- Gastrointestinal Surgery, The First Hospital of Shanxi Medical University, Taiyuan, Shanxi, China
Background: Dysregulation of the long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been linked to some oncogenic pathways that induce cancer initiation and progression. This meta-analysis was conducted to specifically summarize the most recent research on MALAT1 function in human gastric cancer (GC).
Methods: The eligible studies were first identified by searching HowNet, Web of Science, PubMed, The Cochrane Library, Embase, and Nature databases for studies published as of April 1, 2023. The meta-analysis included 14 studies assessing MALAT1 expression and presenting clinical parameters and survival outcomes.
Results: The results illustrated that high MALAT1 expression is predictive of lymph node metastasis (pooled odds ratio [OR] = 2.99, 95% confidence interval [CI] = 1.97–4.54, P < 0.001) and distant metastasis in GC (OR = 3.11, 95% CI = 1.68–5.75, P < 0.001). In addition, MALAT1 was associated with GC tumor invasion (T3/T4 vs. T1/T2: OR = 2.90, 95% CI = 1.90- 4.41, P <0.001) and TNM stage (III/IV vs I/II: OR = 2.93, 95% CI: 1.80-4.77, P <0.001). Additionally, higher MALAT-1 expression predicted poorer overall survival in patients with GC (hazard ratio = 1.64, 95% CI = 1.20–2.09, P < 0.001).
Conclusions: The current findings suggest that the high MALAT1 expression is an adverse biomarker for prognostic outcomes, lymph node metastasis, TNM stage, and distant metastasis in GC and MALAT1 could be a prognostic biomarker for GC.
1 Introduction
Gastric cancer (GC) is the second most common cause of cancer-related death globally (1). Although the incidence of GC has declined in most regions, its impact on the global disease burden and public health persists (2). Number of deaths caused by GC in 2020, both sexes, all ages, is 768793, about 7.7% of all cancers (Source: Globocan 2020). GC is a heterogeneous disease with a complex pathogenesis and geographic variation. The prognosis of GC is difficult to predict (3, 4). Further research on various aspects of GC is required to better identify the pathogenesis of the disease and uncover more practical biomarkers that predict prognosis and response to therapy.
Long non-coding RNAs (lncRNAs) are functional RNA molecules with a length of more than 200 nucleotides. Most of them have a 5′ cap and 3′ poly-A tails. LncRNAs exert their biological roles by binding to DNA, RNA, or proteins. They can act as intracellular competitive endogenous RNAs to regulate gene expression by directly interacting with miRNAs. They can also modulate target gene expression by altering the binding of transcription factors to promoters. In addition, they can facilitate target genes’ activation or silencing by forming scaffolding complexes with effector molecules (5, 6). lncRNAs were revealed to be closely associated with changes in oncogenic phenotypes including cell proliferation, cell differentiation, invasion, apoptosis, and metastasis (7–10). Based on the published evidence, cancer-associated lncRNAs might represent candidate biomarkers for providing precise diagnoses, assessing individualized prognosis, evaluating targeted therapy, and predicting tumor differentiation (11–13).
The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear transcript 2, which has been mapped to human chromosome 11q13, is more than 8000 nucleotides in length (14, 15). Unlike other lncRNAs, MALAT1 is evolutionarily conserved and widely expressed. MALAT1 was originally identified as a metastasis-associated gene and prognostic marker that can be used to identify patients with metastatic early-stage non-small cell lung cancer (NSCLC) who are at high risk of developing metastatic exacerbation (16).
MALAT1 plays significant roles in numerous human cancers, such as regulating pre-messenger RNA splicing, transcription factors, and histone-modifying enzymes (17). Knockdown of MALAT1 in animal experiments inhibited cell migration and proliferation and resulted in reduced expression of genes such as enhancer of zeste homolog 2 (EZH2), Lin28, β-catenin, octamer-binding transcription factor 4 (OCT4), and epithelial–mesenchymal transition (EMT) (18). MALAT1 silencing might inversely regulate miR-129-5p to block triple-negative breast cancer cell migration, proliferation, and invasion (19). As a competing endogenous RNA (ceRNA), MALAT1 regulates Zinc finger E-box-binding homeobox 1 (ZEB1) expression by sponging miR-143-3p. Hepatocellular carcinoma cell proliferation and invasion can be inhibited by MALAT1 inhibition (20). Oncogenic phenotypic transition caused by MALAT1 was detected in GC (21). The MALAT1 gene is overexpressed in GC and related to its occurrence and growth (22). By serving as a ceRNA for miR-23b-3p in chemotherapy-resistant GC cells, MALAT1 silencing was demonstrated to decrease chemotherapy-induced autophagy (23). In conclusion, accumulated data indicate a connection between MATAT1 dysregulation and the emergence of GC.
Over the past decade, increasing numbers of studies have proved the effect of MALAT1 expression on the prognostic outcomes and clinicopathological parameters of GC. Nevertheless, these studies produced inconsistent or controversial conclusions (22, 23). To resolve this issue, we performed a systematic review and meta-analysis to clarify the association of MALAT1 with GC prognostic or clinical features and recapitulate its tumorigenicity in GC.
2 Materials and methods
2.1 Document retrieval
We searched HowNet, Web of Science, PubMed, The Cochrane Library, Embase, and Nature databases for studies published through April 1, 2023. The following keyword combinations were used in this search: (“long noncoding RNA MALAT1” or “lncRNA MALAT1” or “MALAT1”) and (“gastric cancer” or “stomach tumor”). In addition, the reference lists of all qualified studies were retrieved to ensure that all eligible studies were included in the meta-analysis.
2.2 Study selection
The eligibility criteria were as follows: (1) randomized controlled trials (RCTs) or observational tests; (2) examination of the expression of MALAT1 in patients with GC; (3) separation of patients into high- and low-expression groups along with the expression level of MALAT1; (4) inclusion of overall survival (OS), recurrence-free survival (RFS), disease-free survival (DFS), or other clinical parameters with hazard ratios (HRs) and 95% confidence intervals (CIs) or data that can be used to calculate DFS or OS; and (5) the availability of sufficient data. The following studies were excluded from this study: (1) case reports, reviews, meta-analyses, letters, conference presentations, and editorials; (2) duplicated publications; (3) studies with animal experiments or pure cell experiments; and (4) papers with insufficient data.
2.3 Data extraction and quality assessment
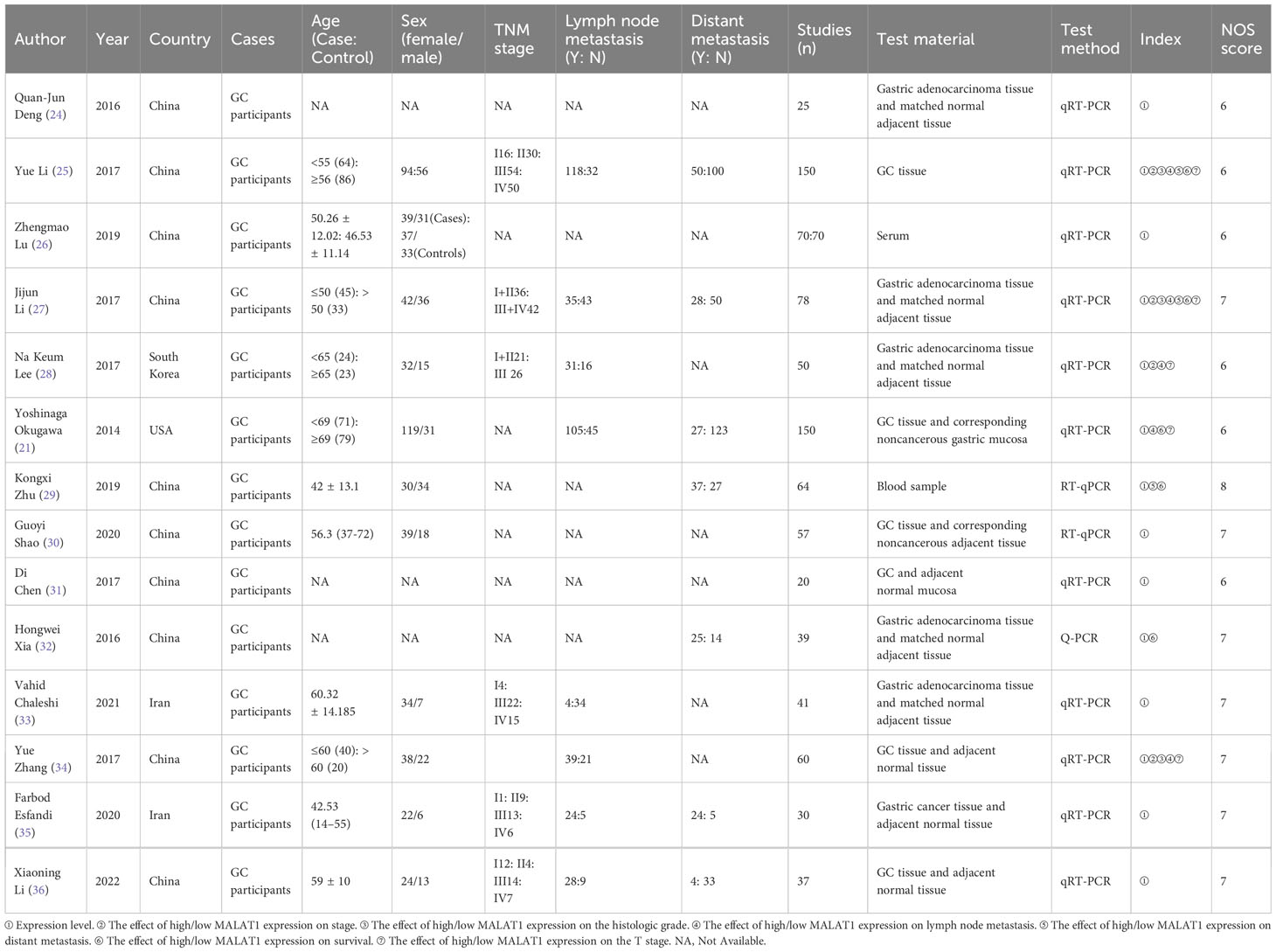
Two authors independently completed data extraction and quality assessment. Any disagreements in the process were resolved by discussion with the third author. The following information was extracted from every article included in this study: name of the first author, year of publication, sex of patients, detection method, number of patients, and source (eg, tissue, serum) of lncRNA MALAT1, indicators in the article (eg, OS, DFS, RFS), and clinical parameters (eg, tumor size, TNM stage, metastasis). In terms of prognostic parameters, HRs and their corresponding 95% CIs were extracted from the article. If an article failed to report the HR or its 95% CI, this information was obtained indirectly, such as by extracting data from survival curves. In addition, the Newcastle–Ottawa Scale (NOS) was used to assess the quality of the included studies. First, some HRs were obtained directly from the article. Second, HRs were estimated using the formula [P0/(1 − P0)]/[P1/(1 − P1)], where P0 is the 5-year survival rate in the low MALAT1 expression group and P1 is the 5-year survival rate in the high MALAT1 expression group. The 95% CI was calculated using the formula exp(lnHR ± 1.96 × SE), where exp is the exponential function, lnHR is the natural logarithm of HR, and SE is the standard error of the HR. Finally, the data were obtained using the survival curves. If these data were analyzed by both univariate and multivariate methods, multivariate analysis was preferred because it has greater precision in explaining confounders. Study quality was assessed using NOS. NOS scores ranged from 0 to 9, and scores ≥ 6 denoted high study quality.
2.4 Statistical analysis
All analyses were performed using Stata 16.0. For prognostic values, such as OS, HRs and the corresponding 95% CIs were used to detect the overall effect. For clinical parameters such as TNM stage, lymph node metastasis, and tumor differentiation, odds ratios (ORs) and their corresponding 95% CIs were used. The I2 test was used to assess the heterogeneity across studies. I2 ≤ 50% indicated the absence of significant heterogeneity among these included studies, and the fixed-effects model was used in the analysis. Otherwise, the random-effects model was used. Funnel plots were made to distinguish bias among included studies. Publication bias in OS meta-analyses was also assessed by Egger’s test.
3 Results
3.1 Identification of the included studies
As presented in Figure 1, 1401 studies were primarily retrieved. After excluding 77 duplicate publications, 1324 papers were retained. Subsequently, 1286 papers were directly excluded after reading the title or abstract, including 466 reviews, 632 irrelevant articles, 188 letters or books, and cell or animal experiments articles. Hence, the full text of 38 papers was reviewed, after which 24 papers were excluded, mainly because of incomplete data or incompatible topics. Ultimately, 14 studies were included in this study.
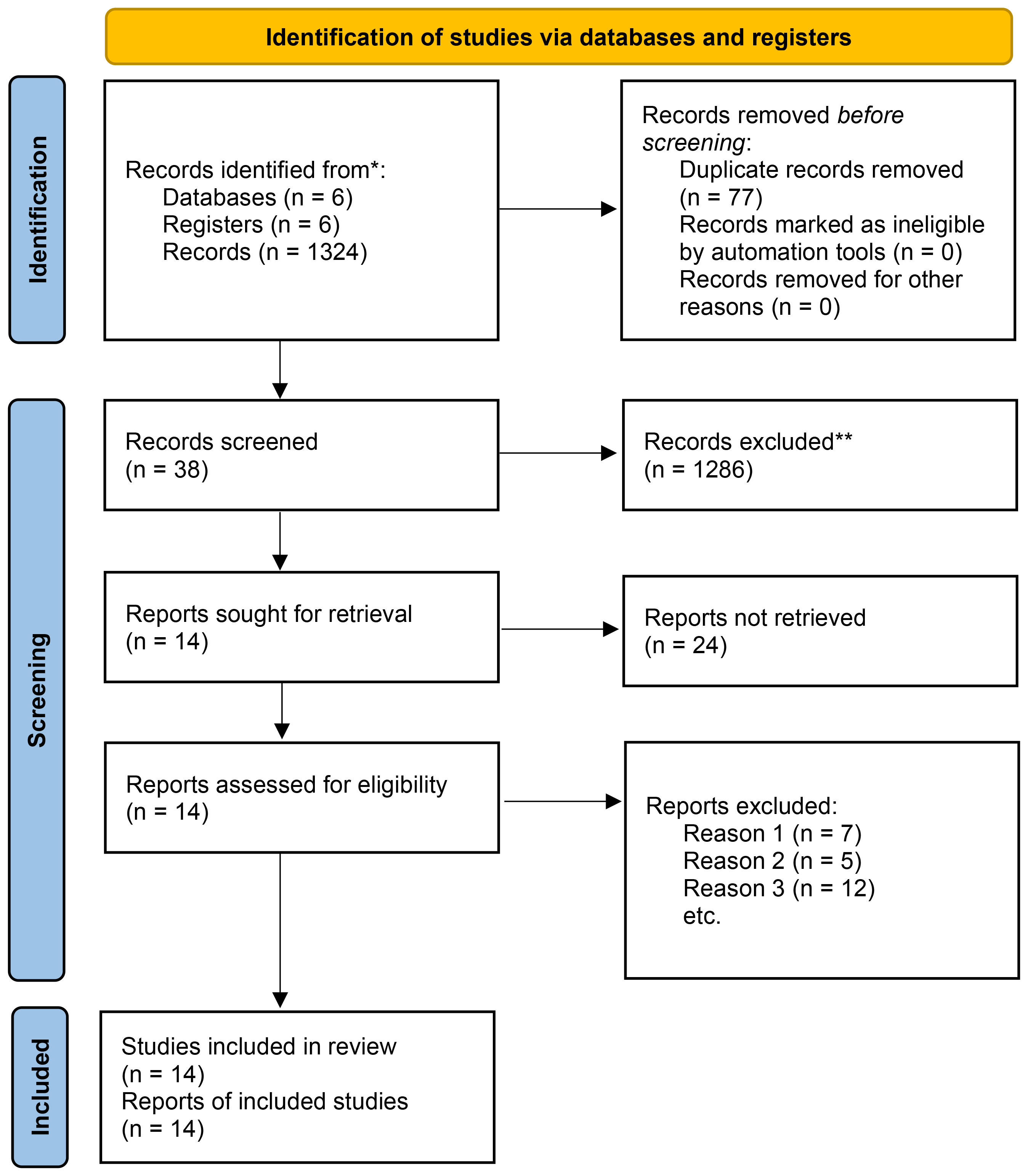
Figure 1 Flowchart of the articles and study selection process. *Consider, if feasible to do so, reporting the number of records identified from each database or register searched (rather than the total number across all databases/registers). **If automation tools were used, indicate how many records were excluded by a human and how many were excluded by automation tools.
3.2 General information
The main characteristics of the included studies are presented in Table 1. The 14 included studies included 10, 2, 1, and 1 study from China, Iran, South Korea, and the United States, respectively, and the studies were published between 2014 and 2023. MALAT1 expression was determined in all studies by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The effect of high/low MALAT1 expression on stage, histologic grade, lymph node metastasis, distant metastasis, survival, and T stage were also analyzed. All studies included the comparison of MALAT1 expression between patients with GC and healthy controls, and MALAT1 expression was higher in patients with GC in all studies. Five studies reported OS data. Of these, three studies presented HRs and 95% CIs, and the needed data were calculated using Kaplan–Meier survival curves for two studies. The NOS score was ≥6 in all studies, indicating high study quality.
3.3 Meta-analysis of clinical parameters
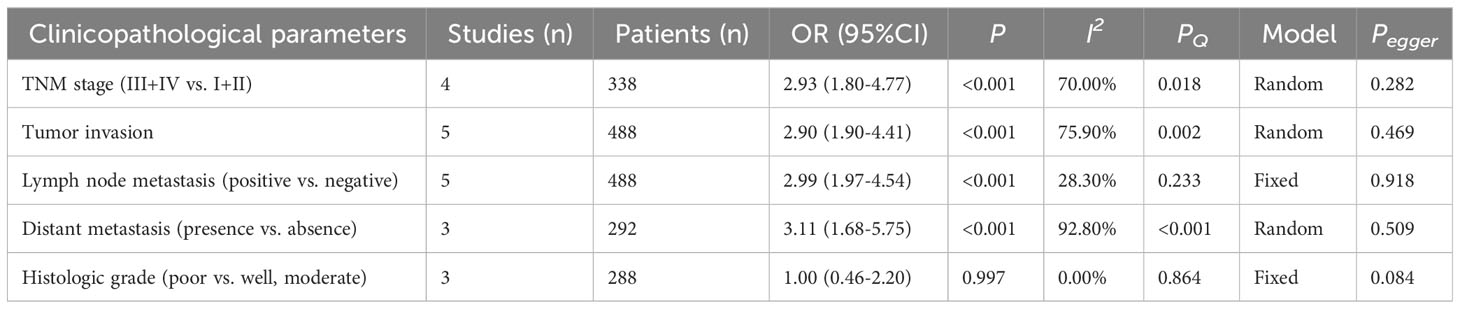
As presented in Table 2 and Figures 2, 3, seven studies were included in the meta-analysis of clinical parameters. Four studies reported the TNM stage (Figure 2A), and high MALAT1 expression predicted an advanced TNM stage (III/IV vs. I/II: OR = 2.93, 95% CI = 1.80–4.77, P < 0.001). Meanwhile, heterogeneity was high among the studies (I2 = 70.00%, PQ = 0.018). Five studies included the stage of tumor invasion (Figure 2B), and high MALAT1 expression was significantly associated with tumor invasion (T3/T4 vs. T1/T2 OR = 2.90, 95% CI = 1.90–4.41, P < 0.001). Heterogeneity was high among the studies (I2 = 74.90%, PQ = 0.002). Five studies included data on lymph node metastasis (Figure 2C), and high MALAT1 expression was significantly related to the presence of lymph node metastasis (positive vs. negative: pooled OR = 2.99, 95% CI = 1.97–4.54, P < 0.001). No significant heterogeneity was detected among the studies (I2 = 28.30%, PQ = 0.788). In addition, three studies reported data on distant metastasis (Figure 2D), and high MALAT1 expression was significantly associated with distant metastasis (positive vs. negative: OR = 3.11, 95% CI = 1.68–5.75, P < 0.001). Significant heterogeneity was detected across the studies (I2 = 92.8%, PQ < 0.001). Three studies reported the tumor histologic grade (Figure 2E). High MALAT1 expression was not associated with the tumor histologic grade (poorly differentiated vs. well-differentiated: OR = 1.00, 95% CI = 0.46–2.20, P = 0.997), and no significant heterogeneity was noted across the studies (I2 = 0.0%, PQ = 0.864). Egger’s test was performed to assess publication bias in the studies, and no significant publication bias was detected (P > 0.05). The symmetrical funnel plot revealed symmetry among clinicopathological outcomes (Figure 3).
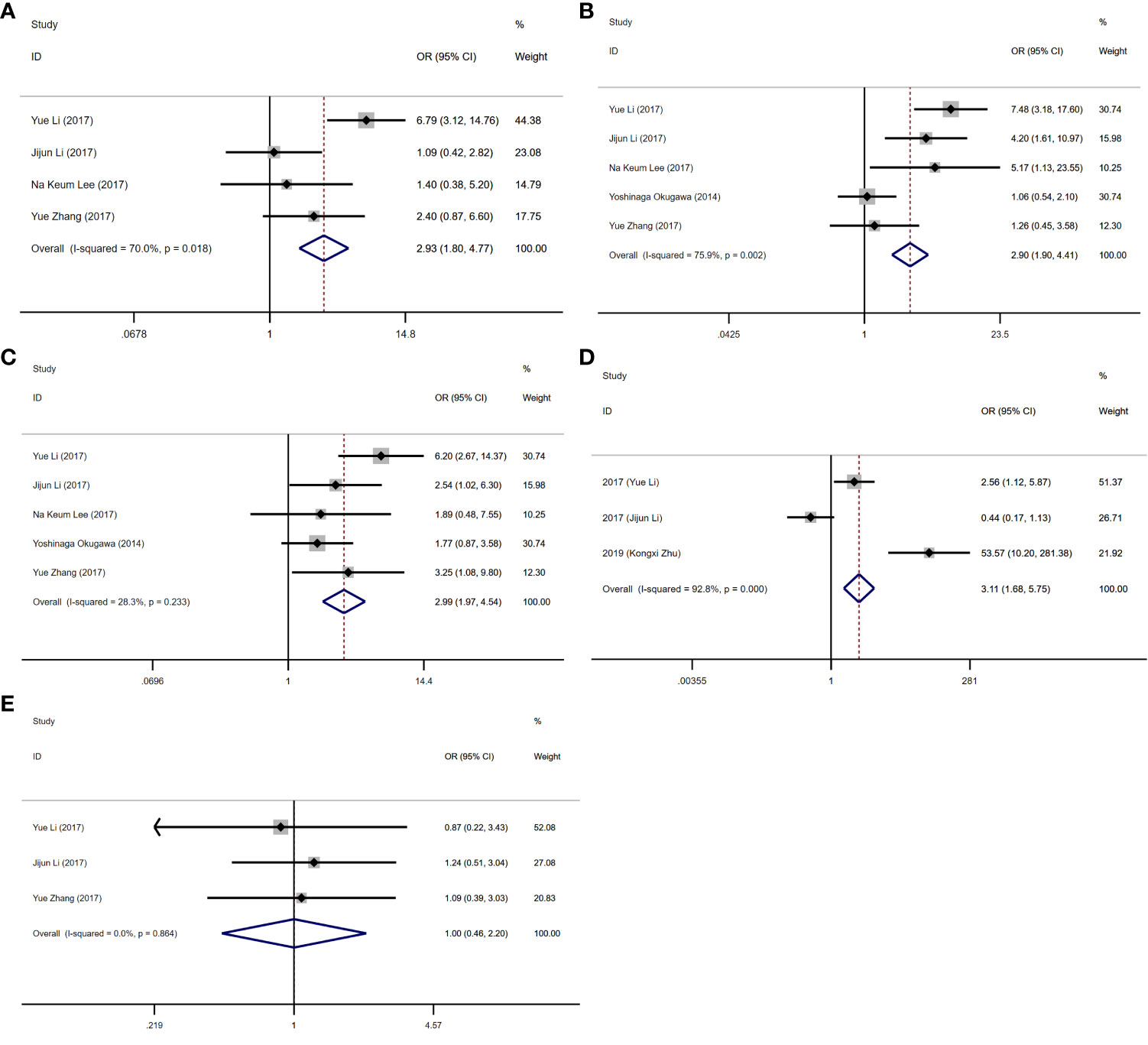
Figure 2 Forest plots of clinicopathological parameters. (A) Forest plot of TNM stage. (B) Forest plot of tumor invasion. (C) Forest plot of lymph node metastasis. (D) Forest plot of distant metastasis. (E) Forest plot of the histologic grade.
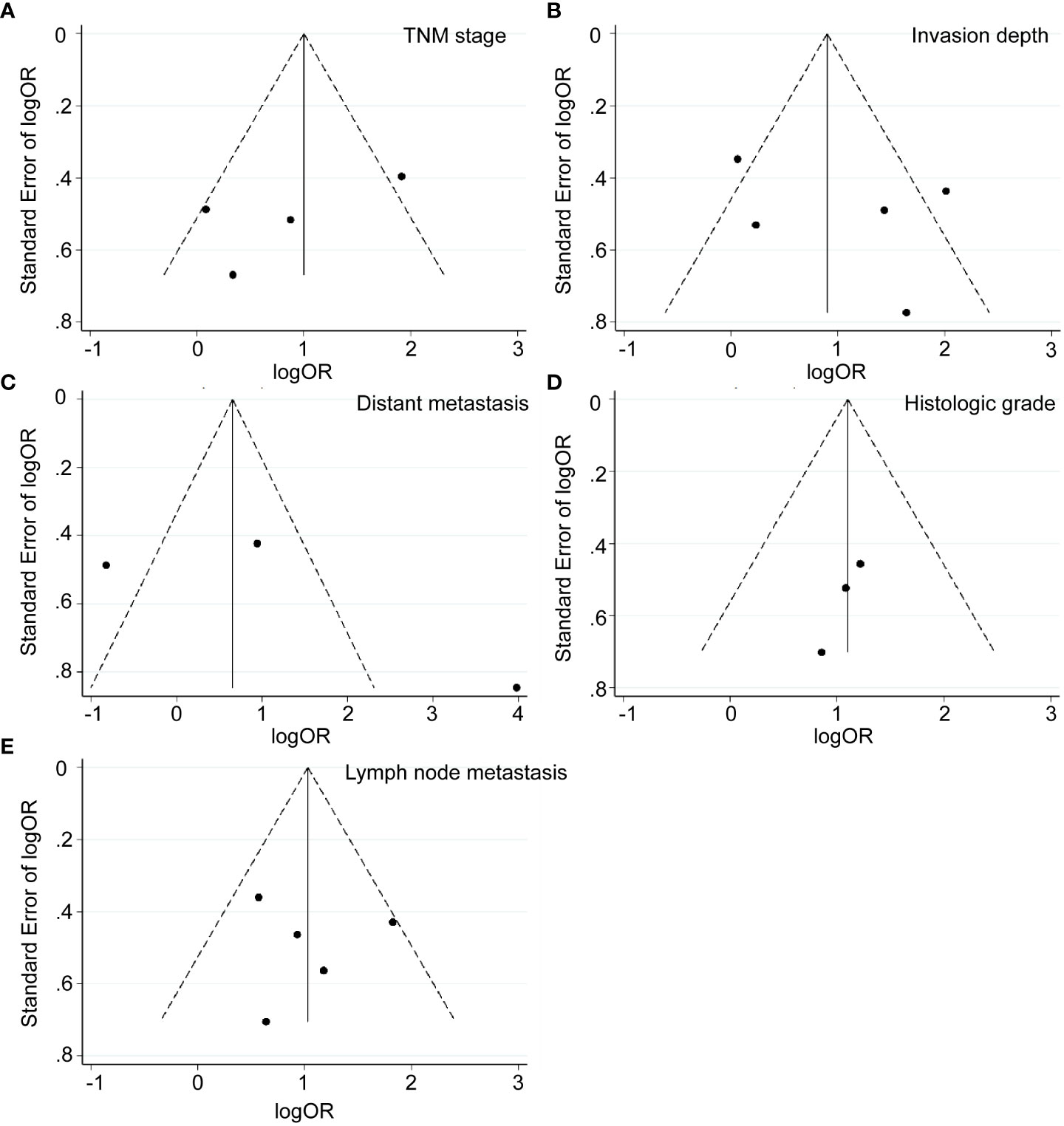
Figure 3 Funnel plots of clinicopathological parameters. (A) TNM stage (stage III/IV vs. Stage I/II). (B) Invasion depth (T3/T4 vs. T1/T2). (C) Distant metastasis (positive vs. negative). (D) Histologic grade (poorly differentiated vs. well-differentiated). (E) Lymph node metastasis (positive vs. negative).
3.4 Association of MALAT1 with survival outcomes
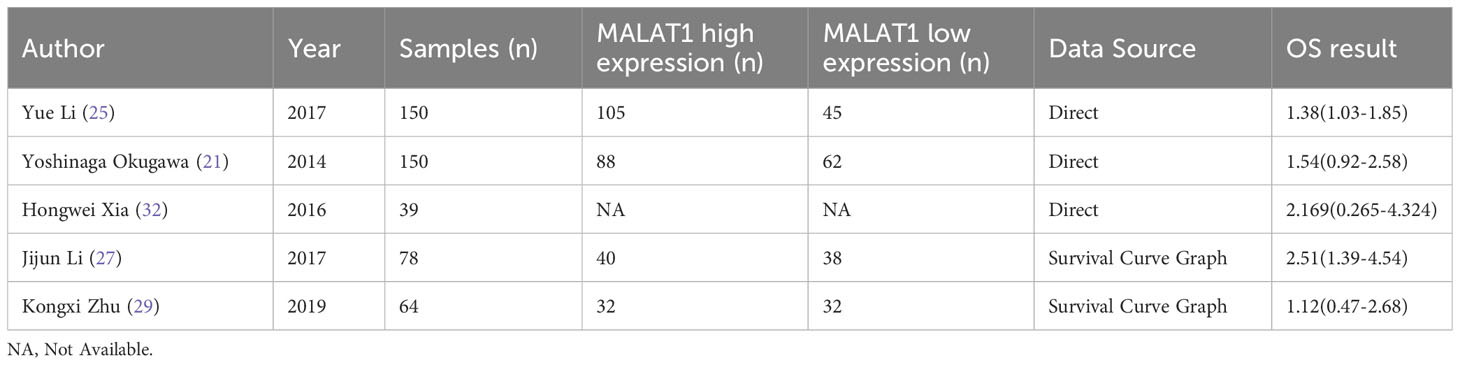
Five eligible studies reported OS according to MALAT1 expression. Because of the lack of heterogeneity across studies (I2 = 0.0%, PQ = 0.417), pooled results were estimated using the fixed-effects model. As presented in Table 3 and Figure 4, elevated MALAT1 expression predicted worse OS in patients with GC (HR = 1.64, 95% CI = 1.20–2.09, P < 0.001). Funnel plots were used to assess publication bias for OS (Figure 5), and the shape of the funnel plots indicated no evidence of asymmetry. In addition, Egger’s results revealed no apparent publication bias for OS (P = 0.70).
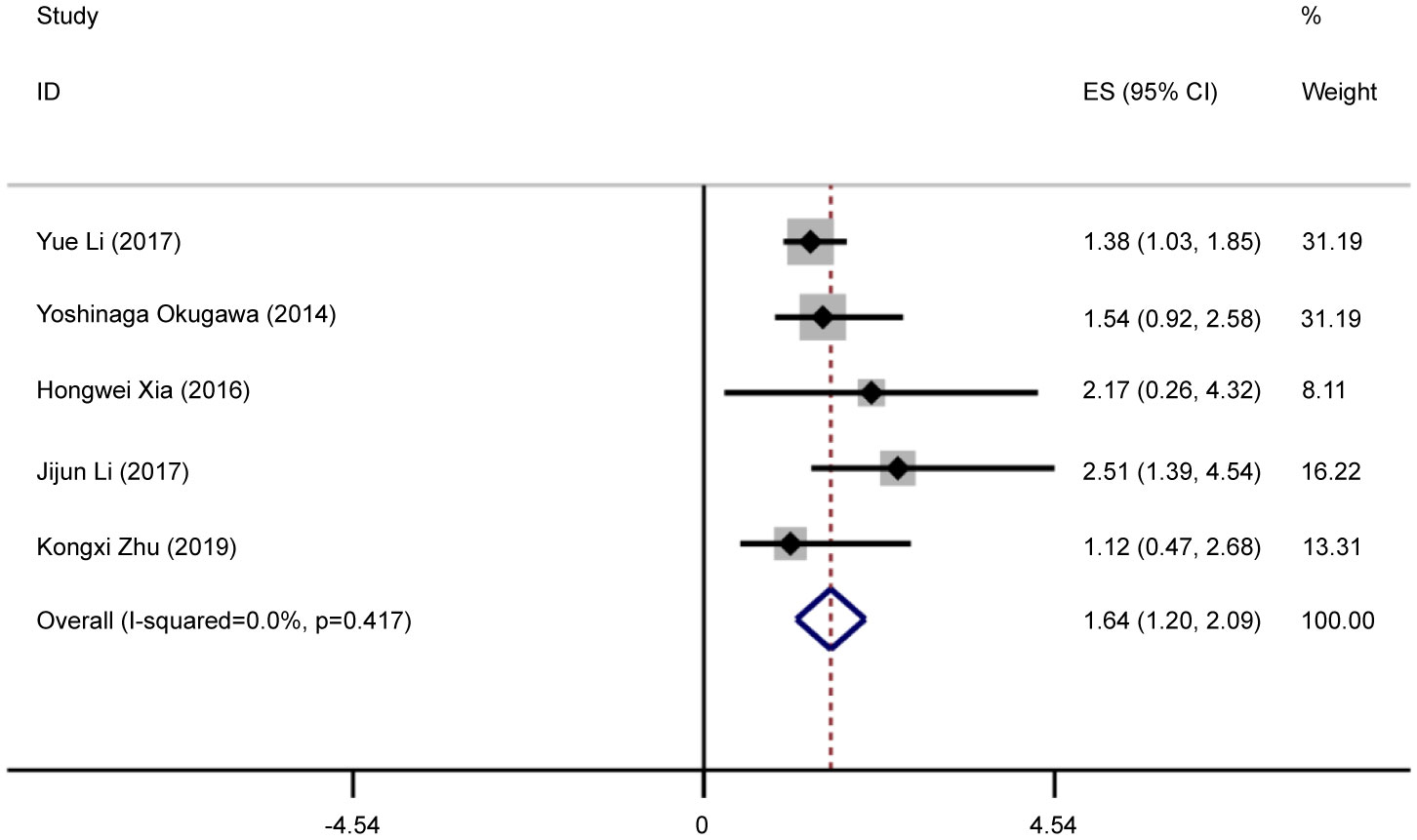
Figure 4 Forest plots of pooled HRs of OS. Elevated MALAT1 expression predicted worse OS in patients with GC (HR = 1.64, 95% CI = 1.20–2.09, P < 0.001).
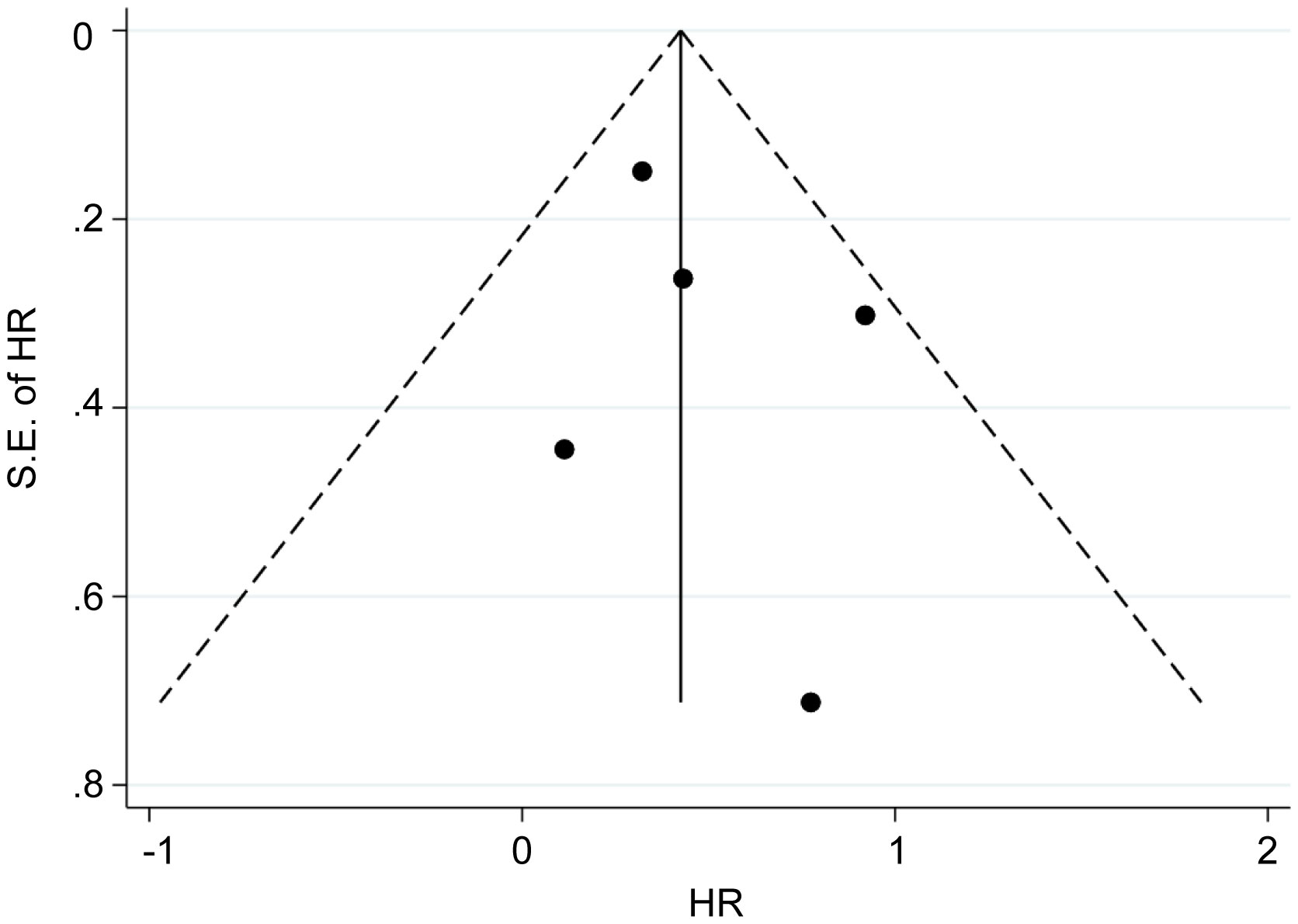
Figure 5 Funnel plots of OS. Funnel plots were used to assess publication bias for OS. The shape of the funnel plots indicated no evidence of asymmetry.
4 Discussion
Despite advances in life and medical sciences, GC remains a global public health problem (37). Therefore, it is necessary to discover the molecular mechanisms of GC progression to prevent tumorigenesis and improve survival. There is increasing evidence that abnormally expressed lncRNAs are involved in progression and tumorigenesis in GC. These lncRNAs participate in many cellular signaling pathways and function as oncogenes or tumor suppressors (38–40). Previous research illustrated that lncRNAs, including MALAT1, are valid predictors of survival outcomes (41, 42).
Several biological roles and abnormal MALAT1 expression have been linked to several cancer types (43–45). In HepG2 cells, MALAT1 inhibition reduced the expression of the transcription factor Oct4, suggesting that MALAT1 can promote stem-like properties in hepatoma cells (46). However, the impact of MALAT1 on GC progression and prognostic outcomes remains controversial. Therefore, to evaluate the clinical and prognostic relevance of MALAT1 in GC, we examined recently published research by meta-analysis.
First, eligible studies were pooled to conduct the meta-analysis. The true link between MALAT1 expression and GC might be more clearly revealed using pooled results because of the correction of confounders that are implicated in many clinical variables. The results illustrated that elevated MALAT1 expression is an effective predictor of GC prognosis, including tumor invasion (T3/T4 vs. T1/T2: OR = 2.90, 95% CI = 1.90- 4.41, P <0.001), lymph node metastasis (pooled odds ratio [OR] = 2.99, 95% confidence interval [CI] = 1.97–4.54, P < 0.001), TNM stage (III/IV vs I/II: OR = 2.93, 95% CI: 1.80-4.77, P < 0.001), and distant metastasis (OR = 3.11, 95% CI = 1.68–5.75, P < 0.001). Furthermore, our study found that patients with high MALAT1 expression had worse OS (HR = 1.64, 95% CI = 1.20–2.09, P < 0.001). Therefore, MALAT1 could be a prognostic biomarker for GC. The findings of this study were similar to those of most prior studies, suggesting that MALAT1 is related to poor prognosis in malignant cancers (47–49). High levels of MALAT1 expression in gastric cancer have been shown to encourage metastasis and development (21). According to a recent clinical investigation, MALAT1 has been linked to colorectal cancer, and patients with stage II/III CRC may have a poorer prognosis if their expression of the gene is high (50). Additionally, it has been demonstrated that MALAT1 is overexpressed in hepatocellular carcinoma, which raises the chance of tumor recurrence following liver transplantation (51).
Nevertheless, because of the limited published data, the study had several limitations. First, the use of different qRT-PCR primer sets could have resulted in heterogeneity across studies. Second, studies might have used different cutoffs for low and high MALAT1 expression. Third, some original studies provided incomplete data. Fourth, confounders such as race can cause significant heterogeneity. Finally, the possibility of a “small study effect” could not be dismissed (52, 53). Thus, larger-scale studies are required to confirm these findings. To our knowledge, this is the first meta-analysis to specifically and systematically evaluate the associations of MALAT1 with lymph node metastasis, distant metastasis, and TNM stage in GC even though some studies partly demonstrated the clinicopathological and prognostic significance of MALAT1 in human cancers (54–57).
In summary, this study suggested that high MALAT1 expression is an adverse biomarker for prognostic outcomes, lymph node metastasis, TNM stage, and distant metastasis in GC. MALAT1 might play a key role in GC tumorigenesis. However, before applying MALAT1 in the treatment and management of GC, additionally high-quality, multi-ethnic, and large-scale studies are required to discover the prognostic value and oncogenic function of MALAT1.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
SB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. JG: Formal analysis, Visualization, Writing – original draft. HZ: Formal analysis, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Natural Science Foundation of Shanxi Province (General Program, No. 201901D111349).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1257120/full#supplementary-material
References
1. Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci (2020) 21(11):4012. doi: 10.3390/ijms21114012
2. Moradzadeh R, Anoushirvani AA. Trend of gastric cancer incidence in an area located in the center of Iran: 2009–2014. J Gastrointestinal Cancer (2020) 51(1):159–64. doi: 10.1007/s12029-019-00227-8
3. La Vecchia C, Conte P. Cancer control in central and eastern Europe. oncologist (2016) 21(10):1161–2. doi: 10.1634/theoncologist.2016-0230
4. Riquelme I, Ili C, Roa JC, Brebi P. Long non-coding RNAs in gastric cancer: mechanisms and potential applications. Oncotarget (2016) 5. doi: 10.18632/oncotarget.9396
5. Xiang A, Wei D, Xiaoge L, Qingling X, Xinhui C, Xuehao Z, et al. Non-coding RNAs regulating mitochondrial function in cardiovascular diseases. J Mol Med (2023) 101(5):501–26. doi: 10.1007/s00109-023-02305-8
6. Liu Y, Ding W, Wang J, Ao X, Xue J. Non-coding RNA-mediated modulation of ferroptosis in cardiovascular diseases. Biomed Pharmacother (2023) 164:114993. doi: 10.1016/j.biopha.2023.114993
7. Spizzo R, MIe A, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene (2012) 31(43):4577–87. doi: 10.1038/onc.2011.621
8. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
9. Xu Y, Qiu M, Chen Y, Wang J, Xia W, Mao Q, et al. tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus (2016) 29(8):950–8. doi: 10.1111/dote.12436
10. Fu D, Zhang Y, Cui H. Long noncoding RNA CCAT2 is activated by E2F1 and exerts oncogenic properties by interacting with PTTG1 in pituitary adenomas. Am J Cancer Res (2018) 8(2):245.
11. Zhang M, Zhao Y, Zhang Y, Wang D, Gu S, Feng W, et al. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochim Biophys Acta (BBA)-Molecular Basis Disease (2018) 1864(5):1770–82. doi: 10.1016/j.bbadis.2018.03.005
12. Zhu Q, Luo Z, Lu G, Gui F, Wu J, Li F, et al. LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to hepatocellular carcinoma cell proliferation, migration and invasion. Biochem Biophys Res Commun (2018) 498(3):551–8. doi: 10.1016/j.bbrc.2018.03.017
13. Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem (2018) 45(5):1904–14. doi: 10.1159/000487966
14. Zhang X, Hamblin MH, Yin K-J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol (2017) 14(12):1705–14. doi: 10.1080/15476286.2017.1358347
15. Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep (2012) 2(1):111–23. doi: 10.1016/j.celrep.2012.06.003
16. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene (2003) 22(39):8031–41. doi: 10.1038/sj.onc.1206928
17. Chen Q, Zhu C, Jin Y. The oncogenic and tumor suppressive functions of the long noncoding RNA MALAT1: an emerging controversy. Front Genet (2020) 11:93. doi: 10.3389/fgene.2020.00093
18. Wang W, Zhu Y, Li S, Chen X, Jiang G, Shen Z, et al. Long noncoding RNA MALAT1 promotes Malignant development of esophageal squamous cell carcinoma by targeting β-catenin via Ezh2. Oncotarget (2016) 7(18):25668. doi: 10.18632/oncotarget.8257
19. Zuo Y, Li Y, Zhou Z, Ma M, Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother (2017) 95:922–8. doi: 10.1016/j.biopha.2017.09.005
20. Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem (2017) 118(12):4836–43. doi: 10.1002/jcb.26158
21. Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis (2014) 35(12):2731–9. doi: 10.1093/carcin/bgu200
22. Guo F, Guo L, Li Y, Zhou Q, Li Z. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol (2015) 8(12):15903.
23. Zhang Y-F, Li C-S, Zhou Y, Lu X-H. Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci (2020) 244:117280. doi: 10.1016/j.lfs.2020.117280
24. Deng QJ, Xie LQ, Li H. Overexpressed MALAT1 promotes invasion and metastasis of gastric cancer cells via increasing EGFL7 expression. Life Sci (2016) 157:38–44. doi: 10.1016/j.lfs.2016.05.041
25. Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett (2017) 395:31–44. doi: 10.1016/j.canlet.2017.02.035
26. Lu Z, Luo T, Pang T, Du Z, Yin X, Cui H, et al. MALAT1 promotes gastric adenocarcinoma through the MALAT1/miR-181a-5p/AKT3 axis. Open Biol (2019) 9(9):190095. doi: 10.1098/rsob.190095
27. Li J, Gao J, Tian W, Li Y, Zhang J. Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int (2017) 17:44. doi: 10.1186/s12935-017-0408-8
28. Lee NK, Lee JH, Ivan C, Ling H, Zhang X, Park CH, et al. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer (2017) 17(1):46. doi: 10.1186/s12885-016-2988-4
29. Zhu K, Ren Q, Zhao Y. lncRNA MALAT1 overexpression promotes proliferation, migration and invasion of gastric cancer by activating the PI3K/AKT pathway. Oncol Lett (2019) 17(6):5335–42. doi: 10.3892/ol.2019.10253
30. Shao G, Zhao Z, Zhao W, Hu G, Zhang L, Li W, et al. Long non-coding RNA MALAT1 activates autophagy and promotes cell proliferation by downregulating microRNA-204 expression in gastric cancer. Oncol Lett (2020) 19(1):805–12.
31. Chen D, Liu L, Wang K, Yu H, Wang Y, Liu J, et al. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand J Gastroenterol (2017) 52(6-7):790–6. doi: 10.1080/00365521.2017.1280531
32. Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget (2016) 7(35):56209–18. doi: 10.18632/oncotarget.10941
33. Chaleshi V, Asadzadeh Aghdaei H, Nourian M, Iravani S, Jalaeikhoo H, Rajaeinejad M, et al. Association of MALAT1 expression in gastric carcinoma and the significance of its clinicopathologic features in an Iranian patient. Gastroenterol Hepatol Bed Bench (2021) 14(2):108–14. doi: 10.18632/oncotarget.10941
34. Zhang Y, Chen Z, Li MJ, Guo HY, Jing NC. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. BioMed Pharmacother (2017) 85:264–71. doi: 10.1016/j.biopha.2016.11.014
35. Esfandi F, Salehnezhad T, Taheri M, Afsharpad M, Hafez AA, Oskooei VK, et al. Expression assessment of a panel of long non-coding RNAs in gastric Malignancy. Exp Mol Pathol (2020) 113:104383. doi: 10.1016/j.yexmp.2020.104383
36. Li X, Zhao J, Zhang H, Cai J. Silencing of lncRNA metastasis-associated lung adenocarcinoma transcript 1 inhibits the proliferation and promotes the apoptosis of gastric cancer cells through regulating microRNA-22-3p-mediated Erbb3. Onco Targets Ther (2020) 13:559–71. doi: 10.2147/OTT.S222375
37. Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol (2020) 18(3):534–42. doi: 10.1016/j.cgh.2019.07.045
38. Yuan L, Xu Z-Y, Ruan S-M, Mo S, Qin J-J, Cheng X-D. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol cancer (2020) 19(1):1–22. doi: 10.1186/s12943-020-01219-0
39. Tan H, Zhang S, Zhang J, Zhu L, Chen Y, Yang H, et al. Long non-coding RNAs in gastric cancer: new emerging biological functions and therapeutic implications. Theranostics (2020) 10(19):8880. doi: 10.7150/thno.47548
40. Wei L, Sun J, Zhang N, Zheng Y, Wang X, Lv L, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer (2020) 19(1):1–17. doi: 10.1186/s12943-020-01185-7
41. Shuai P, Zhou Y, Gong B, Jiang Z, Yang C, Yang H, et al. Long noncoding RNA MALAT1 can serve as a valuable biomarker for prognosis and lymph node metastasis in various cancers: a meta-analysis. Springerplus (2016) 5(1):1–8. doi: 10.1186/s40064-016-3342-7
42. Lin Y, Fang Z, Lin Z, Li Z, Zhao J, Luo Y, et al. The prognostic impact of long noncoding RNA HOTAIR in leukemia and lymphoma: a meta-analysis. Hematology (2018) 23(9):600–7. doi: 10.1080/10245332.2018.1446572
43. Chen Y, Huang W, Sun W, Zheng B, Wang C, Luo Z, et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem (2018) 51(3):1313–26. doi: 10.1159/000495550
44. Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: A long non−coding RNA highly associated with human cancers. Oncol Lett (2018) 16(1):19–26. doi: 10.3892/ol.2018.8613
45. Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp Mol Pathol (2020) 115:104448. doi: 10.1016/j.yexmp.2020.104448
46. Zhao L, Lou G, Li A, Liu Y. lncRNA MALAT1 modulates cancer stem cell properties of liver cancer cells by regulating YAP1 expression via miR−375 sponging. Mol Med Rep (2020) 22(2):1449–57. doi: 10.3892/mmr.2020.11196
47. Jin C, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumor Biol (2016) 37(6):7383–94. doi: 10.1007/s13277-015-4605-6
48. Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumor Biol (2015) 36(3):1477–86. doi: 10.1007/s13277-014-2631-4
49. Luo F, Sun B, Li H, Xu Y, Liu Y, Liu X, et al. MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget (2016) 7(5):5769–87. doi: 10.18632/oncotarget.6806
50. Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, et al. High expression of lncRNA MALAT1 suggests biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol (2014) 15:7:3174–81.
51. Lai MC, Yang Z, Zhou L, QQ Z, HY X, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol (2012) 29:1810–6. doi: 10.1007/s12032-011-0004-z
52. Serghiou S, Kyriakopoulou A, Ioannidis J. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer (2016) 15(1):1–14. doi: 10.1186/s12943-016-0535-1
53. Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. Jama (2011) 305(21):2200–10. doi: 10.1001/jama.2011.713
54. Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open (2015) 5(9):e008653. doi: 10.1136/bmjopen-2015-008653
55. Zhu M, Wang Y, Liu X, Wen X, Liang C, Tu J. LncRNAs act as prognostic biomarkers in gastric cancer: A systematic review and meta-analysis. Front Lab Med (2017) 1(2):59–68. doi: 10.1016/j.flm.2017.05.003
56. Gao S, Zhao ZY, Wu R, Zhang Y, Zhang ZY. Prognostic value of long noncoding RNAs in gastric cancer: a meta-analysis. Onco Targets Ther (2018) 11:4877–91. doi: 10.2147/OTT.S169823
Keywords: meta-analysis, lncRNA, MALAT1, gastric cancer, prognosis
Citation: Bai S, Guo J and Zhang H (2024) A meta-analysis of the clinicopathological significance of the lncRNA MALAT1 in human gastric cancer. Front. Oncol. 13:1257120. doi: 10.3389/fonc.2023.1257120
Received: 12 July 2023; Accepted: 22 November 2023;
Published: 04 January 2024.
Edited by:
Hou-Qun Ying, Second Affiliated Hospital of Nanchang University, ChinaReviewed by:
Sina Azadnajafabad, Tehran University of Medical Sciences, IranMohamed Hassan, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Ying Liu, Qingdao University, China
Copyright © 2024 Bai, Guo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoxiong Bai, YmFpc2hhb3hpb25nXzA5MTVAMTI2LmNvbQ==
 Shaoxiong Bai
Shaoxiong Bai Jiansheng Guo
Jiansheng Guo