- Department of Pathology, University of Iowa Hospitals & Clinics, Iowa City, IA, United States
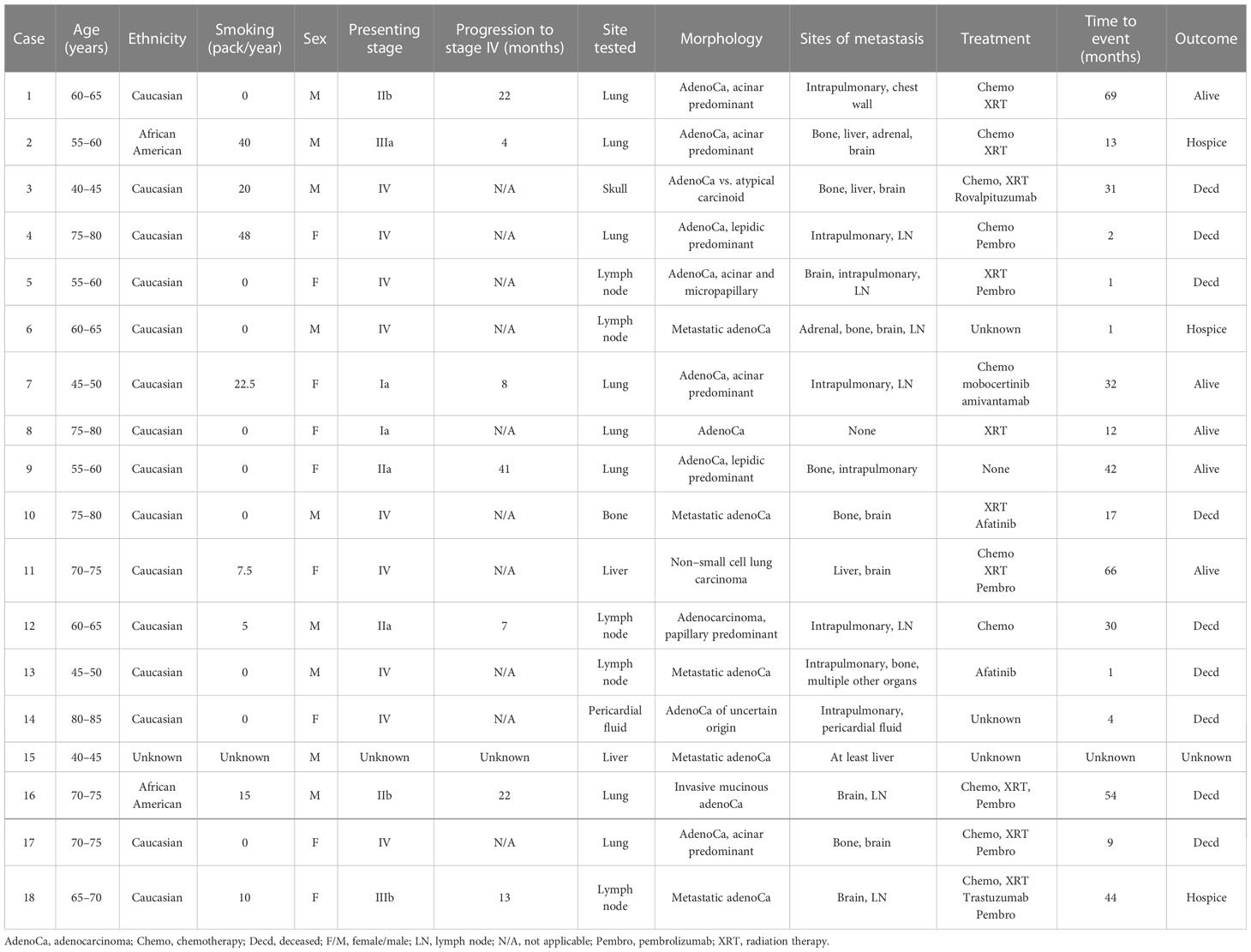
Background: Exon 20 (ex20) in-frame insertions or duplications (ins/dup) in epidermal growth factor receptor (EGFR) and its analog erb-b2 receptor tyrosine kinase 2 (ERBB2) are each detected in 1.5% of non–small cell lung cancer (NSCLC). Unlike EGFR p.L858R or ex19 deletions, ex20 ins/dup is associated with de novo resistance to classic EGFR inhibitors, lack of response to immune checkpoint inhibitors, and poor prognosis. US Food and Drug Administration has approved mobocertinib and amivantamab for targeting tumors with this aberration, but the number of comprehensive studies on ex20 ins/dup NSCLC is limited. We identified 18 cases of NSCLCs with EGFR/ERBB2 ex20 ins/dup and correlated the findings with clinical and morphologic information including programed death-ligand 1 (PD-L1) expression.
Methods: A total of 536 NSCLC cases tested at our institution between 2014 and 2023 were reviewed. A custom-designed 214-gene next-generation sequencing panel was used for detecting DNA variants, and the FusionPlex CTL panel (ArcherDx) was used for the detection of fusion transcripts from formalin-fixed, paraffin-embedded tissue. Immunohistochemistry (IHC)for PD-L1 was performed using 22C3 or E1L3N clones.
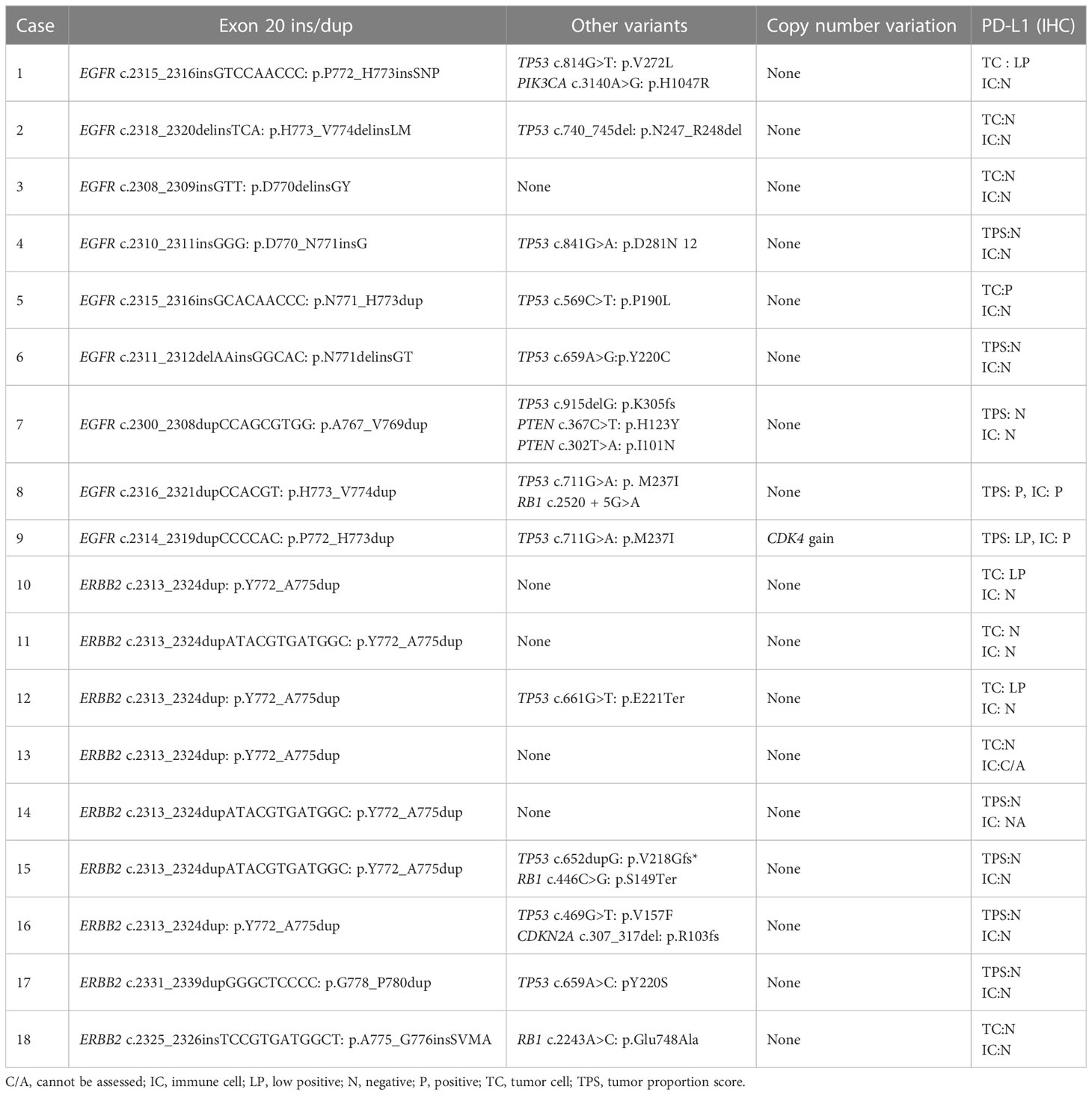
Results: Nine EGFR and nine ERBB2 ex20 ins/dup variants were identified from an equal number of men and women, 14 were non- or light smokers, and 15 had stage IV disease. All 18 cases were adenocarcinomas. Seven of the 11 cases with available primary tumors had acinar predominant pattern, two had lepidic predominant pattern, and the remainder had papillary (one case) and mucinous (one case) patterns. Ex20 ins/dup variants were heterogenous in-frame one to four amino acids spanning A767–V774 in EGFR and Y772–P780 in ERBB2 and were clustered in the loop following the C-helix and α C-helix. Twelve cases (67%) had co-existing TP53 variants. Copy number variation in CDK4 amplification was identified in one case. No fusion or microsatellite instability was identified in any case. PD-L1 was positive in two cases, low positive in four cases, and negative in 11 cases.
Conclusions: NSCLCs harboring EGFR/ERBB2 ex20 ins/dup are rare and tend to be acinar predominant, negative for PD-L1, more frequent in non- or light smokers, and mutually exclusive with other driver mutations in NSCLC. The correlation of different EGFR/ERBB2 ex20 ins/dup variants and co-existing mutations with response to targeted therapy and the possibility of developing resistant mutations after mobocertinib treatment warrants further investigation.
1 Introduction
Lung cancer is the second most common cancer in both men and women and the leading cause of cancer death, accounting for about one in five of all cancer deaths in the United States (https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html). Non–small cell lung cancer (NSCLC) accounts for 82% of lung cancer. In patients with lung adenocarcinoma, epidermal growth factor receptor (EGFR) mutations are present in 10%–20% of Caucasian and in 40%–60% of South-East Asian population (1). Up to 90% EGFR mutations are exon 19 in-frame deletions or the p.L858R hotspot mutation in exon 21 (1). Ex20 insertion/duplication (ins/dup) is the third most common EGFR mutation and detected in approximately 1.5% of NSCLC (2–5).
Erb-b2 receptor tyrosine kinase 2 (ERBB2) encodes for ERBB2, also called human epidermal growth factor receptor 2 (HER2), which is another member of the ERBB family of receptor tyrosine kinases. EGFR and erb-b2 receptor tyrosine kinase 2 (ERBB2) share structural and sequence similarities. Both have an extracellular ligand-binding domain, a transmembrane domain, and a tyrosine kinase domain (6). ERBB2 ex20 ins/dup has also been identified in NSCLCs and shows a similar mutation frequency of 1.5% (3, 7).
Previous studies have shown that similar to other EGFR variants, EGFR/ERBB2 ex20 ins/dup is predominantly found in adenocarcinoma, non-smokers, and women (1). In comparison to the more common and other uncommon EGFR mutations, ex20 ins/dup was associated with poor prognosis and lower overall survival (OS) (8).
Unlike tumors with EGFR p.L858R and ex19 deletion that are sensitive to first- and second-generation tyrosine kinase inhibitors (TKIs), ex20 ins/dup has de novo resistance to classic EGFR TKIs including the third-generation TKI such as osimertinib that targets the p.T790M in ex20 of the EGFR (9–11) and no response to immune checkpoint inhibitors (ICIs) (4). The US Food and Drug Administration (FDA) approved mobocertinib and amivantamab that specifically target EGFR and ERBB2 ex20 ins/dup. Targeted therapy for patients with this aberration showed sustained response (12, 13).
Because of the low incidence of the ex20 ins/dup in NSCLC, there have been a limited number of comprehensive studies published, and about half of the existing studies were performed in the Asian population with a focus on EGFR ex20 ins/dup only. In this study, we identified 18 cases of NSCLCs with EGFR/ERBB2 ex20 ins/dup (nine cases each) from 536 cases tested at our institution using next-generation sequencing (NGS) assays and performed comprehensive molecular profiling of these cases. The molecular findings were correlated with clinical information and morphology. PD-L1 expression and survival data were also evaluated.
2 Methods
2.1 Case selection
This study was approved by the Institutional Review Board. Pathology archives were searched for NSCLC cases that had undergone NGS testing between July 2014 and January 2023. Five hundred thirty-six cases were identified, of which 18 were positive for either EGFR or ERRB2 ex20 ins/dup. For EGFR/ERBB2 ex20 ins/dup-positive cases, additional clinical information was obtained by chart review including patient’s age, gender, smoking history, histologic subtypes, tumor stage, PD-L1 status, progression-free survival (PFS) (time from diagnosis to first metastasis), treatment received, and survival time after initial diagnosis.
Light smokers are those with a smoking history of 1 to 20 packs per year.
2.2 Extraction of nucleic acid
Each case was reviewed by two pathologists, and the optimal formalin-fixed, paraffin-embedded (FFPE) block was selected for testing. The minimal percentage of tumor content was 10%. One hematoxylin and eosin (H&E)–stained slide along with 10 unstained sections (6 µm in thickness) was cut. Areas of interest were circled on the H&E slide, and corresponding areas from the unstained slides were manually scraped using a razor blade. After deparaffinization with xylene and ethanol wash of the pellet, the total nucleic acid was extracted using the RNeasy FFPE mini kit (Qiagen, Valencia, CA) excluding the DNAase treatment step. The concentration of the DNA and RNA was determined using Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA).
2.3 Next-generation sequencing analysis
A custom-designed DNA-based 214-gene NGS panel that covers the full coding sequence of 94 genes and hotspot regions of 120 genes was used for the detection of single-nucleotide variant (SNV), small deletion/duplication, copy number variants (CNV) in 49 genes, and microsatellite instability (MSI) status. DNA (40 ng) was used to generate NGS libraries, and sequencing was performed on the NextSeq (Illumina, Inc., San Diego, CA). Data were analyzed using the Burrows-Wheeler Aligner (BWA) and the Pisces variant caller v.2.1. This assay has a limit of detection of 2.5% for SNV and 6.8% for insertions/deletions. A copy number ratio of 1.9 and 0.5 combined with a z-score ≥ 5.0 was considered as a gene copy gain and loss, respectively.
The RNA-based Comprehensive Thyroid and Lung (CTL) FusionPlex Assay (IDT Technologies, Inc., Coralville, IA) was used for the detection of gene fusions following the manufacturer’s protocol. Briefly, total RNA (250 ng) was reverse-transcribed to cDNA, which was subsequently processed with end repair and dA tailing, followed by ligation with a half-functional adapter that allows amplification from the gene-specific primer (GSP) in one direction only. Two rounds of PCR were performed using GSPs for target enrichment. Libraries were pooled, typically eight at a time, in equimolar concentrations and sequenced using the MiSeq instrument (Illumina Inc., San Diego, CA). A denatured PhiX library was added to each run as a sequencing quality control. The sequence data were analyzed using the CTL Target Region File and vendor-supplied software (Archer Analysis versions 5.0 and 6.0). A minimum of five reads with three or more unique sequencing start sites that cross the breakpoints were set as the cutoff to call for strong evidence of fusions. The Target Mutation file (a text file in variant caller format (VCF) that lists the specific variants of interest) was created and used for targeted variant analysis.
2.4 Immunohistochemistry studies for PD-L1
PD-L1 immunohistochemistry (IHC) study was performed using the standard protocols in the clinical laboratory. Slides (3 μm in thickness) were deparaffinized, followed by rehydration, and heat-induced epitope retrieval in an ethylenediaminetetraacetic acid (EDTA) buffer at pH 9.0. Antibody 22C3 or E1L3N clones were used. Tumor proportion score (TPS) and immune cell (IC) staining score were used for 22C3 clone, and tumor cell (TC) (4) and IC staining score were used for the E1L3N clone. For TCs and TPSs, >1% was considered low positive, ≥50% was considered positive, and ≥5% tumor area occupied by ICs was considered positive.
3 Results
3.1 Patients characteristics
A total of 536 NSCLCs were tested; among them 18 (3.3%) harbored EGFR or ERBB2 ex20 ins/dup. There were an equal number of men and women. The mean age at the time of diagnosis was 63 years and ranged from 41 to 83 years. There were 15 Caucasian (88%, n = 17), two African Americans, and one patient with unknown ethnicity. Fourteen patients were never (nine) or light (five) smokers, three were heavy smokers, and one had an unknown smoking history. Among the 15 patients with available treatment information, seven received chemoradiation therapy; one each had chemotherapy or radiation therapy only, and one did not receive any treatment. Six patients received pembrolizumab: five with chemotherapy, and one with radiation therapy. Two patients were treated with afatinib. One patient received both mobocertinib and amivantamab. Three patients had unknown treatment history.
With an average follow-up time of 24 months (ranging from 0 to 69 months) after initial diagnosis, 17 of the 18 patients developed distant metastases (stage IV). One patient had mediastinal lymphadenopathy but no known distant metastasis after 12 months (Case #8). The most frequent site of metastasis was the brain (9 of 17; 52.9%) followed by the bone and intrapulmonary site (each 7 of 17; 41.1%). Additional sites of metastasis, which were biopsy-proven or clinically suspected by imaging, include the lymph node, liver, and adrenal gland (Table 1).
3.2 EGFR/ERBB2 ex20 ins/dup variants and co-occurring genomic alternations
The variants detected by the 214-gene NGS panel from 536 patients are summarized in Figure 1. Seventy-nine patients had EGFR or ERBB2 aberrations; among them 18 patients had ex20 ins/dup (Figure 1A). The most frequently mutated gene was TP53, followed by KMT2D and KRAS (Figure 1B). Genes that had the most frequent pathogenic/likely pathogenic variants were TP53, KRAS, and EGFR (Figure 1C).
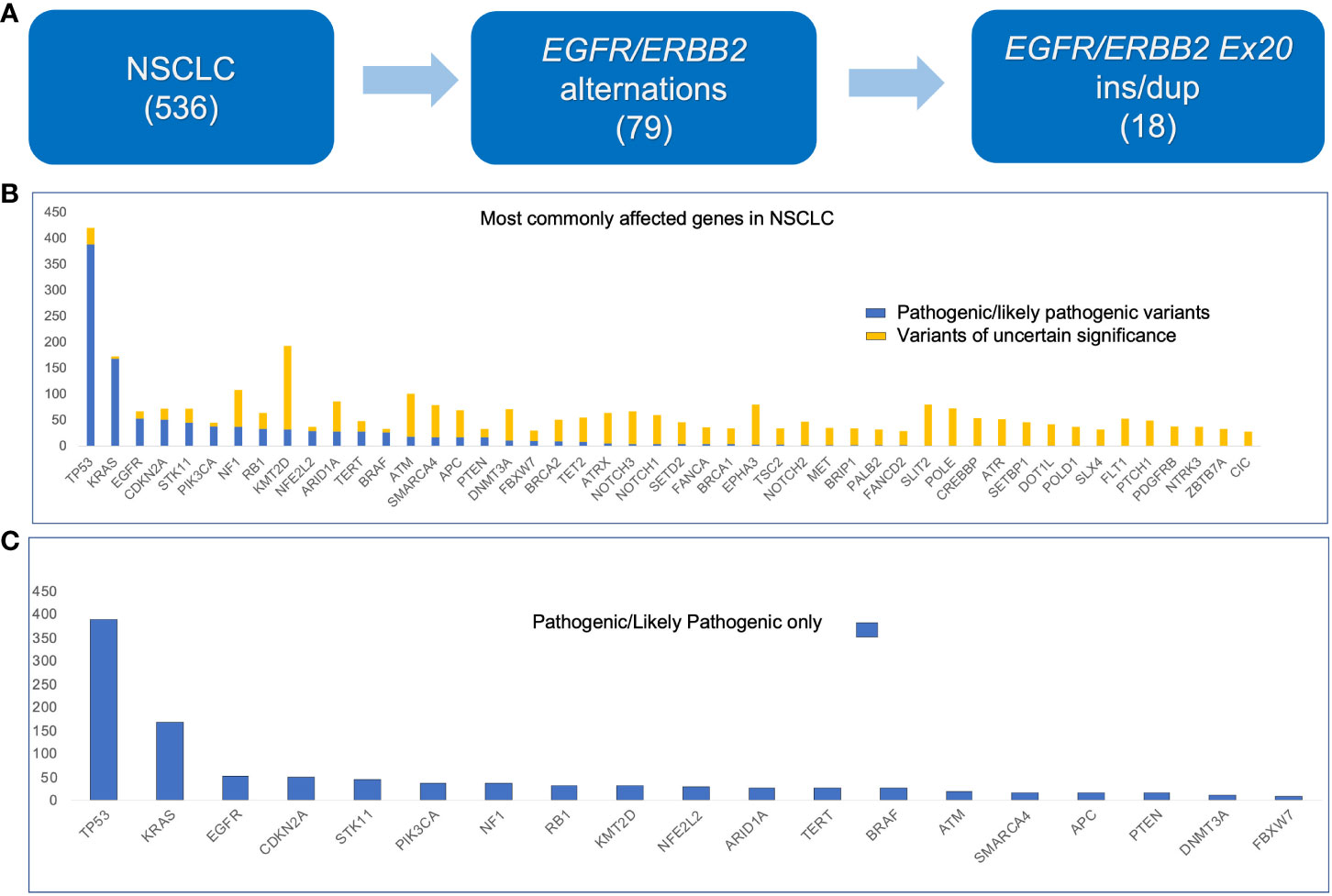
Figure 1 Histogram showing genes with mutations detected in non–small cell lung carcinoma (536 cases) using the 214-gene targeted panel. (A) Number of samples used in this study. (B) All variants. (C) Pathogenic and likely pathogenic variants only.
Nine of each EGFR and ERBB2 ex20 ins/dup variants were identified, which were heterogenous in-frame insertion/duplication of one to four amino acids spanning A767−V774 in EGFR and Y772−P780 in ERBB2. All the EFGR ex20 ins/dup variants were clustered in the loop following the alpha C-helix. Seven of the nine (77.8%) ERBB2 ex20 dup were duplication of four amino acids (p.Y772_A775dup) in the alpha C-helix domain, and only two (p.A775_G776 insSVMA and p.G778_P780Y) were located in the loop following the alpha C-helix domain (Figure 2).
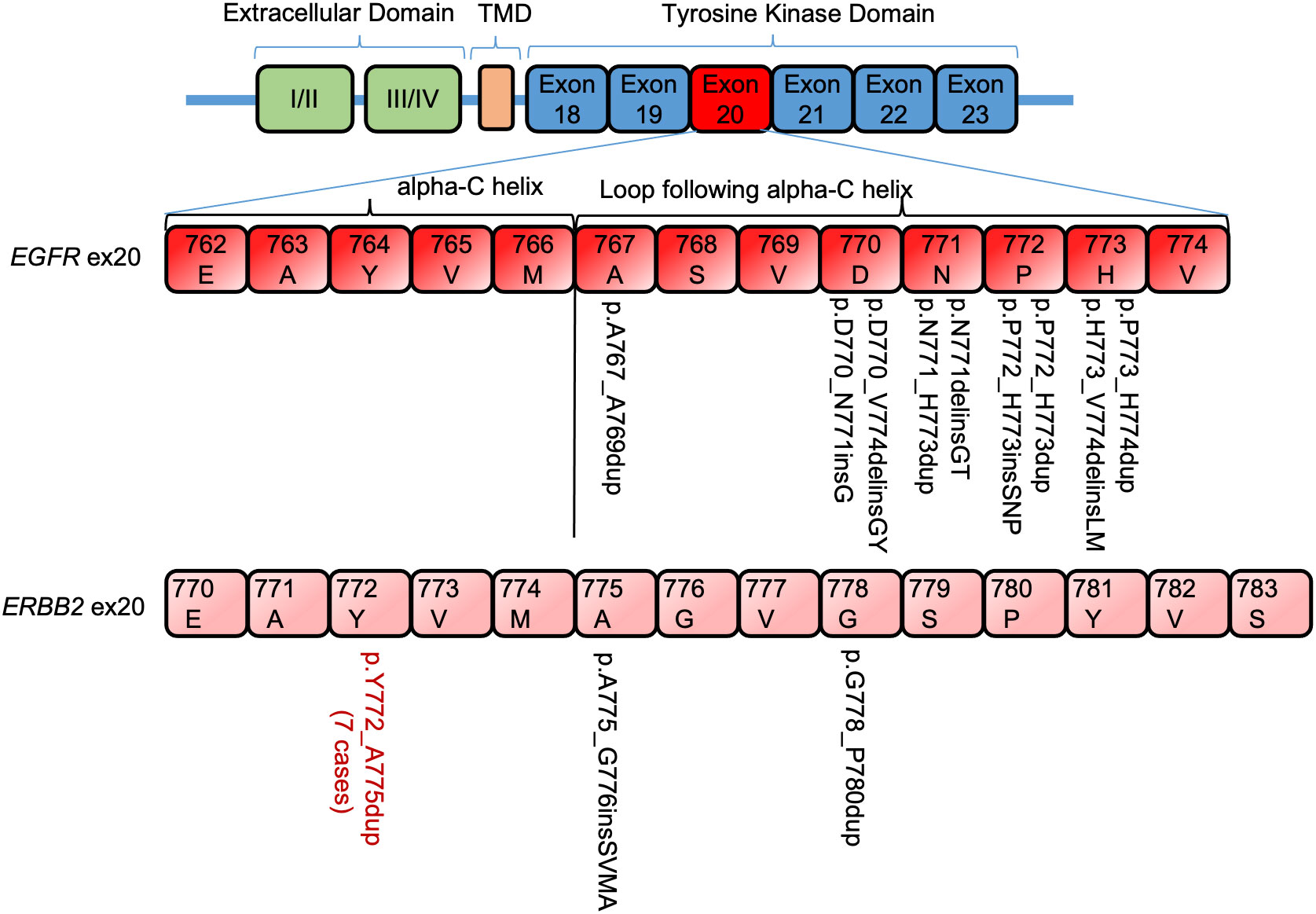
Figure 2 Distribution of EGFR/ERBB2 exon 20 insertion/duplication variants in non–small cell lung carcinoma. TMD, transmembrane domain.
Co-existing variants were identified in 13 (72.2%) EGFR/ERBB2 ex20 ins/dup-positive cases. TP53 mutations were the most common co-occurring variants (12 of 18, 67%), followed by RB1 (3 of 18, 17%). Pathogenic/likely pathogenic variants in PIK3CA, PTEN, and CDKN2A were detected in one case each. In five cases, EGFR/ERBB2 ex20 ins/dup was the only pathogenic variant. Only Case #9 had a low level of CDK4 amplification. All other cases had no CNV. No fusion transcript was detected in any cases, and all cases were microsatellite stable (Table 2).
Tumor mutation burden (TMB) information was available for four patients who had molecular tests performed at other facilities. Of these, three cases had low TMB (<10 mutations/mega bases). The only case that had likely high TMB (16 mutations/mega bases, Case #17) was from a patient who was a non-smoker and had negative TPS and IC scores in the tumor.
3.3 Molecular and morphologic correlations
Primary tumors were available in 11 EGFR/ERBB2 ex20 ins/dup-positive cases. Seven cases (64%) had an acinar predominant growth pattern, two (18%) were of the lepidic predominant pattern, and one each had a papillary or mucinous predominant pattern (9%).
3.4 PD-L1 immunohistochemistry and correlation with molecular findings
PD-L1 IHC was performed on all ex20 ins/dup-positive cases. TPS and IC were positive in two cases (including one with low positive TPS) and negative in six cases (22C3 clone). TC was positive in one case, low positive in three cases, and negative in six cases. All cases were IC negative (E1L3N clone) (Table 1). Among the cases with positive TPS or TC scores, only the one patient with a high TC score was treated with ICI (pembrolizumab) in addition to radiotherapy. This patient had an OS of 1 month (Case #5). Mobocertinib and amivantamab were used as part of the treatment regime for Patient #7, and the patient is still alive 35 months after initial diagnosis.
4 Discussion
In-frame insertion/duplication in EGFR ex20 comprises 4%–12% of all EGFR mutations in NSCLC (14–16) and represents a distinct subset of EGFR-mutated NSCLC. Ex20 ins/dup confers resistance to the conventional EGFR TKIs as well as other commonly available immunotherapies (17) and is associated with poor prognosis. The FDA approval of mobocertinib and amivantamab has changed the course for patients whose tumors harbor EGFR/ERBB2 ex20 ins/dup. However, because of the low frequency of ex20 ins/dup in NSCLC, this subgroup of tumors has not been well characterized. Here, we presented findings from the comprehensive genomic profiling of 18 EGFR and ERBB2 ex20 ins/dup-positive NSCLCs including co-existing pathogenic/pathogenic variants, CNV, MSI, and PD-L1 status and correlation of the genomic findings with clinicopathologic features.
Similar to the classic EGFR variants, EGFR ex20 ins/dup was found more common in Asian women (18), non- or light smokers, and adenocarcinomas (2). In our cohort, 15 patients were Caucasian and two were African American. No Asian ethnicity was identified. All 18 EGFR/ERBB2 ex20 ins/dup-positive cases were adenocarcinomas and most frequently had the acinar predominant growth pattern (64%, n = 11). Other growth patterns were also identified in ex20 ins/dup-positive cases including two lepidic and one each papillary and mucinous predominant pattern. Fourteen of the 18 patients (78%) were either non-smokers (50%) or light smokers (28%). The most common metastatic site in our patients was the brain (52.9%, n = 18) followed by the lung and bone (33% each), similar to the previous reports (5). In our study, the distribution of EGFR/ERBB2 ex20 ins/dup variants showed no gender preference. The predominance in Caucasian patients could be due to the bias in our patient population.
EGFR and ERBB2 ex20 ins/dup was each identified in 1.7% of 536 NSCLCs analyzed, similar to the incidence reported in the literature (4, 7). EGFR ex20 ins/dup is known to be mostly in-frame insertion/duplication of one to seven amino acids between codons A767 and V774 (8). The V769_D770 and D770_N771 were the two most commonly affected locations and together accounted for approximately 40% of all ex20 ins/dup (4). All the EGFR ex20 ins/dup identified in our patients were one to three amino acids ins/dup and located in A767–V774 (Table 2). These variants had notable heterogeneity, and each case had a different ex20 ins/dup variant. The p.P772_H773insSNP detected in a male non-smoker patient is novel (Cosmic Database). p.D770delinsGY was reported to have a more favorable response to EGFR TKIs (19). This variant was detected in Patient #3, a 41-year-old man who presented with stage IV disease. He did not receive any EGFR-specific TKI and was deceased 31 months after diagnosis.
ERBB2 and EGFR both belong to the ErbB family of receptor tyrosine kinase. The structural analog of ex20 ins/dup that promotes ligand-independent activation of EGFR signaling pathway has also been shown in ERBB2 (14, 19). Similar to the classic EGFR TKIs, trastuzumab did not show any clinical benefit against NSCLCs with ERBB2 ex20 ins/dup (20). ERBB2 ex20 ins/dup, although uncommon, represents the most common ERBB2 mutations in NSCLC (14). The most common ERBB2 ex20 ins/dup was reported to be the p.A775_G776insYVMA in the loop following the alpha C-helix domain (3). Only one patient in our cohort had a variant in this location (p.A775_G776 insSVMA), another case had a variant in the adjacent codon (p.G778_P780Y), and the remaining seven (78%, n = 9) were duplication of four amino acids (p.Y772_A775dup) in the alpha C-helix domain (Figure 2).
It has been determined that the mechanism underlying the TKI resistance of EGFR ex20 ins/dup is through inducing structural changes (21). Ex20 ins/dup variants are clustered in the alpha C-helix and the P-loop of the EGFR protein that are the key regulatory regions for EGFR activation status (22). Structural alterations in these regions prevent binding of the reversible TKIs to EGFR protein resulting in resistance. Mobocertinib irreversibly binds to EGFR with exon 20 insertion mutation, preventing EGFR-mediated signaling and leading to cell death. ERBB2 ex20 ins/dup was targeted similarly (10, 21, 23, 24). The correlation of different variants with metastasis, prognosis, and therapy response was controversial (8, 15, 25).
Almost all EGFR/ERBB2 ex20 ins/dup variants were mutually exclusive with other known oncogenic drivers in NSCLC such as KRAS, ALK, or ROS1 fusions (2). Co-occurring mutations have been reported in NSCLCs with EGFR ex20 ins/dup and predominantly were alternations in tumor suppressors such as TP53 (up to 65%) and RB1 (11%) and cell cycle inhibitors (cyclin-dependent kinase inhibitor 2A and 2B, 22% and 16%, respectively) (4). Five of the 18 patients with EGFR/ERBB2 ex20 ins/dup had no other mutations providing more evidence that EGFR/ERBB2 ex20 ins/dup was the oncogenic driver in this molecular subtype of NSCLC. Mutations in TP53 (67%) and RB1 (17%) were the most frequently co-occurring aberrations in our cohort of ex20 ins/dup-positive cases. PTEN and CDKN2A mutation was identified in one case each. TP53 co-mutation decreased the EGFR TKI efficacy in patients with non-ex20 ins/dup mutated NSCLC (26). The effect of TP53/RB1 mutation in ex20 ins/dup-positive tumors is uncertain, but the high incidence of co-existing mutations in tumor suppressors may contribute to chemoradiation resistance and poor prognosis of this subgroup of NSCLC.
Riess et al. (2) reported a high incidence of co-occurring EGFR amplification in EGFR ex20 ins/dup-positive NSCLCs (58 of 263, 22%). In our cohort, only one case had a low level CDK4 amplification (Case #9). The discrepancy in EGFR CNV could be due to the relatively small number of cases in our study.
Immune checkpoint inhibitors have been shown to be ineffective against NSCLC with EGFR ex20 ins (2, 4). The study performed by Reiss et al. (2) was the largest and likely the only study that evaluated TMB in EGFR ex20 ins/dup-positive cases. They found that EGFR ex20 ins-positive NSCLCs with high TMB, which is more common in smoking-associated cases, had a higher response rate to ICIs, potentially due to increased T-cell activity against neoantigens generated by tumor mutations. Less than 4% (n = 263) of their cases had intermediate TMB and only 0.7% (2/263) had high TMB (2). Four of our patients had TMB information available (three low and one likely high). Our patients showed a limited response to ICI even when there was positive PD-L1. One patient (Case #17) has a likely high TMB. The patient received therapy including ICI and had an OS of 9 months. Another patient who had an NSCLC with high TC score was treated with ICI (pembrolizumab) in addition to radiotherapy and had an OS of 1 month (Case #5). It seems that TMB status and the high TC score did not provide significant benefits for these two patients. Large-scale studies are needed to further evaluate immunotherapy response in EGFR/ERBB2 ex20 ins/dup-positive patients.
Ex20 ins/dup is known to be associated with a worse prognosis compared to other EGFR mutations. Seventeen of the 18 patients in our study developed stage IV disease during follow-up (average 24 months). We only had 17 patients (one patient with no clinical information available), and some of the patients are still alive. The small number of patients may not generate meaningful/reliable survival data due to limited statistical power. Therefore, a larger cohort, The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center (MSKCC) data, was used for survival analysis. Although both ex19 del and p.L858R are associated with better outcome in comparison to ex20 ins/dup, further stratification of the two classic variants showed that patients with NSCLC harboring the p.L858R variant had worse OS compared to patients with ex19 del (27–29). We therefore compared the differences between NSCLC cases with EGFR/ERBB2 ex20 ins/dup and p.L858R mutation using the TCGA and MSKCC data in cBioPortal. Of 2,743 patients with NSCLC, 573 had EGFR or ERBB2 alterations; of which 73 have EGFR/ERBB ex20 ins/dup. Survival analyses showed that patients with EGFR/ERBB2 ex20 ins/dup had a poor OS compared to patients with EGFR p.L858R mutation (p = 0.017). The median OS of patients with ex20 ins/dup was 17.7 months, whereas, in patients with EGFR p.L858R and other variants, the OS was 37.7 and 35.4 months, respectively. Ex20 ins/dup was also associated with decreased PSF that was 2.33 months in patients with ex20 ins/dup, 2.63 months in patients with p.L858R, and 3.5 months in patients with other EGFR/ERBB2 mutations (p = 0.057) (Figure 3). The lack of statistical significance in PFS could be due to the small number of patients in each group.

Figure 3 Comparison of overall survival and progression-free survival curves in patients with NSCLC with EGFR/ERBB2 p.L858R, ex20 ins/dup, and other EGFR/ERBB2 aberrations using TCGA and MSKCC data in cBioPortal. (A) Overall survival. (B) Progression-free survival.
In conclusion, we performed extensive studies of NSCLCs harboring EGFR/ERBB2 ex20 ins/dup. No ethnicity or gender preference was observed. Ex20 ins/dup was frequently associated with non- or light smoking history, adenocarcinomas with acinar predominant growth pattern, negativity for PD-L1, and advanced disease. Our data also suggested that ex20 ins/dup could be sufficient to drive oncogenesis in this molecular subset of NSCLC. Given the notable heterogeneity of these variants and the high rate of co-occurring TP53/RB1 mutations, additional studies are needed to evaluate the correlation of different EGFR/ERBB2 ex20 ins/dup variants and co-mutation with targeted therapy. The possible development of resistant mutation after mobocertinib treatment also warrants further investigation.
Data availability statement
The datasets presented in this article are not readily available because of privacy restrictions. Requests to access the datasets should be directed to the corresponding author.
Ethics statement
The studies involving human/animal participants were reviewed and approved by the Institutional Review Board on Human Subjects Office of the University of Iowa. Written informed consent from the patients was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
RS performed data search/collection and analysis, put Figures 1 and 3 together, and revised the manuscript. BD participated in the data collection and drafting of the manuscript, performed chart review, and put the tables together. NG provided technique support for next-generation sequencing and put Figure 2 together. ADB provided critical review and edits of the manuscript. DM designed the project and participated in drafting, revising, and finalizing the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank all the members of the Molecular Pathology Laboratory at University of Iowa Hospitals and Clinics for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res (2015) 5(9):2892–911.
2. Riess JW, Gandara DR, Frampton GM, Madison R, Peled N, Bufill JA, et al. Diverse EGFR exon 20 insertions and Co-occurring molecular alterations identified by comprehensive genomic profiling of NSCLC. J Thorac Oncol (2018) 13(10):1560–8. doi: 10.1016/j.jtho.2018.06.019
3. Wang Y, Jiang T, Qin Z, Jiang J, Wang Q, Yang S, et al. HER2 exon 20 insertions in non-small-cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann Oncol (2019) 30(3):447–55. doi: 10.1093/annonc/mdy542
4. Leal JL, Alexander M, Itchins M, Wright GM, Kao S, Hughes BGM, et al. EGFR exon 20 insertion mutations: clinicopathological characteristics and treatment outcomes in advanced non-small cell lung cancer. Clin Lung Cancer. (2021) 22(6):e859–e69. doi: 10.1016/j.cllc.2021.04.009
5. Fang W, Huang Y, Hong S, Zhang Z, Wang M, Gan J, et al. EGFR exon 20 insertion mutations and response to osimertinib in non-small-cell lung cancer. BMC Cancer. (2019) 19(1):595. doi: 10.1186/s12885-019-5820-0
6. Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol (2001) 2(2):127–37. doi: 10.1038/35052073
7. Liu Z, Wu L, Cao J, Yang Z, Zhou C, Cao L, et al. Clinical characterization of ERBB2 exon 20 insertions and heterogeneity of outcomes responding to afatinib in Chinese lung cancer patients. Onco Targets Ther (2018) 11:7323–31. doi: 10.2147/OTT.S173391
8. Remon J, Hendriks LEL, Cardona AF, Besse B. EGFR exon 20 insertions in advanced non-small cell lung cancer: a new history begins. Cancer Treat Rev (2020) 90:102105. doi: 10.1016/j.ctrv.2020.102105
9. Zeng Y, Yu D, Tian W, Wu F. Resistance mechanisms to osimertinib and emerging therapeutic strategies in nonsmall cell lung cancer. Curr Opin Oncol (2022) 34(1):54–65. doi: 10.1097/CCO.0000000000000805
10. Harrison PT, Vyse S, Huang PH. Rare epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer. Semin Cancer Biol (2020) 61:167–79. doi: 10.1016/j.semcancer.2019.09.015
11. Riely GJ, Ahn MJ, Felip E, Ramalingam SS, Smit EF, Tsao AS, et al. Encorafenib plus binimetinib in patients with BRAF(V600)-mutant non-small cell lung cancer: phase II PHAROS study design. Future Oncol (2022) 18(7):781–91. doi: 10.2217/fon-2021-1250
12. Riely GJ, Neal JW, Camidge DR, Spira AI, Piotrowska Z, Costa DB, et al. Activity and safety of mobocertinib (TAK-788) in previously treated non-small cell lung cancer with EGFR exon 20 insertion mutations from a phase I/II trial. Cancer Discovery (2021) 11(7):1688–99. doi: 10.1158/2159-8290.CD-20-1598
13. Yun J, Lee SH, Kim SY, Jeong SY, Kim JH, Pyo KH, et al. Antitumor activity of amivantamab (JNJ-61186372), an EGFR-MET bispecific antibody, in diverse models of EGFR exon 20 insertion-driven NSCLC. Cancer Discovery (2020) 10(8):1194–209. doi: 10.1158/2159-8290.CD-20-0116
14. Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res (2012) 18(18):4910–8. doi: 10.1158/1078-0432.CCR-12-0912
15. Yang G, Li J, Xu H, Yang Y, Yang L, Xu F, et al. EGFR exon 20 insertion mutations in Chinese advanced non-small cell lung cancer patients: molecular heterogeneity and treatment outcome from nationwide real-world study. Lung Cancer. (2020) 145:186–94. doi: 10.1016/j.lungcan.2020.03.014
16. Oxnard GR, Lo PC, Nishino M, Dahlberg SE, Lindeman NI, Butaney M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol (2013) 8(2):179–84. doi: 10.1097/JTO.0b013e3182779d18
17. Wang J, Lam D, Yang J, Hu L. Discovery of mobocertinib, a new irreversible tyrosine kinase inhibitor indicated for the treatment of non-small-cell lung cancer harboring EGFR exon 20 insertion mutations. Med Chem Res (2022) 31(10):1647–62. doi: 10.1007/s00044-022-02952-5
18. Byeon S, Kim Y, Lim SW, Cho JH, Park S, Lee J, et al. Clinical outcomes of EGFR exon 20 insertion mutations in advanced non-small cell lung cancer in Korea. Cancer Res Treat (2019) 51(2):623–31. doi: 10.4143/crt.2018.151
19. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res (2004) 64(24):8919–23. doi: 10.1158/0008-5472.CAN-04-2818
20. Halliday PR, Blakely CM, Bivona TG. Emerging targeted therapies for the treatment of non-small cell lung cancer. Curr Oncol Rep (2019) 21(3):21. doi: 10.1007/s11912-019-0770-x
21. Yasuda H, Park E, Yun CH, Sng NJ, Lucena-Araujo AR, Yeo WL, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med (2013) 5(216):216ra177. doi: 10.1126/scitranslmed.3007205
22. Tamirat MZ, Kurppa KJ, Elenius K, Johnson MS. Structural basis for the functional changes by EGFR exon 20 insertion mutations. Cancers (Basel). (2021) 13(5). doi: 10.3390/cancers13051120
23. Vyse S, Huang PH. Targeting EGFR exon 20 insertion mutations in non-small cell lung cancer. Signal Transduct Target Ther (2019) 4:5. doi: 10.1038/s41392-019-0038-9
24. Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med (2018) 24(5):638–46. doi: 10.1038/s41591-018-0007-9
25. Cardona AF, Rojas L, Zatarain-Barron ZL, Freitas HC, Granados ST, Castillo O, et al. EGFR exon 20 insertion in lung adenocarcinomas among hispanics (geno1. 2-CLICaP). Lung Cancer (2018) 125:265–72. doi: 10.1016/j.lungcan.2018.10.007
26. Aggarwal C, Davis CW, Mick R, Thompson JC, Ahmed S, Jeffries S, et al. Influence of TP53 mutation on survival in patients with advanced EGFR-mutant non-Small-Cell lung cancer. JCO Precis Oncol (2018) 2018. doi: 10.1200/PO.18.00107
27. Gijtenbeek RGP, Damhuis RAM, van der Wekken AJ, Hendriks LEL, Groen HJM, van Geffen WH. Overall survival in advanced epidermal growth factor receptor mutated non-small cell lung cancer using different tyrosine kinase inhibitors in the Netherlands: a retrospective, nationwide registry study. Lancet Reg Health Eur (2023) 27:100592. doi: 10.1016/j.lanepe.2023.100592
28. Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res (2006) 12(3 Pt 1):839–44. doi: 10.1158/1078-0432.CCR-05-1846
29. Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res (2006) 12(13):3908–14. doi: 10.1158/1078-0432.CCR-06-0462
Keywords: EGFR/ERBB2 exon20 insertion/duplication, non-small cell lung cancer, ERBB2, clinicopathologic features, PD-L1
Citation: Sompallae RR, Dundar B, Guseva NV, Bossler AD and Ma D (2023) EGFR and ERBB2 exon 20 insertion/duplication in advanced non–small cell lung cancer: genomic profiling and clinicopathologic features. Front. Oncol. 13:1163485. doi: 10.3389/fonc.2023.1163485
Received: 10 February 2023; Accepted: 02 May 2023;
Published: 22 May 2023.
Edited by:
Giorgio Scagliotti, University of Torino, ItalyReviewed by:
Prabhat Singh Malik, All India Institute of Medical Sciences, IndiaHushan Zhang, Fudan University, China
Copyright © 2023 Sompallae, Dundar, Guseva, Bossler and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deqin Ma, ZGVxaW4tbWFAdWlvd2EuZWR1
†Present address: Aaron D. Bossler
Department of Molecular Pathology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, United States
‡These authors have contributed equally to this work
 Ramakrishna R. Sompallae
Ramakrishna R. Sompallae Bilge Dundar
Bilge Dundar Natalya V. Guseva
Natalya V. Guseva Aaron D. Bossler
Aaron D. Bossler Deqin Ma
Deqin Ma