
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Oncol., 19 May 2023
Sec. Cancer Metabolism
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1186539
This article is part of the Research TopicMetabolic Barriers in Cancer and Cancer TherapyView all 10 articles
Arginine, glutamine, and the branched chain amino acids (BCAAs) are a focus of increased interest in the field of oncology due to their importance in the metabolic reprogramming of cancer cells. In the tumor microenvironment (TME), these amino acids serve to support the elevated biosynthetic and energy demands of cancer cells, while simultaneously maintaining the growth, homeostasis, and effector function of tumor-infiltrating immune cells. To escape immune destruction, cancer cells utilize a variety of mechanisms to suppress the cytotoxic activity of effector T cells, facilitating T cell exhaustion. One such mechanism is the ability of cancer cells to overexpress metabolic enzymes specializing in the catabolism of arginine, glutamine, and the BCAAs in the TME. The action of such enzymes supplies cancer cells with metabolic intermediates that feed into the TCA cycle, supporting energy generation, or providing precursors for purine, pyrimidine, and polyamine biosynthesis. Armed with substantial metabolic flexibility, cancer cells redirect amino acids from the TME for their own advantage and growth, while leaving the local infiltrating effector T cells deprived of essential nutrients. This review addresses the metabolic pressure that cancer cells exert over immune cells in the TME by up-regulating amino acid metabolism, while discussing opportunities for targeting amino acid metabolism for therapeutic intervention. Special emphasis is given to the crosstalk between arginine, glutamine, and BCAA metabolism in affording cancer cells with metabolic dominance in the TME.
Recent advances in our understanding of the interactions between cancer and immune cells strongly suggest the outcome of the anti-tumor T cell response is dictated by the nutrient availability and the flexibility of cancer and T cell metabolism (1–3). Cancer cells remodel their metabolism to escape immune surveillance in the TME creating nutrient-depleted TME with dysfunctional and exhausted T cells (4, 5). Amino acid deprivation is one of the signatures of nutrient-deprived TME.
Arginine, glutamine, and the BCAAs are needed to support the increased biosynthetic and bioenergetic demands of the growing tumor and the incoming tumor infiltrating lymphocytes (TILs) (6–8). These amino acids interconnect at several metabolic steps. Breakdown of BCAAs to branched chain keto acids (BCKAs) releases glutamate, which is the precursor for glutamine (9). Glutamine is converted into ornithine, which is the precursor of arginine (10). Arginine and ornithine are precursors for polyamine synthesis, which is upregulated in cancer and immune cells (11). The depletion of glutamine, arginine or the BCAAs in the TME, alone or in combination, may impact the ability of TILs to eliminate cancer cells. However, TILs and cancer cells share similar requirements for these amino acids, creating a practical conundrum regarding nutrient-based cancer treatments (12, 13). This review provides an overview of glutamine, arginine and the BCAAs based on recent discoveries in the context of TME and the challenges associated with future therapeutic approaches.
Dietary intake and protein degradation are the main sources of arginine for growing children. Postnatally, humans synthesize arginine via the intestinal-renal axis. This interorgan process includes the synthesis of citrulline by the small intestines and its absorption by the kidneys where citrulline is converted to arginine by argininosuccinate synthase 1 (ASS1) and lyase (ASL) (14). Once released in the circulation, arginine enters cells preferentially via cationic amino acid transporters (CATs) existing in eight different isoforms, each with different tissue distribution (Figure 1) (15). Inside the cells, arginine is incorporated into new protein, or used for polyamine and collagen synthesis, or as an activator of the mammalian target of rapamycin (mTOR) (Figure 1) (16). Thus, arginine availability is crucial for maintaining physiological cell function.
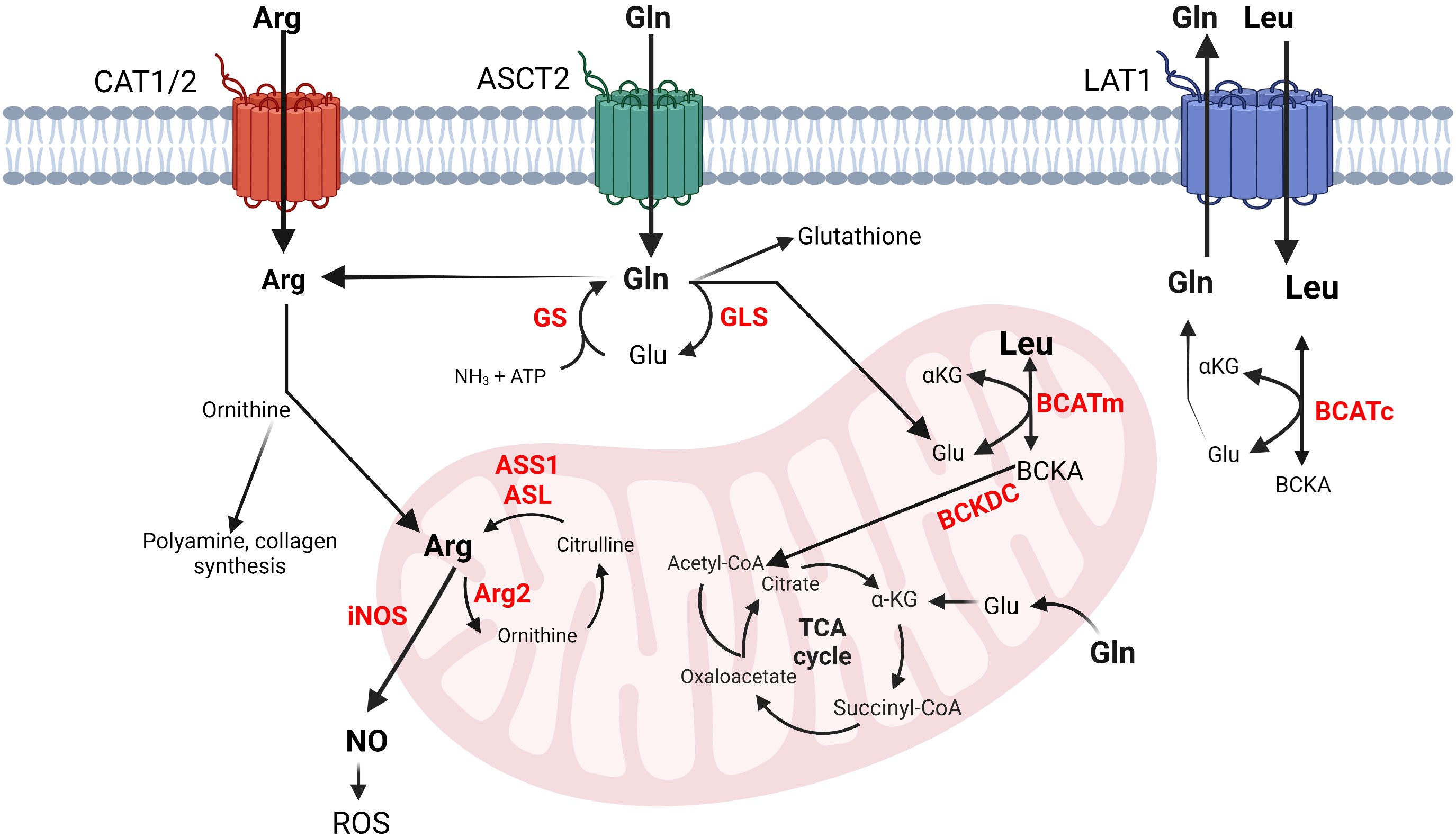
Figure 1 Simplified schematics of glutamine, arginine and BCAA metabolic interconnections in cancer and immune cells. Left to right: Arginine transportation is assisted by CAT. Arginine can be converted into ornithine, polyamines, collagen, or nitric oxide (NO). Glutamine enters the cells via ASCT2 and is converted into glutamate, glutathione, arginine, or nucleotides (not shown) or it may exit the cells via LAT1, which transfers leucine in exchange for glutamine. Leucine is converted into its corresponding BCKA in the cytosol or in the mitochondria. Arginine and glutamine (not shown) can be also synthesized in mitochondria. Glutamine and the BCAAs contribute to energy production by feeding into the TCA cycle. The metabolism of BCAAs is illustrated with leucine. The enzyme names are given in red. arginine, Arg, glutamine; Gln, glutamate; Glu, glutamine synthase; GS, glutaminase; GLS, arginosuccinate synthase 1; ASS1, arginosuccinate lyase; ASL, inducible nitric oxide synthase; iNOS, arginase 2; Arg2, leucine; Leu, α-ketoglutarate; αKG, cytosolic and mitochondrial branched chain aminotransferase BCATc and BCATm, branched chain keto acids; BCKAs, reactive oxygen species; ROS.
Arginine catabolism includes the urea cycle and nitric oxide (NO) production. The urea cycle comprises five enzymatic reactions that occur within the liver. Carbamoyl phosphate synthetase 1 (CPS1) incorporates ammonia into carbamoyl phosphate followed by formation of citrulline by ornithine transcarbamoylase (OTC), and argininosuccinate by ASS1. Arginine is then produced by ASL followed by hydrolysis by arginase 1 (Arg1) to urea and ornithine (16). Arg1 is a cytosolic enzyme expressed in the liver; however, humans express mitochondrial arginase, Arg2, in most tissues (17). During NO synthesis, nitric oxide synthases (NOS) catalyze the oxidation of arginine to NO and citrulline (Figure 1) (18). Mammals have three NOS isoforms, NOS1-3. NOS2 is the inducible and prevalent isoform in immune cells (iNOS) (19). The mononuclear myeloid-derived suppressor cells (M-MDSCs) rely on iNOS to drive immunosuppression (20). High expression of iNOS in M-MDSCs cells releases NO, which is converted into reactive oxygen species (ROS) causing DNA damage and promoting tumor growth (21).
Arginine is conditionally essential in patients with severe trauma, compromised immune system, or cancer cachexia. Under these disease states, the demand for arginine exceeds its endogenous production (22, 23).
Defective arginine synthesis (arginine auxotrophy) is a common occurrence in cancer cells. It primarily associates with a deficiency in ASS1 (24). To persist in the TME, CD4+ and CD8+ T cells must maintain adequate arginine concentrations. Arg2-deficient CD8+ T cells display enhanced cytotoxic activity against murine melanoma B16-OVA and colon adenocarcinoma MC38-OVA (25). The Arg2-deficent CD8+ T cells have improved effector function as seen by increased perforin, granzyme, IFN-γ and IL-2 (25). Alternatively, Arg2-specific human CD8+ T cells recognize Arg2-expressing regulatory T cells (Tregs), suggesting a naturally existing immunomodulatory potential of CD8+ T cells to remove immune suppression by targeting Tregs with high Arg2 expression (26). Similarly, Arg1-specific T cells target Arg1-expressing myeloid cells (27). In another study, bone marrow derived dendritic cells (BMDCs) and peritoneal macrophages synthesize arginine via ASL and ASS1 and supply CD4+ T cells with arginine (28). Studies with colorectal cancer patients failed to support the hypothesis that supplementation with arginine reduces the frequency of immunosuppressive M-MDSCs and polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) but increases the frequency of CD4+ T cells. Thus, while arginine deficiency contributes to immunosuppression, systemic arginine supplementation alone does not restore immune system activity (29).
The rest of the urea cycle enzymes, CPS1 and OTC are studied to a lesser extent in cancer and immune cells (30). Cancer cells upregulate CPS1 to prevent ammonia buildup. A small-molecule inhibitor of CPS1 (H3B-120) that blocks CPS1 activity in human hepatocytes might be valuable for future therapeutic approaches (31). In contrast to CPS1, OTC is downregulated in cancer cells leading to accumulation of ammonia. Cancer cells can recycle ammonia for amino and nucleic acid synthesis (32). Lastly, a virus-induced metabolic reprogramming of mouse liver, results in transcriptional repression of the OTC and ASS1 genes leading to decreased arginine but increased ornithine concentrations in the circulation, which in turn suppresses virus-specific CD8+ T cells (33).
Glutamine is the most abundant non-essential amino acid within human plasma. It contributes to nucleic acid (34) and protein synthesis (35), cellular response to ROS (36), and energy production through the TCA cycle (37). It is conditionally essential for proliferating cells during high demand, where endogenous synthesis is insufficient to support cellular homeostasis (38). Glutamine synthase (GS) generates glutamine from glutamate and ammonia (34). This reaction facilitates interorgan ammonium and glutamate transport, prevents toxic encephalopathy and blood acidification (35). Glutamine hydrolysis to glutamate and ammonia is facilitated, in part, by glutaminase-1 (GLS-1) in the kidney and glutaminase-2 (GLS-2) in the liver. Different transport systems specialize in assisting glutamine import and export by the cells. Among them are the sodium-dependent transporter ASCT2 (Solute Carrier 1a5, Slc1a5) and the sodium-independent antiporter Slc3a2 that work together with Slc7a5 (also known as L-type amino acid transporter 1, LAT1) to exchange glutamine for leucine (Figure 1). These transporters have vast tissue distribution, but most notably they are overexpressed in immune and cancer cells (12, 36, 37).
Cancer reliance on glutamine is established in tumors throughout the body, including pancreatic (39), prostate (40), breast (41), and liver (42) cancers. Increased expression of ASCT2 and GLS are found in squamous cell carcinoma, adenocarcinoma, and neuroendocrine lung tumors (43). Such increases in the expression of ASCT2 and GLS are linked to tumors with aberrant oncogene c-MYC (40, 44). With a growing dependence on exogenous glutamine, tumor cells exhibit “glutamine addiction”. Glutamine addiction prevents cells from relying on endogenous glutamine synthesis and leads to cell death in glutamine free environments (45).
Similarly, immune cells rely on glutamine to sustain homeostasis and execute proper functions. A blockage of glutamine metabolism by DON (6-diazo-5-oxy-L-norleucine), or its modified prodrug JHU-083, causes a shift of CD8+ T cells towards a long-lived memory state and increases their tumor infiltration potential and survival in the TME (46–48). A loss of GLS halts Th17 differentiation but promotes the expression of Tbet and stimulates Th1 and CD8+ T cells. A long-term loss of GLS correlates with an impaired Th17 immune response, yet a transient loss of GLS promotes Th17, but restricts Th1 and CD8+ T cell effector differentiation (49). In a glutamine-depleted environment, activated CD8+ T cells produce significantly less IFN-γ and TNF-α (50). Selective GLS inhibition by CB-839, Telaglenastat, impairs the clonal expansion and activation of CD8+ T cells in the context of combinatorial anti-PD-1 treatment (51). In glutamine-addicted clear cell renal cell carcinoma (ccRCC), tumor-associated macrophages (TAMs) shift to M2 (immunosuppressive phenotype) promoting a pro-tumor environment. Such TAMs produce IL-23 in the context of hypoxia (HIF-α activation), activating Tregs (52). Taken together, glutamine metabolism plays an important role in T cell activation and function.
The BCAAs (leucine, isoleucine, and valine) are supplemented through the diet to mammalian cells. BCAAs make up ~35% of the essential amino acids in the blood (53). The BCAAs are important nutrients under physiological and pathological conditions (54). They are nitrogen donors to glutamate and alanine and stimulate protein synthesis in the muscle (55). In the brain, the BCAAs maintain the glutamate-glutamine interconversions by engaging in “glutamate-BCAA” cycles between neurons and astrocytes (56). BCAAs trigger insulin release from the pancreatic β-islets; however, chronically elevated plasma BCAAs are a common clinical finding in patients with Type 2 Diabetes and Cardiovascular Disease (56, 57).
BCAAs travel across the plasma membranes utilizing the heterodimeric transporter Slc7a5/Slc3a2. As stated earlier, this transporter works in antiport with Slc1a5 where glutamine efflux proceeds BCAA influx (6, 58). Once inside the cells, BCAAs are incorporated into protein or subjected to degradation by the cytosolic branched chain aminotransferase, BCATc (6). Alternately, the BCAAs enter the mitochondria, assisted by the Scl24a44 transporter, to become subjected to degradation by the mitochondrial BCATm (59). BCATc and BCATm catalyze the reversible transamination of the BCAAs to their corresponding BCKAs, which are subjected to irreversible oxidative decarboxylation by the mitochondrial branched chain alpha-ketoacid dehydrogenase complex (BCKDC). Following this step, each BCAA commits to their unique degradation pathways releasing propionyl-CoA, acetoacetate, or acetyl-CoA that feed into the TCA cycle or other pathways (Figure 1) (60).
BCAAs are important for sustainable tumor growth. The growing tumor obtains BCAAs from the circulation or the tissues surrounding it. Positive association between elevated plasma BCAAs and the risk of colorectal adenoma and pancreatic adenocarcinoma are reported in human patients but controversial in animal studies (61–65). High plasma concentrations of BCAAs, due to disruption in BCAA metabolism, or dietary supplementation with BCAAs, are associated with delayed onset of lymphoma, or suppression of breast cancer in mice (63, 64). In contrast, mice subjected to a diet high in BCAAs, have increased incidences of pancreatic ductal adenocarcinoma (PDAC) (66). Elevated BCAA metabolism at the BCAT step is implicated in the onset of many cancers including glioblastoma (53) myeloid leukemia (54) lymphoma (50) lung (55), gastric (56), pancreatic (57) and breast cancers (58).
To a lesser extent, BCAAs and their metabolism are studied in immune cells. Leucine is indispensable for T cell activation as insufficient leucine prevents clonal expansion to Th1, Th17 and CD8+ T cells (37). Mice deficient of Slc3a2 in Foxp3+ Tregs, generate a low number of Foxp3+ Tregs and fail to suppress intestinal inflammation (67). CD4+ T cells, deficient in BCATc or BCATm, have higher glycolytic capacity, improved oxygen consumption and increased capacity to secrete IFNγ (6, 68). Studies with BCATc in human macrophages identified a non-catalytic role for BCATc in the metabolic events associated with fragmented TCA cycle (69, 70). It remains to be further established whether the non-enzymatic function of BCATc represents a universal mechanism to regulate cellular metabolism.
Rapidly dividing cancer cells are forced to reprogram their metabolism to ensure long term survival and metastatic growth. Their major opponents, the effector Th1 and CD8+ T cells, must also reprogram metabolism to embrace the harsh TME. However, these functionally unrelated cells have similar demands for nutrients, including amino acids (71, 72).
Numerous reports have demonstrated uptake of glutamine, arginine, and the BCAAs is upregulated in cancer and activated Th1 and CD8+ T cells (6, 73–75). There is a high redundancy in transport preference for these amino acids, making current approaches to target amino acid uptake particularly challenging (6, 76). Ovarian cancer cells, CD4+ and CD8+ memory T cells, and M0 macrophages overexpress the arginine CAT1 transporter. Silencing CAT1 in the ovarian cancer cells significantly reduces the concentration of arginine but lowers the concentrations of BCAAs (77). The uptake of glutamine by human breast HCC1806 cancer cells, deficient in ASCT2, is sensitive to the inhibition of leucine uptake when LAT1 is targeted by JPH203 (78). This suggests that LAT1 plays a role as a rescue transporter for glutamine. In breast cancer biopsies, high LAT1 expression is associated with invasive breast cancer where LAT1 overexpression positively correlates with the expression of the estrogen receptor (ER) and the programmed death ligand-1 (PD-L1) (79). LAT1 is highly expressed in malignant skin lesions (80) and in cells from patients with skin disorders (81). Increased LAT1 expression is observed in keratinocytes and dermal infiltrating lymphocytes of patients with psoriasis, where LAT1 expression is upregulated by IL-23 and IL-1b (81). Thus, scientific evidence exists to support the notion of high reliance of malignant and non-malignant cells on amino acid transporters specializing in the uptake of arginine, glutamine, and the BCAAs. Because these transporters exert overlapping functions, their targeting may impact the uptake of more than one amino acid in clinical trials.
Most of arginine, glutamine and the BCAAs are delivered to the TME for incorporation in new protein. However, 20-25% are degraded or used to stimulate signal transduction cascades, such as mTOR pathway (72). Such distribution is necessary to supply the cells with fuel and precursor metabolites for purine, pyrimidine, or polyamine biosynthesis (Figure 2) (82–85). The intracellular concentrations of these amino acids, however, fluctuate based on shared metabolic precursors and enzymatic reactions. A global deletion of BCATm leads to a reduction in lymphoma burden, which correlates with elevated concentrations of BCAAs, but reduced concentrations of glutamine (64). In a non-small cell lung carcinoma (NSCLC), nitrogen derived from BCAA transamination supports glutamine and nucleotide synthesis via the glutamine-purine-pyrimidine axis. However, such reliance on nitrogen from BCAAs is not observed in PDAC (86). In contrast, BCATc selective inhibition, but not changes in the BCAAs, results in upregulation of genes involved in the transport of glutamate and the conversion of glutamate into glutathione in human macrophages (70). Similarly, mouse embryonic fibroblasts, grown in a glutamine-depleted environment, show a significant increase in arginine, but not in BCAAs. The arginine levels balance off, while the levels of BCAAs increase when TP53 is deleted. The authors thus identified the tumor suppressor p53 as an important transcriptional regulator of arginine uptake during a pro-survival response to glutamine-induced metabolic stress (87). In the TME, restricting glutamine or glutamine-dependent purine and pyrimidine synthesis shifts CD4+ T cells toward Tregs but this shift is abolished if GS is inhibited. GS is described as de-repressed under low glutamine, or nucleotide starvation (88). Arginine is a precursor of polyamines and targeting enzymes such as Arg1, can impact the synthesis of polyamines in the TME. Polyamines exert immunosuppressive effects, promoting tumor growth (83). Arg1 is overexpressed in dendritic cells and represents one of the immune checkpoints in the TME (11). Dendritic cells may deprive the TME of arginine causing T cell exhaustion (83).
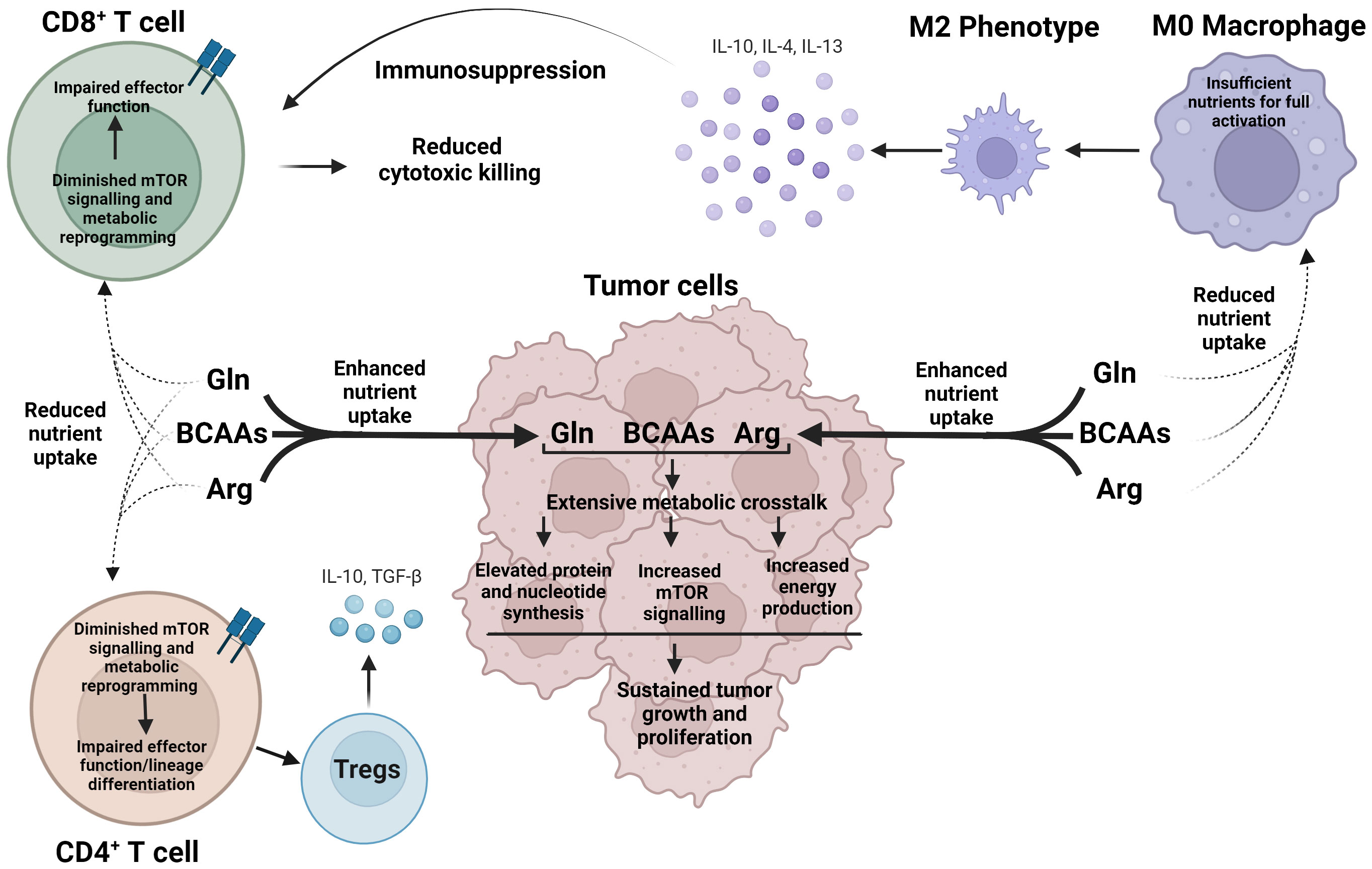
Figure 2 Cancer cells exert metabolic dominance over immune cells within the TME to avoid detection and destruction. In a nutrient-depleted TME, cancer cells preferentially uptake arginine, glutamine, and BCAAs, which undergo vast, interconnected metabolic pathways to produce essential biosynthetic precursors to support rapid cancer growth, as well as activate mTOR signaling. Oppositely, reduced nutrient uptake of arginine, glutamine, and the BCAAs voids CD4+ and CD8+ T cells of essential nutrients and diminishes mTOR signaling leading to impaired effector function and aberrant lineage commitment. As a result, immune cells, such as M2 macrophages and Tregs cells, are generated, which in turn release immunosuppressive cytokines, promoting an environment for cancer growth. Arg, arginine; Gln, glutamine; BCAAs Branched chain amino acids.
Lastly, arginine, glutamine and the BCAAs activate complex 1 of mTOR in cancer and immune cells. Nutrient sensing via mTOR is essential for growth and survival; however, in the context of TME, this is yet another mechanism cancer and immune cells exploit to compete for nutrients (Figure 2). mTOR signaling is dysregulated in cancer cells, while T cell function requires upregulation of mTOR (89–91). While leucine is the most potent activator of mTOR as reviewed in (6), glutamine and arginine are other stimulators of mTOR signaling. mTOR sensing may occur via Rag-GTPase-dependent and independent pathways and may engage different protein targets (92, 93). Leucine-driven activation of mTOR includes GATOR1-2, Sestrin2, and SAR1B and follows the Rag-GTPase dependent mechanism (94, 95). Arginine cannot bind Sestrin 2 or SAR1B but requires a lysosomal membrane protein SLC38A9 (96). Glutamine synergizes asparagine to activate mTOR signaling via Rag-GTPase independent mechanism (93). In summary, cancer and immune cells co-exist in the TME in a bidirectional metabolic relationship, influenced by the fluctuations in arginine, glutamine and the BCAAs.
Because arginine, glutamine and the BCAAs are required for growth of cancer and immune cells, targeted deprivation or supplementation of these amino acids may lead to undesirable therapeutic effects (97). Selectively limiting the availability of these amino acids in tumor cells while supplying them to immune cells may help overcome this obstacle. Indeed, pharmacological inhibition of glutamine uptake by the ASCT2 inhibitor, V-9302, blocks glutamine uptake in triple negative breast cancer cells but not in CD8+ T cells. The CD8+ T cells adapt by upregulating a Na+/Cl-dependent neutral and cationic amino acid transporter ATB0,+ (98). A similar approach is used in keratinocytes from patients with psoriasis, where deleting LAT1 controls skin inflammation, while CD4+ T cells use alternative amino acid transporters (LAT2 and LAT3) (81). Lastly, pro-drugs, such as DRP104, target GLS-1 in tumors and cause CD8+ T cell-dependent tumor regression (94) Such approaches could potentially unleash the immune cells in destroying cancer cells in the TME.
The endurance of the chimeric antigen receptor T (CAR-T) cells in hematological and solid malignancies can be affected by amino-acid depleted TME. Induced expression of ASS1 in re-engineered CAR-T cells increases their proliferation without compromising their function (99).
A combinatorial therapy including multivesicular liposome technology, designed to supply arginine to melanoma tumors, and selective suppression of the CAT2 transporter, leads to arginine starvation of tumor cells but promotes the infiltration of CD8+ T cells in the TME (100). Similarly, a local therapy using nanoparticles to deliver poly(L-arginine) and hyaluronic acid to tumor-associated macrophages successfully induces tumor-suppressive M1 phenotype and leads to an increased iNOS expression in these cells (101).
Although still in their infancy, nanomaterials or liposome-based technologies could be expanded to deliver glutamine and BCAAs to CD8+ and CD4+T cells in the TME. In addition, new generations of CAR-T cells could be designed to competitively intake arginine, glutamine and BCAAs from the TME. Under such a scenario, systemic side effects should be minimal and can address the low therapeutic efficacy of the conventional cancer therapies.
TW and EA designed the mini review. TW prepared the glutamine overview and Figures 1, 2. SE prepared the arginine overview. LW and LF prepared the BCAA overview. EA compiled and edit the different sections. All authors contributed to the article and approved the submitted version.
The digital illustrations in Figure 1 and Figure 2 were created using Biorender.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Delgoffe GM. Filling the tank: keeping antitumor T cells metabolically fit for the long haul. Cancer Immunol Res (2016) 4(12):1001–6. doi: 10.1158/2326-6066.CIR-16-0244
2. DePeaux K, Delgoffe GM. Metabolic barriers to cancer immunotherapy. Nat Rev Immunol (2021) 21(12):785–97. doi: 10.1038/s41577-021-00541-y
3. Lim AR, Rathmell WK, Rathmell JC. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. Elife. (2020) 9. doi: 10.7554/eLife.55185
4. Kim SK, Cho SW. The evasion mechanisms of cancer immunity and drug intervention in the tumor microenvironment. Front Pharmacol (2022) 13:868695. doi: 10.3389/fphar.2022.868695
5. Aktar N, Yueting C, Abbas M, Zafar H, Paiva-Santos AC, Zhang Q, et al. Understanding of immune escape mechanisms and advances in cancer immunotherapy. J Oncol (2022) 2022:8901326. doi: 10.1155/2022/8901326
6. Wetzel TJ, Erfan SC, Ananieva EA. The emerging role of the branched chain aminotransferases, BCATc and BCATm, for anti-tumor T-cell immunity. Immunometabolism (2023) 5(1):e00014. doi: 10.1097/IN9.0000000000000014
7. Endicott M, Jones M, Hull J. Amino acid metabolism as a therapeutic target in cancer: a review. Amino Acids (2021) 53(8):1169–79. doi: 10.1007/s00726-021-03052-1
8. Lieu EL, Nguyen T, Rhyne S, Kim J. Amino acids in cancer. Exp Mol Med (2020) 52(1):15–30. doi: 10.1038/s12276-020-0375-3
9. Holeček M. Branched-chain amino acids in health and disease: metabolism, alterations in blood plasma, and as supplements. Nutr Metab (2018) 15:33. doi: 10.1186/s12986-018-0271-1
10. Watford M. Glutamine metabolism and function in relation to proline synthesis and the safety of glutamine and proline supplementation. J Nutr (2008) 138(10):2003s–7s. doi: 10.1093/jn/138.10.2003S
11. Martí ILAA, Reith W. Arginine-dependent immune responses. Cell Mol Life Sci (2021) 78(13):5303–24. doi: 10.1007/s00018-021-03828-4
12. Wei Z, Liu X, Cheng C, Yu W, Yi P. Metabolism of amino acids in cancer. Front Cell Dev Biol (2020) 8:603837. doi: 10.3389/fcell.2020.603837
13. Kelly B, Pearce EL. Amino assets: how amino acids support immunity. Cell Metab (2020) 32(2):154–75. doi: 10.1016/j.cmet.2020.06.010
14. Marini JC, Agarwal U, Robinson JL, Yuan Y, Didelija IC, Stoll B, et al. The intestinal-renal axis for arginine synthesis is present and functional in the neonatal pig. Am J Physiol Endocrinol Metab (2017) 313(2):E233–e42. doi: 10.1152/ajpendo.00055.2017
15. Werner A, Pieh D, Echchannaoui H, Rupp J, Rajalingam K, Theobald M, et al. Cationic amino acid transporter-1-Mediated arginine uptake is essential for chronic lymphocytic leukemia cell proliferation and viability. Front Oncol (2019) 9:1268. doi: 10.3389/fonc.2019.01268
16. Morris SM Jr. Arginine metabolism revisited J Nutr (2016) 146(12):2579s–86s doi: 10.3945/jn.115.226621
17. Grzywa TM, Sosnowska A, Matryba P, Rydzynska Z, Jasinski M, Nowis D, et al. Myeloid cell-derived arginase in cancer immune response. Front Immunol (2020) 11:938. doi: 10.3389/fimmu.2020.00938
18. Stuehr DJ. Enzymes of the l-arginine to nitric oxide pathway. J Nutr (2004) 134(10 Suppl):2748S–51S. doi: 10.1093/jn/134.10.2748S
19. Morris SM Jr. Arginine: master and commander in innate immune responses Sci Signal (2010) 3(135):pe27 doi: 10.1126/scisignal.3135pe27
20. Ekmekcioglu S, Grimm EA, Roszik J. Targeting iNOS to increase efficacy of immunotherapies. Hum Vaccin Immunother. (2017) 13(5):1105–8. doi: 10.1080/21645515.2016.1276682
21. Wu Y, Yi M, Niu M, Mei Q, Wu K. Myeloid-derived suppressor cells: an emerging target for anticancer immunotherapy. Mol Cancer. (2022) 21(1):184. doi: 10.1186/s12943-022-01657-y
22. Tabe Y, Lorenzi PL, Konopleva M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood. (2019) 134(13):1014–23. doi: 10.1182/blood.2019001034
23. Arribas-López E, Zand N, Ojo O, Snowden MJ, Kochhar T. The effect of amino acids on wound healing: a systematic review and meta-analysis on arginine and glutamine. Nutrients. (2021) 13(8):2498. doi: 10.3390/nu13082498
24. Delage B, Fennell DA, Nicholson L, McNeish I, Lemoine NR, Crook T, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J Cancer. (2010) 126(12):2762–72. doi: 10.1002/ijc.25202
25. Martí i Líndez AA, Dunand-Sauthier I, Conti M, Gobet F, Núñez N, Hannich JT, et al. Mitochondrial arginase-2 is a cell−autonomous regulator of CD8+ T cell function and antitumor efficacy. JCI Insight (2019) 4(24):e132975. doi: 10.1172/jci.insight.132975
26. Perez-Penco M, Weis-Banke SE, Schina A, Siersbæk M, Hübbe ML, Jørgensen MA, et al. TGFβ-derived immune modulatory vaccine: targeting the immunosuppressive and fibrotic tumor microenvironment in a murine model of pancreatic cancer. J Immunother Cancer (2022) 10(12):e005491. doi: 10.1136/jitc-2022-005491
27. Martinenaite E, Ahmad SM, Bendtsen SK, Jørgensen MA, Weis-Banke SE, Svane IM, et al. Arginase-1-based vaccination against the tumor microenvironment: the identification of an optimal T-cell epitope. Cancer Immunol Immunother. (2019) 68(11):1901–7. doi: 10.1007/s00262-019-02425-6
28. Crowther RR, Schmidt SM, Lange SM, McKell MC, Robillard MC, Zhao J, et al. Cutting edge: l-arginine transfer from antigen-presenting cells sustains CD4(+) T cell viability and proliferation. J Immunol (2022) 208(4):793–8. doi: 10.4049/jimmunol.2100652
29. Szefel J, Ślebioda T, Walczak J, Kruszewski WJ, Szajewski M, Ciesielski M, et al. The effect of l-arginine supplementation and surgical trauma on the frequency of myeloid-derived suppressor cells and T lymphocytes in tumour and blood of colorectal cancer patients. Adv Med Sci (2022) 67(1):66–78. doi: 10.1016/j.advms.2021.12.005
30. Hajaj E, Sciacovelli M, Frezza C, Erez A. The context-specific roles of urea cycle enzymes in tumorigenesis. Mol Cell (2021) 81(18):3749–59. doi: 10.1016/j.molcel.2021.08.005
31. Yao S, Nguyen TV, Rolfe A, Agrawal AA, Ke J, Peng S, et al. Small molecule inhibition of CPS1 activity through an allosteric pocket. Cell Chem Biol (2020) 27(3):259–68.e5. doi: 10.1016/j.chembiol.2020.01.009
32. Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. (2017) 358(6365):941–6. doi: 10.1126/science.aam9305
33. Lercher A, Bhattacharya A, Popa AM, Caldera M, Schlapansky MF, Baazim H, et al. Type I interferon signaling disrupts the hepatic urea cycle and alters systemic metabolism to suppress T cell function. Immunity. (2019) 51(6):1074–87.e9. doi: 10.1016/j.immuni.2019.10.014
34. Cruzat V, Macedo Rogero M, Noel Keane K, Curi R, Newsholme P. Glutamine: metabolism and immune function, supplementation and clinical translation. Nutrients. (2018) 10(11):1564. doi: 10.3390/nu10111564
35. Hakvoort TB, He Y, Kulik W, Vermeulen JL, Duijst S, Ruijter JM, et al. Pivotal role of glutamine synthetase in ammonia detoxification. Hepatology. (2017) 65(1):281–93. doi: 10.1002/hep.28852
36. Scalise M, Pochini L, Galluccio M, Console L, Indiveri C. Glutamine transport and mitochondrial metabolism in cancer cell growth. Front Oncol (2017) 7:306. doi: 10.3389/fonc.2017.00306
37. Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol (2013) 14(5):500–8. doi: 10.1038/ni.2556
38. Yang WH, Qiu Y, Stamatatos O, Janowitz T, Lukey MJ. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer. (2021) 7(8):790–804. doi: 10.1016/j.trecan.2021.04.003
39. Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. (2013) 496(7443):101–5. doi: 10.1038/nature12040
40. Mukha A, Kahya U, Linge A, Chen O, Löck S, Lukiyanchuk V, et al. GLS-driven glutamine catabolism contributes to prostate cancer radiosensitivity by regulating the redox state, stemness and ATG5-mediated autophagy. Theranostics. (2021) 11(16):7844–68. doi: 10.7150/thno.58655
41. Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther (2014) 13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870
42. Jin H, Wang S, Zaal EA, Wang C, Wu H, Bosma A, et al. A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. Elife (2020) 9:e56749. doi: 10.7554/eLife.56749
43. Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H, et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res (2013) 19(3):560–70. doi: 10.1158/1078-0432.CCR-12-2334
44. Nagarajan A, Malvi P, Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. (2016) 2(7):365–77. doi: 10.1016/j.trecan.2016.06.002
45. Wang Y, Bai C, Ruan Y, Liu M, Chu Q, Qiu L, et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat Commun (2019) 10(1):201. doi: 10.1038/s41467-018-08033-9
46. Oh MH, Sun IH, Zhao L, Leone RD, Sun IM, Xu W, et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J Clin Invest. (2020) 130(7):3865–84. doi: 10.1172/JCI131859
47. Nabe S, Yamada T, Suzuki J, Toriyama K, Yasuoka T, Kuwahara M, et al. Reinforce the antitumor activity of CD8(+) T cells via glutamine restriction. Cancer Sci (2018) 109(12):3737–50. doi: 10.1111/cas.13827
48. Leone RD, Zhao L, Englert JM, Sun IM, Oh MH, Sun IH, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. (2019) 366(6468):1013–21. doi: 10.1126/science.aav2588
49. Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. (2018) 175(7):1780–95.e19. doi: 10.1016/j.cell.2018.10.001
50. Presnell SR, Spear HK, Durham J, Riddle T, Applegate A, Lutz CT. Differential fuel requirements of human NK cells and human CD8 T cells: glutamine regulates glucose uptake in strongly activated CD8 T cells. Immunohorizons. (2020) 4(5):231–44. doi: 10.4049/immunohorizons.2000020
51. Best SA, Gubser PM, Sethumadhavan S, Kersbergen A, Negrón Abril YL, Goldford J, et al. Glutaminase inhibition impairs CD8 T cell activation in STK11-/Lkb1-deficient lung cancer. Cell Metab (2022) 34(6):874–87.e6. doi: 10.1016/j.cmet.2022.04.003
52. Fu Q, Xu L, Wang Y, Jiang Q, Liu Z, Zhang J, et al. Tumor-associated macrophage-derived interleukin-23 interlinks kidney cancer glutamine addiction with immune evasion. Eur Urol. (2019) 75(5):752–63. doi: 10.1016/j.eururo.2018.09.030
53. Blair MC, Neinast MD, Arany Z. Whole-body metabolic fate of branched-chain amino acids. Biochem J (2021) 478(4):765–76. doi: 10.1042/BCJ20200686
54. Cuomo P, Capparelli R, Iannelli A, Iannelli D. Role of branched-chain amino acid metabolism in type 2 diabetes, obesity, cardiovascular disease and non-alcoholic fatty liver disease. Int J Mol Sci (2022) 23(8):4325. doi: 10.3390/ijms23084325
55. Holeček M. Branched-chain amino acid supplementation in treatment of liver cirrhosis: updated views on how to attenuate their harmful effects on cataplerosis and ammonia formation. Nutrition. (2017) 41:80–5. doi: 10.1016/j.nut.2017.04.003
56. Yudkoff M. Interactions in the metabolism of glutamate and the branched-chain amino acids and ketoacids in the CNS. Neurochem Res (2017) 42(1):10–8. doi: 10.1007/s11064-016-2057-z
57. Siddik MAB, Shin AC. Recent progress on branched-chain amino acids in obesity, diabetes, and beyond. Endocrinol Metab (2019) 34(3):234–46. doi: 10.3803/EnM.2019.34.3.234
58. Wang W, Zou W. Amino acids and their transporters in T cell immunity and cancer therapy. Mol Cell (2020) 80(3):384–95. doi: 10.1016/j.molcel.2020.09.006
59. Yoneshiro T, Wang Q, Tajima K, Matsushita M, Maki H, Igarashi K, et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature. (2019) 572(7771):614–9. doi: 10.1038/s41586-019-1503-x
60. Neinast M, Murashige D, Arany Z. Branched chain amino acids. Annu Rev Physiol (2019) 81:139–64. doi: 10.1146/annurev-physiol-020518-114455
61. Mayers JR, Wu C, Clish CB, Kraft P, Torrence ME, Fiske BP, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med (2014) 20(10):1193–8. doi: 10.1038/nm.3686
62. Lee K, Blanton C. The effect of branched-chain amino acid supplementation on cancer treatment. Nutr Health (2023), 2601060231153428. doi: 10.1177/02601060231153428
63. Chi R, Yao C, Chen S, Liu Y, He Y, Zhang J, et al. Elevated BCAA suppresses the development and metastasis of breast cancer. Front Oncol (2022) 12:887257. doi: 10.3389/fonc.2022.887257
64. Ananieva EA, Bostic JN, Torres AA, Glanz HR, McNitt SM, Brenner MK, et al. Mice deficient in the mitochondrial branched-chain aminotransferase (BCATm) respond with delayed tumour growth to a challenge with EL-4 lymphoma. Br J Cancer. (2018) 119(8):1009–17. doi: 10.1038/s41416-018-0283-7
65. Budhathoki S, Iwasaki M, Yamaji T, Yamamoto H, Kato Y, Tsugane S. Association of plasma concentrations of branched-chain amino acids with risk of colorectal adenoma in a large Japanese population. Ann Oncol (2017) 28(4):818–23. doi: 10.1093/annonc/mdw680
66. Li JT, Li KY, Su Y, Shen Y, Lei MZ, Zhang F, et al. Diet high in branched-chain amino acid promotes PDAC development by USP1-mediated BCAT2 stabilization. Natl Sci Rev (2022) 9(5):nwab212. doi: 10.1093/nsr/nwab212
67. Ikeda K, Kinoshita M, Kayama H, Nagamori S, Kongpracha P, Umemoto E, et al. Slc3a2 mediates branched-chain amino-Acid-Dependent maintenance of regulatory T cells. Cell Rep (2017) 21(7):1824–38. doi: 10.1016/j.celrep.2017.10.082
68. Ananieva EA, Patel CH, Drake CH, Powell JD, Hutson SM. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. J Biol Chem (2014) 289(27):18793–804. doi: 10.1074/jbc.M114.554113
69. Papathanassiu AE, Ko JH, Imprialou M, Bagnati M, Srivastava PK, Vu HA, et al. BCAT1 controls metabolic reprogramming in activated human macrophages and is associated with inflammatory diseases. Nat Commun (2017) 8:16040. doi: 10.1038/ncomms16040
70. Ko J-H, Olona A, Papathanassiu AE, Buang N, Park K-S, Costa ASH, et al. BCAT1 affects mitochondrial metabolism independently of leucine transamination in activated human macrophages. J Cell Sci (2020) 133(22). doi: 10.1242/jcs.247957
71. Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell (2020) 78(6):1019–33. doi: 10.1016/j.molcel.2020.05.034
72. Vettore L, Westbrook RL, Tennant DA. New aspects of amino acid metabolism in cancer. Br J Cancer. (2020) 122(2):150–6. doi: 10.1038/s41416-019-0620-5
73. Ren W, Liu G, Yin J, Tan B, Wu G, Bazer FW, et al. Amino-acid transporters in T-cell activation and differentiation. Cell Death Dis (2017) 8(3):e2655. doi: 10.1111/jpi.12394
74. Lopes C, Pereira C, Medeiros R. ASCT2 and LAT1 contribution to the hallmarks of cancer: from a molecular perspective to clinical translation. Cancers (Basel). (2021) 13(2):203. doi: 10.3390/cancers13020203
75. Nachef M, Ali AK, Almutairi SM, Lee SH. Targeting SLC1A5 and SLC3A2/SLC7A5 as a potential strategy to strengthen anti-tumor immunity in the tumor microenvironment. Front Immunol (2021) 12:624324. doi: 10.3389/fimmu.2021.624324
76. Häfliger P, Charles RP. The l-type amino acid transporter LAT1-an emerging target in cancer. Int J Mol Sci (2019) 20(10):2428. doi: 10.3390/ijms20102428
77. You S, Zhu X, Yang Y, Du X, Song K, Zheng Q, et al. SLC7A1 overexpression is involved in energy metabolism reprogramming to induce tumor progression in epithelial ovarian cancer and is associated with immune-infiltrating cells. J Oncol (2022) 2022:5864826. doi: 10.1155/2022/5864826
78. Bröer A, Gauthier-Coles G, Rahimi F, van Geldermalsen M, Dorsch D, Wegener A, et al. Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells. J Biol Chem (2019) 294(11):4012–26. doi: 10.1074/jbc.RA118.006378
79. Kurozumi S, Kaira K, Matsumoto H, Kurosumi M, Yokobori T, Kanai Y, et al. Association of l-type amino acid transporter 1 (LAT1) with the immune system and prognosis in invasive breast cancer. Sci Rep (2022) 12(1):2742. doi: 10.1038/s41598-022-06615-8
80. Shimizu A, Kaira K, Kato M, Yasuda M, Takahashi A, Tominaga H, et al. Prognostic significance of l-type amino acid transporter 1 (LAT1) expression in cutaneous melanoma. Melanoma Res (2015) 25(5):399–405. doi: 10.1097/CMR.0000000000000181
81. Cibrian D, Castillo-González R, Fernández-Gallego N, de la Fuente H, Jorge I, Saiz ML, et al. Targeting l-type amino acid transporter 1 in innate and adaptive T cells efficiently controls skin inflammation. J Allergy Clin Immunol (2020) 145(1):199–214.e11. doi: 10.1016/j.jaci.2019.09.025
82. Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev BioMed Eng. (2017) 19:163–94. doi: 10.1146/annurev-bioeng-071516-044546
83. Lian J, Liang Y, Zhang H, Lan M, Ye Z, Lin B, et al. The role of polyamine metabolism in remodeling immune responses and blocking therapy within the tumor immune microenvironment. Front Immunol (2022) 13:912279. doi: 10.3389/fimmu.2022.912279
84. Kuo MT, Chen HHW, Feun LG, Savaraj N. Targeting the proline-Glutamine-Asparagine-Arginine metabolic axis in amino acid starvation cancer therapy. Pharmaceuticals (2021) 14(1):72. doi: 10.3390/ph14010072
85. Kurmi K, Haigis MC. Nitrogen metabolism in cancer and immunity. Trends Cell Biol (2020) 30(5):408–24. doi: 10.1016/j.tcb.2020.02.005
86. Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR, et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant kras-driven cancers. Science. (2016) 353(6304):1161–5. doi: 10.1126/science.aaf5171
87. Lowman XH, Hanse EA, Yang Y, Ishak Gabra MB, Tran TQ, Li H, et al. p53 promotes cancer cell adaptation to glutamine deprivation by upregulating Slc7a3 to increase arginine uptake. Cell Rep (2019) 26(11):3051–60.e4. doi: 10.1016/j.celrep.2019.02.037
88. Metzler B, Gfeller P, Guinet E. Restricting glutamine or glutamine-dependent purine and pyrimidine syntheses promotes human T cells with high FOXP3 expression and regulatory properties. J Immunol (2016) 196(9):3618–30. doi: 10.4049/jimmunol.1501756
89. Chen CL, Hsu SC, Ann DK, Yen Y, Kung HJ. Arginine signaling and cancer metabolism. Cancers (Basel). (2021) 13(14):3541. doi: 10.3390/cancers13143541
90. Kim J, Guan KL. mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol (2019) 21(1):63–71. doi: 10.1038/s41556-018-0205-1
91. Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev (2012) 249(1):43–58. doi: 10.1111/j.1600-065X.2012.01152.x
92. Bodineau C, Tomé M, Murdoch PDS, Durán RV. Glutamine, MTOR and autophagy: a multiconnection relationship. Autophagy. (2022) 18(11):2749–50. doi: 10.1080/15548627.2022.2062875
93. Meng D, Yang Q, Wang H, Melick CH, Navlani R, Frank AR, et al. Glutamine and asparagine activate mTORC1 independently of rag GTPases. J Biol Chem (2020) 295(10):2890–9. doi: 10.1074/jbc.AC119.011578
94. Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. (2016) 351(6268):53–8. doi: 10.1126/science.aad2087
95. Chen J, Ou Y, Luo R, Wang J, Wang D, Guan J, et al. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature. (2021) 596(7871):281–4. doi: 10.1038/s41586-021-03768-w
96. Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. (2017) 171(3):642–54.e12. doi: 10.1016/j.cell.2017.09.046
97. Lemberg KM, Vornov JJ, Rais R, Slusher BS. We're not "DON" yet: optimal dosing and prodrug delivery of 6-Diazo-5-oxo-L-norleucine. Mol Cancer Ther (2018) 17(9):1824–32. doi: 10.1158/1535-7163.MCT-17-1148
98. Edwards DN, Ngwa VM, Raybuck AL, Wang S, Hwang Y, Kim LC, et al. Selective glutamine metabolism inhibition in tumor cells improves antitumor T lymphocyte activity in triple-negative breast cancer. J Clin Invest (2021) 131(4):e140100. doi: 10.1172/JCI140100
99. Fultang L, Booth S, Yogev O, Martins da Costa B, Tubb V, Panetti S, et al. Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood. (2020) 136(10):1155–60. doi: 10.1182/blood.2019004500
100. Zhang J, Wang S, Guo X, Lu Y, Liu X, Jiang M, et al. Arginine supplementation targeting tumor-killing immune cells reconstructs the tumor microenvironment and enhances the antitumor immune response. ACS Nano. (2022) 16(8):12964–78. doi: 10.1021/acsnano.2c05408
101. Lim JW, Son HY, Huh YM, Haam S. Cationic poly(amino acid) surface functionalized manganese nanoparticles for nitric oxide-based immunotherapy and magnetic resonance imaging. J Mater Chem B (2022) 10(28):5402–9. doi: 10.1039/D2TB00794K
Keywords: glutamine, arginine, leucine, isoleucine, valine, TME, metabolism
Citation: Wetzel TJ, Erfan SC, Figueroa LD, Wheeler LM and Ananieva EA (2023) Crosstalk between arginine, glutamine, and the branched chain amino acid metabolism in the tumor microenvironment. Front. Oncol. 13:1186539. doi: 10.3389/fonc.2023.1186539
Received: 14 March 2023; Accepted: 03 May 2023;
Published: 19 May 2023.
Edited by:
Benny Abraham Kaipparettu, Baylor College of Medicine, United StatesReviewed by:
Luigi Ippolito, University of Florence, ItalyCopyright © 2023 Wetzel, Erfan, Figueroa, Wheeler and Ananieva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elitsa A. Ananieva, ZWxpdHNhLmFuYW5pZXZhLXN0b3lhbm92YUBkbXUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.