- 1Department of Pathology, GROW-School for Oncology and Reproduction, Maastricht University, Medical Centre+, Maastricht, Netherlands
- 2Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Al-Baha University, Al-Baha, Saudi Arabia
- 3Department of Medical Laboratories Technology, Faculty of Applied Medical Sciences, Jazan University, Jazan, Saudi Arabia
- 4Department of Urology, Maastricht University, Medical Centre+, Maastricht, Netherlands
- 5Pathology Department, Faculty of Medicine, Jazan University, Jazan, Saudi Arabia
Objective: Urothelial cell carcinoma (UCC) is the most common type of urinary bladder. JCPyV and BKPyV have been detected in the urine and tissue of urothelial cell carcinomas (UCC) in immunocompetent patients. Here, we investigated the presence of several HPyVs in UCC samples using diverse molecular techniques to study the prevalence of HPyVs in UCC.
Methods: A large single-institution database of urine cytology specimens (UCS; n = 22.867 UCS) has previously been searched for decoy cells (n = 30), suggesting polyomavirus infection. The available urine sediments and formalin-fixed paraffin-embedded (FFPE) tissue samples of UCC patients were tested for the presence of JCPyV-LTAg expression by immunohistochemistry (IHC) labeled with SV40-LTAg antibody (clone: PAb416) and subsequent PCR followed by sequencing. In addition, the presence of the oncogenic Merkel cell polyomavirus (MCPyV) and the presence of human polyomavirus 6 (HPyV6) and 7 (HPyV7) DNA were tested with DNA PCR or IHC.
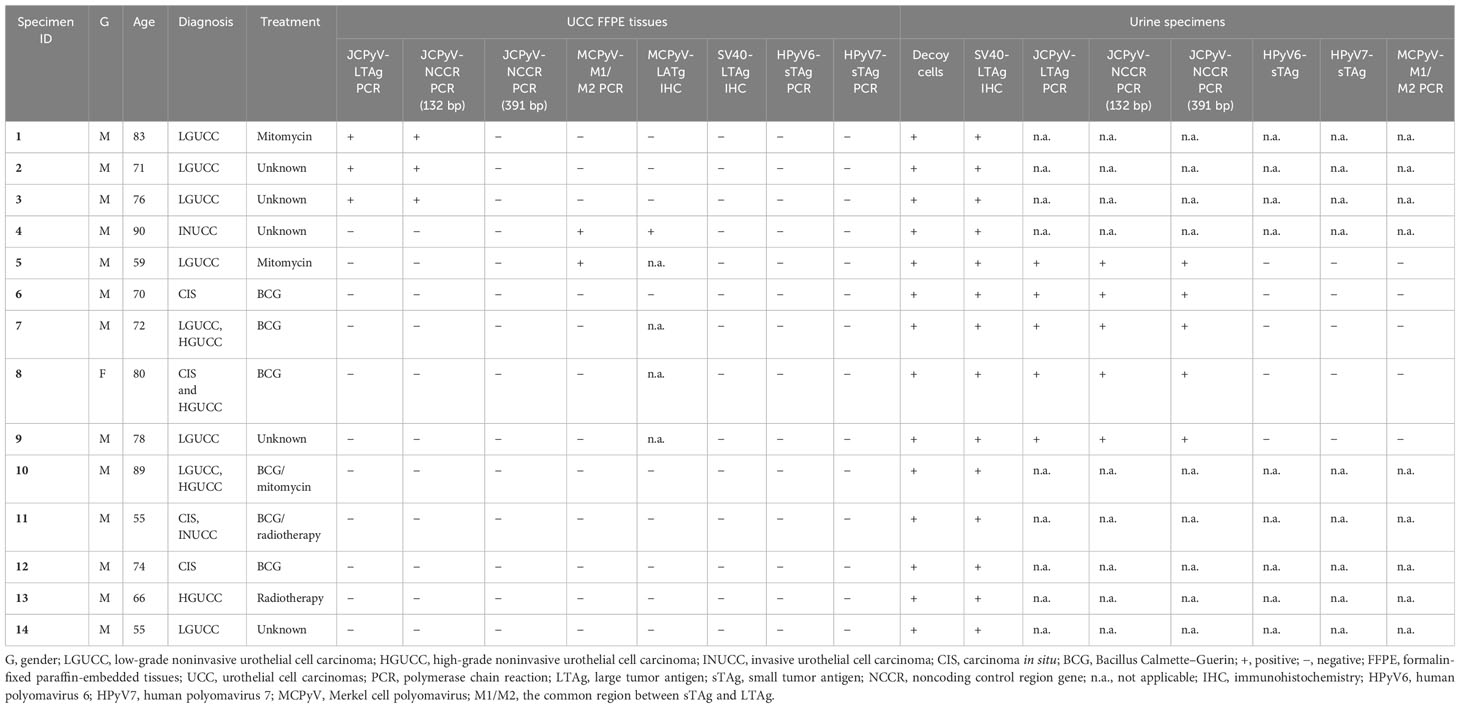
Results: Of the 30 patients harboring decoy cells, 14 were diagnosed with UCC of the urinary bladder (14/30; 46.6%) before presenting with decoy cells in the urine. The SV40-LTAg IHC was positive in all 14 UCC urine sediments and negative in the FFPE tissues. JCPyV-DNA was identified in all five available UCS and in three FFPE samples of UCC (three of 14; 21.4%). Two UCC cases were positive for MCPyV-DNA (two of 14; 14.3%), and one of them showed protein expression by IHC (one of 14; 7.1%). All specimens were HPyV6 and HPyV7 negative.
Conclusion: Our findings show the presence of JCPyV in the urine and UCC of immunocompetent patients. Moreover, MCPyV was detected in two UCC cases. In total, five UCC cases showed the presence of either JCPyV or MCPyV. The evidence here supports the hypothesis that these viruses might sporadically be associated with UCC. Further studies are needed to confirm the relevance of JCPyV or MCPyV as a possible risk factor for UCC development.
Introduction
Urothelial cell carcinoma (UCC) is the most common type of bladder cancer (BC) (1, 2). Several factors are associated with an increased risk of BC development, including but not limited to schistosomiasis infection, smoking, and genetic predisposition (3). To date, it is estimated that up to 15% of the world’s cancer case burden is attributed to viral infections (4, 5). JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV) are small, unenveloped, circular, and double-stranded DNA viruses, and both were the first human polyomaviruses isolated from patients in 1971 (6, 7). Recently, the urotheliotropic BKPyV has been increasingly well-studied and has been discussed as a possible oncogenic virus in the development of UCC in immunocompromised populations (8). JCPyV (HPyV2) is a neurotropic virus with a genome that shares about 75% homology with BKPyV (9). It has been identified as the etiological agent in the development of progressive multifocal leukoencephalopathy (PML), most commonly in immunocompromised individuals (9).
JCPyV (HPyV2) is acquired during early childhood, has shown about 80% seropositivity in humans, and has been described as a possible oncogenic virus in immunocompromised patients (10–12). In addition, JCPyV is known to remain latent or persistent in tubular epithelial cells of the kidney and urothelial tissue after primary infection (13). JCPyV large tumor antigen (LTAg) is located in the early region and has shown the capability to bind specifically with the p53 protein and retinoblastoma (pRB) protein, as known for some of the human polyomaviruses (HPyVs) (14–16). The presence of JCPyV-DNA was previously reported in many different tumor tissues, such as cervical, colorectal, gastric, lung, breast, brain, and urothelial cancers (17–23). The current number of members of HPyVs detected in various types of cancers has recently risen. Among them, Merkel cell polyomavirus (MCPyV) is the most recently identified human DNA tumor virus, which is clonally integrated into the majority of Merkel cell carcinoma (MCC) (24–26). MCPyV (HPyV5) has also been detected in the fresh frozen tissues of bladder cancer (27). Additionally, Husseiny and colleagues reported low levels of MCPyV viral load in urine specimens from immunosuppressed patients who were prospectively enrolled in kidney transplants (28). However, the potential role of HPyVs in UCC tumorigenesis is not fully elucidated.
Urine cytology using Papanicolaou staining is well accepted to reliably detect virally infected epithelial cells in the voided urine also known as “decoy cells”. These cells are characterized by enlarged nuclei and intranuclear inclusion cellular changes, as noticed in UCC neoplasia (29). The presence of decoy cells in urine has been used mostly as a marker to predict BKPyV and JCPyV reactivation (30, 31). In addition, immunohistochemical (IHC) immunolabeled with mouse anti-SV40 large T antigen monoclonal antibody, which is known to cross-react with both JCPyV and BKPyV LTAg, is used as an alternative marker to detect the reactivation of both respected viruses in urothelial cells in voided urine (10, 32, 33).
Recently, we studied the association between the BKPyV infection and UCC in patients with urine cytology positive for decoy cells (30). However, in our patient cohort, both primary and recurrent UCC tissues tested negative for BKPyV by PCR and IHC (30). Recent studies have investigated the role of JCPyV (HPyV2) in UCC tumorigenesis (34, 35). The aims of this study were to evaluate the presence of JCPyV, HPyV6, HPyV7, and MCPyV (HPyV5) in the UCC samples and in the voided urine of the patients diagnosed with UCC and with decoy cells in urine cytology.
Materials and methods
Patients and sample collection
Thirty urine cytology specimens (UCS) containing decoy cells were previously identified from a retrospective cohort study, including a total of 22,867 voided urine cytology specimens retrieved from the files of the Department of Pathology, MUMC+, The Netherlands, between January 2004 and December 2019, as reported previously (Figures 1, 2A) (30). Clinicopathologic data were collected from the medical record, as described previously (Figure 1) (30). The SV40-LTAg IHC and bladder formalin-fixed paraffin-embedded (FFPE) specimens were collected from the archive of the Department of Pathology, MUMC+. All the slides were reviewed by two pathologists (IVS and AzH). The study was approved by the Medical Ethics Review Committee of the Maastricht University Medical Centre+, The Netherlands (2019-0977). All specimens were collected and studied in accordance with the protocol of the Dutch Code of Conduct for Observational Research with Personal Data (2004) and Tissue (36).
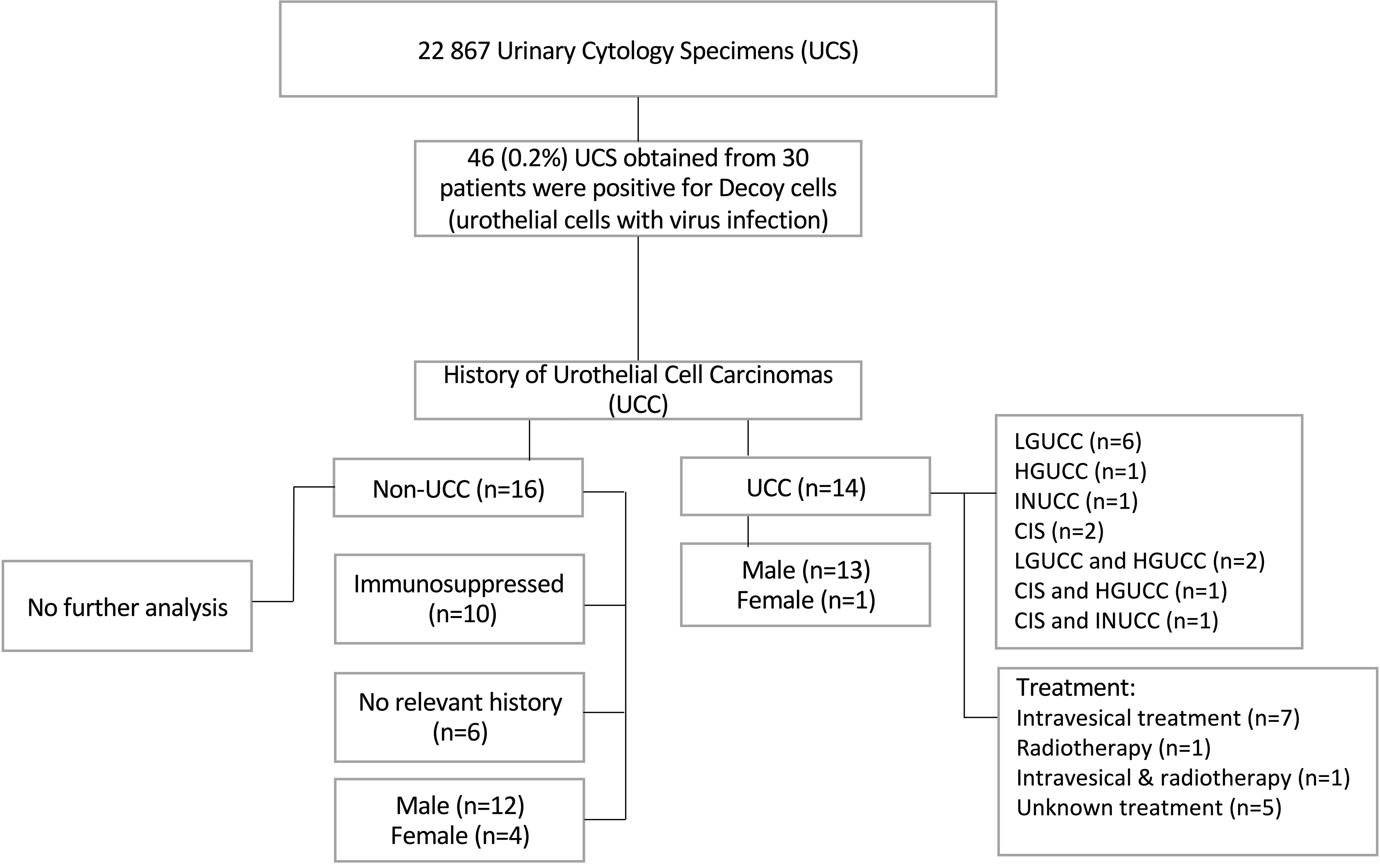
Figure 1 Flow chart summarizing the cohort clinicopathological data and urine cytology results. LGUCC, low-grade noninvasive urothelial cell carcinoma; HGUCC, high-grade noninvasive urothelial cell carcinoma; INUCC, invasive urothelial cell carcinoma; CIS, carcinoma in situ.

Figure 2 (A) Represents the decoy cells in urine cytology stained with Papanicolaou staining. (B) SV40-LTAg immunohistochemistry for UCC urine sediments. (C) H&E of low-grade noninvasive urothelial cell carcinoma (LGUCC). (D) H&E of high-grade noninvasive urothelial cell carcinoma (HGUCC). (E) SV40-LTAg immunohistochemistry for LGUCC FFPE tissues (left corner box is the positive control). (F) SV40-LTAg immunohistochemistry for HGUCC FFPE tissues (left corner box is the positive control). (G) Positive IHC for MCPyV, specific nuclear expression (brown) of MCPyV (CM2B4 antibody) in the nuclei of UCC epithelial (left corner box is the positive MKL1 control). (H) Negative IHC for MCPyV (left corner box is the positive MKL1 control).
Cytology, histology, and immunohistochemistry
Two slides were prepared for each urine sediment sample using the previously published Cytospin protocol (37). The first slide was stained with Papanicolaou stain, and the other slide was used for immunoperoxidase IHC using SV40-LTAg antibody (clone: PAb416, dilution 1:500, Calbiochem Inc, San Diego, CA, USA) with a Dako’s autostainer according to the routine diagnostic pathology department protocol.
The urinary bladder FFPE sections were stained with routine H&E staining for light microscopy. Additionally, the UCC FFPE were immunolabeled with SV40-LTAg antibody (clone: PAb416, dilution 1:500, Calbiochem Inc, San Diego, CA, USA) as described previously (30). Furthermore, 10 UCC FFPE tissues were tested for the expression of MCPyV large tumor antigen (LTAg) protein using the CM2B4 monoclonal antibody (clone: CM2B4, dilution 1:50; Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA), and the MCC cell line MKL-1, which is positive for MCPyV, was used as a positive control for the CM2B4 antibody. Briefly, 5-μm-thick FFPE sections were immunolabeled with a Dako’s autostainer Link 48 according to the EnVision FLEX Visualization manufacturer’s kit (K8008, Dako, Carpinteria, CA, USA), and protein expression in the nucleus was interpreted as a positive. Next, all the IHC slide photo results were obtained by digital scanning with the VENTANA iScan-HT slide scanner (Roche Diagnostics Inc, Tucson, AZ, USA).
DNA extraction and specific HPyV-DNA PCR
DNA was extracted from the available five urine sediments obtained from UCC patients and 14 UCC FFPE tissue blocks according to the protocol of genomic DNA isolation using NucleoSpin® Tissue (Macherey–Nagel GmbH & Co, Düren, Germany). As previously described in our study, the DNA concentration, integrity, and quality were evaluated using a spectrophotometer (NanoDrop 2000, Thermo Scientific, Wilmington, DE, USA) and using multiplex primers [specimen control size (SCS)] as described previously (30, 38, 39).
JCPyV-DNA PCR was conducted by adding a total of 125 ng of DNA to the PCR master mix. Three sets of primers were used; the first set of primers was a previously published targeted conserved region of JCPyV and BKPyV LTAg using the following sequences: (FW: AAGTCTTTAGGGTCTTCTAC) (RV: GTGCCAACCTATGGAACAGA) (40). Another previously published set of primers was also used to amplify 391 bp of JCPyV noncoding control region gene (NCCR) (FW: TTCCTCCCTATTCAGCACTT) (RV: AAAACAGCTCTGGCTCGCAA) NCCR gene (41). In addition, a third set of primers designed by our group (FW: GCTCATACCTAGGGAGCCAA) (RV: CTGCTTTCCACTTCCCCTTGT) to target the JCPyV-NCCR gene were used to amplify 132 bp. The two sets of primers targeting JCPyV-NCCR were employed in order to distinguish between JCPyV strains, either archetype or Mad strains. To validate the PCR analysis, a known JCPyV-positive specimen was used as a positive control. In addition, DNA PCR targeting HpyV6, HpyV7, and MCPyV were performed using protocols as recently described (42, 43). DNA-PCR were used to amplify the small tumor antigen (sTAg) of HPyV6 and HPyV7, targeting the common region between sTAg and LTAg of MCPyV. HPyV6 and HPyV7 plasmids with inserted histidine tag and the MCPyV-positive MCC cell line (MKL-1) were used as positive controls. Bioperformance-certified water was used as a nontemplate negative control. In positive cases, all the amplified PCR products were purified, subsequently submitted for sequencing, and analyzed with the consensus sequences of the National Center for Biotechnology Information (NCBI).
Results
SV40-LTAg and MCPyV-LTAg IHC in FFPE tissues of UCC
No viral histomorphological cytopathic changes were seen in the UCC specimens of all 14 patients (Figures 2C, D). We previously assessed the expression of SV40-LTAg in all 14 UCC FFPE specimens and revealed no expression of SV40-LTAg in both invasive and in situ UCC samples (Table 1; Figures 2E, F) (30). UCC samples were assessed for MCPyV-LTAg by IHC, and one UCC sample tested positive (one of 10; 10%; Table 1; Figure 2G). The expression of MCPyV-LTAg revealed weak to moderate nuclear immunostaining in less than 60% of the tumor cells, and no cytoplasmic expression was observed in the neoplastic urothelium (Table 1; Figures 2G, H).
JCPyV-, HPyV6-, HPyV7-, and MCPyV-specific PCRs in FFPE tissues and urine of UCC
JCPyV- (LTAg and NCCR) DNA-PCR was conducted on all 14 UCC FFPE specimens, and the results revealed that three samples (three of 14; 21.4%) tested positive using both JCPyV-LTAg and JCPyV-NCCR primer sets (primary and recurrent UCC) (Table 1; Figure 3). Sequencing analyses of the PCR amplicons confirmed that all PCR products were 96% to 98% identical to JCPyV. Interestingly, all three UCC specimens, positive for JCPyV-LTAg and JCPyV-NCCR PCR, were low-grade noninvasive urothelial cell carcinoma (LGUCC) (Table 1). The other five patients with UCC, initially negative for JCPyV LTAg and NCCR, showed positivity for this virus in their urine sediments (Table 1). Of note and important to remember, urine cytology was taken in a follow-up period after the initial UCC resection, and four of these patients received an intravesical treatment (BCG or mitomycin). The JCPyV-LTAg and JCPyV-NCCR were amplified in all five specimens and confirmed by sequencing with homology that ranges from 97% to 99% identical to JCPyV (Table 1; Figure 3). Furthermore, using JCPyV-NCCR PCR revealed that the urine and tissue specimens were positive for the JCPyV harbor archetype strain. Additionally, HPyV6-, HPyV7-, and MCPyV-specific PCR were conducted on all 14 UCC FFPE samples and five urine sediments. Two UCC samples (two of 14; 14.3%) were positive for MCPyV PCR targeting the common region of LTAg and sTAg. One patient presented with MCPyV in the UCC FFPE specimen and JCPyV-LTAg in the urine sediment (ID5, Table 1). Neither HPyV6- nor HPyV7-DNA could be amplified by PCR (Table 1).
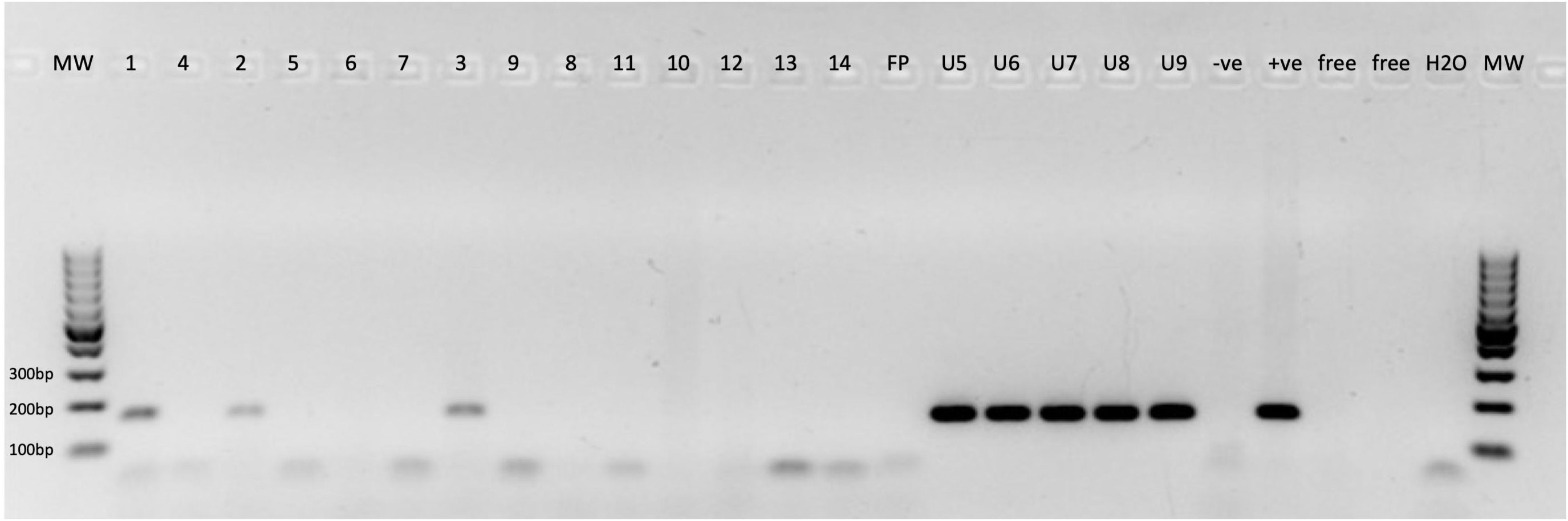
Figure 3 Specific JCPyV-DNA-PCR. 1–14, JCPyV-DNA PCR for urothelial cell carcinoma (UCC) FFPE tissue specimens; FP, free paraffin as negative control; U10–U14, JCPyV-DNA PCR for urine sediments. −ve, clinical specimen negative for JCPyV; +ve, clinical specimen positive for JCPyV; Free, empty slot (no master mix added to the gel); H2O, water nontemplate; MW, molecular weight marker.
Discussion
We recently reported that we did not find an association between BKPyV detection in the urine sediments of patients with prior resected urothelial cell carcinomas. BKPyV reactivation was not restricted to immunosuppression and was possibly related to intravesical BCG or mitomycin treatment in immunocompetent patients (30). JCPyV is closely related to BKPyV, and the large- and small-tumor antigens of both viruses were found to be capable of transforming cells in vitro (44–47). In contrast to the relationship of BKPyV with UCC, limited information is available regarding the role of JCPyV in UCC tumorigenesis, especially in immunocompetent cases (34, 35).
We analyzed the presence of JCPyV on the protein level in combination with DNA-PCR in FFPE tissues and voided urine sediments obtained from UCC immunocompetent patients. Previously, we demonstrated that the detection of decoy cells was infrequent, present in only 0.2% of a large urine specimen cohort from our institution (30). The detection of decoy cells in urine cytology is used in a diagnostic setting for identifying possible polyomavirus infections (48). However, the decoy cells were identified in the urine cytology of all 14 UCC cases, which suggests polyomavirus reactivation (30). Indeed, SV40-LTAg expression was detectable in the urine sediments, which confirmed polyomavirus reactivation in all 14 UCC patients (Figure 2B). Although the SV40-LTAg monoclonal antibody is commonly used in diagnostic pathology, it is known to cross-react with the closely related human polyomaviruses BKPyV and JCPyV (49). We confirmed the presence of JCPyV-LTAg by PCR among the five available urine sediments obtained from five UCC patients. Also, we previously reported the detection of BKPyV-LTAg by PCR in all of the five urine sediments (30). Thus, it can be speculated that the tubular epithelial cells are coinfected with both JCPyV and BKPyV, and both may contribute to the presence of the decoy cells.
Here, we performed JCPyV-LTAg and JCPyV-NCCR PCR in UCC FFPE samples obtained from patients who were diagnosed with decoy cells in their urine. It is important to mention that the patients were diagnosed with UCC, and urine cytology was collected during the postoperative follow-up. The main finding in our data is that 21.4% of UCC FFPE tissues tested positive using JCPyV LTAg and NCCR PCR, as confirmed by sequencing. Furthermore, the JCPyV-NCCR positive tissues and urine specimens were identical to the archetype strain, as confirmed by the presence of the 64 and 23 bp sequences that are absent in the Mad strains, which have been associated with human brain tumors. However, there is a study that reported the detection of archetype strain sequences in two cases of oligodendroglioma (50). The detection of JCPyV-DNA was previously reported in many different tumors, such as cervical, colorectal, gastric, lung, breast, brain, and urothelial cancers (17–23). Infection with JCPyV has been suggested to be associated with bladder carcinoma, which still remains a controversial hypothesis (34, 35). Results from a study that screened for JCPyV among urothelial bladder cancer patients in the UK concluded that 0.9% of bladder tumors were positive for JCPyV (34). In contrast, Fioriti et al. suggested that JCPyV might play a role in bladder cancer and reported the detection of JCPyV-DNA in 19% of bladder urothelial carcinoma (35).
JCPyV is known to remain latent in the kidney after infection and reactivate in immunosuppressed patients (11). However, in our study, JCPyV-PCR positivity in urine sediments was seen in immunocompetent UCC patients; most of the patients were known to have received either mitomycin or intravesical BCG treatment. Thus, our results suggest that the reactivation of latent JCPyV could be related to the locally administered UCC intravesical treatment since the patients were diagnosed with UCC of the urinary bladder before presenting with decoy cells in the urine cytology.
MCPyV is an oncogenic virus discovered in 2008 and has been linked to the pathogenesis of the majority of MCC. MCC are rare, highly aggressive neuroendocrine nonmelanoma skin cancers (24–26). In contrast, there is yet no evidence established regarding the role of MCPyV in bladder carcinoma. One study reported the presence of MCPyV-DNA in voided urine specimens of transitional cell carcinomas of the bladder and revealed 2.7% positivity by PCR (51). Also, Loyo et al. reported the detection of MCPyV-DNA in 75% of bladder cancer tissues with a low viral load (< 0.001 copies per genome) (27). Another study revealed that MCPyV was detected in 30% of the urine specimens from prospectively enrolled immunosuppressed kidney transplant patients by using qPCR with a relatively low viral load (10 to 3.7 × 102 genome copies/mL) (28). In our study, MCPyV was not detectable in the urine sediments of UCC, while MCPyV-DNA was positive in two 14.3% UCC FFPE tissues, and specific nuclear MCPyV-LTAg expression was seen in the neoplastic cells of one UCC tissue. Indeed, it is very interesting that MCPyV immunoreactivity was found to be abundantly present in UCC tumor cells. However, a direct contributing role of MCPyV to UCC carcinogenesis seems unlikely, at least based on our findings unlikely (24–26). The observed pattern of MCPyV protein expression in UCC tissues possibly might imply an indirect role for MCPyV in the development of UCC by, e.g., inflammation as it is known for hepatotropic viruses (e.g., hepatitis B and C viruses) (52). However, further investigations are needed to identify any oncogenic mutations or clonal integration of MCPyV LTAg or sTAg into the genome of UCC.
Noteworthy, the mouse anti-SV40-LTAg monoclonal antibody (clone PAb416) is commonly used in clinical pathology settings and is well known to cross-react with both JCPyV and BKPyV LTAg (53). However, Toptan et al. reported that anti-SV40-LTAg (PAb416) is not restricted to JCPyV and BKPyV. Of note, PAb416 also detects LTAg proteins of KIV, WUV, HPyV6, HPyV7, TSV, HPyV10, and HPyV11 (54). HPyV6 and HPyV7 share approximately 68% genome sequence identity and reveal high seropositivity in healthy populations (55–57). Since anti-SV40-LTAg showed the ability to detect both HPyV6 and HPyV7, we have tested all 14 UCC tissues and the five urine sediments for the presence of HPyV6- and HPyV7-DNA. Our results showed that all UCC specimens were negative for both HPyV6 and HPyV7, and to our knowledge, this is the first study to test UCC specimens for HPyV6 and HPyV7.
Some limitations of this study need to be mentioned. First, the size of the study cohort is relatively small, which partially reflects the rarity of UCC and the number of UCC patients admitted to our hospital. Second, obtaining healthy control tissues is practically impossible. In addition, parallel screening for HPyV-DNA in blood specimens would have been an interesting add-on to our study; however, we were unable to carry out these experiments due to the unavailability of blood.
To the best of our knowledge, this is the first study to investigate the presence of several HPyVs in UCC samples using diverse molecular techniques and the possible contribution of HPyVs in immunocompetent UCC etiopathogenesis. In total, five UCC cases showed the presence of either JCPyV or MCPyV. Both HPyV6 and HPyV7 were not detected. Further studies are warranted to confirm the relevance of JCPyV or MCPyV as a risk factor for developing UCC and to identify any oncogenic mutations or clonal integration into the genome of urothelial cell carcinoma.
Conclusions
JCPyV-DNA is detected in the urine and urothelial cells, and MCPyV was detected in urothelial cell carcinoma. Since there is inadequate evidence of a role for JCPyV in carcinogenicity in UCC, these findings support the hypothesis that JCPyV infection could contribute to urothelial carcinoma tumorigenesis. Moving forward, it is important to define whether or not both JCPyV and MCPyV are involved in UCC tumorigenesis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The study was approved by the Medical Ethics Review Committee of the Maastricht University Medical Centre+, The Netherlands (2019-0977). All specimens were collected and studied in accordance with the protocol of the Dutch Code of Conduct for Observational Research with Personal Data (2004) and Tissue. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
All authors have contributed substantially to conceptualizing the research design planning and continued supervision of the work (FK, GM, AH, IS), aacquisition of data (FK, GM, SS, TM, RA, MM, JR), performed and processed the experimental data and analysis (FK GM SS RA, SA), analysis and interpretation of data (FK, GM, TM, MM, SA, AH, IS), drafting the manuscript (FK, GM, AH, IS), and revising it critically (JR, AH, IS). All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: Cancer J Clin (2015) 65(1):5–29. doi: 10.3322/caac.21254
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
3. Chang SS, Bochner BH, Chou R, Dreicer R, Kamat AM, Lerner SP, et al. Treatment of non-metastatic muscle-invasive bladder cancer: AUA/ASCO/ASTRO/SUO guideline. J urol (2017) 198(3):552–9. doi: 10.1016/j.juro.2017.04.086
4. Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer (2010) 10(12):878–89. doi: 10.1038/nrc2961
5. Ahuja R, Jamal A, Nosrati N, Pandey V, Rajput P, Saxena N, et al. Human oncogenic viruses and cancer. Curr Sci (2014) 107(5):768–85.
6. Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet (1971) 1(7712):1253–7.
7. Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet (1971) 1(7712):1257–60. doi: 10.1016/S0140-6736(71)91777-6
8. Starrett GJ, Buck CB. The case for BK polyomavirus as a cause of bladder cancer. Curr Opin virol (2019) 39:8–15. doi: 10.1016/j.coviro.2019.06.009
9. Pinto M, Dobson S. BK and JC virus: a review. J Infect (2014) 68(Suppl 1):S2–8. doi: 10.1016/j.jinf.2013.09.009
10. Roberts IS, Besarani D, Mason P, Turner G, Friend PJ, Newton R. Polyoma virus infection and urothelial carcinoma of the bladder following renal transplantation. Br J cancer (2008) 99(9):1383–6. doi: 10.1038/sj.bjc.6604711
11. De Caprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiol (2013) 11(4):264–76. doi: 10.1038/nrmicro2992
12. Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PloS pathogens (2009) 5(3):e1000363. doi: 10.1371/journal.ppat.1000363
13. Shen CH, Wu JD, Hsu CD, Jou YC, Lin CT, Wang M, et al. The high incidence of JC virus infection in urothelial carcinoma tissue in Taiwan. J Med virol (2011) 83(12):2191–9. doi: 10.1002/jmv.22240
14. Dyson N, Bernards R, Friend SH, Gooding LR, Hassell JA, Major EO, et al. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J virol (1990) 64(3):1353–6. doi: 10.1128/jvi.64.3.1353-1356.1990
15. Bollag B, Prins C, Snyder EL, Frisque RJ. Purified JC virus T and T' proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology (2000) 274(1):165–78. doi: 10.1006/viro.2000.0451
16. Sharma AK, Kumar G. A 53 kDa protein binds to the negative regulatory region of JC virus early promoter. FEBS Lett (1991) 281(1-2):272–4. doi: 10.1016/0014-5793(91)80409-v.
17. Zheng HC, Xue H, Zhang CY. The oncogenic roles of JC polyomavirus in cancer. Front Oncol (2022) 12:976577. doi: 10.3389/fonc.2022.976577
18. Hori R, Murai Y, Tsuneyama K, Abdel-Aziz HO, Nomoto K, Takahashi H, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Archiv an Int J pathol (2005) 447(4):723–30. doi: 10.1007/s00428-005-0014-3
19. Ksiaa F, Ziadi S, Mokni M, Korbi S, Trimeche M. The presence of JC virus in gastric carcinomas correlates with patient's age, intestinal histological type and aberrant methylation of tumor suppressor genes. Modern Pathol (2010) 23(4):522–30. doi: 10.1038/modpathol.2009.184
20. Abdel-Aziz HO, Murai Y, Hong M, Kutsuna T, Takahashi H, Nomoto K, et al. Detection of the JC virus genome in lung cancers: possible role of the T-antigen in lung oncogenesis. Appl immunohistochem Mol morphol AIMM (2007) 15(4):394–400. doi: 10.1097/01.pai.0000213126.96590.64
21. Zheng HC EY, Cui ZG, Zhao S, Zhang Y. The oncogenic roles of JC virus T antigen in breast carcinogenesis. Front Mol Biosci (2021) 8:687444. doi: 10.3389/fmolb.2021.687444
22. Rastogi A, Gulati N, Bihari C, Chaudhary A, Bansal K, Sasturkar S, et al. JC virus-related progressive multifocal leukoencephalopathy after living-donor liver transplant: A rare case. Exp Clin Transplant (2019) 17(3):414–7. doi: 10.6002/ect.2016.0242
23. Knöll A, Stoehr R, Jilg W, Hartmann A. Low frequency of human polyomavirus BKV and JCV DNA in urothelial carcinomas of the renal pelvis and renal cell carcinomas. Oncol Rep (2003) 10(2):487–91.
24. Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Sci (New York NY) (2008) 319(5866):1096–100. doi: 10.1126/science.1152586
25. Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, et al. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res (2008) 68(13):5009–13. doi: 10.1158/0008-5472.CAN-08-0949
26. Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci United States America (2008) 105(42):16272–7. doi: 10.1073/pnas.0806526105
27. Loyo M, Guerrero-Preston R, Brait M, Hoque MO, Chuang A, Kim MS, et al. Quantitative detection of Merkel cell virus in human tissues and possible mode of transmission. Int J cancer (2010) 126(12):2991–6. doi: 10.1002/ijc.24737
28. Husseiny MI, Anastasi B, Singer J, Lacey SF. A comparative study of Merkel cell, BK and JC polyomavirus infections in renal transplant recipients and healthy subjects. J Clin Virol (2010) 49(2):137–40. doi: 10.1016/j.jcv.2010.06.017
29. Ranzi AD, Prolla JC, Keitel E, Brackmann R, Kist R, dos Santos G, et al. The role of urine cytology for 'decoy cells' as a screening tool in renal transplant recipients. Acta cytologica (2012) 56(5):543–7. doi: 10.1159/000341425
30. Klufah F, Mobaraki G, Hausen AZ, Samarska IV. Reactivation of BK polyomavirus in urine cytology is not associated with urothelial cell carcinoma. Viruses (2020) 12(12):1412. doi: 10.3390/v12121412
31. Sekito T, Araki M, Yoshinaga K, Maruyama Y, Sadahira T, Nishimura S, et al. Presence of decoy cells for 6 months on urine cytology efficiently predicts BK virus nephropathy in renal transplant recipients. Int J Urol (2021) 28(12):1240–6. doi: 10.1111/iju.14679
32. Morace R, Kumar T, Tantisattamo E, Gibson J, Britton S, Li W, et al. Feasibility of BK virus real-time PCR testing in renal graft biopsies with negative SV40 staining. Transplant Proc (2017) 49(6):1294–300. doi: 10.1016/j.transproceed.2017.03.095
33. Yamada Y, Tsuchiya T, Inagaki I, Seishima M, Deguchi T. Prediction of early BK virus infection in kidney transplant recipients by the number of cells with intranuclear inclusion bodies (Decoy cells). Transplant direct (2018) 4(2):e340. doi: 10.1097/txd.0000000000000759
34. Llewellyn MA, Gordon NS, Abbotts B, James ND, Zeegers MP, Cheng KK, et al. Defining the frequency of human papillomavirus and polyomavirus infection in urothelial bladder tumours. Sci Rep (2018) 8(1):11290. doi: 10.1038/s41598-018-29438-y
35. Fioriti D, Pietropaolo V, Dal Forno S, Laurenti C, Chiarini F, Degener AM. Urothelial bladder carcinoma and viral infections: different association with human polyomaviruses and papilloma viruses. Int J Immunopathol Pharmacol (2003) 16(3):283–8. doi: 10.1177/039463200301600315
36. Fedra. Human Tissue and Medical Research: Code of Conduct for responsible use. (2011). www.federa.org, ISBN/EAN 978-90-817510-0-1, The Netherlands.
37. Qamar I, Rehman S, Mehdi G, Maheshwari V, Ansari HA, Chauhan S. Utility of cytospin and cell block technology in evaluation of body fluids and urine samples: A comparative study. J cytol (2018) 35(2):79–82. doi: 10.4103/joc.Joc_240_16
38. Klufah F, Mobaraki G, Chteinberg E, Alharbi RA, Winnepenninckx V, Speel EJM, et al. High prevalence of human polyomavirus 7 in cholangiocarcinomas and adjacent peritumoral hepatocytes: preliminary findings. Microorganisms (2020) 8(8):1125. doi: 10.3390/microorganisms8081125
39. van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia (2003) 17(12):2257–317. doi: 10.1038/sj.leu.2403202
40. Nickeleit V, Klimkait T, Binet IF, Dalquen P, Del Zenero V, Thiel G, et al. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. New Engl J Med (2000) 342(18):1309–15. doi: 10.1056/nejm200005043421802
41. Del Valle L, Khalili K. Induction of brain tumors by the archetype strain of human neurotropic JCPyV in a transgenic mouse model. Viruses (2021) 13(2):162. doi: 10.3390/v13020162
42. Rennspiess D, Pujari S, Keijzers M, Abdul-Hamid MA, Hochstenbag M, Dingemans AM, et al. Detection of human polyomavirus 7 in human thymic epithelial tumors. J Thorac Oncol (2015) 10(2):360–6. doi: 10.1097/JTO.0000000000000390
43. Chteinberg E, Klufah F, Rennspiess D, Mannheims MF, Abdul-Hamid MA, Losen M, et al. Low prevalence of Merkel cell polyomavirus in human epithelial thymic tumors. Thorac Cancer (2019) 10(3):445–51. doi: 10.1111/1759-7714.12953
44. Bollag B, Chuke WF, Frisque RJ. Hybrid genomes of the polyomaviruses JC virus, BK virus, and simian virus 40: identification of sequences important for efficient transformation. J virol (1989) 63(2):863–72. doi: 10.1128/jvi.63.2.863-872.1989
45. Del Valle L, Gordon J, Ferrante P, Khalili K. JC virus in experimental and clinical brain tumorigenesis. Hum Polyomaviruses (2001), 409–30. doi: 10.1002/0471221945.ch15
46. Shivakumar CV, Das GC. Interaction of human polyomavirus BK with the tumor-suppressor protein p53. Oncogene (1996) 13(2):323–32.
47. Haukland HH, Vonen B, Traavik T. Transformed rat pancreatic islet-cell lines established by BK virus infection in vitro. Int J Cancer (1992) 51(1):79–83. doi: 10.1002/ijc.2910510116
48. Geetha V, Rao L, Monappa V, Susmitha M, Prabhu R. Decoy cells in urine cytology: A useful clue to post-transplant polyoma virus infection. J cytol (2012) 29(2):133–4. doi: 10.4103/0970-9371.97157
49. Pelletier DJ, Czeczok TW, Bellizzi AM. A monoclonal antibody against SV40 large T antigen (PAb416) does not label Merkel cell carcinoma. Histopathology (2018) 73(1):162–6. doi: 10.1111/his.13483
50. Del Valle L, Enam S, Lara C, Ortiz-Hidalgo C, Katsetos CD, Khalili K. Detection of JC polyomavirus DNA sequences and cellular localization of T-antigen and agnoprotein in oligodendrogliomas. Clin Cancer Res (2002) 8(11):3332–40.
51. Polesel J, Gheit T, Talamini R, Shahzad N, Lenardon O, Sylla B, et al. Urinary human polyomavirus and papillomavirus infection and bladder cancer risk. Br J cancer (2012) 106(1):222–6. doi: 10.1038/bjc.2011.519
52. Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology (2011) 54(1):173–84. doi: 10.1002/hep.24351
53. Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J virol (1981) 39(3):861–9. doi: 10.1128/jvi.39.3.861-869.1981
54. Toptan T, Yousem SA, Ho J, Matsushima Y, Stabile LP, Fernández-Figueras M-T, et al. Survey for human polyomaviruses in cancer. (2016) 1(2):e85562. doi: 10.1172/jci.insight.85562
55. Fava P, Merlino C, Novelli M, Ponti R, Galliano I, Montanari P, et al. HPyV6, HPyV7 and TSPyV DNA sequences detection in skin disease patients and healthy subjects. J Eur Acad Dermatol Venereol JEADV (2016) 30(4):624–7. doi: 10.1111/jdv.13094
56. Sroller V, Hamsikova E, Ludvikova V, Musil J, Nemeckova S, Salakova M. Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors. J Med virol (2016) 88(7):1254–61. doi: 10.1002/jmv.24440
Keywords: JCPyV, bladder cancer, polyomavirus, tumorigenesis, decoy cells, HPyV6, HPyV7, MCPyV
Citation: Klufah F, Mobaraki G, Shi S, Marcelissen T, Alharbi RA, Mobarki M, Almalki SSR, van Roermund J, zur Hausen A and Samarska I (2023) Human polyomaviruses JCPyV and MCPyV in urothelial cell carcinoma: a single institution experience. Front. Oncol. 13:1251244. doi: 10.3389/fonc.2023.1251244
Received: 01 July 2023; Accepted: 27 November 2023;
Published: 13 December 2023.
Edited by:
Fernanda Martini, University of Ferrara, ItalyReviewed by:
Luis Del Valle, Louisiana State University, United StatesJohn Charles Rotondo, University of Ferrara, Italy
Copyright © 2023 Klufah, Mobaraki, Shi, Marcelissen, Alharbi, Mobarki, Almalki, van Roermund, zur Hausen and Samarska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iryna Samarska, SXJ5bmEuc2FtYXJza2FAbXVtYy5ubA==; Faisal Klufah, ZmtsdWZhaEBidS5lZHUuc2E=
†These authors share first authorship
 Faisal Klufah
Faisal Klufah Ghalib Mobaraki
Ghalib Mobaraki Shuai Shi
Shuai Shi Tom Marcelissen4
Tom Marcelissen4 Axel zur Hausen
Axel zur Hausen Iryna Samarska
Iryna Samarska