
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Oncol., 01 November 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1276695
8p11 myeloproliferative syndrome is a rare hematological malignancy with aggressive course caused by the various translocation of FGFR1. In this study, a novel FGFR1 fusion was identified by RNA sequencing in a 28-year-old male patient with acute B-lymphoblastic leukemia. The patient harbors an in-frame fusion between KIF5B exon 15 and FGFR1 exon 10. The FGFR1 fusion and its protein expression was validated by Sanger sequencing and Western blot. Meanwhile, cytogenetic analysis reported a normal karyotype and targeted DNA sequencing identified no driver mutations, respectively. Despite he achieved complete remission after induction regimen, a relapse occurred and he became refractory to chemotherapy, and salvage haploidentical hematopoietic stem cell transplantation failed to control the progressive disease. In conclusion, we present the first case of KIF5B-FGFR1 fusion in hematological malignancy. These findings extend the spectrum of translocation in 8p11 myeloproliferative syndrome, and demonstrate the great prospect of RNA sequencing in clinical practice again.
Myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions (MLN-TK) is a special category of hematologic malignancy driven by rearrangements of a subset of tyrosine kinase genes (1, 2). These rearrangements lead to fusion proteins in which the kinase domain is constitutively activated, with resulting oncogenic properties. 8p11 myeloproliferative syndrome (EMS) is a specific subtype of MLN-TK caused by a translocation involving FGFR1 gene. Seventeen FGFR1 fusions has been reported in EMS so far, of which the most common are ZNF198-FGFR1, BCR-FGFR1 and CEP110-FGFR1 (3). Most EMS patients are diagnosed with CML (chronic myeloid leukemia), aCML (atypical chronic myeloid leukemia), T-LBL (T-cell lymphoblastic lymphoma), or AML (acute myeloid leukemia). With aggressive clinical course and rapid progression, most EMS patients are resistant to traditional chemotherapeutic agents and most tyrosine kinase inhibitors (TKIs). Herein, we describe an in-frame KIF5B-FGFR1 fusion gene firstly identified by RNA sequencing in a patient with acute B- lymphoblastic leukemia (B-ALL).
A 28-year-old man was referred to our hospital, complaining of petechia, abdominal distension and hemosputum in July 2017. Sternum tenderness, lymphadenopathy of left neck and splenomegaly were found by physical examination. Complete blood count revealed that leukocytosis (125.55×109/L), anemia (86 g/L) and thrombocytopenia (8 ×109/L). At the same time, peripheral blood smear showed 67% blasts, 1% eosinophils, 5% neutrophils and 26% lymphocytes. Bone marrow (BM) aspirates revealed hypercellular with 64% blasts (Figure 1A). Immunophenotypic analysis showed a large subset of (76.4%) leukemic cells positive for CD10 and CD19 (Figure 1B). Cytogenetic analysis of the BM showed a normal karyotype: 46, XY (20/20 cells) (Figure 1C). Meanwhile, a fluorescence in situ hybridization (FISH) test for the detection of BCR-ABL1 rearrangement and a targeted next-generation DNA sequencing using a panel of 172 genes (Table S1) were negative. Besides, a negative result was detected by a multiplex reverse transcription-polymerase chain reaction (RT-PCR) analysis for 43 acute leukemia-related fusion genes.
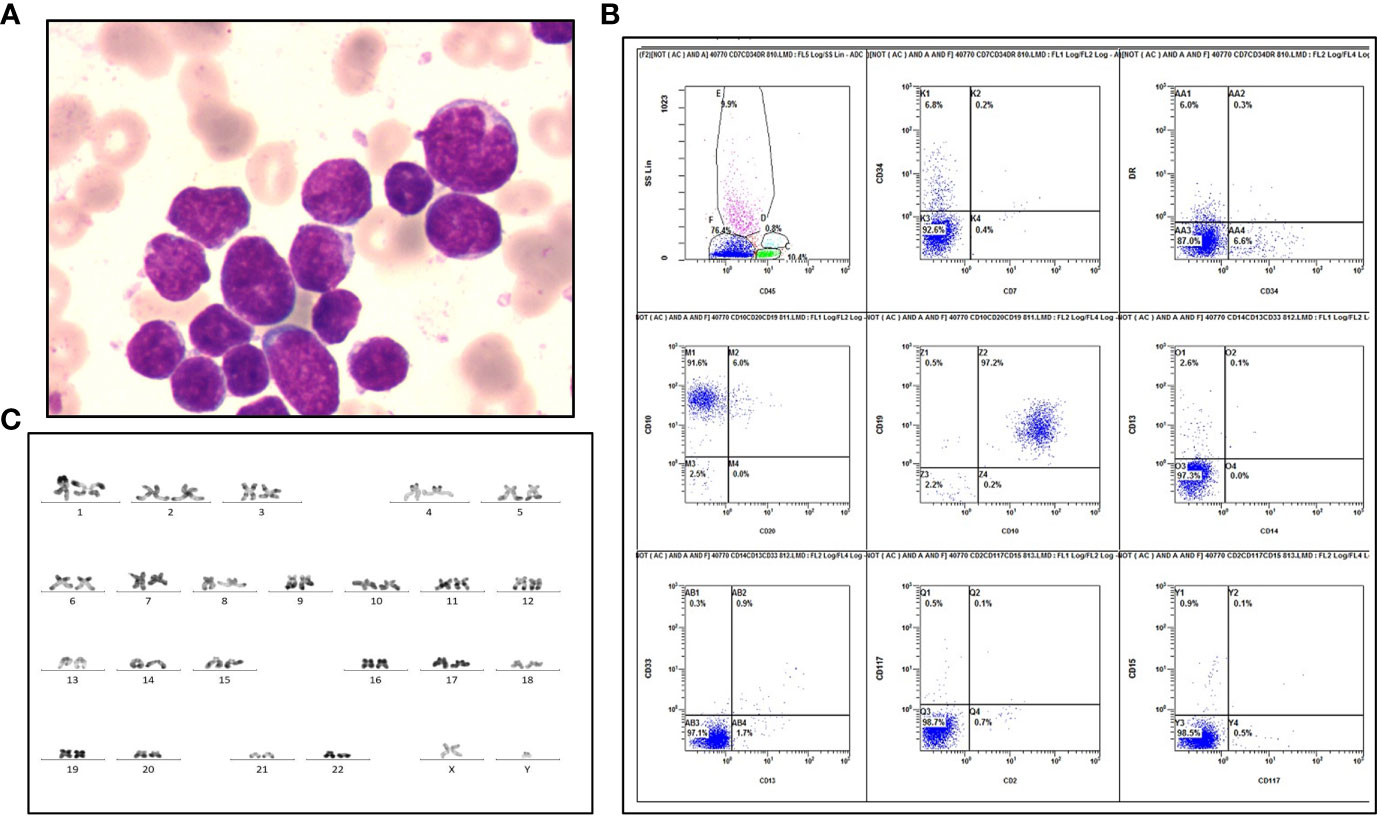
Figure 1 Laboratory examinations of the patient diagnosed as B-ALL. (A) Wright-Giemsa staining of blasts at diagnosis. (B) Flow cytometric immunophenotyping reveals a large CD45-negative blast population positive for CD10 and CD19, and negative for HLA-DR, CD2, CD7, CD13, CD14, CD15, CD20, CD33, CD34 and CD117. (C) R-band karyotyping shows 46, XY [20] in this patient.
However, RNA sequencing revealed a fusion involving exon1-15 of KIF5B (NM_004521.3) and exon10-18 of FGFR1 (NM_023110.3), which mapped to chromosome 10p11 and 8p11, respectively (Figure 2A). The KIF5B-FGFR1 mRNA was reverse-transcribed into cDNA using master mix for RT- PCR (Takara, Japan), and sequential PCR was performed using the following primers: KIF5B F: 5’-CAGGAGGAGCTTTTGGCATCT-3’, and FGFR1 R: 5’-TGCGTCCGACTTCAACATCT-3’, which is the same as described previously (4). As a result, a 616-bp product was specifically amplified only from the patient’s cDNA (Figure 2B), which confirmed the presence of this fusion gene. Moreover, Sanger sequencing analysis of the fusion transcript revealed that KIF5B exon 15 was fused in-frame to FGFR1 exon 10 (Figure 2C). Western blot was performed using the BM mononuclear lysates from the patient with a rabbit anti-FGFR1 antibody (Cell Signaling Technology, USA). As shown in Figure 2D, a protein band corresponding to the KIF5B fused FGFR1 was detected, which demonstrated the KIF5B-FGFR1 fusion gene can lead to the formation of fusion protein.
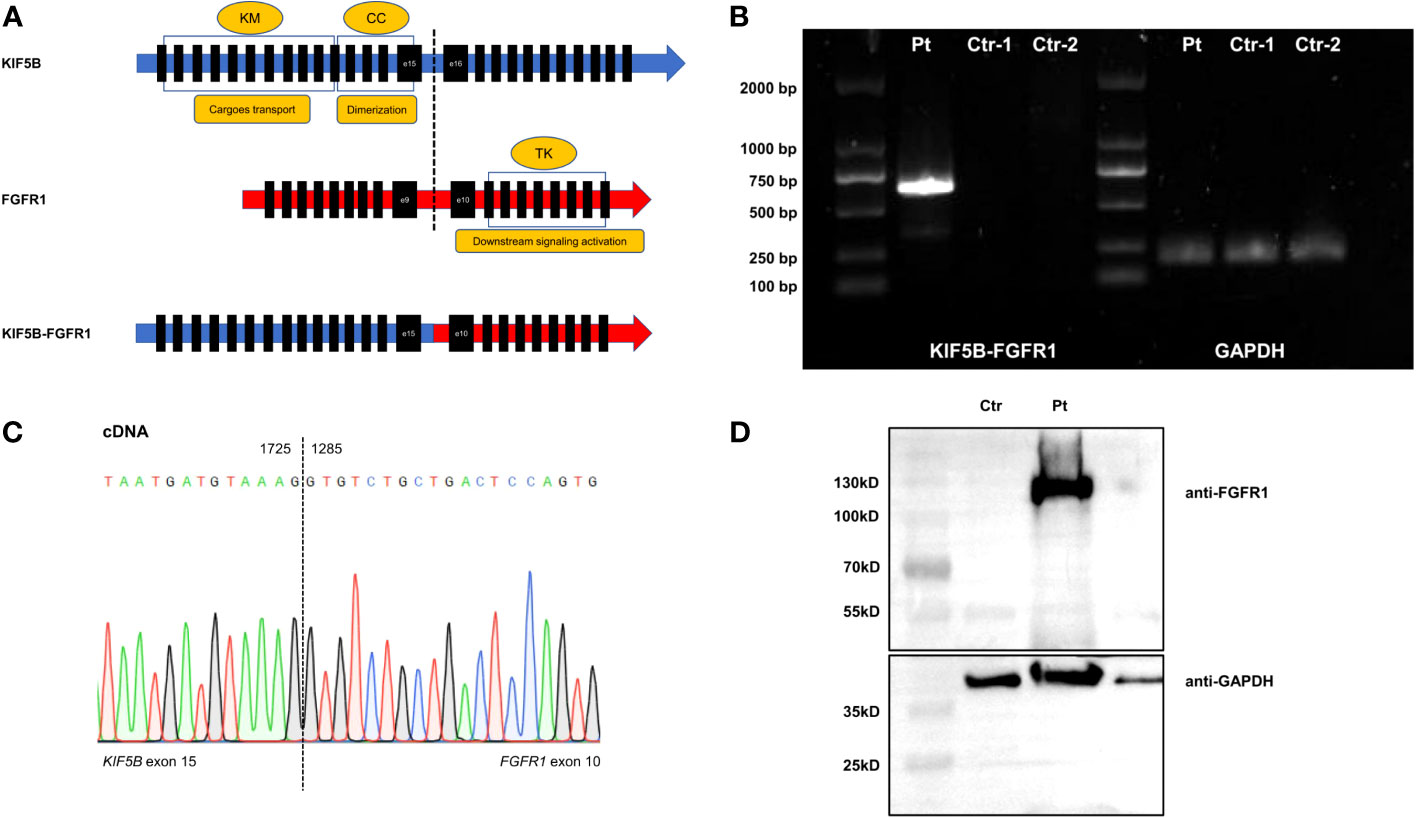
Figure 2 (A) The structure diagram of wildtype KIF5B, FGFR1, and KIF5B-FGFR1 fusion transcript. (B) Electrophoresis of RT-PCR products. Showing the positive KIF5B-FGFR1 fusion transcript in the patient’s bone marrow sample. Pt was the diagnostic sample. Ctr-1 and Ctr-2 were the negative control. (C) Sanger sequencing of the PCR product confirmed the fusion between exon 15 of the KIF5B gene and exon 10 of the FGFR1 gene. (D) Western blot analysis of fusion protein. Detecting one protein containing FGFR1 with a size between 100kD and 130 kD in the patient’s bone marrow sample. Pt was the diagnostic sample. Ctr was the negative control. KM, kinesin motor; CC, coiled coin; TK, tyrosine kinase.
According to above tests, the patient was diagnosed as B-ALL with rearrangement of FGFR1, which is a subtype of BCR-ABL1 like ALL. After supporting treatment, he received a course of induction chemotherapy using IVP regimen (idarubicin, vindesine and dexamethasone) and achieved complete remission (CR) with a detectable minimal residual disease (MRD), which was consistent with previous reports that BCR-ABL1 like ALL correlated with MRD persistence. Consolidated with 2 courses of hyper-CVAD regimen (cyclophosphamide, vincristine, doxorubicin, and dexamethasone), the patient relapsed and was treated by a course of FLAG-IDA regimen (fludarabine, cytarabine, idarubicin, and granulocyte colony-stimulating factor). Without any improvement, he underwent haploidentical hematopoietic stem cell transplantation (HSCT) from his brother and achieved CR. However, the MRD become positive two months after transplantation and donor lymphocyte infusion was administered. Unfortunately, an early relapse finally occurred about one month later with pancytopenia, decreased short tandem repeats chimerism (86.5%) and 12% blasts detected in BM smears. Immunosuppressants were stopped and MOpAD regimen (methotrexate, vincristine, asparaginase, and dexamethasone) combined with DLI was used to control the disease, but he failed to achieve remission. Then the patient transferred to another hospital for economic reasons and died of progressive disease five months later.
B-ALL with FGFR1 rearrangements belongs to BCR-ABL1 like ALL, which is a poor-risk subset and shares a similar gene expression profile to that of ALL with BCR-ABL1 (5, 6). In fact, there is some overlap between the BCR-ABL1 like ALL and MLN-TK diagnostic criterion, and some Ph-like ALL cases could also be diagnosed as MLN-TK, for MLN-TK include a broad range of histologic types and have complex clinical manifestations. Although the latest International Consensus Classification (ICC) of myeloid neoplasms and acute leukemias distinguishes BCR-ABL1 like ALL from MLN-TK presenting as B-ALL by identifying involvement of TK gene fusion in the background myeloid cells in addition to o lymphoblasts (7), the 5th edition of the World Health Organization (WHO) Classification of hematolymphoid tumors does not emphasize the difference between these two groups and holds the opinion that the diagnosis of MLN-TK supersedes other myeloid and lymphoid types from a diagnostic hierarchy standpoint (1). The clinical characteristics (young male, absence of eosinophilia, no MPN history, poor outcome) of our case support a diagnosis of BCR-ABL1 like ALL rather than typical MLN-TK according to ICC. However, we preferred a diagnosis of EMS (MLN-TK) according to the 5th WHO classification, because it could better specify the underlying molecular genetic aberrations of such cases, considering FGFR1 rearrangements are quite uncommon in BCR-ABL1 like ALL (5, 8–11).
EMS is a rare subtype of hematologic neoplasms with fewer than 120 reported cases worldwide (3). The disease could occur at any age with protean and variable clinical manifestations, which makes it easy to ignore or misdiagnose as other hematologic malignancies. Apart from a male predominance, eosinophilia and monocytosis were noted in a substantial proportion of patients with EMS (12). According to previous literature, ZNF198 (also called ZMYM2), located 13q11-12, is the most frequent gene partner of FGFR1 in EMS. Most patients with ZNF198-FGFR1 are diagnosed with T-LBL/T-lymphoma, with the presence of eosinophilia and lymphadenopathy (13). In addition, the t(8;22) (p11;q11), which generates BCR-FGFR1, is the second common translocation in patients with EMS. Patients with BCR-FGFR1 have clinical manifestations resembling CML and tend to develop B-ALL or lymphoma presenting with basophilia (14). Another common translocation in EMS is t(8;9)(p11-12;q32-34), which results in CEP110-FGFR1 fusion gene. Of note, patients with CEP110-FGFR1 often present with both eosinophilia and monocytosis (15). Besides, lymphadenopathy, purpura and skin lesions are also common symptoms at the time of diagnosis. Meanwhile, the initial diagnosis of patients with CEP110-FGFR1 were diverse, including AML, T-LBL, and aCML. Besides, other recurrent fusion genes identified in EMS are FOP1-FGFR1, HERVK-FGFR1, NUP98-FGFR1, FOP2-FGFR1, TIF1-FGFR1, MYO18A-FGFR1, TPR-FGFR1 and NUP358-FGFR1 (3). It should be noted that G-banding karyotype analysis of our case failed to identify translocation between chromosome 10p11 and 8p11 at diagnosis. To better identify the patients with balanced reciprocal translocations, it is necessary to detect FGFR1 rearrangements by a combination of karyotype analysis, FISH and next-generation sequencing of molecular genetic techniques.
Aside from complex clinical presentation and molecular aberrations, most EMS patients are resistant to traditional treatment and thus have a very unfavorable prognosis. Previous studies showed that the CR rate and 1-year overall survival of EMS patients was merely 27% and 43.1%, respectively (12, 16). At present, allogeneic HSCT is the only method to improve the survival of EMS patients (17). Although non-selective TKIs such as imatinib, nilotinib, and dasatinib have showed limited efficacy on MLN with FGFR1 rearrangements (18), pemigatinib, a selective and potent inhibitor of FGFR1-3, has been approved for relapsed or refractory MLN with FGFR1 rearrangements due to promising results in the FIGHT-203 (NCT03011372) study (19), which may offer a long-term treatment option for EMS patients. Additionally, another selective FGFR inhibitor, infigratinib, has showed inhibitory effect on FOP2-FGFR1 and TPR-FGFR1 in vitro.
To our knowledge, the fusion KIF5B-FGFR1 has been only reported in a case of multicentric reticulohistiocytosis (MRH) (4). Our case is the first to identify the presence of this fusion gene in hematologic malignancies. Consistent with our patient, the MRH patient with KIF5B-FGFR1 carried no identifiable driver point mutations. Furthermore, bioinformatic analysis of RNA sequencing in Murakami et al. revealed that KIF5B-FGFR1 was associated with marked activated tyrosine kinase activity. Notably, the coiled-coil domain derived from KIF5B, which was contained by the fusion protein, could involve in protein dimerization, and then phosphorylate and constitutively activate the kinase domains domains (20). These findings suggest that the fusion protein might dimerize and activate FGFR1 tyrosine kinase to promote proliferation and survival of neoplasms without help from additional driver mutations, which needs to be further investigated in disease models.
In summary, we described an in-frame KIF5B-FGFR1 fusion gene that consists of KIF5B exon 15 and FGFR1 exon 10, which was firstly reported in hematologic malignancy. Similar to most EMS or BCR-ABL1 like ALL cases, the patient suffered from disease relapse and then became refractory to traditional treatment. Future studies are needed to define the underlying mechanisms of this fusion gene on contributing to leukemogenesis, and new agents targeting the FGFR-related signaling pathway may provide a potential therapeutic option for hematopoietic neoplasms with FGFR1 rearrangements. More importantly, RNA sequencing could improve detection rate of rare and novel fusion genes, and its future widespread application in clinical practice will provide a deeper understanding of disease biology.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by Ethics Committee of the First Affiliated Hospital of Soochow University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article. All procedures performed in studies involving human participants were in accordance with the ethical standards of the First Affiliated Hospital of Soochow University committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
ZBZ: Writing – original draft, Formal Analysis, Visualization. YZ: Data curation, Writing – review & editing. ZW: Data curation, Writing – review & editing. ZZ: Validation, Writing – review & editing. LW: Methodology, Supervision, Writing – review & editing. LZ: Project administration, Validation, Writing – review & editing. SC: Funding acquisition, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grant from the National Key R&D Program of China (2019YFA0111004, 2022YFC2502701), the National Natural Science Foundation of China (82170158, 81970142, 82100175), the Translational Research Grant of NCRCH (2021WSB01, 2020WSB03, 2020WSB11, 2020WSB13), the Open Project of Jiangsu Biobank of Clinical Resources (SBK202003001, SBK202003003).
All the samples were from Jiangsu Biobank of Clinical Resources.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1276695/full#supplementary-material
1. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia (2022) 36(7):1703–19. doi: 10.1038/s41375-022-01613-1
2. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: integrating morphologic, clinical, and genomic data. Blood (2022) 140(11):1200–28. doi: 10.1182/blood.2022015850
3. Li T, Zhang G, Zhang X, Lin H, Liu Q. The 8p11 myeloproliferative syndrome: Genotypic and phenotypic classification and targeted therapy. Front Oncol (2022) 12:1015792. doi: 10.3389/fonc.2022.1015792
4. Murakami N, Sakai T, Arai E, Muramatsu H, Ichikawa D, Asai S, et al. Targetable driver mutations in multicentric reticulohistiocytosis. Haematologica (2020) 105(2):e61–e4. doi: 10.3324/haematol.2019.218735
5. Tasian SK, Loh ML, Hunger SP. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood (2017) 130(19):2064–72. doi: 10.1182/blood-2017-06-743252
6. Conant JL, Czuchlewski DR. BCR-ABL1-like B-lymphoblastic leukemia/lymphoma: Review of the entity and detection methodologies. Int J Lab Hematol (2019) 41:126–30. doi: 10.1111/ijlh.13012
7. Wang SA, Orazi A, Gotlib J, Reiter A, Tzankov A, Hasserjian RP, et al. The international consensus classification of eosinophilic disorders and systemic mastocytosis. Am J Hematol (2023) 98(8):1286–306. doi: 10.1002/ajh.26966
8. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in ph-like acute lymphoblastic leukemia. N Engl J Med (2014) 371(11):1005–15. doi: 10.1056/NEJMoa1403088
9. Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, et al. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children's Oncology Group. Blood (2017) 129(25):3352–61. doi: 10.1182/blood-2016-12-758979
10. Roberts KG, Reshmi SC, Harvey RC, Chen IM, Patel K, Stonerock E, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children's Oncology Group. Blood (2018) 132(8):815–24. doi: 10.1182/blood-2018-04-841676
11. Paietta E, Roberts KG, Wang V, Gu ZH, Buck GAN, Pei DQ, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood (2021) 138(11):948–58. doi: 10.1182/blood.2020010144
12. Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol (2010) 41(4):461–76. doi: 10.1016/j.humpath.2009.11.003
13. Urrea Pineda LY, Perilla O, Santiago-Pacheco V, Trujillo Montoya S. Myeloproliferative syndrome with eosinophilia associated with translocation t(8; 13) and T-cell lymphoblastic lymphoma: A case report and review of the literature. Cureus (2022) 14(3):e22734. doi: 10.7759/cureus.22734
14. Montenegro-Garreaud X, Miranda RN, Reynolds A, Tang G, Wang SA, Yabe M, et al. Myeloproliferative neoplasms with t(8;22)(p11.2;q11.2)/BCR-FGFR1: a meta-analysis of 20 cases shows cytogenetic progression with B-lymphoid blast phase. Hum Pathol (2017) 65:147–56. doi: 10.1016/j.humpath.2017.05.008
15. Chen MY, Wang K, Cai XH, Zhang XW, Chao HY, Chen SN, et al. Myeloid/lymphoid neoplasm with CEP110-FGFR1 fusion: An analysis of 16 cases show common features and poor prognosis. Hematology (2021) 26(1):153–9. doi: 10.1080/16078454.2020.1854493
16. Umino K, Fujiwara SI, Ikeda T, Toda Y, Ito S, Mashima K, et al. Clinical outcomes of myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement. Hematology (2018) 23(8):470–7. doi: 10.1080/10245332.2018.1446279
17. Konishi Y, Kondo T, Nakao K, Asagoe K, Otsuka Y, Nishikori M, et al. Allogeneic hematopoietic stem cell transplantation for 8p11 myeloproliferative syndrome with BCR-FGFR1 gene rearrangement: a case report and literature review. Bone Marrow Transplant (2019) 54(2):326–9. doi: 10.1038/s41409-018-0287-1
18. Kreil S, Adès L, Bommer M, Stegelmann F, Ethell ME, Lubking A, et al. Limited efficacy of ponatinib in myeloproliferative neoplasms associated with FGFR1 fusion genes. Blood (2015) 126(23):2812. doi: 10.1182/blood.V126.23.2812.2812
19. Gotlib J, Kiladjian J-J, Vannucchi A, Rambaldi A, Reiter A, Shomali W, et al. A phase 2 study of pemigatinib (FIGHT-203; INCB054828) in patients with myeloid/lymphoid neoplasms (MLNs) with fibroblast growth factor receptor 1 (FGFR1) rearrangement (MLN FGFR1). Blood (2021) 138(Supplement 1):385. doi: 10.1182/blood-2021-148103
Keywords: acute lymphoblastic leukemia, Kif5B, FGFR1, rearrangement, 8p11 myeloproliferative
Citation: Zhang Z, Zhu Y, Wang Z, Zeng Z, Wen L, Zhang L and Chen S (2023) Case Report: A novel FGFR1 fusion in acute B-lymphoblastic leukemia identified by RNA sequencing. Front. Oncol. 13:1276695. doi: 10.3389/fonc.2023.1276695
Received: 12 August 2023; Accepted: 12 October 2023;
Published: 01 November 2023.
Edited by:
Liren Qian, Fifth Medical Center of the PLA General Hospital, ChinaReviewed by:
Cosimo Cumbo, University of Bari Aldo Moro, ItalyCopyright © 2023 Zhang, Zhu, Wang, Zeng, Wen, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Zhang, TFoyMDE4NDAzMjAxMEAxNjMuY29t; Lijun Wen, c3VkYXdsakAxNjMuY29t; Suning Chen, Y2hlbnN1bmluZ0BzdWRhLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.