- 1Department of Neurosurgery, Children’s Hospital of Nanjing Medical University, Nanjing, Jiangsu, China
- 2Department of Neurosurgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, China
- 3Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu, China
Background: Increasing evidence supports a relationship between E twenty-six variant transcription factor 4 (ETV4) and several cancers, but no pan-cancer analysis has been reported.
Methods: The present study surveyed the effects of ETV4 on cancer using RNA sequencing data obtained from The Cancer Genome Atlas and GTEx, and further explored its role in drug sensitivity using data from Cellminer. Differential expression analyses were conducted for multiple cancers using R software. Cox regression and survival analysis were employed to calculate correlations between ETV4 levels and survival outcomes in multiple cancers using the online tool Sangerbox. ETV4 expression was also compared with immunity, heterogeneity, stemness, mismatch repair genes, and DNA methylation among different cancers.
Results: ETV4 was found to be significantly upregulated in 28 tumors. Upregulation of ETV4 was associated with poor overall survival, progression free interval, disease-free-interval, and disease specific survival in several cancer types. Expression of ETV4 was also remarkably correlated with immune cell infiltration, tumor heterogeneity, mismatch repair gene expression, DNA methylation, and tumor stemness. Furthermore, ETV4 expression seemed to affect sensitivity to a number of anticancer drugs.
Conclusions: These results suggest that ETV4 may be useful as a prognostic factor and therapeutic target.
Introduction
Cancer is a major public health problem that causes remarkable morbidity and mortality. By 2040, the global cancer rate is expected to exceed 29 million cases annually and account for one out of every six deaths (1, 2). As medical technology continuously improves, so too do cancer screening and management. However, only a few malignancies are able to be cured due to the complexity of tumorigenesis, progression, and therapy resistance. Big data may be leveraged to more efficiently investigate the potential molecular mechanisms of tumorigenesis, progression, and guide clinicians to select cancer-sensitivity drugs according to the characteristics of patient’s tumor cells. Pan-cancer analysis is a recent and critical development, useful for uncovering mutations and other genetic abnormalities that are common to many different cancers. The present study investigated the role of E twenty-six variant transcription factor 4 (ETV4) among different cancers via pan-cancer analysis.
ETV4 is a member of the E26 transcription factor superfamily, which contains the ETS domain and binds to the core GGA(A/T) sequence (3–5). It plays critical roles in embryonic development, including neurogenesis (4, 6), lung branching (7, 8), spermatogenesis, and limb bud formation (9). Recent studies revealed that ETV4 is aberrantly expressed in many types of tumors, and its overexpression is related to poor prognosis of cancer patients, including breast (10, 11), prostate (12), lung (13), and gastric (10, 14) cancers. Additionally, increasing studies have identified that ETV4 promotes cancer growth, invasion, metastasis, and drug resistance (15). ETV4 may promote cancer metastasis by triggering transcription of ZEB1 and SNAIL1 (16). However, it remains uncertain whether ETV4 contributes to the etiology of various cancers through common molecular processes.
In the current study we investigated ETV4 expression among various tumors and its relationship with patient prognosis, mismatch repair genes, mRNA methylation, immunity, heterogeneity, and stemness using RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA). We also surveyed the impact of ETV4 levels on drug tolerance and sensitivity using data from CellMiner. Our findings showed that ETV4 may be useful as an oncogenic prognostic factor and therapy target.
Methods
Data collection
RNA-seq data and clinical parameters of different cancers and matched normal tissues were collected from TCGA database. The RNA-seq data were then normalized via the UCSC Xena tool (https://xena.ucsc.edu/). Genetic profiles of 31 human normal tissues were obtained from GTEx (https://commonfund.nih.gov/GTEx). Duplicated data were excluded.
Expressional levels and survival outcome analysis of ETV4
The prognostic value of ETV4 in various types of cancers was analyzed using univariate Cox analysis and Kaplan-Meier survival analysis through the online tool Sangerbox (http://vip.sangerbox.com/). Data for expression analysis were from TCGA and GTEx database. Each expression value was transformed via log2(x+1) and single cancer with less than three samples was removed. Unpaired Student’s t-Tests were used for differential significance analysis. The clinical parameters used for survival analysis were obtained from TCGA database. Cox regression and Kaplan-Meier survival analysis were used for survival outcome analysis.
Immunohistochemistry staining
Immunohistochemistry images of ETV4 in tumors and non-tumor specimens were acquired from the Human Protein Atlas (http://www.proteinatlas.org/).
Immunological correlation analysis
Correlations between ETV4 expression levels and immune checkpoints, ImmuneScore, StromalScore, and ESTIMATEScore were calculated using the online tool Sangerbox with Pearson’s analysis. Data used for the analysis were downloaded from TCGA database.The relationship between ETV4 levels and immune-infiltrating cells (CD4+ T cells, CD8+ T cells, B cells, neutrophils, and dendritic cells) was calculated using the online tool Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) with Pearson’s analysis. A p-value <0.05 was regarded as statistically significant.
Correlation between ETV4 levels and tumor stemness and heterogeneity
The relationships between ETV4 expression and tumor heterogeneity and stemness were assessed among all TCGA tumor specimens using the online tool Sangerbox with Pearson’s correlation analysis. mRNA and DNA methylation profiles were used to calculate the six stemness scores (ss): RNA expression-based (RNAss), epigenetically regulated RNA expression-based (EREG.EXPss), DNA methylation-based (DNAss), epigenetically regulated DNA methylation-based (EREGMETHss), differentially methylated probes-based (DMPss), and enhancer elements/DNA methylation-based (ENHss) according to previous research (17). These stemness indices range from 0 to 1, where 0 means that the tumor cells are less similar to stem cells, and 1 means that the tumor cells are more similar to stem cells.
Correlations between ETV4 expression and mismatch repair genes and DNA methyltransferases
Expression data for five MMR genes (PMS2, MLH1, EPCAM, MSH2, and MSH6) were obtained from TCGA database and correlated with ETV4 expression using Spearman’s correlation. A similar method was also employed to assess the correlation between ETV4 levels and four DNA methyltransferases (DNMT1, DNMT2, DNMT3A, DNMT3B).
Drug sensitivity evaluation
To evaluate the impact of ETV4 levels on drug sensitivity, drug sensitivity and gene expression data were downloaded from the CellMiner database (https://discover.nci.nih.gov/cellminer/) which is a large database for collecting, processing, and integrating molecular data on NCI-60 and other tumor cells. Cell sensitivity to a drug was indicated by the z-score in the compound activity profile; the drug’s anticancer activity increased as the z-score value increased. Data were analyzed using Pearson’s correlation using R (4.2.1) software. Drugs were only included if they were currently in a clinical trial or had been approved by the FDA. A p-value <0.01 was regarded as statistically significant. Findings were visualized using “ggplot2” and “ggpubr” packages.
Results
Pan-cancer analysis of ETV4 expression
To investigate the differential expression of ETV4 between normal tissues and tumor specimens, mRNA levels were first compared between tissue types using RNA-seq data from TCGA database. As shown in Figure 1A and Supplementary Table 1, in comparison with normal tissues, ETV4 expression was remarkably elevated in 17 tumors such as glioblastoma multiforme (GBM), lung adenocarcinoma (LUAD), colon adenocarcinoma (COAD), esophageal carcinoma, and colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma (COADREAD). In some tumors, ETV4 expression was lower than in normal tissues: kidney renal papillary cell carcinoma (KIRP), prostate adenocarcinoma (PRAD), pan-kidney cohort (KIPAN), kidney renal clear cell carcinoma (KIRC), and pheochromocytoma and paraganglioma (PCPG). Because TCGA dataset comprises mainly tumor tissue and very little normal tissue, ETV4 expression was reanalyzed using data from both TCGA and GTEx. As shown in Figure 1B, the consolidated result showed that ETV4 was markedly elevated in 28 tumors (including GBM and LUAD) and still downregulated in KIRP, KIPAN, PRAD, KIRC, and PCPG. Similar results were also observed for the immunohistochemical tissue assay (Figure 1C). These data reveal that over-expression of ETV4 in most cancer tissues compared to normal tissues.
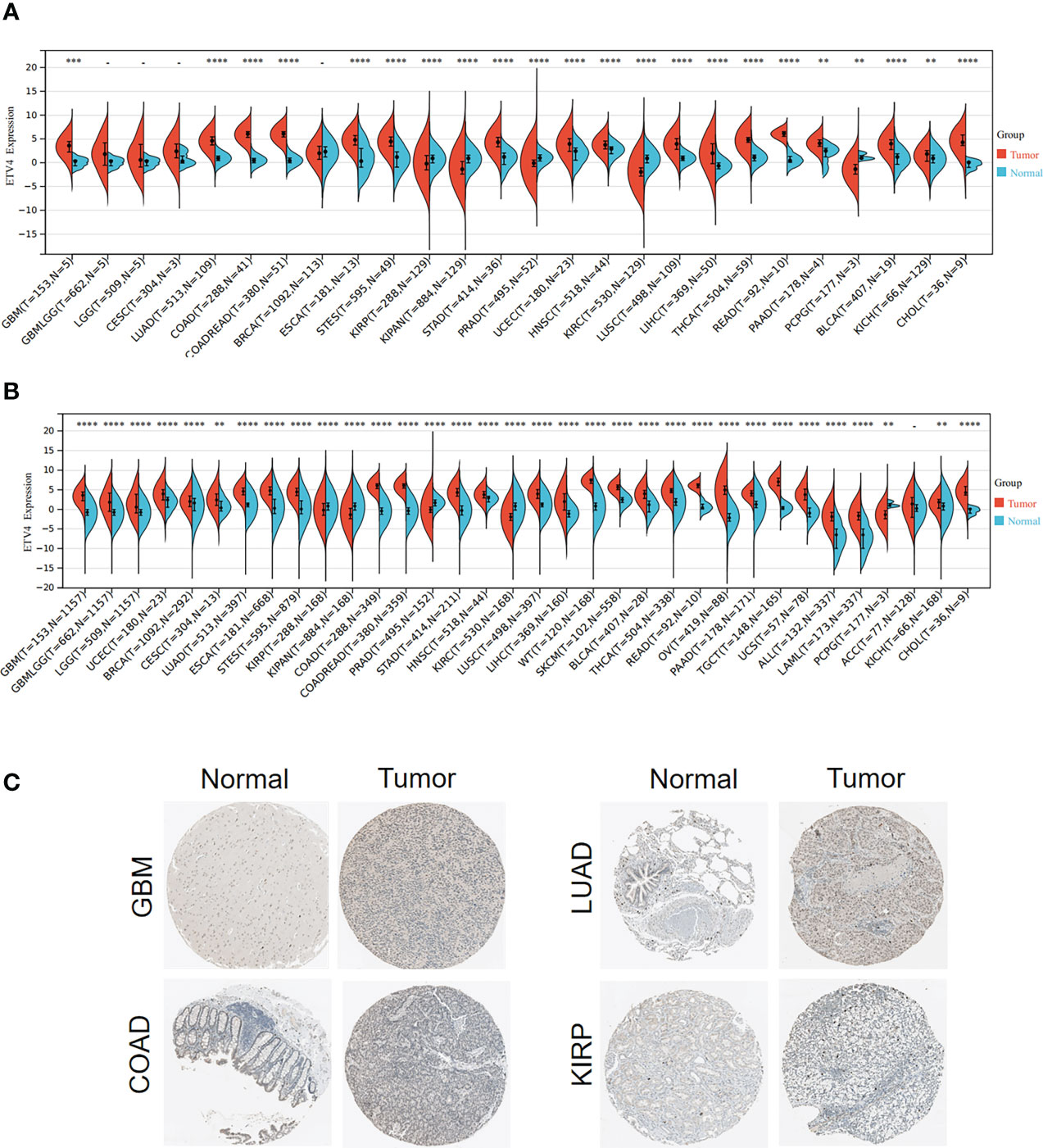
Figure 1 ETV4 expression in various human tumors. (A) mRNA levels of ETV4 among different tumors and adjacent normal specimens using TCGA data. (B) mRNA levels of ETV4 in tumors and normal tissues using the combined data from TCGA and GTEx. (C) Protein expression levels of ETV4 in tumors and their corresponding normal tissues based on The Human Protein Atlas. "-" :no statistic difference **p < 0.01; ***p < 0.001; ****p < 0.0001.
Prognostic value of ETV4 among various cancers
To better understand the prognostic value of ETV4 in different cancer types, we investigated correlations between ETV4 expression and survival outcomes for each cancer using Cox regression and Kaplan-Meier survival analysis. As seen in Figures 2A, 3A, patients suffering glioma (GBMLGG), brain low grade glioma (LGG), sarcoma, kidney renal papillary cell carcinoma, head and neck squamous cell carcinoma, GBM, KIRC, mesothelioma (MESO), liver hepatocellular carcinoma (LIHC), and adrenocortical carcinoma (ACC) with high expression of ETV4 had worse overall survival. Association analysis of disease-free-interval showed that ETV4 expression was a hazard factor in lung squamous cell carcinoma, COADREAD, COAD, and PCPG, but was a favorable factor in esophageal carcinoma (Figure 2B and Figure 3B). High expression of ETV4 predicted poorer progression-free interval for GBMLGG, ACC, MESO, PCPG, LGG, and cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), and better progression-free interval for ovarian serous cystadenocarcinoma (Figures 2C, 3C). High expression of ETV4 was remarkably correlated with poor disease-specific survival in GBMLGG, ACC, LGG, KIRC, GBM, LIHC, KIRP, MESO, and sarcoma, and with improved prognosis in thyroid carcinoma (THCA) (Figures 2D, 3D). The above data indicate that ETV4 may take a pivotal role in the survival of patients with GBMLGG, ACC, LGG, KIRC, GBM, LIHC, KIRP, MESO, sarcoma and THCA.
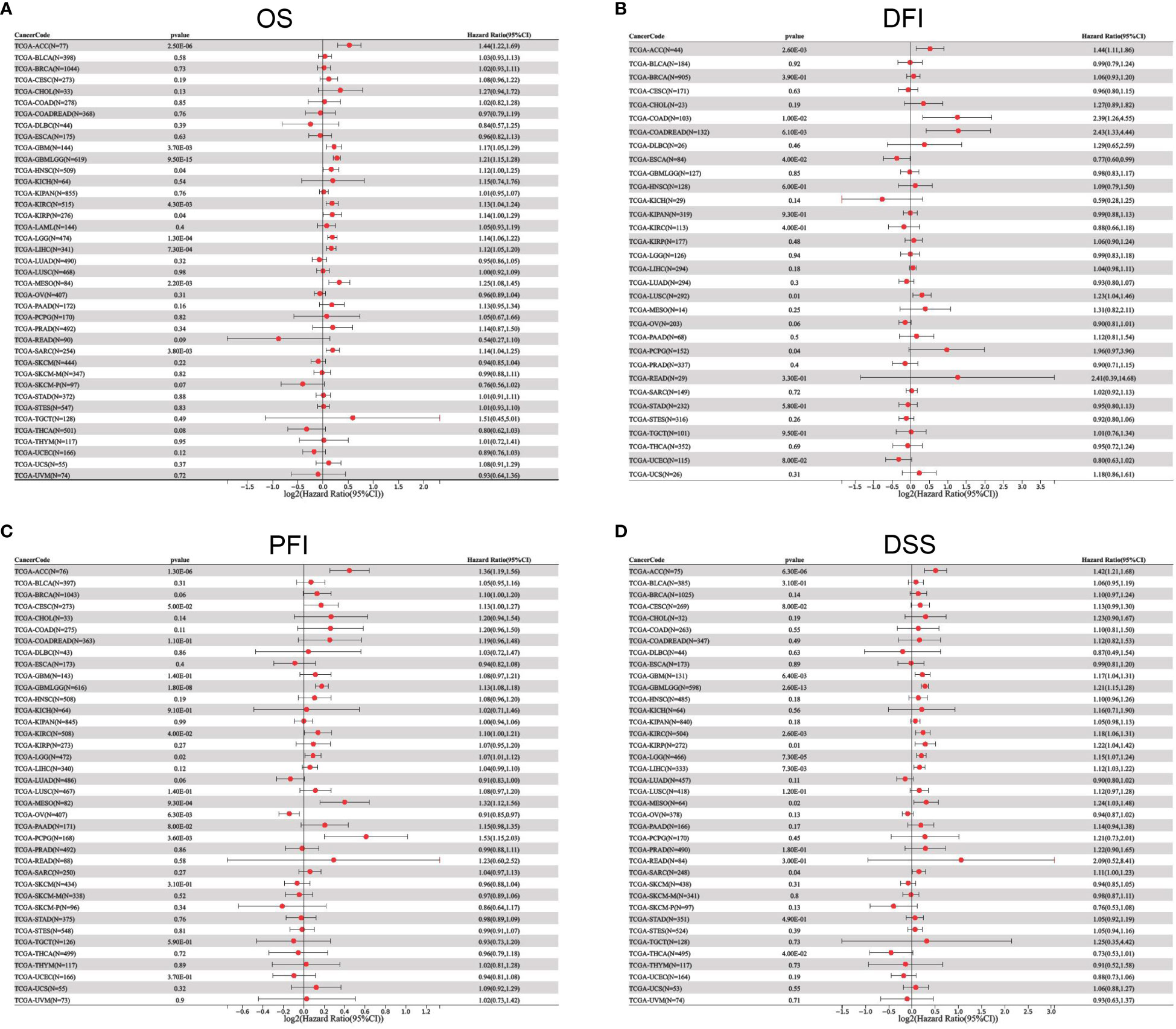
Figure 2 Univariate Cox regression analyses for ETV4 expression in different types of cancers. (A) overall survival, (B) disease-free interval, (C) progression-free interval, (D) disease-specific survival. Hazard ratio (HR) values < 1 represent favorable factors, whereas HR values >1 represent risk factors.
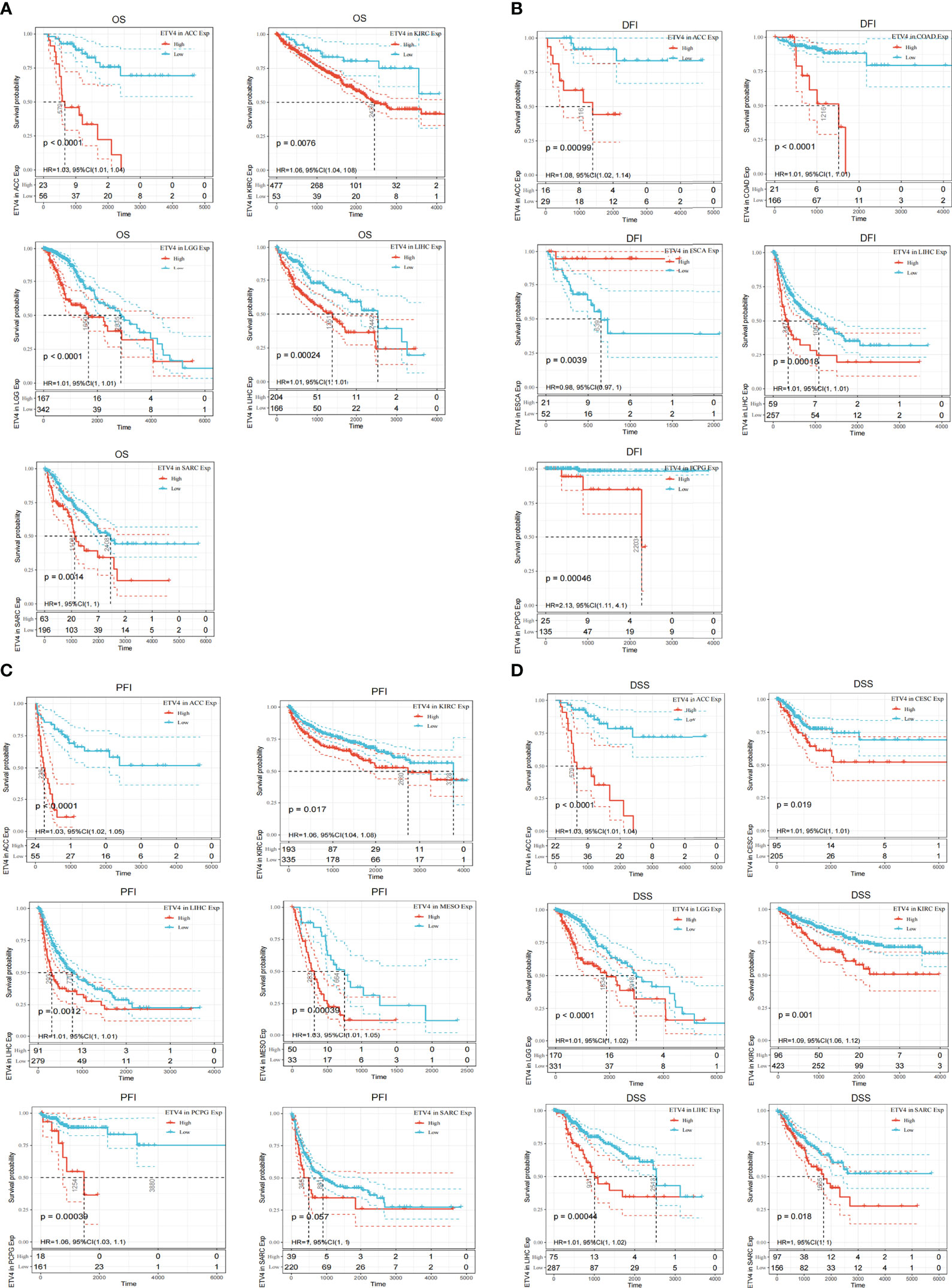
Figure 3 Kaplan-Meier analyses of the correlation between ETV4 expression and overall survival. (A),disease-free interval (B), progression-free interval (C), and disease-specific survival (D) in various cancer types.
Analysis of the relationship between ETV4 expression and immunity
The tumor microenvironment plays an important role in tumor initiation, metastasis, progression, and therapeutic outcomes (18, 19) and it has been confirmed that balance of immune checkpoints and immune cells in the TME is critical to suppress tumor development and progression (20, 21). To investigate the function of ETV4 in the tumor microenvironment, we calculated the correlation between ETV4 levels and 60 common immune checkpoint (ICP) genes (36 stimulatory and 24 inhibitory). As shown in Figure 4A, the relationship between ETV4 and ICP genes varied among different cancer types. ETV4 expression was positively associated with majority of ICP genes in many cancers, such as ovarian cancer and PCPG, but negatively correlated in KIPAN and COAD.This finding implied that ETV4 might promote tumorigenesis and progression via breaking the balance of ICP. Next, relationship between ETV4 levels and immune cell infiltration (comprising B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) was also investigated. As shown in Figure 4B, ETV4 expression was significantly negative correlated with most of immune cell infiltration in cancers. Additionally, ImmuneScore and StromalScore were also employed to evaluate the relationship between ETV4 expression and immune cell infiltration among cancers. As shown in Supplementary Figure S1, ETV4 expression was positively related to StromalScore in GBMLGG, PRAD, thymoma, and PCPG, and negatively related to StromalScore in 19 other tumors. ETV4 expression was associated positively with ImmuneScore and ESTIMATEScore in PRAD and PCPG, and negatively in GBM, LGG, and CESC. The three tumors most strongly correlated with ETV4 expression were PRAD, COAD, stomach adenocarcinoma (STAD) (StromalScore) (Figure 5A); PRAD,COAD, STAD (ImmuneScore) (Figure 5B); and PRAD, COAD, STAD (ESTIMATEScore) (Figure 5C). These results reveal that ETV4 may promote cancer progression via regulating immune checkpoint genes expression and immune cells infiltration.
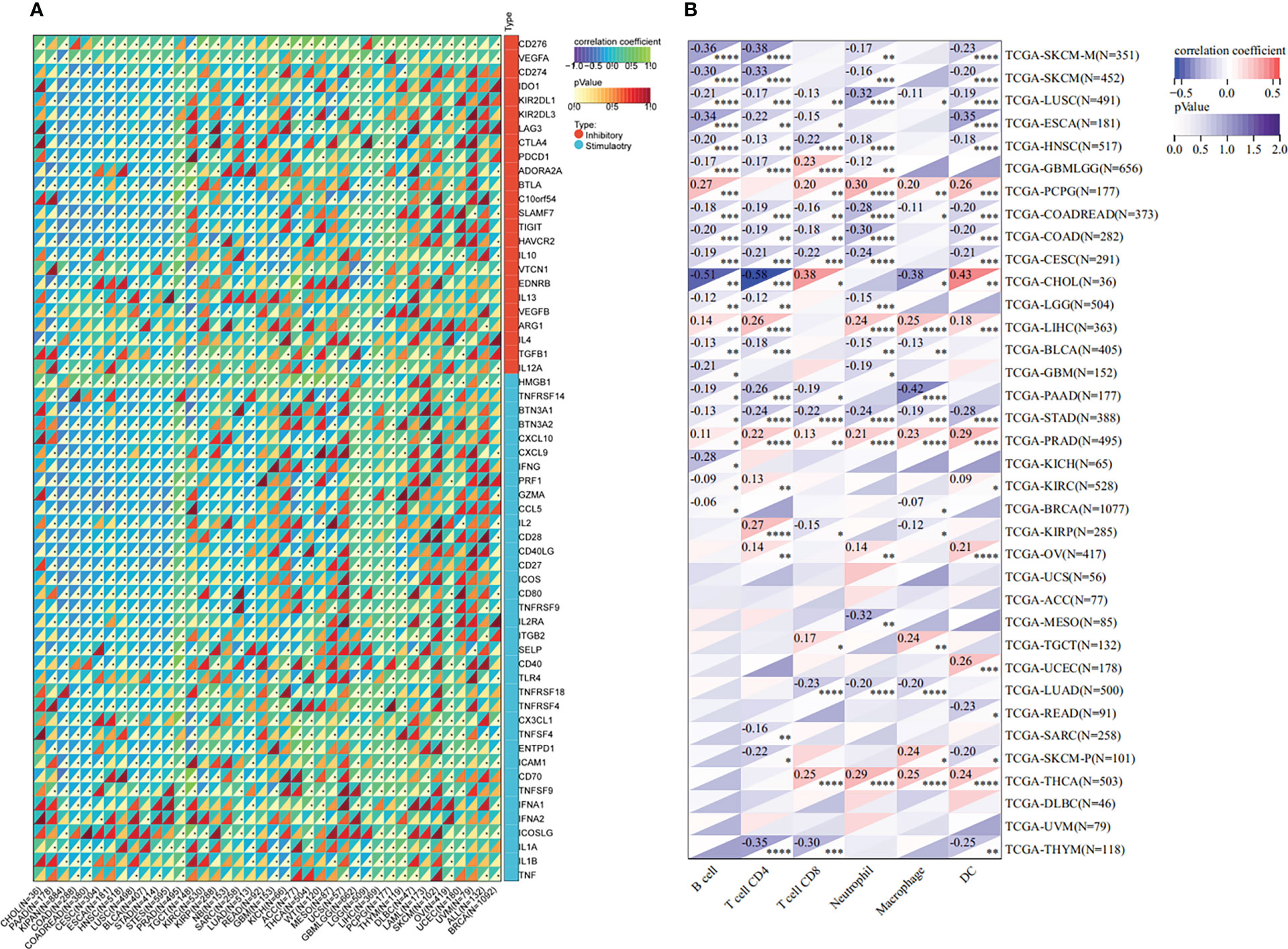
Figure 4 Immunological association analyses. (A) The association between ETV4 expression and immune checkpoint genes.The upper triangle of each square represented the magnitude of the p value of the correlation coefficient, and the lower triangle represented the magnitude of correlation test. The star dots in the lower triangle represent that difference is significant. (B) The association between ETV4 expression and immune infiltration cells. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
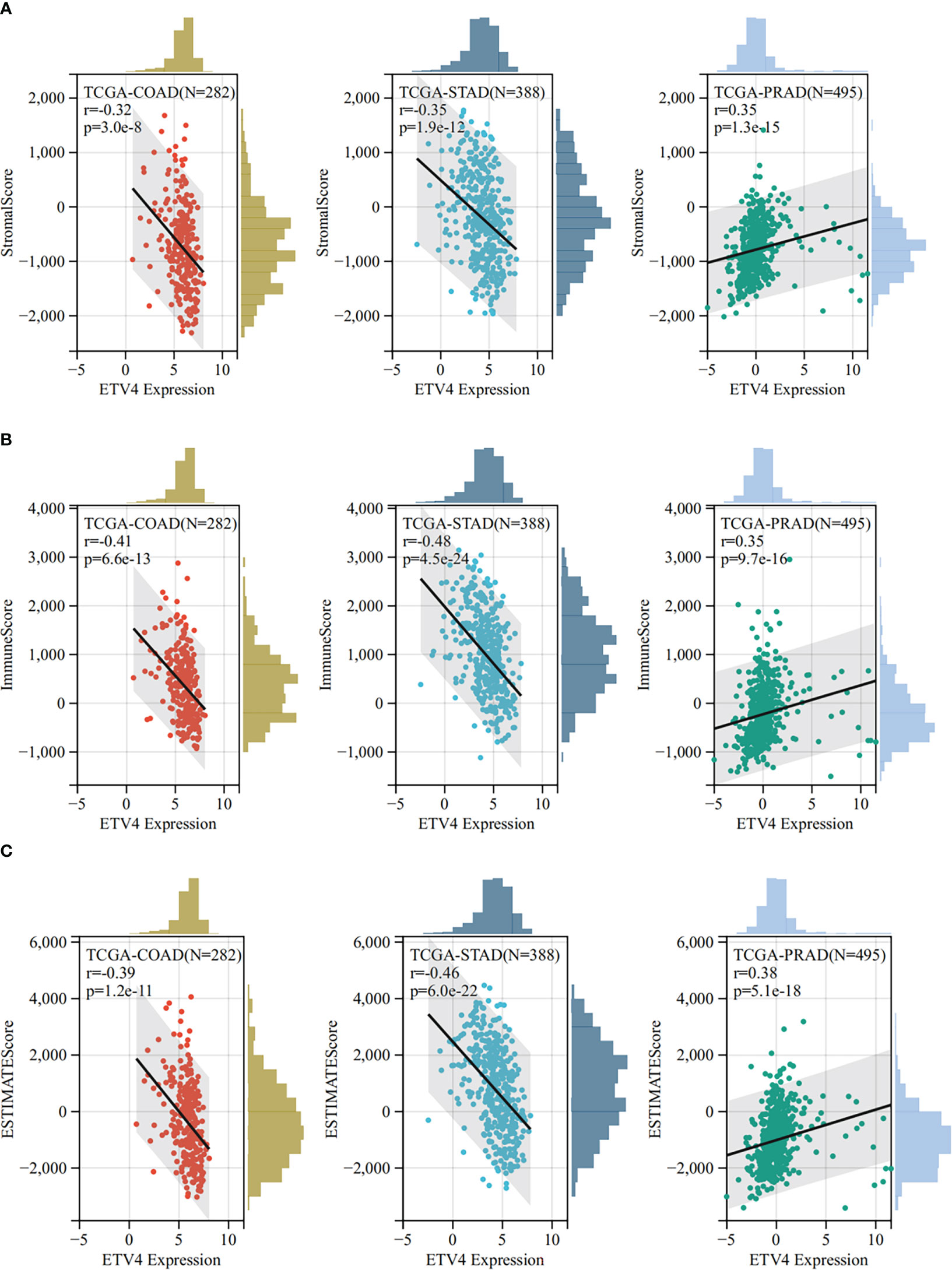
Figure 5 The Sangerbox online tool was used to estimate immune and stromal ratios of various cancers. (A) The relationship between ETV4 expression and StromalScore in prostate adenocarcinoma, colon adenocarcinoma, and kidney renal clear cell carcinoma. (B) The relationship between ETV4 expression and ImmuneScore in colon adenocarcinoma, prostate adenocarcinoma, and stomach adenocarcinoma. (C) The correlation between ETV4 expression and ESTIMATEScore.
Analysis of the relationship between ETV4 expression and heterogeneity
Heterogeneity is a distinct feature of malignancies. After several divisions and proliferation, their descendants acquire genes or altered molecular biology that lead to further differences in proliferation rates, capacity for invasion and metastasis, or drug resistance. Tumor heterogeneity can be assessed by microsatellite instability (MSI), tumor mutational burden (TMB), homologous recombination deficiency (HRD), tumor purity, neoantigen (NEO), loss of heterozygosity (LOH), tumor ploidy, and mutant-allele tumor heterogeneity (MATH). RNA-seq data from TCGA were used to assess the relationship between ETV4 expression and cancer heterogeneity. As shown in Figure 6, ETV4 was positively associated with TMB in MESO and ACC and negatively related to TMB in LUAD, COAD, COADREAD, KIRP, and CHOL. ETV4 was positively associated with MATH in 14 tumors and negatively associated with MATH in GBMLGG, LGG, and skin cutaneous melanoma. ETV4 was positively associated with MSI in CESC, KIPAN, MESO, skin cutaneous melanoma and kidney chromophobe and negatively associated with MSI in COAD, COADREAD, stomach and esophageal carcinoma, STAD and GBMLGG. ETV4 was negatively associated with NEO in LUAD, COAD, COADREAD, testicular germ cell tumors, and CHOL. It was positively associated with tumor purity in 19 tumors and negatively associated with purity in PRAD, LIHC, testicular germ cell tumors, and PCPG. ETV4 was positively associated with ploidy in nine tumors and negatively associated with ploidy in GBMLGG, LGG, KIPAN, THCA, and ovarian serous cystadenocarcinoma. It was related positively to HRD in 14 tumors and negatively in GBMLGG and LGG. ETV4 was positively correlated with LOH in 18 tumors and negatively correlated with LOH in LUAD and THCA. Together these results indicate that ETV4 may accelerate tumor genesis and prognosis by regulating tumor heterogeneity.

Figure 6 Relationship between ETV4 and tumor heterogeneity in various cancer types. (A) tumor mutational burden, (B) mutant-allele tumor heterogeneity, (C) microsatellite instability, (D) neoantigen, (E) tumor purity, (F) ploidy, (G) homologous recombination deficiency, and (H) loss of heterozygosity were calculated using data from TCGA datasets.
Relationship of ETV4 with tumor stemness
The acquisition of stem cell-like features is a crucial driver of tumor progression (22). Stemness indices rely on DNA methylation and mRNA expression are calculated to reflect the resemblance between cancer cells and stem cells (17). The indices include RNAss, EREG EXPss, DNAss, EREG METHss, DMPss, and ENHss, each of which varies from zero (least resemblance to stem cells) to one (most resemblance to stem cells). ETV4 expression was compared to stemness indices across various tumor types. As shown in Figure 7, ETV4 levels were significantly correlate with RNAss index in 18 cancers (positively associated in 14cancers), EREG EXPss in 20cancers (positively associated in 14cancers), DNAss in 17 cancers (positively associated in16 cancers), EREG METHss in 17 cancers (positively associated in 16 cancers), DMPss in 16 cancers (positively associated in 13 cancers), and ENHss in 15 cancers (positively associated in 14cancers) respectively. These results suggest that ETV4 may be related to the genesis of tumor stemness in most cancers.
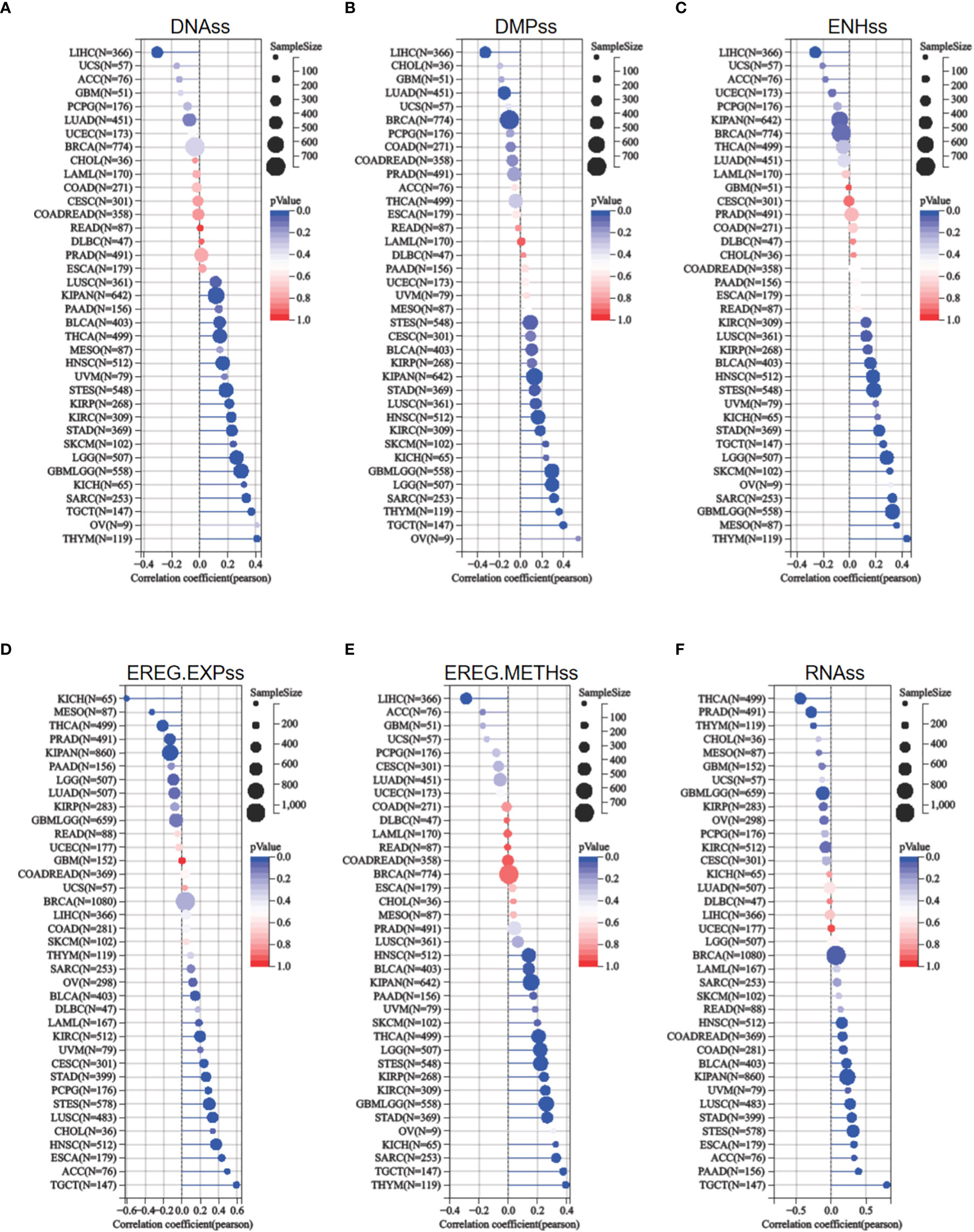
Figure 7 Relationship between ETV4 and cancer stemness in various cancer types. (A) DNA expression-based stemness score (DNAss), (B) Differentially methylated probes-based stemness score(DMPss), (C) Enhancer elements/DNA methylation-based stemness score (ENHss), (D) Epigenetically regulated RNA expression-based stemness score (EREG.EXPss), (E) Epigenetically regulated DNA methylation-based stemness score(EREG.METHss),and (F) RNA expression-based stemness score(RNAss) were calculated using the TCGA datasets.
Correlation of ETV4 expression with MMR genes and DNA methyltransferases
Disorder of MMR genes and DNA methyltransferases plays a pivotal role in tumorigenesis and prognosis (23, 24). To investigate the role of ETV4 in tumorigenesis, ETV4 was correlated with five different MMR genes. As shown in Figure 8A, ETV4 was significantly associated with MMR genes in 26 tumors (notably, it was not associated with MMR genes in acute myeloid leukemia, ovarian serous cystadenocarcinoma, PRAD, READ, thymoma, uterine carcinosarcoma, and uveal melanoma). ETV4 was then correlated with four DNA methyltransferases. As shown in Figure 8B, ETV4 expression was highly associated with these four methyltransferases in many cancers. These results suggest that ETV4 may promote tumorigenesis and poor prognosis by modulating DNA repair genes and DNA methyltransferases.
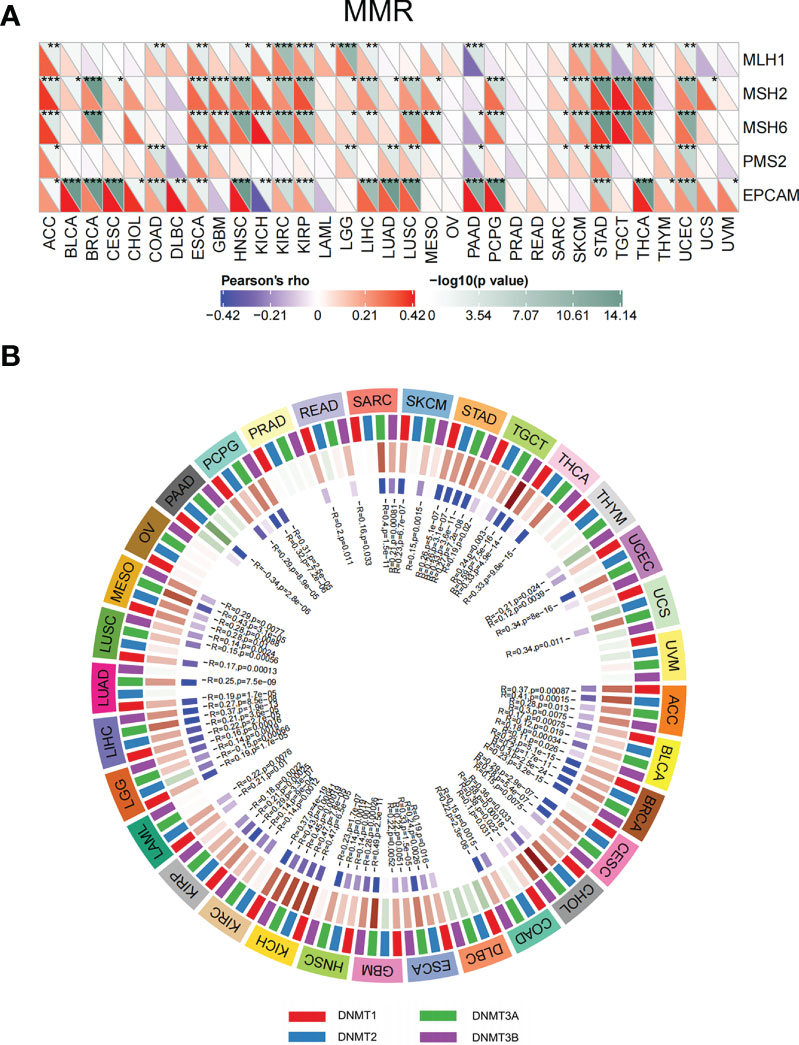
Figure 8 Correlations between ETV4 expression, mismatch repair genes, and DNA methyltransferases among different cancers. (A) Relationship between ETV4 expression and mismatch repair genes. The upper triangle of each square represented the magnitude of the p value of the correlation test, and the lower triangle represented the magnitude of correlation coefficient. (B) Correlation between ETV4 expression and DNA methyltransferases (Red/green in the third layer referring to correlation coefficient and blue/purple in the forth layer referring to p-value).*p < 0.05, **p < 0.01, ***p < 0.001.
Relationship between ETV4 levels and drug sensitivity
The relationship between ETV4 levels and drug sensitivity was assessed using data from CellMiner. The 16 drugs with the strongest correlations are shown in Figure 9. Notably, ETV4 levels were negatively correlated with drug sensitivity of GSK-60903, AZD-2858, Futibatinib, ON-123300, LY-2090314 and positively associated with sensitivity of BGB-283, TAK-632, MLN-2480, AZ-628, LXH-254, LY-3009120, Dabrafenib, Refametinib, Selumetinib, PLX-4720, and PLX-8394. These findings suggest that ETV4 levels can be used for anticancer drug selection.
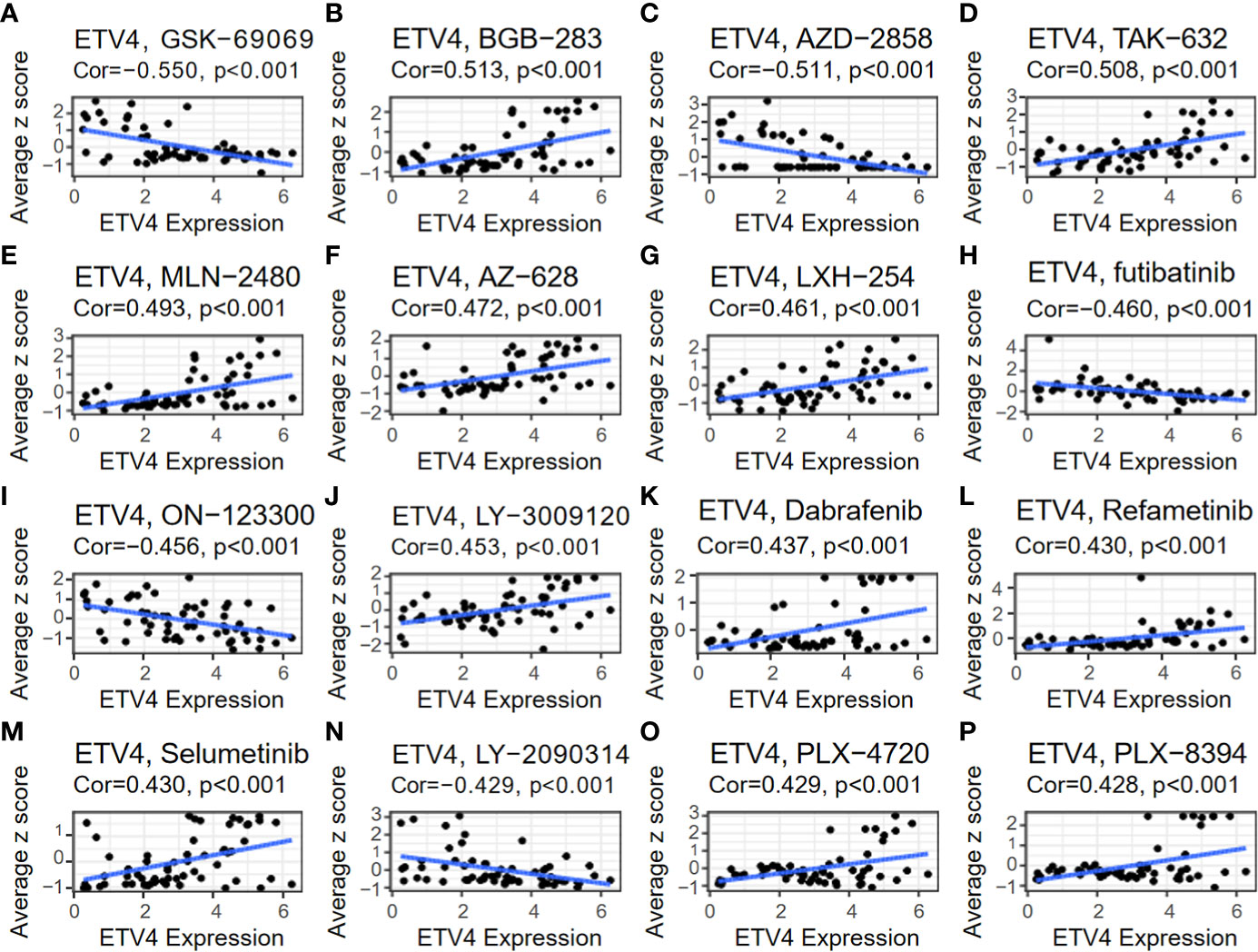
Figure 9 Relationship between ETV4 levels and drug sensitivity. The expression level of ETV4 was correlated with the drug sensitivity of (A) GSK-690693, (B) BGB-283, (C) AZD-2858, (D) TAK-632, (E) MLN-2480, (F) AZ-628, (G) LXH-254, (H) Futibatinib, (I) ON-123300, (J) LY-3009120, (K) Dabrafenib, (L) Refametinib, (M) Selumetinib, (N) LY-2090314, (O) PLX-4720, and (P) PLX-8394.
Discussion
ETV4 is not only an essential factor for embryonic development (4, 8, 9), but also influences the occurrence and prognosis of many diseases, particularly malignant cancers (10, 11, 13). However, it remains unclear whether ETV4 affects various cancers through common molecular processes.
In the present study the potential function of ETV4 in various cancers was investigated using pan-cancer analysis. Through bioinformatics analyses and immunohistochemistry arrays of tumor tissue, ETV4 expression was found to be elevated in multiple cancers. For many different cancers, patients with higher ETV4 expression had worse disease prognosis. These data were in accordance with previous studies (25–27). Many studies have confirmed that disordered immunity, MMR genes, and DNA methyltransferases contribute to cancer progression and therapeutic outcome (28–30). The present study showed that ETV4 levels were not only positively correlated with ICP genes and immune cell infiltration but also significantly associated with MMR genes and methyltransferases, implying that ETV4 may be useful as an immunotherapy target.
Tumor heterogeneity is defined as the diverse pool of cells within and around tumors that express distinct genotypes and phenotypes and express different signature molecules (31–33). It is a major factor in tumor prognosis, therapy resistance, and recurrence (31, 34). Previous research confirmed that abnormal expression of ETV4 was associated with melanoma heterogeneity (35). To examine whether this effect of ETV4 on tumor heterogeneity is a common phenomenon, we calculated the association between ETV4 and TMB, tumor purity, MSI, NEO, HRD, MATH, LOH, and tumor ploidy among multiple types of cancers. Indeed, the expression of ETV4 was positively associated with tumor heterogeneity in the majority of cancers studied.
Stemness is the ability to self-renew and differentiate from origin cells. Cancer stem cells not only have the ability to self-renew but also to produce heterogeneous tumor cells (36, 37). Accumulating evidence has shown that cancer stem cells play a critical role in tumorigenesis, metastasis (38), drug resistance, and recurrence (39, 40). Tao Zhu and colleagues reported that ETV4 promotes breast cancer cell stemness by activating glycolysis and CXCR4-mediated sonic hedgehog signaling (11). The current study confirmed that ETV4 expression was positively correlated with indices of tumor stemness in most tumor types, suggesting that elevated ETV4 expression may enhance tumor proliferation, metastasis, therapy resistance, and recurrence in general.
Finally, effects of ETV4 on drug sensitivity were investigated. ETV4 expression was positively correlated with many drugs (BGB-283, TAK-632, MLN-2480, AZ-628, LXH-254, LY-3009120, Dabrafenib, Refametinib, Selumetinib, PLX-4720, and PLX-8394) and negatively correlated with others (GSK-60903, AZD-2858, Futibutinib, ON-123300, and LY-2090314). These results indicate that ETV4 expression could be used to guide the selection of chemotherapeutic agents.
Taken together, the present results demonstrate that ETV4 is upregulated in multiple cancers and that high expression of ETV4 is correlated with cancer progression and undesirable survival outcomes. Furthermore, ETV4 expression was correlated with immunity, tumor heterogeneity, MMR genes, DNA methyltransferase genes, and tumor stemness in multiple cancers. Finally, ETV4 expression was associated with drug sensitivity. Together these results suggest that ETV4 may be useful as a prognostic factor and therapeutic target for multiple cancers.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: data for expression analysis were obtained from the UCSC database (https://xenabrowser.net/), TCGA Pan-Cancer (PANCAN,N=10535,G=60499) and TCGA TARGET GTEx (PANCAN,N=19131,G=60499). Any further inquiries can be directed to the corresponding author/s..
Author contributions
NL and JH contributed to the study design and conception. RZ, KY, JQ, XW, WL, TJ, YZ, ZG, YP, DQ, and GW performed data analysis and wrote the main manuscript text. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81602212), Natural Science Foundation of Jiangsu Province (BK20161119), Key Project supported by Medical Science and Technique Development Foundation (YKK15139), and Nanjing Medical Science and Technique Development Foundation (QRX17167).
Acknowledgments
We thank Medjaden Inc. for scientific editing of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1121258/full#supplementary-material
Supplementary Figure 1 | The relationship between ETV4 expression and three different immune scores in various cancers. (A) StromalScore, (B) ImmuneScore, (C) ESTIMATEScore.
Supplementary Table 1 | The abbreviations of cancers.
References
1. Deo SVS, Sharma J, Kumar S. Globocan 2020 report on global cancer burden: Challenges and opportunities for surgical oncologists. Ann Surg Oncol (2022) 29(11):6497–500. doi: 10.1245/s10434-022-12151-6
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
3. Oh S, Shin S, Janknecht R. Etv1, 4 and 5: An oncogenic subfamily of ets transcription factors. Biochim Biophys Acta (2012) 1826(1):1–12. doi: 10.1016/j.bbcan.2012.02.002
4. Fontanet PA, Rios AS, Alsina FC, Paratcha G, Ledda F. Pea3 transcription factors, Etv4 and Etv5, are required for proper hippocampal dendrite development and plasticity. Cereb Cortex (2018) 28(1):236–49. doi: 10.1093/cercor/bhw372
5. Xin JH, Cowie A, Lachance P, Hassell JA. Molecular cloning and characterization of Pea3, a new member of the ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev (1992) 6(3):481–96. doi: 10.1101/gad.6.3.481
6. Kandemir B, Gulfidan G, Arga KY, Yilmaz B, Kurnaz IA. Transcriptomic profile of Pea3 family members reveal regulatory codes for axon outgrowth and neuronal connection specificity. Sci Rep (2020) 10(1):18162. doi: 10.1038/s41598-020-75089-3
7. Jones MR, Lingampally A, Dilai S, Shrestha A, Stripp B, Helmbacher F, et al. Characterization of Tg(Etv4-gfp) and Etv5 (Rfp) reporter lines in the context of fibroblast growth factor 10 signaling during mouse embryonic lung development. Front Genet (2019) 10:178. doi: 10.3389/fgene.2019.00178
8. Herriges JC, Verheyden JM, Zhang Z, Sui P, Zhang Y, Anderson MJ, et al. Fgf-regulated etv transcription factors control fgf-shh feedback loop in lung branching. Dev Cell (2015) 35(3):322–32. doi: 10.1016/j.devcel.2015.10.006
9. Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of sonic hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell (2009) 16(4):600–6. doi: 10.1016/j.devcel.2009.02.005
10. Keld R, Guo B, Downey P, Cummins R, Gulmann C, Ang YS, et al. Pea3/Etv4-related transcription factors coupled with active erk signalling are associated with poor prognosis in gastric adenocarcinoma. Br J Cancer (2011) 105(1):124–30. doi: 10.1038/bjc.2011.187
11. Zhu T, Zheng J, Zhuo W, Pan P, Li M, Zhang W, et al. Etv4 promotes breast cancer cell stemness by activating glycolysis and Cxcr4-mediated sonic hedgehog signaling. Cell Death Discovery (2021) 7(1):126. doi: 10.1038/s41420-021-00508-x
12. Qian C, Li D, Chen Y. Ets factors in prostate cancer. Cancer Lett (2022) 530:181–9. doi: 10.1016/j.canlet.2022.01.009
13. Cheng T, Zhang Z, Cheng Y, Zhang J, Tang J, Tan Z, et al. Etv4 promotes proliferation and invasion of lung adenocarcinoma by transcriptionally upregulating Msi2. Biochem Biophys Res Commun (2019) 516(1):278–84. doi: 10.1016/j.bbrc.2019.06.115
14. Okimoto RA, Breitenbuecher F, Olivas VR, Wu W, Gini B, Hofree M, et al. Inactivation of capicua drives cancer metastasis. Nat Genet (2017) 49(1):87–96. doi: 10.1038/ng.3728
15. Jiang W, Xu Y, Chen X, Pan S, Zhu X. E26 transformation-specific variant 4 as a tumor promotor in human cancers through specific molecular mechanisms. Mol Ther Oncolytics (2021) 22:518–27. doi: 10.1016/j.omto.2021.07.012
16. Chen Y, Sumardika IW, Tomonobu N, Kinoshita R, Inoue Y, Iioka H, et al. Critical role of the mcam-Etv4 axis triggered by extracellular S100a8/A9 in breast cancer aggressiveness. Neoplasia (2019) 21(7):627–40. doi: 10.1016/j.neo.2019.04.006
17. Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell (2018) 173(2):338–54.e15. doi: 10.1016/j.cell.2018.03.034
18. De Visser KE, Joyce JA. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell (2023) 41(3):374–403. doi: 10.1016/j.ccell.2023.02.016
19. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
20. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol (2020) 30(16):R921–R5. doi: 10.1016/j.cub.2020.06.081
21. Khosravi N, Mokhtarzadeh A, Baghbanzadeh A, Hajiasgharzadeh K, Shahgoli VK, Hemmat N, et al. Immune checkpoints in tumor microenvironment and their relevance to the development of cancer stem cells. Life Sci (2020) 256:118005. doi: 10.1016/j.lfs.2020.118005
22. Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics (2020) 10(19):8721–43. doi: 10.7150/thno.41648
23. He Y, Zhang L, Zhou R, Wang Y, Chen H. The role of DNA mismatch repair in immunotherapy of human cancer. Int J Biol Sci (2022) 18(7):2821–32. doi: 10.7150/ijbs.71714
24. Zhang W, Xu J. DNA Methyltransferases and their roles in tumorigenesis. biomark Res (2017) 5:1. doi: 10.1186/s40364-017-0081-z
25. Cai LS, Chen Q-X, Fang S-Y, Lian M-Q, Lian M-J, Cai M-Z. Etv4 promotes the progression of gastric cancer through regulating Kdm5d. Eur Rev Med Pharmacol Sci (2020) 24(5):2442–51. doi: 10.26355/eurrev_202003_20511
26. Qi M, Liu Z, Shen C, Wang L, Zeng J, Wang C, et al. Overexpression of Etv4 is associated with poor prognosis in prostate cancer: Involvement of Upa/Upar and mmps. Tumour Biol (2015) 36(5):3565–72. doi: 10.1007/s13277-014-2993-7
27. Fonseca AS, Ramão A, Bürger MC, de Souza JES, Zanette DL, de Molfetta GA, et al. Etv4 plays a role on the primary events during the adenoma-adenocarcinoma progression in colorectal cancer. BMC Cancer (2021) 21(1):207. doi: 10.1186/s12885-021-07857-x
28. Taieb J, Svrcek M, Cohen R, Basile D, Tougeron D, Phelip JM. Deficient mismatch Repair/Microsatellite unstable colorectal cancer: Diagnosis, prognosis and treatment. Eur J Cancer (2022) 175:136–57. doi: 10.1016/j.ejca.2022.07.020
29. Darvin P, Toor SM, Sasidharan Nair V, Elkord EA-O. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp Mol Med (2018) 50(12):1–11. doi: 10.1038/s12276-018-0191-1
30. Győrffy B, Bottai G, Fleischer T, Munkácsy G, Budczies J, Paladini L, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer (2016) 138(1):87–97. doi: 10.1002/ijc.29684
31. Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol (2019) 12(1):134. doi: 10.1186/s13045-019-0818-2
32. Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol (2021) 18(2):79–92. doi: 10.1038/s41585-020-00400-w
33. Pe'er D, Ogawa S, Elhanani O, Keren L, Oliver TG, Wedge D. Tumor heterogeneity. Cancer Cell (2021) 39(8):1015–7. doi: 10.1016/j.ccell.2021.07.009
34. Vitale I, Shema EA-OX, Loi SA-O, Galluzzi LA-O. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med (2021) 27(2):212–24. doi: 10.1038/s41591-021-01233-9
35. Parisi F, Ariyan S, Narayan D, Bacchiocchi A, Hoyt K, Cheng E, et al. Detecting copy number status and uncovering subclonal markers in heterogeneous tumor biopsies. BMC Genomics (2011) 12:230. doi: 10.1186/1471-2164-12-230
36. Prasetyanti PR, Medema JA-O. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer (2017) 16(1):41. doi: 10.1186/s12943-017-0600-4
37. Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature (2013) 501(7467):328–37. doi: 10.1038/nature12624
38. Nandy SB, Lakshmanaswamy R. Cancer stem cells and metastasis. Prog Mol Biol Transl Sci (2017) 151:137–76. doi: 10.1016/bs.pmbts.2017.07.007
39. Dzobo K, Ganz C, Thomford NE, Senthebane DA. Cancer stem cell markers in relation to patient survival outcomes: Lessons for integrative diagnostics and next-generation anticancer drug development. OMICS (2021) 25(2):81–92. doi: 10.1089/omi.2020.0185
Keywords: ETV4, pancancer, microenvironment, immunity, prognostic, drug sensitivity
Citation: Zhang R, Peng Y, Gao Z, Qian J, Yang K, Wang X, Lu W, Zhu Y, Qiu D, Jin T, Wang G, He J and Liu N (2023) Oncogenic role and drug sensitivity of ETV4 in human tumors: a pan-cancer analysis. Front. Oncol. 13:1121258. doi: 10.3389/fonc.2023.1121258
Received: 11 December 2022; Accepted: 22 March 2023;
Published: 02 May 2023.
Edited by:
Boheng Li, Southwest University, ChinaReviewed by:
Andrea Lapucci, University of Florence, ItalyPeter Hollenhorst, Indiana University, United States
Stephanie Metcalf, Indiana University School of Medicine, United States, in collaboration with reviewer PH
Copyright © 2023 Zhang, Peng, Gao, Qian, Yang, Wang, Lu, Zhu, Qiu, Jin, Wang, He and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junping He, SGVqdW5waW5nMzU5QDE2My5jb20=; Ning Liu, TGl1bmluZzA4NTNAMTI2LmNvbQ==
 Rui Zhang
Rui Zhang Yanfang Peng3
Yanfang Peng3 Ning Liu
Ning Liu