- 1Department of Pancreatobiliary Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China
- 2Department of Anesthesiology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China
Hepatocellular carcinoma (HCC) is a highly malignant tumor that carries a significant risk of morbidity and mortality. This type of cancer is prevalent in Asia due to the widespread presence of risk factors. Unfortunately, HCC often goes undetected until it has reached an advanced stage, making early detection and treatment critical for better outcomes. Alpha-fetoprotein (AFP) is commonly used in clinical practice for diagnosing HCC, but its sensitivity and specificity are limited. While surgery and liver transplantation are the main radical treatments, drug therapy and local interventions are better options for patients with advanced HCC. Accurately assessing treatment efficacy and adjusting plans in a timely manner can significantly improve the prognosis of HCC. Non-coding RNA gene transcription products cannot participate in protein production, but they can regulate gene expression and protein function through the regulation of transcription and translation processes. These non-coding RNAs have been found to be associated with tumor development in various types of tumors. Noncoding RNA released by tumor or blood cells can circulate in the blood and serve as a biomarker for diagnosis, prognosis, and efficacy assessment. This article explores the unique role of circulating noncoding RNA in HCC from various perspectives.
1 Introduction
Hepatocellular carcinoma (HCC) is the most prevalent type of cancer that originates in the liver. It ranks fifth in terms of overall cancer occurrences worldwide and is second in terms of fatality among malignant tumors (1). Unfortunately, the incidence of HCC continues to increase as its primary risk factors, including non-alcoholic steatohepatitis, hepatitis B virus infection, and hepatitis C virus infection, remain widespread (2). While current research is focused on examining the molecular mechanism of HCC, there has been no substantial improvement in the prognosis of advanced cases. This is due to the aggressive nature of HCC and its tendency to recur, resulting in an overall survival rate that falls short of satisfactory (3). Early diagnosis plays a critical role in improving the survival time and rate of patients with HCC. Currently, the primary means of clinical diagnosis includes assessing serum alpha-fetoprotein (AFP) levels and utilizing imaging examinations like liver ultrasound, CT, and MRI (4). Although alpha-fetoprotein is a serum biomarker, its specificity is not strong enough. Therefore, current research aims to discover a biomarker that boasts high diagnostic accuracy and specificity. Drug therapy has limited effectiveness, and for patients with early-stage HCC, surgical resection or liver transplantation remains a promising standard of treatment (5). At the same time, in addition to drug therapy and surgery, local radiofrequency therapy and transcatheter arterial chemoembolization (TACE) have gradually been accepted clinically as treatment methods for HCC. At present, drug resistance in HCC has gradually emerged, which has become the main bottleneck limiting the effect of drugs. By continuously improving these treatments, patients with HCC can attain a more favorable prognosis.
Biomarkers play an important role in the field of oncology. For example, alpha-fetoprotein (AFP) is a widely accepted biomarker for the detection of HCC, while CA-199 is sensitive to pancreatic cancer (6), and carcinoembryonic antigen (CEA) is commonly used for colon cancer patients (7). These are just a few examples of the many biomarkers in use today. Biomarkers can: 1) classify patients according to risk; 2) Diagnosis and detection of disease development; 3) Effectively derive patient prognosis and adjust treatment (8). Biomarker tests known as “liquid biopsies” are extracted from body fluids such as blood and urine. This method is less invasive and less painful compared to traditional biopsies. Additionally, it provides relatively good insight into the heterogeneity of tumor molecules and is highly sensitive (9). This liquid biopsy offers numerous advantages over tissue biopsy. Firstly, it inflicts less trauma, making it more appealing for patients. Additionally, it is convenient and fast, offering a simpler and quicker alternative. Moreover, this method is much more accurate and precise. Finally, it also reduces the likelihood of tumor metastasis.
2 Non-coding RNA
Since the discovery of non-coding RNA in the 1990s, researchers have continuously delved deeper into its study, leading to a greater understanding of how gene expression is regulated (10). Non-coding RNA is a byproduct of gene transcription, which cannot be used for protein production. Nonetheless, it has the ability to regulate gene expression and protein function by controlling both transcription and translation processes (11). Through these basic biological functions, it participates in the process of cell growth, migration, autophagy, apoptosis and differentiation, and is widely involved in biological behavior (12). Non-coding RNAs are implicated in the development of numerous diseases, including cancer, heart disease, and immune disorders. As a result, some researchers are now exploring their potential as biomarkers for early diagnosis and treatment evaluation in patients with hepatocellular carcinoma. However, the use of non-coding RNA for tissue assessment may cause discomfort to patients as it requires invasive procedures, unlike using serum AFP. The concentration level of circulating non-coding RNA is relatively stable because of the protection provided by exosomes, making it an effective tool for distinguishing tumor patients from normal individuals (13). Circulating non-coding RNAs primarily result from the discharge of intracellular non-coding RNAs into the bloodstream due to tissue damage or from the discharge of cells as exosomes (14) (Figure 1). The utilization of circulating noncoding RNAs as a means of identifying biomarkers has shown considerable promise in various types of cancer, including lung, prostate, and gastric cancers (15). Non-coding RNA found in the circulation offers significant potential as a non-invasive biomarker for HCC, leaving ample room for research opportunities. Employing non-coding RNA as a biomarker could bring considerable enhancements to the diagnosis, prognosis, and treatment efficacy of HCC. The main components of circulating ncRNA are circulating microRNA and circulating long non-coding RNA (lncRNA). Unfortunately, there are currently limited studies conducted on circulating circular RNA (circRNA). Additionally, no obvious correlation with tumors has been found, so it will not be discussed further in this paper. Therefore, this article will primarily focus on the first two aspects: circulating microRNA and circulating lncRNA.
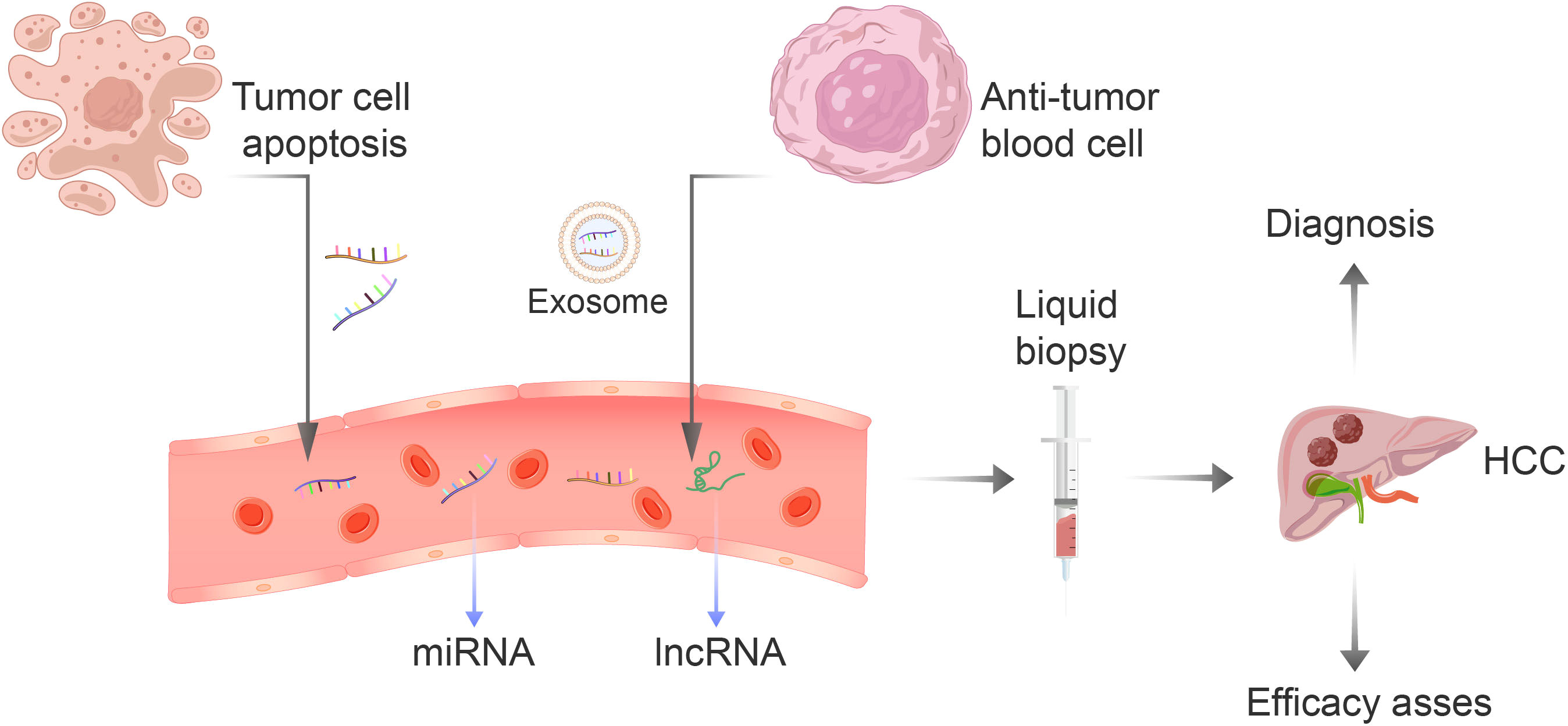
Figure 1 The non-coding RNA present in blood is primarily derived from two sources: the apoptosis of tumor cells or the secretion of blood cells that have anti-tumor properties. This RNA includes miRNA-122, let-7 family, miRNA-34a, and lncRNA. The liquid biopsy technique is an efficient, convenient, and non-invasive method for early detection and evaluation of the effectiveness of treatment for liver cancer.
3 Circulating microRNA
MicroRNA is a type of small non-coding RNA that consists of approximately 18-24 nucleotides in length. Once matured, microRNA joins the RNA-induced silencing complex located in the cytoplasm. From there, it combines with the 3’untranslated region (3’-UTR) of the mRNA to facilitate degradation and translational repression. The primary goal of this process is to regulate gene expression (16, 17). Abnormal regulation of microRNA often leads to the development of tumors. MicroRNA can bind to proteins present in different body fluids, or it can be enveloped in bilayer lipids to attain a stable state. The stability of microRNA is the highest when it is situated in exosomes. Thus, to assess its potential as a biomarker, basic quantitative polymerase chain reaction (qPCR) and other methods can be employed (18). The microRNAs that only exist in serum are derived from non-blood cells, some of which are derived from tumor cell apoptosis or secretion (19). Most of the microRNAs in the circulation mainly come from blood cells that exert anti-tumor functions (20). Furthermore, the genetic and molecular characteristics of a tumor can be revealed by analyzing circulating microRNA. This method is especially effective at detecting the changes that occur in response to tumors. In fact, the levels of serum microRNA are typically 1.6 times higher in tumor patients compared to those of healthy individuals (21). Abnormal expression of miRNA in patients with HCC is closely related to the occurrence and development of tumors, and the let-7 family (22) and miRNA-122 expression are downregulated widely reported in HCC (23), and miRNA-221, miRNA-222 and miRNA-224 are upregulated (24, 25). According to research, miRNA-122, miRNA-34a, and miRNA-196a-5p are known to suppress tumors, while the let-7 family of miRNAs have been found to promote cancer growth. Compared to other malignant tumors, the abnormal miRNA profile of HCC is more distinct in terms of diagnosis, treatment, and prognosis (26). Incorporating miRNA as a biomarker and utilizing it in clinical practice can significantly enhance clinicians’ ability to gain a more precise understanding of their patients’ conditions, ultimately leading to more accurate medical decisions.
3.1 Circulating microRNAs and HCC diagnosis
Currently, the incidence of HCC is consistently increasing each year. Due to its deceptive symptoms and rapid progression, it is commonly detected in the advanced stages of tumor development during diagnosis (27). At present, the primary method for early screening of liver cancer is measuring the serum AFP levels. However, this method’s low sensitivity has led to debates surrounding its diagnostic efficacy (28). The AFP level is found to be elevated not only in patients with HCC, but also in those with chronic hepatitis B and hepatitis C infections (29). Recent studies have suggested that the combination of miRNA and AFP could markedly enhance early detection of HCC by improving both sensitivity and specificity (30). Among them, many studies have been published on the role of circulating miRNA-122 (31), let-7 family (32) and other miRNAs in the diagnosis of HCC (Figure 2).
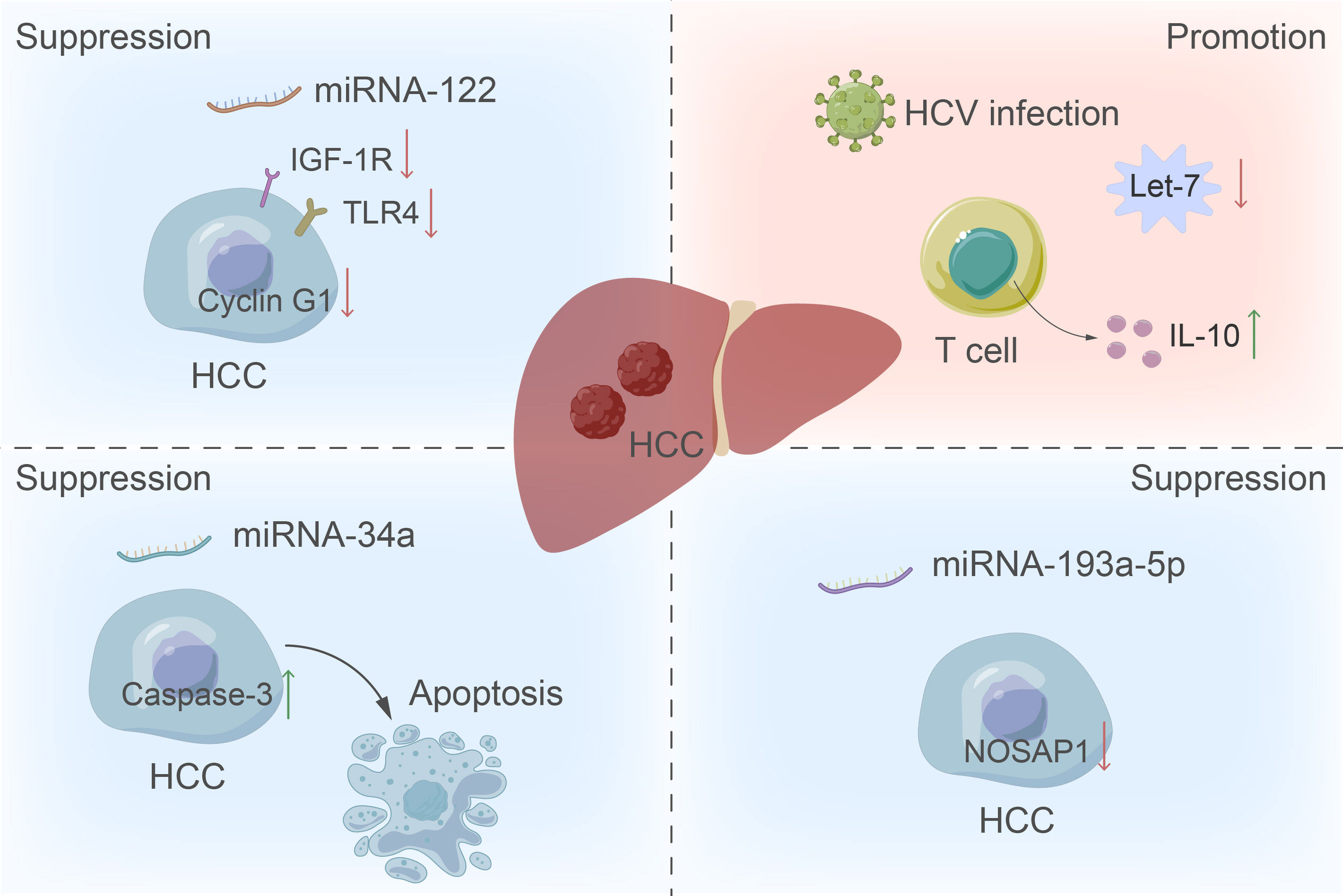
Figure 2 The role of miRNA in the development of HCC. The up-regulation of miRNA-122 inhibits liver cancer by inhibiting cyclin G1, the insulin-like growth factor 1 receptor pathway, and the expression of toll-like receptor 4 in tumor cells. Conversely, downregulation of the let-7 family increases T cell production of IL-10, providing the virus with a survival advantage by manipulating the host immune response and promoting tumor formation. Additionally, miRNA-34a functions as a tumor suppressor by inducing apoptosis through increased activity of caspase-3.
3.1.1 miRNA-122
miRNA-122 is a major microRNA in the liver, accounting for approximately 70% of all liver microRNAs (33), and is involved in the development and pathogenesis of HCC (34). The expression of miRNA-122 is regulated by factors such as enhancer-binding protein α, hepatocyte nuclear factor (HNF) 1α, HNF3β, and HNF4α (35),which explains why miRNA-122 is a liver-specific microRNA. miRNA-122 can inhibit the expression of p53 by down-regulating the cell cycle G1-p53 complex to inhibit the RNA replication of hepatitis B virus, and hepatitis B virus can regulate miRNA-122, reduce its expression to promote virus infection, and eventually lead to tumorigenesis (36). Previous studies have shown that the absence of miRNA-122 will promote liver steatosis and liver fibrosis, and the reduction of miRNA-122 levels has been observed in people with non-alcoholic steatohepatitis and liver cirrhosis (37). Liver tumors appeared successively in mice knocked out of the miRNA-122 gene, and liver tumors in mice were suppressed when microRNA levels were restored (38). miRNA-122 exerts its tumor suppressive effect by inhibiting cyclin G1, inhibiting the insulin-like growth factor 1 receptor pathway, inhibiting the expression of toll-like receptor 4 in tumor cells, etc. (37).
Serum miRNA-122 levels in the HCC group were found to be significantly elevated in early-stage HCC patients compared with healthy controls (31). When miR-122-5p is detected in the circulation, it indicates the early occurrence of liver cancer (39). However, there are similar problems with AFP in the diagnosis of liver cancer. At present, only HCC patients and healthy people are compared, and the serum level in non-HCC patients such as chronic hepatitis B virus infection and liver cirrhosis is still unclear. A recent meta-analysis study showed that (40), the level of miRNA-122 in serum was significantly different between patients with HCC and healthy people or patients with chronic hepatitis B virus infection. However, for patients with liver cirrhosis, the sensitivity of judging whether there is HCC is not high, and auxiliary examinations such as imaging studies are still needed. And in a survey of European population, it was found that serum miRNA-122 was significantly increased in patients with acute liver injury, and it was positively correlated with transaminase levels and alpha-fetoprotein levels (41). Although the sensitivity of miRNA-122 in the diagnosis of liver cancer is better than that of AFP (42), it still has its shortcomings. Some researchers proposed to analyze the binding of circulating miRNA-122 to telomerase reverse transcriptase in circulating cell DNA to evaluate its diagnostic efficacy as a biomarker. The differential diagnosis of HCC, cirrhosis and non- HCC is the highest (43). And miRNA-122 is a kind of tumor suppressor factor, and the serum concentration rises when HCC occurs, which may be secreted from blood cells that play an anti-tumor effect.
3.1.2 let-7 family
The let-7 family consists of 12 miRNAs that function as microRNAs. They play a crucial role in negatively regulating oncogenes and cell cycle factors (44). Relevant studies have pointed out that the circulating let-7 family can be mainly divided into three clusters according to clustering, and let7 b/c/g are the representatives of the three clusters (45). The let-7 family mainly targets hepatitis C virus infection, and interferon α increases the production of let-7s through the signaling pathway to inhibit the replication of hepatitis C virus (46). Moreover, the level of let-7 in HCC tissues caused by hepatitis C virus infection is lower than that in healthy people, and its circulating level is also negatively correlated with the degree of liver cirrhosis (47), indicating that the decreased level of let-7 family in the circulation should be related to HCV infection. Downregulation of the let-7 family leads to increased production of IL-10 by T cells, which in turn confers an important survival advantage to the virus by manipulating the host immune response (48). Subsequent studies have also found that the level of let-7i in the circulation after antiviral therapy can be used as a biomarker for the progression of HCC (32). Circulating miRNAs have good diagnostic efficacy for hepatitis C viral HCC developed from liver cirrhosis and chronic hepatitis C virus, and when used as non-invasive disease biomarkers together with alpha-fetoprotein, the diagnostic efficacy have significantly improved (49). By detecting the level of the let-7 family in the circulation and determining the presence of hepatitis C virus infection, it is possible to identify the condition of patients with hepatitis C virus infection in a timely manner and detect HCC at an early stage.
3.1.3 miRNA-34a
MiRNA-34a has been confirmed as a tumor suppressor (50), that is, patients with tumors with high expression levels of miRNA-34a have a better prognosis. In hepatocellular carcinoma, abnormal expression of miRNA-34a promotes tumor proliferation and metastasis through cell cycle and p53 signaling pathway (51). Recent studies have found that miRNA-34a can significantly downregulate the expression of transcriptional activators E2F1 and E2F3, both of which are upregulated in HCC patients and are associated with the degree of malignancy of tumors (52). However, miRNA-34a induces apoptosis through caspase-3 (53). It was indeed observed in subsequent in vitro experiments that miRNA-34a significantly increased the activity of caspase-3 (52). The presence of suppressed tumor suppressors in the tissues of HCC patients should also be at lower levels secreted by tumor cells into the circulation via exosomes. miRNA-34a is a specific and sensitive indicator for the diagnosis of hepatocellular carcinoma (54). After investigation, it was found that the serum miRNA-34a level of patients with HCC was much lower than that of healthy people, and the circulating level of patients before surgery was also lower than that of patients after surgery. Moreover, the decrease of miRNA-34a level in the circulation is related to the clinical characteristics of the tumor, such as TNM stage, vascular invasion and lymph node metastasis, etc. (55) Upregulation of miR-34a-5p levels in serum can predict the onset of cirrhosis (56), which can also point to early changes in HCC (39).
3.1.4 Other microRNAs
Furthermore, in addition to the aforementioned three microRNAs, there are numerous additional microRNAs that are currently being studied for their potential in aiding the diagnosis of HCC. For instance, the miRNA-193a-5p has shown promise as a tumor suppressor, as evidenced by its reduced expression in patients with HCC (57). This microRNA appears to hinder tumor growth by lowering levels of nucleoli and spindle-associated protein 1, and its low expression levels are significantly linked to unfavorable prognoses (58). Through comparison of the circulating miRNA-193a-5p levels of postoperative HCC patients with those of healthy individuals, it was discovered that patients with HCC exhibited elevated circulating levels of this particular miRNA. Moreover, those who had higher levels of miRNA-193a-5p prior to surgery had generally poorer survival outcomes. This increased level of miRNA-193a-5p in circulation is likely attributable to tumor cell apoptosis, which results in greater secretion of this miRNA into the bloodstream (59).. In patients with hepatocellular carcinoma, the level of miRNA-223-3p in circulation is significantly reduced, and the diagnostic accuracy of alpha-fetoprotein can reach 100% for intermediate and advanced hepatocellular carcinoma. miRNA-223-3p is an independent prognostic factor for patients with HCC (60). Research indicates that the detection of miR-100-5p, miR-34a-5p, and miR-365-5p in the bloodstream can signify initial alterations prior to the development of HCC (39). In this study, we found that serum miRNA-16 levels were significantly higher in patients with early-stage HCC compared to the control group, after amplifying the circulating samples. This was confirmed by ROC curves, which showed that miRNA-16 had high diagnostic accuracy. Furthermore, analyzing miRNA-16 and alpha-fetoprotein together could further increase its accuracy (42). In this study, we discovered that miRNA-518d-5p expression levels were elevated in both tumor and healthy individuals, as well as in both tumor and non-tumorous specimens of HCC patients. Furthermore, we observed that higher levels of this miRNA in serum samples were correlated with poorer treatment outcomes and shorter treatment times (61). Elevated levels of circulating MiRNA-107 can indicate the early stages of HCC development and have been linked to a poorer prognosis (62).
3.2 Circulating microRNAs and HCC efficacy assessment
Currently, there are three main approaches used to treat HCC: drug treatment, surgical treatment, and local interventional treatment. Drug treatment options include chemotherapy, targeted therapy, and immunotherapy. Surgical treatment encompasses radical resection and liver transplantation. On the other hand, local intervention mainly employs radiofrequency therapy and hepatic arterial chemoembolization as its mainstay procedures. Although treatments exist for hepatocellular carcinoma (HCC), these options are primarily effective for patients in the early stage of the disease, which only represents a small fraction of HCC cases (63). Unfortunately, HCC is notorious for its inconspicuous symptoms, and it is typically diagnosed when the tumor has reached an advanced stage. Consequently, surgical treatment offers only limited benefits. As such, drug therapy and local interventional procedures are the predominant methods of treating patients with advanced HCC (64). Hepatocellular carcinoma exhibits heterogeneity both between and within individuals. The former is largely attributable to differences in risk factors and genomic environments, while the latter can be attributed to clonal evolution of cancer cells (65, 66). Heterogeneity can have varying causes, all of which can lead to decreased effectiveness of targeted drugs. A major hurdle in drug treatment is the emergence of drug resistance, which greatly limits efficacy. Unfortunately, there is a current lack of biomarkers that are sensitive to drug resistance. Despite being a potentially effective treatment for HCC, local interventional therapy is not consistently successful in cases of heterogeneous HCC. Consequently, surgeons require a biomarker that can aid in evaluating a patient’s response to treatment in order to address this pressing issue. MicroRNAs can reflect some characteristics of tumors (67), and it is generally believed that processes such as apoptosis caused by treatment are accompanied by the secretion of cytokines, growth factors, and microRNAs (68). As a result, certain studies are focused on exploring whether microRNAs can function as biological indicators for evaluating the efficacy of treatments.
3.2.1 Radiofrequency therapy and microRNA
Radiofrequency ablation is a safe and effective treatment option for patients with early-stage, unresectable, and non-transplantable HCC nodules that are less than 3 cm in diameter (69). Studies comparing local radiofrequency therapy and surgical resection have found no significant difference in progression-free survival (RFS) or overall survival (OS) between the two treatments (70). However, a significant proportion of patients experience tumor recurrence (71). To effectively monitor recurrence after radiofrequency therapy, it is crucial to have a non-invasive biomarker. The Let-7 family, as previously mentioned, is particularly useful for detecting HCC in its early stages, particularly in patients with risk factors for hepatitis C infection. Let-7c is a member of the let-7 family that has been found to have a cancer-suppressing effect. It achieves this by inhibiting tumor cell proliferation and promoting apoptosis. This is achieved through its ability to target CD25A, PI3K/Akt/FoxO, and Wnt signaling (72, 73). It is worth noting that Let-7c is down-regulated in HCC tissues. Research has demonstrated that the levels of circulating let-7 can indicate early recurrence following treatment for HCC. Additionally, overexpression of let-7 has been linked to portal vein invasion, changes in TNM stage, and overall survival post-treatment (74). Similarly, studies have shown that patients treated with local radiofrequency ablation and who have elevated levels of circulating let-7 are more likely to experience a recurrence of HCC after treatment (71). Radiofrequency therapy primarily relies on the direct cautery of tumor tissue through high temperatures. As a result, the peak of circulating microRNA typically occurs approximately 60-90 minutes after the radiofrequency treatment (75). While let-7 is known to act as a tumor suppressor during tumorigenesis, recent findings suggest that microRNAs may have varying roles, and even exhibit opposing patterns, within the tumor environment. Although the exact mechanism remains unclear, it is possible that circulating blood cell secretion may be involved. In a study of patients who underwent radiofrequency therapy, it was found that levels of circulating miRNA-210 and miRNA-200a increased in those who experienced early progression (75). MiRNA-210 has been shown to play a role in tumor progression and metastasis by regulating various cellular processes such as mitochondrial metabolism, angiogenesis, cell proliferation, and apoptosis (76). The function of miRNA-200a in tumorigenesis and progression remains unclear, although it is widely believed to act as a tumor suppressor. Evidence suggests that elevated levels of miRNA-200a in circulation are associated with a negative prognosis in patients with colorectal cancer (77). Clinicians can improve their assessment of a patient’s treatment by analyzing the fluctuations in microRNA levels present in circulation. This allows for the adjustment of treatment strategies that are appropriate for the patient’s needs.
3.2.2 Transarterial chemoembolization therapy and microRNA
For patients with advanced stages of cancer and larger tumors, transarterial chemoembolization is a highly recommended treatment option (78). This treatment primarily involves injecting chemotherapy drugs, such as cisplatin, into the arteries. This is then followed by the embolization of the tumor’s corresponding blood vessels using embolic agents (79). Under high concentrations of embolism, tumor cells experience hypoxia and a cytostatic effect. Certain tumors may not respond to arterial injection of cisplatin, which highlights the importance of identifying a reliable biomarker that can effectively assess the effectiveness of treatment. The study revealed that patients who responded well to TACE had higher baseline levels of miR-106b, miR-107, and miR-133b compared to non-responders. On the other hand, it was found that non-responders exhibited elevated levels of miR-26a and miR-31 compared to responders. Notably, miR-133b and miR-26a demonstrated the most promising response to TACE treatment, with miR-133b proving particularly effective in distinguishing between complete response, partial response, and no response (80). In other tumors, miR-133b has been discovered to increase sensitivity to chemotherapy drugs by inhibiting ABCC1 and MDR1. Additionally, it has been identified as a tumor suppressor factor (81, 82). Researchers have discovered that combining miR-26a with other microRNAs can accurately predict the effectiveness of treatment within one year after TACE surgery for HCC (83). Following TACE treatment, the levels of circulating microRNA will reach their highest point after 24 hours. This is due to the fact that TACE treatment is a slower ischemic process, which means that it takes longer to reach its peak compared to radiofrequency therapy (75). TACE therapy has been found to increase the levels of miRNA-210 in circulation. However, there is a lack of studies that have investigated the correlation between this increase and patient prognosis.
3.2.3 Chemotherapy and microRNA
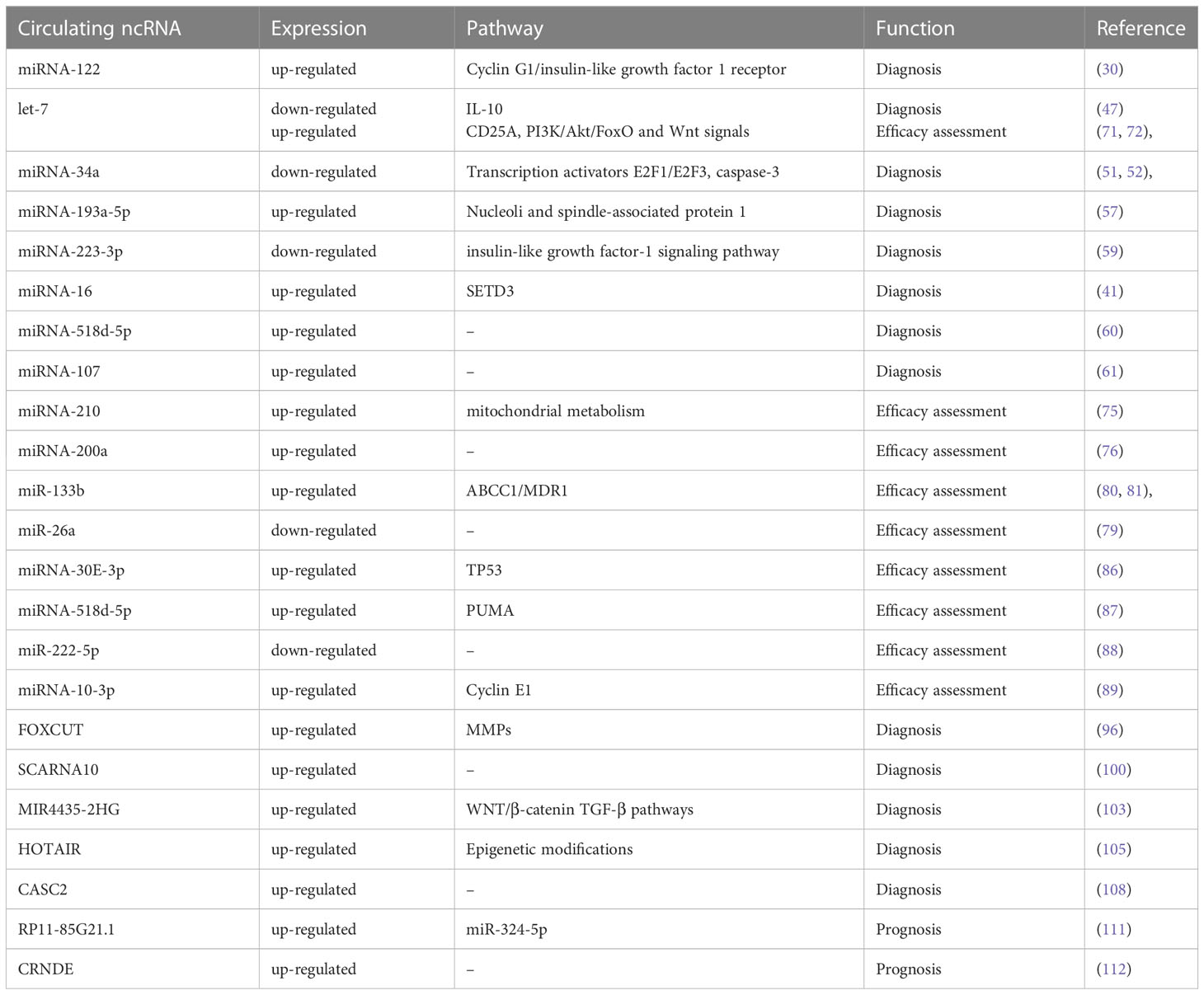
Chemotherapy drugs for HCC are typically categorized into three main types: chemotherapy, targeted therapy, and immunotherapy. While immunotherapy drugs have been approved for widespread clinical use (84), they may not be suitable for patients with poor liver function or a history of autoimmune diseases. In such cases, targeted drugs like sorafenib are the optimal solution for drug treatment. One of the primary challenges in using sorafenib for clinical purposes is the development of drug resistance that occurs during the course of treatment. If clinicians can identify a biomarker that indicates resistance to therapeutic drugs, it could greatly improve the prognosis of patients receiving advanced drug therapy. Serum exosomes found in patients with HCC have been found to inhibit apoptosis through the HGF/c-Met/Akt pathway. This leads to resistance to sorafenib, a common treatment for HCC (85). This demonstrates that circulating microRNAs play a role in the development of drug resistance. Studies have demonstrated that miR-31-5p, miR-221, miR-30e-3p, and miR-30d are linked to sorafenib resistance in hepatoma cells that are cultured in vitro (86, 87). High levels of miRNA-30E-3p in the bloodstream are frequently indicative of the onset of sorafenib resistance. However, the precise mechanism behind this phenomenon remains unclear. It is possible that sorafenib prompts TP53 to enhance extracytocrine, which could be a contributing factor. Targeting MDM2 primarily results in inhibiting tumor growth when TP53 is present, whereas targeting PTEN and p27 under TP53 regulation promotes tumor development (87). High levels of miRNA-518d-5p in serum correlate with survival time during treatment with sorafenib (61). The process in question may be linked to PUMA, which is the primary regulator involved in the apoptosis process. PUMA is mainly expressed in the mitochondria and works in conjunction with sorafenib to enhance tumor suppression (88). A clinical cohort study on sorafenib treatment revealed that miR-200c-3p serum levels were upregulated post-treatment. Furthermore, the study found that patients with HCC who had elevated levels of miR-200c-3p had a lower mortality rate. After sorafenib treatment, both miR-222-5p and miR-512-3p levels in circulation were found to be downregulated. Furthermore, the clinical survival data indicated that patients with a decreased level had a poorer outcome (89). Elevated levels of miRNA-10-3p in serum suggest that sorafenib treatment may be effective, while higher concentrations indicate that the drug is actively impacting tumor cells. This is supported by the fact that Cyclin E1, a promoter associated with sorafenib resistance, is a downstream target of miRNA-10-3p. These findings underscore the clinical relevance of miRNA-10-3p as a biomarker for predicting the efficacy of sorafenib treatment (90). By analyzing these serum markers, clinicians are able to assess the effectiveness of a patient’s drug treatment and ultimately enhance their prognosis.
4 Long non-coding RNA
Long noncoding RNA (lncRNA) is a type of linear RNA that exceeds 200 nucleotides in length. Unlike other RNA molecules, lncRNA does not encode proteins or peptides. However, its secondary and three-dimensional structure allows it to perform functions similar to both RNA and proteins (91). LncRNA is primarily found in the nucleus (92), but it can also operate in body fluids by being secreted through exosomes (93).
In comparison to microRNAs, lncRNAs exhibit a wider range of behavioral patterns, which have been validated in both pathophysiological and pathological contexts. It interferes with mRNA editing or other independent signaling pathways, affecting the transcription and translation of genes. (LncRNAs) have various functions, which can be categorized into three main types: cis-action, trans-action, and regulation of adjacent gene expression. Cis-acting LncRNAs can either activate or inhibit chromatin. Meanwhile, LncRNAs can affect the expression of adjacent genes by modulating the location of RNA polymerase II (94). It has been established that dysregulated lncRNAs can play a role in tumor formation by affecting various cellular processes, such as cell proliferation, migration, invasion, epithelial to mesenchymal transformation, apoptosis, and anti-tumor resistance (95, 96). Currently, there are studies that examine the relationship between the level of detection in circulation and the development and spread of tumors. Specifically, lncRNA has been found to play a role in the occurrence and development of hepatocellular carcinoma. Furthermore, early changes in lncRNA levels in circulation can serve as targets for early detection of tumors (97). LncRNA is utilized as a biomarker for tumors, which improves the sensitivity of diagnosing HCC and the prognosis of tumors.
4.1 lncRNA and HCC diagnosis
The lncRNA molecules NBAT-1 and FOXCUT have been found to have increased serum levels in patients with hepatitis C virus-induced hepatocellular carcinoma (HCC). Furthermore, higher levels of these molecules appear to be associated with more positive clinical outcomes in patients with HCC (98). The NBAT-1 gene is known to suppress tumors and is typically down-regulated in various types of cancer. Its primary function is to hinder cell proliferation and migration by controlling cancer-causing microRNAs and transcription factors (99). The reason for the upregulation of NBAT-1 in the serum of patients with HCC is primarily the body’s natural defense mechanism against the growth of tumors. FOXCUT is a tumor suppressor gene that plays a crucial role in inhibiting cell proliferation, migration, and invasion. It also induces cell cycle arrest and apoptosis. Moreover, the induction of FOXCUT significantly reduces the expression of matrix metalloproteinases (MMPs). These endopeptides are responsible for mediating extracellular matrix degradation, which promotes tumor growth (100). Based on the collation of data from the database, researchers have identified three lncRNAs, namely AC005332.5, ELF3-AS1, and LINC00665, that can serve as effective markers for the diagnosis of hepatocellular carcinoma. These lncRNAs have demonstrated promising responses in the model (101). In patients with HCC, the serum level of lncRNA SCARNA10 was found to be significantly higher than that in both the benign liver disease and control groups. Furthermore, there was a positive correlation between the level of lncRNA SCARNA10 and the degree of tumor malignancy. When combined with AFP, the ROC curve showed higher sensitivity (102). The biomarkers MIR4435-2HG and lnc-POLD3-2, as well as their combinations, have been demonstrated to be effective in distinguishing between hepatocellular carcinoma and non-hepatocellular carcinoma. The ROC curves indicate that these biomarkers have higher sensitivity in patients with normal alpha-fetoprotein levels (103). In a recent study (104), it was demonstrated that circulating lncRNA is consistent with the tumor microenvironment, and that levels of circulating lncRNA can reflect immune infiltration within tumors. The study also found that the expression of MIR4435-2HG and lnc-POLD3-2 in circulation could potentially serve as biomarkers for HCC (103). The lncRNA MIR4435-2HG plays a role in the induction of HCC through the WNT/β-catenin and transforming growth factor-β pathways (105). Its upregulation in serum levels is indicative of poor tumor biological behavior. The lncRNA HOTAIR (homeobox transcript antisense intergenic RNA) plays a crucial role in regulating epigenetic modifications among multiple genes. It is also implicated in the metastasis and drug resistance of various tumors (106). The lncRNA BRM (associated with Brahma) and lncRNA ICR (long noncoding RNA related to ICAM-1) are both found to be highly expressed in liver malignancies (107). Elevated levels of serum lncRNA HOTAIR, BRM, and ICR have been identified as accurate diagnostic markers for hepatocellular carcinoma (108). In patients with HCC, the concentration of LINC01793 in whole blood was significantly higher compared to healthy individuals or those with chronic hepatitis. The diagnostic efficacy was found to be highest when combined with AFP (109). The lncRNA cancer susceptibility candidate gene 2 (CASC2) acts as a tumor suppressor gene in endometrial cancer by being expressed in a down-regulated manner. On the other hand, the lncRNA taurine up-regulated gene 1 (TUG1) has been found to contribute to tumor formation in various types of cancers. In patients with hepatitis C-induced HCC, the expression of CASC2 was found to decrease, while the expression of TUG1 increased in healthy individuals (110). This suggests a potential antagonistic relationship between the two, although the exact mechanism remains unclear. The NEAT1 lncRNA is a significant constituent of parameningeal proteins and has been linked to various types of cancer. The analysis of the TCGA database revealed that NEAT1 is expressed at high levels in patients with hepatitis C virus-induced HCC. Furthermore, there was no significant decrease in serum NEAT1 levels after antiviral treatment (111). This suggests that NEAT1 may play a role in triggering liver malignancy and could serve as a useful biomarker for early HCC diagnosis.
4.2 lncRNA and HCC prognosis
In both HCC stem cells and mice, the expression of lncRNA-H19 increased as the disease progressed. The researchers also discovered that patients who exhibited high levels of lncRNA-H19 after treatment often experienced tumor recurrence (112). In their study, Huang et al. found that a recently identified lncRNA, RP11-85G21.1 (Lnc85), plays a significant role in promoting the proliferation and migration of hepatoma cells. This is achieved through its binding and regulation of miR-324-5p, ultimately impacting patient prognosis. The lncRNA CRNDE has been shown to increase the migration, activity, and invasion of hepatoma cells (113). In this study, we found that the expression level of lncRNA CRNDE in serum is positively correlated with tumor volume and negatively correlated with degree of differentiation. Furthermore, our analysis revealed that lncRNA CRNDE is an independent predictor of overall survival time in patients with HCC. Including lncRNA CRNDE as a marker can accurately predict the prognosis of HCC (114). LINC00161 is an oncogene that is involved in promoting the migration and invasion of hepatocellular carcinoma (HCC). LINC00161 has been identified as a biomarker for HCC in various studies. It has been found to have elevated levels in the serum of HCC patients. Additionally, LINC00161 has been linked to lower survival rates in HCC patients (115).
5 Conclusions and perspectives
Hepatocellular carcinoma is the primary form of liver cancer, with hepatitis viruses - particularly hepatitis B and C - being endemic in Asia. This has resulted in a high incidence and mortality rate of HCC in the region. Non-alcoholic fatty cirrhosis and alcoholic cirrhosis are both considered high-risk factors for HCC. The onset of HCC is often insidious, and the tumor is frequently discovered in its early stages. However, by the time a liver mass lesion is identified, the tumor may have already advanced and spread to other areas, such as distant metastasis or portal vein cancer thrombus. There is an urgent need for an effective biological marker that can detect and diagnose hepatocellular carcinoma in its early stages. Currently, the diagnostic criteria for HCC rely on imaging combined with serum marker AFP. However, imaging is not very effective in detecting early lesions, and the specificity and sensitivity of serum AFP are low. As a result, it is imperative to establish a more dependable diagnostic approach. Recent experiments have demonstrated the involvement of non-coding RNA in tumor development and progression. Additionally, non-coding RNA can be released into the bloodstream via tumor cell death or the circulation of exosomes containing tumor and blood cells. Thus, the significance of non-coding RNA in cancer diagnosis cannot be overstated. The non-invasive extraction of biomarkers is commonly referred to as “liquid biopsy”. In recent times, numerous studies have demonstrated the effectiveness of circulating microRNAs (such as miRNA-122, let-7 family, miRNA-34a, and other circulating microRNAs) and lncRNA in the early detection of HCC (Table 1). Additionally, many studies have suggested that combining circulating non-coding RNA with serum AFP can lead to even higher diagnostic efficiency. In the treatment of HCC, surgical radical resection and liver transplantation are considered the most effective methods. However, it is important to note that surgical treatment may have limited benefits for patients with advanced HCC. This has led to the development of alternative treatment options such as transarterial chemoembolization, radiofrequency ablation, and drug therapy for patients with HCC. Currently, targeted therapy is the primary drug treatment for HCC. However, the use of drugs can lead to the development of drug resistance in tumors. Prior research has examined the alteration of serum noncoding RNA levels in patients before and after surgery. By utilizing a specific biomarker to assess the patient’s condition following treatment, the prognosis for HCC treatment can be enhanced. In the context of TACE and radiofrequency therapy, circulating microRNA has been identified as a crucial factor in treatment evaluation and early postoperative recurrence. Similarly, in patients receiving sorafenib, certain molecules such as miR-31-5p, miR-221, miR-30e-3p, and miR-30 in circulation have been found to indicate drug resistance over time. Additionally, doctors can use lncRNA in serum to determine the prognosis of patients. In summary, HCC patients have a higher number of non-coding RNAs (ncRNAs) which are highly sensitive to HCC. These ncRNAs are secreted into the bloodstream, providing clinicians with valuable information about tumors in a minimally invasive manner. If this technology is applied in clinical settings, it has the potential to revolutionize the treatment strategy of HCC and significantly improve patient prognosis.
Author contributions
All authors participated in conception and design. JY wrote the manuscript. YX and YC revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Hong Kong Scholars Program (Grant No. XJ2020012); the National Natural Science Foundation of China (Grant No. 81902431); the Excellent Youth Project of Natural Science Foundation of Heilongjiang (Grant No. YQ2019H007); the Special Project of China Postdoctoral Science Foundation (Grant No. 2019T120279); the Special Project of Heilongjiang Postdoctoral Science Foundation (Grant No. LBHTZ1016); the China Postdoctoral Science Foundation (Grant No. 2018M641849 and 2018M640311); the Heilongjiang Postdoctoral Science Foundation (Grant No. LBH-Z18107 and LBH-Z18112). This work also received funding from Marshal Initiative Funding of Harbin Medical University (HMUMIF-22008); Natural Science Foundation of Heilongjiang Province (LH2023H043); Thematic Research Support Scheme of State Key Laboratory of Liver Research, The University of Hong Kong (SKLLR/TRSS/2022/08); Beijing Xisike Clinical Oncology Research Foundation (Y-Young2022-0188); Chen Xiaoping Foundation for the Development of Science and Technology of Hubei Province; Medjaden Academy & Research Foundation for Young Scientists (MJR20220903, MJR202309101); Opening Project of State Key Laboratory of Analytical Chemistry for Life Science (SKLACLS2302); Opening Project of State Key Laboratory of Chemical Oncogenomics; Opening Project of Key Laboratory of Basic Pharmacology of Ministry of Education, Zunyi Medicial University (2022-449); Opening Project of Key Laboratory of Gastrointestinal Cancer, Fujian Medical University, Ministry of Education (FMUGIC-202203); Opening Project of Key Laboratory of Environment and Health, Ministry of Education (2022GWKFJJ01); Opening Project of Key Laboratory of Tumor Immunology and Pathology, Army Medical University, Ministry of Education (2022jsz801); Opening Project of Anhui Province Key Laboratory of Translational Cancer Research, Bengbu Medical College (KFKT202301); Opening Project of Key Laboratory of Functional and Clinical Translational Medicine, Fujian Province University (XMMC-FCTM202205); Opening Project of Guangxi Laboratory of Enhanced Recovery after Surgery for Gastrointestinal Cancer (GXEKL202204); Opening Project of Key Laboratory of Biomarkers and In Vitro Diagnosis Translation of Zhejiang Province (KFJJ-2022002); Opening Project of Jiangsu Province Engineering Research Center of Tumor Targeted Nano Diagnostic and Therapeutic Materials (JETNM202210); Opening Project of Key Laboratory of Intelligent Pharmacy and Individualized Therapy of Huzhou & Changxing Anti-cancer Association (NZKF-20230203); Opening Project of Jiangsu Province Engineering Research Center of Tumor Targeted Nano Diagnostic and Therapeutic Materials (JETNM202210); and Opening Project of Key Laboratory of Intelligent Pharmacy and Individualized Therapy of Huzhou & Changxing Anti-cancer Association (NZKF-20230203).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The funding bodies did not play any role in writing the manuscript. The content is solely the responsibility of the authors.
References
1. Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol (2020) 72:250–61. doi: 10.1016/j.jhep.2019.08.025
2. Straś W, Małkowski P, Tronina O. Hepatocellular carcinoma in patients with non-alcoholic steatohepatitis - epidemiology, risk factors, clinical implications and treatment. Clin Exp Hepatol (2020) 6:170–5. doi: 10.5114/ceh.2020.99506
3. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology (2019) 156:477–491.e471. doi: 10.1053/j.gastro.2018.08.065
4. Shariff MI, Cox IJ, Gomaa AI, Khan SA. Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol (2009) 3:353–67. doi: 10.1586/egh.09.35
5. Feng M, Pan Y, Kong R, Shu S. Therapy of primary liver cancer. Innovation (Camb) (2020) 1:100032. doi: 10.1016/j.xinn.2020.100032
6. Luo G, Jin K, Deng S, Cheng H, Fan Z, Gong Y, et al. Roles of CA19-9 in pancreatic cancer: biomarker, predictor and promoter. Biochim Biophys Acta Rev Cancer (2021) 1875:188409. doi: 10.1016/j.bbcan.2020.188409
7. Liu Y, Chen J. Expression levels and clinical significance of serum miR-497, CEA, CA24-2, and HBsAg in patients with colorectal cancer. BioMed Res Int (2022) 2022:3541403. doi: 10.1155/2022/3541403
8. Viereck J, Thum T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res (2017) 120:381–99. doi: 10.1161/circresaha.116.308434
9. Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol (2021) 18:79–92. doi: 10.1038/s41585-020-00400-w
10. Mohapatra S, Pioppini C, Ozpolat B, Calin GA. Non-coding RNAs regulation of macrophage polarization in cancer. Mol Cancer (2021) 20:24. doi: 10.1186/s12943-021-01313-x
11. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov (2021) 20:629–51. doi: 10.1038/s41573-021-00219-z
12. Mirzaei S, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Ranjbar A, et al. Regulation of nuclear factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: inhibiting or promoting carcinogenesis? Cancer Lett (2021) 509:63–80. doi: 10.1016/j.canlet.2021.03.025
13. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
14. Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer (2015) 113:275–81. doi: 10.1038/bjc.2015.201
15. Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev (2015) 81:75–93. doi: 10.1016/j.addr.2014.09.001
16. Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol (2010) 64:123–41. doi: 10.1146/annurev.micro.112408.134243
17. Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut (2021) 70:784–95. doi: 10.1136/gutjnl-2020-322526
18. Usó M, Jantus-Lewintre E, Sirera R, Bremnes RM, Camps C. miRNA detection methods and clinical implications in lung cancer. Future Oncol (2014) 10:2279–92. doi: 10.2217/fon.14.93
19. Pascut D, Krmac H, Gilardi F, Patti R, Calligaris R, Crocè LS, et al. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci Rep (2019) 9:8265. doi: 10.1038/s41598-019-44580-x
20. Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PloS One (2012) 7:e46045. doi: 10.1371/journal.pone.0046045
21. Babayan A, Neumann MHD, Herdean A, Shaffer JM, Janning M, Kobus F, et al. Multicenter evaluation of independent high-throughput and RT-qPCR technologies for the development of analytical workflows for circulating miRNA analysis. Cancers (Basel) (2020) 12:1166. doi: 10.3390/cancers12051166
22. Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev (2015) 81:62–74. doi: 10.1016/j.addr.2014.10.029
23. Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol (2010) 6:402. doi: 10.1038/msb.2010.58
24. Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA (2010) 107:264–9. doi: 10.1073/pnas.0907904107
25. Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem (2008) 283:13205–15. doi: 10.1074/jbc.M707629200
26. Oura K, Morishita A, Masaki T. Molecular and functional roles of MicroRNAs in the progression of hepatocellular carcinoma-a review. Int J Mol Sci (2020) 21:8362. doi: 10.3390/ijms21218362
27. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
28. Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr (2020) 9:452–63. doi: 10.21037/hbsn-20-480
29. Harris PS, Hansen RM, Gray ME, Massoud OI, McGuire BM, Shoreibah MG. Hepatocellular carcinoma surveillance: an evidence-based approach. World J Gastroenterol (2019) 25:1550–9. doi: 10.3748/wjg.v25.i13.1550
30. Peng C, Ye Y, Wang Z, Guan L, Bao S, Li B, et al. Circulating microRNAs for the diagnosis of hepatocellular carcinoma. Dig Liver Dis (2019) 51:621–31. doi: 10.1016/j.dld.2018.12.011
31. El-Garem H, Ammer A, Shehab H, Shaker O, Anwer M, El-Akel W, et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis c related hepatocellular carcinoma. World J Hepatol (2014) 6:818–24. doi: 10.4254/wjh.v6.i11.818
32. Tsai YS, Huang CI, Tsai PC, Yeh ML, Huang CF, Hsieh MH, et al. Circulating let-7 family members as non-invasive biomarkers for predicting hepatocellular carcinoma risk after antiviral treatment among chronic hepatitis c patients. Cancers (Basel) (2022) 14:2023. doi: 10.3390/cancers14082023
33. Mattis AN, Song G, Hitchner K, Kim RY, Lee AY, Sharma AD, et al. A screen in mice uncovers repression of lipoprotein lipase by microRNA-29a as a mechanism for lipid distribution away from the liver. Hepatology (2015) 61:141–52. doi: 10.1002/hep.27379
34. Parizadeh SM, Jafarzadeh-Esfehani R, Ghandehari M, Goldani F, Parizadeh SMR, Hassanian SM, et al. MicroRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Curr Drug Targets (2019) 20:1129–40. doi: 10.2174/1389450120666190307095720
35. Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, et al. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology (2010) 52:1431–42. doi: 10.1002/hep.23818
36. Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X, et al. Hepatitis b virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol (2016) 64:278–91. doi: 10.1016/j.jhep.2015.09.013
37. Chun KH. Molecular targets and signaling pathways of microRNA-122 in hepatocellular carcinoma. Pharmaceutics (2022) 14:1380. doi: 10.3390/pharmaceutics14071380
38. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122–a key factor and therapeutic target in liver disease. J Hepatol (2015) 62:448–57. doi: 10.1016/j.jhep.2014.10.004
39. Ding LH, Fallgren CM, Yu Y, McCarthy M, Edmondson EF, Ullrich RL, et al. Orthologs of human circulating miRNAs associated with hepatocellular carcinoma are elevated in mouse plasma months before tumour detection. Sci Rep (2022) 12:10927. doi: 10.1038/s41598-022-15061-5
40. Zhao XF, Li N, Lin DD, Sun LB. Circulating MicroRNA-122 for the diagnosis of hepatocellular carcinoma: a meta-analysis. BioMed Res Int (2020) 2020:5353695. doi: 10.1155/2020/5353695
41. Franck M, Schütte K, Malfertheiner P, Link A. Prognostic value of serum microRNA-122 in hepatocellular carcinoma is dependent on coexisting clinical and laboratory factors. World J Gastroenterol (2020) 26:86–96. doi: 10.3748/wjg.v26.i1.86
42. Fang Y, Yan D, Wang L, Zhang J, He Q. Circulating microRNAs (miR-16, miR-22, miR-122) expression and early diagnosis of hepatocellular carcinoma. J Clin Lab Anal (2022) 36:e24541. doi: 10.1002/jcla.24541
43. Trung NT, Hoan NX, Trung PQ, Binh MT, Van Tong H, Toan NL, et al. Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci Rep (2020) 10:8181. doi: 10.1038/s41598-020-65213-8
44. Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA (2008) 105:3903–8. doi: 10.1073/pnas.0712321105
45. Tsai YS, Yeh ML, Tsai PC, Huang CI, Huang CF, Hsieh MH, et al. Clusters of circulating let-7 family tumor suppressors are associated with clinical characteristics of chronic hepatitis c. Int J Mol Sci (2020) 21:4945. doi: 10.3390/ijms21144945
46. Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, et al. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis c virus activity by targeting IGF2BP1. J Virol (2013) 87:9707–18. doi: 10.1128/jvi.00802-13
47. Matsuura K, De Giorgi V, Schechterly C, Wang RY, Farci P, Tanaka Y, et al. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis c. Hepatology (2016) 64:732–45. doi: 10.1002/hep.28660
48. Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, et al. Differential regulation of the let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol (2012) 188:6238–46. doi: 10.4049/jimmunol.1101196
49. Huang Y, Chen Y, Tu S, Zhang J, Qiu Y, Yu W. Diagnostic accuracy of circulating microRNAs for hepatitis c virus-associated hepatocellular carcinoma: a systematic review and meta-analysis. BMC Infect Dis (2022) 22:323. doi: 10.1186/s12879-022-07292-8
50. Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog (2009) 48:479–87. doi: 10.1002/mc.20484
51. Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang Y, et al. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res (2014) 74:6236–47. doi: 10.1158/0008-5472.Can-14-0855
52. Han R, Chen X, Li Y, Zhang S, Li R, Lu L. MicroRNA-34a suppresses aggressiveness of hepatocellular carcinoma by modulating E2F1, E2F3, and caspase-3. Cancer Manag Res (2019) 11:2963–76. doi: 10.2147/cmar.S202664
53. Ding N, Wu H, Tao T, Peng E. NEAT1 regulates cell proliferation and apoptosis of ovarian cancer by miR-34a-5p/BCL2. Onco Targets Ther (2017) 10:4905–15. doi: 10.2147/ott.S142446
54. Bharali D, Jebur HB, Baishya D, Kumar S, Sarma MP, Masroor M, et al. Expression analysis of serum microRNA-34a and microRNA-183 in hepatocellular carcinoma. Asian Pac J Cancer Prev (2018) 19:2561–8. doi: 10.22034/apjcp.2018.19.9.2561
55. Chen S, Mao Y, Chen W, Liu C, Wu H, Zhang J, et al. Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J Cancer (2022) 13:1410–7. doi: 10.7150/jca.57205
56. Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep (2019) 9:10464. doi: 10.1038/s41598-019-46872-8
57. Roy S, Hooiveld GJ, Seehawer M, Caruso S, Heinzmann F, Schneider AT, et al. microRNA 193a-5p regulates levels of nucleolar- and spindle-associated protein 1 to suppress hepatocarcinogenesis. Gastroenterology (2018) 155:1951–1966.e1926. doi: 10.1053/j.gastro.2018.08.032
58. Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, et al. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis b- and c-related-hepatocellular-carcinoma. World J Gastroenterol (2016) 22:3907–36. doi: 10.3748/wjg.v22.i15.3907
59. Loosen SH, Wirtz TH, Roy S, Vucur M, Castoldi M, Schneider AT, et al. Circulating levels of microRNA193a-5p predict outcome in early stage hepatocellular carcinoma. PloS One (2020) 15:e0239386. doi: 10.1371/journal.pone.0239386
60. Pratedrat P, Chuaypen N, Nimsamer P, Payungporn S, Pinjaroen N, Sirichindakul B, et al. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis b virus-related hepatocellular carcinoma. PloS One (2020) 15:e0232211. doi: 10.1371/journal.pone.0232211
61. Fernández-Tussy P, Rodríguez-Agudo R, Fernández-Ramos D, Barbier-Torres L, Zubiete-Franco I, Davalillo SL, et al. Anti-miR-518d-5p overcomes liver tumor cell death resistance through mitochondrial activity. Cell Death Dis (2021) 12:555. doi: 10.1038/s41419-021-03827-0
62. Loosen SH, Castoldi M, Jördens MS, Roy S, Vucur M, Kandler J, et al. Serum levels of circulating microRNA-107 are elevated in patients with early-stage HCC. PloS One (2021) 16:e0247917. doi: 10.1371/journal.pone.0247917
63. European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol (2018) 69:182–236. doi: 10.1016/j.jhep.2018.03.019
64. Tao S, Liang S, Zeng T, Yin D. Epigenetic modification-related mechanisms of hepatocellular carcinoma resistance to immune checkpoint inhibition. Front Immunol (2022) 13:1043667. doi: 10.3389/fimmu.2022.1043667
65. Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol (2015) 12:408–24. doi: 10.1038/nrclinonc.2015.103
66. Colombo F, Baldan F, Mazzucchelli S, Martin-Padura I, Marighetti P, Cattaneo A, et al. Evidence of distinct tumour-propagating cell populations with different properties in primary human hepatocellular carcinoma. PloS One (2011) 6:e21369. doi: 10.1371/journal.pone.0021369
67. Cui M, Wang H, Yao X, Zhang D, Xie Y, Cui R, et al. Circulating MicroRNAs in cancer: potential and challenge. Front Genet (2019) 10:626. doi: 10.3389/fgene.2019.00626
68. Ahmed M, Kumar G, Gourevitch S, Levchenko T, Galun E, Torchilin V, et al. Radiofrequency ablation (RFA)-induced systemic tumor growth can be reduced by suppression of resultant heat shock proteins. Int J Hyperthermia (2018) 34:934–42. doi: 10.1080/02656736.2018.1462535
69. Gao J, Wang SH, Ding XM, Sun WB, Li XL, Xin ZH, et al. Radiofrequency ablation for single hepatocellular carcinoma 3 cm or less as first-line treatment. World J Gastroenterol (2015) 21:5287–94. doi: 10.3748/wjg.v21.i17.5287
70. Li JK, Liu XH, Cui H, Xie XH. Radiofrequency ablation vs. surgical resection for resectable hepatocellular carcinoma: a systematic review and meta-analysis. Mol Clin Oncol (2020) 12:15–22. doi: 10.3892/mco.2019.1941
71. Canale M, Foschi FG, Andreone P, Ercolani G, Marisi G, Conti F, et al. Role of circulating microRNAs to predict hepatocellular carcinoma recurrence in patients treated with radiofrequency ablation or surgery. HPB (Oxford) (2022) 24:244–54. doi: 10.1016/j.hpb.2021.06.421
72. Li Y, Li P, Wang N. Effect of let-7c on the PI3K/Akt/FoxO signaling pathway in hepatocellular carcinoma. Oncol Lett (2021) 21:96. doi: 10.3892/ol.2020.12357
73. Jin B, Wang W, Meng XX, Du G, Li J, Zhang SZ, et al. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the wnt signaling pathway. BMC Cancer (2016) 16:863. doi: 10.1186/s12885-016-2904-y
74. Shi W, Zhang Z, Yang B, Guo H, Jing L, Liu T, et al. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Med (Baltimore) (2017) 96:e7764. doi: 10.1097/md.0000000000007764
75. Andrasina T, Juracek J, Zavadil J, Cechova B, Rohan T, Vesela P, et al. Thermal ablation and transarterial chemoembolization are characterized by changing dynamics of circulating MicroRNAs. J Vasc Interv Radiol (2021) 32:403–11. doi: 10.1016/j.jvir.2020.10.024
76. Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, et al. MiRNA-210: a current overview. Anticancer Res (2017) 37:6511–21. doi: 10.21873/anticanres.12107
77. O'Brien SJ, Carter JV, Burton JF, Oxford BG, Schmidt MN, Hallion JC, et al. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: a systematic review. Int J Cancer (2018) 142:2501–11. doi: 10.1002/ijc.31282
78. Llovet JM, Real MI, Montaña X, Planas R, Coll S, Aponte J, et al. Arterial embolisation or chemoembolisation versus symptomatic treatment in patients with unresectable hepatocellular carcinoma: a randomised controlled trial. Lancet (2002) 359:1734–9. doi: 10.1016/s0140-6736(02)08649-x
79. Gomes AS, Monteleone PA, Sayre JW, Finn RS, Sadeghi S, Tong MJ, et al. Comparison of triple-drug transcatheter arterial chemoembolization (TACE) with single-drug TACE using doxorubicin-eluting beads: long-term survival in 313 patients. AJR Am J Roentgenol (2017) 209:722–32. doi: 10.2214/ajr.17.18219
80. Ali HEA, Emam AA, Zeeneldin AA, Srour R, Tabashy R, El-Desouky ED, et al. Circulating miR-26a, miR-106b, miR-107 and miR-133b stratify hepatocellular carcinoma patients according to their response to transarterial chemoembolization. Clin Biochem (2019) 65:45–52. doi: 10.1016/j.clinbiochem.2019.01.002
81. Chen M, Li D, Gong N, Wu H, Su C, Xie C, et al. miR-133b down-regulates ABCC1 and enhances the sensitivity of CRC to anti-tumor drugs. Oncotarget (2017) 8:52983–94. doi: 10.18632/oncotarget.17677
82. Chen S, Jiao JW, Sun KX, Zong ZH, Zhao Y. MicroRNA-133b targets glutathione s-transferase π expression to increase ovarian cancer cell sensitivity to chemotherapy drugs. Drug Des Devel Ther (2015) 9:5225–35. doi: 10.2147/dddt.S87526
83. Kim SS, Cho HJ, Nam JS, Kim HJ, Kang DR, Won JH, et al. Plasma MicroRNA-21, 26a, and 29a-3p as predictive markers for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Korean Med Sci (2018) 33:e6. doi: 10.3346/jkms.2018.33.e6
84. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med (2020) 382:1894–905. doi: 10.1056/NEJMoa1915745
85. Qu Z, Wu J, Wu J, Luo D, Jiang C, Ding Y. Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res (2016) 35:159. doi: 10.1186/s13046-016-0430-z
86. He J, He J, Min L, He Y, Guan H, Wang J, et al. Extracellular vesicles transmitted miR-31-5p promotes sorafenib resistance by targeting MLH1 in renal cell carcinoma. Int J Cancer (2020) 146:1052–63. doi: 10.1002/ijc.32543
87. Gramantieri L, Pollutri D, Gagliardi M, Giovannini C, Quarta S, Ferracin M, et al. MiR-30e-3p influences tumor phenotype through MDM2/TP53 axis and predicts sorafenib resistance in hepatocellular carcinoma. Cancer Res (2020) 80:1720–34. doi: 10.1158/0008-5472.Can-19-0472
88. Dudgeon C, Peng R, Wang P, Sebastiani A, Yu J, Zhang L. Inhibiting oncogenic signaling by sorafenib activates PUMA via GSK3β and NF-κB to suppress tumor cell growth. Oncogene (2012) 31:4848–58. doi: 10.1038/onc.2011.644
89. de la Cruz-Ojeda P, Schmid T, Boix L, Moreno M, Sapena V, Praena-Fernández JM, et al. miR-200c-3p, miR-222-5p, and miR-512-3p constitute a biomarker signature of sorafenib effectiveness in advanced hepatocellular carcinoma. Cells (2022) 11:2673. doi: 10.3390/cells11172673
90. Shao YY, Chen PS, Lin LI, Lee BS, Ling A, Cheng AL, et al. Low miR-10b-3p associated with sorafenib resistance in hepatocellular carcinoma. Br J Cancer (2022) 126:1806–14. doi: 10.1038/s41416-022-01759-w
91. Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci (2013) 14:23672–84. doi: 10.3390/ijms141223672
92. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi: 10.1101/gr.132159.111
93. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell (2016) 29:653–68. doi: 10.1016/j.ccell.2016.03.004
94. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
95. Xu Y, Zhang X, Hu X, Zhou W, Zhang P, Zhang J, et al. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med (2018) 24:52. doi: 10.1186/s10020-018-0050-5
96. Liu M, Zhang H, Li Y, Wang R, Li Y, Zhang H, et al. HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer. Cancer Sci (2018) 109:2717–33. doi: 10.1111/cas.13745
97. Kim JU, Shariff MI, Crossey MM, Gomez-Romero M, Holmes E, Cox IJ, et al. Hepatocellular carcinoma: review of disease and tumor biomarkers. World J Hepatol (2016) 8:471–84. doi: 10.4254/wjh.v8.i10.471
98. Ali MA, Shaker OG, Ezzat EM, Eid HM, Ali DY, Hassan EA, et al. Serum lncRNAs, NBAT-1, and FOXCUT signature in hepatocellular carcinoma developed on top of chronic hepatitis c. Mol Carcinog (2023) 62:319–31. doi: 10.1002/mc.23488
99. Yang C, Wang G, Yang J, Wang L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am J Cancer Res (2017) 7:2009–19.
100. Wang X, Hu Y, Cui J, Zhou Y, Chen L. Coordinated targeting of MMP-2/MMP-9 by miR-296-3p/FOXCUT exerts tumor-suppressing effects in choroidal malignant melanoma. Mol Cell Biochem (2018) 445:25–33. doi: 10.1007/s11010-017-3248-x
101. Fu P, Gong B, Li H, Luo Q, Huang Z, Shan R, et al. Combined identification of three lncRNAs in serum as effective diagnostic and prognostic biomarkers for hepatitis b virus-related hepatocellular carcinoma. Int J Cancer (2022) 151:1824–34. doi: 10.1002/ijc.34201
102. Han Y, Jiang W, Wang Y, Zhao M, Li Y, Ren L. Serum long non-coding RNA SCARNA10 serves as a potential diagnostic biomarker for hepatocellular carcinoma. BMC Cancer (2022) 22:431. doi: 10.1186/s12885-022-09530-3
103. Kunadirek P, Pinjaroen N, Nookaew I, Tangkijvanich P, Chuaypen N. Transcriptomic analyses reveal long non-coding RNA in peripheral blood mononuclear cells as a novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. Int J Mol Sci (2022) 23:7882. doi: 10.3390/ijms23147882
104. Shaath H, Toor SM, Nair VS, Elkord E, Alajez NM. Transcriptomic analyses revealed systemic alterations in gene expression in circulation and tumor microenvironment of colorectal cancer patients. Cancers (Basel) (2019) 11:1994. doi: 10.3390/cancers11121994
105. Farzaneh Z, Vosough M, Agarwal T, Farzaneh M. Critical signaling pathways governing hepatocellular carcinoma behavior; small molecule-based approaches. Cancer Cell Int (2021) 21:208. doi: 10.1186/s12935-021-01924-w
106. Qu X, Alsager S, Zhuo Y, Shan B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett (2019) 454:90–7. doi: 10.1016/j.canlet.2019.04.016
107. Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun (2016) 7:13608. doi: 10.1038/ncomms13608
108. Lou ZH, Xu KY, Qiao L, Su XQ, Ou-Yang Y, Miao LB, et al. Diagnostic potential of the serum lncRNAs HOTAIR, BRM and ICR for hepatocellular carcinoma. Front Biosci (Landmark Ed) (2022) 27:264. doi: 10.31083/j.fbl2709264
109. Mo C, Wu J, Sui J, Deng Y, Li M, Cao Z, et al. Long non-coding RNA LINC01793 as a potential diagnostic biomarker of hepatitis b virus-related hepatocellular carcinoma. Clin Biochem (2022) 108:56–62. doi: 10.1016/j.clinbiochem.2022.06.006
110. Refai NS, Louka ML, Halim HY, Montasser I. Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: clinical significance. J Gene Med (2019) 21:e3112. doi: 10.1002/jgm.3112
111. Tripathi SK, Pal A, Ghosh S, Goel A, Aggarwal R, Banerjee S, et al. LncRNA NEAT1 regulates HCV-induced hepatocellular carcinoma by modulating the miR-9-BGH3 axis. J Gen Virol (2022) 103:10. doi: 10.1099/jgv.0.001809
112. Rojas Á., Gil-Gómez A, de la Cruz-Ojeda P, Muñoz-Hernández R, Sánchez-Torrijos Y, Gallego-Durán R, et al. Long non-coding RNA H19 as a biomarker for hepatocellular carcinoma. Liver Int (2022) 42:1410–22. doi: 10.1111/liv.15230
113. Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, et al. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis (2020) 11:676. doi: 10.1038/s41419-020-02853-8
114. Wang T, Zhu H, Xiao M, Zhou S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J Clin Lab Anal (2021) 35:e23959. doi: 10.1002/jcla.23959
Keywords: circulating noncoding RNA, hepatocellular carcinoma, biomarker, diagnosis, treatment
Citation: You J, Xia H, Huang Z, He R, Zhao X, Chen J, Liu S, Xu Y and Cui Y (2023) Research progress of circulating non-coding RNA in diagnosis and treatment of hepatocellular carcinoma. Front. Oncol. 13:1204715. doi: 10.3389/fonc.2023.1204715
Received: 12 April 2023; Accepted: 28 June 2023;
Published: 20 July 2023.
Edited by:
Amudha Ganapathy, University of Illinois Chicago, United StatesReviewed by:
Deyu Zhang, Second Military Medical University, ChinaDebanjali Dasgupta, Mayo Clinic, United States
Copyright © 2023 You, Xia, Huang, He, Zhao, Chen, Liu, Xu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunfu Cui, eWZjdWk3QDE2My5jb20=; Yi Xu, eHV5aWhtdUAxNjMuY29t
 Junqi You
Junqi You Haoming Xia
Haoming Xia Ziyue Huang
Ziyue Huang Risheng He1
Risheng He1 Yi Xu
Yi Xu Yunfu Cui
Yunfu Cui