- 1School of Basic Medical Sciences & The First Affiliated Hospital, Zhengzhou University, Zhengzhou, Henan, China
- 2Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu, China
- 3Department of Medical Oncology, National Cancer Center, National Clinical Research Center for Cancer, Cancer Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 4BGI College, Zhengzhou University, Zhengzhou, Henan, China
- 5Henan Institute of Medical and Pharmaceutical Sciences & Henan Key Medical Laboratory of Tumor Molecular Biomarkers, Zhengzhou University, Zhengzhou, Henan, China
- 6Department of Clinical Laboratory in the First Affiliated Hospital & Key Clinical Laboratory of Henan Province, Zhengzhou University, Zhengzhou, Henan, China
- 7Department of Clinical Laboratory, Jiangsu Province Hospital of Chinese Medicine, Nanjing, Jiangsu, China
Purpose: Previous studies have shown that DNA methylation in peripheral blood may be associated with breast cancer (BC). To explore the association between the methylation level of the Cathepsin Z (CTSZ) gene in peripheral blood and BC, we conducted a case–control study in the Chinese population.
Methods: Peripheral blood samples were collected from 567 BC cases, 635 healthy controls, and 303 benign breast disease (BBD) cases. DNA extraction and bisulfite-specific PCR amplification were performed for all samples. The methylation levels of seven sites of the CTSZ gene were quantitatively determined by Mass spectrometry. The odds ratios (ORs) of CpG sites were evaluated for BC risk using per 10% reduction and quartiles analyses by logistic regression.
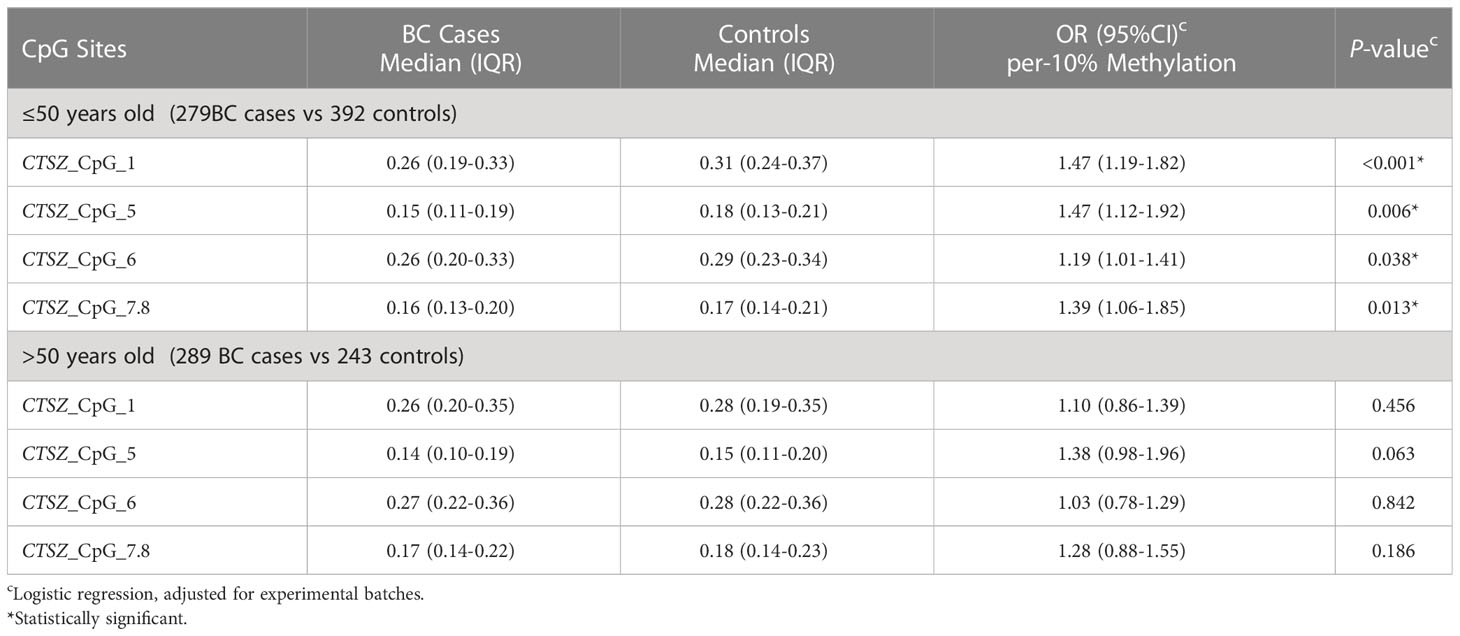
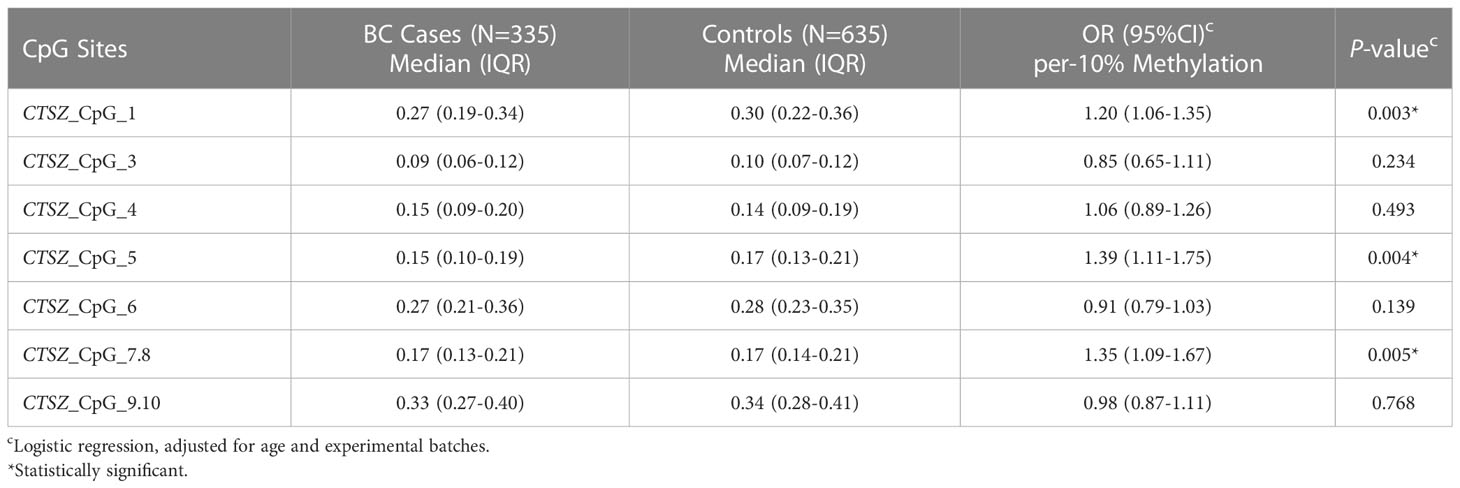
Results: Our analysis showed that five out of the seven CpG sites exhibited significant associations with hypomethylation of CTSZ and BC, compared to healthy controls. The highest OR was for Q2 of CTSZ_CpG_1 (OR: 1.62, P=0.006), particularly for early-stage breast cancer in the case of per 10% reduction of CTSZ_CpG_1 (OR: 1.20, P=0.003). We also found that per 10% reduction of CTSZ_CpG_5 (OR: 1.39, P=0.004) and CTSZ_CpG_7,8 (OR: 1.35, P=0.005) were associated with increased BC risk. Our study also revealed that four out of seven CpG sites were linked to increased BC risk in women under 50 years of age, compared to healthy controls. The highest OR was for per 10% reduction of CTSZ_CpG_1 (OR: 1.47, P<0.001). Additionally, we found that BC exhibited lower methylation levels than BBD at CTSZ_CpG_4 (OR for Q1: 2.18, P<0.001) and CTSZ_CpG_7,8 (OR for Q1: 2.01, P=0.001). Furthermore, we observed a correlation between methylation levels and tumor stage, ER, and HER2 status in BC patients.
Conclusion: Overall, our findings suggest that altered CTSZ methylation levels in peripheral blood may be associated with breast cancer, particularly in young women, and may serve as a potential biomarker for early-stage BC.
1 Introduction
In 2020, Global cancer statistics reported that the leading cause of disability and mortality among women worldwide is breast cancer (BC) (1). As early as 2018, the global cancer statistics showed that BC accounts for 15% of all types of female cancer deaths (2). BC is a complex and heterogeneous disease. Previous research has suggested there is a significant correlation between the incidence of BC and race and ethnicity, which is also normally associated with a range of risk factors including exposure to hormones, diet, and lifestyles (3). Despite the advance in the prevention and treatment that can improve the survival rates for BC patients, there is still a lack of effective early diagnosis methods (4). One study demonstrated that early detection of breast cancer could reduce overall mortality by 37% (5). Recently, a large body of compelling evidence indicates that the potential of epigenetic modifications for the prevention, diagnosis, and treatments of BC has an important impact on the development and progression (6).
Epigenetics refers to changes in gene expression levels that are not based on changes in gene sequence. Epigenetics includes DNA methylation, chromatin remodeling, histone modifications, and non-coding RNA, which can produce heritable variations without changing the DNA sequence (7). DNA methylation is a covalent modification. DNA methylation occurs exclusively on cytosine nucleotides and almost always in the context of CpG. As an important epigenetic mechanism, DNA methylation usually refers to the transfer of methyl groups to the C5 position of the cytosine to form a 5-methylcytosine under the action of DNA methyltransferase (DNMT). 5-methylcytosine, the methylated form of DNA base cytosine, is the main epigenetic marker in mammalian nuclear DNA and may be involved in gene transcription regulation, posttranscriptional modification to affect the maturation, stability, and translation of mRNA molecules. It is also one of the indispensable epigenetic adaptations that affect cell proliferation, apoptosis, differentiation, cell cycle, and transformation (8). The transcriptional start sites of many genes encoding tumor suppressors, such as retinoblastoma-associated protein 1 (RB1), MLH1, p16 and BRCA1, among others, lie within CGIs (CpG islands). In somatic cells, gene body methylation is a major cause of cancer gene mutations in tumor suppressor genes. In general, the tumor genome is hypomethylated, yet the potential role of methylation of CpG-poor promoters, enhancers, insulators, repetitive elements and gene bodies in cancer is almost completely unknown (9). With regards to tissue samples, hypermethylation in the promoter regions of tumor suppressor genes often leads to transcriptional repression and decreased gene expression (10, 11). However, genome-wide DNA hypomethylation may promote tumor formation by increasing chromosomal instability. Therefore, the identification of methylation changes in DNA from peripheral blood and their association with breast cancer have been of great interest for early disease diagnosis and treatment (12–14).
Cathepsin Z (CTSZ) is a differentiated member of the cathepsins. CTSZ has only mono- and dipeptidyl carboxypeptidase activity and being responsible for cleavage. CTSZ also activates some molecules, such as the integrin called lymphocyte function associated antigen (LFA-1), the focal adhesion kinase (FAK), the SRC kinase, and others. CTSZ is expressed in several primary tumor types, such as PCa (prostate cancer), colorectal, gastric, liver, melanoma, and pancreatic neuroendocrine tumors (15). In some of these studies, CTSZ was found in tumor-associated macrophages. It indicated that this protease played a role in the antitumor immune response (16). So far, there have been no studies about the correlation between CTSZ methylation and breast cancer. Thus, we hypothesized that methylation level on the site of CTSZ may effect breast cancer. It has been reported that DNA methylation from tissue or circulating blood tumor DNA may be a promising biomarker and could predicted and distinguished different subtypes of BC (17). To provide evidence for the selection of early markers for breast cancer, we explored the methylation levels between the CTSZ gene and breast cancer among Chinese women through a case-control design in this study.
2 Materials and methods
2.1 Study populations
This study was approved by the ethics committees of Nanjing Medical University, the Chinese Academy of Medical Sciences and the First Affiliated hospital of Zhengzhou University. The written informed consent was obtained from each recruited participants before the peripheral blood samples collection. All the individuals were Chinese women. BC was confirmed by postoperative pathological analyses in each case.
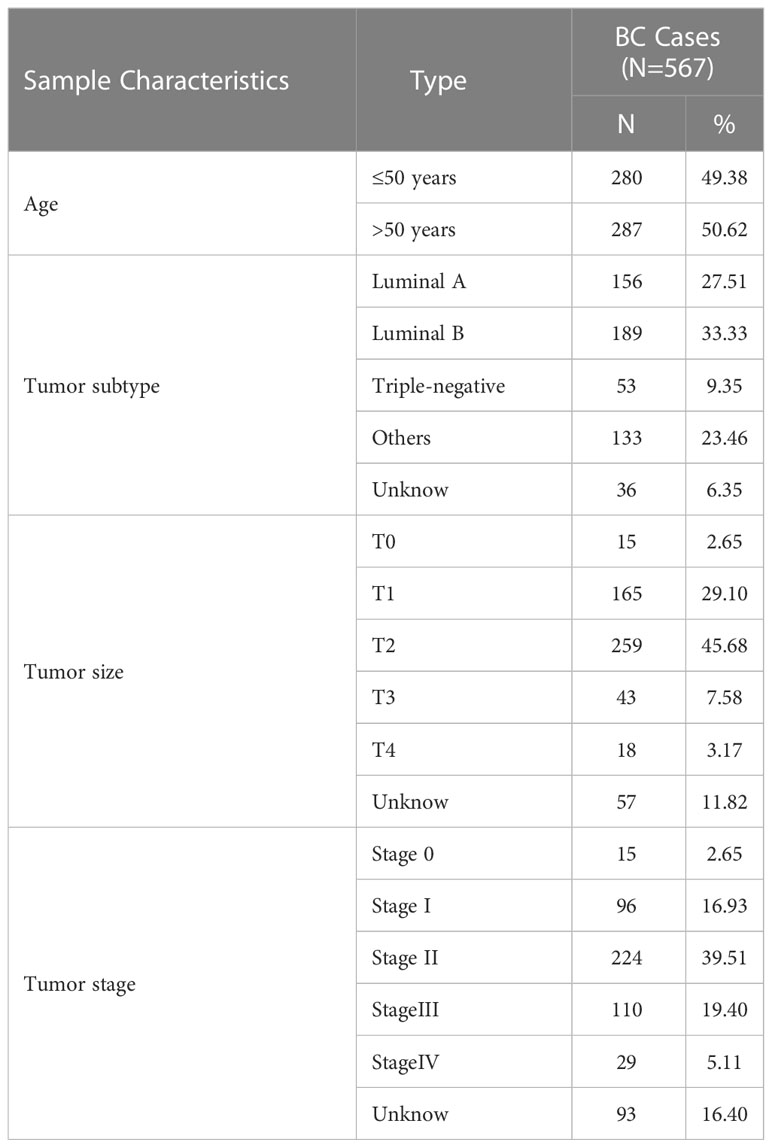
In this study, we collected 1505 whole blood samples, all from peripheral blood. 567 BC patients with the mean age of 50.5 years old (range: 22.0-85.0 years old) were from the Cancer Hospital of the Chinese Academy of Medical Sciences and the First Affiliated Hospital of Zhengzhou University. The criteria for breast cancer patients were those who had been diagnosed with cancer for the first time without any treatment. A total of 635 healthy female control group were enrolled by the Health Center in Nanjing Hospital of Chinese Medicine. The mean age was 47.5 years old (range: 24.0-94.0 years old). The physical examination report was normal, no tumor history, no family history of tumor disease. In addition, a total of 303 patients with BBD (Benign breast disease) were collected from the Cancer Hospital of the Chinese Academy of Medical Sciences and the First Affiliated Hospital of Zhengzhou University. The mean age was 48.0 years old (range: 24.0-78.0 years old). The criteria for the recruitment of BBD were women who had two or more biopsies within six months and the results were benign and no tumor history. Additional clinical characteristics information is described in detail in Table 1.
2.2 Sample collecting and processing
Peripheral blood was collected using Ethylene Diamine Tetraacetic Acid (EDTA) tubes and stored at −80°C. Whole-genome DNA was extracted from the whole blood sample according to the instructions of the DNA extraction kit (TANTICA, China). The above-described extracted DNA was treated with bisulfite using the EZ-96 DNA Methylation Gold Kit (Zymo Research, USA) kit according to the above instructions.
2.3 MALDI-TOF mass spectrometry
Agena matrix-assisted laser desorption/ionization (MALDI)- time-of-flight (TOF) mass spectrometry (Agena Bioscience, San Diego, California, United States) described by Yang et al. was used to determine the levels of DNA methylation semiquantitatively (18, 19). In short, the bisulfite-converted DNA was amplified by bisulfite-specific primers (Figure 1).
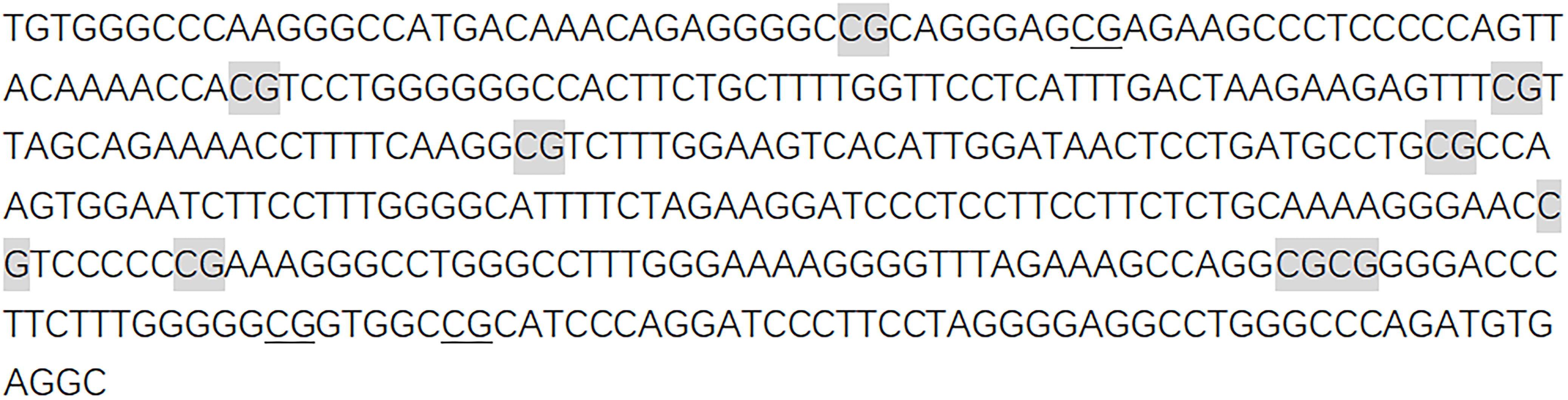
Figure 1 Sequences of CTSZ amplicon for MassARRAY methylation analysis. The sequence of the CTSZ amplicon examined by the EpiTyper assay. The EpiTyper assay determined the methylation levels of 12 CpGs in this amplicon, and yielded 7 distinguishable peaks. The 9 measurable CpG sites are highlighted, and the undetectable CpG sites are underlined.
Forward primer: aggaagagagTGTGGGTTTAAGGGTTATGATAAAT, reverse primer: cagtaatacgactcactatagggagaaggctACCTCACATCTAAACCCAAACCT. Upper case letters represent specific regions of the sequence, while lower case letters are used for non-specific tags. Neither the primers nor the amplicons were overlapped with any known SNPs. Most of the mass peaks detected by this method have only one CpG site and a few contain two. For example, CTSZ_CpG_7 and CTSZ_CpG_8 after the EpiTyper treatment are located in the same fragment, and thus the mass peak shows the average methylation level of CTSZ_CpG_7 and CTSZ_CpG_8 (expressed as CTSZ_CpG_7,8 in the manuscript). This is applied to CTSZ_CpG_9,10 as well. The polymerase chain reaction (PCR) products were processed according to the standard protocol of Agena EpiTyper Assay. Then, it is cleaned with resin and dispensed to 384 SpectroCHIP via Nanodispenser. MassARRAY system was used for data collection and the EpiTYPER v1.2 software was used for calculation. In all the processing and analyses, BC cases and controls were handled in parallel.
2.4 Statistical analyses
SPSS version 26.0 was used for all the statistical analysis. Age and batche adjusted logistic regression was used to assess the association between BC risk and the methylation levels of CTSZ, as well as between BC and BBD cases. The odds ratios (ORs) with 95% confidence interval (CI) of CpG sites were evaluated for BC risk using per 10% reduction and quartiles analyses by logistic regression. OR>1 indicates that CTSZ methylation level is a risk factor for BC. Tests for linear trend across quartiles of methylation levels of each CpG site in CTSZ were conducted using the logistic regression model. The group with the highest methylation level (Q4) was used as the reference group, and the other three groups were set as dummy variables to enter the regression model. The signifcant trend indicated that the relative risk of breast cancer showed a rising trend as the methylation level decreased.The interquartile analysis was performed by calculating the quartiles of methylation levels at each site in BC case and controls. We used Kruskal–Wallis test and Mann–Whitney U test to analyze the significant correlation between CTSZ methylation level and the different clinical characteristics. P <0.05 was considered statistical significance, it indicates that there is a correlation between two groups. All statistical tests were two-sided.
3 Results
3.1 Association between hypomethylation of CTSZ in peripheral blood and BC
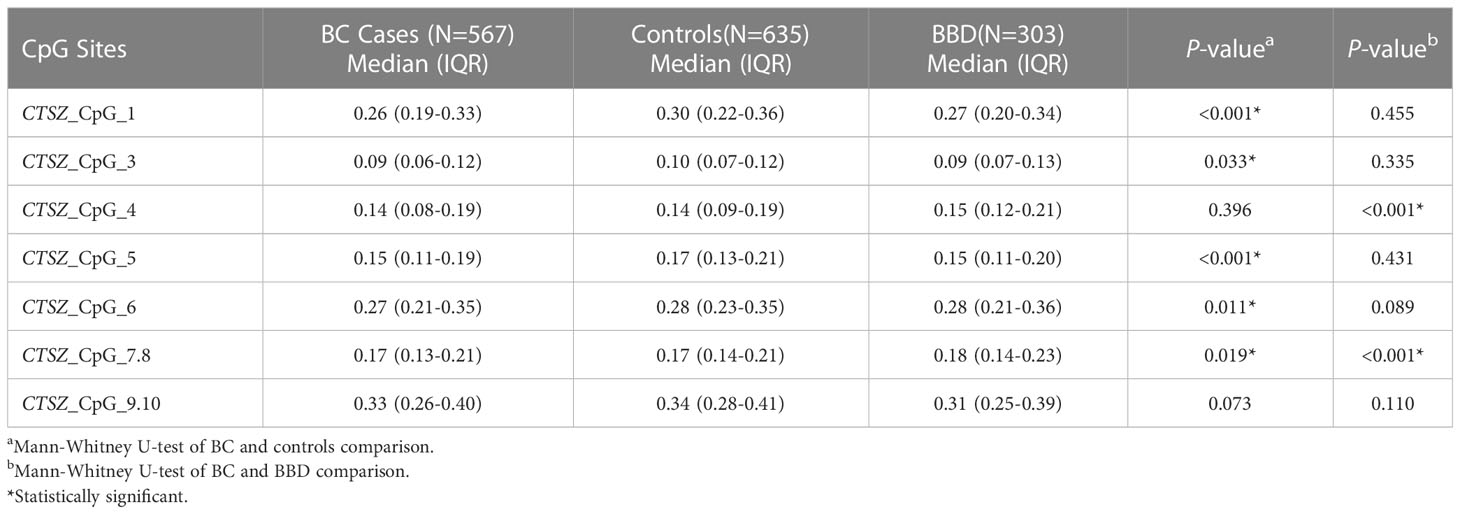
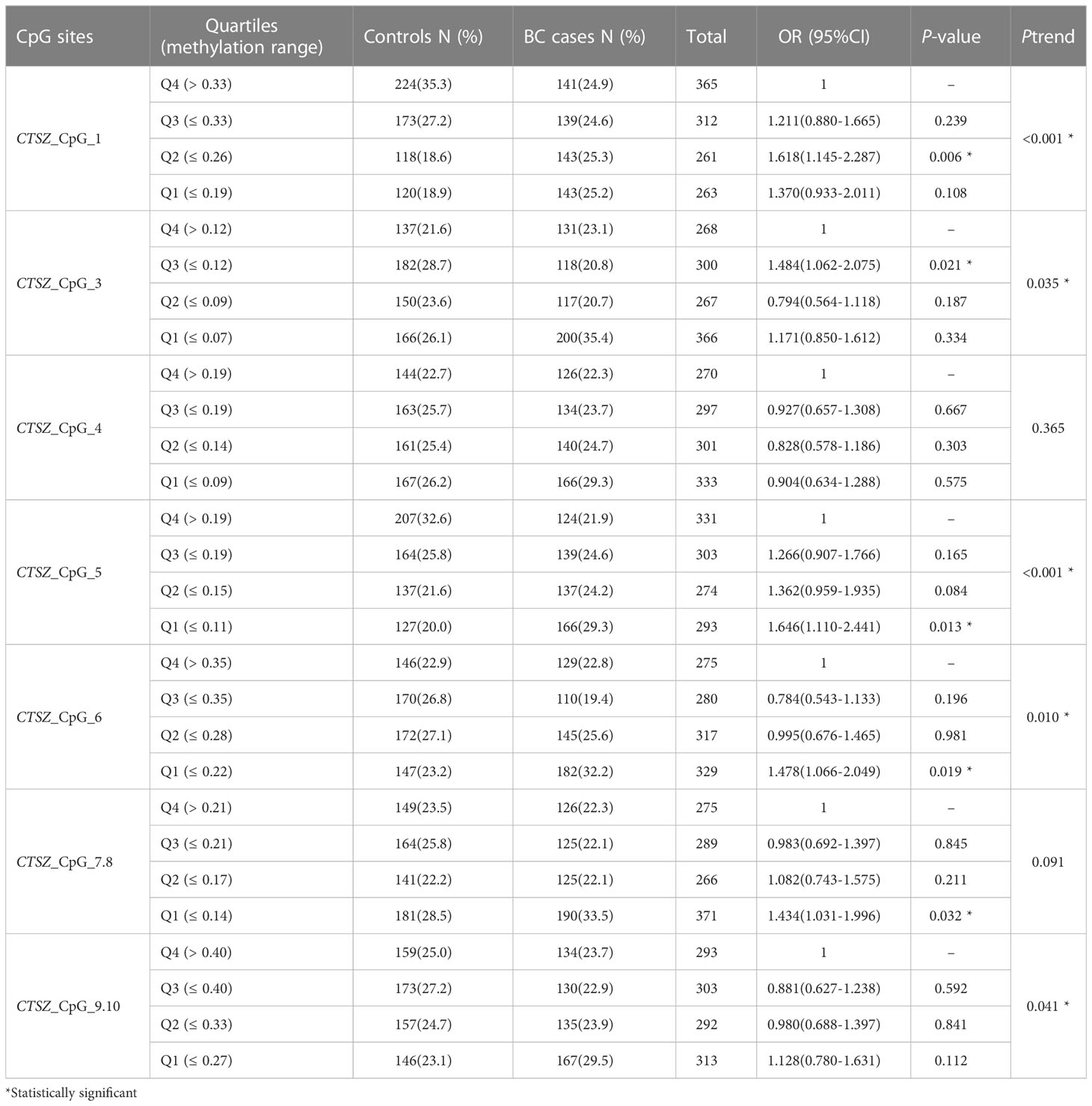
To investigate the association between CTSZ methylation and BC, a case-control study with 567 BC cases and 635 age matched control groups was conducted. As shown in Figure 2, there was a significant association between BC and controls on the site of CTSZ_CpG_1, CTSZ_CpG_3, CTSZ_CpG_5, CTSZ_CpG_6 and CTSZ_CpG_7,8. Meanwhile, BC had lower methylation level compared to controls (Table 2). Interquartile analyses were also performed to assess the ORs of methylation levels in all the CTSZ CpG sites to the risk of BC. Comparing to the highest methylation level (Q4), CTSZ_CpG_1, CTSZ_CpG_3, CTSZ_CpG_5 CTSZ_CpG_6 and CTSZ_CpG_7,8 showed significant association between hypomethylation of CTSZ and BC compared to controls with the highest OR for Q1 of CTSZ_CpG_5 (OR:1.646, P=0.013, Table 3). The effects of methylation in CTSZ_CpG_1, CTSZ_CpG_3, CTSZ_CpG_5 and CTSZ_CpG_6 were also enhanced along with lower interquartiles (Ptrend for CTSZ_CpG_1<0.001, Ptrend for CTSZ_CpG_3 = 0.035, Ptrend for CTSZ_CpG_5 < 0.001, Ptrend for CTSZ_CpG_6 = 0.010). In addition, a significant result was found at CTSZ_CpG_7,8 when comparing Q1 and Q4(OR=1.434, 95%CI:1.031-1.996, P=0.032), but no significant Ptrend existed. Although we did not find a correlation between CTSZ_CpG_9,10 and BC, we did find that OR tended to increase as the interquartile decrease (Ptrend for CTSZ_CpG_9,10 = 0.041; Table 3).
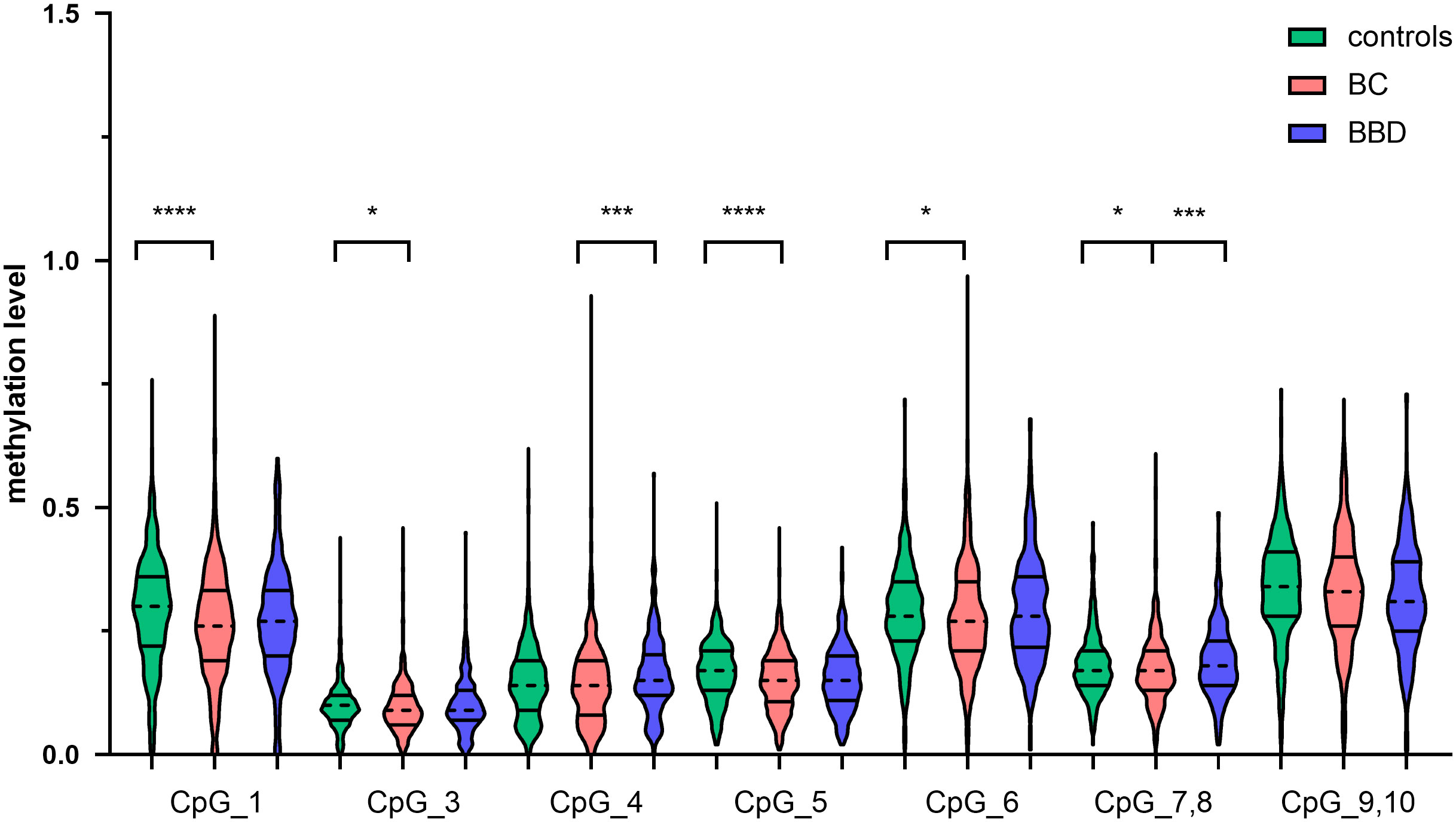
Figure 2 The CTSZ methylation levels of BC, BBD and controls on each site and their correlation. DNA extraction and bisulfite-specific PCR amplification were performed for 567 BC cases, 303 BBD cases and 635 controls. The methylation levels of seven sites of the CTSZ gene were quantitatively determined by Mass spectrometry. Mann-Whitney U test was used to analyze the methylation levels (median±IQR) between two groups. *P<0.05, ***P<0.001, ****P<0.0001.
3.2 The methylation differences of CTSZ between BC and BBD
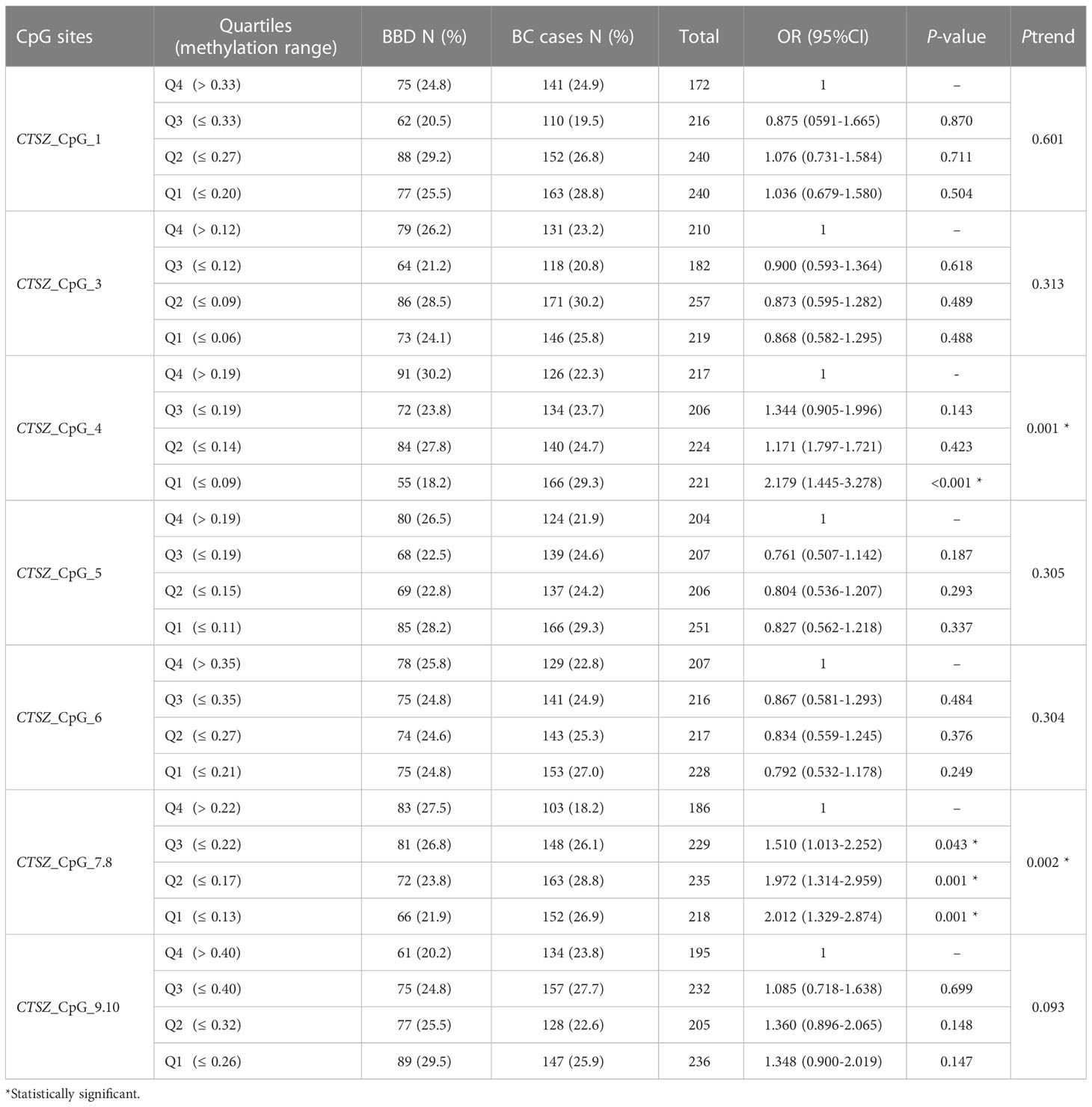
To evaluate the methylation differences of CTSZ between BC and BBD. We conducted a case-control study, which had 567 BC cases and 303 age matched BBD cases. As shown in Figure 2, there was a significant association between BC and BBD on the sites of CTSZ_CpG_4 and CTSZ_CpG_7,8. We found a significant association between hypomethylation of CTSZ and BC compared to BBD (Table 2). However, we further carried out an interquartile analysis, we found that BC had lower methylation level compared to BBD on the site of CTSZ_CpG_4 (OR(95%CI) for Q1 of CTSZ_CpG_4: 2.179 (1.445-3.278), P<0.001) and CTSZ_CpG_7,8 (OR (95%CI) for Q1 of CTSZ_CpG_7,8: 2.012 (1.329-2.874), P=0.001, Table 4). Moreover, with the lower interquartile of methylation levels, the higher risk of BC on site of CTSZ_CpG_4, CTSZ_CpG_7,8 (Ptrend for CTSZ_CpG_4 = 0.001, Ptrend for CTSZ_CpG_5 = 0.002) were be found.
3.3 Association between CTSZ methylation and BC by age
Because age plays an important role in BC risk, we calculated the association between BC and methylation levels by age (20). Here, we stratified the subjects by the age of 50 years. Then we conducted interaction analysis between CTSZ methylation and age, and found that there were significant difference on site of CTSZ_CpG_1, CTSZ_CpG_5, CTSZ_CpG_6, CTSZ_CpG_7,8. In the group of under the age of 50 (including 50 years), there were 279 BC cases and 392 controls. We conducted a binary logistic regression analysis and controlled for experimental batches, we found that hypomethylation of CTSZ_CpG_1, CTSZ_CpG_5, CTSZ_CpG_6, CTSZ_CpG_7,8 was associated with BC, while the highest OR for per 10% reduction of CTSZ_CpG_1 was 1.47(95%CI:1.19-1.82, P<0.001, Table 5). In the contrast, in the group of age>50 including 289 BC cases and 243 controls, there was none of the CpG sites significantly correlated with BC risk.
3.4 Association between CTSZ methylation levels and BC at early stage
To calculate the association between methylation levels and BC at early stage (stage0-stageII), we conducted a binary logistic regression analysis in a case-control study, which had 335 BC cases and 635 age matched controls. When adjusted for age and experimental batches, there was a significant association between hypomethylation of CTSZ and BC cases compared to controls for per 10% reduction of CTSZ_CpG_1 (OR:1.20; P=0.003); CTSZ_CpG_5 (OR:1.39; P=0.004); CTSZ_CpG_7,8 (OR:1.35; P=0.005) in Table 6. Meanwhile, we further carried out an interquartile analyses and found that there were association between BC and CTSZ methylation on the site of CTSZ_CpG_1, CTSZ_CpG_5 and CTSZ_CpG_7,8 where the highest OR of CTSZ_CpG_5 was 1.815(95%CI:1.217-2.707, P=0.003, Table 7). Moreover, with the lower interquartile of methylation levels, the higher risk of BC on site of CTSZ_CpG_1, CTSZ_CpG_3, CTSZ_CpG_5, CTSZ_CpG_7,8 were found (Ptrend for CTSZ_CpG_1<0.001, Ptrend for CTSZ_CpG_3 = 0.010, Ptrend for CTSZ_CpG_5 < 0.001, Ptrend for CTSZ_CpG_7,8 = 0.045; Table 7).
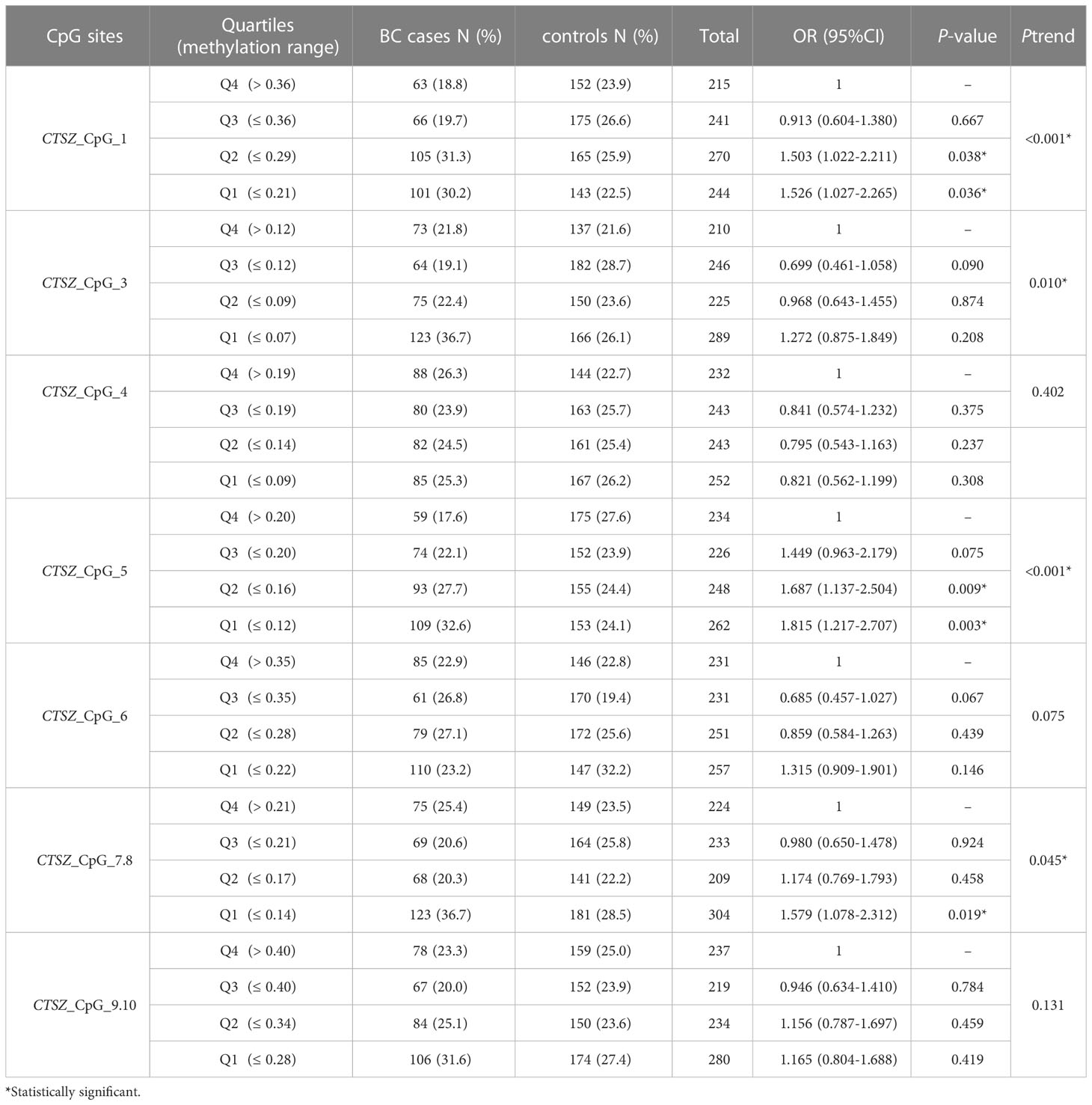
Table 7 Inter-quartile analysis for CTSZ methylation level between BC cases and controls at early stage.
3.5 CTSZ methylation levels and the clinical characteristics of BC patients
The relationship between CTSZ methylation level and the clinical characteristics of BC patients was investigated in 567 BC cases. Patients with higher tumor stage had lower methylation levels in CTSZ_CpG_4 (P<0.05, Table 8). Meanwhile, the negative status of ER was correlated with lower methylation levels at CTSZ_CpG_7,8 (P=0.028) while the hypermethylation level at CTSZ_CpG_7,8 was related to the HER2 positive status(P=0.020). However, the other six sites did not show this trend. In addition, our results showed no correlation of methylation level of CTSZ with tumor size, tumor subtype and the level of Ki67 (Table 8).
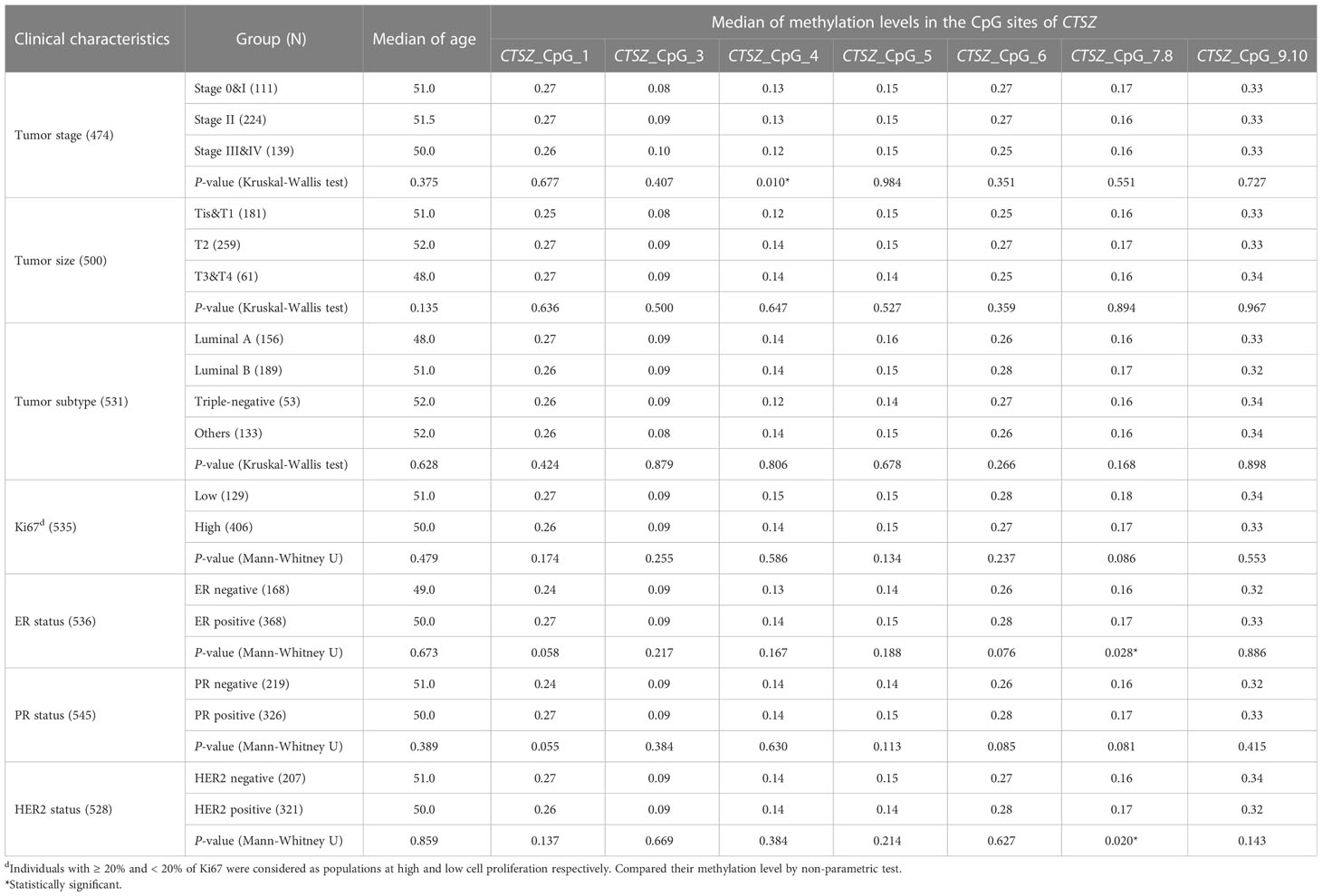
Table 8 The association between CTSZ methylation level and the clinical characteristics of BC patients.
4 Discussion
Immune cells, such as monocytes, macrophages, and dendritic cells regularly express CTSZ (16). According to the above, we found a negative correlation between CTSZ methylation and BC in whole blood with more than 1500 subjects. In our study, four out of seven CpG sites showed significant correlation between BC and controls where BC cases had lower methylation level compared to controls in all sites. Case–control studies concluded that such genomic hypomethylation continuum can be evident at blood DNA level and may identify women at high-risk of BC (21). Some retrospective case–control studies also reported that breast cancer patients have lower global methylation levels compared to control groups in blood DNA (22). In conclusion, our results are consistent with those previous studies. Although there are no reports about the association between BC and BBD, it is worth doing some research. In our study, we found that there was a significant correlation between BC and BBD cases in the two out of seven CpG sites, meanwhile, BC cases had lower methylation than BBD cases in the six out of seven sites. Therefore, the methylation level of CTSZ in BBD cases is a potential indicator to assess the risk for BC. It can also be used as a reference to distinguish benign and malignant breast nodules.
Aging is a natural and irreversible process in human life. To date, it is clear that epigenetic changes, particularly DNA methylation, are associated with the aging process (23). Prospective studies reported that DNA methylation may serve as a measure of aging because of its strong association with age (24). In our study, we not only found that hypomethylation was associated with BC by age, but also validated the correlation between BC and methylation levels was younger than 50 years. Meanwhile, we found that BC cases had lower methylation levels compared to the control groups in each site. The mean age and menopause for Asian women is about 49 years old, and perimenopause refers to 3 to 4 years before menopause, which is 45-49 years old (25). Since estrogen levels decrease with age in Chinese women, we divided the age of the subjects into 50 years. Age is an important factor in alteration of DNA methylation levels (26). However, there have been rarely reports on the correlation between CTSZ methylation levels and age to date. According to the above, we inferred that CTSZ methylation levels in women who have breast cancer and younger than 50 (mostly premenopausal) may be related to age.
We further analyzed the blood-based CTSZ methylation and the clinical characteristics in 567 BC cases and 635 controls. The available clinical features of our study included tumor stage, tumor size, tumor subtype, Ki67 levels, estrogen receptor (ER) status, progesterone receptor (PR) status and human epidermal growth factor receptor 2 (HER2) status. We found patients with higher tumor stage had lower levels of methylation. This suggested that the methylation levels of CTSZ may be a means to assess early BC. In addition, we also found an association between ER status, HER2 status and BC on CTSZ_CpG_7,8 sites. Previous study have found that demethylated state of several matrix metalloproteinase promoter regions in tumors is associated with low expression of HER2 (27). In our study, CTSZ methylation was also significantly lower in HER2 negative BC cases. ER negative also had lower methylation levels compared to ER positive in BC cases. We propose that the expression of HER2 and ER status may influence methylation levels of CTSZ. In previous studies, HER2 positive BC has been associated with a higher incidence of DNA methylation in several genes, including PGR (which codes for PR), HSD17B4 (28). Alternatively, HER2 negative BC was associated with reduced methylation of PGR and HSD17B4. Our findings suggested that aberrant methylation of CTSZ in blood might be an important predictor of BC development and might be a biomarker for the prognosis of BC. Thus, we can hypothesize that there might be a significant association between CTSZ methylation and BC in younger women with higher level of estrogen and HER2 positive BC carriers.
5 Conclusion
Our study found associations between altered methylation and breast cancer in a large sample size of Chinese women. Specifically, we discovered that hypomethylation of the CTSZ gene is associated with early-stage breast cancer in premenopausal Chinese women. This finding may lead to the identification of potential biomarkers, such as CTSZ methylation, which could help improve early detection and treatment of breast cancer. We also identified a correlation between CTSZ methylation level in BC and tumor stage, ER status and HER2 status. Nevertheless, our findings can only suggest the blood-based DNA methylation markers as an early diagnostic marker for BC due to the limitations of the retrospective study. To determine whether these biomarkers could be constituted as early diagnostic markers for BC, prospective multicenter clinical studies with population-based cohorts should be mandatory for future studies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Nanjing Medical University Chinese Academy of Medical Sciences Zhengzhou University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
RY, FM, and LD contributed to the study conception and design. XZ, LL, WG, JQL, JYL, JJL, and JZ performed material preparation. Data collection was performed by JYL, ZW, JJL, and YZ, and JQL and YZ performed analysis. The first draft of the manuscript was written by JYL and all authors commented on previous versions. All authors contributed to the article and approved the submitted version.
Acknowledgments
We are grateful to Feifei Di, Jin Zhang and Jun Wang from Nanjing TANTICA Biotechnology Co. Ltd for their contribution to the sample processing. We also gratefully acknowledge Henan Key Laboratory of Pharmacology for Liver Diseases for providing platform support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 68:394–424. doi: 10.3322/caac.21492
3. Momenimovahed Z, Salehiniya H. Epidemiological characteristics of 577and risk factors for breast cancer in the world. Breast Cancer (2019) 11:151–64. doi: 10.2147/BCTT.S176070
4. Francies FZ, Hull R, Khanyile R, Dlamini Z. Breast cancer in low-middle income countries: abnormality in splicing and lack of targeted treatment options. Am J Cancer Res (2020) 10:1568–91.
5. Plevritis SK, Munoz D, Kurian AW, Stout NK, Alagoz O, Near AM, et al. Association of screening and treatment with breast cancer mortality by molecular subtype in US women 2000–2012. JAMA (2018) 319:154–64. doi: 10.1001/jama.2017.19130
6. Bhat SA, Majid S, Wani HA, Rashid S. Diagnostic utility of epigenetics in breast cancer–a review. Cancer Treat Res Commun (2019) 19:100125. doi: 10.1016/j.ctarc.2019.100125
7. Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol (2016) 8(9):a019505. doi: 10.1101/cshperspect.a019505
8. Pan Y, Liu G, Zhou F, Su B, Li Y. DNA Methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med (2018) 18:1–14. doi: 10.1007/s10238-017-0467-0
9. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet (2012) 13(7):484–92. doi: 10.1038/nrg3230
10. Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenetics (2016) 8(1):115. doi: 10.1186/s13148-016-0282-6
11. Kim SY, Han YK, Song JM, Lee CH, Kang K, Yi JM, et al. Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC). Clin Epigenetics (2019) 11(1):1–12. doi: 10.1186/s13148-019-0715-0
12. Basse C, Arock M. The increasing roles of epigenetics in breast cancer: implications for pathogenicity, biomarkers, prevention and treatment. Int J Cancer (2015) 137(12):2785–94. doi: 10.1002/ijc.29347
13. Min HL, Kim J, Kim WH, Jang BG, Kim MA. Epigenetic silencing of the putative tumor suppressor gene GLDC (Glycine dehydrogenase) in gastric carcinoma. Anticancer Res (2016) 36(1):179–87.
14. Chopra-Tandon N, Wu H, Arcaro KF, Sturgeon SR. Relationships between global DNA methylation in circulating white blood cells and breast cancer risk factors. J Cancer Epidemiol (2017) 1):2705860. doi: 10.1155/2017/2705860
15. Olson OC, Joyce JA. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat Rev Cancer (2015) 15:712–29. doi: 10.1038/nrc4027
16. Jechorek D, Votapek J, Meyer F, Kandulski A, Roessner A, Franke S. Characterization of cathepsin X in colorectal cancer development and progression. Pathol Res Pract (2014) 210:822–9. doi: 10.1016/j.prp.2014.08.014
17. Gyorffy B, Bottai G, Fleischer T, Munkacsy G, Budczies J, Paladini L, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer (2016) 138:87–97. doi: 10.1002/ijc.29684
18. Yang R, Stocker S, Schott S, Heil J, Marme F, Cuk K, et al. The association between breast cancer and S100P methylation in peripheral blood by multicenter case-control studies. Carcinogenesis (2017) 38:312–20. doi: 10.1093/carcin/bgx004
19. Yin Q, Yang X, Li L, Xu T, Zhou W, Gu W, et al. The association between breast cancer and blood-based methylation of S100P and HYAL2 in the Chinese population. Front Genet (2020) 11:977. doi: 10.3389/fgene.2020.00977
20. Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc (2018) 77(4):412–22. doi: 10.1017/S0029665118000150
21. van Veldhoven K, Polidoro S, Baglietto L, Severi G, Sacerdote C, Panico S, et al. Epigenome-wide association study reveals decreased average methylation levels years before breast cancer diagnosis. Clin Epigenetics (2015) 7(1):67. doi: 10.1186/s13148-015-0104-2
22. Severi G, Southey MC, English DR, Jung CH, Lonie A, McLean C, et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat (2014) 148(3):665–73. doi: 10.1007/s10549-014-3209-y
23. Kane AE, Sinclair DA. Epigenetic changes during aging and their reprogramming potential. Crit Rev Biochem Mol Biol (2019) 54:61–83. doi: 10.1080/10409238.2019.1570075
24. Dugue PA, Bassett JK, Joo JE, Jung CH, Ming WE, MorenoBetancur M, et al. DNA Methylation-based biological aging and cancer risk and survival: pooled analysis of seven prospective studies. Int J Cancer (2018) 142:1611–9. doi: 10.1002/ijc.31189
25. Dunneram Y, Greenwood DC, Cade JE. Diet, menopause and the risk of ovarian, endometrial and breast cancer. Proc Nutr Soc (2019) 78:438–48. doi: 10.1017/S0029665118002884
26. Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol Ther (2019) 195:172–85. doi: 10.1016/j.pharmthera.2018.11.001
27. Simonova OA, Kuznetsova EB, Tanas AS, Rudenko VV, Poddubskaya EV, Kekeeva TV, et al. Abnormal hypermethylation of CpG dinucleotides in promoter regions of matrix metalloproteinases genes in breast cancer and its relation to epigenomic subtypes and HER2 overexpression. Biomedicines (2020) 2020:8. doi: 10.3390/biomedicines8050116
Keywords: CTSZ, DNA methylation, breast cancer, case-control study, Chinese women
Citation: Li J, Zhou X, Li L, Ji L, Li J, Qu Y, Wang Z, Zhao Y, Zhang J, Liang F, Liu J, Gu W, Yang R, Ma F and Dai L (2023) The association between CTSZ methylation in peripheral blood and breast cancer in Chinese women. Front. Oncol. 13:1148635. doi: 10.3389/fonc.2023.1148635
Received: 20 January 2023; Accepted: 02 May 2023;
Published: 18 May 2023.
Edited by:
Bin Fang, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, ChinaReviewed by:
Chenshen Huang, Fujian Provincial Hospital, ChinaShaokun Shu, Peking University, China
Jing Xie, Peter MacCallum Cancer Centre, Australia
Copyright © 2023 Li, Zhou, Li, Ji, Li, Qu, Wang, Zhao, Zhang, Liang, Liu, Gu, Yang, Ma and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Dai, bHBkYWlAenp1LmVkdS5jbg==; Fei Ma, ZHJtYWZlaUAxMjYuY29t; Rongxi Yang, cm9uZ3hpeWFuZ0Buam11LmVkdS5jbg==
 Jinyu Li1
Jinyu Li1 Lixi Li
Lixi Li Wanjian Gu
Wanjian Gu Rongxi Yang
Rongxi Yang Fei Ma
Fei Ma Liping Dai
Liping Dai