
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 18 August 2022
Sec. Cancer Immunity and Immunotherapy
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.979569
This article is part of the Research TopicTranscriptional Control in Normal and Malignant B-LymphocytesView all 8 articles
 Catharina Muylaert
Catharina Muylaert Lien Ann Van Hemelrijck
Lien Ann Van Hemelrijck Anke Maes
Anke Maes Kim De Veirman
Kim De Veirman Eline Menu
Eline Menu Karin Vanderkerken
Karin Vanderkerken Elke De Bruyne*
Elke De Bruyne*Drug resistance (DR) of cancer cells leading to relapse is a huge problem nowadays to achieve long-lasting cures for cancer patients. This also holds true for the incurable hematological malignancy multiple myeloma (MM), which is characterized by the accumulation of malignant plasma cells in the bone marrow (BM). Although new treatment approaches combining immunomodulatory drugs, corticosteroids, proteasome inhibitors, alkylating agents, and monoclonal antibodies have significantly improved median life expectancy, MM remains incurable due to the development of DR, with the underlying mechanisms remaining largely ill-defined. It is well-known that MM is a heterogeneous disease, encompassing both genetic and epigenetic aberrations. In normal circumstances, epigenetic modifications, including DNA methylation and posttranslational histone modifications, play an important role in proper chromatin structure and transcriptional regulation. However, in MM, numerous epigenetic defects or so-called ‘epimutations’ have been observed and this especially at the level of DNA methylation. These include genome-wide DNA hypomethylation, locus specific hypermethylation and somatic mutations, copy number variations and/or deregulated expression patterns in DNA methylation modifiers and regulators. The aberrant DNA methylation patterns lead to reduced gene expression of tumor suppressor genes, genomic instability, DR, disease progression, and high-risk disease. In addition, the frequency of somatic mutations in the DNA methylation modifiers seems increased in relapsed patients, again suggesting a role in DR and relapse. In this review, we discuss the recent advances in understanding the involvement of aberrant DNA methylation patterns and/or DNA methylation modifiers in MM development, progression, and relapse. In addition, we discuss their involvement in MM cell plasticity, driving myeloma cells to a cancer stem cell state characterized by a more immature and drug-resistant phenotype. Finally, we briefly touch upon the potential of DNA methyltransferase inhibitors to prevent relapse after treatment with the current standard of care agents and/or new, promising (immuno) therapies.
Multiple myeloma (MM) is an incurable B-cell malignancy characterized by the uncontrolled proliferation and accumulation of malignant plasma cells (PC) in the bone marrow (BM) (1). MM accounts for 1% of all cancers and around 10% of all hematological cancers, being the second most common hematological malignancy (2). In 2020, 176,404 new cases of MM and 117,077 deaths caused by MM (accounting for 1.2% of all cancer deaths) were observed worldwide (3). Age is the most significant risk factor, as MM occurs most frequently in the elderly population with the median age of diagnosis being 65. Consequently, due to an aging population in western countries, the global burden of MM is expected to further increase in the following years (4).
MM is often preceded by a premalignant condition called MGUS or monoclonal gammopathy of unknown significance. An abnormal increase of one type (clone) of PCs can be observed in individuals presenting with MGUS, resulting in the excessive production of one specific immunoglobulin (monoclonal antibody/mAb) termed the M-protein or M-component (5). MGUS progresses to MM at a rate of 1% a year (6, 7). A minor group of the elderly population also suffers from smoldering multiple myeloma (SMM), which is an intermediate phenotype between MGUS and MM. SMM has a 10% risk of developing into MM within the first five years, 5% per year for the following five years, and 1% per year after ten years (8). Patients evolving from MGUS and SMM to MM start to exhibit specific symptoms. In general, MM is diagnosed based on the presence of ≥10% clonal BM plasma cells (BMPC) and the presence of at least one of the MM defining events (MDE). MDE include a serum free light chain (FLC) ratio of ≥ 100, a proportion of clonal BMPC of ≥60%, the detection of one or more focal lesions through MRI, and the presence of one or more CRAB symptoms, including hypercalcemia, renal failure, anemia, and lytic bone lesions (9, 10).
Although MM remains an incurable disease, new treatments developed over the last years have significantly increased the median life expectancy by six to ten years. There are two major lines of treatment options based on transplantation eligibility (age and comorbidities) and risk stratification (11). For patients that are fit enough, first line treatment consists of an induction therapy combining two or three standard of care (SoC) agents, including immunomodulatory drugs (IMiDs; lenalidomide), corticosteroids (dexamethasone), proteasome inhibitors (PI; bortezomib; Bz), and/or monoclonal antibodies (daratumumab, elotuzumab) (10). This induction therapy is then followed by a high dose melphalan treatment combined with an autologous stem cell transplantation (SCT) (12). The second group, people that are not eligible for the SCT, will only be treated with different combinations of the previously mentioned drugs. Although most patients initially respond very well to treatment, most of them will eventually relapse and with each new round of therapy they will respond less until they become completely refractory. Therefore, new therapies are still being developed, like chimeric antigen receptor (CAR) T-cell therapy, antibody drug conjugates (ADC), and T-cell engagers, which are currently being tested in clinical trials (13–16). Although clinical trials showed encouraging results, patients are still relapsing. Hence, there is still no definite treatment to cure MM.
One of the most important reasons for relapse is the development of drug resistance (DR) against all SoC agents. DR can be established on different levels. The close interaction between MM cells and the BM niche is one of the major mechanisms playing a role in the development of DR. Two categories of BM-related DR exist, namely cell adhesion mediated DR (CAM-DR), caused through the interactions between the MM cells and the cellular compartment (BM stromal cells, endothelial cells, osteoblasts, osteoclasts, and immune cells) and/or extracellular matrix, and soluble factor mediated DR (SFM-DR), caused through the interactions between the MM cells and the non-cellular compartment (cytokines, growth factors, chemokines, and exosomes) (17, 18). In addition, changes in the MM cells themselves also lead to DR. These changes can be due to multiple causes: genetic and epigenetic abnormalities, disruptions in intracellular signalization pathways, aberrant metabolism, and aberrant drug transport (19–21).
On a genetic level, a large intra- and interpatient clonal heterogeneity is observed. Patients can roughly be divided into two groups, namely the hyperdiploid group and the non-hyperdiploid group. The hyperdiploid group is characterized by odd-numbered chromosome trisomies involving chromosomes 3, 5, 7, 9, 11, 15, 19, and 21, while the non-hyperdiploid group is characterized by primary translocations involving chromosomes t(11,14)(q13;q32), t(4,14)(p16;q32), t(6,14)(p21;q32), t(14,16)(q32;q23), and t(14,20)(q32;q12) (Figure 1) (22, 23). Patients in the hyperdiploid group and patients harboring the t(6,14)(p21;q32) and t(11,14)(q13;q32) translocation have a more favorable prognosis, while patients harboring one of the other primary translocations have a poor prognosis (23). On top, many non-recurrent secondary translocations and mutations are acquired during disease progression, including the translocation of the MYC gene, gain-of-function mutations in several oncogenes (NRAS, KRAS, BRAF, and CCND1), and loss-of-function mutations in tumor suppressor genes such as p15, p16, and P53 (Figure 1) (24–28).
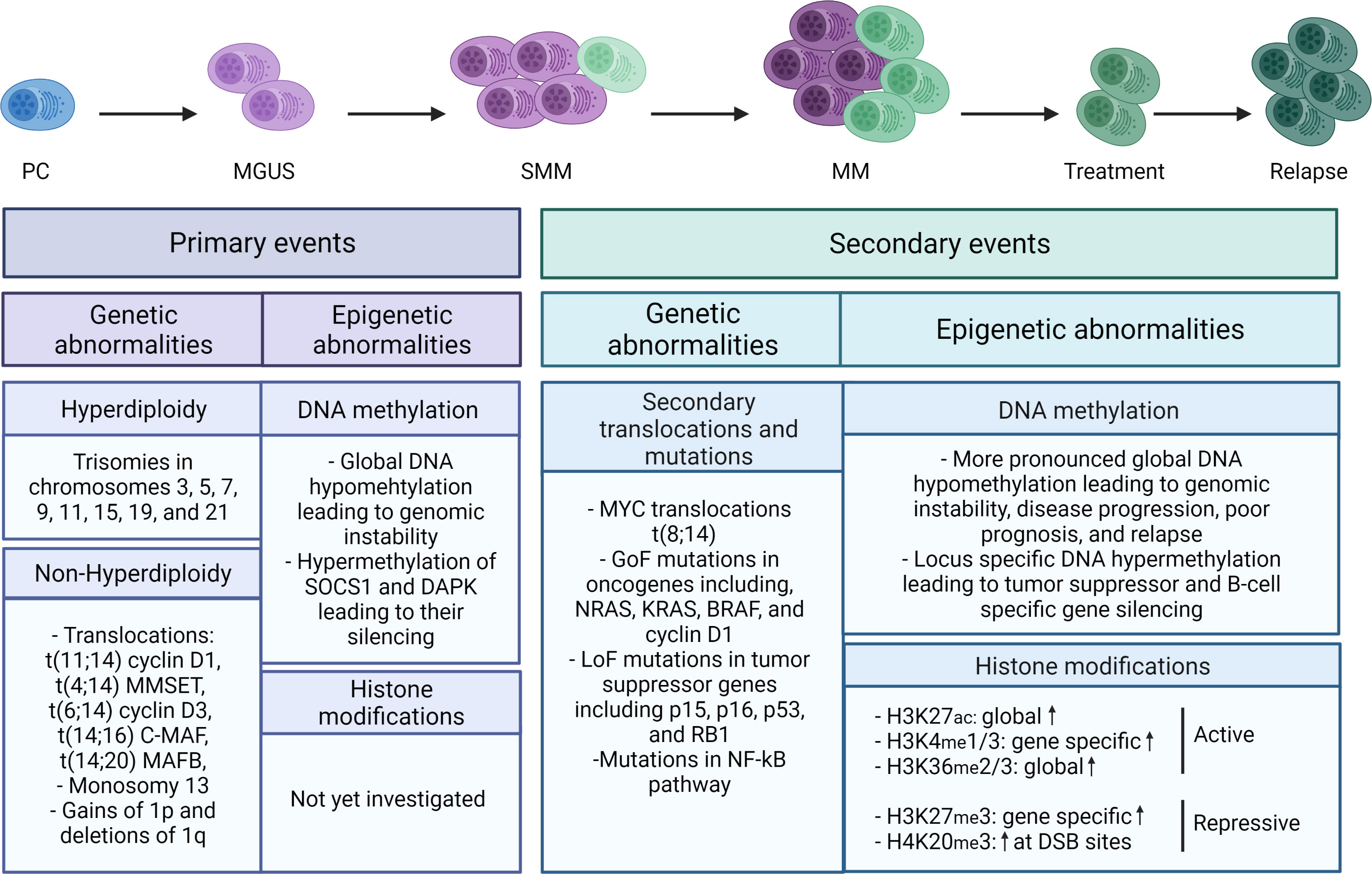
Figure 1 Schematic overview of the genetic and epigenetic abnormalities in MM. Primary (epi)genetic events are driving MM onset, while the secondary events are fostering MM progression and relapse. Primary genetic events include both hyperdiploid and non-hyperdiploid abnormalities, while primary epigenetic events are mainly characterized by global DNA hypomethylation and promotor specific hypermethylation of the tumor suppressors SOCS1 and DAPK. Secondary genetic events include non-recurrent secondary translocations, gain-of-function (GoF) mutations in oncogenes, and loss-of-function (LoF) mutations in tumor suppressor genes, while on the epigenetic level more pronounced global DNA hypomethylation and locus specific hypermethylation are observed together with increased (both global and locus specific) histone mark levels, including H3K27Ac, H3K4me1/3, H3K36me2/3, H3K27me3, and H4K20me3. The blue cell represents a normal plasma cell, while the purple and green cells represent respectively MM cells with primary and secondary events. The color gradation represents the increase in abnormalities in each stage. DSB, double strand breaks.
Apart from being a genetic disease, MM is also deregulated on an epigenetic level (11). In MM, sequencing and gene expression profiling studies have identified numerous epigenetic defects (also called ‘epimutations’) including defects in DNA methylation and posttranslational histone modifications and aberrant expression of miRNAs, probably resulting from genetic defects and/or deregulated expression of the epigenetic modifiers (writer, reader, and eraser proteins) (Figure 1) (29). These epimutations play an important role in disrupting critical regulatory networks in MM, such as the Wnt/β-catenin, JAK/STAT, Cyclin/CDK/Rb, and DAPK/p14ARF/p53 pathway amongst others (30). Hence, these epimutations are well-known to contribute to genomic instability, disease progression, and high-risk disease in MM (11, 31). More recently, increasing evidence has been provided that epiplayers also play an important role in MM cell DR (11, 31). In this review, we discuss recent advances in our understanding of the role of aberrant DNA methylation and DNA methylation modifiers in MM pathogenesis. Furthermore, we describe the role of these modifiers in MM cell DR and disease progression and the potential of combining DNA methylation modifier inhibitors with SoC or novel agents to avoid relapse.
DNA methylation, one of the most studied epigenetic modifications so far, is a vital process for mammalian development and plays an essential role in many biological processes, including cellular differentiation and tissue-specific gene expression, X-chromosome inactivation, genomic imprinting, and silencing of transposable elements (30). Methylation of the DNA is a non-random process in which a methyl group from the methyl donor S-adenosyl-L-methionine (SAM) is covalently added on the fifth carbon position of cytosine residues (5mC) that are directly followed by guanine in the 5’ to 3’ direction, the so-called CpG dinucleotide (32). Methylation of the DNA is generally accepted to result in increased nucleosome compactness and thus gene silencing. However, transcriptional activation upon DNA methylation resulting from, in all likelihood, the recruitment of transcription factors (TF) that preferentially bind methylated DNA (such as RFX) has also been observed and this especially upon 3’ methylation in embryonic stem cells (ESC) (33). In healthy cells, CpG dinucleotides are mainly found in intergenic regions, gene bodies, and repetitive elements and 70-80% of these CpG dinucleotides are methylated, leading to transcriptional inactivation of e.g. transposable and viral elements, which is necessary to preserve the normal integrity of the genome (Figure 2) (34, 35). In addition, 10% of the CpGs can be found in a large number of consecutive CpG dinucleotides, called CpG islands. These CpG islands are mainly located at the transcription start sites of promotors and it is estimated that 50-60% of the gene promotors contain CpG islands (Figure 2) (36). Although these CpG islands are most often unmethylated in healthy cells thus permitting gene expression, methylated CpG islands can also be found and this especially in imprinted genes and in multiple X-chromosome genes that are inactivated in females (37). In contrast, in cancer cells, these normal DNA methylation patterns are often completely disturbed, with a general shift towards global hypomethylation in the intergenic regions and locus specific hypermethylation of the CpG island in the promotor region of tumor suppressor genes. The abnormal DNA methylation patterns in MM will be discussed in more depth in the following chapter (Chapter 3).
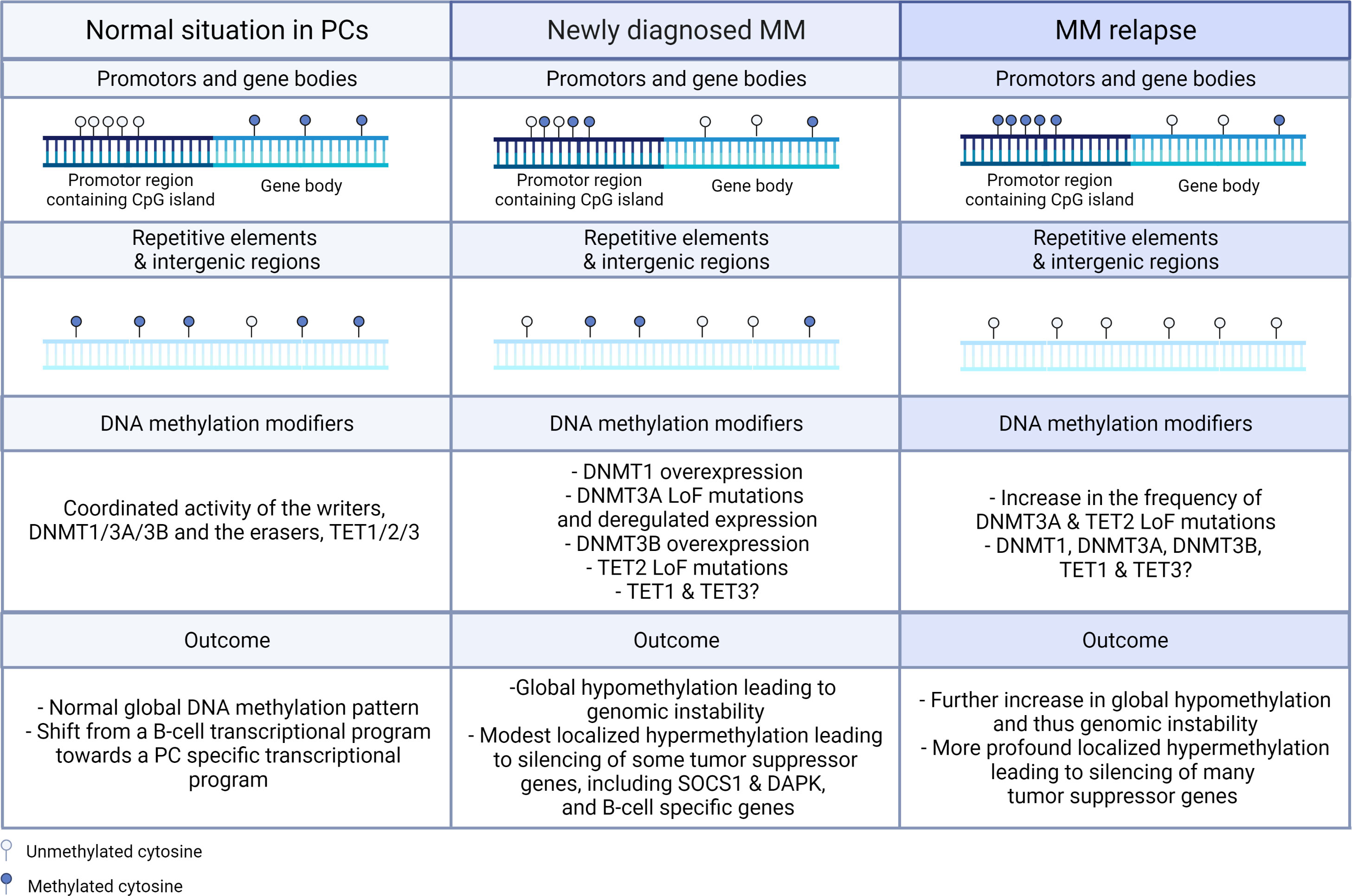
Figure 2 Schematic representation of the changes in global and gene specific DNA methylation patterns and DNA methylation modifiers in newly diagnosed and relapsed MM. In normal circumstances, CpG islands found in the promotor regions are in general not methylated, while the CpG dinucleotide in gene bodies, intergenic regions, and repetitive elements are mostly methylated. In MM, global hypomethylation is found in gene bodies, intergenic regions, and repetitive elements, while hypermethylation is observed in the CpG islands found in the promotor regions of tumor suppressor genes. In the relapsed settings, these events are even more pronounced. On the levels of the DNMT and TET enzymes, overexpression of DNMT1 and DNMT3B and loss-of-function (LoF) mutations in DNMT3A and TET2 are observed in newly diagnosed patients and the frequency of the LoF mutations in DNMT3A and TET2 are even further increased in the relapse setting. In healthy PCs, normal global methylation patterns are observed with a shift from the B-cell transcriptional program towards the PC specific transcriptional program, while in MM cells global hypomethylation leading to genomic instability and localized hypermethylation leading to silencing of tumor suppressor and B-cell specific genes are observed. PC, plasma cell.
The DNA (de)methylation process is mediated by three types of chromatin modifying enzymes, namely the DNA methylation writers, readers, and erasers. The writers, involving the DNA methyltransferase (DNMT) family, are responsible for establishing the 5mC mark. The readers recognize and bind the methylated DNA and influence chromatin compactness and gene expression by recruiting several activator or repressor complexes. Modification and removal of the 5mC marks are established by the erasers, involving mainly the TET enzymes.
The DNA methyltransferase family consist out of 5 DNMTs, encompassing DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L. However, only DNMT1, DNMT3A, and DNMT3B are able to methylate the DNA. DNMT1, DNMT3A, and DNMT3B all have a regulatory N-terminal domain and a catalytic C-terminal domain as illustrated in Figure 3, with the N-terminal domain being responsible for their distinct activity. The N-terminal domain of DNMT1 contains several (sub)domains, including a DNA methyltransferase 1-associated protein (DMAP)-binding domain, which binds DMAP and can consequently interact with the histone deacetylase HDAC2; a nuclear localization signal (NLS); a replication foci-targeting sequence (RFTS), which localizes DNMT1 to the DNA replication fork; a Zn finger CXXC-domain, which recognizes unmethylated CpG containing DNA; 2 bromo-adjacent homology (BAH) domains with unknown function; and a glycine-lysine (GK) repeat, which links the N-region to the C-region. In contrast, the N-terminal domain of DNMT3A and DNMT3B both consist out of a proline-tryptophan-tryptophan-proline (PWWP) domain needed for heterochromatin localization and an ATRX-DNMT3A/B-DNMT3L (ADD) domain that is necessary for the interaction of DNMT3A/B with unmethylated H3K4 (histone 3 lysine 4) (38). The C-terminal domain is quite similar for DNMT1, DNMT3A, and DNMT3B and is responsible for the deposition of the methylation mark in the presence of SAM.
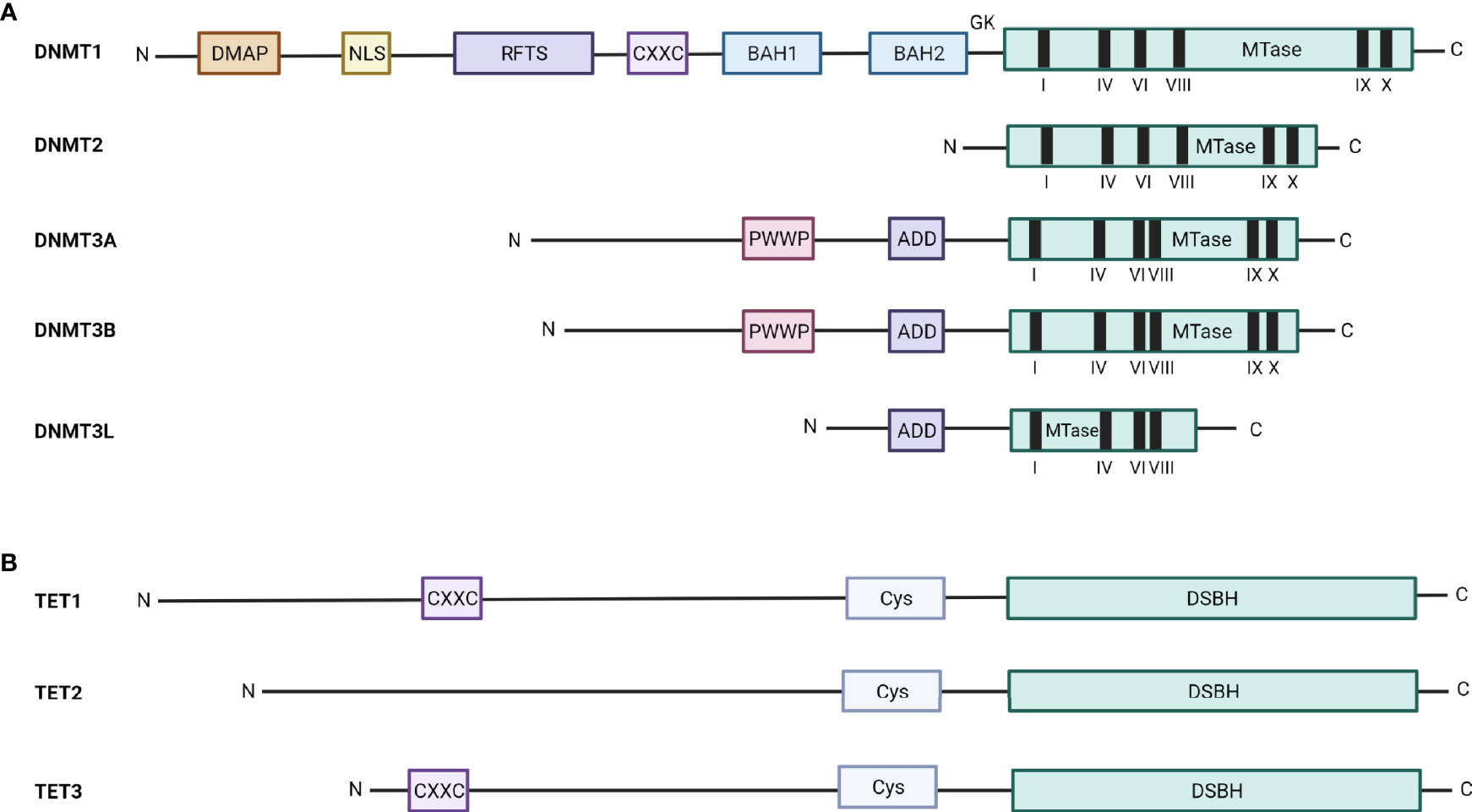
Figure 3 Schematic overview of the structure of the DNMT and TET enzymes. The structure and different domains of (A) the five DNMT family members, DNMT1, DNMT2, DNMT3A, DNMT3B, and DNMT3L, and (B) the three TET family members, TET1, TET2, and TET3, are depicted. DMAP, DNA methyltransferase 1-associated protein binding domain; NLS, nuclear localization signal; RFTS, replication foci-targeting sequence; CXXC, Zn finger CXXC-domain; BAH, bromo-adjacent homology; GK, glycine-lysine repeat; MTase, methyl transferase; PWWP, proline-tryptophan-tryptophan-proline; ADD, ATRX-DNMT3A/B-DNMT3L; Cys, cysteine-rich domain; DSBH, double-stranded β-helix domain.
The DNMTs are recruited to the DNA in both a gene/locus specific and a non-specific manner, which is regulated through the binding of the DNMTs with many different binding partners, which, in turn, regulate their activity. Specific recruitment is regulated by polycomb (PcG) proteins and TF, but is also observed upon DNA repair (DNMT1 recruitment via PCNA), while the unspecific recruitment involves the cooperation with heterochromatin readers or replication associated proteins (39).
In general, DNMT1 is considered a maintenance methyltransferase, meaning that it is responsible for maintaining CpG methylation after DNA replication, as it preferentially recognizes hemimethylated DNA (40). DNMT1 is thus mainly responsible for the maintenance of DNA methylation patterns upon completion of the embryonic cell fate specification and is strongly expressed in almost all adult tissues (41, 42). Knock-out (KO) of Dnmt1 in mice showed that Dnmt1 is necessary for embryonic development, gene imprinting, and X-chromosome inactivation (43). Furthermore, homozygous mutations in Dnmt1 in mice resulted in a delayed development with mice dying before birth, thus (further) showing that DNMT1 is essential for mammalian development and normal cell function (44). In contrast, DNMT3A and DNMT3B are mainly seen as de novo DNA methyltransferases, that are able to recognize unmethylated DNA and establish new DNA methylation patterns during embryogenesis (32, 45, 46). More recently, however, DNMT3A and DNMT3B have been suggested to contribute to the maintenance of the DNA methylation patterns in differentiated cells as well, although their catalytic activity appears to be 20x lower than that of DNMT1 (43). Both DNMT3A and DNMT3B are highly expressed in undifferentiated embryonic stem cells and the expression declines after birth. Yet, distinct functions between DNMT3A and DNMT3B are observed. DNMT3B is highly expressed at E7.5 embryonic development, thus playing a critical role early in the development, while DNMT3A is highly expressed at E8.5 and E9.5 playing a role later in the development and even after birth (32, 45, 46). In line, DNMT3B knockdown (KD) mice died before birth, while DNMT3A KD mice died four weeks after birth. Moreover, DNMT3A is important for methylation of imprinted genes, while DNMT3B is important for methylation of centromeric, pericentromeric, and subtelomeric regions (39, 47). This difference in function can be explained by the fact that there is only a 28% homology in the N-terminal domain between DNMT3A and DNMT3B. Importantly, DNMT3A and DNMT3B not only play a role during embryogenesis, but also during the normal life span (48). In adults, DNMT3A is expressed in almost all tissues, while high DNMT3B levels are mainly observed in the testis, thyroid, and the bone marrow (49). Within the bone marrow, DNMT3B expression levels vary among the different cell types, with the CD34+ cells (the hematopoietic stem cells) showing high DNMT3B levels compared to the more differentiated cell types (41).
Importantly, as a result of alternative splicing, multiple isoforms with distinct activities can be found for DNMT1, DNMT3A, and DNMT3B (Table 1) (61). In mice, three different Dnmt1 isoforms are reported, namely Dnmt1s, Dnmt1o, and Dnmt1p. These isoforms are the result of alternative splicing of the N-terminal exons, with each of the isoforms having their own specificity. DNMT1s is specific to somatic cells, while Dnmt1o and Dnmt1p are specific to oocytes and pachytene spermatocytes respectively (50). However, these DNMT1 isoforms have not yet been identified in humans. For DNMT3A, two isoforms have been described, namely DNMT3A1 and DNMT3A2. DNMT3A1 is the full-length protein, while DNMT3A2 lacks 223 amino acids at the N-terminal end (51). Although both isoforms have the PWWP, ADD domain, and the identical catalytic domain, a difference in DNA methylation activity is observed; with DNMT3A1 having a higher DNA-binding and DNA methylation activity compared to DNMT3A2 (62). Moreover, DNMT3A1 also shows a higher affinity for heterochromatin, while DNMT3A2 has more affinity for euchromatin (51, 52). For DNMT3B, more than forty different isoforms have been reported and are known to be expressed in a tissue specific manner. However, so far, only nine isoforms have been described in literature in more detail (42, 49, 54–60). The activity of these isoforms is determined by the preservation of the highly conserved catalytic domain. DNMT3B1 and DNMT3B2 are both catalytically active isoforms, while the DNMT3B4 and DNMT3B5 isoforms are catalytically inactive due to a frameshift mutation, leading to an early stop codon causing disruption of the catalytic domain (49, 55). The catalytic inactive isoforms are thought to regulate the activity of the active DNMT3A and DNMT3B isoforms by physical interaction (53). Although alternative splicing is most commonly observed in the C-terminal catalytic region of DNMT3B, it can also be observed in the N-terminal region. DNMT3B isoforms lacking the N-terminal region are called delta (Δ) DNMT3B. At the moment, at least seven different ΔDNMT3B are known (63). As to our knowledge, no difference in activity has been described among these different ΔDNMT3B (64).
The two other members of the DNMT family, DNMT2 and DNMT3L, are not able to methylate the DNA. DNMT2 only contains the catalytic C-terminal domain and plays an important role in the methylation of a specific cytosine in tRNAAsp. In contrast, DNMT3L lacks both the N-terminal PWWP domain and the catalytic activity of the C-terminal domain and is thus seen as a truncated version of DNMT3B. DNMT3L appears to play an important role as cofactor for DNMT3A/B and is known to increase their methyltransferase activity (65).
For a long time, it was believed that DNA methylation resulted in a stable and permanent repression of the genes, but new evidence showed that the DNA methylation marks can be removed making DNA methylation a reversible process. There are two forms of DNA demethylation, namely active and passive demethylation. Passive demethylation is induced upon cell division, when a lower DNMT1 methylation activity is observed. In contrast, active demethylation is initiated by the Ten-eleven translocation (TET) enzymes (66, 67). There are three TET enzymes (TET1-3) with different activity and structure as shown in Figure 3. TET1 and TET3 have a N-terminal domain, which contains a CXXC-domain, and a catalytic C-terminal domain, consisting out of cysteine-rich (Cys) and double-stranded β-helix (DSBH) domains. TET2 has the same catalytic C-terminal domain, but lacks the N-terminal CXXC-domain (68). For TET1 and TET3, the N-terminal domain recognizes and recruits them to the methylated DNA, while TET2 recruitment to methylated DNA is facilitated through the interaction with DNA binding proteins, including the tissue-specific TF early B-cell factor 1 (EBF1) (69). The TET enzymes oxidize the 5mC to 5hmC (5-hydroxymethylcytosine), 5fC (5-formylcytosine) and 5caC (5-carboxylcytosine). Final active demethylation is then mediated by the thymine-DNA-glycosylase, which recognizes and excises 5fC and 5caC, followed by base excision repair (BER)-mediated replacement of the modified cytosine by an unmodified one. While all three TET enzymes have similar activity, TET1 and TET2 play a role in the demethylation at specific loci in the primordial germ cells (PGCs), whereas TET3 plays an important role in erasing paternal methylation marks in the male pronuclei of zygotes (38).
Isoforms of the TET enzymes, resulting from alternative splicing, have also been observed; with each isoform having a distinct expression pattern and function. TET1 isoforms encompass TET1 Fl (full length) and TET1s (short) isoform, with TET1s lacking the N-terminal side containing the CXXC-domain. TET1 Fl is observed in early embryos, ESCs, and PGCs, while TET1s is observed in somatic cells. TET2 has two different isoforms, namely TET2 Fl and TET2n, an N-terminus isoform which lacks the C-terminus and thus shows no enzymatic activity. For TET3, three different isoforms exist, namely TET3 Fl, TET3s, and TET3o (ovarian), with TET3s and TET3o both lacking the CXXC-domain. TET3 Fl is specific for neuronal differentiation, while TET3s is highly expressed in neurons and the retina and TET3o is expressed in oocytes and zygotes (70, 71).
The methylated cytosines will be recognized and bound by the methyl binding proteins. There are three families of methylated DNA binding proteins, namely the methyl-CpG-binding domain (MBD) family, the SET and RING finger associated (SRA) domain protein family, and the methyl CpG binding Zinc finger family. Members of the MBD family include, MeCP2 and MBD1-4. MeCP2, MBD1, MBD2, and MBD4 all contain a methyl-CpG-binding domain (MBD), allowing to recognize and bind methylated DNA. In addition, MBD4 also contains a glycosylase domain, which appears to have a function in DNA mismatch repair. In contrast, MBD3 contains a mutated MBD domain, making it unable to bind to methylated DNA on its own. Instead, MBD3 localization to the DNA is possible since MBD3 is a subunit of the Nucleosome Remodeling Deacetylase (NuRD) complex, which also contains the histone deacetylases 1 and/or 2 (HDAC1/2) amongst others. MeCP2 and MBD1-2 also contain a transcription repressing domain (TRD), which allows the interaction with different repressor complexes and chromatin modifying enzymes, such as HDAC1/2 or the histone methyltransferases (HMT) SUV39h1/HP1 and SETDB1, resulting in further transcriptional repression (38, 72–74) and thus a self-reinforcing loop of silencing (75). The SRA domain proteins include the ubiquitin-like with PHD and Ring Finger Domains (UHRF) 1 and 2. UHRF1 plays a role in the localization of DNMT1 to DNA replication foci and also recognizes histone modifications such as H3K9me3, while UHRF2 preferentially binds to 5hmC (76, 77). The methyl CpG binding Zinc finger family includes ZBTB33, ZBTB38, ZBTB4, Zfp57, Klf4 (Krüppel-like factor 4), WT1 (Wilms tumor protein 1), Egr1 (growth response protein 1), and CTCF (CCCTC-binding factor), which are reviewed in depth by Hudson et al. (78).
In MM cells, just like cancer cells in general, a disturbed DNA methylation landscape is observed with global hypomethylation leading to genomic instability and localized hypermethylation contributing to silencing of tumor suppressor and B-cell specific genes (Figure 2). In general, the changes in DNA methylation patterns are disease-stage specific, with global hypomethylation occurring already in the early stages of the disease, whereas locus specific hypermethylation is more frequent in the more advanced disease stages (79, 80). In line, Walker et al. showed that the determination of the methylation pattern is capable of distinguishing the premalignant (MGUS) from the malignant (MM) conditions (80).
Global DNA hypomethylation is already present in MGUS and newly diagnosed (ND) MM patients and correlates with disease progression and poor prognosis (Table 2) (81, 82). In 2009, Bollati et al. found that global hypomethylation of the repetitive elements LINE1, Alu, and SAT alpha in MM is linked with chromosomal instability, with lower methylation levels of LINE1 and SAT alpha being observed in the non-hyperdiploid group compared to the hyperdiploid group. Furthermore, lower levels of methylated Alu and SAT alpha were observed in the t(4,14) group, a group with a worse prognosis compared to the other myeloma cytogenetic subgroups (81). Walker et al. confirmed these findings by showing that the t(4,14) translocation is associated with the strongest heterogeneity in DNA methylation profiles, characterized by the highest amount of hypomethylated regions on the one side and the highest amount of hypermethylated genes on the other side (80). Moreover, they also showed a further decrease in global methylation levels upon disease progression (80). More recently, genome-wide hypomethylation was also observed in patients from the MMRF CoMMpass study, which represents the largest molecular profiling initiative in MM patients, with median global CpG methylation levels of 41% compared to normal PC and B-cells with respectively 71% and 89% of global CpG methylation levels (90).
In contrast, DNA hypermethylation is mainly observed in the later stages of MM and this mainly in the CpG islands present in the promotor regions of tumor suppressor genes [including RASSF4, p15, p16, p73, TP53, SOCS1, DAPK, SFRP1, SFRP2, VHL, and EGLN3 (Table 3)] and enhancer regions of B-cell specific genes (including BCL11A, BATF, EBF1, and PAX5), causing inactivation of these genes (93–118). Importantly, the promotor specific hypermethylation appears to further increase from the ND stage to the relapse stage, with the highest promotor methylation levels being observed in the plasma cell leukemia (PCL) stage. Hypermethylation of SOCS1 and DAPK promotor regions has been identified as early events in the MM pathogenesis, since hypermethylation of these promotor regions has already been observed in the MGUS stage (Figure 2). In contrast, hypermethylation of p16, SHP1, and E-CAD are only first observed in the MM stage, thus contributing to disease progression (94, 102, 118). Furthermore, when patients from an early disease stage developed to a later disease stage, the promotor region of seventy-seven (tumor suppressor) genes was found to become hypermethylated, with the hypermethylation of some of these genes, including p16, DAPK, BCL2/BNIP3, and CDH1 (E-CAD) (Table 3) correlating with a shorter overall survival (OS) (79, 80). Apart from gene specific hypermethylation, gene specific hypomethylation has also been observed in the oncogenes JAG2 and ABC transporter, thus leading to their aberrant expression (Table 3) (91, 92). Furthermore, although it was long assumed that aberrant hypermethylation patterns are primarily present in promotor regions of genes, increasing evidence is now showing that these patterns are also present in gene bodies (Notch1 & GATA3) and intergenic regions (32). In line, Agirre et al. reported profound hypermethylation outside the CpG rich promotors, in the intronic enhancers overlapping with binding sites of B-cell specific TF such as BCL11A, BATF, EBF1, and PAX5 (119).
More recently, with the availability of enhanced reduced representation bisulfite sequencing (eRRBS), it has become apparent that the MM epigenome is not only characterized by global hypomethylation and focal hypermethylation of CpG islands, but also by a high degree of intratumoral epigenetic methylation heterogeneity (83). Moreover, when comparing the epiallele composition changes (= epiallele shifts) between normal PCs and MM patients at diagnosis, the extent of these epiallele shifts was found to be highly variable between patients and appeared associated with poor OS when acquired at the time of diagnosis (and this independently of high-risk genetic lesions) (Table 2). In addition, comparison of the epiallele shift between matched newly diagnosed and relapsed patients revealed that upon relapse, 42% of the patients showed substantial accumulation of stochastic methylation. Importantly, these stochastic methylation gains were mainly found in bivalent promotors of developmental genes, which are in normal circumstances tightly regulated by the balance between the transcriptionally repressive histone mark H3K27me3 and the active histone mark H3K4me3 and generally free from DNA methylation. This allows flexible regulation of the expression of these developmental genes. However, in B-cell tumors, epigenetic switching from H3K27me3 to DNA methylation is observed at some loci. This switch is also referred to as “Polycomb repression-associated DNA methylator phenotype” or PRAMP. PRAMP decreases the flexibility between the repressive and active state, leading to a more permanent “silenced” state of key regulatory genes (120). Together, these findings suggest that the enhanced stochastic methylation variation makes it possible for MM cells to adapt to their environment (including treatment pressure) to survive (83).
The exact reason for the disturbed DNA methylation landscape in cancer cells is currently still unknown, but the most likely explanation is the aberrant expression and/or activity of the DNA methylation modifiers (31, 93). In this review, we will focus on the role of the writers and erasers in MM disease. Overexpression of DNMT1, DNMT3A, and DNMT3B has been observed in many solid and hematological cancers, including MM (121, 122). Moreover, mutations in the DNA methylation modifiers are also observed in MM, correlating with a shorter OS when grouped all together (Table 2). Although mutations in DNA methylation modifiers are only observed in 4% of the patients at diagnosis, their frequency appears significantly increased in the relapsed setting and this especially for DNMT3A and TET2 mutations (87). In contrast, mutations in DNMT1/3B and TET1/3 are less frequent in MM.
Increased expression of DNMT1 is often found in several solid and hematological cancers and has been linked with a poor prognosis, especially in solid cancers (32). DNMT1 is crucial for faithfully maintaining methylation patterns in human cancer cells and DNMT1 KO leads to severe mitotic defects and even cell death (40). In MM, DNMT1 is also found overexpressed and the levels further increase as the disease progresses, indicating a role for DNMT1 in disease progression (81). In line, KD of DNMT1 using siRNA was shown to decrease MM cell proliferation, due to a G1-phase block and re-expression of SOCS1 and p16, and to increase apoptosis due to cleavage of caspase 3 and PARP (123, 124). A recent study in MM also showed that DNMT1 is responsible for the hypermethylation of tight junction protein 1 (TJP1), a negative regulator of the epithelial-to-mesenchymal transition (EMT), leading to decreased TJP1 levels and poor OS (125). The exact mechanisms behind the aberrant expression of DNMT1 are currently unknown, but studies in other cancers are indicating that the DNMT1 levels could be regulated by miR-148a (126). In addition, some proteins such as the fatty acid-binding protein 4 (FABP4) and nucleolin were also found to regulate DNMT1 levels in hematological cancers (127–130). Inhibition of these proteins resulted in the downregulation of DNMT1 and a decreased clonogenic potential of cancers cells in acute myeloid leukemia (AML) and chronic myeloid leukemia (CML) (128–130).
Mutations of DNMT3A are frequently observed in different hematological malignancies, including AML (both adult and pediatric), T-cell acute lymphoblastic leukemia (T-ALL), myelodysplastic syndrome (MDS), and T-cell lymphomas (TCL) (131). In AML, DNA sequencing showed that 22.1% of the newly diagnosed patients have mutations in DNMT3A. The most common mutations found in AML patients are missense mutations, especially those observed at the amino acid R882 located in the methyltransferase domain, resulting in a dominant negative effect. Other mutations such as deletions, frameshift, nonsense, and splice-site mutations are also described. Most of these mutations are consistently associated with loss-of-function, leading to genome-wide hypomethylation and correlated with a shorter OS (132). Mutations at R882 are also observed in the DNMT3A isoforms. Overexpression of mutated DNMT3A1 or DNMT3A2V (a DNMT3A2 variant lacking 68 base pairs) in AML cells resulted in increased proliferation rates, while overexpression of normal DNMT3A1 or DNMT3A2 significantly reduced cell proliferation, thus strengthening the hypothesis that DNMT3A has a tumor suppressive role (133). In line with this tumor suppressive role of DNMT3A, a recent study also reported upregulation of DNMT3A in AML patients, correlating with a better leukemia-free and OS (134). Currently, there are only few studies that have focused on DNMT3A in MM. Mutations in the DNMT3A gene are less frequently observed in MM compared to the other types of leukemia (87, 135). Nevertheless, Walker et al. found that DNMT3A is one of the sixty-three identified mutated driver genes in early MM development (136). In line with this study, a very recent study also reported the presence of a pathogenic DNMT3A mutation in the MGUS stage (137). Furthermore, the mutation frequency in DNMT3A was shown to increase upon relapse (87). On the transcriptional level, two studies demonstrated DNMT3A downregulation in MM and PCL, correlating with poor OS (Table 2) (79, 81). In contrast, Amodio et al. reported increased DNMT3A levels in MM and PCL patients compared to healthy controls. In addition, they provided evidence that miR-29b is a direct regulator of DNMT3A and that targeting DNMT3A using miR-29b mimics reduces MM cell growth (138). In line with the study of Amodio et al, Walker et al. also showed upregulation of DNMT3A in MM. This DNMT3A upregulation was particularly observed in the t(4,14) subgroup of patients and was the result of hypomethylation (80). Furthermore, the recent study from Luzna et al. also showed increased DNMT3A levels in MM patients compared to healthy controls and a trend to increased DNMT3A levels in ND and relapsed MM patients (the active phase of MM) compared to those in remission, thus showing its potential as a biomarker of MM progression (Table 2) (88). However, so far, the exact role of DNMT3A in MM biology has not yet been identified.
An oncogenic role for DNMT3B is observed in several hematological cancers, including T-ALL, AML, and Burkitt and diffuse large B-cell lymphoma (DLBCL), where increased DNMT3B levels are reported to be mostly the result of increased MYC-levels. Moreover, in AML, DNMT3A/B was also shown to be a direct target of miR-29b and forced miR-29b overexpression resulted in a decrease in DNMT3B and global methylation levels, resulting in the re-expression of some tumor suppressor genes including p15 (139). In T-ALL, silencing DNMT3B using DNMT3B shRNA reduced cell viability and cell growth, as evidenced by a decrease in the number of cells in the S-phase and an increase in the levels of CDKN1A (p21CIP1), CDKN2B (p15INK4b), CDKN2A (p16INK4a), and CDKN2D (p19INK4d) (140). Moreover, both in AML and DLBCL, high DNMT3B levels are correlated with a bad prognosis and a more aggressive disease (141, 142). However, while most studies indicate that DNMT3B has oncogenic properties in hematological cancers, some studies have also reported the opposite (143–145). For example, Dnmt3b deletion in the MLL-AF9 driven AML mouse model led to accelerated progression (146). Furthermore, Dnmt3b haploinsufficiency in mice resulted in the development of various hematologic malignancies, including TCL (145). In MM, Amodio et al. reported an inverse correlation between miR-29b and DNMT3B levels and showed that targeting DNMT3A/B using miR-29b mimics reduces MM cell growth, indicating that DNMT3A/B has an oncogenic role in MM (138). Furthermore, the enhanced stemness of MM cells observed upon coculturing them with granulocytic myeloid-derived suppressor cells (G-MDSC), was recently suggested to be the result of increased piRNA-823 and DNMT3B levels (147). However, to the best of our knowledge, the exact role of DNMT3B in MM biology and disease progression has not yet been thoroughly investigated.
TET2 is by far the most investigated TET family member, as it is the one found most frequently mutated. TET2 mutations can either be heterozygous or homozygous and appear to be very heterogeneous, encompassing nonsense and missense mutations, frame shift mutations, and in-frame deletions. Most of these mutations are loss-of-function mutations, leading to a reduced dioxygenase activity and thus a significant decrease in global 5hmC levels (148). These inactivating TET2 mutations suggest a tumor suppressive role for the TET2 protein, which is confirmed by the spontaneous development of myeloid, T-cell, and B-cell malignancies in a TET2 KO mouse model (149). TET2 mutations are frequently observed in hematological cancers, like myeloproliferative neoplasms (MPNs), chronic myelomonocytic leukemia (CMML), DLBCL, and AML (150–152). In contrast, in MM, TET2 mutations were only observed in about 1% of the patients from the Myeloma XI clinical trial (MyXI) (87). Nevertheless, Walker et al. identified TET2 as one of the sixty-three identified mutated driver genes, indicating that TET2 mutations are also an early event in MM development (136). Furthermore, the mutation frequency in TET2 was shown to increase upon relapse (115). Importantly, the prognostic value of the TET2 mutations remains controversial. In MDS, one study showed a correlation between TET2 mutations and a more favorable prognosis, while other studies showed no impact of TET2 mutations on the prognosis in MDS and AML (153–155) or even an inverse correlation (156). For MM, no prognostic potential was shown for TET2 mutations on its own, but Pawlyn et al. found that mutations in any of the DNA methylation modifiers (TET1/2/3, IDH1/2 or DNMT1/3A/B) correlated with a shorter OS (87). Of note, TET2 mutations are often found in combination with several other mutations, like NPM1, FLT3-ITD, FLT3-TKD, RUNX1, CEBPA, CBL, and KRAS (156). The differential combination of TET2 mutations with these other mutations might be a possible explanation as to why outcomes differ and why the prognostic potential remains controversial. Apart from the loss-of-function mutations, aberrant expression levels of TET2, although less frequent, have also been observed in hematological cancers. In MDS, downregulation of TET2, irrespective of the presence or absence of TET2 mutations, has been reported, while in AML patients, TET2 expression levels are significantly increased (157, 158). For MM, an increase in TET2 levels correlating with a better OS has also been observed (Table 2) (89).
Although mutations in TET1 and TET3 have been observed in some hematological malignancies (like AML, T-ALL, and chronic lymphocytic leukemia (CLL) for TET1 and MDS, myeloproliferative neoplasms, CMML, and B-cell acute lymphoblastic leukemia (B-ALL) for TET3) and were reported to be enriched in relapsed B-ALL cases, TET1 and TET3 mutations are in general quite infrequent (150, 159, 160). In contrast, TET1 and TET3 aberrant expression are more frequently observed in hematological malignancies. For TET1, overexpression has been reported in both MLL-rearranged leukemia and cytogenetically normal AML patients and was found to be correlated with a poor OS in the latter group, thus suggesting a pro-oncogenic role for TET1 in hematological cancers (161, 162). In contrast, Zhang et al. showed that TET1 expression is significantly reduced in AML patients (158). Moreover, hypermethylation and thus transcriptional silencing of TET1 is also observed in B-cell lymphoma, where its tumor suppressor activity was demonstrated in vivo, as KO of TET1 resulted in decreased survival of the mice (163). For TET3, a downregulation in TET3 levels was observed in CLL, but no significant correlation with OS was found although TET3 low expressers tended to have a worse OS (164). In contrast, TET3 overexpression was observed in AML and was associated with a longer disease-free and OS compared to patients with low TET3 expression (158). Furthermore, TET3 levels positively correlated with CDKN2B, ZIC2, and miR-196a levels, which appear to have anti-leukemic effects, thus suggesting a tumor suppressive role for TET3 in AML (158). However, more recently, TET3 overexpression in AML cell lines was shown to promote AML growth, suggesting rather an oncogenic role for TET3 (165). Of interest, a recent study in MDS showed an inverse correlation between TET2 and TET3. The authors suggest that the TET3 levels increase as a kind of compensation mechanism for the loss of TET2 expression and a lack in TET3 compensation correlated with high-risk features (increase in percentage of bone marrow blasts) and a poor outcome (157). In contrast, a recent study in AML patients showed a positive correlation between TET2 and TET3 and KO of both TET2 and TET3 in hematopoietic precursor cells in mice resulted in an almost complete loss of 5hmC and the emergence of myeloid leukemia (158, 166). Thus, it appears that the role of both TET1 and TET3 in hematological cancers (either oncogenic or tumor suppressive) is disease/context dependent and remains to be identified for MM.
It is well-known that the epigenetic machinery tightly regulates the differentiation and maturation of hematopoietic stem cells (HSC) to mature B-cells and PCs. Dysregulation of the epigenetic machinery during this normal PC differentiation process is therefore linked to various B-cell related disorders. In MM, the tumor population is composed out of different subpopulations that differ in maturation stage, clonogenic capacity, and drug sensitivity (29, 167, 168). Importantly, it is believed that there exists a certain degree of ‘epigenetic plasticity’ between these different subclones, allowing the reprogramming/dedifferentiation of the terminally differentiated MM cells into the more immature and resilient subclones and vice versa upon treatment pressure (167, 169). Below we will briefly discuss the reported role of the DNA methylation modifiers in normal PC differentiation and MM cell plasticity and stemness.
The HSC present in the BM give rise to pro-B-cells and mature B-cells, which will then, upon antigen encounter further differentiate and mature into memory B-cells, expressing CD19 but not CD38, and plasmablasts, expressing both CD19 and CD38. The plasmablasts will then re-enter the BM to undergo terminal differentiation towards mature, non-dividing and immunoglobulin-secreting PCs. These long-living mature PCs are CD19-/CD38+/CD138+/Xbp1s+ (170). During the normal PC differentiation process, many epigenetic changes will take place. A prerequisite for the terminal differentiation of mature B-cells into fully mature PCs, is the shift from a B-cell transcriptional program, that maintains the B-cell phenotype (PAX5 and BCL-6), towards a PC specific transcriptional program (IRF4, Blimp-1, and Xbp1); with Blimp-1 as the master TF in PC generation (31, 171, 172). Blimp-1 shuts down the B-cell expression program by silencing over more than 250 B-cell specific genes, including PAX5 and BCL-6, by recruiting various co-repressors of the epigenetic machinery. Moreover, a recent study also indicated the importance of TET2/3 in PC differentiation, through demethylation of the IRF4 locus resulting in high IRF4 levels (173). In addition, DNMT3A/B also proved to play an important role in the repression of the B-cell expression program that is necessary for the B-cell activation and PC differentiation. DNMT3A/B deficient B-cells showed significantly less DNA methylation upon PC maturation compared to their normal counterparts due to failure of de novo DNA methylation. This lack of de novo DNA methylation resulted in increased chromatin accessibility at both B-cell and PC factors, including respectively PU.1, and IRF4 and E2A (174). In addition, changes in the expression levels of several of the DNA methylation modifiers have been observed during normal B-cell and PC differentiation. For example, DNMT1 and DNMT3B levels are upregulated and DNMT3A levels are downregulated during the transition from the naive to the germinal center (GC) B-cell stage and return to normal again in the post-GC memory B-cell. During the transition from memory B-cells to PCs, the DNMT3A levels will then decrease again, while the DNMT3B levels are increasing, although to a lesser extent than in the transition from the naive to the GC B-cell stage (175). Furthermore, using RNA sequencing analysis, upregulation of DNMT3B, TET1, IDH1/2, MBD1, and ZBTB38 during the transition of memory B-cells into PCs has been documented (176). In line with the changes in the expression levels of the DNA methylation modifiers, a global shift towards hypomethylation is observed during B-cell differentiation and this especially at the later stages, including the memory B-cell and the PC stage (177, 178). However, although the PC and the memory B-cells have a similar methylome, they appear to have a very different transcriptional program (175). This indicates that, as mentioned above, other epigenetic modifiers are also at play in the differentiation of memory B-cells to fully mature PCs. Indeed, several histone deacetylases (HDAC) and histone methyl transferases (HMTs), such as G9a, Enhancer of Zeste Homolog 2 (EZH2) and LSD1, have been shown to be involved in normal PC differentiation as well (176, 179).
As mentioned earlier, the MM cell population is composed out of different subpopulations with each subpopulation differing in the level of maturation, response to treatment (drug sensitivity), transcriptional profiles, and clonogenic potential. In general, four co-existing MM subpopulations have been suggested, namely plasmablasts, which are CD19+; pre-PC, which are CD19- and CD138-; CD138low PCs; and the more mature CD19-/CD138+ PCs, which make up the bulk of the MM cells (170). Importantly, Chaidos et al. proposed a bidirectional transition between the pre-PCs and the mature PCs in vivo in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice, with the pre-PC showing an enrichment in genes encoding for epiplayers, such as histone acetyltransferases (HATs), HDACs, HMTs, histone demethylases (HDMs), and the methylation reader CCCTC-binding factor (CTCF) compared to the PC. These findings are suggestive of an epigenetic plasticity controlling the reversible and bidirectional transition between the more mature MM cells and the less mature myeloma cells, resulting in a PC/pre-PC equilibrium. Importantly, the authors also showed that the pre-PC are significantly less sensitive to chemotherapy combined with the PI Bz than the PC, suggesting that the mechanisms involved in this epigenetic plasticity are playing an important role in developing DR against the current MM therapies (167).
Moreover, it is strongly believed that the subpopulations of less mature MM cells have stem-like properties and are responsible for MM tumor initiation and propagation. Hence, these less mature subpopulations are thought to comprise the so-called MM stem cells (169, 180). Cancer stem cells (CSC) are well-known for their quiescence, self-renewal, and drug-resistant capacity, contributing to tumor aggressiveness, treatment resistance, and tumor recurrence (181). However, the exact nature of this MM stem cell population, including the surface markers, is still largely debatable (182). Increasing evidence is indicating that dysregulation of the epigenetic machinery, including aberrant expression and/or mutations in DNA methylation modifiers, is playing a role in CSC maintenance in both solid and hematological cancers (183). In MM, the enhancer regions of B-cell specific genes such as PAX5 and BATF are hypermethylated in MM cells compared to their normal counterparts. Importantly, these enhancers are also highly methylated in stem cells. This suggests that the MM cells (or at least a subfraction of them) either retain or regain stem cell features via epigenetic mechanisms. In line, a recent study suggested a role for DNMT3B in MM cell stemness. In short, granulocytic-myeloid-derived suppressor cells (G-MDSCs) cocultured with MM cells were shown to increase DNMT3B expression through piRNA-823 in the myeloma cells and increase their tumorigenic potential. Silencing of piRNA-823 led to decreased DNMT3B levels and a decreased stemness potential. The later was also supported by the decreased levels of CSC related genes, including NANOG, OCT4, and SOX2 (147). Moreover, both in MM cell lines and primary MM cells, an increase in clonogenic potential was observed for the residual cells following pomalidomide treatment. Further analysis revealed increased SOX2 and decreased levels of the methylation reader MBD3 in this population of residual myeloma cells, together with a clear deregulation of embryonal stem cell pathways. In line, MBD3 silencing by siRNA resulted in an increase in the clonogenic potential in myeloma cell lines (184). Finally, treatment of MM cells with berberine, a naturally occurring isoquinoline alkaloid, was recently shown to reduce the clonogenic potential of human myeloma cell lines (HMCLs) by inducing degradation of the DNA methylation reader UHRF1. In line, silencing UHRF1 in HMCLs using siRNA was also able to reduce their colony formation ability (185).
Since epigenetic modifications are reversible, they represent interesting targets to (partially) reprogram the MM cells back to their normal counterparts. Over the past two decades, several epigenetic modulating agents (EMAs) have been developed and investigated for their anti-myeloma activity. Below, we summarize the current and upcoming epigenetic therapies, focussing on the DNA methyltransferase inhibitors (DNMTi), alone or in combination with SoC agents and/or novel promising agents. Two main classes of DNMTi are available, namely the nucleoside analogs and the non-nucleoside analogs.
By far the most clinical advanced DNMTi are the nucleoside analogs azacytidine (AZA) and decitabine (DAC). At the moment, AZA and DAC are already approved by the Food and Drug Administration for the treatment of MDS and other leukemias such as AML (186). Both AZA and DAC are incorporated into the genome upon DNA replication. In general, AZA and DAC have two modes of actions. First, upon treatment with relatively low concentrations, the DNMTs are trapped by the incorporated cytosine analogs, leading to their degradation. This process results in the depletion of DNMTs in the nucleus, leading to genome wide DNA hypomethylation and re-expression of tumor suppressor genes, which in turn reduces tumor growth and survival (187). Second, upon treatment with high AZA and DAC concentrations, massive protein-DNA cross-links are formed, leading to DNA replication fork stalling and the induction of a DNA damage response followed by cell death. In MM, AZA treatment led to p16 re-expression, a G0/G1 phase arrest and caspase-mediated apoptosis. In addition, AZA also suppressed the IL6 and NFkB signaling pathways (Table 4) (188). On the other hand, DAC treatment led to the re-expression of p15, p27, and p21 and the phosphorylation of p38 MAP kinase, which consequently led to both a G0/G1 and a G2/M phase arrest (11, 189, 190). We showed that, at high concentrations, DAC also induces DNA damage in MM cell lines. However, some of the cell lines were able to partially repair the DNA lesions because of their increased homologous recombination (HR) and/or non-homologous end joining (NHEJ) activity (as evidenced by the increased RAD51 and 53BP1 foci formation upon DAC treatment), thus resulting in attenuated cytotoxicity (Table 4) (190). In addition, we also demonstrated potent in vivo anti-MM activity in the 5T33MM mouse model, as evidenced by the significant higher survival rates in the DAC treated mice (190). Finally, a recent study also showed that DAC targets the monocytic myeloid derived suppressor cells (M-MDSC) and this both in vitro and in vivo. In line, combination of DAC treatment with MDSC targeting resulted in a stronger decrease in in vivo tumor growth compared to both single agents (202). However, despite these promising preclinical results, clinical trials so far showed a lack of efficacy when AZA or DAC were used as monotherapy in relapsed MM patients (Table 5). This can be partially explained by the fact that AZA and DAC are both quite unstable in aqueous solutions. Therefore, attempts were made to improve the efficacy of these two compounds (203).
A more stable version of DAC is a dinucleotide of DAC called guadecitabine (SGI-110). Guadecitabine is resistant to cytidine deaminase and has been tested in a phase 2 clinical trial of 107 newly diagnosed AML patients and a phase 2 clinical trial of fifty-five adult relapsed/refractory AML and MDS patients. The first study showed a complete response rate of more than 50%, while the second study showed responses in 14.3% of the patients resulting in prolonged OS. Other phase 1 and 2 clinical trials that are using guadecitabine in AML are currently ongoing (143). In MM, treatment of human cell lines with guadecitabine resulted in increased expression of miR-375, which is well-known to target 3-phosphoinositide-dependent protein kinase 1 (PDPK1). In line, guadecitabine also reduced PDPK1 levels (Table 4) (191). However, the authors did not report about the functional outcome. A second, more stable nucleoside analog that has been evaluated in MM is zebularine, which appears to be less toxic compared to AZA and DAC (204). In MM, zebularine reduced both DNMT3A and DNMT3B levels, resulting in reduced DNA methylation levels and reduced MM cell viability (Table 4) (192). However, so far, these improved versions of the nucleoside analogs have only been tested in vitro in MM and part of the reason for this might be the limited bioavailability observed in different species (205). Other more stable DNMTi such as CP4200, 5,6dihydroazacytidine, and 5-Fluoro-2’-Deoxycytidine (5F-CdR) have also been described, but have so far not been tested yet in MM (Table 4) (65, 193).
A second major drawback of all the nucleoside analogs is that cell cycle progression is needed for the incorporation of these cytosine analogs into the DNA. However, myeloma cells are well-known for their relatively low proliferation rates, thus implying that only a minor fraction of the myeloma cells will be affected at a given time (190, 206). This shows a need for DNMTi that are not dependent on incorporation into the DNA for their activity. Currently, a small number of DNMTi have been described that block the catalytic activity of the DNMTs by directly binding the DNMTs or, in some cases, by binding CpG rich sequences. Apart from the fact that these so-called non-nucleoside DNMTi do not require incorporation in the DNA to exert their catalytic activity, another major advantage of these non-nucleoside analogs is that, since no formation of DNA-DNMT adducts leading to DNA damage takes place, lower levels of cytotoxicity are expected, thus resulting in a larger therapeutic window compared to the nucleoside analogs. Below, we will zoom in on the most interesting non-nucleoside analogs that have been currently tested in MM.
A first non-nucleoside DNMTi that was tested in MM is mithramycin A. Treatment of MM cell lines with mithramycin A resulted in an arrest of the cells at the G1/S transition point. Moreover, mithramycin A was shown to exert anti-angiogenic effects both in vitro, using the endothelial cell migration assay and the rat aortic ring assay, and in vivo, using the 5TGM1 MM mouse model (Table 4) (194). However, it is still under debate whether mithramycin A reversibly binds CpG-rich DNA sequences or whether it binds the catalytic domain of DNMT1, leading to its depletion. Another novel DNA hypomethylating drug that falls under the class of non-nucleoside analogs is Nanaomycin A. Kuck et al. showed that this antibiotic of the anthracycline group is a DNMT3B specific inhibitor that reduces global DNA methylation levels and induces cytotoxicity in human cancer cell lines (Table 4) (195). In addition, as mentioned above, a more recent study showed that Nanaomycin A significantly reduces the amount of MM CSC, thus suggesting that inhibition of DNMT3B using Nanaomycin A would be effective in targeting the CSC in MM (147). Another non-nucleoside analog is (–)-epigallocatechin-3-gallate (EGCG). This compound is well-known to have anti-MM effects as evidenced by decreased proliferation and increased apoptosis of MM cells upon EGCG treatment (196, 197). EGCG has already been tested in a phase 2 clinical trial in MGUS and SMM patients, but was terminated early due to low patient enrolment (Table 5). Although EGCG has been shown to inhibit the HMT EZH2 in MM, so far, no study has yet investigated the effect of EGCG treatment on the activity of the DNMT enzymes in MM (Table 4) (196, 198). Other non-nucleoside inhibitors that might be interesting to test for the treatment of hematological cancers, including MM, are RG108, procaine, SGI-1027, NSC 14778, and NSC 106084 (Table 4) (65, 199). For a detailed description of these non-nucleoside analogs we refer to the review written by Foulks et al. (65).
Given the presumed role for epigenetic modifications in MM cell plasticity, epigenetic alterations have also been suggested to play a role in developing DR against current MM therapies. This provides the rationale for combining one or more epigenetic modulating agents (EMA) with SoC agents to overcome or even prevent relapse. Here, we will focus on the combination of DNMTi with SoC agents, other EMAs and/or new (immuno)therapies that have already been tested in MM.
As mentioned earlier, we previously showed that the increased HR and NHEJ activity in MM cells partially protects the MM cells from DAC-mediated cytotoxicity. However, we also showed that the histone deacetylase inhibitor (HDACi) JNJ-585 (also known as quisinostat) strongly enhances the in vitro and in vivo anti-MM activity of DAC by decreasing HR DNA repair (190, 207). Moreover, using gene expression profiling of DNMTi and/or HDACi treated MM cells, we later on constructed a gene-expression based score to predict patient outcome and MM sensitivity toward HDACi/DNMTi combination treatment. Patients with a low combo score were characterized by a mature BMPC gene signature, whereas patients with a high combo score were characterized by a proliferation and MYC-associated gene signature and a worse OS (high-risk patients) (Table 2) (84). Nevertheless, the MM cells from this high-risk group showed a higher sensitivity towards the combination of the HDACi quisinostat or trichostatin A (TSA) and the DNMTi DAC. Mechanistically, we showed that the HDACi/DNMTi combination resulted in the reprogramming of the MM cells by strongly downregulating IRF4 and MYC and inducing a normal BMPC gene expression profile. Importantly, this strong conjoined downregulation of MYC and IRF4 expression was only observed after the combination treatment and not after DNMTi or HDACi treatment alone (208). Together, these findings provide a strong rationale for the targeting of MM cells with at least two different EMA classes. In line, a more recent study designed epigenetic compounds simultaneously targeting HDACs and DNMT1 (compound 12a) on the one hand and HDACs, DNMT1, and the methyltransferase G9a (compound 9a) on the other hand. Both the dual and triple epigenetic inhibitor resulted in reduced proliferation of MM cells. Furthermore, both inhibitors led to an increase in H3K9ac levels and to increased hypomethylation, whereas compound 9a also led to a significant reduction in the levels of the H3K9me2 mark. Compound 12a was also found to significantly reduce tumor growth in the MM1.S xenograft mouse model. Unfortunately, compound 9a could not be tested in vivo as 9a proved to be lethal for the mice (209).
As IMiDs are well-known to target IRF4 and MYC, our findings also suggest that the combination of IMiDs with DNMTi/HDACi combo treatment could be of therapeutic interest for high-risk MM patients. In line, Dimopoulos et al. recently showed that dual inhibition of DNMTs and the HMT EZH2 using AZA and EPZ-6438 respectively overcomes both intrinsic and acquired IMiD resistance and this independently of cereblon (CRBN). Importantly, the IMiD resistant MM cells were found characterized by increased genome-wide DNA methylation levels and reduced chromatin accessibility and thus reduced gene expression levels. Combination of AZA with EPZ-6438 re-sensitized the IMiD resistant MM cells to both lenalidomide and pomalidomide by reversing the reduced chromatin accessibility (210). A recent study also found that hypermethylation of an active intronic CRBN enhancer was more pronounced in IMiD-refractory MM. Treatment of two MM cell lines with DNMTi resulted in the demethylation of this CRBN enhancer region, thus resulting in increased sensitivity against lenalidomide (211). Together, these results provide evidence that IMiD-acquired resistance in MM is, next to genetic mutations in CRBN, also driven by epigenetic mechanisms and that one or more EMAs together with IMiDs can restore sensitivity.
Apart from (re)boosting IMiD activity, DNMTi have also been shown to potentiate the activity of other SOC and novel agents. A pre-clinical study using the MM cell line RPMI 8226 showed that the DNMTi DAC also enhances the anti-myeloma activity of Bz, as evidenced by a stronger reduction in proliferation and stronger increase in apoptosis compared to both single agents (212). Moreover, a more recent study showed that the combinatory effect observed upon combining DAC and Bz is regulated, at least in part, by the Wnt/β-catenin signaling pathway, as re-expression of the Wnt antagonists SFRP3 and DKK1 was observed upon DAC treatment (213). In addition, DAC has also been shown to restore drug sensitivity in the dexamethasone resistant OPM1 cell line, by inducing re-expression of the tumor suppressor gene RASD1 (115). Furthermore, both AZA and DAC were only very recently shown to restore sensitivity to the monoclonal anti-CD38 Ab daratumumab in MM cell lines, by reverting epigenetic silencing of CD38 upon daratumumab treatment (214). Finally, DAC treatment was recently also shown to sensitize myeloma cells to CAR T therapy. DAC treatment resulted in global hypomethylation and increased expression of the cancer testis antigen NY-ESO-1, thereby enhancing cell lysis upon DAC and NY-ESO-1-specific CAR combination treatment (215). These recent results suggest that (pre-)treatment with DAC might sensitize the myeloma cells to daratumumab and CAR T-cell therapy.
So far, clinical trials have mainly investigated the efficacy of AZA in combination with lenalidomide and/or dexamethasone (Table 5). Reu et al. found that low-dose AZA (subcutaneously) in combination with lenalidomide and/or dexamethasone yielded response rates of about 23% in patients with relapsed or refractory MM, but with the cost of grade 3/4 toxicities in about half of the patients (216). In addition, another study investigating the combination of oral AZA together with lenalidomide and/or dexamethasone in relapsed or refractory MM patients with a history of lenalidomide failure showed overall response rates of about 37.5% without significant toxicities. The author suggested that these superior results, compared to the study from Reu et al, were the result of the extended exposure to AZA (217). Finally, a recent case report described the treatment of a MM patient with a combination of AZA and lenalidomide followed by a combination of AZA and daratumumab upon relapse to be successful (218). Furthermore, a clinical trial investigating a combination therapy combining AZA with e.g., daratumumab and dexamethasone is currently ongoing (Table 5). Clinical trials combining DNMTi with Bz or CAR T-cells in MM are still awaited.
It has become apparent that epigenetic defects, including global DNA hypomethylation and locus specific DNA hypermethylation together with aberrant expression and/or mutations in the DNA methylation modifiers, play an important role in MM onset, progression, and relapse. Moreover, it is becoming increasingly apparent that aberrant DNA methylation patterns/modifiers also play a role in acquiring a heterogeneous MM cell population, encompassing both the mature MM cells and the more immature MM cells characterized by their drug-resistant capacity, and allows for the bidirectional transition between these two states. This epigenetic plasticity allows the MM cells to adapt to their environment, including treatment pressure, and thus escape from all currently available MM therapies. However, although the aberrant DNA methylation patterns and modifiers are clearly major obstacles for curing MM, they also represent great opportunities. Firstly, DNA methylation modifiers represent attractive targets to overcome or delay (acquired) MM cell DR. Indeed, (pre)clinical studies have repeatedly shown that combining DNMTi with either lenalidomide, dexamethasone, Bz, or HDACi is promising for (re)sensitizing the MM cells. Furthermore, DNMTi were recently also shown to upregulate the epigenetically silenced surface proteins used as targets in immunotherapies, including daratumumab and CAR T therapy. These findings advocate for the careful consideration of incorporating DNMTi in the current MM therapies. Moreover, it would also be interesting to find out whether DNMTi could also be of benefit to increase the efficacy of the new, emerging immunotherapies such as ADC and T-cell engagers. Secondly, it is well-known that DNA methylation patterns, as a consequence transcriptional patterns, change upon MM progression. Characterization of the DNA methylation patterns and/or methylation-regulated gene expression in MM patients might, on the one hand, predict response to treatment and OS and, on the other hand, identify those patients that might benefit most from DNMTi therapy. In fact, Moreaux et al. and we developed gene expression-based scores that predict not only patient outcome, but also primary MM cell sensitivity to HDACi and/or DNMTi treatment (84–86). Furthermore, a DNA methylation inference framework called MethSig was recently developed that was demonstrated to be superior in distinguishing stochastic DNA methylation changes, with no biological consequence, from likely driver DNA methylation changes in CLL and MM. When developing a prediction score based on the identified candidate DNA methylation drivers, a higher DNA methylation driver risk score was found associated with an adverse outcome in CLL patients (219). Currently, this DNA methylation driver risk score has not yet been constructed for MM. Hence, it will be interesting to further apply this MethSig tool on MM DNA methylation sequencing cohorts to further identify the oncogenic DNA methylation drivers in MM and to construct and test a MM DNA methylation driver risk score.
However, in order to seize these opportunities, some challenges must be tackled first. Since only a tip of the iceberg concerning the role of the different DNA methylation modifiers in MM disease progression and especially in relapse has thus far been revealed, it is of utmost importance to further investigate the role of these DNA methylation modifiers. In this regard, it will be important to, apart from the DNA methylation writers and erasers, not lose sight of the DNA methylation readers; a group of modifiers that has until now completely been neglected in MM. Recent evidence however points out that DNA methylation readers including readers from the methyl binding domain (MBD) family (MBD3), the SET and RING finger associated (SRA) domain protein family (UHRF1), and the zinc finger family (EGR1 and KLF4) are also involved in cancer pathogenesis. Moreover, in order to make it even more complicated, it is well-known that many DNA methylation modifiers have multiple splice variants/isoforms, with apparently each of these isoforms having distinct activities. At the moment, the role of these different isoforms and their consequences in MM disease is not yet explored. As these isoforms can regulate the activity of the default DNA methylation modifiers, it will be interesting to investigate the role of both catalytic active and inactive isoforms in MM disease. Furthermore, although preclinical (combination) studies showed promising results, clinical studies demonstrated that the pan-DNMTi are subject to a lack of efficacy and high toxicity profiles complicating the broad application of these agents in the clinic. Hence, more specific DNA hypomethylating drugs are urgently needed. Different novel DNMTi, encompassing both the newer nucleoside and non-nucleoside inhibitors, have already been developed and this number is only increasing. However, till today, only a small number of these inhibitors has been tested in MM. Moreover, when looking in the direction of the erasers, no TET inhibitors (TETi) have so far been tested in MM. Two promising TET inhibitors (TETi) have only very recently been identified, C35 compound and TETi76. Both compounds specifically target all three TET members (200, 201). Although one of the two compounds, namely TETi76, has already been tested in AML and was shown to restrict colony formation, neither compounds have yet been tested thoroughly in cancer. Further research is needed to provide insights into the applicability of TET inhibitors for the treatment of MM.
In conclusion, it is clear that the true potential of DNA methylation patterns and modifiers for MM patient stratification and therapeutic targeting is only starting to unfold itself. Although many challenges still need to be overcome, we envision that in time DNA methylation sequencing and the use of DNA methylation modulating agents will become common practice for the follow-up and treatment of MM.
CM and EDB developed the design and arguments for the paper and drafted the manuscript. CM, LH, and EDB designed the figures. LH, AM, KV, EM, and KV revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Fonds voor Wetenschappelijk Onderzoek (FWO), Wetenschappelijk Fonds Willy Gepts (WFWG), and Strategic Research Programme (SRP48), K. De Veirman is a postdoctoral fellow of FWO (12I0921N).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Robak P, Drozdz I, Szemraj J, Robak T. Drug resistance in multiple myeloma. Cancer Treat Rev (2018) 70:199–208. doi: 10.1016/j.ctrv.2018.09.001
2. Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol (2014) 15(12):E538–48. doi: 10.1016/s1470-2045(14)70442-5
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
4. Zhou LH, Yu Q, Wei GQ, Wang LQ, Huang Y, Hu KJ, et al. Measuring the global, regional, and national burden of multiple myeloma from 1990 to 2019. BMC Cancer (2021) 21(1):606. doi: 10.1186/s12885-021-08280-y
5. Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood (2009) 113(22):5418–22. doi: 10.1182/blood-2008-12-195008
6. Kyle RA, Larson DR, Therneau TM, Dispenzieri A, Kumar S, Cerhan JR, et al. Long-term follow-up of monoclonal gammopathy of undetermined significance. New Engl J Med (2018) 378(3):241–9. doi: 10.1056/NEJMoa1709974
7. Kyle RA, Therneau TM, Rajkumar SV, Offord JR, Larson DR, Plevak MF, et al. A long-term study of prognosis in monoclonal gammopathy of undetermined significance. New Engl J Med (2002) 346(8):564–9. doi: 10.1056/NEJMoa01133202
8. Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Primers 3 (2017) 3:17046. doi: 10.1038/nrdp.2017.46
9. Gerecke C, Fuhrmann S, Strifler S, Schmidt-Hieber M, Einsele H, Knop S. The diagnosis and treatment of multiple myeloma. Deutsches Arzteblatt Int (2016) 113(27-28):470–+. doi: 10.3238/arztebl.2016.0470
10. Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol (2020) 95(5):548–67. doi: 10.1002/ajh.25791
11. Maes K, Menu E, Van Valckenborgh E, Van Riet I, Vanderkerken K, De Bruyne E. Epigenetic modulating agents as a new therapeutic approach in multiple myeloma. Cancers (Basel) (2013) 5(2):430–61. doi: 10.3390/cancers5020430
12. van Rhee F, Giralt S, Barlogie B. The future of autologous stem cell transplantation in myeloma. Blood (2014) 124(3):328–33. doi: 10.1182/blood-2014-03-561985
13. D'Agostino M, Raje N. Anti-BCMA CAR T-cell therapy in multiple myeloma: can we do better? Leukemia (2020) 34(1):21–34. doi: 10.1038/s41375-019-0669-4
14. Brown S, Pawlyn C, Tillotson AL, Sherratt D, Flanagan L, Low E, et al. Bortezomib, vorinostat, and dexamethasone combination therapy in relapsed myeloma: Results of the phase 2 MUK four trial. Clin Lymphoma Myeloma Leukemia (2021) 21(3):154–+. doi: 10.1016/j.clml.2020.11.019
15. Yu B, Liu DL. Antibody-drug conjugates in clinical trials for lymphoid malignancies and multiple myeloma. J Hematol Oncol (2019) 12(1):94. doi: 10.1186/s13045-019-0786-6
16. Shah Z, Malik MN, Batool SS, Kotapati S, Akhtar A, Rehman OU, et al. Bispecific T-cell engager (BiTE) antibody based immunotherapy for treatment of relapsed refractory multiple myeloma (RRMM): A systematic review of preclinical and clinical trials. Blood (2019) 134:5567. doi: 10.1182/blood-2019-129652
17. Di Marzo L, Desantis V, Solimando AG, Ruggieri S, Annese T, Nico B, et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget (2016) 7(37):60698–711. doi: 10.18632/oncotarget.10849
18. Suzuki K, Nishiwaki K, Yano S. Treatment strategies considering micro-environment and clonal evolution in multiple myeloma. Cancers (2021) 13(2):215. doi: 10.3390/cancers13020215
19. Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N, et al. Drug resistance in cancer: An overview. Cancers (2014) 6(3):1769–92. doi: 10.3390/cancers6031769
20. Anreddy N, Hazlehurst LA. Targeting intrinsic and extrinsic vulnerabilities for the treatment of multiple myeloma. J Cell Biochem (2017) 118(1):15–25. doi: 10.1002/jcb.25617
21. Pinto V, Bergantim R, Caires HR, Seca H, Guimaraes JE, Vasconcelos MH. Multiple myeloma: Available therapies and causes of drug resistance. Cancers (2020) 12(2):407. doi: 10.3390/cancers12020407
22. Bianchi G, Munshi NC. Pathogenesis beyond the cancer clone(s) in multiple myeloma. Blood (2015) 125(20):3049–58. doi: 10.1182/blood-2014-11-568881
23. Goldman-Mazur S, Vesole D, Jurczyszyn A. Clinical implications of cytogenetic and molecular aberrations in multiple myeloma. Acta Haematol Polonica (2021) 52(1):18–28. doi: 10.5603/AHP.2021.0004
24. Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the intergroupe francophone du myelome. Blood (2007) 109(8):3489–95. doi: 10.1182/blood-2006-08-040410
>25. Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun (2014) 5:2997. doi: 10.1038/ncomms3997
>26. Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell (2014) 25(1):91–101. doi: 10.1016/j.ccr.2013.12.015
27. Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature (2011) 471(7339):467–72. doi: 10.1038/nature09837
28. Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappa b pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell (2007) 12(2):115–30. doi: 10.1016/j.ccr.2007.07.004
29. Issa ME, Takhsha FS, Chirumamilla CS, Perez-Novo C, Berghe WV, Cuendet M. Epigenetic strategies to reverse drug resistance in heterogeneous multiple myeloma. Clin Epigenet (2017) 9:17. doi: 10.1186/s13148-017-0319-5
30. Dimopoulos K, Gimsing P, Gronbaek K. The role of epigenetics in the biology of multiple myeloma. Blood Cancer J (2014) 4(5):e207. doi: 10.1038/bcj.2014.29
>31. De Smedt E, Lui H, Maes K, De Veirman K, Menu E, Vanderkerken K, et al. The epigenome in multiple myeloma: Impact on tumor cell plasticity and drug response. Front Oncol (2018) 8:566. doi: 10.3389/fonc.2018.00566
32. Zhang W, Xu J. DNA Methyltransferases and their roles in tumorigenesis. biomark Res (2017) 5:1. doi: 10.1186/s40364-017-0081-z
33. Zhu H, Wang GH, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet (2016) 17(9):551–65. doi: 10.1038/nrg.2016.83
34. Suzuki MM, Bird A. DNA Methylation landscapes: provocative insights from epigenomics. Nat Rev Genet (2008) 9(6):465–76. doi: 10.1038/nrg2341
35. Moore LD, Le T, Fan GP. DNA Methylation and its basic function. Neuropsychopharmacology (2013) 38(1):23–38. doi: 10.1038/npp.2012.112
36. Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci (2003) 60(8):1647–58. doi: 10.1007/s00018-003-3088-6
37. Baylin SB, Herman JG. DNA Hypermethylation in tumorigenesis - epigenetics joins genetics. Trends Genet (2000) 16(4):168–74. doi: 10.1016/s0168-9525(99)01971-x
38. Hamidi T, Singh AK, Chen TP. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics (2015) 7(2):247–65. doi: 10.2217/epi.14.80
>39. Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenet (2018) 10:17. doi: 10.1186/s13148-018-0450-y
40. Chen TP, Hevi S, Gay F, Tsujimoto N, He T, Zhang BL, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet (2007) 39(3):391–6. doi: 10.1038/ng1982
>41. Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood (2001) 97(5):1172–9. doi: 10.1182/blood.V97.5.1172
42. Robertson KD, Uzvolgyi E, Liang GN, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res (1999) 27(11):2291–8. doi: 10.1093/nar/27.11.2291
43. Robertson KD. DNA Methylation, methyltransferases, and cancer. Oncogene (2001) 20(24):3139–55. doi: 10.1038/sj.onc.1204341
44. Li E, Bestor TH, Jaenisch R. Targeted mutation of the dna methyltransferase gene results in embryonic lethality. Cell (1992) 69(6):915–26. doi: 10.1016/0092-8674(92)90611-f
45. Okano M, Bell DW, Haber DA, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell (1999) 99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6
46. Watanabe D, Suetake I, Tada T, Tajima S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech Dev (2002) 118(1-2):187–90. doi: 10.1016/s0925-4773(02)00242-3
47. Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature (2004) 429(6994):900–3. doi: 10.1038/nature02633
48. Van Emburgh BO, Robertson KD. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res (2011) 39(12):4984–5002. doi: 10.1093/nar/gkr116
49. Xie SP, Wang ZJ, Okano M, Nogami M, Li Y, He WW, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene (1999) 236(1):87–95. doi: 10.1016/s0378-1119(99)00252-8
50. Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development (1998) 125(5):889–97. doi: 10.1242/dev.125.5.889
51. Chen TP, Ueda Y, Xie SP, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem (2002) 277(41):38746–54. doi: 10.1074/jbc.M205312200
52. Saravanaraman P, Selvam M, Ashok C, Srijyothi L, Baluchamy S. De novo methyltransferases: Potential players in diseases and new directions for targeted therapy. Biochimie (2020) 176:85–102. doi: 10.1016/j.biochi.2020.07.004
53. Gordon CA, Hartono SR, Chedin F. Inactive DNMT3B splice variants modulate De novo DNA methylation. PloS One (2013) 8(7):e69486. doi: 10.1371/journal.pone.0069486
54. Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci United States America (2002) 99(15):10060–5. doi: 10.1073/pnas.152121799
55. Gujar H, Weisenberger DJ, Liang GN. The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes (2019) 10(2):172. doi: 10.3390/genes10020172
56. Ostler K, Davis E, Payne S, Gosalia B, Exposito-Cespedes J, Le Beau MM, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene (2007) 26(38):5553–63. doi: 10.1038/sj.onc.1210351
57. Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res (2010) 70(14):5840–50. doi: 10.1158/0008-5472.can-10-0847
58. Vasanthakumar A, Lepore JB, Zegarek MH, Kocherginsky M, Singh M, Davis EM, et al. Dnmt3b is a haploinsufficient tumor suppressor gene in myc-induced lymphomagenesis. Blood (2013) 121(11):2059–63. doi: 10.1182/blood-2012-04-421065
59. Liao J, Karnik R, Gu HC, Ziller MJ, Clement K, Tsankov AM, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet (2015) 47(5):469–U64. doi: 10.1038/ng.3258
60. Singh P, Sailu S, Palchamy E, Coumar MS, Baluchamy S. Identification of a novel leukemic-specific splice variant of DNMT3B and its stability. Med Oncol (2017) 34(8):145. doi: 10.1007/s12032-017-1008-0
61. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet (2018) 19(2):81–92. doi: 10.1038/nrg.2017.80
62. Suetake I, Mishima Y, Kimura H, Lee YH, Goto Y, Takeshima H, et al. Characterization of DNA-binding activity in the n-terminal domain of the DNA methyltransferase Dnmt3a. Biochem J (2011) 437:141–8. doi: 10.1042/bj20110241
63. Wang J, Walsh G, Liu DD, Lee JJ, Mao L. Expression of delta DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res (2006) 66(17):8361–6. doi: 10.1158/0008-5472.can-06-2031
64. Wang J, Bhutani M, Pathak AK, Lang W, Ren H, Jelinek J, et al. Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res (2007) 67(22):10647–52. doi: 10.1158/0008-5472.can-07-1337
65. Foulks JM, Parnell KM, Nix RN, Chau S, Swierczek K, Saunders M, et al. Epigenetic drug discovery: Targeting DNA methyltransferases. J Biomolecular Screening (2012) 17(1):2–17. doi: 10.1177/1087057111421212
66. Alzrigat M, Parraga AA, Jernberg-Wiklund H. Epigenetics in multiple myeloma: From mechanisms to therapy. Semin Cancer Biol (2018) 51:101–15. doi: 10.1016/j.semcancer.2017.09.007
67. Williams K, Christensen J, Helin K. DNA Methylation: TET proteins-guardians of CpG islands? EMBO Rep (2012) 13(1):28–35. doi: 10.1038/embor.2011.233
68. Wu XJ, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet (2017) 18(9):517–34. doi: 10.1038/nrg.2017.33
69. Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev (2016) 30(7):733–50. doi: 10.1101/gad.276568.115
70. Melamed P, Yosefzon Y, David C, Tsukerman A, Pnueli L. Tet enzymes, variants, and differential effects on function. Front Cell Dev Biol (2018) 6:22. doi: 10.3389/fcell.2018.00022
71. Liu DY, Li GP, Zuo YC. Function determinants of TET proteins: the arrangements of sequence motifs with specific codes. Briefings Bioinf (2019) 20(5):1826–35. doi: 10.1093/bib/bby053
72. Lyko F, Brown R. DNA Methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst (2005) 97(20):1498–506. doi: 10.1093/jnci/dji311
73. Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem (2003) 278(26):24132–8. doi: 10.1074/jbc.M302283200
74. Du Q, Luu PL, Stirzaker C, Clark SJ. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics (2015) 7(6):1051–73. doi: 10.2217/epi.15.39
75. Jabbour E, Issa JP, Garcia-Manero G, Kantarjian H. Evolution of decitabine development - accomplishments, ongoing investigations, and future strategies. Cancer (2008) 112(11):2341–51. doi: 10.1002/cncr.23463
76. >Shimbo T, Wade PA. Proteins that read DNA methylation. DNA Methyltransferases - Role Funct (2016) 945:303–20. doi: 10.1007/978-3-319-43624-1_13
77. Patnaik D, Estève PO, Pradhan S. Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger-containing 1 (UHRF1) for anti-cancer drug development. Oncotarget (2018) 9(40):26243–58. doi: 10.18632/oncotarget.25425
78. Hudson NO, Buck-Koehntop BA. Zinc finger readers of methylated DNA. Molecules (2018) 23(10):2555. doi: 10.3390/molecules23102555
79. Heuck CJ, Mehta J, Bhagat T, Gundabolu K, Yu YT, Khan S, et al. Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis. J Immunol (2013) 190(6):2966–75. doi: 10.4049/jimmunol.1202493
80. Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood (2011) 117(2):553–62. doi: 10.1182/blood-2010-04-279539
81. Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, et al. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis (2009) 30(8):1330–5. doi: 10.1093/carcin/bgp149
82. Sive JI, Feber A, Smith D, Quinn J, Beck S, Yong K. Global hypomethylation in myeloma is associated with poor prognosis. Br J Haematol (2016) 172(3):473–5. doi: 10.1111/bjh.13506
83. Derrien J, Guerin-Charbonnel C, Gaborit V, Campion L, Devic M, Douillard E, et al. The DNA methylation landscape of multiple myeloma shows extensive inter- and intrapatient heterogeneity that fuels transcriptomic variability. Genome Med (2021) 13(1):127. doi: 10.1186/s13073-021-00938-3
84. Moreaux J, Reme T, Leonard W, Veyrune JL, Requirand G, Goldschmidt H, et al. Development of gene expression-based score to predict sensitivity of multiple myeloma cells to DNA methylation inhibitors. Mol Cancer Ther (2012) 11(12):2685–92. doi: 10.1158/1535-7163.mct-12-0721
85. Moreaux J, Bruyer A, Veyrune JL, Goldschmidt H, Hose D, Klein B. DNA Methylation score is predictive of myeloma cell sensitivity to 5-azacitidine. Br J Haematol (2014) 164(4):613–6. doi: 10.1111/bjh.12660
86. Moreaux J, Reme T, Leonard W, Veyrune JL, Requirand G, Goldschmidt H, et al. Gene expression-based prediction of myeloma cell sensitivity to histone deacetylase inhibitors. Br J Cancer (2013) 109(3):676–85. doi: 10.1038/bjc.2013.392
87. Pawlyn C, Kaiser MF, Heuck C, Melchor L, Wardell CP, Murison A, et al. The spectrum and clinical impact of epigenetic modifier mutations in myeloma. Clin Cancer Res (2016) 22(23):5783–94. doi: 10.1158/1078-0432.ccr-15-1790
88. Luzna P, Drozdkova DW, Flodrova P, Ondruskova K, Uberall I, Minarik J, et al. Global DNA methylation and increased DNMT3A expression in multiple myeloma patients. Biomed Papers-Olomouc doi. doi: 10.5507/bp.2022.006
89. Yang T, Liu XB, Kumar SK, Jin FY, Dai Y. Decoding DNA methylation in epigenetics of multiple myeloma. Blood Rev (2022) 51:100872. doi: 10.1016/j.blre.2021.100872
90. Barwick BG, Powell DR, Penaherrera D, Skerget S, Keats JJ, Auclair D, et al. Whole genome DNA methylation analysis of multiple myeloma identifies pervasive hypomethylation and biomarkers of survival. Cancer Res (2019) 79(13):839. doi: 10.1158/1538-7445.am2019-839
91. Houde C, Li YL, Song L, Barton K, Zhang Q, Godwin J, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood (2004) 104(12):3697–704. doi: 10.1182/blood-2003-12-4114
92. Turner JG, Gump JL, Zhang CC, Cook JM, Marchion D, Hazlehurst L, et al. ABCG2 expression, function, and promoter methylation in human multiple myeloma. Blood (2006) 108(12):3881–9. doi: 10.1182/blood-2005-10-009084
93. Galm O, Wilop S, Reichelt J, Jost E, Gehbauer G, Herman JG, et al. DNA Methylation changes in multiple myeloma. Leukemia (2004) 18(10):1687–92. doi: 10.1038/sj.leu.2403434
94. Chim CS, Liang R, Leung MH, Kwong YL. Aberrant gene methylation implicated in the progression of monoclonal gammopathy of undetermined significance to multiple myeloma. J Clin Pathol (2007) 60(1):104–6. doi: 10.1136/jcp.2006.036715
95. Braggio E, Maiolino A, Gouveia ME, Magalhaes R, Souto JT, Garnica M, et al. Methylation status of nine tumor suppressor genes in multiple myeloma. Int J Hematol (2010) 91(1):87–96. doi: 10.1007/s12185-009-0459-2
96. Chim CS, Fung TK, Liang R. Disruption of INK4/CDK/Rb cell cycle pathway by gene hypermethylation in multiple myeloma and MGUS. Leukemia (2003) 17(12):2533–5. doi: 10.1038/sj.leu.2403133
97. Seidl S, Ackermann J, Kaufmann H, Keck A, Nosslinger T, Zielinski CC, et al. DNA-Methylation analysis identifies the e-cadherin gene as a potential marker of disease progression in patients with monoclonal gammopathies. Cancer (2004) 100(12):2598–606. doi: 10.1002/cncr.20295
98. Hodge DR, Peng B, Cherry JC, Hurt EM, Fox SD, Kelley JA, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promoter methylation. Cancer Res (2005) 65(11):4673–82. doi: 10.1158/0008-5472.can-04-3589
99. Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet (1999) 21(2):163–7. doi: 10.1038/5947
100. Laird PW, Jaenisch R. Dna methylation and cancer. Hum Mol Genet (1994) 3:1487–95. doi: 10.1093/hmg/3.suppl_1.1487
101. Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res (1988) 48(5):1159–61.
102. Reddy J, Shivapurkar N, Takahashi T, Parikh G, Stastny V, Echebiri C, et al. Differential methylation of genes that regulate cytokine signaling in lymphoid and hematopoietic tumors. Oncogene (2005) 24(4):732–6. doi: 10.1038/sj.onc.1208032
103. Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood (2003) 101(7):2784–8. doi: 10.1182/blood-2002-06-1735
104. Heller G, Schmidt WM, Ziegler B, Holzer S, Mullauer L, Bilban M, et al. Genome-wide transcriptional response to 5-aza-2 '-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res (2008) 68(1):44–54. doi: 10.1158/0008-5472.can-07-2531
105. Chim CS, Liang R, Fung TK, Choi CL, Kwong YL. Epigenetic dysregulation of the death-associated protein kinase/p14/HDM2/p53/Apaf-1 apoptosis pathway in multiple myeloma. J Clin Pathol (2007) 60(6):664–9. doi: 10.1136/jcp.2006.038331
106. Chen GH, Wang Y, Huang HW, Lin FR, Wu DP, Sun AN, et al. Combination of DNA methylation inhibitor 5-azacytidine and arsenic trioxide has synergistic activity in myeloma. Eur J Haematol (2009) 82(3):176–83. doi: 10.1111/j.1600-0609.2008.01189.x
107. de Carvalho F, Colleoni GWB, Almeida MSS, Carvalho AL, Vettore AL. TGF beta R2 aberrant methylation is a potential prognostic marker and therapeutic target in multiple myeloma. Int J Cancer (2009) 125(8):1985–91. doi: 10.1002/ijc.24431
108. De Smedt E, Maes K, Verhulst S, Lui H, Kassambara A, Maes A, et al. Loss of RASSF4 expression in multiple myeloma promotes RAS-driven malignant progression. Cancer Res (2018) 78(5):1155–68. doi: 10.1158/0008-5472.can-17-1544
109. Hatzimichael E, Dranitsaris G, Dasoula A, Benetatos L, Stebbing J, Crook T, et al. Von hippel-lindau methylation status in patients with multiple myeloma: A potential predictive factor for the development of bone disease. Clin Lymphoma Myeloma (2009) 9(3):239–42. doi: 10.3816/CLM.2009.n.047
110. Hatzimichael E, Dasoula A, Shah R, Syed N, Papoudou-Bai A, Coley HM, et al. The prolyl-hydroxylase EGLN3 and not EGLN1 is inactivated by methylation in plasma cell neoplasia. Eur J Haematol (2010) 84(1):47–51. doi: 10.1111/j.1600-0609.2009.01344.x
111. Tshuikina M, Jernberg-Wiklund H, Nilsson K, Oberg F. Epigenetic silencing of the interferon regulatory factor ICSBP/IRF8 in human multiple myeloma. Exp Hematol (2008) 36(12):1673–81. doi: 10.1016/j.exphem.2008.08.001
112. Kaiser MF, Johnson DC, Wu P, Walker BA, Brioli A, Mirabella F, et al. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood (2013) 122(2):219–26. doi: 10.1182/blood-2013-03-487884
113. Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, et al. High-frequency promoter hypermethylation of the deleted in liver cancer-1 gene in multiple myeloma. J Clin Pathol (2006) 59(9):947–51. doi: 10.1136/jcp.2005.031377
114. De Bruyne E, Bos TJ, Asosingh K, Broek IV, Menu E, Van Valckenborgh E, et al. Epigenetic silencing of the tetraspanin CD9 during disease progression in multiple myeloma cells and correlation with survival. Clin Cancer Res (2008) 14(10):2918–26. doi: 10.1158/1078-0432.ccr-07-4489
115. Nojima M, Maruyama R, Yasui H, Suzuki H, Maruyama Y, Tarasawa I, et al. Genomic screening for genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clin Cancer Res (2009) 15(13):4356–64. doi: 10.1158/1078-0432.ccr-08-3336
116. Ng MHL, Chung YF, Lo KW, Wickham NWR, Lee JCK, Huang DP. Frequent hypermethylation of p16 and p15 genes in multiple myeloma. Blood (1997) 89(7):2500–6. doi: 10.1182/blood.V89.7.2500
117. Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of wnt signaling pathway in multiple myeloma. Leukemia (2007) 21(12):2527–36. doi: 10.1038/sj.leu.2404939
118. Chim CS, Fung TK, Cheung WC, Liang R, Kwong YL. SOCS1 and SHP1 hypermethylation in multiple myeloma: implications for epigenetic activation of the Jak/STAT pathway. Blood (2004) 103(12):4630–5. doi: 10.1182/blood-2003-06-2007
119. Agirre X, Castellano G, Pascual M, Heath S, Kulis M, Segura V, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of b cell-specific enhancers. Genome Res (2015) 25(4):478–87. doi: 10.1101/gr.180240.114
120. Oakes CC, Martin-Subero JI. Insight into origins, mechanisms, and utility of DNA methylation in b-cell malignancies. Blood (2018) 132(10):999–1006. doi: 10.1182/blood-2018-02-692970
121. Celik H, Kramer A, Challen GA. DNA Methylation in normal and malignant hematopoiesis. Int J Hematol (2016) 103(6):617–26. doi: 10.1007/s12185-016-1957-7
122. Gagliardi M, Strazzullo M, Matarazzo MR. DNMT3B functions: Novel insights from human disease. Front Cell Dev Biol (2018) 6:140. doi: 10.3389/fcell.2018.00140
123. Zhou WW, Chen HY, Hong XL, Niu XQ, Lu QY. Knockdown of DNA methyltransferase-1 inhibits proliferation and derepresses tumor suppressor genes in myeloma cells. Oncol Lett (2014) 8(5):2130–4. doi: 10.3892/ol.2014.2481
124. Harada T, Ohguchi H, Grondin Y, Kikuchi S, Sagawa M, Tai YT, et al. HDAC3 regulates DNMT1 expression in multiple myeloma: therapeutic implications. Leukemia (2017) 31(12):2670–7. doi: 10.1038/leu.2017.144
125. Li M, Qi L, Xu JB, Zhong LY, Chan S, Chen SN, et al. Methylation of the promoter region of the tight junction protein-1 by DNMT1 induces EMT-like features in multiple myeloma. Mol Ther-Oncolytics (2020) 19:197–207. doi: 10.1016/j.omto.2020.10.004
126. Lombard AP, Mooso BA, Libertini SJ, Lim RM, Nakagawa RM, Vidallo KD, et al. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol Carcinogenesis (2016) 55(5):757–67. doi: 10.1002/mc.22319
127. Shen N, Yan F, Pang JX, Wu LC, Al-Kali A, Litzow MR, et al. A nucleolin-DNMT1 regulatory axis in acute myeloid leukemogenesis. Oncotarget (2014) 5(14):5494–509. doi: 10.18632/oncotarget.2131
128. Tomasello L, Vezzalini M, Boni C, Bonifacio M, Scaffidi L, Yassin M, et al. Regulative loop between beta-catenin and protein tyrosine receptor type gamma in chronic myeloid leukemia. Int J Mol Sci (2020) 21(7):2298. doi: 10.3390/ijms21072298
129. Shen N, Yan F, Pang JX, Zhao N, Gangat N, Wu LC, et al. Inactivation of receptor tyrosine kinases reverts aberrant DNA methylation in acute myeloid leukemia. Clin Cancer Res (2017) 23(20):6254–66. doi: 10.1158/1078-0432.ccr-17-0235
130. Yan F, Shen N, Pang JX, Zhao N, Zhang YW, Bode AM, et al. A vicious loop of fatty acid-binding protein 4 and DNA methyltransferase 1 promotes acute myeloid leukemia and acts as a therapeutic target. Leukemia (2018) 32(4):865–73. doi: 10.1038/leu.2017.307
131. Brunetti L, Gundry MC, Goodell MA. DNMT3A in leukemia. Cold Spring Harbor Perspect Med (2017) 7(2):a030320. doi: 10.1101/cshperspect.a030320
132. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. New Engl J Med (2010) 363(25):2424–33. doi: 10.1056/NEJMoa1005143
133. Lin N, Fu W, Zhao C, Li BX, Yan XJ, Li Y. Biologico-clinical significance of DNMT3A variants expression in acute myeloid leukemia. Biochem Biophys Res Commun (2017) 494(1-2):270–7. doi: 10.1016/j.bbrc.2017.10.041
134. Zhang TJ, Zhang LC, Xu ZJ, Zhou JD. Expression and prognosis analysis of DNMT family in acute myeloid leukemia. Aging-Us (2020) 12(14):14677–90. doi: 10.18632/aging.103520
135. Kim MS, Kim YR, Yoo NJ, Lee SH. Mutational analysis of DNMT3A gene in acute leukemias and common solid cancers. Apmis (2013) 121(2):85–94. doi: 10.1111/j.1600-0463.2012.02940.x
136. Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies FE, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood (2018) 132(6):587–97. doi: 10.1182/blood-2018-03-840132
137. Jirabanditsakul C, Dakeng S, Kunacheewa C, U-pratya Y, Owattanapanich W. Comparison of clinical characteristics and genetic aberrations of plasma cell disorders in Thailand population. Technol Cancer Res Treat (2022) 21:15330338221111228. doi: 10.1177/15330338221111228
138. Amodio N, Leotta M, Bellizzi D, Di Martino MT, D'Aquila P, Lionetti M, et al. DNA-Demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget (2012) 3(10):1246–58. doi: 10.18632/oncotarget.675
139. Garzon R, Liu SJ, Fabbri M, Liu ZF, Heaphy CEA, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood (2009) 113(25):6411–8. doi: 10.1182/blood-2008-07-170589
140. Poole CJ, Zheng WL, Lodh A, Yevtodiyenko A, Liefwalker D, Li HL, et al. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and burkitt's lymphoma. Oncotarget (2017) 8(44):76898–920. doi: 10.18632/oncotarget.20176
141. Hayette S, Thomas X, Jallades L, Chabane K, Charlot C, Tigaud I, et al. High DNA methyltransferase DNMT3B levels: A poor prognostic marker in acute myeloid leukemia. PloS One (2012) 7(12):e51527. doi: 10.1371/journal.pone.0051527
142. Amara K, Ziadi S, Hachana M, Soltani N, Korbi S, Trimeche M. DNA Methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large b-cell lymphomas. Cancer Sci (2010) 101(7):1722–30. doi: 10.1111/j.1349-7006.2010.01569.x
143. Wong KK, Lawrie CH, Green TM. Oncogenic roles and inhibitors of DNMT1, DNMT3A, a DNMT3B in acute myeloid leukaemia. biomark Insights 14 (2019) 14:1177271919846454. doi: 10.1177/1177271919846454
144. Lopusna K, Nowialis P, Opavska J, Abraham A, Riva A, Opavsky R. Dnmt3b catalytic activity is critical for its tumour suppressor function in lymphomagenesis and is associated with c-met oncogenic signalling. Ebiomedicine (2021) 63:103191. doi: 10.1016/j.ebiom.2020.103191
145. Lopusna K, Nowialis P, Opavska J, Abraham A, Riva A, Haney SL, et al. Decreases in different Dnmt3b activities drive distinct development of hematologic malignancies in mice. J Biol Chem (2021) 296:100285. doi: 10.1016/j.jbc.2021.100285
146. Zheng Y, Zhang H, Wang Y, Li X, Lu P, Dong F, et al. Loss of Dnmt3b accelerates MLL-AF9 leukemia progression. Leukemia (2016) 30(12):2373–84. doi: 10.1038/leu.2016.112
147. Ai LS, Mu SD, Sun CY, Fan FJ, Yan H, Qin Y, et al. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer (2019) 18:88. doi: 10.1186/s12943-019-1011-5
148. Feng YM, Li XP, Cassady K, Zou ZM, Zhang X. TET2 function in hematopoietic malignancies, immune regulation, and DNA repair. Front Oncol (2019) 9:210. doi: 10.3389/fonc.2019.00210
149. Li Z, Cai XQ, Cai CL, Wang JP, Zhang WY, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood (2011) 118(17):4509–18. doi: 10.1182/blood-2010-12-325241
150. Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood (2009) 114(1):144–7. doi: 10.1182/blood-2009-03-210039
151. Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, Nielsen AB, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large b-cell lymphoma. Haematologica (2013) 98(12):1912–20. doi: 10.3324/haematol.2013.088740
152. Seethy A, Pethusamy K, Chattopadhyay I, Sah R, Chopra A, Dhar R, et al. TETology: Epigenetic mastermind in action. Appl Biochem Biotechnol (2021) 193(6):1701–26. doi: 10.1007/s12010-021-03537-5
153. Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood (2009) 114(15):3285–91. doi: 10.1182/blood-2009-04-215814
154. Nibourel O, Kosmider O, Cheok M, Boissel N, Renneville A, Philippe N, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood (2010) 116(7):1132–5. doi: 10.1182/blood-2009-07-234484
155. Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet (2009) 41(7):838–U102. doi: 10.1038/ng.391
156. Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia (2012) 26(5):934–42. doi: 10.1038/leu.2011.326
157. Gurnari C, Pagliuca S, Guan YH, Adema V, Hershberger CE, Ni Y, et al. TET2 mutations as a part of DNA dioxygenase deficiency in myelodysplastic syndromes. Blood Adv (2022) 6(1):100–7. doi: 10.1182/bloodadvances.2021005418
158. Zhang TJ, Zhao YL, Zhao YJ, Zhou JD. Expression and prognosis analysis of TET family in acute myeloid leukemia. Aging-Us (2020) 12(6):5031–47. doi: 10.18632/aging.102928
159. Lasho TL, Vallapureddy R, Finke CM, Mangaonkar A, Gangat N, Ketterling R, et al. Infrequent occurrence of TET1, TET3, and ASXL2 mutations in myelodysplastic/myeloproliferative neoplasms. Blood Cancer J (2018) 8:32. doi: 10.1038/s41408-018-0057-8
160. Waanders E, Gu ZH, Dobson SM, Crawford JC, Ma XT, Edmonson MN, et al. Mutational landscape and patterns of clonal evolution in relapsed pediatric acute lymphoblastic leukemia. Blood Cancer Discovery (2020) 1(1):96–111. doi: 10.1158/2643-3249.bcd-19-0041
161. Wang JH, Li FL, Ma ZX, Yu MX, Guo Q, Huang JS, et al. High expression of TET1 predicts poor survival in cytogenetically normal acute myeloid leukemia from two cohorts. Ebiomedicine (2018) 28:90–6. doi: 10.1016/j.ebiom.2018.01.031
162. Huang H, Jiang X, Li ZJ, Li YY, Song CX, He CJ, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci United States America (2013) 110(29):11994–9. doi: 10.1073/pnas.1310656110
163. Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu YT, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol (2015) 16(6):653–+. doi: 10.1038/ni.3148
164. Van Damme M, Crompot E, Meuleman N, Maerevoet M, Mineur P, Bron D, et al. Characterization of TET and IDH gene expression in chronic lymphocytic leukemia: comparison with normal b cells and prognostic significance. Clin Epigenet (2016) 8:132. doi: 10.1186/s13148-016-0298-y
165. Pulikkottil AJ, Bamezai S, Ammer T, Mohr F, Feder K, Vegi NM, et al. TET3 promotes AML growth and epigenetically regulates glucose metabolism and leukemic stem cell associated pathways. Leukemia (2022) 36(2):416–25. doi: 10.1038/s41375-021-01390-3
166. An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun (2015) 6:10071. doi: 10.1038/ncomms10071
167. Chaidos A, Barnes CP, Cowan G, May PC, Melo V, Hatjiharissi E, et al. Clinical drug resistance linked to interconvertible phenotypic and functional states of tumor-propagating cells in multiple myeloma. Blood (2013) 121(2):318–28. doi: 10.1182/blood-2012-06-436220
168. Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, et al. Xbp1s-negative tumor b cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell (2015) 28(4):541–2. doi: 10.1016/j.ccell.2015.09.010
169. Yaccoby S. The phenotypic plasticity of myeloma plasma cells as expressed by dedifferentiation into an immature, resilient, and apoptosis-resistant phenotype. Clin Cancer Res (2005) 11(21):7599–606. doi: 10.1158/1078-0432.ccr-05-0523
170. Karadimitris A, Chaidos A, Caputo V, Goudevenou K, Ponnusamy K, Xiao XL. Myeloma propagating cells, drug resistance and relapse. Stem Cells (2015) 33(11):3205–11. doi: 10.1002/stem.2199
171. Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol (2011) 23(5):341–9. doi: 10.1016/j.smim.2011.08.010
172. Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK, et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol (2016) 17(3):323–30. doi: 10.1038/ni.3348
173. Fujii K, Tanaka S, Hasegawa T, Narazaki M, Kumanogoh A, Koseki H, et al. Tet DNA demethylase is required for plasma cell differentiation by controlling expression levels of IRF4. Int Immunol (2020) 32(10):683–90. doi: 10.1093/intimm/dxaa042
174. Barwick BG, Scharer CD, Martinez RJ, Price MJ, Wein AN, Haines RR, et al. B cell activation and plasma cell differentiation are inhibited by de novo DNA methylation. Nat Commun (2018) 9(1):1900. doi: 10.1038/s41467-018-04234-4
175. Lai AY, Mav D, Shah R, Grimm SA, Phadke D, Hatzi K, et al. DNA Methylation profiling in human b cells reveals immune regulatory elements and epigenetic plasticity at alu elements during b-cell activation. Genome Res (2013) 23(12):2030–41. doi: 10.1101/gr.155473.113
176. Kassambara A, Herviou L, Ovejero S, Jourdan M, Thibaut C, Vikova V, et al. RNA-Sequencing data-driven dissection of human plasma cell differentiation reveals new potential transcription regulators. Leukemia (2021) 35(5):1451–62. doi: 10.1038/s41375-021-01234-0
177. Lee ST, Xiao YY, Muench MO, Xiao JQ, Fomin ME, Wiencke JK, et al. A global DNA methylation and gene expression analysis of early human b-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res (2012) 40(22):11339–51. doi: 10.1093/nar/gks957
178. Kulis M, Merkel A, Heath S, Queiros AC, Schuyler RP, Castellano G, et al. Whole-genome fingerprint of the DNA methylome during human b cell differentiation. Nat Genet (2015) 47(7):746–+. doi: 10.1038/ng.3291
179. Nadeau S, Martins GA. Conserved and unique functions of Blimp1 in immune cells. Front Immunol (2022) 12:805260. doi: 10.3389/fimmu.2021.805260
180. Matsui W, Huff CA, Wang QJ, Malehorn MT, Barber J, Tanhehco Y, et al. Characterization of clonogenic multiple myeloma cells. Blood (2004) 103(6):2332–6. doi: 10.1182/blood-2003-09-3064
181. Bao B, Ahmad A, Azmi AS, Ali S, Sarkar FH. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: implications for cancer therapy. Curr Protoc Pharmacol (2013) 61(1):14.25. doi: 10.1002/0471141755.ph1425s61. Chapter 14, Unit 14.25.
182. Guo WC, Wang HQ, Chen P, Shen XK, Zhang BX, Liu J, et al. Identification and characterization of multiple myeloma stem cell-like cells. Cancers (2021) 13(14):3523. doi: 10.3390/cancers13143523
183. Kumar VE, Nambiar R, De Souza C, Nguyen A, Chien J, Lam KS. Targeting epigenetic modifiers of tumor plasticity and cancer stem cell behavior. Cells (2022) 11(9):1403. doi: 10.3390/cells11091403
184. Verma R, Branagan AR, Xu ML, Flavell RA, Dhodapkar KM, Dhodapkar MV. Role of MBD3-SOX2 axis in residual myeloma following pomalidomide. Leukemia (2021) 35(11):3319–23. doi: 10.1038/s41375-021-01145-0
185. Gu CM, Yin Z, Nie H, Liu YJ, Yang JH, Huang GP, et al. Identification of berberine as a novel drug for the treatment of multiple myeloma via targeting UHRF1. BMC Biol (2020) 18(1):33. doi: 10.1186/s12915-020-00766-8
186. Issa JP. Optimizing therapy with methylation inhibitors in myelodysplastic syndromes: dose, duration, and patient selection. Nat Clin Pract Oncol (2005) 2 Suppl 1:S24–9. doi: 10.1038/ncponc0355
187. Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer (2008) 123(1):8–13. doi: 10.1002/ijc.23607
188. Khong T, Sharkey J, Spencer A. The effect of azacitidine on interleukin-6 signaling and nuclear factor-kappa b activation and its in vitro and in vivo activity against multiple myeloma. Haematologica (2008) 93(6):860–9. doi: 10.3324/haematol.12261
189. Lavelle D, DeSimone J, Hankewych M, Kousnetzova T, Chen YH. Decitabine induces cell cycle arrest at the G1 phase via p21(WAF1) and the G2/M phase via the p38 MAP kinase pathway. Leukemia Res (2003) 27(11):999–1007. doi: 10.1016/s0145-2126(03)00068-7
190. Maes K, De Smedt E, Lemaire M, De Raeve H, Menu E, Van Valckenborgh E, et al. The role of DNA damage and repair in decitabine-mediated apoptosis in multiple myeloma. Oncotarget (2014) 5(10):3115–29. doi: 10.18632/oncotarget.1821
191. Tatekawa S, Chinen Y, Ri M, Narita T, Shimura Y, Matsumura-Kimoto Y, et al. Epigenetic repression of miR-375 is the dominant mechanism for constitutive activation of the PDPK1/RPS6KA3 signalling axis in multiple myeloma. Br J Haematol (2017) 178(4):534–46. doi: 10.1111/bjh.14707
192. Krzeminski P, Garcia-Sanz R, Gutierrez NC. Zebularine-induced myeloma cell death is accompanied by decreased c-myc expression. Cell Oncol (2020) 43(4):743–50. doi: 10.1007/s13402-020-00516-6
193. Brueckner B, Rius M, Markelova MR, Fichtner I, Hals PA, Sandvold ML, et al. Delivery of 5-azacytidine to human cancer cells by elaidic acid esterification increases therapeutic drug efficacy. Mol Cancer Ther (2010) 9(5):1256–64. doi: 10.1158/1535-7163.mct-09-1202
194. Otjacques E, Binsfeld M, Rocks N, Blacher S, Vanderkerken K, Noel A, et al. Mithramycin exerts an anti-myeloma effect and displays anti-angiogenic effects through up-regulation of anti-angiogenic factors. PloS One (2013) 8(5):e62818. doi: 10.1371/journal.pone.0062818
195. Kuck D, Caulfield T, Lyko F, Medina-Franco JL. Nanaomycin a selectively inhibits DNMT3B and reactivates silenced tumor suppressor genes in human cancer cells. Mol Cancer Ther (2010) 9(11):3015–23. doi: 10.1158/1535-7163.mct-10-0609
196. Zhou CG, Hui LM, Luo JM. Epigallocatechin gallate inhibits the proliferation and induces apoptosis of multiple myeloma cells via inactivating EZH2. Eur Rev Med Pharmacol Sci (2018) 22(7):2093–8. doi: 10.26355/eurrev_201804_14742
197. Wang Q, Li J, Gu JL, Huang BH, Zhao Y, Zheng D, et al. Potentiation of (-)-epigallocatechin-3-gallate-induced apoptosis by bortezomib in multiple myeloma cells. Acta Biochim Et Biophys Sin (2009) 41(12):1018–26. doi: 10.1093/abbs/gmp094
198. Almatroodi SA, Almatroudi A, Khan AA, Alhumaydhi FA, Alsahli MA, Rahmani AH. Potential therapeutic targets of epigallocatechin gallate (EGCG), the most abundant catechin in green tea, and its role in the therapy of various types of cancer. Molecules (2020) 25(14):3146. doi: 10.3390/molecules25143146
199. Kuck D, Singh N, Lyko F, Medina-Franco JL. Novel and selective DNA methyltransferase inhibitors: Docking-based virtual screening and experimental evaluation. Bioorganic Med Chem (2010) 18(2):822–9. doi: 10.1016/j.bmc.2009.11.050
200. Singh AK, Zhao B, Liu XH, Wang X, Li HZ, Qin HJ, et al. Selective targeting of TET catalytic domain promotes somatic cell reprogramming. Proc Natl Acad Sci United States America (2020) 117(7):3621–6. doi: 10.1073/pnas.1910702117
201. Guan YH, Tiwari AD, Phillips JG, Hasipek M, Grabowski DR, Pagliuca S, et al. A therapeutic strategy for preferential targeting of TET2-mutant and TET dioxygenase-deficient cells in myeloid neoplasms. Blood Cancer Discovery (2021) 2(2):146–61. doi: 10.1158/2643-3230.bcd-20-0173
202. Zhou JH, Shen Q, Lin HQ, Hu LN, Li GQ, Zhang XY. Decitabine shows potent anti-myeloma activity by depleting monocytic myeloid-derived suppressor cells in the myeloma microenvironment. J Cancer Res Clin Oncol (2019) 145(2):329–36. doi: 10.1007/s00432-018-2790-6
203. Shen H, Laird PW. In epigenetic therapy, less is more. Cell Stem Cell (2012) 10(4):353–4. doi: 10.1016/j.stem.2012.03.012
204. Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: A novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol (2002) 321(4):591–9. doi: 10.1016/s0022-2836(02)00676-9
205. Holleran JL, Parise RA, Joseph E, Eiseman JL, Covey JM, Glaze ER, et al. Plasma pharmacokinetics, oral bioavailability, and interspecies scaling of the DNA methyltransferase inhibitor, zebularine. Clin Cancer Res (2005) 11(10):3862–8. doi: 10.1158/1078-0432.ccr-04-2406
206. Hose D, Reme T, Hielscher T, Moreaux J, Messner T, Seckinger A, et al. Proliferation is a central independent prognostic factor and target for personalized and risk-adapted treatment in multiple myeloma. Haematologica-the Hematol J (2011) 96(1):87–95. doi: 10.3324/haematol.2010.030296
207. De Beck L, Melhaoui S, De Veirman K, Menu E, De Bruyne E, Vanderkerken K, et al. Epigenetic treatment of multiple myeloma mediates tumor intrinsic and extrinsic immunomodulatory effects. Oncoimmunology (2018) 7(10):e1484981. doi: 10.1080/2162402x.2018.1484981
208. Bruyer A, Maes K, Herviou L, Kassambara A, Seckinger A, Cartron G, et al. DNMTi/HDACi combined epigenetic targeted treatment induces reprogramming of myeloma cells in the direction of normal plasma cells. Br J Cancer (2018) 118(8):1062–73. doi: 10.1038/s41416-018-0025-x
209. Rabal O, San Jose-Eneriz E, Agirre X, Sanchez-Arias JA, de Miguel I, Ordonez R, et al. Design and synthesis of novel epigenetic inhibitors targeting histone deacetylases, DNA methyltransferase 1, and lysine methyltransferase G9a with In vivo efficacy in multiple myeloma. J Med Chem (2021) 64(6):3392–426. doi: 10.1021/acs.jmedchem.0c02255
210. Dimopoulos K, Helbo AS, Munch-Petersen HF, Sjo L, Christensen J, Kristensen LS, et al. Dual inhibition of DNMTs and EZH2 can overcome both intrinsic and acquired resistance of myeloma cells to IMiDs in a cereblon-independent manner. Mol Oncol (2018) 12(2):180–95. doi: 10.1002/1878-0261.12157
211. Haertle L, Barrio S, Munawar U, Han SB, Zhou X, Vogt C, et al. Cereblon enhancer methylation and IMiD resistance in multiple myeloma. Blood (2021) 138(18):1721–6. doi: 10.1182/blood.2020010452
212. Cao Y, Qiu GQ, Wu HQ, Wang ZL, Lin Y, Wu W, et al. Decitabine enhances bortezomib treatment in RPMI 8226 multiple myeloma cells. Mol Med Rep (2016) 14(4):3469–75. doi: 10.3892/mmr.2016.5658
213. Jin YL, Xu L, Wu XD, Feng J, Shu MM, Gu HT, et al. Synergistic efficacy of the demethylation agent decitabine in combination with the protease inhibitor bortezomib for treating multiple myeloma through the wnt/beta-catenin pathway. Oncol Res (2019) 27(6):729–37. doi: 10.3727/096504018x15443011011637
214. Choudhry P, Mariano MC, Geng HM, Martin TG, Wolf JL, Wong SW, et al. DNA Methyltransferase inhibitors upregulate CD38 protein expression and enhance daratumumab efficacy in multiple myeloma. Leukemia (2020) 34(3):938–41. doi: 10.1038/s41375-019-0587-5
215. Klar AS, Gopinadh J, Kleber S, Wadle A, Renner C. Treatment with 5-Aza-2 '-deoxycytidine induces expression of NY-ESO-1 and facilitates cytotoxic T lymphocyte-mediated tumor cell killing. PloS One (2015) 10(10):e0139221. doi: 10.1371/journal.pone.0139221
216. Dimopoulos K, Gronbaek K. Epigenetic therapy in hematological cancers. Apmis (2019) 127(5):316–28. doi: 10.1111/apm.12906
217. Kalff A, Khong T, Mithraprabhu S, Bergin K, Reynolds J, Bowen KM, et al. Oral azacitidine (CC-486) in combination with lenalidomide and dexamethasone in advanced, lenalidomide-refractory multiple myeloma (ROAR study). Leukemia Lymphoma (2019) 60(9):2143–51. doi: 10.1080/10428194.2019.1571201
218. Berthon C, Nudel M, Boyle EM, Goursaud L, Boyer T, Marceau A, et al. Acute myeloid leukemia synchronous with multiple myeloma successfully treated by azacytidine/lenalidomide and daratumumab without a decrease in myeloid clone size. Leuk Res Rep (2020) 13:100202. doi: 10.1016/j.lrr.2020.100202
Keywords: multiple myeloma, epigenetics, DNA methylation modifiers, MM cell plasticity, DNMTi
Citation: Muylaert C, Van Hemelrijck LA, Maes A, De Veirman K, Menu E, Vanderkerken K and De Bruyne E (2022) Aberrant DNA methylation in multiple myeloma: A major obstacle or an opportunity? Front. Oncol. 12:979569. doi: 10.3389/fonc.2022.979569
Received: 27 June 2022; Accepted: 22 July 2022;
Published: 18 August 2022.
Edited by:
Alexander A. Shtil, Russian Cancer Research Center NN Blokhin, RussiaReviewed by:
Tiziana Vaisitti, University of Turin, ItalyCopyright © 2022 Muylaert, Van Hemelrijck, Maes, De Veirman, Menu, Vanderkerken and De Bruyne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elke De Bruyne, RWxrZS5EZS5CcnV5bmVAdnViLmJl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.