- Cancer Biology Research Center, Cancer Institute, Tehran University of Medical Sciences, Tehran, Iran
Autophagy regenerates cellular nutrients, recycles metabolites, and maintains hemostasis through multistep signaling pathways, in conjunction with lysosomal degradation mechanisms. In tumor cells, autophagy has been shown to play a dual role as both tumor suppressor and tumor promoter, leading to the discovery of new therapeutic strategies for cancer. Therefore, regulation of autophagy is essential during cancer progression. In this regard, the use of nanoparticles (NPs) is a promising technique in the clinic to modulate autophagy pathways. Here, we summarized the importance of breast cancer worldwide, and we discussed its classification, current treatment strategies, and the strengths and weaknesses of available treatments. We have also described the application of NPs and nanocarriers (NCs) in breast cancer treatment and their capability to modulate autophagy. Then the advantages and disadvantaged of NPs in cancer therapy along with future applications will be disscussed. The purpose of this review is to provide up-to-date information on NPs used in breast cancer treatment and their impacts on autophagy pathways for researchers.
1 Introduction
Despite the advances in breast cancer diagnosis and treatment, the incidence and mortality rates of breast cancer are still increasing especially in poorly developed regions of the world (1). The results of many studies represent the fact that a decline in infection-associated cancers is offset by the rising number of cancer cases being highly related to dietary, hormonal, and reproductive factors in countries with fast transitions in economic and communal issues. Hence, fundamental strategies for prevention, diagnosis in early stages, and novel targeted therapies can result in a decrease in expected cancer incidence (2). Most breast cancer survivors have encountered aggressive relapse. It is mainly explained by the fact that breast cancer tumors are highly heterogeneous and difficult to target (3). In anticancer therapies, the primary objective is the specific inhibition of the malignant function of cancerous cells while unaffected cells remain healthy. Ordinary treatments for cancers are encountered with inevitable side effects and drug resistance, which are discussed in the body of this article (4). In this regard, an intracellular mechanism such as autophagy targeting may be a good approach for breast cancer-targeted therapy. Autophagy as a conserved intracellular mechanism prepares the primary materials for the synthesis of vital macromolecules such as proteins by removing the unwanted molecules or organelles. Furthermore, autophagy preserves normal cells from intrinsic and extrinsic stressors promoting DNA mutations and instability that mainly lead to pre-neoplastic changes and propagation (3). Deficiency in autophagy function has led to diseases such as different malignancies. In cancer cells, autophagy has a dual role in homeostasis maintenance as a tumor suppressor in early stages and a tumor promoter at advanced stages. This important function of autophagy in cancer has led to more investigations in the field of targeting autophagy, which both inhibits and induces it in different cancers with different characteristics being critical. Many investigations in mammary cancer models revealed that the inhibition of autophagy genes can weaken the initial growth in tumors while paradoxically can result in overt metastasis and outgrowth in cancerous cells. In breast cancer, the genetic inhibition of autophagy in both early and late stages leads to the spread of tumor cells, which offers differentiation in pro-metastatic basal epithelial cells (5). In cancer initiation of mammary glands, the autophagy-related genes confer a suppressive role; however, this function is lost throughout breast cancer development, and impaired autophagy results in cancer progression (3). Of note, autophagy is detected as one of the main causes of chemotherapy resistance in breast cancer patients in a way that the inhibition of autophagy dramatically enhances the sensitivity of cancer cells to anticancer medicines (6). However, autophagy inducers could be effective in such situations and push the cells to apoptosis. Generally, efficient and genuine progress in this treatable disorder is not achieved unless a persistent and interconnected effort toward innovative procedures in cancer treatment (1). Most of the autophagy modulators that are currently available have low specificity, as they do not preferentially target a single cell type. Nanoparticles (NPs) can improve the efficacy of drugs through their high power of permeability and reduce their toxicity due to nanosized properties. They lead to more effective targeting of tissue, cells, or organelles and enhance the pharmaceutical properties of drugs such as stability, solubility, plasma half-life, and tumor accumulation. The connection between NPs and their impact on autophagy has shown that they can impact autophagy through both its induction and inhibition (7). In this review article, we aim to describe the application of different NPs and nanocarriers (NCs) in breast cancer treatment and their capability to modulate autophagy. We then discuss the advantages and disadvantages of NPs in cancer therapy, along with their future applications. The purpose of this review is to provide researchers with up-to-date information on NPs used in breast cancer treatment and their impacts on autophagy pathways.
2 Breast cancer
Breast cancer is one of the main causes of death in most parts of the world, especially in low-income areas (1). The incidence of breast cancer has increased in Western countries from the 1980s to 1990s and then has been stable due to better detection and the use of modern treatment. In 2020, almost 2.3 million women were diagnosed with breast cancer worldwide, and approximately 700,000 individuals died (8). According to the World Health Organization (WHO) prediction, by 2040, newly diagnosed breast cancer cases are expected to rise by almost 40% every year. A dramatic increase in the trend of breast cancer will be observed especially in countries with a low human development index (HDI), where the number of new cases and the mortality rate is projected to be doubled by 2040 (9). In Iran, breast cancer was determined to have the highest incidence rate per 100,000 women and the second-highest mortality rate after colorectal cancer in 2020 (10).
The risk factors for breast cancer are categorized as reproductive and non-reproductive factors, based on economic issues. Major reproductive factors include age, age of menarche, age at first pregnancy, some indicators of ovarian activity, age at natural menopause, duration of breastfeeding, history of breast disease, genetic status, nulliparity, and familial history of breast cancer (11). However, non-reproductive risk factors comprise obesity, lifestyle, being overweight during a post-menopausal state, and drinking level (2, 12). In Iran, other factors such as hormone replacement therapy, passive smoking, advanced maternal age at pregnancy, abortion, high levels of sugar consumption, and the genotype of Arg/Arg could also increase the risk of breast cancer development, whereas late menarche, breastfeeding for at least 13–24 months, routine exercise, and having vegetables in diet reduce the risk of breast cancer. The relationship between the polymorphism in codon 72 of the p53 gene and the risk of breast cancer shows that although the genotype of Arg/Pro was not related to the development of breast cancer, the genotype of Arg/Arg raised the risk of breast cancer significantly (13).
2.1 Breast cancer staging
In the 1940s and 1950s, the first staging of breast cancer was reported by Pierre De-noix, which was mainly based on the anatomic shape of the breast (14, 15). Lately, an extensively detailed system of staging cancers has been developed by the American Joint Committee on Cancer (AJCC) and has been updated eight times. In the breast cancer chapter, levels of estrogen receptor (ER) and progesterone receptor (PR) expression, human epidermal growth factor receptor 2 (HER2) or erythroblastic oncogene B (ERBB2) expression, histologic grade, regional lymph node involvement, distant metastases, and prognostic biomarkers are included in order to confer precise prognosis and guide treatment decisions (14, 16). This system is named TNM staging, which shows the status of tumors (T), lymph node involvement (N), and the level of tumor metastasis (M).
The combination of immune histological data such as the tumor grade, the determination of hormone receptor status, and a multi-gene panel with anatomic information results in a more explicit prognosis. However, tumors with lower-graded ER− and PR− tend to be more common among populations; therefore, multi-gene panels should be performed for further information (17).
2.2 Biomarkers in breast cancer
The biomarkers can be extremely helpful in the determination of breast cancer, especially in the early stage, to achieve a better prescription. In general, they are classified into tissue, genetic, and serum markers. The tissue markers or hormone receptors, namely, ER and PR, are critical for identifying patients that should be treated with hormone therapy. ER and PR should be tested for all patients with a breast cancer diagnosis. Nevertheless, ER alone is a weak prognostic factor (18). HER2 measurement is essential for all patients with invasive breast cancer and is the indicator for the patients who may be treated with trastuzumab and who may benefit from anthracycline-based adjuvant chemotherapy (19).
The most important genetic markers of breast cancer are defined as breast cancer gene 1 (BRCA1) and breast cancer gene 2 (BRCA2) mutation genes, which are the most powerful predictive tools to identify patients suffering from breast and ovarian cancer with a 40%–80% risk of developing cancer (20). In primary tumors, DNA ploidy is found to be an independent prognostic tool for surgical cases. Aneuploidy implies genetic abnormalities like single-nucleotide point mutations and changes in chromosome structure. Diploid tumors have a slightly longer survival time than those diagnosed with aneuploid tumors (21). The use of circulating tumor DNA (ctDNA), as a representative of tumorous DNA in the plasma or other body fluids of patients with cancer, is also important in detecting tumor cells. Detecting ctDNA reduces the need for tumor biopsy and helps a physician to determine the effective treatment, and a decrease in the levels of ctDNA in the process of treatment indicates a successful therapy (22).
The serum biomarkers are of great significance in breast cancer management therapy protocol, as they may help determine early diagnosis, prognosis, and response to therapies. They consist of carcinoembryonic antigen (CEA), the MUC-1 family agents (CA15-3, BR 27.29, MCA, and CA549), the serum cytokeratins [e.g., tissue polypeptide antigen (TPA) and tissue polypeptide-specific antigen (TPS)], and the serum oncoproteins (e.g., HER2/c-erbB-2). The approved serum markers designed for breast cancer are CA15-3 and CEA. Increased levels of CA15-3 (e.g., to 150 U/ml) and/or CEA (e.g., to 120 ng/ml) in a patient indicate a non-metastatic tumor (23). The measurement of two parameters such as urokinase-type plasminogen activator (UPA) and plasminogen activator inhibitor (PAI-1) is mostly performed in lymph node-negative patients, for whom adjuvant chemotherapy would not be helpful. Low levels of both the UPA and PAI-1 factors in such patients show a low risk of disease relapse (24).
2.3 Breast cancer treatment
According to the classification of breast cancer into ER and PR expression and HER2/ERBB2 gene amplification, different treatment strategies are applied (25). For ER- or PR-positive breast cancers, the first choice is the use of endocrine agents to downregulate ER signaling. For those with HER2 or HER2/neu positive, ERBB2-targeted therapy is suggested, including anti-ERBB2 antibodies (e.g., trastuzumab and pertuzumab) and small-molecule tyrosine kinase inhibitors (such as lapatinib and neratinib) (26). The main purpose of therapy for non-metastatic breast cancers is the removal of all the tumors or axillary lymph nodes by surgery, followed by postoperative radiation. Systemic therapy can also be performed preoperatively (neoadjuvant), postoperatively (adjuvant), or both. For metastatic breast cancers, the same basic categories of systemic therapy are used as neoadjuvant/adjuvant approaches (27).
For patients undergoing refractory metastatic breast cancer with a diagnosed deletion in BRCA1/2 genes, treatment with poly adenosine diphosphate-ribose polymerase (PARP) inhibitors like olaparib and talazoparib is approved by the US Food and Drug Administration (FDA) (28). Conventional chemotherapy for breast cancer targets the cells non-selectively and inhibits the proliferation of both normal and cancerous cells. Therefore, gastrointestinal problems and hair loss are some of their important side effects. The other major problem is drug resistance (4), which is discussed in the following paragraph. Figure 1 illustrates the treatment protocol for different breast cancers.
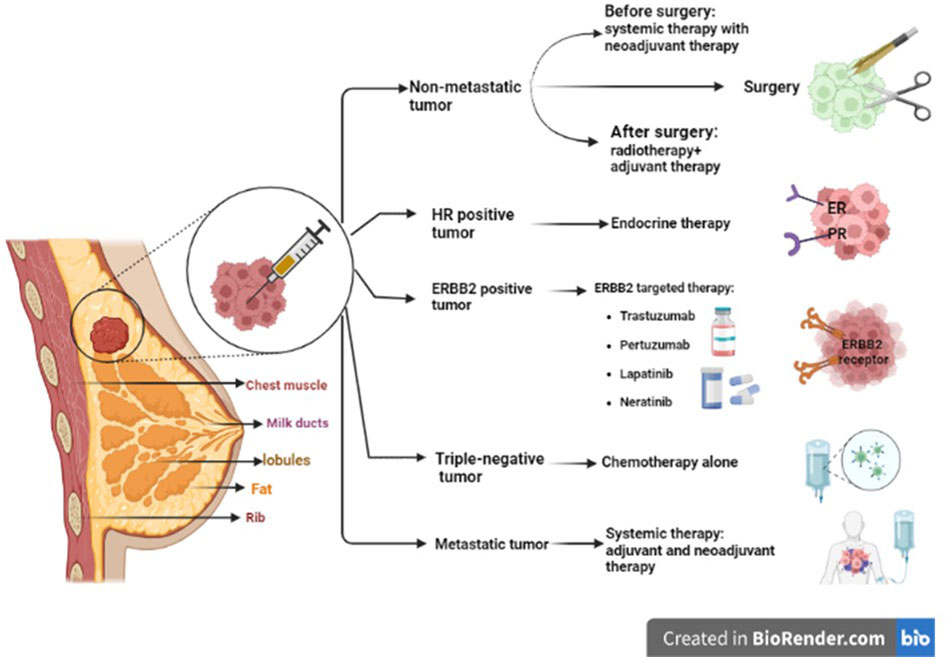
Figure 1 Different strategies to treat different types of breast cancer. ER/PR-positive breast cancers are much more likely to respond to hormone therapy. For HER2 or HER2/neu-positive patients, targeted therapy including anti-ERBB2 antibodies (trastuzumab and pertuzumab) and small-molecule tyrosine kinase inhibitors (such as lapatinib and neratinib) are used. For non-metastatic breast cancer, complete removal of the tumor or axillary lymph nodes by surgery and postoperative radiation is recommended. For metastatic breast cancer, systemic therapy is prescribed, such as neoadjuvant/adjuvant therapies. ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; ERBB2, erythroblastic oncogene B.
2.4 Breast cancer and drug resistance
Multidrug resistance (MDR) is the main reason for the relapse of breast cancer. The development of MDR is due to various molecular mechanisms, including increased expression of efflux transporters, epithelial-to-mesenchymal transition (EMT), and stem cell drug resistance (29). Cancer cells prevent the accumulation of chemical drugs inside the tumor and hinder their effectiveness via different mechanisms. Drug efflux transporters or efflux pumps are defensive mechanisms of cancer cells that direct anticancer drugs out of tumors. They are members of the ATP-binding cassette (ABC) superfamily involved in transporting substances into and out of cells with ATP hydrolysis (29).
Another mechanism is defined as EMT, a process by which epithelial cells lose their intercellular adhesion and apical-basal polarity properties and migrate into the mesenchyme. These cells gain new features that lead to increased drug efflux and resistance to apoptosis (30).
Breast cancer stem cells (BCSCs) are one of the major problems in drug resistance, metastasis, and cancer recurrence. Due to properties such as self-renewal, long lifespan, slow proliferation, high drug transport capacity, DNA repair mechanisms, and anti-apoptotic properties, BCSCs show intrinsic resistance to conventional therapies, and their removal is considered to increase drug susceptibility. One reasonable approach is targeting self-renewal pathways, and the other plan attacks the BCSC microenvironment. At present, some novel techniques are being discovered, including the use of cyclin-dependent kinase (CDK) inhibitors, factors that induce stem cell differentiation, and targeted immunotherapies (31).
Cancer stem cells (CSCs) also play a role in the induction of the mitochondrial-expressed genes involved in oxidative stress response, tumor survival, and drug resistance (32).
Under stressful conditions, such as reactive oxygen species (ROS) production or nutrient starvation, mitochondria undergo membrane potential depolarization and sequester into autophagosomes, under the process named mitophagy. During severe and prolonged stress, mitophagy is suppressed, and cell death pathways are activated (33). Indeed, mitophagy is a cytoprotective process during tumor progression and has a critical role in drug resistance and maintenance of stemness and self-renewal of CSCs. However, reports point to the role of mitophagy as a cellular tumor suppressor. Thus, both induction and inhibition of mitophagy contribute to the drug sensitivity of tumor cells (34). Another cause of MDR is intracellular mechanisms such as autophagy activation, discussed later in detail, which can potentially be modulated in cancer therapy.
3 Autophagy
Autophagy is defined as a “self-degradative” pathway that manages cellular homeostasis to provide precursors such as amino acids for the assembly of vital cellular components via catabolic pathways (35). In other words, autophagy helps the cells eliminate the malfunctioned organelles or macromolecules and then return the primary building materials to the manufacturing cycles of the cell, to reestablish cellular homeostasis. Autophagy is induced by various cellular stresses such as hypoxia, starvation, and infection (36). This phenomenon was indicated with studies in yeast in the 1990s by detecting autophagy-related genes (ATGs). Autophagy is classified into three types: microautophagy, macroautophagy, and chaperone-mediated autophagy (CMA) (37). Although all three types take the misfolded proteins to the lysosome, their mechanisms and morphologies are different. In microautophagy, cytoplasmic materials are directly absorbed into the lysosomal lumen through the invagination of the lysosomal membrane (38). In CMA, soluble proteins in the cytosol are selectively recognized by a 70-kDa heat shock protein (hsp70), unfolded, and translocated to lysosomes. These cytoplasmic proteins have special recognition motifs called pentapeptides. The CMA targeting motif is recognized as KFERQ and varies at different residues. For example, at “K” and “R” positions, up to two positively charged amino acids [e.g., arginine (R) or lysine (K)]; at position “F”, two hydrophobic residues [e.g., isoleucine (I), leucine (L), phenylalanine (F), or valine (V)]; and at “E” position, single negatively charged [e.g., glutamate (E) or aspartate (D)] can be placed. The “Q” can be at the N- or C-terminus of the pentapeptide and is flexible for negatively or positively charged or hydrophobic amino acids. This variation of proteins exists in approximately one-third of all the soluble cytosolic proteins. This protein–chaperone complex binds to a receptor of the lysosome and lysosome-associated membrane protein type 2A (LAMP-2A), and the substrate is released. Then, the unwanted proteins are degraded by lysosomal enzymes and reused in the next protein synthesis process (39).
Macroautophagy initiates with special double-membrane vesicles, known as autophagosomes, that progressively isolate autophagic cargo and deliver them to lysosomes by membrane fusion. An organelle that results from the fusion of an autophagosome and a lysosome is often referred to as the autophagolysosome. The digestion of cargo prepares nutrients for cell survival. Macroautophagy is directly related to autophagy (40) and is mainly considered a non-selective or general process. Nevertheless, it is noteworthy that in many cases the selective autophagy pathway has been also observed, which turns out to be essential only for cellular health. Deficiency in autophagy function can lead to diseases such as susceptibility to infections and inflammation, metabolic disorders, cardiovascular and liver problems, cancer, neurodegeneration, and acute brain damage. In tumorigenicity, autophagy performs a two-faced role of tumor suppressor and pre-oncogene (5). Because of these dual roles, finding a new targeted therapy for both activating and inhibiting autophagy pathways is under investigation (38) and will be discussed in the following sections.
3.1 Autophagy machinery
Autophagy contains complex steps that lead to the generation of autophagosomes and their fusion into lysosomes. The process includes several phases, namely, initiation, elongation, lysosomal fusion, and degradation, which are mediated by ATGs (3).
3.1.1 Induction and initiation
In mammalian cells, the autophagy is induced by unc-51-like kinase family (ULK1/ULK2) complex (homolog of the Atg1 complex in yeast), ATG13, and RB1-inducible coiled-coil 1 (RB1CC1/FIP200). Then, C12orf44/ATG101 protein binds directly to ATG13, which occurs in mammals and not in yeast. ULK1 complex induces membrane formation in the phagophore assembly site (PAS). Accordingly, ATG2 transfers phospholipids from the endoplasmic reticulum, Golgi, and mitochondria membrane to PAS, and ATG9 transfers them to the luminal layer of the autophagosome membrane (41). As the phagophore expands, the membrane wraps around the substrate and forms a spherical autophagosome.
Autophagy is immensely regulated by two main kinases, including the mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK), which affect ULK1 complex formation (42). mTORC1 associates with the ULK1/ULK2 complex in autophagosome under rich nutrients and inactivates the complex by its phosphorylation. However, under rapamycin treatment or starvation, mTORC1 dissociates from the complex and induces autophagy. In other words, in the abundance of amino acids and nutrients, mTORC1 inhibits autophagy through the phosphorylation of ULK1 and ATG13 (43). However, the AMPK (the autophagy inducer) can sense the AMP : ATP ratio caused by energy deprivation and be activated to start autophagy by phosphorylating several sites in the central intrinsically disordered region (IDR) in ULK1. Furthermore, the AMPK is an indirect inducer of autophagy by phosphorylation of the regulatory-associated protein of mTOR (RAP-TOR), which leads to mTOR inhibition (44).
3.1.2 Nucleation
In this step, ATG14-containing class III phosphatidylinositol 3-kinase (PtdIns3K) complex binds to the autophagosome. The PtdIns3K generates the PtdIns3P complex, which is involved in the nucleation of the phagophore and consists of PIK3C3/VPS34, PIK3R4/p150, and BECN1. Some reports suggest the association of this complex with ATG14 and UVRAG is necessary for autophagosome formation. Regulation of the PtdIns3K complex is mediated through the proteins that interact with BECN1 such as BCL2. As the BCL2 binds to BECN1, it inhibits the interaction of BECN1 with PIK3C3 and inactivates autophagy (45).
3.1.3 Elongation
Phagophore expands with the formation of the ATG5, ATG12, and ATG16L1 complex. As the phagophore is completed, it dissociates from the complex. The ATG8/LC3 system is also involved in the phagophore expansion. In mammals, there are several Atg8-like proteins that are the LC3 and GABARAP subfamilies. The LC3 forms LC3-1 by ATG4 and its phosphatidylethanolamine (PE)-conjugated or lipidation form, called LC3-II. The form of LC3-II is activated under starvation or other autophagy stressors. ATG9 is a transmembrane protein involved in phagophore expansion and membrane recruitment (46).
3.1.4 Autophagosome fusion with lysosome
The structure of autolysosome is formed by the fusion of the outer membrane of the mature autophagosome with the lysosome (47). There are three main machinery protein families playing a crucial role in the regulation of the fusion process, including membrane-tethering factors (such as HOPS and EPG5), soluble N-ethylmaleimide-sensitive factor attachment proteins (SNAREs), and Rab GTPases, which are situated on the membrane. They hire the tethering complex to bridge to the facing lipid bilayer; therefore, the tethering complex employs SNARE proteins and promotes them to move autophagosomes toward the lysosome and facilitate the fusion process (48). Recently, a growing number of studies have conferred the great role of ATG8 family members in driving autophagosomes near the lysosome. In addition, the role of these members has been suggested as probable hubs in the final fusion stages. In mammals, amphisomes emerge if the autophagosome fuses with the endosome before reaching the lysosome. Microtubules are also involved in driving the autophagosomes to the lysosomes (49). Being able to assist with the UVRAG PtdIns3K complex, the UVRAG activates GTPase RAB7, which promotes autophagosome–lysosome fusion (50). Recent studies have suggested other components of the SNARE machinery system that play role in the fusion process like VAM9 and VAM7 (51).
One of the mechanisms that macroautophagy uses to selectively identify cargo is ubiquitination. The ubiquitin-binding protein SQSTM1/p62 targets the ubiquitinated proteins and then interacts with LC3-II to clear these aggregate proteins from the cytosol and move them to the lysosome (52).
3.1.5 Degradation
Once the autophagosome formation is completed, its outer membrane merges with the lysosomal membrane; then, the inner layer and cargo will be degraded by hydrolases and subsequently with permeases to be competent for cell biosynthetic processes and the generation of energy (53). The product of autophagosome and the lysosome fusion is called “autophagolysosome” or “outolysosome”. The heterophagic (no-self) materials are needed in autolysosomes since approximately all lysosomes take constant flow made by endocytic pathway (54). All the autophagy steps are shown in Figure 2.
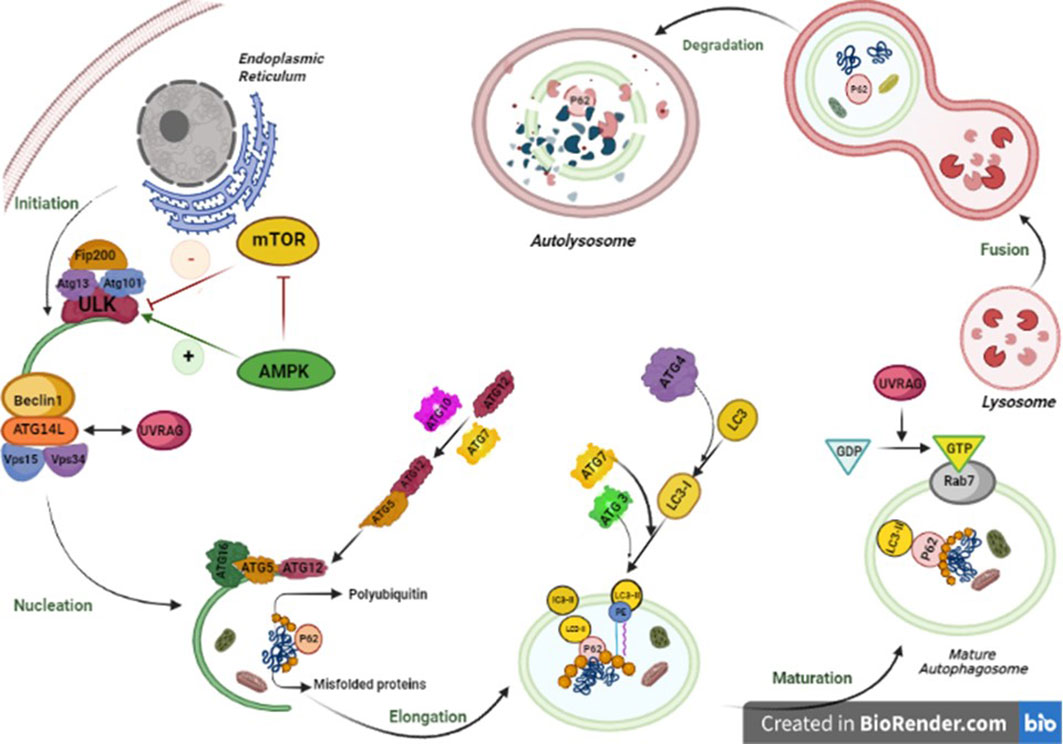
Figure 2 Autophagy pathway is performed in five steps from initiation to fusion with lysosome. In mammalian cells, the initiation phase of autophagy consists of autophagosome formation, which is significantly dependent on the stable complex known as ULK1-Atg13-FIP200-Atg101. The activation of the ULK1 kinase launches the activity of Beclin1 (BECN1)–VPS34 complex including BECN1, VPS34, and Beclin1 regulator 1 (AMBRA-1). In the elongation phase, the WIPI2B scaffold binds to PI3P. The mentioned complex is highly essential for employing two main proteins, ATG7 and ATG10, that can couple ATG5 to ATG12, which makes a complex with ATG16L. The Atg12–Atg5–Atg16 complex positions on the outer membrane due to ATG 5 available binding sites. The next step is the fusion of autophagosome–lysosome and includes two main phases. First, the autophagosome migration to lysosomes, which is implemented by the cytoskeleton in eukaryotic cells by Rab7 and guanosine triphosphate (GTP)-binding protein to microtubules, and second, the fusion of lysosomes, with a single bilayer membrane, and mature autophagosomes, with two lipid bilayer membranes. The last step is degradation of all components in the lysosome. The lysosomal enzymes degrade autophagic cargo.
3.2 Autophagy and cancer
Autophagy has two completely different behaviors in cancer, depending on the severity of cellular stress and the state of the immune system. During tumor initiation, autophagy prevents carcinogenesis, and in the advanced stages of tumors, it aids cancer progression. In fact, autophagy acts as a tumor suppressor in the early stages of carcinogenesis by maintaining genomic stability and paradoxically promotes tumor progression in established cells by providing nutrients (36, 55). It is noteworthy that in cancer cells, autophagy suppresses the immune system by affecting T cells, cytokines, and other immune system cells. In fact, autophagy helps the immune system by removing damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs). Thus, the immune system is prevented from functioning properly in cancerous health conditions due to the increase in autophagy and removal of excessive DAMPs and PAMPs. In other words, the function of T cells, NK cells, cytokines, and other vital parts of the immune system is overshadowed by complicated processes of autophagy (56, 57).
In preclinical breast cancer models, autophagy has performed a strong role in the survival of quiescent disseminated cells (58). Compared to normal tissues, breast cancer cells hold a low level of Beclin1 proteins (59). Human BECN1 monoallelic deletions are reported in up to 50% of breast cancers and 75% of ovarian cancers (60, 61). Numerous studies in the area of breast cancer survivors on the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) dataset demonstrated that a lower expression of BECN1 is associated with a worse probability of survival (61).
The defective activity of any autophagic genes may affect carcinogenesis. It is detected that the mono-allelically deleted Beclin-1, as a haploid tumor suppressor, has an insufficient function in different cancers such as human hepatocellular carcinoma and breast, ovarian, and prostate cancers (62). Notably, many studies on ATG genes have demonstrated that ATG2B, ATG5, ATG9B, ATG12, and ATG16L1 are also oncologically linked to tumorigenesis. Furthermore, the somatic mutations in the ATG5 gene ruin the binding sites of ATG5 and Atg16L1, resulting in an impaired conjugation site for ATG12, and autophagy failure (63).
Evidence shows that in some situations, inhibition of autophagy and, in some cases, induction of autophagy has been beneficial for the treatment of cancers, depending on the tumor type or the stage of disease (64). Autophagy-dependent genes and proteins are valuable markers in tissue samples of patients for better diagnostics (65).
3.3 Autophagy and drug resistance
Several studies indicate that autophagy has a critical role in the survival of tumors and drug resistance. Obviously, autophagy is induced by chemotherapeutic drugs and then prepares the tumor cells for the nutrients by degradation of unwanted proteins or organelles. Accordingly, autophagy prevents DNA damage and enhances drug resistance in tumors (66). Although the exact mechanism of drug resistance with autophagy induction is not fully understood, some reports have suggested the role of autophagy in the induction of DNA damage response and the increase of drug efflux pumps in tumor cells (67). In this regard, autophagy may be involved in apoptosis inhibition by the inactivation of pro-apoptotic factors or the activation of anti-apoptotic agents. In this case, autophagy induction is used to overcome drug resistance. Prolonged autophagy promotes autophagic cell death (type II cell death), which is independent of apoptosis (type I cell death) or necrosis (type III cell death). In tumors, when the cells resist apoptosis, induction of autophagic cell death is an approach to trigger drug resistance. There are some drugs (like bortezomib, rapamycin, and butyrate) that induce autophagosome formation (68). However, considering the dual role of autophagy in cancer, another approach to target tumor drug resistance is autophagy inhibition. The use of gene silencing of autophagy-related genes or chemical inhibitors such as bafilomycin A1, 3-methyladenine (3-MA), and chloroquine (CQ) is commonly used to inhibit autophagosome formation (69).
3.4 Autophagy modulation
Enhanced autophagy in tumors, as a survival mechanism and a cause of MDR, leads to the use of autophagy inhibitors to suppress tumor cell proliferation and induce cell death. The application of autophagy inhibitors in combination with antitumor drugs sensitizes the cancerous cells to chemotherapy (70). For example, treatment of HER-positive breast cancer cells with depletion of Atg5, Atg7, or beclin1 resulted in enhanced effectiveness of tamoxifen or the combination therapy of 3-MA, as an autophagy inhibitor, and trastuzumab, as a chemotherapeutic drug, increasing the effectiveness of chemotherapy in HER2-positive breast cancer cells as compared to monotherapy (71).
However, the use of anticancer drugs induces autophagy as a survival mechanism. However, therapies involving both excessive autophagy induction upon cytotoxic drug treatment or using autophagy inducers may also lead to autophagic cell death (72, 73). Accordingly, there are some autophagy inducers that have been used as anticancer treatments. For example, proteasome inhibitors (PIs) have also been shown to stimulate autophagy. Bortezomib, a PI used in the treatment of multiple myeloma and mantle cell lymphoma, has been shown to increase the early formation of autophagosomes and LC3-II, demonstrating its inducing effects on autophagy (74). A well-known class of autophagy inductors includes analogs of the mTOR inhibitor rapamycin, such as temsirolimus and everolimus. These compounds, which can be used alone or in combination with chemotherapy drugs, show an anti-proliferative effect in mantle cell lymphoma and acute lymphoblastic leukemia by overstimulating autophagy, which might cause tumor cell death. A large number of ongoing trials demonstrate that autophagy modulation in combinatory treatments could successfully overcome the resistance to existing anticancer drugs. For instance, RUBCN (Rubicon autophagy regulator), as a negative regulator of autophagosome biogenesis, leads to the inhibition of basal differentiation and attenuates metastatic growth in vivo (5, 73). There are many FDA-approved and unapproved autophagy modulatory drugs that are introduced for cancer treatment. For example, CQ and hydroxychloroquine (HCQ) are determined as autophagy inhibitors, rapamycin and metformin are used as autophagy inducers in various cancers, and CQ and metformin are mainly used in breast cancer. The mechanism of CQ is suppressing the fusion of autophagosome and lysosome, while metformin activates AMPK (74) through ROS production in the mitochondria and inhibition of the mitochondrial respiratory chain complex 1 (75).
3.5 Breast cancer treatment by autophagy modulation
Since autophagy has a significant role in the survival of cancer cells, its modulation is considered a hopeful strategy for cancer treatment. Previous studies have shown that autophagy is the main reason for resistance to chemotherapy in breast cancer and has resulted in decreased sensitivity to chemotherapy with doxorubicin (DOX). Experiments indicated that the use of autophagy inhibitors can reverse doxorubicin resistance and enhance its efficacy in triple-negative breast cancer MDA-MB-231 cells (76). In addition, the cotreatment of DOX and 3-MA resulted in a necroptotic form of cell death in breast cancer (76). Using CQ is another approach to return DOX sensitivity in breast cancer MCF-7 cells (77).
Overexpression and accumulation of LAMP2A were observed in breast cancer tissue samples, and its inhibition conferred sensitivity to DOX in MCF-7 and T47D breast cancer cells (78). A novel adipokine, resistin, is highly stimulated in patients with breast cancer and facilitates cell proliferation, metastasis, and breast cancer cell migration. Resistin can activate AMPK/mTOR/ULK1 and JNK signaling pathways. Interestingly, the blockage of the two mentioned pathways reduces the ratio of LC3-II/LC3-I, which grants increasing apoptosis in breast cancer cells induced by DOX (79). In addition, in DOX-resistant breast cancer cells, psammaplin A (a natural product isolated from marine sponges with anticancer effects) can stimulate overexpression of damage-regulated autophagy modulator (DRAM) that is induced by p53 protein (80). In estrogen receptor-positive breast cancer, attenuated autophagy sensitized resistant tumors to tamoxifen-induced cell death (81). The summary of some autophagy inhibitors and inducers that are used in breast cancer is listed in Tables 1, 2 respectively.
4 Autophagy modulations by nanoparticles and nanocarriers
Most of the autophagy modulators that are currently available have low specificity, as they do not preferentially target a single cell type. In addition, multiple autophagy mediators contribute to several cellular processes and are involved in autophagy-independent functions. For instance, rapamycin, as a well-known autophagy inducer, has an impact on the inhibition of T-cell proliferation, which results in strong immunosuppression (99). These challenges lead to limitations in the use of autophagy regulators in cancer treatment. In this regard, NPs have multiple advantages to overcome these issues. They can improve the efficacy of drugs by their high power of permeability and reduce their toxicity due to nanosized properties. They lead to more effective targeting of tissue, cells, or organelles and enhance the pharmaceutical properties of drugs such as stability, solubility, plasma half-life, and tumor accumulation (7). NPs can be used alone or in combination with drugs. However, NC is a nanomaterial that is used as a transport mediator for other substances, such as a drug. Generally, NCs are categorized as micelles, polymers, carbon-based materials, liposomes, and other materials and are used in drug delivery, especially in chemotherapy (100).
The connection between NPs and their impact on autophagy has shown that they can impact autophagy through both its induction and inhibition. The induction of autophagy is mainly mediated through oxidative stress activation. Under external stress, the phagocytosis of NPs or NCs is enhanced, and mitochondrial respiration is also increased. Hence, the accumulation of a large number of incompletely reduced oxygen atoms results in the generation of a large number of ROS, which leads to programmed cell death (101). Autophagy induction by NPs may be a defensive mechanism against external signals. The use of CQ and NPs enhances the effectiveness of chemotherapy by the accumulation of NPs in the tumor and decreases the immunological clearance of NPs. The NPs can directly reach the tumor cells; therefore, they can reduce the non-specific functions of autophagy regulators, enhance the accumulation of drugs at tumor sites, and consequently enhance the antitumor efficacy (89).
Although NPs have several advantages in biomedical applications, they have shown some cytotoxic effects on cancerous cells. For example, inflammation, ROS production, apoptosis, or necrosis occurs following the toxicity of NPs, limiting their application. To overcome these barriers, some modifications are necessary in order to increase the efficacy of NPs. For example, changing the dose of treatment or modifying their shape or formulation could reduce the toxicity level. The dose of NPs used in treatment is important in their aggregation and distribution. Moreover, the direct administration of NPs on the skin is less toxic than intravenous injection. Consequently, conducting an accurate protocol for using NPs in biomedicine is critical (90).
The different effects of NPs on autophagy, induction, or inhibition, in different situations, lead to antitumor activity or an increase in cell death (89). In practice, the choice between inhibition and promotion of autophagy is controversial, as it may depend on the role of autophagy in tumor development. As long as autophagy exerts a positive effect on the treatment of certain cancers, strategies that promote autophagy remain desirable. However, when autophagy adversely affects cancer treatment, inhibition of autophagy is the appropriate strategy. Depending on the type of cancer, therapy should involve an appropriate treatment in combination with autophagy modulation (72). Figure 3 illustrates the role of autophagy in cancer, drug resistance, and the modulation of autophagy in breast cancer.
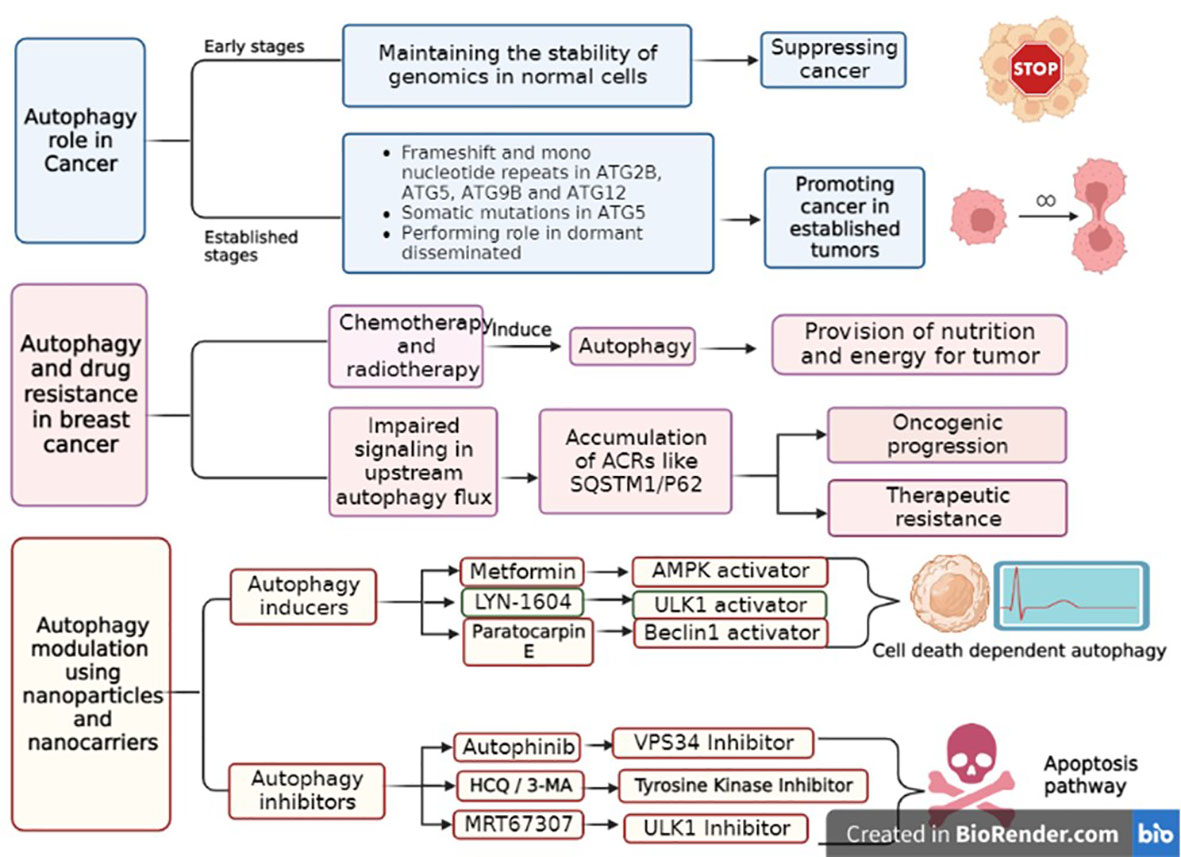
Figure 3 The role of autophagy in breast cancer, its drug resistance and the approaches of autophagy pathway modulation. Autophagy plays a dual role in cancers, according to the stage of tumors. In general, it acts as a tumor suppressor in early stages, while in established tumors, it promotes tumor progression. During chemotherapy or radiotherapy, autophagy is activated to provide nutrients for tumors and induce drug resistance. Autophagy inducers lead to autophagy activation, which finally promotes cell death in an autophagy-dependent manner. In contrast, autophagy inhibitors suppress the nutrient pathway for tumors, ultimately leading to apoptotic cell death.
4.1 Nanocarriers
NCs, as a specific type of drug delivery system, can target tumor cells through passive and active targeting methods and accumulate more in cancerous tissue, compared to normal tissues. Passive targeting means the direct permeation of drugs into tumor tissue, which depends on the action of the enhanced retention system or enhanced permeation system (EPS) effect. As a tumor develops in size and shape, an increased demand for oxygen and nutrients leads to the requirement for new blood vessels. These vessels are often not properly developed and are, therefore, permeable to some particles of certain sizes, usually below 700 nm (90). To function properly, NCs should be less than 100 nm in diameter and have a naturally hydrophilic surface, indicatively to avoid macrophage clearance and adhesion to plasma proteins. Polyethylene glycol, polysaccharides, poloxamines, poloxamers, and amphiphilic copolymers are some NCs with passive targeting ability (102). In active targeting, NCs are coated with specific ligands that can recognize their targets on the surface of cancer cells (91). NCs can be transferred directly to the cancer cells mainly through the following mechanisms. First, they can be delivered directly to the cancer cells through carbohydrate targeting, as the tumor cells have more carbohydrates in their membranes than normal cells. The second approach to the delivery of NCs is through receptor targeting. Some of the NCs are equipped with ligands of specific tumor cell receptors to recognize the tumor cells. In this method, after connecting the NCs to the tumor receptors, the drug is dissociated from the carrier and transferred to the target sites (92).
There are structurally different types of NCs explored for the effective treatment of breast cancer, including polymeric micelles, dendrimers, nanoliposomes, carbon nanotubes, solid lipid NPs (SLNs), nanostructured lipid carriers (NLCs), nanoemulsions (NEs), gold-based NPs (AuNPs), protein nano cargoes, and aptamers (93). In Table 3, we discussed the structure and properties of each nano-drug delivery system in the breast cancer experimental models (94).
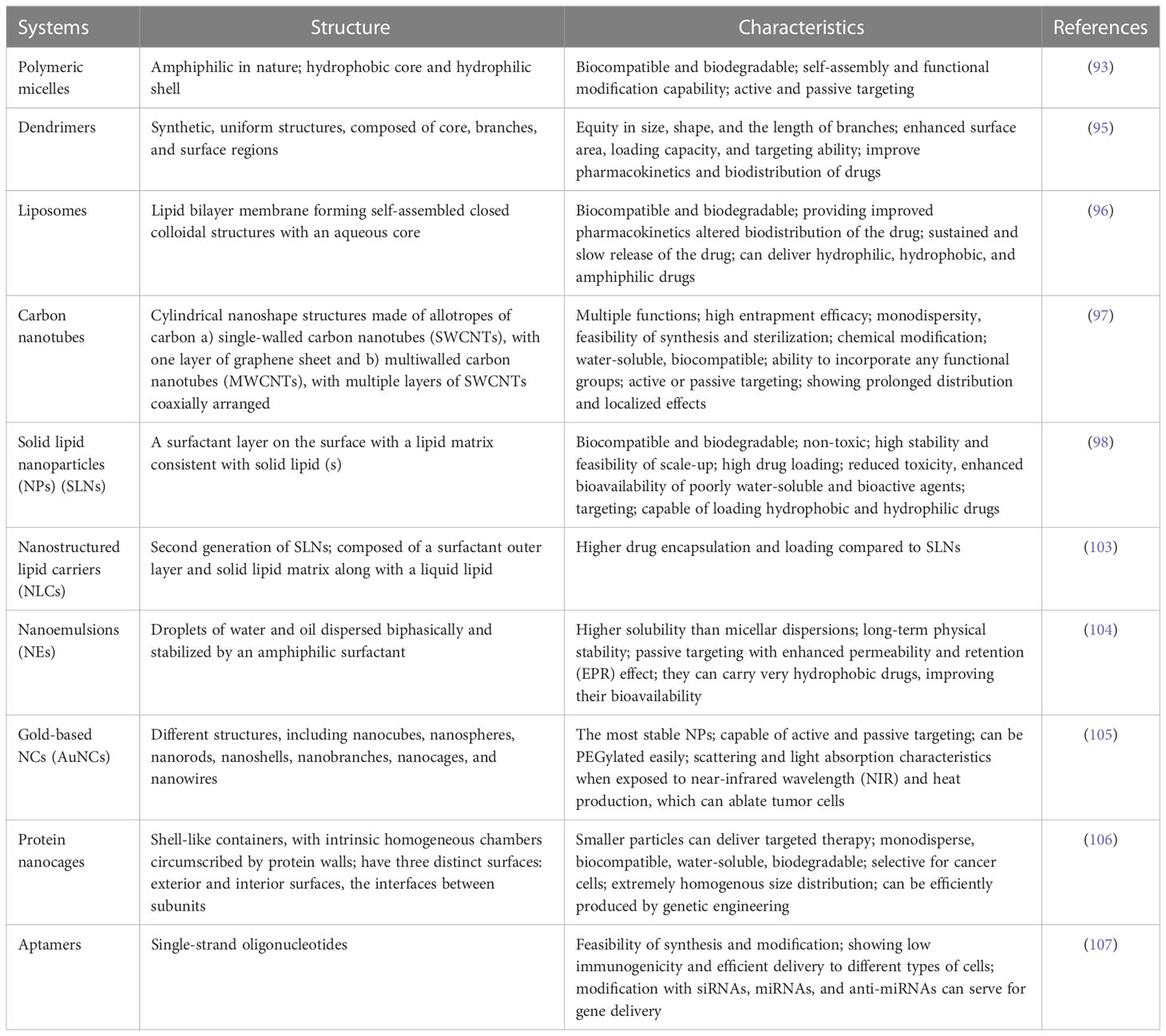
Table 3 The application of different types of nanocarriers (NCs) with their structures and characteristics in breast cancer experimental models.
4.2 Nanoparticles
NPs are commonly described as particulate subjects at least smaller than 100 nm in one dimension (108). NPs are structurally different from each other and include liposomal NPs composed of phospholipids (109); polymeric NPs synthesized from natural products like gelatin, albumin, or artificial polymers (e.g., polylactides); poly alkyl cyanoacrylates; inorganic NPs involving silica NPs; quantum dots; and metal NPs. In addition, other properties such as size, shape, surface charge, and inflexibility are important when choosing NPs as treatment tools. These properties impact the NPs’ cellular uptake through reticuloendothelial systems, targeting the right cells and tissue distribution (110). In the following, the physiochemical properties of NPs are explained in detail (109).
4.2.1 Size of NPs
The size of NPs is a significant factor in drug delivery and activation of the immune system and also modulates the pharmacokinetic actions of therapeutic agents, such as cell uptake, biodistribution, and body fluid half-life (111, 112). For example, gold-based NPs with a size of 10 nm have a longer blood circulation time and a reduced accumulation in the liver and spleen, compared to a size of 20 nm. Large NPs with a diameter between 100 and 200 nm are needed to deliver enough medication to disease sites (111). NPs from 30 to 50 nm exhibit efficient cellular absorption due to increased specific surface area and membrane encapsulation process. Phagocytic activity is considered to be the main pathway by which larger NPs from 250 to 3000 nm in size are internalized into the cells. Glomerular capillary walls have physiological pores between 4.5 and 5 nm; therefore, NPs smaller than 6 nm are effectively filtered, while those larger than 8 nm cannot be eliminated by glomerular filtration (113).
The 100–200-nm NPs, including microparticles, are useful for drug loading, considering their high capacity. However, these large particles undergo active opsonization, readily trigger immune responses, and accumulate rapidly in the liver and spleen, thereby exhibiting poor systemic circulation (114). Small NPs lower than 5 nm should be considered not only because of their low carrying capacity but also because of their fast rate of renal clearance. However, many studies have found that small NPs are advantageous in several respects compared to their larger counterparts, including minimal immunogenicity, long systemic circulation, and easy entry and aggregation in the tumor, as well as independence for caveolin-dependent cell pathways uptake (114). Because of their excellent absorption efficiency and significant tumor accumulation, NPs with a size of 30–200 nm have been widely used in numerous investigations (113).
4.2.2 Shape of NPs
NPs can be used in a variety of nanomaterials, including liposomes, micelles, dendrimers, and metal NPs. The ability of NPs to interact with cell membranes depends on their shape. For instance, non-spherical NPs might yield multivalent interactions with cell surface receptors, whereas spherical ones only interact with a small number of binding site receptors (115).
4.2.3 Surface charge
The plasma membranes of the cells show a moderate negative voltage difference, varying from −90 to −20 mV from cell to cell. The pharmacokinetics and blood circulation time of NPs are influenced by the electrostatic interactions between NPs and the proteins (115). For instance, cationic polymeric NPs encapsulating tetrandrine had superior anticancer activity in A549 cells and a higher cellular absorption efficiency than anionic NPs (116).
4.2.4 Elasticity
It has been thoroughly investigated whether the elasticity of NPs plays a significant physicochemical role in determining their pharmacokinetics and biodistribution. According to certain studies, the flexibility of NPs affects how they interact with tissues and immune cells (117). Additionally, the rigidity or softness of NPs is an important factor in their distribution. The uptake of the stiffer NPs is shown to disrupt the architecture of the cell, while soft NPs can enter the cells through micropinocytosis easily (118). The absorption process is thought to have changed from fusion, which has a low energy dependence, to endocytosis, which has a high energy dependence due to the differential uptake of soft and stiff NPs by cells (117). Soft NPs have a longer half-life in the bloodstream due to the lower absorption by macrophage cells, which is also reliant on their elasticity (117). Commonly, a significant factor that affects the pharmacokinetic behavior of NP therapies is their elastic modulus. It has been demonstrated that elastic modulus optimization affects the interactions of NPs with different cells and their circulating half-life, tumor targeting, and aggregation effectiveness (119).
4.3 NPs and autophagy modulation
Considering the aforementioned data, autophagy modulation may be a promising treatment, alone or in a combination with other therapeutic methods to combat resistance and ensure a successful treatment of breast cancer. However, most current autophagy regulators suffer from low specificity, poor targeting, and bioavailability, as they are insufficiently solubilized in aqueous media and cannot specifically target tumor tissues (120). Leveraging nano-drug delivery systems reduces toxicity and increases drug efficacy through more appropriate targeting (121). The NPs being studied for drug delivery can be divided into different categories considering the type of therapy in which they are implemented. Three major therapeutic NPs are chemotherapeutic, phototherapeutic (including photodynamic therapy, and photothermal therapy), and immuno-therapeutic NPs, along with other categories such as sonodynamic therapeutic NPs. Each of the categories induces cytotoxicity via different intracellular mechanisms such as inhibition or induction of autophagy (72, 121, 122). NPs are divided into three categories: metal, organic polymer, and inorganic non-metallic NPs (115). The NP-based drug delivery into the cell is schematically illustrated in Figure 4 and is discussed in detail in the following section.

Figure 4 Different nanoparticle-mediated autophagy. (A) AuNP delivery system enters the cells, accumulates in the lysosome, and changes its pH. The impairment of lysosome function leads to accumulation of autophagosomes in cells without fusion with lysosomes. Then, apoptosis is induced. Another study has shown the effect of NPs on the Bax/Bcl2 activation that leads to apoptosis. (B) Polymeric/micelle-like NPs can encapsulate the autophagy inducer that disturbs the pH of lysosome leading to cell death. (C) ZnO can conjugate with drugs and impact calcium pathway, which ultimately leads to autophagy. (D) AgNPs induce autophagy through activation of Atg5 expression, which can result in cell death through autophagy or apoptosis induction. (E) Silica NPs can encapsulate chemotherapeutic drugs, such as doxorubicin, and induce intrinsic apoptosis through mitochondria. (F) The effect of photothermal therapy of IONPs in breast cancer cells has been observed through autophagy induction, independently of apoptosis. The use of autophagy inhibitors and IONPs under laser therapy causes cell death. AuNP, gold-based nanoparticles; AgNPs, silver-based nanoparticles; IONPs, iron oxide nanoparticles.
4.3.1 Silver-based NPs
Silver-based NPs (AgNPs) have been shown to induce autophagy in breast cancer cells, ultimately leading to apoptosis. One experiment that used the AgNPs embedded in specific polysaccharides showed good entrance to the cells, generation of ROS, and induction of endoplasmic reticulum stress, leading to cell death through autophagy or apoptosis (123). The effect of AgNPs on cytotoxicity in breast cancer cells also confirmed that regardless of the shape and structure of the AgNPs, they are cytotoxic for triple-negative breast cancer (TNBC) in vitro and xenograft model but have no effect on non-malignant cells. Hence, the application and development of AgNPs in breast cancer treatment should be safe (124).
4.3.2 Gold-based NPs
The effect of gold-based NP (AuNP) cytotoxicity on cells has a different intracellular mechanism. AuNPs enter the cells by endocytosis, accumulate in the lysosome, and change the pH value. Impaired lysosome function has implications for the autophagy pathway. Indeed, AuNPs induce the accumulation of autophagy in cells; however, the formation of autophagy flux is blocked due to an impairment of lysosome degradation (125). Another research showed that induction of apoptosis in AuNP-treated breast cancer cells occurred through p53 and bax/bcl-2 activation (126).
4.3.3 Zinc oxide NPs
The lethal toxicity of zinc oxide NPs (ZnO NPs) on cells could be modified by NP stabilization. An experiment with the conjugation of ZnNPs with porphyrin (MTCP) yielded a cytotoxic effect of NPs in breast cancer cell lines through the calcium signaling pathway, which regulates lysosomal-dependent autophagy and apoptotic cell death (127).
4.3.4 Iron oxide NPs
The effect of photothermal therapy of iron oxide NPs (IONPs) in breast cancer cells was observed by inducing autophagy but not leading to apoptosis. The use of autophagy inhibitors and IONPs under laser therapy causes cell death in both MCF-7 and xenograft breast cancer models (128).
4.3.5 Copper oxide NPs
The cytotoxicity of using copper oxide NPs (CuONPs) in MCF3 breast cancer cells was observed via oxidative stress and autophagy induction. In these cells, autophagy is activated in order to survive the tumor cells. Therefore, the cotreatment of autophagy inhibitors and CuONPs could be a potent treatment against breast cancer cells (129).
4.3.6 Silica-based NPs
The use of nano-drug delivery in cancer cells has been demonstrated to improve biodistribution and increase the sensitivity of the drugs. For instance, the use of DOX-loaded mesoporous silica nanoparticle (MSN) in breast cancer treatment has shown an increased level of apoptosis, in comparison with DOX treatment alone in both in vitro and in vivo experiments. The intracellular mechanisms also confirmed the induction of the pro-death autophagy signaling pathway through inhibition of the AKT-mTOR-p70S6K signaling pathway (130). Another study has shown the cytotoxic effects of silica-based NPs (SiNPs) in breast cancer cell lines through the mitochondrial apoptotic pathway. The NPs with 5–15 nm could induce caspase-9 and caspase-3 activities (131).
4.3.7 Polymeric NPs
Synthetic pH-sensitive polymeric NPs have been used to increase chemical stability and specific biodistribution. For instance, the Bec-1 peptide, as an autophagy inducer, was engineered into the pH-sensitive polymers and then self-assembled with polyethylene glycol to form micellar-like NPs. This structure of NPs alters the acidic status of lysosomes and prevents autophagosome–lysosome fusion. Accumulation of autophagosomes leads to cell death, not tumor survival (132). In Table 4, we have summarized several studies looking at the implementation of an autophagy modulator with a nano-drug delivery system, some of which were also combined with chemotherapeutic agents. In addition, it is shown that NCs are able to modulate autophagy, simultaneously targeting cancer cells and delivering therapeutic agents.
5 The advantages and disadvantages of using nanomaterials in breast cancer treatment
NPs are an ideal drug delivery vehicle due to their nanosized structure, good biodistribution, and cost-saving properties (151), Furthermore, it is widely established that NCs aid in boosting the solubility, bioavailability, and stability of a variety of pharmacological drugs (152). Additionally, by improving cancer cell targeting, NPs could reduce the harmful effect of active cancer drugs. Finally, a safe and efficient agent for combination therapy involving autophagy regulation is one of the nano-drug delivery system advantages (153). Despite their small size, NPs can be easily and specifically delivered to the cells and absorbed by the tissue of interest. NPs may be used in conjugation with target drugs specific to cancer cells without disrupting normal cells’ structure or function. They have specific qualities, including high encapsulation efficiency, biocompatibility, reduced toxicity, and ease of manufacture, making them useful for medications (153). Moreover, NP-based therapy is useful for personalized medicine and biomarker identification. NPs are easily engineered to be pH or temperature sensitive in different situations to transport and release the drug to their specific targets. Despite all these advantages of NPs, not all of them enter the clinical phase because there are some problems like biological and technological difficulties in manufacturing NPs (154). The biological issues, including the route of administration (e.g., oral, intravenous, or skin injected), biological barriers, the toxicity of NPs, and their degradation are essential factors in NP efficacy. To overcome these challenges, some modifications are necessary to improve the NP application. For example, the use of low concentrations, which may decrease the toxicity effects, or the production of NPs with more biocompatible materials, such as chitosan, are beneficial (155). Technological challenges including the scale-up synthesis of NPs and in vitro or in vivo experimental models are not enough to directly use NPs in the clinical study due to little supporting data. In this way, using computational models or organs-on-chip can be a good solution alongside the laboratory results (154).
6 The future of NPs in breast cancer
NPs have a high potential to be used in the future treatments of breast cancer. They overcome the challenges of current treatments and combat drug resistance. They help increase the sensitivity of drugs (65, 156). In addition to therapy, nanomedicine may assist patients in some other aspects, such as diagnostics in the early stages and tumor imaging. Nanotechnology offers potential solutions to the problems that have made breast cancer so challenging to treat. These problems include the diversity of cancer and the rapid evolutionary changes in patients, the multiple pathways driving disease progression simultaneously, the emergence of tolerance, creating therapeutic cocktails, distant breast cancer metastasis, and the severity of side effects of therapies and very poor biodistribution of the injected medications in the body (72, 157). However, nanomedicine has some important challenges and controversies that should be resolved. The major issue is the toxicity of nanomaterials. For instance, ROS production leads to DNA damage and protein denaturation. By standard protocols, deep toxicological studies prior to the application of NPs for human therapy are suggested.
Among many nanomaterials in biomedicine, only a few of them have reached clinical trials and obtained FDA approval. The process by which a new drug passes all the preclinical phases and clinical trials to obtain the license for use in humans is estimated to be approximately 10–15 years (158). The preclinical phases are usually cellular testing, animal studies, toxicology control, safety, efficacy, and dose ranges of the new drugs (159). The clinical trials involve three phases, named I (testing dose and toxicity), II (safety and testing in a larger group), and III (randomized multicenter testing), that should be passed before introducing the new drugs to the market (158). Nanomedicine needs phase IV of post-marketing to consider the limitation or benefits of the application. In this regard, many studies are under investigation to obtain a license in order to enter clinical trials (160). According to clinical trials (www.ClinicalTrials.gov), there are currently 39 studies available that use different shapes and structures of NPs in breast cancer treatment, confirming the importance of using NPs in cancer treatment. Here, we summarize the 10 top studies in Table 5.
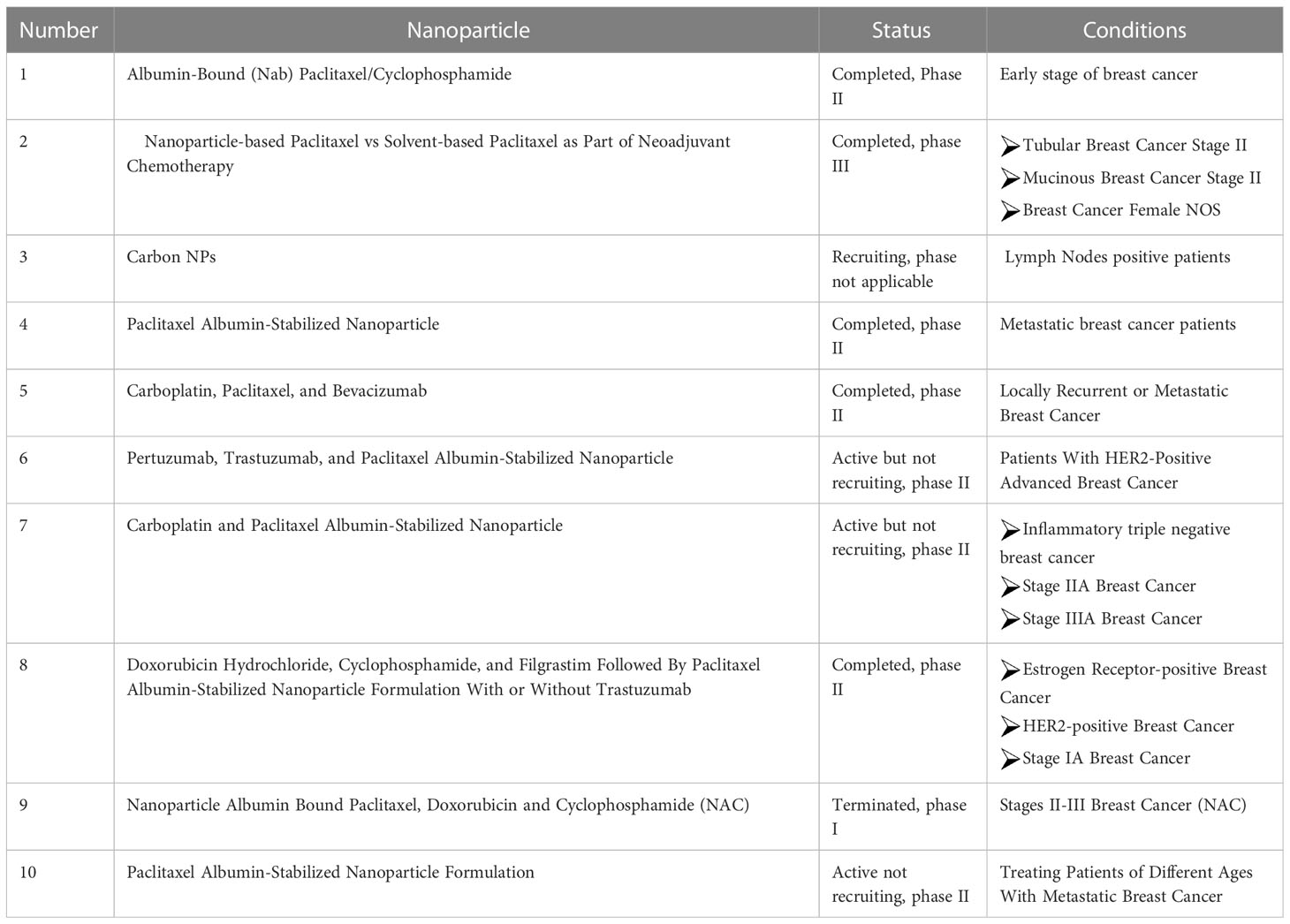
Table 5 Clinical trial of nanoparticles (NP)-based treatment strategies in breast cancer (https://clinicaltrials.gov)
7 Conclusion
NPs are an ideal drug delivery vehicle due to their nanosized structure, good biodistribution, and cost-saving properties. Additionally, by improving cancer cell targeting, NPs could reduce the harmful effect of active cancer drugs. It is challenging to modulate autophagy in breast cancer treatment utilizing NPs due to the different behaviors of autophagy in tumorigenicity. To deal with this problem, a large number of studies are recommended. The different effects of NPs on autophagy, induction, or inhibition, in different situations, lead to antitumor activity or an increase in cell death. In practice, the choice between inhibition and promotion of autophagy is controversial, as it may depend on the role of autophagy in tumor development. As long as autophagy exerts a positive effect on the treatment of certain cancers, strategies that promote autophagy remain desirable. However, when autophagy adversely affects cancer treatment, inhibition of autophagy is the appropriate strategy. Depending on the type of cancer, therapy should involve an appropriate treatment in combination with autophagy modulation. However, NPs have some disadvantages and cannot be used directly in clinical trials. To improve their lethal properties in cancerous cells, more modifications on NPs are needed, or cotreatment with targeted drugs is proposed. Despite some difficulties in the usage of NPs in medicine, their future application is promising and could be a new path for cancer treatment.
Author contributions
AG has contributed as co-author for sections on breast cancer and autophagy mechanisms. She also designs the figures of the manuscript. DS has written the section on nanocarriers. AY has contributed and written the nanoparticles sections. FB has written the autophagy section and prepared the abbreviation section. MR has been invited by the editorial board of the Frontiers in Oncology, proposed the topic, and is a corresponding author of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol (2022) 95:20211033. doi: 10.1259/bjr.20211033
2. Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the human development index (2008-2030): a population-based study. Lancet Oncol (2012) 13:790–801. doi: 10.1016/S1470-2045(12)70211-5
3. Niklaus NJ, Tokarchuk I, Zbinden M, Schläfli AM, Maycotte P, Tschan MP. The multifaceted functions of autophagy in breast cancer development and treatment. Cells (2021) 10(6):1447. doi: 10.3390/cells10061447
4. Gyanani V, Haley JC, Goswami R. Challenges of current anticancer treatment approaches with focus on liposomal drug delivery systems. Pharmaceuticals (2021) 14:835. doi: 10.3390/ph14090835
5. Marsh T, Debnath J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy (2020) 16:1164–5. doi: 10.1080/15548627.2020.1753001
6. Dyczynski M, Yu Y, Otrocka M, Parpal S, Braga T, Henley AB, et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to sunitinib. Cancer Lett (2018) 435:32–43. doi: 10.1016/j.canlet.2018.07.028
7. Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer (2017) 17:20–37. doi: 10.1038/nrc.2016.108
8. Cariolou M, Abar L, Aune D, Balducci K, Becerra-Tomás N, Greenwood DC, et al. Postdiagnosis recreational physical activity and breast cancer prognosis: global cancer update programme (CUP global) systematic literature review and meta-analysis. Int J Cancer (2023) 152:600–15. doi: 10.1002/ijc.34324
9. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast (2022) 66:15–23. doi: 10.1016/j.breast.2022.08.010
10. Zendehdel K. Cancer statistics in I.R. Iran in 2020. Basic Clin Cancer Res (2021) 12(4):159–65. doi: 10.18502/bccr.v12i4.7985
11. Macmahon B, Cole P, Brown J. Etiology of human breast cancer: a review. JNCI J Natl Cancer Inst (1973) 50:21–42. doi: 10.1093/jnci/50.1.21
12. Key T, Appleby P, Barnes I, Reeves G, Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst (2002) 94:606–16. doi: 10.1093/jnci/94.8.606
13. Shamshirian A, Heydari K, Shams Z, Aref AR, Shamshirian D, Tamtaji OR, et al. Breast cancer risk factors in Iran: a systematic review & meta-analysis. Horm Mol Biol Clin Investig (2020) 41(4). doi: 10.1515/hmbci-2020-0021
14. Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin (2017) 67(2):93–9. doi: 10.3322/caac.21388
15. Amin M. American Joint committee on cancer and American cancer society AJCC cancer staging manual. 8th Ed. Am Jt. Commun Cancer (2017)
16. Hortobagyi GN, Edge SB, Giuliano A. New and important changes in the TNM staging system for breast cancer. Am Soc Clin Oncol Educ book Am Soc Clin Oncol Annu Meet (2018) 38:457–67. doi: 10.1200/EDBK_201313
17. Giuliano AE, Connolly JL, Edge SB, Mittendorf EA, Rugo HS, Solin LJ, et al. Breast cancer-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin (2017) 67:290–303. doi: 10.3322/caac.21393
18. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet (London England) (2005) 365:1687–717. doi: 10.1016/S0140-6736(05)66544-0
19. Bernardo G, Palumbo R, Bernardo A, Villani G, Melazzini M, Poggi G, et al. Final results of a phase II study of weekly trastuzumab and vinorelbine in chemonaive patients with HER2-overexpressing metastatic breast cance. J Clin Oncol (2004) 22:731–1. doi: 10.1200/jco.2004.22.90140.731
20. Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther (2014) 10:506–11. doi: 10.4103/0973-1482.137927
21. Cufer T, Lamovec J, Bracko M, Lindtner J, Us-Krasovec M. Prognostic value of DNA ploidy in breast cancer stage I-II. Neoplasma (1997) 44:127–32.
22. Tong Y-K, Lo YMD. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta (2006) 363:187–96. doi: 10.1016/j.cccn.2005.05.048
23. Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem (2006) 52:345–51. doi: 10.1373/clinchem.2005.059832
24. Look MP, van Putten WLJ, Duffy MJ, Harbeck N, Christensen IJ, Thomssen C, et al. Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J Natl Cancer Inst (2002) 94:116–28. doi: 10.1093/jnci/94.2.116
26. Yin L, Duan J-J, Bian X-W, Yu S-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res (2020) 22:61. doi: 10.1186/s13058-020-01296-5
27. Pasha N, Turner NC. Understanding and overcoming tumor heterogeneity in metastatic breast cancer treatment. Nat Cancer (2021) 2:680–92. doi: 10.1038/s43018-021-00229-1
28. Cortes-Ciriano I, Lee S, Park W-Y, Kim T-M, Park PJ. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun (2017) 8:15180. doi: 10.1038/ncomms15180
29. Gote V, Nookala AR, Bolla PK, Pal D. Drug resistance in metastatic breast cancer: tumor targeted nanomedicine to the rescue. Int J Mol Sci (2021) 22:4673. doi: 10.3390/ijms22094673
30. Du B, Shim J. Targeting epithelial–mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules (2016) 21:965. doi: 10.3390/molecules21070965
31. Zeng X, Liu C, Yao J, Wan H, Wan G, Li Y, et al. Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol Res (2021) 163:105320. doi: 10.1016/j.phrs.2020.105320
32. Genovese I, Vezzani B, Danese A, Modesti L, Vitto VAM, Corazzi V, et al. Mitochondria as the decision makers for cancer cell fate: from signaling pathways to therapeutic strategies. Cell Calcium (2020) 92:102308. doi: 10.1016/j.ceca.2020.102308
33. East DA, Campanella M. Ca 2+ in quality control. Autophagy (2013) 9:1710–9. doi: 10.4161/auto.25367
34. Yan C, Li TS. Dual role of mitophagy in cancer drug resistance. Anticancer Res (2018) 38(2):617–21. doi: 10.21873/anticanres.12266
35. Lin P-W, Chu M-L, Liu H-S. Autophagy and metabolism. Kaohsiung J Med Sci (2021) 37:12–9. doi: 10.1002/kjm2.12299
36. Huang F, Wang B-R, Wang Y-G. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol (2018) 24:4643–51. doi: 10.3748/wjg.v24.i41.4643
37. Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer (2020) 19:12. doi: 10.1186/s12943-020-1138-4
38. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov (2017) 16:487–511. doi: 10.1038/nrd.2017.22
39. Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res (2014) 24:92–104. doi: 10.1038/cr.2013.153
40. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res (2014) 24:24–41. doi: 10.1038/cr.2013.168
41. Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell (2014) 55:238–52. doi: 10.1016/j.molcel.2014.05.021
42. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet (2009) 43:67–93. doi: 10.1146/annurev-genet-102808-114910
43. Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol (2011) 13:132–41. doi: 10.1038/ncb2152
44. Tamboli IY, Tien NT, Walter J. Sphingolipid storage impairs autophagic clearance of Alzheimer-associated proteins. Autophagy (2011) 7:645–6. doi: 10.4161/auto.7.6.15122
45. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxidants Redox Signal (2014) 20:460–73. doi: 10.1089/ars.2013.5371
46. Young ARJ, Chan EYW, Hu XW, Köchl R, SG C, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci (2006) 119:3888–900. doi: 10.1242/jcs.03172
47. Ganley IG. Autophagosome maturation and lysosomal fusion. Essays Biochem (2013) 55:65–78. doi: 10.1042/bse0550065
48. Nakamura S, Yoshimori T. New insights into autophagosome–lysosome fusion. J Cell Sci (2017) 130(7):1209–16. doi: 10.1242/jcs.196352
49. Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev (2009) 84:431–48. doi: 10.1111/j.1469-185X.2009.00082.x
50. Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci (2004) 117:4837–48. doi: 10.1242/jcs.01370
51. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with Endosomes/Lysosomes. Cell (2012) 151:1256–69. doi: 10.1016/j.cell.2012.11.001
52. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol (2005) 171:603–14. doi: 10.1083/jcb.200507002
53. Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ (2005) 12:1542–52. doi: 10.1038/sj.cdd.4401765
54. Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun (1988) 151:40–7. doi: 10.1016/0006-291X(88)90556-6
55. Liu B, Wen X, Cheng Y. Survival or death: disequilibrating the oncogenic and tumor suppressive autophagy in cancer. Cell Death Dis (2013) 4:e892. doi: 10.1038/cddis.2013.422
56. Pan H, Chen L, Xu Y, Han W, Lou F, Fei W, et al. Autophagy-associated immune responses and cancer immunotherapy. Oncotarget (2016) 7:21235–46. doi: 10.18632/oncotarget.6908
57. Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy (2018) 14:243–51. doi: 10.1080/15548627.2017.1402992
58. Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun (2018) 9:1944. doi: 10.1038/s41467-018-04070-6
59. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest (2003) 112:1809–20. doi: 10.1172/JCI20039
60. Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, et al. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy (2014) 10:2036–52. doi: 10.4161/auto.34398
61. Delaney JR, Patel CB, Bapat J, Jones CM, Ramos-Zapatero M, Ortell KK, et al. Autophagy gene haploinsufficiency drives chromosome instability, increases migration, and promotes early ovarian tumors. PloS Genet (2020) 16:e1008558. doi: 10.1371/journal.pgen.1008558
62. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature (1999) 402:672–6. doi: 10.1038/45257
63. Kharaziha P, Panaretakis T. Dynamics of Atg5-Atg12-Atg16L1 aggregation and deaggregation. Methods Enzymol (2017) 587:247–55. doi: 10.1016/bs.mie.2016.09.059
64. Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal (2011) 14:2201–14. doi: 10.1089/ars.2010.3482
65. Adibzadeh R, Golhin MS, Sari S, Mohammadpour H, Kheirbakhsh R, Muhammadnejad A, et al. Combination therapy with TiO2 nanoparticles and cisplatin enhances chemotherapy response in murine melanoma models. Clin Transl Oncol (2021) 23:738–49. doi: 10.1007/s12094-020-02463-y
66. Yoon J-H, Ahn S-G, Lee B-H, Jung S-H, Oh S-H. Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA–PKcs and PARP-1. Biochem Pharmacol (2012) 83:747–57. doi: 10.1016/j.bcp.2011.12.029
67. Zhang L, Yang A, Wang M, Liu W, Wang C, Xie X, et al. Enhanced autophagy reveals vulnerability of p-gp mediated epirubicin resistance in triple negative breast cancer cells. Apoptosis (2016) 21:473–88. doi: 10.1007/s10495-016-1214-9
68. Chang H, Zou Z. Targeting autophagy to overcome drug resistance: further developments. J Hematol Oncol (2020) 13:159. doi: 10.1186/s13045-020-01000-2
69. Chang Z, Shi G, Jin J, Guo H, Guo X, Luo F, et al. Dual PI3K/mTOR inhibitor NVP-BEZ235-induced apoptosis of hepatocellular carcinoma cell lines is enhanced by inhibitors of autophagy. Int J Mol Med (2013) 31:1449–56. doi: 10.3892/ijmm.2013.1351
70. Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, et al. Dual role of autophagy in hallmarks of cancer. Oncogene (2018) 37:1142–58. doi: 10.1038/s41388-017-0046-6
71. Jain K, Paranandi KS, Sridharan S, Basu A. Autophagy in breast cancer and its implications for therapy. Am J Cancer Res (2013) 3(3):251–65.
72. Cordani M, Somoza Á. Targeting autophagy using metallic nanoparticles: a promising strategy for cancer treatment. Cell Mol Life Sci (2019) 76:1215–42. doi: 10.1007/s00018-018-2973-y
73. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer (2017) 17:528–42. doi: 10.1038/nrc.2017.53
74. Fauzi YR, Nakahata S, Chilmi S, Ichikawa T, Nueangphuet P, Yamaguchi R, et al. Antitumor effects of chloroquine/hydroxychloroquine mediated by inhibition of the NF-κB signaling pathway through abrogation of autophagic p47 degradation in adult T-cell leukemia/lymphoma cells. PloS One (2021) 16:e0256320. doi: 10.1371/journal.pone.0256320
75. Feng J, Wang X, Ye X, Ares I, Lopez-Torres B, Martínez M, et al. Mitochondria as an important target of metformin: the mechanism of action, toxic and side effects, and new therapeutic applications. Pharmacol Res (2022) 177:106114. doi: 10.1016/j.phrs.2022.106114
76. Aydinlik S, Erkisa M, Cevatemre B, Sarimahmut M, Dere E, Ari F, et al. Enhanced cytotoxic activity of doxorubicin through the inhibition of autophagy in triple negative breast cancer cell line. Biochim Biophys Acta - Gen Subj (2017) 1861:49–57. doi: 10.1016/j.bbagen.2016.11.013
77. Guo B, Tam A, Santi SA, Parissenti AM. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in MCF-7 cells. BMC Cancer (2016) 16:762. doi: 10.1186/s12885-016-2790-3
78. Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy (2012) 8:1643–56. doi: 10.4161/auto.21654
79. Kang J-H, Yu B-Y, Youn D-S. Relationship of serum adiponectin and resistin levels with breast cancer risk. J Korean Med Sci (2007) 22:117. doi: 10.3346/jkms.2007.22.1.117
80. Kim TH, Kim HS, Kang YJ, Yoon S, Lee J, Choi WS, et al. Psammaplin a induces sirtuin 1-dependent autophagic cell death in doxorubicin-resistant MCF-7/adr human breast cancer cells and xenografts. Biochim Biophys Acta (2015) 1850:401–10. doi: 10.1016/j.bbagen.2014.11.007
81. Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat (2008) 112:389–403. doi: 10.1007/s10549-007-9873-4
82. Liu Z, He K, Ma Q, Yu Q, Liu C, Ndege I, et al. Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PloS One (2017) 12:e0177694. doi: 10.1371/journal.pone.0177694
83. Liu PF, Tsai KL, Hsu CJ, Tsai WL, Cheng JS, Chang HW, et al. Drug repurposing screening identifies tioconazole as an ATG4 inhibitor that suppresses autophagy and sensitizes cancer cells to chemotherapy. Theranostics (2018) 8:830–45. doi: 10.7150/thno.22012
84. Sheng Y, Sun B, Guo W-T, Zhang Y-H, Liu X, Xing Y, et al. 3-methyladenine induces cell death and its interaction with chemotherapeutic drugs is independent of autophagy. Biochem Biophys Res Commun (2013) 432:5–9. doi: 10.1016/j.bbrc.2013.01.106
85. Pasquier B, El-Ahmad Y, Filoche-Rommé B, Dureuil C, Fassy F, Abecassis PY, et al. Discovery of (2S)-8-[(3R)-3-methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: a novel potent and selective inhibitor of Vps34 for the treatment of solid tumors. J Med Chem (2015) 58:376–400. doi: 10.1021/jm5013352
86. Sharma N, Thomas S, Golden EB, Hofman FM, Chen TC, Petasis NA, et al. Inhibition of autophagy and induction of breast cancer cell death by mefloquine, an antimalarial agent. Cancer Lett (2012) 326:143–54. doi: 10.1016/j.canlet.2012.07.029
87. Guntuku L, Gangasani JK, Thummuri D, Borkar RM, Manavathi B, Ragampeta S, et al. IITZ-01, a novel potent lysosomotropic autophagy inhibitor, has single-agent antitumor efficacy in triple-negative breast cancer in vitro and in vivo. Oncogene (2019) 38:581–95. doi: 10.1038/s41388-018-0446-2
88. Yang W, Hosford SR, Traphagen NA, Shee K, Demidenko E, Liu S, et al. Autophagy promotes escape from phosphatidylinositol 3-kinase inhibition in estrogen receptor-positive breast cancer. FASEB J (2018) 32:1222–35. doi: 10.1096/fj.201700477R
89. Pelt J, Busatto S, Ferrari M, Thompson EA, Mody K, Wolfram J. Chloroquine and nanoparticle drug delivery: a promising combination. Pharmacol Ther (2018) 191:43–9. doi: 10.1016/j.pharmthera.2018.06.007
90. Raj R, Mongia P, Kumar Sahu S, Ram A. Nanocarriers based anticancer drugs: current scenario and future perceptions. Curr Drug Targets (2016) 17:206–28. doi: 10.2174/1389450116666150722141607
91. Nogueira E, Gomes AC, Preto A, Cavaco-Paulo A. Design of liposomal formulations for cell targeting. Colloids Surf B Biointerfaces (2015) 136:514–26. doi: 10.1016/j.colsurfb.2015.09.034
92. Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res (2006) 66:6732–40. doi: 10.1158/0008-5472.CAN-05-4199
93. Tanaka T, Decuzzi P, Cristofanilli M, Sakamoto JH, Tasciotti E, Robertson FM, et al. Nanotechnology for breast cancer therapy. BioMed Microdevices (2009) 11:49–63. doi: 10.1007/s10544-008-9209-0
94. Kumar L, Baldi A, Verma S, Utreja P. Exploring therapeutic potential of nanocarrier systems against breast cancer. Pharm Nanotechnol (2018) 6:94–110. doi: 10.2174/2211738506666180604101920
95. Jain NK, Gupta U. Application of dendrimer-drug complexation in the enhancement of drug solubility and bioavailability. Expert Opin Drug Metab Toxicol (2008) 4:1035–52. doi: 10.1517/17425255.4.8.1035
96. Condello M, Mancini G, Meschini S. The exploitation of liposomes in the inhibition of autophagy to defeat drug resistance. Front Pharmacol (2020) 66(13):6732–40. doi: 10.3389/fphar.2020.00787
97. Sahoo NG, Bao H, Pan Y, Pal M, Kakran M, Cheng HKF, et al. Functionalized carbon nanomaterials as nanocarriers for loading and delivery of a poorly water-soluble anticancer drug: a comparative study. Chem Commun (Camb) (2011) 47:5235–7. doi: 10.1039/c1cc00075f
98. Müller RH, Mäder K, Gohla S. Solid lipid nanoparticles (SLN) for controlled drug delivery - a review of the state of the art. Eur J Pharm Biopharm (2000) 50:161–77. doi: 10.1016/S0939-6411(00)00087-4
99. Calne RY, Lim S, Samaan A, Collier DSJ, Pollard SG, White DJG, et al. Rapamycin for immunosuppression in organ allografting. Lancet (1989) 334:227. doi: 10.1016/S0140-6736(89)90417-0
100. Wang S, Sun, Zhu, Du, Liu, Qian W-Y. pH-sensitive strontium carbonate nanoparticles as new anticancer vehicles for controlled etoposide release. Int J Nanomedicine (2012) 7:5781. doi: 10.2147/IJN.S34773
101. Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy (2010) 6:838–54. doi: 10.4161/auto.6.7.12113
102. Moghimi SM, Hunter AC. Poloxamers and poloxamines in nanoparticle engineering and experimental medicine. Trends Biotechnol (2000) 18:412–20. doi: 10.1016/S0167-7799(00)01485-2
103. Zhang X-G, Miao J, Dai Y-Q, Du Y-Z, Yuan H, Hu F-Q. Reversal activity of nanostructured lipid carriers loading cytotoxic drug in multi-drug resistant cancer cells. Int J Pharm (2008) 361:239–44. doi: 10.1016/j.ijpharm.2008.06.002
104. Singh Y, Meher JG, Raval K, Khan FA, Chaurasia M, Jain NK, et al. Nanoemulsion: concepts, development and applications in drug delivery. J Control Release (2017) 252:28–49. doi: 10.1016/j.jconrel.2017.03.008
105. Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Gold nanoparticles: interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomedicine (Lond) (2007) 2:681–93. doi: 10.2217/17435889.2.5.681
106. Chen H, Tan X, Fu Y, Dai H, Wang H, Zhao G, et al. The development of natural and designed protein nanocages for encapsulation and delivery of active compounds. Food Hydrocoll (2021) 121:107004. doi: 10.1016/j.foodhyd.2021.107004
107. Kim MW, Jeong HY, Kang SJ, Jeong IH, Choi MJ, You YM, et al. Anti-EGF receptor aptamer-guided Co-delivery of anti-cancer siRNAs and quantum dots for theranostics of triple-negative breast cancer. Theranostics (2019) 9:837–52. doi: 10.7150/thno.30228
108. Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arab J Chem (2019) 12:908–31. doi: 10.1016/j.arabjc.2017.05.011
109. Han S-M, Na Y-G, Lee H-S, Son G-H, Jeon S-H, Bang K-H, et al. Improvement of cellular uptake of hydrophilic molecule, calcein, formulated by liposome. J Pharm Investig (2018) 48:595–601. doi: 10.1007/s40005-017-0358-0
110. Jo DH, Kim JH, Lee TG, Kim JH. Size, surface charge, and shape determine therapeutic effects of nanoparticles on brain and retinal diseases. Nanomedicine Nanotechnology Biol Med (2015) 11:1603–11. doi: 10.1016/j.nano.2015.04.015
111. Liu Y, Hardie J, Zhang X, Rotello VM. Effects of engineered nanoparticles on the innate immune system. Semin Immunol (2017) 34:25–32. doi: 10.1016/j.smim.2017.09.011
112. Rahmati M, Amanpour S, Mohammadpour H. The role of nanoparticles in cancer therapy through apoptosis induction. In: Nanoparticle Drug Deliv. Syst. Cancer Treat. London: Jenny Stanford Publishing, CorporateTaylor & Francis Group (2020). p. 45–74.
113. Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, et al. Renal clearance of quantum dots. Nat Biotechnol (2007) 25:1165–70. doi: 10.1038/nbt1340
114. Su Y-L, Fang J-H, Liao C-Y, Lin C-T, Li Y-T, Hu S-H. Targeted mesoporous iron oxide nanoparticles-encapsulated perfluorohexane and a hydrophobic drug for deep tumor penetration and therapy. Theranostics (2015) 5:1233–48. doi: 10.7150/thno.12843
115. Kiio TM, Park S. Physical properties of nanoparticles do matter. J Pharm Investig (2021) 51:35–51. doi: 10.1007/s40005-020-00504-w
116. Meng R, Li K, Chen Z, Shi C. Multilayer coating of tetrandrine-loaded PLGA nanoparticles: effect of surface charges on cellular uptake rate and drug release profile. J Huazhong Univ Sci Technolog Med Sci (2016) 36:14–20. doi: 10.1007/s11596-016-1535-5
117. Anselmo AC, Zhang M, Kumar S, Vogus DR, Menegatti S, Helgeson ME, et al. Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano (2015) 9:3169–77. doi: 10.1021/acsnano.5b00147
118. Hartmann R, Weidenbach M, Neubauer M, Fery A, Parak WJ. Stiffness-dependent in vitro uptake and lysosomal acidification of colloidal particles. Angew Chem Int Ed Engl (2015) 54:1365–8. doi: 10.1002/anie.201409693
119. Guo P, Liu D, Subramanyam K, Wang B, Yang J, Huang J, et al. Nanoparticle elasticity directs tumor uptake. Nat Commun (2018) 9:130. doi: 10.1038/s41467-017-02588-9
120. Rahmati M, Ebrahim S, Hashemi S, Motamedi M, Moosavi MA. New insights on the role of autophagy in the pathogenesis and treatment of melanoma. Mol Biol Rep (2020) 47:9021–32. doi: 10.1007/s11033-020-05886-6
121. Mohammadinejad R, Moosavi MA, Tavakol S, Vardar DÖ, Hosseini A, Rahmati M, et al. Klionsky DJ %J a (2019) necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 15(1):4–33. doi: 10.1080/15548627.2018.1509171
122. Azimee S, Rahmati M, Fahimi H, Moosavi MA. TiO2 nanoparticles enhance the chemotherapeutic effects of 5-fluorouracil in human AGS gastric cancer cells via autophagy blockade. Life Sci (2020) 248:117466. doi: 10.1016/j.lfs.2020.117466
123. Buttacavoli M, Albanese NN, Di Cara G, Alduina R, Faleri C, Gallo M, et al. Anticancer activity of biogenerated silver nanoparticles: an integrated proteomic investigation. Oncotarget (2018) 9:9685–705. doi: 10.18632/oncotarget.23859
124. Swanner J, Fahrenholtz CD, Tenvooren I, Bernish BW, Sears JJ, Hooker A, et al. Silver nanoparticles selectively treat triple-negative breast cancer cells without affecting non-malignant breast epithelial cells in vitro and in vivo. FASEB BioAdvances (2019) 1:639–60. doi: 10.1096/fba.2019-00021
125. Ma X, Wu Y, Jin S, Tian Y, Zhang X, Zhao Y, et al. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano (2011) 5:8629–39. doi: 10.1021/nn202155y
126. Selim ME, Hendi AA. Gold nanoparticles induce apoptosis in MCF-7 human breast cancer cells. Asian Pacific J Cancer Prev (2012) 13:1617–20. doi: 10.7314/APJCP.2012.13.4.1617
127. Mozdoori N, Safarian S, Sheibani N. Augmentation of the cytotoxic effects of zinc oxide nanoparticles by MTCP conjugation: non-canonical apoptosis and autophagy induction in human adenocarcinoma breast cancer cell lines. Mater Sci Eng C Mater Biol Appl (2017) 78:949–59. doi: 10.1016/j.msec.2017.03.300
128. Ren X, Chen Y, Peng H, Fang X, Zhang X, Chen Q, et al. Blocking autophagic flux enhances iron oxide nanoparticle photothermal therapeutic efficiency in cancer treatment. ACS Appl Mater Interfaces (2018) 10:27701–11. doi: 10.1021/acsami.8b10167
129. Laha D, Pramanik A, Maity J, Mukherjee A, Pramanik P, Laskar A, et al. Interplay between autophagy and apoptosis mediated by copper oxide nanoparticles in human breast cancer cells MCF7. Biochim Biophys Acta - Gen Subj (2014) 1840:1–9. doi: 10.1016/j.bbagen.2013.08.011
130. Duo Y, Li Y, Chen C, Liu B, Wang X, Zeng X, et al. DOX-loaded pH-sensitive mesoporous silica nanoparticles coated with PDA and PEG induce pro-death autophagy in breast cancer. RSC Adv (2017) 7:39641–50. doi: 10.1039/C7RA05135B
131. Krętowski R, Jabłońska-Trypuć A, Cechowska-Pasko M. The effect of silica nanoparticles (SiNPs) on cytotoxicity, induction of oxidative stress and apoptosis in breast cancer cell lines. Int J Mol Sci (2023) 24:2037. doi: 10.3390/ijms24032037
132. Wang Y, Lin Y-X, Qiao Z-Y, An H-W, Qiao S-L, Wang L, et al. Self-assembled autophagy-inducing polymeric nanoparticles for breast cancer interference in-vivo. Adv Mater (2015) 27:2627–34. doi: 10.1002/adma.201405926
133. Sun R, Shen S, Zhang Y-J, Xu C-F, Cao Z-T, Wen L-P, et al. Nanoparticle-facilitated autophagy inhibition promotes the efficacy of chemotherapeutics against breast cancer stem cells. Biomaterials (2016) 103:44–55. doi: 10.1016/j.biomaterials.2016.06.038
134. Joshi P, Chakraborti S, Ramirez-Vick JE, Ansari ZA, Shanker V, Chakrabarti P, et al. The anticancer activity of chloroquine-gold nanoparticles against MCF-7 breast cancer cells. Colloids Surf B Biointerfaces (2012) 95:195–200. doi: 10.1016/j.colsurfb.2012.02.039
135. Feng Q, Yang X, Hao Y, Wang N, Feng X, Hou L, et al. Cancer cell membrane-biomimetic nanoplatform for enhanced sonodynamic therapy on breast cancer via autophagy regulation strategy. ACS Appl Mater Interfaces (2019) 11:32729–38. doi: 10.1021/acsami.9b10948
136. Tang S, Yin Q, Su J, Sun H, Meng Q, Chen Y, et al. Inhibition of metastasis and growth of breast cancer by pH-sensitive poly (β-amino ester) nanoparticles co-delivering two siRNA and paclitaxel. Biomaterials (2015) 48:1–15. doi: 10.1016/j.biomaterials.2015.01.049
137. Wu X, Wu Y, Wang Z, Liu L, Sun C, Chen Y, et al. A cascade-targeting nanocapsule for enhanced photothermal tumor therapy with aid of autophagy inhibition. Adv Healthc Mater (2018) 7:1800121. doi: 10.1002/adhm.201800121
138. Chiu CF, Fu RH, Hsu S, Yu YA, Yang SF, Tsao TC, et al. Delivery capacity and anticancer ability of the berberine-loaded gold nanoparticles to promote the apoptosis effect in breast cancer. Cancers (Basel) (2021) 13:5317. doi: 10.3390/cancers13215317
139. He D, Wu B, Du J, Li L, Zhao J. Synergistic inhibition of the growth of MDA−MB−231 cells in triple−negative breast cancer by salinomycin combined with 17−AAG and its mechanism. Oncol Lett (2022) 23:138. doi: 10.3892/ol.2022.13258
140. Markowska A, Kaysiewicz J, Markowska J, Huczyński A. Doxycycline, salinomycin, monensin and ivermectin repositioned as cancer drugs. Bioorg Med Chem Lett (2019) 29:1549–54. doi: 10.1016/j.bmcl.2019.04.045
141. Hu Y-J, Zhang J-Y, Luo Q, Xu J-R, Yan Y, Mu L-M, et al. Nanostructured dihydroartemisinin plus epirubicin liposomes enhance treatment efficacy of breast cancer by inducing autophagy and apoptosis. Nanomaterials (2018) 8:804. doi: 10.3390/nano8100804
142. Qiu L, Gao M, Xu Y. Enhanced combination therapy effect on paclitaxel-resistant carcinoma by chloroquine co-delivery via liposomes. Int J Nanomedicine (2015) 10:6615–32. doi: 10.2147/IJN.S91463
143. Zafar S, Akhter S, Ahmad I, Hafeez Z, Alam Rizvi MM, Jain GK, et al. Improved chemotherapeutic efficacy against resistant human breast cancer cells with co-delivery of docetaxel and thymoquinone by chitosan grafted lipid nanocapsules: formulation optimization, in vitro and in vivo studies. Colloids Surf B Biointerfaces (2020) 186:110603. doi: 10.1016/j.colsurfb.2019.110603
144. Jing M, Li Y, Wang M, Zhang H, Wei P, Zhou Y, et al. Photoresponsive PAMAM-assembled nanocarrier loaded with autophagy inhibitor for synergistic cancer therapy. Small (2021) 17:e2102295. doi: 10.1002/smll.202102295
145. Wang H, Zheng Y, Sun Q, Zhang Z, Zhao M, Peng C, et al. Ginsenosides emerging as both bifunctional drugs and nanocarriers for enhanced antitumor therapies. J Nanobiotechnology (2021) 19:322. doi: 10.1186/s12951-021-01062-5
146. Zajdel A, Wilczok A, Jelonek K, Kaps A, Musiał-Kulik M, Kasperczyk J. Cytotoxic effect of targeted biodegradable epothilone b and rapamycin co-loaded nanocarriers on breast cancer cells. J BioMed Mater Res A (2021) 109:1693–700. doi: 10.1002/jbm.a.37164
147. Alkhatib MH, Bawadud RS, Gashlan HM. Incorporation of docetaxel and thymoquinone in borage nanoemulsion potentiates their antineoplastic activity in breast cancer cells. Sci Rep (2020) 10:18124. doi: 10.1038/s41598-020-75017-5
148. Zhou X, Suo F, Haslinger K, Quax WJ. Artemisinin-type drugs in tumor cell death: mechanisms, combination treatment with biologics and nanoparticle delivery. Pharmaceutics (2022) 14:395. doi: 10.3390/pharmaceutics14020395
149. Stagni V, Kaminari A, Sideratou Z, Sakellis E, Vlahopoulos SA, Tsiourvas D. Targeting breast cancer stem-like cells using chloroquine encapsulated by a triphenylphosphonium-functionalized hyperbranched polymer. Int J Pharm (2020) 585:119465. doi: 10.1016/j.ijpharm.2020.119465
150. Wang J, Dang MN, Day ES. Inhibition of wnt signaling by Frizzled7 antibody-coated nanoshells sensitizes triple-negative breast cancer cells to the autophagy regulator chloroquine. Nano Res (2020) 13:1693–703. doi: 10.1007/s12274-020-2795-8
151. Nie S, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annu Rev BioMed Eng (2007) 9:257–88. doi: 10.1146/annurev.bioeng.9.060906.152025
152. Kalota A, Shetzline SE, Gewirtz AM. Progress in the development of nucleic acid therapeutics for cancer. Cancer Biol Ther (2004) 3:4–12. doi: 10.4161/cbt.3.1.517
153. Jain V, Kumar H, Anod HV, Chand P, Gupta NV, Dey S, et al. A review of nanotechnology-based approaches for breast cancer and triple-negative breast cancer. J Control Release (2020) 326:628–47. doi: 10.1016/j.jconrel.2020.07.003
154. Gavas S, Quazi S, Karpiński TM. Nanoparticles for cancer therapy: current progress and challenges. Nanoscale Res Lett (2021) 16:173. doi: 10.1186/s11671-021-03628-6
155. Awasthi R, Pant I T, Kulkarni G, Satiko Kikuchi I, de Jesus Andreoli Pinto T, Dua K, et al. Opportunities and challenges in nano-structure mediated drug delivery: where do we stand? Curr Nanomedicine (2016) 6:78–104. doi: 10.2174/2468187306666160808160330
156. Avitabile E, Bedognetti D, Ciofani G, Bianco A, Delogu LG. How can nanotechnology help the fight against breast cancer? Nanoscale (2018) 10:11719–31. doi: 10.1039/C8NR02796J
157. Anselmo AC, Mitragotri S. Nanoparticles in the clinic: an update. Bioeng Transl Med (2019) 4(3):e10143. doi: 10.1002/btm2.10143
158. Bobo D, Robinson KJ, Islam J, Thurecht KJ, Corrie SR. Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm Res (2016) 33:2373–87. doi: 10.1007/s11095-016-1958-5
159. Dobrovolskaia MA. Pre-clinical immunotoxicity studies of nanotechnology-formulated drugs: challenges, considerations and strategy. J Control Release (2015) 220:571–83. doi: 10.1016/j.jconrel.2015.08.056
Keywords: autophagy, breast cancer, drug resistance, nanomaterial, nanoparticles
Citation: Gharoonpour A, Simiyari D, Yousefzadeh A, Badragheh F and Rahmati M (2023) Autophagy modulation in breast cancer utilizing nanomaterials and nanoparticles. Front. Oncol. 13:1150492. doi: 10.3389/fonc.2023.1150492
Received: 24 January 2023; Accepted: 19 April 2023;
Published: 05 May 2023.
Edited by:
Marco Cordani, Faculty of Biology (UCM), SpainReviewed by:
Ilaria Genovese, CNLS_IIT (Istituto Italiano Tecnologia), ItalyChandra Sekhar Bhol, National Institute of Technology Rourkela, India
Md. Rizwanullah, Jamia Hamdard University, India
Kokkarachedu Varaprasad, San Sebastián University, Chile
Copyright © 2023 Gharoonpour, Simiyari, Yousefzadeh, Badragheh and Rahmati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marveh Rahmati, bV9yYWhtYXRpQHNpbmEudHVtcy5hYy5pcg==
 Azar Gharoonpour
Azar Gharoonpour Dorsa Simiyari
Dorsa Simiyari Ali Yousefzadeh
Ali Yousefzadeh Fatemeh Badragheh
Fatemeh Badragheh Marveh Rahmati
Marveh Rahmati