- 1Department of Clinical Laboratory, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
- 2Department of Reproductive Medicine, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
- 3Medical Research Center, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Esophageal squamous cell carcinoma (ESCC) is the most prevalent histological esophageal cancer characterized by advanced diagnosis, metastasis, resistance to treatment, and frequent recurrence. In recent years, numerous human disorders such as ESCC, have been linked to abnormal expression of circular RNAs (circRNAs), suggesting that they are fundamental to the intricate system of gene regulation that governs ESCC formation. The tumor microenvironment (TME), referring to the area surrounding the tumor cells, is composed of multiple components, including stromal cells, immune cells, the vascular system, extracellular matrix (ECM), and numerous signaling molecules. In this review, we briefly described the biological purposes and mechanisms of aberrant circRNA expression in the TME of ESCC, including the immune microenvironment, angiogenesis, epithelial-to-mesenchymal transition, hypoxia, metabolism, and radiotherapy resistance. As in-depth research into the processes of circRNAs in the TME of ESCC continues, circRNAs are promising therapeutic targets or delivery systems for cancer therapy and diagnostic and prognostic indicators for ESCC.
1 Introduction
As one of the most prevalent malignant tumors, esophageal squamous cell carcinoma (ESCC) is associated with a high mortality rate worldwide (1). In 2020, there were 604,100 new esophageal cancer (EC) cases and 544,076 EC-related deaths, ranking seventh and sixth in cancer morbidity and mortality, respectively (2, 3). ESCC accounts for approximately 85% of all EC cases worldwide (4). Currently, available treatment strategies for ESCC include surgical resection, chemotherapy, radiotherapy, molecular targeted therapy, and their combinations (5). However, due to the high probability of recurrence, early metastasis, and extremely low five-year survival rate, the patient prognosis continues to be poor (6, 7).
CircRNAs have been identified and represent unusual canonical and non-canonical RNA splicing errors (8). However, high-throughput sequencing and specialized computational pipelines have led to recognizing circular RNAs (circRNAs) as a class of endogenous non-coding RNA existing objectively in multiple species without a 5’ end cap and a 3’ end poly(A) tail, forming a circular structure with covalent bonds rather than “transcriptional noise.” (9, 10) Such a unique loop structure makes circRNAs more stable than linear RNAs because they are resistant to RNA exonucleases and RNase R nucleic acid exonucleases (11). CircRNAs have been recently discovered to be involved in cancer growth, metastasis, recurrence, and therapy resistance by regulating sponging miRNAs, binding with proteins, and encoding proteins and peptides (12–16).
The communication between ESCCs and other cells and the interaction among different cell types in the tumor microenvironment (TME) determines ESCC development and progression (6, 17). To establish a TME, tumor cells change their normal developing environment as cancer progresses (18). Through interaction with tumor cells, the TME, constituting the region around the tumor during development, is vital in cancer-related disorders (19). The TME plays a significant role in carcinoma progression, including proliferation, metastasis, immune evasion, and chemoradiation resistance (20). In addition, extracellular metabolites, such as exosomes, which are typically communication signals between various cellular compartments, frequently play a role in the complicated interactions between tumor cells, normal cells, their microenvironment, and the accompanying stroma (18). CircRNAs primarily contribute to and control ESCC development by affecting the TME immunological microenvironment, metabolism, hypoxia, angiogenesis, and epithelial-to-mesenchymal transition (EMT) (19, 21, 22). The development of more potent ESCC therapies may be facilitated by establishing a circRNA-involved TME network, with tailored therapy being an option based on the interaction with circRNAs (23–25).
In this review, we explored the irreplaceable role of circRNAs in regulating cancer progression in the TME of ESCC. In summary, circRNAs exhibit good development potential despite the TME studies on them being in their early stages. CircRNA research translation into clinical applications is required.
2 Biogenesis regulation, degradation, and function of circRNA
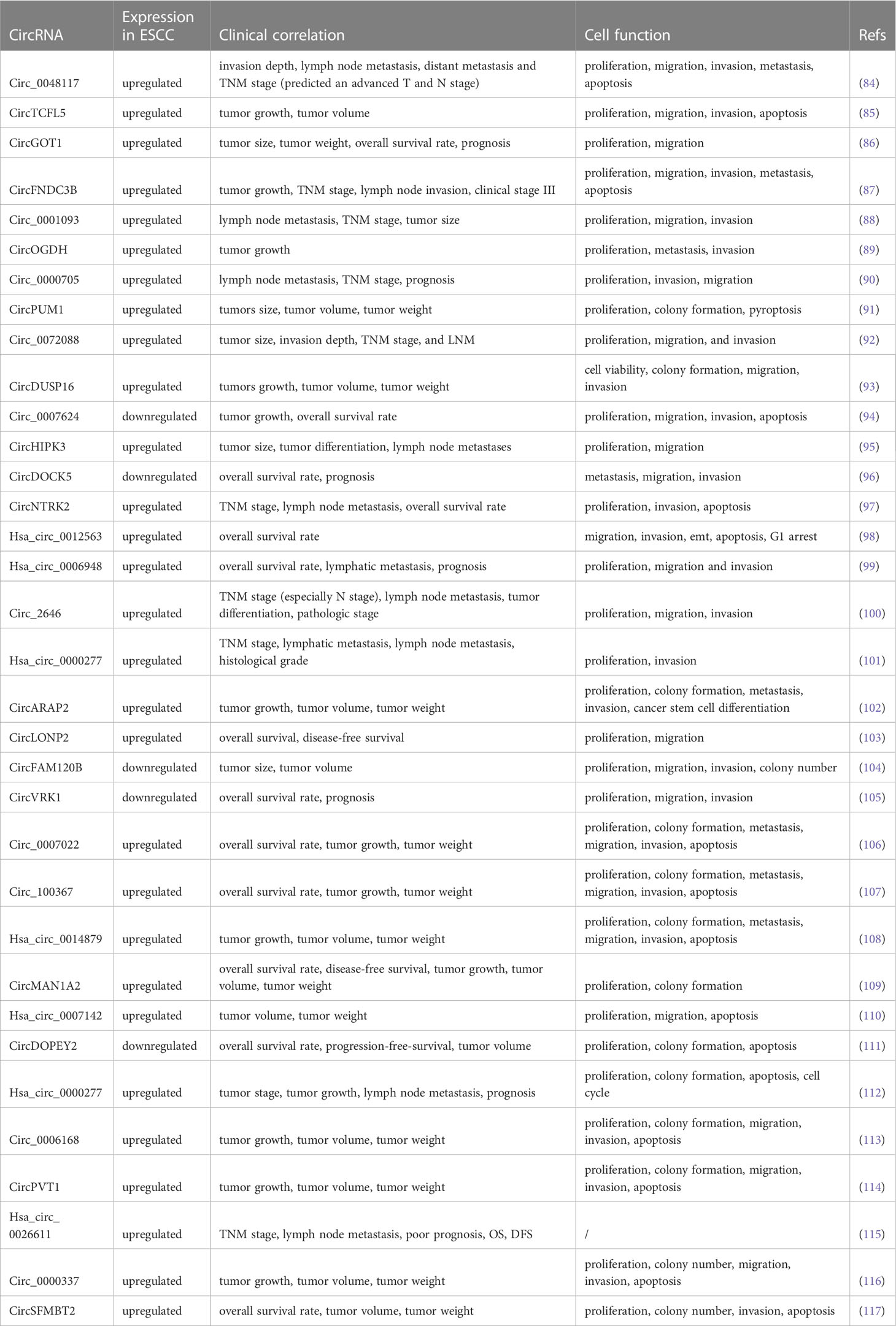
For a long time, most circRNAs were considered “splicing noise” or “byproducts of RNA processing.” (26) In recent years, circRNAs have been increasingly studied and observed to play essential roles in normal cell differentiation, tissue homeostasis, and disease development. Their expression is usually independent of host gene expression (27). CircRNAs are stable byproducts of mRNA splicing and products of newly regulated selective splicing, which have distinctive molecular mechanisms (28). This circular structure endows circRNAs with many distinct features: sequence conservation, evolutionary species conservation, tissue-specific expression patterns, and high abundance and stability, demonstrating that circRNAs have vital non-coding functions (29–31). We will focus later on the regulation, degradation and function of circRNA (Figure 1).
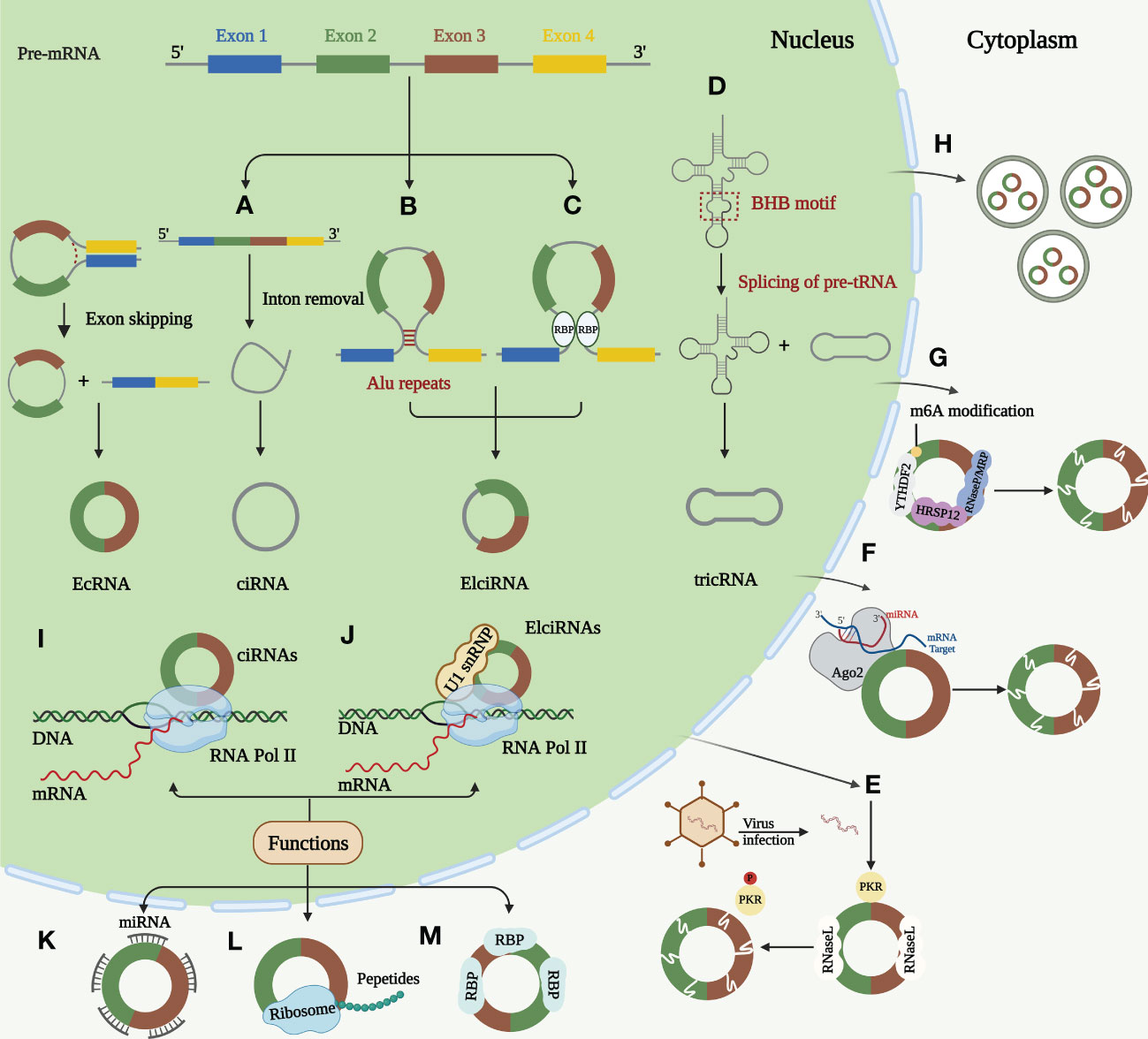
Figure 1 Biogenesis, degradation, and functions of circRNAs. (A) Lariat-driven circularization. The skipped exon and intron sequences are spliced from the previous mRNA. The 5 ‘splice receptor and 3’ splice donor are connected to form a lariat structure. Larch further undergoes internal splicing to remove the intron sequence, thus generating EIciRNA or EcRNA. Some spliced introns can be cyclized to form ciRNA. (B) Intron paring-driven circularization. This circularization depends on the complementary base pairing of different fan introns to form a hairpin structure. The complementary sequence in the flanking intron of the exon is spliced after splicing, so that the downstream 5 ‘splice site is close to the upstream 3’ splice site for cycle. Most intron pairing patterns are promoted by ALU duplication. (C) RBPs-driven circularization. RBP binds to the RBP binding sites on the sector sequence, and these two RBPs can facilitate the cycle by bringing the splicing sites closer together. (D) Formation of tricRNAs. Introns carrying pre-tRNA are cut at the characteristic bulge helix bulge (BHB) motif by the tRNA splicing endonuclease (TSEN) complex. Ligase links the RNA that is generated at the end of the tron to form stabilized tricRNA. (E) Active RNase L triggers circRNA degradation upon virus infection, which relieves the suppression of PKR. (F) miRNAs can bind to circRNAs through base pairing and guide Ago2 dependent cleavage. (G) CircRNA containing m6A is recognized by YTHD F2 which can interact with RNase P/MRP bridged by HRSP12 and then the complex initiating the nuclear degradation of circRNAs. (H) CircRNAs can be excreted to the extracellular space by exosomes. Function of circRNAs. (I) Transcriptional regulation. ciRNAs regulates the transcription of host genes by promoting the extension activity of RNA Pol II. (J) Transcriptional regulation. ElciRNAs can combine with U1 snRNP and then interact with RNA Pol II to regulate host gene transcription in nucleus. (K) miRNA sponging. CircRNAs act as miRNA sponge to regulate the function of miRNA target mRNAs. (L) Coding of proteins and peptides. CircRNAs can perform regulatory functions by encoding proteins and peptides. (M) RNA binding proteins. CircRNAs can combine with RBPs by different mechanisms.
2.1 Biogenesis, regulation, and degradation of circRNA
Most circRNAs are produced by reverse splicing of precursor mRNAs (pre-mRNAs), creating a single-stranded covalent closed-loop structure by connecting the upstream 3′ splice acceptor site to the downstream 5′ splice donor site containing a specific junction site. This is followed by removing all or part of the intron by the spliceosome and ligating the remaining sequences, producing different circRNAs (32, 33). This procedure was performed under the regulation of cis-acting elements and trans-acting factors (34). CircRNAs can be divided into three main categories according to their source sequences: exonic circRNAs (EcRNAs), intronic circRNAs (ciRNAs), and exon-intron circRNAs (EIciRNA) (35–37). The back-splicing hypothesis models currently proposed to describe circRNA generation are Lariat-driven circularization, intron pairing-driven circularization, and RNA-binding protein (RBP)-driven circularization. The three models have different mechanisms (11, 28, 32).
Lariat-driven circularization occurs while removing introns or exons from pre-mRNAs (38, 39). Intron pairing-driven circularization: The basis of this type of circulation is the complementary base pairing of various fan introns, and complementary sequences—short repeat elements (e.g., ALU repeats) or non-repeat elements—in introns flanking post-snap exons are important cis-acting elements of circRNA biogenesis (40). Regarding RBP-driven circularization, fan sequences contain RBP binding sites. By bringing the splice sites close, both RBPs can enhance recycling (41). Certain RBPs are trans-acting regulators of circRNA synthesis (42). In contrast, some RBPs may prevent circRNA synthesis by preventing proper matching of intron elements (43, 44). In addition to pre-mRNA production, a minor portion of intron-derived circRNAs is formed by precursor tRNA (pre-tRNA) splicing (45). RNA synthesized at the intron end is ligated by ligase into a stable circRNA called tRNA intronic circular RNA (tricRNA) (45, 46).
The high circRNA expression in cancer is often closely associated with poor prognosis. Therefore, understanding circRNA degradation and inactivation mechanisms is crucial for emerging targeted therapies (12). A study revealed that circRNAs can be degraded by RNase L. Endogenous circRNAs frequently form defective double complexes, preventing protein kinase (PKR) activation by double-stranded RNA (dsRNA)-activated PKR. Their reduction triggers abnormal PKR activation and autoimmune responses (47). Argonaute 2 (Ago2)-mediated RNA degradation is the most commonly reported mode of circRNA degradation (48, 49). Ago2 is a nucleic acid endonuclease that uses endogenous guide RNAs, such as miRNAs (50). However, Ago2-dependent circRNA degradation also has some drawbacks, as it is inapplicable to circRNAs without specific miRNA targets (51). The most frequent RNA modification is m6A (52). Recent studies have revealed that YTHDF2 recognizes circRNAs containing m6A, and HRSP12 links YTHDF2 and RNase P or MRP, enabling RNase P/MRP to initiate the intranuclear degradation of YTHDF 2-bound circRNAs (53). According to recent research, all circRNAs that possess an open reading frame (ORF) were predicted to possess m6A sites (54). Besides intracellular pathways, circRNAs can be further exported to extracellular vesicles contributing to their clearance (55). Cancer progression caused by circRNA dysregulation promises new clinical therapeutic advances as the biogenesis and degradation mechanisms of circRNAs are further investigated.
2.2 Biological functions of circRNAs
The biological roles of circRNAs have also been thoroughly explained. According to recent studies, circRNAs perform their tasks using four primary methods: miRNA sponging, transcription regulation, protein and peptide coding, and RNA-binding protein incorporation (27, 28, 56). CircRNAs can function as miRNA sponges: Most circRNAs in this review can affect the ESCC TME by acting as miRNA sponges to regulate downstream targets. Moreso, circRNAs act as competing endogenous RNAs (ceRNAs) and microRNA (miRNA) sponges by binding to target mRNAs in a base-paired pattern, causing mRNA cleavage or limiting mRNA translation to regulate miRNA activity on other target genes (10, 56, 57). According to previous reports, circRNAs simultaneously regulate several miRNAs and have multiple miRNA-binding sites. These evolutionarily conserved binding sites guarantee the target efficacy (58). Notably, only a few circRNAs have multiple miRNA-binding sites (59). For instance, Sun et al. have found that a total nine miRNAs and nine candidate mRNA were predicted to have an interaction with has_circ_0000520 (60). CircRNAs regulate transcription: In addition, circRNAs play a significant role in post-transcriptional or transcriptional regulation of gene expression (61, 62). This role is mainly mediated by EIciRNAs and ciRNAs expressed in the nucleus during parental gene regulation (63, 64). The promoter region of the parental gene is bound by a complex formed by specific RNA-RNA interactions between EIciRNA, U1 small nuclear ribonucleoprotein (U1snRNP), and U1 small ribonucleic acid (snRNA). This complex then interacts with RNA Pol II to enhance and promote the transcription of the parental gene (63, 65). ciRNAs regulate their parental function by boosting RNA Pol II elongation (66). CircRNAs can encode proteins and peptides: Despite circRNAs being non-coding RNAs, they also regulate by encoding proteins and polypeptides (67–69). Studies have revealed a connection between circRNAs and polymers, a portion containing the initiation codon AUG and a putative ORF of favorable length, indicating that circRNAs have protein-coding potential (70, 71). Although circRNAs are classified as lncRNAs, they can encode some short peptides. The coding circRNA contains an internal ribosomal entry site (IRES) that allows translation of some of the coding sequences in the circRNA (72). Proteomics application has provided the most visual evidence that ncRNAs can encode regulatory peptides (71). Pamudurti et al. demonstrated that circMbl is capable of producing proteins of approximately 10kDa size in Drosophila by establishing intron-exon-intron minigenes (73). Studies have revealed that some proteins or peptides encoded by circRNAs play irreplaceable regulatory roles in tumorigenesis and progression (74–78). CircRNAs can interact with RNA-binding proteins: In addition to the functions mentioned above, circRNAs combine with RBPs through different mechanisms. RBPs are a broad category of proteins involved in the transcription and translocation of genes and can serve as fundamental elements of circRNA function (41). Three primary modes of action have been described for circRNA binding to RBPs: formation of protein complexes (79), inhibition of protein function (protein decoys) (80), and interactions between different proteins (81). The first is the formation of protein complexes: circRNAs with multiple protein-binding sites can serve as dynamic scaffolding for assembling massive RNA-protein complexes by modulating protein-protein interactions (82). The second category is protein decoys, where circRNA-encoded proteins are in competing with their homologous linear splice protein isoforms for binding molecules, thereby inhibiting normal isoform function (80). Finally, circRNAs bind and chelate specific proteins and regulate certain protein-protein and protein-RNA interactions (81, 83). CircRNAs can form distinct circRNA-protein complexes (circRNPs) by interacting with various proteins, changing the related proteins’ mode.
3 CircRNA dysregulation modulates the clinical characteristics and biological processes of ESCC
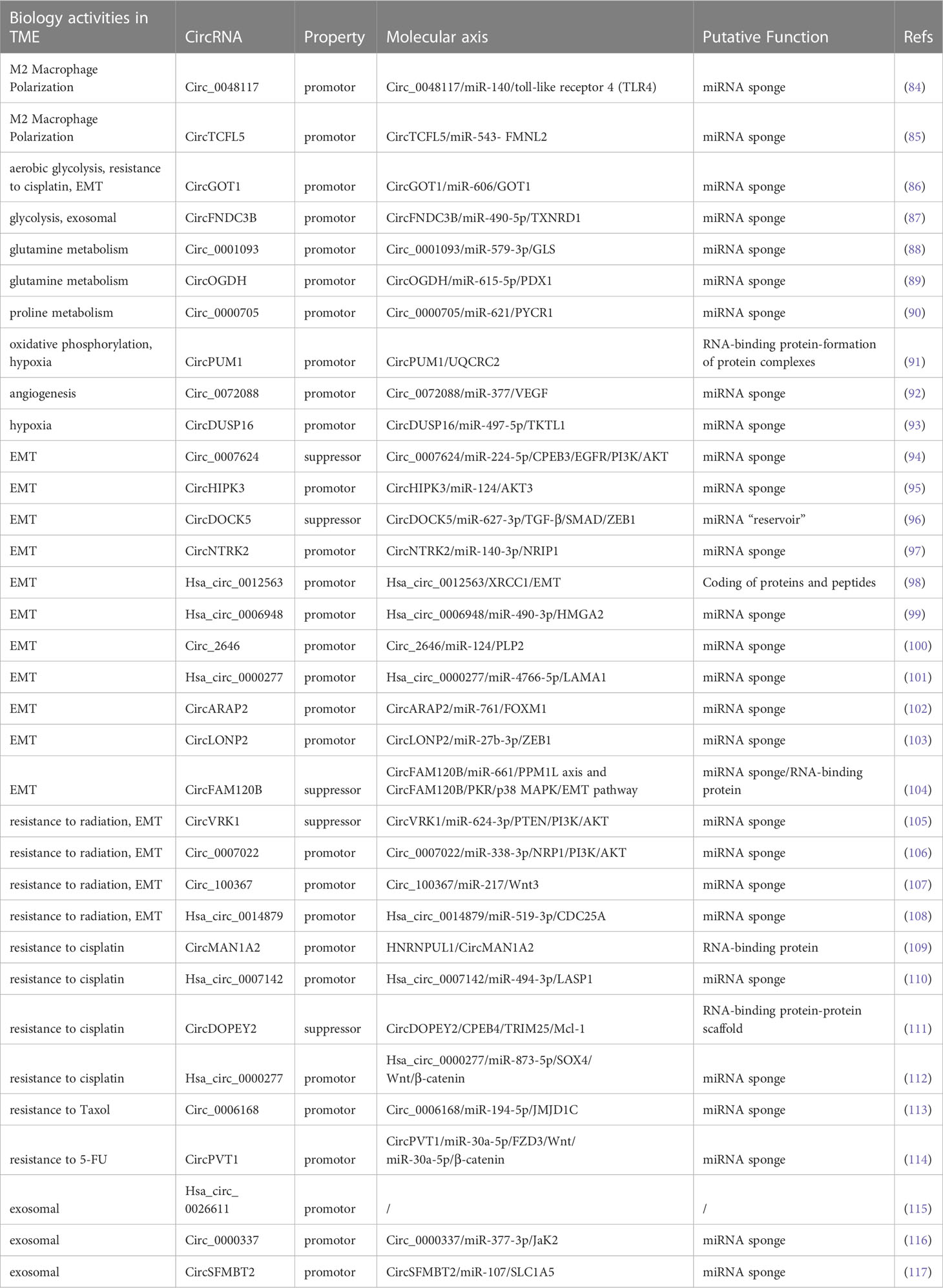
A few aberrantly produced circRNAs were recently revealed to control the proliferation, invasion, migration, and apoptosis of ESCC in TME, thereby affecting ESCC progression. This could lead to new insights into circRNAs as therapeutics. Here, we summarize the biological functions and clinicopathological features of the aberrantly expressed circRNAs in ESCC (Table 1).
ESCC mainly carries out distant metastasis through the lymphatic system, causing patients to lose the opportunity for early surgery, and is directly related to poor prognosis (118–120). High expression of circ_0048117, circ_0001093, circ_0000705, and circHIPK3 are closely related to lymph node metastasis and the TNM stage of ESCC (84, 88, 90, 95). Conversely, as a tumor suppressor, low circDOCK5 expression was positively correlated with overall survival (96). In addition, the highly expressed circNTRK2, hsa_circ_0006948, circ_2646, and hsa_circ_0000277 were closely associated with lymph node metastasis and overall survival time and tumor differentiation (97, 99–101). Cheng et al. revealed that upregulated hsa_circ_0000277 is associated with lymph node metastasis and recurrence in patients with ESCC (112). Liu et al. demonstrated that hsa_circ_0026611 was in the serum of patients with ESCC in the form of serum exosomes and could identify whether ESCC was complicated by lymph node metastasis. In addition, hsa_circ_0026611 was closely related to TNM staging. Thus, hsa_circ_0026611 could be a biomarker in clinical practice (115).
The TNM stage of patients with ESCC is directly related to prognosis and is closely associated with surgery and chemotherapy (121). CircFNDC3B and circ_0072088 are highly expressed in ESCC and are positively correlated with the TNM stage and poor survival (87, 92). In addition, circRNAs are related to tumor volume and weight (85, 86, 89, 91). CircDUSP16, circARAP2, circFAM120B, and hsa_circ_0014879 promote an increased tumor volume by promoting tumor cell proliferation and inhibiting apoptosis (93, 102, 104, 108). Circ_0007142 was upregulated in ESCC tissues and cells and correlated with cisplatin resistance. Mechanistically, circ0007142 increased cell survival by increasing the resistance of tumor cells to cisplatin, leading to increased tumor volume and weight (110). Circ_0006168 resists chemotherapy by reducing paclitaxel sensitivity in resistant cells, increasing tumor volume (113). CircPVT1 is vital in maintaining ESCC chemoresistance to 5-FU through ferroptosis and the Wnt/β pathway. In vivo, circPVT1 reduced the sensitivity of tumor cells to 5-FU and increased the tumor volume under chemotherapy (114). As an oncogenic factor, circ_0000337 knockdown can improve the sensitivity of ESCCs to cisplatin, thereby reducing tumor volume and weight in vivo (116).
4 CircRNAs as a novel regulator of tumor microenvironment in ESCC
The TME is a vital factor in determining cancer progression at all stages and is a complex ecosystem in which cancer cells and the stroma around them coexist (122). Recruitment and conversion of tumor cells into nearby normal cells are essential for cancer advancement (123). The TME consists of stromal cells, immune cells, the vascular system, and the extracellular matrix (ECM). Crosstalk among TME components is essential for influencing cancer progression (124–126). As mentioned above, abnormal expression circRNAs levels in ESCC are caused by various mechanisms. The functional mechanisms of circRNAs in ESCC TME are discussed in the following sections. (Figure 2; Table 2).
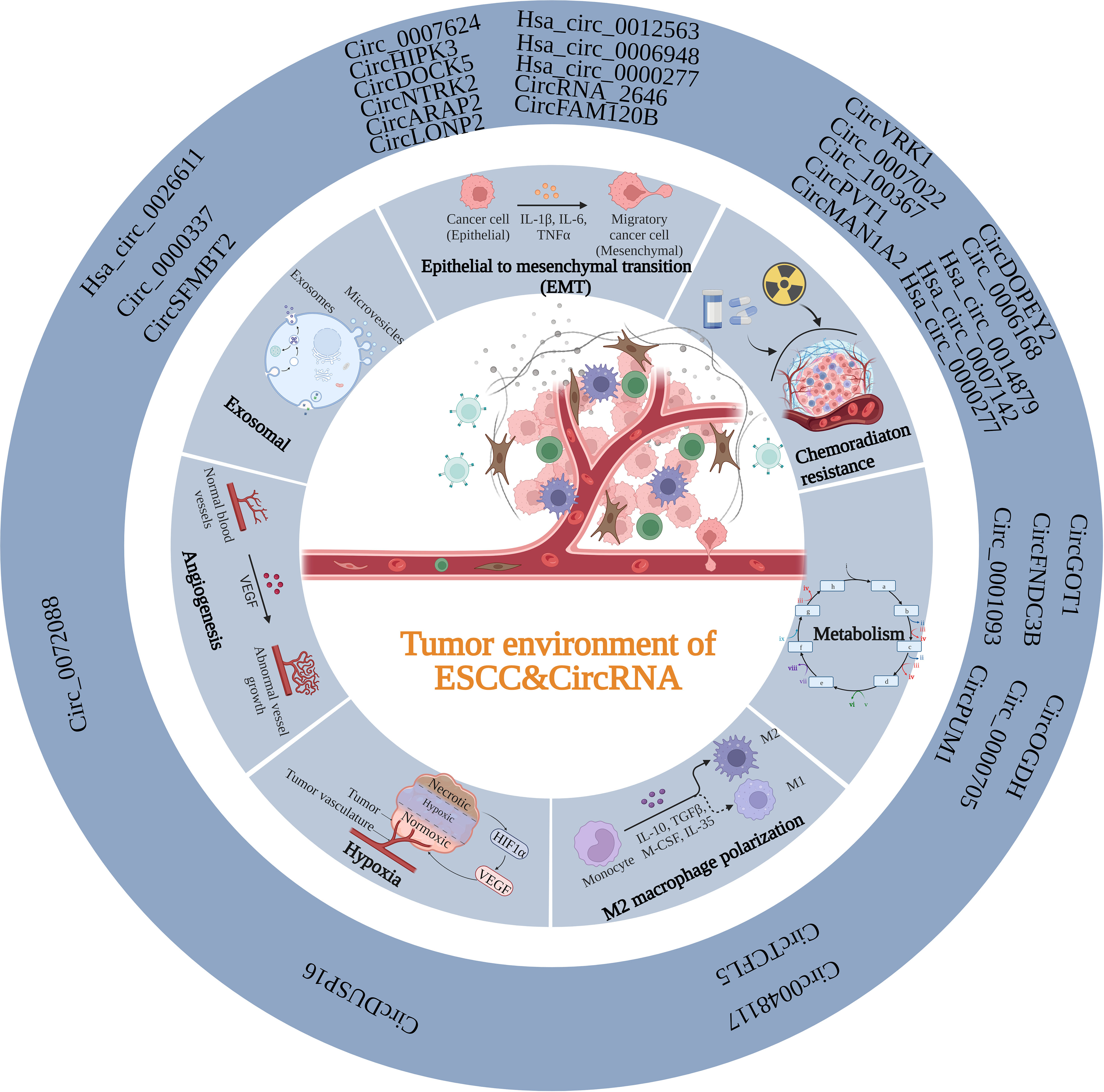
Figure 2 Summary of the function of circRNAs in the TME of ESCC. CircRNAs can participate in the TME of ESCC through various mechanisms, including EMT, exosomal, angiogenesis, hypoxia, M2 polarization, metabolism and chemoradiation resistance.
4.1 Modulating the immune microenvironment in ESCC
The term “macrophage polarization” describes the macrophage activation state at a specific moment (127). Activated macrophages are in two groups based on the guidance of various microenvironmental signals: classically activated macrophages (M1) and alternatively activated macrophages (M2) (128). The polarization of M1/M2 macrophages is critical for tumor progression. Historically, M1 macrophages have been thought to have antitumor activities, whereas M2-polarized macrophages, also known as tumor-associated macrophages (TAMs), have anti-inflammatory and pro-tumor actions (129, 130). In contrast to M1’s antitumor properties, M2-polarized macrophages are incredibly adaptable and multifunctional cells, which also significantly contribute to many pro-tumorigenic outcomes through tumorigenesis, together with angiogenic and lymphangiogenic regulation, immune suppression, hypoxia induction, and stemness (131). CircRNAs are generally associated with M2 polarization (132–134); however, the specific mechanism between circRNAs and M2 polarization is unclear. In recent studies, Lu et al. observed that hypoxia encouraged ESCC cells to produce more exosomes rich in hsa_circ_0048117, which might induce TAMs to differentiate into the M2 type via the miR-140/toll-like receptor 4 (TLR4) pathway. Thus, hsa_circ_0048117 can be seen as a macrophage miR-140 “sponge” which directly competes with TLR4 to bind miRNA. In addition, the ability of ESCC cells to invade and migrate may be enhanced by M2 macrophages (84). M2 macrophages can release Arg1, IL-10, and TGF-α which boost the tumor cells’ capacity for invasion and migration and aid in ESCC metastasis. Compared to healthy participants, serum exosomal hsa_circ_0048117 levels were considerably higher in patients with ESCC. Higher exosomal levels of circ_0048117 were strongly linked to TNM grade and predicted an advanced T and N stage (84). Similarly, in another research, Lin et al. discovered that circTCFL5 was markedly increased in ESCC and could speed up the disease evolution by upregulating FMNL2 by sponging miR-543 in vitro and in vivo. Mechanistically, circTCFL5 accelerates esophageal cancer development by modifying M2 macrophage polarization through the miR-543/FMNL2 axis (85).
Previous studies have revealed that circRNAs can interact with the tumor immune microenvironment by participating in multiple approaches (19). In addition to participating in M2 polarization and interacting with macrophages, circRNAs mediate tumor immune surveillance (135, 136), regulate immune escape via PD-L1 (137–140), regulate natural killer cells’ cytotoxicity (141–143), and regulate neutrophils, myeloid-derived suppressor cells, and cancer-associated fibroblast (144). However, whether circRNA affects ESCC TME through the aforementioned effects, except M2 polarization, has not yet been reported.
4.2 Energy metabolism regulation in the TME
Tumors caused by genomic instability frequently exhibit energy metabolism reprogramming (30). Deregulation of metabolism, common in tumor development, is a crucial source of growth and division. Proteins and non-coding RNAs (ncRNAs) are involved in orchestrating these changes in energy metabolism (145). CircRNAs, a novel class of regulatory molecules, regulate cancer metabolism. CircRNAs regulate the energy metabolism of ESCC through different targets.
Glucose is a vital carbon substrate for generating energy and metabolic intermediates (146). Cancer cells frequently rely on glycolytic metabolism for maintenance; nonetheless, cellular reprogramming of metabolic processes frequently occurs in developing tumors when blood perfusion is constrained (147). For instance, circGOT1 functions as an oncogene in ESCC, which promotes aerobic glycolysis through the miR-606/GOT1 axis, acting as an mRNA sponge (86). CircGOT1 knockdown reduces ESCC cell motility, proliferation, and chemoresistance to cisplatin (86). Additionally, circGOT1 inhibition decreased lactate synthesis, glucose consumption, and ATP levels (86). Further, circFNDC3B was upregulated in ESCC tissues and cells, and circFNDC3B knockdown caused apoptosis, decreased cell glycolysis, and inhibited xenograft tumor formation (87). Sponging miR-490-5p and upregulating TXNRD1 expression resulted in the aforementioned inhibitory effects (87). TXNRD1 overexpression reduced the repressive function by enhancing cell proliferation, migration, and glycolysis and lowering apoptosis in ESCC (87). In addition, tumors from patients with clinical stage III tumors expressed more circFNDC3B than tissues from individuals with clinical stage I + II (87).
Amino acid metabolism is involved in cancer progression. Cancer cells exert worsened metabolic activity to maintain their increased proliferation and microenvironmental adaptation, to survive in nutrient-poor environments. Tumors exhibit increased energy-producing processes such as glycolysis, glutaminolysis, and fatty acid production (148). Since the 1950s, researchers have discovered that glutamine is the amino acid most tumor cells use. The fundamental biological activities require glutamine, which glutaminase transforms into glutamate. Glutamate can also provide nitrogen for purines and pyrimidines production or participate in the TCA cycle via conversion to α-ketoglutarate (145). Glutamate is another source of glutathione, a vital cellular antioxidant. Glutamine’s catabolism creates an anaplerotic pathway that feeds the Krebs cycle; therefore, some cancer cells depend highly on it (149). Thus, glutamine is crucial for tumor development. Recently, Qian et al. discovered that circ_0001093 is overexpressed in ESCC and functions as an mRNA sponge for miR-579-3p to promote GLS expression, boosting glutamine metabolism and malignant phenotype of ESCC (88). Similarly, miR-615-5p was sponged by circOGDH to release PDX1, which increased glutamine metabolism and promoted tumor progression in ESCC (89). Further studies have revealed that elevated miR-615-5p reduces ATP content, α-KG synthesis, and glutamine consumption, which are suppressed by PDX1 (89).
In addition to glutamate metabolism, circRNAs influence ESCC progression by participating in other amino acid metabolism pathways. Proline metabolism is essential to metabolic reprogramming in some malignancies, producing ATP. In cancerous cells, proline metabolism is associated with ATP synthesis, protein and nucleotide production, and redox balance (150–153). Recent studies have demonstrated that proline metabolism is critical for metabolic reprogramming, promoting cell proliferation, preventing cell apoptosis, and generating metabolites that support cancer cell survival in various stressful environments (153–157). Circ_0000705 knockdown boosted ROS generation, decreasing proline and PYCR1 expression and ATP synthesis in ESCC cells (90). Additionally, the inhibitory effects of circ_0000705 knockdown on the aforementioned biological functions in ESCC were reversed by miR-621 suppression or PYCR1 expression restoration (90). Therefore, circ_0000705 might accelerate ESCC development by targeting the miR621/PYCR1 axis and promoting proline metabolism (90).
In addition, increased oxidative phosphorylation may contribute to hypoxia in cancerous cells; however, the exact mechanism underlying this phenomenon is currently being investigated. There are few reports on the potential role of circRNAs in the oxidative phosphorylation of tumor cells (158–160). CircPUM1 is essential for maintaining mitochondrial complex III integrity (91). In ESCC, circPUM1 serves as a scaffold for UQCRC1 binding to UQCRC2 (91). Lower intracellular oxygen levels, significantly suppressed oxidative phosphorylation, decreased mitochondrial membrane potential, and increased ROS production are effects of circPUM1 knockdown (91). Mitochondrial complex III becomes dysfunctional because of circPUM1 deficiency (91). In addition, circPUM1 disruption causes pyroptosis, which initiates ESCC cell death by altering the intracellular ATP levels (91).
Several circRNAs are tightly linked to glycolysis, glutamine metabolism, proline metabolism, and oxidative phosphorylation. In addition, circRNAs may affect ESCC progression through other mechanisms, such as serine and lipid metabolism (145). We could uncover the mystery of ESCC development with further studies in this area.
4.3 Abnormal regulation of angiogenesis
Angiogenesis is a crucial process in growth and development and has been the focus of research in recent years. Endothelial cells (ECs) proliferate, differentiate, and migrate during angiogenesis to form new blood vessels based on pre-existing capillaries or venules (161, 162). Additionally, angiogenesis mainly involves cell migration, proliferation, and vascular endothelial growth factor (VEGF) (163–165). A characteristic of cancer is angiogenesis induction (166). The significance of malignant cell neovasculature in tumor biology and nutrient and oxygen delivery to growing tumors is crucial, including tumor metastasis/dissemination (167), metabolic dysfunction (168), and cancer stem cell maintenance (169, 170). CircRNAs within the TME can influence various physiological and pathological processes, including tumor angiogenesis (171, 172). Circ_0072088 is involved in tumor biological processes, such as proliferation, migration, invasion, and apoptosis in different cancers (173–179). Fang et al. verified that circ_0072088 upregulation considerably enhanced the protein and mRNA expression of VEGF in ESCC by sponging miR-377 (92). According to previous studies, VEGF upregulation by circ_0072088 can be reversed by overexpressing miRNA-377 (92). Thus, circ_0072088 can lessen its inhibitory effect on VEGF expression by modulating the miR-377/VEGF axis (92). This could increase angiogenesis regulation, which would encourage ESCC progression.
4.4 Regulation of hypoxia in the TME
Oxygen homeostasis is crucial in human physiology and metabolism. Oxygen is required by the machinery for energy production and by many cofactors and substrates for enzymes (180). Hypoxia is a vital component of physiological and pathological processes. Understanding the mechanisms underlying angiogenesis, glucose metabolism, and cell proliferation is particularly important (181). Hypoxia is a typical TME characteristic of various malignancies. The rapid proliferation of cancer cells does not match a lack of oxygen. Thus, several adaptive cellular responses and biological changes are initiated when cancer cells sense a drop in oxygen levels, causing them to swiftly adapt to the hypoxic environment and accelerate cancer progression (182). Recent studies have revealed that circRNAs play crucial roles in developing hypoxic microenvironments and exhibit variable expression in response to hypoxia (183, 184). According to previous studies, miR-497-5p is targeted by PRKAA1 and contributes to ESCC cell proliferation and motility control by being minimally expressed in ESCC (185). In addition, low miR-497-5p expression regulates the radiosensitivity of ESCC by directly targeting the 3’-UTR of CDC25A (186). However, Ma et al. illustrated that miR-497-5p overexpression, which is sponged by circDUSP16, could inhibit the ESCC cell progression under hypoxic conditions (93). In hypoxic conditions, ESCC cells had higher levels of circDUSP16 (93). By inhibiting miR-497-5p and increasing TKTL1, circDUSP16 knockdown restored hypoxia-stimulated malignant tendencies in ESCC cells (93). Under hypoxic conditions, circDUSP16 knockdown clearly hindered ESCC survival, colony formation, and invasion (93). Furthermore, the reduction in glucose intake, lactate generation, and HK2 and LDHA levels following circDUSP16 knockdown demonstrated that circDUSP16 silencing caused an apparent inhibition of glycolysis in ESCCs under hypoxic conditions (93). Additionally, circDUSP16 knockdown prevents ESCC carcinogenesis in vivo (93). These findings suggest that circDUSP16 knockdown partially alters the biologically malignant responses of ESCCs to hypoxia (93).
Hypoxia significantly influences tumor progression, metastasis, invasion, angiogenesis, epigenetic reprogramming, metabolic reprogramming, immune evasion, and glycolysis in ESCC (187–190). Further proof of the possible applications of circRNAs may be provided by understanding the mechanisms of circRNAs in hypoxia.
4.5 Inducing epithelial-to-mesenchymal transition and tumor cell migration
The complex program of cell plasticity, EMT, is abnormally reactivated in cancer. The interaction between cancerous cells and TME is crucial for EMT and tumorigenesis (191). CircRNA-related signaling pathways, such as the PI3K/Akt and TGF- signaling axes, are involved in the EMT process. For instance, the PI3K/Akt pathway is affected by circ_0007624 (94). This circRNA functions as a ceRNA to sponge miR-224-5p, increasing CPEB3 expression and inactivating the EGFR/PI3K/AKT axis, which could prevent cell proliferation, metastasis, and EMT in ESCC (94). CircHIPK3 is another circRNA that directly modulates AKT3 expression, acting as an miR-124 sponge in regulating ESCC progression (95). Vimentin and E-cadherin expression were drastically reduced by circHIPK3 expression, indicating that circHIPK3 prevented EMT (95). CircDOCK5 acts as a miR-627-3p reservoir in the TGF-β signaling pathway by stabilizing miR-627-3p to impede TGFB2 and repressing secreted TGF-β, which then dysregulates ZEB1 expression and inhibits EMT (96). In other words, circDOCK5 prevents EMT in ESCC by affecting the miR-627-3p/TGF-β/SMAD/ZEB1 pathway (96).
Despite this, emerging research has revealed that circRNAs are involved in EMT regulation and tumor cell migration in ESCC via other pathways. CircNTRK2 altered NRIP1 function as miR-140-3p sponging, which helped promote the malignant biological cell behaviors (EMT included) of ESCC (97). Zhang et al. observed that hsa_circ_0012563 upregulation in ESCC could enhance cell migration and invasion by modulating the XRCC1/EMT axis. Furthermore, its expression is tightly linked to tumor pathogenesis and metastasis (98). Hsa_circ 0006948 can directly bind to miR-490-3p, which regulates ESCC progression by targeting the 3’-UTR of the oncogene HMGA2 (99). By binding to miR-124, circ_2646 facilitates ESCC cell proliferation, migration, invasion, and EMT (100). MiR-124 can slow ESCC development by suppressing PLP2 expression (100). Zhou et al. observed that circPDE3B functions as a ceRNA and promotes LAMA1-mediated EMT, metastasis, and proliferation in ESCC by acting as an miR-4766-5p sponge (101). Rescue experiments revealed that co-transfection of the circPDE3B vector or miR-4766-5p inhibitor partly reduced the inhibitory effect (101). Similarly, circARAP2 influences EMT and cancer stem cell differentiation by regulating miR-761/FOXM1 (102). Zhu et al. explained that circLONP2 facilitated ESCC proliferation and migration via miR-27b-3p sponging and governed its target gene—ZEB1—expression; consequently, circLONP2/miR-27b-3p-ZEB1 axis involvement might be an efficient strategy for ESCC treatment (103). CircFAM120B, also known as hsa_circ_0001666, is typically dysregulated in ESCC and is favorably correlated with overall survival (104). Essentially, circFAM120B significantly reduced the biological functions of ESCC by disrupting PKR to affect the p38/MAPK/EMT axis or sponging miR-661 to restore PPM1L expression (104).
The aforementioned data continuously verified the crucial role of circRNAs in malignant tumor invasion and metastasis, demonstrating that circRNAs are collectively involved in the EMT process by participating in modulating EMT-related pathways.
5 Clinical application of circRNA in ESCC
With the deepening of circRNA studies, various emerging targets have been identified and verified to be involved in the occurrence and development of ESCC (68, 192). Furthermore, current studies have revealed that circRNAs can be used as biomarkers. Due to their unique resistance to chemotherapy medications, targeted therapy can be carried out for patients with clinical chemotherapy resistance (Figure 3) (31, 193).
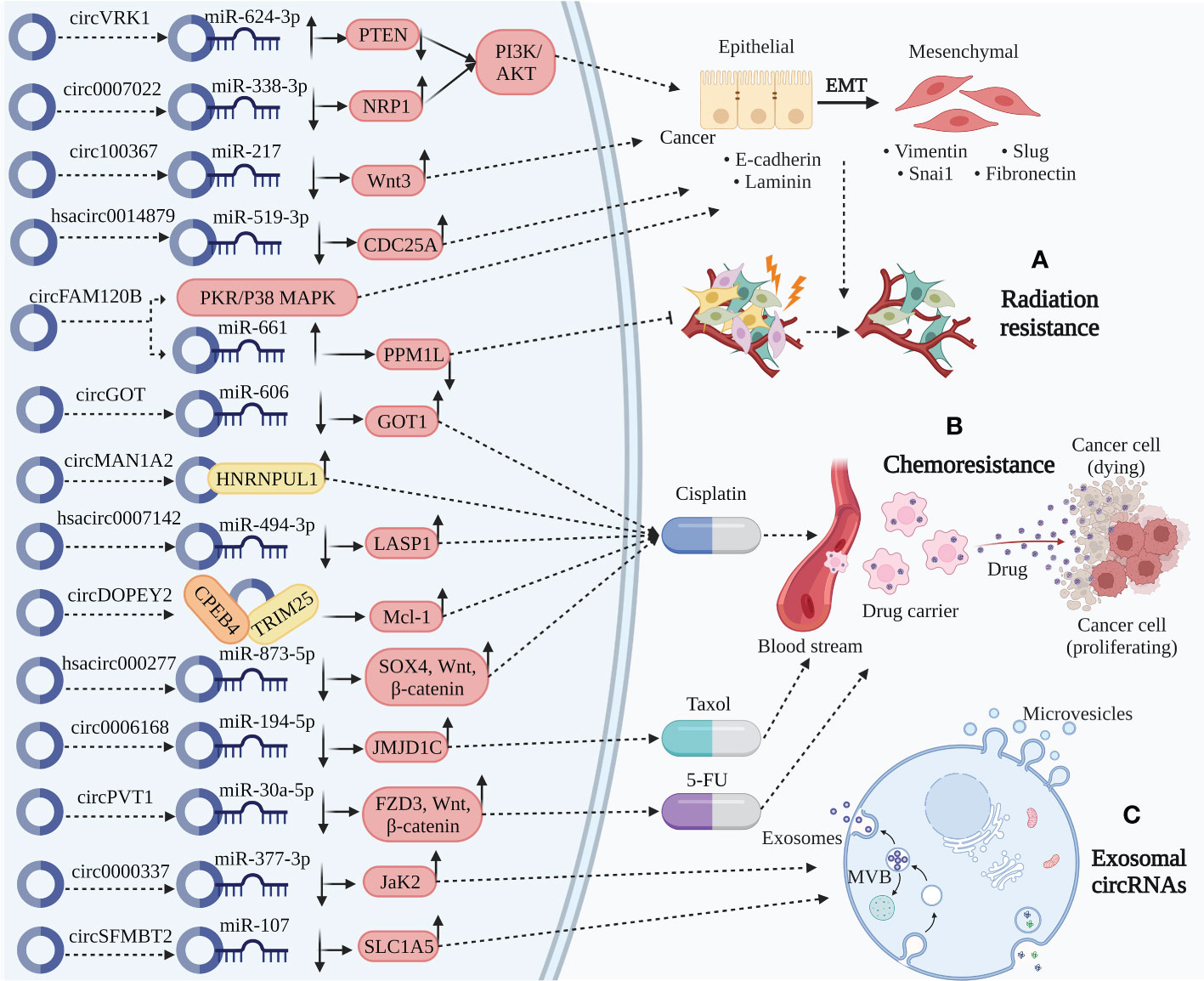
Figure 3 Clinical application of circRNA in ESCC. (A) Radiation resistance. CircVRK1 downregulate PETN by sponging miR-624-3p and inactivate the PI3K/AKT pathway and EMT. Circ_0007022 upregulate NRP1 by sponging miR-338-3p activating the PI3K/AKT pathway and EMT. Circ_100367 and hsa_circ_0014879 upregulate Wnt3 and CDC25A by sponging miR-217 and miR-519-3p, thereby promoting EMT. CircFAM120B significantly promote EMT by disrupting PKR to affect the p38/MAPK axis. Besides, circFAM120B downregulate PPM1L by sponging miR-661 directly functioning on radiation resistance. The role of circRNAs above participate in radiation resistance to affect the progression of ESCC. (B) Chemoresistance. CircMAN1A2 binding with HNRNPUL1 related to cisplatin resistance. CircDOPEY2 functions as a protein scaffold binding with CPEB4 and TRIM25, upregulating Mcl-1 translation and increasing cisplatin resistance. CircGOT1, hsa_circ_0007142, and hsa_circ_0000277 upregulate GOT1, LASP1, and SOX4/Wnt/β-catenin by sponging miR-606, miR-494-2p, and miR-873-5p increasing cisplatin resistance. Circ_0006168 upregulate JMJD1C by sponging miR-194-5p increasing Taxol resistance. CircPVT1 increase 5-FU resistance through miR-30a-5p/FZD3/Wnt/β-catenin axis. (C) Exosomal circRNAs. Circ_0000337 and circSFMBT2, serum exosomal circRNAs, upregulate JaK2 and SLC1A5 by sponging miR-377-3p and miR-107, thereby promoting carcinogenic activity of ESCC.
5.1 Regulating chemoradiation resistance in ESCC
Malignant tumors are often accompanied by metastasis postoperatively; therefore, radiotherapy and chemotherapy still need to be administered locally or systemically (194). However, due to the emergence of resistance, some people with cancer still experience local recurrence and lymph node metastases (195). Thus, the discovery of different targets to enhance treatments can be accomplished by understanding the regulatory processes of circRNAs implicated in radiation and chemotherapeutic resistance (196). The three major concerns associated with anticancer agents are drug resistance, metastasis, and EMT. According to available data, drug resistance and EMT are associated in many ways. Cancerous cells that are resistant to drugs might have higher EMT and invasive capabilities (197, 198). Accumulating evidence has clearly revealed that circRNAs play a significant role in EMT regulation and radiation resistance in ESCC. For instance, increased circVRK1 levels decreased the migration potential, reversed EMT progression, and drastically increased ESCC radiosensitivity by modulating the miR-624-3p/PETN axis and inactivating the PI3K/AKT signaling pathway (105). Zhang et al. verified that circ_0007022 can function as a miRNA sponge to regulate the miR-338-3p/NRP1 axis, activating EMT and the PI3K/AKT pathway to generate radiation resistance in ESCC (106). Circ_100367, an oncogenic circRNA in ESCC cells with a higher potency of EMT coordinated with miR-217, extraordinarily regulates the radioresistance of ESCC through the Wnt3 signaling pathway (107). Liu et al. demonstrated that hsa_circ_0014879 knockdown enhanced ESCC radiosensitivity and reduced ESCC migration and EMT via the miR-519-3p/CDC25A regulatory axis (108).
One of the most commonly used chemotherapeutic drugs is platinum, which is a complex barrier in cancer therapy (199). Albeit the mechanisms of platinum drug resistance have been the subject of numerous investigations, few efficient, targeted treatments are available (200–202). Mounting evidence suggests that circRNAs are involved in multiple cellular processes and malignancies, including ESCC (203, 204). Understanding the underlying causes of chemoresistance is crucial for developing effective ESCC treatments and preventing recurrence. Zhou et al. unveiled that circGOT1 functions as an oncogene in ESCC, stimulating cell proliferation, migration, glycolytic metabolism, and reducing apoptosis by acting as an miR-606 sponge to induce GOT1 expression (86). CircGOT1 knockdown restored the cisplatin resistant ESCC sensitivity to cisplatin (86). In patients with ESCC undergoing platinum-based chemotherapy postoperatively, increased HNRNPUL1 expression is linked to a poor prognosis with a greater likelihood of recurrence and shorter disease-free survival (DFS) (109). By controlling circMAN1A2 production, HNRNPUL1 inhibition reduces the cisplatin-chemoresistance of ESCCs (109). Chang et al. uncovered that circ_0007142 depletion can modulate the miR-494-3p/LASP1 axis, leading to cisplatin resistance in ESCC (110). Moreover, multiple chemotherapeutic drug resistance-associated proteins produce GST-π, which can reduce the cytotoxicity of chemotherapy medications (205). P-gp is an energy-dependent transporter that may combine with drugs and ATP to decrease medication concentration and supply energy for chemotherapeutic drug excretion (206). Remarkably, cisplatin resistant ESCCs with hsa_circ_0007142 silencing had lower P-gp and GST-π levels (110). Additionally, hsa_circ_0007142 knockdown significantly reduced cisplatin resistance in ESCC cells (110). Apoptosis is also closely associated with carcinogenesis. CircRNAs can tip the balance between cell death signaling and may improve chemoresistance efficacy (207). A BCL-2 family member that has been the subject of the most research is MCL-1, an anti-apoptotic oncoprotein (208). Mcl-1 is crucial for malignant biological functions because it mediates the apoptotic signaling pathway (209). It is also fundamentally altered, upregulated, and largely related to chemoresistance in several cancer (210). Liu et al. observed that in ESCC cells, circDOPEY2 functions as a protein scaffold to enhance CPEB4 ubiquitination and degradation in a TRIM25-dependent manner, increasing cisplatin-induced apoptosis by inhibiting CPEB4’s promotion of Mcl-1 translation and reducing cisplatin resistance (111). Another ESCC study revealed that hsa_circ_0000277 can modulate the miR-873-5p/SOX4/Wnt/β-catenin axis by acting as an miR-873-5p sponge to accelerate progression and chemoresistance in ESCC (112). JMJD1C facilitates tumor growth in various malignancies by promoting cancer cell growth and preventing apoptosis (211, 212). A study demonstrated that inhibiting circ_0006168 decreased tumor growth in vivo by sponging miR-194-5p and modulating JMJD1C, and in vitro by reducing cell proliferation, invasion, and migration, and Taxol-resistant ESCC apoptosis (113). Functionally, circPVT1 modulates the miR-30a-5p/FZD3/Wnt/β-catenin axis and ferroptosis, and circPVT1 knockdown can drastically increase the expression of ferroptosis-associated parameters MDA/Fe, which can improve chemoresistance in ESCC (114).
With accumulating evidence for circRNAs elucidated by research on chemoradiation resistance in ESCC as a new biomarker, circRNA has excellent potential in predicting the efficacy and prognosis of chemoradiotherapy or interfering with chemoradiation resistance and can be a target for clinical cancer therapy.
5.2 Exosomal circRNAs in ESCC TME
Exosomes are discoidal vesicles of 30–150 nm diameter that carry cargo derived from the host cell, such as proteins, lipids, DNA, and RNA. Exosomes carry their cargo to recipient cells, allowing immune and cancerous cells to communicate through them (213, 214). Cancer invasion and metastasis may be facilitated by exosome-mediated interactions between cancerous cells and the TME, encouraging EMT, angiogenesis, and immune escape (215). The TME is essential for intercellular information transmission and oncogenesis. Numerous studies have reported extensive and sustainable circRNAs in the exosomes (216–221). In a recent study, Liu et al. identified that serum exosomal hsa_circ_0026611 expression is markedly increased in ESCC, indicating that it can be utilized as a valuable indicator to distinguish lymph from non-lymph node-metastatic ESCC (115). Additionally, elevated hsa_circ_0026611 expression is strongly associated with poor overall survival (OS) and DFS (115). In another study, Zang et al. demonstrated that circ_0000337 was highly upregulated in ESCC (116). In addition, exosomes released by cisplatin-resistant ESCC cells that contain abundant circ_0000337 enhance the ability of ESCC cells to resist cisplatin and promote cell proliferation and metastasis (116). Xenotransplantation supported this hypothesis (116). Thus, exosomal circ_0000337 drastically affects cisplatin resistance in ESCC by modulating the miR-377-3p/Jak2 axis (116). Exosomal circSFMBT2 has been linked to a poor prognosis and is upregulated in ESCC (117). CircSFMBT2 silencing may slow cancer progression by preventing ESCC proliferation, invasion, and glutamine metabolism (117). Mechanistically, circSFMBT2 leads to SLC1A5 upregulation by sponging miR-107, thereby promoting carcinogenic activity (117). CircSFMBT2 exosomes may be an effective new therapeutic indicator for ESCC because they facilitate circSFMBT2 metastasis in ESCC cells, which causes the cells to exhibit the aforementioned malignant behaviors (117). Exo-circRNAs are defined by a transferrable target-specific capability, in addition to the initial physiological functions of circRNAs, since circRNAs are detected in exosomes (19). Continued studies may further reveal the involvement and promising effects of exo-circRNAs in the TME; thus, they require confirmation.
6 Discussion
As well as being affected by abnormal mutations in malignant cells, cancer is also associated with the composition of the TME. The TME is characterized by heterogeneous composition according to a large amount of research. Nevertheless, nearly all types of cancer can be treated with specific cells and mediators to improve patient health. There is increasing evidence that circRNAs play a crucial role in TME regulation. The exact physiological and pathological roles of circRNA in TME, as well as the underlying mechanisms, remain largely unknown. Currently, the interaction mechanisms between circRNAs and TME elements have received considerable attention. Establishing TME networks involving circRNAs may be an emerging direction for targeted therapies based on their interactions with circRNAs. Current findings on circRNAs have primarily concentrated on how they affect the bioactivity of malignancy, while systematic summaries for specific solid tumor microenvironments are lacking.
In this review, we discussed the biogenesis/degradation mechanisms and biological functions of circRNA occurrence. We systematically summarized the biological functions and clinical features of ESCC affected by circRNA dysregulation and its regulatory mechanisms in TME. Especially, in TME-related immune microenvironment, energy metabolism regulation, angiogenesis, hypoxia, epithelial-to-mesenchymal transition and tumor cell migration. Finally, we also discussed the mechanisms by which circRNAs drive chemoradiation resistance in ESCC and the role of the emerging exosomal circRNA in ESCC TME, thus providing insight into the future role of circRNA as a potential diagnostic indicator of ESCC. Nevertheless, there still has some relevant potential mechanisms that have not yet been elucidated in studies of circRNA in ESCC TME. In the immune microenvironment we focus on the relationship between circRNA and macrophage M2 polarization, the expression of other immune cells associated with TME such as T cells, NK cells, and the surface immune checkpoint molecules PD-L1/PD-1 in tumor cells, and the expression of important components of TME: stromal cells such as cancer-associated fibroblasts, endothelial cells, and pericytes, for which more studies are needed to explore their potential regulation. Researches on circRNA in ESCC TME regarding energy metabolism, angiogenesis, EMT, and hypoxia mechanisms is increasing and becoming more detailed. However, current studies on metabolic mechanisms are mainly focused on glycolysis, glutamine metabolism, proline metabolism, and oxidative phosphorylation. Other metabolic-related mechanisms, such as serine and lipid metabolism, need to be explored.
Clinical applications related to the role of circRNA in ESCC TME also need to be further explored, in addition to some of the above mechanistic studies to be elucidated. Despite the exploration of animal models to validate circRNA in chemoradiation resistance in ESCC, the applicability of circRNA-targeted therapy for ESCC in clinical trials remains to be investigated. Moreover, exosomal circRNAs play a crucial role in ESCC, providing new therapeutic targets and promising biomarkers for exosome-loaded small molecules that could be designed for ESCC diagnosis. We believe that with the investigation of circRNA function and mechanism in ESCC TME, it will definitely contribute to the developments of new strategies for ESCC treatment.
Author contributions
JM, YS, and BZ designed and guided the review. JL wrote and edited the manuscript. HC helped with reference collection. All authors contributed to the article and approved the submitted version.
Funding
The present study was supported by Henan Province Medical Science and Technology Research Program (co-supported by ministry and province) (Project number: SBGJ202102176), the key research and development and promotion projects (tackling of key scientific and technical problems) (Project number: 212102310048), Science and Technology Department of Henan Province, China and Independent Innovation Project for Postgraduate Students of Medical Science Academy of Zhengzhou University.
Acknowledgments
Figures in this manuscript were created with BioRender.com
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology (2018) 154:360–73. doi: 10.1053/j.gastro.2017.08.023
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Rogers JE, Sewastjanow-Silva M, Waters RE, Ajani JA. Esophageal cancer: emerging therapeutics. Expert Opin Ther Targets (2022) 26:107–17. doi: 10.1080/14728222.2022.2036718
4. Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol (2021) 18:432–43. doi: 10.1038/s41575-021-00419-3
5. He S, Xu J, Liu X, Zhen Y. Advances and challenges in the treatment of esophageal cancer. Acta Pharm Sin B (2021) 11:3379–92. doi: 10.1016/j.apsb.2021.03.008
6. Zhang X, Peng L, Luo Y, Zhang S, Pu Y, Chen Y, et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat Commun (2021) 12:5291. doi: 10.1038/s41467-021-25539-x
7. Li Y, Xu F, Chen F, Chen Y, Ge D, Zhang S, et al. Transcriptomics based multi-dimensional characterization and drug screen in esophageal squamous cell carcinoma. EBioMedicine (2021) 70:103510. doi: 10.1016/j.ebiom.2021.103510
8. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
9. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495:333–8. doi: 10.1038/nature11928
10. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
11. Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform (2021) 22:1706–28. doi: 10.1093/bib/bbaa001
12. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol (2022) 19:188–206. doi: 10.1038/s41571-021-00585-y
13. Zhang M, Bai X, Zeng X, Liu J, Liu F, Zhang Z. circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta (2021) 523:120–30. doi: 10.1016/j.cca.2021.09.013
14. Chen J, Gu J, Tang M, Liao Z, Tang R, Zhou L, et al. Regulation of cancer progression by circRNA and functional proteins. J Cell Physiol (2022) 237:373–88. doi: 10.1002/jcp.30608
15. Singh D, Kesharwani P, Alhakamy NA, Siddique HR. Accentuating CircRNA-miRNA-Transcription factors axis: a conundrum in cancer research. Front Pharmacol (2021) 12:784801. doi: 10.3389/fphar.2021.784801
16. van Zonneveld AJ, Kölling M, Bijkerk R, Lorenzen JM. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol (2021) 17:814–26. doi: 10.1038/s41581-021-00465-9
17. Chen Z, Zhao M, Liang J, Hu Z, Huang Y, Li M, et al. Dissecting the single-cell transcriptome network underlying esophagus non-malignant tissues and esophageal squamous cell carcinoma. EBioMedicine (2021) 69:103459. doi: 10.1016/j.ebiom.2021.103459
18. Wu Y, Niu D, Deng S, Lei X, Xie Z, Yang X. Tumor-derived or non-tumor-derived exosomal noncodingRNAs and signaling pathways in tumor microenvironment. Int Immunopharmacol (2022) 106:108626. doi: 10.1016/j.intimp.2022.108626
19. Zhou X, Liu J, Meng A, Zhang L, Wang M, Fan H, et al. Gastric juice piR-1245: a promising prognostic biomarker for gastric cancer. J Clin Lab Anal (2020) 34:e23131. doi: 10.1002/jcla.23131
20. Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med (2015) 13:45. doi: 10.1186/s12916-015-0278-7
21. Shao Y, Lu B. The crosstalk between circular RNAs and the tumor microenvironment in cancer metastasis. Cancer Cell Int (2020) 20:448. doi: 10.1186/s12935-020-01532-0
22. Baba Y, Nomoto D, Okadome K, Ishimoto T, Iwatsuki M, Miyamoto Y, et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci (2020) 111:3132–41. doi: 10.1111/cas.14541
23. Ma Z, Shuai Y, Gao X, Wen X, Ji J. Circular RNAs in the tumour microenvironment. Mol Cancer (2020) 19:8. doi: 10.1186/s12943-019-1113-0
24. Viralippurath Ashraf J, Sasidharan Nair V, Saleh R, Elkord E. Role of circular RNAs in colorectal tumor microenvironment. BioMed Pharmacother (2021) 137:111351. doi: 10.1016/j.biopha.2021.111351
25. Zhang LX, Gao J, Long X, Zhang PF, Yang X, Zhu SQ, et al. The circular RNA circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung adenocarcinomas and squamous cell carcinomas via the miR-181a-5p/CARM1 axis. Mol Cancer (2022) 21:110. doi: 10.1186/s12943-022-01586-w
26. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell (2018) 71:428–42. doi: 10.1016/j.molcel.2018.06.034
27. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
28. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol (2020) 21:475–90. doi: 10.1038/s41580-020-0243-y
29. Li Z, Cheng Y, Wu F, Wu L, Cao H, Wang Q, et al. The emerging landscape of circular RNAs in immunity: breakthroughs and challenges. biomark Res (2020) 8:25. doi: 10.1186/s40364-020-00204-5
30. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer (2020) 19:172. doi: 10.1186/s12943-020-01286-3
31. Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell (2022) 185:2016–34. doi: 10.1016/j.cell.2022.04.021
32. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol (2015) 12:381–8. doi: 10.1080/15476286.2015.1020271
33. Shen T, Han M, Wei G, Ni T. An intriguing RNA species–perspectives of circularized RNA. Protein Cell (2015) 6:871–80. doi: 10.1007/s13238-015-0202-0
34. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett (2015) 365:141–8. doi: 10.1016/j.canlet.2015.06.003
35. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
36. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
37. Ali SA, Peffers MJ, Ormseth MJ, Jurisica I, Kapoor M. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol (2021) 17:692–705. doi: 10.1038/s41584-021-00687-y
38. Wilkinson ME, Charenton C, Nagai K. RNA Splicing by the spliceosome. Annu Rev Biochem (2020) 89:359–88. doi: 10.1146/annurev-biochem-091719-064225
39. Wang S, Latallo MJ, Zhang Z, Huang B, Bobrovnikov DG, Dong D, et al. Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD. Nat Commun (2021) 12:4908. doi: 10.1038/s41467-021-25082-9
40. Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev (2014) 28:2233–47. doi: 10.1101/gad.251926.114
41. Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res (2020) 98:87–97. doi: 10.1002/jnr.24356
42. Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J (2021) 19:910–28. doi: 10.1016/j.csbj.2021.01.018
43. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
44. Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the alu invasion of the human genome. Nature (2017) 544:115–9. doi: 10.1038/nature21715
45. Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res (2019) 47:6452–65. doi: 10.1093/nar/gkz311
46. Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, et al. Metazoan tRNA introns generate stable circular RNAs in vivo. Rna (2015) 21:1554–65. doi: 10.1261/rna.052944.115
47. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell (2019) 177:865–880.e21. doi: 10.1016/j.cell.2019.03.046
48. Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang Q, et al. MicroRNA-1224 splicing CircularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J Neurosci (2019) 39:2125–43. doi: 10.1523/jneurosci.1631-18.2018
49. Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H, et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit gliomagenesis. Mol Cancer (2020) 19:138. doi: 10.1186/s12943-020-01253-y
50. Pantazopoulou VI, Delis AD, Georgiou S, Pagakis SN, Filippa V, Dragona E, et al. AGO2 localizes to cytokinetic protrusions in a p38-dependent manner and is needed for accurate cell division. Commun Biol (2021) 4:726. doi: 10.1038/s42003-021-02130-0
51. Marzec M. New insights into the function of mammalian Argonaute2. PloS Genet (2020) 16:e1009058. doi: 10.1371/journal.pgen.1009058
52. Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep (2020) 31:107641. doi: 10.1016/j.celrep.2020.107641
53. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res (2017) 27:626–41. doi: 10.1038/cr.2017.31
54. Papatsirou M, Diamantopoulos MA, Katsaraki K, Kletsas D, Kontos CK, Scorilas A. Identification of novel circular RNAs of the human protein arginine methyltransferase 1 (PRMT1) gene, expressed in breast cancer cells. Genes (Basel) (2022) 13:1133. doi: 10.3390/genes13071133
55. Lasda E, Parker R. Circular RNAs Co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PloS One (2016) 11:e0148407. doi: 10.1371/journal.pone.0148407
56. Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ (2022) 29:481–91. doi: 10.1038/s41418-022-00948-7
57. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol (2018) 1087:67–79. doi: 10.1007/978-981-13-1426-1_6
58. Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics (2014) 30:2243–6. doi: 10.1093/bioinformatics/btu257
59. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol (2014) 15:409. doi: 10.1186/s13059-014-0409-z
60. Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer biomark (2018) 21:299–306. doi: 10.3233/cbm-170379
61. Chen Q, Wang H, Li Z, Li F, Liang L, Zou Y, et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol (2022) 76:135–47. doi: 10.1016/j.jhep.2021.08.027
62. Chen Z, Lu T, Huang L, Wang Z, Yan Z, Guan Y, et al. Circular RNA cia-MAF drives self-renewal and metastasis of liver tumor-initiating cells via transcription factor MAFF. J Clin Invest (2021) 131:e148020. doi: 10.1172/jci148020
63. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22:256–64. doi: 10.1038/nsmb.2959
64. Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics (2009) 93:291–8. doi: 10.1016/j.ygeno.2008.11.009
65. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2017) 24:194. doi: 10.1038/nsmb0217-194a
66. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
67. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer (2020) 19:30. doi: 10.1186/s12943-020-1135-7
68. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer (2020) 19:22. doi: 10.1186/s12943-020-1147-3
69. Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther (2019) 27:1718–25. doi: 10.1016/j.ymthe.2019.09.001
70. Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol (2017) 14:1007–17. doi: 10.1080/15476286.2016.1227905
71. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep (2016) 6:34985. doi: 10.1038/srep34985
72. Dawoud A, Ihab Zakaria Z, Hisham Rashwan H, Braoudaki M, Youness RA. Circular RNAs: new layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res (2023) 8:60–74. doi: 10.1016/j.ncrna.2022.09.011
73. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
74. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene (2018) 37:1805–14. doi: 10.1038/s41388-017-0019-9
75. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst (2018) 110:304–15. doi: 10.1093/jnci/djx166
76. Chen H, Liu Y, Li P, Zhu D. RE: novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst (2019) 111:435. doi: 10.1093/jnci/djy116
77. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun (2018) 9:4475. doi: 10.1038/s41467-018-06862-2
78. Mo Y, Wang Y, Xiong F, Ge X, Li Z, Li X, et al. Proteomic analysis of the molecular mechanism of lovastatin inhibiting the growth of nasopharyngeal carcinoma cells. J Cancer (2019) 10:2342–9. doi: 10.7150/jca.30454
79. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res (2016) 44:2846–58. doi: 10.1093/nar/gkw027
80. Zhao H, Zhou Q, Li X. Protein bait hypothesis: circRNA-encoded proteins competitively inhibit cognate functional isoforms. Trends Genet (2021) 37:616–24. doi: 10.1016/j.tig.2021.04.002
81. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics (2020) 10:3503–17. doi: 10.7150/thno.42174
82. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer (2020) 6:319–36. doi: 10.1016/j.trecan.2020.01.012
83. Luo J, Liu H, Luan S, Li Z. Guidance of circular RNAs to proteins’ behavior as binding partners. Cell Mol Life Sci (2019) 76:4233–43. doi: 10.1007/s00018-019-03216-z
84. Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther (2020) 13:11883–97. doi: 10.2147/ott.S284192
85. Lin C, Xi Y, Yu H, Wang Z, Chen X, Shen W. circRNA TCFL5 promote esophageal cancer progression by modulating M2 macrophage polarization via the miR-543-FMNL2 axis. J Oncol (2022) 2022:5075615. doi: 10.1155/2022/5075615
86. Zhou S, Guo Z, Lv X, Zhang X. CircGOT1 promotes cell proliferation, mobility, and glycolysis-mediated cisplatin resistance via inhibiting its host gene GOT1 in esophageal squamous cell cancer. Cell Cycle (2022) 21:247–60. doi: 10.1080/15384101.2021.2015671
87. Tang B, Zhang Q, Liu K, Huang Y. Exosomal circRNA FNDC3B promotes the progression of esophageal squamous cell carcinoma by sponging miR-490-5p and regulating thioredoxin reductase 1 expression. Bioengineered (2022) 13:13829–48. doi: 10.1080/21655979.2022.2084484
88. Qian CJ, Tong YY, Wang YC, Teng XS, Yao J. Circ_0001093 promotes glutamine metabolism and cancer progression of esophageal squamous cell carcinoma by targeting miR-579-3p/glutaminase axis. J Bioenerg Biomembr (2022) 54:119–34. doi: 10.1007/s10863-022-09935-6
89. Liang Z, Zhao B, Hou J, Zheng J, Xin G. CircRNA circ-OGDH (hsa_circ_0003340) acts as a ceRNA to regulate glutamine metabolism and esophageal squamous cell carcinoma progression by the miR-615-5p/PDX1 axis. Cancer Manag Res (2021) 13:3041–53. doi: 10.2147/cmar.S290088
90. Qian CJ, Tong YY, Wu LK, Wang YC, Teng XS, Yao J. Circ_0000705 facilitates proline metabolism of esophageal squamous cell carcinoma cells by targeting miR-621/PYCR1 axis. Discov Oncol (2022) 13:50. doi: 10.1007/s12672-022-00513-1
91. Gong W, Xu J, Wang Y, Min Q, Chen X, Zhang W, et al. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct Target Ther (2022) 7:40. doi: 10.1038/s41392-021-00865-0
92. Fang N, Shi Y, Fan Y, Long T, Shu Y, Zhou J. Circ_0072088 promotes proliferation, migration, and invasion of esophageal squamous cell cancer by absorbing miR-377. J Oncol (2020) 2020:8967126. doi: 10.1155/2020/8967126
93. Ma L, Li H, Lin Y, Wang G, Xu Q, Chen Y, et al. CircDUSP16 contributes to cell development in esophageal squamous cell carcinoma by regulating miR-497-5p/TKTL1 axis. J Surg Res (2021) 260:64–75. doi: 10.1016/j.jss.2020.11.052
94. Cheng J, Ma H, Yan M, Zhang Z, Xing W. Circ_0007624 suppresses the development of esophageal squamous cell carcinoma via targeting miR-224-5p/CPEB3 to inactivate the EGFR/PI3K/AKT signaling. Cell Signal (2022) 99:110448. doi: 10.1016/j.cellsig.2022.110448
95. Yao D, Lin S, Chen S, Wang Z. circHIPK3 regulates cell proliferation and migration by sponging microRNA-124 and regulating serine/threonine kinase 3 expression in esophageal squamous cell carcinoma. Bioengineered (2022) 13:9767–80. doi: 10.1080/21655979.2022.2060776
96. Meng L, Zheng Y, Liu S, Ju Y, Ren S, Sang Y, et al. ZEB1 represses biogenesis of circ-DOCK5 to facilitate metastasis in esophageal squamous cell carcinoma via a positive feedback loop with TGF-β. Cancer Lett (2021) 519:117–29. doi: 10.1016/j.canlet.2021.06.026
97. Chen X, Jiang J, Zhao Y, Wang X, Zhang C, Zhuan L, et al. Circular RNA circNTRK2 facilitates the progression of esophageal squamous cell carcinoma through up-regulating NRIP1 expression via miR-140-3p. J Exp Clin Cancer Res (2020) 39:133. doi: 10.1186/s13046-020-01640-9
98. Zhang Z, Li X, Xiong F, Ren Z, Han Y. Hsa_circ_0012563 promotes migration and invasion of esophageal squamous cell carcinoma by regulating XRCC1/EMT pathway. J Clin Lab Anal (2020) 34:e23308. doi: 10.1002/jcla.23308
99. Pan Z, Lin J, Wu D, He X, Wang W, Hu X, et al. Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY) (2019) 11:11937–54. doi: 10.18632/aging.102519
100. Zeng B, Liu Z, Zhu H, Zhang X, Yang W, Li X, et al. CircRNA_2646 functions as a ceRNA to promote progression of esophageal squamous cell carcinoma via inhibiting miR-124/PLP2 signaling pathway. Cell Death Discov (2021) 7:99. doi: 10.1038/s41420-021-00461-9
101. Zhou PL, Wu Z, Zhang W, Xu M, Ren J, Zhang Q, et al. Circular RNA hsa_circ_0000277 sequesters miR-4766-5p to upregulate LAMA1 and promote esophageal carcinoma progression. Cell Death Dis (2021) 12:676. doi: 10.1038/s41419-021-03911-5
102. Xu P, Wang L, Liu Q, Gao P, Hu F, Xie X, et al. The abnormal expression of circ-ARAP2 promotes ESCC progression through regulating miR-761/FOXM1 axis-mediated stemness and the endothelial-mesenchymal transition. J Transl Med (2022) 20:318. doi: 10.1186/s12967-022-03507-3
103. Zhu C, Bi W, Li H, Wang W. CircLONP2 accelerates esophageal squamous cell carcinoma progression via direct MiR-27b-3p-ZEB1 axis. Front Oncol (2022) 12:822839. doi: 10.3389/fonc.2022.822839
104. Song H, Tian D, Sun J, Mao X, Kong W, Xu D, et al. circFAM120B functions as a tumor suppressor in esophageal squamous cell carcinoma via the miR-661/PPM1L axis and the PKR/p38 MAPK/EMT pathway. Cell Death Dis (2022) 13:361. doi: 10.1038/s41419-022-04818-5
105. He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol (2019) 125:116–23. doi: 10.1016/j.ijbiomac.2018.11.273
106. Zhang J, Yu Y, Yin X, Feng L, Li Z, Liu X, et al. A circ-0007022/miR-338-3p/Neuropilin-1 axis reduces the radiosensitivity of esophageal squamous cell carcinoma by activating epithelial-To-Mesenchymal transition and PI3K/AKT pathway. Front Genet (2022) 13:854097. doi: 10.3389/fgene.2022.854097
107. Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY) (2019) 11:12412–27. doi: 10.18632/aging.102580
108. Liu Z, Lu X, Wen L, You C, Jin X, Liu J. Hsa_circ_0014879 regulates the radiosensitivity of esophageal squamous cell carcinoma through miR-519-3p/CDC25A axis. Anticancer Drugs (2022) 33:e349–61. doi: 10.1097/cad.0000000000001213
109. Li J, Sang M, Zheng Y, Meng L, Gu L, Li Z, et al. HNRNPUL1 inhibits cisplatin sensitivity of esophageal squamous cell carcinoma through regulating the formation of circMAN1A2. Exp Cell Res (2021) 409:112891. doi: 10.1016/j.yexcr.2021.112891
110. Chang N, Ge N, Zhao Y, Yang L, Qin W, Cui Y. Hsa_circ_0007142 contributes to cisplatin resistance in esophageal squamous cell carcinoma via miR-494-3p/LASP1 axis. J Clin Lab Anal (2022) 36:e24304. doi: 10.1002/jcla.24304
111. Liu Z, Gu S, Wu K, Li L, Dong C, Wang W, et al. CircRNA-DOPEY2 enhances the chemosensitivity of esophageal cancer cells by inhibiting CPEB4-mediated mcl-1 translation. J Exp Clin Cancer Res (2021) 40:361. doi: 10.1186/s13046-021-02149-5
112. Cheng J, Zhang R, Yan M, Li Y. Circular RNA hsa_circ_0000277 promotes tumor progression and DDP resistance in esophageal squamous cell carcinoma. BMC Cancer (2022) 22:238. doi: 10.1186/s12885-022-09241-9
113. Qu F, Wang L, Wang C, Yu L, Zhao K, Zhong H. Circular RNA circ_0006168 enhances taxol resistance in esophageal squamous cell carcinoma by regulating miR-194-5p/JMJD1C axis. Cancer Cell Int (2021) 21:273. doi: 10.1186/s12935-021-01984-y
114. Yao W, Wang J, Meng F, Zhu Z, Jia X, Xu L, et al. Circular RNA CircPVT1 inhibits 5-fluorouracil chemosensitivity by regulating ferroptosis through MiR-30a-5p/FZD3 axis in esophageal cancer cells. Front Oncol (2021) 11:780938. doi: 10.3389/fonc.2021.780938
115. Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin Y, et al. Upregulated expression of serum exosomal hsa_circ_0026611 is associated with lymph node metastasis and poor prognosis of esophageal squamous cell carcinoma. J Cancer (2021) 12:918–26. doi: 10.7150/jca.50548
116. Zang R, Qiu X, Song Y, Wang Y. Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p. Front Cell Dev Biol (2021) 9:673237. doi: 10.3389/fcell.2021.673237
117. Chang Z, Fu Y, Jia Y, Gao M, Song L, Zhang W, et al. Circ-SFMBT2 drives the malignant phenotypes of esophageal cancer by the miR-107-dependent regulation of SLC1A5. Cancer Cell Int (2021) 21:495. doi: 10.1186/s12935-021-02156-8
118. Li YL, Hung WC. Reprogramming of sentinel lymph node microenvironment during tumor metastasis. J Biomed Sci (2022) 29:84. doi: 10.1186/s12929-022-00868-1
119. Wang Z, Mao Y, Gao S, Li Y, Tan L, Daiko H, et al. Lymph node dissection and recurrent laryngeal nerve protection in minimally invasive esophagectomy. Ann New York Acad Sci (2020) 1481:20–9. doi: 10.1111/nyas.14427
120. Yang H, Hu B. Recent advances in early esophageal cancer: diagnosis and treatment based on endoscopy. Postgraduate Med (2021) 133:665–73. doi: 10.1080/00325481.2021.1934495
121. Sudo N, Ichikawa H, Muneoka Y, Hanyu T, Kano Y, Ishikawa T, et al. Clinical utility of ypTNM stage grouping in the 8th edition of the American joint committee on cancer TNM staging system for esophageal squamous cell carcinoma. Ann Surg Oncol (2021) 28:650–60. doi: 10.1245/s10434-020-09181-3
122. Belli C, Trapani D, Viale G, D’Amico P, Duso BA, Della Vigna P, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev (2018) 65:22–32. doi: 10.1016/j.ctrv.2018.02.004
123. Jia Z, Jia J, Yao L, Li Z. Crosstalk of exosomal non-coding RNAs in the tumor microenvironment: novel frontiers. Front Immunol (2022) 13:900155. doi: 10.3389/fimmu.2022.900155
124. Mahadevan NR, Zanetti M. Tumor stress inside out: cell-extrinsic effects of the unfolded protein response in tumor cells modulate the immunological landscape of the tumor microenvironment. J Immunol (2011) 187:4403–9. doi: 10.4049/jimmunol.1101531
125. Khan SU, Khan MU, Khan MI, Fadahunsi AA, Khan A, Gao S, et al. Role of circular RNAs in disease progression and diagnosis of cancers: an overview of recent advanced insights. Int J Biol Macromol (2022) 220:973–84. doi: 10.1016/j.ijbiomac.2022.08.085
126. Xue C, Gu X, Bao Z, Su Y, Lu J, Li L. The mechanism underlying the ncRNA dysregulation pattern in hepatocellular carcinoma and its tumor microenvironment. Front Immunol (2022) 13:847728. doi: 10.3389/fimmu.2022.847728
127. Wang C, Ma C, Gong L, Guo Y, Fu K, Zhang Y, et al. Macrophage polarization and its role in liver disease. Front Immunol (2021) 12:803037. doi: 10.3389/fimmu.2021.803037
128. Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol (2020) 877:173090. doi: 10.1016/j.ejphar.2020.173090
129. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol (2002) 23:549–55. doi: 10.1016/s1471-4906(02)02302-5
130. Najafi M, Hashemi Goradel N, Farhood B, Salehi E, Nashtaei MS, Khanlarkhani N, et al. Macrophage polarity in cancer: a review. J Cell Biochem (2019) 120:2756–65. doi: 10.1002/jcb.27646
131. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci (2021) 22:6995. doi: 10.3390/ijms22136995
132. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology (2020) 72:906–22. doi: 10.1002/hep.31068
133. Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut (2017) 66:157–67. doi: 10.1136/gutjnl-2015-310514
134. Wang Y, Li C, Zhao R, Qiu Z, Shen C, Wang Z, et al. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics (2021) 11:6315–33. doi: 10.7150/thno.52843
135. Ou ZL, Luo Z, Wei W, Liang S, Gao TL, Lu YB. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol (2019) 16:1592–603. doi: 10.1080/15476286.2019.1649585
136. Katopodi T, Petanidis S, Domvri K, Zarogoulidis P, Anestakis D, Charalampidis C, et al. Kras-driven intratumoral heterogeneity triggers infiltration of M2 polarized macrophages via the circHIPK3/PTK2 immunosuppressive circuit. Sci Rep (2021) 11:15455. doi: 10.1038/s41598-021-94671-x
137. Xu YJ, Zhao JM, Gao C, Ni XF, Wang W, Hu WW, et al. Hsa_circ_0136666 activates treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell Signal (2021) 86:110095. doi: 10.1016/j.cellsig.2021.110095
138. Huang L, Ma J, Cui M. Circular RNA hsa_circ_0001598 promotes programmed death-ligand-1-mediated immune escape and trastuzumab resistance via sponging miR-1184 in breast cancer cells. Immunol Res (2021) 69:558–67. doi: 10.1007/s12026-021-09237-w
139. Zheng Y, Ren S, Zhang Y, Liu S, Meng L, Liu F, et al. Circular RNA circWWC3 augments breast cancer progression through promoting M2 macrophage polarization and tumor immune escape via regulating the expression and secretion of IL-4. Cancer Cell Int (2022) 22:264. doi: 10.1186/s12935-022-02686-9
140. Tian Q, Wu T, Zhang X, Xu K, Yin X, Wang X, et al. Immunomodulatory functions of the circ_001678/miRNA-326/ZEB1 axis in non-small cell lung cancer via the regulation of PD-1/PD-L1 pathway. Hum Mol Genet (2022) 31:4094–106. doi: 10.1093/hmg/ddac155
141. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett (2019) 17:388–97. doi: 10.3892/ol.2018.9606
142. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis (2021) 12:94. doi: 10.1038/s41419-020-03334-8
143. Li S, Chen Z, Zhou R, Wang S, Wang W, Liu D, et al. Hsa_circ_0048674 facilitates hepatocellular carcinoma progression and natural killer cell exhaustion depending on the regulation of miR-223-3p/PDL1. Histol Histopathol (2022) 37:1185–99. doi: 10.14670/hh-18-440
144. Guan L, Hao Q, Shi F, Gao B, Wang M, Zhou X, et al. Regulation of the tumor immune microenvironment by cancer-derived circular RNAs. Cell Death Dis (2023) 14:132. doi: 10.1038/s41419-023-05647-w
145. Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol (2019) 12:90. doi: 10.1186/s13045-019-0776-8
146. DePeaux K, Delgoffe GM. Metabolic barriers to cancer immunotherapy. Nat Rev Immunol (2021) 21:785–97. doi: 10.1038/s41577-021-00541-y
147. Jones JC, Bodenstine TM. Connexins and glucose metabolism in cancer. Int J Mol Sci (2022) 23:10172. doi: 10.3390/ijms231710172
148. Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer (2021) 21:669–80. doi: 10.1038/s41568-021-00378-6
149. Ortiz-Pedraza Y, Muñoz-Bello JO, Olmedo-Nieva L, Contreras-Paredes A, Martínez-Ramírez I, Langley E, et al. Non-coding RNAs as key regulators of glutaminolysis in cancer. Int J Mol Sci (2020) 21:2872. doi: 10.3390/ijms21082872
150. Geng P, Qin W, Xu G. Proline metabolism in cancer. Amino Acids (2021) 53:1769–77. doi: 10.1007/s00726-021-03060-1
151. Liu N, Shi F, Yang L, Liao W, Cao Y. Oncogenic viral infection and amino acid metabolism in cancer progression: molecular insights and clinical implications. Biochim Biophys Acta Rev Cancer (2022) 1877:188724. doi: 10.1016/j.bbcan.2022.188724
152. Vettore LA, Westbrook RL, Tennant DA. Proline metabolism and redox; maintaining a balance in health and disease. Amino Acids (2021) 53:1779–88. doi: 10.1007/s00726-021-03051-2
153. Alaqbi SS, Burke L, Guterman I, Green C, West K, Palacios-Gallego R, et al. Increased mitochondrial proline metabolism sustains proliferation and survival of colorectal cancer cells. PloS One (2022) 17:e0262364. doi: 10.1371/journal.pone.0262364
154. Patriarca EJ, Cermola F, D’Aniello C, Fico A, Guardiola O, De Cesare D, et al. The multifaceted roles of proline in cell behavior. Front Cell Dev Biol (2021) 9:728576. doi: 10.3389/fcell.2021.728576
155. Kay EJ, Paterson K, Riera-Domingo C, Sumpton D, Däbritz JHM, Tardito S, et al. Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat Metab (2022) 4:693–710. doi: 10.1038/s42255-022-00582-0
156. Kazberuk A, Chalecka M, Palka J, Bielawska K, Surazynski A. NSAIDs induce proline Dehydrogenase/Proline oxidase-dependent and independent apoptosis in MCF7 breast cancer cells. Int J Mol Sci (2022) 23:3813. doi: 10.3390/ijms23073813
157. Schcolnik-Cabrera A, Juárez-López D. Dual contribution of the mTOR pathway and of the metabolism of amino acids in prostate cancer. Cell Oncol (Dordr) (2022) 45:831–59. doi: 10.1007/s13402-022-00706-4
158. Tang Y, Zhang Z, Chen Y, Qin S, Zhou L, Gao W, et al. Metabolic adaptation-mediated cancer survival and progression in oxidative stress. Antioxid (Basel) (2022) 11:1324. doi: 10.3390/antiox11071324
159. Sica V, Bravo-San Pedro JM, Stoll G, Kroemer G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int J Cancer (2020) 146:10–7. doi: 10.1002/ijc.32616
160. Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res (2018) 24:2482–90. doi: 10.1158/1078-0432.Ccr-17-3070
161. Herbert SP, Stainier DY. Molecular control of endothelial cell behaviour during blood vessel morphogenesis. Nat Rev Mol Cell Biol (2011) 12:551–64. doi: 10.1038/nrm3176
162. Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell (1996) 86:353–64. doi: 10.1016/s0092-8674(00)80108-7
163. Vimalraj S. A concise review of VEGF, PDGF, FGF, notch, angiopoietin, and HGF signalling in tumor angiogenesis with a focus on alternative approaches and future directions. Int J Biol Macromol (2022) 221:1428–38. doi: 10.1016/j.ijbiomac.2022.09.129
164. Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell (2019) 176:1248–64. doi: 10.1016/j.cell.2019.01.021
165. Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis (2017) 20:409–26. doi: 10.1007/s10456-017-9562-9
166. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci (2020) 77:1745–70. doi: 10.1007/s00018-019-03351-7
167. Deryugina EI, Quigley JP. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol (2015) 44-46:94–112. doi: 10.1016/j.matbio.2015.04.004
168. Zhang W, Zhang X, Huang S, Chen J, Ding P, Wang Q, et al. FOXM1D potentiates PKM2-mediated tumor glycolysis and angiogenesis. Mol Oncol (2021) 15:1466–85. doi: 10.1002/1878-0261.12879
169. Jin X, Yin J, Kim SH, Sohn YW, Beck S, Lim YC, et al. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res (2011) 71:7125–34. doi: 10.1158/0008-5472.Can-11-1330
170. Yan B, Liu L, Zhao Y, Xiu LJ, Sun DZ, Liu X, et al. Xiaotan sanjie decoction attenuates tumor angiogenesis by manipulating notch-1-regulated proliferation of gastric cancer stem-like cells. World J Gastroenterol (2014) 20:13105–18. doi: 10.3748/wjg.v20.i36.13105
171. Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer (2018) 17:22. doi: 10.1186/s12943-018-0766-4
172. Jiang S, Fu R, Shi J, Wu H, Mai J, Hua X, et al. CircRNA-mediated regulation of angiogenesis: a new chapter in cancer biology. Front Oncol (2021) 11:553706. doi: 10.3389/fonc.2021.553706
173. Li L, Xiao C, He K, Xiang G. Circ_0072088 promotes progression of hepatocellular carcinoma by activating JAK2/STAT3 signaling pathway via miR-375. IUBMB Life (2021) 73:1153–65. doi: 10.1002/iub.2520
174. Tan Z, Cao F, Jia B, Xia L. Circ_0072088 promotes the development of non-small cell lung cancer via the miR-377-5p/NOVA2 axis. Thorac Cancer (2020) 11:2224–36. doi: 10.1111/1759-7714.13529
175. Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor cell-derived exosomal circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front Cell Dev Biol (2021) 9:726323. doi: 10.3389/fcell.2021.726323
176. Cao F, Liu S, Li Z, Meng L, Sang M, Shan B. Activation of circ_0072088/miR-1261/PIK3CA pathway accelerates lung adenocarcinoma progression. Thorac Cancer (2022) 13:1548–57. doi: 10.1111/1759-7714.14369
177. Sun H, Liu F, Zhang H. Circ_0072008, an oncogene in pancreatic ductal adenocarcinoma, contributes to tumour cell malignant progression and glycolysis by regulating miR-545-3p/SLC7A11 axis. Autoimmunity (2022) 55:203–13. doi: 10.1080/08916934.2022.2027919
178. Zhou M, Yang Z, Wang D, Chen P, Zhang Y. The circular RNA circZFR phosphorylates Rb promoting cervical cancer progression by regulating the SSBP1/CDK2/cyclin E1 complex. J Exp Clin Cancer Res (2021) 40:48. doi: 10.1186/s13046-021-01849-2
179. Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun (2018) 505:346–52. doi: 10.1016/j.bbrc.2018.09.073
180. Wicks EE, Semenza GL. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest (2022) 132:e159839. doi: 10.1172/jci159839
181. Della Rocca Y, Fonticoli L, Rajan TS, Trubiani O, Caputi S, Diomede F, et al. Hypoxia: molecular pathophysiological mechanisms in human diseases. J Physiol Biochem (2022) 78:739–52. doi: 10.1007/s13105-022-00912-6
182. Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol (2020) 21:268–83. doi: 10.1038/s41580-020-0227-y
183. Islam S, Mukherjee C. Molecular regulation of hypoxia through the lenses of noncoding RNAs and epitranscriptome. Wiley Interdiscip Rev RNA (2022) 14:e1750. doi: 10.1002/wrna.1750
184. Huang Q, Yang J, Goh RMW, You M, Wang L, Ma Z. Hypoxia-induced circRNAs in human diseases: from mechanisms to potential applications. Cells (2022) 11:1381. doi: 10.3390/cells11091381
185. He Z. LINC00473/miR-497-5p regulates esophageal squamous cell carcinoma progression through targeting PRKAA1. Cancer Biother Radiopharm (2019) 34:650–9. doi: 10.1089/cbr.2019.2875
186. Jiang H, Liu H, Jiang B. Long non-coding RNA FALEC promotes colorectal cancer progression via regulating miR-2116-3p-targeted PIWIL1. Cancer Biol Ther (2020) 21:1025–32. doi: 10.1080/15384047.2020.1824514
187. Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer (2019) 5:632–53. doi: 10.1016/j.trecan.2019.08.005
188. Infantino V, Santarsiero A, Convertini P, Todisco S, Iacobazzi V. Cancer cell metabolism in hypoxia: role of HIF-1 as key regulator and therapeutic target. Int J Mol Sci (2021) 22:5703. doi: 10.3390/ijms22115703
189. Romero Y, Aquino-Gálvez A. Hypoxia in cancer and fibrosis: part of the problem and part of the solution. Int J Mol Sci (2021) 22:8335. doi: 10.3390/ijms22158335
190. You L, Wu W, Wang X, Fang L, Adam V, Nepovimova E, et al. The role of hypoxia-inducible factor 1 in tumor immune evasion. Med Res Rev (2021) 41:1622–43. doi: 10.1002/med.21771
191. Amicone L, Marchetti A, Cicchini C. Exosome-associated circRNAs as key regulators of EMT in cancer. Cells (2022) 11:1716. doi: 10.3390/cells11101716
192. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer (2019) 18:116. doi: 10.1186/s12943-019-1041-z
193. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol (2020) 14:539–55. doi: 10.1002/1878-0261.12629
194. D’Alterio C, Scala S, Sozzi G, Roz L, Bertolini G. Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin Cancer Biol (2020) 60:351–61. doi: 10.1016/j.semcancer.2019.08.019
195. Weiss F, Lauffenburger D, Friedl P. Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat Rev Cancer (2022) 22:157–73. doi: 10.1038/s41568-021-00427-0
196. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sinica B (2021) 11:2783–97. doi: 10.1016/j.apsb.2021.01.001
197. Kichi ZA, Soltani M, Rezaei M, Shirvani-Farsani Z, Rojhannezhad M. The emerging role of EMT-related lncRNAs in therapy resistance and their applications as biomarkers. Curr Med Chem (2022) 29:4574–601. doi: 10.2174/0929867329666220329203032
198. Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu Y, et al. The epithelial to mesenchymal transition (EMT) and cancer stem cells: implication for treatment resistance in pancreatic cancer. Mol Cancer (2017) 16:52. doi: 10.1186/s12943-017-0624-9
199. Rezayatmand H, Razmkhah M, Razeghian-Jahromi I. Drug resistance in cancer therapy: the pandora’s box of cancer stem cells. Stem Cell Res Ther (2022) 13:181. doi: 10.1186/s13287-022-02856-6
200. Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y, et al. Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer (2021) 20:162. doi: 10.1186/s12943-021-01455-y
201. Cai R, Wang P, Zhao X, Lu X, Deng R, Wang X, et al. Reticulocalbin3: a Ca(2+) homeostasis regulator that promotes esophageal squamous cell carcinoma progression and cisplatin resistance. Cancer Sci (2022) 113:3593–607. doi: 10.1111/cas.15487
202. Fujita K, Motoyama S, Sato Y, Wakita A, Nagaki Y, Minamiya Y, et al. Effects of SLC31A1 and ATP7B polymorphisms on platinum resistance in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiotherapy. Med Oncol (2021) 38:6. doi: 10.1007/s12032-020-01450-1
203. Wei L, Sun J, Zhang N, Shen Y, Wang T, Li Z, et al. Novel implications of MicroRNAs, long non-coding RNAs and circular RNAs in drug resistance of esophageal cancer. Front Cell Dev Biol (2021) 9:764313. doi: 10.3389/fcell.2021.764313
204. Wang S, Qian L, Cao T, Xu L, Jin Y, Hu H, et al. Advances in the study of CircRNAs in tumor drug resistance. Front Oncol (2022) 12:868363. doi: 10.3389/fonc.2022.868363
205. Kulaksiz-Erkmen G, Dalmizrak O, Dincsoy-Tuna G, Dogan A, Ogus IH, Ozer N. Amitriptyline may have a supportive role in cancer treatment by inhibiting glutathione s-transferase pi (GST-π) and alpha (GST-α). J Enzyme Inhib Med Chem (2013) 28:131–6. doi: 10.3109/14756366.2011.639017
206. Chen YL, Yang TY, Chen KC, Wu CL, Hsu SL, Hsueh CM. Hypoxia can impair doxorubicin resistance of non-small cell lung cancer cells by inhibiting MRP1 and p-gp expression and boosting the chemosensitizing effects of MRP1 and p-gp blockers. Cell Oncol (Dordr) (2016) 39:411–33. doi: 10.1007/s13402-016-0285-5
207. Wang M, Yu F, Zhang Y, Zhang L, Chang W, Wang K. The emerging roles of circular RNAs in the chemoresistance of gastrointestinal cancer. Front Cell Dev Biol (2022) 10:821609. doi: 10.3389/fcell.2022.821609
208. Mittal P, Singh S, Sinha R, Shrivastava A, Singh A, Singh IK. Myeloid cell leukemia 1 (MCL-1): structural characteristics and application in cancer therapy. Int J Biol Macromol (2021) 187:999–1018. doi: 10.1016/j.ijbiomac.2021.07.166
209. Fu D, Pfannenstiel L, Demelash A, Phoon YP, Mayell C, Cabrera C, et al. MCL1 nuclear translocation induces chemoresistance in colorectal carcinoma. Cell Death Dis (2022) 13:63. doi: 10.1038/s41419-021-04334-y
210. Abdel-Magid AF. Myeloid cell leukemia-1 inhibitors as emerging cancer treatment. ACS Med Chem Lett (2021) 12:334–6. doi: 10.1021/acsmedchemlett.1c00099
211. Chen C, Aihemaiti M, Zhang X, Qu H, Sun QL, He QS, et al. Downregulation of histone demethylase JMJD1C inhibits colorectal cancer metastasis through targeting ATF2. Am J Cancer Res (2018) 8:852–65.
212. Sui Y, Gu R, Janknecht R. Crucial functions of the JMJD1/KDM3 epigenetic regulators in cancer. Mol Cancer Res (2021) 19:3–13. doi: 10.1158/1541-7786.Mcr-20-0404
213. Ashekyan O, Abdallah S, Shoukari AA, Chamandi G, Choubassy H, Itani ARS, et al. Spotlight on exosomal non-coding RNAs in breast cancer: an in silico analysis to identify potential lncRNA/circRNA-miRNA-Target axis. Int J Mol Sci (2022) 23:8351. doi: 10.3390/ijms23158351
214. Wei X, Shi Y, Dai Z, Wang P, Meng X, Yin B. Underlying metastasis mechanism and clinical application of exosomal circular RNA in tumors (Review). Int J Oncol (2021) 58:289–97. doi: 10.3892/ijo.2021.5179
215. Seimiya T, Otsuka M, Iwata T, Shibata C, Tanaka E, Suzuki T, et al. Emerging roles of exosomal circular RNAs in cancer. Front Cell Dev Biol (2020) 8:568366. doi: 10.3389/fcell.2020.568366
216. Li C, Ni YQ, Xu H, Xiang QY, Zhao Y, Zhan JK, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther (2021) 6:383. doi: 10.1038/s41392-021-00779-x
217. Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong XW, et al. Exosomal ncRNAs: novel therapeutic target and biomarker for diabetic complications. Pharmacol Res (2022) 178:106135. doi: 10.1016/j.phrs.2022.106135
218. Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer (2022) 21:49. doi: 10.1186/s12943-021-01471-y
219. Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer (2021) 20:144. doi: 10.1186/s12943-021-01448-x
220. Zhou H, He X, He Y, Ou C, Cao P. Exosomal circRNAs: emerging players in tumor metastasis. Front Cell Dev Biol (2021) 9:786224. doi: 10.3389/fcell.2021.786224
Keywords: circRNA, esophageal squamous cell carcinoma, tumor microenvironment, biological mechanism, chemoradiation resistance
Citation: Li J, Song Y, Cai H, Zhou B and Ma J (2023) Roles of circRNA dysregulation in esophageal squamous cell carcinoma tumor microenvironment. Front. Oncol. 13:1153207. doi: 10.3389/fonc.2023.1153207
Received: 29 January 2023; Accepted: 30 May 2023;
Published: 13 June 2023.
Edited by:
Christos K. Kontos, National and Kapodistrian University of Athens, GreeceReviewed by:
Maria Papatsirou, National and Kapodistrian University of Athens, GreeceRicardo Marques, University of Coimbra, Portugal
Marcelo R. S. Briones, Federal University of São Paulo, Brazil
Copyright © 2023 Li, Song, Cai, Zhou and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Zhou, emhvdWJvQHp6dS5lZHUuY24=; Jun Ma, bWFqdW5Aenp1LmVkdS5jbg==
 Jingyi Li
Jingyi Li Yuxia Song2
Yuxia Song2 Huihong Cai
Huihong Cai Jun Ma
Jun Ma