- 1Institute of Human Genetics, Jena University Hospital, Jena, Germany
- 2Urology and Nephrology Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Men’s Health and Reproductive Health Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Department of Clinical Analysis, College of Pharmacy, Hawler Medical University, Erbil, Kurdistan, Iraq
- 5Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Long non-coding RNAs (lncRNAs) are regulatory transcripts with essential roles in the pathogenesis of almost all types of cancers, including prostate cancer. They can act as either oncogenic lncRNAs or tumor suppressor ones in prostate cancer. Small nucleolar RNA host genes are among the mostly assessed oncogenic lncRNAs in this cancer. PCA3 is an example of oncogenic lncRNAs that has been approved as a diagnostic marker in prostate cancer. A number of well-known oncogenic lncRNAs in other cancers such as DANCR, MALAT1, CCAT1, PVT1, TUG1 and NEAT1 have also been shown to act as oncogenes in prostate cancer. On the other hand, LINC00893, LINC01679, MIR22HG, RP1-59D14.5, MAGI2-AS3, NXTAR, FGF14-AS2 and ADAMTS9-AS1 are among lncRNAs that act as tumor suppressors in prostate cancer. LncRNAs can contribute to the pathogenesis of prostate cancer via modulation of androgen receptor (AR) signaling, ubiquitin–proteasome degradation process of AR or other important signaling pathways. The current review summarizes the role of lncRNAs in the evolution of prostate cancer with an especial focus on their importance in design of novel biomarker panels and therapeutic targets.
Introduction
Prostate cancer is the most commonly diagnosed cancer among males being responsible for 27% of all diagnosed cases (1). It also accounts for the greatest number of deaths from cancer among men after lung cancer (1). A number of risk factors have been identified for prostate cancer among them are age, ethnicity, genetics, family history, obesity, and smoking (2, 3). Prostate cancer is developed via a multistep process, starting from prostatic intraepithelial neoplasia and being evolved to localized, advanced prostate cancer with local invasion and metastatic prostate cancer, respectively (4). The aggressiveness of prostate cancer is best described by the Gleason grading system (5). The hormone responsiveness is an important feature in this cancer resulting in tumor regression following castration (6). Therefore, androgen deprivation therapy has been suggested as the regular therapeutic regimen for prostate cancer. However, resistance to this therapeutic modality can develop (4).
Identification of the underlying cause of initiation and progression of prostate cancer is an imperative step in development of novel therapies for this kind of malignancy. Moreover, it can facilitate design of novel biomarkers for early detection of cancers. Long non-coding RNAs (lncRNAs) are promising transcripts for both purposes (7–9). These transcripts have sizes more than 200 nucleotides and are responsible for a variety of regulatory mechanisms at different levels of gene expression regulation (10). Aberrations in the expression of lncRNAs might be representative of certain phases of cancer progression, and can be used to predict early progression of cancer or induction of cancer‐related signaling pathways (11, 12). Therefore, these transcripts have attained much attention during recent years for their contribution in the pathogenesis of almost all kinds of cancers, including prostate cancer. The current review summarized the role of lncRNAs in the evolution of prostate cancer with an especial focus on their importance in design of novel biomarker panels and therapeutic targets. We used PubMed and Google Scholar databases with the key words “lncRNA” or “long non-coding RNA” and “prostate cancer”. Then, we screened the obtained articles and included the relevant ones in the manuscript. Finally, we tabulated the data obtained from these articles for the purpose of better classification of the data.
Up-regulated lncRNAs in prostate cancer
Using quantitative real time PCR method, several lncRNAs have been shown to be over-expressed in prostate cancer tissues compared with adjacent non-cancerous tissues or benign prostate hyperplasia (BPH) samples, representing an oncogenic role for these transcripts in the progression of prostate cancer (Table 1). Small nucleolar RNA host genes (SNHGs) are among the mostly assessed lncRNAs in this field. A number of well-known oncogenic lncRNAs in other cancers such as DANCR, MALAT1, CCAT1, PVT1, TUG1 and NEAT1 have also been shown to act as oncogenes in prostate cancer. For instance, DANCR has been found to contribute to the taxol resistance of in this type of cancer via modulation of miR-33b-5p/LDHA axis (44). Expression of this lncRNA has been up-regulated in serum samples of prostate cancer patients, parallel with down-regulation of miR-214-5p. Notably, DANCR expression has been correlated with PSA level, Gleason score and T stage in these patients. DANCR expression not only can be used for prostate cancer diagnosis, but also can predict poor prognosis of this type of cancer with high diagnostic value. Mechanistically, up-regulation of DANCR or down-regulation of miR-214-5p could enhance proliferation and migration, preclude apoptosis, and induce activity of TGF-β signaling (45). DANCR can also target miR-185-5p to increase expression of LIM and SH3 protein 1 promoting prostate cancer through the FAK/PI3K/AKT/GSK3β/snail axis (46).

Table 1 Summary of function of up-regulated lncRNAs in prostate cancer (Official HUGO Gene Nomenclature symbols are used).
In addition, MALAT1 has been found to regulate glucose metabolism through modulation of MYBL2/mTOR axis (47). Moreover, in vitro and in vivo studies have shown the importance of MALAT1/miR-140/BIRC6 axis in the progression of prostate cancer (48). In fact, MALAT1 acts as a molecular sponge for miR-140 to enhance expression of the anti-apoptotic protein BIRC6 (48). In turn, expression and activity of MALAT1 have been shown to be regulated by miR-423-5p, a miRNA that impedes activity of MALAT1 in enhancement of proliferation, migration, and invasiveness of prostate cancer cells (49). Most importantly, up-regulation of miR-423-5p could enhance survival and decrease metastasis formation in a xenograft model of prostate cancer (49). In addition, MALAT1 has a possible diagnostic value in prostate cancer. Expression levels of PCA3 and MALAT1 in urinary exosomes have been shown to be superior to the currently used clinical parameters in detection of prostate cancer, particularly high-grade ones (51).
NEAT1 has also been shown to regulate aerobic glycolysis to affect tumor immunosurveillance by T cells in this type of cancer (13). It can also promote progression of prostate cancer through modulation of miR-766-5p/E2F3 axis (54).
CTBP1-AS is reported as the antisense-RNA transcript positively regulated by androgen and promotes castration-resistant prostate cancer tumor growth (123). This lncRNA is localized in the nucleus and its levels are mostly increased in prostate cancer. It enhances both hormone-dependent and castration-resistant tumor growth. From a mechanistical point of view, CTBP1-AS suppresses the expression of CTBP1 through recruitment of PSF and histone deacetylases. It also exerts androgen-dependent function through inhibition of tumor-suppressor genes and enhancement of cell cycle progression (123).
Epigenetic repression of AR corepressor is an important mechanism for AR activation. ARLNC1 is also regulated by androgen and upregulates AR mRNA stability by binding to the 3’-UTR. In line with this, ARLNC1 silencing leads to inhibition of AR expression and suppression of AR signaling as well as of growth of prostate cancer. In fact, ARLNC1 has a role in the preservation of a positive feedback loop that induces AR signaling in the course of prostate cancer progression (124). In addition to these lncRNAs, several CRPC-specific AR-regulated lncRNAs are important for overexpression of AR and its variant. These AR-regulated lncRNAs are over-expressed in CRPC tissues. An experiment in these cells has shown that knock-down of PRKAG2-AS1 and HOXC-AS1 leads to suppression of CRPC tumor growth in addition to inhibition of expression of AR and AR variant. Mechanistically, PRKAG2-AS1 modulates the subcellular localization of the splicing factor, U2AF2. This splicing factor is involved in the AR splicing system (125).
SChLAP1 is another up-regulated lncRNA in prostate cancer whose up-regulation is associated with poor patient outcomes, such as metastases and prostate cancer specific mortality. It has a critical role in invasiveness and metastasis. Functionally, SChLAP1 influences the localization and regulatory function of the SWI/SNF complex (126).
PCAT-1 is another up-regulated lncRNA in prostate cancer which enhances cell proliferation through cMyc. Mechanistically, PCAT-1–associated proliferation depends on stabilization of cMyc protein. Moreover, cMyc has an essential role in a number of PCAT-1–induced expression alterations (127).
HOTAIR as regarded as an AR-repressed lncRNA is upregulated after androgen deprivation therapy and in CRPC. Mechanistically, HOTAIR binds to the AR protein to inhibit its interactions with the E3 ubiquitin ligase MDM2, thus suppressing AR ubiquitination and its degradation. Therefore, HOTAIR induces androgen-independent AR activation and drives the AR-mediated transcriptional program in the absence of androgen (128). Another study has shown that NEAT1 induces oncogenic growth in prostate tissue through changing the epigenetic marks in the target genes promoters to induce their transcription (129). Moreover, PCGEM1 and PRNCR1 bind to AR and enhance selective looping of AR-bound enhancers to target gene promoters (130). Similarly, SOCS2-AS1 interacts with AR for co-factor interaction (131).
The importance of other up-regulated lncRNAs in prostate cancer is summarized in Figure 1 and Table 1.
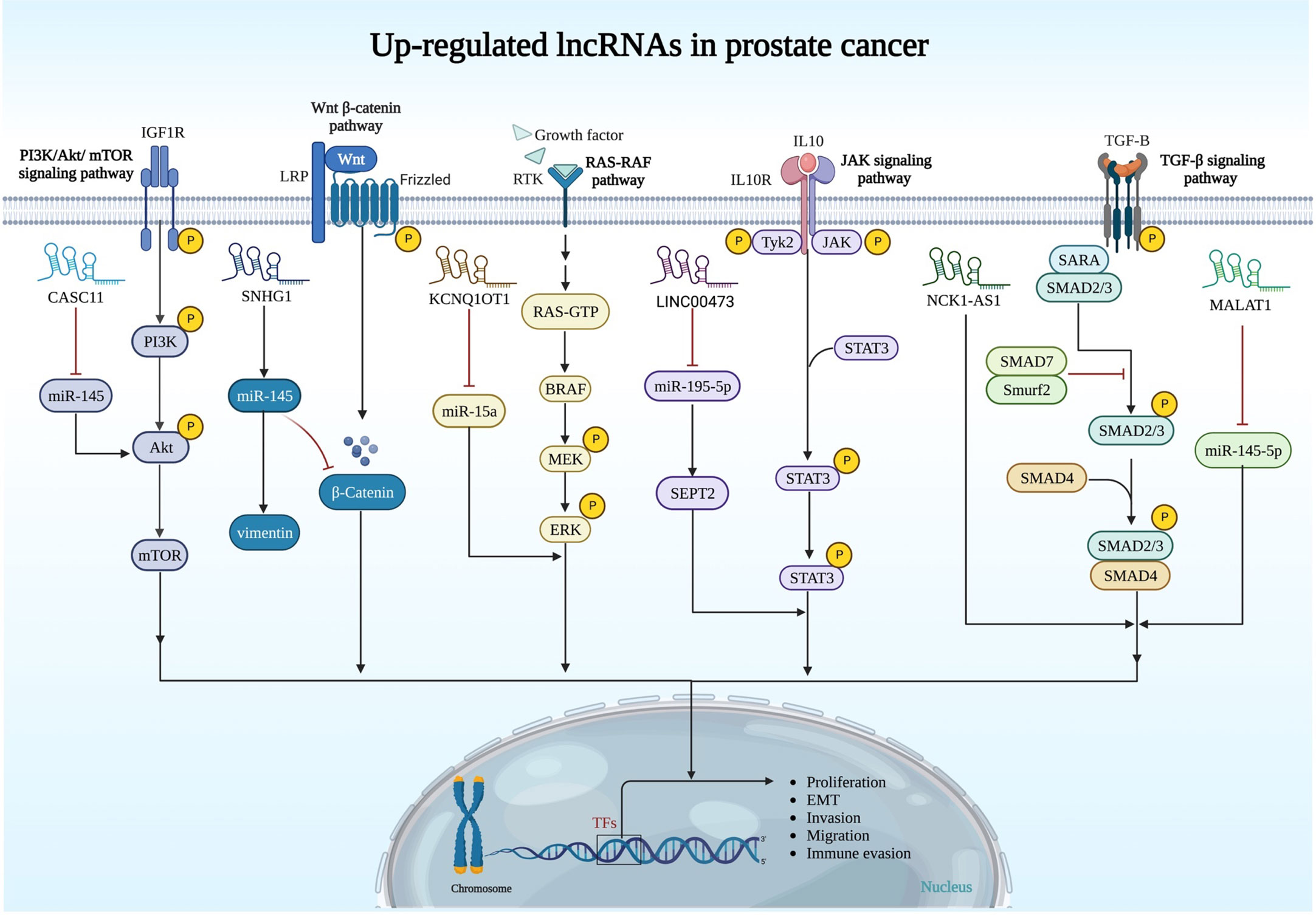
Figure 1 Upregulation of oncogenic lncRNAs and their relation with signaling pathways in prostate cancer. PI3K/AKT/mTOR, Wnt/β-catenin, RAS/RAF, JAK and TGF-β pathways are regulated by oncogenic lncRNAs in prostate cancer.
Down-regulated lncRNAs in prostate cancer
A number of other lncRNAs have been found to act as tumor suppressors in prostate cancer (Table 2). For instance, LINC00893 can inhibit progression of this type of cancer via modulation of miR-3173-5p/SOCS3/JAK2/STAT3 axis (132). Similarly, the sponging effect of LINC01679 on miR-3150a-3p has a role in inhibition of progression of prostate cancer through affecting expression of SLC17A9 (133). MIR22HG is another tumor suppressor lncRNA that acts as a molecular sponge for miR-9-3p (134). The tumor suppressor role of RP1-59D14.5 in prostate cancer is mediated through activation of the Hippo signaling and enhancement of autophagy (135). Moreover, MAGI2-AS3 has been shown to inactivate STAT3 signaling and suppress proliferation of prostate cancer cells through acting as a miR-424-5p sponge (136). NXTAR is another tumor suppressor lncRNA that modulates expression of androgen receptor (AR) and resistance to enzalutamide (137). Totally, the number of identified tumor suppressor lncRNAs in prostate cancer is far below that of oncogenic lncRNAs (Figure 2). Table 2 summarizes the information about tumor suppressor lncRNAs in prostate cancer.
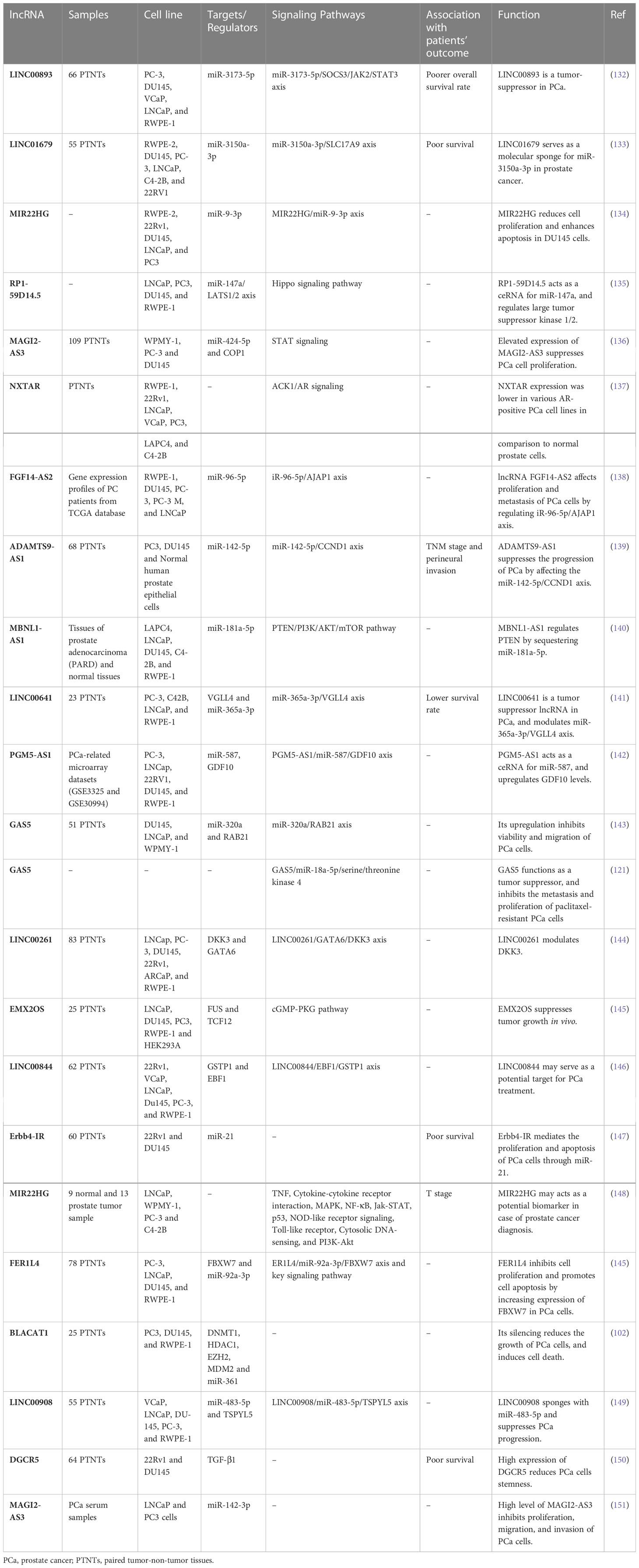
Table 2 Summary of function of down-regulated lncRNAs in prostate cancer (Official HUGO Gene Nomenclature symbols are used).
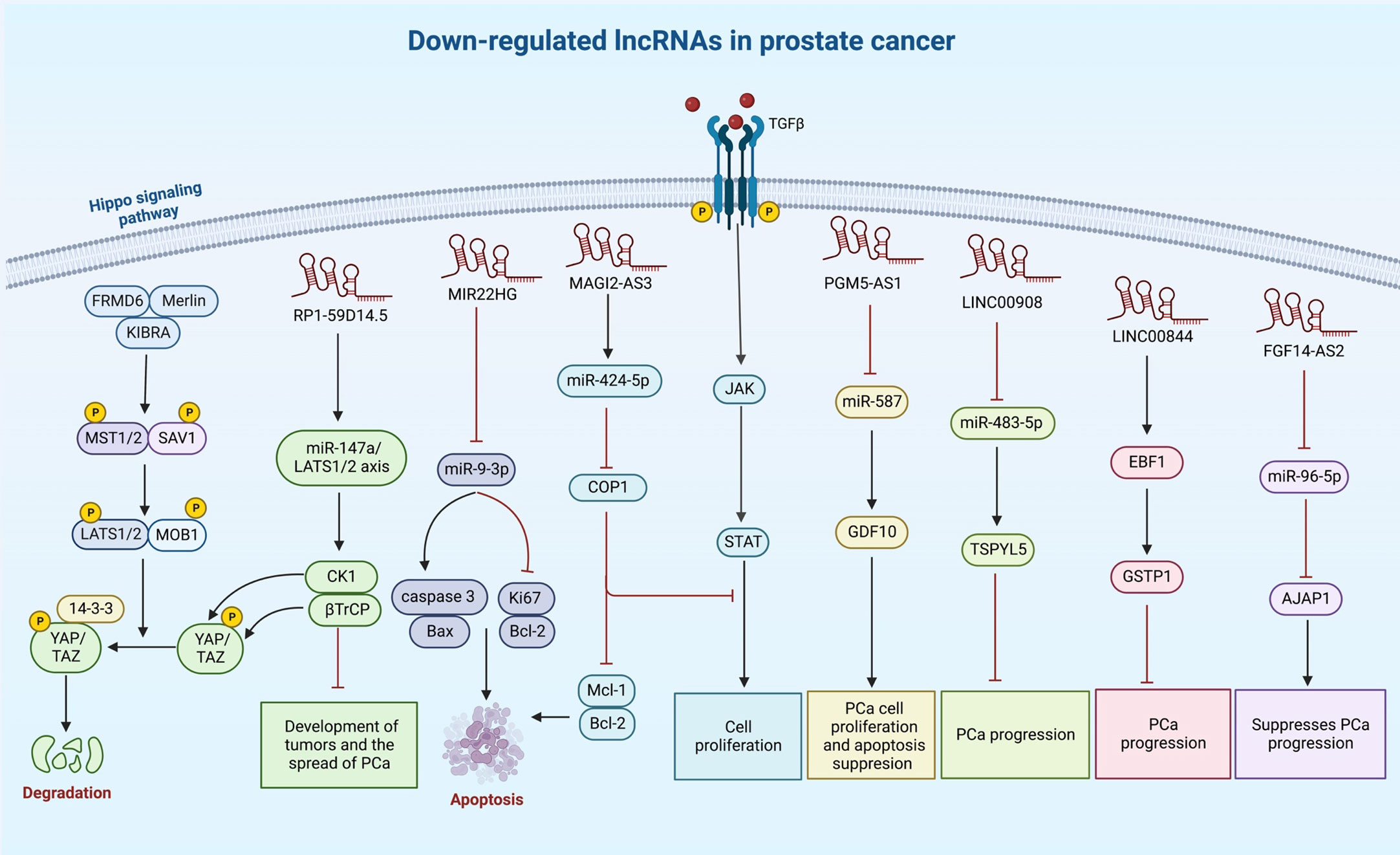
Figure 2 A synopsis of the known roles of lncRNA tumor suppressors in prostate cancer. Several lncRNAs can reduce cell proliferation and invasiveness of prostate cancer cells, particularly through sponging oncogenic miRNAs.
Contribution of lncRNAs variants in prostate cancer
Contribution of single nucleotide polymorphisms (SNPs) within GAS5, POLR2E, MEG3, MALAT1 and HOTAIR in the risk of prostate cancer has been assessed in different ethnic groups (Table 3). Three SNPs within GAS5 have been the subject of these investigations. First, rs145204276 (delCAAGG) is located within the promoter region of GAS5. Compared with subjects carrying ins/ins genotype, cases with ins/del or del/del genotype of this polymorphism have shown decreased risk of pathological lymph node metastasis (152). The rs17359906 in GAS5 is another SNP whose A allele has been shown to be a risk allele for prostate cancer. Similarly, A allele of rs1951625 SNP within GAS5 has been associated with higher risk of this cancer. Both rs17359906 G > A and rs1951625 G > A have been associated with high plasma level of PSA. Most importantly, the recurrence-free survival of patients with prostate cancer has been lowest in patients having AA genotype of rs17359906 and highest in those having GG genotype. Similar findings have been reported for the rs1951625 (153).
A systematic review and meta-analysis of 5 studies on the role of rs3787016 within POLR2E has revealed increased susceptibility to prostate cancer for carriers of T allele in all genotype models (154). The results of other studies on contribution of lncRNAs SNPs in prostate cancer are shown in Table 3.
Importance of lncRNAs as prognostic factors in prostate cancer
Several studies have indicated the importance of dysregulation of lncRNAs in the prediction of survival times of patients with prostate cancer (Table 4). Overall, up-regulation of oncogenic lncRNAs is predictive of lower survival time of patients in terms of overall survival or progression-free survival. For tumor suppressor lncRNAs, an opposite effect has been seen.
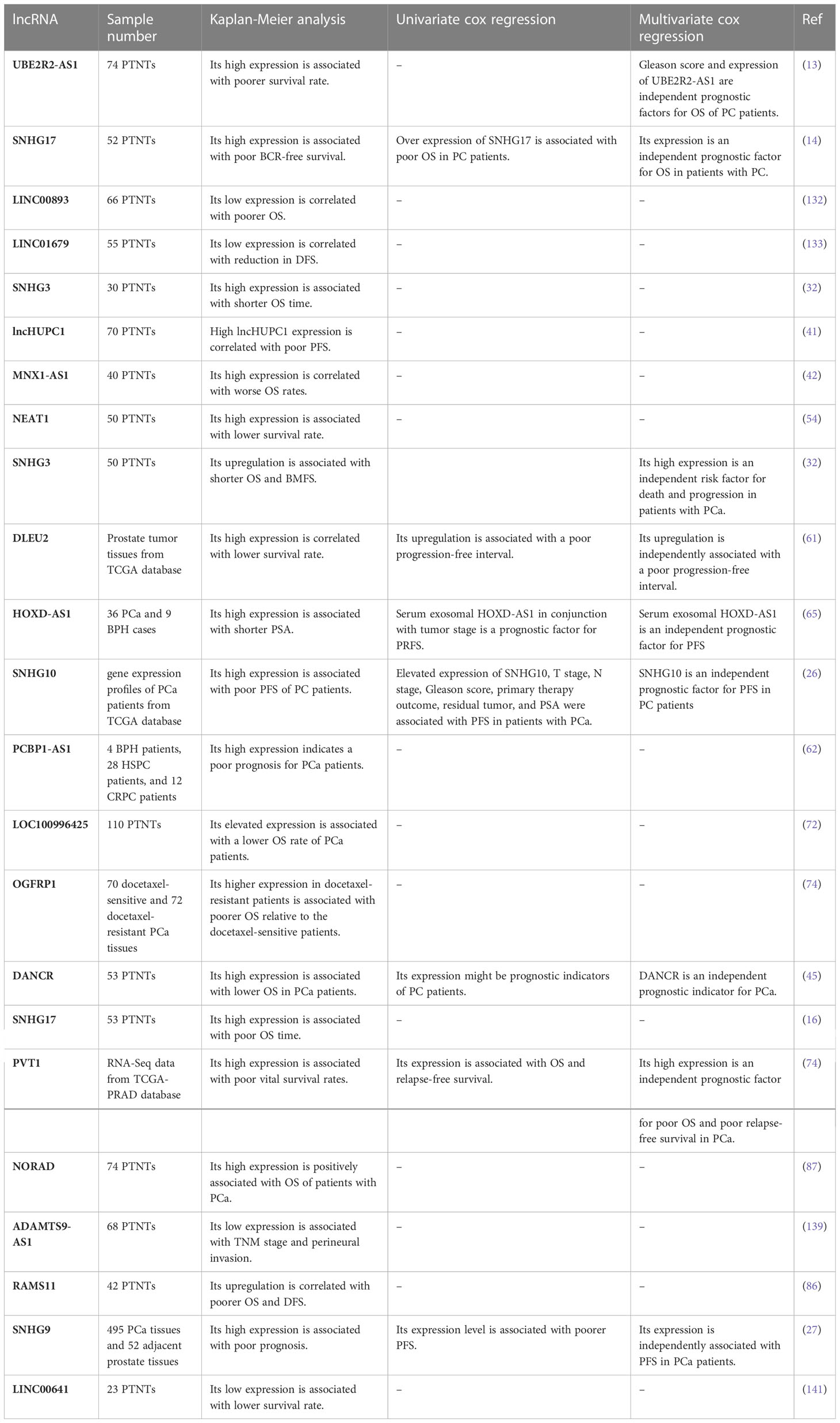
Table 4 Importance of lncRNAs as prognostic factors in prostate cancer (PTNTs, paired tumor-non-tumor tissues; PCa, prostate cancer; OS, overall survival; PFS, progression-free survival).
Discussion
Several lncRNAs have been shown to contribute to the pathogenesis of prostate cancer via modulation of AR signaling, ubiquitin–proteasome degradation process of AR or other important signaling pathways. Some of them such as PCA3 are highly specific for this kind of cancer, representing an appropriate biomarker for prostate cancer (151). Others might be over-/under-expressed in a variatey of cancers, being therapeutic targets for a wide range of human malignnacies. The observed differences in expression of some lncRNAs between castration-resistant prostate cancer and androgen deprivation therapy-responsive cases imply the importance of these transcripts in defining response of patients to this therapeutic modality and represent these transcripts as targets for management of resistance to this therapy.
Although numerous prostate cancer-specific or prostate cancer-associated lncRNAs have been recognized, few lncRNAs have been verified in independent patient cohorts or approved for using in clinical settings. The most important milestone in the field of lncRNA research is probably approval of urinary PCA3 as a biomarker for detection of prostate cancer by the United States Food and Drug Administration (158). This lncRNA is a promising factor for urine test for prostate cancer and has a superior performance compared with PSA in urinary detection of this disorder. Further reseraches are needed to find other appropriate lncRNA biomarkers for this kind of cancer.
LncRNA profiles can also been used to identify prostae cancer patients that benefit from radiotherapy. For instance, UCA1 has beens shwon to mediate radiosensitivity in prostate cancer cell lines and therefore might be a marker to predict response to radiotherapy in these patients. This lncRNA affects radiosensitivity through influencing cell cycle progression (159).
The importance of lncRNAs in the mediation of cell proliferation, invasiveness and metastasis has potentiated them as therapeutic targets for prostate cancer. The results of animal studies have been promising particularly for some AR-regulated lncRNAs. However, clinical studies are missing in this field.
Notably, LncRNAs are also involved in drug resistance in prostate cancer cells, thus they are proper candidates for therapeutic targeting (160). For instance, HORAS5 up-regulation can trigger taxane resistance in CRPC cells through upregulation of BCL2A1. HORAS5 silencing can reduce resistance of prostate cancer cells to cabazitaxel and enhance the efficacy of chemotherapy (161).
PI3K/AKT/mTOR, Wnt/β-catenin, TGF-β, p53, FAK/PI3K/AKT/GSK3β/Snail, STAT3, FAK/AKT/β catenin, Ras/ERK, NF-κB and FOXO signaling pathways are among signaling pathways that are modulated by lncRNAs in the context of prostate cancer. Moreover, several lncRNAs have been shown to act as molecular sponges for miRNAs to regulated expression of miRNA targets. miR-145/IGF1R, miR-23a/OTUB1, miR-339-5p/STAT5A/SNORA71B, miR-144/CD51, miR-5590-3p/YY1, miR-195/CCNE1, miR-184/IGF, miR-152-3p/SLC7A11, miR-214-3p/TGF-β, miR‐577/SMURF1, miR-377-3p/AKT2, miR-133b/SDCCAG3, miR-2113/MDM2, miR-16-5p/HMGA2, miR-140/BIRC6 axis, miR-145-5p-SMAD3/TGFBR2, miR-129-5p/CDT1 axis, miR-766-5p/E2F3, miR-1182/AKT3, miR-582-5p/SGK1, miR-361-5p/FOXM1, miR-24-3p/JPT1, miR-509-3p/PBX3, miR-370-3p/DDX3X, miR-212‐5p/FZD5, miR-3167/YWHAZ, miR-490-3p/FRAT1, miR-24-3p/FSCN1, miR-149-5p/IL-6, miR-1245b-5p/CASK, miR-628-5p/FOXP2, miR-326/Hnrnpa2b1, miR-195-5p/FKBP1A, miR-15b/IGF1R, miR-494-3p/STAT3, miR-486-5p/GOLPH3, miR-15a-5p/KIF23 and miR-101/Rap1A are among putative miRNA/mRNA axes that are modulated by oncogenic lncRNAs in the context of prostate cancer.
Although expression profile of lncRNAs have been comprhensively assessed in tumoral tissues of patients with prostate cancer, less effort has been made for analysis of their expression in urine or serum samples. Based on the availability of these sources for non-invasive diagnostic procedures, future studies should focus on these biofluids to facilitate early detection of prostate cancer via non-invasive methods.
Taken together, lncRNAs have been found to contribute to the pathogenesis of prostate cancer through various mechanisms. These transcripts can be used as targets for therapeutic interventions in this kind of cancer.
Author contributions
MT and AB designed and supervised the study. SG-F wrote the draft and revised it. EB, BH, and AK collected the data and designed the figures and tables. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistic. CA Cancer J Clin (2022) 72:7–33. doi: 10.3322/caac.21708
3. Huncharek M, Haddock KS, Reid R, Kupelnick B. Smoking as a risk factor for prostate cancer: A meta-analysis of 24 prospective cohort studies. Am J Public Health (2010) 100:693–701. doi: 10.2105/AJPH.2008.150508
4. Wang G, Zhao D, Spring DJ, Depinho RA. Genetics and biology of prostate cancer. Genes Dev (2018) 32:1105–40. doi: 10.1101/gad.315739.118
5. Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 international society of urological pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol (2016) 40:244–52. doi: 10.1097/PAS.0000000000000530
6. Huggins C, Hodges CV. Studies on prostatic cancer. Cancer Res (1941) 1:293–7. doi: 10.1001/archsurg.1941.01210140043004
7. Misawa A, Takayama KI, Inoue S. Long non-coding RNAs and prostate cancer. Cancer Sci (2017) 108:2107–14. doi: 10.1111/cas.13352
8. Taheri M, Pouresmaeili F, Omrani MD, Habibi M, Sarrafzadeh S, Noroozi R, et al. Association of ANRIL gene polymorphisms with prostate cancer and benign prostatic hyperplasia in an Iranian population. Biomarkers Med (2017) 11:413–22. doi: 10.2217/bmm-2016-0378
9. Taheri M, Khoshbakht T, Jamali E, Kallenbach J, Ghafouri-Fard S, Baniahmad A. Interaction between non-coding RNAs and androgen receptor with an especial focus on prostate cancer. Cells (2021) 10. doi: 10.3390/cells10113198
10. Ghafouri-Fard S, Dashti S, Hussen BM, Farsi M, Taheri M. BCYRN1: An oncogenic lncRNA in diverse cancers. Pathol - Res Pract (2021) 220:153385. doi: 10.1016/j.prp.2021.153385
11. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov (2011) 1:391–407. doi: 10.1158/2159-8290.CD-11-0209
12. Hussen BM, Azimi T, Abak A, Hidayat HJ, Taheri M, Ghafouri-Fard S. Role of lncRNA BANCR in human cancers: An updated review. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.689992
13. Xia KG, Wang CM, Shen DY, Song XY, Mu XY, Zhou JW, et al. LncRNA NEAT1-associated aerobic glycolysis blunts tumor immunosurveillance by T cells in prostate cancer. Neoplasma (2022) 69:594–602. doi: 10.4149/neo_2022_211021N1497
14. Sun X, Xin S, Zhang Y, Jin L, Liu X, Zhang J, et al. Long non−coding RNA CASC11 interacts with YBX1 to promote prostate cancer progression by suppressing the p53 pathway. Int J Oncol (2022) 61.
15. Capik O, Sanli F, Kurt A, Ceylan O, Suer I, Kaya M, et al. CASC11 promotes aggressiveness of prostate cancer cells through miR-145/IGF1R axis. Prostate Cancer Prostatic Dis (2021) 24:891–902. doi: 10.1038/s41391-021-00353-0
16. Zhao H, Dong H, Wang P, Zhu H. Long non-coding RNA SNHG17 enhances the aggressiveness of C4-2 human prostate cancer cells in association with β-catenin signaling. Oncol Lett (2021) 21:472. doi: 10.3892/ol.2021.12733
17. Wu G, Hao C, Qi X, Nie J, Zhou W, Huang J, et al. LncRNA SNHG17 aggravated prostate cancer progression through regulating its homolog SNORA71B via a positive feedback loop. Cell Death Dis (2020) 11:393. doi: 10.1038/s41419-020-2569-y
18. Bai M, Lei Y, Wang M, Ma J, Yang P, Mou X, et al. Long non-coding RNA SNHG17 promotes cell proliferation and invasion in castration-resistant prostate cancer by targeting the miR-144/CD51 axis. Front Genet (2020) 11:274. doi: 10.3389/fgene.2020.00274
19. Weng W, Liu C, Li G, Ruan Q, Li H, Lin N, et al. Long non-−coding RNA SNHG16 functions as a tumor activator by sponging miR−373−3p to regulate the TGF−β−R2/SMAD pathway in prostate cancer. Mol Med Rep (2021) 24.
20. Shao M, Yu Z, Zou J. LncRNA-SNHG16 silencing inhibits prostate carcinoma cell growth, downregulate GLUT1 expression and reduce glucose uptake. Cancer Manag Res (2020) 12:1751–7. doi: 10.2147/CMAR.S231370
21. Luo ZF, Peng Y, Liu FH, Ma JS, Hu G, Lai SL, et al. Long noncoding RNA SNHG14 promotes malignancy of prostate cancer by regulating with miR-5590-3p/YY1 axis. Eur Rev Med Pharmacol Sci (2020) 24:4697–709.
22. Chen Z, Qi T, Qin XP, Wang J, Huang ZS, Hu XY, et al. Long noncoding RNA SNHG12 promotes prostate tumor occurrence and progression via AKT regulation. BioMed Res Int (2020) 2020:8812923. doi: 10.1155/2020/8812923
23. Wang X, He C, Yang Z, Li S, Qiao L, Fang L. Dysregulation of long non-coding RNA SNHG12 alters the viability, apoptosis, and autophagy of prostate cancer cells by regulating miR-195/CCNE1 axis. Int J Clin Exp Pathol (2019) 12:1272–83.
24. Li Y, Pan B, Guo X, Meng X, Tian X. Prognostic value of long noncoding RNA SNHG11 in patients with prostate cancer. Horm Metab Res (2022) 54:187–93. doi: 10.1055/a-1745-8952
25. Xie Q, Zhao S, Kang R, Wang X. lncRNA SNHG11 facilitates prostate cancer progression through the upregulation of IGF−1R expression and by sponging miR−184. Int J Mol Med (2021) 48.
26. Chen Q, Yang X, Gong B, Xie W, Ma M, Fu S, et al. SNHG10 is a prognostic biomarker correlated with immune infiltrates in prostate cancer. Front Cell Dev Biol (2021) 9:731042. doi: 10.3389/fcell.2021.731042
27. Li C, Hu J, Hu X, Zhao C, Mo M, Zu X, et al. LncRNA SNHG9 is a prognostic biomarker and correlated with immune infiltrates in prostate cancer. Transl Androl Urol (2021) 10:215–26. doi: 10.21037/tau-20-1134
28. Shi Z, Zhang H, Jie S, Yang X, Huang Q, Mao Y, et al. Long non-coding RNA SNHG8 promotes prostate cancer progression through repressing miR-384 and up-regulating HOXB7. J Gene Med (2021) 23:e3309. doi: 10.1002/jgm.3309
29. Liu J, Yuan JF, Wang YZ. METTL3-stabilized lncRNA SNHG7 accelerates glycolysis in prostate cancer via SRSF1/c-myc axis. Exp Cell Res (2022) 416:113149. doi: 10.1016/j.yexcr.2022.113149
30. Xia Q, Li J, Yang Z, Zhang D, Tian J, Gu B. Long non-coding RNA small nucleolar RNA host gene 7 expression level in prostate cancer tissues predicts the prognosis of patients with prostate cancer. Med (Baltimore) (2020) 99:e18993. doi: 10.1097/MD.0000000000018993
31. Cao C, Sun G, Liu C. Long non-coding RNA SNHG6 regulates the sensitivity of prostate cancer cells to paclitaxel by sponging miR-186. Cancer Cell Int (2020) 20:381. doi: 10.1186/s12935-020-01462-x
32. Xi X, Hu Z, Wu Q, Hu K, Cao Z, Zhou J, et al. High expression of small nucleolar RNA host gene 3 predicts poor prognosis and promotes bone metastasis in prostate cancer by activating transforming growth factor-beta signaling. Bioengineered (2022) 13:1895–907. doi: 10.1080/21655979.2021.2020393
33. Wang X, Song Y, Shi Y, Yang D, Li J, Yin B. SNHG3 could promote prostate cancer progression through reducing methionine dependence of PCa cells. Cell Mol Biol Lett (2022) 27:13. doi: 10.1186/s11658-022-00313-z
34. Yu L, Ren Y. Long noncoding RNA small nucleolar RNA host gene 3 mediates prostate cancer migration, invasion, and epithelial-mesenchymal transition by sponging miR-487a-3p to regulate TRIM25. Cancer Biother Radiopharm (2022) 37:451–65. doi: 10.1089/cbr.2020.3988
35. Li T, Xing Y, Yang F, Sun Y, Zhang S, Wang Q, et al. LncRNA SNHG3 sponges miR-577 to up-regulate SMURF1 expression in prostate cancer. Cancer Med (2020) 9:3862–52. doi: 10.1002/cam4.2992
36. Tan X, Chen WB, Lv DJ, Yang TW, Wu KH, Zou LB, et al. LncRNA SNHG1 and RNA binding protein hnRNPL form a complex and coregulate CDH1 to boost the growth and metastasis of prostate cancer. Cell Death Dis (2021) 12:138. doi: 10.1038/s41419-021-03413-4
37. Xiong H, Shen J, Chen Z, Yang J, Xie B, Jia Y, et al. H19/let−7/Lin28 ceRNA network mediates autophagy inhibiting epithelial−mesenchymal transition in breast cancer. Int J Oncol (2020) 56:794–806.
38. Tang Q, Li Z, Han W, Cheng S, Wang Y. High expression of lncRNA SNHG1 in prostate cancer patients and inhibition of SNHG1 suppresses cell proliferation and promotes apoptosis. Indian J Pathol Microbiol (2020) 63:575–80. doi: 10.4103/IJPM.IJPM_612_19
39. Meng XF, Liu AD, Li SL. SNHG1 promotes proliferation, invasion and EMT of prostate cancer cells through miR-195-5p. Eur Rev Med Pharmacol Sci (2020) 24:9880–8.
40. Xie M, Zhang Z, Cui Y. Long noncoding RNA SNHG1 contributes to the promotion of prostate cancer cells through regulating miR-377-3p/AKT2 axis. Cancer Biother Radiopharm (2020) 35:109–19. doi: 10.1089/cbr.2019.3177
41. Ge Y, Liu L, Luo L, Fang Y, Ni T. MIR22HG aggravates oxygen-glucose deprivation and reoxygenation-induced cardiomyocyte injury through the miR-9-3p/SH2B3 axis. Cardiovasc Ther (2022) 2022:7332298. doi: 10.1155/2022/7332298
42. Liang D, Tian C, Zhang X. lncRNA MNX1−AS1 promotes prostate cancer progression through regulating miR−2113/MDM2 axis. Mol Med Rep (2022) 26.
43. Huang F, Zhou LQ. Effect and mechanism of lncRNA CERS6-AS1 on the biological behavior of prostate cancer cell. Appl Bionics Biomech (2022) 2022:9292538. doi: 10.1155/2022/9292538
44. Wang YY, Chen C. lncRNA-DANCR promotes taxol resistance of prostate cancer cells through modulating the miR-33b-5p-LDHA axis. Dis Markers (2022) 2022:9516774. doi: 10.1155/2022/9516774
45. Deng H, Zhu B, Dong Z, Jiang H, Zhao X, Wu S. miR-214-5p targeted by LncRNA DANCR mediates TGF-β signaling pathway to accelerate proliferation, migration and inhibit apoptosis of prostate cancer cells. Am J Transl Res (2021) 13:2224–40.
46. Sun W, Zu S, Shao G, Wang W, Gong F. Long non-coding DANCR targets miR-185-5p to upregulate LIM and SH3 protein 1 promoting prostate cancer via the FAK/PI3K/AKT/GSK3β/snail pathway. J Gene Med (2021) 23:e3344. doi: 10.1002/jgm.3344
47. Mu X, Shen Z, Lin Y, Xiao J, Xia K, Xu C, et al. LncRNA-MALAT1 regulates cancer glucose metabolism in prostate cancer via MYBL2/mTOR axis. Oxid Med Cell Longev (2022) 2022:8693259. doi: 10.1155/2022/8693259
48. Hao T, Wang Z, Yang J, Zhang Y, Shang Y, Sun J. MALAT1 knockdown inhibits prostate cancer progression by regulating miR-140/BIRC6 axis. BioMed Pharmacother (2020) 123:109666. doi: 10.1016/j.biopha.2019.109666
49. Ferri C, Di Biase A, Bocchetti M, Zappavigna S, Wagner S, Le Vu P, et al. MiR-423-5p prevents MALAT1-mediated proliferation and metastasis in prostate cancer. J Exp Clin Cancer Res (2022) 41:20. doi: 10.1186/s13046-021-02233-w
50. Zhang D, Fang C, Li H, Lu C, Huang J, Pan J, et al. Long ncRNA MALAT1 promotes cell proliferation, migration, and invasion in prostate cancer via sponging miR-145. Transl Androl Urol (2021) 10:2307–19. doi: 10.21037/tau-20-1526
51. Li Y, Ji J, Lyu J, Jin X, He X, Mo S, et al. A novel urine exosomal lncRNA assay to improve the detection of prostate cancer at initial biopsy: A retrospective multicenter diagnostic feasibility study. Cancers (Basel) (2021) 13. doi: 10.3390/cancers13164075
52. Liu H, He X, Li T, Qu Y, Xu L, Hou Y, et al. PCGEM1 promotes proliferation, migration and invasion in prostate cancer by sponging miR-506 to upregulate TRIAP1. BMC Urol (2022) 22:14. doi: 10.1186/s12894-022-00969-x
53. Bai J, Huang G. Role of long non-coding RNA NEAT1 in the prognosis of prostate cancer patients. Med (Baltimore) (2020) 99:e20204. doi: 10.1097/MD.0000000000020204
54. Zhao W, Zhu X, Jin Q, Lin B, Ji R. The lncRNA NEAT1/miRNA-766-5p/E2F3 regulatory axis promotes prostate cancer progression. J Oncol (2022) 2022:1866972. doi: 10.1155/2022/1866972
55. Nitusca D, Marcu A, Dema A, Balacescu L, Balacescu O, Bardan R, et al. Long noncoding RNA NEAT1 as a potential candidate biomarker for prostate cancer. Life (Basel) (2021) 11. doi: 10.3390/life11040320
56. Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer (2020) 19:171. doi: 10.1186/s12943-020-01293-4
57. Zhou J, Li J, Qian C, Qiu F, Shen Q, Tong R, et al. LINC00624/TEX10/NF-κB axis promotes proliferation and migration of human prostate cancer cells. Biochem Biophys Res Commun (2022) 601:1–8. doi: 10.1016/j.bbrc.2022.02.078
58. Arslan A, Batar B, Temiz E, Tozkir H, Koyuncu I, Bozgeyik E. Silencing of TP73-AS1 impairs prostate cancer cell proliferation and induces apoptosis via regulation of TP73. Mol Biol Rep (2022) 49:6859–69. doi: 10.1007/s11033-022-07141-6
59. Qin D, Ni C, Tan B, Huang S, Deng B, Huang Z. LINC01207 promotes prostate cancer progression by sponging miR-1182 to upregulate AKT3. Oncol Lett (2022) 23:57. doi: 10.3892/ol.2021.13175
60. Yan Y, Liu J, Xu Z, Ye M, Li J. lncRNA PCAT14 is a diagnostic marker for prostate cancer and is associated with immune cell infiltration. Dis Markers (2021) 2021:9494619. doi: 10.1155/2021/9494619
61. Li P, Xu H, Yang L, Zhan M, Shi Y, Zhang C, et al. E2F transcription factor 2-activated DLEU2 contributes to prostate tumorigenesis by upregulating serum and glucocorticoid-induced protein kinase 1. Cell Death Dis (2022) 13:77. doi: 10.1038/s41419-022-04525-1
62. Cai Z, Wu Y, Ju G, Wang G, Liu B. Role of BCAR4 in prostate cancer cell autophagy. Transl Androl Urol (2021) 10:4253–61. doi: 10.21037/tau-21-929
63. Ye C, Qin S, Guo F, Yang Y, Wang H, Zhang C, et al. LncRNA EIF3J-AS1 functions as an oncogene by regulating MAFG to promote prostate cancer progression. J Cancer (2022) 13:146–52. doi: 10.7150/jca.60676
64. Ahani M, Ghaderian SMH, Mehr Azma M, Kamali K, Naghavi Gargari B, Bahramali G, et al. Differential gene expression of BCL-2, ZEB2-AS1 and BALR-2 in prostate cancer and benign prostatic hyperplasia. Andrologia (2022) 54:e14344. doi: 10.1111/and.14344
65. Jiang Y, Zhao H, Chen Y, Li K, Li T, Chen J, et al. Exosomal long noncoding RNA HOXD-AS1 promotes prostate cancer metastasis via miR-361-5p/FOXM1 axis. Cell Death Dis (2021) 12:1129. doi: 10.1038/s41419-021-04421-0
66. Cheng Y, Xiong HY, Li YM, Zuo HR, Liu Y, Liao GL. LncRNA HOXA11-AS promotes cell growth by sponging miR-24-3p to regulate JPT1 in prostate cancer. Eur Rev Med Pharmacol Sci (2021) 25:4668–77.
67. Yang C, Shen S, Zheng X, Ye K, Ge H, Sun Y, et al. Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. FASEB J (2020) 34. doi: 10.1096/fj.201900731RR
68. Pan X, Chen G, Hu W. lncRNA HLA complex group 18 (HCG18) facilitated cell proliferation, invasion, and migration of prostate cancer through modulating miR-370-3p/DDX3X axis. Reprod Sci (2021) 28:3406–16. doi: 10.1007/s43032-021-00614-2
69. Peng N, Zhang Z, Wang Y, Yang M, Fan J, Wang Q, et al. Down-regulated LINC00115 inhibits prostate cancer cell proliferation and invasion via targeting miR-212-5p/FZD5/Wnt/β-catenin axis. J Cell Mol Med (2021) 25:10627–10637. doi: 10.1111/jcmm.17000
70. Qu W, Wei X, Zhang H, Hou J. FOXD1-AS1 promotes malignant behaviours of prostate cancer cells via the miR-3167/YWHAZ axis. Andrologia (2022) 54:e14263. doi: 10.1111/and.14263
71. Fu C, Xin J, Zhang W, Lai J, Huang Z. LINC00992 exerts oncogenic activities in prostate cancer via regulation of SOX4. Exp Cell Res (2021) 408:112855. doi: 10.1016/j.yexcr.2021.112855
72. Cai X, Dai Y, Gao P, Ren G, Cheng D, Wang B, et al. LncRNA CCAT1 promotes prostate cancer cells proliferation, migration, and invasion through regulation of miR-490-3p/FRAT1 axis. Aging (Albany NY) (2021) 13:18527–44. doi: 10.18632/aging.203300
73. Li X, Han X, Wei P, Yang J, Sun J. Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol Ther (2020) 21:452–62. doi: 10.1080/15384047.2020.1727700
74. Liu J, Li Y, Zhang Q, Lv C, Wang M, Jiao Y, et al. PVT1 expression is a predictor for poor survival of prostate cancer patients. Technol Cancer Res Treat (2021) 20:1533033820971610. doi: 10.1177/1533033820971610
75. Zhao X, Liu Y, Luo C, Zuo Y. AGAP2-AS1/miR-628-5p/FOXP2 feedback loop facilitates the growth of prostate cancer via activating WNT pathway. Carcinogenesis (2021) 42:1270–80. doi: 10.1093/carcin/bgab062
76. Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z, et al. m(6) a modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med (2021) 11:e426. doi: 10.1002/ctm2.426
77. Leng W, Liu Q, Zhang S, Sun D, Guo Y. LncRNA AFAP1-AS1 modulates the sensitivity of paclitaxel-resistant prostate cancer cells to paclitaxel via miR-195-5p/FKBP1A axis. Cancer Biol Ther (2020) 21:1072–80. doi: 10.1080/15384047.2020.1829266
78. Jiang H, Deng W, Zhu K, Zeng Z, Hu B, Zhou Z, et al. LINC00467 promotes prostate cancer progression via M2 macrophage polarization and the miR-494-3p/STAT3 axis. Front Oncol (2021) 11:661431. doi: 10.3389/fonc.2021.661431
79. Song HR, Guo XB, Duan Y, Meng HY, Wang ZY. PAX5-induced upregulation of LINC01194 exerts oncogenic properties by regulating GOLPH3 expression via miR-486-5p in prostate cancer. Eur Rev Med Pharmacol Sci (2021) 25:2528–41.
80. Cui Z, Gao H, Yan N, Dai Y, Wang H, Wang M, et al. LncRNA PlncRNA-1 accelerates the progression of prostate cancer by regulating PTEN/Akt axis. Aging (Albany NY) (2021) 13:12113–28. doi: 10.18632/aging.202919
81. Xing P, Wang Y, Zhang L, Ma C, Lu J. Knockdown of lncRNA MIR4435−2HG and ST8SIA1 expression inhibits the proliferation, invasion and migration of prostate cancer cells in vitro and in vivo by blocking the activation of the FAK/AKT/β−catenin signaling pathway. Int J Mol Med (2021) 47.
82. Sun F, Wu K, Yao Z, Mu X, Zheng Z, Sun M, et al. Long noncoding RNA PVT1 promotes prostate cancer metastasis by increasing NOP2 expression via targeting tumor suppressor MicroRNAs. Onco Targets Ther (2020) 13:6755–65. doi: 10.2147/OTT.S242441
83. Pan Z, Wu C, Li Y, Li H, An Y, Wang G, et al. LncRNA DANCR silence inhibits SOX5-medicated progression and autophagy in osteosarcoma via regulating miR-216a-5p. Biomed Pharmacother (2020) 122:109707. doi: 10.1016/j.biopha.2019.109707
84. Yu S, Yu H, Zhang Y, Liu C, Zhang W, Zhang Y. Long non-coding RNA LINC01116 acts as an oncogene in prostate cancer cells through regulation of miR-744-5p/UBE2L3 axis. Cancer Cell Int (2021) 21:168. doi: 10.1186/s12935-021-01843-w
85. Hasan MF, Ganapathy K, Sun J, Khatib A, Andl T, Soulakova JN, et al. LncRNA PAINT is associated with aggressive prostate cancer and dysregulation of cancer hallmark genes. Int J Cancer (2021) 149. doi: 10.1101/2020.10.29.361105
86. Zheng Z, Qiu K, Huang W. Long non-coding RNA (lncRNA) RAMS11 promotes metastatis and cell growth of prostate cancer by CBX4 complex binding to Top2α. Cancer Manag Res (2021) 13:913–23. doi: 10.2147/CMAR.S270144
87. Hu CY, Chen J, Qin XH, You P, Ma J, Zhang J, et al. Long non-coding RNA NORAD promotes the prostate cancer cell extracellular vesicle release via microRNA-541-3p-regulated PKM2 to induce bone metastasis of prostate cancer. J Exp Clin Cancer Res (2021) 40:98. doi: 10.1186/s13046-021-01891-0
88. Zhang Y, Li Y. Long non-coding RNA NORAD contributes to the proliferation, invasion and EMT progression of prostate cancer via the miR-30a-5p/RAB11A/WNT/β-catenin pathway. Cancer Cell Int (2020) 20:571. doi: 10.1186/s12935-020-01665-2
89. Chen QH, Li B, Liu DG, Zhang B, Yang X, Tu YL. LncRNA KCNQ1OT1 sponges miR-15a to promote immune evasion and malignant progression of prostate cancer via up-regulating PD-L1. Cancer Cell Int (2020) 20:394. doi: 10.1186/s12935-020-01481-8
90. Xie H, Zhao J, Wan J, Zhao J, Wang Q, Yang X, et al. Long non−coding RNA AC245100.4 promotes prostate cancer tumorigenesis via the microRNA−145−5p/RBBP5 axis. Oncol Rep (2021) 45:619–29. doi: 10.3892/or.2020.7894
91. Liao B, Chen S, Li Y, Yang Z, Yang Y, Deng X, et al. LncRNA BLACAT1 promotes proliferation, migration and invasion of prostate cancer cells via regulating miR-29a-3p/DVL3 axis. Technol Cancer Res Treat (2021) 20:1533033820972342. doi: 10.1177/1533033820972342
92. Xiu D, Liu L, Cheng M, Sun X, Ma X. Knockdown of lncRNA TUG1 enhances radiosensitivity of prostate cancer via the TUG1/miR-139-5p/SMC1A axis. Onco Targets Ther (2020) 13:2319–31. doi: 10.2147/OTT.S236860
93. Yao ZF, Pan ZY, Yao YW, Chen JF. [Long noncoding RNA Linc00662 promotes the tumorigenesis of prostate cancer cells]. Zhonghua Nan Ke Xue (2020) 26:588–94.
94. Wu J, Lv Y, Li Y, Jiang Y, Wang L, Zhang X, et al. MCM3AP-AS1/miR-876-5p/WNT5A axis regulates the proliferation of prostate cancer cells. Cancer Cell Int (2020) 20:307. doi: 10.1186/s12935-020-01365-x
95. Dong W, Liu X, Yang C, Wang D, Xue Y, Ruan X, et al. Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target. Clin Transl Med (2021) 11:e411. doi: 10.1002/ctm2.411
96. Yi S, Li G, Sun B. Overexpression of LINC00852 promotes prostate cancer cell proliferation and metastasis. Asia Pac J Clin Oncol (2021) 17:435–41. doi: 10.1111/ajco.13418
97. Cui R, Liu C, Lin P, Xie H, Wang W, Zhao J, et al. LncRNA AC245100.4 binds HSP90 to promote the proliferation of prostate cancer. Epigenomics (2020) 12:1257–71. doi: 10.2217/epi-2020-0270
98. Zeng C, Fan D, Xu Y, Li X, Yuan J, Yang Q, et al. Curcumol enhances the sensitivity of doxorubicin in triple-negative breast cancer via regulating the miR-181b-2-3p-ABCC3 axis. Biochem Pharmacol (2020) 174:113795. doi: 10.1016/j.bcp.2020.113795
99. Zhu X, Ma X, Zhao S, Cao Z. DLX6-AS1 accelerates cell proliferation through regulating miR-497-5p/SNCG pathway in prostate cancer. Environ Toxicol (2021) 36:308–19. doi: 10.1002/tox.23036
100. Wen Y, Gong X, Dong Y, Tang C. Long non coding RNA SNHG16 facilitates proliferation, migration, invasion and autophagy of neuroblastoma cells via sponging miR-542-3p and upregulating ATG5 expression. OncoTargets Ther (2020) 13:263. doi: 10.2147/OTT.S226915
101. Yao C, Cheng X, Guo X, Lu X, Bu F, Xu Y. NNT-AS1 modulates prostate cancer cell proliferation, apoptosis and migration through miR-496/DDIT4 axis. Cancer Cell Int (2020) 20:463. doi: 10.1186/s12935-020-01505-3
102. Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol (2020) 22:187–99. doi: 10.1038/s41556-019-0450-y
103. Wu S, Ding L, Xu H, Gao J, Shao Y, Zhang S, et al. The long non-coding RNA IDH1-AS1 promotes prostate cancer progression by enhancing IDH1 enzyme activity. Onco Targets Ther (2020) 13:7897–906. doi: 10.2147/OTT.S251915
104. Yao L, Yang L, Song H, Liu T, Yan H. Silencing of lncRNA XIST suppresses proliferation and autophagy and enhances vincristine sensitivity in retinoblastoma cells by sponging miR-204-5p. Eur Rev Med Pharmacol Sci (2020) 24:3526–37.
105. Song X, Wang H, Wu J, Sun Y. Long noncoding RNA SOX2-OT knockdown inhibits proliferation and metastasis of prostate cancer cells through modulating the miR-452-5p/HMGB3 axis and inactivating wnt/β-catenin pathway. Cancer Biother Radiopharm (2020) 35:682–95. doi: 10.1089/cbr.2019.3479
106. Angeles AK, Heckmann D, Flosdorf N, Duensing S, Sültmann H. The ERG-regulated LINC00920 promotes prostate cancer cell survival via the 14-3-3ϵ-FOXO pathway. Mol Cancer Res (2020) 18:1545–59. doi: 10.1158/1541-7786.MCR-20-0021
107. Meng L, Li Z, Chen Y, Liu D, Liu Z. LINC00689 promotes prostate cancer progression via regulating miR-496/CTNNB1 to activate wnt pathway. Cancer Cell Int (2020) 20:215. doi: 10.1186/s12935-020-01280-1
108. Xing Z, Li S, Liu Z, Zhang C, Meng M, Bai Z. The long non-coding RNA LINC00473 contributes to cell proliferation via JAK-STAT3 signaling pathway by regulating miR-195-5p/SEPT2 axis in prostate cancer. Biosci Rep (2020) 40. doi: 10.1042/BSR20191850
109. Xie Y, Gu J, Qin Z, Ren Z, Wang Y, Shi H, et al. Long non-coding RNA FAM66C is associated with clinical progression and promotes cell proliferation by inhibiting proteasome pathway in prostate cancer. Cell Biochem Funct (2020) 38:1006–16. doi: 10.1002/cbf.3531
110. Ma T, Chen H, Wang P, Yang N, Bao J. Downregulation of lncRNA ZEB1-AS1 represses cell proliferation, migration, and invasion through mediating PI3K/AKT/mTOR signaling by miR-342-3p/CUL4B axis in prostate cancer. Cancer Biother Radiopharm (2020) 35:661–72. doi: 10.1089/cbr.2019.3123
111. Li G, Yang J, Chong T, Huang Y, Liu Y, Li H. TUG1 knockdown inhibits the tumorigenesis and progression of prostate cancer by regulating microRNA-496/Wnt/β-catenin pathway. Anticancer Drugs (2020) 31:592–600. doi: 10.1097/CAD.0000000000000882
112. Hao SD, Ma JX, Liu Y, Liu PJ, Qin Y. Long non-coding TUG1 accelerates prostate cancer progression through regulating miR-128-3p/YES1 axis. Eur Rev Med Pharmacol Sci (2020) 24:619–32.
113. Chen W, Yu Z, Huang W, Yang Y, Wang F, Huang H. LncRNA LINC00665 promotes prostate cancer progression via miR-1224-5p/SND1 axis. Onco Targets Ther (2020) 13:2527–35. doi: 10.2147/OTT.S241578
114. Tan SF, Ni JX, Xiong H. LncRNA UNC5B-AS1 promotes malignant progression of prostate cancer by competitive binding to caspase-9. Eur Rev Med Pharmacol Sci (2020) 24:2271–80.
115. Pan J, Xu X, Wang G. lncRNA ZFAS1 is involved in the proliferation, invasion and metastasis of prostate cancer cells through competitively binding to miR-135a-5p. Cancer Manag Res (2020) 12:1135–49. doi: 10.2147/CMAR.S237439
116. Fan L, Li H, Wang W. Long non-coding RNA PRRT3-AS1 silencing inhibits prostate cancer cell proliferation and promotes apoptosis and autophagy. Exp Physiol (2020) 105:793–808. doi: 10.1113/EP088011
117. Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long non-coding RNA LINC00673 silencing inhibits proliferation and drug resistance of prostate cancer cells via decreasing KLF4 promoter methylation. J Cell Mol Med (2020) 24:1878–92. doi: 10.1111/jcmm.14883
118. Guan Z, Song Y, Ma J, Li F, Zhao X, Liang G, et al. Altered expression of lncRNA NCK1-AS1 distinguished patients with prostate cancer from those with benign prostatic hyperplasia. Oncol Lett (2019) 18:6379–84. doi: 10.3892/ol.2019.11039
119. Zhang Y, Zhang J, Liang S, Lang G, Liu G, Liu P, et al. Long non-coding RNA VIM-AS1 promotes prostate cancer growth and invasion by regulating epithelial-mesenchymal transition. J buon (2019) 24:2090–8.
120. Mao Y, Li W, Weng Y, Hua B, Gu X, Lu C, et al. METTL3-mediated m(6)A modification of lncRNA MALAT1 facilitates prostate cancer growth by activation of PI3K/AKT signaling. Cell Transplant (2022) 31:9636897221122997. doi: 10.1177/09636897221122997
121. Lu TT, Tao X, Li HL, Gai L, Huang H, Li F. LncRNA GAS5 enhances tumor stem cell-like medicated sensitivity of paclitaxel and inhibits epithelial-to-mesenchymal transition by targeting the miR-18a-5p/STK4 pathway in prostate cancer. Asian J Androl (2022) 24. doi: 10.21203/rs.3.rs-218133/v1
122. Zeng Q, Liu J, Wu Q, Song R, Miao W, Ma Y, et al. Long non-coding RNA AC008972.1 as a novel therapeutic target for prostate cancer. Cancer Biother Radiopharm. (2022). doi: 10.1089/cbr.2022.0031
123. Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J (2013) 32:1665–80. doi: 10.1038/emboj.2013.99
124. Zhang Y, Pitchiaya S, Cieślik M, Niknafs YS, Tien JC, Hosono Y, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet (2018) 50:814–24. doi: 10.1038/s41588-018-0120-1
125. Takayama KI, Fujimura T, Suzuki Y, Inoue S. Identification of long non-coding RNAs in advanced prostate cancer associated with androgen receptor splicing factors. Commun Biol (2020) 3:393. doi: 10.1038/s42003-020-01120-y
126. Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet (2013) 45:1392–8. doi: 10.1038/ng.2771
127. Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia (2014) 16:900–8. doi: 10.1016/j.neo.2014.09.001
128. Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, et al. LncRNA HOTAIR enhances the androgen-Receptor-Mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep (2015) 13:209–21. doi: 10.1016/j.celrep.2015.08.069
129. Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun (2014) 5:5383. doi: 10.1038/ncomms6383
130. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature (2013) 500:598–602. doi: 10.1038/nature12451
131. Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem (2016) 291:17861–80. doi: 10.1074/jbc.M116.718536
132. Yu C, Fan Y, Zhang Y, Liu L, Guo G. LINC00893 inhibits the progression of prostate cancer through miR-3173-5p/SOCS3/JAK2/STAT3 pathway. Cancer Cell Int (2022) 22:228. doi: 10.1186/s12935-022-02637-4
133. Mi YY, Sun CY, Zhang LF, Wang J, Shao HB, Qin F, et al. Long non-coding RNAs LINC01679 as a competitive endogenous RNAs inhibits the development and progression of prostate cancer via regulating the miR-3150a-3p/SLC17A9 axis. Front Cell Dev Biol (2021) 9:737812. doi: 10.3389/fcell.2021.737812
134. Zhang W, Shi C, Xu Q, Chen X, Zhu H, Zheng B. Long non-coding RNA MIR22HG suppresses cell proliferation and promotes apoptosis in prostate cancer cells by sponging microRNA-9-3p. Bioengineered (2022) 13:13108–17. doi: 10.1080/21655979.2022.2079244
135. Zhong B, Zhao Z, Jiang X. RP1-59D14.5 triggers autophagy and represses tumorigenesis and progression of prostate cancer via activation of the hippo signaling pathway. Cell Death Dis (2022) 13:458. doi: 10.1038/s41419-022-04865-y
136. Wei X, Hou Y, Zhang Y, Zhang H, Sun Z, Meng X, et al. Long non-coding RNA MAGI2-AS3 inactivates STAT3 pathway to inhibit prostate cancer cell proliferation via acting as a microRNA-424-5p sponge. J Cancer (2022) 13:343–53. doi: 10.7150/jca.60749
137. Ghildiyal R, Sawant M, Renganathan A, Mahajan K, Kim EH, Luo J, et al. Loss of long noncoding RNA NXTAR in prostate cancer augments androgen receptor expression and enzalutamide resistance. Cancer Res (2022) 82:155–68. doi: 10.1158/0008-5472.CAN-20-3845
138. Li R, Chen Y, Wu J, Cui X, Zheng S, Yan H, et al. LncRNA FGF14-AS2 represses growth of prostate carcinoma cells via modulating miR-96-5p/AJAP1 axis. J Clin Lab Anal (2021) 35:e24012. doi: 10.1002/jcla.24012
139. Zhou Z, Wu X, Zhou Y, Yan W. Long non-coding RNA ADAMTS9-AS1 inhibits the progression of prostate cancer by modulating the miR-142-5p/CCND1 axis. J Gene Med (2021) 23:e3331. doi: 10.1002/jgm.3331
140. Ding X, Xu X, He XF, Yuan Y, Chen C, Shen XY, et al. Muscleblind-like 1 antisense RNA 1 inhibits cell proliferation, invasion, and migration of prostate cancer by sponging miR-181a-5p and regulating PTEN/PI3K/AKT/mTOR signaling. Bioengineered (2021) 12:803–14. doi: 10.1080/21655979.2021.1890383
141. Liu WH, Lu JJ, Yu RK, Zhou L, Yu Q, Li DF, et al. LINC00641 regulates prostate cancer cell growth and apoptosis via the miR-365a-3p/VGLL4 axis. Eur Rev Med Pharmacol Sci (2021) 25:108–15.
142. Du L, Gao Y. PGM5-AS1 impairs miR-587-mediated GDF10 inhibition and abrogates progression of prostate cancer. J Transl Med (2021) 19:12. doi: 10.1186/s12967-020-02572-w
143. Ma X, Wang Z, Ren H, Bao X, Zhang Y, Wang B, et al. Long non-coding RNA GAS5 suppresses tumor progression and enhances the radiosensitivity of prostate cancer through the miR-320a/RAB21 axis. Cancer Manag Res (2020) 12:8833–45. doi: 10.2147/CMAR.S244123
144. Li Y, Li H, Wei X. Long noncoding RNA LINC00261 suppresses prostate cancer tumorigenesis through upregulation of GATA6-mediated DKK3. Cancer Cell Int (2020) 20:474. doi: 10.1186/s12935-020-01484-5
145. Huo W, Qi F, Wang K. Long non-coding RNA FER1L4 inhibits prostate cancer progression via sponging miR-92a-3p and upregulation of FBXW7. Cancer Cell Int (2020) 20:64. doi: 10.1186/s12935-020-1143-0
146. Qiu K, Zheng Z, Huang Y. Long intergenic noncoding RNA 00844 promotes apoptosis and represses proliferation of prostate cancer cells through upregulating GSTP1 by recruiting EBF1. J Cell Physiol (2020) 235:8472–85. doi: 10.1002/jcp.29690
147. Zhou J, Song Q, Liu X, Ye H, Wang Y, Zhang L, et al. lncRNA Erbb4-IR is downregulated in prostate carcinoma and predicts prognosis. Oncol Lett (2020) 19:3425–30. doi: 10.3892/ol.2020.11464
148. Shen H, Weng XD, Yang D, Wang L, Liu XH. Long noncoding RNA MIR22HG is down-regulated in prostate cancer. Math Biosci Eng (2019) 17:1776–86. doi: 10.3934/mbe.2020093
149. Fan L, Li H, Zhang Y. LINC00908 negatively regulates microRNA-483-5p to increase TSPYL5 expression and inhibit the development of prostate cancer. Cancer Cell Int (2020) 20:10. doi: 10.1186/s12935-019-1073-x
150. Li B, Guo Z, Liang Q, Zhou H, Luo Y, He S, et al. lncRNA DGCR5 up-regulates TGF-β1, increases cancer cell stemness and predicts survival of prostate cancer patients. Cancer Manag Res (2019) 11:10657–63. doi: 10.2147/CMAR.S231112
151. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Baniahmad A, Taheri M, Rashnoo F. A review on the role of PCA3 lncRNA in carcinogenesis with an especial focus on prostate cancer. Pathol-Res Pract (2022) 231:153800. doi: 10.1016/j.prp.2022.153800
152. Lin CY, Wang SS, Yang CK, Li JR, Chen CS, Hung SC, et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. Int J Med Sci (2019) 16:1424–9. doi: 10.7150/ijms.38080
153. Zhao L, Zheng W, Li C. Association of long-chain non-coding RNA GAS5 gene polymorphisms with prostate cancer risk and prognosis in Chinese han population. Med (Baltimore) (2020) 99:e21790. doi: 10.1097/MD.0000000000021790
154. Huang S, Cui H, Lou Z, Wang X, Chen L, Xie Z, et al. Association of rs3787016 in long non-coding RNAs POLR2E and rs2910164 in MiRNA-146a with prostate cancer: A systematic review and meta-analysis. Iran J Public Health (2018) 47:623–32.
155. Xu B, Zhang M, Liu C, Wang C, You Z, Wang Y, et al. Association of long non-coding RNA MEG3 polymorphisms and risk of prostate cancer in Chinese han population. Urol J (2020) 18:176–80.
156. Hu JC, Wang SS, Chou YE, Chiu KY, Li JR, Chen CS, et al. Associations between LncRNA MALAT1 polymorphisms and lymph node metastasis in prostate cancer. Diagn (Basel) (2021) 11. doi: 10.3390/diagnostics11091692
157. Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, et al. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene (2017) 613:20–4. doi: 10.1016/j.gene.2017.02.031
158. Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem (2006) 52:1089–95. doi: 10.1373/clinchem.2005.063289
159. Fotouhi Ghiam A, Taeb S, Huang X, Huang V, Ray J, Scarcello S, et al. Long non-coding RNA urothelial carcinoma associated 1 (UCA1) mediates radiation response in prostate cancer. Oncotarget (2017) 8:4668–89. doi: 10.18632/oncotarget.13576
160. Mirzaei S, Paskeh MDA, Okina E, Gholami MH, Hushmandi K, Hashemi M, et al. Molecular landscape of LncRNAs in prostate cancer: A focus on pathways and therapeutic targets for intervention. J Exp Clin Cancer Res (2022) 41:214. doi: 10.1186/s13046-022-02406-1
Keywords: lncRNA, prostate cancer, biomarker, expression, diagnostic
Citation: Taheri M, Badrlou E, Hussen BM, Kashi AH, Ghafouri-Fard S and Baniahmad A (2023) Importance of long non-coding RNAs in the pathogenesis, diagnosis, and treatment of prostate cancer. Front. Oncol. 13:1123101. doi: 10.3389/fonc.2023.1123101
Received: 13 December 2022; Accepted: 07 March 2023;
Published: 21 March 2023.
Edited by:
Yafeng Ma, Ingham Institute of Applied Medical Research, AustraliaReviewed by:
Kenichi Takayama, Tokyo Metropolitan Institute of Gerontology, JapanNathan J. Bowen, Clark Atlanta University, United States
Lin Ye, Tongji University, China
Tao Liu, Zhongnan Hospital, Wuhan University, China
Copyright © 2023 Taheri, Badrlou, Hussen, Kashi, Ghafouri-Fard and Baniahmad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soudeh Ghafouri-Fard, cy5naGFmb3VyaWZhcmRAc2JtdS5hYy5pcg==; Aria Baniahmad, YXJpYS5iYW5pYWhtYWRAbWVkLnVuaS1qZW5hLmRl
 Mohammad Taheri
Mohammad Taheri Elham Badrlou
Elham Badrlou Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Amir Hossein Kashi
Amir Hossein Kashi Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Aria Baniahmad
Aria Baniahmad