
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 08 September 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1256537
This article is part of the Research Topicnon-coding RNAs: Master Messengers in the Tumor Microenvironment and BeyondView all 5 articles
Lung cancer (LC) is a heterogeneous disease with high malignant degree, rapid growth, and early metastasis. The clinical outcomes of LC patients are generally poor due to the insufficient elucidation of pathological mechanisms, low efficiency of detection and assessment methods, and lack of individualized therapeutic strategies. Non-coding RNAs (ncRNAs), including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA), are endogenous regulators that are widely involved in the modulation of almost all aspects of life activities, from organogenesis and aging to immunity and cancer. They commonly play vital roles in various biological processes by regulating gene expression via their interactions with DNA, RNA, or protein. An increasing amount of studies have demonstrated that ncRNAs are closely correlated with the initiation and development of LC. Their dysregulation promotes the progression of LC via distinct mechanisms, such as influencing protein activity, activating oncogenic signaling pathways, or altering specific gene expression. Furthermore, some ncRNAs present certain clinical values as biomarker candidates and therapeutic targets for LC patients. A complete understanding of their mechanisms in LC progression may be highly beneficial to developing ncRNA-based therapeutics for LC patients. This review mainly focuses on the intricate mechanisms of miRNA, lncRNA, and circRNA involved in LC progression and discuss their underlying applications in LC treatment.
Lung cancer (LC) is considered a major obstacle to increasing life expectancy worldwide (1). Globally, LC cases and deaths are rising rapidly. In 2020, GLOBOCAN estimated more than 2.2 million new LC cases occurred (2). LC has become a serious global health concern, bringing significant pain and economic burdens to patients and their families. According to pathological characteristics, LC is mainly classified into two subtypes: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (3, 4). The clinical outcomes of LC patients are commonly poor due to their unobvious early symptom, lack of efficient prognostic evaluation method, and insufficient understanding of pathogenesis (5, 6). Therefore, elucidating the regulatory mechanisms of LC progression may greatly benefit patients in the adjustment of therapeutic strategies and the identification of valuable biomarkers or targets.
Non-coding RNAs (ncRNAs) are functional RNA transcripts that have no protein-coding capacity (7–9). According to their biological functions, ncRNAs are mainly grouped into housekeeping ncRNA and regulatory ncRNA (10). Housekeeping ncRNAs (e.g., ribosomal RNA) are stably expressed in eukaryotic cells. Their products are essential for maintaining the basic life activity of cells (11). Regulatory ncRNAs are key players in almost all biological processes (12). Based on their structural features, ncRNAs are further categorized into microRNA (miRNA) (13), long non-coding RNA (lncRNA) (14), circular RNA (circRNA) (15), small interfering RNA (16), and PIWI-interacting RNA (17). They participate in the regulation of various biological processes, including transcription, development, and immunity, by altering specific gene expression (18–20). Therefore, their dysregulation is closely correlated with various diseases, such as brain disease, diabetes, and cancer (21–27) NcRNA dysregulation has been reported to contribute to almost all aspects of LC development, including apoptosis, cell cycle, metastasis, and autophagy, as well as cell stemness (28–31). However, investigations of LC-related ncRNAs are still lacking.
In this review, we mainly present the modes of action of miRNA, lncRNA, and circRNA and their regulatory mechanisms involved in the initiation and development of LC. We also explore the underlying utilization of these ncRNAs in LC clinical treatment.
NcRNAs (e.g., miRNAs, lncRNAs, and circRNAs) are essential regulators in various physiological and pathological processes, such as regeneration, development, immunopathogenesis, intracerebral hemorrhage, and LC (32–36).
MiRNAs are well-studied small ncRNAs, with a single-stranded structure of 19–25 nucleotides (37). Approximately 2300 miRNAs are found in human cells, and they can serve as post-transcriptional regulators to modulate over 60% of the protein-coding genes (38, 39). The canonical function of miRNAs is to regulate specific gene expression by influencing messenger RNA (mRNA) stability (40). In general, miRNAs suppress gene expression by directly interacting with partially complementary sequences in their target mRNAs (41). The method of gene inhibition relies on the complementary extent between miRNA and target mRNA. Exact matching commonly results in mRNA degradation, whereas partial matching induces translational suppression (42). Moreover, miRNAs recognize and mediate mRNA degradation and/or translational inhibition by recruiting the miRNA-induced silencing complex consisting of Argonaute proteins and GW182 (43). The near-seed or non-seed regions of miRNAs are also required for miRNA-mediated modulation of gene expression (44). In addition, nuclear miRNAs are found to mediate the silencing or activation of transcriptional genes (45–47).
LncRNAs are the largest type of ncRNAs and comprise 81.8% of the total ncRNAs (48, 49). They exhibit highly specific lineage, spatiotemporal, and tissue/cell-dependent patterns, but their abundance, stability, and conservation are less than mRNA (50, 51). LncRNAs are essential modulators that participate in almost every step of gene expression (52, 53). Their canonical mechanism of action is to inhibit target gene expression by binding to miRNA and imposing an additional post-transcriptional regulation level. LncRNAs can also induce transcription factors (TFs) away from chromatin by serving as molecular sinks, thereby altering gene expression (54). Furthermore, some studies suggest that they function as scaffolds to form scaffolding complexes with effectors, resulting in the alteration of gene expression (55). LncRNAs can also guide the ribonucleoprotein complex to the promoters of downstream target genes, thereby altering the transcriptional activity of genes (56). In addition, lncRNAs also alter gene expression through influencing mRNA processing, maturation, and stability (53).
CircRNAs are single-stranded ncRNA molecules generated from the pre-mRNA back-splicing process and possess a covalently closed-loop structure (57). The closed ring structure can protect circRNAs from exonuclease-mediated degradation, resulting in their stable existence in various subcellular structures (58). CircRNAs are key modulators in many biological processes, including gene transcription, protein translation, immune response, and carcinogenesis, as well as chemoresistance (59–61). The most widely investigated role of circRNAs is to weaken their effect on target mRNAs by serving as miRNA sponges, ultimately resulting in the alteration of correlated gene expression. These circRNAs commonly possess multiple miRNA response elements (62, 63). CircRNAs also participate in biological processes by influencing the functions of proteins (64–66). Furthermore, EIciRNAs are found to facilitate the RNA polymerase II-mediated transcription of their parental genes by binding to U1 small nuclear ribonucleoproteins (33). CircURI1 regulates the AS of multiple migration-related genes by directly interacting with hnRNPM, leading to the inhibition of gastric cancer metastasis (67). In addition, a small part of endogenous circRNAs, which contain open reading frames, have been shown to translate into peptides or proteins (68). However, their potential functions are still unclear.
Differentially expressed ncRNAs play crucial roles in LC occurrence and development (69). Zhang et al. revealed 190 differentially expressed miRNAs between pleural effusion induced by lung adenocarcinoma (LUAD) and pleural effusion induced by tuberculosis, including 99 highly expressed miRNAs and 91 low expression miRNAs. These miRNAs probably influenced the production of pleural effusion via tumor immune response (70). In another study, Zeng et al. distinguished 24 aberrantly expressed miRNAs between NSCLC patients with tumor shrinkage of ≤30% after radiotherapy and patients with tumor shrinkage of 30%–50%, 11 (6 upregulated and 5 downregulated) between patients with tumor shrinkage of ≤30% and patients with tumor shrinkage of ≥50%, and 35 between patients with tumor shrinkage of 30%–50% and patients with tumor shrinkage of ≥50% (71). Furthermore, by comparing the plasma of LUAD patients with benign pulmonary nodule patients, Tong et al. confirmed 1762 differentially expressed lncRNAs in LUAD patients, 946 in lung squamous cell carcinoma patients, and 298 in SCLC patients (72). Huang et al. revealed 177 highly expressed lncRNAs and 215 low expression lncRNAs in the exosomes of LUAD pleural effusion compared with that of benign pleural effusion (73). In addition, Cai et al. performed high-throughput sequencing and identified 598 differentially expressed circRNAs between LUAD patients with bone metastasis and patients without bone metastasis, among which 238 were upregulated and 360 were downregulated (74).
Recent studies suggest that the crosstalk between ncRNA and oncogenic signaling pathway is involved in LC initiation and development (75–77) (Figure 1). A better understanding of ncRNA action in targeting cancer-related signaling pathways may be of great benefit to the prevention and treatment of LC.
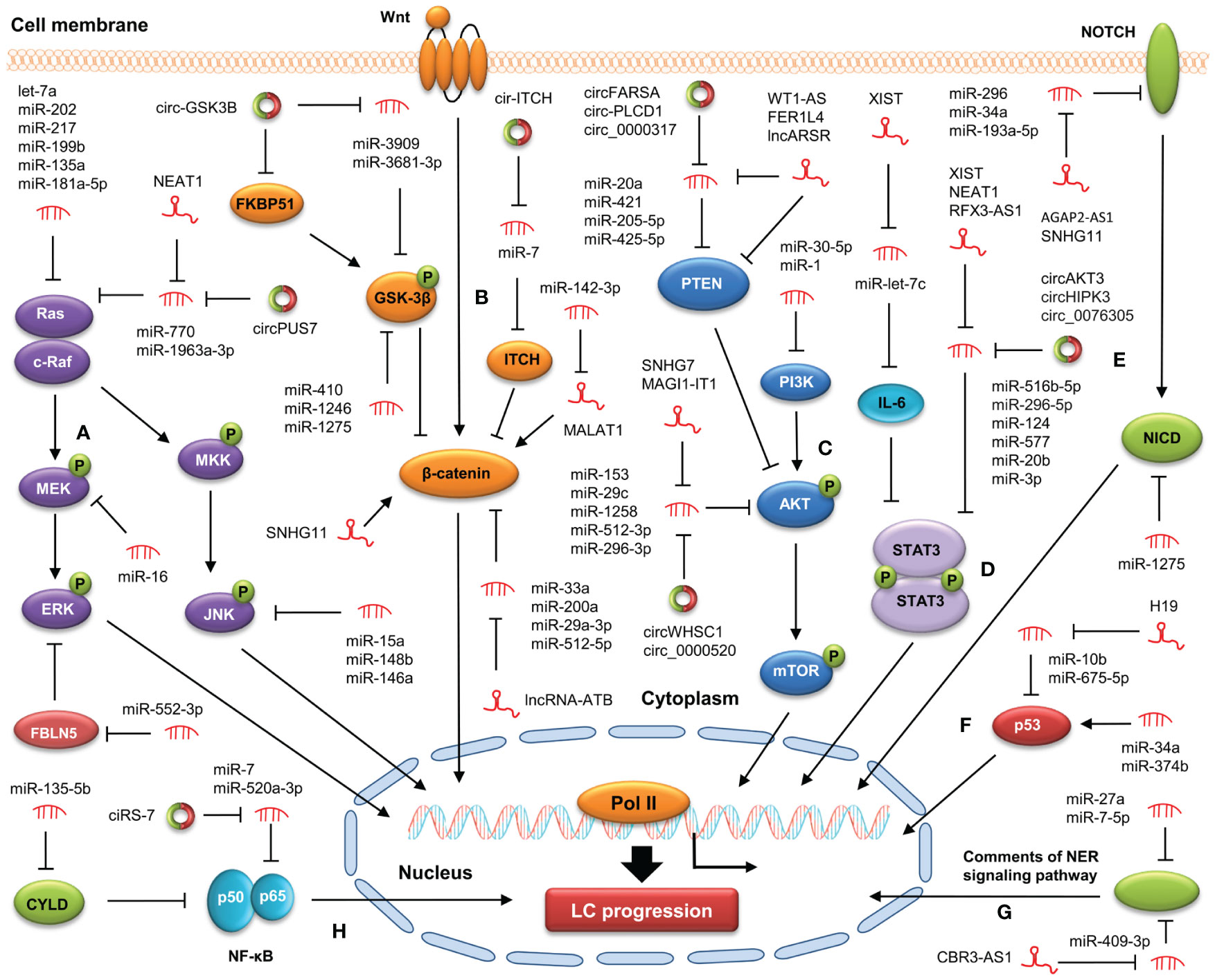
Figure 1 Modulation of ncRNAs on cancer-related signaling pathways in LC. NcRNAs are involved in LC progression by targeting the MAPK (A), WNT/β-catenin (B), PI3K/AKT (C), STAT3 (D), Notch (E), p53 (F), NER (G), NF-κB (H) signaling pathways.
The PI3K/AKT signaling pathway is a conserved signaling cascade involved in various biological processes, including growth, differentiation, metabolism, and survival. The aberrant activation of this pathway contributes to LC progression (78). MiRNAs play vital roles in LC progression by targeting the PI3K/AKT signaling pathway. For example, in a study by Niu et al., miRNA let-7c-3p was found to inactivate the PI3K/AKT signaling pathway through downregulating PIK3CA, thereby suppressing proliferation and migration in NSCLC cell lines H460 and A549 (79). Furthermore, Shi et al. showed that miR-514b-5p facilitated NSCLC progression by downregulating SGTB and enhancing the PI3K/AKT signaling pathway (80). LncRNAs and circRNAs are also key regulators of the PI3K/AKT signaling pathway during LC progression. For example, high expression of LASTR was observed in both LUAD and lung squamous cell carcinoma (LUSC) samples. LASTR overexpression increased the levels of transforming growth factor alpha by sponging miR-137, thereby activating the PI3K/AKT signaling pathway and ultimately leading to the facilitation of LC progression (81). Liu et al. demonstrated that circGRAMD1B enhanced the activity of the PI3K/AKT pathway by increasing SOX4 levels via sequestering miR-4428, resulting in the facilitation of LUAD progression (82).
The MAPK signaling pathway is a highly conserved pathway that plays an important role in maintaining cellular behaviors and processes, including epithelial-to-mesenchymal transition (EMT), apoptosis, and migration (83). NcRNA dysregulation has been demonstrated to participate in LC progression by modulating the MAPK signaling pathway. For example, exosomal miRNA let-7c-5p and miR-181b-5p was found to repress EMT in bronchial epithelial cells (BEAS-2B) by suppressing the MAPK signaling pathway, thereby inhibiting the invasion of BEAS-2B cells (84). Besides, Shi et al. showed that miRNA let-7a overexpression significantly suppressed the activity of the MAPK signaling pathway by downregulating Ras, p-Raf1/Raf1, and p-MEK1/MEK1 via targeting Rsf-1 in LC cells, resulting in the repression of cell proliferation after radiotherapy (85). Furthermore, Zhu et al. demonstrated that the levels of LINC00649 expression were remarkably increased in LUSC cells, and its upregulation facilitated the occurrence and development of LUSC. Mechanistically, LINC00649 activated the MAPK signaling pathway by enhancing the transcription and stability of MAPK6 via recruiting TATA-box binding protein associated factor 15 in LUSC cells, resulting in the promotion of LUSC progression (86). Wang et al. discovered that lncRNA PCAT19 increased MAP2K4 levels by binding to miR-25-3p, thereby repressing the MAPK signaling pathway and LC progression (87). Moreover, our previous study discovered that circ-ZKSCAN1 increased the levels of FAM83A via sequestering miR-330-5p, leading to the inactivation of the MAPK signaling pathway and subsequent facilitation of NSCLC progression (88). In addition, Zhuang et al. revealed that circ-RAD23B increased MAP4K3 levels by sequestering miR-142-3p, thereby enhancing the activity of the MAPK signaling pathway and promoting NSCLC progression (89). Hu et al. discovered that circCNN2 activated the MAPK signaling pathway through upregulating E2F TF 1 via sponging miR-184, thereby promoting LUSC progression (90).
The Wnt/β-catenin signaling pathway governs various physiological processes, such as embryo development and tissue homeostasis, and its aberrant activation is tightly linked with cancer progression (91). Some ncRNAs (e.g., miR-1275, miR-199, MIR4435-2HG, and FLVCR1-AS1) have been demonstrated to contribute to LC progression through the Wnt/β-catenin signaling pathway (92–97). Furthermore, multiple lncRNAs and circRNAs can regulate the Wnt/β-catenin signaling pathway by serving as competing endogenous RNA (ceRNA) for miRNAs in LC. Liu et al. showed that lncRNA RP11-79H23.3 knockdown enhanced the activity of the Wnt/β-catenin signaling pathway in NSCLC cells by sequestering miR-29c, resulting in the facilitation of LC progression (98). Yang et al. revealed that circ_0017109 knockdown decreased FZD4 levels by releasing miR-671-5p, resulting in the enhancement of the Wnt/β-catenin signaling pathway and subsequent facilitation of NSCLC progression (99). In addition, circCDR1 was found to activate the Wnt/β-catenin signaling pathway by interacting with SRSF1, thereby facilitating PM2.5-induced LC development (100).
The Notch signaling pathway is involved in the regulation of multiple biological processes, such as cell fate determination, embryo formation, and organism homeostasis (101). The dysregulation of the Notch signaling pathway contribute to many aspects of LC progression, including uncontrolled proliferation, cancer cell stemness, and TME (102). Ji et al. found that miR-34a inactivated the Notch signaling pathway by downregulating Hes-1, Notch-1, and Survivin, resulting in the suppression of cell growth and invasiveness and facilitation of apoptosis in NSCLC cells (103). Xue et al. showed that miR-200 modulated the crosstalk of LUAD cells with adjacent cancer-associated fibroblasts (CAFs) by targeting Jagged1 and Jagged2 (Notch ligands), thereby activating the Notch signaling pathway in CAFs and subsequently repressing LUAD metastasis (104). Furthermore, exosomal AGAP2-AS1 activated the Notch signaling pathway in LC cells by upregulating Notch2 via sequestering miR-296, leading to the enhancement of cell malignant behaviors (105). SNHG11 upregulated Notch3 by sponging miR-193a-5p, thereby activating the Notch signaling pathway and subsequently facilitating LUAD progression (106). In addition, circ_0000190 was found to counteract the repression of luteolin on LC progression by activating the Notch-1 signaling pathway via sponging miR-130a-3p (107).
NcRNAs can also play a role in regulating LC progression through other signaling pathways. Multiple ncRNAs (e.g., miR-520a-3p, lncRNA MIR503HG, and circ_cMras) have been shown to suppress LC development by inactivating the NF-κB pathway, whereas some other ncRNAs (e.g., miR-135b and lncRNA SNHG5) can promote LC progression by enhancing the NF-κB pathway (108–112). Furthermore, lncRNA H19 enhanced the STAT3 signaling pathway by increasing STAT3 levels via sequestering miR-17, thereby promoting the progression of NSCLC (113). Hsa_circ_0002874 repressed the p53 signaling pathway by upregulating MDM2 (the E3 ubiquitin ligase of p53) via sponging miR-1273f, thereby enhancing the PTX resistance of NSCLC cells (114). In addition, hsa_circ_0001946 suppressed cisplatin resistance in NSCLC cells by modulating the NER signaling pathway (115).
Uncontrolled proliferation and escape from apoptosis are the most defining characteristics of tumor cells. However, the regulatory network involved in proliferation and apoptosis remains unclear and need to be further clarified (116). NcRNAs has been demonstrated to be vital regulators of the two cellular processes in LC. Luo et al. discovered that the levels miRNA-144-5p were significantly increased in LUAD, and its upregulation suppressed proliferation and promoted apoptosis in LUAD cells by targeting CDCA3 (117). Han et al. showed that miR-4491 was remarkably upregulated in NSCLC cells. Its overexpression facilitated proliferation and repressed apoptosis in NCI-H1650 cells through targeting TRIM7 (118). LncRNAs and circRNAs can also modulate LC proliferation and apoptosis via serving as ceRNAs for miRNA. For example, lncRNA-UCA1 was found to upregulate VEGF-A via sequestering miR-383, thereby facilitating proliferation and inhibiting apoptosis in HCC-78 cells (119). Circ_0000520 increased the levels of breast cancer-overexpressed gene 1 via sequestering miR-512-5p, resulting in the facilitation of proliferation and induction of apoptosis in LC cells (120).
Some cell cycle-related proteins are identified as downstream targets of ncRNAs (121), indicating that ncRNAs may participate in the modulation of the proliferation and apoptosis in LC through influencing cell cycle process. For instance, Huang et al. discovered that let-7c-5p overexpression arrested cells in G0/G1 phase by targeting cell division cycle 25A, resulting in the repression of proliferation and the promotion of apoptosis in LUAD cells (122). Wang et al. revealed that lnc-TMEM132D-AS1 induced M2/G-phase cell cycle arrest, facilitated proliferation, and repressed apoptosis in NSCLC cells by upregulating CD39 via sponging miR-766-5p (123). In addition, circPIM3 was found to increase TNFAIP8 levels by sponging miR-338-3p, thereby repressing apoptosis and promoting cell cycle progress and proliferation in taxol-resistant A549 and PC9 cells (124). Taken together, these findings strongly suggest that ncRNAs play pleiotropic roles in LC progression.
The invasion and metastasis of tumor cells are major causes of cancer recurrence and mortality (125, 126). Therefore, elucidating the potential mechanisms involved in invasion and metastasis is essential for developing therapeutic strategies to ameliorate prognosis for LC patients. NcRNAs have been proven to serve as key modulators that mediate invasion and metastasis in LC. For example, miR-96-5p was found to activate the MAPK signaling pathway by targeting domain-binding protein 2, leading to the repression of invasion and metastasis in LC cells (127). MiR-520a-3p decreased NF-κB p65 levels by targeting AKT1, thereby inactivating the NF-κB pathway and ultimately suppressing cell invasion and metastasis in NSCLC cells (128). Moreover, lncRNA TEX41 suppressed the PI3K/AKT signaling pathway by increasing Runx2 expression, leading to the facilitation of invasion, metastasis, and autophagy in LUAD cells (129). Circ_0000376 silencing decreased PDPK1 expression by releasing miR-545-3p, thereby repressing invasion and metastasis in NSCLC cells (130).
EMT is a biological process of cellular morphological alterations in which epithelial cells obtain mesenchymal characteristics. Recent studies suggest that ncRNA dysregulation endows LC cells with invasive and metastatic characteristics by altering EMT (84, 131, 132). Exosomal miR-181b-5p and let-7c-5p was found to inhibit the EMT process in BEAS-2B cells by modulating the MAPK signaling pathway, resulting in the enhancement of migratory and invasive ability in BEAS-2B cells (84). Moreover, Yang et al. showed that lncRNA PCAT6 significantly repressed the migration, invasion, and EMT of A549 and H1975 cells by increasing EGFR expression via sequestering miR-545-3p (131). Liu et al. revealed that circSCN8A suppressed invasion, metastasis, and EMT in NSCLC cell lines. Mechanistically, circSCN8A increased the levels of ACSL4 by sponging miR-1290, thereby repressing NSCLC progression (132). Collectively, as the key modulators of invasion and metastasis during LC progression, ncRNAs have presented great value as target candidates in LC treatment.
Angiogenesis denotes the development of new vessels from existing ones, by which tumor cells acquire sufficient material supplement for their growth (133). Targeting angiogenesis is considered a promising strategy in cancer treatment. NcRNAs are modulators of angiogenesis in LC. Gan et al. found that let-7d-5p expression was remarkably increased in LC cells treated with Trametes robiniophila, and its upregulation inhibited angiogenesis and tumor growth in LC by targeting NAP1L1 (134). Chang et al. demonstrated that exosomal miR-197-3p from LUAD cells could facilitate the angiogenesis of HUVECs by directly downregulating TIMP2/3 (135). Furthermore, Pan et al. discovered that LANCL1-AS1 upregulation dramatically repressed the angiogenesis of NSCLC cells by upregulating glia maturation factor gamma via sponging miR-3680-3p (136). Wang et al. revealed that ZNRD1-AS1 upregulation increased tensin 1 levels by sponging miR-942, thereby suppressing LC angiogenesis (137). In addition, circ_0043256 upregulation remarkably repressed angiogenesis in LC cells by upregulating KLF2 via absorbing miR-1206 (138). In-depth investigations are needed to further clarify the ncRNA action in angiogenesis, which may bring significant advantage for the development of theoretical basis in LC treatment.
Tumor microenvironment (TME) is a highly complicated ecosystem that contains tumor cells, nontumoral cells, and various cytokines and chemokines generated by them. The continuous interaction between tumor cells and TME contributes to carcinogenesis, metastasis, and drug resistance (139). NcRNAs are involved in LC development through targeting the cellular components of TME, such as CAFs and tumor-associated macrophages (TAMs) (140–145). Liu et al. discovered that CAF-derived exosomal miR-200 inhibited morphological and metastatic characteristics of NSCLC cells by downregulating ZEB1 (140). Enukashvily et al. showed that satellite lncRNA knockdown reduced cell aging and attenuated inflammatory CAF phenotype in human lung fibroblasts (141). Furthermore, Li et al. revealed that LINC01798 remarkably increased ITGA8 levels through absorbing miR-17-5p, resulting in the alteration of TME and stemness in LUAD cells (142). As the main immune cell population in the TME, TAMs play vital roles in shaping the TME (143). NSCLC cell-derived exosomal miR-181b was found to enhance TAM M2 polarization through the activation of the STAT3 signaling pathway (144). Moreover, Wu et al. demonstrated that LINC01094 activated the transcription of CCL7 by facilitating the shuttling of SPI1 from cytoplasm to nucleus, resulting in the accumulation of M2 TAMs and the dissemination of LUAD cells (145). In addition, exosomal circFARSA was found to polarize TAMs to an M2 phenotype by enhancing the activation of the PI3K/AKT pathway. NSCLC cells co-cultured with TAMs transfected with circFARSA exhibited enhanced EMT and metastasis (146).
Cancer stem cell (CSC) belongs to a specific type of self-renewal cells, which is considered the major factor contributing to metastasis, chemoresistance, and recurrence in cancer (147). Elucidating the detailed mechanism involved in the modulation of CSC functions may bring significant benefit to the development of individualized treatment of LC patients. NcRNAs are key regulators of stemness in LC cells (148–151). Moro et al. discovered that miR-486-5p facilitated apoptosis and decreased viability in CD133+ lung CSCs by inactivating the PI3K/AKT pathway, leading to the inhibition of the tumor-initiating roles of these cells (148). Liu et al. demonstrated that miR-1246 knockdown attenuated the stemness of LC cells by directly targeting TRIM17 (149). Furthermore, the overexpression of ADAMTS9-AS1 was found to significantly increase NPNT expression by sequestering miR-5009-3p, thereby repressing the stemness of LUDA-CSCs (150). Lu et al. revealed that TDRG1 was remarkably increased in lung CSCs compared with parental LC cells. TDRG1 overexpression enhanced the stemness of lung CSCs by upregulating Sox2 (stemness marker) via binding to its mRNA (151). In addition, circRACGAP1 enhanced stemness and metastasis in NSCLC cells via promoting SIRT3-mediated RIF1 deacetylation (152). Collectively, these studies suggest that ncRNAs are key modulators of stemness in LC cells. However, their regulatory mechanisms remain not fully understood, which need to be further elucidated.
Chemotherapy is a well-established treatment method for distinct cancer types and can significantly extend patients’ life spans, but the development of chemoresistance limits its further utilization and ultimately results in patients’ death (153). NcRNA dysregulation is closely correlated with the emergency of chemoresistance in LC treatment (154). Our previous study showed that miR-608 was remarkably downregulated in NSCLC samples. MiR-608 overexpression in NSCLC cells facilitated doxorubicin-induced apoptosis by targeting TFAP4 (155). Vinciguerra et al. discovered that miR-301a was dramatically decreased in cisplatin-resistant NSCLC cells. The overexpression of miR-301a downregulated GLIPR1 by targeting Fra-2, thereby improving cisplatin resistance in NSCLC cells (156). Besides, miR-936 was significantly downregulated in NSCLC cells, and its overexpression inactivated the Galphaq Rho GTPase pathway by targeting GPR78, resulting in the repression of cisplatin resistance in NSCLC cells (157). Furthermore, Yu et al. demonstrated that lncRNA LOC85009 inhibited ATG5-induced autophagy by decreasing the stability of upstream TF 1 via sequestering ubiquitin-specific proteinase 5, thereby triggering cell apoptosis and suppressing docetaxel resistance in LUAD cells. Interestingly, exosomal LOC85009 derived from LUAD cells enhanced docetaxel sensitivity in docetaxel-resistant cells (158). Liu et al. discovered that DDX11-AS1A was remarkably increased in LUAD, and its upregulation attenuated paclitaxel sensitivity in LUAD cells through promoting DNA damage repair (159). In addition, CircPIM3 decreased taxol sensitivity and inhibit apoptosis in taxol-resistant NSCLC cells. Mechanistically, circPIM3 upregulated tumor necrosis factor-alpha-induced protein-8 via absorbing miR-338-3p, thereby enhancing taxol resistance in NSCLC cells (124).
Tobacco smoking is considered the major risk factor of LC that generates long-lasting and progressive impairment to the lung tissue (160). Some ingredients in tobacco have been shown to contribute to tumorigenesis and progression of LC by promoting malignant behaviors of cancer cells and inducing chronic inflammation (161, 162). However, the exact mechanisms of tobacco smoking in LC remain largely unknown. Recent studies suggest that ncRNAs play vital roles in pathogenesis of tobacco smoking-induced LC (163–165). Tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a well-studied strong carcinogen. Kalscheuer et al. discovered that the levels of miR-101, miR-126*, miR-199 and miR-34 were significantly downregulated in male rats treated with NNK, indicating the potential value of these miRNAs as diagnostic biomarker for early LC development. Functional analysis revealed that NNK exerted its oncogenic role by increasing cytochrome P450 (CYP) 2A3 levels via downregulating miR-126* (163). Chen et al. showed that NNK treatment decreased lncRNA AC007255.8 expression by inducing its promoter hypermethylation, resulting in the promotion of proliferation and the suppression of apoptosis in human bronchial epithelial Beas-2B cells (164). Furthermore, Hua et al. demonstrated that circ_0035266 regulated the inflammatory responses of Beas-2B cells to NNK and lipopolysaccharide (LPS) by altering the secretion of IL-6 and IL-8. Mechanistically, circ_0035266 overexpression upregulated DDX3X by sponging miR-181d-5p, thereby facilitating IL-6 and IL-8 secretion and ultimately resulting in the enhancement of inflammatory responses of cells to NNK and LPS (165). They also found that circ_0035266 knockdown significantly repressed the proliferation, cell cycle process, and migration of Beas-2B cells treated with NNK and LPS (166). Nicotine is a primary alkaloid derived from tobacco plants. Liu et al. revealed that the levels of miR-218 were remarkably decreased in NSCLC cells treated with nicotine and its downregulation facilitated the expression of CDK6, leading to the promotion of cell proliferation (167). Zhao et al. showed that the nicotine-induced upregulation of LINC00460 promoted the proliferation and migration of NSCLC cells and the inhibition of cell apoptosis (161). In addition, Zong et al. demonstrated that lncRNA CCAT1 was significantly upregulated in human bronchial epithelial (HBE) cells treated with cigarette smoke extract (CSE). CCAT1 overexpression activated the ERK signaling pathway by sponging miR-152-3p, resulting in the enhancement of inflammation in CSE-treated HBE cells (168). Collectively, these findings strongly suggest that ncRNAs are key regulators in tobacco smoking-induced LC progression. Understanding their exact mechanisms in tobacco smoking -associated LC may provide novel insights in the development of individualized treatment of tobacco-using patients with LC.
In summary, ncRNAs can play pleiotropic roles in almost all aspects of LC occurrence and development, such as EMT, apoptosis, angiogenesis, TME, stemness, and chemoresistance (Figure 2). The key functions of ncRNAs in the development of LC malignant characteristics endow them with great clinical application value in LC treatment.
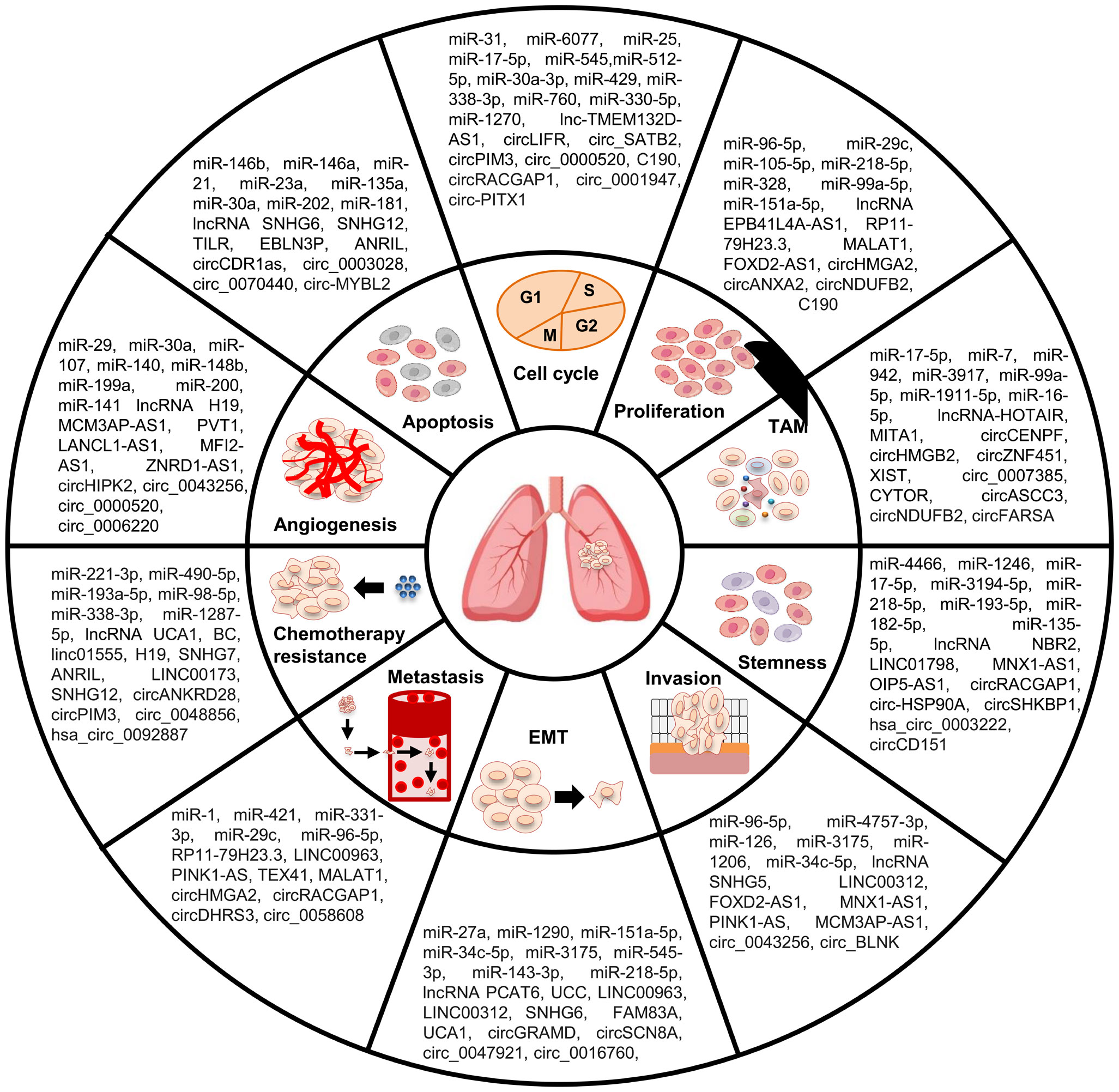
Figure 2 Role of ncRNAs in malignant behaviors of LC cells. NcRNAs participate in the regulation of LC malignant behaviors, including cell apoptosis, proliferation, cell cycle, invasion, metastasis, EMT, TAM, stemness, angiogenesis, and chemoresistance.
Currently, most LC patients have poor clinical outcomes because of the lack of effective early diagnosis and prognostic assessment means (33). Multiple proteins (e.g., PSA, CA-125, and CYFRA 21-1) have been applied in LC treatment by serving as biomarkers. However the unsatisfactory accuracy and reliability restrict their further utilization (169). NcRNAs have exhibited differently expressed patterns, high stability and specificity, and detectability (170–172). These specific features endow them with great value as noninvasive biomarkers for LC patients (Tables 1, 2). Wang et al. performed an in-depth meta-analysis and discovered that the area under the curve (AUC) value for miR-21 in distinguishing LC was 0.87, with 77% sensitivity and 86% specificity. Moreover, high miR-21 levels were remarkably associated with overall survival (OS) in LC patient (170). Wu et al., found that miR-340 was dramatically downregulated in plasma from NSCLC patients, whereas miR-450b-5p were upregulated. The AUC values for circulating miR-340 and miR-450b-5p in distinguishing NSCLC were 0.740 and 0.808, respectively. Furthermore, lower miR-340 and higher miR-450b-5p were significantly correlated with prognosis in NSCLC (171).
LncRNAs and circRNAs have also been utilized in LC clinical research. Yuan et al. discovered that the plasma levels of CRNDE and TA73-AS1 were significantly increased in NSCLC tissues. Their AUC values in distinguishing NSCLC were 0.822 and 0.815, separately. Moreover, their plasma levels were also closely correlated with poor tumor-free survival in NSCLC (172). Zhang et al. showed that NPSR1-AS1 was much higher in LUAD samples compared with benign samples. The AUC value for high NPSR1-AS1 in the diagnosis of LUAD was 0.904, with a 95% CI ranging from 0.881 to 0.927. Furthermore, the levels of NPSR1-AS1 exhibited positive correlation with OS in LUAD patients (213). Zou et al. demosntrated that AUC for serum circERBB2 in distinguishing NSCLC was 0.871, which was higher than CYFRA21-1 (0.693) and CEA (0.861). Moreover, LC patients with low circERBB2 levels had higher 36-month cumulative survival rate than patients with high circERBB2 levels (p < 0.05) (214). In addition, Li et al. showed that high levels of hsa_circ_001010 and hsa_circ-ZNF609 was negatively correlated with OS and DFS, whereas low levels of hsa_circ-CRIMI1, hsa_circ-EPB41L2, and hsa_circ_0072309 was positively associated with OS and DFS in LUAD. The four circRNAs also exhibits great potential in distinguishing LUAD (215).
As the key regulators of LC progression, ncRNAs have displayed huge therapeutic potential (133). Targeting oncogenic ncRNAs represents a highly feasible solution for patients to improve LC interventions. Chu et al. showed that miR-96-5p levels were dramatically increased in LC tissues, and its upregulation altered the expression of Bax, MMP9, and Bcl-2 through downregulating domain-binding protein 2, thereby facilitating invasion and proliferation in H1299 cell lines (127). In another study by Lv et al., lncRNA MNX1-AS1 was remarkably increased in LC samples, and its downregulation repressed the proliferation, migration, invasion, and sphere-forming abilities of LC CSCs by activating myosin IG (216). Furthermore, Sun et al. revealed that circ_0000376 knockdown decreased PDPK1 levels by releasing miR-545-3p, thereby suppressing NSCLC progression (130). Upregulating tumor-suppressive ncRNAs in cancer cells could be another effective strategy in LC treatment. For example, miR-1 overexpression repressed cell growth and oncogenic signaling in SCLC cells by targeting CXCR4. Consistent with this, intracardiac injection of miR-1 SCLC cells in mice exhibited a reduction in distant tissue metastasis (217). Furthermore, Gao et al. demonstrated that lncRNA FAM138B suppressed cell proliferation and invasion by targeting miR-105-5p in NSCLC cells (218). Song et al. revealed that circANKRD28 overexpression enhanced cisplatin sensitivity in NSCLC cells through increasing SOCS3 levels via absorbing miR-221-3p (219). To summarize, therapeutic strategies that directly target ncRNAs or use ncRNAs will bring significant benefit to the development of individualized treatment of LC patients.
LC, the most commonly diagnosed type of cancer in respiratory system, severely shortens patients’ life expectancy. The pathogenesis of LC is very complex and still unclear. Clarifying the regulatory mechanisms involved in LC progression is extremely urgent for developing efficient therapies of LC patients. Recent studies have identified a large amount of differently expressed ncRNAs (e.g., miRNAs, lncRNAs, and circRNAs) in LC. These ncRNAs play vital roles in LC progression by influencing almost all biological processes, such as cell invasion, autophagy, CSCs, and chemoresistance (28–30). Moreover, the differently expressed ncRNAs are easily examined in body fluids (e.g., serum and lymph) of LC patients, and their differentiated expression patterns are also closely correlated with some pathological characteristics, including tumor-free survival, OS, and DFS (172, 215). These unique characteristics mean that ncRNAs are valuable candidates of non-invasive biomarker and target in LC treatment (Figure 3). However, some challenges (e.g., ununified standardization strategies, unknown side effects, and insufficient patient size) still exist, which should be addressed before applying ncRNAs in LC clinical treatment. Nevertheless, recent studies strongly suggest that ncRNAs are effective biomarkers and promising targets for LC patients. Future investigations should focus on elucidating the exact functions of ncRNA in LC pathogenesis and developing novel ncRNA-based therapeutic strategies.
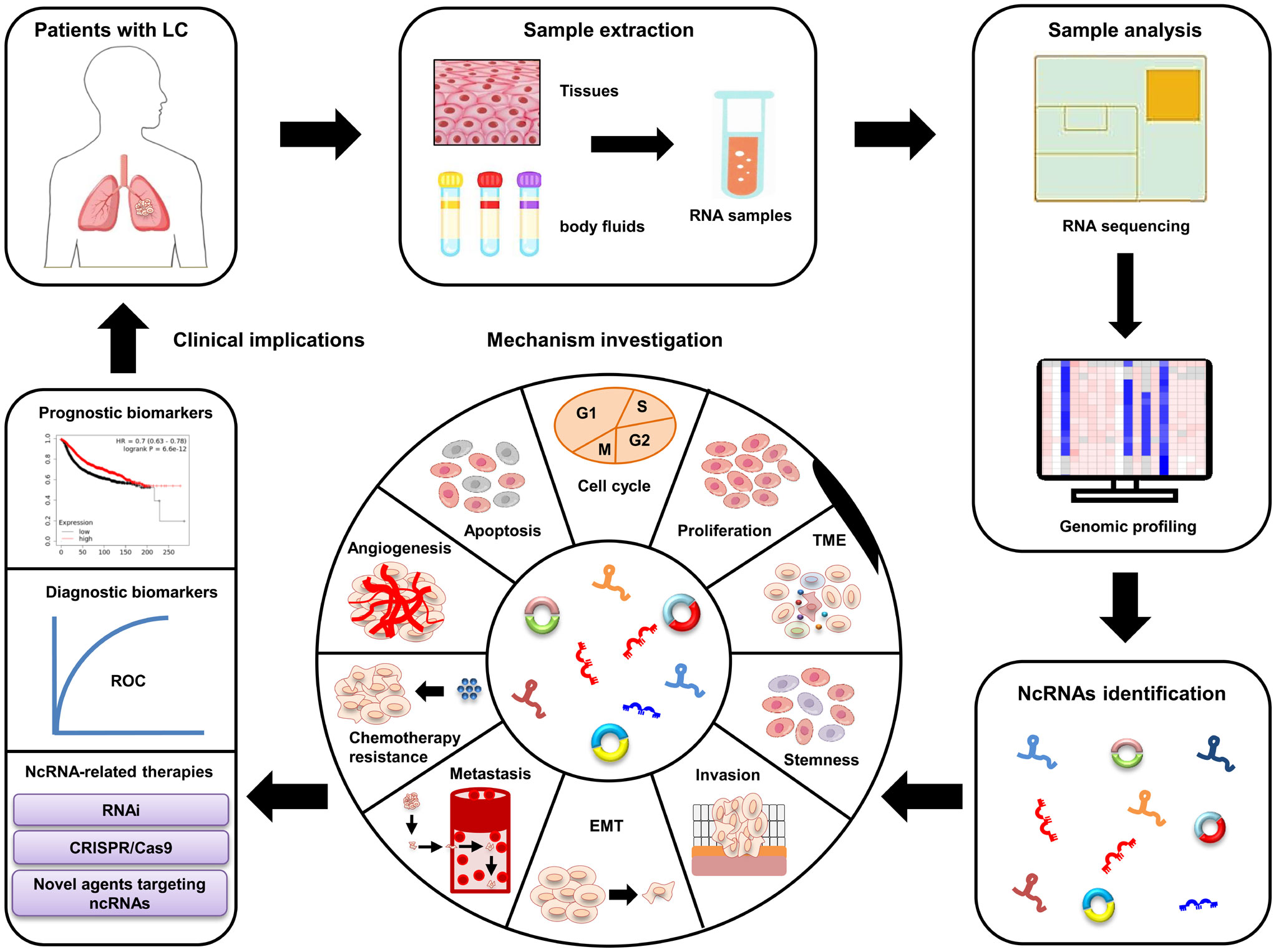
Figure 3 Clinical applications of ncRNAs in LC. The dysregulated ncRNAs are identified from LC patient samples using RNA sequencing and bioinformatics. In vitro and in vivo studies are performed to further clarify the underlying mechanisms of these ncRNAs involved in LC progression. Large patient cohorts are used to validate their potential as diagnostic and prognostic biomarkers. Novel ncRNA-based therapeutic strategies are developed for LC patients.
YL: Original draft preparation, writing—review and editing. WD: Data curation. JW: Data curation. XA: Original draft preparation, funding acquisition. JX: Writing—conceptualization, original draft preparation; writing—review and editing. All authors contributed to the article and approved the submitted version.
All authors are supported by Qingdao Medical College, Qingdao University. This work was funded by the China Postdoctoral Science Foundation (2018M642607).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lahiri A, Maji A, Potdar PD, Singh N, Parikh P, Bisht B, et al. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer (2023) 22(1):40. doi: 10.1186/s12943-023-01740-y
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol (2015) 16(4):e165–72. doi: 10.1016/S1470-2045(14)71180-5
4. Zhang S, Li M, Ji H, Fang Z. Landscape of transcriptional deregulation in lung cancer. BMC Genomics (2018) 19(1):435. doi: 10.1186/s12864-018-4828-1
5. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin (2018) 68(1):7–30. doi: 10.3322/caac.21442
6. Adams SJ, Stone E, Baldwin DR, Vliegenthart R, Lee P, Fintelmann FJ. Lung cancer screening. Lancet (2023) 401(10374):390–408. doi: 10.1016/S0140-6736(22)01694-4
7. Wang M, Zhang Y, Chang W, Zhang L, Syrigos KN, Li P. Noncoding RNA-mediated regulation of pyroptotic cell death in cancer. Front Oncol (2022) 12:1015587. doi: 10.3389/fonc.2022.1015587
8. Wang ZY, Wen ZJ, Xu HM, Zhang Y, Zhang YF. Exosomal noncoding RNAs in central nervous system diseases: biological functions and potential clinical applications. Front Mol Neurosci (2022) 15:1004221. doi: 10.3389/fnmol.2022.1004221
9. Chang W, Li P. Bone marrow mesenchymal stromal cell-derived small extracellular vesicles: A novel therapeutic agent in ischemic heart diseases. Front Pharmacol (2022) 13:1098634. doi: 10.3389/fphar.2022.1098634
10. Ao X, Ding W, Li X, Xu Q, Chen X, Zhou X, et al. Non-coding RNAs regulating mitochondrial function in cardiovascular diseases. J Mol Med (Berl) (2023) 101(5):501–26. doi: 10.1007/s00109-023-02305-8
11. Leng S, Qu H, Lv X, Liu X. Role of ncRNA in multiple myeloma. biomark Med (2022) 16(16):1181–91. doi: 10.2217/bmm-2022-0349
12. Zhou Z, Wang Z, Gao J, Lin Z, Wang Y, Shan P, et al. Noncoding RNA-mediated macrophage and cancer cell crosstalk in hepatocellular carcinoma. Mol Ther Oncolytics (2022) 25:98–120. doi: 10.1016/j.omto.2022.03.002
13. Zhang L, Zhang Y, Yu F, Li X, Gao H, Li P. The circRNA-miRNA/RBP regulatory network in myocardial infarction. Front Pharmacol (2022) 13:941123. doi: 10.3389/fphar.2022.941123
14. Zhou Z, Cao Q, Diao Y, Wang Y, Long L, Wang S, et al. Non-coding RNA-related antitumor mechanisms of marine-derived agents. Front Pharmacol (2022) 13:1053556. doi: 10.3389/fphar.2022.1053556
15. Zhang L, Zhang Y, Wang Y, Zhao Y, Ding H, Li P. Circular RNAs: functions and clinical significance in cardiovascular disease. Front Cell Dev Biol (2020) 8:584051. doi: 10.3389/fcell.2020.584051
16. Traber GM, Yu AM. RNAi-based therapeutics and novel RNA bioengineering technologies. J Pharmacol Exp Ther (2023) 384(1):133–54. doi: 10.1124/jpet.122.001234
17. Jia DD, Jiang H, Zhang YF, Zhang Y, Qian LL, Zhang YF. The regulatory function of piRNA/PIWI complex in cancer and other human diseases: The role of DNA methylation. Int J Biol Sci (2022) 18(8):3358–73. doi: 10.7150/ijbs.68221
18. Wang M, Chen X, Yu F, Zhang L, Zhang Y, Chang W. The targeting of noncoding RNAs by quercetin in cancer prevention and therapy. Oxid Med Cell Longev (2022) 2022:4330681. doi: 10.1155/2022/4330681
19. Zuo YB, Zhang YF, Zhang R, Tian JW, Lv XB, Li R, et al. Ferroptosis in cancer progression: role of noncoding RNAs. Int J Biol Sci (2022) 18(5):1829–43. doi: 10.7150/ijbs.66917
20. Zhang Y, Yu W, Chang W, Wang M, Zhang L, Yu F. Light chain amyloidosis-induced autophagy is mediated by the foxo3a/beclin-1 pathway in cardiomyocytes. Lab Invest (2023) 103(2):100001. doi: 10.1016/j.labinv.2022.100001
21. Zhou Z, Gong Q, Wang Y, Li M, Wang L, Ding H, et al. The biological function and clinical significance of SF3B1 mutations in cancer. biomark Res (2020) 8:38. doi: 10.1186/s40364-020-00220-5
22. Wang M, Yu F, Zhang Y, Chang W, Zhou M. The effects and mechanisms of flavonoids on cancer prevention and therapy: focus on gut microbiota. Int J Biol Sci (2022) 18(4):1451–75. doi: 10.7150/ijbs.68170
23. Zhang Y, Zhang L, Wang M, Li P. The applications of nanozymes in neurological diseases: From mechanism to design. Theranostics (2023) 13(8):2492–514. doi: 10.7150/thno.83370
24. Zhang Y, Yu W, Chen M, Zhang B, Zhang L, Li P. The applications of nanozymes in cancer therapy: based on regulating pyroptosis, ferroptosis and autophagy of tumor cells. Nanoscale (2023) 15(29):12137–56. doi: 10.1039/d3nr01722b
25. Xiao D, Chang W. Phosphatidylserine in diabetes research. Mol Pharm (2023) 20(1):82–9. doi: 10.1021/acs.molpharmaceut.2c00707
26. Zhang Y, Yu W, Flynn C, Chang W, Zhang L, Wang M, et al. Interplay between gut microbiota and NLRP3 inflammasome in intracerebral hemorrhage. Nutrients (2022) 14(24):5251. doi: 10.3390/nu14245251
27. Ding W, Zhang X, Xiao D, Chang W. Decreased in n-3 DHA enriched triacylglycerol in small extracellular vesicles of diabetic patients with cardiac dysfunction. J Diabetes (2023). doi: 10.1111/1753-0407.13457
28. Rajakumar S, Jamespaulraj S, Shah Y, Kejamurthy P, Jaganathan MK, Mahalingam G, et al. Long non-coding RNAs: an overview on miRNA sponging and its co-regulation in lung cancer. Mol Biol Rep (2023) 50(2):1727–41. doi: 10.1007/s11033-022-07995-w
29. Hua J, Liu J, Ma M, Xie L, Tian J. MicroRNA in the diagnosis of lung cancer: An overview of ten systematic reviews. Ann Clin Biochem (2023) 60(1):6–13. doi: 10.1177/00045632221128684
30. Sufianov A, Begliarzade S, Beilerli A, Liang Y, Ilyasova T, Beylerli O. Circular RNAs as biomarkers for lung cancer. Noncoding RNA Res (2023) 8(1):83–8. doi: 10.1016/j.ncrna.2022.11.002
31. Kielbowski K, Ptaszynski K, Wojcik J, Wojtys ME. The role of selected non-coding RNAs in the biology of non-small cell lung cancer. Adv Med Sci (2023) 68(1):121–37. doi: 10.1016/j.advms.2023.02.004
32. Loganathan T, Doss CG. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genomics (2023) 23(1):33. doi: 10.1007/s10142-022-00947-4
33. Liu Y, Ao X, Yu W, Zhang Y, Wang J. Biogenesis, functions, and clinical implications of circular RNAs in non-small cell lung cancer. Mol Ther - Nucleic Acids (2022) 27:50–72. doi: 10.1016/j.omtn.2021.11.013
34. Zhang Y, Yu W, Liu Y, Chang W, Wang M, Zhang L. Regulation of nuclear factor erythroid-2-related factor 2 as a potential therapeutic target in intracerebral hemorrhage. Front Mol Neurosci (2022) 15:995518. doi: 10.3389/fnmol.2022.995518
35. Zhang Y, Wei YJ, Zhang YF, Liu HW, Zhang YF. Emerging functions and clinical applications of exosomal ncRNAs in ovarian cancer. Front Oncol (2021) 11:765458. doi: 10.3389/fonc.2021.765458
36. He B, Xu HM, Liu HW, Zhang YF. Unique regulatory roles of ncRNAs changed by PM(2.5) in human diseases. Ecotoxicol Environ Saf (2023) 255:114812. doi: 10.1016/j.ecoenv.2023.114812
37. Zhang L, Zhang Y, Zhao Y, Wang Y, Ding H, Xue S, et al. Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert Opin Ther Pat (2018) 28(8):591–601. doi: 10.1080/13543776.2018.1503650
38. Diener C, Keller A, Meese E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends Genet (2022) 38(6):613–26. doi: 10.1016/j.tig.2022.02.006
39. Liu Y, Ao X, Jia Y, Li X, Wang Y, Wang J. The FOXO family of transcription factors: key molecular players in gastric cancer. J Mol Med (Berl) (2022) 100(7):997–1015. doi: 10.1007/s00109-022-02219-x
40. Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci (2019) 20(24):6249. doi: 10.3390/ijms20246249
41. Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci (2019) 76(3):441–51. doi: 10.1007/s00018-018-2940-7
42. van den Berg A, Mols J, Han J. RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta (2008) 1779(11):668–77. doi: 10.1016/j.bbagrm.2008.07.005
43. Jungers CF, Djuranovic S. Modulation of miRISC-mediated gene silencing in eukaryotes. Front Mol Biosci (2022) 9:832916. doi: 10.3389/fmolb.2022.832916
44. Seok H, Ham J, Jang ES, Chi SW. MicroRNA target recognition: insights from transcriptome-wide non-canonical interactions. Mol Cells (2016) 39(5):375–81. doi: 10.14348/molcells.2016.0013
45. Miao L, Yao H, Li C, Pu M, Yao X, Yang H, et al. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta (2016) 1859(4):650–62. doi: 10.1016/j.bbagrm.2016.02.016
46. Roberts TC. The microRNA biology of the mamMalian nucleus. Mol Ther Nucleic Acids (2014) 3(8):e188. doi: 10.1038/mtna.2014.40
47. Zhou Z, Gong Q, Lin Z, Wang Y, Li M, Wang L, et al. Emerging roles of SRSF3 as a therapeutic target for cancer. Front Oncol (2020) 10:577636. doi: 10.3389/fonc.2020.577636
48. Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci (2021) 64(1):22–50. doi: 10.1007/s11427-020-1700-9
49. Liu Y, Ao X, Wang Y, Li X, Wang J. Long non-coding RNA in gastric cancer: mechanisms and clinical implications for drug resistance. Front Oncol (2022) 12:841411. doi: 10.3389/fonc.2022.841411
50. Nunez-Martinez HN, Recillas-Targa F. Emerging Functions of lncRNA Loci beyond the Transcript Itself. Int J Mol Sci (2022) 23(11):6258. doi: 10.3390/ijms23116258
51. Giannakakis A, Zhang J, Jenjaroenpun P, Nama S, Zainolabidin N, Aau MY, et al. Contrasting expression patterns of coding and noncoding parts of the human genome upon oxidative stress. Sci Rep (2015) 5:9737. doi: 10.1038/srep09737
52. Zhang Q, Wang C, Yang Y, Xu R, Li Z. LncRNA and its role in gastric cancer immunotherapy. Front Cell Dev Biol (2023) 11:1052942. doi: 10.3389/fcell.2023.1052942
53. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol (2023) 24(6):430–47. doi: 10.1038/s41580-022-00566-8
54. Liu Y, Ding W, Yu W, Zhang Y, Ao X, Wang J. Long non-coding RNAs: Biogenesis, functions, and clinical significance in gastric cancer. Mol Ther Oncolytics (2021) 23:458–76. doi: 10.1016/j.omto.2021.11.005
55. Greco S, Gorospe M, Martelli F. Noncoding RNA in age-related cardiovascular diseases. J Mol Cell Cardiol (2015) 83:142–55. doi: 10.1016/j.yjmcc.2015.01.011
56. Lv N, Shen S, Chen Q, Tong J. Long noncoding RNAs: glycolysis regulators in gynaecologic cancers. Cancer Cell Int (2023) 23(1):4. doi: 10.1186/s12935-023-02849-2
57. Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in cancer. Mol Ther Nucleic Acids (2020) 21:367–83. doi: 10.1016/j.omtn.2020.06.008
58. Zhang L, Wang Y, Zhang Y, Zhao Y, Li P. Pathogenic mechanisms and the potential clinical value of circFoxo3 in cancers. Mol Ther Nucleic Acids (2021) 23:908–17. doi: 10.1016/j.omtn.2021.01.010
59. Wen ZJ, Xin H, Wang YC, Liu HW, Gao YY, Zhang YF. Emerging roles of circRNAs in the pathological process of myocardial infarction. Mol Ther Nucleic Acids (2021) 26:828–48. doi: 10.1016/j.omtn.2021.10.002
60. Zhang Y, Jia DD, Zhang YF, Cheng MD, Zhu WX, Li PF, et al. The emerging function and clinical significance of circRNAs in Thyroid Cancer and Autoimmune Thyroid Diseases. Int J Biol Sci (2021) 17(7):1731–41. doi: 10.7150/ijbs.55381
61. Wang M, Yu F, Zhang Y, Zhang L, Chang W, Wang K. The emerging roles of circular RNAs in the chemoresistance of gastrointestinal cancer. Front Cell Dev Biol (2022) 10:821609. doi: 10.3389/fcell.2022.821609
62. Shoda K, Kuwano Y, Ichikawa D, Masuda K. circRNA: A new biomarker and therapeutic target for esophageal cancer. Biomedicines (2022) 10(7):1643. doi: 10.3390/biomedicines10071643
63. Liu R, Zhang L, Zhao X, Liu J, Chang W, Zhou L, et al. circRNA: Regulatory factors and potential therapeutic targets in inflammatory dermatoses. J Cell Mol Med (2022) 26(16):4389–400. doi: 10.1111/jcmm.17473
64. Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol (2020) 186:101746. doi: 10.1016/j.pneurobio.2020.101746
65. Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J (2017) 38(18):1402–12. doi: 10.1093/eurheartj/ehw001
66. Wang L, Wu S, He H, Ai K, Xu R, Zhang L, et al. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab Invest (2022) 102(12):1323–34. doi: 10.1038/s41374-022-00826-3
67. Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci U.S.A. (2021) 118(33):e2012881118. doi: 10.1073/pnas.2012881118
68. Liu C, Wu X, Gokulnath P, Li G, Xiao J. The functions and mechanisms of translatable circular RNAs. J Pharmacol Exp Ther (2023) 384(1):52–60. doi: 10.1124/jpet.122.001085
69. Sun Y, He P, Li L, Ding X. The significance of the crosstalk between ubiquitination or deubiquitination and ncRNAs in non-small cell lung cancer. Front Oncol (2022) 12:969032. doi: 10.3389/fonc.2022.969032
70. Zhang X, Bao L, Yu G, Wang H. Exosomal miRNA-profiling of pleural effusion in lung adenocarcinoma and tuberculosis. Front Surg (2022) 9:1050242. doi: 10.3389/fsurg.2022.1050242
71. Zeng L, Zeng G, Ye Z. Bioinformatics analysis for identifying differentially expressed microRNAs derived from plasma exosomes associated with radiotherapy resistance in non-small-cell lung cancer. Appl Bionics Biomech (2022) 2022:9268206. doi: 10.1155/2022/9268206
72. Zhang H, Ma RR, Zhang G, Dong Y, Duan M, Sun Y, et al. Long noncoding RNA lnc-LEMGC combines with DNA-PKcs to suppress gastric cancer metastasis. Cancer Lett (2022) 524:82–90. doi: 10.1016/j.canlet.2021.09.042
73. Huang X, Zhou H, Yang X, Shi W, Hu L, Wang J, et al. Construction and analysis of expression profile of exosomal lncRNAs in pleural effusion in lung adenocarcinoma. J Clin Lab Anal (2022) 36(12):e24777. doi: 10.1002/jcla.24777
74. Cai Y, Zhu C, Wang Y, Jiang Y, Zhu Z. Comprehensive circular RNA expression profile of lung adenocarcinoma with bone metastasis: Identification of potential biomarkers. Front Genet (2022) 13:961668. doi: 10.3389/fgene.2022.961668
75. Asghariazar V, Sakhinia E, Mansoori B, Mohammadi A, Baradaran B. Tumor suppressor microRNAs in lung cancer: An insight to signaling pathways and drug resistance. J Cell Biochem (2019) 120(12):19274–89. doi: 10.1002/jcb.29295
76. Doghish AS, Ismail A, Elrebehy MA, Elbadry AMM, Mahmoud HH, Farouk SM, et al. A study of miRNAs as cornerstone in lung cancer pathogenesis and therapeutic resistance: A focus on signaling pathways interplay. Pathol Res Pract (2022) 237:154053. doi: 10.1016/j.prp.2022.154053
77. Diao MN, Zhang XJ, Zhang YF. The critical roles of m6A RNA methylation in lung cancer: from mechanism to prognosis and therapy. Br J Cancer (2023) 129(1):8–23. doi: 10.1038/s41416-023-02246-6
78. Ghareghomi S, Atabaki V, Abdollahzadeh N, Ahmadian S, Hafez Ghoran S. Bioactive PI3-kinase/akt/mTOR inhibitors in targeted lung cancer therapy. Adv Pharm Bull (2023) 13(1):24–35. doi: 10.34172/apb.2023.003
79. Niu H, Wang D, Wen T, Liu H, Jie J, Song L, et al. Anwuligan inhibits the progression of non-small cell lung cancer via let-7c-3p/PI3K/AKT/mTOR axis. Cancer Med (2023) 12(5):5908–25. doi: 10.1002/cam4.5382
80. Shi L, Kan J, Zhuo L, Wang S, Chen S, Zhang B, et al. Bioinformatics identification of miR-514b-5p promotes NSCLC progression and induces PI3K/AKT and p38 pathways by targeting small glutamine-rich tetratricopeptide repeat-containing protein beta. FEBS J (2023) 290(4):1134–50. doi: 10.1111/febs.16639
81. Xia M, Zhu W, Tao C, Lu Y, Gao F. LncRNA LASTR promote lung cancer progression through the miR-137/TGFA/PI3K/AKT axis through integration analysis. J Cancer (2022) 13(4):1086–96. doi: 10.7150/jca.66067
82. Liu X, Wang Y, Zhou G, Zhou J, Tian Z, Xu J. circGRAMD1B contributes to migration, invasion and epithelial-mesenchymal transition of lung adenocarcinoma cells via modulating the expression of SOX4. Funct Integr Genomics (2023) 23(1):75. doi: 10.1007/s10142-023-00972-x
83. Liu Y, Ding W, Wang J, Ao X, Xue J. Non-coding RNA-mediated modulation of ferroptosis in cardiovascular diseases. BioMed Pharmacother (2023) 164:114993. doi: 10.1016/j.biopha.2023.114993
84. Liu Y, Su CY, Yan YY, Wang J, Li JJ, Fu JJ, et al. Exosomes of A549 Cells Induced Migration, Invasion, and EMT of BEAS-2B Cells Related to let-7c-5p and miR-181b-5p. Front Endocrinol (Lausanne) (2022) 13:926769. doi: 10.3389/fendo.2022.926769
85. Shi Z, Liu J, Sun D. Let-7a targets Rsf-1 to modulate radiotherapy response of non-small cell lung cancer cells through Ras-MAPK pathway. J BUON (2021) 26(4):1422–31.
86. Zhu H, Liu Q, Yang X, Ding C, Wang Q, Xiong Y. LncRNA LINC00649 recruits TAF15 and enhances MAPK6 expression to promote the development of lung squamous cell carcinoma via activating MAPK signaling pathway. Cancer Gene Ther (2022) 29(8-9):1285–95. doi: 10.1038/s41417-021-00410-9
87. Wang B, Yang S, Jia Y, Yang J, Du K, Luo Y, et al. PCAT19 Regulates the Proliferation and Apoptosis of Lung Cancer Cells by Inhibiting miR-25-3p via Targeting the MAP2K4 Signal Axis. Dis Markers (2022) 2022:2442094. doi: 10.1155/2022/2442094
88. Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y, et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl Lung Cancer Res (2019) 8(6):862–75. doi: 10.21037/tlcr.2019.11.04
89. Zhuang Q, Huang Z, Zhuang W, Hong Y, Huang Y. Knockdown of circ-RAD23B inhibits non-small cell lung cancer progression via the miR-142-3p/MAP4K3 axis. Thorac Cancer (2022) 13(5):750–60. doi: 10.1111/1759-7714.14319
90. Hu S, Cao P, Kong K, Han P, Yue J, Deng Y, et al. circCNN2 Accelerates Cell Proliferation and Invasion in Lung Squamous Cell Carcinoma via Regulating miR-184/E2F1 and Activating MAPK Signaling Pathway. Dis Markers (2022) 2022:6329097. doi: 10.1155/2022/6329097
91. Liu Y, Li Y, Du C, Kuang S, Zhou X, Zhang J, et al. Underlying mechanisms of epithelial splicing regulatory proteins in cancer progression. J Mol Med (Berl) (2022) 100(11):1539–56. doi: 10.1007/s00109-022-02257-5
92. Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei Y, et al. HIF-1a-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/beta-catenin and Notch signaling. Theranostics (2020) 10(6):2553–70. doi: 10.7150/thno.41120
93. Su WZ, Ren LF. MiRNA-199 inhibits Malignant progression of lung cancer through mediating RGS17. Eur Rev Med Pharmacol Sci (2019) 23(8):3390–400. doi: 10.26355/eurrev_201904_17703
94. Qian H, Chen L, Huang J, Wang X, Ma S, Cui F, et al. The lncRNA MIR4435-2HG promotes lung cancer progression by activating beta-catenin signalling. J Mol Med (Berl) (2018) 96(8):753–64. doi: 10.1007/s00109-018-1654-5
95. Lin H, Shangguan Z, Zhu M, Bao L, Zhang Q, Pan S. lncRNA FLVCR1-AS1 silencing inhibits lung cancer cell proliferation, migration, and invasion by inhibiting the activity of the Wnt/beta-catenin signaling pathway. J Cell Biochem (2019) 120(6):10625–32. doi: 10.1002/jcb.28352
96. Li XY, Liu YR, Zhou JH, Li W, Guo HH, Ma HP. Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non-small cell lung cancer by modulating Wnt/beta-catenin pathway. Eur Rev Med Pharmacol Sci (2019) 23(8):3366–74. doi: 10.26355/eurrev_201904_17700
97. Ding L, Yao W, Lu J, Gong J, Zhang X. Upregulation of circ_001569 predicts poor prognosis and promotes cell proliferation in non-small cell lung cancer by regulating the Wnt/beta-catenin pathway. Oncol Lett (2018) 16(1):453–8. doi: 10.3892/ol.2018.8673
98. Liu M, Liu C, Li X, Li S. RP11-79H23.3 inhibits the proliferation and metastasis of non-small-cell lung cancer through promoting miR-29c. Biochem Genet (2023) 61(2):506–20. doi: 10.1007/s10528-022-10263-y
99. Yang B, Zhang B, Qi Q, Wang C. CircRNA has_circ_0017109 promotes lung tumor progression via activation of Wnt/beta-catenin signaling due to modulating miR-671-5p/FZD4 axis. BMC Pulm Med (2022) 22(1):443. doi: 10.1186/s12890-022-02209-2
100. Xu J, Huang L, Bao T, Duan K, Cheng Y, Zhang H, et al. CircCDR1as mediates PM(2.5)-induced lung cancer progression by binding to SRSF1. Ecotoxicol Environ Saf (2023) 249:114367. doi: 10.1016/j.ecoenv.2022.114367
101. Xia R, Xu M, Yang J, Ma X. The role of Hedgehog and Notch signaling pathway in cancer. Mol BioMed (2022) 3(1):44. doi: 10.1186/s43556-022-00099-8
102. Zhang H, Yang Y, Li X, Yuan X, Chu Q. Targeting the Notch signaling pathway and the Notch ligand, DLL3, in small cell lung cancer. BioMed Pharmacother (2023) 159:114248. doi: 10.1016/j.biopha.2023.114248
103. Ji X, Wang Z, Geamanu A, Goja A, Sarkar FH, Gupta SV. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells. Int J Cancer (2012) 131(11):2668–77. doi: 10.1002/ijc.27549
104. Xue B, Chuang CH, Prosser HM, Fuziwara CS, Chan C, Sahasrabudhe N, et al. miR-200 deficiency promotes lung cancer metastasis by activating Notch signaling in cancer-associated fibroblasts. Genes Dev (2021) 35(15-16):1109–22. doi: 10.1101/gad.347344.120
105. Zhang F, Sang Y, Chen D, Wu X, Wang X, Yang W, et al. M2 macrophage-derived exosomal long non-coding RNA AGAP2-AS1 enhances radiotherapy immunity in lung cancer by reducing microRNA-296 and elevating NOTCH2. Cell Death Dis (2021) 12(5):467. doi: 10.1038/s41419-021-03700-0
106. Deng Y, Zhang L. LncRNA SNHG11 accelerates the progression of lung adenocarcinoma via activating Notch pathways. Pathol Res Pract (2022) 234:153849. doi: 10.1016/j.prp.2022.153849
107. Zheng H, Zhu X, Gong E, Lv Y, Li Y, Cai X. Luteolin suppresses lung cancer progression through targeting the circ_0000190/miR-130a-3p/notch-1 signaling pathway. J Chemother (2022) 35(4):1–13. doi: 10.1080/1120009X.2022.2102303
108. Mirzaei S, Saghari S, Bassiri F, Raesi R, Zarrabi A, Hushmandi K, et al. NF-kappaB as a regulator of cancer metastasis and therapy response: A focus on epithelial-mesenchymal transition. J Cell Physiol (2022) 237(7):2770–95. doi: 10.1002/jcp.30759
109. Wang Y, Liu F, Chen L, Fang C, Li S, Yuan S, et al. Neutrophil extracellular traps (NETs) promote non-small cell lung cancer metastasis by suppressing lncRNA MIR503HG to activate the NF-kappaB/NLRP3 inflammasome pathway. Front Immunol (2022) 13:867516. doi: 10.3389/fimmu.2022.867516
110. Zhou Q, Sun Y. Circular RNA cMras suppresses the progression of lung adenocarcinoma through ABHD5/ATGL axis using NF-kappaB signaling pathway. Cancer Biother Radiopharm (2020) 38(5):336–46. doi: 10.1089/cbr.2020.3709
111. Zhao J, Wang X, Mi Z, Jiang X, Sun L, Zheng B, et al. STAT3/miR-135b/NF-kappaB axis confers aggressiveness and unfavorable prognosis in non-small-cell lung cancer. Cell Death Dis (2021) 12(5):493. doi: 10.1038/s41419-021-03773-x
112. Kang S, Ou C, Yan A, Zhu K, Xue R, Zhang Y, et al. Long noncoding RNA SNHG5 induces the NF-kappaB pathway by regulating miR-181c-5p/CBX4 axis to promote the progression of non-small cell lung cancer. Arch Bronconeumol (2023) 59(1):10–8. doi: 10.1016/j.arbres.2022.07.001
113. Huang Z, Lei W, Hu HB, Zhang H, Zhu Y. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol (2018) 233(10):6768–76. doi: 10.1002/jcp.26530
114. Xu J, Ni L, Zhao F, Dai X, Tao J, Pan J, et al. Overexpression of hsa_circ_0002874 promotes resistance of non-small cell lung cancer to paclitaxel by modulating miR-1273f/MDM2/p53 pathway. Aging (Albany NY) (2021) 13(4):5986–6009. doi: 10.18632/aging.202521
115. Huang MS, Liu JY, Xia XB, Liu YZ, Li X, Yin JY, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol (2019) 9:508. doi: 10.3389/fonc.2019.00508
116. Liu Y, Ao X, Zhou X, Du C, Kuang S. The regulation of PBXs and their emerging role in cancer. J Cell Mol Med (2022) 26(5):1363–79. doi: 10.1111/jcmm.17196
117. Luo J, Xia L, Zhang L, Zhao K, Li C. MiRNA-144-5p down-modulates CDCA3 to regulate proliferation and apoptosis of lung adenocarcinoma cells. Mutat Res (2022) 825:111798. doi: 10.1016/j.mrfmmm.2022.111798
118. Han F, Chen G, Guo Y, Li B, Sun Y, Qi X, et al. MicroRNA-4491 enhances cell proliferation and inhibits cell apoptosis in non-small cell lung cancer via targeting TRIM7. Oncol Lett (2021) 22(2):591. doi: 10.3892/ol.2021.12852
119. Tang L, Wang S, Wang Y, Li K, Li Q. LncRNA-UCA1 regulates lung adenocarcinoma progression through competitive binding to miR-383. Cell Cycle (2023) 22(2):213–28. doi: 10.1080/15384101.2022.2111929
120. Wang L, Jin W, Wu X, Liu Y, Gu W. Circ_0000520 interacts with miR-512-5p to upregulate KIAA0100 to promote Malignant behaviors in lung cancer. Histol Histopathol (2023) 38(1):73–89. doi: 10.14670/HH-18-498
121. Sati I, Parhar I. MicroRNAs regulate cell cycle and cell death pathways in glioblastoma. Int J Mol Sci (2021) 22(24):13550. doi: 10.3390/ijms222413550
122. Huang L, Lou K, Wang K, Liang L, Chen Y, Zhang J. Let-7c-5p represses cisplatin resistance of lung adenocarcinoma cells by targeting CDC25A. Appl Biochem Biotechnol (2023) 195(3):1644–55. doi: 10.1007/s12010-022-04219-6
123. Wang N, Zhao Q, Huang Y, Wen C, Li Y, Bao M, et al. Lnc-TMEM132D-AS1 as a potential therapeutic target for acquired resistance to osimertinib in non-small-cell lung cancer. Mol Omics (2023) 19(3):238–51. doi: 10.1039/d2mo00261b
124. Du L, Guo D, Sun C, Yan X, Lin S, Xu S. CircPIM3 regulates taxol resistance in non-small cell lung cancer via miR-338-3p/TNFAIP8 axis. Anticancer Drugs (2023) 34(1):115–25. doi: 10.1097/CAD.0000000000001347
125. Ao X, Ding W, Zhang Y, Ding D, Liu Y. TCF21: a critical transcription factor in health and cancer. J Mol Med (Berl) (2020) 98(8):1055–68. doi: 10.1007/s00109-020-01934-7
126. Wang M, Yu F. Research progress on the anticancer activities and mechanisms of polysaccharides from ganoderma. Front Pharmacol (2022) 13:891171. doi: 10.3389/fphar.2022.891171
127. Chu F, Xu X, Zhang Y, Cai H, Peng J, Li Y, et al. LIM-domain binding protein 2 was down-regulated by miRNA-96-5p inhibited the proliferation, invasion and metastasis of lung cancer H1299 cells. Clinics (Sao Paulo) (2023) 78:100145. doi: 10.1016/j.clinsp.2022.100145
128. Fang X, Shi H, Sun F. The microRNA-520a-3p inhibits invasion and metastasis by targeting NF-kappaB signaling pathway in non-small cell lung cancer. Clin Transl Oncol (2022) 24(8):1569–79. doi: 10.1007/s12094-022-02797-9
129. Li R, Lin Y, Hu F, Liao Y, Tang J, Shen Y, et al. LncRNA TEX41 regulates autophagy by increasing Runx2 expression in lung adenocarcinoma bone metastasis. Am J Transl Res (2023) 15(2):949–66.
130. Sun C, Guan H, Li J, Gu Y. circ_0000376 knockdown suppresses non-small cell lung cancer cell tumor properties by the miR-545-3p/PDPK1 pathway. Open Med (Wars) (2023) 18(1):20230641. doi: 10.1515/med-2023-0641
131. Yang C, Huang H, Li Y, Zhuo T, Zhu L, Luo C, et al. LncRNA PCAT6 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of lung adenocarcinoma cell by targeting miR-545-3p. Mol Biol Rep (2023) 50(4):3557–68. doi: 10.1007/s11033-023-08259-x
132. Liu B, Ma H, Liu X, Xing W. CircSCN8A suppresses Malignant progression and induces ferroptosis in non-small cell lung cancer by regulating miR-1290/ACSL4 axis. Cell Cycle (2023) 22(7):758–76. doi: 10.1080/15384101.2022.2154543
133. Liao Y, Wu X, Wu M, Fang Y, Li J, Tang W. Non-coding RNAs in lung cancer: emerging regulators of angiogenesis. J Transl Med (2022) 20(1):349. doi: 10.1186/s12967-022-03553-x
134. Gan H, Xu X, Bai Y. Trametes robiniophila represses angiogenesis and tumor growth of lung cancer via strengthening let-7d-5p and targeting NAP1L1. Bioengineered (2022) 13(3):6698–710. doi: 10.1080/21655979.2021.2012619
135. Chang RM, Fu Y, Zeng J, Zhu XY, Gao Y. Cancer-derived exosomal miR-197-3p confers angiogenesis via targeting TIMP2/3 in lung adenocarcinoma metastasis. Cell Death Dis (2022) 13(12):1032. doi: 10.1038/s41419-022-05420-5
136. Pan H, Peng J, Qiao X, Gao H. Upregulation of lncRNA LANCL1-AS1 inhibits the progression of non-small-cell lung cancer via the miR-3680-3p/GMFG axis. Open Med (Wars) (2023) 18(1):20230666. doi: 10.1515/med-2023-0666
137. Wang J, Tan L, Yu X, Cao X, Jia B, Chen R, et al. lncRNA ZNRD1-AS1 promotes Malignant lung cell proliferation, migration, and angiogenesis via the miR-942/TNS1 axis and is positively regulated by the m(6)A reader YTHDC2. Mol Cancer (2022) 21(1):229. doi: 10.1186/s12943-022-01705-7
138. Zhou Y, Liu H, Wang R, Zhang M. Circ_0043256 upregulates KLF2 expression by absorbing miR-1206 to suppress the tumorigenesis of lung cancer. Thorac Cancer (2023) 14(7):683–99. doi: 10.1111/1759-7714.14794
139. Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther (2021) 221:107753. doi: 10.1016/j.pharmthera.2020.107753
140. Liu J, Cao L, Li Y, Deng P, Pan P, Hu C, et al. Pirfenidone promotes the levels of exosomal miR-200 to down-regulate ZEB1 and represses the epithelial-mesenchymal transition of non-small cell lung cancer cells. Hum Cell (2022) 35(6):1813–23. doi: 10.1007/s13577-022-00766-6
141. Enukashvily NI, Ponomartsev NV, Ketkar A, Suezov R, Chubar AV, Prjibelski AD, et al. Pericentromeric satellite lncRNAs are induced in cancer-associated fibroblasts and regulate their functions in lung tumorigenesis. Cell Death Dis (2023) 14(1):19. doi: 10.1038/s41419-023-05553-1
142. Li X, Zhu G, Li Y, Huang H, Chen C, Wu D, et al. LINC01798/miR-17-5p axis regulates ITGA8 and causes changes in tumor microenvironment and stemness in lung adenocarcinoma. Front Immunol (2023) 14:1096818. doi: 10.3389/fimmu.2023.1096818
143. Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature (2021) 595(7868):578–84. doi: 10.1038/s41586-021-03651-8
144. Ma J, Chen S, Liu Y, Han H, Gong M, Song Y. The role of exosomal miR-181b in the crosstalk between NSCLC cells and tumor-associated macrophages. Genes Genomics (2022) 44(10):1243–58. doi: 10.1007/s13258-022-01227-y
145. Wu Z, Bai X, Lu Z, Liu S, Jiang H. LINC01094/SPI1/CCL7 axis promotes macrophage accumulation in lung adenocarcinoma and tumor cell dissemination. J Immunol Res (2022) 2022:6450721. doi: 10.1155/2022/6450721
146. Chen T, Liu Y, Li C, Xu C, Ding C, Chen J, et al. Tumor-derived exosomal circFARSA mediates M2 macrophage polarization via the PTEN/PI3K/AKT pathway to promote non-small cell lung cancer metastasis. Cancer Treat Res Commun (2021) 28:100412. doi: 10.1016/j.ctarc.2021.100412
147. Zhou H, Tan L, Liu B, Guan XY. Cancer stem cells: Recent insights and therapies. Biochem Pharmacol (2023) 209:115441. doi: 10.1016/j.bcp.2023.115441
148. Moro M, Fortunato O, Bertolini G, Mensah M, Borzi C, Centonze G, et al. MiR-486-5p Targets CD133+ Lung Cancer Stem Cells through the p85/AKT Pathway. Pharm (Basel) (2022) 15(3):297. doi: 10.3390/ph15030297
149. Liu Z, Zhao W, Yang R. MiR-1246 is responsible for lung cancer cells-derived exosomes-mediated promoting effects on lung cancer stemness via targeting TRIM17. Environ Toxicol (2022) 37(11):2651–9. doi: 10.1002/tox.23625
150. Wang P, Zhang Y, Lv X, Zhou J, Cang S, Song Y. LncRNA ADAMTS9-AS1 inhibits the stemness of lung adenocarcinoma cells by regulating miR-5009-3p/NPNT axis. Genomics (2023) 115(3):110596. doi: 10.1016/j.ygeno.2023.110596
151. Lu Y, Cheng J, Mao Q, Wang Z, Wei Q. Long non-coding RNA TDRG1 aggravates lung cancer stemness by binding to Sox2 mRNA. Environ Toxicol (2023) 38(3):645–53. doi: 10.1002/tox.23714
152. Xiong H, Liu B, Liu XY, Xia ZK, Lu M, Hu CH, et al. circ_rac GTPase-activating protein 1 facilitates stemness and metastasis of non-small cell lung cancer via polypyrimidine tract-binding protein 1 recruitment to promote sirtuin-3-mediated replication timing regulatory factor 1 deacetylation. Lab Invest (2023) 103(1):100010. doi: 10.1016/j.labinv.2022.100010
153. Liu Y, Wang Y, Li X, Jia Y, Wang J, Ao X. FOXO3a in cancer drug resistance. Cancer Lett (2022) 540:215724. doi: 10.1016/j.canlet.2022.215724
154. Zhou X, Ao X, Jia Z, Li Y, Kuang S, Du C, et al. Non-coding RNA in cancer drug resistance: Underlying mechanisms and clinical applications. Front Oncol (2022) 12:951864. doi: 10.3389/fonc.2022.951864
155. Wang YF, Ao X, Liu Y, Ding D, Jiao WJ, Yu Z, et al. MicroRNA-608 promotes apoptosis in non-small cell lung cancer cells treated with doxorubicin through the inhibition of TFAP4. Front Genet (2019) 10:809. doi: 10.3389/fgene.2019.00809
156. Rampioni Vinciguerra GL, Capece M, Distefano R, Nigita G, Vecchione A, Lovat F, et al. Role of the miR-301a/Fra-2/GLIPR1 axis in lung cancer cisplatin resistance. Signal Transduct Target Ther (2023) 8(1):37. doi: 10.1038/s41392-022-01228-z
157. Huang X, Wang Y, Chen M, Li G. MiR-936 targets GPR78 and regulates chemotherapy resistance in non-small cell lung cancer by activating the galphaq Rho GTPase pathway. Altern Ther Health Med (2023) 29(2):58–63.
158. Yu Z, Tang H, Chen S, Xie Y, Shi L, Xia S, et al. Exosomal LOC85009 inhibits docetaxel resistance in lung adenocarcinoma through regulating ATG5-induced autophagy. Drug Resist Update (2023) 67:100915. doi: 10.1016/j.drup.2022.100915
159. Liu J, Yang X, Gao S, Wen M, Yu Q. DDX11-AS1 modulates DNA damage repair to enhance paclitaxel resistance of lung adenocarcinoma cells. Pharmacogenomics (2023) 24(3):163–72. doi: 10.2217/pgs-2022-0121
160. Warren GW, Cummings KM. Tobacco and lung cancer: risks, trends, and outcomes in patients with cancer. Am Soc Clin Oncol Educ Book (2013), 359–64. doi: 10.14694/EdBook_AM.2013.33.359
161. Zhao H, Wang Y, Ren X. Nicotine promotes the development of non-small cell lung cancer through activating LINC00460 and PI3K/Akt signaling. Biosci Rep (2019) 39(6):BSR20182443. doi: 10.1042/BSR20182443
162. Liu CH, Chen Z, Chen K, Liao FT, Chung CE, Liu X, et al. Lipopolysaccharide-mediated chronic inflammation promotes tobacco carcinogen-induced lung cancer and determines the efficacy of immunotherapy. Cancer Res (2021) 81(1):144–57. doi: 10.1158/0008-5472.CAN-20-1994
163. Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis (2008) 29(12):2394–9. doi: 10.1093/carcin/bgn209
164. Chen E, Zhou J, Xu E, Zhang C, Liu J, Zhou J, et al. A genome-wide screen for differentially methylated long noncoding RNAs identified that lncAC007255.8 is regulated by promoter DNA methylation in Beas-2B cells Malignantly transformed by NNK. Toxicol Lett (2021) 346:34–46. doi: 10.1016/j.toxlet.2021.04.013
165. Hua Q, Liu Y, Li M, Chen Y, Diao Q, Zeng H, et al. Tobacco-related exposure upregulates circ_0035266 to exacerbate inflammatory responses in human bronchial epithelial cells. Toxicol Sci (2021) 179(1):70–83. doi: 10.1093/toxsci/kfaa163
166. Hua Q, Liu Y, Li M, Li X, Chen W, Diao Q, et al. Upregulation of circ_0035266 contributes to the Malignant progression of inflammation-associated Malignant transformed cells induced by tobacco-specific carcinogen NNK. Toxicol Sci (2022) 189(2):203–15. doi: 10.1093/toxsci/kfac072
167. Liu Z, Lu C, Zhao G, Han X, Dong K, Wang C, et al. Downregulation of miR-218 by nicotine promotes cell proliferation through targeting CDK6 in non-small cell lung cancer. J Cell Biochem (2019) 120(10):18370–7. doi: 10.1002/jcb.29148
168. Zong D, Liu X, Li J, Long Y, Ouyang R, Chen Y. LncRNA-CCAT1/miR-152-3p is involved in CSE-induced inflammation in HBE cells via regulating ERK signaling pathway. Int Immunopharmacol (2022) 109:108818. doi: 10.1016/j.intimp.2022.108818
169. Boyle P, Chapman CJ, Holdenrieder S, Murray A, Robertson C, Wood WC, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol (2011) 22(2):383–9. doi: 10.1093/annonc/mdq361
170. Wang W, Li X, Liu C, Zhang X, Wu Y, Diao M, et al. MicroRNA-21 as a diagnostic and prognostic biomarker of lung cancer: a systematic review and meta-analysis. Biosci Rep (2022) 42(5):BSR20211653. doi: 10.1042/BSR20211653
171. Wu Y, Jing H, Zhang J. MicroRNA-340 and microRNA-450b-5p: plasma biomarkers for detection of non-small-cell lung cancer. J Environ Public Health (2022) 2022:8024700. doi: 10.1155/2022/8024700
172. Yuan RX, Dai CH, Chen P, Lv MJ, Shu Y, Wang ZP, et al. Circulating TP73-AS1 and CRNDE serve as diagnostic and prognostic biomarkers for non-small cell lung cancer. Cancer Med (2023) 12(2):1655–72. doi: 10.1002/cam4.5013
173. Alexandre D, Teixeira B, Rico A, Valente S, Craveiro A, Baptista PV, et al. Molecular beacon for detection miRNA-21 as a biomarker of lung cancer. Int J Mol Sci (2022) 23(6):3330. doi: 10.3390/ijms23063330
174. Sui J. MiRNA-30 play an important role in non-small cell lung cancer (NSCLC). Clin Lab (2020) 66(4). doi: 10.7754/Clin.Lab.2019.190712
175. Liu S, Wang W, Ning Y, Zheng H, Zhan Y, Wang H, et al. Exosome-mediated miR-7-5p delivery enhances the anticancer effect of Everolimus via blocking MNK/eIF4E axis in non-small cell lung cancer. Cell Death Dis (2022) 13(2):129. doi: 10.1038/s41419-022-04565-7
176. Hanafi AR, Jayusman AM, Alfasunu S, Sadewa AH, Pramono D, Heriyanto DS, et al. Serum miRNA as predictive and prognosis biomarker in advanced stage non-small cell lung cancer in Indonesia. Zhongguo Fei Ai Za Zhi (2020) 23(5):321–32. doi: 10.3779/j.issn.1009-3419.2020.104.02
177. D'Antona P, Cattoni M, Dominioni L, Poli A, Moretti F, Cinquetti R, et al. Serum miR-223: A validated biomarker for detection of early-stage non-small cell lung cancer. Cancer Epidemiol Biomarkers Prev (2019) 28(11):1926–33. doi: 10.1158/1055-9965.EPI-19-0626
178. Trakunram K, Chaniad P, Geater SL, Keeratichananont W, Chittithavorn V, Uttayamakul S, et al. Serum miR-339-3p as a potential diagnostic marker for non-small cell lung cancer. Cancer Biol Med (2020) 17(3):652–63. doi: 10.20892/j.issn.2095-3941.2020.0063
179. Hetta HF, Zahran AM, Shafik EA, El-Mahdy RI, Mohamed NA, Nabil EE, et al. Circulating miRNA-21 and miRNA-23a Expression Signature as Potential Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. Microrna (2019) 8(3):206–15. doi: 10.2174/1573399815666190115151500
180. Liu F, Li T, Hu P, Dai L. Upregulation of serum miR-629 predicts poor prognosis for non-small-cell lung cancer. Dis Markers (2021) 2021:8819934. doi: 10.1155/2021/8819934
181. Huang YF, Liu MW, Xia HB, He R. Expression of miR-92a is associated with the prognosis in non-small cell lung cancer: An observation study. Med (Baltimore) (2022) 101(41):e30970. doi: 10.1097/MD.0000000000030970
182. Huang X, Huang M, Chen M, Chen X. lncRNA SLC9A3-AS1 Promotes Oncogenesis of NSCLC via Sponging microRNA-760 and May Serve as a Prognosis Predictor of NSCLC Patients. Cancer Manag Res (2022) 14:1087–98. doi: 10.2147/CMAR.S352308
183. Yao X, Wang T, Sun MY, Yuming Y, Guixin D, Liu J. Diagnostic value of lncRNA HOTAIR as a biomarker for detecting and staging of non-small cell lung cancer. Biomarkers (2022) 27(6):526–33. doi: 10.1080/1354750X.2022.2085799
184. Min L, Zhu T, Lv B, An T, Zhang Q, Shang Y, et al. Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int J Clin Oncol (2022) 27(6):1013–24. doi: 10.1007/s10147-022-02129-5
185. Yang B, Miao S. lncRNA ELFN1-AS1 predicts poor prognosis and promotes tumor progression of non-small cell lung cancer by sponging miR-497. Cancer biomark (2022) 34(4):637–46. doi: 10.3233/CBM-210393
186. Luo Y, Zhang Q, Lv B, Shang Y, Li J, Yang L, et al. CircFOXP1: A novel serum diagnostic biomarker for non-small cell lung cancer. Int J Biol Markers (2022) 37(1):58–65. doi: 10.1177/17246008211073151
187. Chen Y, Lou C, Ma X, Zhou C, Zhao X, Li N, et al. Serum exosomal hsa_circ_0069313 has a potential to diagnose more aggressive non-small cell lung cancer. Clin Biochem (2022) 102:56–64. doi: 10.1016/j.clinbiochem.2022.01.005
188. Zhang Q, Qin S, Peng C, Liu Y, Huang Y, Ju S. Circulating circular RNA hsa_circ_0023179 acts as a diagnostic biomarker for non-small-cell lung cancer detection. J Cancer Res Clin Oncol (2022) 149(7):3649–60. doi: 10.1007/s00432-022-04254-0
189. Huang Y, Qin S, Gu X, Zheng M, Zhang Q, Liu Y, et al. Comprehensive assessment of serum hsa_circ_0070354 as a novel diagnostic and predictive biomarker in non-small cell lung cancer. Front Genet (2021) 12:796776. doi: 10.3389/fgene.2021.796776
190. Yang F, Ma C, Qiu J, Feng X, Yang K. Identification of circRNA_001846 as putative non-small cell lung cancer biomarker. Bioengineered (2021) 12(1):8690–7. doi: 10.1080/21655979.2021.1991161
191. Yu Y, Zuo J, Tan Q, Zar Thin K, Li P, Zhu M, et al. Plasma miR-92a-2 as a biomarker for small cell lung cancer. Cancer biomark (2017) 18(3):319–27. doi: 10.3233/CBM-160254
192. Li M, Shan W, Hong B, Zou J, Li H, Han D, et al. Circulating miR-92b and miR-375 for monitoring the chemoresistance and prognosis of small cell lung cancer. Sci Rep (2020) 10(1):12705. doi: 10.1038/s41598-020-69615-6
193. Ranade AR, Cherba D, Sridhar S, Richardson P, Webb C, Paripati A, et al. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol (2010) 5(8):1273–8. doi: 10.1097/JTO.0b013e3181dea6be
194. Li D, Tong Q, Lian Y, Chen Z, Zhu Y, Huang W, et al. Inhibition of lncRNA KCNQ1OT1 improves apoptosis and chemotherapy drug response in small cell lung cancer by TGF-beta1 mediated epithelial-to-mesenchymal transition. Cancer Res Treat (2021) 53(4):1042–56. doi: 10.4143/crt.2020.1208
195. Chen S, Wu H, Lv N, Wang H, Wang Y, Tang Q, et al. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. BioMed Pharmacother (2016) 82:583–8. doi: 10.1016/j.biopha.2016.05.017
196. Liu B, Liu X, Yang Y, Ge J, Li J. [Expression of lncRNA AK09398 and relationship with prognosis in patients with small cell lung cancer]. Zhongguo Fei Ai Za Zhi (2016) 19(11):754–9. doi: 10.3779/j.issn.1009-3419.2016.11.06
197. Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ (2020) 27(5):1709–27. doi: 10.1038/s41418-019-0455-x
198. Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, et al. FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res (2019) 25(4):1302–17. doi: 10.1158/1078-0432.CCR-18-1447
199. Shimada Y, Yoshioka Y, Kudo Y, Mimae T, Miyata Y, Adachi H, et al. Extracellular vesicle-associated microRNA signatures related to lymphovascular invasion in early-stage lung adenocarcinoma. Sci Rep (2023) 13(1):4823. doi: 10.1038/s41598-023-32041-5
200. Jiang X, Yuan Y, Tang L, Wang J, Zhang D, Cho WC, et al. Identification and validation prognostic impact of miRNA-30a-5p in lung adenocarcinoma. Front Oncol (2022) 12:831997. doi: 10.3389/fonc.2022.831997
201. Tang L, Yuan Y, Zhai H, Wang J, Zhang D, Liang H, et al. MicroRNA-125b-5p correlates with prognosis and lung adenocarcinoma progression. Front Mol Biosci (2021) 8:788690. doi: 10.3389/fmolb.2021.788690
202. Gu X, Zhang Y, Xiong H. Correlation between plasma lncRNA CASC11 and Malignancy in lung adenocarcinoma patients and the prognostic value of lncRNA CASC11. Per Med (2023) 20(2):167–73. doi: 10.2217/pme-2022-0104
203. Liu Z, Jiang H, Zhao R, Quan Q, Huang X. Long noncoding RNA IPW is a novel diagnostic and predictive biomarker in lung adenocarcinoma. Genet Test Mol Biomarkers (2023) 27(1):18–26. doi: 10.1089/gtmb.2022.0173
204. Shen X, Zeng Y, Yang C, Jiang L, Chen S, Chen F, et al. The diagnostic and prognostic value of pseudogene SIGLEC17P in lung adenocarcinoma and a preliminary functional study. Cell Biol Int (2023) 47(1):86–97. doi: 10.1002/cbin.11919
205. Wang S, Luo Z, Yuan L, Lin X, Tang Y, Yin L, et al. tRNA-derived small RNAs: novel insights into the pathogenesis and treatment of cardiovascular diseases. J Cardiovasc Transl Res (2022) 16(2):300–9. doi: 10.1007/s12265-022-10322-0
206. Kang Y, You J, Gan Y, Chen Q, Huang C, Chen F, et al. Serum and Serum Exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as Diagnostic Biomarkers for Lung Adenocarcinoma. Front Oncol (2022) 12:912246. doi: 10.3389/fonc.2022.912246
207. Chen SW, Lu HP, Chen G, Yang J, Huang WY, Wang XM, et al. Downregulation of miRNA-126-3p is associated with progression of and poor prognosis for lung squamous cell carcinoma. FEBS Open Bio (2020) 10(8):1624–41. doi: 10.1002/2211-5463.12920
208. Yang S, Sui J, Liu T, Wu W, Xu S, Yin L, et al. Expression of miR-486-5p and its signi fi cance in lung squamous cell carcinoma. J Cell Biochem (2019) 120(8):13912–23. doi: 10.1002/jcb.28665
209. Li X, Qin M, Huang J, Ma J, Hu X. Clinical significance of miRNA−1 and its potential target gene network in lung squamous cell carcinoma. Mol Med Rep (2019) 19(6):5063–78. doi: 10.3892/mmr.2019.10171
210. Hu J, Xu L, Shou T, Chen Q. Systematic analysis identifies three-lncRNA signature as a potentially prognostic biomarker for lung squamous cell carcinoma using bioinformatics strategy. Transl Lung Cancer Res (2019) 8(5):614–35. doi: 10.21037/tlcr.2019.09.13
211. Wu KL, Tsai YM, Huang YC, Wu YY, Chang CY, Liu YW, et al. LINC02323 facilitates development of lung squamous cell carcinoma by miRNA sponge and RBP dysregulation and links to poor prognosis. Thorac Cancer (2023) 14(4):407–18. doi: 10.1111/1759-7714.14760
212. Wang Y, Zhang H, Wang J, Li B, Wang X. Circular RNA expression profile of lung squamous cell carcinoma: identification of potential biomarkers and therapeutic targets. Biosci Rep (2020) 40(4):BSR20194512. doi: 10.1042/BSR20194512
213. Zhang H, Yuan J, Xiang Y, Liu Y. Comprehensive analysis of NPSR1-AS1 as a novel diagnostic and prognostic biomarker involved in immune infiltrates in lung adenocarcinoma. J Oncol (2022) 2022:2099327. doi: 10.1155/2022/2099327
214. Zou G, Lu W, Dai X. Diagnostic and prognostic value of serum CircERBB2 level in NSCLC and its correlation with clinicopathological features in NSCLC patients. Am J Transl Res (2023) 15(2):1215–22.
215. Li Z, Ma G, Pan Y. Five circRNAs serve as potential diagnostic and prognostic biomarkers in lung adenocarcinoma. Clin Lab (2021) 67(8). doi: 10.7754/Clin.Lab.2020.200523
216. Lv D, Wang Y, Li S, Shao X, Jin Q. Activation of MYO1G by lncRNA MNX1-AS1 drives the progression in lung cancer. Mol Biotechnol (2023) 65(1):72–83. doi: 10.1007/s12033-022-00531-y
217. Khan P, Siddiqui JA, Kshirsagar PG, Venkata RC, Maurya SK, Mirzapoiazova T, et al. MicroRNA-1 attenuates the growth and metastasis of small cell lung cancer through CXCR4/FOXM1/RRM2 axis. Mol Cancer (2023) 22(1):1. doi: 10.1186/s12943-022-01695-6
218. Gao J, Pan T, Wang H, Wang S, Chai J, Jin C. LncRNA FAM138B inhibits the progression of non-small cell lung cancer through miR-105-5p. Cell Cycle (2023) 22(7):808–17. doi: 10.1080/15384101.2022.2154556
Keywords: lung cancer, microRNAs, long non-coding RNAs, circular RNAs, biomarker, therapeutic target
Citation: Liu Y, Ding W, Wang J, Ao X and Xue J (2023) Non-coding RNAs in lung cancer: molecular mechanisms and clinical applications. Front. Oncol. 13:1256537. doi: 10.3389/fonc.2023.1256537
Received: 11 July 2023; Accepted: 24 August 2023;
Published: 08 September 2023.
Edited by:
Catalin Marian, Victor Babes University of Medicine and Pharmacy, RomaniaReviewed by:
Patricia Silveyra, Indiana University Bloomington, United StatesCopyright © 2023 Liu, Ding, Wang, Ao and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Ao, eGlhbmdhbzIwMTZAMTYzLmNvbQ==; Junqiang Xue, eHVlanFAcWR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.