- 1Department of Clinical Medicine, Fenyang College of Shanxi Medical University, Fenyang, China
- 2Department of Medical Laboratory Science, Fenyang College of Shanxi Medical University, Fenyang, China
- 3Key Laboratory of Lvliang for Clinical Molecular Diagnostics, Fenyang, China
- 4Department of Clinical Laboratory, Fenyang Hospital of Shanxi Province, Fenyang, China
The eukaryotic translation initiation factor eIF4E can specifically bind to the cap structure of an mRNA 5' end, mainly regulating translation initiation and preferentially enhancing the translation of carcinogenesis related mRNAs. The expression of eIF4E is closely related to a variety of malignant tumors. In tumor cells, eIF4E activity is abnormally increased, which stimulates cell growth, metastasis and translation of related proteins. The main factors affecting eIF4E activity include intranuclear regulation, phosphorylation of 4EBPs, and phosphorylation and sumoylation of eIF4E. In this review, we summarize the biological functions and the research progress of eIF4E, the main influencing factors of eIF4E activity, and the recent progress of drugs targeting eIF4E, in the hope of providing new insights for the treatment of multiple malignancies and development of targeted drugs.
1 Introduction
Translation initiation is the major regulatory stage of protein synthesis (1), which is an important rate-limiting link in protein translation (2). The first step in eukaryotic translation initiation that is dependent on the 5'-end cap structure of mRNAs is the assembly of the translation initiation complex eIF4F on the cap structure (3), and this complex recruits the ribosome to the 5'-end of the mRNAs (4). The eIF4F complex consists of the cap binding protein eIF4E, the large scaffolding protein eIF4G and the DEAD-box protein eIF4A. EIF4E can bind to the cap structure (5), recruiting eIF4G and eIF4A (2, 3, 6, 7).
EIF4E and eIF4G are oncogenes whose overexpression promotes cell transformation. Studies reported that eIF4E, eIF4G and eIF4A genes showed increased amplification or transcription in various human cancers (4). The overexpression of eIF4E precisely elevates the translation of mRNAs associated with tumor growth and invasion (8), making it an important target for tumor therapy. Xu et al. showed that reducing the levels of these translation factors, which have long been considered housekeeping genes, does not interfere with normal development or cell physiology, but is significantly associated with carcinogenesis, supporting the idea that eIF4E activity is critically involved in cancer development (4). Recent studies have shown that eIF4E can inhibit obesity and fatty liver caused by high-fat food. Therefore, the relationship among obesity, canceration and eIF4E may become a new direction for the study of eIF4E.
The action mechanism of eIF4E in promoting malignant transformation of cells has triggered wide attention, and some progress has been made so far. In this paper, the biological functions, activity influencing factors, and the research status of eIF4E in the treatment of tumors are reviewed.
2 Structures of eIF4E
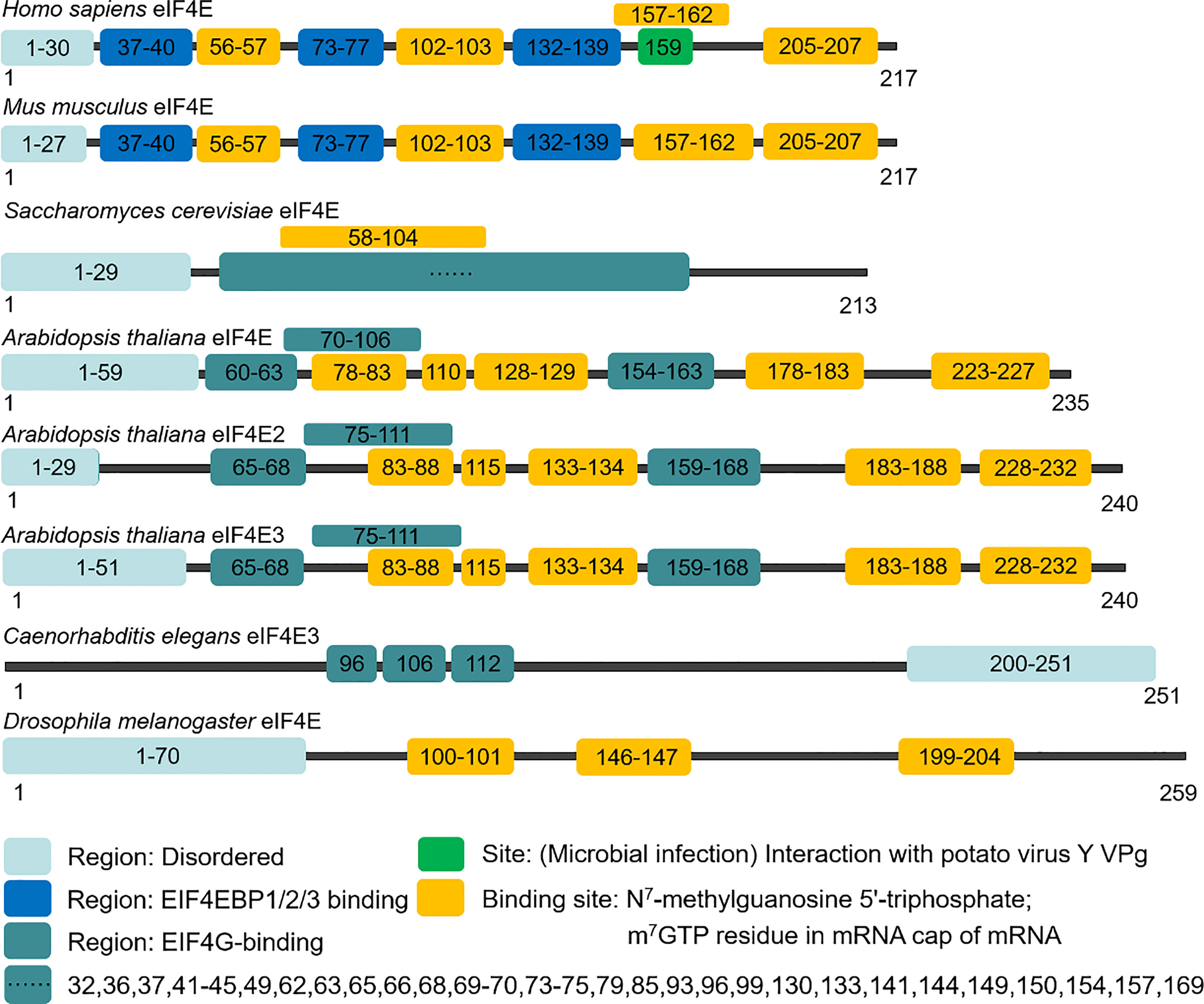
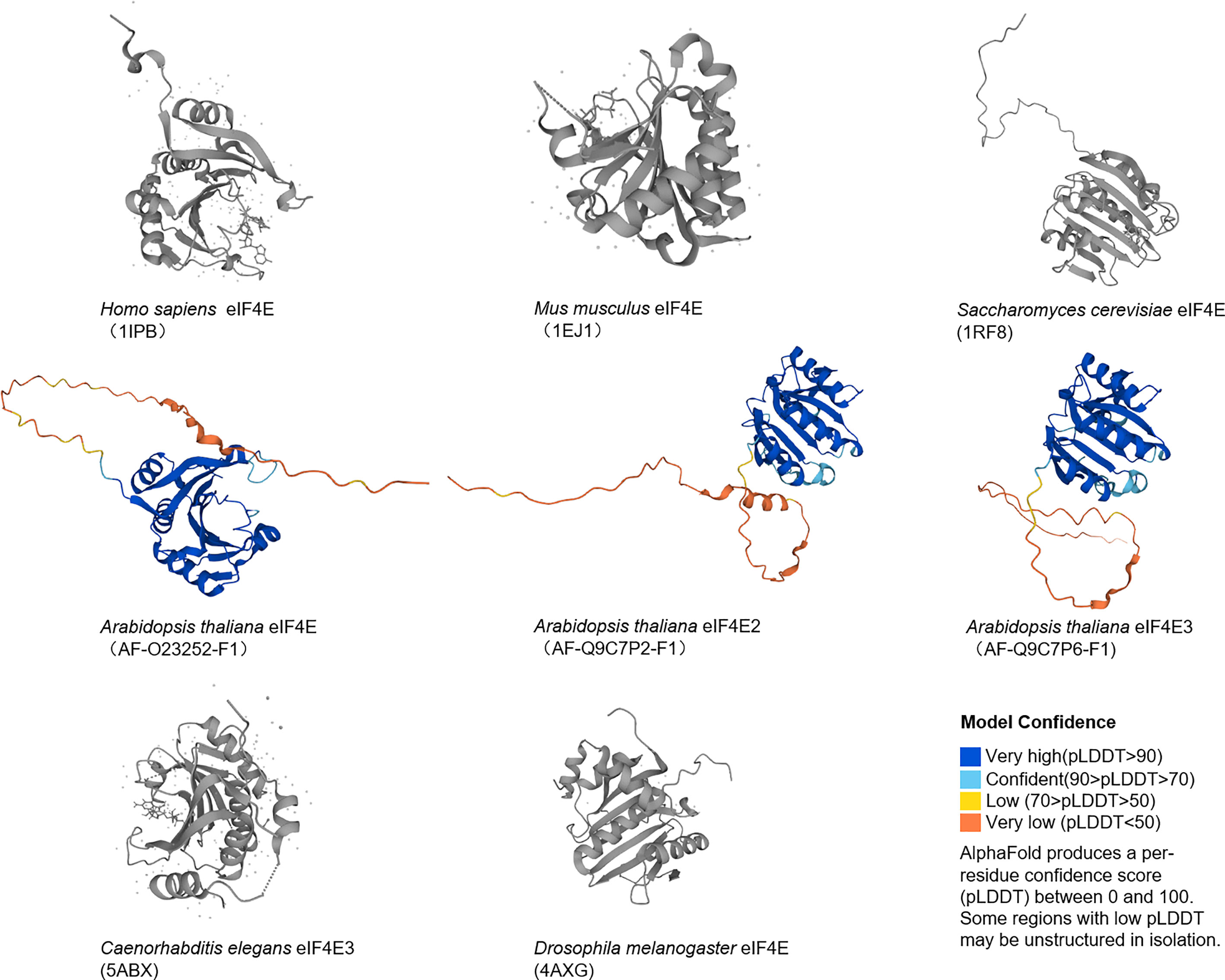
The eIF4E gene, located on chromosome 4q21.25, encodes a conserved protein with a molecular weight of 25 kDa that can bind to the 5' end cap structure of mRNAs. In mammals, there are three members in the eIF4E family, eIF4E1, eIF4E2 and eIF4E3, which differ in structures, functions and expression patterns. EIF4E1 (eIF4E) is present in all eukaryotes (Figures 1, 2), and orthologs of eIF4E2 appears to exist only in metazoans, while those of eIF4E3 are found only in chordates. EIF4E2 is ubiquitously expressed, with the highest levels in testis, whereas eIF4E3 is expressed only in skeletal muscle, heart, spleen and lung. Compared with eIF4E1, eIF4E2 and eIF4E3 respectively have some functions of eIF4E1. EIF4E2 cannot combine with eIF4G to initiate translation, and eIF4E3 cannot combine with 4EBPs (9). Three-dimensional structural analysis showed that there are two main binding sites on the surface of eIF4E: cap binding site and eIF4G (or 4EBPs) binding site. The concave face of eIF4E contains a concave hydrophobic pocket assembled by eight β-sheets and three α-helices. The cap binding site is on the basal face of the pocket, and the cap structure is stacked between two highly conserved tryptophan residues. EIF4G or 4EBPs specifically bind to the backside bulge of eIF4E, both of which share and competitively bind to this region (10, 11).
3 Biological functions and associated interacting proteins of eIF4E
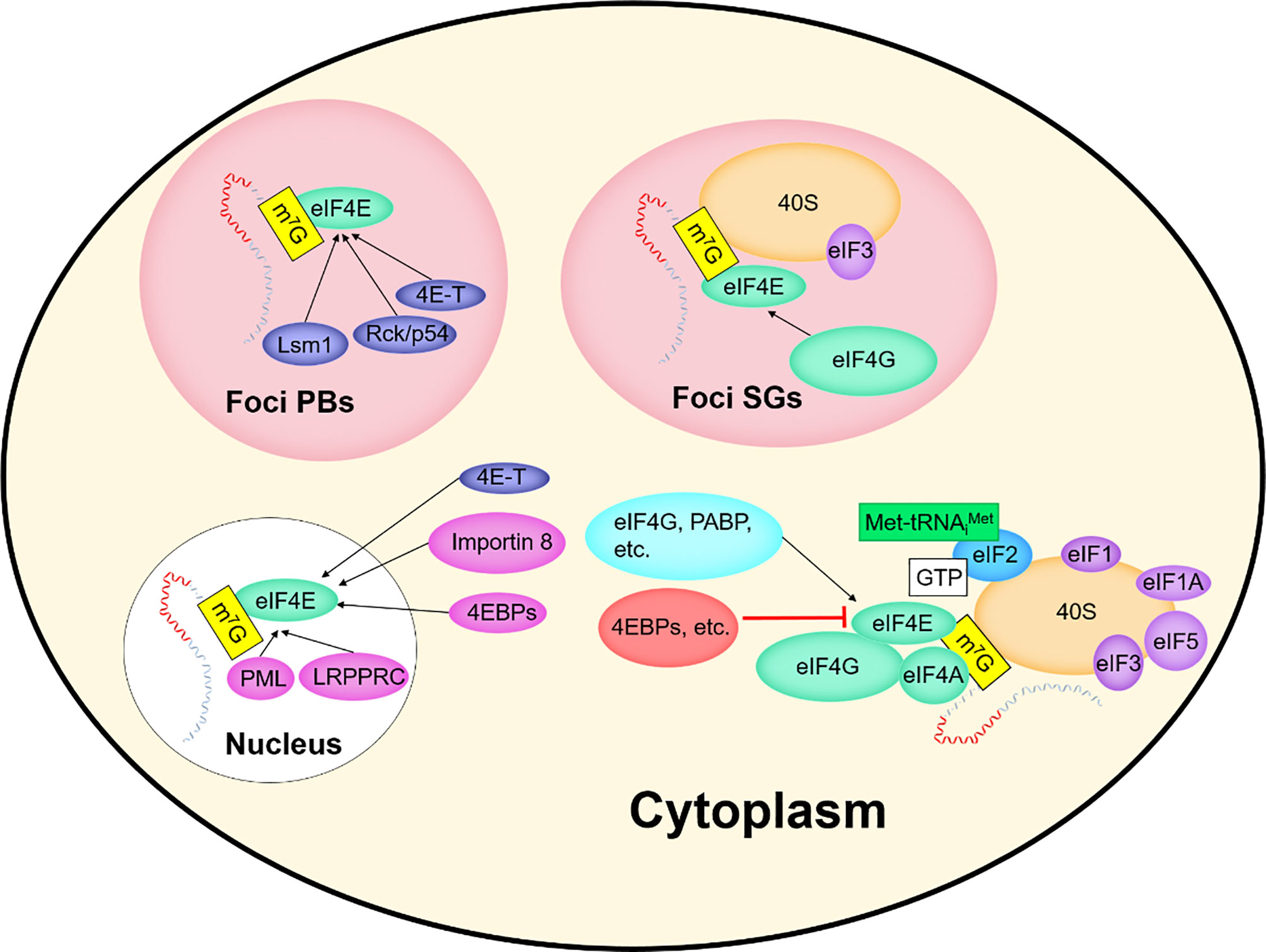
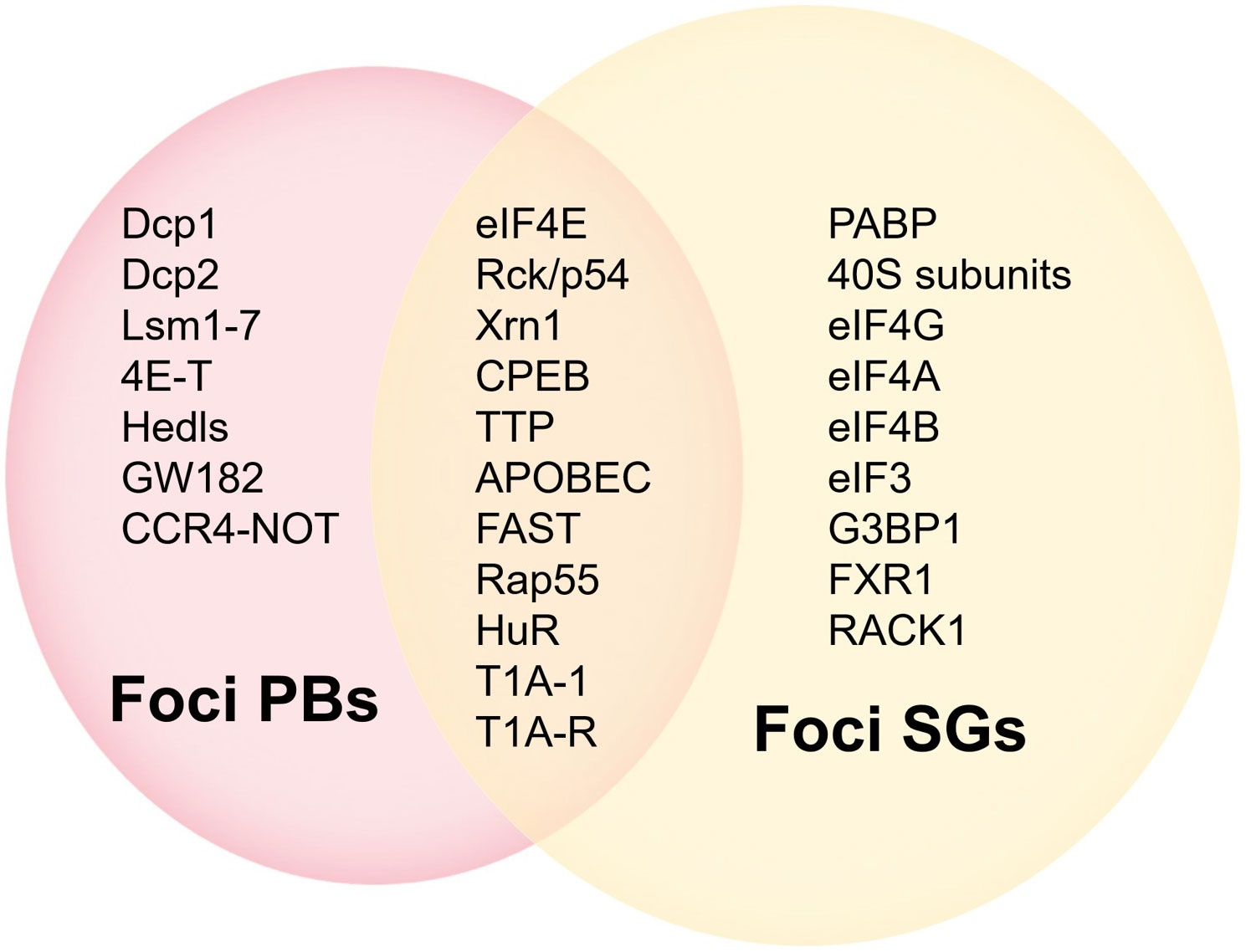
EIF4E plays a key role in mRNAs translation. In this paper, we associate eIF4E with the interacting protein 4E-IPs and focus on some functions of eIF4E: improving the translation efficiency of some mRNAs in the cytoplasm, mediating the nuclear export of mRNAs containing specific elements, and playing a role in the cytoplasmic foci. The activity and biological specificity of eIF4E depend on its interaction with different 4E-IPs. Over the years, about 47 different 4E-IPs have been found (12), which play an auxiliary role in the different functions of eIF4E (Figure 3).
3.1 EIF4E and nucleocytoplasmic transport
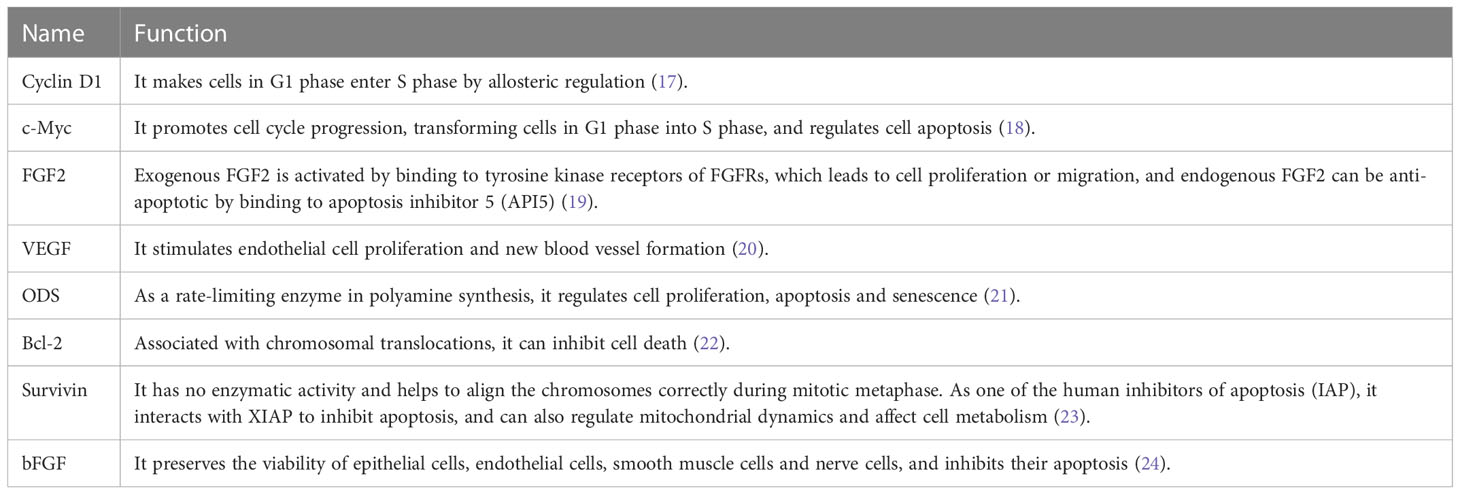
EIF4E is expressed in both nucleus and cytoplasm, with up to 68% of eIF4E distributed in the nucleus (13). In the nucleus, eIF4E binds the m7GpppN cap structure of transcripts and is solely responsible for regulating the nuclear export of eIF4E--dependent mRNAs, a class of mRNAs which mainly encode proteins related with proliferation and survival, such as Cyclin D1 (13–16) (Table 1), thus increasing their cytoplasmic concentration and protein production (14, 16, 25). The 3'UTR of this class of mRNAs have a structure of approximately 50 nucleotide units in length, which is termed the eIF4E sensitive element (4ESE) (14, 15). These eIF4E-dependent mRNAs are termed weak mRNAs after they are transported to the cytoplasm. Overexpression of eIF4E can alter the composition of the nuclear pore complex (NPC), increasing nuclear export of eIF4E-dependent mRNAs (26).
The transport mechanism of eIF4E-dependent mRNAs is different from that of general mRNAs. mRNPs exported by eIF4E differ from general mRNPs in the export pathway, as eIF4E and eIF4E sensitive mRNAs(eIF4E-dependent mRNAs) are not associated with general export factors such as TAP/NXF1 or REF/Aly (27, 28). Cofactors involved in eIF4E export in mammalian nuclei include LRPPRC proteins and the export receptor CRM1 (25). mRNPs exported by eIF4E consist of eIF4E, mRNA containing the 4ESE element, LRPPRC, and other components, but not REF/Aly (26).
CRM1 mediates the nuclear export of hundreds of different functional proteins, including many tumor suppressors, cell cycle regulators, and many mRNAs. Inhibition of CRM1 with Leptomycin B may abrogate the export of eIF4E-dependent mRNAs, suggesting that eIF4E may interact with CRM1 and eIF4E-dependent mRNAs transport is mediated by CRM1 (25, 29). GST pull-down assay proved that CRM1 could directly bind LRPPRC and act as an export receptor to form LRPPRC-eIF4E-mRNA-CRM1 complex (25). LRPPRC could immunoprecipitate eIF4E, and knockdown of LRPPRC reduced the ability of eIF4E to immunoprecipitate with 4ESE RNA in the nucleus, which directly suggested that eIF4E could interact with LRPPRC (25, 27).
3.2 EIF4E and translation
3.2.1 The mechanism of eIF4E in translation
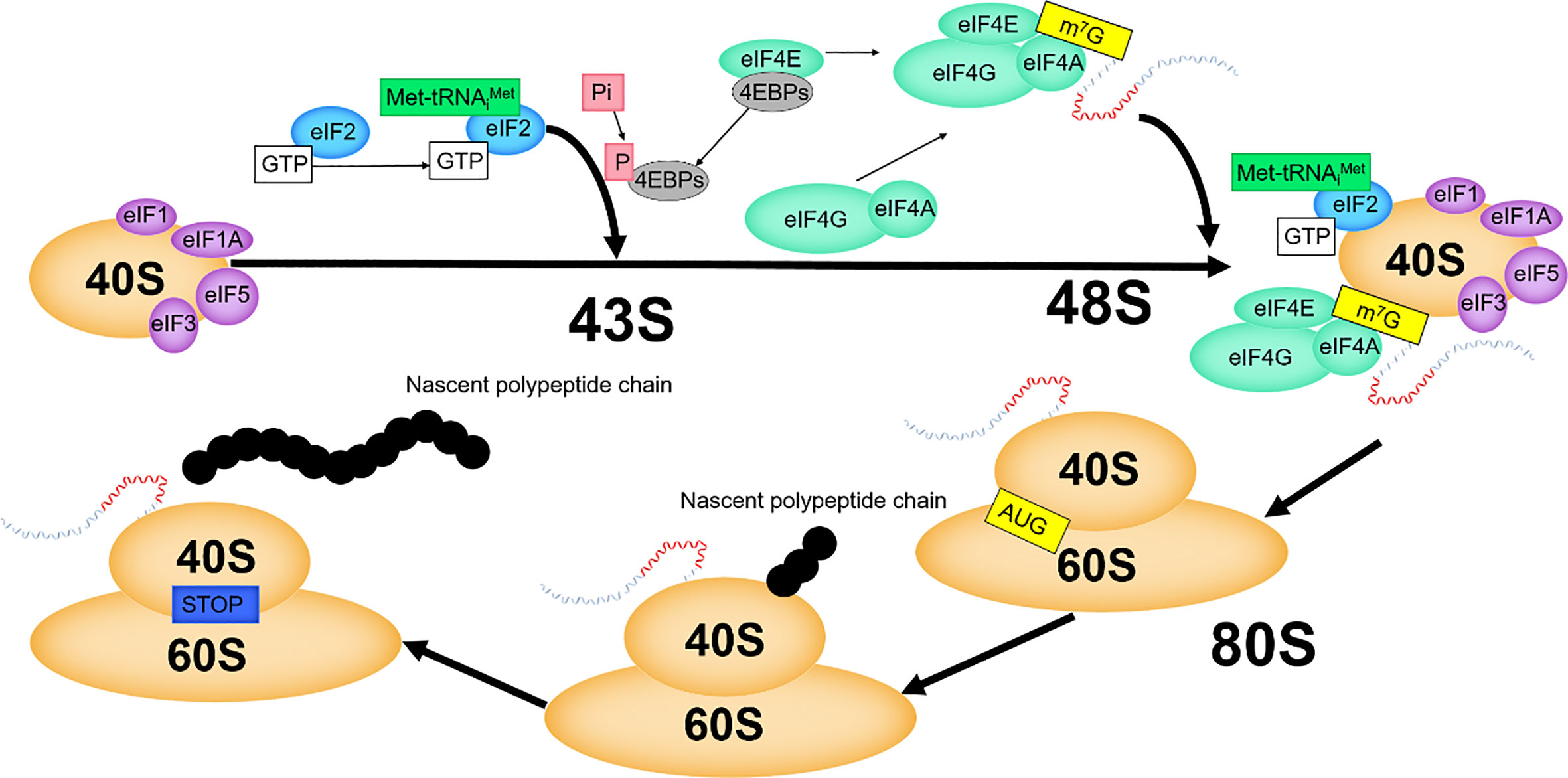
In eukaryotes, translation is initiated by decoding AUG in mRNAs, which is executed by the 48S pre-initiation complex (PIC) composed of the mRNAs, eIF2/GTP/Met-tRNAi ternary complex (TC), the small subunit of the ribosome, and the eIF4F complex. Then, 48S PIC combines with the large ribosomal subunit and assembles into the initiation complex. Subsequently, translation enters the elongation cycle (Figure 4).
EIF1, eIF1A, eIF3 and eIF5 promote the recruitment of TC to the small ribosomal subunit, and these factors then assemble to form 43S PIC (30, 31). In eukaryotes, eIF4F binds cap structure to activate mRNAs, which is crucial (30, 31). EIF4F promotes mRNAs to combine with the preassembled 43S PIC, forming 48S PIC (30, 31). By binding with the cap structure, eIF4E brings eIF4A to the 5' end of mRNAs, and makes eIF4A exert helicase activity to open the secondary structure of the 5' end of mRNAs. The interaction of eIF4G with eIF1, eIF3 and eIF5 plays an important role in facilitating the recognition of the 5' end of mRNAs by 43S PIC. The helicase activity of eIF4A is increased by the interaction of eIF4G with eIF4A (32). The eIF4E-eIF4G interaction is a key step in mRNAs recruitment, and this node is regulated by 4EBPs (33). After scanning from the 5' end to the 3' end of the mRNAs and positioning the start codon AUG (6), the 48S PIC changes from an “open” to a “closed” conformation to prevent scanning and binds to the 60S large subunit, which, with the release of initiation factors, eventually generates the 80S initiation complex with translation elongation capacity (34).
3.2.2 EIF4E improves translation efficiency
Due to the limited expression of eIF4E and its binding to inhibitory 4EBPs, the active of eIF4F complexes is normally limited. Therefore, intracellular mRNAs must compete for binding to eIF4F to initiate translation (35).
The “availability” of mRNAs in the cytoplasm varies considerably, with the length and structure of its 5'UTR significantly affecting translation efficiency (36). According to the “availability”, mRNAs can be divided into strong mRNA and weak mRNA. The 5'UTR of weak mRNAs are long and complex, with stable secondary structures, which hinders the association between the 43S PIC and mRNAs, and also the scanning for the 5'UTR. Weak mRNAs require RNA helicases (such as eIF4A) to open the secondary structure, so the translation efficiency is low (37). Proteins such as VEGF, FGF2, c-Myc, ODC, Cyclin D1, Bcl2 and Survivin are translated from weak mRNAs (35, 36, 38), and most of them can promote the development of cancer. Weak mRNAs are more susceptible to regulation by eIF4E than other mRNAs and are known as eIF4E sensitive genes (13), and they are less translationally competent when eIF4F complex formation is restricted (36). The 5'UTR of strong mRNAs are short and simple, with low GC contents and less stable secondary structures, which can be effectively scanned to initiate start codon recognition. For example, β-actin and GAPDH encoded by strong mRNAs (35, 36), are efficiently translated even when eIF4F is limiting.
EIF4E in the cytoplasm is a translation enhancer of weak mRNAs, especially during cellular stress and proliferation (15, 16). When the levels of eIF4E are limited, the effect on the translation of strong mRNAs is minimal (36). When the levels of eIF4E are increased, the translation of weak mRNAs is disproportionately increased, there is a nonlinear relationship between the increase of eIF4E and the increase rate of weak mRNAs translation. With the increase of eIF4E, the increase rate of weak mRNAs translation is variable rather than constant (35, 36). Numerous studies have shown that the elevation of eIF4E levels preferentially enhances the translation of weak mRNAs (35), leading to tumorigenesis and metastasis.
3.3 EIF4E and cytoplasmic foci
The processing, transportation, degradation and translation of mRNAs are essential for gene expression regulation (39, 40), and these processes are controlled by specific RNA-binding proteins (RBPs) (39, 40). Many RBPs bind to mRNAs and assemble into mRNPs (41). After being exported from the nucleus, some mRNPs are transported to specific regions of subcellular localization, cytoplasmic foci (40, 42). Non-membrane-enclosed cytoplasmic foci, such as Stress Granules (SGs) and processing bodies (P-Bodies, PBs), dynamically sequester untranslatable mRNPs into compartments distinct from the surrounding cytoplasm. Both cytoplasmic foci are associated with translation events that affect cell survival (40). Representatives of the various enzymes required for the breakdown of mRNAs into their constituent parts including a deadenylase, a decapping enzyme, and an exonuclease, are concentrated in the cytoplasmic foci (43).
PBs differ from SGs in composition and function (Figures 3, 5) (44, 45), but share some components and attributes, including eIF4E, mRNAs, and RBPs. Several components contained in PBs, such as eIF4E-T, Lsm1 and Rck/p54, are required for eIF4E accumulation in PBs. However, PBs lack SGs-associated eIF3, PABP, small ribosomal subunits, and many signaling proteins, and eIF4G accumulates only in SGs (40, 46, 47). Tryptophan residues needed for recognizing mRNA cap structure are not essential for the recruitment of eIF4E to PBs or SGs, and those required for protein-protein interactions are critical for eIF4E accumulation in cytoplasmic foci (46).
PBs are RNPs aggregates that contain a variety of proteins associated with mRNAs degradation (46, 48). PBs are associated with miRNA induced translation silence and siRNA induced mRNA degradation (43, 46). PBs can also act as a “storage depot” for mRNAs, where the stored mRNAs can be recycled under certain conditions (48). Among the new studies, researchers have identified two decapping activators, Dhh1p and Pat1p. Approximately only 10% of cells with a deletion of both proteins can form PBs, rendering the cells unable to normally inhibit the translation of some of the proteins that should not be expressed. The human homologue of Dhh1p is Rck/p54, which functions to inhibit translation in vitro (49). However, eIF4E can contact with 4E-T and Rck/p54 molecules in PBs in vivo, and this implies that the role played by eIF4E in cytoplasmic foci is also relevant to translation. PBs are rich in mRNAs degrading enzymes and contain eIF4E but not eIF4G, suggesting that eIF4E may play an early role in the transition from active translation to degradation of mRNAs. Moreover, due to the protection of the cap structure by eIF4E, some mRNAs, although their translation were repressed, may not be immediately targeted for degradation in the 5' to 3' direction (47).
Under certain non-physiological conditions in higher eukaryotes, such as heat or chemical stress, inactive mRNPs accumulate in the cytoplasm and SGs appear. SGs contain most of the components of 48S PIC but lack eIF2, which are called “abnormal aggregates of 48S PIC” (46, 47). SGs affect translation and stability of mRNAs and are associated with apoptotic. Disrupting eIF4F complexes, such as interfering with eIF4E activity, can lead to SGs formation.
The above studies suggest a novel role of eIF4E in mRNAs translation in cytoplasmic foci.
3.4 EIF4E interacting proteins
3.4.1 LRPPRC and importin 8
LRPPRC is a leucine rich pentapeptide repeat protein implicated in the nuclear export of eIF4E. LRPPRC acts as a specific factor to recruit mRNAs containing 4ESE in the nucleus, and can directly bind eIF4E and 4ESE-containing mRNA through the N and C termini respectively, and binds CRM1 at the same time, thereby transporting the complex to the cytoplasm through the nuclear pore. LRPPRC is a central component of this mRNAs export pathway and is the first factor to function in eIF4E-sensitive mRNAs selection (25, 27). LRPPRC mediated nuclear output requires the integrity of its binding site with eIF4E (27).
EIF4E nuclear import is directly mediated by importin 8 and is competitively regulated by the m7G cap structure and importin 8 (50).
3.4.2 4EBPs and 4E-T
4EBPs, widely distributed in the nucleus and cytoplasm, are eIF4E binding proteins and usually inhibit the role of eIF4E in translation. 4EBPs share and competitively bind to a common eIF4E binding site with eIF4G (10, 11), preventing their assembly into eIF4F. 4EBP1 and 4EBP2 are expressed in most tissues, while 4EBP3 shows a more limited expression pattern (51). Binding of 4EBPs to eIF4E is regulated by their phosphorylation status. Non-phosphorylated 4EBPs (active) bind to eIF4E with high affinity and phosphorylated 4EBPs (inactive) can not bind to eIF4E (52–54), and the expression of phosphorylated 4EBPs is associated with malignant progression and poor prognosis. Deletion mutations of 4EBPs genes are frequently found in patients with pancreatic cancer and head and neck cancer, suggesting a tumor-suppressive role of 4EBPs in some cancers (4).
4E transporter (4E-T), an eIF4E binding protein (4EBPs) present in PBs, represses translation and promotes mRNAs degradation and interacts with factors including DDX6, Lsm14 and the Lsm1-7-PAT1 complex (55, 56).
3.4.3 PRH
Proline-rich homeodomain proteins (PRH) are composed of a proline-rich N-terminal domain, a central homeodomain that binds to specific DNA sequences, and an acidic C-terminal domain. This protein is encoded by a haematopoietically expressed homeobox (HHEX) gene (57), and acts on eIF4E in the nucleus. PRH is a regulator of transcription and translation, and plays an important role in the control of cell proliferation and differentiation (58), early embryonic patterning and hematopoietic processes (59), and is essential for forebrain, liver, and thyroid development. PRH is highly expressed in pluripotent erythromyeloid and B cell progenitors and is downregulated during differentiation of most hematopoietic lineages (60). In addition to hematopoietic cells, PRH is expressed in a limited set of tissues in the adult, including myeloid cells, lung, thyroid, and liver tissues, and has been localized as a tissue-specific regulator of eIF4E. Proline-rich N-terminal domain is essential for the inhibition of myeloid cell proliferation and cell transformation (58). PRH can directly interact with eIF4E through a conserved binding site and inhibit the transformation and growth promotion function of eIF4E by inhibiting its mRNA transport activity (61).
3.4.4 PML
The promyelocytic leukemiaprotein, PML, acts on eIF4E in the nucleus. The integrity of PML nuclear body depends to some extent on the integrity of eIF4E nuclear body, whereas the integrity of eIF4E nuclear body is not related to PML (62, 63). PML contains three cysteine-rich zinc-binding domains, called RING and B-box (B1 and B2), which are required for the transformation suppressive and pro-apoptotic functions of PML (64). PML binds to the back of eIF4E, including Trp73, using a region around the first zinc binding site of the RING domain. The RING is essential for some physiological functions of PML, including nuclear bodies formation, growth inhibition and apoptosis (62, 65). Mechanism of PML binding to eIF4E as detailed later in “intranuclear regulation of eIF4E”.
3.4.5 EIF4G and eIF4A
EIF4G is a large scaffold protein associated with DEAD-box proteins, with domains that bind to mRNA, eIF4E, eIF4A, poly (A)-binding protein (PABP), eIF3, eIF1, and eIF5 (2, 3, 5, 6).
EIF4A is a protein with a molecular weight of 44kDa and an ATP-dependent helicase belonging to the DEAD-box protein family, which is thought to unwind the secondary structure of mRNAs (4), facilitate scanning and exposure of the start codon of mRNAs (30, 36, 66).
4 Regulation of eIF4E expression
EIF4E is often highly expressed in tumor cells, and few studies have investigated how the levels and activity of eIF4E are elevated in malignant cells.
4.1 Increased gene copy number
Sorrells and Haydon et al. investigated eIF4E gene copy number by using western blot and PCR. Overexpression of eIF4E in breast cancer samples was associated with increased gene copy number, which was not detected in benign breast tissue (67). EIF4E gene copy number is increased in invasive carcinomas compared to normal tissues and benign tumors. The extent of eIF4E gene copy number increase is variable within each tissue category, and progression of malignant phenotype appears to be related to the extent of eIF4E gene copy number increase, which may be one of the mechanisms of eIF4E oncoprotein overexpression (68).
4.2 Increased stability of eIF4E mRNA
The changes in the stability of eIF4E mRNA may be related to the expression of eIF4E. EIF4E 3'UTR contains a unique AU-rich elements (AREs), which is responsible for binding eIF4E transcriptional stability regulators. HuR (69) and AUF1 p42 isoform are two factors that regulate transcriptional stability of eIF4E and competitively bind to the mRNA 3'UTR of eIF4E. In malignant tumor specimens with high expression of eIF4E, HuR is up-regulated, and its deletion leads to a decrease in eIF4E levels. With the increase of HuR level, the stability of eIF4E mRNA in cancer cells is improved. HuR and eIF4E regulate common transcripts involved in cell proliferation (Cyclin D1 and c-Myc) and neovascularization (VEGF). The p42 isoform of AUF1 interacts with the 3'UTR of eIF4E mRNA and decreases its stability (70).
4.3 Epigenetic modification
4.3.1 Methylation
Protein arginine methyltransferase 5 (PRMT5) expression is significantly upregulated in a variety of cancer cells (71). Knockdown of PRMT5 decreases the methylation levels of H3 and H4 on promoter of eIF4E, thus reducing eIF4E expression (72–74). In colorectal cancer (CRC) tissues, the expression of eIF4E is positively correlated with PRMT5, silencing PRMT5 leads to decreased eIF4E expression (75). This helps to gain a deeper understanding of the overexpression of eIF4E in tumor cells.
4.3.2 MiRNAs regulation
MiRNAs expression is frequently dysregulated in many diseases such as cancers and immune disorders, which is associated with the development of many pathological conditions and diseases (76). Down-regulation of miR-141 increases the expression of eIF4E, VEGF and c-Myc in H1299 or H2009 cells (77, 78). MiRNA-455-3p directly targets eIF4E and inhibits cap-dependent translation (79). On the other hand, miRNAs can recruit eIF4E2 to compete with eIF4E for binding to mRNAs, induce the dissociation of eIF4A from target mRNAs, block the assembly of the translation initiation complex, and thus inhibit mRNAs translation (80).
5 EIF4E and metabolic reprogramming
In 2011, Hanahan and Weinberg summarized immune evasion and metabolic reprogramming as the main characteristics of tumor cells (81). The overexpression of eIF4E plays an up-regulated role in both lipid metabolism and glycolysis, which suggests that overexpression of eIF4E can promote cancer by affecting metabolism.
5.1 EIF4E and glycolysis
Activation of mTORC1/eIF4E pathway promotes the translation of HIF-1α and up-regulation of HIF-1α can increase the expression levels of several glycolytic enzymes to drive glycolysis and produce more ATP (82, 83).
5.2 EIF4E and lipid metabolism
In 2012, about 3.9% (544300 cases) of cancers worldwide were related to obesity. In 2016, the International Agency for Research on Cancer concluded that there was a causal relationship between obesity and 13 cancers (84). Fang et al. reported that BMI is significantly positively correlated with endometrial cancer, esophageal adenocarcinoma and kidney cancer, while it is significantly negatively correlated with oral cavity cancer, lung cancer, premenopausal breast cancer and localized prostate cancer (85). Davide et al. showed that eIF4E single copy knockout mice gained only half the weight of normal mice under the same feeding conditions, suggesting that eIF4E is associated with obesity and that reducing eIF4E expression levels can enhance lipid metabolism and suppress obesity (86). Specifically, mRNAs involved in lipid storage pathways are translated in an eIF4E-dependent manner. Knockdown of eIF4E, which renders these mRNAs unable to be upregulated, leads to increased fatty acid oxidation and energy expenditure. Inhibition of eIF4E phosphorylation can suppress weight gain after intake of a high-fat diet. In conclusion, the translation control of eIF4E may be the driving factor of weight gain induced by high-fat diet and eIF4E can be used as a pharmacological target for obesity treatment (86).
6 Main factors affecting eIF4E activity
6.1 Intranuclear regulation of eIF4E
Both cytoplasmic and nuclear functions of eIF4E depend on its ability to bind to mRNA. Intranuclear regulation of eIF4E activity is mainly achieved by two proteins: PRH and PML, which are negative regulators of eIF4E-dependent mRNAs export (13, 87, 88).
PRH directly interacts with eIF4E through a conservative binding site, and inhibits the growth-promoting function of eIF4E by inhibiting its mRNAs transport function (61), and this is similar to that observed in the proline rich N-terminal region of eIF4G and 4EBP1 (62). PRH plays a major role in myelopoiesis, acting as both a tumor suppressor and an oncogene (60, 87). Alterations in the expression and intracellular localization of PRH are associated with breast, liver, thyroid cancers and certain leukemia subtypes. PRH is up-regulated in certain types of lymphoid leukemia (60), while it is down-regulated and eIF4E is up-regulated in some types of myeloid leukemia (AML, CML) (60, 87). In addition, PRH is a tissue-specific inhibitor of eIF4E-dependent Cyclin D1 mRNA transport (61). The level of Cyclin D1 is increased in leukemic specimens (87). Multiple PRH target genes including the genes encoding VEGF and VEGF receptors, are known to be important in the control of cell proliferation and cell survival (57). These findings suggest that the transport disorder of eIF4E mediated by PRH may be related to hematological malignancies.
PML is a potent inhibitor of eIF4E-dependent mRNAs export (62, 89), and is the first factor reported to modulate nuclear eIF4E functions (64). PML was shown to co-localize and co-immunoprecipitate with nuclear eIF4E (13). The RING domain of PML regulates the activity of eIF4E by directly interacting with the backside of eIF4E to drastically reduce (over 100 fold) the affinity of eIF4E for the 5' end cap structure of mRNAs (64, 88, 90–92). The back side of eIF4E is also the binding region for eIF4G and 4EBPs, suggesting that this region contributes to the positive and negative regulations of eIF4E activity (65). Overexpression of PML can inhibit AKT activity, leading to decreased phosphorylation levels of 4EBP1, which in turn affects cell survival, so PML may regulate eIF4E by inhibiting AKT activation (90).
Unlike the limited expression of PRH, PML is expressed in all mammalian cells and almost exclusively distributed in the nucleus, whereas eIF4E and PRH have a punctate distribution in the nucleus and the cytoplasm (61), but transport of eIF4E-dependent mRNAs is only inhibited by the nuclear part of PRH (87).
6.2 Phosphorylation of 4EBPs
The PI3K-AKT-mTOR signal transduction system significantly affects mRNAs translation by regulating the phosphorylation of its downstream targets. Growth factors, mitogens, and hormones can activate the PI3K-AKT-mTOR signaling pathway (93). Activated mTOR promotes the phosphorylation of 4EBPs (94). mTOR can form two distinct complexes, mTORC1 and mTORC2, which differ in their compositions, downstream targets, and sensitivity to rapamycin (95). The mTORC1 signaling pathway controls many cellular biological processes including mRNAs translation, cell growth and proliferation, and its downstream substrates mainly include 4EBPs and S6K (96). 4EBPs can regulate cell proliferation and S6K can regulate cell growth (97). mTORC1 sequentially phosphorylates 4EBP1 at T70 and S65, enabling the release of eIF4E to allow eIF4F assembly (54). Activated S6K can affect multiple factors involved in eIF4F assembly, promoting the recruitment of eIF4B to the pre-translational initiation complex for binding to eIF4A (98). mTORC2 controls cell survival by regulating the activity of AGC kinases, such as AKT, SGK1, and regulates the stability of nascent peptide chains. Studies have shown that more than 70% of cancer patients have excessive activation of mTOR (96).
A variety of factors can affect the phosphorylation of 4EBPs, such as endotoxin (LPS), insulin-like growth factor-1 protein (IGF-1) and branched-chain amino acids (99). LPS can transiently decrease phosphorylated 4EBP1 and increase the binding of 4EBP1 to eIF4E, leading to downregulation of IGF-1 expression (100), and 4EBP1 is inactivated by hyperphosphorylation in response to IGF-1 stimulation (101). In addition, O-glucoylation of 4EBP1 may regulate interaction between eIF4E and eIF4G, for example, when the level of O-glucoylation increases the binding of eIF4E to eIF4G decreases (102).
6.3 Phosphorylation and sumoylation of eIF4E
6.3.1 Phosphorylation of eIF4E
The only kinases that phosphorylate eIF4E are mitogen-activated protein kinase interaction kinases MNK1 and MNK2 (103). The phosphorylation of eIF4E can stimulate the translation of tumor-promoting mRNAs, thus enhancing its carcinogenic characteristics (104).
RAS-RAF-MEK-ERK-MNK and ASK1-MKK3/6-p38-MNK cascade reactions activate transcription factors and regulate gene expression. The cascade involves three upstream kinases, namely mitogen-activated protein kinase (MAPK), MAPK kinase (MAPKKs/MEK/MKK) and MAPKK kinase (MAPKKKs). The three kinases integrate upstream signals and transmit them to a series of downstream effectors. MAPK pathway has four main branches, namely extracellular signal-regulated protein kinase (ERK), p38 mitogen-activated protein kinase (p38 MAPK), c-Jun N-terminal kinase (JNK) and extracellular signal-regulated protein kinase 5 (ERK5) (105). MEK1 and MEK2 are activators of ERK1/2, MEK5 is an activator of ERK5, MKK4 and MKK7 are activators of JNK, and MKK3 and MKK6 are activators of p38 MAPK.
6.3.1.1 RAS-RAF-MEK-ERK-MNK pathway
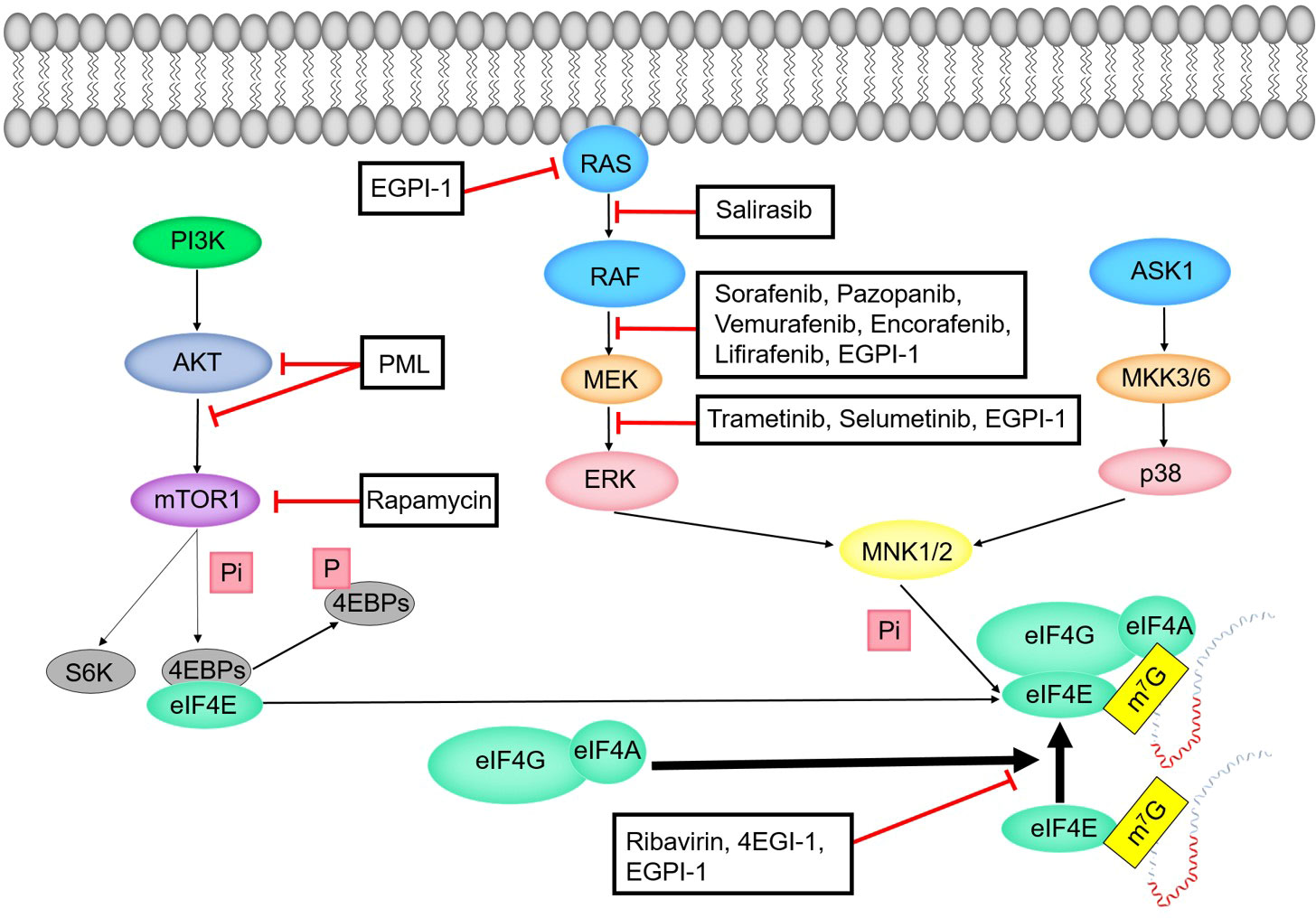
The process of phosphorylation of eIF4E by RAS-RAF-MEK-ERK-MNK pathway can be divided into five stages (Figure 6). In the first stage, RAS protein recruits and activates protein kinase RAF. RAS protein is related to many physiological processes such as cell metabolism, cell proliferation and apoptosis (106). RAS is often activated in a variety of human cancers (106). About 19% of cancer patients have RAS mutations (107). The RAS mutation that causes the constitutive activation of MAPK pathway is the most common mutation in human cancers (108). In the second stage, RAF, as MAPKKK, starts the activity of MEK by phosphorylating the activated fragment of MEK. In the third stage, MEK transmits signals by phosphorylating the activated fragment of ERK1/2. ERK1 and ERK2 belong to the mitogen activated protein kinase family. MAPK/ERK pathway is related to cell proliferation, differentiation, migration, aging and apoptosis (105). In the fourth stage, activated ERK1/2 is transferred to the nucleus to phosphorylate a variety of substrates including transcription factors (108–110). In the fifth stage, MNK1 and MNK2 phosphorylate the conserved site S209 of eIF4E after they were activated by ERK and p38 (103, 111). When eIF4E is combined with the N-terminal of eIF4G, MNK1/MNK2 is combined with the C-terminal of eIF4G. In other words, MNK phosphorylates S209 of eIF4E with eIF4G as the docking site (112, 113).
6.3.1.2 ASK1-MKK3/6-p38-MNK pathway
The process of phosphorylation of eIF4E by ASK1-MKK3/6-p38-MNK pathway can be divided into four stages (Figure 6). In the first stage, apoptosis signal regulated kinase 1 (ASK1) is the MAPKKK of JNK and p38 MAPK pathways. ASK1 is preferentially activated by various cytotoxic stressors and plays a key role in various cellular responses (114). In the second stage, ASK1 activates two different subgroups of MAPKK, SEK1 (or MKK4) and MKK3/MKK6. In the third stage, MKK3/MKK6 activates the p38 subgroup of MAPK (115, 116). In the fourth stage, p38 MAPK activates MNK to phosphorylate eIF4E.
6.3.2 Sumoylation of eIF4E
Many targets related to sumoylation are linked to transcription, DNA repair and nucleocytoplasmic transport. Sumoylation of eIF4E can enhance its translation activity (117) and promote the formation of active eIF4F, and its destruction can inhibit the translation of eIF4E-dependent mRNAs. More than 20% of eIF4E combined with cap structure of mRNAs are sumoylated, which indicates that sumoylation may be crucial to the functions of eIF4E at the initiation of translation. Sumoylation does not directly affect the cap binding activity of eIF4E, but promotes the dissociation of eIF4E with 4EBP1. At the molecular level, sumoylation may regulate the function of proteins by changing their conformations. The conformational change of eIF4E may increase its affinity with proteins that have a certain conformation (such as eIF4G), and decrease its affinity with 4EBP1 and other proteins. Phosphorylation of S209 is necessary for eIF4E sumoylation (117, 118), while phosphorylation of eIF4E does not require its sumoylation. Histone deacetylases (HDACs) induce the formation of active eIF4F complexes by stimulating eIF4E sumoylation (119).
7 EIF4E and tumorigenesis
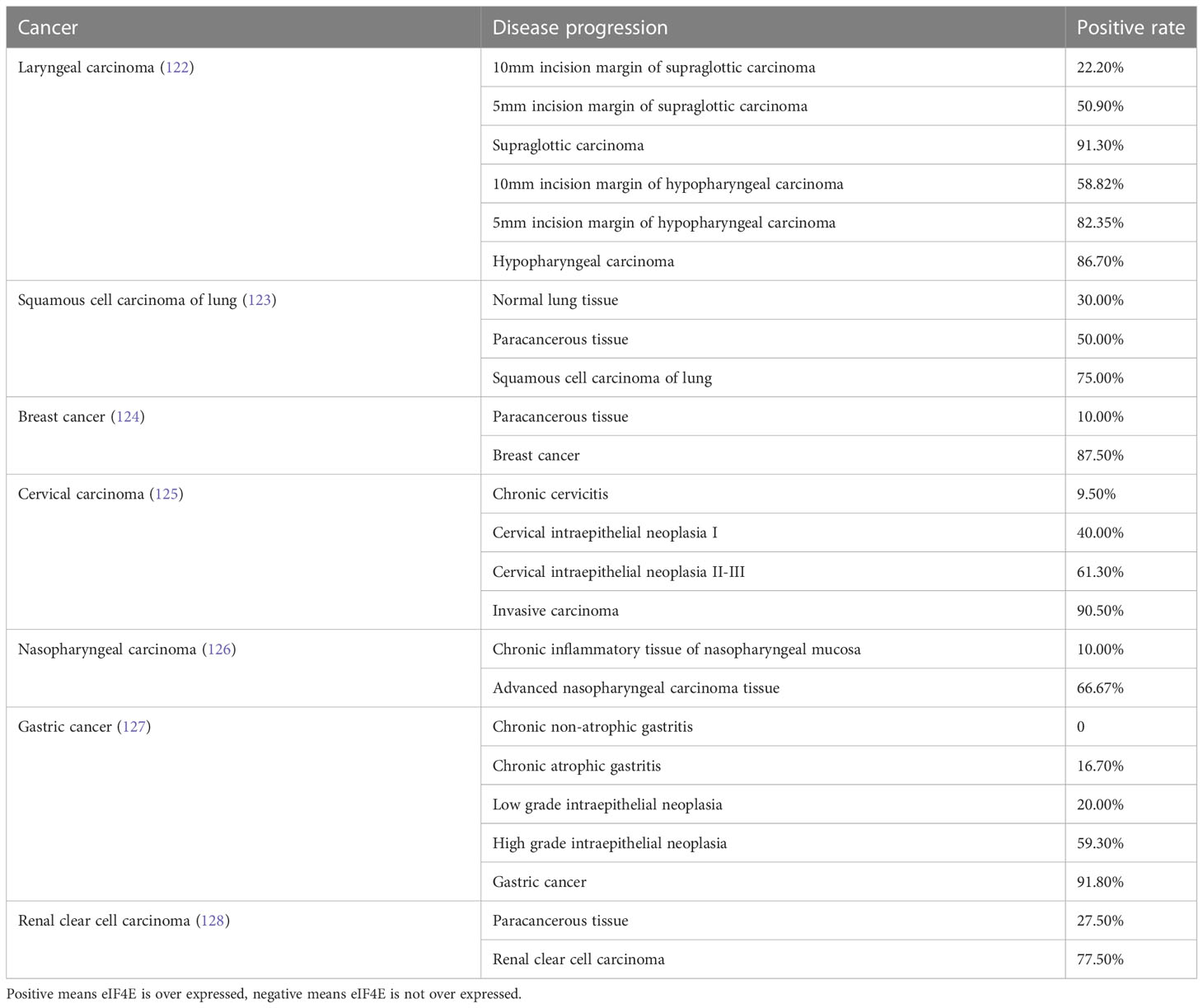
EIF4E plays a crucial role in the malignant transformation, progression and drug resistance of many human solid tumors (120). When tissue cells are stimulated by growth factors, mitogens, et al., multiple pathways work together to increase the expression and activity of eIF4E. Overexpression of eIF4E can selectively enhance the translation of weak mRNA that is not expressed or weakly expressed in normal cells, resulting in the abnormal expression of these genes related to cell cycle, vascular growth and cell survival. Then the translation of multiple essential proteins in the process of tumor occurrence and development is increased, promoting cell proliferation and malignant transformation, increasing the malignant degree of tumor tissues, facilitating the formation of tumor blood vessels and thus providing more nutrition for tumor tissues, which is conducive to the growth, invasion and metastasis of tumor cells. There is a common mechanism for eIF4E in different cancers: eIF4E mediates normal cell proliferation, but induces tumorigenesis when it is dysregulated and over expressed (121). The expression of eIF4E in tumor tissue is significantly higher than that in normal tissue. With the progress of cancer, the expression of eIF4E is increasing. In addition, overexpression of eIF4E will increase the probability of cancer recurrence. Therefore, eIF4E is a marker of tumor progression, malignant transformation, metastasis, and poor prognosis. It is also a sensitive molecular marker in tumor detection (Table 2).
7.1 Expression of eIF4E in laryngeal carcinoma
Franklin et al. showed that eIF4E in the tumor area of patients was over expressed, and the expression of FGF2 and VEGF was increased. The overexpression of eIF4E at the incisal edge would increase the probability of tumor recurrence (129). Liang et al. found that eIF4E was over expressed in laryngeal squamous cell carcinoma samples, but not in vocal cord polyps samples. Overexpression of eIF4E led to up regulation of bFGF. EIF4E and bFGF played a synergistic role in the genesis, development, invasion and metastasis of laryngeal squamous cell carcinoma (130). Yi et al. detected tumor markers such as Cyclin D1, p53 and eIF4E at the surgical margin, and the results showed that eIF4E was the most sensitive molecular indicator among these molecules (131).
7.2 Expression of eIF4E in lung cancer
Chan et al. found that the overexpression of 4EBP1 led to the decrease of eIF4E, and the expression of FGF2 and VEGF was inhibited, which could inhibit the proliferation of K-RASLA1 mouse lung cancer cells (132). According to Dong et al., the expression of eIF4E in the serum of NSCLC patients was significantly higher than that in healthy individuals, and eIF4E was an independent prognostic factor for shortening overall survival and progression free survival (133). According to Qi et al., eIF4E was highly expressed in multiple lung cancer cell lines, and siRNA-eIF4E could significantly inhibit lung cancer cell proliferation (134). Yoshizawa et al. showed that the overexpression rate of p-eIF4E in 300 NSCLC tissues was 39.9%, suggesting that the activity of eIF4E in NSCLC was increased (135).
7.3 Expression of eIF4E in breast cancer
Kerekatte et al. showed that eIF4E was overexpressed in breast carcinomas, but not in normal breast tissue and benign breast lesions (hyperplasia or inflammation) (136). Derek et al. believed that the expression level of eIF4E was related to tumor recurrence, and eIF4E might increase the risk of recurrence of breast cancer (137). Avdulov et al. found that the level of p-4EBP1 in breast cancer cells was increased, and the assembly ability of eIF4F was enhanced (53). Yang et al. showed that the overexpression of eIF4E was related to the formation of various human malignant tumors, including breast cancer (138). RNA interference system driven by the Survivin promoter efficiently and specifically downregulated eIF4E expression in human breast cancer cells but not in normal human breast epithelial cells. Therefore, RNAi driven by Survivin promoter targeting eIF4E could be used as an adjuvant therapy tool for human breast cancer, with tumor specificity and efficiency (120).
7.4 Expression of eIF4E in head and neck squamous cell carcinoma
Haydon et al. found that the amplification and overexpression of eIF4E gene gradually increased from normal head and neck tissues to benign tumors, and ultimately to invasive cancer cells in the head and neck. This expression characteristic of eIF4E makes it a potential tumor detection indicator (68). Patients with head and neck squamous cell carcinoma (HNSCC) have a high risk of metastasis and recurrence, and aberrant activation of PI3K/AKT/mTOR signaling occurs in approximately 80% of HNSCC, which has been suggested as a prognostic biomarker for patients with recurrence or metastasis (139). The expression of eIF4E was elevated in HNSCC, and its overexpression made VEGF and FGF2 preferentially up-regulated. Inhibition of eIF4E expression by antisense RNA could reduce the tumorigenicity and angiogenesis of cells. Antisense RNA therapy of eIF4E might be used as an adjuvant treatment for head and neck cancer, especially when eIF4E was found to be elevated at the surgical margin (140). The expression of eIF4E and Cyclin D1 increases in HNSCC, both of which could stimulate cell cycle progression and transform squamous epithelial cells (141). In HNSCC, eIF4E seems to be a more significant prognostic indicator than p53 (142). The expression of eIF4E relative to 4EBP1 is a more precise predictor of HNSCC and its progression (143).
7.5 EIF4E and colorectal carcinoma
Niu et al. showed that eIF4E was an indicator of tumor progression and poor prognosis in colon cancer patients (144). Gao et al. found that eIF4E was up-regulated in CRC, and its expression frequency (EF) in cancer tissues was higher than that in normal adjacent tissues (145). Ruan et al. showed that eIF4E could significantly affect CRC organoid growth (146).
7.6 EIF4E and leukemia
The elevation of eIF4E level will lead to the imbalance of eIF4E-dependent mRNAs transport, which will hinder the differentiation of granulocytes and monocytes, especially in myeloid leukemia and may contribute to the occurrence of leukemia. This abnormality of eIF4E is caused by the imbalance of PRH (87).
In acute myeloid leukemia (AML), the increased expression and phosphorylation of eIF4E are associated with poor prognosis, and phosphorylated eIF4E can be used as a prognostic indicator and potential anti-cancer target for biological therapy of AML (147, 148). In chronic myeloid leukemia (CML), the expression and phosphorylation level of eIF4E are up-regulated, and the activity of eIF4E in patients with advanced CML is significantly increased. In addition, abnormal activation of eIF4E in leukemic stem cells of CML in blast phase significantly increases the synthesis of β-Catenin (149).
Changes of many signal pathways have been found in acute lymphoblastic leukemia (ALL), including RAS-RAF-MEK-ERK and PI3K-AKT-mTOR pathways (150). Inhibition of eIF4E may be a new method to treat pre-B cell leukemia while preserving the development and function of normal B cells (7).
8 Targeted therapy of eIF4E
EIF4E specific antisense oligonucleotide (4EASO) is a new drug that can reduce the level of eIF4E and the formation of eIF4F complex (151, 152). 4EASO can effectively down-regulate the expression of eIF4E protein in breast and prostate tumor xenografts (153), significantly inhibits tumor growth by binding to eIF4E mRNA, triggering RNA degradation mediated by RNA enzyme H (153). The early generation of ASO lacks nuclease resistance and tissue stability, resulting in a relatively short half-life and significantly reduced efficacy. In contrast, the efficacy, nuclease resistance and tissue half-life of the second generation ASO were significantly improved (154). LY2275796 is a second-generation antisense anticancer drug (Figure 7 and Table 3).
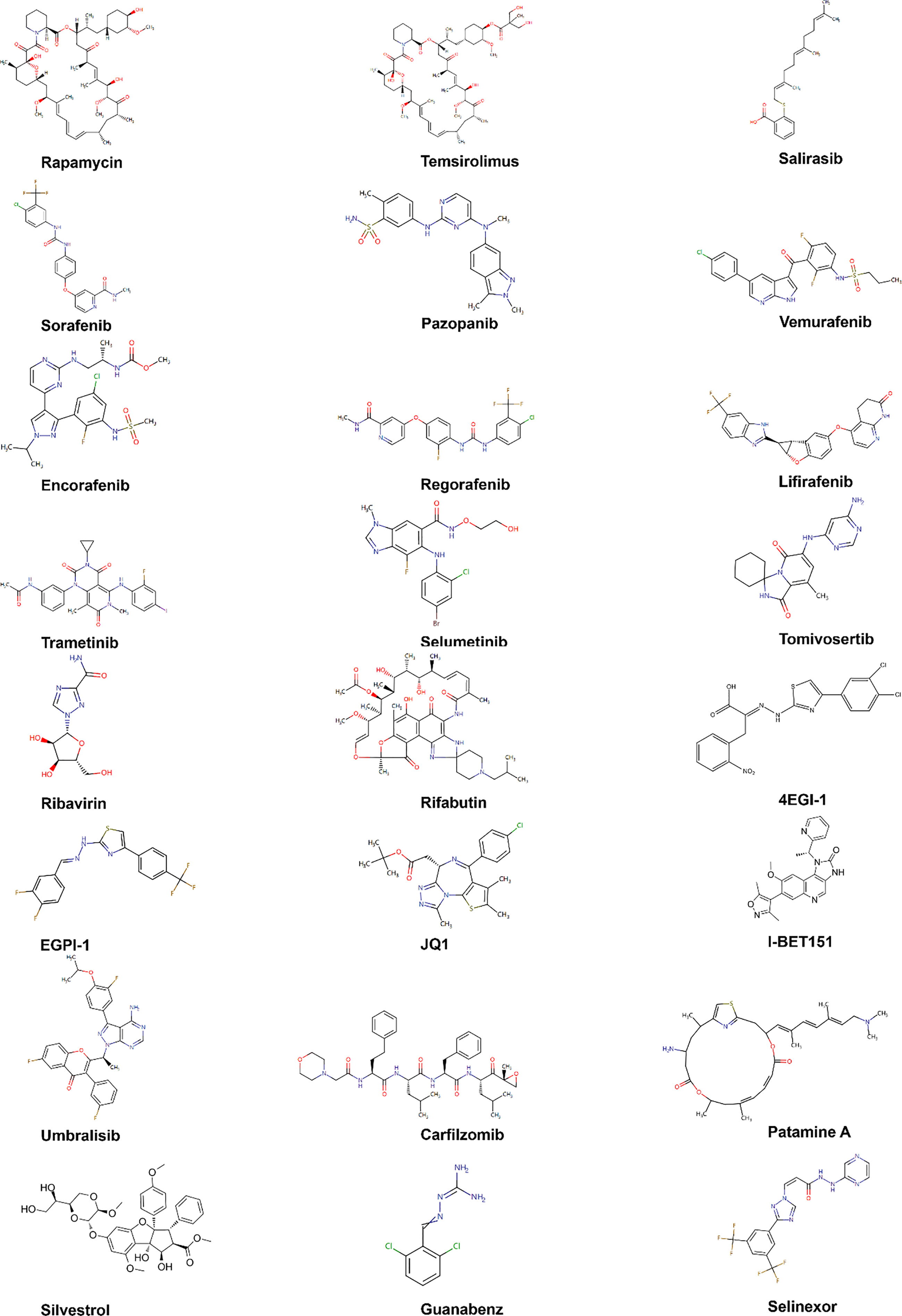
Figure 7 Chemical structures of inhibitors. Chemical structures are sourced from the DrugBank database (https://go.drugbank.com/).
Rapamycin is an FDA approved antibiotic and immunosuppressant against mTOR, which can inhibit the kinase activity of mTOR1. Unlike rapamycin analogues, ATP competitive mTOR inhibitors target ATP binding sites (179), weakening the activity of mTORC1 and mTORC2 (179, 180). At present, many clinical trials of ATP competitive mTOR inhibitors for malignant tumors are under way (179).
Trametinib is a clinically approved anti MEK drug, but its efficiency is low, which may be due to the rapid development of drug-resistant diseases. Trametinib’s transience is attributed to negative feedback loop inhibition, which reactivates MAPK pathway through various compensatory mechanisms (181). For some xenografts, the combined treatment of trametinib and pazopanib resulted in a continuous reduction of tumor volume by 50% or more (182).
The anti-cancer drug eFT508 (tomivosertib) developed by Davide et al. selectively blocked the phosphorylation of eIF4E by targeting inhibition of MNK1/2, thus inhibiting the weight gain after eating a high-fat diet. Studies showed that eFT508 alone can enhance anti-tumor immunity. This also supports the statement that there is a connection between eIF4E, obesity and cancer development (86, 183).
Ribavirin competitively inhibits the binding of eIF4E to the 5' end cap of mRNAs, as well as has an anti-proliferation effect. It can down regulate the phosphorylation levels of AKT, mTOR, 4EBP1 and eIF4E proteins in the mTOR-eIF4E signal pathway, as well as the phosphorylation levels of MEK, ERK, MNK1 and eIF4E proteins in the ERK-MNK1-eIF4E signal pathway. Ribavirin significantly increased the binding of eIF4E to 4EBP1, and decreased the binding of eIF4E to eIF4G. Ribavirin effectively targets eIF4E of leukemia patients with poor prognosis, leading to significant clinical reactions including complete remission and partial remission (148). The combination of ribavirin and imatinib can enhance the anti-leukemia effect (184).
EIF4E and β-Catenin are key regulators of the growth and survival of lung cancer cells, and their pharmacological inhibition may have therapeutic effects on lung cancer (185). Phosphorylation of eIF4E S209 can activate β-Catenin (186). Rifabutin inhibits eIF4E phosphorylation, leading to the decreased phosphorylation of β-Catenin and its subsequent transcriptional activity. The absence of eIF4E eliminates the effect of rifabutin on the inhibition of β-Catenin activity, which further confirms that rifabutin plays a role in lung cancer cells by targeting eIF4E and β-Catenin. The overexpression of β-Catenin reverses the inhibition of rifabutin on cell growth and survival.
EIF4E regulates gene translation and has been proved to play an important role in the progression of lung cancer. BET protein can regulate gene transcription. BET inhibitors JQ1 and I-BET151 inhibit the growth of NSCLC and down regulate the expression of eIF4E. When BRD4, a member of the BET family, was knocked out by siRNA, the growth of NSCLC was inhibited and the level of eIF4E protein was reduced. In addition, overexpression of eIF4E partially eliminated the growth inhibition function of JQ1, while knockout of eIF4E enhanced the growth inhibition function of JQ1. This indicates that JQ1 and I-BET151 inhibit the growth of NSCLC by inhibiting BET to reduce the transcription and subsequent protein expression of eIF4E (176, 177).
4EGI-1, a small molecular compound targeting to inhibit eIF4E-eIF4G binding, can reduce the expression of c-Myc without affecting the expression of β-actin (173) and has been proved to play an anti-cancer role in human cancer cells (187) without obvious toxicity (175).
EGPI-1 is a small molecule compound that can induce the down-regulation of eIF4E downstream proteins such as c-Myc. Further studies showed that EGPI-1 could significantly inhibit the proliferation of various lung cancer cells, such as A549, NCIL-H460, NCI-H1650 and 95D, but did not affect the proliferation of HUVEC cells (134). EGPI-1 showed good safety and pharmacokinetics characteristics in vivo. These results indicate that EGPI-1 can be used as an excellent lead compound to develop new anti-cancer drugs targeting eIF4E-eIF4G interface, and also as a chemical genetic probe to study the mechanism of eIF4E in biological processes and human diseases (188).
9 Outlook
This review mainly introduces eIF4E from its structure, biological function, main factors affecting its activity, and targeted drugs. However, there is still a lot of content not covered in this paper.
EIF4E mediates the proliferation of normal cells, but induces tumorigenesis when over expressed. Maintaining the level of eIF4E below its cancer promoting threshold is an important anti-cancer measure for normal cells (189). First of all, for future clinical trials, we should focus on setting up prospective randomized controlled studies to analyze the levels of patients' eIF4E and 4EBPs as indicators of disease detection, so as to formulate effective treatments at the early stage of the disease. In addition, the exact mechanism of tumor occurrence, development, and metastasis mediated by eIF4E is still poorly understood, and the relationship between eIF4E and tumors should be paid attention to and further studied. The sensitivities of different tumors to the same drug are also different, and the reasons for this difference deserve further in-depth study in order to achieve efficient treatment and reduce side effects. Finally, using eIF4E as a molecular target for tumor therapy, establishing models that multiple anti-tumor drugs are used in combination may provide new ideas for improving the efficacy and reducing side effects. For example, trametinib and pazopanib, umbilisib and carfilzomab, as well as patamine A and silvestrol can be used in combination to achieve anti-tumor effects.
To sum up, eIF4E will become an important target for the treatment of tumors in the future, and as a highly sensitive biomarker in tumor detection, it will be used for early diagnosis and evaluation of prognosis, opening up broad prospects for the treatment of malignant tumors.
Author contributions
Conception: LL and BX. Writing-original draft: XC, LL, and BX. Methodology, investigation and revising: YA, MT, and DX. Writing-Reviewing and Editing: XC, LL, and BX. All authors listed have made substantial, direct and intellectual contributions to this work, and approved it for publication.
Funding
This study was supported by Fundamental Research Program of Shanxi Province (Grant no. 20210302123397; 202203021212351), Science and Technology Innovation Project of Colleges and Universities in Shanxi Province (Grant no. 2020L0749), Key R&D Projects of Introducing High-Level Scientific and Technological Talents in Lvliang City (Grant no. 2021RC-1-4), the Project of Lvliang City Science and Technology Program (Grant no. 2020SHFZ29), the National College Students’ Innovation and Entrepreneurship Training Program (Grant no. 20221569), the Key Projects of Innovation and Entrepreneurship Training for College Students in Shanxi Province (Grant no. 20221577), Projects of Innovation and Entrepreneurship Training Program for College Students of Fenyang College of Shanxi Medical University (Grant no. FDC202209; FDC202215; FDC202214), and Special Fund for Key Disciplines of Fenyang College of Shanxi Medical University (Grant no. 2022B14).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol (2010) 11(2):113–27. doi: 10.1038/nrm2838
2. Jan E, Mohr I, Walsh D. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu Rev Virol (2016) 3(1):283–307. doi: 10.1146/annurev-virology-100114-055014
3. Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA (2011) 2(2):277–98. doi: 10.1002/wrna.52
4. Xu Y, Ruggero D. The role of translation control in tumorigenesis and its therapeutic implications. Annu Rev Cancer Biol (2020) 4(1):437–57. doi: 10.1146/annurev-cancerbio-030419-033420
5. Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol (2012) 4(12):a011528. doi: 10.1101/cshperspect.a011528
6. Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem (2014) 83:779–812. doi: 10.1146/annurev-biochem-060713-035802
7. Chiu H, Buono R, Jackson LV, Herzog LO, Mallya S, Conn CS, et al. Reduced eIF4E function impairs B-cell leukemia without altering normal B-lymphocyte function. iScience (2021) 24(7):102748. doi: 10.1016/j.isci.2021.102748
8. Yang X, Zang J, Pan X, Yin J, Xiang Q, Yu J, et al. miR-503 inhibits proliferation making human hepatocellular carcinoma cells susceptible to 5-fluorouracil by targeting EIF4E. Oncol Rep (2017) 37(1):563–70. doi: 10.3892/or.2016.5220
9. Joshi B, Cameron A, Jagus R. Characterization of mammalian eIF4E-family members. Eur J Biochem (2004) 271(11):2189–203. doi: 10.1111/j.1432-1033.2004.04149.x
10. Tettweiler G, Kowanda M, Lasko P, Sonenberg N, Hernández G. The distribution of eIF4E-family members across insecta. Comp Funct Genomics (2012) 2012:960420. doi: 10.1155/2012/960420
11. Jin H, Xu W, Rahman R, Na D, Fieldsend A, Song W, et al. TRIBE editing reveals specific mRNA targets of eIF4E-BP in Drosophila and in mammals. Sci Adv (2020) 6(33):eabb8771. doi: 10.1126/sciadv.abb8771
12. Hernández G. The versatile relationships between eIF4E and eIF4E-interacting proteins. Trends Genet (2022) 38(8):801–4. doi: 10.1016/j.tig.2022.04.003
13. Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol (2006) 175(3):415–26. doi: 10.1083/jcb.200607020
14. Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle (2007) 6(1):65–9. doi: 10.4161/cc.6.1.3688
15. Culjkovic-Kraljacic B, Skrabanek L, Revuelta MV, Gasiorek J, Cowling VH, Cerchietti L, et al. The eukaryotic translation initiation factor eIF4E elevates steady-state m7G capping of coding and noncoding transcripts. Proc Natl Acad Sci USA (2020) 117(43):26773–83. doi: 10.1073/pnas.2002360117
16. Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res (2013) 33(5):227–38. doi: 10.1089/jir.2012.0142
17. Montalto FI, De Amicis F. Cyclin D1 in cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells (2020) 9(12):2648. doi: 10.3390/cells9122648
18. Thompson EB. The many roles of c-Myc in apotosis. Annu Rev Physiol (1998) 60:575–600. doi: 10.1146/annurev.physiol.60.1.575
19. Kostas M, Lampart A, Bober J, Wiedlocha A, Tomala J, Krowarsch D, et al. Translocation of exogenous FGF1 and FGF2 protects the cell against apoptosis independently of receptor activation. J Mol Biol (2018) 430(21):4087–101. doi: 10.1016/j.jmb.2018.08.004
20. Shibuya M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes Cancer (2011) 2(12):1097–105. doi: 10.1177/1947601911423031
21. Niu C, Zhang S, Mo G, Jiang Y, Li L, Xu H, et al. Effects of ODC on polyamine metabolism, hormone levels, cell proliferation and apoptosis in goose ovarian granulosa cells. Poult Sci (2021) 100(8):101226. doi: 10.1016/j.psj.2021.101226
22. Bruckheimer EM, Cho SH, Sarkiss M, Herrmann J, McDonnell TJ. The Bcl-2 gene family and apoptosis. Adv Biochem Eng Biotechnol (1998) 62:75–105. doi: 10.1007/BFb0102306
23. Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci (2019) 132(7):jcs223826. doi: 10.1242/jcs.223826
24. Peluso JJ. Basic fibroblast growth factor (bFGF) regulation of the plasma membrane calcium ATPase (PMCA) as part of an anti-apoptotic mechanism of action. Biochem Pharmacol (2003) 66(8):1363–9. doi: 10.1016/s0006-2952(03)00486-6
25. Volpon L, Culjkovic-Kraljacic B, Sohn HS, Blanchet-Cohen A, Osborne MJ, Borden KLB. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA (2017) 23(6):927–37. doi: 10.1261/rna.060137.116
26. Culjkovic-Kraljacic B, Baguet A, Volpon L, Amri A, Borden KL. The oncogene eIF4E reprograms the nuclear pore complex to promote mRNA export and oncogenic transformation. Cell Rep (2012) 2(2):207–15. doi: 10.1016/j.celrep.2012.07.007
27. Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J (2009) 28(8):1087–98. doi: 10.1038/emboj.2009.53
28. Bong SM, Bae SH, Song B, Gwak H, Yang SW, Kim S, et al. Regulation of mRNA export through API5 and nuclear FGF2 interaction. Nucleic Acids Res (2020) 48(11):6340–52. doi: 10.1093/nar/gkaa335
29. Baumhardt JM, Walker JS, Lee Y, Shakya B, Brautigam CA, Lapalombella R, et al. Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation. Mol Biol Cell (2020) 31(17):1879–91. doi: 10.1091/mbc.E20-04-0233
30. Gulay S, Gupta N, Lorsch JR, Hinnebusch AG. Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo. Elife (2020) 9:e58243. doi: 10.7554/eLife.58243
31. Mishra RK, Datey A, Hussain T. mRNA recruiting eIF4 factors involved in protein synthesis and its regulation. Biochemistry (2020) 59(1):34–46. doi: 10.1021/acs.biochem.9b00788
32. Park E-H, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1•PABP mRNPs in vivo. EMBO J (2011) 30(2):302–16. doi: 10.1038/emboj.2010.312
33. Huggins HP, Subash JS, Stoffel H, Henderson MA, Hoffman JL, Buckner DS, et al. Distinct roles of two eIF4E isoforms in the germline of Caenorhabditis elegans. J Cell Sci (2020) 133(6):jcs.237990. doi: 10.1242/jcs.237990
34. Haimov O, Sehrawat U, Tamarkin-Ben Harush A, Bahat A, Uzonyi A, Will A, et al. Dynamic interaction of eukaryotic initiation factor 4G1 (eIF4G1) with eIF4E and eIF1 underlies scanning-dependent and -independent translation. Mol Cell Biol (2018) 38(18):e00139–18. doi: 10.1128/MCB.00139-18
35. De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene (2004) 23(18):3189–99. doi: 10.1038/sj.onc.1207545
36. Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res (2008) 68(3):631–4. doi: 10.1158/0008-5472.CAN-07-5635
37. Gaikwad S, Ghobakhlou F, Young DJ, Visweswaraiah J, Zhang H, Hinnebusch AG. Reprogramming of translation in yeast cells impaired for ribosome recycling favors short, efficiently translated mRNAs. Elife (2021) 10:e64283. doi: 10.7554/eLife.64283
38. Teng B, Ma P, Yu C, Zhang X, Feng Q, Wen L, et al. Upconversion nanoparticles loaded with eIF4E siRNA and platinum(iv) prodrug to sensitize platinum based chemotherapy for laryngeal cancer and bioimaging. J Mater Chem B (2017) 5(2):307–17. doi: 10.1039/c6tb02360f
39. Gehring NH, Wahle E, Fischer U. Deciphering the mRNP code: RNA-bound determinants of post-transcriptional gene regulation. Trends Biochem Sci (2017) 42(5):369–82. doi: 10.1016/j.tibs.2017.02.004
40. Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol (2019) 11(5):a032813. doi: 10.1101/cshperspect.a032813
41. Zarnack K, Balasubramanian S, Gantier MP, Kunetsky V, Kracht M, Schmitz ML, et al. Dynamic mRNP remodeling in response to internal and external stimuli. Biomolecules (2020) 10(9):1310. doi: 10.3390/biom10091310
42. Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell (2014) 54(4):547–58. doi: 10.1016/j.molcel.2014.04.033
44. Suzuki Y, Minami M, Suzuki M, Abe K, Zenno S, Tsujimoto M, et al. The Hsp90 inhibitor geldanamycin abrogates colocalization of eIF4E and eIF4E-transporter into stress granules and association of eIF4E with eIF4G. J Biol Chem (2009) 284(51):35597–604. doi: 10.1074/jbc.M109.036285
45. Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell (2009) 36(6):932–41. doi: 10.1016/j.molcel.2009.11.020
46. Ferrero PV, Layana C, Paulucci E, Gutiérrez P, Hernández G, Rivera-Pomar RV. Cap binding-independent recruitment of eIF4E to cytoplasmic foci. Biochim Biophys Acta (2012) 1823(7):1217–24. doi: 10.1016/j.bbamcr.2012.03.013
47. Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA (2005) 11(5):717–27. doi: 10.1261/rna.2340405
48. Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (2003) 300(5620):805–8. doi: 10.1126/science.1082320
49. Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell (2005) 122(6):875–86. doi: 10.1016/j.cell.2005.07.012
50. Volpon L, Culjkovic-Kraljacic B, Osborne MJ, Ramteke A, Sun Q, Niesman A, et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci USA (2016) 113(19):5263–8. doi: 10.1073/pnas.1524291113
51. Rong L, Livingstone M, Sukarieh R, Petroulakis E, Gingras AC, Crosby K, et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA (2008) 14(7):1318–27. doi: 10.1261/rna.950608
52. Igreja C, Peter D, Weiler C, Izaurralde E. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat Commun (2014) 5:4790. doi: 10.1038/ncomms5790
53. Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell (2004) 5(6):553–63. doi: 10.1016/j.ccr.2004.05.024
54. Githaka JM, Tripathi N, Kirschenman R, Patel N, Pandya V, Kramer DA, et al. BAD regulates mammary gland morphogenesis by 4E-BP1-mediated control of localized translation in mouse and human models. Nat Commun (2021) 12(1):2939. doi: 10.1038/s41467-021-23269-8
55. Räsch F, Weber R, Izaurralde E, Igreja C. 4E-T-bound mRNAs are stored in a silenced and deadenylated form. Genes Dev (2020) 34(11-12):847–60. doi: 10.1101/gad.336073.119
56. Nishimura T, Padamsi Z, Fakim H, Milette S, Dunham WH, Gingras AC, et al. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5' and 3' termini of target mRNAs. Cell Rep (2015) 11(9):1425–36. doi: 10.1016/j.celrep.2015.04.065
57. Gaston K, Tsitsilianos MA, Wadey K, Jayaraman PS. Misregulation of the proline rich homeodomain (PRH/HHEX) protein in cancer cells and its consequences for tumour growth and invasion. Cell Biosci (2016) 6:12. doi: 10.1186/s13578-016-0077-7
58. Soufi A, Gaston K, Jayaraman PS. Purification and characterisation of the PRH homeodomain: Removal of the N-terminal domain of PRH increases the PRH homeodomain-DNA interaction. Int J Biol Macromol (2006) 39(1-3):45–50. doi: 10.1016/j.ijbiomac.2006.01.004
59. Soufi A, Smith C, Clarke AR, Gaston K, Jayaraman PS. Oligomerisation of the developmental regulator proline rich homeodomain (PRH/Hex) is mediated by a novel proline-rich dimerisation domain. J Mol Biol (2006) 358(4):943–62. doi: 10.1016/j.jmb.2006.02.020
60. Ploski JE, Topisirovic I, Park KW, Borden KL, Radu A. A mechanism of nucleocytoplasmic trafficking for the homeodomain protein PRH. Mol Cell Biochem (2009) 332(1-2):173–81. doi: 10.1007/s11010-009-0188-0
61. Topisirovic I, Culjkovic B, Cohen N, Perez JM, Skrabanek L, Borden KLB. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J (2003) 22(3):689–703. doi: 10.1093/emboj/cdg069
62. Piserà A, Campo A, Campo S. Structure and functions of the translation initiation factor eIF4E and its role in cancer development and treatment. J Genet Genomics (2018) 45(1):13–24. doi: 10.1016/j.jgg.2018.01.003
63. Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ (2003) 10(12):1290–9. doi: 10.1038/sj.cdd.4401313
64. Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J (2001) 20(16):4547–59. doi: 10.1093/emboj/20.16.4547
65. Strudwick S, Borden KL. Finding a role for PML in APL pathogenesis: a critical assessment of potential PML activities. Leukemia (2002) 16(10):1906–17. doi: 10.1038/sj.leu.2402724
66. Jiang C, Tang Y, Ding L, Tan R, Li X, Lu J, et al. Targeting the N terminus of eIF4AI for inhibition of its catalytic recycling. Cell Chem Biol (2019) 26(10):1417-26.e5. doi: 10.1016/j.chembiol.2019.07.010
67. Sorrells DL, Black DR, Meschonat C, Rhoads R, De Benedetti A, Gao M, et al. Detection of eIF4E gene amplification in breast cancer by competitive PCR. Ann Surg Oncol (1998) 5(3):232–7. doi: 10.1007/BF02303778
68. Haydon MS, Googe JD, Sorrells DS, Ghali GE, Li BD. Progression of eIF4e gene amplification and overexpression in benign and malignant tumors of the head and neck. Cancer (2000) 88(12):2803–10. doi: 10.1002/1097-0142(20000615)88:12<2803::aid-cncr20>3.0.co;2-5
69. Chang SH, Elemento O, Zhang J, Zhuang ZW, Simons M, Hla T. ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc Natl Acad Sci USA (2014) 111(51):18309–14. doi: 10.1073/pnas.1412172111
70. Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol (2009) 29(5):1152–62. doi: 10.1128/MCB.01532-08
71. Yang L, Ma DW, Cao YP, Li DZ, Zhou X, Feng JF, et al. PRMT5 functionally associates with EZH2 to promote colorectal cancer progression through epigenetically repressing CDKN2B expression. Theranostics (2021) 11(8):3742–59. doi: 10.7150/thno.53023
72. Powers MA, Fay MM, Factor RE, Welm AL, Ullman KS. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res (2011) 71(16):5579–87. doi: 10.1158/0008-5472.CAN-11-0458
73. Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res (2009) 37(15):4965–76. doi: 10.1093/nar/gkp516
74. Gu Z, Gao S, Zhang F, Wang Z, Ma W, Davis RE, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J (2012) 446(2):235–41. doi: 10.1042/BJ20120768
75. Zhang B, Dong S, Li Z, Lu L, Zhang S, Chen X, et al. Targeting protein arginine methyltransferase 5 inhibits human hepatocellular carcinoma growth via the downregulation of beta-catenin. J Transl Med (2015) 13:349. doi: 10.1186/s12967-015-0721-8
76. Stavast CJ, Erkeland SJ. The non-canonical aspects of MicroRNAs: Many roads to gene regulation. Cells (2019) 8(11):1465. doi: 10.3390/cells8111465
77. Tejero R, Navarro A, Campayo M, Viñolas N, Marrades RM, Cordeiro A, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PloS One (2014) 9(7):e101899. doi: 10.1371/journal.pone.0101899
78. Wang D, Ma J, Ji X, Xu F, Wei Y. miR-141 regulation of EIF4E expression affects docetaxel chemoresistance of non-small cell lung cancer. Oncol Rep (2017) 37(1):608–16. doi: 10.3892/or.2016.5214
79. Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li Y, et al. MicroRNA-455-3p functions as a tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep (2017) 37(4):2449–58. doi: 10.3892/or.2017.5502
80. Chen S, Gao G. MicroRNAs recruit eIF4E2 to repress translation of target mRNAs. Protein Cell (2017) 8(10):750–61. doi: 10.1007/s13238-017-0444-0
81. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
82. Lin J, Fan L, Han Y, Guo J, Hao Z, Cao L, et al. The mTORC1/eIF4E/HIF-1α pathway mediates glycolysis to support brain hypoxia resistance in the gansu zokor, Eospalax cansus. Front Physiol (2021) 12:626240. doi: 10.3389/fphys.2021.626240
83. Chi PI, Huang WR, Chiu HC, Li JY, Nielsen BL, Liu HJ. Avian reovirus σA-modulated suppression of lactate dehydrogenase and upregulation of glutaminolysis and the mTOC1/eIF4E/HIF-1α pathway to enhance glycolysis and the TCA cycle for virus replication. Cell Microbiol (2018) 20(12):e12946. doi: 10.1111/cmi.12946
84. Sung H, Siegel RL, Torre LA, Pearson-Stuttard J, Islami F, Fedewa SA, et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J Clin (2019) 69(2):88–112. doi: 10.3322/caac.21499
85. Fang X, Wei J, He X, Lian J, Han D, An P, et al. Quantitative association between body mass index and the risk of cancer: A global Meta-analysis of prospective cohort studies. Int J Cancer (2018) 143(7):1595–603. doi: 10.1002/ijc.31553
86. Conn CS, Yang H, Tom HJ, Ikeda K, Oses-Prieto JA, Vu H, et al. The major cap-binding protein eIF4E regulates lipid homeostasis and diet-induced obesity. Nat Metab (2021) 3(2):244–57. doi: 10.1038/s42255-021-00349-z
87. Topisirovic I, Guzman ML, McConnell MJ, Licht JD, Culjkovic B, Neering SJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol (2003) 23(24):8992–9002. doi: 10.1128/MCB.23.24.8992-9002.2003
88. von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol (2004) 11(6):503–11. doi: 10.1038/nsmb779
89. Lai HK, Borden KL. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene (2000) 19(13):1623–34. doi: 10.1038/sj.onc.1203473
90. Culjkovic B, Tan K, Orolicki S, Amri A, Meloche S, Borden KL. The eIF4E RNA regulon promotes the Akt signaling pathway. J Cell Biol (2008) 181(1):51–63. doi: 10.1083/jcb.200707018
91. Prabhu SA, Moussa O, Miller WH Jr, Del Rincón SV. The MNK1/2-eIF4E axis as a potential therapeutic target in melanoma. Int J Mol Sci (2020) 21(11):4055. doi: 10.3390/ijms21114055
92. Strudwick S, Borden KLB. The emerging roles of translation factor eIF4E in the nucleus. Differentiation (2002) 70(1):10–22. doi: 10.1046/j.1432-0436.2002.700102.x
93. Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene (2006) 25(48):6416–22. doi: 10.1038/sj.onc.1209888
94. Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue P, Fu H, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res (2005) 65(16):7052–8. doi: 10.1158/0008-5472.CAN-05-0917
95. Tsukumo Y, Alain T, Fonseca BD, Nadon R, Sonenberg N. Translation control during prolonged mTORC1 inhibition mediated by 4E-BP3. Nat Commun (2016) 7:11776. doi: 10.1038/ncomms11776
96. Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res (2012) 72(24):6468–76. doi: 10.1158/0008-5472.CAN-12-2395
97. So L, Lee J, Palafox M, Mallya S, Woxland CG, Arguello M, et al. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci Signal (2016) 9(430):ra57. doi: 10.1126/scisignal.aad8463
98. Cho S, Lee G, Pickering BF, Jang C, Park JH, He L, et al. mTORC1 promotes cell growth via m6A-dependent mRNA degradation. Mol Cell (2021) 81(10):2064–75.e8. doi: 10.1016/j.molcel.2021.03.010
99. Ishiguro H, Katano Y, Nakano I, Ishigami M, Hayashi K, Honda T, et al. Clofibrate treatment promotes branched-chain amino acid catabolism and decreases the phosphorylation state of mTOR, eIF4E-BP1, and S6K1 in rat liver. Life Sci (2006) 79(8):737–43. doi: 10.1016/j.lfs.2006.02.037
100. Lang CH, Frost RA, Jefferson LS, Kimball SR, Vary TC. Endotoxin-induced decrease in muscle protein synthesis is associated with changes in eIF2B, eIF4E, and IGF-I. Am J Physiol Endocrinol Metab (2000) 278(6):E1133–43. doi: 10.1152/ajpendo.2000.278.6.E1133
101. Jacobson BA, De A, Kratzke MG, Patel MR, Jay-Dixon J, Whitson BA, et al. Activated 4E-BP1 represses tumourigenesis and IGF-I-mediated activation of the eIF4F complex in mesothelioma. Br J Cancer (2009) 101(3):424–31. doi: 10.1038/sj.bjc.6605184
102. Shu XE, Swanda RV, Qian SB. Nutrient control of mRNA translation. Annu Rev Nutr (2020) 40:51–75. doi: 10.1146/annurev-nutr-120919-041411
103. Guo C, Hou Y, Yu X, Zhang F, Wu X, Wu X, et al. The ERK-MNK-eIF4F signaling pathway mediates TPDHT-induced A549 cell death in vitro and in vivo. Food Chem Toxicol (2020) 137:111158. doi: 10.1016/j.fct.2020.111158
104. Furic L, Beardsley E, Topisirovic I. eIF4E phosphorylation downstream of MAPK pathway. In: Translation and its regulation in cancer biology and medicine (Dordrecht: Springer) (2014). p. 363–74.
105. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res (2015) 35(6):600–4. doi: 10.3109/10799893.2015.1030412
106. ur Rasool R, Rah B, Amin H, Nayak D, Chakraborty S, Rawoof A, et al. Dual modulation of Ras-Mnk and PI3K-AKT-mTOR pathways: A novel c-FLIP inhibitory mechanism of 3-AWA mediated translational attenuation through dephosphorylation of eIF4E. Sci Rep (2016) 6:18800. doi: 10.1038/srep18800
107. Chen K, Zhang Y, Qian L, Wang P. Emerging strategies to target RAS signaling in human cancer therapy. J Hematol Oncol (2021) 14(1):116. doi: 10.1186/s13045-021-01127-w
108. Tran TH, Chan AH, Young LC, Bindu L, Neale C, Messing S, et al. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat Commun (2021) 12(1):1176. doi: 10.1038/s41467-021-21422-x
109. Lavoie H, Gagnon J, Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol (2020) 21(10):607–32. doi: 10.1038/s41580-020-0255-7
110. Li L, Zhao GD, Shi Z, Qi LL, Zhou LY, Fu ZX. The Ras/Raf/MEK/ERK signaling pathway and its role in the occurrence and development of HCC. Oncol Lett (2016) 12(5):3045–50. doi: 10.3892/ol.2016.5110
111. Hay N. Mnk earmarks eIF4E for cancer therapy. Proc Natl Acad Sci USA (2010) 107(32):13975–6. doi: 10.1073/pnas.1008908107
112. Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J (1999) 18(1):270–9. doi: 10.1093/emboj/18.1.270
113. Yang X, Zhong W, Cao R. Phosphorylation of the mRNA cap-binding protein eIF4E and cancer. Cell Signal (2020) 73:109689. doi: 10.1016/j.cellsig.2020.109689
114. Takeda K, Noguchi T, Naguro I, Ichijo H. Apoptosis signal-regulating kinase 1 in stress and immune response. Annu Rev Pharmacol Toxicol (2008) 48:199–225. doi: 10.1146/annurev.pharmtox.48.113006.094606
115. Tobiume K. Characterization of mouse apoptosis signal-regulating kinase 1. Kokubyo Gakkai Zasshi (1998) 65(1):42–52. doi: 10.5357/koubyou.65.42
116. Barros-Miñones L, Orejana L, Goñi-Allo B, Suquía V, Hervías I, Aguirre N, et al. Modulation of the ASK1-MKK3/6-p38/MAPK signalling pathway mediates sildenafil protection against chemical hypoxia caused by malonate. Br J Pharmacol (2013) 168(8):1820–34. doi: 10.1111/bph.12071
117. Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. Sumoylation of eIF4E activates mRNA translation. EMBO Rep (2010) 11(4):299–304. doi: 10.1038/embor.2010.18
118. Gu Y, Zhou H, Gan Y, Zhang J, Chen J, Gan X, et al. Small-molecule induction of phospho-eIF4E sumoylation and degradation via targeting its phosphorylated serine 209 residue. Oncotarget (2015) 6(17):15111–21. doi: 10.18632/ontarget.3615
119. Xu X, Vatsyayan J, Gao C, Bakkenist CJ, Hu J. HDAC2 promotes eIF4E sumoylation and activates mRNA translation gene specifically. J Biol Chem (2010) 285(24):18139–43. doi: 10.1074/jbc.C110.131599
120. Dong K, Wang R, Wang X, Lin F, Shen JJ, Gao P, et al. Tumor-specific RNAi targeting eIF4E suppresses tumor growth, induces apoptosis and enhances cisplatin cytotoxicity in human breast carcinoma cells. Breast Cancer Res Treat (2009) 113(3):443–56. doi: 10.1007/s10549-008-9956-x
121. Bitterman PB, Polunovsky VA. eIF4E-mediated translational control of cancer incidence. Biochim Biophys Acta (2015) 1849(7):774–80. doi: 10.1016/j.bbagrm.2014.09.007
122. Sun S, Liu Q, Zhang A, He E. Expression and clinical value of eukaryotic initiation factor 4E and matrix metalloproteinase-9 in supraglottic and hypopharyngeal carcinoma and metastasic lymph. J Clin Otorhinolaryngol (China) (2006) 20(24):1116–9. doi: 10.3969/j.issn.1001-1781.2006.24.005
123. Hu S. The expression and clinical significance of mTOR、 eIF4E in lung squamous cell cancer. [Master's thesis] (Zhengzhou: Zhengzhou University) (2010). doi: 10.7666/d.y1834160
124. Xie M, Huang Y, Yang L, Zhang W, Zhao J, Huang Q. The expressions of survivin, eIF4E protein in breast invasive ductal carcinoma and their significance. China Journal of Modern Medicine (2013) 23(04):30–3. doi: 10.3969/j.issn.1005-8982.2013.04.008
125. Gao M. HPV E6/E7 enhances transcriptional expression of eIF4E to promote proliferation and migration of cervical carcinoma. [Master's thesis] (Guangdong: Guangdong Medical College) (2011). doi: 10.7666/d.y2013377
126. Yu Y, Fei Z, Lu L. Relationship between eIF4E gene expression and radiochemotherapy sensitivity in advanced nasopharyngeal carcinoma. Guangdong Medical Journal (2019) 40(1):128–31. doi: 10.13820/j.cnki.gdyx.20177677
127. Zhang Y, Fang X, He X, Zhang F. Expression and significance of eIF4E, C-myc and CDK4 in gastric carcinoma and premalignant conditions. Chin J Clin Gastroenterol (2020) 32(02):102–7. doi: 10.3870/lcxh.j.issn.1005-541X.2020.02.08
128. Li G, Liu Y, Chong T, Wang Z. Expression and role of eukaryotic translation initiation factor 4E in the tissues of renal clear cell carcinoma. Journal of Xi’an Jiaotong University(Medical Sciences) (2019) 40(01):106–110+125. doi: 10.7652/jdyxb201901020
129. Franklin S, Pho T, Abreo FW, Nassar R, De Benedetti A, Stucker FJ, et al. Detection of the proto-oncogene eIF4E in larynx and hypopharynx cancers. Arch Otolaryngol Head Neck Surg (1999) 125(2):177–82. doi: 10.1001/archotol.125.2.177
130. Liang Z, Lei T, LuYing Z, YuPing G. The expression of proto-oncogene eIF4E in laryngeal squamous cell carcinoma. Laryngoscope (2003) 113(7):1238–43. doi: 10.1097/00005537-200307000-00024
131. Yi HJ, Zhang BQ, Guo W, Zhao LD, Yang SM. The role of molecular margins as prognostic factors in laryngeal carcinoma in Chinese patients. Acta Otolaryngol (2012) 132(8):874–8. doi: 10.3109/00016489.2012.663503
132. Chang SH, Kim JE, Lee JH, Minai-Tehrani A, Han K, Chae C, et al. Aerosol delivery of eukaryotic translation initiation factor 4E-binding protein 1 effectively suppresses lung tumorigenesis in K-rasLA1 mice. Cancer Gene Ther (2013) 20(6):331–5. doi: 10.1038/cgt.2013.24
133. Dong Q, Dong L, Liu S, Kong Y, Zhang M, Wang X. Tumor-derived exosomal eIF4E as a biomarker for survival prediction in patients with non-small cell lung cancer. Med Sci Monit (2020) 26:e923210. doi: 10.12659/MSM.923210
134. Qi X, Zhang S, Chen Z, Wang L, Zhu W, Yin C, et al. EGPI-1, a novel eIF4E/eIF4G interaction inhibitor, inhibits lung cancer cell growth and angiogenesis through Ras/MNK/ERK/eIF4E signaling pathway. Chem Biol Interact (2022) 352:109773. doi: 10.1016/j.cbi.2021.109773
135. Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res (2010) 16(1):240–8. doi: 10.1158/1078-0432.CCR-09-0986
136. Kerekatte V, Smiley K, Hu B, Smith A, Gelder F, Benedetti AD. The proto-oncogene/translation factor eIF4E: a survey of its expression in breast carcinomas. Int J Cancer (1995) 64(1):27–31. doi: 10.1002/ijc.2910640107
137. McClusky DR, Chu Q, Yu H, Debenedetti A, Johnson LW, Meschonat C, et al. A prospective trial on initiation factor 4E (eIF4E) overexpression and cancer recurrence in node-positive breast cancer. Ann Surg (2005) 242(4):584–90. doi: 10.1097/01.sla.0000184224.55949.90
138. Yang H, Li LW, Shi M, Wang JH, Xiao F, Zhou B, et al. In vivo study of breast carcinoma radiosensitization by targeting eIF4E. Biochem Biophys Res Commun (2012) 423(4):878–83. doi: 10.1016/j.bbrc.2012.06.064
139. Huang CI, Wang CC, Tai TS, Hwang TZ, Yang CC, Hsu CM, et al. eIF4E and 4EBP1 are prognostic markers of head and neck squamous cell carcinoma recurrence after definitive surgery and adjuvant radiotherapy. PloS One (2019) 14(11):e0225537. doi: 10.1371/journal.pone.0225537
140. DeFatta RJ, Nathan CA, Benedetti AD. Antisense RNA to eIF4E suppresses oncogenic properties of a head and neck squamous cell carcinoma cell line. Laryngoscope (2000) 110(6):928–33. doi: 10.1097/00005537-200006000-00007
141. Oridate N, Kim HJ, Xu X, Lotan R. Growth inhibition of head and neck squamous carcinoma cells by small interfering RNAs targeting eIF4E or cyclin D1 alone or combined with cisplatin. Cancer Biol Ther (2005) 4(3):318–23. doi: 10.4161/cbt.4.3.1504
142. Singh J, Jayaraj R, Baxi S, Mileva M, Skinner J, Dhand NK, et al. Immunohistochemical expression levels of p53 and eIF4E markers in histologically negative surgical margins, and their association with the clinical outcome of patients with head and neck squamous cell carcinoma. Mol Clin Oncol (2016) 4(2):166–72. doi: 10.3892/mco.2015.689
143. Sunavala-Dossabhoy G, Palaniyandi S, Clark C, Nathan CO, Abreo FW, Caldito G. Analysis of eIF4E and 4EBP1 mRNAs in head and neck cancer. Laryngoscope (2011) 121(10):2136–41. doi: 10.1002/lary.22144
144. Niu Z, Wang J, Muhammad S, Niu W, Liu E, Peng C, et al. Protein expression of eIF4E and integrin αvβ6 in colon cancer can predict clinical significance, reveal their correlation and imply possible mechanism of interaction. Cell Biosci (2014) 4:23. doi: 10.1186/2045-3701-4-23
145. Gao M, Zhang X, Li D, He P, Tian W, Zeng B. Expression analysis and clinical significance of eVEGF-C, E-cadherin and MMP-2 in colorectal adenocarcinoma. Oncotarget (2016) 7(51):85502–14. doi: 10.18632/oncotarget.13453
146. Ruan H, Li X, Xu X, Leibowitz BJ, Tong J, Chen L, et al. eIF4E S209 phosphorylation licenses myc- and stress-driven oncogenesis. Elife (2020) 9:e60151. doi: 10.7554/eLife.60151
147. Zhou H, Xu RZ, Gu Y, Shi PF, Qian S. Targeting of phospho-eIF4E by homoharringtonine eradicates a distinct subset of human acute myeloid leukemia. Leuk Lymphoma (2020) 61(5):1084–96. doi: 10.1080/10428194.2017.1390229
148. Culjkovic B, Borden KL. Understanding and targeting the eukaryotic translation initiation factor eIF4E in head and neck cancer. J Oncol (2009) 2009:981679. doi: 10.1155/2009/981679
149. Cheng R, Huang Y, Fang Y, Wang Q, Yan M, Ge Y. Cryptotanshinone enhances the efficacy of Bcr-Abl tyrosine kinase inhibitors via inhibiting STAT3 and eIF4E signalling pathways in chronic myeloid leukaemia. Pharm Biol (2021) 59(1):893–903. doi: 10.1080/13880209.2021.1944224
150. Barrett D, Brown VI, Grupp SA, Teachey DT. Targeting the PI3K/AKT/mTOR signaling axis in children with hematologic malignancies. Paediatr Drugs (2012) 14(5):299–316. doi: 10.2165/11594740-000000000-00000
151. Jacobson BA, Thumma SC, Jay-Dixon J, Patel MR, Dubear Kroening K, Kratzke MG, et al. Targeting eukaryotic translation in mesothelioma cells with an eIF4E-specific antisense oligonucleotide. PloS One (2013) 8(11):e81669. doi: 10.1371/journal.pone.0081669
152. Duffy AG, Makarova-Rusher OV, Ulahannan SV, Rahma OE, Fioravanti S, Walker M, et al. Modulation of tumor eIF4E by antisense inhibition: A phase I/II translational clinical trial of ISIS 183750-an antisense oligonucleotide against eIF4E-in combination with irinotecan in solid tumors and irinotecan-refractory colorectal cancer. Int J Cancer (2016) 139(7):1648–57. doi: 10.1002/ijc.30199
153. Thumma SC, Jacobson BA, Patel MR, Konicek BW, Franklin MJ, Jay-Dixon J, et al. Antisense oligonucleotide targeting eukaryotic translation initiation factor 4E reduces growth and enhances chemosensitivity of non-small-cell lung cancer cells. Cancer Gene Ther (2015) 22(8):396–401. doi: 10.1038/cgt.2015.34
154. Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest (2007) 117(9):2638–48. doi: 10.1172/JCI32044
155. Pal I, Safari M, Jovanovic M, Bates SE, Deng C. Targeting translation of mRNA as a therapeutic strategy in cancer. Curr Hematol Malig Rep (2019) 14(4):219–27. doi: 10.1007/s11899-019-00530-y
156. Bustinza-Linares E, Kurzrock R, Tsimberidou A-M. Salirasib in the treatment of pancreatic cancer. Future Oncol (2010) 6(6):885–91. doi: 10.2217/fon.10.71
157. Wolfson E, Schmukler E, Schokoroy ST, Kloog Y, Pinkas-Kramarski R. Enhancing FTS (Salirasib) efficiency via combinatorial treatment. Biol Cell (2015) 107(5):130–43. doi: 10.1111/boc.201400087
158. Rotblat B, Ehrlich M, Haklai R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid (Salirasib) disrupts the spatiotemporal localization of active Ras: a potential treatment for cancer. Methods Enzymol (2008) 439:467–89. doi: 10.1016/S0076-6879(07)00432-6
159. Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res (2006) 66(24):11851–8. doi: 10.1158/0008-5472.CAN-06-1377
160. Matsuda Y, Fukumoto M. Sorafenib: complexities of Raf-dependent and Raf-independent signaling are now unveiled. Med Mol Morphol (2011) 44(4):183–9. doi: 10.1007/s00795-011-0558-z
161. Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther (2008) 7(10):3129–40. doi: 10.1158/1535-7163.MCT-08-0013
162. Gril B, Palmieri D, Qian Y, Smart D, Ileva L, Liewehr DJ, et al. Pazopanib reveals a role for tumor cell B-Raf in the prevention of HER2+ breast cancer brain metastasis. Clin Cancer Res (2011) 17(1):142–53. doi: 10.1158/1078-0432.CCR-10-1603
163. Gril B, Palmieri D, Qian Y, Anwar T, Ileva L, Bernardo M, et al. The B-Raf status of tumor cells may be a significant determinant of both antitumor and anti-angiogenic effects of pazopanib in xenograft tumor models. PloS One (2011) 6(10):e25625. doi: 10.1371/journal.pone.0025625
164. Jordan EJ, Kelly CM. Vemurafenib for the treatment of melanoma. Expert Opin Pharmacother (2012) 13(17):2533–43. doi: 10.1517/14656566.2012.737780
165. Hartsough E, Shao Y, Aplin AE. Resistance to RAF inhibitors revisited. J Invest Dermatol (2014) 134(2):319–25. doi: 10.1038/jid.2013.358
166. Dornan MH, Petrenyov D, Simard JM, Boudjemeline M, Mititelu R, DaSilva JN, et al. Synthesis of a 11C-isotopologue of the B-Raf-selective inhibitor encorafenib using in-loop [11C]CO2 fixation. ACS Omega (2020) 5(33):20960–6. doi: 10.1021/acsomega.0c02419
167. Davis J, Wayman M. Encorafenib and binimetinib combination therapy in metastatic melanoma. J Adv Pract Oncol (2022) 13(4):450–5. doi: 10.6004/jadpro.2022.13.4.7
168. Crona DJ, Keisler MD, Walko CM. Regorafenib: a novel multitargeted tyrosine kinase inhibitor for colorectal cancer and gastrointestinal stromal tumors. Ann Pharmacother (2013) 47(12):1685–96. doi: 10.1177/1060028013509792
169. Yuan X, Tang Z, Du R, Yao Z, Cheung SH, Zhang X, et al. RAF dimer inhibition enhances the antitumor activity of MEK inhibitors in K-RAS mutant tumors. Mol Oncol (2020) 14(8):1833–49. doi: 10.1002/1878-0261.12698
170. Desai J, Gan H, Barrow C, Jameson M, Atkinson V, Haydon A, et al. Phase I, open-label, dose-escalation/dose-expansion study of lifirafenib (BGB-283), an RAF family kinase inhibitor, in patients with solid tumors. J Clin Oncol (2020) 38(19):2140–50. doi: 10.1200/JCO.19.02654
171. Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med (2019) 381(7):626–36. doi: 10.1056/NEJMoa1904059
172. Urtishak KA, Wang LS, Culjkovic-Kraljacic B, Davenport JW, Porazzi P, Vincent TL, et al. Targeting EIF4E signaling with ribavirin in infant acute lymphoblastic leukemia. Oncogene (2019) 38(13):2241–62. doi: 10.1038/s41388-018-0567-7
173. De A, Jacobson BA, Peterson MS, Jay-Dixon J, Kratzke MG, Sadiq AA, et al. 4EGI-1 represses cap-dependent translation and regulates genome-wide translation in malignant pleural mesothelioma. Invest New Drugs (2018) 36(2):217–29. doi: 10.1007/s10637-017-0535-z
174. Takrouri K, Chen T, Papadopoulos E, Sahoo R, Kabha E, Chen H, et al. Structure-activity relationship study of 4EGI-1, small molecule eIF4E/eIF4G protein-protein interaction inhibitors. Eur J Med Chem (2014) 77:361–77. doi: 10.1016/j.ejmech.2014.03.034
175. Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem Soc Trans (2015) 43(5):763–72. doi: 10.1042/BST20150126
176. Gao Z, Yuan T, Zhou X, Ni P, Sun G, Li P, et al. Targeting BRD4 proteins suppresses the growth of NSCLC through downregulation of eIF4E expression. Cancer Biol Ther (2018) 19(5):407–15. doi: 10.1080/15384047.2018.1423923
177. Pham TND, Kumar K, DeCant BT, Shang M, Munshi SZ, Matsangou M, et al. Induction of MNK kinase-dependent eIF4E phosphorylation by inhibitors targeting BET proteins limits efficacy of BET inhibitors. Mol Cancer Ther (2019) 18(2):235–44. doi: 10.1158/1535-7163.MCT-18-0768
178. Hao P, Yu J, Ward R, Liu Y, Hao Q, An S, et al. Eukaryotic translation initiation factors as promising targets in cancer therapy. Cell Commun Signal (2020) 18(1):175. doi: 10.1186/s12964-020-00607-9
179. Niu P, Li J, Chen H, Zhu Y, Zhou J, Shi D. Anti-proliferative effect of cardamonin on mTOR inhibitor-resistant cancer cells. Mol Med Rep (2020) 21(3):1399–407. doi: 10.3892/mmr.2019.10898
180. Badawi M, Kim J, Dauki A, Sutaria D, Motiwala T, Reyes R, et al. CD44 positive and sorafenib insensitive hepatocellular carcinomas respond to the ATP-competitive mTOR inhibitor INK128. Oncotarget (2018) 9(40):26032–45. doi: 10.18632/oncotarget.25430
181. Yang H, Liang SQ, Xu D, Yang Z, Marti TM, Gao Y, et al. HSP90/AXL/eIF4E-regulated unfolded protein response as an acquired vulnerability in drug-resistant KRAS-mutant lung cancer. Oncogenesis (2019) 8(9):45. doi: 10.1038/s41389-019-0158-7
182. Ball DW, Jin N, Xue P, Bhan S, Ahmed SR, Rosen DM, et al. Trametinib with and without pazopanib has potent preclinical activity in thyroid cancer. Oncol Rep (2015) 34(5):2319–24. doi: 10.3892/or.2015.4225
183. Fabbri L, Chakraborty A, Robert C, Vagner S. The plasticity of mRNA translation during cancer progression and therapy resistance. Nat Rev Cancer (2021) 21(9):558–77. doi: 10.1038/s41568-021-00380-y
184. Shi F, Len Y, Gong Y, Shi R, Yang X, Naren D, et al. Ribavirin inhibits the activity of mTOR/eIF4E, ERK/Mnk1/eIF4E signaling pathway and synergizes with tyrosine kinase inhibitor imatinib to impair Bcr-Abl mediated proliferation and apoptosis in ph+ leukemia. PloS One (2015) 10(8):e0136746. doi: 10.1371/journal.pone.0136746
185. Li J, Huang Y, Gao Y, Wu H, Dong W, Liu L. Antibiotic drug rifabutin is effective against lung cancer cells by targeting the eIF4E-β-catenin axis. Biochem Biophys Res Commun (2016) 472(2):299–305 doi: 10.1016/j.bbrc.2016.02.120
186. Lim S, Saw TY, Zhang M, Janes MR, Nacro K, Hill J, et al. Targeting of the MNK-eIF4E axis in blast crisis chronic myeloid leukemia inhibits leukemia stem cell function. Proc Natl Acad Sci USA (2013) 110(25):E2298–307. doi: 10.1073/pnas.1301838110
187. Yang X, Dong QF, Li LW, Huo JL, Li PQ, Fei Z, et al. The cap-translation inhibitor 4EGI-1 induces mitochondrial dysfunction via regulation of mitochondrial dynamic proteins in human glioma U251 cells. Neurochem Int (2015) 90:98–106. doi: 10.1016/j.neuint.2015.07.019
188. Wang L, Guo C, Li X, Yu X, Li X, Xu K, et al. Design, synthesis and biological evaluation of bromophenol-thiazolylhydrazone hybrids inhibiting the interaction of translation initiation factors eIF4E/eIF4G as multifunctional agents for cancer treatment. Eur J Med Chem (2019) 177:153–70. doi: 10.1016/j.ejmech.2019.05.044
Keywords: eIF4E, translation initiation, biological function, malignant tumor, antineoplastic drugs
Citation: Chen X, An Y, Tan M, Xie D, Liu L and Xu B (2023) Biological functions and research progress of eIF4E. Front. Oncol. 13:1076855. doi: 10.3389/fonc.2023.1076855
Received: 04 November 2022; Accepted: 30 January 2023;
Published: 03 August 2023.
Edited by:
Ui Soon Khoo, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Greco Hernández, National Institute of Cancerology (INCAN), MexicoHo Tsoi, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2023 Chen, An, Tan, Xie, Liu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Liu, bGw3NzJ4QHN4bXUuZWR1LmNu; Benjin Xu, YmVuamluX2licHJuYWxhYkBzaW5hLmNvbQ==
†These authors share first authorship
 Xiaocong Chen
Xiaocong Chen Yang An
Yang An Mengsi Tan
Mengsi Tan Dongrui Xie
Dongrui Xie Ling Liu2,3,4*
Ling Liu2,3,4* Benjin Xu
Benjin Xu