
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 09 September 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.985897
This article is part of the Research TopicReviews in Molecular and Cellular OncologyView all 47 articles
miRNAs play a crucial role in regulating genes involved in cancer progression. Recently, miR-153 has been mainly well-known as a tumor suppressive miRNA modulating genes in proliferation, metastasis, EMT, angiogenesis and drug resistance ability of a variety types of cancer. Mechanistic activity of miR-153 in tumorigenicity has not been fully reviewed. This manuscript presents a comprehensive review on the tumor suppressive activity of miR-153 as well as introducing the controversial role of miR-153 as an oncogenic miRNA in cancer. Furthermore, it summarizes all potential non-coding RNAs such as long non-coding RNAs (LncRNAs), transcribed ultra-conserved regions (T-UCRs) and circular RNAs (CircRNAs) targeting and sponging miR-153. Understanding the critical role of miR-153 in cell growth, metastasis, angiogenesis and drug resistance ability of cancer cells, suggests miR-153 as a potential prognostic biomarker for detecting cancer as well as providing a novel treatment strategy to combat with several types of cancer.
Cancer is one of the most critical diseases with an increasing prevalence rate and high mortality. Cancer is a type of cell transformation and loss of cell differentiation with uncontrolled cell growth. It is also a genetic and epigenetic disorder, having much heterogeneity in both tissue and cellular levels. It is characterized by different properties such as proliferation, migration and invasion, metastasis, angiogenesis and chemo/radio resistance abilities. There are many oncogenes, tumor suppressor and regulatory genes which modulate cancer characterizations through various mechanistic pathways (1, 2).
miRNAs are non-coding small RNAs (∼20–24 nucleotides) which regulate target genes negatively, by binding to complementary sites of 3´ UTR in a specific gene’s mRNA, resulting in mRNA degradation or translation inhibition based on complete or partial complementary, respectively (3). Recently, the role of microRNAs (miRNAs) as oncogenes or tumor suppressor genes, has been confirmed in regulating genes involved in the progression of various types of malignancies. The expression level of numerous miRNAs has modified in different types of cancer cells (4–6). Dysregulation of miRNAs is associated with the development and progression of cancer. Therefore, miRNAs have been proposed for using as prognostic biomarkers, predicting and detecting cancer as well as usage as novel strategy for treatment of cancer (7). miR-153, one of the miRNAs involved in cancer progression and development, has recently been reported in several studies. The two miRNAs genes, miR-153-1 (miR-153-5p) and miR-153-2 (miR-153-3p) with a conserved sequence are evolutionarily-conserved genes in Eukaryotes, Vertebrata, Mammalia, Primates, Hominidae, and Homo located on chromosome 2q35 (Chromosome 2 - NC_000002.12) with 90 bp (https://www.ncbi.nlm.nih.gov/gene/406944) and chromosome 7q36.3 (Chromosome 7-NC_000007.14) with 87 bp (https://ncbi.nlm.nih.gov/gene/406945) Evaluation of genomic sequence of miRNA-153-1 and miRNA-153-2 has been revealed that these miRNAs as intragenic miRNAs, are embedded within the 19th intron of IA-2 (also known as PTPRN) and IA-2β (also known as PTPRN2), the conserved host genes, respectively (8).
Recently, dysregulation of miR-153 has been verified in different types of cancer cells such as cervical, lung, neuroblastoma and gastric cancer (9–12). It is implicated in cancer pathophysiological processes, including proliferation, apoptosis, invasion, EMT and metastasis, angiogenesis and chemo/radiotherapy resistance (13). Most importantly, miR-153 acts as a tumor suppressive miRNA to reduce stemness phenotypes of cancer cells (14, 15). Both 5′-arm and 3′-arm of the miR-153 precursor suppress tumor initiation and progression through modulating expression of their target genes (16). Besides anti-tumorigenicity activity of miR-153, several studies have reported the oncogenic role of miR-153 in some types of cancer cells (16–18). It recommends a controversial role of miR-153 as both a tumor suppressor gene and an oncogene.
Critical role of miR-153 in different types of cancers and especially the controversial role of miR-153 in tumorigenicity of cancer cells made the author put forth this review, which is the first comprehensive review with details about miR-153, its transcriptional regulation and mechanistic effects on cancer. This review presents a study on the mechanistic roles of miR-153 as either a tumor suppressor gene or an oncogene in proliferation, apoptosis, migration and invasion, metastasis, angiogenesis and chemo/radio resistance ability of cancer cells. Moreover, it introduces a number of non-coding RNAs (ncRNAs) which regulate the expression of miR-153 in different mechanistic pathways of malignancy. Furthermore, it summarizes some suggested strategies for miR-153-related therapy as well as suggesting as a prognostic biomarker that is associated with some pathological parameters. Recognition and understanding the molecular mechanisms modulated by miR-153 in cancer cells, can be effective for discovering novel treatment strategies as well as usage as a specific prognostic biomarker for the prognosis of different types of cancer.
Primarily, transcriptional regulation of miRNAs is dependent on genomic stability, activity of transcription factors, epigenetic regulation and miRNA biogenesis pathway. There are some potential mechanisms that deregulate miRNAs in cancers including the amplification or deletion of miRNA genes, aberrant activity of transcription factors, epigenetic dysregulation and defects in the miRNA biogenesis pathway (19). miRNAs is regulated by several transcription activators, co-activators or suppressors, so dysregulation of some key transcription factors results in abnormal expression of miRNAs in tumors. Search to discover the potential transcription factors for transcriptional regulation of miR-153 in TRANSFAC database has exhibited the predicted binding sites for transcription factors such as CREB-binding protein (CBP)/p300, cAMP-response element binding protein (CREB), C/EBPß, and ATF4 at promoter sequence of miR-153 (20). Both CBP and p300 interact with CREB/ATF4 in a pathway dependent on the cAMP-dependent protein kinase A and may perform an important role in alteration in gene expression and the regulation of signaling pathways (21). Furthermore, it has been reported that KLF4 may also act as a transcription factor of miR-153 (22). Therefore, the regulation of miR-153 may be mediated by a group of transcription factors and dysregulation of these transcription factors may result in abnormal expression of miR-153 in tumors. Also, it has been reported that the intragenic cryptic promoter of miR-153 may be regulated epigenetically with a site of enrichment for H3K4me3 in hippocampal neurons (20). There is no more evidence on transcriptional regulation of miR-153 in cancer. Nevertheless, further studies are needed to discover the exact mechanism of miR-153 dysregulation in cancer.
miRNA-153 is transcribed by RNA polymerase II as a primary transcripts (pri-miRNAs). The primary transcript is cleaved by the Drosha ribonuclease III enzyme to produce an approximately 70-nt stem-loop precursor miRNA (pre-miRNA), which is further cleaved by the cytoplasmic Dicer ribonuclease to generate the mature miRNA-153 (https://ncbi.nlm.nih.gov/gene/406945). The mature miRNA-153 is incorporated into a RNA-induced silencing complex (RISC), which recognizes target mRNAs through base pairing with the miRNA and results in translational inhibition or destabilization of the target mRNAs (8).
Proliferation of cancer cells is mediated by different genes and mechanistic pathways. miR-153, as a tumor suppressor gene, is able to regulate genes related to proliferation (Table 1). Down-regulation of miR-153 has been verified in proliferation of various types of cancer, whereas, up-regulation of miR-153 inhibits proliferation through targeting molecules involved in survival and proliferation and modulating some suggested mechanisms (75). FYN, a tyrosine-protein kinase, implicated in cell proliferation and metastasis, is downregulated by miR-153. Downregulation of miR-153 results in overexpression of FYN, promoting proliferation and metastasis of esophageal squamous cell carcinoma (62). Also, the expression level of AKT increases in some types of cancer such as lung cancer, ovarian cancer and glioblastoma. The studies have revealed that an increase in AKT expression can be contributed to downregulation of miR-153. However, overexpression of miR-153 induces cell cycle arrest in G0/G1 and G2/M phases as well as inhibiting proliferation by regulating the expression of AKT, CDK1 through binding to and degrading their mRNAs (3, 23, 24, 26). Moreover, miR-153 suppresses the AKT signaling pathway in glioblastoma through reduction in expression of insulin receptor substrate-2 (Irs-2) which acts as a molecular adaptor, mediating influences of insulin and insulin-like growth factor 1 (IGF-1) on cell proliferation (Figure 1) (27). miR-153 inhibits the proliferation of glioma cells by targeting AKT which is also well known as a mTOR complex 2 (mTORC2) indicator (25). In addition, high expression levels of an essential subunit of mTORC2, Rictor, along with overexpression of mTORC2 have been reported in some cancer cells such as glioma cells, whereas miR-153 as an anti-Rictor is downregulated in these cells. Therefore, modulating the mTOR pathway contributes to anti-tumorigenicity activity of miR-153. Furthermore, ribosomal protein S6 kinase B1 (RPS6KB1) is a serine/threonine kinase in downstream of mTOR pathway, implicated in cell viability and inhibition of apoptosis through phosphorylating BAD protein in thyroid carcinoma. miR-153 is involved in inhibiting proliferation and development of thyroid carcinoma by targeting RPS6KB1 in the mTOR-dependent pathway (28). Furthermore, miR-153 regulates the expression level of an E3 ubiquitin ligase, Zinc and ring finger 2 (ZNRF2), which is implicated in the activity of PI3K/Akt/mTOR pathway in papillary thyroid cancer. It recommends the miR-153/ZNRF2 axis, as one of the most important mechanistic pathways in suppressing the PI3K/Akt/mTOR (29).
Also, anti-proliferative activity of miR-153 is mediated by targeting Kruppel-like factor 5 (KLF5) in breast and gastric cancers. KLF5 is a transcription activator of genes involved in proliferation that is activated by the MEK/ERK1/2 signaling pathway (12, 30). Moreover, upregulation of other transcription factors such as Zinc finger and BTB domain-containing protein 2 (ZBTB2) and RUNX2 due to downregulation of miR-153 may also affect tumorigenicity of gastric cancer. ZBTB2 is a novel partner of the nucleosome remodeling and deacetylase (NuRD) complex which is positively correlated with cancer cell proliferation and metastasis (33). Also, overexpression of RUNX2, a transcription factor involved in regulation of genes related to EMT, metastasis and development of cancer cells, is associated with downregulation of miR-153 in breast cancer cells (60). It suggests that ZBTB2 and RUNX2 may be considered as targets of miR-153, respectively. On the other hand, it has also been confirmed that miR-153 decreases proliferation and promotes cell cycle arrest by targeting E2F3, resulting in downregulation of E2F family transcription factors and Ki67 in thyroid cancer (31).
The other target gene of miR-153, TGFβ, is up-regulated in cancer cells such as osteosarcoma, resulting in overexpression of p-SMAD2, p-SMAD3, EGFR, IGF binding protein-3 (IGFBP-3) which are the crucial proteins in downstream of TGFβ signaling pathway. Therefore, upregulation of miR-153 suppresses proliferation of cancer cells through modulating TGFβ, negatively, which leads to low expression of its downstream proteins (32). It shows that miR-153 acts as an anti-proliferative factor which is downregulated in various types of cancer. Totally, it may be introduced as a novel biomarker to detect cancer as well as usage as a novel strategy for cancer therapy.
Recently, the studies have shown the pro-apoptotic role of miR-153 in cancer cells (Table 1) (14, 76). It has been reported that the low rate of apoptosis in some cancer cells, including glioblastoma, ovarian cancer, CML and AML, may be contributed to downregulation of miR-153 which targets three antiapoptotic genes, Bcl-2, Mcl-1 and XIAP through binding to 3´UTR of their mRNAs (34, 36, 39). Recently, it has been shown that Mcl-1 is also well known as a miR-153 target gene in inhibiting metastasis in some types of cancer, including oral and liver cancer (57, 59). Thus, besides the anti-apoptotic role, MCL-1 plays an important activity in migration, metastasis and development of a variety of cancer cells. In addition, it has been confirmed that miR-153 has a critical role in cell viability reduction and apoptosis induction through activating caspase 3, 9 and rising Bax/Bcl2 ratio, which are mediated by targeting TGFβ/Samd2 axis in nasopharyngeal cancer cells (37). On the other hand, miR-153 is considered as a negative regulator of homologous to the E6-associated protein carboxyl terminus domain containing 3 (HECTD3), an E3 ubiquitin ligase. HECTD3 promotes ubiquitination of some types of caspases, including caspase 8 and 9, resulting in apoptosis inhibition and cell survival promotion in cancer cells such as breast cancer. Thus, modulating HECTD3 is implicated in miR-153-induced apoptosis in these cancer cells (38). Besides apoptotic activity, miR-153 regulates autophagy in CML and osteosarcoma. miR-153 inhibits autophagy by downregulating Bcl2 as well as targeting ATG5 (35, 40). Therefore, it can be recommended as a therapy strategy through stimulating apoptosis and suppressing autophagy.
miR-153 regulates invasion, metastasis and progression of different types of cancer through some suggested mechanisms. Numerous studies have verified that downregulation of miR-153 increases migration and metastasis of some types of cancer cells, whereas upregulation of miR-153 suppresses invasion and metastatic phenotypes of cancer cells (Table 1) (41, 43, 46, 54, 74). In addition, the role of miR-153 has been shown in inhibiting stemness phenotypes of cancer stem cells (CSCs) such as self-renewal, proliferation and tumorigenicity of lung cancer cells (42). Jagged1/Notch, one of the signaling pathways involved in CSCs, is regulated by miR-153 through targeting Jagged 1, Notch1 ligand in non-small cell lung cancer (42). Thus, Jagged1/Notch can be considered as targets for lung cancer treatment modulated by miR-153.
On the other hand, epithelial mesenchymal transition (EMT), which is a tumor progression process in malignant cancer cells and CSCs, is regulated by miR-153. Downregulation of miR-153 leads to the development of EMT through decrease in E-cadherin, the epithelial marker, as well as an increase in expression levels of mesenchymal markers, including vimentin, Snail and fibronectin. Overexpression of mesenchymal markers is mediated by upregulation of SNAI1, one of the other miR-153 target genes in various types of cancer cells such as melanoma, esophageal squamous cell carcinoma, oral cancer, osteosarcoma, human epithelial cancer, pancreatic adenocarcinoma and laryngeal squamous cell carcinoma (48, 50, 54, 56, 57, 77). miR-153 suppresses SNAI1-induced metastasis through negative regulation of SNAI1 in some types of cancer (44, 46, 47, 49). Also, WNT1-inducible signaling pathway protein-1 (WISP1) inhibits miR-153-induced downregulation of Snail mRNA translation and promotes migration, metastasis and EMT of oral squamous cell carcinoma cells (Figure 2) (55). Therefore, it demonstrates WISP1/miR-153/Snail axis acts as an important pathway in EMT and metastasis of malignancies.
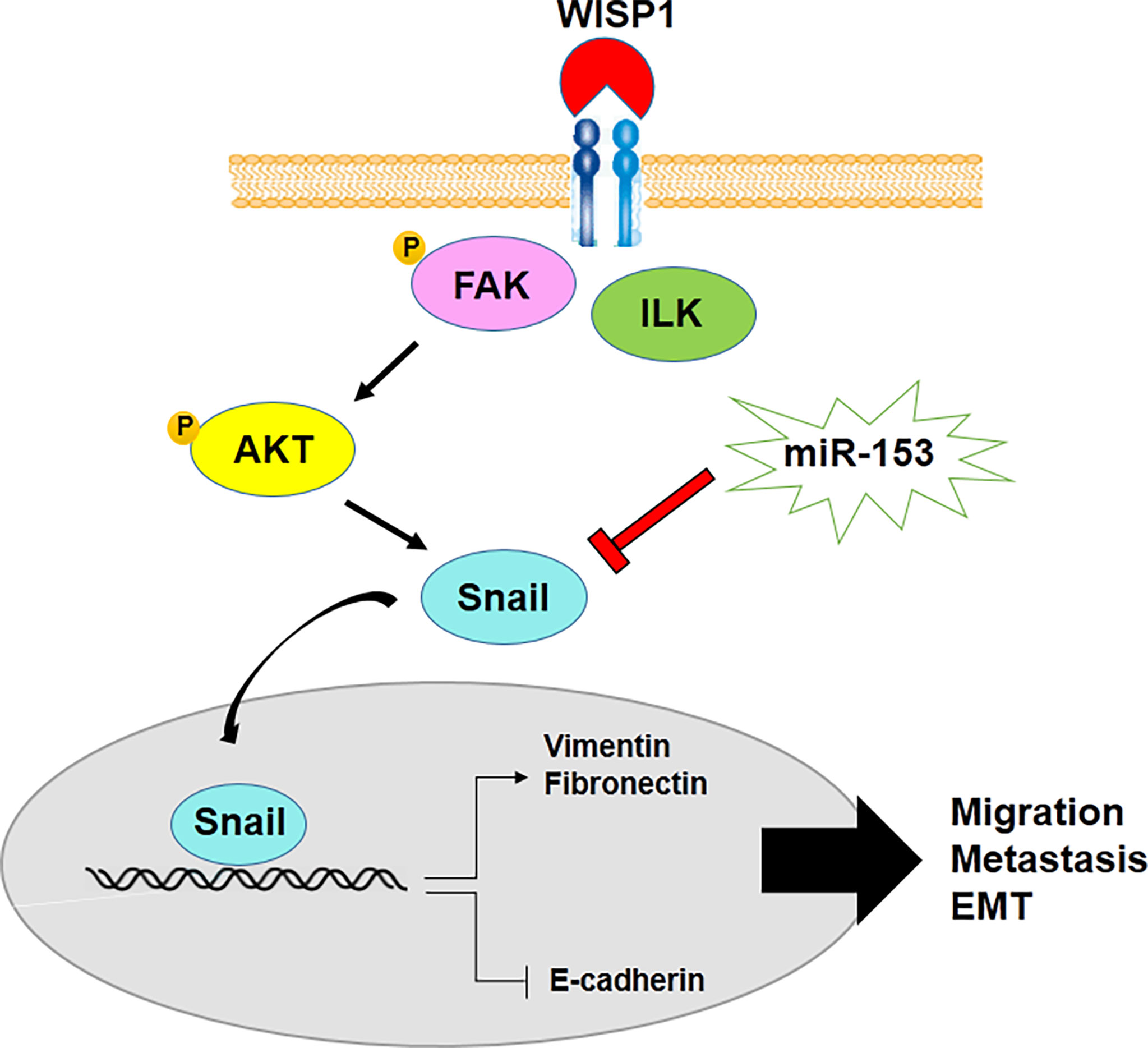
Figure 2 miR-153 regulates metastasis and EMT of cancer cells through decrease in E-cadherin and increase in expression levels of mesenchymal markers.
One of the other suggested molecular mechanisms involved in anti-metatstatic activity of miR-153 is mediated by targeting TGFβ receptor 2 (TGFβR2) that leads to inhibit invasion, migration and EMT in breast cancer (41). In addition, metadherin (MTDH), a new target of miR-153, is an oncogene inducing EMT through activating PI3K/Akt, Wnt/β-catenin, MAPK and NFκB signaling pathways. Therefore, these signaling pathways may be recognized as indirect targets of miR-153, which are over activated by downregulation of miR-153 in cancer cells (64, 78). It recommends the miR-153/MTDH axis as a prognostic biomarker and a novel targeted therapy for treatment of breast cancer. Also, it has been shown that expression of miR-153 is closely correlated with decreased expression of p-Akt in the PI3K signaling pathway regulating invasion and metastasis of glioma cells (65).
Furthermore, anti-tumorigenicity activity of miR-153 contributes to aberrant expression of a transcriptional repressor of E-cadherin coding gene, ZEB2, which plays a critical role in metastasis and EMT (43, 44). In addition, the expression of RhoGTPase activating protein 18 (ARHGAP18), Rab-like protein 3 (Rabl3) and Rho associated coiled-coil containing protein kinase 1 (ROCK1), which play a vital role in migration and metastasis of cancer cells, are negatively regulated by miR-153 in hepatocellular carcinoma and breast cancer (51–53). Kinesin-like protein (KIF20A) is another target of miR-153 which is responsible for intracellular organelle transport and cell migration. miR-153 promotes downreulation of KIF20A and inhibits migration and invasion of cervical cancer cells (61). Also, S100A14, a calcium dependent regulatory protein with a phosphorylation site by protein kinase C (PKC), is one of the other miR-153 targets which is closely related to proliferation and metastasis of non-small cell lung cancer cells (58). Furthermore, degradation of extracellular matrix proteins by A Disintegrin and Metalloproteinase (ADAM) proteolytic enzymes is a key step to migration and metastasis of cancer cells. Downregulation of miR-153 is correlated with overexpression of ADMA19, proposing ADMA19 as another miR-153 target gene in non-small cell lung cancer (63). Totally, miR-153 suppresses migration, invasion, EMT and metastasis of cancer cells through some suggested pathways, resulting in introducing miR-153 as a prognosis marker and therapy target for malignancies.
Angiogenesis is a vital process in the development of tumors that is modulated by hypoxia and thus, hypoxia-induced HIF1 mediates angiogenesis in cancer cells. HIF1α promotes the expression of some angiogenic factors, including vascular endothelial growth factor (VEGF) and angiopoietin 1 (ANG1) by binding to HIF1 response element (HRE) (79). Recently, it has been verified that miR-153, as a tumor suppressor gene, regulates angiogenesis by modulating VEGF and ANG1 in glioma and breast cancer, respectively (Table 1). miR-153 suppresses angiogenesis through targeting HIF1α as well as negative regulation of ANG1 and inhibition of endothelial cell tube formation and migration in breast cancer (Figure 3) (67, 80, 81). On the other hand, miR-153 declines the expression of angiogenesis promoting factors such as VEGFA and cdc42 by binding to 3´UTR of their mRNAs in glioma (68). Therefore, it indicates that miR-153 regulates HIF1α/VEGF axis in angiogenesis, proposing usage as an anti-angiogenesis therapy in cancer. In addition, the insulin-like growth factor1 receptor (IGF-1R) mediates proliferation of vascular smooth muscle cells (VSMCs) through promoting the PI3K/Akt signaling pathway in hyperplasia. miR-153 suppresses proliferation of VSMCs by targeting and downregulating IGF-1R as well as inhibiting PI3K/Akt signaling pathway (69).
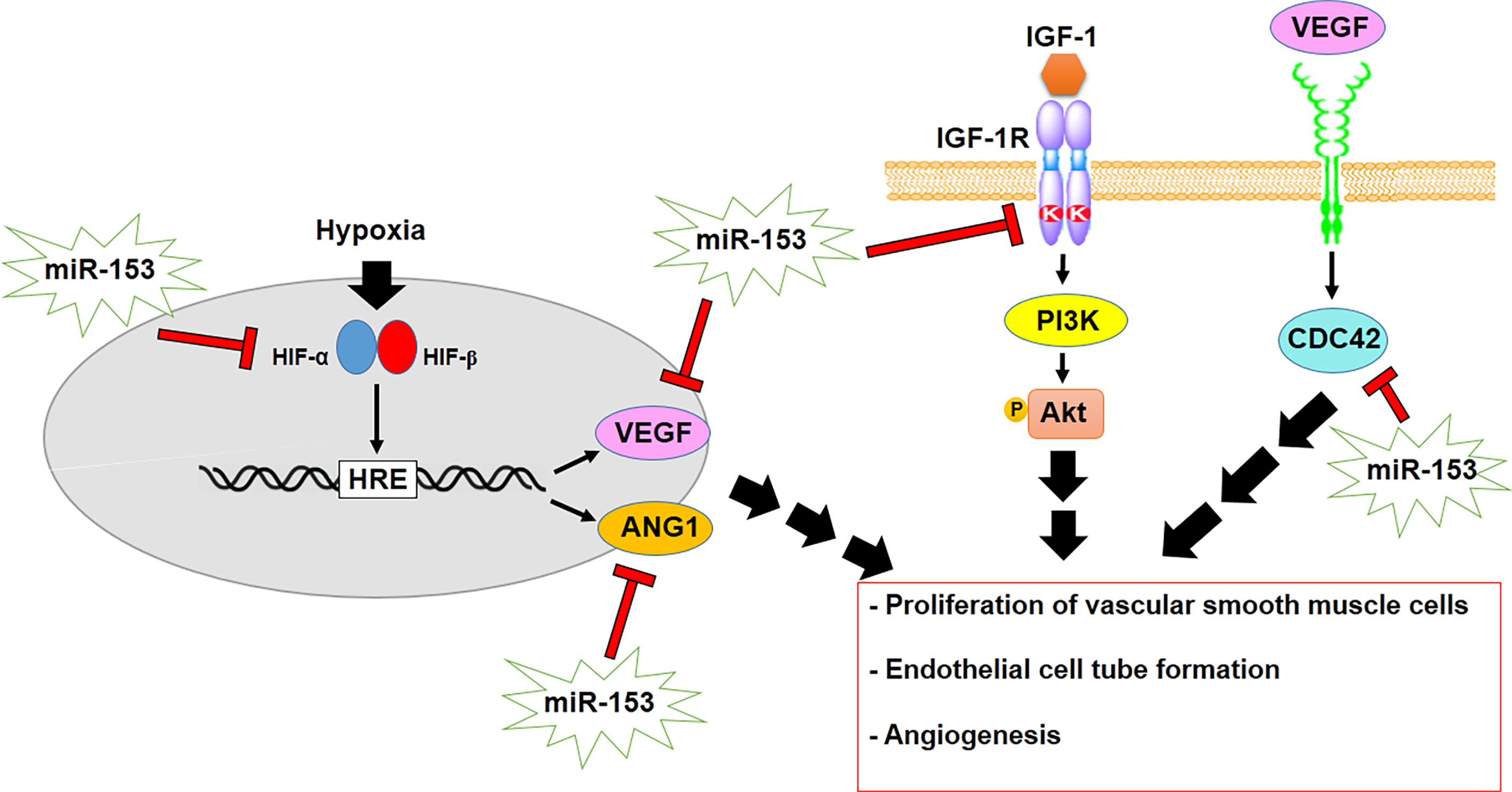
Figure 3 miR-153 regulates angiogenesis and proliferation of VSMCs by modulating HIF1α/VEGF/CDC42 axis as well as inhibiting PI3K/Akt signaling.
Furthermore, high metabolism in cancer cells is related to malignancy of cancer cells. Increased metabolism of tryptophan has been observed in angiogenesis. Metabolism of tryptophan is mediated by indoleamine 2,3-dioxygenase 1 (IDO1), which is downregulated by miR-153 in bladder cancer. Therefore, miR-153 can reduce tryptophan metabolism and also modulate angiogenesis through suppressing the IL6/STAT3/VEGF signaling pathway (66). The studies have confirmed that miR-153 may be used as a prognostic marker for angiogenesis and applied as a target for treatment of a variety of malignancies.
Some potential mechanisms are implicated in radio/chemotherapy resistance ability of cancer cells and CSCs mediating recurrence in some types of cancer (82). Recently, numerous studies have reported the role of miRNAs, in particular, miR-153, in regulating radio and drug resistance ability of cancer cells (Table 1) (70, 71, 83, 84). The expression of miR-153 is dysregulated in radiotherapy resistant cancer cells. However, high expression level of miR-153 increases sensitivity of cancer cells to radiotherapy, which may be mediated by targeting Snail and Jagged canonical notch ligand1 (JAG1) in pancreatic cancer (70, 71). Moreover, downregulation of miR-153 has been reported in drug resistant cancer cells, whereas miR-153 overexpression enhances the sensitivity of cancer cells to chemotherapy (26, 35, 76). One of the suggested mechanisms of drug resistance of cancer cells is upregulation of ABCE1 due to downregulation of miR-153 in lung cancer cells (72). Therefore, ABCE1 is suggested as miR-153 target gene modulating drug resistance of cancer cells. In addition, miR-153 regulates drug resistance by modulating expression of Cbp/p300-interacting transactivator 2 (CITED2), which is implicated in drug resistance of gastric cancer cells (Figure 4). CITED2 regulates the TGF-β signaling pathway through its association with the SMAD/p300/CBP-mediated transcriptional coactivator complex (73). Also, miR-153 is able to regulate the nuclear factor erythroid 2-related factor 2 (NRF2), a transcription factor upregulated in antioxidant responses of radio/chemotherapy resistant cancer cells. NRF2 overexpression controls the expression of glutathione peroxidase 1 (GPX1) and reduces ROS, which leads to enhancement of chemotherapy resistance (15). However, aberrant expression of NRF2 as a result of upregulation of miR-153, increases sensibility of cancer cells and CSCs to radio/chemotherapy in some types of cancer cells including esophageal squamous cell carcinoma and glioma stem cells (15, 74). In addition, miR-153 decreases drug resistance of AML through targeting XIAP, which is responsible for protecting cells from apoptosis by blocking the function of caspases (39). Recently, miR-153 has been recommended as an adjuvant in treatment of various types of leukemias. Acute promyelocytic leukemia (APL) and Chronic myeloid leukemia (CML) show drug resistance to As2O3, which is well-known as an effective drug in the treatment of APL, CML and other leukemias (85). The expression of miR-153 reduces in As2O3-resistant CML, whereas overexpression of miR-153 induces greater apoptosis in combination with As2O3 in CML (76). Also, downregulation of miR-153 has been observed in drug resistance ability of CML to imatinib. However, miR-153 upregulation increases sensitivity of CML to chemotherapy through downregulating Bcl2, resulting in autophagy reduction in drug resistant CML (35). It indicates that miR-153 overexpression in combination with chemotherapy may be used as a novel strategy to combat with leukemias. Totally, the studies suggest that miR-153 can be used as a molecular target in order to reduce the drug resistance of cancer cells.
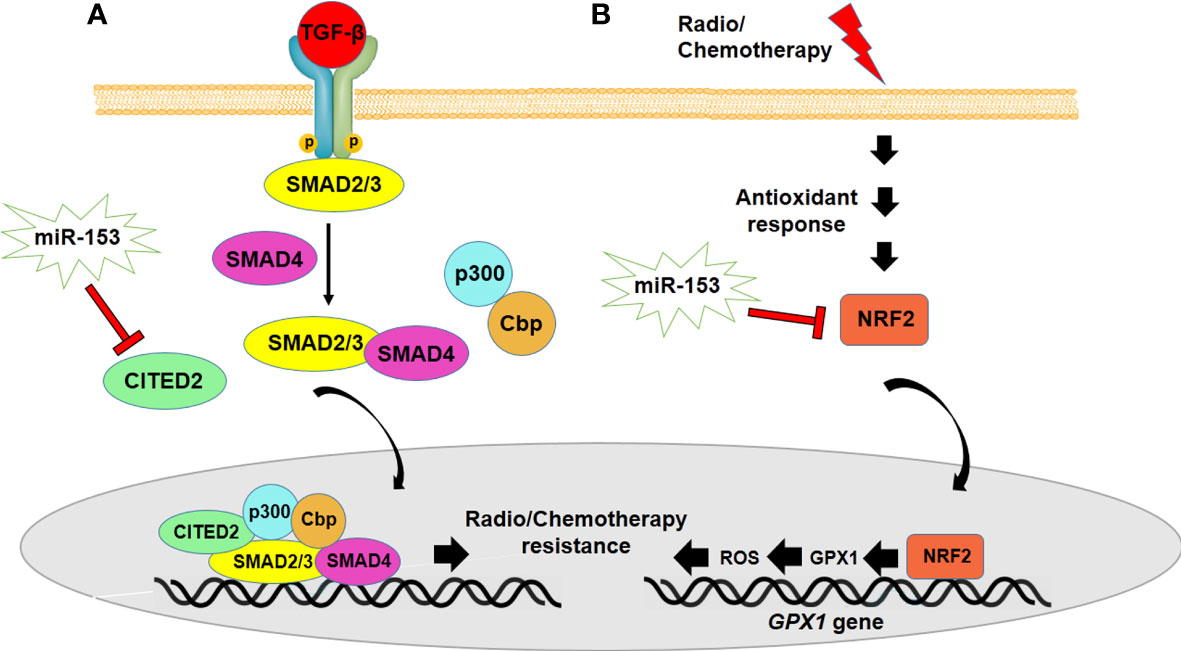
Figure 4 miR-153 regulates radio/chemotherapy resistance by modulating expression of CITED2 (A) and NRF2 (B). (A) miR-153 targets CITED2 which is implicated in drug resistance of cancer cells through regulating TGF-β signaling. (B) NRF2 overexpression controls the expression of glutathione peroxidase 1 (GPX1) and reduces ROS which leads to enhancement of chemotherapy resistance.
Besides tumor suppressor activity of miR-153 in various types of cancer, the oncogenic role of miR-153 has been confirmed in some studies (16, 17, 86). There is a positive correlation between the overexpression of miR-153 and aggressive phenotypes of cancer cells including, proliferation, lymph node metastasis, EMT and drug resistance that can be mediated by targeting some suggested target genes (Table 2). Proliferative and metastatic activity of miR-153 is mediated by activating the PI3K/Akt signaling pathway. One of the suggested mechanisms involved in the oncogenic role of miR-153 is downregulating PTEN, which is responsible for suppressing the PI3K/Akt signaling pathway in prostate cancer cells (17). miR-153 promotes proliferation of prostate cancer cells through enhancing G1/S phase transition, cyclin D expression as well as inhibiting CDK inhibitor (p21) that are mediated by targeting PTEN. Aberrant expression of PTEN results in activation of PI3K/Akt signaling and downregulation of Forkhead box class O 1 (FOXO1) in these types of cancer cells (86, 87). Also, the expression of Argonaute RISC catalytic component 1 (AGO1), a miRNA processing protein in RISC complex, is downregulated in renal cell carcinoma due to upregulation of miR-153. The miR-153/AGO1 axis induces progression of renal cell carcinoma through activating PI3K/Akt signaling pathway. It recommends both AGO1 and miR-153 as potential prognostic markers for detecting this type of cancer (16). One of the other signaling pathways involved in tumorigenicity activity of miR-153 is Wnt/β catenin signaling pathway which is activated by miR-153 targeting WWOX, a tumor suppressor and inhibitor of Wnt/β catenin in hepatocellular carcinoma (88). Moreover, it has been reported that miR-153 is upregulated in triple negative breast cancer and may exhibit an oncogenic role in breast cancer development and progression. miR-153 inhibitor suppresses proliferation and induces apoptosis through enhancing caspase 3/7 activity in triple negative breast cancer (89). The oncogenic role of miR-153 is associated with an increase in anti-apoptotic proteins such as surviving and BCL2, as well as a decrease in p21. miR-153 promotes proliferation and metastasis of cancer cells such as hepatocellular carcinoma through suppressing the efficiency of chemical and targeted drugs including Etoposide, Paclitaxel and Sorafenib (86). miR-153-induced dug resistance of cancer cells is promoted by production of matrix metalloproteinase (MMP9) as well as inhibiting transcription factor Forkhead box class O 3a (FOXO3a), a tumor suppressor protein in colorectal cancer (18). Totally, miR-153 can be considered as an oncogene and potential prognostic marker for detecting some types of cancer that may be suggested as a novel therapy target.
It has been confirmed that miR-153 acts as both tumor suppressor gene and oncogene which may be dependent on type of cancer cells with different gene expression profiles (17). It proposes a tumor suppressive role of miR-153 which is downregulated in most types of tumors including non-small cell lung cancer, liver cancer, glioblastoma, thyroid carcinoma, gastric cancer, osteosarcoma, melanoma, oral cancer, pancreatic adenocarcinoma, bladder cancer, cervical cancer and leukemias, and an oncogenic activity of miR-153 which is upregulated in prostate, colorectal and renal cell carcinoma. In renal cell carcinoma, the signaling pathway involved in oncogenic effect of miR-153 is mainly PI3K/AKT signaling pathway which has a critical role in cell cycle progression and proliferation by upregulation of cyclin D1 as well as EMT-mediated metastasis by downregulation of E-cadherin and upregulation of Snai1. Also, PI3K/AKT signaling pathway is suppressed by AGO1, a tumor suppressor gene and one of the miR-153 targets which is implicated in gene silencing and translational inhibition of mRNAs (16). miR-153 has also an oncogenic role in prostate cancer. One of the signaling pathways promoting prostate cancer is PI3K/AKT signaling pathway which is regulated by PTEN, a PI3K/AKT signaling inhibitor. PTEN is frequently downregulated in prostate tumors that may be mediated by upregulation of miR-153. miR-153 targets PTEN to activate PI3K/AKT signaling pathway, resulting in cell cycle progression and proliferation through upregulation of cyclin D1 and downregulation of p21, a CDK inhibitor (87). Moreover, in colorectal cancer, higher expression of miR-153 has been detected in primary cancer cells compared with normal colonic epithelial cells and in advanced stage of tumor compared with early stages. It has been revealed that miR-153 supports colorectal cancer via pleiotropic effects that enhance progression and chemotherapeutic resistance (18). Compared to early stages of colorectal cancer, overexpression of MMP9 is strongly associated with more advanced stages. It has been confirmed that miR-153 upregulates MMP9 through targeting some predicted transcription factors that may suggest mechanism for this process (18). In addition, it is assumed that miR-153 has a controversial role as both tumor suppressive and oncogenic miRNAs in hepatocellular carcinoma and breast cancer in a manner dependent on heterogeneity and tumor stage. In early clinical stages, miR-153, as an oncogenic miRNA, promotes proliferation and tumor growth of hepatocellular carcinoma through modulating β-catenin signaling pathway (88). On the other hand, it acts as tumor suppressor miRNA in metastatic cancer and in an advanced clinical stage of hepatocellular carcinoma by targeting ARHGAP18, Rabl3 and snail which are up-regulated and implicated in migration and metastasis of this type of cancer cells (51, 52, 56). Moreover, abnormal downregulation of miR-153 can also be associated with advanced clinical stage of breast cancer as detected in human ovarian tumor (89, 90). Also, the expression of a specific miRNA may be different due to diverse histological subtypes in a certain cancer (91). For example, data from The Encyclopedia of RNA Interactomes (ENCORI) showed that the expression levels of miR-153 in kidney chromophob is higher than kidney renal papillary cell carcinoma and is lower than kidney renal clear cell carcinoma. The expression levels of miR-153 is also higher in lung adenocarcinoma compared with lung squamous cell carcinoma (Figure 5). Further investigation is recommended to detect miRNA expression in different histological subtypes of other cancers.
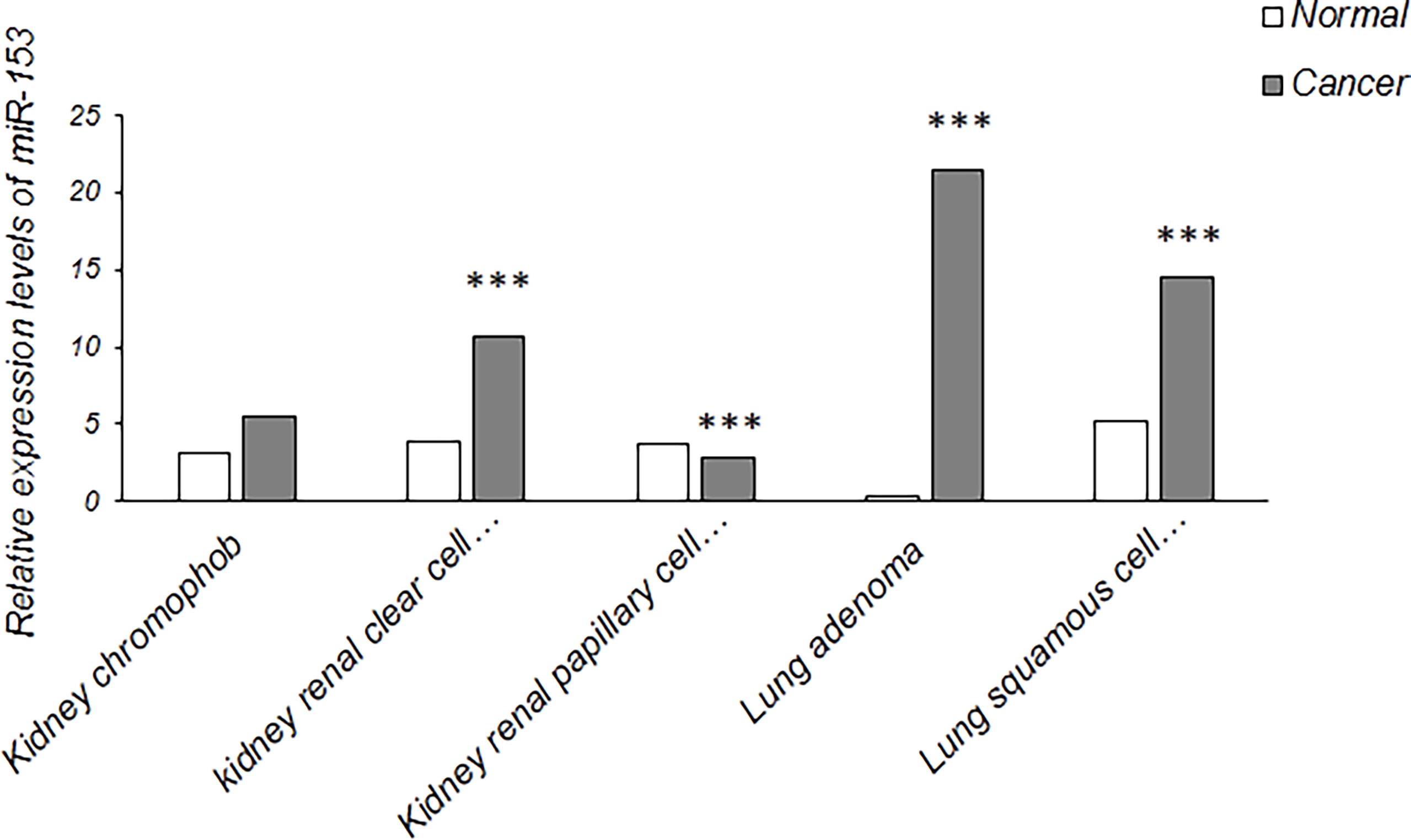
Figure 5 Relative expression levels of miR-153 in some cancers with diverse histological subtypes (The data were obtained from ENCORI). Kidney chromophob (N = 65) against normal samples (N = 24), kidney renal clear cell carcinoma (N = 517) against normal samples (N = 71), kidney renal papillary cell carcinoma (N = 289) against normal samples (N = 32), lung adenocarcinoma (N = 512) against normal samples (N = 20) and lung squamous cell carcinoma (N = 475) against normal samples (N = 38). N indicates the number of tissue samples. ***P < 0.001.
In addition, miRNA concentration affect gene silencing. Some target genes require lower miRNA concentration and some others need higher miRNA abundance for silencing that depends on miRNA-mRNA complementary base pairing (92). It is suggested that the concentration of miR-153 is one of the other reasons causing the different effects of miR-153. Because oncogenes are more abundant targets of miR-153, they require low concentrations of miRNA to be silenced. On the other hand, tumor suppressor genes are less abundant targets of miR-153, they require high concentrations of miRNA to be silenced. Therefore, miR-153 has the tumor suppressive and oncogenic roles in the low and high concentrations, respectively (Figure 6).
To date, many miR-153 target genes have been confirmed, and there are numerous other unverified genes that can be regulated by miR-153 and affect several signaling pathways, simultaneously. Also, a specific target can be regulated by various miRNAs. Therefore, it influences the ultimate function of miRNA, just as the expression level of the target genes also affects the function of miRNA. In a study, tumor suppressive and oncogenic effects of miR-204 have been confirmed through targeting XRN1 in prostatic adenocarcinoma cells and neuroendocrine-like prostate cancer cells, respectively (93). In another report, inhibition of all miRNAs targeting FOXO1 in endometrial cancer cells with low levels of FOXO1 expression (Ishikawa cells) induced cell cycle arrest, while no significant changes were observed in endometrial cancer cells with high levels of FOXO1 (HEC-1B cells) (94). Therefore, contradictory role of miRNAs in cancer may be due to the variable expression levels of the target genes in cancer cells. It has been shown that miR-153 induces apoptosis through targeting HECTD3 in BT-549 breast cancer cells, whereas in another report miR-153 inhibits apoptosis through indirect targeting of caspase 3/7 activity in triple negative breast cancer (38, 89). Ultimately, the function of miRNA is determined by the network of these target genes with different expression and functions (92). The abundance of these targets with opposite function may be contributed to the dual role of miR-153 in cancers.
Furthermore, the dual role of miR-153 in cancer may also be contributed to single nucleotide polymorphisms (SNPs) in miRNAs or miRNA-binding sites (92). SNPs can affect binding affinity of miRNAs to their target genes. For example, it has been reported that there is a significant association between SNP-rs66461782 in miR-186 and breast cancer. On the other hand, rs1062577 on its target gene, ESR1, decreased the binding affinity of miR-186 to ESR1 mRNA due to alteration in the number of hydrogen bonds (92). Several SNPs in pre-miR-153-5p, pre-miR-153-3p and three SNPs on the seed site of miR-153-3p (rs747400621, rs1478975893 and rs1034944138) have been recognized (http://bioinfo.life.hust.edu.cn/miRNASNP/#!/) that may affect the biogenesis and function of miR-153 on its target genes. There is still no evidence that has revealed the association between these SNPs with various cancers. Furthermore, there are numerous SNPs on the 3´UTR of target genes of miR-153 that may create or disturb negative regulatory effects of miR-153 on target mRNAs. For example, there is a significant association between 3′UTR WWOX SNP (rs73569323) and risk of hepatocellular carcinoma, as it has shown that T allele decreases risk of hepatocellular carcinoma with an unknown mechanism (95). Also, it has been reported that 3′UTR polymorphisms of amyloid precursor protein (APP) (APP-118C/A and APP-534G/A) may affect the binding affinity of miR-153 and the regulation of APP expression by this miRNA in Alzheimer’s disease (96). In the other research, it has also been verified that the expression of C/T allele variants (rs6932603) in CCDC170 may be regulated by miR-153. miR-153 significantly downregulated rs6932603-T expression in bone osteosarcoma epithelial cells (97). Therefore, studies can be extended to identify the association between SNPs in miR-153 and its target genes with different types of cancers as well as detecting effects of these SNPs on the miR-153 binding to its target genes in cancers.
There are various types of non-coding RNAs, including long non-coding RNAs (LncRNAs), transcribed ultra-conserved regions (T-UCRs) and circular RNAs (CircRNAs) regulating miR-153, negatively. Several types of non-coding RNAs are upregulated in various types of cancer cells that results in inhibiting anti-cancerous effects of miR-153. A number of LncRNAs, T-UCRs and CircRNAs play an oncogenic role in proliferation, invasion, metastasis, angiogenesis, and drug resistance ability of cancer cells by targeting and sponging miR-153 through some suggested mechanisms (Table 3). Figure 7 also presents a summary for non-coding RNAs-related regulation of miR-153 and an overall schematic representation of miR153 in cancer. LncRNA CASC15 promotes proliferation of breast cancer cells through sponging miR-153 targeting KLF5 (98). It has been confirmed that LncRNA TTN-AS1 and LncRNA OIP5-AS1 increase proliferation of papillary thyroid cancer and gastric cancer through miR-153/ZNRF2 axis and miR-153/ZBTB2 axis, respectively (29, 33). LncRNA CDKN2BAS and LncRNA LINC00858 induce migration, invasion and metastasis of hepatocellular carcinoma through suppressing miR-153 and upregulating ARHGAP18 and Rabl3 (51, 52). Also, LncRNA HIF1A-AS2 and LncRNA NEAT1 promote migration, invasion and metastasis of non-small cell lung cancer in a manner dependent on the miR-153/S100A14 axis and the miR-153/Wnt signaling axis (58, 78). Moreover, oncogenic activity of LncRNAs including LncRNA-XIST, LncRNA FGD5-AS1, LncRNA LINC00152 is mediated by overexpressing SNAI1, MCL1, FYN in osteosarcoma, oral cancer, esophageal carcinoma, respectively (48, 59, 62).
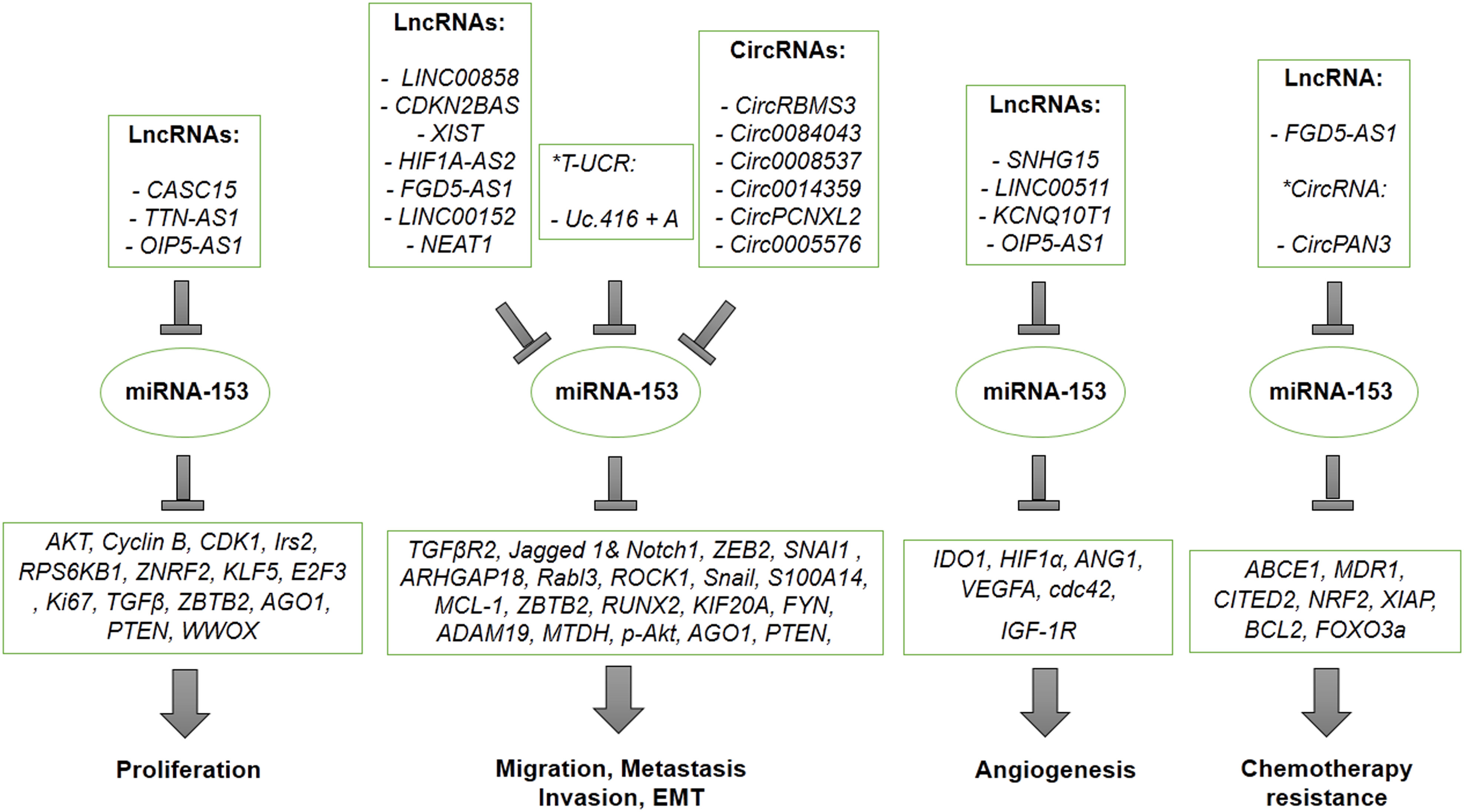
Figure 7 A summary figure for lncRNA-related regulation of miR-153 and an overall schematic representation of miR153 in cancer.
On the other hand, angiogenic activity of LncRNAs has been confirmed by some studies. It has been reported that angiogenic activity of LncRNA SNHG15 is regulated by suppressing miR-153, which targets VEGFA and cdc42 in glioma (68). Furthermore, LncRNAs including LncRNA LINC00511 and LncRNA KCNQ10T1 promote angiogenesis by overexpressing HIF1 in a manner dependent on LncRNAs/miR-153/HIF1 in colorectal cancer and retinoblastoma (80, 81). HIF1 is considered as a potential transcription factor regulating the expression of LncRNA LINC00511. Therefore, it suggests positive feedback for LncRNA LINC00511 as miR-153/HIF1/LINC00511 (80). Also, aberrant upregulation of lncRNA OIP5-AS1 regulates angiogenesis and autophagy of osteosarcoma through down-regulating miR-153, which targets ATG (40).
In addition, resistance to chemotherapy may be mediated by Lnc-RNA FGD5-AS1 suppressing miR-153 and upregulating CITED2, a gene target of miR-153, in gastric cancer (73).
T-UCR ncRNA Uc.416 + A is a non-coding RNA which is well-known as a crucial factor for EMT of cancer cells such as renal carcinoma. Tumorigenic activity of T-UCR ncRNA Uc.416 + A contributed to upregulating vimentin and snail through sponging miR-153 (77).
CircRNAs, the other regulating RNAs, have been recognized as non-coding RNAs which have an important role in tumorigenicity of cancer cells by modulating miRNAs in particular miR-153. CircRNAs such as CircRBMS3, CircRNA-0084043 and Hsa-Circ-0008537 promote cancer cell migration, invasion and metastasis through upregulating SNAI1 and snail as a result of sponging miR-153 in melanoma, gastric cancer and liver cancer (54, 57, 99). One of the other CircRNAs contributing to migration, invasion and metastasis of cancer cells by modulating miR-153 is Has-Circ-0014359, which is upregulated in glioma cells and leads to an increase in the PI3K signaling pathway (65). Also, miR-153 can be targeted by CircPCNXL2, which overexpresses ZEB2 in renal carcinoma in a manner dependent on the CircPCNXL2/miR-153/ZEB2 axis (100). Circ-0005576 is also overexpressed in cervical cancer resulting in upregulation of KIF20A, one of the other target genes of miR-153 (61). Furthermore, one of the other CircRNAs sponging miR-153, is CircPAN3, which induces chemotherapy resistance of AML through suppressing miR-153 and overexpressing XIAP (39).
LncRNAs can also regulate the expression of transcription factors related to miR-153 expression. For instance, LncRNA Taurine upregulated1 (TUG1) downregulates miR-153 indirectly in colorectal cancer through targeting KLF4, which is well-known as a miR-153 transcription factor (22). It demonstrates non-coding RNAs as molecular targets in order to discover a novel strategy for treatment of a variety of cancers.
Some research have examined anti-cancerous activity of several potential therapeutic drugs and regents that are able to modulate the expression of miR-153 in vitro models of some cancers including glioblastoma and breast cancer. 4-phenylbutyric acid, 5-aza-2´-deoxycytidine and Olea europaea leaf extract induce apoptosis through upregulation of miR-153 targeting Bcl-2, Mcl-1 and Irs2 in glioblastoma (27, 101). Also, Mifepristone and Mifepristone derivative FZU-00,003 inhibit cell cycle progression and proliferation of breast cancer cells as well as inducing apoptosis through suppressing KLF5 expression which is mediated by overexpression of miR-153 (30, 102). Therefore, it can be suggested that regents and drugs regulating the expression of miR-153 can be extended in the experiment for a variety of tumor cells.
In addition, expanding prospective trials with larger sample sizes and further validation by independent studies on different types of tumors, miR-153 could be entered the clinical trial process and recommended as a novel treatment strategy in the future. For this purpose, there are some suggested miRNA treatment strategies including miRNA mimics and inhibitors, polymer based delivery system, nanoparticles, viral and non-viral approaches. For oncogenic miRNAs, miRNAs inhibitors, antagomiRs, anti-miR oligonucleotides, single-stranded antisense, locked nucleic acid anti-miRs, miRNA sponges prevent miRNA biogenesis or miRNA-mRNA interaction. On the other hand, for tumor suppressive miRNAs, synthetic miRNA mimics can be applied to compensate for downregulated miRNA (103).
Being unstable in the body due to numerous ribonucleases, not uptaking in the cell due to their negative charge, acting tumor microenvironment related immune cells, miRNA mimics and inhibitors have some problems with delivering in cancer cells (103). To overcome these limitations, researchers have applied several approaches to deliver therapeutic miRNA inhibitors and mimics. Using a local delivery system such as intratumoral injection of miRNA mimics or inhibitors is followed by minimal nonspecific uptake which provides effective delivery (104). Recently, some studies have been reported that local delivery of miRNAs using a polymer based delivery system, polyethylenimine and nanoparticles could achieve a significant anti-tumor effects (104–106). Mostly, local delivery systems can be applied to accessible solid tumors, thus improvement of other delivery approaches is essential to include metastatic tumors and other types of cancers. Systemic miRNA delivery systems have shown developed delivery of miRNA mimics and inhibitors (104).
In addition, using viral vectors, lentivirus, adenovirus and adeno-associated virus (AAV) have effectively delivered miRNA mimics and inhibitors. Nevertheless, the immunogenic response of viral vectors may provide serious concern in therapeutic applications (107). Therefore, improved non-viral approaches with chemical modifications is recommended for clinical applications (103). Non-viral systems are categorized into nanoparticles, lipid-based vectors (neutral/cationic), polymeric vectors (polyethylenimine, polylactic-co-glycolic acid/PLGA, chitosans, collagen and gelatin), biomaterials, and inorganic materials (gold, diamond, silica, and ferric oxide). Nanoparticles such as lipid-based systems with biocompatibility, flexibility, low immunogenicity, and versatility are the most common used approaches which constitute sphere-shaped structures composed of phospholipid bilayers in delivering exogenous nucleic acids. Furthermore, exosomes are biological nanoparticles with lipid-bilayer membrane naturally secreted from cells to mediate cell communication through intercellular transmission of nucleic acids and proteins (103, 106, 107).
miR-153 is predominantly well-known as a tumor suppressive miRNA which negatively regulates genes related to cancer development. Down-regulation of miR-153 has been associated with development and progression of cancer. Expression level of miR-153 is nearly twice or more in normal tissues compared with different tumors. It can target numerous oncogenes such as AKT, CDK1, Bcl2, Mcl1, SNAI1, Snail and multiple other genes involved in proliferation, invasion, metastasis, angiogenesis and chemo/radiotherapy resistance of several types of cancer cells. Besides being as tumor suppressor, oncogenic role of miR-153 has been confirmed by some research. It can downregulate tumor suppressive genes, including PTEN, AGO1, FOXO3a and WWOX in different types of cancer. It recommends a controversial role of miR-153 as both tumor suppressor gene and oncogene which may be dependent on type and stage of cancer cells with different gene expression profiles (16–18, 88). Nevertheless, these data should be further validated by independent studies on different types and stages of tumors. On the other hand, several LncRNAs, T-UCRs and CircRNAs are introduced as oncogenic non-coding RNAs targeting and sponging miR-153 in some suggested mechanisms. Up-regulation or down-regulation of miR-153 is associated with prognostic parameters such as lymph node involvement, distant metastasis, stage and differential grade but not associated with age, gender and size of tumor in different types of cancer (Table 4). Ultimately, it recommends miR-153 as a potential diagnostic biomarker for detecting cancer as well as providing a novel treatment strategy to combat with several types of cancer. In conclusion, further studies, cohorts and prospective trials on a larger scale should be applied to achieve this value.
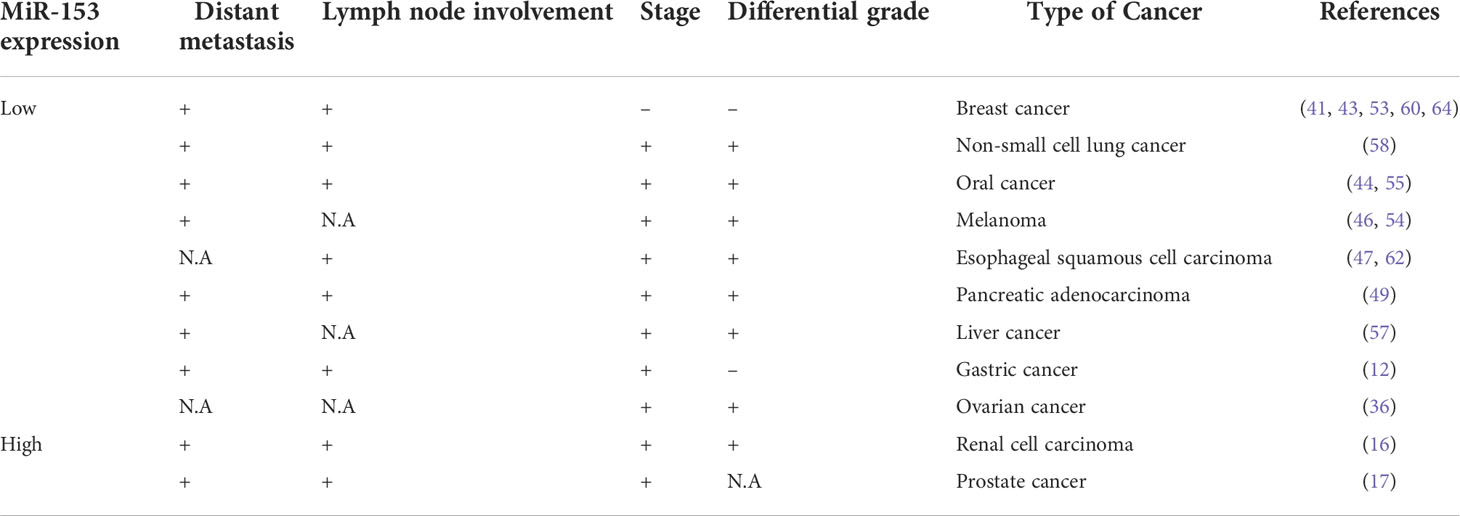
Table 4 Low or high expression level of miRNA-153 is associated with the prognostic parameters in different types of cancer.
SY, conception, providing the data and design, and manuscript writing.
I am sincere to my colleagues at University of Isfahan for their valuable discussions.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Grandér D. How do mutated oncogenes and tumor suppressor genes cause cancer? Med Oncol (1998) 15:20–6. doi: 10.1007/BF02787340
2. Lee EY, Muller WJ. Oncogenes and tumor suppressor genes. Cold Spring Harbor Perspect Biol (2010) 2:a003236. doi: 10.1101/cshperspect.a003236
3. Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med (2019) 70:3–20. doi: 10.1016/j.mam.2018.07.003
4. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci (2020) 21:1723. doi: 10.3390/ijms21051723
5. Khan S, Ayub H, Khan T, Wahid F. MicroRNA biogenesis, gene silencing mechanisms and role in breast, ovarian and prostate cancer. Biochimie (2019) 167:12–24. doi: 10.1016/j.biochi.2019.09.001
6. Palanichamy JK, Rao DS. miRNA dysregulation in cancer: towards a mechanistic understanding. Front Genet (2014) 5:54. doi: 10.3389/fgene.2014.00054
7. Chakraborty C, Sharma AR, Sharma G, Sarkar BK, Lee S-S. The novel strategies for next-generation cancer treatment: miRNA combined with chemotherapeutic agents for the treatment of cancer. Oncotarget (2018) 9:10164. doi: 10.18632/oncotarget.24309
8. Mandemakers W, Abuhatzira L, Xu H, Caromile L, Hébert S, Snellinx A, et al. Co-Regulation of intragenic microRNA miR-153 and its host gene ia-2 β: Identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia (2013) 56:1547–56. doi: 10.1007/s00125-013-2901-5
9. Chen W-J, Zhang E-N, Zhong Z-K, Jiang M-Z, Yang X-F, Zhou D-M, et al. MicroRNA-153 expression and prognosis in non-small cell lung cancer. Int J Clin Exp Pathol (2015) 8:8671.
10. Jiang J, Liu Y, Zhao Y, Tian F, Wang G. miR-153-3p suppresses inhibitor of growth protein 2 expression to function as tumor suppressor in acute lymphoblastic leukemia. Technol Cancer Res Treat (2019) 18:1533033819852990. doi: 10.1177/1533033819852990
11. Patil KS, Basak I, Pal R, Ho H-P, Alves G, Chang EJ, et al. A proteomics approach to investigate miR-153-3p and miR-205-5p targets in neuroblastoma cells. PloS One (2015) 10:e0143969. doi: 10.1371/journal.pone.0143969
12. Ouyang Y, Yuan W, Qiu S. MicroRNA−153 functions as a tumor suppressor in gastric cancer via targeting kruppel−like factor 5. Exp Ther Med (2018) 16:473–82. doi: 10.3892/etm.2018.6226
13. Ning W, Li S, Yang W, Yang B, Xin C, Ping X, et al. Blocking exosomal miRNA-153-3p derived from bone marrow mesenchymal stem cells ameliorates hypoxia-induced myocardial and microvascular damage by targeting the ANGPT1-mediated VEGF/PI3k/Akt/eNOS pathway. Cell Signalling (2021) 77:109812. doi: 10.1016/j.cellsig.2020.109812
14. Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, et al. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep (2013) 40:2789–98. doi: 10.1007/s11033-012-2278-4
15. Yang W, Shen Y, Wei J, Liu F. MicroRNA-153/Nrf-2/GPx1 pathway regulates radiosensitivity and stemness of glioma stem cells via reactive oxygen species. Oncotarget (2015) 6:22006. doi: 10.18632/oncotarget.4292
16. Li Z, Zhao S, Zhu S, Fan Y. MicroRNA-153-5p promotes the proliferation and metastasis of renal cell carcinoma via direct targeting of AGO1. Cell Death Dis (2021) 12:1–13. doi: 10.1038/s41419-020-03306-y
17. Bi C-w, Zhang G-y, Bai Y, Zhao B, Yang H. Increased expression of miR-153 predicts poor prognosis for patients with prostate cancer. Medicine (2019) 98(36):e16705. doi: 10.1097/MD.0000000000016705
18. Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res (2013) 73:6435–47. doi: 10.1158/0008-5472.CAN-12-3308
19. Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: A signature for cancer progression. Biomed Pharmacother (2021) 138:111528. doi: 10.1016/j.biopha.2021.111528
20. Mathew RS, Tatarakis A, Rudenko A, Johnson-Venkatesh EM, Yang YJ, Murphy EA, et al. A microRNA negative feedback loop downregulates vesicle transport and inhibits fear memory. Elife (2016) 5:e22467. doi: 10.7554/eLife.22467
21. Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, et al. Co-Condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol Cell (2021) 81:1682–97. e7. doi: 10.1016/j.molcel.2021.01.031
22. Shao H, Dong D, Shao F. Long non-coding RNA TUG1-mediated down-regulation of KLF4 contributes to metastasis and the epithelial-to-mesenchymal transition of colorectal cancer by miR-153-1. Cancer Manage Res (2019) 11:8699. doi: 10.2147/CMAR.S208508
23. Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang Y, et al. Suppression of AKT expression by mi r-153 produced anti-tumor activity in lung cancer. Int J Cancer (2015) 136:1333–40. doi: 10.1002/ijc.29103
24. Li W, Wang M, Meng B, Yu J, Chen Q, Li H, et al. MicroRNA-153 regulated AKT1 expression and suppressed cell proliferation of epithelial ovarian cancer cells. Int J Clin Exp Pathol (2017) 10:7417.
25. Cui Y, Zhao J, Yi L, Jiang Y. microRNA-153 targets mTORC2 component rictor to inhibit glioma cells. PloS One (2016) 11:e0156915. doi: 10.1371/journal.pone.0156915
26. Wang Y, Wu N, Zhang J, Wang H, Men X. MiR-153-5p enhances the sensitivity of triple-negative breast cancer cells to paclitaxel by inducing G2M phase arrest. OncoTargets Ther (2020) 13:4089. doi: 10.2147/OTT.S241640
27. Xu J, Liao X, Lu N, Liu W, Wong CW. Chromatin-modifying drugs induce miRNA-153 expression to suppress irs-2 in glioblastoma cell lines. Int J Cancer (2011) 129:2527–31. doi: 10.1002/ijc.25917
28. Ghafouri-Fard S, Farsani ZS, Taheri M. The role of micrornas in the pathogenesis of thyroid cancer. Non-Coding RNA Res (2020) 5(2020):88–98. doi: 10.1016/j.yexmp.2019.104332
29. Cui Z, Luo Z, Lin Z, Shi L, Hong Y, Yan C. Long non-coding RNA TTN-AS1 facilitates tumorigenesis of papillary thyroid cancer through modulating the miR-153-3p/ZNRF2 axis. J Gene Med (2019) 21:e3083. doi: 10.1002/jgm.3083
30. Liu R, Chen H, Zhao P, Chen C-H, Liang H, Yang C, et al. Mifepristone derivative FZU-00,003 suppresses triple-negative breast cancer cell growth partially via miR-153-KLF5 axis. Int J Biol Sci (2020) 16:611. doi: 10.7150/ijbs.39491
31. Deng X, Guo B, Fan Y. MiR-153-3p suppresses cell proliferation, invasion and glycolysis of thyroid cancer through inhibiting E3F3 expression. OncoTargets Ther (2021) 14:519. doi: 10.2147/OTT.S267887
32. Niu G, Li B, Sun L, An C. MicroRNA-153 inhibits osteosarcoma cells proliferation and invasion by targeting TGF-β2. PloS One (2015) 10:e0119225. doi: 10.1371/journal.pone.0119225
33. Zhi X, Jiang K, Ma Y, Zhou L. OIP5-AS1 promotes the progression of gastric cancer cells via the miR-153-3p/ZBTB2 axis. Eur Rev Med Pharmacol Sci (2020) 24:2428–41. doi: 10.26355/eurrev_202003_20510
34. Xu J, Liao X, Wong C. Downregulations of b-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer (2010) 126:1029–35. doi: 10.1002/ijc.24823
35. Li Y-L, Tang J-M, Chen X-Y, Luo B, Liang G-H, Qu Q, et al. MicroRNA-153-3p enhances the sensitivity of chronic myeloid leukemia cells to imatinib by inhibiting b-cell lymphoma-2-mediated autophagy. Hum Cell (2020) 33:610–8. doi: 10.1007/s13577-020-00367-1
36. Li C, Zhang Y, Zhao W, Cui S, Song Y. miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J Cell Biochem (2019) 120:19147–58. doi: 10.1002/jcb.29244
37. Guo G, Zhang Y, Hu L, Bian X. MicroRNA−153 affects nasopharyngeal cancer cell viability by targeting TGF−β2. Oncol Lett (2019) 17:646–51. doi: 10.3892/ol.2018.9570
38. Wu X, Li L, Li Y, Liu Z. MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res (2016) 6:1563.
39. Shang J, Chen W-M, Wang Z-H, Wei T-N, Chen Z-Z, Wu W-B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p–XIAP axis. Exp Hematol (2019) 70:42–54. e3. doi: 10.1016/j.exphem.2018.10.011
40. Li Y, Lin S, Xie X, Zhu H, Fan T, Wang S. Highly enriched exosomal lncRNA OIP5-AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR-153 and ATG5. Am J Trans Res (2021) 13:4211.
41. Wang J, Liang S, Duan X. Molecular mechanism of miR-153 inhibiting migration, invasion and epithelial-mesenchymal transition of breast cancer by regulating transforming growth factor beta (TGF-β) signaling pathway. J Cell Biochem (2019) 120:9539–46. doi: 10.1002/jcb.28230
42. Zhao G, Zhang Y, Zhao Z, Cai H, Zhao X, Yang T, et al. MiR-153 reduces stem cell-like phenotype and tumor growth of lung adenocarcinoma by targeting Jagged1. Stem Cell Res Ther (2020) 11:1–12. doi: 10.1186/s13287-020-01679-7
43. Shi D, Li Y, Fan L, Zhao Q, Tan B, Cui G. Upregulation of miR-153 inhibits triple-negative breast cancer progression by targeting ZEB2-mediated EMT and contributes to better prognosis. OncoTargets Ther (2019) 12:9611. doi: 10.2147/OTT.S223598
44. Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis (2013) 34:539–49. doi: 10.1093/carcin/bgs374
45. Zhou J, Xie M, Shi Y, Luo B, Gong G, Li J, et al. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol Rep (2015) 34:111–20. doi: 10.3892/or.2015.3952
46. Zeng HF, Yan S, Wu SF. MicroRNA-153-3p suppress cell proliferation and invasion by targeting SNAI1 in melanoma. Biochem Biophys Res Commun (2017) 487:140–5. doi: 10.1016/j.bbrc.2017.04.032
47. Zuo J, Wang D, Shen H, Liu F, Han J, Zhang X. MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumor Biol (2016) 37:16135–40. doi: 10.1007/s13277-016-5427-x
48. Wen JF, Jiang YQ, Li C, Dai XK, Wu T, Yin WZ. LncRNA-XIST promotes the oxidative stress-induced migration, invasion, and epithelial-to-mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biol Int (2020) 44:1991–2001. doi: 10.1002/cbin.11405
49. Bai Z, Sun J, Wang X, Wang H, Pei H, Zhang Z. MicroRNA-153 is a prognostic marker and inhibits cell migration and invasion by targeting SNAI1 in human pancreatic ductal adenocarcinoma. Oncol Rep (2015) 34:595–602. doi: 10.3892/or.2015.4051
50. Zhang B, Fu T, Zhang L. MicroRNA−153 suppresses human laryngeal squamous cell carcinoma migration and invasion by targeting the SNAI1 gene. Oncol Lett (2018) 16:5075–83. doi: 10.3892/ol.2018.9302
51. Chen J, Huang X, Wang W, Xie H, Li J, Hu Z, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY) (2018) 10:3371. doi: 10.18632/aging.101645
52. Qi W, Mao X, He Y, Xiao C. Long non-coding RNA LINC00858 promotes cells proliferation and invasion through the miR-153-3p/Rabl3 axis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci (2020) 24:9343–52. doi: 10.26355/eurrev_202009_23017
53. Sun L, Wang H, Jiang J, Bi X. miR-153-3p inhibits proliferation and migration of breast cancer cells via down-regulating ROCK1. Xi bao yu fen zi mian yi xue za zhi= Chin J Cell Mol Immunol (2020) 36:138–44.
54. Luan W, Shi Y, Zhou Z, Xia Y, Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun (2018) 502:22–9. doi: 10.1016/j.bbrc.2018.05.114
55. Chang A-C, Lien M-Y, Tsai M-H, Hua C-H, Tang C-H. WISP-1 promotes epithelial-mesenchymal transition in oral squamous cell carcinoma cells via the miR-153-3p/Snail axis. Cancers (2019) 11:1903. doi: 10.3390/cancers11121903
56. Xia W, Ma X, Li X, Dong H, Yi J, Zeng W, et al. miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting snail. Oncol Rep (2015) 34:655–62. doi: 10.3892/or.2015.4008
57. Yang G, Li X, Liu J, Huang S, Weng Y, Zhu J, et al. Hsa_circ_0008537 facilitates liver carcinogenesis by upregulating MCL1 and Snail1 expression via miR−153−3p. Oncol Rep (2021) 45:1072–82. doi: 10.3892/or.2021.7941
58. Zhang W, Liu K, Pei Y, Tan J, Ma J, Zhao J. Long noncoding RNA HIF1A-AS2 promotes non-small cell lung cancer progression by the miR-153-5p/S100A14 axis. OncoTargets Ther (2020) 13:8715. doi: 10.2147/OTT.S262293
59. Ge C, Dong J, Chu Y, Cao S, Zhang J, Wei J. LncRNA FGD5-AS1 promotes tumor growth by regulating MCL1 via sponging miR-153-3p in oral cancer. Aging (Albany NY) (2020) 12:14355. doi: 10.18632/aging.103476
60. Zuo Z, Ye F, Liu Z, Huang J, Gong Y. MicroRNA−153 inhibits cell proliferation, migration, invasion and epithelial−mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp Ther Med (2019) 17:4693–702. doi: 10.3892/etm.2019.7470
61. Ma H, Tian T, Liu X, Xia M, Chen C, Mai L, et al. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed Pharmacother (2019) 118:109311. doi: 10.1016/j.biopha.2019.109311
62. Liu D, Gao M, Wu K, Zhu D, Yang Y, Zhao S. LINC00152 facilitates tumorigenesis in esophageal squamous cell carcinoma via miR-153-3p/FYN axis. Biomed Pharmacother (2019) 112:108654. doi: 10.1016/j.biopha.2019.108654
63. Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun (2015) 456:385–91. doi: 10.1016/j.bbrc.2014.11.093
64. Li W, Zhai L, Zhao C, Lv S. miR-153 inhibits epithelial–mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat (2015) 150:501–9. doi: 10.1007/s10549-015-3346-y
65. Shi F, Shi Z, Zhao Y, Tian J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun (2019) 510:614–20. doi: 10.1016/j.bbrc.2019.02.019
66. Zhang W, Mao S, Shi D, Zhang J, Zhang Z, Guo Y, et al. MicroRNA-153 decreases tryptophan catabolism and inhibits angiogenesis in bladder cancer by targeting indoleamine 2, 3-dioxygenase 1. Front Oncol (2019) 9:619. doi: 10.3389/fonc.2019.00619
67. Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, et al. miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis (2018) 21:849–60. doi: 10.1007/s10456-018-9630-9
68. Ma Y, Xue Y, Liu X, Qu C, Cai H, Wang P, et al. SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol Rep (2017) 38:3265–77. doi: 10.3892/or.2017.5985
69. Song L, Duan P, Guo P, Li D, Li S, Xu Y, et al. Downregulation of miR-223 and miR-153 mediates mechanical stretch-stimulated proliferation of venous smooth muscle cells via activation of the insulin-like growth factor-1 receptor. Arch Biochem biophys (2012) 528:204–11. doi: 10.1016/j.abb.2012.08.015
70. Liu F, Liu B, Qian J, Wu G, Li J, Ma Z. miR-153 enhances the therapeutic effect of gemcitabine by targeting snail in pancreatic cancer. Acta Biochim Biophys Sin (2017) 49:520–9. doi: 10.1093/abbs/gmx039
71. Zhao Z, Shen X, Zhang D, Xiao H, Kong H, Yang B, et al. miR-153 enhances the therapeutic effect of radiotherapy by targeting JAG1 in pancreatic cancer cells. Oncol Lett (2021) 21:1–8. doi: 10.3892/ol.2020.12303
72. Wang L, Lv X, Fu X, Su L, Yang T, Xu P. MiR-153 inhibits the resistance of lung cancer to gefitinib via modulating expression of ABCE1. Cancer Biomarkers (2019) 25:361–9. doi: 10.3233/CBM-190094
73. Gao Y, Xie M, Guo Y, Yang Q, Hu S, Li Z. Long non-coding RNA FGD5-AS1 regulates cancer cell proliferation and chemoresistance in gastric cancer through miR-153-3p/CITED2 axis. Front Genet (2020) 11. doi: 10.3389/fgene.2020.00715
74. Zuo J, Zhao M, Fan Z, Liu B, Wang Y, Li Y, et al. MicroRNA-153-3p regulates cell proliferation and cisplatin resistance via nrf-2 in esophageal squamous cell carcinoma. Thorac Cancer (2020) 11:738–47. doi: 10.1111/1759-7714.13326
75. Liu L, Lai X, Yuan C, Lv X, Yu T, He W, et al. Aberrant expression of miR−153 is associated with the poor prognosis of cervical cancer. Oncol Lett (2018) 15:9183–7. doi: 10.3892/ol.2018.8475
76. Liu L, Chen R, Huang S, Wu Y, Li G, Zhang B, et al. miR-153 sensitized the K562 cells to As2O3-induced apoptosis. Med Oncol (2012) 29:243–7. doi: 10.1007/s12032-010-9807-6
77. Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Quoc TP, et al. Uc. 416+ a promotes epithelial-to-mesenchymal transition through miR-153 in renal cell carcinoma. BMC Cancer (2018) 18:1–10. doi: 10.1186/s12885-018-4863-y
78. Zhao L, Bi M, Zhang H, Shi M. Downregulation of NEAT1 suppresses cell proliferation, migration, and invasion in NSCLC Via sponging miR-153-3p. Cancer Biother Radiopharmaceuticals (2020) 35:362–70. doi: 10.1089/cbr.2019.3119
79. Liang H, Xiao J, Zhou Z, Wu J, Ge F, Li Z, et al. Hypoxia induces miR-153 through the IRE1α-XBP1 pathway to fine tune the HIF1α/VEGFA axis in breast cancer angiogenesis. Oncogene (2018) 37:1961–75. doi: 10.1038/s41388-017-0089-8
80. Sun S, Xia C, Xu Y. HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed Pharmacother (2020) 125:110014. doi: 10.1016/j.biopha.2020.110014
81. Wang Y, Wang J, Hao H, Luo X. lncRNA KCNQ1OT1 promotes the proliferation, migration and invasion of retinoblastoma cells by upregulating HIF-1α via sponging miR-153-3p. J Invest Med (2020) 68:1349–56. doi: 10.1136/jim-2020-001431
82. Yousefnia S, Seyed Forootan F, Seyed Forootan S, Nasr Esfahani MH, Gure AO, Ghaedi K. Mechanistic pathways of malignancy in breast cancer stem cells. Front Oncol (2020) 10:452. doi: 10.3389/fonc.2020.00452
83. Sun D, Mu Y, Piao H. MicroRNA-153-3p enhances cell radiosensitivity by targeting BCL2 in human glioma. Biol Res (2018) 51:1–10. doi: 10.1186/s40659-018-0203-6
84. He Y, Zhang L, Tan F, Wang L-F, Liu D-H, Wang R-J, et al. MiR-153-5p promotes sensibility of colorectal cancer cells to oxaliplatin via targeting bcl-2-mediated autophagy pathway. Biosci biotechnol Biochem (2020) 84:1645–51. doi: 10.1080/09168451.2020.1760784
85. Yousefnia S. Mechanistic effects of arsenic trioxide on acute promyelocytic leukemia and other types of leukemias. Cell Biol Int (2021) 45(6):1148–57. doi: 10.1002/cbin.11563
86. Chen Y, Feng F, Gao X, Wang C, Sun H, Zhang C, et al. MiRNA153 reduces effects of chemotherapeutic agents or small molecular kinase inhibitor in HCC cells. Curr Cancer Drug Targets (2015) 15:176–87. doi: 10.2174/1568009615666150225122635
87. Wu Z, He B, He J, Mao X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate (2013) 73:596–604. doi: 10.1002/pros.22600
88. Hua H-W, Jiang F, Huang Q, Liao Z, Ding G. MicroRNA-153 promotes wnt/β-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget (2015) 6:3840. doi: 10.18632/oncotarget.2927
89. Anaya-Ruiz M, Cebada J, Delgado-López G, Sanchez-Vazquez ML, Perez-Santos JLM. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pacific J Cancer Prev (2013) 14:2983–6. doi: 10.7314/APJCP.2013.14.5.2983
90. Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, et al. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology (2010) 57:734–43. doi: 10.1111/j.1365-2559.2010.03686.x
91. Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer (2016) 16:1–8. doi: 10.1186/s12885-016-2135-2
92. Xiang Y, Tian Q, Guan L, Niu S-s. The dual role of miR-186 in cancers: oncomir battling with tumor suppressor miRNA. Front Oncol (2020) 10:233. doi: 10.3389/fonc.2020.00233
93. Ding M, Lin B, Li T, Liu Y, Li Y, Zhou X, et al. A dual yet opposite growth-regulating function of miR-204 and its target XRN1 in prostate adenocarcinoma cells and neuroendocrine-like prostate cancer cells. Oncotarget (2015) 6:7686. doi: 10.18632/oncotarget.3480
94. Myatt SS, Wang J, Monteiro LJ, Christian M, Ho K-K, Fusi L, et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res (2010) 70:367–77. doi: 10.1158/0008-5472.CAN-09-1891
95. Lee H-L, Cheng H-L, Liu Y-F, Chou M-C, Yang S-F, Chou Y-E. Functional genetic variant of WW domain-containing oxidoreductase (WWOX) gene is associated with hepatocellular carcinoma risk. PloS One (2017) 12:e0176141. doi: 10.1371/journal.pone.0176141
96. Zhou Q, Luo L, Wang X, Li X. Relationship between single nucleotide polymorphisms in the 3′ UTR of amyloid precursor protein and risk of alzheimer’s disease and its mechanism. Biosci Rep (2019) 39(5):BSR20182485. doi: 10.1042/BSR20182485
97. Liu X, Li Y-G, Tan F, Liu J, Yi R, Zhao X. Three functional polymorphisms in CCDC170 were associated with osteoporosis phenotype. Biol Open (2021) 10:bio050930. doi: 10.1242/bio.050930
98. Yu L, Xu Q, Yu W, Duan J, Dai G. LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153–3p/KLF5 positive feedback loop. Biochem Biophys Res Commun (2018) 506:819–25. doi: 10.1016/j.bbrc.2018.10.131
99. Li G, Xue M, Yang F, Jin Y, Fan Y, Li W. CircRBMS3 promotes gastric cancer tumorigenesis by regulating miR-153–SNAI1 axis. J Cell Physiol (2019) 234:3020–8. doi: 10.1002/jcp.27122
100. Zhou B, Zheng P, Li Z, Li H, Wang X, Shi Z, et al. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle (2018) 17:2644–54. doi: 10.1080/15384101.2018.1553354
101. Tezcan G, Aksoy S, Tunca B, Bekar A, Mutlu M, Cecener G, et al. Oleuropein modulates glioblastoma miRNA pattern different from olea europaea leaf extract. Hum Exp Toxicol (2019) 38:1102–10. doi: 10.1177/0960327119855123
102. Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics (2016) 6:533. doi: 10.7150/thno.14315
103. Forterre A, Komuro H, Aminova S, Harada M. A comprehensive review of cancer MicroRNA therapeutic delivery strategies. Cancers (2020) 12:1852. doi: 10.3390/cancers12071852
104. Fujita Y, Kuwano K, Ochiya T. Development of small RNA delivery systems for lung cancer therapy. Int J Mol Sci (2015) 16:5254–70. doi: 10.3390/ijms16035254
105. Manikkath J, Jishnu PV, Wich PR, Manikkath A, Radhakrishnan R. Nanoparticulate strategies for the delivery of miRNA mimics and inhibitors in anticancer therapy and its potential utility in oral submucous fibrosis. Nanomedicine (2022) 17:181–95. doi: 10.2217/nnm-2021-0381
106. Ban E, Kwon T-H, Kim A. Delivery of therapeutic miRNA using polymer-based formulation. Drug Deliv Trans Res (2019) 9:1043–56. doi: 10.1007/s13346-019-00645-y
107. Yang N. An overview of viral and nonviral delivery systems for microRNA. Int J Pharm Invest (2015) 5:179. doi: 10.4103/2230-973X.167646

Keywords: miR-153, tumorigenicity, non-coding RNAs, metastasis, chemo/radiotherapy resistance
Citation: Yousefnia S (2022) A comprehensive review on miR-153: Mechanistic and controversial roles of miR-153 in tumorigenicity of cancer cells. Front. Oncol. 12:985897. doi: 10.3389/fonc.2022.985897
Received: 04 July 2022; Accepted: 22 August 2022;
Published: 09 September 2022.
Edited by:
Zhi-qian Zhang, Southern University of Science and Technology, ChinaReviewed by:
Arun Renganathan, Washington University in St. Louis, United StatesCopyright © 2022 Yousefnia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saghar Yousefnia, c2FnaGFyX3lvdXNlZm5pYUB5YWhvby5jb20=; cy55b3VzZWZuaWFAc2NpLnVpLmFjLmly
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.