
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 29 September 2023
Sec. Gastrointestinal Cancers: Gastric and Esophageal Cancers
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1224669
This article is part of the Research Topic Probiotics and Their Metabolites in Cancer Therapy View all 6 articles
 Yiwen Wang1,2
Yiwen Wang1,2 Wenjie Han1,2
Wenjie Han1,2 Na Wang1,2
Na Wang1,2 Mengzhen Han1,2
Mengzhen Han1,2 Meng Ban3
Meng Ban3 Jianying Dai4
Jianying Dai4 Yuesheng Dong4
Yuesheng Dong4 Tao Sun1,5
Tao Sun1,5 Junnan Xu1,2,5*
Junnan Xu1,2,5*The stomach was once considered a sterile organ until the discovery of Helicobacter pylori (HP). With the application of high-throughput sequencing technology and macrogenomics, researchers have identified fungi and fivemajor bacterial phyla within the stomachs of healthy individuals. These microbial communities exert regulatory influence over various physiological functions, including energy metabolism and immune responses. HP is a well-recognized risk factor for gastric cancer, significantly altering the stomach’s native microecology. Currently, numerous studies are centered on the mechanisms by which HP contributes to gastric cancer development, primarily involving the CagA oncoprotein. However, aside from exogenous infections such as HP and EBV, certain endogenous dysbiosis can also lead to gastric cancer through multiple mechanisms. Additionally, gut microbiota and its metabolites significantly impact the development of gastric cancer. The role of microbial therapies, including diet, phages, probiotics and fecal microbiota transplantation, in treating gastric cancer should not be underestimated. This review aims to study the mechanisms involved in the roles of exogenous pathogen infection and endogenous microbiota dysbiosis in the development of gastric cancer. Also, we describe the application of microbiota therapy in the treatment and prognosis of gastric cancer.
Gastric cancer is the fifth most common cancer and the third leading cause of cancer-related deaths worldwide (1). Gastric cancer rarely detects in its early stage, and the lack of effective biomarkers can accurately diagnose early gastric cancer and monitor prognosis during the adjuvant stage, which results in higher mortality rates, with only one in five patients surviving more than five years after being diagnosed with gastric cancer (2). Consequently, the development of more accurate and accessible biomarkers is necessary to aid in the early-stage diagnosis of gastric cancer and to monitor relapse.
The microbiota in the human body comprises viruses, fungi and bacteria. High-fat diets, antibiotics, exogenous microbial infections and host genetics can lead to dysbiosis (3–5). The definition of dysbiosis is a change in the composition and function of the microbiota (5). A healthy and stable microbiota inhibits cancer development, whereas a dysbiotic microbiota has a limited protective effect on the host and promotes cancer development. Gastric cancer is the result of the interaction between the dysbiosis of the gastric microbiota and the host gastric epithelial cells (6). Moreover, as gastric cancer progresses, damage to the gastric mucosal barrier and persistent chronic inflammation will further promote dysbiosis (7).
Helicobacter pylori (HP) infection is one of the riskiest factors for gastric cancer (8). The majority of intestinal-type gastric cancers are associated with HP. HP infection accelerates the progression from atrophic gastritis (AG) to intestinal metaplasia (IM) and eventually to gastric cancer. Furthermore, individuals with HP infection have a 3-6 fold higher risk of developing gastric cancer than those without HP infection (2, 9). In recent years, viruses such as Epstein-Barr virus (EBV), Hepatitis B virus (HBV), and John Cunningham virus (JCV) have also attracted attention for their role in gastric cancer carcinogenesis. In addition to exogenous pathogen infections, some dysbiotic endogenous microbiota such as Lactobacillus, F. nucleatum, and Candida albicans can also be involved in developing gastric cancer through various mechanisms. Metabolites from the gut microbiota influence gastric cancer progression and improve anticancer therapy for gastric cancer patients. Microbiota therapy will be an essential target for future cancer treatment. In this review, the direct and indirect mechanisms by which exogenous infections and endogenous dysbiosis contribute to gastric cancer development are examined first. Next, the effects of oral and gut microbiota on gastric cancer are explored. Finally, the application of microbiota therapy in treating gastric cancer, including diet, bacteriophages, probiotics, and fecal microbiota transplantation (FMT), is discussed. Our goal is to investigate the impact of microbiota on gastric cancer, which could provide new insights for early screening and subsequent treatment of gastric cancer.
Due to its high acidity, the stomach was once regarded as a sterile organ and unsuitable for microbial colonization. However, the discovery of HP led to the realization that some microbes could indeed colonize the stomach (2). Advancements in high-throughput sequencing technologies in microbiology have facilitated the discovery of an increasing number of exogenous and endogenous microbiota. The normal human stomach harbors a diverse microbiota, with over 130 lineages representing 7 to 13 bacterial phyla, of which 5 are the most dominant, including Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria (10, 11). Fungi, a common yet easily overlooked component of gastric microbiota, have also been identified. Investigations of the gastric environment in healthy individuals, patients with chronic gastritis (CG), and gastric ulcers have detected hundreds of fungal strains in the stomach. Fungi may possess an effective acid tolerance mechanism, enabling them to proliferate more efficiently in a gastric environment with a pH of only 1.4 compared to bacteria. Researchers have detected fungal species, such as Candida tropicales and Candida lusitanae, in addition to the most common Candida albicans. Fungi usually colonize areas like the gastric mucosa and stomach contents (12, 13).
Both bacteria and fungi in the human stomach play crucial roles in various physiological activities, including energy metabolism, nutrient absorption, immune response, and nervous system regulation (14). However, some studies have observed differences in the flora of the gastric mucosa and gastric juice (8, 10, 11, 15, 16). Due to swallowing in the oral cavity and reflux in the duodenum, the gastric juice may contain more bacteria than the gastric mucosa (17). Although most current gastric flora studies have sampled the gastric mucosa, bacteria in the gastric juice should not be overlooked. These bacteria can contribute to the proper functioning of the gastrointestinal system but may also cause some adverse reactions.
The results are diverse for changes in the diversity and abundance of microbiota in gastric cancer. Some studies showed that the abundance and diversity of the gastric microbiota did not change gradually with the development of gastric cancer (18, 19). Other studies have taken the opposite view, suggesting that the abundance and diversity of the microbiota decrease as gastric cancer progresses, which is what most studies have concluded (20–22). Another study has shown a significant increase in the abundance and diversity of the microbiota in cancerous tissues compared to paracancerous tissues. They also found that oral bacteria (such as Peptostreptococcus, Streptococcus, and Fusobacterium) dominated the cancerous tissues, and lactic acid-producing bacteria (such as Lactococcus lactis and Lactobacillus brevis) were enriched in the paracancerous tissues (23). However, Liu et al. found that Prevotella melanogenic, Streptococcus anginosus and Propionibacterium acnes were enriched in cancerous tissues compared to normal and paracancerous tissues. In contrast, the abundance of HP, Prevotella copri and Bacteroides uniformis was significantly reduced in cancerous tissues (24).
Many current studies exist on the microbiota of gastric cancer patients and controls. However, their results varied widely, and some were even contradictory, possibly due to different sample types, study populations, sequencing technologies and analysis methods (Table 1). However, it is undeniable that there is a significant difference in the microbial composition of gastric cancer patients compared to controls. In general, the microbiota of gastric cancer patients shows a decreased abundance of HP, a significant enrichment of the oral bacteria, and reduced diversity and abundance of the microbiota.
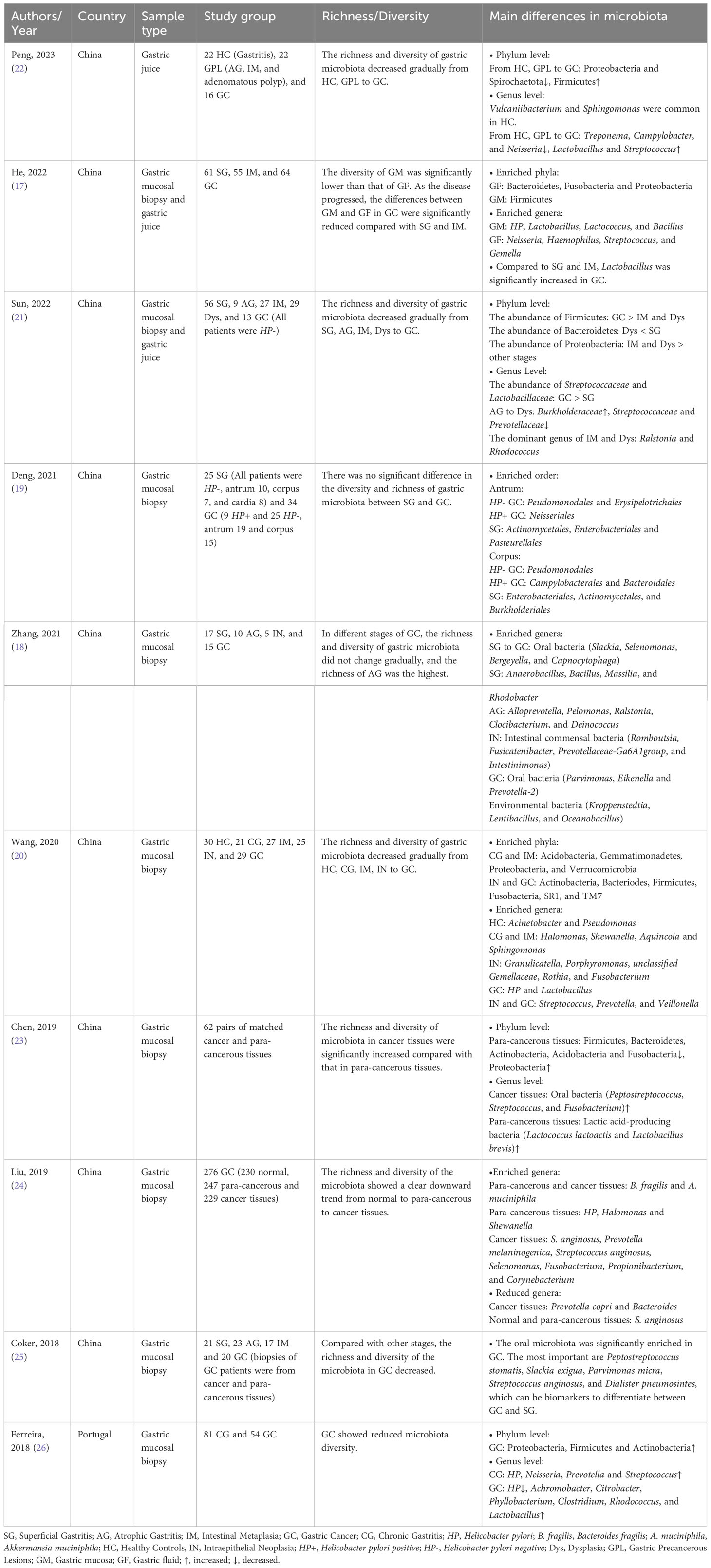
Table 1 Studies on the difference in gastric microbiota composition between patients with gastric cancer and controls.
HP is a gram-negative bacterium that parasitizes the human stomach. The colonization of HP can lead to dysbiosis. The diversity and abundance of the gastric microbiota decrease after infection with HP (27). HP-positive gastric cancers demonstrate an abundance of Lactobacillus, Achromobacter, Citrobacter, Clostridium, and Rhodococcus but lack HP and Neisseria compared to other gastric diseases (26). A recent meta-analysis suggests that eradicating HP can restore the diversity of the gastric microbiota and reduce the risk of gastric cancer (28). However, some studies contradict this result by suggesting that eradicating HP does not entirely prevent the development of gastric cancer, which indicates that other microorganisms in the stomach besides HP are also associated with gastric cancer carcinogenesis (29). Lertpiriyapong et al. demonstrated this hypothesis with INS-GAS mice. They found that HP mice colonized by complex or restricted microbiota were more susceptible to HP-induced gastric cancer than germ-free mice (30). This result suggests that after HP infection of the gastric epithelium causes dysbiosis, HP acts synergistically with the dysbiotic microbiota to promote gastric carcinogenesis.
Although HP is recognized as a type 1 carcinogen by the World Health Organization (WHO), not all gastric cancers are caused by HP infection. HP-negative gastric cancers also account for a percentage ranging from 0.7% to 47.8% of gastric cancer patients (31). The role of microbiota in the development of HP-negative gastric cancer has attracted increasing attention. In HP-negative gastric cancer, Kim et al. found there was a significant increase in the abundance and diversity of Lactobacillaceae, Streptococcaceae, and Prevotellaceae and a decrease in the numbers of Burkholderiaceae, Haemophilus, and Campylobacter (32) (Figure 1). These findings are inconsistent in related studies, but the increased abundance of Lactobacillaceae was detected in almost all patients with gastric cancer. Lactobacillaceae is a nitrosating bacterium, and Haemophilus is a nitrate-reducing bacterium. An increase in the abundance of Lactobacillaceae and a decrease in the abundance of Haemophilus can increase the formation of N-nitroso compounds, which can cause persistent inflammation and promote the development of gastric cancer (32). Ding et al. also concluded that HP-negative gastric cancer occurs due to disruption of the normal microbiota, with elevated levels of some bacteria with pro-cancer activity and decreased levels of cancer-suppressing bacteria (33). In other words, it is not a certain kind of bacteria that causes gastric cancer alone. Instead, the microbiota in the stomach undergoes dysbiosis, and the various dysbiotic microbiota work together to participate in the development of gastric cancer.
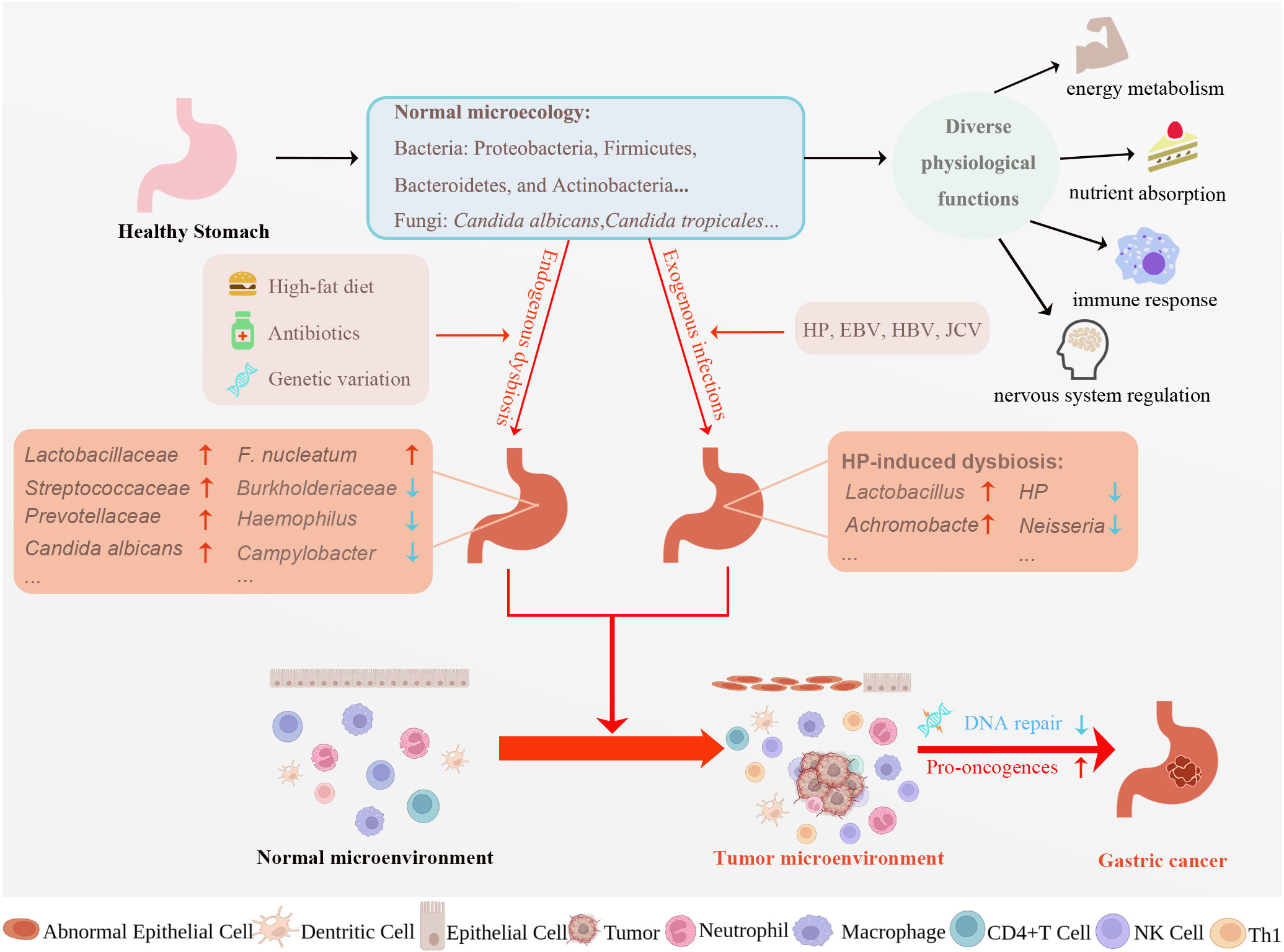
Figure 1 Factors affecting gastric microecological dysbiosis and potential mechanisms underlying microecological dysbiosis causing gastric carcinogenesis. A multitude of microorganisms resides in the stomach, creating normal gastric microecology. Normal microecology plays a vital role in various physiological activities such as energy metabolism, nutrient absorption, immune response, and nervous system regulation. High-fat diet, antibiotics, genetic variation, and exogenous infections can all dysregulate normal gastric microecology, turning the normal microenvironment into the tumor microenvironment and eventually leading to gastric cancer development. Exploring the mechanism of gastric cancer progression from the perspective of gastric microecology is essential for early diagnosis, treatment, and prognosis of gastric cancer. HP, Helicobacter pylori; EBV, Epstein-Barr virus; HBV, Hepatitis B virus; JCV, John Cunningham virus, F. nucleatum, Fusobacterium nucleatum; Th, helper T cell; CTL, Cytotoxic T lymphocyte. Created with BioRender.com. (accessed on 17 May 2023).
Numerous studies have found a connection between the development of gastric cancer and exogenous infections, such as HP and EBV, as well as endogenous dysbiosis. A shared mechanism among these factors is the induction of chronic inflammation, which heightens the risk of cancer (34). Chronic inflammation is vital in all three stages of gastric cancer development, from AG to IM and finally to gastric cancer. Chronic inflammation leads to the accumulation of mutations and the destabilization of the host genome by accelerating DNA replication rates and diminishing DNA damage repair capabilities. The result is pro-oncogenes’ activation and tumor suppressor genes’ (TSG) inactivation, ultimately leading to gastric cancer development (35).
HP, a representative exogenous bacteria in the stomach’s microecology, can be transmitted through saliva, vomit, and feces. The carcinogenic mechanism of HP is closely linked to its virulence factors, which include cytotoxin-associated gene A (CagA), vesicular cytotoxin A (VacA), neutrophil-activating protein (NAP) and outer membrane proteins (OMPs) (3) (Table 2). These factors damage DNA in gastric epithelial cells and cause the destabilization of the host genome.
CagA, an oncoprotein, interacts with various signaling pathways upon entering the cytoplasm of gastric epithelial cells, disrupts intercellular junctions, and activates pro-inflammatory and oncogenic signaling pathways. This process leads to sustained inflammatory responses and uncontrollable cell proliferation. CagA can be transfected into the cytoplasm of gastric epithelial cells via the type 4 secretion system (T4SS) encoded by the Cag pathogenicity island (CagPAI) (2). CagA can regulate signal transduction through both phosphorylated and non-phosphorylated pathways. Phosphorylated CagA can directly bind or recruit the phosphatase SHP2 to activate the MAPK/ERK pathway. SHP2 is a key protein in CagA pathogenesis and can regulate ERK signaling in a non-Ras-dependent manner (36). Non-phosphorylated CagA, when combined with E-cadherin, decreases E-cadherin expression and promotes epithelial-mesenchymal transition (EMT), accelerating gastric cancer progression (37).
VacA possesses selective anion channel properties that facilitate the release of bicarbonate and organic anions into the cytoplasm, contributing to HP colonization in gastric epithelial cells and leading to apoptosis (36). Moreover, HP infection can induce gastric carcinogenesis through hypermethylation silencing of multiple TSGs, such as miR-124a-1, miR-124a-2, and miR-124a-3 (60). Abnormal alterations in the methylation levels of the CpG island of TSG promoters following HP infection affect the transcription of downstream genes, causing irreversible TSG inactivation (61).
NAP is the main pro-inflammatory factor in HP infection. The protein was initially named for its ability to activate neutrophils to produce Reactive Oxygen Species (ROS) and promote neutrophils’ adhesion to gastric epithelial cells (62). In addition to neutrophils, NAP activates monocytes and mast cells to release a variety of pro-inflammatory chemokines such as IL-8, IL-6, TNF-α, MIP-1α, MIP-1β, and β-hexosaminidase (63). The release of these factors causes damage to the gastric mucosa and accelerates the process of chronic gastritis. In addition, NAP has been found to play an essential role in bacterial protection. NAP is a DNA-binding protein belonging to the Dps family and has structural similarity to bacterial ferritin. Therefore, NAP can isolate free iron and bind DNA, which can protect HP DNA from oxidative damage and promote the growth and colonization of HP (40).
The most studied adhesins among HP outer membrane proteins are blood group antigen-binding adhesin (BabA) and sialic acid-binding adhesion (SabA). They can bind to receptors on the surface of gastric epithelial cells, and this binding contributes to HP adhesion and colonization, increases HP pathogenicity, and promotes persistent infection (64). BabA binds to Leb and enhances CagA translocation by promoting T4SS activity. CagA induces the production of massive pro-inflammatory factors, leading to IM and precancerous lesions (49). SabA binds to the sialyl-Lex antigen and promotes neutrophil activation and infiltration, inducing oxidative damage and causing a persistent inflammatory response (51). The expression of BabA and SabA is closely associated with the development and prognosis of various gastrointestinal diseases, such as chronic gastritis and gastric cancer (50, 65). Therefore, they may be potential targets for preventing and treating HP-related diseases.
Additionally, HP infection may significantly diminish the effectiveness of immunotherapy for gastric cancer. Firstly, HP can decrease the activity of CD4+ T cells, dendritic cells (DCs), macrophages, and NKT cells, while also increasing the activity of myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Treg cells) (66). Secondly, HP elevates the expression of PD-L1 in gastric epithelial cells and induces CD4+T cell apoptosis (67) (Figure 2). This finding implies that HP infection leads to non-specific suppression of T cells and reduced immune checkpoint inhibitors (ICIs) efficacy. Finally, Oster et al. discovered that HP infection could reduce the effectiveness of anti-CTLA-4 and anti-PD-L1 therapy, decrease the potency of cancer vaccines, and inhibit in situ tumor immunotherapy (66, 68).
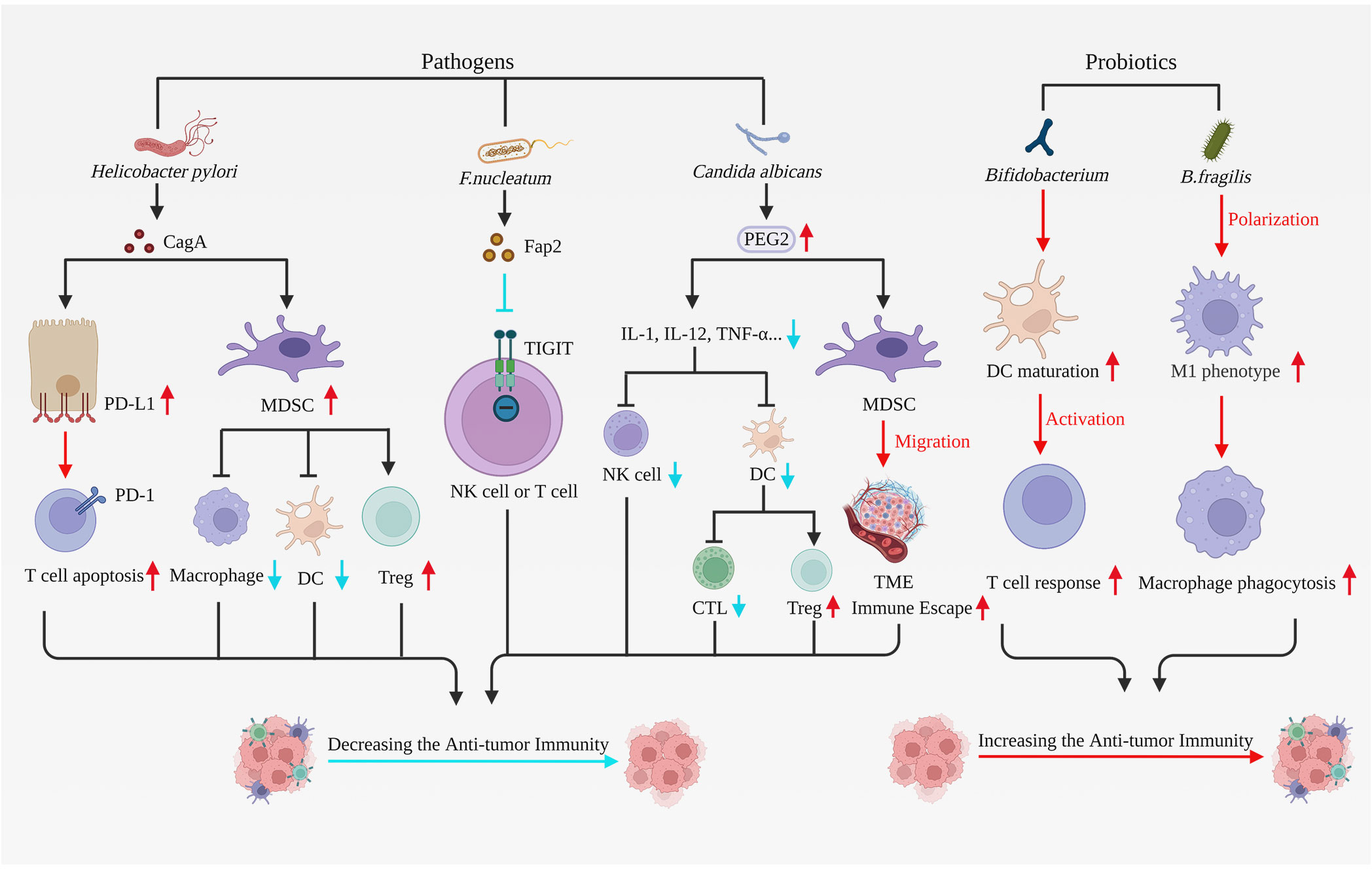
Figure 2 Pathogens and probiotics have effects on anti-tumor immunity. Pathogens correlated with gastric carcinogenesis comprise Helicobacter pylori, F. nucleatum, and Candida albicans, all of which can suppress immune cell function, stimulate tumor cell proliferation and hinder apoptosis, thereby fostering tumor advancement. Helicobacter pylori upregulate PD-L1 expression in gastric epithelial cells, consequently inducing T cell apoptosis. Helicobacter pylori additionally activate MDSCs, reducing DCs and macrophage function while enhancing Treg cell activity. The F. nucleatum virulence factor Fap2 obstructs tumor elimination by immune cells via binding and interacting with TIGIT on NK cells or lymphocytes. Candida albicans infection leads to heightened expression of the inflammatory factor PGE2. PGE2 diminishes cytokine levels such as IL-1, IL-12, and TNF-α, which restrain DC and NK cell activity and function, obstruct CTL activation, and promote Treg cell maturation, ultimately suppressing tumor immunity. Furthermore, PGE2 facilitates MDSC migration to TME, thus promoting tumor immune escape. Probiotics like Bifidobacterium and B. fragilis elicit robust immune responses, enhance anti-tumor immunity, and impede tumor progression. Bifidobacterium directly stimulates DC maturation and fosters immune response production by T cells. B. fragilis induces macrophage polarization toward the M1 phenotype and enhances macrophage phagocytic capacity. In summary, microbial pathogens associated with gastric carcinogenesis suppress anti-tumor immunity. However, probiotics can potentially enhance anti-tumor immunity, which has significant implications for the treatment and prognosis of gastric cancer. F. nucleatum, Fusobacterium nucleatum; B. fragilis, Bacteroides fragilis; MDSCs, myeloid-derived suppressor cells; Treg cells, regulatory T cells; TIGIT, T cell immunoreceptors with IG and ITIM domains; DC, Dendritic cell; TME, Tumor microenvironment. Created with BioRender.com. (accessed on 17 May 2023).
EBV, first isolated from Burkitt’s lymphoma cell line in 1964, was among the initial viruses identified to be linked to human malignancy. EBV infection also elevates the risk of developing gastric cancer by more than 18-fold, with EBV-associated gastric cancer constituting approximately 10% of all gastric cancers (69, 70).
Following EBV infection, viral genes such as EBERs, EBNA-1, and BARTs are expressed and can interact with host proteins to promote cell proliferation and inhibit apoptosis. The DNA damage and cell proliferation resulting from this process contribute to gastric cancer development. Additionally, EBV can disrupt the host’s epigenetic machinery and induce malignant transformation of the epigenome. EBV upregulates host DNA Methyltransferases (DNMTs) through the expression of Latent Membrane Protein 1 (LMP1) and Latent Membrane Protein 2A (LMP2A) viral oncoproteins (13, 71). Consequently, most EBV-associated gastric cancers exhibit CpG island hypermethylation at TSGs, such as PTEN, leading to the inactivation of TSGs and immune escape. Plasma EBV DNA load is a valid marker for assessing the efficacy and prognosis of EBV-associated gastric cancers (72). It has been discovered that EBV and HP may synergistically impact the development of gastric cancer. HP-positive patients have a higher EBV DNA load. EBV infection can enhance the oncogenic potential of CagA. Gastric cancers co-infected with EBV and HP display numerous methylated genes. EBV and HP can synergistically promote chronic inflammation and increase tissue damage during the early stages of the gastric carcinogenesis process (2, 73). Immunotherapy is typically employed to treat patients with EBV-associated gastric cancer because EBV subtypes are positively associated with PD-L1 overexpression (35). Su et al. observed an increased cellular immune response and cytotoxicity in EBV-associated gastric cancer xenograft model mice through CRISPR/Cas9 system-mediated disruption of PD-1 on T cells (74).
Several studies have suggested that other viruses, such as JCV and HBV, may also play a crucial role in gastric cancer development (75). The oncogenic effect of JCV is primarily achieved through its encoded oncoprotein T-Ag. T-Ag can interact with tumor suppressor proteins like p53 and pRB and participate in gastric carcinogenesis through critical signaling pathways, such as Wnt/β-Catenin (76). HBV infection results in a 1.29-fold increased risk of gastric cancer, with the hepatitis B virus X protein (HBx) potentially playing a vital role in this process (77). Cui et al. discovered that HBV could induce gastric epithelial cell carcinogenesis through HBx (78). Moreover, chronic inflammation caused by HBV infection heightens the risk of gastric carcinogenesis (79).
Although Lactobacillus is widely regarded as a probiotic in the intestine, it is often abundantly enriched in gastric cancer, particularly in advanced gastric cancer (32, 80). Several factors contribute to Lactobacillus causing gastric cancer. First, lactate, produced by Lactobacillus, plays a massive role in the Warburg effect of tumors (81). Second, Lactobacillus can upregulate pro-inflammatory genes, such as Ptger4 and Tgf-β, and oncogenic genes, such as Nos2, Tnf-α, Cxcl1 (Kc), and Ccl2 (Mcp-1) to induce gastric carcinogenesis (30). Third, Lactobacillus can reduce nitrate to nitrites and eventually form nitrosamines (82). Furthermore, Lactobacillus is a potent ROS inducer, leading to DNA damage (83). Lastly, Lactobacillus can convert human fibroblasts into multipotent cells, directly promoting gastric cancer development (84). These factors may jointly contribute to gastric cancer development, but the exact mechanism requires further investigation.
Fusobacterium nucleatum (F. nucleatum) is most widely known for its carcinogenic effect on colorectal cancer. However, in recent years, F. nucleatum has also been recognized as a significant cause of gastric cancer development. F. nucleatum is more abundant in gastric cancer than normal tissues, and F. nucleatum positivity is associated with a worse prognosis and shorter overall survival in patients with diffuse-type gastric cancer (85, 86). F. nucleatum binds to E-cadherin in gastric epithelial cells via the characteristic virulence factor FadA, increasing endothelial permeability, promoting self-colonization, propagation, and immune escape (87), and activating the Wnt/β-catenin signaling pathway to enhance tumor cell proliferation (88). Familial adenomatous polyposis 2 (Fap2) is another virulence factor of F. nucleatum. Fap2, through binding and interaction with T cell immunoreceptors with IG and ITIM domains (TIGIT), can inhibit the killing of NK cells and lymphocytes to tumors, thereby protecting tumors and promoting the formation of an inflammatory microenvironment (89, 90). F. nucleatum also produces an induced inflammatory response through NF-κB-mediated cytokines such as IL-6, IL-1β, IL-17, IFN-γ, and TNF-α (87, 91).
Zhong et al. conducted ITS rDNA gene analysis on cancerous and non-cancerous tissues from 45 gastric cancer patients and discovered an imbalance of fungal communities in gastric cancer. After statistical analysis, they determined that Candida albicans were abundantly enriched in gastric cancer. Ultimately, it was demonstrated that Candida albicans could serve as a fungal biomarker for diagnosing gastric cancer. Candida albicans may promote gastric cancer development by reducing the diversity and abundance of fungi in the stomach, such as Candida glabrata, Aspergillus montevidensis, Saitozyma podzolica, and Penicillium arenicola (92).
Candida albicans may possess multiple carcinogenic mechanisms, including damage to mucosal epithelial cells, induction of chronic inflammation, and production of carcinogens leading to cancer.
Upon invasion of the mucosal epithelium, Candida albicans cause apoptosis and necrosis, which disrupts the immune barrier of the epithelium and leads to structural changes in the epithelium (93). Long-term colonization and infestation of Candida albicans in the mucosal epithelium can cause chronic inflammation. During this process, the expression of the inflammatory factor prostaglandin E2 (PGE2) increases. PGE2 downregulates macrophage cytokines, inhibits DC and NK cell activity and function, blocks cytotoxic T cell activation, and promotes maturation and suppressive activity of Treg cells, thereby suppressing tumor immunity. PGE2 aids in the migration of MDSCs to the tumor microenvironment, proliferation of malignant cells, and inhibition of apoptosis, directly promoting tumor progression. PGE2 can also promote tumorigenesis through ROS production, activation of oncogenic transcription factors, and promotion of angiogenesis (94).
Candida albicans-produced nitrosamines and acetaldehyde can contribute to cancer development to some extent. Acetaldehyde impacts DNA replication and can cause point mutations of genes and chromosomal abnormalities. Meanwhile, acetaldehyde interferes with enzymes involved in cytosine methylation and DNA damage repair, resulting in proto-oncogene activation and cell cycle abnormalities, thereby promoting cancer development. Additionally, acetaldehyde can cause mitochondrial damage and induce apoptosis (93).
In conclusion, Candida albicans dysbiosis can increase the likelihood of cancer occurrence and promote cancer progression.
The oral and gastric environments are linked through swallowed saliva and ingested food. The oral microbiota is a major source of gastric microbiota (95). Many studies have found that dysbiosis of the oral microbiota is associated with gastric carcinogenesis. Oral bacteria such as Parvimonas, Eikenella and Prevotella-2 were significantly enriched in gastric cancer compared to other precancerous stages (18). The abundance of oral bacteria such as Peptostreptococcus, Streptococcus, and Fusobacterium was higher in cancerous tissues than in paracancerous tissues (23). Network analysis revealed that oral microorganisms such as Peptostreptococcus stomatitis, Slackia exigua, Parvimonas micra, Streptococcus anginosus, and Dialister pneumosintes might play critical roles in the development of gastric cancer (25). Under normal conditions, most of the oral microbiota entering the stomach is either destroyed by gastric acid or protected by the mucus-bicarbonate barrier from invading the gastric epithelium. However, certain disease states, such as gastric ulcers and gastric cancer, can damage the gastric mucosa and neutralize gastric acid, allowing oral bacteria to colonize the gastric mucosa from gastric juices (17). Recently, some studies have also been on the interaction between oral microbiota and HP. On the one hand, HP infection can disrupt the homeostasis of the oral microbiota (96). On the other hand, ectopic colonization on the gastric mucosa by the oral microbiota may, in turn, be necessary for HP-induced ecological dysbiosis (95).
Since the pH in the intestine is more conducive to microbial colonization, intestinal bacteria are nearly 107 times more abundant than those in the stomach (16, 97). Approximately 99% of the 1014 microorganisms that comprise the human microbiome reside in the intestine (98). Numerous studies have demonstrated that intestinal flora can directly and indirectly affect the development, treatment, and prognosis of gastric cancer.
In a study conducted in Shanxi province, researchers investigated changes in the gut microbiota of 116 patients with gastric cancer and 88 healthy controls. They observed an increase in the abundance of intestinal flora, a decrease in butyrate-producing bacteria, and a significant enrichment of Lactobacillus, Escherichia, and Klebsiella in patients with gastric cancer (99). According to Sarhadi et al., Enterobacteriaceae were abundant in stool samples from patients with all types of gastric cancer (100). Enterobacteriaceae may serve as a potential biomarker in the diagnosis of gastric cancer.
Certain metabolites of intestinal bacteria, such as acetic acid and butyrate, also influence the development of gastric cancer. Acetic acid and butyrate are the major short-chain fatty acids, and intestinal bacteria such as Eubacterium, Clostridium, Ruminococcus, and Coprococcus can produce butyrate (35). Increasingly, research has found that butyrate plays a crucial role in inhibiting gastric cancer development. First, butyrate inhibits the Warburg effect of gastric cancer by binding pyruvate kinase M2 (PKM2), increasing the content of glucose intermediates in mitochondria, preventing the conversion of tricarboxylic acid cycle intermediates to ATP, and ultimately depriving tumor growth of sufficient energy, thus inhibiting gastric cancer development. Second, butyrate can directly interfere with the mitochondria of gastric cancer cells, upregulate oxidative stress, and significantly increase the level of ROS. Moreover, butyrate can promote caspase 9 production and inhibit BCL-2 synthesis, leading to the apoptosis of gastric cancer cells (101). Lastly, a recent study discovered that butyrate could inhibit the growth, migration, and invasion of gastric cancer cells and aerobic glycolysis by blocking the Wnt/β-catenin/c-Myc signaling pathway (102).
Alterations in the microbiota may directly impact gastric cancer immunotherapy (103, 104). Matson et al. discovered that higher proportions of Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium led to more effective anti-PD-L1 therapy (105). A meta-analysis revealed that Bacteroides caccae was enriched in all types of immune checkpoint inhibitor therapies. Faecalibacterium prausnitzii, Bacteroides thetaiotamicron, and Holdemania filiformis were enriched in responders to anti-CTLA-4 and anti-PD-1 ICIs treatments (106). Based on these findings, researchers have proposed microbiota modification as a means to enhance the efficacy of immunotherapy for gastric cancer. Current approaches to microbiota modification include the use of probiotics and FMT.
Probiotics, such as Bifidobacterium, Lactobacillus, and Saccharomyces, are active microbes that benefit the host’s health (107, 108). There is growing evidence that probiotics may inhibit the growth of gastric cancer to some extent. Probiotics can significantly reduce inflammatory responses, enhance the immune system, promote tumor cell apoptosis, restore gut microbiota homeostasis, and inhibit cancer signaling pathways (35, 109, 110). Lactobacillus can inhibit the development of gastric cancer by reducing the expression of NF-κB and the phosphorylation of the PI3K/Akt/mTOR signaling pathway (111). Bifidobacterium can directly induce DC maturation and cytokine (IFNγ, TNFα, IL-10, IL-17) production, thus promoting anti-tumor immunity and anti-PD-L1 efficacy and almost inhibiting tumor progression after combination therapy with PD-L1 monoclonal antibody (mAb) (104). Bacteroides thetaotaomicron and Bacteroides fragilis (B. fragilis) are also potential probiotics. Vétizou et al. found that the antitumor and immunotherapeutic effects of CTLA-4 blockade were associated with Bacteroides thetaotaomicron and B. fragilis. Oral administration of Bacteroides thetaotaomicron or B. fragilis to germ-free mice enhances the antitumor effects of CTLA-4 blockers (112). B. fragilis also increase macrophage phagocytosis, prompting them to polarize towards an M1 phenotype (113).
Probiotics have become a prominent research topic due to their beneficial effects on gastric cancer. Immunotherapeutic probiotics could be developed to improve the efficacy of immunotherapy (114). However, contrary to the aforementioned findings, it is relatively easy to identify inconsistencies. The role of Lactobacillus in gastric cancer remains unclear. In addition to Lactobacillus, there are inconsistent findings on the effects of other probiotics on gastric cancer. Based on human microbiome studies and animal models, researchers have cautioned against the direct use of Lactobacillus in treating patients with gastric cancer (81).
FMT is transplanting fecal material from a healthy donor into a recipient’s intestine, ultimately altering the recipient’s intestinal flora to match the donor’s microbial profile (115). Several methods of fecal microbiota transplantation exist, including oral, nasal, and rectal administration. Rectal administration is preferred, as nasal administration may cause pulmonary or gastrointestinal complications, and although oral administration is convenient, careful avoidance of first-pass effects is necessary (116, 117). Given FMT’s numerous benefits, there is growing interest in its potential for enhancing cancer immunotherapy.
Routy et al. discovered that the aberrant composition of intestinal flora results in primary resistance to immune checkpoint inhibitors (ICIs) in patients with late-stage cancer and that antibiotics interfere with the therapeutic efficacy of ICIs in patients. Their findings revealed that the anti-tumor effects of PD-1 inhibitors are more effective in germ-free mice or those not treated with antibiotics after FMT from cancer patients who respond to ICIs. However, this was not the case in the FMT of patients who did not respond to ICIs. They also analyzed the metagenomics of fecal material from patients and found a correlation between the efficacy of ICIs and the abundance of Akkermansia muciniphila (A. muciniphila). After FMT, patients who did not respond were given A. muciniphila can restore the efficacy of PD-1 inhibitors (103). Fecal material from immunotherapy responders was transplanted into germ-free mice, resulting in slower tumor growth and increased immunotherapy efficacy (105). In summary, FMT can enhance patient responses to immunotherapy.
Since FMT has a more substantial impact on intestinal flora than gastric flora, most current research focuses on the effect of FMT on intestinal diseases. However, researchers should also consider FMT as a treatment for gastric cancer and precancerous lesions in the stomach. As a safe and effective therapy, FMT may treat gastric cancer in the future by interfering with the composition of the intestinal flora. However, we need further studies to evaluate FMT’s safety and reduce the risk of side effects.
In addition to the FMT and probiotics mentioned above, many potential microbial interventions have been used to treat gastric cancer, including diet, prebiotic, and bacteriophage-based strategies, all pointing to the promising future of microbial therapies.
Bacteriophages are viruses that selectively infect bacteria (118) and are more than 100 times more abundant in the gut than in bacteria and human cells (119). Given that most bacteriophages are particular to specific pathogenic bacteria but do not disrupt the homeostasis of normal flora (120), many studies have constructed bacteriophage-based biotic–abiotic hybrid nanosystems to accurately remove tumor microorganisms (121).
In order to precisely regulate intestinal flora to treat colorectal cancer, Zheng et al. loaded irinotecan inside dextran nanoparticles (122). And through bioorthogonal reaction, it was covalently linked with azide-modified phages that could specifically lyse F. nucleatum, which could not only effectively inhibit the proliferation of F. nucleatum, but also reduce the toxic side effects caused by the non-targeted release of conventional chemotherapeutic drugs. Moreover, phages can also remodel TME. F. nucleatum selectively amplifies MDSCs, thereby blocking the body’s anti-tumor immune response. Through phage display technology, Dong et al. screened M13 phage that can specifically bind F. nucleatum (123). They further assembled silver nanoparticles on the surface capsid protein of the M13 phage (M13@Ag). Subsequently, it was also demonstrated that M13@Ag could specifically clear F. nucleatum and inhibit the proliferation of MDSCs, reversing the immunosuppressive TME. In addition, the M13 phage utilizes its coat protein to activate antigen-presenting cells, which further activates the host immune response and inhibits CRC. Animal experiments demonstrated that M13@Ag combined with immune checkpoint inhibitors or chemotherapeutic agents significantly prolonged the survival of CRC mice. A recent study on phage strategies targeting HP may open new avenues for treating HP-positive gastric cancer (124).
Diet has an important influence on the composition of the gut microbiota. Different dietary habits can lead to differences in the composition of the gut microbiota (125). Compared to omnivores, the intestinal flora of strict vegetarians was characterized by a higher abundance of Bacteroidetes and a lower abundance of Firmicutes (126). However, the opposite alteration of the intestine flora was observed in people on chronic high-fat diets (127). Therefore, the composition of the intestinal flora can be adjusted by dietary interventions, thereby creating a more favourable microecological environment for the host. Dietary interventions such as fasting and calorie restriction have been used in clinical trials for various cancers, and the results have shown that dietary interventions can alter the systemic metabolism of cancer patients, improve anti-tumor immunity (128), and inhibit tumor growth. For example, in an ongoing clinical trial (NCT01642953), researchers restrict patients’ diets after gastric cancer surgery to assess whether fasting promotes recovery and reduces mortality and adverse events (129). Dietary interventions can also inhibit inflammation, improve anti-cancer immune surveillance (130) and prevent chemotherapy-induced toxic side effects (131).
WHO defines the concept of prebiotics as “an inactive food ingredient that promotes the health of the organism by modulating the composition of the microbiota (107).” Common prebiotics are fructooligosaccharides (FOS), galactooligosaccharides (GOS) and inulin (132). Prebiotics can help the body resist pathogenic infections, maintain stable gastrointestinal function, promote the production of short-chain fatty acids by beneficial bacteria, regulate immunity and energy metabolism, and increase the absorption of micronutrients, thus improving the efficacy of cancer treatment (132–134). To demonstrate the role of prebiotics in cancer treatment, many preclinical studies and clinical trials are underway or have been completed. One clinical trial in the United States (NCT04682665) is investigating eicosapentaenoic acid’s role in treating liver metastases from colorectal cancer (135). Furthermore, It has been shown that a high intake of Ganoderma spp (136). Polysaccharides and raffinose (137) potentially reduce the risk of gastric cancer, but more extensive independent studies and clinical trials are needed to confirm this.
The microbiota has dual effects on gastric cancer. On the one hand, pathogenic bacterial infections and dysbiosis can promote the development of gastric cancer and is detrimental to the treatment and prognosis of gastric cancer. On the other hand, microbial therapies that optimize the microbiota through dietary changes, the use of bacteriophages and probiotics, and even transplantation of the fecal microbiota hold promise as potential treatments for gastric cancer. Furthermore, specific microorganisms and metabolites in gastric and intestinal flora could potentially serve as markers for diagnosing and monitoring the prognosis of gastric cancer, thereby improving early detection and treatment. Studying the microbiota helps us further understand the mechanisms of gastric cancer development and may provide new ideas for targeting microbiota interventions to treat cancer. However, there are still some problems in the current research on the microbiota of gastric cancer. Firstly, the mechanism by which some microbes promote gastric cancer has not been conclusively demonstrated and requires further investigation. Second, due to the differences in sequencing technology, analysis method or study population, the results obtained by many studies are quite different, and the interference of these factors should be excluded as far as possible. Third, there is currently a lack of more definitive biomarkers for gastric cancer other than prevention or eradication of HP infection, which hinders the early diagnosis of gastric cancer. In the future, more convenient and efficient biomarkers should be explored in larger sample sizes. In conclusion, further exploration is needed to modify the microbiota and ultimately improve the prognosis and effectiveness of gastric cancer treatment.
JX conceived and designed the review. YW and NW wrote the draft of the paper. WH and MH collected the information. YW and MB drew the picture. YD, JD, TS, and JX participated in the manuscript revision. All authors read and approved the final manuscript and submission of this manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Nature Science Foundation of China (82373113, JX) Shenyang Breast Cancer Clinical Medical Research Center (2020-48-3-1, TS), Liaoning Cancer Hospital Yangtse River Scholars Project (TS, JX), LiaoNing Revitalization Talents Program (XLYC1907160, JX), Beijing Medical Award Foundation (YXJL-2020-0941-0752, TS), Wu Jieping Medical Foundation (320.6750.2020-12-21,320.6750.2020-6-30, TS) and the Fundamental Research Funds for the Central Universities (202229, ST; 202230, JX).
MB is employed by Kanghui Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
HP, Helicobacter pylori; EBV, Epstein-Barr virus; HBV, Hepatitis B virus; JCV, John Cunningham virus; AG, atrophic gastritis; IM, intestinal metaplasia; FMT, fecal microbiota transplantation; CG, chronic gastritis; TSG, tumor suppressor genes; CagA, cytotoxin-associated gene A; VacA, vesicular cytotoxin A; T4SS, type 4 secretion system; CagPAI, Cag pathogenicity island; EMT, epithelial-mesenchymal transition; MDSCs, myeloid-derived suppressor cells; Treg cells, regulatory T cells; ICIs, immune checkpoint inhibitors; DNMTs, DNA Methyltransferases; LMP1, Latent Membrane Protein 1 (LMP1); LMP2A, Latent Membrane Protein 2A; HBx, hepatitis B virus X protein; F. nucleatum, Fusobacterium nucleatum; TIGIT, T cell immunoreceptors with IG and ITIM domains; PGE2, prostaglandin E2; PKM2, pyruvate kinase M2; mAb, monoclonal antibody; B. fragilis, Bacteroides fragilis; A. muciniphila, Akkermansia muciniphila; NAP, Neutrophil-Activating Protein; HtrA, High Temperature Requirement A; gGT, γ-Glutamyl Transpeptidase; BabA, Blood Group Antigen-Binding Adhesin; SabA, Sialic Acid-Binding Adhesin; OipA, Outer Inflammatory Protein; Hop, Helicobacter pylori Outer Membrane Protein; ROS, Reactive Oxygen Species; DC, Dendritic cell, WHO, World Health Organization.
1. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. (2020) 396(10251):635–48. doi: 10.1016/S0140-6736(20)31288-5
2. Bessède E, Mégraud F. Microbiota and gastric cancer. Semin Cancer Biol (2022) 86:11–7. doi: 10.1016/j.semcancer.2022.05.001
3. Chattopadhyay I, Gundamaraju R, Rajeev A. Diversification and deleterious role of microbiome in gastric cancer. Cancer Rep (Hoboken) (2023), e1878. doi: 10.1002/cnr2.1878
4. Yang J, Zhou X, Liu X, Ling Z, Ji F. Role of the gastric microbiome in gastric cancer: from carcinogenesis to treatment. Front Microbiol (2021) 12:641322. doi: 10.3389/fmicb.2021.641322
5. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17(4):219–32. doi: 10.1038/nri.2017.7
6. Garrett WS. Cancer and the microbiota. Science. (2015) 348(6230):80–6. doi: 10.1126/science.aaa4972
7. Weng MT, Chiu YT, Wei PY, Chiang CW, Fang HL, Wei SC. Microbiota and gastrointestinal cancer. J Formos Med Assoc (2019) 118 Suppl 1:S32–41. doi: 10.1016/j.jfma.2019.01.002
8. Cao L, Yu J. Effect of helicobacter pylori infection on the composition of gastric microbiota in the development of gastric cancer. Gastrointest Tumors. (2015) 2(1):14–25. doi: 10.1159/000380893
9. Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes (2020) 11(5):1220–30. doi: 10.1080/19490976.2020.1762520
10. Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. (2006) 103(3):732–7. doi: 10.1073/pnas.0506655103
11. Li XX, Wong GLH, To KF, Wong VWS, Lai LH, Chow DKL, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PloS One (2009) 4(11):e7985. doi: 10.1371/journal.pone.0007985
12. Zwolinska-Wcisło M, Budak A, Bogdał J, Trojanowska D, Stachura J. Fungal colonization of gastric mucosa and its clinical relevance. Med Sci Monit (2001) 7(5):982–8.
13. Zhang C, Powell SE, Betel D, Shah MA. The gastric microbiome and its influence on gastric carcinogenesis: current knowledge and ongoing research. Hematol Oncol Clin North Am (2017) 31(3):389–408. doi: 10.1016/j.hoc.2017.01.002
14. Zhang S, Shi D, Li M, Li Y, Wang X, Li W. The relationship between gastric microbiota and gastric disease. Scand J Gastroenterol (2019) 54(4):391–6. doi: 10.1080/00365521.2019.1591499
15. Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One (2008) 3(7):e2836. doi: 10.1371/journal.pone.0002836
16. Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. Participation of microbiota in the development of gastric cancer. World J Gastroenterol (2014) 20(17):4948–52. doi: 10.3748/wjg.v20.i17.4948
17. He C, Peng C, Shu X, Wang H, Zhu Z, Ouyang Y, et al. Convergent dysbiosis of gastric mucosa and fluid microbiome during stomach carcinogenesis. Gastric Cancer. (2022) 25(5):837–49. doi: 10.1007/s10120-022-01302-z
18. Zhang X, Li C, Cao W, Zhang Z. Alterations of gastric microbiota in gastric cancer and precancerous stages. Front Cell Infect Microbiol (2021) 11:559148. doi: 10.3389/fcimb.2021.559148
19. Deng Y, Ding X, Song Q, Zhao G, Han L, Ding B, et al. Alterations in mucosa-associated microbiota in the stomach of patients with gastric cancer. Cell Oncol (Dordr). (2021) 44(3):701–14. doi: 10.1007/s13402-021-00596-y
20. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the gastric mucosal microbiome associated with histological stages of gastric carcinogenesis. Front Microbiol (2020) 11:997. doi: 10.3389/fmicb.2020.00997
21. Sun QH, Zhang J, Shi YY, Zhang J, Fu WW, Ding SG. Microbiome changes in the gastric mucosa and gastric juice in different histological stages of Helicobacter pylori-negative gastric cancers. World J Gastroenterol (2022) 28(3):365–80. doi: 10.3748/wjg.v28.i3.365
22. Peng X, Yao S, Huang J, Zhao Y, Chen H, Chen L, et al. Alterations in bacterial community dynamics from noncancerous to Gastric cancer. Front Microbiol (2023) 14:1138928. doi: 10.3389/fmicb.2023.1138928
23. Chen XH, Wang A, Chu AN, Gong YH, Yuan Y. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol (2019) 10:1261. doi: 10.3389/fmicb.2019.01261
24. Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. (2019) 40:336–48. doi: 10.1016/j.ebiom.2018.12.034
25. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. (2018) 67(6):1024–32. doi: 10.1136/gutjnl-2017-314281
26. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, MaChado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. (2018) 67(2):226–36. doi: 10.1136/gutjnl-2017-314205
27. Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. Alterations in gastric microbiota after H. Pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep (2017) 7:44935. doi: 10.1038/srep44935
28. Li Y, Hu Y, Zhan X, Song Y, Xu M, Wang S, et al. Meta-analysis reveals Helicobacter pylori mutual exclusivity and reproducible gastric microbiome alterations during gastric carcinoma progression. Gut Microbes (2023) 15(1):2197835. doi: 10.1080/19490976.2023.2197835
29. Cheung KS, Leung WK. Risk of gastric cancer development after eradication of Helicobacter pylori. World J Gastrointest Oncol (2018) 10(5):115–23. doi: 10.4251/wjgo.v10.i5.115
30. Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. (2014) 63(1):54–63. doi: 10.1136/gutjnl-2013-305178
31. Yoon H, Kim N, Lee HS, Shin CM, Park YS, Lee DH, et al. Helicobacter pylori-negative gastric cancer in South Korea: incidence and clinicopathologic characteristics. Helicobacter. (2011) 16(5):382–8. doi: 10.1111/j.1523-5378.2011.00859.x
32. Kim HN, Kim MJ, Jacobs JP, Yang HJ. Altered gastric microbiota and inflammatory cytokine responses in patients with helicobacter pylori-negative gastric cancer. Nutrients. (2022) 14(23):4981. doi: 10.3390/nu14234981
33. Ding J, Man YG, Deng X, Chen T. Differences in community structure of gastrointestinal tract between Helicobacter pylori positive patients and negative patients with gastric cancer. J Cancer. (2022) 13(6):1905–13. doi: 10.7150/jca.69873
34. Sahan AZ, Hazra TK, Das S. The pivotal role of DNA repair in infection mediated-inflammation and cancer. Front Microbiol (2018) 9:663. doi: 10.3389/fmicb.2018.00663
35. Mathebela P, Damane BP, Mulaudzi TV, Mkhize-Khwitshana ZL, Gaudji GR, Dlamini Z. Influence of the microbiome metagenomics and epigenomics on gastric cancer. Int J Mol Sci (2022) 23(22):13750. doi: 10.3390/ijms232213750
36. Salvatori S, Marafini I, Laudisi F, Monteleone G, Stolfi C. Helicobacter pylori and gastric cancer: pathogenetic mechanisms. Int J Mol Sci (2023) 24(3):2895. doi: 10.3390/ijms24032895
37. Ansari S, Yamaoka Y. Helicobacter pylori virulence factor cytotoxin-associated gene A (CagA)-mediated gastric pathogenicity. Int J Mol Sci (2020) 21(19):7430. doi: 10.3390/ijms21197430
38. Szabò I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, et al. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J (1999) 18(20):5517–27. doi: 10.1093/emboj/18.20.5517
39. Zhu P, Xue J, Zhang ZJ, Jia YP, Tong YN, Han D, et al. Helicobacter pylori VacA induces autophagic cell death in gastric epithelial cells via the endoplasmic reticulum stress pathway. Cell Death Dis (2017) 8(12):3207. doi: 10.1038/s41419-017-0011-x
40. Yokoyama H, Fujii S. Structures and metal-binding properties of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center. Biomolecules. (2014) 4(3):600–15. doi: 10.3390/biom4030600
41. Fu HW. Helicobacter pylori neutrophil-activating protein: from molecular pathogenesis to clinical applications. World J Gastroenterol (2014) 20(18):5294–301. doi: 10.3748/wjg.v20.i18.5294
42. Polenghi A, Bossi F, Fischetti F, Durigutto P, Cabrelle A, Tamassia N, et al. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J Immunol (2007) 178(3):1312–20. doi: 10.4049/jimmunol.178.3.1312
43. Nishioka H, Baesso I, Semenzato G, Trentin L, Rappuoli R, Del Giudice G, et al. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) activates the MAPK pathway in human neutrophils. Eur J Immunol (2003) 33(4):840–9. doi: 10.1002/eji.200323726
44. Harrer A, Boehm M, Backert S, Tegtmeyer N. Overexpression of serine protease HtrA enhances disruption of adherens junctions, paracellular transmigration and type IV secretion of CagA by Helicobacter pylori. Gut Pathog (2017) 9:40. doi: 10.1186/s13099-017-0189-6
45. Hoy B, Löwer M, Weydig C, Carra G, Tegtmeyer N, Geppert T, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep (2010) 11(10):798–804. doi: 10.1038/embor.2010.114
46. Ricci V, Giannouli M, ROmano M, Zarrilli R. Helicobacter pylori gamma-glutamyl transpeptidase and its pathogenic role. World J Gastroenterol (2014) 20(3):630–8. doi: 10.3748/wjg.v20.i3.630
47. Oertli M, Noben M, Engler DB, Semper RP, Reuter S, Maxeiner J, et al. Helicobacter pylori γ-glutamyl transpeptidase and vacuolating cytotoxin promote gastric persistence and immune tolerance. Proc Natl Acad Sci U S A. (2013) 110(8):3047–52. doi: 10.1073/pnas.1211248110
48. Ling SSM, Yeoh KG, Ho B. Helicobacter pylori γ-glutamyl transpeptidase: a formidable virulence factor. World J Gastroenterol (2013) 19(45):8203–10. doi: 10.3748/wjg.v19.i45.8203
49. Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, et al. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem (2011) 286(28):25256–64. doi: 10.1074/jbc.M111.233601
50. Gerhard M, Lehn N, Neumayer N, Borén T, Rad R, Schepp W, et al. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A. (1999) 96(22):12778–83. doi: 10.1073/pnas.96.22.12778
51. Unemo M, Aspholm-Hurtig M, Ilver D, Bergström J, Borén T, Danielsson D, et al. The sialic acid binding SabA adhesin of Helicobacter pylori is essential for nonopsonic activation of human neutrophils. J Biol Chem (2005) 280(15):15390–7. doi: 10.1074/jbc.M412725200
52. Horridge DN, Begley AA, Kim J, Aravindan N, Fan K, Forsyth MH. Outer inflammatory protein a (OipA) of Helicobacter pylori is regulated by host cell contact and mediates CagA translocation and interleukin-8 response only in the presence of a functional cag pathogenicity island type IV secretion system. Pathog Dis (2017) 75(8):ftx113. doi: 10.1093/femspd/ftx113
53. Teymournejad O, Mobarez AM, Hassan ZM, Moazzeni SM, Ahmadabad HN. In vitro suppression of dendritic cells by Helicobacter pylori OipA. Helicobacter. (2014) 19(2):136–43. doi: 10.1111/hel.12107
54. Teymournejad O, Mobarez AM, Hassan ZM, Talebi Bezmin Abadi A. Binding of the Helicobacter pylori OipA causes apoptosis of host cells via modulation of Bax/Bcl-2 levels. Sci Rep (2017) 7(1):8036. doi: 10.1038/s41598-017-08176-7
55. Behrens IK, Busch B, Ishikawa-Ankerhold H, Palamides P, Shively JE, Stanners C, et al. The hopQ-CEACAM interaction controls cagA translocation, phosphorylation, and phagocytosis of helicobacter pylori in neutrophils. mBio. (2020) 11(1):e03256–19. doi: 10.1128/mBio.03256-19
56. Gur C, Maalouf N, Gerhard M, Singer BB, Emgård J, Temper V, et al. The Helicobacter pylori HopQ outermembrane protein inhibits immune cell activities. Oncoimmunology. (2019) 8(4):e1553487. doi: 10.1080/2162402X.2018.1553487
57. Nguyen QA, Schmitt L, Mejías-Luque R, Gerhard M. Effects of Helicobacter pylori adhesin HopQ binding to CEACAM receptors in the human stomach. Front Immunol (2023) 14:1113478. doi: 10.3389/fimmu.2023.1113478
58. Kennemann L, Brenneke B, Andres S, Engstrand L, Meyer TF, Aebischer T, et al. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect Immun (2012) 80(12):4364–73. doi: 10.1128/IAI.00977-12
59. Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res (1999) 27(16):3325–33. doi: 10.1093/nar/27.16.3325
60. Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. (2009) 124(10):2367–74. doi: 10.1002/ijc.24219
61. Muhammad JS, Eladl MA, Khoder G. Helicobacter pylori-induced DNA methylation as an epigenetic modulator of gastric cancer: recent outcomes and future direction. Pathogens. (2019) 8(1):23. doi: 10.3390/pathogens8010023
62. Fu HW, Lai YC. The Role of Helicobacter pylori Neutrophil-Activating Protein in the Pathogenesis of H. pylori and Beyond: from a virulence factor to therapeutic targets and therapeutic agents. Int J Mol Sci (2022) 24(1):91. doi: 10.3390/ijms24010091
63. Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, et al. Helicobacter pylori virulence factors-mechanisms of bacterial pathogenicity in the gastric microenvironment. Cells. (2020) 10(1):27. doi: 10.3390/cells10010027
64. Ansari S, Yamaoka Y. Helicobacter pylori Virulence Factors Exploiting Gastric Colonization and its Pathogenicity. Toxins (Basel). (2019) 11(11):677. doi: 10.3390/toxins11110677
65. Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, et al. Helicobacter pylori sabA adhesin in persistent infection and chronic inflammation. Science. (2002) 297(5581):573–8. doi: 10.1126/science.1069076
66. Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer C, et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut. (2022) 71(3):457–66. doi: 10.1136/gutjnl-2020-323392
67. Das S, Suarez G, Beswick EJ, Sierra JC, Graham DY, Reyes VE. Expression of B7-H1 on gastric epithelial cells: its potential role in regulating T cells during Helicobacter pylori infection. J Immunol (2006) 176(5):3000–9. doi: 10.4049/jimmunol.176.5.3000
68. Oster P, Vaillant L, McMillan B, Velin D. The efficacy of cancer immunotherapies is compromised by helicobacter pylori infection. Front Immunol (2022) 13:899161. doi: 10.3389/fimmu.2022.899161
69. Naseem M, Barzi A, Brezden-Masley C, Puccini A, Berger MD, Tokunaga R, et al. Outlooks on Epstein-Barr virus associated gastric cancer. Cancer Treat Rev (2018) 66:15–22. doi: 10.1016/j.ctrv.2018.03.006
70. Tavakoli A, Monavari SH, Solaymani Mohammadi F, Kiani SJ, Armat S, Farahmand M. Association between Epstein-Barr virus infection and gastric cancer: a systematic review and meta-analysis. BMC Cancer. (2020) 20(1):493. doi: 10.1186/s12885-020-07013-x
71. Kusano M, Toyota M, Suzuki H, Akino K, Aoki F, Fujita M, et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. (2006) 106(7):1467–79. doi: 10.1002/cncr.21789
72. Qiu M, He C, Lu S, Guan W, Wang F, Wang X, et al. Prospective observation: Clinical utility of plasma Epstein–Barr virus DNA load in EBV-associated gastric carcinoma patients. Int J Cancer. (2020) 146(1):272–80. doi: 10.1002/ijc.32490
73. Singh S, Jha HC. Status of epstein-barr virus coinfection with helicobacter pylori in gastric cancer. J Oncol (2017) 2017:3456264. doi: 10.1155/2017/3456264
74. Su S, Zou Z, Chen F, Ding N, Du J, Shao J, et al. CRISPR-Cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer. Oncoimmunology. (2017) 6(1):e1249558. doi: 10.1080/2162402X.2016.1249558
75. Wang H, Chen XL, Liu K, Bai D, Zhang WH, Chen XZ, et al. Associations between gastric cancer risk and virus infection other than epstein-barr virus: A systematic review and meta-analysis based on epidemiological studies. Clin Transl Gastroenterol (2020) 11(7):e00201. doi: 10.14309/ctg.0000000000000201
76. Mirzaei H, Goudarzi H, Eslami G, Faghihloo E. Role of viruses in gastrointestinal cancer. J Cell Physiol (2018) 233(5):4000–14. doi: 10.1002/jcp.26194
77. Li M, Wu S, Luo H, Niu J, Yan Y, Fang Y, et al. Serological and molecular characterization of hepatitis B virus infection in gastric cancer. Front Cell Infect Microbiol (2022) 12:894836. doi: 10.3389/fcimb.2022.894836
78. Cui H, Jin Y, Chen F, Ni H, Hu C, Xu Y, et al. Clinicopathological evidence of hepatitis B virus infection in the development of gastric adenocarcinoma. J Med Virol (2020) 92(1):71–7. doi: 10.1002/jmv.25584
79. Song C, Lv J, Liu Y, Chen JG, Ge Z, Zhu J, et al. Associations between hepatitis B virus infection and risk of all cancer types. JAMA Netw Open (2019) 2(6):e195718. doi: 10.1001/jamanetworkopen.2019.5718
80. Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, et al. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharmacol Ther (2020) 51(8):770–80. doi: 10.1111/apt.15675
81. Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. (2019) 1872(2):188309. doi: 10.1016/j.bbcan.2019.07.004
82. Li ZP, Liu JX, Lu LL, Wang LL, Xu L, Guo ZH, et al. Overgrowth of Lactobacillus in gastric cancer. World J Gastrointest Oncol (2021) 13(9):1099–108. doi: 10.4251/wjgo.v13.i9.1099
83. Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem (2012) 19(10):1519–29. doi: 10.2174/092986712799828283
84. Ohta K, Kawano R, Ito N. Lactic acid bacteria convert human fibroblasts to multipotent cells. PloS One (2012) 7(12):e51866. doi: 10.1371/journal.pone.0051866
85. Boehm ET, Thon C, Kupcinskas J, Steponaitiene R, Skieceviciene J, Canbay A, et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci Rep (2020) 10(1):16240. doi: 10.1038/s41598-020-73448-8
86. Nie S, Wang A, Yuan Y. Comparison of clinicopathological parameters, prognosis, micro-ecological environment and metabolic function of Gastric Cancer with or without Fusobacterium sp. Infection. J Cancer. (2021) 12(4):1023–32. doi: 10.7150/jca.50918
87. Stasiewicz M, Karpiński TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol (2022) 86(Pt 3):633–42. doi: 10.1016/j.semcancer.2021.11.002
88. Hatta MNA, Mohamad Hanif EA, Chin SF, Neoh HM. Pathogens and carcinogenesis: A review. Biol (Basel). (2021) 10(6):533. doi: 10.3390/biology10060533
89. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
90. Sun Z, Xiong C, Teh SW, Lim JCW, Kumar S, Thilakavathy K. Mechanisms of oral bacterial virulence factors in pancreatic cancer. Front Cell Infect Microbiol (2019) 9:412. doi: 10.3389/fcimb.2019.00412
91. Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes (2020) 11(3):511–25. doi: 10.1080/19490976.2019.1695494
92. Zhong M, Xiong Y, Zhao J, Gao Z, Ma J, Wu Z, et al. Candida albicans disorder is associated with gastric carcinogenesis. Theranostics. (2021) 11(10):4945–56. doi: 10.7150/thno.55209
93. Yu D, Liu Z. The research progress in the interaction between Candida albicans and cancers. Front Microbiol (2022) 13:988734. doi: 10.3389/fmicb.2022.988734
94. Nasry WHS, Rodriguez-Lecompte JC, Martin CK. Role of COX-2/PGE2 mediated inflammation in oral squamous cell carcinoma. Cancers (Basel). (2018) 10(10):348. doi: 10.3390/cancers10100348
95. Wu ZF, Zou K, Xiang CJ, Jin ZJ, Ding HH, Xu S, et al. Helicobacter pylori infection is associated with the co-occurrence of bacteria in the oral cavity and the gastric mucosa. Helicobacter. (2021) 26(2):e12786. doi: 10.1111/hel.12786
96. Chen X, Wang N, Wang J, Liao B, Cheng L, Ren B. The interactions between oral-gut axis microbiota and Helicobacter pylori. Front Cell Infect Microbiol (2022) 12:914418. doi: 10.3389/fcimb.2022.914418
97. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533
98. Panebianco C, Potenza A, Andriulli A, Pazienza V. Exploring the microbiota to better understand gastrointestinal cancers physiology. Clin Chem Lab Med (2018) 56(9):1400–12. doi: 10.1515/cclm-2017-1163
99. Qi YF, Sun JN, Ren LF, Cao XL, Dong JH, Tao K, et al. Intestinal microbiota is altered in patients with gastric cancer from Shanxi province, China. Dig Dis Sci (2019) 64(5):1193–203. doi: 10.1007/s10620-018-5411-y
100. Sarhadi V, Mathew B, Kokkola A, Karla T, Tikkanen M, Rautelin H, et al. Gut microbiota of patients with different subtypes of gastric cancer and gastrointestinal stromal tumors. Gut Pathog (2021) 13(1):11. doi: 10.1186/s13099-021-00403-x
101. Zhang K, Ji X, Song Z, Wu F, Qu Y, Jin X, et al. Butyrate inhibits gastric cancer cells by inducing mitochondria-mediated apoptosis. Comb Chem High Throughput Screen (2023) 26(3):630–8. doi: 10.2174/1386207325666220720114642
102. Liang Y, Rao Z, Du D, Wang Y, Fang T. Butyrate prevents the migration and invasion, and aerobic glycolysis in gastric cancer via inhibiting Wnt/β-catenin/c-Myc signaling. Drug Dev Res (2023) 84(3):532–41. doi: 10.1002/ddr.22043
103. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
104. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
105. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
106. Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. (2017) 19(10):848–55. doi: 10.1016/j.neo.2017.08.004
107. Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinf (2018) 16(1):33–49. doi: 10.1016/j.gpb.2017.06.002
108. Xu X, Ying J. Gut microbiota and immunotherapy. Front Microbiol (2022) 13:945887. doi: 10.3389/fmicb.2022.945887
109. Barra WF, Sarquis DP, Khayat AS, Khayat BCM, Demachki S, Anaissi AKM, et al. Gastric cancer microbiome. Pathobiology. (2021) 88(2):156–69. doi: 10.1159/000512833
110. van Vliet MJ, Harmsen HJM, de Bont ESJM, Tissing WJE. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PloS Pathog (2010) 6(5):e1000879. doi: 10.1371/journal.ppat.1000879
111. Kumar R, Sharma A, Gupta M, Padwad Y, Sharma R. Cell-free culture supernatant of probiotic lactobacillus fermentum protects against H2O2-induced premature senescence by suppressing ROS-akt-mTOR axis in murine preadipocytes. Probiotics Antimicrob Proteins. (2020) 12(2):563–76. doi: 10.1007/s12602-019-09576-z
112. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
113. Deng H, Li Z, Tan Y, Guo Z, Liu Y, Wang Y, et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci Rep (2016) 6:29401. doi: 10.1038/srep29401
114. Dai Z, Zhang J, Wu Q, Fang H, Shi C, Li Z, et al. Intestinal microbiota: a new force in cancer immunotherapy. Cell Commun Signal (2020) 18(1):90. doi: 10.1186/s12964-020-00599-6
115. Fakharian F, Asgari B, Nabavi-Rad A, Sadeghi A, Soleimani N, Yadegar A, et al. The interplay between Helicobacter pylori and the gut microbiota: An emerging driver influencing the immune system homeostasis and gastric carcinogenesis. Front Cell Infect Microbiol (2022) 12:953718. doi: 10.3389/fcimb.2022.953718
116. Antushevich H. Fecal microbiota transplantation in disease therapy. Clin Chim Acta (2020) 503:90–8. doi: 10.1016/j.cca.2019.12.010
117. Wang JW, Kuo CH, Kuo FC, Wang YK, Hsu WH, Yu FJ, et al. Fecal microbiota transplantation: Review and update. J Formos Med Assoc (2019) 118 Suppl 1:S23–31. doi: 10.1016/j.jfma.2018.08.011
118. Łusiak-Szelachowska M, Weber-Dąbrowska B, Jończyk-Matysiak E, Wojciechowska R, Górski A. Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog (2017) 9:44. doi: 10.1186/s13099-017-0196-7
119. Nandi D, Parida S, Sharma D. The gut microbiota in breast cancer development and treatment: the good, the bad, and the useful! Gut Microbes (2023) 15(1):2221452. doi: 10.1080/19490976.2023.2221452
120. El Tekle G, Garrett WS. Bacteria in cancer initiation, promotion and progression. Nat Rev Cancer (2023) 23(9):600–18. doi: 10.1038/s41568-023-00594-2
121. Hussain S, Joo J, Kang J, Kim B, Braun GB, She ZG, et al. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat BioMed Eng. (2018) 2(2):95–103. doi: 10.1038/s41551-017-0187-5
122. Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat BioMed Eng. (2019) 3(9):717–28. doi: 10.1038/s41551-019-0423-2
123. Dong X, Pan P, Zheng DW, Bao P, Zeng X, Zhang XZ. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci Adv (2020) 6(20):eaba1590. doi: 10.1126/sciadv.aba1590
124. Cuomo P, Papaianni M, Fulgione A, Guerra F, Capparelli R, Medaglia C. An innovative approach to control H. pylori-induced persistent inflammation and colonization. Microorganisms (2020) 8(8):1214. doi: 10.3390/microorganisms8081214
125. Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. (2018) 362(6416):776–80. doi: 10.1126/science.aau5812
126. Franco-de-Moraes AC, de Almeida-Pititto B, da Rocha Fernandes G, Gomes EP, da Costa Pereira A, Ferreira SRG. Worse inflammatory profile in omnivores than in vegetarians associates with the gut microbiota composition. Diabetol Metab Syndr (2017) 9:62. doi: 10.1186/s13098-017-0261-x
127. Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J (2012) 6(10):1848–57. doi: 10.1038/ismej.2012.27
128. Vernieri C, Fucà G, Ligorio F, Huber V, Vingiani A, Iannelli F, et al. Fasting-mimicking diet is safe and reshapes metabolism and antitumor immunity in patients with cancer. Cancer Discovery (2022) 12(1):90–107. doi: 10.1158/2159-8290.CD-21-0030
129. Hur H. Prospective clinical study for early recovery after gastric cancer surgery. clinicaltrials.gov (2012). Report No.: NCT01642953.
130. Rubio-Patiño C, Bossowski JP, Donatis GMD, Mondragón L, Villa E, Aira LE, et al. Low-protein diet induces IRE1α-dependent anticancer immunosurveillance. Cell Metab (2018) 27(4):828–842.e7. doi: 10.1016/j.cmet.2018.02.009
131. Zhang XY, Yang KL, Li Y, Zhao Y, Jiang KW, Wang Q, et al. Can dietary nutrients prevent cancer chemotherapy-induced cardiotoxicity? An evidence mapping of human studies and animal models. Front Cardiovasc Med (2022) 9:921609. doi: 10.3389/fcvm.2022.921609
132. Kaźmierczak-Siedlecka K, Daca A, Fic M, van de Wetering T, Folwarski M, Makarewicz W. Therapeutic methods of gut microbiota modification in colorectal cancer management – fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes (2020) 11(6):1518–30. doi: 10.1080/19490976.2020.1764309
133. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol (2019) 16(10):605–16. doi: 10.1038/s41575-019-0173-3
134. Aguilar-Toalá JE, Hall FG, Urbizo-Reyes UC, Garcia HS, Vallejo-Cordoba B, González-Córdova AF, et al. In silico prediction and in vitro assessment of multifunctional properties of postbiotics obtained from two probiotic bacteria. Probiotics Antimicrob Proteins. (2020) 12(2):608–22. doi: 10.1007/s12602-019-09568-z
135. Hull MA. Biospecimen collection for:Prebiotic effect of eicosapentaenoic acid treatment for colorectal cancer liver metastases. clinicaltrials.gov (2021). Report No.: NCT04682665.
136. Zheng M, Pi X, Li H, Cheng S, Su Y, Zhang Y, et al. Ganoderma spp. polysaccharides are potential prebiotics: a review. Crit Rev Food Sci Nutr (2022), 1–19. doi: 10.1080/10408398.2022.2110035
Keywords: gastric cancer, microbiota, Helicobacter pylori (HP), Epstein-Barr virus (EBV), immunotherapy
Citation: Wang Y, Han W, Wang N, Han M, Ban M, Dai J, Dong Y, Sun T and Xu J (2023) The role of microbiota in the development and treatment of gastric cancer. Front. Oncol. 13:1224669. doi: 10.3389/fonc.2023.1224669
Received: 18 May 2023; Accepted: 28 August 2023;
Published: 29 September 2023.
Edited by:
Mingzhou Guo, People’s Liberation Army General Hospital, ChinaReviewed by:
Li Zhang, University of Minnesota Twin Cities, United StatesCopyright © 2023 Wang, Han, Wang, Han, Ban, Dai, Dong, Sun and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junnan Xu, eGpuMDAyQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.