- Department of General Surgery, China-Japan Friendship Hospital, Beijing, China
Introduction: Breast cancer has a strong genetic predisposition, and its genetic architecture is not fully understood thus far. In this study, we aimed to perform a meta-analysis to evaluate the association of genetic alterations in LEP and ADIPOQ genes, as well as their receptor-encoded genes with risk for breast cancer.
Methods: Only published studies conducted in humans and written in English were identified by searching PubMed, SCOPUS, CINAHIL and Embase from their inception to October 2022. Eligibility assessment and data collection were completed independently by two researchers. Statistical analyses were done using the STATA software.
Results: After literature search, 33 publications were eligible for inclusion. Overall, LEP gene rs7799039-G allele (odds ratio [OR]: 0.78, 95% confidence interval [CI]: 0.62 to 0.98) and ADIPOQ gene rs1501299-T allele (OR: 1.41, 95% CI: 1.06 to 1.88) were associated with the significant risk of breast cancer. In subgroup analyses, differences in menopausal status, obesity, race, study design, diagnosis of breast cancer, genotyping method and sample size might account for the divergent observations of individual studies. Circulating leptin levels were comparable across genotypes of LEP gene rs7799039, as well as that of LEPR gene rs1137101 (P>0.05). Begg’s funnel plots seemed symmetrical, with the exception of LEPR gene rs1137100 and ADIPOQ gene rs1501299.
Discussion: Taken together, we found, in this meta-analysis, that LEP gene rs7799039 and ADIPOQ gene rs1501299 were two promising candidate loci in predisposition to breast cancer risk.
Introduction
Breast cancer is a leading cause of death in women (1). Global statistics show that the incidence rate of breast cancer was annually increased by 0.5% during the period from 2010 to 2019 (2). Breast cancer is a multifactorial malignancy that has a genetic predisposition (3). Prior studies have demonstrated that the development of mammary carcinoma in the opposite breast of familial patients with unilateral disease was three times higher than that in sporadic patients (4). Recently, a growing number of genome-wide association studies have been conducted to decipher the genetic architecture of breast cancer worldwide (5–9). In spite of great endeavors, deciphering genetic codes of breast cancer is still in its infancy. Evaluating genes with definitive biological function and direct implications in breast carcinogenesis represents a good alternative. Echoing this claim, obesity-related cytokines such as leptin and adiponectin are increasingly recognized as promising candidates in the development of breast cancer (10).
It is widely recognized that obesity is linked to an enhanced risk of tumorigenesis (11). Leptin as an inducer of epithelial-mesenchymal transition was found to promote tumor progression and metastasis (11). Experimental data supported that leptin can influence mammary tumor growth and progression through regulation of autocrine/paracrine factors and by modulating the extracellular matrix composition (12). Clinical evidence showed that women with breast cancer had increased levels of circulating leptin and its receptor (13). Another important obesity-related cytokine, adiponectin, was found to be capable to induce autophagic cell death in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis (14). There is evidence that circulating adiponectin levels were lower in women with breast cancer than in healthy controls, especially in postmenopausal women (15). Grossmann and Cleary have written an excellent review and highlighted the balance between leptin and adiponectin in the control of mammary tumorigenesis (16). Specifically, imbalance in leptin-adiponectin levels and leptin receptor expression was found to precipitate the progression of triple negative breast cancer (17). Above data collectively support the contributory roles of leptin and adiponectin in the pathogenesis of breast cancer. We thereby hypothesize that genes coding leptin (LEP) and adiponectin (ADIPOQ) and their receptors are promising candidates in predisposition to breast cancer risk.
To test this hypothesis, we conducted a meta-analysis on genetic alterations in LEP and ADIPOQ genes as well as their receptor-encoded genes by pooling published summary data, aiming to evaluate their association with risk for breast cancer, as well as circulating leptin and adiponectin levels.
Methods
Meta-analysis guideline
The conduct of this meta-analysis complied with the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guideline (18).
Search strategy
Only peer-reviewed published studies were retrieved in this meta-analysis by searching PubMed, SCOPUS, CINAHIL and Embase electronic datasets from their inception to October 2022. The key words used for indexing studies in above datasets were formulated from the MeSH (Medical Subject Headings) database, and they are expressed in logistic relations, that is, (“breast cancer” or “breast neoplasm” or “breast tumor” or “breast carcinoma” or “cancer or breast” or “mammary cancer”) and (“leptin” or “lep” or “leptin receptor” or “adiponectin” or “ADIPOQ” or “adiponectin receptor” or “ADIPOQR”) and (“polymorphism” or “variant” or “mutation” or “mutant” or “SNP” or “allele” or “genotype”). The search process was completed by two researchers (X.L. and C.L.) independently. Search results from different datasets were managed by the ENDNOTE software version X9.3.3, and duplicate records were deleted.
In addition, potential missing studies were complemented by checking the references of reviews, meta-analyses and major original articles in search results.
Eligibility criteria
Eligible studies were expected to meet all five inclusion criteria (1): breast cancer as clinical outcome (2); involvement of both breast cancer patients and control participants (3); complete genetic data (genotypes or alleles or effect sizes) of any genetic alteration in LEP or ADIPOQ genes or their receptor-encoded genes between patients with breast cancer and controls or mean or median values of circulating leptin or adiponectin levels for single genotypes or their combination (4); publication using English language (5); valid diagnosis of breast cancer.
Meanwhile, other forms of publications such as comment, editorial, perspective, letter to the editor and case report/series were not covered.
Data collection
Collection of necessary data from eligible studies was independently conducted by two researchers (X.L. and C.L.). Items of data covered surname of first author, year of publication, study design, race, country where study participants resided in, menopause status, source of control participants, factors matched between patients and controls, genotyping method, diagnostic criteria of breast cancer, sample size, chronological age, age at menarche, age of first delivery, nulliparous, percentage of ER, PR and Her-2 of patients with breast cancer, height, weight, body mass index, cigarette smoking, alcohol drinking, family history of breast cancer and genotypes of genetic alteration between patients and controls.
If there was disagreement between the two researchers, original article was assessed, and if necessary, a third researcher (W.P.) was involved.
Data analyses
Summary data from identified eligible studies were pooled by the Stata software version 15.0. To derive a sufficient power to detect significance, a minimum number of eligible studies was set at 3 for genetic alterations analyzed in this study. The association between genetic alterations and breast cancer was expressed as odds ratio (OR) and 95% confidence interval (95% CI). The association between genetic alterations and circulating leptin or adiponectin levels was expressed as standardized mean difference (SMD) and its 95% CI. Effect-size estimates were generated using the DerSimonian-Laird method and under the random-effects model. Heterogeneity between studies was justified by using a percent, inconsistency index (I2), and I2 over 50% or the probability of associated χ2 test less than 0.1 was indicative of statistical significance. Exploring sources of heterogeneity was implemented by using subgroup analyses according to categorical items of interest. Contribution of each study to overall OR was illustrated by sensitivity analyses.
Publication bias was judged by the Begg’s funnel plot and Egger’s linear regression tests from visual and statistical aspects, respectively. In the case of evident publication bias, the trim-and-fill method was used to take theoretically missing studies into consideration when estimating effect-size estimates.
Results
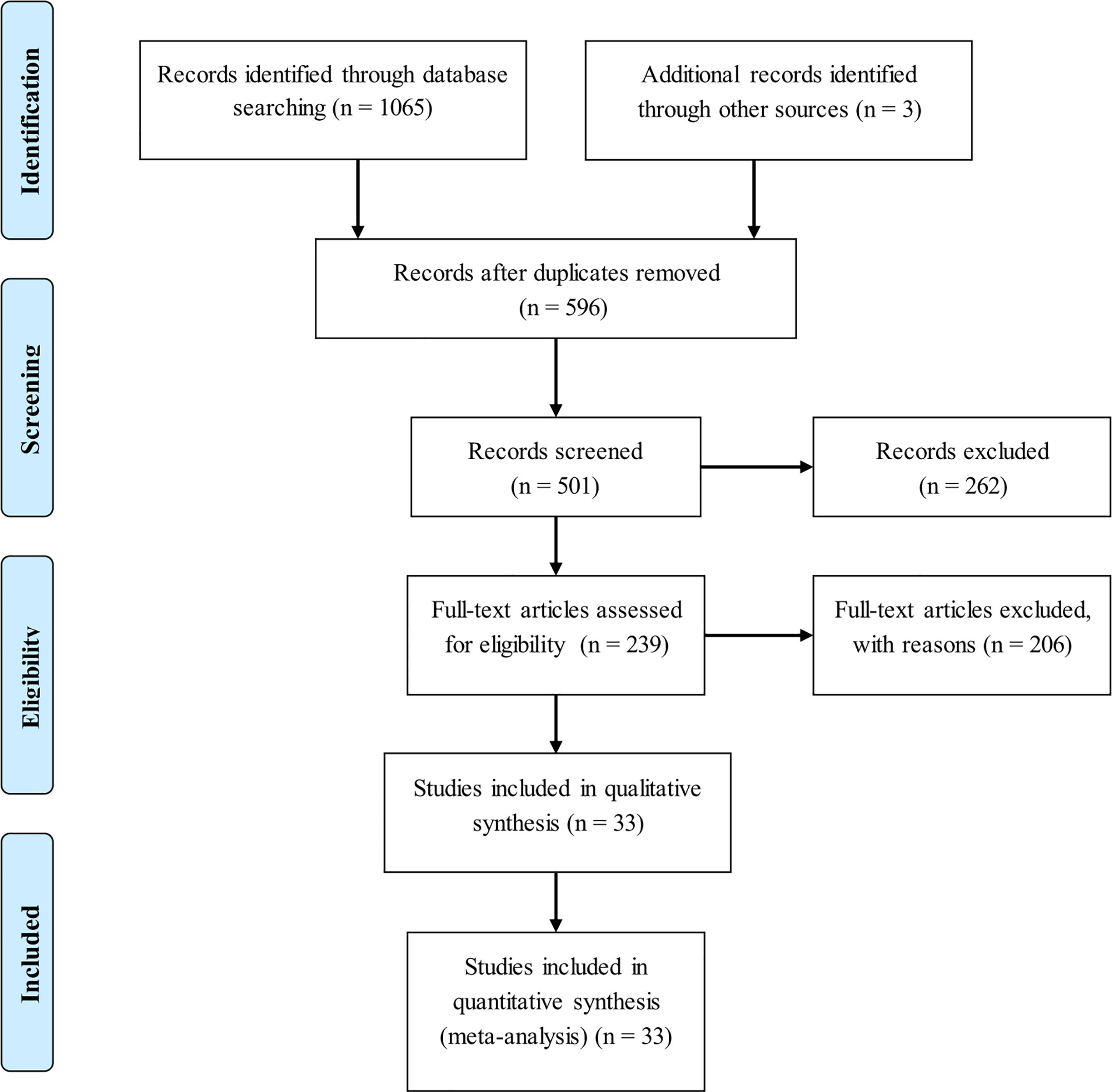
Eligible studies
Initial search of 4 public datasets identified a total of 596 publications after deleting duplicates. Only 33 of these publications were eligible for inclusion (10, 19–50). The selection process of eligible articles was displayed in the form of flow diagram (Figure 1). In the case of publications containing more than one group, each group was treated separately. Finally, 55 studies were meta-analyzed for the association between 5 genetic alterations in 3 genes (LEP, leptin receptor [LEPR] and ADIPOQ) and breast cancer risk, 4 studies for the association between rs7799039 genotypes and circulating leptin levels, and 8 studies for the association between rs1137101 genotypes and circulating leptin levels.
The baseline characteristics of 55 studies in this meta-analysis are presented in Table 1.
Overall association analyses
The association of 5 genetic alterations with breast cancer risk was displayed in the form of forest plots under allele mode of inheritance (Figure 2). Overall, LEP gene rs7799039-G allele and LEPR gene rs1137100-A allele were associated with reduced breast cancer risk relative to the corresponding reference alleles, and the risk was close to statistical significance. By contrast, ADIPOQ gene rs1501299-T allele increased breast cancer risk significantly by 26% (OR: 1.26, 95% CI: 1.00 to 1.59) relative to the corresponding G allele. The I2 ranged from 62% to 85.4%, denoting the moderate-strong evidence of heterogeneity between studies.

Figure 2 Pooled estimates of 5 genetic alterations associated with breast cancer under allele mode of inheritance.
Besides allele mode, pooled estimates under dominant and genotype modes of inheritance are shown in Supplementary Figure 1 and Supplementary Figure 2, respectively. Under dominant mode, the protective effects of LEP gene rs7799039 GG plus GA genotypes and LEPR gene rs1137100 AA plus AG genotypes on breast cancer risk dwindled, and the risk conferred by ADIPOQ gene rs1501299 TT plus TG genotypes was enhanced, with OR of 1.41 (95% CI: 1.06 to 1.88). Under genotype mode, LEP gene rs7799039-GG was associated with a 22% reduced risk of breast cancer significantly (OR: 0.78, 95% CI: 0.62 to 0.98), and no significance was detected for the other comparisons.
Subgroup analyses by menopause and obesity
The association of 5 genetic alterations with breast cancer stratified by menopause and obesity is summarized in Table 2.
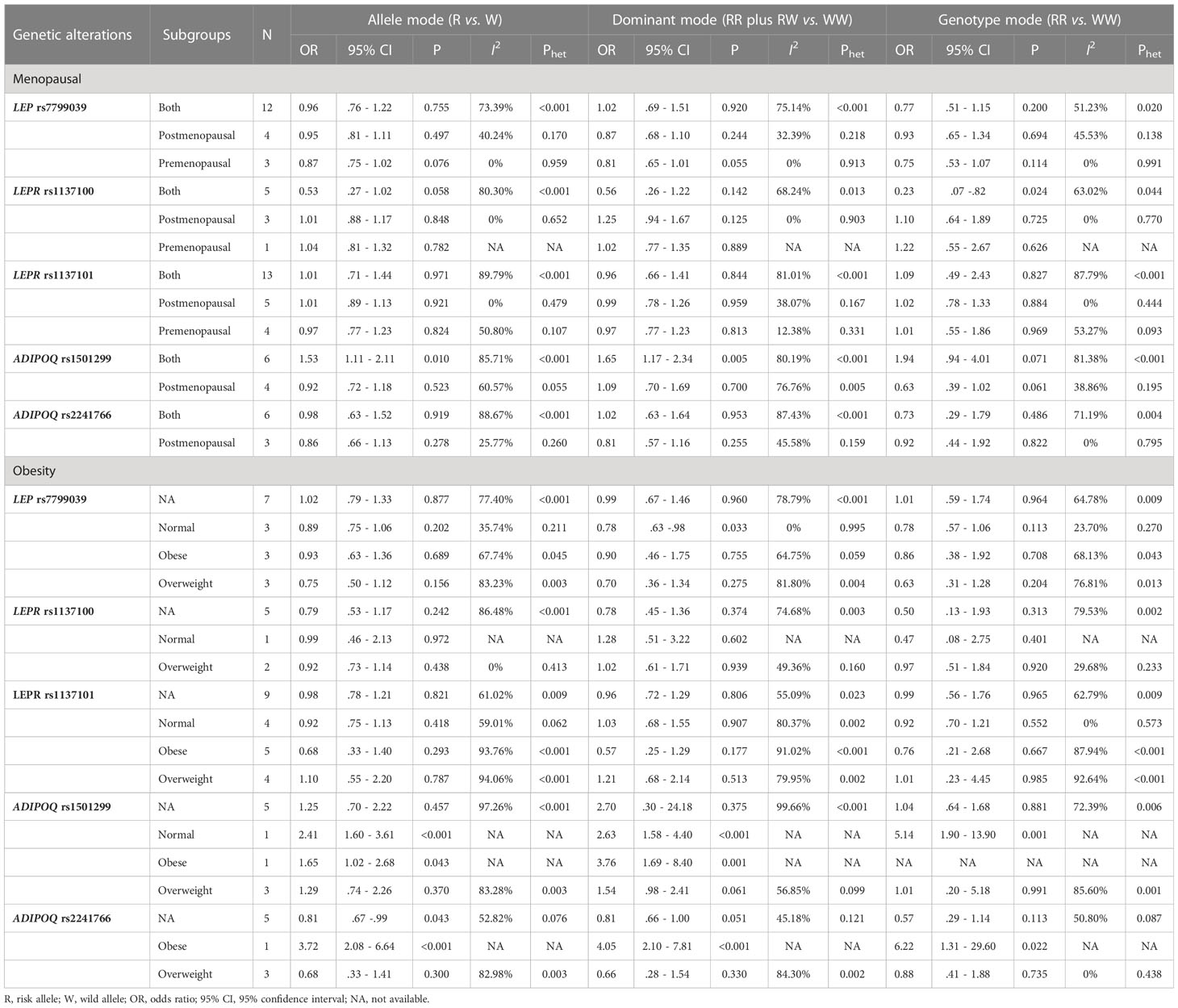
Table 2 Subgroup association analyses of 5 genetic alterations with breast cancer by menopause and obesity.
By menopausal status, LEPR gene rs1137100-AA genotype carriers conferred a significantly reduced risk of breast cancer compared with GG genotype carriers (OR: 0.23, 95% CI: 0.07 to 0.82) in both premenopausal and postmenopausal women. For ADIPOQ gene rs1501299, the risk for breast cancer was significant under both allele (OR: 1.53, 95% CI: 1.11 to 2.11) and dominant (OR: 1.65, 95% CI: 1.17 to 2.34) modes of inheritance in both premenopausal and postmenopausal women.
By obesity, the association of LEP gene rs7799039 GG plus GA genotypes with breast cancer was substantiated in normal-weight women (OR: 0.78, 95% CI: 0.63 to 0.98).
Subgroup analyses by other features
Table 3 shows the subgroup association of 5 genetic alterations with breast cancer stratified by other features of interest. For LEP rs7799039, the association was significant in studies with prospective design, with histologically-confirmed breast cancer, and involving sample size exceeding 300 under three genetic modes of inheritance. For ADIPOQ rs1501299, significance was noticed in women from East Asia, in studies involving hospital-sourced controls, in studies adopting RFLP technique, and in studies with histologically-confirmed breast cancer under both allele and dominant modes.
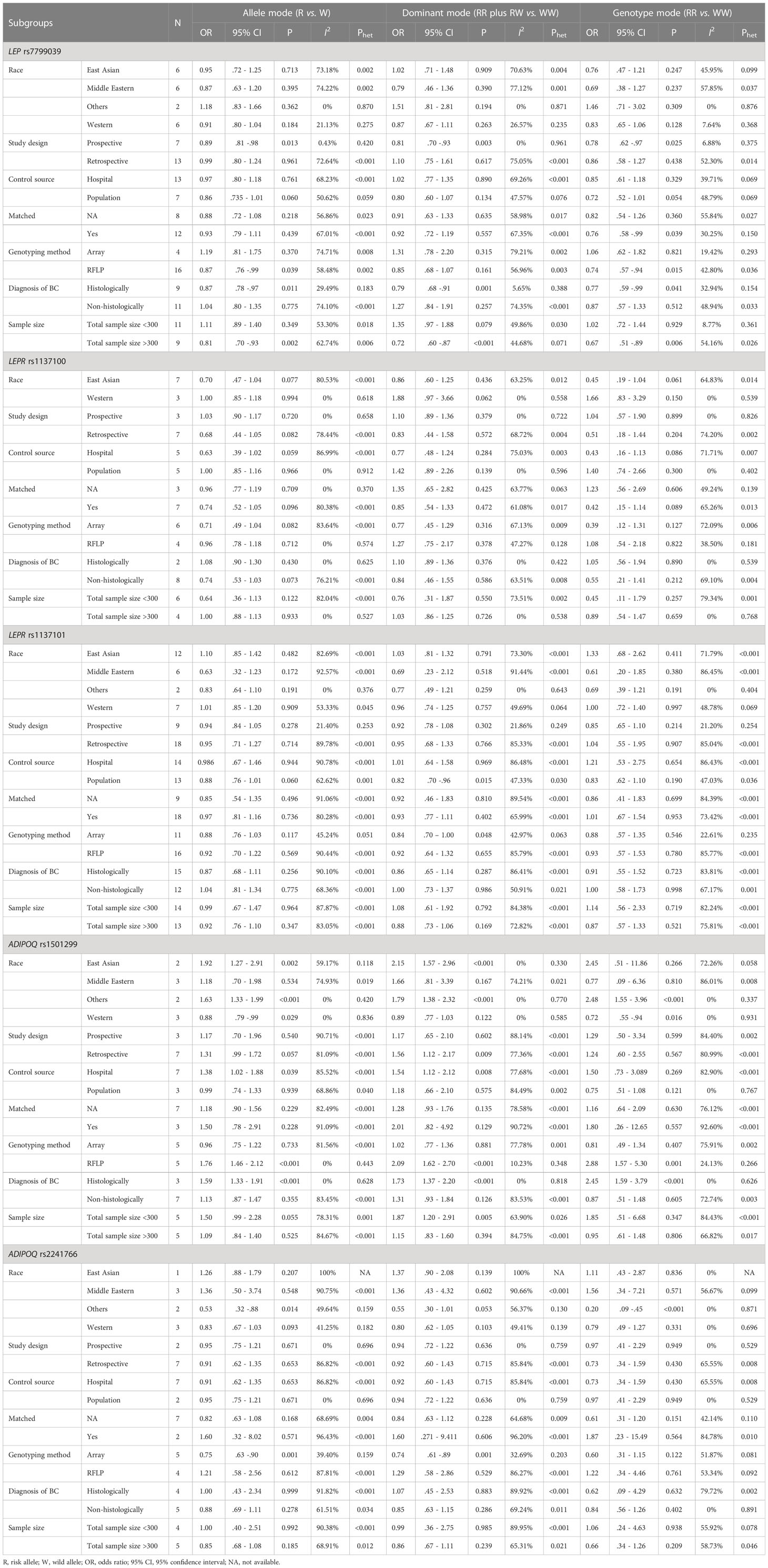
Table 3 Subgroup association analyses of 5 genetic alterations with breast cancer by other features.
Sensitivity analyses
Sensitivity analyses were performed for 5 genetic alterations associated with breast cancer under allele mode of inheritance, respectively (Supplementary Figure 3). There was no observably significant impact of any individual studies on overall effect-size estimates for 5 genetic alterations evaluated.
Publication bias
Publication bias was assessed in the form of funnel plots and regression tests. As shown in Figure 3, Begg’s funnel plots seemed symmetrical, with the exception of LEPR gene rs1137100 and ADIPOQ gene rs1501299, which was confirmed by the Egger’s regression tests (P: 0.075 and 0.077, respectively). Filled funnel plots revealed that two studies and one study were theoretically missing for LEPR gene rs1137100 and ADIPOQ gene rs1501299, respectively. After taking these missing studies into consideration, effect-size estimates were changed slightly.
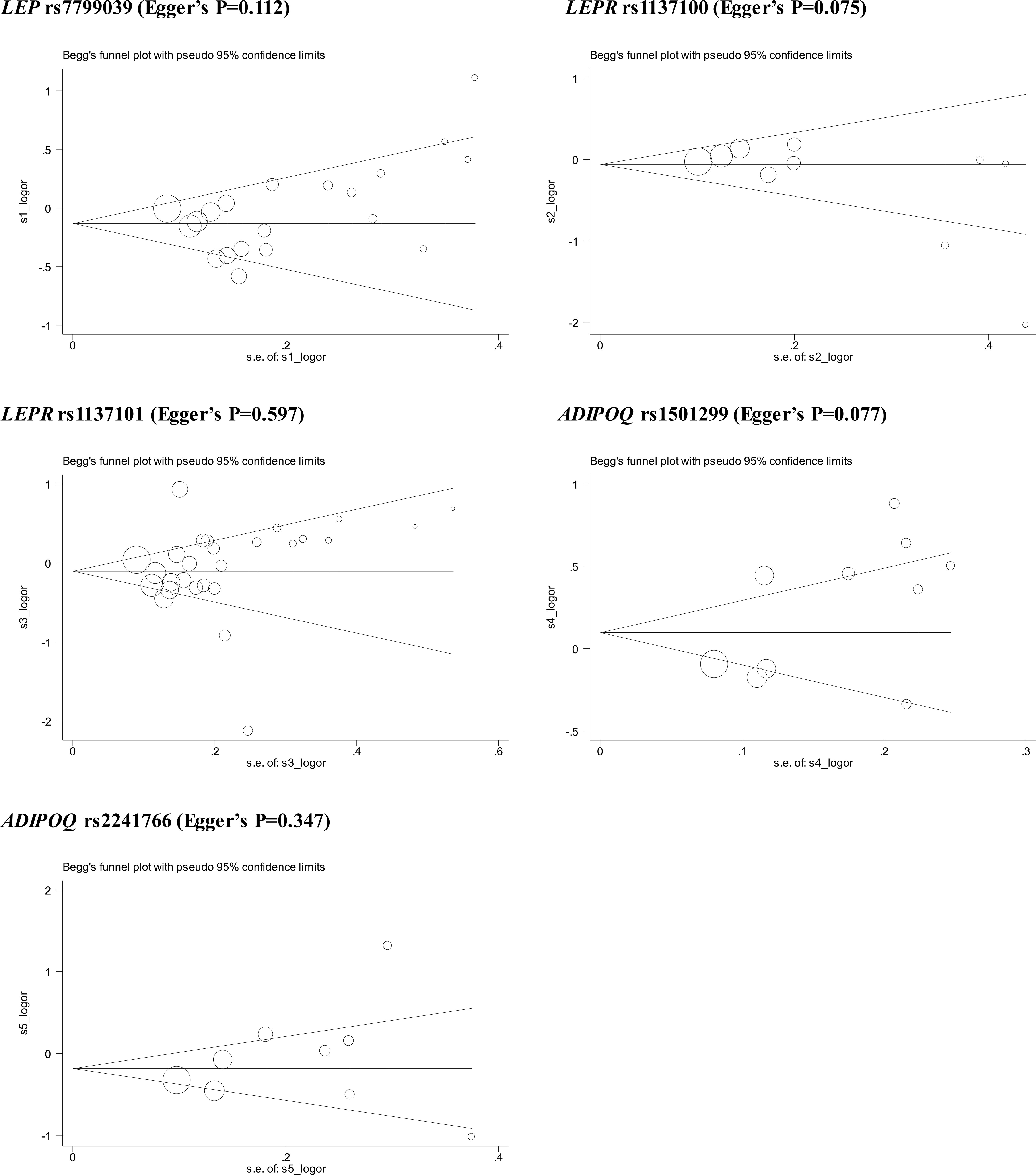
Figure 3 Begg’s funnel plots of 5 genetic alterations associated with breast cancer under allele mode of inheritance.
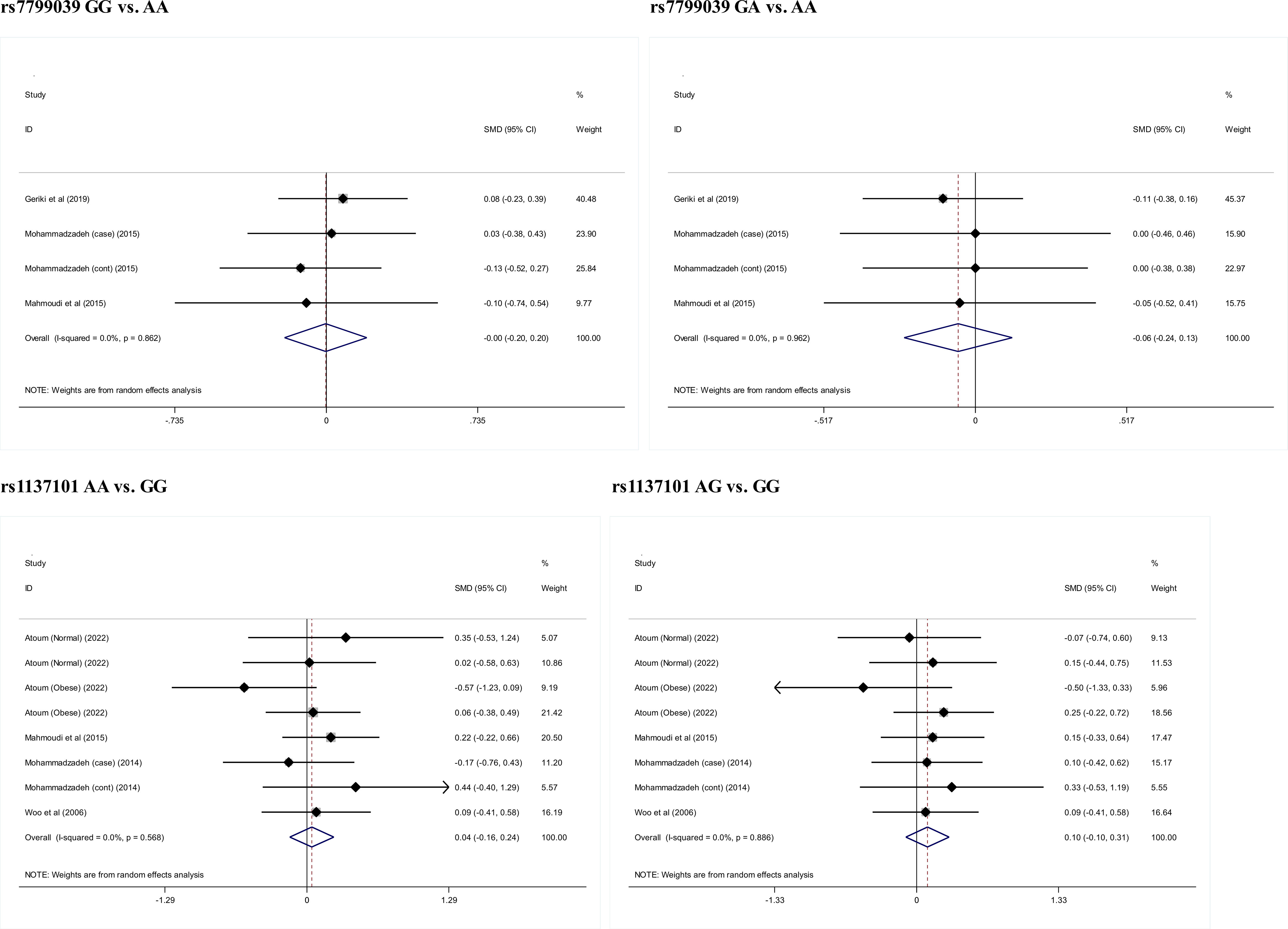
Circulating leptin levels
Figure 4 presents the comparison of circulating leptin levels between genotypes of LEP gene rs7799039 and LEPR gene rs1137101. There was no noticeable difference for all comparisons (P>0.05).
Discussion
The aim of this meta-analysis was to examine the association of 5 genetic alterations in LEP and ADIPOQ genes, as well as their receptor-encoded genes, with breast cancer risk and circulating leptin levels. Importantly, we found that LEP gene rs7799039 and ADIPOQ gene rs1501299 were two promising candidate loci in predisposition to breast cancer risk. Additionally, we found that differences in menopausal status, obesity, race, study design, diagnosis of breast cancer, genotyping method and sample size might account for the divergent results of previous studies in the literature. To the best of our knowledge, this meta-analysis is thus far the most comprehensive on the susceptibility of LEP and ADIPOQ genes to breast cancer.
Breast cancer has a strong genetic predisposition, and the heritability among first degree relatives is estimated to be around 35% (51, 52). To unravel the genetic linings of breast cancer, a large panel of studies have been conducted, and many genes were identified to be susceptible to breast cancer, such as breast cancer susceptibility gene (BRCA) (53). However, for the majority of identified genes, uncertainty still exists over which gene is actually involved in the pathogenesis of breast cancer. One of the biggest hurdles is lack of reproducibility across single studies. The reasons for this irreproducibility are mainly attributed to insufficient power to detect significance, discrepant sampling criteria of participants and varying characteristics of participants. Taking these possible reasons into consideration, we in this meta-analysis tested the hypothesis that genes encoding leptin and adiponectin and their receptors are potential candidates to breast cancer. Our findings supported this hypothesis by showing that LEP gene rs7799039 and ADIPOQ gene rs1501299 were two promising breast cancer-susceptibility loci. By contrast, a recent meta-analysis by Sayad and coworkers did not support the association of LEP gene rs7799039 and LEPR gene rs1137100 with breast cancer (54). It is possibly because of the differing number of eligible studies involved between the meta-analysis by Sayad and coworkers (54) and the present meta-analysis. As far as we know, we, for the first time, meta-analyzed the association between ADIPOQ gene and breast cancer.
To seek possible reasons behind the irreproducible findings of previous studies, we further conducted subgroup analyses for the association of 5 genetic alterations with breast cancer under three genetic modes of inheritance. It is of importance to see that menopausal status, obesity and race were potential attributes responsible for this irreproducibility. The impact of menopausal status and obesity on breast carcinogenesis has been well established, with evidence from both clinical and experimental aspects (55–57). The attribute race merited special discussion, as it is not uncommon to notice that a genetic alteration is associated with a disease in one racial group but not in another (58, 59). Given that linkage disequilibrium and genetic sequences may not be identical across races, it is a wise choice to establish candidate genes and genetic alterations within each race or ethnicity group.
Although the significant association between LEP, LEPR and ADIPOQ genes and breast cancer risk, we did not notice remarkable differences in circulating leptin levels across genotypes of their genetic alterations. The possibility for this phenomenon might be the limited number of studies measuring and comparing circulating leptin levels across genotypes. Another possibility is that studies for the association with breast cancer and circulating leptin levels are not identical, and differences in study designs, sample sizes and participant characteristics may matter. Practically, it is expected to validate the association with circulating leptin levels by large, well-designed cohorts in the future.
Limitations
Some possible limitations needed to be addressed for this meta-analysis. The first is the probability of selection bias. This meta-analysis merely retrieved published studies in English, and studies written the other languages known as “grey” literature were not covered. The second limitation is the cross-sectional nature of all retrieved studies, and the association derived in this meta-analysis cannot imply the cause-and-effect relationship, calling for further investigations to fill this gap in knowledge. The third limitation is the insufficient power in most subgroup association analyses. The fourth limitation is that this meta-analysis is based on summary estimates, instead of individual participant data, which made the statistical correction for some confounding factors such as menopausal status and body mass index impractical. The fifth limitation is the possibility of publication bias for two of five genetic alterations assessed in this meta-analysis; however, incorporation of adding theoretically missing studies did not materially change our effect-size estimates.
In conclusion, through a comprehensive analysis of 33 publications, we found that LEP gene rs7799039 and ADIPOQ gene rs1501299 were two promising candidate loci in predisposition to breast cancer risk. Additionally, we found that differences in menopausal status, obesity, race, study design, diagnosis of breast cancer, genotyping method and sample size might account for the divergent results of previous studies in the literature. We agree that further investigations from genetic and experimental points of view are necessary to ascertain the implication of these genes in the pathogenesis of breast cancer, which might shed more light on knowledge and preferences toward breast cancer screening for high-risk women.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
W-zP conceived the study; XL and C-fL conducted the literature search. XL and C-fL extracted the required data. XL and C-fL performed data analysis and interpretation. W-zP and JZ did statistical analyses. W-zP and XL drafted the manuscript. All authors contributed to the writings and approved the final version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1125189/full#supplementary-material
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
2. Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast cancer statistics, 2022. CA Cancer J Clin (2022) 72(6):524–41. doi: 10.3322/caac.21754
3. Criscitiello C, Corti C. Breast cancer genetics: diagnostics and treatment. Genes (Basel) (2022) 13(9). doi: 10.3390/genes13091593
4. Lynch HT, Harris RE, Organ CH Jr., Lynch JF. Management of familial breast cancer. i. biostatistical-genetic aspects and their limitations as derived from a familial breast cancer resource. Arch Surg (1978) 113(9):1053–8. doi: 10.1001/archsurg.1978.01370210035004
5. Morton LM, Sampson JN, Armstrong GT, Chen TH, Hudson MM, Karlins E, et al. Genome-wide association study to identify susceptibility loci that modify radiation-related risk for breast cancer after childhood cancer. J Natl Cancer Inst (2017) 109(11). doi: 10.1093/jnci/djx058
6. Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet (2015) 47(4):373–80. doi: 10.1038/ng.3242
7. Cai Q, Zhang B, Sung H, Low SK, Kweon SS, Lu W, et al. Genome-wide association analysis in East asians identifies breast cancer susceptibility loci at 1q32. 1 5q14.3 15q26.1. Nat Genet (2014) 46(8):886–90. doi: 10.1038/ng.3041
8. Orr N, Lemnrau A, Cooke R, Fletcher O, Tomczyk K, Jones M, et al. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet (2012) 44(11):1182–4. doi: 10.1038/ng.2417
9. Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet (2010) 42(6):504–7. doi: 10.1038/ng.586
10. Geriki S, Bitla AR, SrinivasaRao P, Hulikal N, Yootla M, Sachan A, et al. Association of single nucleotide polymorphisms of adiponectin and leptin genes with breast cancer. Mol Biol Rep (2019) 46(6):6287–97. doi: 10.1007/s11033-019-05070-5
11. Sabol RA, Bowles AC, Cote A, Wise R, O'Donnell B, Matossian MD, et al. Leptin produced by obesity-altered adipose stem cells promotes metastasis but not tumorigenesis of triple-negative breast cancer in orthotopic xenograft and patient-derived xenograft models. Breast Cancer Res (2019) 21(1):67. doi: 10.1186/s13058-019-1153-9
12. Perera CN, Spalding HS, Mohammed SI, Camarillo IG. Identification of proteins secreted from leptin stimulated MCF-7 breast cancer cells: a dual proteomic approach. Exp Biol Med (Maywood). (2008) 233(6):708–20. doi: 10.3181/0710-RM-281
13. Romero-Figueroa Mdel S, Garduno-Garcia Jde J, Duarte-Mote J, Matute-Gonzalez G, Gomez-Villanueva A, de la Cruz-Vargas J. Insulin and leptin levels in obese patients with and without breast cancer. Clin Breast Cancer (2013) 13(6):482–5. doi: 10.1016/j.clbc.2013.08.001
14. Chung SJ, Nagaraju GP, Nagalingam A, Muniraj N, Kuppusamy P, Walker A, et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy (2017) 13(8):1386–403. doi: 10.1080/15548627.2017.1332565
15. Bielawski K, Rhone P, Bulsa M, Ruszkowska-Ciastek B. Pre-operative combination of normal BMI with elevated YKL-40 and leptin but lower adiponectin level is linked to a higher risk of breast cancer relapse: a report of four-year follow-up study. J Clin Med (2020) 9(6). doi: 10.3390/jcm9061742
16. Grossmann ME, Cleary MP. The balance between leptin and adiponectin in the control of carcinogenesis - focus on mammary tumorigenesis. Biochimie (2012) 94(10):2164–71. doi: 10.1016/j.biochi.2012.06.013
17. Sultana R, Kataki AC, Borthakur BB, Basumatary TK, Bose S. Imbalance in leptin-adiponectin levels and leptin receptor expression as chief contributors to triple negative breast cancer progression in northeast India. Gene (2017) 621:51–8. doi: 10.1016/j.gene.2017.04.021
18. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PloS Med (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
19. Li L, Meng X, Liu L, Xiang Y, Wang F, Yu L, et al. Single-nucleotide polymorphisms in LEP and LEPR associated with breast cancer risk: results from a multicenter case-control study in Chinese females. Front Oncol (2022) 12:809570. doi: 10.3389/fonc.2022.809570
20. Atoum MF, Hamaid Alparrey AA. Association of leptin receptor Q223R gene polymorphism and breast cancer patients: a case control study. Asian Pac J Cancer Prev (2022) 23(1):177–82. doi: 10.31557/APJCP.2022.23.1.177
21. Özgöz A, Mutlu Içduygu F, Yükseltürk A, Samli H, Hekimler Öztürk K, Baskan Z, et al. Postmenopausal estrogen receptor positive breast cancer and obesity associated gene variants. Excli J (2021) 20:1133–44. doi: 10.17179/excli2020-2860
22. Hołysz H, Paszel-Jaworska A, Romaniuk-Drapała A, Grodecka-Gazdecka S, Rubiś B. LEP (-2548G>A LEP) and LEPR (223Gln>Arg, 109Lys>Arg) polymorphisms as breast cancer risk factors in the polish female population. Mol Biol Rep (2021) 48(4):3237–44. doi: 10.1007/s11033-021-06328-7
23. Mahmoud EH, Fawzy A, El-Din WM, Shafik NF. Diagnostic value of adiponectin gene polymorphism and serum level in postmenopausal obese patients with breast cancer. J Cancer Res Ther (2020) 16(6):1269–73. doi: 10.4103/jcrt.JCRT_1091_19
24. Cerda-Flores RM, Camarillo-Cárdenas KP, Gutiérrez-Orozco G, Villarreal-Vela MP, Garza-Guajardo R, Ponce-Camacho MA, et al. ADIPOQ single nucleotide polymorphisms and breast cancer in northeastern Mexican women. BMC Med Genet (2020) 21(1):187. doi: 10.1186/s12881-020-01125-8
25. Pasha HF, Mohamed RH, Toam MM, Yehia AM. Genetic and epigenetic modifications of adiponectin gene: potential association with breast cancer risk. J Gene Med (2019) 21(10):e3120. doi: 10.1002/jgm.3120
26. Macias-Gomez NM, Hernandez-Terrones MC, Ramirez-Guerrero AA, Leal-Ugarte E, Gutierrez-Angulo M, Peregrina-Sandoval J. ADIPOQ rs2241766 SNP as protective marker against DIBC development in Mexican population. PloS One (2019) 14(3):e0214080. doi: 10.1371/journal.pone.0214080
27. Liu CR, Li Q, Hou C, Li H, Shuai P, Zhao M, et al. Changes in body mass index, leptin, and leptin receptor polymorphisms and breast cancer risk. DNA Cell Biol (2018) 37(3):182–8. doi: 10.1089/dna.2017.4047
28. Rodrigo C, Tennekoon KH, Karunanayake EH, De Silva K, Amarasinghe I, Wijayasiri A. Circulating leptin, soluble leptin receptor, free leptin index, visfatin and selected leptin and leptin receptor gene polymorphisms in sporadic breast cancer. Endocr J (2017) 64(4):393–401. doi: 10.1507/endocrj.EJ16-0448
29. El-Hussiny MA, Atwa MA, Rashad WE, Shaheen DA, Elkady NM. Leptin receptor Q223R polymorphism in Egyptian female patients with breast cancer. Contemp Oncol (Pozn) (2017) 21(1):42–7. doi: 10.5114/wo.2017.66655
30. Khandouzi M, Deka M, Narayan Baruah M, Rozati R, Kordafshari M, Kashyap P, et al. Genetic variation in adiponectin, leptin and leptin receptors with reference to risk of breast cancer in northeast obese women in India. BIOSCIENCE Biotechnol Res Commun (2016) 9(2):163–70. doi: 10.21786/bbrc/9.2/1
31. Erbay B, Yılmaz TU, Eraldemir C, Üren N, Tiryaki Ç, Ergül E, et al. The relationship between adiponectin and breast cancer. J Breast Health (2016) 12(2):67–71. doi: 10.5152/tjbh.2016.2881
32. Rostami S, Kohan L, Mohammadianpanah M. The LEP G-2548A gene polymorphism is associated with age at menarche and breast cancer susceptibility. Gene (2015) 557(2):154–7. doi: 10.1016/j.gene.2014.12.021
33. Mohammadzadeh G, Ghaffari MA, Bafandeh A, Hosseini SM, Ahmadi B. The relationship between -2548 G/A leptin gene polymorphism and risk of breast cancer and serum leptin levels in ahvazian women. Iran J Cancer Prev (2015) 8(2):100–8.
34. Mahmoudi R, Noori Alavicheh B, Nazer Mozaffari MA, Fararouei M, Nikseresht M. Polymorphisms of leptin (-2548 G/A) and leptin receptor (Q223R) genes in Iranian women with breast cancer. Int J Genomics (2015) 2015:132720. doi: 10.1155/2015/132720
35. Karakus N, Kara N, Ulusoy AN, Ozaslan C, Tural S, Okan I. Evaluation of CYP17A1 and LEP gene polymorphisms in breast cancer. Oncol Res Treat (2015) 38(9):418–22. doi: 10.1159/000438940
36. Mohammadzadeh G, Ghaffari MA, Bafandeh A, Hosseini SM. Effect of leptin receptor Q223R polymorphism on breast cancer risk. Iran J Basic Med Sci (2014) 17(8):588–94.
37. Robles MJG. The LEP G-2548A polymorphism is not associated with breast cancer susceptibility in obese Western Mexican women. J Clin Cell Immunol (2013) 04(01). doi: 10.4172/2155-9899.1000133
38. Kaklamani VG, Hoffmann TJ, Thornton TA, Hayes G, Chlebowski R, Van Horn L, et al. Adiponectin pathway polymorphisms and risk of breast cancer in African americans and hispanics in the women's health initiative. Breast Cancer Res Treat (2013) 139(2):461–8. doi: 10.1007/s10549-013-2546-6
39. Kim KZ, Shin A, Lee YS, Kim SY, Kim Y, Lee ES. Polymorphisms in adiposity-related genes are associated with age at menarche and menopause in breast cancer patients and healthy women. Hum Reprod (2012) 27(7):2193–200. doi: 10.1093/humrep/des147
40. Gu F, Kraft P, Rice M, Michels KB. Leptin and leptin receptor genes in relation to premenopausal breast cancer incidence and grade in Caucasian women. Breast Cancer Res Treat (2012) 131(1):17–25. doi: 10.1007/s10549-011-1778-6
41. Nyante SJ, Gammon MD, Kaufman JS, Bensen JT, Lin DY, Barnholtz-Sloan JS, et al. Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes. Breast Cancer Res Treat (2011) 129(2):593–606. doi: 10.1007/s10549-011-1517-z
42. Cleveland RJ, Gammon MD, Long CM, Gaudet MM, Eng SM, Teitelbaum SL, et al. Common genetic variations in the LEP and LEPR genes, obesity and breast cancer incidence and survival. Breast Cancer Res Treat (2010) 120(3):745–52. doi: 10.1007/s10549-009-0503-1
43. Teras LR, Goodman M, Patel AV, Bouzyk M, Tang W, Diver WR, et al. No association between polymorphisms in LEP, LEPR, ADIPOQ, ADIPOR1, or ADIPOR2 and postmenopausal breast cancer risk. Cancer Epidemiol Biomarkers Prev (2009) 18(9):2553–7. doi: 10.1158/1055-9965.EPI-09-0542
44. Okobia MN, Bunker CH, Garte SJ, Zmuda JM, Ezeome ER, Anyanwu SN, et al. Leptin receptor Gln223Arg polymorphism and breast cancer risk in Nigerian women: a case control study. BMC Cancer (2008) 8:338. doi: 10.1186/1471-2407-8-338
45. Kaklamani VG, Sadim M, Hsi A, Offit K, Oddoux C, Ostrer H, et al. Variants of the adiponectin and adiponectin receptor 1 genes and breast cancer risk. Cancer Res (2008) 68(9):3178–84. doi: 10.1158/0008-5472.CAN-08-0533
46. Han CZ, Du LL, Jing JX, Zhao XW, Tian FG, Shi J, et al. Associations among lipids, leptin, and leptin receptor gene Gin223Arg polymorphisms and breast cancer in China. Biol Trace Elem Res (2008) 126(1-3):38–48. doi: 10.1007/s12011-008-8182-z
47. Liu CL, Chang YC, Cheng SP, Chern SR, Yang TL, Lee JJ, et al. The roles of serum leptin concentration and polymorphism in leptin receptor gene at codon 109 in breast cancer. Oncology (2007) 72(1-2):75–81. doi: 10.1159/000111097
48. Gallicchio L, McSorley MA, Newschaffer CJ, Huang HY, Thuita LW, Hoffman SC, et al. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect Prev (2007) 31(2):95–101. doi: 10.1016/j.cdp.2007.02.004
49. Woo HY, Park H, Ki CS, Park YL, Bae WG. Relationships among serum leptin, leptin receptor gene polymorphisms, and breast cancer in Korea. Cancer Lett (2006) 237(1):137–42. doi: 10.1016/j.canlet.2005.05.041
50. Snoussi K, Strosberg AD, Bouaouina N, Ben Ahmed S, Helal AN, Chouchane L. Leptin and leptin receptor polymorphisms are associated with increased risk and poor prognosis of breast carcinoma. BMC Cancer. (2006) 6:38. doi: 10.1186/1471-2407-6-38
51. Ju W, Wang J, Li B, Li Z. An epidemiology and molecular genetic study on breast cancer susceptibility. Chin Med Sci J (2000) 15(4):231–7.
52. Calvo Chozas A, Mahjani B, Ronnegard L. Family history of breast cancer is associated with elevated risk of prostate cancer: evidence for shared genetic risks. Hum Hered (2021). doi: 10.1159/000521215
53. Osorio A, de la Hoya M, Rodriguez-Lopez R, Martinez-Ramirez A, Cazorla A, Granizo JJ, et al. Loss of heterozygosity analysis at the BRCA loci in tumor samples from patients with familial breast cancer. Int J Cancer (2002) 99(2):305–9. doi: 10.1002/ijc.10337
54. Sayad S, Dastgheib SA, Farbod M, Asadian F, Karimi-Zarchi M, Salari S, et al. Association of PON1, LEP and LEPR polymorphisms with susceptibility to breast cancer: a meta-analysis. Asian Pac J Cancer Prev (2021) 22(8):2323–34. doi: 10.31557/APJCP.2021.22.8.2323
55. Crujeiras AB, Diaz-Lagares A, Stefansson OA, Macias-Gonzalez M, Sandoval J, Cueva J, et al. Obesity and menopause modify the epigenomic profile of breast cancer. Endocr Relat Cancer (2017) 24(7):351–63. doi: 10.1530/ERC-16-0565
56. Katoh A, Watzlaf VJ, D'Amico F. An examination of obesity and breast cancer survival in post-menopausal women. Br J Cancer (1994) 70(5):928–33. doi: 10.1038/bjc.1994.422
57. Garcia-Estevez L, Cortes J, Perez S, Calvo I, Gallegos I, Moreno-Bueno G. Obesity and breast cancer: a paradoxical and controversial relationship influenced by menopausal status. Front Oncol (2021) 11:705911. doi: 10.3389/fonc.2021.705911
58. Sweeney C, Bernard PS, Factor RE, Kwan ML, Habel LA, Quesenberry CP Jr., et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev (2014) 23(5):714–24. doi: 10.1158/1055-9965.EPI-13-1023
Keywords: breast cancer, genetic alternation, meta-analysis, risk, association
Citation: Peng W-z, Liu X, Li C-f and Zhao J (2023) Genetic alterations in LEP and ADIPOQ genes and risk for breast cancer: a meta-analysis. Front. Oncol. 13:1125189. doi: 10.3389/fonc.2023.1125189
Received: 19 December 2022; Accepted: 10 May 2023;
Published: 19 May 2023.
Edited by:
Ricardo Ribeiro, Universidade do Porto, PortugalReviewed by:
Manish Charan, The Ohio State University, United StatesHortensia Moreno-Macias, Universidad Autonoma Metropolitana, Mexico
Kyoung Sik Park, Konkuk University Medical Center, Republic of Korea
Copyright © 2023 Peng, Liu, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-zhao Peng, cHd5Xzc1QDE2My5jb20=
 Wei-zhao Peng
Wei-zhao Peng Xin Liu
Xin Liu Chao-feng Li
Chao-feng Li