- 1Microbiology and Immunology, College of Medicine and Health Sciences, United Arab Emirates University, Abu Dhabi, United Arab Emirates
- 2Zayed Bin Sultan Center for Health Sciences, College of Medicine and Health Sciences, United Arab Emirates University, Abu Dhabi, United Arab Emirates
- 3Institute of Public Health, College of Medicine and Health Sciences, United Arab Emirates University, Abu Dhabi, United Arab Emirates
- 4General Surgery Division, Surgery Department, Tawam Hospital, Abu Dhabi, United Arab Emirates
Background: Blastocystis is an anaerobic intestinal protozoan. Nine Blastocystis subtypes (STs) were detected in humans. A subtype-dependent association between Blastocystis and different cancer types has been debated in many studies. Thus, this study aims to assess the possible association between Blastocystis infection and cancer, especially colorectal cancer (CRC). We also screened the presence of gut fungi and their association with Blastocystis.
Methods: We used a case-control design; cancer patients and cancer-free (CF) participants. The cancer group was further sub-group into CRC group and cancers outside the gastrointestinal tract (COGT) group. Macroscopic and microscopic examinations were performed to identify intestinal parasites in participants’ stool samples. Molecular and phylogenetic analyses were conducted to identify and subtype Blastocystis. Furthermore, gut fungi were investigated molecularly.
Results: 104 stool samples were collected and matched between CF (n=52) and cancer patients (n=52); CRC (n=15) and COGT (n=37). As anticipated, Blastocystis prevalence was significantly higher among CRC patients (60%, P=0.002) and insignificant in COGT patients (32.4%, P=0.161) compared to CF group (17.3%). The most common subtypes were ST2 among cancer group and ST3 in the CF group.
Conclusion: Cancer patients have a higher risk of Blastocystis infection compared to CF individuals (OR=2.98, P=0.022). Increased risk of Blastocystis infection was associated with CRC patients (OR=5.66, P=0.009). Nevertheless, further studies are required to understand the underlying mechanisms of Blastocystis and cancer association.
1 Introduction
Blastocystis is an anaerobic intestinal protozoan found in humans and a wide range of animals. Morphological forms of Blastocystis include vacuolar, granular, amoeboid, and cyst forms, the vacuolar form is predominant in fresh stool samples and laboratory cultures (1). The Blastocystis prevalence rate is 60% in developing countries, due to poor hygiene and close contact with animals, compared to developed countries (5-20%) (2). More than 17 subtypes (STs) of Blastocystis spp. are known, and only nine subtypes are found in humans (3, 4). Blastocystis was considered a commensal parasite and caused asymptomatic infections at most. However, there is an increasing number of studies investigating the role and pathogenicity of Blastocystis in the gut (3, 4).
Some studies showed that Blastocystis infections contributed to the severity of multiple conditions, such as AIDs, cancer, IBD, and gut microbiota dysbiosis (5–7). Conversely, other studies have shown that the presence of Blastocystis promotes high diversity of gut microbiota in healthy individuals preventing intestinal disorders (8, 9). Blastocystis culture filtrates did not affect the growth of cancer cell lines in one study (10). These conflicting findings are probably attributed to the genetic diversity of Blastocystis, inter and intra-subtype variations (7, 11, 12).
Globally, cancer is regarded as one of the leading causes of death with an estimation of 10 million deaths in 2020 (13). The most common cancers in order are breast, lung, colorectal, prostate, skin, and stomach cancers according to WHO in 2020 (13). Some of risk factors include age, family history, obesity, diet, and infectious pathogens (13, 14). Since 30-50% of cancer is preventable by avoiding its risk factors and 30% of cancer are caused by infectious pathogens, it is important to identify all possible cancer-promoting infections to limit progression of existing cases and emergence of new cases (13). Colorectal cancer (CRC) is the third most common cancer, and the second leading cause of cancer-related deaths worldwide (2020) (15). CRC cases are detected in males more than females. The majority of CRC cases are diagnosed in the late stages of the disease, partially due to its non-specific symptoms (16–19). One of the risk factors of CRC is the gut microbiota dysbiosis (17–20). Several studies have investigated the role of microbiota in different cancers initiation or progression (21–27). However, previous studies focused on the bacterial content overlooking other micro-organisms such as protozoa and fungi (28).
Since Blastocystis spp. is considered a normal intestinal flora, investigating Blastocystis association with cancer, focusing on CRC, is essential to understanding the gut microbiota effect on tumor initiation and progression (8, 29). Many studies investigated the interaction between Blastocystis spp. and gut microbiota, and one study reported an association between Blastocystis with increased levels of five gut fungi (Mycobiota) (7–9, 30). Other studies associated gut mycobiota with carcinogenesis, initiation, and development of different cancers (20, 31–33). Thus, this study aimed to assess the possible association between Blastocystis infection and fungi in cancer patients locally. Then, compare Blastocystis infection in different cancers’ patients to cancer-free controls (CF).
2 Methods
2.1 Study design
In this observational study, a matched case-control study design was used. The case patients affected by cancer were gender and age-group-matched with a CF control group. The study participants were recruited from March 2020 to April 2022. This study followed the STROBE guidelines (34).
2.2 Study population and variables
All participants signed informed consent (n=104), and the study was performed per the Declaration of Helsinki. Participants of both genders were 18 years old and above. Age was categorized into three groups: Youth (18-24 years old), adults (25-59 years old), and elderly (≥60 years old). Nationalities were classified into six regions of origin: Africa, Americas, South-East Asia, Europe, Eastern Mediterranean, and Western Pacific. Patients who took any anti-parasitic drug in the last six months, did not provide informed consent, or were unwilling to give a stool sample were excluded from this study. The consumption of antibiotics, gastrointestinal surgery history in the past two years, hospitalization, cancer therapy, and the number of therapy cycles were extracted from patients’ medical records.
2.3 Case patients
Patients referred to the Oncology Services at Tawam Hospital, United Arab Emirates, with confirmed cancer (n=52) cases histopathologically, in any stage, and undergoing any treatment were included in this study. The cases were further sub-categorized into two groups: CRC (n=15) and cancers outside the gastrointestinal tract (COGT) (n=37) patients.
2.4 Control participants
Community-based control participants (n=52) were gender- and age-group matched subjects recruited. Excluding subjects with intestinal disorders, immunological or neoplastic disorders, and antibiotics users in the last six months before recruitment.
2.5 Sample collection and processing
Stool samples were collected using a commercial stool collection kit (alpha laboratories, UK). All stool samples were transported for analysis to the Microbiology Laboratory, College of Medicine and Health Sciences (CMHS), United Arab Emirates University (UAEU). Stool samples were each split into two parts; one was stored at 4°C and processed within one to two days for macroscopy, and microscopy, while the second was put in Eppendorf tubes and stored at -20°C for molecular work.
2.6 Macroscopic and microscopic investigation
As mentioned previously (35), macroscopic and microscopic examinations were performed to define the types of organisms and check for blood and mucus. Stool samples were stained with Wheatley Trichrome for Blastocystis detection, following the manufacturer’s instructions. Then, smears were examined microscopically under ×40 and ×100 magnification objectives. Two microbiologists examined all stool slides independently.
2.7 Molecular investigation
Genomic DNA was extracted from stool samples as previously published (35). Primers and PCR conditions used are listed in Table S1. Blastocystis spp. and gut fungi DNAs were amplified by Polymerase Chain Reaction (PCR). A mix of 5µl of 5x Q-Solution, 2.5µl of 10x CoralLoad PCR buffer, 1µl of each primer (10mM), 0.5µl of QIAGEN Taq DNA Polymerase (250U),1µl of dNTP Blend (100mM) (Applied Biosystems, USA), and 2µl of stool genomic DNA diluted in free-nuclease water to reach final volume of 25µl. For the ITS reaction mixtures, an extra 0.5µl of MgCl2 was added.
2.8 Sequencing and phylogenetic analysis
Gel and PCR Clean-Up System (Promega, Madison, Wisconsin, USA) was used following the manufacturer’s instructions. Barcode region and ITS primers were used to sequence the purified products in both directions via capillary electrophoresis using a Big DyeTM Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) in ABI PRISM 3130xI Genetic Analyzer. The confirmed Blastocystis-positive sequences were assigned to the best-matched Blastocystis subtype by aligning them to reference sequences in the GenBank database using nBLAST program (36). Query cover and per identity of ≥97% were used to determine subtypes matches. The ClustalW algorithm of MEGA-X was used to align the sequences (37). Acquired sequences were submitted to PubMLST database to confirm subtypes and identify relevant alleles (38). The established fungi-positive sequences were assigned to the best-match fungi species by aligning them to reference sequences in the GenBank database. Query cover of ≥80% and 97-100% per identity were used to determine the most probable match.
Blastocystis samples and reference sequences along with an outgroup species (Proteromonas lacertae, GenBank accession no. U37108) went through phylogenetic analysis. The best substitution models were determined via the Bayesian information criterion. The maximum Likelihood (ML) method and Bayesian Inference (BI) were used to construct the tree. ML tree was constructed using MEGA-X. Tamura 3-parameter with gamma distribution was used, and Bootstrapping analysis with 1000 replicates was performed. For the BI method, Jmodeltest v2.1.10 and MrBayes v3.2.7 were used (39, 40). Hasegawa-Kishino-Yano model with gamma distribution was used, and four Markov chains were run for 5 million generations, with a sampling frequency of 100 and a 25% burn-in. Tree Graph 2 combined the two constructed trees (41).
2.9 Statistical analysis
Data were analyzed by IBM SPSS Statistics v26.0. Qualitative variables were expressed as numbers and percentages, while quantitative variables were expressed as means and medians. The chi-square, Fisher’s Exact, and Fisher-Freeman-Halton tests for categorical variables. Logistic regression was used to predict factors associated with Blastocystis prevalence, and a p-value of ≤ 0.05 indicated statistical significance.
2.10 Ethical approval
The study was approved by the Tawam Human Research Ethics Committee (T-HREC) of Tawam Hospital, Al Ain, Abu Dhabi, UAE (THREC-678).
3 Results
In this case-control study, a total of 104 matched participants were recruited. The controls were CF participants (n=52), and the cases were cancer patients (n=52) (Table S2). The study participants consisted of 44 males (42.3%) and 60 females (57.7%). CF participants’ mean age was 43.3 (median: 41.5, range: 23-87). Cancer patients’ mean age was 49.3 (median: 51, range: 22-75). There was no statistical association between Blastocystis infection and gender (P=0.147) nor Blastocystis infection and age groups (P=0.277). The prevalence of Blastocystis among regions differed but was statistically insignificant (P=0.056).
Of the cancer patients, 37 patients were COGT, while 15 individuals were CRC patients. COGT patients were of 19 different cancer types reclassified into 8 categories based on tumor site (Table S2). The majority of patients had breast cancer (n=16, 15.4%), and hematologic cancer (n=9, 8.7%). The Blastocystis prevalence within cancer types was statistically insignificant (P=0.440). Also, antibiotics usage, GIT surgery history, number of chemotherapy cycles and hospitalization were statistically insignificant to Blastocystis infection (P=0.721, P=0.07, P=0.705, and P=0.241) in cancer group (Table S3).
3.1 Stool analysis
Stool samples macroscopic parameters had an insignificant association with Blastocystis infection. Thirteen samples were positive for Blastocystis via microscopy (Figure 1). Moreover, eight other protozoans and three helminths were identified in participants’ stool samples, and the majority of infections were identified in CF group. The most common protozoa found were Entamoeba, Cryptosporidium, and Retortamonas intestinalis (Table S4), and the most common helminths infection were Enterobius vermicularis and Ascaris lumbricoides (Table S4). Blastocystis co-infection was studied; however, there was no significant association with other intestinal parasites.
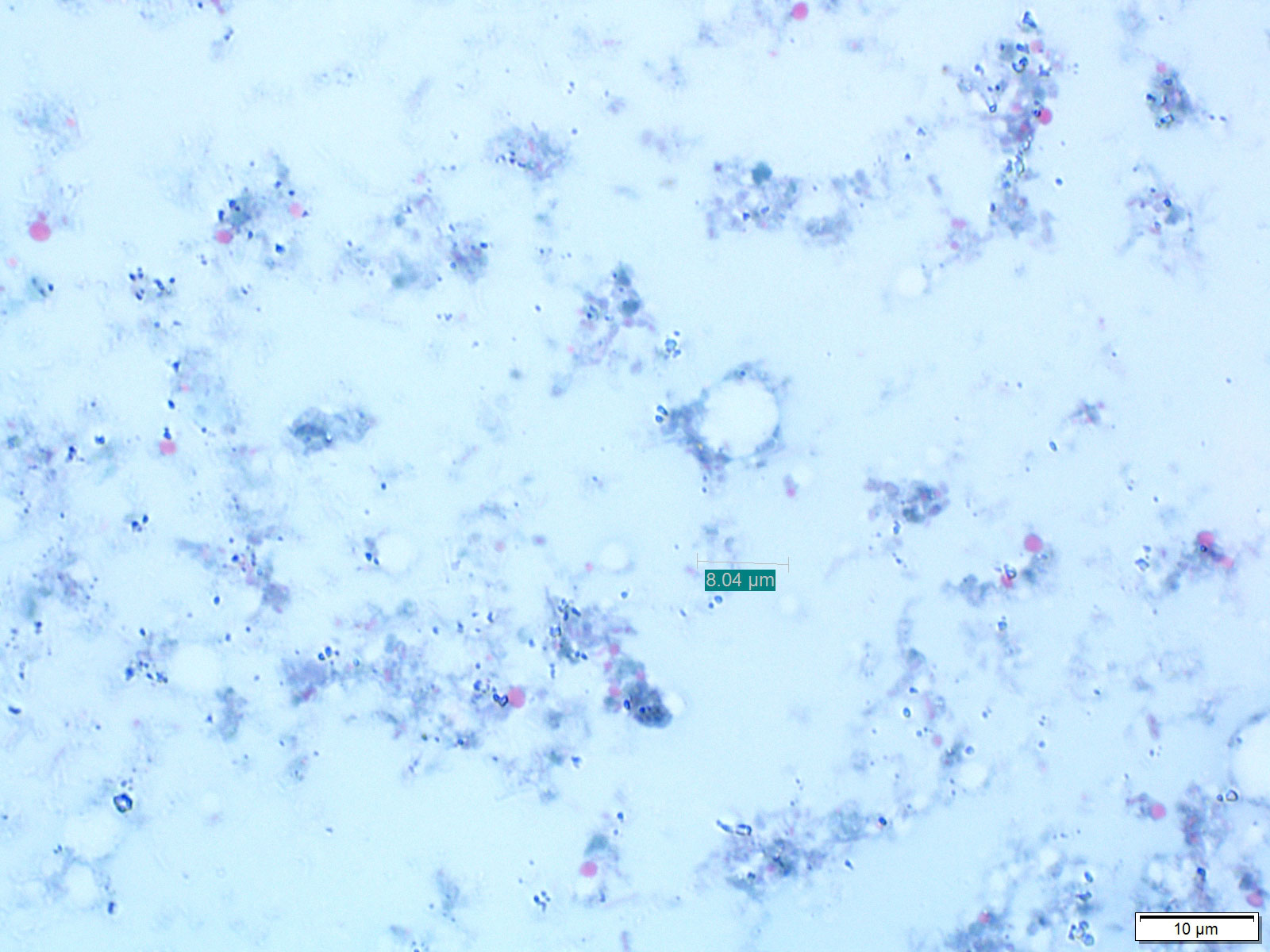
Figure 1 Blastocystis cyst (black arrow) in stool sample stained with Trichrome stain at 100x magnification objectives using light microscopy.
3.2 Molecular investigation: Blastocystis infection
Twenty-four samples were Blastocystis-positive via PCR and 13 via microscopy (Table S5). In total, 30 samples were Blastocystis-positive regardless of the detection method (Table 1). As anticipated, Blastocystis prevalence was significantly higher among all cancer types (40.4%) compared to CF group (17.3%) (P=0.009). Also, Blastocystis prevalence was significantly higher in CRC sub-group (60%) compared to CF group (P=0.002). However, COGT sub-group (32.4%) was insignificant compared to CF group (P=0.161).
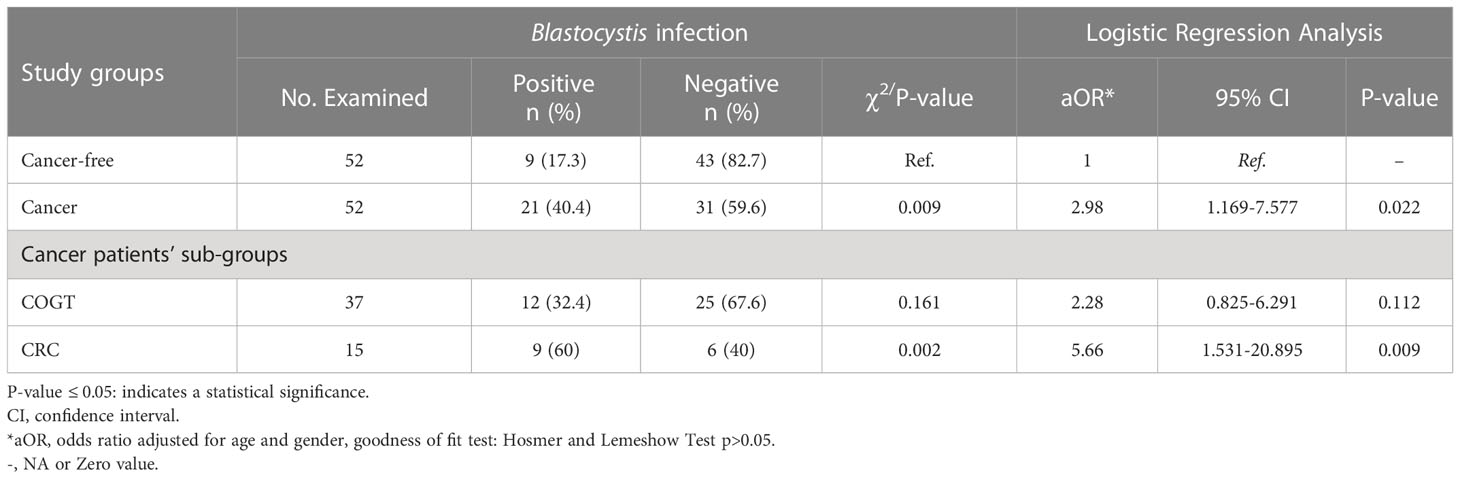
Table 1 Logistic regression and Pearson chi-square test of Blastocystis infection among the study population (n = 104).
The odds of Blastocystis infection were almost threefold higher in cancer group than in the CF group (OR=2.98, P=0.022) and more than fivefold higher in the CRC group (OR= 5.66, P=0.009). In contrast, the Blastocystis infection odds in COGT to CF group were insignificant (Table 1). Blastocystis fold change between CRC and COGT groups was insignificant (OR=2.35, P=0.209), even though the prevalence of Blastocystis spp. in CRC group alone was high (60%) (Table 2).
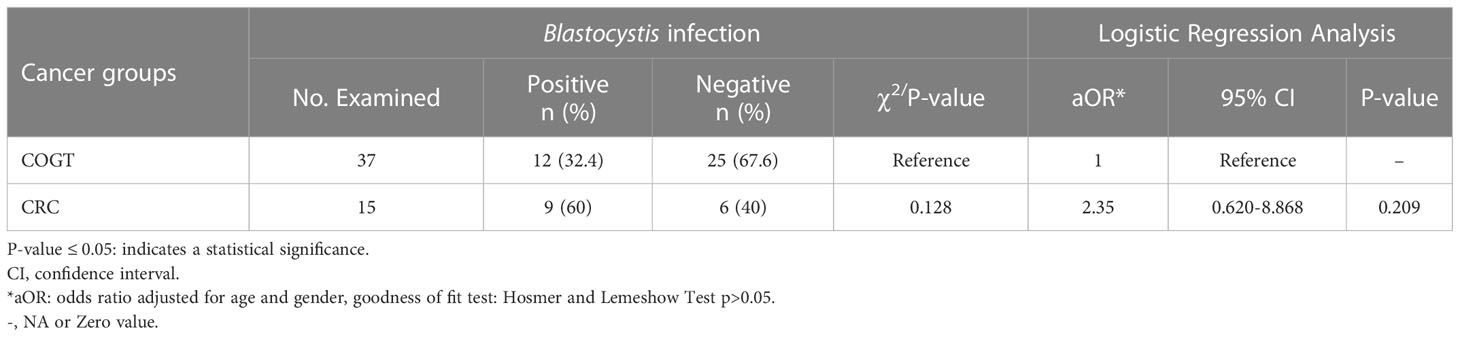
Table 2 Logistic regression and Pearson chi-square test of Blastocystis infection among cases, COGT vs. CRC group.
3.3 Molecular investigation: Gut fungi
Via gel electrophoresis, amplicon size of ~450-800bp using ITS primers was considered positive for gut fungi. Sixty individuals (57.7%) were tested positive for gut fungi, of which 30 were from CF group (57.7%), and 30 were from the cancer group (57.7%). Twenty-two individuals (59.5%) of the COGT subgroup had gut fungi, and 8 (53.3%) of the CRC subgroup.
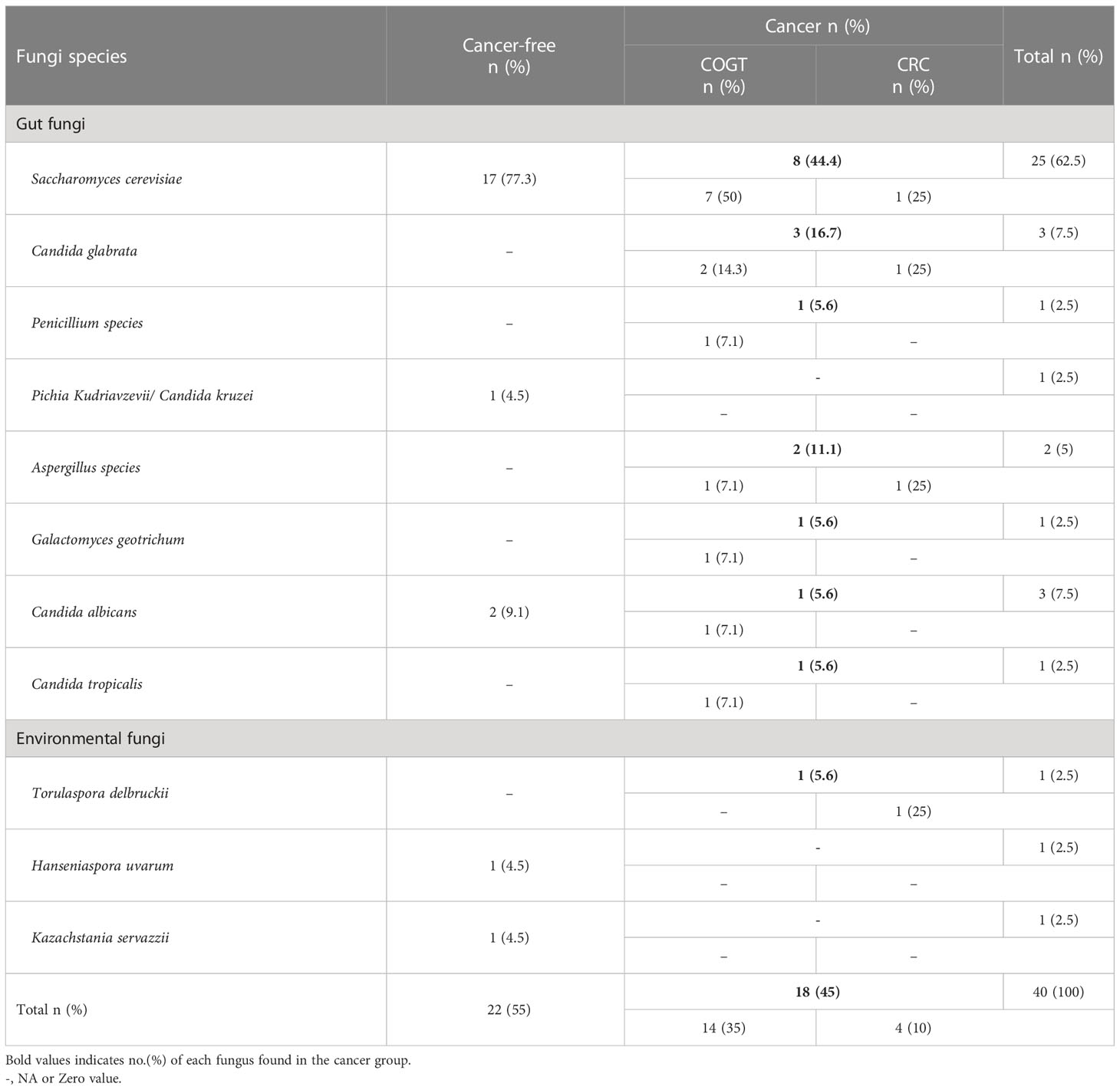
Thirty-nine fungi-positive samples were sequenced. Eleven types of fungi were identified; eight were gut mycobiome, and three were environmental fungal species (Table 3). Saccharomyces cerevisiae was the most common gut fungi (n=25, 62.5%). S. cerevisiae was detected in 17 samples (77.3%) from CF group and 8 samples (44.4%) from the cancer group. However, S. cerevisiae prevalence was not significantly associated with cancer (P=0.193) nor Blastocystis spp.(P=0.478).
3.4 Blastocystis subtyping and phylogenetic analysis
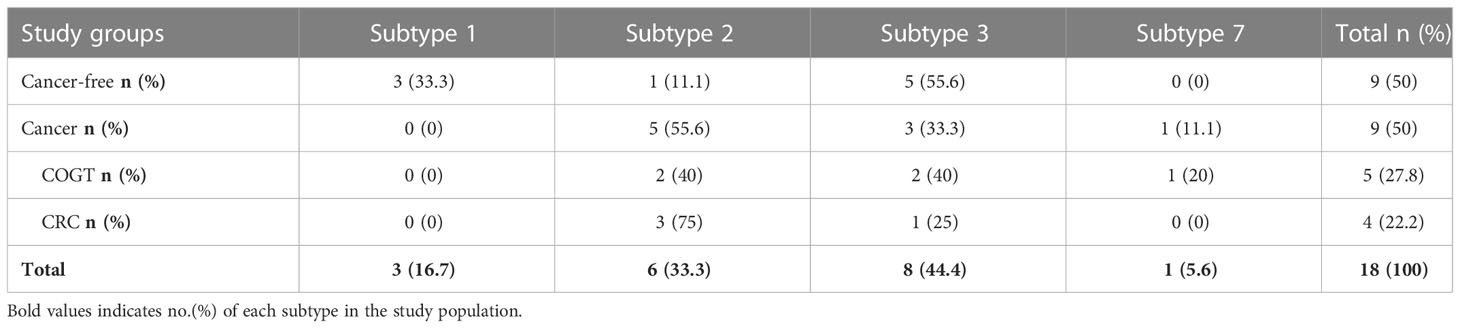
Eighteen samples were Blastocystis-subtyped via sequencing. The most common Blastocystis subtype was ST3 (n=8, 44.4%), then ST2 (n=6, 33.3%) (Table 4), In our study, the most common subtype was ST3 (n=5, 55.6%) in CF group and ST2 (n=5, 55.6%) in the cancer group, and only one ST7 was detected in a breast cancer patient.
Fifteen Blastocystis-subtyped samples were assigned an accession number via GenBank. Allele sequence analyses showed allele 4 in all ST1-positive samples, allele 15 in three ST2-positive samples, and allele 12 in one ST2-positive sample (Table S6). In ST3-positive samples, allele 34 (n=2) and allele 36 (n=3) were identified. Allele 137 was identified in the ST7-positive sample.
The ML and BI trees include 15 sample sequences submitted to NCBI GenBank with the accession numbers (OM478515-OM478518, OM478527-OM478529, OM976632, OM976635-OM976638, and ON185813-ON185815) (Figure 2). The topologies between the original reconstructed trees with ML (Figure S1) and BI (Figure S2) were broadly consistent. Figure S3 shows the branch lengths of the combined tree.
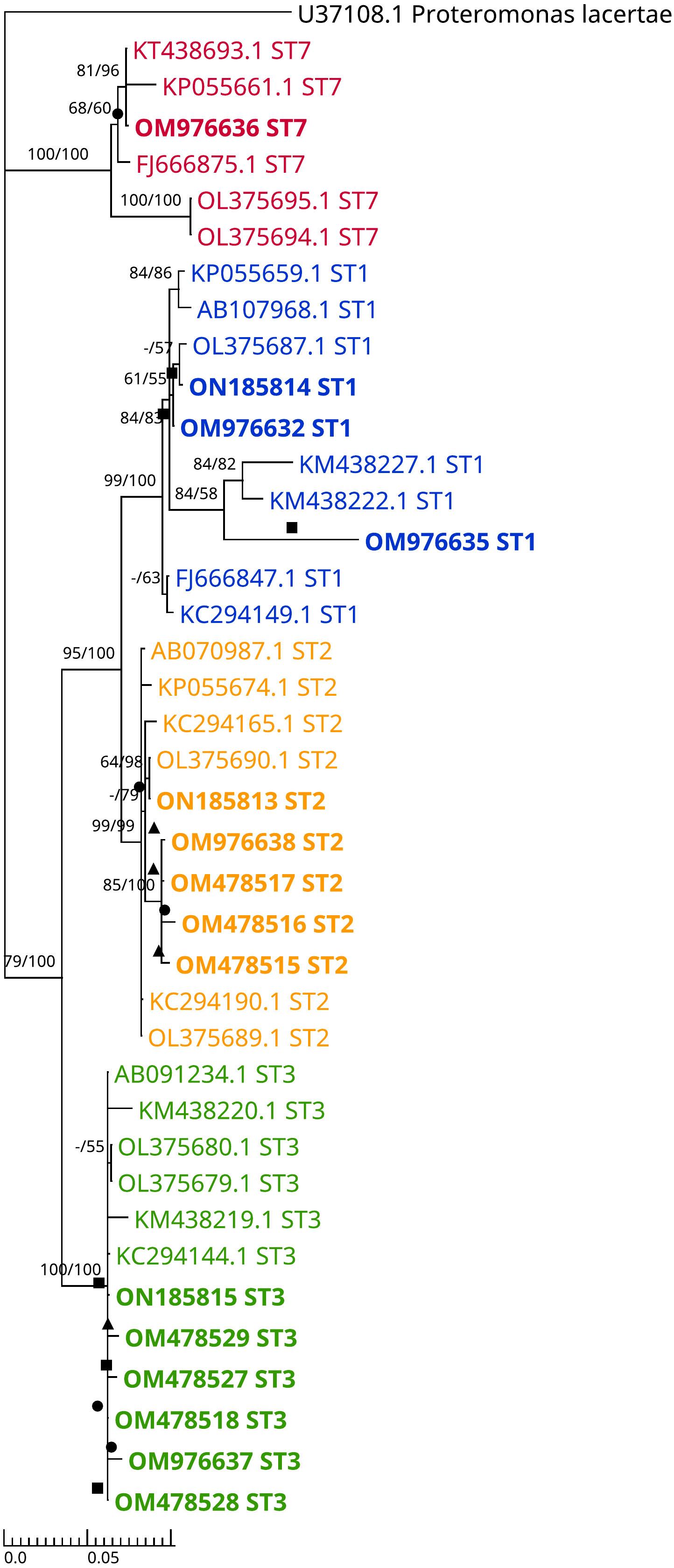
Figure 2 Dendrogram representing combined (ML+BI) phylogenetic tree inferred using the Barcode region of SSU rRNA gene sequences. This tree includes 15 isolated samples, 19 reference sequences from the GenBank (isolates from Cancer-free participants and CRC patients), 5 sequences of different alleles from PubMLST (AB107968.1, KT438693.1, AB070987.1, AB091234.1, and AB107965.1), and an outgroup (Proteromonas lacertae). The best tree of ML analysis was with Tamura 3-parameter model, while Bayesian tree best model was Hasegawa-Kishino-Yano model. Bootstrapping Proportions of more than 50% are shown on the left side of the branch. Bayesian posterior probability was also performed (ngen=5 million, samplefreq=100 and Burn-in 25%) and values of more than 50% are shown on the right side of the branch. A solid triangle indicates an isolate from a CRC patient, a solid circle indicates an isolate from a COGT patient and a solid square indicates an isolate from a cancer-free patient.
4 Discussion
Cancer is one of the most common causes of death worldwide (15, 42). The majority of new cases in the UAE are among women compared to men. (42). Colorectal, skin, and prostate cancers are the most prevalent in men, whereas breast, thyroid, and colorectal cancers are the most common in women. (42). In our study, Blastocystis spp. was significantly higher in cancer patients (OR=2.98). Similarly, a regional study from the Kingdom of Saudi Arabia (KSA) reported a significant association between Blastocystis infection and patients with cancer (OR=2.15) (43).
CRC is among the most diagnosed malignancies and mortalities globally (15). Most incident cancers among Emirati males were related to CRC (16). In the current study, approximately 60% of CRC patients were infected with Blastocystis, which was higher than in previous studies in Iran (23.9%), KSA (29.7%), Egypt (52%), Turkey (7.5%), and Poland (12%) (43–47). The disparities might be attributed to the Blastocystis detection method or the population’s diversity (48). Consistent with the results of other studies, our study reported significantly high odds of Blastocystis in CRC patients (43–46).
Blastocystis spp. prevalence in the COGT group (32.4%) was insignificant compared to CF group (17.3%), similar to previous studies (43, 49). On the other hand, Taşova et al. reported a significant difference in Blastocystis prevalence between hematologic cancer patients (13%) and the non-cancer GIT patients group (1%) (50).
Blastocystis prevalence did not significantly differ within the cancer types included in our study (P=0.440). Furthermore, Blastocystis prevalence between CRC (60%) and COGT (32.4%) groups was insignificant (P=0.128), in agreement with previous studies on cancer groups (47, 51, 52). While Yersal and colleagues reported significantly higher Blastocystis prevalence in lung cancer (38.1%) compared to other cancer types (7.2-8.9%) (47).
In this study, patients’ medical records were assessed to identify potential risk factors associated with Blastocystis in cancer patients. Blastocystis prevalence association with risk factors tested was found insignificant (Table S3), as reported in other studies (44, 52).
Except for two patients (breast and hematologic/blood malignancies), all cancer patients in our study had received cancer therapy. Twenty-four cancer patients received chemotherapy, of which 18 (75%) were COGT, and 6 (25%) were CRC. The potential association of the number of completed chemotherapy cycles with Blastocystis prevalence was insignificant (P=0.705). In contrast, other studies reported that patients receiving at least eight chemotherapy cycles had a significantly higher Blastocystis prevalence than patients receiving fewer cycles (47, 53). Generally, Blastocystis infections were detected consistently in patients receiving ≥8 chemotherapy cycles compared to earlier cycles, and compared to patients who did not start treatment (47, 53, 54). Interestingly, current study findings and previous studies suggest that CRC patients had a significantly higher Blastocystis prevalence than healthy participants, regardless of their treatment status (43, 45, 46). Also, COGT patients receiving multiple chemotherapy cycles presented a significantly higher Blastocystis infection than healthy participants (43, 50). These findings may help in understanding the complex relationship between Blastocystis and cancer and the effects of chemotherapy on Blastocystis prevalence.
We have detected more Blastocystis infection in males (36.4%) compared to females (23.3%), in agreement with previous studies which have reported that Blastocystis was higher in males (11.8-53.8%) versus females (0-46.2%) (2, 44, 45, 47, 53). Ali et al. linked the previous findings to more outdoor activities and exposure to infection sources in male patients (44).
Reportedly, the gut fungi Aspergillus flavus, Debaryomyces hansenii, Mucor mucedo, Mucor racemosus, and Issatchenkia terricola were significantly higher in the presence of Blastocystis (30). Therefore, we investigated the relationship between Blastocystis and gut fungi in cancer patients. Fungal sequences were identified via nBLAST and confirmed as gut fungi via the human gut mycobiome database published previously (28, 36, 55, 56). However, none of the aforementioned fungi were detected in our fungi-positive samples. We have detected S. cerevisiae in most gut fungi-positive samples 25 (62.5%). Various studies saw S. cerevisiae to inhibit CRC progression and metastasis and stimulate apoptosis of cancer cells (57, 58). S. cerevisiae was more prevalent in CF group (77.3%) compared to cancer patients (44.4%) with an insignificant difference (P=0.193). The latter observation was consistent with previous studies, except the association was statistically significant (59, 60).
The predominant Blastocystis subtype in this study was ST3 (n=5) in CF group and ST2 (n=5) in cancer patients. In accordance with studies from Egypt and the UAE, where ST3 was the most common subtype in control participants (44, 61). On the other hand, studies in KSA and France showed ST2 and ST4 are the most common subtypes in a healthy population (43, 62). Moreover, ST2 was the most common sub-type in cancer group (n=5) and CRC patients(n=3), unlike other studies where ST3 and ST1 were the most predominant sub-type in cancer patients (43, 44, 46, 47, 51, 53).
In our study, the predominant Blastocystis subtypes are ST2 and ST3 in COGT group, with the same percentage (40%). Similarly, in Mohamed et al. work, Blastocystis ST2 was predominantly seen in COGT patients (43.7%) (43). Also, we found ST7 in one breast cancer patient while Ali et al. and Poirier et al. found ST7 in two CRC patients and one hematologic cancer patient, respectively (44, 62). Those difference in subtyping is possibly due to detection methods variation (43, 47, 63).
To our knowledge, this is the first study in the UAE to investigate Blastocystis intra-subtypes (alleles) variations in cancer patients. Few studies examined Blastocystis alleles prevalence in healthy individuals (61, 64). In our research, Blastocystis ST3 alleles (34 and 36) were identified in 3 cancer patients and 2 CF participants (Table S6). These alleles were also identified in studies conducted on healthy individuals (61, 64). We detected Blastocystis ST1 allele 4 in CF participants, and ST2 alleles 15 and 12 in cancer patients. While AbuOdeh et al., in the UAE, reported ST1 allele 4, and ST2 allele 9 in healthy subjects (61). We identified Blastocystis ST7 allele 137, a similar finding was reported in Rezaei et al. study (65). A study from Turkey reported alleles 2, 4, and 88 of ST1, and alleles 34 and 36 of ST3 in cancer patients (66).
We submitted Blastocystis allele sequences OM478515 ST2 and OM478527 ST3 to PubMLST and 15 of the first UAE Blastocystis isolates. In this study, Blastocystis-ST1-positive controls (n=4) were all of allele 4, ST2-positive controls (n=2) were of allele 9, and ST3-positive controls (n=2) were of allele 34 and allele 36.
In conclusion, Blastocystis infection was significantly associated with cancer, with ST2 being the most common subtype. Furthermore, CRC patients had a higher risk of Blastocystis infection than CF. Since Blastocystis infection is more common among cancer patients than CF individuals, further studies are needed to understand the association between Blastocystis infection and cancer in general, and CRC in particular. Thus, routine Blastocystis infection screening in cancer patients might be a useful tool to be added to the usual patient’s care in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM478529 https://www.ncbi.nlm.nih.gov/genbank/, OM478517 https://www.ncbi.nlm.nih.gov/genbank/, OM478518 https://www.ncbi.nlm.nih.gov/genbank/, OM478516 https://www.ncbi.nlm.nih.gov/genbank/, OM478528 https://www.ncbi.nlm.nih.gov/genbank/, OM478526 https://www.ncbi.nlm.nih.gov/genbank/, OM478525 https://www.ncbi.nlm.nih.gov/genbank/, OM478521 https://www.ncbi.nlm.nih.gov/genbank/, OM478519 https://www.ncbi.nlm.nih.gov/genbank/, OM478513 https://www.ncbi.nlm.nih.gov/genbank/, OM478520 https://www.ncbi.nlm.nih.gov/genbank/, OM478514 https://www.ncbi.nlm.nih.gov/genbank/, OM478512 https://www.ncbi.nlm.nih.gov/genbank/, OM478511 https://www.ncbi.nlm.nih.gov/genbank/, OM976635 https://www.ncbi.nlm.nih.gov/genbank/, OM976632 https://www.ncbi.nlm.nih.gov/genbank/, OM976639 https://www.ncbi.nlm.nih.gov/genbank/, ON185815 https://www.ncbi.nlm.nih.gov/genbank/, ON185814 https://www.ncbi.nlm.nih.gov/genbank/, OM976638 https://www.ncbi.nlm.nih.gov/genbank/, OM976637 https://www.ncbi.nlm.nih.gov/genbank/, OM976636 https://www.ncbi.nlm.nih.gov/genbank/, ON185813 https://www.ncbi.nlm.nih.gov/genbank/, OM976633 https://www.ncbi.nlm.nih.gov/genbank/, OM976634.
Ethics statement
The studies involving human participants were reviewed and approved by Tawam Human Research Ethics Committee (T-HREC) of Tawam Hospital, Al Ain, Abu Dhabi, UAE (THREC-678). The patients/participants provided their written informed consent to participate in this study.
Author contributions
LL: Methodology, data curation, analysis, interpretation, visualization, writing, and editing. SZ: Conception and design, methodology, data analysis, interpretation, revision, and writing. SA: Methodology, interpretation, writing, revision, and editing. MO: Methodology, analysis, and revision. SS: Study design and methodology. ZR: Conception and design, supervision, funding acquisition, methodology, data analysis, interpretation, revision, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work received a grant from the College of Medicine and Health Sciences, United Arab Emirates University (Fund number:12M081)
Acknowledgments
The authors thank Dr. Khalid Balaraj, and Mr. Khaled Al Qawasmeh from Tawam Oncology Center; Dr. Ziad Peerwani, Dr. Timothy Anthony Collyns, Ms. Shiekha Awad Al Kaabi, and Mr. Edmendo Garcia from Tawam Laboratory Divisions; for their enormous help in samples and data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1115835/full#supplementary-material
References
1. Wawrzyniak I, Poirier P, Viscogliosi E, Dionigia M, Texier C, Delbac F, et al. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther Adv Infect Dis (2013) 1(5):167–78. doi: 10.1177/2049936113504754
2. Salehi M, Mardaneh J, Niazkar HR, Minooeianhaghighi M, Arshad E, Soleimani F, et al. Prevalence and subtype analysis of blastocystis hominis isolated from patients in the northeast of Iran. J Parasitol Res (2021) 2021:e8821885. doi: 10.1155/2021/8821885
3. Kumarasamy V, Anbazhagan D, Subramaniyan V, Vellasamy S. Blastocystis sp., parasite associated with gastrointestinal disorders: An overview of its pathogenesis, immune modulation and therapeutic strategies. Curr Pharm Des (2018) 24(27):3172–5. doi: 10.2174/1381612824666180807101536
4. Partida-Rodríguez O, Serrano-Vázquez A, Nieves-Ramírez ME, Moran P, Rojas L, Portillo T, et al. Human intestinal microbiota: Interaction between parasites and the host immune response. Arch Med Res (2017) 48(8):690–700. doi: 10.1016/j.arcmed.2017.11.015
5. Kumarasamy V, Kuppusamy UR, Jayalakshmi P, Samudi C, Ragavan ND, Kumar S. Exacerbation of colon carcinogenesis by blastocystis sp. PloS One (2017) 12(8):e0183097. doi: 10.1371/journal.pone.0183097
6. Kesuma Y, Firmansyah A, Bardosono S, Sari IP, Kurniawan A. Blastocystis ST-1 is associated with irritable bowel syndrome-diarrhoea (IBS-d) in Indonesian adolescences. Parasite Epidemiol Control (2019) 6:e00112. doi: 10.1016/j.parepi.2019.e00112
7. Yason JA. Interactions between a pathogenic blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome (2019) 7:30. doi: 10.1186/s40168-019-0644-3
8. Billy V, Lhotská Z, Jirků M, Kadlecová O, Frgelecová L, Parfrey LW, et al. Blastocystis colonization alters the gut microbiome and, in some cases, promotes faster recovery from induced colitis. Front Microbiol (2021) 12:641483. doi: 10.3389/fmicb.2021.641483
9. Kodio A, Coulibaly D, Koné AK, Konaté S, Doumbo S, Guindo A, et al. Blastocystis colonization is associated with increased diversity and altered gut bacterial communities in healthy malian children. Microorganisms (2019) 7(12):649. doi: 10.3390/microorganisms7120649
10. Long H, Handschack A, König W, Ambrosch A. Blastocystis hominis modulates immune responses and cytokine release in colonic epithelial cells. Parasitol Res (2001) 87(12):1029–30. doi: 10.1007/s004360100494
11. Deng L, Wojciech L, Gascoigne NRJ, Peng G, Tan KSW. New insights into the interactions between blastocystis, the gut microbiota, and host immunity. PloS Pathog (2021) 17(2):e1009253. doi: 10.1371/journal.ppat.1009253
12. Qi M. Genetic diversity of blastocystis in kindergarten children in southern xinjiang, China. Parasites and Vectors (2020) 13:15. doi: 10.1186/s13071-020-3890-0
13. WHO W. Cancer [Internet] (2022) [cited 2022 May 21]. Available at: https://www.who.int/news-room/fact-sheets/detail/cancer.
14. Understanding Cancer Risk [Internet]. Cancer.Net. (2010) [cited 2023 Jan 25]. Available at: https://www.cancer.net/navigating-cancer-care/prevention-and-healthy-living/understanding-cancer-risk.
15. Cancer (IARC) TIA for R onGlobal Cancer Observatory [Internet] (2021) [cited 2022 Apr 4]. Available at: https://gco.iarc.fr/.
16. Abancens M, Bustos V, Harvey H, McBryan J, Harvey BJ. Sexual dimorphism in colon cancer. Front Oncol (2020) 10:607909. doi: 10.3389/fonc.2020.607909
17. Cleveland Clinic Abu Dhabi LLC. Colorectal cancer program [Internet]. Cleveland clinic Abu Dhabi (2021). Available at: https://www.clevelandclinicabudhabi.ae/en/institutes-and-specialties/digestive-disease-institute/pages/colorectal-cancer.aspx.
18. ADPHC T. ADPHC revises recommendations for early colorectal (2022). Available at: https://www.doh.gov.ae/en/news/ADPHC-revises-recommendations-for-early-colorectal-cancer-screening.
19. HAAD T. Department of health –HAAD fact: 62% of colorectal cancer cases are men (2022). Available at: https://www.doh.gov.ae/en/news/haad-fact-62-of-colorectal-cancer-cases-are-men.
20. Rebersek M. Gut microbiome and its role in colorectal cancer. BMC Cancer (2021) 21(1):1–13. doi: 10.1186/s12885-021-09054-2
21. Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol (2016) 22(2):501–18. doi: 10.3748/wjg.v22.i2.501
22. Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis (2017) 36(5):757–69. doi: 10.1007/s10096-016-2881-8
23. Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human Gut Microbiota and Gastrointestinal Cancer. Genomics Proteomics Bioinformatics [Internet] (2018) 16(1):33–49. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6000254/.
24. Plaza-Díaz J, Álvarez-Mercado AI, Ruiz-Marín CM, Reina-Pérez I, Pérez-Alonso AJ, Sánchez-Andujar MB, et al. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer [Internet] (2019) 19:495. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6534876/.
25. Zhang J, Xia Y, Sun J. Breast and gut microbiome in health and cancer. Genes Dis [Internet] (2020) 8(5):581–9. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8278537/.
26. Zhao Y, Liu Y, Li S, Peng Z, Liu X, Chen J, et al. Role of lung and gut microbiota on lung cancer pathogenesis. J Cancer Res Clin Oncol [Internet] (2021) 147(8):2177–86. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8236441/.
27. Georgiou K, Marinov B, Farooqi AA, Gazouli M. Gut Microbiota in Lung Cancer: Where Do We Stand? Int J Mol Sci [Internet] (2021) 22(19):10429. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8508914/.
28. Raimondi S, Amaretti A, Gozzoli C, Simone M, Righini L, Candeliere F, et al. Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping, and colonization. Front Microbiol (2019) 10:1575. doi: 10.3389/fmicb.2019.01575
29. Kaya S, Sesl E, Arikan S, Dem M. Pathogenicity of blastocystis hominis, a clinical reevaluation. Turkiye Parazitol Derg (2007) 31(3):184-7.
30. Nieves-Ramírez ME, Partida-Rodríguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, et al. Asymptomatic intestinal colonization with protist blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems (2018) 3(3):e00007–18. doi: 10.1128/mSystems.00007-18
31. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8
32. Dohlman AB, Klug J, Mesko M, Gao IH, Lipkin SM, Shen X, et al. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell (2022) 185(20):3807–3822.e12.
33. Zhang L, Chai D, Chen C, Li C, Qiu Z, Kuang T, et al. Mycobiota and C-Type Lectin Receptors in Cancers: Know thy Neighbors. Front Microbiol [Internet] (2022) 13(20):946995. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9326027/.
34. Cuschieri S. The STROBE guidelines. Saudi J Anaesth (2019) 13(Suppl 1):S31–4. doi: 10.4103/sja.SJA_543_18
35. Al-Rifai RH, Loney T, Sheek-Hussein M, Zoughbor S, Ajab S, Olanda M, et al. Prevalence of, and factors associated with intestinal parasites in multinational expatriate workers in Al ain city, united Arab Emirates: An occupational cross-sectional study. J Immigr Minor Health (2020) 22(2):359–74. doi: 10.1007/s10903-019-00903-8
36. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol (1990) 215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2
37. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol (2018) 35(6):1547–9. doi: 10.1093/molbev/msy096
38. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res (2018) 3:124. doi: 10.12688/wellcomeopenres.14826.1
39. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinforma Oxf Engl (2001) 17(8):754–5. doi: 10.1093/bioinformatics/17.8.754
40. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods (2012) 9(8):772–2. doi: 10.1038/nmeth.2109
41. Stöver BC, Müller KF. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf (2010) 11(1):7. doi: 10.1186/1471-2105-11-7
42. National Cancer Registry (UAE) [Internet]. Available at: https://smartapps.moh.gov.ae/ords/f?p=105:511.
43. Mohamed AM, Ahmed MA, Ahmed SA, Al-Semany SA, Alghamdi SS, Zaglool DA. Predominance and association risk of blastocystis hominis subtype I in colorectal cancer: a case control study. Infect Agent Cancer (2017) 12(1):21. doi: 10.1186/s13027-017-0131-z
44. Ali SH, Ismail MAM, El-Badry AA, Abu-Sarea EY, Dewidar AM, Hamdy DA. An association between blastocystis subtypes and colorectal cancer patients: A significant different profile from non-cancer individuals. Acta Parasitol (2022) 67:752–63. doi: 10.1007/s11686-021-00508-y
45. Mahmoudvand H, Sepahvand A, Badparva E, Khatami M, Niazi M, Moayyedkazemi A. Possible association and risk factors of blastocystis infection and colorectal cancers in Western Iran. Arch Clin Infect Dis (2021) 16(1). doi: 10.5812/archcid.90861
46. Sulżyc-Bielicka V, Kołodziejczyk L, Adamska M, Skotarczak B, Jaczewska S, Safranow K, et al. Colorectal cancer and blastocystis sp. infection. Parasit Vectors (2021) 14(1):200. doi: 10.1186/s13071-021-04681-x
47. Yersal O, Malatyali E, Ertabaklar H, Oktay E, Barutca S, Ertug S. Blastocystis subtypes in cancer patients: Analysis of possible risk factors and clinical characteristics. Parasitol Int (2016) 65(6 Pt B):792–6. doi: 10.1016/j.parint.2016.02.010
48. UNESCO. Awareness and promotion of cultural diversity values. In: Diversidad de las expresiones culturales (2020) UNESCO Diversity of Cultural Expressions. Available at: https://es.unesco.org/creativity/policy-monitoring-platform/awareness-promotion-cultural.
49. Zaki N. Intestinal protozoan infections among Egyptian neutropenic patients with acute leukemia. Trop Biomed (2021) 38:50–6. doi: 10.47665/tb.38.1.009
50. Taşova Y, Sahin B, Koltaş S, Paydaş S. Clinical significance and frequency of blastocystis hominis in Turkish patients with hematological malignancy. Acta Med Okayama. (2000) 54(3):133–6. doi: 10.18926/AMO/32298
51. Zhang W, Ren G, Zhao W, Yang Z, Shen Y, Sun Y, et al. Frontiers | genotyping of enterocytozoon bieneusi and subtyping of blastocystis in cancer patients: Relationship to diarrhea and assessment of zoonotic transmission. Microbiology (2017) 8:1835. doi: 10.3389/fmicb.2017.01835/full
52. Mülayim S, Dalkılıç S, Akbulut HH, Aksoy A, Kaplan M. Investigation of the relationship between lymphocyte subsets and intestinal parasites. Acta Trop (2022) 225:106221. doi: 10.1016/j.actatropica.2021.106221
53. Asghari A, Zare M, Hatam G, Shahabi S, Gholizadeh F, Motazedian M. Molecular identification and subtypes distribution of blastocystis sp. isolated from children and adolescent with cancer in Iran: evaluation of possible risk factors and clinical features. Acta Parasitol (2020) 65(2):462–73. doi: 10.2478/s11686-020-00186-2
54. Chandramathi S, Suresh K, Anita ZB, Kuppusamy UR. Infections of blastocystis hominis and microsporidia in cancer patients: are they opportunistic? Trans R Soc Trop Med Hyg (2012) 106(4):267–9. doi: 10.1016/j.trstmh.2011.12.008
55. Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martínez I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol (2015) 15:9–17. doi: 10.1016/j.funeco.2015.01.006
56. Suhr MJ, Banjara N, Hallen-Adams HE. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol (2015) 62(3):209–15. doi: 10.1111/lam.12539
57. Abedi J, Vakili Saatloo M, Nejati V, Hobbenaghi R, Tukmechi A, Nami Y, et al. Selenium-enriched saccharomyces cerevisiae reduces the progression of colorectal cancer. (Iran: SpringerLink) (2018) 185:424–32. doi: 10.1007/s12011-018-1270-9
58. Sambrani R, Abdolalizadeh J, Kohan L, Jafari B. Saccharomyces cerevisiae inhibits growth and metastasis and stimulates apoptosis in HT-29 colorectal cancer cell line. (Iran:SpringerLink) (2018) 28:985–95. doi: 10.1007/s00580-018-2855-6
59. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut (2019) 68(4):654–62. doi: 10.1136/gutjnl-2018-317178
60. Li JQ, Li JL, Xie YH, Wang Y, Shen XN, Qian Y, et al. Saccharomyces cerevisiae may serve as a probiotic in colorectal cancer by promoting cancer cell apoptosis. J Dig Dis (2020) 21(10):571–82. doi: 10.1111/1751-2980.12930
61. AbuOdeh R, Ezzedine S, Samie A, Stensvold CR, ElBakri A. Prevalence and subtype distribution of blastocystis in healthy individuals in sharjah, united Arab Emirates. Infect Genet Evol (2016) 37:158–62. doi: 10.1016/j.meegid.2015.11.021
62. Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. Development and evaluation of a real-time PCR assay for detection and quantification of blastocystis parasites in human stool samples: Prospective study of patients with hematological malignancies▿. J Clin Microbiol (2011) 49(3):975–83. doi: 10.1128/JCM.01392-10
63. Stensvold CR. Comparison of sequencing (Barcode region) and sequence-Tagged-Site PCR for blastocystis subtyping. J Clin Microbiol (2013) 51(1):190–4. doi: 10.1128/JCM.02541-12
64. Pandey PK, Verma P, Marathe N, Shetty S, Bavdekar A, Patole MS, et al. Prevalence and subtype analysis of blastocystis in healthy Indian individuals. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis (2015) 31:296–9. doi: 10.1016/j.meegid.2015.02.012
65. Rezaei Riabi T, Mirjalali H, Haghighi A, Rostami Nejad M, Pourhoseingholi MA, Poirier P, et al. Genetic diversity analysis of blastocystis subtypes from both symptomatic and asymptomatic subjects using a barcoding region from the 18S rRNA gene. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis (2018) 61:119–26. doi: 10.1016/j.meegid.2018.03.026
Keywords: colorectal cancer, ST subtypes, phylogenetic analysis, UAE, Blastocystis infection, cancer, Fungi
Citation: Labania L, Zoughbor S, Ajab S, Olanda M, Shantour SNM and Al Rasbi Z (2023) The associated risk of Blastocystis infection in cancer: A case control study. Front. Oncol. 13:1115835. doi: 10.3389/fonc.2023.1115835
Received: 12 December 2022; Accepted: 27 January 2023;
Published: 20 February 2023.
Edited by:
Michael Linnebacher, University Medical Center Rostock, GermanyReviewed by:
Muhammad Jameel, George Washington University, United StatesKatie Lynn Summers, USDA, United States
Copyright © 2023 Labania, Zoughbor, Ajab, Olanda, Shantour and Al Rasbi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zakeya Al Rasbi, cmFzYmlAdWFldS5hYy5hZQ==
†These authors have contributed equally to this work
 Lena Labania
Lena Labania Sumaya Zoughbor
Sumaya Zoughbor Suad Ajab
Suad Ajab Marie Olanda1,2
Marie Olanda1,2 Zakeya Al Rasbi
Zakeya Al Rasbi