
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 24 February 2023
Sec. Molecular and Cellular Oncology
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1130472
This article is part of the Research TopicPredictive and Prognostic Value of Liquid Biopsy Biomarkers in Metastatic Cancers: from Basic Science, across High Throughput Profiling up to Clinical PracticeView all 8 articles
Cancer consists of a group of diseases with the salient properties of an uncontrolled cell cycle, metastasis, and evasion of the immune response, mainly driven by the genomic instability of somatic cells and the physicochemical environment. Long noncoding RNAs (lncRNAs) are defined as noncoding RNAs with a length of more than 200 nucleotides. LncRNA dysregulation participates in diverse disease types and is tightly associated with patient clinical features, such as age, disease stage, and prognosis. In addition, an increasing number of lncRNAs have been confirmed to regulate a series of biological and pathological processes through numerous mechanisms. The lncRNA epidermal growth factor receptor antisense RNA 1 (EGFR-AS1) was recently discovered to be aberrantly expressed in many types of diseases, particularly in cancers. A high level of EGFR-AS1 was demonstrated to correlate with multiple patient clinical characteristics. More importantly, EGFR-AS1 was found to be involved in the mediation of various cellular activities, including cell proliferation, invasion, migration, chemosensitivity, and stemness. Therefore, EGFR-AS1 is a promising marker for cancer management. In this review, we introduce the expression profile, molecular mechanisms, biological functions, and clinical value of EGFR-AS1 in cancers.
Cancer has been a leading cause of disease-related mortality worldwide in the last decade, with high morbidity and poor functional outcomes (1–5). Despite the existence of accurate biopsy-based diagnosis methods and advances in anticancer drug development, cancer recurrence and poor prognosis still cannot be avoided (6–9). The molecular pathogenesis of various cancers has been persistently investigated and our understanding has expanded, and the related findings help to accurately diagnose cancer and personalize treatment (10–13).
Increasing evidences have shown that noncoding RNAs (ncRNAs) play essential roles in tumor progression (14, 15). Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs with a length of over 200 nucleotides (16–18). An increasing number of lncRNA structures and functions have been discovered in recent years (19, 20). Various lncRNAs have been discovered to be abnormally expressed in many human diseases, especially cancers (21–24). Moreover, emerging studies have revealed lncRNA involvement in various cellular processes, including the cell cycle, differentiation, and chromatin modification (25–28). Functional experiments confirmed that lncRNAs drive their roles through diverse molecular mechanisms, including interactions with proteins, binding to miRNAs, and combining with chromatin-modifying complexes (29–33). In view of their diverse signatures, lncRNAs exhibit a specific advantage in classifying different types of disease as biomarkers for diagnosis, prognosis, and treatment response (34–36).
As a 2.821-kb transcript from the antisense chain of epidermal growth factor receptor (EGFR), lncRNA EGFR antisense RNA 1 (EGFR-AS1) belongs to the family of receptor tyrosine kinases ErbB and is located on human chromosome 7p11.2 (chr7:55179750-55188934 according to hg38) (Figure 1A) (Images in Figure 1A from the GeneCards website, https://www.genecards.org/cgi-bin/carddisp.pl?gene=EGFR-AS1&keywords=EGFR-AS1). Numerous articles have pointed out that the aberrant expression of EGFR-AS1 is involved in the progression of diverse diseases, such as lung cancer (37–40), cervical cancer (41, 42), glioma (43, 44), bladder cancer (45, 46), kidney cancer (47, 48), head and neck cancer (49–51), gastric cancer (52), colorectal cancer (53), liver cancer (54), uterine cancer (55), preeclampsia (56), gestational diabetes mellitus (57), and cryptorchidism (58). EGFR-AS1 is also crucial in physiological processes, including cell migration, invasion, multiplication, and drug sensitivity. High EGFR-AS1 is closely correlated with a poor prognosis and adverse clinical features, such as malignant lymphatic metastasis status, advanced tumor stage, and large tumor size. Moreover, EGFR-AS1 was recommended to be utilized to evaluate cancer prognosis and the efficacy of chemotherapies and has promising diagnostic performance in different diseases. This review mainly focused on the correlation of EGFR-AS1 expression with the clinicopathological features and prognosis of patients, its functions in regulating cellular processes, and its promising clinical applications.
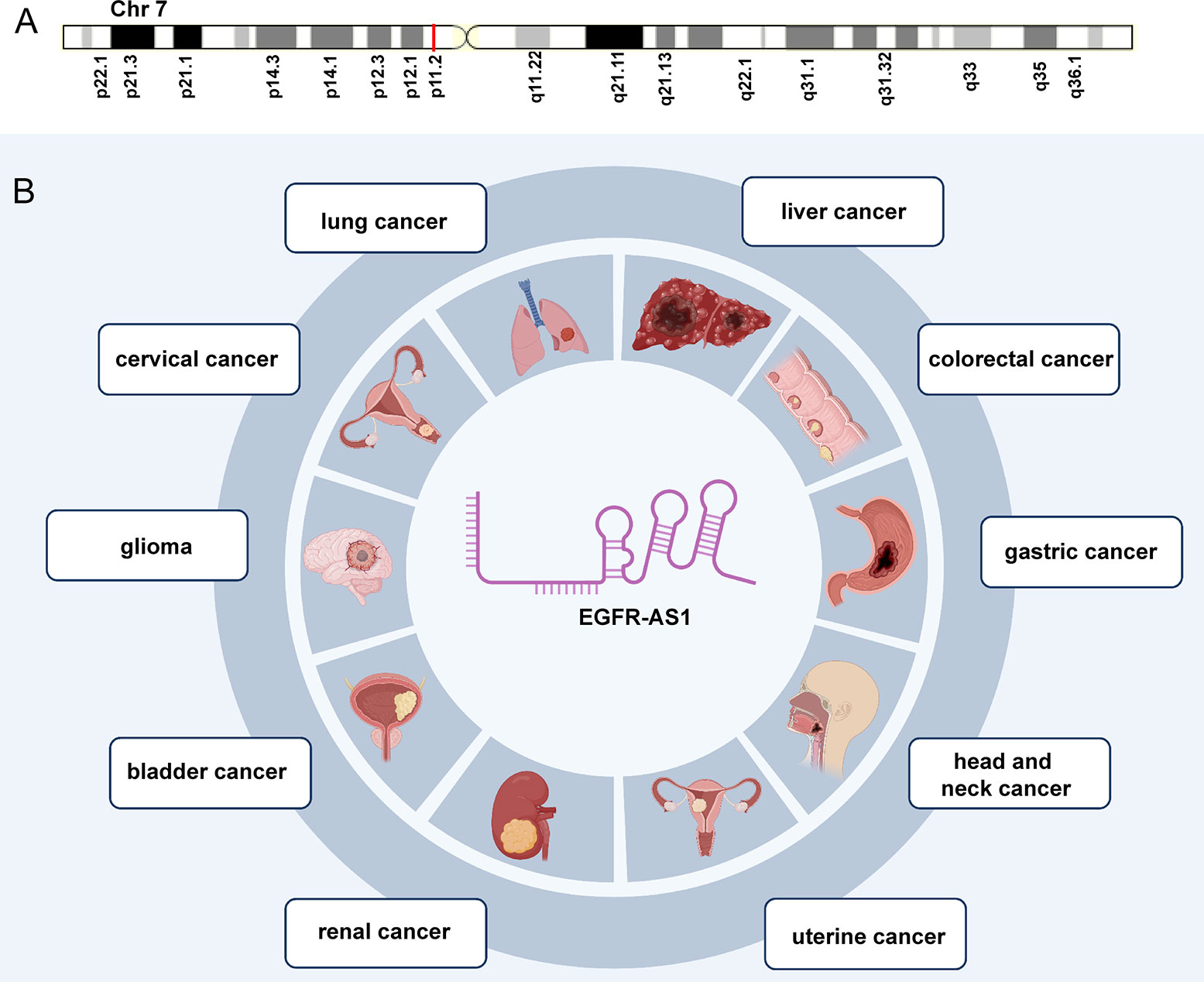
Figure 1 The expression and potential roles in cancer types. (A) Chromosomal location of EGFR-AS1. (B) EGFR-AS1 was overexpressed in lung cancer, cervical cancer, glioma, bladder cancer, kidney cancer, head and neck cancer, gastric cancer, colorectal cancer, liver cancer, and uterine cancer.
EGFR-AS1 was recently shown to be upregulated in diverse types of cancers, including lung cancer (37–40), cervical cancer (41, 42), glioma (43, 44), bladder cancer (45, 46), kidney cancer (47, 48), head and neck cancer (49–51), gastric cancer (52), colorectal cancer (53), liver cancer (54), and uterine cancer (55) (Figure 1B). Numerous studies have indicated that high EGFR-AS1 expression is closely correlated with multiple clinical characteristics, including tumor size, clinical stage, vascular invasion, portal vein thrombosis, cumulative recurrence (CR), lymph node metastasis, overall survival (OS), disease-free survival (DFS), and recurrence-free survival (RFS) (Table 1). More importantly, EGFR-AS1 participates in cancer progression via regulation of biological processes, such as cell proliferation, migration, invasion, and even drug response (Table 2). In this review, we mainly illustrated the understanding of EGFR-AS1 in terms of its expression, correlation with clinicopathological characteristics, biological roles, and relevant mechanisms.
Several studies reported that EGFR-AS1 was overexpressed in lung tissues and cell lines (A549, NCI-H460, NCI-H1299, NCI-H358, HCC827, NCH-H23, and NCI-H1650). High expressed EGFR-AS1 was strongly associated with poor CR and OS, large tumor size, and advanced clinical stage. In addition, EGFR-AS1 was functionally proven to enhance the proliferation, chemotherapy resistance, invasion, and stemness abilities of lung cancer cells and the tumor growth of mouse xenografts (37–40).
EGFR-AS1 expression was increased in cervical cancer tissues and different cell lines. Previous research indicated that EGFR-AS1 contributes to the progression of cervical cancer by promoting the proliferation, and invasion of SiHa and CaSki cells (41, 42).
In glioma, EGFR-AS1 expression was elevated in tissues and U87, U251, MG, and T98 G cells. EGFR-AS1 exerts its oncogenic roles through the promotion of cell migration, invasion, apoptosis, and drug resistance in U87 and U251 cells and through the promotion of xenograft growth (43, 44).
High EGFR-AS1 levels were observed in bladder cancer HT-1197, 5637, and T24 cells as well as tissues. Increased EGFR-AS1 expression indicated a poor patient prognosis and unfavorable features in terms of tumor size, stages, grades, and lymph node metastasis. Functionally, EGFR-AS1 facilitates cell proliferation and invasion and tumor growth and metastasis, thus promoting bladder cancer progression (45, 46).
EGFR-AS1 overexpression was also discovered in renal cancer tissues and 786O, OSRC-2, RCC4, ACHN, A498, and KETR-3 cells. A high level of EGFR-AS1 was related to unfavorable features in terms of tumor size, Fuhrman grade, TNM stage, erlotinib resistance, distant metastasis, OS, RFS, and DFS. Additionally, EGFR-AS1 stimulated the proliferation, erlotinib resistance, and invasion of 786-O and A498 cells and lung metastasis of renal cancer cells in mice (47, 48).
Multiple studies have revealed the upregulation of EGFR-AS1 in the tumor tissues and cells of head and neck cancer, including oral squamous cell carcinoma and esophageal squamous cell carcinoma (ESCC) (49–51). EGFR-AS1 was also demonstrated to promote the invasion and migration of esophageal squamous cell carcinoma cells (KYSE-30 and EC109) and thus achieved its pro-oncogenic effect (49).
In addition, EGFR-AS1 expression was upregulated in gastric cancer tissues and SGC7901, BGC823, MGC803, and MKN-28 cells. High EGFR-AS1 expression was remarkably associated with larger tumor size and promoted MGC803 and SGC-7901 cell proliferation as well as mouse tumor growth (52). In colorectal cancer, increased EGFR-AS1 levels reflect adverse patient features in terms of tumor grade, tumor status, lymph node metastasis, vascular invasion, and outcomes (53). EGFR-AS1 overexpression was also discovered in liver cancer tissues and a various of HCC cell lines and was closely correlated with lymph metastasis (54). In vivo and in vitro experiments validated that EGFR-AS1 promotes the invasion and proliferation of Huh-7 liver cells, resulting in the development of liver cancer (54). Additionally, EGFR-AS1 was upregulated in uterine cancer SK-LMS-1 and SK-UT-1 cells as well as tissues. EGFR-AS1 exhibited an oncogenic effect in uterine cancer by suppressing T-cell infiltration and motivating immune escape and tumor growth (55).
Multiple mechanistic studies have reported that lncRNAs function as tumor promoters through the regulation of a series of biological processes of cancers, such as cell proliferation, apoptosis, invasion and migration (22, 24, 28, 59, 60). In this section, we summarize the major biological mechanisms of EGFR-AS1 during cancer progression.
Uncontrolled cell proliferation causes malignant transformation and ultimately tumorigenesis, which has long been a hotspot in cancer research (61–65). Additionally, aberrantly activated cell migration and invasion are responsible for tumor expansion into the adjacent tissues, which accounts for more than 90% of cancer-related deaths (66–69). The understanding of the molecular mechanisms underlying dysregulated cellular processes may shed light on the improvement of cancer management (70–74). As revealed by multiple studies, EGFR-AS1 participates in the mediation of diverse cellular activities through interactions with its target molecules. In lung cancer, EGFR-AS1 inhibited miR-524-5p and rescued DRAM1 expression and therefore promoted the invasion of HCC827 and NCI-H1650 cells (37). EGFR-AS1 was also indicated to enhance the proliferation and chemotherapy resistance of NCI-H1299 and NCI-H358 cells via the miR-223/IGF1R/AKT signaling pathway (39). In addition, previous research reported that nicotine-derived nitrosamine ketone (NNK) downregulated EGFR-AS1 levels and then elevated HIF2A and FOXP3 expression and promoted Notch1-mediated enhancement of cancer cell stemness (40) (Figure 2). In cervical cancer, H3K27 acetylation-activated EGFR-AS1 interacts with miR-2355-5p and activates the ACTN4-mediated WNT pathway to promote the proliferation, migration, and invasion of SiHa and CaSki cells (42). It was also shown in glioma U87 and U251 cells that EGFR-AS1 promotes cell migration and invasion by sponging miR-133b to facilitate RACK1 expression (43). In bladder cancer, EGFR-AS1 accelerates cell proliferation and invasion in the T24 and 5637 cell lines by increasing the expression of EGFR (45). EGFR-AS1 also sponges microRNA-381 to elevate ROCK2 levels, which promotes bladder cancer HT-1197 cell invasion and migration (46). Moreover, EGFR-AS1 enhances the cell proliferation and invasion abilities of renal cancer 786-O and A498 cells through the enhancement of HuR-mediated EGFR expression (48). In esophageal squamous cell carcinoma, EGFR-AS1 combines with miR-145 and increases ROCK1 expression to increase the rates of KYSE-30 and EC109 cell invasion and migration (49). EGFR-AS1 was also proven to stabilize EGFR expression via the EGFR-dependent PI3K/AKT pathway and to facilitate the proliferation of MGC803 and SGC-7901 gastric cancer cells (52). In colorectal cancer, EGFR-AS1 exerts a pro-oncogenic effect through the miRNA-133b/EGFR/STAT3 axis (53). EGFR-AS1 also decreases EGFR levels to enhance the invasion and proliferation abilities of Huh-7 liver cancer cells (54). Moreover, EGFR-AS1 combines with EGFR to stimulate MYC and subsequent PD-1 expression, achieving uterine cancer cell proliferation (55).
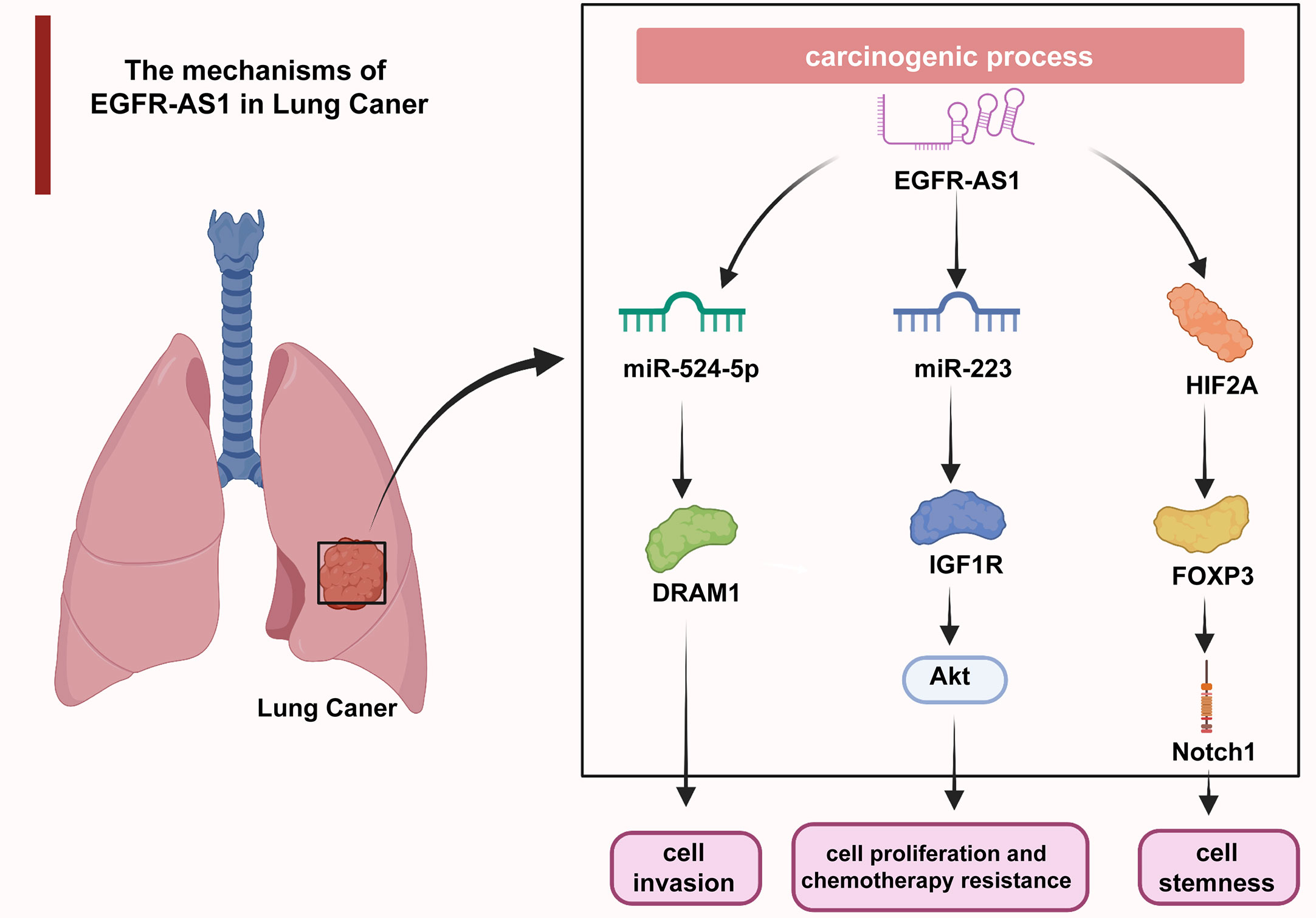
Figure 2 In lung cancer, EGFR-AS1 enhanced cell proliferation, chemotherapy resistance, invasion, and stemness via the miR-524-5p/DRAM1 axis, miR-223/IGF1R/AKT signaling pathway, and HIF2A/FOXP3/Notch1 axis.
Mounting evidence indicates that lncRNAs play pivotal roles in the different stages of cancer progression and possess considerable potential for clinical diagnosis, prognosis evaluation, and even treatment (23, 75–77) (Figure 3). Concerning its broad participation in the regulation of diverse cellular processes, EGFR-AS1 cloud be a promising novel biomarker for disease diagnosis, treatment and prognosis prediction. For example, given the overexpression of EGFR-AS1 in lung cancer patient plasma, plasma EGFR-AS1 was regarded as a noninvasive marker for cancer diagnosis as well as an independent prognostic predictor for the CR and OS of lung cancer patients (39). Xu et al. further indicated that EGFR-AS1 expression has an inverse relationship with the response to cisplatin and gemcitabine (39). EGFR-AS1 was also experimentally validated by Nath A et al. as a predictive marker of lung cancer patient response to anti-EGFR drugs such as erlotinib (38). Similarly, high EGFR-AS1 levels could differentiate bladder cancer tissue from adjacent normal tissues with a high AUC value of 0.845. Kaplan–Meier analysis also suggested that increasing EGFR-AS1 expression in bladder cancer was markedly associated with poor outcomes in terms of factors such as DFS and OS (45). As an independent prognostic factor for renal cancer patients, EGFR-AS1 was demonstrated to regulate sensitivity to the EGFR inhibitor erlotinib/Tarceva (ERLO) (47, 48). Survival curves for esophageal squamous cell carcinoma showed that the 5-year survival rate of patients in the EGFR-AS1 overexpression group was distinctly decreased (49). In oral squamous cell carcinoma, EGFR-AS1 knockdown was proven to reverse patient resistance to tyrosine kinase inhibitors (50). Similar results regarding the potential of EGFR-AS1 for disease diagnosis and prognosis were found in colorectal and liver cancer (53, 54).
EGFR-AS1 was found to be overexpressed in diverse cancers, including lung cancer, cervical cancer, glioma, bladder cancer, kidney cancer, head and neck cancer, gastric cancer, colorectal cancer, liver cancer, and uterine cancer. Upregulation of EGFR-AS1 was reported to be closely correlated with unfavorable clinicopathological characteristics such as tumor stage and distant metastasis as well as a poor prognosis in patients with multiple cancers. In vitro and in vivo research elucidated the molecular mechanisms of EGFR-AS1 in cancer-related biological processes, including cell proliferation, invasion, and migration, which mainly involve interaction with its target molecules. Regarding the exploration of its clinical value, EGFR-AS1 was validated to function as a sensitive indicator of cancer diagnosis, prognosis, and treatment response in several cancer types. Further studies clarifying the in-depth mechanisms of EGFR-AS1 in cancer progression and validating the efficacy and safety of EGFR-AS1 application in cancer management are warranted.
LL and KS designed and guided the study. DZ and XO wrote and edited the manuscript. YZ and XY helped with reference collection. All authors contributed to the article and approved the submitted version.
This study was funded by the National Key Research and Development Program of China (2021YFC230180, 2019YFC0840600, and 2019YFC0840609) and the Fundamental Research Funds for the Central Universities (2022ZFJH003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. GBD 2017 Causes of Death Collaborators Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: A systematic analysis for the global burden of disease study 2017. Lancet (2018) 392:1736–88. doi: 10.1016/s0140-6736(18)32203-7
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
3. Maruthappu M, Head MG, Zhou CD, Gilbert BJ, El-Harasis MA, Raine R, et al. Investments in cancer research awarded to UK institutions and the global burden of cancer 2000-2013: a systematic analysis. BMJ Open (2017) 7:e013936. doi: 10.1136/bmjopen-2016-013936
4. Yabroff KR, Lund J, Kepka D, Mariotto A. Economic burden of cancer in the united states: estimates, projections, and future research. Cancer Epidemiol Biomarkers Prev (2011) 20:2006–14. doi: 10.1158/1055-9965.Epi-11-0650
5. Chaker L, Falla A, van der Lee SJ, Muka T, Imo D, Jaspers L, et al. The global impact of non-communicable diseases on macro-economic productivity: A systematic review. Eur J Epidemiol (2015) 30:357–95. doi: 10.1007/s10654-015-0026-5
6. Lau MT, Ghazanfar S, Parkin A, Chou A, Rouaen JR, Littleboy JB, et al. Systematic functional identification of cancer multi-drug resistance genes. Genome Biol (2020) 21:27. doi: 10.1186/s13059-020-1940-8
7. Szymczyk J, Sluzalska KD, Materla I, Opalinski L, Otlewski J, Zakrzewska M. FGF/FGFR-dependent molecular mechanisms underlying anti-cancer drug resistance. Cancers (Basel) (2021) 13:5796. doi: 10.3390/cancers13225796
8. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin (2019) 69:363–85. doi: 10.3322/caac.21565
9. Simard S, Thewes B, Humphris G, Dixon M, Hayden C, Mireskandari S, et al. Fear of cancer recurrence in adult cancer survivors: a systematic review of quantitative studies. J Cancer Surviv (2013) 7:300–22. doi: 10.1007/s11764-013-0272-z
10. Corona RI, Seo JH, Lin X, Hazelett DJ, Reddy J, Fonseca MAS, et al. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat Commun (2020) 11:2020. doi: 10.1038/s41467-020-15951-0
11. Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood (2011) 117:2022–32. doi: 10.1182/blood-2010-04-280750
12. Chinn SB, Myers JN. Oral cavity carcinoma: Current management, controversies, and future directions. J Clin Oncol (2015) 33:3269–76. doi: 10.1200/jco.2015.61.2929
13. Kloetgen A, Thandapani P, Ntziachristos P, Ghebrechristos Y, Nomikou S, Lazaris C, et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat Genet (2020) 52:388–400. doi: 10.1038/s41588-020-0602-9
14. Xue C, Li G, Lu J, Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct Target Ther (2021) 6:400. doi: 10.1038/s41392-021-00788-w
15. Xue C, Li G, Zheng Q, Gu X, Bao Z, Lu J, et al. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer (2022) 21:108. doi: 10.1186/s12943-022-01582-0
16. Xing YH, Bai Z, Liu CX, Hu SB, Ruan M, Chen LL. Research progress of long noncoding RNA in China. IUBMB Life (2016) 68:887–93. doi: 10.1002/iub.1564
17. Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell (2013) 154:996–1009. doi: 10.1016/j.cell.2013.07.047
18. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell (2007) 129:823–37. doi: 10.1016/j.cell.2007.05.009
19. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. doi: 10.1101/gr.132159.111
20. Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer (2016) 15:43. doi: 10.1186/s12943-016-0530-6
21. Flippot R, Malouf GG, Su X, Mouawad R, Spano JP, Khayat D. Cancer subtypes classification using long non-coding RNA. Oncotarget (2016) 7:54082–93. doi: 10.18632/oncotarget.10213
22. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene (2017) 36:5661–7. doi: 10.1038/onc.2017.184
23. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res (2017) 77:3965–81. doi: 10.1158/0008-5472.Can-16-2634
24. McCabe EM, Rasmussen TP. lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol (2021) 75:38–48. doi: 10.1016/j.semcancer.2020.12.012
25. Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep (2012) 13:971–83. doi: 10.1038/embor.2012.145
26. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev (2009) 23:1494–504. doi: 10.1101/gad.1800909
27. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet (2009) 10:155–9. doi: 10.1038/nrg2521
28. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) (2021) 41:109–20. doi: 10.1002/cac2.12108
29. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature (2011) 472:120–4. doi: 10.1038/nature09819
30. Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci (2013) 56:876–85. doi: 10.1007/s11427-013-4553-6
31. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature (2012) 491:454–7. doi: 10.1038/nature11508
32. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell (2013) 154:26–46. doi: 10.1016/j.cell.2013.06.020
33. Chen L, Zhou Y, Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res (2018) 257:25–32. doi: 10.1016/j.virusres.2018.08.018
34. Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer (2016) 15:39. doi: 10.1186/s12943-016-0524-4
35. Bolha L, Ravnik-Glavač M, Glavač D. Long noncoding RNAs as biomarkers in cancer. Dis Markers (2017) 2017:7243968. doi: 10.1155/2017/7243968
36. Yang Z, Jiang S, Shang J, Jiang Y, Dai Y, Xu B, et al. LncRNA: Shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res Rev (2019) 52:17–31. doi: 10.1016/j.arr.2019.04.001
37. Xue Y, Zhang J, Hou J, Wang X. EGFR-AS1 promotes nonsmall cell lung cancer (NSCLC) progression via downregulating the miR-524-5p/DRAM1 axis and inhibiting autophagic lysosomal degradation. J Oncol (2022) 2022:4402536. doi: 10.1155/2022/4402536
38. Nath A, Lau EYT, Lee AM, Geeleher P, Cho WCS, Huang RS. Discovering long noncoding RNA predictors of anticancer drug sensitivity beyond protein-coding genes. Proc Natl Acad Sci U S A (2019) 116:22020–9. doi: 10.1073/pnas.1909998116
39. Xu YH, Tu JR, Zhao TT, Xie SG, Tang SB. Overexpression of lncRNA EGFR−AS1 is associated with a poor prognosis and promotes chemotherapy resistance in non small cell lung cancer. Int J Oncol (2019) 54:295–305. doi: 10.3892/ijo.2018.4629
40. Qi H, Wang S, Wu J, Yang S, Gray S, Ng CSH, et al. EGFR-AS1/HIF2A regulates the expression of FOXP3 to impact the cancer stemness of smoking-related non-small cell lung cancer. Ther Adv Med Oncol (2019) 11:1758835919855228. doi: 10.1177/1758835919855228
41. Yao H, Jiang X, Fu H, Yang Y, Jin Q, Zhang W, et al. Exploration of the immune-related long noncoding RNA prognostic signature and inflammatory microenvironment for cervical cancer. Front Pharmacol (2022) 13:870221. doi: 10.3389/fphar.2022.870221
42. Li J, Wang H. H3K27ac-activated EGFR-AS1 promotes cell growth in cervical cancer through ACTN4-mediated WNT pathway. Biol Direct (2022) 17:3. doi: 10.1186/s13062-021-00315-5
43. Dong ZQ, Guo ZY, Xie J. The lncRNA EGFR-AS1 is linked to migration, invasion and apoptosis in glioma cells by targeting miR-133b/RACK1. BioMed Pharmacother (2019) 118:109292. doi: 10.1016/j.biopha.2019.109292
44. Kadioglu O, Saeed MEM, Mahmoud N, Azawi S, Mrasek K, Liehr T, et al. Identification of novel drug resistance mechanisms by genomic and transcriptomic profiling of glioblastoma cells with mutation-activated EGFR. Life Sci (2021) 284:119601. doi: 10.1016/j.lfs.2021.119601
45. Wang A, Jiang A, Gan X, Wang Z, Huang J, Dong K, et al. EGFR-AS1 promotes bladder cancer progression by upregulating EGFR. BioMed Res Int (2020) 2020:6665974. doi: 10.1155/2020/6665974
46. Yuan S, Luan X, Chen H, Shi X, Zhang X. Long non-coding RNA EGFR-AS1 sponges micorRNA-381 to upregulate ROCK2 in bladder cancer. Oncol Lett (2020) 19:1899–905. doi: 10.3892/ol.2020.11283
47. Grépin R, Guyot M, Dumond A, Durivault J, Ambrosetti D, Roussel JF, et al. The combination of bevacizumab/Avastin and erlotinib/Tarceva is relevant for the treatment of metastatic renal cell carcinoma: The role of a synonymous mutation of the EGFR receptor. Theranostics (2020) 10:1107–21. doi: 10.7150/thno.38346
48. Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi J, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis (2019) 10:154. doi: 10.1038/s41419-019-1331-9
49. Feng Z, Li X, Qiu M, Luo R, Lin J, Liu B. LncRNA EGFR-AS1 upregulates ROCK1 by sponging miR-145 to promote esophageal squamous cell carcinoma cell invasion and migration. Cancer Biother Radiopharm (2020) 35:66–71. doi: 10.1089/cbr.2019.2926
50. Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med (2017) 23:1167–75. doi: 10.1038/nm.4401
51. Dhamodharan S, Rose MM, Chakkarappan SR, Umadharshini KV, Arulmurugan R, Subbiah S, et al. Genetic variant rs10251977 (G>A) in EGFR-AS1 modulates the expression of EGFR isoforms a and d. Sci Rep (2021) 11:8808. doi: 10.1038/s41598-021-88161-3
52. Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y, et al. Long noncoding RNA EGFR-AS1 promotes cell proliferation by increasing EGFR mRNA stability in gastric cancer. Cell Physiol Biochem (2018) 49:322–34. doi: 10.1159/000492883
53. Atef MM, Amer AI, Hafez YM, Elsebaey MA, Saber SA, Abd El-Khalik SR. Long non-coding RNA EGFR-AS1 in colorectal cancer: potential role in tumorigenesis and survival via miRNA-133b sponge and EGFR/STAT3 axis regulation. Br J BioMed Sci (2021) 78:122–9. doi: 10.1080/09674845.2020.1853913
54. Qi HL, Li CS, Qian CW, Xiao YS, Yuan YF, Liu QY, et al. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol (2016) 37:1079–89. doi: 10.1007/s13277-015-3887-z
55. Chen ML, Fan L, Huang GR, Sun ZF. lncRNA EGFR-AS1 facilitates leiomyosarcoma progression and immune escape via the EGFR-MYC-PD-L1 axis. Int Immunol (2022) 34:365–77. doi: 10.1093/intimm/dxac014
56. Zhao ZM, Jiang J. Lowly expressed EGFR-AS1 promotes the progression of preeclampsia by inhibiting the EGFR-JAK/STAT signaling pathway. Eur Rev Med Pharmacol Sci (2018) 22:6190–7. doi: 10.26355/eurrev_201810_16024
57. Li T, Hu D, Gong Y. Identification of potential lncRNAs and co-expressed mRNAs in gestational diabetes mellitus by RNA sequencing. J Matern Fetal Neonatal Med (2021) 35:5125–39. doi: 10.1080/14767058.2021.1875432
58. Hadziselimovic F, Verkauskas G, Vincel B, Stadler MB. Testicular expression of long non-coding RNAs is affected by curative GnRHa treatment of cryptorchidism. Basic Clin Androl (2019) 29:18. doi: 10.1186/s12610-019-0097-3
59. Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett (2018) 419:27–9. doi: 10.1016/j.canlet.2018.01.008
60. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinf (2016) 14:42–54. doi: 10.1016/j.gpb.2015.09.006
61. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
62. de Oliveira JC, Oliveira LC, Mathias C, Pedroso GA, Lemos DS, Salviano-Silva A, et al. Long non-coding RNAs in cancer: Another layer of complexity. J Gene Med (2019) 21:e3065. doi: 10.1002/jgm.3065
63. Ramundo V, Giribaldi G, Aldieri E. Transforming growth factor-β and oxidative stress in cancer: A crosstalk in driving tumor transformation. Cancers (Basel) (2021) 13(12):3093. doi: 10.3390/cancers13123093
64. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science (2013) 339:1546–58. doi: 10.1126/science.1235122
65. Castillo JJ, Mull N, Reagan JL, Nemr S, Mitri J. Increased incidence of non-Hodgkin lymphoma, leukemia, and myeloma in patients with diabetes mellitus type 2: A meta-analysis of observational studies. Blood (2012) 119:4845–50. doi: 10.1182/blood-2011-06-362830
66. Kreis NN, Friemel A, Ritter A, Roth S, Rolle U, Louwen F, et al. Function of p21 (Cip1/Waf1/CDKN1A) in migration and invasion of cancer and trophoblastic cells. Cancers (Basel) (2019) 11:989. doi: 10.3390/cancers11070989
67. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science (2011) 331:1559–64. doi: 10.1126/science.1203543
68. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell (2017) 168:670–91. doi: 10.1016/j.cell.2016.11.037
69. McGregor AL, Hsia CR, Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr Opin Cell Biol (2016) 40:32–40. doi: 10.1016/j.ceb.2016.01.011
70. Zheng K, Chen Z, Feng H, Chen Y, Zhang C, Yu J, et al. Sphingomyelin synthase 2 promotes an aggressive breast cancer phenotype by disrupting the homoeostasis of ceramide and sphingomyelin. Cell Death Dis (2019) 10:157. doi: 10.1038/s41419-019-1303-0
71. Wang S, Li GX, Tan CC, He R, Kang LJ, Lu JT, et al. FOXF2 reprograms breast cancer cells into bone metastasis seeds. Nat Commun (2019) 10:2707. doi: 10.1038/s41467-019-10379-7
72. Ling A, Löfgren-Burström A, Larsson P, Li X, Wikberg ML, Öberg ÅChecktae, et al. TAP1 down-regulation elicits immune escape and poor prognosis in colorectal cancer. Oncoimmunology (2017) 6:e1356143. doi: 10.1080/2162402x.2017.1356143
73. Saha SK, Yin Y, Chae HS, Cho SG. Opposing regulation of cancer properties via KRT19-mediated differential modulation of wnt/β-Catenin/Notch signaling in breast and colon cancers. Cancers (Basel) (2019) 11:99. doi: 10.3390/cancers11010099
74. Ramis-Zaldivar JE, Gonzalez-Farré B, Balagué O, Celis V, Nadeu F, Salmerón-Villalobos J, et al. Distinct molecular profile of IRF4-rearranged large b-cell lymphoma. Blood (2020) 135:274–86. doi: 10.1182/blood.2019002699
75. Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci (2019) 20:5758. doi: 10.3390/ijms20225758
76. Tang R, Wu Z, Rong Z, Xu J, Wang W, Zhang B, et al. Ferroptosis-related lncRNA pairs to predict the clinical outcome and molecular characteristics of pancreatic ductal adenocarcinoma. Brief Bioinform (2022) 23:bbab388. doi: 10.1093/bib/bbab388
Keywords: lncRNA, EGFR-AS1, expression, functions, clinical values
Citation: Zhu D, Ouyang X, Zhang Y, Yu X, Su K and Li L (2023) A promising new cancer marker: Long noncoding RNA EGFR-AS1. Front. Oncol. 13:1130472. doi: 10.3389/fonc.2023.1130472
Received: 23 December 2022; Accepted: 03 February 2023;
Published: 24 February 2023.
Edited by:
Yubing Zhou, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Yoshimitsu Akiyama, Tokyo Medical and Dental University, JapanCopyright © 2023 Zhu, Ouyang, Zhang, Yu, Su and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanjuan Li, bGpsaUB6anUuZWR1LmNu; Kunkai Su, a3N1QHpqdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.