
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 09 August 2023
Sec. Genitourinary Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1175183
This article is part of the Research TopicNovel Insights from Tumor Immune Microenvironments in the Prognosis Prediction and Treatment Guidance of Bladder CancerView all 8 articles
Background: The role of Eph receptors and related ephrin (EFN) ligands (as the largest family of transmembrane-bound RTKs) in immunomodulation in many types of cancer, especially bladder cancer (BLCA), is scarcely known.
Methods: A pan-cancer dataset was retrieved from The Cancer Genome Atlas (TCGA) to explore the relation between Eph receptor/EFN ligand family genes and immunomodulators and tumor-infiltrated immune cells (TIICs). Local BLCA, GSE32894, and GSE31684 cohorts were applied to validate. The IMvigor210 cohort was employed to explore the relationship between EPHB6 and immunotherapy response. Moreover, association between EPHB6 and molecular subtype was investigated to explore potential therapeutic strategies. Immunohistochemical staining of CD8 and CD68 was performed to validate the correlation between EPHB6 and TIICs.
Results: The pan-cancer analysis revealed variations in the immunological effects of Eph receptor/EFN ligand family genes across different types of cancer. EPHB6 expression negatively correlated with the expression of the majority of immunomodulators (including HLA and immune checkpoints), and CD8 T cells and macrophages in both the TCGA-BLCA and validation BLCA cohorts, shaping a cold immune microenvironment with inhibited immunity. In the IMvigor210 cohort, patients with high-EPHB6 highly correlated with a non-inflamed, low PD-L1 expression immune phenotype, and correspondingly, with less responders to immunotherapy. The high-EPHB6 group, enriched with the basal subtype, presented significantly fewer TP53 and more FGFR3 genomic alterations. Finally, a novel EPHB6-related Genes signature, with reliable and robust ability in prognosis prediction, was constructed.
Conclusions: This study comprehensively investigated the immunological effects of Eph receptor/EFN ligand family genes pan-cancer, and specially identified the immunosuppressive role of EPHB6 in BLCA. Furthermore, EPHB6 may predict the molecular subtype and prognosis of BLCA, and serve as a novel therapeutic target to improve the sensitivity of immunotherapy.
Since the discovery of the first EphA1 in 1987, a growing body of evidence has revealed that Eph receptors and their ephrin (EFN) ligands are involved in various cell processes, including cell communication, proliferation and migration, and tissue homeostasis (1). Eph receptors, the largest family of transmembrane-bound receptor tyrosine kinases (RTKs), play crucial roles in angiogenesis, lymphangiogenesis, carcinogenesis, and metastasis across various types of cancer (2, 3). Based on sequence homology analysis, Eph receptors can be categorized into two classes: nine Eph A and five Eph B members, which differ primarily in the EFN ligand binding site. This difference is what determines the binding specificity between Eph receptors and EFNs (4).. Eph A and B receptors promiscuously bind to these five EFN A and three EFN B ligand members, respectively; however, cross interactions do occur between groups (5, 6). Eph receptors were found to signal through cross-talk with RTKs and other factors, that is, ligand-independent signaling (7).
Recently, the role of Eph receptors and EFN ligands in the immune system has been garnering attention, and Eph receptors especially have great potential to become immunotherapeutic targets. In addition to the ubiquitous expression in tumor cells, Eph receptors and EFN ligands are also widely expressed in immune cell subsets, such as monocytes, platelets, macrophages, dendritic cells, B cells, and T cells (8). EphA3, the first receptor found as a tumor-associated antigen, can be recognized by a CD4+ T cell clone in melanoma and this process stimulates selective immunoreactivity (9). EphA2-derived epitopes can induce the elevated immunoreactivity of CD4+ and CD8+ T cells in the EphA2-positive renal cell carcinoma (10), and dendritic cell vaccination with EphA2 peptides has been applied in the clinical trial of glioblastoma multiforme (11). Furthermore, several studies have revealed that the expression of Eph receptors in tumor or tumor-infiltrating immune cells is particularly with immunosuppressive response (8). Intrinsic EphA2 expression in pancreatic cancer cells correlated with the suppression of immune response because it retains the exclusion of T cells and causes a low infiltrating level of T cells; whereas, the knockout of EphA2 gene can reverse T cell exclusion, increase the abundance of CD4+ and CD8+ T cells, and improve the sensitivity to immunotherapy (12). EphA10 expression in breast cancer cells positively correlated with the expression of PD-L1 (13), indicating that more cancer cells can escape immune surveillance. Eph receptors have also been identified in many tumor infiltrating cells; for example, EphA3 is extensively expressed in stromal fibroblasts in many solid tumors, which may promote tumor progression and invasiveness, and also inhibit the anti-cancer immunity (14). Additionally, the expression of EFN ligands on monocytes and macrophages is closely associated with inflammation, and EFNB1-3 ligands may affect T cell receptor-mediated signaling and T cell co-stimulation (15).
BLCA is a significant genitourinary disease, with approximately 573,278 new cases and over 212,536 related deaths reported annually (16). Due to the frequent recurrence of BLCA in patients after transurethral resection of bladder tumors and the need for repeat surgeries, it is the most expensive cancer to treat on a per-patient basis (17). Currently, immune checkpoint inhibitors (ICIs) targeted at PD-1 or PD-L1 are becoming a novel and effective treatment option for BLCA, with durable antitumor efficacy. However, only about one-fifth of unselected patients respond to ICIs, and three prospective trials have shown that ICIs do not improve overall survival (OS) compared to chemotherapy alone (18–20). The limitations in improving the overall response rate for ICIs in BLCA therapy mainly stem from the individual heterogeneity in genetic and tumor immune microenvironment, and thus, the lack of robust predictive biomarkers to precisely select which patients will or will not benefit from the therapy (21–23). Therefore, it is necessary to identify novel biomarkers that can accurately predict immunotherapy response in BLCA patients. Eph receptors and EFN ligands prevalently correlated with the activation or suppression of innate and adaptive immune response in different types of cancer. Remarkably, patients with metastatic BLCA treated with combination therapy of EFNB2 inhibition and pembrolizumab had better OS and improved treatment responses than those receiving anti-PD-1/PD-L1 monotherapy (24). While the inhibition of PD-1/PD-L1 and Ephrin-related pathways has demonstrated promising outcomes in preclinical models (25), the expression pattern of Eph receptors and EFN ligands in most tumors, particularly BLCA, remains unclear, and their immunological roles are not well understood.
In this study, a pan-cancer analysis was initially performed to reveal the relationship between the Eph receptors and EFN ligands expression and immunoregulatory factors and identified inactive kinase EPHB6 which shaped a cold immune microenvironment and promoted the immune escape in BLCA. Previous studies have suggested that EPHB6 may have both oncogene (26) and tumor suppressor (27, 28) roles in different types of cancer. However, its specific function in BLCA remains unknown. Thereafter, the role of EPHB6 in the immune microenvironment, anti-cancer immunity, and as correlate of immunotherapy response of BLCA was investigated. Briefly, our study elucidated the expression patterns of Eph receptors and EFN ligands pan-cancer, and identified a novel biomarker of EPHB6 as a potential effective therapeutic target to improve the immunotherapy response of BLCA.
The transcriptomic data, mutational profile, copy number variant (CNV), methylation, and related clinical information of pan-cancer cohort, including 32 types of cancer, were retrieved from The Cancer Genome Atlas (TCGA). RNA-seq data were log2 (x+0.001) transformed, whereas TMB level was calculated using VarScan2 package. Based on the pan-cancer cohort, 406 patients with BLCA (all belonged to the muscle-invasive BLCA subtype, namely MIBC) were filtered and constituted as the TCGA-BLCA cohort. In addition, four other independent datasets of GSE32548, GSE13507, GSE188715, and GSE32894 from the Gene Expression Omnibus (GEO) were downloaded to verify the novel constructed prognostic signature.
Twenty-eight MIBC patients were enrolled in this study, and we have received their written informed consent. This study was approved by the Ethics Committee of Ningbo First Hospital. Tumor and matched normal tissues were collected from the patients in the local BLCA cohort after the surgical resection. A FastPure® Cell/Tissue Total RNA Isolation Kit V2 (Vazyme, Jiangsu, China) was used to extract total RNA, which was quantified and qualified using the Qubit (ThermoFisher Scientific, MA, the United States) and Agilent 2100 bioanalyzer (Agilent Technologies, CA, United States), respectively. The NEBNext® Ultra™ RNA Library Prep Kit (NEB, MA, US) was subsequently used for the construction of RNA library. The prepared RNA library was finally sequenced on the Illumina Novaseq-6000 machine (Illumina, MA, US).
In this study, several indices including immunomodulator gene expression, tumor immune cell infiltration, cancer immunity cycle, and inhibitory immune checkpoints-related gene expression were employed for the evaluation of the immune-related features, anti-cancer immunity, and microenvironmental status in BLCA. A total of 122 immunomodulators (including chemokines, paired receptors, MHC molecules, and immunostimulators), which have been reported previously, were collected to estimate the immunomodulation of TME in BLCA (29). The cancer immunity cycle was reviewed to represent the anti-cancer immunity, and the following contained seven steps. Step 1: release of cancer cell antigens, step 2: cancer antigen presentation, step 3: priming and activation, step 4: trafficking of immune cells to cancer cells, step 5: immune cell infiltration into tumor, step 6: recognition of cancer cells by T cell, and step 7: killing of cancer cell (30). These steps were performed using a single sample gene set enrichment analysis (ssGSEA), based on the transcriptomic data of each sample (31). To decrease the errors, several algorithms, including TIMER, CIBERSORT, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms, were performed to assess the infiltrating levels of tumor immune cells (mainly CD8+, Macrophage, Dendritic cells, Natural Killer (NK) cells, and Th1 cells). The ESTIMATE algorithm was also employed to evaluate the immunological status of TME in BLCA. Furthermore, the profile of inhibitory immune checkpoints was obtained from a study by Auslander (32). As described, a pan-cancer T cell inflamed score (an 18 genes signature (33),) associated with pre-existing cancer immunity, which could predict the response of immunotherapy.
The transcriptomic data and related clinical information of 348 patients with urothelial cancer (UC) in the IMvigor210 cohort were downloaded from: http://research-pub.gene.com/IMvigor210CoreBiologies/. These patients with UC reportedly received an immunotherapy via anti-PD-L1/PD-1 antibodies. Accordingly, the therapy response was defined similarly as the published criteria (34): CR: complete response, PR: partial response, SD: stable disease, and PD: progressive disease; and CR/PR and SD/PD were defined as the binary response groups. In the IMvigor210 cohort, BLCA samples were categorized into three immune phenotypes, namely deserted, excluded, and inflamed microenvironment, according to the distribution, status, and infiltrating level of CD8+ T cells. In addition, IC0, IC1, and IC2+ phenotypes were characterized as the lowest, medium, and the highest PD-1 expression, respectively. Thus, these known parameters were employed in this study to determine the role of EPHB6 during immunotherapy. Eventually, a majority of immunotherapy-predicted pathways were performed using the ssGSEA algorithm, and we further explored the association between EPHB6 and the activities of these immunotherapy-predicted pathways.
The Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Hallmark pathway enrichment analysis in this study was conducted to explore the EPHB6-associated signaling pathways. First, the differentially expressed genes (DEGs; log2 (Fold change)>1.5; p<0.01) between EPHB6-high and -low expression groups were acquired. Then, a functional enrichment analysis of these DEGs was conducted using the fgsea package to calculate the enrichment score. In the GO enrichment analysis, three categories of BP were delineated: biological process (GOBP), CC: Cellular component (GOCC), and MF: molecular function (GOCC). In addition, the whole genome sequencing data in the TCGA-BLCA cohort were downloaded to visualize the overall mutational profile in BLCA and the differences in the genomic landscape between EPHB6-high and -low expression groups using maftools package. Genomic alteration enrichment analysis was performed to directly display the prevalent genes in different groups.
In addition to the TMB, microsatellite instability, and inhibitory immune checkpoints, several studies have revealed that molecular subtype may predict the response of some therapies in BLCA (35–38). In this study, several molecular subtyping strategies were used, including Consensus, CIT, Lund, MDA, TCGA, Baylor, and UNC subtypes. Accordingly, some pathways involved with urothelial differentiation, Ta pathway, cell cycle activity, mitochondria, basal differentiation, myofibroblasts, interferon (IFN) response, B and T lymphocytes, smooth muscle, and neuroendocrine differentiation were used for evaluation of each molecular subtype, and ssGSEA was conducted to calculate the enrichment score. The receiver operating characteristic (ROC) curve and the value of area under ROC curve (AUC) were used to evaluate the accuracy of EPHB6 in predicting the molecular subtype. As reported, RB1 (39), ATM (40), ERBB2 (41), ERCC2 (42), and FANCC (43) alterations highly correlated with the response of adjuvant or neoadjuvant chemotherapy. In addition, the Drugbank database was used to predict the potential target drugs against BLCA.
We collected specimens from a local cohort of 21 BLCA patients to analyze the expression level of EPHB6 and its correlation with TME feature. These patients were classified into three groups: NMIBC-low grade (n=7), NMIBC-high grade (n=7), and MIBC-high grade (n=7). Tissue samples (4-µm-thick sections) were subjected to immunohistochemical staining for EPHB6, CD8, CD68 using EPHB6 (ab217542, 1:100, Abcam) CD8 (ab217344, 1:100 Abcam) and CD68 (ab955, 1:50, Abcam) antibodies, respectively. The immunohistochemical staining method was performed as described previously by Sun et al. (44). The staining score for EPHB6 was classified as high (staining in more than 50% of malignant cells), low (staining in less than 25% of malignant cells), or medium (staining in 25%-50% of malignant cells). The staining of CD8 and CD68 was only evaluated in intratumoral infiltrating leukocytes. The score was calculated independently by two pathologists from five high magnification (200) visual fields using CaseViewer2.2 (Thermo Fisher).
Finally, the prognostic value of Eph receptor/EFN ligand family genes were explored via univariate Cox regression analysis. Notably, EPHB6 markedly correlated with the prognosis of BLCA, indicating that EPHB6 also can be a potential prognostic biomarker. To identify EPHB6-related differentially expressed genes (DEGs), we employed the limma R package to compare tumor vs. normal tissues and high- vs. low-EPHB6 expression groups in the TCGA-BLCA dataset. The criteria for determining differential DEGs were adjusted P-value < 0.01 and |log(fold change)|>1. We then used the VennDiagram R package to collect and analyze the overlapped genes. Based on the DEGs analysis, 35 prognosis-related genes were screened out for the construction of a EBGs prognostic signature. In this study, seven kinds of machine learning algorithms were applied, including RF, Ridge, stepwise Cox, Enet, Boost, multivariate Cox, and LASSO, of which two methods were randomly selected to discover the most robust EBGs signature, as the standard of the highest C-index in the training and four independent validation cohorts. The Kaplan–Meier curve analysis was conducted to compare the OS between high- and low- EBGs score groups. The ROC curve and corresponding AUC value were used to evaluate the reliability and robustness of EBGs signature. Moreover, the predictive performance of EBGs signature was further verified in four other independent cohorts: GSE32548, GSE13507, GSE188715, and GSE32894 from the GEO database.
In this study, all statistical analyses were conducted using the R studio. Correlation analyses between variables were performed using Pearson coefficients. Categorical variables analysis was conducted using chi-square and Fisher’s exact tests. In addition, Mann–Whitney U test was performed for continuous variables. In the Kaplan–Meier curve analysis, the log-rank test was performed. The data was considered statistically significant at p<0.05. All applied software and version information was provided in the Supplementary Table 1.
The immunological correlation between Eph receptors and EFN ligands in the TCGA pan-cancer cohorts was investigated. For most types of cancer, a positive and significant association was noted between most of Eph receptors/EFN ligands and immunomodulatory genes at the transcriptomic level (Supplementary Figure 1). Furthermore, a notable heterogeneity was observed in the relationship between Eph receptors/EFN ligands and main checkpoints (CD274, PDCD1, CTLA4, and LAG3) in different types of cancer (Figure 1A). For the first time, it was identified that EPHB6, a previously recognized inactive kinase receptor, had a significantly relation with immunomodulatory genes expression in most types of cancer (Figure 1B). Consistent with the result exhibited in Figure 1A, EPHB6 expression was found to have a significantly negative correlation with immunomodulatory genes expression in mesonephric adenocarcinoma (MESO), thyroid carcinoma (THCA), thymoma (THYM), and BLCA. In BLCA, EPHB6 demonstrated a different immunological profile compared to other Eph receptor/EFN ligand family genes, which showed the most negative connection with the majority of immunomodulators (Figure 1C).

Figure 1 Immunological correlation between Eph receptors and EFN ligands in the pan-cancer cohort. (A) Correlation matrix showing the relationship between the expression levels of all the Eph receptors and EFN ligands and LAG3, CTLA4, PDCD1 and CD274 in TCGA pan-cancer cohorts. Dots labeled with yellow color indicated a negative correlation between genes and checkpoints in a tumor type; whereas, those with purple indicated a positive correlation. (B) Heatmap revealing the association between immunomodulatory genes and all the Eph receptors and EFN ligands in TCGA pan-cancer cohorts. (C) Correlation analysis showing the relationship between all the Eph receptors and EFN ligands and immunomodulatory genes in TCGA-bladder cancer (BLCA) cohort alone. *p<0.05.
In the local BLCA cohort, most Eph receptor genes (EphA3, EphA4, EphA5, EphA6, EphA7, EphA8, EphA10, EphB3, EphB4, and EPHB6) and several EFN ligand genes (EFNA3, EFNA4, and EFNB2) expressions were significantly dysregulated (p<0.05; Figure 2A). This result suggested that these Eph receptor/EFN ligand family genes may be also correlated with the prognosis of BLCA. Subsequently, univariate Cox regression analysis revealed that EPHB6 and EphA10 expressions were positively correlated with OS, DSS, and PFI; whereas, EphB4, EphB3, EphB1, EphA6, EphA5, and EphA3 expressions were negatively correlated with OS, DSS, and PFI (p<0.05; Figure 2B). Based on these features, the following experiments mainly focused on investigating the role of EPHB6 and tumor immunology in BLCA.

Figure 2 Expression and prognostic role of Eph receptors and EFN ligands in bladder cancer. (A) Difference in the mRNA expression level between bladder cancer tumor tissues and normal tissues in local bladder cancer cohort. (B) Forest plot for the association between Eph receptors and EFN ligands and survival by median cutoff in the TCGA-BLCA cohort; data were presented with HR and 95% CI. * p<0.05. OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval; HR, hazard ratio. *p<0.05; **p<0.01; ***p<0.001; ns: non-significant.
Subsequently, the immunological effects regulated by EPHB6 expression were deeply investigated in the TCGA-BLCA cohort. As demonstrated, the expressions of most chemokines and their paired receptors (CXCL9, CXCL10, CXCL11, CXCL13, CCL3, CCL4, CCL8, CCR2, CCR3, CCR4, CCR5, CCR6, CCR8, CXCR3, and CXCR6) were significantly lower in the EPHB6-high expression group when compared to those in the EPHB6-low expression group (p<0.05; Figure 3A). Most immunostimulators (including TNFSF4, TNFSF14, TNFRSF9, TNFSF13B, TNFRSF17, CD27, CD28, CD48, and CD80) were also downregulated in the EPHB6-high expression group. Furthermore, expressions of several MHC genes (HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1, HLA-A, HLA-B, HLA-E, and HLA-F) were negatively correlated with EPHB6 expression. In summary, EPHB6 may shape a cold immune microenvironment in BLCA. Correspondingly, EPHB6 expression negatively correlated with most steps engaged in cancer immunity cycle, including the step 1 of cancer cell antigen release and step 4 of basophil, CD8+ T cell, dendritic cell, eosinophil, macrophage, MDSC, neutrophil, NK cell, Th1 cell, Th17 cell, and Th22 cell recruiting (p<0.05; Figure 3B). However, step 6 activity of recognition of tumor cells by T cells was significantly downregulated in the EPHB6-low expression group (p<0.001), probably owing to the high level of inhibitory immune checkpoints stimulating immune escape. The step 7 activity of killing of cancer cells was significantly upregulated in the EPHB6-low expression group (p<0.05; Figure 3B). Notably, it was consistent that EPHB6 expression negatively correlated with macrophages, dendritic cells, NK cells, and CD8+ T cells infiltrating levels in different algorithms, including TIMER, CIBERSORT, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC (Figure 3C). Similarly, the effector genes expression of these immune cells was downregulated in the EPHB6-high expression group (Supplementary Figure 2A). Furthermore, EPHB6 was found to have a negative relation with nearly all known inhibitory immune checkpoints at the transcriptomic level, including PD-L1, PD-1, CTLA4, LAG3, TIGHT, IDO1, and IDO2 (Supplementary Figure 2B). Notably, in the TCGA-BLCA cohort, EPHB6 expression was negatively correlated with the enrichment scores of most immunotherapy predicted pathways, such as IFN-γ signaling, DNA damage repair (DDR) signaling, cell cycle, and DNA replication related signaling (p<0.05; Figure 3D). As previously reported, T cell inflamed score is associated with immunotherapy response; therefore, the relation between EPHB6 expression and T cell inflamed score was evaluated, demonstrating that EPHB6 expression had a remarkably negative correlation with pan-cancer T cell inflamed score (R = -0.31, p<0.0001; Figure 3E). EPHB6 expression also negatively correlated with each individual T cell inflamed signature-related gene expression (Figure 3F).
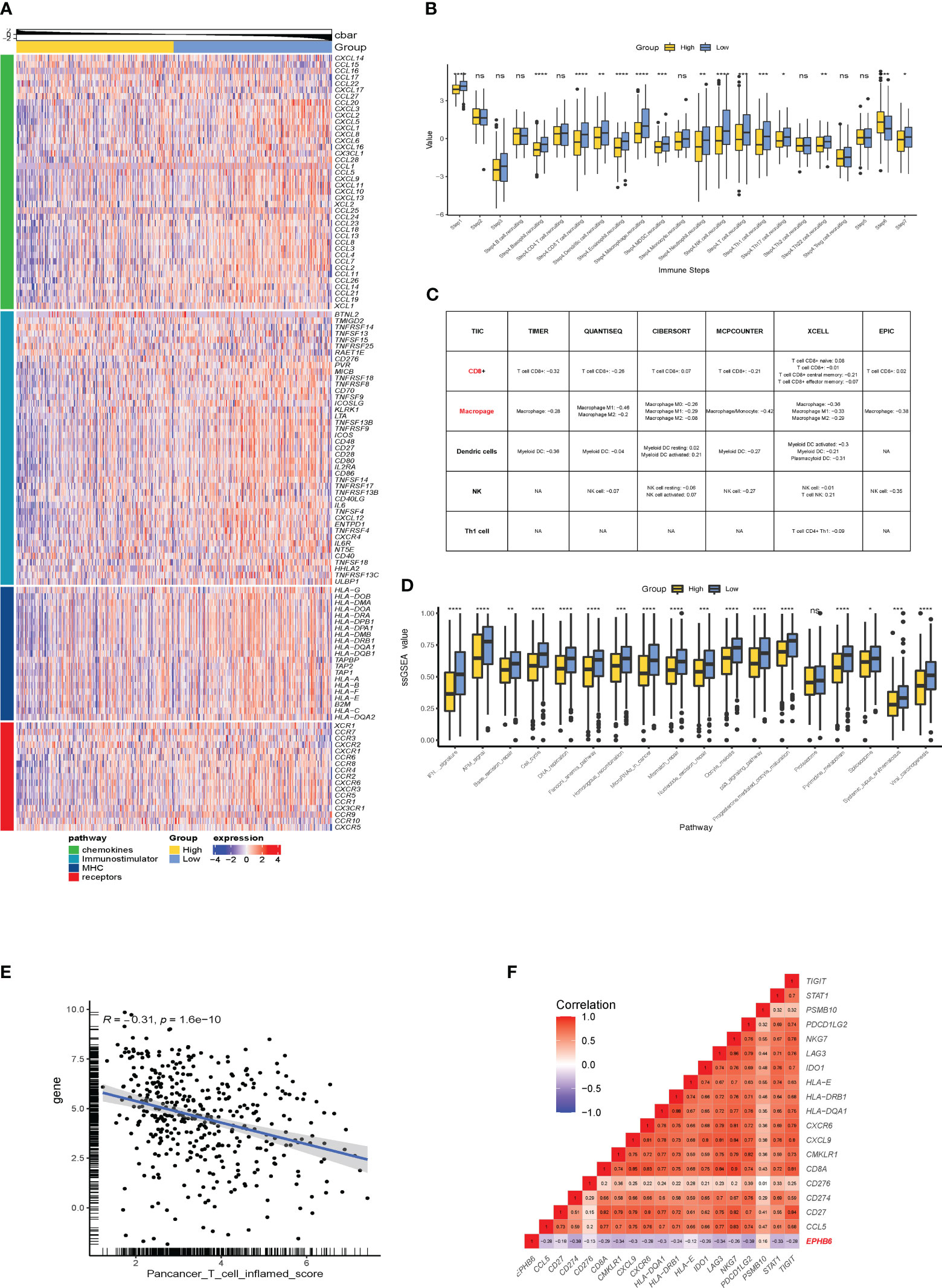
Figure 3 EPHB6 is associated with a cold immune microenvironment in bladder cancer. (A) Difference in the expression levels of immunomodulatory genes between the high- and low-EPHB6 groups in the TCGA-BLCA cohort. (B) Bar plots showing the difference in the activities of seven immune steps between the high- and low-EPHB6 groups in the TCGA-BLCA cohort. (C) Correlation between EPHB6 expression levels and tumor-infiltrated immune cells (TIICs) in bladder cancer. The levels of TIICs were analyzed by multiple algorithms, including TIMER, CIBERSOFT, QUANTISEQ, MCPCOUNTER, XCELL and EPIC. (D) Bar plots revealing the difference in the immune-related signal pathways between the high- and low-EPHB6 groups. (E) Correlation between EPHB6 expression level and T cell inflamed score. (F) Correlation between EPHB6 and each individual T cell inflamed signature-related gene expression. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, non-significant.
In the local BLCA and GSE32894 cohorts, we observed a significant negative correlation between the expressions of most HLA-family members and immune checkpoints and the mRNA expression level of EPHB6 (Figure 4A). However, HLA-DQB2, CD40, CD44, and TNFRSF14 were exceptions to this correlation. These findings were further supported by similar results obtained in the GSE31684 cohort (Supplementary Figure 3), which confirmed that EPHB6 may contribute to the development of a cold immune microenvironment in BLCA. Considering the consistent negative correlation between EPHB6 mRNA expression level and the abundance of CD8 T cells and macrophages, we conducted immunochemical staining of EPHB6, CD8, and CD68 to validate this correlation. Firstly, we observed heterogeneity in the protein expression level of EPHB6 among BLCA patients. However, no significant difference in the protein expression level of EPHB6 was found among patients with NMIBC-low grade, NMIBC-high grade, and MIBC in the local cohort (Figures 4B, C). Furthermore, there was a noticeable difference in the staining of CD8 and CD68 between BLCA patients with high and low EPHB6 expression levels (Figures 4D, E). Subsequently, we found a significant negative correlation between EPHB6 and CD8 (r=-0.64, p<0.01, Figure 4F). Although the correlation between EPHB6 and CD68 was also negative (r=-0.22), this difference was not statistically significant in our protein analysis (Figure 4G). Nevertheless, the negative correlation between the protein expression profile of EPHB6 and CD8 or CD68 was consistent with the mRNA expression correlation observed in multiple datasets (Supplementary Figure 4).
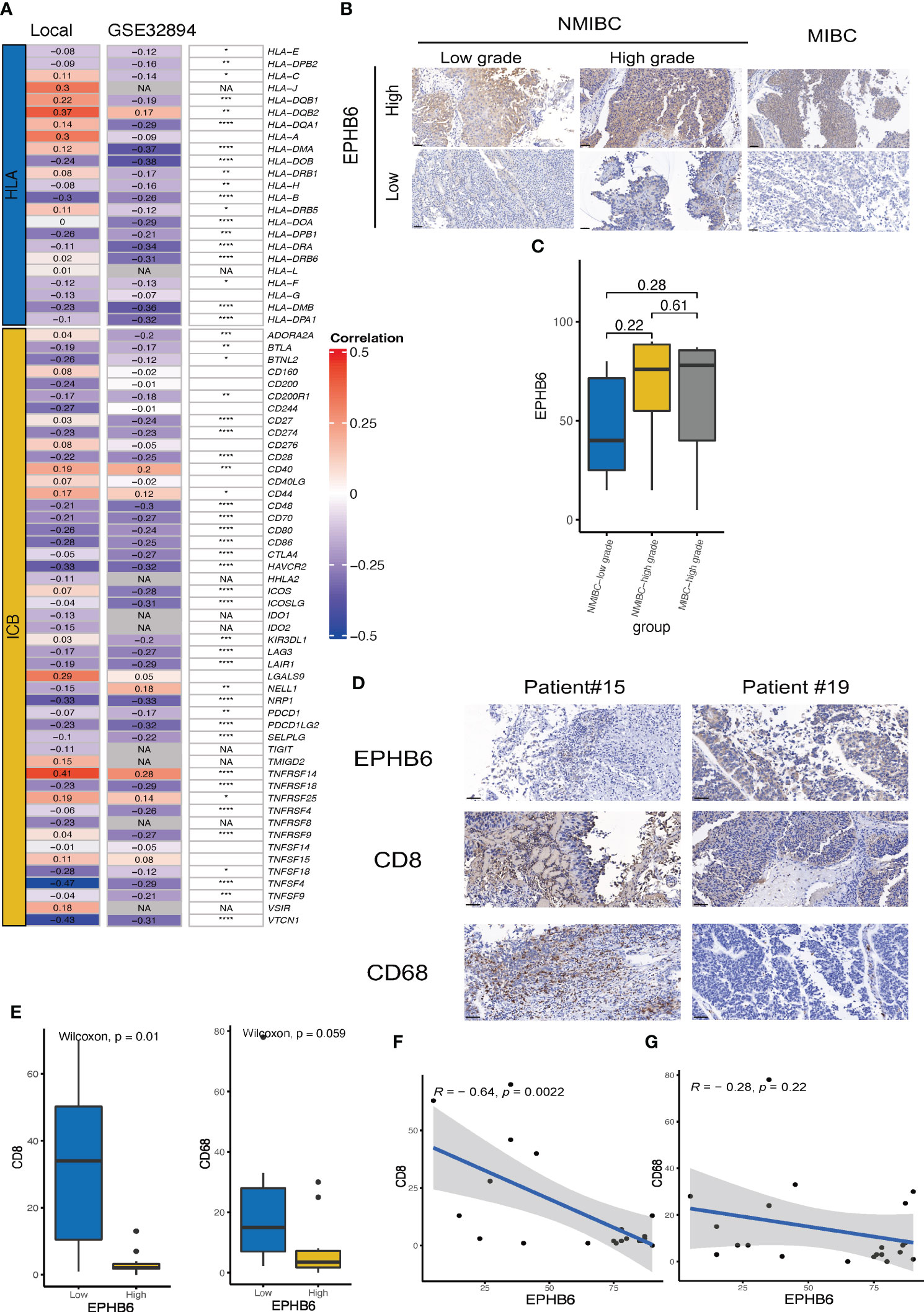
Figure 4 Validation of the association between EPHB6 and immunology feature in local and other bladder cancer datasets. (A) Correlation analysis of the mRNA expression levels of immunomodulatory genes (HLA and ICBs) and EPHB6 in local bladder cancer (BLCA) and GSE32894 datasets. (B) Immunohistochemical staining of EPHB6 in patients with NMIBC-low grade (left panel), NMIBC-high grade (middle panel), and MIBC (right panel) from local cohort. (C) Bar plot showing the difference in EPHB6 protein expression level in patients with NMIBC-low grade, NMIBC-high grade, and MIBC from local cohort. The images were captured at a magnification of 200, and a scale bar indicating 50 μm was included in the left bottom corner of each image. (D) Representative image of immunohistochemical staining of EPHB6, CD8 and CD68 in patients with high- and low-EPHB6 protein expression level. The images were captured at a magnification of 200, and a scale bar indicating 50 μm was included in each image’s left bottom corner. (E) Bar plot showing the difference in the CD8 and CD68 protein expression levels in patients with high- and low-EPHB6 protein expression level. Correlation analysis of the protein expression levels of EPHB6 and CD8 (F) or CD68 (G) in BLCA patients from local cohort. HLA, human leukocyte antigens; ICB, immune checkpoint; NMIBC, non-muscle invasive bladder cancer; MIBC, muscle invasive bladder cancer. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; NA: not applicable.
Furthermore, the immunological role of EPHB6 was evaluated in the IMvigor210 cohort. Patients with UC in the EPHB6-low expression group had a markedly higher level of immunomodulators, T cell inflamed signature-related genes, immune checkpoints, and immunotherapy predicted pathways which was consistent with the cold immune feature as previously mentioned (Figure 5A). Based on the classification of three immune phenotypes (deserted, excluded, and inflamed) in the IMvigor210 cohort, deserted and inflamed phenotype groups were found to have the highest and lowest EPHB6 expression, respectively (p<0.05; Figure 5B). According to the PD-1 expression status, higher PD-L1 expressed immune cells group (IC2+, with the highest PD-1 expression) had the lowest EPHB6 expression than that in the IC0 and IC1 phenotype groups; conversely, the highest EPHB6 expression was observed in the IC0 phenotype group (with the lowest PD-1 expression) when compared to the IC1 and IC2+ phenotype groups (p<0.05; Figure 5C). Moreover, TC2+ phenotype group (with the highest PD-L1 expression on tumor cells) also had the lowest EPHB6 expression (p<0.05); however, no significant difference was noted in EPHB6 expression between TC0 and TC1 phenotype groups (Figure 5D). Briefly, EPHB6 expression had a strongly negative correlation with the cold immune microenvironment in UC. Patients with high-EPHB6 in the IMvigor210 cohort had an inferior OS (Figure 5E). Remarkably, in the IMvigor210 cohort, patients with UC who showed a complete or partial response (CR/PR) to immunotherapy had the significantly downregulated expression of EPHB6, compared with those with a progressive or stable disease (PD/SD) (p<0.01; Figure 5F). Significantly higher proportion of responders to immunotherapy were present in the low-EPHB6 group. Notably, after both TP53 mutation and EPHB6 stratification, we found that TP53mtEPHB6low group and TP53mtEPHB6high had the highest and lowest proportion of responders, respectively (Supplementary Figure 5). Meanwhile, differences between high- and low-EPHB6 group were nearly insignificant when TP53 was not mutated.
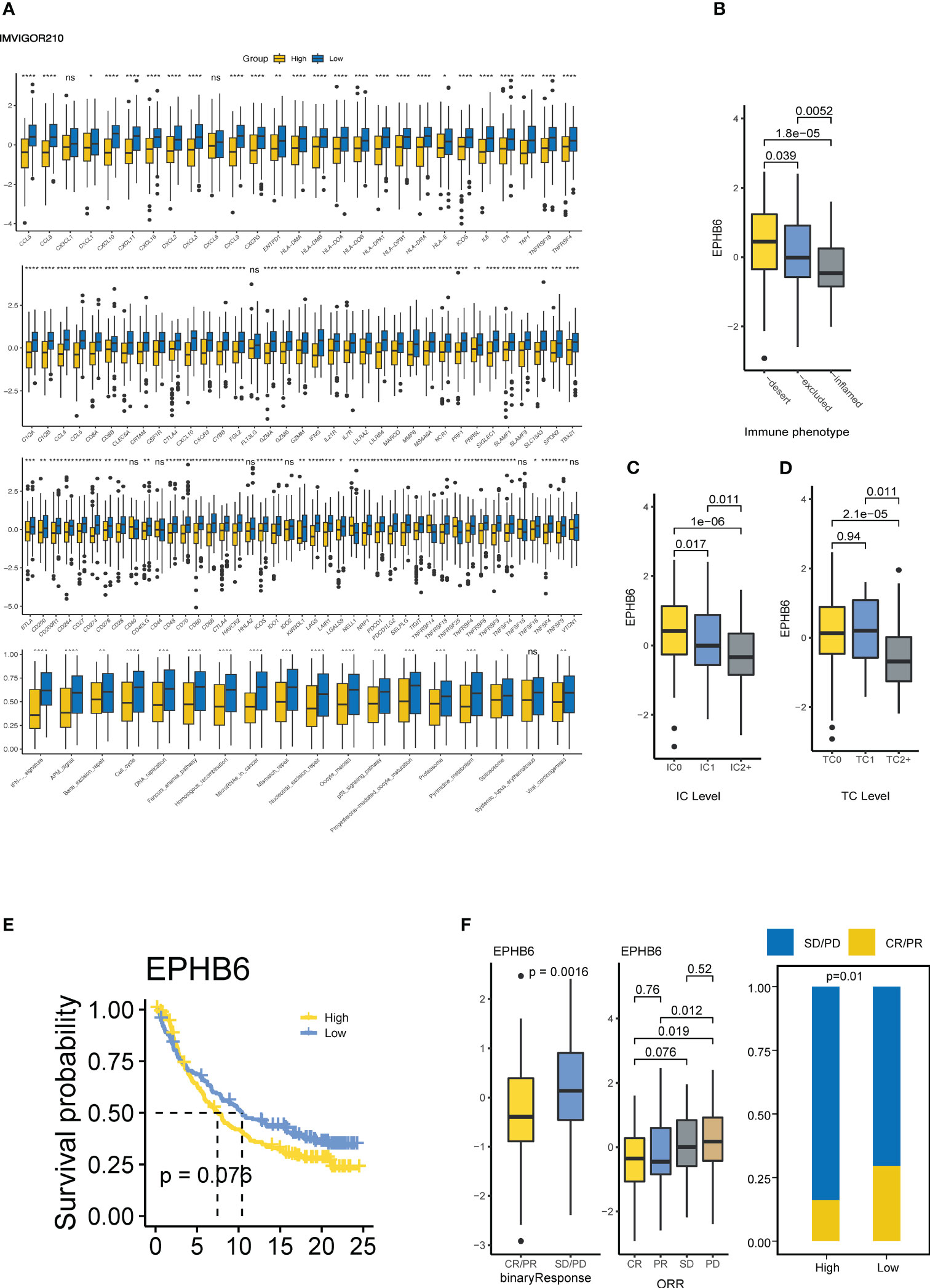
Figure 5 EPHB6 is associated with immunotherapy response in bladder cancer. (A) Difference in the expression levels of immunomodulatory genes between high- and low-EPHB6 groups in Imvigor210 cohort. Difference in the EPHB6 mRNA expression level between groups with various immune phenotypes (B), PD-L1 expression in immune cells (C) PD-L1 expression in tumor cells (D). (E) Kaplan-Meyer plot comparing patients with high- or low-EPHB6 expression levels treated with immunotherapy. (F) Difference in the EPHB6 mRNA expression level between responders (CR/PR) and non-responders (SD/PD) treated with immunotherapy. IC: PD-L1 immunohistochemistry levels in immune cells: IC0 (<1%); IC1(≥1% and <5%); IC2+ (≥5%). TC: PD-L1 immunohistochemistry levels in tumor cells TC0 (<1%); TC1 (≥1% and <5%); TC2+ (≥5%); IC0 (<1%); IC1 (≥1% and <5%); IC2+ (≥5%); CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, non-significant.
Underlying EPHB6 expression-related DEGs, GOBP of antimicrobial humoral immune response mediated by antimicrobial peptide, B cell mediated immunity, antigen receptor-mediated signaling pathway, and activation of immune response; GOCC of immunoglobulin complex, collagen-containing extracellular matrix, and receptor complex; and GOMF of immunoglobulin receptor-binding, antigen-binding, immune receptor activity, and cytokine receptor activity were significantly enriched (Figures 6A-C). KEGG pathway enrichment analysis further revealed that cytokine–cytokine receptor interaction, metabolism of xenobiotics by cytochrome P450, and chemokine signaling pathway were differentially regulated between the EPHB6-high and -low expression groups (p<0.0001; Figure 6D). Hallmark pathway enrichment analysis displayed the enrichment of E2F targets, G2M checkpoint, epithelial–mesenchymal transition, allograft rejection, inflammatory response, IL6-JAK-STAT3 signaling, IFN-α response, IFN-γ response, complement, and TNFα signaling via NFκB (p<0.0001; Figure 6E). Biological enrichment analysis verified that EPHB6 expression was markedly associated with the regulation of immune-related pathways.
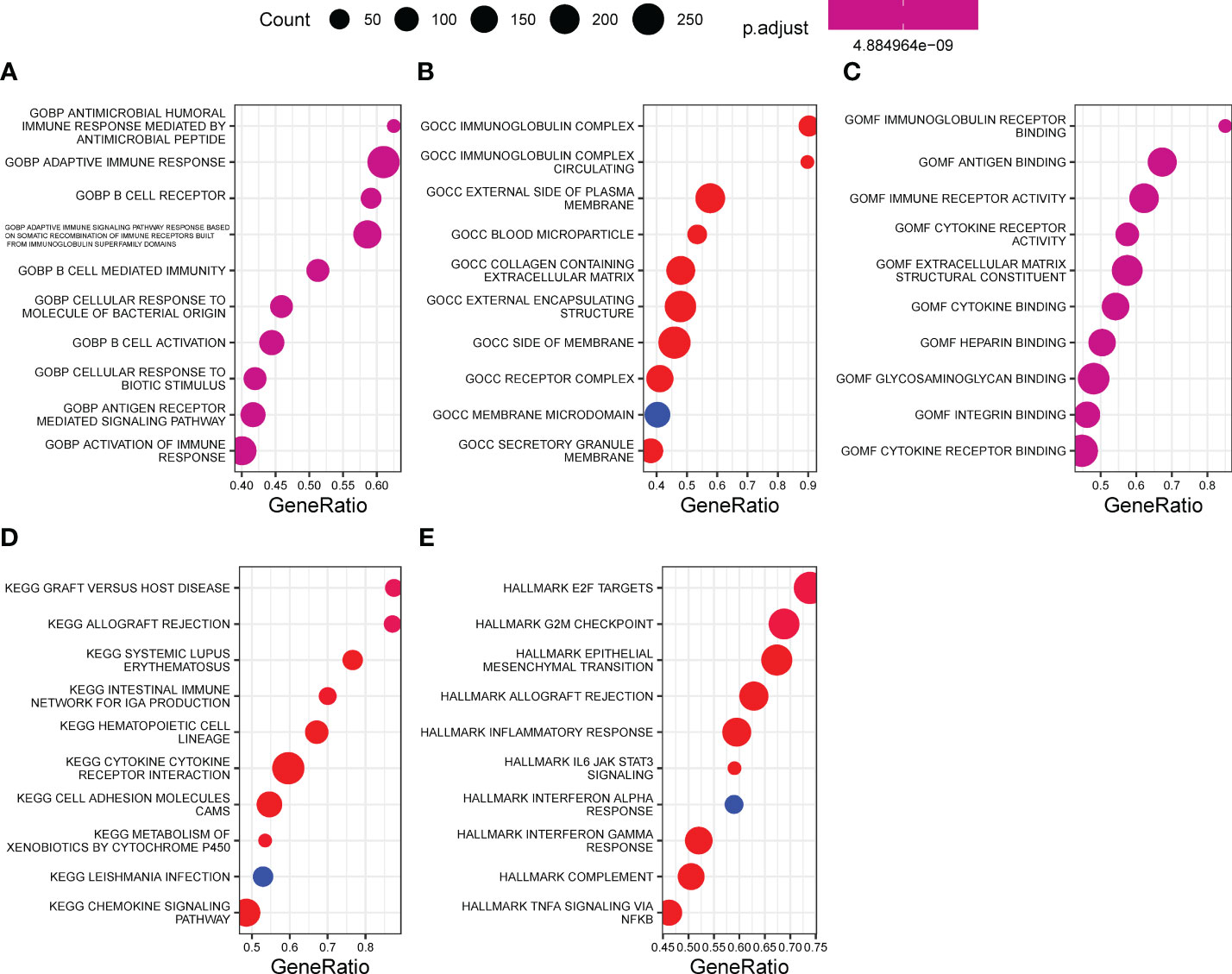
Figure 6 Biological enrichment underlying EPHB6. Function enrichment of EPHB6 expression related DEGs by GO (A-C), KEGG (D) and hallmark (E). DEG, differentially expressed genes; GOBP, gene ontology biological process; GOCC, gene ontology cellular component; GOMF, gene ontology molecular function; KEGG, kyoto encyclopedia of genes and genomes.
Underlying EPHB6 expression, we further deeply investigated the differences in genomic characteristics between EPHB6-high and -low expression groups. In spite of no statistical significance existing in the TMB level between EPHB6-high and -low expression groups (p=0.34; Figure 7A), the comprehensive genomic characterization revealed notable discrepancies in the genomic alteration landscape (Figures 7B, C). As shown in Figure 7D, TP53, ARID1A, and FAT4 alterations were significantly enriched in the EPHB6-low expression group; whereas, FGFR3, STAG2, and TENM3 alterations were more prevalent in the EPHB6-high expression group (p<0.05). Meanwhile, the specific DDR alteration landscape was also investigated; only BRCA2 alterations (Figure 7E) were found to be significantly more frequent in the EPHB6-high expression group. When comparing alterations in the signaling pathway, alterations in RTK/RAS signaling were discovered to be more frequent in the EPHB6-high expression group, whereas alterations in TP53 signaling pathway were more prevalent in the EPHB6-low expression group (Figure 7F).
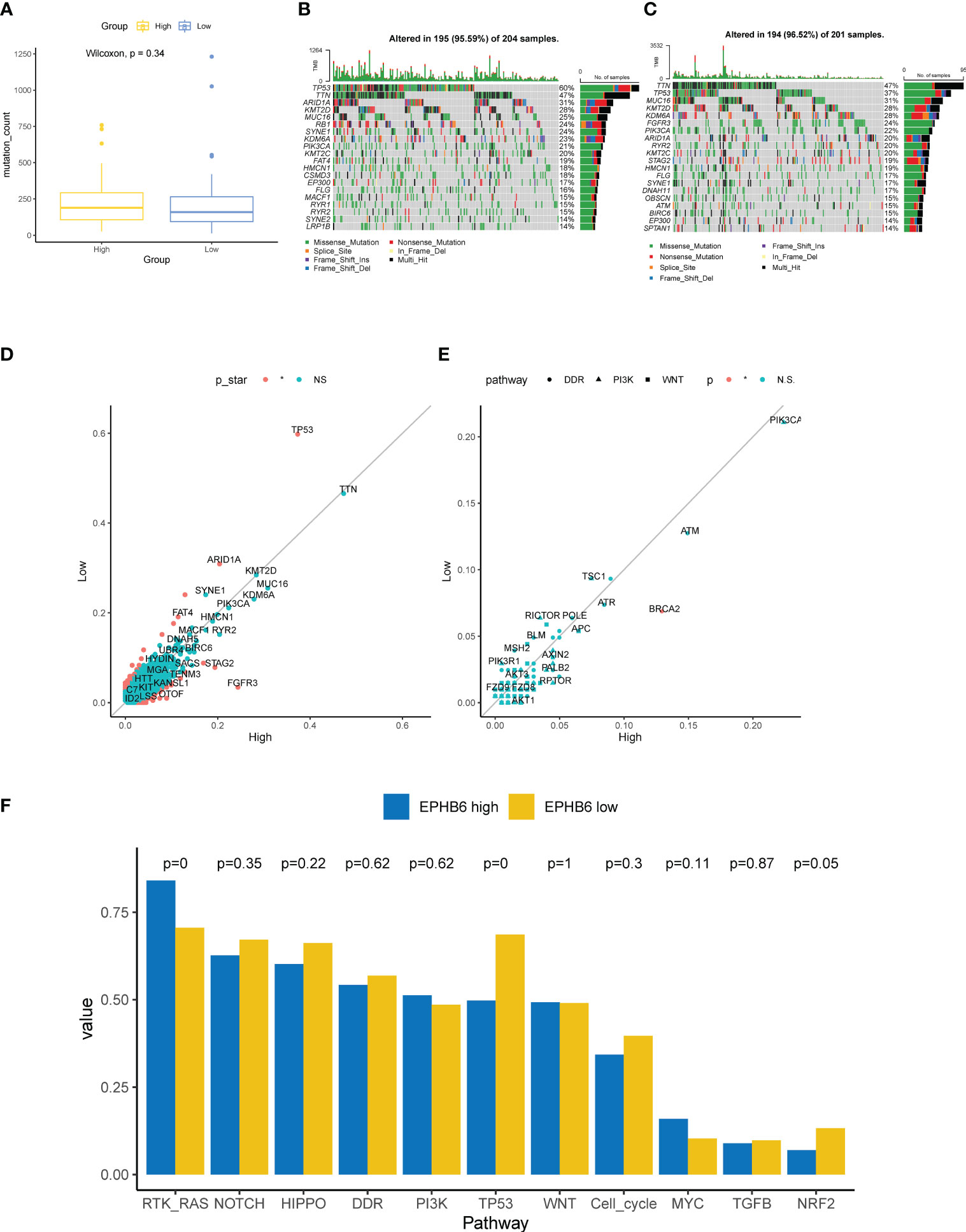
Figure 7 Genomic characteristics associated with EPHB6. (A) Difference in the mutation counts level between high- and low-EPHB6 group. Prevalent mutated genes in high- (B) and low-EPHB6 group (C). (D) Difference in the prevalence of mutated genes between high- and low-EPHB6 group. Genes colored in red were those significantly differed (p<0.05). (E) Difference in the prevalence of genes involved in DDR, PI3K or WNT signaling pathways between high- and low-EPHB6 group. (F) Difference in the mutated signaling pathways between high- and low-EPHB6 group.
According to the seven molecular subtyping systems, a markedly higher proportion of luminal (papillary) subtype was observed to always occur in the EPHB6-high expression group; in comparison, patients with basal subtype were more abundantly enriched in the EPHB6-low expression group (Figure 8A). The enrichment scores of urothelial differentiation and Ta pathway were apparently higher in the EPHB6-high expression group, whereas the enrichment scores of myofibroblasts and IFN response seemed to be higher in the EPHB6-low expression group. Regarding cell cycle activity, mitochondria, basal differentiation, B and T lymphocytes, smooth muscle, and neuroendocrine differentiation, their enrichment scores between EPHB6-high and -low expression groups were nearly equivalent. Furthermore, the AUC values of EPHB6 predicting the molecular subtype were all > 0.70, except in the Baylor subtyping system (Figure 8B). These findings reveal that EPHB6 expression was highly correlated with molecular subtype accompanied with some specific molecular characteristics, which guided significance for the therapy selection to some extent. The chemotherapy-associated mutational profiling demonstrated that ARID1A (31% vs. 20%) and RB1 (24% vs. 13%) alterations were more prevalent in the EPHB6-low expression group (p<0.05; Figures 8C-E). In addition, the Drugbank database analysis demonstrated a significantly higher response to most chemotherapeutic and immunotherapeutic target drugs in the EPHB6-low expression group (Figure 8F).
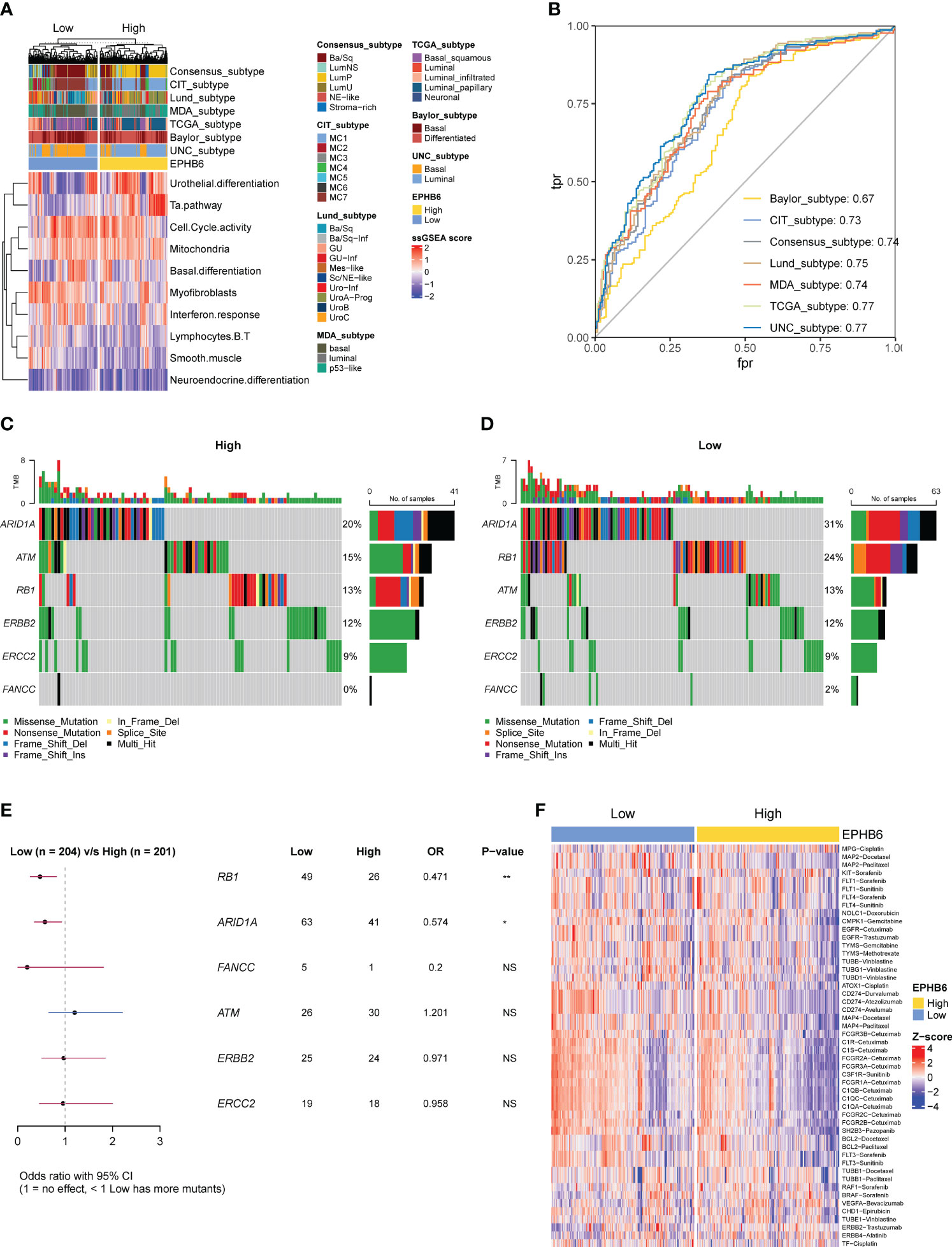
Figure 8 EPHB6 related molecular subtype and therapeutic strategies in bladder cancer. (A) Difference in the bladder cancer signatures between high- and low-EPHB6 group. (B) ROC curves showing the accuracy of EPHB6 for predicting molecular subtypes. (C-E) Difference in the prevalence of neoadjuvant chemotherapy-related genes between high- and low-EPHB6 group. (F) Difference in the enrichment scores of different therapeutic signatures between high- and low-EPHB6 group. Each therapeutic signature was indicated as target- regimen.
Ninety-one overlapped genes between the DEGs of BLCA vs. normal tissues (Figure 9A) and EPHB6-related DEGs (Figure 9B) were acquired (Figure 9C; Supplementary Table 2). The univariate Cox regression analysis found 35 genes to be significantly associated with the prognosis of BLCA (Supplementary Table 2). After calculation, the combinational algorithms of RF and Ridge exhibited the best performance (Figure 9D), identifying a total of 21 genes enrolled in the EPHB6-related Genes signature (EBGs; Supplementary Table 2). According to the median cut-off value of EBGs score, patients with BLCA were divided into high- and low-EBGs score groups. In the TCGA-BLCA cohort, high EBGs score group had worse clinical outcome. Moreover, the performance of EBGs signature was validated in other four independent BLCA cohorts, and consistently higher EBGs score indicated an inferior OS. All the AUC values of EBGs signature in predicting prognosis for patients with BLCA were approximately 0.65 (Figure 9E).
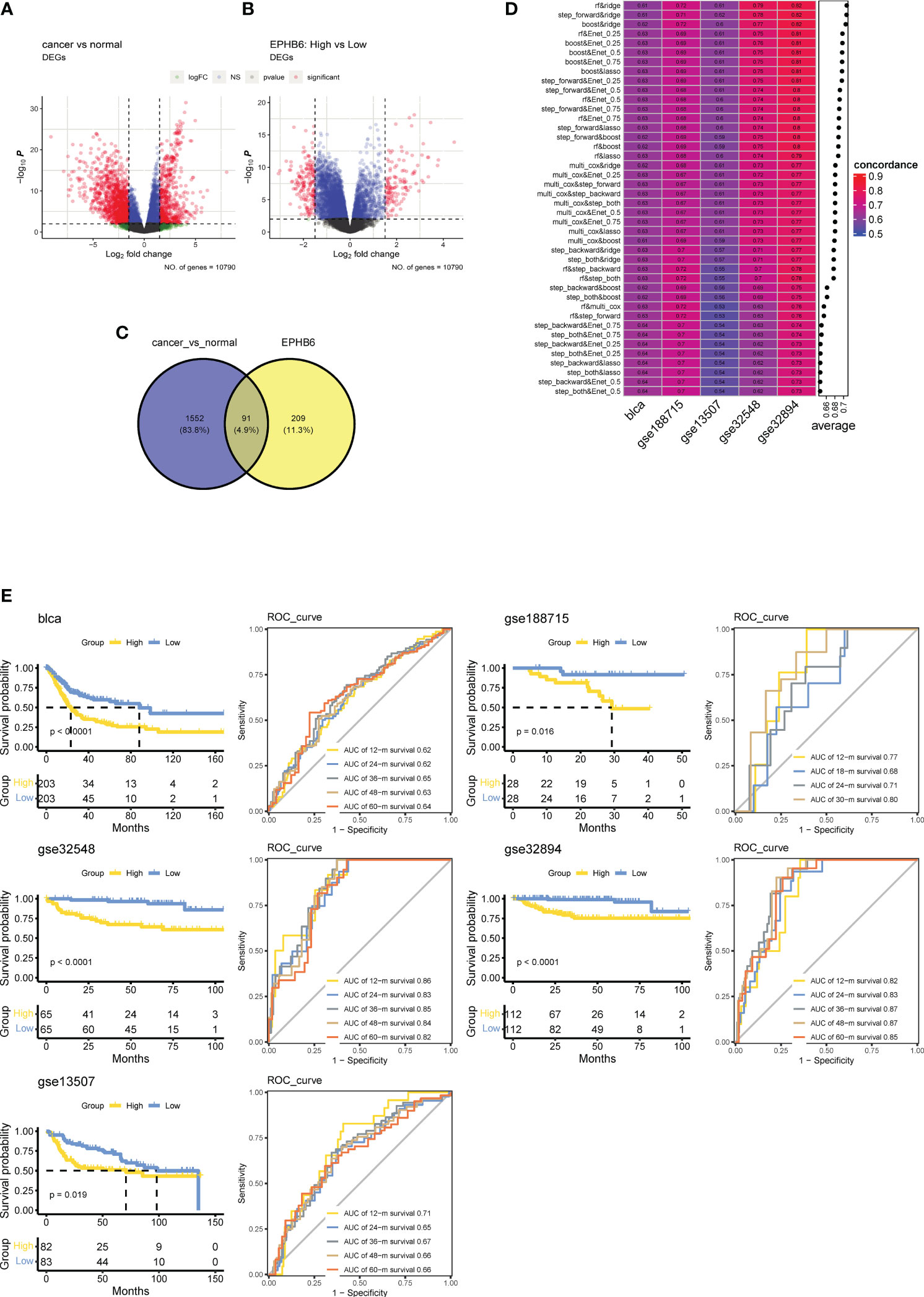
Figure 9 Establish a prognostic signature based on EPHB6 expression feature in bladder cancer. (A) Differentially expressed genes (DEGs) between tumor and normal bladder tissues as showing by the volcano plot. (B) Volcano plot showing the DEGs between high- and low-EPHB6 group. (C) Veen plot showing the shared genes. (D) Combination of machine learning algorithms for establishing EPHB6 related genes (EBGs) signatures. The C-index of each model was calculated across bladder cancer datasets, including TCGA-BLCA, GSE188715, GSE13507, GSE 32548 and GSE32894. (E) Kaplan-Meyer plots and time-dependent ROC curve for predicting survivals at 1- to 5-years in TCGA-BLCA, GSE188715, GSE13507, GSE 32548 and GSE32894 datasets.
Globally, BLCA is one of the most prevalent malignant urinary carcinomas with approximately 550,000 new cases diagnosed and 170,000 deaths annually (45). Generally, BLCA can develop to NMIBC and MIBC tumors, and each subtype has specific clinicopathological and molecular features (46). Currently, surgical resection remains the major therapeutic strategy for the localized BLCA (47). For patients with NMIBC, intravesical therapies, such as Bacillus Calmette-Guérin or alternative therapies after resection are frequently used to prevent disease progression and recurrence. More aggressive therapies, including radical cystectomy and urinary diversion with chemotherapy, radiotherapy, neoadjuvant chemotherapy, or immunotherapy are mainly proposed for patients with MIBC (48). Despite that, patients with advanced or metastatic BLCA still have a poor prognosis, with an estimated 5-year survival rate of < 30%, especially with the difficulty of distinguishing patients who will and will not respond to therapy. Therefore, it is urgent to discover more novel biomarkers that can become effective therapeutic targets for BLCA.
In this study, we conducted the first pan-cancer analysis revealing the correlation between the largest family of RTKs and immunomodulation. Eph receptor/EFN ligand family genes expression was highly correlated with immunomodulation of TME in 32 types of cancer, and different Eph receptor/EFN ligand genes had distinct immunological effects in different types of cancer. BLCA is an immunogenic tumor type (49), and an in-depth investigation of Eph receptor/EFN ligand family genes in BLCA identified that inactive kinase EPHB6 may be a potential therapeutic target. Evidence has revealed that EPHB6 could shape a cold immune microenvironment in BLCA. Furthermore, lower expression of EPHB6 in BLCA indicated a higher T cell inflamed score, which can efficiently quantify T cell inflamed microenvironment, and such a score was confirmed to positively correlate with the response to immune checkpoints blockade (33). Moreover, IMvigor210 cohort analysis showed that lower EPHB6 expression may assist in predicting the inflamed, IC2+, and TC2+ phenotypes, which has been confirmed to be markedly correlated with immunotherapy response of patients with UC treated with atezolizumab (anti-PD-L1 agent) (34). In addition, in the IMvigor210 cohort, the patients with UC showing a complete or partial response had a lower EPHB6 expression, directly indicating that anti-EPHB6 immunotherapy may be a novel and reliable therapeutic strategy and could greatly help to promote the response to immunotherapy and improve clinical outcomes for patients with BLCA. Notably, pan-cancer analysis showed that EPHB6 could not exert the similarly cold immune effects in most other types of cancer; hence, anti-EPHB6 immunotherapy might be suitable for patients with BLCA only, but it requires further clinical investigations.
Immunomodulators, cancer immunity, tumor immune cells infiltration, and immune checkpoints have been frequently used to evaluate the immunological status of TME (50). Most chemokines and paired receptors, particularly CXCL9, CXCL10, CXCL11, CXCL13, CCL3, CCR2, CCR3, and CXCR3, stimulate the recruitment of CD8+ T cells in different types of cancer in humans (29, 51–53). As reported, the downregulated chemokine/receptors in the EPHB6-high expression group may result in a reduced activity of anti-cancer immunity. In addition, downregulation of immunostimulators may also cause a cold immune microenvironment in the EPHB6-high expression group. Meanwhile, the reduced expression of MHC molecules would also attenuate the ability of antigen presentation and processing (29). Comprehensively, the complex functions and interactions of these immunomodulators could be integrated and reflected by seven major steps, directly representing the anti-cancer immunity to tumor cells (30). In the EPHB6-high expression group, the activities of steps 1, 4, and 7 in the cancer immunity cycle were weakened from the perspective of molecular functions, which implies that the activities of cancer cell antigen release and immune cells tracking tumor cells were restricted. Owing to the reduced level of immunomodulators, the infiltrating levels of several effector cells including macrophages, dendritic cells, NK cells, and CD8+ T cells were all expectedly decreased, which was also validated in our local BLCA cohort. Collectively, EPHB6 shaped a cold immune microenvironment in BLCA. Although no drugs have been directly targeted at EPHB6, related pathway-regulating drugs could potentially affect its expression and/or activity and then regulate tumor cell biology. In vitro analysis has shown that drugs that inhibit PI3K could reactivate EPHB6, thereby suppressing lung cell metastasis and proliferation (54). Moreover, enzalutamide, a widely applied anti-androgen therapy in advanced prostate cancer, has been found to suppress EPHB6 expression by binding to the androgen-response-element on the EPHB6 promoter to stimulate tumor cell metastasis (55). Therefore, given that EPHB6 has been identified as a protective prognostic factor in BLCA, monotherapy using its related targeted therapy could potentially lead to a worse clinical outcome. However, for patients with advanced or metastatic BLCA, EPHB6-directed therapy could potentially serve as a stimulator to immune checkpoint inhibitors (ICIs), reprogramming a cold immune tumor microenvironment into a hot one, which merited further study and investigation.
The comprehensive functional characterization subsequently displayed the enrichment of immunological processes, irrespective of initial (antimicrobial humoral immune response mediated by antimicrobial peptide) and adaptive immune responses, which further confirmed that EPHB6 expression was remarkably associated with immunomodulation in BLCA. Especially, IFN-γ (the only member belonging to the type II interferon family), as the uppermost cytokine involved in the anti-cancer immunity, may function to inhibit angiogenesis, induce apoptosis of cancer cells and T regulatory (Treg) cells, and further activate M1 macrophages to hinder tumor progression (56). However, some researchers have reported that IFN-γ may contribute to the development and progression of tumor (57–60). In this study, the findings revealed that IFN-γ signature enrichment score was markedly higher in the EPHB6-low expression group compared to that in the EPHB6-high group. The higher level of IFN-γ signaling likely contributed to the excessively inflamed microenvironment, consequently causing the inferior prognosis (OS, DSS, and PFI) in the EPHB6-low expression. From another perspective, IFN-γ is widely believed to be a critical factor influencing the response of immunotherapy (61). Ayers et al. has reported that patients with metastatic cancer (gastric cancer, head and neck squamous cell carcinoma, and melanoma) that responded to anti-PD-L1/PD-1 immunotherapy had a higher expression level of IFN-γ signaling-related genes (IFNG, CXCL9, CXCL10, HLA-DRA, STAT1, and IDO1) compared with the non-responders (33). A similar result was obtained in urothelial, melanoma, and non-small cell lung carcinoma in which IFN-γ signaling-related genes, including IFNG, CD274, LAG3, and CXCL9, could well predict about who could benefit from the immunotherapy via anti-PD-L1/PD-1 antibody (62, 63). Mechanistically, IFN-γ could activate the integrative inflammatory response and related immunological signaling to achieve the enhanced immunotherapy efficiency. Moreover, EPHB6 expression was negatively correlated with the enrichment scores of immunotherapy-predicted pathways.
Remarkably, mutational spectrum analysis further showed that the EPHB6-low expression group had a higher enrichment of TP53 genomic alterations, whereas especially FGFR3 alterations were abundantly enriched in the EPHB6-high expression group. Altered TP53 is known to prevalently associate with the poor clinical outcome in BLCA, and generally, TP53 alteration frequently occurred in patients with advanced or metastatic BLCA (64). In contrast, FGFR3 alterations were significantly correlated with lower pT stage, lower tumor grade, and expectedly longer survival in BLCA (65). From these molecular characteristics, EPHB6 was verified to be a potential prognostic biomarker. Additionally, EPHB6 expression may help identify the patients with altered FGFR3. Erdafitinib, as a later-line therapeutic regimen targeting FGFR3, recently achieved great advances in treating patients with UC having FGFR3 alterations (66). In addition, FGFR3-altered BLCA tumors had a lower expression of a fibroblast TGF-β response signature and downregulation of epithelial–mesenchymal transition signature (67). Accordingly, higher expression of EPHB6 was associated with the lower immune and stromal score in both TCGA-BLCA and local BLCA cohorts, and two other independent BLCA datasets. TP53 alterations were found to be positively correlated with the immune-promoting microenvironment; however, FGFR3 alterations might contribute to the cold immune microenvironment in BLCA. Furthermore, FGFR3 alterations were prevalent in the luminal papillary MIBC subtype (68); as expected, the EPHB6-high group had the higher proportion of luminal (papillary) subtype. Based on the molecular subtype classification, the EPHB6-low group showed a notable enrichment of basal subtype. Correspondingly, the enrichment scores of urothelial differentiation and Ta pathway were observed to be higher in the EPHB6-high expression group. As reported, urothelial differentiation is upregulated in the luminal subtype (69), and Ta pathway is positively correlated with FGFR3 alterations (70). Therefore, not only does EPHB6 have the prediction ability in prognosis in BLCA but also in molecular subtype.
At the molecular level, different molecular subtypes were noted to be correlated with distinct responses to chemotherapy, radiotherapy, neoadjuvant chemotherapy, and immunotherapy (71–74). A previous study regarding the consensus subtype of MIBC reported that receiving immunotherapy was more suitable for basal subtype tumors (38). Moreover, PURE-01 (NCT02736266) study also demonstrated that basal type tumors had the highest infiltrating level of tumor immune cells and pathological response rate to the neoadjuvant pembrolizumab (75). As previously described, RB1 (76) and ARID1A (77) alterations highly correlated with the response of chemotherapy. Both RB1 and ARID1A alterations were prevalently altered in the EPHB6-low expression (enriched basal subtype) group suggesting that patients with low expression of EPHB6 were more likely to be sensitive to the chemotherapeutic drugs. The Drugbank database analysis predicted that the EPHB6-low expression group had the higher response to most chemotherapeutic and immunotherapeutic regimens. Overall, adjuvant or neoadjuvant chemotherapy and immunotherapy (either monotherapy or combination therapy) could be considered for patients with BLCA having low expression of EPHB6. Based on the EPHB6-determined transcriptomic profile, a novel EBGs signature was eventually established which performed more robustly in the prognosis prediction.
In this study, the role of Eph receptor/EFN ligand family genes was investigated in a pan-cancer cohort for the first time. EPHB6 exhibited unique cold immune effects on BLCA tumors. From multiple perspectives, it was established that anti-EPHB6 immunotherapy might be a novel therapeutic regimen for patients with BLCA, which would greatly improve the clinical outcomes of BLCA. However, there are some limitations in the present study. First, number of patients in our local BLCA cohort was small; therefore, it is necessary to validate these results in a larger cohort of patients with BLCA. Second, the performance of EPHB6 in predicting the response of immunotherapy was only verified in a IMvigor210 cohort, which should be further investigated. Third, experimental explorations and clinical trials should be proposed to testify the efficacy and efficiency of anti-EPHB6 immunotherapy.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by The ethics committee of Ningbo First Hospital. The patients/participants provided their written informed consent to participate in this study.
XJ, JJ, and LX contributed to conception and design of the study. DZ and CZ organized the database. DZ performed the statistical analysis. XJ wrote the first draft of the manuscript. ZY, and ZJ wrote sections of the manuscript. XJ, DZ, CZ contributed equally to this work. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the following funding: (1) Medical and Health Science and Technology Plan Project of Zhejiang Province, No.2019KY575, No.2020ZH007, No.2021KY995); (2) Zhejiang Natural Science Fund (LTGY23H050002); (3) Ningbo Top Medical and Health Research Program (No.2022020203)
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1175183/full#supplementary-material
Supplementary Figure 1 | Correlation matrix showing the relationship between the expression levels of all the Eph receptors and EFN ligands and immunomodulatory genes a in pan-cancers.
Supplementary Figure 2 | EPHB6 is associated with a cold immune microenvironment in Bladder cancer. (A) Heatmap showing the elevated in the expression level of marker genes of CD8 T cells, dendric cells, macrophages, NK cells and Th1 cells. (B) Correlation between the expression levels of immune checkpoints and EPHB6 in TCGA-BLCA cohort.
Supplementary Figure 3 | Difference in the expression levels of immunomodulatory genes between the high- and low- EPHB6 groups in GSE31684 dataset.
Supplementary Figure 4 | Correlation analysis of the mRNA expression level of EPHB6 and CD8A or CD68 in TCGA-BLCA, Imvigor210 and Local BLCA datasets.
Supplementary Figure 5 | Binary response stratified in IMvigor210 cohort by TP53 mutation (mt vs wt) and EPHB6 expression level (high vs low). Mt: mutated; wt: wildtype.
1. Taylor H, Campbell J, Nobes CD. Ephs and ephrins. Curr Biol (2017) 27(3):R90–5. doi: 10.1016/j.cub.2017.01.003
2. Kullander K, Klein R. Mechanisms and functions of eph and ephrin signalling. Nat Rev Mol Cell Biol (2002) 3(7):475–86. doi: 10.1038/nrm856
3. Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer (2010) 10(3):165–80. doi: 10.1038/nrc2806
4. Pasquale EB. The eph family of receptors. Curr Opin Cell Biol (1997) 9(5):608–15. doi: 10.1016/S0955-0674(97)80113-5
5. Himanen J-P, Henkemeyer M, Nikolov DB. Crystal structure of the ligand-binding domain of the receptor tyrosine kinase EphB2. Nature (1998) 396(6710):486–91. doi: 10.1038/24904
6. Dai D, Huang Q, Nussinov R, Ma B. Promiscuous and specific recognition among ephrins and eph receptors. Biochim Biophys Acta (BBA) - Proteins Proteomics (2014) 1844(10):1729–40. doi: 10.1016/j.bbapap.2014.07.002
7. Arvanitis D, Davy A. Eph/ephrin signaling: networks. Genes Dev (2008) 22(4):416–29. doi: 10.1101/gad.1630408
8. Janes PW, Vail ME, Ernst M, Scott AM. Eph receptors in the immunosuppressive tumor microenvironment. Cancer Res (2021) 81(4):801–5. doi: 10.1158/0008-5472.Can-20-3047
9. Shiuan E, Chen J. Eph receptor tyrosine kinases in tumor immunity. Cancer Res (2016) 76(22):6452–7. doi: 10.1158/0008-5472.Can-16-1521
10. Tatsumi T, Herrem CJ, Olson WC, Finke JH, Bukowski RM, Kinch MS, et al. Disease stage variation in CD4+ and CD8+ t-cell reactivity to the receptor tyrosine kinase EphA2 in patients with renal cell carcinoma. Cancer Res (2003) 63(15):4481–9.
11. Rapp M, Grauer OM, Kamp M, Sevens N, Zotz N, Sabel M, et al. A randomized controlled phase II trial of vaccination with lysate-loaded, mature dendritic cells integrated into standard radiochemotherapy of newly diagnosed glioblastoma (GlioVax): study protocol for a randomized controlled trial. Trials (2018) 19(1):293. doi: 10.1186/s13063-018-2659-7
12. Markosyan N, Li J, Sun YH, Richman LP, Lin JH, Yan F, et al. Tumor cell–intrinsic EPHA2 suppresses antitumor immunity by regulating PTGS2 (COX-2). J Clin Invest (2019) 129(9):3594–609. doi: 10.1172/JCI127755
13. Yang W-H, Cha J-H, Xia W, Lee H-H, Chan L-C, Wang Y-N, et al. Juxtacrine signaling inhibits antitumor immunity by upregulating PD-L1 expression. Cancer Res (2018) 78(14):3761–8. doi: 10.1158/0008-5472.Can-18-0040
14. Vail ME, Murone C, Tan A, Hii L, Abebe D, Janes PW, et al. Targeting EphA3 inhibits cancer growth by disrupting the tumor stromal microenvironment. Cancer Res (2014) 74(16):4470–81. doi: 10.1158/0008-5472.Can-14-0218
15. Darling TK, Lamb TJ. Emerging roles for eph receptors and ephrin ligands in immunity. Front Immunol (2019) 10. doi: 10.10.3389/fimmu.2019.01473
16. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
17. Teoh JY-C, Kamat AM, Black PC, Grivas P, Shariat SF, Babjuk M. Recurrence mechanisms of non-muscleinvasive bladder cancer — a clinical perspective. Nat Rev Urol (2022) 19(5):280–94. doi: 10.1038/s41585-022-00578-1
18. Powles T, Csőszi T, Özgüroğlu M, Matsubara N, Géczi L, Cheng SYS, et al. Pembrolizumab alone or combined with chemotherapy versus chemotherapy as first-line therapy for advanced urothelial carcinoma (KEYNOTE-361): a randomised, open-label, phase 3 trial. Lancet Oncol (2021) 22(7):931–45. doi: 10.1016/s1470-2045(21)00152-2
19. Powles T, van der Heijden MS, Castellano D, Galsky MD, Loriot Y, Petrylak DP, et al. Durvalumab alone and durvalumab plus tremelimumab versus chemotherapy in previously untreated patients with unresectable, locally advanced or metastatic urothelial carcinoma (DANUBE): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol (2020) 21(12):1574–88. doi: 10.1016/s1470-2045(20)30541-6
20. Davis ID, Galsky MD, Garcia del Muro X, Park SH, De Giorgi U, Alekseev B, et al. Abstract CT040: Updated overall survival (OS) analysis of atezolizumab (atezo) monotherapy vs chemotherapy in untreated locally advanced or metastatic urothelial carcinoma (mUC) in IMvigor130. Cancer Res (2021). doi: 10.1158/1538-7445.am2021-ct040
21. Deshpande RP, Sharma S, Watabe K. The confounders of cancer immunotherapy: Roles of lifestyle, metabolic disorders and sociological factors. Cancers (2020) 12(10). doi: 10.3390/cancers12102983
22. Desrichard A, Kuo F, Chowell D, Lee KW, Riaz N, Wong RJ, et al. Tobacco smoking-associated alterations in the immune microenvironment of squamous cell carcinomas. J Natl Cancer Institute (2018) 110(12):1386–92. doi: 10.1093/jnci/djy060
23. Barr T, Girke T, Sureshchandra S, Nguyen C, Grant K, Messaoudi I. Alcohol consumption modulates host defense in rhesus macaques by altering gene expression in circulating leukocytes. J Immunol (Baltimore Md 1950) (2016) 196(1):182–95. doi: 10.4049/jimmunol.1501527
24. Sadeghi S, Quinn D, Dorff T, Pal S, Groshen S, Tsao-Wei D, et al. EphrinB2 inhibition and pembrolizumab in metastatic urothelial carcinoma. J Clin Oncol (2023) 41(3):640–50. doi: 10.1200/JCO.21.02923
25. Sadeghi S, Hether T, Yeon J, Mangio R, Reeves J, Liang Y, et al. 455 impact of EphB4 and PD-1 treatment on immune infiltrate in advanced bladder cancer. J ImmunoTherapy Cancer (2020) 8(Suppl 3):A276–6. doi: 10.1136/jitc-2020-SITC2020.0455
26. Toosi BM, El Zawily A, Truitt L, Shannon M, Allonby O, Babu M, et al. EPHB6 augments both development and drug sensitivity of triple-negative breast cancer tumours. Oncogene (2018) 37(30):4073–93. doi: 10.1038/s41388-018-0228-x
27. Manuela Z, Patrizia R, Probir C, Colin DHR, Steven H, Martina D, et al. EphB6 regulates TFEB-lysosomal pathway and survival of disseminated indolent breast cancer cells. Cancers (2021) 13(5). doi: 10.3390/cancers13051079
28. Yu H, Qin X-K, Yin K-W, Li Z-M, Ni E-D, Yang J-M, et al. EphB6 deficiency in intestinal neurons promotes tumor growth in colorectal cancer by neurotransmitter GABA signaling. Carcinogenesis (2023). doi: 10.1093/carcin/bgad041
29. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep (2017) 18(1):248–62. doi: 10.1016/j.celrep.2016.12.019
30. Chen Daniel S, Mellman I. Oncology meets immunology: The cancer-immunity cycle. Immunity (2013) 39(1):1–10. doi: 10.1016/j.immuni.2013.07.012
31. Xu L, Deng C, Pang B, Zhang X, Liu W, Liao G, et al. TIP: A web server for resolving tumor immunophenotype profiling. Cancer Res (2018) 78(23):6575–80. doi: 10.1158/0008-5472.Can-18-0689
32. Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med (2018) 24(10):1545–9. doi: 10.1038/s41591-018-0157-9
33. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest (2017) 127(8):2930–40. doi: 10.1172/JCI91190
34. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of t cells. Nature (2018) 554(7693):544–8. doi: 10.1038/nature25501
35. Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell (2014) 25(2):152–65. doi: 10.1016/j.ccr.2014.01.009
36. Rebouissou S, Bernard-Pierrot I, de Reyniès A, Lepage ML, Krucker C, Chapeaublanc E, et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci Transl Med (2014) 6(244):244ra291. doi: 10.1126/scitranslmed.3008970
37. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell (2017) 171(3):540–556.e525. doi: 10.1016/j.cell.2017.09.007
38. Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol (2020) 77(4):420–33. doi: 10.1016/j.eururo.2019.09.006
39. Rubio C, Martínez-Fernández M, Segovia C, Lodewijk I, Suarez-Cabrera C, Segrelles C, et al. CDK4/6 inhibitor as a novel therapeutic approach for advanced bladder cancer independently of RB1 status. Clin Cancer Res (2019) 25(1):390–402. doi: 10.1158/1078-0432.Ccr-18-0685
40. Gil-Jimenez A, van Dorp J, Contreras-Sanz A, van der Vos K, Vis DJ, Braaf L, et al. Assessment of predictive genomic biomarkers for response to cisplatin-based neoadjuvant chemotherapy in bladder cancer. Eur Urol (2022) 83(4):313–7. doi: 10.1016/j.eururo.2022.07.023
41. Groenendijk FH, de Jong J, Fransen van de Putte EE, Michaut M, Schlicker A, Peters D, et al. ERBB2 mutations characterize a subgroup of muscle-invasive bladder cancers with excellent response to neoadjuvant chemotherapy. Eur Urol (2016) 69(3):384–8. doi: 10.1016/j.eururo.2015.01.014
42. Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discovery (2014) 4(10):1140–53. doi: 10.1158/2159-8290.cd-14-0623
43. Tse J, Ghandour R, Singla N, Lotan Y. Molecular predictors of complete response following neoadjuvant chemotherapy in urothelial carcinoma of the bladder and upper tracts. Int J Mol Sci (2019) 20(4). doi: 10.3390/ijms20040793
44. Sun S, Pan X, Zhao L, Zhou J, Wang H, Sun Y. The expression and relationship of CD68-Tumor-Associated macrophages and microvascular density with the prognosis of patients with laryngeal squamous cell carcinoma. Clin Exp Otorhinolaryngology (2016) 9(3):270–7. doi: 10.21053/ceo.2015.01305
45. Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA: A Cancer J Clin (2020) 70(5):404–23. doi: 10.3322/caac.21631
46. Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, et al. Bladder cancer. Nat Rev Dis Primers (2017) 3(1):17022. doi: 10.1038/nrdp.2017.22
47. Witjes JA, Bruins HM, Cathomas R, Compérat EM, Cowan NC, Gakis G, et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: Summary of the 2020 guidelines. Eur Urol (2021) 79(1):82–104. doi: 10.1016/j.eururo.2020.03.055
48. Lenis AT, Lec PM, Chamie K, MSHS M. Bladder cancer: A review. JAMA (2020) 324(19):1980–91. doi: 10.1001/jama.2020.17598
49. Grivas P, Agarwal N, Pal S, Kalebasty AR, Sridhar SS, Smith J, et al. Avelumab first-line maintenance in locally advanced or metastatic urothelial carcinoma: Applying clinical trial findings to clinical practice. Cancer Treat Rev (2021) 97:102187. doi: 10.1016/j.ctrv.2021.102187
50. Hu J, Yu A, Othmane B, Qiu D, Li H, Li C, et al. Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics (2021) 11(7):3089–108. doi: 10.7150/thno.53649
51. Gordon-Alonso M, Hirsch T, Wildmann C, van der Bruggen P. Galectin-3 captures interferon-gamma in the tumor matrix reducing chemokine gradient production and t-cell tumor infiltration. Nat Commun (2017) 8(1):793. doi: 10.1038/s41467-017-00925-6
52. Li J, Ybarra R, Mak J, Herault A, De Almeida P, Arrazate A, et al. IFNγ-induced chemokines are required for CXCR3-mediated t-cell recruitment and antitumor efficacy of anti-HER2/CD3 bispecific antibody. Clin Cancer Res (2018) 24(24):6447–58. doi: 10.1158/1078-0432.Ccr-18-1139
53. Workel HH, Lubbers JM, Arnold R, Prins TM, van der Vlies P, de Lange K, et al. A transcriptionally distinct CXCL13+CD103+CD8+ t-cell population is associated with b-cell recruitment and neoantigen load in human cancer. Cancer Immunol Res (2019) 7(5):784–96. doi: 10.1158/2326-6066.Cir-18-0517
54. Fan Y-H, Ding H-W, Kim D, Liu J-Y, Hong J-Y, Xu Y-N, et al. The PI3Kα inhibitor DFX24 suppresses tumor growth and metastasis in non-small cell lung cancer via ERK inhibition and EPHB6 reactivation. Pharmacol Res (2020) 160:105147. doi: 10.1016/j.phrs.2020.105147
55. Chen J, Li L, Yang Z, Luo J, Yeh S, Chang C. Androgen-deprivation therapy with enzalutamide enhances prostate cancer metastasis via decreasing the EPHB6 suppressor expression. Cancer Lett (2017) . 408:155–63. doi: 10.1016/j.canlet.2017.08.014
56. Hu KH, Krummel MF. Carpet-bombing tumors with IFN-γ. Nat Cancer (2020) 1(3):270–2. doi: 10.1038/s43018-020-0042-6
57. Kelly SA, Gschmeissner S, East N, Balkwill FR. Enhancement of metastatic potential by gamma-interferon. Cancer Res (1991) 51(15):4020–7.
58. Andrianifahanana M, Singh AP, Nemos C, Ponnusamy MP, Moniaux N, Mehta PP, et al. IFN-gamma-induced expression of MUC4 in pancreatic cancer cells is mediated by STAT-1 upregulation: a novel mechanism for IFN-gamma response. Oncogene (2007) 26(51):7251–61. doi: 10.1038/sj.onc.1210532
59. Chapela PJ, Broaddus RR, Hawkins SM, Lessey BA, Carson DD. Cytokine stimulation of MUC4 expression in human female reproductive tissue carcinoma cell lines and endometrial cancer. J Cell Biochem (2015) 116(11):2649–57. doi: 10.1002/jcb.25213
60. Singh S, Kumar S, Srivastava RK, Nandi A, Thacker G, Murali H, et al. Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-γ signalling. Nat Cell Biol (2020) 22(5):591–602. doi: 10.1038/s41556-020-0495-y
61. Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-γ in tumor progression and regression: a review. biomark Res (2020) 8(1):49. doi: 10.1186/s40364-020-00228-x
62. Higgs BW, Morehouse CA, Streicher K, Brohawn PZ, Pilataxi F, Gupta A, et al. Interferon gamma messenger RNA signature in tumor biopsies predicts outcomes in patients with non–small cell lung carcinoma or urothelial cancer treated with durvalumab. Clin Cancer Res (2018) 24(16):3857–66. doi: 10.1158/1078-0432.Ccr-17-3451
63. Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol (2018) 10:1758834017749748. doi: 10.1177/1758834017749748
64. Kaubryte J, Lai AG. Pan-cancer prognostic genetic mutations and clinicopathological factors associated with survival outcomes: a systematic review. NPJ Precis Oncol (2022) 6(1):27. doi: 10.1038/s41698-022-00269-5
65. van Rhijn BWG, Mertens LS, Mayr R, Bostrom PJ, Real FX, Zwarthoff EC, et al. FGFR3 mutation status and FGFR3 expression in a large bladder cancer cohort treated by radical cystectomy: Implications for anti- FGFR3 treatment?†. Eur Urol (2020) 78(5):682–7. doi: 10.1016/j.eururo.2020.07.002
66. Cathomas R, Lorch A, Bruins HM, Compérat EM, Cowan NC, Efstathiou JA, et al. The 2021 updated european association of urology guidelines on metastatic urothelial carcinoma. Eur Urol (2022) 81(1):95–103. doi: 10.1016/j.eururo.2021.09.026
67. Wang L, Gong Y, Saci A, Szabo PM, Martini A, Necchi A, et al. Fibroblast growth factor receptor 3 alterations and response to PD-1/PD-L1 blockade in patients with metastatic urothelial cancer. Eur Urol (2019) 76(5):599–603. doi: 10.1016/j.eururo.2019.06.025
68. Shi M-J, Fontugne J, Moreno-Vega A, Meng X-Y, Groeneveld C, Dufour F, et al. FGFR3 mutational activation can induce luminal-like papillary bladder tumor formation and favors a male sex bias. Eur Urol (2022) 83(1):70–81. doi: 10.1016/j.eururo.2022.09.030
69. Kim J, Akbani R, Creighton CJ, Lerner SP, Weinstein JN, Getz G, et al. Invasive bladder cancer: Genomic insights and therapeutic promise. Clin Cancer Res (2015) 21(20):4514–24. doi: 10.1158/1078-0432.Ccr-14-1215
70. Neuzillet Y, Paoletti X, Ouerhani S, Mongiat-Artus P, Soliman H, de The H, et al. A meta-analysis of the relationship between FGFR3 and TP53 mutations in bladder cancer. PloS One (2012) 7(12):e48993. doi: 10.1371/journal.pone.0048993
71. McConkey DJ, Choi W, Shen Y, Lee IL, Porten S, Matin SF, et al. A prognostic gene expression signature in the molecular classification of chemotherapy-naïve urothelial cancer is predictive of clinical outcomes from neoadjuvant chemotherapy: A phase 2 trial of dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin with bevacizumab in urothelial cancer. Eur Urol (2016) 69(5):855–62. doi: 10.1016/j.eururo.2015.08.034
72. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell (2018) 174(4):1033. doi: 10.1016/j.cell.2018.07.036
73. Nadal R, Bellmunt J. Management of metastatic bladder cancer. Cancer Treat Rev (2019) 76:10–21. doi: 10.1016/j.ctrv.2019.04.002
74. Singla N, Ghandour RA, Raj GV. Biomarkers for platinum sensitivity in bladder cancer: are we there yet? Trans Andrology Urol (2019) 8:S236–9. doi: 10.21037/tau.2019.01.10
75. Necchi A, Raggi D, Gallina A, Ross JS, Farè E, Giannatempo P, et al. Impact of molecular subtyping and immune infiltration on pathological response and outcome following neoadjuvant pembrolizumab in muscle-invasive bladder cancer. Eur Urol (2020) 77(6):701–10. doi: 10.1016/j.eururo.2020.02.028
76. Pietzak EJ, Zabor EC, Bagrodia A, Armenia J, Hu W, Zehir A, et al. Genomic differences between “Primary” and “Secondary” muscle-invasive bladder cancer as a basis for disparate outcomes to cisplatin-based neoadjuvant chemotherapy. Eur Urol (2019) 75(2):231–9. doi: 10.1016/j.eururo.2018.09.002
Keywords: EPHB6, bladder cancer, immunomodulator, tumor microenvironment, immunotherapy, molecular subtype, prognostic signature
Citation: Jia X, Zhang D, Zhou C, Yan Z, Jiang Z, Xie L and Jiang J (2023) Eph receptor B6 shapes a cold immune microenvironment, inhibiting anti-cancer immunity and immunotherapy response in bladder cancer. Front. Oncol. 13:1175183. doi: 10.3389/fonc.2023.1175183
Received: 27 February 2023; Accepted: 10 July 2023;
Published: 09 August 2023.
Edited by:
Sergei Kusmartsev, University of Florida, United StatesReviewed by:
Song Wu, Shenzhen University, ChinaCopyright © 2023 Jia, Zhang, Zhou, Yan, Jiang, Xie and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Xie, eGllbHBAemp1LmVkdS5jbg==; Junhui Jiang, SmlhbmdqaDIwMDUwOUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.