- State Key Laboratory of Experimental Hematology, National Clinical Research Center for Blood Diseases, Haihe Laboratory of Cell Ecosystem, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin, China
Copy number variations (CNVs) are widespread in both pediatric and adult cases of B-cell acute lymphoblastic leukemia (B-ALL); however, their clinical significance remains unclear. This review primarily discusses the most prevalent CNVs in B-ALL to elucidate their clinical value and further personalized management of this population. The discovery of the molecular mechanism of gene deletion and the development of targeted drugs will further enhance the clinical prognosis of B-ALL.
Introduction
B cell acute lymphoblastic leukemia (B-ALL) is a heterogeneous and invasive hematological malignancy with the accretion of genetic lesions (1, 2). Recent research has comprehensively investigated the genetic landscape of both adult and pediatric B-ALL (3–5). Over 90% of pediatric patients with B-ALL can attain complete remission (CR), 20% relapse, and 10% remain incurable (6). The conventional approach for pediatric B-ALL remission-induction chemotherapy drugs mainly consists of glucocorticoid, vincristine, asparaginase and/or anthracycline (7). With the first course of induction therapy administration for 4-6 weeks, the CR rate population of pediatric B-ALL may reach 98% (7).
The genomic pattern of adult B-ALL might differ from pediatric cases, accompanied by more devastating clinical outcomes (8). However, chances of newly emerged drugs, chimeric antigen receptor T cell therapy, and hematopoietic stem cell transplantation (HSCT) improved the clinical response of specific subtypes of B-ALL patients remarkably (9–12). Nevertheless, 40% of adult patients with B-ALL relapsed at a median duration of 13 months (28 days to 12 years) (13). In this population, around 30%-40% of relapsed and refractory B-ALL cases can attain complete remission by first salvage chemotherapy. Besides, the long-term survival, that is, the 5-year survival rate, of patients with B-ALL remains at 20% only (14, 15). Hence, it is imperative to find a novel biomarker that could help determine the characteristics and prognosis of newly diagnosed B-ALL (16, 17).
Copy number variations (CNV; a.k.a. copy number aberrations [CNAs]) are a specific type of genetic abnormality with a high incidence in B-ALL (1, 14, 18), ranging from 1 Kb to less than 5 Mb (19). CNVs denote the deletion, insertion, replication, and multipoint variants of DNA fragments. Previously, the initial cognition of CNV was found in healthy people and correlated with neuropsychiatric disorders. Today, CNV is broadly recognized as a major cause of various solid tumors (20) and acute myeloid leukemia (21). This review primarily focuses on the CNV biomarker analysis in B-ALL and their prognostic significance.
CNV detection method
As CNVs are challenging to detect by karyotype analysis, fluorescence in situ hybridization (FISH), and PCR amplification; besides, their research and application are limited to some extent (22, 23). Indeed, FISH is traditionally used in CNV research but is limited to the imbalance design of both satisfying multi-genes location and the FISH gene-specific probes. With the advent of various sequencing technologies, array-based CNV analysis was commonly used for detecting genomic DNA fragments. For example, CNV can be recognized by array comparative genomic hybridization and single-nucleotide polymorphism arrays; however, the high cost and complex process of these techniques hinder their widespread use in clinical practice. In 2002, Schouten established multiplex ligation-dependent probe amplification (MLPA) assay to analyze the CNV spectrum; this technology is a fast and reliable gene CNV detection method that can detect the copy number changes of 45 gene probes simultaneously with high specificity and at a low cost (24). Kiss R et al. (25) proposed the digital MLPA-based approach based on the next-generation sequencing technology to detect hundreds of exon-positions CNV panels at the same time. The next-generation sequencing method can simultaneously detect sequence variation of a single base, insertion, or deletion of short fragments and CNV (19).
To date, many studies have investigated various software projects to examine copy number changes (26). Zhou B et al. (23) compared different sequencing depths (1×, 3×, and 5× coverages) using whole-genome sequencing by different sequencing libraries (short/3 kb/5 kb); they recommended that the gold standard for CNV detection was under the large library and low sequencing depth. Optical genome mapping is a new whole-genome sequencing method in which each DNA molecule is linearized and unfolded by nano-microfluidic CHIP with high-resolution fluorescence imaging (27, 28).
All structural variations and CNVs can be detected by providing original DNA information for downstream applications of genomics. Unlike other traditional cytogenetic methods, optical genome mapping has a full coverage of all types of mutations, detects small tumor-related mutations, and has high consistency in detecting hematological malignancies–related chromosomal and DNA abnormalities. In addition, LüHMANN JL et al. (29) established that optical genome mapping was superior to any other traditional method in the area of detecting the classical gene deletions (e.g., IKZF1) and gene losses that were previously undetected (e.g., SETD2). Owing to the insensitivity of whole-genome sequencing hybridization and capture, the reads captured in an exon fragment vary markedly from sample to sample. Thus, new technologies emerged gradually, such as noninvasive prenatal testing technology, which could detect CNVs in tumor circulating free DNA of 7 MB size with >95% sensitivity and specificity (30).
Reportedly, RNA-seq is limited to detect CNVs in ALL as a result of mismatching B-allele frequency. BAŘINKA et.al (31). developed a robust tool RNAseqCNV package based on the normalized gene expression and minor allele frequency to classify arm-level CNVs. In addition, InferCNV was applied widely to identify large-scale chromosomal CNVs in tumor single-cell RNA sequencing (scRNA-seq) data. The basic idea is to compare the gene expression of each tumor cell with the average expression or “normal” reference cell gene expression in the whole genome to determine its expression intensity (32). However, the genomic location of specific CNVs is not available to precisely classify tumor and normal cells copy number spectrum. Considering the critical need for distinguishing normal cell types from malignant cells in the tumor microenvironment, copy number karyotype of tuments (CopyKAT), as an integrated Bayesian segmentation method, was developed to estimate the CNV spectrum, with an average genome resolution of 5 MB from the reading depth of high-throughput scRNA-seq data (33).
CNV prevalence in B-ALL
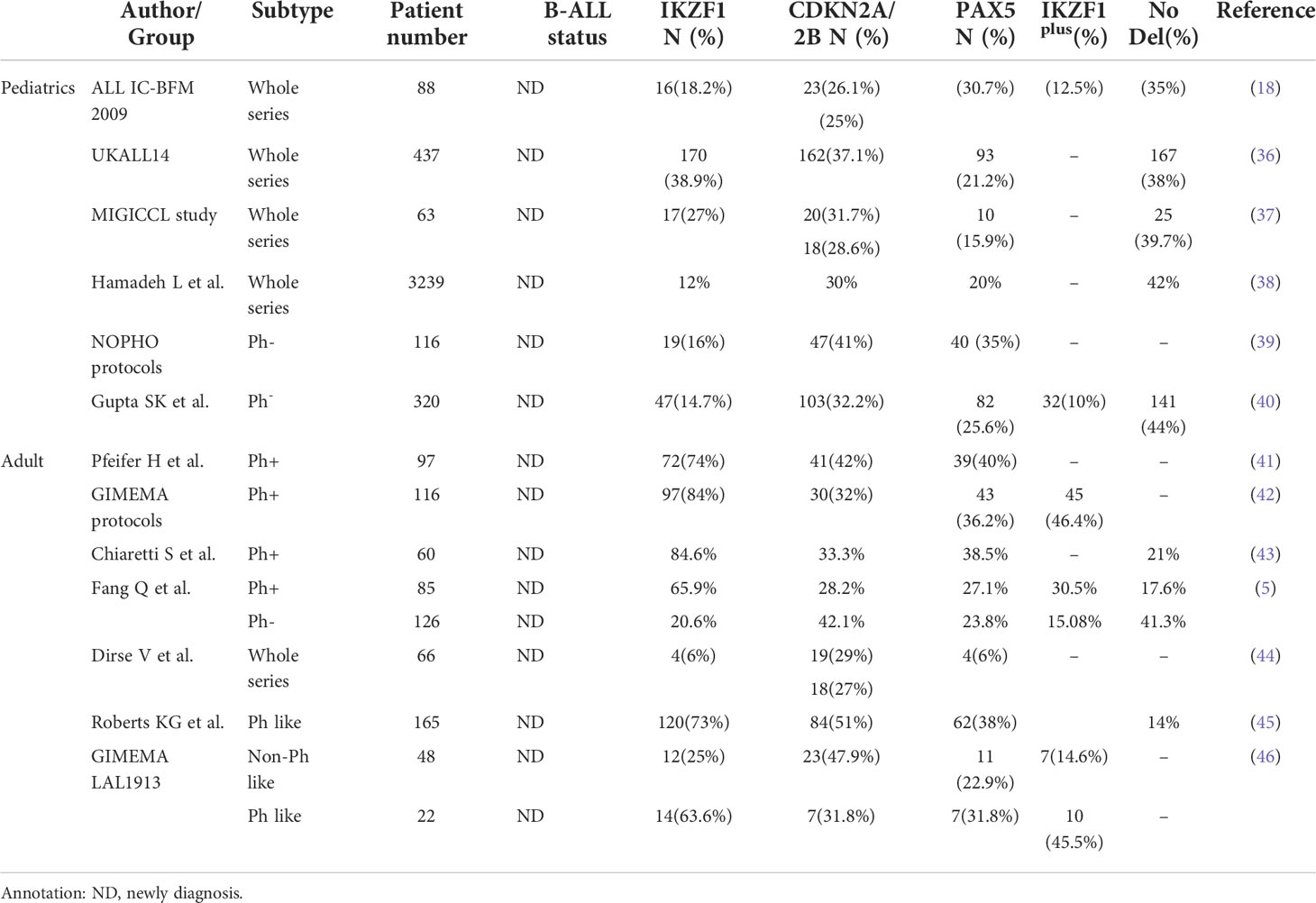
CNVs are frequently detected in B-ALL with considerable heterogeneity distribution (34). Overall, about 40%–49% of B-ALL carried gene CNVs that regulate early B-line cell differentiation and development-related genes (e.g., PAX5, IKZF1, and EBF1) and about 60% carry deletions of cell-cycle regulatory genes (e.g., CDKN2A/B and RB1) (5, 14). Broadly, CNVs occurred in 65% of pediatric B-ALL cases (35). Table 1 summarizes the incidence of common CNVs (including IKZF1, CDKN2A/B, and PAX5 genes) detected using MLPA from multiple cohorts (5, 18, 36–46); these occurred in the order of IKZF1, CDKN2A/B, and PAX5. In adult B-ALL cases, deletions of these genes were markedly enriched in the Philadelphia chromosome-positive (Ph+) B-ALL group than in the Ph− B-ALL group (82.4% vs. 58.7%, P<0.01) (18). Furthermore, Ribera J et al. (47) detected CNAs of 12 genetic regions in 142 adolescents and adults with de-novo precursor B-ALL using MLPA; CDKN2A/B deletion occurred in 59/142 (42%) cases, while IKZF1 deletion occurred in 49/142 (35%) cases.
Nevertheless, the research on CNV clones in relapsed B-ALL is limited. Despite being the preferred and widely used method for detecting CNVs in the related literature, MLPA might not be able to detect CNVs in samples presenting a low leukemia burden (carried <25% CNV clone). Moreover, CNVs in relapsed B-ALL remain unclear owing to limited paired B-ALL (newly diagnosed and relapsed) samples. The CNVs of relapsed B-ALL evolved from the diagnosis for examining specific gene content and clone size. By comparing the first-relapsed B-ALL to the newly diagnosed stage. RIBERA J et al. (48) established that CDKN2A/B, PAX5, and IKZF1 deletions were more frequent at relapse. Mullighan CG et al. (49) performed the genome-wide CNV and LOH analyses on matched diagnostic and relapse bone marrow samples from 61 pediatric patients with ALL, and identified a mean of 10.8 somatic CNV per B-ALL case and 7.1 CNVs per T-ALL case at diagnosis. In addition, they observed a significant increase in the mean number of CNVs per case in relapsed B-ALL samples (10.8 at diagnosis vs. 14.0 at relapse, P = 0.0005); however, no significant changes were observed in the lesion frequency in T-ALL. The majority (88.5%) of relapse samples harbored at least some of the CNAs present in the matched diagnosis sample, suggesting a common clonal origin, although 91.8% of samples showed a change in the pattern of CNVs from diagnosis to relapse. Of these cases, 34% acquired new CNVs, 12% exhibited loss of lesions present at diagnosis, and 46% both acquired new lesions and lost lesions present at diagnosis. Moreover, Ribera (48) compared CNVs at diagnosis and relapse, observing the trend to acquire homozygous CDKN2A/B deletions and a considerable increase in CNVs from diagnosis to the first relapse. Besides, evolution from an ancestral clone was the main pattern of clonal evolution. When focusing on the acquired CNVs in relapsed clones, gene alterations mostly correlated with proliferation and drug resistance.
Clinical significance of recurrent CNV genes in B-ALL
IKZF1 gene deletions
The Ikaros Zinc Finger 1 (IKZF1) gene, located at 7p12.2, encodes 519 amino acids by 8 full-length exons (50). Exons are essential for Ikaros gene functions, except for exon 1 (which does not participate in transcription) and exons 2, 3, and 7 (undetermined significance). IKZF1 deletions in both coding and noncoding regions might interfere with the gene activity and promote B-ALL progression through specific targets. For example, EBF1, MSH2, and MCL1 genes, as the target genes of IKZF1, play a vital role in affecting B-cell differentiation (EBF1 gene), DNA repair (MSH2 gene), and anti-apoptosis (MCL1 gene). The primary functions of the IKZF1 gene include B-cell differentiation blocking, metabolic reprogramming, leukemia microenvironment adhesion, disease relapse, and drug resistance (51).
Increasing evidence indicated that IKZF1 deletions mediate cellular drug resistance and relapse. For example, Rogers et.al (52) established that the IKZF1 deletion was resistant to dexamethasone, asparaginase, and daunorubicin by upregulating the JAK/STAT pathway. In addition, the IKZF1 deletion affects sensitivity to cytarabine by downregulating the SAMHD1 pathway (52); STEEGHS et. al (14) suggested that the loss of IKZF1 caused prednisolone resistance by elevating intracellular ATP and glucose levels, whereas drug sensitivity was recovered by inhibition of glycolysis. Moreover, IKZF1 deletion events, accompanied by CREBBP deletion or mutation, were common in relapsed pediatric B-ALL patients, which could correlate with the selective pressure of chemotherapeutic drugs on tumor cells (8).
Notably, IKZF1 gene deletions comprise localized large fragment deletions, single exon deletions, and other nonlocalized deletions, among which localized large fragment deletions are the most common. The loss of IKZF1 can be separated depending on its functional effect. While IK1–IK3 is considered a functional subtype, other subtypes are dominant-negative isoforms (DN isoforms), that is, functional defect subtype. In addition, IK6, often located in the cytoplasm, is a functional defect subtype with the complete loss of N-terminal zinc finger structure due to exon 4–7 deletion. IK6 functions as DN effects by isolating normal cytoplasmic proteins (53). Loss-of-function was designated as the total allelic inactivation. The loss of haploid dysfunction due to exon 2 deletion can decrease the Ikaros protein level.
Some studies reported IKZF1 deletions in around 15% of pediatric B-ALL cases and 30%–40% of adult B-ALL cases (40, 54). Perhaps, IKZF1 deletions in pediatric B-ALL are a hallmark of high-risk stratification and relapse independently carried by 70% of high-risk pediatric B-ALL (45, 55, 56).
In adult B-ALL, IKZF1 deletions were detected about 70% of Ph+ B-ALL cases (4, 5), around 15%–30% of Ph− B-ALL (53), and 40% of Ph-like B-ALL cases (13). In a study, IKZF1 deletions were mostly enriched in the adult Ph+ B-ALL group than in the Ph− B-ALL group (65.9% vs. 20.6%, P < 0.01) (5). Ribera reported that IKZF1 deletions were more prevalent in Ph+ B-ALL (52%) and correlated with advanced age and high white blood cell count (47, 57). Another study reported that IKZF1 deletions correlated with the CALF2 gene overexpression (P = 0.001), particularly in DN isoforms (P = 0.006), regardless of age (54). Furthermore, IKZF1 deletions with CRLF2 overexpression indicated a poor prognosis in both adult and pediatric B-ALL patients (54).
The prognostic impact of IKZF1 alterations in B-ALL remains debatable (58).
Kobitzsch (53) reported that loss-of-function not DN intragenic IKZF1 deletions correlated with an adverse prognosis in adult BCR-ABL-negative ABL. Yeoh AEJ et al. (59) compared the 5-year cumulative incidence of relapse (CIR) of Malaysia–Singapore MS2003 (n = 507) and MS2010 (n = 316) of pediatric B-ALL; the findings revealed that the loss of IKZF1 strongly correlated with a higher 5-year CIR (20.5% vs. 8.0%, P = 0.01) in MS2003. However, the treatment of IKZF1 deletion patients was intensified in MS2010, and the 5-year CIR presented no more significant difference in pediatric Ph− B-ALL (11.4% vs. 4.4%, P = 0.09). In addition, Ribera reported that IKZF1 deletions conferred a higher relapse incidence (40% vs. 58%, P = 0.048) and worse 5-year overall survival (OS; 29% vs. 50%, P = 0.023) than IKZF1 undeleted in Ph− B-ALL (47). Zhang W et al. (58) conducted a meta-analysis of the correlation between IKZF1 deletion and survival; IKZF1 lesions could independently predict unfavorable OS (hazard ratio [HR] 1.60, 95% confidence interval [CI] 1.25–2.06) and event-free survival (EFS; HR 1.67, 95% CI: 1.28–2.17) in Ph− B-ALL. In the EsPhALL cohort (pediatric BCR-ABL1-positive), IKZF1 deletions correlated with an unfavorable prognosis (4-year Disease Free Survival [DFS] of 51.9% ± 8.8% for IKZF1-deleted vs. 78.6% ± 13.9% for IKZF1 wild-type; P = 0.03). The massive analysis of IKZF1-loss patients demonstrated that it played a crucial role in Ph-like B-ALL. In ALL-BFM protocols, IKZF1 deletions acted as an independent risk factor, with the lower 5-year EFS than wild-type IKZF1 (0.69% vs. 0.85%, P < 0.0001) (51). Furthermore, IKZF1 deletions in Ph-like B-ALL multivariate models could precast EFS and OS (60, 61).
The response of early chemotherapy induction in patients with IKZF1 deletions was disappointing over the whole series. Several studies established that patients with IKZF1 lesions exhibited a high minimal residual disease (MRD) level (51, 60, 62). Reportedly, these patients could benefit more from intensive/alternate therapy than standard ones (4). Reportedly, the combination of vincristine and steroids in patients with IKZF1 deletions during maintenance treatment could be an effective and reasonable approach to prevent relapse. Dhédin N et al. (63) demonstrated that patients with IKZF1 deletions were likely to benefit from allogeneic HSCT (allo-HSCT) in terms of EFS (HR 0.42, 95% CI: 0.18–1.07, P = 0.025) and OS (HR 0.35, 95% CI: 0.16–0.75, P = 0.007), compared with non-IKZF1 alteration groups in adult Ph− B-ALL populations. However, whether the poor prognosis of IKZF1 overcame by stem cell transplantation warrants further investigation.
CDKN2A/CDKN2B gene deletion
Cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) is a common deletion in pediatric and adult B-ALL CNV profiles (1, 57, 64), as well as a major proposition of E2A-PBX1–positive B-ALL, but limited in MLL-rearranged patients (P = 0.005) (65). CDKN2A/B deletion is the major suppressor gene CNV in chromosome 9p21 (66). Compared with children, the CDKN2A/B incidence rate is marginally higher in adults (P = 0.002) (67). Moreover, 24.6% (14/57) of Ph-like patients present with enriched biallelic loss of CDKN2A/B (68). Reportedly, this lesion was highly representative of high white blood cell count, older age at initial diagnosis, and often accompanied by IKZF1 deletions (called I&C) (36, 69). Remarkably, clones with CDKN2A/B deletions detected in the initial diagnosis always persisted in relapse cases. Furthermore, CDKN2A/B presented a notable increase in the CNVs of relapse B-ALL (48).
In some studies, pediatric B-ALL patients with CDKN2A/B deletions exhibited a trend of shorter relapse time and EFS (35, 67), although the OS rate remains debatable. Kathiravan et. al (35) indicated that the 28-month EFS of CDKN2A/B lesions in ICICLE (Indian adaption of UKMRC2007 protocol) was notably decreased (42% vs. 90%, P = 0.0004) compared with non-CDKN2A/B deletions. Moreover, Braun M et al. (69) proved that CDKN2A deletions decreased the RFS significantly (HR 2.21, P = 0.028). No evidence indicates that loss of CDKN2A/B affected the prognosis in pediatric EORTC trials (70). Conversely, Feng J et al. (71) suggested that CDKN2A/B deletions inferred the 3-year EFS rate (69.8% vs. 89.2%, P = 0.000) and 3-year OS rate (89.4% vs. 94.7%, P = 0.037).
The frequency of adult CDKN2A/B deletions in the Ph-B-ALL group was much higher than in the Ph+ B-ALL group (39.7% vs. 24.7%, P = 0.041) (5). The prognostic value of CDKN2A/B in adults has been debated previously (35, 41, 44). Most studies emphasized that CDKN2A/B did not affect EFS and OS of adult patients with B-ALL. Only a few studies emphasized that CDKN2A/B adversely affected adult patients with B-ALL. Fang et. al (72). reported that CDKN2A/B is the vital relapsing and inferior prognostic marker for adult Ph− B-ALL (2-year OS: 38.2% vs. 80.3%, P = 0.002; 2-year RFS: 44% vs. 88.9%, P = 0.006). Messina M et al. (73) enrolled B-ALL-negative patients for BCR-ABL1 (Ph− B-ALL) population, including children, adolescents, and adults; the CDKN2A/B/RB1 deletion was reported as the negative prognostic factor (HR 2.12, P = 0.048) regardless of age. Pfeifer H et al. (41) suggested that CDKN2A/B deletions played an independent prognostic role in predicting the risk of relapse (DFS HR 2.621, P = 0.0054) and OS (HR 2.162, P = 0.014) in the adult Ph+ B-ALL population. Moreover, Dirse et. al (44). reported that CDKN2A/B decreased the EFS (multivariate HR 2.607, P = 0.034) in the whole series of adult B-ALL.
PAX5 gene deletion
The transcription factor paired box domain gene 5 (PAX5) was considered to regulate B-cell lineage differentiation and contribute to leukemogenesis in B-ALL (74, 75). PAX5 acts on the downstream transcription factors E2A and EBF1 and is crucial for B-line differentiation (76). In PAX5-deficient mice, the development of B cells in the bone marrow was blocked in the early Pro-B stage (77). The alterations of PAX5 comprise partial exon deletion on chromosome 9 (14%) and amplification of exon 2 or 5, resulting in frameshift mutation (7%). PAX5 deletions might increase genetic instability. Consequently, the probability of a secondary strike markedly increases and induces the recurrence and development of leukemia. In a study, PAX5 deletions decreased leukemia cell viability by inducing apoptotic cell death using a new ribozyme-derived isotype-specific knockdown system in the B-ALL cell model (77). Furthermore, transplantation experiments and exhaustive sequencing validated that PAX5 deletion made it sensitive to malignant transformation by forming an abnormal progenitor cell population (78).
As shown in Table 1, PAX5 deletions occurred in 15.9%–31.7% of pediatric Ph− B-ALL, 33% of pediatric Ph+ B-ALL (14), 27.1%–40% of adult Ph+ B-ALL, and 22.9%–23.8% of adult Ph-B-ALL (31.8%–38% Ph-like ALL) cases. No statistical difference has been reported between adult Ph− B-ALL and Ph+ B-ALL (27.1% vs. 27.8%, P = 0.549) cases (5). Most PAX5 deletions coexisted with CDKN2A/B deletions (83.3% of children and 100.0% of adults) and were commonly deleted in ETV6-RUNX1 B-ALL. The prognostic significance of PAX5 deletions in adult B-ALL also remains debatable. BHANDARI P et al. (64) claimed that PAX5 deletions were unsuitable for an independent prognostic marker for predicting prognosis because of no significant influence of RFS among B-ALL subgroups (P = 0.6839). Moreover, Iacobucci I et al. (79) reported no correlation between PAX5 deletions and OS (P = 0.3294) or DFS (P = 0.9249) in adult Ph+ B-ALL. In contrast, FEDULLO AL et al. (42) suggested that adult Ph+ B-ALL with PAX5 deletions showed shortened DFS (24.9% vs. 43.3%; P = 0.026).
In pediatric B-ALL groups, the prognosis of PAX5 deletion was strongly dependent on IKZF1 codeletion (61, 80). However, no significant prognostic correlation was observed in PAX5 deletions alone in children (74). In other words, the PAX5 -loss group presented no relapsing risk after excluding IKZF1 deletions. Indeed, double deletion of PAX5 and IKZF1 was improved by treatment intensification in MS2010, with 0% 5-year CIR than 80.0% in MS2003 (P = 0.05).
Prognostic relevance of integrated CNV profiling
Extensive research integrated gene CNV profile into pediatric B-ALL risk stratification (17). Moorman AV et al. (34) identified an 8-gene CNV panel, including IKZF1, CDKN2A/B, PAR1, BTG1, EBF1, PAX5, ETV6, and RB1, for stratifying the pediatric B-ALL risk level known as the UKALL-CNV classifier (Table 2). This tool has robust decision-making ability in intermediate-risk cytogenetics subgroups and even patients with different leukemia protocols baseline (37, 38). Besides, the UKALL-CNV classifier can refine the established cytogenetic risk groups.
Based on the Moorman’s criteria, Gupta SK et al. (83) subgrouped the MRD-negative intermediate-risk pediatric Ph− B-ALL into two subgroups with different EFS (77% vs. 38%, P = 0.045) and OS (90% vs. 30%, P = 0.037), whereas the criteria had no classifying power in MRD-positive groups (OS 75% vs. 57%, P = 0.293). A total of 3239 pediatric B-ALL cases were applied to validate the UKALL classifier (38). By integrating CNV and cytogenetic data, Hamadeh revised the overall genetic classification by defining four risk groups with distinct EFS rates (P < 0.001)—very good (91%), good (81%), intermediate (73%), and poor (54%). Stanulla M et al. (81) proposed a very-poor prognostic subtype defined as IKZF1plus subtype: IKZF1 occurred with additional mutations, containing CDKN2A, CDKN2B, PAX5, or PAR1 deletions simultaneously but without ERG deletions (Table 2). Besides, the IKZF1plus 5-year EFS rate in pediatrics was 53% ± 6% compared with 79% ± 5% in adults (P < 0.001).
In adult Ph+ B-ALL, IKZF1plus negatively affected the survival outcome than IKZF1 alone (DFS: 43.3% vs. 24.9%, P = 0.026; OS: 62.6% vs. 40.2%, P = 0.02) (42). Reportedly, IKZF1plus patients had been under similar conditions in the GIMEMA LAL2116 cohort (DFS: 84.5% vs. 54.5%, P = 0.026) and GIMEMA LAL1509 protocol (DFS: 0% vs. 60%, P = 0.0008; OS: 20% vs. 69.5%, P = 0.0068) (43, 82). However, the prognostic significance of IKZF1 plus in adult Ph+ B-ALL was not detected (5). Likewise, Chiaretti S et al. (46) reported no statistical correlation between IKZF1plus in adult Ph-like B-ALL (HR 1.869, 95% CI: 0.49–6.67, P = 0.339). In addition, GUPTA SK et al. (40) proposed the “MRplus” risk score system by integrating IKZF1plus and the UKALL-CNV classifier subtyping to better classify pediatric Ph− B-ALL prognosis. The 0, 1, 2 groups defined by MR plus system markedly discriminated postinduction remission response and 4-year OS (Table 2).
Considering the primary chromosomal abnormalities significantly correlate with the CNV frequency, Moorman AV et al. (36) revised the stratification by adding cytogenetic risk factors, like KMT2A fusions, complex karyotype and low hypodiploidy/near-triploidy. The new risk system could predict the 3-year OS (64% vs. 47%; HR 1.65 95% CI: 1.27–2.12, P < 0.001).
Future perspectives
Many studies have proved that CNV is a common molecular abnormality in the development of B-ALL (48). Current evidence suggests that the CNV pattern of adult and pediatric B-ALL has a different cytogenetic abnormality and pathological significances. Moreover, growing evidence indicates that high number and diverse CNVs observed are acquired in the process of disease relapsing (37). This study mainly discussed the clinical significance of the CNV spectrum, which has been well recognized in patients with B-ALL. Among them, IKZF1, CDKN2A/B, and PAX5 are the leading prevalent gene alterations in B-ALL (47). Moreover, these CNVs in Ph-like and Ph+ B-ALL remain equally frequent (68). However, some research of gene prognostic value is inconsistent, which could be because of difference in enrolled patients and treatment regimen. Undoubtedly, CNVs guided the risk of relapsing and survival outcome of both pediatric and adult B-ALL (84). Intensive chemotherapy combined with allo-HSCT is expected to overcome the adverse impact of CNVs. Perhaps, the combination of intensive chemotherapy and allo-HSCT could overcome the adverse impact of CNVs.
Typically, the risk stratification of B-ALL based on the CNV profiles is largely limited to the pediatric population (36). Currently, the IKZF1plus and UKALL-CNV classifier are broadly promoted in the adult B-ALL classification (5, 37, 43). Considering the different cytogenetic patterns of adults and children, the risk system in adults warrants revision in future. With the new exploration of new targets of rearrangement in B-ALL (e.g., DUX4, ZNF384, and MEF2D), the survival risk stratification system will be consistently updated in the future. Besides, further research will help identify new prognostic indicators and potential therapeutic targets.
In conclusion, this review characterizes B-ALL–related copy number events, which is valuable for precise patient subgroup stratification. In addition, this study provides insights into the new immunotherapy-based approaches and tailored treatment strategies for patients with B-ALL. Nevertheless, additional multicenter survival data will be needed for further verification in the future.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author contributions
YS drafted the manuscript. QF and YM revised the manuscript. All authors read and approved the final manuscript.
Funding
Basic Scientific Research Project of National Universities (332021059).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Patel S, Mason CC, Glenn MJ, Paxton CN, South ST, Cessna MH, et al. Genomic analysis of adult b-ALL identifies potential markers of shorter survival. Leukemia Res (2017) 56:44–51. doi: 10.1016/j.leukres.2017.01.034
2. Lejman M, Chałupnik A, Chilimoniuk Z, Dobosz M. Genetic biomarkers and their clinical implications in b-cell acute lymphoblastic leukemia in children. Int J Mol Sci (2022) 23(5):2755. doi: 10.3390/ijms23052755
3. Ueno H, Yoshida K, Shiozawa Y, Nannya Y, Iijima-Yamashita Y, Kiyokawa N, et al. Landscape of driver mutations and their clinical impacts in pediatric b-cell precursor acute lymphoblastic leukemia. Blood Adv (2020) 4(20):5165–73. doi: 10.1182/bloodadvances.2019001307
4. Mitchell RJ, Kirkwood AA, Barretta E, Clifton-Hadley L, Lawrie E, Lee S, et al. IKZF1 alterations are not associated with outcome in 498 adults with b-precursor ALL enrolled in the UKALL14 trial. Blood Adv (2021) 5(17):3322–32. doi: 10.1182/bloodadvances.2021004430
5. Fang Q, Song Y, Gong X, Wang J, Li Q, Liu K, et al. Gene deletions and prognostic values in b-linage acute lymphoblastic leukemia. Front Oncol (2021) 11:677034. doi: 10.3389/fonc.2021.677034
6. O'Brien MM, Ji L, Shah NN, Rheingold SR, Bhojwani D, Yuan CM, et al. Phase II trial of inotuzumab ozogamicin in children and adolescents with relapsed or refractory b-cell acute lymphoblastic leukemia: Children's oncology group protocol AALL1621. J Clin Oncol (2022) 40(9):956–67. doi: 10.1200/JCO.21.01693
7. Inaba H, Mullighan CG. Pediatric acute lymphoblastic leukemia. Haematol (2020) 105(11):2524–39. doi: 10.3324/haematol.2020.247031
8. Sayyab S, Lundmark A, Larsson M, Ringnér M, Nystedt S, Marincevic-Zuniga Y, et al. Mutational patterns and clonal evolution from diagnosis to relapse in pediatric acute lymphoblastic leukemia. Sci Rep (2021) 11(1):15988. doi: 10.1038/s41598-021-95109-0
9. Jabbour E, Short NJ, Ravandi F, Huang X, Daver N, DiNardo CD, et al. Combination of hyper-CVAD with ponatinib as first-line therapy for patients with Philadelphia chromosome-positive acute lymphoblastic leukaemia: long-term follow-up of a single-centre, phase 2 study. Lancet Haematol (2018) 5(12):e618–e27. doi: 10.1016/S2352-3026(18)30176-5
10. Locatelli F, Zugmaier G, Rizzari C, Morris JD, Gruhn B, Klingebiel T, et al. Effect of blinatumomab vs chemotherapy on event-free survival among children with high-risk first-relapse b-cell acute lymphoblastic leukemia: A randomized clinical trial. JAMA (2021) 325(9):843–54. doi: 10.1001/jama.2021.0987
11. Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in children and young adults with b-cell lymphoblastic leukemia. N Engl J Med (2018) 378(5):439–48. doi: 10.1056/NEJMoa1709866
12. Ling Y, Xu N, Zhao K, Han L, Zhang Q, Fan Z, et al. Allogeneic hematopoietic cell transplant overcomes the poor prognostic value of CDKN2 deletion in adult b-lineage acute lymphoblastic leukemia. Cancer Lett (2021) 510:59–66. doi: 10.1016/j.canlet.2021.04.009
13. Paietta E, Roberts KG, Wang V, Gu Z, Buck GAN, Pei D, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative b-ALL. Blood (2021) 138(11):948–58. doi: 10.1182/blood.2020010144
14. Steeghs EMP, Boer JM, Hoogkamer AQ, Boeree A, de Haas V, de Groot-Kruseman HA, et al. Copy number alterations in b-cell development genes, drug resistance, and clinical outcome in pediatric b-cell precursor acute lymphoblastic leukemia. Sci Rep (2019) 9(1):4634. doi: 10.1038/s41598-019-41078-4
15. Gökbuget N, Dombret H, Ribera JM, Fielding AK, Advani A, Bassan R, et al. International reference analysis of outcomes in adults with b-precursor ph-negative relapsed/refractory acute lymphoblastic leukemia. Haematologica (2016) 101(12):1524–33. doi: 10.3324/haematol.2016.144311
16. Gregory S. Adult acute lymphoblastic leukemia: Treatment and management updates. Semin Oncol Nurs (2019) 35(6):150951. doi: 10.1016/j.soncn.2019.150951
17. Erbilgin Y, Firtina S, Mercan S, Hatirnaz Ng O, Karaman S, Tasar O, et al. Prognostic gene alterations and clonal changes in childhood b-ALL. Leukemia Res (2019) 83:106159. doi: 10.1016/j.leukres.2019.05.009
18. Crepinsek K, Marinsek G, Kavcic M, Prelog T, Kitanovski L, Jazbec J, et al. Clinical impacts of copy number variations in b-cell differentiation and cell cycle control genes in pediatric b-cell acute lymphoblastic leukemia: a single centre experience. Radiol Oncol (2021) 56(1):92–101. doi: 10.2478/raon-2021-0050
19. Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinf (2013) 14 Suppl 11(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1
20. Bartlett J, Amemiya Y, Arts H, Bayani J, Eng B, Grafodatskaya D, et al. Multisite verification of the accuracy of a multi-gene next generation sequencing panel for detection of mutations and copy number alterations in solid tumours. PLoS One (2021) 16(10):e0258188. doi: 10.1371/journal.pone.0258188
21. Nibourel O, Guihard S, Roumier C, Pottier N, Terre C, Paquet A, et al. Copy-number analysis identified new prognostic marker in acute myeloid leukemia. Leukemia (2017) 31(3):555–64. doi: 10.1038/leu.2016.265
22. Henrichsen CN, Chaignat E, Reymond A. Copy number variants, diseases and gene expression. Hum Mol Genet (2009) 18(R1):R1–8. doi: 10.1093/hmg/ddp011
23. Zhou B, Ho SS, Zhang X, Pattni R, Haraksingh RR, Urban AE. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J Med Genet (2018) 55(11):735–43. doi: 10.1136/jmedgenet-2018-105272
24. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res (2002) 30(12):e57. doi: 10.1093/nar/gnf056
25. Kiss R, Gángó A, Benard-Slagter A, Egyed B, Haltrich I, Hegyi L, et al. Comprehensive profiling of disease-relevant copy number aberrations for advanced clinical diagnostics of pediatric acute lymphoblastic leukemia. Mod Pathol (2020) 33(5):812–24. doi: 10.1038/s41379-019-0423-5
26. Suvakov M, Panda A, Diesh C, Holmes I, Abyzov A. CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing. Gigascience (2021) 10(11):1–9. doi: 10.1093/gigascience/giab074
27. Rack K, De Bie J, Ameye G, Gielen O, Demeyer S, Cools J, et al. Optimizing the diagnostic workflow for acute lymphoblastic leukemia by optical genome mapping. Am J Hematol (2022) 97(5):548–61. doi: 10.1002/ajh.26487
28. Gerding WM, Tembrink M, Nilius-Eliliwi V, Mika T, Dimopoulos F, Ladigan-Badura S, et al. Optical genome mapping reveals additional prognostic information compared to conventional cytogenetics in AML/MDS patients. Int J Cancer (2022) 150(12):1998–2011. doi: 10.1002/ijc.33942
29. Lühmann JL, Stelter M, Wolter M, Kater J, Lentes J, Bergmann AK, et al. The clinical utility of optical genome mapping for the assessment of genomic aberrations in acute lymphoblastic leukemia. Cancers (Basel) (2021) 13(17):4388. doi: 10.3390/cancers13174388
30. Molparia B, Nichani E, Torkamani A. Assessment of circulating copy number variant detection for cancer screening. PLoS One (2017) 12(7):e0180647. doi: 10.1371/journal.pone.0180647
31. Bařinka J, Hu Z, Wang L, Wheeler DA, Rahbarinia D, McLeod C, et al. RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data. Leukemia (2022) 36:1492–8. doi: 10.1038/s41375-022-01547-8
32. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (2014) 344(6190):1396–401. doi: 10.1126/science.1254257
33. Gao R, Bai S, Henderson YC, Lin Y, Schalck A, Yan Y, et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol (2021) 39(5):599–608. doi: 10.1038/s41587-020-00795-2
34. Moorman AV, Enshaei A, Schwab C, Wade R, Chilton L, Elliott A, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood (2014) 124(9):1434–44. doi: 10.1182/blood-2014-03-562918
35. Kathiravan M, Singh M, Bhatia P, Trehan A, Varma N, Sachdeva MS, et al. Deletion of CDKN2A/B is associated with inferior relapse free survival in pediatric b cell acute lymphoblastic leukemia. Leuk Lymphoma (2019) 60(2):433–41. doi: 10.1080/10428194.2018.1482542
36. Moorman AV, Barretta E, Butler ER, Ward EJ, Twentyman K, Kirkwood AA, et al. Prognostic impact of chromosomal abnormalities and copy number alterations in adult b-cell precursor acute lymphoblastic leukaemia: a UKALL14 study. Leukemia (2022) 36(3):625–36. doi: 10.1038/s41375-021-01448-2
37. Rosales-Rodríguez B, Núñez-Enríquez JC, Velázquez-Wong AC, González-Torres C, Gaytán-Cervantes J, Jiménez-Hernández E, et al. Copy number alterations are associated with the risk of very early relapse in pediatric b-lineage acute lymphoblastic leukemia: A nested case-control MIGICCL study. Arch Med Res (2021) 52(4):414–22. doi: 10.1016/j.arcmed.2020.12.013
38. Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, et al. Validation of the united kingdom copy-number alteration classifier in 3239 children with b-cell precursor ALL. Blood Advances (2019) 3(2):148–57. doi: 10.1182/bloodadvances.2018025718
39. Ofverholm I, Tran AN, Heyman M, Zachariadis V, Nordenskjöld M, Nordgren A, et al. Impact of IKZF1 deletions and PAX5 amplifications in pediatric b-cell precursor ALL treated according to NOPHO protocols. Leukemia (2013) 27(9):1936–9. doi: 10.1038/leu.2013.92
40. Gupta SK, Bakhshi S, Kamal VK, Gupta R, Sharma P, Pushpam D, et al. Proposal and clinical application of molecular genetic risk scoring system, "MRplus", for BCR-ABL1 negative pediatric b-cell acute lymphoblastic leukemia- report from a single centre. Leuk Res (2021) 111:106683. doi: 10.1016/j.leukres.2021.106683
41. Pfeifer H, Raum K, Markovic S, Nowak V, Fey S, Obländer J, et al. Genomic CDKN2A/2B deletions in adult ph(+) ALL are adverse despite allogeneic stem cell transplantation. Blood (2018) 131(13):1464–75. doi: 10.1182/blood-2017-07-796862
42. Fedullo AL, Messina M, Elia L, Piciocchi A, Gianfelici V, Lauretti A, et al. Prognostic implications of additional genomic lesions in adult Philadelphia chromosome-positive acute lymphoblastic leukemia. Haematologica (2019) 104(2):312–8. doi: 10.3324/haematol.2018.196055
43. Chiaretti S, Ansuinelli M, Vitale A, Elia L, Matarazzo M, Piciocchi A, et al. A multicenter total therapy strategy for de novo adult Philadelphia chromosome positive acute lymphoblastic leukemia patients: final results of the GIMEMA LAL1509 protocol. Haematologica (2021) 106(7):1828–38. doi: 10.3324/haematol.2020.260935
44. Dirse V, Bertasiute A, Gineikiene E, Zvirblis T, Dambrauskiene R, Gerbutavicius R, et al. A population-based single nucleotide polymorphism array analysis of genomic aberrations in younger adult acute lymphoblastic leukemia patients. Genes Chromosomes Cancer (2015) 54(5):326–33. doi: 10.1002/gcc.22246
45. Roberts KG, Gu Z, Payne-Turner D, McCastlain K, Harvey RC, Chen IM, et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J Clin Oncol (2017) 35(4):394–401. doi: 10.1200/JCO.2016.69.0073
46. Chiaretti S, Messina M, Della Starza I, Piciocchi A, Cafforio L, Cavalli M, et al. Philadelphia-Like acute lymphoblastic leukemia is associated with minimal residual disease persistence and poor outcome. first report of the minimal residual disease-oriented GIMEMA LAL1913. Haematologica (2021) 106(6):1559–68. doi: 10.3324/haematol.2020.247973
47. Ribera J, Morgades M, Zamora L, Montesinos P, Gómez-Seguí I, Pratcorona M, et al. Prognostic significance of copy number alterations in adolescent and adult patients with precursor b acute lymphoblastic leukemia enrolled in PETHEMA protocols. Cancer (2015) 121(21):3809–17. doi: 10.1002/cncr.29579
48. Ribera J, Zamora L, Morgades M, Mallo M, Solanes N, Batlle M, et al. Copy number profiling of adult relapsed b-cell precursor acute lymphoblastic leukemia reveals potential leukemia progression mechanisms. Genes Chromosomes Cancer (2017) 56(11):810–20. doi: 10.1002/gcc.22486
49. Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science (2008) 322(5906):1377–80. doi: 10.1126/science.1164266
50. Yamashita M, Morio T. Inborn errors of IKAROS and AIOLOS. Curr Opin Immunol (2021) 72:239–48. doi: 10.1016/j.coi.2021.06.010
51. Dörge P, Meissner B, Zimmermann M, Möricke A, Schrauder A, Bouquin JP, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica (2013) 98(3):428–32. doi: 10.3324/haematol.2011.056135
52. Rogers JH, Gupta R, Reyes JM, Gundry MC, Medrano G, Guzman A, et al. Modeling IKZF1 lesions in b-ALL reveals distinct chemosensitivity patterns and potential therapeutic vulnerabilities. Blood Adv (2021) 5(19):3876–90. doi: 10.1182/bloodadvances.2020002408
53. Kobitzsch B, Gökbuget N, Schwartz S, Reinhardt R, Brüggemann M, Viardot A, et al. Loss-of-function but not dominant-negative intragenic IKZF1 deletions are associated with an adverse prognosis in adult BCR-ABL-negative acute lymphoblastic leukemia. Haematologica (2017) 102(10):1739–47. doi: 10.3324/haematol.2016.161273
54. Maciel ALT, Barbosa TDC, Blunck CB, Wolch K, AdAL M, da Costa ES, et al. IKZF1 deletions associate with CRLF2 overexpression leading to a poor prognosis in b-cell precursor acute lymphoblastic leukaemia. Trans Oncol (2022) 15(1):101291. doi: 10.1016/j.tranon.2021.101291
55. Clappier E, Grardel N, Bakkus M, Rapion J, De Moerloose B, Kastner P, et al. IKZF1 deletion is an independent prognostic marker in childhood b-cell precursor acute lymphoblastic leukemia, and distinguishes patients benefiting from pulses during maintenance therapy: results of the EORTC children's leukemia group study 58951. Leukemia (2015) 29(11):2154–61. doi: 10.1038/leu.2015.134
56. van der Veer A, Zaliova M, Mottadelli F, De Lorenzo P, Te Kronnie G, Harrison CJ, et al. IKZF1 status as a prognostic feature in BCR-ABL1-positive childhood ALL. Blood (2014) 123(11):1691–8. doi: 10.1182/blood-2013-06-509794
57. Gupta SK, Bakhshi S, Kumar L, Kamal VK, Kumar R. Gene copy number alteration profile and its clinical correlation in b-cell acute lymphoblastic leukemia. Leuk Lymphoma (2017) 58(2):333–42. doi: 10.1080/10428194.2016.1193855
58. Zhang W, Kuang P, Li H, Wang F, Wang Y. Prognostic significance of IKZF1 deletion in adult b cell acute lymphoblastic leukemia: a meta-analysis. Ann Hematol (2017) 96(2):215–25. doi: 10.1007/s00277-016-2869-6
59. Yeoh AEJ, Lu Y, Chin WHN, Chiew EKH, Lim EH, Li Z, et al. Intensifying treatment of childhood b-lymphoblastic leukemia with IKZF1 deletion reduces relapse and improves overall survival: Results of Malaysia-Singapore ALL 2010 study. J Clin Oncol (2018) 36(26):2726–35. doi: 10.1200/JCO.2018.78.3050
60. van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with b-cell precursor ALL. Blood (2013) 122(15):2622–9. doi: 10.1182/blood-2012-10-462358
61. Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med (2009) 360(5):470–80. doi: 10.1056/NEJMoa0808253
62. Zhang J, Xu XJ, Liu L, Song H, Shen H, Xu W, et al. Clinical and genetic characteristics of IKZF1 mutation in Chinese children with b-cell acute lymphoblastic leukemia. Front Genet (2022) 13:822832. doi: 10.3389/fgene.2022.822832
63. Dhédin N, Huynh A, Maury S, Tabrizi R, Beldjord K, Asnafi V, et al. Role of allogeneic stem cell transplantation in adult patients with ph-negative acute lymphoblastic leukemia. Blood (2015) 125(16):2486–96. doi: 10.1182/blood-2014-09-599894
64. Bhandari P, Ahmad F, Das BR. Molecular profiling of gene copy number abnormalities in key regulatory genes in high-risk b-lineage acute lymphoblastic leukemia: frequency and their association with clinicopathological findings in Indian patients. Med Oncol (2017) 34(5):92. doi: 10.1007/s12032-017-0940-3
65. Zhou B, Chu X, Tian H, Liu T, Liu H, Gao W, et al. The clinical outcomes and genomic landscapes of acute lymphoblastic leukemia patients with E2A-PBX1: A 10-year retrospective study. Am J Hematol (2021) 96(11):1461–71. doi: 10.1002/ajh.26324
66. Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S, et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1–positive acute lymphoblastic leukemia patients. Clin Cancer Res (2011) 17(23):7413–23. doi: 10.1158/1078-0432.CCR-11-1227
67. Agarwal M, Bakhshi S, Dwivedi SN, Kabra M, Shukla R, Seth R. Cyclin dependent kinase inhibitor 2A/B gene deletions are markers of poor prognosis in Indian children with acute lymphoblastic leukemia. Pediatr Blood Cancer (2018) 65(6):e27001. doi: 10.1002/pbc.27001
68. Hrabovsky S, Vrzalova Z, Stika J, Jelinkova H, Jarosova M, Navrkalova V, et al. Genomic landscape of b-other acute lymphoblastic leukemia in an adult retrospective cohort with a focus on BCR-ABL1-like subtype. Acta Oncol (2021) 60(6):760–70. doi: 10.1080/0284186X.2021.1900908
69. Braun M, Pastorczak A, Fendler W, Madzio J, Tomasik B, Taha J, et al. Biallelic loss of CDKN2A is associated with poor response to treatment in pediatric acute lymphoblastic leukemia. Leuk Lymphoma (2017) 58(5):1162–71. doi: 10.1080/10428194.2016.1228925
70. Mirebeau D, Acquaviva C, Suciu S, Bertin R, Dastugue N, Robert A, et al. The prognostic significance of CDKN2A, CDKN2B and MTAP inactivation in b-lineage acute lymphoblastic leukemia of childhood. Results of the EORTC studies 58881 and 58951. Haematologica (2006) 91(7):881–5. doi: 10.1038/sj.leu.2404282
71. Feng J, Guo Y, Yang W, Zou Y, Zhang L, Chen Y, et al. Childhood acute b-lineage lymphoblastic leukemia with CDKN2A/B deletion is a distinct entity with adverse genetic features and poor clinical outcomes. Front Oncol (2022) 12:878098. doi: 10.3389/fonc.2022.878098
72. Fang Q, Yuan T, Li Y, Feng J, Gong X, Li Q, et al. Prognostic significance of copy number alterations detected by multi-link probe amplification of multiple genes in adult acute lymphoblastic leukemia. Oncol Lett (2018) 15(4):5359–67. doi: 10.3892/ol.2018.7985
73. Messina M, Chiaretti S, Fedullo AL, Piciocchi A, Puzzolo MC, Lauretti A, et al. Clinical significance of recurrent copy number aberrations in b-lineage acute lymphoblastic leukaemia without recurrent fusion genes across age cohorts. Br J Haematol (2017) 178(4):583–7. doi: 10.1111/bjh.14721
74. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of b-progenitor acute lymphoblastic leukemia. Nat Genet (2019) 51(2):296–307. doi: 10.1038/s41588-018-0315-5
75. Shahjahani M, Norozi F, Ahmadzadeh A, Shahrabi S, Tavakoli F, Asnafi AA, et al. The role of Pax5 in leukemia: diagnosis and prognosis significance. Med Oncol (2015) 32(1):360. doi: 10.1007/s12032-014-0360-6
76. Cobaleda C, Jochum W, Busslinger M. Conversion of mature b cells into T cells by dedifferentiation to uncommitted progenitors. Nature (2007) 449(7161):473–7. doi: 10.1038/nature06159
77. Cozma D, Yu D, Hodawadekar S, Azvolinsky A, Grande S, Tobias JW, et al. B cell activator PAX5 promotes lymphomagenesis through stimulation of b cell receptor signaling. J Clin Invest (2007) 117(9):2602–10. doi: 10.1172/JCI30842
78. Martín-Lorenzo A, Hauer J, Vicente-Dueñas C, Auer F, González-Herrero I, García-Ramírez I, et al. Infection exposure is a causal factor in b-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discovery (2015) 5(12):1328–43. doi: 10.1158/2159-8290.CD-15-0892
79. Iacobucci I, Lonetti A, Paoloni F, Papayannidis C, Ferrari A, Storlazzi CT, et al. The PAX5 gene is frequently rearranged in BCR-ABL1-positive acute lymphoblastic leukemia but is not associated with outcome. a report on behalf of the GIMEMA acute leukemia working party. Haematologica (2010) 95(10):1683–90. doi: 10.3324/haematol.2009.020792
80. Li Z, Lee SHR, Chin WHN, Lu Y, Jiang N, Lim EH, et al. Distinct clinical characteristics of DUX4- and PAX5-altered childhood b-lymphoblastic leukemia. Blood Advances (2021) 5(23):5226–38. doi: 10.1182/bloodadvances.2021004895
81. Stanulla M, Dagdan E, Zaliova M, Möricke A, Palmi C, Cazzaniga G, et al. IKZF1plus defines a new minimal residual disease–dependent very-poor prognostic profile in pediatric b-cell precursor acute lymphoblastic leukemia. J Clin Oncol (2018) 36(12):1240–9. doi: 10.1200/JCO.2017.74.3617
82. Foà R, Bassan R, Vitale A, Elia L, Piciocchi A, Puzzolo MC, et al. Dasatinib-blinatumomab for ph-positive acute lymphoblastic leukemia in adults. N Engl J Med (2020) 383(17):1613–23. doi: 10.1056/NEJMoa2016272
83. Gupta SK, Bakhshi S, Chopra A, Kamal VK. Molecular genetic profile in BCR-ABL1 negative pediatric b-cell acute lymphoblastic leukemia can further refine outcome prediction in addition to that by end-induction minimal residual disease detection. Leuk Lymphoma (2018) 59(8):1899–904. doi: 10.1080/10428194.2017.1408087
Keywords: gene deletion, copy number variation, acute lymphoblastic leukemia, CDKN2A/2B deletion, IKZF1 deletion, PAX5 deletion, prognosis
Citation: Song Y, Fang Q and Mi Y (2022) Prognostic significance of copy number variation in B-cell acute lymphoblastic leukemia. Front. Oncol. 12:981036. doi: 10.3389/fonc.2022.981036
Received: 29 June 2022; Accepted: 11 July 2022;
Published: 04 August 2022.
Edited by:
Kevin J. Ni, St. George Hospital Cancer Care Centre, AustraliaReviewed by:
Yinghua Li, Harbin Medical University, ChinaChunyan Ji, Qilu Hospital, Shandong University, China
Copyright © 2022 Song, Fang and Mi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiuyun Fang, ZmFuZ3FpdXl1bkBpaGNhbXMuYWMuY24=; Yingchang Mi, eWNobWlAaWhjYW1zLmFjLmNu
 Yang Song
Yang Song Qiuyun Fang
Qiuyun Fang Yingchang Mi
Yingchang Mi