- 1Department of Forensic Science, Guru Ghasidas Vishwavidyalaya, Bilaspur, India
- 2Department of Biotechnology, Guru Ghasidas Vishwavidyalaya, Bilaspur, India
- 3Department of Immunology and Microbiology, University of Missouri, Columbia, SC, United States
- 4Center for Disease Biology and Integrative Medicine, Faculty of Medicine, The University of Tokyo, Bunkyo, Japan
- 5Department of Pharmacy, Guru Ghasidas Vishwavidyalaya, Bilaspur, India
- 6Department of Microbiology, RK University, Rajkot, India
- 7Department of Zoology, Udayana Charya (UR) College, Lalit Narayan Mithila University, Darbhanga, India
- 8Department of Zoology, Veer Kunwar Singh University, Arrah, India
T cells are an important component of adaptive immunity and T-cell-derived lymphomas are very complex due to many functional sub-types and functional elasticity of T-cells. As with other tumors, tissues specific factors are crucial in the development of T-cell lymphomas. In addition to neoplastic cells, T- cell lymphomas consist of a tumor micro-environment composed of normal cells and stroma. Numerous studies established the qualitative and quantitative differences between the tumor microenvironment and normal cell surroundings. Interaction between the various component of the tumor microenvironment is crucial since tumor cells can change the microenvironment and vice versa. In normal T-cell development, T-cells must respond to various stimulants deferentially and during these courses of adaptation. T-cells undergo various metabolic alterations. From the stage of quiescence to attention of fully active form T-cells undergoes various stage in terms of metabolic activity. Predominantly quiescent T-cells have ATP-generating metabolism while during the proliferative stage, their metabolism tilted towards the growth-promoting pathways. In addition to this, a functionally different subset of T-cells requires to activate the different metabolic pathways, and consequently, this regulation of the metabolic pathway control activation and function of T-cells. So, it is obvious that dynamic, and well-regulated metabolic pathways are important for the normal functioning of T-cells and their interaction with the microenvironment. There are various cell signaling mechanisms of metabolism are involved in this regulation and more and more studies have suggested the involvement of additional signaling in the development of the overall metabolic phenotype of T cells. These important signaling mediators include cytokines and hormones. The impact and role of these mediators especially the cytokines on the interplay between T-cell metabolism and the interaction of T-cells with their micro-environments in the context of T-cells lymphomas are discussed in this review article.
1 Introduction
T-cell lymphoma (TCL) is a subclass of non-Hodgkin lymphoma (NHL) (1). The diagnosis and identification of risk factors associated with TCL are challenging due to the lower number of cases. TCLs cover a wide range of conditions that differ in disease progression and clinical symptoms. Broadly, TCLs are classified into peripheral T-cell lymphoma (PTCL) and cutaneous T-cell lymphoma (CTCL) (1) (Table 1). TCLs account for only one-sixth of all NHLs, and most of them are of PTCL type (4). The basis of differentiation between PTCL and CTCL may be phenotypic, diagnostic, or prognostic (5). Over the years, this classification has been updated by the Revised European American Lymphoma (REAL) classification and the European Organization of Research and Treatment of Cancer (EORTC) (6). REAL is a WHO project which recently updated the categorization of tumors of lymphoid and hematopoietic origin (7). This updated classification identifies around 30 different types of PTCLs (7, 8). CTCL lymphomas are present on the skin with an absence of any subcutaneous symptoms at the time of diagnosis (6, 9). Clinically, CTCLs may have a slow or very aggressive mode of prognosis (6, 9, 10). Treatment and diagnosis of TCLs require a multidisciplinary approach inclusive of short-term or long treatment goals (9, 10).
With the recent advancement in the field of metabolomics or lipidomics, the understanding of cancer metabolism has increased. Developing tumors requires high energy and the macromolecule building block to support not only their proliferation but also the adaptation to changing environments during the process of metastasis (11–13). The Warburg effect along with increased glucose uptake is the characteristic metabolic feature of tumor cells (11–13). To fulfill these requirements, the cancer cell metabolism must be reprogrammed (13, 14). The activated T-cell metabolism resembles cancer cell metabolism up on activation through T-cell receptor signaling accompanied by co-stimulation through CD28 induction (15). To sustain this active state, IL-2-mediated signaling is a must (16). Similar to tumor cells, post-activation T cells divide rapidly much faster than any somatic cell (15, 17). Metabolically active T cells reprogram their metabolism to increase the uptake and utilization of glucose accompanied by changes in calcium flux with upregulated lactate production (18, 19). T-cell receptor (TCR)-mediated signaling starts glucose transport upregulation, but to attain the maximum metabolic reprogramming, CD28-mediated signaling must be activated (19, 20). In addition to TCR and costimulatory signaling, cytokine-mediated signaling is of utmost importance for the development of effector T cells, which resembles T-lymphoma cells metabolically (15, 21).
Irrespective of these metabolic similarities, the activated T cells rarely become cancerous, and once the immune challenges are over, the activated T cell either goes to apoptosis or attains the quiescent memory T-cell state (22–24). In the case of TCLs or other lymphomas, instead of going to apoptosis or resting memory cell state, the T lymphocyte remains in the state of hyperactivity due to gain-of-function mutation in the oncogene or loss of function of the tumor-suppressor gene (22, 25). This continuous state of proliferation is associated with altered metabolism in comparison with normal T cells (19, 25). Various reports associated the different genetic alternations in genes related to cytokine signaling with the pathogenesis of TCLs (25–30). Interactions between lymphoma cells and their microenvironment mediated by the altered cytokine profile of the tumor microenvironment (TME) disturb tissue homeostasis and thus favor lymphoma development and progression (31–34). The various aspects of T-cell lymphoma development like metastasis, migration, invasion apoptosis, and epithelial–mesenchymal transition (EMT) are regulated by the altered metabolic phenotype of T lymphoma cells and their abnormal interaction with other non-neoplastic cellular components of the TME (22, 24, 31). As with other solid and hematologic cancers, TCL progenesis depends upon the nature of tumor-infiltrating lymphocytes (TiLs). These TiLs are different from the original lymphoid organ immune cell and are predominantly T lymphocytes. It has been postulated that the interaction between TiLs and T lymphoma cells dictates the subsequent disease progression (35, 36). The TME is ever changing due to the entry of TiLs and subsequent inflammation associated with it (35–37). It has been reported that these TiLs induce inflammation and help in tumor cell survival (35).
For the proper functioning of the immune system, intercellular communication between immune cells is crucial. The important mediator of this immune communication is cytokine, a protein with low molecular weight. Cytokines are mainly secreted by immune cells and stromal cells and have a regulatory role in immune cell activation and subsequent differentiation, proliferation, and apoptosis (38). The role of cytokine in TCL progression is not very clear and depends upon the TME. The effect of cytokine may be either antitumor or malignancy promoting, which depends on the ratio of pro- and anti-inflammatory cytokines along with the level of expression of cytokine receptors present in the TME (39, 40). Since cytokines are a very important cog in the wheel of cellular communication, this review is aiming to discuss their role in the metabolism of T-cell lymphomas and the subsequent development of the tumor microenvironment.
2 Tumor microenvironment of T-cell lymphoma and the role of cytokine and other signaling pathways in cross-talk between its cellular constituents
The tumor cell microenvironment (TME) is a heterogenous body constituted of tumor cells, normal resident cells, infiltrating immune cells, extracellular matrix, and a variety of secreted mediators (31–34, 41, 42). The normal cells of the TME help in tumor promotion, and the intercellular communication inside the TME is mediated through the interwoven complex network involving various cytokines and enzymes, along with a plethora of growth factors (43–47). To understand the cancer disease mechanism, elucidation of this intercellular signaling is of utmost importance. T-cell lymphomas are generally highly proliferative and resistant to apoptosis and accumulate in the nodal and particular extranodal sites (48–51). Despite the general resistance to chemotherapy, culturing T-cell lymphoma cell lines for long is difficult because these cell lines are very prone to spontaneous apoptosis in in vitro conditions (52). These observations established the role of the TME in the pathogenesis of T-cell lymphomas (35–37). The TME is the main source of extrinsic growth, and the survival signal provided by the TME’s non-neoplastic cells including different hematopoietic (myeloid, lymphoid) and non-hematopoietic cells is essential for T-lymphoma survival (35–37) (Figure 1). The source T-cell variant from which the tumorigenic T cell derives significantly dictates the composition of the TME (39, 42, 53, 54). As in the case of AITL (angio-immunoblastic T-cell lymphoma), the overexpression of follicular T helper cells (Tfh) associated with cell surface receptors (ICOS, PD-1, and CXCR-5) along with BCL-6 established that AITL tumorigenic cells originated from Tfh cells. The cytokines (CXCL13, LT-beta, and IL-21) influence the recruitment, functional polarization, and expansion of cells within the TME of AITL (49, 55–57). While B-cell recruitment in the TME is promoted by CXCL-13 and IL-21, recruitment of follicular dendrite cells has been promoted by LT-β (58–60). The unique expression portfolio of different subsets of T-lymphoma cells for the cell surface ligand, cytokines, and chemokines provides uniqueness to its respective TME (25, 37, 61). The uniqueness of the TME of different subtypes of TCL is regulated by unique transcription factors specific for each subset of T cells (36, 61–63).
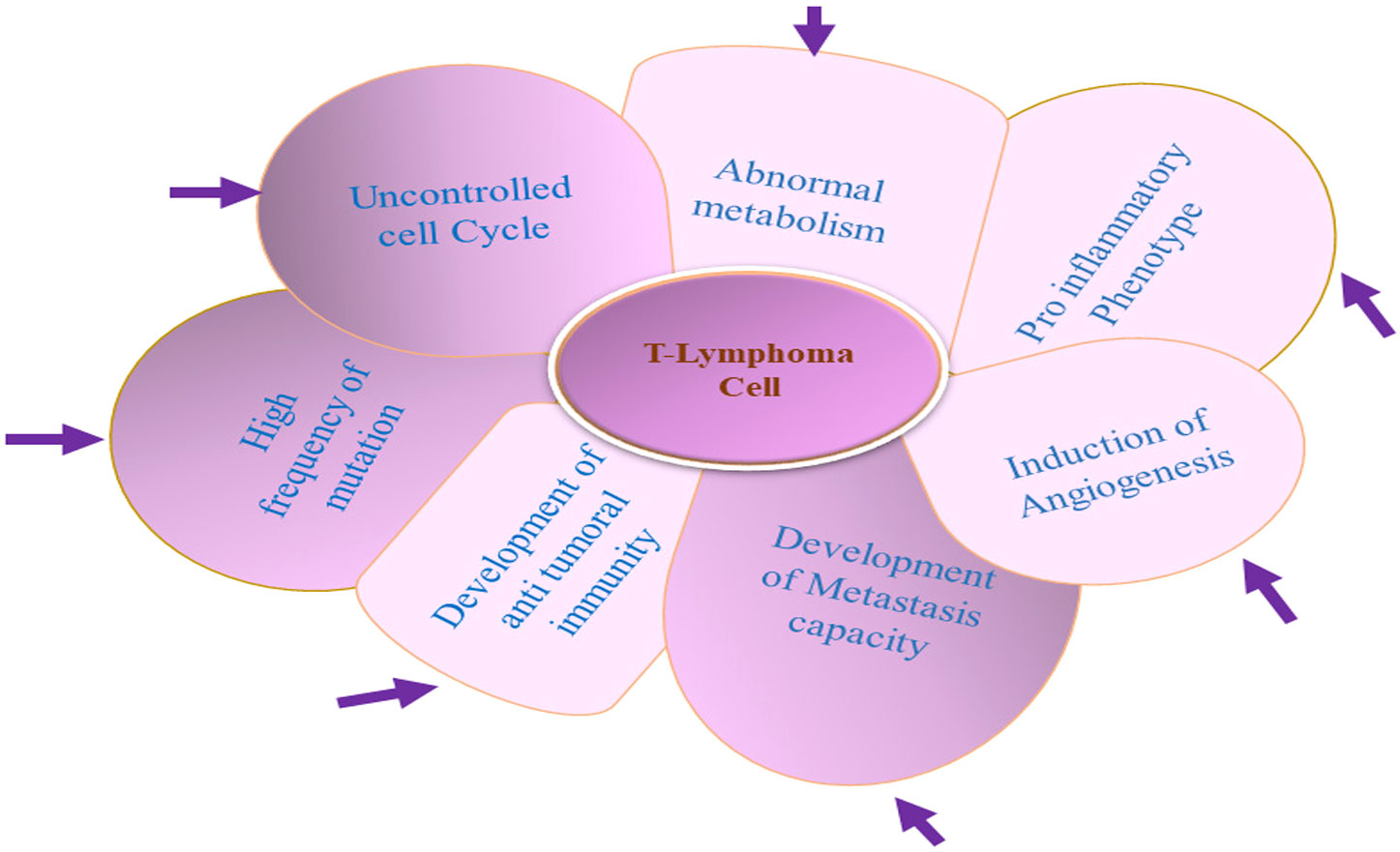
Figure 1 Unusual properties of T-lymphoma cell: T-lymphoma cell’s unusual properties is dictated by an interwoven complex signaling network between T lymphoma cell and other normal constituent cells of the tumor microenvironment (TME).
Recently, it has been found that in non-Tfh-derived T-cell lymphomas like PTCL-NOS, gene regulators like t-bet, GATA-3, and FoxP3 regulate the differential expression of their target genes including those cytokines that regulate the constituents of the TME (64–66). These observations suggested that the cell of origin is a major factor behind the composition of the TME in developing T-cell lymphomas and established the importance of TCR-mediated signaling aided by co-stimulatory and cytokine signaling in transcriptional and posttranscriptional regulations of these master transcription factors (38). These signaling alternations subsequently result in the development of a specific TME, which is evident by the homology between the receptor profile of T-lymphoma cells and the T cells’ source of origin subtypes (53, 54). Subsequently, the constituents of the TME in turn influence the availability of ligands and cytokines which drive the T-cell lymphoma growth proliferation and survival.
2.1 Cytokine and cell signaling in the tumor microenvironment of cutaneous T-cell lymphomas
As discussed in the above section, the nature and composition of the TME depends upon various inter- and intracellular signaling pathways. The TME associated with hematologic tumors vastly differs from the solid tumor microenvironment (67). Mycosis fungoides (MF) is the most frequent type of CTCL, whereas the leukemic variant of CTCL, Sezary syndrome (SS), is the rarest (67). Apart from T-cell transformation progression, MF depends on the interactions between T cells with other cellular components of its TME like antigen-presenting cells (68). It has been established that the interaction between T lymphomas and various components of the microenvironment is regulated by specific cell signaling, and to understand the T-cell abnormalities in CTCL (38), deciphering the mechanism of these signaling pathways is crucial. Alterations in these signaling networks promote malignant T-cell proliferation, survival, T-cell migration, inflammation, and suppression of immune regulation of malignant T cells, resulting in the development of a skin microenvironment that allows disease progression (69, 70). Targeting key proteins like ITK, PI3K, SYK, and mTOR of signaling pathways which are under clinical trials in other disease conditions may open the way for the development of new therapeutic options for CTCL treatment (38). Analyzing the signals responsible for the interaction of T-lymphoma cells with other cells and the extracellular matrix of the microenvironment is important for the identification of new molecular therapeutics molecules (31, 35).
With the help of recently developed high-throughput technologies, various important genetic modulations in cytokine signaling-associated genes have been identified in MF (71–73). The overexpression of downstream signaling components like MAPK of cytokine receptors is due to gain-of-function mutation in B-Raf (p. Asp594Asn) and ERK-1 (p. Glu322Ala, p. Glu322lys) in MF subjects (74). In stage IV subjects of MF, mutations were identified in KRAS (Kristen Rat Sarcoma Viral oncogene homolog) and NRAS (Neuroblastoma RAS Viral oncogene homolog) genes (74–76). Immunohistochemistry studies revealed that in the nucleus of 53% cells of MF lesion, ERK1/2 gets phosphorylated, which has been associated with phospho-4E-BP1 (Eukaryotic Translation Initiation Factor 4E Binding Protein 1) (p-4E-BP1) upregulation. Phospho-4E-BP1 (p-4E-BP1) is an important component of PI3K/Akt downstream signaling (69, 77, 78), and the PI3/Akt pathway is important to support metabolic alternation in the TME. Phosphorylation of Akt, an important component associated with cytokine downstream signaling, is a clear indicator of poor patient survival, and the level of phosphorylated PTEN (Phosphate and TENsin homolog) (inhibitor of PI3K-downstream signaling) is inversely associated with the later clinical phase of MF (69, 78–80). Alternation in PI3K-related genes involved in TCR-CD28 signaling along with deletion in PTEN was found through whole-exome sequencing of MF and SS subjects in comparison with healthy skin tissue (81, 82). Mutation in PTEN regulatory proteins such as p. Arg297Cys in PREX2 has also been reported in MF (69, 83). It has also been reported that upregulation of 14 miR-122 occurred in advanced-stage MF, which has been correlated with a decrease in chemotherapy response via Akt activation along with p53.1 inhibition (84).
It has been reported that mutation altering the gene of MAPK and the PI3K/Akt/mTOR axis is crucial for cell metabolism and rarely results in alternation in the amino acid sequence of the protein (85, 86). Therefore, the reason behind the upregulation in these signaling pathways may be attributed to changes in the TME of CTCL. Activation of T-cell lymphoma may produce ligands that can activate this signaling pathway. It has been reported that IL-2 levels in the TME of CTCL get upregulated, which is inducive for the ERK1/2 pathway (87, 88). The exosomes containing proteins of signaling pathway RAS-MEK secreted in response to T-cell activation may further induce ERK-1/2 activation in a paracrine way (47, 69, 89). In addition to these, triggering other signaling pathways downstream of cytokine receptors like NF-κ and JAK/STAT can result in the activation of MAPK and PI3K/Akt signaling in the TME of CTCL (69, 90–92). It has been established that these pathways are crucial for metabolic alternation to support TCL progression. The ERK-1/2 and PI3K/Akt/mTOR pathways downstream of cytokine signaling cross-talk among themselves and may work together synergistically in the TME of CTCL (38, 69, 92–95). It has been reported that simultaneous induction of nuclear p-ERK, P-Akt, and p-mTOR takes place in 27% of MF lesions (69). Various genetical studies reported that there are changes mainly in the T-cell receptor (TCR) and tumor necrosis factor receptor (TNFR) which have been correlated with the survival and proliferation of tumorous T cells (38). Genetic alternation in these receptors invariably affects TCR-mediated signaling, TCR complex assembly formation, and the level of various proteins such as TCR-associated enzymes, along with various transcription factors. All these are important for the proper execution of downstream signaling mediated through these receptors (8, 26, 26, 38, 50, 76, 83, 96, 97). It has been found that enhanced TCR activation through gain-of-function mutation in the extracellular domain of CD28 and the translocation of CTLA4-CD28 occurred in CTCL (98, 99).
In general, tumor cells bring immune suppression in the TME through the programmed cell death protein 1 (PD-1)-mediated signaling pathway induced by its ligands, programmed death ligands 1 and 2 (PD-L1 and PD-L2) (100). Expression of PD-1 occurs predominantly on activated immune cells like T cells, dendritic cells, natural killer cells, and B cells as well as various types of tumor cells including TCL-expressed PD-1 ligand PD-L1 (82, 101–103). The anti-PD-L1 antibody has shown antitumor response in cancer patients with high levels of PD-L1 expression on tumor-infiltrating immune cells, and with some difference in results, a high expression of PD-1 has been observed in CTCL too (82, 104, 105). This high level of expression of PD-1 is associated with CTCL progression as the level of expression of PD-1 and PD-L1 is higher in an advanced stage in comparison with better-managed CTCL (82, 103). It has been reported that PD-1/PD-L1 inhibition through specific antibodies downregulates tumor progression in mice with enhanced IFN-γ production. The same study reported the objective response rate (ORR) of 15 and 38% in MF when treated with nivolumab in MF and pembrolizumab, respectively (106, 107). It has been reported that transcription factor NF-κ, which is important for TCR-mediated immune response, gets activated in CTCL (108, 109). Various kinds of mutations such as somatic, splice site, heterozygous deletions, and truncations have been reported in genes like NF-κ 2 and CARD11, which are crucial for NF-κ signaling (98, 108, 109). Another possible mechanism for activation of the NF-κ pathway is through tumor necrosis factor receptor 2 (TNFR2, encoded by TNFRSF1B) (38, 110, 111). The ligand for TNFR2 is TNF-α, and in the case of CTCL, point mutation in TNFRSF1B was observed along with amplification of the related gene (38, 111). An inhibitor of TNF-α-induced NF-κ is TNF-α-induced protein 3 (TNFAIP3), which has been downregulated in some SS subjects (69, 112). Kinases like TAK1 are the main regulator of NF-κ, and the level of TAK1 has been reported to be upregulated in CTCL, but inhibition of TAK1 hampers the NF-κ pathway, resulting in the induction of apoptosis in the CTCL cell line in both in vitro and in vivo conditions (69, 113, 114). Toll-like receptors (TLRs) are a class of pattern-recognition receptors comprising more than 10 surface receptors important for pathogen recognition. The signaling mediated through TLRs and subsequent induction of cytokine release are important in various skin pathologies including MF (115, 116). Treatment with TLR9 stimulators such as cytosine-phosphate-guanine (CpG) and oligodeoxynucleotides induces Th1-inducing cytokines, which have been proven to be clinically significant in MF and SS subjects (115, 117, 118). In addition to that, the agonist of TLR7/8 is very useful in clearing MF-associated psoralen and ultraviolet A-resistant plaques (119, 120). In an independent trial with patients of MF, imiquimod, an immunomodulatory agent, had given positive results (121).
UV-induced transition plays an important role in the development of MF, and transitions have been represented by both C>T and CC>>TT mutations (122–124). There are three specific mutations associated with UV exposure that have been reported in MF, and furthermore, the most frequent (41%) exotic mutations are C to T transitions in MF (125–127). This finding indicates a link between exposure to UV light and genetic changes in the MF microenvironment. These exposures seem to be a gain-of-function mutation in proto-oncogenes along with loss of function in tumor suppressors and an unusual expression of cytokines and adhesion proteins (122–127). The C>T transition in the P53 gene is a UV signature mutation (128) that is very crucial for tumor progression and is the most frequent gene mutation associated with CTCL, but this mutation is more frequent in the later stage of cancer than the early tumor stage in MF (125, 126). Mutations in important cell-cycle regulators like RB1 and CDKN1B/CDKN2A are also common in CTCL (69, 129). A sufficient supply of oxygen and nutrient is of utmost importance to sustain the growing mass of the tumor (11, 15, 19, 20). To combat this hypoxic condition, the cells of the TME activate the hypoxia-inducible factor (HIF-1) signaling (130). The expression of HIF-1α on CD4+ T cells along with different cytokine receptors in MF patients has been associated with disease progression (131). The role of HIF-1 in the regulation of the vascular endothelial growth factor-A (VEGF-A) and, subsequently, in angiogenesis is well established (132). In the MF, the level of angiogenesis is directly linked with tumor progression, which provides suitable conditions in the TME of MF for tumor growth (133). It has been observed by analyzing the expression of CD34 that there are an increased number of microvessels in MF in comparison with normal skin tissue (134, 135). The lesioned skin at different stages of MF, viz., IB, IIB, and IVA1, have higher VEGF-A mRNA levels in correlation with CCL27 mRNA levels (136, 137). Another evidence for the correlation between tumor progression and angiogenic markers came from the finding that overexpression of placental growth factor (PlGF), another member of the VEGF family in stages IIB and IVA1 lesional skin of MF, has been linked with CCL27 and IL-4 expression (136). It has been reported that in stage IIB, the serum PlGF levels are upregulated along with serum CCL17, Ang-2, and IL-10 levels (136). Alternation induced in mitochondrial DNA (mtDNA) due to frequent exposure to UVA radiation results in impairment of mitochondrial function and upregulation in metalloproteinase MMP-1 expression (138, 139). In the case of the advanced MF stage, the overexpression of metalloproteinases MMP-2 and MMP-9 occurs in microvascular endothelial cells along with that in stroma fibroblasts (136, 140, 141). Therefore, it is clear that TCL aptly subverts the signaling pathway including the cytokine-mediated pathway to adapt to TME stress conditions and may be a point of focus to avoid tumor development in CTCL.
2.2 Tumor microenvironment in peripheral T-cell lymphomas and cytokine-mediated signaling
The disease pathogenesis of PTCL is quite different from CTL, and it has been noticed that PTCL varieties of TCL can be predicted for their developmental organ site based on special gene expression signatures which are organ-specific (142). This observation indicates the importance of the TME in developing the specific type of tumor (143). As with other tumor cells, the TME of PTCLs constitutes various non-neoplastic cells that are the basis of characterization as in the case of angioimmunoblastic T-cell lymphoma (AITL) in which vascularization patterns and contents of immunoblast are considered as defining features (55, 144). Similarly, in the case of NOS, the follicular variant of PTCLs, the growth pattern of lymphoma cells, and their association with follicular dendritic cells are the distinguished features (145, 146). The lymphoepithelial variants of PTCLs are characterized by the presence of numerous histolytic infiltrates in the TME (142, 147). It has also been observed that the composition and nature of the TME vary from patient to patient in the case of PTCLs (142).
Although the exact nature and interaction between the individual component in the TME of PTCLs are still not well understood, it is clear that the TME influences the tumor progression in the case of PTCLs (55), as in the case of AITL which had been named angioimmunoblastic lymphadenopathy initially but has later been renamed as immunoblastic T-cell lymphomas due to a better understanding of its TME and its intercellular interactions that occur in the TME (148). Initially, it was believed that the reason behind the disease is an abnormal hyperimmune B-cell system but the elucidation of clonal T-cell receptor (TCR) gene rearrangements and identification of clonal cytogenic abnormalities confirmed the neoplastic nature of immunoblastic T-cell lymphoma (49). AITL is unique because in this TCL, neoplastic cells migrate from the lymph node to many different tissues and anatomical sites to develop appropriate conditions to support their growth (149).
Flowcytometry analysis has shown that lymphocytosis is uncommon in AITL, but a variable number of immune cells with abnormal immune-surface phenotype occur and are based on the morphology of these cells in the TME (150). The AITL can be referred to as patterns I, II, and, III. Pattern III with the diffuse proliferation of lymphocytes is more frequent than pattern I (hyperplastic follicle pattern) and pattern II (with depleted follicles) (151, 152). The elucidation of the fact that AITL originated from T-follicular helper (Tfh) cells enhances the understanding of the pathophysiology of AITL (145, 146, 149) (Figure 2). The basis of this is the expression of the Tfh marker specially CXCL13, which has been further validated by a study of genome-wide transcription markers (145). The role of Tfh cells is critical as its interaction with B cells at the germinal center (GC) is essential for B-cell survival, somatic hypermutation, and immunoglobulin class switching (153). These interactions of Tfh cells with B cells at the germinal center ultimately led to the development of effector plasma cells and memory B cells (58, 153, 154). The expression of chemokines CXCL13 and cytokine IL-21 is very crucial for the recruitment of B cells in the germinal center (155). The expression of CXCR5 (receptor of CXCL13) is responsible for Tfh localization in the GC (156). At the cellular level, tumor cells of AITL vary in size (small to medium) and are mature CD4+ and CD8+ T cells (157, 158). Since these AITL cells are derived from Tfh cells, they continue to express various Tfh-associated proteins like BCL6, ICOS, C-MAF, CD200, CXCL13, and SAP (157, 159). In the AITL neoplastic cells, neutral endo-peptidase CD10 is also observed in variable proportions (160). In conclusion, these observations indicated that CD10 and Tfh markers highlighted the source of origin of this tumor cell which resides among non-neoplastic reactive T cells (CD4+ and CD8+) in the TME of AITL (144, 157, 160). It is a fact that in the TME of AITL, the non-neoplastic cells often outnumbered the neoplastic cells, and clinically, dysregulated inflammatory and immune responses are observed in AITL manifestation instead of tumor growth-related complications, which support the theory of para-neoplastic immunological dysfunction (49).
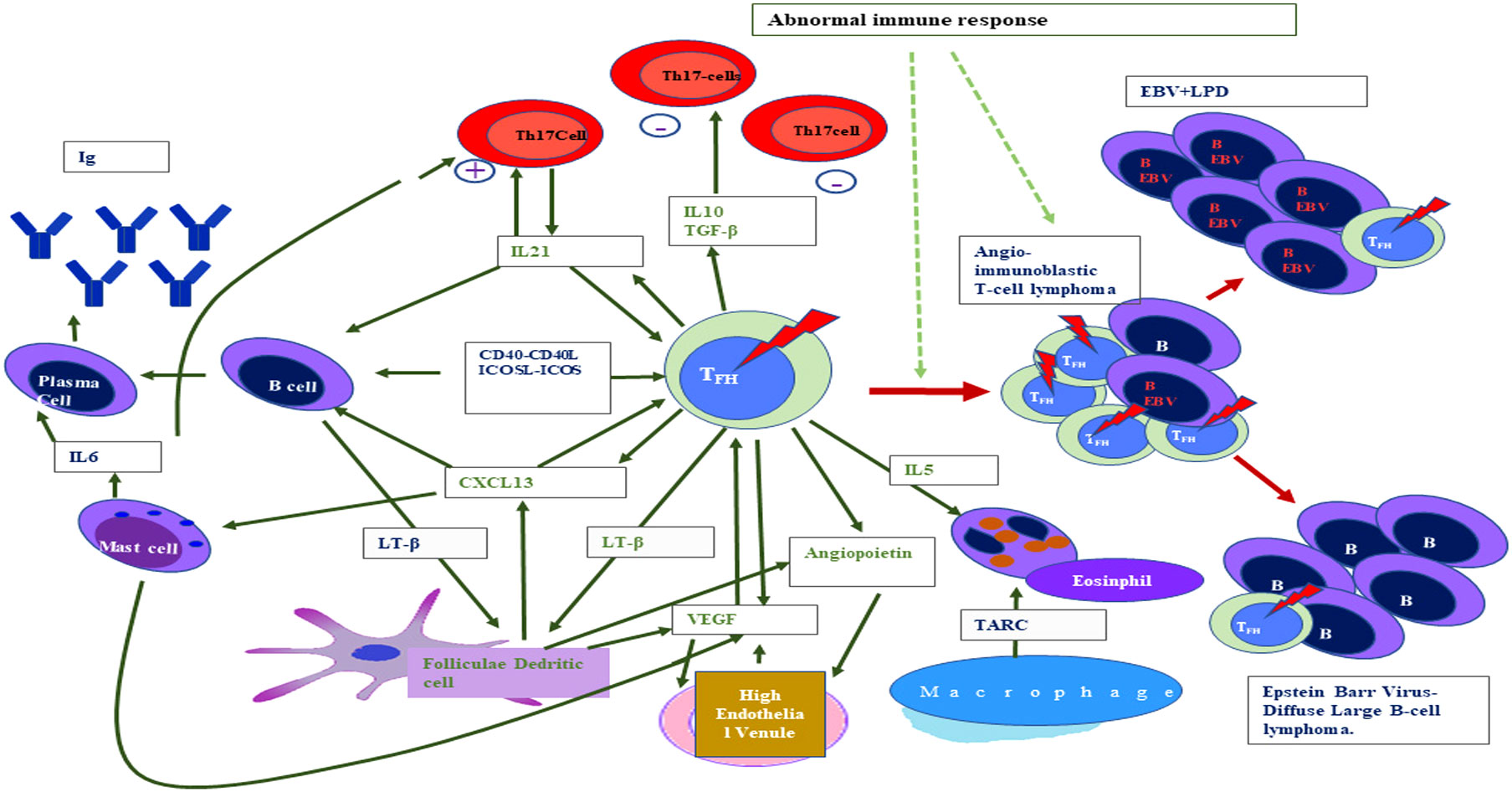
Figure 2 Interaction between different cellular non-neoplastic components of the tumor microenvironment and T-lymphoma cells in the case of angioimmunoblastic T-cell lymphoma (standard abbreviation used).
The molecular profiling of AITL gene expression clearly indicated the influence of the TME, which is evident by overexpression of B cells and follicular dendritic cell (FDC) genes such as various immunoglobulins, clusterins, cytokines, and cytokine receptors along with genes related to the extracellular matrix (161). EBV-infected large B-cell blasts are often found in the TME of the majority of AITL (162). In the TME of AITL, the contact interaction occurs between B cells and Tfh cells and neoplastic T-cell rosettes are generally found around large B-cell blasts (55, 144). In the TME, these interactions occur via the ligand–receptor binding like cytokine binding with its respective receptor on the surface of the interacting cells like CD4-CD40L and ICOS-ICOL (163, 164). The role of Tfh in the TME is very crucial because it secretes different soluble factors such as cytokine which act in an autocrine manner to support TCL (151, 157, 165). CXCL13 is the most crucial immuno-mediator secreted by Tfh which subsequently promotes B-cell expansion along with plasmocytic differentiation, which ultimately leads to abnormal immune manifestations associated with AITL (55, 144–147). This dysimmune function in AITL is also promoted by cytokines released from other cells of the TME (166). For example, IL-6 released from mast cells and neoplastic Tfh cells promote plasmacytosis of IL-21, another soluble factor released from Tfh cells that work in an autocrine fashion and subsequently exert a positive effect on B cells resulting in increased antibody production by plasma cells (58, 150, 151, 153). It has been reported that lymphotoxin released by B cells after induction by CXCL13 is also expressed in AITL tumor cells (167, 168). This lymphotoxin is suspected to be involved in the induction of FDC proliferation, and these FDC of the TME may show an abnormal expression of FDC-specific markers such as CD23 and CD21, but a direct link between abnormal expression of CD21 along with CD-23 with development of FDC neoplasm has not been reported (58, 151, 153, 168). The AITL overexpression of the vascular endothelial growth factor (VEGF) is associated with an increase in the vascularization evident in the disease and immunostaining of neoplastic cells and endothelial cells of the TME, revealing that there is some paracrine or autocrine loop as evident by the expression of both VEGFs to sustain the angiogenesis (49, 157, 158, 161, 163, 166). Since angioprotein-I is also expressed by AITL and FDC, the role of angioprotein-I may also be crucial in increased angiogenesis (157).
Eosinophils are always present in the TME of AITL albeit in differential numbers, as evidenced by blood hypereosinophilia in around 30%–50% of AITL subjects (169). It has also been reported that bone marrow hyperplasia along with eosinophilia is common in AITL (159). The link between eosinophilic infiltrates present in the tumor biopsies and CCL17/TRAC mRNA expression on mononuclear cells present in the TME has been reported (170). It has also been observed that the neoplastic cell of the TME expresses IL-5 but not CCL11/eotoxin1 (171). The CD-3+ T cells isolated from AITL overexpressed IL-4, IL-5, and IL-13 than the normal lymphoma, which may be associated with eosinophilia observed in the AITL subjects (148, 171–173). However, it is still not clear about the role of eosinophilia in the disease pathology of AITL. In an epithelioid variant of AITL, the TME contains macrophages in variable numbers with numerous epithelioid cells in clusters, which creates mistakes in diagnosis and can be misinterpreted as a granulomatous disease (152). The macrophages present in the TME of AITL are of both M1 and M2 phenotypes (2, 174). The role of T-cell mediated response is not clear, but CD8+ T cells are suspected to play an immunosuppressive role; usually, the TME of AITL is immunosuppressive which is evident by the low number of Treg cells together with M2 macrophage expansion and Th17 cell accumulation (142, 148, 152, 175, 176). Cytokines such as IL-10 and TGF- produced by Tfh inhibit T-cell immune response by suppressing the proper functioning of normal CD4+ T helper cells (177, 178). In the TME, the FDC differentiation is induced by TGF-β in an autocrine manner and this immunosuppression in the TME reactivates EBV, which further induces Tfh-cell and B-cell expansion (179, 180). During the disease progression, the TME cytokine milieu of AITL changes in favor of neoplastic cells with a simultaneous decrease in the proportion of non-neoplastic cells (179). An experiment conducted on a mouse model of NOG in which serial transplantation of AITL had been done ended in the subsequent reduction in an immuno-active components like the B cells and CD8+T cells (161). These observations suggested that the TME microenvironment is dynamic and the neoplastic cell growth becomes independent from the influences of the TME with time. In T-cell lymphoma especially in PTCL, the tumor development is dependent upon the supporting environment of the TME and the alternation in cytokine profile of immune cells and the TME is interdependent, as evidenced by the various studies discussed in this section especially in the context of PTCL.
3 Alternation in cell signaling and subsequent reprogramming of T-cell lymphoma metabolism
As reviewed in the previous section, the role of various signaling pathways in the alternation of the TME especially cytokine-mediated signaling is crucial and these alternations result in the hypoxic condition and acidic environment in the TME, which is the hallmark of any cancer (11, 18). To overcome this hostile microenvironment, the TME normalizes oxygen level and ensures adequate nutrient supply by promoting angiogenesis through co-ordination between its various intrinsic and extrinsic factors (33, 34). The upregulation of aerobic glycolysis in cancer cells is one example of an intrinsic factor that resulted in the “Warburg effect”. The “Warburg effect” results in the upregulation of glycolysis along with enhanced lactate production even in inadequate availability of oxygen (11, 13, 15). This increased level of lactate, in turn, alters the extrinsic factors resulting in alteration in the extracellular matrix (ECM) and angiogenesis along with tumor invasion (18). It has been shown that oncogenes like C-Myc, KRAS, and PI3K regulated the uplifting of glycolysis rate by upregulating the expression of various proteins of the glycolytic pathway (32–34).
These oncogenes are also responsible for increased glutaminolysis, a hallmark of tumor metabolism which resulted in both ATP generation and synthesis of precursors for the biosynthesis of other macromolecules such as nucleic acid, proteins, and lipids (11, 18). Previously, due to misinterpretation of the “Warburg effect”, it had been postulated that in cancer cells, the mitochondrial oxidative phosphorylation gets defective but now the importance of the mitochondrial electron transport chain in tumor progression has been accepted as evident by intra-aperitive 13C tracing experiments in human suffering from brain and lung cancer (32, 33, 37, 181). In various cancer types, enzyme mutation is another important intrinsic factor (15). In acute myeloid leukemia, lower-level gliomas and secondary glioblastomas are associated with somatic mutation of isocitrate dehydrogenase-1 and -2 (IDH1 and IDH2), which results in the conversion of alpha-ketoglutarate (αKG) to D-2-hydroxyglutarate (D-2HG), subsequently altering the various alpha-KG-dependent deoxygenase (11, 13, 15, 182). In addition to that, tumor progression is supported by a mutation in either succinate dehydrogenase complex (SDH) or fumarate hydratase (FH) enzymes which have antitumor effects in hereditary tumor syndromes (183–185). Fumarate also triggers cell migration in endometrial cancer (185).
Naive T cells are metabolically similar to other cells and need IL-7 for the maintenance of their basal required metabolism (19). The naïve T cells are smaller in size and generally do not divide. Metabolically, they are highly oxidative and depend completely on the efficient metabolic breakdown of glucose and glutamine (13, 19, 20). In humans, these naive T cells have a comparatively longer life span ranging from months to years. This longer life of T cells is attributed to the continuous induction of IL-7 (22) and alteration in anti-apoptotic protein BCL-2 (B-cell lymphoma-2) which result in the prevention of apoptosis initiation. Upon activation, the metabolic portfolio of a T cell gets a very significant transformation (19, 20, 24). Once they are activated through the TCR and co-stimulatory signal, T cells require continuous IL-2-mediated signaling to maintain their activated state (38). All these signaling mechanisms like TCR-mediated, CD-28-mediated, or IL-2-mediated trigger different pathways to support the upregulated metabolism (19, 24, 38). To support the highly activated and proliferative state, the activated T cells increase the glucose uptake, and upon activation, changes in calcium flux and lactate production occur instantly in these immune cells (19, 24).
Although TCR signaling is needed for initiation of upregulation in glucose transportation, for maximum glucose uptake, CD-28-mediated signaling along with AKT pathway initiation is a must (186, 187). IL-2 produced by activated T cells maintains this upregulation in glucose transformation in an autocrine fashion (188–190). This increased uptake of glucose is accompanied by upregulated oxygen consumption, which is quite similar to the cancer cells (18, 19, 33, 188). Increased glucose uptake results in ATP generation through glycolysis as well as the production of precursors of nucleic acid, lipids, and antioxidants through the pentose phosphate pathway (PPP) (15, 20). These metabolic changes are dependent on cytokine stimulators (191). It has been reported that T regulatory cells are less glucose-dependent metabolically, and rather than this, they depend on fatty acid oxidation to support their metabolism (192, 193). It has been demonstrated that the naive cells activated without glucose do not develop into effectors cells but are easily differentiated into T cells (19). Recently, the role of hypoxia-inducible factor (HIF-1) in the development of Th-17 cells has been documented (194, 195). This underlying difference in metabolism between effector T cells and T-reg cells is very crucial to understanding TCL metabolism in the context of normal cell metabolism (15, 38). JAK-STAT and P13K-AKT downstream of the cytokine signaling pathway of T-cell activation are very crucial to maintaining the growth and proliferation of activated T cells (19, 191, 196). STAT is associated with the IL-2 receptor, and upon IL-2R binding with IL-2, it upregulates some proteins such as BCL-2, C-Myc, and cyclin D (188, 189). The P13K-AKT pathway is associated with the CD-28 co-stimulatory receptor and cytokine receptor in almost all cancer cells including TCL, and this pathway is activated in an inappropriate way (22, 38, 191). Phosphorylated AKT in TCL activates the mammalian target of rapamycin complex-1 (mTORC-1) to bring upregulation in protein translocation (197). The activated mTORC-1 deactivates protein synthesis inhibitor eIF4E binding protein to allow cap-dependent protein synthesis; furthermore, mTORC-I activates S-6 kinase resulting in Sb-ribosomal protein activation (198). The availability of free amino acid is critical to continuing the activity of mTORC-I, and this continued activation of mTORC-I ultimately results in the overrepresentation of glucose transporter-I (Glut-I) along with C-Myc and HIF-I alpha in TCL cells (199). The activation of Akt downstream to CD28 maintains the Glut-I expression on the cell surface and also helps in the upregulation of hexokinases for maximum utilization of glucose by the cell along with the activation of PPP to facilitate the synthesis of other macromolecule precursors (15, 20, 196, 200). This metabolic phenotype of activated T cells is different from other normal cells but similar to the TCL cells (93).
Mitochondrial membrane permeabilization is a crucial step to initiating apoptosis, and Akt activation through cytokine signaling has the ability of hexokinase localization which may hinder mitochondrial membrane permeabilization (201, 202). It has also been reported that activated Akt promotes ubiquitination of tumor-suppressive P53 and subsequent degradation of P53, which results in suppression of synthesis of the proapoptotic P53-upregulated mediator of apoptosis (PUMA) protein (203, 204). A significant decrease in Puma accumulation inside the activated T cell, as well as T lymphoma cell, is associated with upregulation in glucose uptake and activation of Akt (201, 204). The changes in metabolic phenotype brought by upregulation in glucose uptake are also associated with the stability of anti-apoptotic protein myeloid cell leukemia-I inside the activated T cell and T lymphoma cell (205). This observation indicates that Akt and other signaling pathways inside activated T cells may result in anti-apoptotic signaling alternation (93).
As discussed above, the metabolic alternation observed in the activated T cells is very similar to T lymphoma cells (Figure 3). It has been reported that many cancer cells including TCL upregulate the expression of the embryonic M2 isoform of pyruvate kinase (PKM 2) which results in the promotion of aerobic glycolysis and lactate production, bypassing the TCA cycle (206). An irregular expression of C-MYC or HIF-I alpha in tumorigenic T cells inactivates pyruvate dehydrogenase and upregulates lactate dehydrogenase A (LDHA), which promote the conversion of pyruvate to lactate (194). Recently, it has been reported that tumor cells like many activated lymphocytes use extracellular glutamine as a carbon source as well as a primary energy source, and this is linked to the inappropriate expression of C-MYC in mice (93, 207). Although T lymphoma cells are very similar to activated T cells, tumor cells have mutations in enzymes of the TCA cycle which is not the case with normal activated T lymphocytes (93). Examples are the mutations observed in isocitrate dehydrogenase (IDH) I and IDH 2 in gliomas and leukemias (208, 209). IDH is an important enzyme that regulates the conversion of isocitrate to alpha-keto-glutarate, and after mutation, they convert alpha-ketoglutarate to 2-hydroxyglutarate, a novel metabolite which subsequently alters the epigenome of the cell (208, 210).
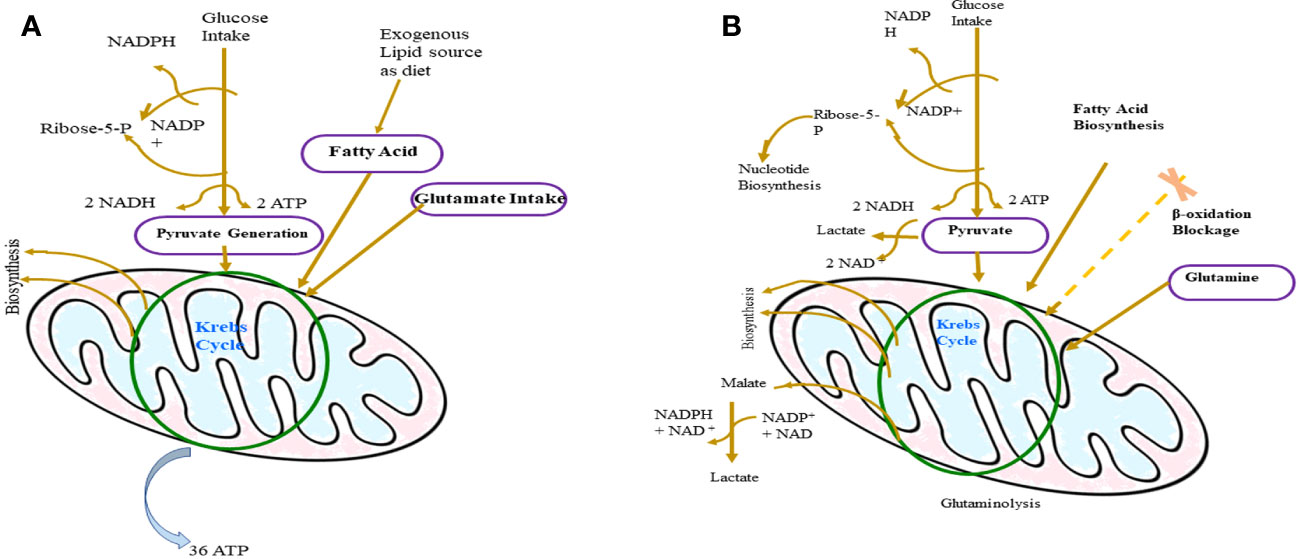
Figure 3 Difference between the metabolism of normal cells (A) and T-lymphoma cell (B). In T-lymphoma cells, glucose is the backbone of its growth as it fuels the biosynthesis of other important molecules like protein and nucleic acid. Glutamine is the main source of lactate production in tumor cells and in addition to that glutamine supports tumor growth by synthesizing Kreb’s cycle and NADPH regeneration intermediates.
The T lymphoma cell expresses muted versions of growth factor receptors and thus mimic the activated lymphocyte for their survival (93, 211). This is evident by the observation that in more than 50% of patients with T-cell acute lymphoblastic leukemia, proteins of the Notch pathway were mutated (212). The Notch pathway is very critical for the development and lineage selection of lymphocytes (213). As discussed, the expression of C-Myc is associated with upregulated glucose metabolism and mutations resulting in the overexpression of C-Myc have been reported in various lymphomas like Burkitt’s lymphoma (196). It can be concluded that these alternations in the signaling pattern of cancer cells including T lymphoma cells are the chief reason behind the Warburg phenotype and constitutive expression of Akt, along with C-Myc results in upregulation of aerobic glycolysis, inhibition of the TCA cycle (through the activation of proto-oncogene), and suppression of apoptotic genes. It has been clear that the metabolic reprogramming of tumor cells is essential for their survival and development. As discussed in this section, T-lymphoma cells mimic activated T cells metabolically, but the T-lymphoma cell differs from normally activated T cells because it expresses various oncogenes and inhibits tumor-suppressor genes. Due to this difference, whereas normally activated T cells either go through apoptosis or attain the state of anergy, T-lymphoma cells continue to survive and divide. To support this continuous growth, metabolic reprogramming is essential, which is also termed as the “Warburg effect”. For the effectiveness of the Warburg effect, signaling mediated by various cytokines is essential, as highlighted in this section. Various signaling pathways downstream of cytokine receptor-mediated signaling pathways like P13K-AKT and JAK-STAT are responsible for the pro-tumorigenic metabolic alternation. Cytokines like IL-7 and IL-2 have proven roles in the maintenance of basal metabolic requirements, and as discussed, their expression level is directly linked with metabolic reprogramming. Therefore, on the basis of the findings discussed in this section, it may be easily concluded that cell signaling alternations especially cytokine mediated are crucial for T-lymphoma cells’ metabolic reprogramming.
4 Cell signaling and cytokine in the context of T-cell lymphoma development
In the earlier section of this review, it has been discussed that the role of the cytokine in the TME development of the T-lymphoma cell and subsequent metabolic reprogramming to support the demanding metabolic requirement is crucial. The recent whole-exome signaling and gene expression profiling studies have shown that in T-cell lymphomas, all three signals (TCR signaling, co-stimulating signaling, and cytokine signaling) get altered (214, 215). In signal-1 in normal T cells, their T-cell receptor recognizes MHC-bound protein which subsequently activates the T cell through the downstream signaling via CD3 (186, 187). This interaction of TCR with the MHC-bonded peptide is very specific and sensitive. Mainly the kinases like LCK (lymphocyte-specific protein tyrosine kinase) and FYN are involved in the autophosphorylation of conserved immune-receptor tyrosine-based activation motifs (ITAMs) located in the cytoplasmic domain of the CD3 (216). These T-cell activation kinase domains get autophosphorylation by the SRC family kinase (SKFs), and dephosphorylation is mediated through several phosphates like phosphates of cytosol, protein tyrosine phosphatase non-receptor-6 (PTPN-6), PPTN22, CD45, and DUSP22 (dual-specificity phosphatase22) (216, 217). The inhibitory signal to the activation of this pathway is mediated by inhibitory C-terminal tyrosine which gets phosphorylated by CSK (C-terminal SRC kinase) and dephosphorylated by CD45 (217). LCK remains continuously active to maintain the signal required for the survival of naïve T cells (186). LCK activity along with ζ-chain activation via phosphorylation recruits ZAP70 (zeta-chain-associated protein kinase 70) and SYK family kinase, which results in the attainment of ZAP70 to its active conformation. This activation of ZAP70 results in the phosphorylation of adopter proteins like LAT (linker for activation of T cells) and SLP-76 (an SH2 domain-containing leucocyte protein of 76 kDa (186, 216) (Figure 4). Thus, TCR-dependent signaling of T-cell activation is rather complex involving multiprotein signalosomes, which result in the activation of multiple signaling networks essential for T-cell activation. TCR expression on T lymphoma cells is intact with some exceptions (218, 219). It has been reported that in peripheral T-cell lymphomas not otherwise specified (PTCL, NOS), the elements of the TCR complex were intact in more than 85% of cases, along with a normal expression of ZAP-70, LAT, and ITK. These observations were also similar for angioimmunoblastic T cells which use this TCR-mediated signaling for their growth and survival (220). The required engagement with MHC/peptide to activate TCR-mediated signaling in T lymphoma cells may be provided by the TME of tumorigenic T cells (53). To support this, it has been observed that immature DCs and macrophage favor the T-cell tumor development in vitro, which can be inhibited by MHC-blocking antibodies or clonotypic TCR (108). It has not been clear whether antigen-independent TCR signaling occurs in T-cell lymphoma, but the role of TCR-mediated signaling is established through the study on mouse models. Wang et al. have shown that 100% PTCL is developed in mouse models if the TCR remains intact and functional (108, 221). The importance of TCR signaling in TCL is further supported by studies on the role of antigen drug-associated neo-antigen in the development of T-cell lymphoma (38, 220).
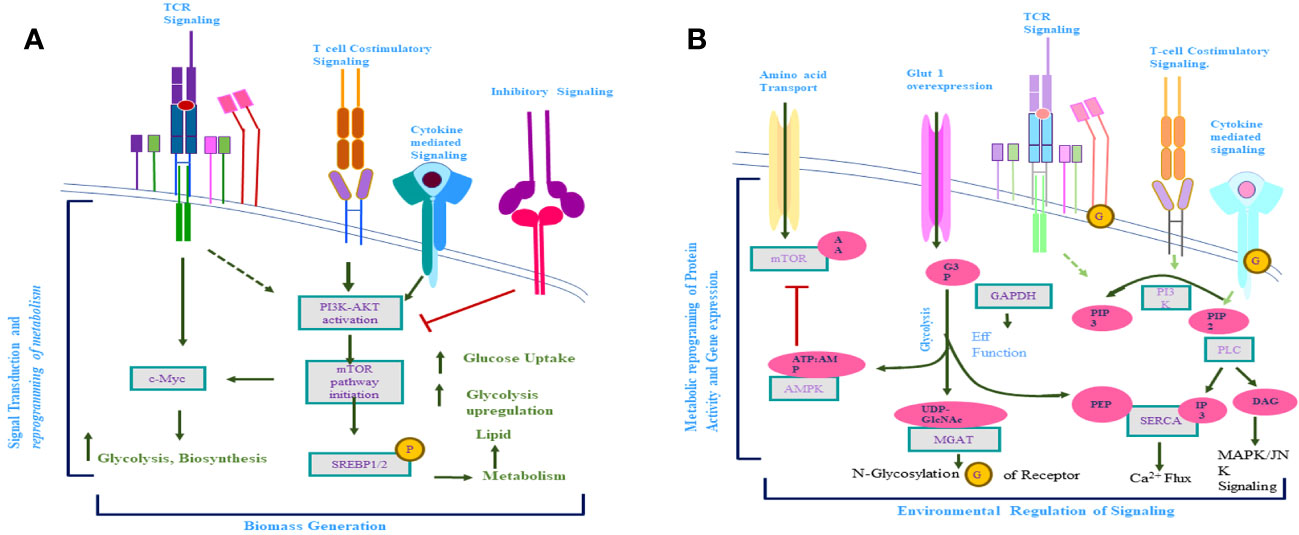
Figure 4 (A) Activation of T-cell receptor (TCR)-mediated signaling induction of costimulatory and cytokine receptor-mediated signaling takes place. Activation of these signaling pathways results in the downstream activation of mTOR and C-Myc and the reprogramming of activated T-cell metabolism. (B) Metabolite produced inside the cell regulates the signaling pathway of activated T cells. Afterward, the activation-activated T cells increase the uptake of glucose and amino acids, which results in a significant increase in the metabolite concern with these biomolecules. This increased level of metabolites influences the signaling pathways (TCR mediated, costimulatory, cytokine mediated) of activated T cells, which further alter the metabolism of activated T cells.
Although the role of CD3-TCR-mediated signaling in the development of T-cell lymphoma is universal, in many subtypes of PTCL, there are various alterations that result in TCR complex independent activation of downstream TCR signaling. The case of systemic anaplastic large cell lymphomas (ALCL), in which NPM-ALK (nucleophosmin-anaplastic lymphoma kinase) translocation fuses the catalytic domain of ALK with the dimerization domain of NPM, results in autophosphorylation of ALK resulting in activation of the pathways which are either redundant with RAS/RAF/ERK and P13K/STAT-3 pathway or complimentary to the JAK3/STAT3 pathway (38, 216, 222, 223). This ALK-dependent pathway may work as a substitute for TCR-independent downstream signaling activation (223). Although the mechanism is still not well elucidated, it has been reported that translocation involving the interferon regulatory factor (IRF)-4-DUSp22 (dual-specificity phosphatase 22) results in low expression of DUSP22 which can lead to the uninhibited activation of LCK and ERK and lead to the sustaining of downstream signaling induced by the TCR complex (224, 225). In approx. 20% of PTCL NOS, a translocation is observed between ITK and ZAP70 homologue SYK (226). This translocation results in the fusion of the pleckstrin and T homologous domain of ITK with the SYK kinase domain resulting in a catalytic fusion protein that can phosphorylate PLC-, SLP-76, and LAT1 along with endogenous SYK (227). This unique ITK-SYK fusion protein induces SRC family kinase-independent mimics of TCR-mediated signaling cascades (224, 227). This fusion protein is reported to be an oncogenic and transgenic expression, causing lymphoproliferative disorder similar to the human PTCL in mouse models (226, 227). Mutation in the gene-specific TCR signaling has been reported in many T-cell lymphomas. One of the examples is a recurrent mutation in RHOA, a small GTPase protein that has been observed in approx. 70% AITL and around 20% of PTCL and NOS. This RHOA (RAS family protein) binds active GTP, and the mutation (G17V) in the GTP binding site of RHOA in T-cell progenitors present in the thymus results in the development of T-cell lymphoblast leukemia in mouse models (228–231). It has been reported that when dominant-negative RHOA (Gi7V) is transduced in the cell line, it results in altered F-actin stress fiber development (230–232). This actin cytoskeleton is crucial for proper TCR activation since it maintains the spatiotemporal integrity of the cell membrane (233, 234). Other important proteins crucial for regulating the actin cytoskeleton are E2H2 (polycomb group protein) and Vav1 (A guanine exchange factor) (233).
In addition to TCR-mediated signaling, additional non-MHC signaling is essential for the proper activation of T cells. Various co-stimulatory signaling receptors have been reported to belong to either the Ig superfamily or the TNF receptor superfamily (TNRSF) (235, 236). Along with this activator co-stimulatory receptor, there are numerous homologous receptors with inhibitory effects present on T cells enabling the maintenance of fine-tuning of T-cell-mediated immune response (235). These co-stimulatory receptors mediate the activation of different signaling pathways like P13K, MAPK, NFAT, and NF- (236). It has been observed that CD28 engagement induced the proliferation of Sézary cells (237). Recently, a recurrent mutation in both the intracellular and extracellular domains of the CD28 receptor is reported in some PTCL resulting in increased affinity of receptor CD28 to its corresponding ligands (238). These observations are suggestive of the role of CD28 co-stimulation in T-cell lymphoma pathogenesis. Furthermore, in Sézary syndrome patients, the fusion of the transmembrane and extracellular domains of CTLA-4 with the intracytoplasmic domain of CD28 has been reported, which results in upregulation in CD28 signaling (239). The abundant availability of CD28 ligands in the TME supports the growth and development of T-cell lymphomas (34).
Following the engagement of TCR and CD28, the expression of ICOS (inducible T-cell co-stimulator) was induced. A knockout experiment in mouse models indicated the important role of P13K in ICOS-dependent proliferation, differentiation, and activation of T cells (38, 240). Although ICOS is expressed in various types of T cells, it is present in the germinal center and essential for GC homeostasis (241). It has been reported that AITL which has been originated from Tfh cells highly expresses ICOS (242). Another co-stimulatory receptor is CD134 (ox40) which belongs to TNRSF that is induced after TCR engagement but is not expressed by either naive or memory T cells (243). The ligand of CD134 is expressed by professional antigen-presenting cells, active T cells, and some non-hematopoietic cells (244). After activation through its ligand, CD134 trimerizes and its recruitment in the lipid raft is followed by association with TNFR-associated factor (TRAF) proteins 2, 3, and 5 through a conserved motif present within its cytoplasmic tail (245, 246). CD134 is expressed in more than 95% of AITL, 17% in PTCL, and NOS and absent in anaplastic large cell lymphomas (ALCL) (247). Similarly, TNFRSF8 (CD30) is expressed in a variable manner in PTCL (248). Although the role of TNFRSF in T-cell lymphoma pathogenesis is poorly understood, the importance of TNFSFs in T-cell lymphoma is indicated by the fact that TNFRSFS homologous to B cells plays an important role in B-cell lymphoma pathogenesis (249).
Although MHC-peptide-dependent and co-stimulatory signals are crucial for the activation of naive T cells, cytokine-dependent signaling is essential for effective T-cell response. Various studies reported the important role of IL-12 and IFN-alpha/beta in CD8 T-cell maintenance and activation (38, 250). The signaling pathways of most of the cytokine receptors are induced by respective cytokine binding to its receptor followed by activation of a family of Janus kinases (JAK1, JAK2, JAK3, and tyrosine kinase-2), which is associated with the cytokine receptor cytoplasmic tail. Cytokine binding to its receptor results in conformational changes in JAKs which subsequently phosphorylate signal transducers and activators of transcription (STAT) monomer (38, 251). Activated STAT dimerizes and after translocation to the nucleus regulates the gene expression (251). In T-cell lymphoma, any cytokine that suppresses host anti-tumor activity is considered a tumor-promoting cytokine. It has been reported that various inflammatory and homeostasis cytokines have a role in T-cell lymphoma pathogenesis (38, 87). Transcription factor GATA-3 is a master regulator of Th2 differentiation since it binds to the locus of Th2-specific cytokines. Two dominant subclasses of cytokines have been reported in the case of PTCL: one is GATA-3 enriched (IL-4, IL-5, IL-13), and another is t-bet enriched (IFN-gamma and IFN-gamma inducible gene). In the PTCL, NOS and the GATA-3+ subclass of T-cell lymphomas resembled CTCL on a molecular level in which type 2 cytokines have a direct role in the proliferation and survival of tumor cells. IL-5-dependent hypereosinophilia has been observed in the GATA 3+ subclass of PTCL (64, 252). Recent whole-genome and whole-exome studies have reported that recurrent mutations in JAK and STAT genes are frequent in some classes of T-cell lymphomas (70). For example, in T-cell polympholytic leukemia (T-PLL), recurring mutation in gamma-chain, JAK1, JAK3, and STAT has been reported in more than 75% of cases (253–255). Similarly, STAT5b mutation is frequent in gamma F-PTCL and NK-cell-derived lymphomas (255). Although STAT3 mutation is not very prevalent, it has an important role in indolent T-cell lymphoproliferative disorders (256). It has also been reported that the PTPRK (receptor type tyrosine-protein phosphatase K) gene located at chromosome 6q is mostly deleted in extranodal NK-cell lymphoma and T-cell lymphoma, which results in STAT3 dephosphorylation and subsequent activation of STAT3 in approx. 50% of NK/T-cell lymphoma (257). It has been observed that JAK/STAT mutation is more prevalent in TCL than recurring mutation in its cytokine receptors (258). Mutation in chemokine receptor CCR4 is an exception, which is more frequent in TCL (259). The c-terminal truncating recurring mutation in CCR4 is reported in more than 25% of ATLL cases (260). This mutation in CCR4 hinders its internalization, a kind of gain-of-function mutation, and it was reported to be associated with increased PI3K/Akt activation which in turn plays a crucial role in metabolic reprogramming in TCL (260, 261).
It is easy to describe the role of these different T-cell signaling pathways in TCL pathogenesis in isolation, but in actuality, all these signaling pathways are interwoven and regulate the TCL pathogenesis in a coordinated manner. The cross-talk between these signaling receptors has been observed, and their dependency on each other in TCL development and progression has been reported. However, it is a fact that alternation in TCR-mediated signaling is crucial to bringing required changes in other receptor signaling pathways for the survival of T-lymphoma cells. Through the earlier works discussed in this section, it has been clear that in addition to metabolic alternation, cytokine-mediated signaling is also important for the overall development of T-lymphoma cells. Signaling mediated by these cytokines regulates the different stages of development, i.e., from naive T-cell to activated T-cell. In fact, all three T-cell signaling pathways, i.e., TCR mediated, co-stimulatory, and cytokine-mediated, play a crucial role in TCL, which helps in the differentiation of T-lymphoma cells to attain its tumorigenic morphology and physiology. Cytokine-mediated signaling depends upon the various intrinsic and extrinsic factors, and it may be easily concluded that cytokine plays an important role in deciding the fate of the developing T-cell lymphoma in the tumor microenvironment.
5 Interrelation of cytokine and metabolism in T-cell lymphoma
Metabolism and immune response are interlinked, and as discussed in earlier sections, metabolic alternation is the hallmark of the tumor cell and this metabolic reprogramming in mammalian immune cells is a crucial immunological adaption to mounting a proper immune response (22). This association of metabolism with immune response has been known for the last 60 years (19, 23) (Figure 5). It has been reported that IL-6, a pro-inflammatory cytokine, can regulate glucose as well as lipid metabolism (262). It is a fact that increased uptake of amino acid is crucial to support continuous mTOR activity and, along with C-Myc expression, IL-2 signaling plays a significant role in this (191, 198). It has been reported that IL-2 signaling induces amino acid transporter SLC7A5 and it redirects T-cell metabolic reprogramming through pyruvate dehydrogenase kinase (PDK)-mediated upregulation of mTOR, which does not require P13K/AKT signaling in CD8+ T cells (191, 198, 263). The role of IL-2 in Treg-cell metabolism regulation is evident by the high expression of the IL-2 receptor (IL-2R), which is accompanied by the phenotypic and functionally active state of Treg cells (189, 264). IL-2-mediated signaling is crucial for T-cell metabolism as it co-ordinates P13K/AKT/mTOR activity, upregulates SLC7A5 expression, and upregulates C-Myc expression in CD4+T cells and CD8+ T cells (60, 189, 198). To support functionally active Treg cell metabolism, a continuous level of glycolysis and lipid biosynthesis along with the proper mitochondrial function is required and IL-2 has been reported to play an important role in all of these metabolic requirements of Treg cells (198, 265, 266). Another important cytokine for the maintenance of T-cell metabolic homeostasis is IL-7. It has been reported that IL-7-mediated signaling plays a crucial role in sustaining glycolysis in T cells through STAT-5-mediated AKT induction. It also induces GLUT1 expression in CD8+ memory T cells and IL-7 involved in triglyceride synthesis through increasing the rate of glycol uptake (198, 267).
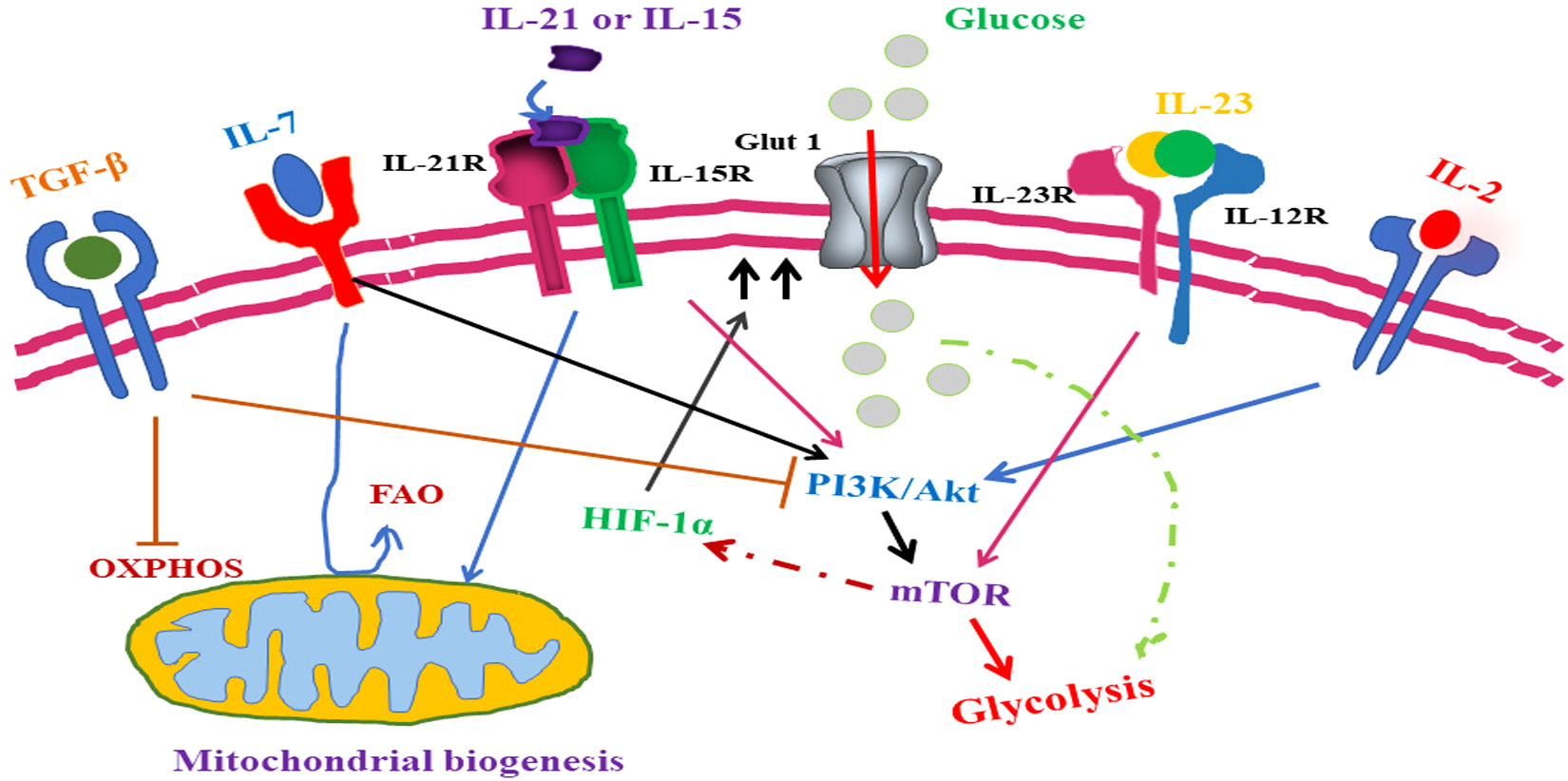
Figure 5 Role of cytokine-mediated signaling in T-cell lymphoma metabolic reprogramming. TGF-β, an anti-inflammatory cytokine, inhibits T lymphoma cell glycolysis whereas IL-7 signaling promotes glycolysis through upregulation of Glut1 expression. With a similar role in antioxidant production, IL-15- and IL-21-mediated signaling plays an important role in oxidative phosphorylation along with mitochondrial biogenesis inside the T-lymphoma cells. IL-15- and IL-21-mediated signaling induces the mTOR pathway and subsequent alternation of T-lymphoma cell metabolism. In addition to promoting the growth of Th17 helper cells, IL-1β also induces the mTOR pathway. Although IL-2 promotes mTOR activity in both CD4+T cell and CD8+ T cell through different pathways, in CD8+ T cell, IL-2 induces mTOR through PDK whereas in CD4+ T cell, IL-2 induces mTOR through activation of the PI3/Akt pathway. Upregulation in amino acid intake by T-cell lymphoma cells is promoted through overexpression of SLC7A5.
IL-15 is essential for memory T-cell development, which alters the metabolism of these developing cells through the remodeling of cristae, a fusion of mitochondria, and upregulation in mitochondrial spare respiratory capacity (SRC) (268). IL-15 also supports fatty acid oxidation (FAO) through the upregulation of carnitine O-palmitoyltransferase-1 (CPT1a) which facilitate the transfer of the acyl group of long-chain fatty acyl-CoA from coenzyme A to 1-carnitine. It has been reported that IFN-α, which is an inducer of IL-15, can further upregulate transcription of antioxidant molecules like glutathione reductase, thioredoxin reductase, superoxide dismutase, and peroxiredoxin resulting in the long life of these memory cells (189, 191, 268, 269). This effect of IL-15 may be due to the neutralization of a high level of RO generated through the hyperoxidative phosphorylation activity of mitochondria by these antioxidant molecules. IL-15 plays an important role in the development of the tissue-resident population of T cells by promoting the mTOR activity in memory CD8+ T cells resulting in fastened activation and proliferation of CD8+ T cells in case of reinfection (191, 199, 268, 270). It has been reported that IL-21 has an important role in T-cell metabolism since it promotes fatty acid oxidation, antioxidant production, and mitochondrial biogenesis. It has also been reported that IL-21 has a role in promoting mTOR activity in Treg cells. It has been shown that priming of T cells with IL-21 and IL-15 before using in cancer T-cell therapy prolongs the life of these therapeutic T cells in a cancer mouse model (271–274). Studies have reported that by upregulating mitochondrial activity and reduction in oxidative stress in T cells, both IL-15 and IL-21 delayed the expression of apoptosis markers such as PD1 and LAG-3 (274, 275).
Although the role of inflammatory cytokines in T-cell metabolism has not been investigated much, evidence is growing which is suggestive of the roles of inflammatory cytokines in the alteration of T-cell metabolism (191) (Table 2). A study has shown that IFN-γ can sustain the production of effector CD8+ T cells despite of a weak TCR signal (301). This effect of IFN-γ is through the activation of mTOR via STAT-1. The roles of IL-1β and IL-23 are crucial for the shifting of the Treg/Th17 axis toward Th17 through the promotion of human Th17 CD4+T cells. IL-1β-mediated signaling triggers HIF-1α expression along with upregulation of glycolysis-related genes like GLUT1 and HK2 (302–304). This further shift analysis suggested that IL-1β upregulates Th17 cell glycolysis. IL-23 has a synergistic effect on Th-17 cell differentiation and IL-23 is considered as the chief architect behind the Th17 cell phenotype (303). There are various phosphorylation targets on IL-23 identified through phosphor-proteomics analysis including an important glycolytic enzyme PKM2 whose upregulation results in hyperphosphorylation of STAT3 resulting in metabolic alternations (304, 305). It has also been reported that PKM2 is crucial for the homeostasis of the metabolism of Th17 cells (304). It has been observed that these effects of IL-23 can be inhibited by CD28 signaling, which also upregulates SIGIR expression in differentiating Th17 cells, resulting in the limitation of mTOR signaling (306, 307).
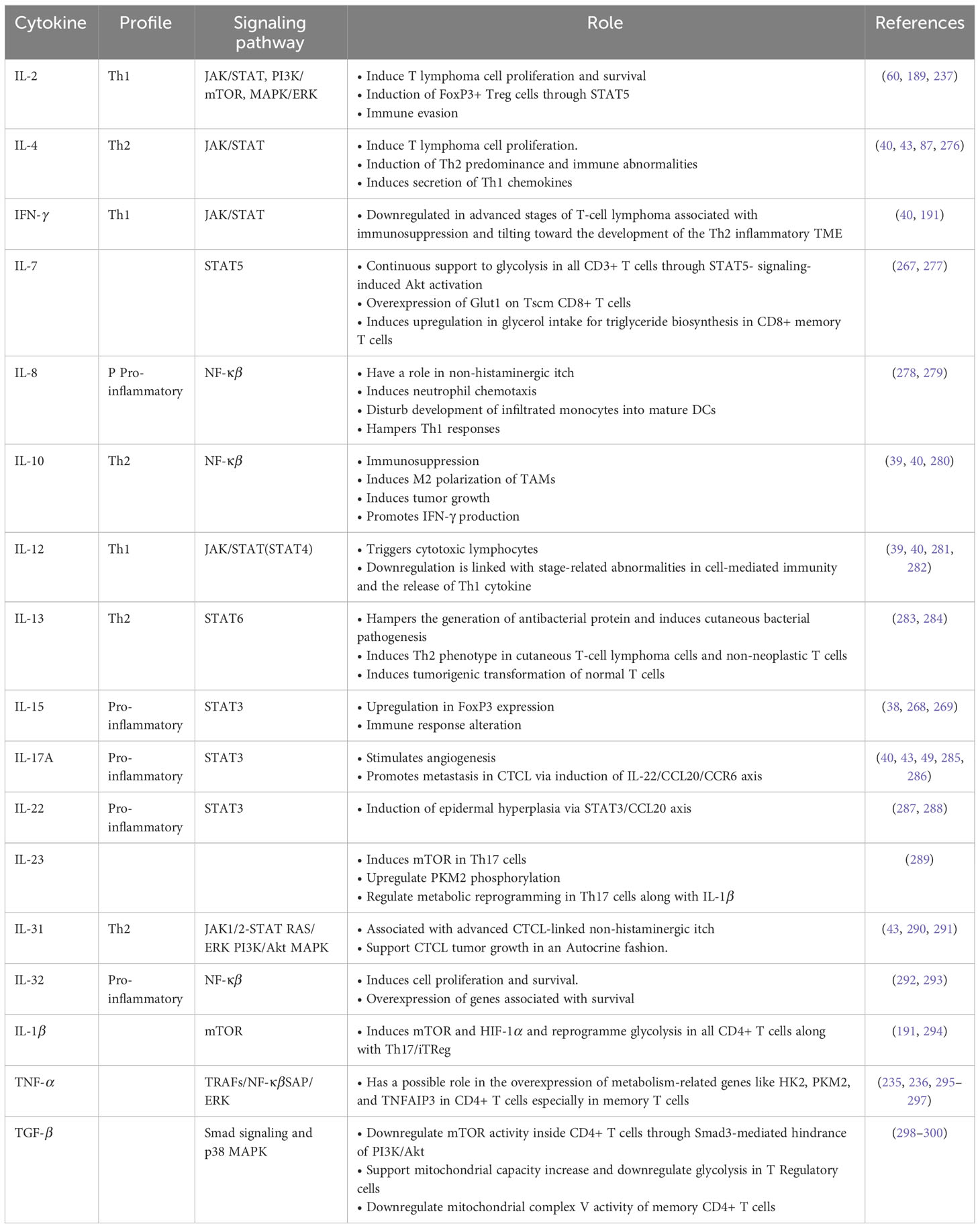
Table 2 List of Cytokines crucial in T-cell lymphoma pathogenesis and metabolism (Source reference: 87, 191).
The TNF-α-mediated signaling inhibits CD4+ T-cell maturation and supports those cells which express anti-inflammatory cytokine IL-10 (308). Various studies have shown the role of TNF-α in calcium signaling which control T-cell metabolism. There is a recent report on the impact of TNF-α on the metabolic profile of CD4+ T-cells, but further studies are needed to fully elucidate the role of TNF-α in cellular metabolism in general and of T cells in particular (308–310). Although IL-10 has been reported to have a role in B-cell and macrophage metabolism alternation, evidence for the role of this anti-inflammatory cytokine in T-cell metabolism is few (280). TGF-β induced Smad-3-mediated inhibition of PI3K/Akt signaling in CD4+T cells, which results in the downregulation of important components of glycolysis like GIUT1 and HK2 (280, 298). Due to its suppressive effect on mTOR signaling, TGF-β favors T cells toward the inducible Treg phenotype. A study has reported that circulating Treg treated with TGF-β reduced the suppressive capacity of these Tregs through inhibition of glycolysis via the PI3K/AKT mTOR pathway (311). One in vitro study has shown that the presence of TGF-β in the tumor microenvironment can reduce OXPHOS in effector CD4+ T cells (299). Although there is growing evidence that cytokines play an important role in the reprogramming of T-cell metabolism, in turn, this metabolic alternation changes the cytokine profile of T-lymphoma cells as well as of the TME. Further studies are required to understand the mechanism behind cytokine-mediated metabolic reprogramming. As reported and summarized in this section, the cytokine-mediated signaling is important for metabolic reprogramming, and in turn, this metabolic alternation further changes the cytokine milieu of the tumor cell microenvironment in favor of T-lymphoma cell development and survival.
6 Future investigation and challenges to make cytokine a diagnostic marker
This is a matter of fact that cytokine receptor-mediated signaling is very crucial to the development and survival of tumor cells. With the advancement in tools and techniques, various novel biomarkers have been identified for the early diagnosis and prognosis of cancer. These biomarkers are a different class of biomolecules, and usually, cancer biomarkers are the molecules derived from the tumor cell and an ideal cancer biomarker should help in screening, prognosis, and cancer stage prediction as well as monitoring of treatment efficacy (312, 313). Another important criterion to be a good biomarker is that techniques based upon these biomarkers should be easy to handle, cost-effective, and reproducible (313). The currently approved biomarker for cancer has limited utility due to its low specificity and sensitivity (314). As cytokines have been associated with every stage of cancer development, these signaling mediators have great potential as a cancer biomarker (315, 316). Usually, a low amount of cytokines exists locally, but the upregulation in the cytokine level is observed in various abnormal conditions including tumors. The potential of cytokine as a biomarker is good because in tumors, the level of cytokines is related with tumor progression and changes in their expression level can be easily detected in body fluid mostly in the blood (317). The level of cytokines such as IL-6, IL-10, IL-8, TNF-, and VEGF can be co-related with disease progression in ovarian, breast, and cervical carcinoma (317, 318). As discussed in the various sections of this review, the level of cytokine is clearly associated with T-lymphoma progression and metabolic rewiring required for the survival of T-lymphoma cells. As mentioned in Tables 1, 2, various cytokines and their expression level influence the metabolic alternation in T-lymphoma cells. Some more obvious cytokines such as IL-2, IL-6, IL-7, and IL-15 have a direct link with T-cell lymphoma progression, and these cytokines’ expression levels have the potential to be used as biomarkers especially to predict the developmental stage of T-lymphoma tumor cells.
Despite of this clear co-relation, the main hindrance in using cytokines as a biomarker is that cytokines are mechanism specific, not tumor specific. In recent times, numerous works have been reported which evaluated the potential of cytokines as a cancer biomarker, but mostly, these studies evaluate cytokines in isolation; however, in reality, the cytokine expression depends upon various interwoven factors (319–322). The potential of cytokine as an independent biomarker will be realized through the elucidation of the interwoven factors affecting the cytokine level in specific conditions. Another problem in using cytokine as a biomarker is to decide the cutoff value, which is essential to using any biomarker as a diagnostic tool. In the case of cytokine, it is really a huge challenge to differentiate between normal levels and abnormal levels of cytokine because only few studies have been done under any particular condition to decide the base-level value of any particular cytokine to use as a biomarker for any cancer including T-cell lymphoma. Due to the advent of multiplex arrays nowadays, it is possible to monitor the level of different cytokines simultaneously and then use these cytokines as a biomarker group. The receiver operating characteristics analysis (ROC analysis) which is based upon the plot of the true positive rate versus false positive rate of a particular disease at various cutoff may be helpful in deciding the cutoff value of cytokine to enable them to be used as a biomarker (323). In addition, in infectious disease where the roles of cytokines are more clear, the diagnostic potential of cytokines has been established in certain chronic conditions like Alzheimer’s disease (AD) and tumors. A recent study conducted on Malaysian people reveals the clear-cut link between blood IP-10 (interferon gamma inducible protein 10) along with IL-13. The level of these anti-inflammatory mediators was found to be on the lower side in AD subjects in comparison with healthy control significantly (324). Similarly, cytokines such as IL-6 and IL-8 are associated with poor neurological outcomes in individuals who experienced cardiac arrest. The cutoff value to predict poor neurological progress is 1423 pg/ml for IL-8 and 2708 pg/ml for IL-6 with 100% sensitivity and 86% specificity (325). The 1.97-pg/ml serum level of IL-6 with 81.8% sensitivity and 66.7% specificity is found to be the optimal diagnostic cutoff for gastric cancer (326). Two cutoff values, i.e., 400 pg/ml (92% sensitivity and 60% specificity) and 1,200 pg/ml (84% sensitivity and 87%), for IL-6 has been evaluated for stage I and stage II ovarian cancer subjects. The VEGF diagnostic cutoff was found to be 90% sensitive and 80% specific at the serum level value of 400 pg/ml (327). Many studies have been reported with a moderate rate of success to establish the cytokine as a diagnostic or prognostic biomarker. It is clear that to establish the cytokine as a biomarker for T-cell lymphoma development, a particular cutoff value is needed to be identified, and for this, elucidation of complex cytokine biology is needed. The ROC analysis will help in this regard, but the sample size needs to be sufficiently high to draw any conclusion. However, due to a limited number of cases, it is really difficult to find a cutoff value of cytokines to be established as a diagnostic or prognostic marker. The development of an animal model is an alternative to solve this problem and requires a high degree of attention of researchers working in the area. However, despite these limitations, cytokine has certain usefulness as a supportive biomarker to predict the T-lymphoma cell progression and diagnosis. These cytokines may be used as a corroborative marker along with other clinical symptoms and outcomes to estimate the treatment responsiveness and disease progression, but certainly, it warrants more attention from clinicians and researchers.
7 Conclusion
As of other cancer cells, the classical T-cell lymphoma TME constitutes various immune-active non-neoplastic cells like lymphocytes, mast cells, macrophages, and eosinophils. These cells may even outnumber the neoplastic cells (31–33). Although the biology of real interaction between the T-lymphoma cells and other cells of the TME is not fully elucidated, the role of cytokine-mediated signaling is getting much attention (35). T-lymphoma cells undergo metabolic alternation to fulfill the ever-increasing demand of ATP, NADH, and NADPH bio-synthesis to support their growth and proliferation. It has been established that in T-cell lymphomas as with other cancers, all the neoplastic cells are not in the same state metabolically (31, 39). The ever-changing TME exerts selective pressure on T-lymphoma cells. Due to the Warburg effect, tumor cells release an increasing amount of lactate in the tumor microenvironment resulting in acidosis and hypoxia (328). Cytokines play an important role in the development of the TME and metabolic reprogramming in T-cell lymphoma (40, 43, 87). The cytokine milieu of the TME dictates the tumor progression, and mutation in cytokine signaling protein significantly favors the tumor development. In addition, TCR- and CD28-mediated signaling and cytokine receptor signaling are very crucial for proper T-cell activation and proliferation. Cytokines like IL-2 and IL-7 are well-established mediators of T-cell lymphoma metabolic reprogramming (38, 60, 277). Alternation in downstream signaling of cytokine receptors has been reported in various types of T-cell lymphomas, which has been discussed in the various sections of the review. The specific mutation in the AKT-P13K and JAK/STAT pathways associated with cytokine receptors has been reported. These alternations ultimately lead to much-needed changes in metabolism to sustain tumor growth and proliferation. Apart from metabolic reprogramming, cytokine-mediated signaling plays an important role in the overall cellular compositions of the TME, as discussed in various sections of this review. T-cell lymphoma development depends upon various extrinsic as well as intrinsic factors, and cytokines have certainly an important role in coordinating these extrinsic and intrinsic factors resulting in T-cell lymphoma development and progression. The cytokines released from different tumor-infiltrating lymphocytes have an important role in the cross-talk between T-lymphomas and their corresponding TME (35). Although T-lymphoma cells resemble metabolically activated T cells, the mutation in the genes related to cytokine signaling has been observed in the development of almost all subtypes of T-cell lymphomas. Thus, the importance of cytokine milieus of the TME in T-lymphoma progression is further needed to be explored for developing a new therapeutic target against T-cell lymphomas. Elucidation of the role of cytokine in tumor development will help in finding T-lymphoma-specific biomarkers for early diagnosis of T-lymphoma and to monitor the disease progression along with in follow-up to the treatment response.
Author contributions
Corresponding and joint first authors from Guru Ghasidas Vishwavidyalaya wrote and edited the manuscript. All other co-authors contributed intellectually to the designing, editing, and proofreading of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors are supported by the University Grants Commission (UGC), New Delhi, India, Guru Ghasidas Vishwavidyalaya (GGV), Bilaspur, India, The University of Tokyo, Japan, University of Missouri, Columbia, USA, Veer Kunwar Singh University, Arrah, India, Lalit Narayan Mithila University, Darbhanga, India, and RK University, Rajkot, India.
Acknowledgments
The authors acknowledge institutional and administrative support of the Department of Forensic Science, Guru Ghasidas Vishwavidyalaya (GGV). The authors also acknowledge the research support of our collaborators and their host institutions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Peng F, Qin Y, Mu S, Li J, Ai L, Hu Y. Prognostic role of regulatory T cells in lymphoma: a systematic review and meta-analysis. J Cancer Res Clin Oncol (2020) 146:3123–35. doi: 10.1007/s00432-020-03398-1
2. Gaulard P, de Leval L. The microenvironment in T-cell lymphomas: emerging themes. Semin Cancer Biol (2014) 24:49–60. Academic Press. doi: 10.1016/j.semcancer.2013.11.004
3. Satou A, Bennani NN, Feldman AL. Update on the classification of T-cell lymphomas, Hodgkin lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev Hematol (2019) 12(10):833–43. doi: 10.1080/17474086.2019.1647777
4. Macapagal SC, Bennani NN. Nodal peripheral T-cell lymphoma: Chemotherapy-free management, are we there yet? Blood Rev (2023) 60:101071. doi: 10.1016/j.blre.2023.101071
5. Pytlik R, Polgarova K, Karolova J, Klener P. Current immunotherapy approaches in non-Hodgkin lymphomas. Vaccines (2020) 8(4):708. doi: 10.3390/vaccines8040708
6. Willemze R, Cerroni L, Kempf W, Berti E, Facchetti F, Swerdlow SH, et al. The 2018 update of the WHO-EORTC classification for primary cutaneous lymphomas. Blood J Am Soc Hematol (2019) 133(16):1703–14. doi: 10.1182/blood-2018-11-881268
7. de Leval L, Jaffe ES. Lymphoma classification. Cancer J (2020) 26(3):176–85. doi: 10.1097/PPO.0000000000000451
8. Sandell RF, Boddicker RL, Feldman AL. Genetic landscape and classification of peripheral T cell lymphomas. Curr Oncol Rep (2017) 19:1–10. doi: 10.1007/s11912-017-0582-9
9. Levy R, Pope E. Primary cutaneous lymphoma. In: Harper’s textbook of pediatric dermatology (Wiley Online) (2019). p. 1044–62.
10. Reinhard D, Vermeer MH, Scarisbrick JJ, Kim YH, Connor S, Tensen CP, et al. Cutaneous T cell lymphoma (Primer). Nat Reviews: Dis Primers (2021) 7(1). doi: 10.1038/s41572-021-00296-9
11. Altea-Manzano P, Cuadros AM, Broadfield LA, Fendt SM. Nutrient metabolism and cancer in the in vivo context: a metabolic game of give and take. EMBO Rep (2020) 21(10):e50635. doi: 10.15252/embr.202050635
12. Ghosh P, Vidal C, Dey S, Zhang L. Mitochondria targeting as an effective strategy for cancer therapy. Int J Mol Sci (2020) 21(9):3363. doi: 10.3390/ijms21093363
13. Sneeggen M, Guadagno NA, Progida C. Intracellular transport in cancer metabolic reprogramming. Front Cell Dev Biol (2020) 8:597608. doi: 10.3389/fcell.2020.597608
14. Ahmad F, Cherukuri MK, Choyke PL. Metabolic reprogramming in prostate cancer. Br J Cancer (2021) 125(9):1185–96. doi: 10.1038/s41416-021-01435-5
15. Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol (2010) 22(3):314–20. doi: 10.1016/j.coi.2010.01.018
16. Sockolosky JT, Trotta E, Parisi G, Picton L, Su LL, Le AC, et al. Selective targeting of engineered T cells using orthogonal IL-2 cytokine-receptor complexes. Science (2018) 359(6379):1037–42. doi: 10.1126/science.aar3246
17. Stopnicki B. Glycogen dynamics in proliferating human helper T cells (Doctoral dissertation, concordia university) (Quebec, Canada: University of Montreal) (2019).
18. Herbel C, Patsoukis N, Bardhan K, Seth P, Weaver JD, Boussiotis VA. Clinical significance of T cell metabolic reprogramming in cancer. Clin Trans Med (2016) 5:1–23. doi: 10.1186/s40169-016-0110-9
19. Chen H, Yang T, Zhu L, Zhao Y. Cellular metabolism on T-cell development and function. Int Rev Immunol (2015) 34(1):19–33. doi: 10.3109/08830185.2014.902452
20. Palmer CS, Hussain T, Duette G, Weller TJ, Ostrowski M, Sada-Ovalle I, et al. Regulators of glucose metabolism in CD4+ and CD8+ T cells. Int Rev Immunol (2016) 35(6):477–88. doi: 10.3109/08830185.2015.1082178
21. Kohlgruber AC, Donado CA, LaMarche NM, Brenner MB, Brennan PJ. Activation strategies for invariant natural killer T cells. Immunogenetics (2016) 68:649–63. doi: 10.1007/s00251-016-0944-8
22. Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science (2013) 342(6155):1242454. doi: 10.1126/science.1242454
23. Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity (2013) 38(4):633–43. doi: 10.1016/j.immuni.2013.04.005
24. Gaber T, Chen Y, Krauß PL, Buttgereit F. Metabolism of T lymphocytes in health and disease. Int Rev Cell Mol Biol (2019) 342:95–148. doi: 10.1016/bs.ircmb.2018.06.002
25. Durinck K, Goossens S, Peirs S, Wallaert A, Van Loocke W, Matthijssens F, et al. Novel biological insights in T-cell acute lymphoblastic leukemia. Exp Hematol (2015) 43(8):625–39. doi: 10.1016/j.exphem.2015.05.017
26. Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet (2015) 47(12):1426–34. doi: 10.1038/ng.3444
27. McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood J Am Soc Hematol (2015) 126(4):508–19. doi: 10.1182/blood-2014-11-611194
28. Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RSK, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science (1993) 262(5134):747–50. doi: 10.1126/science.8235596
29. Crescenzo R, Abate F, Lasorsa E, Gaudiano M, Chiesa N, Di Giacomo F, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell (2015) 27(4):516–32. doi: 10.1016/j.ccell.2015.03.006
30. Travert M, Huang Y, De Leval L, Martin-Garcia N, Delfau-Larue MH, Berger F, et al. Molecular features of hepatosplenic T-cell lymphoma unravels potential novel therapeutic targets. Blood J Am Soc Hematol (2012) 119(24):5795–806. doi: 10.1182/blood-2011-12-396150
31. Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med (2015) 13(1):1–14. doi: 10.1186/s12916-015-0278-7
32. Bejarano L, Jordāo MJ, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov (2021) 11(4):933–59. doi: 10.1158/2159-8290.CD-20-1808
33. Wang JJ, Lei KF, Han F. J. E. R. M. P. S. Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci (2018) 22(12):3855–64. doi: 10.26355/eurrev_201806_15270
34. Pereira JF, Jordan P, Matos P. A signaling view into the inflammatory tumor microenvironment. Immuno (2021) 1(2):91–118. doi: 10.3390/immuno1020007
35. Yu P, Fu YX. Tumor-infiltrating T lymphocytes: friends or foes? Lab Invest (2006) 86(3):231–45. doi: 10.1038/labinvest.3700389
36. Liu X, Jin S, Hu S, Li R, Pan H, Liu Y, et al. Single-cell transcriptomics links Malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma. Nat Commun (2022) 13(1):1158. doi: 10.1038/s41467-022-28799-3
37. Galli F, Aguilera JV, Palermo B, Markovic SN, Nisticò P, Signore A. Relevance of immune cell and tumor microenvironment imaging in the new era of immunotherapy. J Exp Clin Cancer Res (2020) 39(1):1–21. doi: 10.1186/s13046-020-01586-y
38. Wilcox RA. A three-signal model of T-cell lymphoma pathogenesis. Am J Hematol (2016) 91(1):113–22. doi: 10.1002/ajh.24203
39. Fanok MH, Sun A, Fogli LK, Narendran V, Eckstein M, Kannan K, et al. Role of dysregulated cytokine signaling and bacterial triggers in the pathogenesis of cutaneous T-cell lymphoma. J Invest Dermatol (2018) 138(5):1116–25. doi: 10.1016/j.jid.2017.10.028
40. Li L, Yu R, Cai T, Chen Z, Lan M, Zou T, et al. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int Immunopharmacol (2020) 88:106939. doi: 10.1016/j.intimp.2020.106939
41. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol (2020) 30(16):R921–5. doi: 10.1016/j.cub.2020.06.081
42. Ansell SM, Vonderheide RH. Cellular composition of the tumor microenvironment. Am Soc Clin Oncol Educ Book (2013) 33(1):e91–7. doi: 10.14694/EdBook_AM.2013.33.e91
43. Odenthal J, Takes R, Friedl P. Plasticity of tumor cell invasion: governance by growth factors and cytokines. Carcinogenesis (2016) 37(12):1117–28. doi: 10.1093/carcin/bgw098
44. Antonyak MA, Cerione RA. Microvesicles as mediators of intercellular communication in cancer. Cancer Cell Signaling: Methods Protoc (2014) 1165:147–73. doi: 10.1007/978-1-4939-0856-1_11
45. Brücher BL, Jamall IS. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol Biochem (2014) 34(2):213–43. doi: 10.1159/000362978
46. Maacha S, Bhat AA, Jimenez L, Raza A, Haris M, Uddin S, et al. Extracellular vesicles-mediated intercellular communication: roles in the tumor microenvironment and anti-cancer drug resistance. Mol Cancer (2019) 18:1–16. doi: 10.1186/s12943-019-0965-7
47. Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev Biol (2018) 6:18. doi: 10.3389/fcell.2018.00018
48. Jaffe ES. Pathobiology of peripheral T-cell lymphomas. ASH Educ Program Book (2006) 2006(1):317–22. doi: 10.1182/asheducation-2006.1.317
49. Chiba S, Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia (2020) 34(10):2592–606. doi: 10.1038/s41375-020-0990-y
50. Kim H, Ko YH. The pathologic and genetic characteristics of extranodal NK/T-Cell lymphoma. Life (2022) 12(1):73. doi: 10.3390/life12010073
51. Ma H, Abdul-Hay M. T-cell lymphomas, a challenging disease: types, treatments, and future. Int J Clin Oncol (2017) 22:18–51. doi: 10.1007/s10147-016-1045-2
52. Eray M, Postila V, Eeva J, Ripatti A, Karjalainen-Lindsberg ML, Knuutila S, et al. Follicular lymphoma cell lines, an in vitro model for antigenic selection and cytokine-mediated growth regulation of germinal centre B cells. Scandinavian J Immunol (2003) 57(6):545–55. doi: 10.1046/j.1365-3083.2003.01264.x
53. Anderson KG, Stromnes IM, Greenberg PD. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell (2017) 31(3):311–25. doi: 10.1016/j.ccell.2017.02.008
54. Yamagishi M. The role of epigenetics in T-cell lymphoma. Int J Hematol (2022) 116(6):828–36. doi: 10.1007/s12185-022-03470-1
55. Pritchett JC, Yang ZZ, Kim HJ, Villasboas JC, Tang X, Jalali S, et al. High-dimensional and single-cell transcriptome analysis of the tumor microenvironment in angioimmunoblastic T cell lymphoma (AITL). Leukemia (2022) 36(1):165–76. doi: 10.1038/s41375-021-01321-2
56. Leca J, Lemonnier F, Meydan C, Foox J, El Ghamrasni S, Mboumba DL, et al. IDH2 and TET2 mutations synergize to modulate T Follicular Helper cell functional interaction with the AITL microenvironment. Cancer Cell (2023) 41(2):323–39. doi: 10.1016/j.ccell.2023.01.003
57. Grogg KL, Attygalle AD, Macon WR, Remstein ED, Kurtin PJ, Dogan A. Angioimmunoblastic T-cell lymphoma: a neoplasm of germinal-center T-helper cells? Blood (2005) 106(4):1501. doi: 10.1182/blood-2005-03-1083
58. Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol Rev (2010) 237(1):72–89. doi: 10.1111/j.1600-065X.2010.00937.x
59. Rubio AJ, Porter T, Zhong X. Duality of B cell-CXCL13 axis in tumor immunology. Front Immunol (2020) 11:521110. doi: 10.3389/fimmu.2020.521110
60. Chabab G, Bonnefoy N, Lafont V. IL-21 signaling in the tumor microenvironment. In: Tumor microenvironment: the role of interleukins–part A (Springer) (2020). p. 73–82.
61. Lin YW, Aplan PD. Gene expression profiling of precursor T-cell lymphoblastic leukemia/lymphoma identifies oncogenic pathways that are potential therapeutic targets. Leukemia (2007) 21(6):1276–84. doi: 10.1038/sj.leu.2404685
62. WeNiger MA, Küppers R. Molecular biology of Hodgkin lymphoma. Leukemia (2021) 35(4):968–81. doi: 10.1038/s41375-021-01204-6
63. Gaydosik AM, Queen DS, Trager MH, Akilov OE, Geskin LJ, Fuschiotti P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood (2020) 136(15):1748–59. doi: 10.1182/blood.2019004725
64. Wang T, Feldman AL, Wada DA, Lu Y, Polk A, Briski R, et al. GATA-3 expression identifies a high-risk subset of PTCL, NOS with distinct molecular and clinical features. Blood J Am Soc Hematol (2014) 123(19):3007–15. doi: 10.1182/blood-2013-12-544809
65. Weiss J, Reneau J, Wilcox RA. PTCL, NOS: An update on classification, risk-stratification, and treatment. Front Oncol (2023) 13. doi: 10.3389/fonc.2023.1101441
66. Bennani NN, Ansell SM. Peripheral T-cell lymphoma not otherwise specified. In: The peripheral T-cell lymphomas. (Wiley Online) (2021). p. 105–14.
67. Chao Y, Liu Z. Biomaterials tools to modulate the tumour microenvironment in immunotherapy. Nat Rev Bioeng (2023) 1(2):125–38. doi: 10.1038/s44222-022-00004-6
68. Pulitzer MP, Horna P, Almeida J. Sézary syndrome and mycosis fungoides: An overview, including the role of immunophenotyping. Cytometry Part B: Clin Cytometry (2021) 100(2):132–8. doi: 10.1002/cyto.b.21888
69. Rendón-Serna N, Correa-Londoño LA, Velásquez-Lopera MM, Bermudez-Muñoz M. Cell signaling in cutaneous T-cell lymphoma microenvironment: promising targets for molecular-specific treatment. Int J Dermatol (2021) 60(12):1462–80. doi: 10.1111/ijd.15451
70. Van Arnam JS, Lim MS, Elenitoba-Johnson KS. Novel insights into the pathogenesis of T-cell lymphomas. Blood J Am Soc Hematol (2018) 131(21):2320–30. doi: 10.1182/blood-2017-11-764357
71. Cortes JR, Patrone CC, Quinn SA, Gu Y, Sanchez-Martin M, Mackey A, et al. Jak-STAT inhibition mediates romidepsin and mechlorethamine synergism in cutaneous T-cell lymphoma. J Invest Dermatol (2021) 141(12):2908–20. doi: 10.1016/j.jid.2021.04.023
72. Junkins-Hopkins JM, Kim EJ. Aggressive cytotoxic primary cutaneous T-cell lymphomas. In: Hematopathology of the skin: clinical & Pathological approach (Wolters Kluwer) (2023).
73. Sequeira C, Ozer H. Immunology of the lymphomas. In: Neoplastic diseases of the blood (Springer) (2018). p. 827–52.
74. Yanagi T, Nishihara H, Fujii K, Nishimura M, Narahira A, Takahashi K, et al. Comprehensive cancer-related gene analysis reveals that active KRAS mutation is a prognostic mutation in mycosis fungoides. J Dermatol Sci (2017) 88(3):367–70. doi: 10.1016/j.jdermsci.2017.07.013
75. Wobser M, Roth S, Appenzeller S, Houben R, Schrama D, Goebeler M, et al. Targeted deep sequencing of mycosis fungoides reveals intracellular signaling pathways associated with aggressiveness and large cell transformation. Cancers (2021) 13(21):5512. doi: 10.3390/cancers13215512
76. Khadzhieva MB, Zakharova ES, Kalinina EV, Abramov DS, Rumyantsev AG, Larin SS. Molecular genetic features of cutaneous T-cell lymphomas development on example of mycosis fungoides and Sezary syndrome. Oncohematology (2022) 17(1):65–74. doi: 10.17650/1818-8346-2022-17-1-65-74
77. Zhang XH, Nam S, Wu J, Chen CH, Liu X, Li H, et al. Multi-kinase inhibitor with anti-p38γ Activity in cutaneous T-cell lymphoma. J Invest Dermatol (2018) 138(11):2377–87. doi: 10.1016/j.jid.2018.04.030
78. Bresin A, Cristofoletti C, Caprini E, Cantonetti M, Monopoli A, Russo G, et al. Preclinical evidence for targeting PI3K/mTOR signaling with dual-inhibitors as a therapeutic strategy against cutaneous T-cell lymphoma. J Invest Dermatol (2020) 140(5):1045–53. doi: 10.1016/j.jid.2019.08.454
79. Karagianni F, Pavlidis A, Malakou LS, Piperi C, Papadavid E. Predominant role of mTOR signaling in skin diseases with therapeutic potential. Int J Mol Sci (2022) 23(3):1693. doi: 10.3390/ijms23031693
80. Sun Z, Yao X, Ding X, Li X, Tian X. MicroRNAs and their signaling pathway in mycosis fungoides. Medicine (2022) 101(25):e29248. doi: 10.1097/MD.0000000000029248
81. Elenitoba-Johnson KS, Wilcox R. A new molecular paradigm in mycosis fungoides and Sezary syndrome. Semin Diagn Pathol (2017) 34(1):15–21. WB Saunders. doi: 10.1053/j.semdp.2016.11.002
82. Wartewig T, Ruland J. PD-1 tumor suppressor signaling in T cell lymphomas. Trends Immunol (2019) 40(5):403–14. doi: 10.1016/j.it.2019.03.005
83. da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen CP, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet (2015) 47(12):1465–70. doi: 10.1038/ng.3442
84. Manfè V, Biskup E, Rosbjerg A, Kamstrup M, Skov AG, Lerche CM, et al. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PloS One (2012) 7(1):e29541. doi: 10.1371/journal.pone.0029541
85. Yu L, Wei J, Liu P. Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. Semin Cancer Biol (2022) 85:69–94. Academic Press. doi: 10.1016/j.semcancer.2021.06.019
86. Murugan AK. mTOR: Role in cancer, metastasis and drug resistance. Semin Cancer Biol (2019) 59:92–111. Academic Press. doi: 10.1016/j.semcancer.2019.07.003
87. Patil K, Kuttikrishnan S, Khan AQ, Ahmad F, Alam M, Buddenkotte J, et al. Molecular pathogenesis of Cutaneous T cell Lymphoma: Role of chemokines, cytokines, and dysregulated signaling pathways. Semin Cancer Biol (2022) 86:382–99. Academic Press. doi: 10.1016/j.semcancer.2021.12.003
88. Ikeda S, Kitadate A, Ito M, Abe F, Nara M, Watanabe A, et al. Disruption of CCL20-CCR6 interaction inhibits metastasis of advanced cutaneous T-cell lymphoma. Oncotarget (2016) 7(12):13563. doi: 10.18632/oncotarget.6916
89. Salaroglio IC, Mungo E, Gazzano E, Kopecka J, Riganti C. ERK is a pivotal player of chemo-immune-resistance in cancer. Int J Mol Sci (2019) 20(10):2505. doi: 10.3390/ijms20102505
90. Patil K, Sher G, Kuttikrishnan S, Moton S, Alam M, Buddenkotte J, et al. The cross-talk between miRNAs and JAK/STAT pathway in cutaneous T cell lymphoma: Emphasis on therapeutic opportunities. Semin Cell Dev Biol (2022). Academic Press. doi: 10.1016/j.semcdb.2022.09.015
91. Isabelle C, Boles A, Chakravarti N, Porcu P, Brammer J, Mishra A. Cytokines in the pathogenesis of large granular lymphocytic leukemia. Front Oncol (2022) 712. doi: 10.3389/fonc.2022.849917
92. Redell MS, Tweardy DJ. Targeting transcription factors for cancer therapy. Curr Pharm design (2005) 11(22):2873–87. doi: 10.2174/1381612054546699
93. Altman BJ, Dang CV. Normal and cancer cell metabolism: lymphocytes and lymphoma. FEBS J (2012) 279(15):2598–609. doi: 10.1111/j.1742-4658.2012.08651.x
94. Kittipongdaja W, Wu X, Garner J, Liu X, Komas SM, Hwang ST, et al. Rapamycin suppresses tumor growth and alters the metabolic phenotype in T-cell lymphoma. J Invest Dermatol (2015) 135(9):2301–8. doi: 10.1038/jid.2015.153
95. Han TJ, Xu HZ, Li JS, Geng LY, Li XY, Zhou XX, et al. Leptin and its receptor in glucose metabolism of T‐cell lymphoma. Oncol Lett (2018) 16(5):5838–46. doi: 10.3892/ol.2018.9356
96. Bertness V, Kirsch I, Hollis G, Johnson B, Bunn PA Jr. T-cell receptor gene rearrangements as clinical markers of human T-cell lymphomas. N Engl J Med (1985) 313(9):534–8. doi: 10.1056/NEJM198508293130902
97. Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga JI, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet (2015) 47(11):1304–15. doi: 10.1038/ng.3415
98. Damsky WE, Choi J. Genetics of cutaneous T cell lymphoma: from bench to bedside. Curr Treat options Oncol (2016) 17:1–14. doi: 10.1007/s11864-016-0410-8
99. Yoo HY, Kim P, Kim WS, Lee SH, Kim S, Kang SY, et al. Frequent CTLA4-CD28 gene fusion in diverse types of T-cell lymphoma. Haematologica (2016) 101(6):757. doi: 10.3324/haematol.2015.139253
100. Han Y, Liu D, Li L. PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res (2020) 10(3):727.
101. Dong Y, Sun Q, Zhang X. PD-1 and its ligands are important immune checkpoints in cancer. Oncotarget (2017) 8(2):2171. doi: 10.18632/oncotarget.13895
102. Dai S, Jia R, Zhang X, Fang Q, Huang L. The PD-1/PD-Ls pathway and autoimmune diseases. Cell Immunol (2014) 290(1):72–9. doi: 10.1016/j.cellimm.2014.05.006
103. Phillips D, Matusiak M, Gutierrez BR, Bhate SS, Barlow GL, Jiang S, et al. Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nat Commun (2021) 12(1):6726. doi: 10.1038/s41467-021-26974-6
104. Hamanishi J, Mandai M, Matsumura N, Abiko K, Baba T, Konishi I. PD-1/PD-L1 blockade in cancer treatment: perspectives and issues. Int J Clin Oncol (2016) 21:462–73. doi: 10.1007/s10147-016-0959-z
105. Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat (2018) 167:671–86. doi: 10.1007/s10549-017-4537-5
106. Elsayad K, Stadler R, Steinbrink K, Eich HT. Combined total skin radiotherapy and immune checkpoint inhibitors: A promising potential treatment for mycosis fungoides and Sezary syndrome. JDDG: J der Deutschen Dermatologischen Gesellschaft (2020) 18(3):193–7. doi: 10.1111/ddg.14044
107. Khodadoust MS, Rook AH, Porcu P, Foss F, Moskowitz AJ, Shustov A, et al. Pembrolizumab in relapsed and refractory mycosis fungoides and Sézary syndrome: a multicenter phase II study. J Clin Oncol (2020) 38(1):20. doi: 10.1200/JCO.19.01056
108. Wang T, Lu Y, Polk A, Chowdhury P, Zamalloa CM, Fujiwara H, et al. T-cell receptor signaling activates an ITK/NF-κB/GATA-3 axis in T-cell lymphomas facilitating resistance to chemotherapy. Clin Cancer Res (2017) 23(10):2506–15. doi: 10.1158/1078-0432.CCR-16-1996
109. Durgin JS, Weiner DM, Wysocka M, Rook AH. The immunopathogenesis and immunotherapy of cutaneous T cell lymphoma: Pathways and targets for immune restoration and tumor eradication. J Am Academy (2021) 84(3):587–95. doi: 10.1016/j.jaad.2020.12.027
110. Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet (2015) 47(9):1056–60. doi: 10.1038/ng.3370
111. Vanamee É.S, Faustman DL. TNFR2: a novel target for cancer immunotherapy. Trends Mol Med (2017) 23(11):1037–46. doi: 10.1016/j.molmed.2017.09.007
112. Braun FCM, Grabarczyk P, Möbs M, Braun FK, Eberle J, Beyer M, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sezary syndrome. Leukemia (2011) 25(9):1494–501. doi: 10.1038/leu.2011.101
113. Prasad S, Ravindran J, Aggarwal BB. NF-κB and cancer: how intimate is this relationship. Mol Cell Biochem (2010) 336:25–37. doi: 10.1007/s11010-009-0267-2
114. Grondona P, Bucher P, Schulze-Osthoff K, Hailfinger S, Schmitt A. NF-κB activation in lymphoid Malignancies: genetics, signaling, and targeted therapy. Biomedicines (2018) 6(2):38. doi: 10.3390/biomedicines6020038
115. Huen AO, Rook AH. Toll receptor agonist therapy of skin cancer and cutaneous T-cell lymphoma. Curr Opin Oncol (2014) 26(2):237–44. doi: 10.1097/CCO.0000000000000048
116. El Tawdy AM, Amin IM, Abdel Hay RM, Hassan AS, Gad ZS, Rashed LA. Toll-like receptor (TLR) 7 expression in mycosis fungoides and psoriasis: a case–control study. Clin Exp Dermatol (2017) 42(2):172–7. doi: 10.1111/ced.13008
117. Harsini S, Beigy M, Akhavan-Sabbagh M, Rezaei N. Toll-like receptors in lymphoid Malignancies: double-edged sword. Crit Rev Oncol/Hematol (2014) 89(2):262–83. doi: 10.1016/j.critrevonc.2013.08.010
118. Kunz W. The role of the chemokine CCL22 in the interaction of dendritic cells and regulatory T cells (Doctoral dissertation, lmu) (Germany: Ludwig Maximilian University of Munich) (2017).
119. Gao D, Li W, Wang W, Cai Y, Wang Y, Luo X, et al. Synergy of purine-scaffold TLR7 agonist with doxorubicin on systemic inhibition of lymphoma in mouse model. J Cancer (2017) 8(16):3183. doi: 10.7150/jca.20015
120. Zhu J, He S, Du J, Wang Z, Li W, Chen X, et al. Local administration of a novel Toll-like receptor 7 agonist in combination with doxorubicin induces durable tumouricidal effects in a murine model of T cell lymphoma. J Hematol Oncol (2015) 8(1):1–10. doi: 10.1186/s13045-015-0121-9
121. Bubna AK. Imiquimod-Its role in the treatment of cutaneous Malignancies. Indian J Pharmacol (2015) 47(4):354. doi: 10.4103/0253-7613.161249
122. Hussein MR. Ultraviolet radiation and skin cancer: molecular mechanisms. J cutaneous Pathol (2005) 32(3):191–205. doi: 10.1111/j.0303-6987.2005.00281.x
123. Tsai KY, Tsao H. The genetics of skin cancer. Am J Med Genet Part C: Semin Med Genet (2004) 131(1):82–92. Hoboken: Wiley Subscription Services, Inc., A Wiley Company. doi: 10.1002/ajmg.c.30037
124. Greinert R. Skin cancer: new markers for better prevention. Pathobiology (2009) 76(2):64–81. doi: 10.1159/000201675
125. Jones CL, Degasperi A, Grandi V, Amarante TD, Mitchell TJ, Nik-Zainal S, et al. Spectrum of mutational signatures in T-cell lymphoma reveals a key role for UV radiation in cutaneous T-cell lymphoma. Sci Rep (2021) 11(1):1–13. doi: 10.1038/s41598-021-83352-4
126. Yumeen S, Girardi M. Focus: skin: insights into the molecular and cellular underpinnings of cutaneous T cell lymphoma. Yale J Biol Med (2020) 93(1):111.
127. Torres AB, Najidh S, Tensen CP, Vermeer MH. Molecular advances in cutaneous T-cell lymphoma. Semin cutaneous Med Surg (2018) 37(1):81–5. doi: 10.12788/j.sder.2018.007
128. Brash DE. UV signature mutations. Photochem Photobiol (2015) 91(1):15–26. doi: 10.1111/php.12377
129. Gallardo F, Pujol RM. Genetics abnorMalities with clinical impact in primary cutaneous lymphomas. Cancers (2022) 14(20):4972. doi: 10.3390/cancers14204972
130. Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis (2018) 7(1):10. doi: 10.1038/s41389-017-0011-9
131. Alcántara-Hernández M, Torres-Zárate C, Pérez-Montesinos G, Jurado-Santacruz F, Domínguez-Gómez MA, Peniche-Castellanos A, et al. Overexpression of hypoxia-inducible factor 1 alpha impacts FoxP3 levels in mycosis fungoides—Cutaneous T-cell lymphoma: Clinical implications. Int J Cancer (2014) 134(9):2136–45. doi: 10.1002/ijc.28546
132. Geindreau M, Ghiringhelli F, Bruchard M. Vascular endothelial growth factor, a key modulator of the anti-tumor immune response. Int J Mol Sci (2021) 22(9):4871. doi: 10.3390/ijms22094871
133. Pileri A, Guglielmo A, Grandi V, Violetti SA, Fanoni D, Fava P, et al. The microenvironment’s role in mycosis fungoides and Sézary syndrome: from progression to therapeutic implications. Cells (2021) 10(10):2780. doi: 10.3390/cells10102780
134. Jankowska-Konsur A, Maj J, Woźniak Z, Baran E. Angiogenesis assessment in patients with mycosis fungoides. Adv Dermatol Allergology/Postępy Dermatologii i Alergologii (2009) 26(4):186–9.
135. Bagherani N, Smoller BR. An overview of cutaneous T cell lymphomas. F1000Research (2016) 5:F1000 Faculty Rev-1882. doi: 10.12688/f1000research.8829.1
136. Miyagaki T, Sugaya M, Oka T, Takahashi N, Kawaguchi M, Suga H, et al. Placental growth factor and vascular endothelial growth factor together regulate tumour progression via increased vasculature in cutaneous T-cell lymphoma. Acta Dermato-Venereologica (2017) 97(5):586–92. doi: 10.2340/00015555-2623
137. Serrano L, Martinez-Escala ME, Zhou XA, Guitart J. Pruritus in cutaneous T-cell lymphoma and its management. Dermatologic Clinics (2018) 36(3):245–58. doi: 10.1016/j.det.2018.02.011
138. Birch-Machin MA, Russell EV, Latimer JA. Mitochondrial DNA damage as a biomarker for ultraviolet radiation exposure and oxidative stress. Br J Dermatol (2013) 169(s2):9–14. doi: 10.1111/bjd.12207
139. He H, Xiong L, Jian L, Li L, Wu Y, Qiao S. Role of mitochondria on UV-induced skin damage and molecular mechanisms of active chemical compounds targeting mitochondria. J Photochem Photobiol B: Biol (2022) 232:112464. doi: 10.1016/j.jphotobiol.2022.112464
140. Vacca A, Moretti S, Ribatti D, Pellegrino A, Pimpinelli N, Bianchi B, et al. Progression of mycosis fungoides is associated with changes in angiogenesis and expression of the matrix metalloproteinases 2 and 9. Eur J Cancer (1997) 33(10):1685–92. doi: 10.1016/S0959-8049(97)00186-X
141. Aronovich A, Moyal L, Gorovitz B, Amitay-Laish I, Naveh HP, Forer Y, et al. Cancer-associated fibroblasts in mycosis fungoides promote tumor cell migration and drug resistance through CXCL12/CXCR4. J Invest Dermatol (2021) 141(3):619–27. doi: 10.1016/j.jid.2020.06.034
142. Pizzi M, Margolskee E, Inghirami G. Pathogenesis of peripheral T cell lymphoma. Annu Rev Pathology: Mech Dis (2018) 13:293–320. doi: 10.1146/annurev-pathol-020117-043821
143. Chen X, Wu W, Wei W, Zou L. Immune checkpoint inhibitors in peripheral T-cell lymphoma. Front Pharmacol (2022) 13:1482. doi: 10.3389/fphar.2022.869488
144. Lemonnier F, Mak TW. Angioimmunoblastic T-cell lymphoma: more than a disease of T follicular helper cells. J Pathol (2017) 242(4):387–90. doi: 10.1002/path.4920
145. Agostinelli C, Hartmann S, Klapper W, Korkolopoulou P, Righi S, Marafioti T, et al. Peripheral T cell lymphomas with follicular T helper phenotype: a new basket or a distinct entity? Revising Karl Lennert’s personal archive. Histopathology (2011) 59(4):679–91. doi: 10.1111/j.1365-2559.2011.03981.x
146. Pileri SA. Follicular helper T-cell–related lymphomas. Blood J Am Soc Hematol (2015) 126(15):1733–4. doi: 10.1182/blood-2015-08-665075
147. O’Leary HM, Savage KJ. Update on the World Health Organization classification of peripheral T-cell lymphomas. Curr Hematol Malig Rep (2009) 4(4):227–35. doi: 10.1007/s11899-009-0030-5
148. Lunning MA, Vose JM. Angioimmunoblastic T-cell lymphoma: the many-faced lymphoma. Blood J Am Soc Hematol (2017) 129(9):1095–102. doi: 10.1182/blood-2016-09-692541
149. Yabe M, Dogan A, Horwitz SM, Moskowitz AJ. Angioimmunoblastic T-cell lymphoma. T-Cell NK-Cell Lymphomas: From Biol to Novel Therapies (2019) 176:99–126. doi: 10.1007/978-3-319-99716-2_5
150. Liu HL, Liao CK, Weng SW, Shih LY, Chou CC, You HL, et al. Flow cytometry analysis reveals a wide cytologic and immunophenotypic spectrum of peripheral B lymphocytes in angioimmunoblastic T-cell lymphoma. Pathobiology (2022) 90:1–12. doi: 10.1159/000526284
151. Gaulard P, De Leval L. Follicular helper T cells: implications in neoplastic hematopathology. Semin Diagn Pathol (2011) 28(3):202–13. WB Saunders. doi: 10.1053/j.semdp.2011.03.003
152. De Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic T-cell lymphoma. Br J Haematol (2010) 148(5):673–89. doi: 10.1111/j.1365-2141.2009.08003.x
153. Nutt SL, Tarlinton DM. Germinal center B and follicular helper T cells: siblings, cousins or just good friends? Nat Immunol (2011) 12(6):472–7. doi: 10.1038/ni.2019
154. Biram A, Davidzohn N, Shulman Z. T cell interactions with B cells during germinal center formation, a three-step model. Immunol Rev (2019) 288(1):37–48. doi: 10.1111/imr.12737
155. Rao DA. T cells that help B cells in chronically inflamed tissues. Front Immunol (2018) 9:1924. doi: 10.3389/fimmu.2018.01924
156. Song W, Craft J. T follicular helper cell heterogeneity: time, space, and function. Immunol Rev (2019) 288(1):85–96. doi: 10.1111/imr.12740
157. de Leval L, Gaulard P. Pathology and biology of peripheral T-cell lymphomas. Histopathology (2011) 58(1):49–68. doi: 10.1111/j.1365-2559.2010.03704.x
158. Gaulard P, de Leval L. Pathology of peripheral T-cell lymphomas: where do we stand? Semin Hematol (2014) 51(1):5–16. WB Saunders. doi: 10.1053/j.seminhematol.2013.11.003
159. Gaulard P, Hoeller S. Misleading features of bone marrow involvement by peripheral T-cell lymphomas. In: Bone marrow lymphoid infiltrates: diagnosis and clinical impact (London: Springer) (2012). p. 253–70.
160. Attygalle A, Al-Jehani R, Diss TC, Munson P, Liu H, Du MQ, et al. Neoplastic T cells in angioimmunoblastic T-cell lymphoma express CD10. Blood J Am Soc Hematol (2002) 99(2):627–33. doi: 10.1182/blood.V99.2.627
161. Witalis M. Immune mechanisms controlling angioimmunoblastic T cell lymphoma progression (Quebec, Canada: University of Montreal) (2021).
162. Delfau-Larue MH, de Leval L, Joly B, Plonquet A, Challine D, Parrens M, et al. Targeting intratumoral B cells with rituximab in addition to CHOP in angioimmunoblastic T-cell lymphoma. A clinicobiological study of the GELA. haematologica (2012) 97(10):1594. doi: 10.3324/haematol.2011.061507
163. Verma NK, Wong BHS, Poh ZS, Udayakumar A, Verma R, Goh RKJ, et al. Obstacles for T-lymphocytes in the tumour microenvironment: Therapeutic challenges, advances and opportunities beyond immune checkpoint. EBioMedicine (2022) 83:104216. doi: 10.1016/j.ebiom.2022.104216
164. Shimizu K, Iyoda T, Okada M, Yamasaki S, Fujii SI. Immune suppression and reversal of the suppressive tumor microenvironment. Int Immunol (2018) 30(10):445–55. doi: 10.1093/intimm/dxy042
165. Peña-Romero AC, Orenes-Piñero E. Dual effect of immune cells within tumour microenvironment: pro-and anti-tumour effects and their triggers. Cancers (2022) 14:1681. doi: 10.3390/cancers14071681
166. Basnet A. Angioimmunoblastic T-cell lymphoma: an immunological masquerade. QJM: Int J Med (2023) 116(7):577–8. doi: 10.1093/qjmed/hcad057
167. Hussain M, Adah D, Tariq M, Lu Y, Zhang J, Liu J. CXCL13/CXCR5 signaling axis in cancer. Life Sci (2019) 227:175–86. doi: 10.1016/j.lfs.2019.04.053
168. Grogg KL, Attygale AD, Macon WR, Remstein ED, Kurtin PJ, Dogan A. Expression of CXCL13, a chemokine highly upregulated in germinal center T-helper cells, distinguishes angioimmunoblastic T-cell lymphoma from peripheral T-cell lymphoma, unspecified. Modern Pathol (2006) 19(8):1101–7. doi: 10.1038/modpathol.3800625
169. Kaffenberger B, Haverkos B, Tyler K, Wong H, Porcu P, Gru AA. Extranodal Marginal Zone Lymphoma (MALT)–Like Presentations of Angioimmunoblastic T-cell lymphoma: A T-cell lymphoma masquerading as a B-cell lymphoproliferative disorder. Am J Dermatopathol (2015) 37(8):604. doi: 10.1097%2FDAD.0000000000000266
170. Chang CJ, Cheng JH, Lin MS, Dai YC, Hsiue TR. Eosinophilic pleural effusion as the first presentation of angioimmunoblastic T cell lymphoma. J Formosan Med Assoc (2007) 106(2):156–60. doi: 10.1016/S0929-6646(09)60232-1
171. Timmins MA, Wagner SD, Ahearne MJ. The new biology of PTCL-NOS and AITL: current status and future clinical impact. Br J Haematol (2020) 189(1):54–66. doi: 10.1111/bjh.16428
172. Fajardo Despaigne JE. Functional role of follicular helper T cells in chronic lymphocytic leukemia (Master’s thesis) (Winnipeg, Canada: University of Manitoba) (2020).
173. Willard-Gallo KE, Badran BM, Ravoet M, Zerghe A, Burny A, Martiat P, et al. Defective CD3γ gene transcription is associated with NFATc2 overexpression in the lymphocytic variant of hypereosinophilic syndrome. Exp Hematol (2005) 33(10):1147–59. doi: 10.1016/j.exphem.2005.06.027
174. de Leval L, Gaulard P. Mature T-cell lymphomas. Diagn Histopathol (2015) 21(10):408–20. doi: 10.1016/j.mpdhp.2015.09.005
175. Tse E, Kwong YL. T-cell lymphoma: microenvironment-related biomarkers. Semin Cancer Biol (2015) 34:46–51. Academic Press. doi: 10.1016/j.semcancer.2015.06.001
176. Tripodo C, Gri G, Piccaluga PP, Frossi B, Guarnotta C, Piconese S, et al. Mast cells and Th17 cells contribute to the lymphoma-associated pro-inflammatory microenvironment of angioimmunoblastic T-cell lymphoma. Am J Pathol (2010) 177(2):792–802. doi: 10.2353/ajpath.2010.091286
177. Eivazi S, Bagheri S, Hashemzadeh MS, Ghalavand M, Qamsari ES, Dorostkar R, et al. Development of T follicular helper cells and their role in disease and immune system. Biomedicine pharmacotherapy (2016) 84:1668–78. doi: 10.1016/j.biopha.2016.10.083
178. Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res (2010) 20(1):4–12. doi: 10.1038/cr.2009.138
179. Facchetti F, Lorenzi L. Follicular dendritic cells and related sarcoma. Semin Diagn Pathol (2016) 33(5):262–76. WB Saunders. doi: 10.1053/j.semdp.2016.05.002
180. Dolcetti R, Dal Col J, Martorelli D, Carbone A, Klein E. Interplay among viral antigens, cellular pathways and tumor microenvironment in the pathogenesis of EBV-driven lymphomas. Semin Cancer Biol (2013) 23(6):441–56. Academic Press. doi: 10.1016/j.semcancer.2013.07.005
181. Fan TW, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, et al. Altered regulation of metabolic pathways in human lung cancer discerned by 13C stable isotope-resolved metabolomics (SIRM). Mol Cancer (2009) 8(1):1–19. doi: 10.1186/1476-4598-8-41
182. Chen WL, Wang JH, Zhao AH, Xu X, Wang YH, Chen TL, et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood J Am Soc Hematol (2014) 124(10):1645–54. doi: 10.1182/blood-2014-02-554204
183. Kancherla P, Daneshvar M, Sager RA, Mollapour M, Bratslavsky G. Fumarate hydratase as a therapeutic target in renal cancer. Expert Opin Ther Targets (2020) 24(9):923–36. doi: 10.1080/14728222.2020.1804862
184. Sulkowski PL, Sundaram RK, Oeck S, Corso CD, Liu Y, Noorbakhsh S, et al. Krebs-cycle-deficient hereditary cancer syndromes are defined by defects in homologous-recombination DNA repair. Nat Genet (2018) 50(8):1086–92. doi: 10.1038/s41588-018-0170-4
185. Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta (BBA)-Bioenergetics (2011) 1807(11):1432–43. doi: 10.1016/j.bbabio.2011.07.003
186. Chisolm DA, Weinmann AS. TCR-signaling events in cellular metabolism and specialization. Front Immunol (2015) 6:292. doi: 10.3389/fimmu.2015.00292
187. Daniels MA, Teixeiro E. TCR signaling in T cell memory. Front Immunol (2015) 6:617. doi: 10.3389/fimmu.2015.00617
188. Klein Geltink RI, Kyle RL, Pearce EL. Unraveling the complex interplay between T cell metabolism and function. Annu Rev Immunol (2018) 36:461–88. doi: 10.1146/annurev-immunol-042617-053019
189. Cornish GH, Sinclair LV, Cantrell DA. Differential regulation of T-cell growth by IL-2 and IL-15. Blood (2006) 108(2):600–8. doi: 10.1182/blood-2005-12-4827
190. Ross SH, Cantrell DA. Signaling and function of interleukin-2 in T lymphocytes. Annu Rev Immunol (2018) 36:411–33. doi: 10.1146/annurev-immunol-042617-053352
191. Bishop EL, Gudgeon N, Dimeloe S. Control of T cell metabolism by cytokines and hormones. Front Immunol (2021) 12:653605. doi: 10.3389/fimmu.2021.653605
192. Lochner M, Berod L, Sparwasser T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol (2015) 36(2):81–91. doi: 10.1016/j.it.2014.12.005
193. Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol (2020) 20(1):55–70. doi: 10.1038/s41577-019-0203-y
194. Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell (2011) 146(5):772–84. doi: 10.1016/j.cell.2011.07.033
195. Kheshtchin N, Hadjati J. Targeting hypoxia and hypoxia-inducible factor-1 in the tumor microenvironment for optimal cancer immunotherapy. J Cell Physiol (2022) 237(2):1285–98. doi: 10.1002/jcp.30643
196. Shyer JA, Flavell RA, Bailis W. Metabolic signaling in T cells. Cell Res (2020) 30(8):649–59. doi: 10.1038/s41422-020-0379-5
197. Kawata T, Tada K, Kobayashi M, Sakamoto T, Takiuchi Y, Iwai F, et al. Dual inhibition of the mTORC 1 and mTORC 2 signaling pathways is a promising therapeutic target for adult T-cell leukemia. Cancer Sci (2018) 109(1):103–11. doi: 10.1111/cas.13431
198. Linke M, Fritsch SD, Sukhbaatar N, Hengstschläger M, Weichhart T. mTORC 1 and mTORC 2 as regulators of cell metabolism in immunity. FEBS Lett (2017) 591(19):3089–103. doi: 10.1002/1873-3468.12711
199. Liu C, Chapman NM, Karmaus PW, Zeng H, Chi H. mTOR and metabolic regulation of conventional and regulatory T cells. J leukocyte Biol (2015) 97(5):837–47. doi: 10.1189/jlb.2RI0814-408R
200. Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity (2002) 16(6):769–77. doi: 10.1016/S1074-7613(02)00323-0
201. Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K, et al. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell (2004) 16(5):819–30. doi: 10.1016/j.molcel.2004.11.014
202. Bonora M, Giorgi C, Pinton P. Molecular mechanisms and consequences of mitochondrial permeability transition. Nat Rev Mol Cell Biol (2022) 23(4):266–85. doi: 10.1038/s41580-021-00433-y
203. Liu Y, Gu W. The complexity of p53-mediated metabolic regulation in tumor suppression. Semin Cancer Biol (2022) 85:4–32. Academic Press. doi: 10.1016/j.semcancer.2021.03.010
204. Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol (2013) 23(12):620–33. doi: 10.1016/j.tcb.2013.07.006
205. Karjalainen K. Candidate therapeutic targets against acute myeloid leukemia identified via screening combinatorial peptide and chemical libraries. (Finland: University of Helsinki) (2015).
206. Rahman M, Hasan MR. Cancer metabolism and drug resistance. Metabolites (2015) 5(4):571–600. doi: 10.3390/metabo5040571
207. Jin J, Byun J-K, Choi Y-K, Park. K-G. Targeting glutamine metabolism as a therapeutic strategy for cancer. Exp Mol Med (2023) 55:1–10. doi: 10.1038/s12276-023-00971-9
208. Schaap FG, French PJ, Bovée JV. Mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in tumors. Adv Anat Pathol (2013) 20(1):32–8. doi: 10.1097/PAP.0b013e31827b654d
209. McKenney AS, Levine RL. Isocitrate dehydrogenase mutations in leukemia. J Clin Invest (2013) 123(9):3672–7. doi: 10.1172/JCI67266
210. Grabacka M, Plonka PM. Mitochondrial sirtuins at the crossroads of energy metabolism and oncogenic transformation. In: Sirtuin biology in cancer and metabolic disease. Elsevier, United States: Academic Press (2021). p. 103–26.
211. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
212. Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer (2006) 6(5):347–59. doi: 10.1038/nrc1880
213. Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol (2007) 8(5):451–6. doi: 10.1038/ni1453
214. Huang Y, De Reyniès A, De Leval L, Ghazi B, Martin-Garcia N, Travert M, et al. Gene expression profiling identifies emerging oncogenic pathways operating in extranodal NK/T-cell lymphoma, nasal type. Blood J Am Soc Hematol (2010) 115(6):1226–37. doi: 10.1182/blood-2009-05-221275
215. de Leval L, Rickman DS, Thielen C, Reynies AD, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood J Am Soc Hematol (2007) 109(11):4952–63. doi: 10.1182/blood-2006-10-055145
216. Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol (2014) 15(9):798–807. doi: 10.1038/ni.2940
217. Courtney AH, Lo WL, Weiss A. TCR signaling: mechanisms of initiation and propagation. Trends Biochem Sci (2018) 43(2):108–23. doi: 10.1016/j.tibs.2017.11.008
218. Agostinelli C, Rizvi H, Paterson J, Shende V, Akarca AU, Agostini E, et al. Intracellular TCR-signaling pathway: novel markers for lymphoma diagnosis and potential therapeutic targets. Am J Surg Pathol (2014) 38(10):1349–59. doi: 10.1097/PAS.0000000000000309
219. Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduct Target Ther (2021) 6(1):412. doi: 10.1038/s41392-021-00823-w
220. Warner K, Weit N, Crispatzu G, AdmIrand J, Jones D, Herling M. T-cell receptor signaling in peripheral T-cell lymphoma–a review of patterns of alterations in a central growth regulatory pathway. Curr Hematol Malig Rep (2013) 8:163–72. doi: 10.1007/s11899-013-0165-2
221. Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood J Am Soc Hematol (2019) 133(15):1664–76. doi: 10.1182/blood-2018-09-872549
222. Kinney MC, Higgins RA, Medina EA. Anaplastic large cell lymphoma: twenty-five years of discovery. Arch Pathol Lab Med (2011) 135(1):19–43. doi: 10.5858/2010-0507-RAR.1
223. Webb TR, Slavish J, George RE, Look AT, Xue L, Jiang Q, et al. Anaplastic lymphoma kinase: role in cancer pathogenesis and small-molecule inhibitor development for therapy. Expert Rev Anticancer Ther (2009) 9(3):331–56. doi: 10.1586/14737140.9.3.331
224. Li JP, Yang CY, Chuang HC, Lan JL, Chen DY, Chen YM, et al. The phosphatase JKAP/DUSP22 inhibits T-cell receptor signalling and autoimmunity by inactivating Lck. Nat Commun (2014) 5(1):1–13. doi: 10.1038/ncomms4618
225. Gaud G, Lesourne R, Love PE. Regulatory mechanisms in T cell receptor signalling. Nat Rev Immunol (2018) 18(8):485–97. doi: 10.1038/s41577-018-0020-8
226. Streubel B, Vinatzer U, Willheim M, Raderer M, Chott A. Novel t (5, 9)(q33; q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia (2006) 20(2):313–8. doi: 10.1038/sj.leu.2404045
227. Krisenko MO, Geahlen RL. Calling in SYK: SYK’s dual role as a tumor promoter and tumor suppressor in cancer. Biochim Biophys Acta (BBA)-Molecular Cell Res (2015) 1853(1):254–63. doi: 10.1016/j.bbamcr.2014.10.022
228. Cools J. RHOA mutations in peripheral T cell lymphoma. Nat Genet (2014) 46(4):320–1. doi: 10.1038/ng.2937
229. Kim JG, Islam R, Cho JY, Jeong H, Cap KC, Park Y, et al. Regulation of RhoA GTPase and various transcription factors in the RhoA pathway. J Cell Physiol (2018) 233(9):6381–92. doi: 10.1002/jcp.26487
230. Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet (2014) 46(4):371–5. doi: 10.1038/ng.2916
231. Zain JM, Hanona P. Aggressive T-cell lymphomas: 2021 Updates on diagnosis, risk stratification and management. Am J Hematol (2021) 96(8):1027–46. doi: 10.1002/ajh.26270
232. Heemskerk N, Schimmel L, Oort C, Van Rijssel J, Yin T, Ma B, et al. F-actin-rich contractile endothelial pores prevent vascular leakage during leukocyte diapedesis through local RhoA signalling. Nat Commun (2016) 7(1):10493. doi: 10.1038/ncomms10493
233. Stricker J, Falzone T, Gardel ML. Mechanics of the F-actin cytoskeleton. J Biomech (2010) 43(1):9–14. doi: 10.1016/j.jbiomech.2009.09.003
234. Calvo V, Izquierdo M. Role of actin cytoskeleton reorganization in polarized secretory traffic at the immunological synapse. Front Cell Dev Biol (2021) 9:629097. doi: 10.3389/fcell.2021.629097
235. Dostert C, Grusdat M, Letellier E, Brenner D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol Rev (2019) 99(1):115–60. doi: 10.1152/physrev.00045.2017
236. Müller D. Targeting co-stimulatory receptors of the TNF superfamily for cancer immunotherapy. BioDrugs (2023) 37(1):21–33. doi: 10.1007/s40259-022-00573-3
237. Picozza M, Cristofoletti C, Bresin A, Fioretti M, Sambucci M, Scala E, et al. Genetically driven CD39 expression affects sezary cell viability and IL-2 production and detects two patient subsets with distinct prognosis. J Invest Dermatol (2022) 142(11):3009–19. doi: 10.1016/j.jid.2022.04.017
238. Rohr J, Guo S, Huo J, Bouska A, Lachel C, Li Y, et al. Recurrent activating mutations of CD28 in peripheral T-cell lymphomas. Leukemia (2016) 30(5):1062–70. doi: 10.1038/leu.2015.357
239. Sekulic A, Liang WS, Tembe W, Izatt T, Kruglyak S, Kiefer JA, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA 4: CD 28 fusion. Mol Genet genomic Med (2015) 3(2):130–6. doi: 10.1002/mgg3.121
240. Zheng J, Chan PL, Liu Y, Qin G, Xiang Z, Lam KT, et al. ICOS regulates the generation and function of human CD4+ Treg in a CTLA-4 dependent manner. PloS One (2013) 8(12):e82203. doi: 10.1371/journal.pone.0082203
241. Wikenheiser DJ, Stumhofer JS. ICOS co-stimulation: friend or foe? Front Immunol (2016) 7:304. doi: 10.3389/fimmu.2016.00304
242. Amatore F, Gorvel L, Olive D. Role of Inducible Co-Stimulator (ICOS) in cancer immunotherapy. Expert Opin Biol Ther (2020) 20(2):141–50. doi: 10.1080/14712598.2020.1693540
243. Bansal-Pakala P, Gebre-Hiwot Jember A, Croft M. Signaling through OX40 (CD134) breaks peripheral T-cell tolerance. Nat Med (2001) 7(8):907–12. doi: 10.1038/90942
244. Deng J, Zhao S, Zhang X, Jia K, Wang H, Zhou C, et al. OX40 (CD134) and OX40 ligand, important immune checkpoints in cancer. OncoTargets Ther (2019) 12:7347. doi: 10.2147/OTT.S214211
245. Croft M. Control of immunity by the TNFR-related molecule OX40 (CD134). Annu Rev Immunol (2009) 28:57–78. doi: 10.1146/annurev-immunol-030409-101243
246. Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev (2009) 229(1):173–91. doi: 10.1111/j.1600-065X.2009.00766.x
247. Tsuchiya T, Ohshima K, Karube K, Yamaguchi T, Suefuji H, Hamasaki M, et al. Th1, Th2, and activated T-cell marker and clinical prognosis in peripheral T-cell lymphoma, unspecified: comparison with AILD, ALCL, lymphoblastic lymphoma, and ATLL. Blood (2004) 103(1):236–41. doi: 10.1182/blood-2002-05-1352
248. Bossard C, Dobay MP, Parrens M, Lamant L, Missiaglia E, Haioun C, et al. Immunohistochemistry as a valuable tool to assess CD30 expression in peripheral T-cell lymphomas: high correlation with mRNA levels. Blood J Am Soc Hematol (2014) 124(19):2983–6. doi: 10.1182/blood-2014-07-584953
249. Dillon SR, Gross JA, Ansell SM, Novak AJ. An APRIL to remember: novel TNF ligands as therapeutic targets. Nat Rev Drug Discov (2006) 5(3):235–46. doi: 10.1038/nrd1982
250. Freeman BE, Hammarlund E, Raué HP, Slifka MK. Regulation of innate CD8+ T-cell activation mediated by cytokines. Proc Natl Acad Sci (2012) 109(25):9971–6. doi: 10.1073/pnas.1203543109
251. Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, et al. STATs shape the active enhancer landscape of T cell populations. Cell (2012) 151(5):981–93. doi: 10.1016/j.cell.2012.09.044
252. Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood J Am Soc Hematol (2014) 123(19):2915–23. doi: 10.1182/blood-2013-11-536359
253. Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood J Am Soc Hematol (2014) 124(9):1460–72. doi: 10.1182/blood-2014-03-559542
254. Bergmann AK, Schneppenheim S, Seifert M, Betts MJ, Haake A, Lopez C, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosomes Cancer (2014) 53(4):309–16. doi: 10.1002/gcc.22141
255. Nairismägi ML, Gerritsen ME, Li ZM, Wijaya GC, Chia BKH, Laurensia Y, et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia (2018) 32(5):1147–56. doi: 10.1038/s41375-017-0004-x
256. Jerez A, Clemente MJ, Makishima H, Koskela H, LeBlanc F, Peng Ng K, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood J Am Soc Hematol (2012) 120(15):3048–57. doi: 10.1182/blood-2012-06-435297
257. Chen YW, Guo T, Shen L, Wong KY, Tao Q, Choi WW, et al. Receptor-type tyrosine-protein phosphatase κ directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood J Am Soc Hematol (2015) 125(10):1589–600. doi: 10.1182/blood-2014-07-588970
258. Han JJ, O’byrne M, Stenson MJ, Maurer MJ, Wellik LE, Feldman AL, et al. Prognostic and therapeutic significance of phosphorylated STAT3 and protein tyrosine phosphatase-6 in peripheral-T cell lymphoma. Blood Cancer J (2018) 8(11):110. doi: 10.1038/s41408-018-0138-8
259. Iżykowska K, Rassek K, Korsak D, Przybylski GK. Novel targeted therapies of T cell lymphomas. J Hematol Oncol (2020) 13(1):1–38. doi: 10.1186/s13045-020-01006-w
260. Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood J Am Soc Hematol (2014) 123(9):1293–6. doi: 10.1182/blood-2013-10-531509
261. Velusamy T, Kiel MJ, Sahasrabuddhe AA, Rolland D, Dixon CA, Bailey NG, et al. A novel recurrent NPM1-TYK2 gene fusion in cutaneous CD30-positive lymphoproliferative disorders. Blood J Am Soc Hematol (2014) 124(25):3768–71. doi: 10.1182/blood-2014-07-588434
262. Lehrskov LL, Christensen RH. The role of interleukin-6 in glucose homeostasis and lipid metabolism. Semin Immunopathology (2019) 41(4):491–9. Berlin/Heidelberg: Springer Berlin Heidelberg. doi: 10.1007/s00281-019-00747-2
263. Andrejeva G, Rathmell JC. Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metab (2017) 26(1):49–70. doi: 10.1016/j.cmet.2017.06.004
264. Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity (2009) 31(3):457–68. doi: 10.1016/j.immuni.2009.07.002
265. Soto-Heredero G, Gomez de las Heras MM, Gabandé-Rodríguez E, Oller J, Mittelbrunn M. Glycolysis–A key player in the inflammatory response. FEBS J (2020) 287(16):3350–69. doi: 10.1111/febs.15327
266. Pacella I, Procaccini C, Focaccetti C, Miacci S, Timperi E, Faicchia D, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci (2018) 115(28):E6546–55. doi: 10.1073/pnas.1720113115
267. Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood J Am Soc Hematol (2008) 111(4):2101–11. doi: 10.1182/blood-2007-06-096297
268. Marchetti P, Fovez Q, Germain N, Khamari R, Kluza J. Mitochondrial spare respiratory capacity: mechanisms, regulation, and significance in non-transformed and cancer cells. FASEB J (2020) 34(10):13106–24. doi: 10.1096/fj.202000767R
269. Coppola C, Hopkins B, Huhn S, Du Z, Huang Z, Kelly WJ. Investigation of the impact from IL-2, IL-7, and IL-15 on the growth and signaling of activated CD4+ T cells. Int J Mol Sci (2020) 21(21):7814. doi: 10.3390/ijms21217814
270. Argilés JM, Stemmler B, López-Soriano FJ, Busquets S. Inter-tissue communication in cancer cachexia. Nat Rev Endocrinol (2019) 15(1):9–20. doi: 10.1038/s41574-018-0123-0
271. Zhou X, Zhu X, Zeng H. Fatty acid metabolism in adaptive immunity. FEBS J (2023) 290(3):584–99. doi: 10.1111/febs.16296
272. Tian Y, Zajac AJ. IL-21 and T cell differentiation: consider the context. Trends Immunol (2016) 37(8):557–68. doi: 10.1016/j.it.2016.06.001
273. Hermans D, Gautam S, García-Cañaveras JC, Gromer D, Mitra S, Spolski R, et al. Lactate dehydrogenase inhibition synergizes with IL-21 to promote CD8+ T cell stemness and antitumor immunity. Proc Natl Acad Sci (2020) 117(11):6047–55. doi: 10.1073/pnas.1920413117
274. Loschinski R, Böttcher M, Stoll A, Bruns H, Mackensen A, Mougiakakos D. IL-21 modulates memory and exhaustion phenotype of T-cells in a fatty acid oxidation-dependent manner. Oncotarget (2018) 9(17):13125. doi: 10.18632/oncotarget.24442
275. Alizadeh D, Wong RA, Yang X, Wang D, Pecoraro JR, Kuo CF, et al. IL15 enhances CAR-T cell antitumor activity by reducing mTORC1 activity and preserving their stem cell memory phenotypeSuperior antitumor activity of CAR-T cells cultured in IL15. Cancer Immunol Res (2019) 7(5):759–72. doi: 10.1158/2326-6066.CIR-18-0466
276. Lin YJ, Goretzki A, Schülke S. Immune metabolism of IL-4-activated B cells and Th2 cells in the context of allergic diseases. Front Immunol (2021) 12:790658. doi: 10.3389/fimmu.2021.790658
277. Jacobs SR, Michalek RD, Rathmell JC. IL-7 is essential for homeostatic control of T cell metabolism in vivo. J Immunol (2010) 184(7):3461–9. doi: 10.4049/jimmunol.0902593
278. Liao P, Chang N, Xu B, Qiu Y, Wang S, Zhou L, et al. Amino acid metabolism: challenges and opportunities for the therapeutic treatment of leukemia and lymphoma. Immunol Cell Biol (2022) 100(7):507–28. doi: 10.1111/imcb.12557
279. Hasan A, Akhter N, Al-Roub A, Thomas R, Kochumon S, Wilson A, et al. TNF-α in combination with palmitate enhances IL-8 production via the MyD88-independent TLR4 signaling pathway: potential relevance to metabolic inflammation. Int J Mol Sci (2019) 20(17):4112. doi: 10.3390/ijms20174112
280. Komai T, Inoue M, Okamura T, Morita K, Iwasaki Y, Sumitomo S, et al. Transforming growth factor-β and interleukin-10 synergistically regulate humoral immunity via modulating metabolic signals. Front Immunol (2018) 9:1364. doi: 10.3389/fimmu.2018.01364
281. Vacaflores A, Chapman NM, Harty JT, Richer MJ, Houtman JC. Exposure of human CD4 T cells to IL-12 results in enhanced TCR-induced cytokine production, altered TCR signaling, and increased oxidative metabolism. PloS One (2016) 11(6):e0157175. doi: 10.1371/journal.pone.0157175
282. Athie-Morales V, Smits HH, Cantrell DA, Hilkens CM. Sustained IL-12 signaling is required for Th1 development. J Immunol (2004) 172(1):61–9. doi: 10.4049/jimmunol.172.1.61
283. McCormick SM, Heller NM. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine (2015) 75(1):38–50. doi: 10.1016/j.cyto.2015.05.023
284. Karpathiou G, Papoudou-Bai A, Ferrand E, Dumollard JM, Peoc’h M. STAT6: a review of a signaling pathway implicated in various diseases with a special emphasis in its usefulness in pathology. Pathology-Research Pract (2021) 223:153477. doi: 10.1016/j.prp.2021.153477
285. Chen XW, Zhou SF. Inflammation, cytokines, the IL-17/IL-6/STAT3/NF-κB axis, and tumorigenesis. Drug Design Dev Ther (2015) 9:2941. doi: 10.2147/DDDT.S86396
286. Sundrud MS, Trivigno C. Identity crisis of Th17 cells: many forms, many functions, many questions. Semin Immunol (2013) 25(4):263–72. Academic Press. doi: 10.1016/j.smim.2013.10.021
287. Lim C, Savan R. The role of the IL-22/IL-22R1 axis in cancer. Cytokine Growth factor Rev (2014) 25(3):257–71. doi: 10.1016/j.cytogfr.2014.04.005
288. Jiang R, Sun B. IL-22 signaling in the tumor microenvironment. In: Tumor microenvironment: the role of interleukins–part B (Springer, Cham) (2021). p. 81–8.
289. Whitley SK, Li M, Kashem SW, Hirai T, Igyártó BZ, Knizner K, et al. Local IL-23 is required for proliferation and retention of skin-resident memory TH17 cells. Sci Immunol (2022) 7(77):eabq3254. doi: 10.1126/sciimmunol.abq325
290. Singer EM, Nattkemper LA, Benoit BM, Klein RS, Shin DB, Didigu CA, et al. IL-31 is produced by the Malignant T cell population in cutaneous T-cell lymphoma and its expression correlates with pruritus. J Invest Dermatol (2013) 133:S159. doi: 10.1038/jid.2013.227
291. Kunimura K, Fukui Y. The molecular basis for IL-31 production and IL-31-mediated itch transmission: from biology to drug development. Int Immunol (2021) 33(12):731–6. doi: 10.1093/intimm/dxab065
292. Jiang W, Lian J, Yue Y, Zhang Y. IL-33/ST2 as a potential target for tumor immunotherapy. Eur J Immunol (2021) 51(8):1943–55. doi: 10.1002/eji.202149175
293. Allegra A, Innao V, Tartarisco G, Pioggia G, Casciaro M, Musolino C, et al. The ST2/interleukin-33 axis in hematologic Malignancies: the IL-33 paradox. Int J Mol Sci (2019) 20(20):5226. doi: 10.3390/ijms20205226
294. Voronov E, Dotan S, Krelin Y, Song X, Elkabets M, Carmi Y, et al. Unique versus redundant functions of IL-1α and IL-1β in the tumor microenvironment. Front Immunol (2013) 4:177. doi: 10.3389/fimmu.2013.00177
295. Sole Chimenti M, Tucci P, Candi E, Perricone R, Melino G, Willis A. Metabolic profiling of human CD4+ cells following treatment with methotrexate and anti-TNF-α infliximab. Cell Cycle (2013) 12(18):3025–36. doi: 10.4161/cc.26067
296. Guilbaud E, Gautier EL, Yvan-Charvet L. Macrophage origin, metabolic reprogramming and IL-1β signaling: Promises and pitfalls in lung cancer. Cancers (2019) 11(3):298. doi: 10.3390/cancers11030298
297. Urbano PC, Aguirre-Gamboa R, Ashikov A, van Heeswijk B, Krippner-Heidenreich A, Tijssen H, et al. TNF-α–induced protein 3 (TNFAIP3)/A20 acts as a master switch in TNF-α blockade–driven IL-17A expression. J Allergy Clin Immunol (2018) 142(2):517–29. TNF. doi: 10.1016/j.jaci.2017.11.024
298. Priyadharshini B, Loschi M, Newton RH, Zhang JW, Finn KK, Gerriets VA, et al. Cutting edge: TGF-β and phosphatidylinositol 3-kinase signals modulate distinct metabolism of regulatory T cell subsets. J Immunol (2018) 201(8):2215–9. doi: 10.4049/jimmunol.1800311
299. Dimeloe S, Gubser P, Loeliger J, Frick C, Develioglu L, Fischer M, et al. Tumor-derived TGF-β inhibits mitochondrial respiration to suppress IFN-γ production by human CD4+ T cells. Sci Signaling (2019) 12(599):eaav3334. doi: 10.1126/scisignal.aav3334
300. Delisle JS, Giroux M, Boucher G, Landry JR, Hardy MP, Lemieux S, et al. The TGF-β-Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes Immun (2013) 14(2):115–26. doi: 10.1038/gene.2012.63
301. Stoycheva D, Deiser K, Stärck L, Nishanth G, Schlüter D, Uckert W, et al. IFN-γ regulates CD8+ memory T cell differentiation and survival in response to weak, but not strong, TCR signals. J Immunol (2015) 194(2):553–9. doi: 10.4049/jimmunol.1402058
302. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol (2015) 6:639. doi: 10.3389/fimmu.2015.00639
303. Diller ML, KudChadkar RR, Delman KA, Lawson DH, Ford ML. Balancing inflammation: the link between Th17 and regulatory T cells. Mediators Inflamm (2016) 2016. doi: 10.1155/2016/6309219
304. Zhang Y, Liu Z, Tian M, Hu X, Wang L, Ji J, et al. The altered PD-1/PD-L1 pathway delivers the ‘one-two punch’effects to promote the Treg/Th17 imbalance in pre-eclampsia. Cell Mol Immunol (2018) 15(7):710–23. doi: 10.1038/cmi.2017.70
305. Kono M. New insights into the metabolism of Th17 cells. Immunol Med (2023) 46(1):15–24. doi: 10.1080/25785826.2022.2140503
306. Pastor-Fernández G, Mariblanca IR, Navarro MN. Decoding IL-23 signaling cascade for new therapeutic opportunities. Cells (2020) 9(9):2044. doi: 10.3390/cells9092044
307. Revu S, Wu J, Henkel M, Rittenhouse N, Menk A, Delgoffe GM, et al. IL-23 and IL-1β drive human Th17 cell differentiation and metabolic reprogramming in absence of CD28 costimulation. Cell Rep (2018) 22(10):2642–53. doi: 10.1016/j.celrep.2018.02.044
308. Povoleri GA, Lalnunhlimi S, Steel KJ, Agrawal S, O’Byrne AM, Ridley M, et al. Anti-TNF treatment negatively regulates human CD4+ T-cell activation and maturation in vitro, but does not confer an anergic or suppressive phenotype. Eur J Immunol (2020) 50(3):445–58. doi: 10.1002/eji.201948190
309. Carruthers DM, Arrol HP, Bacon PA, Young SP. Dysregulated intracellular Ca2+ stores and Ca2+ signaling in synovial fluid T lymphocytes from patients with chronic inflammatory arthritis. Arthritis Rheumatism: Off J Am Coll Rheumatol (2000) 43(6):1257–65. doi: 10.1002/1529-0131(200006)43:6<1257::AID-ANR8>3.0.CO;2-Q
310. Ali M, Ponchel F, Wilson KE, Francis MJ, Wu X, Verhoef A, et al. Rheumatoid arthritis synovial T cells regulate transcription of several genes associated with antigen-induced anergy. J Clin Invest (2001) 107(4):519–28. doi: 10.1172/JCI8027
311. Mjösberg J, Svensson J, Johansson E, Hellström L, Casas R, Jenmalm, M. C., et al. Systemic reduction of functionally suppressive CD4dimCD25highFoxp3+ Tregs in human second trimester pregnancy is induced by progesterone and 17β-estradiol. J Immunol (2009) 183(1):759–69. doi: 10.4049/jimmunol.0803654
312. Sarhadi VK, Armengol G. Molecular biomarkers in cancer. Biomolecules (2022) 12(8):1021. doi: 10.3390/biom12081021
313. O’connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol (2017) 14(3):169–86. doi: 10.1038/nrclinonc.2016.162
314. Füzéry AK, Levin J, Chan MM, Chan DW. Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin Proteomics (2013) 10(1):1–14. doi: 10.1186/1559-0275-10-13
315. Loo SW, Pui TS. Cytokine and cancer biomarkers detection: the dawn of electrochemical paper-based biosensor. Sensors (2020) 20(7):1854. doi: 10.3390/s20071854
316. Monastero RN, Pentyala S. Cytokines as biomarkers and their respective clinical cutoff levels. Int J Inflamm (2017) 2017. doi: 10.1155/2017/4309485
317. Sánchez-Zauco N, Torres J, Gómez A, Camorlinga-Ponce M, Muñoz-Pérez L, Herrera-Goepfert R, et al. Circulating blood levels of IL-6, IFN-γ, and IL-10 as potential diagnostic biomarkers in gastric cancer: a controlled study. BMC Cancer (2017) 17(1):1–10. doi: 10.1186/s12885-017-3310-9
318. Modi U, Makwana P, Vasita R. Molecular insights of metastasis and cancer progression derived using 3D cancer spheroid co-culture in vitro platform. Crit Rev Oncol/Hematol (2021) 168:103511. doi: 10.1016/j.critrevonc.2021.103511
319. Chiamulera MMA, Zancan CB, Remor AP, Cordeiro MF, Gleber-Netto FO, Baptistella AR. Salivary cytokines as biomarkers of oral cancer: A systematic review and meta-analysis. BMC Cancer (2021) 21(1):1–16. doi: 10.1186/s12885-021-07932-3
320. Scheede-Bergdahl C, Watt HL, Trutschnigg B, Kilgour RD, Haggarty A, Lucar E, et al. Is IL-6 the best pro-inflammatory biomarker of clinical outcomes of cancer cachexia? Clin Nutr (2012) 31(1):85–8. doi: 10.1016/j.clnu.2011.07.010
321. Hardy-Werbin M, Rocha P, Arpi O, Taus Á., Nonell L, Durán X, et al. Serum cytokine levels as predictive biomarkers of benefit from ipilimumab in small cell lung cancer. Oncoimmunology (2019) 8(6):e1593810. doi: 10.1080/2162402X.2019.1593810
322. Lee LT, Wong YK, Hsiao HY, Wang YW, Chan MY, Chang KW. Evaluation of saliva and plasma cytokine biomarkers in patients with oral squamous cell carcinoma. Int J Oral Maxillofac Surg (2018) 47(6):699–707. doi: 10.1016/j.ijom.2017.09.016
323. Fan J, Upadhye S, Worster A. Understanding receiver operating characteristic (ROC) curves. Can J Emergency Med (2006) 8(1):19–20. doi: 10.1017/S1481803500013336
324. Dayana SMH, Lim SM, Tan MP, Chin AV, Poi PJH, Kamaruzzaman SB, et al. 115 IP-10 AND IL-13 AS POTENTIALLY NEW, NON-CLASSICAL BLOOD-BASED CYTOKINE BIOMARKERS FOR ALZHEIMER’S DISEASE. Age Ageing (2014) 43(suppl_1):i32–2. doi: 10.1093/ageing/afu045.2
325. Oda Y, Tsuruta R, Kasaoka S, Inoue T, Maekawa T. The cutoff values of intrathecal interleukin 8 and 6 for predicting the neurological outcome in cardiac arrest victims. Resuscitation (2009) 80(2):189–93. doi: 10.1016/j.resuscitation.2008.10.001
326. Ashizawa T, Okada R, Suzuki Y, Takagi M, Yamazaki T, Sumi T, et al. Clinical significance of interleukin-6 (IL-6) in the spread of gastric cancer: role of IL-6 as a prognostic factor. Gastric Cancer (2005) 8:124–31. doi: 10.1007/s10120-005-0315-x
327. Gorelik E, Landsittel DP, Marrangoni AM, Modugno F, Velikokhatnaya L, Winans MT, et al. Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol Biomarkers Prev (2005) 14(4):981–7. doi: 10.1158/1055-9965.EPI-04-0404
Keywords: T cell lymphoma, metabolism, cytokine, tumor micro environment, cell signaling
Citation: Yadav M, Uikey BN, Rathore SS, Gupta P, Kashyap D, Kumar C, Shukla D, Vijayamahantesh, Chandel AS, Ahirwar B, Singh AK, Suman SS, Priyadarshi A and Amit A (2023) Role of cytokine in malignant T-cell metabolism and subsequent alternation in T-cell tumor microenvironment. Front. Oncol. 13:1235711. doi: 10.3389/fonc.2023.1235711
Received: 06 June 2023; Accepted: 14 August 2023;
Published: 07 September 2023.
Edited by:
Ajay Kumar, Banaras Hindu University, IndiaReviewed by:
Aklank Jain, Central University of Punjab, IndiaVirendra Singh, Jawaharlal Nehru University, India
Copyright © 2023 Yadav, Uikey, Rathore, Gupta, Kashyap, Kumar, Shukla, Vijayamahantesh, Chandel, Ahirwar, Singh, Suman, Priyadarshi and Amit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ajay Amit, YWpheTJhbWl0QGdtYWlsLmNvbQ==
†These authors share first authorship
 Megha Yadav
Megha Yadav Blessi N. Uikey
Blessi N. Uikey Shantnu Singh Rathore1
Shantnu Singh Rathore1 Priyanka Gupta
Priyanka Gupta Chanchal Kumar
Chanchal Kumar Dhananjay Shukla
Dhananjay Shukla Arvind Singh Chandel
Arvind Singh Chandel Bharti Ahirwar
Bharti Ahirwar Ashish Kumar Singh
Ashish Kumar Singh Ajay Amit
Ajay Amit