- 1Department of Biotechnology, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi, India
- 2Department of Zoology, Deshbandhu College, University of Delhi, Delhi, India
Introduction: Artemisia absinthium (wormwood) exhibits anticancer properties by inhibiting proliferation and causing cell death in breast cancer. Targeted drug delivery of A. absinthium nanoformulation using N-isopropyl acrylamide, N-vinyl pyrrolidone, and acrylic acid-based polymeric nanoparticles (NVA-AA NPs) was ensured by utilizing features of the tumor microenvironment, although their mechanism of action involved in cytotoxicity remains unknown.
Methods: The present study employed nano LC-MS/MS to identify differences in secretory protein expression associated with the treatment of breast cancer cell lines (MCF-7; MDA-MB-231) by NVA-AA NPs for the determination of affected pathways and easily accessible therapeutic targets. Different bioinformatics tools were used to identify signature differentially expressed proteins (DEPs) using survival analysis by GENT2 and correlation analysis between their mRNA expressions and sensitivity toward small-molecule drugs as well as immune cell infiltration by GSCA.
Results: Analysis by GENT2 revealed 22 signature DEPs with the most significant change in their expression regulation, namely, gelsolin, alpha-fetoprotein, complement component C3, C7, histone H2B type 1-K, histone H2A.Z, H2AX, heat shock cognate 71 kDa protein, heat shock 70 kDa protein 1-like, cytochrome c somatic, GTP-binding nuclear protein Ran, tubulin beta chain, tubulin alpha-1B chain, tubulin alpha-1C chain, phosphoglycerate mutase 1, kininogen 1, carboxypeptidase N catalytic chain, fibulin-1, peroxiredoxins 4, lactate dehydrogenase C, SPARC, and SPARC-like protein 1. Correlation analysis between their mRNA expressions versus immune cell infiltrates showed a positive correlation with antitumor immune response elicited by these NPs as well as a correlation with drug response shown by the GDSC and CTRP drugs in different cancer cells.
Discussion: Our results suggest that NVA-AA NPs were able to invade the tumor microenvironment; transformed the communication network between the cancer cells; affected potential drivers of microtubular integrity, nucleosome assembly, and cell cycle; and eventually caused cell death.
1 Introduction
Breast cancer is one of the most common and leading causes of mortality among women worldwide. According to an estimation by the American Cancer Society, 1.9 million new cancer cases were diagnosed with 609,360 cancer deaths in the United States in 2022 (1). Several conventional therapeutic interventions are available for breast cancer that mainly include chemotherapy, radiotherapy, mastectomy, hormone therapy, immunotherapy, or a combination of any of these (2). These treatment modalities such as chemotherapy are often associated with acute side effects such as hair fall, nausea, vomiting, cytopenia, cardiotoxicity, ovarian failure, chances of tumor relapse, and drug resistance among patients. Radiotherapy has proven to be effective with low chances of recurrence and high overall survival; however, radiation dermatitis is reported in patients (3). Mastectomy impacts the psychological well-being of patients and their quality of life (4). Similarly, hormone therapy also poses challenges among patients that include hypertension, osteoporosis, thromboembolic events, endometrial cancer, and dyspareunia (5). Different immune therapeutic strategies of immunotherapy targeted against different breast cancer subtypes have been developed that mainly deal with the recruitment of targeted antibodies or immune checkpoint inhibitors (ICI) aimed at triggering or restoring the immune responses against tumor cells. Different clinical subtypes of breast cancer vary in their immunological characteristics, comprising of tumor-infiltrating lymphocytes (TILs CD4, CD8), expression of programmed death ligand-1 (PD-L1), leukemia inhibitory factor (LIF), tumor-associated antigen, and random gene mutations (6). The monoclonal antibodies target molecular cell surface receptors of tumor cells enabling blockade of proliferative pathways (e.g., HER2, PD-L1) or tumor-infiltrating immune cell subsets in the tumor microenvironment hampering the activation of immune cells by tumor cells and abolishing the tumor. Already existing antitumor immune response elicited by TILs in the tumor microenvironment was associated with good responsiveness toward chemotherapy and prognosis (7). Similarly, other immunomarkers (PD-L1 or LIF) are also predictive indicators of the clinical outcome of ICI therapy. A number of drugs are clinically approved for targeted therapies against HER2+, HR+/HER2−, TNBC, and gBRCAm that include trastuzumab, neratinib, everolimus, abemaciclib, pembrolizumab, olaparib, and talazoparib (8). However, different targeted drugs in combination with chemotherapy for varying breast cancer subtypes showed a limited success rate and significant discrepancies in immune-modulatory response, efficacy, safety, and prognosis (9).
Sonodynamic therapy (SDT) is a new therapeutic strategy employing ultrasound for the activation of sonosensitizers that produce reactive oxygen species which could destroy tumor cells; however, a hypoxic tumor microenvironment poses certain limitations to SDT. Nanoplatforms are a widely explored biocompatible delivery system for treating different types of cancer that could considerably increase the efficiency of existing treatment by utilizing the tumor microenvironment. Recently, ultrasmall titanium nitride nanodots (TiNs) and titanium dioxide (TiO2)-based hollow nanoshells have been developed which act as sonosensitizers and carriers of free radical generators or hypoxia-selective cytotoxin, synergistically increasing the therapeutic efficiency of SDT against solid tumors (10–12). Similarly, photothermal therapy (PPT) is another strategy that uses near-infrared waves to eliminate solid tumors. Ultrasmall zirconium carbide nanodots (NDs) and mesoporous NiS2 nanospheres (mNiS2 NSs) have been designed that significantly improved the PTT effect by inhibiting tumor growth in vitro and in vivo (13, 14).
It is estimated that approximately 75% of anticancer drugs are designed and prepared from plant-derived natural products (15). Artemisia absinthium L. (wormwood) is a perennial herb that belongs to the family Asteraceae and is found all over the world such as in the Indian subcontinent. This plant has been used in ethnic medicine due to its several medicinal properties such as antiseptic, antibacterial, antifungal, antiparasitic, antihelminthic, and anti-inflammatory. The anticancer effect of A. absinthium methanolic extract has been studied in-vitro on human breast carcinoma cell lines MDA-MB-231 and MCF-7, depicting its antiproliferative effect mediated via the apoptotic pathway (16). Previously, our research group evaluated the therapeutic potential of A. absinthium ethanolic extract-loaded polymeric nanoparticles (NVA-AA) on breast cancer cell lines (MCF-7 and MDA-MB-231) targeting the tumor microenvironment. Targeted drug delivery involves the simultaneous administration of anticancer agents or phytoconstituents with various routes of action in order to combat the aforementioned drawbacks of the majority of anticancer medications. Temperature- and pH-sensitive, self-assembled polymeric nanoparticles, made of N-isopropyl acrylamide (NIPAAM), N-vinyl pyrrolidone (VP), and acrylic acid (AA), are biocompatible and have been used in the past with several drugs (e.g., ketorolac, riluzole) for site-specific delivery of different formulations (17, 18). Free radical polymerization was used to prepare NIPAAM, VP, and AA-based polymeric nanoparticles (NVAs). In our previous study, the cytotoxic potential of A. absinthium extract-loaded polymeric nanoparticles (NVA-AAs) was explored against breast cancer with a proteomic analysis approach to find out their mechanism of action. These NPs were able to cause significant cell proliferation inhibition, cell death, and cell cycle arrest at the G0/G1 phase in MCF-7 as well as MDA-MB-231 cell lines (19).
Secretome is the group of proteins secreted in the extracellular space by a cell, tissue, organ, or organism within the local environment which comprises an extracellular matrix (ECM) and is mainly responsible for cell–cell communication. Altered patterns of secretion are closely associated with the growth and progression of cancer as these transformed signals aid the cancer cells to metastasize. Altered cancer secretome tends to have an important role in tumor microenvironment communication as well as the development of chemoresistance (20). Moreover, the secretome is considered a reservoir of potential cancer biomarkers as well as drug and therapeutic targets involved in the molecular cascade of drug development (21). Therefore, the investigation of proteins secreted in the medium by in-vitro-cultured cancer cells will provide insight into the interaction of NVA-AA with the tumor microenvironment, unraveling easily accessible biomarkers and potential therapeutic targets.
Secretome profiling of the cell lines through the proteomics approach helps in the identification and selection of secreted proteins into the extracellular milieu to be utilized as biomarkers or drug targets for therapeutic application. The nano liquid chromatography-tandem mass spectrometry (nano LC-MS/MS) approach has been used previously for secretome profiling of different cell lines that helps in the quantification of hundreds to thousands of secretory proteins present in the media (22). In the present study, the nano LC-MS/MS proteomic approach was employed to compare the secretome of NVA-AA-treated MCF-7 and MDA-MB-231 cell lines with the corresponding untreated cell lines as a control to identify potential therapeutic targets among differentially expressed secretory proteins. A total of 208 and 194 specific proteins were identified in the MCF-7 and MDA-MB-231 cell lines, among which 65 and 156 proteins were significantly differentially expressed in the MCF-7 and MDA-MB-231 cell lines, respectively. In MCF-7, some of the upregulated proteins are reported to be involved in apoptosis, vesicular trafficking, inhibition of angiogenesis, cell proliferation, and metastasis. Some tumor suppressor genes were also highly upregulated in NVA-AA-treated cells. At the same time, a few proteins involved in tumorigenesis were downregulated by NVA-AA treatment in MCF-7 cells. Similarly, proteins involved in vesicular trafficking and inhibition of cancer invasion were highly upregulated in NVA-AA NP-treated MDA-MB-231 cells, while proteins involved in proliferation, tumorigenesis, proteasomal degradation, cell adhesion, and migration were downregulated after NVA-AA NP treatment.
2 Materials and methods
2.1 Chemicals
N-isopropylacrylamide (NIPAAM) was procured from Sigma-Aldrich, USA and recrystallized with N-hexane overnight. N-vinyl 2-pyrrolidone (VP) and acrylic acid (AA) were purchased from Across Organics (USA). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), antibiotic solution (100×), trypsin (with 0.5% EDTA), and ammonium persulfate (APS) were bought from HiMedia, USA, and ferrous ammonium sulfate (FAS) was from SRL Pvt Ltd. India MCF-7 and MDA-MB-231 cell lines were procured from the National Centre for Cell Science (NCCS), Pune, India.
2.2 Preparation of Artemisia absinthium ethanolic extract-loaded NIPAAM-VP-AA polymeric nanoparticles
NIPAAM-VP-AA (NVA-AA) polymeric nanoparticles were synthesized based on previous studies (19, 23, 24). The monomers N-isopropyl acrylamide (NIPAAM), N-vinylpyrrolidone (VP), and acrylic acid (AA) were employed in the molar ratio of 90:10:5 for free radical polymerization. N,N′-methylene-bisacrylamide (MBA) was used as a cross-linker, FAS as an activator, and APS as an initiator. One hundred eighty milligrams of NIPAAM, 20 µl of VP, and 10 µl of AA were dissolved in 20 ml of double distilled water accompanied by vigorous vortexing. One hundred microliters of MBA (0.049 g/ml), 60 µl of FAS (5 mg/ml), and 100 µl of APS (saturated) were added to trigger the polymerization reaction, and nitrogen gas was passed to maintain an inert environment for 24 h at 32°C. The final solution was dialyzed through a cellulose dialyzing membrane (cutoff 12 kDa) and characterized for the determination of the average size distribution and morphology of the synthesized NPs. The uniform, monodisperse, and spherical-shaped NPs were characterized by dynamic scattering light (DLS) spectroscopy and transmission electron microscopy (TEM). Artemisia absinthium ethanolic extract was added slowly to the aqueous solution of NPs with continuous vortexing and mild sonication to assist its physical entrapment into the hydrophobic core of NPs.
2.3 Cell culture
The selection of relevant breast cancer cell lines for the translational study was done as per the a) molecular subtype, b) metastatic status, c) best or worst prognosis, and d) response rate to different therapies. Two subtypes were further chosen out of the four existing breast cancer subtypes: the first is luminal A subtype which is ER and/or PR+, HER2−, slow growing with a low-grade, high response rate, and the finest prognosis, and the second was triple-negative breast cancer (TNBC) subtype which is ER−, PR−, and HER2−, aggressive in nature with a low response rate and a variable prognosis. Therefore, MCF-7 and MDA-MB-231 were selected for the luminal A and the TNBC subtype, respectively. MCF-7 and MDA-MB-231 cells were grown in DMEM supplemented with 10% FBS at 37°C and 5% CO2. Cells (10,000 cells per well) were grown onto 96-well plates and treated with an IC50 concentration of A. absinthium ethanolic extract-loaded NIPAAM-VP-AA (NVA-AA) polymeric nanoparticles at 176.83 ± 11.8 μg/ml for MCF-7 and 181.39 ± 23.2 μg/ml for MDA-MB-231 (19). After 24 h, the conditioned media were collected and centrifuged for removing debris. The supernatants were concentrated by lyophilization and the protein was quantified by Bradford’s method.
2.4 Sample preparation
Samples were dissolved in 6 M of Gn-HCl (50 mM Tris, pH 8.8) buffer. A 50-µg sample was reduced with 5 mM of TCEP and further alkylated with 50 mM of iodoacetamide. It was further digested with trypsin (1:50, trypsin/lysate ratio) for 16 h at 37°C. Digests were cleaned using a C18 silica cartridge to remove the salt and dried using a speed vac. The dried pellet was resuspended in buffer A (2% acetonitrile, 0.1% formic acid).
2.5 Nano LC-MS/MS
All the experiments were performed quantitatively using the RSLC Nano system (Thermo Fisher Scientific, USA) coupled to Thermo Fisher-QExactive Plus equipped with a nano electrospray ion source. One microgram was loaded on a C18 column 50 cm, 3.0 μm EASY-Spray column (Thermo Fisher Scientific). Peptides were eluted with a 0%–40% gradient of buffer B (80% acetonitrile, 0.1% formic acid) at a flow rate of 300 nl/min and injected for mass spectrometry analysis. LC gradients were run for 100 min. MS1 spectra were acquired in the Orbitrap at 70,000 resolution. Dynamic exclusion was employed during data-dependent acquisition (DDA) for 10 s excluding all charge states for a given precursor. This method was chosen to ensure that each fragment obtained can be related back to a specific precursor, as the resulting MS/MS spectrum contains only fragments from the precursor. Moreover, at the same pressure of He gas, the ion trap performs both ion collection and fragmentation in the used dynamic exclusion method. MS2 spectra were acquired at 17,500 resolution. All the processed samples were subjected to mass spectrometry run in technical replicates. Raw files containing mass/charge values were produced for each of the samples. Then, these raw files were analyzed through Thermo Proteome Discoverer (v2.2) against the UniProt Human database. For the SEQUEST and Amanda search, the precursor and fragment mass tolerance were set at 10 ppm and 0.5 Da, respectively. The protease used to generate peptides, i.e., enzyme specificity, was set for trypsin/P (cleavage at the C terminus of “K/R: unless followed by “P”) along with a maximum missed cleavage value of 2. Carbamidomethyl on cysteine was considered as fixed modification, while oxidation of methionine and acetylation of N terminus were both considered as variable modifications for database search. Both peptide spectrum match and protein false discovery rate were set to 0.01 false discovery rate (FDR). Based on UniProt accession number, Pfam, KEGG pathways, and GO annotations were assigned for the list of identified proteins. Differentially expressed proteins (DEPs) were identified by calculating their abundance ratio (treated/untreated) as log2 of fold change.
2.6 Bioinformatics analysis
2.6.1 Selection of signature differentially expressed proteins
A web-based gene expression database called the Gene Expression database of Normal and Tumor tissues 2 (GENT2) (http://gent2.appex.kr/gent2/) generated by the Affymetrix U133A or U133Plus2 microarray platform [GPL570 platform (HG-U133_Plus_2)] was utilized to find out the signature secretory proteins out of the total identified DEPs from NVA-AA-treated MCF-7 and MDA-MB-231 cell lines (25). Briefly, the tissue-wide expression pattern of each identified DEP was studied in normal and breast cancer patient samples (N = 1,246) to filter out the most important key proteins (DEPs) affected by the treatment of MCF-7 and MDA-MB-231 cells in comparison to untreated cell lines. Two-sample t-tests were applied, and filtered-out proteins were further verified for their distinct expression pattern across different cancer subtypes varying in estrogen receptor (ER) status. Similarly, survival analysis for the key proteins was further carried out, and Kaplan–Meier (KM) plots with median cutoff were produced to investigate the impact of their gene expression on overall survival (OS) of breast cancer patients, as well as their clinical significance and prognostic value. As MCF-7 and MDA-MB-231 represent luminal A and triple-negative breast cancer (TNBC) subtypes, respectively, therefore, the identified key proteins from MCF-7 and MDA-MB-231 were further compared with the existing expression profile across luminal A and TNBC subtype using GENT2. P-value <0.05 was considered statistically significant for expression as well as survival analysis.
2.6.2 Correlation of gene expression with drug sensitivity and immune cell infiltration in GSCA
An integrated platform, Gene Set Cancer Analysis (GSCA) (http://bioinfo.life.hust.edu.cn/GSCA/#/) (26), was used for Pearson correlation analysis between mRNA expression of selected signature genes and drug sensitivity (IC50) of 265 small molecules and 481 compounds toward breast cancer, provided by the Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/) and Cancer Therapeutics Response Portal (CTRP) (https://portals.broadinstitute.org/ctrp/) datasets, respectively. P-value was adjusted by FDR. Red and blue colors specify positive and negative correlations, respectively, such that a darker color represents a stronger correlation (*: P-value < 0.05; #: FRD < 0.05). These analyses would help to predict the sensitivity of MCF-7 and MDA-MB-231 toward NVA-AA NP treatments, which are shared with the list of anticancer drugs provided by the GDSC and CTRP. A positive correlation indicates that a high expression of genes provides a high resistance toward the drug and vice versa.
GSCA also provides multidimensional genomic data across 33 cancer types from the Cancer Genome Atlas (TCGA). Spearman correlation analysis was applied to estimate the association between signature gene mRNA expression and 24 immune cell infiltrates in BRCA through the ImmuCellAI module of GSCA (27). Immune infiltration and the GSVA score module of GSCA were utilized to study the correlation between signature gene expression and immune cell infiltrates in the BRCA cancer type. P-value was adjusted by FDR. Red and blue colors indicate positive and negative correlations, respectively, with a darker color representing a stronger correlation (*: P-value < 0.05; #: FDR < 0.05).
2.6.3 Gene annotation and KEGG analysis by GeneCodis
Singular and modular enrichment analysis (SEA and MEA) of the selected signature DEPs was done using an online web tool, GeneCodis (https://genecodis.genyo.es/). This tool uses standard hypergeometric or Wallenius’ non-central hypergeometric distribution for enrichment analysis, where hypergeometric or Wallenius tested the raw P-values and corrected them via Benjamini/Hochberg FDR. Different modules, namely, KEGG, Reactome, gene annotation for cellular components, PharmGKB, and DisGeNET, were used to determine the association of these signature genes with other metabolic pathways, cellular components, pharmacogenomics information about drug responses, and other human diseases, respectively. The bar charts were generated, where bar size signifies −log10(Pval Adj) and the red intensity signifies the number of genes present (28).
2.6.4 Protein–protein interaction and PANTHER analysis
The protein–protein interaction (PPI) among significantly differentially expressed signature proteins selected after nano LC-MS/MS analysis was built using the STRING module of Cytoscape version 3.7.1. Proteins of interest were enlisted into the STRING database using their UniProt IDs to generate the interaction between these proteins to highlight the mechanism involved in cytotoxicity caused by NVA-AA treatment and identify secretory protein targets. All the identified proteins were also classified based on their molecular function, biological roles, pathways, cellular components, and protein classes using PANTHER 17.0 (http://www.pantherdb.org/) online software.
2.7 Real-time PCR
Validation of the expression of selected altered signature DEPs, namely, gelsolin (GSN) (actin-binding protein and tumor suppressor), complement component C3 (C3) (component of innate immune response), and cytochrome c somatic (CYCS) (apoptotic biomarker) due to NVA-AA treatment of MCF-7 and MDA-MB-231 cell lines, was done by quantitation of the corresponding mRNA transcripts using quantitative PCR. The total RNA from the untreated and treated MCF-7 and MDA-MB-231 cell lines was isolated using the RNASure® Fusion RNA Mini Kit. The quantity and quality of RNA were assessed using NanoDrop 2000c spectrophotometer. The cDNA synthesis was done using normalized RNA samples by RevertAid H Minus Reverse Transcriptase (Thermo Scientific) Kit. Quantitative PCR of synthesized cDNA was performed on the Bio-Rad real-time PCR cycler using SYBR Green chemistry. The total reaction volume was made 10 μl which included 5 μl of 2× Fermentas Maxima SYBR Green qPCR Master Mix (Thermo Scientific), 0.25 μl of each primer (20 μM) (Supplementary Table S1), 2 μl of cDNA, and 2.5 μl of nuclease-free water. All reactions were performed in triplicates with the following temperature conditions: i) 96°C for 5 min (initial denaturation) and ii) 34 cycles at 96°C for 1 min, 56°C for 1 min, and 72°C for 5 min, followed by iii) melt curve analysis at a temperature range of 55°C–95°C. The raw data were analyzed with the CFX Maestro software (version 2.3). The Ct values were obtained using a constant threshold value for all the genes assayed. The relative gene expression was quantitated by applying a comparative ΔCt (2−ΔΔCT) method. The untreated sample was used as the calibrator/control. HPRT1 served as the normalizer gene.
2.8 Statistical analysis
Data were analyzed through unpaired t-test for the identification of differentially expressed secretory proteins among NVA-AA NP-treated and untreated MCF-7 and MDA-MB-231 cell lines using GraphPad Prism software (version 7.04, GraphPad software). P-value <0.05 was considered statistically significant.
3 Results
In this study, polymeric nanoparticles were synthesized using NIPAAM, VP, and AA with the average size of 131.4 nm ± 19.7 nm (by DLS) and 110 nm ± 12.6 nm (by TEM) (19). Artemisia absinthium ethanolic extract was encapsulated in the hydrophobic core of these polymeric nanoparticles (NVA-AA NPs). Nano LC-MS/MS was employed to quantify the unique proteins secreted in the media of NVA-AA-treated MCF-7 and MDA-MB-231 cell lines. Raw data files obtained from nano LC-MS/MS were analyzed against the human subset of the RefSeq database from NCBI, and the master list of proteins was prepared with their relative quantification. Unique proteins were selected out of the total protein groups and peptide groups identified in Thermo Proteome Discoverer against the Human database based on abundance count (=1). Among these, 65 and 156 proteins were significantly differentially expressed (P < 0.05) in MCF-7 and MDA-MB-231 cell lines (as shown in Supplementary Figures S1A, B), respectively. The lists of a few important significant DEPs in MCF-7 and MDA-MB-231 are shown in Tables 1, 2, respectively.
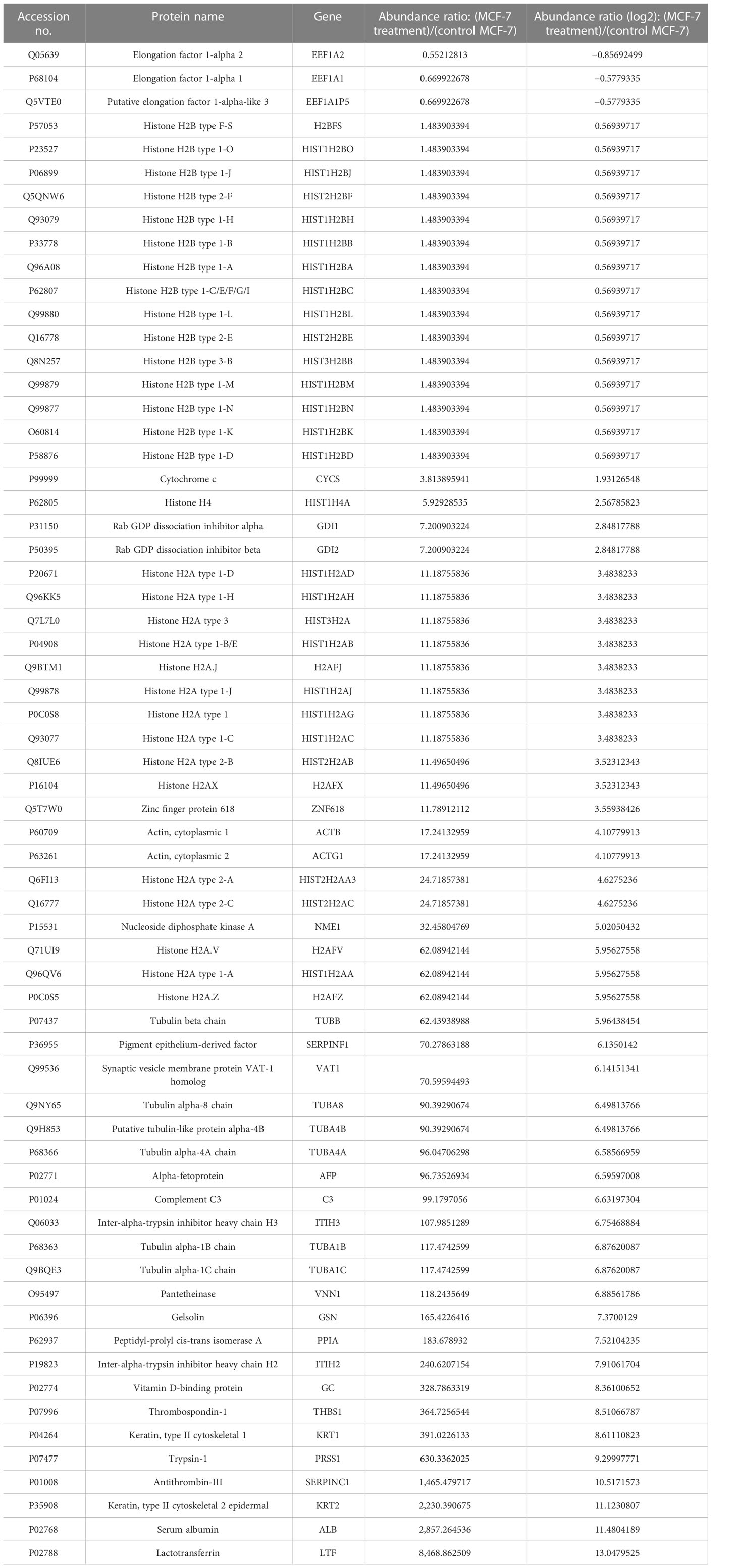
Table 1 List of some significantly differentially expressed proteins in NVA-AA NP-treated MCF-7 cell line with their abundance ratio and abundance ratio (log2).
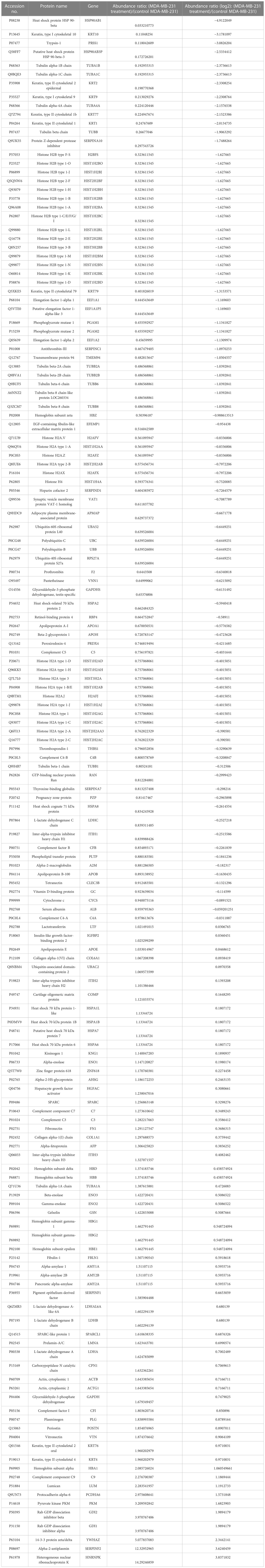
Table 2 List of some significantly differentially expressed proteins in NVA-AA NP-treated MDA-MB-231 cell line with their abundance ratio and abundance ratio (log2).
3.1 Secretome profiling of MCF-7 and MDA-MB-231 after NVA-AA NP treatment
A comparative analysis of the untreated and NVA-AA-treated MCF-7 and MDA-MB-231 cell lines revealed alteration in key secretory proteins involved in apoptosis, cell proliferation, tumorigenesis, metastasis, cancer progression, and suppression. In MCF-7, out of 65 proteins, some of the upregulated proteins (fold change ≥ 1) are involved in apoptosis, e.g., cytochrome c (CYCS), histone H2AX (H2AFX), nucleoside diphosphate kinase A (NME1), and lactotransferrin (LTF); a few are involved in vesicular trafficking, e.g., Rab GDP dissociation inhibitor beta (GDI2), while others are involved in the inhibition of angiogenesis, e.g., pigment epithelium-derived factor (SERPINF1) and thrombospondin-1 (THBS1). Some tumor suppressor genes are also highly upregulated in NVA-AA-treated cells, e.g., inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3) and gelsolin (GSN), while proteins involved in the inhibition of cell proliferation and metastasis are also upregulated, e.g., vitamin D-binding protein (GC). However, a few proteins involved in tumorigenesis were downregulated (fold change ≤ −1), e.g., elongation factor 1-alpha 2 (EEF1A2) and elongation factor 1-alpha 1 (EEF1A1). A few proteins belonging to the histone family (HIST1H4A, HIST1H2AD, H2AFX, HIST2H2AC, H2AFZ) had upregulated expression, suggesting that NVA-AA NPs affected the genome integrity of treated MCF-7 cells through nucleosome disassembly as compared with untreated cells. Increased expression of tubulin isoforms (TUBB, TUBA8, TUBA4A, TUBA1B) indicates the potential of NVA-AA to hamper the dynamics of microtubule assembly in MCF-7 cells. This could be linked with the downregulation of EEF1A2 and EEF1A1 in the treated cells that are required for cytoskeletal protein rearrangement comprised of actin and tubulins.
Similarly, proteins involved in the inhibition of cancer invasion are highly upregulated (fold change ≥ 1) in NVA-AA-treated MDA-MB-231 cells, e.g., lumican (LUM), along with proteins involved in vesicular trafficking, e.g., Rab GDP dissociation inhibitor beta (GDI2). Moreover, proteins involved in proliferation and tumorigenesis were downregulated (fold change ≤ −1) after treatment, e.g., elongation factor 1-alpha 1 (EEF1A1), elongation factor 1-alpha 2 (EEF1A2), peroxiredoxin-4 (PRDX4), GTP-binding nuclear protein Ran (RAN), and alpha-enolase (ENO1), as well as proteins involved in proteosomal degradation, cell adhesion, and migration, e.g., ubiquitin-60S ribosomal protein L40 (UBA52), fibronectin (FN1), and periostin (POSTN). The downregulation of tubulin family genes, TUBA1B, TUBA4A, TUBB, and TUBB8, after NVA-AA NP treatment in MDA-MB-231 suggests a disturbance in tubulin assembly. Heat shock protein HSP 90-beta (HSP90AB1), which is associated with the survival of certain oncogenes, was also significantly downregulated by NVA-AA NPs. Proteins belonging to the family of keratins (KRT1, KRT10, KRT2, KRT77) were also downregulated in our study.
3.2 Bioinformatic analysis
3.2.1 Identification of signature DEPs
Out of the total proteins, analysis by GENT2 revealed 22 signature DEPs having the most significant change in their expression level after NVA-AA treatment of MCF-7 and MDA-MB-231. Differential expression levels, fold change (log2FC), and the P-values of these proteins in breast cancer tissues are represented along with their expression after treatment in the affected cell lines in Table 3. The expression profile of selected signature DEPs across cancer subtypes differing in ER status is represented in Supplementary Figure S2 and their survival analysis is represented in Figure 1. Fold change and the expression profile of suitable signature genes from MCF-7 and MDA-MB-231 were again compared across luminal A and TNBC subtypes as shown in Supplementary Figure S3 and Supplementary Table S2.
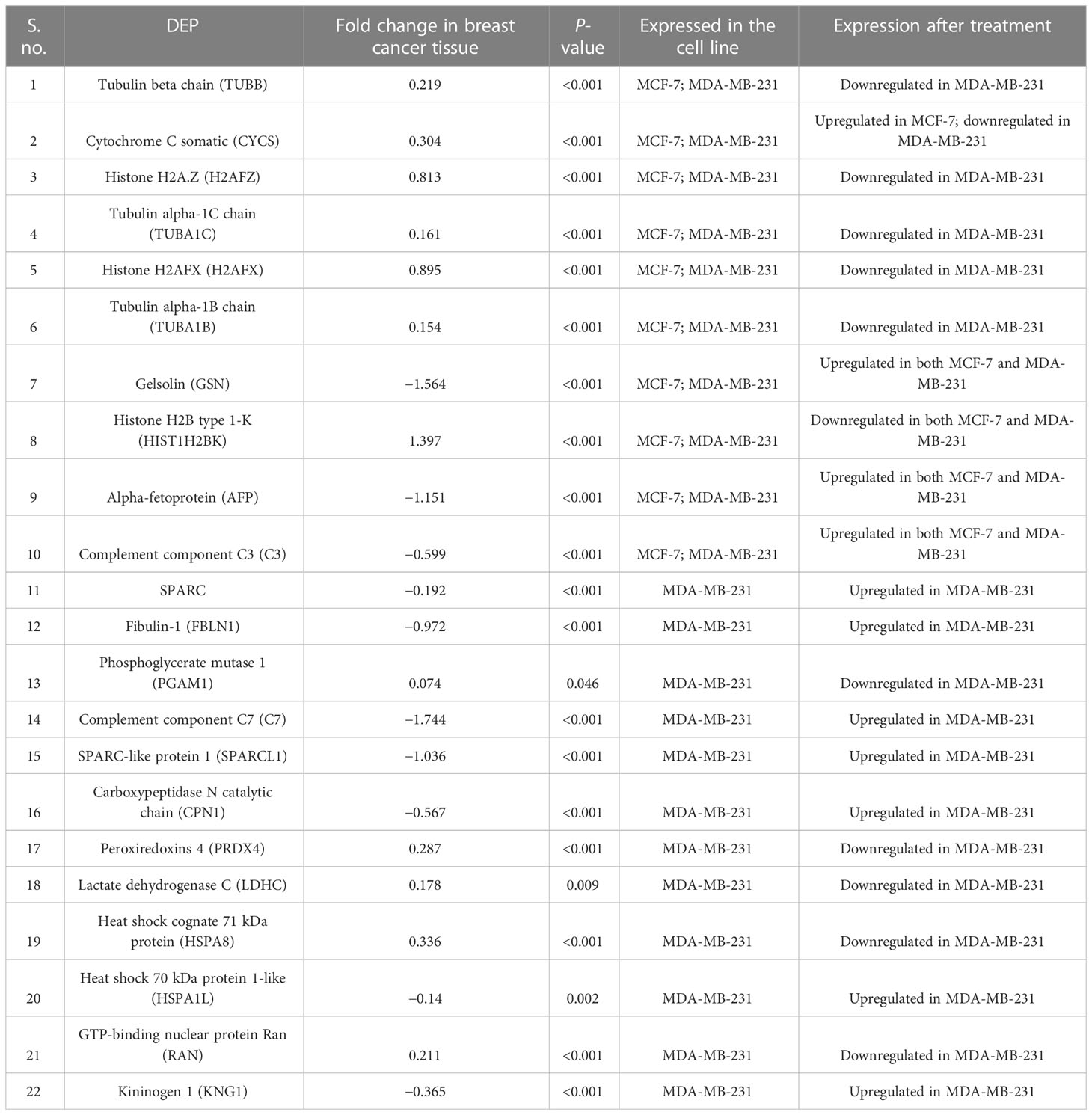
Table 3 List of 22 signature differentially expressed proteins (DEPs) in NVA-AA NP-treated MCF-7 and MDA-MB-231 cell lines obtained from the analysis by the GENT2 online database whose expression was found to be most altered.
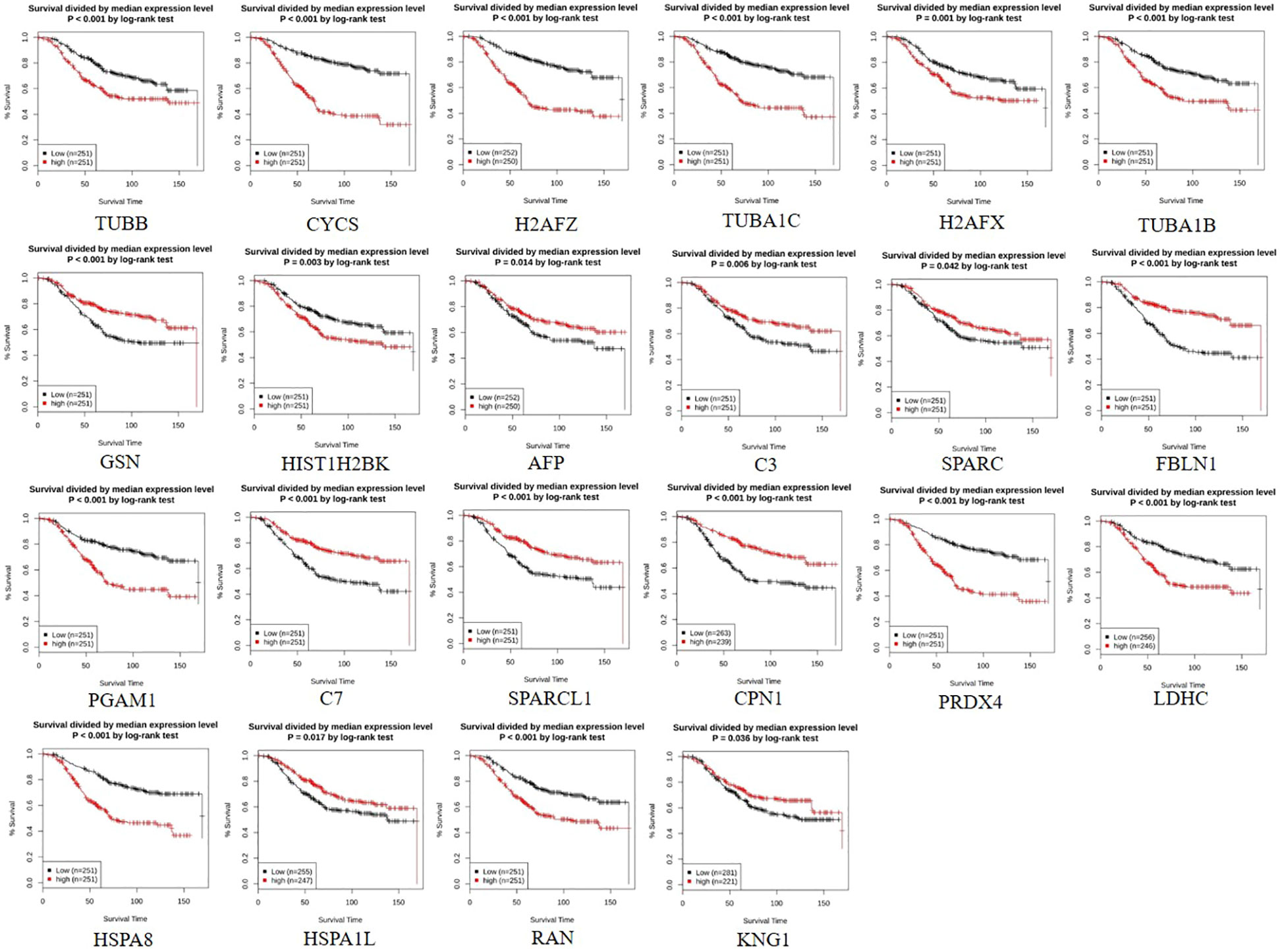
Figure 1 Survival curve of 22 signature differentially expressed proteins (DEPs) obtained from N-isopropyl acrylamide, N-vinyl pyrrolidone, and acrylic acid-based polymeric nanoparticle (NVA-AA NP)-treated MCF-7 and MDA-MB-231 cell lines showing their clinical significance and prognostic value in breast cancer as generated by GENT2 employed for overall survival (OS) analysis. The low expression of TUBB, CYCS, H2AFZ, TUBA1C, H2AFX, TUBA1B, HIST1H2BK, PGAM1, PRDX4, LDHC, HSPA8, and RAN, which are considered favorable for prognosis in breast cancer patients, coincides with their downregulation in NVA-AA NP-treated cell lines indicating a favorable outcome of NVA-AA NP treatment. Similarly, the high expression of GSN, AFP, C3, SPARC, FBLN1, C7, SPARCL1, CPN1, HSPA1L, and KNG1 correlates with their upregulation in NVA-AA NP-treated cell lines and justifies the favorable outcome of the treatment.
3.2.2 Correlation analysis between mRNA of signature DEP and drug sensitivity by GSCA
Bubble plots were obtained from GSCA showing the correlation between mRNA of selected signature DEPs and drug response as shown in Figure 2. Inputted gene having an association with at least one drug and vice versa was obtained. Bubble size is positively correlated with the FDR significance, where the blue bubble indicates a negative correlation, the red bubble indicates a positive correlation, and the darker color indicates a higher correlation. As per the analysis, out of the 22 signature DEPs, 17 were found to be correlated with 6 GDSC drugs (Figure 2A). A CDK7 inhibitor, THZ-2-102-1, was found to be highly positively correlated with the expression of GSN (FDR <= 0.0001) and C3 (FDR = 0.001); moderately positively correlated with FBLN1, TUBA1C, and SPARC (FDR = 0.01); least positively correlated with KNG1, AFP, and PRDX4 (FDR = 0.05); and negatively correlated with RAN (FDR = 0.01). YM155 which is a survivin suppressant showed a positive correlation with KNG1 and AFP (FDR = 0.01). A JNK1 inhibitor, ZG-10, had a positive correlation with AFP (FDR = 0.05) and a negative correlation with TUBB and TUBA1B (FDR = 0.05). WH-4-023, an Lck and Src inhibitor, was positively correlated with RAN (FDR = 0.05) while negatively correlated with AFP, SPARC, GSN, and C3 (FDR = 0.05). A membrane permeability inhibitor of PI3K p110β subunit, TGX221, was highly positively correlated with RAN (FDR = 0.001) and least positively correlated with HSPA8 (FDR = 0.05), while it was highly negatively correlated with SPARC (FDR <= 0.0001), moderately negatively correlated with AFP, GSN, and C3 (FDR = 0.001), and least negatively correlated with PRDX4 and CPN1 (FDR = 0.05). Pazopanib, which is a tyrosine kinase inhibitor, displayed a moderate negative correlation with SPARC (FDR = 0.001) and a low negative correlation with GSN, AFP, FBLN1, and KNG1 (FDR = 0.05).
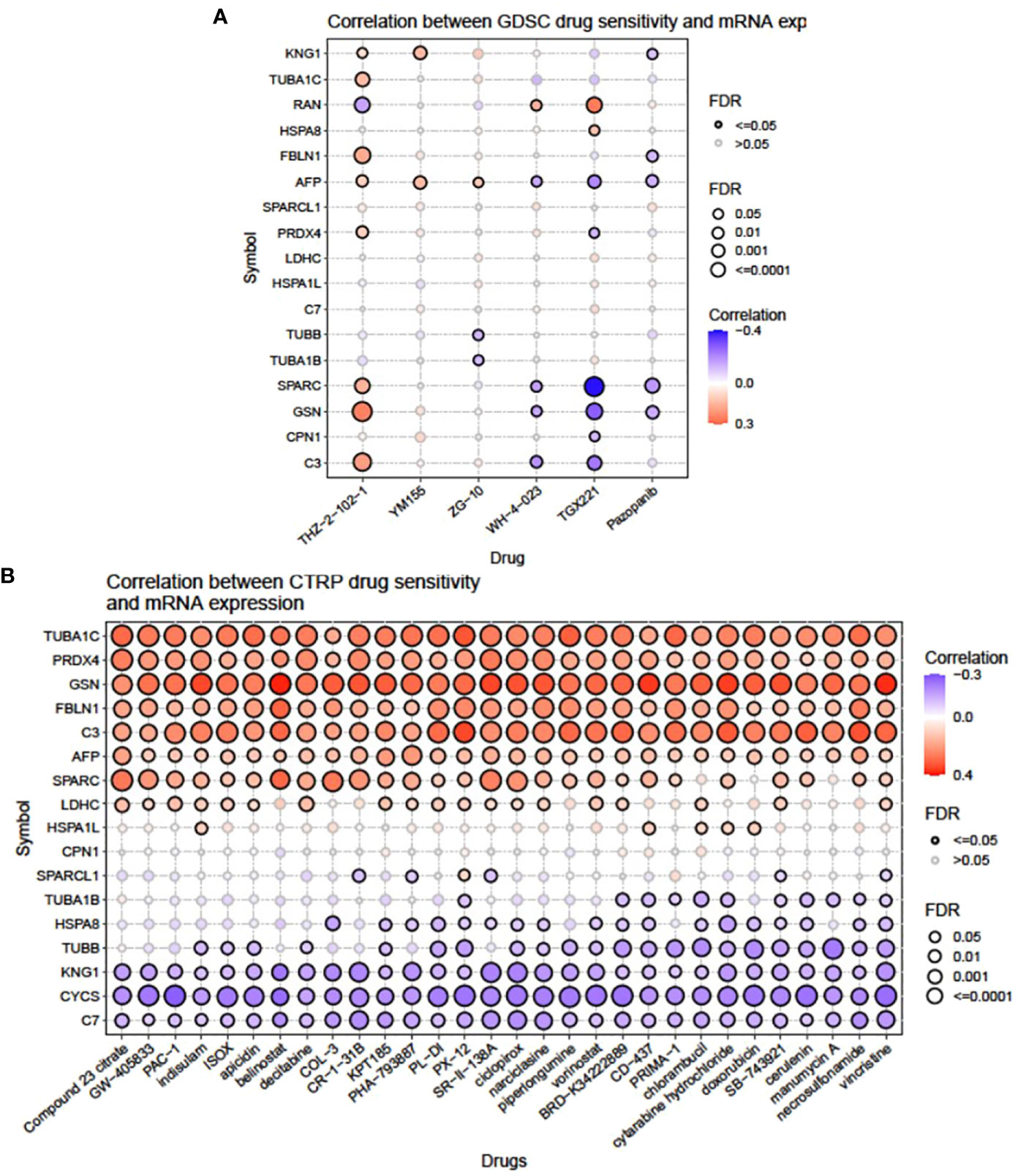
Figure 2 Illustration showing the correlation between mRNA expression of individual signature genes and drug sensitivity from the (A) GDSC and (B) CTRP portal.
Thirty CTRP drugs acting as inhibitors of different signaling pathways, namely, compound 23 citrate, GW-405833, PAC-1, indisulam, ISOX, apicidin, belinostat, decitabine, COL-3, CR-1-31B, KPT185, PHA-793887, PL-DI, PX-12, SR-II-138A, ciclopirox, narciclasine, piperlongumine, vorinostat, BRD-K34222889, CD-437, PRIMA-1, chlorambucil, cytarabine hydrochloride, doxorubicin, SB-743921, cerulenin, manumycin A, necrosulfonamide, and vincristine, showed a variable degree of correlation with the mRNA expression of selected signature DEPs (Figure 2B). Out of these, TUBA1C, PRDX4, GSN, FBLN1, C3, and AFP exhibited the highest range of positive correlation (FDR <= 0.0001; 0.001) with all the 30 drugs. SPARC, LDHC, and HSPA1L showed moderate to low levels of positive correlation (FDR = 0.01; 0.05) with many drugs. However, KNG1, CYCS, and C7 had the highest negative correlation (FDR <= 0.0001; 0.001) with all the 30 drugs. TUBB, HSPA8, and TUBA1B showed moderate to low levels of negative correlation (FDR = 0.01; 0.05) with many drugs.
3.2.3 Correlation analysis between mRNA expression of signature DEP and immune cell infiltrates by GSCA
A bubble plot was generated indicating a correlation between mRNA expression of signature DEPs and 24 immune cell infiltrates (Figure 3A), where C3, TUBB, PRDX4, TUBA1B, RAN, PGAM1, HSPA8, and TUBA1C presented the highest significant positive correlation with infiltration score (FDR <= 0.0001), whereas KNG1, FBLN1, CYCS, GSN, and SPARC had moderate to low positive correlation with infiltration score (FDR = 0.001; 0.01). AFP displayed the highest negative correlation with infiltration score (FDR <= 0.0001), and SPARCL1 and HSPA1L showed moderate and low negative correlation (FDR=0.001; 0.01). Upregulated expression of KNG1 showed a positive correlation with infiltrates of cytotoxic, CD8_T, Exhausted, macrophage, Th1, iTreg, B cell, Tr1, central_memory, NK, and Tfh and a negative correlation with NKT, MAIT, Th17, and neutrophil cell types. The FBLN1-altered expression had the highest positive correlation with cytotoxic, NK, Tfh, CD4_T, Gamma_delta, and NKT and negative correlation with effector_memory, nTreg, and neutrophils. Enhanced expression of C3 in both cell lines correlated positively with cytotoxic, CD8_T, Exhausted, Th1, iTreg, B cell, Th2, NK, Tfh, CD4_T, Gamma_delta, and NKT and negatively with monocyte, CD8_naive, Th17, neutrophil, and CD4_naive. TUBB had a positive correlation with cytotoxic, CD8_T, Exhausted, macrophage, Th1, iTreg, B cell, Tr1, monocyte, DC, effector_memory, and nTreg and a negative correlation with CD4_T, CD4_naive, Gamma_delta, NKT, MAIT, CD8_naive, Th17, and neutrophil. SPARC correlated positively with macrophage, Tr1, monocyte, NKT, and CD8_naive and negatively with B cell and neutrophil. Similarly, a change in the expression of PRDX4, SPARCL1, AFP, GSN, TUBA1C, CYCS, HSPA8, PGAM1, RAN, TUBA1B, C7, and HSPA1L also showed a positive and negative correlation with the maximum number of different infiltrate cell types with a variable degree of FDR.
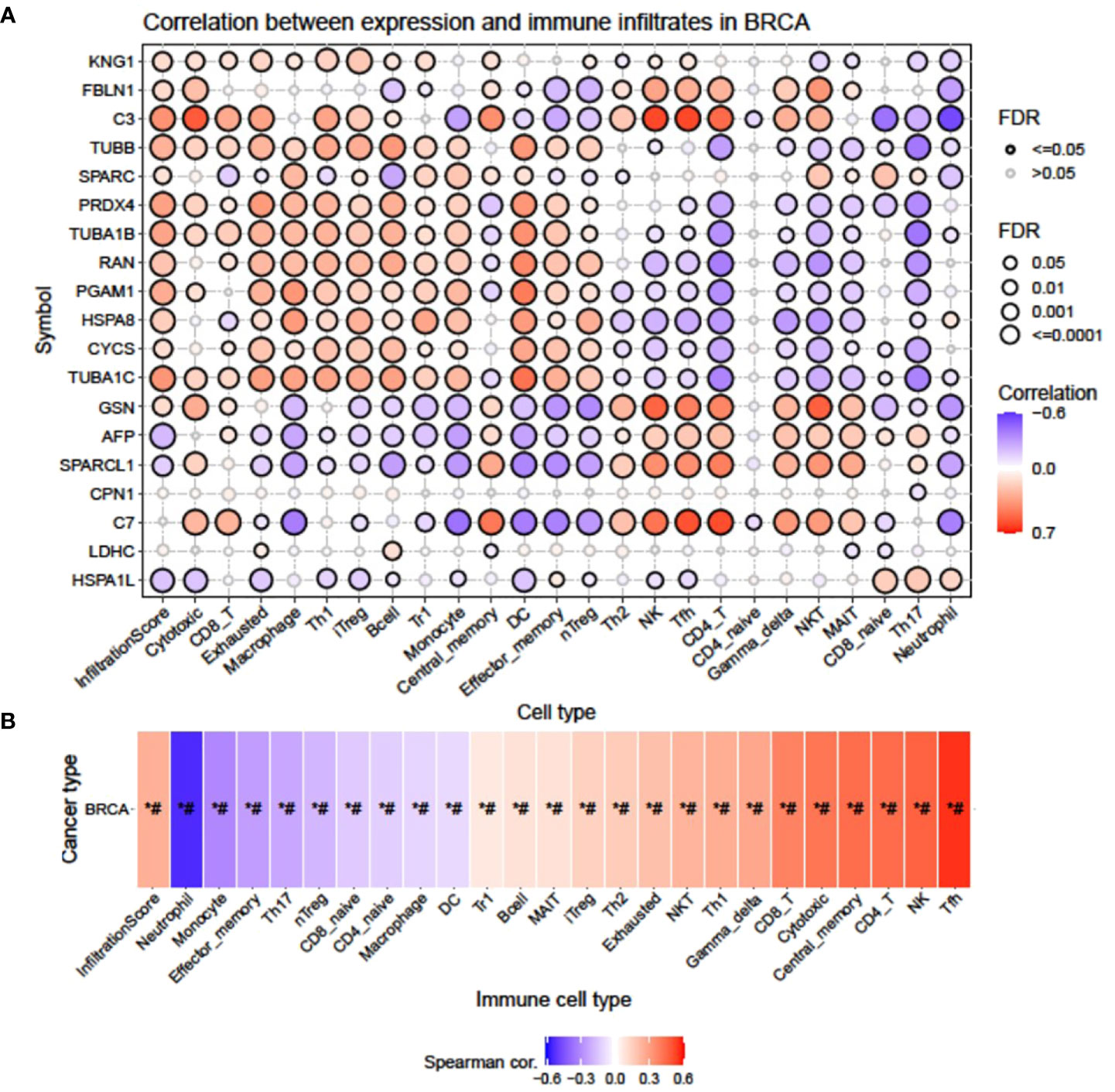
Figure 3 Correlation between mRNA expression of signature DEPs and 24 immune cell infiltrates by GSCA. (A) Bubble plot showing the correlation of individual genes with different immune cell infiltrates and (B) heatmap showing the cumulative correlation of a set of genes with individual immune cell infiltrates.
The heatmap (Figure 3B) obtained from the Spearman correlation analysis showed the association of expression of our signature gene set with 24 immune cell infiltrates on the horizontal axis. P-value was adjusted by FDR. Red and blue colors indicate positive and negative correlations, respectively, with the darker color representing the stronger correlation (*: P-value < 0.05; #: FDR < 0.05). It was observed that the expression pattern of our signature genes was positively correlated (P-value ≤ 0.05; #: FDR ≤ 0.05) with Tfh, NK, CD4_T, central_memory, cytotoxic, CD8_T, Gamma_delta, NKT, Th1, Exhausted, Th2, iTreg, MAIT, B cell, and Tr1 and negatively correlated (P-value ≤ 0.05; #: FDR ≤ 0.05) with neutrophil, monocyte, effector_memory, Th17, nTreg, CD8_naive, CD4_naive, macrophage, and DC.
3.2.4 Gene annotation and KEGG analysis by GeneCodis
All the signature DEPs selected in NVA-AA-treated MCF-7 and MDA-MB-231 were categorized based on KEGG molecular pathways, PharmGKB, DisGeNET, cellular component, and Reactome modules. The results of this analysis are shown in Figure 4. KEGG enrichment analysis illustrated that these genes were associated with the prion disease (C7, CYCS, HSPA1L, TUBA1B, TUBB), legionellosis (C3, CYCS, HSPA1L, HSPA8), systemic lupus erythematosus (C3, C7, H2AFX, H2AFZ HIST1H2BK), phagosome (C3, TUBA1B, TUBA1C, TUBB), gap junction (C3, TUBA1B, TUBA1C, TUBB), complement and coagulation cascades (KNG1, C3, C7), and neutrophil extracellular trap formation (C3, H2AFX, H2AFZ, HIST1H2BK). PharmGKB analysis revealed an association of these genes with responses toward different drugs, namely, hormonal contraceptives for systemic use (KNG1), eculizumab (C3), abacavir (HSPA1L), antineoplastic agents (SPARC), carbamazepine (HSPA1L), ranibizumab (C3), leucovorin (SPARC), bevacizumab (C3), capecitabine (SPARC), and docetaxel (PRDX4). The DisGeNET module showed the relationship of different signature genes with other human diseases like acute coronary syndrome (HSPA8, GSN, TUBA1C); transient ischemic attack (CYCS, C3); crescendo transient ischemic attack (CYCS, C3); paratuberculosis (C3, GSN); brain stem ischemia, transient (CYCS, C3); carotid circulation transient ischemic attack (CYCS, C3); posterior circulation transient ischemic attack (CYCS, C3), transient ischemic attack; vertebrobasilar circulation (CYCS, C3), transient ischemic attack; anterior circulation (CYCS, C3); and transient cerebral ischemia (CYCS, C3). Reactome analysis by GeneCodis revealed the association of the signature gene with other biological pathways like M phase (H2AFX, H2AFZ, HIST1H2BK, RAN, TUBA1B, TUBA1C, TUBB); cellular responses to stress (CYCS, H2AFX, H2AFZ, HIST1H2BK, HSPA1L, HSPA8, TUBA1B, TUBA1C); cellular responses to stimuli (CYCS, H2AFX, H2AFZ, HIST1H2BK, HSPA1L, HSPA8, TUBA1B, TUBA1C), cell cycle; mitotic (H2AFX, H2AFZ, HIST1H2BK, RAN, TUBA1B, TUBA1C, TUBB); HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand (HSPA1L, HSPA8, TUBA1B, TUBA1C); post-translational protein phosphorylation (AFP, C3, KNG1, SPARCL1); transcriptional regulation by small RNAs (H2AFX, H2AFZ, HIST1H2BK, RAN); and regulation of complement cascade (CPN1, C3, C7). Gene annotation for cellular components related to these signature genes by GeneCodis showed that C3, C7, FBLN1, GSN, H2AFX, H2AFZ, HSPA8, KNG1, LDHC, PGAM1, PRDX4, RAN, and TUBB belong to the extracellular exosome, whereas AFP, C3, C7, CPN1, FBLN1, GSN, HSPA8, KNG1, PGAM1, PRDX4, SPARC, SPARCL1, and TUBB were part of the extracellular region. Similarly, AFP, C3, CPN1, FBLN1, GSN, HIST1H2BK, HSPA8, KNG1, SPARC, and SPARCL1 belong to components of the extracellular space, and C3, GSN, HSPA8, PGAM1, and PRDX4 were found to be components of secretory granule lumen.
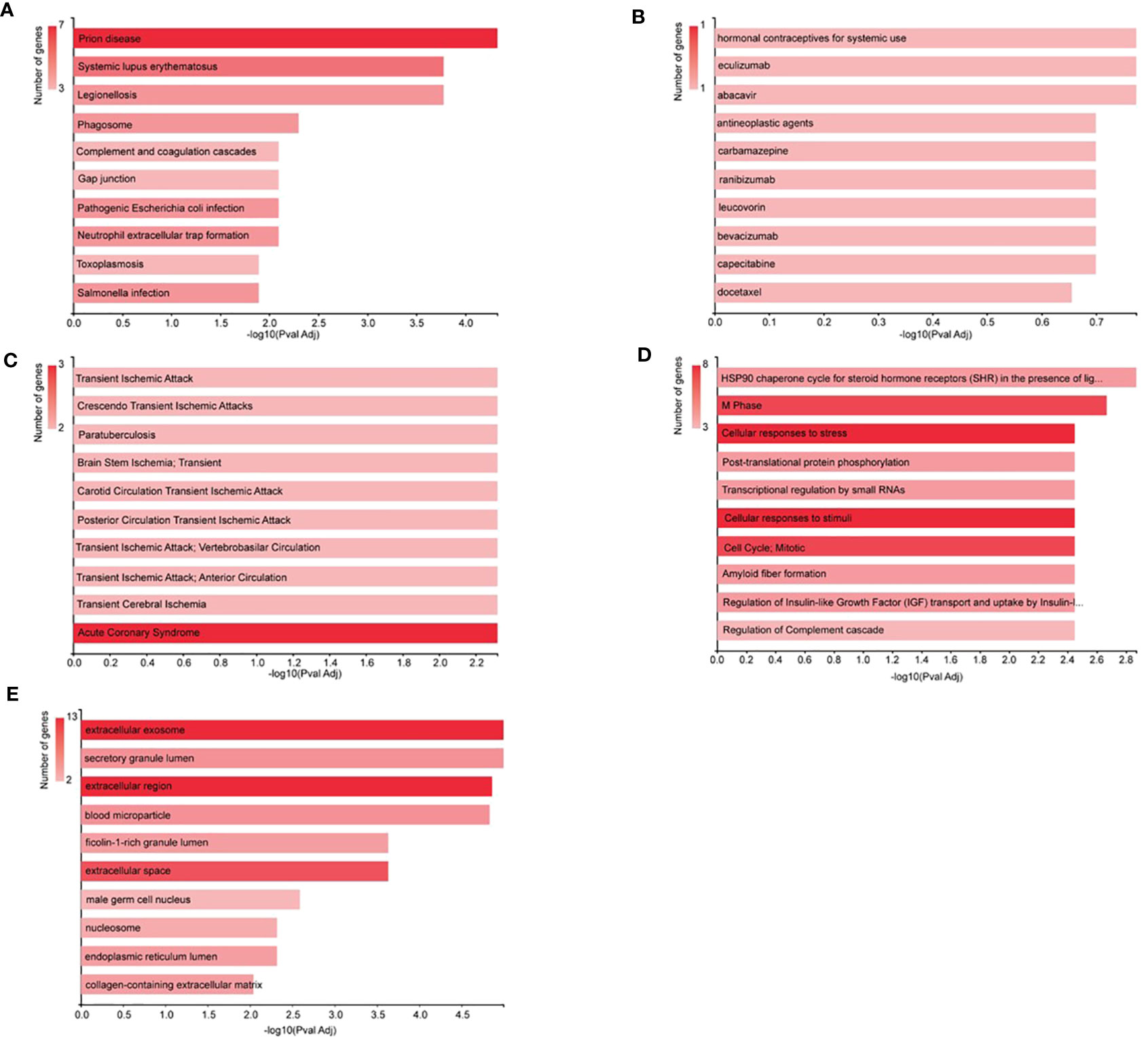
Figure 4 Analysis by GeneCodis. All the signature DEPs from NVA-AA-treated MCF-7 and MDA-MB-231 were categorized by (A) KEGG molecular pathways, (B) PharmGKB, (C) DisGeNET, (D) cellular component, and (E) Reactome modules.
3.2.5 Pathway analysis by Cytoscape
Protein–protein interaction of signature DEPs was generated by the STRING module of Cytoscape software as shown in Figure 5. Out of 22 signature DEPs, 18 showed interaction with each other. Interacting proteins belong to different biological processes including regulation of complement cascade (CPN1, C3, C7), cell cycle (H2AFX, H2AFZ, HIST1H2BK, RAN, TUBA1B, TUBA1C, TUBB), cellular responses to stimuli (CYCS, H2AFX, H2AFZ, HIST1H2BK, HSPA1L, HSPA8, TUBA1B, TUBA1C), innate immunity (C3, C7, CPN1, GSN, HSPA8, PGAM1, PRDX4, TUBB), and neutrophil degranulation (C3, GSN, HSPA8, PGAM1, PRDX4, TUBB).
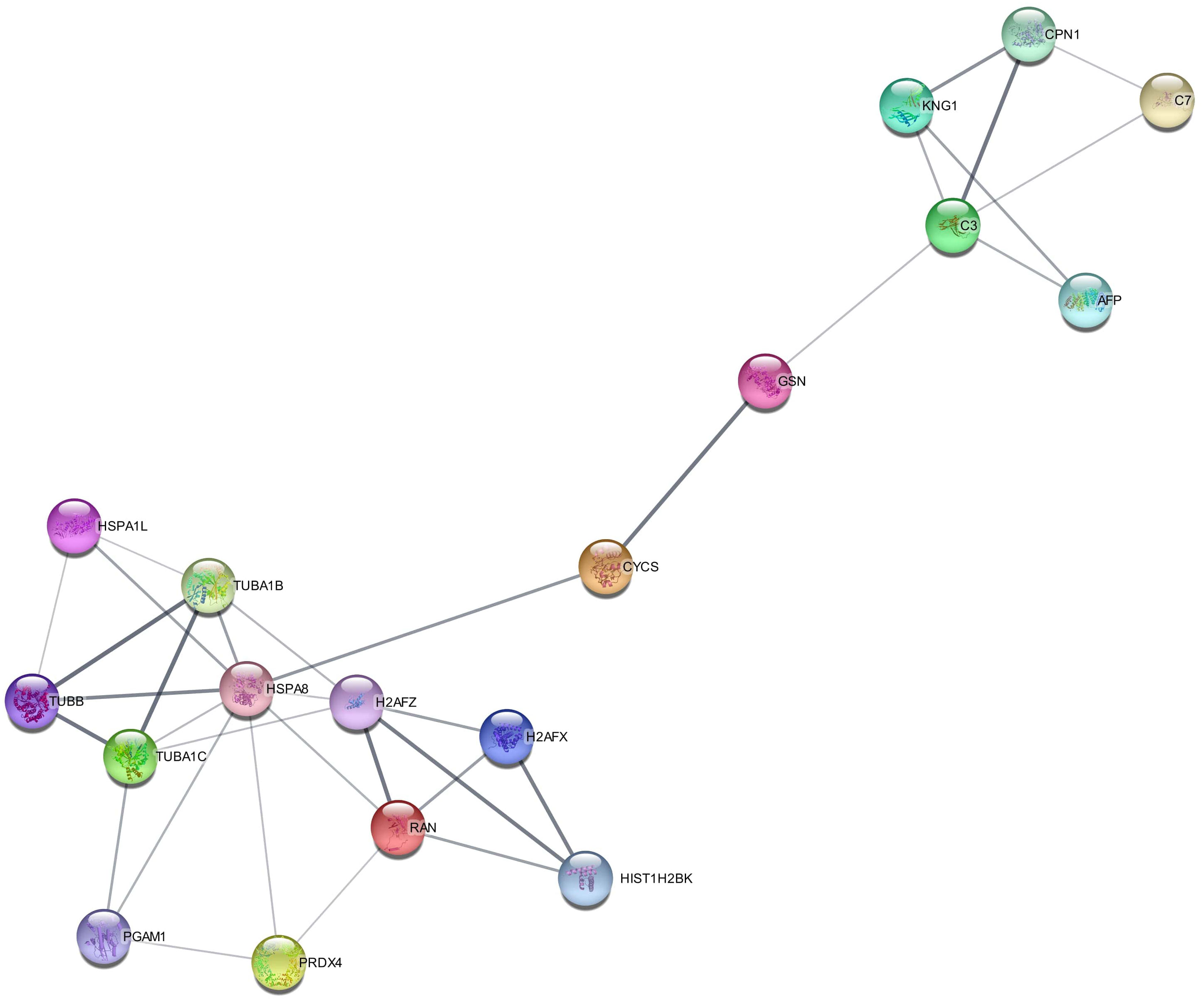
Figure 5 Protein–protein interaction (PPI) of signature differentially expressed proteins (DEPs) in the secretome of Artemisia absinthium whole-plant ethanolic extract-loaded NIPAAM-VP-AA (NVA-AA) nanoparticle-treated MCF-7 and MDA-MB-231 cell lines. This interactome was generated by the STRING module of Cytoscape software in which 18 out of 22 DEPs showed interaction with each other.
3.2.6 Functional annotation analysis by PANTHER
All the significant DEPs identified in NVA-treated MCF-7 and MDA-MB-231 were categorized based on their protein class, biological role, molecular functions, cellular component, and pathways involved. The results of this analysis are shown in Supplementary Figure S4. The majority of the proteins in MCF-7 and MDA-MB-231 are clustered as DNA binding, ATP-dependent activity, and functions corresponding to catalytic and regulatory activity. The biological classification of these proteins identified in MCF-7 and MDA-MB-231 revealed that these are majorly involved in cellular and metabolic processes as well as localization, signaling, biological adhesion, and immune system processes. In MCF-7 and MDA-MB-231, a large portion of proteins belong to the class of proteins that participate in chromatin binding or regulatory protein, while other proteins were cell adhesion molecules, chaperones, cytoskeleton proteins, transcriptional regulators, protein-binding activity modulators, and carrier proteins. A few additional proteins in MDA-MB-231 were categorized under the class of proteins responsible for RNA metabolism, extracellular matrix proteins, immunity system, and intercellular signal molecule. Pathway analysis in both cell lines shows that most of the proteins were involved in ATP synthesis, apoptosis signaling pathway, p53 pathway, blood coagulation, cadherin signaling, FAS signaling pathway, inflammation mediated by chemokine, cytokine, vitamin D metabolism, and Wnt signaling pathway. However, few proteins involved in EGF-receptor signaling, glycolysis, plasminogen activating cascade, and PI3 kinase pathway were additionally found in MDA-MB-231.
3.3 Real-time PCR
The gene expression analysis of GSN, C3, and CYCS in treated and untreated cell lines was conducted using quantitative real-time PCR (Figure 6). There was a significant upregulation of the GSN (P < 0.01, P < 0.001) in both NVA-AA NP-treated MCF-7 and MDA-MB-231 cell lines, indicating inhibition of cell cycle regulation; however, C3 (P < 0.01) showed a significant upregulation in NVA-AA NP-treated MCF-7 and a non-significant upregulation in NVA-AA NP-treated MDA-MB-231 cell lines, suggesting stimulation of innate immune response by NVA-AA NPs in MCF-7 and MDA-MB-231 cell lines. Moreover, CYCS was upregulated in treated MCF-7 and significantly downregulated (P < 0.05) in treated MDA-MB-231 suggesting induction of caspase-dependent and caspase-independent cell death by NVA-AA NPs.
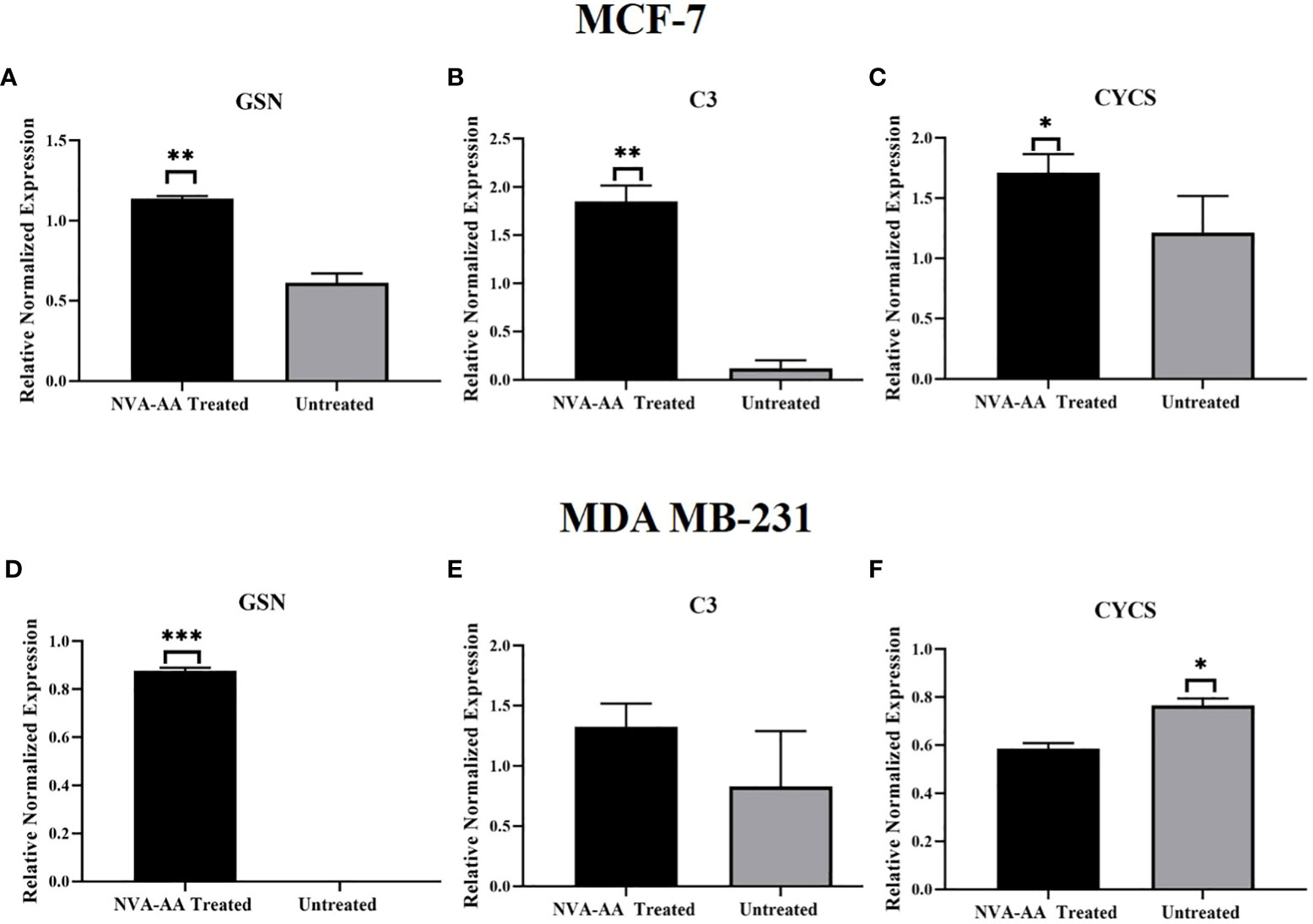
Figure 6 Quantitative real-time PCR analysis showing the relative expression of the mRNA transcripts of signature differentially expressed proteins (DEPs) belonging to cell cycle regulation (A, D) GSN, innate immune response (B, E) C3, and apoptosis (C, F) CYCS in MCF-7 and MDA-MB-231. The values are expressed as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001 when NVA-AA-treated cell lines were compared with untreated cell lines using unpaired t-test).
4 Discussion
Temperature- and pH-responsive polymeric nanoparticles were previously utilized by our research group for site-specific drug delivery targeted against the tumor microenvironment comprised of low pH and high temperature accompanied by inflammation. We have reported the cytotoxic and anticancer potential of A. absinthium whole-plant ethanolic extract-loaded NIPAAM-VP-AA (NVA-AA) against the breast cancer cell lines MCF-7 and MDA-MB-231. NVA-AA NPs significantly inhibited cell proliferation and cell viability by inducing apoptosis in the treated cell lines (19). In the present study, we investigated differentially expressed secretory proteins in the media of NVA-AA NP-treated MCF-7 and MDA-MB-231 cell lines by employing nano LC-MS/MS to identify key pathways and easily accessible potential therapeutic protein targets affected by the cytotoxic action of NVA-AA NPs targeted against the tumor microenvironment. Our study revealed an altered secretome profile of A. absinthium whole-plant extract-loaded NVA-AA polymeric nanoparticle-treated MCF-7 and MDA-MB-231 cell lines. Several proteins involved in different biological functions and pathways were found to be significantly altered. Twenty-two signature DEPs were selected out of the total proteins which were most significantly altered due to the treatment. The selection of proteins was done by utilizing TCGA datasets available at GENT2 and GSCA.
Protein–protein interaction among 22 signature secretory proteins demonstrated that most affected signaling pathways accompanied by apoptosis induction and cell death by NVA-AA NPs in MCF-7 and MDA-MB-231 involved the regulation of complement cascade [carboxypeptidase N catalytic chain, complement components (C3 and C7)], modulation of the cell cycle [histone proteins (H2AFX, H2AFZ, HIST1H2BK), GTP-binding nuclear protein (RAN), tubulin (TUBA1B, TUBA1C, and TUBB)], cellular responses to stimuli [cytochrome c, histone (H2AFX, H2AFZ, HIST1H2BK), heat shock proteins (HSPA1L, HSPA8), tubulins (TUBA1B, TUBA1C)], triggering of innate immune response [complement components (C3, C7), CPN1, gelsolin, heat shock proteins (HSPA8), phosphoglycerate mutase 1, perioxidin-4, tubulin (TUBB)], and neutrophil degranulation [complement C3, gelsolin, heat shock proteins (HSPA8), phosphoglycerate mutase 1, perioxidin-4, tubulin (TUBB)].
Gelsolin is an actin-binding protein that causes conformational changes in actin monomers, facilitating filament polymerization. It also regulates the length of actin filament and other cellular functions including cell motility, division, and apoptosis by participating in different signaling pathways. It is a known tumor-suppressor gene that is downregulated in breast cancer due to epigenetic changes including histone acetylation during chromatin structural configuration (29). Previously, overexpression of gelsolin using recombinant adenovirus encoding wild-type gelsolin (Ad-GSN) arrested cell cycle at the G2/M phase, reduced cell division in bladder cancer cells (KU-7 and UMUC-2), and inhibited tumor growth in the orthotopic bladder cancer nude mouse model (30). Gelsolin reduced tumorigenicity in a human lung cancer cell line (PC10) via inhibiting phospholipases C (PLC)/protein kinase C (PKC) signal transduction pathway (31). Similarly, highly expressed gelsolin significantly inhibited invasion and metastasis in human colon carcinoma (CC) cells in vitro (32). Thus, these findings were similar to our results, i.e., upregulation of gelsolin due to NVA-AA treatment indicates that these NPs could cause cell death by impeding microtubule organization and cell division, irrespective of the breast cancer subtype. Similarly, cytochrome c somatic (CYCS) is an apoptotic biomarker whose expression was different in both treated cell lines, i.e., elevated in MCF-7 and reduced in MDA-MB-231. ROS-mediated programmed cell death is associated with the release of pro-apoptotic proteins like cytochrome c into the cytosol, which in turn, activates the caspase-dependent cascade of apoptosis (33). However, there are increasing studies demonstrating that different cytotoxic drugs are capable of inducing caspase-independent programmed cell death as a defense mechanism against external stimuli or stress (34). Apart from cytochrome c, apoptosis-inducing factor (AIF) is another protein with oxidoreductase activity and caspase-independent apoptogenic function, in which high ROS level triggers activation of PARP, translocation of AIF into the nucleus which in turn causes chromatin condensation, DNA fragmentation, and subsequent cell death (35). Increased level of CYCS in the extracellular space of treated MCF-7 in our study indicates the release of cytochrome c from the mitochondrial membrane associated with the induction of apoptosis by NVA-AA NPs. However, a reduced level of CYCS in treated MDA-MB-231 with significant cell death proposes the presence of cytotoxic drug-mediated caspase-independent cell death involving perturbation in ROS stability causing depolarization of mitochondrial membrane potential, the release of AIF, and finally cell death (36). Therefore, the anticancer effect of NVA-AA NPs causes differential apoptotic gene expression changes in MCF-7 and MDA-MB-231, due to differences in cell genotype.
Alpha-fetoprotein (AFP) is a glycoprotein that functions as a carrier protein for various ligands as well as the regulator of cellular growth of different cell types, which was overexpressed in NVA-AA NP-treated MCF-7 as well as MDA-MB-231 cell lines in our study. According to a study by Sierralta et al., the addition of an active site of AFP consisting of cyclized 9-amino acid peptide (cP) to MCF-7 breast cancer cells and estrogen-dependent ZR75-1 cells inhibited cell proliferation (37). Therefore, the upregulation of AFP along with cell proliferation inhibition induced by NVA-AA NPs proposes the possibility of these NPs interfering with the cell cycle regulation.
Immune cells are the key players whose pathological implications vary with the heterogeneous pattern of gene expression in cancer initiation, progression, and response to cytotoxic drugs. Complement components are one of the crucial factors in cancer that specifically binds to cells at immune surveillance facilitating the removal of apoptotic cells by macrophages (38). On the contrary, complement components can also contribute to tumor growth by supporting angiogenesis and causing chronic inflammation (39). Among several components, a high level of C3 has been considered a diagnostic marker in breast and lung cancer (40). Previously, upregulation of C3 due to docetaxel/epirubicin-based chemotherapy suggested it as a good predictive marker of therapeutic response in breast cancer (41). Thus, complement pathway-mediated regulation of tumor growth varies among different tumor types. Monteran et al. reported increased expression of C3 and C7 in lung fibroblasts of doxorubicin-treated mice and proposed their involvement in the formation of the immunosuppressive tumor microenvironment (42). In the present study, increased levels of two isoforms of complement components, i.e., complement component C3 (in MCF-7 and MDA-MB-231) and C7 (MDA-MB-231) by NVA-AA NPs, were seen in both cell lines indicating activation of the complement pathway via triggering the innate immune system. Thus, our data suggest that cytotoxicity caused by NVA-AA NPs elicited activation of complement cascade due to induced apoptosis.
Tubulins are cytoskeletal proteins whose alpha and beta subunits heterodimerize for the microtubule assembly required for cell division. Agents affecting tubulin dynamics have been utilized as an ideal approach for chemotherapeutics (43, 44). NVA-AA NPs resulted in reduced expression of signature DEPs from the tubulin family including TUBB, TUBA1B, and TUBA1C, confirming its ability to hinder the formation of microtubules by binding to tubulins, eventually causing cell death through apoptosis. Heat shock protein HSP 90-beta (HSP90AB1) was the most significantly downregulated secretory protein in MDA-MB-231 after the NVA-AA NP treatment. Therefore, our results were consistent with the study by Rasouliha et al. that showed the downregulation of tubulin genes and HSP90AB1 due to doxorubicin treatment in MCF-7 cells (45). Therefore, this decrease in tubulin levels would have reduced the rate of cell division in MDA-MB-231. On the contrary, a few tubulin genes (TUBB, TUBA1B) were overexpressed in NVA-AA NP-treated MCF-7 cell lines. Cytoskeletal-interacting agents can either stabilize or destabilize the microtubule organization, i.e., enhanced tubulin expression represents an increase in tubulin monomer required for polymerization due to its depletion, whereas reduced tubulin expression demonstrates the increase in the tubulin monomer due to inhibition of polymerization (46). Thus, we anticipate that these differences between both cell lines could be due to the difference in the nature of interaction of these NPs with different breast cancer subtypes.
Histone variants including H2AFZ and H2AFX were downregulated in MDA-MB-231, whereas HIST1H2BK was downregulated in both MCF-7 and MDA-MB-231 cell lines after NVA-AA NP treatment. Histones are critical drivers of DNA packaging into the nucleosome which is differentially regulated throughout the cell cycle. H2AFZ is an oncogene that was overexpressed in breast cancer patients with adverse clinical outcomes (47). Similarly, the H2AFX variant was also found to be associated with the initiation and progression of breast cancer (48). Previously, upregulation of HIST1H2BK in TNBC MDA-MB-231 was found to be correlated with chemoresistance induced by doxorubicin (49). HIST1H2BK was one of the most significantly downregulated differentially expressed histone proteins due to Canady Helios Cold Plasma™ (CHCP) treatment of breast cancer cells where CHCP mainly degrades histone proteins during the early S phase of the cell cycle (50). Therefore, apparent cell death associated with histone regulation by NVA-AA NPs in our study determines its ability to cause chromatin instability and aberrant cell cycle regulation, subsequently affecting cell survival.
Heat shock proteins (HSPs) are stress-responsive molecules that are required for the stabilization of oncogenes, their transcriptional regulation, protein folding, and cell cycle maintenance (51). Two signature HSPs were altered by NVA-AA NPs in MDA-MB-231, where the expression of heat shock cognate 71 kDa protein (HSPA8) was reduced and heat shock 70 kDa protein 1-like (HSPA1L) was elevated. Sporadic breast cancer is found in association with a deletion mutation in HSPA8, located on chromosome 11q23.3 (52). Overexpressed HSPA8 was also considered a poor prognosis marker of breast cancer (53). HSPA1L was one of the downregulated genes after gemcitabine treatment used against breast cancer (54). In our study, NVA-AA NPs stimulated altered expression of these HSPs as a consequence of modifications in gene transcriptional regulation.
Phosphoglycerate mutase 1 (PGAM1) is a key metabolic enzyme in the glycolysis pathway whose overexpression is associated with breast cancer. However, Zhang et al. demonstrated the non-metabolic function of PGAM1, which is involved in modulating actin filament organization, cell motility, migration, and enhancing cancer progression (55). Previously, methotrexate (MTX), a chemotherapeutic drug, has shown downregulation of PGAM1 in the MCF-7 cell line (56). In the present study, PGAM1 was also significantly downregulated, which can be corroborated by the reduction in cell proliferation and viability in MDA-MB-231 caused by NVA-AA NPs.
GTP-binding nuclear protein RAN is a regulatory protein mainly involved in the exchange of molecules across the nucleus membrane by undergoing GTP/GDP-bound conformation changes. It also plays an important role during the cycle by controlling the assembly of the mitotic spindle, cell cycle checkpoint, and nuclear envelope formation. Upregulation of RAN has been found in many tumor types, including breast tumors, contributing to cancer invasion and poor prognosis (57). An in-vitro investigation by Sheng et al. showed inhibition of cell proliferation, motility, and cell cycle arrest in MDA-MB-231 cell lines by treatment with si-RNA directed against RAN (RAN-si-RNA) (58). Therefore, our findings with significant cell death accompanied by downregulation of RAN in NVA-AA NP-treated MDA-MB-231 implicated the potential of RAN to be considered as a therapeutic target.
Kininogen 1 (KNG1) is a crucial component of the blood coagulation system whose expression is lowered in a variety of cancers including breast, ovarian, and prostate. It is known as a metastasis inhibitor, capable of inhibiting angiogenesis, invasion, and migration of human prostate cancer (59). Downregulated KNG1 was also identified in metastatic tumor-draining lymph from metastatic mammary carcinoma through LC/MS proteomic analysis (60). To our knowledge, this is the first study reporting the upregulation of KNG1 in MDA-MB-231 after NVA-AA NP treatment, representing it as a promising therapeutic target for inhibiting cancer invasion and metastasis. Carboxypeptidase N catalytic chain (CPN1) exhibits peptidase activity by cleaving the substrate peptide, KNG1, at the carboxy-terminal arginine residue. A low level of CPN1 was observed in MDA-MB-231 and MCF-7 than in non-tumorigenic MCF-10A cell lines (61). CPN1 was considered a tumor biomarker for assessing invasion and metastasis status in breast cancer patients (62). Similarly, protein–protein interaction in our study also demonstrated a direct interaction between upregulated KNG1 and CPN1 supporting the existing data.
Fibulin-1 (FBLN1) is an ECM-associated glycoprotein, localized in the basement membrane and interacting with other components like laminin. FBLN1 has a critical role in epithelial-to-mesenchymal transition (EMT) during cancer due to its capability of regulating cell differentiation, adhesion, migration, and proliferation. High as well as aberrant expression of FBLN1 was assessed in breast carcinoma (63). However, the elevated expression level of FBLN1 in fibrosarcoma-derived cells has demonstrated inhibition of tumor formation in nude mice as well as invasion in the gels of reconstituted basement membrane extracts (64). Fibulin-1 has previously shown the ability to inhibit cell migration, invasion, and motility in melanoma, epidermoid carcinoma, and breast carcinoma cell lines through inhibition of ERK activation based on fibronectin-specific mechanisms (65). Silencing of fibulin-1 has shown an increment in cell proliferation in MCF-7 in vitro, and a low proliferation index was found in fibulin-1-expressing breast cancer tissue samples (66). Thus, the role of fibulin-1 in cancer progression may vary with cancer type. Therefore, our study corroborates with these studies as the overexpression of fibulin-1 in the MDA-MB-231 cell line was associated with the inhibition of cell proliferation and migration by destabilizing ECM structure and integrity.
Cellular damage caused by oxidative stress and redox status imbalance is closely associated with cancer cell proliferation and survival. Peroxiredoxins 4 (PRDX4) is one of the antioxidant enzymes specifically located in the endoplasmic reticulum (ER) and secreted in the extracellular space. It has been suggested that PRDX4 has a potential role in tumor initiation, progression, chemoresistance against docetaxel, and disease recurrence. High expression of PRDX4 has been reported in a variety of cancers including breast cancer; on the other hand, increased tissue level of PRDX4 was found to be correlated with a better survival rate in breast cancer patients (67). However, our study reporting downregulation of the antioxidant enzyme PRDX4 after treatment is in agreement with the OS analysis provided by TCGA datasets in GETN2 representing the association of low expression with better survival, thus helping us to postulate that oxidative stress burden was reduced due to the antioxidant potential of A. absinthium extract encapsulated in NVA NPs.
Secreted protein acidic and rich in cysteine (SPARC) binds with ECM components and growth factors mediating cell–ECM interaction, modulating different cellular processes, cell microenvironment, adhesion, and drug resistance (68). Contradictory regulation of SPARC has been documented in the initiation and progression of cancer, although the majority of existing literature reveals that downregulation of SPARC is closely associated with the aggressive phenotype of breast cancer progression (69, 70). In a study by Nagai et al., negative SPARC immunostaining of tumors was associated with poor prognosis in luminal A and triple-negative breast cancer patients (71). Previously, SPARC has been proposed as a tumor-suppressor protein that can induce apoptosis and inhibit angiogenesis by reducing VEGF expression (72, 73). Therefore, overexpression of SPARC by NVA-AA NPs supports the previous findings reporting the induction of apoptosis associated with high expression of SPARC in breast cancer. Similarly, secreted protein acidic and rich in cysteine-like protein 1 (SPARCL1) was another ECM-glycoprotein whose reduced expression was reported in human breast cancer tissues, and NVA-AA NP treatment caused an increase in the SPARCL1 expression (74). SPARCL1 is 62% identical to SPARC and contains conservative structural domains with functional similarity (75). It has also been considered a tumor suppressor as well as an oncogene (76, 77); thus, altered regulation of SPARCL1 after treatment can be useful in implicating it as a therapeutic target in breast cancer.
Lactate dehydrogenase C (LDHC) is an isoform of the lactate dehydrogenase (LDH) family whose elevated expression was observed in serum-derived exosomes as well as tissue of breast cancer patients and associated with adverse clinical outcomes, poor survival, and remission (78). It is also considered a tumor-associated antigen due to its immunogenic nature promoting cytotoxic immune response in breast cancer (79). In a study by Naik et al., LDHC silencing led to the abrupt progression of the cell cycle with substantial expression alteration in cell cycle checkpoint components, disassembly of nuclear and microtubule components, and DNA damage regulators in breast cancer (80). Downregulation of LDHC in our study along with cell cycle arrest at the G0/G1 phase corroborates with the observations of the previous study by Naik et al., further confirming LDHC to be used as a potential therapeutic target modulating DNA integrity.
Angiogenesis in cancer progression is highly regulated through interactions of ECM constituting the tumor microenvironment. In the present study, THBS1 was highly expressed in NVA-AA-treated MCF-7 cell lines, which is known as a potential inhibitor of angiogenesis (81). THBS1 is a glycoprotein that can strongly bind to ECM, affecting their structure and function via modulating direct as well as indirect interactions between other secretory factors (82, 83). THBS1 could inhibit angiogenesis by modifying the assembly of the actin cytoskeleton and focal adhesion of endothelial cells which would lead to the inhibition of cellular migration and invasion (84). A few genes belonging to the family of keratins (KRT1, KRT10, KRT2, KRT77) were downregulated in our study, which are filament proteins required for the maintenance of epithelial cells and have a role in cell motility, protein synthesis and regulation of signaling pathways.
Evaluation of the correlation between the mRNA expressions of 22 signature DEPs in MCF-7 and MDA-MB-231 cell lines versus the sensitivity of cancer cells toward small-molecule drugs of GDSC and CTRP was done. The dysregulated proteins in NVA-AA-treated MCF-7 and MDA-MB-231 cells, viz., KNG1, TUBA1C, FBLN1, AFP, PRDX4, SPARC, GSN, and C3, were found to have a positive correlation of their mRNA expressions with the GDSC drug THZ-2-102-1. Similarly, KNG1 and AFP with YM156, AFP with ZG-10, RAN with WH-4-023, and RAN and HSPA8 with TGX221 were found to be positively correlated. Since downregulation of TUBA1C and PRDX4 proteins was observed only in NVA-AA-treated MDA-MB-231 cells, their downregulated expressions might make MDA-MB-231 sensitive to NVA-AA treatment in the same fashion as shown by cancer cells to THZ-2-102-1 treatments. However, overexpression of GSN, C3, and AFP due to NVA-AA NPs in both cell lines (MCF-7 and MDA-MB-231) displayed negative correlations with WH-4-023, TGX221, and pazopanib. Thus, their overexpression might increase the sensitivity to NVA-AA treatment similar to WH-4-023, TGX211, and pazapanib treatments to other cancer cells. Moreover, the change in the expression of TUBA1C, PRDX4, LDHC, C7, and KNG1 due to NVA-AA treatment corroborated with the sensitivity provided by CTRP drugs to cancer cells (85). Thus, differential expression of proteins by NVA-AA NPs imparting sensitivity to the two cancer cells could be correlated to their sensitivities with the GDSC and CTRP drugs in different cancer cells.
Immune cell infiltration (Tfh, NK, CD4_T, CD8_T, NKT, B cell) showed positive correlations with the expressions of the dysregulated signature genes/proteins in NVA-AA-treated MCF-7 and MDA-MB-231 cell lines. These immune cells were reported to be involved in antitumor immune responses and also considered important targets in immunotherapy (86–90). However, immune cells, viz., neutrophil, Th17, nTreg, and macrophage, showed negative correlations of their infiltrations with the expressions of the dysregulated signature genes/proteins in both NVA-AA-treated cell lines. These immune cells were mainly found to be involved in angiogenesis, cancer promotion, metastasis, poor prognosis, and relapse of tumor cells, which are the hallmarks of cancer manifestations and progression (91–95).
The limitations of our in-vitro study include further experimental validation at the in-vivo level due to variation in the tumor microenvironment, the presence of certain biological barriers at the cell line and organism level, and bioinformatics analysis findings, which may further be comprehended through in-vitro and in-vivo experiments. Remarkably, validating and establishing A. absinthium extract-loaded NVA-AA NPs as the drug carrier ensuring site-specific drug delivery would increase the therapeutic effectiveness and lessen side effects related to the existing anticancer payload associated with currently available treatments. However, the clinical application of NVA-AA NPs against breast cancer requires more vigorous preclinical studies to ensure its efficacy and safety at an individual level. Also, it will be interesting to investigate the effect of these NPs on other cancer types as well.
This study has a fundamentally exploratory aim which was focused on the secretome profiling to unravel the altered processes and pathways in MCF-7 and MDA-MB-231 breast cancer cell lines caused due to the cytotoxicity of A. absinthium whole-plant extract-loaded NVA-AA polymeric nanoparticles. We can conclude that cytotoxicity by NVA-AA NPs was mainly associated with their capacity to interfere with the cytoskeletal dynamics, cell cycle regulators, cell–cell interaction, intracellular trafficking, cell polarization, and migration of cancer cells, irrespective of the breast cancer subtype.
Data availability statement
The datasets presented in this study can be found in online repository named “PRIDE database”, with accession number “PXD042365”.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
SK did the investigation, data curation, formal analysis, methodology, visualization, validation, writing—original draft, and writing—review and editing. MM was involved in data curation, methodology, validation, and writing—review and editing. IM did the investigation, visualization, and writing—review and editing. SS was involved in the methodology and writing—review and editing. SW did the conceptualization, methodology, supervision, project administration, funding acquisition, validation, visualization, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
The authors acknowledge the financial assistance by CSIR (Council of Scientific and Industrial Research), India, by providing fellowship to SK (Award letter no. 09/591(0158)/2018-EMR-I) and to SS (Award letter no. 09/591(0159)/2018-EMR-I) and the Indian Council of Medical Research (ICMR), Govt. of India, to MM under the Research Associateship (RA) scheme (45/25/2018-NAN/BMS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1209168/full#supplementary-material
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
2. Van den Berg MM, Winkels RM, de Kruif JT, Van Laarhoven HW, Visser M, De Vries JH, et al. Weight change during chemotherapy in breast cancer patients: a meta-analysis. BMC Cancer (2017) 17(1):1–3. doi: 10.1186/s12885-017-3242-4
3. Yee C, Wang K, Asthana R, Drost L, Lam H, Lee J, et al. Radiation-induced skin toxicity in breast cancer patients: a systematic review of randomized trials. Clin Breast Cancer (2018) 18(5):e825–40. doi: 10.1016/j.clbc.2018.06.015
4. Ng ET, Ang RZ, Tran BX, Ho CS, Zhang Z, Tan W, et al. Comparing quality of life in breast cancer patients who underwent mastectomy versus breast-conserving surgery: a meta-analysis. Int J Environ Res Public Health (2019) 16(24):4970. doi: 10.3390/ijerph16244970
5. Visvanathan K, Fabian CJ, Bantug E, Brewster AM, Davidson NE, DeCensi A, et al. Use of endocrine therapy for breast cancer risk reduction: ASCO clinical practice guideline update. J Clin Oncol (2019) 37(33):3152–65. doi: 10.1200/JCO.19.01472
6. Franzoi MA, ROmano E, Piccart M. Immunotherapy for early breast cancer: too soon, too superficial, or just right? Ann Oncol (2021) 32:323–6. doi: 10.1016/j.annonc.2020.11.022
7. Savas P, Salgado R, Denkert C, Sotiriou CC, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol (2016) 13(4):228–41. doi: 10.1038/nrclinonc.2015.215
8. Jacobs AT, Castaneda-Cruz DM, Rose MM, Connelly L. Targeted therapy for breast cancer: An overview of drug classes and outcomes. Biochem Pharmacol (2022) 14:115209. doi: 10.1016/j.bcp.2022.115209
9. Semiglazov V, Tseluiko A, Kudaybergenova A, Artemyeva A, Krivorotko P, Donskih R. Immunology and immunotherapy in breast cancer. Cancer Biol Med (2022) 19(5):609. doi: 10.20892/j.issn.2095-3941.2021.0597
10. Wang X, Wang X, Yue Q, Xu H, Zhong X, Sun L, et al. Liquid exfoliation of TiN nanodots as novel sonosensitizers for photothermal-enhanced sonodynamic therapy against cancer. Nano Today (2021) 39:101170. doi: 10.1016/j.nantod.2021.101170
11. Ning S, Dai X, Tang W, Guo Q, Lyu M, Zhu D, et al. Cancer cell membrane-coated C-TiO2 hollow nanoshells for combined sonodynamic and hypoxia-activated chemotherapy. Acta Biomaterialia (2022) 152:562–74. doi: 10.1016/j.actbio.2022.08.067
12. Guo W, Wang T, Huang C, Ning S, Guo Q, Zhang W, et al. Platelet membrane-coated C-TiO2 hollow nanospheres for combined sonodynamic and alkyl-radical cancer therapy. Nano Res (2023) 16(1):782–91. doi: 10.1007/s12274-022-4646-2
13. He G, Ma Y, Zhou H, Sun S, Wang X, Qian H, et al. Mesoporous NiS 2 nanospheres as a hydrophobic anticancer drug delivery vehicle for synergistic photothermal–chemotherapy. J Materials Chem B (2019) 7(1):143–9. doi: 10.1039/C8TB02473A
14. Liu D, Dai X, Zhang W, Zhu X, Zha Z, Qian H, et al. Liquid exfoliation of ultrasmall zirconium carbide nanodots as a noninflammatory photothermal agent in the treatment of glioma. Biomaterials (2023) 292:121917. doi: 10.1016/j.biomaterials.2022.121917
15. Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Natural Products (2020) 83(3):770–803. doi: 10.1021/acs.jnatprod.9b01285
16. Shafi G, Hasan TN, Syed NA, Al-Hazzani AA, Alshatwi AA, Jyothi A, et al. Artemisia absinthium (AA): a novel potential complementary and alternative medicine for breast cancer. Mol Biol Rep (2012) 39:7373–9. doi: 10.1007/s11033-012-1569-0
17. Gupta AK, Madan S, Majumdar DK, Maitra A. Ketorolac entrapped in polymeric micelles: preparation, characterisation and ocular anti-inflammatory studies. Int J Pharmaceutics (2000) 209(1-2):1–4. doi: 10.1016/S0378-5173(00)00508-1
18. Ahmad N, Umar S, Ashafaq M, Akhtar M, Iqbal Z, Samim M, et al. A comparative study of PNIPAM nanoparticles of curcumin, demethoxycurcumin, and bisdemethoxycurcumin and their effects on oxidative stress markers in experimental stroke. Protoplasma (2013) 250:1327–38. doi: 10.1007/s00709-013-0516-9
19. Mughees M, Wajid S, Samim M. Cytotoxic potential of Artemisia absinthium extract loaded polymeric nanoparticles against breast cancer cells: Insight into the protein targets. Int J Pharmaceutics (2020) 586:119583. doi: 10.1016/j.ijpharm.2020.119583
20. Madden EC, Gorman AM, Logue SE, SaMali A. Tumour cell secretome in chemoresistance and tumour recurrence. Trends Cancer (2020) 6(6):489–505. doi: 10.1016/j.trecan.2020.02.020
21. Bonin-Debs AL, Boche I, Gille H, Brinkmann U. Development of secreted proteins as biotherapeutic agents. Expert Opin Biol Ther (2004) 4(4):551–8. doi: 10.1517/14712598.4.4.551
22. Chen ST, Pan TL, Juan HF, Chen TY, Lin YS, Huang CM. Breast tumor microenvironment: proteomics highlights the treatments targeting secretome. J Proteome Res (2008) 7(4):1379–87. doi: 10.1021/pr700745n
23. Swami S, Mughees M, Kauser S, Wajid S. Evaluation of acute oral toxicity of Ipomoea turpethum extract loaded polymeric nanoparticles in Wistar rats. Front Pharmacol (2023) 14:1086581. doi: 10.3389/fphar.2023.1086581
24. Kauser S, Mughees M, Swami S, Wajid S. Pre-clinical toxicity assessment of Artemisia absinthium loaded Polymeric nanoparticles associated with their oral administration. Front Pharmacol (2023) 14:1196842. doi: 10.3389/fphar.2023.1196842
25. Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics (2019) 12(5):1–8. doi: 10.1186/s12920-019-0514-7
26. Liu CJ, Hu FF, Xie GY, Miao YR, Li XW, Zeng Y, et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Briefings Bioinf (2023) 24(1):bbac558. doi: 10.1093/bib/bbac558
27. Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Advanced Sci (2020) 7(7):1902880. doi: 10.1002/advs.201902880
28. García-Moreno A, López-Domínguez R, Ramirez-Mena A, Pascual-Montano A, Aparicio-Puerta E, Hackenberg M, et al. GeneCodis 4: Expanding the modular enrichment analysis to regulatory elements. bioRxiv (2021) 15:2021–04. doi: 10.1101/2021.04.15.439962
29. Mielnicki LM, Ying AM, Head KL, Asch HL, Asch BB. Epigenetic regulation of gelsolin expression in human breast cancer cells. Exp Cell Res (1999) 249(1):161–76. doi: 10.1006/excr.1999.4461
30. Sazawa A, Watanabe T, Tanaka M, Haga K, Fujita H, Harabayashi T, et al. Adenovirus mediated gelsolin gene therapy for orthotopic human bladder cancer in nude mice. J Urol (2002) 168(3):1182–7. doi: 10.1016/S0022-5347(05)64622-8
31. Sagawa N, Fujita H, Banno Y, Nozawa Y, Katoh H, Kuzumaki N. Gelsolin suppresses tumorigenicity through inhibiting PKC activation in a human lung cancer cell line, PC10. Br J Cancer (2003) 88(4):606–12. doi: 10.1038/sj.bjc.6600739
32. Li WX, Yang MX, Hong XQ, Dong TG, Yi T, Lin SL, et al. Overexpression of gelsolin reduces the proliferation and invasion of colon carcinoma cells. Mol Med Rep (2016) 14(4):3059–65. doi: 10.3892/mmr.2016.5652
33. Cohen GM. Caspases: the executioners of apoptosis. Biochem J (1997) 326(1):1–6. doi: 10.1042/bj3260001
34. Broker LE, Kruyt FA, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res (2005) 11(9):3155–62. doi: 10.1158/1078-0432.CCR-04-2223
35. Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene (2004) 23(16):2785–96. doi: 10.1038/sj.onc.1207517
36. Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci (2004) 25(5):259–64. doi: 10.1016/j.tips.2004.03.005
37. Sierralta WD, Epuñan MJ, Reyes JM, Valladares LE, Andersen TT, Bennett JA, et al. A peptide derived from alpha-fetoprotein inhibits the proliferation induced by estradiol in mammary tumor cells in culture. Oncol Rep (2008) 19(1):229–35. doi: 10.3892/or.19.1.229
38. Païdassi H, Tacnet-Delorme P, Garlatti V, Darnault C, Ghebrehiwet B, Gaboriaud C, et al. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J Immunol (2008) 180(4):2329–38. doi: 10.4049/jimmunol.180.4.2329
39. Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, et al. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J Immunol (2012) 189(9):4674–83. doi: 10.4049/jimmunol.1201654
40. Opstal-van Winden AW, Vermeulen RC, Peeters PH, Beijnen JH, van Gils CH. Early diagnostic protein biomarkers for breast cancer: How far have we come? Breast Cancer Res Treat (2012) 134(1):1–12. doi: 10.1007/s10549-011-1907-2
41. Michlmayr A, Bachleitner-Hofmann T, Baumann S, Marchetti-Deschmann M, Rech-Weichselbraun I, Burghuber C, et al. Modulation of plasma complement by the initial dose of epirubicin/docetaxel therapy in breast cancer and its predictive value. Br J Cancer (2010) 103(8):1201–8. doi: 10.1038/sj.bjc.6605909
42. Monteran L, Ershaid N, Doron H, Zait Y, Scharff YE, Ben-Yosef S, et al. Chemotherapy-induced complement signaling modulates immunosuppression and metastatic relapse in breast cancer. Nat Commun (2022) 13(1):5797. doi: 10.1038/s41467-022-33598-x
43. Jordan A, Hadfield JA, Lawrence NJ, McGown AT. Tubulin as a target for anticancer drugs: agents which interact with the mitotic spindle. Medicinal Res Rev (1998) 18(4):259–96. doi: 10.1002/(SICI)1098-1128(199807)18:4<259::AID-MED3>3.0.CO;2-U
44. Islam M, Iskander MN. Microtubulin binding sites as target for developing anticancer agents. Mini Rev Medicinal Chem (2004) 4(10):1077–104. doi: 10.2174/1389557043402946
45. Rasouliha BH, Zhou Y, Chen WN. iTRAQ-coupled 2D LC-MS/MS analysis of protein profile in MCF-7 human breast cancer cells inCubated with doxorubicin: potential role of heat shock protein 90. J Med Imaging Health Informatics (2011) 1(2):193–5. doi: 10.1166/jmihi.2011.1026
46. Stargell LA, Heruth DP, Gaertig JA, Gorovsky MA. Drugs affecting microtubule dynamics increase alpha-tubulin mRNA accumulation via transcription in Tetrahymena thermophila. Mol Cell Biol (1992) 12(4):1443–50. doi: 10.1128/MCB.12.4.1443
47. Qi L, Zhou B, Chen J, Hu W, Bai R, Ye C, et al. Significant prognostic values of differentially expressed-aberrantly methylated hub genes in breast cancer. J Cancer (2019) 10(26):6618. doi: 10.7150/2Fjca.33433
48. Podralska M, Ziółkowska-Suchanek I, Żurawek M, Dzikiewicz-Krawczyk A, Słomski R, Nowak J, et al. Genetic variants in ATM, H2AFX and MRE11 genes and susceptibility to breast cancer in the polish population. BMC Cancer (2018) 18(1):1–1. doi: 10.1186/s12885-018-4360-3
49. Han J, Lim W, You D, Jeong Y, Kim S, Lee JE, et al. Chemoresistance in the human triple-negative breast cancer cell line MDA-MB-231 induced by doxorubicin gradient is associated with epigenetic alterations in histone deacetylase. J Oncol (2019). doi: 10.1155/2019/1345026
50. Cheng X, Murthy SR, Zhuang T, Ly L, Jones O, Basadonna G, et al. Canady helios cold plasma induces breast cancer cell death by oxidation of histone mRNA. Int J Mol Sci (2021) 22(17):9578. doi: 10.3390/ijms22179578
51. Zajac M, Gomez G, Benitez J, Martínez-Delgado B. Molecular signature of response and potential pathways related to resistance to the HSP90 inhibitor, 17AAG, in breast cancer. BMC Med Genomics (2010) 3(1):1–3. doi: 10.1186/1755-8794-3-44
52. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med (2017) 377(6):523–33. doi: 10.1056/NEJMoa1706450
53. Cava C, Sabetian S, Castiglioni I. Patient-specific network for personalized breast cancer therapy with multi-omics data. Entropy (2021) 23(2):225. doi: 10.3390/e23020225
54. Hernández-Vargas H, Rodríguez-Pinilla SM, Julián-Tendero M, Sánchez-Rovira P, Cuevas C, Anton A, et al. Gene expression profiling of breast cancer cells in response to gemcitabine: NF-κB pathway activation as a potential mechanism of resistance. Breast Cancer Res Treat (2007) 102:157–72. doi: 10.1007/s10549-006-9322-9
55. Zhang D, Jin N, Sun W, Li X, Liu B, Xie Z, et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene (2017) 36(20):2900–9. doi: 10.1038/onc.2016.446
56. Chen S, Cai J, Zhang W, Zheng X, Hu S, Lu J, et al. Proteomic identification of differentially expressed proteins associated with the multiple drug resistance in methotrexate-resistant human breast cancer cells. Int J Oncol (2014) 45(1):448–58. doi: 10.3892/ijo.2014.2389
57. Matchett KB, McFarlane S, Hamilton SE, Eltuhamy YS, Davidson MA, Murray JT, et al. Ran GTPase in nuclear envelope formation and cancer metastasis. Cancer Biol Nucl Envelope: Recent Adv May Elucidate Past Paradoxes (2014), 323–51. doi: 10.1007/978-1-4899-8032-8_15
58. Sheng C, Qiu J, Wang Y, He Z, Wang H, Wang Q, et al. Knockdown of Ran GTPase expression inhibits the proliferation and migration of breast cancer cells. Mol Med Rep (2018) 18(1):157–68. doi: 10.3892/mmr.2018.8952
59. Colman RW. Role of the light chain of high molecular weight kininogen in adhesion, cell-associated proteolysis and angiogenesis. Biol Chem (2001) 382(1):65–70. doi: 10.1515/BC.2001.011
60. Mohammed SI, Torres-Luquis O, Zhou W, Lanman NA, Espina V, Liotta L. Tumor-draining lymph secretome en route to the regional lymph node in breast cancer metastasis. Breast Cancer: Targets Ther (2020) 25:57–67. doi: 10.2147/BCTT.S236168
61. Fan J, Tea MK, Yang C, Ma L, Meng QH, Hu TY, et al. Profiling of cross-functional peptidases regulated circulating peptides in BRCA1 mutant breast cancer. J Proteome Res (2016) 15(5):1534–45. doi: 10.1021/acs.jproteome.6b00010
62. Cui R, Wang C, Zhao Q, Wang Y, Li Y. Serum carboxypeptidase N1 serves as a potential biomarker complementing CA15-3 for breast cancer. Anti-Cancer Agents Medicinal Chem (2020) 20(17):2053–65. doi: 10.2174/1871520620666200703191135
63. Greene LM, Twal WO, Duffy MJ, McDermott EW, Hill AD, O’Higgins NJ, et al. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br J cancer. (2003) 88(6):871–8. doi: 10.1038/sj.bjc.6600802
64. Qing J, Maher VM, Tran H, Argraves WS, Dunstan RW, McCormick JJ. Suppression of anchorage-independent growth and matrigel invasion and delayed tumor formation by elevated expression of fibulin-1D in human fibrosarcoma-derived cell lines. Oncogene (1997) 15(18):2159–68. doi: 10.1038/sj.onc.1201385
65. Twal WO, Czirok A, Hegedus B, Knaak C, Chintalapudi MR, Okagawa H, et al. Fibulin-1 suppression of fibronectin-regulated cell adhesion and motility. J Cell science. (2001) 114(24):4587–98. doi: 10.1242/jcs.114.24.4587
66. Marano F, Zunino V, Frairia R, Arvat E, Castellano I, Bosco O, et al. Fibulin-1 interacts with sex hormone binding globulin and is linked to less aggressive estrogen-dependent breast cancers. Life Sci (2018) 207:372–80. doi: 10.1016/j.lfs.2018.06.024
67. Jia L, Waxman DJ. Thrombospondin-1 and pigment epithelium-derived factor enhance responsiveness of KM12 colon tumor to metronomic cyclophosphamide but have disparate effects on tumor metastasis. Cancer Letters (2013) 330(2):241–9. doi: 10.1016/j.canlet.2012.11.055
68. Sung SY, Hsieh CL, Wu D, Chung LW, Johnstone PA. Tumor microenvironment promotes cancer progression, metastasis, and therapeutic resistance. Curr Problems Cancer (2007) 31(2):36–100. doi: 10.1016/j.currproblcancer.2006.12.002
69. Beck AH, Espinosa I, Gilks CB, Van De Rijn M, West RB. The fibromatosis signature defines a robust stromal response in breast carcinoma. Lab Invest (2008) 88(6):591–601. doi: 10.1038/labinvest.2008.31
70. Wong SY, Crowley D, Bronson RT, Hynes RO. Analyses of the role of endogenous SPARC in mouse models of prostate and breast cancer. Clin Exp Metastasis (2008) 25:109–18. doi: 10.1007/s10585-007-9126-2
71. Nagai MA, Gerhard R, Fregnani JH, Nonogaki S, Rierger RB, Netto MM, et al. Prognostic value of NDRG1 and SPARC protein expression in breast cancer patients. Breast Cancer Res Treat (2011) 126:1–4. doi: 10.1007/s10549-010-0867-2
72. Yunker CK, Golembieski W, Lemke N, Schultz CR, Cazacu S, Brodie C, et al. SPARC-induced increase in glioma matrix and decrease in vascularity are associated with reduced VEGF expression and secretion. Int J Cancer (2008) 122(12):2735–43. doi: 10.1002/ijc.23450
73. Yiu GK, Chan WY, Ng SW, Chan PS, Cheung KK, Berkowitz RS, et al. SPARC (secreted protein acidic and rich in cysteine) induces apoptosis in ovarian cancer cells. Am J Pathol (2001) 159(2):609–22. doi: 10.1016/S0002-9440(10)61732-4
74. Cao F, Wang K, Zhu R, Hu YW, Fang WZ, Ding HZ. Clinicopathological significance of reduced SPARCL1 expression in human breast cancer. Asian Pacific J Cancer Prev (2013) 14(1):195–200. doi: 10.7314/APJCP.2013.14.1.195
75. Girard JP, Springer TA. Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity (1995) 2(1):113–23. doi: 10.1016/1074-7613(95)90083-7
76. Claeskens A, Ongenae N, Neefs JM, Cheyns P, Kaijen P, Cools M, et al. Hevin is down-regulated in many cancers and is a negative regulator of cell growth and proliferation. Br J Cancer (2000) 82(6):1123–30. doi: 10.1054/bjoc.1999.1051
77. Lau CY, Poon RP, Cheung ST, Yu WC, Fan ST. SPARC and Hevin expression correlate with tumour angiogenesis in hepatocellular carcinoma. J Pathol (2006) 210(4):459–68. doi: 10.1002/path.2068
78. Cui Z, Chen Y, Hu M, Lin Y, Zhang S, Kong L, et al. Diagnostic and prognostic value of the cancer-testis antigen lactate dehydrogenase C4 in breast cancer. Clinica Chimica Acta (2020) 503:203–9. doi: 10.1016/j.cca.2019.11.032
79. Thomas R, Shaath H, Naik A, Toor SM, Elkord E, Decock J. Identification of two HLA-A* 0201 immunogenic epitopes of lactate dehydrogenase C (LDHC): potential novel targets for cancer immunotherapy. Cancer immunol Immunother (2020) 69:449–63. doi: 10.1007/s00262-020-02480-4
80. Naik A, Decock J. Targeting of lactate dehydrogenase C dysregulates the cell cycle and sensitizes breast cancer cells to DNA damage response targeted therapy. Mol Oncol (2022) 16(4):885–903. doi: 10.1002/1878-0261.13024
81. Nör JE, Mitra RS, Sutorik MM, Mooney DJ, Castle VP, Polverini PJ. Thrombospondin-1 induces endothelial cell apoptosis and inhibits angiogenesis by activating the caspase death pathway. J Vasc Res (2000) 37(3):209–18. doi: 10.1159/000025733
82. Dawson DW, Volpert OV, Pearce SF, Schneider AJ, Silverstein RL, Henkin J, et al. Three distinct D-amino acid substitutions confer potent antiangiogenic activity on an inactive peptide derived from a thrombospondin-1 type 1 repeat. Mol Pharmacol (1999) 55(2):332–8. doi: 10.1124/mol.55.2.332
83. Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res Treat (2009) 114:85–96. doi: 10.1007/s10549-008-9992-6
84. Sage EH, Bornstein P. Extracellular proteins that modulate cell-matrix interactions. SPARC, tenascin, and thrombospondin. J Biol Chem (1991) 266(23):14831–4. doi: 10.1016/S0021-9258(18)98545-5
85. Cai D, Liang J, Cai XD, Yang Y, Liu G, Zhou F, et al. Identification of six hub genes and analysis of their correlation with drug sensitivity in acute myeloid leukemia through bioinformatics. Trans Cancer Res (2021) 10(1):126. doi: 10.21037/tcr-20-2712
86. Lin X, Ye L, Wang X, Liao Z, Dong J, Yang Y, et al. Follicular helper T cells remodel the immune microenvironment of pancreatic cancer via secreting CXCL13 and IL-21. Cancers (2021) 13(15):3678. doi: 10.3390/cancers13153678
87. Chan IS, Ewald AJ. The changing role of natural killer cells in cancer metastasis. J Clin Invest (2022) 132(6). doi: 10.1172/JCI143762
88. Webb TJ, Yuan W, Meyer E, Dellabona P. NKT Cells in cancer immunotherapy. Front Immunol (2020) 11:1314. doi: 10.3389/fimmu.2020.01314
89. Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy—new insights into old paradigms. Cancer Gene Ther (2021) 28(1-2):5–17. doi: 10.1038/s41417-020-0183-x
90. Maeda Y, Wada H, Sugiyama D, Saito T, Irie T, Itahashi K, et al. Depletion of central memory CD8+ T cells might impede the antitumor therapeutic effect of Mogamulizumab. Nat Commun (2021) 12(1):7280. doi: 10.1038/s41467-021-27574-0
91. Xiong S, Dong L, Cheng L. Neutrophils in cancer carcinogenesis and metastasis. J Hematol Oncol (2021) 14(1):1–7. doi: 10.1186/s13045-021-01187-y
92. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell (2019) 35(4):588–602. doi: 10.1016/j.ccell.2019.02.009
93. Asadzadeh Z, Mohammadi H, Safarzadeh E, Hemmatzadeh M, Mahdian-Shakib A, Jadidi-Niaragh F, et al. The paradox of Th17 cell functions in tumor immunity. Cell Immunol (2017) 322:15–25. doi: 10.1016/j.cellimm.2017.10.015
94. Paluskievicz CM, Cao X, Abdi R, Zheng P, Liu Y, Bromberg JS. T regulatory cells and priming the suppressive tumor microenvironment. Front Immunol (2019) 10:2453. doi: 10.3389/fimmu.2019.02453
Keywords: Artemisia absinthium, nano LC-MS/MS, GENT2, GSCA, GDSC, CTRP
Citation: Kauser S, Mughees M, Mangangcha IR, Swami S and Wajid S (2023) Secretome profiling of Artemisia absinthium extract-loaded polymeric nanoparticle-treated MCF-7 and MDA-MB-231 revealed perturbation in microtubule assembly and cell migration. Front. Oncol. 13:1209168. doi: 10.3389/fonc.2023.1209168
Received: 20 April 2023; Accepted: 04 August 2023;
Published: 31 August 2023.
Edited by:
Antonio Macciò, Ospedale Oncologico Armando Businco, ItalyReviewed by:
Marta Canel, University of Edinburgh, United KingdomAleksandra Slawinska, University of Edinburgh, United Kingdom, in collaboration with reviewer MC
Xianwen Wang, Anhui Medical University, China
Copyright © 2023 Kauser, Mughees, Mangangcha, Swami and Wajid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saima Wajid, c3dhamlkQGphbWlhaGFtZGFyZC5hYy5pbg==; d2FqaWQuc2FpbWFAZ21haWwuY29t
†Present address: Mohd Mughees, Department of Breast Medical Oncology, University of Texas MD Anderson Cancer Center, Houston, TX, United States
‡These authors have contributed equally to this work and share first authorship
§ORCID: Saima Wajid, orcid.org/0000-0003-1558-8233
 Sana Kauser
Sana Kauser Mohd Mughees
Mohd Mughees Irengbam Rocky Mangangcha
Irengbam Rocky Mangangcha Sanskriti Swami
Sanskriti Swami Saima Wajid
Saima Wajid