
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Oncol. , 12 May 2023
Sec. Hematologic Malignancies
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1167143
 Peifang Xiao1†
Peifang Xiao1† Nan Chen2†
Nan Chen2† Tingting Shao2
Tingting Shao2 Xinni Bian1
Xinni Bian1 Jie Miao1
Jie Miao1 Jiajia Zheng1
Jiajia Zheng1 Xingping Lang2
Xingping Lang2 Yiting Wang2
Yiting Wang2 Xiaojun Chen2
Xiaojun Chen2 Liqin Jin2
Liqin Jin2 Shaoyan Hu1*
Shaoyan Hu1* Sheng Xiao3*
Sheng Xiao3*The synuclein family, consisting of α-, β-, and γ-synuclein, is primarily expressed in neurons. Mutations of α- and β-synuclein have been linked to Parkinson’s disease and dementia with Lewy bodies, respectively. Recent studies have shown that synucleins are upregulated in various tumors, including breast, ovarian, meningioma, and melanoma, and high synuclein expression is associated with poor prognosis and drug resistance. We report a novel rearrangement of β-synuclein in a pediatric T-cell acute lymphoblastic leukemia (T-ALL) case, where β-synuclein (SNCB) is fused in-frame with ETS variant transcription factor 6 (ETV6), a gene frequently rearranged in acute leukemia including acute myeloid leukemia (AML), B-cell acute lymphoblastic leukemia (B-ALL), and T-ALL. An additional case of β-synuclein rearrangement was identified in a squamous cell carcinoma of the lung through analysis of the public TCGA database. Both rearrangements involve the C-terminal of β-synuclein. Since β-synuclein shares extensive amino acid similarities with α-synuclein and α-synuclein binds to 14-3-3, an important regulator of apoptosis, the rearranged β-synuclein may contribute to tumorigenesis by deregulating apoptosis. In addition, overexpression of synucleins has been shown to increase cell proliferation, suggesting that the rearranged β-synuclein may also deregulate the cell cycle.
Synuclein is a group of three small acidic proteins, α-synuclein (140 amino acids), β-synuclein (134 amino acids), and γ-synuclein (127 amino acids). These proteins contain an N-terminal lysine-rich amphipathic α-helix domain, a central hydrophobic non-amyloid β component (NAC) domain, and a C-terminal structureless acidic region. The N-terminal domain is positively charged and capable of lipid binding to cell membranes, while the NAC domain, which is an antiparallel β-sheet structure, is involved in fibril formation and aggregation. The structureless acidic region interacts with the N-terminal domain to prevent synuclein aggregation. The C-terminal region is also responsible for protein interaction. α-synuclein and β-synuclein were first purified from the human brain and are both enriched in presynaptic terminals of neurons. γ-synuclein was first identified as the breast cancer-specific gene 1 due to its overexpression in breast tumors and is expressed in the nervous system, the endocrine tissues, and the kidneys and urinary tract.
The synucleins are mainly known for their role in synucleinopathies, a group of neurodegenerative disorders. Mutations of SNCA, encoding α-synuclein, or increased copies of the wild-type SNCA lead to early-onset autosomal dominant Parkinson’s disease, characterized by the loss of dopaminergic neurons in the substantia nigra. Two mutations of SNCB, encoding β-synuclein, the P123H and the V70M, are associated with dementia with Lewy bodies, a disorder closely related to Parkinson’s disease. Lewy bodies are protein aggregates in neurons and are pathological hallmarks in synucleinopathies, with α-synuclein being a major component.
The role of synucleins in cancer is supported by their overexpression and links to both disease progression and therapeutic response. α-synuclein is overexpressed in various types of cancer, including meningioma, pancreatic cancer, glioblastoma, ovarian cancer, colorectal cancer, and melanoma (1, 2). The expression of α-synuclein is associated with disease progression, as it is only expressed in meningiomas of grades 2 and 3, but not grade 1 (3). In pancreatic cancer, α-synuclein expression is correlated with neurotropism, a hallmark of tumor progression (4). There is limited evidence of β-synuclein’s involvement in cancer, but it is expressed in certain types of brain tumors, such as ependymoma, astrocytoma, oligodendroglioma, and medulloblastoma (5). γ-synuclein is overexpressed in many tumors, including breast, ovary, colon, liver, and cervical cancer, and its expression is higher in more advanced cancers (6, 7). In vitro assays and animal models suggest that γ-synuclein plays a crucial role in promoting tumor invasion and metastasis. Moreover, recent studies indicate that tumors with γ-synuclein overexpression are more resistant to chemotherapy and radiotherapy (8, 9).
We present a novel discovery of a β-synuclein rearrangement in a pediatric T-ALL tumor, where SNCB is fused to the well-known leukemic gene ETV6. We also found another case of SNCB rearrangement in a case of squamous lung cancer by searching the TCGA database. These are the first reported cases of tumors with SNCB rearrangements.
Immunohistochemistry (IHC) was performed on 5-μm tissue sections using the following protocol: (1) the slides were baked at 60 °C for 1 hour, (2) deparaffinized and rehydrated with 100% xylene, 100% ethanol, 70% ethanol, and running water, (3) blocked in a solution of 10% normal serum and 1% bovine serum albumin (BSA) in Tris-buffered saline, (4) incubated with primary antibodies for 2 hours, (5) endogenous peroxidase was blocked with 0.3% hydrogen peroxide, and (6) slides were incubated with horseradish peroxidase-labeled polymer (DAKO) according to the manufacturer’s instructions. The tissue sections were developed using 3,3’-diaminobenzidine (DAKO) as the chromogen and counterstained with Mayer’s hematoxylin.
The bone marrow specimens were processed as follows: (1) cells were collected in sodium heparin and counted using a TC-20 Automated Cell Counter (Bio-Rad, Hercules, CA), (2) 5 × 106 cells were cultured overnight and treated with ethidium bromide (5 µg/mL) for 1.5 hours and Colcemid (0.5 µg/mL) for 20 minutes, (3) cells were incubated in a hypotonic solution, fixed, and metaphase spreading slides were manually prepared, (4) slides were baked at 70°C for 1 hour, (5) chromosomes were Giemsa-Trypsin banded and analyzed. For fluorescence in situ hybridization (FISH), a two-colored split apart probe for ETV6 was added to metaphase slides, co-denatured, and hybridized at 37°C overnight. Post-hybridization wash was performed in 0.4 × SSC/0.3% NP-40 at 73°C for 3 minutes, and slides were counterstained with DAPI.
Total RNA was isolated from the bone marrow aspirate using the TRIZOL reagent (ThermoFisher, Waltham, MA) according to the manufacturer’s instructions. The reverse transcription, end repairing, dA-tailing, and adaptor ligation were performed following standard NGS protocols (NEB, Cat E7771 and E6111, Ipswich, MA, USA). A group of 75 genes commonly involved in hematological malignancies was targeted using PCR enrichment with primers specific to these genes. The PCR products were sequenced using an Illumina NovaSeq sequencer (San Diego, CA, USA). The sequencing results were analyzed using SeqNext software (JSI, Ettenheim, Germany) and laboratory-developed pipelines (Sano Medical Laboratories, China).
cDNA synthesis from total RNA was performed using random priming and the SuperScript™ IV reverse transcriptase (ThermoFisher). PCR amplification was then carried out using specific primers for ETV6::SNCB (forward primer ETV6: 5’-TCTTAAATGACCGCGTCTGGC and reverse primer SNCB: 3’-GACAGAATTGTGCTGCTGGTG, nested PCR forward primer ETV6: 5’-TTGGGGAGAGGAAAGGAAAGTG and reverse primer SNCB: 3’-CATACTCCTGATATTCCTCCTGGG) and ETV6::SNCB::GPRIN1 (forward primer ETV6: 5’-TTCTTAAATGACCGCGTCTGGC and reverse primer GPRIN1: 3’-ATCCTGTGGGCAGAAGAAGG, nested PCR forward primer ETV6: 5’-TGGGGAGAGGAAAGGAAAGTG and reverse primer GPRIN1: 3’-GGGCTGGAATCCTTTTGAAGC). The PCR protocol consisted of one cycle at 94°C for 2 minutes followed by 30 cycles at 94°C for 30 seconds, 68°C for 30 seconds, and 72°C for 30 seconds, one cycle at 72°C for 1 minute, and final holding at 4°C. The 1st PCR product (0.5 µl) was used as template DNA for the nested PCR. The PCR products were then subjected to Sanger sequencing.
A 5-year-old girl presented with a neck mass for 2 months. Physical examination revealed lymphadenopathy on both sides of her neck. Complete blood counts showed WBC 1.39 x 109/L, Hb 91 g/L, PLT 111 x 109/L, 31.7% neutrophils, 65% lymphocytes, 0% monocytes, 0% eosinophils, and 0% basophils. A neck lymph node biopsy showed an effaced node with diffuse infiltration of blasts positive for CD3 (dim), CD7, TdT, CD99, PAX5, Ki-67 (40%) and negative for MPO, CD20, CD21 (Figure 1A). The bone marrow smear revealed that 65% of the cells were blasts of various sizes with small amounts of blue-grey cytoplasm, dispersed nuclear chromatin, and inconspicuous nucleoli. Cytochemistry was positive for periodic acid-Schiff (PAS) and negative for myeloperoxidase. The bone marrow flow cytometry showed 58% of CD45+ cells with cCD3+, CD5+, CD7+, CD33dim, CD34dim, CD99+, CD1a-, CD2-, sCD3-, CD4-, CD8-, CD13-, CD19-, cCD79a-, MPO-, TdT-. Based on these results, a diagnosis of T-ALL was made. The karyotype analysis of her bone marrow showed a complex karyotype with 45,XX, t(5;12)(q35;p13), t(7;9)(q34;q22), add(12)(p13), der(13) del(13)(q14q32)t(12;13) (p13;q34), -21[16]/46, XX[4] (Figure 1B). Notably, both copies of chromosome 12p13 (ETV6 locus) were rearranged. The FISH assays revealed that one locus of the ETV6 was rearranged and the other was partially deleted at the 3’ end of ETV6. The rearranged 5’ ETV6 was found at the 5q terminal, which was consistent with the translocation between chromosomes 5 and 12 (Figure 1C). To identify the ETV6 fusion partner gene, a targeted RNA-NGS assay was conducted. This showed the presence of two in-frame fusion transcripts: the first was made up of the first exon of ETV6 and the last two exons (exon 5 and 6) of SNCB, while the second contained sequences from three genes, including the first exon of ETV6, exon 5 of SNCB, and the last exon (exon 2) of GPRIN1. As the GPRIN1 is located immediately 3’ to SNCB, the three-gene fusion transcript is believed to be a read-through product of the ETV6::SNCB rearrangement. The presence of both fusion transcripts was confirmed by RT-PCR with gene-specific primers and subsequent Sanger sequencing (Figures 2A–E). We believe this is the first fusion transcript involving 3 genes observed in the tumor.
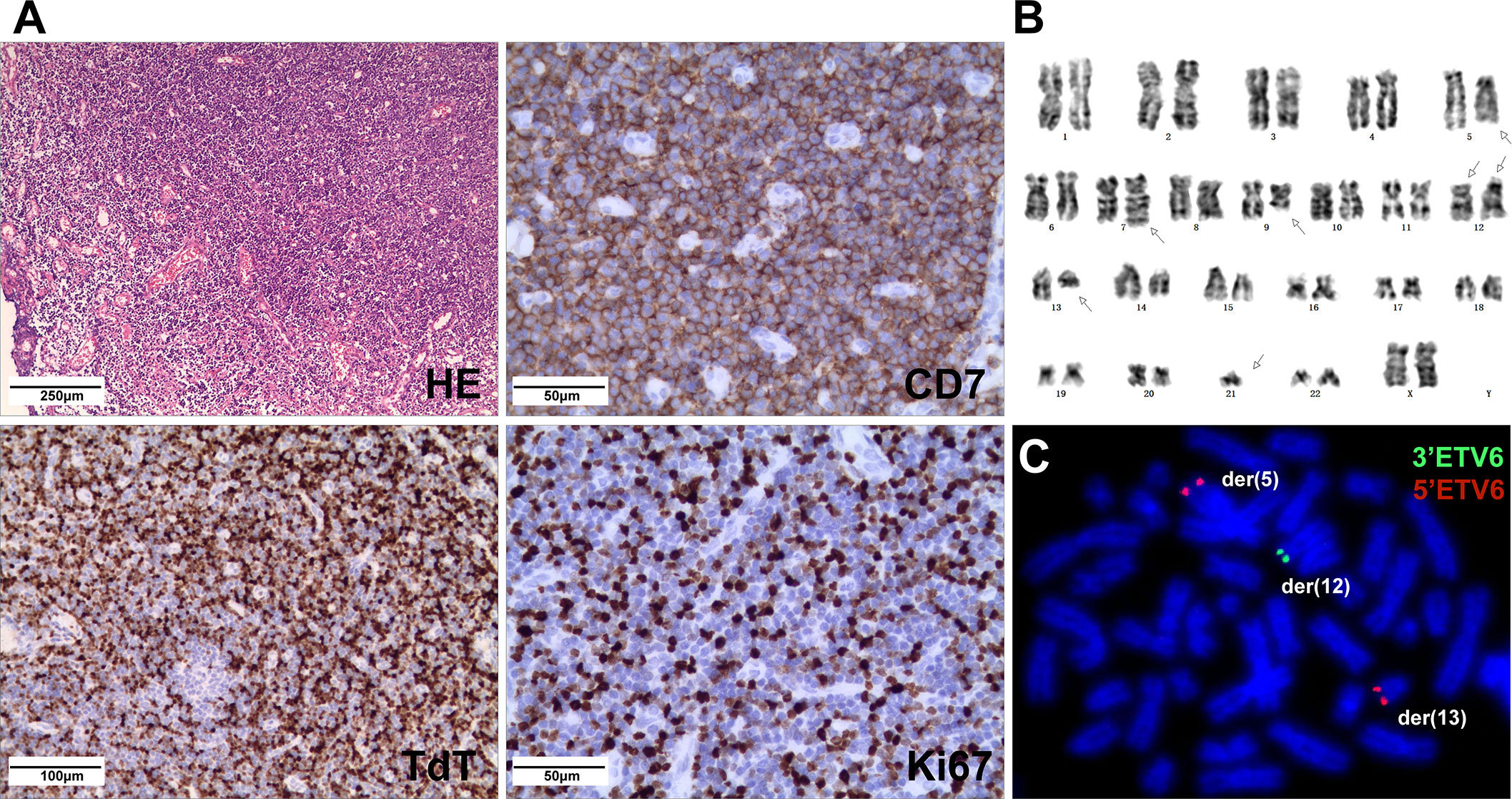
Figure 1 (A) Neck lymph node biopsy showed complete effacement of architecture with small lymphoblasts (HE stain) and were positive for CD7, TdT, and Ki67 (40%) by IHC. (B) The karyotype of the cultured bone marrow showed complex chromosome changes with both copies of chromosome 12p13 rearranged. (C) FISH analysis on abnormal metaphases with the ETV6 break-apart probe revealed an ETV6 rearrangement by the t(5;12), while the other ETV6 allele had a deletion of the 3’ETV6, with the 5’ETV6 translocated to der(13).
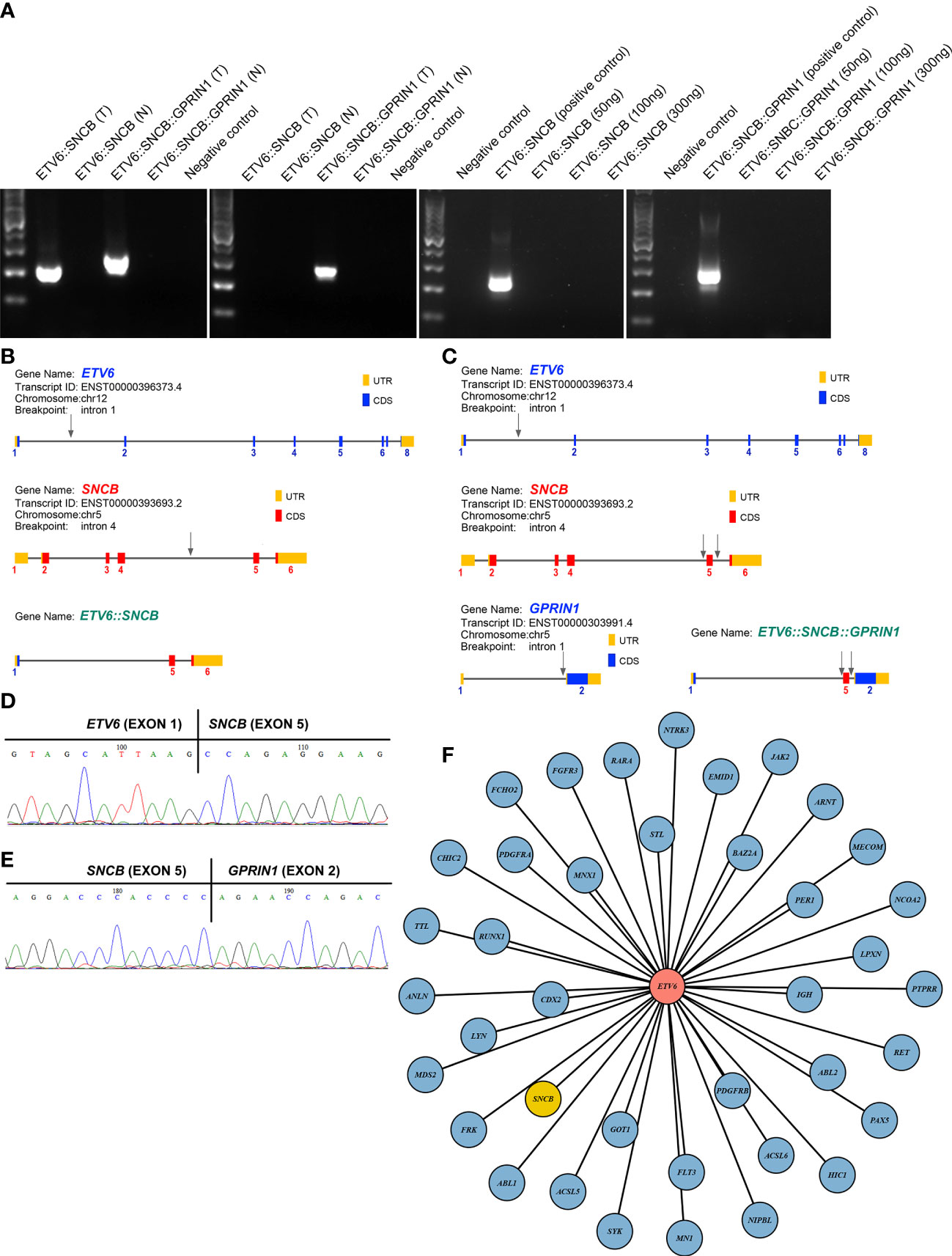
Figure 2 (A) RT-PCR amplified 2 fusion transcripts, ETV6::SNCB and ETV6::SNCB::GPRIN1, from the diagnostic bone marrow specimen, consistent with the RNA NGS finding (left panel). At day 19, only ETV6::SNCB::GPRIN1 was observed, consistent with MRD (next to the left panel). At day 49, both fusion transcripts were absent from the patient’s bone marrow (right 2 panels). T: tumor bone marrow; N: normal bone marrow; Negative control: reactions without cDNA template; Positive control: tumor materials from the initial diagnostic specimen; (B) Exons and breakpoints (arrows) of the ETV6::SNCB; (C) Exons and breakpoints (arrows) of the ETV6::SNCB::GPRIN1; (D) Sanger sequencing of the RT-PCR for ETV6::SNCB; (E) Sanger sequencing of the ETV6::SNCB::GPRIN1; (F) Total 39 fusion partners of the ETV6 in cancer.
The patient was enrolled in a clinical trial conducted by the Chinese Children’s Cancer Group (CCCG) and was treated with the CCCG-2020 protocol. This protocol involved a four-day pre-treatment phase with dexamethasone, followed by induction therapy combining dexamethasone, vincristine, daunorubicin, peg-asparaginase, cyclophosphamide, cytarabine, and 6-mercaptopurine. The patient also underwent additional early intensification therapy consisting of cyclophosphamide, cytarabine, 6-mercaptopurine, vincristine, and peg-asparaginase, followed by consolidation therapy with high-dose methotrexate, triple intrathecal therapy every other week, and daily 6-mercaptopurine for four courses. After one year, the patient remains in remission.
The therapeutic response was monitored using both ETV6::SNCB and ETV6::SNCB::GPRIN1. On day 19 of the treatment, the ETV6::SNCB disappeared but the ETV6::SNCB::GPRIN1 remained positive in the bone marrow, which is consistent with minimal residual disease (MRD). However, on day 46 of the treatment, both ETV6::SNCB and ETV6::SNCB::GPRIN1 had disappeared (as shown in Figure 2A). These results, obtained through RT-PCR, were consistent with the patient’s clinical remission, negative histology findings, and negative flow cytometry.
In a search of 10,565 cancer genomes from the TCGA database, a lung squamous cell carcinoma with the presence of LDLRAD3::SNCB fusion (TCGA-85-A50M-01A) was identified, among a total of 423 cases of lung squamous cell carcinoma. The patient was a 46-year-old male with a 15-year smoking history. The rearrangement involves the first four exons (exons 1-4) of LDLRAD3, which encodes low-density lipoprotein receptor class A domain containing 3, and the last four exons of SNCB (exons 3-7). The reading frame is intact, which encodes the domains 1-3 (D1-3) of the LDLRAD3 and the partial N-terminal, the NAC domain, and the C-terminal of the β-synuclein (Figure 3). The role of LDLRAD3 in cancer has not been extensively studied, however, LDLRAD3 is overexpressed in several types of cancers including breast cancer, glioma, prostate adenocarcinoma, skin cutaneous melanoma, testicular germ cell tumor, and pancreatic cancer (TCGA, data not shown). Similarly, circular RNA circ-LDLRAD3 is upregulated in pancreatic cancer, lung adenocarcinoma, gastric cancer and is significantly associated with venous invasion, lymphatic invasion, and metastasis in pancreatic cancer (10).
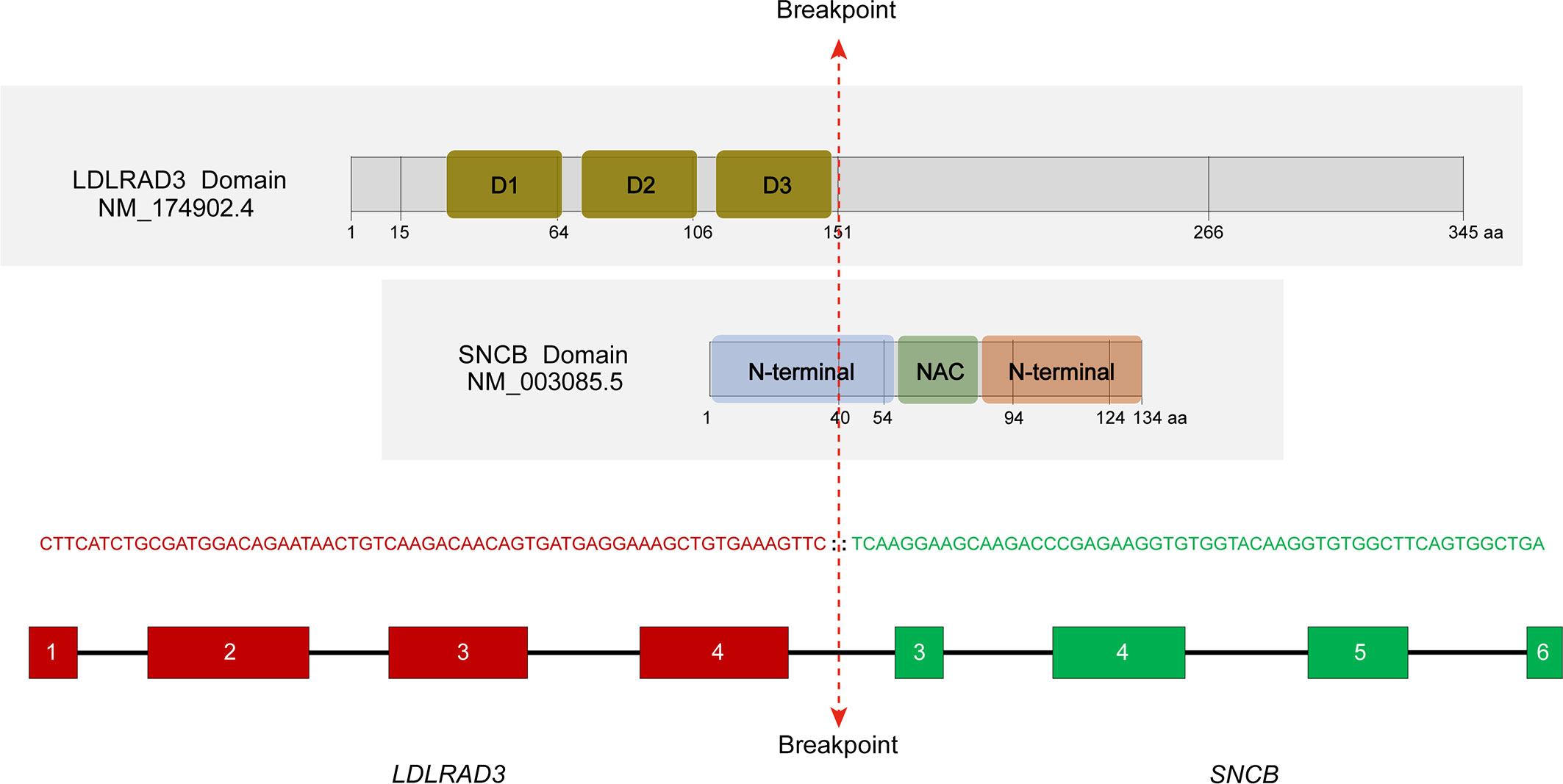
Figure 3 Characterization of the LDLRAD3::SNCB rearrangement. The location of breakpoints of corresponding proteins was marked. D1-3, domains 1-3; NAC, non-amyloid β component domain. RNA sequencing showed an in-frame fusion between LDLRAD3 exon 4 and SNCB exon 3.
ETV6 is a promiscuity gene that plays a role in the development of various tumors, such as leukemia, lymphoma, secretory carcinoma of the breast, infantile fibrosarcoma, congenital mesoblastic nephroma, and thyroid cancer. Our literature review uncovered 38 fusion partners of ETV6, with the SNCB being the 39th fusion partner as shown in Figure 2F. ETV6 rearrangements can lead to oncogenesis through various mechanisms (11). One of the most common rearrangements, ETV6::RUNX1, is found in around 25% of pediatric B-ALL and leads to the dysregulation of genome-wide gene expression. The wild-type ETV6 protein has the ability to form dimers with ETV6::RUNX1, which reduces the transforming activity of ETV6::RUNX1 and mitigates the malignant process. However, in many tumors, the wild-type ETV6 is lost, which exacerbates the malignant process. In our case with the ETV6::SNCB rearrangement, we observed a similar loss of wild-type ETV6, which likely promotes cancer progression through a similar mechanism.
14-3-3s are a group of small proteins that play a crucial role in programmed cell death. α-synuclein binds to 14-3-3s, and both are present in Lewy bodies in patients with Parkinson’s disease. The interaction between α-synuclein and 14-3-3s can have both positive and negative effects on the fate of neurons. 14-3-3s serve as chaperones to reduce α-synuclein aggregation, providing neuroprotective activity, but α-synuclein can also sequester 14-3-3s and release the apoptotic BAD and BAX, leading to apoptosis. The potential oncogenic mechanism of the ETV6::SNCB or LDLRAD3::SNCB may involve the overexpression of the C-terminal synuclein driven by its fusion with either ETV6 or LDLRAD3, both of which are highly expressed genes. This overexpression may bind and sequester 14-3-3s, compromising their anti-apoptotic signaling. Other mechanisms, including p53-dependent and Akt-related apoptotic pathways, may also be involved (12, 13).
The involvement of synucleins in cell proliferation is primarily seen in γ-synuclein. Overexpression of γ-synuclein leads to increased cell proliferation in H175 (squamous cell carcinoma) by binding and activating the AKT. γ-synuclein is a nicotine-responsive protein and is essential for nicotine-induced cancer cell proliferation (14). Diabetes is also a risk factor for lung cancer, and glucose-induced lung cancer cell proliferation is mediated by γ-synuclein (15). In ovarian cancer cell lines A2780 and OVCAR5, forced expression of γ-synuclein increased cell proliferation by activating the MAPK signaling pathway and attenuated the chemotherapeutic drug-induced apoptosis by blocking the c-Jun N-terminal kinase (JNK) signaling (8). The ETV6::SNCB only retains the C-terminal acidic region of β-synuclein, while the LDLRAD3::SNCB contains the partial N-terminal α-helix domain, NAC domain, and C-terminal acidic region of β-synuclein. Further investigation is needed to determine if these fusion proteins have different effects on cell proliferation and/or apoptosis.
In conclusion, we have reported the first cases of synuclein rearrangement in tumors. Further study of these chimeric proteins may provide insights into not only tumorigenesis but also the physiological function of β-synuclein, which is crucial to understand synucleinopathies.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Project design: PX, SH and SX. Clinical studies: PX, SH, XB, JM and JZ. Laboratory work: NC, TS, XL, YW, XC and LJ. Result interpretation: PX, SH, XL, NC and SX. Draft manuscript: NC and SX. All authors contributed to the article and approved the submitted version.
This study was supported by funds #2022YFC2502700 and #ZD2022056
Authors NC, TS, XL, YW, XC and LJ were employed by Suzhou Sano Precision Medicine Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CC declared a shared parent affiliation with the author SX to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rodríguez-Losada N, de la Rosa J, Larriva M, Wendelbo R, Aguirre JA, Castresana JS, et al. Overexpression of alpha-synuclein promotes both cell proliferation and cell toxicity in human sh-Sy5y neuroblastoma cells. J Adv Res (2020) 23:37–45. doi: 10.1016/j.jare.2020.01.009
2. Ryskalin L, Biagioni F, Morucci G, Busceti CL, Frati A, Puglisi-Allegra S, et al. Spreading of alpha synuclein from glioblastoma cells towards astrocytes correlates with stem-like properties. Cancers (Basel) (2022) 14(6):1417. doi: 10.3390/cancers14061417
3. Ge Y, Xu K. Alpha-synuclein contributes to malignant progression of human meningioma Via the Akt/Mtor pathway. Cancer Cell Int (2016) 16:86. doi: 10.1186/s12935-016-0361-y
4. Bianchini M, Giambelluca M, Scavuzzo MC, Di Franco G, Guadagni S, Palmeri M, et al. In pancreatic adenocarcinoma alpha-synuclein increases and marks peri-neural infiltration. Int J Mol Sci (2022) 23(7):3775. doi: 10.3390/ijms23073775
5. Fung KM, Rorke LB, Giasson B, Lee VM, Trojanowski JQ. Expression of alpha-, beta-, and gamma-synuclein in glial tumors and medulloblastomas. Acta Neuropathol (2003) 106(2):167–75. doi: 10.1007/s00401-003-0718-x
6. Yanagawa N, Tamura G, Honda T, Endoh M, Nishizuka S, Motoyama T. Demethylation of the synuclein gamma gene cpg island in primary gastric cancers and gastric cancer cell lines. Clin Cancer Res (2004) 10(7):2447–51. doi: 10.1158/1078-0432.ccr-03-0107
7. Tastekin D, Kargin S, Karabulut M, Yaldız N, Tambas M, Gurdal N, et al. Synuclein-gamma predicts poor clinical outcome in esophageal cancer patients. Tumour Biol (2014) 35(12):11871–7. doi: 10.1007/s13277-014-2429-4
8. Pan Z-Z, Bruening W, Giasson BI, Lee VMY, Godwin AK. Γ-synuclein promotes cancer cell survival and inhibits stress- and chemotherapy drug-induced apoptosis by modulating mapk pathways*. J Biol Chem (2002) 277(38):35050–60. doi: 10.1074/jbc.M201650200
9. Tian L, Zhao Y, Truong MJ, Lagadec C, Bourette RP. Synuclein gamma expression enhances radiation resistance of breast cancer cells. Oncotarget (2018) 9(44):27435–47. doi: 10.18632/oncotarget.25415
10. Yang F, Liu D-Y, Guo J-T, Ge N, Zhu P, Liu X, et al. Circular rna circ-Ldlrad3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol (2017) 23(47):8345. doi: 10.3748/wjg.v23.i47.8345
11. De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. Etv6 fusion genes in hematological malignancies: a review. Leuk Res (2012) 36(8):945–61. doi: 10.1016/j.leukres.2012.04.010
12. Brockhaus K, Böhm MRR, Melkonyan H, Thanos S. Age-related beta-synuclein alters the P53/Mdm2 pathway and induces the apoptosis of brain microvascular endothelial cells in vitro. Cell Transplant (2018) 27(5):796–813. doi: 10.1177/0963689718755706
13. Hashimoto M, Bar-On P, Ho G, Takenouchi T, Rockenstein E, Crews L, et al. Beta-synuclein regulates akt activity in neuronal cells. a possible mechanism for neuroprotection in parkinson’s disease. J Biol Chem (2004) 279(22):23622–9. doi: 10.1074/jbc.M313784200
14. Hsu CC, Su YF, Tsai KY, Kuo FC, Chiang CF, Chien CY, et al. Gamma synuclein is a novel nicotine responsive protein in oral cancer malignancy. Cancer Cell Int (2020) 20:300. doi: 10.1186/s12935-020-01401-w
Keywords: SNCB, ETV6, GPRIN1, T-ALL, synuclein
Citation: Xiao P, Chen N, Shao T, Bian X, Miao J, Zheng J, Lang X, Wang Y, Chen X, Jin L, Hu S and Xiao S (2023) Intragenic β-synuclein rearrangements in malignancy. Front. Oncol. 13:1167143. doi: 10.3389/fonc.2023.1167143
Received: 16 February 2023; Accepted: 27 April 2023;
Published: 12 May 2023.
Edited by:
Guru Prasad Maiti, Oklahoma Medical Research Foundation, United StatesReviewed by:
Sabira Mohammed Jazir, University of Oklahoma Health Sciences Center, United StatesCopyright © 2023 Xiao, Chen, Shao, Bian, Miao, Zheng, Lang, Wang, Chen, Jin, Hu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoyan Hu, aHN5MTM5QDEyNi5jb20=; Sheng Xiao, c3hpYW9AcGFydG5lcnMub3Jn
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.