
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 16 January 2023
Sec. Thoracic Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.969032
This article is part of the Research Topic Targeting Ubiquitination to Overcome Cancer Drug Resistance View all 5 articles
Lung cancer (LC) remains the leading cause of cancer-related deaths worldwide, with extremely high morbidity and mortality rates. Non-small cell lung cancer (NSCLC) is the most critical type of LC. It seriously threatens the life and health of patients because of its early metastasis, late clinical symptoms, limited early screening methods, and poor treatment outcomes. Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), participate in cell proliferation, metastasis, and chemoresistance. Several previous studies have proven that ncRNAs are vital regulators of tumorigenesis. Ubiquitination plays the most crucial role in protein post-translational modification (PTM). Deubiquitination and ubiquitination form a homeostasis. In summary, ubiquitination and deubiquitination play essential roles in mediating the degradation or overexpression of a range of crucial proteins in various cancers. A growing number of researchers have found that interactions between ncRNAs and ubiquitination (or deubiquitination) play a crucial role in NSCLC. This review presents several typical examples of the important effects of ncRNAs and ubiquitination (or deubiquitination) in NSCLC, aiming to provide more creative ideas for exploring the diagnosis and treatment of NSCLC.
Many well-known international studies have confirmed that cancer is the primary cause of premature death. Lung cancer still ranks first in terms of fatality rate (18.0%) and is the second most diagnosed cancer (11.4%) (1).Based on histopathological characteristics, lung cancer can be divided into small-cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC accounts for up to 85% of lung cancer (2). The latest advances in diagnostic tools and systematic treatment methods have improved the therapeutic effect of NSCLC, with a 5-year survival rate of 33% for regional-stage disease and 60% for localized-stage disease. However, owing to the characteristics of early metastasis and late clinical symptoms, the 5-year survival rate of patients with distant metastasis is only 6% (3). Therefore, early detection, diagnosis, and treatment are essential for prolonging the survival time of NSCLC patients.
The Human Genome Project found that genes encoding proteins account for only 2% of the human genome, and most of the remaining human transcriptomes are ncRNAs (4). The vast number of ncRNAs was once considered non-functional “garbage” until the ENCODE project showed that the non-protein-coding parts of genes could be copied into thousands of RNA molecules, which regulate fundamental biological processes and play vital roles in the entire human disease spectrum. However, the boom in ncRNA research has only begun (5). There are many types of ncRNAs, and current research mainly focuses on microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs).
MiRNAs are small RNAs of approximately 22 nucleotides that regulate the expression of specific genes by pairing them completely or incompletely with the 3’-UTR region of the mRNA of the target genes to degrade mRNAs or inhibit their post-transcriptional translation (6). MiRNAs are typically processed in the nucleus by RNA polymerase II (polII) and Drosha as precursor miRNAs (pri-miRNAs), which are then exported to the cytoplasm and cleaved into double-stranded RNA by the RNase III enzyme, Dier. One of the strands was then selected and incorporated into a miRNA-induced silencing complex (miRISC). Complementary miRNA and mRNA usually lead to the degradation of the target mRNA. However, when not entirely complementary, miRNAs usually prevent the expression of target genes at the protein level without influencing mRNA stability (7). MiRNAs are often heavily dysregulated and function as oncogenes or tumor suppressors in cancer cells. MiRNAs can act as diagnostic and prognostic markers, which bring vast attention to cancer diagnosis and treatment (8, 9).
LncRNAs are non-coding RNAs that are longer than 200 nucleotides and have 5’-modified caps and 3’-polyadenylated tails. LncRNAs are closely related to their cellular localization and play essential roles in almost all phases of gene regulation (10). LncRNAs affect chromatin and transcription levels in the nucleus, leading to post-transcriptional regulation in the cytoplasm. In addition, they regulate fine-tuning of the translation process and RNA molecules directly or indirectly by affecting the expression of upstream or downstream genes (11). In terms of mechanisms of action, decoy lncRNAs can bind proteins or RNAs, resulting in the negative regulation of protein expression. In addition, guide lncRNAs direct protein localization by binding to proteins and signal lncRNAs to interact with transcription factors or chromatin-modifying enzymes, resulting in the regulation of transcription and signaling pathways. Lastly, scaffold lncRNAs act as organizing structures where molecules can bind and interact with each other more efficiently (12). Aberrant expression of numerous lncRNAs participates in several types of carcinogenesis.
CircRNAs are a class of endogenous RNAs that regulate gene expression, and the 3’ and 5’ ends of circRNAs are covalently bound by trans-splicing to form a closed circular structure. Unlike linear RNA structures, circRNAs do not have 5’-3’ polarity or polyadenylated tails, which makes them more stable (13). As a result, circRNAs can resist the decomposition of RNase enzymes and have higher sequence conservation, abundance, and tissue specificity (14). CircRNAs mainly work through four molecular mechanisms. First, circRNAs can act as competing endogenous RNAs (ceRNAs) of miRNAs. Second, circRNAs interact with RNA-binding proteins (RBPs) to regulate the mRNA of many genes. Third, a balance exists between linear RNAs and circRNAs through competitive complementary pairing. Fourth, although circRNAs are non-coding RNAs, some circRNAs can perform regulatory functions through translation (15). CircRNAs affect all aspects of the occurrence and development of different cancers.
The ubiquitin-proteasome system (UPS) is a critical PTM of proteins that regulates protein degradation and maintains protein homeostasis (16). The UPS consists of many key components, including ubiquitin(Ub), a highly conserved 76-amino-acid protein that conjugates other cellular proteins and modifies them (17). In addition, the UPS consists of deubiquitinating enzymes (DUBs) and a three-enzyme cascade involving ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s) (18). Deubiquitination can remove mono Ub or the whole polyubiquitin chains on proteins to resist the protein’s degradation or the alteration of the subcellular position (19). Also included in the UPS is the 26S proteasome, which mainly consists of two parts: a 20S core particle (CP) complex, where protein degradation mainly occurs, and a 19S regulatory particle (RP), responsible for the substrate choosing the right degradation site (20). Usually, E1 activates Ub in an ATP-dependent manner. The activated Ub is then moved to E2 through a transthiolation reaction. The E3 and substrate protein complex acquire Ub from E2, and Ub targets the protein. Alternatively, Ub is moved to E3 through a transthiolation reaction, E3 recognizes the protein specifically, and the C-terminal of Ub is attached to the Lys residue of the target protein (21). In humans, two E1s, 38 E2s, and approximately 600–1000 E3s have been found so far (22). Studies have found that ubiquitination and deubiquitination reactions are responsible for developing various tumors and play key roles in cancer treatment (23). The ubiquitinization and deubiquitination process is shown in the Figure 1.
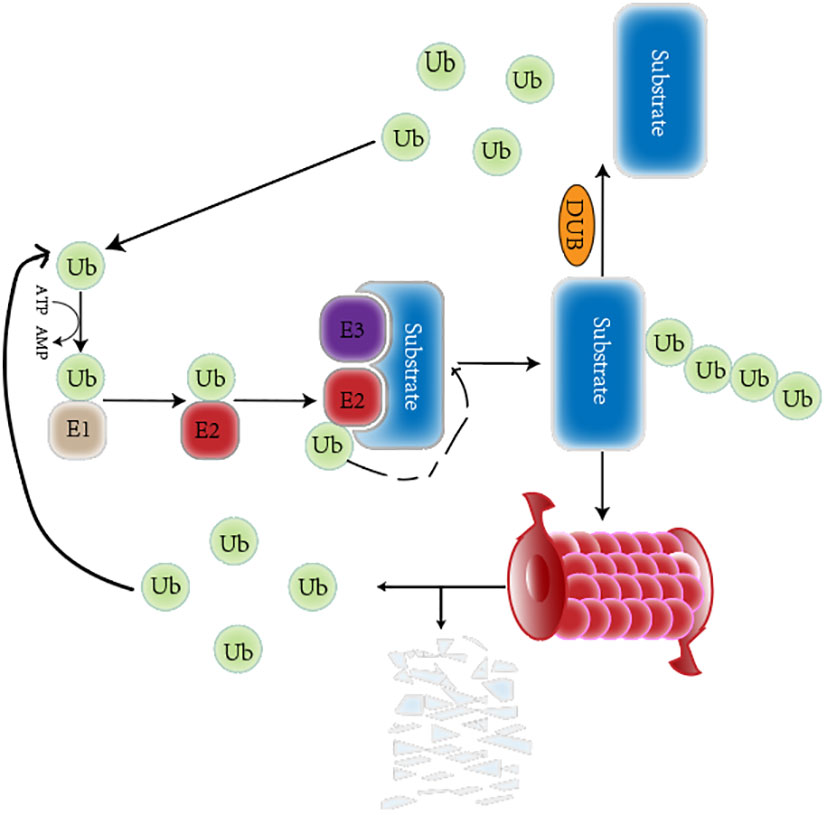
Figure 1 A brief overview of the ubiquitination and deubiquitination circulation. The Ub is respectively activated by the E1 Ub-activating enzyme, conjugated to the E2 Ub-conjugating enzyme, and attached to a specific substrate selected by E3 Ub ligase. The ubiquitinated proteins can be degraded by the proteasome. The deubiquitinase can remove Ub from substrate. Those released Ub participates in ubiquitination again.
NcRNAs participate in all aspects of NSCLC occurrence and development and play an essential role in NSCLC diagnosis and treatment. Parts of the crucial ncRNAs are enumerated in this review to explain their critical role in NSCLC.
Dysregulation of miRNAs causes abnormal expression of target genes and influences every aspect of NSCLC. Numerous studies have verified that miRNAs influence NSCLC through cell growth, cell cycle, apoptosis, metastasis, immune escape, and drug resistance (24).
In 2004, let-7 was reported to have low expression in NSCLC causing cell growth inhibition, and was associated with shortened postoperative survival (25). The let-7 family inhibits tumor growth and metastasis in lung adenocarcinoma through the MAPK/ERK and Wnt/β-catenin pathways (26, 27). let-7 has a strong relationship with drug resistance by targeting different proteins or regulating other factors. Cancer stem cells (CSCs) have stem cell-like characteristics that enable the growth of tumor cells, which is closely related to tumor treatment resistance (28). Lin28 is a highly conserved RNA-binding protein that induces pluripotent stem cell differentiation. The double negative feedback loop established by let-7 and Lin28 significantly influences NSCLC chemotherapy and radiation therapy resistance (29, 30). A recent study found that metformin could increase mature let-7b expression by mediating m6A formation on pri-let-7b and enforce osimertinib sensitivity by decreasing the expansion of stem cell groups (31).
MiR-21 is one of cancer research’s first identified “miRNA enhancers” (32). All cells in the human body can produce extracellular vesicles (EVs), which are membranous vesicles released into the extracellular matrix. Exosomes are EVs with diameters ranging between 30 and 150 nm. The molecular content of exosomes, including nucleic acids, proteins, and lipids, has an important influence on various cells. Therefore, exosomal ncRNAs and their pathophysiological roles significantly impact NSCLC (33). MiR-21 was significantly higher in sputum and plasma samples from patients with NSCLC than healthy donors. Thus, in addition to promoting growth and invasion phenotypes, miR-21 is a non-invasive biomarker for NSCLC diagnosis (32, 34). Recent studies have focused on the combined effects of let-7 and miR-21. Bai et al. found that the simultaneous downregulation of miR-21 and upregulation of let-7 could inhibit NSCLC development. This finding indicates a feedback regulation loop between miR-21 and let-7, with K-Ras as the target gene (35). This study also looks at the combined effects of miRNAs, which could be significant in NSCLC research.
The characteristics of early metastasis make NSCLC survival rates extremely low. Epithelial-mesenchymal transition (EMT) is a typical change in the invasiveness and migratory abilities of tumor cells. Accompanied by converting epithelial cells to mesenchymal cells, epithelial cells lose cell-cell adhesion, have increased motility, and have altered the expression of some key genes (36). The miR-200 family includes five conserved miRNAs: miR-200a, miR-200b, miR-200c, miR-141, and miR-429. MiR-200 is a well-known tumor suppressor that inhibits EMT in various cancers. Previous studies have found that the typical EMT regulator, miR-200/ZEB1 axis, regulates ECM-dependent β1-integrin/FAK signaling in NSCLC through CRKL. In addition, previous studies further explored the intracellular signaling pathways in NSCLC responsible for tumor cell invasion and metastasis through the EMT activation (37). At the same time, a novel double-negative feedback loop between FOXF2 and miR-200 was discovered, revealing a parallel axis to miR-200/ZEB1 that controls cancer cell invasion and migration through the EMT (38). In addition to influencing invasion and metastasis, the EMT can also mediate drug resistance. A study found that Cathepsin L (CTSL) and miRNA-200c suppress each other and mediate paclitaxel resistance through EMT changes (39). Moreover, new research found that miRNA-200c could downregulate Lin28B and improve EMT-related EGFR tyrosine kinase inhibitor (TKI) resistance in NSCLC (40). MiR-200s and let-7s have multiple members that regulate overlapping target sets.
Three typical miRNAs in NSCLC and three research suggestions regarding the correlation between miRNAs and NSCLC are provided. First, early NSCLC diagnosis is critical for mortality reduction because more miRNAs function as exosomes. Second, miRNAs are prone to develop drug resistance during NSCLC treatment, as they can modulate drug resistance through many methods. Therefore, research should improve the efficiency of NSCLC treatment. Last but not least, miRNAs can be the enhancer or suppressor of NSCLC. In addition, several miRNAs have the same target genes. Thus, the overlapping effects of these miRNAs in NSCLC, which play a leading role, are expected to provide new insights into future research on NSCLC.
To date, numerous lncRNAs have been identified in NSCLC. Similarly, the effects of lncRNAs on the various stages of NSCLC development have received widespread attention. This paper clarifies the reciprocal of lncRNAs and NSCLC and future research directions by enumerating classic lncRNAs that have been identified.
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched abundant transcript 2 (NEAT2), is the first lncRNA to be studied in NSCLC. It is upregulated in tumor tissues of NSCLC and is considered a specific marker for NSCLC (41). LncRNAs can regulate the biological functions of NSCLC through many mechanisms; the most common regulating method is by acting as endogenous miRNA sponges. Several miRNAs are sponged by MALAT1 and regulate functions, including cell proliferation, differentiation, and development, through competing with mRNAs. For example, miR-1297 binding to MALAT1 was predicted using software and confirmed by luciferase reporter assays. P300 is the downstream target molecule of MALAT1 and miR-1297. The P300/β-catenin complex is essential for activating the Wnt pathway during transcriptional activity (42). In addition, the Wnt signaling pathway is closely related to drug resistance. Therefore, through inhibiting MALAT1/miR-1297/p300/β-catenin/Wnt signaling, the A549/DDP cells were re-sensitized. Asiatic acid can achieve this regulation (43). Similarly, the MALAT1/miR-27a-5p/PBOV1 axis was recently found to enhance gemcitabine resistance in NSCLC cells (44). In addition to being a ceRNA, MALAT1 is also involved in regulating NSCLC through other mechanisms. IGF2BP2 promotes MALAT1 stability via m6A modification and promotes the proliferation of NSCLC cells through the MALAT1/ATG12 axis (45). Furthermore, MALAT1 is an important circulating diagnostic biomarker for diagnosing NSCLC (46). The single nucleotide polymorphism (SNP) in MALAT1, rs3200401, has been associated with NSCLC susceptibility, which is essential for identifying early screening populations for NSCLC (47).
LncRNA H19 (referred to as H19) is another classic lncRNA that can influence NSCLC. LncRNA H19 is encoded by H19 protein and is highly expressed in NSCLC tissues and cells. FOXF2 has been certified as one of the promoters to accelerate H19 transcription. Upregulated H19 can promote the proliferation and migration of NSCLC through the EZH2/PTEN axis (48). H19 decreased the overall survival of NSCLC patients, the combination of H19 and miR-21 played important roles in diagnostic and treatments value in NSCLC (49). In addition, H19 can also act as a sponge for several microRNAs (miRNAs). For example, downregulating H19 can significantly inhibit EMT progression through the upregulation of miR-203 in NSCLC (50). As for drug resistance, related research found that H19 could be secreted into exosomes assisted by heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) and induce gefitinib resistance in NSCLC cells (51). The latest study confirmed that exosomal H19 promotes erlotinib resistance via the miR-615-3p/ATG7 axis (52). H19 can also regulate the radiosensitivity of NSCLC cells. Radiotherapy (RT) is a crucial treatment modality. Theoretically, higher doses of radiation have a better effect on radiotherapy; however, related side effects may gradually appear. Therefore, it is essential to enhance radiosensitivity. Zhao et al. found that the H19 sponge miR-130a-3p increased the expression of WNK lysine deficient protein kinase 3 (WNK3) and elevated radiosensitivity to X-rays in NSCLC cells (53). Moreover, SNPs in H19 were associated with NSCLC susceptibility (54).
LncRNA-MEG3 is a representative inhibitory lncRNA in NSCLC. Tumor-suppressive lncRNAs inhibit carcinogenesis and progression by enhancing apoptosis via several methods. MEG3 induces apoptosis and inhibits proliferation by activating p53 (55). The upregulation of MEG3 inhibits stem cell-like characteristics and prevents metastasis in NSCLC through miR-650/SLC34A2 (56). As for therapeutic aspects, MEG3 might enhance cisplatin sensitivity via different signaling pathways (57, 58). Autophagy refers to a lysosome-mediated catabolic process and is a double-edged sword for NSCLC that can regulate the development of NSCLC through different proteins and signaling pathways. On the one hand, autophagy restrains tumorigenesis by removing harmful cytotoxic agents to reduce stress injury, prevent genome damage, and maintain cellular integrity (59). Wang et al. found that MEG3 influences autophagy by regulating the miR-543/IDO axis (60). On the other hand, the MEG3 rs4081134 polymorphism has been associated with NSCLC susceptibility in the Chinese population (61).
Considering the current research on lncRNAs and NSCLC, it can be concluded that lncRNAs regulate the progression of NSCLC through multiple mechanisms. Therefore, lncRNAs play a key role in the diagnosis and treatment outcomes of NSCLC. In addition, SNPs in a few classic lncRNAs are closely related to NSCLC susceptibility, which is of great significance for early lung cancer screening.
Extensive research was only conducted on circRNAs after lncRNAs and miRNAs. However, due to their more stable characteristics, research on NSCLC and circRNAs is very prosperous, and the related mechanistic studies are more detailed and in-depth.
As mentioned above, exosomes can be used as diagnostic markers to regulate drug resistance and can be key factors in promoting the progression of NSCLC. More than 1000 exosomal circRNAs have been discovered in human serum, much more than linear RNAs. These exosomal circRNAs have a significant effect on NSCLC. For example, three circRNAs, circ_0047921, circ_0056285, and circ_0007761, were found as exosomal circRNAs in NSCLC patients’ serum exosomes, and could perfectly distinguish early-stage NSCLC and other lung diseases, such as pulmonary tuberculosis, in healthy people (62). Therefore, circRNAs have become excellent non-invasive diagnostic biomarkers for NSCLC. Moreover, several exosomal circRNAs have been found to regulate resistance to chemotherapy and radiotherapy in NSCLC. Exosomal circ_0001658 has been found to sponge miR-409-3p and increase TWIST1 expression to promote gefitinib resistance in NSCLC (63). The exosomal circUSP7 modulates the miR-934/SHP2 axis to induce anti-PD1 resistance and promote immune escape in NSCLC (64). Controlling tumor cell escape is crucial for improving radiotherapy sensitivity.
The tumor microenvironment (TME) includes a variety of cells and structures surrounding tumor cells. TME cells undergo metabolic modifications and regulate drug resistance (65). Recent studies have revealed the relationship between circRNAs and altered metabolism, which could assist in addressing the progression and drug insensitivity of NSCLC. The expression of circ_0008797 is low in NSCLC tissues and cell lines, which could attenuate proliferation, metastasis, and aerobic glycolysis by regulating miR-301a-3p/SOCS2 (66). The downregulation of circ_0011298 enhances the Taxol sensitivity of Taxol-resistant NSCLC cells by decreasing cell growth, metastasis, and glycolysis and promoting apoptosis and cell cycle arrest via the miR-486-3p/CRABP2 axis (67). In addition, circSLC25A16 and circPIP5K1A can induce hypoxia-inducible factor (HIF)-1a-dependent glycolysis by sponging miRNAs (68, 69).
The induction of programmed cell death (PCD) is the primary mechanism leading to tumor cell death. Pyroptosis, autophagy, and ferroptosis are novel and essential methods for inducing PCD. CircRNAs have an intimate relationship with PCD (70). A current study found that circDTL functions as an oncogene and the knockdown of circDTL could improve the efficacy of chemotherapy drugs, ferroptosis, and apoptosis of NSCLC cells through the miR-1287-5p/GPX4 axis (71). CircRNAs can regulate autophagy by directly targeting key proteins or signaling pathways related to autophagy. Hsa_circ_0085131 functions as a ceRNA and enhances cisplatin resistance of NSCLC cells by upregulating the autophagy-associated factor, ATG7, leading to autophagy of tumor cells (72). Similarly, circ_FOXM1 can lead to autophagy by sequestering miR-149-5p and upregulating ATG5 (73). In addition, some bioinformatic predictions have identified six circRNAs that regulate autophagy proteins by sponging miRNAs and enhancing the sensitivity of NSCLC to radiotherapy (74). Future experiments are expected to verify these predictions.
This paper did not enumerate the classic circRNAs to clarify their relationship with NSCLC. Although the research on circRNA is limited, many circRNAs have been found to influence NSCLC by regulating factors, including metabolism, TME, and autophagy. These are novel and popular NSCLC mechanism studies. Therefore, circRNAs are bound to be excellent research objects for diagnosing and treating NSCLC or exploring new mechanism targets and networks.
During ubiquitination and deubiquitination in NSCLC, the E3 ligases and the DUBs play crucial roles in regulating protein stability that influences the progression of NSCLC. Therefore, E3 ligases and DUBs illustrated the relationship between ubiquitination, deubiquitination, and NSCLC.
The superfamily of tripartite motif proteins (TRIM) are proteins containing an N-terminal RING finger, a coiled-coil (CC) domain, and one or two B-boxes (75). The RING domain is responsible for conjugations with ubiquitin or ubiquitin-like molecule, including interferon-stimulated gene15 (ISG15) and small ubiquitin-like modifier (SUMO). The C-terminal domains can categorize TRIM proteins and recognize and regulate substrate proteins (76). Many TRIM proteins have been found to regulate the development and treatment of NSCLC by targeting different proteins and signaling pathways (77). TRIM59 expression in NSCLC patients was two to three fold higher than in normal patients; these patients had worse survival outcomes suggesting that TRIM59 might be a novel biomarker for NSCLC diagnosis (78).
Similarly, DNA hypermethylation of TRIM58 was upregulated in NSCLC tissues and presented promising results in Area Under Curve (AUC) when discriminating between NSCLC and control groups (79). The TRIM family also affects drug resistance in NSCLC. TRIM46 promotes cell proliferation, glycolysis, and DDP in NSCLC by upregulating HK2 via AKT phosphorylation and PHLPP2 ubiquitination by interaction with TRIM46 is the key method for inducing p-AKT (80). TRIM23 was highly expressed in DDP-resistant lung adenocarcinoma (LUAD) cells and tissues. A mechanistic study suggested that TRIM23 could ubiquitinate proteins to activate the NF‐κB pathway and further regulate glucose metabolism in DDP cells (81). In addition to the TRIM family, many other classic E3 ligases participate in regulating NSCLC. Ubiquitin-conjugating enzyme E2O (UBE2O), an E2/E3 hybrid ubiquitin-protein ligase, targets MAX-interacting protein 1 (Mxi1) for ubiquitination and degradation at the K46 residue, which can suppress the occurrence of NSCLC and enhance radiosensitivity (82). Another E3 ligase, MIB1, stimulates the degradation of the antioxidant transcription factor, nuclear factor erythroid 2-related factor 2 (Nrf2), in a ubiquitin manner and induces them to be more sensitive to ferroptosis (83).
Ubiquitin-specific proteases (USPs) are the most versatile class of known deubiquitinases with the most diverse structures that remove Ub from proteins. Among currently available research, USPs regulate multiple known NSCLC-related pathways (84). Knocking out the USP1 could lead to hypersensitivity DNA damage. Recent studies have found that inhibiting the activity of the USP1/UAF1 complex can improve cisplatin sensitivity in NSCLC (85). USP18 removes the conjugate of interferon-induced Ubl ISG15 and inhibits 14-3-3ζ acetylation by ISG15 to accelerate NSCLC metastasis (86). Additionally, USP35 targeted ferroportin and induced ferroptosis in NSCLC (87). USP5 increased PD-L1 levels by cleaving and stabilizing the polyubiquitin chain and maintaining stability. Thus, USP5 knockdown might prevent immune response and drug resistance by reducing PD-L1 protein (88). OTU deubiquitinase 3 (OTUD3) binds to the ovarian tumor-related protease family, PTEN protector. OTUD3 is a potential tumor suppressor in NSCLC, a deubiquitylase of GRP78, and promotes tumorigenesis of NSCLC (89). Ubiquitin C-terminal hydrolase-L3 (UCHL3) belongs to the ubiquitin COOH-terminal hydrolase family, and its high expression is associated with poor survival in LUAD. In addition, UCHL3 promotes the proliferation and stem cell traits of NSCLC cells by deubiquitinating Aryl hydrocarbon receptor (AhR) (90).
The number of E3 ligases and DUBs is enormous. They target key proteins in NSCLC by inducing ubiquitination or deubiquitination of these proteins or further regulating post-modifications of other proteins to regulate the progression of NSCLC. Therefore, the balance between ubiquitination and deubiquitination plays a crucial role in proliferation, metastasis, apoptosis, the cell cycle, immune escape, and drug resistance in NSCLC.
The ncRNA and the UPS systems have huge members and diverse control methods, which is why they became key regulators of NSCLC. Many studies focused on the interactions between ncRNAs and the UPS in NSCLC. However, compared to the vast number of these two systems, interaction research was fairly small. Recent studies have shown that ncRNAs could act on critical Ub-enzymes, other essential proteins, and pathways to regulate the development of NSCLC. Furthermore, certain ubiquitination and deubiquitination processes can conversely regulate ncRNAs. Such crosstalk between ncRNAs and ubiquitination or deubiquitination is significant and may even lead to breakthroughs in further NSCLC studies. Next, the necessity of research on crosstalk is illustrated by providing classic examples.
Several studies have been conducted on the let-7 family and the UPS. Histones, H2A and H2B, have monoubiquitination sites at their lysines, and K120 is the only site of H2B monoubiquitination that promotes the occurrence of H2Bub1. H2Bub1 functions as a tumor suppressor in lung cancer and can be catalyzed by the E3 ubiquitin-ligase complex, RNF20/RNF40, and erased by various DUBs (91). Ambra et al. performed a bioinformatic screen to identify the effects of miRNAs on H2Bub1 homeostasis. The results showed that let-7b targeted USP42, USP44, and ATXN7L3. USP42 and USP44 were reported to act on H2Bub1, while ATXN7L3 could remove H2Bub1 through the activity of USP22. Further experimental data showed that let-7b and let-7c significantly reduced the steady-state mRNAs and ATXN7L3 and USP42 protein levels but did not influence RNF20 protein levels. The results showed that the let-7 family members, especially let-7b and let-7c, could positively regulate H2Bub1. Further, RNA pull-down assays strongly supported that ATXN7L3, USP44, and USP42 were enriched by pulling down let-7b and were the direct let-7b targets. Altogether, these findings suggest that the let-7 family could prevent the ubiquitination of H2B by directly regulating DUBs to prevent the migration of NSCLC (92). Epidermal growth factor receptor (EGFR)-TKIs can induce drug resistance by elevating autophagic flux (93). One study found that miR-4487 expression increased in NSCLC cells treated with gefitinib. Subsequently, miR-4487 targeted USP37 directly and downregulated USP37 expression. These results led to whole-cell ubiquitination and increased autophagic flux, which also allowed the observation of miRNAs’ effects on NSCLC resistance by regulating deubiquitination and autophagy (94). Previous studies have suggested that KRAS mutations might promote tumor cell growth by inducing DNA damage and genotoxic stress. As a highly conserved recombinase, RAD51 may repair DNA damage caused by KRAS-MT and improve resistance to radiation (95). MiR-376a-5p acts as an upstream regulator of TRIM36; knockdown of miR-376a-5p upregulates TRIM36. Elevated levels of TRIM36 can inhibit DNA repair and promote radiosensitivity (96). Cullin 4B (CUL4B) belongs to the cullin family, which can assemble DNA damage-binding protein 1 (DDB1) and its substrate to form a new RING-based E3 ubiquitin ligase, cullin4B-Ring E3 ligase complex (CRL4B). MiR-194 can target CUL4B directly, inhibiting its translation and functions. On the other hand, CUL4B was important for H2AK119ub1 and EZH2 recruitment and the consequent H3K27me3 in miR-194. EZH2 repressed miR-194. This study described the role played by the negative feedback loop, including miR-194 and CUL4B. Negative feedback plays an essential role in NSCLC (97). More information on the effects of miRNAs interacting with the UPS in NSCLC is shown in Table 1.
Some points regarding the interactions between miRNAs and the UPS can be concluded from the examples provided. Based on current research, miRNAs usually regulate the ubiquitination degradation or deubiquitination stabilization of key proteins through E3s or DUBs, which are the main methods that affect NSCLC development. Other studies have also revealed that E3s or DUBs could regulate miRNAs, partially forming a negative feedback loop. Such reaction networks play a critical role in the tumorigenesis, diagnosis, and treatment of NSCLC. Some miRNAs regulate the expression levels of target proteins and change their locations through E3s or DUBs, thus promoting or inhibiting the subsequent reactions. Studies on drug resistance and radiotherapy sensitivity of NSCLC influenced by miRNAs and the UPS are equally popular. Further miRNAs and E3s or DUBs interactions have been discovered, but further mechanistic research is required (107, 108). Thus, further research should lead to more spillovers in diagnosing and treating NSCLC.
The interactions between lncRNAs and E3s or DUBs are more complex than the interactions between miRNAs and the UPS. Tumor-associated macrophages (TAMs) are closely related to the TME and can be classified into M1 and M2 macrophages. M1 macrophages function as tumor suppressors, whereas M2 macrophages have the opposite effect. M2 polarization occurs more easily in the TME (109). In addition, exosomes and M2 macrophages can interact with each other to promote tumorigenesis (110). MiR-19b-3p could work as an exosomal miRNA and play an oncogenic role in NSCLC. On the one hand, exosomal miR‐19b‐3p promotes M2 polarization. Further, M2-polarized macrophages secrete exosomal LINC00273, which recruits neural precursor cell expressed developmentally downregulated 4 (NEDD4) and induces large tumor suppressor kinase 2 (LATS2) ubiquitination. NEDD4 is an E3 ligase that functions as an oncogene by promoting the ubiquitination of tumor-suppressive proteins (111). Therefore, miR‐19b‐3p regulates the Hippo/YAP pathway through LATS2 ubiquitination and promotes tumor progression. On the other hand, LINC00273 could also increase the RBMX level through Hippo/Yes associated transcriptional regulator (YAP). X-linked RNA-binding motif protein (RBMX) is an hnRNP that functions as an RBP to adjust the packaging of exosomal miRNAs. Therefore, RBMX helped package miR-19b-3p into LUAD cell-derived exosomes (112). This closed-loop plays an essential role in M2 polarization and LUAD development. This paper elucidates the crosstalk between TAMs and LUAD cells mediated by exosomes, ncRNAs, crucial pathways, and proteins, which provides new possibilities for LUAD diagnosis and treatment. The expression of UCHL3 was upregulated in NSCLC tissues and cells and is related to an unfavorable prognosis. Overexpression of LINC00665 and silencing of miR-582-5p enhanced the resistance of NSCLC cells to radiotherapy by upregulating UCHL3 and PD-L1 and stabilizing AhR to promote immune escape (113). Another study confirmed that lncRNA CCAT1 could activate and stabilize the PI3K/AKT/mTOR pathway. This stabilization is done by translocating fatty acid binding protein 5 (FABP5) into the nucleus to induce the PPAR‐RXR complex and pyruvate dehydrogenase kinase 1 (PDK1) translation. Further, it binds to USP49 and deubiquitinates FABP5, thereby binding to RAPTOR to induce AKT phosphorylation. CCAT1 participates in reprogramming FA metabolism and enhances the malignant phenotype of NSCLC (114). More information on the effects of lncRNA interactions with the UPS on NSCLC is shown in Table 2.
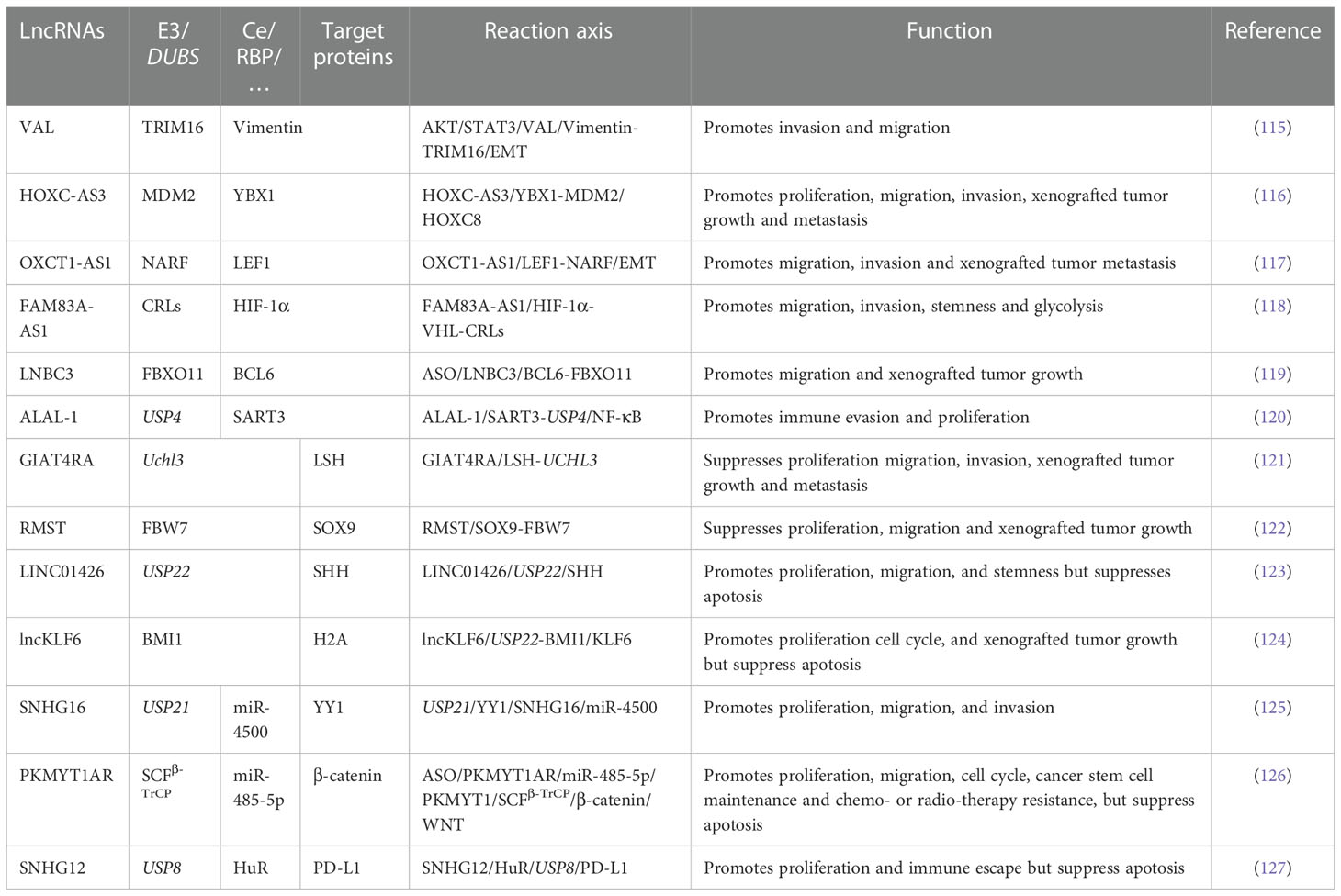
Table 2 The interaction between lncRNAs and UPS in NSCLC(when a protein acts as two roles, the two charts were merged).
Current research shows that the reactions between lncRNAs and the UPS are exceptionally complicated. In some studies, RBPs remained the target genes of the UPS. LncRNAs regulate ubiquitination or deubiquitination by competing with the binding sites of proteins or destroying the combination of proteins and E3s or DUBs. Some E3s or DUBs are RBPs, and lncRNAs can regulate enzymes directly, thus adjusting further reactions. Other lncRNAs regulate E3s or DUBs through their long miRNAs or RBPs. Some lncRNAs may influence multiple ubiquitination and deubiquitination processes. A few lncRNAs can alter the position of E3s and DUBs to regulate their effects. In conclusion, the diversity of ubiquitination or deubiquitination regulation by lncRNAs considerably impacts the occurrence and development of all stages of NSCLC. Whether it is TME, metabolism, SCS, immune escape, or other popular NSCLC research fields, crosstalk reactions can be seen everywhere. Therefore, further related studies should be conducted in the future.
The number of studies related to the regulation of circRNAs and the UPS is insufficient. Some studies have involved the regulation between E3s, DUBs, and circRNAs, but more profound mechanistic studies have not yet been conducted. Such research is relatively new and lacking, but it is crucial for the new direction of NSCLC diagnosis and treatment. CircIGF2BP3, a circRNA derived from the back-splicing of IGF2BP3, suppresses CD8T cell infiltration in NSCLC. Further, circIGF2BP3 is overexpressed and compromises anti-tumor immunity in NSCLC. METTL3 and METTL14 are composed of a stable methyltransferase complex (MTC) in a 1:1 ratio and exerts a methylation function (128). Promoting m6A levels in the circIGF2BP3 transcript in a METTL3-dependent manner could enhance the circularization of circIGF2BP3. In addition, circIGF2BP3 upregulates PKP3 expression in NSCLC cells by binding to miR-3173-5p or miR-328-3p. Plakophilin 3 (PKP3) belongs to the armadillo protein family, which can mediate PD-L1 expression and rescue it from proteasomal degradation. OTUB1 functions as a downstream effector of PKP3 and inhibits its degradation, thus further avoiding the killing effects of T cells and leading to immune escape by upregulating the PD-1 and PD-L1 complex (129). In this study, circIGF2BP3 and DUB became the PKP3 upstream and downstream, respectively, and the indirect effects of these two factors had an essential impact on the occurrence and development of NSCLC. Insulin-like growth factor-2 mRNA-binding proteins (IGF2BPs), IGF2BP1 and IGF2BP3, are evolutionarily conserved families of RNA-binding proteins that regulate important parts of cancer cells and promote the development of cancers (130). CircNDUFB2 has a length of 249 nucleotides and is generated from NDUFB2. Functionally, circNDUFB2 serves as a suppressor of NSCLC development. CircNDUFB2 enhances the interactions between TRIM25 and IGF2BPs and subsequently boosts the IGF2BPs ubiquitination by TRIM25. Interestingly, circNDUFB2 exerts a tumor-suppressive role by promoting IGF2BPs ubiquitination and eliciting immune responses in NSCLC cells. Retinoic acid-inducible gene I (RIG-I) is a member of the RIG-I-like receptor (RLR) family, which recognizes viral RNAs and induces innate immune responses against viral infections. When lacking an RNA ligand, RIG-I adopts an auto-repressed conformation that prevents the N-terminal caspase recruitment domains (CARDs) from signaling. CircNDUFB2 activates RIG-I by destabilizing the interaction between CARDs and its helicase domain, thereby inducing the activation of RIG-I-MAVS signaling cascades (131). In addition, circNDUFB2 promotes IGF2BPs ubiquitination through the circNDUFB2/TRIM25/IGF2BPs signaling pathway and causes cellular immune responses by activating RIG-I. CircNDUFB2 inhibited tumorigenesis by two mechanisms, which prompted a focus on the effects of circRNAs on tumors from multiple perspectives.
Most circRNAs regulated the E3s or DUBs by sponging miRNAs. Therefore, it can be said that the research on the crosstalk between the circRNAs and the UPS is in its infancy. However, with further development, crosstalk studies between circRNAs and the UPS are bound to bring more remarkable progress in diagnosing and treating NSCLC.
This article reviews the current research progress on the effects of interactions between ncRNAs and ubiquitination (and deubiquitination) in NSCLC. The intrinsic correlation between regulatory ncRNAs and UPS in NSCLC has been increasingly studied in recent years. It is worth noting that the interactions between ncRNAs and the UPS have been found to influence the progression of NSCLC, which makes them critical therapeutic targets. However, there is limited research that uncovers the precise mechanisms. There are many hurdles to overcome in studying the interplay between regulatory ncRNAs and the UPS in NSCLC. Nevertheless, with the continuous innovations of experimental methods and techniques, interactions between ncRNAs and ubiquitination and deubiquitination to innovate diagnosis methods and improve treatment efficiency in NSCLC are foreseeable.
YS wrote the draft and revised it. PH designed and supervised the study. LL and XD collected the data and designed the figures. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc (2008) 83(5):584–94. doi: 10.1016/S0025-6196(11)60735-0
3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin (2022) 72(1):7–33. doi: 10.3322/caac.21708
4. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
5. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. (2017) 127(3):761–71. doi: 10.1172/JCI84424
6. Gebert L, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol (2019) 20(1):21–37. doi: 10.1038/s41580-018-0045-7
7. Qu J, Lin Z. Autophagy regulation by crosstalk between miRNAs and ubiquitination system. Int J Mol Sci (2021) 22(21). doi: 10.3390/ijms222111912
8. Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol (2020) 72(12):1732–49. doi: 10.1111/jphp.13351
9. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
10. Lin C, Yang L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol (2018) 28(4):287–301. doi: 10.1016/j.tcb.2017.11.008
11. Balakittnen J, Weeramange CE, Wallace DF, Duijf PHG, Cristino AS, Kenny L, et al. Noncoding RNAs in oral cancer. Wiley Interdiscip Rev RNA (2022):e1754. doi: 10.1002/wrna.1754
12. Hull R, Mbita Z, Dlamini Z. Long non-coding RNAs (LncRNAs), viral oncogenomics, and aberrant splicing events: therapeutics implications. Am J Cancer Res (2021) 11(3):866–83.
13. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
14. Wilusz JE. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA. (2018) 9(4):e1478. doi: 10.1002/wrna.1478
15. Xue C, Li G, Zheng Q, Gu X, Bao Z, Lu J, et al. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer. (2022) 21(1):108. doi: 10.1186/s12943-022-01582-0
16. Mevissen T, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem (2017) 86:159–92. doi: 10.1146/annurev-biochem-061516-044916
17. Swatek KN, Komander D. Ubiquitin modifications. Cell Res (2016) 26(4):399–422. doi: 10.1038/cr.2016.39
18. Jin JO, Puranik N, Bui QT, Yadav D, Lee PC. The ubiquitin system: An emerging therapeutic target for lung cancer. Int J Mol Sci (2021) 22(17). doi: 10.3390/ijms22179629
19. Farshi P, Deshmukh RR, Nwankwo JO, Arkwright RT, Cvek B, Liu J, et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat. (2015) 25(10):1191–208. doi: 10.1517/13543776.2015.1056737
20. Lobaina DP, Tarazi R, Castorino T, Vaslin M. The ubiquitin-proteasome system (UPS) and viral infection in plants. Plants (Basel) (2022) 11(19). doi: 10.3390/plants11192476
21. Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol (2018) 19(1):59–70. doi: 10.1038/nrm.2017.83
22. Morreale FE, Walden H. Types of ubiquitin ligases. Cell (2016) 165(1):248–248.e1. doi: 10.1016/j.cell.2016.03.003
23. Liu J, Leung CT, Liang L, Wang Y, Chen J, Lai KP, et al. Deubiquitinases in cancers: Aspects of proliferation, metastasis, and apoptosis. Cancers (Basel) (2022) 14(14). doi: 10.3390/cancers14143547
24. Zhu X, Kudo M, Huang X, Sui H, Tian H, Croce CM, et al. Frontiers of MicroRNA signature in non-small cell lung cancer. Front Cell Dev Biol (2021) 9:643942. doi: 10.3389/fcell.2021.643942
25. Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res (2004) 64(11):3753–6. doi: 10.1158/0008-5472.CAN-04-0637
26. Li Y, Dong R, Lu M, Cheng C, Feng Z, Zhao R, et al. Let-7b-3p inhibits tumor growth and metastasis by targeting the BRF2-mediated MAPK/ERK pathway in human lung adenocarcinoma. Transl Lung Cancer Res (2021) 10(4):1841–56. doi: 10.21037/tlcr-21-299
27. He XY, Chen JX, Zhang Z, Li CL, Peng QL, Peng HM. The let-7a microRNA protects from growth of lung carcinoma by suppression of k-ras and c-myc in nude mice. J Cancer Res Clin Oncol (2010) 136(7):1023–8. doi: 10.1007/s00432-009-0747-5
28. Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol (2018) 53:156–67. doi: 10.1016/j.semcancer.2018.11.006
29. Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 pathway in cancer. Front Genet (2017) 8:31. doi: 10.3389/fgene.2017.00031
30. Yin J, Zhao J, Hu W, Yang G, Yu H, Wang R, et al. Disturbance of the let-7/LIN28 double-negative feedback loop is associated with radio- and chemo-resistance in non-small cell lung cancer. PloS One (2017) 12(2):e0172787. doi: 10.1371/journal.pone.0172787
31. Li K, Gao S, Ma L, Sun Y, Peng ZY, Wu J, et al. Stimulation of let-7 maturation by metformin improved the response to tyrosine kinase inhibitor therapy in an m6A dependent manner. Front Oncol (2021) 11:731561. doi: 10.3389/fonc.2021.731561
32. Rhim J, Baek W, Seo Y, Kim JH. From molecular mechanisms to therapeutics: Understanding MicroRNA-21 in cancer. Cells (2022) 11(18). doi: 10.3390/cells11182791
33. Paramanantham A, Asfiya R, Das S, McCully G, Srivastava A. Extracellular vesicle (EVs) associated non-coding RNAs in lung cancer and therapeutics. Int J Mol Sci (2022) 23(21). doi: 10.3390/ijms232113637
34. Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci (2018) 75(19):3539–51. doi: 10.1007/s00018-018-2877-x
35. Bai J, Shi Z, Wang S, Pan H, Zhang T. MiR-21 and let-7 cooperation in the regulation of lung cancer. Front Oncol (2022) 12:950043. doi: 10.3389/fonc.2022.950043
36. Nam MW, Kim CW, Choi KC. Epithelial-mesenchymal transition-inducing factors involved in the progression of lung cancers. Biomol Ther (Seoul). (2022) 30(3):213–20. doi: 10.4062/biomolther.2021.178
37. Ungewiss C, Rizvi ZH, Roybal JD, Peng DH, Gold KA, Shin DH, et al. The microRNA-200/Zeb1 axis regulates ECM-dependent β1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci Rep (2016) 6:18652. doi: 10.1038/srep18652
38. Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, et al. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene (2016) 35(2):173–86. doi: 10.1038/onc.2015.71
39. Zhao YF, Han ML, Xiong YJ, Wang L, Fei Y, Shen X, et al. A miRNA-200c/cathepsin l feedback loop determines paclitaxel resistance in human lung cancer A549 cells in vitro through regulating epithelial-mesenchymal transition. Acta Pharmacol Sin (2018) 39(6):1034–47. doi: 10.1038/aps.2017.164
40. Sato H, Shien K, Tomida S, Okayasu K, Suzawa K, Hashida S, et al. Targeting the miR-200c/LIN28B axis in acquired EGFR-TKI resistance non-small cell lung cancer cells harboring EMT features. Sci Rep (2017) 7:40847. doi: 10.1038/srep40847
41. Cao B, Bray F, Ilbawi A, Soerjomataram I. Effect on longevity of one-third reduction in premature mortality from non-communicable diseases by 2030: a global analysis of the sustainable development goal health target. Lancet Glob Health (2018) 6(12):e1288–96. doi: 10.1016/S2214-109X(18)30411-X
42. Malleske DT, Hayes D Jr, Lallier SW, Hill CL, Reynolds SD. Regulation of human airway epithelial tissue stem cell differentiation by β-catenin, P300, and CBP. Stem Cells (2018) 36(12):1905–16. doi: 10.1002/stem.2906
43. Cheng Q, Zhang S, Zhong B, Chen Z, Peng F. Asiatic Acid re-sensitizes multidrug-resistant A549/DDP cells to cisplatin by down regulating long non-coding RNA metastasis associated lung adenocarcinoma transcript 1/β-catenin signaling. Bioengineered (2022) 13(5):12972–84. doi: 10.1080/21655979.2022.2079302
44. Chen W, Tan X, Yang Q, Fang Z, Xu Y. MALAT1 enhances gemcitabine resistance in non-small cell lung cancer cells by directly affecting miR-27a-5p/PBOV1 axis. Cell Signal (2022) 94:110326. doi: 10.1016/j.cellsig.2022.110326
45. Han L, Lei G, Chen Z, Zhang Y, Huang C, Chen W. IGF2BP2 regulates MALAT1 by serving as an N6-methyladenosine reader to promote NSCLC proliferation. Front Mol Biosci (2021) 8:780089. doi: 10.3389/fmolb.2021.780089
46. Yuan S, Xiang Y, Guo X, Zhang Y, Li C, Xie W, et al. Circulating long noncoding RNAs act as diagnostic biomarkers in non-small cell lung cancer. Front Oncol (2020) 10:537120. doi: 10.3389/fonc.2020.537120
47. Tong G, Tong W, He R, Cui Z, Li S, Zhou B, et al. MALAT1 polymorphisms and lung cancer susceptibility in a Chinese northeast han population. Int J Med Sci (2022) 19(8):1300–6. doi: 10.7150/ijms.73026
48. Xu JL, Hua T, Ding J, Fan Y, Liu ZJ, Lian JW. FOXF2 aggravates the progression of non-small cell lung cancer through targeting lncRNA H19 to downregulate PTEN. Eur Rev Med Pharmacol Sci (2019) 23(24):10796–802. doi: 10.26355/eurrev_201912_19782
49. Zhou Y, Sheng B, Xia Q, Guan X, Zhang Y. Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther (2017) 24(8):317–24. doi: 10.1038/cgt.2017.20
50. Ge XJ, Zheng LM, Feng ZX, Li MY, Liu L, Zhao YJ, et al. H19 contributes to poor clinical features in NSCLC patients and leads to enhanced invasion in A549 cells through regulating miRNA-203-mediated epithelial-mesenchymal transition. Oncol Lett (2018) 16(4):4480–8. doi: 10.3892/ol.2018.9187
51. Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumor−released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non−small cell lung cancer. Oncol Rep (2018) 40(6):3438–46. doi: 10.3892/or.2018.6762
52. Pan R, Zhou H. Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 axis. Cancer Manag Res (2020) 12:4283–97. doi: 10.2147/CMAR.S241095
53. Zhao X, Jin X, Zhang Q, Luo H, Yang Z, Geng Y, et al. Silencing of the lncRNA H19 enhances sensitivity to X-ray and carbon-ions through the miR-130a-3p /WNK3 signaling axis in NSCLC cells. Cancer Cell Int (2021) 21(1):644. doi: 10.1186/s12935-021-02268-1
54. Li L, Guo G, Zhang H, Zhou B, Bai L, Chen H, et al. Association between H19 SNP rs217727 and lung cancer risk in a Chinese population: a case control study. BMC Med Genet (2018) 19(1):136. doi: 10.1186/s12881-018-0573-1
55. Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. (2013) 13:461. doi: 10.1186/1471-2407-13-461
56. Zhao Y, Zhu Z, Shi S, Wang J, Li N. Long non-coding RNA MEG3 regulates migration and invasion of lung cancer stem cells via miR-650/SLC34A2 axis. BioMed Pharmacother. (2019) 120:109457. doi: 10.1016/j.biopha.2019.109457
57. Wang P, Chen D, Ma H, Li Y. LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Onco Targets Ther (2017) 10:5137–49. doi: 10.2147/OTT.S146423
58. Xia Y, He Z, Liu B, Wang P, Chen Y. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/β-catenin signaling pathway. Mol Med Rep (2015) 12(3):4530–7. doi: 10.3892/mmr.2015.3897
59. Marinković M, Šprung M, Buljubašić M, Novak I. Autophagy modulation in cancer: Current knowledge on action and therapy. Oxid Med Cell Longev (2018) 2018:8023821. doi: 10.1155/2018/8023821
60. Wang C, Tao X, Wei J. Effects of LncRNA MEG3 on immunity and autophagy of non-small cell lung carcinoma through IDO signaling pathway. World J Surg Oncol (2021) 19(1):244. doi: 10.1186/s12957-021-02346-8
61. Yang Z, Li H, Li J, Lv X, Gao M, Bi Y, et al. Association between long noncoding RNA MEG3 polymorphisms and lung cancer susceptibility in Chinese northeast population. DNA Cell Biol (2018) 37(10):812–20. doi: 10.1089/dna.2018.4277
62. Xian J, Su W, Liu L, Rao B, Lin M, Feng Y, et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-Small-Cell lung cancer in the Chinese population. J Mol Diagn. (2020) 22(8):1096–108. doi: 10.1016/j.jmoldx.2020.05.011
63. Yu X, Fu X, Zhang X, Bai C, Wang Y. Circ_0001658 regulates gefitinib resistance of non-small cell lung cancer through miR-409-3p/TWIST1 axis. Anticancer Drugs (2022) 33(2):158–66. doi: 10.1097/CAD.0000000000001257
64. Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. (2021) 20(1):144. doi: 10.1186/s12943-020-01292-5
65. Fridman ES, Ginini L, Gil Z. The role of extracellular vesicles in metabolic reprogramming of the tumor microenvironment. Cells (2022) 11(9). doi: 10.3390/cells11091433
66. Abuduwaili K, Zhu X, Shen Y, Lu S, Liu C. circ_0008797 attenuates non-small cell lung cancer proliferation, metastasis, and aerobic glycolysis by sponging miR-301a-3p/SOCS2. Environ Toxicol (2022) 37(7):1697–710. doi: 10.1002/tox.23518
67. Wu Y, Xie J, Wang H, Hou S, Feng J. Circular RNA hsa_circ_0011298 enhances taxol resistance of non-small cell lung cancer by regulating miR-486-3p/CRABP2 axis. J Clin Lab Anal (2022) 36(5):e24408. doi: 10.1002/jcla.24408
68. Shangguan H, Feng H, Lv D, Wang J, Tian T, Wang X. Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis (2020) 11(6):437. doi: 10.1038/s41419-020-2635-5
69. Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem (2019) 120(11):19019–30. doi: 10.1002/jcb.29225
70. Luo Y, Li J, Yu P, Sun J, Hu Y, Meng X, et al. Targeting lncRNAs in programmed cell death as a therapeutic strategy for non-small cell lung cancer. Cell Death Discovery (2022) 8(1):159. doi: 10.1038/s41420-022-00982-x
71. Shanshan W, Hongying M, Jingjing F, Yiming Y, Yu R, Rui Y. CircDTL functions as an oncogene and regulates both apoptosis and ferroptosis in non-small cell lung cancer cells. Front Genet (2021) 12:743505. doi: 10.3389/fgene.2021.743505
72. Kong R. Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int (2020) 44(9):1945–56. doi: 10.1002/cbin.11401
73. Wei H, Li L, Zhang H, Xu F, Chen L, Che G, et al. Circ-FOXM1 knockdown suppresses non-small cell lung cancer development by regulating the miR-149-5p/ATG5 axis. Cell Cycle (2021) 20(2):166–78. doi: 10.1080/15384101.2020.1867780
74. Fan L, Li B, Li Z, Sun L. Identification of autophagy related circRNA-miRNA-mRNA-Subtypes network with radiotherapy responses and tumor immune microenvironment in non-small cell lung cancer. Front Genet (2021) 12:730003. doi: 10.3389/fgene.2021.730003
75. Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem (2017) 161(2):135–44. doi: 10.1093/jb/mvw087
76. Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, Fimia GM. TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ (2020) 27(3):887–902. doi: 10.1038/s41418-020-0495-2
77. Zhan W, Zhang S. TRIM proteins in lung cancer: Mechanisms, biomarkers and therapeutic targets. Life Sci (2021) 268:118985. doi: 10.1016/j.lfs.2020.118985
78. Lou M, Gao Z, Zhu T, Mao X, Wang Y, Yuan K, et al. TRIM59 as a novel molecular biomarker to predict the prognosis of patients with NSCLC. Oncol Lett (2020) 19(2):1400–8. doi: 10.3892/ol.2019.11199
79. Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, Saigi M, Pajares MJ, Garcia D, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res (2016) 22(13):3361–71. doi: 10.1158/1078-0432.CCR-15-2346
80. Tantai J, Pan X, Chen Y, Shen Y, Ji C. TRIM46 activates AKT/HK2 signaling by modifying PHLPP2 ubiquitylation to promote glycolysis and chemoresistance of lung cancer cells. Cell Death Dis (2022) 13(3):285. doi: 10.1038/s41419-022-04727-7
81. Zhang Y, Du H, Li Y, Yuan Y, Chen B, Sun S. Elevated TRIM23 expression predicts cisplatin resistance in lung adenocarcinoma. Cancer Sci (2020) 111(2):637–46. doi: 10.1111/cas.14226
82. Huang Y, Yang X, Lu Y, Zhao Y, Meng R, Zhang S, et al. UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance. Cell Death Differ (2021) 28(2):671–84. doi: 10.1038/s41418-020-00616-8
83. Wang H, Huang Q, Xia J, Cheng S, Pei D, Zhang X, et al. The E3 ligase MIB1 promotes proteasomal degradation of NRF2 and sensitizes lung cancer cells to ferroptosis. Mol Cancer Res (2022) 20(2):253–64. doi: 10.1158/1541-7786.MCR-21-0342
84. Chen S, Liu Y, Zhou H. Advances in the development ubiquitin-specific peptidase (USP) inhibitors. Int J Mol Sci (2021) 22(9). doi: 10.3390/ijms22094546
85. Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, et al. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol (2011) 18(11):1390–400. doi: 10.1016/j.chembiol.2011.08.014
86. Chen Z, Zheng L, Chen Y, Liu X, Kawakami M, Mustachio LM, et al. Loss of ubiquitin-specific peptidase 18 destabilizes 14-3-3ζ protein and represses lung cancer metastasis. Cancer Biol Ther (2022) 23(1):265–80. doi: 10.1080/15384047.2022.2054242
87. Tang Z, Jiang W, Mao M, Zhao J, Chen J, Cheng N. Deubiquitinase USP35 modulates ferroptosis in lung cancer via targeting ferroportin. Clin Transl Med (2021) 11(4):e390. doi: 10.1002/ctm2.390
88. Pan J, Qiao Y, Chen C, Zang H, Zhang X, Qi F, et al. USP5 facilitates non-small cell lung cancer progression through stabilization of PD-L1. Cell Death Dis (2021) 12(11):1051. doi: 10.1038/s41419-021-04356-6
89. Du T, Li H, Fan Y, Yuan L, Guo X, Zhu Q, et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat Commun (2019) 10(1):2914. doi: 10.1038/s41467-019-10824-7
90. Ouyang L, Yan B, Liu Y, Mao C, Wang M, Liu N, et al. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct Target Ther (2020) 5(1):78. doi: 10.1038/s41392-020-0181-3
91. Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr Relat Cancer. (2015) 22(1):T19–33. doi: 10.1530/ERC-14-0185
92. Spolverini A, Fuchs G, Bublik DR, Oren M. Let-7b and let-7c microRNAs promote histone H2B ubiquitylation and inhibit cell migration by targeting multiple components of the H2B deubiquitylation machinery. Oncogene (2017) 36(42):5819–28. doi: 10.1038/onc.2017.187
93. Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PloS One (2011) 6(6):e18691. doi: 10.1371/journal.pone.0018691
94. Kim MS, Kim SH, Yang SH, Kim MS. miR-4487 enhances gefitinib-mediated ubiquitination and autophagic degradation of EGFR in non-small cell lung cancer cells by targeting USP37. Cancer Res Treat (2022) 54(2):445–57. doi: 10.4143/crt.2021.622
95. Hu J, Zhang Z, Zhao L, Li L, Zuo W, Han L. High expression of RAD51 promotes DNA damage repair and survival in KRAS-mutant lung cancer cells. BMB Rep (2019) 52(2):151–6. doi: 10.5483/BMBRep.2019.52.2.213
96. Yu S, Li W, Liu X, Zhang H, Liu X, Zhang LW. TRIM36 enhances lung adenocarcinoma radiosensitivity and inhibits tumorigenesis through promoting RAD51 ubiquitination and antagonizing hsa-miR-376a-5p. Biochem Biophys Res Commun (2022) 628:1–10. doi: 10.1016/j.bbrc.2022.08.053
97. Mi J, Zou Y, Lin X, Lu J, Liu X, Zhao H, et al. Dysregulation of the miR-194-CUL4B negative feedback loop drives tumorigenesis in non-small-cell lung carcinoma. Mol Oncol (2017) 11(3):305–19. doi: 10.1002/1878-0261.12038
98. Pan X, Chen Y, Shen Y, Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis (2019) 10(6):429. doi: 10.1038/s41419-019-1660-8
99. Chae DK, Park J, Cho M, Ban E, Jang M, Yoo YS, et al. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β receptor I ubiquitination. Mol Oncol (2019) 13(12):2663–78. doi: 10.1002/1878-0261.12581
100. Liu X, Liu J, Zhang X, Tong Y, Gan X. MiR-520b promotes the progression of non-small cell lung cancer through activating hedgehog pathway. J Cell Mol Med (2019) 23(1):205–15. doi: 10.1111/jcmm.13909
101. Zhao J, Wang X, Mi Z, Jiang X, Sun L, Zheng B, et al. STAT3/miR-135b/NF-κB axis confers aggressiveness and unfavorable prognosis in non-small-cell lung cancer. Cell Death Dis (2021) 12(5):493. doi: 10.1038/s41419-021-03773-x
102. Park J, Cho M, Cho J, Kim EE, Song EJ. MicroRNA-101-3p suppresses cancer cell growth by inhibiting the USP47-induced deubiquitination of RPL11. Cancers (Basel) (2022) 14(4). doi: 10.3390/cancers14040964
103. Hua X, Chu H, Wang C, Shi X, Wang A, Zhang Z. Targeting USP22 with miR−30−5p to inhibit the hypoxia−induced expression of PD−L1 in lung adenocarcinoma cells. Oncol Rep (2021) 46(4). doi: 10.3892/or.2021.8166
104. Li W, Huang K, Wen F, Cui G, Guo H, He Z, et al. Intermittent hypoxia-induced downregulation of microRNA-320b promotes lung cancer tumorigenesis by increasing CDT1 via USP37. Mol Ther Nucleic Acids (2021) 24:528–41. doi: 10.1016/j.omtn.2020.12.023
105. Wang Y, Zhang S, Bao H, Mu S, Zhang B, Ma H, et al. MicroRNA-365 promotes lung carcinogenesis by downregulating the USP33/SLIT2/ROBO1 signalling pathway. Cancer Cell Int (2018) 18:64. doi: 10.1186/s12935-018-0563-6
106. Zhang P, Li L, Wang B, Ran X, Yang S, Luo Y, et al. miR-489-3p promotes malignant progression of non-small cell lung cancer through the inactivation of wnt/β-catenin signaling pathway via regulating USP48. Respir Res (2022) 23(1):93. doi: 10.1186/s12931-022-01988-w
107. Zhao H, Zheng C, Wang Y, Hou K, Yang X, Cheng Y, et al. miR-1323 promotes cell migration in lung adenocarcinoma by targeting cbl-b and is an early prognostic biomarker. Front Oncol (2020) 10:181. doi: 10.3389/fonc.2020.00181
108. Cui F, Luo P, Wu R, Meng J. miR-381 inhibits proliferation and invasion of non-Small-Cell cancer cells by targeting USP39. Dis Markers. (2022) 2022:2195393. doi: 10.1155/2022/2195393
109. Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res (2006) 66(2):605–12. doi: 10.1158/0008-5472.CAN-05-4005
110. Pritchard A, Tousif S, Wang Y, Hough K, Khan S, Strenkowski J, et al. Lung tumor cell-derived exosomes promote M2 macrophage polarization. Cells (2020) 9(5). doi: 10.3390/cells9051303
111. Huang X, Chen J, Cao W, Yang L, Chen Q, He J, et al. The many substrates and functions of NEDD4-1. Cell Death Dis (2019) 10(12):904. doi: 10.1038/s41419-019-2142-8
112. Chen J, Zhang K, Zhi Y, Wu Y, Chen B, Bai J, et al. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via hippo pathway. Clin Transl Med (2021) 11(9):e478. doi: 10.1002/ctm2.478
113. Xu LM, Yuan YJ, Yu H, Wang S, Wang P. LINC00665 knockdown confers sensitivity in irradiated non-small cell lung cancer cells through the miR-582-5p/UCHL3/AhR axis. J Transl Med (2022) 20(1):350. doi: 10.1186/s12967-022-03516-2
114. Chen J, Alduais Y, Zhang K, Zhu X, Chen B. CCAT1/FABP5 promotes tumour progression through mediating fatty acid metabolism and stabilizing PI3K/AKT/mTOR signalling in lung adenocarcinoma. J Cell Mol Med (2021) 25(19):9199–213. doi: 10.1111/jcmm.16815
115. Tian H, Lian R, Li Y, Liu C, Liang S, Li W, et al. AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent vimentin degradation. Nat Commun (2020) 11(1):5127. doi: 10.1038/s41467-020-18929-0
116. Su H, Fan G, Huang J, Qiu X. LncRNA HOXC-AS3 promotes non-small-cell lung cancer growth and metastasis through upregulation of YBX1. Cell Death Dis (2022) 13(4):307. doi: 10.1038/s41419-022-04723-x
117. Li B, Zhu L, Li L, Ma R. lncRNA OXCT1-AS1 promotes metastasis in non-Small-Cell lung cancer by stabilizing LEF1, In vitro and in vivo. BioMed Res Int (2021) 2021:4959381. doi: 10.1155/2021/4959381
118. Chen Z, Hu Z, Sui Q, Huang Y, Zhao M, Li M, et al. LncRNA FAM83A-AS1 facilitates tumor proliferation and the migration via the HIF-1α/ glycolysis axis in lung adenocarcinoma. Int J Biol Sci (2022) 18(2):522–35. doi: 10.7150/ijbs.67556
119. Shen J, Ma J, Li J, Wang X, Wang Y, Ma J. A long non-coding RNA LNBC3 facilitates non-small cell lung cancer progression by stabilizing BCL6. J Clin Lab Anal (2020) 34(4):e23122. doi: 10.1002/jcla.23122
120. Athie A, Marchese FP, González J, Lozano T, Raimondi I, Juvvuna PK, et al. Analysis of copy number alterations reveals the lncRNA ALAL-1 as a regulator of lung cancer immune evasion. J Cell Biol (2020) 219(9). doi: 10.1083/jcb.201908078
121. Yang R, Liu N, Chen L, Jiang Y, Shi Y, Mao C, et al. GIAT4RA functions as a tumor suppressor in non-small cell lung cancer by counteracting Uchl3-mediated deubiquitination of LSH. Oncogene (2019) 38(46):7133–45. doi: 10.1038/s41388-019-0909-0
122. Pei Y, Zhou B, Liu X. The long non-coding RNA rhabdomyosarcoma 2-associated transcript exerts anti-tumor effects on lung adenocarcinoma via ubiquitination of SOX9. Ann Transl Med (2022) 10(1):10. doi: 10.21037/atm-21-6052
123. Liu X, Yin Z, Xu L, Liu H, Jiang L, Liu S, et al. Upregulation of LINC01426 promotes the progression and stemness in lung adenocarcinoma by enhancing the level of SHH protein to activate the hedgehog pathway. Cell Death Dis (2021) 12(2):173. doi: 10.1038/s41419-021-03435-y
124. Li F, Zhang Q, Gong Y, Yu J. The lncKLF6/KLF6 feedback loop regulates the growth of non-small cell lung cancer. Am J Cancer Res (2018) 8(8):1427–39.
125. Xu P, Xiao H, Yang Q, Hu R, Jiang L, Bi R, et al. The USP21/YY1/SNHG16 axis contributes to tumor proliferation, migration, and invasion of non-small-cell lung cancer. Exp Mol Med (2020) 52(1):41–55. doi: 10.1038/s12276-019-0356-6
126. He Y, Jiang X, Duan L, Xiong Q, Yuan Y, Liu P, et al. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating wnt signaling pathway. Mol Cancer. (2021) 20(1):156. doi: 10.1186/s12943-021-01469-6
127. Huang Y, Xia L, Tan X, Zhang J, Zeng W, Tan B, et al. Molecular mechanism of lncRNA SNHG12 in immune escape of non-small cell lung cancer through the HuR/PD-L1/USP8 axis. Cell Mol Biol Lett (2022) 27(1):43. doi: 10.1186/s11658-022-00343-7
128. Wu X, Ye W, Gong Y. The role of RNA methyltransferase METTL3 in normal and malignant hematopoiesis. Front Oncol (2022) 12:873903. doi: 10.3389/fonc.2022.873903
129. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. (2021) 20(1):105. doi: 10.1186/s12943-021-01398-4
130. Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression. Cell Mol Life Sci (2013) 70(15):2657–75. doi: 10.1007/s00018-012-1186-z
Keywords: miRNA, lncRNA, circRNA, ubiquitination, deubiquitination, NSCLC
Citation: Sun Y, He P, Li L and Ding X (2023) The significance of the crosstalk between ubiquitination or deubiquitination and ncRNAs in non-small cell lung cancer. Front. Oncol. 12:969032. doi: 10.3389/fonc.2022.969032
Received: 14 June 2022; Accepted: 28 December 2022;
Published: 16 January 2023.
Edited by:
Mohammad Taheri, University Hospital Jena, GermanyCopyright © 2023 Sun, He, Li and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping He, aGVwQHNqLWhvc3BpdGFsLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.