
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 11 October 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1222575
This article is part of the Research TopicCancer Stem Cells in Adenocarcinoma as Therapeutic Targets: From Basic Research to Clinical ApplicationView all 5 articles
The role of tumor interaction with stromal components during carcinogenesis is crucial for the design of efficient cancer treatment approaches. It is widely admitted that tumor hypoxic stress is associated with tumor aggressiveness and thus impacts susceptibility and resistance to different types of treatments. Notable biological processes that hypoxia functions in include its regulation of tumor heterogeneity and plasticity. While hypoxia has been reported as a major player in tumor survival and dissemination regulation, the significance of hypoxia inducible factors in cancer stem cell development remains poorly understood. Several reports indicate that the emergence of cancer stem cells in addition to their phenotype and function within a hypoxic tumor microenvironment impacts cancer progression. In this respect, evidence showed that cancer stem cells are key elements of intratumoral heterogeneity and more importantly are responsible for tumor relapse and escape to treatments. This paper briefly reviews our current knowledge of the interaction between tumor hypoxic stress and its role in stemness acquisition and maintenance. Our review extensively covers the influence of hypoxia on the formation and maintenance of cancer stem cells and discusses the potential of targeting hypoxia-induced alterations in the expression and function of the so far known stem cell markers in cancer therapy approaches. We believe that a better and integrated understanding of the effect of hypoxia on stemness during carcinogenesis might lead to new strategies for exploiting hypoxia-associated pathways and their targeting in the clinical setting in order to overcome resistance mechanisms. More importantly, at the present time, efforts are oriented towards the design of innovative therapeutical approaches that specifically target cancer stem cells.
Cancer remains one of the leading causes of death worldwide. The mortality rate associated with cancer is high because subpopulations of cancer cells exhibit metastasis. Indeed, metastatic invasion involves complex overlapping processes in which cells undergo multiple steps of reprogramming to promote mechanisms of repair, resistance to cell death, adaptation to changes in metabolism, acquisition of stem cell-like properties, and ultimately, survival. In addition to tumor plasticity and heterogeneity, these subpopulations are further equipped with the capacity to resist therapeutic strategies (1). Tumor heterogeneity refers to the variations observed among tumors of the same type in different patients. This diversity serves as the foundation for the development of personalized treatment approaches aimed at maximizing effectiveness. Furthermore, within a tumor there exists a cellular heterogeneity due to the variable microenvironment shaping the tumor. Hypoxia within solid tumors contributes to this heterogeneity and shapes the behavior of a cell population. As a result, different subpopulations of cancer cells may have differential responses to the same therapy; therefore, the more heterogenous the tumor is, the more likely it will resist therapy and be defined as a treatment-resistant tumor. Within the tumor microenvironment, cancer stem cells (CSCs) are supported by hypoxia and are known to be resistant to therapy. These cells are key to cancer progression and cancer recurrence.
In this review, we examine the existing understanding of the effects of hypoxic stress within the tumor microenvironment (TME) on tumor heterogeneity, plasticity, and resistance. We address the mechanisms that lead to the generation of CSCs, focusing on the potential role of hypoxia in stemness acquisition and maintenance. Identifying CSCs populations within a tumor necessitates a clear understanding of their molecular characteristics, as such we have compiled a set of CSCs markers that are currently recognized as an identifying factor and hence a possible target for therapy. We also discuss the resistant mechanisms that these cells adapt in response to therapy and accordingly, the putative therapeutic strategies for targeting these multifaceted interactions of CSC with TME components.
The tumor microenvironment (TME) comprises tumor cells, immune cells, signaling molecules, blood vessels and the extracellular matrix (ECM) components (2). An important feature of the TME of solid tumors is hypoxia that arises when oxygen decreases below the level required to maintain tissue homeostasis. The oxygen percentage varies depending on tumor type and has been reported to be as low as 0.3% in pancreatic cancer (6.8% normal tissue), and 1.9% in lung cancer (5.6% normal tissue) (3). Hypoxia ensues due to a decrease in blood oxygen content, or to the non-availability or improper structure of the blood vessels, that could result from increasing proliferative rate of cancer cells (4, 5). The plastic nature of the TME, therefore, results in variations in the severity (mild or severe) and duration of hypoxia (chronic or intermittent).
HIF transcription factors are the main sensors for hypoxia. HIF are comprised of HIF1, HIF2 and HIF3 and include three oxygen sensitive alpha (α) subunits and three nuclear beta (β) subunits. HIFs form a heterodimer of the two subunits α and β. Under normal oxygen tension, the HIF-α subunit is inhibited by Factor Inhibiting HIF (FIH). FIH is an asparagine hydroxylase that hydroxylates HIF and blocks its association with transcriptional co-activators, thus inhibiting its transcriptional activity (6–8). HIF is also degraded following hydroxylation by prolyl hydroxylase (PHD). However, when exposed to low oxygen concentrations, PHD and FIH are down-regulated resulting in the stabilization of HIF-α. HIF-α subsequently translocates to the nucleus, dimerizes with the β subunit and activates the expression of genes that promote carcinogenesis.
HIF-1α is ubiquitously expressed and is degraded under normal oxygen tension, whereas HIF-1β is a stable constitutively expressed nuclear protein. Both proteins have similar DNA sequence specificity, but they differ in their transactivation domains, suggesting that each subunit has distinct roles. This distinct role for HIF-1α and HIF-1β was demonstrated through deletion experiments conducted in mice that showed that HIF-1α and HIF-1β signaling in breast tumors control tumor dissemination in a site-specific manner (9). HIF-1α binds to specific hypoxia responsive elements (HRE) on target genes, it induces NFκB activation resulting in the expression of targets including MIP-2/CXCL2/3, CXCL1 and TNFα (10). Together these induce proliferation of pre-cancerous lesions, thereby facilitating tumorigenesis (11), including invasion of tissues, cell survival, recurrence of tumors, and the formation of CSCs (12, 13).
HIF-2α is highly homologous to HIF-1α and is regulated in a similar fashion through ubiquitin mediated proteasomal degradation, however the expression pattern of the two proteins is distinct: HIF-2α being expressed mainly in vascularized cells, several evidence points to its role in mediating the remodeling and recruitment of vasculature (14). HIF-2α is also found to mediate the chronic hypoxic response (14). In addition to its roles in the induction of EMT (15), and CSCs induction (16), HIF-2α is shown to be essential for T-regs development (17). Furthermore, studies showed that a crosstalk between the expression of HIF-1α and HIF-2α in T-regs contributes to a tumor-suppressive activity (17).
HIF-3α is less studied, and it has been shown to play a role in cancer cell invasion and migration (18). And some research has demonstrated a positive role for HIF-3α in non-small cell lung cancer (NSCLC) (19). However recent work evaluated the expression levels of HIF-3α in various types of cancer, and interestingly found that in contrast to HIF-1α and HIF-2α, an increase in expression levels of HIF-3α correlated with better survival (20). Additional studies are needed to further dissect the role of this protein and its interplay with HIF1 and HIF2 in tumorigenesis of specific cancer types.
Moreover, the hypoxic TME is acidic because HIF1 regulates tumor cells’ metabolic activities, nutrient sensing, and availability (21). In tumor cells glycolysis results from the anaerobic breakdown of glucose due to the lack of oxygen or from aerobic glycolysis (Warburg effect), which further leads to the production of lactate resulting in an acidic microenvironment (21). HIF1 directly plays a role in this process by increasing the expression of pyruvate dehydrogenase kinase, that subsequently inhibits pyruvate dehydrogenase and thereby represses oxygen consumption and redirects pyruvate to be used in glycolysis (21). HIF1 also enhances the expression of glucose transporters and glycolytic enzymes (22). The resultant acidic TME has significant effects on several cells including suppressing the immune response (23).
Finally, the variability in hypoxia directly impacts the behaviors of the tumor vis a vis resistance to therapy and immune escape and renders targeted treatment more challenging (3, 24). HIF protein response to different types of hypoxias plays a role in this by increasing the complexity of the TME. For example, HIF-1α expression is an acute response and it gets degraded in chronic hypoxia, whereas HIF-2α protein levels increase for longer duration. HIF-2α on the other hand is more sensitive to mild hypoxia (5%) compared to HIF-1α (25). In addition, the cyclic nature of hypoxia differentially controls HIFs, HIF-1α increases in cyclic hypoxia but HIF-2α decreases (26) but it is important to note that it is very difficult to monitor and follow the spaciotemporal hypoxia fluctuations in individual tumors. And it is evident that better understanding of the hypoxia response in individual patients would be necessary to initiate effective treatment strategies.
To adapt to hypoxic stress, cells activate several genes that regulate many pathways (27). Hypoxia plays an important role in the induction of epithelial to mesenchymal transition (EMT), where epithelial cells acquire the capacity to migrate and invade neighboring tissues (28). Hypoxia within the TME plays an important role in CSCs initiation and maintenance (29). CSCs drive tumor initiation, recurrence, metastatic potential, residual disease, and therapy resistance. Furthermore, these cells maintain properties of normal stem cells and are unique in that they are capable of self-renewal and remaining undifferentiated (30). Understanding the processes that give rise to and sustain CSCs is therefore crucial as they are present across various cancer types and targeting them could be detrimental to tumor survival.
While cancer occurs as a sporadic event, resultant from environmental factors (carcinogens, chemicals, biological agents, radiation) or as an inherited event, CSCs origins are debated, and evidence supports that they could originate from existing stem cells or develop following tumor formation. Because cancer can develop from the gradual accumulation of mutations in a single specific cell over a prolonged period, stem cells – that could have long lifespans – have the potential to accumulate mutations that initiate cancer. This is supported by early work where pathologists determined that cancer tissues contain cells that exhibit properties of early embryonic dormant tissue (31) that lead to the “embryonic rest” hypothesis of cancer development which suggested that cancer may arise from embryonic dormant cells that persist in developing organs after embryogenesis (32).
In the TME, hypoxia plays a key role in the acquisition of CSCs and this occurs on several fronts. Under hypoxic conditions, signaling pathways regulate stemness, pluripotency and viability of CSC populations. For example, HIF-1α regulates stem cell phenotypes through activation of signaling pathways including Notch, TGF-/SMAD, NF-B, PI3K/Akt/mTOR, STAT3, MAPK/ERK as well as via transcription factors including C/EBPδ, SOX2 and c-MYC (33, 34) (Figure 1). In hypoxic areas, HIF-1α protein has been shown to activate Notch, Wnt, and Hedgehog (Hh) pathways target genes, inhibiting their differentiation and stimulating stem cell self-renewal and multi-potency (39). The Notch receptors, along with the Jagged or Delta ligand family, translocate into the nucleus to create a DNA-binding complex. Cofactors like mastermind-like (MAML), CSL, NICD, and p300, together promote the activation of Notch target genes. This process significantly contributes to the maintenance of the stem cell population (39, 40). Furthermore, hypoxia and HIF factors have also been shown to enhance the propagation of CSCs identified by the upregulation of CSC markers including CD133, Sox2, Oct4, CD44, and ALDH (34) (Figure 1). This suggests a particular role for HIF in cancer cell phenotype and plasticity. Finally, hypoxia through HIF-1α activation induces methylation of genes promoting CSCs. Recent evidence has shown that mutated DNMT3a, in a mechanistic manner, can activate specific enhancers, resulting in localized DNA methylation and histone acetylation changes. These alterations ultimately lead to disruptions in stemness pathways (41). Moreover, epigenetic modifications of the DNA packaging protein Histone H3 (H3Kme3 and H3K27Ac) at the promoter region of IFN-γ increases the expression of IFN stimulated genes, including PD-L1 expression (42). PD-L1 in turn promotes CSC expansion (43).
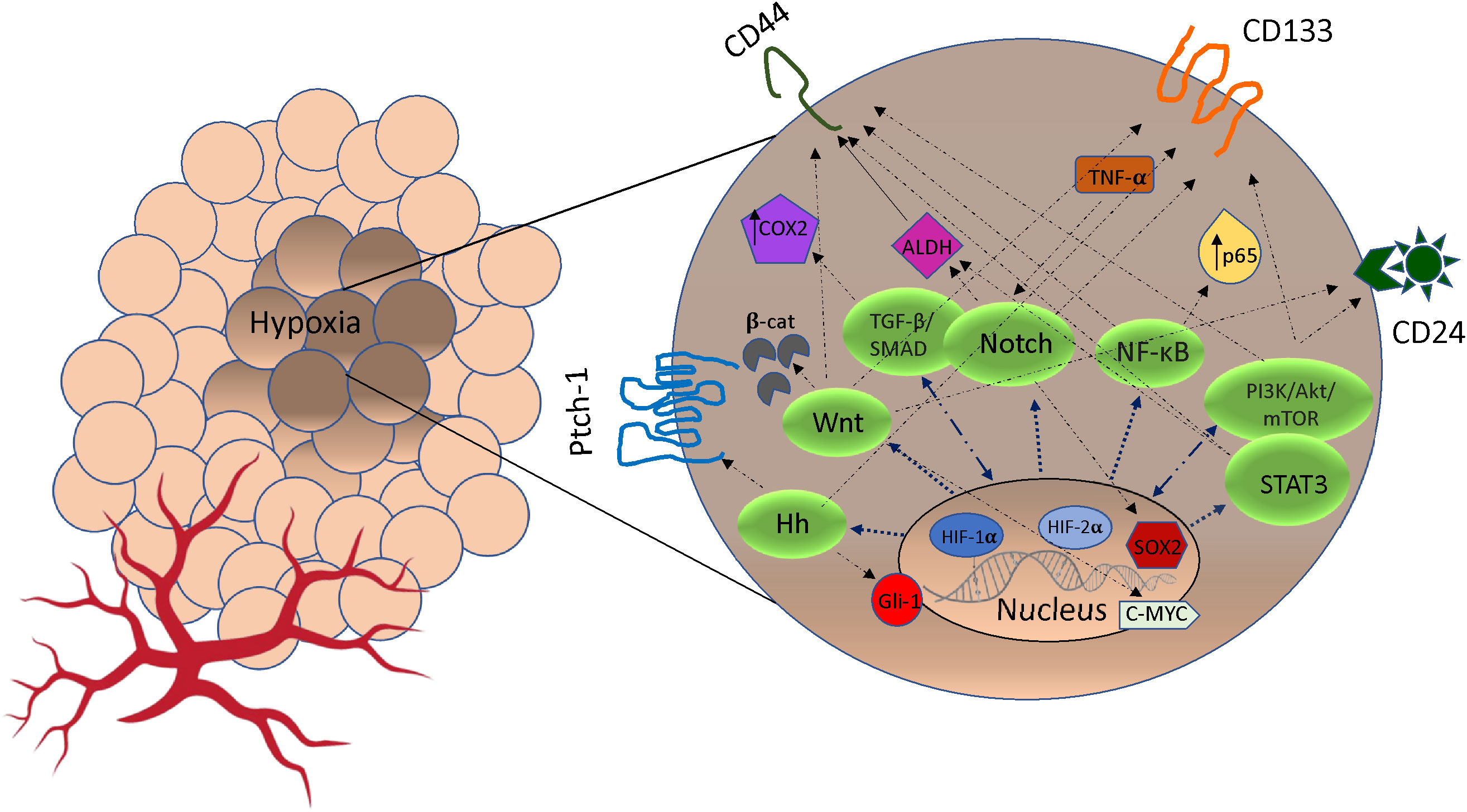
Figure 1 The crosstalk between pathways signaling and markers of cancer stem cells (CSCs) stimulated by hypoxia. Under hypoxic condition, Notch, Wnt, Hedgehog (Hh), NF-κB, TGF-β/SMAD, PI3K/AkT/mTOR and STAT3 pathways can be activated by HIF expression. In some cases, cross-talk between pathways may promote markers expression (CD44, CD24, ALDH1, Patch-1, PROM1, Gli-1, SOX2, c-MYC, p65, TNF-α/β, COX2) that promote phenotypes of CSCs and resistance to cancer therapeutics (35–38). Hh, Hedgehog; TGF-β, Transforming Growth Factor Beta; NF-κB, Nuclear factor-κB; PI3K, intracellular phosphatidylinositol kinase; Akt, serine/threonine kinase; mTOR, mammalian target of rapamycin; STAT3, signal transducer and activator of transcription 3; β-cat, β-catenin; COX2, cytochrome c oxidase subunit 2; ADLH, aldehyde dehydrogenase; TNF-α, Tumor Necrosis Factor Alpha; CD133, Cluster of Differentiation 133; CD24, Cluster of Differentiation 24; CD44, Cluster of Differentiation 44.
Another factor in the TME that contributes to tumor progression and sustains CSCs is the intercellular communication between cancer and stromal cells within the TME. Recent studies demonstrated that non-CSCs could become CSC by processes mediated by secreted factors. Indeed, proteins, cytokines, chemokines, microRNAs, and other substances could be secreted by cancer cells to mediate tumor maintenance and CSCs formation (44–46). It has been reported that IL-6 present in the TME can induce non-CSCs to transform into tumor stem cells (47, 48). Likewise, tumor associated mesenchymal stem cells through direct contact with cancer cells have been shown to promote CSCs features through pathways involving microRNAs (49). Cancer-associated fibroblasts (CAFs) release hepatocyte growth factors (HGF) and annexin A1 which have the ability to revert differentiated tumor cells back to stem cell-like phenotypes (50). An additional example that highlighted the importance of intercellular communications in tumorigenesis came from work that demonstrated that macrophages, could secrete an increased amount of the cytokine osteopontin (OPN) when cocultured with CD44-positive cancer cells, that subsequently promoted tumorigenicity (51). Macrophages secrete oncostatin-M, an IL-6 family cytokine, that can activate the dedifferentiation of non-CSCs into aggressive CSCs (52). Cancer associated fibroblasts can also modulate CSC plasticity through signaling pathways including IGF-II/IGF1R; FAK and c-Met/FRA1/HEY1 (53–55). The activation of NOTCH1 signaling by dermal fibroblasts derived from mesenchymal stem cells was observed to regulate both plasticity and stemness (56). Therefore, the interaction of the microenvironment facilitates the plasticity and stemness of these cells and can be modulated by the secretome.
Several specific markers are established to categorize cancer stem cells, enabling their identification, purification, and potential use for targeted therapies (Table 1). These markers’ expression is regulated by spatial and temporal characteristics, indicating the remarkable adaptability of these cells (152). In 1997, the first evidence of the existence of CSCs surfaced from experiments that demonstrated the existence of a subpopulation of CD34-expressing cells in leukemia. These cells were capable of initiating tumors in NOD/SCID mice that resembled the donor’s tumor (32). Following this, in 2002 it was shown that cancer stem like sphere-forming cells from human gliomas could induce tumors resembling the original tumor when transplanted intracranially in nude mice (153, 154). As the research progressed, additional specific markers of CSCs were identified. The most common markers that are in use for CSC isolation are CD133 (also known as PROM1), CD44, ALDH1A1, CD34, CD24 (155, 156). Indeed, CD133 positive cells isolated from colon carcinomas could grow as tumor spheroids in vitro as well as initiate tumor growth when xenografted in immunodeficient mice (157). Hypoxia and HIF transcription factors play a key role in the expression of these markers, however the molecular mechanisms leading to the increased expression is not yet understood for several of these markers (Table 1). It should be noted that due to the expression of a heterogeneous range of stem cell markers, cancer stem cells that originate from a hypoxic microenvironment may exist in a distinct stem cell states or exhibit variations in the expression of different sets of stem cell markers. Targeting CSCs markers could be effective in treatment strategies. Indeed, downregulation of CD133 using short hairpin RNAs led to slower cell growth of human metastatic melanoma and slowed the spheroid formation and metastatic potential of melanoma (158).
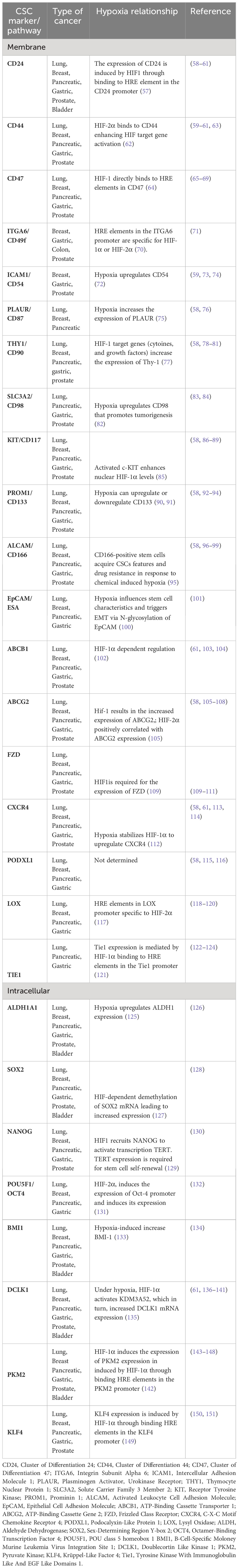
Table 1 Relationship of various cancer stem cell markers with hypoxia in selected solid tumors (Lung, Breast, Pancreatic, Gastric, Prostate, Bladder).
Hypoxia also influences mesenchymal stem cells (MSCs) features including differentiation cell viability, proliferation capacity, migration, and metabolism. It has been reported that the intratumoral MSC (T-MSC) play a key role in tumor progression and immune regulation (159, 160). T-MSC are either Bone Marrow-derived MSC that migrate and infiltrate the TME (161, 162). Alternatively, as demonstrated in the Wilm’s tumor, T-MSC represent the neoplastic mesenchymal tissue that is originated by a common neoplastic stem cell that also generates the blastemal and the epithelial components (163). T-MSC may be also directly generated by particular tumor cell subsets as reported in the neuroblastoma (164) Immunohistochemical analysis and in vitro studies show that T-MSC establish direct cellular crosstalk with the tumor cells (159), and/or pro inflammatory cells such M2 macrophages (165, 166). In addition, several tumor-associated inflammatory markers such as COX-2, nitric oxide synthase and nitrotyrosine, may be detected within the tumor stroma (159, 161, 165). In addition T-MSC (tumor-associated mesenchymal stem cells), possess potent immunosuppressive properties that can affect T cells (161), impair the cytolytic functions of NK cells and induce the polarization of monocytes towards alternatively activated macrophages (M2) (164, 166–169) this may, in turn, further compromise the functions of NK cell (168). Efficient tumor elimination requires a combined action both on tumor cells and stromal components (159, 170). As future perspective, to eliminate or disable T-MSC in vivo could involve targeting of the mesenchymal marker TRC105 with the TRC105 monoclonal antibodies (mAb) exploiting their ability to induce antibody-dependent cell mediated cytotoxicity (ADCC) (167, 170), or inducing their senescence through the application of anti-cancer drugs such as isoalantolactone which has demonstrated effectiveness both in vitro and in vivo.
Hypoxia in the TME induces and maintains CSCs which are equipped with mechanisms to evade treatment modalities. In addition, hypoxia confers radiotherapy resistance through an increase in reactive oxygen species (ROS). Although ROS, induced by radiation therapy or hypoxia, triggers DNA damage and cell death, this effect is mitigated under hypoxic stress by the induction of antioxidant HIF target genes resulting in ROS buffering action (171).
CSCs populations resist therapy and could also increase in response to therapy. Because CSCs are predominantly in the G0 or resting phase of the cell cycle, they escape conventional treatment regimens focused on eradicating proliferating tumor masses (172). Unlike differentiated cells that undergo apoptosis, non-cancerous stem cells do not, and this is important because it enables them to restore and rebuild normal organs following damage. CSCs like normal stem cells have mechanisms of resistance to apoptosis. Indeed, CSCs express high levels of antiapoptotic proteins such as Bcl-2 family proteins and inhibitors of apoptosis (173–176). Overexpression of Bcl-2 protein in the hematopoietic system results in an increase in hematopoietic stem cell number and chemoresistance (177, 178). Furthermore, stem cells possess asynchronous DNA synthesis activity and increased DNA repair activity (179). During asynchronous DNA synthesis, the parental ‘immortal’ DNA strand consistently segregates with the stem cell rather than the differentiating progeny. This segregation process may be regulated by P53 (180). As a result, stem cells gain an advantage by avoiding the accumulation of mutations related to replication and the detrimental effects of DNA-damaging agents and antiapoptotic proteins. Moreover, there are populations of stem cell-like cells found in many tumors that have been shown to express high levels of multidrug efflux pumps (MDR) or transporter proteins/detoxification proteins, such as MDR1, ABCB1, ABCG2 (BCRP) that play a significant role in expelling cytotoxic drugs from cells leading to high resistance to chemotherapeutic agents (179). The overexpression of ABC protein is a critical protective mechanism for CSCs in response to chemotherapy (152, 181). Indeed, numerous studies have reported that CSCs exhibit more resistance to chemotherapy and/or radiotherapy compared to differentiated tumor cells (182–184) in various types of cancer including breast cancer (185), ovarian cancer (186), colon cancer (157, 187) lung cancer (188, 189) and other deadly forms of cancers such as pancreatic cancers (190), myeloma (191, 192) and leukemia (193). In vivo and in vitro studies of common cancers have demonstrated resistance of CSCs to standard chemotherapy agents such as: oxaliplatin and 5-fluorouracil in colorectal cancers (194), cisplatin and paclitaxel in ovarian cancers and docetaxel and doxorubicin in breast cancers (195). These studies demonstrated that these cells are less susceptible to chemotherapy when compared to differentiated cells. A study by Chen et al. in 2012 showed that after treatment with temozolamide (TMZ) there was complete restoration of tumor cell population when CSCs were present (196). An additional study conducted in breast cancer revealed that taxane treatment could increase the generation of CSCs and further contribute to therapy resistance (197). Furthermore, after undergoing standard chemotherapy treatment with docetaxel, doxorubicin, cyclophosphamide and trastuzumab, breast cancer cells that are CD44+ and CD24- were found to exhibit resistance to chemotherapy. Specifically, 12 weeks post-chemotherapy the population of CD44+CD24-/low cells, increased from 4.7% to 13.6% while the proportion of epithelial cancer cells remained relatively unchanged (198). These findings suggest that CD44+CD24-/low cells may play a significant role in mediating chemotherapy resistance in breast cancer.
Another important reason for cancers resistance to chemotherapy drugs like cisplatin, etoposide, fluorouracil, and gefitinib is the overexpression of cytosolic enzyme called aldehyde dehydrogenase (ALDH) that protects cells from the toxic effects of elevated levels of reactive oxygen species (ROS) (199, 200). ALDH proteins scavenge free radicals generated by oxidative stress induced by radiation or drugs (201). However, when the activity of ALDH is inhibited in therapy-resistant CSCs, it results in the accumulation of excessive ROS. This accumulation of ROS leads to DNA damage and triggers apoptosis causing toxic effects on CSCs (199). patients with resectable esophageal cancer who exhibit high expression levels of ALDH1 are predicted to experience a poor response or resistance to preoperative chemotherapy (202, 203). Moreover, the CSC population can present clonal variation as well as distinct CSC-driven clones that differ in their growth rate or resistance to therapy, modeling tumor behavior. Finally, CSCs of various cancers not only develop chemoresistance, but they also develop resistance to radiation therapy leading to failure of treatments (204). Therefore, targeting CSCs utilizing their unique cell surface markers to develop antibodies or antibodies-drug conjugates could be more effective (Tables 1–3) (158).
Autophagy is a self-eating mechanism that is activated in response to stress in order to sustain homeostasis and cell survival. Many studies addressed the role for autophagy in self-renewal, pluripotency, and differentiation of normal stem cells (250, 251). In the context of cancer autophagy could function to either prevent or promote cancer, depending on the cancer stage (252). Furthermore, CSCs within a tumor are characterized by a high level of autophagy. Autophagy is upregulated in mammospheres (representing stem-like cells) compared to parental adherent cells (253). In addition to breast CSCs (253, 254), autophagy has been linked to CSCs in liver (255), pancreatic (256), osteosarcoma (257), ovarian (258) and glioblastoma (259).
Ongoing research is focused on elucidating the mechanisms through which autophagy contributes to the maintenance of stemness, as well as understanding the reliance of stem cells on autophagy (260). Numerous studies have shown that autophagy regulates the maintenance of pluripotency and homeostasis of CSCs under various pathophysiological conditions (260–262). Two important autophagy proteins, Beclin1 and Atg4a, were found to be critical for the maintenance and expansion of breast CSCs as well as tumor development in nude mice (253, 254). In the same line of thought, the suppression of autophagy by knocking down ATG5 and ATG7 drastically decreases the stemness characteristics of colorectal CSCs. This is evident through the decrease of stemness markers such as OCT4, SOX2, and NANOG, the induction of cellular senescence, and the decline of the proliferative capacities of CSCs in tumors (263). In another study, it was shown that inhibition of autophagy by knockdown of ATG7 or BECN1 modified the CD44+/CD24low/- (stem cell phenotype) population of breast cancer cells by regulating CD24 and IL-6 secretion (264). Indeed, IL-6 secretion was crucial for CSC maintenance, mammosphere formation, and conversion of non-CSCs into CSCs in different breast cell lines and a prostate cell line (264, 265). Moreover, autophagy inhibition decreased the secretion of IL-6 (264), most likely via JAK2/STAT3 signaling pathway, which was preferentially active in breast cancer cells compared with other tumor cell types (266). It is worth noting that, STAT3 has been reported to regulate the expression of multiple autophagy genes, including ATG3, BECN1, and BNIP3 (267). In a study involving a mouse model of breast CSCs two distinct signaling pathways were identified (268) (Figure 2). Yeo and colleagues isolated two subpopulations of breast CSCs, one luminal one (ALDH+) and one mesenchymal one (CD29high/CD61+). Intriguingly, stemness markers (ALDH, CD29, CD61) were downregulated in both populations following depletion of FIP200 (a component of ULK1 complex), which was correlated with decreased EGFR/STAT3 and TGF beta/SMAD signaling (268). Taken together, these findings indicate that the activation of STAT3 signaling could play an important role in the development of CSCs.
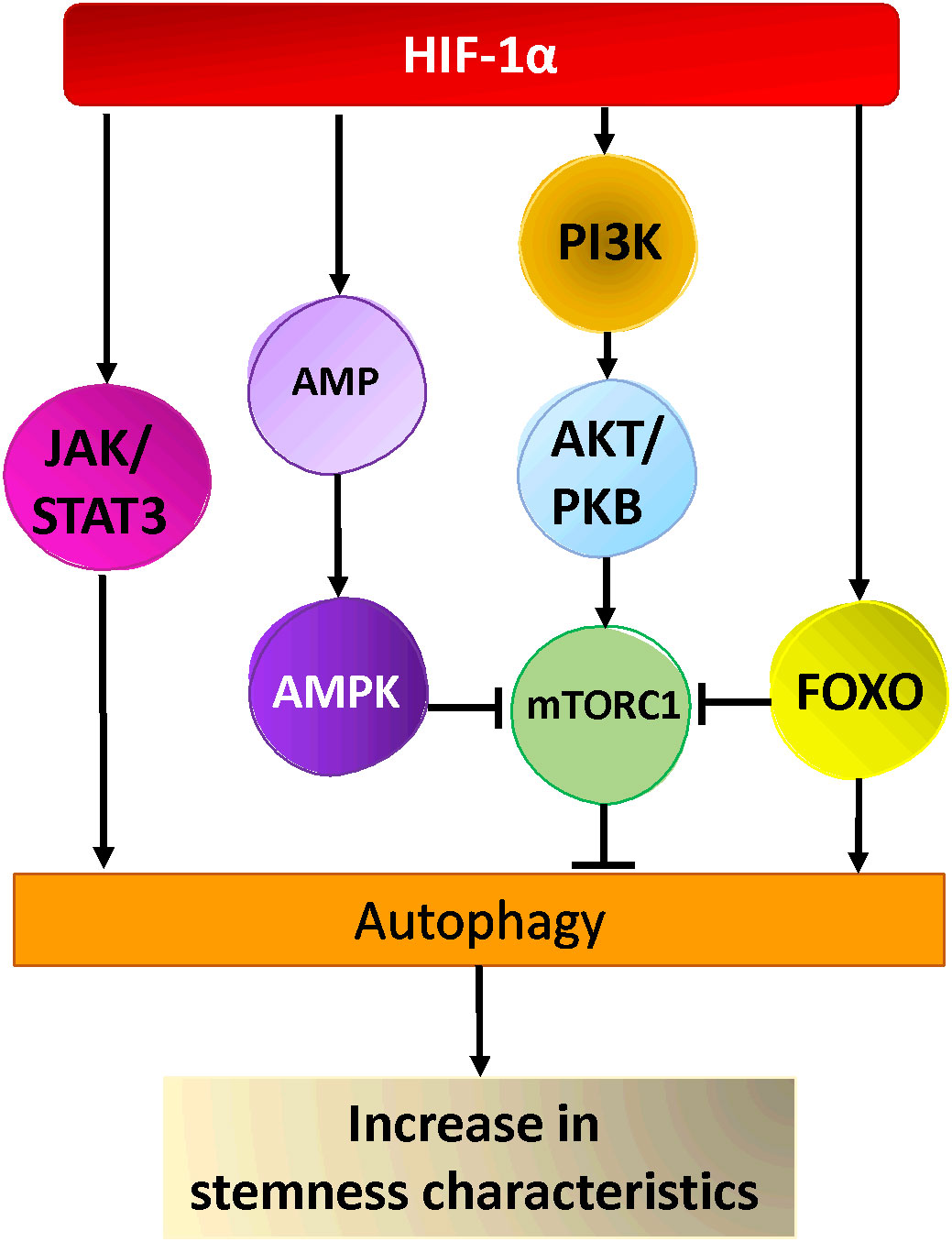
Figure 2 Hypoxia activated pathways in cancer stem cells. HIF-1α activates AKT via PI3-Kinase, leading to the activation of mTOR-C1, AMPK and FOXO activation by HIF-1α leads to inhibition of mTORC1 and autophagy activation. Hypoxia activates the JAK/STAT pathway via HIF-1α resulting in the activation of autophagy and cancer stem cells maintenance/generation.
Other research suggested that the fate of various CSCs could also be regulated by FoxO3a protein, a member of the Forkhead box O protein family. Indeed, the modulation of FoxO3a in breast cancer, affected CSC markers expression and had an impact on the formation of mammospheres as well as breast cancer-initiating potential (269). Moreover, the knockdown of FoxO3a led to an increase in the self-renewal capacity of the prostate, colorectal, and ovarian cancer stem cells as well as their tumorigenic potential (270–272). FoxO3a, which integrates the signals from Akt and Erk pathways, also plays a pivotal role in the control of differentiation and tumorigenicity of glioma CSCs (273). Several studies showed that FoxO transcription factors (including FoxO3a) are able to induce the expression of multiple ATG genes (267). Additionally, cytosolic FoxOs are able to regulate autophagy by interacting directly with cytosolic autophagy protein (267)(Figure 2). Nevertheless, further investigation is necessary to understand how FoxO-dependent autophagy and FoxO-dependent regulation of stemness are interrelated in tumorigenesis. Recently, a new link between autophagy and stemness was discovered, showing that Forkhead box A2 (FOXA2) is highly expressed in ovarian CSCs and modulates autophagy. Inhibition of autophagy induces FOXA2 downregulation and impairment of the self-renewal ability of ovarian CSCs.
In glioblastoma, several regulators of autophagy are highly expressed in tumors with a mesenchymal signature. Notably, the key regulator of selective autophagy p62/SQSTM1 and DNA damage-regulated autophagy modulator 1 (DRAM1) are both highly expressed in glioma CSCs (GSCs). Knockdown of DRAM1 and p62/SQSTM1 in GSCs leads to alteration of cellular bioenergetics and inhibits their migratory and invasive abilities. Moreover, these data suggest that the RAS/MAPK pathway may positively modulate autophagy in GSCs (274). However, other studies indicate that autophagy can regulate the differentiation of GSCs. The enhancement of autophagy promotes differentiation, whereas inhibition of autophagy suppresses differentiation (275, 276). Thus, it is unclear whether autophagy regulates stemness in glioma stem cells and it requires further elucidation.
Interestingly, in hematological malignancies, autophagy’s function could be reversed depending on the type of progenitors and the state of leukemia expansion (tumor initiation vs progression). In chronic myeloid leukemia (CML), inhibition of autophagy by silencing ATG7 or ATG4B curbs the expansion of CML CD34+ stem/progenitor cells (277, 278). Conversely, in acute myeloid leukemia (AML), monoallelic loss of a key autophagy gene Atg5 is sufficient to accelerate the disease progression and aggressiveness in a mouse AML model (279). Altogether, this evidence highlights that CSCs are often characterized by an increase in autophagy that maintains their pluripotency. However, different signaling pathways could be responsible for autophagy-dependent CSCs maintenance. Probably the mechanisms that underlie these activities depend on the cell type or malignancy degree. Therefore, the governing role of autophagy in CSCs is complex, and additional research is necessary.
CSCs are accountable for not only the formation, progression, and spread of tumors but also for resistance to treatment. Therefore, targeting CSCs specifically may be an appropriate approach to combat cancer. Because CSCs are used as a detection index, the availability of assays that allow detection and identification of CSCs after tumor initiation is of high relevance to guide treatment modalities. Furthermore, as chemotherapy or radiotherapy are known to target dividing cells, development of improved screening and targeting strategies could bring new perspectives to cancer exploration and cancer therapy (Table 2). While targeting CSCs holds promise, it also presents significant challenges. The development of therapies that selectively target CSCs without affecting normal stem cells remains a huge challenge. The identification and isolation of CSCs can be complex due to their heterogeneity and dynamic nature. However, the pursuit of CSC-targeted therapies in the context of hypoxic cancer environments presents an exciting avenue for advancing cancer treatment strategies and improving patient outcomes. In hypoxic cancer environments, targeting cancer stem cells (CSCs) has gained considerable attention as a promising approach to cancer treatment due to the following facts:
1. CSCs Contribute to Tumor Progression and Recurrence: Cancer stem cells are thought to be responsible for initiating tumors, driving their growth, and contributing to disease recurrence after treatment. They possess self-renewal and differentiation capabilities, allowing them to regenerate the entire tumor hierarchy. Targeting CSCs aims to disrupt this regenerative potential and halt tumor progression.
2. Resistance to Conventional Therapies: CSCs have been shown to exhibit increased resistance to various conventional cancer treatments, such as chemotherapy and radiation therapy. This resistance is due to their slow-cycling nature, enhanced DNA repair mechanisms, and expression of drug efflux transporters. By targeting CSCs, researchers aim to overcome the limitations posed by treatment-resistant cell populations.
3. Tumor Heterogeneity and Plasticity: Hypoxic environments in tumors can promote genetic and phenotypic heterogeneity, contributing to therapy resistance. CSCs are often associated with this heterogeneity and plasticity, making them a key target for therapy to prevent the emergence of treatment-resistant cell populations.
4. Microenvironmental Adaptation: CSCs are known to adapt to the hypoxic tumor microenvironment by upregulating hypoxia-inducible factors (HIFs) and other survival mechanisms. Targeting these adaptations can sensitize CSCs to therapy and disrupt their ability to survive under adverse conditions.
5. Reducing Relapse: Eliminating CSCs can reduce the likelihood of disease relapse. If CSCs are not effectively targeted, they can remain dormant and later give rise to new tumors, contributing to relapse and metastasis.
6. Long-Term Treatment Efficacy: By targeting CSCs, the goal is to achieve long-term treatment efficacy by eradicating the cell population responsible for initiating and sustaining the disease. This approach could lead to more durable responses and improved patient outcomes.
7. Combination Strategies: Targeting CSCs can be combined with conventional therapies to create synergistic effects. By targeting both the bulk of the tumor and the CSC subpopulation, treatment effectiveness may be enhanced.
8. Personalized Medicine: Understanding the molecular characteristics of CSCs and their responses to hypoxia can facilitate the development of personalized treatment strategies. Tailoring treatments to target CSCs based on individual patient profiles could enhance treatment outcomes.
9. Emerging Therapeutic Approaches: Researchers are actively exploring innovative approaches to target CSCs, including the use of specific antibodies, nanoparticles, gene therapies, and small molecules that inhibit key signaling pathways responsible for CSC maintenance.
10. Advancements in Research: As our understanding of CSC biology and the impact of hypoxia on these cells improves, more precise and effective targeting strategies can be developed.
Shown in Table 2 are examples of approaches developed to target and eradicate cancer stem cells and in Table 3 are current therapies targeting CSC pathways. By inhibiting signaling pathways specific to cancer stem cells researchers can prevent cancer stem cells from surviving and multiplying (223).
Developing immunotherapeutic approaches to target specific markers on cancer stem cells could be highly effective (280–283), however, it is important to note that as CSCs and normal stem cells share common markers, these types of treatments may result in adverse non desirable effects and therefore would have to be addressed prior to execution. Through differentiation therapy CSCs are induced to differentiate into non-tumorigenic cells. By doing so, researchers can prevent cancer stem cells from proliferating and spreading (284–286). The use of natural compounds such as curcumin, resveratrol, and sulforaphane are being investigated for their potential as CSCs targeting agents because they have been shown to have anti-CSCs properties (287). Finally, nanoparticles can be designed to selectively target CSCs for direct delivery of therapeutic agents resulting in improved efficacy and reduced toxicity (288–290).
Another potential strategy to target CSCs is by inducing them to exit their quiescent state, which is a state of dormancy that allows CSCs to resist chemotherapy and radiation and enter the cell cycle (291). By targeting quiescence, it may be possible to sensitize CSCs to conventional therapies and reduce the risk of tumor recurrence. Several approaches have been proposed to target quiescence in CSCs. One strategy is using drugs to target the signaling pathways that regulate quiescence, such as the Notch, Wnt (223), and Hedgehog pathways. Another approach is to use drugs that interfere with the interactions between CSCs and their microenvironment, which play a critical role in maintaining CSC quiescence (292–294). In addition to drug-based approaches, physical and mechanical cues can also be used to target CSC quiescence. For example, mechanical stress or compression can induce CSCs to exit their quiescent state and become more susceptible to chemotherapy. Another approach is to target the unique metabolic features of quiescent CSCs. CSCs generate energy mainly through glycolysis, (Warburg effect) and this results in rapid ATP production, in the presence of abundant glucose. As quiescent cells they have different metabolic requirements than proliferating cells, which can be exploited for therapy. For example, targeting the metabolism of quiescent CSCs with drugs that inhibit mitochondrial metabolism or fatty acid oxidation has been shown to selectively kill these cells (295). Overall, targeting quiescence represents a promising strategy for destroying CSCs and improving the effectiveness of conventional cancer therapies. However, more research is needed to fully understand the mechanisms underlying CSC quiescence and to develop effective strategies for targeting this state. While there is still much to learn about CSC biology and quiescence, ongoing research in this area holds great promise for the future of cancer treatment.
Despite significant efforts and progress made in comprehending cancer over the years, the fact remains that tumors can relapse, metastasize, and recur. While the discovery that tumors possess a heterogeneous population of cells, a subset of which with stem cell-like characteristics, was fascinating and promising, their actual significance in clinical settings was still uncertain. Numerous studies provided increasing evidence for the clinical significance of CSCs and for the important role of hypoxia in supporting stemness. Given CSCs presence in tumors, it is reasonable to assume that targeting them could be the most effective approach to achieve complete cancer elimination. In 2016, Piero and Debashish demonstrated that stem cells make up 4 to 7% of the cells within colorectal tumors, and that patients with this population have a greater likelihood of relapse, even in the second stage of the disease. Furthermore, chemotherapy does not appear to benefit these patients (296). Therefore, developing a more effective method to analyze CSCs within a tumor will be important.
Many pathways that CSCs utilize for their survival have been identified and studied and they present a viable option as cancer therapy targets. Clinical trials are underway to assess the effectiveness of agents targeting stemness pathways, such as Wnt, TGFβ, Notch, Hedgehog, and JAK/STAT, for a broad spectrum of cancers. These agents are being evaluated alone or in combination with conventional therapies, with the goal of completely eradicating the cancer (Tables 2, 3) (297).
The cellular and metabolic TME is currently attracting a lot of interest given its key role in carcinogenesis. In addition, strong evidence has been provided indicating that tumor heterogeneity represents a serious obstacle for therapy. Carcinogenesis and resistance mechanisms seem to be overlapping, carcinogenesis could endow cells with resistance, but resistance may not dictate carcinogenesis, and to determine conclusively if one mechanism contributes solely to carcinogenesis or resistance exclusively would have to be tested in one context excluding the other, which is not usually tested as most research assumes overlap of the two. Hypoxia clearly can exert several effects on the emergence and function of cancer stem cells, that play roles in both carcinogenesis and resistance. In this respect, the microenvironmental hypoxia is able to induce the alteration of gene expression, and HIF proteins can modulate several CSCs characteristics. It has been reported that microenvironmental hypoxia is able to mediate its effects by several potential mechanisms: altering gene expression, the activation of oncogenes, inactivation of tumor suppressor genes, reducing genomic stability and clonal selection. Significant scientific efforts are currently dedicated to understanding the complexity of the crosstalk between the tumor and its hostile hypoxic microenvironment in order to avoid tolerance and attenuate resistance of the tumor cells.
Here we briefly reviewed the current advances in the understanding of hypoxia and its role in stemness acquisition and how tumor hypoxia and its associated pathways may interfere with CSCs plasticity that impacts their phenotype and function and how it may offer a potential target that could be exploited therapeutically. Clearly, CSC are of particular interest because they are believed to be the clonogenic core of the tumor and therefore represent the cell population that drives growth and progression. We believe that a better elucidation of the hypoxia inducing stemness can clearly help the design of more adapted anti-cancer therapies approaches. This review could help the design of innovative therapeutic treatment approaches by considering the interlink between TME and CSC plasticity and may also contribute to further understand the huge complexity of tumor plasticity in response to anti-cancer therapies. Therefore, the putative targeting of the hypoxic CSC niche would be highly effective for controlling tumor metastasis and dormant CSCs. Thus, it would be of paramount importance to identify potential actionable targets. The challenging targeting of tumor stemness in the context of TME complexity remains, however, a considerable challenge.
All authors, ZR, RM, AB, KS, and CS have made substantial, direct, and intellectual contribution to the review, and approved it for publication.
This work was funded by Gulf Medical University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Weiss F, Lauffenburger D, Friedl P. Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat Rev Cancer (2022) 22(3):157–73. doi: 10.1038/s41568-021-00427-0
2. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol (2020) 30(16):R921–5. doi: 10.1016/j.cub.2020.06.081
3. McKeown SR. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radiol (2014) 87(1035):20130676. doi: 10.1259/bjr.20130676
4. Horsman MR, Vaupel P. Pathophysiological basis for the formation of the tumor microenvironment. Front Oncol (2016) 6:66. doi: 10.3389/fonc.2016.00066
5. Span PN, Bussink J. Biology of hypoxia. Semin Nucl Med (2015) 45(2):101–9. doi: 10.1053/j.semnuclmed.2014.10.002
6. Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev (2001) 15(20):2675–86. doi: 10.1101/gad.924501
7. Befani C, Liakos P. The role of hypoxia-inducible factor-2 alpha in angiogenesis. J Cell Physiol (2018) 233(12):9087–98. doi: 10.1002/jcp.26805
8. Yang SL, Wu C, Xiong XF, Fang X. Progress on hypoxia-inducible factor-3: Its structure, gene regulation and biological function (Review). Mol Med Rep (2015) 12(2):2411–2. doi: 10.3892/mmr.2015.3689
9. Todd VM, Vecchi LA, Clements ME, Snow KP, Ontko CD, Himmel L, et al. Hypoxia inducible factor signaling in breast tumors controls spontaneous tumor dissemination in a site-specific manner. Commun Biol (2021) 4(1):1122. doi: 10.1038/s42003-021-02648-3
10. Scortegagna M, Cataisson C, Martin RJ, Hicklin DJ, Schreiber RD, Yuspa SH, et al. HIF-1alpha regulates epithelial inflammation by cell autonomous NFkappaB activation and paracrine stromal remodeling. Blood (2008) 111(7):3343–54. doi: 10.1182/blood-2007-10-115758
11. Korbecki J, Simińska D, Gąssowska-Dobrowolska M, Listos J, Gutowska I, Chlubek D, et al. Chronic and cycling hypoxia: drivers of cancer chronic inflammation through HIF-1 and NF-κB activation: A review of the molecular mechanisms. Int J Mol Sci (2021) 22(19):10701. doi: 10.3390/ijms221910701
12. Bui BP, Nguyen PL, Lee K, Cho J. Hypoxia-inducible factor-1: A novel therapeutic target for the management of cancer, drug resistance, and cancer-related pain. Cancers (Basel) (2022) 14(24):6054. doi: 10.3390/cancers14246054
13. Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol (2014) 9:47–71. doi: 10.1146/annurev-pathol-012513-104720
14. Davis L, Recktenwald M, Hutt E, Fuller S, Briggs M, Goel A, et al. Targeting HIF-2α in the tumor microenvironment: redefining the role of HIF-2α for solid cancer therapy. Cancers (Basel) (2022) 14(5):1259. doi: 10.3390/cancers14051259
15. Yang J, Zhang X, Zhang Y, Zhu D, Zhang L, Li Y, et al. HIF-2α promotes epithelial-mesenchymal transition through regulating Twist2 binding to the promoter of E-cadherin in pancreatic cancer. J Exp Clin Cancer Res (2016) 35:26. doi: 10.1186/s13046-016-0298-y
16. Hallis SP, Kim SK, Lee JH, Kwak MK. Association of NRF2 with HIF-2alpha-induced cancer stem cell phenotypes in chronic hypoxic condition. Redox Biol (2023) 60:102632. doi: 10.1016/j.redox.2023.102632
17. Hsu TS, Lin YL, Wang YA, Mo ST, Chi PY, Lai AC, et al. HIF-2α is indispensable for regulatory T cell function. Nat Commun (2020) 11(1):5005. doi: 10.1038/s41467-020-18731-y
18. Zhou X, Guo X, Chen M, Xie C, Jiang J. HIF-3alpha promotes metastatic phenotypes in pancreatic cancer by transcriptional regulation of the rhoC-ROCK1 signaling pathway. Mol Cancer Res (2018) 16(1):124–34. doi: 10.1158/1541-7786.MCR-17-0256
19. Wei L, Yuan N, Chen Y, Gong P. Aberrant expression of HIF3A in plasma of patients with non-small cell lung cancer and its clinical significance. J Clin Lab Anal (2021) 35(8):e23889. doi: 10.1002/jcla.23889
20. Yazdani B, Sirous H. Expression analysis of HIF-3alpha as a potent prognostic biomarker in various types of human cancers: a case of meta-analysis. Res Pharm Sci (2022) 17(5):508–26. doi: 10.4103/1735-5362.355210
21. Missiaen R, Lesner NP, Simon MC. HIF: a master regulator of nutrient availability and metabolic cross-talk in the tumor microenvironment. EMBO J (2023) 42(6):e112067. doi: 10.15252/embj.2022112067
22. Taylor CT, Scholz CC. The effect of HIF on metabolism and immunity. Nat Rev Nephrol (2022) 18(9):573–87. doi: 10.1038/s41581-022-00587-8
23. Pal S, Sharma A, Mathew SP, Jaganathan BG. Targeting cancer-specific metabolic pathways for developing novel cancer therapeutics. Front Immunol (2022) 13:955476. doi: 10.3389/fimmu.2022.955476
24. Rashid M, Zadeh LR, Baradaran B, Molavi O, Ghesmati Z, Sabzichi M, et al. Up-down regulation of HIF-1α in cancer progression. Gene (2021) 798:145796. doi: 10.1016/j.gene.2021.145796
25. Lofstedt T, Fredlund E, Holmquist-Mengelbier L, Pietras A, Ovenberger M, Poellinger L, et al. Hypoxia inducible factor-2alpha in cancer. Cell Cycle (2007) 6(8):919–26. doi: 10.4161/cc.6.8.4133
26. Bader SB, Dewhirst MW, Hammond EM. Cyclic hypoxia: an update on its characteristics, methods to measure it and biological implications in cancer. Cancers (Basel) (2020) 13(1):23. doi: 10.3390/cancers13010023
27. Abou Khouzam R, Zaarour RF, Brodaczewska K, Azakir B, Venkatesh GH, Thiery J, et al. The effect of hypoxia and hypoxia-associated pathways in the regulation of antitumor response: friends or foes? Front Immunol (2022) 13:828875. doi: 10.3389/fimmu.2022.828875
28. Tirpe AA, Gulei D, Ciortea SM, Crivii C, Berindan-Neagoe I. Hypoxia: overview on hypoxia-mediated mechanisms with a focus on the role of HIF genes. Int J Mol Sci (2019) 20(24):6140. doi: 10.3390/ijms20246140
29. Moreno-Manzano V, Rodríguez-Jiménez FJ, Aceña-Bonilla JL, Fustero-Lardíes S, Erceg S, Dopazo J, et al. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. J Biol Chem (2010) 285(2):1333–42. doi: 10.1074/jbc.M109.008326
30. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med (2017) 23(10):1124–34. doi: 10.1038/nm.4409
31. Ratajczak MZ, Bujko K, Mack A, Kucia M, Ratajczak J. Cancer from the perspective of stem cells and misappropriated tissue regeneration mechanisms. Leukemia (2018) 32(12):2519–26. doi: 10.1038/s41375-018-0294-7
32. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med (1997) 3(7):730–7. doi: 10.1038/nm0797-730
33. Balamurugan K, Mendoza-Villanueva D, Sharan S, Summers GH, Dobrolecki LE, Lewis MT, et al. C/EBPδ links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene (2019) 38(20):3765–80. doi: 10.1038/s41388-018-0516-5
34. Hajizadeh F, Okoye I, Esmaily M, Ghasemi Chaleshtari M, Masjedi A, Azizi G, et al. Hypoxia inducible factors in the tumor microenvironment as therapeutic targets of cancer stem cells. Life Sci (2019) 237:116952. doi: 10.1016/j.lfs.2019.116952
35. Manni W, Min W. Signaling pathways in the regulation of cancer stem cells and associated targeted therapy. MedComm (2020) (2022) 3(4):e176. doi: 10.1002/mco2.176
36. Sun X, Lv X, Yan Y, Zhao Y, Ma R, He M, et al. Hypoxia-mediated cancer stem cell resistance and targeted therapy. BioMed Pharmacother (2020) 130:110623. doi: 10.1016/j.biopha.2020.110623
37. Jeng KS, Chang CF, Sheen IS, Jeng CJ, Wang CH. Cellular and molecular biology of cancer stem cells of hepatocellular carcinoma. Int J Mol Sci (2023) 24(2):1417. doi: 10.3390/ijms24021417
38. Gonzalez-Torres C, Gaytan-Cervantes J, Vazquez-Santillan K, Mandujano-Tinoco EA, Ceballos-Cancino G, Garcia-Venzor A, et al. NF-κB participates in the stem cell phenotype of ovarian cancer cells. Arch Med Res (2017) 48(4):343–51. doi: 10.1016/j.arcmed.2017.08.001
39. Soeda A, Park M, Lee D, Mintz A, Androutsellis-Theotokis A, McKay RD, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene (2009) 28(45):3949–59. doi: 10.1038/onc.2009.252
40. Wilson A, Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett (2006) 580(12):2860–8. doi: 10.1016/j.febslet.2006.03.024
41. Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, et al. Epigenetic perturbations by arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell (2016) 30(1):92–107. doi: 10.1016/j.ccell.2016.05.008
42. Chen Y, Shen X, Tang Y, Weng Y, Yang W, Liu M, et al. The diverse pancreatic tumor cell-intrinsic response to IFNγ is determined by epigenetic heterogeneity. Cancer Lett (2023) 562:216153. doi: 10.1016/j.canlet.2023.216153
43. Fu L, Fan J, Maity S, McFadden G, Shi Y, Kong W. PD-L1 interacts with Frizzled 6 to activate β-catenin and form a positive feedback loop to promote cancer stem cell expansion. Oncogene (2022) 41(8):1100–13. doi: 10.1038/s41388-021-02144-2
44. Xu J, Liao K, Zhou W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells Int (2018) 2018:4837370. doi: 10.1155/2018/4837370
45. Nadesh R, Menon KN, Biswas L, Mony U, Subramania Iyer K, Vijayaraghavan S, et al. Adipose derived mesenchymal stem cell secretome formulation as a biotherapeutic to inhibit growth of drug resistant triple negative breast cancer. Sci Rep (2021) 11(1):23435. doi: 10.1038/s41598-021-01878-z
46. Hass R, von der Ohe J, Ungefroren H. Impact of the tumor microenvironment on tumor heterogeneity and consequences for cancer cell plasticity and stemness. Cancers (Basel) (2020) 12(12):3716. doi: 10.3390/cancers12123716
47. Margreiter C, Aigner F, Resch T, Berenji AK, Oberhuber R, Sucher R, et al. Enteroscopic biopsies in the management of pancreas transplants: a proof of concept study for a novel monitoring tool. Transplantation (2012) 93(2):207–13. doi: 10.1097/TP.0b013e31823cf953
48. Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res (2011) 71(2):614–24. doi: 10.1158/0008-5472.CAN-10-0538
49. Cuiffo BG, Campagne A, Bell GW, Lembo A, Orso F, Lien EC, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell (2014) 15(6):762–74. doi: 10.1016/j.stem.2014.10.001
50. Geary LA, Nash KA, Adisetiyo H, Liang M, Liao CP, Jeong JH, et al. CAF-secreted annexin A1 induces prostate cancer cells to gain stem cell-like features. Mol Cancer Res (2014) 12(4):607–21. doi: 10.1158/1541-7786.MCR-13-0469
51. Rao G, Wang H, Li B, Huang L, Xue D, Wang X, et al. Reciprocal interactions between tumor-associated macrophages and CD44-positive cancer cells via osteopontin/CD44 promote tumorigenicity in colorectal cancer. Clin Cancer Res (2013) 19(4):785–97. doi: 10.1158/1078-0432.CCR-12-2788
52. Doherty MR, Parvani JG, Tamagno I, Junk DJ, Bryson BL, Cheon HJ, et al. The opposing effects of interferon-beta and oncostatin-M as regulators of cancer stem cell plasticity in triple-negative breast cancer. Breast Cancer Res (2019) 21(1):54. doi: 10.1186/s13058-019-1136-x
53. Chen WJ, Ho CC, Chang YL, Chen HY, Lin CA, Ling TY, et al. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat Commun (2014) 5:3472. doi: 10.1038/ncomms4472
54. Begum A, McMillan RH, Chang YT, Penchev VR, Rajeshkumar NV, Maitra A, et al. Direct interactions with cancer-associated fibroblasts lead to enhanced pancreatic cancer stem cell function. Pancreas (2019) 48(3):329–34. doi: 10.1097/MPA.0000000000001249
55. Lau EY, Lo J, Cheng BY, Ma MK, Lee JM, Ng JK, et al. Cancer-Associated Fibroblasts Regulate Tumor-Initiating Cell Plasticity in Hepatocellular Carcinoma through c-Met/FRA1/HEY1 Signaling. Cell Rep (2016) 15(6):1175–89. doi: 10.1016/j.celrep.2016.04.019
56. Du Y, Shao H, Moller M, Prokupets R, Tse YT, Liu ZJ. Intracellular notch1 signaling in cancer-associated fibroblasts dictates the plasticity and stemness of melanoma stem/initiating cells. Stem Cells (2019) 37(7):865–75. doi: 10.1002/stem.3013
57. Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF, et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer Res (2012) 72(21):5600–12. doi: 10.1158/0008-5472.CAN-11-3666
58. Zheng Y, Wang L, Yin L, Yao Z, Tong R, Xue J, et al. Lung cancer stem cell markers as therapeutic targets: an update on signaling pathways and therapies. Front Oncol (2022) 12:873994. doi: 10.3389/fonc.2022.873994
59. Gómez-Gallegos AA, Ramírez-Vidal L, Becerril-Rico J, Pérez-Islas E, Hernandez-Peralta ZJ, Toledo-Guzmán ME, et al. CD24+CD44+CD54+EpCAM+ gastric cancer stem cells predict tumor progression and metastasis: clinical and experimental evidence. Stem Cell Res Ther (2023) 14(1):16. doi: 10.1186/s13287-023-03241-7
60. Ricardo S, Vieira AF, Gerhard R, Leitão D, Pinto R, Cameselle-Teijeiro JF, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol (2011) 64(11):937–46. doi: 10.1136/jcp.2011.090456
61. Gzil A, Zarębska I, Bursiewicz W, Antosik P, Grzanka D, Szylberg Ł. Markers of pancreatic cancer stem cells and their clinical and therapeutic implications. Mol Biol Rep (2019) 46(6):6629–45. doi: 10.1007/s11033-019-05058-1
62. Johansson E, Grassi ES, Pantazopoulou V, Tong B, Lindgren D, Berg TJ, et al. CD44 interacts with HIF-2α to modulate the hypoxic phenotype of perinecrotic and perivascular glioma cells. Cell Rep (2017) 20(7):1641–53. doi: 10.1016/j.celrep.2017.07.049
63. Güler G, Guven U, Oktem G. Characterization of CD133. Analyst (2019) 144(6):2138–49. doi: 10.1039/c9an00093c
64. Zhang H, Lu H, Xiang L, Bullen JW, Zhang C, Samanta D, et al. HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci U.S.A. (2015) 112(45):E6215–23. doi: 10.1073/pnas.1520032112
65. Liu L, Zhang L, Yang L, Li H, Li R, Yu J, et al. Anti-CD47 antibody as a targeted therapeutic agent for human lung cancer and cancer stem cells. Front Immunol (2017) 8:404. doi: 10.3389/fimmu.2017.00404
66. Upton R, Banuelos A, Feng D, Biswas T, Kao K, McKenna K, et al. Combining CD47 blockade with trastuzumab eliminates HER2-positive breast cancer cells and overcomes trastuzumab tolerance. Proc Natl Acad Sci U.S.A. (2021) 118(29):e2026849118. doi: 10.1073/pnas.2026849118
67. Michaels AD, Newhook TE, Adair SJ, Morioka S, Goudreau BJ, Nagdas S, et al. CD47 blockade as an adjuvant immunotherapy for resectable pancreatic cancer. Clin Cancer Res (2018) 24(6):1415–25. doi: 10.1158/1078-0432.CCR-17-2283
68. Yoshida K, Tsujimoto H, Matsumura K, Kinoshita M, Takahata R, Matsumoto Y, et al. CD47 is an adverse prognostic factor and a therapeutic target in gastric cancer. Cancer Med (2015) 4(9):1322–33. doi: 10.1002/cam4.478
69. Semiz HS, Küçük Ü, Kısa E, Keskinkılıç M, Süyün DE, Arayıcı ME, et al. CD47 (don’t eat me signal) expression levels and its relationship with clinicopathologic features in early-stage prostate carcinoma. Prostate (2022) 82(16):1564–71. doi: 10.1002/pros.24432
70. Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer (2016) 15:26. doi: 10.1186/s12943-016-0510-x
71. Schulenburg A, Blatt K, Cerny-Reiterer S, Sadovnik I, Herrmann H, Marian B, et al. Cancer stem cells in basic science and in translational oncology: can we translate into clinical application? J Hematol Oncol (2015) 8:16. doi: 10.1186/s13045-015-0113-9
72. Song X, Zhang Y, Zhang L, Song W, Shi L. Hypoxia enhances indoleamine 2,3-dioxygenase production in dendritic cells. Oncotarget (2018) 9(14):11572–80. doi: 10.18632/oncotarget.24098
73. Li C, Liu S, Yan R, Han N, Wong KK, Li L. CD54-NOTCH1 axis controls tumor initiation and cancer stem cell functions in human prostate cancer. Theranostics (2017) 7(1):67–80. doi: 10.7150/thno.16752
74. Taftaf R, Liu X, Singh S, Jia Y, Dashzeveg NK, Hoffmann AD, et al. ICAM1 initiates CTC cluster formation and trans-endothelial migration in lung metastasis of breast cancer. Nat Commun (2021) 12(1):4867. doi: 10.1038/s41467-021-25189-z
75. Minchenko OH, Kharkova AP, Kubaichuk KI, Minchenko DO, Hlushchak NA, Kovalevska OV. Effect of hypoxia on the expression of CCN2, PLAU, PLAUR, SLURP1, PLAT and ITGB1 genes in ERN1 knockdown U87 glioma cells. Ukr Biochem J (2014) 86(4):79–89. doi: 10.15407/ubj86.04.079
76. Narayanaswamy PB, Baral TK, Haller H, Dumler I, Acharya K, Kiyan Y. Transcriptomic pathway analysis of urokinase receptor silenced breast cancer cells: a microarray study. Oncotarget (2017) 8(60):101572–90. doi: 10.18632/oncotarget.21351
77. Pérez LA, Leyton L, Valdivia A. Thy-1 (CD90), integrins and syndecan 4 are key regulators of skin wound healing. Front Cell Dev Biol (2022) 10:810474. doi: 10.3389/fcell.2022.810474
78. Gao L, Li J, He J, Liang L, He Z, Yue C, et al. CD90 affects the biological behavior and energy metabolism level of gastric cancer cells by targeting the PI3K/AKT/HIF-1α signaling pathway. Oncol Lett (2021) 21(3):191. doi: 10.3892/ol.2021.12451
79. Montanari M, Carbone MR, Coppola L, Giuliano M, Arpino G, Lauria R, et al. Epigenetic silencing of. Mol Cancer Res (2019) 17(2):628–41. doi: 10.1158/1541-7786.MCR-17-0324
80. Zhu J, Thakolwiboon S, Liu X, Zhang M, Lubman DM. Overexpression of CD90 (Thy-1) in pancreatic adenocarcinoma present in the tumor microenvironment. PloS One (2014) 9(12):e115507. doi: 10.1371/journal.pone.0115507
81. True LD, Zhang H, Ye M, Huang CY, Nelson PS, von Haller PD, et al. CD90/THY1 is overexpressed in prostate cancer-associated fibroblasts and could serve as a cancer biomarker. Mod Pathol (2010) 23(10):1346–56. doi: 10.1038/modpathol.2010.122
82. Cormerais Y, Giuliano S, LeFloch R, Front B, Durivault J, Tambutté E, et al. Genetic disruption of the multifunctional CD98/LAT1 complex demonstrates the key role of essential amino acid transport in the control of mTORC1 and tumor growth. Cancer Res (2016) 76(15):4481–92. doi: 10.1158/0008-5472.CAN-15-3376
83. Li Z, Chen S, He X, Gong S, Sun L, Weng L. SLC3A2 promotes tumor-associated macrophage polarization through metabolic reprogramming in lung cancer. Cancer Sci (2023) 114(6):2306–17. doi: 10.1111/cas.15760
84. Yang Y, Toy W, Choong LY, Hou P, Ashktorab H, Smoot DT, et al. Discovery of SLC3A2 cell membrane protein as a potential gastric cancer biomarker: implications in molecular imaging. J Proteome Res (2012) 11(12):5736–47. doi: 10.1021/pr300555y
85. Laugier F, Delyon J, André J, Bensussan A, Dumaz N. Hypoxia and MITF regulate KIT oncogenic properties in melanocytes. Oncogene (2016) 35(38):5070–7. doi: 10.1038/onc.2016.39
86. Tan WJ, Thike AA, Tan SY, Tse GM, Tan MH, Bay BH, et al. CD117 expression in breast phyllodes tumors correlates with adverse pathologic parameters and reduced survival. Mod Pathol (2015) 28(3):352–8. doi: 10.1038/modpathol.2014.111
87. Mattiolo P, Hong SM, Paolino G, Rusev BC, Marchegiani G, Salvia R, et al. CD117 is a specific marker of intraductal papillary mucinous neoplasms (IPMN) of the pancreas, oncocytic subtype. Int J Mol Sci (2020) 21(16):5794. doi: 10.3390/ijms21165794
88. Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M. CD117: a sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol (1998) 11(8):728–34.
89. Harris KS, Shi L, Foster BM, Mobley ME, Elliott PL, Song CJ, et al. CD117/c-kit defines a prostate CSC-like subpopulation driving progression and TKI resistance. Sci Rep (2021) 11(1):1465. doi: 10.1038/s41598-021-81126-6
90. Matsumoto K, Arao T, Tanaka K, Kaneda H, Kudo K, Fujita Y, et al. mTOR signal and hypoxia-inducible factor-1 alpha regulate CD133 expression in cancer cells. Cancer Res (2009) 69(18):7160–4. doi: 10.1158/0008-5472.CAN-09-1289
91. Iida H, Suzuki M, Goitsuka R, Ueno H. Hypoxia induces CD133 expression in human lung cancer cells by up-regulation of OCT3/4 and SOX2. Int J Oncol (2012) 40(1):71–9. doi: 10.3892/ijo.2011.1207
92. Soleimani A, Dadjoo P, Avan A, Soleimanpour S, Rajabian M, Ferns G, et al. Emerging roles of CD133 in the treatment of gastric cancer, a novel stem cell biomarker and beyond. Life Sci (2022) 293:120050. doi: 10.1016/j.lfs.2021.120050
93. Yang J, Aljitawi O, Van Veldhuizen P. Prostate cancer stem cells: the role of CD133. Cancers (Basel) (2022) 14(21):5448. doi: 10.3390/cancers14215448
94. Brugnoli F, Grassilli S, Al-Qassab Y, Capitani S, Bertagnolo V. CD133 in breast cancer cells: more than a stem cell marker. J Oncol (2019) 2019:7512632. doi: 10.1155/2019/7512632
95. Zhao M, Zhang Y, Zhang H, Wang S, Zhang M, Chen X, et al. Hypoxia-induced cell stemness leads to drug resistance and poor prognosis in lung adenocarcinoma. Lung Cancer (2015) 87(2):98–106. doi: 10.1016/j.lungcan.2014.11.017
96. Ni T, Wang H, Zhan D, Tao L, Lv M, Wang W, et al. CD133+/CD166+ human gastric adenocarcinoma cells present the properties of neoplastic stem cells and emerge more Malignant features. Life Sci (2021) 269:119021. doi: 10.1016/j.lfs.2021.119021
97. Burkhardt M, Mayordomo E, Winzer KJ, Fritzsche F, Gansukh T, Pahl S, et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J Clin Pathol (2006) 59(4):403–9. doi: 10.1136/jcp.2005.028209
98. Fujiwara K, Ohuchida K, Sada M, Horioka K, Ulrich CD, Shindo K, et al. CD166/ALCAM expression is characteristic of tumorigenicity and invasive and migratory activities of pancreatic cancer cells. PloS One (2014) 9(9):e107247. doi: 10.1371/journal.pone.0107247
99. Massoner P, Thomm T, Mack B, Untergasser G, Martowicz A, Bobowski K, et al. EpCAM is overexpressed in local and metastatic prostate cancer, suppressed by chemotherapy and modulated by MET-associated miRNA-200c/205. Br J Cancer (2014) 111(5):955–64. doi: 10.1038/bjc.2014.366
100. Zhang D, Yang L, Liu X, Gao J, Liu T, Yan Q, et al. Hypoxia modulates stem cell properties and induces EMT through N-glycosylation of EpCAM in breast cancer cells. J Cell Physiol (2020) 235(4):3626–33. doi: 10.1002/jcp.29252
101. Liu Y, Wang Y, Sun S, Chen Z, Xiang S, Ding Z, et al. Understanding the versatile roles and applications of EpCAM in cancers: from bench to bedside. Exp Hematol Oncol (2022) 11(1):97. doi: 10.1186/s40164-022-00352-4
102. Patak P, Jin F, Schäfer ST, Metzen E, Hermann DM. The ATP-binding cassette transporters ABCB1 and ABCC1 are not regulated by hypoxia in immortalised human brain microvascular endothelial cells. Exp Transl Stroke Med (2011) 3:12. doi: 10.1186/2040-7378-3-12
103. Abd El-Aziz YS, Spillane AJ, Jansson PJ, Sahni S. Role of ABCB1 in mediating chemoresistance of triple-negative breast cancers. Biosci Rep (2021) 41(2):BSR20204092. doi: 10.1042/BSR20204092
104. de Oliveira J, Felipe AV, Neto RA, Oshima CT, de Souza Silva M, Forones NM. Association between ABCB1 immunohistochemical expression and overall survival in gastric cancer patients. Asian Pac J Cancer Prev (2014) 15(16):6935–8. doi: 10.7314/APJCP.2014.15.16.6935
105. Xiang L, Liu ZH, Huan Q, Su P, Du GJ, Wang Y, et al. Hypoxia-inducible factor-2a is associated with ABCG2 expression, histology-grade and Ki67 expression in breast invasive ductal carcinoma. Diagn Pathol (2012) 7:32. doi: 10.1186/1746-1596-7-32
106. He X, Wang J, Wei W, Shi M, Xin B, Zhang T, et al. Hypoxia regulates ABCG2 activity through the activivation of ERK1/2/HIF-1α and contributes to chemoresistance in pancreatic cancer cells. Cancer Biol Ther (2016) 17(2):188–98. doi: 10.1080/15384047.2016.1139228
107. Wang J, Yunyun Z, Wang L, Chen X, Zhu Z. ABCG2 confers promotion in gastric cancer through modulating downstream CRKL in vitro combining with biostatistics mining. Oncotarget (2017) 8(3):5256–67. doi: 10.18632/oncotarget.14128
108. Sabnis NG, Miller A, Titus MA, Huss WJ. The efflux transporter ABCG2 maintains prostate stem cells. Mol Cancer Res (2017) 15(2):128–40. doi: 10.1158/1541-7786.MCR-16-0270-T
109. Liu Z, Sun L, Cai Y, Shen S, Zhang T, Wang N, et al. Hypoxia-induced suppression of alternative splicing of MBD2 promotes breast cancer metastasis via activation of FZD1. Cancer Res (2021) 81(5):1265–78. doi: 10.1158/0008-5472.CAN-20-2876
110. Dong D, Na L, Zhou K, Wang Z, Sun Y, Zheng Q, et al. FZD5 prevents epithelial-mesenchymal transition in gastric cancer. Cell Commun Signal (2021) 19(1):21. doi: 10.1186/s12964-021-00708-z
111. Li Y, Liu Z, Zhang Y. Expression and prognostic impact of FZDs in pancreatic adenocarcinoma. BMC Gastroenterol (2021) 21(1):79. doi: 10.1186/s12876-021-01643-6
112. Schioppa T, Uranchimeg B, Saccani A, Biswas SK, Doni A, Rapisarda A, et al. Regulation of the chemokine receptor CXCR4 by hypoxia. J Exp Med (2003) 198(9):1391–402. doi: 10.1084/jem.20030267
113. Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Devel Ther (2015) 9:4953–64. doi: 10.2147/DDDT.S84932
114. Gu Y, Gu W, Xie R, Chen Z, Xu T, Fei Z. Role of CXCR4 as a prognostic biomarker associated with the tumor immune microenvironment in gastric cancer. Front Cell Dev Biol (2021) 9:654504. doi: 10.3389/fcell.2021.654504
115. He S, Du W, Li M, Yan M, Zheng F. PODXL might be a new prognostic biomarker in various cancers: a meta-analysis and sequential verification with TCGA datasets. BMC Cancer (2020) 20(1):620. doi: 10.1186/s12885-020-07108-5
116. Zhi Q, Chen H, Liu F, Han Y, Wan D, Xu Z, et al. Podocalyxin-like protein promotes gastric cancer progression through interacting with RUN and FYVE domain containing 1 protein. Cancer Sci (2019) 110(1):118–34. doi: 10.1111/cas.13864
117. Wang V, Davis DA, Yarchoan R. Identification of functional hypoxia inducible factor response elements in the human lysyl oxidase gene promoter. Biochem Biophys Res Commun (2017) 490(2):480–5. doi: 10.1016/j.bbrc.2017.06.066
118. Ramos S, Ferreira S, Fernandes AS, Saraiva N. Lysyl oxidases expression and breast cancer progression: A bioinformatic analysis. Front Pharmacol (2022) 13:883998. doi: 10.3389/fphar.2022.883998
119. Murdocca M, De Masi C, Pucci S, Mango R, Novelli G, Di Natale C, et al. LOX-1 and cancer: an indissoluble liaison. Cancer Gene Ther (2021) 28(10-11):1088–98. doi: 10.1038/s41417-020-00279-0
120. Miller BW, Morton JP, Pinese M, Saturno G, Jamieson NB, McGhee E, et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Mol Med (2015) 7(8):1063–76. doi: 10.15252/emmm.201404827
121. Li C, Yang N, Chen Z, Xia N, Shan Q, Wang Z, et al. Hypoxia-induced Tie1 drives stemness and cisplatin resistance in non-small cell lung carcinoma cells. Cancer Cell Int (2021) 21(1):57. doi: 10.1186/s12935-020-01729-3
122. Katsuta E, Qi Q, Peng X, Hochwald SN, Yan L, Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci Rep (2019) 9(1):1310. doi: 10.1038/s41598-018-37909-5
123. Lin W, Kao HW, Robinson D, Kung HJ, Wu CW, Chen HC. Tyrosine kinases and gastric cancer. Oncogene (2000) 19(49):5680–9. doi: 10.1038/sj.onc.1203924
124. Scheutz F. Anxiety and dental fear in a group of parenteral drug addicts. Scand J Dent Res (1986) 94(3):241–7. doi: 10.1111/j.1600-0722.1986.tb01759.x
125. Soehngen E, Schaefer A, Koeritzer J, Huelsmeyer V, Zimmer C, Ringel F, et al. Hypoxia upregulates aldehyde dehydrogenase isoform 1 (ALDH1) expression and induces functional stem cell characteristics in human glioblastoma cells. Brain Tumor Pathol (2014) 31(4):247–56. doi: 10.1007/s10014-013-0170-0
126. Yue H, Hu Z, Hu R, Guo Z, Zheng Y, Wang Y, et al. ALDH1A1 in cancers: bidirectional function, drug resistance, and regulatory mechanism. Front Oncol (2022) 12:918778. doi: 10.3389/fonc.2022.918778
127. Chen G, Liu B, Yin S, Li S, Guo Y, Wang M, et al. Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA. Oncogenesis (2020) 9(9):81. doi: 10.1038/s41389-020-00265-z
128. Novak D, Hüser L, Elton JJ, Umansky V, Altevogt P, Utikal J. SOX2 in development and cancer biology. Semin Cancer Biol (2020) 67(Pt 1):74–82. doi: 10.1016/j.semcancer.2019.08.007
129. Lu H, Lyu Y, Tran L, Lan J, Xie Y, Yang Y, et al. HIF-1 recruits NANOG as a coactivator for TERT gene transcription in hypoxic breast cancer stem cells. Cell Rep (2021) 36(13):109757. doi: 10.1016/j.celrep.2021.109757
130. Vasefifar P, Motafakkerazad R, Maleki LA, Najafi S, Ghrobaninezhad F, Najafzadeh B, et al. Nanog, as a key cancer stem cell marker in tumor progression. Gene (2022) 827:146448. doi: 10.1016/j.gene.2022.146448
131. Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev (2006) 20(5):557–70. doi: 10.1101/gad.1399906
132. Monferrer E, Burgos-Panadero R, Blanquer-Maceiras M, Cañete A, Navarro S, Noguera R. High Oct4 expression: implications in the pathogenesis of neuroblastic tumours. BMC Cancer (2019) 19(1):1. doi: 10.1186/s12885-018-5219-3
133. Cui CP, Wong CC, Kai AK, Ho DW, Lau EY, Tsui YM, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut (2017) 66(12):2149–59. doi: 10.1136/gutjnl-2016-313264
134. Wang MC, Li CL, Cui J, Jiao M, Wu T, Jing LI, et al. BMI-1, a promising therapeutic target for human cancer. Oncol Lett (2015) 10(2):583–8. doi: 10.3892/ol.2015.3361
135. Dandawate P, Ghosh C, Palaniyandi K, Paul S, Rawal S, Pradhan R, et al. The histone demethylase KDM3A, increased in human pancreatic tumors, regulates expression of DCLK1 and promotes tumorigenesis in mice. Gastroenterology (2019) 157(6):1646–1659.e11. doi: 10.1053/j.gastro.2019.08.018
136. Panneerselvam J, Mohandoss P, Patel R, Gillan H, Li M, Kumar K, et al. DCLK1 regulates tumor stemness and cisplatin resistance in non-small cell lung cancer via ABCD-member-4. Mol Ther Oncolytics (2020) 18:24–36. doi: 10.1016/j.omto.2020.05.012
137. Liu H, Wen T, Zhou Y, Fan X, Du T, Gao T, et al. DCLK1 plays a metastatic-promoting role in human breast cancer cells. BioMed Res Int (2019) 2019:1061979. doi: 10.1155/2019/1061979
138. Carli ALE, Afshar-Sterle S, Rai A, Fang H, O'Keefe R, Tse J, et al. Cancer stem cell marker DCLK1 reprograms small extracellular vesicles toward migratory phenotype in gastric cancer cells. Proteomics (2021) 21(13-14):e2000098. doi: 10.1002/pmic.202000098
139. Cao Z, Weygant N, Chandrakesan P, Houchen CW, Peng J, Qu D. Tuft and cancer stem cell marker DCLK1: A new target to enhance anti-tumor immunity in the tumor microenvironment. Cancers (Basel) (2020) 12(12):3801. doi: 10.3390/cancers12123801
140. Shafiei S, Kalantari E, Saeednejad Zanjani L, Abolhasani M, Asadi Lari MH, Madjd Z. Increased expression of DCLK1, a novel putative CSC maker, is associated with tumor aggressiveness and worse disease-specific survival in patients with bladder carcinomas. Exp Mol Pathol (2019) 108:164–72. doi: 10.1016/j.yexmp.2019.04.015
141. Jiang D, Xiao C, Xian T, Wang L, Mao Y, Zhang J, et al. Association of doublecortin-like kinase 1 with tumor aggressiveness and poor biochemical recurrence-free survival in prostate cancer. Onco Targets Ther (2018) 11:1077–86. doi: 10.2147/OTT.S157295
142. Hasan D, Gamen E, Abu Tarboush N, Ismail Y, Pak O, Azab B. PKM2 and HIF-1α regulation in prostate cancer cell lines. PloS One (2018) 13(9):e0203745. doi: 10.1371/journal.pone.0203745
143. Wang C, Zhang S, Liu J, Tian Y, Ma B, Xu S, et al. Secreted pyruvate kinase M2 promotes lung cancer metastasis through activating the integrin beta1/FAK signaling pathway. Cell Rep (2020) 30(6):1780–1797.e6. doi: 10.1016/j.celrep.2020.01.037
144. Li H, Yan M, Wu X, Wang Y, Huang L. Expression and clinical significance of pyruvate kinase M2 in breast cancer: A protocol for meta-analysis and bioinformatics validation analysis. Med (Baltimore) (2021) 100(18):e25545. doi: 10.1097/MD.0000000000025545
145. Kumar Y, Gurusamy K, Pamecha V, Davidson BR. Tumor M2-pyruvate kinase as tumor marker in exocrine pancreatic cancer a meta-analysis. Pancreas (2007) 35(2):114–9. doi: 10.1097/mpa.0b013e3180537237
146. Guo W, Zhang Z, Li G, Lai X, Gu R, Xu W, et al. Pyruvate kinase M2 promotes prostate cancer metastasis through regulating ERK1/2-COX-2 signaling. Front Oncol (2020) 10:544288. doi: 10.3389/fonc.2020.544288
147. Xia Y, Wang X, Liu Y, Shapiro E, Lepor H, Tang MS, et al. PKM2 is essential for bladder cancer growth and maintenance. Cancer Res (2022) 82(4):571–85. doi: 10.1158/0008-5472.CAN-21-0403
148. Li N, Meng D, Xu Y, Gao L, Shen F, Tie X, et al. Pyruvate kinase M2 knockdown suppresses migration, invasion, and epithelial-mesenchymal transition of gastric carcinoma via hypoxia-inducible factor alpha/B-cell lymphoma 6 pathway. BioMed Res Int (2020) 2020:7467104. doi: 10.1155/2020/7467104
149. Shan F, Huang Z, Xiong R, Huang QY, Li J. HIF1α-induced upregulation of KLF4 promotes migration of human vascular smooth muscle cells under hypoxia. J Cell Physiol (2020) 235(1):141–50. doi: 10.1002/jcp.28953
150. Taracha-Wisniewska A, Kotarba G, Dworkin S, Wilanowski T. Recent discoveries on the involvement of krüppel-like factor 4 in the most common cancer types. Int J Mol Sci (2020) 21(22):8843. doi: 10.3390/ijms21228843
151. Yu F, Li J, Chen H, Fu J, Ray S, Huang S, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene (2011) 30(18):2161–72. doi: 10.1038/onc.2010.591
152. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med (2011) 17(3):313–9. doi: 10.1038/nm.2304
153. Sundar SJ, Hsieh JK, Manjila S, Lathia JD, Sloan A. The role of cancer stem cells in glioblastoma. Neurosurg Focus (2014) 37(6):E6. doi: 10.3171/2014.9.FOCUS14494
154. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U.S.A. (2003) 100(7):3983–8. doi: 10.1073/pnas.0530291100
155. Smith LM, Nesterova A, Ryan MC, Duniho S, Jonas M, Anderson M, et al. CD133/prominin-1 is a potential therapeutic target for antibody-drug conjugates in hepatocellular and gastric cancers. Br J Cancer (2008) 99(1):100–9. doi: 10.1038/sj.bjc.6604437
156. Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res (2007) 67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030
157. Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell (2007) 1(4):389–402. doi: 10.1016/j.stem.2007.08.001
158. Rappa G, Fodstad O, Lorico A. The stem cell-associated antigen CD133 (Prominin-1) is a molecular therapeutic target for metastatic melanoma. Stem Cells (2008) 26(12):3008–17. doi: 10.1634/stemcells.2008-0601
159. Pillat MM, Oliveira-Giacomelli Á, das Neves Oliveira M, Andrejew R, Turrini N, Baranova J, et al. Mesenchymal stem cell-glioblastoma interactions mediated via kinin receptors unveiled by cytometry. Cytometry A (2021) 99(2):152–63. doi: 10.1002/cyto.a.24299
160. Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev (2021) 101(1):147–76. doi: 10.1152/physrev.00048.2019
161. Pelizzo G, Veschi V, Mantelli M, Croce S, Di Benedetto V, D'Angelo P, et al. Microenvironment in neuroblastoma: isolation and characterization of tumor-derived mesenchymal stromal cells. BMC Cancer (2018) 18(1):1176. doi: 10.1186/s12885-018-5082-2
162. Borriello L, Nakata R, Sheard MA, Fernandez GE, Sposto R, Malvar J, et al. Cancer-associated fibroblasts share characteristics and protumorigenic activity with mesenchymal stromal cells. Cancer Res (2017) 77(18):5142–57. doi: 10.1158/0008-5472.CAN-16-2586
163. Royer-Pokora B, Busch M, Beier M, Duhme C, de Torres C, Mora J, et al. Wilms tumor cells with WT1 mutations have characteristic features of mesenchymal stem cells and express molecular markers of paraxial mesoderm. Hum Mol Genet (2010) 19(9):1651–68. doi: 10.1093/hmg/ddq042
164. Canzonetta C, Pelosi A, Di Matteo S, Veneziani I, Tumino N, Vacca P, et al. Identification of neuroblastoma cell lines with uncommon TAZ. J Immunother Cancer (2021) 9(1):e001313. doi: 10.1136/jitc-2020-001313
165. Maturu P, Overwijk WW, Hicks J, Ekmekcioglu S, Grimm EA, Huff V. Characterization of the inflammatory microenvironment and identification of potential therapeutic targets in wilms tumors. Transl Oncol (2014) 7(4):484–92. doi: 10.1016/j.tranon.2014.05.008
166. Komohara Y, Takeya M. CAFs and TAMs: maestros of the tumour microenvironment. J Pathol (2017) 241(3):313–5. doi: 10.1002/path.4824
167. Di Matteo S, Avanzini MA, Pelizzo G, Calcaterra V, Croce S, Spaggiari GM, et al. Neuroblastoma tumor-associated mesenchymal stromal cells regulate the cytolytic functions of NK cells. Cancers (Basel) (2022) 15(1):19. doi: 10.3390/cancers15010019
168. Fiore PF, Vacca P, Tumino N, Besi F, Pelosi A, Munari E, et al. Wilms’ Tumor primary cells display potent immunoregulatory properties on NK cells and macrophages. Cancers (Basel) (2021) 13(2):224. doi: 10.3390/cancers13020224
169. Cantoni C, Serra M, Parisi E, Azzarone B, Sementa AR, Nasto LA, et al. Stromal-like Wilms tumor cells induce human Natural Killer cell degranulation and display immunomodulatory properties towards NK cells. Oncoimmunology (2021) 10(1):1879530. doi: 10.1080/2162402X.2021.1879530
170. Wu HW, Sheard MA, Malvar J, Fernandez GE, DeClerck YA, Blavier L, et al. Anti-CD105 antibody eliminates tumor microenvironment cells and enhances anti-GD2 antibody immunotherapy of neuroblastoma with activated natural killer cells. Clin Cancer Res (2019) 25(15):4761–74. doi: 10.1158/1078-0432.CCR-18-3358
171. Wang H, Jiang H, Van De Gucht M, De Ridder M. Hypoxic radioresistance: can ROS be the key to overcome it? Cancers (Basel) (2019) 11(1):112. doi: 10.3390/cancers11010112
172. Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PloS Biol (2004) 2(10):e301. doi: 10.1371/journal.pbio.0020301
173. Wang S, Yang D, Lippman ME. Targeting Bcl-2 and Bcl-XL with nonpeptidic small-molecule antagonists. Semin Oncol (2003) 30(5 Suppl 16):133–42. doi: 10.1053/j.seminoncol.2003.08.015
174. Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell (2011) 147(4):742–58. doi: 10.1016/j.cell.2011.10.033
175. Madjd Z, Mehrjerdi AZ, Sharifi AM, Molanaei S, Shahzadi SZ, Asadi-Lari M. CD44+ cancer cells express higher levels of the anti-apoptotic protein Bcl-2 in breast tumours. Cancer Immun (2009) 9:4.
176. Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ (2006) 13(6):994–1002. doi: 10.1038/sj.cdd.4401908
177. Domen J. The role of apoptosis in regulating hematopoiesis and hematopoietic stem cells. Immunol Res (2000) 22(2-3):83–94. doi: 10.1385/IR:22:2-3:83
178. Domen J, Weissman IL. Self-renewal, differentiation or death: regulation and manipulation of hematopoietic stem cell fate. Mol Med Today (1999) 5(5):201–8. doi: 10.1016/S1357-4310(99)01464-1
179. Gillespie MS, Ward CM, Davies CC. DNA repair and therapeutic strategies in cancer stem cells. Cancers (Basel) (2023) 15(6):1897. doi: 10.3390/cancers15061897
180. Rambhatla L, Ram-Mohan S, Cheng JJ, Sherley JL. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res (2005) 65(8):3155–61. doi: 10.1158/0008-5472.CAN-04-3161
181. McIntosh K, Balch C, Tiwari AK. Tackling multidrug resistance mediated by efflux transporters in tumor-initiating cells. Expert Opin Drug Metab Toxicol (2016) 12(6):633–44. doi: 10.1080/17425255.2016.1179280
182. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer (2008) 8(10):755–68. doi: 10.1038/nrc2499
183. Alison MR, Lim SM, Nicholson LJ. Cancer stem cells: problems for therapy? J Pathol (2011) 223(2):147–61. doi: 10.1002/path.2793
184. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer (2005) 5(4):275–84. doi: 10.1038/nrc1590
185. Pavlopoulou A, Oktay Y, Vougas K, Louka M, Vorgias CE, Georgakilas AG. Determinants of resistance to chemotherapy and ionizing radiation in breast cancer stem cells. Cancer Lett (2016) 380(2):485–93. doi: 10.1016/j.canlet.2016.07.018
186. Meirelles K, Benedict LA, Dombkowski D, Pepin D, Preffer FI, Teixeira J, et al. Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc Natl Acad Sci U.S.A. (2012) 109(7):2358–63. doi: 10.1073/pnas.1120733109
187. Colak S, Zimberlin CD, Fessler E, Hogdal L, Prasetyanti PR, Grandela CM, et al. Decreased mitochondrial priming determines chemoresistance of colon cancer stem cells. Cell Death Differ (2014) 21(7):1170–7. doi: 10.1038/cdd.2014.37
188. Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U.S.A. (2009) 106(38):16281–6. doi: 10.1073/pnas.0905653106
189. Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ (2008) 15(3):504–14. doi: 10.1038/sj.cdd.4402283
190. Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell (2007) 1(3):313–23. doi: 10.1016/j.stem.2007.06.002
191. Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, et al. Characterization of clonogenic multiple myeloma cells. Blood (2004) 103(6):2332–6. doi: 10.1182/blood-2003-09-3064
192. Jones RJ, Matsui WH, Smith BD. Cancer stem cells: are we missing the target? J Natl Cancer Inst (2004) 96(8):583–5. doi: 10.1093/jnci/djh095
193. Guzman ML, Swiderski CF, Howard DS, Grimes BA, Rossi RM, Szilvassy SJ, et al. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci U.S.A. (2002) 99(25):16220–5. doi: 10.1073/pnas.252462599
194. Park M, Sundaramoorthy P, Sim JJ, Jeong KY, Kim HM. Synergistically anti-metastatic effect of 5-flourouracil on colorectal cancer cells via calcium-mediated focal adhesion kinase proteolysis. Anticancer Res (2017) 37(1):103–14. doi: 10.21873/anticanres.11295
196. Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature (2012) 488(7412):522–6. doi: 10.1038/nature11287
197. Deng X, Apple S, Zhao H, Song J, Lee M, Luo W, et al. CD24 Expression and differential resistance to chemotherapy in triple-negative breast cancer. Oncotarget (2017) 8(24):38294–308. doi: 10.18632/oncotarget.16203
198. Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst (2008) 100(9):672–9. doi: 10.1093/jnci/djn123
199. Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, et al. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res (2014) 74(13):3579–90. doi: 10.1158/0008-5472.CAN-13-3456
200. Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle (2011) 10(9):1378–84. doi: 10.4161/cc.10.9.15486
201. Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med (2013) 56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010
202. Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells (2005) 23(6):752–60. doi: 10.1634/stemcells.2004-0292
203. Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev (2010) 19(2):327–37. doi: 10.1158/1055-9965.EPI-09-0865
204. Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer (2008) 8(7):545–54. doi: 10.1038/nrc2419
205. Nguyen CH, Grandits AM, Purton LE, Sill H, Wieser R. All-trans retinoic acid in non-promyelocytic acute myeloid leukemia: driver lesion dependent effects on leukemic stem cells. Cell Cycle (2020) 19(20):2573–88. doi: 10.1080/15384101.2020.1810402
206. Ehata S, Miyazono K. Bone morphogenetic protein signaling in cancer; some topics in the recent 10 years. Front Cell Dev Biol (2022) 10:883523. doi: 10.3389/fcell.2022.883523
207. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev (2018) 62:50–60. doi: 10.1016/j.ctrv.2017.11.002
208. Rechberger JS, Thiele F, Daniels DJ. Status quo and trends of intra-arterial therapy for brain tumors: A bibliometric and clinical trials analysis. Pharmaceutics (2021) 13(11):1885. doi: 10.3390/pharmaceutics13111885
209. Kabakov AE, Gabai VL. HSP70s in breast cancer: promoters of tumorigenesis and potential targets/tools for therapy. Cells (2021) 10(12):3446. doi: 10.3390/cells10123446
210. Wang Z, Zhou L, Xiong Y, Yu S, Li H, Fan J, et al. Salinomycin exerts anti-colorectal cancer activity by targeting the beta-catenin/T-cell factor complex. Br J Pharmacol (2019) 176(17):3390–406. doi: 10.1111/bph.14770
211. Samuel SM, Varghese E, Koklesova L, Liskova A, Kubatka P, Busselberg D. Counteracting chemoresistance with metformin in breast cancers: targeting cancer stem cells. Cancers (Basel) (2020) 12(9):2482. doi: 10.3390/cancers12092482
212. Gutzmer R, Solomon JA. Hedgehog pathway inhibition for the treatment of basal cell carcinoma. Target Oncol (2019) 14(3):253–67. doi: 10.1007/s11523-019-00648-2
213. Froeling FEM, Swamynathan MM, Deschenes A, Chio IIC, Brosnan E, Yao MA, et al. Bioactivation of napabucasin triggers reactive oxygen species-mediated cancer cell death. Clin Cancer Res (2019) 25(23):7162–74. doi: 10.1158/1078-0432.CCR-19-0302
214. Zhang ZW, Teng X, Zhao F, Ma C, Zhang J, Xiao LF, et al. METTL3 regulates m(6)A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma. Cell Rep (2022) 41(4):111530. doi: 10.1016/j.celrep.2022.111530
215. Nishiga Y, Drainas AP, Baron M, Bhattacharya D, Barkal AA, Ahrari Y, et al. Radiotherapy in combination with CD47 blockade elicits a macrophage-mediated abscopal effect. Nat Cancer (2022) 3(11):1351–66. doi: 10.1038/s43018-022-00456-0
216. Marcu LG, Marcu D. In silico modelling of a cancer stem cell-targeting agent and its effects on tumour control during radiotherapy. Sci Rep (2016) 6:32332. doi: 10.1038/srep32332
217. Haslauer T, Greil R, Zaborsky N, Geisberger R. CAR T-cell therapy in hematological Malignancies. Int J Mol Sci (2021) 22(16):8996. doi: 10.3390/ijms22168996
218. Xu Q, Liu G, Yuan X, Xu M, Wang H, Ji J, et al. Antigen-specific T-cell response from dendritic cell vaccination using cancer stem-like cell-associated antigens. Stem Cells (2009) 27(8):1734–40. doi: 10.1002/stem.102
219. Muriithi W, Macharia LW, Heming CP, Echevarria JL, Nyachieo A, Filho PN, et al. ABC transporters and the hallmarks of cancer: roles in cancer aggressiveness beyond multidrug resistance. Cancer Biol Med (2020) 17(2):253–69. doi: 10.20892/j.issn.2095-3941.2019.0284
220. Liu D, Hong Y, Li Y, Hu C, Yip TC, Yu WK, et al. Targeted destruction of cancer stem cells using multifunctional magnetic nanoparticles that enable combined hyperthermia and chemotherapy. Theranostics (2020) 10(3):1181–96. doi: 10.7150/thno.38989
221. Ribas L, Vanezis K, Imues MA, Piferrer F. Treatment with a DNA methyltransferase inhibitor feminizes zebrafish and induces long-term expression changes in the gonads. Epigenet Chromatin (2017) 10(1):59. doi: 10.1186/s13072-017-0168-7
222. Katarzyna R, Lucyna B. Epigenetic therapies in patients with solid tumors: Focus on monotherapy with deoxyribonucleic acid methyltransferase inhibitors and histone deacetylase inhibitors. J Cancer Res Ther (2019) 15(5):961–70. doi: 10.4103/jcrt.JCRT_403_17
223. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther (2020) 5(1):8. doi: 10.1038/s41392-020-0110-5
224. Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, et al. Targeting Notch signalling pathway of cancer stem cells. Stem Cell Investig (2018) 5:5. doi: 10.21037/sci.2018.02.02
225. Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol (2011) 8(2):97–106. doi: 10.1038/nrclinonc.2010.196
226. Merchant AA, Matsui W. Targeting Hedgehog–a cancer stem cell pathway. Clin Cancer Res (2010) 16(12):3130–40. doi: 10.1158/1078-0432.CCR-09-2846
227. Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel) (2016) 8(2):22. doi: 10.3390/cancers8020022
228. Gampala S, Zhang G, Chang CJ, Yang JY. Activation of AMPK sensitizes medulloblastoma to Vismodegib and overcomes Vismodegib-resistance. FASEB Bioadv (2021) 3(6):459–69. doi: 10.1096/fba.2020-00032
229. Carpenter RL, Ray H. Safety and tolerability of sonic hedgehog pathway inhibitors in cancer. Drug Saf (2019) 42(2):263–79. doi: 10.1007/s40264-018-0777-5
230. Mercurio AM. VEGF/neuropilin signaling in cancer stem cells. Int J Mol Sci (2019) 20(3):490. doi: 10.3390/ijms20030490
231. Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang Z, et al. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther (2017) 24(9):386–92. doi: 10.1038/cgt.2017.30
232. Takahashi S. Vascular endothelial growth factor (VEGF), VEGF receptors and their inhibitors for antiangiogenic tumor therapy. Biol Pharm Bull (2011) 34(12):1785–8. doi: 10.1248/bpb.34.1785
233. Raz G, Allen KE, Kingsley C, Cherni I, Arora S, Watanabe A, et al. Hedgehog signaling pathway molecules and ALDH1A1 expression in early-stage non-small cell lung cancer. Lung Cancer (2012) 76(2):191–6. doi: 10.1016/j.lungcan.2011.10.015
234. Wang S, Wang Y, Xun X, Zhang C, Xiang X, Cheng Q, et al. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J Exp Clin Cancer Res (2020) 39(1):22. doi: 10.1186/s13046-020-1523-2
235. Bartucci M, Hussein MS, Huselid E, Flaherty K, Patrizii M, Laddha SV, et al. Synthesis and characterization of novel BMI1 inhibitors targeting cellular self-renewal in hepatocellular carcinoma. Target Oncol (2017) 12(4):449–62. doi: 10.1007/s11523-017-0501-x
236. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther (2021) 6(1):402. doi: 10.1038/s41392-021-00791-1
237. Furqan M, Akinleye A, Mukhi N, Mittal V, Chen Y, Liu D. STAT inhibitors for cancer therapy. J Hematol Oncol (2013) 6:90. doi: 10.1186/1756-8722-6-90
238. Taniguchi H, Masuishi T, Kawazoe A, Muro K, Kadowaki S, Bando H, et al. Phase I study of napabucasin in combination with FOLFIRI + bevacizumab in Japanese patients with metastatic colorectal cancer. Int J Clin Oncol (2021) 26(11):2017–24. doi: 10.1007/s10147-021-01987-9
239. Doheny D, Sirkisoon S, Carpenter RL, Aguayo NR, Regua AT, Anguelov M, et al. Combined inhibition of JAK2-STAT3 and SMO-GLI1/tGLI1 pathways suppresses breast cancer stem cells, tumor growth, and metastasis. Oncogene (2020) 39(42):6589–605. doi: 10.1038/s41388-020-01454-1
240. Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U.S.A. (2013) 110(50):20224–9. doi: 10.1073/pnas.1314239110
241. Liu Y, Qi X, Donnelly L, Elghobashi-Meinhardt N, Long T, Zhou RW, et al. Mechanisms and inhibition of Porcupine-mediated Wnt acylation. Nature (2022) 607(7920):816–22. doi: 10.1038/s41586-022-04952-2
242. Kim JY, Lee HY, Park KK, Choi YK, Nam JS, Hong IS. CWP232228 targets liver cancer stem cells through Wnt/beta-catenin signaling: a novel therapeutic approach for liver cancer treatment. Oncotarget (2016) 7(15):20395–409. doi: 10.18632/oncotarget.7954
243. Shi L, Fei X, Wang Z, You Y. PI3K inhibitor combined with miR-125b inhibitor sensitize TMZ-induced anti-glioma stem cancer effects through inactivation of Wnt/beta-catenin signaling pathway. In Vitro. Cell Dev Biol Anim (2015) 51(10):1047–55. doi: 10.1007/s11626-015-9931-x
244. Neiheisel A, Kaur M, Ma N, Havard P, Shenoy AK. Wnt pathway modulators in cancer therapeutics: An update on completed and ongoing clinical trials. Int J Cancer (2022) 150(5):727–40. doi: 10.1002/ijc.33811
245. Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: Cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review). Int J Oncol (2017) 51(5):1357–69. doi: 10.3892/ijo.2017.4129
246. Wiese M, Hamdan FH, Kubiak K, Diederichs C, Gielen GH, Nussbaumer G, et al. Combined treatment with CBP and BET inhibitors reverses inadvertent activation of detrimental super enhancer programs in DIPG cells. Cell Death Dis (2020) 11(8):673. doi: 10.1038/s41419-020-02800-7
247. Rodon J, Argilés G, Connolly RM, Vaishampayan U, de Jonge M, Garralda E, et al. Phase 1 study of single-agent WNT974, a first-in-class Porcupine inhibitor, in patients with advanced solid tumours. Br J Cancer (2021) 125(1):28–37. doi: 10.1038/s41416-021-01389-8
248. Molczyk C, Singh RK. CXCR1: A cancer stem cell marker and therapeutic target in solid tumors. Biomedicines (2023) 11(2):576. doi: 10.3390/biomedicines11020576
249. Singh JK, Farnie G, Bundred NJ, Simoes BM, Shergill A, Landberg G, et al. Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and -independent mechanisms. Clin Cancer Res (2013) 19(3):643–56. doi: 10.1158/1078-0432.CCR-12-1063
250. Rodolfo C, Di Bartolomeo S, Cecconi F. Autophagy in stem and progenitor cells. Cell Mol Life Sci (2016) 73(3):475–96. doi: 10.1007/s00018-015-2071-3
251. Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol (2007) 23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154
252. Zaarour RF, Azakir B, Hajam EY, Nawafleh H, Zeinelabdin NA, Engelsen AST, et al. Role of hypoxia-mediated autophagy in tumor cell death and survival. Cancers (Basel) (2021) 13(3):533. doi: 10.3390/cancers13030533
253. Gong C, Bauvy C, Tonelli G, Yue W, Delomenie C, Nicolas V, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene (2013) 32(18):2261–72, 2272e 1-11. doi: 10.1038/onc.2012.252
254. Wolf J, Dewi DL, Fredebohm J, Muller-Decker K, Flechtenmacher C, Hoheisel JD, et al. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res (2013) 15(6):R109. doi: 10.1186/bcr3576
255. Song YJ, Zhang SS, Guo XL, Sun K, Han ZP, Li R, et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett (2013) 339(1):70–81. doi: 10.1016/j.canlet.2013.07.021
256. Zhu H, Wang D, Liu Y, Su Z, Zhang L, Chen F, et al. Role of the Hypoxia-inducible factor-1 alpha induced autophagy in the conversion of non-stem pancreatic cancer cells into CD133+ pancreatic cancer stem-like cells. Cancer Cell Int (2013) 13(1):119. doi: 10.1186/1475-2867-13-119
257. Zhang D, Zhao Q, Sun H, Yin L, Wu J, Xu J, et al. Defective autophagy leads to the suppression of stem-like features of CD271. J BioMed Sci (2016) 23(1):82. doi: 10.1186/s12929-016-0297-5
258. Peng Q, Qin J, Zhang Y, Cheng X, Wang X, Lu W, et al. Autophagy maintains the stemness of ovarian cancer stem cells by FOXA2. J Exp Clin Cancer Res (2017) 36(1):171. doi: 10.1186/s13046-017-0644-8
259. Buccarelli M, Marconi M, Pacioni S, De Pascalis I, D'Alessandris QG, Martini M, et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis (2018) 9(8):841. doi: 10.1038/s41419-018-0864-7
260. Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol (2019) 247(5):708–18. doi: 10.1002/path.5222
261. Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F. Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ (2019) 26(4):690–702. doi: 10.1038/s41418-019-0292-y
262. Han Y, Fan S, Qin T, Yang J, Sun Y, Lu Y, et al. Role of autophagy in breast cancer and breast cancer stem cells (Review). Int J Oncol (2018) 52(4):1057–70. doi: 10.3892/ijo.2018.4270
263. Sharif T, Martell E, Dai C, Kennedy BE, Murphy P, Clements DR, et al. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy (2017) 13(2):264–84. doi: 10.1080/15548627.2016.1260808
264. Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res (2015) 13(4):651–8. doi: 10.1158/1541-7786.MCR-14-0487
265. Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U.S.A. (2011) 108(4):1397–402. doi: 10.1073/pnas.1018898108
266. Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24- stem cell-like breast cancer cells in human tumors. J Clin Invest (2011) 121(7):2723–35. doi: 10.1172/JCI44745
267. Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol (2015) 25(6):354–63. doi: 10.1016/j.tcb.2015.02.002
268. Yeo SK, Wen J, Chen S, Guan JL. Autophagy differentially regulates distinct breast cancer stem-like cells in murine models via EGFR/stat3 and tgfβ/smad signaling. Cancer Res (2016) 76(11):3397–410. doi: 10.1158/0008-5472.CAN-15-2946
269. Smit L, Berns K, Spence K, Ryder WD, Zeps N, Madiredjo M, et al. An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget (2016) 7(3):2596–610. doi: 10.18632/oncotarget.6354
270. Ning Y, Luo C, Ren K, Quan M, Cao J. FOXO3a-mediated suppression of the self-renewal capacity of sphere-forming cells derived from the ovarian cancer SKOV3 cell line by 7-difluoromethoxyl-5,4’-di-n-octyl genistein. Mol Med Rep (2014) 9(5):1982–8. doi: 10.3892/mmr.2014.2012
271. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
272. Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, García-Echeverría C, et al. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci U.S.A. (2009) 106(1):268–73. doi: 10.1073/pnas.0810956106
273. Sunayama J, Sato A, Matsuda K, Tachibana K, Watanabe E, Seino S, et al. FoxO3a functions as a key integrator of cellular signals that control glioblastoma stem-like cell differentiation and tumorigenicity. Stem Cells (2011) 29(9):1327–37. doi: 10.1002/stem.696
274. Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene (2013) 32(6):699–712. doi: 10.1038/onc.2012.111
275. Ciechomska IA, Przanowski P, Jackl J, Wojtas B, Kaminska B. BIX01294, an inhibitor of histone methyltransferase, induces autophagy-dependent differentiation of glioma stem-like cells. Sci Rep (2016) 6:38723. doi: 10.1038/srep38723
276. Zhao Y, Huang Q, Yang J, Lou M, Wang A, Dong J, et al. Autophagy impairment inhibits differentiation of glioma stem/progenitor cells. Brain Res (2010) 1313:250–8. doi: 10.1016/j.brainres.2009.12.004
277. Rothe K, Lin H, Lin KB, Leung A, Wang HM, Malekesmaeili M, et al. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood (2014) 123(23):3622–34. doi: 10.1182/blood-2013-07-516807
278. Karvela M, Baquero P, Kuntz EM, Mukhopadhyay A, Mitchell R, Allan EK, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy (2016) 12(6):936–48. doi: 10.1080/15548627.2016.1162359
279. Watson AS, Riffelmacher T, Stranks A, Williams O, De Boer J, Cain K, et al. Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discovery (2015) 1:15008. doi: 10.1038/cddiscovery.2015.8
280. Sullivan RJ, Hamid O, Gonzalez R, Infante JR, Patel MR, Hodi FS, et al. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat Med (2019) 25(6):929–35. doi: 10.1038/s41591-019-0474-7
281. Migden MR, Rischin D, Schmults CD, Guminski A, Hauschild A, Lewis KD, et al. PD-1 blockade with cemiplimab in advanced cutaneous squamous-cell carcinoma. N Engl J Med (2018) 379(4):341–51. doi: 10.1056/NEJMoa1805131
282. Cortese I, Muranski P, Enose-Akahata Y, Ha SK, Smith B, Monaco M, et al. Pembrolizumab treatment for progressive multifocal leukoencephalopathy. N Engl J Med (2019) 380(17):1597–605. doi: 10.1056/NEJMoa1815039
283. Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med (2019) 380(12):1103–15. doi: 10.1056/NEJMoa1816047
284. Lee KS, Choi JS, Cho YW. Reprogramming of cancer stem cells into non-tumorigenic cells using stem cell exosomes for cancer therapy. Biochem Biophys Res Commun (2019) 512(3):511–6. doi: 10.1016/j.bbrc.2019.03.072
285. Arima Y, Nobusue H, Saya H. Targeting of cancer stem cells by differentiation therapy. Cancer Sci (2020) 111(8):2689–95. doi: 10.1111/cas.14504
286. Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell (2012) 21(3):283–96. doi: 10.1016/j.ccr.2012.03.003
287. Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res (2018) 38(7):3797–809. doi: 10.21873/anticanres.12663
288. Ertas YN, Abedi Dorcheh K, Akbari A, Jabbari E. Nanoparticles for targeted drug delivery to cancer stem cells: A review of recent advances. Nanomaterials (Basel) (2021) 11(7):112260. doi: 10.3390/nano11071755
289. Qin W, Huang G, Chen Z, Zhang Y. Nanomaterials in targeting cancer stem cells for cancer therapy. Front Pharmacol (2017) 8:1. doi: 10.3389/fphar.2017.00001
290. He L, Gu J, Lim LY, Yuan ZX, Mo J. Nanomedicine-mediated therapies to target breast cancer stem cells. Front Pharmacol (2016) 7:313. doi: 10.3389/fphar.2016.00313
291. Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R’s of radiobiology revisited. Stem Cells (2010) 28(4):639–48. doi: 10.1002/stem.318
292. Emami Nejad A, Najafgholian S, Rostami A, Sistani A, Shojaeifar S, Esparvarinha M, et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: a novel approach to developing treatment. Cancer Cell Int (2021) 21(1):62. doi: 10.1186/s12935-020-01719-5
293. Sistigu A, Musella M, Galassi C, Vitale I, De Maria R. Tuning cancer fate: tumor microenvironment’s role in cancer stem cell quiescence and reawakening. Front Immunol (2020) 11:2166. doi: 10.3389/fimmu.2020.02166
294. Huang R, Rofstad EK. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget (2017) 8(21):35351–67. doi: 10.18632/oncotarget.10169
295. Mukha A, Dubrovska A. Metabolic targeting of cancer stem cells. Front Oncol (2020) 10:537930. doi: 10.3389/fonc.2020.537930
296. Dalerba P, Sahoo D, Paik S, Guo X, Yothers G, Song N, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med (2016) 374(3):211–22. doi: 10.1056/NEJMoa1506597
Keywords: cancer stem cell (CSC), hypoxia, hypoxia-inducible factor (HIF), resistance, stemness, tumor microenvironment (TME)
Citation: Zaarour R, Ribeiro M, Azzarone B, Kapoor S and Chouaib S (2023) Tumor microenvironment-induced tumor cell plasticity: relationship with hypoxic stress and impact on tumor resistance. Front. Oncol. 13:1222575. doi: 10.3389/fonc.2023.1222575
Received: 14 May 2023; Accepted: 27 September 2023;
Published: 11 October 2023.
Edited by:
Pu Xia, Jinzhou Medical University, ChinaReviewed by:
Ina Kurth, German Cancer Research Center (DKFZ), GermanyCopyright © 2023 Zaarour, Ribeiro, Azzarone, Kapoor and Chouaib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Chouaib, c2FsZW0uY2hvdWFpYkBnbXUuYWMuYWU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.