- 1Department of Experimental Research, Guangxi Medical University Cancer Hospital, Nanning, China
- 2Department of Respiratory Oncology, Guangxi Medical University Cancer Hospital, Nanning, China
- 3Department of Epidemiology and Health Statistics, School of Public Health, Guangxi Medical University, Nanning, China
- 4Department of Clinical Research, Guangxi Medical University Cancer Hospital, Nanning, China
- 5Department of Cancer Prevention and Control, Guangxi Medical University Cancer Hospital, Nanning, China
- 6Department of Disease Process Management, Guangxi Medical University Cancer Hospital, Nanning, China
- 7Department of Scientific Research Dept, Guangxi Medical University Cancer Hospital, Nanning, China
- 8Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Guangxi Medical University, Ministry of Education, Nanning, China
- 9Guangxi Health Commission, Key Cultivated Laboratory of Cancer Molecular Medicine, Guangxi Medical University Cancer Hospital, Nanning, China
Background: Although the sphingolipid metabolism pathway is known to play a significant role in tumor progression, there have been few studies on how genetic variants in the sphingolipid metabolism pathway genes affect the survival of patients with hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Methods: We utilized available genotyping data to conduct multivariate Cox proportional hazards regression model analysis, examining the associations of 12,188 single nucleotide polymorphisms (SNPs) in 86 sphingolipid metabolism pathway genes on the survival of 866 HBV-HCC patients, and the model was also used in additive interaction analysis. We used bioinformatics functional prediction and expression quantitative trait locus (eQTL) analysis to explore the potential functions of SNPs and to evaluate the association of SNPs with the corresponding mRNA expression, respectively. We also used the online database TIMER2.0 (http://timer.comp-genomics.org/) to analyze the relationship between the corresponding mRNA expression levels and immune cell infiltration.
Results: Our study found that GBA2 rs1570247 G>A was significantly associated with elevated survival of HBV-HCC patients [(hazards ratio (HR)=0.74, 95% confidence interval (CI)=0.64-0.86, P<0.001)]. And on an additive scale, a synergistic effect was observed between the GG genotype of rs1570247 and advanced BCLC stage. Among HBV-HCC patients with advanced BCLC stage, those carrying the GBA2 rs1570247 GG genotype exhibited a significantly elevated risk of mortality (HR=3.32, 95%CI=2.45-4.50). Further functional prediction and eQTL analysis revealed that rs1570247 were located in the 5’ untranslated region of the GBA2, the A allele of SNP rs1570247 was associated with higher mRNA expression levels of GBA2 in normal liver tissues (P=0.009). Moreover, we observed a positive correlation between GBA2 mRNA expression and the infiltration level of B lymphocytes cell (R=0.331, P<0.001), while a negative correlation was noted between GBA2 mRNA expression and the infiltration level of macrophage M2 in HCC (R=-0.383, P<0.001).
Conclusion: Our findings suggest that GBA2 rs1570247 G>A in sphingolipid metabolism pathway may be a key factor for survival of HBV-HCC patients by regulating the expression of corresponding genes and affecting the infiltration level of immune cells.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide and represents a major challenge to global healthcare. In 2020, nearly 906,000 people were diagnosed with liver cancer globally, mostly HCC (1). Unfortunately, HCC is also the third highest cause of cancer-related deaths, with a poor 5-year survival rate of around 15% (2). It’s worth noting that most patients with HCC have a background of hepatitis B virus infection (HBV) in Asia, especially in in Guangxi Province, Sorthern China, the infection rate was as high as 87.7% (3).
Despite surgical procedure for operable HCC patients can lead to a better 5-year survival of over 70%, there is still a large part of patients suffering from poor survival because of high recurrence or metastasis rates (4). In patients with operable HCC, their genetic backgrounds are also likely influence their survival (5, 6). Although several survival-predicting genetic variants have identified in genome-wide association studies (7–9), these variants do not provide clear biological relevance. Recent pathway analyses for identifying survival-predicting genetic variants appear to be effective and found several additional variants that are potentially functional (10–12). Therefore, identifying efficient prognostic biomarkers is still urgent for identifying patients with high-death risk and for strategy-decision making of personalized treatment in operable HBV-HCC patients.
Sphingolipids are one of the main categories of lipids in eukaryotic organisms, participating in the composition of the plasma membrane and serving as regulators of cell-cell interactions and cell recognition (13). Bioactive sphingolipids are now recognized as important regulators of cancer cell biology. For example, sphingosine-1-phosphate can promote tumor cell growth through Stat3 and Akt signaling pathways, upregulate Bcl-2/Bcl-xL, and inhibit p53-mediated apoptosis (14–18). Ceramides, on the other hand, promote tumor progression through chronic inflammation and endoplasmic reticulum stress (19, 20). Additionally, sphingolipids are closely associated with tumor immune responses. Sphingosine-1-phosphate and lysophosphatidic acid can modulate the anti-tumor activity of infiltrating lymphocytes, and the acidic tumor microenvironment can activate acid sphingomyelinase and induce matrix metalloproteinase-9, thereby promoting tumor metastasis and immune evasion (21–24). Studies have also reported that sphingolipids in hepatocellular carcinoma (HCC) can influence patient prognosis by mediating the immune microenvironment(25). Specifically, abnormal ceramides produced by liver cells infected with HBV PreS variants can activate NLRP3 inflammasome in hepatic macrophages, promoting the progression of hepatocellular carcinoma (26). Furthermore, elevated plasma ceramide levels in pancreatic cancer can predict the response to radiotherapy in metastatic tumors; similarly, Ceramides6 has emerged as a strong prognostic factor for survival in human colon cancer patients (27, 28). Therefore, potential genetic variations in sphingolipid metabolic pathway genes may serve as promising predictive biomarkers for HCC patient survival.
In the presen study, we hypothesize that genetic variants in sphingolipid metabolism pathway genes are associated with survival of HBV-HCC patients. We tested this hypothesis by using genotyping data from HBV-HCC patients and focusing on SNPs that may alter their gene function or expression levels and are therefore likely to have biological and functional implications.
Materials and methods
Study populations
In this study, a total of 866 HBV-HCC patients were enrolled. These patients were HBsAg seropositive HCC patients with histologically confirmed operable HCC and underwent hepatectomy at the Guangxi Medical University Cancer Hospital between July 2007 and December 2017. The eligibility criteria and clinical characteristics of all patients have been previously described(6). The investigators collected the general information [i.e age, sex, smoking status, drinking status, serum AFP level, cirrhosis, embolus, and the Barcelona Clinic Liver Cancer (BCLC) stage] and follow-up information of patients by reviewing medical records and telephone follow-up. All patients were followed up every three months within the first two years after surgery and every six months in the next year through telephone calls. Overall survival (OS) was defined as the time from surgery to death or to the date of the last follow-up (March 2020). Patients who were lost to follow-up or survived at the last follow-up were censored. Because this was a single institution study, we used an internal study validation design to ensure the accuracy and reliability of the results, the 866 cases were randomly divided into a discovery group and a replication group at a ratio of 1:1. The present study was approved by Guangxi Medical University Cancer Hospital Institutional Review Committee (LW2023109).
Genotyping, gene and SNP selection
The method used for genotyping SNPs, the quality control of raw data and imputation have been described elsewhere (5). In brief, the whole genomic DNA of each HCC patient was extracted by a blood DNA extraction kit (Concert). Genotyping was performed using Illumina Infinium Global Screening Assay (Shanghai, China). Based on the 1000 Genomes Project (Phase 3 v5) reference population information, Minimac3 was used for imputation (https://imputationserver.sph.umich.edu/index.htm)forthe untyped SNPs.
The sphingolipid metabolism pathway-related genes were gathered from the Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/index.jsp) using the keyword “sphingolipid”. After eliminating 37 duplicates and six genes located in the X chromosome, a total of 86 genes were selected as candidates (Supplementary Table S1).
After quality control and imputation, all SNPs in these genes and their ±2 kb flanking regions were extracted by Plink 1.09 (http://pngu.mgh.harvard.edu/purcell/plink/)(29) based on the following criteria: minor allele frequency (MAF) ≥ 0.05, the genotyping success rate was ≥ 95%, and Hardy-Weinberg equilibrium (HWE) was ≥ 1.0 × 10-6. As a result, a total of 12,188 SNPs were obtained from the discovery dataset for further analysis.
Functional annotation and eQTL analysis
Functional prediction of the identified SNPs was carried out using three online tools: SNPinfo (https://snpinfo.niehs.nih.gov/snpinfo/snpfuNc.htm), RegulomeDB (http://www.regulomedb.org/), and VannoPortal (http://www.mulinlab.org/vportal). The linkage disequilibrium (LD) of the selected SNPs was analyzed and LD plots were generated using Haploview software.
To assess the relationships between different genotypes of SNPs and the mRNA expression levels of corresponding genes, an expression quantitative trait locus (eQTL) analysis was performed using the GTEx database (https://www.gtexportal.org/). The relationships between the expression levels of target genes and the infiltration levels of multiple immune cells in the tumor microenvironment were analyzed using the tumor immune cell infiltration database TIMER2.0 (http://timer.comp-genomics.org/). Additionally, the mRNA expression of target genes in HCC tissues and normal tissues and their association with survival of HCC patients were analyzed through the Kaplan-Meier Plotter (https://kmplot.com/) database.
Statistical analysis
To evaluate the association between SNPs and survival in 866 HBV-HCC patients after surgery, a multivariate Cox proportional hazards regression was performed using the GenABEL package in R 3.1.3. Bayesian false discovery probability (BFDP) was used to correct for the incidence of false positive errors (30). The effect of different SNP genotypes on postoperative OS in patients with HBV-HCC was analyzed using the survival package in R 4.0.3, and Kaplan-Meier curves were generated.
In order to test the reliability of the association analysis results, we used the bootstrap method to calculate the hazard ratio (HR) of each time by 1,000 repeated sampling. The Shapiro-Wilk test was utilized to assess the fitting degree and 95% confidence interval (CI) of the normal distribution of HR values, thereby evaluating the reliability of the study results.
Result
Associations of SNPs in the sphingolipid metabolism pathway with survival
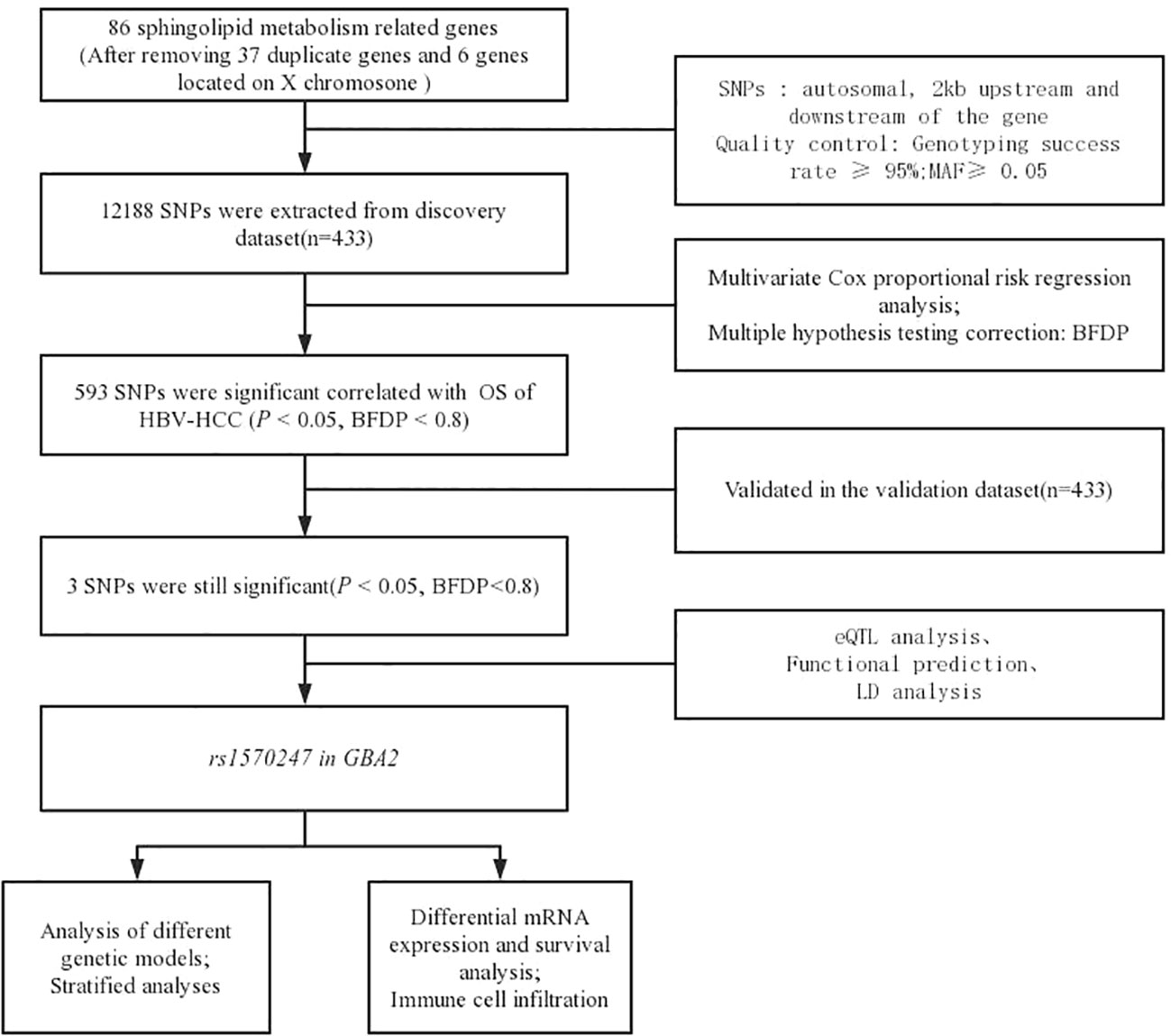
Figure 1 illustrates the flowchart of the study design. The discovery dataset consisted of 12,188 SNPs in 86 genes related to sphingolipid metabolism pathway, among which 593 SNPs were found to be associated with HBV-HCC OS (P<0.05, BFDP<0.8). After verification in the replication dataset, 3 SNPs (rs1570246 G>T, rs1570247 G>A, rs3750434 G>A) located on glucosylceramidase beta 2 (GBA2) remained statistically significant (P<0.05, BFDP<0.8) (Table 1).
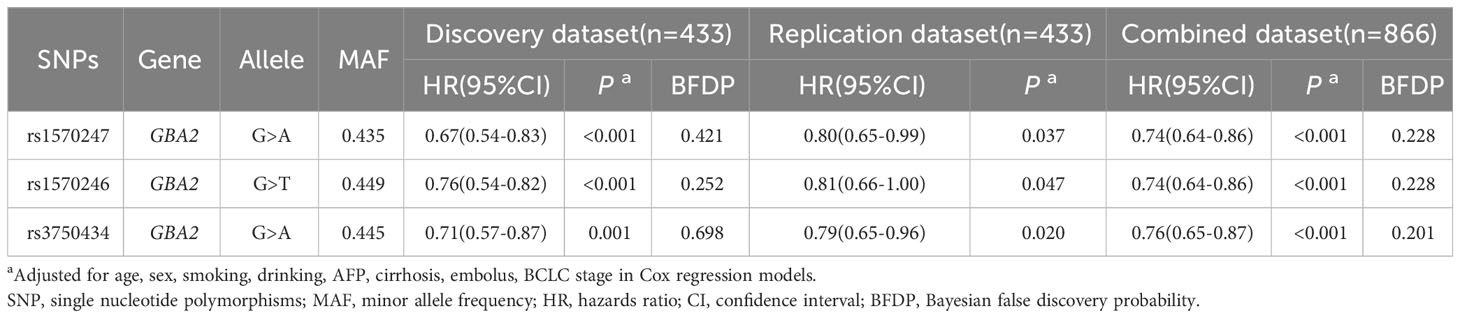
Table 1 Associations of 3 validated significant SNPs with HBV-HCC OS in discovery, validation and combined dataset.
LD analysis and functional annotation
The results of LD analysis revealed high linkage (r2>0.8) between these three SNPs in GBA2 gene (Figure 2). To select a tagSNP with potential functions, we performed functional annotation for these three SNPs using RegulomeDB, SNPinfo, and VannoPortal. As shown in Table 2, rs1570246 and rs1570247 were located in the 5’ untranslated region (UTR) of the GBA2 gene, while rs3750434 was located in the intronic region. Importantly, the rs1570247 with the highest potential functional score and the highest context-dependent priority was located in an active region of chromatin with many histone modifications, indicating a high probability of potential function. Therefore, we selected rs1570247 for subsequent analysis.
Genetic models
To assess the impact of SNPs on the prognosis of HBV-HCC patients, we applied multivariable Cox regression to evaluate the effect of rs1570247 on the risk of death in the combined dataset. The results revealed a dose-response relationship between the A allele amount of rs1570247 and decreased risk of death in patients (Table 3).
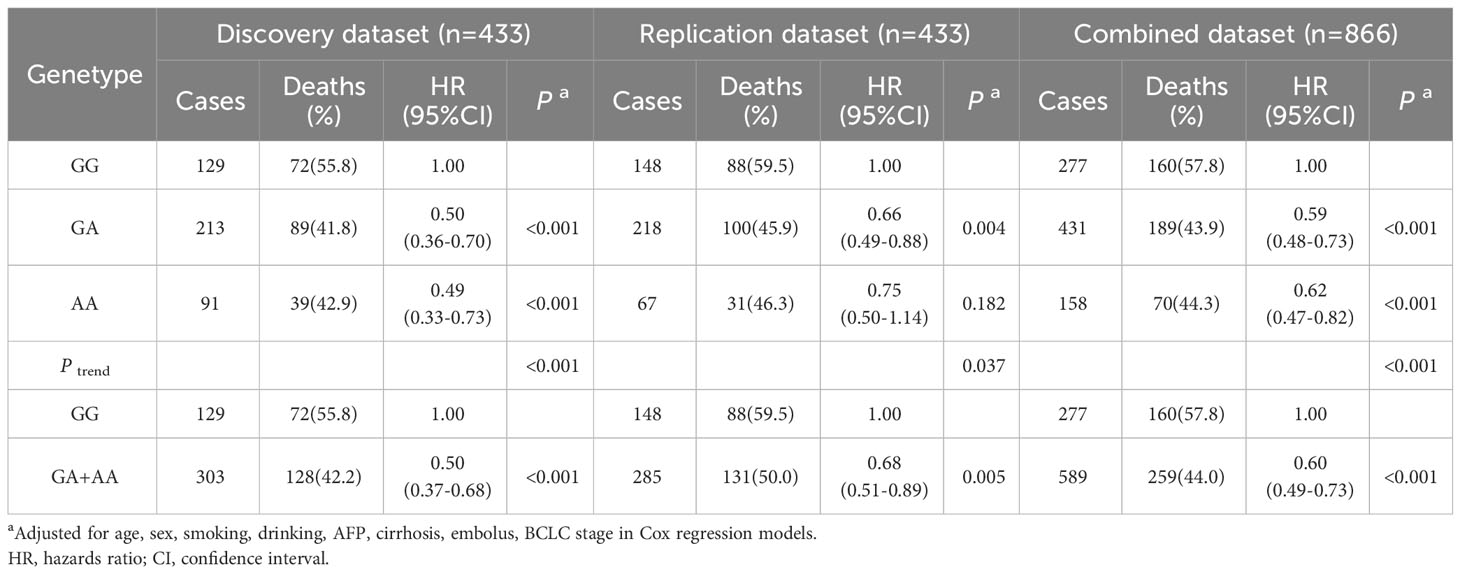
Table 3 Associations between rs1570247 and postoperative OS of HBV-HCC patients in different genetic models of all datasets.
In the combined dataset, the mortality rates for different genotypes of GBA2 rs1570247 were 57.8% for GG, 43.9% for GA, and 44.3% for AA. Patients with the rs1570247 GA or AA genotype had better survival rates than those with the GG genotype (HR=0.60, 95%CI=0.49-0.73, P<0.001). Kaplan-Meier survival curves were generated to illustrate the associations between rs1570247 and OS in patients with HBV-HCC (Figure 3).
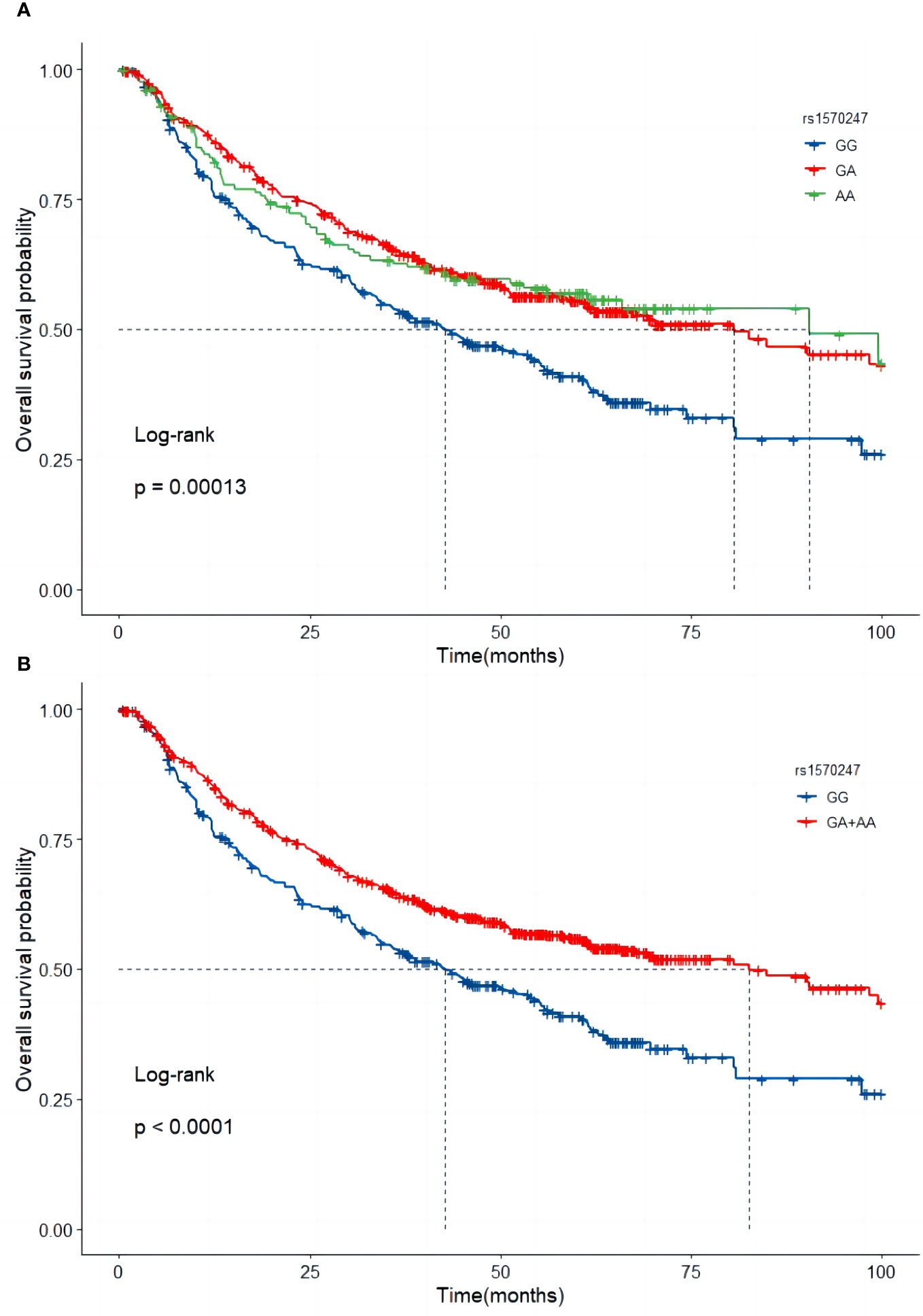
Figure 3 Kaplan-Meier curves of GBA2 rs1570247 in combined dataset. (A) Additive model; (B) Dominant model.
To verify the association analysis results, we conducted 1,000 repeated samplings using the Bootstrap method and performed multivariate Cox regression analysis to calculate HR and 95% CI. The distribution of HR in verification was consistent with a normal distribution (P<0.05). And the HR value for rs1570247 in the survival analysis of HCC patients was within the 95% confidence interval (Figure 4). These observations suggested the overall reliability of association analysis.
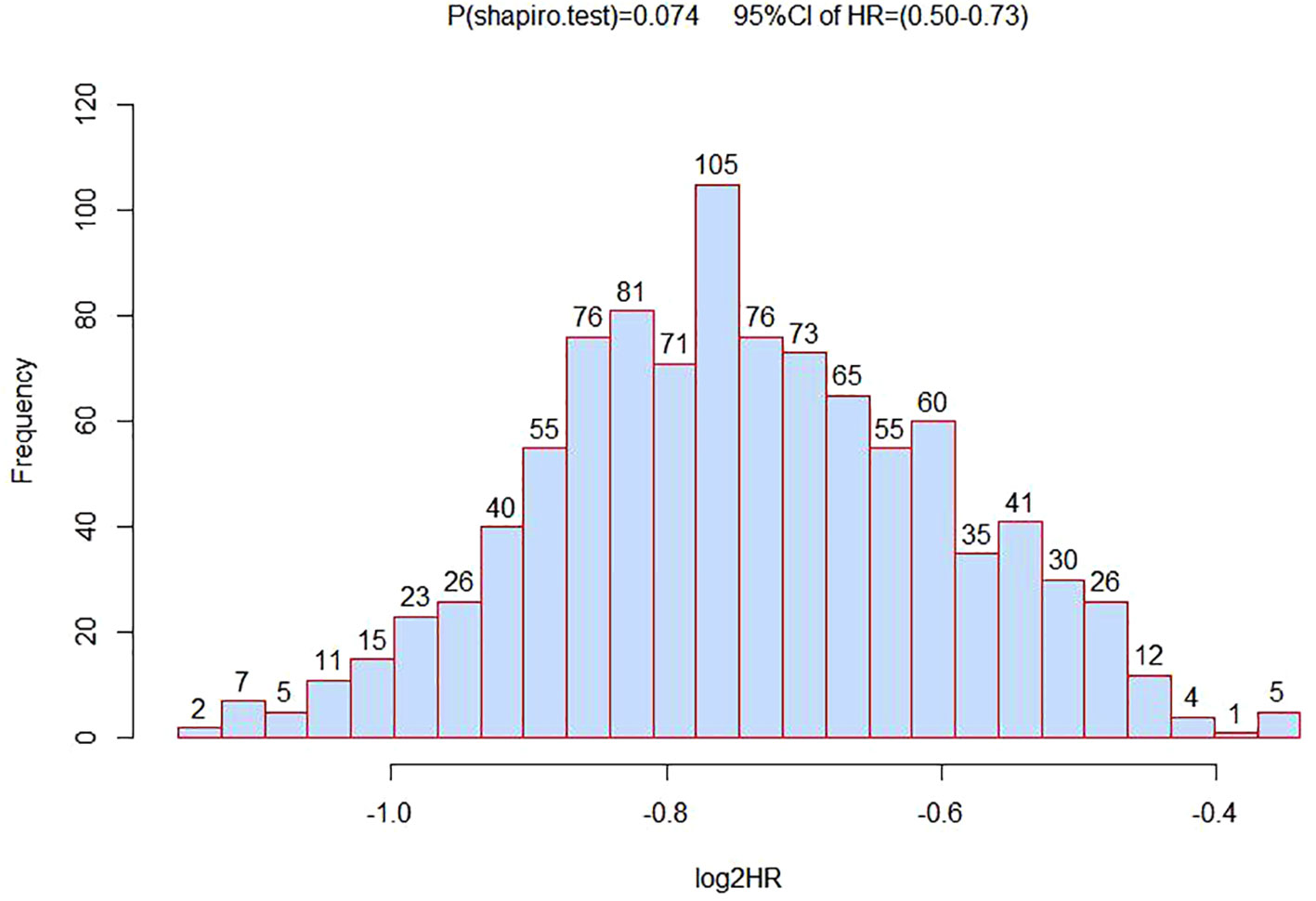
Figure 4 The distribution histogram of 1000 HR values after grouped by bootstrapping for 1000 times.
Stratified analysis
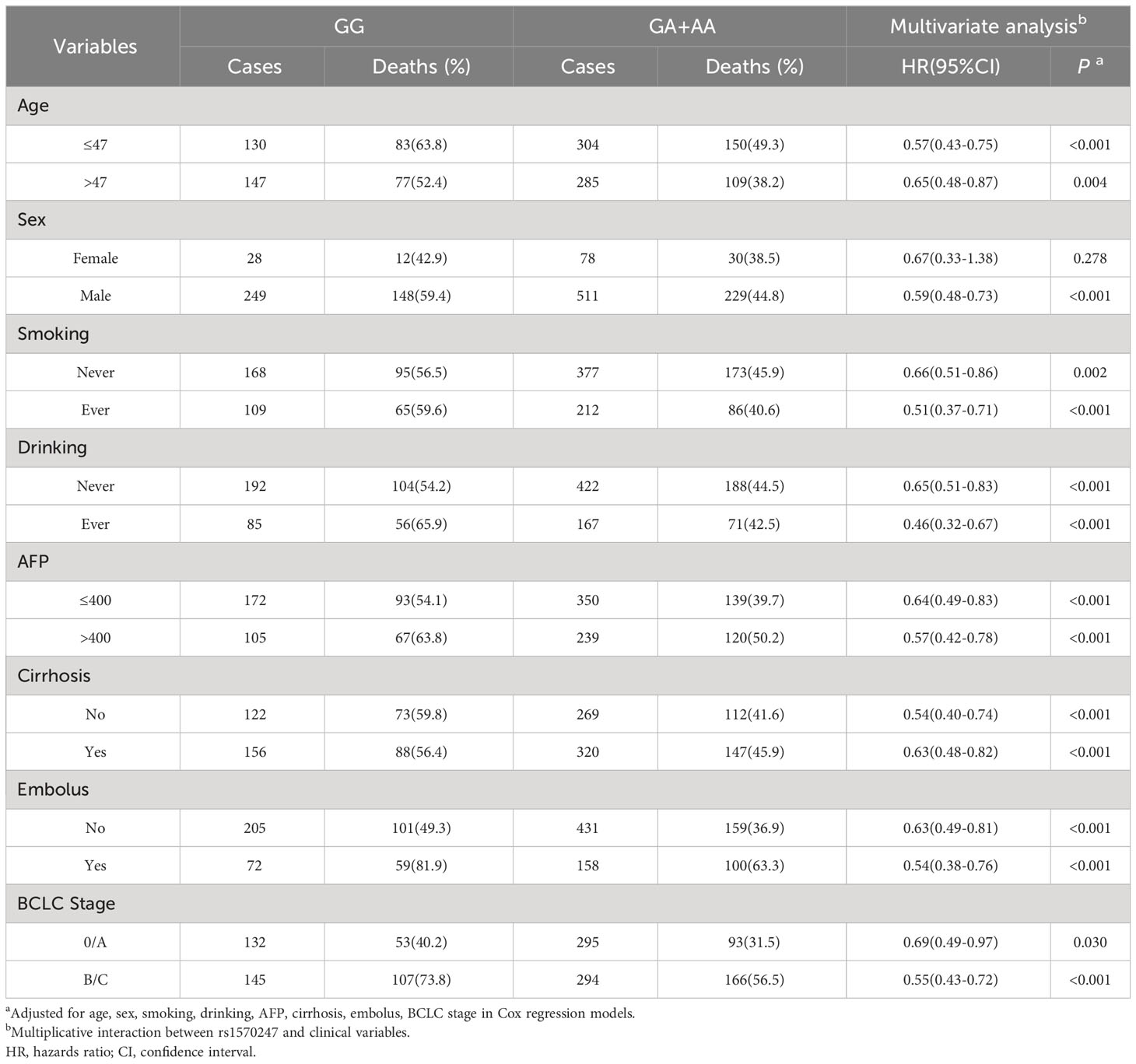
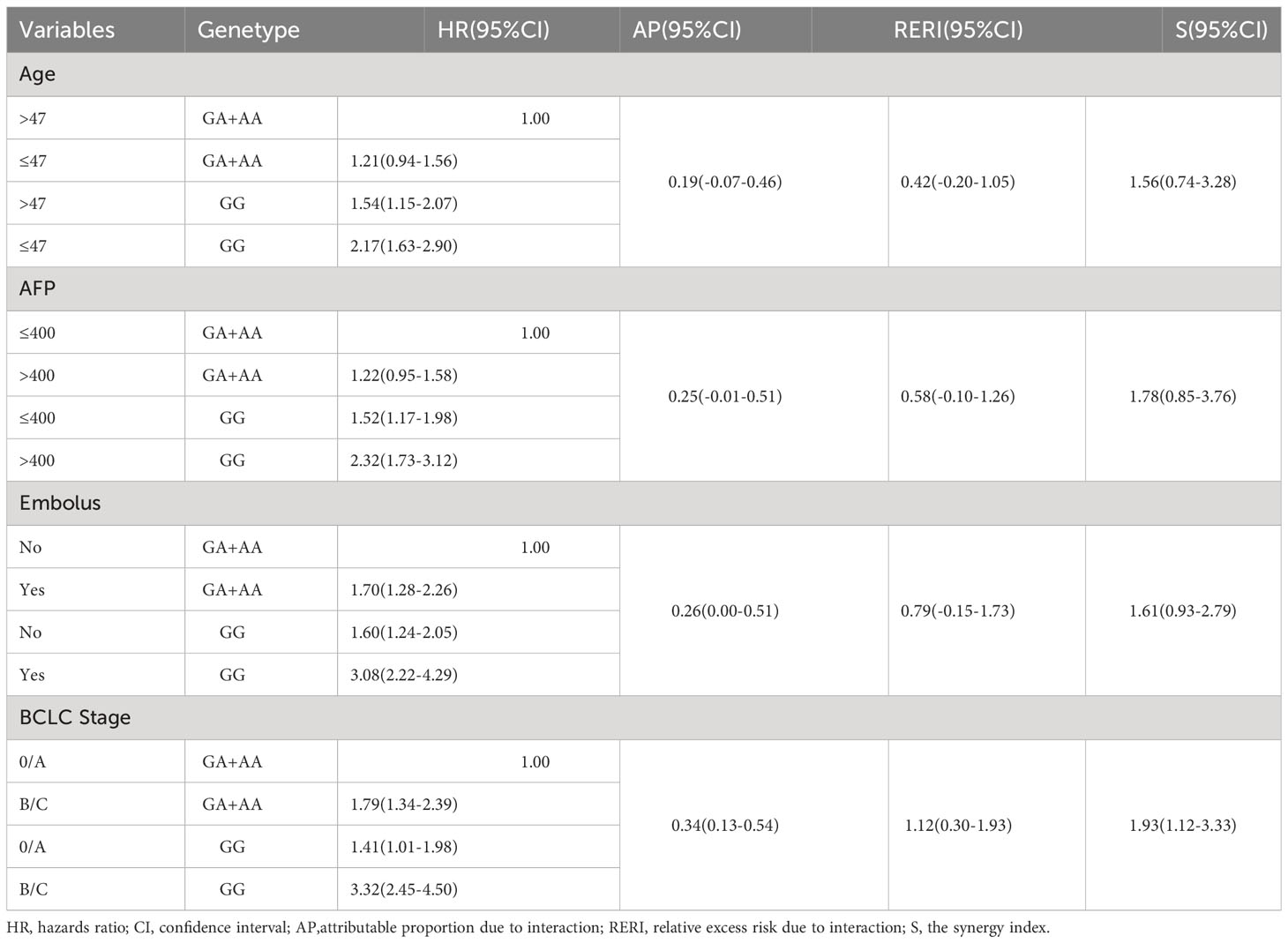
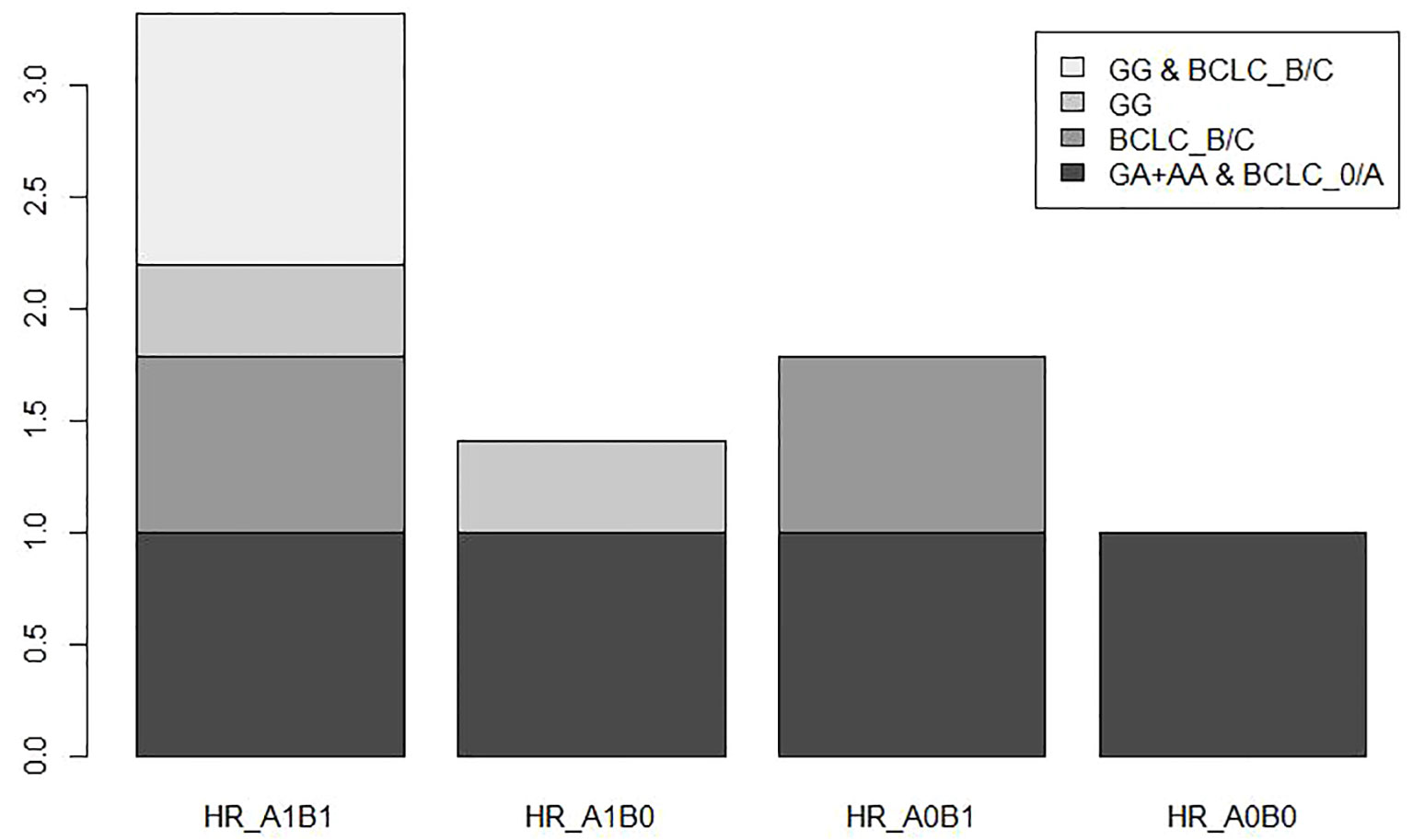
To further investigate the association between different genotypes and OS in the 866 cases, we performed a stratified analysis using covariates such as age, sex, smoking, drinking, AFP, cirrhosis, embolus, and BCLC stage as stratification factors. Obtained results revealed that the GA and AA genotypes of rs1570247 were associated with a significantly reduced risk of postoperative death in multiple subgroups, except for female patients (Table 4). Then, we selected 4 clinical variables that were statistically significant for HBV-HCC survival in our dataset to analyze the interaction with rs1570247 on an additive scale. Our results showed a synergistic effect between rs1570247 GG genotype and HBV-HCC patients with advanced BCLC stage on an additive scale (Table 5). Specifically, HBV-HCC patients with GG genotype in advanced BCLC stage have a higher risk of death(HR=3.32, 95%CI=2.45-4.50). The visualization results were shown in Figure 5.
eQTL analysis, differential mRNA expression analysis and survival analysis
To gain further insight into the role of rs1570247 in HCC, we conducted an eQTL analysis in the GTEx database to investigate the association between rs1570247 genotype and GBA2 mRNA expression levels. Our results showed that the rs1570247 AA genotype was significantly associated with increased GBA2 mRNA expression levels in normal liver tissues (P=0.0087, NES=0.12) (Figure 6).
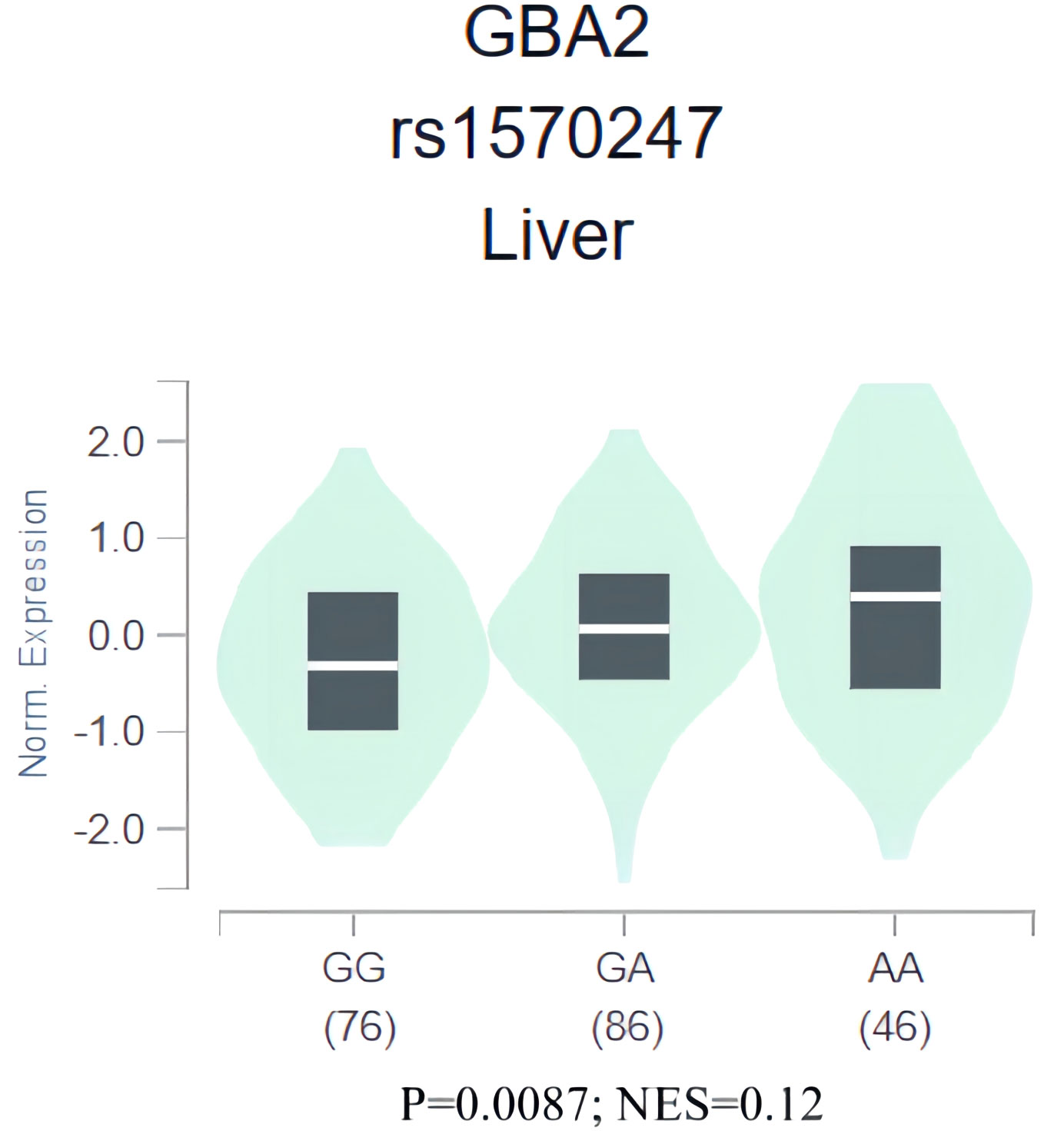
Figure 6 eQTL analysis of rs1570247 and GBA2 mRNA expression levels in liver tissues from GTEx database.
To clarify the effect of GBA2 gene expression in the progression and survival of HCC, we first assessed mRNA expression levels of GBA2 gene in HCC tissues and normal tissues using TNMplot, and analyzed its relationship with OS of HCC patients through Kaplan-Meier Plotter. It was found that the expression level of GBA2 mRNA in HCC was lower than that in normal liver tissues (Figure 7A). Meanwhile, the higher expression levels of GBA2 seem to be associated with better survival probability in liver cancer patients with microvascular invasion or stage III/IV (Figures 7B, C), which is consistent with our additive interaction results.
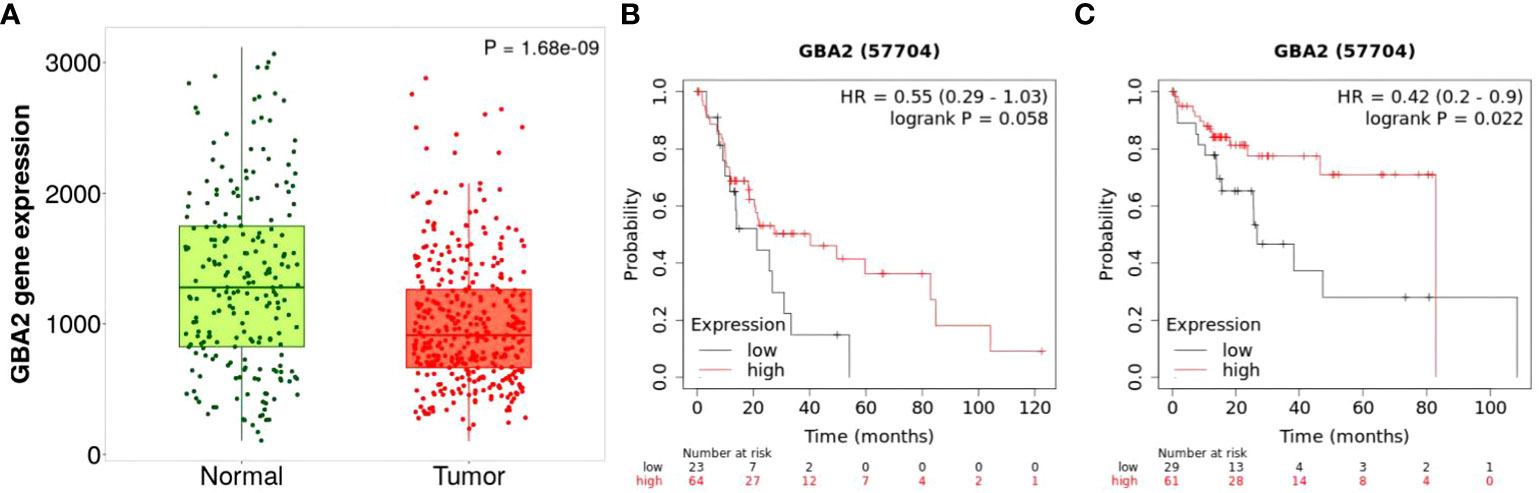
Figure 7 Differential mRNA expression analysis and survival of HCC from the Kaplan-Meier Plotter databases: (A) mRNA expression of GBA2 in HCC tissues and normal liver tissues; (B) mRNA expression of GBA2 relationship with OS in stage 3 + 4 HCC patients; (C) mRNA expression of GBA2 relationship with OS in HCC patients with microvascular invasion.
Relationship between GBA2 and immune cell infiltration in HCC
To gain further insight into the role of GBA2 in HCC, we investigated the relationship between GBA2 and immune cell infiltration in the HCC microenvironment using the TIMER 2.0 database. Our analysis showed that GBA2 expression was positively correlated with B lymphocyte infiltration levels, and negatively correlated with M2 macrophage infiltration levels (B cell: R=0.331, P=2.80×10-10; Macrophage M2: R=-0.383, P=1.67×10-13) (Figure 8). These findings suggest that GBA2 may modulate immune cell infiltration levels in the tumor microenvironment, which could impact the survival of HCC patients.
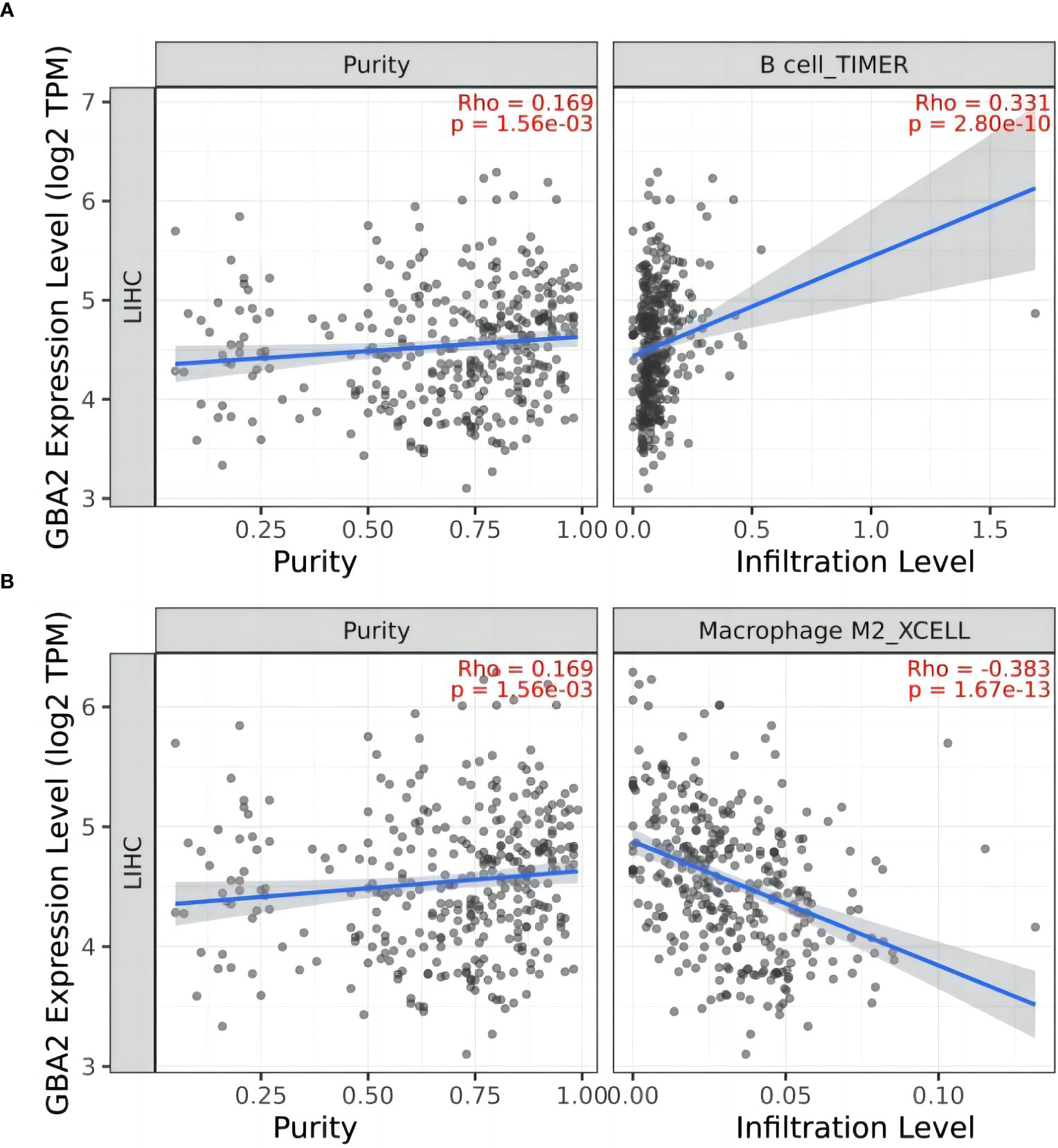
Figure 8 Correlation between GBA2 mRNA expression and immune cells infiltrating level in HCC tissues: (A) B cells; (B) Macrophage M2 cells.
Discussion
In the present study, we evaluated the associations between SNPs in sphingolipid metabolism genes and survival of HBV-HCC patients. Our study identified a novel functional SNP (rs1570247 G>A) located in the 5’UTR of the GBA2 gene as a protective factor for HCC prognosis. Furthermore, eQTL analysis revealed that the GBA2 rs1570247 A allele was associated with increased mRNA expression levels, and GBA2 may play a role in regulating immune cell infiltration in the tumor microenvironment. Overall, our findings suggest that GBA2 rs1570247 G>A in sphingolipid metabolism pathway may have an important impact on survival of HBV-HCC patients, possibly by influencing mRNA expression and affecting the infiltration level of immune cells.
The GBA2 gene is located on the human chromosome 9 and is mapped in position p13.3 (31). GBA2 was first isolated and characterized from the human liver, where it was found to hydrolyze endogenous bile acid 3-O-glucosides (32). GBA2 is a membrane-bound enzyme located in the endoplasmic reticulum(33), and has been found to be present at or near the cell surface(34). It cleaves the β-glucose-sphingosine linkage in glucosylceramide (GlcCer). In addition to its hydrolytic activity on GlcCer, GBA2 also exhibits transglucosylation activity by transferring glucose to cholesterol. This activity allows GBA2 to use the glucose moiety released by the cleavage of GlcCer to form glucocholesterol (GlcChol). Conversely, GlcChol can be deglycosylated by GBA2 to synthesize GlcCer. In this study, we found that GBA2 rs1570247 A allele was a protective factor for survival in HCC patients and was associated with increased GBA2 mRNA expression levels. Similarly, GBA2 mRNA in HCC was lower than that in normal liver tissues, and the higher expression levels of GBA2 seemed to be associated with better survival probability in liver cancer patients with microvascular invasion or stage III/IV. However, since GBA2 can cleave glucosylceramide to produce ceramides, ceramides have been shown to act as tumor suppressors in a variety of tumors (35–37). GBA2 also has been shown to function as a tumor suppressor in melanoma cells and cholangiocarcinoma (38, 39). Meanwhile, based on the role of GBA2 in inflammatory response (40), we analyzed the relationship between GBA2 and immune cell infiltration in the tumor microenvironment, and found that GBA2 was positively correlated with B cells, but negatively correlated with M2 macrophages. B cells arise and mature in the bone marrow and have various functions in immune response (41). Tumor-infiltrating B lymphocytes (TIBs) have been observed in various solid tumors. Studies have shown that TIBs can suppress tumor progression by secreting immunoglobulins, promoting T cell response, and directly killing cancer cells (42). Additionally, TIBs can help induce the infiltration of cytotoxic T lymphocytes (CTL) into tumors by maintaining the structure and function of tertiary lymphoid structures (TLS), which contributes to an effective antitumor response and better patient prognosis (43–45). Macrophages are a crucial component of the tumor microenvironment that facilitate tumor cell migration and invasion, matrix degradation, and angiogenesis. The density of macrophages in the tumor microenvironment has been found to be a prognostic marker of poor outcome for a variety of carcinomas (46–48). M2 macrophages, which are derived from macrophages, have poor antigen presentation ability and produce factors that inhibit the proliferation and activity of T cells. They are typically more suited to promoting angiogenesis, tissue remodeling, and repair (49). Studies have also suggested that M2 macrophages recruited by tumors can facilitate tumor metastasis (50). These findings suggest that GBA2 may affect the survival of HCC patients by influencing the immune response.
Conclusion
In conclusion, our study identified the role of genetic variants in GBA2 (rs1570247 G>A) in HBV-HCC survival. The GBA2 rs1570247 mutant genotypes were associated with better survival of HBV-HCC, possibly by enhancing GBA2 transcription and improving the immune response. However, it is important to note that the datasets used to validate our findings only included Chinese populations. Therefore, our results may not be generalizable to other ethnic groups, and further studies are needed to determine the impact on HCC survival in the general population. Additionally, the molecular mechanism of GBA2 in HCC remains unclear, and further biochemical studies and functional experiments are needed to confirm our findings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving humans were approved by Ethics Committee of the Guangxi Medical University Cancer Hospital (LW2023109). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conception and design, HY; Provision of study materials or patients and data collection, BJ, LQ, JT, SZ, JW, JL, YL, YM and JC; Bioinformation analysis, BJ, ZZ, QL and YJ; Follow-up: XL; Data analysis and interpretation, BJ, and MQ; Writing – Original Draft Preparation, BJ, and MQ; Writing – Review and Editing, HY. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by The Key Project of Guangxi Natural Science Foundation (2018GXNSFDA050012); Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Ministry of Education (GKE-ZZ202104); Key Laboratory of Early Prevention and Treatment for Regional High Frequency Tumor, Ministry of Education (GKE-ZZ202118); the Youth Program of Scientific Research Foundation of Guangxi Medical University Cancer Hospital (2021-10).
Acknowledgments
We would like to thank all study participants, researchers and clinicians who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1252158/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. ‘Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries’, The Lancet. Global Health 6(5) (2018) pp:e555–67. doi: 10.1016/S2214-109X(18)30127-X
3. Lin J, Zhang H, Yu H, Bi X, Zhang W, Yin J, et al. Epidemiological characteristics of primary liver cancer in mainland China from 2003 to 2020: A representative multicenter study. Front Oncol (2022) 12:906778. doi: 10.3389/fonc.2022.906778
4. Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet (2022) 400(10360):1345–62. doi: 10.1016/S0140-6736(22)01200-4
5. Huang Q, Liu Y, Qiu M, Lin Q, Wei X, Zhou Z, et al. Potentially functional variants of MAP3K14 in the NF-κB signaling pathway genes predict survival of HBV-related hepatocellular carcinoma patients. Front Oncol (2022) 12:990160. doi: 10.3389/fonc.2022.990160
6. Lin Q, Qiu M, Wei X, Xiang Z, Zhou Z, Ji I, et al. Genetic variants of SOS2, MAP2K1 and RASGRF2 in the RAS pathway genes predict survival of HBV-related hepatocellular carcinoma patients. Arch Toxicol (2023) 97:1599–611. doi: 10.1007/s00204-023-03469-5
7. Sato Y, Yamamoto N, Kunitoh H, Ohe Y, Minami H, Laird NM, et al. Genome-wide association study on overall survival of advanced non-small cell lung cancer patients treated with carboplatin and paclitaxel. J Thorac Oncol (2011) 6(1):132–8. doi: 10.1097/JTO.0b013e318200f415
8. Wu C, Kraft P, Stolzenberg-Solomon R, Steplowski E, Brotzman M, Xu M, et al. Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut (2014) 63(1):152–60. doi: 10.1136/gutjnl-2012-303477
9. Li W, Middha M, Bicak M, Sjoberg DD, Vertosick E, Dahlin A, et al. Genome-wide scan identifies role for AOX1 in prostate cancer survival. Eur Urol (2018) 74(6):710–9. doi: 10.1016/j.eururo.2018.06.021
10. Sun Z, Chen J, Aakre J, Marks RS, Garces YY, Jiang R, et al. Genetic variation in glutathione metabolism and DNA repair genes predicts survival of small-cell lung cancer patients. Ann Oncol (2010) 21(10):2011–6. doi: 10.1093/annonc/mdq212
11. Cao Y, Lindström S, Schumacher F, Stevens VL, Albanes D, Berndt S, et al. Insulin-like growth factor pathway genetic polymorphisms, circulating IGF1 and IGFBP3, and prostate cancer survival. J Natl Cancer Institute (2014) 106(6):dju085. doi: 10.1093/jnci/dju085
12. Dai W, Liu H, Liu Y, Xu X, Qian D, Luo S, et al. Genetic variants in the folate metabolic pathway genes predict cutaneous melanoma-specific survival. Br J Dermatol (2020) 183(4):719–28. doi: 10.1111/bjd.18878
13. Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol (2018) 19(3):175–91. doi: 10.1038/nrm.2017.107
14. Oskouian B, Sooriyakumaran P, Borowsky AD, Crans A, Dillard-Telm L, Tam YY, et al. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc Natl Acad Sci USA (2006) 103(46):17384–9. doi: 10.1073/pnas.0600050103
15. Colié S, Van Veldhoven PP, Kedjouar B, Bedia C, Albinet V, Sorli S-C, et al. Disruption of sphingosine 1-phosphate lyase confers resistance to chemotherapy and promotes oncogenesis through Bcl-2/Bcl-xL upregulation. Cancer Res (2009) 69(24):9346–53. doi: 10.1158/0008-5472.CAN-09-2198
16. Benesch MGK, Tang X, Dewald J, Dong W-F, Mackey JR, Hemmings DG, et al. Tumor-induced inflammation in mammary adipose tissue stimulates a vicious cycle of autotaxin expression and breast cancer progression. FASEB J (2015) 29(9):3990–4000. doi: 10.1096/fj.15-274480
17. Furuya H, Shimizu Y, Tamashiro PM, Iino K, Bielawski J, Chan OTM, et al. Sphingosine kinase 1 expression enhances colon tumor growth. J Trans Med (2017) 15(1):120. doi: 10.1186/s12967-017-1220-x
18. Magkrioti C, Oikonomou N, Kaffe E, Mouratis M-A, Xylourgidis N, Barbayianni I, et al. The autotaxin-lysophosphatidic acid axis promotes lung carcinogenesis. Cancer Res (2018) 78(13):3634–44. doi: 10.1158/0008-5472.CAN-17-3797
19. Chen W, Wu C, Chen Y, Guo Y, Qiu L, Liu Z, et al. Downregulation of ceramide synthase 1 promotes oral cancer through endoplasmic reticulum stress. Int J Oral Sci (2021) 13(1):10. doi: 10.1038/s41368-021-00118-4
20. El-Hindi K, Brachtendorf S, Hartel JC, Oertel S, Birod K, Merz N, et al. T-cell-specific cerS4 depletion prolonged inflammation and enhanced tumor burden in the AOM/DSS-induced CAC model. Int J Mol Sci (2022) 23(3):1866. doi: 10.3390/ijms23031866
21. Kato Y, Ozawa S, Tsukuda M, Kubota E, Miyazaki K, St-Pierre Y, et al. Acidic extracellular pH increases calcium influx-triggered phospholipase D activity along with acidic sphingomyelinase activation to induce matrix metalloproteinase-9 expression in mouse metastatic melanoma. FEBS J (2007) 274(12):3171–83. doi: 10.1111/j.1742-4658.2007.05848.x
22. Kachler K, Bailer M, Heim L, Schumacher F, Reichel M, Holzinger CD, et al. Enhanced acid sphingomyelinase activity drives immune evasion and tumor growth in non-small cell lung carcinoma. Cancer Res (2017) 77(21):5963–76. doi: 10.1158/0008-5472.CAN-16-3313
23. Chakraborty P, Vaena SG, Thyagarajan K, Chatterjee S, Al-Khami A, Selvam SP, et al. Pro-survival lipid sphingosine-1-phosphate metabolically programs T cells to limit anti-tumor activity. Cell Rep (2019) 28(7):1879–1893.e7. doi: 10.1016/j.celrep.2019.07.044
24. Matas-Rico E, Frijlink E, van der Haar Àvila I, Menegakis A, van Zon M, Morris AJ, et al. Autotaxin impedes anti-tumor immunity by suppressing chemotaxis and tumor infiltration of CD8+ T cells. Cell Rep (2021) 37(7):110013. doi: 10.1016/j.celrep.2021.110013
25. Zhang X, Zhuge J, Liu J, Xia Z, Wang H, Gao Q, et al. Prognostic signatures of sphingolipids: Understanding the immune landscape and predictive role in immunotherapy response and outcomes of hepatocellular carcinoma. Front Immunol (2023) 14:1153423. doi: 10.3389/fimmu.2023.1153423
26. Liu C, Chen K, Zhao F, Xuan L, Wang Y, Xu C, et al. Occult infection with hepatitis B virus PreS variants synergistically promotes hepatocellular carcinoma development in a high-fat diet context by generating abnormal ceramides. BMC Med (2022) 20:279. doi: 10.1186/s12916-022-02481-3
27. Abdul Aziz NA, Mokhtar NM, Harun R, Mollah MMH, Mohamed Rose I, Sagap I, et al. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics (2016) 9(1):58. doi: 10.1186/s12920-016-0218-1
28. Dubois N, Rio E, Ripoche N, Ferchaud-Roucher V, Gaugler M-H, Campion L, et al. Plasma ceramide, a real-time predictive marker of pulmonary and hepatic metastases response to stereotactic body radiation therapy combined with irinotecan. Radiother Oncol (2016) 119(2):229–35. doi: 10.1016/j.radonc.2016.03.014
29. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet (2007) 81(3):559–75. doi: 10.1086/519795
30. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet (2007) 81(2):208–27. doi: 10.1086/519024
31. Matern H, Boermans H, Matern S, Lottspeich F. Molecular cloning and expression of human bile acid β-glucosidase. J Biol Chem (2001) 276(41):37929–33. doi: 10.1074/jbc.M104290200
32. Matern H, Heinemann H, Legler G, Matern S. Purification and characterization of a microsomal bile acid beta-glucosidase from human liver. J Biol Chem (1997) 272(17):11261–7. doi: 10.1074/jbc.272.17.11261
33. Yildiz Y, Matern H, Thompson B, Allegood JC, Warren RL, Ramirez DMO, et al. Mutation of β-glucosidase 2 causes glycolipid storage disease and impaired male fertility. J Clin Invest (2006) 116(11):2985–94. doi: 10.1172/JCI29224
34. Boot RG, Verhoek M, Donker-Koopman W, Strijland A, van Marle J, Overkleeft HS, et al. Identification of the non-lysosomal glucosylceramidase as β-glucosidase 2. J Biol Chem (2007) 282(2):1305–12. doi: 10.1074/jbc.M610544200
35. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer (2018) 18(1):33–50. doi: 10.1038/nrc.2017.96
36. Muñoz-Guardiola P, Casas J, Megías-Roda E, Solé S, Perez-Montoyo H, Yeste-Velasco M, et al. The anti-cancer drug ABTL0812 induces ER stress-mediated cytotoxic autophagy by increasing dihydroceramide levels in cancer cells. Autophagy (2021) 17(6):1349–66. doi: 10.1080/15548627.2020.1761651
37. Lewis AC, Pope VS, Tea MN, Li M, Nwosu GO, Nguyen TM, et al. Ceramide-induced integrated stress response overcomes Bcl-2 inhibitor resistance in acute myeloid leukemia. Blood (2022) 139(26):3737–51. doi: 10.1182/blood.2021013277
38. Sorli S, Colié S, Albinet V, Dubrac A, Touriol C, Guilbaud N, et al. The nonlysosomal β-glucosidase GBA2 promotes endoplasmic reticulum stress and impairs tumorigenicity of human melanoma cells. FASEB J (2013) 27(2):489–98. doi: 10.1096/fj.12-215152
39. Chueakwon P, Jatooratthawichot P, Talabnin K, Ketudat Cairns JR, Talabnin C. Inhibition of ceramide glycosylation enhances cisplatin sensitivity in cholangiocarcinoma by limiting the activation of the ERK signaling pathway. Life (Basel) (2022) 12(3):351. doi: 10.3390/life12030351
40. Loberto N, Tebon M, Lampronti I, Marchetti N, Aureli M, Bassi R, et al. GBA2-encoded β-glucosidase activity is involved in the inflammatory response to Pseudomonas aeruginosa. PLoS One (2014) 9(8):e104763. doi: 10.1371/journal.pone.0104763
41. Laumont CM, Banville AC, Gilardi M, Hollern DP, Nelson BH. Tumour-infiltrating B cells: immunological mechanisms, clinical impact and therapeutic opportunities. Nat Rev Cancer (2022) 22(7):414–30. doi: 10.1038/s41568-022-00466-1
42. Wang S, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol (2019) 16(1):6–18. doi: 10.1038/s41423-018-0027-x
43. Germain C, Gnjatic S, Tamzalit F, Knockaert S, Remark R, Goc J, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med (2014) 189(7):832–44. doi: 10.1164/rccm.201309-1611OC
44. Pitzalis C, Jones GW, Bombardieri M, Jones SA. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol (2014) 14(7):447–62. doi: 10.1038/nri3700
45. Zhu W, Germain C, Liu Z, Sebastian Y, Devi P, Knockaert S, et al. A high density of tertiary lymphoid structure B cells in lung tumors is associated with increased CD4+ T cell receptor repertoire clonality. Oncoimmunology (2015) 4(12):e1051922. doi: 10.1080/2162402X.2015.1051922
46. Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell (2011) 19(4):541–55. doi: 10.1016/j.ccr.2011.02.006
47. Cho HJ, Jung JI, Lim DY, Kwon GT, Her S, Park JH, et al. Bone marrow-derived, alternatively activated macrophages enhance solid tumor growth and lung metastasis of mammary carcinoma cells in a Balb/C mouse orthotopic model. Breast Cancer Res (2012) 14(3):R81. doi: 10.1186/bcr3195
48. Kawata M, Koinuma D, Ogami T, Umezawa K, Iwata C, Watabe T, et al. TGF-β-induced epithelial-mesenchymal transition of A549 lung adenocarcinoma cells is enhanced by pro-inflammatory cytokines derived from RAW 264.7 macrophage cells. J Biochem (2012) 151(2):205–16. doi: 10.1093/jb/mvr136
49. Qian B, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell (2010) 141(1):39–51. doi: 10.1016/j.cell.2010.03.014
Keywords: hepatocellular carcinoma, sphingolipid metabolism, GBA2, single nucleotide polymorphism, overall survival
Citation: Jiang B, Qiu M, Qin L, Tang J, Zhan S, Lin Q, Wei J, Liu Y, Zhou Z, Liang X, Cao J, Lian J, Mai Y, Jiang Y and Yu H (2024) Associations between genetic variants in sphingolipid metabolism pathway genes and hepatitis B virus-related hepatocellular carcinoma survival. Front. Oncol. 13:1252158. doi: 10.3389/fonc.2023.1252158
Received: 31 July 2023; Accepted: 15 December 2023;
Published: 08 January 2024.
Edited by:
Ricardo Ribeiro, Universidade do Porto, PortugalReviewed by:
Artuo Simoni-Nieves, Foundation for Liver Research, United KingdomNghiem Xuan Hoan, 108 Hospital, Vietnam
Copyright © 2024 Jiang, Qiu, Qin, Tang, Zhan, Lin, Wei, Liu, Zhou, Liang, Cao, Lian, Mai, Jiang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongping Yu, eXVob25ncGluZ0BzdHUuZ3htdS5lZHUuY24=
†These authors have contributed equally to this work
 Binbin Jiang
Binbin Jiang Moqin Qiu
Moqin Qiu Liming Qin
Liming Qin Jingmei Tang3
Jingmei Tang3 Yingchun Liu
Yingchun Liu Zihan Zhou
Zihan Zhou Hongping Yu
Hongping Yu