
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 28 September 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.990195
This article is part of the Research Topic365 Days of Progress In Cancer Molecular Targets and TherapeuticsView all 23 articles
Ubiquitin-specific peptidase 10 (USP10) is a member of the ubiquitin-specific protease family that removes the ubiquitin chain from ubiquitin-conjugated protein substrates. We performed a literature search to evaluate the structure and biological activity of USP10, summarize its role in tumorigenesis and tumor progression, and discuss how USP10 may act as a tumor suppressor or a tumor-promoting gene depending on its mechanism of action. Subsequently, we elaborated further on these results through bioinformatics analysis. We demonstrated that abnormal expression of USP10 is related to tumorigenesis in various types of cancer, including liver, lung, ovarian, breast, prostate, and gastric cancers and acute myeloid leukemia. Meanwhile, in certain cancers, increased USP10 expression is associated with tumor suppression. USP10 was downregulated in kidney renal clear cell carcinoma (KIRC) and associated with reduced overall survival in patients with KIRC. In contrast, USP10 upregulation was associated with poor prognosis in head and neck squamous cell carcinoma (HNSC). In addition, we elucidated the novel role of USP10 in the regulation of tumor immunity in KIRC and HNSC through bioinformatics analysis. We identified several signaling pathways to be significantly associated with USP10 expression, such as ferroptosis, PI3K/AKT/mTOR, TGF-β, and G2/M checkpoint. In summary, this review outlines the role of USP10 in various forms of cancer, discusses the relevance of USP10 inhibitors in anti-tumor therapies, and highlights the potential function of USP10 in regulating the immune responses of tumors.
Ubiquitination is an important epigenetic process that regulates transcription factors, histones, and chromatin states (1). A variety of biological processes are affected by ubiquitination in gene transcriptional regulation, including protein degradation (2), cell signaling (3), DNA damage repair (4), stress response (5), and cancer (6). Protein ubiquitination involves the addition of ubiquitin to target proteins through a multi-enzyme cascade, whereas deubiquitination is the removal of ubiquitin from proteins by deubiquitinating enzymes (DUBs) (7). As such, ubiquitination and deubiquitination constitute a dynamic equilibrium in cell biology. DUBs have become important drug targets for various diseases, such as tumors and neurodegenerative diseases (8, 9). Ubiquitin-specific peptidase 10 (USP10) is a member of the USP family of DUBs. It can remove conjugated ubiquitin from target proteins, such as autophagy-regulated gene Beclin1 (BECN1) (10), tumor protein p53 (TP53) (11), cystic fibrosis transmembrane conductance regulator (CFTR) (12), and sorting nexin 3 (SNX3) (13). It is also involved in a tumor necrosis factor receptor-associated factor (TRAF) family member-associated noncanonical nuclear factor-kappa B (NF-κB) activator (TANK)-dependent negative feedback response that attenuates NF-κB activation by deubiquitinating inhibitor of kappa polypeptide gene enhancer in B cells, kinase gamma (IKBKG) or tumor necrosis factor receptor-associated factor 6 (TRAF6) in response to interleukin-1-beta stimulation or DNA damage (14). Additionally, USP10 deubiquitinates and stabilizes T-box transcription factor protein (TBX21) (15). Moreover, it may affect the development of various cancers, including lung cancer (16), liver cancer (17), and acute myeloid leukemia (AML) (18). Interestingly, it also modulates immune-related signaling pathways via deubiquitination of immune-related genes, such as yes-associated protein 1 (YAP1) (19). The relationship between the abnormal expression of USP10 in various tumors and its regulation of immune pathways prompted us to explore the role of USP10 in tumor development and immunity. Here, we review the role of USP10 in tumors and regulation of immune responses and provide new insights into the USP family as potential targets for cancer therapy.
USP10 is located on chromosome 16q24.1 (20) and consists of 798 amino acids (21). Human USP10 contains an Ataxin2C domain and a USP domain (Figure 1). The Ataxin2C domain is approximately 250 residues long, and is located at the C-terminus of eukaryotic ataxin-2. The function of ataxin-2 is unknown; however, the expansion of polyglutamine tracts may cause spinocerebellar ataxia type 2 (SCA2) (22). This expansion disrupts the normal morphology of the Golgi complex, leading to increased cell death (23). The USP domain consists of a catalytic site, protein-protein interaction sites, and localization domains. Since most USP domains require cleavage of the isopeptide bond between two ubiquitin molecules, a minimum of two ubiquitin-binding sites are required (24). The catalytic core of USP consists of six conserved boxes that are present in almost all USP domains. Boxes 1, 5, and 6 contain catalytic Cys, His, and Asp/Asn residues, respectively. Boxes 3 and 4 contain a Cys-X-X-Cys motif that functions as a zinc-binding motif. This zinc-binding motif facilitates the folding of the USP core, which facilitates the interaction between motifs separated by hundreds of residues (24).
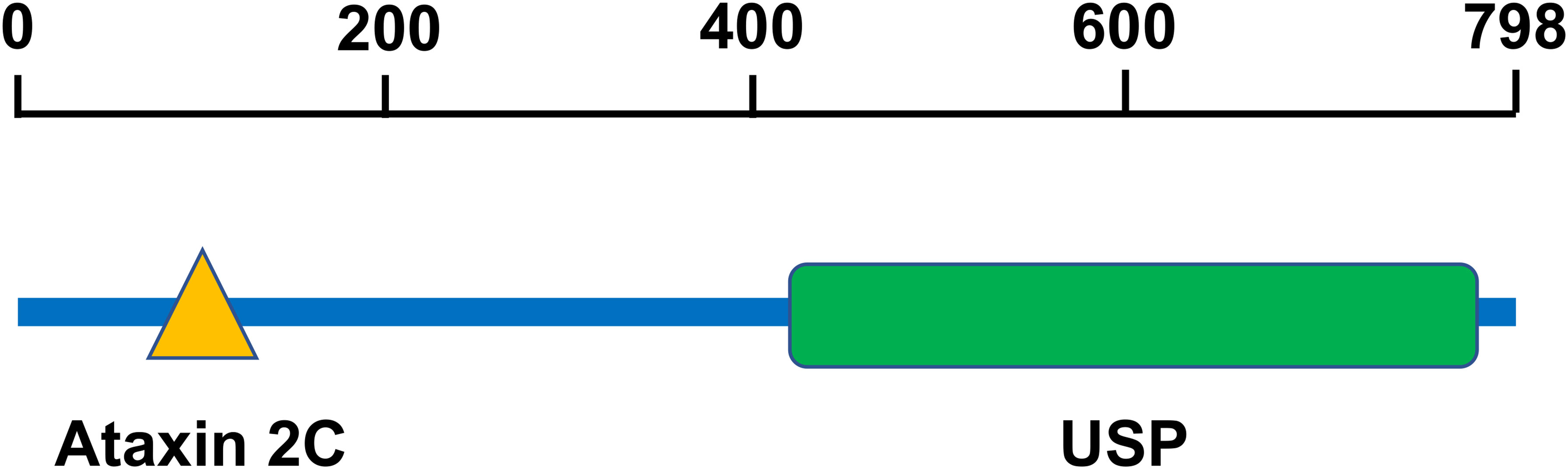
Figure 1 The structure of USP10. Human USP10 contains an Ataxin 2C domain and a USP domain. The Ataxin2C domain is approximately 250 residues long, and is located at the C-terminus of eukaryotic ataxin-2. The USP domain consists of a catalytic site, protein-protein interaction sites, and localization domains. USP10, Ubiquitin-specific peptidase 10.
USP10 has several molecular functions and characteristics. Firstly, USP10 has cysteine-type endopeptidase activity and can catalyze the hydrolysis of α-peptide bonds inside polypeptide chains through the sulfhydryl group of the cysteine residue in the active center (25). Secondly, USP10 may bind to members of one of the p53 protein families (26) and it may also bind to RNA molecules (27). USP10 also has a thiol-dependent iso-peptidase activity that cleaves ubiquitin from its conjugated target protein (25). Finally, USP10 may bind to transmembrane transporters, proteins, or protein complexes, facilitating the transfer of substances across membranes (28). Thus, USP10 is involved in a wide range of biological processes, including removal ubiquitin groups from proteins (26), negative regulation of the I-κB kinase/NF-κB signaling pathway and cellular responses to interleukin-1 and DNA damage stimuli (14), regulation of the cell cycle regulator phosphoprotein p53 in response to DNA damage (26), and in the regulation of autophagy (25). Furthermore, USP10 may also be involved in trans-lesion synthesis, which is the bypass of DNA lesions to overcome stalled replication at sites of DNA damage in the template strand. This is performed by a specialized DNA polymerase that allows DNA synthesis to continue by inserting a nucleotide at the lesion site (29). Thus, USP10 has a diverse array of relatively well characterized molecular and biological functions. However, its precise role in the development and progression of various forms of cancer remains to be elucidated.
Lung cancer has the highest rates of cancer mortality worldwide, and multiple studies have investigated the role of USP10 in lung cancer. For example, Sun et al. reported that USP10 was significantly downregulated in lung cancer, and USP10 may directly interact with and stabilize the missing phosphatase and tensin homologue on chromosome 10 (PTEN) through deubiquitination, thereby acting as a tumor suppressor (30).
In non-small cell lung cancer (NSCLC), USP10 can deubiquitinate K63-linked polyubiquitination, thereby restoring the phosphatase activity of PTEN, reducing the secondary messenger phosphatidylinositol 3,4,5-triphosphate, which in turn attenuates AKT/mammalian target of rapamycin growth-promoting signaling, ultimately inhibiting NSCLC proliferation (31). Additionally, USP10 stabilizes and deubiquitinates MutS homolog 2 (MSH2), which is the core protein of the MutS homology family and is involved in DNA mismatch repair. These findings also suggest that USP10 downregulation leads to stabilization of the MSH2 protein, and thereby is implicated in the tumorigenesis of NSCLC (32, 33). Moreover, USP10 plays a role in regulating p53 ubiquitination and degradation. Zhao et al. reported that the half-life of p53 protein was significantly reduced in insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3)-overexpressing cells, and co-expression of IGF2BP3 with USP10 promoted the ubiquitination level of p53. Therefore, IGF2BP3 might attenuate the deubiquitination of p53 via USP10 (34).
Additionally, Ko et al. found that c-Myc could promote oncogene-induced senescence through the transcriptional activation of p14 alternate reading frame (p14ARF), followed by the activation of p53. Simultaneously, c-Myc increased the stability of p14ARF by inducing the transcription of USP10. Clinically, patients with NSCLC have significantly reduced overall survival (OS) due to disruption of the c-Myc-USP10-p14ARF axis (35). Hu et al. demonstrated that high levels of USP10 were associated with poor OS in TP53-mutant NSCLC but not in wild-type NSCLC. Likewise, USP10 knockout significantly reduced the growth of p53-mutated lung cancer xenografts and increased their sensitivity to cisplatin in vivo. Thus, USP10 might affect cisplatin resistance by deubiquitinating and thereby stabilizing the oncogenic protein histone deacetylase 6 (HDAC6) (36, 37). Another study showed that USP10 plays an important regulatory role in NSCLC via deubiquitinating and stabilizing histone deacetylase 7 (HDAC7), and that USP10 inhibition could significantly accelerate HDAC7 degradation and impair NSCLC proliferation and migration (38).
HCC has become one of the most common life-threatening cancers, due to its susceptibility to metastasis. Yuan et al. indicated that metastasis of advanced HCC might be closely related to the persistent activation of transforming growth factor-β (TGF-β) and Smad4. Yuan and colleagues found that USP10 deubiquitinates Smad4, maintains its protein expression level, and activates TGF-β signaling, thereby promoting HCC metastasis (39). Another study showed that the long non-coding RNA growth arrest associated lncRNA 1 (GASAL1), which may promote HCC progression, can upregulate USP10 expression by competitively binding to miR-193b-5p. Importantly, USP10 can also enhance HCC cell proliferation and migration by deubiquitinating proliferating cell nuclear antigen (PCNA) (40). Yes-associated protein (YAP) and its paralogs, transcriptional co-activators with a PDZ-binding motif (TAZ), play important roles in promoting HCC progression. Zhu et al. found that USP10 activated yes-associated protein/transcriptional coactivator with PDZ-binding motif (YAP/TAZ) activating gene and reduced proteolysis of the YAP/TAZ protein (17). In contrast to the above findings, Lu et al. found that USP10 could deubiquitinate and stabilize AMP-activated protein kinase α (AMPKα) and PTEN in HCC cells, leading to inhibition of mTOR Complex1 (mTORC1) and reduced AKT phosphorylation. Therefore, paradoxically, USP10 might also function as a tumor suppressor in HCC (41). USP10 may promote HCC proliferation on the one hand, yet on the other, it may function as a tumor suppressor. More research is required to clarify the precise role of USP10 in HCC.
The 5-year OS rate for AML patients is only 20% (42). In particular, patients with FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) mutations have a poor prognosis, as tumors with these mutations are associated with increased invasiveness and lethality (43). Furthermore, treatment with FLT3 kinase inhibitors has short-term efficacy due to development of drug resistance (44). Weisberg et al. revealed that USP10, a key enzyme for tumor growth in FLT3-ITD-positive AML, might inhibit FLT3 degradation through deubiquitination (18). The non-receptor tyrosine kinase spleen tyrosine kinase (SYK) is activated in FLT3-ITD-positive AML patients. This enzyme is critical for transformation and is associated with resistance to FLT3-targeting tyrosine kinase inhibitors (TKIs). Recent studies have shown that SYK and FLT3 are regulated by ubiquitination and deubiquitination (45–47). Interestingly, small-molecule inhibitors of USP10 have been found to inhibit the activated SYK-driven proliferation of leukemia cells and induce the degradation of SYK protein. USP10 can form a complex with FLT3-ITD and physically bind with SYK. USP10 stabilizes the expression of FLT3 and SYK by deubiquitination (48). Therefore, USP10 may be an effective drug target in kinase inhibitor-resistant AML.
Ovarian cancer (OC) is highly malignant and prevalent worldwide. Despite the high cure rates for early-stage OC, most patients are diagnosed with advanced tumors and the mortality rate is 75% (49). One study showed that reduced USP10 expression or reduced p14ARF/USP10 expression is an effective indicator of poor prognosis in OC patients (50). In addition, Li et al. proposed that GTPase-activating protein-binding protein 1 (G3BP1) could promote the proliferation and invasion of OC cells (51), and that USP10 knockdown could restore the proliferation and invasion of OC cells inhibited by G3BP1 knockdown. Thus, USP10 may act as a tumor suppressor in OC, however, more research is needed to elucidate its precise role.
Prostate cancer (PCa) is a common malignancy among men, in which androgens and the androgen receptor (AR) play key roles in PCa tumorigenesis (52–54). Many patients are resistant to androgen deprivation therapy (ADT) or develop castration resistance (55, 56), therefore, there is an urgent need to find new therapeutic targets. USP10 stabilizes p53 by deubiquitination and thereby regulates AR-induced epigenetic signaling. GTPase-activating protein-binding protein 2 (G3BP2) is an androgen-responsive gene that can be stabilized by USP10-mediated deubiquitination. Therefore, USP10 may have an important oncogenic role in PCa by regulating the p53-G3BP2 complex and AR signaling (11). Recent studies have suggested that abnormal activation of the epidermal growth factor receptor (EGFR) might promote the progression of castration-resistant PCa by inhibition of androgen signaling (57, 58). Spautin-1, an inhibitor of USP10/13 (25), may inhibit EGFR-related signaling pathways, inducing activation of the MKK4/JNK/Bax axis and inactivation of the MEK1/2/ERK/cyclin D1 axis, leading to significant inhibition of PCa proliferation (59).
Breast cancer (BC) is one of the most common cancers, accounting for 30% of all cancer cases in women (60, 61). Yang et al. identified a novel circRNA, circWSB1, which is highly expressed in BC tissues and strongly associated with poor prognosis in patients with BC. Upon direct binding to USP10, circ WSB1 can reduce USP10-mediated p53 stability, resulting in p53 degradation and promoting BC development (62). USP10 may also contribute to BC progression through topoisomerase IIα (TOP2α), which is essential for chromosome condensation, segregation, and genome integrity (63). RING finger protein 168 (RNF168), an E3 ligase, interacts with TOP2α and mediates its ubiquitination. RNF168 deficiency impairs TOP2α activity and promotes mitotic abnormalities and chromosome segregation defects. RNF168 deficiency in human breast cancer cell lines leads to drug resistance, including resistance to the TOP2 inhibitor, etoposide. USP10 may deubiquitinate TOP2α and inhibit chromatin binding. Therefore, interactions between USP10, RNF168, and TOP2α may play a key role in the development of tumor resistance to TOP2 inhibitors (64).
Gastric cancer (GC) is highly malignant, with 700,000 GC-related deaths occurring annually worldwide (65). Zeng et al. demonstrated that the expression level of USP10 in GC tissues and cell lines was lower than that in non-cancerous mucosal tissues and gastric epithelial immortalized cell lines. Moreover, the expression level of USP10 was negatively correlated with invasion and lymph node metastasis in GC. Low USP10 expression was significantly associated with poor prognosis in GC patients. Furthermore, multivariate analysis indicated that USP10 was an independent prognostic factor for OS in patients with GC (20).
Small intestinal adenocarcinoma is rare, accounting for only 2% of gastrointestinal malignancies (66). p14ARF, encoded by a reading frame within the cyclin-dependent kinase inhibitor p16/the p53 regulator p14 (INK4a/ARF) locus on chromosome 9p21 (67), can inhibit the E3 ubiquitin protein ligase mouse double minute 2 (MDM2)-mediated degradation of TP53, thereby increasing TP53 stability and leading to cell cycle arrest and apoptosis (68). Song et al. revealed that USP10 deletion is associated with advanced tumor-related phenotypes, and that co-deletion of USP10 and p14ARF yields poor outcomes in small intestinal adenocarcinoma, indicating that USP10 and p14ARF may be involved in small intestinal adenocarcinoma (69).
Li et al. reported that NACHT, LRR, and PYD domain-containing protein 7 (NLRP7) plays a key oncogenic role in the proliferation and metastasis of colorectal cancer (CRC) and is associated with poor prognosis. USP10-mediated deubiquitination stabilized NLRP7 protein expression and induced polarization of tumor-promoting M2-like macrophages through NF-κB pathway-mediated monocyte chemoattractant protein-1 (MCP-1) secretion (70). In addition, the tumor suppressor sirtuin 6 (SIRT6) is significantly inversely correlated with tumorigenesis (71), and USP10 can inhibit the transcriptional activity of the c-Myc by increasing the protein stability of SIRT6 and p53, ultimately inhibiting the progression of CRC (72). Kim et al. also noted that USP10 expression was absent in 18.6% of CRC tissue and was significantly associated with distant metastasis and lymph vascular invasion. Likewise, USP10 deletion was associated with shorter OS, disease-free survival, and was an independent prognostic factor in CRC patients (73). In contrast, another study showed that USP10 promoted CRC cell proliferation by deubiquitinating and stabilizing the oncogenic factor musashi 2 (MSI2) (74).
Pancreatic cancer is an extremely lethal malignancy with a high mortality rate worldwide (75). The role of USP10 in pancreatic cancer remains controversial. For example, Liu et al. reported that USP10 promoted Cysteine-Rich 61 (Cyr61) expression by inhibiting YAP1 ubiquitination and degradation, thereby favoring immune escape and promoting the proliferation and metastasis of pancreatic cancer (19). However, another study suggested that miR-191 promotes pancreatic cancer cell proliferation by inhibiting USP10 expression (76). Furthermore, it has been reported that miR-103 may also downregulate USP10 in pancreatic cancer cell lines and tissues. Upregulation of miR-103 expression is associated with lymph node metastasis, advanced TNM stage, and poor prognosis (77). Thus, the specific regulatory role of USP10 in pancreatic cancer requires further investigation.
Neurotrophin receptor-interacting MAGE homologue (NRAGE) is generally considered to be a tumor suppressor, yet Yang et al. found that NRAGE significantly promoted esophageal carcinoma, mainly via upregulation of PCNA. Knockdown of NRAGE promoted PCNA K48-linked polyubiquitination, leading to proteasome-dependent degradation of PCNA and cancer proliferation inhibition, and USP10 is a key regulator in this process (78).
In chronic myeloid leukemia (CML), constitutive activation of the tyrosine kinase Bcr-Abl is a major cause of disease development and progression (79). However, acquired resistance to Bcr-Abl-targeted TKIs severely affects the prognosis of patients with advanced CML. Liao et al. reported that as a co-regulator of Bcr-Abl, S-phase kinase-associated protein 2 (SKP2) mediates activation of its K63 linkages. USP10 can further enhance Bcr-Abl activation by promoting the deubiquitination of SKP2 in CML cells, and stabilizing its protein expression. This study demonstrated that targeting the USP10/SKP2/Bcr-Abl axis could reverse imatinib resistance in CML patients (80).
In glioblastoma (GBM), one study showed that Cyclin D1 (CCND1) is involved in cell cycle control and promotes tumor progression (81). In this study, Sun et al. found that USP10 may interact with CCND1 and prevent its K48-linked polyubiquitination, increasing CCND1 stability. Overall, the USP10/CCND1 axis is expected to be an effective target for the treatment of GBM (82).
In thyroid cancer, Cui et al. reported that 3-deazaneplanocin A (DZNep) may cause the accumulation of p53 protein by upregulating USP10 expression, thereby activating the p53 pathway and ultimately inhibiting the growth of TP53 wild-type thyroid cancer cells (83).
The role of USP10 in different tumors, as discussed above, indicate that the deubiquitination substrates of USP10 are diverse and that USP10 targets different signaling pathways (Table 1). Additionally, we found that USP10 acts as an oncogene in some cancers but not in others, due to the different functions of genes deubiquitinated by USP10. For example, if the gene deubiquitinated by USP10 is an oncogene, it may lead to tumor progression, and if the gene deubiquitinated by USP10 is a tumor suppressor gene, it may lead to tumor suppression. In short, USP10 cannot be simply defined as a tumor suppressor gene or an oncogene, and its function may be directly related to the function of its deubiquitinated gene, rather than USP10 itself having oncogenic or tumor suppressor effects. In conclusion, USP10 plays a diverse set of roles in either promoting or inhibiting the progression of various of cancers. Clinically, it may serve as a new tumor biomarker and potential therapeutic target, however, its precise role would be dependent on the cancer type.
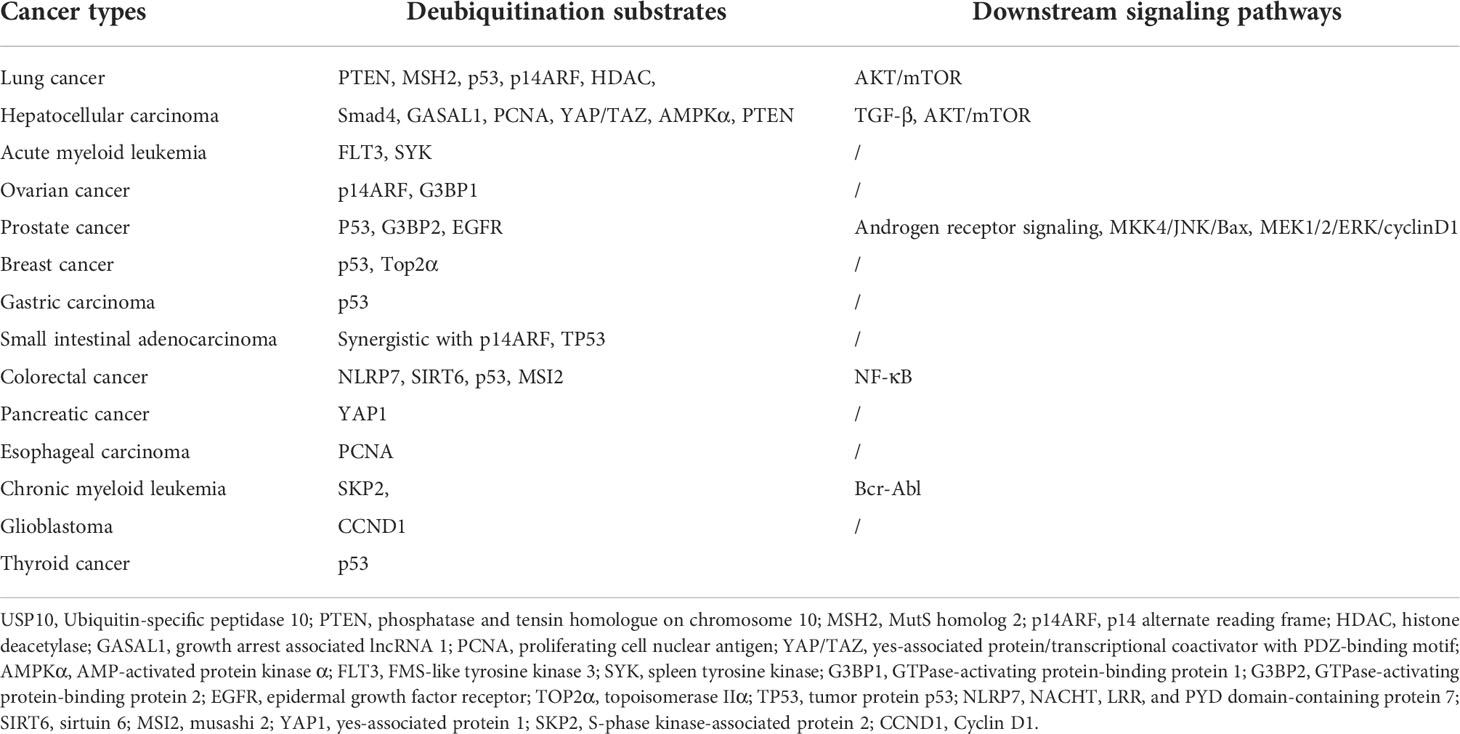
Table 1 The deubiquitination substrates and downstream signaling pathways of USP10 in different cancer types.
The ubiquitin-binding site in USP10 functions as a thiol-dependent isopeptidase, which separates ubiquitin from target proteins, and thereby mediates their stability (84). In this context, USP10 inhibitors that target the binding site might effectively interfere with its oncogenic effects. USP10 expression levels differ widely across various tumors and are significantly correlated with the prognosis of these tumors. Thus, USP10 inhibitors may be clinically useful in cancer settings where overexpression of USP10 is associated with tumor progression or in settings where USP10 serves as an oncogene. To date, only a few USP10 inhibitors have been identified, P22077, HBX19818, and spautin-1 (85). P22077 and HBX19818 were first identified as irreversible USP7 inhibitors. However, one study showed that P22077 and HBX19818 might also inhibit the proliferation of FLT3-ITD-positive tumor cells by inhibiting the deubiquitinase activity of USP10 (18). Spautin-1, a small-molecule inhibitor, has been shown to inhibit the deubiquitinase activities of both USP10 and USP13, leading to increased ubiquitination and degradation of Beclin1 in the Vps34 complex, and an eventual decrease in the level of autophagy in cancer cell lines (25). In addition, spautin-1 inhibition of USP10 significantly attenuates the migration of HCC cells (39). Interestingly, spautin-1 also inhibits the proliferation of prostate cancer, NSCLC, ovarian cancer, and melanoma cells in a USP10-independent manner (86). Spautin-A41 (an analog of spautin-1), was recently found to be more effective than spautin-1 in inhibiting autophagy and inducing microsomal stability (87). However, the mechanism of action of spautin-1 and spautin-A41 in cancer patients still needs to be confirmed. Recently, compound library screening revealed that Wu-5, a novel USP10 inhibitor, enhanced the anti-AML effect of crenolanib and reversed FLT3 inhibitor resistance. Mechanistically, Wu-5 inhibited the proliferation of MV4-11 cells mainly by inhibiting the activity of USP10 and subsequently reducing the expression of the downstream gene AMPKα (88).
There are also several USP10 modulators that indirectly influence USP10 activity. For example, DZNep stabilizes p53 by upregulating USP10 to reduce ubiquitin binding in wild-type GC cells, thereby activating p53 and inhibiting the proliferation of GC cells (89). Quercetin (C15H10O7) is a pentahydroxy flavonoid widely present in vegetables and fruits, and it has been shown that quercetin could reduce USP10 expression, resulting in the downregulation of T-bet expression levels (15). Quercetin inhibits the proliferation of several cancer cells, including colon cancer, breast cancer, and lung cancer (90). However, the mechanism of how quercetin inhibits USP10 remains unclear. Another potential USP10 modulator, Cai’s Neiyi prescription (CNYP), can alleviate inflammation by inhibiting USP10 and promoting apoptosis of endometrial stromal cells, suggesting that CNYP may be an inhibitor of USP10 (91). Unlike other USP10 inhibitors, ubiquitin variant.10.1 (UbV.10.1) is a mixture of peptides and proteins with a high affinity for USP10. UbV.10.1 overexpression can promote the degradation of p53 by inhibiting USP10 (92). However, the antitumor effects of both CNYP and UbV.10.1 have yet to be explored.
No USP10 inhibitors have been considered in clinical trials for cancer treatment. This is likely owing to the low selectivity of most USP10 inhibitors. As such, technologies such as polymer chemistry and selective structural modification may offer strategies to improve both the anti-tumor effects of USP10 inhibitors and their selectivity for tumor cells. Furthermore, it would be necessary to identify suitable patients for USP10 inhibitor treatment based on their cancer type. Considering the immunoregulatory role of USP10, combining USP10 inhibitors with immunotherapy may be a promising future research avenue.
After discussing the literature and knowledge gaps above, we found that the role of USP10 in cancer is conflicting. Therefore, to further examine the expression of USP10 in cancer, we compared the differences in USP10 expression between tumor tissues and adjacent normal tissues by analyzing gene expression data of USP10 in different human tumor samples from The Cancer Genome Atlas (TCGA) database. The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) (93) showed that USP10 gene expression was significantly upregulated in 12 types of tumor samples: breast invasive carcinoma, bladder urothelial carcinoma, cholangiocarcinoma, esophageal carcinoma, colon adenocarcinoma, kidney renal papillary cell carcinoma, head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma, liver hepatocellular carcinoma, lung squamous cell carcinoma, stomach adenocarcinoma, and rectum adenocarcinoma. Conversely, significant downregulation of USP10 gene expression was observed in two types of tumor samples: pheochromocytoma and paraganglioma (PCPG) and uterine corpus endometrial carcinoma (UCEC) (Figure 2A). Based on data from the International Cancer Proteogenome Consortium (ICPC) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC), we analyzed differences in USP10 protein expression between tumor tissues and adjacent normal tissues (94). USP10 protein expression was significantly upregulated in seven types of tumor samples: breast cancer, colon cancer, ovarian cancer, UCEC, lung cancer, HNSC, and glioblastoma. However, significant downregulation of USP10 protein expression was observed in three types of tumor samples: kidney renal clear cell carcinoma (KIRC), pancreatic cancer, and liver cancer (Figure 2B). Interestingly, USP10 was highly expressed in lung cancer samples in the TCGA and CPTAC databases (Figure 2). These results appear to be inconsistent with those reported in the literature. We speculated that it may be due to tumor heterogeneity or the insufficient sample size. Therefore, whether USP10 expression is increased or decreased in lung cancer and whether USP10 plays a role in promoting or suppressing lung cancer require further investigation.
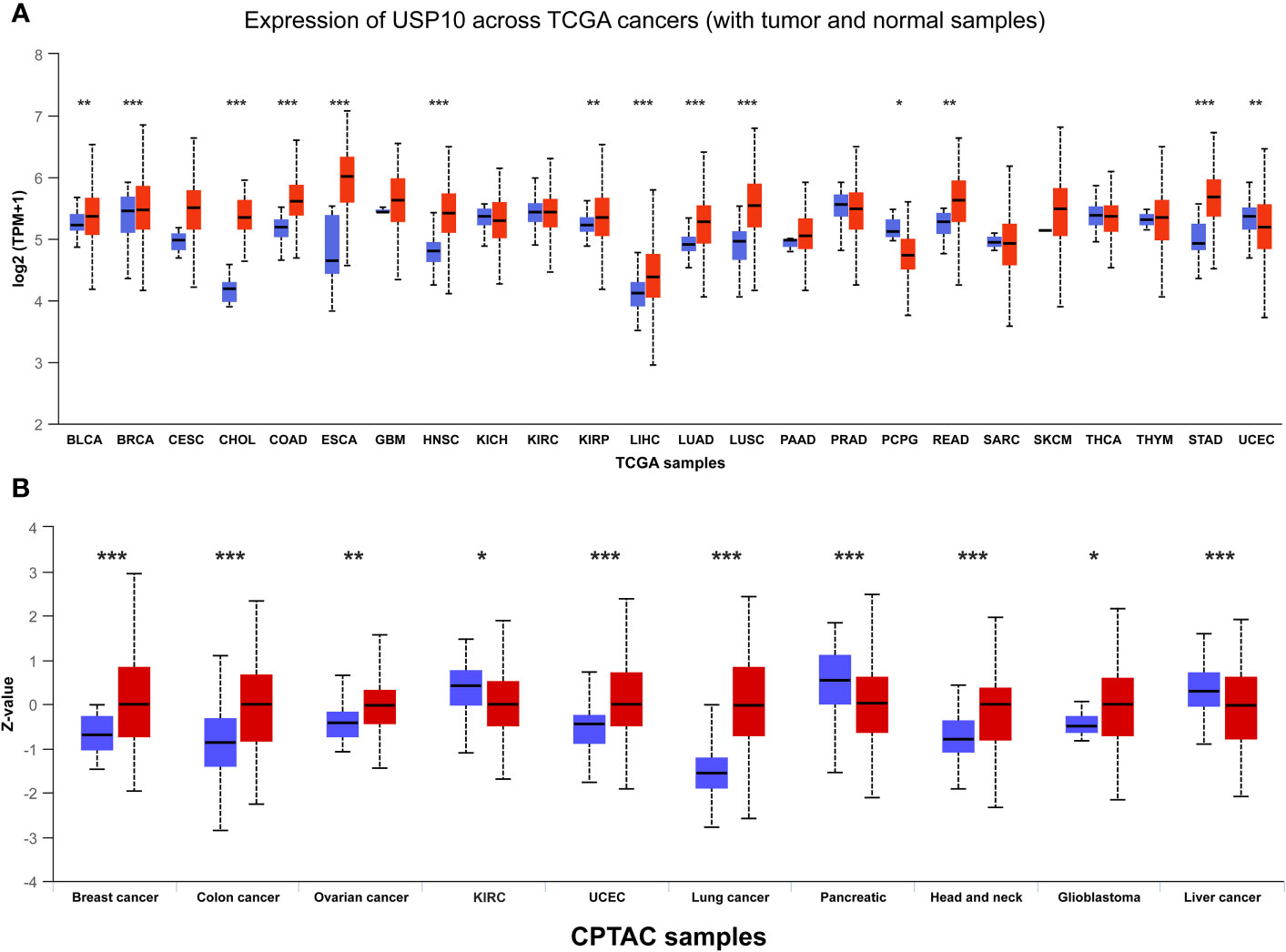
Figure 2 The gene and protein expressions of USP10 in pan-cancers based on TCGA and CPTAC databases. (A) The gene expressions of USP10 in pan-cancers based on TCGA database; (B) The protein expressions of USP10 in pan-cancers based on CPTAC database. Red color: tumor tissues; Blue color: normal tissues. USP10, Ubiquitin-specific peptidase 10; TCGA, The Cancer Genome Atlas; CPTAC, Clinical Proteomic Tumor Analysis Consortium. *P < 0.05; **P < 0.01; ***P < 0.001.
In addition, we investigated whether there was a correlation between USP10 expression and OS or tumor stage. Data collected from the TISIDB website (an integrated repository portal for tumor-immune system interactions) showed that HNSC and prostate adenocarcinoma (PRAD) patients with increased USP10 expression had shorter OS, suggesting that USP10 plays an oncogenic role in HNSC and PRAD (95). Further validation analysis (https://kmplot.com/analysis/) revealed that low levels of USP10 expression was significantly associated with prolonged OS in HNSC patients (Figure 3A). However, USP10 was downregulated in KIRC, and high levels of USP10 was associated with prolonged OS in KIRC patients, suggesting that USP10 may have a tumor suppressor role in KIRC (Figure 3A). In a pan-cancer analysis, we found that high USP10 expression was associated with early tumor stage in KIRC and UCEC patients. However, in testicular germ cell tumor (TGCT) patients, high USP10 expression predicted later tumor stage (Figure 3B). Consistent with these findings, another study also has demonstrated the close relationship between USP10 expression and the progression and prognosis of various cancers, including lung cancer, hepatocellular carcinoma (HCC), ovarian cancer, prostate cancer, and AML (96).
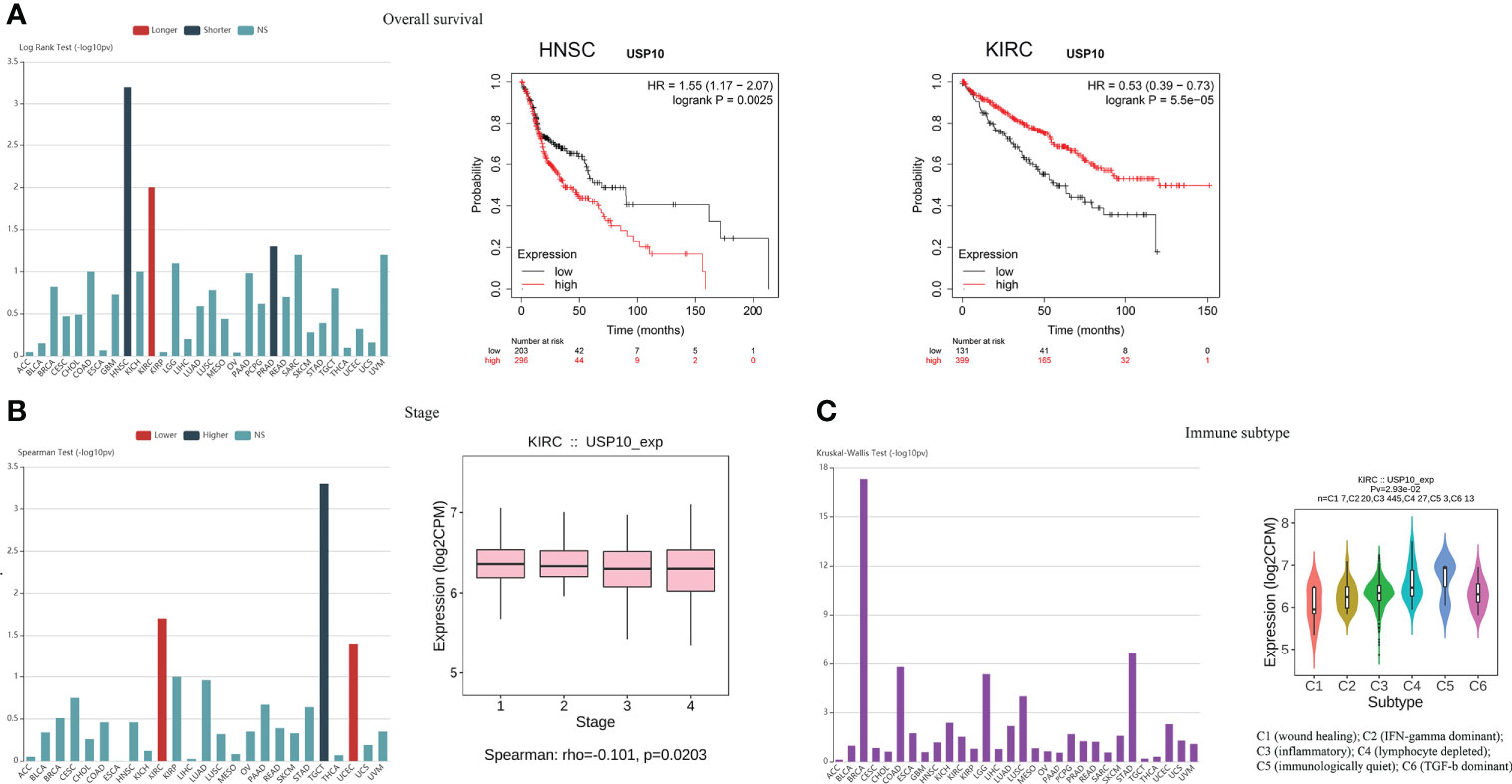
Figure 3 Analysis of the TISIDB and Kaplan-Meier plotter website indicated the role of USP10 in tumors and immune subtype. (A) HNSC patients with increased USP10 expression had lowered OS, however, high levels of USP10 were associated with prolonged OS in KIRC patients. (B) The correlation between USP10 expression and tumor stage in KIRC patients. (C) USP10 was correlated with the immune subtype in KIRC. NS, not significant. USP10, Ubiquitin-specific peptidase 10; HNSC, head and neck squamous cell carcinoma; OS, overall survival; KIRC, kidney renal clear cell carcinoma.
Deubiquitination is a common post-translational modification that modulates multiple cellular functions, including protein stability. Meanwhile, deubiquitination also affects multiple signal transduction pathways, including some immunomodulatory pathways (97). For example, USP10 promotes the deubiquitination of tumor necrosis factor receptor-associated factor 6 (TRAF6) and inhibits NF-κB and the interleukin-1 receptor/Toll-like receptor (IL-1R/TLR) activation (14). TLRs can promote inflammation and immune response in tumors, suggesting a potential role for USP10 in the regulation of immune pathways. Growing evidence suggests that USP10 may be involved in the infiltration of various immune cells in tumors, potentially regulating the infiltration levels of specific immune cells (98); however, its precise role in tumor immune response remains unclear. We further analyzed data from the TISIDB (http://cis.hku.hk/TISIDB/index.php) to evaluate the correlation between USP10 and immunostimulatory molecules, immunosuppressive molecules, major histocompatibility complex (MHC) molecules, chemokines, and MHC receptors, to better understand the immune function of USP10 in cancer regulation.
In HNSC, USP10 was positively correlated with the immunosuppressive molecules transforming growth factor beta receptor 1 (TGFBR1), the immunostimulator tumor necrosis factor receptor superfamily member 14 (TNFRSF14), the chemokine C-X-C motif ligand 8 (CXCL8), the MHC molecule transporter associated with antigen processing 2 (TAP2), lymphocyte T helper type (Th2), and the MHC receptor C-X-C chemokine receptor 1 (CXCR1) (P<0.05, Figure 4). In KIRC, USP10 was correlated with the immune subtype (P=0.029) (Figure 3C). Furthermore, USP10 was positively correlated with the immunosuppressive molecules CD274, immunostimulator TNF receptor superfamily 25 (TNFRSF25), chemokine C-X-C motif ligand 16 (CXCL16), β2-microglobulin (B2M), lymphocyte immature dendritic cells (iDC), and MHC receptor CC Chemokine receptor 1 (CCR1) (P<0.05, Figure 4). Analysis of the TIMER database (http://timer.cistrome.org/) verified the correlation of USP10 with these immune markers (Figures 5, 6). CD274 plays a key role in the induction and maintenance of self-immune tolerance. As a ligand for the inhibitory receptor programmed cell death 1 (PDCD1)/CD279, CD274 modulates the activation threshold of T cells and limits T cell effector responses (99). Tumors utilize PDCD1/CD279-mediated inhibitory pathways to attenuate antitumor immunity and promote tumor survival (100). CXCL16 is a chemotactic agent for immunosuppressive regulatory T cells (Treg), which may be involved in Treg recruitment and tumorigenic functions (101). B2M is an important subunit of MHC class I and plays important biological functions in tumorigenesis and immune control. There is increasing evidence that alterations in B2M genes and proteins contribute to poor responses to cancer immunotherapy by inhibiting antigen presentation (102). CCR1 is a receptor for C-C type chemokines. It may play a role in Treg and tumor immunosuppression (103). TNFRSF25 is a member of the TNF receptor superfamily (TNFRSF) that binds to the TNF-like protein TL1A. Recent studies have demonstrated a role for TNFRSF25 in regulating CD4+ T cell responses. Additionally, TNFRSF25 signaling in CD8+ T cells positively affects the proliferation of CD8+ T cells and their differentiation into CTLs (104). The immature DC phenotype can reduce dendritic cell function, reduce antigen presentation, and affect T cell effector function (105). The above results suggest that USP10 may play an immunosuppressive function by promoting the expression of some immunosuppressive cytokines or by inhibiting the expression of some immune-activating cytokines. However, the specific immune regulation mechanism of USP10 remains to be fully elucidated.
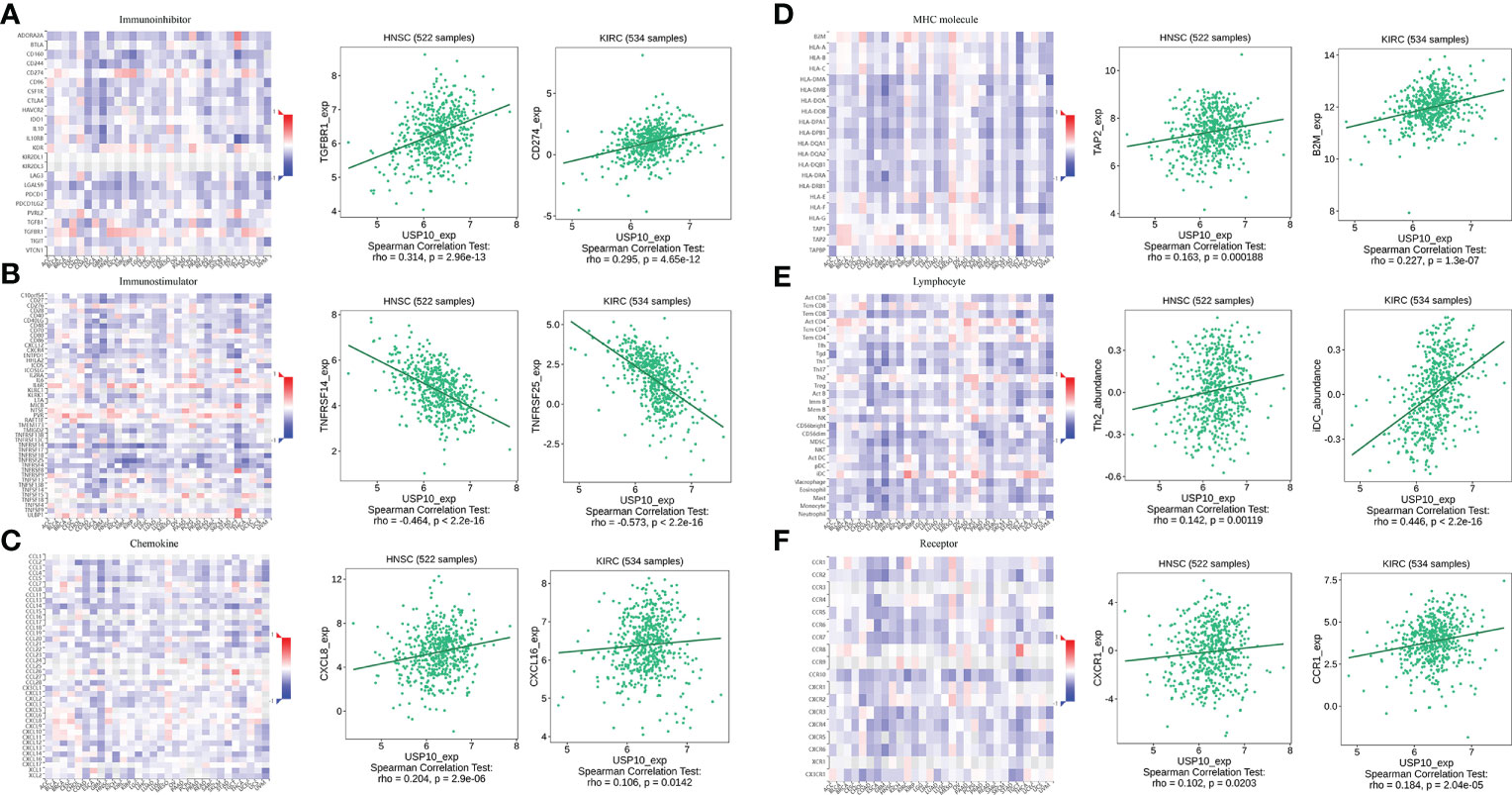
Figure 4 USP10 was positively correlated with immunosuppressive molecules (A), immunostimulatory molecules (B), chemokines (C), MHC molecules (D), lymphocytes (E), and MHC receptors (F). USP10, Ubiquitin-specific peptidase 10; MHC, major histocompatibility complex.
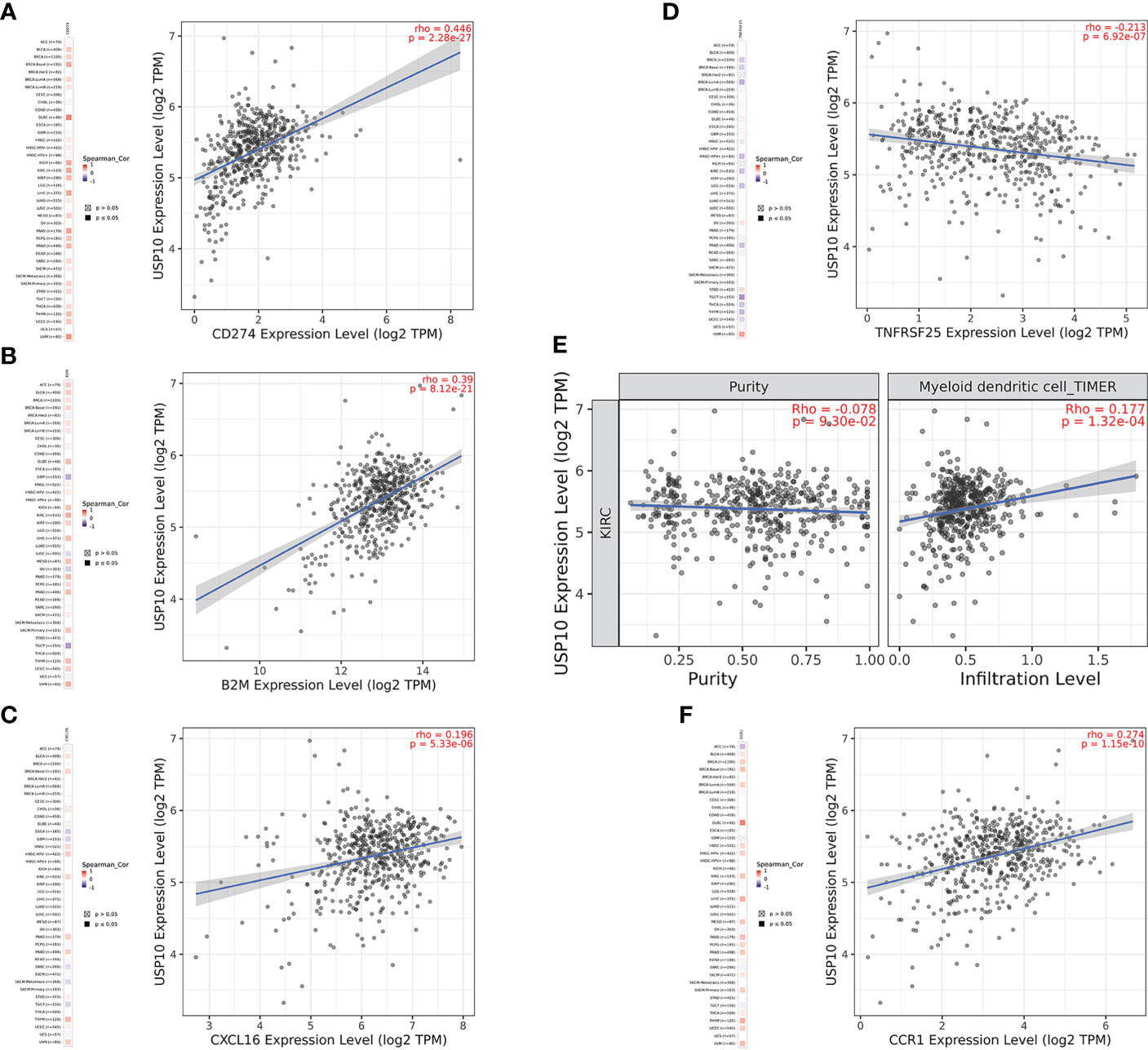
Figure 6 USP10 was positively correlated with CD274 (A), B2M (B), CXCL16 (C), myeloid dendritic cell (E), and CCR1 (F), but negatively correlated with TNFRSF25 (D) in KIRC. USP10, Ubiquitin-specific peptidase 10; TAP2, transporter associated with antigen processing 2; TGFBR1, transforming growth factor beta receptor 1; HNSC, head and neck squamous cell carcinoma; B2M, β2-microglobulin; CXCL16, C-X-C motif ligand 16; CCR1, CC Chemokine receptor 1; TNFRSF25, TNF receptor superfamily 25; KIRC, kidney renal clear cell carcinoma.
We also analyzed the correlations between USP10 and pathway scores by Spearman’s correlation. In HNSC, USP10 was found to be correlated with Epithelial-mesenchymal transition (EMT), extracellular matrix (ECM), angiogenesis, apoptosis, tumor inflammation, G2/M checkpoint, ferroptosis, PI3K/AKT/mTOR, MYC, TGF-β (Figure 7). In KIRC, we found that the signaling pathways significantly related to USP10 included ferroptosis, DNA repair, G2/M checkpoint, inflammatory response, PI3K/AKT/mTOR, p53, c-Myc, TGF-β, and reactive oxygen species (ROS) (Figure 8). In summary, we suggest that USP10 maybe a novel target for cancer immunotherapy; however, the specific mechanisms of USP10 involvement in cancer immunity remain unclear and further research is required.
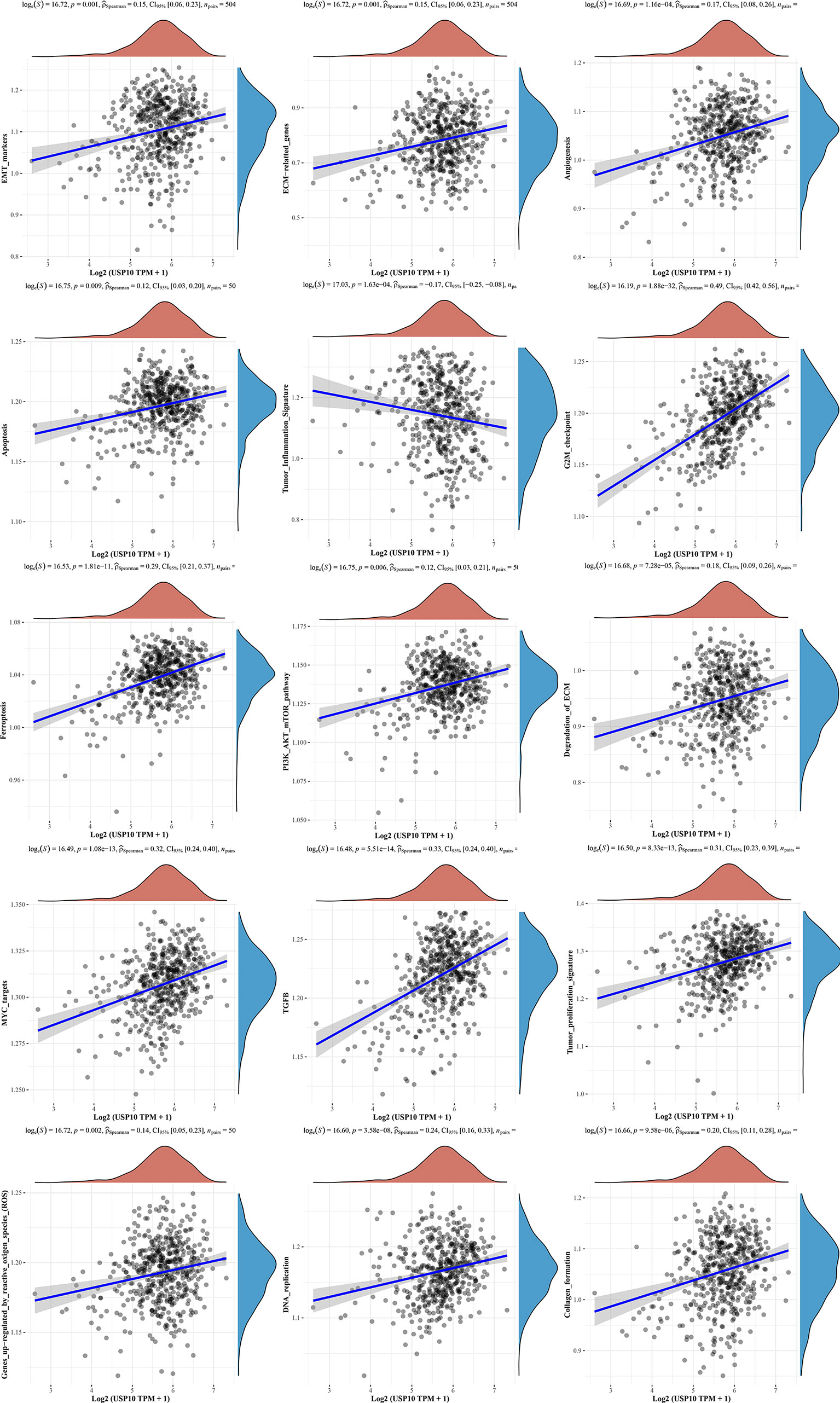
Figure 7 The signaling pathways significantly related to USP10 in HNSC included EMT, ECM, angiogenesis, apoptosis, tumor inflammation, G2/M checkpoint, ferroptosis, PI3K/AKT/mTOR, MYC, TGF-β, tumor proliferation, reaction oxygen species, DNA replication, and collagen formation. USP10, Ubiquitin-specific peptidase 10; HNSC, head and neck squamous cell carcinoma; EMT, Epithelial-mesenchymal transition; ECM, extracellular matrix.
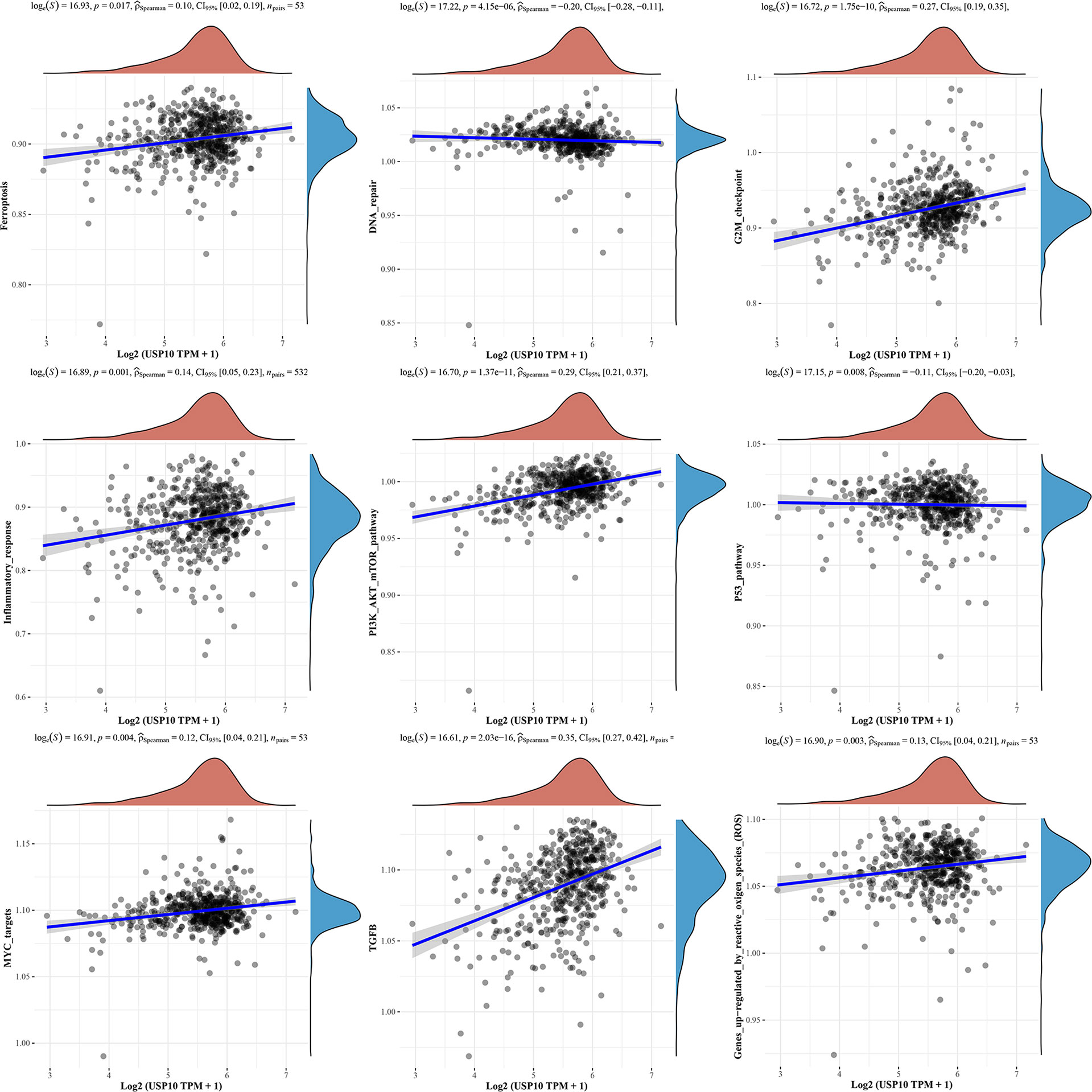
Figure 8 The signaling pathways significantly related to USP10 in KIRC included ferroptosis, DNA repair, G2/M checkpoint, inflammatory response, PI3K/AKT/mTOR, p53, c-Myc, TGF-β, and ROS. USP10, Ubiquitin-specific peptidase 10; KIRC, kidney renal clear cell carcinoma; ROS, reactive oxygen species.
USP10 acts as a tumor suppressor or tumor-promoting gene depending on the target genes modified by deubiquitination. In this paper, we have discussed the biological function of USP10 and its key role in tumor progression and immune response. We have demonstrated that USP10 is a potential drug target for immunotherapy in tumors. In addition, we have summarized the current knowledge regarding USP10 inhibitors. However, it remains unclear how USP10 regulates immunity and whether USP10-mediated deubiquitination may affect tumor response to immunotherapy. Therefore, studying the role and mechanism of USP10 immune regulation will provide a theoretical basis for the clinical application of USP10-oriented therapies in the future.
ZY and ZX designed this study, JC, ZY, and SZ edited the manuscript and contributed to the systematic evaluation of articles and literature. ZX and PH participated in the writing and revision of the manuscript, and all authors approved the final version of the article and agreed to be responsible for all aspects of the work.
This study was supported by the Joint Fund of Zhejiang Provincial Natural Science Foundation (Grant No. LYY21H310005), Natural Science Foundation of Zhejiang Province (Grant Number LYY21H310008), Zhejiang Traditional Chinese Medicine Science and Technology Plan (Grant Number 2022ZQ014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer (2020) 19(1):146. doi: 10.1186/s12943-020-01262-x
2. Chen HF, Chuang HC, Tan TH. Regulation of dual-specificity phosphatase (Dusp) ubiquitination and protein stability. Int J Mol Sci (2019) 20(11):2668. doi: 10.3390/ijms20112668
3. Dumetier B, Zadoroznyj A, Dubrez L. Iap-mediated protein ubiquitination in regulating cell signaling. Cells (2020) 9(5):1118. doi: 10.3390/cells9051118
4. Sharma A, Alswillah T, Singh K, Chatterjee P, Willard B, Venere M, et al. Usp14 regulates DNA damage repair by targeting Rnf168-dependent ubiquitination. Autophagy (2018) 14(11):1976–90. doi: 10.1080/15548627.2018.1496877
5. Shen J, Chen M, Lee D, Law CT, Wei L, Tsang FH, et al. Histone chaperone fact complex mediates oxidative stress response to promote liver cancer progression. Gut (2020) 69(2):329–42. doi: 10.1136/gutjnl-2019-318668
6. Mansour MA. Ubiquitination: Friend and foe in cancer. Int J Biochem Cell Biol (2018) 101:80–93. doi: 10.1016/j.biocel.2018.06.001
7. Mukherjee S, Kumar R, Tsakem Lenou E, Basrur V, Kontoyiannis DL, Ioakeimidis F, et al. Deubiquitination of Nlrp6 inflammasome by cyld critically regulates intestinal inflammation. Nat Immunol (2020) 21(6):626–35. doi: 10.1038/s41590-020-0681-x
8. Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ (2021) 28(2):591–605. doi: 10.1038/s41418-020-00708-5
9. Do HA, Baek KH. Cellular functions regulated by deubiquitinating enzymes in neurodegenerative diseases. Ageing Res Rev (2021) 69:101367. doi: 10.1016/j.arr.2021.101367
10. Yao RQ, Ren C, Xia ZF, Yao YM. Organelle-specific autophagy in inflammatory diseases: A potential therapeutic target underlying the quality control of multiple organelles. Autophagy (2021) 17(2):385–401. doi: 10.1080/15548627.2020.1725377
11. Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of Usp10 with G3bp2 inhibits P53 signaling and contributes to poor outcome in prostate cancer. Mol Cancer Res (2018) 16(5):846–56. doi: 10.1158/1541-7786.MCR-17-0471
12. Bomberger JM, Coutermarsh BA, Barnaby RL, Stanton BA. Arsenic promotes ubiquitinylation and lysosomal degradation of cystic fibrosis transmembrane conductance regulator (Cftr) chloride channels in human airway epithelial cells. J Biol Chem (2012) 287(21):17130–9. doi: 10.1074/jbc.M111.338855
13. Boulkroun S, Ruffieux-Daidie D, Vitagliano JJ, Poirot O, Charles RP, Lagnaz D, et al. Vasopressin-inducible ubiquitin-specific protease 10 increases enac cell surface expression by deubiquitylating and stabilizing sorting nexin 3. Am J Physiol Renal Physiol (2008) 295(4):F889–900. doi: 10.1152/ajprenal.00001.2008
14. Wang W, Huang X, Xin HB, Fu M, Xue A, Wu ZH. Traf family member-associated nf-kappab activator (Tank) inhibits genotoxic nuclear factor kappab activation by facilitating deubiquitinase Usp10-dependent deubiquitination of Traf6 ligase. J Biol Chem (2015) 290(21):13372–85. doi: 10.1074/jbc.M115.643767
15. Pan L, Chen Z, Wang L, Chen C, Li D, Wan H, et al. Deubiquitination and stabilization of T-bet by Usp10. Biochem Biophys Res Commun (2014) 449(3):289–94. doi: 10.1016/j.bbrc.2014.05.037
16. Wang X, Xia S, Li H, Wang X, Li C, Chao Y, et al. The deubiquitinase Usp10 regulates Klf4 stability and suppresses lung tumorigenesis. Cell Death Differ (2020) 27(6):1747–64. doi: 10.1038/s41418-019-0458-7
17. Zhu H, Yan F, Yuan T, Qian M, Zhou T, Dai X, et al. Usp10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing Yap/Taz. Cancer Res (2020) 80(11):2204–16. doi: 10.1158/0008-5472.Can-19-2388
18. Weisberg EL, Schauer NJ, Yang J, Lamberto I, Doherty L, Bhatt S, et al. Inhibition of Usp10 induces degradation of oncogenic Flt3. Nat Chem Biol (2017) 13(12):1207–15. doi: 10.1038/nchembio.2486
19. Liu X, Chen B, Chen J, Su Z, Sun S. Deubiquitinase ubiquitin-specific peptidase 10 maintains cysteine rich angiogenic inducer 61 expression via Yes1 associated transcriptional regulator to augment immune escape and metastasis of pancreatic adenocarcinoma. Cancer Sci (2022) 113(5):1868–79. doi: 10.1111/cas.15326
20. Zeng Z, Wu HX, Zhan N, Huang YB, Wang ZS, Yang GF, et al. Prognostic significance of Usp10 as a tumor-associated marker in gastric carcinoma. Tumour Biol (2014) 35(4):3845–53. doi: 10.1007/s13277-013-1509-1
21. Soncini C, Berdo I, Draetta G. Ras-gap Sh3 domain binding protein (G3bp) is a modulator of Usp10, a novel human ubiquitin specific protease. Oncogene (2001) 20(29):3869–79. doi: 10.1038/sj.onc.1204553
22. Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the cag repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology (1997) 49(4):1009–13. doi: 10.1212/wnl.49.4.1009
23. Huynh DP, Yang HT, Vakharia H, Nguyen D, Pulst SM. Expansion of the polyq repeat in ataxin-2 alters its golgi localization, disrupts the golgi complex and causes cell death. Hum Mol Genet (2003) 12(13):1485–96. doi: 10.1093/hmg/ddg175
24. Ye Y, Scheel H, Hofmann K, Komander D. Dissection of usp catalytic domains reveals five common insertion points. Mol Biosyst (2009) 5(12):1797–808. doi: 10.1039/b907669g
25. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of P53 by regulating the deubiquitination activity of Usp10 and Usp13. Cell (2011) 147(1):223–34. doi: 10.1016/j.cell.2011.08.037
26. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. Usp10 regulates P53 localization and stability by deubiquitinating P53. Cell (2010) 140(3):384–96. doi: 10.1016/j.cell.2009.12.032
27. Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, et al. The mrna-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell (2012) 46(5):674–90. doi: 10.1016/j.molcel.2012.05.021
28. Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme Usp10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem (2009) 284(28):18778–89. doi: 10.1074/jbc.M109.001685
29. Park JM, Yang SW, Yu KR, Ka SH, Lee SW, Seol JH, et al. Modification of pcna by Isg15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol Cell (2014) 54(4):626–38. doi: 10.1016/j.molcel.2014.03.031
30. Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, et al. Usp10 inhibits lung cancer cell growth and invasion by upregulating pten. Mol Cell Biochem (2018) 441(1-2):1–7. doi: 10.1007/s11010-017-3170-2
31. He Y, Jiang S, Mao C, Zheng H, Cao B, Zhang Z, et al. The deubiquitinase Usp10 restores pten activity and inhibits non-small cell lung cancer cell proliferation. J Biol Chem (2021) 297(3):101088. doi: 10.1016/j.jbc.2021.101088
32. Zeng Z, Li D, Yu T, Huang Y, Yan H, Gu L, et al. Association and clinical implication of the Usp10 and Msh2 proteins in non-small cell lung cancer. Oncol Lett (2019) 17(1):1128–38. doi: 10.3892/ol.2018.9702
33. Zhang M, Hu C, Tong D, Xiang S, Williams K, Bai W, et al. Ubiquitin-specific peptidase 10 (Usp10) deubiquitinates and stabilizes muts homolog 2 (Msh2) to regulate cellular sensitivity to DNA damage. J Biol Chem (2016) 291(20):10783–91. doi: 10.1074/jbc.M115.700047
34. Zhao W, Lu D, Liu L, Cai J, Zhou Y, Yang Y, et al. Insulin-like growth factor 2 mrna binding protein 3 (Igf2bp3) promotes lung tumorigenesis via attenuating P53 stability. Oncotarget (2017) 8(55):93672–87. doi: 10.18632/oncotarget.21280
35. Ko A, Han SY, Choi CH, Cho H, Lee MS, Kim SY, et al. Oncogene-induced senescence mediated by c-myc requires Usp10 dependent deubiquitination and stabilization of P14arf. Cell Death Differ (2018) 25(6):1050–62. doi: 10.1038/s41418-018-0072-0
36. Zhuo ZJ, Hua RX, Chen Z, Zhu J, Wang M, Yang Z, et al. Wtap gene variants confer hepatoblastoma susceptibility: A seven-center case-control study. Mol Ther Oncolytic (2020) 18:118–25. doi: 10.1016/j.omto.2020.06.007
37. Zhang XM, Gavande N, Parajuli P, Bepler G. Implications of the Usp10-Hdac6 axis in lung cancer - a path to precision medicine. J Cancer Biol (2021) 2(1). doi: 10.46439/cancerbiology.2.015
38. Guo K, Ma Z, Zhang Y, Han L, Shao C, Feng Y, et al. Hdac7 promotes nsclc proliferation and metastasis via stabilization by deubiquitinase Usp10 and activation of beta-Catenin-Fgf18 pathway. J Exp Clin Cancer Res (2022) 41(1):91. doi: 10.1186/s13046-022-02266-9
39. Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S, et al. Deubiquitinating enzyme Usp10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol (2020) 14(1):197–210. doi: 10.1002/1878-0261.12596
40. Shen C, Li J, Zhang Q, Tao Y, Li R, Ma Z, et al. Lncrna Gasal1 promotes hepatocellular carcinoma progression by up-regulating Usp10-stabilized pcna. Exp Cell Res (2022) 415(1):112973. doi: 10.1016/j.yexcr.2021.112973
41. Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, et al. Usp10 suppresses tumor progression by inhibiting mtor activation in hepatocellular carcinoma. Cancer Lett (2018) 436:139–48. doi: 10.1016/j.canlet.2018.07.032
42. De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: A comprehensive review and 2016 update’. Blood Cancer J (2016) 6(7):e441. doi: 10.1038/bcj.2016.50
43. Martelli MP, Sportoletti P, Tiacci E, Martelli MF, Falini B. Mutational landscape of aml with normal cytogenetics: Biological and clinical implications. Blood Rev (2013) 27(1):13–22. doi: 10.1016/j.blre.2012.11.001
44. Weisberg E, Choi HG, Barrett R, Zhou W, Zhang J, Ray A, et al. Discovery and characterization of novel mutant Flt3 kinase inhibitors. Mol Cancer Ther (2010) 9(9):2468–77. doi: 10.1158/1535-7163.MCT-10-0232
45. Katkere B, Rosa S, Drake JR. The syk-binding ubiquitin ligase c-cbl mediates signaling-dependent b cell receptor ubiquitination and b cell receptor-mediated antigen processing and presentation. J Biol Chem (2012) 287(20):16636–44. doi: 10.1074/jbc.M112.357640
46. Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of cbl-family ubiquitin ligases. Biochim Biophys Acta (2013) 1833(1):122–39. doi: 10.1016/j.bbamcr.2012.10.010
47. Taylor SJ, Thien CB, Dagger SA, Duyvestyn JM, Grove CS, Lee BH, et al. Loss of c-cbl E3 ubiquitin ligase activity enhances the development of myeloid leukemia in Flt3-itd mutant mice. Exp Hematol (2015) 43(3):191–206 e1. doi: 10.1016/j.exphem.2014.11.009
48. Yang J, Meng C, Weisberg E, Case A, Lamberto I, Magin RS, et al. Inhibition of the deubiquitinase Usp10 induces degradation of syk. Br J Cancer (2020) 122(8):1175–84. doi: 10.1038/s41416-020-0731-z
49. Peres LC, Cushing-Haugen KL, Kobel M, Harris HR, Berchuck A, Rossing MA, et al. Invasive epithelial ovarian cancer survival by histotype and disease stage. J Natl Cancer Inst (2019) 111(1):60–8. doi: 10.1093/jnci/djy071
50. Han GH, Chay DB, Yi JM, Cho H, Chung JY, Kim JH. Loss of both Usp10 and P14arf protein expression is an independent prognostic biomarker for poor prognosis in patients with epithelial ovarian cancer. Cancer Genomics Proteomics (2019) 16(6):553–62. doi: 10.21873/cgp.20157
51. Li M, Tang Y, Zuo X, Meng S, Yi P. Loss of ras gtpase-activating protein Sh3 domain-binding protein 1 (G3bp1) inhibits the progression of ovarian cancer in coordination with ubiquitin-specific protease 10 (Usp10). Bioengineered (2022) 13(1):721–34. doi: 10.1080/21655979.2021.2012624
52. Cai C, Yuan X, Balk SP. Androgen receptor epigenetics. Transl Androl Urol (2013) 2(3):148–57. doi: 10.3978/j.issn.2223-4683.2013.09.02
53. Dehm SM, Tindall DJ. Molecular regulation of androgen action in prostate cancer. J Cell Biochem (2006) 99(2):333–44. doi: 10.1002/jcb.20794
54. Debes JD, Tindall DJ. The role of androgens and the androgen receptor in prostate cancer. Cancer Lett (2002) 187(1-2):1–7. doi: 10.1016/s0304-3835(02)00413-5
55. Martin PL, Yin JJ, Seng V, Casey O, Corey E, Morrissey C, et al. Androgen deprivation leads to increased carbohydrate metabolism and hexokinase 2-mediated survival in Pten/Tp53-deficient prostate cancer. Oncogene (2017) 36(4):525–33. doi: 10.1038/onc.2016.223
56. Xiao L, Tien JC, Vo J, Tan M, Parolia A, Zhang Y, et al. Epigenetic reprogramming with antisense oligonucleotides enhances the effectiveness of androgen receptor inhibition in castration-resistant prostate cancer. Cancer Res (2018) 78(20):5731–40. doi: 10.1158/0008-5472.CAN-18-0941
57. Day KC, Lorenzatti Hiles G, Kozminsky M, Dawsey SJ, Paul A, Broses LJ, et al. Her2 and egfr overexpression support metastatic progression of prostate cancer to bone. Cancer Res (2017) 77(1):74–85. doi: 10.1158/0008-5472.CAN-16-1656
58. Shah RB, Ghosh D, Elder JT. Epidermal growth factor receptor (Erbb1) expression in prostate cancer progression: Correlation with androgen independence. Prostate (2006) 66(13):1437–44. doi: 10.1002/pros.20460
59. Liao Y, Guo Z, Xia X, Liu Y, Huang C, Jiang L, et al. Inhibition of egfr signaling with spautin-1 represents a novel therapeutics for prostate cancer. J Exp Clin Cancer Res (2019) 38(1):157. doi: 10.1186/s13046-019-1165-4
60. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
61. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
62. Yang R, Chen H, Xing L, Wang B, Hu M, Ou X, et al. Hypoxia-induced Circwsb1 promotes breast cancer progression through destabilizing P53 by interacting with Usp10. Mol Cancer (2022) 21(1):88. doi: 10.1186/s12943-022-01567-z
63. Nitiss JL. DNA Topoisomerase ii and its growing repertoire of biological functions. Nat Rev Cancer (2009) 9(5):327–37. doi: 10.1038/nrc2608
64. Guturi KKN, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, et al. Rnf168 and Usp10 regulate topoisomerase iialpha function via opposing effects on its ubiquitylation. Nat Commun (2016) 7:12638. doi: 10.1038/ncomms12638
65. Ilic M, Ilic I. Epidemiology of stomach cancer. World J Gastroenterol (2022) 28(12):1187–203. doi: 10.3748/wjg.v28.i12.1187
66. Chang HK, Yu E, Kim J, Bae YK, Jang KT, Jung ES, et al. Adenocarcinoma of the small intestine: A multi-institutional study of 197 surgically resected cases. Hum Pathol (2010) 41(8):1087–96. doi: 10.1016/j.humpath.2010.01.006
67. Sherr CJ. Tumor surveillance via the arf-P53 pathway. Genes Dev (1998) 12(19):2984–91. doi: 10.1101/gad.12.19.2984
68. Zhang Y, Xiong Y, Yarbrough WG. Arf promotes Mdm2 degradation and stabilizes P53: Arf-Ink4a locus deletion impairs both the Rb and P53 tumor suppression pathways. Cell (1998) 92(6):725–34. doi: 10.1016/s0092-8674(00)81401-4
69. Song JS, Yi JM, Cho H, Choi CH, Park Y, Chung EJ, et al. Dual loss of Usp10 and P14arf protein expression is associated with poor prognosis in patients with small intestinal adenocarcinoma. Tumour Biol (2018) 40(10):1010428318808678. doi: 10.1177/1010428318808678
70. Li B, Qi ZP, He DL, Chen ZH, Liu JY, Wong MW, et al. Nlrp7 deubiquitination by Usp10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J Exp Clin Cancer Res (2021) 40(1):126. doi: 10.1186/s13046-021-01920-y
71. Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase Sirt6 is a tumor suppressor that controls cancer metabolism. Cell (2012) 151(6):1185–99. doi: 10.1016/j.cell.2012.10.047
72. Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, et al. Usp10 antagonizes c-myc transcriptional activation through Sirt6 stabilization to suppress tumor formation. Cell Rep (2013) 5(6):1639–49. doi: 10.1016/j.celrep.2013.11.029
73. Kim K, Huh T, Park Y, Koo DH, Kim H, Hwang I, et al. Prognostic significance of Usp10 and P14arf expression in patients with colorectal cancer. Pathol Res Pract (2020) 216(6):152988. doi: 10.1016/j.prp.2020.152988
74. Ouyang SW, Liu TT, Liu XS, Zhu FX, Zhu FM, Liu XN, et al. Usp10 regulates musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett (2019) 593(4):406–13. doi: 10.1002/1873-3468.13323
75. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
76. Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Lu J, et al. Microrna-191 promotes pancreatic cancer progression by targeting Usp10. Tumour Biol (2014) 35(12):12157–63. doi: 10.1007/s13277-014-2521-9
77. Xu L, Yuan X, Ni J, Shen L, Cai M, Jiang D. Gain of microrna-103 triggers metastatic behavior by targeting ubiquitin specific peptidase 10 in pancreatic cancer. Int J Clin Exp Pathol (2019) 12(4):1214–23.
78. Yang Q, Ou C, Liu M, Xiao W, Wen C, Sun F. Nrage promotes cell proliferation by stabilizing pcna in a ubiquitin-proteasome pathway in esophageal carcinomas. Carcinogenesis (2014) 35(7):1643–51. doi: 10.1093/carcin/bgu084
79. Ren R. Mechanisms of bcr-abl in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer (2005) 5(3):172–83. doi: 10.1038/nrc1567
80. Liao Y, Liu N, Xia X, Guo Z, Li Y, Jiang L, et al. Usp10 modulates the Skp2/Bcr-abl axis via stabilizing Skp2 in chronic myeloid leukemia. Cell Discovery (2019) 5:24. doi: 10.1038/s41421-019-0092-z
81. Mahzouni P, Taheri F. An immunohistochemical study of cyclin D1 expression in astrocytic tumors and its correlation with tumor grade. Iran J Pathol (2019) 14(3):252–7. doi: 10.30699/ijp.2019.82024.1771
82. Sun T, Xu YJ, Jiang SY, Xu Z, Cao BY, Sethi G, et al. Suppression of the Usp10/Ccnd1 axis induces glioblastoma cell apoptosis. Acta Pharmacol Sin (2021) 42(8):1338–46. doi: 10.1038/s41401-020-00551-x
83. Cui B, Yang Q, Guan H, Shi B, Hou P, Ji M. Prima-1, a mutant P53 reactivator, restores the sensitivity of Tp53 mutant-type thyroid cancer cells to the histone methylation inhibitor 3-deazaneplanocin a. J Clin Endocrinol Metab (2014) 99(6):E962–70. doi: 10.1210/jc.2013-3147
84. Clague MJ, Urbe S, Komander D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol (2019) 20(6):338–52. doi: 10.1038/s41580-019-0099-1
85. Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H, et al. Inhibition of ubiquitin-specific proteases as a novel anticancer therapeutic strategy. Front Pharmacol (2018) 9:1080. doi: 10.3389/fphar.2018.01080
86. Correa RJ, Valdes YR, Peart TM, Fazio EN, Bertrand M, McGee J, et al. Combination of akt inhibition with autophagy blockade effectively reduces ascites-derived ovarian cancer cell viability. Carcinogenesis (2014) 35(9):1951–61. doi: 10.1093/carcin/bgu049
87. Dong K, Chen X, Xie L, Yu L, Shen M, Wang Y, et al. Spautin-A41 attenuates cerulein-induced acute pancreatitis through inhibition of dysregulated autophagy. Biol Pharm Bull (2019) 42(11):1789–98. doi: 10.1248/bpb.b19-00132
88. Yu M, Fang ZX, Wang WW, Zhang Y, Bu ZL, Liu M, et al. Wu-5, a novel Usp10 inhibitor, enhances crenolanib-induced Flt3-Itd-Positive aml cell death via inhibiting Flt3 and ampk pathways. Acta Pharmacol Sin (2021) 42(4):604–12. doi: 10.1038/s41401-020-0455-x
89. Cheng LL, Itahana Y, Lei ZD, Chia NY, Wu Y, Yu Y, et al. Tp53 genomic status regulates sensitivity of gastric cancer cells to the histone methylation inhibitor 3-deazaneplanocin a (Dznep). Clin Cancer Res (2012) 18(15):4201–12. doi: 10.1158/1078-0432.Ccr-12-0036
90. Reyes-Farias M, Carrasco-Pozo C. The anti-cancer effect of quercetin: Molecular implications in cancer metabolism. Int J Mol Sci (2019) 20(13):3177. doi: 10.3390/ijms20133177
91. Li S, Zhu Y, Zhang T, Hang Y, Chen Q, Jin Y. Cai’s neiyi prescription promotes apoptosis and inhibits inflammation in endometrial stromal cells with endometriosis through inhibiting Usp10. Biotechnol Appl Biochem (2019) 66(2):231–9. doi: 10.1002/bab.1715
92. Zhang W, Sartori MA, Makhnevych T, Federowicz KE, Dong X, Liu L, et al. Generation and validation of intracellular ubiquitin variant inhibitors for Usp7 and Usp10. J Mol Biol (2017) 429(22):3546–60. doi: 10.1016/j.jmb.2017.05.025
93. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. Ualcan: An update to the integrated cancer data analysis platform. Neoplasia (2022) 25:18–27. doi: 10.1016/j.neo.2022.01.001
94. Zhang Y, Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun (2022) 13(1):2669. doi: 10.1038/s41467-022-30342-3
95. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. Tisidb: An integrated repository portal for tumor-immune system interactions. Bioinformatics (2019) 35(20):4200–2. doi: 10.1093/bioinformatics/btz210
96. Bhattacharya U, Neizer-Ashun F, Mukherjee P, Bhattacharya R. When the chains do not break: The role of Usp10 in physiology and pathology. Cell Death Dis (2020) 11(12):1033. doi: 10.1038/s41419-020-03246-7
97. Zhang Q, Liu YJ, Li JP, Zeng SH, Shen H, Han M, et al. Usp35 is a potential immunosuppressive factor in skin cutaneous melanoma. J Inflammation Res (2022) 15:3065–82. doi: 10.2147/JIR.S362619
98. Gao D, Zhang Z, Xu R, He Z, Li F, Hu Y, et al. The prognostic value and immune infiltration of Usp10 in pan-cancer: A potential therapeutic target. Front Oncol (2022) 12:829705. doi: 10.3389/fonc.2022.829705
99. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the pd-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med (2000) 192(7):1027–34. doi: 10.1084/jem.192.7.1027
100. Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of Cmtm6 and Cmtm4 as pd-L1 protein regulators. Nature (2017) 549(7670):106–10. doi: 10.1038/nature23669
101. Korbecki J, Bajdak-Rusinek K, Kupnicka P, Kapczuk P, Siminska D, Chlubek D, et al. The role of Cxcl16 in the pathogenesis of cancer and other diseases. Int J Mol Sci (2021) 22(7):3490. doi: 10.3390/ijms22073490
102. Wang H, Liu B, Wei J. Beta2-Microglobulin(B2m) in cancer immunotherapies: Biological function, resistance and remedy. Cancer Lett (2021) 517:96–104. doi: 10.1016/j.canlet.2021.06.008
103. Wang X, Fu X, Zhang J, Xiong C, Zhang S, Lv Y. Identification and validation of M(6)a rna methylation regulators with clinical prognostic value in papillary thyroid cancer. Cancer Cell Int (2020) 20:203. doi: 10.1186/s12935-020-01283-y
104. Slebioda TJ, Rowley TF, Ferdinand JR, Willoughby JE, Buchan SL, Taraban VY, et al. Triggering of Tnfrsf25 promotes Cd8(+) T-cell responses and anti-tumor immunity. Eur J Immunol (2011) 41(9):2606–11. doi: 10.1002/eji.201141477
Keywords: deubiquitination, epigenetic modification, tumorigenesis, immune response, USP10 inhibitors
Citation: Ye Z, Chen J, Huang P, Xuan Z and Zheng S (2022) Ubiquitin-specific peptidase 10, a deubiquitinating enzyme: Assessing its role in tumor prognosis and immune response. Front. Oncol. 12:990195. doi: 10.3389/fonc.2022.990195
Received: 09 July 2022; Accepted: 09 September 2022;
Published: 28 September 2022.
Edited by:
Giuseppe Giaccone, Cornell University, United StatesReviewed by:
Zhi Zeng, Department of Pathology, Wuhan University, ChinaCopyright © 2022 Ye, Chen, Huang, Xuan and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zixue Xuan, eHVhbnppeHVlMDIyMkAxNjMuY29t; Shuilian Zheng, c2h1aWxpYW56QDE2My5jb20=
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.