- 1Department of Radiation Oncology, Dr. B. R. Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, India
- 2Integrated translational Molecular Biology laboratory, Department of Rog Nidan and Vikriti vigyan (Pathology), All India Institute of Ayurveda (AIIA), New Delhi, India
- 3Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India
- 4Department of Kaya Chikitsa (Internal Medicine), All India Institute of Ayurveda (AIIA), New Delhi, India
With a high mortality rate that accounts for millions of cancer-related deaths each year, breast cancer is the second most common malignancy in women. Chemotherapy has significant potential in the prevention and spreading of breast cancer; however, drug resistance often hinders therapy in breast cancer patients. The identification and the use of novel molecular biomarkers, which can predict response to chemotherapy, might lead to tailoring breast cancer treatment. In this context, accumulating research has reported microRNAs (miRNAs) as potential biomarkers for early cancer detection, and are conducive to designing a more specific treatment plan by helping analyze drug resistance and sensitivity in breast cancer treatment. In this review, miRNAs are discussed in two alternative ways-as tumor suppressors to be used in miRNA replacement therapy to reduce oncogenesis and as oncomirs to lessen the translation of the target miRNA. Different miRNAs like miR-638, miR-17, miR-20b, miR-342, miR-484, miR-21, miR-24, miR-27, miR-23 and miR-200 are involved in the regulation of chemoresistance through diverse genetic targets. For instance, tumor-suppressing miRNAs like miR-342, miR-16, miR-214, and miR-128 and tumor-promoting miRNAs like miR101 and miR-106-25 cluster regulate the cell cycle, apoptosis, epithelial to mesenchymal transition and other pathways to impart breast cancer drug resistance. Hence, in this review, we have discussed the significance of miRNA biomarkers that could assist in providing novel therapeutic targets to overcome potential chemotherapy resistance to systemic therapy and further facilitate the design of tailored therapy for enhanced efficacy against breast cancer.
Highlights
1. Drug resistance is the major obstacle in breast cancer chemotherapy
2. miRNA biomarkers could provide novel therapeutic targets to overcome potential chemotherapy resistance to systemic therapy in breast cancer.
3. Targeting miRNAs—either reducing or raising their expression—seems to be an attractive approach for designing novel, more effective, and personalized treatments for breast cancer.
4. A novel approach to treating breast cancer that combines miRNA therapies with conventional chemotherapeutic techniques and drug targets is possible.
1 Introduction
The most common malignancy worldwide is breast cancer (BC). According to the status update on the GLOBOCAN 2020 projections of cancer incidence and mortality, BC is the primary cause of cancer death in women and accounts for 1 in 4 cancer diagnoses among females (1). An estimated 684,996 people died from breast cancer in 2020, with low-resource areas accounting for a disproportionate share of these deaths. According to statistics, the prevalence of BC ranges from 2-6% in Western nations to 10-20% in Asian nations (2), indicating that BC is becoming a global health concern, even in nations with sizable young populations like India. Although breast cancer diagnoses have increased recently (3), the prognosis for the disease has significantly improved, with expected 5-year survival rates rising from 40% to approximately 90% over the last 50 years. With few exceptions, en-bloc radical resections in the form of Halstead mastectomy and axillary clearing were formerly thought to be essential for managing BC (4). Recent advancements in clinical trials have been brought about by a greater understanding of the molecular processes associated with the heterogeneity of breast tumors. This understanding has allowed for more conservative surgical procedures and the personalization of treatment plans to maximize sensitization to the tumor while minimizing unneeded morbidity to the patient. This includes the era of cancer diagnostics, which has recognized BC as a diverse disease and routinely subcategorized these cancers into four genetically distinct, integral subgroups - luminal A breast cancer (LABC), luminal B breast cancer (LBBC), human epidermal growth factor receptor-2 enriched breast cancer (HER2+) and triple-negative breast cancer (TNBC). These subgroups have different clinical behavior, prognosis, treatment approaches, and clinical outcomes in the known treatments (5).
For BC patients, chemotherapy is regarded as the most successful and crucial therapeutic approach. Anthracyclins, Tamoxifen, Taxane, 5-FU and trastuzumab are the major chemotherapeutic drugs which are administered to BC patients (6–12). Doxorubicin, daunorubicin, and epirubicin are some of the anthracycline antibiotics that are frequently used. Anthracyclines can be administered at all BC stages and have demonstrated a crucial function in treating BC (8). Tamoxifen is specifically used for the oestrogen receptor (ER) positive subtype of BC (9). Anthracyclines and taxanes are used as the predominant treatment for TNBC (10). The two most widely used taxanes are- paclitaxel and docetaxel, causing acute hypersensitivity responses (HSRs) in 5% to 10% of patients (11). The human epidermal growth factor receptor 2 (HER-2) is frequently used to categorize BC patients based on its overexpression (also known as HER-2 positive) or lack of expression (also known as HER-2 negative) (13). The likelihood of BC metastases and poor prognosis are strongly correlated with HER-2 overexpression (13). A targeted therapy for HER-2 is trastuzumab (TRS), a humanized monoclonal antibody (12). Despite our efforts to categorize tumors into prognostic categories, tumor behavior and prognosis remains unpredictable, which makes it challenging to develop strategies that would improve disease control while minimizing toxicities to patients. Although, a better understanding of the disease has led to advancements in treatment over the past few decades, but drug resistance remains a challenge and the underlying molecular causes are still largely undefined (14). Drug-resistant cancer cells multiply rapidly and grow more hostile, increasing the likelihood that the tumor may aggressively spread to other organs. Drug resistance can be categorized in two different ways. One is internal resistance or inherited resistance, which occurs when tumors are resistant to treatment even before receiving it, meaning that even early detection and treatment are ineffective. Another form of resistance is received resistance or acquired resistance which occurs following an initial positive response to the therapy (15). Here, the targets and processes associated are a focus of significant research, and the mechanisms of such drug resistance are largely still under investigation (16). For instance, Martz et al. (2014) demonstrated that stimulation of the Notch-1, mitogen-activated protein kinase (RAS-MAPK), phosphoinositide 3-kinase (PI3K) and mammalian target of rapamycin (mTOR), PI3K/AKT and estrogen receptor (ER) signaling pathways resulted in resistance to a variety of drugs (17). It was observed that when Notch-1 is activated, BRAF (V600E) melanoma cells develop acquired resistance to MAPK inhibitors and breast cancer cells also exhibit resistance to tamoxifen (17). Hence, the research group used a Notch-1 inhibitor to restore sensitivity, indicating that Notch-1 knockdown could be a therapeutic strategy in melanomas and drug-resistant breast malignancies (17). Likewise, it seems, resistance to chemotherapy is also related to the epidermal growth factor receptor (EGFR) pathway. Genetically modified murine model (GEMM), human cell lines, and a clinically applicable model of KRAS-mutant colorectal cancer (CRC) have all been used to study EGFR and PI3K/mTOR (18). According to the evidence, PI3K/mTOR and EGFR inhibition boost drug sensitivity and are increasingly used in cancer therapy to combat drug resistance. Additionally, the use of systemic drugs as neoadjuvant enables the production of in-vivo data on tumor sensitivity, which has been shown to have predictive importance for disease survival and recurrence. These contemporary aspects of traditional breast cancer management shed light on the potential value of emerging biomarkers in advancing the current treatment model. There are currently few biomarkers that may reliably predict response and resistance to systemic and targeted therapy and attempts to use non-invasive approaches to collect such biomarkers have largely been ineffective (19). This highlights how important it is for researchers to find new biomarkers that can assess patient response to therapy, predict the prognosis of breast cancer patients, and offer clinicians cutting-edge oncogenesis-targeting therapeutic approaches. In the context of BC chemoresistance monitoring and systemic therapy, this study focuses on the function of microRNA (miRNA) as new clinical biomarkers.
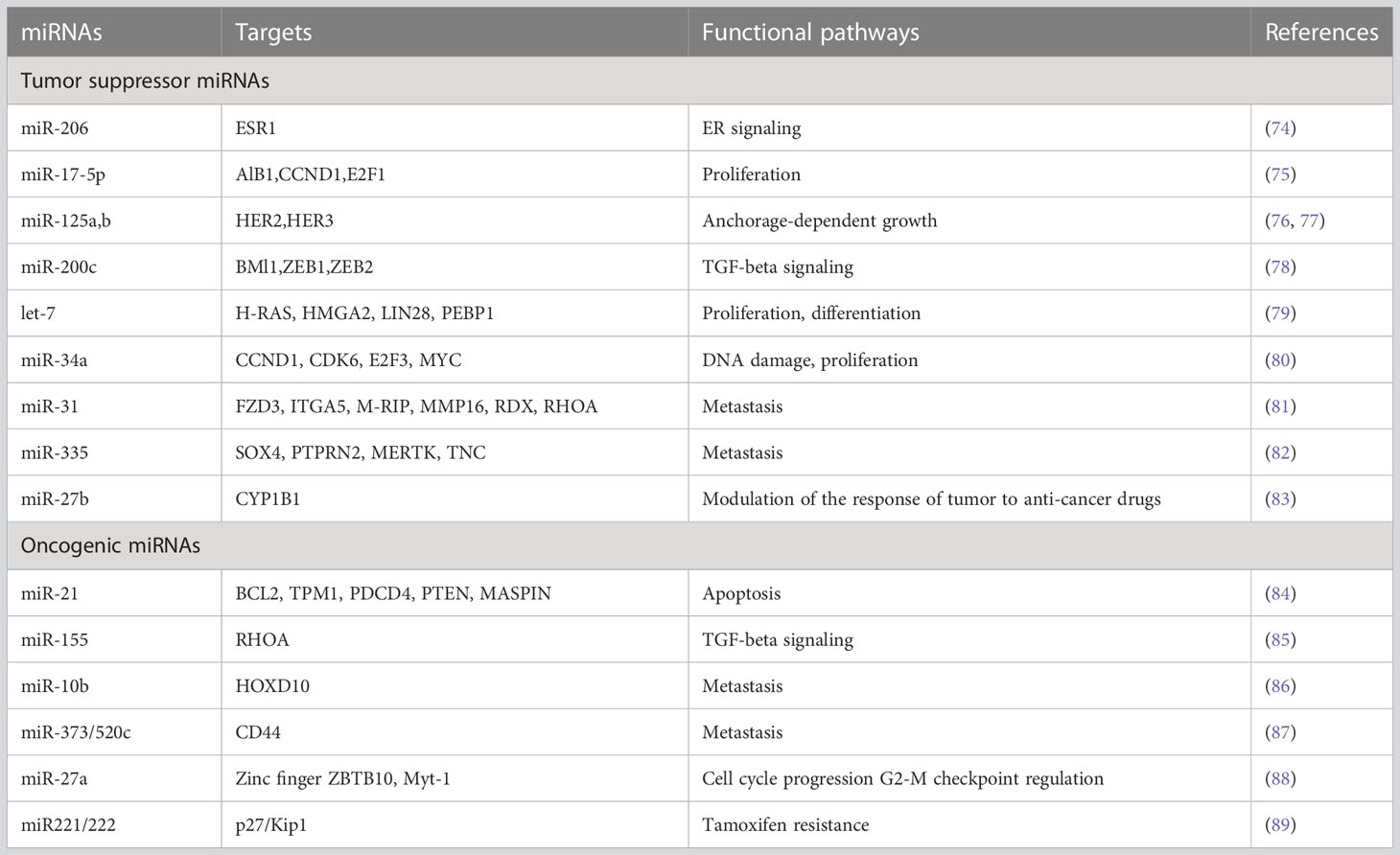
miRNAs are small non-coding RNAs ranging from 19 to 25 nucleotides in size and are involved in a variety of biological activities, including cell cycle, apoptosis, survival, and gene control (20). miRNAs primarily bind to the 3′ or 5′ untranslated region (UTR) of their target mRNAs and, depending on the degree of binding, participate in controlling the translation of proteins or destruction of the mRNA itself (21). A single miRNA may target several mRNAs, while many miRNAs may target single mRNA with varying degrees of efficiency (22). Therefore, changes in miRNA expression levels and gene expression silencing by miRNAs have a significant impact on human health and the emergence of diseases such as cancer, diabetes, neurological disease, and cardiovascular disorders (23–25). In the context of cancer, miRNAs can function as both tumor suppressors and oncogenes/oncomirs (26). In contrast to their counterparts in normal tissue, many miRNAs are reported to be up-or down-regulated in cancer tissues. For instance, practically all cancer types have increased miR-21 expression (27). Numerous B-cell malignancies have been shown to express miR-155 at high levels (28). One of the first miRNAs to be found was let-7 which is essentially missing throughout embryonic stages or tissues, although it is highly expressed in the majority of differentiated tissues (29). Similar to the fall in let-7 expression during development, the decline in let-7 expression in malignancies is more pronounced in cancer cells that are more advanced, less differentiated and have mesenchymal features (29). The generation, biology and function of miRNAs in cancer have been discussed in detail in further sections.
This review focuses primarily on the latest findings about the involvement of miRNAs in breast tumor resistance to chemotherapeutic agents and in the development of systemic therapy. Targeting miRNAs—either reducing or raising their expression—seems to be an attractive approach for designing novel, more effective, and personalized treatments for BC. Boosting drug efficacy by examining the downstream targets/pathways influenced by miRNA targeting and predicting patient response to various therapies can lead to better treatment outcomes for BC patients.
2 Breast cancer chemotherapy
Breast cancer bears 7% of the total number of cancers related deaths in 2020. To date, many strategies have been adapted to combat this disease. Complete surgical removal has usually enabled efficient breast cancer disease management (30). Regardless of the severity of the disease, William Halstead’s radical mastectomy (which required significant removal of all the breast parenchyma, local lymph nodes, and pectoralis major muscle) used to be the cornerstone of breast cancer treatment (4). Cyclophosphamide, methotrexate, and 5-fluorouracil (CMF), the first chemotherapeutic treatment prescribed by Bonadanno et al. in 1976 with the intention of curing breast cancer, significantly decreased breast cancer relapse (94.7% of 207 patients administered chemotherapy vs 76.0% of 179 patients constrained chemotherapy) (31). However, since 1950s, Bernard Fisher and the National Surgical Adjuvant Breast and Bowel Project (NSABP) have hypothesized that aggressive surgery for breast cancer has only limited scientific and biomolecular justification because it is frequently insufficient to achieve complete disease control (32). Fisher’s theory that all breast cancer patients needed systemic therapy (especially with chemotherapy) has, however, been thoroughly refuted.
However, the inherent advantage of treating cancer patients with chemotherapy in the neoadjuvant setting has now been recognized as an oncological practice. Neoadjuvant chemotherapy (NAC) benefits comprised tumor downstaging, expanding patient suitability for breast conservation surgery (BCS), and producing in vivo data related to tumor resistance, which has been shown to hold predictive value for cancer recurrence and overall survival (OS) (33, 34). The Early Breast Cancer Triallist’s Collaborative Group (EBCTCG) recently published data from a meta-analysis of randomized clinical trials showing that locoregional recurrence (LRR) rates are higher following neoadjuvant therapy (21.4% vs. 15.9%), despite the fact that disease-free survival (DFS) and overall survival (OS) results are parallel with those treated in the adjuvant setting (35). Additionally, there is growing proof that people who have a pathological complete response (pCR) with NAC have a higher chance of living longer than those who have a latent disease (34, 36). Nevertheless, the clinical usefulness of NAC has been integrated into best-practice guidelines for HER2+ and Triple-negative breast cancer (TNBC). HER2+ malignancies should be treated with NAC and trastuzumab, with the exception of T1a-T1b N0 disease (37). High-risk LN (lymph node)- patients and those with LN positivity should receive anthracycline- and taxane-based chemotherapy along with trastuzumab (37). Further, until TNBC is identified with cancer stages T1a-T1b N0, patients with TNBC should always be provided with an anthracycline and taxane-based treatment (37). American Society of Clinical Oncology (ASCO) also supports the inclusion of platinum-based chemotherapy in TNBC based on the results of a recent meta-analysis because of a higher likelihood to obtain pCR (52.1% versus 37.0%) (38). Pembrolizumab and NAC significantly increased the pCR rates in the KEYNOTE522 trial’s preliminary findings (pembrolizumab and NAC: 64.8% versus placebo and NAC: 51.2%) (39).
3 Need for miRNA-based therapy
The idea of improving pCR rates, simplifying the de-escalation of adjuvant therapy post pCR, and minimising treatment-related toxicities for patients receiving these neoadjuvant medicines are the main directions for translational research efforts in the future (40). Therefore, numerous clinical trials have focused on practices that improve pCR rates (40). To further improve the pCR, the idea of manipulating treatment with miRNA-based therapies may be helpful in boosting pCR rates to NAC in breast cancer is now popular, and the same has been discussed in depth in this review.
4 Chemoresistance in breast cancer
Various molecular aspects are known to be involved in inducing chemoresistance in cancer cells (Figure 1). Some of them have also been summarized below:
♦ Resistant genes:
4.1 Twist
Twist is a key player in the invasion and metastasis of tumors because it regulates the epithelial-mesenchymal transition (EMT) (41). It has been reported that NF-κB up-regulation of twist-1 is a factor in the chemoresistance (42). Through the downregulation of estrogen receptor alpha (ERα) activity, twist overexpression can also contribute to hormone resistance in breast tumors (43).
4.2 Multidrug resistance gene
MDR1 has a significant impact on breast cancer’s chemoresistance. P-glyocoprotein (P-gp), glutathione S-transferase (GST-π), and P53 are only a few examples of MDR1-encoded proteins that are involved in the chemoresistance cascade (44). In fact, Chen et al., 2022 suggested the synergistic effect of 7-O-geranylquercetin and miR-451 on undoing MDR-1 and P-gp-mediated chemoresistance in breast cancer MCF-7/ADR (Adriamycin) cells (45).
♦ Efflux proteins: Another mechanism of resistance to chemotherapy is mediated by ATP-dependent efflux pumps, which decrease the intracellular concentration of drugs. By using the energy from ATP hydrolysis, or the MDR phenomenon, the ATP-dependent efflux transporters in cancer cells can actively transport a range of substrates outside the cell membrane (46). MDR-associated proteins, such as P-gp, multidrug resistance-associated protein (MRP), ABCC subfamily, and breast cancer resistance protein (BCRP) are examples of ATP-binding cassette (ABC) transporters (47). Downregulation of ABCC4 with miR-124-3p overexpression significantly enhanced the ADR sensitivity in MCF7/ADR cells (48), highlighting the importance of miRNA-based targeting of resistant proteins for improved treatment of breast tumors that are resistant to certain drugs.
♦ Signaling pathways: Resistance to endocrine therapy for breast cancers can be brought on by cell surface receptors like epidermal growth factor receptor (EGFR), human epidermal growth factor receptor-2 (HER-2), and insulin-like growth factor 1 receptor (IGF-1R), and their downstream signaling pathways like PI3K/AKT/mTOR, RAS/MAPK/ERK, and upstream signaling pathways (49). Also, tamoxifen resistance has been linked to EGFR, HER-2/nue, PI3K, and other growth factor signaling pathways (50). Additionally, one of the gefitinib resistance mechanisms in breast cancer may involve EGFR nuclear translocation (51).
♦ Response to DNA damage: Many chemotherapeutic medications work to treat cancer by causing DNA damage (52). Single and double-strand breaks, intra- and inter-strand DNA crosslinks, methylated and oxidized bases, mismatched and protein-DNA adducts are several types of DNA damage. Cancer stem cells (CSCs), in particular, stimulate DNA damage repair (DDR) pathways to counteract DNA damage, explaining why chemotherapy that damages DNA might cause drug resistance (53). The following DDR pathways are found in breast cancer cells: homologous recombination (HR) (54) and non-homologous end joining (NHEJ) (55), involved in the elimination of double-strand breaks; mismatch repair (MMR), in charge of removing incorrect bases (56); nucleotide excision repair (NER) (57) and base excision repair (BER) (58), involved in the repair of single-strand breaks. In addition, these pathways contain a variety of components that are involved in the repair procedure, including the DNA repair genes - Excision Repair Cross-Complementation group (ERCC1/2/5) (59, 60), the BRCA1/2 (61), the Meiotic Recombination 11 (MRE11) and RAD50/51 (60, 62), and others. Understanding DNA repair mechanisms can aid in reversing breast cancer resistance to therapy.
♦ Cancer Stem Cells (CSCs): Chemoresistance in breast cancer is significantly influenced by cancer stem cells (63). CSCs overexpress several ABC transporters, such as ABCB5, ABCC1, ABCG2, and P-gp. Additionally, CSCs can cause drug resistance through increased anti-apoptotic levels and DNA repair activity (64). Another significant factor contributing to CSCs’ drug resistance is Aldehyde dehydrogenase 1 (ALDH1) upregulation (65). Drug resistance of CSCs may also be influenced by signal transduction pathways such as Notch, Hedgehog, and Wnt/-catenin which are involved in the self-renewal and maintenance of stem cells (66). Expression of Let-7, a tumor suppressor miRNA, was shown to be considerably reduced in breast cancer stem cells compared to non-cancer stem cells. However, the upregulation of let-7 microRNA in breast cancer stem cells can encourage CSCs to enter the differentiation phase from the stationary phase, and hence increases the sensitivity of CSCs to the chemotherapeutics (67).
♦ Drug detoxification: Cell detoxification proteins such as ALDH, DNA topoisomerase, protein kinase C, dihydrofolate reductase, glutathione (68) and glutathione S-transferases (GST) are some of the key enzymes that contribute to MDR in cancer cells. These agents have the potential to enhance the transformation and catabolism of anti-neoplastic drugs, shorten the term of effective concentration of chemotherapeutic drugs in tumor cells, decrease drug accumulation in target areas, and ultimately limit drug efficacy (69). For example, GST-π can be employed as a separate index to direct a clinical treatment against BC as its expression in breast cancer patients was associated with the histological grade, the number of lymphatic metastases, and the age of the patients (70).
5 MicroRNAs
MicroRNAs are a class of small noncoding RNAs (ncRNAs), which function in post-transcriptional regulation of gene expression and are powerful regulators of various cellular activities and have been linked to many diseases (20). RNA polymerase II (Pol II), which produces the main transcripts, participates in several stages of microRNA synthesis (pri-miRNA). The pri-miRNA are split up into precursor miRNAs (pre-miRNAs) by the RNase III Drosha (71). The pre-miRNAs are subsequently moved from the nucleus into the cytoplasm by Exportin-5 (Exp5), where they are further split by Dicer into a mature single-stranded miRNA. The miRNA is induced to either degrade or suppress the translation of mRNA targets when the mature miRNA is removed from the pre-miRNA hairpin and attached to the RNA-induced silencing complex (RISC) (72). Figure 2 depicts a pictorial representation of miRNA biogenesis and function. Besides miRNAs, other ncRNAs are long-chain non-coding RNAs (LncRNAs), piRNAs, and circle RNAs (circRNAs), which make up just 1% of the whole genome’s RNA (73).
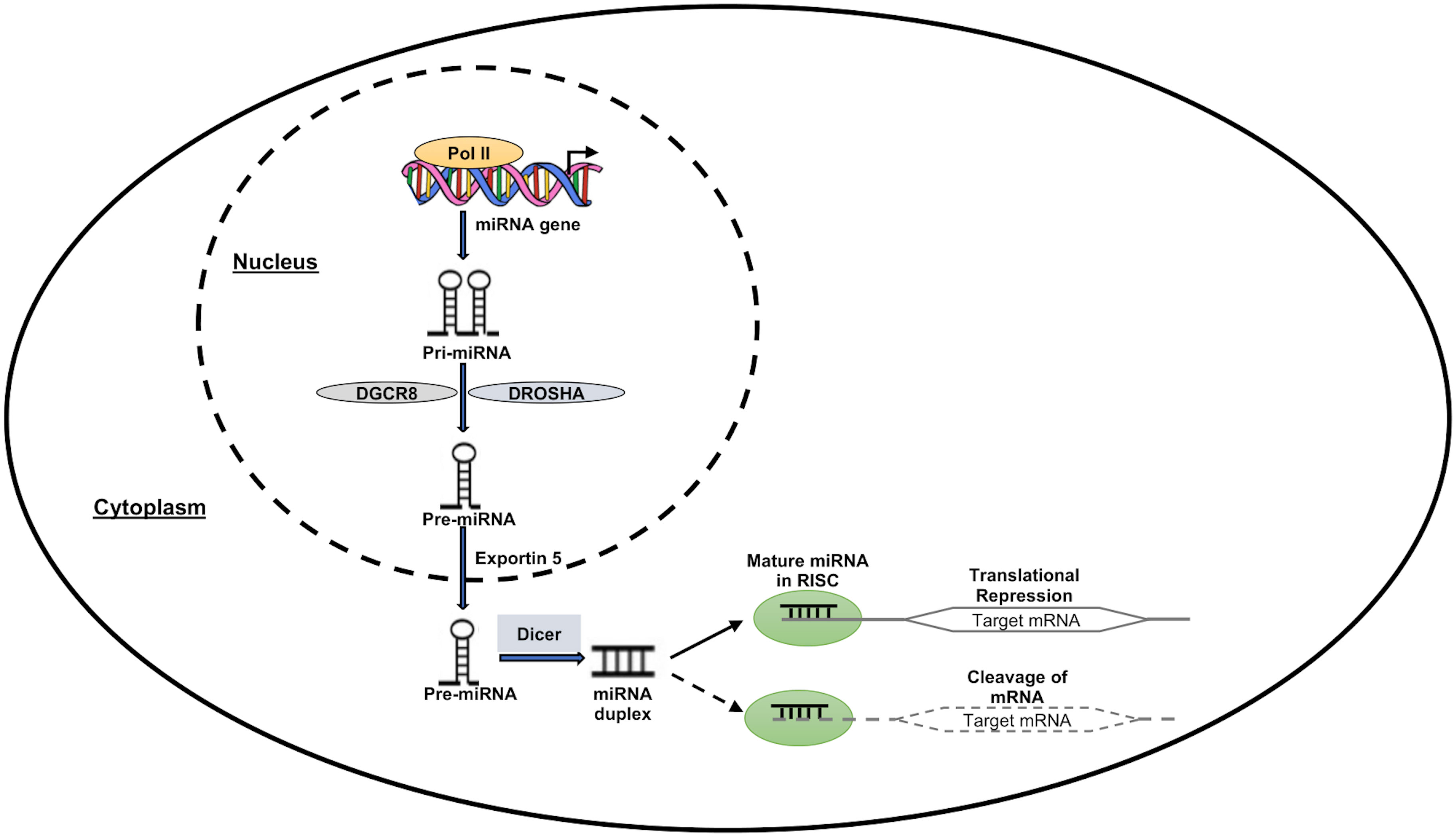
Figure 2 MiRNA expression and function. RNA polymerase II enzyme in the nucleus transcribes the miRNA-encoding genes, forming the “Pri-miRNA” hairpin-shaped molecule. DROSHA and DGCR8 molecules work together to transform the “Pri-miRNA” molecule into the “Pre-miRNA” precursor molecule. Pre-miRNA then travels to the nuclear export receptor “Exportin 5” and reaches the cytoplasm. This precursor is cleaved by Dicer complex in the cytoplasm to create a double-stranded molecule called “miRNA duplex.” One of these two strands is left active after this process, and it has the ability to suppress or even activate the target downstream genes at the transcriptional or translational level.
The molecular revolution enables us to design strategies to maximize patient outcomes, reduce toxicity, and control disease with less strenuous and more focused therapies. The development of chemotherapeutic response biomarkers is necessary in the future to speed up the removal of tumors and reduce the need for extended and excessive treatments. The utility of detecting miRNA expression (both in tumors and in the blood) is now being discussed in the scientific community. Doing so may help doctors prescribe medicines that are suitably targeted, address early relapse, or even enable miRNA-directed therapies. In general, miRNAs can be either tumor suppressors (tumor suppressor miRNA) or oncomirs, and they can affect the development of cancers in either way. Numerous miRNAs, along with their downstream targets, have been shown to be differentially expressed in breast cancer patients when compared to healthy controls (either circulating or in tumors) (Table 1).
6 miRNAs in BC subtyping
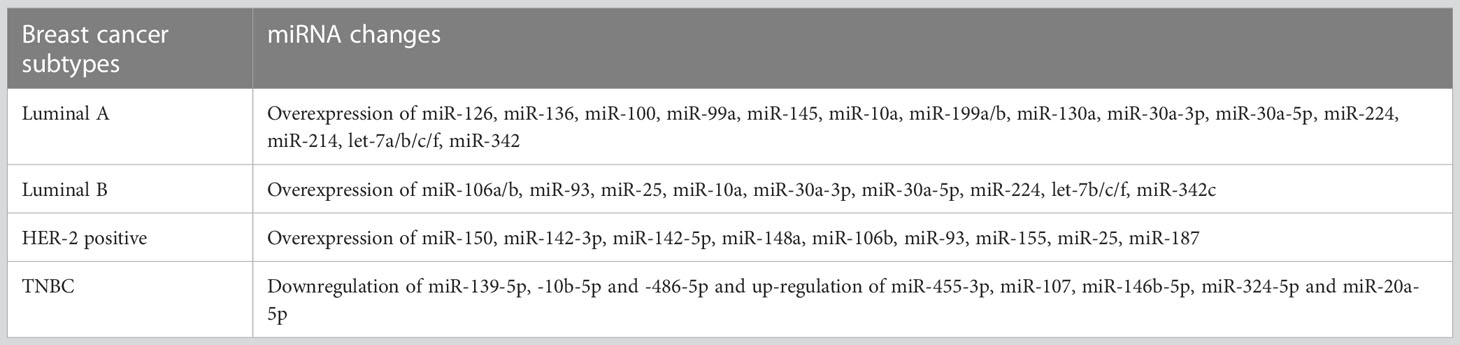
The three main subtypes of breast cancer that exist are (1) Positive ER and PR (2); HER-2 positive; and (3) (ER, PR, and HER-2 negative) Triple negative. However, this subtyping is expanded to a more precise one using the microarray approach for identifying miRNA profiles, including (1):Luminal A (ER-positive with low grade) (2); Luminal B (ER-positive with high grade) (3); HER-2 positive (4); Basal-like (Almost equal to triple-negative condition).
There are several miRNAs that are used for the breast cancer subgrouping as shown in Table 2 (87). Currently, it is possible to use miRNA profiling for the subgrouping of breast tumors. This capability can therefore aid in the selection of cancer patients who will get adjuvant therapy. In addition, miRNA profiling can be successful in identifying new therapeutic targets by revealing the genetic underpinnings of distinct subgroups of breast cancer.
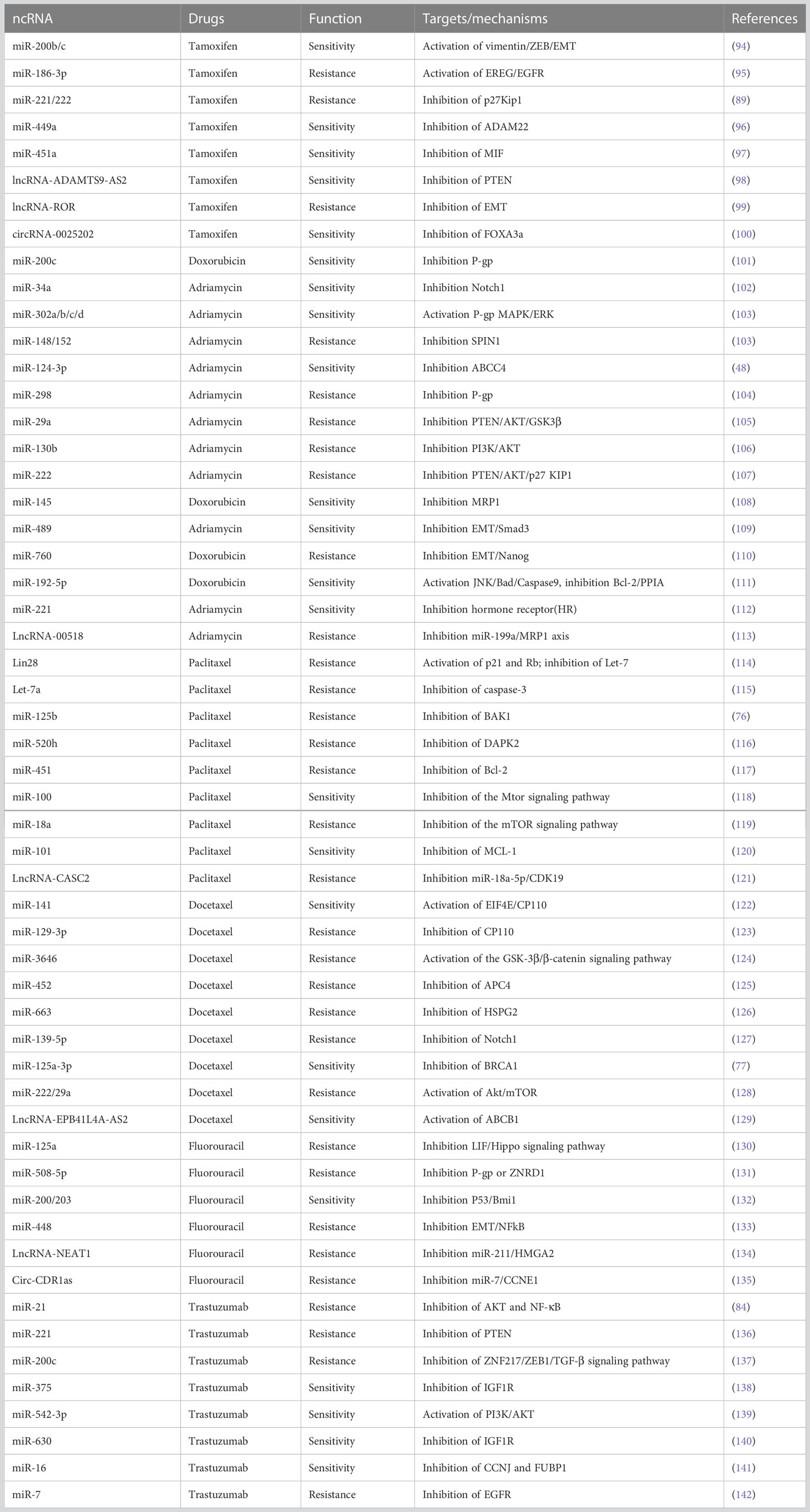
7 Role of miRNA in BC drug resistance
It is well known that miRNAs can regulate drug resistance to traditional chemotherapeutic medicines, endocrine hormone treatments, and radiotherapies in cancer cells (90–93). It has been shown that miRNA expression can influence a breast cancer patient’s ability to respond to or reject systemic treatment as shown in Table 3. The miRNAs along with other ncRNAs significantly reverse the BC cell drug resistance by suppressing signaling pathways such as Wnt/β-catenin, Hippo, AKT, TGF-β, or mTOR signaling pathways. A summary of molecules which can participate in target diversity in miRNA interactions leading to drug resistance using different chemotherapeutic drugs is mentioned in Table 3. According to various reports, there are scientific explanations and processes for chemotherapeutic resistance, including altered drug-target interactions, lower active drug doses, and increased tumor tissue survival (143). Numerous miRNA expression profiles have been linked to the prediction of chemoresistance and their regulatory role in influencing chemoresistance to chemotherapeutics. For example, reduced expression of miR-18a, miR-1207-5p, and miR-5195-3p in TNBC has recently been linked to translational research studies that predict resistance to paclitaxel or docetaxel in TNBC (144, 145). Similarly, Wu et al., 2019 discovered that by downregulating the expression of dCMP deaminase (DCTD) in TNBC, upregulation of miR-620 improves tumor resistance to gemcitabine-based chemotherapies (146). Further, finding higher levels of circulating miR-125b in 56 patients with invasive ductal carcinoma receiving curative treatment was associated with chemoresistance (p = 0.008) (147). Hypoxia-inducible factor-1 (HIF-1) pathway-dependent upregulation of cell resilience to hypoxia and inhibition of chemotherapy-induced apoptosis are two mechanisms through which miR-24 has been demonstrated to promote chemoresistance in early breast cancer (148). miR-155 has shown to be linked to drug resistance and cancer development (149) via modulation of FOXO3a signaling, the interruption of TGF-beta, and the induction of drug resistance through RhoA signaling. Likewise, in 25 breast cancer samples, miR-221 has been shown to alter the PTEN/Akt/mTOR signaling pathway, which promotes breast cancer resistance to Adriamycin (150).
The role of miRNAs in BC chemoresistance has been attributed to some of the following molecular mechanisms (Figure 3):
♦ miRNAs and cell cycle: Cell cycle deregulation is an established hallmark of cancer, and it has been linked to both drug resistance and poor prognosis when it is aberrantly activated. Various miRNAs have been reported to target genes linked to cell cycle regulation, resulting in either drug sensitivity or resistance such as miR-93, involved in G1/S phase arrest, was reported to be downregulated in paclitaxel-resistant BC samples compared to responder patients (Figure 3A) (151). Direct targets of this miRNA were discovered to include CCND1 and the E2F transcription factor 1 (E2F1), which upon downregulation resulted in cell cycle arrest in G1 phase and increased apoptosis via inhibiting AKT phosphorylation (p-AKT), and BCL-2 expression, and increasing the expression levels of BCL-2-associated X, apoptosis regulator (BAX), which could increase the paclitaxel sensitivity.
♦ miRNAs and DNA repair machinery: As mentioned above, most chemotherapy drugs used today to treat breast cancer cause either direct or indirect DNA damage. To counteract DNA damage, however, CSCs activate DDR pathways, explaining why chemotherapy that destroys DNA could result in drug resistance (Figure 3B). One such DDR pathway involves BRCA1 which is engaged in different cellular processes that maintain genomic stability like DNA damage repair, DNA damage-induced cell cycle checkpoint activation, chromatin remodelling, protein ubiquitination, transcriptional control, and cell death [43]. Drug sensitivity is thus impacted by its miRNA influenced control, for instance miR-182 inhibits BRCA1 expression to induce drug sensitization. Furthermore, it has been shown that overexpressing miR-182 makes BC cells more susceptible to PARPi (PADP-ribose polymerase 1 inhibitor). In contrast, miR-182 suppression raises BRCA1 levels and results in PARPi resistance (152).
♦ miRNAs and cell death: The interests of investigators are growing in drug-miRNA combination anticancer therapy since miRNAs can influence cell death (Figure 3C). Examples include miR-125b, which confers paclitaxel resistance by inhibiting the expression of BAK1 (BAX and/or BCL-2 Antagonist/Killer 1), which causes the release of cytochrome C from mitochondria to the cytoplasm, where it binds Apoptotic peptidase Activating Factor 1 (APAF-1) and triggers caspase activation (76). Similar findings were also made from miR-149-5p, whose overexpression was shown to boost BAX expression (153), and from miR-663b, which imparts tamoxifen resistance by indirectly upregulating BAX (154).
♦ miRNAs, CSCs and epithelial to mesenchymal transition (EMT): Breast cancer stem cells (BCSCs) are a small population of cells with a high ability for tumorigenesis and are involved in therapy resistance (155). The modulation of BCSCs’ phenotype is mediated by several molecular mechanisms, the most significant of which is EMT. This process takes place as cancer develops, and it involves a decrease in the expression of molecules associated with epithelial growth, such as E-cadherin, and a rise in molecules associated with mesenchymal development, such as N-cadherin, vimentin (VIM), and fibronectin (FN1) (156). Thus, the cells become more capable of invasion and migration (155) and can nest in various tissues where they can multiply and create new tumors through a process called metastasis (155). In this context, miRNAs play a significant role in controlling stemness and EMT by targeting a few genes implicated in these two pathways (Figure 3D). Among those engaged in the control of EMT, the miR-200 family has received the greatest research attention. Five different miRNAs make up this family: miR-141, miR200a, miR200b, miR200c, and miR-429 (157), which can inhibit the expression of ZEB1 and ZEB2 (Zinc finger E-Box-binding homeobox genes) (157). As a result, it has been demonstrated that overexpressing miR-200 in many cancer cell lines can reverse EMT (158). Another factor contributing to stemness in BC is the Wnt/-catenin signaling pathway. It has been shown that several miRNAs, including miR-105 and miR-93-3p, regulate this pathway. The Wnt/-catenin signaling pathway suppressor Secreted Frizzled Related Protein 1 (SPFR1) is the target of those miRNAs. This led Li et al. to show that those miRNAs encourage cisplatin resistance (159).
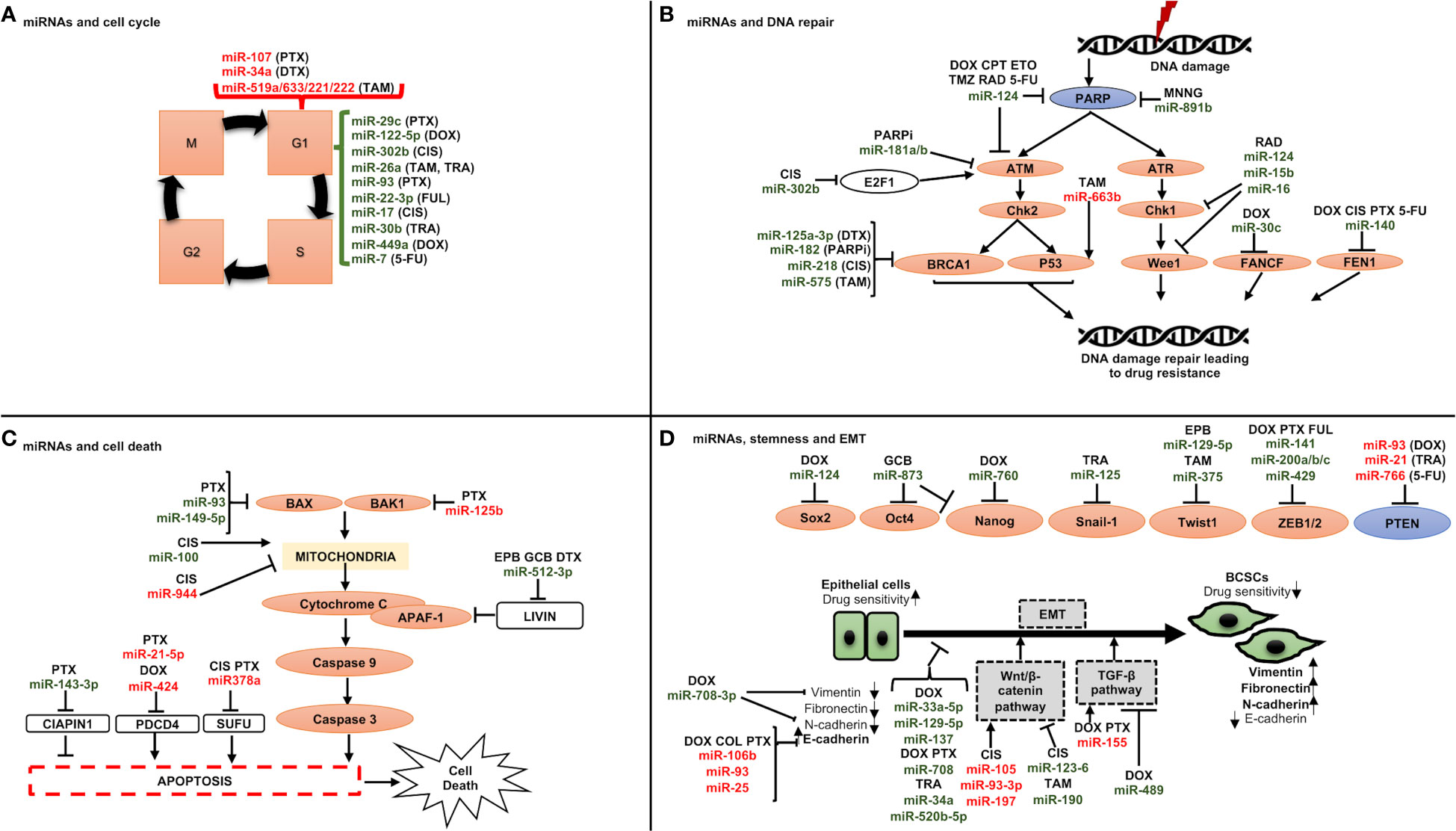
Figure 3 Schematic representation of miRNAs involved in drug resistance through regulating (A) cell cycle, (B) DNA repair checkpoints, (C) cell death, and (D) stemness and epithelial to mesenchymal transition. A line with a perpendicular line at the end designates inhibition and arrows designate activation. miRNAs increasing drug resistance are shown in red, and miRNAs increasing drug sensitivity are shown in green. CIS, cisplatin; DOX, doxorubicin; DTX, docetaxel; FUL, fulvestrant; PTX, paclitaxel; TAM, tamoxifen; TRA, trastuzumab; 5-FU, 5-fuorouracil; CPT, camptothecin; ETO, etoposide; MNNG, N-methylN′-nitro-N-nitrosoguanidine; PARPi, PARP inhibitors; RAD, radiation; TMZ, temozolomide; EPB, epirubicin; GCB, gemcitabine.
Four circulating miRNA patterns linked to pCR were recently identified using profiling of circulating miRNA (ct miRNA detected in plasma) to categorize NAC responders (from non-responders) in Her2+ patients (160). These results demonstrate the potential of miRNA signatures as prognostic and predictive biomarkers that could individualize breast cancer treatments and enhance patient sampling techniques for current therapies, including traditional cytotoxic chemotherapies. The following is a discussion of a few of them:
♦ miR-638 – miR-638 was shown to be downregulated in cases with BC chemoresistance in a microarray analysis (161). A minimal patient-derived xenograft (MiniPDXTM) was also developed by the researchers to assess the chemosensitivity of various drugs. The results of this study demonstrated that in patients, who received 5-FU, miR-638 levels were relatively low in the 5-FU-resistant group compared to the 5-FU-sensitive group. So, according to the MiniPDX™ model, MDA-MB-231 BC cells overexpressing miR-638 were more susceptible to 5-FU treatment in vivo.
♦ miR-17/20 – The serine-threonine kinase Akt1 has been linked to the regulation of cellular homeostasis, proliferation, and growth as well as hyperactivation in human malignancies (162). It is known that the miR-17/20 cluster blocks the proliferation of breast cancer cells by causing the G1/S cell cycle arrest by anchoring to the 3’UTR of cyclin D1 (163). Yu et al., 2014 demonstrated a unique mechanism by which miR17/20 controls p53 and Akt, which further control breast cancer cell apoptosis (163). Additionally, they have demonstrated that miR-17/20 overexpression, via Akt1, makes MCF-7 cells more susceptible to apoptosis brought on by either doxorubicin or UV irradiation. In brief, the apoptosis-inducing miR-17/20 increases Akt breakdown by upregulating p53 expression.
♦ miR-342 – The mesenchymal stem cell-derived exosome (MSCs-Exo) carrying microRNAs has been proven to regulate tumor biological activities (164, 165). Yu et al., 2022 demonstrated the regulatory function of miR-342-3p in MSCs-Exo on the BC (166). They revealed considerably decreased levels of miR-342-3p in patients with metastatic illnesses (166). Additionally, miR-342-3p was found to target the Inhibitor of Differentiation 4 (ID4) and operate as a possible tumor suppressor by preventing the metastasis and chemoresistance of breast cancer cells (166).
♦ miR-484 –By controlling cyclin E-CDK2 signaling, cytosine deaminase (CDA), a crucial chemoresistance axis, inhibits the advancement of the cell cycle (167). In a breast cancer model that is resistant to gemcitabine, miR-484 controls the CDA (167). CDA expression was found to be downregulated and inversely linked with miR-484 expression in clinical samples of BC (167). Additionally, in the same cohort, greater expression of CDA was linked to extended disease-free survival. Collectively, the findings of Ye et al., 2015 established that miR-484-modulated CDA promotes chemoresistance while inhibiting cell proliferation in breast cancer, highlighting the pathophysiological exchange that arises because of chemoresistance in this cancerous condition (167).
♦ miR-451 – Gu et al. (2016) looked at the possible use of miR-451, which is prevalent in the serum of BC patients, to forecast NAC resistance (168). Here, qRT-PCR was used to determine the expression levels of miR-451 in the MCF-7 BC cell line, the docetaxel-resistant MCF-7 BC cell line (MCF-7/DTX), the epirubicin-resistant MCF-7 BC cell line (MCF-7/EPB), normal controls, NAC-sensitive group, and NAC-resistant group. The findings of this investigation confirmed the hypothesis that miR-451 expression was differentially expressed between NAC-sensitive and -resistant BC patients. Additionally, the research team noticed that miR-451 expression was much lower in the MCF-7/EPB and MCF-7/DTX cell lines than it was in the MCF-7 cell lines, indicating that miR-451 may be functionally crucial in predicting NAC resistance in breast cancer patients.
♦ miR-222 and miR-29a – In a 2013 study by Zhong et al., it was discovered that the changed expression pattern of miR-222 and miR-29a contributed to the development of DTX and adriamycin (ADR) resistance in breast cancer MCF-7 cells (128). The research team found that targeting Phosphatase and TENsin homolog (PTEN) with miR-222 and miR-29a mimics and inhibitors partially altered the treatment resistance of breast cancer cells.
♦ miR-141 – About 50% of patients develop resistance to the chemotherapeutic medication docetaxel used to treat BC. The role of miR-141 expression in BC cells with acquired docetaxel resistance was studied by Yao et al., 2015 (122). Docetaxel-resistant cells (MCF-7/DTX and MDA-MB-231/DTX, respectively) were more responsive to the drug when miR-141 was inhibited, but docetaxel-sensitive cells were more resistant when miR-141 was overexpressed (MCF-7 and MDA-MB-231, respectively). This research team showed that miR-141 operates on genes required for drug-induced apoptosis, leaving the cells drug resistant, through direct interaction with eukaryotic translation initiation factor 4E (EIF4E).
♦ miR-140-5p – In a training set, conducted by Di et al., 2019, starting from 51 circulating (ct)-miRNAs linked with pCR, four signatures were validated in the testing set: lapatinib at T0 and T1 [AUC 0.86; 95% confidence interval (CI), 0.73–0.98 and 0.71 (0.55–0.86)], respectively; trastuzumab at T1 (0.81; 0.70–0.92); lapatinib + trastuzumab at T1 (0.67; 0.51–0.83) (160). Although the levels of ct-miR-140-5p, which is a component of the trastuzumab signature, were linked to EFS (HR 0.43; 95% CI, 0.22-0.84), ct-miRNA signatures could not predict event-free survival (EFS). Patients with and without pCR after neoadjuvant lapatinib- and/or trastuzumab-based therapy can be distinguished by ct-miRNAs. To help to de-escalate treatment plans, ct-miRNAs at week two may be useful in identifying individuals who respond to trastuzumab and preventing needless combinations with other anti-HER2 medications.
♦ miR-34a – A link between enhanced miR-34a expression and docetaxel resistance has also been established (80). Kastl et al., 2012 confirmed that B-cell leukemia/lymphoma 2 protein (BCL-2) and cyclin D1 protein (CCND1), both of which are targeted by miR-34a, were shown to be expressed at lower levels in docetaxel-resistant cells (80). It was found that overexpressing miR-34a resulted in resistance in MCF-7 docetaxel-sensitive cells, but miR-34a inhibition improved responsiveness to docetaxel in MCF-7 docetaxel-resistant cells. To propose a prospective therapeutic target for the treatment of docetaxel-resistant breast cancer, this work described a pathway of acquired docetaxel resistance in these cells, presumably involving direct interactions with BCL-2 and CCND1.
♦ miR-23, 24 and 27 – Recent research has shown that the extracted exosomes (D/exo) from the docetaxel-resistant breast cancer cells MCF-7 (MCF-7/Doc) were linked to the genetic cargo’s contribution to resistance transmission (169, 170). The significance of D/exo during exposure to DRβ-H (d Rhamnose -hederin), an active ingredient obtained from the traditional Chinese medicine plant Clematis ganpiniana, was discovered by Chen et al., 2018, in MCF-7/DTX cells (171). Herein, the investigators have found that DRβ-H could reduce the expression of a few of the most common miRNAs (miR-23a, miR-24, and miR-27a) transported by D/exo. Target gene prediction and pathway analysis showed the relevance of these selected miRNAs in pathways related to disease relapse.
♦ miR-200 –The miR-200 family of microRNAs have recently been revealed to be dysregulated in a variety of malignancies, and it has been shown that this family of miRNAs is crucial for tumor development, maintenance, tumor metastasis, and chemotherapy tolerance (172). MiR-200s are currently recognized as master EMT regulators, inhibiting cancer invasion and metastasis by focusing on a number of key inducers of the EMT, such as ZEB1, ZEB2, and SLUG (172). By playing critical and pleiotropic roles in malignancies, miR-200s are promising targets for cancer therapy. However, a recent study revealed that miR-200s play a role in breast cancer metastasis promotion, hence cautious evaluation should be done prior to treatment modalities using miR-200s as therapeutic targets (172).
8 miRNAs in neoadjuvant chemotherapies: predicting response
As already said, breast oncology research has advanced recently to realize that treating patients with chemotherapy in the neoadjuvant setting is both rational and beneficial (173, 174). Although conventional clinicopathological traits have been shown to correlate with response to NAC (33), it is still difficult for oncologists to identify patients who are likely to experience such reactions since success rates are frequently unpredictable. The latest research has linked miRNA expression profiles with breast cancer patients’ responses to NAC therapy. For example, Xing et al., 2021 found that decreased expression of miR-638 and miR-451a and elevated expression of miR-200c-3p, miR-23a-3p and miR-214-3p correlated to chemoresistance (Miller–Payne grade 1) (175). In their analysis of 114 breast cancer patients participating in the Clinical Trials Ireland All-Ireland Cooperative Oncology Research Group (CTRIAL-IE ICORG) 10/11 prospective, multicenter translational trial, McGuire et al., 2020 emphasize the innate value of miR-21 expression as a factor associated to response to conventional NAC (176). Further, a study conducted by Liu et al., 2019 supported the findings of the CTRIAL-IE ICORG 10/11 trial by showing decreased miR-21 expression levels in responders (vs non-responders) following cycle 2 of NAC (177). Di Cosimo et al., 2020 described the clinical utility of venous sampling for miR-140a-5p, miR-148a-3p, and miR-374a-5p, and their predictive value in determining response to subsequent neoadjuvant therapy, with an enhanced combined predictive capability of 54% in determining pCR to trastuzumab in HER2+ illness, compared with 0% in cases of poor expression (178). In their series of 435 patients with either early-stage HER2+ or TNBC illness, Stevic et al. (2018) explained how the overexpression of miR-199a in patient plasma was indicative of pCR to NAC in the GeparSixto study (179). MiR-34a expression levels were shown to accurately distinguish between responders and non-responders in 39 patients receiving treatment for locally advanced breast cancer according to promising findings from Kassem et al., 2019 (area under the curve (AUC): 0.995, sensitivity: 97.4%, specificity: 100%) (180). In patients who successfully achieved a pCR to NAC, Garcia et al., 2019 showed lower miR-145-5p expression levels (AUC: 0.790, p = 0.003) (181). Table 4 shows systematic trials examining the function of miRNAs in determining how patients would respond to neoadjuvant therapy and lists the miRNAs that are important in this setting (160, 176, 178, 179, 183, 184, 186–190). Using miRNA expression profiles to assess response to adjuvant chemotherapy is substantially more difficult. It is quite challenging to measure whether medication improved oncological outcomes for patients who were most likely to succumb to recurrence, estimate the timing of miRNA sampling, and analyze treatment response rates in a crude way. Therefore, it is not surprising that most research evaluates miRNA expression patterns using metrics that indicate response to NAC rather than adjuvant chemotherapy (e.g., RECIST, Miller-Payne grade, Sataloff score, etc.).
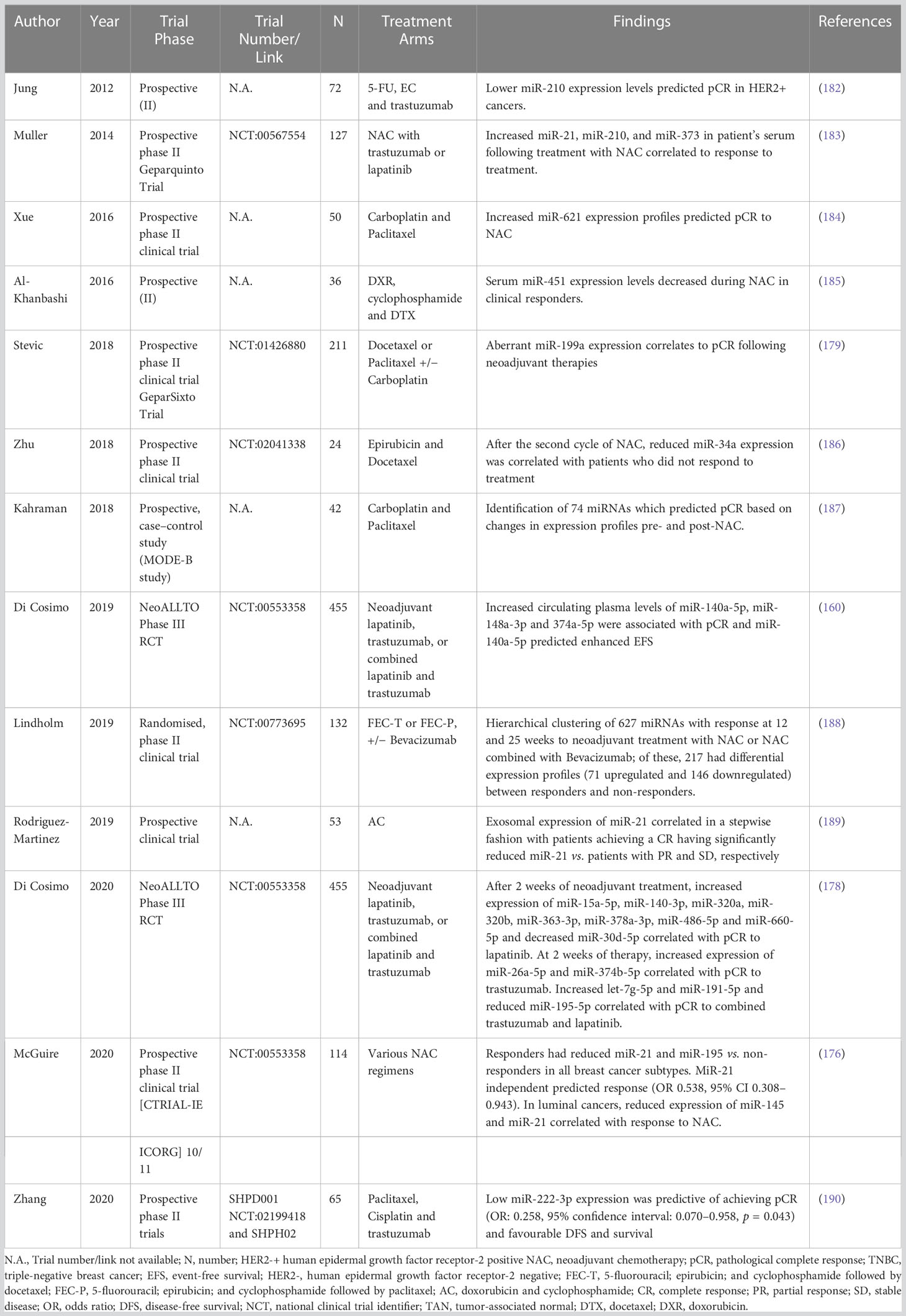
Table 4 Table illustrating prospective trials evaluating the role of miRNAs in predicting response to neoadjuvant therapies.
9 MicroRNAs for therapeutic use in breast cancer
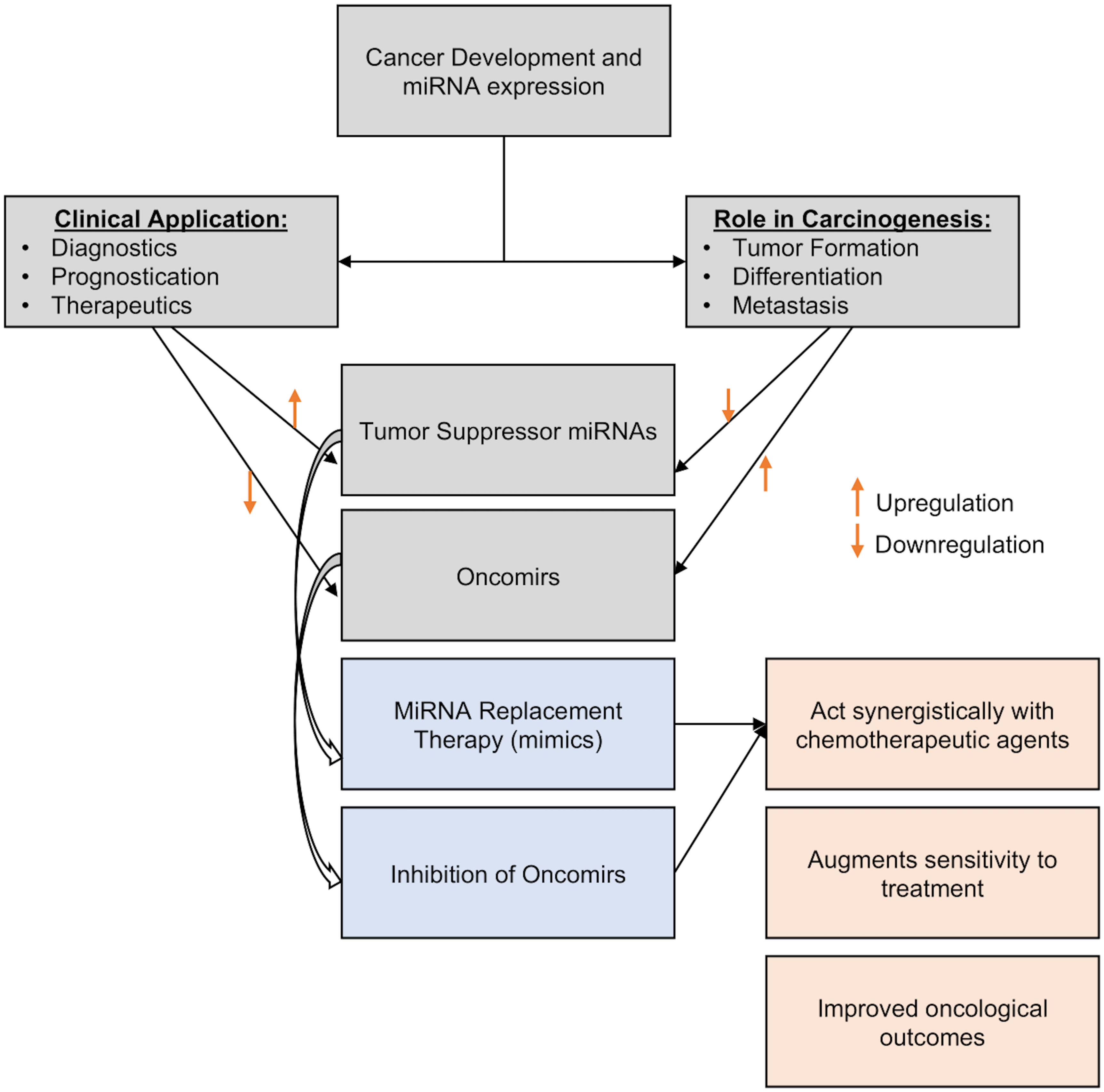
The use of miRNAs for the development of novel treatment approaches has been made easier by the current molecular technology. These entail the administration of carefully chosen miRNAs into the tumor microenvironment for therapeutic purposes or to improve the efficacy of currently available therapeutic modalities employed in standard clinical practice, such as systemic chemotherapy (143, 191). miRNAs can act as tumor suppressors or oncomirs, so there are two possible methods for using them as therapeutics (1): miRNA replacement therapy, which involves inducing and overexpressing specific miRNA to reduce oncogenesis or increasing sensitivity to systemic treatment, or (2) oncomir inhibition, which involves lowering targeted miRNA expression characteristics (i.e., miRNA silencing) by incorporating inhibitory miRNA to lessen the translation of the target miRNA (Figure 4).
♦ miRNA Replacement Therapy – By inhibiting oncogenes and the genes that regulate cell proliferation and death, tumor suppressor miRNAs can prevent the development of cancer (192). MiRNA replacement treatment includes reintroducing tumor-suppressing miRNA (or mimics) into the tumor microenvironment in order to inhibit tumor growth and restrain the spread of malignancy (193). They might be delivered into the cytoplasm of cancer cells through a variety of transporters, such as chemicals, electroporation, and modelling of the endogenous miRNA (192). Park et al., 2014 discussed the possible significance of overexpression of miR-34a in MCF7 cells in reducing cancer stem cell characteristics and increasing sensitivity to doxorubicin treatment by specifically targeting NOTCH1 (194). In studies involving animals and MDA-MB-231 and MDA-MB-549 chemoresistant breast cancer cell lines, Yu et al., 2007 and 2010 and Cochrane et al., 2009 show the value of gradually introducing and increasing the expression levels of let-7a, miR-30, and miR-200c to minimize oncogenesis and increase therapeutic index (195–197). Additionally, Kalinowski et al. (2014) analyzed how miR-7 replacement therapy can improve the efficacy of the traditional breast cancer chemotherapy currently being used to treat breast cancer (198).
♦ Oncomir Inhibition – Generally, oncomirs are elevated in the cancer (191). Anti-miRNA oligonucleotides, targeted miRNA silencing agents (antagomirs), and locked nucleic acids (LNA) can all be used to limit the action of oncogenic miRNAs (199). In various pre-clinical studies, such inhibitor mechanisms have been shown to increase the sensitivity of breast cancer cells to chemotherapeutic agents: For example, in MCF-7/ADR cell lines, miR-3609 was successfully transfected to increase the tumor cells’ susceptibility to adriamycin-based chemotherapy (200). Similarly, Lin et al., 2021 successfully enhanced cell sensitivity to chemotherapy in 65 BC patients by inducing miR-133 into cisplatin-resistant TNBC cells from these patients (201). Li et al., 2021 also successfully overcome paclitaxel resistance in previously resistant breast cancer cells by transfecting miR-155-5p into tumor cells (85). Finally, Mei et al., 2010 report that downregulating miR-21 increased the susceptibility of MCF-7 BC cell lines to docetaxel treatment (202).
9.1 miRNA delivery strategies used for cancer therapy
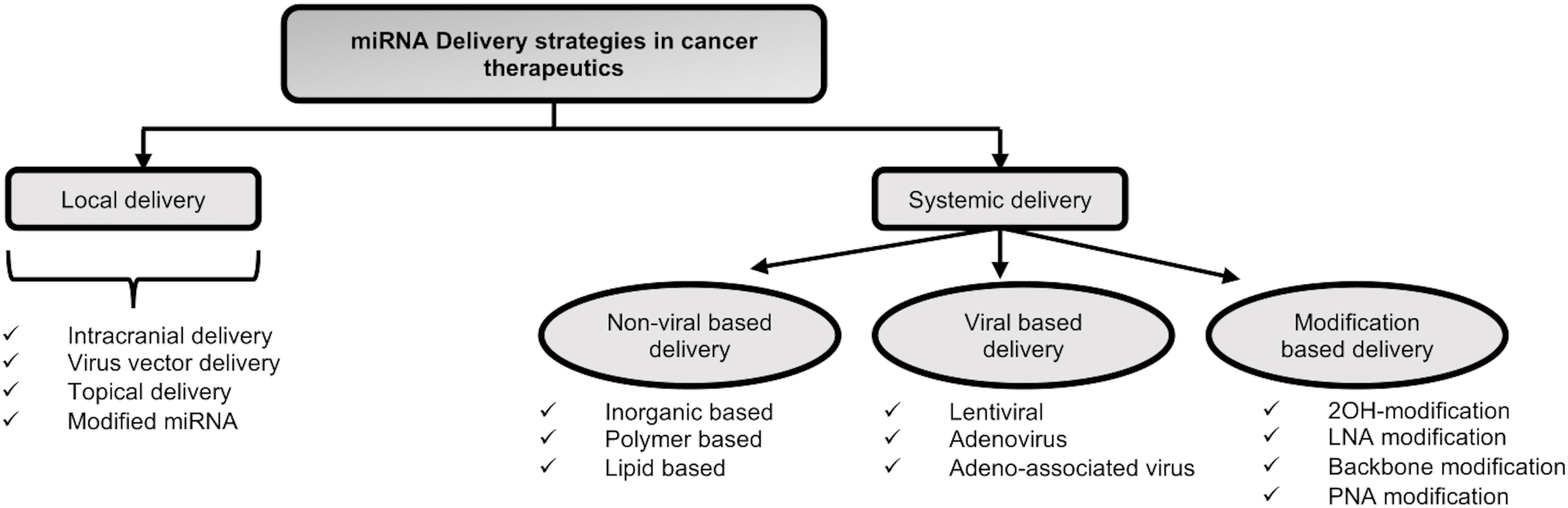
miRNAs can be introduced therapeutically into cancer cells through a variety of methods. These approaches are typically divided into two categories of local and systemic delivery, which are thoroughly discussed below and in Figure 5:
Local delivery of miRNAs: Target gene suppression with less toxicity may result from the local delivery of miRNAs as opposed to the systemic delivery of miRNAs. According to Møller et al., 2013, the aforementioned strategy has been examined mainly for primary tumors including melanoma, breast, and cervical cancers (203). Recently, different local delivery techniques, such as the direct injection of miRNA vectors into the tumor site and the nanoparticles (NP) formulation with surface modifications, have been devised. For instance, glioblastoma multiform was treated using the intracranial miRNA delivery approach (203). In a study by Trang et al., 2010, let-7 was introduced into non-small-cell lung cancer using viral vectors, which inhibited the growth of KRAS-dependent tumors (204). The topical distribution approach is an additional technique for treating skin conditions. The target region is more accessible with fewer adverse effects when topical administration is used (205). Moreover, the local delivery system makes the use of modified miRNAs. For instance, astro-cyte elevated gene-1 (AEG-1) was the target of intratumoral miR-375 mimics in cholesterol-conjugated 2′-O-methyl modified form, which significantly suppressed tumor growth in in-vivo models of hepatoma xenograft (206).
However, as the local delivery system employs direct injection or local application of miRNAs with or without carriers, it cannot be recommended as a good strategy for treating late-stage metastatic disease. Therefore, developing a systemic delivery strategy is essential to provide efficient miRNA cancer therapy.
Systemic delivery of miRNAs: The systemic miRNA delivery technique represents a significant advancement in the effort to increase the effectiveness of cancer therapy and get over the drawback of miRNA delivery in vivo. Different systemic delivery strategies have been devised up until this point. A few of them are covered below:
♦ Modification-based delivery - Systemic distribution of miRNAs into tumor cells was mostly accomplished through chemical alterations (207). The altered oligonucleotides show a stronger propensity for the target molecule. However, there are some restrictions in this regard. For instance, a targeting moiety is necessary for the intracellular uptake of modified miRNAs. Additionally, there might be a sign of short half-lives and uneven biodistribution because of quick renal and hepatic clearance. The stability of miRNA modulators and their resistance to nuclease degradation inside the blood circulation may both be improved by increasing the systemic delivery effect with various chemical modifications (208). These changes lessen the off-target effects of miRNAs and aid to overcome immune responses and low miRNA stability (207). The typical chemical modifications are 2′‐OH group modification, LNA modifications, passenger strand modifications, Phosphorothioate modification and peptide nucleic acid (PNA) modifications.
♦ Viral delivery of miRNAs – miRNAs can be transmitted by being encoded in several types of vectors, including viral and non-viral vectors. In this regard, viral delivery is an advantageous strategy. One of its benefits include low off-target rate, resulting from given miRNAs being translated by tumor cells. Lentiviruses, adenoviruses, and adenoassociated viruses (AAVs) are among the viruses that have been identified as delivery vectors for miRNAs. As a result, targeting components were added to the viral capsid to strengthen the affinity between viral vectors and cancer-specific receptors, allowing for better transportation into tumors (209). However, due to the immunological reaction they cause and the difficulty of scaling up the production process in comparison to nonviral delivery systems, there are still some significant challenges to overcome. Additionally, the potential for a virus with replication competence may raise the risk of the pathogenic condition. For instance, some retroviruses can cause the start of a CNS illness as a result of their active reproduction (210).
♦ Non-viral delivery of miRNAs – The use of non-viral vectors is a beneficial strategy for miRNA delivery. In this approach, site-specific delivery, system optimization, or polyethylene glycol (PEG) molecule augmentation could be employed to achieve targeted ligands or lengthen circulation times. Additionally, nanocarriers are produced in a secure and straightforward manner, and they are distinguished by their affordability, minimal immunogenicity, and adaptability. Non-viral delivery vectors can be divided into three primary categories: inorganic materials, lipid-based carriers, and polymeric carriers (211, 212).
However, non-viral-based approaches to miRNA delivery have their own shortcomings such as lower loading efficiency, lack of cargo protection, lower endosomal escape, nonspecific interaction with target cells and nucleic acids, etc (193).
10 Discussion
Considering that drug resistance continues to be a major obstacle in the clinical context, causing relapse and metastatic spread in many cancer types, novel treatment approaches are of the utmost importance. The discovery of miRNAs has provided a novel perspective on the molecular processes behind cancer, increasing the possibility of creating novel and more potent therapeutic approaches. This review is centered on new findings pertaining to the significance of miRNAs in breast cancer chemoresistance. miRNAs regulate numerous signaling pathways and regulatory networks, therefore even little changes in miRNA expression can have a big impact on the development and the progression of the disease. Targeting miRNAs—either reducing or enhancing their expression—seems promising to develop novel, more effective, and customized treatments, boost therapeutic efficacy, and predict patient response to various treatments. However, to fully explain all the miRNAs that are altered in tumors based on profiling data would be beyond the scope of this review. Numerous organizations are exploring the use of microRNAs as potential therapeutics. In vivo and translational investigations are currently the focus of increased research. Evidence exists that points to miRNAs as possible therapeutic agents, particularly when used in conjunction with anti-cancer chemotherapeutics. This could take the form of mimics that support miRNA function and expression or antagonists that block miRNA expression. By affecting the expression of endogenous microRNAs in cancer cells, miRNA mimics or anti-miRNAs can potentially change chemotherapy’s efficacy. Two clinical investigations have shown the potential therapeutic impact of miRNAs in the future. Among these is a phase 2a clinical trial with Miravirsen in 26 patients who had chronic hepatitis C virus (HCV) genotype 1 infection. Miravirsen is a nucleic acid–modified DNA phosphorothioate antisense oligonucleotide that encases mature miR-122 in a heteroduplex and inhibits its function. No side effects related to the experiment have been reported yet (213). Another phase 1 clinical trial including individuals with liver cancer or metastatic cancer with liver problems is MRX34 (a mimic of the tumor suppressor miR-34). Healthy volunteers and patients with advanced or metastatic liver cancer (hepatocellular carcinoma) are being tested for the safety and effectiveness of MRX34 in this study (214). Future possibilities for these novel medicines are encouraging given the encouraging preliminary findings from both trials.
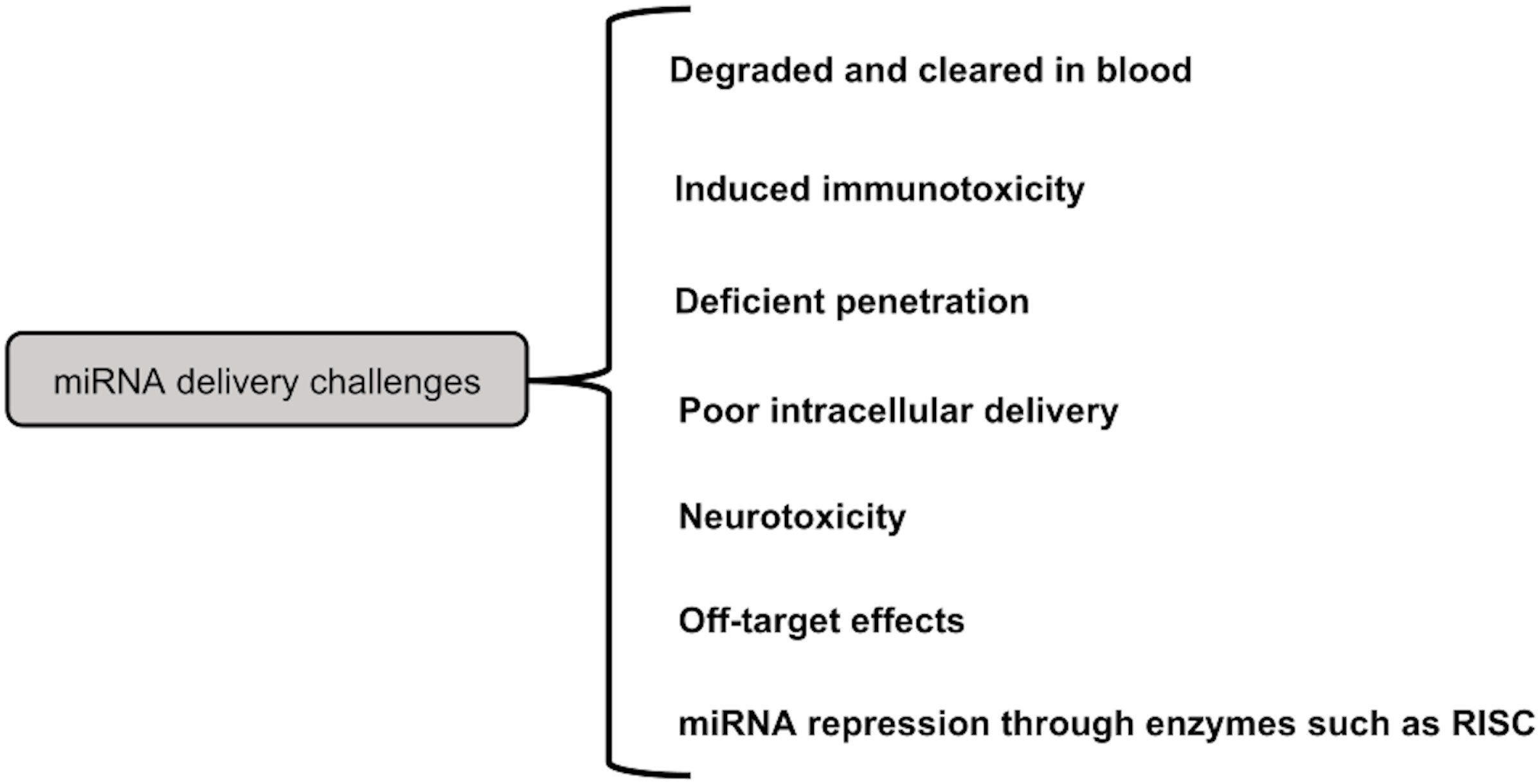
Challenges in the field of miRNA therapy-As mentioned above, the miRNAs can be delivered by either local or systemic approaches. It might not be a suitable strategy, for advanced cancer. However, miRNA cancer therapy works well with systemic delivery. Figure 6 summarizes the various constraints to miRNA delivery. For instance, poor miRNA penetration is caused by the leaky nature of aberrant tumor vasculature (215). The rapid cleavage of naked miRNAs by serum nucleases of the RNase A type poses another challenge (216). Additionally, there is a rapid renal clearance, notably for naked miRNA (217). When utilizing big NPs (>100 nm), reticuloendothelial system (RES) clearance would rise in the liver, spleen, lung, and bone marrow, leading to nonspecific absorption by innate immune cells such monocytes and macrophages (218).
Additionally, the systemic miRNA distribution triggered the innate immune system, as with other nucleic acid types, which resulted in undesired toxicities. Immune system activation includes the release of inflammatory cytokines and Type I IFNs via Toll-like receptors (TLRs) (219). Anti-inflammatory miRNA treatment, however, may prevent the activation of inflammatory pathways (220). On the other hand, some miRNAs work through TLRs to trigger neurodegeneration. For instance, Lehmann et al. (2012) demonstrated that miRNA let7b can cause neurotoxicity by activating TLR7 signaling in neurons (221). Therefore, a significant issue for miRNA systemic cancer therapy is the incidence of miRNA-related neurotoxicity. Additionally, increased miRNA uptake in cancer cells is a problem, and methods to address this issue include increasing endosomal escape and releasing miRNA payloads into the cytoplasm.
Off-target effects brought on by the miRNA mode of action are yet another challenge for miRNA delivery systems. These compounds may have undesirable side effects since they may bind to the 3′-UTR of a number of genes and decrease their expression (222). A developed method to lessen these adverse effects is the use of multifunctional co-delivering systems (223). Furthermore, it was demonstrated that under specific circumstances, such as hypoxia, the activity of miRNA processing enzymes such RISC reduces, which lowers the expression of tumor suppressor miRNAs (224). De Carvalho Vicentini et al. (2013) suggest that altering the expression or activity of these enzymes is another method for suppressing miRNA (225).
11 Conclusion
The discovery, development, and enhancement of miRNAs as potential medicines for the treatment of breast cancer patients have received significant funding, yet this branch of translational research is still in its infancy. Numerous attempts have been made to tailor cancer therapies using miRNA, but little progress has been made in improving clinic-oncological outcomes using miRNA targeting. miRNA therapies are now facing several developmental obstacles. This study is constrained by the fact that most of the research done thus far provides information about in-vitro trials, with very few studies coming from sources other than animal or breast cancer cell lines. Clinical trials assessing the clinical effectiveness, risk profiles, and premium benefit are necessary for addition to the generally accepted scientific technique to support the initial findings of these recent investigations. An in-depth discussion of how clinical trial research has transformed BC patient care over the last four decades is provided in the current review. This research has produced novel, individualized therapeutic approaches, minimally invasive surgical techniques for the breast and axilla, and improved clinico-oncological results for patients who might otherwise have died from their disease in earlier times. The personalization of BC patient care appears to be closer than ever thanks to ongoing trials evaluating cutting-edge targeted therapies like immune checkpoint modulation (39, 226) and the use of poly(adenosine diphosphate-ribose) polymerase inhibitors (or PARP inhibitors) in the treatment of early-stage breast cancer in BRCA mutation carriers (227).
Hence, before we can use miRNAs in the therapeutic setting, numerous obstacles remain to be overcome. The delivery method is the key impediment. We might be able to get over this obstacle with the use of chemical alterations, viral vectors, or nanoparticles. Despite these delivery issues, it is possible that miRNAs will play a significant role in cancer therapy, including BC, in the future. A novel approach to treating breast cancer that combines miRNA therapies with conventional chemotherapeutic techniques and drug targets is possible, but further study is needed before this promising paradigm can be implemented in the clinic. Thus, this review emphasizes how important it is to prioritize clinical trials and therapeutic interventions to advance the precision oncology movement’s goal of “curing” breast cancer.
Author contributions
SS, RT conceived the study. SS drafted the manuscript. SS, HS, AS, SG, VH, RT revised the manuscript critically for important intellectual content. RT, HS provided important comments on the manuscript. All authors approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by the Indian Council of Medical Research (ICMR) project (5/13/58/2020/NCD-III)
Acknowledgments
SS thanks ICMR project (5/13/58/2020/NCD-III) for providing Research Associate fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
miRNAs: MicroRNAs
BC: breast cancer
LABC: Luminal A Breast Cancer
LBBC: Luminal B Breast Cancer
HER2+: Human Epidermal Growth Factor Receptor-2 Enriched Breast Cancer
TNBC: Triple-Negative Breast Cancer
MAPK: mitogen-activated protein kinase
PI3K: Phosphoinositide 3-Kinase
mTOR: Mammalian Target of Rapamycin
ER: Estrogen Receptor
Pol II: RNA polymerase II
pre-miRNAs: Precursor miRNAs
Exp5: Exportin-5
RISC: RNA-induced silencing complex
CMF: Cyclophosphamide
methotrexate: and 5-fluorouracil
NAC: Neoadjuvant chemotherapy
BCS: Breast Conservation Surgery
EBCTCG: The Early Breast Cancer Triallist’s Collaborative Group
LRR: Locoregional Recurrence
DFS: Disease-Free Survival
OS: Overall Survival
pCR: Pathological Complete Response
LN: Lymph node
ASCO: American Society of Clinical Oncology
EMT: Epithelial-Mesenchymal Transition
MiniPDXTM: Minimal Patient-Derived Xenograft
PTEN: Phosphatase and TENsin homolog
5-FU: 5-fluorouracil
MSCs-Exo: Mesenchymal Stem Cell-Derived Exosome
ID4: Inhibitor of Differentiation 4
CDA: Cytosine Deaminase
MCF-7/DTX: Docetaxel-Resistant MCF-7 BC cell line
DOX: Doxorubicin
MCF-7/EPB: Epirubicin-Resistant MCF-7 BC cell line
ADR: Adriamycin
EIF4E: Eukaryotic Translation Initiation Factor 4E
EFS: Event-Free Survival
BCL-2: B-cell Leukemia/Lymphoma 2 Protein
CCND1: Cyclin D1 protein
DCTD: dCMP Deaminase
HIF-1: Hypoxia-inducible factor-1
DTX: Docetaxel
EPB: Epirubicin
GCB: Gemcitabine
ct-miRNA: circulating miRNA
EMT: epithelial-mesenchymal transition
MDR1: Multidrug Resistance gene
P-gp: P-glyocoprotein
GST-π: glutathione S-transferase
MRP: multidrug resistance-associated protein
ABC: ATP-binding cassette transporters
BCRP: breast cancer resistance protein
EGFR: epidermal growth factor receptor
CSCs: Cancer stem cells
DDR: DNA damage repair
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Bajpai J, Ventrapati P, Joshi S, Wadasadawala T, Rath S, Pathak R, et al. Unique challenges and outcomes of young women with breast cancers from a tertiary care cancer centre in India. Breast (2021) 60:177–84. doi: 10.1016/j.breast.2021.09.008
3. DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA Cancer J Clin (2019) 69(6):438–51. doi: 10.3322/caac.21583
4. Sakorafas GH, Safioleas M. Breast cancer surgery: an historical narrative. part II. 18th and 19th centuries. Eur J Cancer Care (Engl) (2010) 19(1):6–29. doi: 10.1111/j.1365-2354.2008.01060.x
5. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol (2013) 24(9):2206–23. doi: 10.1093/annonc/mdt303
6. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol (2019) 21(4):498–510. doi: 10.1038/s41556-019-0299-0
7. Wei Y, Yang P, Cao S, Zhao L. The combination of curcumin and 5-fluorouracil in cancer therapy. Arch Pharm Res (2018) 41(1):1–13. doi: 10.1007/s12272-017-0979-x
8. Shah AN, Gradishar WJ. Adjuvant anthracyclines in breast cancer: what is their role? Oncologist (2018) 23(10):1153–61. doi: 10.1634/theoncologist.2017-0672
9. Chang M. Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) (2012) 20(3):256–67. doi: 10.4062/biomolther.2012.20.3.256
10. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res (2010) 12(5):R68. doi: 10.1186/bcr2635
11. Picard M. Management of hypersensitivity reactions to taxanes. Immunol Allergy Clin North Am (2017) 37(4):679–93. doi: 10.1016/j.iac.2017.07.004
12. Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med (2011) 365(14):1273–83. doi: 10.1056/NEJMoa0910383
13. Łukasiewicz S, Czeczelewski M, Forma A, Baj J, Sitarz R, Stanisławek A. Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancers (Basel) (2021) 13(17). doi: 10.3390/cancers13174287
14. Nwabo Kamdje AH, Seke Etet PF, Vecchio L, Muller JM, Krampera M, Lukong KE. Signaling pathways in breast cancer: therapeutic targeting of the microenvironment. Cell Signal (2014) 26(12):2843–56. doi: 10.1016/j.cellsig.2014.07.034
15. Rebucci M, Michiels C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem Pharmacol (2013) 85(9):1219–26. doi: 10.1016/j.bcp.2013.02.017
16. Haider T, Pandey V, Banjare N, Gupta PN, Soni V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol Rep (2020) 72(5):1125–51. doi: 10.1007/s43440-020-00138-7
17. Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal (2014) 7(357):ra121. doi: 10.1126/scisignal.aaa1877
18. Belmont PJ, Jiang P, McKee TD, Xie T, Isaacson J, Baryla NE, et al. Resistance to dual blockade of the kinases PI3K and mTOR in KRAS-mutant colorectal cancer models results in combined sensitivity to inhibition of the receptor tyrosine kinase EGFR. Sci Signal (2014) 7(351):ra107. doi: 10.1126/scisignal.2005516
19. Toss A, Venturelli M, Peterle C, Piacentini F, Cascinu S, Cortesi L. Molecular biomarkers for prediction of targeted therapy response in metastatic breast cancer: trick or treat? Int J Mol Sci (2017) 18(1). doi: 10.3390/ijms18010085
20. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech (2021) 14(4). doi: 10.1242/dmm.047662
21. Deng X, Liu Y, Luo M, Wu J, Ma R, Wan Q. Circulating miRNA-24 and its target YKL-40 as potential biomarkers in patients with coronary heart disease and type 2 diabetes mellitus. Oncotarget (2017) 8(38):63038–46. doi: 10.18632/oncotarget.18593
22. Ni WJ, Leng XM. Dynamic miRNA-mRNA paradigms: new faces of miRNAs. Biochem Biophys Rep (2015) 4:337–41. doi: 10.1016/j.bbrep.2015.10.011
23. Teng M, Yu ZH, Zhao P, Zhuang GQ, Wu ZX, Dang L, et al. Putative roles as oncogene or tumour suppressor of the mid-clustered microRNAs in gallid alphaherpesvirus 2 (GaHV2) induced marek's disease lymphomagenesis. J Gen Virol (2017) 98(5):1097–112. doi: 10.1099/jgv.0.000786
24. Snowhite IV, Allende G, Sosenko J, Pastori RL, Messinger Cayetano S, Pugliese A. Association of serum microRNAs with islet autoimmunity, disease progression and metabolic impairment in relatives at risk of type 1 diabetes. Diabetologia (2017) 60(8):1409–22. doi: 10.1007/s00125-017-4294-3
25. Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans (2018) 46(1):11–21. doi: 10.1042/BST20170037
26. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci (2020) 21(5). doi: 10.3390/ijms21051723
27. Singh A, Singh AK, Giri R, Kumar D, Sharma R, Valis M, et al. The role of microRNA-21 in the onset and progression of cancer. Future Med Chem (2021) 13(21):1885–906. doi: 10.4155/fmc-2021-0096
28. Due H, Svendsen P, Bødker JS, Schmitz A, Bøgsted M, Johnsen HE, et al. miR-155 as a biomarker in b-cell malignancies. BioMed Res Int (2016) 2016:9513037. doi: 10.1155/2016/9513037
29. Ma Y, Shen N, Wicha MS, Luo M. The roles of the let-7 family of MicroRNAs in the regulation of cancer stemness. Cells (2021) 10(9). doi: 10.3390/cells10092415
30. McVeigh TP, Boland MR, Lowery AJ. The impact of the biomolecular era on breast cancer surgery. Surgeon (2017) 15(3):169–81. doi: 10.1016/j.surge.2016.09.007
31. Bonadonna G, Brusamolino E, Valagussa P, Rossi A, Brugnatelli L, Brambilla C, et al. Combination chemotherapy as an adjuvant treatment in operable breast cancer. N Engl J Med (1976) 294(8):405–10. doi: 10.1056/NEJM197602192940801
32. Fisher B. Biological research in the evolution of cancer surgery: a personal perspective. Cancer Res (2008) 68(24):10007–20. doi: 10.1158/0008-5472.CAN-08-0186
33. Davey MG, Kerin E, O'Flaherty C, Maher E, Richard V, McAnena P, et al. Clinicopathological response to neoadjuvant therapies and pathological complete response as a biomarker of survival in human epidermal growth factor receptor-2 enriched breast cancer - a retrospective cohort study. Breast (2021) 59:67–75. doi: 10.1016/j.breast.2021.06.005
34. Spring LM, Fell G, Arfe A, Sharma C, Greenup R, Reynolds KL, et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res (2020) 26(12):2838–48. doi: 10.1158/1078-0432.CCR-19-3492
35. (EBCTCG) EBCTCG. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol (2018) 19(1):27–39. doi: 10.1016/S1470-2045(17)30777-5
36. Boughey JC, Ballman KV, McCall LM, Mittendorf EA, Symmans WF, Julian TB, et al. Tumor biology and response to chemotherapy impact breast cancer-specific survival in node-positive breast cancer patients treated with neoadjuvant chemotherapy: long-term follow-up from ACOSOG Z1071 (Alliance). Ann Surg (2017) 266(4):667–76. doi: 10.1097/SLA.0000000000002373
37. Korde LA, Somerfield MR, Carey LA, Crews JR, Denduluri N, Hwang ES, et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J Clin Oncol (2021) 39(13):1485–505. doi: 10.1200/JCO.20.03399
38. Li ZY, Zhang Z, Cao XZ, Feng Y, Ren SS. Platinum-based neoadjuvant chemotherapy for triple-negative breast cancer: a systematic review and meta-analysis. J Int Med Res (2020) 48(10):300060520964340. doi: 10.1177/0300060520964340
39. Schmid P, Cortes J, Pusztai L, McArthur H, Kümmel S, Bergh J, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med (2020) 382(9):810–21. doi: 10.1056/NEJMoa1910549
40. Bartsch R, Bergen E, Galid A. Current concepts and future directions in neoadjuvant chemotherapy of breast cancer. Memo (2018) 11(3):199–203. doi: 10.1007/s12254-018-0421-1
41. Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature (2015) 527(7579):525–30. doi: 10.1038/nature16064
42. Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, et al. Upregulation of twist-1 by NF-kappaB blocks cytotoxicity induced by chemotherapeutic drugs. Mol Cell Biol (2007) 27(11):3920–35. doi: 10.1128/MCB.01219-06
43. Vesuna F, Lisok A, Kimble B, Domek J, Kato Y, van der Groep P, et al. Twist contributes to hormone resistance in breast cancer by downregulating estrogen receptor-α. Oncogene (2012) 31(27):3223–34. doi: 10.1038/onc.2011.483
44. Gao L, Yang Y, Song S, Hong H, Zhao X, Li D. The association between genetic variant of MDR1 gene and breast cancer risk factors in Chinese women. Int Immunopharmacol (2013) 17(1):88–91. doi: 10.1016/j.intimp.2013.05.025
45. Chen Y, Li X, Shi L, Ma P, Wang W, Wu N, et al. Combination of 7-. Aging (Albany NY) (2022) 14(17):7156–69. doi: 10.18632/aging.204287
46. Ajith AK, Subramani S, Manickam AH, Ramasamy S. Chemotherapeutic resistance genes of breast cancer patients - an overview. Adv Pharm Bull (2022) 12(4):649–57. doi: 10.34172/apb.2022.048
47. Kort A, Durmus S, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Brain and testis accumulation of regorafenib is restricted by breast cancer resistance protein (BCRP/ABCG2) and p-glycoprotein (P-GP/ABCB1). Pharm Res (2015) 32(7):2205–16. doi: 10.1007/s11095-014-1609-7
48. Hu D, Li M, Su J, Miao K, Qiu X. Dual-targeting of miR-124-3p and ABCC4 promotes sensitivity to adriamycin in breast cancer cells. Genet Test Mol Biomarkers (2019) 23(3):156–65. doi: 10.1089/gtmb.2018.0259
49. Levin ER. Extranuclear estrogen receptor's roles in physiology: lessons from mouse models. Am J Physiol Endocrinol Metab (2014) 307(2):E133–40. doi: 10.1152/ajpendo.00626.2013
50. Ziauddin MF, Hua D, Tang SC. Emerging strategies to overcome resistance to endocrine therapy for breast cancer. Cancer Metastasis Rev (2014) 33(2-3):791–807. doi: 10.1007/s10555-014-9504-6
51. Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, et al. Nuclear translocation of epidermal growth factor receptor by akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem (2011) 286(23):20558–68. doi: 10.1074/jbc.M111.240796
52. Kitao H, Iimori M, Kataoka Y, Wakasa T, Tokunaga E, Saeki H, et al. DNA Replication stress and cancer chemotherapy. Cancer Sci (2018) 109(2):264–71. doi: 10.1111/cas.13455
53. Nikitaki Z, Michalopoulos I, Georgakilas AG. Molecular inhibitors of DNA repair: searching for the ultimate tumor killing weapon. Future Med Chem (2015) 7(12):1543–58. doi: 10.4155/fmc.15.95
54. Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol (2015) 7(4):a016600. doi: 10.1101/cshperspect.a016600
55. Martin JH, Bromfield EG, Aitken RJ, Lord T, Nixon B. Double strand break DNA repair occurs via non-homologous end-joining in mouse MII oocytes. Sci Rep (2018) 8(1):9685. doi: 10.1038/s41598-018-27892-2
56. Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet (2014) 30(8):326–39. doi: 10.1016/j.tig.2014.06.003
57. Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol (2014) 15(7):465–81. doi: 10.1038/nrm3822
58. Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol (2013) 5(4):a012583. doi: 10.1101/cshperspect.a012583
59. Li W, Melton DW. Cisplatin regulates the MAPK kinase pathway to induce increased expression of DNA repair gene ERCC1 and increase melanoma chemoresistance. Oncogene (2012) 31(19):2412–22. doi: 10.1038/onc.2011.426
60. Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, et al. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer Epidemiol (2010) 34(1):85–92. doi: 10.1016/j.canep.2009.11.002
61. Aleskandarany M, Caracappa D, Nolan CC, Macmillan RD, Ellis IO, Rakha EA, et al. DNA Damage response markers are differentially expressed in BRCA-mutated breast cancers. Breast Cancer Res Treat (2015) 150(1):81–90. doi: 10.1007/s10549-015-3306-6
62. Altan B, Yokobori T, Ide M, Bai T, Yanoma T, Kimura A, et al. High expression of MRE11-RAD50-NBS1 is associated with poor prognosis and chemoresistance in gastric cancer. Anticancer Res (2016) 36(10):5237–47. doi: 10.21873/anticanres.11094
63. Pavlopoulou A, Oktay Y, Vougas K, Louka M, Vorgias CE, Georgakilas AG. Determinants of resistance to chemotherapy and ionizing radiation in breast cancer stem cells. Cancer Lett (2016) 380(2):485–93. doi: 10.1016/j.canlet.2016.07.018
64. Zhou ZY, Wan LL, Yang QJ, Han YL, Li D, Lu J, et al. Nilotinib reverses ABCB1/P-glycoprotein-mediated multidrug resistance but increases cardiotoxicity of doxorubicin in a MDR xenograft model. Toxicol Lett (2016) 259:124–32. doi: 10.1016/j.toxlet.2016.07.710
65. Attia YM, El-Kersh DM, Ammar RA, Adel A, Khalil A, Walid H, et al. Inhibition of aldehyde dehydrogenase-1 and p-glycoprotein-mediated multidrug resistance by curcumin and vitamin D3 increases sensitivity to paclitaxel in breast cancer. Chem Biol Interact (2020) 315:108865. doi: 10.1016/j.cbi.2019.108865
66. Angius A, Scanu AM, Arru C, Muroni MR, Rallo V, Deiana G, et al. Portrait of cancer stem cells on colorectal cancer: molecular biomarkers, signaling pathways and miRNAome. Int J Mol Sci (2021) 22(4). doi: 10.3390/ijms22041603
67. Sun X, Qin S, Fan C, Xu C, Du N, Ren H. Let-7: a regulator of the ERα signaling pathway in human breast tumors and breast cancer stem cells. Oncol Rep (2013) 29(5):2079–87. doi: 10.3892/or.2013.2330
68. Hazarika M, Chuk MK, Theoret MR, Mushti S, He K, Weis SL, et al. U.S. FDA approval summary: nivolumab for treatment of unresectable or metastatic melanoma following progression on ipilimumab. Clin Cancer Res (2017) 23(14):3484–8. doi: 10.1158/1078-0432.CCR-16-0712
69. Al-Harras MF, Houssen ME, Shaker ME, Farag K, Farouk O, Monir R, et al. Polymorphisms of glutathione s-transferase π 1 and toll-like receptors 2 and 9: association with breast cancer susceptibility. Oncol Lett (2016) 11(3):2182–8. doi: 10.3892/ol.2016.4159
70. Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol (2014) 15(7):e279–89. doi: 10.1016/S1470-2045(13)70567-9
71. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature (2003) 425(6956):415–9. doi: 10.1038/nature01957
72. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5
73. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
74. Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol (2007) 21(5):1132–47. doi: 10.1210/me.2007-0022
75. Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol (2006) 26(21):8191–201. doi: 10.1128/MCB.00242-06
76. Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem (2010) 285(28):21496–507. doi: 10.1074/jbc.M109.083337
77. Xu X, Lv YG, Yan CY, Yi J, Ling R. Enforced expression of hsa-miR-125a-3p in breast cancer cells potentiates docetaxel sensitivity via modulation of BRCA1 signaling. Biochem Biophys Res Commun (2016) 479(4):893–900. doi: 10.1016/j.bbrc.2016.09.087
78. Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther (2009) 8(5):1055–66. doi: 10.1158/1535-7163.MCT-08-1046
79. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell (2007) 131(6):1109–23. doi: 10.1016/j.cell.2007.10.054
80. Kastl L, Brown I, Schofield AC. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat (2012) 131(2):445–54. doi: 10.1007/s10549-011-1424-3
81. Schmittgen TD. miR-31: a master regulator of metastasis? Future Oncol (2010) 6(1):17–20. doi: 10.2217/fon.09.150
82. Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature (2008) 451(7175):147–52. doi: 10.1038/nature06487
83. Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res (2006) 66(18):9090–8. doi: 10.1158/0008-5472.CAN-06-1403
84. Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem (2011) 286(21):19127–37. doi: 10.1074/jbc.M110.216887
85. Li Y, Zhang L, Dong Z, Xu H, Yan L, Wang W, et al. MicroRNA-155-5p promotes tumor progression and contributes to paclitaxel resistance via TP53INP1 in human breast cancer. Pathol Res Pract (2021) 220:153405. doi: 10.1016/j.prp.2021.153405
86. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature (2007) 449(7163):682–8. doi: 10.1038/nature06174
87. Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol (2008) 10(2):202–10. doi: 10.1038/ncb1681
88. Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-m checkpoint in MDA-MB-231 breast cancer cells. Cancer Res (2007) 67(22):11001–11. doi: 10.1158/0008-5472.CAN-07-2416
89. Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem (2008) 283(44):29897–903. doi: 10.1074/jbc.M804612200
90. Li H, Yang BB. Friend or foe: the role of microRNA in chemotherapy resistance. Acta Pharmacol Sin (2013) 34(7):870–9. doi: 10.1038/aps.2013.35
91. Magee P, Shi L, Garofalo M. Role of microRNAs in chemoresistance. Ann Transl Med (2015) 3(21):332. doi: 10.3978/j.issn.2305-5839.2015.11.32
92. Griñán-Lisón C, Olivares-Urbano MA, Jiménez G, López-Ruiz E, Del Val C, Morata-Tarifa C, et al. miRNAs as radio-response biomarkers for breast cancer stem cells. Mol Oncol (2020) 14(3):556–70. doi: 10.1002/1878-0261.12635
93. Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer (2015) 22(5):R279–300. doi: 10.1530/ERC-15-0355
94. Gao Y, Zhang W, Liu C, Li G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Sci Rep (2019) 9(1):18844. doi: 10.1038/s41598-019-54289-6
95. He M, Jin Q, Chen C, Liu Y, Ye X, Jiang Y, et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene (2019) 38(28):5551–65. doi: 10.1038/s41388-019-0817-3
96. Li J, Lu M, Jin J, Lu X, Xu T, Jin S. miR-449a suppresses tamoxifen resistance in human breast cancer cells by targeting ADAM22. Cell Physiol Biochem (2018) 50(1):136–49. doi: 10.1159/000493964
97. Liu ZR, Song Y, Wan LH, Zhang YY, Zhou LM. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci (2016) 149:104–13. doi: 10.1016/j.lfs.2016.02.059
98. Shi YF, Lu H, Wang HB. Downregulated lncRNA ADAMTS9-AS2 in breast cancer enhances tamoxifen resistance by activating microRNA-130a-5p. Eur Rev Med Pharmacol Sci (2019) 23(4):1563–73. doi: 10.26355/eurrev_201902_17115
99. Zhang HY, Liang F, Zhang JW, Wang F, Wang L, Kang XG. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother Pharmacol (2017) 79(2):327–37. doi: 10.1007/s00280-016-3208-2
100. Sang Y, Chen B, Song X, Li Y, Liang Y, Han D, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther (2019) 27(9):1638–52. doi: 10.1016/j.ymthe.2019.05.011
101. Kopp F, Oak PS, Wagner E, Roidl A. miR-200c sensitizes breast cancer cells to doxorubicin treatment by decreasing TrkB and Bmi1 expression. PloS One (2012) 7(11):e50469. doi: 10.1371/journal.pone.0050469
102. Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, et al. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch Med Res (2012) 43(7):514–21. doi: 10.1016/j.arcmed.2012.09.007
103. Zhao L, Wang Y, Jiang L, He M, Bai X, Yu L, et al. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing p-glycoprotein(P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res (2016) 35:25. doi: 10.1186/s13046-016-0300-8
104. Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of p-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathol (2012) 180(6):2490–503. doi: 10.1016/j.ajpath.2012.02.024
105. Shen H, Li L, Yang S, Wang D, Zhong S, Zhao J, et al. MicroRNA-29a contributes to drug-resistance of breast cancer cells to adriamycin through PTEN/AKT/GSK3β signaling pathway. Gene (2016) 593(1):84–90. doi: 10.1016/j.gene.2016.08.016
106. Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou H, et al. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep (2017) 7:41942. doi: 10.1038/srep41942
107. Wang DD, Yang SJ, Chen X, Shen HY, Luo LJ, Zhang XH, et al. miR-222 induces adriamycin resistance in breast cancer through PTEN/Akt/p27. Tumour Biol (2016) 37(11):15315–24. doi: 10.1007/s13277-016-5341-2
108. Gao M, Miao L, Liu M, Li C, Yu C, Yan H, et al. miR-145 sensitizes breast cancer to doxorubicin by targeting multidrug resistance-associated protein-1. Oncotarget (2016) 7(37):59714–26. doi: 10.18632/oncotarget.10845
109. Jiang L, He D, Yang D, Chen Z, Pan Q, Mao A, et al. MiR-489 regulates chemoresistance in breast cancer via epithelial mesenchymal transition pathway. FEBS Lett (2014) 588(11):2009–15. doi: 10.1016/j.febslet.2014.04.024
110. Hu SH, Wang CH, Huang ZJ, Liu F, Xu CW, Li XL, et al. miR-760 mediates chemoresistance through inhibition of epithelial mesenchymal transition in breast cancer cells. Eur Rev Med Pharmacol Sci (2016) 20(23):5002–8.
111. Zhang Y, He Y, Lu LL, Zhou ZY, Wan NB, Li GP, et al. miRNA-192-5p impacts the sensitivity of breast cancer cells to doxorubicin via targeting peptidylprolyl isomerase a. Kaohsiung J Med Sci (2019) 35(1):17–23. doi: 10.1002/kjm2.12004
112. Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, et al. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie (2011) 34(12):675–80. doi: 10.1159/000334552
113. Chang L, Hu Z, Zhou Z, Zhang H. Linc00518 contributes to multidrug resistance through regulating the MiR-199a/MRP1 axis in breast cancer. Cell Physiol Biochem (2018) 48(1):16–28. doi: 10.1159/000491659
114. Lv K, Liu L, Wang L, Yu J, Liu X, Cheng Y, et al. Lin28 mediates paclitaxel resistance by modulating p21, Rb and let-7a miRNA in breast cancer cells. PloS One (2012) 7(7):e40008. doi: 10.1371/journal.pone.0040008
115. Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis (2008) 13(10):1215–22. doi: 10.1007/s10495-008-0256-z
116. Su CM, Wang MY, Hong CC, Chen HA, Su YH, Wu CH, et al. miR-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene (2016) 35(9):1134–42. doi: 10.1038/onc.2015.168
117. Gu X, Li JY, Guo J, Li PS, Zhang WH. Influence of MiR-451 on drug resistances of paclitaxel-resistant breast cancer cell line. Med Sci Monit (2015) 21:3291–7. doi: 10.12659/MSM.894475
118. Zhang B, Zhao R, He Y, Fu X, Fu L, Zhu Z, et al. MicroRNA 100 sensitizes luminal a breast cancer cells to paclitaxel treatment in part by targeting mTOR. Oncotarget (2016) 7(5):5702–14. doi: 10.18632/oncotarget.6790
119. Sha LY, Zhang Y, Wang W, Sui X, Liu SK, Wang T, et al. MiR-18a upregulation decreases dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur Rev Med Pharmacol Sci (2016) 20(11):2201–8.
120. Liu X, Tang H, Chen J, Song C, Yang L, Liu P, et al. MicroRNA-101 inhibits cell progression and increases paclitaxel sensitivity by suppressing MCL-1 expression in human triple-negative breast cancer. Oncotarget (2015) 6(24):20070–83. doi: 10.18632/oncotarget.4039
121. Zheng P, Dong L, Zhang B, Dai J, Zhang Y, Wang Y, et al. Long noncoding RNA CASC2 promotes paclitaxel resistance in breast cancer through regulation of miR-18a-5p/CDK19. Histochem Cell Biol (2019) 152(4):281–91. doi: 10.1007/s00418-019-01794-4
122. Yao YS, Qiu WS, Yao RY, Zhang Q, Zhuang LK, Zhou F, et al. miR-141 confers docetaxel chemoresistance of breast cancer cells via regulation of EIF4E expression. Oncol Rep (2015) 33(5):2504–12. doi: 10.3892/or.2015.3866
123. Zhang Y, Wang Y, Wei Y, Li M, Yu S, Ye M, et al. MiR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci Rep (2015) 5:15424. doi: 10.1038/srep15424
124. Zhang X, Zhong S, Xu Y, Yu D, Ma T, Chen L, et al. MicroRNA-3646 contributes to docetaxel resistance in human breast cancer cells by GSK-3β/β-Catenin signaling pathway. PloS One (2016) 11(4):e0153194. doi: 10.1371/journal.pone.0153194
125. Hu Q, Chen WX, Zhong SL, Zhang JY, Ma TF, Ji H, et al. MicroRNA-452 contributes to the docetaxel resistance of breast cancer cells. Tumour Biol (2014) 35(7):6327–34. doi: 10.1007/s13277-014-1834-z
126. Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2). J Biol Chem (2013) 288(16):10973–85. doi: 10.1074/jbc.M112.434340
127. Zhang HD, Sun DW, Mao L, Zhang J, Jiang LH, Li J, et al. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun (2015) 465(4):702–13. doi: 10.1016/j.bbrc.2015.08.053
128. Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene (2013) 531(1):8–14. doi: 10.1016/j.gene.2013.08.062
129. Huang P, Li F, Li L, You Y, Luo S, Dong Z, et al. lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci Rep (2018) 8(1):17970. doi: 10.1038/s41598-018-36231-4
130. Nandy SB, Arumugam A, Subramani R, Pedroza D, Hernandez K, Saltzstein E, et al. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the hippo signaling pathway. Oncotarget (2015) 6(19):17366–78. doi: 10.18632/oncotarget.3953
131. Zhang Y, Qu X, Teng Y, Li Z, Xu L, Liu J, et al. Cbl-b inhibits p-gp transporter function by preventing its translocation into caveolae in multiple drug-resistant gastric and breast cancers. Oncotarget (2015) 6(9):6737–48. doi: 10.18632/oncotarget.3253
132. Yin J, Zheng G, Jia X, Zhang Z, Zhang W, Song Y, et al. A Bmi1-miRNAs cross-talk modulates chemotherapy response to 5-fluorouracil in breast cancer cells. PloS One (2013) 8(9):e73268. doi: 10.1371/journal.pone.0073268
133. Li QQ, Chen ZQ, Cao XX, Xu JD, Xu JW, Chen YY, et al. Involvement of NF-κB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial-mesenchymal transition of breast cancer cells. Cell Death Differ (2011) 18(1):16–25. doi: 10.1038/cdd.2010.103
134. Li X, Wang S, Li Z, Long X, Guo Z, Zhang G, et al. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int J Biol Macromol (2017) 105(Pt 1):346–53. doi: 10.1016/j.ijbiomac.2017.07.053
135. Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou D, et al. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J Cell Mol Med (2019) 23(5):3166–77. doi: 10.1111/jcmm.14171
136. Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang L, et al. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep (2014) 47(5):268–73. doi: 10.5483/BMBRep.2014.47.5.165
137. Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, Huang X, et al. MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Cancer (2014) 135(6):1356–68. doi: 10.1002/ijc.28782
138. Ye XM, Zhu HY, Bai WD, Wang T, Wang L, Chen Y, et al. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer (2014) 14:134. doi: 10.1186/1471-2407-14-134
139. Ma T, Yang L, Zhang J. MiRNA−542−3p downregulation promotes trastuzumab resistance in breast cancer cells via AKT activation. Oncol Rep (2015) 33(3):1215–20. doi: 10.3892/or.2015.3713
140. Corcoran C, Rani S, Breslin S, Gogarty M, Ghobrial IM, Crown J, et al. miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol Cancer (2014) 13:71. doi: 10.1186/1476-4598-13-71
141. Venturutti L, Cordo Russo RI, Rivas MA, Mercogliano MF, Izzo F, Oakley RH, et al. MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. Oncogene (2016) 35(48):6189–202. doi: 10.1038/onc.2016.151
142. Huynh FC, Jones FE. MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2Δ16 mediated breast tumorigenesis and reverse trastuzumab resistance. PloS One (2014) 9(12):e114419. doi: 10.1371/journal.pone.0114419
143. Casey MC, Sweeney KJ, Brown JA, Kerin MJ. Exploring circulating micro-RNA in the neoadjuvant treatment of breast cancer. Int J Cancer (2016) 139(1):12–22. doi: 10.1002/ijc.29985
144. Liu M, Gong C, Xu R, Chen Y, Wang X. MicroRNA-5195-3p enhances the chemosensitivity of triple-negative breast cancer to paclitaxel by downregulating EIF4A2. Cell Mol Biol Lett (2019) 24:47. doi: 10.1186/s11658-019-0168-7
145. Hou X, Niu Z, Liu L, Guo Q, Li H, Yang X, et al. miR-1207-5p regulates the sensitivity of triple-negative breast cancer cells to taxol treatment via the suppression of LZTS1 expression. Oncol Lett (2019) 17(1):990–8. doi: 10.3892/ol.2018.9687
146. Wu C, Zhao A, Tan T, Wang Y, Shen Z. Overexpression of microRNA-620 facilitates the resistance of triple negative breast cancer cells to gemcitabine treatment by targeting DCTD. Exp Ther Med (2019) 18(1):550–8. doi: 10.3892/etm.2019.7601
147. Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, et al. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PloS One (2012) 7(4):e34210. doi: 10.1371/journal.pone.0034210
148. Roscigno G, Puoti I, Giordano I, Donnarumma E, Russo V, Affinito A, et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget (2017) 8(12):19507–21. doi: 10.18632/oncotarget.14470
149. Yu DD, Lv MM, Chen WX, Zhong SL, Zhang XH, Chen L, et al. Role of miR-155 in drug resistance of breast cancer. Tumour Biol (2015) 36(3):1395–401. doi: 10.1007/s13277-015-3263-z
150. Yin Y, Wang X, Li T, Ren Q, Li L, Sun X, et al. MicroRNA-221 promotes breast cancer resistance to adriamycin via modulation of PTEN/Akt/mTOR signaling. Cancer Med (2020) 9(4):1544–52. doi: 10.1002/cam4.2817
151. Bao C, Chen J, Chen D, Lu Y, Lou W, Ding B, et al. MiR-93 suppresses tumorigenesis and enhances chemosensitivity of breast cancer via dual targeting E2F1 and CCND1. Cell Death Dis (2020) 11(8):618. doi: 10.1038/s41419-020-02855-6
152. Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell (2011) 41(2):210–20. doi: 10.1016/j.molcel.2010.12.005
153. Xiang F, Fan Y, Ni Z, Liu Q, Zhu Z, Chen Z, et al. Ursolic acid reverses the chemoresistance of breast cancer cells to paclitaxel by targeting MiRNA-149-5p/MyD88. Front Oncol (2019) 9:501. doi: 10.3389/fonc.2019.00501
154. Jiang H, Cheng L, Hu P, Liu R. MicroRNA−663b mediates TAM resistance in breast cancer by modulating TP73 expression. Mol Med Rep (2018) 18(1):1120–6. doi: 10.3892/mmr.2018.9064
155. Dave B, Mittal V, Tan NM, Chang JC. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res (2012) 14(1):202. doi: 10.1186/bcr2938
156. Pinto CA, Widodo E, Waltham M, Thompson EW. Breast cancer stem cells and epithelial mesenchymal plasticity - implications for chemoresistance. Cancer Lett (2013) 341(1):56–62. doi: 10.1016/j.canlet.2013.06.003
157. Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls notch signalling in cancer cells. EMBO J (2011) 30(4):770–82. doi: 10.1038/emboj.2010.349
158. Zhao N, Powell RT, Yuan X, Bae G, Roarty KP, Stossi F, et al. Morphological screening of mesenchymal mammary tumor organoids to identify drugs that reverse epithelial-mesenchymal transition. Nat Commun (2021) 12(1):4262. doi: 10.1038/s41467-021-24545-3
159. Li HY, Liang JL, Kuo YL, Lee HH, Calkins MJ, Chang HT, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res (2017) 19(1):133. doi: 10.1186/s13058-017-0918-2
160. Di Cosimo S, Appierto V, Pizzamiglio S, Tiberio P, Iorio MV, Hilbers F, et al. Plasma miRNA levels for predicting therapeutic response to neoadjuvant treatment in HER2-positive breast cancer: results from the NeoALTTO trial. Clin Cancer Res (2019) 25(13):3887–95. doi: 10.1158/1078-0432.CCR-18-2507
161. Wang B, Wang K, Yu J, Hao XM, Liu YL, Xing AY. miR-638 serves as a biomarker of 5-fluorouracil sensitivity to neoadjuvant chemotherapy in breast cancer. J Breast Cancer (2022) 25(3):193–206. doi: 10.4048/jbc.2022.25.e24
162. Choi JS, Cho YY. Novel wiring of the AKT-RSK2 signaling pathway plays an essential role in cancer cell proliferation via a G. Biochem Biophys Res Commun (2023) 642:66–74. doi: 10.1016/j.bbrc.2022.12.048
163. Yu Z, Xu Z, Disante G, Wright J, Wang M, Li Y, et al. miR-17/20 sensitization of breast cancer cells to chemotherapy-induced apoptosis requires Akt1. Oncotarget (2014) 5(4):1083–90. doi: 10.18632/oncotarget.1804
164. Zhang F, Lu Y, Wang M, Zhu J, Li J, Zhang P, et al. Exosomes derived from human bone marrow mesenchymal stem cells transfer miR-222-3p to suppress acute myeloid leukemia cell proliferation by targeting IRF2/INPP4B. Mol Cell Probes (2020) 51:101513. doi: 10.1016/j.mcp.2020.101513
165. Naseri Z, Oskuee RK, Jaafari MR, Forouzandeh Moghadam M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int J Nanomed (2018) 13:7727–47. doi: 10.2147/IJN.S182384
166. Yu S, Zhou Y, Niu L, Qiao Y, Yan Y. Mesenchymal stem cell-derived exosome mir-342-3p inhibits metastasis and chemo-resistance of breast cancer through regulating ID4. Genes Genomics (2022) 44(5):539–50. doi: 10.1007/s13258-021-01200-1
167. Ye FG, Song CG, Cao ZG, Xia C, Chen DN, Chen L, et al. Cytidine deaminase axis modulated by miR-484 differentially regulates cell proliferation and chemoresistance in breast cancer. Cancer Res (2015) 75(7):1504–15. doi: 10.1158/0008-5472.CAN-14-2341
168. Gu X, Xue JQ, Han SJ, Qian SY, Zhang WH. Circulating microRNA-451 as a predictor of resistance to neoadjuvant chemotherapy in breast cancer. Cancer biomark (2016) 16(3):395–403. doi: 10.3233/CBM-160578
169. Chin AR, Wang SE. Cancer-derived extracellular vesicles: the 'soil conditioner' in breast cancer metastasis? Cancer Metastasis Rev (2016) 35(4):669–76. doi: 10.1007/s10555-016-9639-8
170. Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol (2015) 77:13–27. doi: 10.1146/annurev-physiol-021014-071641
171. Chen WX, Xu LY, Qian Q, He X, Peng WT, Fan WQ, et al. D rhamnose β-hederin reverses chemoresistance of breast cancer cells by regulating exosome-mediated resistance transmission. Biosci Rep (2018) 38(5). doi: 10.1042/BSR20180110
172. Zhang HF, Xu LY, Li EM. A family of pleiotropically acting microRNAs in cancer progression, miR-200: potential cancer therapeutic targets. Curr Pharm Des (2014) 20(11):1896–903. doi: 10.2174/13816128113199990519
173. Yau C, Osdoit M, van der Noordaa M, Shad S, Wei J, de Croze D, et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol (2022) 23(1):149–60. doi: 10.1016/S1470-2045(21)00589-1
174. Provenzano E. Neoadjuvant chemotherapy for breast cancer: moving beyond pathological complete response in the molecular age. Acta Med Acad (2021) 50(1):88–109. doi: 10.5644/ama2006-124.328
175. Xing AY, Wang B, Li YH, Chen X, Wang YW, Liu HT, et al. Identification of miRNA signature in breast cancer to predict neoadjuvant chemotherapy response. Pathol Oncol Res (2021) 27:1609753. doi: 10.3389/pore.2021.1609753
176. McGuire A, Casey MC, Waldron RM, Heneghan H, Kalinina O, Holian E, et al. Prospective assessment of systemic MicroRNAs as markers of response to neoadjuvant chemotherapy in breast cancer. Cancers (Basel) (2020) 12(7). doi: 10.3390/cancers12071820
177. Liu B, Su F, Lv X, Zhang W, Shang X, Zhang Y, et al. Serum microRNA-21 predicted treatment outcome and survival in HER2-positive breast cancer patients receiving neoadjuvant chemotherapy combined with trastuzumab. Cancer Chemother Pharmacol (2019) 84(5):1039–49. doi: 10.1007/s00280-019-03937-9
178. Di Cosimo S, Appierto V, Pizzamiglio S, Silvestri M, Baselga J, Piccart M, et al. Early modulation of circulating MicroRNAs levels in HER2-positive breast cancer patients treated with trastuzumab-based neoadjuvant therapy. Int J Mol Sci (2020) 21(4). doi: 10.3390/ijms21041386
179. Stevic I, Müller V, Weber K, Fasching PA, Karn T, Marmé F, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med (2018) 16(1):179. doi: 10.1186/s12916-018-1163-y
180. Kassem NM, Makar WS, Kassem HA, Talima S, Tarek M, Hesham H, et al. Circulating miR-34a and miR-125b as promising non invasive biomarkers in Egyptian locally advanced breast cancer patients. Asian Pac J Cancer Prev (2019) 20(9):2749–55. doi: 10.31557/APJCP.2019.20.9.2749
181. García-García F, Salinas-Vera YM, García-Vázquez R, Marchat LA, Rodríguez-Cuevas S, López-González JS, et al. miR−145−5p is associated with pathological complete response to neoadjuvant chemotherapy and impairs cell proliferation by targeting TGFβR2 in breast cancer. Oncol Rep (2019) 41(6):3527–34. doi: 10.3892/or.2019.7102
182. Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer (2012) 118(10):2603–14. doi: 10.1002/cncr.26565
183. Müller V, Gade S, Steinbach B, Loibl S, von Minckwitz G, Untch M, et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the geparquinto trial. Breast Cancer Res Treat (2014) 147(1):61–8. doi: 10.1007/s10549-014-3079-3
184. Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene (2016) 35(4):448–58. doi: 10.1038/onc.2015.96
185. Al-Khanbashi M, Caramuta S, Alajmi AM, Al-Haddabi I, Al-Riyami M, Lui WO, et al. Tissue and serum miRNA profile in locally advanced breast cancer (LABC) in response to neo-adjuvant chemotherapy (NAC) treatment. PloS One (2016) 11(4):e0152032. doi: 10.1371/journal.pone.0152032
186. Zhu W, Liu M, Fan Y, Ma F, Xu N, Xu B. Dynamics of circulating microRNAs as a novel indicator of clinical response to neoadjuvant chemotherapy in breast cancer. Cancer Med (2018) 7(9):4420–33. doi: 10.1002/cam4.1723
187. Kahraman M, Röske A, Laufer T, Fehlmann T, Backes C, Kern F, et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci Rep (2018) 8(1):11584. doi: 10.1038/s41598-018-29917-2
188. Lindholm EM, Ragle Aure M, Haugen MH, Kleivi Sahlberg K, Kristensen VN, Nebdal D, et al. miRNA expression changes during the course of neoadjuvant bevacizumab and chemotherapy treatment in breast cancer. Mol Oncol (2019) 13(10):2278–96. doi: 10.1002/1878-0261.12561
189. Rodríguez-Martínez A, de Miguel-Pérez D, Ortega FG, García-Puche JL, Robles-Fernández I, Exposito J, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res (2019) 21(1):21. doi: 10.1186/s13058-019-1109-0
190. Zhang S, Wang Y, Peng J, Yuan C, Zhou L, Xu S, et al. Serum miR-222-3p as a double-edged sword in predicting efficacy and trastuzumab-induced cardiotoxicity for HER2-positive breast cancer patients receiving neoadjuvant target therapy. Front Oncol (2020) 10:631. doi: 10.3389/fonc.2020.00631
191. Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? the duplicity of MicroRNAs in cancer. Cancer Res (2016) 76(13):3666–70. doi: 10.1158/0008-5472.CAN-16-0359
192. Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: a promising therapeutic target in cancer. Int J Mol Sci (2022) 23(19). doi: 10.3390/ijms231911502
193. Mollaei H, Safaralizadeh R, Rostami Z. MicroRNA replacement therapy in cancer. J Cell Physiol (2019) 234(8):12369–84. doi: 10.1002/jcp.28058
194. Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res (2014) 74(24):7573–82. doi: 10.1158/0008-5472.CAN-14-1140
195. Elghoroury EA, ElDine HG, Kamel SA, Abdelrahman AH, Mohammed A, Kamel MM, et al. Evaluation of miRNA-21 and miRNA let-7 as prognostic markers in patients with breast cancer. Clin Breast Cancer (2018) 18(4):e721–e6. doi: 10.1016/j.clbc.2017.11.022
196. Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene (2010) 29(29):4194–204. doi: 10.1038/onc.2010.167
197. Mutlu M, Raza U, Saatci Ö, Eyüpoğlu E, Yurdusev E, Şahin Ö. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J Mol Med (Berl) (2016) 94(6):629–44. doi: 10.1007/s00109-016-1420-5
198. Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J, et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol (2014) 54:312–7. doi: 10.1016/j.biocel.2014.05.040
199. Langer C, Rücker FG, Buske C, Döhner H, Kuchenbauer F. Targeted therapies through microRNAs: pulp or fiction? Ther Adv Hematol (2012) 3(2):97–104. doi: 10.1177/2040620711432582
200. Li D, Wang X, Yang M, Kan Q, Duan Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp Cell Res (2019) 380(1):20–8. doi: 10.1016/j.yexcr.2019.03.025
201. Lin Y, Lin F, Anuchapreeda S, Chaiwongsa R, Duangmano S, Ran B, et al. Effect of miR-133b on progression and cisplatin resistance of triple-negative breast cancer through FGFR1-wnt-β-catenin axis. Am J Transl Res (2021) 13(6):5969–84.
202. Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang GX, et al. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat (2010) 9(1):77–86. doi: 10.1177/153303461000900109
203. Møller HG, Rasmussen AP, Andersen HH, Johnsen KB, Henriksen M, Duroux M. A systematic review of microRNA in glioblastoma multiforme: micro-modulators in the mesenchymal mode of migration and invasion. Mol Neurobiol (2013) 47(1):131–44. doi: 10.1007/s12035-012-8349-7
204. Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther (2011) 19(6):1116–22. doi: 10.1038/mt.2011.48
205. Schneider MR. MicroRNAs as novel players in skin development, homeostasis and disease. Br J Dermatol (2012) 166(1):22–8. doi: 10.1111/j.1365-2133.2011.10568.x
206. He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene (2012) 31(28):3357–69. doi: 10.1038/onc.2011.500
207. Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther (2011) 18(12):1111–20. doi: 10.1038/gt.2011.100
208. Zhou LY, Qin Z, Zhu YH, He ZY, Xu T. Current RNA-based therapeutics in clinical trials. Curr Gene Ther (2019) 19(3):172–96. doi: 10.2174/1566523219666190719100526
209. Kasar S, Salerno E, Yuan Y, Underbayev C, Vollenweider D, Laurindo MF, et al. Systemic in vivo lentiviral delivery of miR-15a/16 reduces malignancy in the NZB de novo mouse model of chronic lymphocytic leukemia. Genes Immun (2012) 13(2):109–19. doi: 10.1038/gene.2011.58
210. Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet (2014) 15(8):541–55. doi: 10.1038/nrg3763
211. Fernandez-Piñeiro I, Badiola I, Sanchez A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol Adv (2017) 35(3):350–60. doi: 10.1016/j.biotechadv.2017.03.002
212. Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming nonviral gene delivery barriers: perspective and future. Mol Pharm (2013) 10(11):4082–98. doi: 10.1021/mp400467x
213. Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med (2013) 368(18):1685–94. doi: 10.1056/NEJMoa1209026
214. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discovery (2013) 12(11):847–65. doi: 10.1038/nrd4140
215. Stylianopoulos T, Jain RK. Combining two strategies to improve perfusion and drug delivery in solid tumors. Proc Natl Acad Sci U S A (2013) 110(46):18632–7. doi: 10.1073/pnas.1318415110
216. Raemdonck K, Vandenbroucke RE, Demeester J, Sanders NN, De Smedt SC. Maintaining the silence: reflections on long-term RNAi. Drug Discovery Today (2008) 13(21-22):917–31. doi: 10.1016/j.drudis.2008.06.008
217. Yu B, Zhao X, Lee LJ, Lee RJ. Targeted delivery systems for oligonucleotide therapeutics. AAPS J (2009) 11(1):195–203. doi: 10.1208/s12248-009-9096-1
218. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discovery (2010) 9(10):775–89. doi: 10.1038/nrd3179
219. Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol (2005) 23(4):457–62. doi: 10.1038/nbt1081
220. Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A (2009) 106(8):2735–40. doi: 10.1073/pnas.0811073106
221. Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, et al. An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci (2012) 15(6):827–35. doi: 10.1038/nn.3113
222. van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods (2008) 5(12):1023–5. doi: 10.1038/nmeth.1267
223. Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics (2010) 11(7):537–61. doi: 10.2174/138920210793175895
224. Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, et al. Functional importance of dicer protein in the adaptive cellular response to hypoxia. J Biol Chem (2012) 287(34):29003–20. doi: 10.1074/jbc.M112.373365
225. Vicentini FT, Borgheti-Cardoso LN, Depieri LV, de Macedo Mano D, Abelha TF, Petrilli R, et al. Delivery systems and local administration routes for therapeutic siRNA. Pharm Res (2013) 30(4):915–31. doi: 10.1007/s11095-013-0971-1
226. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med (2018) 379(22):2108–21. doi: 10.1056/NEJMoa1809615
Keywords: micro RNA, breast cancer, chemoresistance, neoadjuvant (chemo)radiotherapy, systemic therapies
Citation: Singh S, Saini H, Sharma A, Gupta S, Huddar VG and Tripathi R (2023) Breast cancer: miRNAs monitoring chemoresistance and systemic therapy. Front. Oncol. 13:1155254. doi: 10.3389/fonc.2023.1155254
Received: 31 January 2023; Accepted: 05 June 2023;
Published: 16 June 2023.
Edited by:
Zhi-Gang Zhuang, Shanghai First Maternity and Infant Hospital, ChinaReviewed by:
Macrina Beatriz Silva Cázares, Autonomous University of San Luis Potosí, MexicoBruce Alan Bunnell, University of North Texas Health Science Center, United States
Subhayan Das, Indian Institute of Technology Kharagpur, India
Copyright © 2023 Singh, Saini, Sharma, Gupta, Huddar and Tripathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richa Tripathi, cmljaGEudHJwdGhzQGdtYWlsLmNvbQ==
 Shivam Singh
Shivam Singh Heena Saini
Heena Saini Ashok Sharma
Ashok Sharma Subhash Gupta
Subhash Gupta V. G. Huddar
V. G. Huddar Richa Tripathi
Richa Tripathi