
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 06 October 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.986674
 Yin Guan1†
Yin Guan1† Yutong Wang2†
Yutong Wang2† Hongxia Li3†
Hongxia Li3† Jing Meng4†
Jing Meng4† Xia You5,6,7
Xia You5,6,7 Xiaofeng Zhu5,6,7
Xiaofeng Zhu5,6,7 Qin Zhang5,6,7
Qin Zhang5,6,7 Tingting Sun5,6,7
Tingting Sun5,6,7 Chuang Qi5,6,7
Chuang Qi5,6,7 Guangyu An1*
Guangyu An1* Ying Fan8*
Ying Fan8* Binghe Xu8*
Binghe Xu8*ERBB2 amplification is one of the most important and mature targets for HER2-targeted drug therapy. Somatic mutations of ERBB2 in the tyrosine kinase domain have been studied extensively, and play a role in response to anti-HER2 therapy among different cancer types. However, ERBB2 fusion has not been got attention and its relevance to HER2-targeted therapy is unclear. We comprehensively characterized ERBB2 fusions from next-generation sequencing (NGS) data between May 2018 and October 2021 in 32,131 various solid tumors. Among the tumors, 0.28% harbored ERBB2 fusions, which occurred more commonly in gastroesophageal junction cancer (3.12%; 3/96), breast cancer (1.89%; 8/422), urothelial carcinoma (1.72%; 1/58), and gastric cancer (1.60%; 23/1,437). Our population presented with a median age of 65 years (range 28 to 88 years), a high proportion of men (55 men vs 34 women; 61.80%). Among the patients with ERBB2 fusions, TP53 (82%), APC (18%), and CDK4 (15%) were the top3 co-mutant genes. What’s more, most patients with ERBB2 fusion also had ERBB2 amplification (75.28%; 67/89), which was similar to the data in the TCGA database (88.00%; 44/50). Furthermore, TCGA database shows that patients with ERBB2 fusions in pan-cancer had a worse prognosis than those without ERBB2 fusions, as well as in breast cancer. Besides, ERBB2 amplification combined with ERBB2 fusion had worse prognosis than those with only ERBB2 amplification. ERBB2 fusion may interfere the effect of anti-HER2-targeted antibody drugs and influence the prognosis of patients with ERBB2 amplification. Prospective clinical trials are warranted to confirm the results in the future.
Human epidermal growth factor receptor 2 (HER2), encoded by ERBB2, is an important member of the receptor tyrosine kinase (RTK) family. The HER2 receptor is activated by forming homodimers or heterodimers with other ERBB family receptors; particularly, it forms the most stable heterodimer with EGFR (1). Therefore, HER2 can enhance EGFR signaling and promote the continuous differentiation and proliferation of tumor cells (2). The oncogenic activation of HER2 can be caused by HER2 protein over-expression, gene amplification, or gene mutation and occur in various malignant tumors, including breast (3, 4), gastric (5), non-small cell lung (6), bladder (7), ovarian (8), and pancreatic cancers (9).
ERBB2 amplification is the most common mechanism leading to increased HER2 protein over-expression. Among all the cancers related to ERBB2 amplification and HER2 over-expression, breast cancer is most widely studied. ERBB2, amplified in 20%–30% of the breast cancer cases, is associated with aggressive tumor behavior (10). HER2 is also over-expressed in patients with other solid tumors, such as gastric cancer (11), biliary tract (12), colorectal (13), non-small cell lung (14), and bladder cancer (15).
Studies have suggested that the ERBB2 mutations play an important role in the pathogenesis, development, and resistance to anti-HER2-targeted breast cancer drugs (16). ERBB2 mutations are also found in other common cancers (17), including lung (18) and colorectal (19). ERBB2’s exon 20 mutations (4.83%) is a relatively frequent primary oncogenic driver in non-small cell lung cancer (NSCLC), especially lung adenocarcinoma (LUAD) (20). Therefore, anti-HER2 therapies such as trastuzumab, lapatinib, afatinib, and masatinib are effective in heavily pretreated ERBB2-mutated NSCLC (18).
In addition to amplification and SNV, there are various, albeit less common, fusion forms of ERBB2. For example, ERBB2 fusions, representing a different mechanism of HER2 activation, have been described in gastric cancer. ZNF207-ERBB2 and MDK-ERBB2 fusion variants can activate HER2 signaling in a similar manner as wild-type HER2 (21). Meanwhile, ERBB2 fusion has been found in colorectal cancer (22) and breast cancer (23), which may be related to response to HER2-targeted drugs. The mechanism of ERBB2 fusion in tumors remains unclear and ERBB2 fusion could be a potential target. Therefore, systematic research on ERBB2 fusions has clinical significance. However, ERBB2 fusions have not been systematically described in pan-cancer. Here, we retrospectively analyzed the genomic profiling of the ERBB2 fusions from 32,131 Chinese patients with solid tumors. Our results comprehensively revealed enrichment of ERBB2 fusions in certain histologic subtypes, providing a new insights of responses to therapies targeting ERBB2.
Between May 2018 and October 2021, 32,131 consecutive clinical samples of primarily relapsed and refractory solid tumors in the database of Simcere Diagnostics, Co. Ltd. (Nanjing, China) were evaluated retrospectively to search for ERBB2 gene fusions. All the patients included in the study were informed consent concerning genetic testing and research. We identified patients with ERBB2 fusions in the laboratory information management system (LIMS) database using a natural language search program. For those cases, relevant demographic and clinical data were extracted from the database, including age, gender, date of diagnosis, histology type, pathological stage, and evaluation of treatment responses per the reports of the clinical investigators. For tumor tissue samples, the pathologic diagnosis and tumor content for each case was confirmed by pathologists. We analyzed only those patients who had ERBB2 fusion testing, among these cases, 66 cases were analyzed using the same panel containing 69 genes, which were used in co-occurring gene alterations analysis. Other patients chose some matched panels containing ERBB2 fusion detection due to their conditions and needs.
Three commercial kits were used for DNA extraction. Genomic DNA (gDNA) of formalin-fixed and paraffin-embedded (FFPE) tissues and fresh tissues was extracted using the Tissue sample DNA extraction kit (Kai Shuo). Genomic DNA of leucocytes was extracted using MagMAXTM DNA Multi-Sample Ultra Kit (Thermo). Cell-free DNA (cfDNA) of plasma was extracted using MagMAXTM Cell-Free DNA Isolation Kit (Thermo). All of the extraction procedures were performed following the manufacturer’s instructions. DNA was quantified on Qubit Fluorometer with Qubit dsDNA HS Assay kit (Thermo) and its quality was evaluated by Agilent 4200 TapeStation (Agilent).
The probe hybridization capture method was used for library construction. Commercial reagents and customized probes were used for library construction and hybridization capture. In brief, 15 ng-200 ng gDNA was sheared into 200~350 bp by fragmentation enzymes. Indexed paired-end adaptors for the Illumina platform were self-developed and customized (SimcereDx). End repair, A-tailing, and adaptor ligation of sheared DNA and cfDNA was respectively performed by KAPA HyperPlus DNA Library Prep kit (Roche Diagnostics) and VAHTSTM Universal DNA Library Prep Kit for Illumina® (Vazyme). Unligated adaptors were removed by the size selection function of Agencourt AMPure XP beads (Beckman Coulter). The ligation products were PCR amplified to form a pre-library for hybridization. The final library was quantified on Qubit Fluorometer with Qubit dsDNA HS Assay kit (Thermo Fisher) and its quality was evaluated by Agilent 4200 TapeStation (Agilent).
The qualified DNA libraries were sequenced on the Illumina NovaSeq6000 platform (Illumina, San Diego, CA) and generated 150 bp paired-end reads. Base calls from Illumina NovaSeq6000 were conducted to FASTQ files. The software fastp (v.2.20.0) was used for adapter trimming and filtering of low-quality bases (24). The BWA-MEM (v.0.7.17) algorithm was performed to align to the reference genome (UCSC’s hg19 GRCh37) (25). Duplicate reads from PCR were excluded using Dedup with Error Correct. SNVs/InDels were called and annotated via VarDict (v.1.5.7) (26) and InterVar (27), then the variants were filtered against the common SNPs in the public database including 1000 Genome Project (Aug 2015) and Exome Aggregation Consortium (ExAC) Browser28 (v.0.3). CNVs and fusions were analyzed by CNVkit (dx1.1) (28) and factera (v1.4.4) (29), respectively.
The analysis of CNV difference between ERBB2 amplification and ERBB2 amplification combined with fusion using Wilcoxon test. All reported p-values were two-tailed, and p <0.05 was considered statistically significant. Statistical analyses were performed using R package ggpubr v. 0.4.0 (https://cran.r-project.org/package=ggpubr).
In total, eighty-nine (0.28%) of the 32,131 samples harbored 110 ERBB2 fusions (Figure 1; Table 1). The median age of the 89 patients whose tumors harbored ERBB2 fusions was 65 with a range of 28–88; 34 (38.20%) of the patients were female and 55 (61.80%) were male. Most patients had a clinical stage IV disease when initially diagnosed (Table 1). ERBB2 fusions were distributed across different tumor types, including lung cancer (0.14%; 29/20,670), gastric cancer (1.60%; 23/1,437), colorectal cancer (0.30%; 11/3,613), breast cancer (1.89%; 8/422), gastroesophageal junction cancer (3.12%; 3/96), biliary tract cancer (0.40%; 3/745), glioma (0.37%; 3/814), liver cancer (0.07%; 1/1,406), ovarian cancer (0.35%, 1/285), esophageal cancer (0.40%; 1/249), gastrointestinal stromal tumor (0.61%; 1/163), endometrial cancer (1.37%, 1/73), urothelial carcinoma (1.72%; 1/58), and cancer with unknown primary sites (0.14%; 3/2100) (Figures 2A, B and Table 2). 75.28% (67/89) of the ERBB2 fusion-positive patients also harbored ERBB2 amplification, which was similar to 88.00% in TCGA cohort (Supplementary Tables 1, 2).
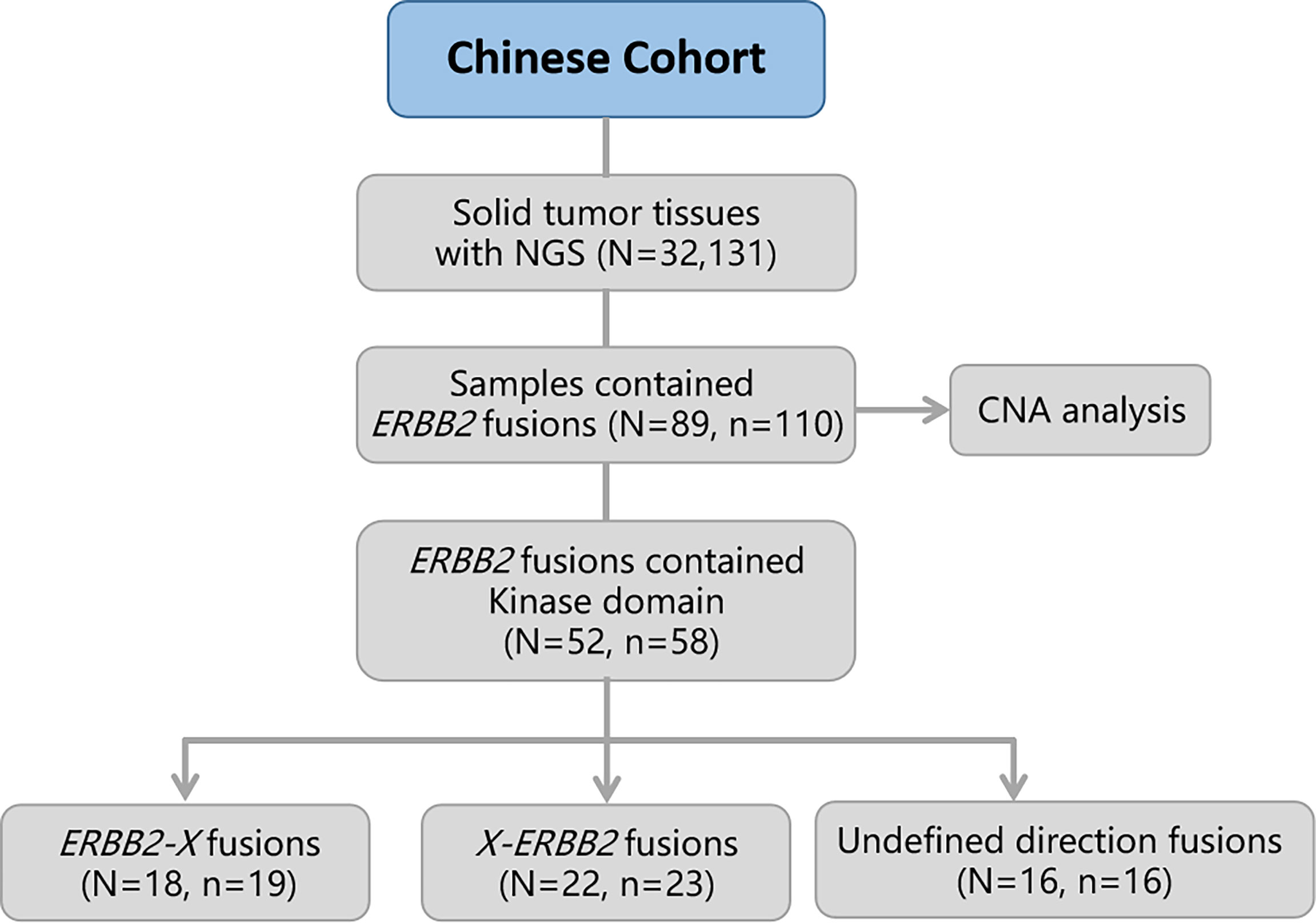
Figure 1 Flow diagram of the study. OS, overall survival; CNA, copy number alteration. N, number of patients; n, number of ERBB2 fusions.

Table 1 A summary of ERBB2 fusion tumor patients’ demographic and clinical characteristics in Chinese cohort.
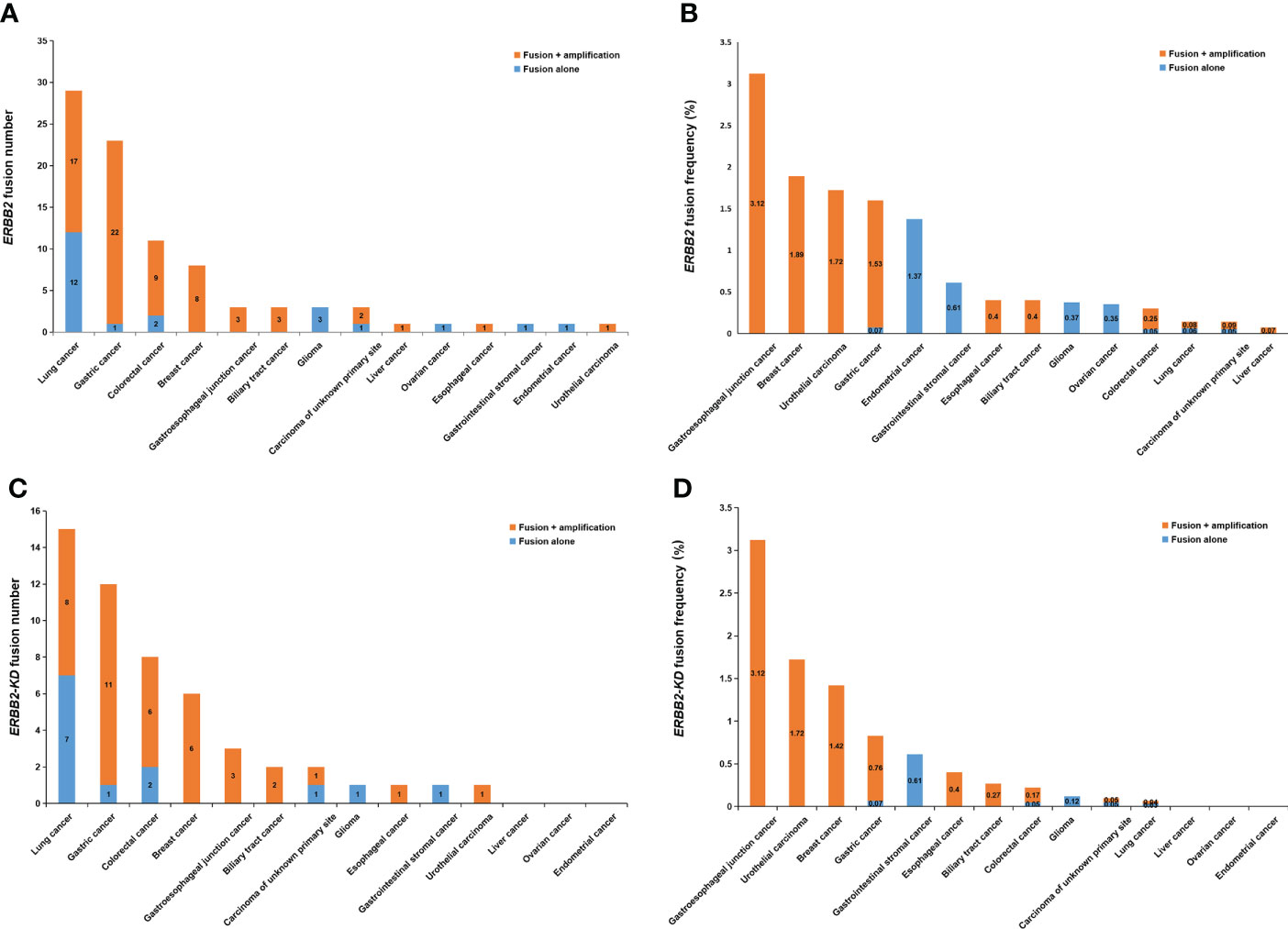
Figure 2 ERBB2 fusions in solid tumors. (A) The number of ERBB2 fusions in different cancer types. (B) The frequency of ERBB2 fusions in different cancer types. (C) The number of ERBB2-KD fusions in different cancer types. (D) The frequency of ERBB2-KD fusions in different cancer types.
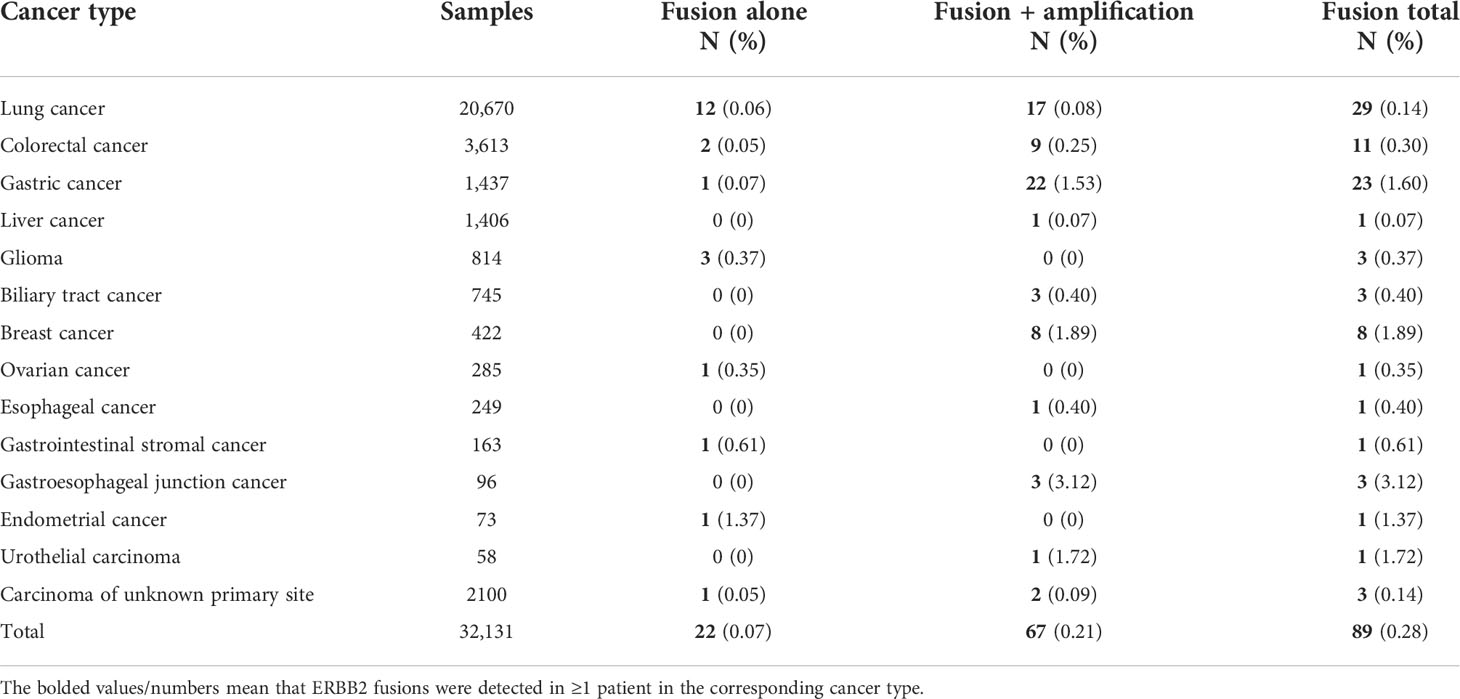
Table 2 Frequencies of ERBB2 alternations and relative distribution of alternation types in 89 ERBB2 fusion solid tumors from Chinese cohort.
Considering the characteristics of the receptor tyrosine kinase, we focused on the ERBB2 fusions which contain the kinase domain. Fifty-two of which can produce an intact HER2 kinase domain (ERBB2-KD fusion) and were collected to conduct follow-up analysis (Figure 1). The median age of the 52 patients whose tumors harbored ERBB2-KD fusions was 63 with a range of 28–88; 22 (42.31%) of the patients were female and 30 (57.69%) were male. Most patients had a clinical stage IV disease when initially diagnosed (Table 3). ERBB2-KD fusions occurred more commonly in gastroesophageal junction cancer (3.12%; 3/96), urothelial carcinoma (1.72%; 1/58), breast cancer (1.42%; 6/422), and gastric cancer (0.83%; 12/1,437) (Figures 2C, D and Table 4).
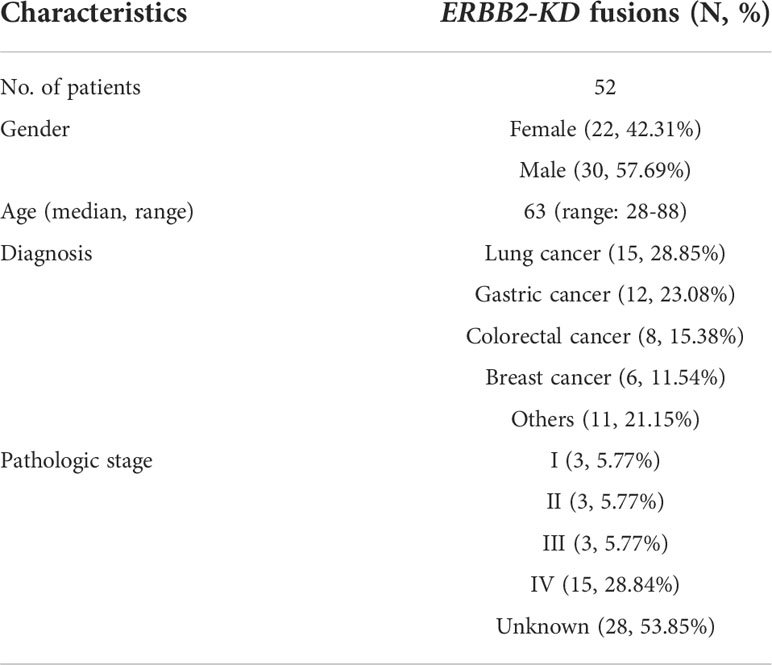
Table 3 A summary of ERBB2-KD fusion tumor patients’ demographic and clinical characteristics in Chinese cohort.
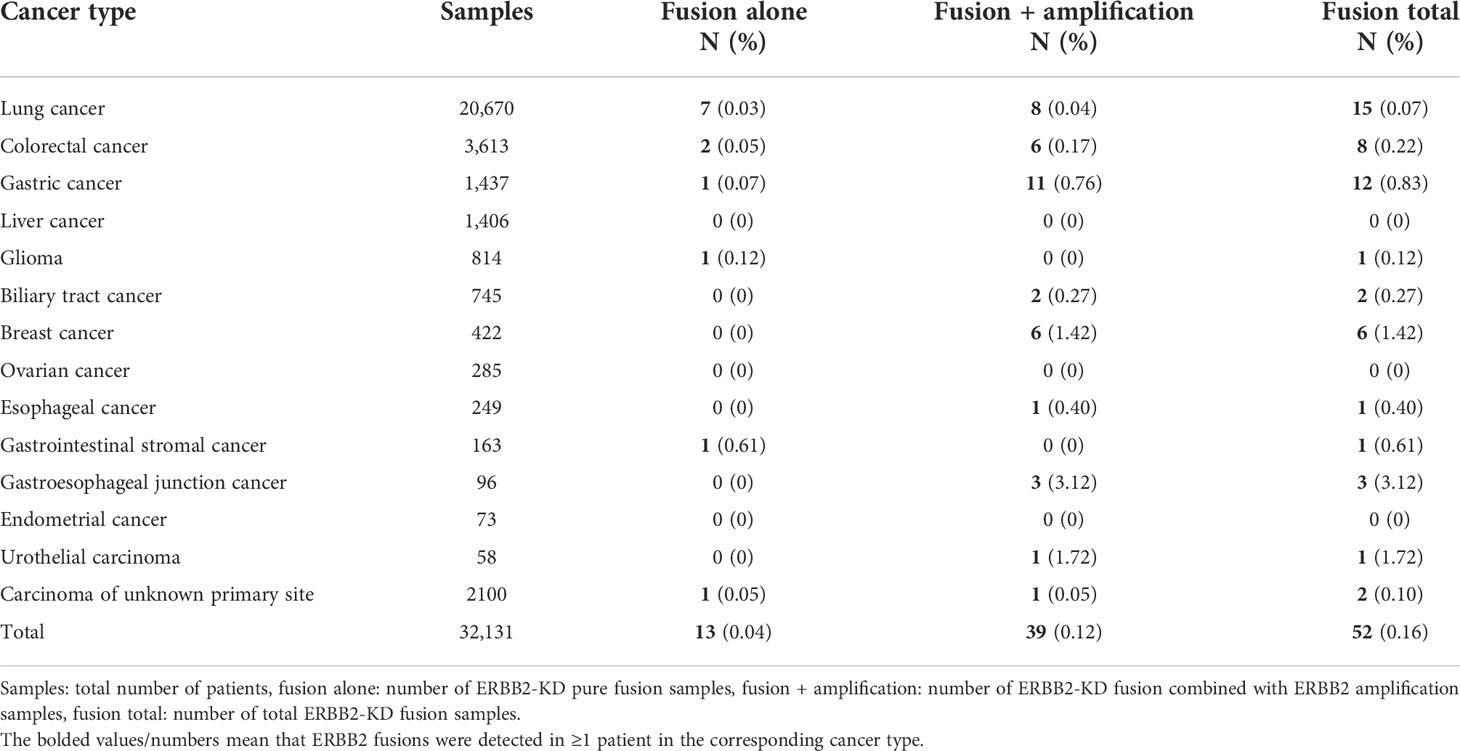
Table 4 Frequencies of ERBB2 alternations and relative distribution of alternation types in 52 ERBB2-KD fusion solid tumors from Chinese cohort.
Of the exon composition of the ERBB2-KD fusions identified, all 58 ERBB2-KD fusions from 52 patients contained intact ERBB2 kinase domain encoded by exons 18–24 (Supplementary Materials). Fusion of ERBB2 gene was more common in exons 17, 25, 26, and 27 (Figure 3A). Two breast cancer patients and two colorectal cancer patients had 2 different ERBB2-KD fusions each, and 1 colorectal cancer patient had 3 different ERBB2-KD fusions. The remaining 47 patients had one ERBB2-KD fusion each. MIEN1 was the most frequent fusion partner identified in 5 cases, followed by IKZF3 with 4 fusions, and CDK12 and TCAP with 2 fusions. All the other fusions were identified in only 1 case (Figure 3B).
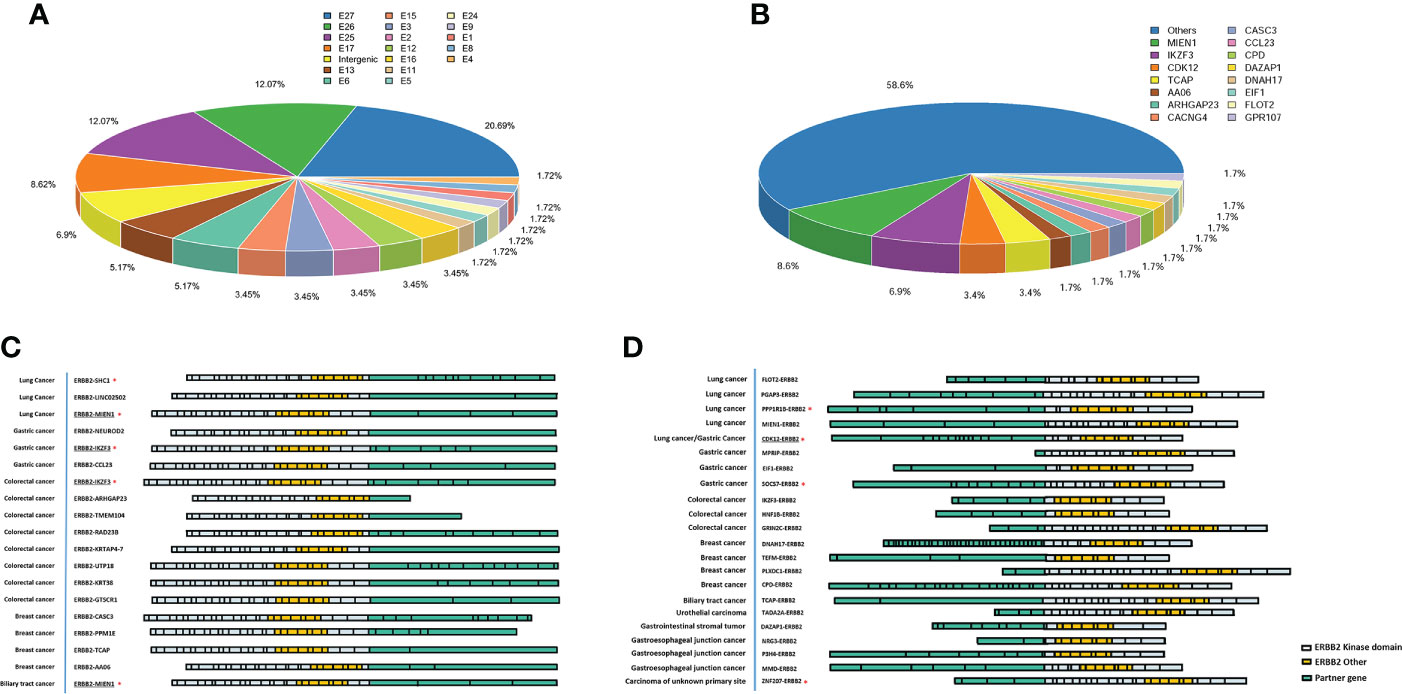
Figure 3 ERBB2-KD rearrangements in solid tumors. (A) Frequency of fusion between different exons of ERBB2. (B) Frequency of ERBB2-KD fusion variants’ partners. Schematic of all ERBB2-X fusions (C) and X-ERBB2 fusions (D) identified, at scale with exons represented by individual boxes. The partners is colored pink, with ERBB2 kinase domain colored orange and other ERBB2 exons colored white. The reported ERBB2 fusions marked with the red asterisk, others were novel ERBB2 fusions. The underlined marks are the same fusion gene with different break point positions.
Consider the binding mechanism of HER2-targeted drugs, we divided the ERBB2-KD fusion into two classes for analysis. Those ERBB2-X fusions which were intact from the extracellular domain to the kinase domain, while those X-ERBB2 fusions only retained kinase domain. In our study, 19 ERBB2-X and 23 X-ERBB2 fusions were identified (Figures 3C, D), and the other ERBB2-KD fusions’ direction was uncertain. In ERBB2-X fusions (Figure 3C), there were 17 different fusion partners, of which 3 have been previously reported, and 14 were previously unreported in public databases (COSMIC and TCGA) or published literature (PubMed). In addition, the ERBB2-IKZF3 fusion was appeared in two patients each, the same as the ERBB2-MIEN1 fusion. The 23 X-ERBB2 fusions detected from 22 patients all could encode an intact HER2 kinase domain. One breast cancer patient had 2 different ERBB2-KD fusions, and the remaining 21 patients had 1 ERBB2-KD fusion. Two CDK12-ERBB2 fusions were identified in gastric tumor and lung cancer. All the other fusions were recognized in only one case. The X-ERBB2 fusions had 22 different fusion partners, of which 4 have been previously reported, and 18 were novel (Figure 3D). All 23 ERBB2 fusions were in-frame with breakpoints that were distributed in different introns and exons of ERBB2 (Supplementary Table 3).
Many patients with ERBB2 fusions also had ERBB2 amplification (75.28%; 67/89) and ERBB2 SNV (8.99%; 8/89). In contrast, patients with only ERBB2 fusions were less common (24.72%; 22/89) (Table 2 and Figure 4A). ERBB2 fusion combined with amplification was found in patients with lung, colorectal, gastric, liver, biliary tract, breast, esophageal, and gastroesophageal junction cancers and urothelial carcinoma. ERBB2 fusion alone was found in patients with lung, colorectal, gastric, glioma, ovarian, endometrial, and gastrointestinal stromal tumors.
Figure 4 Frequency and distribution of ERBB2 variations in solid tumors with ERBB2 fusion. (A) Distribution of ERBB2 variations in three cancer types with ERBB2 fusion, lung cancer, gastric cancer, colorectal cancer, and breast cancer. For each cancer type, the frequency of ERBB2 variations was described as the percentage of all cancer cases analyzed, and the distribution and type of variations were normalized to 100%. (B) Landscape of genomic aberrations of ERBB2 fusion in solid tumors.
We then analyzed the co-mutations of 69 tumor driver genes (listed in the Supplementary Table 4) with ERBB2 in 66 patients with ERBB2 fusions. The top 5 co-mutated genes included TP53 (86%), EGFR (21%), APC (20%), CDK4 (15%), PIK3CA (15%) (Figure 4B). Among them, TP53 and PIK3CA were mainly concentrated on missense mutations, EGFR and CDK4 were mainly amplification, while APC was missense and nonsense mutations. In the ERBB2 fusion alone, the co-mutated genes were mainly missense mutations of TP53, KRAS, NF1, BRAF, APC, PTEN, ERBB4, ROS1 and amplifications of FGFR2, MET, CDK4, and CDK6. The most concurrently-mutated genes with ERBB2 fusions were in the MAP kinase, RTK, cytoskeletal regulation and vesicle trafficking, PI3K/Akt/mTOR, and DNA damage/repair signaling pathways.
Since more than 75% of ERBB2 fusions were accompanied by ERBB2 amplification, we also analyzed the difference in CNV values between the pure ERBB2 amplification (ERBB2 Amp) and ERBB2 amplification combined with fusion (ERBB2 Fusion + Amp) in different cancer types. At the same time, since ERBB2 amplification mainly occurs in gastric cancer, colorectal cancer, and breast cancer, we focused on these cancer types in our work. The results showed that the CNV of ERBB2 Amp was significantly higher than ERBB2 Fusion + Amp in pan-cancer (Figure 5A), gastric cancer (Figure 5B), colorectal cancer (Figure 5C), and breast cancer (Figure 5D) (P<0.01).
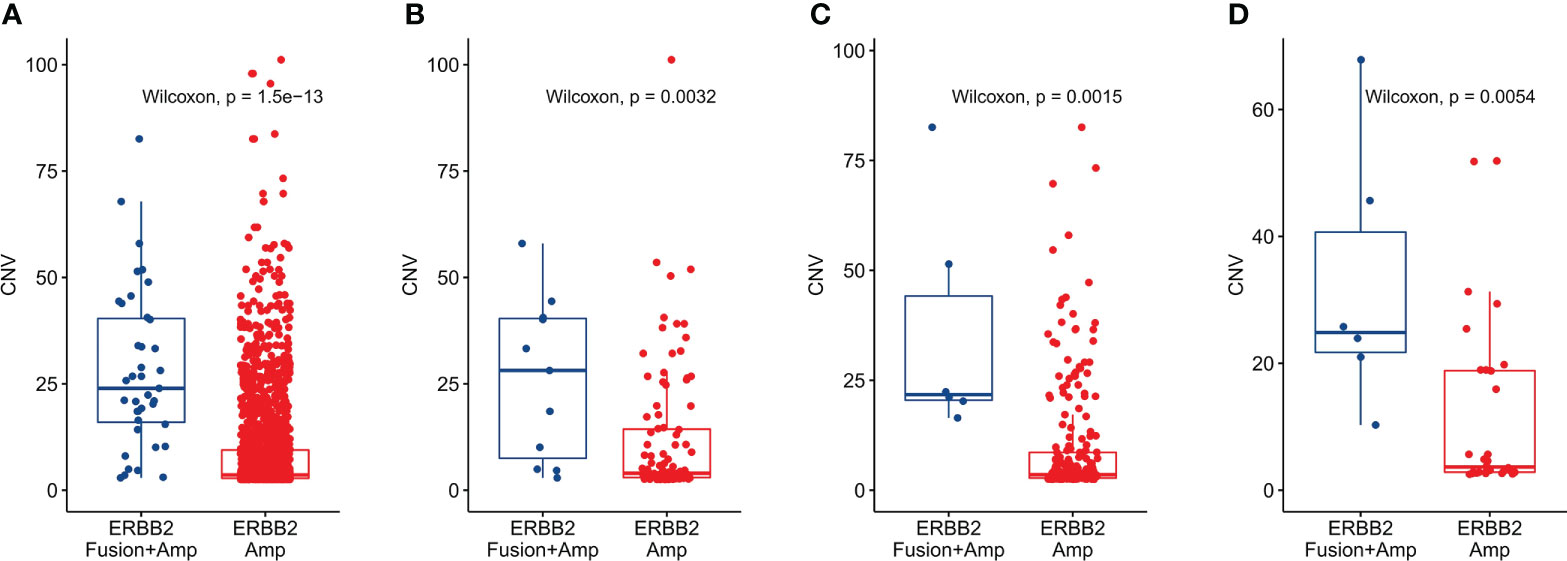
Figure 5 CNV difference between solid tumors with only ERBB2 amplification and tumors with ERBB2 fusion combined with amplification. (A) CNV difference analysis classified by ERBB2 fusion status in pan-cancer, (B) CNV difference analysis classified by ERBB2 fusion status in gastric cancer. (C) CNV difference analysis classified by ERBB2 fusion status in colorectal cancer. (D) CNV difference analysis classified by ERBB2 fusion status in breast cancer.
ERBB2 is one of the most common oncogenic driver genes; its amplification alterations predominantly occur in about 30% of breast cancer (30) and 20% of gastric cancer cases (31). Previously, most studies focused on ERBB2 amplifications. Therefore, there is a need for comprehensive studies on other alterations of ERBB2, such as ERBB2 fusions.
This study identified 110 ERBB2 fusions from 32,131 samples with solid tumors using NGS. Although ERBB2 fusions were rare, they occurred more frequently in gastroesophageal junction cancer (3.12%), breast cancer (1.89%), and urothelial carcinoma (1.72%). Considering the characteristics of the receptor tyrosine kinase, ERBB2 fusions containing the kinase domain were screened. There were 58 ERBB2 fusions containing a full kinase domain (ERBB2-KD fusions) in 52 patients. The ERBB2-KD fusion partners were diverse. Among the 42 fusions with certain directions, 32 fusions were first reported by us. TP53, EGFR, APC, CDK4, and PI3KCA were relatively frequent mutant genes in patients with ERBB2 fusions, and most of the concurrently-mutated genes were identified in the RTK, MAP Kinase, DNA damage/repair signaling, PI3K/Akt/mTOR, and cell cycle pathways. Interestingly, we found that most ERBB2 fusions accompanied ERBB2 amplification, which was the main target of anti-HER2 therapy, while ERBB2 fusions rarely occur by themselves.
The HER2-targeting drugs were divided into two categories, trastuzumab, pertuzumab, and TDM-1, which binding to the proximal membrane domain of HER2, while lapatinib and neratinib binding to the intracellular kinase domain. Therefore, we distinguished the different fusions by the position of ERBB2 in the fusion protein and divided them into ERBB2-X fusions and X-ERBB2 fusions. Given the presence of the HER2 dimerization domain (32, 33), all of ERBB2-X and X-ERBB2 fusion variants were likely to form homodimers in a manner similar to that of amplified wild-type HER2. Meanwhile, they all retained the kinase domain. Therefore, we speculated that these fusions were meaningful. The ERBB2-X fusions retained the intact extracellular domain and can bind to ligands and monoclonal antibodies theoretically. Patients with these ERBB2-X fusions might benefit from HER2 antibodies therapy. While X-ERBB2 fusion proteins, due to the deletion of the signal peptide and transmembrane sequence, were most likely retained in the cytoplasm, which might be effective with HER2-TKIs and might also lead to HER2 antibodies’ resistance in patients with ERBB2 amplification.
As described in the result, most ERBB2 fusions accompanied ERBB2 amplification. The current clinical protocol for selecting HER2-positive patients is based on FISH positivity or an IHC score of 3+, while an IHC score of 2+ requires confirmation using FISH. The FISH probe covers the entire ERBB2 (34). Meanwhile, the antibody used in IHC recognizes the HER2 epitope located at HER2’s intracellular site (35–37); it also covers all X-ERBB2 fusion proteins in this study. Therefore, only the special probes design for detection fusions can distinguish the ERBB2 amplification and fusion by FISH, but NGS could be a feasible, high-throughput method to detect ERBB2 fusions.
A limitation of this study was the lack of a large sample of ERBB2 fusion patients’ prognosis in the real world. However, given the importance of clinical prognosis, we attempted a simple exploratory prognostic analysis of ERBB2 fusions in the TCGA database. The results indicated that the prognosis of the patients with ERBB2 fusions was significantly worse than those without ERBB2 fusions in pan-cancer and breast cancer (Supplementary Figures 1A, B). Besides, the prognosis of these patients harbored ERBB2 amplification only was better than those with ERBB2 fusions combination with amplification in pan-cancer (Supplementary Figure 1C). In addition, in breast cancer, the OS of the patients with ERBB2 amplification was significantly longer than those with ERBB2 fusions with amplification (Supplementary Figure 1D). Since there was no specific CNV value in TCGA cohort, we could not compare the difference of CNV between the two groups. The CNV was significantly higher in ERBB2 amplification combined with fusion than pure ERBB2 amplification in Chinese cohort, we speculate that this phenomenon may also exists in TCGA cohort. High CNV had been revealed as an independent risk factor predicted unfavorable prognosis in HER2-positive gastric adenocarcinoma patients (38), and HER2-positive metastatic breast cancer patients with high HER2 CNV in plasma had worse prognosis after trastuzumab-based therapy (39). Based on our founding and other study results, there were two possibilities. On one hand, the impact of ERBB2 fusions on clinical outcomes might be caused by high CNV. On the other hand, the impact of high CNV on clinical outcomes might be due to the presence of ERBB2 fusions, or some other possibility. Which one is right? We do not have a definite answer to this question at present, and further basic research and mechanism research are needed.
The important thing is that ERBB2 fusion seems not meaningless “passenger fusion” without function. Previous in vitro studies showed that the cells expressing ZNF207 (exon 2)-ERBB2 (exon18) fusion gene lost the ability to bind to T-DM1, which is trastuzumab conjugated with emtansine (DM1), an antimitotic agent. Furthermore, in vivo efficacy study indicated that trastuzumab did not inhibit tumor growth in xenografts expressing the ZNF207-ERBB2 fusion; this finding supported the resistant mechanism to trastuzumab (21). ZNF207 is a kinetochore- and microtubule-binding protein that plays a key role in spindle assembly. It localize in cytoskeleton, kinetochore, and nucleus. We speculate that since this protein does not have a signal peptide, when it is fused with HER2, this ZNF207-HER2 fusion protein cannot localize on the cell membrane, so it is not sensitive to trastuzumab or T-DM1, instead the HER2-TKIs may be effective.
Regarding the effect of ERBB2 fusions on the efficacy of targeted drugs, there are also related reports on colorectal cancer and breast cancer. For example, in patients with colorectal carcinoma, ERBB2-GRB7 fusion is insensitive to HER2 inhibitors (22). Lesurf et al. (23) retrospectively analyzed the correlation between the molecular characteristics of 48 patients in the clinical trial of ACOSOG Z1041 and the efficacy of trastuzumab combined with chemotherapy, i.e., neoadjuvant therapy, for breast cancer and found that 2 patients with ERBB2 fusions (ERBB2 (exon19)-IKZF3, ERBB2 (exon1)-TBC1D3P1-DHX40P1) did not achieve pathologic complete response (pCR). HER2-TBC1D3P1-DHX40P1 only retains one exon region of HER2, so it is not sensitive to targeted drugs. The kinase domain of HER2 protein covers exon 18 to exon 24, while HER2-IKZF3 fusion proteins do not retain the complete kinase domain, and we speculate that this may be the reason for the patient insensitive to trastuzumab. Therefore, we conclude that ERBB2 fusion partners and breakpoint locations play a vital role in HER2-targeted therapy.
In addition to antibody drugs, several small-molecule HER2 kinase inhibitors, including pyrotinib and lapatinib, are available approved agents for patients with HER2-positive breast cancer. Besides, pyrotinib has demonstrated a good potential to treat advanced NSCLC with ERBB2 mutations, especially the ERBB2 exon 20 insertions, in the phase II studies (40, 41). In view of their drug mechanisms which bind to the HER2 kinase domain, pyrotinib or lapatinib can be a potential option for cancer patients with ERBB2 fusions, especially those X-ERBB2 fusions, which needs to be prospectively explored.
Our study is the first comprehensive analysis of a large group of Chinese patients with pan-cancer, it has extended our understanding of ERBB2 fusions in solid tumors significantly. However, there were several limitations in our study. First, we couldn’t collect complete and detailed clinical pathological characteristics and treatment details including survival status of all patients with ERBB2 fusion. To analyze the prognosis of ERBB2 fusion, we utilized the TCGA database and found patients with ERBB2 fusion had a worse prognosis than those without ERBB2 fusion. In addition, the patients harboring ERBB2 fusion and ERBB2 amplification showed a worse prognosis than patients with pure ERBB2 amplification, or ERBB2 amplification without fusion. Second, the function of this novel ERBB2 fusion proteins and the potential effect of ERBB2 fusion to anti-HER2 therapy have not been studied and described clearly. By analyzing the protein structure and functional sequence of HER2, we screened out some potentially meaningful ERBB2 fusion forms. Basic experiments carrying out are exploring the function of the fusion protein, followed by responsiveness to various anti-HER2 drugs. Despite its limitations, this study is the first comprehensive analysis of ERBB2 fusions in a larger group of Chinese patients with various carcinomas, it has extended our understanding of ERBB2 fusions in solid tumors significantly and may provide more inspiration in the upcoming clinical trials.
This study analyzed the ERBB2 fusions profiles in more than 30,000 patients with different solid carcinomas. The prevalence of this rare mutation differs obviously and is relatively higher in upper digestive system tumor. Most ERBB2 fusions are accompanied by ERBB2 amplification and may play a role in the prognosis of pan-cancer, especially BRCA. Further well-designed prospective researches are expected to confirm the role of ERBB2 fusion and to identify the patients who will benefit more from anti-HER2 treatment.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Beijing Chao-Yang Hospital, Capital Medical University. The patients/participants provided their written informed consent to participate in this study.
YG and YF performed the research, collected and analyzed the data, interpreted the results and helped revise the manuscript. YW, HL, and JM prepared samples, gathered detailed clinical information for the study. XY collected and analyzed the data, and wrote the initial draft of the paper. XZ and QZ were responsible for bioinformatics investigation. TS and CQ were responsible for statistical review and participated in review. GA and BX conceived the study, directed and supervised research, and revised the manuscript. The work reported in the paper has been performed by the authors unless clearly specified in the text. All authors contributed to the article and approved the submitted version.
We appreciate the effort of the physicians for enrolling patients and thank all the patients involved for allowing us to analyze their clinical data. Besides, we are grateful to Qianru He and Qianqian Duan for their help in the study.
Authors XY, QZ, XZ, TS, and CQ were employed by Jiangsu Simcere Diagnostics Co., Ltd, Nanjing Simcere Medical Laboratory Science Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.986674/full#supplementary-material
Amp, Amplification; BRCA, Breast invasive carcinoma; CNV, Copy number variation; FISH, Fluorescent in situ hybridization; HER2, Human epidermal growth factor receptor 2; IHC, Immunohistochemical; KD, Kinase domain; MAP kinase, Mitogen Activated Protein Kinases; NGS, Next-generation sequencing;NSCLC, Non-Small Cell Lung Cancer; OS, Overall survival; PD, Progressive disease;RTK, Receptor Tyrosine Kinase; SNV, Single-nucleotide variant; TKI, Tyrosine kinase inhibitors; T-DM1, Trastuzumab emtansine
1. Ménard S, Pupa SM, Campiglio M, Tagliabue E. Biologic and therapeutic role of HER2 in cancer. Oncogene (2003) 22:6570–8. doi: 10.1038/sj.onc.1206779
2. Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene (2007) 26:6469–87. doi: 10.1038/sj.onc.1210477
3. Krishnamurti U, Silverman JF. HER2 in breast cancer: A review and update. Adv Anat Pathol (2014) 21:100–7. doi: 10.1097/PAP.0000000000000015
4. Yarden Y. Biology of HER2 and its importance in breast cancer. Oncology (2001) 61 Suppl 2:1–13. doi: 10.1159/000055396
5. Boku N. HER2-positive gastric cancer. Gastric Cancer (2014) 17:1–12. doi: 10.1007/s10120-013-0252-z
6. Swanton C, Futreal A, Eisen T. Her2-targeted therapies in non-small cell lung cancer. Clin Cancer Res (2006) 12:4377s–83s. doi: 10.1158/1078-0432.CCR-06-0115
7. Laé M, Couturier J, Oudard S, Radvanyi F, Beuzeboc P, Vieillefond A. Assessing HER2 gene amplification as a potential target for therapy in invasive urothelial bladder cancer with a standardized methodology: results in 1005 patients. Ann Oncol (2010) 21:815–9. doi: 10.1093/annonc/mdp488
8. Chung YW, Kim S, Hong JH, Lee JK, Lee NW, Lee YS, et al. Overexpression of HER2/HER3 and clinical feature of ovarian cancer. J Gynecol Oncol (2019) 30:e75. doi: 10.3802/jgo.2019.30.e75
9. Chou A, Waddell N, Cowley MJ, Gill AJ, Chang DK, Patch AM, et al. Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med (2013) 5:78. doi: 10.1186/gm482
10. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science (1989) 244:707–12. doi: 10.1126/science.2470152
11. Meric-Bernstam F, Johnson AM, Dumbrava EEI, Raghav K, Balaji K, Bhatt M, et al. Advances in HER2-targeted therapy: Novel agents and opportunities beyond breast and gastric cancer. Clin Cancer Res (2019) 25:2033–41. doi: 10.1158/1078-0432.CCR-18-2275
12. Javle M, Borad MJ, Azad NS, Kurzrock R, Abou-Alfa GK, George B, et al. Pertuzumab and trastuzumab for HER2-positive, metastatic biliary tract cancer (MyPathway): a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol (2021) 22:1290–300. doi: 10.1016/S1470-2045(21)00336-3
13. Siena S, Di Bartolomeo M, Raghav K, Masuishi T, Loupakis F, Kawakami H, et al. Trastuzumab deruxtecan (DS-8201) in patients with HER2-expressing metastatic colorectal cancer (DESTINY-CRC01): a multicentre, open-label, phase 2 trial. Lancet Oncol (2021) 22:779–89. doi: 10.1016/S1470-2045(21)00086-3
14. Cox G, Vyberg M, Melgaard B, Askaa J, Oster A, O'Byrne KJ. Herceptest: HER2 expression and gene amplification in non-small cell lung cancer. Int J Cancer (2001) 92:480–3. doi: 10.1002/ijc.1214
15. Oh DY, Bang YJ. HER2-targeted therapies - a role beyond breast cancer. Nat Rev Clin Oncol (2020) 17:33–48. doi: 10.1038/s41571-019-0268-3
16. Gaibar M, Beltrán L, Romero-Lorca A, Fernández-Santander A, Novillo A. Somatic mutations in HER2 and implications for current treatment paradigms in HER2-positive breast cancer. J Oncol (2020) 2020:6375956. doi: 10.1155/2020/6375956
17. Connell CM, Doherty GJ. Activating HER2 mutations as emerging targets in multiple solid cancers. ESMO Open (2017) 2:e000279. doi: 10.1136/esmoopen-2017-000279
18. Mazières J, Peters S, Lepage B, Cortot AB, Barlesi F, Beau-Faller M, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol (2013) 31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095
19. Siena S, Sartore-Bianchi A, Marsoni S, Hurwitz HI, McCall SJ, Penault-Llorca F, et al. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann Oncol (2018) 29:1108–19. doi: 10.1093/annonc/mdy100
20. Mehta A, Nathany S, Tripathi R, Sharma SK, Saifi M, Batra U. Non-amplification genetic alterations of HER2 gene in non-small cell lung carcinoma. J Clin Pathol (2021) 74:106–10. doi: 10.1136/jclinpath-2020-206730
21. Yu DH, Tang L, Dong H, Dong Z, Zhang L, Fu J, et al. Oncogenic HER2 fusions in gastric cancer. J Transl Med (2015) 13:116. doi: 10.1186/s12967-015-0476-2
22. Hechtman JF, Zehir A, Yaeger R, Wang L, Middha S, Zheng T, et al. Identification of targetable kinase alterations in patients with colorectal carcinoma that are preferentially associated with wild-type RAS/RAF. Mol Cancer Res (2016) 14:296–301. doi: 10.1158/1541-7786.MCR-15-0392-T
23. Lesurf R, Griffith OL, Griffith M, Hundal J, Trani L, Watson MA, et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann Oncol (2017) 28:1070–7. doi: 10.1093/annonc/mdx048
24. Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
25. Hwang KB, Lee IH, Li H, Won DG, Hernandez-Ferrer C, Negron JA, et al. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci Rep (2019) 9:3219. doi: 10.1038/s41598-019-39108-2
26. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res (2016) 44:e108. doi: 10.1093/nar/gkw227
27. Li Q, Wang K. InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet (2017) 100:267–80. doi: 10.1016/j.ajhg.2017.01.004
28. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-wide copy number detection and visualization from targeted DNA sequencing. PloS Comput Biol (2016) 12:e1004873. doi: 10.1371/journal.pcbi.1004873
29. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics (2014) 30:3390–3. doi: 10.1093/bioinformatics/btu549
30. King CR, Kraus MH, Aaronson SA. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science (1985) 229:974–6. doi: 10.1126/science.2992089
31. Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol (2008) 19:1523–9. doi: 10.1093/annonc/mdn169
32. Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci USA (2002) 99:15937–40. doi: 10.1073/pnas.252640799
33. Matsushita C, Tamagaki H, Miyazawa Y, Aimoto S, Smith SO, Sato T. Transmembrane helix orientation influences membrane binding of the intracellular juxtamembrane domain in neu receptor peptides. Proc Natl Acad Sci USA (2013) 110:1646–51. doi: 10.1073/pnas.1215207110
34. Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol (1997) 15:2894–904. doi: 10.1200/JCO.1997.15.8.2894
35. Corbett IP, Henry JA, Angus B, Watchorn CJ, Wilkinson L, Hennessy C, et al. NCL-CB11, a new monoclonal antibody recognizing the internal domain of the c-erbB-2 oncogene protein effective for use on formalin-fixed, paraffin-embedded tissue. J Pathol (1990) 161:15–25. doi: 10.1002/path.1711610105
36. Powell WC, Hicks DG, Prescott N, Tarr SM, Laniauskas S, Williams T, et al. A new rabbit monoclonal antibody (4B5) for the immunohistochemical (IHC) determination of the HER2 status in breast cancer: comparison with CB11, fluorescence in situ hybridization (FISH), and interlaboratory reproducibility. Appl Immunohistochem Mol Morphol (2007) 15:94–102. doi: 10.1097/pai.0b013e31802ced25
37. Schrohl AS, Pedersen HC, Jensen SS, Nielsen SL, Brünner N. Human epidermal growth factor receptor 2 (HER2) immunoreactivity: specificity of three pharmacodiagnostic antibodies. Histopathology (2011) 59:975–83. doi: 10.1111/j.1365-2559.2011.04034.x
38. Liu Z, Shi M, Li X, Song S, Liu N, Du H, et al. HER2 copy number as predictor of disease-free survival in HER2-positive resectable gastric adenocarcinoma. J Cancer Res Clin Oncol (2021) 147:1315–24. doi: 10.1007/s00432-021-03522-9
39. Ran R, Huang W, Liu Y, Shao L, Liu X, Niu Y, et al. Prognostic value of plasma HER2 gene copy number in HER2-positive metastatic breast cancer treated with first-line trastuzumab. Onco Targets Ther (2020) 13:4385–95. doi: 10.2147/OTT.S240990
40. Wang Y, Jiang T, Qin Z, Jiang J, Wang Q, Yangn S, et al. HER2 exon 20 insertions in non-small-cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann Oncol (2019) 30:447–55. doi: 10.1093/annonc/mdy542
Keywords: next generation sequencing, ERBB2 fusion, HER2, trastuzumab, clinical impact
Citation: Guan Y, Wang Y, Li H, Meng J, You X, Zhu X, Zhang Q, Sun T, Qi C, An G, Fan Y and Xu B (2022) Molecular and clinicopathological characteristics of ERBB2 gene fusions in 32,131 Chinese patients with solid tumors. Front. Oncol. 12:986674. doi: 10.3389/fonc.2022.986674
Received: 05 July 2022; Accepted: 15 September 2022;
Published: 06 October 2022.
Edited by:
Shoumin Zhu, University of Miami Health System, United StatesReviewed by:
Sara Pizzamiglio, Fondazione IRCCS Istituto Nazionale dei Tumori, ItalyCopyright © 2022 Guan, Wang, Li, Meng, You, Zhu, Zhang, Sun, Qi, An, Fan and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyu An, YW5ndWFuZ3l1QG1haWwuY2NtdS5lZHUuY24=; Binghe Xu, eHViaW5naGVAY3Njby5vcmcuY24=; Ying Fan, ZmFueWluZ2Z5QG1lZG1haWwuY29tLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.