- 1Oncogenic Signalling Laboratory, Telethon Kids Institute, Nedlands, WA, Australia
- 2School of Biomedical Sciences, University of Western Australia, Crawley, WA, Australia
- 3Division of Paediatrics/Centre for Child Health Research, University of Western Australia, Crawley, WA, Australia
The epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases (RTKs) consists of EGFR, ErbB2, ErbB3, and ErbB4. These receptors play key roles in cell proliferation, angiogenesis, cell migration, and in some cases, tumor promotion. ErbB4 is a unique member of the EGFR family, implicated not only in pro-tumorigenic mechanisms, such as cell proliferation and migration, but also in anti-tumorigenic activities, including cell differentiation and apoptosis. ErbB4 is differentially expressed in a wide variety of tissues, and interestingly, as different isoforms that result in vastly different signalling outcomes. Most studies have either ignored the presence of these isoforms or used overexpression models that may mask the true function of ErbB4. ErbB4 is widely expressed throughout the body with significant expression in skeletal tissue, mammary glands, heart, and brain. Knockout models have demonstrated embryonic lethality due to disrupted heart and brain development. Despite high expression in the brain and a critical role in brain development, remarkably little is known about the potential signalling activity of ErbB4 in brain cancer.This review focuses on the unique biology of ErbB4 in the brain, and in particular, highlights brain cancer research findings. We end the review with a focus on high grade gliomas, primarily glioblastoma, a disease that has been shown to involve EGFR and its mutant forms. The role of the different ErbB4 isotypes in high grade gliomas is still unclear and future research will hopefully shed some light on this question.
Overview of ErbB4 biology
The epidermal growth factor receptor (EGFR) family of receptor tyrosine kinases (RTKs), also known as the ErbB family of receptors, comprises four members: EGFR/ErbB1/HER1; ErbB2/Neu/HER2; ErbB3/HER3; and ErbB4/HER4 (1). These receptors play key roles in cellular proliferation, differentiation, migration, angiogenesis, and apoptosis (1). Therefore, unsurprisingly, aberrant signalling through these receptors has been implicated in the pathogenesis of numerous tumor types.
The ErbB receptors share similar domain organization, including an extracellular ligand-binding domain, a single membrane-spanning region, an intracellular tyrosine kinase domain, and a C-terminal region containing various tyrosine residues (Figure 1) (2). Following ligand engagement of these receptors at the cell surface, hetero- or homodimerization occurs, leading to trans-autophosphorylation of tyrosine residues within their cytoplasmic tails. This creates binding or docking sites for adaptor and signalling proteins containing SH2 and PTB domains, such as Grb2, Shc, and PI3-Kinase (PI3-K) (3, 4).
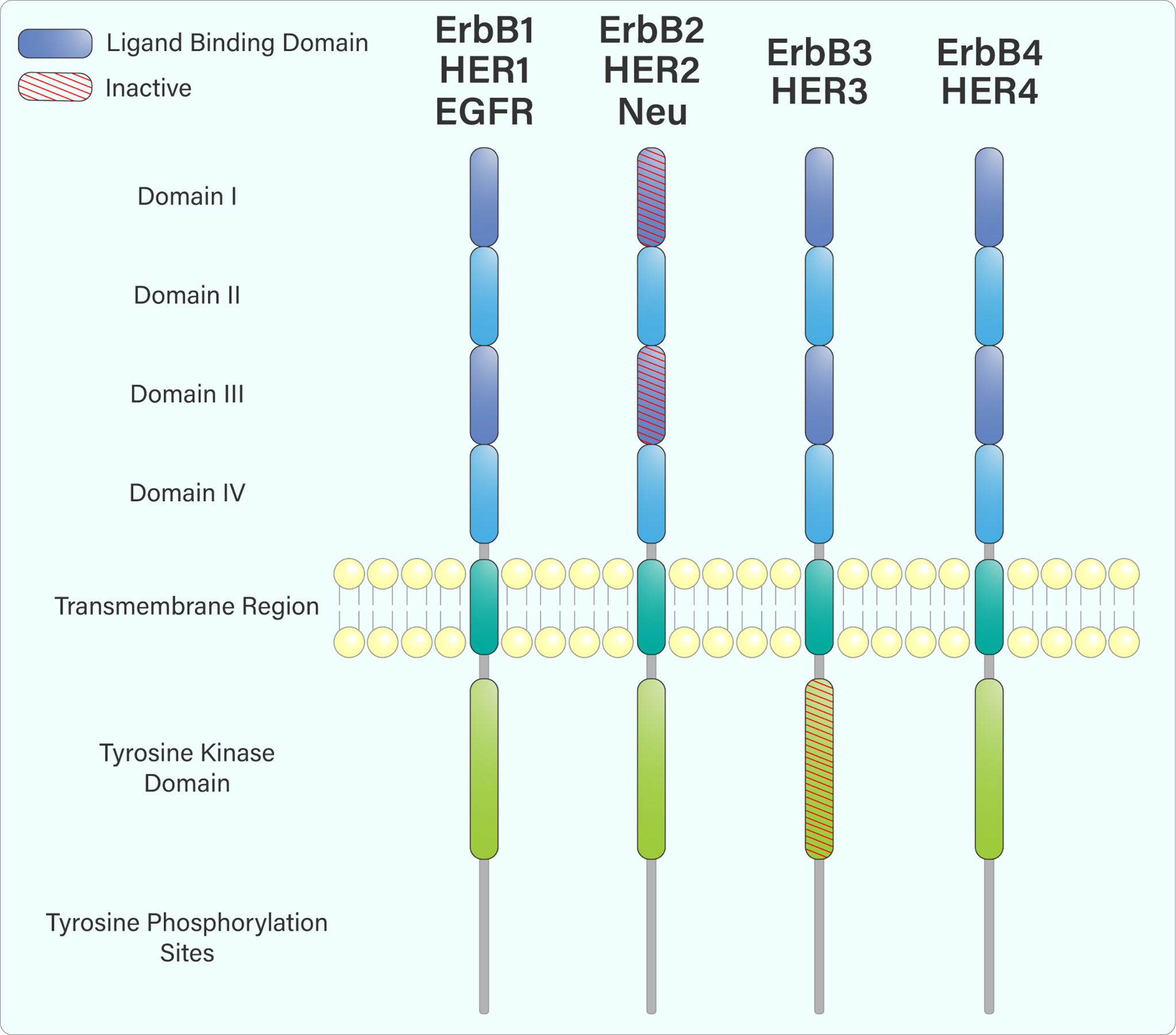
Figure 1 The domain arrangement of the ErbB receptor family. The extracellular region is composed of 4 domains: I, II, III & IV. Domains I and III (purple) form an extracellular ligand-binding domain that facilitates ligand attachment at the cell surface. ErbB2, while containing a ligand-binding domain, does not bind any ligands. Upon ligand engagement, the ErbB family members hetero- or homodimerize with each other, leading to trans-autophosphorylation of tyrosine residues within their cytoplasmic tails through their intracellular tyrosine kinase domain. ErbB3 does not have an active tyrosine kinase domain and must heterodimerize with another ErbB receptor to be phosphorylated.
Several ligands are capable of binding to and activating the ErbB family. Each ErbB-binding ligand contains an EGF-like domain, which helps to confer binding specificity, and allows the ligand to preferentially or specifically bind to a particular ErbB family member (1). Whilst ErbB1, 3, and 4 bind several ligands, ErbB2 does not appear to bind any known ligands (Figure 2) (5). However, ErbB2 dimerizes with other ErbB members, which helps to stabilize ligand interactions. Furthermore, it appears that ErbB2 is the preferred dimerization partner for other ErbB family members (5).
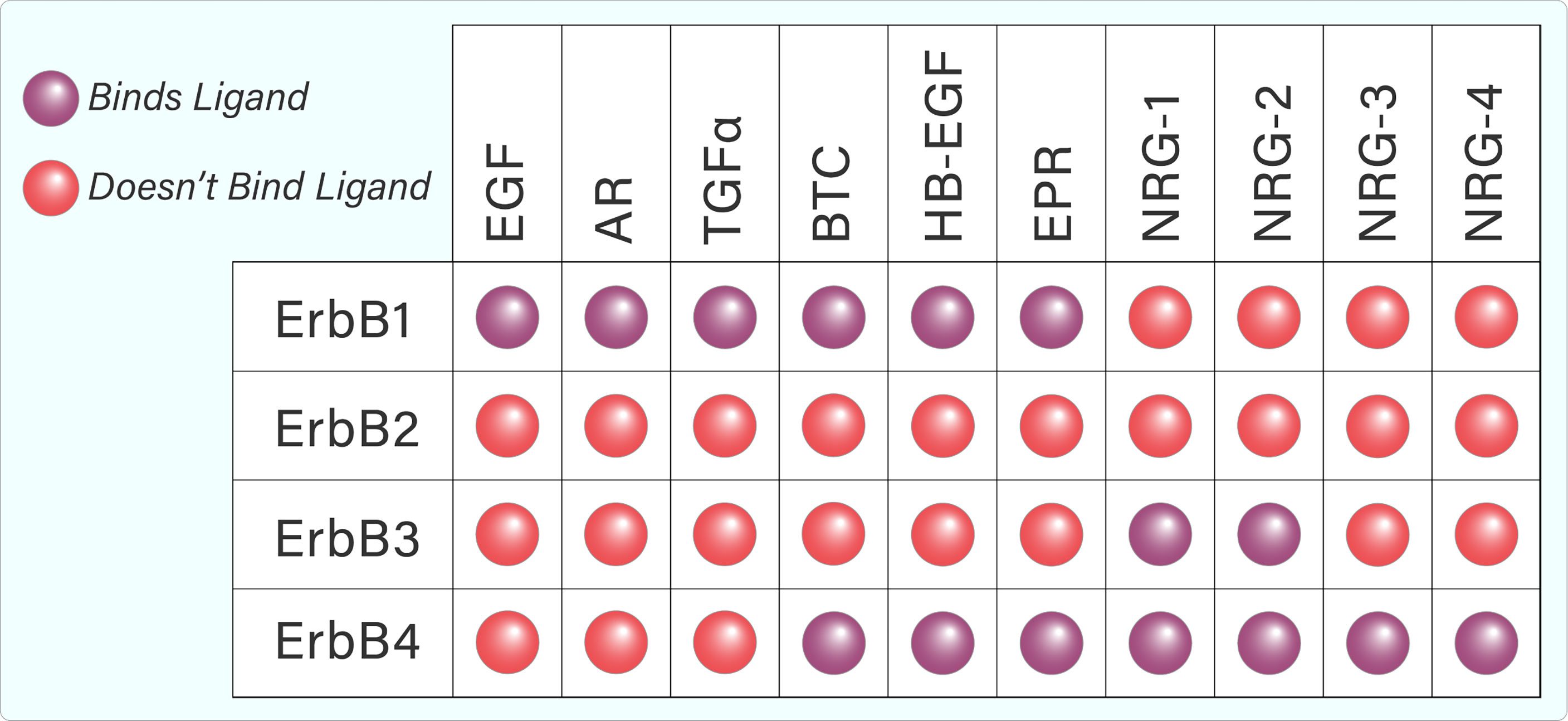
Figure 2 Tabular representation of ErbB ligand binding. ErbB1, 3, and 4 are capable of binding several ligands while ErbB2 does not appear to bind any known ligands.
ErbB4 was cloned in 1993 and shows similar domain organization to the other, previously identified, members of the ErbB family (6, 7). Specifically, the ecto- and cytoplasmic domains are separated by a single transmembrane domain (8). The extracellular domain contains two cysteine-rich regions and displays high levels of similarity to the extracellular domain of ErbB3 (7, 8). The cytoplasmic domain is more homologous to the cytoplasmic domains of EGFR and ErbB2 and is composed of a juxtamembrane (JM) region, a tyrosine kinase domain, and a carboxy terminal tail (7, 8).
ErbB4 domain structure
ErbB4 structure and signalling is complicated by the existence of different splice variants (9). Specifically, exon splicing occurs within the ERBB4 JM domain to produce the splice variants JM-a, JM-b, JM-c, and JM-d (Figure 3) (9–11). A cleavage site for the tumor necrosis factor-alpha converting enzyme (TACE) is encoded within exon 16 of ERBB4 (9). The JM-a and JM-d variants both contain exon 16, and therefore, contain the cleavage site (9). In contrast, the ErbB4 JM-b and JM-c variants do not contain exon 16 (9). Additional splicing results in alterations to the cytoplasmic domain (towards the C-terminal tail), producing the CYT-1 and CYT-2 variants by the inclusion or exclusion of exon 26 (9). This results in the respective presence or absence of a 16-amino acid sequence (Ser1046 through Gly1061) (11–13).
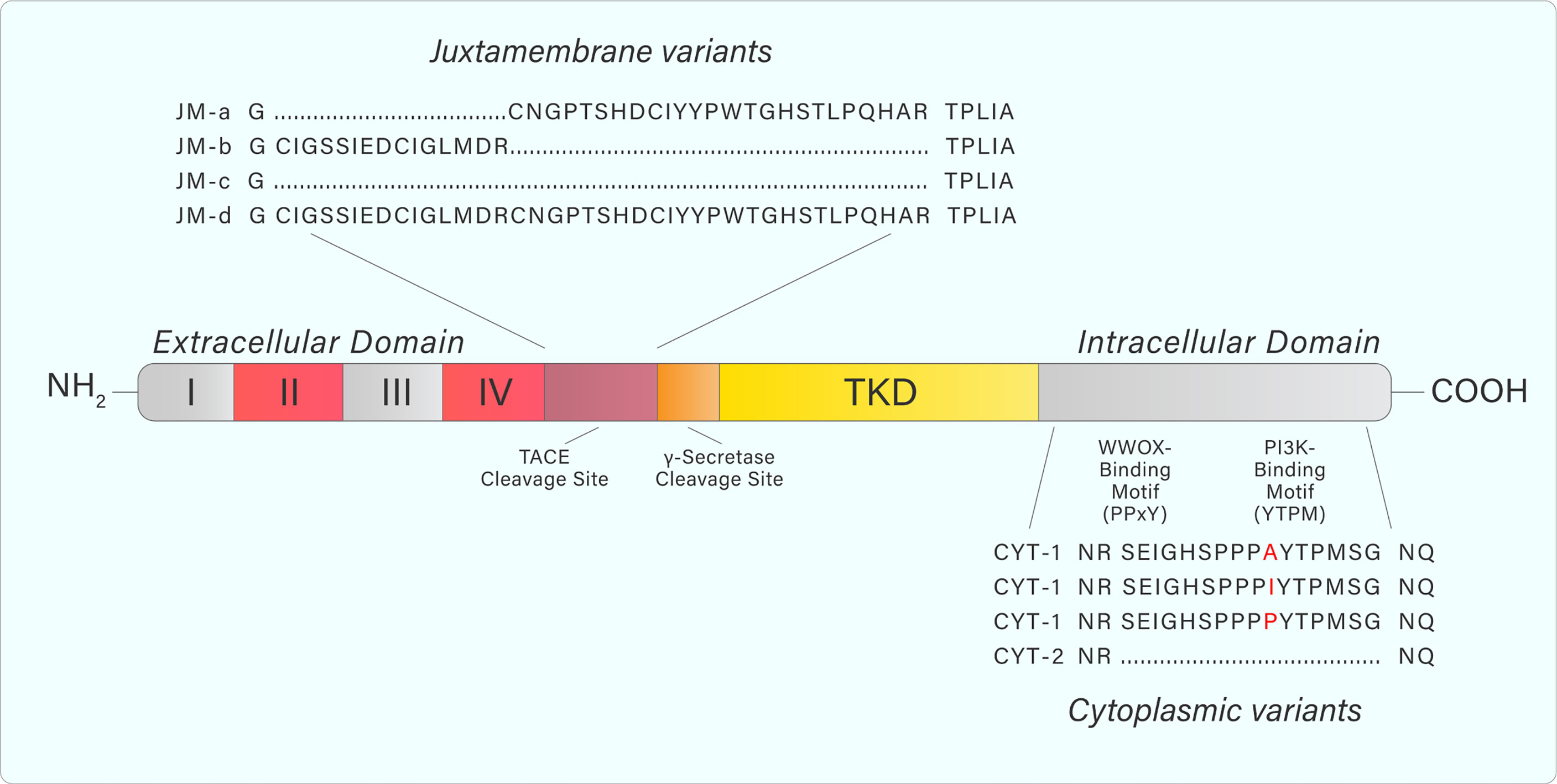
Figure 3 Adapted from Donoghue et al. Graphic representation of ERBB4 juxtamembrane (JM-a, b, c, d) and cytoplasmic (CYT-1, 2) variant sequences. JM-a- and JM-d variants contain tumor necrosis factor converting enzyme (TACE) and γ-secretase cleavage sites causing release of the extracellular domain and subsequent release of a soluble intracellular fragment. Variants containing JM-b and JM-c do not undergo cleavage and remain as a full-length receptor. CYT-1-containing variants include a 16-amino acid sequence that contains WW domain-containing oxidoreductase (WWOX)- and PI3K-binding motifs. These binding motifs are absent from CYT-2-containing variants. Amino acid sequences were generated from the NCBI Nucleotide database.
The different ErbB4 isoforms allow for various signalling outcomes. Subsequent to ligand engagement, cleavage of protease-sensitive ErbB4 isoforms (JM-a, JM-d) within the JM domain (between His-651 and Ser-652) by TACE releases the extracellular domain and leaves a membrane-bound intracellular domain (ICD), known as m80. This triggers a secondary cleavage event by γ-secretase, releasing an intracellular domain fragment (s80), which can then translocate to the nucleus and interact with transcriptional regulators (14, 15). Moreover, ErbB4 contains a Nuclear Localization Sequence, providing further evidence for its role as a transcriptional regulator, which is unique within the ErbB receptor family (16, 17). The 16 additional amino acids present within the CYT-1 isoform contain docking sites for SH2 domains and WW-domain-containing proteins (15). This allows, for example, CYT-1 to signal through the PI3-Kinase pathway, whereas CYT-2 lacks this ability (Figure 4) (12).
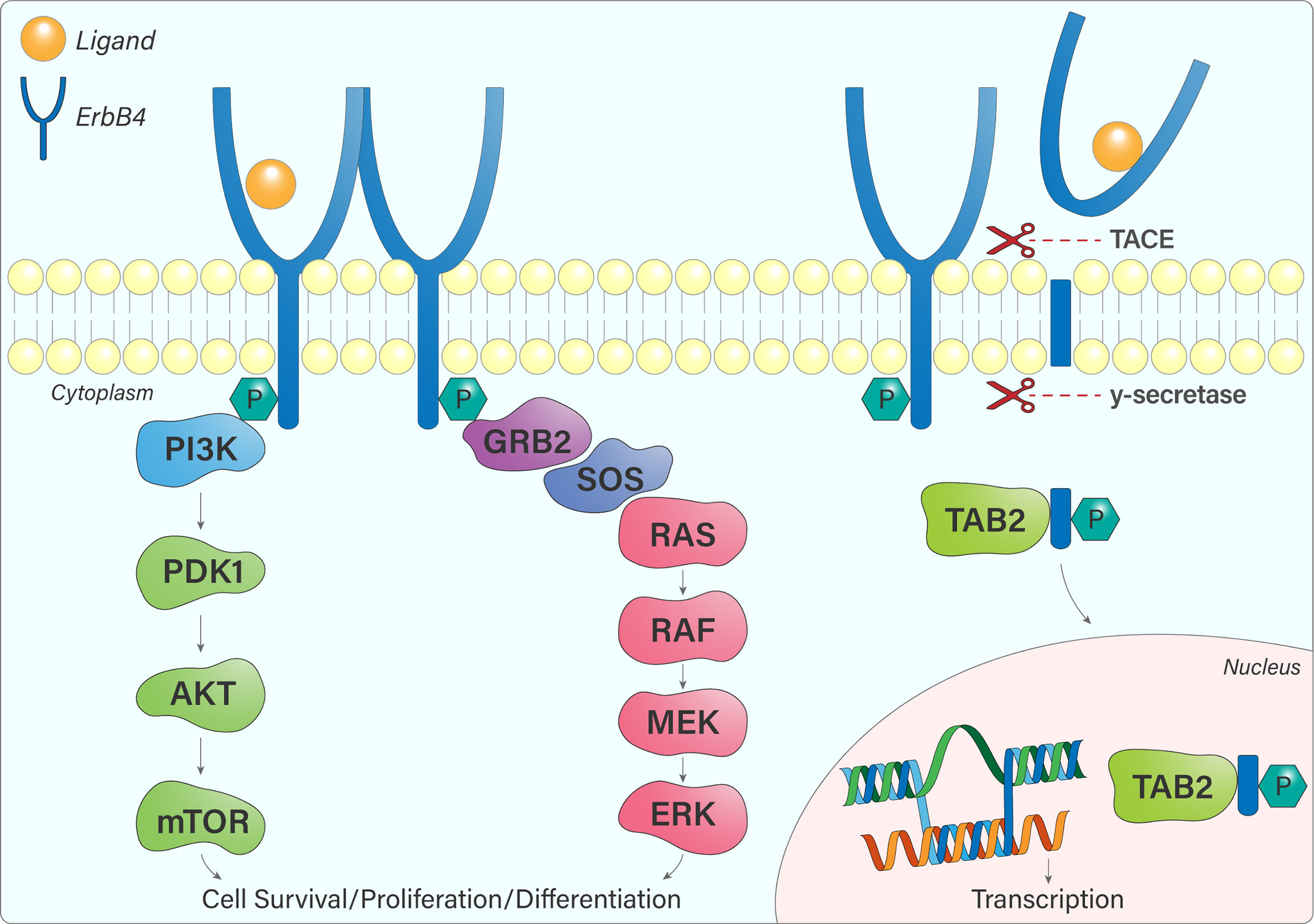
Figure 4 ErbB4 signalling pathways. Once ErbB4 is phosphorylated, signalling cascades become activated, notably the PI3K-AKT and Ras-MAPK pathways that influence cell survival, proliferation, and differentiation. JM-a or JM-d-containing ErbB4 variants are cleaved by TACE and γ-secretase, resulting in the release of a soluble intracellular fragment that forms a complex with TAB2 and undergoes nuclear translocation. In the nucleus, ErbB4 regulates gene transcription.
Ligand binding
The ErbB family is activated by ligands that share an EGF-like motif of 45–55 amino acids (18). Binding specificity is conferred by loops that form within this motif due to covalent interactions between six cysteine residues (18). The ligands can be separated into three groups – those that specifically bind EGFR, those that bind ErbB4 and EGFR (epiregulin, betacellulin, heparin-binding EGF) and the neuregulins (NRG), which specifically bind ErbB4 and ErbB3 (Figure 2) (19). Neuregulin (denoted as Heregulin in humans) belongs to a family of EGF-like polypeptides that are widely expressed. They are encoded by at least six genes (NRG-1–6), each of which have multiple splice forms (19). NRG-1 was the first member of the family to be discovered and is the best-characterized (19, 20). The neuregulins can again be divided into two groups, depending on their ability to bind ErbB3 and ErbB4 (NRG-1, NRG-2), or ErbB4 only (NRG-3, NRG-4). Early studies found that NRG-1 is a preferred ligand for ErbB4 (7, 21). Binding analyses showed that ErbB4/ErbB2 heterodimers have greatly increased affinity for heregulin, well surpassing that of ErbB3/ErbB4 heterodimers or ErbB4 homodimers (22). The ability of ErbB4 to bind a variety of ligands from both the EGF-like and NRG families contributes to its uniqueness in the ErbB family and has the potential to diversify signalling outcomes even further.
Role of ErbB4 in brain development
An RNA analysis study demonstrated that ErbB4 is highly expressed in brain, heart, and skeletal muscle (7). A subsequent study showed that ErbB4 mRNA is expressed in the hindbrain, mid-brain, and ventral forebrain at E9.5 in developing mouse embryos and the protein is detectable in brain tissue of both embryonic and adult mice (23).
Studies addressing specific variant expression in mice have shown that the mouse kidney solely expresses the JM-a variant and the heart solely expresses the JM-b variant, whilst the cerebellum expresses both variants (6). With regards to the cytoplasmic domain variants, CYT-1 is mainly expressed in mouse heart, mammary gland, and skin, while CYT-2 is the main variant in kidney and neural tissues (12). This tissue-specific expression of ErbB4 juxtamembrane and cytoplasmic variants further complicates signalling outcomes.
An essential role for ErbB4 in the central nervous system (CNS) has been demonstrated in mice by the creation of ErbB4 null mutants, which are terminal at E10.5 due to aborted development of myocardial ventricular trabeculae and disruptions to innervation of the hindbrain (23). In the CNS, ErbB4 is the primary receptor for NRG-1 (24, 25). This NRG-1-ErbB pathway is crucial for the proliferation, differentiation, and survival of glial cells, as well as the development of Schwann cells in the peripheral nervous system (26). This pathway also has an important role in inducing receptors for neurotransmitters such as acetylcholine, gamma-aminobutyric acid (GABA), and N-Methyl-D-aspartic acid, as well as regulating dendritogenesis in the cerebellum (27–29). Studies supporting these crucial roles have reported expression of both ErbB4 and NRG-1–4 in the brain (7, 19).
The different NRG isoforms are the products of alternative splicing of a single gene, which is regulated by neuronal activity (30, 31). There are more than 30 isoforms of NRG-1 alone, and these isoforms are categorized into six groups (32). In rats, mRNA analysis has shown that at different ages, each group has a distinctive expression pattern in the brain (30). This is consistent with the previously established role of NRG-1 in neural development. However, there are significantly fewer studies focusing on the NRG-2–4 isoforms. NRG-1 is highly expressed in the embryonic and fetal brain and expression declines during postnatal development. In contrast, NRG-2 levels significantly increase soon after birth (33, 34). This indicates that NRG-1 is vital in prenatal development while NRG-2 has an essential role in the postnatal and adult brain. NRG-2, -3, and -4 stimulate ErbB4 tyrosine phosphorylation (35). Furthermore, NRG-2 and -3 stimulation couples ErbB4 activation to biological responses and NRG-3 is a critical mediator in cortical inhibitory circuit assembly (35, 36). Until recently, NRG-4 was not demonstrated to have a role in neural development (37). However, it has since been shown to have an important role in the regulation of dendritic arborization in the developing cerebral cortex (37). Additionally, NRGs and ErbB4 are essential for synapse formation, interneuron migration, and axon and dendrite development, all of which are necessary for the GABAergic circuitry assembly that controls neural activity in the cerebral cortex (25).
As defective heart development is the reason for lethality in ErbB4 KO mice, one group expressed ErbB4 under a cardiac-specific myosin promoter in these mice, allowing them to reach adulthood (38). Consequently, the potential developmental role of ErbB4 in the brain and other tissues could be studied. Authors observed aberrant cranial neural crest cell migration, abnormal cranial nerve architecture, and significantly more interneurons in the cerebellum of the heart-rescued ErbB4 KO mice (38).
In the developing rat brain, Yau and colleagues analyzed ErbB4 immunoreactivity and showed that ErbB4 is preferentially expressed in interneurons migrating tangentially from the ventral to the dorsal telencephalon (25). ErbB4 mRNA was also detected at high levels in the cerebral cortex from prenatal day 18–20, peaking at postnatal day 0 (day of birth), hinting at the regulatory role of ErbB4 on neural development. Comparable results were reported by Fox and Kornblum, who found high ErbB4 expression in the germinal zones surrounding the lateral ventricles of the mouse forebrain only from prenatal day 17 through to postnatal day 1 (39).
Intriguingly, the high expression of the JM-a isoform in the brain (6) suggests ErbB4 cleavage and nuclear activity could play an important role in this tissue (40). The ability of ErbB4 to translocate into the nucleus and alter transcription has been associated with diverse outcomes, including poor prognostic outcomes in breast cancer (41). In the brain, ErbB4 signalling (specifically, the nuclear translocation of the ErbB4 ICD) is thought to be critical for the timing of astrogenesis in the developing brain (40). Interaction of the ErbB4 ICD with the signalling protein TAB2 and the corepressor N-CoR leads to nuclear translocation of a complex that can repress glial gene transcription and prevent neural precursor cells from differentiating into astrocytes (40).
Role of ErbB4 in brain injury, neurodegenerative diseases, and psychological disorders
ErbB4 has been implicated in neuroprotection from brain ischemia, via a signalling mechanism involving NRG-1β activation that protects against oxygen-glucose deprivation-induced neuronal death (42). Experiments in rodent models have also linked ErbB4 signalling to neuronal protection after brain injury and hemorrhage (43–45). Multiple studies have found ErbB4 to be critical for the assembly of the GABAergic (inhibitory) system (19). Alterations in this circuitry have been associated with the development of neurodevelopmental diseases, neurological and psychological disorders, and neuronal plasticity (46, 47). Furthermore, ErbB4 is expressed at the presynaptic terminals of GABAergic interneurons (48, 49), with detailed analysis narrowing this down to parvalbumin-expressing interneurons (50). Although an extensive coverage of the literature in this field is beyond the scope of this review, we will briefly summarize some of the research addressing the role of ErbB4 in schizophrenia, Alzheimer’s disease (AD), and Parkinson’s disease (PD).
Several studies have identified intronic variants of ErbB4 that are associated with schizophrenia, AD, and PD (51–54). The ERBB4 and NRG-1 genes have been implicated as risk genes for the development of schizophrenia. Indeed, the Neuregulin-ErbB4 signalling pathway plays a critical role in the development of inhibitory circuits in the mammalian cortex (50, 55). Mice hypomorphic for NRG-1 or ERBB4 demonstrate similar behavioral abnormalities and anti-psychotic drug treatment of the NRG-1 hypomorphs is able to partially reverse the behavioral phenotype (56).
Fifteen ErbB4 sequence variants have been associated with schizophrenia. Moreover, one of these variants is associated with an increased risk of schizophrenia in people carrying the Icelandic NRG-1 risk haplotype. Two ErbB4 alleles are significantly overexpressed in Ashkenazi schizophrenia patients, compared with matched controls (57). Other studies have identified additional genetic variants of the ErbB4 gene associated with this disease (58–60). Neuregulin signalling through ErbB4 selectively decreases fast synaptic GABAA currents on hippocampal interneurons which may contribute to the pathophysiology of epilepsy and neuropsychiatric disorders (61).
A 2010 study found significantly higher ErbB4 immunoreactivity in apoptotic hippocampal pyramidal neurons in AD patients, compared with healthy controls (62). These results, the authors argue, reflect a link between ErbB4 up-regulation and the progression of AD pathology (62). In a subsequent study, the same group found increased ErbB4 expression in neurons from the cortico-medial nucleus amygdala, human basal forebrain, and superior frontal gyrus of patients with AD (63). Additionally, ErbB4 is found at high levels surrounding the neuritic plaques that are characteristic of AD and potential allelic variants of ErbB4 have been associated with AD (64). Similarly, there is elevated ErbB4 expression in midbrain tissue sections of PD patients, compared with healthy controls.
The role of ErbB4 in cancer – oncogene, tumor suppressor, or both? the influence of isoforms
The different ErbB4 isoforms and their variable tissue expression may help to explain seemingly opposing signalling outcomes. Non-cleavable isoforms may dimerize with other ErbB members and recruit signalling proteins to the plasma membrane (65–67). Alternatively, proteolytic cleavage may result in translocation of the 80kDa intracellular domain to the nucleus, where it can participate as a transcriptional regulator (Figure 3) (14, 16, 65).
The ErbB4 isoform JM-a/CYT-2 is capable of oncogenic activity as a result of autoactivation, independent of the ligand binding that is required by other isoforms (68, 69). This ligand-independent autophosphorylation can increase cell proliferation and decrease apoptosis (52). Furthermore, the JM-a/CYT-2 isoform is known to promote survival and proliferation of breast cancer cells (68, 70). Specifically, overexpression of the JM-a/CYT-2 isoform combination promotes the proliferation of breast cancer cells, even in the absence of ligand (68). Conversely, expression of CYT-1 in breast cancer cell lines has tumor-suppressor effects by producing ligand-dependent growth inhibition and differentiation (71, 72). Results of such studies indicate isoform-specific roles of ErbB4 in cancer.
Interestingly, cytosolic localization of ErbB4 has been associated with better breast cancer prognosis, whilst nuclear localization is associated with worse prognosis (41). Real-time reverse transcription-PCR (RT-PCR) analysis of breast cancer samples determined that expression of the cleavable JM-a isoform is associated with estrogen receptor-α expression and a high histologic grade of differentiation (73). ErbB4 nuclear immunoreactivity is also associated with poor patient survival, compared with women whose cancer cells had membranous ErbB4 staining (73).
Mutations targeting the ErbB4 gene have been functionally characterized in some non-brain cancers, like non-small cell lung cancer (74). Kurppa and colleagues identified four specific mutations, located in the dimerization interface of the extracellular domain and in the tyrosine kinase domain (74). These mutations were functionally analyzed and found to increase basal and ligand-induced ErbB4 phosphorylation (74).
Similarly, novel ErbB4 mutations are present in 19% of melanoma patients and seven missense mutations in ERBB4 have been found to increase transformation ability and kinase activity (75). ErbB4 gain-of-function mutations are believed to be used by melanoma cells to activate the PI3K pathway (76). Furthermore, mutant ErbB4 in melanoma cells is associated with increased cell growth and this growth is reduced when these cells are treated with the ErbB inhibitor lapatinib (75).Silencing endogenous ErbB4 in ErbB4 mutant-expressing melanoma cells leads to an arrest in AKT phosphorylation and cellular proliferation (75).
However, not all ErbB4 mutations are linked to cancer. For example, a recent study by Jones and colleagues analyzed data from the Cancer Cell Line Encyclopedia (CCLE) and the Cancer Genome Atlas (TCGA), focusing on copy number mutations in ErbB4 (77). The study found that a region in Intron 1 of the ERBB4 gene was deleted in 69.1% of tumor samples harboring ERBB4 copy number loss (77). However, the same deletion was found at a similar frequency in matched normal tissue samples from these glioblastoma patients and in the general population (77).
The lack of isoform-specific and cleavage-specific reagents
Whilst ErbB4 is activated by a specific subset of ligands, once ligand-bound, it can homodimerize with other ErbB4 receptor molecules or heterodimerize with other ErbB family members. This complicates the study of ErbB4-induced signalling events (13). Compounding this, the lack of ErbB4-specific reagents can make it challenging to study its biological role. For example, the assessment of ErbB4 phosphorylation status can be difficult due to the lack of specific antibodies that are able to detect phosphorylation of specific tyrosine residues. A report from Gallo and colleagues found that an antibody directed against Tyr 1056, found within the ErbB4 CYT1 isoform but not the CYT2 isoform, produced a signal in mutant ErbB4 cells lacking this site (31). The authors showed that it not only cross-reacted with other ErbB4 phosphorylation sites, but also with phosphorylated EGFR (55). This highlights the importance of using immunoprecipitation in certain cases to verify the identity of phosphorylated ErbB4.
The existence of different ErbB4 isoforms also makes it inherently complicated to study the biological role of this receptor. It has been demonstrated in many publications that different ErbB4 isoforms produce varying signalling and biological outcomes. Furthermore, the generation of the ErbB4 intracellular domain (4ICD) can result in different subcellular localizations. Although there are currently no antibodies that specifically detect the different ErbB4 isoforms, antibodies have been developed that recognize the N-terminal or C-terminal of the receptor. A study by Tovey and colleagues compared antibody detection of ErbB4 in estrogen receptor-positive breast cancer patients (78). They used two different antibodies: the HFR1 antibody, which recognizes both the intact receptor and cleaved ICD, and the H4.77.16 antibody, which recognizes the extracellular domain of ErbB4, and thus only stains the full length receptor (78). The authors reported that patients demonstrating nuclear staining with the H4.77.16 antibody only had poorer survival (78). No such survival correlation was observed with the HFR1 antibody, suggesting that HFR1 may select for cytoplasmic and nuclear ErbB4 ICDs whilst H4.77.16 selects for membranous ErbB4 (79). These findings suggest that ErbB4 may be recycled in the cytoplasm or nucleus, however, further study is required for confirmation (80).
Due to the well-documented roles of EGFR and ErbB2 as oncogenes, much research has been directed at developing specific inhibitors against these family members, such as Gefitinib (EGFR inhibitor) and CP-724714 (ErbB2 inhibitor) (81, 82). Some of these have shown promise in the clinic, including the EGFR inhibitor Afatinib as a treatment for advanced non-small-cell lung cancer and the ErbB2 inhibitor Lapatinib in the treatment of advanced breast cancer (83, 84).
To date, there are no specific ErbB4 inhibitors. This makes it challenging to study the role of endogenous ErbB4. Many studies have used overexpression models to study the biological role of ErbB4. However, overexpression models, while helpful, have limitations and may not be truly representative of normal cellular function or the role of endogenous ErbB4 (85). Furthermore, many studies looking at expression do not take into account the different isoforms (85). In addition, the use of different ErbB4 antibodies can generate conflicting results (79).
The role of ErbB4 in brain cancers, including high-grade glioma
Whilst there is a wealth of literature on the potential role of ErbB4 in breast cancer, melanoma, and lung cancer, relatively little is known about its potential role in brain cancers. This is surprising, given its high expression in this tissue and the essential role it plays in brain growth and development. Figure 5 highlights some of the available data on ErbB4 expression and/or activation in various brain cancers.

Figure 5 ErbB4 expression and activation in various brain cancers. ErbB4 activation and mutations have only been identified in Glioblastoma. ErbB4 overexpression is observed in medulloblastoma and ependymoma. Isoform expression has been analyzed in all four cancers displayed however only glioblastoma and pilocytic astrocytoma showed a change in the isoform expression pattern.
ErbB4 expression and activity is ubiquitous during brain development. As would be expected, dysfunctions of ErbB4 isoforms have been implicated in different pathologies, including malignancies. Overexpression of ErbB4 has been identified in medulloblastomas, pilocytic astrocytomas, ependymomas, and glioblastomas (GBMs) (Figure 5) (86–89). In most benign meningiomas, however, ErbB4 is underexpressed and co-expression of ErbB4 with ErbB2 has been associated with poor prognosis in medulloblastoma and with increased proliferation index in ependymomas (86–88, 90).
Medulloblastomas, characterized as Grade IV lesions and ranked as the most common solid tumors in childhood, arise from abnormal development of the cerebellum (91). Overexpression of ErbB4 in conjunction with ErbB2 correlates with poor prognosis in medulloblastoma patients (87) (92). Recent analysis of the relative RNA levels of ErbB4 isoforms has shed some light on the differential expression patterns present within medulloblastoma.
A paper by Zeng and colleagues analyzed the relative levels of the different JM and CYT isoforms of ERBB4 in two pediatric brain tumor types: highly malignant medulloblastoma and the relatively benign pilocytic astrocytoma (86). They found different ratios of expression of the JM isoforms, with predominantly more of the cleavable JM-a isoform than the non-cleavable JM-b isoform in medulloblastoma and pilocytic astrocytoma, whereas the opposite pattern occurred in normal brain. Furthermore, rare isoforms (JM-c and JM-d) were differentially expressed in the two tumor types, with both expressing JM-c and only medulloblastoma expressing the JM-d variant. Neither of these rare isoforms were detected in normal brain (86).
Ependymoma is the third most common brain tumors in children (88). Histological grading for these tumors is controversial, hence there is much interest in identifying molecular targets that can be used as prognostic markers (93). Next generation sequencing of an ependymoma tumor found previously unknown mutations for this cancer, including an ERRB4 variant that has been detected in lung adenocarcinoma (93). A study of a large cohort of pediatric ependymoma patients found that co-expression of ErbB4 with ErbB2 occurred in over 70% of cases and was associated with higher proliferative index and poorer patient survival (88). The JM-a/CYT-1 and JM-a/CYT-2 isoforms were most highly expressed, with very little JM-b detected (88).
High grade gliomas (HGGs) are one of the most common forms of cancer affecting the central nervous system (94). HGG commonly targets adults, but as many as 12% of children diagnosed with primary CNS tumors suffer from HGG (95). HGG typically originates in non-neuronal cells, including glial cells, oligodendrocytes, and ependymal cells, in the brain and spinal cord (94). As expected, given the extensive role it plays during brain development, studies have identified ErbB4 as a potential target in HGG (10, 96, 97).
The most studied HGG is GBM, the most common and aggressive primary brain tumor in adults (98). Reports on the expression of ErbB4 in this disease are contradictory, with some reports suggesting that expression levels may influence whether ErbB4 plays an oncogenic or tumor suppressor role (99). ErbB4 mRNA and protein is expressed in the GBM cell lines SKMG‐3, SF767, U87, T98, and LN-229, as well as in primary GBM cells (10, 100–103). However, quantitative RT-PCR analysis of GBM patient samples found that whilst EGFR mRNA was overexpressed, ERBB4 mRNA remained at a level similar to normal brain (80). Moreover, a study by Andersson and colleagues showed that ErbB4 mRNA and protein expression was highest in low-grade gliomas, compared with higher grade tumors, suggesting that ErbB4 may act as a tumor suppressor (100). However, the results of this study were variable, and some GBM samples expressed comparatively high levels of ErbB4, both at the mRNA and protein level (100). Also, this study did not use isoform-specific antibodies and qPCR probes, and therefore, did not assess expression of the different ErbB4 isoforms (100). Comparison of ErbB4 isoform-specific expression in low versus high-grade glioma may be more pertinent. An isoform-specific RT-qPCR analysis of patient-derived GBM cell lines found JM-a/CYT-2 to be the predominantly expressed ErbB4 isoform, which is different from the usual JM-b predominance in neural tissue (10, 73). As previously stated, the JM-a/CYT-2 isoform has the capacity to autophosphorylate without ligand binding and thereby increase cell proliferation and decrease apoptosis (52, 68, 69). It is, therefore, possible that the JM-a/CYT-2 isoform performs these oncogenic functions in GBM, contributing to the aggressiveness of this disease. A recent report using patient-derived GBM samples and xenograft models of GBM also found significant associations between ErbB4 mRNA levels and tumorigenicity, proliferation, angiogenesis, and therapeutic response (10). Patients with high levels of ErbB4 activation had significantly shorter survival than those with little or no ErbB4 activation (10). These findings support the potential role of ErbB4 as a prognostic and therapeutic target in GBM.
Immunohistochemical studies of GBM patient samples have also attempted to relate ErbB family expression patterns to survival outcomes, with conflicting results. As outlined above, the 2004 study by Andersson and colleagues found variable expression of ErbB4 protein in GBM samples (100). Another study examined expression profiles of the ErbB family members in a panel of nine GBM patient samples (104). Whilst ErbB4 was expressed in the vast majority of GBM samples, expression levels were lower in GBM samples than in controls. Moreover, in GBM patient samples, ErbB4 was mainly detected in neuronal-like elements and occasionally in hypertrophic astrocytes (104).
Other studies report more significant ErbB4 expression levels in GBM. Using immunohistochemistry, Bodey and colleagues reported elevated ErbB4 expression in pediatric HGG samples compared with normal brain controls, with ‘strong immunoreactivity’ in GBM samples (97). Nabika and colleagues examined expression profiles of ErbB1–4 and the Cyclin-Dependent Kinases p21 and p27 in high grade (III and IV) gliomas (105). High ErbB1/EGFR and ErbB4 expression, along with low p27 expression, was associated with poor patient outcome (105). In addition, high ErbB4 expression was an independent indicator for poor survival (105). Further studies are required to understand these conflicting results and to clearly define the role that ErbB4 plays in GBM.
In addition to isoform-specific expression levels, mutations may influence ErbB4 function in GBM. Circulating tumor cell clusters in GBM were first identified in 2018 and sequencing analysis identified a structural variant of the ERBB4 gene within these clusters (106). Moreover, targeted next generation sequencing identified ERBB4 mutations in a cohort of 228 primary GBM patients (107). Although these mutations are of unknown significance, in this cohort they were found to contribute to overall survival (107). Therefore, the authors speculated that, based on other data supporting a role for ERBB4 in GBM progression, these may be inactivating mutations (10, 107).
Molecular interactions can also affect ErbB4 signalling in GBM. The WWOX tumor suppressor protein can interact with full-length ErbB4 or with the ICD only, with distinct results (108). WWOX interaction with full-length ErbB4 stabilizes the ErbB4 receptor in the cellular membrane (108). In contrast, when interacting with the ICD only, WWOX prevents the ICD from reaching the nucleus (108). In 2019, Chen and colleagues reported a novel regulatory pathway involving ErbB4 and circular RNA, which is upregulated in glioma (GM) cells (109). The circular RNA, circ_0074026 was highly expressed in HGG samples and correlated with ERBB4 mRNA expression (109). This circular RNA targets miRNA-1304, which appears to modulate ERBB4 expression and possibly contribute to glioma progression (109).
Diffuse midline glioma (DMG), otherwise known as diffuse intrinsic pontine glioma (DIPG), is considered the pediatric counterpart of GBM (110). DMG tumors are rarely surgically removed or biopsied, and consequently, they have not been molecularly characterized to the same level as many other primary brain tumors, such as GBM (111, 112). However, FISH analysis of genetic expression profiles comparing DIPG samples with non-DIPG pediatric GBM found recurrent focal gains for ERBB4 (96).
As previously discussed, ErbB4 can heterodimerize with the other ErbB family members, with each dimer having unique signalling properties. For example, since EGFR is dysregulated in approximately 70% of GBMs, the EGFR/ErbB4 heterodimer would almost certainly be present in tumors and may even be the dominant signalling moiety (113). Likewise, the presence of ErbB2 and ErbB3 has the capacity to alter the function of ErbB4 in tumors. Thus, additional studies are required to determine how the co-expression of ErbB family members may influence the function of ErbB4 in patients.
In summary, research is limited regarding individual ErbB4 isoform expression in brain tumors. However, analysis of ErbB4 isoform differential expression in medulloblastoma and ependymoma has highlighted the association between JM-a isoform expression and higher proliferative indices, translating to poorer survival rates (87, 88). Comparison of isoform-specific expression in low- versus high-grade glioma has not been reported, however the JM-a isoform appears to be the dominant isoform in GBM, and this may contribute to tumor aggressiveness (10). Future mechanistic studies are required to clarify the role of the JM-a isoform in GBM progression.
ErbB4 and possible therapeutic implications in high grade glioma
Currently there are no small molecule inhibitors that specifically inhibit ErbB4. There are, however, inhibitors that block the activity of ErbB4 in conjunction with other ErbB family members. We have demonstrated that the pan-ErbB inhibitor dacomitinib could inhibit the growth of GBM and pediatric medulloblastoma in orthotopic xenograft models (114). Similar results have been obtained with a second pan-ErbB inhibitor, NT113, in models of adult GBM (115). However, it remains unknown whether the inhibition of ErbB4 enhances or impedes the efficacy of these drugs. Another possible approach might be the repurposing of ibrutinib, which was designed to target Bruton’s tyrosine kinase. A recent report demonstrated that it inhibited ErbB4 at low concentrations and showed some preferential efficacy towards high-expressing ErbB4 cell lines (116).
The effective use of agents that inhibit ErB4 in brain cancer will require research that identifies pro-tumorigenic forms of ErbB4; targeting tumor suppressor forms of ErbB4 with drugs could obviously be detrimental to patients. As previously discussed, mutations of ErbB4 have been described in a range of brain cancers and the presence of such activating mutations could identify patients who may benefit from ErbB4 therapeutics. We recently demonstrated that the presence of highly phosphorylated ErbB4 in GBM, independent of phosphorylated EGFR, was linked to shorter patient survival compared to its absence (10). Additionally, in xenograft models, increased ErbB4 activation in GBM cells was associated with increased tumorigenicity, proliferation, and angiogenesis, as well as reduced sensitivity to anti-EGFR treatments (10). This strongly suggests that in GBM, activation of ErbB4 is pro-tumorigenic and thus phosphorylated ErbB4 could be a second biomarker for predicting response to ErbB4 targeting drugs. The mechanisms by which ErbB4 activation increases high-grade glioma aggressiveness are not yet understood. Indeed, there may be other viable targets within this pathway.
No specific ErbB4 inhibitors are currently available. Furthermore, the implications of inhibiting ErbB4 function in the brain, particularly the developing child brain, must be considered. As previously established, ErbB4 has important roles in normal brain physiology and vital roles in brain development (25). Thus, it is reasonable to speculate that negative side effects could occur in patients administered an ErbB4 inhibitor. It may be possible to limit side effects by developing isoform-specific inhibitors that target only the oncogenic ErbB4 isoform JM-a/CYT-2. Using approaches such as CRISPR in patient-derived cell lines will allow us to determine the role of the different isoforms, such as JM-a/CYT-2, in the growth and survival of brain cancer cells. These are the critical studies required before moving ErbB4 therapeutic forward into the clinic. Developing inhibitors for specific ErbB family members has proven a difficult task and may take a significant amount of time and resources to achieve. It is therefore imperative to elucidate ErbB4 signalling mechanisms in HGG and establish the impact of this pathway on disease progression and therapeutic resistance before developing such inhibitors.
We have shown that the interaction between c-MET and EGFR leads to the transactivation of both receptors in GBM (117). It is likely that ErbB4 can also be activated by other receptors expressed in brain cancers. Indeed, as recently extensively reviewed, many G-protein-coupled receptors can activate members of the ErbB family, including ErbB4 (118). Therefore, when determining the role of ErbB4 in brain cancers, other mechanisms of activation apart from ligand engagement must be considered. Identifying the presence of activated ErbB4, rather than the simple presence of ErbB4 protein, may be the most important approach to determining if ErbB4 is a driver in patient tumors (10).
Lessons should be learnt from the development and trial of EGFR-specific inhibitors in GBM treatment. While promising in pre-clinical models, EGFR inhibitors have had inconsistent results in clinical trials (119). This is believed to be due to the highly plastic and heterogenous nature of high-grade gliomas (119). Ultimately, a single target may not be enough to treat these cancers. Perhaps a multi-faceted approach with both ErbB4 inhibitors and EGFR inhibitors could prove more consistently effective against high-grade gliomas.
Future directions
The ‘uniqueness’ of ErbB4 in the ErbB receptor family, epitomized by its isoform variations, tissue- and disease state-specific expression, vast array of activating ligands (and yet, a seemingly selective subset of downstream targets), and large number of tyrosine phosphorylation sites makes it an attractive potential target to treat diseases such as GBM. However, these unique characteristics, in conjunction with the lack of inhibitors and other high-quality reagents for detecting its activation, and its ability to homo- and heterodimerize with other members of the ErbB receptor family, also make it inherently complicated to study.
Clearly, more mechanistic studies are required to further elucidate the role that ErbB4 may play in the progression of GBM. Current therapies do not adequately address the diffuse nature of this tumor or the plasticity of the signalling pathways that underlie its progression. Therefore, further insights into the role of ErbB4 in this disease are warranted.
Author contributions
The draft of this manuscript was prepared equally by J-LP and NA. All authors edited, reviewed, and approved the final version of the manuscript.
Funding
The research work related to the review in the author’s lab is funded by the Robert Connor Dawes Foundation in association with the Pirate Ship Foundation. At the time of writing, the first author of this review was receiving the support of the Australian Government Research Training Program Scholarship at The University of Western Australia.
Acknowledgments
The authors thank Leah Cannon, PhD, for editing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J (2000) 19(13):3159–67. doi: 10.1093/emboj/19.13.3159
2. Fan YX, Wong L, Johnson GR. EGFR kinase possesses a broad specificity for ErbB phosphorylation sites, and ligand increases catalytic-centre activity without affecting substrate binding affinity. Biochem J (2005) 392(Pt 3):417–23. doi: 10.1042/BJ20051122
3. Burden S, Yarden Y. Neuregulins and their receptors: A versatile signaling module in organogenesis and oncogenesis. Neuron (1997) 18(6):847–55. doi: 10.1016/S0896-6273(00)80324-4
4. Kainulainen V, Sundvall M, Määttä JA, Santiestevan E, Klagsbrun M, Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J Biol Chem (2000) 275(12):8641–9. doi: 10.1074/jbc.275.12.8641
5. Garrett TPJ, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, et al. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell (2003) 11(2):495–505. doi: 10.1016/S1097-2765(03)00048-0
6. Elenius K, Corfas G, Paul S, Choi CJ, Rio C, Plowman GD, et al. A novel juxtamembrane domain isoform of HER4/ErbB4.Isoform-specific tissue distribution and differential processing in response to phorbol ester. J Biol Chem (1997) 272(42):26761–8. doi: 10.1074/jbc.272.42.26761
7. Plowman GD, Culouscou JM, Whitney GS, Green JM, Carlton GW, Foy L, et al. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci U.S.A. (1993) 90(5):1746–50. doi: 10.1073/pnas.90.5.1746
8. Carpenter G. ErbB-4: Mechanism of action and biology. Exp Cell Res (2003) 284(1):66–77. doi: 10.1016/S0014-4827(02)00100-3
9. Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle (2011) 10(16):2647–57. doi: 10.4161/cc.10.16.17194
10. Donoghue JF, Kerr LT, Alexander NW, Greenall SA, Longano AB, Gottardo NG, et al. Activation of ERBB4 in glioblastoma can contribute to increased tumorigenicity and influence therapeutic response. Cancers (Basel) (2018) 10(8):243. doi: 10.3390/cancers10080243
11. Tan W, Dean M, Law AJ. Molecular cloning and characterization of the human ErbB4 gene: Identification of novel splice isoforms in the developing and adult brain. PloS One (2010) 5(9):e12924. doi: 10.1371/journal.pone.0012924
12. Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M. Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene (1999) 18(16):2607–15. doi: 10.1038/sj.onc.1202612
13. Gallo RM, Riese DJ 2nd. The antibody sc-33040-R fails to specifically recognize phosphorylation of ErbB4 on tyrosine1056. Growth Factors (2007) 25(5):329–33. doi: 10.1080/08977190701804008
14. Ni CY, Murphy MP, Golde TE, Carpenter G. Gamma -secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science (2001) 294(5549):2179–81. doi: 10.1126/science.1065412
15. Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem (2003) 278(35):33334–41. doi: 10.1074/jbc.M305597200
16. Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, et al. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J Cell Biol (2004) 167(3):469–78. doi: 10.1083/jcb.200403155
17. Long W, Wagner K-U, Lloyd KC, Binart N, Shillingford JM, Hennighausen L, et al. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development (2003) 130(21):5257–68. doi: 10.1242/dev.00715
18. Klapper LN, Kirschbaum MH, Seta M, Yarden Y. Biochemical and clinical implications of the ErbB/HER signaling network of growth factor receptors. Adv Cancer Res (2000) 77:25–79. doi: 10.1016/S0065-230X(08)60784-8
19. Mei L, Nave KA. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron (2014) 83(1):27–49. doi: 10.1016/j.neuron.2014.06.007
20. Birchmeier C. ErbB receptors and the development of the nervous system. Exp Cell Res (2009) 315(4):611–8. doi: 10.1016/j.yexcr.2008.10.035
21. Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, Lavi S, et al. ErbB-3 and ErbB-4 function as the respective low and high affinity receptors of all neu differentiation factor/heregulin isoforms. J Biol Chem (1994) 269(40):25226–33. doi: 10.1016/S0021-9258(17)31521-1
22. Fitzpatrick VD, Pisacane PI, Vandlen RL, Sliwkowski MX. Formation of a high affinity heregulin binding site using the soluble extracellular domains of ErbB2 with ErbB3 or ErbB4. FEBS Lett (1998) 431(1):102–6. doi: 10.1016/S0014-5793(98)00737-6
23. Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature (1995) 378(6555):390–4. doi: 10.1038/378390a0
24. Gerecke KM, Wyss JM, Karavanova I, Buonanno A, Carroll SL. ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J Comp Neurol (2001) 433(1):86–100. doi: 10.1002/cne.1127
25. Yau H-J, Wang H-F, Lai C, Liu F-C. Neural development of the neuregulin receptor ErbB4 in the cerebral cortex and the hippocampus: Preferential expression by interneurons tangentially migrating from the ganglionic eminences. Cereb Cortex (2003) 13(3):252–64. doi: 10.1093/cercor/13.3.252
26. Canoll PD, Musacchio JM, Hardy R, Reynolds R, Marchionni MA, Salzer JL. GGF/neuregulin is a neuronal signal that promotes the proliferation and survival and inhibits the differentiation of oligodendrocyte progenitors. Neuron (1996) 17(2):229–43. doi: 10.1016/S0896-6273(00)80155-5
27. Ozaki M, Sasner M, Yano R, Lu HS, Buonanno A. Neuregulin-beta induces expression of an NMDA-receptor subunit. Nature (1997) 390(6661):691–4. doi: 10.1038/37795
28. Rieff HI, Raetzman LT, Sapp DW, Yeh HH, Siegel RE, Corfas G. Neuregulin induces GABA(A) receptor subunit expression and neurite outgrowth in cerebellar granule cells. J Neurosci (1999) 19(24):10757–66. doi: 10.1523/JNEUROSCI.19-24-10757.1999
29. Liu Y, Ford B, Mann MA, Fischbach GD. Neuregulins increase alpha7 nicotinic acetylcholine receptors and enhance excitatory synaptic transmission in GABAergic interneurons of the hippocampus. J Neurosci (2001) 21(15):5660–9. doi: 10.1523/JNEUROSCI.21-15-05660.2001
30. Liu X, Bates R, Yin D-M, Shen C, Wang F, Su N, et al. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci (2011) 31(23):8491–501. doi: 10.1523/JNEUROSCI.5317-10.2011
31. Marchionni MA, Goodearl ADJ, Chen MS, Bermingham-McDonogh O, Kirk C, Hendricks M, et al. Glial growth factors are alternatively spliced erbB2 ligands expressed in the nervous system. Nature (1993) 362(6418):312–8. doi: 10.1038/362312a0
32. Falls DL. Neuregulins: Functions, forms, and signaling strategies. Exp Cell Res (2003) 284(1):14–30. doi: 10.1016/S0014-4827(02)00102-7
33. Corfas G, Rosen KM, Aratake H, Krauss R, Fischbach GD. Differential expression of ARIA isoforms in the rat brain. Neuron (1995) 14(1):103–15. doi: 10.1016/0896-6273(95)90244-9
34. Longart M, Liu Y, Karavanova I, Buonanno A. Neuregulin-2 is developmentally regulated and targeted to dendrites of central neurons. J Comp Neurol (2004) 472(2):156–72. doi: 10.1002/cne.20016
35. Hobbs SS, Coffing SL, Le ATD, Cameron EM, Williams EE, Andrew M, et al. Neuregulin isoforms exhibit distinct patterns of ErbB family receptor activation. Oncogene (2002) 21(55):8442–52. doi: 10.1038/sj.onc.1205960
36. Bartolini G, Sánchez-Alcañiz JA, Osório C, Valiente M, García-Frigola C, Marín O. Neuregulin 3 mediates cortical plate invasion and laminar allocation of GABAergic interneurons. Cell Rep (2017) 18(5):1157–70. doi: 10.1016/j.celrep.2016.12.089
37. Paramo B, Wyatt S, Davies AM. An essential role for neuregulin-4 in the growth and elaboration of developing neocortical pyramidal dendrites. Exp Neurol (2018) 302:85–92. doi: 10.1016/j.expneurol.2018.01.002
38. Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci U.S.A. (2003) 100(14):8281–6. doi: 10.1073/pnas.1436402100
39. Fox IJ, Kornblum HI. Developmental profile of ErbB receptors in murine central nervous system: implications for functional interactions. J Neurosci Res (2005) 79(5):584–97. doi: 10.1002/jnr.20381
40. Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell (2006) 127(1):185–97. doi: 10.1016/j.cell.2006.07.037
41. Thor AD, Edgerton SM, Jones FE. Subcellular localization of the HER4 intracellular domain, 4ICD, identifies distinct prognostic outcomes for breast cancer patients. Am J Pathol (2009) 175(5):1802–9. doi: 10.2353/ajpath.2009.090204
42. Lu Y-M, Gao Y-P, Tao R-R, Liao M-H, Huang J-Y, Wu G, et al. Calpain-dependent erbb4 cleavage is involved in brain ischemia-induced neuronal death. Mol Neurobiol (2016) 53(4):2600–9. doi: 10.1007/s12035-015-9275-2
43. Yan F, Tan X, Wan W, Dixon BJ, Fan R, Enkhjargal B, et al. ErbB4 protects against neuronal apoptosis via activation of YAP/PIK3CB signaling pathway in a rat model of subarachnoid hemorrhage. Exp Neurol (2017) 297:92–100. doi: 10.1016/j.expneurol.2017.07.014
44. Qian H, Dou Z, Ruan W, He P, Zhang JH, Yan F, et al. ErbB4 preserves blood-brain barrier integrity via the yap/pik3cb pathway after subarachnoid hemorrhage in rats. Front Neurosci (2018) 12:492. doi: 10.3389/fnins.2018.00492
45. Deng W, Luo F, Li B-M, Mei L. NRG1-ErbB4 signaling promotes functional recovery in a murine model of traumatic brain injury via regulation of GABA release. Exp Brain Res (2019) 237(12):3351–62. doi: 10.1007/s00221-019-05680-2
46. Chattopadhyaya B, Cristo GD. GABAergic circuit dysfunctions in neurodevelopmental disorders. Front Psychiatry (2012) 3:51. doi: 10.3389/fpsyt.2012.00051
47. Coyle JT. The GABA-glutamate connection in schizophrenia: which is the proximate cause? Biochem Pharmacol (2004) 68(8):1507–14. doi: 10.1016/j.bcp.2004.07.034
48. Woo R-S, Li X-M, Tao Y, Carpenter-Hyland E, Huang YZ, Weber J, et al. Neuregulin-1 enhances depolarization-induced GABA release. Neuron (2007) 54(4):599–610. doi: 10.1016/j.neuron.2007.04.009
49. Longart M, Chatani-Hinze M, Gonzalez CM, Vullhorst D, Buonanno A. Regulation of ErbB-4 endocytosis by neuregulin in GABAergic hippocampal interneurons. Brain Res Bull (2007) 73(4-6):210–9. doi: 10.1016/j.brainresbull.2007.02.014
50. Fazzari P, Paternain AV, Valiente M, Pla R, Luján R, Lloyd K, et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature (2010) 464(7293):1376–80. doi: 10.1038/nature08928
51. Banerjee A, MacDonald ML, Borgmann-Winter KE, Hahn C-G. Neuregulin 1-erbB4 pathway in schizophrenia: From genes to an interactome. Brain Res Bull (2010) 83(3-4):132–9. doi: 10.1016/j.brainresbull.2010.04.011
52. Iwakura Y, Nawa H. ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: Pathological implications in schizophrenia and parkinson’s disease. Front Cell Neurosci (2013) 7:4. doi: 10.3389/fncel.2013.00004
53. Norton N, Moskovina V, Morris DW, Bray NJ, Zammit S, Williams NM, et al. Evidence that interaction between neuregulin 1 and its receptor erbB4 increases susceptibility to schizophrenia. Am J Med Genet B Neuropsychiatr Genet (2006) 141B(1):96–101. doi: 10.1002/ajmg.b.30236
54. Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science (2008) 320(5875):539–43. doi: 10.1126/science.1155174
55. Marenco S, Geramita M, van der Veen JW, Barnett AS, Kolachana B, Shen J, et al. Genetic association of ErbB4 and human cortical GABA levels in vivo. J Neurosci (2011) 31(32):11628–32. doi: 10.1523/JNEUROSCI.1529-11.2011
56. Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet (2002) 71(4):877–92. doi: 10.1086/342734
57. Silberberg G, Darvasi A, Pinkas-Kramarski R, Navon R. The involvement of ErbB4 with schizophrenia: Association and expression studies. Am J Med Genet B Neuropsychiatr Genet (2006) 141B(2):142–8. doi: 10.1002/ajmg.b.30275
58. Law AJ, Lipska BK, Weickert CS, Hyde TM, Straub RE, Hashimoto R, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5’ SNPs associated with the disease. Proc Natl Acad Sci U.S.A. (2006) 103(17):6747–52. doi: 10.1073/pnas.0602002103
59. Li W, Zhou Y, Jentsch JD, Brown RAM, Tian X, Ehninger D, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U.S.A. (2007) 104(46):18280–5. doi: 10.1073/pnas.0706900104
60. Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci (2008) 9(6):437–52. doi: 10.1038/nrn2392
61. Mitchell RM, Janssen MJ, Karavanova I, Vullhorst D, Furth K, Makusky A, et al. ErbB4 reduces synaptic GABAA currents independent of its receptor tyrosine kinase activity. Proc Natl Acad Sci U.S.A. (2013) 110(48):19603–8. doi: 10.1073/pnas.1312791110
62. Woo R-S, Lee J-H, Yu H-N, Song D-Y, Baik T-K. Expression of ErbB4 in the apoptotic neurons of alzheimer’s disease brain. Anat Cell Biol (2010) 43(4):332–9. doi: 10.5115/acb.2010.43.4.332
63. Woo R-S, Lee J-H, Yu H-N, Song D-Y, Baik T-K. Expression of ErbB4 in the neurons of alzheimer’s disease brain and APP/PS1 mice, a model of alzheimer’s disease. Anat Cell Biol (2011) 44(2):116–27. doi: 10.5115/acb.2011.44.2.116
64. Chaudhury AR, Gerecke KM, Wyss M, Morgan DG, Gordon MN, Carroll SL. Neuregulin-1 and ErbB4 immunoreactivity is associated with neuritic plaques in alzheimer disease brain and in a transgenic model of alzheimer disease. J Neuropathol Exp Neurol (2003) 62(1):42–54. doi: 10.1093/jnen/62.1.42
65. Kaushansky A, Gordus A, Budnik BA, Lane WS, Rush J, MacBeath G. System-wide investigation of ErbB4 reveals 19 sites of tyr phosphorylation that are unusually selective in their recruitment properties. Chem Biol (2008) 15(8):808–17. doi: 10.1016/j.chembiol.2008.07.006
66. Cohen BD, Kiener PA, Green JM, Foy L, Fell HP, Zhang K. The relationship between human epidermal growth-like factor receptor expression and cellular transformation in NIH3T3 cells. J Biol Chem (1996) 271(48):30897–903. doi: 10.1074/jbc.271.48.30897
67. Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett (1999) 447(2-3):227–31. doi: 10.1016/S0014-5793(99)00283-5
68. Määttä JA, Sundvall M, Junttila TT, Peri L, Laine VJO, Isola J, et al. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol Biol Cell (2006) 17(1):67–79. doi: 10.1091/mbc.e05-05-0402
69. Tal-Or P, Erlich S, Porat-Shliom N, Goldshmit Y, Ben-Baruch G, Shaharabani E, et al. Ligand-independent regulation of ErbB4 receptor phosphorylation by activated ras. J Cell Biochem (2006) 98(6):1482–94. doi: 10.1002/jcb.20815
70. Junttila TT, Sundvall M, Lundin M, Lundin J, Tanner M, Härkönen P, et al. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res (2005) 65(4):1384–93. doi: 10.1158/0008-5472.CAN-04-3150
71. Sartor CI, Zhou H, Kozlowska E, Guttridge K, Kawata E, Caskey L, et al. Her4 mediates ligand-dependent antiproliferative and differentiation responses in human breast cancer cells. Mol Cell Biol (2001) 21(13):4265–75. doi: 10.1128/MCB.21.13.4265-4275.2001
72. Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, et al. The ERBB4/HER4 intracellular domain 4icd is a bh3-only protein promoting apoptosis of breast cancer cells. Cancer Res (2006) 66(12):6412–20. doi: 10.1158/0008-5472.CAN-05-2368
73. Junttila TT, Sundvall M, Lundin M, Lundin J, Tanner M, Härkönen P, et al. Cleavable ErbB4 isoform in estrogen receptor–regulated growth of breast cancer cells. Cancer Res (2005) 65(4):1384–93. doi: 10.1158/0008-5472.CAN-04-3150
74. Kurppa KJ, Denessiouk K, Johnson MS, Elenius K. Activating ERBB4 mutations in non-small cell lung cancer. Oncogene (2016) 35(10):1283–91. doi: 10.1038/onc.2015.185
75. Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet (2009) 41(10):1127–32. doi: 10.1038/ng.438
76. Kurppa K, Elenius K. Mutated ERBB4: A novel drug target in metastatic melanoma? Pigment Cell Melanoma Res (2009) 22(6):708–10. doi: 10.1111/j.1755-148X.2009.00635.x
77. Jones DC, Scanteianu A, DiStefano M, Bouhaddou M, Birtwistle MR. Analysis of copy number loss of the ErbB4 receptor tyrosine kinase in glioblastoma. PloS One (2018) 13(1):e0190664. doi: 10.1371/journal.pone.0190664
78. Tovey SM, Witton CJ, Bartlett JMS, Stanton PD, Reeves JR, Cooke TG. Outcome and human epidermal growth factor receptor (HER) 1-4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res (2004) 6(3):R246–51. doi: 10.1186/bcr783
79. Tovey SM, Dunne B, Witton CJ, Cooke TG, Bartlett JMS. HER4 in breast cancer: comparison of antibodies against intra- and extra-cellular domains of HER4. Breast Cancer Res (2006) 8(2):R19. doi: 10.1186/bcr1394
80. Yoshimoto K, Ma X, Guan Y, Mizoguchi M, Nakamizo A, Amano T, et al. Expression of stem cell marker and receptor kinase genes in glioblastoma tissue quantified by real-time RT-PCR. Brain Tumor Pathol (2011) 28(4):291–6. doi: 10.1007/s10014-011-0046-0
81. Herbst RS, Fukuoka M, Baselga J. Gefitinib — a novel targeted approach to treating cancer. Nat Rev Cancer (2004) 4(12):956–65. doi: 10.1038/nrc1506
82. Jani JP, Finn RS, Campbell M, Coleman KG, Connell RD, Currier N, et al. Discovery and pharmacologic characterization of CP-724,714, a selective ErbB2 tyrosine kinase inhibitor. Cancer Res (2007) 67(20):9887–93. doi: 10.1158/0008-5472.CAN-06-3559
83. Liang W, Wu X, Fang W, Zhao Y, Yang Y, Hu Z, et al. Network meta-analysis of erlotinib, gefitinib, afatinib and icotinib in patients with advanced non-small-cell lung cancer harboring EGFR mutations. PloS One (2014) 9(2):e85245. doi: 10.1371/journal.pone.0085245
84. Schlam I, Swain SM. HER2-positive breast cancer and tyrosine kinase inhibitors: the time is now. NPJ Breast Cancer (2021) 7(1):56. doi: 10.1038/s41523-021-00265-1
85. Prelich G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics (2012) 190(3):841–54. doi: 10.1534/genetics.111.136911
86. Zeng N, Liu L, McCabe MG, Jones DTW, Ichimura K, Collins VP. Real-time quantitative polymerase chain reaction (qPCR) analysis with fluorescence resonance energy transfer (FRET) probes reveals differential expression of the four ERBB4 juxtamembrane region variants between medulloblastoma and pilocytic astrocytoma. Neuropathol Appl Neurobiol (2009) 35(4):353–66. doi: 10.1111/j.1365-2990.2008.01001.x
87. Gilbertson RJ, Perry RH, Kelly PJ, Pearson AD, Lunec J. Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res (1997) 57(15):3272–80.
88. Gilbertson RJ, Bentley L, Hernan R, Junttila TT, Frank AJ, Haapasalo H, et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res (2002) 8(10):3054–64.
89. Torp SH, Gulati S, Johannessen E, Dalen A. Coexpression of c-erbB 1-4 receptor proteins in human glioblastomas. an immunohistochemical study. J Exp Clin Cancer Res (2007) 26(3):353–9.
90. Laurendeau I, Ferrer M, Garrido D, D'Haene N, Ciavarelli P, Basso A, et al. Gene expression profiling of ErbB receptors and ligands in human meningiomas. Cancer Invest (2009) 27(6):691–8. doi: 10.1080/07357900802709175
91. Aldaregia J, Errarte P, Olazagoitia-Garmendia A, Gimeno M, Uriz JJ, Gershon TR, et al. Erbb4 is required for cerebellar developmentand malignant phenotype of medulloblastoma. Cancers (Basel) (2020) 12(4):997. doi: 10.3390/cancers12040997
92. Bal MM, Das Rodotra B, Srinivasan R, Sharma SC. Expression of c-erbB-4 in medulloblastoma and its correlation with prognosis. Histopathology (2006) 49(1):92–3. doi: 10.1111/j.1365-2559.2006.02337.x
93. Butt E, Alyami S, Nageeti T, Saeed M, AlQuthami K, Bouazzaoui A, et al. Mutation profiling of anaplastic ependymoma grade III by Ion Proton next generation DNA sequencing. F1000Res (2019) 8:613. doi: 10.12688/f1000research.18721.1
94. Sim HW, Morgan ER, Mason WP. Profiling of anaplastic ependymoma grade iii by ion proton next generation dna sequencing contemporary management of high-grade gliomas. CNS Oncol (2018) 7(1):51–65. doi: 10.2217/cns-2017-0026
95. El-Ayadi M, Ansari M, Sturm D, Gielen GH, Warmuth-Metz M, Kramm CM, et al. High-grade glioma in very young children: A rare and particular patient population. Oncotarget (2017) 8(38):64564–78. doi: 10.18632/oncotarget.18478
96. Paugh BS, Broniscer A, Qu C, Miller CP, Zhang J, Tatevossian RG, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol (2011) 29(30):3999–4006. doi: 10.1200/JCO.2011.35.5677
97. Bodey B, Kaiser HE, Siegel SE. Epidermal growth factor receptor (EGFR) expression in childhood brain tumors. In Vivo (2005) 19(5):931–41.
98. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
99. Lucas LM, Dwivedi V, Senfeld JI, Cullum RL, Mill CP, Piazza JT, et al. The yin and yang of ERBB4: Tumor suppressor and oncoprotein. Pharmacol Rev (2022) 74(1):18–47. doi: 10.1124/pharmrev.121.000381
100. Andersson U, Guo D, Malmer B, Bergenheim AT, Brännström T, Hedman H, et al. Epidermal growth factor receptor family (EGFR, ErbB2-4) in gliomas and meningiomas. Acta Neuropathol (2004) 108(2):135–42. doi: 10.1007/s00401-004-0875-6
101. Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science (2007) 318(5848):287–90. doi: 10.1126/science.1142946
102. Carrasco-García E, Saceda M, Grasso S, Rocamora-Reverte L, Conde M, Gómez-Martínez A, et al. Small tyrosine kinase inhibitors interrupt EGFR signaling by interacting with erbB3 and erbB4 in glioblastoma cell lines. Exp Cell Res (2011) 317(10):1476–89. doi: 10.1016/j.yexcr.2011.03.015
103. Thomas CY, Chouinard M, Cox M, Parsons S, Stallings-Mann M, Garcia R, et al. Spontaneous activation and signaling by overexpressed epidermal growth factor receptors in glioblastoma cells. Int J Cancer (2003) 104(1):19–27. doi: 10.1002/ijc.10880
104. Duhem-Tonnelle V, Bièche I, Vacher S, Loyens A, Maurage C-A, Collier F, et al. Differential distribution of erbB receptors in human glioblastoma multiforme: Expression of erbB3 in CD133-positive putative cancer stem cells. J Neuropathol Exp Neurol (2010) 69(6):606–22. doi: 10.1097/NEN.0b013e3181e00579
105. Nabika S, Kiya K, Satoh H, Mizoue T, Kondo H, Katagiri M, et al. Prognostic significance of expression patterns of EGFR family, p21 and p27 in high-grade astrocytoma. Hiroshima J Med Sci (2010) 59(4):65–70.
106. Krol I, et al. Detection of circulating tumour cell clusters in human glioblastoma. Br J Cancer (2018) 119(4):487–91. doi: 10.1038/s41416-018-0186-7
107. Ji MS, Eldred BSC, Liu R, Pianka ST, Molaie D, Kevan B, et al. Targeted next-generation sequencing of 565 neuro-oncology patients at UCLA: A single-institution experience. Neurooncol Adv (2020) 2(1):vdaa009. doi: 10.1093/noajnl/vdaa009
108. Kosla K, Pluciennik E, Kurzyk A, Jesionek-Kupnicka D, Kordek R, Potemski P, et al. Molecular analysis of WWOX expression correlation with proliferation and apoptosis in glioblastoma multiforme. J Neurooncol (2011) 101(2):207–13. doi: 10.1007/s11060-010-0254-1
109. Chen M, Liu X, Xie P, Wang P, Liu M, Zhan Y, et al. Circular RNA circ_0074026 indicates unfavorable prognosis for patients with glioma and facilitates oncogenesis of tumor cells by targeting miR-1304 to modulate ERBB4 expression. J Cell Physiol (2020) 235(5):4688–97. doi: 10.1002/jcp.29347
110. Kluiver TA, Alieva M, van Vuurden DG, Wehrens EJ, Rios AC. Invaders exposed: Understanding and targeting tumor cell invasion in diffuse intrinsic pontine glioma. Front Oncol (2020) 10:92. doi: 10.3389/fonc.2020.00092
111. Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer (2014) 14(10):651–61. doi: 10.1038/nrc3811
112. Mrugala MM. Advances and challenges in the treatment of glioblastoma: A clinician’s perspective. Discovery Med (2013) 15(83):221–30.
113. Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell (2013) 155(2):462–77. doi: 10.1016/j.cell.2013.09.034
114. Endersby R, Whitehouse J, Hii H, Greenall SA, Johns TG, Gottardo NG. A pre-clinical assessment of the pan-erbb inhibitor dacomitinib in pediatric and adult brain tumors. Neoplasia (2018) 20(5):432–42. doi: 10.1016/j.neo.2018.02.004
115. Yoshida Y, Ozawa T, Yao T-W, Shen W, Brown D, Parsa AT, et al. NT113, a pan-ERBB inhibitor with high brain penetrance, inhibits the growth of glioblastoma xenografts with EGFR amplification. Mol Cancer Ther (2014) 13(12):2919–29. doi: 10.1158/1535-7163.MCT-14-0306
116. Rauf F, Festa F, Park JG, Magee M, Eaton S, Rinaldi C, et al. Ibrutinib inhibition of ERBB4 reduces cell growth in a WNT5A-dependent manner. Oncogene (2018) 37(17):2237–50. doi: 10.1038/s41388-017-0079-x
117. Greenall SA, Donoghue JF, Van Sinderen M, Dubljevic V, Budiman S, Devlin M, et al. EGFRvIII-mediated transactivation of receptor tyrosine kinases in glioma: mechanism and therapeutic implications. Oncogene (2015) 34(41):5277–87. doi: 10.1038/onc.2014.448
118. Palanisamy S, Xue C, Ishiyama S, Prasad SV, Gabrielson K. GPCR-ErbB transactivation pathways and clinical implications. Cell Signalling (2021) 86:110092. doi: 10.1016/j.cellsig.2021.110092
Keywords: ErbB4, EGFR, receptor tyrosine kinase, high grade glioma, diffuse midline glioma
Citation: Pitcher J-L, Alexander N, Miranda PJ and Johns TG (2022) ErbB4 in the brain: Focus on high grade glioma. Front. Oncol. 12:983514. doi: 10.3389/fonc.2022.983514
Received: 01 July 2022; Accepted: 28 July 2022;
Published: 31 August 2022.
Edited by:
Yonehiro Kanemura, Osaka National Hospital (NHO), JapanReviewed by:
Shigeo Ohba, Fujita Health University, JapanJasmin Lalonde, University of Guelph, Canada
Copyright © 2022 Pitcher, Alexander, Miranda and Johns. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jamie-Lee Pitcher, SmFtaWUtbGVlLnBpdGNoZXJAdGVsdGhvbmtpZHMub3JnLmF1
†These authors have contributed equally to this work and share first authorship
 Jamie-Lee Pitcher
Jamie-Lee Pitcher Naomi Alexander1†
Naomi Alexander1† Terrance G. Johns
Terrance G. Johns