
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Oncol. , 24 October 2023
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1276138
 Andrea Alberti1*
Andrea Alberti1* Gertraud Stocker2
Gertraud Stocker2 Florian Lordick2
Florian Lordick2 Ulrich T. Hacker2
Ulrich T. Hacker2 Benjamin Kobitzsch2
Benjamin Kobitzsch2 Ivonne Haffner2
Ivonne Haffner2 Gian Luca Baiocchi3
Gian Luca Baiocchi3 Manuel Zamparini1
Manuel Zamparini1 Guido A. M. Tiberio4
Guido A. M. Tiberio4 Carla Baronchelli5
Carla Baronchelli5 Arnaldo Caruso6
Arnaldo Caruso6 Paolo Bossi7
Paolo Bossi7 Alfredo Berruti1
Alfredo Berruti1Purpose: The Cancer Genome Atlas Research Network identified Epstein-Barr-Virus (EBV)-positive gastric cancer as a distinct molecular subtype. The prevalence is 8-9% and the histological examination shows pronounced lymphocytic infiltration, elevated levels of IFN-γ and consequently overexpression of PD-L1. The role of plasma EBV DNA load as a prognostic factor in patients with this cancer subtype is still to be defined.
Methods and analysis: The present multicenter prospective observational study “EBV PRESAGE”, involving German and Italian cancer centers, aims to evaluate the prognostic role of plasma EBV DNA in EBV-related gastric cancer (GC). The objective is to study the association between plasma EBV DNA load at different consecutive time points and the patient’s prognosis. Every patient with a new diagnosis of gastric cancer (including gastroesophageal junction adenocarcinoma) will be screened for Epstein-Barr encoded small Region (EBER) on tissue biopsies using in situ hybridization (ISH). If EBER ISH is positive, blood analysis for plasma EBV DNA will be conducted. The plasma EBV quantitative analysis will be centralized, and extraction, detection, and quantification of EBV DNA in plasma samples will be performed using real-time PCR.
Discussion: We hypothesized that plasma EBV DNA represents a non-invasive tool for monitoring EBV-related GC and might be valuable as a prognostic marker.
Gastric cancer (GC) is the 4th most common cancer worldwide and the second leading cause of cancer-related death with 700,000 deaths reported annually (1). The 5-year survival rate in stages I-II is 92-96%, however, resectable disease is detected in only 20-30%, and the recurrence rate after resection is high (1). For advanced disease, the reported 1-year survival is approximately 30% (2). The Cancer Genome Atlas Research Network (TCGA) has identified four distinct molecular subtypes of GC: EBV-positive, microsatellite instable, genomically stable and chromosomal instability; 9% of tumors were identified as EBV-positive (3). A meta-analysis involving 9,738 patients from 48 studies revealed an EBV-positive rate of 8.8% in GC (4). Another pooled analysis included 4,599 GC patients from 13 studies and found an 8.2% EBV positivity rate (5). The EBV subtype is more frequent in males and is usually localized in the gastric fundus or body. The histological examination shows pronounced lymphocytic infiltration, elevated levels of IFN-γ and consequently overexpression of PD-L1 (3, 6). EBV positive GCs have a higher prevalence of DNA hypermethylation than any other GC subtype. In particular, they display CDKN2A (p16INK4A) promoter hypermethylation but lack the MLH1 hypermethylation characteristic of MSI-associated CpG island methylator phenotype (3, 6).
Studies on the prognostic significance of EBV in GC are limited by the small number of EBV-positive tumors; results have been inconsistent, with some reporting a significant survival advantage for EBV-positive tumors (7–12), while others report a non-significant increased risk of death (13–15). Liu et al. conducted a meta-analysis on this aspect and concluded that patients with EBV-positive GC have a significantly better survival than those with EBV-negative cancers (HR 0.67, CI 055-0.79) (16). The pooled analysis by Camargo et al. came to consistent conclusions (HR 0.72; 95% CI 0.61-0.86), and although the stage was inversely associated with EBV positivity, the prognostic impact is preserved in early GC as in advanced GC (5).
Serum tumor markers, like carcinoembryonic antigen and cancer antigen 19-9, are of limited utility for the management of GC. They may be elevated in some patients and a drop in blood levels may correlate with response to preoperative therapy (17). However, clinical decisions are rarely taken based on tumor marker changes alone (17). Likewise, in many (but not all) studies, preoperative elevation in serum tumor markers are an independent indicator of adverse prognosis (17–19). While tumor markers should not be used to exclude a patient from surgical approach. Accordingly, recommendations for preoperative evaluation and staging of GC from the NCCN and ESMO do not include an assay of any tumor marker (20, 21).
Plasma EBV-DNA load has been well documented as an effective marker for distinguishing infectious mononucleosis from the presence of EBV in benign B lymphocytes, as well as for diagnostic screening and monitoring of EBV-associated diseases such as lymphoma, post-transplantation lymph proliferative disease and nasopharyngeal carcinoma (22–25). Especially in nasopharyngeal cancer patients, the quantification of plasma EBV DNA can be used to monitor recurrence and predict prognosis (13). Little has been done to evaluate the role of plasma EBV-DNA load in the diagnosis and monitoring of GC. Shoda et al. evaluated the clinical utility of plasma EBV-DNA in EBV-associated GC and found that the sensitivity and specificity of detection were 71.4% and 97.1%, respectively (26). An Asian prospective cohort study from a single cancer center in China identified 140/2760 (5.1%) EBER-positive GC. This study confirmed that EBV-associated GC had better overall survival and suggested that elevated dynamic detection of plasma EBV-DNA load offered an easily accessible biomarker to monitor EBV-associated GC (27). However, there are few data on the role of plasma EBV DNA load as a prognostic or predictive biomarker for EBV-associated GC in a non-Asian population.
Herein, we describe the design and methodology of the EBV PRESAGE study, an international, prospective observational trial, evaluating the prognostic role of plasma EBV DNA in EBV-related GC.
The primary hypothesis is to identify the association between pre-treatment plasma EBV DNA load and the patient’s prognosis in a series of consecutive, prospective early EBV-related GC (stage Ib-IIIc). Secondary objective is to evaluate the role of EBV DNA load after surgery as a prognostic factor for disease-free survival. Moreover, descriptive objectives are to evaluate the possibility to test circulating plasmatic EBV DNA at baseline and at consecutive time points during follow-up and during systemic treatments in advanced (stage IV) EBV-related GC.
The study population includes female and male patients, age ≥ 18 years, diagnosed with early (stage Ib-IIIc) or advanced (stage IV) GC (including gastroesophageal junction adenocarcinoma). The study enrolls patients who are going to start a preoperative or perioperative or a 1st – 2nd line systemic therapy, as well as those who are only candidates for surgical treatment.
Every patient will be screened for EBV by EBER-ISH on tissue biopsy. If EBER ISH is positive, blood analysis for plasma EBV DNA will be conducted. Patients enrolled will be divided according to the type of treatment proposed in operative, perioperative, and palliative categories. Those undergoing surgical treatment with or without any peri-operative treatment will be evaluated for disease free survival analysis. The blood samples will be collected in these categories before the start of any treatment. Moreover, plasma will be collected after surgery and it will be evaluated as a prognostic factor for disease-free survival. In order to describe the changes of plasma EBV DNA levels and correlate with the radiological results, blood samples will be collected during follow-up, approximately every 6 months for the first two years and at the end of the third year. In the advanced setting, this is an exploratory study to describe the trend of plasma EBV DNA during the first and consecutive lines of therapy. Plasma will be collected at the second week after the start of treatment to evaluate early changes and then every 12 weeks to evaluate the clearance rate until progression. Figure 1 shows the study design.
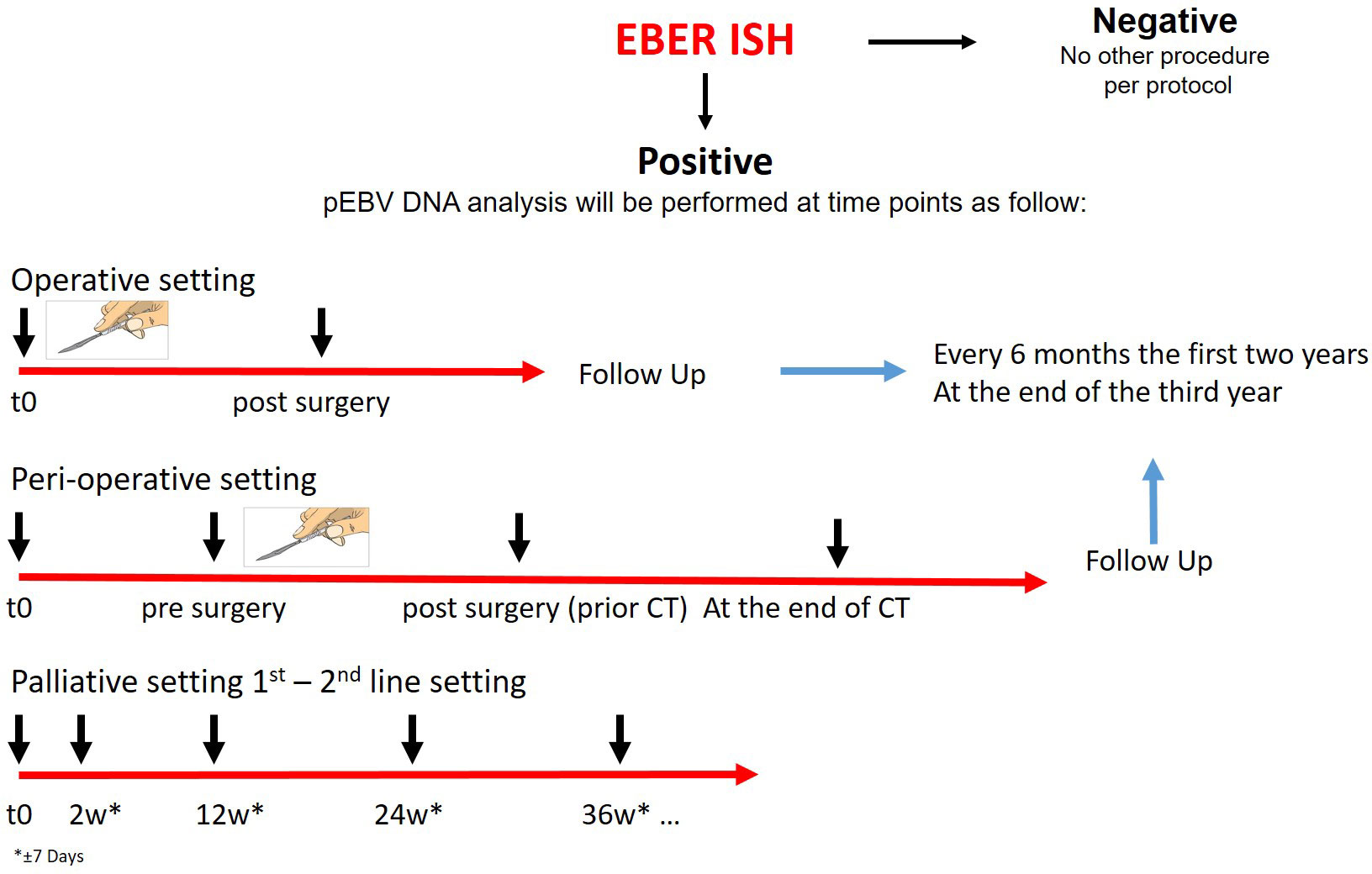
Figure 1 Study design. CT chemotherapy, EBV Epstein Barr virus, EBER Epstein-Barr Encoding Region, ISH in situ hybridization. Patients with GC will be screened with EBER ISH. If EBER ISH is positive, blood analysis for plasma EBV DNA will be conducted. Plasma EBV DNA analysis will be performed according to the setting at different times. During follow up blood samples will be collected every 6 months the first two years and at the end of the third year.
Archival tumor tissue, obtained for clinical practice, will be used for EBER ISH. The test will be performed locally, at each pathological anatomy department of participant centers. EBER detection will be performed on FFPE sample by an ISH kit according to the manufacturer’s specification.
The plasma EBV quantitative analysis will be centralized at the Laboratory of Microbiology of Spedali Civili of Brescia. Extraction, detection and quantification of EBV DNA in plasma samples will be performed using a completely automatized platform based on RT PCR technology (EBV ELITe MGB® Kit). Targets of RT PCR are region of Epstein-Barr virus nuclear antigen 1 (EBNA 1) gene and human beta-globin gene as an internal control.
To assess the prognostic value of plasma EBV DNA load at baseline (pre-treatment) and post-treatment for disease free survival in early EBV-related GC we will use a Cox model. Assuming a log(copies/ml) standard deviation for EBV-DNA of approximately 8.5, an R2 with the other variables (covariates) of 0.2, a baseline 3 years survival of 50% and a HR >= 1.1 for a unit increase in log(EBV-DNA), alpha=5% and power 80%, we would have N=31 patients. Considering a drop-out rate of 10%, 35 patients will be enrolled. The enrollment period is from January 2023 to January 2026; the planned follow-up period is 3 years, so the study duration is 6 years.
During follow-up and in the advanced setting EBV DNA load measured at different time intervals will be described as mean (sd) and median (range), and to represent the trend of plasma EBV DNA in the different time points (descriptive objectives) we will use the raw and percentages variations, testing their difference from 0 through one sample t-test or alternatively the non-parametric sign test, after checking variables distributions. All analyses will be done with the software SAS, SPSS, and R.
It is known that 5-9% of GC are EBV-related. The EBV subtype has a distinct molecular profile, a favourable survival, and the EBV-association seems to be a predictive biomarker for immunotherapy (3, 5, 16, 27, 28). In an Asian population it was shown that dynamic detection of EBV DNA plasma load offers an easily accessible biomarker to monitor GC (26, 27). However, knowledge about the plasma EBV DNA in gastric cancer is still limited and prospective data from Non-Asian populations are lacking. Therefore, we designed the EBV PRESAGE study to investigate the role of plasma EBV DNA in a non-Asian EBV-related GC population.
We hypothesize that like in nasopharyngeal cancer, also in EBV-related GC, there could be an association between plasma EBV DNA levels and survival. Such a prognostic factor could help stratify treatment according to the expected prognosis. Subsequent EBV DNA assessments could also inform current practice to quantify the response to peri-operative treatment and to stratify according to the risk of recurrence. In the future, this information could be used to personalize peri-operative management and integrate plasma EBV DNA into clinical follow-up. In the metastatic setting, this biomarker could help in evaluating treatment response and anticipate disease progression. In conclusion, we hypothesize that data from this study support the evidence that plasma EBV DNA represents a non-invasive tool for monitoring EBV-related GC.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The institutional review board at each participating site has approved the EBV PRESAGE study protocol and relevant supplementary information. The trial is performed in accordance with ethical principles that have their origin in the Declaration of Helsinki and are consistent with the International Council of Harmonisation/Good Clinical Practice and applicable regulatory requirements. Written informed consent will be obtained from each trial participant. The results of this study will be published in international, peer-reviewed journals, and presented at national and international conferences.
AA: Writing – original draft. GS: Writing – review & editing. FL: Writing – review & editing. UH: Writing – review & editing. BK: Writing – review & editing. IH: Writing – review & editing. GB: Writing – review & editing. MZ: Writing – review & editing. GT: Writing – review & editing. CB: Writing – review & editing. AC: Writing – review & editing. PB: Writing – review & editing. AB: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project is supported by the nonprofit foundation RicerChiAmo Onlus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that PB and AB were both associate editors and author AA was a review editor and were members of Frontiers editorial board, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012: Global Cancer Statistics, 2012. CA: A Cancer J Clin (2015) 65:87–108. doi: 10.3322/caac.21262
2. AJCC Cancer Staging Manual. Eighth Edition. New York City: Springer International Publishing (2017).
3. The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature (2014) 513:202–9. doi: 10.1038/nature13480
4. Lee JH, Kim SH, Han SH, An JS, Lee ES, Kim YS. Clinicopathological and molecular characteristics of Epstein-Barr virus-associated gastric carcinoma: A meta-analysis. J Gastroenterol Hepatol (2009) 24:354–65. doi: 10.1111/j.1440-1746.2009.05775.x
5. Camargo MC, Kim WH, Chiaravalli AM, Kim KM, Corvalan AH, Matsuo K, et al. Improved survival of gastric cancer with tumour Epstein–Barr virus positivity: an international pooled analysis. Gut (2014) 63:236–43. doi: 10.1136/gutjnl-2013-304531
6. Roh CK, Choi YY, Choi S, Seo WJ, Cho M, Jang E, et al. Single patient classifier assay, microsatellite instability, and epstein-barr virus status predict clinical outcomes in stage II/III gastric cancer: results from CLASSIC trial. Yonsei Med J (2019) 60:132. doi: 10.3349/ymj.2019.60.2.132
7. Chiaravalli AM, Feltri M, Bertolini V, Bagnoli E, Furlan D, Cerutti R, et al. Intratumour T cells, their activation status and survival in gastric carcinomas characterised for microsatellite instability and Epstein-Barr virus infection. Virchows Arch (2006) 448:344–53. doi: 10.1007/s00428-005-0066-4
8. Park ES, Do IG, Park CK, Kang WK, Noh JH, Sohn TS, et al. Cyclooxygenase-2 is an independent prognostic factor in gastric carcinoma patients receiving adjuvant chemotherapy and is not associated with EBV infection. Clin Cancer Res (2009) 15:291–8. doi: 10.1158/1078-0432.CCR-08-0848
9. Chang MS, Lee HS, Kim CW, Kim YI, Kim WH. Clinicopathologic characteristics of Epstein-Barr virus-incorporated gastric cancers in Korea. Pathol Res Pract (2001) 197:395–400. doi: 10.1078/0344-0338-00052
10. Song HJ, Srivastava A, Lee J, Kim YS, Kim KM, Ki Kang W, et al. Host inflammatory response predicts survival of patients with Epstein-Barr virus-associated gastric carcinoma. Gastroenterology (2010) 139:84–92.e2. doi: 10.1053/j.gastro.2010.04.002
11. van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJH, Middeldorp JM, van den Brule AJC, et al. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol (2004) 22:664–70. doi: 10.1200/JCO.2004.08.061
12. Shibata D, Hawes D, Stemmermann GN, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma among Japanese Americans in Hawaii. Cancer Epidemiol Biomarkers Prev (1993) 2:213–7.
13. Kijima Y, Ishigami S, Hokita S, Koriyama C, Akiba S, Eizuru Y, et al. The comparison of the prognosis between Epstein-Barr virus (EBV)-positive gastric carcinomas and EBV-negative ones. Cancer Lett (2003) 200:33–40. doi: 10.1016/S0304-3835(03)00410-5
14. Koriyama C, Akiba S, Itoh T, Kijima Y, Sueyoshi K, Corvalan A, et al. Prognostic significance of Epstein-Barr virus involvement in gastric carcinoma in Japan. Int J Mol Med (2002) 10:635–9.
15. Truong CD, Feng W, Li W, Khoury T, Li Q, Alrawi S, et al. Characteristics of Epstein-Barr virus-associated gastric cancer: a study of 235 cases at a comprehensive cancer center in U.S.A. J Exp Clin Cancer Res (2009) 28:14. doi: 10.1186/1756-9966-28-14
16. Liu X, Liu J, Qiu H, Kong P, Chen S, Li W, et al. Prognostic significance of Epstein-Barr virus infection in gastric cancer: a meta-analysis. BMC Cancer (2015) 15:782. doi: 10.1186/s12885-015-1813-9
17. Horie Y, Miura K, Matsui K, Yukimasa A, Ohi S, Hamamoto T, et al. Marked elevation of plasma carcinoembryonic antigen and stomach carcinoma. Cancer (1996) 77:1991–7. doi: 10.1002/(SICI)1097-0142(19960515)77:10<1991::AID-CNCR5>3.0.CO;2-K
18. Mihmanli M, Dilege E, Demir U, Coskun H, Eroglu T, Uysalol MD. The use of tumor markers as predictors of prognosis in gastric cancer. Hepatogastroenterology (2004) 51:1544–7.
19. Marrelli D, Pinto E, De Stefano A, Farnetani M, Garosi L, Roviello F. Clinical utility of CEA, CA 19-9, and CA 72-4 in the follow-up of patients with resectable gastric cancer. Am J Surg (2001) 181:16–9. doi: 10.1016/S0002-9610(00)00549-3
20. National Comprehensive Cancer Network (NCCN). NCCN clinical practice guidelines in oncology. Available at: https://www.nccn.org/professionals/physician_gls.
21. Lordick F, Carneiro F, Cascinu S, Fleitas T, Haustermans K, Piessen G, et al. Gastric cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol (2022) 33:1005–20. doi: 10.1016/j.annonc.2022.07.004
22. Zhang Y, Li WF, Mao YP, Guo R, Tang LL, Peng H, et al. Risk stratification based on change in plasma Epstein-Barr virus DNA load after treatment in nasopharyngeal carcinoma. Oncotarget (2016) 7:9576–85. doi: 10.18632/oncotarget.7083
23. Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, et al. Quantification of plasma epstein–barr virus DNA in patients with advanced nasopharyngeal carcinoma. N Engl J Med (2004) 350:2461–70. doi: 10.1056/NEJMoa032260
24. Leung SF, Chan KCA, Ma BB, Hui EP, Mo F, Chow KCK, et al. Plasma Epstein–Barr viral DNA load at midpoint of radiotherapy course predicts outcome in advanced-stage nasopharyngeal carcinoma. Ann Oncol (2014) 25:1204–8. doi: 10.1093/annonc/mdu117
25. Hohaus S, Santangelo R, Giachelia M, Vannata B, Massini G, Cuccaro A, et al. The viral load of epstein–barr virus (EBV) DNA in peripheral blood predicts for biological and clinical characteristics in hodgkin lymphoma. Clin Cancer Res (2011) 17:2885–92. doi: 10.1158/1078-0432.CCR-10-3327
26. Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, et al. Clinical utility of circulating cell-free Epstein-Barr virus DNA in patients with gastric cancer. Oncotarget (2017) 8:28796–804. doi: 10.18632/oncotarget.15675
27. Qiu M, He C, Lu S, Guan W, Wang F, Wang X, et al. Prospective observation: Clinical utility of plasma Epstein–Barr virus DNA load in EBV-associated gastric carcinoma patients. Int J Cancer (2020) 146:272–80. doi: 10.1002/ijc.32490
Keywords: gastric and gastroesophageal junction adenocarcinoma, epstein barr virus DNA, prognostic factor, disease-free survival, prospective multicenter study
Citation: Alberti A, Stocker G, Lordick F, Hacker UT, Kobitzsch B, Haffner I, Baiocchi GL, Zamparini M, Tiberio GAM, Baronchelli C, Caruso A, Bossi P and Berruti A (2023) Plasma EBV DNA as a prognostic factor in EBV associated gastric cancer: a multicenter, prospective study (EBV PRESAGE study). Front. Oncol. 13:1276138. doi: 10.3389/fonc.2023.1276138
Received: 11 August 2023; Accepted: 05 October 2023;
Published: 24 October 2023.
Edited by:
Dakeun Lee, Ajou University, Republic of KoreaReviewed by:
Maria Raffaella Ambrosio, University of Siena, ItalyCopyright © 2023 Alberti, Stocker, Lordick, Hacker, Kobitzsch, Haffner, Baiocchi, Zamparini, Tiberio, Baronchelli, Caruso, Bossi and Berruti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Alberti, YS5hbGJlcnRpMDE1QHVuaWJzLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.