- 1Institute for Urology and Reproductive Health, I.M. Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
- 2I.M. Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
- 3Department of Urology, The First Hospital of Jilin University (Lequn Branch), Changchun, Jilin, China
- 4Department of Radiology, The Second University Clinic, I.M. Sechenov First Moscow State Medical University (Sechenov University), Moscow, Russia
- 5Regional State Budgetary Health Care Institution, Kostroma Regional Clinical Hospital named after Korolev E.I. Avenue Mira, Kostroma, Russia
- 6School of Biomedical Engineering, Faculty of Medicine, Dalian University of Technology, Dalian, Liaoning, China
Multiparametric magnetic resonance imaging (mpMRI) has emerged as a first-line screening and diagnostic tool for prostate cancer, aiding in treatment selection and noninvasive radiotherapy guidance. However, the manual interpretation of MRI data is challenging and time-consuming, which may impact sensitivity and specificity. With recent technological advances, artificial intelligence (AI) in the form of computer-aided diagnosis (CAD) based on MRI data has been applied to prostate cancer diagnosis and treatment. Among AI techniques, deep learning involving convolutional neural networks contributes to detection, segmentation, scoring, grading, and prognostic evaluation of prostate cancer. CAD systems have automatic operation, rapid processing, and accuracy, incorporating multiple sequences of multiparametric MRI data of the prostate gland into the deep learning model. Thus, they have become a research direction of great interest, especially in smart healthcare. This review highlights the current progress of deep learning technology in MRI-based diagnosis and treatment of prostate cancer. The key elements of deep learning-based MRI image processing in CAD systems and radiotherapy of prostate cancer are briefly described, making it understandable not only for radiologists but also for general physicians without specialized imaging interpretation training. Deep learning technology enables lesion identification, detection, and segmentation, grading and scoring of prostate cancer, and prediction of postoperative recurrence and prognostic outcomes. The diagnostic accuracy of deep learning can be improved by optimizing models and algorithms, expanding medical database resources, and combining multi-omics data and comprehensive analysis of various morphological data. Deep learning has the potential to become the key diagnostic method in prostate cancer diagnosis and treatment in the future.
1 Introduction
Prostate cancer (PCa) is a commonly occurring urological malignancy among middle-aged and older men, with its incidence on the rise. In 2020, there were approximately 1.4 million new PCa cases reported globally, resulting in around 375,000 deaths, making it the second most common cancer among men, following lung cancer, and the fifth leading cause of cancer-related deaths among men (1). Although digital rectal examination and prostate-specific antigen (PSA) test are routinely conducted for the diagnosis of PCa, transrectal ultrasound (TRUS) has been the primary imaging technique for clinical suspicion and diagnosis of PCa in the past (2, 3). However, due to its low sensitivity and specificity, particularly for detecting lesions present in the transitional zone (TZ), mpMRI has replaced TRUS as the first-line radiological screening modality for clinical suspicion of PCa (4–6). Compared with other imaging examinations, MRI of the prostate provides a higher soft-tissue resolution and multiple imaging data parameters non-invasively, which facilitates better understanding of the complete prostate gland and its relationship with the surrounding environment and also provides improved guidance for PCa staging (7, 8). Therefore, MRI has become the preferable imaging tool for patients with suspected PCa or those at risk of PCa (9–11). The prostate imaging-reporting and data system (PI-RADS) provides a comprehensive set of standards for scanning, interpreting, and reporting mpMRI (12). Combining mpMRI with PI-RADS scaling results in more precise PCa diagnosis and staging, as well as improved guidance for later biopsies, and has contributed significantly to reducing overdiagnosis (13–15). Although mpMRI is a valuable technique in PCa diagnosis, manual interpretation of mpMRI data is complex, time-consuming, and challenging due to low sensitivity and specificity of the interpreting results (16–18). Deep learning (DL) technology has the capability to mine various features from medical images that are difficult to identify and distinguish using the naked eye in the macroscopic view (19, 20). DL technology can guide clinicians in medical diagnosis and help reduce diagnostic accuracy issues caused by factors mentioned above, providing physicians with accurate disease information (21, 22). Over the past few years, several computer-aided diagnosis (CAD) systems have been applied to PCa diagnosis with positive outcomes (23–26). CAD systems can be classified into two categories: computer-aided detection (CADe) and computer-aided diagnosis (CADx) (27, 28). CADe can determine if a patient has PCa and localize the possible PCa lesion based on the entire mpMRI data. CADx can evaluate a series of manually or automatically selected tumor-suspected areas by radiologists or CADe systems, followed by assessing and evaluating the aggressiveness of PCa (24, 29–31). This review presents a cross-disciplinary summary of research progress in PCa using DL-based CADs to make the artificial intelligence (AI) process understandable to not only radiologists but also general physicians who lack systematic and specialized imaging interpretation training. We briefly describe the application of DL techniques based on prostate MRI data and provide possible research ideas for future studies.
2 Overview of deep learning techniques
According to the latest Prostate Imaging-Reporting and Data System Version 2.1 (PI-RADS 2.1) recommendations, mpMRI image sequences for PCa detection and diagnosis typically consist of T2-weighted imaging (T2W), diffusion-weighted imaging (DWI), average diffusion coefficient (ADC) maps, and dynamic contrast-enhanced (DCE) imaging (12). DWI and ADC sequences are primarily employed to detect peripheral zone lesions, while T2W focuses on detecting transition zone lesions (32, 33). The PI-RADS score, calculated from mpMRI data, ranges from 1 (low likelihood of clinically significant PCa) to 5 (high likelihood of clinically significant PCa) and serves as a crucial diagnostic measure to determine the necessity of a biopsy (34, 35). Due to its significant clinical value, PI-RADS recommendations have been updated for standardizing prostate MRI scanning and interpretation processes. However, accurately interpreting mpMRI data requires a high level of expertise and skill. Furthermore, inter-observer and intra-observer variability values, which pertain to different radiologists interpreting the same MRI results and a single radiologist interpreting the same MRI results multiple times, respectively, tend to exhibit high variability (36, 37). This affects the broader utilization of mpMRI in PCa diagnosis. Consequently, to reduce interpretation time, enhance image interpretation quality, and minimize the risk of overtreatment, DL has emerged as the predominant AI method within machine learning (ML) technology (38, 39). Inspired by human learning patterns, ML can be broadly categorized into supervised learning, which utilizes well-labeled training data examples with fully controlled data input and output, and unsupervised learning, which operates on unlabeled datasets and aims to identify correlations within the dataset (38–40). Semi-supervised learning, a hybrid mode between supervised and unsupervised learning, uses partially labeled training data while the remainder stays unlabeled (39, 41, 42). Another renowned learning framework, reinforcement learning, obtains feedback based on each action’s response in the environment, modifying model parameters to maximize anticipated benefits (39, 43). DL technology, introduced by Hinton in 2006 (44), is an ML subset sharing similar working principles but featuring an advanced multilayer neural network that mimics human biological neural networks for data representation learning (39, 45). A key distinction between ML and DL lies in feature extraction methods, where conventional ML relies on hand-crafted approaches by expert specialists, and DL automatically extracts features within network layers (45, 46). As hand-crafted feature design demands significant effort and considerable workload, a growing number of DL algorithm (DLA) are emerging, tending to replace ML in medical image processing (20, 45, 47). However, hand-crafted approaches persist in situations with limited annotated data, and several studies have demonstrated promising results by fusing models with classic hand-crafted features and DL-extracted features (46, 48, 49). Typically, the majority of CAD systems for PCa involve processing steps such as imaging alignment, prostate localization and segmentation, feature-based lesion detection, and task-based classification (16, 24). With the increasing volume of labeled imaging data, supervised learning models represented by convolutional neural networks (CNNs) and non-supervised learning frameworks, including generative adversarial networks (GANs), have been incorporated into various medical imaging processes (50–53). In general, DLA-based analysis constitutes a new form of computer-aided diagnosis that facilitates the automatic acquisition of data-related features and identification of lesions without the requirement of manual segmentation, provided that the dataset is sufficiently large. This approach is innovative in that it allows for spontaneous learning of features, resulting in improved efficiency and accuracy of lesion detection (16, 20, 45, 54) (Figure 1).
Figure 1 Stratification of artificial intelligence. Created with BioRender.com.
2.1 Main working principles of DLA (Figure 2A)
In summary, within the medical imaging domain, DLAs perform tasks such as image classification, object detection, and semantic segmentation. Image classification determines benignity or malignancy, tumor types, grading, and staging from medical images; object detection localizes tumors and extracts their information from images; and semantic segmentation outlines tumors or adjacent organs in the images (54–57). Among the various DL-based models, deep CNNs have garnered significant attention due to their promising performance in medical imaging (52, 58). The multilayer neural network structure of DLAs, inspired by the human visual system, has the potential to process convolution operations (38, 52, 59). CNN structures primarily consist of a convolutional layer, max-pooling layer, and fully connected layer (20, 38, 58–61). These layers pertain to specific calculation methods or functions that receive, compute, and output relevant data. The convolution layer, the core of CNNs, extracts image features by constructing multiple convolution kernels (60). The max-pooling layer, also known as the down-sampling layer, reduces computational effort by consolidating data within a certain range. The fully connected layer, used as a classifier, integrates all local information acquired from the previous max-pooling or convolutional layer that is class-distinctive, ultimately producing the desired class predictions (58, 61, 62). In brief, the convolutional layer of a CNN functions as a feature extractor, while the fully connected layer serves as a classifier (63) (Figure 2B). The trained CNN forms its network structure and weight files, which are the foundation for predicting the same type of unknown data. Function-dependent networks based on CNN have been designed for specific computer vision tasks, such as AlexNet and ResNet for image classification (64, 65), YOLO and Faster R-CNN for object detection (66, 67), and U-Net and Mask R-CNN for semantic segmentation (58, 61, 62, 68). Another valuable deep learning network, the GAN, has demonstrated effectiveness for semi-supervised learning (69), supervised learning (70), and reinforcement learning (71), despite its initial proposal for unsupervised learning. GAN can be simply described as a deep learning model used to create alternative imaging data similar to the target data, but with improved quality and reduced noise (51, 53, 72). GAN primarily consists of two separate but interdependent neural networks, functioning as the generator and the discriminator. Generated data (false) from the generator, using random variables as input, or target data (true) are then input into the discriminator (53, 72). These two networks are trained competitively and adversarially, with the aim of making the discriminator model strictly capable of distinguishing between the synthesized data generated by the generator and the true data, while the generator intends to create data as realistic as possible compared to the target data (53, 72, 73). The primary objective of GAN is to render the discriminator network incapable of differentiating between the output data generated by the generator network and real data (Figure 2C). More recently, deep convolutional GANs (DCGANs) have emerged by combining CNNs and GANs to achieve better performance and effectiveness, resulting in their increasing popularity for designing various computer-aided diagnosis (CADx) models (53, 74–76).
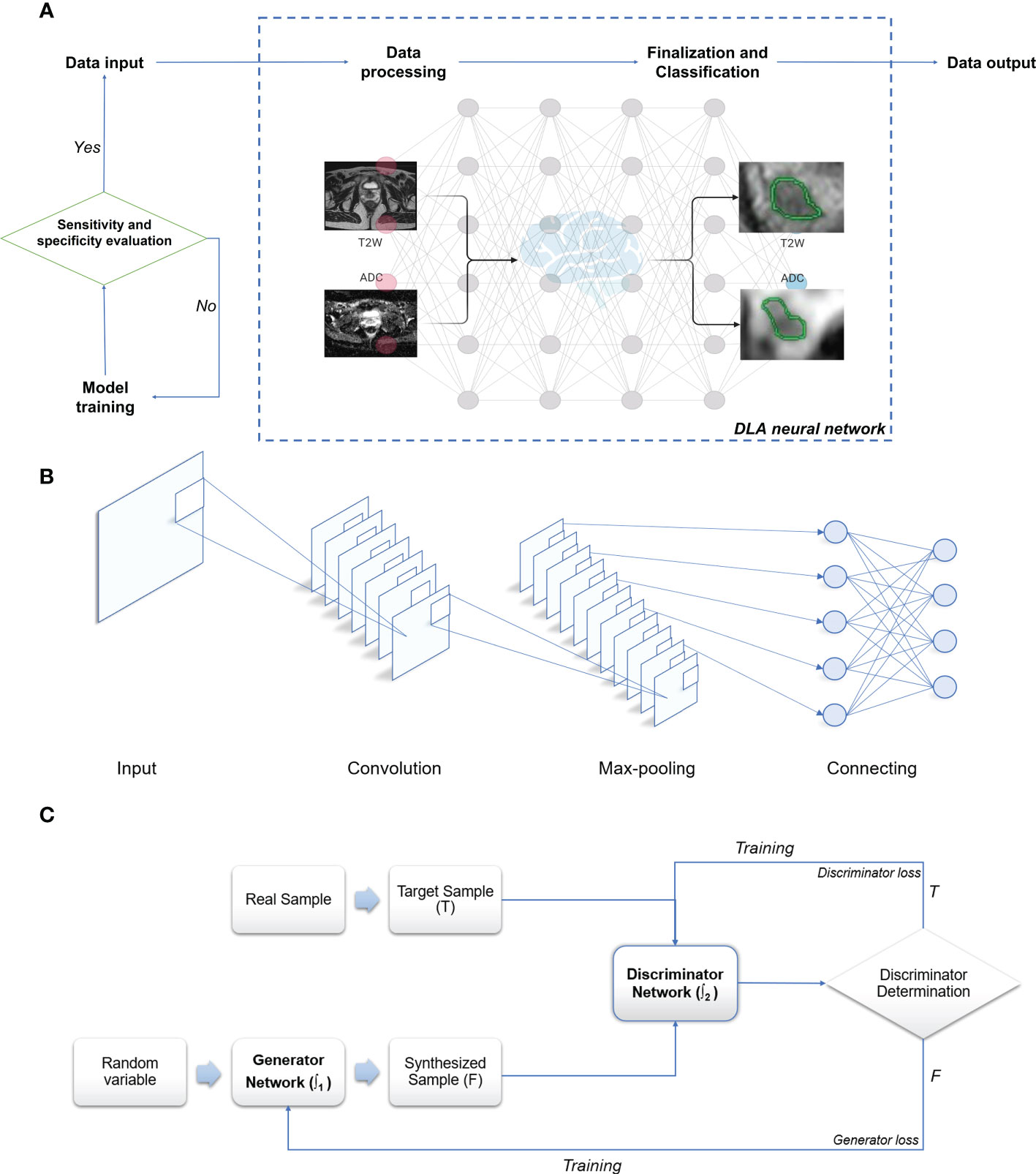
Figure 2 Schematic illustration of deep learning algorithm. (A) Working algorithm of image processing. (B) Principle and architecture of CNN. (C) Principle and architecture of GAN: T, True data; F, False data; ∫1 - Function 1 of the generator network; ∫2 - Function 2 of the discriminator network. Created with BioRender.com.
2.2 Evaluation metrics of DLA
Evaluation metrics for DLA in medical imaging applications encompass accuracy, specificity, sensitivity, dice similarity coefficient (DSC), Jaccard index, receiver operating characteristic (ROC) curve, and area under the ROC curve (AUC) (77–81). While general clinicians need not fully grasp the complex equations and processes involved in these evaluation metrics, a basic understanding of their core principles is crucial for accurately interpreting the performance of pertinent DL models. Sensitivity denotes the likelihood of detecting a positive sample within a positive population (1.2.1), while specificity refers to the probability of identifying a negative result in a negative population (1.2.2) (77). DSC (1.2.3) and the Jaccard index are promising evaluation metrics for assessing segmentation quality. They are typically employed to calculate the similarity between two samples, with values ranging from 0 to 1. A value closer to 1 indicates a better model performance (78, 82). The ROC curve is used to evaluate the diagnostic accuracy and performance of various models. A curve closer to the upper left corner signifies higher diagnostic value, and a larger AUC corresponds to greater application value (79–81). AUC serves as a criterion for determining the quality of classification models, referring to the likelihood of positive examples ranking higher than negative examples in prediction outcomes. An AUC between 0.5 and 1 implies that the model possesses predictive value, and a value closer to 1 signifies superior model performance (79–81).
3 Application of mpMRI-based DLA on PCa
3.1 Diagnosis of PCa
3.1.1 Detection and classification of PCa
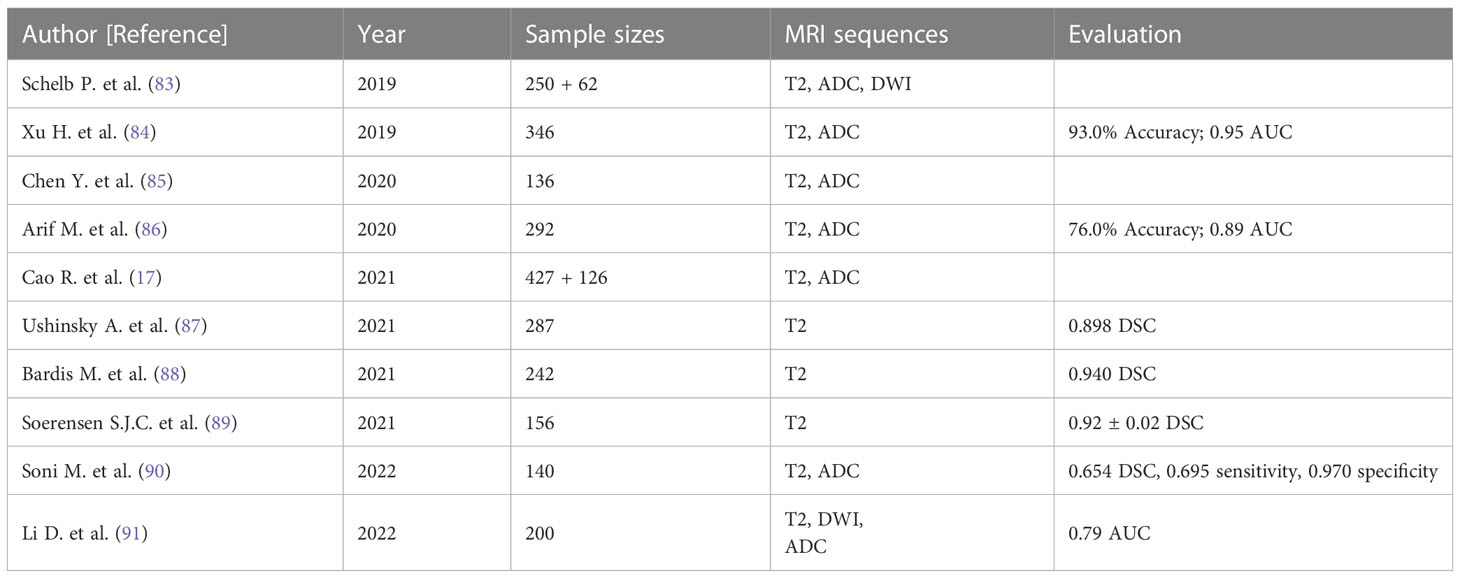
In the clinical management of PCa, accurately distinguishing between low-risk and high-risk cases is crucial to prevent overdiagnosis or delayed treatment (92). For patients with low-risk PCa, mpMRI serves as the primary imaging technique to determine if the lesion has grown or metastasized and to assess disease progression during active surveillance (93). Therefore, a reliable noninvasive assessment system is of significant importance. Fusco et al. (94) performed a systematic literature review, reporting that MRI holds considerable clinical value in localizing and staging PCa. Vente et al. (95) developed a multitasking U-Net model using T2W and DWI sequences of MRI, capable of simultaneously detecting and grading PCa with excellent diagnostic outcomes. Wang et al. (96) designed an end-to-end CNN comprising two sub-networks: one for aligning apparent DWI and T2W, and the other as a convolutional neural classification network. The end-to-end CNN model was trained and assessed on 360 patients using a fivefold cross-validation method, ultimately exhibiting a sensitivity of 0.89 for identifying high-risk PCa cases. Ishioka et al. (97) developed a fully automated PCa detection system using patients’ T2W sequence data, combining two distinct algorithms and demonstrating an AUC of 0.793. Wang et al. (98) compared the detection capabilities of DLAs and non-DLAs in differentiating PCa, using T2W sequences from prostate MRI findings of 172 patients, which included 79 patients with PCa and 93 with benign prostatic hyperplasia (BPH). The final ROC curve value was 0.84 for the DL model, compared to 0.70 for the non-DL model. Sanford et al. (99) conducted PI-RADS scoring with a CNN trained on T2W/ADC/high-b values, confirming that DLAs possess a PCa assessment potential comparable to clinical PI-RADS scoring. Yang et al. (31) collected T2W and DWI sequences from prostate MRI findings of 160 patients and built two parallel deep CNNs. The final features extracted by these two CNNs were input into a classifier based on the support vector machine algorithm, ultimately achieving spontaneous identification of PCa (Table 1).
3.1.2 Segmentation of the prostate gland
The clinical measurement of prostate-specific antigen density (PSA-D) is closely related to prostate volume (PV), and PSA-D serves as an indicator of prostate cancer (PCa), with higher PSA-D values suggesting a greater likelihood of clinically significant PCa (100–102). PV is employed to diagnose BPH in clinical settings and assists urologists in selecting suitable surgical procedures and medication strategies for BPH patients (103–105). TRUS is the most common imaging method for calculating PV in clinical practice (106), but it is susceptible to significant measurement errors when the prostate has an irregular shape. Computing PV based on pixel size and layer thickness, in which the prostate gland is segmented on each MRI image, may be more accurate. In a clinical setting, determinization of the type of surgery, such as prostate tissue-preserving surgery and fascial-sparing surgery requires precise differentiation of prostate gland boundaries. Preservation of the neurovascular bundle for performing the nerve-sparing radical prostatectomy (107) to save the erectile function and sparing of the pelvic fascia for fascial-sparing radical prostatectomy (108) to prevent positive surgical margins followed by high risk of clinical recurrences rely on preoperative imaging guidance. In the case of PCa radiotherapy (discussed in more detail later), precise MRI-guided segmentation in radiotherapy significantly improves target accuracy, effectively prevents damage to normal prostate tissues surrounding the tumor, and reduces toxic side effects (109). Hence, accurate, robust, and efficient MRI-guided segmentation of the prostate gland is crucial for evaluating PCa tumors, calculating PV, selecting surgical options for prostate abnormalities, outlining target areas for radiation planning, and monitoring progressive changes in tumor lesions. However, due to heterogeneity in MRI imaging quality and signal intensity, as well as interference from periprostatic tissues and organs like the bladder or rectum, prostate segmentation remains highly challenging (110, 111). Applying DL for accurate prostate gland segmentation on MRI images could facilitate more precise and easy determination of PV and prostate boundaries. CNNs do not require complex feature extraction and are widely utilized for medical image segmentation (112, 113). Zhu et al. (114) developed a three-dimensional (3D) deep learning model containing dense blocks to segment the prostate gland. The 3D structures enable the network to fully exploit the relationship between adjacent images, and the dense blocks make complete use of both shallow and deep information, achieving a DSC of 0.82. Yan et al. (115) proposed a backpropagation neural network that integrates the optimal combination selected from multi-level feature extraction into a single model for prostate MRI image segmentation, achieving a DSC of 0.84, an average increase of 3.19% compared to traditional ML segmentation algorithms based on random forests. To et al. (116) segmented MRI images and identified PCa using a 3D deep dense multipath CNN constructed from T2W and ADC sequences, achieving DSCs of 0.95 and 0.89 in two independent test sets, respectively. Dai et al. (117) developed a mask region-based CNN for prostate gland and intraprostatic lesion segmentation, showing that this end-to-end DL model could automatically segment the prostate gland and identify suspicious lesions directly from prostate MRI images without manual intervention, demonstrating its potential to guide clinicians in tumor delineation.
3.2 Advanced radiotherapy of PCa
Radiotherapy is a vital component of PCa treatment and relies on a complex series of multimodal medical imaging techniques, such as computed tomography (CT), MRI, cone-beam CT, and positron emission tomography, to localize tumors, establish radiotherapy treatment plans, and assess radiotherapy efficacy (118). Radiotherapy is an indispensable treatment modality for cancer patients, either as neoadjuvant or postoperative therapy, in combination with chemotherapy (119, 120). The main goal of radiotherapy is to maximize the therapeutic gain ratio by delivering an effective radiation dose to the planned target volume (PTV) and avoiding unnecessary radiation exposure to adjacent healthy tissues and organs at risk (OARs) (121, 122). However, manual segmentation of the prostate gland, which is necessary for accurate mapping of PTV and OARs, is prone to errors and can result in less accurate and sensitive outcomes than those desired clinically. In addition, respiratory movements, setup errors, and fluctuations in body weight can lead to displacement of PTV and OARs, potentially resulting in under-measurement of the radiation dose received by the PTV or over-measurement of the radiation dose delivered to OARs (123). To achieve precise tumor localization and appropriate treatment, continuous technological advances have led to the development of precision therapy, such as intensity-modulated radiotherapy (IMRT) and 3D conformal radiation therapy, which aim to provide personalized, precise anticancer treatment by setting an appropriate radiation dose according to the tumor shape while avoiding radiation exposure to OARs as much as possible (124–126). Although precision therapy has improved the accuracy of radiotherapy to some extent, further optimization is necessary to achieve the desired efficacy. Therefore, image-guided adaptive radiotherapy (ART) has emerged as a potential solution to overcome PTV and OAR displacement caused by various factors (123, 127, 128).
3.2.1 ART technology
ART technology allows for systematic monitoring of target lesions and changes in adjacent tissues based on imaging features to optimize radiotherapy plans further (123, 128, 129). ART enables the acquisition of feedback and tracking of target area-related information primarily through offline, online, and real-time modes (129, 130). For instance, offline ART involves measuring setup errors on MRI images obtained during the patient’s initial few treatments, after which the clinical target volume (CTV) coverage is adjusted, and both the dose and treatment plan for subsequent fractions are modified (130, 131). Online ART calculates the necessary data based on the patient’s anatomical imaging information acquired at the time, allowing for modifications to the radiotherapy plan that are directly applied to the current treatment (129–131). Real-time ART involves intra-fraction and inter-beam reprogramming and automatic adjustment of the radiotherapy plan during treatment execution based on dynamic tracking of radiation dose and anatomical details of the target area without manual intervention (129–131). Since the anatomical and geometric variations of PCa are influenced by the degree of bladder and rectal filling, the morphology, location, and volume of PTV and OARs may differ between treatments. Consequently, offline ART is not flexible enough to accommodate these changes (131). While CTV expansion is often used clinically to compensate for these limitations, it can result in increased post-radiotherapy toxicities (132). Although online ART offers improved accuracy compared to offline ART, its time-consuming nature limits its clinical application to some extent. Real-time ART overcomes the drawbacks of both offline and online ART and has been implemented in clinical practice (133), but its safety and robustness require validation due to the lack of a sufficiently comprehensive database for model adaptation and training (134). As mentioned in Section 3.1.2, significant progress has been made in DL-based automatic segmentation of the prostate gland. However, research on developing subsequent radiotherapy systems remains underexplored. Developing an accurate and efficient automated radiotherapy delivery system using DL technology to enhance radiotherapy outcomes has far-reaching clinical implications. Sprouts et al. (135) developed a virtual treatment planner (VTP) based on deep reinforcement learning (DRL) to implement a treatment planning system. The VTP, based on the Q-learning framework, was evaluated using 50 samples, achieving a mean ProKnow plan score of 8.14 ± 1.27 (standard deviation), indicating its potential for IMRT planning in PCa. The application of the conventional ϵ-greedy algorithm for training VTP is time-consuming, restricting its clinical use. Shen et al. (136) introduced a knowledge-guided DRL to adjust treatment plan parameters to enhance VTP training efficiency, achieving a plan quality score of 8.82 ( ± 0.29). Lempart et al. (137) proposed a densely connected DL model based on a modified U-Net, trained on a triplet of 160 patients to predict dose distribution for volumetric-modulated arc therapy. The model maintained the mean percentage error within 1.9% for both CTV and PTV and within 2.6% for OAR, demonstrating its capacity to partially automate the radiotherapy planning process and accelerate treatment progress.
3.2.2 MRI-only radiotherapy
Although MRI offers excellent soft-tissue contrast and facilitates relatively precise tumor segmentation, it does not provide the electron density map or Hounsfield units needed for radiation dose calculation. Consequently, it is essential to map relative regions such as CTV, PTV, and OARs on MRI, after which the outlined contours are mapped to CT via image alignment for clinical radiotherapy planning (134, 138, 139). Combining MRI and CT simulations in PCa radiotherapy plans may reduce acute urogenital toxicity (140). However, the labor-intensive process of CT and MRI alignment and the challenges in achieving full alignment can result in systematic errors, potentially leading to dose distribution issues in the target region and diminishing radiotherapy effectiveness (141). To address these problems, recently developed MRI simulators enable the conversion of MRI data to synthetic CT (sCT), allowing radiation dose measurements to directly contribute to radiotherapy planning and establishing MRI-only radiotherapy (138, 139, 142) (Figure 3). Common approaches for converting MRI data to sCT include bulk density assignment–based methods, voxel-based methods, and atlas-based methods (143, 144). Currently, a bulk density assignment–based system called magnetic resonance for calculating attenuation (MRCAT™ by Philips) and an atlas-based system called MriPlanner™ (by Spectronic Medical) have been employed for automatic generation of pelvic sCT in clinical practice (145, 146). MriPlanner™ has demonstrated promising performance, as evaluated in the MR-OPERA and MR-PROTECT studies (147, 148). In contrast to sCT generation methods, MRCAT™, due to its bulk density assignment–based nature, requires multiple MRI sequences, such as air, liquid, and bone, each assigned to corresponding electron density or Hounsfield unit values necessary for creating a CT image (144, 149) (Table 2). As illustrated previously, the conventional approach to radiotherapy planning, which involves the use of both CT and MRI, necessitates manual intervention for aligning and fusing CT and MRI images, as well as determining the CTV, PTV, and OARs. This manual intervention considerably reduces both accuracy and efficiency. However, by employing DL, which automatically extracts informative features from a large number of training samples to establish a nonlinear mapping from MRI to CT (150), a trained model can swiftly generate highly precise synthetic CT (sCT) images in just a few seconds. These sCT images provide more accurate guidance for ART (151). Fu et al. (152) utilized two-dimensional (2D) and three-dimensional (3D) fully connected CNN based on U-Net to generate pelvic region sCT, with results indicating that accurate sCT was effectively executed using DLA. Conditional GAN, developed by adding a discriminator to U-Net, enable the generated sCT to provide more details, enhancing sCT accuracy and robustness and allowing for more precise radiotherapy planning (153, 154). CycleGAN (cGAN) is a modified adversarial network based on GAN, with additional generators and discriminators incorporated for improved unpaired training data (155, 156). Liu et al. (157) proposed a multi-CycleGAN network and designed a new generator, Z-Net, to improve anatomical details. This approach exhibited lower mean error and mean absolute error and higher dose accuracy of the sCT (Table 3).
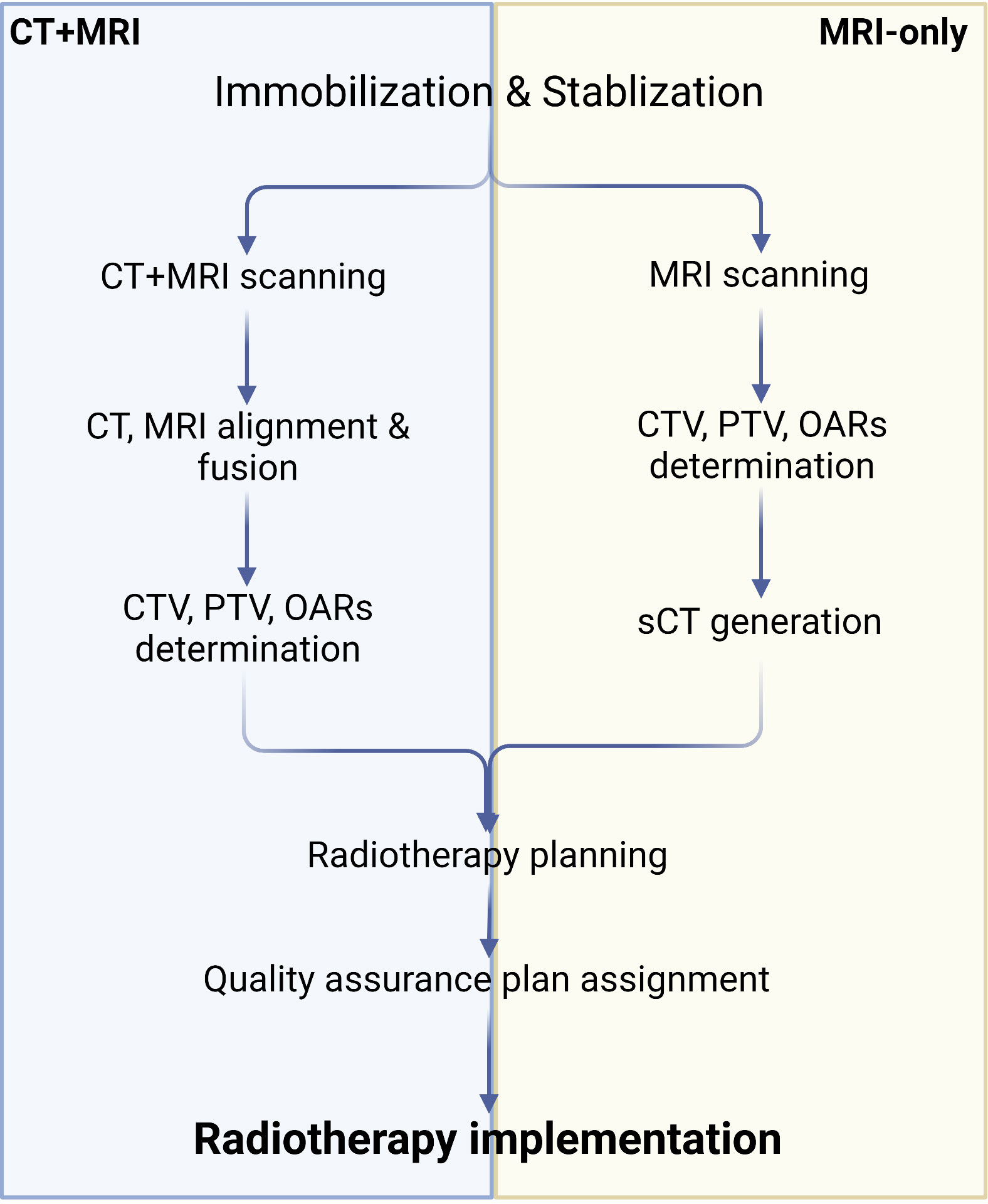
Figure 3 Algorithm of CT+MRI and MRI-only radiotherapy technique. Created with BioRender.com.
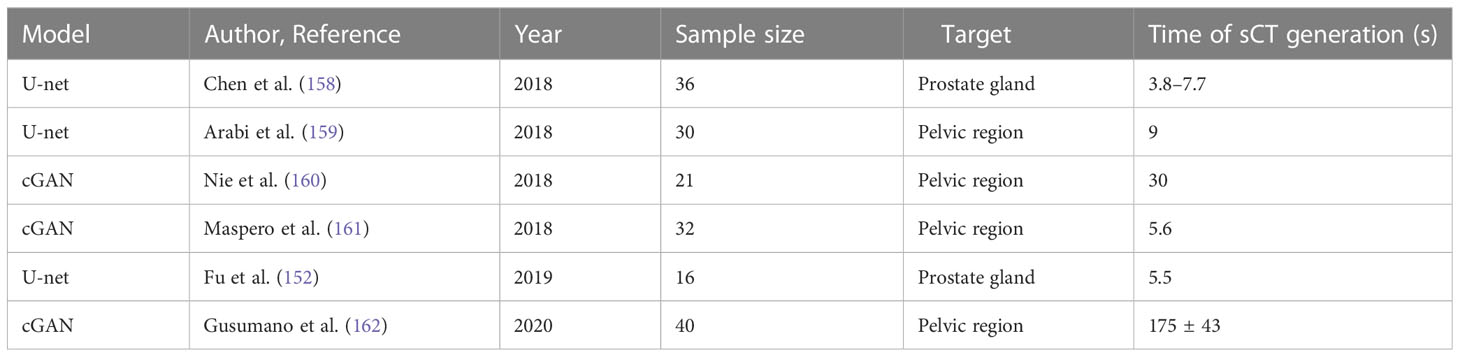
Table 3 Recent designed models of deep learning–based synthetic CT generation in the prostate gland or pelvic region.
3.3 Prognostic assessment of PCa
To improve prognosis monitoring of PCa and reduce mortality, it is essential to consider patients at low risk during active surveillance and those who have undergone radical prostatectomy (163). The European Association of Urology guidelines widely recognize PSA as the primary metric for assessing BCR in clinical practice (164). To mitigate diagnostic bias, leveraging the precise anatomical information provided by MRI is invaluable, as it offers non-invasive insights. This is particularly crucial since PSA levels can fluctuate and be influenced by various factors (165, 166). Furthermore, the role of mpMRI in assessing PCa recurrence has gained importance (167), underscoring the need for comprehensive investigation into MRI-based PCa recurrence prediction. Yan et al. (168) conducted a multicenter study using a DL technique and a novel model called deep radiomic signature for BCR prediction. They combined quantitative features and radiomics extracted from prostate MRI with DL-based survival analysis. The performance of the model was evaluated using data from approximately 600 patients who underwent radical prostatectomy, achieving maximum AUC values of 0.85 and 0.88 for BCR-free survival prediction at 3 years and 5 years, respectively. In addition to recurrence prediction, there should be a significant focus on monitoring metastasis, particularly considering the high occurrence rate of bone metastases in over 80% of patients with advanced PCa (169). The accuracy and sensitivity of conventional bone scintigraphy for detecting skeletal metastases have been questioned (170). Therefore, of the potential in detecting earlier PCa metastasis using PSMA PET-CT and MRI has been established (170, 171). As part of a routine radiological examination for suspected PCa, Liu et al. successfully detected and segmented pelvic bone metastases using dual 3D U-net DLAs rely on T1-weighted imaging and diffusion-weighted imaging sequences (172). Through two rounds of evaluation, they achieved a mean DSC value above 0.85 for pelvic bone segmentation and a maximum AUC of 0.85 for metastasis detection, demonstrating accurate detection and segmentation of pelvic bone metastases.
4 Discussion
Prostate MRI holds significant potential as a first-line diagnostic and therapeutic approach for prostate gland abnormalities. However, the broader application of prostate MRI is currently limited for various reasons. CNN-based DL models have been employed for fully automatic target segmentation. More importantly, DLA can be easily applied to large-scale samples, making them suitable for real-world clinical practice. In addition to their utility in detecting and segmenting lesions on prostate MRI, DLA have a wide range of applications. Presently, some studies have demonstrated the potential applications of DL in multiple areas. A few studies have employed DL to predict the Gleason score of PCa by using DLAs to assess pathological sections, which demonstrated diagnostic power equal to that of pathologists (173–175). DL has also yielded satisfactory results in prostate gland segmentation on TRUS images (176–178). In radiotherapy, prostate gland and adjacent organ contouring based on DL auto-contouring algorithms may reduce workload and inter-observer variability, as evidenced by several clinical evaluations conducted at different radiotherapy centers (179, 180). Additionally, using DLA for detecting and tracking marker seeds during PCa treatment enhances precise target dosage delivery and minimizes radiation-induced adverse events in normal tissues surrounding the tumor (181, 182). Remarkably, DL has been employed for PI-RADS scoring based on mpMRI of the prostate gland in real-world settings, yielding results similar to PI-RADS scores determined by radiologist experts (83). In recent times, there has been notable progress in integrating DL with nomograms. This integration enables the inclusion of crucial variables such as PSA, PV, patient age, free/total PSA ratio, and PSA-D into the diagnostic process of PCa using MRI data (183, 184).
4.1 Limitations and outlook
The research examined in this review highlights the significant potential and wide-ranging prospects of DL applications. Future studies should concentrate on employing DL in prostate MRI for in-depth understanding. Firstly, there is a strong demand for 3D information processing. Presently, most available DLAs rely on 2D images for feature extraction and analysis, indicating that these DLAs may not be suitable for extracting 3D spatial anatomical information from clinically obtained patient images. Although DL has been employed for segmenting 3D medical images of the liver and cardiovascular system (185), there is a scarcity of research on using DL for segmenting 3D images of the prostate gland, necessitating further evaluation. Developing computational segmentation methods appropriate for 3D medical images while preserving the high performance of DL models for PCa detection and diagnosis remains highly challenging. In addition, future research should continually extend to multimodal and multisequence data analyses. Currently, most prostate MRI studies include only T2W and DWI sequences. Despite the diminishing role of DCE according to recent PI-RADS guidelines (186), incorporating ADC into the analysis and fusing multiple modalities of feature descriptions for 3D tumor image segmentation may further enhance the accuracy of CNN in identifying PCa. Furthermore, the effectiveness of using DCE sequence in detecting PCa is still debated, given its time-consuming nature and the associated risk of nephrogenic systemic fibrosis (187, 188). Therefore, focusing on biparametric MRI, which assesses only T2W and DWI sequences, should be prioritized for rapid screening. Improving CNN architectures may also enhance the computational capabilities of DLAs. Based on cumulative findings, we propose that parallelizing sub-networks analyzing different sequences and then inputting the final result into the classifier or connecting various sub-networks in series and generating the final result directly could yield promising outcomes. Utilizing diverse DLAs, developed by modifying neural network architectures, can further improve detection effectiveness. One primary limitation of DL, not only in medical image processing but also in other professional fields, is the incomprehensibility and lack of interpretability of predictions and decisions made by DLAs (189–191). This becomes critically important in cases where DL-based decisions can result in significant consequences, particularly in medical and biological contexts (189, 191). To prevent misdiagnosis and mistreatment that may lead to life-threatening conditions, the rationale and evidence for DL-provided conclusions must be clarified. Developing “explainable AI” (XAI) for accurate predictions with understandable assessment criteria should be further investigated as a future direction. More ambitiously, comprehending complex biological contexts, such as molecular mechanisms, genetic expression, and cellular microenvironments, is crucial for developing novel biomarkers, discovering disease pathogenesis, proposing new treatment strategies, and evaluating analytical approach performance (192, 193). All these advancements necessitate updating CNN architectures to not only make predictions based on data-driven DL approaches but also learn the biological mechanisms behind the data by integrating biological knowledge into the learning process (193, 194). A new concept called digital biopsy involves analyzing digital images and identifying characteristic features focusing on tumor heterogeneity rather than its contour, using computer power from multi-omics to aid in diagnosing or predicting various diseases (195). Investigating DL-based digital biopsy techniques will significantly contribute to assessing and predicting diseases non-invasively, making it a valuable tool in clinical settings. Digital biopsy holds considerable potential to become the “next-generation biopsy” for patients with low risk PCa, substantially benefiting healthcare.
5 Conclusion
DLAs have shown promising results in tumor identification and detection, lesion segmentation, PCa grading and scoring, as well as postoperative recurrence and prognostic outcome prediction, making them gain significant attention and play important roles in urology. However, the diagnostic accuracy of DL models still has room for improvement, and the amount of annotated sample data used is relatively limited. Therefore, optimization of models and algorithms, expansion of medical database resources, and combination of multi-omics data and comprehensive analysis of various morphological data will enhance the usefulness of DL for the diagnosis and treatment of urological diseases. Additionally, continued exploration in developing explainable AI will bring greater transparency and trustworthiness to DL. Undoubtedly, DL has shown a steep learning curve in the interpretation of prostate MRI (196), and its advent will benefit not only radiologists but also general physicians who lack systematic and specialized imaging interpretation training in terms of imaging evaluation of prostatic diseases. We believe that with advancements in technology and research, a significant leap in DLA development would occur, which would be beneficial in PCa diagnosis and treatment.
Author contributions
MH performed the literature search regarding the available databases and drafted the manuscript. YC, XY and AK helped in consulting the relevant literature. RR, SW assisted in implementing images. OM, LZ and GY polished the manuscript. CC evaluated and reinfored the technical background. KH, ME contributed to editing the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Song JM, Kim CB, Chung HC, Kane RL. Prostate-specific antigen, digital rectal examination and transrectal ultrasonography: a meta-analysis for this diagnostic triad of prostate cancer in symptomatic korean men. Yonsei Med J (2005) 46(3):414–24. doi: 10.3349/ymj.2005.46.3.414
3. Moe A, Hayne D. Transrectal ultrasound biopsy of the prostate: does it still have a role in prostate cancer diagnosis? Trans Androl Urol (2020) 9(6):3018–24. doi: 10.21037/tau.2019.09.37
4. Numao N, Yoshida S, Komai Y, Ishii C, Kagawa M, Kijima T, et al. Usefulness of pre-biopsy multiparametric magnetic resonance imaging and clinical variables to reduce initial prostate biopsy in men with suspected clinically localized prostate cancer. J Urol (2013) 190(2):502–8. doi: 10.1016/j.juro.2013.02.3197
5. Borghesi M, Bianchi L, Barbaresi U, Vagnoni V, Corcioni B, Gaudiano C, et al. Diagnostic performance of MRI/TRUS fusion-guided biopsies vs. systematic prostate biopsies in biopsy-naïve, previous negative biopsy patients and men undergoing active surveillance. Nephrology (2021) 73(3):357–66.
6. Chen FK, de Castro Abreu AL, Palmer SL. Utility of ultrasound in the diagnosis, treatment, and follow-up of prostate cancer: state of the art. J Nucl Med (2016) 57(Suppl 3):13s–8s. doi: 10.2967/jnumed.116.177196
7. Pecoraro M, Messina E, Bicchetti M, Carnicelli G, Del Monte M, Iorio B, et al. The future direction of imaging in prostate cancer: MRI with or without contrast injection. Andrology (2021) 9(5):1429–43. doi: 10.1111/andr.13041
8. Bittencourt LK, Hausmann D, Sabaneeff N, Gasparetto EL, Barentsz JO. Multiparametric magnetic resonance imaging of the prostate: current concepts. Radiologia brasileira (2014) 47(5):292–300. doi: 10.1590/0100-3984.2013.1863
9. Würnschimmel C, Chandrasekar T, Hahn L, Esen T, Shariat SF, Tilki D. MRI As a screening tool for prostate cancer: current evidence and future challenges. World J Urol (2023) 41(4):921–928. doi: 10.1007/s00345-022-03947-y
10. Rouvière O, Puech P, Renard-Penna R, Claudon M, Roy C, Mège-Lechevallier F, et al. Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy-naive patients (MRI-FIRST): a prospective, multicentre, paired diagnostic study. Lancet Oncol (2019) 20(1):100–9. doi: 10.1016/s1470-2045(18)30569-2
11. Mastrogiacomo S, Dou W, Jansen JA, Walboomers XF. Magnetic resonance imaging of hard tissues and hard tissue engineered bio-substitutes. Mol Imaging Biol (2019) 21(6):1003–19. doi: 10.1007/s11307-019-01345-2
12. Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol (2019) 76(3):340–51. doi: 10.1016/j.eururo.2019.02.033
13. van der Leest M, Cornel E, Israël B, Hendriks R, Padhani AR, Hoogenboom M, et al. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naïve men with elevated prostate-specific antigen: a Large prospective multicenter clinical study. Eur Urol (2019) 75(4):570–8. doi: 10.1016/j.eururo.2018.11.023
14. Gupta RT, Mehta KA, Turkbey B, Verma S. PI-RADS: past, present, and future. J Magn Reson Imaging: JMRI (2020) 52(1):33–53. doi: 10.1002/jmri.26896
15. Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet (London England) (2017) 389(10071):815–22. doi: 10.1016/s0140-6736(16)32401-1
16. Song Y, Zhang YD, Yan X, Liu H, Zhou M, Hu B, et al. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J Magn Reson Imaging: JMRI (2018) 48(6):1570–7. doi: 10.1002/jmri.26047
17. Cao R, Zhong X, Afshari S, Felker E, Suvannarerg V, Tubtawee T, et al. Performance of deep learning and genitourinary radiologists in detection of prostate cancer using 3-T multiparametric magnetic resonance imaging. J Magn Reson Imaging: JMRI (2021) 54(2):474–83. doi: 10.1002/jmri.27595
18. Sanders JW, Mok H, Hanania AN, Venkatesan AM, Tang C, Bruno TL, et al. Computer-aided segmentation on MRI for prostate radiotherapy, part I: quantifying human interobserver variability of the prostate and organs at risk and its impact on radiation dosimetry. Radiother oncol: J Eur Soc Ther Radiol Oncol (2022) 169:124–31. doi: 10.1016/j.radonc.2021.12.011
19. Ahsan MM, Luna SA, Siddique Z. Machine-Learning-Based disease diagnosis: a comprehensive review. Healthcare (Basel Switzerland) (2022) 10(3):541. doi: 10.3390/healthcare10030541
20. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Engineering (2017) 19:221–48. doi: 10.1146/annurev-bioeng-071516-044442
21. Kumar Y, Koul A, Singla R, Ijaz MF. Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. J Ambient Intell Humaniz Comput (2022) 13:1–28. doi: 10.1007/s12652-021-03612-z
22. Sebastian AM, Peter D. Artificial intelligence in cancer research: trends, challenges and future directions. Life (Basel Switzerland) (2022) 12(12):1991. doi: 10.3390/life12121991
23. Lemaitre G. Computer-aided diagnosis for prostate cancer using multi-parametric magnetic resonance imaging [Doctoral dissertation]. France: Universitat de Girona (2016). Available at: https://www.researchgate.net/profile/Guillaume-Lemaitre-2/publication/312063690_Computer-Aided_Diagnosis_for_Prostate_Cancer_using_Multi-Parametric_Magnetic_Resonance_Imaging/links/58710e0108ae8fce491ede41/Computer-Aided-Diagnosis-for-Prostate-Cancer-using-Multi-Parametric-Magnetic-Resonance-Imaging.pdf.
24. Wang S, Burtt K, Turkbey B, Choyke P, Summers RM. Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research. BioMed Res Int (2014) 2014:789561. doi: 10.1155/2014/789561
25. Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci USA (2015) 112(46):E6265–73. doi: 10.1073/pnas.1505935112
26. Lemaitre G, Marti R, Rastgoo M, Meriaudeau F. (2017). Computer-aided detection for prostate cancer detection based on multi-parametric magnetic resonance imaging. Annu Int Conf IEEE Eng Med Biol Soc (2017) 2017:3138–41. doi: 10.1109/embc.2017.8037522
27. Firmino M, Angelo G, Morais H, Dantas MR, Valentim R. Computer-aided detection (CADe) and diagnosis (CADx) system for lung cancer with likelihood of malignancy. Biomed Eng Online (2016) 15(1):2. doi: 10.1186/s12938-015-0120-7
28. Chan HP, Hadjiiski LM, Samala RK. Computer-aided diagnosis in the era of deep learning. Med Physics (2020) 47(5):e218–27. doi: 10.1002/mp.13764
29. Liu L, Tian Z, Zhang Z, Fei B. Computer-aided detection of prostate cancer with MRI: technology and applications. Acad Radiol (2016) 23(8):1024–46. doi: 10.1016/j.acra.2016.03.010
30. Telecan T, Andras I, Crisan N, Giurgiu L, Căta ED, Caraiani C, et al. More than meets the eye: using textural analysis and artificial intelligence as decision support tools in prostate cancer Diagnosis—A systematic review. J Pers Med (2022) 12(6):983. doi: 10.3390/jpm12060983
31. Yang X, Liu C, Wang Z, Yang J, Min HL, Wang L, et al. Co-Trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. Med Image Anal (2017) 42:212–27. doi: 10.1016/j.media.2017.08.006
32. Hassanzadeh E, Glazer DI, Dunne RM, Fennessy FM, Harisinghani MG, Tempany CM. Prostate imaging reporting and data system version 2 (PI-RADS v2): a pictorial review. Abdominal Radiol (New York) (2017) 42(1):278–89. doi: 10.1007/s00261-016-0871-z
33. Barentsz JO, Weinreb JC, Verma S, Thoeny HC, Tempany CM, Shtern F, et al. Synopsis of the PI-RADS v2 guidelines for multiparametric prostate magnetic resonance imaging and recommendations for use. Eur Urol (2016) 69(1):41–9. doi: 10.1016/j.eururo.2015.08.038
34. Park BK, Park SY. New biopsy techniques and imaging features of transrectal ultrasound for targeting PI-RADS 4 and 5 lesions. J Clin Med (2020) 9(2):530. doi: 10.3390/jcm9020530
35. Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. And data system: 2015, version 2. Eur Urol (2016) 69(1):16–40. doi: 10.1016/j.eururo.2015.08.052
36. Brembilla G, Dell’Oglio P, Stabile A, Damascelli A, Brunetti L, Ravelli S, et al. Interreader variability in prostate MRI reporting using prostate imaging reporting and data system version 2.1. Eur Radiol (2020) 30(6):3383–92. doi: 10.1007/s00330-019-06654-2
37. Smith CP, Harmon SA, Barrett T, Bittencourt LK, Law YM, Shebel H, et al. Intra- and interreader reproducibility of PI-RADSv2: a multireader study. J Magn Reson Imaging: JMRI (2019) 49(6):1694–703. doi: 10.1002/jmri.26555
38. Barragán-Montero A, Javaid U, Valdés G, Nguyen D, Desbordes P, Macq B, et al. Artificial intelligence and machine learning for medical imaging: a technology review. Physica medica: PM: an Int J devoted to Appl Phys to Med biol: Off J Ital Assoc Biomed Phys (AIFB) (2021) 83:242–56. doi: 10.1016/j.ejmp.2021.04.016
39. Yang R, Yu Y. Artificial convolutional neural network in object detection and semantic segmentation for medical imaging analysis. Front Oncol (2021) 11:638182. doi: 10.3389/fonc.2021.638182
40. Jiang T, Gradus JL, Rosellini AJ. Supervised machine learning: a brief primer. Behav Ther (2020) 51(5):675–87. doi: 10.1016/j.beth.2020.05.002
41. Wallin E, Svensson L, Kahl F, Hammarstrand L. DoubleMatch: improving semi-supervised learning with self-supervision. 26th International Conference on Pattern Recognition (ICPR). (2022) 2871–7.
42. Chapelle O, Schölkopf B, Zien A. Semi-supervised learning. IEEE Transactions on Neural Networks (2009) 20(3):542.
43. Hu J, Niu H, Carrasco J, Lennox B, Arvin F. Voronoi-based multi-robot autonomous exploration in unknown environments via deep reinforcement learning. IEEE Trans Vehicular Technol (2020) 69(12):14413–23. doi: 10.1109/TVT.2020.3034800
44. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural computation (2006) 18(7):1527–54. doi: 10.1162/neco.2006.18.7.1527
45. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature (2015) 521(7553):436–44. doi: 10.1038/nature14539
46. Huang X, Li Z, Zhang M, Gao S. Fusing hand-crafted and deep-learning features in a convolutional neural network model to identify prostate cancer in pathology images. Front Oncol (2022) 12:994950. doi: 10.3389/fonc.2022.994950
47. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
48. Lee H, Hong H, Kim J, Jung DC. Deep feature classification of angiomyolipoma without visible fat and renal cell carcinoma in abdominal contrast-enhanced CT images with texture image patches and hand-crafted feature concatenation. Med Physics (2018) 45(4):1550–61. doi: 10.1002/mp.12828
49. Li Z, Wu F, Hong F, Gai X, Cao W, Zhang Z, et al. Computer-aided diagnosis of spinal tuberculosis from CT images based on deep learning with multimodal feature fusion. Front Microbiol (2022) 13:823324. doi: 10.3389/fmicb.2022.823324
50. Zeng X, Huang R, Zhong Y, Xu Z, Liu Z, Wang Y. A reciprocal learning strategy for semisupervised medical image segmentation. Med Physics (2023) 50(1):163–77. doi: 10.1002/mp.15923
51. Iglesias G, Talavera E, Díaz-Álvarez A. A survey on GANs for computer vision: recent research, analysis and taxonomy. Comput Sci Rev (2023) 48:100553. doi: 10.1016/j.cosrev.2023.100553
52. Sarıgül M, Ozyildirim BM, Avci M. Differential convolutional neural network. Neural Networks (2019) 116:279–87. doi: 10.1016/j.neunet.2019.04.025
53. Koshino K, Werner RA, Pomper MG, Bundschuh RA, Toriumi F, Higuchi T, et al. Narrative review of generative adversarial networks in medical and molecular imaging. Ann Trans Med (2021) 9(9):821. doi: 10.21037/atm-20-6325
54. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer (2018) 18(8):500–10. doi: 10.1038/s41568-018-0016-5
55. Cuocolo R, Cipullo MB, Stanzione A, Ugga L, Romeo V, Radice L, et al. Machine learning applications in prostate cancer magnetic resonance imaging. Eur Radiol Experimental (2019) 3(1):35. doi: 10.1186/s41747-019-0109-2
56. Rickman J, Struyk G, Simpson B, Byun BC, Papanikolopoulos N. The growing role for semantic segmentation in urology. Eur Urol Focus (2021) 7(4):692–5. doi: 10.1016/j.euf.2021.07.017
57. Ahmed M, Hashmi KA, Pagani A, Liwicki M, Stricker D, Afzal MZ. And performance analysis of deep learning based object detection in challenging environments. Sensors (Basel Switzerland) (2021) 21(15):5116. doi: 10.3390/s21155116
58. Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Systems (2018) 42(11):226. doi: 10.1007/s10916-018-1088-1
59. Puttagunta M, Ravi S. Medical image analysis based on deep learning approach. Multimedia Tools Appl (2021) 80(16):24365–98. doi: 10.1007/s11042-021-10707-4
60. Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights into Imaging (2018) 9(4):611–29. doi: 10.1007/s13244-018-0639-9
61. Alzubaidi L, Zhang J, Humaidi AJ, Al-Dujaili A, Duan Y, Al-Shamma O, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data (2021) 8(1):53. doi: 10.1186/s40537-021-00444-8
62. Min S, Lee B, Yoon S. Deep learning in bioinformatics. Briefings Bioinf (2017) 18(5):851–69. doi: 10.1093/bib/bbw068
63. Michaely HJ, Aringhieri G, Cioni D, Neri E. Current value of biparametric prostate MRI with machine-learning or deep-learning in the detection, grading, and characterization of prostate cancer: a systematic review. Diagnostics (Basel Switzerland) (2022) 12(4):799. doi: 10.3390/diagnostics12040799
64. Yi Z. Researches advanced in image recognition based on deep learning. Highlights Science Eng Technol (2023) 39:1309–16. doi: 10.54097/hset.v39i.6760
65. Li Z, Wang Y, Zhang N, Zhang Y, Zhao Z, Xu D, et al. Deep learning-based object detection techniques for remote sensing images: a survey. Remote Sens (2022) 14(10):2385. doi: 10.3390/rs14102385
66. Li M, Zhang Z, Lei L, Wang X, Guo X. Agricultural greenhouses detection in high-resolution satellite images based on convolutional neural networks: comparison of faster r-CNN, YOLO v3 and SSD. Sensors (Basel Switzerland) (2020) ;20(17):4938. doi: 10.3390/s20174938
67. Sumit SS, Watada J, Roy A, Rambli DRA. In object detection deep learning methods, YOLO shows supremum to mask r-CNN. J Physics: Conf Ser (2020) 1529(4). doi: 10.1088/1742-6596/1529/4/042086
68. Zanaty E, Abdel-Aty MMJI. Comparing U-net convolutional network with mask r-CNN in nuclei segmentation. IJCSNS Int J Comput Sci Netw Secur (2022) 22(3):273.
69. Li C, Liu H. Medical image segmentation with generative adversarial semi-supervised network. Phys Med Biol (2021) 66(24). doi: 10.1088/1361-6560/ac3d15
70. Isola P, Zhu J-Y, Zhou T, Efros A. Image-to-Image translation with conditional adversarial networks. arXiv (2017), 5967. doi: 10.48550/arXiv.1611.07004
71. Ho J, Ermon S. Generative adversarial imitation learning. Advances in neural information processing systems (2016) 29.
72. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. NIPS (2014).
73. Mohammadi B, Sabokrou M. End-to-End adversarial learning for intrusion detection in computer networks. (2019). doi: 10.1109/LCN44214.2019.8990759
74. Laddha S, Kumar V. DGCNN: deep convolutional generative adversarial network based convolutional neural network for diagnosis of COVID-19. Multimedia Tools Appl (2022) 81(22):31201–18. doi: 10.1007/s11042-022-12640-6
75. Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial network. sarXiv. (2015). doi: 10.48550/arXiv.1511.06434
76. Wang Y, Zhou L, Wang M, Shao C, Shi L, Yang S, et al. Combination of generative adversarial network and convolutional neural network for automatic subcentimeter pulmonary adenocarcinoma classification. Quantitative Imaging Med surgery (2020) 10(6):1249–64. doi: 10.21037/qims-19-982
77. Shreffler J, Huecker MR. Diagnostic testing accuracy: sensitivity, specificity, predictive values and likelihood ratios. In: StatPearls. Treasure Island (FL: StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC (2023).
78. Eelbode T, Bertels J, Berman M, Vandermeulen D, Maes F, Bisschops R, et al. Optimization for medical image segmentation: theory and practice when evaluating with dice score or jaccard index. IEEE Trans Med Imaging (2020) 39(11):3679–90. doi: 10.1109/tmi.2020.3002417
79. Park SH, Goo JM, Jo CH. Receiver operating characteristic (ROC) curve: a practical review for radiologists. Korean J Radiol (2004) 5(1):11–8. doi: 10.3348/kjr.2004.5.1.11
80. Nahm FS. Receiver operating characteristic curve: overview and practical use for clinicians. Korean J anesthesiol (2022) 75(1):25–36. doi: 10.4097/kja.21209
81. Li F, He H. Assessing the accuracy of diagnostic tests. Shanghai Arch Psychiatry (2018) 30(3):207–12. doi: 10.11919/j.issn.1002-0829.218052
82. Yeung M, Rundo L, Nan Y, Sala E, Schönlieb C-B, Yang G. Calibrating the dice loss to handle neural network overconfidence for biomedical image segmentation. J Digital Imaging (2023) 36(2):739–52. doi: 10.1007/s10278-022-00735-3
83. Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, et al. Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology (2019) 293(3):607–17. doi: 10.1148/radiol.2019190938
84. Xu H, Baxter JSH, Akin O, Cantor-Rivera D. Prostate cancer detection using residual networks. Int J Comput assisted Radiol surgery (2019) 14(10):1647–50. doi: 10.1007/s11548-019-01967-5
85. Chen Y, Xing L, Yu L, Bagshaw HP, Buyyounouski MK, Han B. Automatic intraprostatic lesion segmentation in multiparametric magnetic resonance images with proposed multiple branch UNet. Med Physics (2020) 47(12):6421–9. doi: 10.1002/mp.14517
86. Arif M, Schoots IG, Castillo Tovar J, Bangma CH, Krestin GP, Roobol MJ, et al. Clinically significant prostate cancer detection and segmentation in low-risk patients using a convolutional neural network on multi-parametric MRI. Eur Radiol (2020) 30(12):6582–92. doi: 10.1007/s00330-020-07008-z
87. Ushinsky A, Bardis M, Glavis-Bloom J, Uchio E, Chantaduly C, Nguyentat M, et al. A 3D-2D hybrid U-net convolutional neural network approach to prostate organ segmentation of multiparametric MRI. AJR Am J Roentgenol (2021) 216(1):111–6. doi: 10.2214/ajr.19.22168
88. Bardis M, Houshyar R, Chantaduly C, Tran-Harding K, Ushinsky A, Chahine C, et al. Segmentation of the prostate transition zone and peripheral zone on MR images with deep learning. Radiol Imaging Cancer (2021) 3(3):e200024. doi: 10.1148/rycan.2021200024
89. Soerensen SJC, Fan RE, Seetharaman A, Chen L, Shao W, Bhattacharya I, et al. Deep learning improves speed and accuracy of prostate gland segmentations on magnetic resonance imaging for targeted biopsy. J Urol (2021) 206(3):604–12. doi: 10.1097/ju.0000000000001783
90. Soni M, Khan IR, Babu KS, Nasrullah S, Madduri A, Rahin SA. Light weighted healthcare CNN model to detect prostate cancer on multiparametric MRI. Comput Intell Neurosci (2022) 2022:5497120. doi: 10.1155/2022/5497120
91. Li D, Han X, Gao J, Zhang Q, Yang H, Liao S, et al. Deep learning in prostate cancer diagnosis using multiparametric magnetic resonance imaging with whole-mount histopathology referenced delineations. Front Med (2021) 8:810995. doi: 10.3389/fmed.2021.810995
92. Boehm K, Borgmann H, Ebert T, Höfner T, Khaljani E, Schmid M, et al. Stage and grade migration in prostate cancer treated with radical prostatectomy in a Large German multicenter cohort. Clin Genitourinary Cancer (2021) 19(2):162–166.e1. doi: 10.1016/j.clgc.2020.12.004
93. Moore CM, Giganti F, Albertsen P, Allen C, Bangma C, Briganti A, et al. Reporting magnetic resonance imaging in men on active surveillance for prostate cancer: the PRECISE recommendations-a report of a European school of oncology task force. Eur Urol (2017) 71(4):648–55. doi: 10.1016/j.eururo.2016.06.011
94. Fusco R, Sansone M, Granata V, Setola SV, Petrillo A. A systematic review on multiparametric MR imaging in prostate cancer detection. Infect Agents Cancer (2017) 12:57. doi: 10.1186/s13027-017-0168-z
95. Vente C, Vos P, Hosseinzadeh M, Pluim J, Veta M. Deep learning regression for prostate cancer detection and grading in bi-parametric MRI. IEEE Trans Bio-medical Engineering (2021) 68(2):374–83. doi: 10.1109/tbme.2020.2993528
96. Wang Z, Liu C, Cheng D, Wang L, Yang X, Cheng KT. Automated detection of clinically significant prostate cancer in mp-MRI images based on an end-to-End deep neural network. IEEE Trans Med Imaging (2018) 37(5):1127–39. doi: 10.1109/tmi.2017.2789181
97. Ishioka J, Matsuoka Y, Uehara S, Yasuda Y, Kijima T, Yoshida S, et al. Computer-aided diagnosis of prostate cancer on magnetic resonance imaging using a convolutional neural network algorithm. BJU Int (2018) 122(3):411–7. doi: 10.1111/bju.14397
98. Wang X, Yang W, Weinreb J, Han J, Li Q, Kong X, et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep (2017) 7(1):15415. doi: 10.1038/s41598-017-15720-y
99. Sanford T, Harmon SA, Turkbey EB, Kesani D, Tuncer S, Madariaga M, et al. Deep-Learning-Based artificial intelligence for PI-RADS classification to assist multiparametric prostate MRI interpretation: a development study. J Magn Reson Imaging: JMRI (2020) 52(5):1499–507. doi: 10.1002/jmri.27204
100. Omri N, Kamil M, Alexander K, Alexander K, Edmond S, Ariel Z, et al. Association between PSA density and pathologically significant prostate cancer: the impact of prostate volume. Prostate (2020) 80(16):1444–9. doi: 10.1002/pros.24078
101. Feng ZJ, Xue C, Wen JM, Li Y, Wang M, Zhang N. PSAD test in the diagnosis of prostate cancer: a meta-analysis. Clin Laboratory (2017) 63(1):147–55. doi: 10.7754/Clin.Lab.2016.160727
102. Jue JS, Barboza MP, Prakash NS, Venkatramani V, Sinha VR, Pavan N, et al. Re-examining prostate-specific antigen (PSA) density: defining the optimal PSA range and patients for using PSA density to predict prostate cancer using extended template biopsy. Urology (2017) 105:123–8. doi: 10.1016/j.urology.2017.04.015
103. Zoltan E, Lee R, Staskin DR, Te AE, Kaplan SA. Combination therapy for benign prostatic hyperplasia: does size matter? Curr Bladd Dysfunct Rep (2008) 3(2):102–8. doi: 10.1007/s11884-008-0016-5
104. Milonas D, Trumbeckas D, Juska P. The importance of prostatic measuring by transrectal ultrasound in surgical management of patients with clinically benign prostatic hyperplasia. Medicina (Kaunas Lithuania) (2003) 39(9):860–6.
105. Garvey B, Türkbey B, Truong H, Bernardo M, Periaswamy S, Choyke PL. Clinical value of prostate segmentation and volume determination on MRI in benign prostatic hyperplasia. Diagn Interventional Radiol (Ankara Turkey) (2014) 20(3):229–33. doi: 10.5152/dir.2014.13322
106. Harvey CJ, Pilcher J, Richenberg J, Patel U, Frauscher F. Applications of transrectal ultrasound in prostate cancer. Br J Radiol (2012) 85 Spec No 1(Spec Iss 1):S3–17. doi: 10.1259/bjr/56357549
107. Walz J, Epstein JI, Ganzer R, Graefen M, Guazzoni G, Kaouk J, et al. A critical analysis of the current knowledge of surgical anatomy of the prostate related to optimisation of cancer control and preservation of continence and erection in candidates for radical prostatectomy: An update. Eur Urol (2016) 70(2):301–11. doi: 10.1016/j.eururo.2016.01.026
108. Hoeh B, Wenzel M, Hohenhorst L, Köllermann J, Graefen M, Haese A, et al. Anatomical fundamentals and current surgical knowledge of prostate anatomy related to functional and oncological outcomes for robotic-assisted radical prostatectomy. Front Surg (2022) 8:825183. doi: 10.3389/fsurg.2021.825183.
109. Munck Af Rosenschold P, Zelefsky MJ, Apte AP, Jackson A, Oh JH, Shulman E, et al. Image-guided radiotherapy reduces the risk of under-dosing high-risk prostate cancer extra-capsular disease and improves biochemical control. Radiat Oncol (London England) (2018) 13(1):64. doi: 10.1186/s13014-018-0978-1
110. Murphy G, Haider M, Ghai S, Sreeharsha B. The expanding role of MRI in prostate cancer. AJR Am J Roentgenol (2013) 201(6):1229–38. doi: 10.2214/ajr.12.10178
111. Jia H, Xia Y, Song Y, Cai W, Fulham M, Feng DD. Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing (2018) 275:1358–69. doi: 10.1016/j.neucom.2017.09.084
112. Bibault JE, Giraud P, Burgun A. Big data and machine learning in radiation oncology: state of the art and future prospects. Cancer Letters (2016) 382(1):110–7. doi: 10.1016/j.canlet.2016.05.033
113. Mehralivand S, Harmon SA, Shih JH, Smith CP, Lay N, Argun B, et al. Multicenter multireader evaluation of an artificial intelligence-based attention mapping system for the detection of prostate cancer with multiparametric MRI. AJR Am J roentgenol (2020) 215(4):903–12. doi: 10.2214/ajr.19.22573
114. Zhu Q, Du B, Wu J, Yan P. “A deep learning health data analysis approach: automatic 3D prostate MR segmentation with densely-connected volumetric ConvNets,” 2018 International Joint Conference on Neural Networks (IJCNN), (Rio de Janeiro, Brazil) (2018), pp. 1–6. doi: 10.1109/IJCNN.2018.8489136
115. Yan K, Wang X, Kim J, Khadra M, Fulham M, Feng D. A propagation-DNN: deep combination learning of multi-level features for MR prostate segmentation. Comput Methods Programs Biomed (2019) 170:11–21. doi: 10.1016/j.cmpb.2018.12.031
116. To MNN, Vu DQ, Turkbey B, Choyke PL, Kwak JT. Deep dense multi-path neural network for prostate segmentation in magnetic resonance imaging. Int J Comput Assisted Radiol Surgery (2018) 13(11):1687–96. doi: 10.1007/s11548-018-1841-4
117. Dai Z, Carver E, Liu C, Lee J, Feldman A, Zong W, et al. Segmentation of the prostatic gland and the intraprostatic lesions on multiparametic magnetic resonance imaging using mask region-based convolutional neural networks. Adv Radiat Oncol (2020) 5(3):473–81. doi: 10.1016/j.adro.2020.01.005
118. Wang C, Zhu X, Hong JC, Zheng D. Artificial intelligence in radiotherapy treatment planning: present and future. Technol Cancer Res Treat (2019) 18:1533033819873922. doi: 10.1177/1533033819873922
119. Thoms J, Goda JS, Zlotta AR, Fleshner NE, van der Kwast TH, Supiot S, et al. Neoadjuvant radiotherapy for locally advanced and high-risk prostate cancer. Nat Rev Clin Oncol (2011) 8(2):107–13. doi: 10.1038/nrclinonc.2010.207
120. Podder TK, Fredman ET, Ellis RJ. Advances in radiotherapy for prostate cancer treatment. Adv Exp Med Biol (2018) 1096:31–47. doi: 10.1007/978-3-319-99286-0_2
121. Muralidhar A, Potluri HK, Jaiswal T, McNeel DG. Targeted radiation and immune therapies-advances and opportunities for the treatment of prostate cancer. Pharmaceutics (2023) 15(1):252. doi: 10.3390/pharmaceutics15010252
122. Thariat J, Hannoun-Levi JM, Sun Myint A, Vuong T, Gérard JP. Past, present, and future of radiotherapy for the benefit of patients. Nat Rev Clin Oncol (2013) 10(1):52–60. doi: 10.1038/nrclinonc.2012.203
123. Sonke JJ, Aznar M, Rasch C. Adaptive radiotherapy for anatomical changes. Semin Radiat Oncol (2019) 29(3):245–57. doi: 10.1016/j.semradonc.2019.02.007
124. Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci (2012) 9(3):193–9. doi: 10.7150/ijms.3635
125. Alterio D, Marvaso G, Ferrari A, Volpe S, Orecchia R, Jereczek-Fossa BA. Modern radiotherapy for head and neck cancer. Semin Oncol (2019) 46(3):233–45. doi: 10.1053/j.seminoncol.2019.07.002
126. Yang WC, Hsu FM, Yang PC. Precision radiotherapy for non-small cell lung cancer. J Biomed sci (2020) 27(1):82. doi: 10.1186/s12929-020-00676-5
127. Zhang Y, Liang Y, Ding J, Amjad A, Paulson E, Ahunbay E, et al. A prior knowledge-guided, deep learning-based semiautomatic segmentation for complex anatomy on magnetic resonance imaging. Int J Radiat Oncol Biol Phys (2022) 114(2):349–59. doi: 10.1016/j.ijrobp.2022.05.039
128. Wang M, Zhang Q, Lam S, Cai J, Yang R. A review on application of deep learning algorithms in external beam radiotherapy automated treatment planning. Front Oncol (2020) 10:580919. doi: 10.3389/fonc.2020.580919
129. Hunt A, Hansen VN, Oelfke U, Nill S, Hafeez S. Adaptive radiotherapy enabled by MRI guidance. Clin Oncol (Royal Coll Radiologists (Great Britain)) (2018) 30(11):711–9. doi: 10.1016/j.clon.2018.08.001
130. Paganetti H, Botas P, Sharp GC, Winey B. Adaptive proton therapy. Phys Med Biol (2021) 66(22):10.1088/1361-6560/ac344f. doi: 10.1088/1361-6560/ac344f
131. Glide-Hurst CK, Lee P, Yock AD, Olsen JR, Cao M, Siddiqui F, et al. Adaptive radiation therapy (ART) strategies and technical considerations: a state of the ART review from NRG oncology. Int J Radiat Oncol Biol Phys (2021) 109(4):1054–75. doi: 10.1016/j.ijrobp.2020.10.021
132. Jing X, Meng X, Sun X, Yu J. Delineation of clinical target volume for postoperative radiotherapy in stage IIIA-pN2 non-small-cell lung cancer. OncoTargets Ther (2016) 9:823–31. doi: 10.2147/ott.S98765
133. Keall PJ, Nguyen DT, O’Brien R, Caillet V, Hewson E, Poulsen PR, et al. The first clinical implementation of real-time image-guided adaptive radiotherapy using a standard linear accelerator. Radiother Oncol: J Eur Soc Ther Radiol Oncol (2018) 127(1):6–11. doi: 10.1016/j.radonc.2018.01.001
134. Thorwarth D, Low DA. Technical challenges of real-time adaptive MR-guided radiotherapy. Front Oncol (2021) 11:634507. doi: 10.3389/fonc.2021.634507
135. Sprouts D, Gao Y, Wang C, Jia X, Shen C, Chi Y. The development of a deep reinforcement learning network for dose-volume-constrained treatment planning in prostate cancer intensity modulated radiotherapy. Biomed Phys Eng Express (2022) 8(4):10.1088/2057-1976/ac6d82. doi: 10.1088/2057-1976/ac6d82
136. Shen C, Chen L, Gonzalez Y, Jia X. Improving efficiency of training a virtual treatment planner network via knowledge-guided deep reinforcement learning for intelligent automatic treatment planning of radiotherapy. Med Physics (2021) 48(4):1909–20. doi: 10.1002/mp.14712
137. Lempart M, Benedek H, Jamtheim Gustafsson C, Nilsson M, Eliasson N, Bäck S, et al. Volumetric modulated arc therapy dose prediction and deliverable treatment plan generation for prostate cancer patients using a densely connected deep learning model. Phys Imaging Radiat Oncol (2021) 19:112–9. doi: 10.1016/j.phro.2021.07.008
138. Owrangi AM, Greer PB, Glide-Hurst CK. MRI-Only treatment planning: benefits and challenges. Phys Med Biol (2018) 63(5):05tr01. doi: 10.1088/1361-6560/aaaca4
139. Hsu SH, Han Z, Leeman JE, Hu YH, Mak RH, Sudhyadhom A. Synthetic CT generation for MRI-guided adaptive radiotherapy in prostate cancer. Front Oncol (2022) 12:969463. doi: 10.3389/fonc.2022.969463
140. Lachance C, McCormack S. Magnetic resonance imaging simulators for simulation and treatment for patients requiring radiation therapy: a review of the clinical effectiveness, cost-effectiveness, and guidelines. In: CADTH rapid response reports. Ottawa (ON: Canadian Agency for Drugs and Technologies in Health (2019).
141. Chandarana H, Wang H, Tijssen RHN, Das IJ. Emerging role of MRI in radiation therapy. J Magn Reson Imaging: JMRI (2018) 48(6):1468–78. doi: 10.1002/jmri.26271
142. Das IJ, McGee KP, Tyagi N, Wang H. Role and future of MRI in radiation oncology. Br J Radiol (2019) 92(1094):20180505. doi: 10.1259/bjr.20180505
143. Li X, Yadav P, McMillan AB. Synthetic computed tomography generation from 0.35T magnetic resonance images for magnetic resonance-only radiation therapy planning using perceptual loss models. Pract Radiat Oncol (2022) 12(1):e40–8. doi: 10.1016/j.prro.2021.08.007
144. Li X, Yadav P, McMillan AB. Synthetic computed tomography generation from 0.35T magnetic resonance images for magnetic resonance–only radiation therapy planning using perceptual loss models. Pract Radiat Oncol (2022) 12(1):e40–e48. doi: 10.1016/j.prro.2021.08.007
145. Depauw N, Keyriläinen J, Suilamo S, Warner L, Bzdusek K, Olsen C, et al. MRI-Based IMPT planning for prostate cancer. Radiother oncol: J Eur Soc Ther Radiol Oncol (2020) 144:79–85. doi: 10.1016/j.radonc.2019.10.010
146. Palmér E, Persson E, Ambolt P, Gustafsson C, Gunnlaugsson A, Olsson LE. Cone beam CT for QA of synthetic CT in MRI only for prostate patients. J Appl Clin Med Physics (2018) 19(6):44–52. doi: 10.1002/acm2.12429
147. Persson E, Jamtheim Gustafsson C, Ambolt P, Engelholm S, Ceberg S, Bäck S, et al. MR-PROTECT: clinical feasibility of a prostate MRI-only radiotherapy treatment workflow and investigation of acceptance criteria. Radiat Oncol (2020) 15(1):77. doi: 10.1186/s13014-020-01513-7
148. Persson E, Gustafsson C, Nordström F, Sohlin M, Gunnlaugsson A, Petruson K, et al. MR-OPERA: a Multicenter/Multivendor validation of magnetic resonance imaging–only prostate treatment planning using synthetic computed tomography images. Int J Radiat Oncol Biol Phys (2017) 99(3):692–700. doi: 10.1016/j.ijrobp.2017.06.006
149. Johnstone E, Wyatt JJ, Henry AM, Short SC, Sebag-Montefiore D, Murray L, et al. Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging-only radiation therapy. Int J Radiat Oncol Biol Phys (2018) 100(1):199–217. doi: 10.1016/j.ijrobp.2017.08.043
150. Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. Deep Learn Data labeling Med applications: First Int Workshop LABELS 2016 Second Int Workshop DLMIA 2016 held conjunction MICCAI 2016 Athens Greece October 21 (2016) 2016:170–8. doi: 10.1007/978-3-319-46976-8_18
151. Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, et al. MRI-Only brain radiotherapy: assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother Oncol (2019) 136:56–63. doi: 10.1016/j.radonc.2019.03.026
152. Fu J, Yang Y, Singhrao K, Ruan D, Chu FI, Low DA, et al. Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med Physics (2019) 46(9):3788–98. doi: 10.1002/mp.13672
153. Zhang Y, Ding SG, Gong XC, Yuan XX, Lin JF, Chen Q, et al. Generating synthesized computed tomography from CBCT using a conditional generative adversarial network for head and neck cancer patients. Technol Cancer Res Treat (2022) 21:15330338221085358. doi: 10.1177/15330338221085358
154. Schonfeld E, Schiele B, Khoreva A. Available at: https://openaccess.thecvf.com/content_CVPR_2020/html/Schonfeld_A_U-Net_Based_Discriminator_for_Generative_Adversarial_Networks_CVPR_2020_paper.html.
155. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I, et al. Deep MR to CT synthesis using unpaired data. international workshop on simulation and synthesis in medical Imaging Vol. vol. 10557). . Québec City, QC, Canada: Springer (2017) p. 14–23 p.
156. Yang H, Sun J, Carass A, Zhao C, Lee J, Prince JL, et al. Unsupervised MR-to-CT synthesis using structure-constrained CycleGAN. IEEE Trans Med Imaging (2020) 39(12):4249–61. doi: 10.1109/tmi.2020.3015379
157. Liu Y, Chen A, Shi H, Huang S, Zheng W, Liu Z, et al. CT synthesis from MRI using multi-cycle GAN for head-and-neck radiation therapy. Computerized Med Imaging graphics: Off J Computerized Med Imaging Society (2021) 91:101953. doi: 10.1016/j.compmedimag.2021.101953
158. Chen S, Qin A, Zhou D, Yan D. Technical note: U-net-generated synthetic CT images for magnetic resonance imaging-only prostate intensity-modulated radiation therapy treatment planning. Med Physics (2018) 45(12):5659–65. doi: 10.1002/mp.13247
159. Arabi H, Dowling JA, Burgos N, Han X, Greer PB, Koutsouvelis N, et al. Comparative study of algorithms for synthetic CT generation from MRI: consequences for MRI-guided radiation planning in the pelvic region. Med Physics (2018) 45(11):5218–33. doi: 10.1002/mp.13187
160. Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, et al. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Bio-medical Engineering (2018) 65(12):2720–30. doi: 10.1109/tbme.2018.2814538
161. Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk-Schulz IM, et al. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol (2018) 63(18):185001. doi: 10.1088/1361-6560/aada6d
162. Cusumano D, Lenkowicz J, Votta C, Boldrini L, Placidi L, Catucci F, et al. A deep learning approach to generate synthetic CT in low field MR-guided adaptive radiotherapy for abdominal and pelvic cases. Radiother Oncol: J Eur Soc Ther Radiol Oncol (2020) 153:205–12. doi: 10.1016/j.radonc.2020.10.018
163. Dall’Era MA, Albertsen PC, Bangma C, Carroll PR, Carter HB, Cooperberg MR, et al. Active surveillance for prostate cancer: a systematic review of the literature. Eur Urol (2012) 62(6):976–83. doi: 10.1016/j.eururo.2012.05.072
164. Van den Broeck T, van den Bergh RCN, Briers E, Cornford P, Cumberbatch M, Tilki D, et al. Biochemical recurrence in prostate cancer: the European association of urology prostate cancer guidelines panel recommendations. Eur Urol Focus (2020) 6(2):231–4. doi: 10.1016/j.euf.2019.06.004
165. David MK, Leslie SW. Prostate specific antigen. StatPearls. Treasure Island FL: StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC (2023).
166. Kobayashi M, Kijima T, Yashi M, Kamai T. Prostate-specific antigen kinetics contributes to decision making for biopsy referral: the predictive implication for PSA retest in patients with elevated PSA levels. Prostate Int (2023) 11(1):27–33. doi: 10.1016/j.prnil.2022.08.001
167. Bergaglio C, Giasotto V, Marcenaro M, Barra S, Turazzi M, Bauckneht M, et al. The role of mpMRI in the assessment of prostate cancer recurrence using the PI-RR system: diagnostic accuracy and interobserver agreement in readers with different expertise. Diagnostics (Basel) (2023) 13(3):387. doi: 10.3390/diagnostics13030387
168. Yan Y, Shao L, Liu Z, He W, Yang G, Liu J, et al. Deep learning with quantitative features of magnetic resonance images to predict biochemical recurrence of radical prostatectomy: a multi-center study. Cancers (2021) 21:13(12). doi: 10.3390/cancers13123098
169. Hensel J, Thalmann GN. Biology of bone metastases in prostate cancer. Urology (2016) 92:6–13. doi: 10.1016/j.urology.2015.12.039
170. Padhani AR, Lecouvet FE, Tunariu N, Koh DM, De Keyzer F, Collins DJ, et al. Rationale for modernising imaging in advanced prostate cancer. Eur Urol Focus (2017) 3(2-3):223–39. doi: 10.1016/j.euf.2016.06.018
171. Combes AD, Palma CA, Calopedos R, Wen L, Woo H, Fulham M, et al. PSMA PET-CT in the diagnosis and staging of prostate cancer. Diagnostics (Basel Switzerland) (2022) 12(11):2594. doi: 10.3390/diagnostics12112594
172. Liu X, Han C, Cui Y, Xie T, Zhang X, Wang X. Detection and segmentation of pelvic bones metastases in MRI images for patients with prostate cancer based on deep learning. Front Oncol (2021) 11:773299. doi: 10.3389/fonc.2021.773299
173. Kott O, Linsley D, Amin A, Karagounis A, Jeffers C, Golijanin D, et al. Development of a deep learning algorithm for the histopathologic diagnosis and Gleason grading of prostate cancer biopsies: a pilot study. Eur Urol Focus (2021) 7(2):347–51. doi: 10.1016/j.euf.2019.11.003
174. Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep (2018) 8(1):12054. doi: 10.1038/s41598-018-30535-1
175. Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol (2020) 21(2):233–41. doi: 10.1016/s1470-2045(19)30739-9
176. van Sloun RJG, Wildeboer RR, Mannaerts CK, Postema AW, Gayet M, Beerlage HP, et al. Deep learning for real-time, automatic, and scanner-adapted prostate (Zone) segmentation of transrectal ultrasound, for example, magnetic resonance imaging-transrectal ultrasound fusion prostate biopsy. Eur Urol Focus (2021) 7(1):78–85. doi: 10.1016/j.euf.2019.04.009
177. Karimi D, Zeng Q, Mathur P, Avinash A, Mahdavi S, Spadinger I, et al. Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images. Med Image Analysis (2019) 57:186–96. doi: 10.1016/j.media.2019.07.005
178. Feng Y, Yang F, Zhou X, Guo Y, Tang F, Ren F, et al. A deep learning approach for targeted contrast-enhanced ultrasound based prostate cancer detection. IEEE/ACM Trans Comput Biol Bioinf (2019) 16(6):1794–801. doi: 10.1109/tcbb.2018.2835444
179. Walker Z, Bartley G, Hague C, Kelly D, Navarro C, Rogers J, et al. Evaluating the effectiveness of deep learning contouring across multiple radiotherapy centres. Phys Imaging Radiat Oncol (2022) 24:121–8. doi: 10.1016/j.phro.2022.11.003
180. Zabel WJ, Conway JL, Gladwish A, Skliarenko J, Didiodato G, Goorts-Matthews L, et al. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract Radiat Oncol (2021) 11(1):e80–9. doi: 10.1016/j.prro.2020.05.013
181. Amarsee K, Ramachandran P, Fielding A, Lehman M, Noble C, Perrett B, et al. Automatic detection and tracking of marker seeds implanted in prostate cancer patients using a deep learning algorithm. J Med Physics (2021) 46(2):80–7. doi: 10.4103/jmp.JMP_117_20
182. Motley R, Ramachandran P, Fielding A. A feasibility study on the development and use of a deep learning model to automate real-time monitoring of tumor position and assessment of interfraction fiducial marker migration in prostate radiotherapy patients(). Biomed Phys Eng Express (2022) 8(3):10.1088/2057-1976/ac34da. doi: 10.1088/2057-1976/ac34da
183. Hiremath A, Shiradkar R, Fu P, Mahran A, Rastinehad AR, Tewari A, et al. An integrated nomogram combining deep learning, prostate imaging-reporting and data system (PI-RADS) scoring, and clinical variables for identification of clinically significant prostate cancer on parametri MRI: a retrospective multicentre study. Lancet Digital Health (2021) 3(7):e445–54. doi: 10.1016/s2589-7500(21)00082-0
184. Zhou Z, Liang Z, Zuo Y, Zhou Y, Yan W, Wu X, et al. Development of a nomogram combining multiparametric magnetic resonance imaging and PSA-related parameters to enhance the detection of clinically significant cancer across different region. Prostate (2022) 82(5):556–65. doi: 10.1002/pros.24302
185. Dou Q, Yu L, Chen H, Jin Y, Yang X, Qin J, et al. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Analysis (2017) 41:40–54. doi: 10.1016/j.media.2017.05.001
186. Tamada T, Kido A, Yamamoto A, Takeuchi M, Miyaji Y, Moriya T, et al. Comparison of biparametric and multiparametric MRI for clinically significant prostate cancer detection with PI-RADS version 2.1. J Magn Reson Imaging: JMRI (2021) 53(1):283–91. doi: 10.1002/jmri.27283
187. Xu L, Zhang G, Shi B, Liu Y, Zou T, Yan W, et al. Comparison of biparametric and multiparametric MRI in the diagnosis of prostate cancer. Cancer imaging: Off Publ Int Cancer Imaging Soc (2019) 19(1):90. doi: 10.1186/s40644-019-0274-9
188. Scialpi M, Prosperi E, D’ANDREA A, Martorana E, Malaspina C, Palumbo B, et al. Biparametric versus multiparametric MRI with non-endorectal coil at 3T in the detection and localization of prostate cancer. Anticancer Res (2017) 37(3):1263–71. doi: 10.21873/anticanres.11443
189. Gohel P, Singh P, MJapa M. Explainable AI: current status and future directions. arXiv. (2021). doi: 10.48550/arXiv.2107.07045
190. Danilevsky M, Qian K, Aharonov R, Katsis Y, Kawas B, PJapa S. A survey of the state of explainable AI for natural language processing. arXiv. (2020). doi: 10.48550/arXiv.2010.00711
191. Chaddad A, Peng J, Xu J, Bouridane A. Survey of Explainable AI Techniques in Healthcare. Sensors (Basel) (2023) 23(2):634. doi: 10.3390/s23020634
192. Zimmermann MT. The importance of biologic knowledge and gene expression context for genomic data interpretation. Front Genet. (2018) 9:670. doi: 10.3389/fgene.2018.00670
193. Sidak D, Schwarzerová J, Weckwerth W, Waldherr S. Interpretable machine learning methods for predictions in systems biology from omics data. Front Mol Biosci. (2022) 9:926623. doi: 10.3389/fmolb.2022.926623
194. Elmarakeby HA, Hwang J, Arafeh R, Crowdis J, Gang S, Liu D, et al. Biologically informed deep neural network for prostate cancer discovery. Nature (2021) 598(7880):348–52. doi: 10.1038/s41586-021-03922-4
195. Echegaray S, Nair V, Kadoch M, Leung A, Rubin D, Gevaert O, et al. A rapid segmentation-insensitive “Digital biopsy” method for radiomic feature extraction: method and pilot study using CT images of non-small cell lung cancer. Tomography (Ann Arbor Mich) (2016) 2(4):283–94. doi: 10.18383/j.tom.2016.00163
196. Gaziev G, Wadhwa K, Barrett T, Koo BC, Gallagher FA, Serrao E, et al. Defining the learning curve for multiparametric magnetic resonance imaging (MRI) of the prostate using MRI-transrectal ultrasonography (TRUS) fusion-guided transperineal prostate biopsies as a validation tool. BJU Int (2016) 117(1):80–6. doi: 10.1111/bju.12892
Keywords: deep learning, machine learning, computer-aided diagnosis, prostate cancer, radiotherapy, precision therapy
Citation: He M, Cao Y, Chi C, Yang X, Ramin R, Wang S, Yang G, Mukhtorov O, Zhang L, Kazantsev A, Enikeev M and Hu K (2023) Research progress on deep learning in magnetic resonance imaging–based diagnosis and treatment of prostate cancer: a review on the current status and perspectives. Front. Oncol. 13:1189370. doi: 10.3389/fonc.2023.1189370
Received: 19 March 2023; Accepted: 30 May 2023;
Published: 13 June 2023.
Edited by:
Marco Fusella, Abano Terme Hospital, ItalyReviewed by:
Martina Murr, University of Tübingen, GermanyRakesh Shiradkar, Emory University, United States
Copyright © 2023 He, Cao, Chi, Yang, Ramin, Wang, Yang, Mukhtorov, Zhang, Kazantsev, Enikeev and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kebang Hu, aHVrYkBqbHUuZWR1LmNu; Mikhail Enikeev, ZW5pa2Vldl9tX2VAc3RhZmYuc2VjaGVub3YucnU=
 Mingze He
Mingze He Yu Cao2
Yu Cao2 Kebang Hu
Kebang Hu