- 1Blood Diseases and Cell Therapies unit, Bone Marrow Transplant Unit, “ASST-Spedali Civili” Hospital of Brescia, Department of Clinical and Experimental Sciences, University of Brescia, Brescia, Italy
- 2Stem Cell Laboratory, Section of Hematology and Blood Coagulation, Clinical Chemistry Laboratory, Diagnostics Department, ASST Spedali Civili of Brescia, Brescia, Italy
- 3Centro di Ricerca Emato-oncologico AIL (CREA) , “ASST-Spedali Civili” Hospital of Brescia, Brescia, Italy
- 4Department of Hematology, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, University of Milan, Milan, Italy
- 5Department of Laboratory Diagnostics, ASST Spedali Civili, Brescia, Italy
Background: Minimal residual disease (MRD) monitoring is an important tool to optimally address post-transplant management of acute myeloid leukemia (AML) patients.
Methods: We retrospectively analyzed the impact of bone marrow CD34+ molecular chimerism and WT1 on the outcome of a consecutive series of 168 AML patients submitted to allogeneic stem cell transplantation.
Results: The cumulative incidence of relapse (CIR) was significantly lower in patients with donor chimerism on CD34+ cells ≥ 97.5% and WT1 < 213 copies/ABL x 10^4 both at 1st month (p=0.008 and p<0.001) and at 3rd month (p<0.001 for both). By combining chimerism and WT1 at 3rd month, 13 patients with chimerism < 97.5% or WT1 > 213 showed intermediate prognosis. 12 of these patients fell in this category because of molecular chimerism < 97.5% at a time-point in which WT1 was < 213.
Conclusions: Our results confirm that lineage-specific molecular chimerism and WT1 after allo-SCT (1st and 3rd month) are useful MRD markers. When considered together at 3rd month, CD34+ molecular chimerism could represent an earlier predictor of relapse compared to WT1. Further studies are necessary to confirm this preliminary observation.
Introduction
Minimal residual disease (MRD) monitoring is crucial for the management of patients with acute myeloid leukemia (AML) (1–3). Two assays are currently available: multiparametric flow cytometry (MFC) on the leukemia associated immunophenotype (LAIP) and quantitative RT-qPCR on genes known to be mutated or over-expressed in a subgroup of AML (e.g. FlLT3-ITD, NPM1 mutation, CBF-fusion transcripts, WT1 gene,…) (4, 5). Each of these two assays is associated with different specificity, sensitivity and accuracy, and, with the exception of RT-qPCR on NPM1 mutation, no conclusive data are available on the superiority of one test over the other (4, 6). Nevertheless, several studies confirmed the role of MRD monitoring after induction/consolidation, irrespective of the methods used and the threshold adopted, in order to measure the depth of response during the whole treatment program (3, 4). In particular, it has been suggested that MRD monitoring should be considered as a dynamic event, suggesting that AML risk may be refined during the treatment program (3, 4). Focusing on this issue, we reported how bone marrow (BM) LAIP <0.2% and BM-WT1 < 121 copies/ABLx10^4 after first consolidation were associated with improved outcome; moreover, after 1st intensification cycle, peripheral blood (PB) WT1 < 16 copies/ABLx10^4 was significantly correlated with a better prognosis (3). The issue of MRD monitoring is a crucial step in the path to cure of AML patients, especially in low-intermediate ELN risk categories, for which firstline allogeneic stem cell transplantation (Allo-SCT) in case of MRD persistence is a mainstay of good clinical practice (1–4).
Moreover, MRD detection before allo-SCT is very important to guide the intensity of transplant conditioning regimen (7–10). Then, the issue of MRD detection and monitoring after allo-SCT is particularly relevant, since early detection of residual disease may allow a pre-emptive treatment approach, including not only the early immunosuppression withdrawal and donor lymphocytes infusions (DLI), but also the introduction of new drugs such as hypomethylating agents (HMA), venetoclax, and tyrosine-kinase inhibitors (11–13). Although several studies have explored this topic, the methods and timepoints for the detection of patients at high risk of relapse are still a matter of debate (1–4). In particular, besides their limitations in terms of sensitivity and specificity, and the lack of prospective, controlled data, both MFC and RT-qPCR on selected gene targets are applicable in no more than 30-40% of the patients after allo-SCT (4). As a consequence, WT1 has been suggested as a universal marker of MRD monitoring after allo-SCT, as its expression, although with low specificity, is increased in more than 80% of AML at diagnosis (5, 10, 13).
In this scenario, considering that AML arises from the hematopoietic stem cell, and that more than 90% of AML blasts express CD34 antigen, an option to monitor if allo-SCT has been able to cancel autologous hemopoiesis is the assessment of molecular chimerism on CD34+ cells (14–17). Both short tandem repeat analysis and single nucleotide polymorphism analysis by RT-qPCR have been suggested to be potentially useful tools to measure the degree of donor hematopoiesis. Thus, it may be considered as a surrogate marker of MRD, which can be associated with a high probability of disease recurrence (14–17). Several studies have confirmed that lineage-specific molecular chimerism is a reliable marker of MRD and relapse risk (14–17), but the interplay between CD34+ chimerism and other markers of MRD (e.g., leukemic blasts detection with MFC or WT1) possibly associated with MRD persistence is poorly understood and under-studied (18, 19).
With this background, we analyzed a cohort of 168 AML patients consecutively allotransplanted in our Institution between December 2015 and January 2022, for whom at least one between BM-CD34+ chimerism or BM-WT1 level was available at 1 and 3 months after allo-SCT. The primary endpoint of this retrospective analysis on these two tests was to describe their accuracy in measuring the risk of relapse and their interplay in the definition of patients’ prognosis.
Patients and methods
From December 2015 to January 2022, a total of 191 AML patients were consecutively submitted to allo-SCT in our Institution. 168 out of these transplants (88%) are included in the present analysis, as they represent a consecutive series for which data on lineage specific molecular chimerism (CD34+) and/or molecular monitoring of WT1 gene are available at 1st and/or 3rd month after transplant. All patients included in this analysis provided informed consent for data registration in the PROMISE database, in which clinical and biological data are collected. Additional data were extracted from the revision of the clinical charts of each patient, including both the transplant phase and the subsequent follow up. The study was conducted in compliance with current national and European legislation on clinical trials, in accordance with the Declaration of Helsinki and the principles of good clinical practice.
Lineage-specific chimerism and WT1 monitoring
According to our guidelines, molecular chimerism assessment on BM-CD34+ cells was planned at months 3, 6, 9, 12, 18, and 24 after allo-SCT. From 2020 we implemented another timepoint of assessment at day +30 after allo-SCT.
CD34+ cells were isolated from bone marrow using CD34 MicroBeads human (Miltenyi Biotec, Bergisch Gladbach, Germany) following manufacturer protocol. Briefly, cells were incubated with 100 µL of CD34 MicroBeads and 100 µL of FcR Blocking Reagent for 30 minutes at 4°C, washed, resuspended in 500 µL buffer and applied onto one-step, semiautomated MACS device, AutoMACS (Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of cellular subsets post-separation was determined by FACS analysis (BD FACSCanto™ II) and BD FACSDiva software (BD Biosciences, San Jose, CA). Genomic DNA obtained after CD34+ selection from BM samples was extracted using mini blood kit (QIAGEN, Valencia, CA), following the manufacturer instructions. Validation of the CD34-enrichment was performed comparing the chimerism percentage of CD34+ and chimerism percentage of MNC between groups by 2-sided Student t test (continuous variables with normal distribution). P<.05 was considered significant. Calculations were conducted in Prism 5 (GraphPad, La Jolla, CA). Comparative statistical analysis showed significant difference (P= .0008) and validation of the method. The AmpFLSTR® Identifiler® Plus PCR Amplification Kit (Life Technologies Inc., Foster City, CA) containing 15 polymorphic STR (short tandem repeat) loci and the amelogenin marker was used to evaluate chimerism status in patients post transplant (20). Genomic DNA obtained after CD34+ selection (Automacs System -Miltenyi) from bone marrow samples was extracted using mini blood kit (QIAGEN, Valencia, CA), following the manufacturer instructions. Serial dilutions were created by mixing DNA samples with standardized mixed chimeric samples, in a range between 0% and 100%. The level of sensitivity of this test was 2.5%. The data were analyzed by GeneMapper®ID v3.2 software calculating the amount of donor’s DNA.
All the patients with AML were evaluated for WT1 expression level at diagnosis. Focusing on this series, the time-points of WT1 evaluation on BM were the same as for chimerism and its assessment was performed by Q-PCR (protocol: Ipsogen WT1 ProfileQuant) according to the ELN method as previously published (21). The cut-off for positive samples, according to the sensitivity of our platform and available literature, was ≥ 213 WT1 copies/ABL1x10^4 on BM (21).
Statistical analysis
Descriptive statistics was employed for summarizing patients characteristics. Categorical variables were presented as numbers and percentages, continuous variables as median and range, respectively. Chi-Squared or Fisher’s Exact test and the Wilcoxon Rank-sum or Kruskal-Wallis tests were used to test differences among subgroups, as appropriate. Median survival with 95% confidence interval (95%CI) was calculated according to reverse Kaplan-Meier method. Overall survival (OS) was measured from the time of transplant to the date of last follow-up or death, cumulative incidence of relapse (CIR), considering non-relapse mortality (NRM) as a competitive event, was carried out according to the Fine-Gray model. Log-rank and Gray tests were employed to verify differences among the different groups. One-month and 3-month landmark analyses were conducted in order to evaluate association between WT1 (cut-off 213 copies/ABL1x10^4) and donor chimerism (cut-off 97.5%) on subsequent CIR. Sensitivity, specificity and diagnostic accuracy of post-transplant chimerism and WT1 values in predicting relapse occurrence were also measured. Statistical analysis was performed with EZR (version 1.61), as previously described (22).
Results
Table 1 reports the most important clinical and transplant characteristics of these patients. The median age at transplant was 56.5 years (23.8-74.1), and patients were equally distributed between sexes. The disease risk index (DRI) was intermediate/high in two thirds of the cases, and 49.4% of the patients received the transplant in first complete remission (CR) following a myeloablative conditioning in 55.4% of the cases. Peripheral blood stem cells (PBSC) were used in the 76.2% of the cases, and donor was other than a sibling in more than 50% of the transplants (matched unrelated donor in 45.8% and haploidentical in 18.5% of the cases). No significant differences were detected comparing the same characteristics, dividing patients according to the percentage of donor chimerism on CD34+ cells (above or below 97.5%) and WT1 levels (above or below 213 copies/ABL1x10^4) both at 1st and 3rd month (data not shown).
Overall, the total number of patients for whom WT1 could be considered for MRD monitoring (at diagnosis > 213) was 125/168 (74%). Molecular chimerism on CD34+ cells and WT1 at 1st month was available on 36 (21%) and 45 patients (27%), respectively. In 72.2% of the cases (26/36) the percentage of donor CD34+ cells was above 97.5%. Focusing on WT1, its level was < 213 copies/ABL1x10^4 in 24.4% of the cases.
Moving to the 3rd month, molecular chimerism on CD34+ cells and WT1 were available on 99 (53%) and 125 patients (67%), respectively. Donor chimerism ≥ 97.5% was detected in 63 patients (65.6%) and WT1 levels < 213 copies/ABL1x10^4 in 103 cases (82.4%).
Additional molecular markers of disease persistence during follow up were FLT3-ITD (2 cases at 1st month and 4 cases at 3rd month) and NPM1A (2 cases at 1st month and 2 cases at 3rd month). All the patients with positive FlLT3-ITD MRD had mixed chimerism on CD34+ and WT1 level above 213 copies/ABL1x10^4, experienced hematological relapse and did not survive. The 2 patients with NPM1A positive residual disease showed complete donor chimerism and WT1 level < 213 copies/ABL1x10^4 and are alive in continuous complete remission at last follow up.
Cumulative incidence of relapse and overall survival
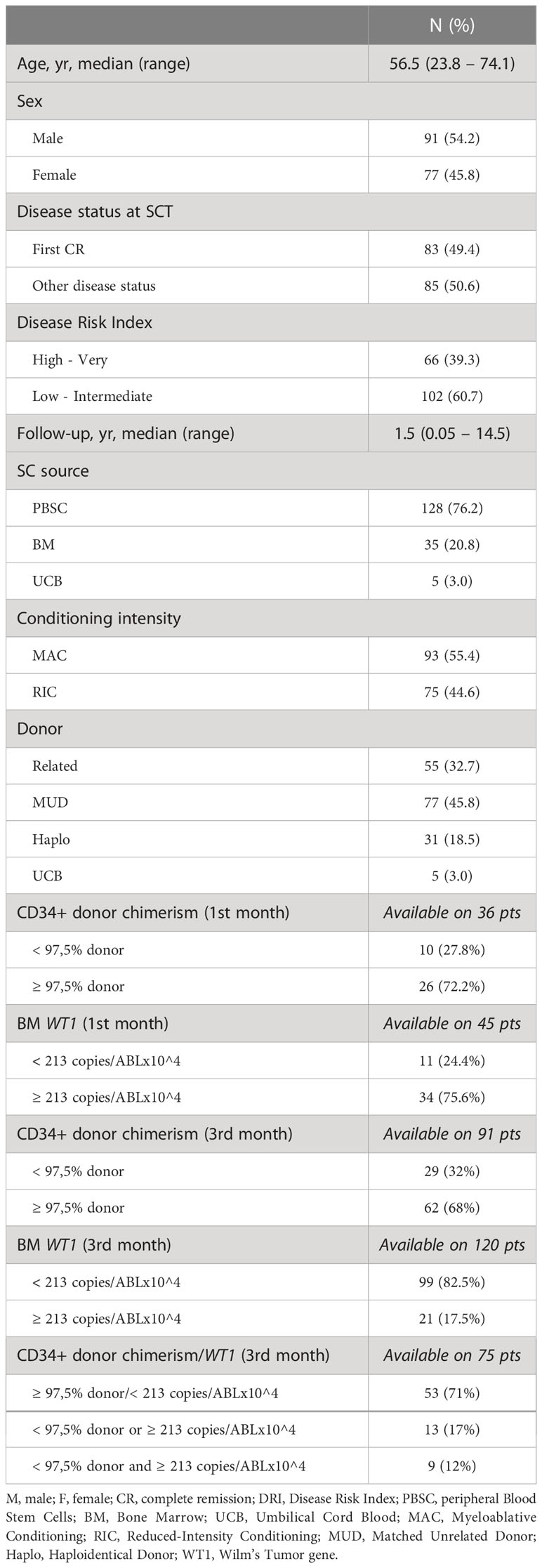
After a median follow up of 4.5 years (range 3,5-5,0), the 1, 3, and 5 years cumulative incidence of relapse (CIR) was 26.9% (95% CI 20.3-34.0), 46.8% (95% CI 38.6-54.4) and 50.8% (95% CI 42.2-58.9), respectively (Figure 1A). This translated into an overall survival (OS) at 1,3 and 5 years of 67.3% (95% CI 59.4-74), 50.9% (95% CI 42.6-58.6), and 43.2% (95% CI 34.8-51.3), respectively (Figure 1B).
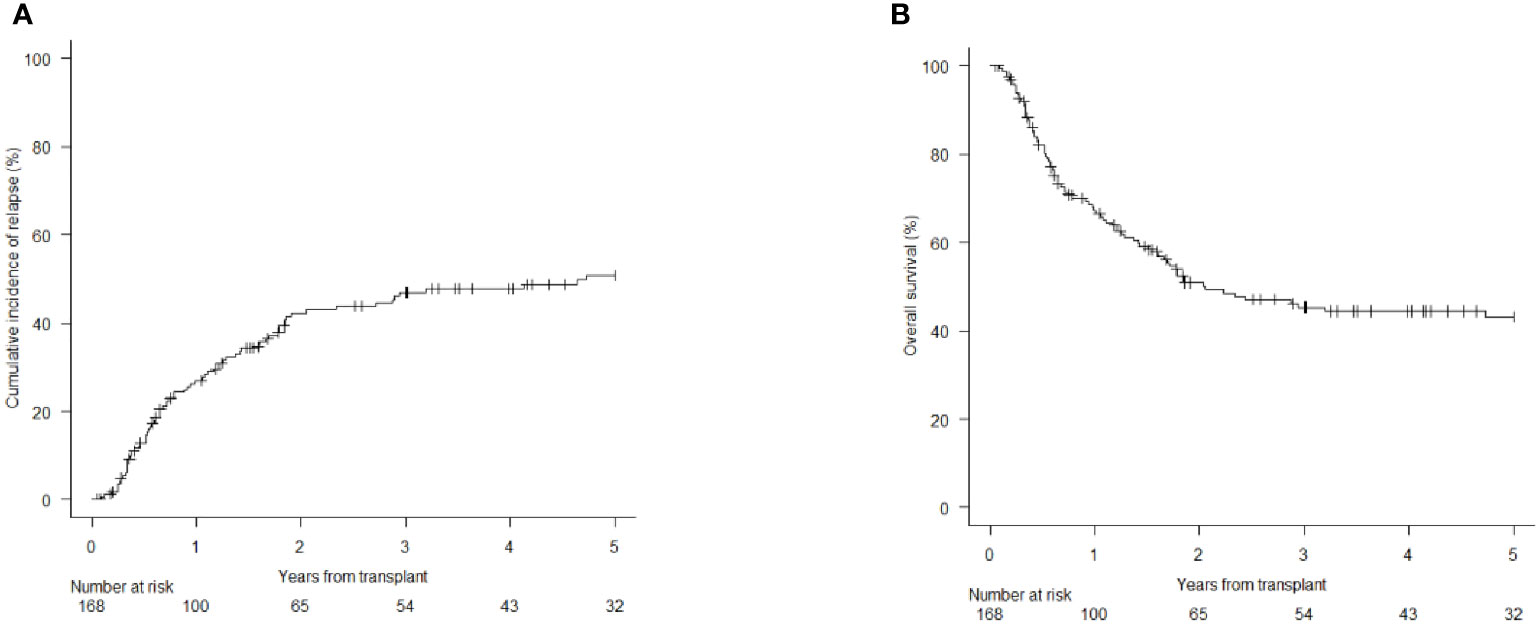
Figure 1 Cumulative Incidence of Relapse (CIR) and Overall Survival (OS) of the 168 AML patients included in this analysis. [CIR at 1, 3 and 5 years; 26.9% (95% CI 20.3-34.0), 46.8% (95% CI 38.6-54.4) and 50.8% (95% CI 42.2-58.9) (A); OS at 1, 3 and 5 years: 67.3% (95% CI 59.4-74), 50.9% (95% CI 42.6-58.6) and 43.2% (95% CI 34.8-51.3) (B)].
At 1st month, both donor chimerism on CD34+ cells ≥ 97.5% and WT1 levels below 213 copies/ABL1x10^4 significantly correlated with CIR (chimerism: 13% vs 70% at 1 year; p=0.008 – Figure 2A; WT1: 31.8% vs 81.8%; p=0.03 – Figure 2B) and OS (chimerism: 81.8% vs 9.5% at 1 year; p<0.001 – Figure 2C; WT1: 54.3% vs 18.2%; p<0.05 – Figure 2D).
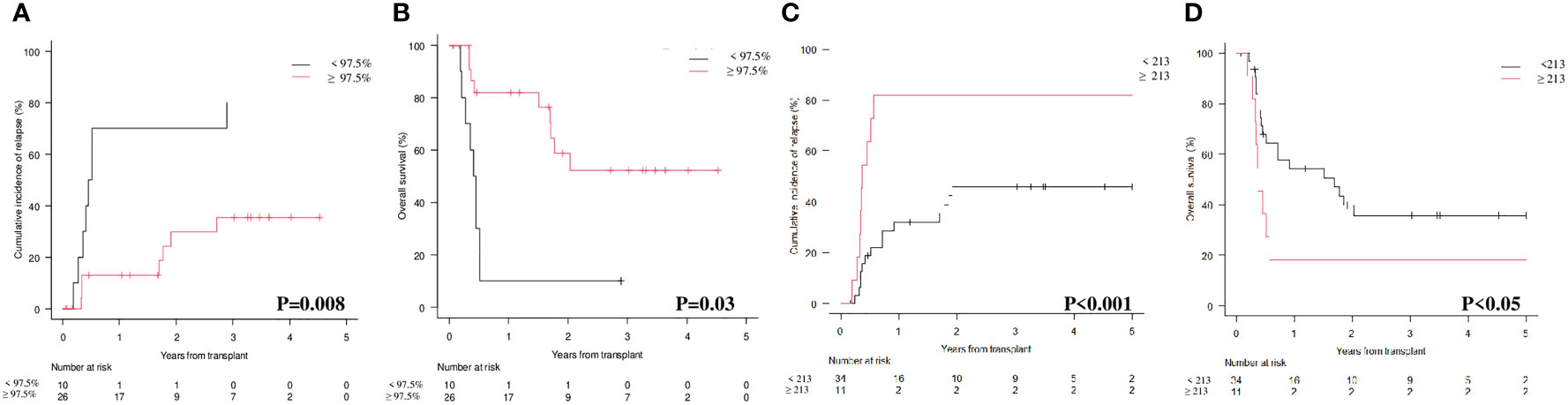
Figure 2 Cumulative Incidence of Relapse (CIR) and Overall Survival (OS) according to molecular chimerism on CD34+ cells and WT1 levels at 1st month. (A) CIR at 1 year CD34+≥97.5% vs <97.5% donor: 13% (95% CI 3.3-29.7) vs 70% (95% CI 32.9.89.2). (B) OS at 1 year CD34+≥97.5% vs <97.5% donor: 81.8% (95% CI 58.5-92.8) vs 9.5% (95% CI 5-35.8). (C) CIR at 1 year WT1 < 213 copies/ABL1×10^4≥213 copies/ABL1×10^4: 31.8% (95%CI 16.7-48.2) vs 81.8% (95%CI 44.7-95.1). (D) OS at 1 year WT1 < 213 copies/ABL1×10^4≥213 copies/ABL1×10^4: 54.3% (95%CI 35.3-69.9) vs 18.2% (95%CI 2.8-44.2).
As reported in Figures 3A, D, the results at 3rd month confirmed the predictive value of the two markers on CIR and OS. In particular, the 1 and 2 years CIR for patients with donor chimerism on CD34+ cells ≥ 97.5% vs those with donor chimerism < 97.5% was 5.3% and 26% vs 61% and 74%, respectively (Figure 3A; p<0.001). This translated into a 1 and 2 years OS of 93% and 72.4% vs 44.2 and 25.4%, respectively (Figure 3B; p<0.001). Moving to WT1 at 3rd month and comparing patients with a level below or above 213 copies/ABL1x10^4, we observed that the CIR at 1 and 2 years was 12.6% and 28.6% vs 80.9% and 97.3%, respectively (Figure 3C; p<0.001). As expected, 1 and 2 years OS was 83.3% and 65.8% vs 20.3% and 6.7% (Figure 3D; p<0.001).
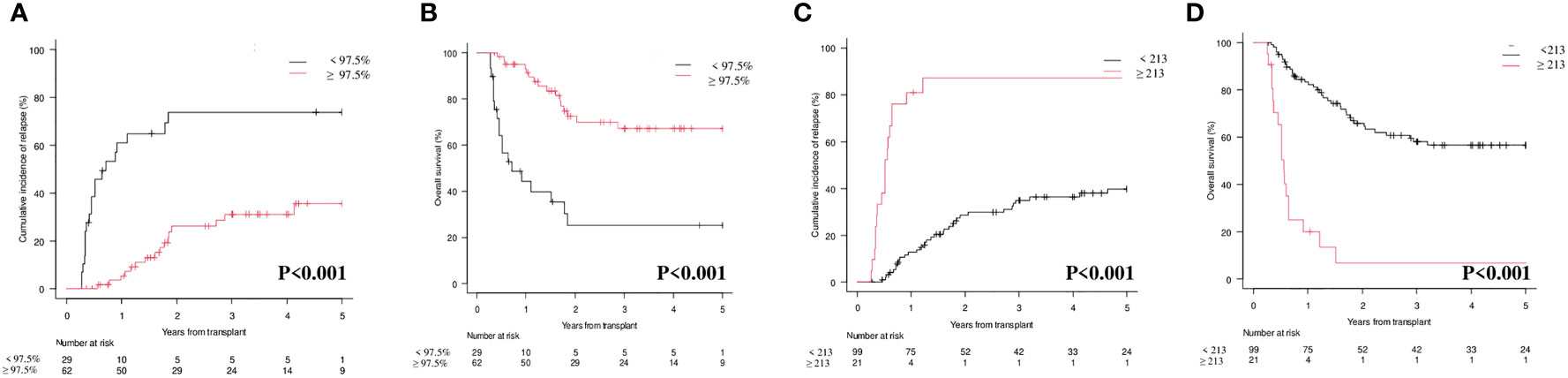
Figure 3 Cumulative Incidence of Relapse (CIR) and Overall Survival (OS) according to molecular chimerism on CD34+ cells and WT1 levels at 3rd month. (A) CIR at 1 year CD34+≥97.5% vs <97.5% donor: 5.3% (95% CI 1.4-13.4) vs 61% (95% CI 40.3-76.4). (B) OS at 1 year CD34+≥97.5% vs <97.5% donor: 93.1% (95% CI 82.6-97.3) vs 44.2% (95% CI 25-61.9). (C) CIR at 1 year WT1 < 213 copies/ABL1×10^4≥213 copies/ABL1×10^4: 12.6% (95%CI 6.9-20.2) vs 80.9% (95%CI 56.9-92-4). (D) OS at 1 year WT1 < 213 copies/ABL1×10^4≥213 copies/ABL1×10^4: 83.3% (95%CI 74.2-89.4) vs 20.1% (95%CI 6.2-39.5).
The sensitivity, specificity, and accuracy of molecular chimerism on CD34+ cells at 3rd month was 53.3% (95% CI 34.3-71.7), 83.7% (95% CI 70.3-92.7), and 72.2% (85% CI 60.9-81.7). For WT1 at 3rd month we observed a sensitivity of 33.3% (95% CI 17.3-52.8), a specificity of 98% (95% CI 89.1-99.9) and an accuracy of 73.4% (95% CI 62.3-82.7). We then looked at the sensitivity, specificity and accuracy of the combination of donor chimerism and WT1 levels at 3rd month and we found that they were 53.3% (95% CI 34.3-71.7), 81.6% (95% CI 68.0-91.2) and 70.9% (95% CI 59.6-80.6).
Interestingly, by combining CD34+ donor chimerism </≥ 97.5% and WT1 </≥ 213 copies/ABL1x10^4, three categories could be identified with significantly different prognosis both on CIR (p<0.001; Figure 3A) and on OS (p<0.001; Figure 3B): (i) donor chimerism ≥ 97.5% and WT1 < 213 (53 patients) [CIR at 1 year 4.1% (95% CI 0.8-12.4) and OS at 1 year 94.2% (95% CI 83.0-98.1)]; (ii) donor chimerism < 97.5 or WT1 ≥ 213 (13 patients) [CIR at 1 year 30.7% (95% CI 9.5-55.4) and OS at 1 year 76.9% (95% CI 44.2-91.9)]; (iii) donor chimerism < 97.5% and WT1 ≥ 213 (9 patients) [CIR at 1 year 100% (95% CI NA) and OS at 1 year 0% (95% CI NA)]. Moreover, 12/13 patients included in the “intermediate” group (donor chimerism < 97.5% or WT1 ≥ 213) fell in this category because of donor chimerism < 97.5% and only 1 patient because of WT1 ≥ 213 copies/ABL1x10^4.
Pre-emptive treatment following the detection of CD34+ donor chimerism < 97.5% and/or WT1 ≥ 200 copies/ABL1x10^4
Overall, 43 and 66 patients had at least one detection of donor CD34+ chimerism < 97.5% and/or WT1 levels ≥ 213 copies/ABL1x10^4 at 1st and/or 3rd month. Whenever clinically possible (no graft versus host disease and no active infections) these patients were managed with early tapering of immunosuppression. If clinical and hematological conditions were permissive, additional pre-emptive therapy was administered (11 patients). Results in the different subgroups are reported in Supplementary Table 1.
Discussion
Minimal Residual Disease (MRD) monitoring is crucial in the management of AML patients, and is a dynamic process during all the treatment plan, including the post-transplant phase (1–17, 23, 24).
Our study clearly shows that both lineage-specific molecular chimerism and WT1 levels are useful markers for MRD detection and monitoring after allo-SCT in AML, either alone or in combination. At 1st month after allo-SCT, lineage-specific molecular chimerism (Figure 2A; p=0.008) and WT1 levels (Figure 2C; p<0.001) were significantly correlated with the CIR. This was also confirmed at 3rd month (Figure 3A; p<0.001 and Figure 3B; p<0.001). Interestingly, by combining molecular chimerism and WT1 at 3rd month, we identified three categories of patients with different prognosis: (i) donor chimerism ≥ 97.5% and WT1 < 213 (53 patients); (ii) donor chimerism < 97.5 or WT1 ≥ 213 (13 patients); (iii) donor chimerism < 97.5% and WT1 ≥ 213 (9 patients). The lowest CIR and the longest OS were observed in patients with donor CD34+ ≥ 97.5% and WT1 < 213 copies/ABL1x10^4 (Figure 4A; p<0.001 and Figure 4B; p< 0.001). This strongly reinforces the significance of these two tests for MRD monitoring after allo-SCT. Notably, focusing on the intermediate category (donor CD34+ chimerism < 97.5% or WT1 ≥ 213 copies/ABL1x10^4), we observed that nearly all of these patients (12/13) were included in this group because of mixed donor chimerism, at a timepoint in which WT1 levels were still within the normal range. This suggests that molecular chimerism may detect persistence of MRD earlier than WT1. In other words, once WT1 gets positive, disease relapse is highly likely to occur in a very short time-frame. Even if numbers are small to be conclusive, we think that these results reinforces the usefulness of both methods for MRD monitoring after allo-SCT and suggests that lineage-specific molecular chimerism may an earlier predictor of relapse than WT1. On the other hands, a recently published paper (18) suggests that day +100 MRD positivity is a stronger predictor of relapse after allo-SCT compared to mixed chimerism. Notably, the series published by Klyuchinov and Colleagues includes intermediate-risk AML only, and MRD monitoring was performed with MFC and RT-qPCR on NPM1A, of which at least NPM1A is extremely disease-specific as a marker of MRD. These two aspects may be responsible for the different results.

Figure 4 Cumulative Incidence of Relapse (CIR) and Overall Survival (OS) according to combination of molecular chimerism on CD34+ cells and WT1 levels at 3rd month. (A) CIR at 1 year CD34+≥97.5% and WT1 < 213 copies/ABL1×10^4 vs CD34+ <97.5% WT1 ≥ 213 copies/ABL1×10^4 vs CD34+ <97.5% or WT1 ≥ 213 copies/ABL1×10^4: 4.1% (95% CI 0.8-12.4) vs 30.7% (95% CI 9.5-55.4) vs 100% (95% CI NA). (B) OS at 1 year CD34+≥97.5% and WT1 < 213 copies/ABL1×10^4 vs CD34+ <97.5% or WT1 ≥ 213 copies/ABL1×10^4 vs CD34+ <97.5% and WT1 ≥ 213 copies/ABL1×10^4: 94.2% (95% CI 83.0-98.1) vs 76.9% (95% CI 44.91.9)] vs 0% (95% CI NA).
We then looked at the use of pre-emptive therapy guided by one or both of the MRD markers (chimerism and/or WT1). Pre-emptive treatment was administered in a minority of patients (n=11). As a consequence results are anecdotal and no conclusions can be drawn. Interestingly, the higher response rate (in terms of conversion to full donor chimerism or increase in the percentage of donor CD34+ cells) was observed in patients with mixed chimerism at 3rd months (n=29). In this group, 7 patients (24%) received a pre-emptive approach with either HMA alone or in combination with venetoclax/DLI or DLI alone, 4/7 (57%) patients achieved a response and at the last follow up 9/29 (31%) patients were alive.
The relatively small number of patients included in our analysis may hamper drawing final conclusions. Nevertheless, our study confirms the prognostic value of lineage-specific chimerism at very early timepoints (1st and 3rd month) and suggests that patients at high risk of relapse may show mixed chimerism before positivity of WT1 as a marker of MRD. The aim of this study was not to compare lineage-specific molecular chimerism and WT1, but our results indirectly suggest that chimerism on CD34+ cells could be an earlier predictor of relapse. The small number of cases with available data at day +30 depends on the fact that early assessment of chimerism and MRD monitoring were implemented only from 2020 in our Institution and suggests caution both in results interpretation and conclusion drawing.
Overall, our data are in line with other published papers, highlighting the role of both lineage specific molecular chimerism and WT1 as markers of MRD after allo-SCT (14–19, 24–28). The issue of the superiority of molecular chimerism on CD34+ cells over other methods for leukemia relapse prediction is still unsolved. Some data suggest that WT1 could be more sensitive than lineage specific molecular chimerism (29) or that the two methods are concordant (30), also when analyzed in specific cellular sub-types, such as CD3 negative mononuclear cells (31). On the other hand, in the study by Rossi and Colleagues a higher concordance between positive results from MFC and WT1 was detected among patients with mixed rather than complete chimerism (32). Several issues are still open, such as the role of the source used for the detection of both chimerism and MRD. If it is true that PB may be used for MRD monitoring in AML and has some advantages over bone marrow (13), there are no conclusive data regarding this issue when we consider lineage-specific molecular chimerism and different sources are used in the published papers, according to each Center’s guideline (15, 26–32). Interestingly, as suggested by Gambacorta and Colleagues, PB may allow a tighter follow up of the patients and may allow higher specificity in case of positive samples. The Authors give an intriguing explanation for this, speculating that BM detects a significant “background noise” possibly related to the aspiration of host stromal cells (15). Moreover, new technologies such as digital PCR (dPCR) or next generation sequencing (NGS) may be a useful tool to increase both the specificity and sensitivity of lineage-specific molecular chimerism. Further prospective studies are thus warranted in order to clarify if lineage-specific molecular chimerism is superior to WT1 to identify imminent relapse, which time-points are more reliable for an optimal prediction of disease recurrence, if PB should be preferred to BM and if new technologies may increase the power of molecular chimerism for relapse prevention.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consent to participate in this study.
Author contributions
MM, NP, ArLa, DR designed the study. EM, VR, EB, SB, FR, DaBo and AlLe collected the data. ArLa, AB, FB, MC performed chimerism analysis. DuBr performed molecular analysis on WT1. MM, NP, AlLe, ArLa, SB, FR analyzed the data. MM, NP, ArLa and DR wrote the Manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
Special thanks to Studio Moretto for English revision.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1133418/full#supplementary-material
References
1. Venditti A, Piciocchi A, Candoni A, Melillo L, Calafiore V, Cairoli R, et al. GIMEMA AML1310 trial of risk-adapted, MRD-directed therapy for young adults with newly diagnosed acute myeloid leukemia. Blood (2019) 134(12):935–45. doi: 10.1182/blood.2018886960
2. Walter RB, Ofran Y, Wierzbowska A, Ravandi F, Hourigan CS, Ngai LL, et al. Measurable residual disease as a biomarker in acute myeloid leukemia: theoretical and practical considerations. Leukemia (2021) 35(6):1529–38. doi: 10.1038/s41375-021-01230-4
3. Malagola M, Skert C, Borlenghi E, Chiarini M, Cattaneo C, Morello E, et al. Postremission sequential monitoring of minimal residual disease by WT1 q-PCR and multiparametric flow cytometry assessment predicts relapse and may help to address risk-adapted therapy in acute myeloid leukemia patients. Cancer Med (2016) 5(2):265–74. doi: 10.1002/cam4.593
4. Heuser M, Freeman SD, Ossenkoppele GJ, Buccisano F, Hourigan CS, Ngai LL, et al. 2021 update on MRD in acute myeloid leukemia: a consensus document from the European LeukemiaNet MRD working party. Blood (2021) 138(26):2753–67. doi: 10.1182/blood.2021013626
5. Lazzarotto D, Candoni A. The role of wilms' tumor gene (WT1) expression as a marker of minimal residual disease in acute myeloid leukemia. J Clin Med (2022) 11(12):3306. doi: 10.3390/jcm11123306
6. Gao MG, Ruan GR, Chang YJ, Liu YR, Qin YZ, Jiang Q, et al. The predictive value of minimal residual disease when facing the inconsistent results detected by real-time quantitative PCR and flow cytometry in NPM1-mutated acute myeloid leukemia. Ann Hematol (2020) 99(1):73–82. doi: 10.1007/s00277-019-03861-1
7. Buckley SA, Wood BL, Othus M, Hourigan CS, Ustun C, Linden MA, et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: a meta-analysis. Haematologica (2017) 102(5):865–73. doi: 10.3324/haematol.2016.159343
8. Maffini E, Labopin M, Beelen DW, Kroeger N, Arat M, Wilson KMO, et al. Measurable residual disease (MRD) status before allogeneic hematopoietic cell transplantation impact on secondary acute myeloid leukemia outcome. a study from the acute leukemia working party (ALWP) of the European society for blood and marrow transplantation (EBMT). Bone Marrow Transplant (2022) 57(10):1556–63. doi: 10.1038/s41409-022-01748-w
9. Loke J, Buka R, Craddock C. Allogeneic stem cell transplantation for acute myeloid leukemia: Who, when, and how? Front Immunol (2021) 12:659595. doi: 10.3389/fimmu.2021.659595
10. Guolo F, Di Grazia C, Minetto P, Raiola AM, Clavio M, Miglino M, et al. Pre-transplant minimal residual disease assessment and transplant-related factors predict the outcome of acute myeloid leukemia patients undergoing allogeneic stem cell transplantation. Eur J Haematol (2021) 107(5):573–82. doi: 10.1111/ejh.13694
11. Malagola M, Greco R, Peccatori J, Isidori A, Romee R, Mohty M, et al. Editorial: Strengths and challenges of allo-SCT in the modern era. Front Oncol (2022) 12:850403. doi: 10.3389/fonc.2022.850403
12. Leotta S, Condorelli A, Sciortino R, Milone GA, Bellofiore C, Garibaldi B, et al. Prevention and treatment of acute myeloid leukemia relapse after hematopoietic stem cell transplantation: The state of the art and future perspectives. J Clin Med (2022) 11(1):253. doi: 10.3390/jcm11010253
13. Malagola M, Skert C, Ruggeri G, Turra A, Ribolla R, Cancelli V, et al. Peripheral blood WT1 expression predicts relapse in AML patients undergoing allogeneic stem cell transplantation. BioMed Res Int (2014) 2014:123079. doi: 10.1155/2014/123079
14. Bacher U, Haferlach T, Fehse B, Schnittger S, Kröger N. Minimal residual disease diagnostics and chimerism in the post-transplant period in acute myeloid leukemia. ScientificWorldJournal (2011) 11:310–9. doi: 10.1100/tsw.2011.16
15. Gambacorta V, Parolini R, Xue E, Greco R, Bouwmans EE, Toffalori C, et al. Quantitative PCR-based chimerism in bone marrow or peripheral blood to predict acute myeloid leukemia relapse in high-risk patients: results from the KIM-PB prospective study. Haematologica (2020) 106(5):1480–3. doi: 10.3324/haematol.2019.238543
16. Lindahl H, Vonlanthen S, Valentini D, Björklund AT, Sundin M, Mielke S, et al. Lineage-specific early complete donor chimerism and risk of relapse after allogeneic hematopoietic stem cell transplantation for acute myeloid leukemia. Bone Marrow Transplant (2022) 57(5):753–9. doi: 10.1038/s41409-022-01615-8
17. Ciurea SO, Kothari A, Sana S, Al Malki MM. The mythological chimera and new era of relapse prediction post-transplant. Blood Rev (2022) 30:100997. doi: 10.1016/j.blre.2022.100997
18. Klyuchnikov E, Badbaran A, Massoud R, Fritsche-Friedland U, Janson D, Ayuk F, et al. Post-transplantation multicolored flow cytometry-minimal residual disease status on day 100 predicts outcomes for patients with refractory acute myeloid leukemia. Transplant Cell Ther (2022) 28(5):267.e1–7. doi: 10.1016/j.jtct.2022.01.014
19. Chiusolo P, Metafuni E, Minnella G, Giammarco S, Bellesi S, Rossi M, et al. Day +60 WT1 assessment on CD34 selected bone marrow better predicts relapse and mortality after allogeneic stem cell transplantation in acute myeloid leukemia patients. Front Oncol (2022) 12:994366. doi: 10.3389/fonc.2022.994366
20. Porta F, Comini M, Soncini E, Carracchia G, Maffeis M, Pintabona V, et al. CD34+ stem cell selection and CD3+ T cell add-back from matched unrelated adult donors in children with primary immunodeficiencies and hematological diseases. Transplant Cell Ther (2021) 27(5):426.e1–9. doi: 10.1016/j.jtct.2021.01.020
21. Cilloni D, Renneville A, Hermitte F, Hills RK, Daly S, Jovanovic JV, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol (2009) 27(31):5195–201. doi: 10.1200/JCO.2009.22.4865
22. Polverelli N, Tura P, Battipaglia G, Malagola M, Bernardi S, Gandolfi L, et al. Multidimensional geriatric assessment for elderly hematological patients (≥60 years) submitted to allogeneic stem cell transplantation. a French-Italian 10-year experience on 228 patients. Bone Marrow Transplant (2020) 55(12):2224–33. doi: 10.1038/s41409-020-0934-1
23. Cilloni D, Gottardi E, De Micheli D, Serra A, Volpe G, Messa F, et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia (2002) 16(10):2115–21. doi: 10.1038/sj.leu.2402675
24. Georgi JA, Stasik S, Bornhäuser M, Platzbecker U, Thiede C. Analysis of subset chimerism for MRD-detection and pre-emptive treatment in AML. Front Oncol (2022) 12:841608. doi: 10.3389/fonc.2022.841608
25. Le Bris Y, Costes D, Bourgade R, Guillaume T, Peterlin P, Garnier A, et al. Impact on outcomes of mixed chimerism of bone marrow CD34+ sorted cells after matched or haploidentical allogeneic stem cell transplantation for myeloid malignancies. Bone Marrow Transplant (2022) 57(9):1435–41. doi: 10.1038/s41409-022-01747-x
26. Bornhäuser M, Oelschlaegel U, Platzbecker U, Bug G, Lutterbeck K, Kiehl MG, et al. Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica (2009) 94(11):1613–7. doi: 10.3324/haematol.2009.007765
27. Bendjelloul M, Usureau C, Etancelin P, Saidak Z, Lebon D, Garçon L, et al. Utility of assessing CD3+ cell chimerism within the first months after allogeneic hematopoietic stem-cell transplantation for acute myeloid leukemia. HLA (2022) 100(1):18–23. doi: 10.1111/tan.14557
28. Hoffmann JC, Stabla K, Burchert A, Volkmann T, Bornhäuser M, Thiede C, et al. Monitoring of acute myeloid leukemia patients after allogeneic stem cell transplantation employing semi-automated CD34+ donor cell chimerism analysis. Ann Hematol (2014) 93(2):279–85. doi: 10.1007/s00277-013-1961-4
29. Rautenberg C, Pechtel S, Hildebrandt B, Betz B, Dienst A, Nachtkamp K, et al. Wilms' tumor 1 gene expression using a standardized European LeukemiaNet-certified assay compared to other methods for detection of minimal residual disease in myelodysplastic syndrome and acute myelogenous leukemia after allogeneic blood stem cell transplantation. Biol Blood Marrow Transplant (2018) 24(11):2337–43. doi: 10.1016/j.bbmt.2018.05.011
30. Candoni A, Toffoletti E, Gallina R, Simeone E, Chiozzotto M, Volpetti S, et al. Monitoring of minimal residual disease by quantitative WT1 gene expression following reduced intensity conditioning allogeneic stem cell transplantation in acute myeloid leukemia. Clin Transplant (2011) 25(2):308–16. doi: 10.1111/j.1399-0012.2010.01251.x
31. Bouvier A, Riou J, Thépot S, Sutra Del Galy A, François S, Schmidt A, et al. Quantitative chimerism in CD3-negative mononuclear cells predicts prognosis in acute myeloid leukemia patients after hematopoietic stem cell transplantation. Leukemia (2020) 34(5):1342–53. doi: 10.1038/s41375-019-0624-4
32. Rossi G, Carella AM, Minervini MM, Savino L, Fontana A, Pellegrini F, et al. Minimal residual disease after allogeneic stem cell transplant: a comparison among multiparametric flow cytometry, wilms tumor 1 expression and chimerism status (Complete chimerism versus low level mixed chimerism) in acute leukemia. Leuk Lymphoma (2013) 54(12):2660–6. doi: 10.3109/10428194.2013.789508
Keywords: WT1, allogeneic stem cell transplantation, minimal residual disease (MRD), lineage specific molecular chimerism, pre-emptive therapy
Citation: Malagola M, Polverelli N, Beghin A, Bolda F, Comini M, Farina M, Morello E, Radici V, Accorsi Buttini E, Bernardi S, Re F, Leoni A, Bonometti D, Brugnoni D, Lanfranchi A and Russo D (2023) Bone marrow CD34+ molecular chimerism as an early predictor of relapse after allogeneic stem cell transplantation in patients with acute myeloid leukemia. Front. Oncol. 13:1133418. doi: 10.3389/fonc.2023.1133418
Received: 28 December 2022; Accepted: 20 February 2023;
Published: 06 March 2023.
Edited by:
Francesco Buccisano, University of Rome Tor Vergata, ItalyReviewed by:
Raffaele Palmieri, University of Rome Tor Vergata, ItalyChristian Thiede, Technical University Dresden, Germany
Daniela Cilloni, University of Turin, Italy
Copyright © 2023 Malagola, Polverelli, Beghin, Bolda, Comini, Farina, Morello, Radici, Accorsi Buttini, Bernardi, Re, Leoni, Bonometti, Brugnoni, Lanfranchi and Russo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele Malagola, bWljaGVsZS5tYWxhZ29sYUB1bmlicy5pdA==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Michele Malagola
Michele Malagola Nicola Polverelli
Nicola Polverelli Alessandra Beghin2
Alessandra Beghin2 Mirko Farina
Mirko Farina Enrico Morello
Enrico Morello Vera Radici
Vera Radici Eugenia Accorsi Buttini
Eugenia Accorsi Buttini Simona Bernardi
Simona Bernardi Federica Re
Federica Re Domenico Russo
Domenico Russo