
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 06 March 2023
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1138481
This article is part of the Research Topic Emerging Mutations in Colorectal Cancer Development and Progression View all 13 articles
Colorectal cancer (CRC) is a leading cause of cancer-related death worldwide due to the lack of effective diagnosis and prognosis biomarkers and therapeutic targets, resulting in poor patient survival rates. Circular RNA (circRNA) is a type of endogenous non-coding RNA (ncRNA) with a closed-loop structure that plays a crucial role in physiological processes and pathological diseases. Recent studies indicate that circRNAs are involved in the diagnosis, prognosis, drug resistance, and development of tumors, particularly in CRC. Therefore, circRNA could be a potential new target for improving CRC diagnosis, prognosis, and treatment. This review focuses on the origin and biological functions of circRNA, summarizes recent research on circRNA’s role in CRC, and discusses the potential use of circRNAs as clinical biomarkers for cancer diagnosis and prognosis, as well as therapeutic targets for CRC treatment.
Colorectal cancer (CRC) is a prevalent malignant tumor worldwide, with the third-highest incidence and the second-highest mortality rate globally (1). The incidence of CRC is increasing worldwide, with over 2.2 million new cases and 1.1 million deaths projected to occur by 2030, according to a recent survey. Developed countries have a higher incidence rate, while developing countries have seen a significant rise in mortality rates (2). The exact mechanism underlying the occurrence and development of CRC is unclear. However, risk factors closely linked to CRC include heredity, diet, smoking, alcohol consumption, physical inactivity, and older age, with age>50 being a particularly significant risk factor. Nonetheless, recent evidence suggests a gradual increase in the incidence of CRC among young people (3). The disease has multifactorial characteristics, and there are no detectable precursors that are universally applicable. Furthermore, there is currently no clinically effective non-invasive screening test to facilitate early diagnosis of CRC (4). There are four subtypes of CRC: CMS1-CMS4, each with different clinical and biological characteristics (5). High-grade CRC tumors are associated with poor prognosis, high metastatic potential, and resistance to conventional therapies, which poses a significant challenge in their treatment. Although our understanding of CRC-related signaling pathways has advanced in recent years, chemotherapy and radiotherapy resistance due to specific molecules in these pathways continues to be a major obstacle to effective treatment (4). Here’s a possible revision: “Non-coding RNAs (ncRNAs) are known to have significant roles in regulating the chemical and radioresistance of CRC, making them promising targets for the development of new anti-drug and anti-radiation therapy strategies (6, 7). These ncRNAs make up the majority (~90%) of the transcribed human genome and include RNAs of varying lengths (8). Numerous studies have demonstrated the involvement of ncRNAs in various physiological and pathological processes, such as diabetes, cardiovascular disease, and cancer (9–11). Short ncRNAs consist of microRNAs (miRNAs), small interfering RNAs (siRNAs), and short piwi-interacting RNAs (piRNAs), whereas both linear lncRNAs (long non-coding RNAs) and circRNAs(Circular RNAs) belong to the category of long non-coding RNAs (12).
CircRNA is a relatively new subtype of ncRNA that was first discovered as a viroid in 1976 (13), and later in eukaryotes (14–16). CircRNA is a single-stranded, covalently closed loop that exhibits strong resistance to exonuclease (17). CircRNA was originally thought to be a non-functional by-product of pre-mRNA splicing (18). However, in recent years, the development and application of high-throughput RNA sequencing (RNA-seq) technology and bioinformatics have identified a large number of circRNAs that play key roles in physiological and developmental processes (19). Moreover, mounting evidence suggests that CircRNA plays a crucial role in tumorigenesis, proliferation, progression, and drug resistance (20, 21). Given its high stability, tissue-specificity, and presence in exosomes and body fluids, circRNA holds great promise as a biomarker or therapeutic target for malignant tumors (21). To elucidate the characteristics and mechanisms of circRNA in CRC, an increasing number of studies are focused on developing new accurate medical biomarkers to identify potential therapeutic targets. Thus, gaining a comprehensive understanding of circRNAs and their roles in CRC signal transduction and molecular mechanisms may aid in identifying more effective treatments. This article aims to introduce the processes and signaling pathways that may be affected by each circRNA during the entire evolution of CRC, with the goal of aiding in early diagnosis, pathological grading, targeted therapy, and prognosis evaluation of CRC.
CircRNA is synthesized during post-transcriptional splicing of pre-mRNA, where mature mRNA is generated after removal of introns by standard splicing. CircRNAs can be classified into four subgroups based on their sequence and domain binding: exonic circRNAs (which are the most common), intronic circRNAs, exonic intron ciRNAs, and tRNA intron ciRNAs (22). Various types of circRNA are produced through different regulation mechanisms. Exon circRNA is produced through both lariat-driven cyclization and reverse splicing (22). In the first pathway, all introns are removed from the lasso, and the remaining exons bind to the 5 ‘ -3 ‘ phosphodiester bond (23).In the second pathway, RNA-binding proteins attached to two introns form eCircRNAs (24, 25). The Alu complementary sequence causes the attachment of two introns during base pairing-driven cyclization (22). Subsequently, reverse splicing leads to the production of EIciRNA (26). Specific circRNAs avoid degradation by linking elements rich in 7 nt GU and elements rich in 11 nt C (27). CiRNA and EIciRNA, located in the nucleus, are important for gene transcription, whereas eCircRNA is widely present in the cytoplasm and does not participate in transcription (22) (Figure 1).
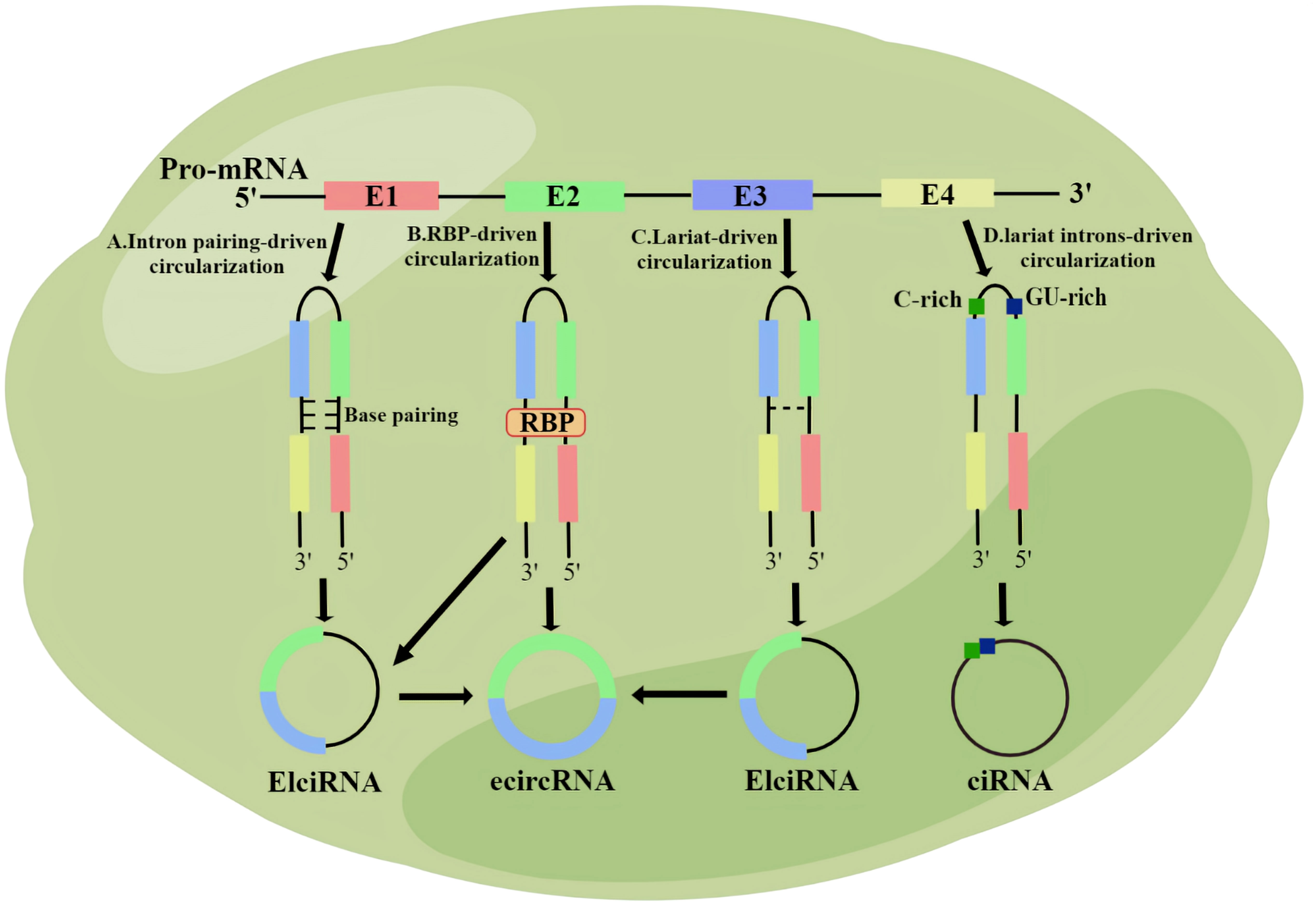
Figure 1 The biological origin of circRNA. (A) Intron pairing-driven circularization; (B) RNA binding protein (RBP)-driven circularization; (C) Lariat-driven circularization; (D) Lariat introns-driven circularization.
CircRNA can regulate parental genes through competition for splicing sites with linear isoforms. For example, CircMbl, derived from the second exon of the splicing factor muscleblind (MBL), competes with linear MBL mRNA for splicing. Due to the presence of the CircMbl binding site in MBL protein, MBL can tightly bind to CircMbl, thereby promoting CircMbl production. As a result, the expression of CircMbl can be upregulated by MBL expression, leading to a reduction in the production of linear mRNA (28). CircRNA can bind to proteins to regulate the transcription of its locus genes. For example, two eiCircRNAs, Circ-EIF3J and Circ-PAIP2, can form an EIciRNA-U1 complex with U1 small ribonucleoprotein (snRNP), which further targets polymerase II (Pol II) in the promoter region of the host gene and enhances the expression of its parental genes (29). Similarly, ciRNAs such as CircAankrd52 and ci-sirt7 can regulate the transcription of their parental genes through cis-regulation of RNA Pol II (27) (Figure 2).
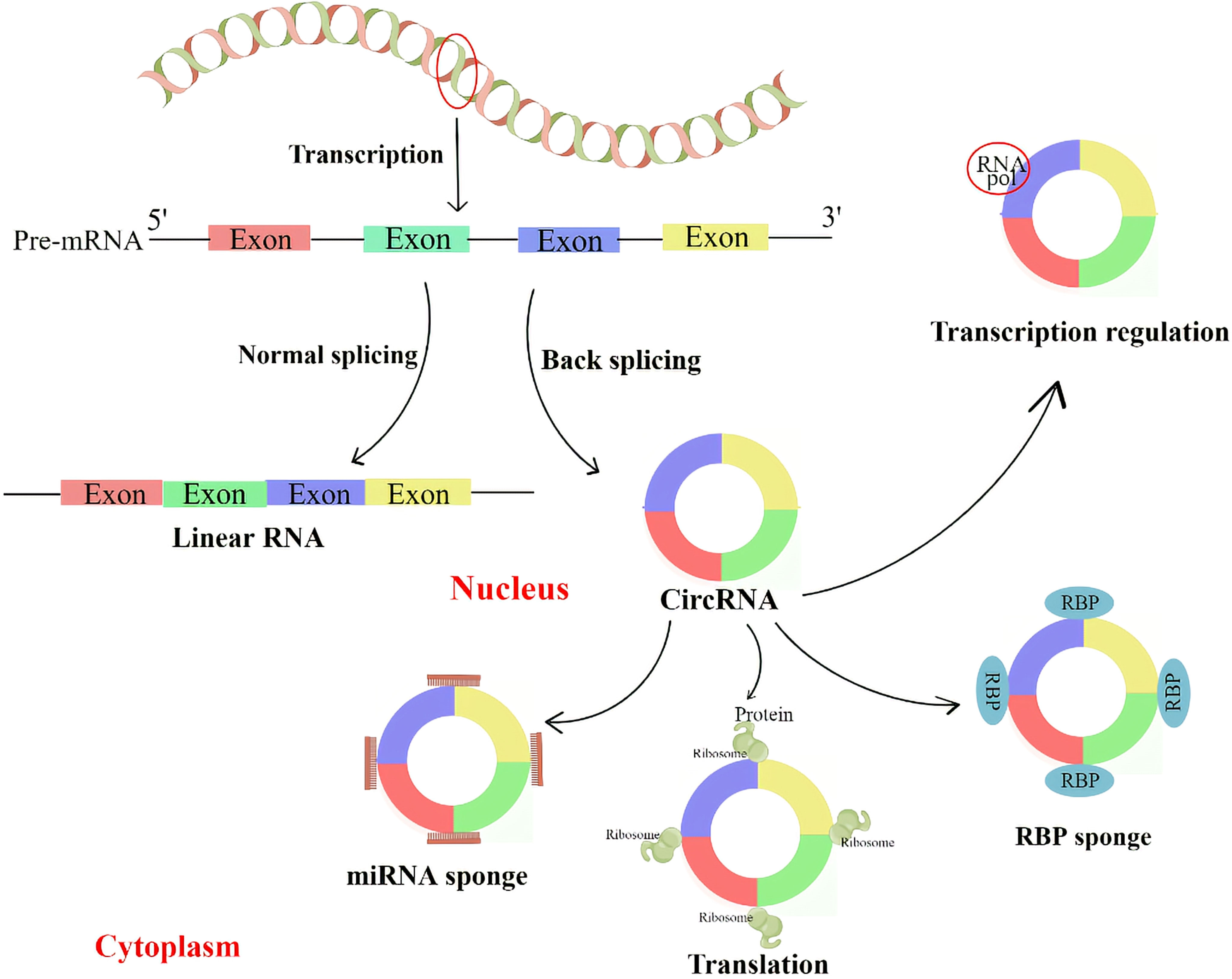
Figure 2 CircRNA regulates the transcription and splicing of parental genes by interacting with RNA polymerase II or transcription-related factors. CircRNA with miRNA binding sites can prevent miRNA from attaching to its target mRNA, thereby blocking the inhibitory effect of miRNA on target proteins. CircRNA can also interact with RNA binding protein (RBP) to regulate protein activity. Some circRNAs can be translated into proteins.
Increasing evidence suggests that some circRNAs are enriched with miRNA response elements (MREs) and can function as miRNAs by competitively binding to other miRNAs. These circRNAs are referred to as competitive endogenous RNAs (ceRNAs) and can sequester miRNAs, thereby relieving their inhibitory effects on target genes (30). MiRNA is another subtype of ncRNA with a length of about 22 nt, and it plays an important role in post-transcriptional regulation by binding to specific sites in the 3’ untranslated regions of mRNA (31, 32). Several lines of evidence have demonstrated that some circRNAs can act as miRNA sponges, including ciRS-7 (33),circSRY (30), circHIPK3 (34), and many other circRNAs (Figure 2).
CircRNAs, similar to miRNA sponges, can interact with RNA-binding proteins (RBPs) and act as protein decoys or antagonists. For example, CircRNA derived from Foxo3 (circFoxo3) interacts with cell cycle protein-dependent kinase 2 (CDK2) and cell cycle protein kinase inhibitor 1 (p21), resulting in anticancer and cell cycle blocking effects (35). Other examples, such as CircDNMT1 (36) and circ-NOL10 (37), mainly function as RNA binding protein (RBP) sponges. Recent studies by Okholm et al. (38) have shown that some RBPs preferentially bind to circRNAs rather than their linear counterparts, and this interaction occurs in a cell-type specific manner. They also discovered that circCDYL interacts with IGFBP1 and IGFBP2 in bladder cancer cell lines. Depletion of either circCDYL or these RBPs is indicative of a cancer gene set, and knockdown of this circRNA affects the expression of two important genes involved in tumorigenesis and progression, TP53 and MYC. However, the specific mechanism by which RBPs regulate circRNA requires further investigation (Figure 2).
Initially, circRNAs were thought to be ncRNAs because they lack a 5’ cap, a 3’ poly-A tail, and a translation initiation structure (39). However, convincing evidence has shown that some endogenous circRNAs can be translated into proteins. The genome of the hepatitis D virus provides the first example of a circRNA that can be translated into a protein. This single-stranded circRNA encodes the hepatitis D antigen (15). Furthermore, Legnini et al. (40) reported that circZNF609 in mice can be translated into a protein that controls myoblast proliferation via the internal ribosome entry site (IRES). Similarly, Pamudurti et al. (41) found that circMbl can be translated into proteins in the head of flies in a cap-independent manner. Additionally, circAKT3 and circFBXW7 (42) were found to encode proteins that inhibit tumorigenesis in human gliomas Figure 2).
Thus far, a cumulative sum of 173 circRNAs have been identified via PubMed and documented as differential expressions in the tumorous and adjacent normal tissues of CRC. Amongst these, 67 circRNAs exhibited an up-regulation, while 23 demonstrated a down-regulation in both cell-culture experiments and animal studies. Tables 1 and 2 depict the prominence of up-regulated circRNAs over their down-regulated counterparts. Throughout the evaluation of these 90 circRNAs, their regulatory impact on fundamental malignant traits has been comprehensively illustrated, encompassing factors such as proliferation, migration, invasion, and apoptosis.
Wang et al. (43) have recently verified that the upregulation of circ_0011385 and MYO6 in both CRC tissues and cells was significant, while the down-regulation of miR-330-3p was observed. Notably, the expression of circ_0011385 in CRC patients exhibited a positive correlation with tumor size and TNM stage. Furthermore, the inhibition of cell proliferation, migration, and invasion and the induction of apoptosis in CRC cells were observed upon silencing of circ_0011385 or down-regulation of MYO6. Remarkably, the inhibitory effect of circ_0011385 silencing on CRC progression was reversed by the down-regulation of miR-330-3p or the overexpression of MYO6. It is noteworthy that circ_0011385 is interrelated with miR-330-3p, and the latter targets MYO6. The in vivo overexpression of Circ_0011385 leads to the promotion of tumor growth through the miR-330-3p/MYO6 axis.
The up-regulation of circ_0006732 has been observed in the tissues of CRC. Further investigation has revealed that circ_0006732 serves as a competitive endogenous RNA (ceRNA), which directly interacts with miR-127-3p, thereby affecting the expression of Ras-related protein Rab-3D (Rab3D). Notably, it has been verified that the down-regulation of circ_0006732 can significantly suppress the proliferation, migration, invasion, and EMT of CRC cells by regulating the miR-127-5p/RAB3D axis. Based on these findings, Circ_0006732 emerges as a prospective therapeutic target for the treatment of CRC (48).
Circ_0056618 and PRRG4 were found to be up-regulated in both CRC tumor tissues and cells, while miR-411-5p exhibited down-regulation. The suppression of CRC cell proliferation, migration, invasion, and EMT was observed following the knockdown of Circ_0056618 or PRRG4. The regulation of PRRG4 expression was positively influenced by Circ_0056618 through the targeting of miR-411-5p. Furthermore, the inhibition of CRC cell proliferation, migration, invasion, and EMT, which were induced by the knockdown of circ_0056618, were rescued by miR-411-5p deletion or PRRG4 overexpression. Animal studies have confirmed the hindrance of tumor growth in vivo due to circ_0056618 knockdown. Therefore, the targeting of the circ_0056618/miR-411-5p/PRRG4 axis could be a promising therapeutic approach for treating CRC (49).
Zou Y et al. (68) reported an abnormal up-regulation of circ_0068464 in both CRC cells and tissues when compared to their normal counterparts. The knockdown of circ_0068464 inhibited the migration and proliferation of CRC cells, induced apoptosis, and decreased the expression of Wnt/β-catenin pathway-related proteins such as β-catenin, cyclin D1, C-myc, and LEF-1. In vivo tumorigenicity experiments in nude mice further confirmed the inhibitory effect of circ_0068464 down-regulation on tumor growth and lung metastasis. Additionally, the interaction between circ_0068464 and miR-383 was established through dual luciferase assay and RNA immunoprecipitation assay. It was found that miR-383 was significantly downregulated in CRC tissues and cells, and the inhibition of miR-383 expression reversed the inhibitory effect of circ_0068464 knockout on CRC cells. Thus, it was concluded that circ_0068464 promotes CRC development by targeting miR-383 to regulate the activation of the Wnt/β-catenin pathway.
In a recent study, Chen B et al. (121) reported the down-regulation of circ_0003215 in CRC, which was negatively correlated with tumor size, TNM stage, and lymph node metastasis. The authors found that the decrease in circ_0003215 level was due to the RNA degradation by m6A read-write protein YTHDF2. Functional assays revealed that Circ_0003215 inhibited cell proliferation, migration, invasion, and metastasis both in vitro and in vivo. The study also demonstrated that Circ_0003215 regulates the expression of DLG4 through miR-663b sponge, thereby inducing metabolic reprogramming in CRC. Mechanistically, DLG4 inhibits the pentose phosphate pathway (PPP) by K48-linked ubiquitination of glucose-6-phosphate dehydrogenase (G6PD).
In CRC tissues and cell lines, a significant downregulation of circ_0007919 and TET1 gene expression was observed, while miR-942-5p exhibited an upregulated expression. Remarkably, in vitro studies demonstrated that the overexpression of circ_0007919 led to a substantial inhibition of both CRC cell growth and migration. Moreover, we discovered that circ_0007919 acts as a competitive inhibitor, binding to miR-942-5p, thereby increasing the expression of the downstream target gene TET1. Altogether, our results reveal that circ_0007919 plays a crucial role in suppressing the oncogenic behavior of CRC cells, functioning through the modulation of the miR-942-5p/TET1 axis. These findings offer a novel therapeutic avenue for the management of CRC (125).
Yan D S et al. (124) have reported a significant reduction in the expression of the circRNA, circ_0065378, in CRC tissues. In vitro experiments demonstrated that the upregulation of circ_0065378 impeded the malignant behavior of CRC cells, as evidenced by the suppression of cellular proliferation, invasion, migration, and epithelial-mesenchymal transition (EMT). At the molecular level, we observed that circ_0065378-mediated tumor suppression was exerted through the sequestration of miR-4701-5p. Remarkably, we found that the knockdown of the TUSC1 gene expression, a direct downstream target of miR-4701-5p, partially abolished the inhibitory effects of either miR-4701-5p inhibitor or circ_0065378 overexpression on the malignant behavior of CRC cells. These findings collectively suggest that the circ_0065378/miR-4701-5p/TUSC1 axis constitutes a promising molecular target for the diagnosis and treatment of CRC.
Given the emerging regulatory role of circRNAs in cancer, their potential as effective therapeutic targets is gaining momentum. However, it has been reported that resistance to certain circRNAs can occur during the treatment of CRC (Table 3).
Circ_0006174 exhibits upregulation in doxorubicin (Dox)-resistant CRC cells and tissues. Notably, the depletion of Circ_0006174 instigated a reduction in Dox resistance, cell proliferation, migration, and invasion of CRC cells. Remarkably, the copious expression of circ_0006174 has been found to emanate from exosomes of Dox-resistant CRC cells. Additionally, Zhang et al. established the targeting association between circ_0006174/miR-1205 or miR-1205/CCND2. The exosomal Circ_0006174 enforces Dox resistance by upregulating CCND2 via miR-1205 mediation. In vivo, circ_0006174 knockdown also enhances tumor sensitivity to Dox by targeting the miR-1205/CCND2 axis. Taken together, these findings signify that exosomes enriched in Circ_0006174 could be utilized as a diagnostic biomarker for chemoresistance in CRC (65).
A mounting body of evidence indicates that exosomes originating from drug-resistant cells can engender resistance in chemosensitive cells. The exosome’s structure contributes to the prevention of RNA degradation, which guarantees an effective concentration of circRNA. The size and membrane structure of exosomes also promote the absorption and fusion of cancer cells. The expression of Circ_0094343 was significantly reduced in CRC tissues, chemoresistant CRC tissues, and metastatic CRC tissues. Furthermore, the exosome-borne Circ_0094343 also impedes HCT116 cell proliferation, clone formation, and glycolysis. Notably, Circ_0094343 augments the chemosensitivity of HCT116 cells to 5-fluorouracil (5-FU), oxaliplatin (L-OHP), and doxorubicin (Dox). Moreover, Circ_0094343 functions as a sponge for miR-766-5p, targeting and regulating TRIM67. The results demonstrate that Circ_0094343 hinders the proliferation, clone formation, and glycolysis of HCT116 cells through the miR-766-5p/TRIM67 axis, thereby instigating chemoresistance (123).
Wang Z F et al. have discovered that circ_0082182 is upregulated in OXA-resistant CRC tissues and cells. Circ_0082182 downregulation impedes drug resistance, proliferation, invasion, and migration of OXA, while promoting apoptosis in OXA-resistant CRC cells. MiR-326 directly targets NFIB, preventing OXA resistance in CRC cells and thwarting cancer development. Circ_0082182 regulates NFIB expression by acting as a sponge for miR-326. In OXA-resistant xenograft tumor models, Circ_0082182 mediates the miR-326/NFIB axis, promoting tumor growth. Consequently, Circ_0082182 boosts NFIB expression by sequestering miR-326, thereby regulating the onset and progression of OXA resistance and CRC. Therefore, Circ_0082182 could be a prospective therapeutic target for OXA-resistant CRC (50).
These findings provide novel insights into our comprehension of the mechanisms behind circRNA-mediated resistance. Their dysregulation propels the evolution of drug resistance in cancer through various mechanisms, including apoptosis inhibition, EMT induction, cell proliferation promotion, and glycolysis suppression (Figure 3). However, the precise mechanism remains incompletely understood. As our comprehension of circRNA deepens, circRNA may hold great clinical promise in the future.
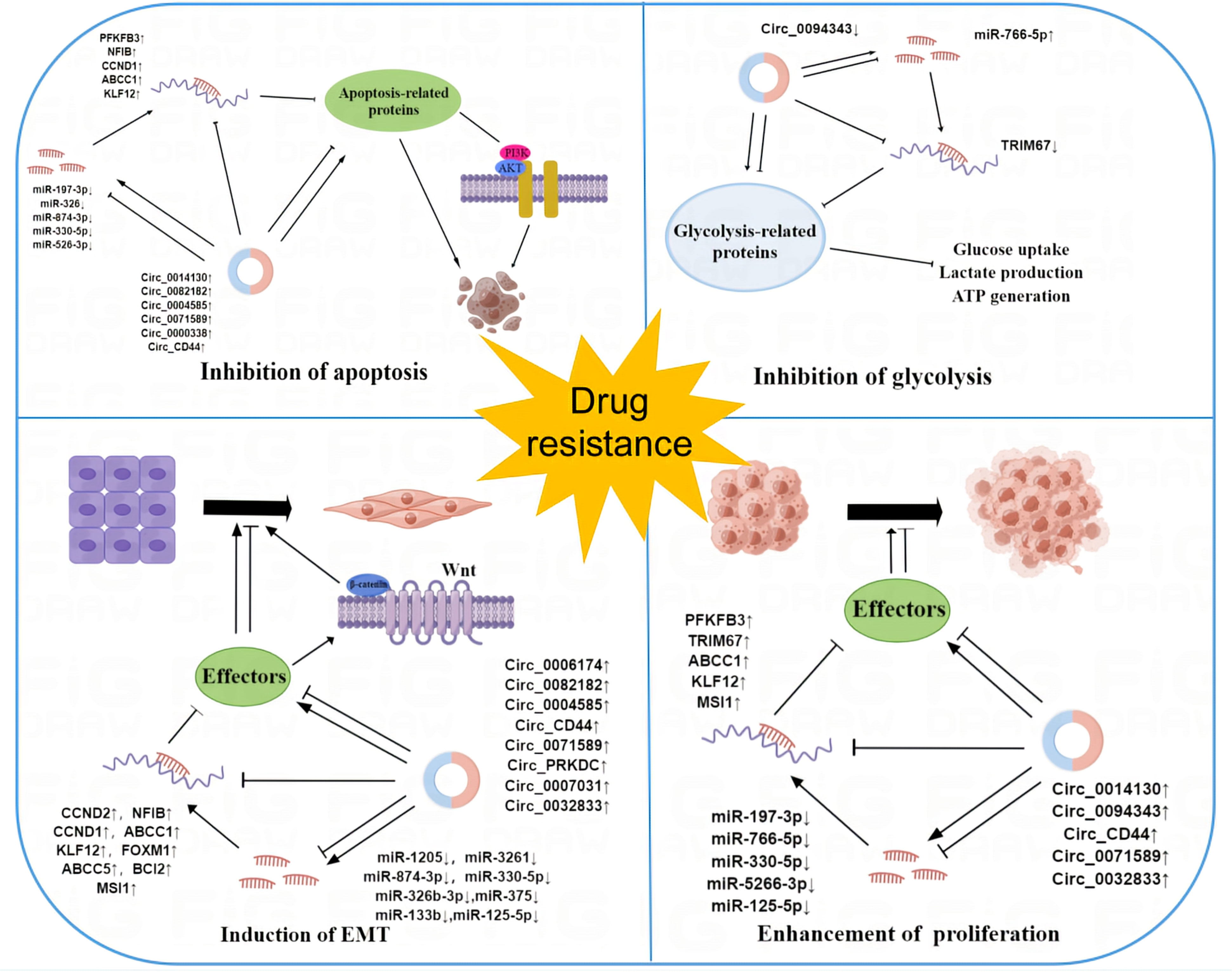
Figure 3 The mechanism of circRNAs in cancer drug resistance. Dysregulation of circRNAs contributes to the development of cancer resistance by regulating a variety of cellular processes in cancer cells, such as apoptosis, EMT, glycolysis, and cell proliferation.
Due to the high mortality rate associated with CRC, there is an urgent need for early diagnostic and prognostic indicators. As circRNAs are abundantly expressed in cancer cells, they represent a promising class of biomarkers for CRC. Indeed, clinical samples, including CRC patient tissues and plasma, have demonstrated the potential of circRNAs as valuable biomarkers for early diagnosis and metastasis prediction of CRC (Table 4, Figure 4).
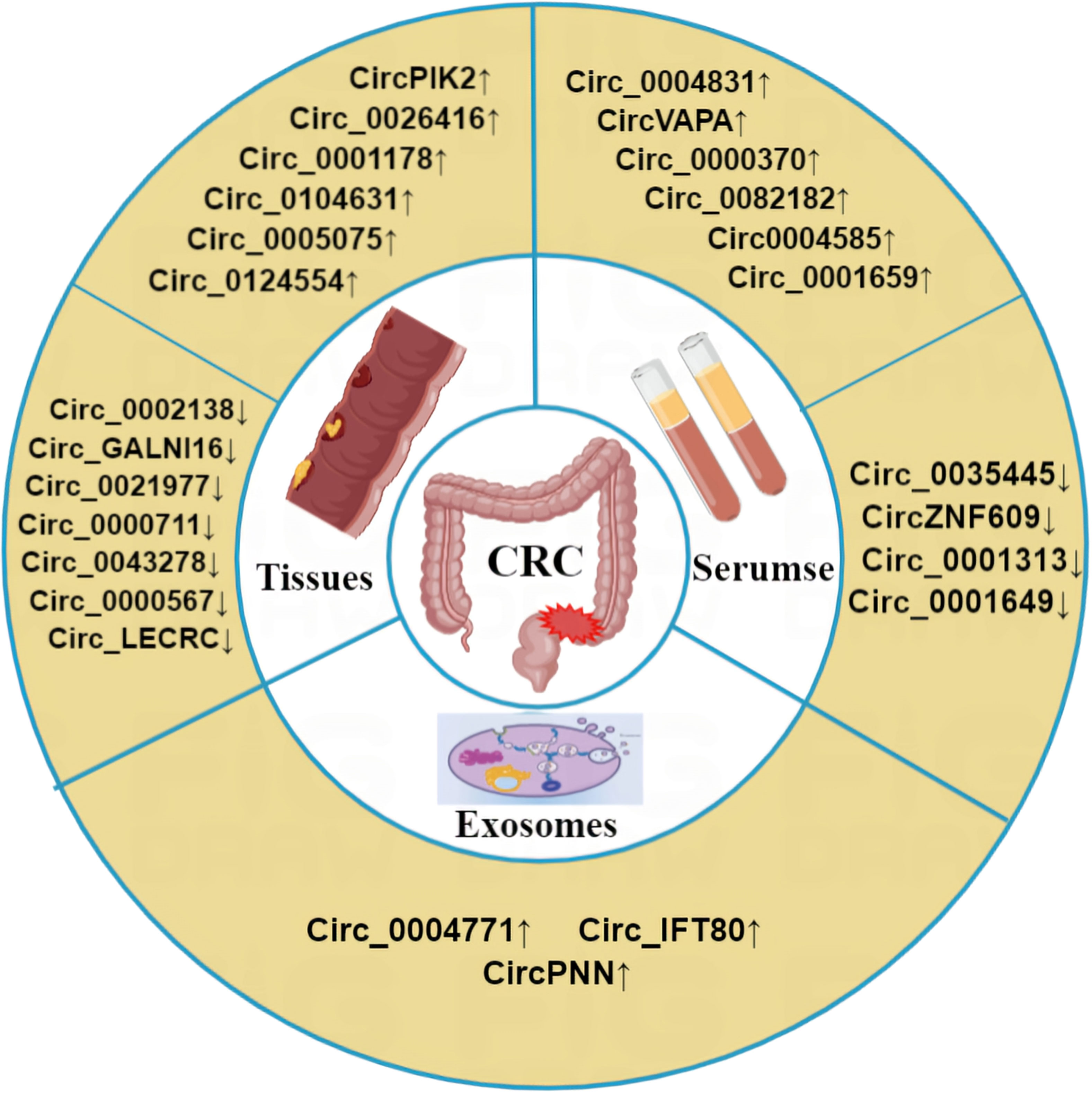
Figure 4 Abnormal expression of circRNA in CRC. Compared with the normal control group, circRNA was differentially expressed in CRC tissues, exosomes and blood of CRC patients. ‘↑’ means up, ‘↓’ means down.
A recent study by Mohammadi et al. has demonstrated that the expression level of circ_0006282 is significantly increased in CRC tissues and plasma samples of CRC patients when compared to healthy controls (p<0.0001). The area under the curve (AUC) was 0.831 (95%CI:0.779-0.883), suggesting that circ_0006282 has the potential to serve as a biomarker for the early detection and metastasis prediction of CRC. Importantly, the expression of circ_0006282 in CRC patients decreased to normal levels after surgery (p<0.0001), indicating its potential role as a monitoring tool. When combined with carcinoembryonic antigens (CEA) and carbohydrate antigen 199 (CA199), the use of circ_0006282 showed high specificity and sensitivity in CRC detection. Furthermore, the study revealed that plasma circ_0006282 can differentiate between patients with colorectal cancer and colitis. Taken together, these findings suggest that plasma circ_0006282 can serve as a promising diagnostic and dynamic monitoring biomarker for CRC (160).
Exosomes represent a critical modulator of intercellular communication and have been the subject of numerous scientific inquiries. These vesicles, originated from cancerous cells, are able to encapsulate circRNAs and release them into the bloodstream (163). Furthermore, there has been a surge of interest in using exosome-derived circRNAs as disease biomarkers (164). Notably, Pan B et al. have observed a marked upregulation of exosomal circ_0004771 in the serum of CRC patients. The use of GW4869, a known inhibitor of exosome secretion, led to a significant reduction in the level of exosomal circ_0004771 in the medium, without causing a noteworthy change in CRC cells. Thus, the presence of circulating exosomal circ_0004771 is a promising candidate as a diagnostic biomarker for early detection of CRC (153).
As a nascent variety of ncRNA, CircRNA participates in diverse physiological processes and pathological conditions. While the precise role of circRNAs is still a matter of debate, mounting evidence supports their function as miRNA sponges, protein decoys, or translational modulators, as well as their role in regulating pre-RNA gene splicing and transcription. Presently, CircRNA represents a frontier in tumor biology and treatment, with no less than hundreds of aberrantly expressed circRNAs detected in CRC tissues, many of which have been demonstrated to modulate CRC cell proliferation, migration, invasion, and apoptosis. These circRNAs have the potential to serve as diagnostic biomarkers and effective therapeutic targets for CRC. Despite novel perspectives on the clinical application of circRNA, experimental and clinical research in CRC lags behind. The molecular mechanisms underlying the cyclization, degradation, and intracellular localization of circRNAs in CRC remain elusive. Furthermore, the biological functions and mechanisms of the majority of circRNAs in CRC are still poorly understood, and alternative functions or mechanisms beyond miRNA sponges are yet to be uncovered. Despite the diagnostic and prognostic potential of certain circRNAs, their clinical utility is limited by low sensitivity and specificity. Therefore, it is crucial to investigate the differential expression of circRNAs in clinically relevant samples, such as blood and urine, in addition to CRC tissues, and to improve their sensitivity and specificity. Liquid biopsy, owing to its minimal or non-invasive nature and widespread acceptance, holds great promise as a potent tool for early screening, diagnosis, and prognosis of cancer. Furthermore, the ability to collect liquid biopsy samples at predetermined intervals facilitates monitoring of cancer treatment efficacy, drug resistance, as well as recurrence and metastasis.
This review comprehensively summarizes the current research progress of circRNA in the diagnosis, prognosis, progression, and drug resistance of CRC, highlighting its immense potential as a novel biomarker for early diagnosis, prognosis, and treatment. However, despite the significant strides that have been made, there is still much to be learned about the regulatory mechanism of circRNAs, and their downstream regulatory networks and clinical correlations. An in-depth understanding of these aspects will undoubtedly enhance our understanding of the role of circRNAs in CRC and pave the way for the development of circRNA-based diagnosis, prognosis, and treatment strategies for CRC. As biological methods and information technology continue to advance, it is expected that an increasing number of circRNAs associated with CRC and their physiological and pathological functions will be discovered. Although the precise mechanisms of circularization, degradation, cell localization, and function of circRNAs in CRC remain largely unknown, the enigma of circRNA will eventually be unraveled. New diagnostic and therapeutic strategies based on circRNAs will undoubtedly find their way into clinical practice in the future.
Study concept and design: GF and YM. Analysis and interpretation of data: GF, DX and TZ. Drafting of the manuscript: GF and DX. Critical revision of the manuscript for important intellectual content: GF, DX, GW, LQ, XG and YM. Obtained funding: GF and YM. Study supervision: TZ and YM. All authors contributed to the article and approved the submitted version.
This study is supported by the “ Six Talent Peaks “ Funding Project in Jiangsu Province, China(WSW-326)and the Graduate Research and Innovation Projects of Bengbu Medical College (Byycx22124).
Thanks are due to Figdraw for assistance with the drawing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912
3. Zheng Y, Hua X, Win AK, MacInnis RJ, Gallinger S, Marchand LL, et al. A new comprehensive colorectal cancer risk prediction model incorporating family history, personal characteristics, and environmental factors. Cancer Epidemiol Biomarkers Prev (2020) 29(3):549–57. doi: 10.1158/1055-9965.Epi-19-0929
4. Al-Joufi FA, Setia A, Salem-Bekhit MM, Sahu RK, Alqahtani FY, Widyowati R, et al. Molecular pathogenesis of colorectal cancer with an emphasis on recent advances in biomarkers, as well as nanotechnology-based diagnostic and therapeutic approaches. Nanomater (Basel) (2022) 12(1). doi: 10.3390/nano12010169
5. Peters NA, Constantinides A, Ubink I, van Kuik J, Bloemendal HJ, van Dodewaard JM, et al. Consensus molecular subtype 4 (CMS4)-targeted therapy in primary colon cancer: A proof-of-concept study. Front Oncol (2022) 12:969855. doi: 10.3389/fonc.2022.969855
6. Chai B, Ma Z, Wang X, Xu L, Li Y. Functions of non-coding RNAs in regulating cancer drug targets. Acta Biochim Biophys Sin (Shanghai) (2022) 54(3):279–91. doi: 10.3724/abbs.2022006
7. Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech (2020) 1863(6):194417. doi: 10.1016/j.bbagrm.2019.194417
8. Pandey GK, Kanduri C. Long non-coding RNAs: Tools for understanding and targeting cancer pathways. Cancers (Basel) (2022) 14(19). doi: 10.3390/cancers14194760
9. Ameli-Mojarad M, Ameli-Mojarad M, Nourbakhsh M, Nazemalhosseini-Mojarad E. Circular RNA hsa_circ_0005046 and hsa_circ_0001791 may become diagnostic biomarkers for breast cancer early detection. J Oncol (2021) 2021:2303946. doi: 10.1155/2021/2303946
10. Tian D, Xiang Y, Tang Y, Ge Z, Li Q, Zhang Y. Circ-ADAM9 targeting PTEN and ATG7 promotes autophagy and apoptosis of diabetic endothelial progenitor cells by sponging mir-20a-5p. Cell Death Dis (2020) 11(7):526. doi: 10.1038/s41419-020-02745-x
11. Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinf (2016) 14(4):216–23. doi: 10.1016/j.gpb.2016.02.003
12. DiStefano JK, Gerhard GS. Long noncoding RNAs and human liver disease. Annu Rev Pathol (2022) 17:1–21. doi: 10.1146/annurev-pathol-042320-115255
13. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U.S.A. (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
14. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature (1979) 280(5720):339–40. doi: 10.1038/280339a0
15. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature (1986) 323(6088):558–60. doi: 10.1038/323558a0
16. Matsumoto Y, Fishel R, Wickner RB. Circular single-stranded RNA replicon in saccharomyces cerevisiae. Proc Natl Acad Sci U.S.A. (1990) 87(19):7628–32. doi: 10.1073/pnas.87.19.7628
17. Beilerli A, Gareev I, Beylerli O, Yang G, Pavlov V, Aliev G, et al. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin Cancer Biol (2022) 83:242–52. doi: 10.1016/j.semcancer.2020.12.026
18. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J (1993) 7(1):155–60. doi: 10.1096/fasebj.7.1.7678559
19. Andralojc KM, Elmelik D, Rasing M, Pater B, Siebers AG, Bekkers R, et al. Targeted RNA next generation sequencing analysis of cervical smears can predict the presence of hrHPV-induced cervical lesions. BMC Med (2022) 20(1):206. doi: 10.1186/s12916-022-02386-1
20. Wang J, Zhang Y, Liu L, Yang T, Song J. Circular RNAs: new biomarkers of chemoresistance in cancer. Cancer Biol Med (2021) 18(2):421–36. doi: 10.20892/j.issn.2095-3941.2020.0312
21. Ding P, Liu P, Wu H, Yang P, Tian Y, Zhao Q. Functional properties of circular RNAs and research progress in gastric cancer. Front Oncol (2022) 12:954637. doi: 10.3389/fonc.2022.954637
22. Zhao X, Cai Y, Xu J. Circular RNAs: Biogenesis, mechanism, and function in human cancers. Int J Mol Sci (2019) 20(16). doi: 10.3390/ijms20163926
23. Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U.S.A. (1996) 93(13):6536–41. doi: 10.1073/pnas.93.13.6536
24. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
25. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
26. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
27. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell (2013) 51(6):792–806. doi: 10.1016/j.molcel.2013.08.017
28. Pamudurti NR, Patop IL, Krishnamoorthy A, Bartok O, Maya R, Lerner N, et al. circMbl functions in cis and in trans to regulate gene expression and physiology in a tissue-specific fashion. Cell Rep (2022) 39(4):110740. doi: 10.1016/j.celrep.2022.110740
29. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol (2015) 22(3):256–64. doi: 10.1038/nsmb.2959
30. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
31. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
32. Trujillo RD, Yue SB, Tang Y, O’Gorman WE, Chen CZ. The potential functions of primary microRNAs in target recognition and repression. EMBO J (2010) 29(19):3272–85. doi: 10.1038/emboj.2010.208
33. Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y, et al. Corrigendum to “Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-a family expression” [Cancer lett. 426 (2018) 37-46]. Cancer Lett (2022) 526:364–5. doi: 10.1016/j.canlet.2021.12.001
34. Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep (2022) 23(11):e56102. doi: 10.15252/embr.202256102
35. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res (2016) 44(6):2846–58. doi: 10.1093/nar/gkw027
36. Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene (2018) 37(44):5829–42. doi: 10.1038/s41388-018-0369-y
37. Cai Y, Zhao X, Chen D, Zhang F, Chen Q, Shao CC, et al. Circ-NOL10 regulated by MTDH/CASC3 inhibits breast cancer progression and metastasis via multiple miRNAs and PDCD4. Mol Ther Nucleic Acids (2021) 26:773–86. doi: 10.1016/j.omtn.2021.09.013
38. Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med (2020) 12(1):112. doi: 10.1186/s13073-020-00812-8
39. Wilusz JE. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA (2018) 9(4):e1478. doi: 10.1002/wrna.1478
40. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell (2017) 66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
41. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell (2017) 66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
42. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal (2021) 35(7):e23866. doi: 10.1002/jcla.23866
43. Wang J, Ke S, Gong Y, Cai Y, Xia L, Shi Z, et al. Circ_0011385 knockdown inhibits cell proliferation, migration and invasion, whereas promotes cell apoptosis by regulating miR-330-3p/MYO6 axis in colorectal cancer. BioMed J (2022). doi: 10.1016/j.bj.2022.01.007
44. Zhang W, Wang B, Lin Y, Yang Y, Zhang Z, Wang Q, et al. hsa_circ_0000231 promotes colorectal cancer cell growth through upregulation of CCND2 by IGF2BP3/miR-375 dual pathway. Cancer Cell Int (2022) 22(1):27. doi: 10.1186/s12935-022-02455-8
45. Gao X, Yin J, Yao Y. hsa_circ_0001955 promotes colorectal cancer progression by regulating miR-583/FGF21 axis. J Oncol (2022) 2022:4288474. doi: 10.1155/2022/4288474
46. Wu R, Tang S, Wang Q, Kong P, Liu F. Hsa_circ_0003602 contributes to the progression of colorectal cancer by mediating the miR-149-5p/SLC38A1 axis. Gut Liver (2022). doi: 10.5009/gnl210542
47. Huang Y, Bai Q, Wang Z, Yu H, Li Y, Lu H, et al. circ_0052184 promotes colorectal cancer progression via targeting miR-604/HOXA9 axis. Anal Cell Pathol (Amst) (2022) 2022:8583382. doi: 10.1155/2022/8583382
48. Yang T, Sun J, Wang W, Li D, Yang X, Jia A, et al. Hsa_circ_0006732 regulates colorectal cancer cell proliferation, invasion and EMT by miR-127-5p/RAB3D axis. Mol Cell Biochem (2022) 477(12):2751–60. doi: 10.1007/s11010-022-04458-5
49. Zhang B, Cao W, Liu Y, Zhao Y, Liu C, Sun B. Circ_0056618 enhances PRRG4 expression by competitively binding to miR-411-5p to promote the malignant progression of colorectal cancer. Mol Cell Biochem (2022). doi: 10.1007/s11010-022-04525-x
50. Wang Z, Liu J, Yang T, Wang Q, Liang R, Tang J. Circ_0082182 upregulates the NFIB level via sponging miR-326 to promote oxaliplatin resistance and malignant progression of colorectal cancer cells. Mol Cell Biochem (2022). doi: 10.1007/s11010-022-04551-9
51. Bai L, Gao Z, Jiang A, Ren S, Wang B. Circ_0001535 facilitates tumor malignant progression by miR-485-5p/LASP1 axis in colorectal cancer. Balkan Med J (2022) 39(6):411–21. doi: 10.4274/balkanmedj.galenos.2022.2022-6-51
52. Ma X, Deng C. Circ_0044556 promotes the progression of colorectal cancer via the miR-665-Dependent expression regulation of diaphanous homolog 1. Dig Dis Sci (2022) 67(9):4458–70. doi: 10.1007/s10620-021-07310-w
53. Zhao X, Cui D, Yan F, Yang L, Zhang M, Huang B. Circ_0006174 promotes the malignancy of colorectal cancer cell via the miR−1205/CCBE1/Wnt pathway. Mol Med Rep (2022) 26(2). doi: 10.3892/mmr.2022.12767
54. Chen J, Xu L, Fang M, Xue Y, Cheng Y, Tang X. Hsa_circ_0060927 participates in the regulation of caudatin on colorectal cancer malignant progression by sponging miR-421/miR-195-5p. J Clin Lab Anal (2022) 36(5):e24393. doi: 10.1002/jcla.24393
55. Hong F, Deng Z, Tie R, Yang S. Hsa_circ_0045932 regulates the progression of colorectal cancer by regulating HK2 through sponging miR-873-5p. J Clin Lab Anal (2022) 36(9):e24641. doi: 10.1002/jcla.24641
56. Xie J, Jin D, Xu J, Yang F, Jin J. Hsa_hsa_circ_0081069 promotes the progression of colorectal cancer through sponging miR-665 and regulating E2F3 expression. J Clin Lab Anal (2022):e24710. doi: 10.1002/jcla.24710
57. Zhao S, Xu F, Ji Y, Wang Y, Wei M, Zhang L. Circular RNA circ-CD44 regulates chemotherapy resistance by targeting the miR-330-5p/ABCC1 axis in colorectal cancer cells. Histol Histopathol (2022), 18516. doi: 10.14670/hh-18-516
58. Zhou Y, Zhang Q, Qiu X, Tian T, Xu Q, Liao B. Hsa_circ_0001550 facilitates colorectal cancer progression through mediating microRNA-4262/nuclear casein kinase and cyclin-dependent kinase substrate 1 cascade. J Clin Lab Anal (2022) 36(7):e24532. doi: 10.1002/jcla.24532
59. Wang S, Cao J, Pei L. Knockdown of circ_0004585 enhances the chemosensitivity of colorectal cancer cells to 5-fluorouracil via the miR-874-3p/CCND1 axis. Histol Histopathol (2022), 18502. doi: 10.14670/hh-18-502
60. Wang Y, Zhang B, Zhu Y, Zhang Y, Li L, Shen T, et al. hsa_circ_0000523/miR-let-7b/METTL3 axis regulates proliferation, apoptosis and metastasis in the HCT116 human colorectal cancer cell line. Oncol Lett (2022) 23(6):186. doi: 10.3892/ol.2022.13306
61. Bai L, Gao Z, Jiang A, Ren S, Wang B. Circular noncoding RNA circ_0007334 sequestrates miR-577 to derepress KLF12 and accelerate colorectal cancer progression. Anticancer Drugs (2022) 33(1):e409–22. doi: 10.1097/cad.0000000000001221
62. Zhang Y, Yu H, Guo Z. Circ_KIAA1199 inhibits MSI1 degradation by targeting miR-34c-5p to drive the malignant cell behaviors and tumor growth of colorectal cancer. Anticancer Drugs (2022) 33(1):e134–44. doi: 10.1097/cad.0000000000001164
63. Ma W, Niu Z, Han D, Wang B, Wang X. Circ-FAT1 up-regulates FOSL2 expression by sponging miR-619-5p to facilitate colorectal cancer progression. Biochem Genet (2022) 60(4):1362–79. doi: 10.1007/s10528-021-10148-6
64. Su S, Lu W, Liu J, Li L, Liu L, Li X, et al. Circ_0007031 silencing inhibits cell proliferation and induces cell apoptosis via downregulating MELK at a miR-485-3p-Dependent way in colorectal cancer. Biochem Genet (2022) 60(2):576–97. doi: 10.1007/s10528-021-10111-5
65. Zhang Y, Tan X, Lu Y. Exosomal transfer of circ_0006174 contributes to the chemoresistance of doxorubicin in colorectal cancer by depending on the miR-1205/CCND2 axis. J Physiol Biochem (2022) 78(1):39–50. doi: 10.1007/s13105-021-00831-y
66. Wang W, Zhou L, Li Z, Lin G. Circ_0014130 is involved in the drug sensitivity of colorectal cancer through miR-197-3p/PFKFB3 axis. J Gastroenterol Hepatol (2022) 37(5):908–18. doi: 10.1111/jgh.15829
67. Song M, Liu J. Circ_0067717 promotes colorectal cancer cell growth, invasion and glutamine metabolism by serving as a miR-497-5p sponge to upregulate SLC7A5. Histol Histopathol (2022), 18494. doi: 10.14670/hh-18-494
68. Zou Y, Liu L, Meng J, Dai M. Circular RNA circ_0068464 combined with microRNA-383 regulates wnt/β-catenin pathway to promote the progression of colorectal cancer. Bioengineered (2022) 13(3):5113–25. doi: 10.1080/21655979.2022.2036905
69. Jin L, Han C, Zhai T, Zhang X, Chen C, Lian L. Circ_0030998 promotes tumor proliferation and angiogenesis by sponging miR-567 to regulate VEGFA in colorectal cancer. Cell Death Discovery (2021) 7(1):160. doi: 10.1038/s41420-021-00544-7
70. Fang G, Chen T, Mao R, Huang X, Ji L. Circular RNA circ_0089153 acts as a competing endogenous RNA to regulate colorectal cancer development by the miR-198/SUMO-specific peptidase 1 (SENP1) axis. Bioengineered (2021) 12(1):5664–78. doi: 10.1080/21655979.2021.1967076
71. Xie L, Pan Z. Circular RNA circ_0000467 regulates colorectal cancer development via miR-382-5p/EN2 axis. Bioengineered (2021) 12(1):886–97. doi: 10.1080/21655979.2021.1889130
72. Jiang Z, Hu H, Hu W, Hou Z, Liu W, Yu Z, et al. Circ-RNF121 regulates tumor progression and glucose metabolism by miR-1224-5p/FOXM1 axis in colorectal cancer. Cancer Cell Int (2021) 21(1):596. doi: 10.1186/s12935-021-02290-3
73. Sun S, Li C, Cui K, Liu B, Zhou M, Cao Y, et al. Hsa_circ_0062682 promotes serine metabolism and tumor growth in colorectal cancer by regulating the miR-940/PHGDH axis. Front Cell Dev Biol (2021) 9:770006. doi: 10.3389/fcell.2021.770006
74. Wang Y, Chen H, Wei X. Circ_0007142 downregulates miR-874-3p-mediated GDPD5 on colorectal cancer cells. Eur J Clin Invest (2021) 51(7):e13541. doi: 10.1111/eci.13541
75. Zhang G, Liu Z, Zhong J, Lin L. Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. Eur J Clin Invest (2021) 51(12):e13607. doi: 10.1111/eci.13607
76. Xu YJ, Zhao JM, Gao C, Ni XF, Wang W, Hu WW, et al. Hsa_circ_0136666 activates treg-mediated immune escape of colorectal cancer via miR-497/PD-L1 pathway. Cell Signal (2021) 86:110095. doi: 10.1016/j.cellsig.2021.110095
77. Liu D, Peng S, Li Y, Guo T. Circ-MFN2 positively regulates the proliferation, metastasis, and radioresistance of colorectal cancer by regulating the miR-574-3p/IGF1R signaling axis. Front Genet (2021) 12:671337. doi: 10.3389/fgene.2021.671337
78. Jiang Z, Hou Z, Liu W, Yu Z, Liang Z, Chen S. Circ-keratin 6c promotes malignant progression and immune evasion of colorectal cancer through microRNA-485-3p/Programmed cell death receptor ligand 1 axis. J Pharmacol Exp Ther (2021) 377(3):358–67. doi: 10.1124/jpet.121.000518
79. Mao G, Zhou B, Xu W, Jiao N, Wu Z, Li J, et al. Hsa_circ_0040809 regulates colorectal cancer development by upregulating methyltransferase DNMT1 via targeting miR-515-5p. J Gene Med (2021) 23(12):e3388. doi: 10.1002/jgm.3388
80. Wang X, Liu S, Xu B, Liu Y, Kong P, Li C, et al. Circ-SIRT1 promotes colorectal cancer proliferation and EMT by recruiting and binding to eIF4A3. Anal Cell Pathol (Amst) (2021) 2021:5739769. doi: 10.1155/2021/5739769
81. Wei J, Lin Y, Wang Z, Liu Y, Guo W. Circ_0006174 accelerates colorectal cancer progression through regulating miR-138-5p/MACC1 axis. Cancer Manag Res (2021) 13:1673–86. doi: 10.2147/cmar.S295833
82. Bai L, Gao Z, Jiang A, Ren S, Wang B. circ_0101802 functions as a sponge of miR-1236-3p to facilitate the proliferation, migration and invasion of colorectal cancer via regulating MACC1. J Pharmacol Sci (2021) 147(1):104–13. doi: 10.1016/j.jphs.2021.06.002
83. Wu H, Tao Y, Zhang W, Wang G, Zhang Q. circ−0000212 promotes cell proliferation of colorectal cancer by sponging miR−491 and modulating FOXP4 expression. Mol Med Rep (2021) 23(4). doi: 10.3892/mmr.2021.11939
84. Zhao K, Cheng X, Ye Z, Li Y, Peng W, Wu Y, et al. Exosome-mediated transfer of circ_0000338 enhances 5-fluorouracil resistance in colorectal cancer through regulating MicroRNA 217 (miR-217) and miR-485-3p. Mol Cell Biol (2021) 41(5). doi: 10.1128/mcb.00517-20
85. Zhang B, Yang S, Wang J. Circ_0084615 is an oncogenic circular RNA in colorectal cancer and promotes DNMT3A expression via repressing miR-599. Pathol Res Pract (2021) 224:153494. doi: 10.1016/j.prp.2021.153494
86. Huang L, Zhu L, Pan S, Xu J, Xie M, Wang W, et al. Circ_0029803 serves as the sponge of miR-216b-5p to promote the progression of colorectal cancer by regulating SKIL expression. World J Surg Oncol (2021) 19(1):268. doi: 10.1186/s12957-021-02368-2
87. Zhang L, Yu R, Li C, Dang Y, Yi X, Wang L. Circ_0026416 downregulation blocks the development of colorectal cancer through depleting MYO6 expression by enriching miR-545-3p. World J Surg Oncol (2021) 19(1):299. doi: 10.1186/s12957-021-02407-y
88. Zhang W, Wang Z, Cai G, Huang P. Circ_DOCK1 regulates USP11 through miR-132-3p to control colorectal cancer progression. World J Surg Oncol (2021) 19(1):67. doi: 10.1186/s12957-021-02173-x
89. Luo M, Peng T, Lin R, Gu L, He Y. Hsa_circ_0031787 promotes cell proliferation and invasion in colorectal cancer. J Clin Lab Anal (2021) 35(7):e23807. doi: 10.1002/jcla.23807
90. Zhang W, Wang Z, Cai G, Huang P. Downregulation of Circ_0071589 suppresses cisplatin resistance in colorectal cancer by regulating the MiR-526b-3p/KLF12 axis. Cancer Manag Res (2021) 13:2717–31. doi: 10.2147/cmar.S294880
91. Huang B, Cui DJ, Yan F, Yang LC, Zhang MM, Zhao X. Circ_0087862 promotes the progression of colorectal cancer by sponging miR-142-3p and up-regulating BACH1 expression. Kaohsiung J Med Sci (2021) 37(12):1048–57. doi: 10.1002/kjm2.12437
92. Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer (2020) 19(1):164. doi: 10.1186/s12943-020-01272-9
93. Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol (2020) 235(3):2881–90. doi: 10.1002/jcp.29193
94. Yang L, Sun H, Liu X, Chen J, Tian Z, Xu J, et al. Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J Cell Physiol (2020). doi: 10.1002/jcp.29484
95. Liang Y, Shi J, He Q, Sun G, Gao L, Ye J, et al. Hsa_circ_0026416 promotes proliferation and migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell Int (2020) 20:494. doi: 10.1186/s12935-020-01593-1
96. Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med (2020) 24(17):9908–24. doi: 10.1111/jcmm.15590
97. Ren C, Zhang Z, Wang S, Zhu W, Zheng P, Wang W. Circular RNA hsa_circ_0001178 facilitates the invasion and metastasis of colorectal cancer through upregulating ZEB1 via sponging multiple miRNAs. Biol Chem (2020) 401(4):487–96. doi: 10.1515/hsz-2019-0350
98. Sun J, Liu J, Zhu Q, Xu F, Kang L, Shi X. Hsa_circ_0001806 acts as a ceRNA to facilitate the stemness of colorectal cancer cells by increasing COL1A1. Onco Targets Ther (2020) 13:6315–27. doi: 10.2147/ott.S255485
99. Wang L, Wu H, Chu F, Zhang L, Xiao X. Knockdown of circ_0000512 inhibits cell proliferation and promotes apoptosis in colorectal cancer by regulating miR-296-5p/RUNX1 axis. Onco Targets Ther (2020) 13:7357–68. doi: 10.2147/ott.S250495
100. Li S, Zheng S. Down-regulation of Circ_0032833 sensitizes colorectal cancer to 5-fluorouracil and oxaliplatin partly depending on the regulation of miR-125-5p and MSI1. Cancer Manag Res (2020) 12:11257–69. doi: 10.2147/cmar.S270123
101. Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco Targets Ther (2020) 13:3689–701. doi: 10.2147/ott.S238338
102. Yu C, Li S, Hu X. Circ_0005576 promotes malignant progression through miR-874/CDK8 axis in colorectal cancer. Onco Targets Ther (2020) 13:7793–805. doi: 10.2147/ott.S249494
103. Lu C, Fu L, Qian X, Dou L, Cang S. Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis. Arch Biochem Biophys (2020) 689:108434. doi: 10.1016/j.abb.2020.108434
104. He X, Ma J, Zhang M, Cui J, Yang H. Circ_0007031 enhances tumor progression and promotes 5-fluorouracil resistance in colorectal cancer through regulating miR-133b/ABCC5 axis. Cancer biomark (2020) 29(4):531–42. doi: 10.3233/cbm-200023
105. Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-a in colorectal cancer. Eur Rev Med Pharmacol Sci (2020) 24(8):4190–202. doi: 10.26355/eurrev_202004_20999
106. Li Y, Zang H, Zhang X, Huang G. circ_0136666 facilitates the progression of colorectal cancer via miR-383/CREB1 axis. Cancer Manag Res (2020) 12:6795–806. doi: 10.2147/cmar.S251952
107. Wang G, Li Y, Zhu H, Huo G, Bai J, Gao Z. Circ-PRKDC facilitates the progression of colorectal cancer through miR-198/DDR1 regulatory axis. Cancer Manag Res (2020) 12:12853–65. doi: 10.2147/cmar.S273484
108. Chen H, Pei L, Xie P, Guo G. Circ-PRKDC contributes to 5-fluorouracil resistance of colorectal cancer cells by regulating miR-375/FOXM1 axis and wnt/β-catenin pathway. Onco Targets Ther (2020) 13:5939–53. doi: 10.2147/ott.S253468
109. Du H, He Z, Feng F, Chen D, Zhang L, Bai J, et al. Hsa_circ_0038646 promotes cell proliferation and migration in colorectal cancer via miR-331-3p/GRIK3. Oncol Lett (2020) 20(1):266–74. doi: 10.3892/ol.2020.11547
110. Chen Z, Ren R, Wan D, Wang Y, Xue X, Jiang M, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene (2019) 38(32):6017–34. doi: 10.1038/s41388-019-0857-8
111. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol (2019) 234(5):7247–56. doi: 10.1002/jcp.27482
112. Zhu CL, Sha X, Wang Y, Li J, Zhang MY, Guo ZY, et al. Circular RNA hsa_circ_0007142 is upregulated and targets miR-103a-2-5p in colorectal cancer. J Oncol (2019) 2019:9836819. doi: 10.1155/2019/9836819
113. Cao JZ, Ma LM, Zhang YL, Guo HC, Niu X, Zhao TB. Circ-0104631 promotes cell proliferation and invasion in colorectal cancer and predicts poor prognosis. Eur Rev Med Pharmacol Sci (2019) 23(11):4730–7. doi: 10.26355/eurrev_201906_18054
114. Jin YD, Ren YR, Gao YX, Zhang L, Ding Z. Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci (2019) 23(8):3311–9. doi: 10.26355/eurrev_201904_17693
115. Lu X, Yu Y, Liao F, Tan S. Homo sapiens circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 axis. Med Sci Monit (2019) 25:6872–83. doi: 10.12659/msm.916064
116. Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. BioMed Pharmacother (2018) 102:1188–94. doi: 10.1016/j.biopha.2018.03.085
117. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res (2017) 23(14):3918–28. doi: 10.1158/1078-0432.Ccr-16-2541
118. Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget (2017) 8(53):91674–83. doi: 10.18632/oncotarget.21748
119. Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int (2017) 41(9):1056–64. doi: 10.1002/cbin.10826
120. An Y, Xu B, Yan G, Wang N, Yang Z, Sun M. YAP derived circ-LECRC functions as a “brake signal” to suppress hyperactivation of oncogenic YAP signalling in colorectal cancer. Cancer Lett (2022) 532:215589. doi: 10.1016/j.canlet.2022.215589
121. Chen B, Hong Y, Gui R, Zheng H, Tian S, Zhai X, et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis (2022) 13(9):804. doi: 10.1038/s41419-022-05245-2
122. Zhang Z, Fan W, Gao Q, Han Y, Ma J, Gao W, et al. Hsa_Circ_0000826 inhibits the proliferation of colorectal cancer by targeting AUF1. J Genet Genomics (2022). doi: 10.1016/j.jgg.2022.07.006
123. Li C, Li X. Exosome-derived Circ_0094343 promotes chemosensitivity of colorectal cancer cells by regulating glycolysis via the miR-766-5p/TRIM67 axis. Contrast Media Mol Imaging (2022) 2022:2878557. doi: 10.1155/2022/2878557
124. Yan D, Liu W, Liu Y, Zhu X. Circular RNA circ_0065378 upregulates tumor suppressor candidate 1 by competitively binding with miR-4701-5p to alleviate colorectal cancer progression. J Gastroenterol Hepatol (2022) 37(6):1107–18. doi: 10.1111/jgh.15862
125. Zhao X, Cui D, Yan F, Yang L, Huang B. Circ_0007919 exerts an anti-tumor role in colorectal cancer through targeting miR-942-5p/TET1 axis. Pathol Res Pract (2022) 229:153704. doi: 10.1016/j.prp.2021.153704
126. Wen C, Feng X, Yuan H, Gong Y, Wang G. Circ_0003266 sponges miR-503-5p to suppress colorectal cancer progression via regulating PDCD4 expression. BMC Cancer (2021) 21(1):284. doi: 10.1186/s12885-021-07997-0
127. Yu C, Li D, Yan Q, Wang Y, Yang X, Zhang S, et al. Circ_0005927 inhibits the progression of colorectal cancer by regulating miR-942-5p/BATF2 axis. Cancer Manag Res (2021) 13:2295–306. doi: 10.2147/cmar.S281377
128. Lu C, Jiang W, Hui B, Rong D, Fu K, Dong C, et al. The circ_0021977/miR-10b-5p/P21 and P53 regulatory axis suppresses proliferation, migration, and invasion in colorectal cancer. J Cell Physiol (2020) 235(3):2273–85. doi: 10.1002/jcp.29135
129. Yang Z, Zhang J, Lu D, Sun Y, Zhao X, Wang X, et al. Hsa_circ_0137008 suppresses the malignant phenotype in colorectal cancer by acting as a microRNA-338-5p sponge. Cancer Cell Int (2020) 20:67. doi: 10.1186/s12935-020-1150-1
130. Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin T, et al. Circ-SMARCA5 suppresses colorectal cancer progression via downregulating miR-39-3p and upregulating ARID4B. Dig Liver Dis (2020) 52(12):1494–502. doi: 10.1016/j.dld.2020.07.019
131. Wang J, Luo J, Liu G, Li X. Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem Biophys Res Commun (2020) 527(2):503–10. doi: 10.1016/j.bbrc.2020.03.165
132. Zhang Y, Zhang Z, Yi Y, Wang Y, Fu J. CircNOL10 acts as a sponge of miR-135a/b-5p in suppressing colorectal cancer progression via regulating KLF9. Onco Targets Ther (2020) 13:5165–76. doi: 10.2147/ott.S242001
133. Li Z, Yao H, Wang S, Li G, Gu X. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res (2020) 39(1):160. doi: 10.1186/s13046-020-01642-7
134. Huang X, Shen X, Peng L, Mai W, Wan Y, Zhang H. CircCSNK1G1 contributes to the development of colorectal cancer by increasing the expression of MYO6 via competitively targeting miR-455-3p. Cancer Manag Res (2020) 12:9563–75. doi: 10.2147/cmar.S262007
135. Wen T, Wu H, Zhang L, Li K, Xiao X, Zhang L, et al. Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer. Anticancer Drugs (2021) 32(1):22–33. doi: 10.1097/cad.0000000000000992
136. Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond) (2019) 133(10):1197–213. doi: 10.1042/cs20190286
137. Shen T, Cheng X, Liu X, Xia C, Zhang H, Pan D, et al. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif Cells Nanomed Biotechnol (2019) 47(1):4038–45. doi: 10.1080/21691401.2019.1669620
138. Yang G, Zhang T, Ye J, Yang J, Chen C, Cai S, et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag Res (2019) 11:6499–509. doi: 10.2147/cmar.S203137
139. Cui W, Dai J, Ma J, Gu H. circCDYL/microRNA-105-5p participates in modulating growth and migration of colon cancer cells. Gen Physiol Biophys (2019) 38(6):485–95. doi: 10.4149/gpb2019037
140. Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. BioMed Pharmacother (2018) 98:775–82. doi: 10.1016/j.biopha.2018.01.015
141. Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun (2018) 503(2):870–5. doi: 10.1016/j.bbrc.2018.06.089
142. Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res (2018) 51(12):e7811. doi: 10.1590/1414-431x20187811
143. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. Cir-ITCH plays an inhibitory role in colorectal cancer by regulating the wnt/β-catenin pathway. PloS One (2015) 10(6):e0131225. doi: 10.1371/journal.pone.0131225
144. Ruan H, Deng X, Dong L, Yang D, Xu Y, Peng H, et al. Circular RNA circ_0002138 is down-regulated and suppresses cell proliferation in colorectal cancer. BioMed Pharmacother (2019) 111:1022–8. doi: 10.1016/j.biopha.2018.12.150
145. Peng C, Tan Y, Yang P, Jin K, Zhang C, Peng W, et al. Circ-GALNT16 restrains colorectal cancer progression by enhancing the SUMOylation of hnRNPK. J Exp Clin Cancer Res (2021) 40(1):272. doi: 10.1186/s13046-021-02074-7
146. Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer (2020) 19(1):13. doi: 10.1186/s12943-020-1139-3
147. Xing L, Xia M, Jiao X, Fan L. Hsa_circ_0004831 serves as a blood-based prognostic biomarker for colorectal cancer and its potentially circRNA-miRNA-mRNA regulatory network construction. Cancer Cell Int (2020) 20(1):557. doi: 10.1186/s12935-020-01651-8
148. Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. BioMed Pharmacother (2019) 112:108611. doi: 10.1016/j.biopha.2019.108611
149. Ye DX, Wang SS, Huang Y, Chi P. A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int (2019) 19:276. doi: 10.1186/s12935-019-0995-7
150. Zhang X, Zhao Y, Kong P, Han M, Li B. Expression of circZNF609 is down-regulated in colorectal cancer tissue and promotes apoptosis in colorectal cancer cells by upregulating p53. Med Sci Monit (2019) 25:5977–85. doi: 10.12659/msm.915926
151. Tu FL, Guo XQ, Wu HX, He ZY, Wang F, Sun AJ, et al. Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am J Transl Res (2020) 12(1):281–91.
152. Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun (2018) 497(1):122–6. doi: 10.1016/j.bbrc.2018.02.036
153. Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet (2019) 10:1096. doi: 10.3389/fgene.2019.01096
154. Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y, et al. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids (2019) 18:375–87. doi: 10.1016/j.omtn.2019.08.024
155. Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol (2020) 10:982. doi: 10.3389/fonc.2020.00982
156. Li J, Ni S, Zhou C, Ye M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res (2018) 10:2777–84. doi: 10.2147/cmar.S172388
157. Tian J, Xi X, Wang J, Yu J, Huang Q, Ma R, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res (2019) 11:5413–23. doi: 10.2147/cmar.S199436
158. Wang J, Wang T, Hu S, Li J, Ni C, Ye M. Hsa_circ_0043278 inhibits tumorigenesis and is downregulated in colorectal cancer. Cancer Manag Res (2021) 13:965–75. doi: 10.2147/cmar.S289775
159. He B, Chao W, Huang Z, Zeng J, Yang J, Luo D, et al. Hsa_circ_001659 serves as a novel diagnostic and prognostic biomarker for colorectal cancer. Biochem Biophys Res Commun (2021) 551:100–6. doi: 10.1016/j.bbrc.2021.02.121
160. Mohammadi D, Zafari Y, Estaki Z, Mehrabi M, Moghbelinejad S. Evaluation of plasma circ_0006282 as a novel diagnostic biomarker in colorectal cancer. J Clin Lab Anal (2022) 36(1):e24147. doi: 10.1002/jcla.24147
161. Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal (2018) 32(5):e22379. doi: 10.1002/jcla.22379
162. Tang J, Zhang C, Huang Y, Wang L, Xu Z, Zhang D, et al. CircRNA circ_0124554 blocked the ubiquitination of AKT promoting the skip lymphovascular invasion on hepatic metastasis in colorectal cancer. Cell Death Dis (2021) 12(3):270. doi: 10.1038/s41419-021-03565-3
163. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids (2019) 16:118–29. doi: 10.1016/j.omtn.2019.02.005
Keywords: circRNAs, colorectal cancer, biomarker, cancer therapy, cancer diagnosis
Citation: Fang G, Xu D, Zhang T, Wang G, Qiu L, Gao X and Miao Y (2023) Biological functions, mechanisms, and clinical significance of circular RNA in colorectal cancer. Front. Oncol. 13:1138481. doi: 10.3389/fonc.2023.1138481
Received: 05 January 2023; Accepted: 20 February 2023;
Published: 06 March 2023.
Edited by:
Paul Ruff, University of the Witwatersrand, South AfricaReviewed by:
Clem Penny, University of the Witwatersrand, South AfricaCopyright © 2023 Fang, Xu, Zhang, Wang, Qiu, Gao and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchang Miao, bHlnZXlteWNAMTYzLmNvbQ==; Xuzhu Gao, YWxleGd3YW5AMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.