- 1Department of Hematology, Nagano Red Cross Hospital, Nagano, Japan
- 2Division of Molecular Therapy, Institute of Medical Science, Advanced Clinical Research Center, The University of Tokyo, Tokyo, Japan
- 3Department of Diagnostic Pathology and Cytology, Osaka International Cancer Institute, Osaka, Japan
- 4Department of Pathology, Nagano Red Cross Hospital, Nagano, Japan
- 5Department of Applied Genomics, Research Hospital, Institute of Medical Science, University of Tokyo, Tokyo, Japan
- 6Molecular Pathology and Genetics Division, Kanagawa Cancer Center Research Institute, Yokohama, Japan
- 7Division of Health Medical Data Science, Health Intelligence Center, Institute of Medical Science, University of Tokyo, Tokyo, Japan
- 8Division of Cancer Systems Biology, Aichi Cancer Center Research Institute, Nagoya, Japan
- 9Department of Integrated Data Science, Medical and Dental Data Science Center, Tokyo Medical and Dental University, Tokyo, Japan
- 10Department of Diagnostic Pathology, IMSUT Hospital, Institute of Medical Science, The University of Tokyo, Tokyo, Japan
- 11Department of Data Science and Faculty Affairs, Tokyo Medical and Dental University, Tokyo, Japan
Langerhans cell histiocytosis (LCH) and acute myeloid leukemia (AML) are distinct entities of blood neoplasms, and the exact developmental origin of both neoplasms are considered be heterogenous among patients. However, reports of concurrent LCH and AML are rare. Herein we report a novel case of concurrent LCH and AML which shared same the driver mutations, strongly suggesting a common clonal origin.An 84-year-old female presented with cervical lymphadenopathy and pruritic skin rash on the face and scalp. Laboratory tests revealed pancytopenia with 13% of blasts, elevated LDH and liver enzymes, in addition to generalised lymphadenopathy and splenomegaly by computed tomography. Bone marrow specimens showed massive infiltration of MPO-positive myeloblasts, whereas S-100 and CD1a positive atypical dendritic cell-like cells accounted for 10% of the atypical cells on bone marrow pathology, suggesting a mixture of LCH and AML. A biopsy specimen from a cervical lymph node and the skin demonstrated the accumulation of atypical cells which were positive for S-100 and CD1a. LCH was found in lymph nodes, skin and bone marrow; AML was found in peripheral blood and bone marrow (AML was predominant compared with LCH in the bone marrow).
Next generation sequencing revealed four somatic driver mutations (NRAS-G13D, IDH2-R140Q, and DNMT3A-F640fs/-I715fs), equally shared by both the lymph node and bone marrow, suggesting a common clonal origin for the concurrent LCH and AML. Prednisolone and vinblastine were initially given with partial response in LCH; peripheral blood blasts also disappeared for 3 months. Salvage chemotherapy with low dose cytarabine and aclarubicin were given for relapse, with partial response in both LCH and AML. She died from pneumonia and septicemia on day 384. Our case demonstrates a common cell of origin for LCH and AML with a common genetic mutation, providing evidence to support the proposal to classify histiocytosis, including LCH, as a myeloid/myeloproliferative malignancy.
Introduction
Langerhans cell histiocytosis (LCH) and acute myeloid leukemia (AML) are distinct entities of blood neoplasms, and the exact developmental origin of both neoplasms is considered to be heterogenous among patients (1–6). However, to our knowledge, only ten cases of concurrent LCH and AML have been reported so far (2, 3, 7–11). Herein we report a novel case of concurrent AML and LCH which shared the same driver mutations, strongly suggesting a common clonal origin.
Case description
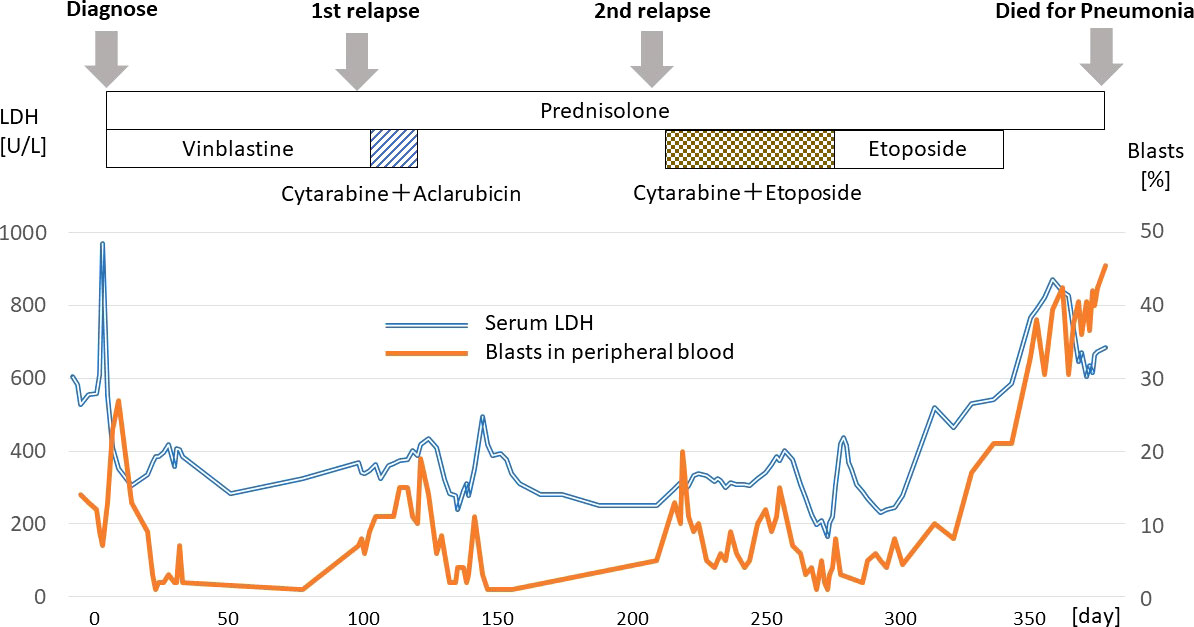
An 84 year-old female presented with cervical lymphadenopathy and pruritic skin rash on the face and scalp. She had no past history or family history of hematological or solid organ malignancies or other treatments. She had no fever but multiple lymph nodes were palpated bilaterally in the neck, axilla, and inguinal region. Laboratory tests revealed pancytopenia (WBC 2,520/μL including 13% blasts, 32% neutrophils, 6% eosinophils, 0% basophils, 3% monocytes, 46% lymphocytes, Hemoglobin (Hb) of 10.9 g/dL, and a platelet count of 92,000/μL) and elevated LDH (604 U/L) and liver enzymes (AST 51 U/L, ALT 39 U/L). Systemic lymphadenopathy and splenomegaly, but no other abnormal findings, were noted on computed tomography. Bone marrow specimens showing a massive infiltration of MPO-positive blasts including 53.5% of myeloblastic cells and 13.8% of monoblastic cells indicated the diagnosis of AML. Whereas, 9.1% of cellular components were occupied by MPO-negative atypical dendritic cell-like cells (Figures 1A–C). On flow cytometric analysis, monoclonal cells were positive for CD13, CD33, CD34, CD38, CD7, and TdT. On bone marrow pathology, S-100 and CD1a-positive atypical cells accounted for 10% of the blasts (Figures 1D, E), suggesting a mixture of AML and LCH. A biopsy specimen from a cervical lymph node demonstrated the accumulation of atypical round or horseshoe-shaped cells with indented or folded nuclei which were positive for S-100 and CD1a, and negative for MPO and CD34, confirming the diagnosis of LCH (Figures 1F, H–J). AML cells were absent not only on pathology, but also on flow cytometry in lymph nodes. Atypical cells were partially positive for langerin (Figure 1G), not suggestive of indeterminate cell histiocytosis (ICH). Around the atypical cells were scattered mononuclear cells positive for CD3, CD20, and CD68 (Figures 1K–M). Simultaneously, skin biopsy demonstrated atypical cells with an immunohistochemical profile similar to that in the lymph nodes, suggesting skin invasion of LCH. The atypical cells were negative for MPO and CD34, suggesting absence of skin invasion by the AML. The G-banding assay revealed a normal karyotype in both the bone marrow and lymph nodes.
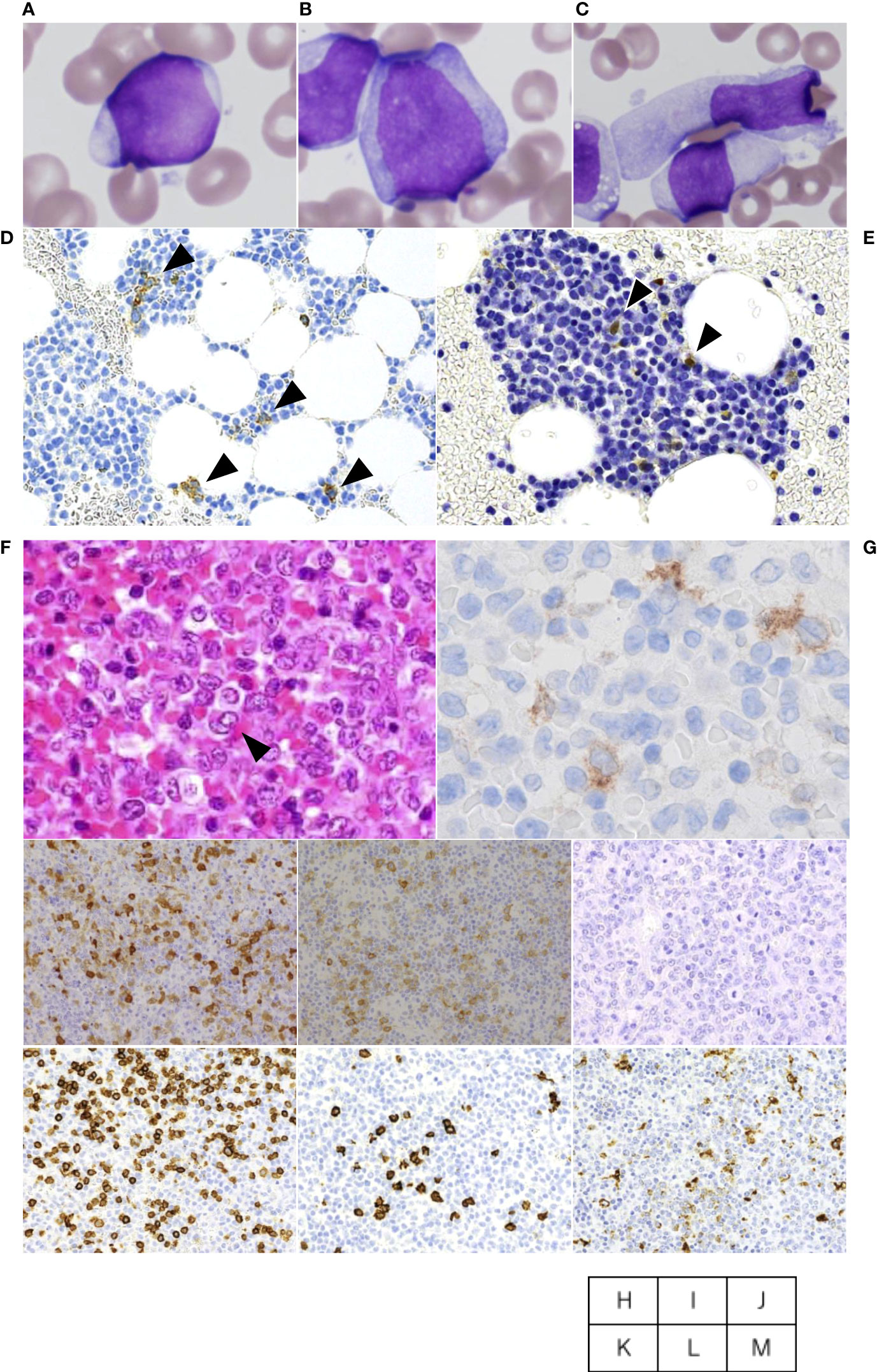
Figure 1 Cervical lymph node biopsy and bone marrow aspiration. The aspiration smears of bone marrow show myeloblastic cells accounted for 53.5% (A) and monoblastic cells accounted for 13.8% (B), whereas dendritic cell-like atypical cells are also found at a frequency of 9.1% (C). On bone marrow pathology, CD1a (D)- and S-100 (E)-positive atypical cells accounted for 10% of the blasts. Cervical lymph node biopsy specimen shows atypical cells with indented or folded nuclei like one indicated by arrowhead or other ones with mild folding (F), and immunostaining was Langerin-partially positive (G), S-100-positive (H), CD1a-positive (I), MPO-negative (J). Around the atypical cells were scattered mononuclear cells positive for CD3 (K), CD20 (L), and CD68 (M) in the periphery.
LCH was found in lymph nodes, skin and bone marrow; AML was found in peripheral blood and bone marrow (AML was predominant compared with LCH in the bone marrow). Elevated liver enzymes and splenomegaly suggested the presence of hepatic and splenic lesions, but which tumor was responsible could not be determined.
Diagnostic assessment
Taken together, the patient was diagnosed with concurrent LCH and AML. Next generation sequencing with the TruSight Myeloid Sequencing Panel (Illumina) revealed four somatic driver mutations (NRAS-G13D, IDH2-R140Q, and DNMT3A-F640fs/-I715fs), equally found in both the lymph node and bone marrow (Table 1). BRAF-V600E was negative, which was also confirmed by immunohistochemistry staining (data not shown). The mutant allele frequencies were 24.6%-33.0% and 27.0%-38.5% in the lymph node and bone marrow, respectively. Assuming each mutant allele to be a single hit, 50-70% of tumor cells harbored each mutation, suggesting a common clonal origin of concurrent LCH and AML.
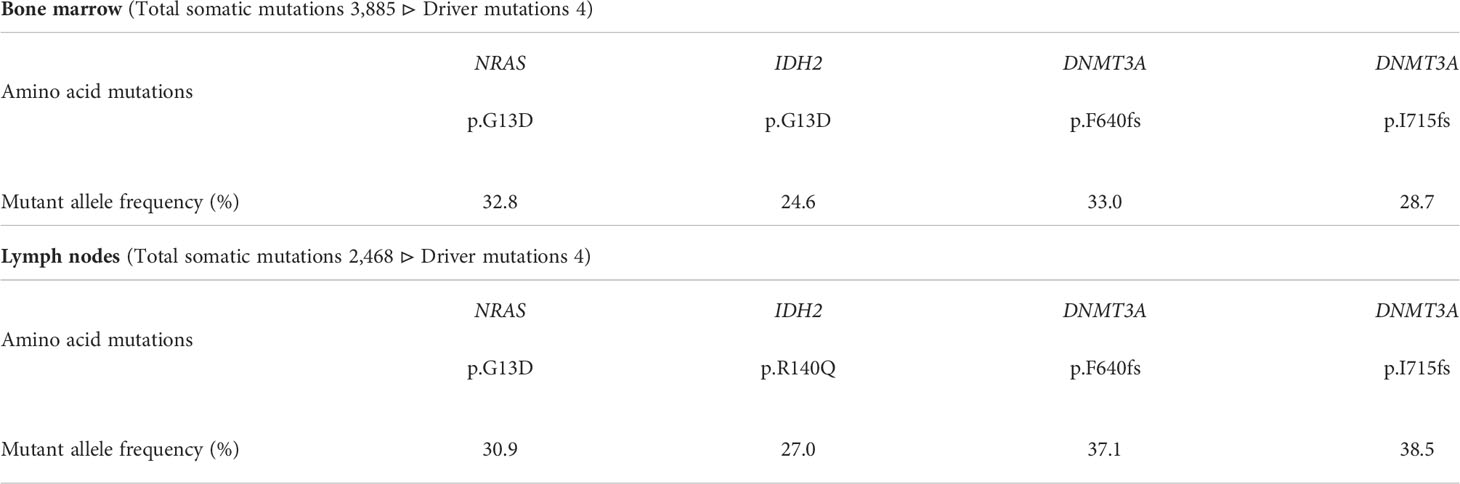
Table 1 Results of genomic analysis of cervical lymph node biopsy and bone marrow aspiration by panel analysis.
Prednisolone and vinblastine were initially given with partial response in LCH, additionally peripheral blood blasts disappeared for 3 months. From day 120, low dose cytarabine and aclarubicin were administered as a salvage chemotherapy with partial response in both LCH and AML. Subsequently, from day 253, the patient received low dose cytarabine and etoposide, which did not give rise to a durable response, and she died from pneumonia and septicemia on day 384 (Figure 2).
Discussion
Histiocytic disorders are a group of rare diseases characterized by organ infiltration by macrophages, dendritic cells, and monocytes (12). Gene mutations have been identified in a number of histiocytoses, including LCH and non-LCH (e.g., Erdheim-Chester disease (ECD), indeterminate cell histiocytosis, and histiocytic sarcoma) (5, 13–20), most of which are mutations in genes encoding proteins in the mitogen-activated protein kinase (MAPK) pathway (21). Therefore, the pathogenesis of histiocytic disorders, including LCH are mainly attributed to unregulated activation of the MAPK pathway (1, 22), and these disorders are clonal neoplastic diseases caused by this (23, 24). Activating mutations of MAPK pathway members are almost mutually exclusive. Among these, BRAF is a major target of mutation, and BRAF-V600E is most commonly observed in LCH (1). In contrast, mutation of NRAS, another member of the MAPK pathway, is rare in LCH, but one report stated that NRAS mutations were present in 40% of pulmonary LCH (25). Furthermore, genetic mutations in the MAPK pathway members including NRAS were identified in 57% of Langerhans cell sarcomas (26). NRAS mutations were more common in AML (10%) than LCH, and, in combination with IDH2, DNMT3A and other mutations, contribute to the pathogenesis of AML (27–29).
Cases of LCH combined with malignant neoplasms are rare and generally the subject of isolated case reports (2, 3). Most reports of AML associated with LCH are treatment-related AML after treatment for previous LCH. Only ten cases of simultaneous diagnosis of LCH and AML have been reported to date, most of which were characterized by generalized LCH, monocytic leukemia as the predominant type of associated AML, and poor prognosis, as in the present case (2, 3, 7–11). On the other hand, a high concomitant rate of myeloid neoplasm has been reported in non-LCH (e.g., 10.1% in ECD) (30, 31).
The exact cell of origin of histiocytic disorders, including LCH, is unknown. Allelic assessment of Langerhans cells in LCH clearly distinguished them from skin Langerhans cells (4), as BRAF-V600E mutations were identified in a subset of dendritic cells, mature monocytes, myeloid progenitor cells, and CD34+ cells in LCH and ECD patients. Therefore, it can be estimated myeloid progenitor cells are the cell of origin for histiocytic disorders (5, 32, 33). However, clinical and experimental evidence is lacking.
Recently, advances in genetic analysis have led to the discovery of commonly mutated genes in cases of histiocytic disorders and myeloid neoplasms, and the existence of a common cellular origin of histiocytic disorders and myeloid neoplasms has been proposed. In a review by Kemps PG et al. (34), mutated genes in histiocytic disorders and myeloid neoplasms have been reported in 30 cases (29, 33–55). A total of 31 cases, including one additional case of a common mutation found in LCH and primary myelofibrosis (39), including the present case are shown in Table 2. There have been five cases, of LCH occurring concurrently with a myeloid neoplasm, including the present case. Gene mutations of the MAPK pathway were frequently detected in LCH and non-LCH (30, 34, 35, 37, 40–44). There have been 5 cases of shared NRAS mutations, and one of them was at the same locus as the present case (NRAS p.G13D) (42). In this case, an adult female was diagnosed with ECD from skin lesions, and bone marrow biopsy revealed chronic myelomonocytic leukemia. The only mutation detected was NRAS p.G13D, which was common to both lesions, but BRAF-V600E was not examined.
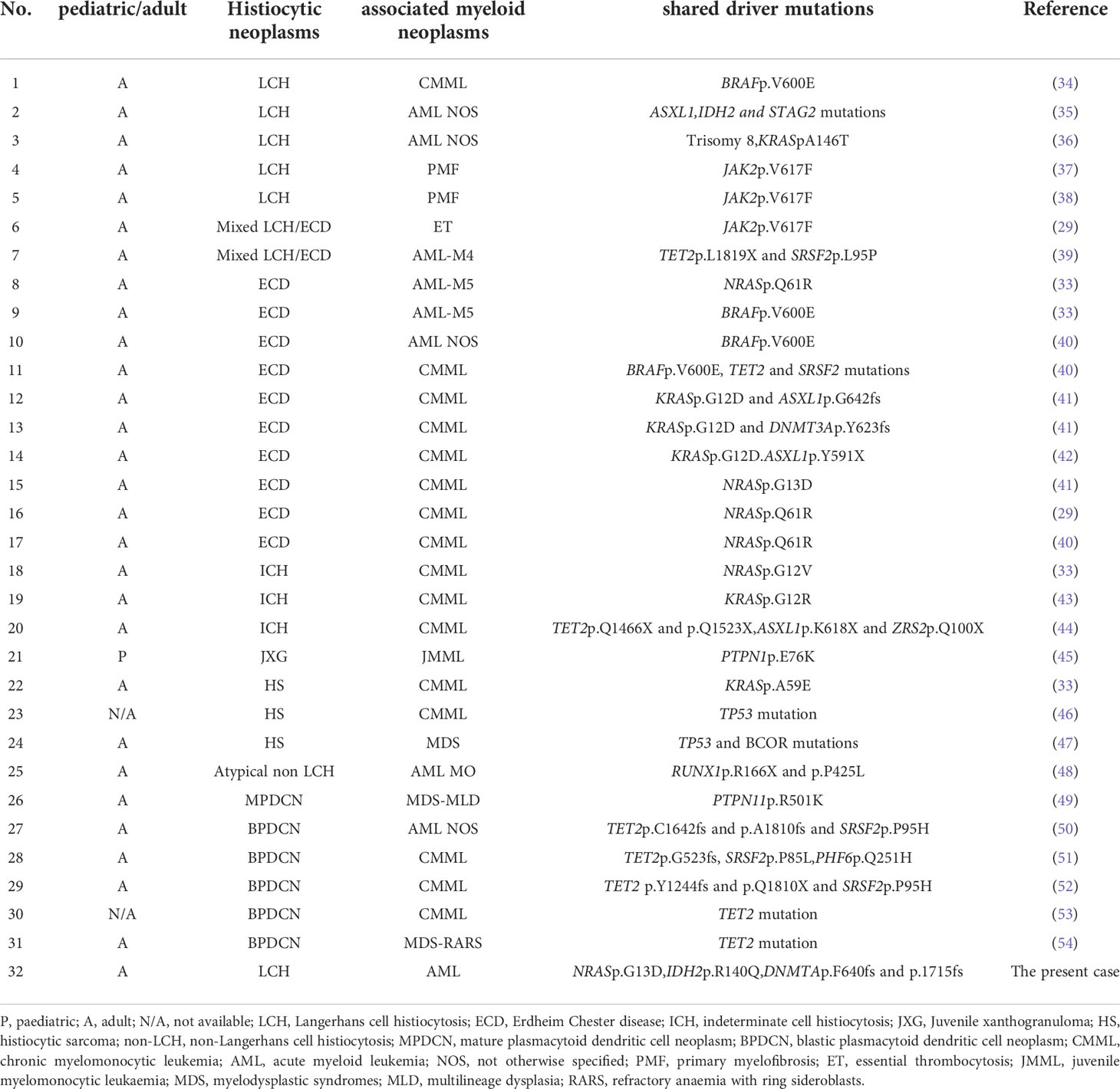
Table 2 Overview of reported cases with histiocytic disorders and additional myeloid neoplasms bearing the same genetic alteration(s).
Five cases of myeloid neoplasms have been suggested to have a common origin with LCH (2 cases of AML, 2 cases of polycrythemia vera, and 1 case of chronic myelomonocytic leukemia), thus, this is the third report of common gene mutation between LCH and AML. In the other two reports of shared mutation between LCH and AML, both adult males were diagnosed with LCH on skin biopsy and AML from bone marrow biopsy. One patient shared ASXL1, IDH2, and STAG2 mutations (36), and the other shared KRAS mutation and trisomy 8 (37). Of note, in both cases, BRAF-V600E mutation was detected only in LCH cells. In the present case, allele frequencies of 4 driver mutations were comparable to each other as well as between LCH (lymph node) and AML (bone marrow), no genetic mutations present in only AML or LCH were detected, making it difficult to dissect the developmental process of LCH and AML. Thus, the present case provides important evidence that myeloid progenitors are the common origin of the two neoplasms, and reaffirms the importance of MAPK pathway activation in the pathogenesis of histiocytic disorders.
On the other hand, a different biological mechanism by which leukemic progenitor cells are misdirected into LCH by environmental factors was considered. This disease is an inflammatory myeloid neoplasm with features of both abnormal reaction processes and neoplastic processes. De Graaf et al. reported that inflammatory cytokines are expressed in LCH lesions (56). Kannourakis et al. extracted and cultured monocytes from the eosinophilic granuloma tissue of LCH patients, and found that such monocytes highly express inflammatory cytokines (57). In addition to a cytokine storm in local lesions, the levels of several pro-inflammatory cytokines in the serum of LCH patients are high, suggesting that cytokines are associated with the pathogenesis of LCH (58). LCH cells in the target tissues of LCH patients are surrounded by lymphocytic infiltrates and multinucleated giant cells, including T cells, macrophages, eosinophils, and B cells. Cytokines derived from LCH cells and T cells are considered to regulate the differentiation, maturation, and migration of myeloid dendritic cell precursors originating from hematopoietic stem cells (59, 60). In our case, the lymph node lesions also exhibited CD3-, CD20-, and CD68-positive mononuclear cell infiltrates around the LCH cells, suggesting that the microenvironment of these inflammatory cells played a role in the induction of LCH. Additionally, LCH lesions in this case were widely distributed in lymph nodes, but organs susceptible to LCH such as bone remained intact. Considering the common AML genotype and atypical LCH phenotype, there is one possible explanation for this case, that leukemic progenitor cells which migrated into lymph nodes could have been misguided toward LCH by environmental factors including the lymph node microenvironment and soluble factors.
According to the 2016 WHO classification, histiocytic disorders, including LCH and ECD, are classified as lymphoid neoplasms (61). This classification is based on several case reports describing individual patients with secondary malignant histiocytosis clonally associated with lymphoid neoplasms (62–66). However, with the recent development of genetic analysis techniques, the number of shared mutations in histiocytosis and myeloid neoplasms has surpassed those in lymphoid neoplasms (33).
Conclusion
In summary, our case demonstrates a common cell of origin for LCH and AML with a common genetic mutation, providing evidence to support the proposal to classify histiocytosis, including LCH, as a myeloid/myeloproliferative malignancy.
Data availability statement
The detailed clinical data and datasets presented in this article are not publicly available due to ethical and privacy restrictions. Requests to access the datasets should be directed to the corresponding authors.
Ethics statement
Written informed consent was obtained from the [individual(s) AND minor(s)’ legal guardian/next of kin] for the publication of any potentially identifiable images or data included in this article.
Author contributions
SK, TU, HKa, HS, MS, II, YT, TD, YO, and HKo were involved in the description of clinical information. KY, NY, ES, RY, SI, SM, and AT were involved in the data analysis. All authors contributed to the article and approved the submitted version.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Allen CE, Parsons DW. Biological and clinical significance of somatic mutations in langerhans cell histiocytosis and histiocytic neoplastic disorders. Hematol Am Soc Hematol Educ Program (2015) 2015:559–64. doi: 10.1182/asheducation-2015.1.559
2. Egeler RM, Neglia JP, Puccetti DM, Brennan CA, Nesbit ME. Association of langerhans cell histiocytosis with malignant neoplasms. Cancer (1993) 71(3):865–73. doi: 10.1002/1097-0142(19930201)71:3<865::aid-cncr2820710334>3.0.co;2-0
3. Egeler RM, Neglia JP, Aricò M, Favara BE, Heitger A, Nesbit ME, et al. The relation of langerhans cell histiocytosis to acute leukemia, lymphomas, and other solid tumors. the LCH-malignancy study group of the histiocyte society. Hematol Oncol Clin North Am (1998) 12(2):369–78. doi: 10.1016/s0889-8588(05)70516-5
4. Allen CE, Li L, Peters TL, Leung HCE, Yu A, Man TK, et al. Cell-specific gene expression in langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal langerhans cells. J Immunol (2010) 184(8):4557–67. doi: 10.4049/jimmunol.0902336
5. Berres ML, Lim KPH, Peters T, Price J, Takizawa H, Salmon H, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med (2014) 211(4):669–83. doi: 10.1084/jem.20130977
6. Allen CE, Merad M, McClain KL. Langerhans-cell histiocytosis. N Engl J Med (2018) 379(9):856–68. doi: 10.1056/NEJMra1607548
7. Egeler RM, Neglia JP, Arico M, Favara BE, Heitger A, Nesbit ME. Acute leukemia in association with langerhans cell histiocytosis. Med Pediatr Oncol (1994) 23(2):81–5. doi: 10.1002/mpo.2950230204
8. Pina-Oviedo S, Torres-Cabala CA, Miranda RN, Tetzlaff MT, Singh S, Rapini RP, et al. Concomitant cutaneous langerhans cell histiocytosis and leukemia cutis. Am J Dermatopathol (2017) 39(5):388–92. doi: 10.1097/DAD.0000000000000775
9. Yohe SL, Chenault CB, Torlakovic EE, Asplund SL, McKenna RW. Langerhans cell histiocytosis in acute leukemias of ambiguous or myeloid lineage in adult patients: Support for a possible clonal relationship. Mod Pathol (2014) 27(5):651–6. doi: 10.1038/modpathol.2013.181
10. Hwang YY, Tsui P, Leung RYY, Kwong YL. Disseminated langerhans cell histiocytosis associated with acute myeloid leukaemia: Complete remission with daunorubicin and cytarabine. Ann Hematol (2013) 92(2):267–8. doi: 10.1007/s00277-012-1555-6
11. Bohn OL, Feldman JL, Heaney ML, Teruya-Feldstein J. Acute myeloid leukemia with t(9;11) (p22;q23) and synchronous langerhans cell histiocytosis. Int J Surg Pathol (2014) 22(2):172–6. doi: 10.1177/1066896913487985
12. Emile JF, Abla O, Fraitag S, Horne A, Haroche J, Donadieu J, et al. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood (2016) 127(22):2672–81. doi: 10.1182/blood-2016-01-690636
13. Durham BH. Molecular characterization of the histiocytoses: neoplasia of dendritic cells and macrophages. Semin Cell Dev Biol (2019) 86:62–76. doi: 10.1016/j.semcdb.2018.03.002
14. Badalian-Very G, Vergilio JA, Degar BA, MacConaill LE, Brandner B, Calicchio ML, et al. Recurrent BRAF mutations in langerhans cell histiocytosis. Blood (2010) 116(11):1919–23. doi: 10.1182/blood-2010-04-279083
15. Haroche J, Charlotte F, Arnaud L, Deimling AV, Hélias-Rodzewicz Z, Hervier B, et al. High prevalence of BRAF V600E mutations in erdheim-Chester disease but not in other non-langerhans cell histiocytoses. Blood (2012) 120(13):2700–3. doi: 10.1182/blood-2012-05-430140
16. Chakraborty R, Hampton OA, Shen X, Simko SJ, Shih A, Abhyankar H, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood (2014) 124(19):3007–15. doi: 10.1182/blood-2014-05-577825
17. Sahm F, Capper D, Preusser M, Meyer J, Stenzinger A, Lasitschka F, et al. BRAFV600E mutant protein is expressed in cells of variable maturation in langerhans cell histiocytosis. Blood (2012) 120(12):e28–34. doi: 10.1182/blood-2012-06-429597
18. Satoh T, Smith A, Sarde A, Lu HC, Mian S, Trouillet C, et al. B-RAF mutant alleles associated with langerhans cell histiocytosis, a granulomatous pediatric disease. PloS One (2012) 7(4):e33891. doi: 10.1371/journal.pone.0033891
19. Brown NA, Furtado LV, Betz BL, Kiel MJ, Weigelin HC, Lim MS, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative langerhans cell histiocytosis. Blood (2014) 124(10):1655–8. doi: 10.1182/blood-2014-05-577361
20. Nelson DS, Quispel W, Badalian-Very G, Halteren AGSV, Bos CVD, Bovée JVMG, et al. Somatic activating ARAF mutations in langerhans cell histiocytosis. Blood (2014) 123(20):3152–5. doi: 10.1182/blood-2013-06-511139
21. Durham BH, Rodrigo EL, Picarsic J, Abramson D, Rotemberg V, Munck SD, et al. Activating mutations in CSF1R and additional receptor tyrosine kinases in histiocytic neoplasms. Nat Med (2019) 25(12):1839–42. doi: 10.1038/s41591-019-0653-6
22. Berres ML, Merad M, Allen CE. Progress in understanding the pathogenesis of langerhans cell histiocytosis: Back to histiocytosis X? Br J Haematol (2015) 169(1):3–13. doi: 10.1111/bjh.13247
23. Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO classification of tumours of soft tissue and bone. 4th ed Vol. Volume 5. . Lyon: IARC (2013).
24. Swerdlow S, Campo E, Harris N, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed Vol. Volume 2. . Lyon: IARC (2017).
25. Mourah S, How-Kit A, Meignin V, Gossot D, Lorillon G, Bugnet E, et al. Recurrent NRAS mutations in pulmonary langerhans cell histiocytosis. Eur Respir J (2016) 47(6):1785–96. doi: 10.1183/13993003.01677-2015
26. Shanmugam V, Griffin GK, Jacobsen ED, Fletcher CDM, Sholl LM, Hornick JL. Identification of diverse activating mutations of the RAS-MAPK pathway in histiocytic sarcoma. Mod Pathol (2019) 32(6):830–43. doi: 10.1038/s41379-018-0200-x
27. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov (2015) 13(11):828–51. doi: 10.1038/nrd4389
28. Wang S, Wu Z, Li T, Li Y, Wang W, Hao Q, et al. Mutational spectrum and prognosis in NRAS‐mutated acute myeloid leukemia. Sci Rep (2020) 10(1):12512. doi: 10.1038/s41598-020-69194-6
29. Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cancer targets in the ras pathway. Cold Spring Harb Symp Quant Biol (2005) 70:461–8. doi: 10.1101/sqb.2005.70.044
30. Papo M, Diamond EL, Cohen-Aubart F, Emile JF, Roos-Weil D, Gupta N, et al. High prevalence of myeloid neoplasms in adults with non-langerhans cell histiocytosis. Blood (2017) 130(8):1007–13. doi: 10.1182/blood-2017-01-761718
31. Kommalapati A, Tella SH, Durkin M, Go RS, Goyal G. Histiocytic sarcoma: a population-based analysis of incidence, demographic disparities, and long-term outcomes. Blood (2018) 131(2):265–8. doi: 10.1182/blood-2017-10-812495
32. Milne P, Bigley V, Bacon CM, Néel A, McGovern N, Bomken S, et al. Hematopoietic origin of langerhans cell histiocytosis and erdheim-Chester disease in adults. Blood (2017) 130(2):167–75. doi: 10.1182/blood-2016-12-757823
33. Durham BH, Roos-Weil D, Baillou C, Cohen-Aubart F, Yoshimi A, Miyara M, et al. Functional evidence for derivation of systemic histiocytic neoplasms from hematopoietic stem/progenitor cells. Blood (2017) 130(2):176–80. doi: 10.1182/blood-2016-12-757377
34. Kemps PG, Hebeda KM, Pals ST, Verdijk RM, Lam KH, Bruggink AH, et al. Spectrum of histiocytic neoplasms associated with diverse hematological malignancies bearing the same oncogenic mutation. J Pathol Clin Res (2021) 7(1):10–26. doi: 10.1002/cjp2.177
35. Konstantinou MP, Lucas P, Uthurriague C, Severino-Freire M, Spenatto N, Gaudin C, et al. Langerhans cell histiocytosis associated with chronic myelomonocytic leukemia both harboring the same BRAF V600E mutation: efficacy of vemurafenib. J Eur Acad Dermatol Venereol (2021) 35(2):e120–1. doi: 10.1111/jdv.16850
36. Khurana S, Sluzevich JC, He R, Reimer DK, Kharfan-Dabaja MA, Foran JM, et al. Association between highgrade myelodysplastic syndrome and cutaneous langerhans cell histiocytosis suggested by next-generation sequencing. JAMA Dermatol (2020) 156(7):817–9. doi: 10.1001/jamadermatol.2020.0544
37. Wang X, Wang Z. Revealing homologous clonality by synchronous trisomy 8 in langerhans cell histiocytosis and acute myeloid leukemia. Blood (2019) 134(1):5040. doi: 10.1182/blood-2019-127883
38. Bonometti A, Bagnoli F, Fanoni D, Venegoni L, Corti L, Bianchi P, et al. JAK2-mutated langerhans cell histiocytosis associated with primary myelofibrosis treated with ruxolitinib. Hum Pathol (2018) 73:171–5. doi: 10.1016/j.humpath.2017.10.017
39. Holst JM, Enemark MB, Plesner TL, Pedersen MB, Ludvigsen M, d'Amore F. Coexisting BRAF-mutated langerhans cell histiocytosis and primary myelofibrosis with shared JAK2 mutation. Case Rep Hematol (2021). doi: 10.1155/2021/6623706
40. Ghobadi A, Miller CA, Li T, O'Laughlin M, Lee YS, Ali M, et al. Shared cell of origin in a patient with erdheim–Chester disease and acute myeloid leukemia. Haematologica (2019) 104(8):e373–5. doi: 10.3324/haematol.2019.217794
41. Tzankov A, Kremer M, Leguit R, Orazi A, Walt JVD, Gianelli U, et al. Histiocytic cell neoplasms involving the bone marrow: summary of the workshop cases submitted to the 18th meeting of the European association for haematopathology (EAHP) organized by the European bone marrow working group, Basel 2016. Ann Hematol (2018) 97(11):2117–28. doi: 10.1007/s00277-018-3436-0
42. Bonnet P, Chasset F, Moguelet P, Abisror N, Itzykson R, Bouaziz JD, et al. Erdheim-Chester disease associated with chronic myelomonocytic leukemia harboring the same clonal mutation. Haematologica (2019) 104(11):e530–3. doi: 10.3324/haematol.2019.223552
43. Goyal G, Liu Y, Ravindran A, Al-Kali A, Go RS, Patnaik MM, et al. Concomitant erdheim-Chester disease and chronic myelomonocytic leukaemia: genomic insights into a common clonal origin. Br J Haematol (2019) 187(2):e51–4. doi: 10.1111/bjh.16177
44. Loghavi S, Curry JL, Garcia-Manero G, Patel KP, Xu J, Khoury JD, et al. Chronic myelomonocytic leukemia masquerading as cutaneous indeterminate dendritic cell tumor: expanding the spectrum of skin lesions in chronic myelomonocytic leukemia. J Cutan Pathol (2017) 44(12):1075–9. doi: 10.1111/cup.13039
45. Santos-Briz A, Medina-Miguelañez M, Moyano-Bueno D, Viñolas-Cuadros A, Martínez TG, Izquierdo MM, et al. Indeterminate dendritic cell tumor as cutaneous involvement of chronic myelomonocytic leukemia successfully treated with phototherapy. Am J Dermatopathol (2020) 42(11):876–80. doi: 10.1097/DAD.0000000000001703
46. Bátai B, Krizsán S, Gángó A, Hegyi L, Csóka M, Erdélyi DJ, et al. Juvenile myelomonocytic leukaemia presentation after preceding juvenile xanthogranuloma harbouring an identical somatic PTPN11 mutation. Pediatr Blood Cancer (2020) 67(9):e28368. doi: 10.1002/pbc.28368
47. Facchetti F, Pileri SA, Lorenzi L, Tabanelli V, Rimsza L, Pittaluga S, et al. Histiocytic and dendritic cell neoplasms: what have we learnt by studying 67 cases. Virchows Arch (2017) 471(4):467–89. doi: 10.1007/s00428-017-2176-1
48. Tashkandi H, Dogan A. Histiocytic sarcoma arising in patient with history of clonally-related germ cell tumour and myelodysplastic syndrome. Br J Haematol (2020) 188(4):482. doi: 10.1111/bjh.16372
49. Mugairi AA, Turki SA, Salama H, Ahmadi KA, Abuelgasim KA, Damlaj M. Isolated bone marrow non-langerhans cell histiocytosis preceding RUNX1-mutated acute myeloid leukemia: Case report and literature review. Am J Clin Pathol (2019) 151(6):638–46. doi: 10.1093/ajcp/aqz018
50. Bodmer A, Menter T, Juskevicius D, Arranto C, Wenzel F, Dirnhofer S, et al. Sharing of a PTPN11 mutation by myelodysplastic bone marrow and a mature plasmacytoid dendritic cell proliferation provides evidence for their common myelomonocytic origin. Virchows Arch (2017) 470(4):469–73. doi: 10.1007/s00428-017-2075-5
51. Luskin MR, Kim AS, Patel SS, Wright K, LeBoeuf NR, Lane AA, et al. Evidence for separate transformation to acute myeloid leukemia and blastic plasmacytoid dendritic cell neoplasm from a shared ancestral hematopoietic clone. Leuk Lymphoma (2020) 61(9):2258–61. doi: 10.1080/10428194.2020.1755856
52. Patnaik MM, Lasho T, Howard M, Finke C, Ketterling RL, Al-Kali A, et al. Biallelic inactivation of the retinoblastoma gene results in transformation of chronic myelomonocytic leukemia to a blastic plasmacytoid dendritic cell neoplasm: Shared clonal origins of two aggressive neoplasms. Blood Cancer J (2018) 8(9):82. doi: 10.1038/s41408-018-0120-5
53. Brunetti L, Battista VD, Venanzi A, Schiavoni G, Martelli MP, Ascani S, et al. Blastic plasmacytoid dendritic cell neoplasm and chronic myelomonocytic leukemia: a shared clonal origin. Leukemia (2017) 31(5):1238–40. doi: 10.1038/leu.2017.38
54. Sukegawa S, Sakata-Yanagimoto M, Matsuoka R, Momose H, Kiyoki Y, Noguchi M, et al. Blastic plasmacytoid dendritic cell neoplasm accompanied by chronic myelomonocytic leukemia successfully treated with azacitidine. Rinsho Ketsueki (2018) 59(12):2567–73. doi: 10.11406/rinketsu.59.2567
55. Krause JR, Baugh L, Swink A, Burchet M. Blastic plasmacytoid dendritic cell neoplasm following acquired erythropoietic protoporphyria. Baylor Univ Med Cent Proc (2017) 30(4):450–1. doi: 10.1080/08998280.2017.11930225
56. Graaf JHD, Tamminga RY, Dam-Meiring A, Kamps WA, Timens W. The presence of cytokines in langerhans’ cell histiocytosis. J Pathol (1996) 180(4):400–6. doi: 10.1002/(SICI)1096-9896(199612)180:4<400::AID-PATH701>3.0.CO;2-W
57. Kannourakis G, Abbas A. The role of cytokines in the pathogenesis of langerhans cell histiocytosis. Br J Cancer Suppl (1994) 23:S37–40.
58. Feng C, Li Y, Ke H, Peng X, Guo H, Zhan L, et al. Immune microenvironment in langerhans cell histiocytosis: Potential prognostic indicators. Front Oncol (2021) 11:631682. doi: 10.3389/fonc.2021.631682
59. Egeler RM, Favara BE, Meurs MV, Laman JD, Claassenet E. Differential In situ cytokine profiles of langerhans-like cells and T cells in langerhans cell histiocytosis: Abundant expression of cytokines relevant to disease and treatment. Blood (1999) 94(12):4195–201.
60. Rizzo FM, Cives M, Simone V, Silvestris F. New insights into the molecular pathogenesis of langerhans cell histiocytosis. Oncologist (2014) 19(2):151–63. doi: 10.1634/theoncologist.2013-0341
61. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the world health organization classification of lymphoid neoplasms. Blood (2016) 127(20):2375–90. doi: 10.1182/blood-2016-01-643569
62. Feldman AL, Arber DA, Pittaluga S, Martinez A, Burke JS, Raffeld M, et al. Clonally related follicular lymphomas and histiocytic/ dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood (2008) 111(12):5433–9. doi: 10.1182/blood-2007-11-124792
63. West DS, Dogan A, Quint PS, Tricker-Klar ML, Porcher JC, Ketterling RP, et al. Clonally related follicular lymphomas and langerhans cell neoplasms: Expanding the spectrum of transdifferentiation. Am J Surg Pathol (2013) 37(7):978–86. doi: 10.1097/PAS.0b013e318283099f
64. Shao H, Xi L, Raffeld M, Feldman AL, Ketterling RP, Knudson R, et al. Clonally related histiocytic/dendritic cell sarcoma and chronic lymphocytic leukemia/small lymphocytic lymphoma: A study of seven cases. Mod Pathol (2011) 24(11):1421–32. doi: 10.1038/modpathol.2011.102
65. Chen W, Jaffe R, Zhang L, Hill C, Block AM, Sait S, et al. Langerhans cell sarcoma arising from chronic lymphocytic lymphoma/small lymphocytic leukemia: Lineage analysis and BRAF V600E mutation study. N Am J Med Sci (2013) 5(6):386–91. doi: 10.4103/1947-2714.114172
Keywords: langerhans cell histiocytosis, acute myeloid leukemia, NRAS, MAPK pathway, histiocytic disorders, dendritic cells, BRAF V600E, inflammatory myeloid neoplasm
Citation: Kazama S, Yokoyama K, Ueki T, Kazumoto H, Satomi H, Sumi M, Ito I, Yusa N, Kasajima R, Shimizu E, Yamaguchi R, Imoto S, Miyano S, Tanaka Y, Denda T, Ota Y, Tojo A and Kobayashi H (2022) Case report: Common clonal origin of concurrent langerhans cell histiocytosis and acute myeloid leukemia. Front. Oncol. 12:974307. doi: 10.3389/fonc.2022.974307
Received: 21 June 2022; Accepted: 18 August 2022;
Published: 16 September 2022.
Edited by:
Jerry L. Spivak, The Johns Hopkins Hospital, Johns Hopkins Medicine, United StatesReviewed by:
Giorgio Alberto Croci, University of Milan, ItalyShih-Sung Chuang, Chi Mei Medical Center, Taiwan
Copyright © 2022 Kazama, Yokoyama, Ueki, Kazumoto, Satomi, Sumi, Ito, Yusa, Kasajima, Shimizu, Yamaguchi, Imoto, Miyano, Tanaka, Denda, Ota, Tojo and Kobayashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuaki Yokoyama, ay15b2tvQGltcy51LXRva3lvLmFjLmpw; Arinobu Tojo, dG9qby5hZG1AdG1kLmFjLmpw
†These authors have contributed equally to this work and share first authorship
 Shintaro Kazama
Shintaro Kazama Kazuaki Yokoyama
Kazuaki Yokoyama Toshimitsu Ueki
Toshimitsu Ueki Hiroko Kazumoto
Hiroko Kazumoto Hidetoshi Satomi
Hidetoshi Satomi Masahiko Sumi1
Masahiko Sumi1 Eigo Shimizu
Eigo Shimizu Rui Yamaguchi
Rui Yamaguchi Arinobu Tojo
Arinobu Tojo