
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 16 October 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1269531
This article is part of the Research TopicNovelties in Acute Myeloid Leukemia: From Biology to Clinical ApplicationsView all 13 articles
SET-CAN/NUP214 fusion is a recurrent event commonly observed in adult male patients diagnosed with T-cell acute lymphoblastic leukemia (T-ALL) and has occasionally been reported in other diseases such as acute myeloid leukemia (AML), myeloid sarcoma (MS), acute undifferentiated leukemia (AUL), chronic myeloid leukemia (CML) and B-cell acute lymphoblastic leukemia (B-ALL). This fusion gene is derived from chromosome del(9)(q34.11;q34.13) or t(9;9)(q34;q34) and may have an inhibitory effect on primitive progenitor differentiation. The prognosis of the reported patients is varied, with these patients often show resistance to chemotherapy regimens that include high doses of glucocorticoids. The optional treatment has not been determined, more cases need to be accumulated and evaluated. The scope of this review is to summarize the general features and prognostic significance in leukemia associated with the SET-CAN/NUP214 fusion gene and to discuss the methods of detection and treatment, aiming at providing some useful references for relevant researchers in the field of blood tumor.
Leukemia is a malignant clonal disease originating from hematopoietic stem and progenitor cells. Leukemia cells with proliferation and survival advantages proliferate and accumulate uncontrollably in the body, gradually replacing normal hematopoiesis and invading other organs and systems, resulting in a series of symptoms such as anemia, hemorrhage, infection and immersion. According to the degree of differentiation and maturation of leukemia cells and the natural course of disease, leukemia can be roughly divided into two categories: acute leukemia and chronic leukemia, and then divided into myelogenic/myeloid and lymphocytic/lymphoblastic according to the cell of origin.
SET-CAN/NUP214 fusion gene is formed by del(9)(q34.11;q34.13) or t(9;9)(q34;q34) and has been identified in the LOUCY cell line of T -ALL and the MEGAL cell line of AML(1, 2). In 1992, Von Lindern et al. first identified the SET-CAN/NUP214 fusion gene in a case of acute undifferentiated leukemia (AUL). Since then, with the development of detection technology and the deepening understanding of leukemia, subsequent cases of AML, MS, AUL, CML, and B-ALL have also been found (3–6). Overall, the disease experienced by most patients carrying SET-CAN/NUP214 is T-ALL.
The NUP214 protein, also known as CAN, is a nucleoporin with FG repeats rich in phenylalanine-glycine. The NUP214 gene is located on band 9q34.1 and it has a total of 36 exons numerically labeled from 1 to 36 (Figure 1). Chromosome abnormality involving NUP214 occur repeatedly in leukemia, in addition to the SET-CAN/NUP214 reviewed here, other chromosome abnormalities were found such as DEK-NUP214, SQSTM1-NUP214 and NUP214-ABL1. DEK-NUP214 [t(6;9)(p22;q34)] was associated with AML, NUP214-ABL1 was identified in T-ALL patients, the rarest leukemia NUP214 fusion protein is SQSTM1-NUP214: to date, only two cases have been reported, one in ALL and the other in AML. The structure of the SQSTM1-NUP214 fusion gene consists of five exons located at the N-terminus of the SQSTM1 gene fused to a portion of the C-terminus of NUP214, including its last 14 FG repeats (7). In eukaryotic cells, nucleo-cytoplasmic transport plays an important role in maintaining the normal function and integrity of cells (8). Molecules with a molecular mass greater than 40kDa cannot move across the nuclear membrane by simple diffusion, but require to be facilitated by nuclear transporter receptors (NTRs) with the help of nuclear pore complexes (NPCs) embedded within the nuclear membrane (9–11). NUP214 interacts with NTRs via the FG repeat region in the cytoplasmic filaments of the nuclear pore complexes (NPCs) to control macromolecule trafficking (12). NUP214 has been shown to interact with exportin-1 (XPO1) and nuclear RNA export factor 1 (NXF1) of NTRs, which are highly mobile in cells (13) and play an important role in the response to NUP214 by nuclear export sequences (NES) protein; Furthermore, NUP214 fusion proteins such as SET-CAN/NUP214 and DEK-NUP214, reduce the mobility of XPO1 and lead to the accumulation of XPO1 cargo within the nucleus, impair nuclear output by sequestering XPO1 in the nucleus, interfere with nuclear-cytoplasmic transport of macromolecules, and potentially affect the transcriptional regulatory function of the NF-κB pathway (14), leading to various blood diseases (15). Moreover, genomic knockout of NUP214 led to embryonic lethality in mice (1).
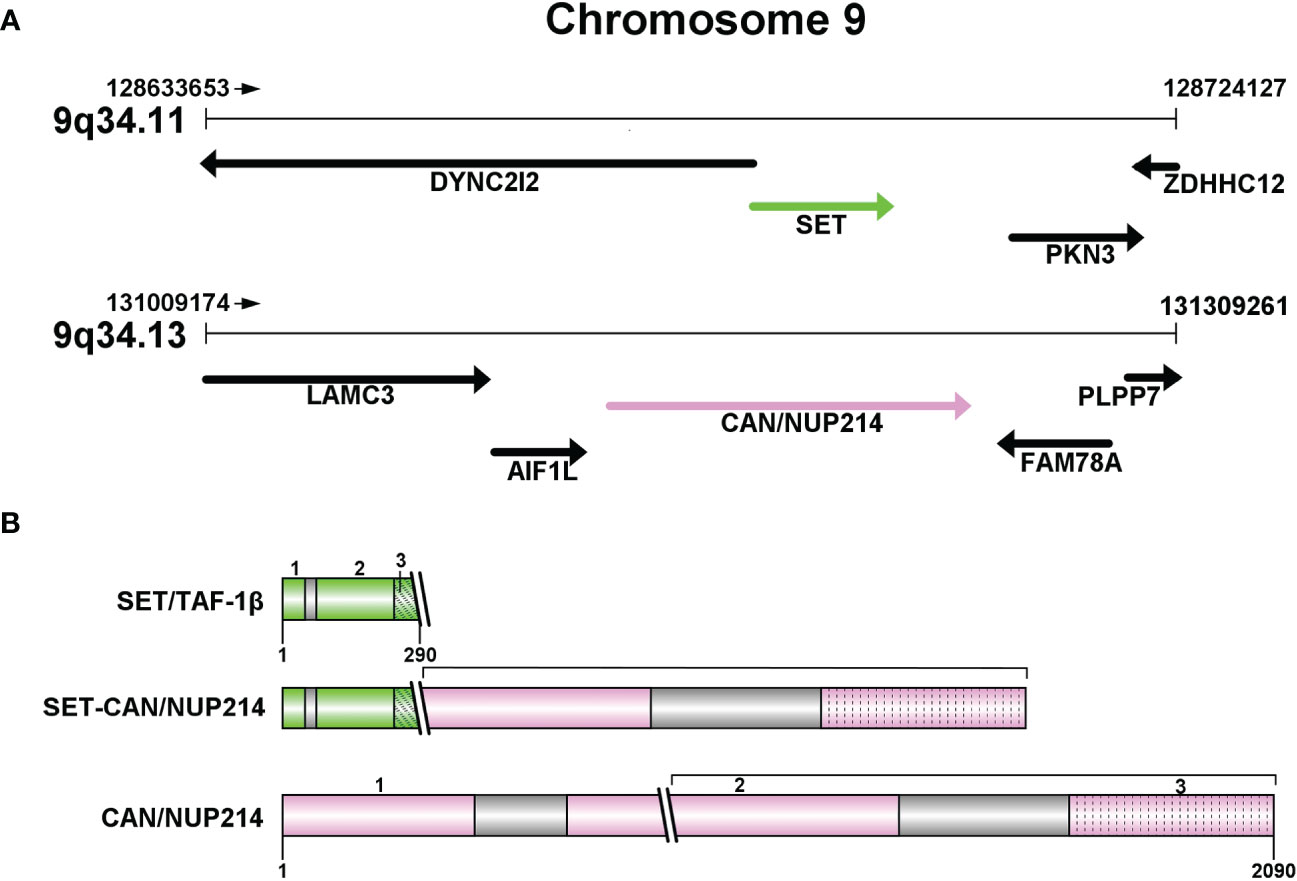
Figure 1 (A) Partial structure of chromosome 9 long arm (9q34): SET at 9q34.11 and CAN/NUP214 at 9q34.13. (B) Protein structures of SET/TAF-1β, CAN/NUP214 and SET-CAN/NUP214. SET/TAF-1β: 1-3: N-terminal dimerization domain; “Earmuff” domain; acidic and negatively charged C-terminal domain. CAN/NUP214: 1-3: β-propeller; coiled-coil region; FG repeats C-terminal region.
SET, also referred to as TATA box binding protein-associated factor 1 (TAF1). SET is a component of the histone acetyltransferase inhibitor (Inhat), which has been reported to be a putative oncogene involved in transcription by regulating chromatin organization (16). SET encodes a protein which can exert an inhibitory effect on apoptosis induced by cytotoxic T lymphocytes (4). In eukaryotic cells, the occurrence of selective splicing in the first two exons of the TAF1 gene results in the formation of two forms of SET expression: the two heterodimeric forms, TAF1-α and TAF1-β (1). Whereas in SET-CAN/NUP214, only the TAF1-β isoform is present (17). The structure of SET/TAF1-β consists of three parts: an N-terminal dimerization domain, a central “Earmuff” domain named for its headphone-like structure, and an acidic and negatively charged C-terminal domain (Figure 1). SET/TAF-Iβ has a variety of different activities, such as inhibiting phosphatase 2A activity, inducing cell transformation and differentiation, and transferring histones to naked DNA. The structural and negative regulatory functions may be related to glucocorticoid resistance (16, 18, 19).
SET-CAN/NUP214 fusion gene encodes a protein containing an almost complete portion of SET fused to the carboxy-terminal two-thirds of CAN, which is a rare gene rearrangement occurs primarily in hematological malignancies (3). The appearance of the fusion gene may be the result of prior cancer therapy, but it may also occur de novo.
SET-CAN/NUP214 positive patients often show resistance to chemotherapy including glucocorticoids, but the mechanism is not completely clear. The optional treatment has not been determined, previous studies have adopted different treatment options with varying prognoses for patients. Some previous studies have shown that SET-CAN/NUP214 fusion gene positive patients have a worse prognosis (3, 7, 20), while clinical studies have shown that there is no significant difference in 3-year event-free survival (EFS) and overall survival (OS) between patients with SET-CAN/NUP214 fusion gene positive and SET-CAN/NUP214 negative patients (21, 22). Conventional techniques such as chromosomal karyotype analysis may have limitations in detecting patients with SET-CAN/NUP214. Due to the emergence of more advanced detection techniques such as fluorescence in situ hybridization (FISH), previously challenging fusion genes like SET-CAN/NUP214 can now be detected with increasing frequency. This necessitates more precise disease classification and optimization of therapeutic regimens. Research shows that HSCT can improve the prognosis, the level of SET-CAN/NUP214 after transplantation can predict recurrence to a certain extent (23), new methods such as CAR-T may be effective for patients and further research is needed (24).
In this review, we summarized the general features and clinical advances of SET-CAN/NUP214 fusion gene in leukemia.
The cases and literature cited and included in this review were retrieved by Jingyu Song and his colleagues using PubMed, Web of Science, Google Scholar, and metstr databases or websites.
The whole screening process is shown in Figure 2. First we exhaustively searched the literature through the databases or websites, and in this step of the search we disregarded the country of publication and time constraints of the literature in order to obtain more comprehensive results. After the search was completed, we performed the exclusion of duplicates and initial screening. Next, by scanning the full-text content, we screened the literature based on its content and excluded incomplete and missing information, leaving behind content that (1) contained complete information and data (2) related to clinical cases, basic research, or reviews of SET-CAN/NUP214.
After completing the screening, we proceeded to the integration of viewpoints and statistics of cases.
We analyzed the statistical case data by SPSS software and performed survival analysis using Kaplan-Meier survival curves.
In the 2022 international consensus classification of acute lymphoblastic leukemia/lymphoma, SET-CAN/NUP214 fusion gene positive has been listed as a subtype of the HOXA gene family in the latest eight temporary entities (25). SET-CAN/NUP214 fusion gene is rare in leukemia patients, and there is no prospective clinical study for such patients. Relevant articles focus on case reports and mechanism studies. This section provides an overview of the general features of patients.
According to a statistic in 2016, a total of 42 SET-CAN/NUP214 positive patients were reported up to that year, including T-ALL(38/42,90.5%), AUL(2/42,4.8%), AML(1/42;2.4%) and B-ALL(1/42;2.4%) (4), another study involving 59 T-ALL patients showed that about 10.3% of T-ALL patients carried SET-CAN/NUP214 fusion gene (20), Ben Abdelli et al. reported that the positive rate of SET-CAN/NUP214 fusion gene in 196 patients with T-ALL was about 5.6% (21), in 2022, Yan C and others first reported two CML patients with positive SET-CAN/NUP214 fusion gene (7). The data revealed that although SET-CAN/NUP214 fusion gene occurs in various types of leukemia, it mainly occurs in T-ALL. This review compiled relevant literature containing more complete patient characteristics published since the emergence of the first SET-CAN/NUP214 fusion gene positive case to date, some articles were not included due to lack of patient information, a total of 81 patients’ information was collected, the overall statistical characteristics of the patients are listed in Table 1, and detailed information on the individual characteristics of the patients are listed in Table 2. Among the 81 patients in Table 2, there are 57(57/81, 70.4%) patients with T-ALL, which is much higher than other types, consistent with the conclusion that the fusion gene is more likely to occur in T-ALL.
Among the fusion gene positive patients counted in this review, there are 59 male and 22 female patients, respectively, with the proportion of male patients reaching more than 70%, suggesting that the SET-CAN/NUP214 fusion gene is more likely to occur in male patients. The number of fusion gene positive T-ALL patients included 40 males and 17 females, with the proportion of males reaching 70.2%. Although there were fewer cases of other types of leukemia, there were still significantly more males than females, which suggests that the type of leukemia in fusion gene positive patients may not be an influencing factor in the proportion of males and females in the disease (4, 7, 20, 21).
There is a large difference in the age of patients at initial diagnosis, the youngest patient is only 8 years old (T-ALL), the oldest patient is 58 years old (T-ALL), the average age is 30.2 years old and the patients are distributed in all age groups (6, 17, 24, 26, 35, 41). Relatively speaking, the probability of fusion gene positive in adult leukemia patients is higher (40). Two CML patients with SET-CAN/NUP214 fusion gene positive were 37 and 42 years old, far from the average age of fusion gene positive patients. However, due to the small number of cases and the older age of CML patients, the relationship between age and fusion gene could not be established.
In previous cases, the patients with fusion gene positive leukemia did not show symptoms different from those with fusion gene negative leukemia, and most remained symptomatic with classic anemia, fever, and lower sternal segment tenderness. However, liver and spleen enlargement, lymph node enlargement, mediastinal involvement, as well as tumor bulk and rapid growth were more common than in fusion gene negative patients (5, 29, 34, 42). Some patients came to see doctors because of liver and spleen enlargement and related symptoms caused by mediastinal mass. Sang-Guk Lee et al. described a 28-year-old patient who complained of dyspnea and chest pain. Physical examination found that multiple lymph nodes in the neck were swollen. Chest CT showed that mediastinal mass compressed the main pulmonary artery with pleural effusion and splenomegaly. Finally, the patient was diagnosed as SET-CAN/NUP214 positive T-ALL (34). Song Y et al. (43) also confirmed that patients often have extramedullary infiltration at the onset of the disease, including areas such as the skin, liver and breast. According to the statistics, the median WBC count of the patients was 18.0×109/L. Based on the collected patient information, the highest WBC count was 604.4 × 109/L (T-ALL) and this patient died 5 months after diagnosis. The median percentage of leukemic blasts in the bone marrow was high (82.0-97.0%), probably reflecting the high proliferation status of fusion gene positive patients (4, 24).
Patients with fusion gene positive may have normal chromosome karyotype or complex karyotype, the existence of a complex karyotype may mask the presence of the fusion gene (34, 44). As a molecular abnormality with low frequency, this is also the reason why SET-CAN/NUP214 patients were not widely concerned at first.
In terms of immunophenotype, the fusion gene positive leukemia cells showed characteristics of extreme immaturity. Flow cytometry showed that their most frequent immunophenotype was CD7, except for the two CML cases mentioned previously (7), only one T-ALL patient and one AML patient reported by Zhang H (6)and Rosati R (29) did not detect CD7+. CD7 was highly frequent in SET-CAN/NUP214 fusion gene positive leukemia, and the other immunophenotypes with higher frequency were cCD3, CD34, CD33 and CD13. The immunophenotypic results suggest that the transformation of fusion gene positive leukemia may occur in the early stage of myeloid or T-lymphocyte differentiation, and it may be related to the inhibition of differentiation of primitive progenitor cells by the fusion gene (6, 7, 35, 45, 46).
Generally, myeloid markers such as CD13 and CD33 are only expressed in about 19% of T-ALL cases. The reason why the fusion gene induces myeloid marker expression remains to be further investigated.
SET-CAN/NUP214 fusion gene impairs the process of hematopoietic differentiation, but it alone is not sufficient to induce leukemia. Additional chromosomal aberrations and molecular events are required to mediate the development of leukemia. Understanding the process is greatly helpful for understanding the disease.
SET-CAN/NUP214 fusion gene may contribute to leukemia through direct and indirect effects. Saito S et al. (47) developed transgenic mice expressing SET-CAN/NUP214, which is active in different groups of hematopoietic cell groups, and the transgenic mice carrying SET-CAN/NUP214 gradually developed symptoms such as anemia, thrombocytopenia and splenomegaly, so that within 6 months, a considerable number of transgenic mice died successively, the course and characteristics of the lesions are more similar to those of leukemias, and the characterization of bone marrow cells in mice during the course of the disease showed that the SET-CAN/NUP214 fusion gene increased the number of immature cells and impaired the hematopoietic differentiation of erythroid, granulocytic, and megakaryocytic lineages (47).
Previous studies have shown that the fusion gene impairs the process of hematopoietic differentiation, but cannot induce the occurrence of leukemia alone. HOXA upregulation may be the key mechanism and play an intermediary role. HOX genes is a kind of gene that specially regulates biological form in organisms. The expression of HOX gene in various organisms is similar, and its sequence is related to its action sequence and action position. Human HOX gene can be divided into four gene clusters: HOXA, HOXB, HOXC, and HOXD, which are located on different chromosomes respectively. The DNA sequence of these gene family members is similar to the protein sequence transcribed. Quantitative and comparative analysis of bone marrow samples from SET-CAN/NUP214 positive patients during initial diagnosis and morphological remission using RT-PCR and other detection methods showed that the expression level of HOXA9 and HOXA10 at initial diagnosis was 3.53 and 4.15 times higher than that during morphological remission, while the expression level of HOXA5 was similar (34). Sang-Guk Lee and others found that the up-regulation of HOXA gene was caused by the interaction of SET-CAN/NUP214 fusion gene with XPO1, hDOT1L and HOXA promoters. The fusion genes with similar mechanism include CALM-AF10 and MLL-AF10 (33), which can lead to the H3K79 hypermethylation of HOXA genes and mediate the occurrence of leukemia. Hypermethylation and subsequent upregulation of HOXA genes play an important role in the pathogenesis of leukemia with positive fusion gene (48). Gorello P et al. detected the overexpression of HOXA7, HOXA9 and HOXA10 in the fusion gene positive patients selected from 256 ALL patients (7, 42). 17 fusion gene positive patients were detected by FISH analysis, and all patients(17/17, 100%) had overexpression of HOXA gene. There were also studies that summarized the up-regulation of HOXA gene with the positive expression of several NUP214 fusion gene subtypes. SET-NUP214, DEK-NUP214 and SQSTM1-NUP214 have the same characteristics, which can lead to the up-regulation of HOXA3, HOXA5, HOXA7, HOXA9, HOXA10 and HOXB in the HOX family (2, 33, 49, 50). These studies also confirmed the relationship between HOXA and SET-CAN/NUP214 fusion gene.
Na Lin et al. (40) evaluated common recurrent mutations in SET-CAN/NUP214 positive T-ALL patients through next-generation sequencing. The results showed that mutations were more common in NOTCH1(23/31,74.2%), PHF6(11/21,52.38%), KRAS(6/14,42.86%), JAK3(4/12,33.33%), CCND3(3/12,25%), JAK1(3/15,20%), STAT5B(2/10,20%), DNM2(2/10,20%) and EED(2/10,20%), these are common recurrent mutations in SET-CAN/NUP214 positive patients in T-ALL and ETP-ALL. The patients with fusion gene positive are accompanied by more molecular events than those with fusion gene negative. These complex molecular events may promote adverse reactions to induction therapy, and may also be one of the factors of poor prognosis (51). As the total number of cases remains low, these issues remain to be explored.
The protein encoded by NOTCH gene is a highly conserved cell surface receptor, which can regulate the development of a variety of biological cells. NOTCH signaling can affect a series of normal life processes of cells, including the differentiation of pluripotent progenitor cells, cell apoptosis, proliferation and cell boundary formation. The abnormality of NOTCH signaling is related to esophageal cancer, gastric cancer, leukemia and other diseases, Among them, abnormal NOTCH1 is most often detected in tumor diseases.
The activation mutation of NOTCH1 or the inactivation mutation of NOTCH1 negative regulatory factor(FBXW7) can be found in about 60% of T-ALL cases. However, the proportion of NOTCH1 mutation seems to be higher in SET-CAN/NUP214 positive leukemia patients. A gene sequencing of 6 SET-CAN/NUP214 positive T-ALL patients by Dai HP et al.(Jiangsu Institute of Hematology, China) showed most T-ALL patients with positive fusion gene have NOTCH1 mutations(5/6,83.3%) and PHF6 mutations(4/6,66.7%) (20). The next-generation sequencing of patients by Na Lin et al. (40) showed that the proportion of NOTCH1 mutations in 31 patients reached 74.2%, similarly, the results of the test performed by Wang Q et al. (52) on the association between 96 fusion gene positive patients and mutations such as NOTCH, JAK1 and others demonstrated a possible positive correlation between NOTCH1 mutations and fusion gene positivity.
The mutations of NOTCH1, PHF6 and JAK1 are closely linked in the process of leukemia, which may be the secondary genetic alterations of SET-CAN/NUP214 fusion gene. PHF6 is a tumor suppressor gene with transcriptional regulation linked to the X sex chromosome in the nucleus. Tumorigenic mutations have a higher incidence rate in T-ALL and can also be seen in AML, most of them occur in male patients. JAK1 plays a key role in initiating reactions related to a variety of major cytokine receptor families. It appears in about 20% of adult T-ALL patients, generally indicating poor prognosis. If the patients with positive fusion gene have co-mutation of NOTCH1 and PHF6, they are more likely to have symptoms such as splenomegaly and lymph node enlargement (2, 22, 52–54). In addition, the existence of SET-CAN/NUP214 fusion gene is related to the up-regulation of the expression level of lymphoblastic leukemia-associated hematopoietic regulator 1(LYL1) and myocyte enhancer 2C(MEF2C) genes (22). Contrary to the common mutations such as NOTCH1, PHF6 and JAK1, the overexpression of CALM-AF10, SIL-TAL, TLX1 or TLX3 is mutually exclusive with the existence of SET-CAN/NUP214 fusion gene. A gene test of 11 fusion gene positive T-ALL patients by Ben et al (21). showed that none of the 11 patients expressed CALM-AF10, SIL-TAL, TLX1 or TLX3(0/11,0%).
In the process of leukemogenesis mediated by SET-CAN/NUP214 fusion gene, it is generally accepted that additional chromosomal aberrations also play a role. Chae H et al. (35) reported del (12)(p13)/ETV6 in 3 of 4 patients, while Ben et al. (21) found this aberration numerous times in their cases. Similarly, the patients in the reports also presented del (6) (q21q23) and del (11) (q22q23) chromosomal aberrations (51, 55). The recurrent chromosomal aberrations in the rare fusion gene positive patients are intriguing and worth pondering.
The prognosis of patients with positive SET-CAN/NUP214 fusion gene is different. Most studies consider that the prognosis is poor. The prognosis of patients may vary due to leukemia classification, concomitant molecular events, treatment plan and the age stage. Patients generally showed delayed response and drug resistance to chemotherapy including glucocorticoids, but studies showed that this drug resistance might not have a negative impact on clinical outcomes (21). Yang Q et al. demonstrated that the prognosis of T-ALL patients with SET-CAN/NUP214 was quite poor, their treatment of three patients with fusion gene positive showed that none of the three patients achieved complete remission(CR) during chemotherapy, and all of them were infected by drug-resistant bacteria such as Candida tropicalis and Pseudomonas aeruginosa. Because of the disease progress and the inability to control the concurrent infection, two patients died during chemotherapy (3). Gorello P et al. also found that the prognosis of fusion gene positive patients was poor. In this study, 6 of the 7 patients received treatment, of which 4 patients died 12 to 24 months after treatment. The main causes of death were refractory disease and leukemia recurrence (7). The treatment results of 6 patients by Dai HP et al. showed that 4 of the 6 patients had recurrence (the median recurrence time was only 7.8 months), and 3 of them died (20). There are also studies show that the positive fusion gene has no effect on the clinical outcome of patients. In the study of Ben et al., the difference between the 3-year total survival rate(3y OS) and event-free survival rate(3y EFS) of fusion gene positive patients and fusion gene negative patients is not statistically significant(3y OS:73% vs 68%; 3y EFS:45% vs 59%) (21, 24), while in the study of Chen B et al, the 3-year overall survival rate(3y OS) and event-free survival rate(3y EFS) of 8 fusion gene positive patients were 87.5% and 70% respectively (22). It can be seen that the outcomes of patients in different clinical trials vary greatly, and finding more effective treatment methods may be beneficial to patients.
Patients with positive SET-CAN/NUP214 fusion gene usually exhibit general resistance to chemotherapy regimens including glucocorticoids in the early stages of induction therapy. Although patients have a delayed response to chemotherapy, the overall CR rate is not affected (40).
The relevant research evaluated patients based on in vitro drug sensitivity screening, monitoring of blasts during induction and MRD results after induction. Compared with the patients with negative fusion gene, the rate of corticosteroid resistance in patients with positive fusion gene(91% of patients had corticosteroid resistance, while the data of patients with negative fusion gene was only 44%) and the rate of early chemotherapy resistance (nearly 100% of patients had early chemotherapy resistance, and only 44% of patients with negative fusion gene) were significantly higher (3, 34, 39, 40).
The anti-inflammatory, immunosuppressive and proapoptotic effects of glucocorticoids play an important role in the treatment of various inflammatory, autoimmune and tumor diseases. In the treatment of leukemia, glucocorticoids are involved in various chemotherapy regimens, especially for ALL. Corticosteroid therapy induced GR target gene transcription is also one of the reference treatment options for ALL (56, 57). The powerful role of glucocorticoids is based on the ubiquitous glucocorticoid receptors(GR) in human cells (58), ligands activate GR and bind with glucocorticoid response elements(GREs) in the nucleus. The transcription process starts under the mediation of “coactivators” such as steroid receptor coactivator 1 (SRC1) and glucocorticoid receptor interaction protein 1 (GRIP1). Under pathological conditions, SET is fused with CAN/NUP214, and the SET subtype mainly exists in SET-CAN/NUP214 is TAF1-β. TAF1-β serves as a component in the INHAT complex, which interacts with a variety of trans-acting factors through TAF1-β to inhibit the transcriptional activity of multiple transcription factors and nuclear receptors. Due to this mechanism, Takamasa Ichijo et al. reported that the potential cause of glucocorticoid resistance in patients with positive SET-CAN/NUP214 fusion gene is the co-precipitation of SET-CAN/NUP214 fusion protein and glucocorticoid response element, which inhibits the transcription activity of glucocorticoid receptor and histone acetylation (56, 59). The in vitro experimental data reported by Yang Q and others also believe that the lack of histone acetylation regulation mediated by SET-CAN/NUP214 may be the cause of glucocorticoid resistance in many patients (3).
Even though nearly 100% of SET-CAN/NUP214 fusion gene positive patients exhibit resistance during the early stages of chemotherapy, studies have shown a high complete response rate(26 of 36 patients,72.22%) (40, 60). The CR rate of the 69 patients counted in Table 1 is also relatively high, reaching 75.4% (52/69). The drug resistance situation and mechanism of the patients still need further research, which may be helpful for the selection of chemotherapy regimen.
The optional treatment method of SET-CAN/NUP214 fusion gene positive leukemia has not been determined. We present patients with clear treatment methods and outcome information reported so far in Table 3 for reference. Analyzing the treatment methods and prognosis of previous cases may provide guidance for the establishment of treatment strategies for such patients.
The SET-CAN/NUP214 fusion gene is mainly found in T-ALL patients. Table 3 contains 49 T-ALL patients, of which 18 patients received chemotherapy and 31 patients received transplantation. Among the patients receiving chemotherapy, 7 patients survived, 11 patients died, and 9 patients relapsed; Among the patients receiving transplantation, 18 patients survived, 13 patients died and 13 patients relapsed.
Most patients developed drug resistance at the initial stage of chemotherapy, but 35 T-ALL patients finally achieved complete remission (CR, 35/49, 71.4%), which was similar to the complete remission rate suggested in previous studies (72.22%) (40, 60). Yang Q et al. (3) reported that CLAG chemotherapy combined with asparaginase might be a potential treatment option for adult SET-CAN/NUP214 fusion gene positive T-ALL patients. They implemented VICP chemotherapy for the first two patients (No.52-53) in the case, but the effect was not obvious. The patients eventually died because of the disease progress and uncontrollable infection of drug-resistant bacteria, for the third patient(No.54), the CLAG chemotherapy regimen combined with asparaginase was used. Surprisingly, the patient’s condition was quickly controlled. Na Lin et al. (40) conducted a drug sensitivity screening tests on the leukemic cells of a refractory fusion gene positive T-ALL patient (No.57) with up to 165 drugs, suggesting that the DAE protocol of “AML like treatment” (daunorubicin+cytarabine+etoposide) showed the highest inhibition rate in vitro. At the same time, they suggested that the induction treatment could adopt a 28-day course of chemotherapy such as used in GRAALL 2003 or 2005. The reason why such “AML like treatment” is effective for patients with fusion gene positive may be related to the frequent occurrence of markers such as CD33 and CD34. Carfilzomib may have a strong inhibitory effect on leukemic cells with positive fusion gene. It can mediate the production of reactive oxygen species as an inducer and synergistically enhance the cytotoxicity of dexamethasone. It is worth noting that in the drug sensitivity screening test, the inhibition rate of single drug treatment of carfilzomib is 37.57%, which shows that carfilzomib may also have potential benefits for patients with refractory SET-CAN/NUP214 fusion gene positive T-ALL (40, 61, 62). Unfortunately, carfilzomib is not currently available in China.
In the treatment of fusion gene positive patients, transplantation may benefit more. A literature based comparison of the treatment methods of patients shows that the average survival time of the chemotherapy group was 22.5 months, the average survival time of the transplantation group was 50.1 months, the average survival time of the chemotherapy group was less than half of that in the transplantation group (24). The statistical analysis shows that hematopoietic stem cell transplantation (HSCT) can significantly improve the survival rate of patients, we can consider that only chemotherapy for patients with fusion gene positive is not enough. The total 3-year overall survival rate (3y OS) of the 9 patients with fusion gene positive T-ALL who received allogeneic hematopoietic stem cell transplantation was 73% (21), which is similar to the outcome of the patients with fusion gene negative after allogeneic hematopoietic stem cell transplantation. This suggests that transplantation can significantly improve the prognosis of patients. It may be a good choice to complete the transplantation at the right time in the first CR.
In this review, we screened 46 effective cases from 49 T-ALL patients in Table 3 (excluding No. 32, No. 34 and No. 56), 30 patients received transplantation, of which 13 died with a median survival of 49 months, 16 patients received chemotherapy, of which 11 died with a median survival of 20 months. The difference between the two groups was tested to be statistically significant (P=0.012). We listed the Kaplan-Meier survival curves of the patients in Figure 3, and it is clear that for T-ALL patients, transplantation can significantly improve the survival status and prolong the overall survival.
CAR-T may play a role in acute leukemia patients with positive fusion gene. The expression frequency of CD7 in previous cases is close to 100%. Research shows that CD7 may play a role in promoting chemoresistance and accelerating disease progression in leukemia (63, 64). Gomes-silva et al. (65) demonstrated that CAR-T targeting CD7 can delay disease progression and prolong patient survival in the mouse model. In the MPAL case reported by Li MY et al. (27) (no. 3), they performed two times of CAR-T cell infusion treatment on the patients who had relapsed after HSCT, which significantly improved the patient’s condition. By the end of follow-up, the patients had survived for more than 42 months. It suggests the application prospect of CAR-T technology in the treatment of fusion gene positive leukemia, which is worthy of further exploration and research.
In this review, 12 patients(12/70) with fusion gene positive AML were included, and the number was second only to T-ALL. Although the chemotherapy regimen of 12 patients was not the same, they all achieved complete remission(CR, 12/12, 100%), all patients received HSCT. Finally, 9 patients survived, 3 patients died and 4 patients relapsed. Chen SM et al. (28) showed that the survival data of SET-CAN/NUP214 fusion gene positive AML patients were similar to those of fusion gene negative patients.
To date, only four patients with fusion gene positive B-ALL have been reported. Similarly, their treatment process was very difficult. The two patients reported by Nowak NJ et al. (32) and Hong HZ et al. (4) were resistant to chemotherapy, and have not achieved complete remission. Unfortunately, the report didn’t mentioned the follow-up of the two patients. One of the two patients(No.20-21) reported by Chen SM et al. (28) achieved complete remission, and both patients received HSCT, but they died of graft-versus-host disease (GVHD) and relapse respectively 9 and 15 months after transplantation. Although the sample of related B-ALL cases is small, we can still speculate that the patients with SET-CAN/NUP214 fusion gene positive B-ALL may have poor prognosis.
Chen Y et al. (5) first reported two rare cases of SET-CAN/NUP214 fusion gene positive CML in 2022 (No.18-19), the two patients detected BCR-ABL1 and SET-CAN/NUP214 fusion transcripts after 7 and 2 years of treatment with tyrosine kinase inhibitor (TKI), one patient (no.18) received chemotherapy and HSCT, and still survived up to the end of follow-up (95.7 months after initial diagnosis, 6.5 months after transplantation), the other patient (No.19) gave up treatment and died 36 months after the initial diagnosis. Retrospective analysis of samples from two patients showed that SET-CAN/NUP214 fusion transcript was present at the initial diagnosis, but not during TKI treatment. The disease progression of CML is slow and typically categorized into three phases. The chronic phase (CP) is often asymptomatic but may include mild fatigue, emaciation, and splenomegaly on physical examination. The accelerated phase (AP) is characterized by fever, progressive splenomegaly, and the appearance of additional chromosomal abnormalities. The acute transformation stage (BP) is marked by the continued deterioration of symptoms and signs. Additional chromosome abnormalities play an important role in the deterioration of CML in chronic phase (CP) and accelerated phase (AP), SET-CAN/NUP214 fusion gene may be used as the main clone in CML to promote disease transformation, and its combination with BCR-ABL1 accelerates disease progression. Similar to the treatment of other fusion genes in CML cases, high intensity TKI chemotherapy and HSCT may be more effective for these patients (5, 66).
SET-CAN/NUP214 fusion gene has also been found in AUL, MS and MPAL. The incidence of AUL is relatively rare. It is considered to be the result of clone expansion and maturation stagnation of undifferentiated hematopoietic cells, and does not express myeloid or lymphoid specific antigen. MS is a limited tumor formed by the proliferation and infiltration of myeloid primitive cells or immature myeloid cells outside the marrow. It may occur in association with various myeloproliferative disorders or in isolation. The lesions are mostly located in a single site, and sometimes multifocal or multiorgan involvement is present (67, 68). In this review, a case of SET-CAN/NUP214 fusion gene positive MS patient (No.2) was included. During the treatment, the patient also suffered from bone marrow compression and pericardial effusion. The incidence of MPAL in acute leukemia is relatively low, accounting for only 2-5% of acute leukemia cases. At present, MPAL lacks a unified treatment option, and the prognosis of patients is usually worse than AML or ALL (69). Li MY et al. (27) treated a 29-year-old SET-CAN/NUP214 fusion gene positive MPAL patient identified by them (Table 3, No.3) with induction and consolidation therapy leading to CR and transplanted the patient, but the patient relapsed six months later, followed by a lymphocyte consumption program based on fludarabine (30 mg/m2, 1-3days) and cyclophosphamide (300 mg/m2, 1-3days) and CAR-T cell therapy. The patient ultimately survived greater than 42 months. Chen SM et al. (28) used the treatment regimen CODLP or VPIA (vincristine+prednisone+daunorubicin+cytarabine) and transplantation for two patients(Table 3, No.4-5) with positive SET-CAN/NUP214 fusion gene positive MPAL who were 22 years old and 34 years old. Both patients ultimately survived to the end of the follow-up period(survival>42 months and>24 months).
Among the 70 patients counted in Table 3, 28 patients relapsed and 30 patients died. Relapse and death are common clinical outcomes in SET-CAN/NUP214 fusion gene positive leukemia. We need to monitor the prognosis of patients with some indicators and detection methods, so as to better evaluate the condition of patients and timely intervene.
Current research shows that the detection of SET-CAN/NUP214 fusion gene may be a minor residual disease (MRD) with early recurrence, or an early indicator of poor prognosis (24). Chen SM et al. carried out a long-term continuous monitoring of SET-CAN/NUP214 gene transcript level in 24 patients, and learned that the expression level of fusion gene was lower than 0.001% continuously, which was a sign of good prognosis. The median time of morphological relapse in patients with expression level higher than 0.001% was only 5 months. Gao MG et al. (41) studied the prognostic significance of fusion gene expression level before and after allogeneic hematopoietic stem cell transplantation for patients. The expression level of fusion gene after transplantation is higher than 0.02%, which is an effective indicator of patients’ relapse. Monitoring the expression level of SET-CAN/NUP214 fusion gene through RQ-PCR is more sensitive than flow cytometry (FCM), its sensitivity for detection of various genetic abnormalities and mutation types can reach 10-5, whereas the sensitivity of FCM is usually at 10-4 (70). 4 of the 5 patients with relapse after transplantation have SET-CAN/NUP214+ before relapse, and their FCM detection results are negative. Previous studies also emphasized the significance of MRD monitoring in transplantation. Positive MRD before transplantation may indicate poor prognosis after transplantation (70, 71).
In summary, SET-CAN/NUP214 fusion gene is relatively rare in leukemia and mainly occurs in adult male T-ALL patients. It has also been reported in AUL, MS, MPAL, AML, CML and B-ALL. Patients are generally resistant to chemotherapy, and the prognosis in different diseases may be different. The clinical symptoms of positive and negative fusion gene patients are relatively similar, and the common immunephenotypes are CD7, cCD3, CD34, CD33 and CD13. The karyotypes may be normal or complex, the concomitant molecular events can become the influencing factors of disease progression and prognosis. HSCT can significantly improve the survival rate of patients, CAR-T is also a potential treatment method. RQ-PCR is an effective monitoring method, and the monitoring of fusion gene may be more sensitive than FCM. Prognosis prediction and recurrence intervention based on the expression level of SET-CAN/NUP214 fusion gene can improve the treatment effect. Further research is needed to evaluate the role of SET-CAN/NUP214 fusion gene in leukemia.
JS: Data curation, Investigation, Methodology, Software, Visualization, Writing – original draft. HL: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. SF: Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (NSFC, 81903966) and the Science Foundation of the First Affiliated Hospital of Harbin Medical University (2021M02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Mendes A, Fahrenkrog B. Nup214 in leukemia: it’s more than transport. Cells (2019) 8(1). doi: 10.3390/cells8010076
2. Zhou MH, Yang QM. Nup214 fusion genes in acute leukemia (Review). Oncol Lett (2014) 8(3):959–62. doi: 10.3892/ol.2014.2263
3. Yang Q, Qian H, Jin Z, Yu Z, Yu K, Zhang S, et al. Set-can fusion gene as poor prognosis predictor in adult T-cell acute lymphoblastic leukemia. Leuk Lymphoma (2020) 61(1):217–20. doi: 10.1080/10428194.2019.1660966
4. Zhu HH, Zhao XS, Qin YZ, Lai YY, Jiang H. B-cell acute lymphoblastic leukemia associated with set-nup214 rearrangement: A case report and review of the literature. Oncol Lett (2016) 11(4):2644–50. doi: 10.3892/ol.2016.4260
5. Chen Y, Wang Q, Cen J, Xu C, Tao TT, Xie J, et al. Blast phase of chronic myeloid leukemia with concurrent bcr::Abl1 and set::Nup214: A report of two cases. Mol Carcinog (2023) 62(2):117–21. doi: 10.1002/mc.23480
6. Zhang H, Zhang L, Yan_ Li, Wang X. Set-can fusion gene in acute leukemia and myeloid neoplasms: report of three cases and a literature review. OncoTargets Ther (2020). doi: 10.2147/OTT.S258365
7. Gorello P, La Starza R, Di Giacomo D, Messina M, Puzzolo MC, Crescenzi B, et al. Sqstm1-nup214: A new gene fusion in adult T-cell acute lymphoblastic leukemia. Haematologica (2010) 95(12):2161–3. doi: 10.3324/haematol.2010.029769
8. Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer (2004) 4(2):106–17. doi: 10.1038/nrc1274
9. Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol (2010) 11(7):490–501. doi: 10.1038/nrm2928
10. Wente SR, Rout MP. The Nuclear Pore Complex and Nuclear Transport. Cold Spring Harb Perspect Biol (2010) 2(10):a000562. doi: 10.1101/cshperspect.a000562
11. Wälde S, Kehlenbach RH. The part and the whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol (2010) 20(8):461–9. doi: 10.1016/j.tcb.2010.05.001
12. Roloff S, Spillner C, Kehlenbach RH. Several phenylalanine-glycine motives in the nucleoporin nup214 are essential for binding of the nuclear export receptor crm1. J Biol Chem (2013) 288(6):3952–63. doi: 10.1074/jbc.M112.433243
13. Daelemans D, Costes SV, Lockett S, Pavlakis GN. Kinetic and molecular analysis of nuclear export factor crm1 association with its cargo in vivo. Mol Cell Biol (2005) 25(2):728–39. doi: 10.1128/MCB.25.2.728-739.2005
14. Fu SC, Huang HC, Horton P, Juan HF. Validness: A database of validated leucine-rich nuclear export signals. Nucleic Acids Res (2013) 41(Database issue):D338–43. doi: 10.1093/nar/gks936
15. Saito S, Cigdem S, Okuwaki M, Nagata K. Leukemia-associated nup214 fusion proteins disturb the xpo1-mediated nuclear-cytoplasmic transport pathway and thereby the nf-kappab signaling pathway. Mol Cell Biol (2016) 36(13):1820–35. doi: 10.1128/MCB.00158-16
16. Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, et al. Relationship between the structure of set/taf-ibeta/inhat and its histone chaperone activity. Proc Natl Acad Sci U.S.A. (2007) 104(11):4285–90. doi: 10.1073/pnas.0603762104
17. Lindern MV, Breems D, Baal SV, Adriaansen H, Grosveld G. Characterization of the translocation breakpoint sequences of two dek-can fusion genes present in T(6;9) acute myeloid leukemia and a set-can fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer (2010) 5(3):227–34. doi: 10.1002/gcc.2870050309
18. Chae YC, Kim KB, Kang JY, Kim SR, Jung HS, Seo SB. Inhibition of foxo1 acetylation by inhat subunit set/taf-ibeta induces P21 transcription. FEBS Lett (2014) 588(17):2867–73. doi: 10.1016/j.febslet.2014.06.053
19. Wang D, Kon N, Lasso G, Jiang L, Leng W, Zhu WG, et al. Acetylation-regulated interaction between P53 and set reveals a widespread regulatory mode. Nature (2016) 538(7623):118–22. doi: 10.1038/nature19759
20. Dai HP, Wang Q, Wu LL, Ping NN, Chen SN. [Expression of set-nup214 fusion gene in patients with T-cell acute lymphoblastic leukemia and its clinical significance]. J Exp Hematol (2012) 20(5):1047–51.
21. Ben Abdelali R, Roggy A, Leguay T, Cieslak A, Renneville A, Touzart A, et al. Set-nup214 is a recurrent gammadelta lineage-specific fusion transcript associated with corticosteroid/chemotherapy resistance in adult T-all. Blood (2014) 123(12):1860–3. doi: 10.1182/blood-2013-08-521518
22. Chen B, Jiang L, Zhong ML, Li JF, Li BS, Peng LJ, et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U.S.A. (2018) 115(2):373–8. doi: 10.1073/pnas.1717125115
23. Ni X, Li Z, Li X, Zhang X, Bai G, Liu Y, et al. Socioeconomic inequalities in cancer incidence and access to health services among children and adolescents in China: A cross-sectional study. Lancet (2022) 400(10357):1020–32. doi: 10.1016/S0140-6736(22)01541-0
24. Wang J, Q-r Z, Lu XX, Zhang LJ, Wang XX, Zhang HY. The characteristics and prognostic significance of the set-can/nup214 fusion gene in hematological Malignancies: A systematic review. Medicine (2022) 101(30). doi: 10.1097/md.0000000000029294
25. Duffield AS, Mullighan CG, Borowitz MJ. International consensus classification of acute lymphoblastic leukemia/lymphoma. Virchows Arch (2023) 482(1):11–26. doi: 10.1007/s00428-022-03448-8
26. Kim J, Lee SG, Song J, Kim SJ, Rha SY, Lee KA, et al. Molecular characterization of alternative set-nup214 fusion transcripts in a case of acute undifferentiated leukemia. Cancer Genet Cytogenet (2010) 201(2):73–80. doi: 10.1016/j.cancergencyto.2010.05.010
27. Li MY, Lin ZH, Hu MM, Kang LQ, Wu XX, Chen QW, et al. Secondary donor-derived humanized cd19-modified car-T cells induce remission in relapsed/refractory mixed phenotype acute leukemia after allogeneic hematopoietic stem cell transplantation: A case report. biomark Res (2020) 8:36. doi: 10.1186/s40364-020-00216-1
28. Chen SM, Song WJ, Qin YZ, Wang Z, Dang H, Shi Y, et al. [Analysis of the clinical characteristics of 24 cases of hematological Malignancies with set-nup214 fusion gene]. Zhonghua Xue Ye Xue Za Zhi (2021) 42(6):459–65. doi: 10.3760/cma.j.issn.0253-2727.2021.06.004
29. Rosati R, Starza RL, Barba G, Gorello P, Mecucci C. Cryptic chromosome 9q34 deletion generates taf-ialpha/can and taf-ibeta/can fusion transcripts in acute myeloid leukemia. Haematologica (2007) 92(2):232–5. doi: 10.3324/haematol.10538
30. Jeong IH, An GD, Lim HH, Woo KS, Kim KH, Kim JM, et al. A rare case of acute myeloid leukemia with set-nup214 fusion and massive hyperdiploidy. Ann Lab Med (2019) 39(4):403–5. doi: 10.3343/alm.2019.39.4.403
31. Zheng YZ, Wen JJ, Wang LY, Zheng H, Hua XL, Li J, et al. [Set-nup214-positive pediatric acute myeloid leukemia: A report of two cases]. Zhonghua Xue Ye Xue Za Zhi (2021) 42(9):769. doi: 10.3760/cma.j.issn.0253-2727.2021.09.011
32. Nowak NJ, Sait SN, Zeidan A, Deeb G, Gaile D, Liu S, et al. Recurrent deletion of 9q34 in adult normal karyotype precursor B-cell acute lymphoblastic leukemia. Cancer Genet Cytogenet (2010) 199(1):15–20. doi: 10.1016/j.cancergencyto.2010.01.014
33. Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, et al. The recurrent set-nup214 fusion as a new hoxa activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood (2008) 111(9):4668–80. doi: 10.1182/blood-2007-09-111872
34. Lee SG, Park TS, Cho SY, Lim G, Park GJ, Oh SH, et al. T-cell acute lymphoblastic leukemia associated with complex karyotype and set-nup214 rearrangement: A case study and review of the literature. Ann Clin Lab Sci (2011) 41(3):267.
35. Chae H, Lim J, Kim M, Park J, Kim Y, Han K, et al. Phenotypic and genetic characterization of adult T-cell acute lymphoblastic leukemia with del(9)(Q34);Set-nup214 rearrangement. Ann Hematol (2012) 91(2):193–201. doi: 10.1007/s00277-011-1289-x
36. Li WJ, Cui L, Gao C, Zhao XX, Liu SG, Xin YP, et al. [Gene rearrangement pattern of immunoglobulin and T-cell receptor (Ig/tr) and its clinical characteristics in children with set-nup214 fusion gene-positive leukemia/lymphoma]. J Exp Hematol (2011) 19(6):1362.
37. Lee EY, Park TS, Kim MJ, Chang MH, Cho EH, Park SJ, et al. Detection of set-nup214 rearrangement using multiplex reverse transcriptase-polymerase chain reaction (Rt-pcr) in acute leukemias: A case report and literature review on a korean case series. Ann Hematol (2012) 91(7):1135–8. doi: 10.1007/s00277-011-1366-1
38. Prokopiou C, Koumas S, Neokleous N, Seimeni O, Barmpouti A. Set-nup214 rearrangement in isolation is insufficient to induce leukemia: A single center experience. Leuk Lymphoma (2016) 57(2):451–2. doi: 10.3109/10428194.2015.1049169
39. Xu X, Zhai Q, Jin H, Yu Y, Han D, Zhang H, et al. Set-nup214 fusion gene involved early T-cell precursor acute lymphoblastic leukemia in adult with B marker expression. Int J Gen Med (2021) 14:659–64. doi: 10.2147/IJGM.S294715
40. Lin N, Liu Z, Li Y, Yan X, Wang L. Determining the appropriate treatment for T-cell acute lymphoblastic leukemia with set-can/nup214 fusion: perspectives from a case report and literature review. Front Oncol (2021) 11:651494. doi: 10.3389/fonc.2021.651494
41. Gao MG, Hong Y, Qin YZ, Chang YJ, Wang Y, Zhang XH, et al. Prognostic significance of set-nup214 fusion gene in acute leukemia after allogeneic hematopoietic stem cell transplantation. Med (Baltimore) (2020) 99(50):e23569. doi: 10.1097/MD.0000000000023569
42. Liu F, Gao L, Jing Y, Xu YY, Ding Y, Zhou MH, et al. Detection and clinical significance of gene rearrangements in chinese patients with adult acute lymphoblastic leukemia. Leuk Lymphoma (2013) 54(7):1521–6. doi: 10.3109/10428194.2012.754888
43. Song Y, Gong XY, Wei SN, Li QH, Zhang GJ, Wang Y, et al. Clinical analysis of set - Nup214 fusion gene positive patients with acute leukemia. J Exp Hematol (2023) 31(2):6. doi: 10.19746/j.cnki.issn1009-2137.2023.02.007
44. Falini B, Mecucci C, Tiacci E, Alcalay M, Martelli MF. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. New Engl J Med (2005) 352(3):254–66. doi: 10.1056/NEJMoa041974
45. Uckun FM, Sather HN, Gaynon PS, Arthur DC, Trigg ME, Tubergen DG, et al. Clinical features and treatment outcome of children with myeloid antigen positive acute lymphoblastic leukemia: A report from the children’s cancer group. Blood (1997) 90(1):28–35.
46. Kandilci A, Mientjes E, Grosveld G. Effects of set and set-can on the differentiation of the human promonocytic cell line U937. Leukemia (2004) 18(2):337–40. doi: 10.1038/sj.leu.2403227
47. Saito S, Nouno K, Shimizu R, Yamamoto M, Nagata K. Impairment of erythroid and megakaryocytic differentiation by a leukemia-associated and T(9;9)-derived fusion gene product, set/taf-ibeta-can/nup214. J Cell Physiol (2008) 214(2):322–33. doi: 10.1002/jcp.21199
48. Fabarius A, Leitner A, Hochhaus A, Muller MC, Hanfstein B, Haferlach C, et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of cml: long-term observation of 1151 patients from the randomized cml study iv. Blood (2011) 118(26):6760–8. doi: 10.1182/blood-2011-08-373902
49. Birthe F. Nucleoporin gene fusions and hematopoietic Malignancies. New J Science,2014,(2014-5-27) (2014) 2014:1–18.
50. Sandahl JD, Coenen EA, Forestier E, Harbott J, Johansson B, Kerndrup G, et al. T(6;9)(P22;Q34)/dek-nup214-rearranged pediatric myeloid leukemia: an international study of 62 patients. Haematologica (2014) 99(5):865–72. doi: 10.3324/haematol.2013.098517
51. Gorello P, La Starza R, Varasano E, Chiaretti S, Elia L, Pierini V, et al. Combined interphase fluorescence in situ hybridization elucidates the genetic heterogeneity of T-cell acute lymphoblastic leukemia in adults. Haematologica (2010) 95(1):79–86. doi: 10.3324/haematol.2009.010413
52. Wang Q, Qiu H, Jiang H, Wu L, Dong S, Pan J, et al. Mutations of phf6 are associated with mutations of notch1, jak1 and rearrangement of set-nup214 in T-cell acute lymphoblastic leukemia. Haematologica (2011) 96(12):1808–14. doi: 10.3324/haematol.2011.043083
53. Weng AP, Ferrando AA, Lee W, JPt M, LB S, Sanchez-Irizarry C, et al. Activating mutations of notch1 in human T cell acute lymphoblastic leukemia. Science (2004) 306(5694):269–71. doi: 10.1126/science.1102160
54. Brown FC, Still E, Koche RP, Yim CY, Takao S, Cifani P, et al. Mef2c phosphorylation is required for chemotherapy resistance in acute myeloid leukemia. Cancer Discovery (2018) 8(4):478–97. doi: 10.1158/2159-8290.CD-17-1271
55. Andreasson P, Johansson B, Arheden K, Billström R, Mitelman F, Höglund M, et al. Deletions of cdkn1b and etv6 in acute myeloid leukemia and myelodysplastic syndromes without cytogenetic evidence of 12p abnormalities. Genes Chromosomes Cancer (1997) 19(2):77–83. doi: 10.1002/(sici)1098-2264(199706)19:2<77::aid-gcc2>3.0.co;2-x
56. Ichijo T, Chrousos GP, Kino T. Activated glucocorticoid receptor interacts with the inhat component set/taf-ibeta and releases it from a glucocorticoid-responsive gene promoter, relieving repression: implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with set-can translocation. Mol Cell Endocrinol (2008) 283(1-2):19–31. doi: 10.1016/j.mce.2007.10.014
57. Inaba H, Pui CH. Glucocorticoid use in acute lymphoblastic leukaemia. Lancet Oncol (2010) 11(11):1096–106. doi: 10.1016/S1470-2045(10)70114-5
58. Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid receptors and associated diseases. Essays Biochem (2004) 40:137–55. doi: 10.1042/bse0400137
59. Mckenna NJ. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev (1999). doi: 10.1210/er.20.3.321
60. Huang ZF, Wang TY, Fu MW, Liu W, Hao M, Qiu LG, et al. [Treatment and prognosis of adult T cell acute lymphoblastic leukemia]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao (2019) 41(4):485–91. doi: 10.3881/j.issn.1000-503X.10807
61. Hosseini MS, Mohammadi MH, Vahabpour Roudsari R, Jafari L, Mashati P, Gharehbaghian A. Proteasome inhibition by carfilzomib induced apotosis and autophagy in a T-cell acute lymphoblastic leukemia cell line. Iran J Pharm Res (2019) 18(Suppl1):132–45. doi: 10.22037/ijpr.2020.112692.13898
62. Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J (2017) 7(6):e577. doi: 10.1038/bcj.2017.53
63. Chang H, Yeung J, Brandwein J, Yi QL. Cd7 expression predicts poor disease free survival and post-remission survival in patients with acute myeloid leukemia and normal karyotype. Leuk Res (2007) 31(2):157–62. doi: 10.1016/j.leukres.2006.06.001
64. Chang H, Salma F, Yi QL, Patterson B, Brien B, Minden MD. Prognostic relevance of immunophenotyping in 379 patients with acute myeloid leukemia. Leuk Res (2004) 28(1):43–8. doi: 10.1016/s0145-2126(03)00180-2
65. Gomes-Silva D, Atilla E, Atilla PA, Mo F, Tashiro H, Srinivasan M, et al. Cd7 car T cells for the therapy of acute myeloid leukemia. Mol Ther (2019) 27(1):272–80. doi: 10.1016/j.ymthe.2018.10.001
66. Salem A, Loghavi S, Tang G, Huh YO, Jabbour EJ, Kantarjian H, et al. Myeloid neoplasms with concurrent bcr-abl1 and cbfb rearrangements: A series of 10 cases of a clinically aggressive neoplasm. Am J Hematol (2017) 92(6):520–8. doi: 10.1002/ajh.24710
67. Claerhout H, Van Aelst S, Melis C, Tousseyn T, Gheysens O, Vandenberghe P, et al. Clinicopathological characteristics of de novo and secondary myeloid sarcoma: A monocentric retrospective study. Eur J Haematology (2018) 100(6):603–12. doi: 10.1111/ejh.13056
68. Podgaetz E, Kriegsmann M, Dincer EH, Allan JS. Myeloid sarcoma: an unusual presentation for acute tracheal stenosis. Clin Respir J (2016) 10(6):800–4. doi: 10.1111/crj.12287
69. Khan M, Siddiqi R, Naqvi K. An update on classification, genetics, and clinical approach to mixed phenotype acute leukemia (Mpal). Ann Hematol (2018) 97(6):945–53. doi: 10.1007/s00277-018-3297-6
70. Mo XD, Lv M, Huang XJ. Preventing relapse after haematopoietic stem cell transplantation for acute leukaemia: the role of post-transplantation minimal residual disease (Mrd) monitoring and mrd-directed intervention. Br J Haematol (2017) 179(2):184–97. doi: 10.1111/bjh.14778
Keywords: SET-CAN/NUP214 fusion gene, leukemia, T-cell acute lymphoblastic leukemia (T-ALL), acute myeloid leukemia (AML), molecular anomaly, treatment, prognosis
Citation: Song J, Li H and Fan S (2023) SET-CAN/NUP214 fusion gene in leukemia: general features and clinical advances. Front. Oncol. 13:1269531. doi: 10.3389/fonc.2023.1269531
Received: 30 July 2023; Accepted: 02 October 2023;
Published: 16 October 2023.
Edited by:
Massimiliano Bonifacio, University of Verona, ItalyReviewed by:
Jessica Heath, University of Vermont Children’s Hospital, United StatesCopyright © 2023 Song, Li and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huibo Li, bGlodWliby0xOTg1QDE2My5jb20=; Shengjin Fan, ZmFuc2pobXVAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.