- 1Department of Pulmonary and Critical Care Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Key Laboratory of Pulmonary Diseases of National Health Commission, Tongji Hospital, Tongji Medical College, Huazhong University of Sciences and Technology, Wuhan, China
Notch signaling is involved in cell fate determination and deregulated in human solid tumors. Hypoxia is an important feature in many solid tumors, which activates hypoxia-induced factors (HIFs) and their downstream targets to promote tumorigenesis and cancer development. Recently, HIFs have been shown to trigger the Notch signaling pathway in a variety of organisms and tissues. In this review, we focus on the pro- and anti-tumorigenic functions of Notch signaling and discuss the crosstalk between Notch signaling and cellular hypoxic response in cancer pathogenesis, including epithelia-mesenchymal transition, angiogenesis, and the maintenance of cancer stem cells. The pharmacological strategies targeting Notch signaling and hypoxia in cancer are also discussed in this review.
1 Introduction
The discovery of Notch signaling dates back to the early 1900s when a specific Drosophila wing phenotype showed notches on the wings which resulted from the mutations in the Notch receptor. Meanwhile, several other mutations have also been identified, such as Delta and Serrate, which similarly turned out to reside in genes encoding ligands related to the Notch pathway (1). Studies of the Notch signaling have flourished since then and the principal components and process of the signaling transduction cascade were identified. As a juxtacrine signaling, Notch signaling relies on the interaction between receptors and ligands expressed on juxtaposed cells to initiate signaling. The Notch signaling has been extensively characterized as a highly conserved pathway involved in cell proliferation, fate, differentiation, and stem cell maintenance (2). It is universally acknowledged that the normal Notch signaling is vital to most developmental decision-making in animals, and that pathway dysfunction is involved in many conditions, including cancer (3).
The Notch signaling pathway plays a critical role in tumor initiation and progression. Notch can function as an oncogene or a tumor suppressor in different cancers. Hypoxia is a common feature in a majority of malignant tumors. Hypoxia triggers a complex signaling network in tumor cells to alter cell metabolism and regulate angiogenesis, epithelia-mesenchymal transition (EMT), and the maintenance and functions of cancer stem cells (CSCs). Hypoxia-induced factors (HIFs), as global regulators of cellular hypoxia responses, can interact with Notch and directly regulate the Notch signaling pathway. This review systematically summarizes the intersection between Notch signaling and the cellular hypoxic response and highlights the underlying molecular mechanisms involved in the cancer pathogenesis, which contributes to the discovery and development of a combinational strategy targeting Notch and hypoxia in cancer treatment.
2 Notch signaling pathway
Notch signaling exerts its effect in a canonical or noncanonical fashion. The specific mechanisms of canonical and non-canonical Notch signaling are described as follows.
2.1 Canonical Notch signaling
Canonical Notch signaling is initiated by γ-secretase-mediated cleavage of the Notch receptor, resulting in the release of the active intracellular domain of Notch, which migrates to the nucleus and interacts with CSL (for CBF1, Suppressor of Hairless, Lag1; also known as RBPJ), leading to the activation of downstream target genes (Figure 1). In mammals, there are four Notch receptors (Notch 1/2/3/4) and five ligands (Delta-like 1/3/4 or Jagged 1/2). The Notch receptors and ligands are structurally related in some ways. They both contain a large number of epidermal growth factor (EGF)-like repeats in their extracellular domains. Briefly, Notch receptors are produced in the endoplasmic reticulum and synthesized as single precursor proteins, which are then trafficked to the Golgi compartment. In the Golgi compartment, Notch receptor precursors undergo S1 cleavage by a furin-like protease, creating the heterodimeric Notch receptor consisting of a Notch extracellular domain (NECD) and a Notch transmembrane and intracellular domain (TMIC). The part of the extracellular domain of Notch receptor consists of 36 EGF-like repeats and a negative regulatory region. EGF-like repeats 11 and 12 function as specific protein binding domains mediating interaction with ligands (4). The ligand-receptor interaction triggers proteolytic cleavages by an ADAM metalloprotease (S2-cleavage). In this process, ligand will be endocytosed after it binds to Notch receptor. Epsin-dependent ligand endocytosis exerts force on the negative regulatory region exposing the S2 site for cleavage (5). Then, the remainder of the receptor subjected to S3 cleavage by the γ-secretase complex releases the Notch intracellular domain (NICD), which translocates into the nucleus. In the nucleus, NICD interacts with a DNA-binding protein CSL, converting CSL from a transcriptional repressor to an activator. The NICD-CSL interaction is stabilized by Mastermind like transcriptional coactivator (MAML), forming a ternary NICD/MAML/CSL complex to activate the transcription of downstream genes including Hes (hairy-enhancer of split), Hey (Hes related to YRPW), and so on (6, 7). Different ligands could generate diverse Notch activity dynamics in signaling receiving cells, inducing different cell fates via activating distinct target gene programs (8).
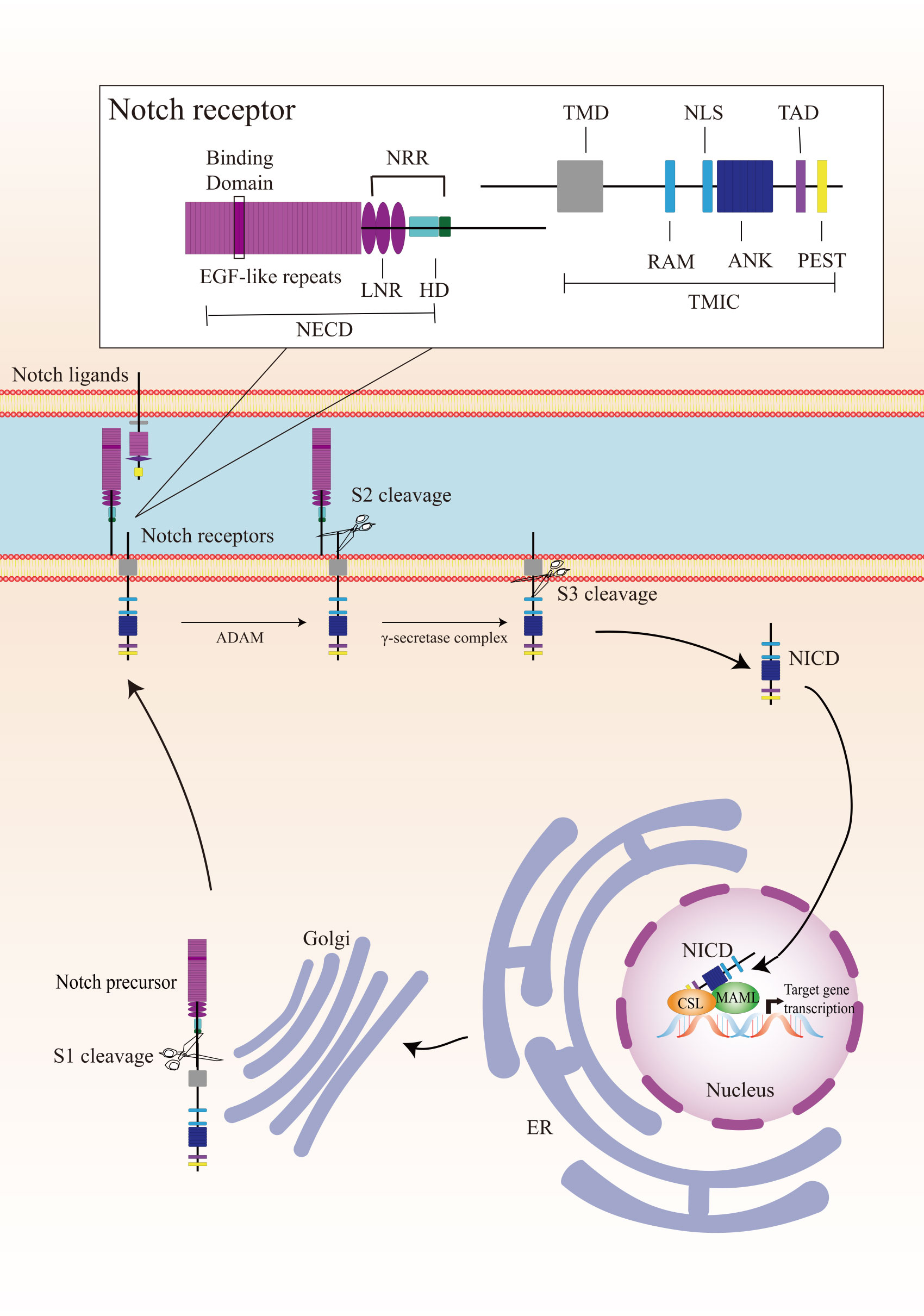
Figure 1 Overview of the Notch signaling pathway. The Notch receptor is produced in the endoplasmic reticulum (ER) and undergoes S1-cleavage in the Golgi compartment. The cleavage results in the formation of a heterodimer receptor, consisting of a Notch extracellular domain (NECD) and a Notch transmembrane and intracellular domain (TMIC), which is then transported to the plasma membrane. Upon interacting with a transmembrane ligand, the Notch receptor undergoes two sequentially cleavage, releases the Notch intracellular domain (NICD), which translocates into the nucleus. In the cell nucleus, NICD forms a ternary complex with the DNA-binding protein CSL and MAML to regulate transcription of downstream genes. A detailed description of the various domains in Notch receptor is presented in the box on the top. Notch receptor consists of a NECD, a transmembrane domain (TMD), and a NICD. NECD consists of epidermal growth factor (EGF) - like repeats domain, and a negative regulatory region (NRR), which including three Lin Notch repeats (LNR) and a heterodimerization (HD) domain. EGF-like repeats 11 and 12 function as specific protein binding domains mediating interaction with ligands. NICD consists of a RBPJ associated molecule (RAM), ankyrin repeats (ANK), a translational active domain (TAD), and a PEST domain.
2.2 Non-canonical Notch signaling
Non-canonical Notch signaling is an important arm of Notch signaling. Notch is proved active in cells where the canonical ligands and downstream effectors were defective, indicating that Notch acts in a second way independently (9). Non-canonical Notch signaling can be initiated by a non-canonical ligand via CSL-independent manner (10–12).
Notch signaling can be elicited by diverse non-canonical ligands, including ligands structurally similar to canonical ligands, structurally unrelated ligands, and secreted proteins (13, 14). Delta like non-canonical Notch ligand 1 is an integral membrane protein containing tandem EGF-like repeats in its extracellular domain but lacking the DSL domain. It can directly interact with Notch1 and act as an antagonist (14). Another structurally similar non-canonical ligand Delta/Notch-like EGF-related receptor functioned as a trans-ligand to affect glial morphological changes (15). A diverse group of structurally unrelated non-canonical ligands have also been identified as Notch activators. F3/contactin1 and NB3/contactin6 interacted with Notch EGF-like repeat distal to the DSL domain binding site to induce oligodendrocyte differentiation (11, 16). In addition, a number of secreted proteins act as non-canonical ligands of Notch. In vertebrates, CCN3 and MAGP-2 can bind to the extracellular domains of Notch receptor, resulting in its cleavage and activation (17, 18). In Drosophila, Scabrous activated transcription of the Notch target gene E(spl)C m3 to regulate eye ommatidia and sensory bristles (19, 20).
In CSL-independent non-canonical Notch signaling, the cleaved NICD interacts with multiple pathways and regulates cell survival. The CSL knockout mice developed breast tumors similar to CSL heterozygous and control mice, indicating that Notch-induced breast tumor development was CSL-independent (21). Interleukin-6 has been identified as a novel Notch target in breast tumor cells. The Notch-mediated interleukin-6 up-regulation required two NF-κB signaling-related proteins and P53 (22). The membrane-tethered NICD inhibited cell apoptosis through interacting with mTOR and Rictor (companion of mTOR) to trigger Akt phosphorylation in activated T cells (23). Notch activated the PI3K-Akt pathway via Deltex1 and played oncogenic functions in cervical cancer (24). In addition, Notch1 was demonstrated to directly regulate vascular barrier function through a flow-mediated, non-canonical, transcription-independent signaling mechanism (25, 26).
3 Notch signaling pathway in cancer
The mutations in the Notch signaling pathway genes and dysregulated Notch signaling pathways exhibit dual biological functions in tumorigenesis and cancer progression (Table 1). Notch1 mutation was first identified in patients with acute T-cell acute lymphoblastic leukemia (T-ALL) and occurs in approximately 50% of T-ALL (27). Oncogenic and gain-of-function mutations of Notch genes have been implicated in chronic lymphocytic leukemia (30), splenic marginal zone B-cell lymphoma (31), squamous cell lung carcinoma (44) and salivary adenoid cystic carcinomas (58). Moreover, aberrant activation of Notch signaling has been found in many solid tumors including prostate (59), breast (60), cervical (61), melanoma (62), and lung cancer (63, 64).
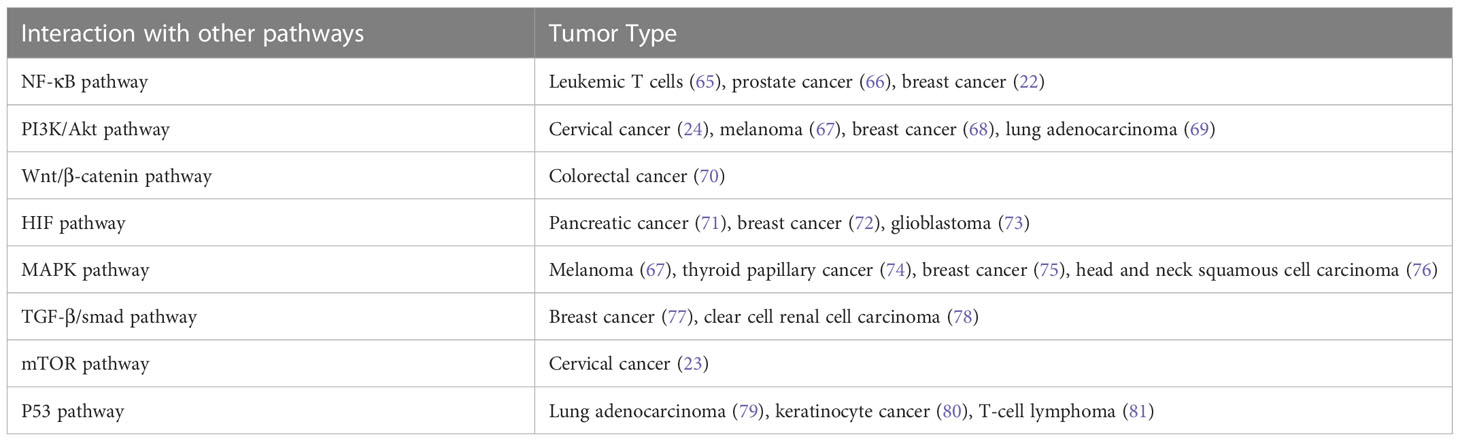
In addition, Notch signaling can interact with other signaling pathways to promote tumorigenesis and cancer progression (Table 2). The Notch signaling contributed to the development of leukemia and breast cancer through interacting with the NF-κB pathway (22, 65, 82). Notch inhibited cervical cancer cell apoptosis via the mTOR–Rictor pathway (23).
In addition to its oncogenic role in human malignancies, Notch also functions as a tumor suppressor (83). Nicolas et al. has demonstrated that Notch1 deficiency in skin resulted in the sustained expression of Gli2 and derepressed β-catenin signaling, causing the development of tumor (84). In addition, Notch was reported to play a suppressive role in B cell ALL (85), human hepatocellular carcinoma (86), small cell lung cancer (41), and neuroendocrine tumors (87). In a word, Notch acts as an oncogene or tumor suppressor in cancer depending on different contexts. To comprehend the full spectrum of Notch effects, efforts were required to identify the specific ligand-receptor interactions, the downstream targets of Notch signaling, and the functions of Notch modifiers (88).
Tumor microenvironment is comprised of a complex network, including stromal cells, immune cells, fibroblasts, blood vessels, and secreted factors (89). The interaction between tumor cells and tumor microenvironment (TME) is interdependent. A normal TME has a potential to suppress tumors. Lim et al. has suggested that tumor-stroma interactions can drive disease progression in squamous cell carcinoma arising in different tissues, indicating that the tumor context defines metastatic progression (90).
Accumulating evidence suggested that Notch signaling plays a role in regulating the immune responses in tumors, which may be associated with the critical role of Notch signaling in hematopoiesis and immune development (88, 91). A single-cell RNA-sequencing analysis has revealed that Jagged1-Notch pathway regulated immune cell homeostasis during minimal residual disease in hematologic neoplasm, which was a potential target to delay tumor recurrence (92). In breast cancer, the Jagged1-Notch pathway regulated tumor-associated macrophage differentiation towards M2 phenotype to induce aromatase inhibitor resistance (93). Activation of the Notch signaling in triple-negative breast cancer resulted in the secretion of pro-inflammatory cytokines and the recruitment of pro-tumoral macrophages to the TME (94). Delta-like 1 (Dll1)-mediated Notch signaling was implicated in the crosstalk between tumor cells and cancer-associated fibroblasts to promote radio-resistance in breast cancer (95). In general, Notch signaling plays a critical role in regulating tumor cells and TME, which may provide new strategies for Notch-targeted cancer therapy.
4 Hypoxia in cancer
Oxygen is indispensable for mammals that maintain intracellular ATP levels and serves as an electron acceptor in a large number of biochemical reactions (96). Hypoxia is a major feature of solid tumor and associated with poor prognosis and resistance to therapy (97–99). Under hypoxic condition, tumor cells undergo various biological processes including cell proliferation, migration, apoptosis, and EMT (100). Hypoxia also triggers multiple signaling pathways to regulate advanced but dysfunctional vascularization in TME (101).
The transcriptional factor HIFs are principal regulators and orchestrate cellular adaptive mechanisms in responses to hypoxia. HIFs contain two different subunits: α and β. The α-subunit protein is regulated by cellular oxygen levels, whereas the β subunit is constitutively expressed (102, 103). HIF-α proteins are oxygen-sensitive that contain an oxygen-dependent degradation domain with target prolyl residues, and a C-terminal transactivation domain which contains the target asparaginyl residue. Under normoxic condition, HIF-α subunits are hydroxylated by prolyl hydroxylases. After hydroxylation, the von-Hippel Lindau tumor suppressor gene interacts with HIF-α and tags it for 26s proteasomal degradation (104, 105). Under hypoxic condition, HIF-α hydroxylation is prevented due to the inactivation of prolyl hydroxylases, resulting in the inhibition of ubiquitin-mediated proteasome degradation of HIF-α. HIF-α is stabilized and form the HIF heterodimer, which then enters the nucleus and combines with hypoxia-response elements to activate the downstream genes (106). Moreover, HIF transcriptional activity is modulated by factor inhibiting HIF-1 (FIH-1), which hydroxylates an asparagine residue in the transactivation domain of HIF-α subunits, thereby blocking its transactivation function (107, 108).
There are three known α subunits (HIF-1α, HIF-2α, and HIF-3α) and three β subunits (HIF-1β, HIF-2β, and HIF-3β). HIF-1α is widely expressed in most human tissues, while HIF-2α and HIF-3α are detected in more restricted tissues, such as lung, kidney, and so on (109, 110). In canonical HIF signaling, hypoxia leads to the stabilization of the labile protein HIF-1α or HIF-2α which complexes with HIF-β, forming heterodimers that bind to hypoxia-response elements in target genes (111). HIF-1α and HIF-2α are structurally closely related and share both common and distinct target genes (112). The role of HIF-3α in the regulation of the HIF pathway is not completely understood and mainly regarded as a negative regulator of HIF-1α and HIF-2α (113).
HIFs are overexpressed and significantly associated with poor prognosis in a variety of cancers (114–117). HIFs-regulated genes encode proteins involved in critical aspects of cancer biology, including energy metabolism, cell survival and invasion, angiogenesis, EMT, and so on. Tumor cells tend to turn metabolism from an oxygen-dependent tricarboxylic acid cycle to glycolysis (118). HIF-1 regulates glycolytic enzymes, including hexokinase 2 and phosphofructokinase 1, which involved in tumor initiation and growth (119, 120). A number of growth factors regulated by HIFs played a role in cell survival, such as transforming growth factor-β, insulin-like growth factor 2, endothelin-1, erythropoietin, and epidermal growth factor receptor (100, 121–123). HIFs mediated angiogenesis via activating the transcription of multiple angiogenic growth factors, including vascular endothelial growth factor (VEGF), placenta-like growth factor, angiopoietin (124, 125). HIF-1 can directly induces the transcription of ZEB1, TWIST, and TCF3, which promote EMT in cancers (126–128). In a word, HIFs play a key role in cancer initiation and progression.
5 Crosstalk between Notch signaling and hypoxia pathway
HIF signaling pathway is the primary regulator in the physiological and pathological response to hypoxia. The Notch signaling pathway plays a critical role in cell fate control, including tumorigenesis and progression. The link between Notch signaling and hypoxia was first described in a transcriptomic analysis, in which the Notch target gene Hes1 was upregulated in hypoxic neuroblastoma cell lines (129). Thereafter, a study of Notch and hypoxia-activated genes in glioblastoma tumor confirmed a combined gene signature of these two pathways and their role in tumor prognosis (130). Gustaffson et al. provided important evidence that hypoxia directly regulated Notch signaling (131). In this study, HIF-1α was recruited to Notch-responsive promoters and interacted with NICD, leading to stabilization of NICD and activation of Notch downstream genes (Hes and Hey). HIF-1α can also be recruited to the Hey-2 promoter in myogenic cell (131). The up-regulation of the Notch ligands (Jagged 2 and Delta-like 4) induced by hypoxia leaded to activation of Notch signaling (132–134). HIF-2α promoted stem phenotype conversion and resistance to Paclitaxel by activating Notch and Wnt pathways in breast cancer cells (72). Besides, HIF-1α was revealed to interact with γ-secretase and upregulate γ-secretase activity to promote cell invasion and metastasis through a novel function independent of transcription factor (135). HIF-1α and HIF-2α synergized with the Notch co-activator MAML1 to potentiate Notch activity in breast cancer cells (136). The indirect regulation of Notch signaling by HIF was reported in lung cancer cells that HIF-mediated miR-1275 up-regulation exerted its tumorigenic effect through co-activating Notch and Wnt/β-catenin signaling pathways (137).
On the other hand, Notch signaling can also regulate hypoxic response. Notch was demonstrated to transcriptionally upregulate the expression of HIF-2α in certain tumor cells via a HIF1α-to-HIF2α switch (138). The γ-secretase inhibitor of Notch decreased the mRNA expression of the HIF-1 target PGK-1 (131).
FIH-1 is involved in the crosstalk between hypoxia and Notch signaling pathways. Both HIF-1α and Notch are substrates for the asparagine hydroxylase FIH-1. Two asparagine residues in the NICD ankyrin repeat domain are hydroxylated by FIH-1, leading to inactivation of Notch signaling. FIH-1 binds to NICD more efficiently than HIF-1α, indicating that NICD sequesters FIH-1 away from HIF-1α, which results in an under-hydroxylation on HIF1α (139, 140). This may shed light on another oxygen-dependent interface that modulates HIF signaling.
To summarize, the crosstalk between Notch signaling and the cellular hypoxic response is extensive and the underlying molecular mechanism is complex (Figure 2). A Notch-hypoxia crosstalk has been involved in a variety of physiological situations and pathological conditions, including vascular diseases and cancers (64, 141).
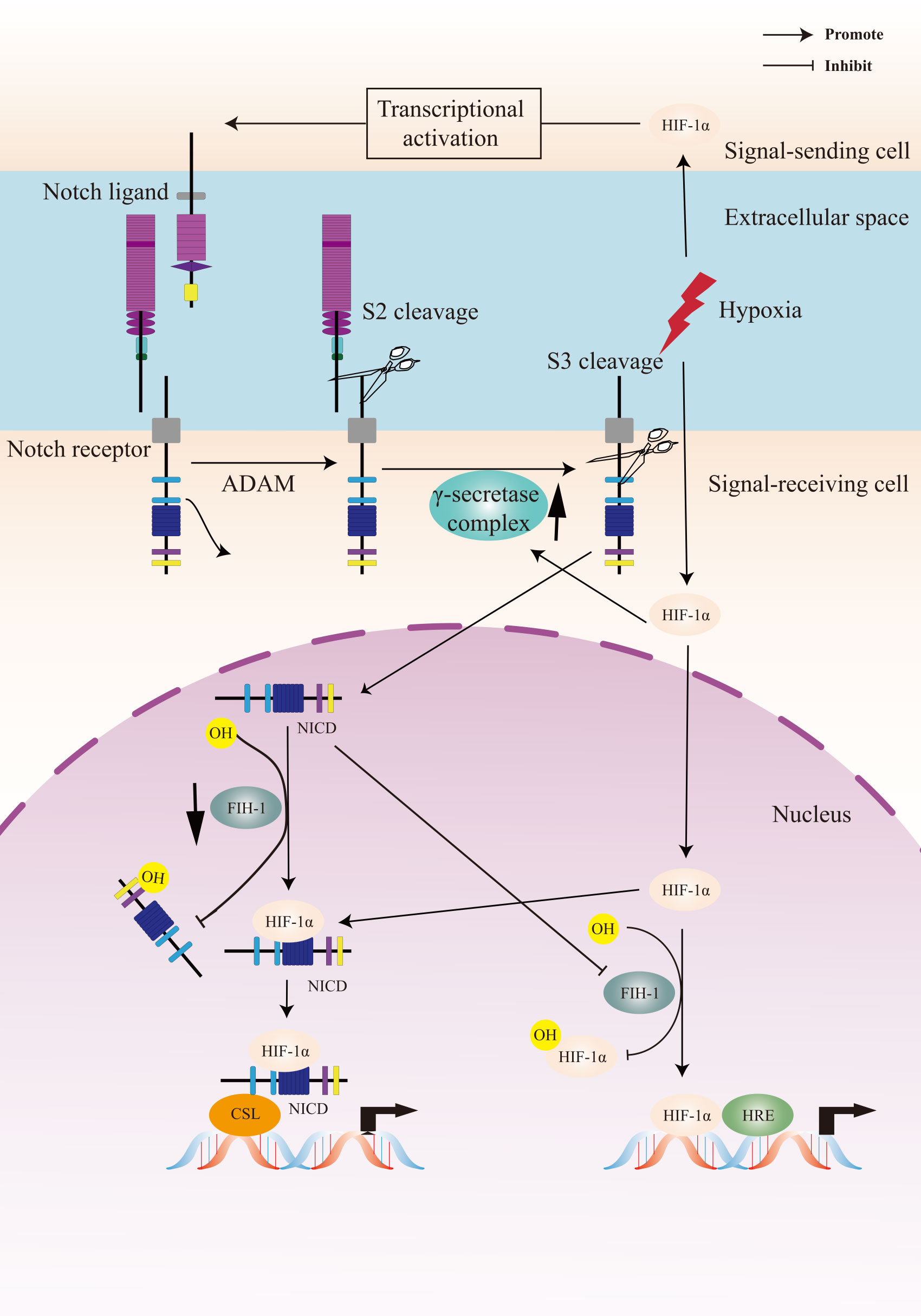
Figure 2 A Crosstalk between Notch signaling and hypoxia pathway. Upon activation of the Notch receptor, the Notch intracellular domain (NICD) accumulates in the cell nucleus and activates target genes. Hypoxia induces the canonical hypoxia response pathway, which involves the activation of hypoxia response element (HRE)-driven target genes. Under hypoxic conditions, hypoxia-induced factors-1α (HIF-1α) potentiates Notch-dependent activation of target genes through interaction with the NICD. Besides, HIF-1α interacts with γ-secretase and upregulated γ-secretase activity. Factor-inhibiting HIF-1 (FIH-1) hydroxylates the asparagine residues of HIF-α and NICD, leading to inactivation of Notch and hypoxia signaling pathways. Hypoxia decreases the activity of FIH-1. In addition, FIH-1 binds NICD more efficiently than HIF-1α. NICD sequesters FIH-1 away from HIF-1α, indirectly resulting in an activation of HRE-driven target genes.
6 Biological processes in cancer regulated by a Notch-hypoxia crosstalk
A functional relationship between hypoxia and Notch signaling pathways has been observed in many types of tumors. Accumulating evidences have revealed that the crosstalk between Notch and the cellular hypoxic response has diverse roles in cancer pathogenesis by regulating several important biological processes, including EMT, angiogenesis, the maintenance of CSCs, and so on.
6.1 A Notch-hypoxia crosstalk in cancer EMT
EMT is one of the critical mechanisms of cancer metastasis (142, 143). The hallmark of EMT is the loss of E-cadherin expression through the up-regulation of its repressors (144, 145). E-cadherin repressors are classified into two groups depending on their effects on the E-cadherin promoter. Snail, Zeb, E47, and KLF8 bind to and repress the activity of the E-cadherin promoter (146, 147), whereas several factors such as Twist, Goosecoid, E2.2, and FoxC2 indirectly repress E-cadherin transcription (148).
HIF-1 was reported to upregulate the expression of Twist to promote EMT (149). A number of studies suggested that hypoxia induced EMT via activating Notch signaling in tumor cells (136, 150–152). Notch can regulate the expression of Snail-1 via two distinct mechanisms in hypoxia. One relied on the transcriptional up-regulation of Snail-1. The other concerned the protein stabilization of Snail-1 via the increase of lysyl oxidase which was transcriptionally regulated by HIF-1α and potentiated by Notch (150). Hypoxia-mediated increase in Snail and Slug required Notch pathway in the initiation of EMT in breast cancer cells (136). HIF-1α can also exert a non-transcriptional function in regulating the expression of NICD and E-cadherin in lung cancer cells (153).
6.2 A Notch-hypoxia crosstalk in angiogenesis
Tumor growth is fed by nearby blood vessels. Hypoxia occurs as the tumor grows. New blood vessels are essential for continued primary tumor growth. The ability of forming vasculature has been termed angiogenesis. Activation of endothelial cells was a key step of angiogenesis and a number of growth factors upregulated by HIF were involved in the process, such as VEGF (154).
Notch signaling was activated and played an important role in the process of angiogenesis (155). The expression of Notch ligand Dll4 was much higher in the endothelium of tumor blood vessels compared to nearby normal blood vessels, indicating that Notch signaling were implicated in tumor angiogenesis (132, 156, 157). Dll4 was upregulated by VEGF as a negative feedback modulator, which prevented VEGF-induced overexuberant angiogenic sprouting and branching via Notch signaling, guaranteeing the formation of a well-differentiated vascular network (158, 159). HIF1α-induced basic fibroblast growth factor and VEGF were reported to play a synergistic role in the regulation of Dll4 in tumor cells (156). Hypoxia-induced up-regulation of Dll4 and Hey repressed COUP-TFII (known as a regulator of vein identity) in endothelial progenitor cells, which may contribute to tumor angiogenesis (160). Another Notch ligand Jagged 2 was transcriptionally activated by HIF-1α, which triggered Notch signaling and activated Hey1 to promote vascular development and angiogenesis (133).
6.3 A Notch-hypoxia crosstalk in the maintenance of CSCs
CSCs represent a discrete subpopulation of cancer cells with stem cell properties, which is responsible for tumor growth. CSCs are self-renewal and can produce more committed progenitor or “transit-amplifying” cells whose progeny differentiate aberrantly to promote the tumorigenesis (161, 162). Stem cell “niches” are considered as particular microenvironments that maintain the combined properties of CSCs self-renewal and multipotency. The Notch signaling is highly conserved and is critical for cell fate decisions and the maintenance of stem cells (163). HIF stabilization in hypoxic tumor cells can promote stem cell properties, including self-renewal and multipotency partly via inducing the expression and activity of the Notch signaling pathway (164–167). Hypoxia-induced the 66-kDa isoform of the SHC gene controlled the expression of Notch3 to regulate the stem cell properties (168). In glioblastomas, HIF-1α played an important role in the hypoxia-mediated maintenance of glioma stem cells via the interaction with NICD (73). A further study suggested that hypoxia can promote glioma stem cells proliferation and maintain the characteristics of stem cells through activating Notch1 and Oct3/4 (169). In addition, HIF-1α was reported to promote pancreatic cancer cell dedifferentiation into stem-like cell phenotypes by activating Notch signaling, revealing a novel regulatory mechanism (71).
7 Strategies for cancer therapy
7.1 Therapeutic targets in the Notch signaling pathway
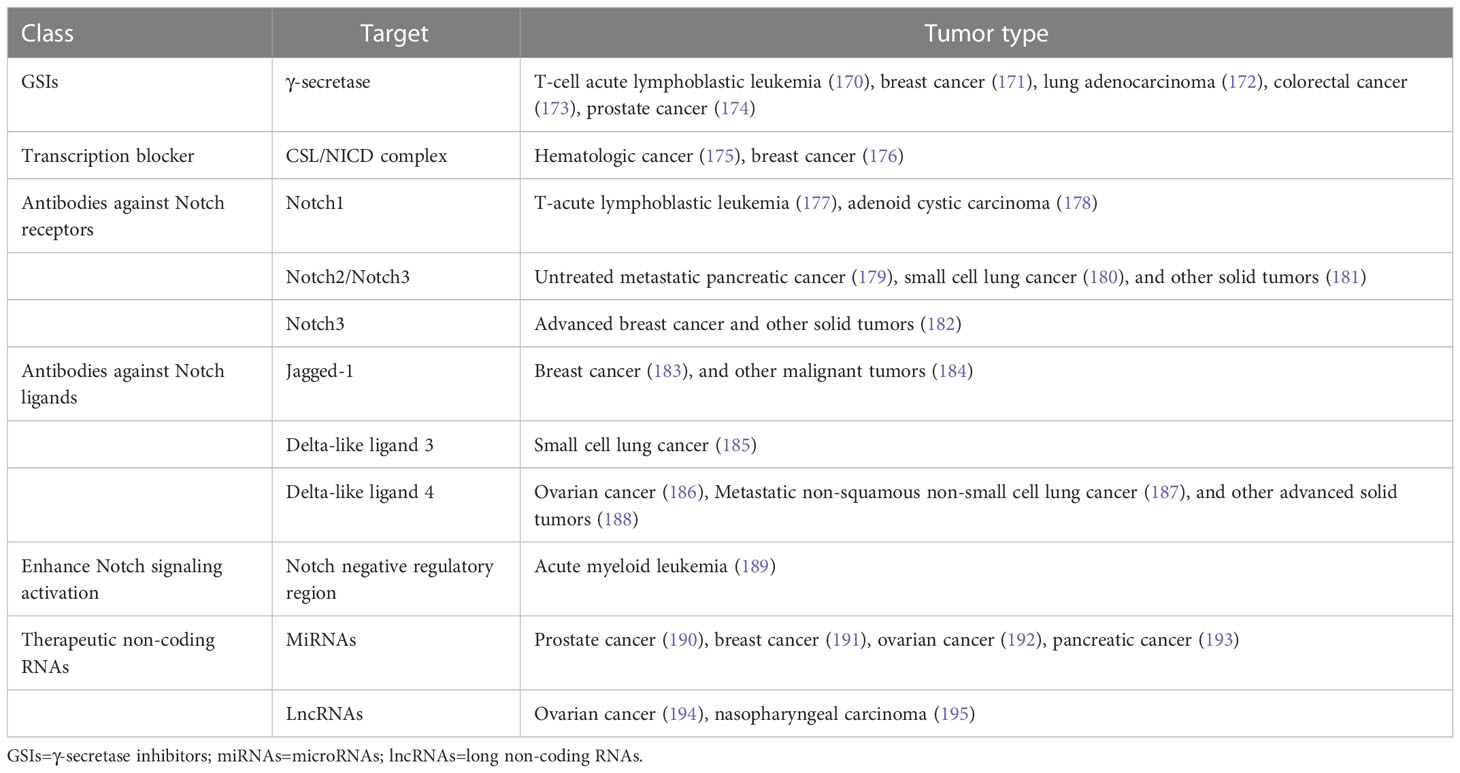
In view of the critical role of Notch signaling in tumor pathogenesis, Notch is regarded as a promising therapeutic target. Numerous approaches have been developed to inhibit different steps of Notch signaling pathway for therapy: γ-secretase inhibitors (GSIs), antibodies targeting ligands or receptors, compounds targeting transcription activation, and so on (Figure 3). The drugs are listed by therapeutic category in Table 3.
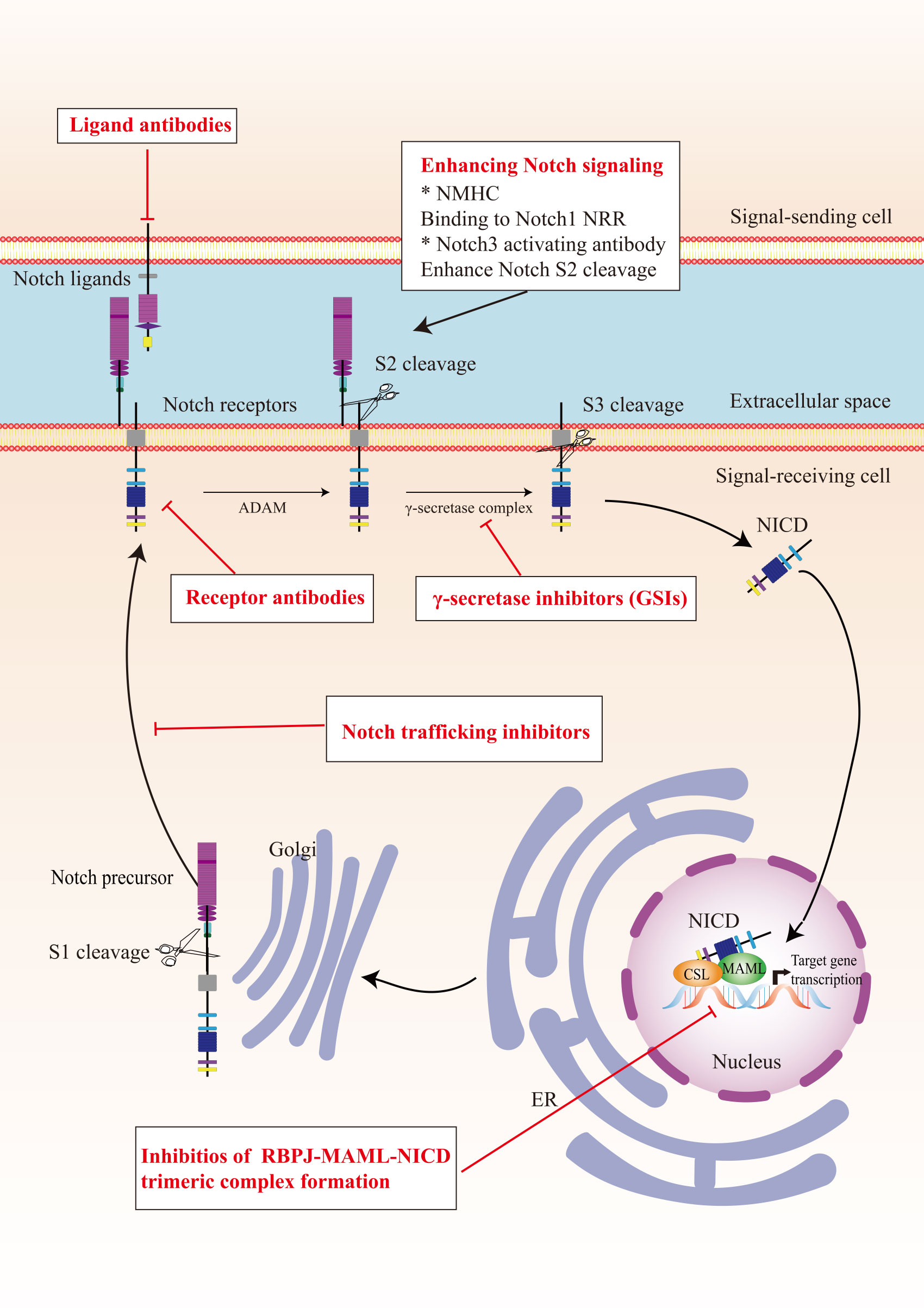
Figure 3 The potential therapeutics targeting Notch signaling pathway. Here are several strategies to modulate Notch signaling pathway: (I) inhibitors of Notch pre-processing, (II) receptor and ligand antibodies blocking ligand-receptor interaction, (III) inhibitors of the trimeric transcriptional complex assembly, (IV) molecules activating Notch signaling. ER, endoplasmic reticulum; NICD, Notch intracellular domain; NRR, negative regulatory region; NMHC, N-methylhemeanthidine chloride; MAML, Mastermind like transcriptional coactivator; GSIs, γ-secretase inhibitors.
GSIs were the first and most extensively studied small-molecule Notch inhibitors. Initially, GSIs were developed for treating Alzheimer’s disease because γ-secretase catalyzed the production of the β-amyloid peptide from amyloid precursor protein (196). The use of GSIs for cancer treatment is based on inhibiting the cleavage of γ-secretase which mediates S3 cleavage to generate NICD, resulting in blocking Notch signaling. However, studies have shown that systemic inhibition of Notch signaling by GSIs results in “on-target” gastrointestinal toxicity because of the accumulation of secretory goblet cells in the intestine. The above observation can be explained by alterations in the differentiation of intestinal stem cells following the dual inhibition of Notch1 and Notch2 (197). Co-administration of glucocorticoid may alleviate the toxicity through inducing transcriptional up-regulation of cyclin D2 and protecting mice from developing the GSIs-induced intestinal goblet cell metaplasia in a preclinical mouse model of T-ALL (198).
Considering the inherent mechanism-based toxicity caused by pan-Notch inhibitor GSIs, novel inhibitors that selectively target individual Notch ligands and receptors have been developed. Selective blocking of Notch1 signaling inhibited cancer cell growth and deregulation of angiogenesis (199). The antibodies against Notch receptors are divided into two classes, one directed against the EGF-like repeat region and the other directed against the Notch negative regulatory region (200). Several potent and selective inhibitors against Notch1, Notch2, and Notch3 have been developed (199, 201, 202). However, there is a lack of inhibitor against Notch 4. The antibodies that selectively target the canonical ligands have also been investigated, such as Jagged antagonism (203).
In the past decades, several molecules targeting Notch trafficking and processing have been developed. The dihydropyridine FLI-06 as the first small molecular chemical compound functioned at an early stage in secretory traffic through disrupting the Golgi apparatus and inhibiting general secretion before exiting from the endoplasmic reticulum (204). FLI-06 was also demonstrated to block Notch activation and decrease the self-renewal ability of tongue CSCs (205). In addition, direct inhibition of the CSL/NICD complex has been reported to treat cancers. SAHM1, as a high-affinity binding of the hydrocarbon-stapled peptide, could prevent the assembly of the active transcriptional complex, resulting in genome-wide suppression of Notch-activated genes for the treatment of leukemia (206). There are other small molecules inhibiting the transcriptional activation complex, which have been investigated, such as IMR-1, CB-103, and RIN1 (175, 176, 207). However, given that loss of CSL derepressed target gene promoter and promoted tumorigenesis, targeting CSL may bring potential problems (208).
As mentioned above, Notch can act as a tumor suppressor in specific contexts, thus enhancing Notch signaling activation is a potential therapeutic strategy for cancer. A study demonstrated that N-methylhemeanthidine chloride, a novel Amaryllidaceae alkaloid, activated the Notch signaling via docking in the hydrophobic cavity within the Notch1 negative regulatory region and promoting Notch1 proteolytic cleavage (189). A monoclonal antibody was reported to enhance Notch3 cleavage and mimic the effects of ligand-induced Notch activation via binding to overlapping epitopes within negative regulatory region (202).
Accumulating evidence demonstrated that the non-coding RNAs’ (ncRNAs) played a critical role in cancer therapy. NcRNAs are a class of RNAs including microRNAs (miRNAs) and long ncRNAs (lncRNAs) and other short ncRNAs. miRNAs and lncRNAs regulated cell fate determination via various signaling pathways (209). miRNA-34 was reported to suppress Notch1 expression, inducing ovarian cancer cell death (210). In contrast, miRNA-223 as an oncogene activated Notch signaling to induce tumor cell proliferation in colorectal cancer (211). The versatility is one of the advantages of miRNA therapeutics, which can suppress or mimic the activity of a miRNA. However, the delivery of miRNA remains an important challenge. LncRNAs mostly act as oncogenes in cancers. LncRNAs can interact with Notch or act as competing endogenous RNAs for miRNAs to indirectly induce Notch signaling in various cancers (212–214). Besides, other therapeutics targeting Notch are currently under investigation, such as natural products, virotherapy, and so on.
7.2 Hypoxia targeting strategies
Considering the critical role of hypoxia in tumor initiation, progression and therapy resistance, a growing number of preclinical and clinical cancer studies targeting hypoxia have been performed. In general, the strategies can be classified into hypoxia activated prodrugs (HAPs) and pharmacological inhibitors of the HIF signaling pathway.
7.2.1 Hypoxia activated prodrugs
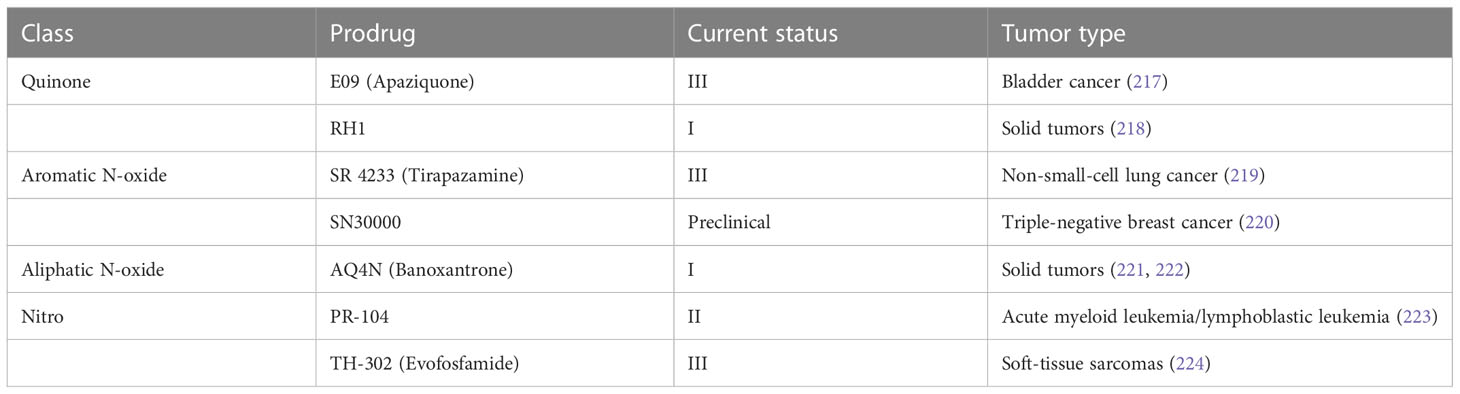
HAPs are bioreductive drugs which are reduced by specific reductases under hypoxic conditions and release cytotoxins to kill cells (215). Five different chemical entities have the potential to target hypoxia based on their enzymatical reductive reaction under hypoxic conditions (216), including nitro groups, quinones, aromatic N-oxides, aliphatic N-oxides and transition metals. To date, several HAPs have been developed, including EO9 (apaziquone), RH1, SR 4233 (tirapazamine), SN30000, AQ4N (banoxantrone), PR-104, and TH-302 (evofosfamide) (Table 4). The effects of HAPs are different depending on the degree of hypoxia and the activity of reductase enzymes. The selection of the appropriate agents in different patients is dependent on the clinical context and requires predictive biomarkers (225).
7.2.2 Inhibitors of HIF signaling
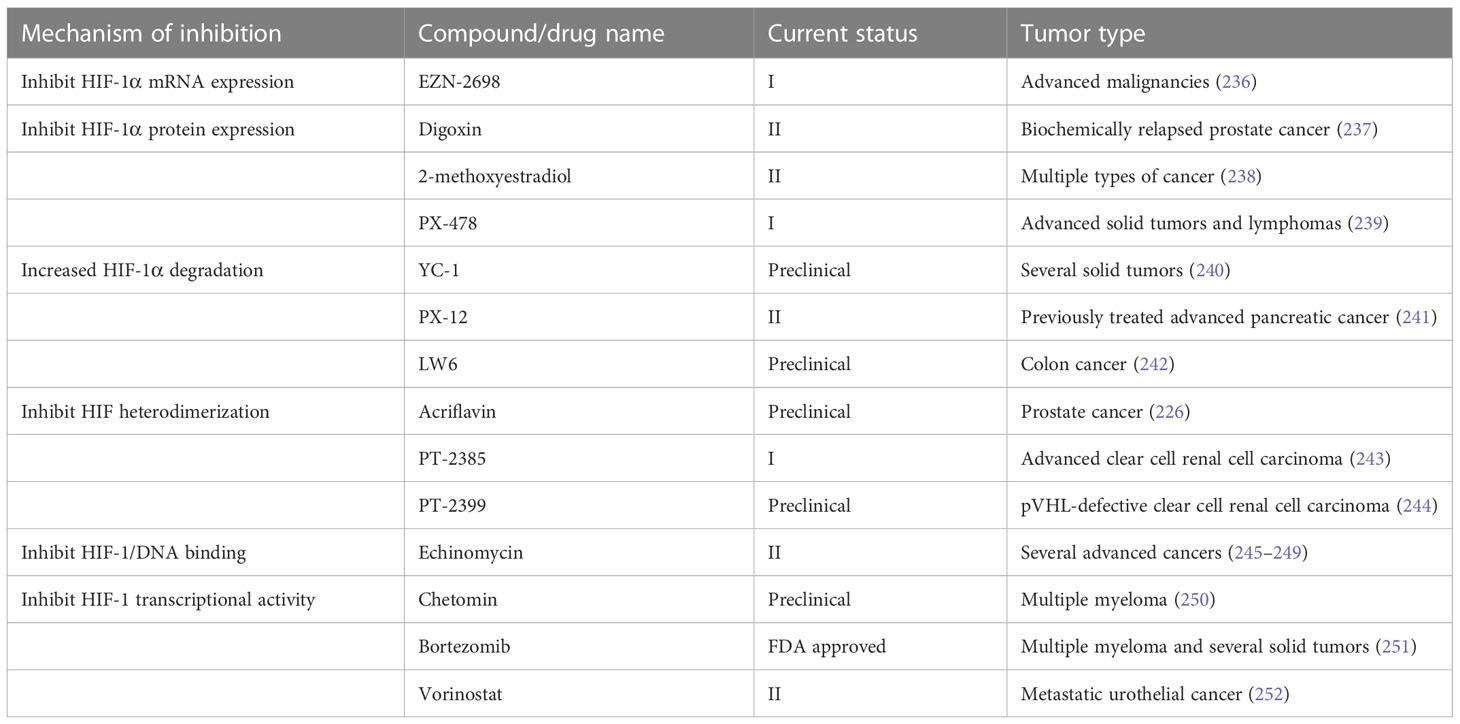
HIF signaling is an attractive target for cancer treatment. Several inhibitors have been developed to directly bind to HIF-1α or HIF-2α, resulting in inhibition of their heterodimerization with HIF-β, such as acriflavine (226), PT2385 (227) and PT2399 (228). Heat shock protein 90 (Hsp90) can bind to HIF-1α and block the VHL-dependent proteasomal degradation of HIF-1α. A number of Hsp90 inhibitors have been developed during the past two decades. Hsp90 was identified as the biological target of the ansamycin class of natural products and derivatives, which has been extensively studied in cancer treatment (229). Hsp90 inhibitors apigenin and radicicol reduced hypoxia-induced VEGF expression to decrease angiogenesis (230, 231). Hsp90 can also modulate the conformation of the HIF-1 heterodimer, increasing its interaction with hypoxia-responsive elements, inducing HIF-1 transcriptional activity (231). Hsp90 can be regulated by posttranslational modifications, including acetylation. The process of histone acetylation is regulated by opposing activities of histone acetyltransferases and histone deacetylases (HDACs). HDAC6 functions as an Hsp90 deacetylase (232). HDAC inhibitor vorinostat was developed to inhibit HIF-1 transcriptional activity via direct Hsp90 acetylation, decreasing Hsp90-HIF-1 affinity and the interaction between HIF and hypoxia-responsive elements (233). Chetomin, a small molecule blocking the transcriptional co-activation of HIF-1 pathway, was evaluated as a promising candidate treatment for several types of cancers (234). Paradoxically, the stabilization of HIF-1α through inhibition of prolyl hydroxylase domain-containing protein 2 has antitumor effects in certain context. The loss of EGLN1 which encodes prolyl hydroxylase domain-containing protein 2 inhibited the proliferation of clear cell ovarian cancer cells (235). In general, anti-HIF agents are classified by different molecular mechanisms, including inhibition of HIF protein synthesis, degradation, and transcriptional activity. A detailed review of experimental chemical compounds and approved drugs directly targeting HIF pathway are presented in Table 5.
Targeting HIF signaling can be performed via interfering with other signaling pathways. PI3K/AKT/mTOR and MAPK/ERK pathways can increase HIF-1α synthesis in a cell type-specific manner (253). PI3K inhibitors LY294002 and wortmannin have been recognized as the synthesis inhibition of HIF-1α protein in the prostate carcinoma-derived cell lines PC-3 and DU145 (254). Temsirolimus, everolimus, and sirolimus as mTOR inhibitors are currently in clinical development for the treatment of solid tumors (255). The phase III clinical trials for temsirolimus and everolimus have been completed and showed a significant gain in survival for patients of metastatic renal cell carcinoma (256).
7.3 Combination therapy
Combination therapy is an important trend in the development of anticancer agents, and targeting hypoxia is critical in the new strategy (225). The anti-hypoxia agents were combined with immune checkpoint inhibitors to enhance the effect of immune checkpoint inhibitors in cancer treatment, which was based on hypoxia-induced expression and activity of immune checkpoints and immune checkpoint ligands on immune-cells and tumor cells (257). A phase II clinical trial of pembrolizumab and HDAC inhibitor vorinostat demonstrated the combination was active for patients with recurrent/metastatic squamous cell carcinomas of the head and neck, and salivary gland cancer (258). In a neuroblastoma xenograft model, the combination of anti-angiogenic drug sunitinib with hypoxia-activated prodrug evofosfamide was demonstrated to improve survival of mice (259).
Hypoxia and cellular interaction between tumor and non-tumor cells are two important TME. There are strong links between these two themes, and hypoxia contributes to TME to adversely affect therapeutic outcomes. Notch signaling plays an important role in regulating the crosstalk between the different compartments of the TME. Therefore, a combination of targeting Notch and hypoxia implies a potential treatment strategy of cancer to alter TME. In addition, hypoxia and Notch signaling have been shown to form a complex web of interaction in cancer, providing new insights into the combination therapeutics. Notch is a key regulator of tumor angiogenesis (260). The anti-angiogenesis drugs aggravated tumor hypoxia (261), indicating that targeting Notch may induce hypoxia. While, hypoxia activated Notch signaling pathway and may reduce the effect of Notch signaling inhibitors. Therefore, the combination of anti-hypoxia and Notch-targeted agents may present a new strategy for addressing the adverse effect of hypoxia.
8 Conclusion
The Notch signaling, as an evolutionarily conserved pathway, is usually activated and extensively involved in tumor initiation and progression. Notch signaling plays a critical role in the interaction between the tumor cells and the surrounding TME, acting as an oncogene or a tumor suppressor. Hypoxia is recognized as a hallmark of TME and the HIF pathway is a master regulator of the cellular hypoxic response. The interaction of Notch and HIF pathways played a key role in multiple biological processes in hypoxic tumor, including EMT, angiogenesis, and the maintenance of CSCs. A broad spectrum of anti-hypoxia agents and Notch signaling inhibitors have been developed during the past decades. The combination therapy has been an important trend of cancer treatment. Considering the complex web of hypoxia and Notch signaling, the combination of them implies a potential treatment strategy of cancer.
Author contributions
XCL and MG conceived the study. MG drafted the manuscript. XCL revised the manuscript critically for important intellectual content. YN, MX, and XSL provided important comments on the manuscript. All authors approved the final version of the manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. All authors contributed to the article and approved the submitted version. The reviewer BZ declared a shared affiliation with the authors to the handling editor at the time of review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HIFs, Hypoxia-induced factors; EMT, Epithelial-mesenchymal transition; CSCs, Cancer stem cells; EGF, Epidermal growth factor; NICD, Notch intracellular domain; MAML, Mastermind like transcriptional coactivator; ALL, Acute lymphoblastic leukemia; TME, Tumor microenvironment; FIH-1, Factor inhibiting HIF-1; VEGF, Vascular endothelial growth factor; Dll4, Delta-like 4; GSIs, γ-secretase inhibitors; NcRNAs, Non-coding RNAs; MiRNAs, MicroRNAs; LncRNAs, Long non-coding RNAs; HAPs, Hypoxia activated prodrugs; Hsp90, Heat shock protein 90; HDACs, Histone deacetylases.
References
1. Mugisha S, Di X, Disoma C, Jiang H, Zhang S. Fringe family genes and their modulation of notch signaling in cancer. Biochim Biophys Acta Rev Cancer (2022) 1877(4):188746. doi: 10.1016/j.bbcan.2022.188746
2. Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev (2017) 97(4):1235–94. doi: 10.1152/physrev.00005.2017
3. Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol (2010) 92:1–29. doi: 10.1016/s0070-2153(10)92001-2
4. Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of notch mediate interactions with delta and serrate: implications for notch as a multifunctional receptor. Cell (1991) 67(4):687–99. doi: 10.1016/0092-8674(91)90064-6
5. Langridge PD, Struhl G. Epsin-dependent ligand endocytosis activates notch by force. Cell (2017) 171(6):1383–96.e12. doi: 10.1016/j.cell.2017.10.048
6. Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol (2016) 17(11):722–35. doi: 10.1038/nrm.2016.94
7. Sprinzak D, Blacklow SC. Biophysics of notch signaling. Annu Rev Biophys (2021) 50:157–89. doi: 10.1146/annurev-biophys-101920-082204
8. Nandagopal N, Santat LA, LeBon L, Sprinzak D, Bronner ME, Elowitz MB. Dynamic ligand discrimination in the notch signaling pathway. Cell (2018) 172(4):869–80.e19. doi: 10.1016/j.cell.2018.01.002
9. Rusconi JC, Corbin V. Evidence for a novel notch pathway required for muscle precursor selection in drosophila. Mech Dev (1998) 79(1-2):39–50. doi: 10.1016/s0925-4773(98)00170-1
10. Ayaz F, Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol (2014) 4:345. doi: 10.3389/fonc.2014.00345
11. Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, et al. F3/contactin acts as a functional ligand for notch during oligodendrocyte maturation. Cell (2003) 115(2):163–75. doi: 10.1016/s0092-8674(03)00810-9
12. Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J (2006) 25(1):129–38. doi: 10.1038/sj.emboj.7600902
13. D'Souza B, Miyamoto A, Weinmaster G. The many facets of notch ligands. Oncogene (2008) 27(38):5148–67. doi: 10.1038/onc.2008.229
14. Traustadóttir G, Jensen CH, Thomassen M, Beck HC, Mortensen SB, Laborda J, et al. Evidence of non-canonical NOTCH signaling: Delta-like 1 homolog (DLK1) directly interacts with the NOTCH1 receptor in mammals. Cell Signal (2016) 28(4):246–54. doi: 10.1016/j.cellsig.2016.01.003
15. Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, et al. DNER acts as a neuron-specific notch ligand during bergmann glial development. Nat Neurosci (2005) 8(7):873–80. doi: 10.1038/nn1492
16. Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, et al. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem (2004) 279(24):25858–65. doi: 10.1074/jbc.M313505200
17. Heath E, Tahri D, Andermarcher E, Schofield P, Fleming S, Boulter CA. Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene. BMC Dev Biol (2008) 8:18. doi: 10.1186/1471-213x-8-18
18. Miyamoto A, Lau R, Hein PW, Shipley JM, Weinmaster G. Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J Biol Chem (2006) 281(15):10089–97. doi: 10.1074/jbc.M600298200
19. Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science (1990) 250(4986):1370–7. doi: 10.1126/science.2175046
20. Powell PA, Wesley C, Spencer S, Cagan RL. Scabrous complexes with notch to mediate boundary formation. Nature (2001) 409(6820):626–30. doi: 10.1038/35054566
21. Raafat A, Lawson S, Bargo S, Klauzinska M, Strizzi L, Goldhar AS, et al. Rbpj conditional knockout reveals distinct functions of Notch4/Int3 in mammary gland development and tumorigenesis. Oncogene (2009) 28(2):219–30. doi: 10.1038/onc.2008.379
22. Jin S, Mutvei AP, Chivukula IV, Andersson ER, Ramsköld D, Sandberg R, et al. Non-canonical notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene (2013) 32(41):4892–902. doi: 10.1038/onc.2012.517
23. Perumalsamy LR, Nagala M, Banerjee P, Sarin A. A hierarchical cascade activated by non-canonical notch signaling and the mTOR-rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ (2009) 16(6):879–89. doi: 10.1038/cdd.2009.20
24. Veeraraghavalu K, Subbaiah VK, Srivastava S, Chakrabarti O, Syal R, Krishna S. Complementation of human papillomavirus type 16 E6 and E7 by Jagged1-specific Notch1-phosphatidylinositol 3-kinase signaling involves pleiotropic oncogenic functions independent of CBF1;Su(H);Lag-1 activation. J Virol (2005) 79(12):7889–98. doi: 10.1128/jvi.79.12.7889-7898.2005
25. Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, et al. A non-canonical notch complex regulates adherens junctions and vascular barrier function. Nature (2017) 552(7684):258–62. doi: 10.1038/nature24998
26. Fischer A, Braga VMM. Vascular permeability: Flow-mediated, non-canonical notch signalling promotes barrier integrity. Curr Biol (2018) 28(3):R119–r121. doi: 10.1016/j.cub.2017.11.065
27. Weng AP, Ferrando AA, Lee W, JPt M, LB S, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science (2004) 306(5694):269–71. doi: 10.1126/science.1102160
28. Bernasconi-Elias P, Hu T, Jenkins D, Firestone B, Gans S, Kurth E, et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene (2016) 35(47):6077–86. doi: 10.1038/onc.2016.133
29. King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell (2013) 153(7):1552–66. doi: 10.1016/j.cell.2013.05.041
30. Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med (2011) 208(7):1389–401. doi: 10.1084/jem.20110921
31. Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med (2012) 209(9):1553–65. doi: 10.1084/jem.20120910
32. Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large b-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U.S.A. (2012) 109(10):3879–84. doi: 10.1073/pnas.1121343109
33. Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large b-cell lymphoma. Cancer Sci (2009) 100(5):920–6. doi: 10.1111/j.1349-7006.2009.01130.x
34. Xie M, Wei S, Wu X, Li X, You Y, He C. Alterations of notch pathway in patients with adenoid cystic carcinoma of the trachea and its impact on survival. Lung Cancer (2018) 121:41–7. doi: 10.1016/j.lungcan.2018.04.020
35. Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest (2013) 123(7):2965–8. doi: 10.1172/jci67201
36. Pappas K, Martin TC, Wolfe AL, Nguyen CB, Su T, Jin J, et al. NOTCH and EZH2 collaborate to repress PTEN expression in breast cancer. Commun Biol (2021) 4(1):312. doi: 10.1038/s42003-021-01825-8
37. Wu D, Wang S, Oliveira DV, Del Gaudio F, Vanlandewijck M, Lebouvier T, et al. The infantile myofibromatosis NOTCH3 L1519P mutation leads to hyperactivated ligand-independent notch signaling and increased PDGFRB expression. Dis Model Mech (2021) 14(2):dmm046300. doi: 10.1242/dmm.046300
38. Mosquera JM, Sboner A, Zhang L, Chen CL, Sung YS, Chen HW, et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosomes Cancer (2013) 52(11):1075–87. doi: 10.1002/gcc.22102
39. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (2011) 333(6046):1154–7. doi: 10.1126/science.1206923
40. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (2011) 333(6046):1157–60. doi: 10.1126/science.1208130
41. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature (2015) 524(7563):47–53. doi: 10.1038/nature14664
42. Maraver A, Fernandez-Marcos PJ, Cash TP, Mendez-Pertuz M, Dueñas M, Maietta P, et al. NOTCH pathway inactivation promotes bladder cancer progression. J Clin Invest (2015) 125(2):824–30. doi: 10.1172/jci78185
43. Rampias T, Vgenopoulou P, Avgeris M, Polyzos A, Stravodimos K, Valavanis C, et al. A new tumor suppressor role for the notch pathway in bladder cancer. Nat Med (2014) 20(10):1199–205. doi: 10.1038/nm.3678
44. Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U.S.A. (2011) 108(43):17761–6. doi: 10.1073/pnas.1114669108
45. Zender S, Nickeleit I, Wuestefeld T, Sörensen I, Dauch D, Bozko P, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell (2013) 23(6):784–95. doi: 10.1016/j.ccr.2013.04.019
46. Gramantieri L, Giovannini C, Lanzi A, Chieco P, Ravaioli M, Venturi A, et al. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int (2007) 27(7):997–1007. doi: 10.1111/j.1478-3231.2007.01544.x
47. Ning L, Wentworth L, Chen H, Weber SM. Down-regulation of Notch1 signaling inhibits tumor growth in human hepatocellular carcinoma. Am J Transl Res (2009) 1(4):358–66.
48. Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res (2003) 63(23):8323–9.
49. Ye J, Wen J, Ning Y, Li Y. Higher notch expression implies poor survival in pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. Pancreatology (2018) 18(8):954–61. doi: 10.1016/j.pan.2018.09.014
50. Hanlon L, Avila JL, Demarest RM, Troutman S, Allen M, Ratti F, et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res (2010) 70(11):4280–6. doi: 10.1158/0008-5472.Can-09-4645
51. Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest (2005) 115(11):3166–76. doi: 10.1172/JCI25001
52. Müller CS. Notch signaling and malignant melanoma. Adv Exp Med Biol (2012) 727:258–64. doi: 10.1007/978-1-4614-0899-4_19
53. Zhang L, Sha J, Yang G, Huang X, Bo J, Huang Y. Activation of notch pathway is linked with epithelial-mesenchymal transition in prostate cancer cells. Cell Cycle (2017) 16(10):999–1007. doi: 10.1080/15384101.2017.1312237
54. Zhu H, Zhou X, Redfield S, Lewin J, Miele L. Elevated jagged-1 and notch-1 expression in high grade and metastatic prostate cancers. Am J Transl Res (2013) 5(3):368–78.
55. Bazzoni R, Bentivegna A. Role of notch signaling pathway in glioblastoma pathogenesis. Cancers (Basel) (2019) 11(3):292. doi: 10.3390/cancers11030292
56. El Hindy N, Keyvani K, Pagenstecher A, Dammann P, Sandalcioglu IE, Sure U, et al. Implications of Dll4-notch signaling activation in primary glioblastoma multiforme. Neuro Oncol (2013) 15(10):1366–78. doi: 10.1093/neuonc/not071
57. Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of notch-1 and its ligands, delta-like-1 and jagged-1, is critical for glioma cell survival and proliferation. Cancer Res (2005) 65(6):2353–63. doi: 10.1158/0008-5472.Can-04-1890
58. Ho AS, Ochoa A, Jayakumaran G, Zehir A, Valero Mayor C, Tepe J, et al. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J Clin Invest (2019) 129(10):4276–89. doi: 10.1172/jci128227
59. Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res (2004) 64(19):6854–7. doi: 10.1158/0008-5472.Can-04-2500
60. Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res (2005) 65(18):8530–7. doi: 10.1158/0008-5472.CAN-05-1069
61. Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U.S.A. (1995) 92(14):6414–8. doi: 10.1073/pnas.92.14.6414
62. Hendrix MJ, Seftor RE, Seftor EA, Gruman LM, Lee LM, Nickoloff BJ, et al. Transendothelial function of human metastatic melanoma cells: role of the microenvironment in cell-fate determination. Cancer Res (2002) 62(3):665–8.
63. Chen Y, De Marco MA, Graziani I, Gazdar AF, Strack PR, Miele L, et al. Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Res (2007) 67(17):7954–9. doi: 10.1158/0008-5472.Can-07-1229
64. Li X, Cao X, Zhao H, Guo M, Fang X, Li K, et al. Hypoxia activates Notch4 via ERK/JNK/P38 MAPK signaling pathways to promote lung adenocarcinoma progression and metastasis. Front Cell Dev Biol (2021) 9:780121. doi: 10.3389/fcell.2021.780121
65. Vacca A, Felli MP, Palermo R, Di Mario G, Calce A, Di Giovine M, et al. Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. EMBO J (2006) 25(5):1000–8. doi: 10.1038/sj.emboj.7600996
66. Zhang J, Kuang Y, Wang Y, Xu Q, Ren Q. Notch-4 silencing inhibits prostate cancer growth and EMT via the NF-κB pathway. Apoptosis (2017) 22(6):877–84. doi: 10.1007/s10495-017-1368-0
67. Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating n-cadherin expression. Cancer Res (2006) 66(8):4182–90. doi: 10.1158/0008-5472.Can-05-3589
68. Li X, Wang S, Deng N, Guo X, Fu M, Ma Y, et al. N-((1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl)-2-methylene-3-oxo-olean-12-en-28-amide induces apoptosis in human breast cancer cells by stimulating oxidative stress and inhibiting the notch-akt signaling pathway. Oxid Med Cell Longev (2022) 2022:8123120. doi: 10.1155/2022/8123120
69. Wang W, Sun R, Zeng L, Chen Y, Zhang N, Cao S, et al. GALNT2 promotes cell proliferation, migration, and invasion by activating the Notch/Hes1-PTEN-PI3K/Akt signaling pathway in lung adenocarcinoma. Life Sci (2021) 276:119439. doi: 10.1016/j.lfs.2021.119439
70. Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/β-catenin pathway in colorectal cancer. Cancer Res (2013) 73(6):2003–13. doi: 10.1158/0008-5472.Can-12-3159
71. Mu R, Zou YK, Tu K, Wang DB, Tang D, Yu Z, et al. Hypoxia promotes pancreatic cancer cell dedifferentiation to stem-like cell phenotypes with high tumorigenic potential by the HIF-1α/Notch signaling pathway. Pancreas (2021) 50(5):756–65. doi: 10.1097/mpa.0000000000001828
72. Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OI, et al. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating wnt and notch pathways. J Exp Clin Cancer Res (2018) 37(1):256. doi: 10.1186/s13046-018-0925-x
73. Qiang L, Wu T, Zhang HW, Lu N, Hu R, Wang YJ, et al. HIF-1α is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating notch signaling pathway. Cell Death Differ (2012) 19(2):284–94. doi: 10.1038/cdd.2011.95
74. Yamashita AS, Geraldo MV, Fuziwara CS, Kulcsar MA, Friguglietti CU, da Costa RB, et al. Notch pathway is activated by MAPK signaling and influences papillary thyroid cancer proliferation. Transl Oncol (2013) 6(2):197–205. doi: 10.1593/tlo.12442
75. Izrailit J, Berman HK, Datti A, Wrana JL, Reedijk M. High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFβ pathways as fundamental notch regulators in breast cancer. Proc Natl Acad Sci U.S.A. (2013) 110(5):1714–9. doi: 10.1073/pnas.1214014110
76. Zeng Q, Li S, Chepeha DB, Giordano TJ, Li J, Zhang H, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of notch signaling. Cancer Cell (2005) 8(1):13–23. doi: 10.1016/j.ccr.2005.06.004
77. Sun Y, Lowther W, Kato K, Bianco C, Kenney N, Strizzi L, et al. Notch4 intracellular domain binding to Smad3 and inhibition of the TGF-beta signaling. Oncogene (2005) 24(34):5365–74. doi: 10.1038/sj.onc.1208528
78. Sjölund J, Boström AK, Lindgren D, Manna S, Moustakas A, Ljungberg B, et al. The notch and TGF-β signaling pathways contribute to the aggressiveness of clear cell renal cell carcinoma. PLoS One (2011) 6(8):e23057. doi: 10.1371/journal.pone.0023057
79. Licciulli S, Avila JL, Hanlon L, Troutman S, Cesaroni M, Kota S, et al. Notch1 is required for kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer Res (2013) 73(19):5974–84. doi: 10.1158/0008-5472.Can-13-1384
80. Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev (2007) 21(5):562–77. doi: 10.1101/gad.1484707
81. Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by notch in lymphomagenesis: implications for initiation and regression. Cancer Res (2005) 65(16):7159–68. doi: 10.1158/0008-5472.Can-05-1664
82. Shin HM, Tilahun ME, Cho OH, Chandiran K, Kuksin CA, Keerthivasan S, et al. NOTCH1 can initiate NF-κB activation via cytosolic interactions with components of the T cell signalosome. Front Immunol (2014) 5:249. doi: 10.3389/fimmu.2014.00249
83. Maillard I, Pear WS. Notch and cancer: best to avoid the ups and downs. Cancer Cell (2003) 3(3):203–5. doi: 10.1016/s1535-6108(03)00052-7
84. Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet (2003) 33(3):416–21. doi: 10.1038/ng1099
85. Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of b-cell malignancies. Blood (2005) 106(12):3898–906. doi: 10.1182/blood-2005-01-0355
86. Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med (2011) 208(10):1963–76. doi: 10.1084/jem.20110198
87. Crabtree JS, Singleton CS, Miele L. Notch signaling in neuroendocrine tumors. Front Oncol (2016) 6:94. doi: 10.3389/fonc.2016.00094
88. Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity (2003) 19(6):781–91. doi: 10.1016/s1074-7613(03)00325-x
89. Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol (2015) 35 Suppl(Suppl):S199–s223. doi: 10.1016/j.semcancer.2015.02.007
90. Lim YZ, South AP. Tumour-stroma crosstalk in the development of squamous cell carcinoma. Int J Biochem Cell Biol (2014) 53:450–8. doi: 10.1016/j.biocel.2014.06.012
91. Maillard I, He Y, Pear WS. From the yolk sac to the spleen: New roles for notch in regulating hematopoiesis. Immunity (2003) 18(5):587–9. doi: 10.1016/s1074-7613(03)00119-5
92. Janghorban M, Yang Y, Zhao N, Hamor C, Nguyen TM, Zhang XH, et al. Single-cell analysis unveils the role of the tumor immune microenvironment and notch signaling in dormant minimal residual disease. Cancer Res (2022) 82(5):885–99. doi: 10.1158/0008-5472.Can-21-1230
93. Liu H, Wang J, Zhang M, Xuan Q, Wang Z, Lian X, et al. Jagged1 promotes aromatase inhibitor resistance by modulating tumor-associated macrophage differentiation in breast cancer patients. Breast Cancer Res Treat (2017) 166(1):95–107. doi: 10.1007/s10549-017-4394-2
94. Jaiswal A, Murakami K, Elia A, Shibahara Y, Done SJ, Wood SA, et al. Therapeutic inhibition of USP9x-mediated notch signaling in triple-negative breast cancer. Proc Natl Acad Sci U.S.A. (2021) 118(38):e2101592118. doi: 10.1073/pnas.2101592118
95. Nandi A, Debnath R, Nayak A, To TKJ, Thacker G, Reilly M, et al. Dll1-mediated notch signaling drives tumor cell cross-talk with cancer-associated fibroblasts to promote radioresistance in breast cancer. Cancer Res (2022) 82(20):3718–33. doi: 10.1158/0008-5472.Can-21-1225
96. Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol (2020) 21(5):268–83. doi: 10.1038/s41580-020-0227-y
97. Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev (2007) 26(2):225–39. doi: 10.1007/s10555-007-9055-1
98. Brahimi-Horn MC, Chiche J, Pouysségur J. Hypoxia and cancer. J Mol Med (Berl) (2007) 85(12):1301–7. doi: 10.1007/s00109-007-0281-3
99. Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the tumor in the hypoxic niche: The emerging concept and associated therapeutic strategies. Trends Pharmacol Sci (2017) 38(8):669–86. doi: 10.1016/j.tips.2017.05.002
100. Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer (2002) 2(1):38–47. doi: 10.1038/nrc704
101. Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) (2015) 3:83–92. doi: 10.2147/hp.S93413
102. Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell (2012) 148(3):399–408. doi: 10.1016/j.cell.2012.01.021
103. Kaelin WG Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell (2008) 30(4):393–402. doi: 10.1016/j.molcel.2008.04.009
104. Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von hippel-lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science (2001) 292(5516):468–72. doi: 10.1126/science.1059796
105. Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, et al. Hypoxia inducible factor-alpha binding and ubiquitylation by the von hippel-lindau tumor suppressor protein. J Biol Chem (2000) 275(33):25733–41. doi: 10.1074/jbc.M002740200
106. Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature (2015) 524(7565):303–8. doi: 10.1038/nature14883
107. Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev (2001) 15(20):2675–86. doi: 10.1101/gad.924501
108. Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev (2002) 16(12):1466–71. doi: 10.1101/gad.991402
109. Yang SL, Wu C, Xiong ZF, Fang X. Progress on hypoxia-inducible factor-3: Its structure, gene regulation and biological function (Review). Mol Med Rep (2015) 12(2):2411–6. doi: 10.3892/mmr.2015.3689
110. Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, et al. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol (2000) 157(2):411–21. doi: 10.1016/s0002-9440(10)64554-3
111. Suzuki N, Gradin K, Poellinger L, Yamamoto M. Regulation of hypoxia-inducible gene expression after HIF activation. Exp Cell Res (2017) 356(2):182–6. doi: 10.1016/j.yexcr.2017.03.013
112. Kenneth NS, Rocha S. Regulation of gene expression by hypoxia. Biochem J (2008) 414(1):19–29. doi: 10.1042/bj20081055
113. Heikkilä M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol Life Sci (2011) 68(23):3885–901. doi: 10.1007/s00018-011-0679-5
114. Frolova O, Samudio I, Benito JM, Jacamo R, Kornblau SM, Markovic A, et al. Regulation of HIF-1α signaling and chemoresistance in acute lymphocytic leukemia under hypoxic conditions of the bone marrow microenvironment. Cancer Biol Ther (2012) 13(10):858–70. doi: 10.4161/cbt.20838
115. Dales JP, Garcia S, Meunier-Carpentier S, Andrac-Meyer L, Haddad O, Lavaut MN, et al. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: retrospective study in a series of 745 patients. Int J Cancer (2005) 116(5):734–9. doi: 10.1002/ijc.20984
116. Rasheed S, Harris AL, Tekkis PP, Turley H, Silver A, McDonald PJ, et al. Hypoxia-inducible factor-1alpha and -2alpha are expressed in most rectal cancers but only hypoxia-inducible factor-1alpha is associated with prognosis. Br J Cancer (2009) 100(10):1666–73. doi: 10.1038/sj.bjc.6605026
117. Schmitz KJ, Müller CI, Reis H, Alakus H, Winde G, Baba HA, et al. Combined analysis of hypoxia-inducible factor 1 alpha and metallothionein indicates an aggressive subtype of colorectal carcinoma. Int J Colorectal Dis (2009) 24(11):1287–96. doi: 10.1007/s00384-009-0753-8
118. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science (2009) 324(5930):1029–33. doi: 10.1126/science.1160809
119. Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med (2011) 208(2):313–26. doi: 10.1084/jem.20101470
120. Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science (2012) 337(6097):975–80. doi: 10.1126/science.1222278
121. Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev (2001) 11(3):293–9. doi: 10.1016/s0959-437x(00)00193-3
122. Grimshaw MJ. Endothelins and hypoxia-inducible factor in cancer. Endocr Relat Cancer (2007) 14(2):233–44. doi: 10.1677/erc-07-0057
123. Smith K, Gunaratnam L, Morley M, Franovic A, Mekhail K, Lee S. Silencing of epidermal growth factor receptor suppresses hypoxia-inducible factor-2-driven VHL-/- renal cancer. Cancer Res (2005) 65(12):5221–30. doi: 10.1158/0008-5472.Can-05-0169
124. Rey S, Semenza GL. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res (2010) 86(2):236–42. doi: 10.1093/cvr/cvq045
125. Kelly BD, Hackett SF, Hirota K, Oshima Y, Cai Z, Berg-Dixon S, et al. Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ Res (2003) 93(11):1074–81. doi: 10.1161/01.Res.0000102937.50486.1b
126. Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, et al. HIF-1α promotes epithelial-mesenchymal transition and metastasis through direct regulation of ZEB1 in colorectal cancer. PLoS One (2015) 10(6):e0129603. doi: 10.1371/journal.pone.0129603
127. Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol (2008) 10(3):295–305. doi: 10.1038/ncb1691
128. Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, et al. Hypoxia-inducible factor-1-dependent repression of e-cadherin in von hippel-lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res (2006) 66(5):2725–31. doi: 10.1158/0008-5472.Can-05-3719
129. Jögi A, Øra I, Nilsson H, Lindeheim A, Makino Y, Poellinger L, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci U.S.A. (2002) 99(10):7021–6. doi: 10.1073/pnas.102660199
130. Irshad K, Mohapatra SK, Srivastava C, Garg H, Mishra S, Dikshit B, et al. A combined gene signature of hypoxia and notch pathway in human glioblastoma and its prognostic relevance. PLoS One (2015) 10(3):e0118201. doi: 10.1371/journal.pone.0118201
131. Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell (2005) 9(5):617–28. doi: 10.1016/j.devcel.2005.09.010
132. Mailhos C, Modlich U, Lewis J, Harris A, Bicknell R, Ish-Horowicz D. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation (2001) 69(2-3):135–44. doi: 10.1046/j.1432-0436.2001.690207.x
133. Pietras A, von Stedingk K, Lindgren D, Påhlman S, Axelson H. JAG2 induction in hypoxic tumor cells alters notch signaling and enhances endothelial cell tube formation. Mol Cancer Res (2011) 9(5):626–36. doi: 10.1158/1541-7786.Mcr-10-0508
134. Lanner F, Lee KL, Ortega GC, Sohl M, Li X, Jin S, et al. Hypoxia-induced arterial differentiation requires adrenomedullin and notch signaling. Stem Cells Dev (2013) 22(9):1360–9. doi: 10.1089/scd.2012.0259
135. Villa JC, Chiu D, Brandes AH, Escorcia FE, Villa CH, Maguire WF, et al. Nontranscriptional role of hif-1α in activation of γ-secretase and notch signaling in breast cancer. Cell Rep (2014) 8(4):1077–92. doi: 10.1016/j.celrep.2014.07.028
136. Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates notch signaling in breast cancer leading to decreased e-cadherin expression and increased cell migration and invasion. Br J Cancer (2010) 102(2):351–60. doi: 10.1038/sj.bjc.6605486
137. Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei Y, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating wnt/β-catenin and notch signaling. Theranostics (2020) 10(6):2553–70. doi: 10.7150/thno.41120
138. Mutvei AP, Landor SK, Fox R, Braune EB, Tsoi YL, Phoon YP, et al. Notch signaling promotes a HIF2α-driven hypoxic response in multiple tumor cell types. Oncogene (2018) 37(46):6083–95. doi: 10.1038/s41388-018-0400-3
139. Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, et al. Asparaginyl hydroxylation of the notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem (2007) 282(33):24027–38. doi: 10.1074/jbc.M704102200
140. Zheng X, Linke S, Dias JM, Zheng X, Gradin K, Wallis TP, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the notch and hypoxia signaling pathways. Proc Natl Acad Sci U.S.A. (2008) 105(9):3368–73. doi: 10.1073/pnas.0711591105
141. Guo M, Zhang M, Cao X, Fang X, Li K, Qin L, et al. Notch4 mediates vascular remodeling via ERK/JNK/P38 MAPK signaling pathways in hypoxic pulmonary hypertension. Respir Res (2022) 23(1):6. doi: 10.1186/s12931-022-01927-9
142. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
143. Lin YT, Wu KJ. Epigenetic regulation of epithelial-mesenchymal transition: focusing on hypoxia and TGF-β signaling. J BioMed Sci (2020) 27(1):39. doi: 10.1186/s12929-020-00632-3
144. Wells A, Yates C, Shepard CR. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis (2008) 25(6):621–8. doi: 10.1007/s10585-008-9167-1
145. Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev (2009) 28(1-2):151–66. doi: 10.1007/s10555-008-9179-y
146. Peinado H, Olmeda D, Cano A. Snail, zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer (2007) 7(6):415–28. doi: 10.1038/nrc2131
147. Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, et al. Krüppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res (2007) 67(15):7184–93. doi: 10.1158/0008-5472.Can-06-4729
148. Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, Portillo F, et al. The class I bHLH factors E2-2A and E2-2B regulate EMT. J Cell Sci (2009) 122(Pt 7):1014–24. doi: 10.1242/jcs.028241
149. Yang MH, Wu KJ. TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle (2008) 7(14):2090–6. doi: 10.4161/cc.7.14.6324
150. Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U.S.A. (2008) 105(17):6392–7. doi: 10.1073/pnas.0802047105
151. Ishida T, Hijioka H, Kume K, Miyawaki A, Nakamura N. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol Lett (2013) 6(5):1201–6. doi: 10.3892/ol.2013.1549
152. Tian Q, Xue Y, Zheng W, Sun R, Ji W, Wang X, et al. Overexpression of hypoxia-inducible factor 1α induces migration and invasion through notch signaling. Int J Oncol (2015) 47(2):728–38. doi: 10.3892/ijo.2015.3056
153. Fujiki K, Inamura H, Miyayama T, Matsuoka M. Involvement of Notch1 signaling in malignant progression of A549 cells subjected to prolonged cadmium exposure. J Biol Chem (2017) 292(19):7942–53. doi: 10.1074/jbc.M116.759134
154. Cook KM, Figg WD. Angiogenesis inhibitors: current strategies and future prospects. CA Cancer J Clin (2010) 60(4):222–43. doi: 10.3322/caac.20075
155. Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer Cell (2005) 8(1):1–3. doi: 10.1016/j.ccr.2005.06.013
156. Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res (2005) 65(19):8690–7. doi: 10.1158/0008-5472.Can-05-1208
157. Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U.S.A. (2004) 101(45):15949–54. doi: 10.1073/pnas.0407290101
158. Hainaud P, Contrerès JO, Villemain A, Liu LX, Plouët J, Tobelem G, et al. The role of the vascular endothelial growth factor-delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions. Cancer Res (2006) 66(17):8501–10. doi: 10.1158/0008-5472.Can-05-4226
159. Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature (2007) 445(7129):776–80. doi: 10.1038/nature05571
160. Diez H, Fischer A, Winkler A, Hu CJ, Hatzopoulos AK, Breier G, et al. Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp Cell Res (2007) 313(1):1–9. doi: 10.1016/j.yexcr.2006.09.009
161. Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer (2005) 5(4):311–21. doi: 10.1038/nrc1592
162. Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell (2007) 129(3):465–72. doi: 10.1016/j.cell.2007.04.019
163. Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol (2010) 92:367–409. doi: 10.1016/s0070-2153(10)92012-7
164. Wang K, Ding R, Ha Y, Jia Y, Liao X, Wang S, et al. Hypoxia-stressed cardiomyocytes promote early cardiac differentiation of cardiac stem cells through HIF-1α/Jagged1/Notch1 signaling. Acta Pharm Sin B (2018) 8(5):795–804. doi: 10.1016/j.apsb.2018.06.003
165. Main H, Lee KL, Yang H, Haapa-Paananen S, Edgren H, Jin S, et al. Interactions between notch- and hypoxia-induced transcriptomes in embryonic stem cells. Exp Cell Res (2010) 316(9):1610–24. doi: 10.1016/j.yexcr.2009.12.012
166. Moriyama H, Moriyama M, Isshi H, Ishihara S, Okura H, Ichinose A, et al. Role of notch signaling in the maintenance of human mesenchymal stem cells under hypoxic conditions. Stem Cells Dev (2014) 23(18):2211–24. doi: 10.1089/scd.2013.0642
167. Zhang S, Chan RWS, Ng EHY, Yeung WSB. Hypoxia regulates the self-renewal of endometrial mesenchymal Stromal/Stem-like cells via notch signaling. Int J Mol Sci (2022) 23(9):4613. doi: 10.3390/ijms23094613
168. Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells (2007) 25(3):807–15. doi: 10.1634/stemcells.2006-0442
169. Zeng F, Chen H, Zhang Z, Yao T, Wang G, Zeng Q, et al. Regulating glioma stem cells by hypoxia through the Notch1 and Oct3/4 signaling pathway. Oncol Lett (2018) 16(5):6315–22. doi: 10.3892/ol.2018.9442
170. Papayannidis C, DeAngelo DJ, Stock W, Huang B, Shaik MN, Cesari R, et al. A phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J (2015) 5(9):e350. doi: 10.1038/bcj.2015.80
171. Sardesai S, Badawi M, Mrozek E, Morgan E, Phelps M, Stephens J, et al. A phase I study of an oral selective gamma secretase (GS) inhibitor RO4929097 in combination with neoadjuvant paclitaxel and carboplatin in triple negative breast cancer. Invest New Drugs (2020) 38(5):1400–10. doi: 10.1007/s10637-020-00895-5
172. Morgan KM, Fischer BS, Lee FY, Shah JJ, Bertino JR, Rosenfeld J, et al. Gamma secretase inhibition by BMS-906024 enhances efficacy of paclitaxel in lung adenocarcinoma. Mol Cancer Ther (2017) 16(12):2759–69. doi: 10.1158/1535-7163.Mct-17-0439
173. Akiyoshi T, Nakamura M, Yanai K, Nagai S, Wada J, Koga K, et al. Gamma-secretase inhibitors enhance taxane-induced mitotic arrest and apoptosis in colon cancer cells. Gastroenterology (2008) 134(1):131–44. doi: 10.1053/j.gastro.2007.10.008
174. Cui D, Dai J, Keller JM, Mizokami A, Xia S, Keller ET. Notch pathway inhibition using PF-03084014, a γ-secretase inhibitor (GSI), enhances the antitumor effect of docetaxel in prostate cancer. Clin Cancer Res (2015) 21(20):4619–29. doi: 10.1158/1078-0432.Ccr-15-0242
175. Hurtado C, Safarova A, Smith M, Chung R, Bruyneel AAN, Gomez-Galeno J, et al. Disruption of NOTCH signaling by a small molecule inhibitor of the transcription factor RBPJ. Sci Rep (2019) 9(1):10811. doi: 10.1038/s41598-019-46948-5
176. Lehal R, Zaric J, Vigolo M, Urech C, Frismantas V, Zangger N, et al. Pharmacological disruption of the notch transcription factor complex. Proc Natl Acad Sci U.S.A. (2020) 117(28):16292–301. doi: 10.1073/pnas.1922606117
177. Agnusdei V, Minuzzo S, Frasson C, Grassi A, Axelrod F, Satyal S, et al. Therapeutic antibody targeting of Notch1 in T-acute lymphoblastic leukemia xenografts. Leukemia (2014) 28(2):278–88. doi: 10.1038/leu.2013.183
178. Ferrarotto R, Eckhardt G, Patnaik A, LoRusso P, Faoro L, Heymach JV, et al. A phase I dose-escalation and dose-expansion study of brontictuzumab in subjects with selected solid tumors. Ann Oncol (2018) 29(7):1561–8. doi: 10.1093/annonc/mdy171
179. Hu ZI, Bendell JC, Bullock A, LoConte NK, Hatoum H, Ritch P, et al. A randomized phase II trial of nab-paclitaxel and gemcitabine with tarextumab or placebo in patients with untreated metastatic pancreatic cancer. Cancer Med (2019) 8(11):5148–57. doi: 10.1002/cam4.2425
180. Lu H, Jiang Z. Advances in antibody therapeutics targeting small-cell lung cancer. Adv Clin Exp Med (2018) 27(9):1317–23. doi: 10.17219/acem/70159
181. Smith DC, Chugh R, Patnaik A, Papadopoulos KP, Wang M, Kapoun AM, et al. A phase 1 dose escalation and expansion study of tarextumab (OMP-59R5) in patients with solid tumors. Invest New Drugs (2019) 37(4):722–30. doi: 10.1007/s10637-018-0714-6
182. Rosen LS, Wesolowski R, Baffa R, Liao KH, Hua SY, Gibson BL, et al. A phase I, dose-escalation study of PF-06650808, an anti-Notch3 antibody-drug conjugate, in patients with breast cancer and other advanced solid tumors. Invest New Drugs (2020) 38(1):120–30. doi: 10.1007/s10637-019-00754-y
183. Pandya K, Wyatt D, Gallagher B, Shah D, Baker A, Bloodworth J, et al. PKCα attenuates jagged-1-Mediated notch signaling in ErbB-2-Positive breast cancer to reverse trastuzumab resistance. Clin Cancer Res (2016) 22(1):175–86. doi: 10.1158/1078-0432.Ccr-15-0179
184. Masiero M, Li D, Whiteman P, Bentley C, Greig J, Hassanali T, et al. Development of therapeutic anti-JAGGED1 antibodies for cancer therapy. Mol Cancer Ther (2019) 18(11):2030–42. doi: 10.1158/1535-7163.Mct-18-1176
185. Saunders LR, Bankovich AJ, Anderson WC, Aujay MA, Bheddah S, Black K, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med (2015) 7(302):302ra136. doi: 10.1126/scitranslmed.aac9459
186. Perez-Fidalgo JA, Ortega B, Simon S, Samartzis EP, Boussios S. NOTCH signalling in ovarian cancer angiogenesis. Ann Transl Med (2020) 8(24):1705. doi: 10.21037/atm-20-4497
187. McKeage MJ, Kotasek D, Markman B, Hidalgo M, Millward MJ, Jameson MB, et al. Phase IB trial of the anti-cancer stem cell DLL4-binding agent demcizumab with pemetrexed and carboplatin as first-line treatment of metastatic non-squamous NSCLC. Target Oncol (2018) 13(1):89–98. doi: 10.1007/s11523-017-0543-0
188. Chiorean EG, LoRusso P, Strother RM, Diamond JR, Younger A, Messersmith WA, et al. A phase I first-in-Human study of enoticumab (REGN421), a fully human delta-like ligand 4 (Dll4) monoclonal antibody in patients with advanced solid tumors. Clin Cancer Res (2015) 21(12):2695–703. doi: 10.1158/1078-0432.Ccr-14-2797
189. Ye Q, Jiang J, Zhan G, Yan W, Huang L, Hu Y, et al. Small molecule activation of NOTCH signaling inhibits acute myeloid leukemia. Sci Rep (2016) 6:26510. doi: 10.1038/srep26510
190. Rios-Colon L, Chijioke J, Niture S, Afzal Z, Qi Q, Srivastava A, et al. Leptin modulated microRNA-628-5p targets jagged-1 and inhibits prostate cancer hallmarks. Sci Rep (2022) 12(1):10073. doi: 10.1038/s41598-022-13279-x
191. Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res (2014) 74(24):7573–82. doi: 10.1158/0008-5472.Can-14-1140
192. Jeong JY, Kang H, Kim TH, Kim G, Heo JH, Kwon AY, et al. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett (2017) 386:168–78. doi: 10.1016/j.canlet.2016.11.017
193. Zhao Z, Shen X, Zhang D, Xiao H, Kong H, Yang B, et al. miR-153 enhances the therapeutic effect of radiotherapy by targeting JAG1 in pancreatic cancer cells. Oncol Lett (2021) 21(4):300. doi: 10.3892/ol.2021.12561
194. Bai L, Wang A, Zhang Y, Xu X, Zhang X. Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp Cell Res (2018) 366(2):161–71. doi: 10.1016/j.yexcr.2018.03.014
195. Shen Z, Wu Y, He G. Long non-coding RNA PTPRG-AS1/microRNA-124-3p regulates radiosensitivity of nasopharyngeal carcinoma via the LIM homeobox 2-dependent notch pathway through competitive endogenous RNA mechanism. Bioengineered (2022) 13(4):8208–25. doi: 10.1080/21655979.2022.2037364
196. Imbimbo BP, Panza F, Frisardi V, Solfrizzi V, D'Onofrio G, Logroscino G, et al. Therapeutic intervention for alzheimer's disease with γ-secretase inhibitors: still a viable option? Expert Opin Investig Drugs (2011) 20(3):325–41. doi: 10.1517/13543784.2011.550572
197. Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep (2008) 9(4):377–83. doi: 10.1038/embor.2008.7
198. Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med (2009) 15(1):50–8. doi: 10.1038/nm.1900
199. Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual notch receptors. Nature (2010) 464(7291):1052–7. doi: 10.1038/nature08878
200. Aste-Amézaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One (2010) 5(2):e9094. doi: 10.1371/journal.pone.0009094
201. Sharma A, Gadkari RA, Ramakanth SV, Padmanabhan K, Madhumathi DS, Devi L, et al. A novel monoclonal antibody against Notch1 targets leukemia-associated mutant Notch1 and depletes therapy resistant cancer stem cells in solid tumors. Sci Rep (2015) 5:11012. doi: 10.1038/srep11012
202. Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, et al. Modulation of notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem (2008) 283(12):8046–54. doi: 10.1074/jbc.M800170200
203. Lafkas D, Shelton A, Chiu C, de Leon Boenig G, Chen Y, Stawicki SS, et al. Therapeutic antibodies reveal notch control of transdifferentiation in the adult lung. Nature (2015) 528(7580):127–31. doi: 10.1038/nature15715
204. Krämer A, Mentrup T, Kleizen B, Rivera-Milla E, Reichenbach D, Enzensperger C, et al. Small molecules intercept notch signaling and the early secretory pathway. Nat Chem Biol (2013) 9(11):731–8. doi: 10.1038/nchembio.1356
205. Gan RH, Lin LS, Xie J, Huang L, Ding LC, Su BH, et al. FLI-06 intercepts notch signaling and suppresses the proliferation and self-renewal of tongue cancer cells. Onco Targets Ther (2019) 12:7663–74. doi: 10.2147/ott.S221231
206. Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature (2009) 462(7270):182–8. doi: 10.1038/nature08543
207. Astudillo L, Da Silva TG, Wang Z, Han X, Jin K, VanWye J, et al. The small molecule IMR-1 inhibits the notch transcriptional activation complex to suppress tumorigenesis. Cancer Res (2016) 76(12):3593–603. doi: 10.1158/0008-5472.Can-16-0061
208. Kulic I, Robertson G, Chang L, Baker JH, Lockwood WW, Mok W, et al. Loss of the notch effector RBPJ promotes tumorigenesis. J Exp Med (2015) 212(1):37–52. doi: 10.1084/jem.20121192
209. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell (2019) 179(5):1033–55. doi: 10.1016/j.cell.2019.10.017
210. Jia Y, Lin R, Jin H, Si L, Jian W, Yu Q, et al. MicroRNA-34 suppresses proliferation of human ovarian cancer cells by triggering autophagy and apoptosis and inhibits cell invasion by targeting notch 1. Biochimie (2019) 160:193–9. doi: 10.1016/j.biochi.2019.03.011
211. Liu Z, Ma T, Duan J, Liu X, Liu L. MicroRNA−223−induced inhibition of the FBXW7 gene affects the proliferation and apoptosis of colorectal cancer cells via the notch and Akt/mTOR pathways. Mol Med Rep (2021) 23(2):154. doi: 10.3892/mmr.2020.11793
212. Pan T, Ding H, Jin L, Zhang S, Wu D, Pan W, et al. DNMT1-mediated demethylation of lncRNA MEG3 promoter suppressed breast cancer progression by repressing Notch1 signaling pathway. Cell Cycle (2022) 21(21):2323–37. doi: 10.1080/15384101.2022.2094662
213. Deng Y, Zhang L. LncRNA SNHG11 accelerates the progression of lung adenocarcinoma via activating notch pathways. Pathol Res Pract (2022) 234:153849. doi: 10.1016/j.prp.2022.153849
214. Jiang H, Li X, Wang W, Dong H. Long non-coding RNA SNHG3 promotes breast cancer cell proliferation and metastasis by binding to microRNA-154-3p and activating the notch signaling pathway. BMC Cancer (2020) 20(1):838. doi: 10.1186/s12885-020-07275-5
215. Phillips RM. Targeting the hypoxic fraction of tumours using hypoxia-activated prodrugs. Cancer Chemother Pharmacol (2016) 77(3):441–57. doi: 10.1007/s00280-015-2920-7
216. Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer (2011) 11(6):393–410. doi: 10.1038/nrc3064
217. Phillips RM, Loadman PM, Reddy G. Inactivation of apaziquone by haematuria: implications for the design of phase III clinical trials against non-muscle invasive bladder cancer. Cancer Chemother Pharmacol (2019) 83(6):1183–9. doi: 10.1007/s00280-019-03812-7
218. Danson SJ, Johnson P, Ward TH, Dawson M, Denneny O, Dickinson G, et al. Phase I pharmacokinetic and pharmacodynamic study of the bioreductive drug RH1. Ann Oncol (2011) 22(7):1653–60. doi: 10.1093/annonc/mdq638
219. Williamson SK, Crowley JJ, Lara PN Jr., McCoy J, Lau DH, Tucker RW, et al. Phase III trial of paclitaxel plus carboplatin with or without tirapazamine in advanced non-small-cell lung cancer: Southwest oncology group trial S0003. J Clin Oncol (2005) 23(36):9097–104. doi: 10.1200/jco.2005.01.3771
220. Hunter FW, Hsu HL, Su J, Pullen SM, Wilson WR, Wang J. Dual targeting of hypoxia and homologous recombination repair dysfunction in triple-negative breast cancer. Mol Cancer Ther (2014) 13(11):2501–14. doi: 10.1158/1535-7163.Mct-14-0476
221. Albertella MR, Loadman PM, Jones PH, Phillips RM, Rampling R, Burnet N, et al. Hypoxia-selective targeting by the bioreductive prodrug AQ4N in patients with solid tumors: results of a phase I study. Clin Cancer Res (2008) 14(4):1096–104. doi: 10.1158/1078-0432.Ccr-07-4020
222. Papadopoulos KP, Goel S, Beeram M, Wong A, Desai K, Haigentz M, et al. A phase 1 open-label, accelerated dose-escalation study of the hypoxia-activated prodrug AQ4N in patients with advanced malignancies. Clin Cancer Res (2008) 14(21):7110–5. doi: 10.1158/1078-0432.Ccr-08-0483
223. Konopleva M, Thall PF, Yi CA, Borthakur G, Coveler A, Bueso-Ramos C, et al. Phase I/II study of the hypoxia-activated prodrug PR104 in refractory/relapsed acute myeloid leukemia and acute lymphoblastic leukemia. Haematologica (2015) 100(7):927–34. doi: 10.3324/haematol.2014.118455
224. Tap WD, Papai Z, Van Tine BA, Attia S, Ganjoo KN, Jones RL, et al. Doxorubicin plus evofosfamide versus doxorubicin alone in locally advanced, unresectable or metastatic soft-tissue sarcoma (TH CR-406/SARC021): an international, multicentre, open-label, randomised phase 3 trial. Lancet Oncol (2017) 18(8):1089–103. doi: 10.1016/s1470-2045(17)30381-9
225. Singleton DC, Macann A, Wilson WR. Therapeutic targeting of the hypoxic tumour microenvironment. Nat Rev Clin Oncol (2021) 18(12):751–72. doi: 10.1038/s41571-021-00539-4
226. Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U.S.A. (2009) 106(42):17910–5. doi: 10.1073/pnas.0909353106
227. Xie C, Gao X, Sun D, Zhang Y, Krausz KW, Qin X, et al. Metabolic profiling of the novel hypoxia-inducible factor 2α inhibitor PT2385 In vivo and in vitro. Drug Metab Dispos (2018) 46(4):336–45. doi: 10.1124/dmd.117.079723
228. Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature (2016) 539(7627):112–7. doi: 10.1038/nature19796
229. Porter JR, Ge J, Lee J, Normant E, West K. Ansamycin inhibitors of Hsp90: nature's prototype for anti-chaperone therapy. Curr Top Med Chem (2009) 9(15):1386–418. doi: 10.2174/156802609789895719
230. Osada M, Imaoka S, Funae Y. Apigenin suppresses the expression of VEGF, an important factor for angiogenesis, in endothelial cells via degradation of HIF-1alpha protein. FEBS Lett (2004) 575(1-3):59–63. doi: 10.1016/j.febslet.2004.08.036
231. Hur E, Kim HH, Choi SM, Kim JH, Yim S, Kwon HJ, et al. Reduction of hypoxia-induced transcription through the repression of hypoxia-inducible factor-1alpha/aryl hydrocarbon receptor nuclear translocator DNA binding by the 90-kDa heat-shock protein inhibitor radicicol. Mol Pharmacol (2002) 62(5):975–82. doi: 10.1124/mol.62.5.975
232. Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell (2005) 18(5):601–7. doi: 10.1016/j.molcel.2005.04.021
233. Zhang C, Yang C, Feldman MJ, Wang H, Pang Y, Maggio DM, et al. Vorinostat suppresses hypoxia signaling by modulating nuclear translocation of hypoxia inducible factor 1 alpha. Oncotarget (2017) 8(34):56110–25. doi: 10.18632/oncotarget.18125
234. Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell (2004) 6(1):33–43. doi: 10.1016/j.ccr.2004.06.009
235. Price C, Gill S, Ho ZV, Davidson SM, Merkel E, McFarland JM, et al. Genome-wide interrogation of human cancers identifies EGLN1 dependency in clear cell ovarian cancers. Cancer Res (2019) 79(10):2564–79. doi: 10.1158/0008-5472.Can-18-2674
236. Greenberger LM, Horak ID, Filpula D, Sapra P, Westergaard M, Frydenlund HF, et al. A RNA antagonist of hypoxia-inducible factor-1alpha, EZN-2968, inhibits tumor cell growth. Mol Cancer Ther (2008) 7(11):3598–608. doi: 10.1158/1535-7163.Mct-08-0510
237. Lin J, Zhan T, Duffy D, Hoffman-Censits J, Kilpatrick D, Trabulsi EJ, et al. A pilot phase II study of digoxin in patients with recurrent prostate cancer as evident by a rising PSA. Am J Cancer Ther Pharmacol (2014) 2(1):21–32.
238. Tinley TL, Leal RM, Randall-Hlubek DA, Cessac JW, Wilkens LR, Rao PN, et al. Novel 2-methoxyestradiol analogues with antitumor activity. Cancer Res (2003) 63(7):1538–49.
239. Lee K, Kim HM. A novel approach to cancer therapy using PX-478 as a HIF-1α inhibitor. Arch Pharm Res (2011) 34(10):1583–5. doi: 10.1007/s12272-011-1021-3
240. Yeo EJ, Chun YS, Cho YS, Kim J, Lee JC, Kim MS, et al. YC-1: a potential anticancer drug targeting hypoxia-inducible factor 1. J Natl Cancer Inst (2003) 95(7):516–25. doi: 10.1093/jnci/95.7.516
241. Ramanathan RK, Abbruzzese J, Dragovich T, Kirkpatrick L, Guillen JM, Baker AF, et al. A randomized phase II study of PX-12, an inhibitor of thioredoxin in patients with advanced cancer of the pancreas following progression after a gemcitabine-containing combination. Cancer Chemother Pharmacol (2011) 67(3):503–9. doi: 10.1007/s00280-010-1343-8
242. Lee K, Kang JE, Park SK, Jin Y, Chung KS, Kim HM, et al. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1alpha via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol (2010) 80(7):982–9. doi: 10.1016/j.bcp.2010.06.018
243. Courtney KD, Infante JR, Lam ET, Figlin RA, Rini BI, Brugarolas J, et al. Phase I dose-escalation trial of PT2385, a first-in-Class hypoxia-inducible factor-2α antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J Clin Oncol (2018) 36(9):867–74. doi: 10.1200/jco.2017.74.2627
244. Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2α antagonist in preclinical kidney cancer models. Nature (2016) 539(7627):107–11. doi: 10.1038/nature19795
245. Taylor SA, Metch B, Balcerzak SP, Hanson KH. Phase II trial of echinomycin in advanced soft tissue sarcomas. a southwest oncology group study. Invest New Drugs (1990) 8(4):381–3. doi: 10.1007/bf00198595
246. Brown TD, Goodman PJ, Fleming TR, Taylor SA, Macdonald JS. Phase II trial of echinomycin in advanced colorectal cancer. a southwest oncology group study. Invest New Drugs (1991) 9(1):113–4. doi: 10.1007/bf00194561
247. Schilsky RL, Faraggi D, Korzun A, Vogelzang N, Ellerton J, Wood W, et al. Phase II study of echinomycin in patients with advanced breast cancer: a report of cancer and leukemia group b protocol 8641. Invest New Drugs (1991) 9(3):269–72. doi: 10.1007/bf00176982
248. Marshall ME, Wolf MK, Crawford ED, Taylor S, Blumenstein B, Flanigan R, et al. Phase II trial of echinomycin for the treatment of advanced renal cell carcinoma. a southwest oncology group study. Invest New Drugs (1993) 11(2-3):207–9. doi: 10.1007/bf00874157
249. Shevrin DH, Lad TE, Guinan P, Kilton LJ, Greenburg A, Johnson P, et al. Phase II trial of echinomycin in advanced hormone-resistant prostate cancer. an Illinois cancer council study. Invest New Drugs (1994) 12(1):65–6. doi: 10.1007/bf00873239
250. Viziteu E, Grandmougin C, Goldschmidt H, Seckinger A, Hose D, Klein B, et al. Chetomin, targeting HIF-1α/p300 complex, exhibits antitumour activity in multiple myeloma. Br J Cancer (2016) 114(5):519–23. doi: 10.1038/bjc.2016.20
251. Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med (2006) 57:33–47. doi: 10.1146/annurev.med.57.042905.122625
252. Quinn DI, Tsao-Wei DD, Twardowski P, Aparicio AM, Frankel P, Chatta G, et al. Phase II study of the histone deacetylase inhibitor vorinostat (Suberoylanilide hydroxamic acid; SAHA) in recurrent or metastatic transitional cell carcinoma of the urothelium - an NCI-CTEP sponsored: California cancer consortium trial, NCI 6879. Invest New Drugs (2021) 39(3):812–20. doi: 10.1007/s10637-020-01038-6
253. Albadari N, Deng S, Li W. The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. Expert Opin Drug Discovery (2019) 14(7):667–82. doi: 10.1080/17460441.2019.1613370
254. Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ (2001) 12(7):363–9.
255. Wang D, Eisen HJ. Mechanistic target of rapamycin (mTOR) inhibitors. Handb Exp Pharmacol (2022) 272:53–72. doi: 10.1007/164_2021_553
256. Ravaud A, Bernhard JC, Gross-Goupil M, Digue L, Ferriere JM. [mTOR inhibitors: temsirolimus and everolimus in the treatment of renal cell carcinoma]. Bull Cancer (2010) 97:45–51. doi: 10.1684/bdc.2010.1069
257. Kopecka J, Salaroglio IC, Perez-Ruiz E, Sarmento-Ribeiro AB, Saponara S, De Las Rivas J, et al. Hypoxia as a driver of resistance to immunotherapy. Drug Resist Update (2021) 59:100787. doi: 10.1016/j.drup.2021.100787
258. Rodriguez CP, Wu QV, Voutsinas J, Fromm JR, Jiang X, Pillarisetty VG, et al. A phase II trial of pembrolizumab and vorinostat in recurrent metastatic head and neck squamous cell carcinomas and salivary gland cancer. Clin Cancer Res (2020) 26(4):837–45. doi: 10.1158/1078-0432.Ccr-19-2214
259. Kumar S, Sun JD, Zhang L, Mokhtari RB, Wu B, Meng F, et al. Hypoxia-targeting drug evofosfamide (TH-302) enhances sunitinib activity in neuroblastoma xenograft models. Transl Oncol (2018) 11(4):911–9. doi: 10.1016/j.tranon.2018.05.004
260. Akil A, Gutiérrez-García AK, Guenter R, Rose JB, Beck AW, Chen H, et al. Notch signaling in vascular endothelial cells, angiogenesis, and tumor progression: An update and prospective. Front Cell Dev Biol (2021) 9:642352. doi: 10.3389/fcell.2021.642352
Keywords: Notch signaling, hypoxia, hypoxia-induced factors pathway, cancer, therapeutics
Citation: Guo M, Niu Y, Xie M, Liu X and Li X (2023) Notch signaling, hypoxia, and cancer. Front. Oncol. 13:1078768. doi: 10.3389/fonc.2023.1078768
Received: 24 October 2022; Accepted: 19 January 2023;
Published: 31 January 2023.
Edited by:
João Pessoa, University of Coimbra, PortugalReviewed by:
Binghan Zhou, Huazhong University of Science and Technology, ChinaOlivier Meurette, Université Claude Bernard Lyon 1, France
Xiawa Mao, Zhejiang University, China
Copyright © 2023 Guo, Niu, Xie, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaochen Li, bGl4aWFvY2hlbjNuMmJAMTYzLmNvbQ==
 Mingzhou Guo
Mingzhou Guo Yang Niu1,2
Yang Niu1,2 Min Xie
Min Xie Xiansheng Liu
Xiansheng Liu Xiaochen Li
Xiaochen Li