
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Oncol., 08 June 2023
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1174577
Background: Circular RNAs (circRNAs) are types of endogenous noncoding RNAs produced by selective splicing that are expressed highly specifically in various organisms and tissues and have numerous clinical implications in the regulation of cancer development and progression. Since circRNA is resistant to digestion by ribonucleases and has a long half-life, there is increasing evidence that circRNA can be used as an ideal candidate biomarker for the early diagnosis and prognosis of tumors. In this study, we aimed to reveal the diagnostic and prognostic value of circRNA in human pancreatic cancer (PC).
Methods: A systematic search for publications from inception to 22 July 2022 was conducted on Embase, PubMed, Web of Science (WOS), and the Cochrane Library databases. Available studies that correlated circRNA expression in tissue or serum with the clinicopathological, diagnostic, and prognostic values of PC patients were enrolled. Odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were used to evaluate clinical pathological characteristics. Area under the curve (AUC), sensitivity, and specificity were adopted to assess diagnostic value. Hazard ratios (HRs) were utilized to assess disease-free survival (DFS) and overall survival (OS).
Results: This meta-analysis enrolled 32 eligible studies, including six on diagnosis and 21 on prognosis, which accounted for 2,396 cases from 245 references. For clinical parameters, high expression of carcinogenic circRNA was significantly associated with degree of differentiation (OR = 1.85, 95% CI = 1.47–2.34), TNM stage (OR = 0.46, 95% CI = 0.35–0.62), lymph node metastasis (OR = 0.39, 95% CI = 0.32–0.48), and distant metastasis (OR = 0.26, 95% CI = 0.13–0.51). As for clinical diagnostic utility, circRNA could discriminate patients with pancreatic cancer from controls, with an AUC of 0.86 (95% CI: 0.82–0.88), a relatively high sensitivity of 84%, and a specificity of 80% in tissue. In terms of prognostic significance, carcinogenic circRNA was correlated with poor OS (HR = 2.00, 95% CI: 1.76–2.26) and DFS (HR = 1.96, 95% CI: 1.47–2.62).
Conclusion: In summary, this study demonstrated that circRNA may act as a significant diagnostic and prognostic biomarker for pancreatic cancer.
Pancreatic cancer (PC), a malignant tumor with a poor prognosis, is the seventh leading cause of cancer-related deaths worldwide, with increasing morbidity and mortality (1). Thus, it remains a burden on the international medical landscape. To our knowledge, personal features, environment, lifestyle behaviors, and related primary diseases are the main risk factors (2). Among them, the risk factors related to personal characteristics mostly include sex, age, race, obesity, smoking, heredity, and so on (3, 4). As for environment and lifestyle behaviors, exposure to trace elements (5), smoking (6), excessive alcohol consumption (7), overweight and obesity (8), red meat, and saturated fat increase the probability of pancreatic cancer. In addition, underlying diseases such as diabetes (9), chronic pancreatitis (10), and allergies (11) can also significantly contribute to the risk of pancreatic cancer.
Traumatic surgery is an option for patients with non-metastatic pancreatic cancer. High-end medical equipment and gradually improved surgical techniques significantly reduced patient postoperative mortality (12). In recent years, preoperative adjuvant therapy has also helped patients with pancreatic cancer who had already metastasized before surgery (13). Nonetheless, the overall 5-year survival rate of pancreatic cancer patients was only 2%–9%, which was quite low when compared to other cancers (3). The situation compelled us to reconsider how to increase the rate of pancreatic cancer diagnosis in the early stages to improve patient prognosis.
The use of imaging technologies such as endoscopy or magnetic resonance imaging (MRI) for screening in high-risk populations can significantly improve pancreatic cancer detection rates (14). However, the screening projects have not been widely implemented in China. Because of its high cost and limited sensitivity, imaging technology may not be suitable for early diagnosis, whereas biomarkers have piqued the interest of researchers due to their non-invasive and cost-effective characteristics (15, 16). Carbohydrate antigen199 (CA199) is currently the biomarker used in the clinical diagnosis of pancreatic cancer (17), but an increase in CA199 does not always indicate the presence of cancer, which could be due to biliary infection, inflammation, obstruction, or other diseases (18). The appearance of false positives will reduce the biomarker’s reliability. Although the use of CA199 alone has limitations (19), some studies have found that combining CA199 with other markers such as MUC5AC or microRNA 196a can improve the accuracy of pancreatic cancer diagnosis while still not showing a better result (20, 21).
Recently, new biomarker research on cancer has been emerging endlessly, including microRNA, circRNA, and other novel biomarkers (22, 23). Circular RNAs are a class of long, non-coding RNA molecules that shape a covalently closed continuous loop that has no 5’–3’ polarity and contains no polyA tail (24, 25). The stability of its function is determined by its closed ring structure (26). Previous studies suggested that circRNA may not have biological functions (27), but recent research evidence showed that it may play an important role in the occurrence and development of cancers (28), including liver cancer (29), colorectal cancer (30), breast cancer (31), and so on. The study found that circRNA can adsorb miRNA like a sponge and then affect biological functions; RNA-binding proteins are widely involved in the transcription and translation of proteins (32). They participate in the whole process of circRNA synthesis. CircRNA can also affect the expression of RNA-binding proteins; circRNA can also serve as a template for translation, a cell protein scaffold, and a protein function enhancer (33, 34). Of course, some research teams are also devoted to exploring the function of circRNA in pancreatic cancer. Researchers have detected a variety of circRNAs through microarrays or chips, but only a small part of them confirmed the critical role of their occurrence and development in pancreatic cancer, and the biological process of pancreatic cancer is still under study. The research was still in its initial stages, which did not prevent circRNA from becoming a more valuable biomarker for the diagnosis, treatment, and prognosis of pancreatic cancer in the future (32).
Although the research on circRNA in cancer has been extensively explored, its value in the diagnosis and prognosis of pancreatic cancer has not yet been comprehensively evaluated. The purpose of this study was to evaluate the diagnostic and prognostic value of circRNA in pancreatic cancer and to provide a basis for later functional mechanism research.
The study implemented comprehensive searching until 22 July 2022 to identify relevant literature. The resources were primarily from the Pubmed, Embase, Web of Science, Cochrane Library, CNKI, VIP, and Wanfang databases. Meanwhile, the study strictly followed the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) checklist. The search terms were used as follows (1): pancreatic (2); neoplasm OR cancer OR tumor OR neoplastic OR neoplasia OR carcinoma OR malignancy OR malignancies (3); circRNAs* OR circular RNA* (4); diagnosis OR diagnostic OR area under the curve OR AUC OR sensitivity OR specificity OR ROC OR receiver operation characteristic curve OR detection OR screen OR screening; and (5) prognosis OR prognostic OR HR OR Hazard Rate.
Enrolled studies were required to meet the following criteria: (1) the subjects of studies were all pancreatic cancer; (2) the study design was case-control or cohort; and (3) the content of studies was associated with circular RNA and diagnosis or prognosis.
The reports were excluded if the studies accorded with one of the following criteria: (1) the topic of studies was not pancreatic cancer or circular RNA; (2) the type of studies was original articles rather than conference papers or books; (3) no valid data could be obtained, such as the sensitivity and specificity of circRNA or HR; and (4) the studies were not written in English.
The data were extracted by two independent individuals. The details were extracted from the included studies as follows: a. first author; b. publish year; c. country; d. ethnicity; e. cancer type; f. circRNA type; g. expression level; h. specimen source; i. no. of patients; j. no. of controls; k. AUC, sensitivity, and specificity (diagnosis); l. follow up time, OS, and HR (prognosis); m. age, gender, tumor size, lymph node metastasis, grade of histology, TNM stage, and distant metastasis (clinical characteristics).
The Quality Assessment for Studies of Diagnostic Accuracy (QUADAS)-2 scale was used to evaluate the quality of the diagnosis studies that were enrolled in this study. Similarly, the Newcastle-Ottawa Scale (NOS) was applied to prognosis studies. The research was considered as high-quality if the scores of the QUADAS scale and NOS all reached higher than six.
Stata 15.1 was used to analyze the data. The effect sizes of clinical characteristics (OR) were shown in a forest plot. SROC (summary receiver operator characteristic), pooled sensitivity, and specificity were adopted to evaluate the diagnostic value of circRNA for pancreatic cancer. A pooled hazard ratio (HR) was used to assess the prognosis ability. The random effect model was applied to the merge data. The I2 test and P-value were used to check the heterogeneity of the merged results. I2 >50% or P <0.05 seemed to indicate strong heterogeneity. Egger’s test was used to examine publication bias quantitatively.
The detailed process of screening literature is shown in Figure 1. The study identified 439 articles through comprehensive searching; however, 154 of them were duplicates. After looking through the titles and abstracts of each article, 195 articles were not considered to be associated with pancreatic cancer or circRNA and were removed. A total of 91 articles were read thoroughly and carefully, and 36 studies were preserved fortunately in the end (35–70). Among them, there are 27 studies related to clinicopathological features, nine studies related to diagnosis, and 24 studies related to prognosis. A total of 2,930 subjects were included, and the publication period of the article was from 2017 to 2022. All studies were carried out in China. Most of the cancer types were pancreatic ductal adenocarcinomas, and the study samples included tissue and serum.
Figures 2 and 3 described the correlation between clinicopathological characters and circRNA expression level. The correlation between age, gender, and tumor size with circRNA expression was not found (Figure 2 and 3). Interestingly, the results showed that the circRNA expression level was associated with lymph node metastasis (OR = 0.39, 95% CI = 0.32–0.48, Figure 3A), degree of differentiation (OR = 1.85, 95% CI = 1.47–2.34, Figure 3B), TNM stage (OR = 0.46, 95% CI = 0.35–0.62, Figure 3C), and distant metastasis (OR = 0.26, 95% CI = 0.13–0.51, Figure 3D). This means that the lower the degree of tumor differentiation, the higher the expression level of circRNA; the occurrence of lymph node metastasis and distant metastasis is also related to the higher expression of circRNA; and the later the TNM stage, the higher the expression level of circRNA. The detailed information on the included studies with clinicopathological characters is in Supplementary Tables 1–7.
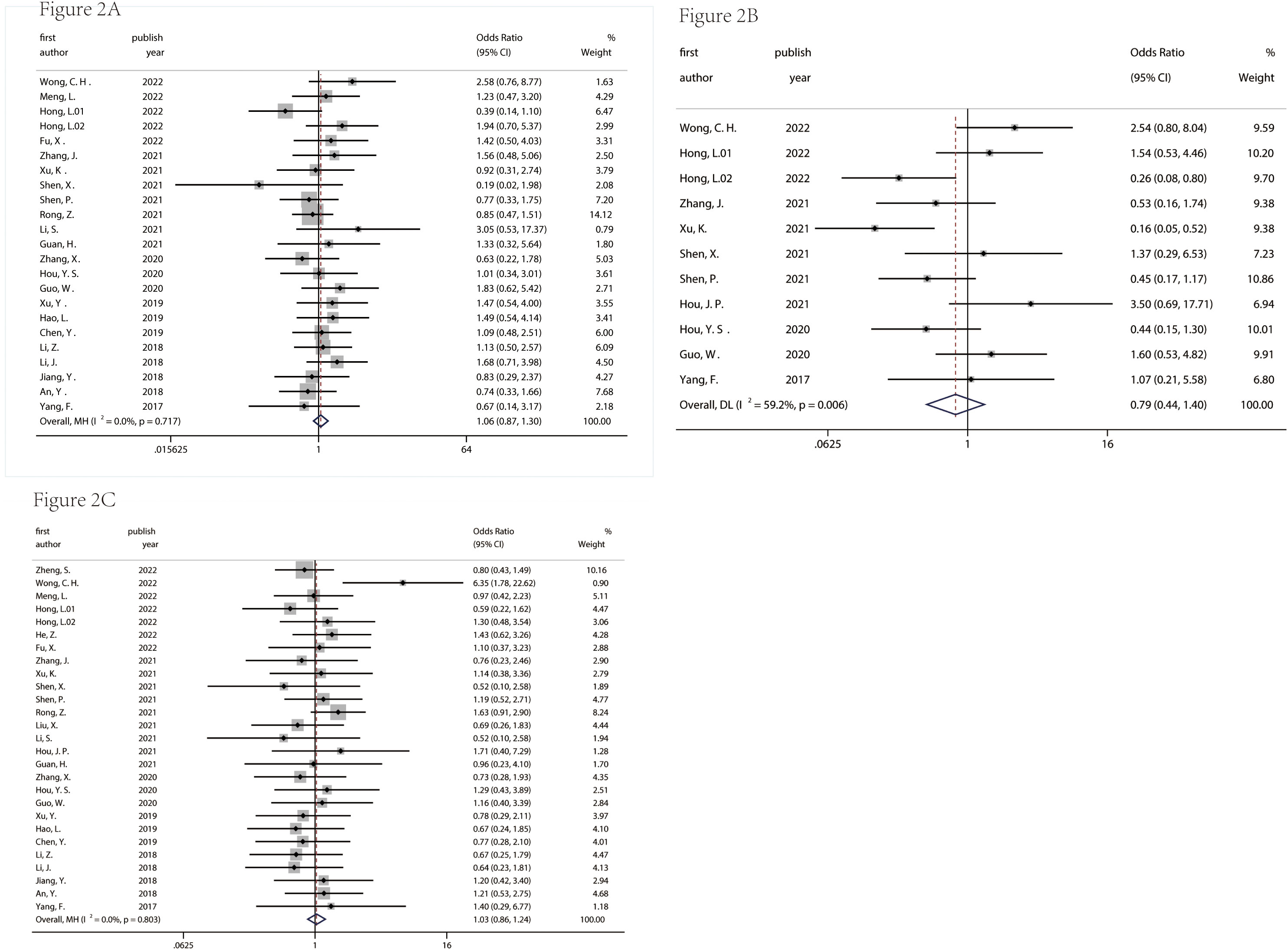
Figure 2 Correlation between age (A), gender (B), tumor size (C) and circRNA expression in pancreatic cancer. age: ≥60 years/<60years; gender: male/female; tumor size: ≥5 cm/<5 cm.
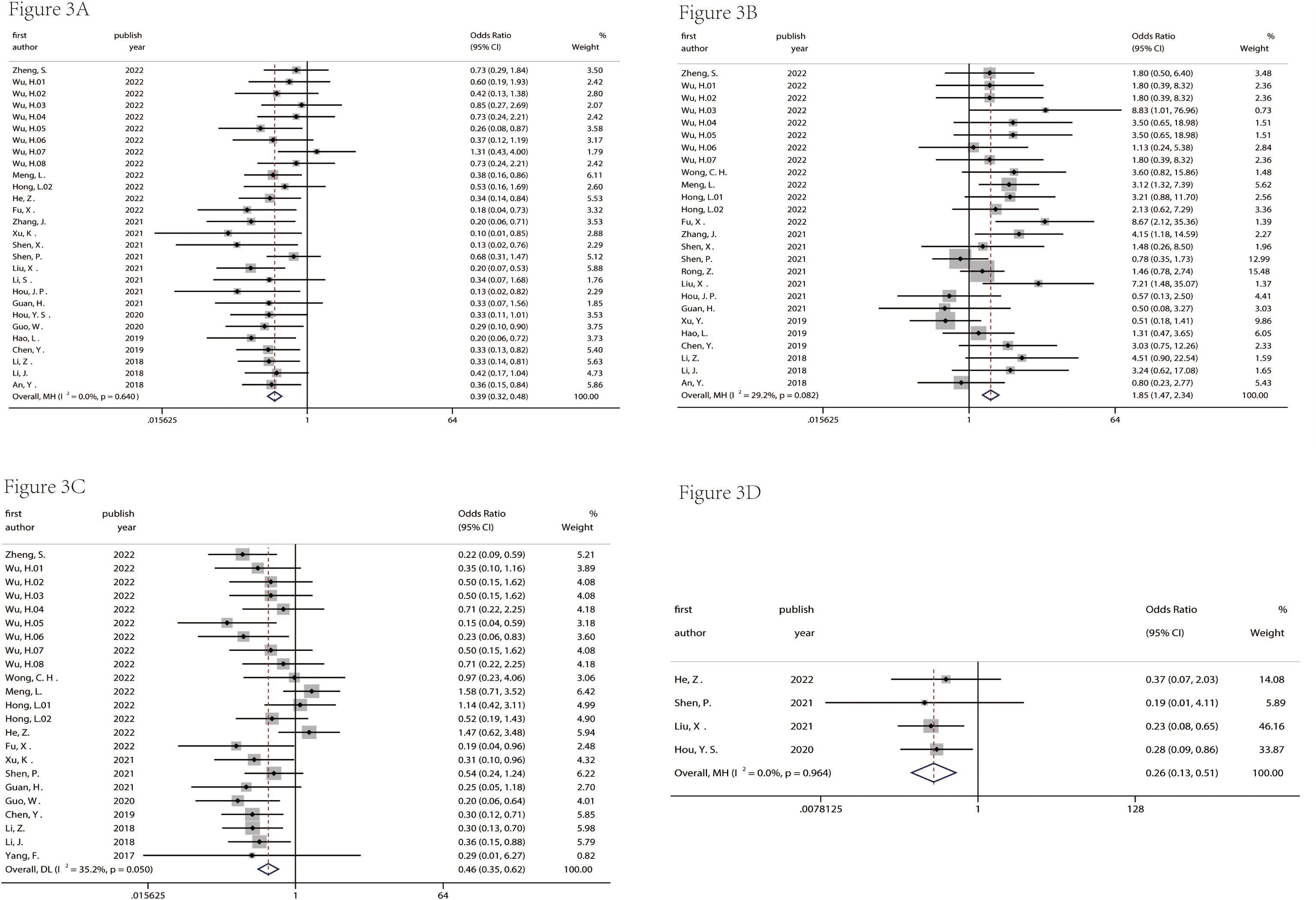
Figure 3 Correlation between lymph node metastasis (A), degree of differentiation (B), TNM stage (C), and distant metastasis (D) and circRNA expression in pancreatic cancer.
There were nine studies used to assess the diagnostic value (Table 1). Table 2 indicated that the included studies of diagnosis were high quality. The merged diagnostic performance of circRNA for pancreatic cancer was 0.83 (95% CI = 0.80–0.86) from the SROC curve in Figure 4A. However, we also found that the pooled sensitivity and specificity were highly heterogeneous (Figure 4B). The bivariate boxplot was used to seek out the sources of heterogeneity. As shown in the bivariate boxplot (Figure 4C), two studies were not involved in the boxplot, including studies 5 and 10. The testing sample source in study 5 was tissue, but in study 10, it was plasma, which may indicate that the specimen source was the main factor in heterogeneity. The results of the subgroup analysis showed that if the sample was from tissue, the diagnostic value reached 0.86 (95% CI = 0.82–0.88), and pooled sensitivity and specificity were 84% and 80%, respectively (Figures 5A, B). But the diagnostic value of the serum sample (AUC = 0.75, 95% CI = 0.72–0.78) was lower than that of the tissue sample; and the pooled sensitivity and specificity were 70% and 72%, respectively (Figures 5C, D). The results of Figure 4D indicated that there was no significant publication bias in the literature included in the diagnostic studies (P = 0.51).
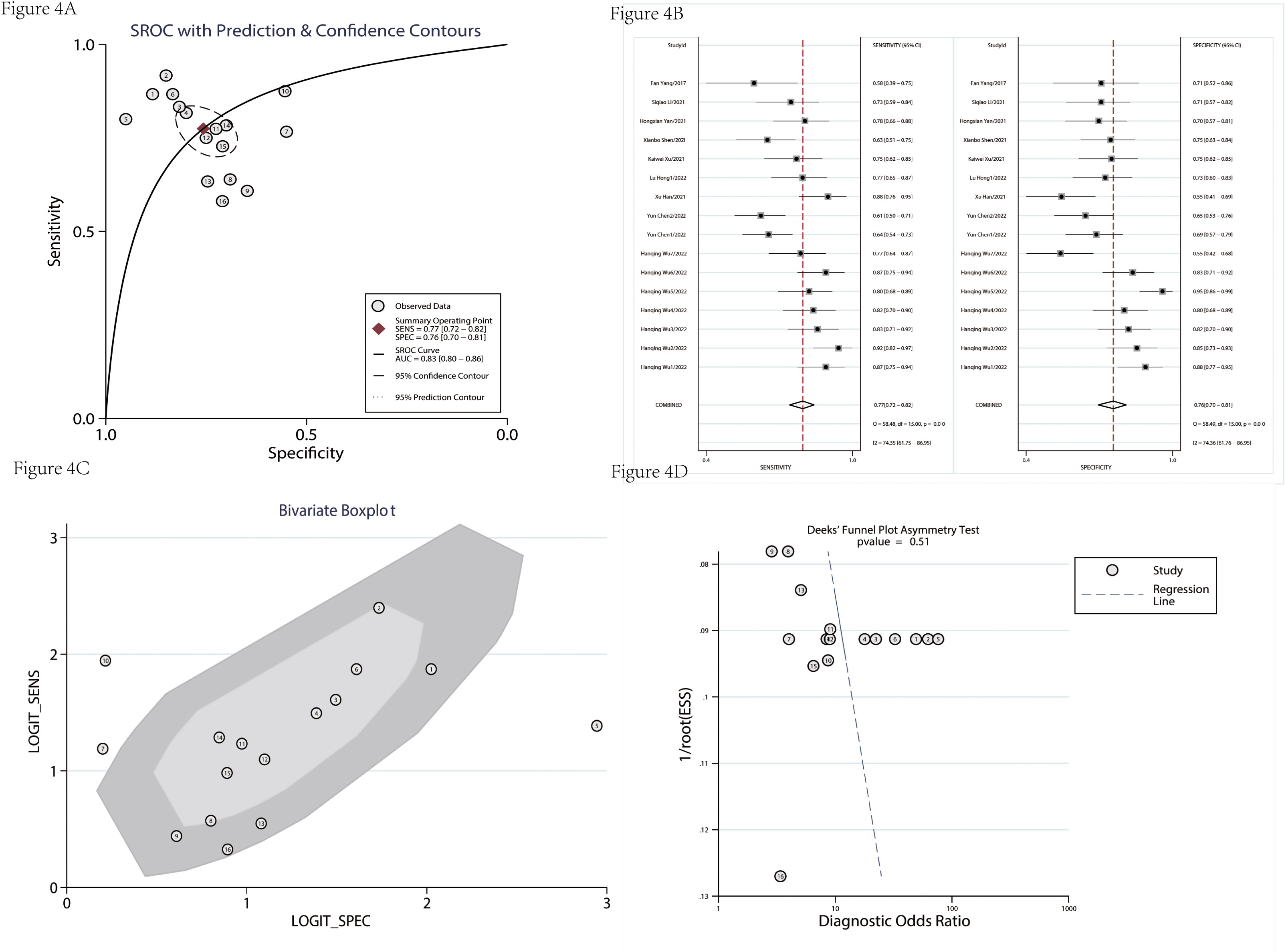
Figure 4 The diagnostic value was evaluated. SROC of enrolled diagnostic studies (A). The pooled sensitivity and specificity of enrolled diagnostic studies (B). The bivariate boxplot of diagnostic studies (C). The funnel plot was used to assess the publication bias of enrolled diagnostic studies (D). SROC, summary receiver operator characteristic.
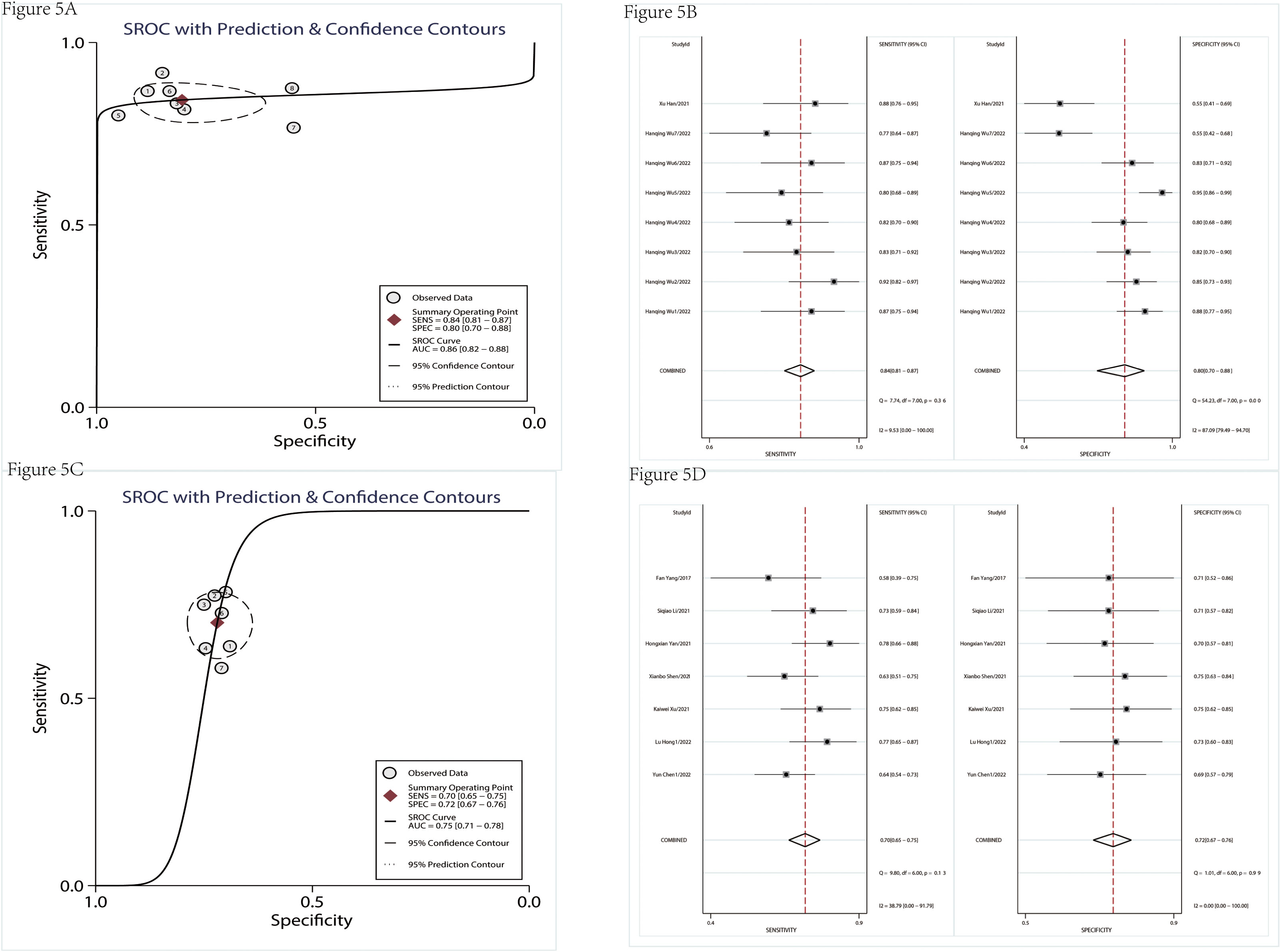
Figure 5 The subgroup analysis of diagnostic studies. SROC of diagnostic studies in tissue (A). The pooled sensitivity and specificity of diagnostic studies in tissue (B). SROC of diagnostic studies in serum (C). The pooled sensitivity and specificity of diagnostic studies in serum (D). SROC, summary receiver operator characteristic.
Table 3 showed the detailed information of prognosis. There was no significant heterogeneity in the combined hazard ratio (HR). The results of Figures 6A, D showed that the high expression level of circRNAs indicated a poor prognosis; the pooled HRs of OS and DFS were 1.98 (95% CI = 1.77–2.21) and 1.82 (95% CI = 1.51–2.20), respectively. The sensitivity analysis showed that no single study could influence results (Figures 6B, E), which suggested that circRNA was a more stable biomarker and has some prognostic implications for patients’ DFS or OS. A funnel plot (Figures 6C, F) and Egger’s test were used to evaluate publication bias. The P-values of Egger’s test were 0.08 for OS and 0.84 for DFS.
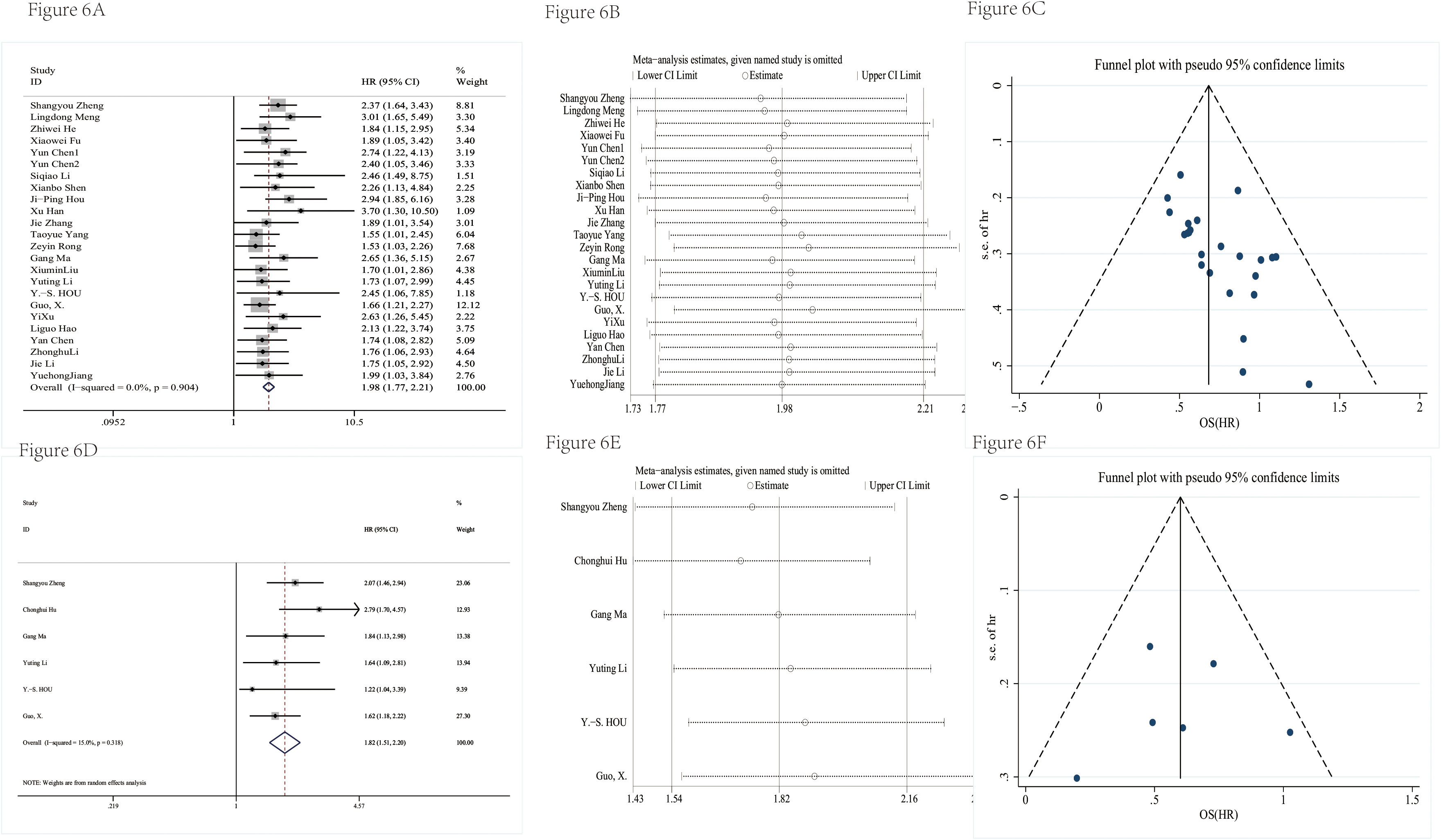
Figure 6 The prognosis value was evaluated. The forest plot had a pooled HR of OS (A). The sensitivity analysis of OS (B). The funnel plot of OS (C). The forest plot had a pooled HR of DFS (D). The sensitivity analysis of DFS (E). The funnel plot of DFS (F).
In this study, a meta-pooling analysis of the 36 included studies showed that circRNA may be a biomarker with great potential for both diagnosis and prognosis of pancreatic cancer, and it was also found that the expression level of circRNA in humans correlated with the degree of tumor differentiation, TNM stage, whether the lymph nodes metastasized, and whether distant metastasis occurred. Using tissues as samples to detect circRNA expression levels may have higher diagnostic power, while using circRNA assays as observational monitoring indicators during the later stages of patient treatment may more positively suggest patient outcomes and long-term prognosis.
The correlation between circRNA expression level in pancreatic cancer and clinicopathological characteristics did not reach a consistent conclusion, so it is necessary to carry out a corresponding analysis. The study found that has_ circ_ 0075829 is highly expressed in pancreatic cancer and is related to tumor size and lymph node metastasis (71). However, the pooled analysis results did not find that tumor size was related to circRNA expression levels. While many studies have shown that the expression of circRNA is related to the occurrence of lymph node metastasis and TNM stage, which is consistent with the conclusion of our study. The high expression of circ BFAR (53) and circ-ASH2L (56) in pancreatic cancer was positively correlated with lymph node metastasis and advanced TNM stage, hsa_ circ_ 0001649 (59) and circ-IARS (58) were related to the degree of tumor differentiation. In the published research, Yang and others found that circ-LDLRAD3 can promote the proliferation, invasion, and migration of tumor cells by acting on the miR-137-3p/PTN axis through functional mechanism research in 2017 (60); in 2018, Li and team also found that circ-PDE8A stimulated the growth of tumor cells by targeting the miR-338/MACC1/MET axis while affecting the liver metastasis of tumor cells (57). The in-depth study of the mechanism also suggested that the high expression of circRNA in tumor tissue would lead to a poor prognosis, which was confirmed by the results of the survival analysis of patients. Zheng found in the 2022 study that the higher the expression level of circCUL2 in tumor tissue, the shorter the disease-free survival and total survival time of PDAC patients (35).
Although the current research results believe that circRNA is a diagnostic tool for pancreatic cancer, there is a greater correlation between the size of the diagnostic efficacy and the samples tested. This study found that the diagnostic ability of tissue samples is significantly higher than that of serum samples, which is consistent with the result that the expression level of circRNA in tissues is higher than that of serum. However, compared with the stability of the biomarkers in the serum, the preoperative tissue samples submitted for examination in clinical practice have greater heterogeneity, which has a great relationship with the technical level of the sending doctors and the tumor location. Previous studies have shown that circRNA is abundant in serum exosomes (72). Therefore, the research on tumor biomarkers in serum is still worth exploring, and the focus is on serum tumor markers with strong diagnostic ability.
This study is the first to comprehensively analyze the correlation between circRNA expression in pancreatic cancer and clinicopathological characteristics, as well as its potential diagnostic and prognostic tool for pancreatic cancer in Chinese population, which is one of the greatest advantages of this study. Secondly, this study conducted a comprehensive analysis of the functions of circRNA in the diagnosis, treatment, and prognosis of pancreatic cancer patients, which may prove that circRNA, as an emerging biomarker, may have greater potential in the future. However, there are also some deficiencies in this study. For example, for diagnostic research, tissue as a sample shows greater diagnostic potential than serum. Although we have tried our best to comprehensively screen high-quality research, the amount of literature that can be included in both is relatively small, which may indicate that the direction of our future research needs to be improved. There was some publication bias in the pooled OS in prognostic studies, which may be because published studies prefer positive results, but there was no doubt about circRNA as a prognostic biomarker for pancreatic cancer, so we may need to include more studies at a later stage to prove the conclusion. In addition, most of the studies included in this study were carried out in Asia, and such studies were carried out in other ethnic groups to observe whether the research results were different from those in Asia, which is worth exploring later.
In conclusion, this study confirmed the important role of circRNA in the diagnosis and prognosis of pancreatic cancer through a systematic and comprehensive search and screening of literature and analysis of data, laying a solid foundation for further basic research. At the same time, we should also realize that researchers’ research on circRNA is still at the basic stage and that there is still a long way to go to realize its clinical value, which is a huge challenge for scientific research.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
RZ and ZH wrote the draft of the manuscript. ZH, HZ, and YX were responsible for extracting data. XBC, XGC, and YX contributed to the conception to the work and the structure of the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1174577/full#supplementary-material
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol (2021) 18(7):493–502. doi: 10.1038/s41575-021-00457-x
3. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol (2018) 24(43):4846–61. doi: 10.3748/wjg.v24.i43.4846
4. Chen F, Childs EJ, Mocci E, Bracci P, Gallinger S, Li D, et al. Analysis of heritability and genetic architecture of pancreatic cancer: a PanC4 study. Cancer epidemiology Biomarkers Prev Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol (2019) 28(7):1238–45. doi: 10.1158/1055-9965.Epi-18-1235
5. Camargo J, Pumarega JA, Alguacil J, Sanz-Gallén P, Gasull M, Delclos GL, et al. Toenail concentrations of trace elements and occupational history in pancreatic cancer. Environ Int (2019) 127:216–25. doi: 10.1016/j.envint.2019.03.037
6. Lynch SM, Vrieling A, Lubin JH, Kraft P, Mendelsohn JB, Hartge P, et al. Cigarette smoking and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Am J Epidemiol (2009) 170(4):403–13. doi: 10.1093/aje/kwp134
7. Lucenteforte E, La Vecchia C, Silverman D, Petersen GM, Bracci PM, Ji BT, et al. Alcohol consumption and pancreatic cancer: a pooled analysis in the international pancreatic cancer case-control consortium (PanC4). Ann Oncol Off J Eur Soc Med Oncol (2012) 23(2):374–82. doi: 10.1093/annonc/mdr120
8. Michaud DS, Giovannucci E, Willett WC, Colditz GA, Stampfer MJ, Fuchs CS. Physical activity, obesity, height, and the risk of pancreatic cancer. Jama (2001) 286(8):921–9. doi: 10.1001/jama.286.8.921
9. Grote VA, Rohrmann S, Nieters A, Dossus L, Tjønneland A, Halkjær J, et al. Diabetes mellitus, glycated haemoglobin and c-peptide levels in relation to pancreatic cancer risk: a study within the European prospective investigation into cancer and nutrition (EPIC) cohort. Diabetologia (2011) 54(12):3037–46. doi: 10.1007/s00125-011-2316-0
10. Duell EJ, Lucenteforte E, Olson SH, Bracci PM, Li D, Risch HA, et al. Pancreatitis and pancreatic cancer risk: a pooled analysis in the international pancreatic cancer case-control consortium (PanC4). Ann Oncol Off J Eur Soc Med Oncol (2012) 23(11):2964–70. doi: 10.1093/annonc/mds140
11. Gandini S, Lowenfels AB, Jaffee EM, Armstrong TD, Maisonneuve P. Allergies and the risk of pancreatic cancer: a meta-analysis with review of epidemiology and biological mechanisms. Cancer epidemiology Biomarkers Prev Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol (2005) 14(8):1908–16. doi: 10.1158/1055-9965.Epi-05-0119
12. Torphy RJ, Fujiwara Y, Schulick RD. Pancreatic cancer treatment: better, but a long way to go. Surg Today (2020) 50(10):1117–25. doi: 10.1007/s00595-020-02028-0
13. Khorana AA, McKernin SE, Berlin J, Hong TS, Maitra A, Moravek C, et al. Potentially curable pancreatic adenocarcinoma: ASCO clinical practice guideline update. J Clin Oncol Off J Am Soc Clin Oncol (2019) 37(23):2082–8. doi: 10.1200/jco.19.00946
14. Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol (2018) 24(19):2047–60. doi: 10.3748/wjg.v24.i19.2047
15. Schiffman JD, Fisher PG, Gibbs P. Early detection of cancer: past, present, and future. Am Soc Clin Oncol Educ book Am Soc Clin Oncol Annu Meeting 2015:57–65. doi: 10.14694/EdBook_AM.2015.35.57
16. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer (2021) 20(1):13. doi: 10.1186/s12943-020-01298-z
17. Marrelli D, Caruso S, Pedrazzani C, Neri A, Fernandes E, Marini M, et al. CA19-9 serum levels in obstructive jaundice: clinical value in benign and malignant conditions. Am J Surg (2009) 198(3):333–9. doi: 10.1016/j.amjsurg.2008.12.031
18. Mujica VR, Barkin JS, Go VL. Acute pancreatitis secondary to pancreatic carcinoma. Study Group Participants. Pancreas (2000) 21(4):329–32. doi: 10.1097/00006676-200011000-00001
19. Safi F, Roscher R, Bittner R, Schenkluhn B, Dopfer HP, Beger HG. High sensitivity and specificity of CA 19-9 for pancreatic carcinoma in comparison to chronic pancreatitis. Serological immunohistochemical findings. Pancreas (1987) 2(4):398–403. doi: 10.1097/00006676-198707000-00006
20. Kaur S, Smith LM, Patel A, Menning M, Watley DC, Malik SS, et al. A combination of MUC5AC and CA19-9 improves the diagnosis of pancreatic cancer: a multicenter study. Am J Gastroenterol (2017) 112(1):172–83. doi: 10.1038/ajg.2016.482
21. Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer (2012) 131(3):683–91. doi: 10.1002/ijc.26422
22. Zhang H-d, Jiang L-h, Sun D-w, Hou J-c, Ji Z-l. CircRNA: a novel type of biomarker for cancer. Breast Cancer (2018) 25(1):1–7. doi: 10.1007/s12282-017-0793-9
23. Rawat M, Kadian K, Gupta Y, Kumar A, Chain PSG, Kovbasnjuk O, et al. MicroRNA in pancreatic cancer: from biology to therapeutic potential. Genes (Basel) (2019) 10(10):752. doi: 10.3390/genes10100752
24. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol (2015) 12(4):381–8. doi: 10.1080/15476286.2015.1020271
25. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol (2014) 32(5):453–61. doi: 10.1038/nbt.2890
26. Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res (2016) 44(3):1370–83. doi: 10.1093/nar/gkv1367
27. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J Off Publ Fed Am Societies Exp Biol (1993) 7(1):155–60. doi: 10.1096/fasebj.7.1.7678559
28. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
29. Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi L, et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol (2021) 14(1):134. doi: 10.1186/s13045-021-01145-8
30. Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer (2021) 20(1):26. doi: 10.1186/s12943-021-01318-6
31. Zhang M, Bai X, Zeng X, Liu J, Liu F, Zhang Z. circRNA-miRNA-mRNA in breast cancer. Clinica chimica acta; Int J Clin Chem (2021) 523:120–30. doi: 10.1016/j.cca.2021.09.013
32. Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J, et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics (2021) 11(6):2755–69. doi: 10.7150/thno.56174
33. Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett (2021) 505:49–57. doi: 10.1016/j.canlet.2021.02.004
34. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol (2022) 19(3):188–206. doi: 10.1038/s41571-021-00585-y
35. Zheng S, Hu C, Lin H, Li G, Xia R, Zhang X, et al. circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-κB signaling pathway. J Exp Clin Cancer Res CR (2022) 41(1):71. doi: 10.1186/s13046-021-02237-6
36. Wu H, Wang B, Wang L, Xue Y. Circular RNAs 0000515 and 0011385 as potential biomarkers for disease monitoring and determining prognosis in pancreatic ductal adenocarcinoma. Oncol Lett (2022) 23(2):56. doi: 10.3892/ol.2021.13174
37. Wong CH, Lou UK, Fung FKC, Tong JHM, Zhang CH, To KF, et al. CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol Cancer (2022) 21(1):10. doi: 10.1186/s12943-021-01481-w
38. Meng L, Zhang Y, Wu P, Li D, Lu Y, Shen P, et al. CircSTX6 promotes pancreatic ductal adenocarcinoma progression by sponging miR-449b-5p and interacting with CUL2. Mol Cancer (2022) 21(1):121. doi: 10.1186/s12943-022-01599-5
39. Hu C, Xia R, Zhang X, Li T, Ye Y, Li G, et al. circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer (2022) 21(1):24. doi: 10.1186/s12943-022-01501-3
40. Hong L, Xu L, Jin L, Xu K, Tang W, Zhu Y, et al. Exosomal circular RNA hsa_circ_0006220, and hsa_circ_0001666 as biomarkers in the diagnosis of pancreatic cancer. J Clin Lab Anal (2022) 36(6):e24447. doi: 10.1002/jcla.24447
41. He Z, Cai K, Zeng Z, Lei S, Cao W, Li X. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis (2022) 13(3):233. doi: 10.1038/s41419-022-04677-0
42. Fu X, Sun G, Tu S, Fang K, Xiong Y, Tu Y, et al. Hsa_circ_0046523 mediates an immunosuppressive tumor microenvironment by regulating MiR-148a-3p/PD-L1 axis in pancreatic cancer. Front Oncol (2022) 12:877376. doi: 10.3389/fonc.2022.877376
43. Zhang J, Zhang Z. Mechanisms of circular RNA circ_0066147 on pancreatic cancer progression. Open Life Sci (2021) 16(1):495–510. doi: 10.1515/biol-2021-0047
44. Yang T, Shen P, Chen Q, Wu P, Yuan H, Ge W, et al. FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res CR (2021) 40(1):261. doi: 10.1186/s13046-021-02063-w
45. Xu K, Qiu Z, Xu L, Qiu X, Hong L, Wang J. Increased levels of circulating circular RNA (hsa_circ_0013587) may serve as a novel biomarker for pancreatic cancer. biomark Med (2021) 15(12):977–85. doi: 10.2217/bmm-2020-0750
46. Shen X, Chen Y, Li J, Huang H, Liu C, Zhou N. Identification of Circ_001569 as a potential biomarker in the diagnosis and prognosis of pancreatic cancer. Technol Cancer Res Treat (2021) 20:1533033820983302. doi: 10.1177/1533033820983302
47. Rong Z, Shi S, Tan Z, Xu J, Meng Q, Hua J, et al. Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol Cancer (2021) 20(1):106. doi: 10.1186/s12943-021-01400-z
48. Ma G, Li G, Fan W, Xu Y, Song S, Guo K, et al. Circ-0005105 activates COL11A1 by targeting miR-20a-3p to promote pancreatic ductal adenocarcinoma progression. Cell Death Dis (2021) 12(7):656. doi: 10.1038/s41419-021-03938-8
49. Liu X, Zhong L, Jiang W, Wen D. Repression of circRNA_000684 inhibits malignant phenotypes of pancreatic ductal adenocarcinoma cells via miR-145-mediated KLF5. Pancreatology (2021) 21(2):406–17. doi: 10.1016/j.pan.2020.12.023
50. Hou JP, Men XB, Yang LY, Han EK, Han CQ, Liu LB. CircCCT3 acts as a sponge of miR-613 to promote tumor growth of pancreatic cancer through regulating VEGFA/VEGFR2 signaling. Balkan Med J (2021) 38(4):229–38. doi: 10.5152/balkanmedj.2021.21145
51. Han X, Fang Y, Chen P, Xu Y, Zhou W, Rong Y, et al. Upregulated circRNA hsa_circ_0071036 promotes tumourigenesis of pancreatic cancer by sponging miR-489 and predicts unfavorable characteristics and prognosis. Cell Cycle (Georgetown Tex) (2021) 20(4):369–82. doi: 10.1080/15384101.2021.1874684
52. Hou YS, Li X. Circ_0005273 induces the aggravation of pancreatic cancer by targeting KLF12. Eur Rev Med Pharmacol Sci (2020) 24(22):11578–86. doi: 10.26355/eurrev_202011_23799
53. Guo X, Zhou Q, Su D, Luo Y, Fu Z, Huang L, et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol Cancer (2020) 19(1):83. doi: 10.1186/s12943-020-01196-4
54. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun (2019) 509(1):138–42. doi: 10.1016/j.bbrc.2018.12.088
55. Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y, et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem (2019) 120(3):3780–9. doi: 10.1002/jcb.27658
56. Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang Y, et al. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res CR (2019) 38(1):466. doi: 10.1186/s13046-019-1436-0
57. Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett (2018) 432:237–50. doi: 10.1016/j.canlet.2018.04.035
58. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res CR (2018) 37(1):177. doi: 10.1186/s13046-018-0822-3
59. Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene (2018) 675:88–93. doi: 10.1016/j.gene.2018.06.099
60. Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol (2017) 23(47):8345–54. doi: 10.3748/wjg.v23.i47.8345
61. Yan HX LH, Liu SP, Bai MH, Zheng YW. Expression of circular RNA chr14∶101402109-101464448c in pancreatic cancer and its effect on proliferation, invasion and metastasis of pancreatic cancer cells. Chin J Exp Surg (2021) 38(10):1947–51. doi: 10.3760/cma.j.cn421213-20201220-01460
62. Chen Y LJ, Yang Z, Zhou N. Peripheral blood circular RNA circ_0141633 and circ_0008234 as potential diagnostic and prognostic biomarkers in pancreatic cancer. J Chin Physician (2022) 24(04):566–72. doi: 10.3760/cma.j.cn431274-20220221-00119
63. Li SQ CQ, Liu HC, Liu SP. Expression and clinical significance of circ-MFN2 in pancreatic cancer. Chin J Bases Clinics Gen Surg (2021) 28(06):762–7.
64. Li YT KY, Luo YM, Zheng HH, Zhu J, Gao BW. Et.al. circular RNA-PCAC1 promotes the metastasis of pancreatic ductal adenocarcinoma. Chin Arch Gen Surg(Electronic Edition) (2020) 14(03):174–9.
65. Shen P, Yang T, Chen Q, Yuan H, Wu P, Cai B, et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and a-to-I RNA-editing. Mol Cancer (2021) 20(1):51. doi: 10.1186/s12943-021-01333-7
66. Li S, Song J, He Y, Huang Y. Expression of circ-RANBP1 in pancreatic cancer tissue and its effect on cell proliferation, migration and invasion. Chin J Gastroenterol (2021) 26(2):76–81. doi: 10.3969/j.issn.1008-7125.2021.02.003
67. Guo W, Zhao L, Wei G, Liu P, Zhang Y, Fu L. Blocking circ_0013912 suppressed cell growth, migration and invasion of pancreatic ductal adenocarcinoma cells in vitro and in vivo partially through sponging miR-7-5p. Cancer Manage Res (2020) 12:7291–303. doi: 10.2147/cmar.S255808
68. An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D, et al. CircZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem (2018) 51(5):2224–36. doi: 10.1159/000495868
69. Wong CH, Lou UK, Tong JHM, To KF, Chen Y. CircRTN4 promotes tumor growth and metastasis of pancreatic ductal adenocarcinoma through circRNA/miRNA/lncRNA axis and stabilizing RAB11FIP1 protein. Pancreas (2020) 49(10):1437. doi: 10.1186/s12943-021-01481-w
70. Xie H, Zhao Q, Yu L, Lu J, Peng K, Xie N, et al. Circular RNA circ_0047744 suppresses the metastasis of pancreatic ductal adenocarcinoma by regulating the miR-21/SOCS5 axis. Biochem Biophys Res Commun (2022) 605:154–61. doi: 10.1016/j.bbrc.2022.03.082
71. Zhang X, Xue C, Cui X, Zhou Z, Fu Y, Yin X, et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR-1287-5p and activating LAMTOR3 signalling. J Cell Mol Med (2020) 24(24):14596–607. doi: 10.1111/jcmm.16089
Keywords: pancreatic cancer, diagnostic, prognostic, circular RNAs, meta-analysis
Citation: Zhao R, Han Z, Zhou H, Xue Y, Chen X and Cao X (2023) Diagnostic and prognostic role of circRNAs in pancreatic cancer: a meta-analysis. Front. Oncol. 13:1174577. doi: 10.3389/fonc.2023.1174577
Received: 03 March 2023; Accepted: 22 May 2023;
Published: 08 June 2023.
Edited by:
John Gibbs, Hackensack Meridian Health, United StatesReviewed by:
Qin Wenxing, Shanghai Changzheng Hospital, ChinaCopyright © 2023 Zhao, Han, Zhou, Xue, Chen and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobing Chen, emx5eWNoZW54YjA4MDdAenp1LmVkdS5jbg==; Xinguang Cao, eGluZ3VhbmdjYW9AMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.