
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 20 July 2023
Sec. Cancer Genetics
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1229168
N6-methyladenosine (m6A) is the most prevalent internal modification found in both mRNA and lncRNA. It exerts reversible regulation over RNA function and affects RNA processing and metabolism in various diseases, especially tumors. The m6A binding protein, hnRNPA2B1, is extensively studied as a member of the heterogeneous nuclear ribonucleoprotein (hnRNP) protein family. It is frequently dysregulated and holds significant importance in multiple types of tumors. By recognizing m6A sites for variable splicing, maintaining RNA stability, and regulating translation and transport, hnRNPA2B1 plays a vital role in various aspects of tumor development, metabolism, and regulation of the immune microenvironment. In this review, we summarized the latest research on the functional roles and underlying molecular mechanisms of hnRNPA2B1. Moreover, we discussed its potential as a target for tumor therapy.
Epigenetics is now widely recognized as the study of stable and heritable alterations that occur in chromosomal regions without changes in the DNA sequence (1). Regulatory mechanisms of epigenetic modifications include DNA methylation, histone modification, noncoding RNA regulation, chromatin remodeling, and nucleosome positioning (2). Among these mechanisms, RNA methylation modification, specifically N6-methyladenosine (m6A), holds paramount importance. m6A is the most abundant form of internal modification found in mRNA and lncRNA, and it has been shown to play a crucial role in various biological functions (3). In coding transcripts, m6A sites are predominantly located near the 3’-untranslated terminal region (UTR) (4) and translated near the 5’-UTR (5), while they can be found anywhere in noncoding transcripts. These modifications affect RNA alternative splicing (6), stability maintenance (7, 8), and regulation of translation and transport (4, 9). Regulators involved in m6A modifications can be categorized into three types: the multicomponent m6A methyltransferase complex (MTC) (10, 11), RNA demethylases (12, 13), and m6A binding proteins (14, 15). The multicomponent m6A methyltransferase complex, comprising METTL3, METTL14, RBM15, WTAP, and KIAA1429, is commonly referred to as the “writers” as it adds m6A modifications to target RNA (16). Conversely, the RNA demethylases FTO and ALKBH5, known as “erasers”, are responsible for removing m6A modifications (17, 18). These m6A sites can be recognized by various m6A binding proteins categorized as “readers”, including the IGF2BP proteins, the YTH domain family of proteins and the HNRNP protein family (19–21). The dynamic regulation of m6A modifications has been shown to play a significant role in various processes related to tumor development and drug susceptibility.
An illustrative example is the abnormal overexpression of ALKBH5 in acute myeloid leukemia (AML), which serves as a prognostic indicator for poor outcomes and plays a pivotal role in the self-renewal of cancer stem cells (22). Additionally, in HIF2α low/− clear cell renal cell carcinoma (ccRCC), FTO displays elevated expression levels, leading to increased sensitivity of ccRCC to BRD9 inhibitors (23).
HnRNPs represent a diverse group of RNA-binding proteins that are widely expressed in human tissues and encompass approximately 20 different protein types (24). Among them, hnRNPA2B1 belongs to the A/B subfamily of hnRNPs. The hnRNPA2B1 pre-mRNA exhibits four splice subtypes: B1, A2, B1b, and A2b (25, 26), although B1b and A2b are typically not expressed in human cells. Distinguishing between the isoforms of hnRNPA2B1 has proven challenging due to a mere 12-amino acid difference near the N-terminus between A2 and B1 (27).
Structurally, hnRNPA2B1 consists of two RNA recognition motifs (RRMs) located at its N-terminus, referred explicitly to as RRM1 and RRM2. Additionally, it contains a glycine-rich low-complexity region (LC) at its C-terminus, which encompasses an RGG box, a core PrLD, and an M9-NLS (28–30) (Figure 1). The RRMs and RGG box are crucial in RNA binding (24, 30). HnRNPA2B1 interacts with specific RNA sequences and is involved in m6A modification through its RRMs (31). While the RGG box significantly influences binding strength, its impact on specific RNA binding is relatively minor (32).
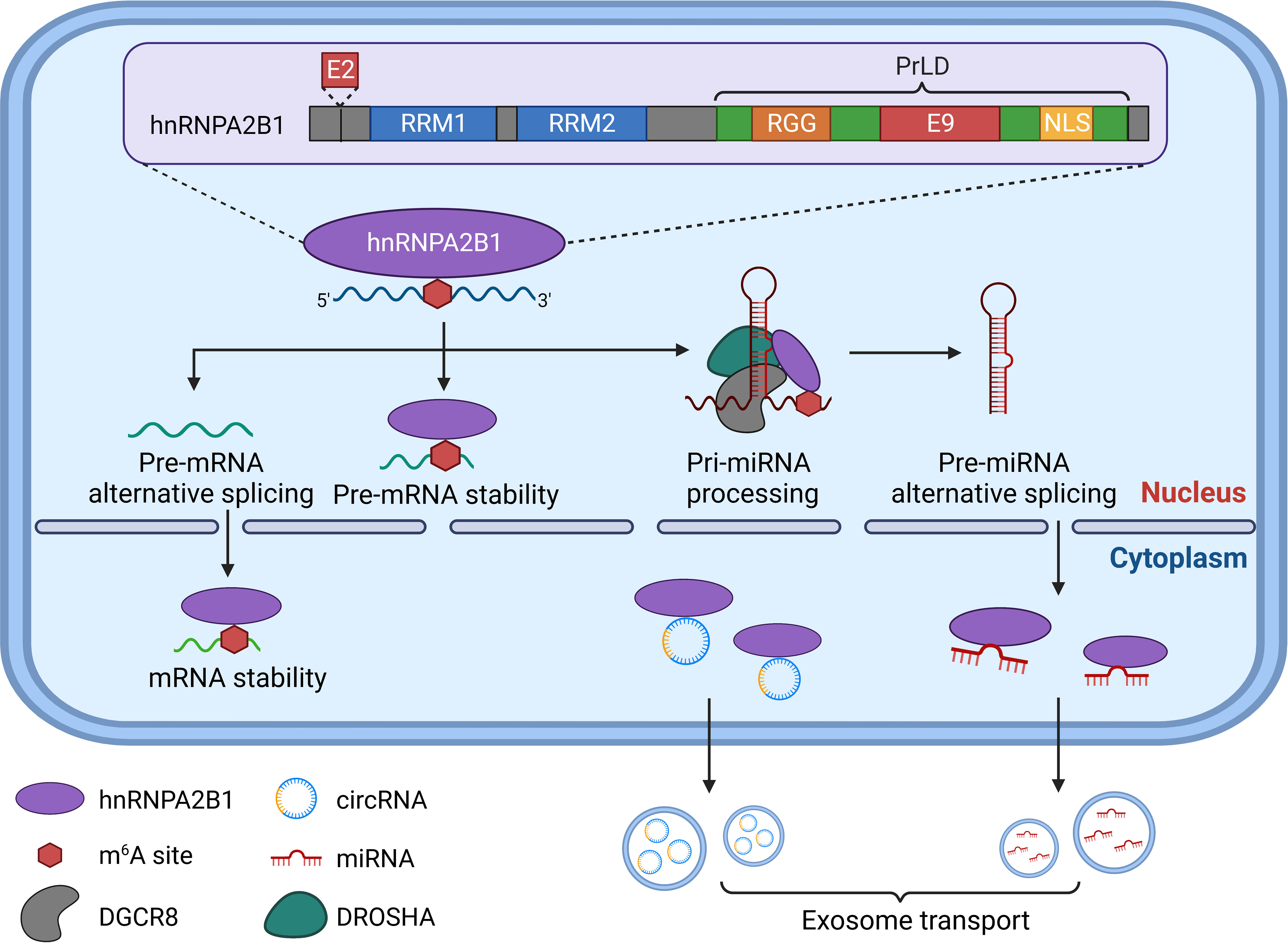
Figure 1 The protein structure and biological function of hnRNPA2B1. RRM1, RRM2 at N-terminal and RGG box, PrLD and LNS at C-terminal constitute the important protein structure of hnRNPA2B1. Differentiating between the A2 and B1 has been challenging due to only a 12-amino acid (Exon 2) near N-termina. For m6A modification-dependent functions, hnRNPA2B1 regulates mRNA or miRNA splicing efficiency by binding m6A-containing pre-mRNA or pre-miRNA. In addition, hnRNPA2B1 regulates mRNA or pre-mRNA stability as a trans-acting RNA-binding protein. HnRNPA2B1 also specifically recognizes and binds to pri-miRNA-containing transcripts and enhances the binding of DGCR8 to pri-miRNA transcripts, promoting pri-miRNA processing. Moreover, hnRNPA2B1 can load circRNA and miRNA into exosomes to mediate their transport.
Presently, two main perspectives exist regarding the various mechanisms by which hnRNPA2B1 contributes to m6A modification regulation. Alarcón et al. have demonstrated that hnRNPA2B1 binds to RNA sites containing the RGAC motif in nuclear transcripts, thereby promoting m6A-dependent pre-miRNA processing events (33). However, another study suggests that hnRNPA2B1 may not directly act on the m6A locus. Instead, it may interact with specific targets containing AGG and UAG motifs to activate an “m6A switch” by binding to the RRM1 and RRM2 domains (30). Through the utilization of m6A markers, hnRNPA2B1 participates in nearly every step of RNA synthesis and processing. It specifically recognizes and binds to transcripts containing pri-miRNAs, enhancing the binding of DGCR8 to pri-miRNA transcripts and facilitating pri-miRNA processing (33). Recent research has also identified a role for hnRNPA2B1 in exosome loading, where sumoylated hnRNPA2B1 can bind and sort miRNAs into exosomes (34). Furthermore, hnRNPA2B1 is involved in regulating mRNA splicing efficiency, potentially through its binding to m6A-containing pre-mRNA (35). Additionally, as a trans-acting RNA-binding protein, hnRNPA2B1 contributes to the regulation of mRNA stability (36).
In addition to affecting RNA methylation modification, hnRNPA2B1 can also change its metabolism through ubiquitination, acetylation, and other post-translational modifications. For example, in liver cancer cells, lncRNA miR503HG promotes the degradation of hnRNPA2B1 through ubiquitin-proteasome pathway (37). HnRNPA2B1 also can be acetylated by p300, a common transcriptional coactivator, and promotes nonsmall cell lung cancer (NSCLC) growth by activating COX-2 signaling (38). Numerous studies have highlighted the frequent dysregulation of hnRNPA2B1 and its crucial role in the biological processes of tumorigenesis and development.
In this review, we focused on recent researches regarding the biological behaviors and underlying mechanisms of hnRNPA2B1 in various aspects, including epithelial-mesenchymal transition (EMT), tumorigenesis, tumor microenvironment (TME), and tumor metabolism. Furthermore, we investigated the potential of targeting hnRNPA2B1 as a therapeutic strategy for tumors.
Tumor growth arises from uncontrolled cell proliferation and invasion, destroying surrounding tissue and distant metastasis. The process of tumor formation and progression is influenced by mutual dynamic crosstalk between the genomic/epigenetic features of tumor cells and tumor microenvironment (TME) components. Extensive studies have demonstrated that hnRNPA2B1 is upregulated in multiple tumors and is associated with poor prognosis (37, 39, 40). It actively regulates RNA processing, translation, transport, and metabolism, thereby influencing EMT, tumorigenesis, TME, and tumor metabolism. (Table 1).
EMT is a biological process where polarized epithelial cells downregulate epithelial characteristics and acquire a mesenchymal phenotype (56). In the context of early lung cancer detection, hnRNPA2B1 has been identified as an early marker of EMT and carcinogenesis using monoclonal antibodies 703D4 (57, 58). The downregulation of E-cadherin and upregulation of vimentin and N-cadherin are early features of EMT initiation (59). Transcription factors, Twist, Snail, Zeb1 and so on, play crucial roles in EMT across various types of cancer (60, 61). In colorectal cancer (CRC), lncRNA H19 binds directly to hnRNPA2B1 and activates Raf/ERK/Snail signaling (46). Another lncRNA RP11 is reported to regulate EMT of CRC as well. It post-translationally degrades Siah1 and Fbxo45 mRNA by binding with hnRNPA2B1, resulting in increased expression of Zeb1 (47). Consistent with this, silencing hnRNPA2B1 can down-regulate the expression of E-cadherin inhibitors, Snai1 and Twist (41). Cancer drug resistance can occur due to the inactivation of chemotherapeutic drugs, altered drug targets, and increased efflux, resulting in reduced drug absorption by cancer cells (62). This complex phenomenon may arise from various mechanisms, such as inhibition of cell death, EMT, cellular heterogeneity, and epigenetic influences (63). In cetuximab resistant CRC, hnRNPA2B1 binds to lncRNA MIR100HG through the RRM2 domain and stabilizes TCF7L2 mRNA, enhancing Wnt/β-catenin signaling (39). The Wnt pathway plays a critical role in regulating EMT. In the context of this study, the Wnt target genes ZEB1 and Slug were found to be significantly regulated when hnRNPA2B1 was overexpressed or knocked down (39). Thus, hnRNPA2B1 served as an essential mediator between lncRNA and its target RNA within the EMT signaling pathway.
Alternative splicing (AS) has the capacity to influence mRNA translation, localization, and regulation of stability. As a consequence, it leads to the generation of distinct protein isoforms that exhibit divergent functions, localizations, and varying degrees of relevance in cellular processes (64). HnRNPA2B1 is known to generate multiple proteins with distinct exon combinations and functions by alternative splicing of a single target pre-mRNA (31). The alternative splicing of pre-mRNA by hnRNPA2B1 may contribute to the pathogenesis of chronic myeloid leukemia (CML). With modifying the splicing of the β1-integrin-responsive non-receptor tyrosine kinase (PYK2) mRNA precursor, the expression of full-length Pyk2 increases in BCR/ABL-containing HSCs (55). Bcl-x is a well-known player in apoptosis, and its AS gives rise to two distinct isoforms: the anti-apoptotic Bcl-xL and the pro-apoptotic Bcl-xS, which exert opposing effects on apoptosis (65). Notably, hnRNPA2B1 competes with Sam68 for binding to the Bcl-x pre-mRNA, while also interacting with the lncRNA BC200. The upregulation of Bcl-xL expression enhances uncontrolled cell proliferation in estrogen-dependent breast cancer, a phenomenon that can be reversed by the knockdown of hnRNPA2B1 (35).
Another important RNA-binding protein, DGCR8, forms a complex with the RNase III enzyme Drosha, resulting in the formation of a microprocessor complex responsible for cleaving primary microRNA transcripts (pri-miRs) (66). In NSCLC, hnRNPA2B1 is regulated by the lncRNA LINC01234 and binds to DGCR8 to participate in the splicing of the miR-106b-5p precursor. Mature miR-106b-5p downregulates CRY2 and upregulates c-Myc. Activated c-MYC, in turn, increases the transcription of LINC01234 and promotes proliferation (42). This mechanism is also observed in multiple myeloma (MM) osteolytic bone disease, where the hnRNPA2B1-DGCR8 complex plays a role in pri-miRNA processing events through m6A modification. Under this mechanism, the generation of miR-92a-2-5p and miR-373-3p is increased, and they are packaged into exosomes and delivered to monocytes or mesenchymal stem cells (MSCs), activating osteoclast formation and inhibiting osteogenesis by suppressing IRF8 or RUNX2 (53). In the study conducted by Yin et al., it has been demonstrated that hnRNPA2B1 plays a role in the upregulation and modulation of cell division protein kinase 6 (CDK6) expression in NSCLC cell lines. CDK6 is a cyclin-dependent kinase active during interphase, regulating the transition from the G1 to S phase of the cell cycle. Aberrant CDK6 activity is frequently observed in various human cancers, contributing to their development and progression (67). Mechanistically, hnRNPA2B1 recruits the RNA helicase DHX9 and binds to the UUAGG motif in the CDK6 3’-UTR, collaborating with miR-506 to suppress the translation of CDK6 (40).
The transcription and degradation of mRNA are dependent on changes in its concentration (68). mRNA stability is determined by mRNA cis-elements and corresponding trans-acting binding proteins (36). Jiang et al. have revealed that hnRNPA2B1 identifies the m6A site of ILF3 mRNA in MM and reduces its fragility, thereby mediating AKT3 to promote MM cell proliferation (54). During migration and invasion in ovarian cancer, hnRNPA2B1 plays a positive regulatory role by interacting with Lin28B mRNA and reducing its degradation (50). However, contrary to the findings in other cancer studies, hnRNPA2B1 shows an inverse correlation with breast cancer (BC) metastasis, and patients with high HNRNPA2B1 expression exhibit better survival and prognosis (49). Based on research findings, hnRNPA2B1 directly binds to the UAGGG region in the 3′-UTR, leading to the degradation of PFN2 mRNA and suppressing the pro-invasive phenotype (49). Consistently, in hepatocellular carcinoma (HCC) cells, hnRNPA2B1 is degraded by miR503HG specifical interaction, which promotes the metabolism of p52 and p65 mRNA, inhibits the NF-κB signaling pathway, and thereby suppresses HCC metastasis (37).
In a word, during the proliferation, apoptosis, invasion, and metastasis of a tumor, hnRNPA2B1 can directly regulate the alternative splicing of pre-mRNA and mRNA stability or cooperate with miRNA on mRNA to regulate downstream signaling.
Metabolic remodeling plays a crucial role in tumor initiation and progression (69). Major metabolic changes in cancer include increased aerobic glycolysis (70), dysregulated lipid metabolism (71), elevated reactive oxygen species (ROS) generation (72), and disturbances in enzyme activities (73). The ATP-binding cassette transporter (ABCA1) facilitates the formation of high-density lipoprotein (HDL) and enhances plasma HDL levels, thereby promoting cholesterol efflux (74). Cholesterol 7a-hydroxylase (CYP7A1) plays a crucial role in converting cholesterol into bile acids (75). The proper expression of both ABCA1 and CYP7A1 is vital for maintaining cholesterol metabolism homeostasis. In hepatocytic cholesterol metabolism, the intranuclear expression level of hnRNPA2B1 is increased by lncRNA lnc-HC, which forms a protein-RNA complex targeting Cyp7a1 and Abca1, resulting in mRNA destabilization and decreased translation levels (76). SREBP1, a master regulator of fatty acid and triglyceride synthesis, increases lipid accumulation upon activation of c-Jun signaling (77). The lncRNA NEAT1, in conjunction with hnRNPA2B1, targets RPRD1B mRNA stability, activating the c-Jun/c-Fos/SREBP1 axis to promote fatty acid metabolism and primary tumor lymph node implantation in gastric cancer (GC) (45). Mitochondrial uncoupling protein 2 (UCP2) sustains the transition from mitochondrial oxidative phosphorylation (OXPHOS) to aerobic glycolysis, known as the Warburg effect (43). Pyruvate kinase subtype M2 (PKM2) is highly enriched in tumor cells and induces the Warburg effect (78). In PC cells, UCP2 induces hnRNPA2B1 generation through its antioxidant function, resulting in the upregulation of GLUT1 and PKM2 and increased lactate secretion (43). Similarly, in ccRCC, hnRNPA2B1 interacts with VHLα. Through the regulation of pyruvate kinase (PKM) splicing and the subsequent increase in PKM2 expression, hnRNPA2B1 exerts a tumor-suppressive effect on cellular glucose metabolism (51). The negative regulation of hnRNPA2B1 occurs through N-terminal-mediated ubiquitination of VHLα. In contrast, its positive regulation forms a feedback loop that regulates downstream abnormal c-myc signaling, ultimately contributing to its tumor-suppressive effect (52). Activation of oncogenes, such as MYC, transcription factors like HIF1α, and deactivation of tumor suppressor genes like p53 lead to increased glucose uptake and inhibition of OXPHOS (79). In CRC, circMYH9 promotes tumor cell proliferation and serine metabolism. circMYH9 recruits nuclear hnRNPA2B1 to inhibit its combination and stabilization with p53 pre-mRNA. Additionally, an amino acid deficiency increases ROS levels and HIF1α expression, which further enhances circMYH9 expression, forming a positive feedback loop (48). Overall, these results indicate that hnRNPA2B1 covers multiple metabolic imbalances in tumor cells.
Recent research has revealed the significant involvement of the m6A family in tumor immunity, drawing extensive attention due to its impact on the effectiveness of immune checkpoint blockade therapy in cancer cells (80, 81). HnRNPA2B1 has been found to be significantly overexpressed in esophageal cancer and associated with the regulation of the TME (82). Overexpression of hnRNPA2B1 leads to the enrichment of tumor-derived extracellular vesicles (EVs) containing miR-378a-3p in prostate cancer bone metastases. Upon uptake by bone marrow macrophages (BMMs), these EVs target and activate the Dyrk1a/Nfatc1/Angptl2 axis, promoting bone metastasis (83). Tumor-associated macrophages (TAMs), consisting of M2 and M1 cells, not only fail to recognize and engulf tumor cells but also contribute to immune evasion (84). Recent research in adrenocortical carcinoma (ACC) has shown that HNRNPA2B1 affects tumor development by regulating infiltration of M0 macrophages into the TME (85). Drug-induced DNA damage in macrophages stimulates the secretion of IFN-β through the hnRNPA2B1-cGAS axis (86). Exosomes, natural carriers known for their role in overcoming chemoresistance, have a significant impact on tumor cell transcriptome changes and immune responses (87). The composition of exosomes, which includes diverse nucleic acids, plays a crucial role in facilitating these processes (88). Elaborate studies have demonstrated that hnRNPA2B1 induces the packaging of circNEIL3 into exosomes, which are then taken up by infiltrating TAMs. This enables the stabilization of oncogenic protein IGF2BP3, conferring immunosuppressive properties and promoting glioma progression (89). Furthermore, hnRNPA2B1 is involved in the polarization of M2 macrophages. In LUAD, hnRNPA2B1 facilitates the delivery of exosomal miR-3153 to activate JNK signaling and induce M2 macrophage polarization (90). Furthermore, in CRC, hnRNPA2B1 binds to the GGAG sequence and facilitates the packaging of miR-934 into exosomes, which are transferred to recipient macrophages. This leads to downregulation of PTEN expression, activation of the PI3K/AKT signaling pathway, and further polarization of macrophages. The resulting M2 macrophages secrete CXCL13, which acts on the CXCR5 receptor, activating downstream NFκB/p65/miR-934 signaling in CRC cells (91). A study of CD8+ T cells found that hnRNPA2B1-dependent secretion of exosomal circCCAR1 leads to its uptake by CD8+ T cells, causing their dysfunction by stabilizing the expression of PD-1 and promoting resistance to anti-PD-1 immunotherapy (92) (Figure 2). The crucial role of hnRNPA2B1-mediated vesicle transport in tumor immunity is evident. Consequently, discovering the novel role of hnRNPA2B1 in the tumor microenvironment opens up new avenues and ideas for future tumor immunotherapy.
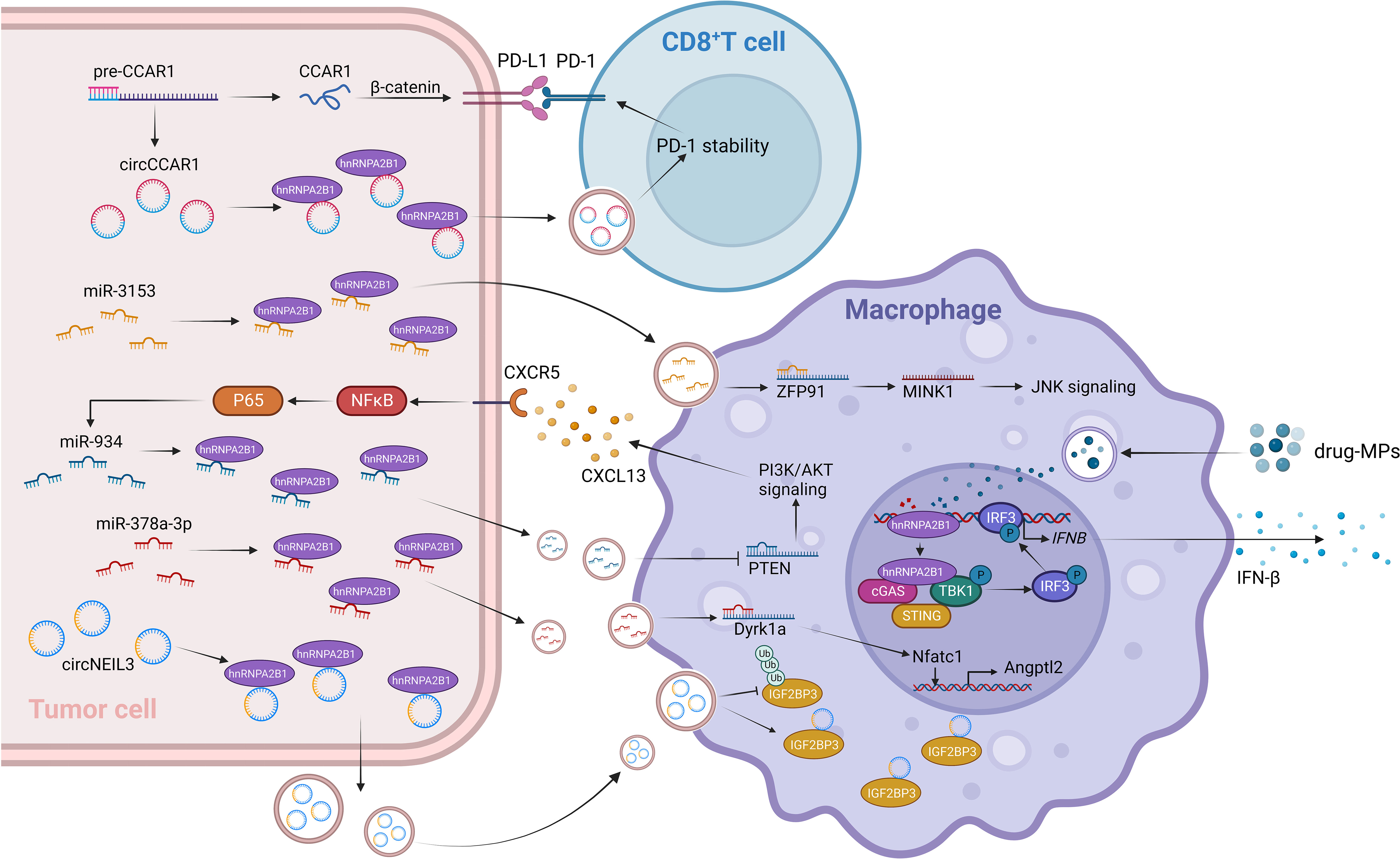
Figure 2 HnRNPA2B1 plays a role in tumor immunity by inhibiting the immune response through the transport of miRNA and circRNA to recipient immune cells. HnRNPA2B1-circNEIL3 exosomes target TAMs and stabilize IGF2BP3, leading to immunosuppressive properties. Moreover, the uptake of hnRNPA2B1-mediated tumor-derived EVs enriched with miR-378a-3p by BMMs activates the Dyrk1a/Nfatc1/Angptl2 axis, promoting prostate cancer bone metastasis. Macrophages exist in two phenotypes: proinflammatory (M1) and anti-inflammatory (M2). JNK signaling and PI3K/AKT signaling pathways are associated with M2 polarization. HnRNPA2B1 mediates the secretion of exosomes containing miR-3153 and miR-934, which respectively activate JNK signaling and PI3K/AKT signaling in macrophages, promoting M2 polarization. Some drugs have been reported to reverse M2 polarization. Drug-induced M1-shifted macrophages increase IFN-β secretion through the hnRNPA2B1-cGAS axis. Furthermore, hnRNPA2B1 contributes to a decrease in the effectiveness of immune checkpoint inhibitors. Exosomal circCCAR1, secreted in a hnRNPA2B1-dependent manner, is taken up by CD8+ T cells and stabilizes the interaction between PD-1 and PD-L1, promoting resistance to anti-PD-1 immunotherapy.
The therapeutic targeting of hnRNPA2B1 shows promise for cancer treatment due to its significant impact on multiple cancers. A series of drugs targeting different functions of hnRNPA2B1 have emerged.
In glioma cells (U251), β-asarone inhibits hnRNPA2B1, leading to increased MMP-9 and p-STAT3 expression and the induction of a malignant phenotype (93). β-asarone also promotes the processing of Bcl-x, shifting the balance from Bcl-xL to Bcl-xS, which may contribute to apoptosis through the mitochondrial pathway (94). Extracts from the South African medicinal plant Cotyledon orbiculata cause the splicing of hnRNPA2B1 to the hnRNPA2 subtype, also increasing the ratio of Bcl-xS/Bcl-xL and activating caspase-3 cleavage and apoptosis (95). Epirubicin treatment induces the secretion of exosomes carrying anti-tumoral miR-503 by preventing hnRNPA2B1 from interacting with miR-503/ANXA2 in endothelial cells (96). The natural product MO-460 binds to the glycine-rich domain of HNRNPA2B1, inhibiting its association with the 3′-UTR of HIF-1 (97). The aptamer C6-8, targeting ROS 17/2.8 in rat osteosarcoma cells, inhibits tumor cell growth by specifically binding to hnRNPA2B1 after conjugation with fluorescent carbon nanodots (CDots) (98). L-norleucine interacts with the RRM1 and RRM2 domains of hnRNPA2B1, inhibiting Twist1 and Snail and increasing E-cadherin, thereby hindering EMT and tumor metastasis (99). Dietary flavones like apigenin have also been considered potential chemo-sensitizers. Apigenin sensitizes triple-negative breast cancer (TNBC) spheroids to doxorubicin through its interaction with hnRNPA2B1, promoting apoptosis induced by ABCC4 and ABCG2 transporters (100) (Figure 3).
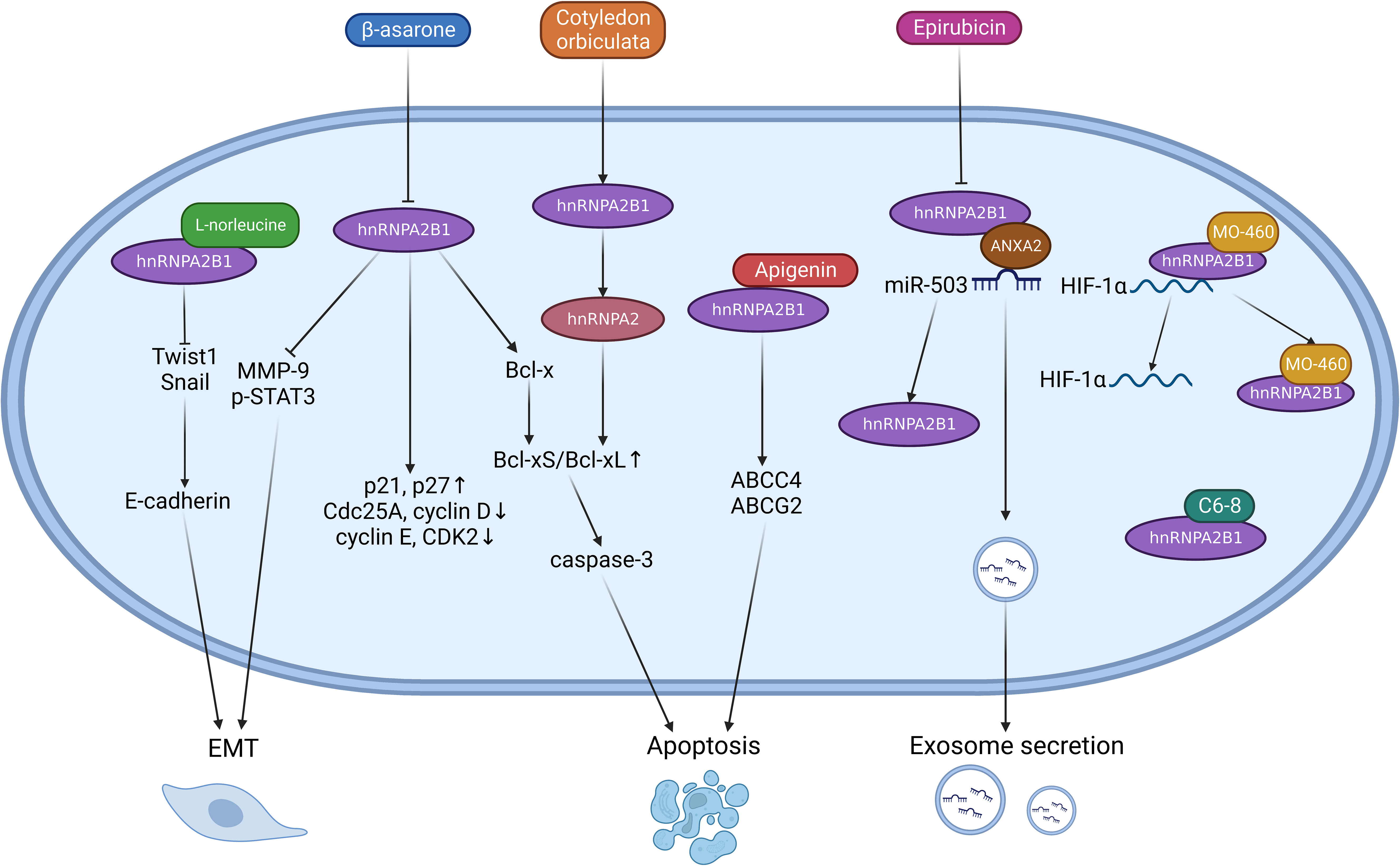
Figure 3 Targeting hnRNPA2B1 for cancer treatment involves the use of certain drugs that act as inhibitors, although the exact mechanisms of action are not fully understood. Inhibition of hnRNPA2B1 by β-asarone leads to reduced expression of MMP-9 and p-STAT3, thereby decreasing EMT. Additionally, β-asarone promotes the splicing of Bcl-xS, which mediates tumor cell apoptosis. Similarly, cotyledon orbiculate induces apoptosis through the same effect. Epirubicin inhibits the interaction between hnRNPA2B1 and the miR503-ANAX2 complex, facilitating exosome transport. Some drugs bind directly to specific regions of hnRNPA2B1. L-norleucine binds to the RRM1 and RRM2 domains of hnRNPA2B1, inhibiting EMT by regulating transcription factors like Twist1 and Snail. Apigenin directly acts on hnRNPA2B1, promoting apoptosis induced by ABCC4 and ABCG2. MO-460 competes with HIF-1α for binding to hnRNPA2B1, leading to reduced translational expression of HIF-1α. C6-8 exert antitumor effects by directly targeting hnRNPA2B1.
As previously mentioned, hnRNPA2B1 exhibits expression in various human tissues, and the potential toxicity associated with targeted therapy utilizing this molecule is evident. Currently, research on hnRNPA2B1 targeted therapy is primarily focused on tumor cells, necessitating further animal experiments to validate the feasibility of this molecular targeted drug.
Furthermore, understanding how hnRNPA2B1 exerts opposing roles in the same cancer type requires careful investigation. It is worth noting that in BC and kidney cancer (49, 101), hnRNPA2B1 plays a tumor-promoting or tumor-suppressing role in different populations, possibly due to variations in research models, microenvironments, and tumor heterogeneity. Comprehensive research is needed to better understand these complexities in order to effectively guide targeted therapy effectively.
Although there are only 12 amino acid differences between hnRNPA2 and B1, studies have shown that they have slightly different recognizable sequences, indicating that hnRNPA2 and B1 have different functions (102). Previous research has demonstrated that hnRNPA2, rather than hnRNPB1, is upregulated in an inflammation-induced mouse liver cancer model. HnRNPA2 activates the Ras/Raf/MAPK/ERK signaling pathway and promotes tumorigenesis through A-Raf splicing (44). Further studies should delve into the functions and mechanisms of various isoforms of hnRNPA2B1 in m6A modification and their implications for targeted therapy.
Proliferation, resistance to growth inhibitory factors, evasion of cell death, replicative immortality, induction of angiogenesis, invasion, and metastasis are considered hallmarks of cancer (103). Recent studies have revealed multiple mechanisms through which hnRNPA2B1 affects cancer. These studies primarily focus on cell proliferation, invasion, metastasis, and dysregulation of metabolism (45, 50, 67). However, emerging evidence suggests that hnRNPA2B1 also plays a crucial role in other biological processes, such as immune microenvironment regulation. For example, Cao et al. have found that hnRNPA2B1 can initiate and amplify the innate immune response to DNA viruses (104). Therefore, hnRNPA2B1 may also mediate antitumor immune responses. Further research is needed to explore the effects and mechanisms of hnRNPA2B1 on each cancer biomarker.
In conclusion, hnRNPA2B1 was identified as a modifier of various RNAs that impacted tumorigenesis and development in an m6A-dependent manner. This modification occurred regardless of its dependence or independence on other members of the m6A methylase family. HnRNPA2B1-targeted therapy is still in its early stages, and ongoing efforts should be directed toward designing and optimizing tumor treatment strategies involving hnRNPA2B1.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by National Natural Science Foundation of China (82172689, 81902386); China Postdoctoral Science Foundation (2021M700543, 2021M700547).
All figures were created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Wu C, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science (2001) 293(5532):1103–5. doi: 10.1126/science.293.5532.1103
2. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell (2012) 150(1):12–27. doi: 10.1016/j.cell.2012.06.013
3. Zaccara S, Jaffrey SR. A unified model for the function of Ythdf proteins in regulating M(6)a-Modified Mrna. Cell (2020) 181(7):1582–95 e18. doi: 10.1016/j.cell.2020.05.012
4. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of Mrna methylation reveals enrichment in 3' Utrs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
5. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' Utr M(6)a promotes cap-independent translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
6. Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vagbo CB, et al. Splice site M(6)a methylation prevents binding of U2af35 to inhibit rna splicing. Cell (2021) 184(12):3125–42 e25. doi: 10.1016/j.cell.2021.03.062
7. He PC, Wei J, Dou X, Harada BT, Zhang Z, Ge R, et al. Exon architecture controls mrna M(6)a suppression and gene expression. Science (2023) 379(6633):677–82. doi: 10.1126/science.abj9090
8. Uzonyi A, Dierks D, Nir R, Kwon OS, Toth U, Barbosa I, et al. Exclusion of M6a from splice-site proximal regions by the exon junction complex dictates M6a topologies and Mrna stability. Mol Cell (2023) 83(2):237–51 e7. doi: 10.1016/j.molcel.2022.12.026
9. Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-bound Mettl3 maintains myeloid leukaemia by M(6)a-dependent translation control. Nature (2017) 552(7683):126–31. doi: 10.1038/nature24678
10. Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, et al. M(6)a Rna methylation regulates the fate of endogenous retroviruses. Nature (2021) 591(7849):312–6. doi: 10.1038/s41586-020-03135-1
11. Zhang T, Ding C, Chen H, Zhao J, Chen Z, Chen B, et al. M(6)a Mrna modification maintains colonic epithelial cell homeostasis Via Nf-Kappab-Mediated antiapoptotic pathway. Sci Adv (2022) 8(12):eabl5723. doi: 10.1126/sciadv.abl5723
12. Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, et al. Fto mediates Line1 M(6)a demethylation and chromatin regulation in mescs and mouse development. Science (2022) 376(6596):968–73. doi: 10.1126/science.abe9582
13. Zhou J, Zhang X, Hu J, Qu R, Yu Z, Xu H, et al. M(6)a demethylase Alkbh5 controls Cd4(+) T cell pathogenicity and promotes autoimmunity. Sci Adv (2021) 7(25):eabg0470. doi: 10.1126/sciadv.abg0470
14. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mrna M(6)a methylation and Ythdf1 in dendritic cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
15. Huang J, Jiang B, Li GW, Zheng D, Li M, Xie X, et al. M(6)a-modified Lincrna Dubr is required for neuronal development by stabilizing Ythdf1/3 and facilitating Mrna translation. Cell Rep (2022) 41(8):111693. doi: 10.1016/j.celrep.2022.111693
16. Huang W, Chen TQ, Fang K, Zeng ZC, Ye H, Chen YQ. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J Hematol Oncol (2021) 14(1):117. doi: 10.1186/s13045-021-01129-8
17. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear Rna is a major substrate of the obesity-associated Fto. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
18. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. Alkbh5 is a mammalian rna demethylase that impacts rna metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
19. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of Rna N(6)-methyladenosine by Igf2bp proteins enhances mrna stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
20. Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, et al. Crystal structure of the Yth domain of Ythdf2 reveals mechanism for recognition of N6-methyladenosine. Cell Res (2014) 24(12):1493–6. doi: 10.1038/cr.2014.152
21. Wang LC, Chen SH, Shen XL, Li DC, Liu HY, Ji YL, et al. M6a Rna methylation regulator Hnrnpc contributes to tumorigenesis and predicts prognosis in glioblastoma multiforme. Front Oncol (2020) 10:536875. doi: 10.3389/fonc.2020.536875
22. Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. Rna demethylase Alkbh5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell (2020) 27(1):64–80 e9. doi: 10.1016/j.stem.2020.04.009
23. Zhang C, Chen L, Lou W, Su J, Huang J, Liu A, et al. Aberrant activation of M6a demethylase fto renders Hif2alpha(Low/-) clear cell renal cell carcinoma sensitive to Brd9 inhibitors. Sci Transl Med (2021) 13(613):eabf6045. doi: 10.1126/scitranslmed.abf6045
24. Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. Hnrnp proteins and the biogenesis of Mrna. Annu Rev Biochem (1993) 62:289–321. doi: 10.1146/annurev.bi.62.070193.001445
25. Kamma H, Horiguchi H, Wan L, Matsui M, Fujiwara M, Fujimoto M, et al. Molecular characterization of the Hnrnp A2/B1 proteins: tissue-specific expression and novel isoforms. Exp Cell Res (1999) 246(2):399–411. doi: 10.1006/excr.1998.4323
26. Hatfield JT, Rothnagel JA, Smith R. Characterization of the mouse hnrnp A2/B1/B0 gene and identification of processed Pseudogenes. Gene (2002) 295(1):33–42. doi: 10.1016/s0378-1119(02)00800-4
27. Burd CG, Swanson MS, Gorlach M, Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of Rna binding proteins is generated by small peptide inserts. Proc Natl Acad Sci U.S.A. (1989) 86(24):9788–92. doi: 10.1073/pnas.86.24.9788
28. Mears WE, Rice SA. The Rgg box motif of the herpes simplex virus Icp27 protein mediates an Rna-binding activity and determines in vivo methylation. J Virol (1996) 70(11):7445–53. doi: 10.1128/JVI.70.11.7445-7453.1996
29. Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in Hnrnpa2b1 and Hnrnpa1 cause multisystem proteinopathy and Als. Nature (2013) 495(7442):467–73. doi: 10.1038/nature11922
30. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of Rna substrates by human hnrnp A2/B1. Nat Commun (2018) 9(1):420. doi: 10.1038/s41467-017-02770-z
31. Liu Y, Shi SL. The roles of hnrnp A2/B1 in Rna biology and disease. Wiley Interdiscip Rev RNA (2021) 12(2):e1612. doi: 10.1002/wrna.1612
32. Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnrnp U protein: binding Rna through Rgg box. EMBO J (1992) 11(7):2655–64. doi: 10.1002/j.1460-2075.1992.tb05331.x
33. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. Hnrnpa2b1 is a mediator of M(6)a-dependent nuclear Rna processing events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
34. Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated Hnrnpa2b1 controls the sorting of mirnas into exosomes through binding to specific motifs. Nat Commun (2013) 4:2980. doi: 10.1038/ncomms3980
35. Singh R, Gupta SC, Peng WX, Zhou N, Pochampally R, Atfi A, et al. Regulation of alternative splicing of Bcl-X by Bc200 contributes to breast cancer pathogenesis. Cell Death Dis (2016) 7(6):e2262. doi: 10.1038/cddis.2016.168
36. Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of Mrna decay. Nat Rev Mol Cell Biol (2007) 8(2):113–26. doi: 10.1038/nrm2104
37. Wang H, Liang L, Dong Q, Huan L, He J, Li B, et al. Long noncoding rna Mir503hg, a prognostic indicator, inhibits tumor metastasis by regulating the Hnrnpa2b1/Nf-kappab pathway in hepatocellular carcinoma. Theranostics (2018) 8(10):2814–29. doi: 10.7150/thno.23012
38. Xuan Y, Wang J, Ban L, Lu JJ, Yi C, Li Z, et al. Hnrnpa2/B1 activates cyclooxygenase-2 and promotes tumor growth in human lung cancers. Mol Oncol (2016) 10(4):610–24. doi: 10.1016/j.molonc.2015.11.010
39. Liu H, Li D, Sun L, Qin H, Fan A, Meng L, et al. Interaction of Lncrna Mir100hg with Hnrnpa2b1 facilitates M(6)a-dependent stabilization of Tcf7l2 mrna and colorectal cancer progression. Mol Cancer (2022) 21(1):74. doi: 10.1186/s12943-022-01555-3
40. Yin M, Cheng M, Liu C, Wu K, Xiong W, Fang J, et al. Hnrnpa2b1 as a trigger of Rna switch modulates the mirna-mediated regulation of Cdk6. iScience (2021) 24(11):103345. doi: 10.1016/j.isci.2021.103345
41. Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. Hnrnp A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res (2010) 70(18):7137–47. doi: 10.1158/0008-5472.CAN-10-0860
42. Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, et al. Integrative analysis of Nsclc identifies Linc01234 as an oncogenic Lncrna that interacts with Hnrnpa2b1 and regulates Mir-106b biogenesis. Mol Ther (2020) 28(6):1479–93. doi: 10.1016/j.ymthe.2020.03.010
43. Brandi J, Cecconi D, Cordani M, Torrens-Mas M, Pacchiana R, Dalla Pozza E, et al. The antioxidant uncoupling protein 2 stimulates Hnrnpa2/B1, Glut1 and Pkm2 expression and sensitizes pancreas cancer cells to glycolysis inhibition. Free Radic Biol Med (2016) 101:305–16. doi: 10.1016/j.freeradbiomed.2016.10.499
44. Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, Rauch J, et al. Splicing factor hnrnp A2 activates the ras-Mapk-Erk pathway by controlling a-raf splicing in hepatocellular carcinoma development. RNA (2014) 20(4):505–15. doi: 10.1261/rna.042259.113
45. Jia Y, Yan Q, Zheng Y, Li L, Zhang B, Chang Z, et al. Long non-coding rna Neat1 mediated Rprd1b stability facilitates fatty acid metabolism and lymph node metastasis Via c-Jun/C-Fos/Srebp1 axis in gastric cancer. J Exp Clin Cancer Res (2022) 41(1):287. doi: 10.1186/s13046-022-02449-4
46. Zhang Y, Huang W, Yuan Y, Li J, Wu J, Yu J, et al. Long non-coding rna H19 promotes colorectal cancer metastasis Via binding to Hnrnpa2b1. J Exp Clin Cancer Res (2020) 39(1):141. doi: 10.1186/s13046-020-01619-6
47. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. M(6)a-induced lncrna Rp11 triggers the dissemination of colorectal cancer cells Via upregulation of Zeb1. Mol Cancer (2019) 18(1):87. doi: 10.1186/s12943-019-1014-2
48. Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. Circmyh9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a P53-dependent manner. Mol Cancer (2021) 20(1):114. doi: 10.1186/s12943-021-01412-9
49. Liu Y, Li H, Liu F, Gao LB, Han R, Chen C, et al. Heterogeneous nuclear ribonucleoprotein A2/B1 is a negative regulator of human breast cancer metastasis by maintaining the balance of multiple genes and pathways. EBioMedicine (2020) 51:102583. doi: 10.1016/j.ebiom.2019.11.044
50. Yang Y, Wei Q, Tang Y, Yuanyuan W, Luo Q, Zhao H, et al. Loss of Hnrnpa2b1 inhibits malignant capability and promotes apoptosis Via down-regulating Lin28b expression in ovarian cancer. Cancer Lett (2020) 475:43–52. doi: 10.1016/j.canlet.2020.01.029
51. Liu Y, Yang H, Li L, Chen S, Zuo F, Chen L. A novel vhlalpha isoform inhibits warburg effect Via modulation of pkm splicing. Tumour Biol (2016) 37(10):13649–57. doi: 10.1007/s13277-016-5191-y
52. Liu Y, Zhang H, Li X, Zhang C, Huang H. Identification of anti-tumoral feedback loop between vhlalpha and Hnrnpa2b1 in renal cancer. Cell Death Dis (2020) 11(8):688. doi: 10.1038/s41419-020-02861-8
53. Liu R, Zhong Y, Chen R, Chu C, Liu G, Zhou Y, et al. M(6)a reader Hnrnpa2b1 drives multiple myeloma osteolytic bone disease. Theranostics (2022) 12(18):7760–74. doi: 10.7150/thno.76852
54. Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, et al. Hnrnpa2b1 promotes multiple myeloma progression by increasing Akt3 expression Via M6a-dependent stabilization of Ilf3 mrna. J Hematol Oncol (2021) 14(1):54. doi: 10.1186/s13045-021-01066-6
55. Salesse S, Dylla SJ, Verfaillie CM. P210bcr/Abl-induced alteration of pre-mrna splicing in primary human Cd34+ hematopoietic progenitor cells. Leukemia (2004) 18(4):727–33. doi: 10.1038/sj.leu.2403310
56. Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol (2020) 21(6):341–52. doi: 10.1038/s41580-020-0237-9
57. Zhou J, Mulshine JL, Unsworth EJ, Scott FM, Avis IM, Vos MD, et al. Purification and characterization of a protein that permits early detection of lung cancer. identification of heterogeneous nuclear ribonucleoprotein-A2/B1 as the antigen for monoclonal antibody 703d4. J Biol Chem (1996) 271(18):10760–6. doi: 10.1074/jbc.271.18.10760
58. Fielding P, Turnbull L, Prime W, Walshaw M, Field JK. Heterogeneous nuclear ribonucleoprotein A2/B1 up-regulation in bronchial lavage specimens: a clinical marker of early lung cancer detection. Clin Cancer Res (1999) 5(12):4048–52.
59. Sisto M, Ribatti D, Lisi S. Cadherin signaling in cancer and autoimmune diseases. Int J Mol Sci (2021) 22(24):13358. doi: 10.3390/ijms222413358
60. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, et al. The emt-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol (2017) 19(5):518–29. doi: 10.1038/ncb3513
61. Mohammadi Ghahhari N, Sznurkowska MK, Hulo N, Bernasconi L, Aceto N, Picard D. Cooperative interaction between eralpha and the emt-inducer Zeb1 reprograms breast cancer cells for bone metastasis. Nat Commun (2022) 13(1):2104. doi: 10.1038/s41467-022-29723-5
62. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature (2019) 575(7782):299–309. doi: 10.1038/s41586-019-1730-1
63. Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: an overview. Cancers (Basel) (2014) 6(3):1769–92. doi: 10.3390/cancers6031769
64. Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol (2017) 18(7):437–51. doi: 10.1038/nrm.2017.27
65. Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, et al. Bcl-X, a bcl-2-Related gene that functions as a dominant regulator of apoptotic cell death. Cell (1993) 74(4):597–608. doi: 10.1016/0092-8674(93)90508-n
66. Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. Dgcr8 is essential for microrna biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet (2007) 39(3):380–5. doi: 10.1038/ng1969
67. Goel S, Bergholz JS, Zhao JJ. Targeting Cdk4 and Cdk6 in cancer. Nat Rev Cancer (2022) 22(6):356–72. doi: 10.1038/s41568-022-00456-3
68. Nouaille S, Mondeil S, Finoux AL, Moulis C, Girbal L, Cocaign-Bousquet M. The stability of an mrna is influenced by its concentration: a potential physical mechanism to regulate gene expression. Nucleic Acids Res (2017) 45(20):11711–24. doi: 10.1093/nar/gkx781
69. Li Y, Su R, Deng X, Chen Y, Chen J. Fto in cancer: functions, molecular mechanisms, and therapeutic implications. Trends Cancer (2022) 8(7):598–614. doi: 10.1016/j.trecan.2022.02.010
70. Liberti MV, Locasale JW. The warburg effect: how does it benefit cancer cells? Trends Biochem Sci (2016) 41(3):211–8. doi: 10.1016/j.tibs.2015.12.001
71. Zaidi N, Lupien L, Kuemmerle NB, Kinlaw WB, Swinnen JV, Smans K. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res (2013) 52(4):585–9. doi: 10.1016/j.plipres.2013.08.005
72. Sharma S, Wang J, Cortes Gomez E, Taggart RT, Baysal BE. Mitochondrial complex ii regulates a distinct oxygen sensing mechanism in monocytes. Hum Mol Genet (2017) 26(7):1328–39. doi: 10.1093/hmg/ddx041
73. Massari F, Ciccarese C, Santoni M, Iacovelli R, Mazzucchelli R, Piva F, et al. Metabolic phenotype of bladder cancer. Cancer Treat Rev (2016) 45:46–57. doi: 10.1016/j.ctrv.2016.03.005
74. Vaisman BL, Lambert G, Amar M, Joyce C, Ito T, Shamburek RD, et al. Abca1 overexpression leads to hyperalphalipoproteinemia and increased biliary cholesterol excretion in transgenic mice. J Clin Invest (2001) 108(2):303–9. doi: 10.1172/JCI12517
75. Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor lxr alpha. Cell (1998) 93(5):693–704. doi: 10.1016/s0092-8674(00)81432-4
76. Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, et al. A novel long noncoding rna lnc-hc binds Hnrnpa2b1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology (2016) 64(1):58–72. doi: 10.1002/hep.28391
77. Cubero FJ, Drvarov O, Trautwein C. C-jun Nh(2)-terminal kinase 1 in hepatocytes: an essential mediator of insulin resistance. Hepatology (2010) 51(6):2221–3. doi: 10.1002/hep.23737
78. Alquraishi M, Puckett DL, Alani DS, Humidat AS, Frankel VD, Donohoe DR, et al. Pyruvate kinase M2: a simple molecule with complex functions. Free Radic Biol Med (2019) 143:176–92. doi: 10.1016/j.freeradbiomed.2019.08.007
79. Yeung SJ, Pan J, Lee MH. Roles of P53, myc and hif-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci (2008) 65(24):3981–99. doi: 10.1007/s00018-008-8224-x
80. Bao Y, Zhai J, Chen H, Wong CC, Liang C, Ding Y, et al. Targeting M(6)a reader Ythdf1 augments antitumour immunity and boosts anti-Pd-1 efficacy in colorectal cancer. Gut (2023) 72:1497–1509. doi: 10.1136/gutjnl-2022-328845
81. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. Alkbh5 regulates anti-Pd-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U.S.A. (2020) 117(33):20159–70. doi: 10.1073/pnas.1918986117
82. Guo W, Tan F, Huai Q, Wang Z, Shao F, Zhang G, et al. Comprehensive analysis of pd-L1 expression, immune infiltrates, and M6a rna methylation regulators in esophageal squamous cell carcinoma. Front Immunol (2021) 12:669750. doi: 10.3389/fimmu.2021.669750
83. Wang J, Du X, Wang X, Xiao H, Jing N, Xue W, et al. Tumor-derived mir-378a-3p-Containing extracellular vesicles promote osteolysis by activating the Dyrk1a/Nfatc1/Angptl2 axis for bone metastasis. Cancer Lett (2022) 526:76–90. doi: 10.1016/j.canlet.2021.11.017
84. Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discovery (2022) 21(11):799–820. doi: 10.1038/s41573-022-00520-5
85. Jin Y, Wang Z, He D, Zhu Y, Hu X, Gong L, et al. Analysis of M6a-related signatures in the tumor immune microenvironment and identification of clinical prognostic regulators in adrenocortical carcinoma. Front Immunol (2021) 12:637933. doi: 10.3389/fimmu.2021.637933
86. Wei K, Zhang H, Yang S, Cui Y, Zhang B, Liu J, et al. Chemo-drugs in cell microparticles reset antitumor activity of macrophages by activating lysosomal P450 and nuclear Hnrnpa2b1. Signal Transduct Target Ther (2023) 8(1):22. doi: 10.1038/s41392-022-01212-7
87. Li S, Yi M, Dong B, Jiao Y, Luo S, Wu K. The roles of exosomes in cancer drug resistance and its therapeutic application. Clin Transl Med (2020) 10(8):e257. doi: 10.1002/ctm2.257
88. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
89. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. Ewsr1-induced Circneil3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization Via stabilizing Igf2bp3. Mol Cancer (2022) 21(1):16. doi: 10.1186/s12943-021-01485-6
90. Xu L, Wang L, Yang R, Li T, Zhu X. Lung adenocarcinoma cell-derived exosomes promote M2 macrophage polarization through transmission of mir-3153 to activate the jnk signaling pathway. Hum Mol Genet (2023) 32(12):2162–2176. doi: 10.1093/hmg/ddad052
91. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-derived exosomal mir-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol (2020) 13(1):156. doi: 10.1186/s13045-020-00991-2
92. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, et al. Exosome-derived Circccar1 promotes Cd8 + T-cell dysfunction and anti-Pd1 resistance in hepatocellular carcinoma. Mol Cancer (2023) 22(1):55. doi: 10.1186/s12943-023-01759-1
93. Li L, Wu M, Wang C, Yu Z, Wang H, Qi H, et al. Beta-asarone inhibits invasion and emt in human glioma U251 cells by suppressing splicing factor hnrnp A2/B1. Molecules (2018) 23(3):671. doi: 10.3390/molecules23030671
94. Li L, Yang Y, Wu M, Yu Z, Wang C, Dou G, et al. Beta-asarone induces apoptosis and cell cycle arrest of human glioma U251 cells via suppression of hnrnp A2/B1-mediated pathway in vitro and in vivo. Molecules (2018) 23(5):1072. doi: 10.3390/molecules23051072
95. Makhafola TJ, Mbele M, Yacqub-Usman K, Hendren A, Haigh DB, Blackley Z, et al. Apoptosis in cancer cells is induced by alternative splicing of Hnrnpa2/B1 through splicing of bcl-X, a mechanism that can be stimulated by an extract of the south African medicinal plant, cotyledon orbiculata. Front Oncol (2020) 10:547392. doi: 10.3389/fonc.2020.547392
96. Perez-Boza J, Boeckx A, Lion M, Dequiedt F, Struman I. Hnrnpa2b1 inhibits the exosomal export of mir-503 in endothelial cells. Cell Mol Life Sci (2020) 77(21):4413–28. doi: 10.1007/s00018-019-03425-6
97. Soung NK, Kim HM, Asami Y, Kim DH, Cho Y, Naik R, et al. Mechanism of the natural product moracin-O derived Mo-460 and its targeting protein Hnrnpa2b1 on hif-1alpha inhibition. Exp Mol Med (2019) 51(2):1–14. doi: 10.1038/s12276-018-0200-4
98. Li H, Guo L, Huang A, Xu H, Liu X, Ding H, et al. Nanoparticle-conjugated aptamer targeting hnrnp A2/B1 can recognize multiple tumor cells and inhibit their proliferation. Biomaterials (2015) 63:168–76. doi: 10.1016/j.biomaterials.2015.06.013
99. He T, Jin M, Xu C, Ma Z, Wu F, Zhang X. The homeostasis-maintaining metabolites from bacterial stress response to bacteriophage infection suppress tumor metastasis. Oncogene (2018) 37(43):5766–79. doi: 10.1038/s41388-018-0376-z
100. Sudhakaran M, Parra MR, Stoub H, Gallo KA, Doseff AI. Apigenin by targeting Hnrnpa2 sensitizes triple-negative breast cancer spheroids to doxorubicin-induced apoptosis and regulates expression of Abcc4 and Abcg2 drug efflux transporters. Biochem Pharmacol (2020) 182:114259. doi: 10.1016/j.bcp.2020.114259
101. Li L, Tao Z, Zhao Y, Li M, Zheng J, Li Z, et al. Prognostic characteristics and immune effects of N(6)-methyladenosine and 5-Methylcytosine-Related regulatory factors in clear cell renal cell carcinoma. Front Genet (2022) 13:864383. doi: 10.3389/fgene.2022.864383
102. Nguyen ED, Balas MM, Griffin AM, Roberts JT, Johnson AM. Global profiling of hnrnp A2/B1-rna binding on chromatin highlights lncrna interactions. RNA Biol (2018) 15(7):901–13. doi: 10.1080/15476286.2018.1474072
103. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
Keywords: epigenetics, m6A, hnRNPA2B1, tumor, biological mechanisms, cancer therapy
Citation: Wu Y, Li A, Chen C, Fang Z, Chen L and Zheng X (2023) Biological function and research progress of N6-methyladenosine binding protein heterogeneous nuclear ribonucleoprotein A2B1 in human cancers. Front. Oncol. 13:1229168. doi: 10.3389/fonc.2023.1229168
Received: 26 May 2023; Accepted: 03 July 2023;
Published: 20 July 2023.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Xiaodan Hao, Qingdao University, ChinaCopyright © 2023 Wu, Li, Chen, Fang, Chen and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Zheng, emhlbmd4aWFvQHN1ZGEuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.