
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol. , 20 June 2023
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1166835
This article is part of the Research Topic Identifying Clinically Relevant Transcriptional Signatures and Methylation Profiles in the Course, Treatment and Outcome of Colorectal Cancer View all 11 articles
 Eva Turyova1*
Eva Turyova1* Peter Mikolajcik2
Peter Mikolajcik2 Marian Grendar3
Marian Grendar3 Eva Kudelova2
Eva Kudelova2 Veronika Holubekova3
Veronika Holubekova3 Michal Kalman4
Michal Kalman4 Juraj Marcinek4
Juraj Marcinek4 Matej Hrnciar5
Matej Hrnciar5 Michal Kovac5
Michal Kovac5 Juraj Miklusica2
Juraj Miklusica2 Ludovit Laca2
Ludovit Laca2 Zora Lasabova1*
Zora Lasabova1*Introduction: Colorectal cancer (CRC) is one of the most common types of cancer worldwide. The carcinogenesis of CRC is indeed complex, and there are many different mechanisms and pathways that contribute to the development of malignancy and the progression from primary to metastatic tumors. The OCT4A, encoded by the POU5F1 gene, is a transcription factor responsible for the phenotype of stem cells, maintaining pluripotency and regulation of differentiation. The POU5F1 gene is made up of five exons that can create numerous isoforms through alternative promoter or alternative splicing. In addition to OCT4A, other isoforms called OCT4B are also translated into protein; however, their role in cells has been unclear. The aim of our work was to investigate the expression patterns of OCT4 isoforms in primary and metastatic CRC, providing us with useful information about their role in the development and progression of CRC.
Methods: Surgical specimens from a total of 78 patients were collected and isolated from primary tumors (n = 47) and metastases (n = 31). The relative gene expression of OCT4 isoforms was investigated using the RT-qPCR method together with the TaqMan probes for particular OCT4 isoforms.
Results: Our results suggest significantly downregulated expression of the OCT4A and OCT4Bs isoforms in both primary (p = 0.0002 and p < 0.0001, respectively) and metastatic tumors (p = 0.0006 and p = 0.00051, respectively) when compared with the control samples. We also observed a correlation between reduced expression of all OCT4 isoforms and both primary and left-sided tumors (p = 0.001 and p = 0.030, respectively). On the other hand, the expression of all OCT4 isoforms was significantly upregulated in metastases compared with primary tumors (p < 0.0001).
Discussion: Unlike previous reports, we found out that the expression of OCT4A, OCT4Bs, and all OCT4 isoforms was significantly reduced in primary tumors and metastases compared with control samples. On the other hand, we supposed that the expression rate of all OCT4 isoforms may be related to the cancer type and side, as well as to liver metastases. However, further studies are required to investigate the detailed expression patterns and significance of individual OCT4 isoforms in carcinogenesis.
Colorectal cancer (CRC) is one of the most common types of cancer worldwide. The global statistics from 2020 show that colorectal cancer creates 10% of all newly diagnosed cases, which means that after breast and lung cancer, it is the third most common type (1). Regardless of the enormous interest in CRC research, the annual incidence and number of CRC-related deaths have been increasing worldwide. The incidence of CRC is higher in developed countries due to unhealthy lifestyle including high consumption of red meat and alcohol, smoking, sedentary lifestyle, and inflammatory bowel diseases (2–4).
The octamer binding transcription factor 4 (OCT4) isoforms are encoded by the POU5F1 gene which is located at the short arm of chromosome 6 (5). Proteins from the POU protein family contain the so-called POU domains which allow them to bind to DNA and influence the gene expression, as well as interact with other transcription factors and cofactors. The POU domain recognizes and binds to the octamer consensus DNA sequence ATGCAAT, and in this way, the OCT4 can regulate gene expression, making it the main regulator of maintaining pluripotency and self-renewal of the stem cells (6–8). The human OCT4 gene can, through alternative transcription initiation or alternative splicing, create numerous different isoforms. These isoforms differ not only in nucleotide sequence but also in subcellular localization and properties. At the RNA level, OCT4 creates four groups of variants, namely, OCT4A, OCT4B, OCT4C, and OCT4D, and each of them uses a unique transcription start site. Furthermore, the mechanism of alternative splicing is responsible for the emergence of other OCT4B and OCT4C isoforms (9, 10). The discovery of individual transcripts and isoforms was gradual, and to date, overall, 10 OCT4 transcripts have been identified (9, 11–16). Not all transcripts have been identified also at the protein level, and there are only assumptions that the length of the protein product would be 164 amino acids (16). On the other hand, there is unequivocal evidence that the OCT4A and OCT4B variants generate distinct protein products that differ in their properties. The OCT4A isoform encodes the longest protein composed of 360 amino acids, which fulfills the role of the transcription factor. In contrast, the OCT4B transcript may be through the mechanism of alternative translation translated into three different proteins with lengths of 265, 190, and 164 amino acids (17). OCT4B proteins differ from OCT4A in DNA-binding properties; as a consequence of the inability of OCT4B proteins to regulate gene expression, their role in the cell remains unknown (18).
The OCT4A is the most known isoform, and its correct and precise expression is necessary for the regulation of the expression of further genes, as well as for the repression of genes involved in differentiation (18). The OCT4A protein plays a pivotal role in pluripotency and its expression was confirmed in embryonic stem (ES) and embryonic cancer (EC) cells, tumor cells, and cell lines (19, 20), but its presence has not yet been detected in non-pluripotent cell types (11, 18). The definite role of OCT4A in pluripotent cells was clarified by Takahashi and Yamanaka who proved that OCT4 is one of the four transcription factors needed for the reprogramming of somatic cells to pluripotent cells (12, 13, 21). In contrast to OCT4A, which is located exclusively in the nucleus, the OCT4B isoform is diffusely localized in both the cytoplasm and nucleus (17, 22). Atlasi et al. demonstrated that OCT4B is expressed in almost all cell types tested, including ES, EC, and somatic cell types (12). Interestingly, another identified OCT4B isoform, called OCT4B1, has properties and expression patterns more similar to the OCT4A than to the OCT4B isoform.
The role of POU5F1, especially the OCT4A isoform in stem cells, emphasizes its potential role in carcinogenesis. The so-called theory of stem cells in carcinogenesis claims that cancer stem cells are capable of inducing tumorigenesis and tumor growth due to their self-renewal ability, as well as giving rise to additional progenitor cells (23). Due to the great diversity of OCT4 isoforms, little is known about their expression patterns and role in primary and metastatic colorectal cancer. In our study, we focused on determining the expression patterns of OCT4A, OCT4Bs, and all OCT4 isoforms in primary and metastatic colorectal cancer, as well as the correlation between their expression and clinical parameters.
In total, 78 CRC patients were registered and underwent CRC resection. Primary tumor samples as well as metastatic samples were collected in collaboration with the University Hospital Martin, the Clinic of General, Visceral and Transplant Surgery, and the Department of Pathological Anatomy (Martin, Slovakia). Control, adjacent non-tumorous samples were obtained from patients with primary tumor (n = 30). The inclusion criteria were confirmation of diagnosis by histopathological examination, TNM classification, and clinical stages I, II, and III for primary tumor samples (n = 47) and confirmation of diagnosis by histopathological examination, TNM classification, and clinical stage IV for samples of liver metastases (n = 31). The pathologist also decided to take a non-cancer control sample from the adjacent tissue as far as possible from the tumor. The exclusion criteria were age less than 40 years or the simultaneous presence of other cancer types or liver metastasis of unknown origin. The histopathological assessment such as staging, grading, and typing of tumors was done by experienced pathologists (MK and JM). The clinicopathological characteristics of the patients included in the study are listed in Table 1. Tumor surgical specimens were collected by the pathologists and on the same day embedded into a solution of Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (10%), and penicillin/streptomycin and stored at 4°C. Immediately, the samples were frozen in RNAlater and then transferred to the Department of Molecular Biology and Genomics for RNA isolation.
Total RNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, treated with DNase, and stored at −80°C. Total RNA was quantified using the Qubit 3.0 Fluorometer (Thermo Fisher, MA, USA), and RNA quality assessment was performed on Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA). Reverse transcription was performed for each sample using a High-Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Thermo Fisher, MA, USA) according to the manufacturer’s instructions in a total volume of 20 µl consisting of 2 µl of 10× RT buffer, 0.8 µl of 2× dNTP mix (100 mM), 2 µl of 10× RT random primers, 1 µl of MultiScribe™ Reverse Transcriptase, 1 µl of RNase inhibitor, 3.2 µl of nuclease-free water, and 10 µl of RNA diluted in nuclease-free water to an overall concentration of 500 ng/µl. Reverse transcription was performed using the Bio-Rad MJ Mini Personal Thermal Cycler (Bio-Rad, Hercules, CA, USA), and the thermal conditions were as follows: 25°C for 10 min, followed by 37°C for 120 min and 85°C for 5 min. The prepared cDNA was stored at −20°C.
For the RT-PCR analysis, 10 µl of Gene Expression Master Mix was mixed with 1 µl of POU5F1 TaqMan assay, 1 µl of GAPDH TaqMan assay, and 1 µl of cDNA. The total volume of 20 µl per reaction was supplemented with nuclease-free water. Expression analysis was performed by real-time quantitative PCR (qPCR) using LightCycler ABI 3500 (Applied Biosystems, Foster City, CA, USA), TaqMan Gene Expression Master Mix, and several TaqMan assays (Thermo Fisher, MA, USA) for specific OCT4 isoforms and endogenous control. We determined the expression of the OCT4A isoform, OCT4B isoforms, and all OCT4 isoforms together using three different TaqMan assays (FAM-MGB) (assay ID Hs01654807_s1, Hs04195369_s1, and Hs04260367_gH, respectively). Individual assays were chosen based on genomic map analysis (Supplementary Figure 1) according to the manufacturer, as well as based on the relevant literature. The chosen assays have already been validated and used in several articles, for instance, Hs04260367_gH (24, 25), Hs01654807_s1 (26, 27), and Hs04195369_s1 (28, 29). The assay used for the detection of the OCT4A isoform did not recognize other OCT4 isoforms due to its exon 1 specificity. The same principle was used in the experiment with assay specific for almost all OCT4B isoforms. Some of the OCT4B and OCT4A isoforms were not recognized by this assay due to the absence of a certain exon. The last assay was used to determine all OCT4 isoforms that share the same last exon (Supplementary Figure 1). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control, and its expression was determined by TaqMan assay (VIC-MGB, assay ID Hs99999905_m1), too. As a reference sample, we used the Total Human RNA Control (Thermo Fisher, MA, USA; Cat. number 4307281) which was also reverse-transcribed into cDNA. The thermal conditions of the reactions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. All reactions were performed in duplicate. Contamination was controlled by using a no-template control without adding cDNA as a template molecule. The relative quantification of OCT4 isoform expression in the primary and metastatic tumor samples but also in adjacent non-tumorous tissue was performed by the ΔΔCt method (30). We calculated the relative expression of OCT4A, OCT4B, and all OCT4 isoforms against the GAPDH expression separately and obtained a fold-change value (log2FC). For statistical analysis, log2(FC) values were used.
Data were explored and analyzed in collaboration with the Biomedical Centre Martin in R ver. 4.0.5, with the aid of different libraries (31–41). Data were summarized as the mean, SD, min, quartiles, and max. Boxplot overlaid with a swarm plot and quantile–quantile plot with the 95% confidence band constructed by bootstrap were used to assess the normality of data. Welch’s t-test was used to test the null hypothesis that the population mean of the log2(FC) is 0. The regression model was used to model the association between log2(FC) and clinical data. Using the Wilkinson–Rogers notation, the full model that we used can be written as log2FC ~ type + side + T + N + M + stage, where type is a factor with levels p (for primary tumor) and t (for metastasis); side is a 0/1 factor where the left side is coded as 1; T, N, and M are the factors, describing the amount and spread of cancer; and stage is the pathological stage. The Akaike information criterion (AIC) was used for model selection. The model selected by the AIC was subjected to standard diagnostic analyses. Adjusted R2 was used to measure the effect size. Marginal means and post hoc pairwise comparisons were performed where the AIC-selected model contained other predictors aside from the intercept. The post hoc p-values were adjusted by the Tukey method.
The STAR-processed RNA sequencing level 1 data were downloaded from the GDC Portal of the NIH National Cancer Institute. Files were generated within the projects TCGA-COAD, TCGA-READ, CPTAC-2, and HCMI-CMDC. The set contained tsv files of 737 samples from 616 individuals, of which 615 were tumor samples, 45 were labeled as normal tissue, and 11 were labeled as metastases. Sixty-six samples with no information of their origin were omitted from further analysis. Given that all files were open access level 1 data, no preprocessing was carried out.
In our study, we focused on determining the expression of different OCT4 isoforms in primary and metastatic colorectal cancer samples. We investigated the expression of the OCT4A isoform, OCT4B isoforms, and all OCT4 isoforms together, as well as the relationship between isoform expression in certain sample types and clinical data. A total of 78 cancer samples (47 primary cancer samples and 31 liver metastases samples) were analyzed in duplicates for each isoform (A, B, and all isoforms). Control samples (n = 30) represented adjacent non-tumor tissues of the intestinal epithelium.
Fold-change values were calculated based on qPCR results for individual isoforms in specific sample types. At first, we compared the expression of the OCT4 isoform in all obtained tumor samples (primary + metastatic) vs. control (adjacent non-tumor tissue). The expression of all tested isoforms (namely, OCT4A, OCT4B, and all OCT4 isoforms) was significantly downregulated in tumor tissues compared with controls. The log2(FC) values for the OCT4A and OCT4B isoforms were −1.25 and −1.53, respectively, which refers to significantly reduced expression in tumor samples compared with controls (p < 0.0001 and p < 0.0001, respectively). In the case of all OCT4 isoforms, the log2(FC) was −1.03 and the expression was lower in tumors compared with control samples (p < 0.0001) (Figure 1). All data are listed in Supplementary Table 1.
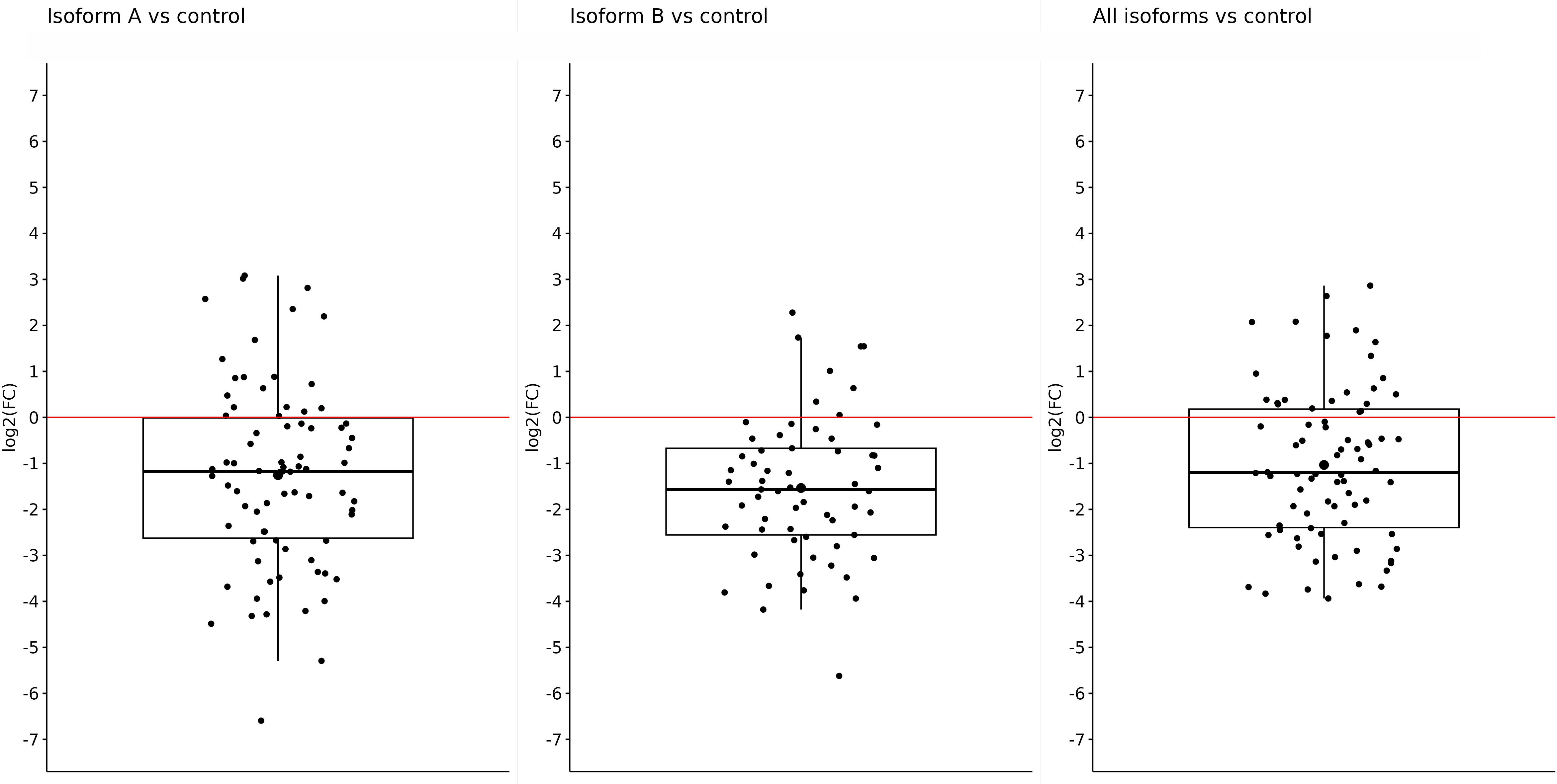
Figure 1 Expression of OCT4A, OCT4B, and all OCT4 isoforms in tumor samples (primary + metastatic) compared with control samples. On the y-axis, we see log2(FC) values for OCT4A (−1.25), OCT4B isoforms (−1.53), and all OCT4 isoforms (−1.03), which means a significantly reduced expression of all mentioned isoforms in primary tumors compared with control.
After getting these results, we tried to determine whether the primary or metastatic samples were responsible for such reduced expression of individual isoforms in tumor samples or if it was a result of a mutual effect. Thus, subsequently, we determined the changes in the expression in primary and metastatic tumors compared with the control separately. In primary tumors, we also observed significantly reduced expression of all tested isoforms. For the OCT4A isoform, the log2(FC) value was −1.08, referring to reduced expression in primary tumors compared with control samples (p = 0.0002). The expression of OCT4B isoforms was the lowest in primary tumors, and compared with the control samples, the log2(FC) value was −1.69 (p < 0.0001). Furthermore, we obtained statistically significant results also for the remaining isoforms. For all OCT4 isoforms, the log2(FC) value was −1.55, which means decreased expression in primary tumors compared with controls (p < 0.0001) (Figure 2).
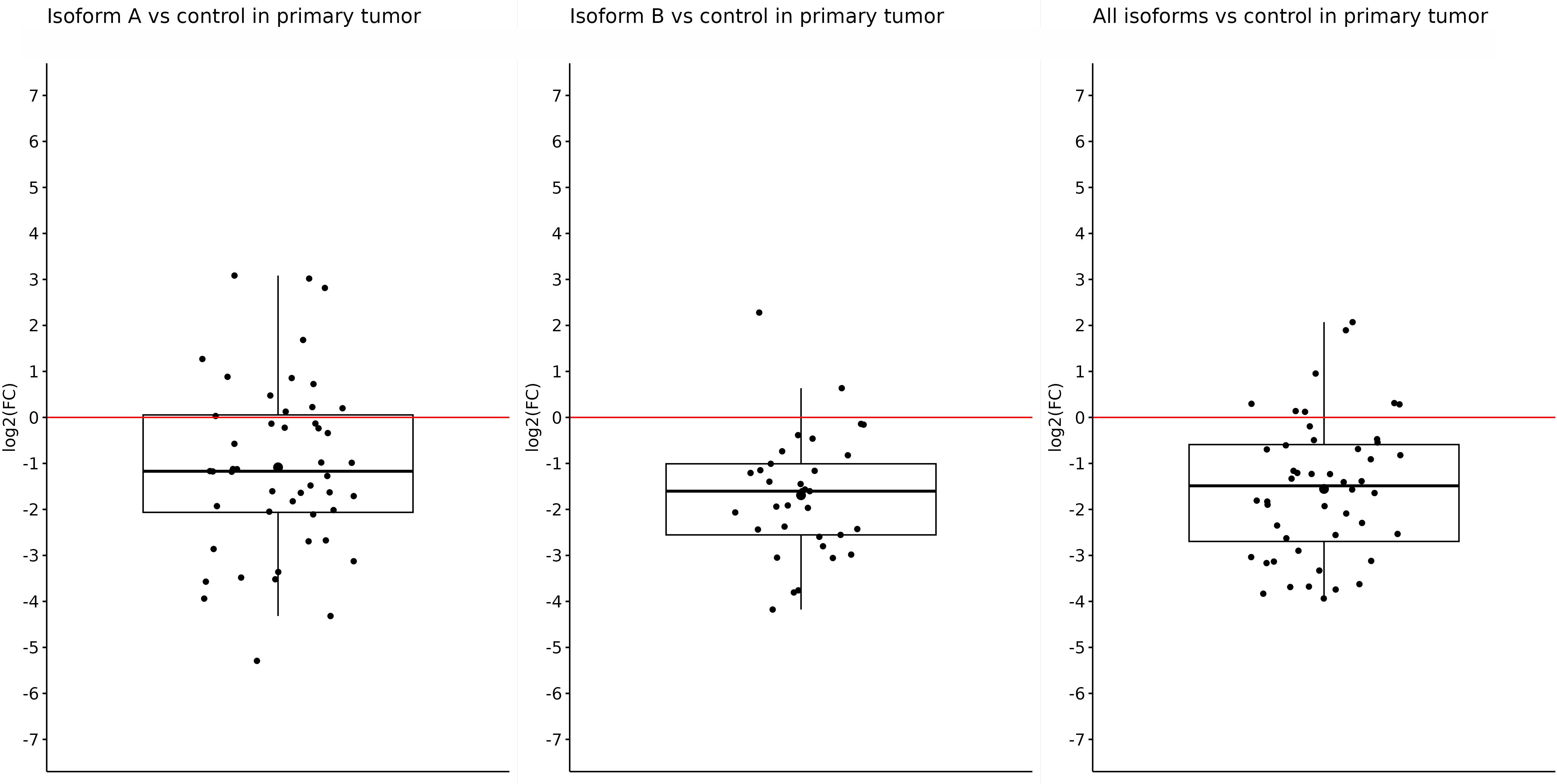
Figure 2 Expression of OCT4A, OCT4B, and all OCT4 isoforms in primary tumors compared with control samples. In the artwork, we can see the significantly downregulated expression of all tested isoforms in primary tumors compared with control samples. The log2(FC) values are −1.08 for OCT4A, −1.69 for OCT4B, and −1.55 for all OCT isoforms.
Similar results were also obtained in metastatic samples; in other words, the expression of OCT4A and OCT4B isoforms was significantly downregulated in tumor samples compared with controls. Both the OCT4A and OCT4B isoforms had statistically significant results. The expression of OCT4A was reduced (log2(FC) = −1.53) in liver metastasis compared with control tissues (p = 0.0006). For OCT4B, the log2(FC) value was −1.35, indicating a lower expression in metastasis (p = 0.00051). On the other hand, although the log2(FC) for all OCT4 isoforms was −0.2 and indicated reduced expression in metastases compared with controls, this difference was not enough to be statistically significant (p = 0.5) (Figure 3).
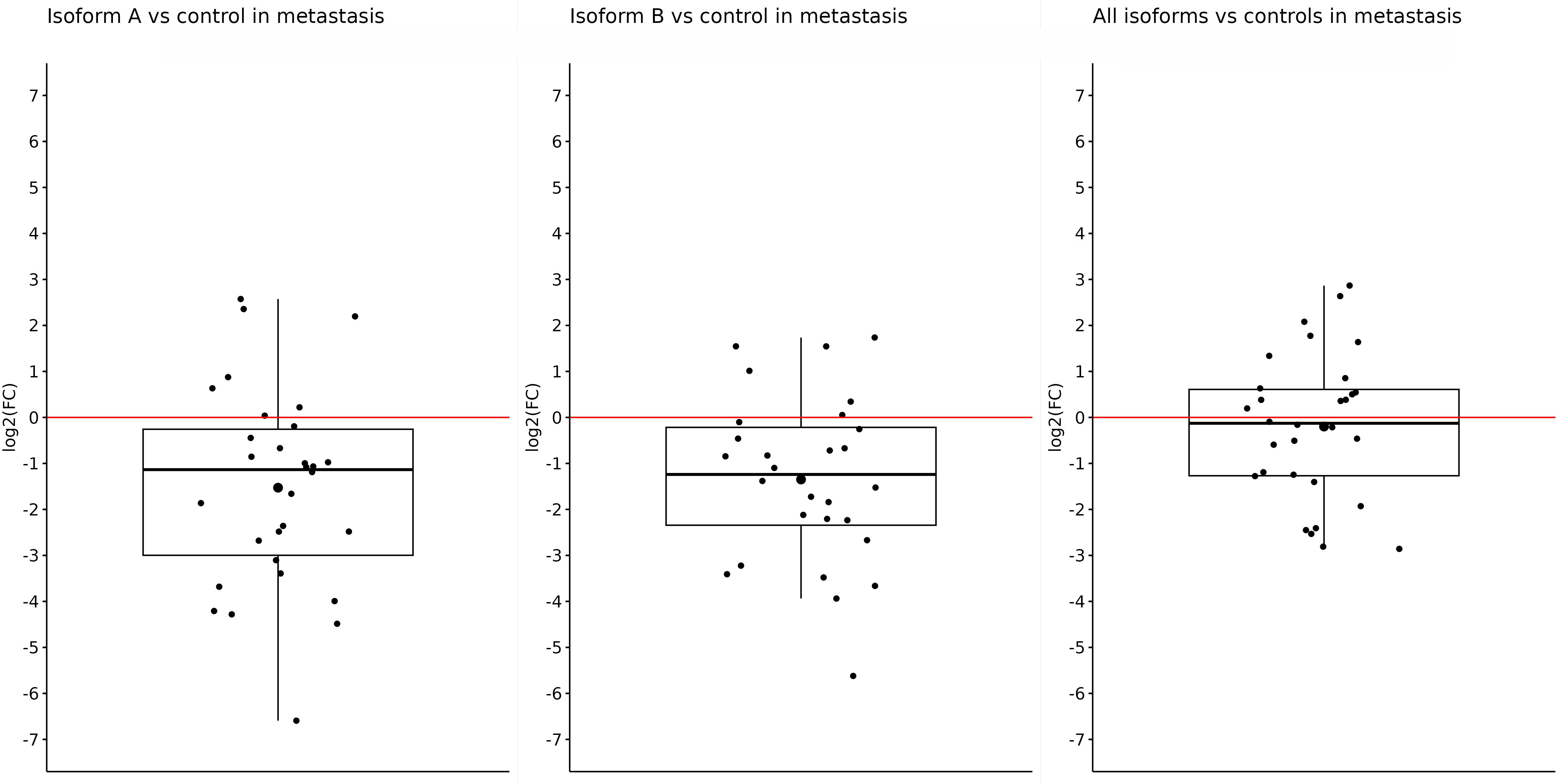
Figure 3 Expression of OCT4A, OCT4B, and all OCT4 isoforms in metastases compared with control samples. When comparing metastases with controls, a significantly reduced expression was observed for only OCT4A (log2(FC) = −1.53) and OCT4B isoforms (log2(FC) = −1.35). For all OCT4 isoforms, the log2(FC) value was the lowest (−0.2) and without statistical significance.
After that, we did an expression comparison in primary and metastatic tumors. Surprisingly, the expression of OCT4A and OCT4B was not significantly different in metastases and primary tumors, and log2(FC) −0.44 and 0.34 did not show statistically significant results (p = 0.27 and p = 0.33, respectively). Nonetheless, the expression of OCT4A was lower in metastases compared with primary tumors. On the contrary, OCT4B isoforms were overexpressed in liver metastases. We obtained the only statistically significant result in the case of all OCT4 isoforms which were significantly overexpressed in metastases compared with primary tumors (log2(FC) = 1.357; p < 0.0001) (Figure 4).
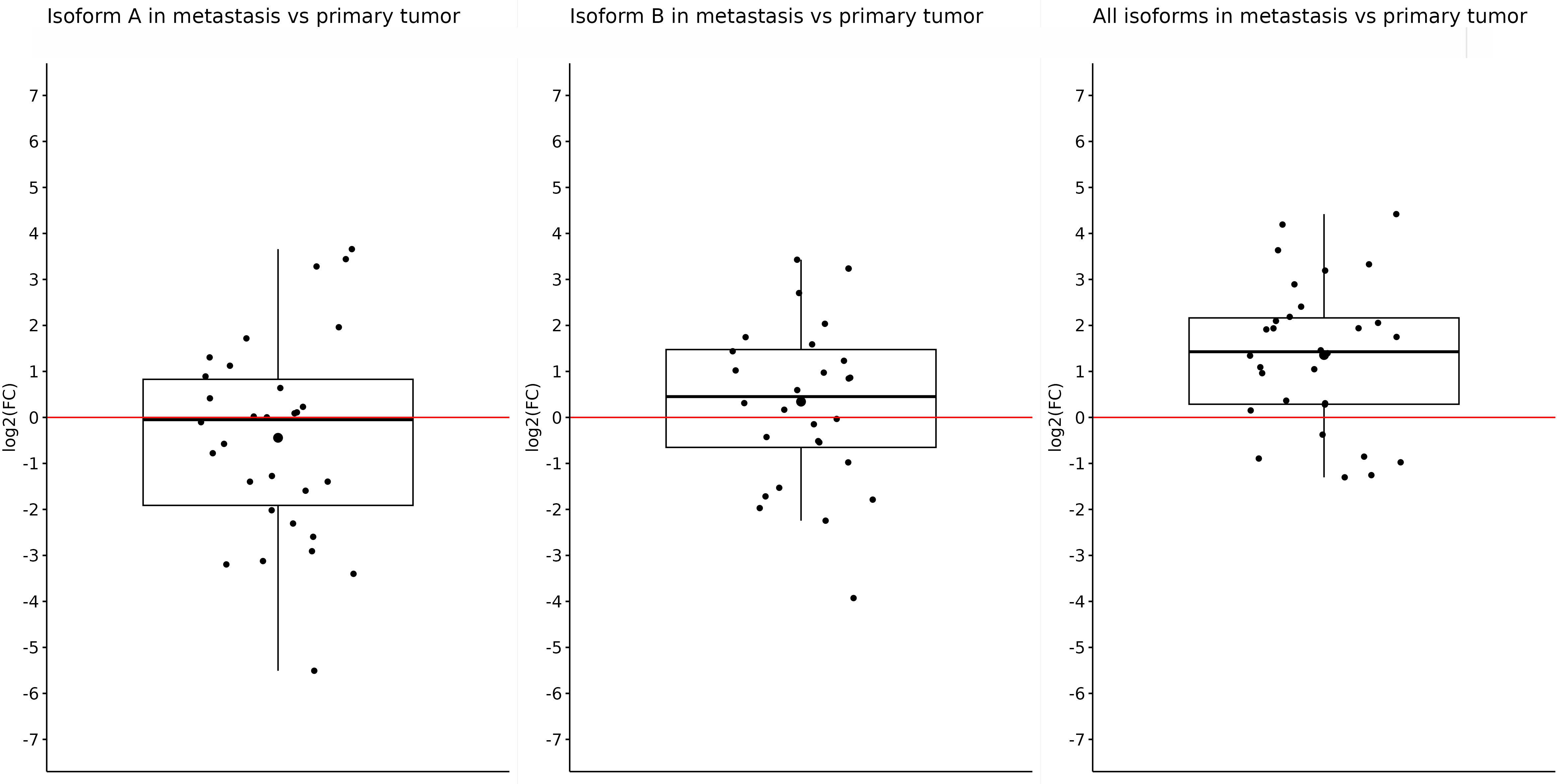
Figure 4 Expression of OCT4A, OCT4B, and all OCT4 isoforms in metastases compared with primary tumor samples. Comparison of the expression in metastases and primary tumors proved only one statistically significant result, which showed us that the expression of all OCT4 isoforms is significantly upregulated in metastasis (log2(FC) = 1.357). For the OCT4A and OCT4B isoforms, there were slight downregulation and upregulation (log2(FC) = −0.44 and 0.34, respectively).
So, in the case of OCT4A, the most downregulated expression was observed in metastatic tumors. Regarding the OCT4B isoforms, the most downregulated expression was also detected in metastases, too. In the case of all OCT4 isoforms, when comparing metastases and primary tumors, the expression was significantly upregulated in metastatic samples. Data are available on data-mendeley.com/datasets (42).
We further evaluated the relationship between the isoform expression and several clinical parameters, for instance, cancer type, tumor side, clinical, and TNM stage. Comparison of the tumor samples (primary + metastatic) with the control showed a correlation between significantly downregulated expression of all OCT4 isoforms and primary tumors (p = 0.001). Regression analysis also revealed the relationship between significantly reduced expression of all OCT4 isoforms and left-sided tumors (p = 0.030). Other correlations regarding the expression of certain isoforms and the type or side of tumors were not observed.
We have downloaded the STAR-processed RNA sequencing level 1 data of 671 samples (colorectal tumors: n = 615, metastases: n = 11, unpaired samples from normal tissue: n = 45) from the Cancer Genome Atlas Consortium Portal and used them to extract transcripts per kilobase million (TPM) counts for POU5F1 (Ensemble ID ENSG00000204531.20). POU class 5 homeobox 1 has seven splice variants, which unfortunately are not discriminated by level 1 data. The level of gene expression in normal, tumor, and metastatic tissues is summarized in Supplementary Table 3 and Supplementary Figure 2. The TCGA results (Supplementary Table 3) indicate a rise in the expression of the POU5F1 gene from non-tumor samples to primary tumors and subsequently to metastases, although without statistical significance. Given the low number of observations for two out of three groups and the skewness toward higher TPM values, it is inconclusive at any significance level to say that there is an increased expression from normal to primary tumors and to metastases, although there might be some trend in this direction. Welch’s t-test was used for pairwise testing of the null hypothesis that the population means of TPM in the normal, tumor, and metastatic groups are the same. Contrary to our previous expectations, in no tested combination did we reach the statistical significance at the level of <0.05 (normal vs. tumor: p = 0.17; normal vs. metastatic: p = 0.05, tumor vs. metastatic tissue: p = 0.06), probably owing to the high number of outliers.
It is well known that the OCT4A isoform is expressed in numerous cell types including ES, EC, and cancer cell lines. Based on very little evidence about its expression in adult tissues (12, 43) and the study of Feldman et al., who found out that OCT4A transcription is turned off during gastrulation due to its promoter and enhancer methylation (44), it was generally thought that OCT4A is not expressed in adult somatic tissues. The OCT4A was shown to be an irreplaceable factor for stem-like cell phenotype, maintaining pluripotency and self-renewal of the stem cells and also for precise embryonic development, making it one of the most popular transcription factors ever (45–48). At present, a large number of articles demonstrate the presence of OCT4A in cancer stem cells (CSCs), and it is thought that OCT4A is responsible for stem-like cell properties of the cancer cells such as self-renewal, resistance, and the possibility to give rise to progenitor cells as well as epithelial–mesenchymal transition (19, 49). These assumptions make it a suitable target for treatment (50). In the past, it was proven that downregulation of OCT4 expression resulted in inhibited tumorigenesis, reduced drug resistance, and induced G2/M phase arrest (51). Furthermore, OCT4 appears to play a role in the angiogenesis and conversion of human fibroblasts to functional endothelial cells (52–54), and its expression was confirmed in all 13 CRC cell lines established from patients with both primary and metastatic tumors (55). Altogether, it appears that OCT4A contributes to tumor initiation, cancer growth, metastasis, and therapy resistance (56).
To date, 10 different OCT4 isoforms have been identified at the RNA level, but not all are translated into protein (15). At the protein level, we can distinguish OCT4A and OCT4B isoforms which differ in their exon composition, nucleotide sequence, cell localization, and properties (11) (Supplementary Table 2). Due to the inability to bind to DNA, the OCT4B isoform is unable to regulate gene expression, and therefore, the role of OCT4B isoforms in cells remains unknown and unclear (18). Even though there are numerous OCT4B transcripts, the existence of only three protein isoforms has been confirmed (57). OCT4B proteins have been shown to play a role in stress response in two different ways. OCT4B-190 protects cells against apoptosis after heat shock (17), and in contrast, OCT4B-265 promotes apoptosis in reaction to genotoxic stress through the p53 signaling pathway (58).
In our study, we demonstrated an expression pattern of different OCT4 isoforms in different sample types, as well as the relationship between isoform expression and several clinical parameters.
Comparing the expression of all determined isoforms (A, B, and all isoforms) in the control and tumor samples, regardless of the type, we observed significant overexpression in the control samples (Figures 1–3). Distinct results were presented by Liu et al. (59) who found out that OCT4 was overexpressed in tumor tissue compared with their matched normal counterparts of CRC. However, the authors did not distinguish primary and metastatic samples nor the expression of individual isoforms because they used primers specific for OCT4A as well as OCT4B isoforms (59). On the other hand, similar results for the expression of all OCT4 isoforms, such as reduced gene expression in tumor tissue compared with the control and regardless of the isoforms, were also obtained in breast cancer (60).Aside from the other isoforms, OCT4A also had a higher expression in the control samples. On the other hand, the most reduced expression was detected in metastases compared with control samples, and the lowest difference was observed in the comparison of primary tumors with control tissues. In earlier investigations, devoted to OCT4A expression, the authors pointed out the possible distortion of the results due to the existence of numerous OCT4 pseudogenes, so we should also consider the fact that our expression data may be influenced by pseudogenes expression (61–63). To date, eight OCT4 pseudogenes have been identified and the transcription of these pseudogenes can have a confusing effect on research and knowledge of the OCT4 gene expression (64). On the contrary, Saha et al. published results in which the expression of several OCT4 pseudogenes has the same trend compared with the OCT4 expression which indicates that pseudogene expression should not have an impact on the overall direction of expression (60). It could be precisely pseudogenes that are responsible for such high OCT4A expression, but even though our results indicate that OCT4A expression is the highest in the control samples, our raw expression data demonstrated that OCT4A expression is not as high as it can be seen in the controls (average Ct approximately 31). We also observed that the expression of OCT4A was mildly decreased in metastases compared with primary tumors. We suppose that it is related to the change in OCT4B expression. Li et al. (65) found out that OCT4B functions as a non-coding RNA, modulating OCT4A expression by competitive binding with microRNAs. This may highlight the role of OCT4B in miRNA regulation of OCT4A expression (65). Hypothetically, this could be the reason why the expression of OCT4A was reduced in primary tumors, and with the mild increase in OCT4B expression, we observed a little but statistically non-significant decrease in OCT4A expression in metastases.
We also observed significantly upregulated expression of all OCT4 isoforms in metastasis compared with primary tumors. Such increased expression may be caused by using a probe that specifically recognizes a mutual exon shared by all of the already identified OCT4 isoforms. Thus, we were probably able to detect the expression of not only the OCT4A and OCT4B isoforms but also fewer known isoforms such as OCT4C and OCT4D, whose existence at the protein level has not yet been confirmed (15, 16). Several research groups have published results that emphasize the role of OCT4 in the aggressive behavior of CRC and its contribution to forming liver metastasis in CRC, especially OCT4 in the high-expression group, which is consistent with our results (66–68). Furthermore, OCT4 expression was denoted as an independent prognostic biomarker for predicting worse disease-specific survival and overall survival in CRC (69). On the other hand, the expression of all OCT4 isoforms in both primary and metastatic samples than in control samples was significantly downregulated. Similar results, such as reduced gene expression of OCT4 in tumor tissue compared with control and regardless of the isoforms, were also obtained from breast cancer (60).
For the OCT4B isoforms, there is a typical expression in various non-pluripotent cell types and differentiated tissues at different levels based on a specific isoform. There is also unequivocal evidence that OCT4A is also expressed in adult human stem cells and differentiated somatic cells, in addition to pluripotent cells, but at a much lower level (14). Surprisingly, OCT4B1 has a similar expression pattern to OCT4A (18). As a result of determining the expression of all OCT4B isoforms together, we were not able to designate which isoform had a higher expression and vice versa. In primary tumors, the expression of the OCT4B isoforms was significantly reduced compared with controls. When compared with metastases, the expression was slightly reduced but without a statistically significant result.
Although we used data from the TCGA and despite our efforts, we were not able to confirm in the TCGA dataset our findings of reduced POU5F1 gene expression in tumor samples compared with non-tumor controls at a statistically significant level. The TCGA results (Supplementary Table 3) indicate a non-significant trend of the increase of the POU5F1 expression from normal tissue to primary tumor and to metastases. Our analyses demonstrate only a significant increase in gene expression solely between the primary and metastatic samples. In contrast, we have observed a decrease in gene expression in tumor samples relative to non-tumor samples. Although it would have been ideal to have paired TCGA data, the number of non-cancerous samples available for comparison was rather limited. To avoid any contradictions in our research, we believe that it is important to conduct further validation studies with a larger sample size and better access to the TCGA data. It will also be necessary to differentiate between individual OCT4 isoforms, which were not distinguished in many previous studies, to obtain more conclusive results on the gene expression of POU5F1 and its isoforms. These efforts will provide a better understanding of the role of POU5F1 in colorectal cancer.
The role of OCT4 expression as a prognostic marker, as well as its role in metastatic CRC, has already been explored. Patients with high OCT4 expression had a poorer prognosis, making it a potential marker for the diagnosis and assessment of the prognosis of CRC. In addition, the results also indicate that OCT4 expression was correlated with clinical stage, tumor grade, metastasis, and TNM stage (69–71). In our study, we did not observe any correlation between OCT4 isoform expression and clinical or TNM tumor stage. The authors of the studies mentioned above did not recognize between individual OCT4 isoforms which may cause inconsistency in the results. Our results also suggest a correlation between reduced expression of all OCT4 isoforms and both primary and left-sided tumors. On the other hand, Talebi et al. did not observe OCT4 expression in any of the tissues tested (normal, polyp, and cancer tissue) and concluded that the diagnostic power of the OCT4 gene is not enough to identify cancer (72). Even though the exact role of OCT4B isoforms in the cell is still under investigation, several studies indicate that in some way they may contribute to the properties of cancer cells, such as invasion, having antiapoptotic properties, and resistance to chemotherapy (73, 74). In addition, Gazouli et al. confirmed the expression of the OCT4B and OCT4B1 isoforms in CRC samples and observed that the level of OCT4B1 mRNA was correlated with poorly and moderately differentiated CRC and with the progression of cancer stage (62). In our study, no correlation was detected between OCT4B expression and clinical data as well as the type or side of the tumor or TNM stage. Another research group showed that OCT4B1 has a potential role in regulating the self-renewal of colorectal CSC through its involvement in epithelial–mesenchymal transition (75). Furthermore, Simó-Riudalbas et al. demonstrated a pro-oncogenic effect of OCT4B1 through its association with protein kinases and subsequent activation of intracellular signaling events as well as cytoskeletal rearrangements (76). All these findings can signify a potential role of OCT4B, especially OCT4B1 as a marker of tumor-initiating cells or CSCs, and not only OCT4A but all OCT4 isoforms might play a significant role in carcinogenesis.
Based on previous results as well as our results, the expression level of OCT4 isoforms could be a useful tool not only for diagnosis, especially metastatic disease, but also for prognosis prediction. As the results of the TCGA analysis and our own analyses are not fully consistent, further studies will be needed to clarify these differences. Thus, these results emphasize the importance of precise characterization of individual OCT4 isoforms whether transcriptional or protein, as well as their expression patterns in colorectal cancer.
Unlike previous reports, we found out that the expression of OCT4A, OCT4Bs, and all OCT4 isoforms is significantly reduced in primary tumors and metastases compared with control samples. On the other hand, the expression of all OCT4 isoforms was significantly upregulated in metastases compared with primary tumors, which was not caused by the upregulation of the OCT4A isoform and only partially by the OCT4B isoform, emphasizing the role of less-known isoforms in metastatic CRC. Furthermore, the reduced expression of all OCT4 isoforms was correlated with primary and left-sided tumors. Based on these results, we supposed that the expression rate of all OCT4 isoforms can be related to the cancer type and side, as well as to liver metastasis. However, further studies are required to investigate the detailed expression patterns and significance of individual OCT4 isoforms in carcinogenesis.
The datasets presented in this study are available on https://data.mendeley.com/datasets/2c5296ytbr (42).
The studies involving human participants were reviewed and approved by the Ethics Committee of the Jessenius Faculty of Medicine in Martin, Comenius University in Bratislava by decision number 1863/2016. The patients/participants provided their written informed consent to participate in this study.
ET was responsible for the experimental design, performed the experiments, and wrote the manuscript. ZL secured the financial funding, was responsible for the experimental design, supervised the data analysis and interpretation, and critically reviewed and revised the manuscript drafts. VH and ZL assisted in conducting the experiments. MG performed the bioinformatic analyses and generated the figures. MH and MKo performed the gene analysis from the TCGA raw reads and wrote the TCGA part of the manuscript. MKa and JMa performed the histopathological evaluation. EK and PM collected the patients’ samples. JMi and LL critically reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from The Ministry of Education, Science, Research and Sport of the Slovak Republic (grant VEGA no. 1/0269/22) and by The Slovak Research and Development Agency (grant APVV no. 16-0066 and no. 21-0448). Michal Kovac received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie COFUND SASPro2 grant agreement no. 2207/02/01.
We would like to thank Mrs. Andrea Vaňochová for her professional technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1166835/full#supplementary-material
Supplementary Figure 1 | Areas within the POU5F1 gene which are recognized by specific TaqMan probe.
Supplementary Figure 2 | Gene expression of POU5F1 based on Transcript Per Kilobase Million (TPM) counts from RNA sequencing experiments. Conformity to the Gaussian distribution was assessed and was subsequently rejected in each group, owing to the substantial enrichment of samples with high expression (Shapiro-Wilk test: normal tissue: P = 2.901e-10, tumor tissue: P < 2.2e-16, metastases: P = 0.0009742).
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin (2021) 7:209–49. doi: 10.3322/caac.21660
2. Chang VC, Cotterchio M, De P, Tinmouth J. Risk factors for early-onset colorectal cancer: a population-based case–control study in Ontario, Canada. Cancer Cause Control (2021) 32:1063–83. doi: 10.1007/s10552-021-01456-8
3. Botteri E, Borroni E, Sloan E, Bagnardi V, Bosetti C, Peveri G, et al. Smoking and colorectal cancer risk, overall and by molecular subtypes: a meta-analysis. Am J Gastroenterol (2020) 115:1940–9. doi: 10.14309/ajg.0000000000000803
4. Gausman V, Dornblaser D, Anand S, Hayes RB, O’Connell K, Du M, et al. Risk factors associated with early-onset colorectal cancer. Clin Gastroenterol Hepatol (2020) 18:2752–9. doi: 10.1016/j.cgh.2019.10.009
5. Guillaudeux T, Mattei MG, Depetris D, Le Bouteiller P, Pontarotti P. In situ hybridization localizes the human OTF3 to chromosome 6p21.3 ?p22 and OTF3L to 12p13. Cytogenet Cell Genet (1993) 63:212–4. doi: 10.1159/000133537
6. Herr W, Sturm RA, Clerc. RG, Corcoran LM, Baltimore D, Sharp PA, et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and caenorhabditis elegans unc-86 gene products. Genes Dev (1988) 2:1513–6. doi: 10.1101/gad.2.12a.1513
7. Verijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating JJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J (1992) 11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x
8. Shi G, Jin Y. Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res Ther (2010) 1:39. doi: 10.1186/scrt39
9. Mehravar M, Ghaemimanesh F, Poursani EM. An overview on the complexity of OCT4: at the level of DNA, RNA and protein. Stem Cell Rev Rep (2021) 17:1121–36. doi: 10.1007/s12015-020-10098-3
10. Poursani EM, Mehravar M, Shahryari A, Mowla SJ, Mohammad Soltani B. Alternative splicing generates different 5′ UTRs in OCT4B variants. Avicenna J Med Biotechnol (2017) 9:201–4.
11. Takeda J, Seino S, Bell GI. Human Oct3 gene family: cDNA sequences, alternative splicing, gene organization, chromosomal location, and expression at low levels in adult tissues. Nucleic Acids Res (1992) 20:4613–20. doi: 10.1093/nar/20.17.4613
12. Atlasi Y, Mowla SJ, Ziaee SAM, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells (2008) 26:3068–74. doi: 10.1634/stemcells.2008-0530
13. Poursani EM, Mehravar M, Soltani BM, Mowla SJ. OCT4B2, a novel alternative spliced variant of OCT4, is significantly upregulated under heat-stress condition and downregulated in differentiated cells. Tumour Biol (2017) 39. doi: 10.1177/1010428317724280
14. Zhao FQ, Misra Y, Li DB, Wadsworth MP, Krag D, Weaver D, et al. Differential expression of Oct3/4 in human breast cancer and normal tissues. Int J Oncol (2018) 52:2069–78. doi: 10.3892/ijo.2018.4341
15. Mehravar M, Poursani EM. Novel variant of OCT4, named OCT4B5, is highly expressed in human pluripotent cells. Stem Cell Rev Rep (2021) 17:1068–73. doi: 10.1007/s12015-020-10093-8
16. Malakootian M, Azad FM, Naeli P, Pakzad M, Fouani Y, Bajgan ET. Novel spliced variants of OCT4, OCT4C and OCT4C1, with distinct expression patterns and functions in pluripotent and tumor cell lines. Eur J Cell Biol (2017) 96:347–55. doi: 10.1016/j.ejcb.2017.03.009
17. Wang X, Zhao Y, Xiao Z, Chen B, Wei Z, Wang B, et al. Alternative translation of OCT4 by an internal ribosome entry site and its novel function in stress response. Stem Cells (2009) 27:1265–75. doi: 10.1002/stem.58
18. Lee J, Kim HK, Rho JY, Han YM, Kim J. The human OCT-4 isoforms differ in their ability to confer self-renewal. J Biol Chem (2006) 281:33554–65. doi: 10.1074/jbc.M603937200
19. Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. (2005) 26:495–502. doi: 10.1093/carcin/bgh321
20. Palmieri SL, Peter W, Hess H, Schöler HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol (1994) 166:259–67. doi: 10.1006/dbio.1994.1312
21. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (2007) 318:1917–20. doi: 10.1126/science.1151526
22. Cauffman G, Liebaers I, Van Steirteghem A, Van de Velde H. POU5F1 isoforms show different expression patterns in human embryonic stem cells and preimplantation embryos. Stem Cells (2006) 24:2685–91. doi: 10.1634/stemcells.2005-0611
23. O´Flaherty JD, Barr N, Fennell D, Richard D, Reynolds J, O´Leary J, et al. The cancer stem-cell hypothesis: its emerging role in lung cancer biology and its relevance for future therapy. J Thorac Oncol (2012) 7:1880–90. doi: 10.1097/JTO.0b013e31826bfbc6
24. Ilieva M, Dufva M. SOX2 and OCT4 mRNA-expressing cells, detected by molecular beacons, localize to the center of neurospheres during differentiation. PLoS One (2013) 8:e73669. doi: 10.1371/journal.pone.0073669
25. Losada-García A, Salido-Guandarrama I, Cortes-Ramirez SA, Cruz-Burgos M, Morales-Pacheco M, Vazquez-Santillan. SFRP1 induced a stem cell phenotype in prostate cancer cells. Front Cell Dev Biol (2023) . 11:1096923. doi: 10.3389/fcell.2023.1096923
26. Yamashita A, Yoshitomi H, Kihara S, Toguchida J, Tsumaki N. Culture substrate-associated YAP inactivation underlies chondrogenic differentiation of human induced pluripotent stem cells. Stem Cells Transl Med (2021) 1:115–27. doi: 10.1002/sctm.20-0058
27. Okuno M, Aoki S, Kawai S, Imataki R, Abe Y, Harada K, et al. Effect of non-thermal atmospheric pressure plasma on differentiation potential of human deciduous dental pulp fibroblast like cells. Appl Sci (2021) . 21:10119. doi: 10.3390/app112110119
28. Gil-Kulik P, Chomik P, Krzyzanowski A, Radzikowska-Buchner E, Maciejewski R, Kwasniewska A, et al. Influence of the type of delivery, use of oxytocin, and maternal age on POU5F1 gene expression in stem cells dervied from wharton´s jelly within the umbilical cord. Oxid Med Cell Longev (2019) 2019:1027106. doi: 10.1155/2019/1027106
29. Timmerman DM, Eleveld TF, Gillis AJM, Friedrichs CC, Hillenius S, Remmers TL, et al. The role of TP53 in cisplatin resistance in mediastinal and testicular germ cell tumors. Int J Mol Sci (2021) . 22:11774. doi: 10.3390/ijms222111774
30. Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat bioinforma bioma (2013) 3:71–85.
31. Gagolewski M. Stringi: fast and portable character string processing in r. j. stat. Software (2022) 103:2. doi: 10.18637/jss.v103.i02
32. Goode K, Rey K. ggResidpanel: panels and interactive versions of diagnostic plots using ´ggplot2´. R package (2022). Available at: https://goodekat.github.io/ggResidpanel/.
33. Kassambara A. Ggpubr: 'ggplot2' based publication ready plots. R package (2020). Available at: https://rpkgs.datanovia.com/ggpubr.
34. Kassambara A. Rstatix: pipe-friendly framework for BasicStatistical tests. R package (2021). Available at: https://CRAN.R-project.org/package=rstatix.
35. Lenth R. Emmeans: estimated marginal means, aka least-SquaresMeans. R package (2022). Available at: https://CRAN.R-project.org/package=emmeans.
36. Lüdecke D. sjPlot: data visualization for statistics in social science. R package (2023). Available at: https://CRAN.R-project.org/package=sjPlot.
37. Ogle DH, Doll JC, Wheeler AP, Dinno A. FSA: Simple FisheriesStock assessment methods. R package (2023). Available at: https://CRAN.R-project.org/package=FSA.
38. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2021). Available at: https://www.r-project.org/.
39. Wickham H. Reshaping data with the reshape package. J Stat Software (2007) 21:12. doi: 10.18637/jss.v021.i12
40. Wickham H. GGplot2: elegant graphics for data analysis (2016). Available at: https://ggplot2.tidyverse.org.
41. Zhu H. kableExtra: construct complex table with ´kable´ andPipe syntax. R package (2021). Available at: https://cran.r-project.org/web/packages/kableExtra/index.html.
42. Turyova E, Mikolajcik P, Grendar M, Kudelova E, Holubekova V, Kalman M, et al. Supplement to expression of OCT4 isoforms is reduced in primary colorectal cancer. Mendeley Data (2023) V1. doi: 10.17632/2c5296ytbr.1
43. Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell (1990) 60:461–72. doi: 10.1016/0092-8674(90)90597-8
44. Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol (2006) 8:188–94. doi: 10.1038/ncb1353
45. Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell (1998) 95:379–91. doi: 10.1016/S0092-8674(00)81769-9
46. Fogarty NME, McCarthy A, Snijders KE, Powell BE, Kubikova N, Blakeley P, et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature (2017) 550:67–73. doi: 10.1038/nature24033
47. Mulas C, Chia G, Jones KA, Hodgson AC, Stirparo GG, Nichols J. Oct4 regulates the embryonic axis and coordinates exit from pluripotency and germ layer specification in the mouse embryo. Development (2018) 145. doi: 10.1242/dev.159103
48. Lo JHH, Edwards M, Langerman J, Sridharan R, Plath K, Smale ST. Oct4:Sox2 binding is essential for establishing but not maintaining active and silent states of dynamically regulated genes in pluripotent cells. Genes Dev (2022) 36:1079–95. doi: 10.1101/gad.350113.122
49. Zhang Q, Han Z, Zhu Y, Chen J, Li W. The role and specific mechanism of OCT4 in cancer stem cells: a review. Int J Stem Cells (2020) 13:312–25. doi: 10.15283/ijsc20097
50. Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest (2010) 120:41–50. doi: 10.1172/JCI41004
51. Ruan Z, Yang X, Cheng W. OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag Res (2018) 11:389–99. doi: 10.2147/CMAR.S180418
52. Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, et al. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol (2013) 33:1366–75. doi: 10.1161/ATVBAHA.112.301167
53. Hess DL, Kelly-Goss MR, Cherepanova OA, Nguyen AT, Baylis RA, Tkachenko S, et al. Perivascular cell-specific knockout of the stem cell pluripotency gene Oct4 inhibits angiogenesis. Nat Commun (2019) 10:967. doi: 10.1038/s41467-019-08811-z
54. Liu HL, ting TH, lin YH, Deng TT, Xu YP, Xu SQ, et al. Oct4 regulates the transition of cancer stem-like cells to tumor endothelial-like cells in human liver cancer. Front Cell Dev Biol (2020) 8:563316. doi: 10.3389/fcell.2020.563316
55. Ku JL, Shin YK, Kim DW, Kim KH, Choi JS, Hong SH, et al. Establishment and characterization of 13 human colorectal carcinoma cell lines: mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis (2010) 31:1003–9. doi: 10.1093/carcin/bgq043
56. Wang YJ, Herlyn M. The emerging roles of Oct4 in tumor-initiating cells. Am J Physiol Cell Physiol (2015) 309:C709–718. doi: 10.1152/ajpcell.00212.2015
57. Gao Y, Wang X, Han J, Xiao Z, Chen B, Su G, et al. The novel OCT4 spliced variant OCT4B1 can generate three protein isoforms by alternative splicing into OCT4B. J Genet Genomics (2010) 7:461–5. doi: 10.1016/S1673-8527(09)60065-5
58. Gao Y, Wei J, Han J, Wang X, Su G, Zhao Y, et al. The novel function of OCT4B isoform-265 in genotoxic stress. Stem Cells (2012) 4:665–72. doi: 10.1002/stem.1034
59. Liu YH, Li Y, Liu XH, Sui HM, Liu YX, Xiao ZQ, et al. A signature for induced pluripotent stem cell-associated genes in colorectal cancer. Med Oncol (2013) 1:426. doi: 10.1007/s12032-012-0426-2
60. Saha SK, Jeong Y, Cho S, Cho SG. Systematic expression alteration analysis of master reprogramming factor OCT4 and its three pseudogenes in human cancer and their prognostic outcomes. Sci Rep (2018) 8:14806. doi: 10.1038/s41598-018-33094-7
61. Suo G, Han J, Wang X, Zhang J, Zhao Y, Zhao Y, et al. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun (2005) 337:1047–51. doi: 10.1016/j.bbrc.2005.09.157
62. Gazouli M, Roubelakis MG, Theodoropoulos GE, Papailiou J, Vaiopoulou A, Pappa KI, et al. OCT4 spliced variant OCT4B1 is expressed in human colorectal cancer. Mol Carcinog (2012) 51:165–73. doi: 10.1002/mc.20773
63. Poursani EM, Mohammad Soltani B, Mowla SJ. Differential expression of OCT4 pseudogenes in pluripotent and tumor cell lines. Cell J (2016) 18:28–36. doi: 10.22074/cellj.2016.3984
64. Pain D, Chirn GW, Strassel C, Kemp DM. Multiple retropseudogenes from pluripotent cell-specific gene expression indicates a potential signature for novel gene identification. J Biol Chem (2005) 280:6265–8. doi: 10.1074/jbc.C400587200
65. Li D, Yang ZK, Bu JY, Xu CY, Sun H, Tang JB, et al. OCT4B modulates OCT4A expression as ceRNA in tumor cells. Oncol Rep (2015) 33:2622–30. doi: 10.3892/or.2015.3862
66. Dai X, Ge J, Wang X, Qian X, Zhang C, Li X. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol Rep (2013) 29:155–60. doi: 10.3892/or.2012.2086
67. Fujino S, Miyoshi N. Oct4 gene expression in primary colorectal cancer promotes liver metastasis. Stem Cells Int (2019) 2019:7896524. doi: 10.1155/2019/7896524
68. Roudi R, Barodabi M, Madjd Z, Roviello G, Corona SP, Panahi M. Expression patterns and clinical significance of the potential cancer stem cell markers OCT4 and NANOG in colorectal cancer patients. Mol Cell Oncol (2020) 7:1788366. doi: 10.1080/23723556.2020.1788366
69. Fang W, Ni M, Zhang M, Chen H. Prognostic value of OCT4 in colorectal cancer: analysis using immunohistochemistry and bioinformatics validation. biomark Med (2020) 14:1473–84. doi: 10.2217/bmm-2020-0069
70. Zhou H, Hu YU, Wang W, Mao Y, Zhu J, Zhou B, et al. Expression of Oct-4 is significantly associated with the development and prognosis of colorectal cancer. Oncol Lett (2015) 2:691–6. doi: 10.3892/ol.2015.3269
71. Miyoshi N, Fujino S, Ohue M, Yasui M, Takahashi Y, Sugimura K, et al. The POU5F1 gene expression in colorectal cancer: a novel prognostic marker. Surg Today (2018) 7:709–15. doi: 10.1007/s00595-018-1644-9
72. Talebi A, Kianersi K, Beiraghdar M. Comparison of gene expression of SOX2 and OCT4 in normal tissue, polyps, and colon adenocarcinoma using immunohistochemical staining. Adv BioMed Res (2015) 4:234. doi: 10.4103/2277-9175.167958
73. Wen KM, Zhang GH, Li J, Chen ZQ, Cheng YL, Su X, et al. OCT4B1 promotes cell growth, migration and invasion suppressing sensitivity to οxaliplatin in colon cancer. Oncol Rep (2015) 34:2943–52. doi: 10.3892/or.2015.4286
74. Wen K, Fu Z, Wu X, Feng J, Chen W, Qian J. Oct-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: effects associated with STAT3/Survivin. Cancer Lett (2013) 333(1):56–65. doi: 10.1016/j.canlet.2013.01.009
75. min ZJ, qing H, Jiang H, lin CY, hong FJ, quan CZ, et al. OCT4B1 promoted EMT and regulated the self-renewal of CSCs in CRC: effects associated with the balance of miR-8064/PLK1. Mol Ther – Oncolytics (2019) 15:7–20. doi: 10.1016/j.omto.2019.08.004
Keywords: colorectal cancer, gene expression, oct4, isoforms, carcinogenesis
Citation: Turyova E, Mikolajcik P, Grendar M, Kudelova E, Holubekova V, Kalman M, Marcinek J, Hrnciar M, Kovac M, Miklusica J, Laca L and Lasabova Z (2023) Expression of OCT4 isoforms is reduced in primary colorectal cancer. Front. Oncol. 13:1166835. doi: 10.3389/fonc.2023.1166835
Received: 15 February 2023; Accepted: 02 May 2023;
Published: 20 June 2023.
Edited by:
Matjaz Rokavec, Ludwig Maximilian University of Munich, GermanyReviewed by:
Roxana Pincheira, University of Concepcion, ChileCopyright © 2023 Turyova, Mikolajcik, Grendar, Kudelova, Holubekova, Kalman, Marcinek, Hrnciar, Kovac, Miklusica, Laca and Lasabova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eva Turyova, dHVyeW92YTEwQHVuaWJhLnNr; Zora Lasabova, em9yYS5sYXNhYm92YUB1bmliYS5zaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.