- 1Department of Oncology, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Clinical School, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 3Operating Room, Sichuan University West China Hospital School of Nursing, Chengdu, China
- 4Department of Gastroenterology, China Academy of Chinese Medical Sciences Guang’anmen Hospital, Beijing, China
Background: Malignant tumors, mainly solid tumors, are a significant obstacle to the improvement of life expectancy at present. Epithelial cell adhesion molecule (EpCAM), a cancer stem cell biomarker, showed widespread expression in most normal epithelial cells and most cancers. Although the clinical significance of EpCAM in various malignant solid tumors has been studied extensively, the latent relationships between EpCAM and pathological and clinical characteristics in solid tumors and differences in the roles of EpCAM among tumors have not been clearly determined. The destination point of this study was to analyze the value of EpCAM in solid tumors in clinicopathological and prognostic dimension using a meta-analysis approach.
Method and materials: A comprehensive and systematic search of the researches published up to March 7th, 2022, in PubMed, EMBASE, Web of Science, Cochrane library and PMC databases was performed. The relationships between EpCAM overexpression, clinicopathological characteristics, and survival outcomes were analyzed. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) and odds ratios (ORs) were estimated as indicators of the degree of correlation. This research was registered on PROSPERO (International prospective register of systematic reviews), ID: CRD42022315070.
Results: In total, 57 articles and 14184 cases were included in this study. High EpCAM expression had a significant coherence with a poorer overall survival (OS) (HR: 1.30, 95% CI: 1.08–1.58, P < 0.01) and a worse disease-free survival (DFS) (HR: 1.58, 95% CI: 1.28–1.95, P < 0.01), especially of gastrointestinal tumors’ OS (HR: 1.50, 95% CI: 1.15–1.95, P < 0.01), and DFS (HR: 1.84, 95% CI: 1.52–2.33, P < 0.01). The DFS of head and neck tumors (HR: 2.33, 95% CI: 1.51–3.61, P < 0.01) was also associated with the overexpression of EpCAM. There were no positive relationships between the overexpression of EpCAM and sex (RR: 1.03, 95% CI: 0.99–1.07, P = 0.141), T classification (RR: 0.93, 95% CI: 0.82–1.06, P = 0.293), lymph node metastasis (RR: 0.85, 95% CI: 0.54–1.32, P = 0.461), distant metastasis (RR: 0.97, 95% CI: 0.84–1.10, P = 0.606), vascular infiltration (RR: 1.05, 95% CI: 0.85–1.29, P = 0.611), and TNM stage (RR: 0.93, 95% CI: 0.83–1.04, P = 0.187). However, the overexpression of EpCAM exhibited a significant association with the histological grades (RR: 0.88, 95% CI: 0.80–0.97, P < 0.01).
Conclusion: Based on pooled HRs, the positive expression of EpCAM was totally correlated to a worse OS and DFS in solid tumors. The expression of EpCAM was related to a worse OS in gastrointestinal tumors and a worse DFS in gastrointestinal tumors and head and neck tumors. Moreover, EpCAM expression was correlated with the histological grade. The results presented pointed out that EpCAM could serve as a prognostic biomarker for gastrointestinal and head and neck tumors.
Systematic review registration: https://www.crd.york.ac.uk/prospero, identifier CRD42022315070.
1 Introduction
Cancer, the first or second principal cause of death in most countries (1), is a significant obstacle to the improvement of life expectancy at present (2). Epithelial cell adhesion molecule (EpCAM), which showed widespread expression in most normal epithelial cells and most cancers, is an epithelial glycoprotein encoded by GA-733-2 (3). The molecule is participated in various physiological processes, such as cell adhesion, proliferation, migration, and mitotic signal transduction (4). EpCAM has been identified as a cancer stem cell (CSC) marker. CSCs have strong self-renewal ability and are directly related to tumor formation (5), accounting for 0.05–3% of the total number of tumor cells (6). CSCs have been a focus of research in recent decades and have been implicated in tumor generation, metastasis, recurrence, heterogeneity, resistance to chemotherapy and radiotherapy, and avoidance of immune surveillance (7, 8). A number of recent meta-analyses have shown that EpCAM expression levels are competent to serve as a significant prognostic marker in stomach (9), hepatic (10), prostate (11), and colorectal malignant tumors (12). Although the clinical significance of EpCAM in various cancers has been widely studied, differences of EpCAM expression in heterogeneous cancers and the relationships between EpCAM and pathological and clinical characteristics in solid tumors have not been determined. We employed a meta-analysis method for this investigation to comprehensively and systematically analyze EpCAM expression in different cancers and its relationship with survival outcomes and clinical characteristics. Furthermore, we conducted subgroup analyses to establish the prognostic and clinical validity of EpCAM in different cancers. The results provide basis for further studies of the applications of EpCAM.
2 Methods and materials
2.1 Search strategy and inclusion criteria
A comprehensive and systematic search of studies published up to March 7, 2022, in PubMed, EMBASE, Web of Science, Cochrane library and PMC databases was conducted. The search terms were as follows: (‘EpCAM’ OR ‘Epithelial Cell Adhesion Molecule’) AND (Tumor OR Neoplasm OR Neoplasia OR Cancer OR Carcinoma OR Malignancy) AND (Prognosis OR outcome OR survival).
Published articles that were in full compliance with following inclusion criteria were considered eligible: (1) published in English; (2) studies with pathologically accurate solid tumor diagnoses, including lung, breast, ovarian, gastric, hepatic, colorectal and pancreatic cancer and etc.; (3) EpCAM levels were detected by immunohistochemistry (IHC), quantitative real time-polymerase chain reaction (qRT-PCR), or enzyme-linked immunosorbent assay (ELISA); (4) studies evaluating the correlation between EpCAM overexpression and overall survival (OS), disease-free survival (DFS), and/or clinicopathological features of solid tumors; (5) hazard ratios (HRs) with 95% confidence intervals (CIs) are reported or data are available to calculate HRs and 95% CIs. Studies on the basis of the following criteria were excluded: (1) articles with overlapping or duplicate results, a lack of information, reviews, animal reports, conference abstracts, expert opinions, case reports, and letters; (2) studies irrelevant to the subjects of interest; (3) studies in which participators were in administration of any kind of anti-cancer treatment, for instance chemotherapy and radiotherapy, prior to surgical pathology or biopsy; (4) studies with a sample size of less than 40 patients.
2.2 Data extraction and quality assessment
Assessments of the abstract and the whole text, data retrieval, and data quality assessment were conducted by two researchers (PW Ding and PY Chen) independently. Key information was extracted into the baseline table. Differences that arose during the retrieval process were unraveled in reference with a third researcher. Extracted basic information contains: first author, year of publication, publication country, sample size, histological type, sampling method, EpCAM detection assay, cut-off value, follow-up time, HR estimation method, and HRs and 95% CIs. Patient characteristics included age, gender, histological grade, TNM stage, the size of tumor, T classification, lymphatic nodes metastasis, vascular invasion, and distant metastasis.
The HRs and 95% CIs of OS and DFS were obtained directly from the articles, when available. For studies that did not present HRs and 95% CIs, Kaplan-Meier survival curves (K-M curves) were used to estimate the results. The Newcastle-Ottawa Scale (NOS) was aimed to assess the qualities of included studies (13). An NOS score of 5 or higher indicated a high quality. Otherwise, the study was defined as low-quality. All processes were in observance of the PRISMA (Preferred Reporting Item for Systems Evaluation and Meta-Analysis) guidelines (14).
2.3 Statistical methods
Predictive capability of EpCAM overexpression for the prognosis of patients with solid tumors were appraised by the HRs and 95% CIs. Engauge Digitizer version 11.1 was applied to extract survival data from K-M curves, and STATA version 12.1 (STATA Corporation, College Station, TX, USA) was used for data processing of the meta-analysis.
When the heterogeneity in the combined studies was significant (I2> 60%), a random effects model was selected; otherwise, a fixed effect model was used. Furthermore,the sources of heterogeneity were determined by subgroup analyses. When estimating HR values, multivariate analyses adjusting for other prognostic factors were prioritized; otherwise, data from univariate analyses were used. K-M curves were used for the calculation of HRs (15, 16). Multivariate HR could better demonstrate the independent effect of high EpCAM expression in predicting the prognosis of patients with solid tumors. If the prognosis of patients with solid tumors with EpCAM over expression is poor, the combined HR should be more than 1.0, and its 95% CI should not overlap 1.0. Pooled odds ratios (ORs) were used to evaluate the relationship between clinicopathological characteristics and EpCAM positive expression. The continual deletion of individual studies was used to conduct a sensitivity analysis (17). To evaluate publication bias, a funnel plot, a Begg’s funnel plot, and the Egger test were utilized.
3 Results
3.1 Study inclusion and characteristics
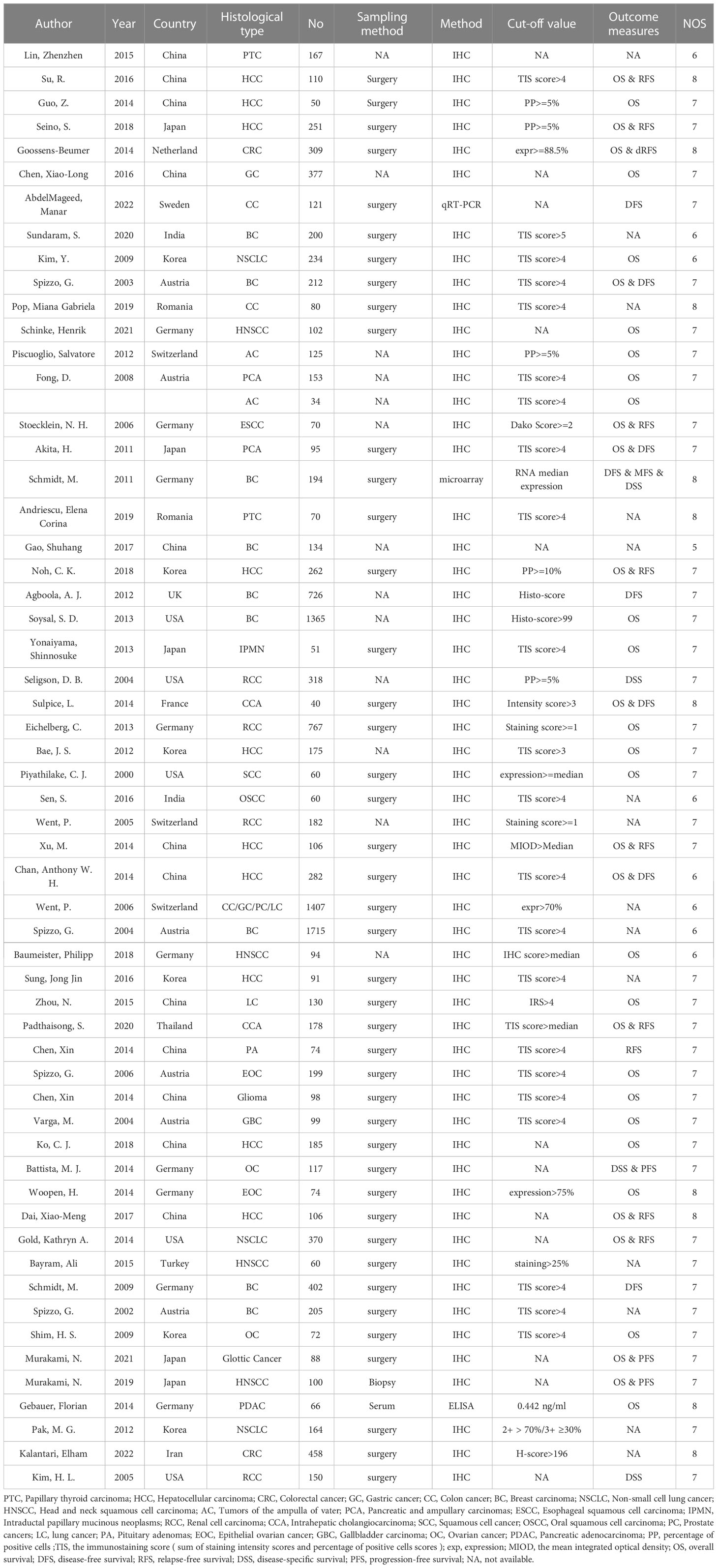
In total, 3747 studies in PubMed, EMBASE, Web of Science, Cochrane library and PMC databases were initially retrieved. After excluding 1069 duplications, 2678 studies were retained. Then, by sifting the titles and abstracts, 2509 studies were excluded. Of 169 full-text records evaluated in detail, 57 were in accordance with the exclusion and inclusion criteria and were selected for the final review (18–75). A flow diagram of researches selection is illustrated in Figure 1. The publication years ranged from 2000–2022. A total of 14184 cases were recruited in the selected studies. In detail, 12 out of 57 studies were in China, nine were in Germany, six each were in South Korea and Australia, five were in both Japan and the USA, and the others were in France, India, Iran, Netherlands, Romania, Sweden, Switzerland, Thailand, Turkey, the UK, and other countries. Additionally, 10 of 57 reports were focused on hepatocellular cancer, nine on breast cancer, four on head and neck squamous cell cancer, four on renal cell cancer, and the rest on lung, ampullary, colorectal, thyroid, cervical, pancreatic, and ovarian cancers (Table 1). There were 13 articles reporting only pathological features and 44 articles reporting both pathological features and prognostic results, including 36 articles targeting OS and 21 articles targeting DFS, RFS, or progression-free survival. Immunohistochemistry was a principal approach for detecting EpCAM, except for three articles using microarray, qRT-PCR, and ELISA as detection methods. Of note, in most articles, the total immunostaining score (TIS), which combines the proportion of staining with staining intensity, was used to divide the expression of EpCAM into high or low levels. Other articles used the positive percentage (pp), median expression, or a single factor, such as the intensity or the staining score as the cut-off value. All included articles were of high quality, with NOS scores of ranging five to eight.
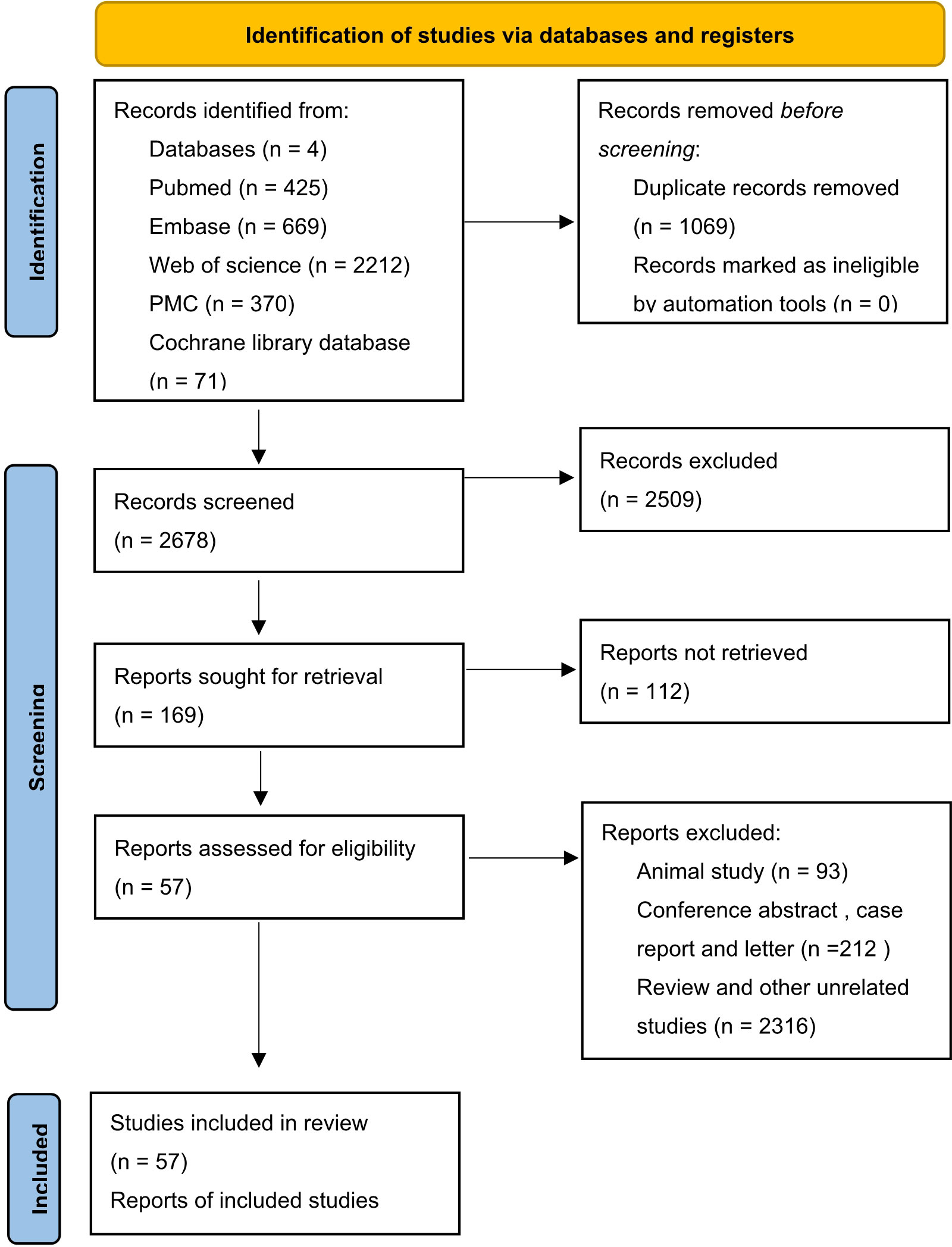
Figure 1 Flow chart of study identification process (14).
3.2 Association between EpCAM and clinicopathological features
3.2.1 EpCAM overexpression and OS
Since there was obvious heterogeneity (76.6%; P < 0.01), the HRs and 95% CIs were evaluated by a random effects model. As shown in Figure 2, high EpCAM expression was significantly associated with a worse OS (HR: 1.30, 95% CI: 1.08–1.58, P < 0.01). In gastrointestinal tumors (HR: 1.50, 95% CI: 1.15–1.95, P < 0.01), the overexpression of EpCAM was significantly related to a worse OS. In thoracic tumors (HR 1.33, 95% CI: 0.93–1.90, P = 0.16) and head and neck tumors (HR: 1.11, 95% CI: 0.58–2.14, P = 0.752), the relationship was not significant. However, in urogenital tumors (HR = 0.70, 95% CI: 0.55–0.89, P = 0.22), the opposite relationship was detected.
3.2.2 EpCAM overexpression and DFS
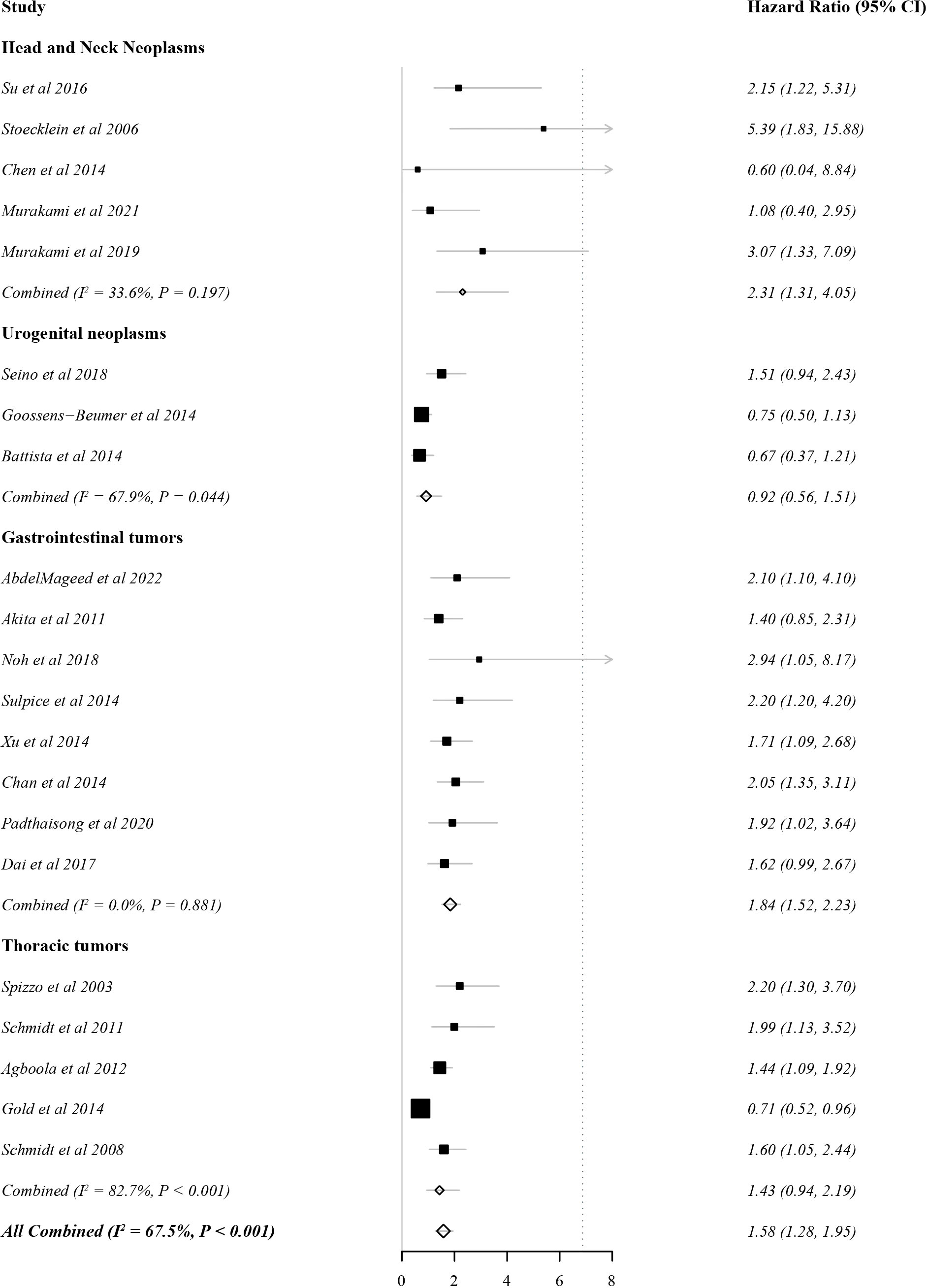
In total, 21 studies evaluated DFS as the outcome indicator. Because heterogeneity was 67.5% (>60%), the HR and 95% CI were analysed by a random effects model. There was a significant correlation between EpCAM expression and a worse DFS (Figure 3, HR: 1.58, 95% CI: 1.28–1.95, P < 0.01). In gastrointestinal tumors (HR: 1.84, 95% CI: 1.52–2.33, P < 0.01) and head and neck tumors (HR: 2.33, 95% CI: 1.51–3.61, P < 0.01), the overexpression of EpCAM was significantly associated with a worse DFS. However, this relationship was unclear in thoracic tumors (HR 1.43, 95% CI: 0.94–2.19, P = 0.10) and urogenital tumors (HR 0.92, 95% CI: 0.56–1.51, P = 0.74).
3.3 EpCAM overexpression: Pathological and clinical characteristics
EpCAM overexpression was not significantly related to sex (RR: 1.03, CI: 0.99–1.07, P = 0.141), T classification (RR: 0.93, CI: 0.82–1.06, P = 0.293), lymph node metastasis (RR: 0.85, CI: 0.54–1.32, P = 0.461), distant metastasis (RR: 0.97, CI: 0.84–1.10, P = 0.606), vascular infiltration (RR: 1.05, CI: 0.85–1.29, P = 0.611), and TNM stage (RR: 0.93, CI: 0.83–1.04, P = 0.187). However, it was significantly correlated with the histological grade (RR: 0.88, CI: 0.80–0.97, P < 0.01). The detailed information is listed in Table 2. Owing to a lack of data, the relationships between the overexpression of EpCAM and other pathological features were not explored.
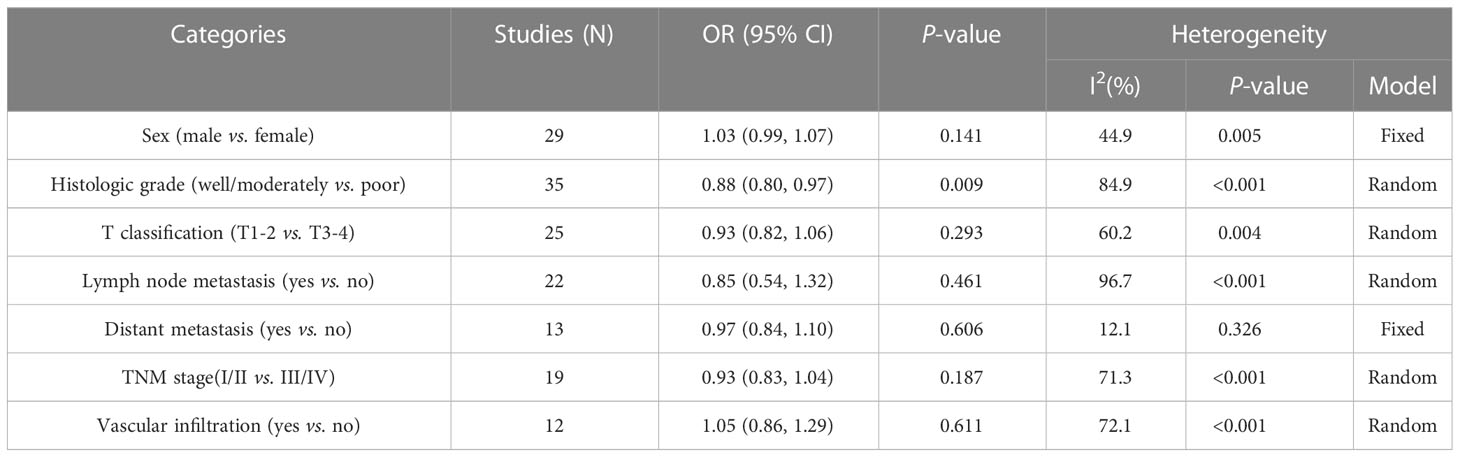
Table 2 Results of the associations of high EpCAM expression with multiple clinicopathological parameters.
3.4 Sensitivity analysis and publication bias
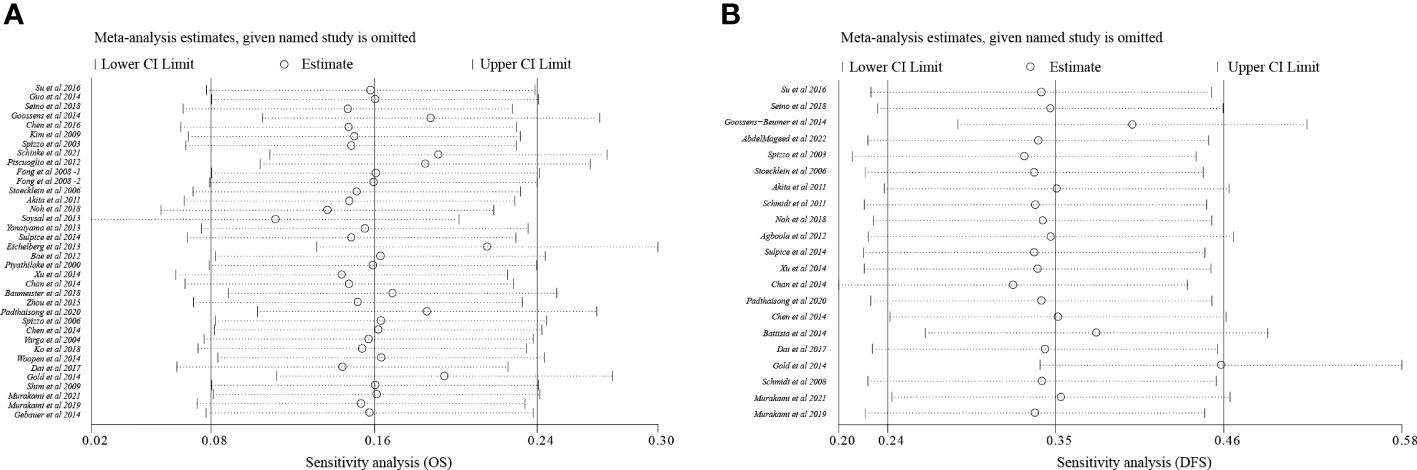
To analyze sensitivity, each study was successively deleted, revealing that the statistical result of relationships between EpCAM overexpression and survival periods were affected by no individual study. These outcomes attested to the validity of the meta-analysis (Figure 4).
Egger’s test, Begg’s test, and funnel plots were performed to make evaluation of publication bias. As illustrated in Figures 4, 5, significant publication bias was not detected for OS (P = 0.84) and DFS (P = 0.10) by Begg’s tests. By Egger’s test, no obvious publication bias for OS (P = 0.25) was found; however, significant publication bias was detected for DFS (P = 0.04). This publication bias might be due to the smaller sample size for DFS than for OS and unpublished research that has not been included also was contributing.
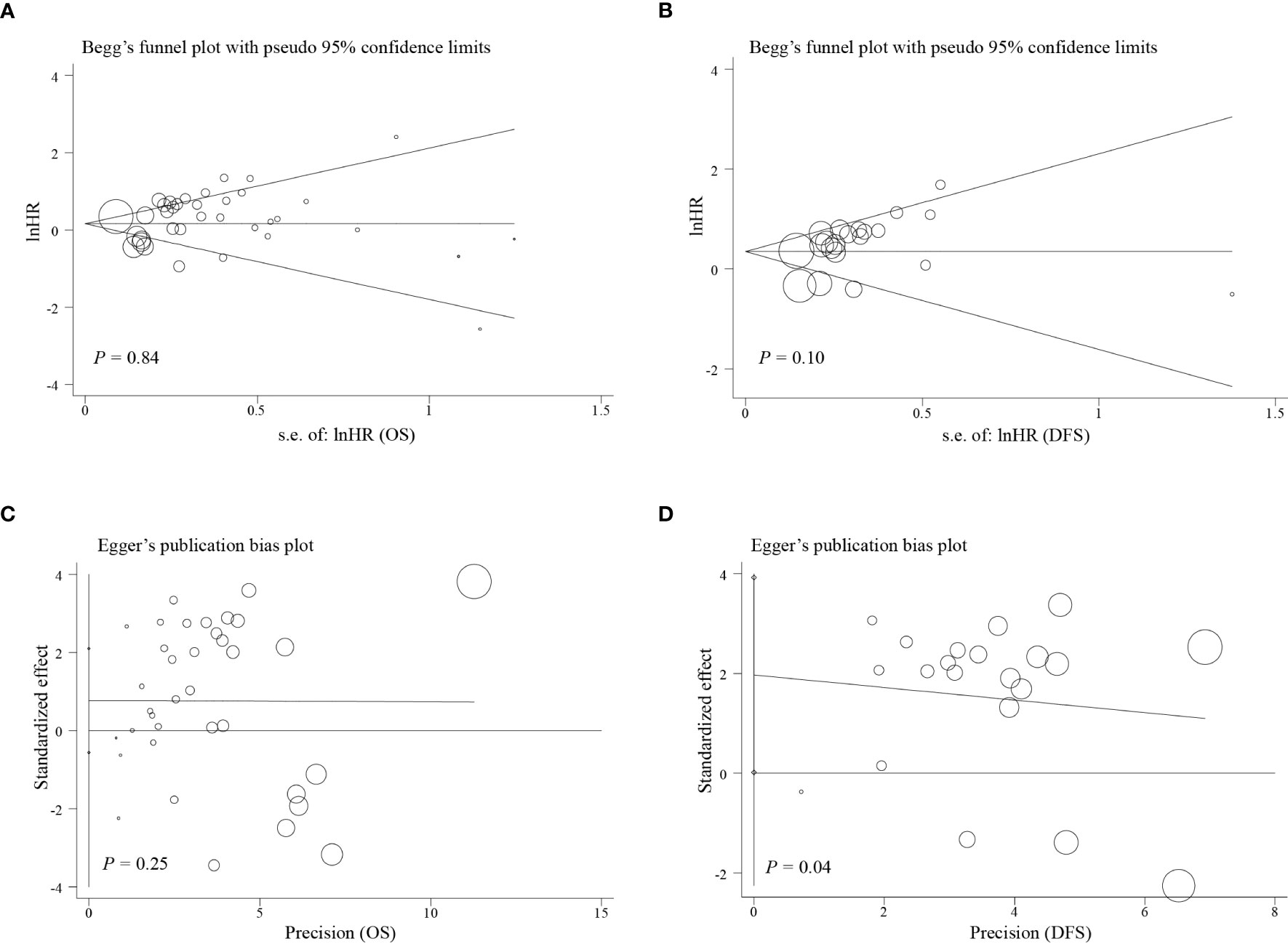
Figure 5 Funnel plot of the analysis on survival. (A) Begg’s funnel plot of the outcome of OS; (B) Begg’s funnel plot of the outcome of DFS; (C) Egger’s publication bias plot of outcome of OS; (D) Egger’s publication bias plot of the outcome of DFS.
4 Discussion
EpCAM, one of the first CSC biomarkers, was first discovered in 1979 as a colorectal cancer secretory antigen recognized by humoral immunity (5). Based on its prominent role in adhesion structure and polarity, EpCAM was considered a cell adhesion molecule initially (76). However, the complexities of EpCAM functions have recently been determined. It is now recognized that EpCAM does not play a significant role in cell adhesion and migration (77) and affects downstream pathways by inhibiting nPKCs (78, 79).
Hematological neoplasms were not included in this study, given the differences in growth and metastasis and the lack of studies focused on hematological neoplasms with EpCAM as a prognostic marker. This study indicated that the overexpression of EpCAM in solid tumors suggests a worse OS and DFS. In a subgroup analysis, high EpCAM expression in gastrointestinal tumors was related to a worse OS, while the opposite relationship was obtained for urogenital tumors. High EpCAM expression levels in gastrointestinal tumors and head and neck tumors suggest a worse DFS. In digestive system tumors, EpCAM was osculate related to a poor prognosis. Elevated EpCAM expression in solid tumors suggests a worse degree of cancer cell differentiation. Previously, it has reported that EpCAM is a predictor of tumor metastasis (80); however, this was not supported by the discoveries of current studies. In the following discussion, we discuss a few key issues relate to our results.
EpCAM, as a humoral immune antigen found in carcinoma of colon cells, is of significantly close relationship to congenital tufting enteropathy, inflammatory bowel disease, and cholestatic liver injury, in addition to cancer (81–84). It has also been reported that EpCAM plays a certain role in the differentiation and regeneration of hepatobiliary cells (85). EpCAM regulates intestinal epithelial homeostasis via various signaling pathways, including ROCK and nPKCs (86, 87). Although there is no laboratory evidence to prove that EpCAM is involved in gastrointestinal cancer, there is some evidence supporting this relationship. EpCAM and claudin-7’s interaction may be answerable to the growth of tumors in colorectal cancer. (88, 89).
It has been claimed that the overexpression of EpCAM can inhibit the migration of ovarian cancer cells stimulated by EGF (90). Direct evidence for the relationship between EpCAM expression and urological tumors, such as renal cell carcinoma, has not been reported. The clinical significance of the overexpression of EpCAM may differ between urogenital tumors and gastrointestinal tumors. Based on the function of EpCAM in separating the mesoderm and endoderm and guiding endoderm differentiation (91), the digestive system generally originates from the endoderm and the genitourinary system primarily originates from the mesoderm; cells of the two embryonic layers have various differences, and these differences may explain why EpCAM has opposite effects in the two distinct cancers. However, comparative analyses of the effects of EpCAM in different embryonic cells are lacking; accordingly, further laboratory research is needed to resolve this issue.
Second, EpCAM was upregulated in undifferentiated P19 cells of mouse embryonic cancer (92), but downregulated in differentiated ones. EpCAM is crucial for preserving the pluripotency of embryonic stem cells. EpICD of EpCAM supports pluripotency by activating the transcription of reprogramming factors (93). However, its mechanism of action in somatic stem cells is unclear (94). EpCAM affected somatic reprogramming by related pathways or by forming complexes with other molecules. For example, EpCAM as well as claudin-7 complexes are essential for somatic reprogramming in both mice and humans. To improve pluripotent reprogramming, EpCAM complexes may promote Oct4 transcription while blocking the p53 and p21 pathways (95). EpEX/EpCAM may also lead to the nuclear translocation of hypoxia inducible factor 1a, via stimulating signal transducer and activator of transcription 3 (STAT3), thus enabling somatic reprogramming. To synthesize human induced pluripotent stem cells, EpEX/EpCAM when combined with Oct3/4 or Kruppel-like factor 4 is adequate (96). Since EpCAM serves as crucial for the maintenance of cell pluripotency, it may be overexpressed in cancer cells with low differentiation.
Finally, as EpCAM was initially recognized as a cell adhesion molecule, the molecule was expected to influence cell adhesion, migration, and other functions (86). However, it showed no significant effects on lymph node and distal metastasis in this study,. Early studies of EpCAM suggested that it could be considered a homophilic cell adhesion molecule because its ectopic expression in mouse fibroblasts and mouse breast cancer cells induced cell aggregation and separation and reduced invasive growth (97). Subsequent studies found that EpCAM disrupted the combination of E-cadherin and cytoskeleton and inhibited E-cadherin-mediated cell aggregation (98, 99). It is also possible that EpCAM is an antagonist of E-cadherin. These contradictory opposite make the functions of EpCAM in terms of adhesion unclear. It had also been pointed out that the adhesion and migration functions of EpCAM are not related to its adhesion functions. Instead, it can inhibit myosin activity by regulating nonclassical nPKCs to produce downstream cascade reactions, thereby avoiding the excessive activation of myosin, leading to unstable adhesion contact and a loss of calcium mucoprotein (86). In terms of migration, there are conflicting results for EpCAM and EMT (90, 91). The view that EMT is critical for cell migration and invasion has been repeatedly challenged in recent years. Current research suggests a dynamic collective invasion mode (100, 101), in which EpCAM plays a complex role. As a single index, EpCAM expression is not a powerful tool to predict tumor progression, recurrence, and metastasis. More research is needed to characterize the multifaceted roles of EpCAM.
This study had the following limitations. First, the object of the study was pan-solid tumors, and there was substantial heterogeneity. Second, the impact of radiotherapy and chemotherapy after surgery or biopsy on survival was not considered, and this is another source of heterogeneity. Third, in addition to differences in detection methods, the cut-off values varied. Most studies adopted a TIS score of >4 as the standard, while others adopted PP ≥ 5%, expression ≥ 88.5%, Dako Score ≥ 2, and other standards. In addition, due to the lack of information or inconsistent classification criteria in the literature, variables such as age and racial pathological type were not able to be combined and analyzed. In each case,heterogeneity was detected. Furthermore, a large part of the studies were retrospective, and more prospective studies are required to establish the causal relationship between EpCAM and prognostic indicators.
To overcome drug resistance in radiotherapy and chemotherapy as well as recurrence and metastasis, destroying CSCs, while shrinking the tumor has become an important strategy (102). They are a key area of tumor research and an important target for future cancer treatment (7). Researches on CSCs and their biomarkers is highly significant for the advancement of precision medicine. Precision medicine refers to treatments that differentiating a particular patient from other individuals exhibiting similar clinical manifestations according to genetic, biomarker, phenotype, or psychosocial characteristics, aimed at the needs of individual patients (103). The development of relevant therapies based on biomarkers targeting CSCs is a promising strategy for precise medical treatment and for reducing radiochemotherapy resistance, recurrence, and metastasis in patients with tumors.
Our results clearly demonstrated that the overexpression of EpCAM is an unfavorable prognostic indicator of OS and DFS in solid tumors, especially in gastrointestinal tumors. And EpCAM overexpression was related to the clinicopathological characteristics of solid tumors, particularly worse differentiation. EpCAM,with complex biological characteristics, serves as a promising candidate molecule for solid tumor detection and therapy. Further experimental and clinical researches are expected to reveal the mechanism by which EpCAM is conducive to the occurrence and development of solid tumors and to apply the biological characteristics of EpCAM to diagnosis and treatment.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
Research conception and design: PD, PC, JO, QL, and SL. Acquisition of data: PD, PC, and JO. Analysis of data: PD and PC. Drafting of the manuscript: PD and PC. Critical revision of the manuscript for important intellectual content: QL and SL. Statistical analysis: PD and PC. Supervision: QL and SL. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer (2021) 127:3029–30. doi: 10.1002/cncr.33587
3. Litvinov SV, Velders MP, Bakker HA, Fleuren GJ, Warnaar SO. Ep-CAM: A human epithelial antigen is a homophilic cell-cell adhesion molecule. J Cell Biol (1994) 125:437–46. doi: 10.1083/jcb.125.2.437
4. Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res (2009) 69:5627–9. doi: 10.1158/0008-5472.Can-09-0654
5. Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc Natl Acad Sci USA (1979) 76:1438–42. doi: 10.1073/pnas.76.3.1438
6. Ingangi V, Minopoli M, Ragone C, Motti ML, Carriero MV. Role of microenvironment on the fate of disseminating cancer stem cells. Front Oncol (2019) 9:82. doi: 10.3389/fonc.2019.00082
7. Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer (2005) 5:899–904. doi: 10.1038/nrc1740
8. Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int (2016) 2016:1740936. doi: 10.1155/2016/1740936
9. Dai M, Yuan F, Fu C, Shen G, Hu S, Shen G. Relationship between epithelial cell adhesion molecule (EpCAM) overexpression and gastric cancer patients: A systematic review and meta-analysis. PloS One (2017) 12:e0175357. doi: 10.1371/journal.pone.0175357
10. Zhou L, Zhu Y. The EpCAM overexpression is associated with clinicopathological significance and prognosis in hepatocellular carcinoma patients: A systematic review and meta-analysis. Int J Surg (2018) 56:274–80. doi: 10.1016/j.ijsu.2018.06.025
11. Hu Y, Wu Q, Gao J, Zhang Y, Wang Y. A meta-analysis and The Cancer Genome Atlas data of prostate cancer risk and prognosis using epithelial cell adhesion molecule (EpCAM) expression. BMC Urol (2019) 19:67. doi: 10.1186/s12894-019-0499-8
12. Han S, Zong S, Shi Q, Li H, Liu S, Yang W, et al. Is ep-CAM expression a diagnostic and prognostic biomarker for colorectal cancer? A systematic meta-analysis. EBioMed (2017) 20:61–9. doi: 10.1016/j.ebiom.2017.05.025
13. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
14. Page MJ, Mckenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ (2021) 372:n71. doi: 10.1136/bmj.n71
15. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med (1998) 17:2815–34. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8
16. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16
17. Kicinski M, Springate DA, Kontopantelis E. Publication bias in meta-analyses from the Cochrane Database of Systematic Reviews. Stat Med (2015) 34:2781–93. doi: 10.1002/sim.6525
18. Piyathilake CJ, Frost AR, Weiss H, Manne U, Heimburger DC, Grizzle WE. The expression of Ep-CAM (17-1A) in squamous cell cancers of the lung. Hum Pathol (2000) 31:482–7. doi: 10.1053/hp.2000.6711
19. Spizzo G, Obrist P, Ensinger C, Theurl I, Dünser M, Ramoni A, et al. Prognostic significance of Ep-CAM AND Her-2/neu overexpression in invasive breast cancer. Int J Cancer (2002) 98:883–8. doi: 10.1002/ijc.10270
20. Spizzo G, Gastl G, Wolf D, Gunsilius E, Steurer M, Fong D, et al. Correlation of COX-2 and Ep-CAM overexpression in human invasive breast cancer and its impact on survival. Br J Cancer (2003) 88:574–8. doi: 10.1038/sj.bjc.6600741
21. Seligson DB, Pantuck AJ, Liu X, Huang Y, Horvath S, Bui MH, et al. Epithelial cell adhesion molecule (KSA) expression: pathobiology and its role as an independent predictor of survival in renal cell carcinoma. Clin Cancer Res (2004) 10:2659–69. doi: 10.1158/1078-0432.ccr-1132-03
22. Spizzo G, Went P, Dirnhofer S, Obrist P, Simon R, Spichtin H, et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res Treat (2004) 86:207–13. doi: 10.1023/b:Brea.0000036787.59816.01
23. Varga M, Obrist P, Schneeberger S, Mühlmann G, Felgel-Farnholz C, Fong D, et al. Overexpression of epithelial cell adhesion molecule antigen in gallbladder carcinoma is an independent marker for poor survival. Clin Cancer Res (2004) 10:3131–6. doi: 10.1158/1078-0432.ccr-03-0528
24. Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, et al. Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. J Urol (2005) 173:1496–501. doi: 10.1097/01.ju.0000154351.37249.f0
25. Went P, Dirnhofer S, Salvisberg T, Amin MB, Lim SD, Diener PA, et al. Expression of epithelial cell adhesion molecule (EpCam) in renal epithelial tumors. Am J Surg Pathol (2005) 29:83–8. doi: 10.1097/01.pas.0000.146028.70868.7a
26. Spizzo G, Went P, Dirnhofer S, Obrist P, Moch H, Baeuerle PA, et al. Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an independent prognostic marker for reduced survival of patients with epithelial ovarian cancer. Gynecol Oncol (2006) 103:483–8. doi: 10.1016/j.ygyno.2006.03.035
27. Stoecklein NH, Siegmund A, Scheunemann P, Luebke AM, Erbersdobler A, Verde PE, et al. Ep-CAM expression in squamous cell carcinoma of the esophagus: a potential therapeutic target and prognostic marker. BMC Cancer (2006) 6:165. doi: 10.1186/1471-2407-6-165
28. Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer (2006) 94:128–35. doi: 10.1038/sj.bjc.6602924
29. Fong D, Steurer M, Obrist P, Barbieri V, Margreiter R, Amberger A, et al. Ep-CAM expression in pancreatic and ampullary carcinomas: frequency and prognostic relevance. J Clin Pathol (2008) 61:31–5. doi: 10.1136/jcp.2006.037333
30. Schmidt M, Hasenclever D, Schaeffer M, Boehm D, Cotarelo C, Steiner E, et al. Prognostic effect of epithelial cell adhesion molecule overexpression in untreated node-negative breast cancer. Clin Cancer Res (2008) 14:5849–55. doi: 10.1158/1078-0432.Ccr-08-0669
31. Kim Y, Kim HS, Cui ZY, Lee HS, Ahn JS, Park CK, et al. Clinicopathological implications of EpCAM expression in adenocarcinoma of the lung. Anticancer Res (2009) 29:1817–22.
32. Shim HS, Yoon BS, Cho NH. Prognostic significance of paired epithelial cell adhesion molecule and E-cadherin in ovarian serous carcinoma. Hum Pathol (2009) 40:693–8. doi: 10.1016/j.humpath.2008.10.013
33. Akita H, Nagano H, Takeda Y, Eguchi H, Wada H, Kobayashi S, et al. Ep-CAM is a significant prognostic factor in pancreatic cancer patients by suppressing cell activity. Oncogene (2011) 30:3468–76. doi: 10.1038/onc.2011.59
34. Schmidt M, Petry IB, Böhm D, Lebrecht A, Von Törne C, Gebhard S, et al. Ep-CAM RNA expression predicts metastasis-free survival in three cohorts of untreated node-negative breast cancer. Breast Cancer Res Treat (2011) 125:637–46. doi: 10.1007/s10549-010-0856-5
35. Agboola AJ, Paish EC, Rakha EA, Powe DG, Macmillan RD, Ellis IO, et al. EpCAM expression is an indicator of recurrence in basal-like breast cancer. Breast Cancer Res Treat (2012) 133:575–82. doi: 10.1007/s10549-011-1813-7
36. Bae JS, Noh SJ, Jang KY, Park HS, Chung MJ, Park CK, et al. Expression and role of epithelial cell adhesion molecule in dysplastic nodule and hepatocellular carcinoma. Int J Oncol (2012) 41:2150–8. doi: 10.3892/ijo.2012.1631
37. Pak MG, Shin DH, Lee CH, Lee MK. Significance of EpCAM and TROP2 expression in non-small cell lung cancer. World J Surg Oncol (2012) 10:53. doi: 10.1186/1477-7819-10-53
38. Piscuoglio S, Lehmann FS, Zlobec I, Tornillo L, Dietmaier W, Hartmann A, et al. Effect of EpCAM, CD44, CD133 and CD166 expression on patient survival in tumours of the ampulla of Vater. J Clin Pathol (2012) 65:140–5. doi: 10.1136/jclinpath-2011-200043
39. Eichelberg C, Chun FK, Bedke J, Heuer R, Adam M, Moch H, et al. Epithelial cell adhesion molecule is an independent prognostic marker in clear cell renal carcinoma. Int J Cancer (2013) 132:2948–55. doi: 10.1002/ijc.27970
40. Soysal SD, Muenst S, Barbie T, Fleming T, Gao F, Spizzo G, et al. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2(+), basal-like, and HER2 intrinsic subtypes of breast cancer. Br J Cancer (2013) 108:1480–7. doi: 10.1038/bjc.2013.80
41. Yonaiyama S, Toyoki Y, Morohashi S, Sakuraba S, Yoshizawa T, Suzuki T, et al. Epithelial cell adhesion molecule (EpCAM) overexpression is correlated with Malignant potentials of intraductal papillary mucinous neoplasms (IPMNs) of the pancreas. Biomed Res (2013) 34:87–95. doi: 10.2220/biomedres.34.87
42. Battista MJ, Cotarelo C, Jakobi S, Steetskamp J, Makris G, Sicking I, et al. Overexpression of epithelial cell adhesion molecule protein is associated with favorable prognosis in an unselected cohort of ovarian cancer patients. J Cancer Res Clin Oncol (2014) 140:1097–102. doi: 10.1007/s00432-014-1672-9
43. Chan AW, Tong JH, Chan SL, Lai PB, To KF. Expression of stemness markers (CD133 and EpCAM) in prognostication of hepatocellular carcinoma. Histopathology (2014) 64:935–50. doi: 10.1111/his.12342
44. Gebauer F, Struck L, Tachezy M, Vashist Y, Wicklein D, Schumacher U, et al. Serum EpCAM expression in pancreatic cancer. Anticancer Res (2014) 34:4741–6.
45. Gold KA, Kim ES, Liu DD, Yuan P, Behrens C, Solis LM, et al. Prediction of survival in resected non-small cell lung cancer using a protein expression-based risk model: implications for personalized chemoprevention and therapy. Clin Cancer Res (2014) 20:1946–54. doi: 10.1158/1078-0432.Ccr-13-1959
46. Goossens-Beumer IJ, Zeestraten EC, Benard A, Christen T, Reimers MS, Keijzer R, et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br J Cancer (2014) 110:2935–44. doi: 10.1038/bjc.2014.226
47. Guo Z, Li LQ, Jiang JH, Ou C, Zeng LX, Xiang BD. Cancer stem cell markers correlate with early recurrence and survival in hepatocellular carcinoma. World J Gastroenterol (2014) 20:2098–106. doi: 10.3748/wjg.v20.i8.2098
48. Sulpice L, Rayar M, Turlin B, Boucher E, Bellaud P, Desille M, et al. Epithelial cell adhesion molecule is a prognosis marker for intrahepatic cholangiocarcinoma. J Surg Res (2014) 192:117–23. doi: 10.1016/j.jss.2014.05.017
49. Woopen H, Pietzner K, Richter R, Fotopoulou C, Joens T, Braicu EI, et al. Overexpression of the epithelial cell adhesion molecule is associated with a more favorable prognosis and response to platinum-based chemotherapy in ovarian cancer. J Gynecol Oncol (2014) 25:221–8. doi: 10.3802/jgo.2014.25.3.221
50. Xu M, Qian G, Xie F, Shi C, Yan L, Yu L, et al. Expression of epithelial cell adhesion molecule associated with elevated ductular reactions in hepatocellar carcinoma. Clin Res Hepatol Gastroenterol (2014) 38:699–705. doi: 10.1016/j.clinre.2014.04.015
51. Chen X, Ma WY, Xu SC, Liang Y, Fu YB, Pang B, et al. The overexpression of epithelial cell adhesion molecule (EpCAM) in glioma. J Neurooncol (2014) 119:39–47. doi: 10.1007/s11060-014-1459-5
52. Chen X, Pang B, Liang Y, Xu SC, Xin T, Fan HT, et al. Overexpression of EpCAM and Trop2 in pituitary adenomas. Int J Clin Exp Pathol (2014) 7:7907–14.
53. Ali B, Yüce İ, Çağlı S, Canöz Ö, Güney E. Predictive value of E-cadherin and Ep-CAM in cervical lymph node metastasis of supraglottic larynx carcinoma. Am J Otolaryngol (2015) 36:736–40. doi: 10.1016/j.amjoto.2015.08.006
54. Lin Z, Lu X, Li W, Sun M, Peng M, Yang H, et al. Association of cancer stem cell markers with aggressive tumor features in papillary thyroid carcinoma. Cancer Control (2015) 22:508–14. doi: 10.1177/107327481502200418
55. Zhou N, Wang H, Liu H, Xue H, Lin F, Meng X, et al. MTA1-upregulated EpCAM is associated with metastatic behaviors and poor prognosis in lung cancer. J Exp Clin Cancer Res (2015) 34:157. doi: 10.1186/s13046-015-0263-1
56. Iliaz R, Akyuz U, Tekin D, Serilmez M, Evirgen S, Cavus B, et al. Role of several cytokines and adhesion molecules in the diagnosis and prediction of survival of hepatocellular carcinoma. Arab J Gastroenterol (2016) 17:164–7. doi: 10.1016/j.ajg.2016.10.002
57. Sen S, Carnelio S. Expression of epithelial cell adhesion molecule (EpCAM) in oral squamous cell carcinoma. Histopathology (2016) 68:897–904. doi: 10.1111/his.12870
58. Su R, Nan H, Guo H, Ruan Z, Jiang L, Song Y, et al. Associations of components of PTEN/AKT/mTOR pathway with cancer stem cell markers and prognostic value of these biomarkers in hepatocellular carcinoma. Hepatol Res (2016) 46:1380–91. doi: 10.1111/hepr.12687
59. Sung JJ, Noh SJ, Bae JS, Park HS, Jang KY, Chung MJ, et al. Immunohistochemical expression and clinical significance of suggested stem cell markers in hepatocellular carcinoma. J Pathol Transl Med (2016) 50:52–7. doi: 10.4132/jptm.2015.10.09
60. Chen XL, Chen XZ, Wang YG, He D, Lu ZH, Liu K, et al. Clinical significance of putative markers of cancer stem cells in gastric cancer: A retrospective cohort study. Oncotarget (2016) 7:62049–69. doi: 10.18632/oncotarget.11384
61. Gao S, Sun Y, Liu X, Zhang D, Yang X. EpCAM and COX−2 expression are positively correlated in human breast cancer. Mol Med Rep (2017) 15:3755–60. doi: 10.3892/mmr.2017.6447
62. Dai XM, Huang T, Yang SL, Zheng XM, Chen GG, Zhang T. Peritumoral epCAM is an independent prognostic marker after curative resection of HBV-related hepatocellular carcinoma. Dis Markers (2017) 2017:8495326. doi: 10.1155/2017/8495326
63. Baumeister P, Hollmann A, Kitz J, Afthonidou A, Simon F, Shakhtour J, et al. High expression of epCAM and sox2 is a positive prognosticator of clinical outcome for head and neck carcinoma. Sci Rep (2018) 8:14582. doi: 10.1038/s41598-018-32178-8
64. Ko CJ, Li CJ, Wu MY, Chu PY. Overexpression of epithelial cell adhesion molecule as a predictor of poor outcome in patients with hepatocellular carcinoma. Exp Ther Med (2018) 16:4810–6. doi: 10.3892/etm.2018.6794
65. Noh CK, Wang HJ, Kim CM, Kim J, Yoon SY, Lee GH, et al. EpCAM as a predictive marker of tumor recurrence and survival in patients who underwent surgical resection for hepatocellular carcinoma. Anticancer Res (2018) 38:4101–9. doi: 10.21873/anticanres.12700
66. Seino S, Tsuchiya A, Watanabe Y, Kawata Y, Kojima Y, Ikarashi S, et al. Clinical outcome of hepatocellular carcinoma can be predicted by the expression of hepatic progenitor cell markers and serum tumour markers. Oncotarget (2018) 9:21844–60. doi: 10.18632/oncotarget.25074
67. Andriescu EC, Giuşcă SE, Ciobanu Apostol DG, Lozneanu L, Căruntu ID. EpCAM (MOC-31) - immunohistochemical profile and clinico-pathological correlations in different histological variants of papillary thyroid carcinoma. Rom J Morphol Embryol (2019) 60:429–36.
68. Murakami N, Mori T, Nakamura S, Yoshimoto S, Honma Y, Ueno T, et al. Prognostic value of the expression of epithelial cell adhesion molecules in head and neck squamous cell carcinoma treated by definitive radiotherapy. J Radiat Res (2019) 60:803–11. doi: 10.1093/jrr/rrz053
69. Pop MG, Bartoş DM, Fiţ AM, Vesa ŞC, Bartoş A, Corpădean AG, et al. Detection of epithelial specific cell adhesion molecules in colon cancer and the correlation with clinical and pathological characteristics EpCAM expression in colon cancer. Ann Ital Chir (2019) 90:318–23.
70. Padthaisong S, Thanee M, Namwat N, Phetcharaburanin J, Klanrit P, Khuntikeo N, et al. Overexpression of a panel of cancer stem cell markers enhances the predictive capability of the progression and recurrence in the early stage cholangiocarcinoma. J Transl Med (2020) 18:64. doi: 10.1186/s12967-020-02243-w
71. Sundaram S, Christian SD, Krishnakumar R, Ramya R, Ramadoss M, Karunagaran D. Clinicopathologic implications of epithelial cell adhesion molecule expression across molecular subtypes of breast carcinoma. J Cancer Res Ther (2020) 16:1354–9. doi: 10.4103/jcrt.JCRT_490_20
72. Abdelmageed M, Ismail HTH, Olsson L, Lindmark G, Hammarström ML, Hammarström S, et al. Clinical significance of stem cell biomarkers epCAM, LGR5 and LGR4 mRNA levels in lymph nodes of colon cancer patients. Int J Mol Sci (2021) 23:403. doi: 10.3390/ijms23010403
73. Murakami N, Mori T, Machida R, Kodaira T, Ito Y, Shikama N, et al. Prognostic value of epithelial cell adhesion molecules in T1-2N0M0 glottic cancer. Laryngoscope (2021) 131:1522–7. doi: 10.1002/lary.29348
74. Schinke H, Heider T, Herkommer T, Simon F, Blancke Soares A, Kranz G, et al. Digital scoring of EpCAM and slug expression as prognostic markers in head and neck squamous cell carcinomas. Mol Oncol (2021) 15:1040–53. doi: 10.1002/1878-0261.12886
75. Kalantari E, Taheri T, Fata S, Abolhasani M, Mehrazma M, Madjd Z, et al. Significant co-expression of putative cancer stem cell markers, EpCAM and CD166, correlates with tumor stage and invasive behavior in colorectal cancer. World J Surg Oncol (2022) 20:15. doi: 10.1186/s12957-021-02469-y
76. Huang L, Yang Y, Yang F, Liu S, Zhu Z, Lei Z, et al. Functions of EpCAM in physiological processes and diseases (Review). Int J Mol Med (2018) 42:1771–85. doi: 10.3892/ijmm.2018.3764
77. Tsaktanis T, Kremling H, Pavšič M, Von Stackelberg R, Mack B, Fukumori A, et al. Cleavage and cell adhesion properties of human epithelial cell adhesion molecule (HEPCAM). J Biol Chem (2015) 290:24574–91. doi: 10.1074/jbc.M115.662700
78. Maghzal N, Vogt E, Reintsch W, Fraser JS, Fagotto F. The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J Cell Biol (2010) 191:645–59. doi: 10.1083/jcb.201004074
79. Maghzal N, Kayali HA, Rohani N, Kajava AV, Fagotto F. EpCAM controls actomyosin contractility and cell adhesion by direct inhibition of PKC. Dev Cell (2013) 27:263–77. doi: 10.1016/j.devcel.2013.10.003
80. Gires O, Pan M, Schinke H, Canis M, Baeuerle PA. Expression and function of epithelial cell adhesion molecule EpCAM: where are we after 40 years? Cancer Metastasis Rev (2020) 39:969–87. doi: 10.1007/s10555-020-09898-3
81. Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell (2001) 104:605–17. doi: 10.1016/s0092-8674(01)00246-x
82. Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, et al. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology (2012) 142:305–15. doi: 10.1053/j.gastro.2011.10.025
83. Tanaka H, Takechi M, Kiyonari H, Shioi G, Tamura A, Tsukita S. Intestinal deletion of Claudin-7 enhances paracellular organic solute flux and initiates colonic inflammation in mice. Gut (2015) 64:1529–38. doi: 10.1136/gutjnl-2014-308419
84. Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule. Hepatology (2017) 66:1183–96. doi: 10.1002/hep.29209
85. Dollé L, Theise ND, Schmelzer E, Boulter L, Gires O, Van Grunsven LA. EpCAM and the biology of hepatic stem/progenitor cells. Am J Physiol Gastrointest Liver Physiol (2015) 308:G233–250. doi: 10.1152/ajpgi.00069.2014
86. Fagotto F, Aslemarz A. EpCAM cellular functions in adhesion and migration, and potential impact on invasion: A critical review. Biochim Biophys Acta Rev Cancer (2020) 1874:188436. doi: 10.1016/j.bbcan.2020.188436
87. Ouchi T, Morimura S, Dow LE, Miyoshi H, Udey MC. EpCAM (CD326) regulates intestinal epithelial integrity and stem cells via rho-associated kinase. Cells (2021) 10:256. doi: 10.3390/cells10020256
88. Kuhn S, Koch M, Nübel T, Ladwein M, Antolovic D, Klingbeil P, et al. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res (2007) 5:553–67. doi: 10.1158/1541-7786.Mcr-06-0384
89. Nübel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, et al. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res (2009) 7:285–99. doi: 10.1158/1541-7786.Mcr-08-0200
90. Fan Q, Cheng JC, Qiu X, Chang HM, Leung PC. EpCAM is up-regulated by EGF via ERK1/2 signaling and suppresses human epithelial ovarian cancer cell migration. Biochem Biophys Res Commun (2015) 457:256–61. doi: 10.1016/j.bbrc.2014.12.097
91. Sarrach S, Huang Y, Niedermeyer S, Hachmeister M, Fischer L, Gille S, et al. Spatiotemporal patterning of EpCAM is important for murine embryonic endo- and mesodermal differentiation. Sci Rep (2018) 8:1801. doi: 10.1038/s41598-018-20131-8
92. Shimazaki T, Okazawa H, Fujii H, Ikeda M, Tamai K, Mckay RD, et al. Hybrid cell extinction and re-expression of Oct-3 function correlates with differentiation potential. EMBO J (1993) 12:4489–98. doi: 10.1002/j.1460-2075.1993.tb06138.x
93. Lu TY, Lu RM, Liao MY, Yu J, Chung CH, Kao CF, et al. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem (2010) 285:8719–32. doi: 10.1074/jbc.M109.077081
94. Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development (2009) 136:1951–60. doi: 10.1242/dev.031369
95. Huang HP, Chen PH, Yu CY, Chuang CY, Stone L, Hsiao WC, et al. Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem (2011) 286:33520–32. doi: 10.1074/jbc.M111.256164
96. Kuan Ii, Liang KH, Wang YP, Kuo TW, Meir YJ, Wu SC, et al. EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci Rep (2017) 7:41852. doi: 10.1038/srep41852
97. Litvinov SV, Bakker HA, Gourevitch MM, Velders MP, Warnaar SO. Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes Commun (1994) 2:417–28. doi: 10.3109/15419069409004452
98. Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-De Bruijn IH, Prins F, et al. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol (1997) 139:1337–48. doi: 10.1083/jcb.139.5.1337
99. Winter MJ, Nagelkerken B, Mertens AE, Rees-Bakker HA, Briaire-De Bruijn IH, Litvinov SV. Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp Cell Res (2003) 285:50–8. doi: 10.1016/s0014-4827(02)00045-9
100. Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol (2012) 14:777–83. doi: 10.1038/ncb2548
101. Campbell K, Casanova J. A common framework for EMT and collective cell migration. Development (2016) 143:4291–300. doi: 10.1242/dev.139071
102. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med (2011) 17:313–9. doi: 10.1038/nm.2304
Keywords: EpCAM, solid tumors, prognosis, cancer stem cells, biomarkers, meta-analysis
Citation: Ding P, Chen P, Ouyang J, Li Q and Li S (2023) Clinicopathological and prognostic value of epithelial cell adhesion molecule in solid tumours: a meta-analysis. Front. Oncol. 13:1242231. doi: 10.3389/fonc.2023.1242231
Received: 18 June 2023; Accepted: 27 July 2023;
Published: 16 August 2023.
Edited by:
Ru Wen, Stanford University, United StatesReviewed by:
Wanrun Lin, University of Texas Southwestern Medical Center, United StatesHao Wu, Exelixis Inc., United States
Sijia Yu, The State University of New Jersey, United States
Copyright © 2023 Ding, Chen, Ouyang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijie Li, Y2hpbmFqZWZAMTI2LmNvbQ==; Qiang Li, bGlxaWFuZzUzMzFAMTYzLmNvbQ==
†These authors share first authorship
‡These authors have contributed equally to this work and share last authorship
 Peiwen Ding
Peiwen Ding Panyu Chen3†
Panyu Chen3† Jiqi Ouyang
Jiqi Ouyang