
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 21 October 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.968884
This article is part of the Research TopicMolecular Drivers in Bone and Soft Tissue SarcomasView all 8 articles
 Marcella Martinelli1*
Marcella Martinelli1* Caterina Mancarella2
Caterina Mancarella2 Luca Scapoli1
Luca Scapoli1 Annalisa Palmieri1
Annalisa Palmieri1 Paola De Sanctis1
Paola De Sanctis1 Cristina Ferrari2
Cristina Ferrari2 Michela Pasello2
Michela Pasello2 Cinzia Zucchini1†
Cinzia Zucchini1† Katia Scotlandi2*†
Katia Scotlandi2*†Ewing sarcoma (EWS), the second most common malignant bone tumor in children and adolescents, occurs abruptly without clear evidence of tumor history or progression. Previous association studies have identified some inherited variants associated with the risk of developing EWS but a common picture of the germline susceptibility to this tumor remains largely unclear. Here, we examine the association between thirty single nucleotide polymorphisms (SNPs) of the IGF2BP3, a gene that codes for an oncofetal RNA-binding protein demonstrated to be important for EWS patient’s risk stratification, and five SNPs of SENCR, a long non-coding RNA shown to regulate IGF2BP3. An association between polymorphisms and EWS susceptibility was observed for three IGF2BP3 SNPs - rs112316332, rs13242065, rs12700421 - and for four SENCR SNPs - rs10893909, rs11221437, rs12420823, rs4526784 -. In addition, IGF2BP3 rs34033684 and SENCR rs10893909 variants increased the risk for female respect to male subgroup when carried together, while IGF2BP3 rs13242065 or rs76983703 variants reduced the probability of a disease later onset (> 14 years). Moreover, the absence of IGF2BP3 rs10488282 variant and the presence of rs199653 or rs35875486 variant were significantly associated with a worse survival in EWS patients with localized disease at diagnosis. Overall, our data provide the first evidence linking genetic variants of IGF2BP3 and its modulator SENCR to the risk of EWS development and to disease progression, thus supporting the concept that heritable factors can influence susceptibility to EWS and may help to predict patient prognosis.
Ewing sarcoma (EWS), the second most common primary tumor of bone in the pediatric population, is a mesenchymal very aggressive cancer with high tendency to form distal metastasis and still unmet clinical needs (1). From a genetic point of view, EWS is characterized by a very low mutational burden (2–4) while its genetic landscape is thought to be driven by the aberrant transcript that derives from the fusion of EWSR1 gene with a member of the ETS family genes, in most of the cases represented by EWSR1-FLI chimera (5). EWS is not considered a heritable cancer but disparity in EWS epidemiological distribution, with higher incidence in European than in Asian and African population (6) together with some reports indicating EWS in siblings or cousins (7, 8) and reports of family aggregation of different malignant tumors between EWS patients and their relatives (9, 10) suggest that genetic susceptibility factors may exist for this tumor. Indeed, in the last decade several evidence of correlation between polymorphic variants and EWS risk has been reported (11–18). Through genome-wide association studies (GWAS), multiple genetic susceptibility loci (1p36.22, 6p25.1, 10q21, 15q15, 20p11.22 and 20p11.23) associated with EWS risk have been identified (16, 17). Most of these loci reside near GGAA repeat sequences and may condition the binding of the aberrant transcriptional factor EWS-FLI1. A noteworthy example is the locus 10q21, in which rs79965208 variant increases the number of consecutive GGAA motifs and the consequent EWS-FLI1-dependent enhancer activity, leading to EGR2 overexpression and favoring EWS susceptibility and aggressiveness (18). Deeply investigation of genes already known to be involved in the pathogenesis and progression of EWS has been performed by several groups as an alternative option to identify predisposing factors for this disease. In particular, analysis of single nucleotide polymorphisms (SNPs) in the EWSR1 gene revealed that the rs4820804 variant in homozygosis may increase the chance of chromosome breakage and occurrence of chromosomal translocation (11). The relationship between polymorphic variants in genes implicated in EWS pathogenesis and progression and their role in EWS susceptibility has been also studied for NROB1 and CAV1, two EWS-FLI1 target genes (19), and for CD99, another hallmark of EWS critically associated with EWS cell differentiation, migration and metastasis (20). Specifically, CD99 rs311059-T allele was found to be associated with early EWS onset, while rs312257-T variant was related with a reduced risk of relapse (12). In this study, we focused on the analysis of genetic polymorphisms of the Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), a gene that encodes for an oncofetal RNA-binding protein that was found to be undetectable in most adult tissues but strongly expressed in embryos and in diverse tumor types (21). In EWS, high levels of IGF2BP3 were found to support cell migration and metastasis formation besides correlating with disease progression and poor patient’s prognosis (22–24). Thirty SNPs mapping on IGF2BP3 were genotyped in a cohort of 73 EWS Italian patients to evaluate the genetic influence of IGF2BP3 polymorphisms on EWS susceptibility and to establish whether a potential link between IGF2BP3 somatic variants and EWS progression exists. In addition, we analyzed five genetic polymorphisms of SENCR, a long non-coding RNA transcribed antisense from the 5’ end of the FLI1 gene, which was shown to regulate IGF2BP3 (25).
A cohort of 73 unrelated Italian patients with localized (58 cases) or disseminated (15 cases) EWS treated at the IRCCS Istituto Ortopedico Rizzoli (Bologna, Italy) was considered. Patients underwent local treatments (surgery; surgery plus radiotherapy; radiotherapy only, when the surgeon considered the lesion inoperable or due to patient refusal) and neo-adjuvant chemotherapy according to protocols that were previously reported in detail (26, 27). For radiotherapy administered in combination with surgery, the doses ranged between 45 Gy and 54 Gy, depending on the individual factors (age, site, size, surgical margins, response to chemotherapy); for radiotherapy administered alone, the doses ranged between 54-60 Gy. The timing of radiation therapy ranged between 4 to 6 weeks (26, 27). Clinical-pathological data are shown in Table 1. Patients with localized EWS were followed-up for 120 months and clinical information updated. Ethical committee approval was obtained from the Comitato Etico di Area Vasta Emilia Centro (Codice CE AVEC 505/2019/Sper/IOR) and written informed consent was obtained. The study was conducted in accordance with the Declaration of Helsinki ethical guidelines. Control sample consisted of three populations among the 1000 Genome Project, i.e Toscani in Italia (TSI), Utah residents with Northern and Western European ancestry (CEU), and Iberian population in Spain (IBS). Genotypes for each polymorphism were obtained from the Ensembl.org genome browser (GRCh37/hg19).
Quality and concentration of DNA obtained from peripheral blood leukocytes or from muscle tissue using standard DNAzol procedure (Thermo Fisher Scientific, Foster City, CA, USA) were evaluated by Nanodrop (Thermo Fisher Scientific). Aliquots of 12 ng/μl DNA from each patient were plated for being processed by the Sequenom MALDI-TOF mass spectrometer MassArray system (as a service at Applied Biomedical Research Center, S. Orsola-Malpighi Polyclinic, Bologna, Italy). The SNPs were selected using the SNPclip tool [https://ldlink.nci.nih.gov/] among the Genome1000 Phase1 Vars (GRCh37/hg19). Caucasian, Italian and Iberian populations (CEU+TSI+IBS dataset) were used to explore the haplotype complexity of each locus considered, applying the thresholds R2 0.8 and MAF 0.07. To a selection of 30 SNPs distributed along the entire IGF2BP3 sequence with the minimal redundancy level (28) were added 5 SNPs of the SENCR gene. Assay design was performed using specific Sequenom software package (Sequenom, San Diego, California, USA). Primers were synthesized at Metabion (Martinsried, Germany) (sequences available upon request). Allele peaks were analyzed with the Sequenom Typer Analysis software.
The distribution of genotypes in patient and control groups was tested for deviations from the Hardy–Weinberg equilibrium using Pearson’s χ2 test. The PLINK software was used to test for allelic association within different sample subsets, defined by patient sex or age at disease onset within an alternate phenotype file (29). Odds ratios were calculated to estimate the level of association of the rare allele carriers, i.e heterozygotes versus non-carriers, as well as homozygotes versus non-carriers. A permutation procedure was used to generate empirical significance levels. Such procedure relaxed assumptions about normality of continuous phenotypes and Hardy-Weinberg equilibrium and faced with rare alleles and small sample sizes. Haplotype association analysis using a likelihood ratio approach was performed with the aid of UNPHASED software v3.1.7 for loci showing nominal evidence of association in allelic association analysis. The full model test was performed to obtain a global P for association. In addition, for the combinations of SNPs that showed a global P < 0.05 in the overall association test, a specific analysis was carried out to evaluate a difference in risk between one haplotype versus all the others pooled together.
Association between IGF2BP3 or SENCR variants and overall survival (OVS) was also estimated. Survival curves for OVS were obtained using the Kaplan–Meier method, while the log-rank test was used to calculate univariate statistical significance. OVS was defined as the time from diagnosis to cancer-related death. Patients who were lost to follow-up were censored at the last contact date. The genetic variants significantly associated with OVS in univariate analysis were entered into a Cox proportional hazards model. Values of 95% confidence intervals (CIs) of hazard ratios (HRs) were provided (30). Analyses were performed with SPSS software, version 22.0. P value ≤ 0.05 were considered significant.
Genotyping was carried out on a cohort of 73 EWS patients whose clinical-pathological features are summarized in Table 1. Among the 35 genotyped polymorphisms, three SNPs (rs17796758, rs62468200 and rs70954368) mapping on IGF2BP3 and one SNP (rs7930515) mapping on SENCR were excluded from any statistical analyses because of a low call rate.
Pairwise association analysis was performed to test the impact of the remaining variants and results are shown in Table 2. In details, the IGF2BP3 rs12700421 variant was found to be significantly less frequent in EWS patients than in the control group. Specifically, heterozygote genotype led to a reduced risk of developing EWS [ORhet = 0.47 (95% CI 0.24-0.91)]. A similar trend was observed for the IGF2BP3 rs13242065 variant [ORhet = 0.29 (95% CI 0.09-0.98)]. Instead, the adjacent rs112316332 variant showed association with increased risk of disease [ORhet = 1.94 (95% CI 1.08-3.49)]. The IGF2BP3 rs146075134 variant also showed a significant association level but it was excluded from further analyses because of a deviation from Hardy-Weinberg equilibrium observed in the control-group (P value < 0.01). The analysis of SENCR polymorphisms evidenced a protective effect of the variant allele at rs11221437, rs12420823, and rs4526784 [ORhet = 0.48 (95% CI 0.27-0.86); ORhet = 0.52 (95% CI 0.3-0.92); ORhet = 0.5 (95% CI 0.28-0.87) respectively]. An opposite role was observed for the SENCR rs10893909 (P = 0.0092), as the variant allele of this marker increased the risk of EWS more than three times when carried in homozygosis [ORhom = 3.33 (95% CI 1.35-8.19)].
To verify if the level of association varies in relation to patient sex or age at disease onset, stratified data were considered (Tables 3, 4). Females were found to be more prone than males to incur in EWS when carrying the IGF2BP3 rs34033684 variant [OR = 3.38 (95% CI 1.44-7.94)]. An increased risk for females [OR = 2.48 (95% CI 1.28-4.82)] was also observed for the SENCR rs10893909 variant allele (Table 3). Of note, when females carry the variant at both rs34033684 and rs10893909, their risk to develop EWS is further increased [OR = 4.43 (95% CI 1.11-19.01)]. In patients with a later onset of EWS (> 14 years) a significantly lower frequency of the rare allele was observed both for IGF2BP3 rs13242065 and rs76983703 [OR = 0.19 (95% CI 0.04-0.78) and OR = 0.29 (95% CI 0.11-0.74), respectively] (Table 4).

Table 3 Association analysis with data stratified by patient sex; only most significant data are reported.
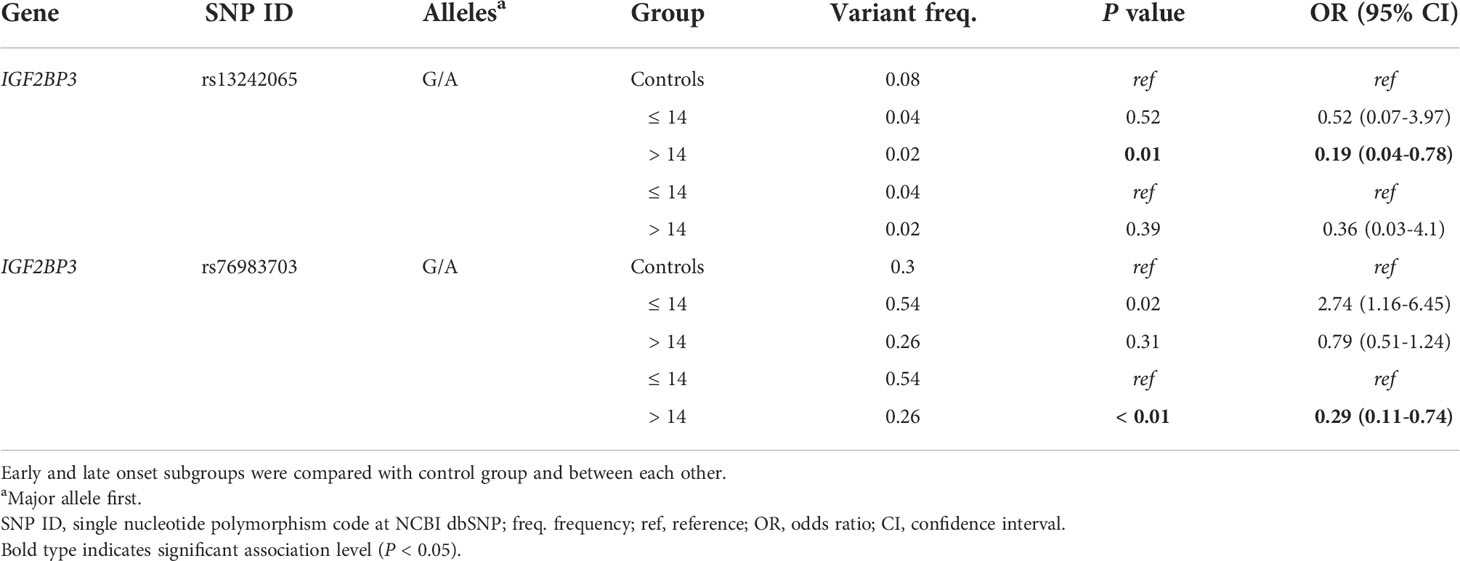
Table 4 Association analysis with data stratified by age at EWS diagnosis; only most significant data are reported.
The multipoint association analysis confirmed the role of the IGF2BP3 genetic region that includes rs112316332 and rs13242065 in influencing the risk of EWS development (Supplementary Table 1). Significant P value levels were also obtained when the IGF2BP3 rs58201821, rs12533936, rs34033684, and rs6953027 SNPs, which map close to the rs112316332 and rs13242065 SNPs, were considered in the haplotype analysis. Both over- or under-represented haplotypes were found in EWS patients. Notably, haplotypes including four, five, or six SNPs had a higher level of association compared to the effect produced by single allele in pairwise analysis.
To search for the possibility that IGF2BP3 or SENCR SNPs impact on the probability for patients to have a different outcome, we stratified patients according to the presence (censored as POS) or absence (censored as NEG) of the variant allele at the 30 previously considered SNPs (26 in IGF2BP3 and 4 in SENCR, respectively). In order to limit possible drawbacks related to the presence of metastasis at diagnosis, an event known to be associated to a worse prognosis (31), we limited our analysis to 58 patients with primary, localized tumor homogeneously treated in a single Institution (Table 1, Dataset II). Kaplan-Meier curves and log-rank test performed on IGF2BP3 polymorphisms showed that the absence of the variant allele at rs10488282 and the presence at rs199653 or rs35875486 were significantly associated with a worse OVS at 120 months (Figure 1). Multivariate analysis was performed for the three variables identified by univariate analysis and confirmed the prognostic value of the absence of the variant allele at IGF2BP3 rs10488282 (Table 5) as an independent factor of worse outcome.
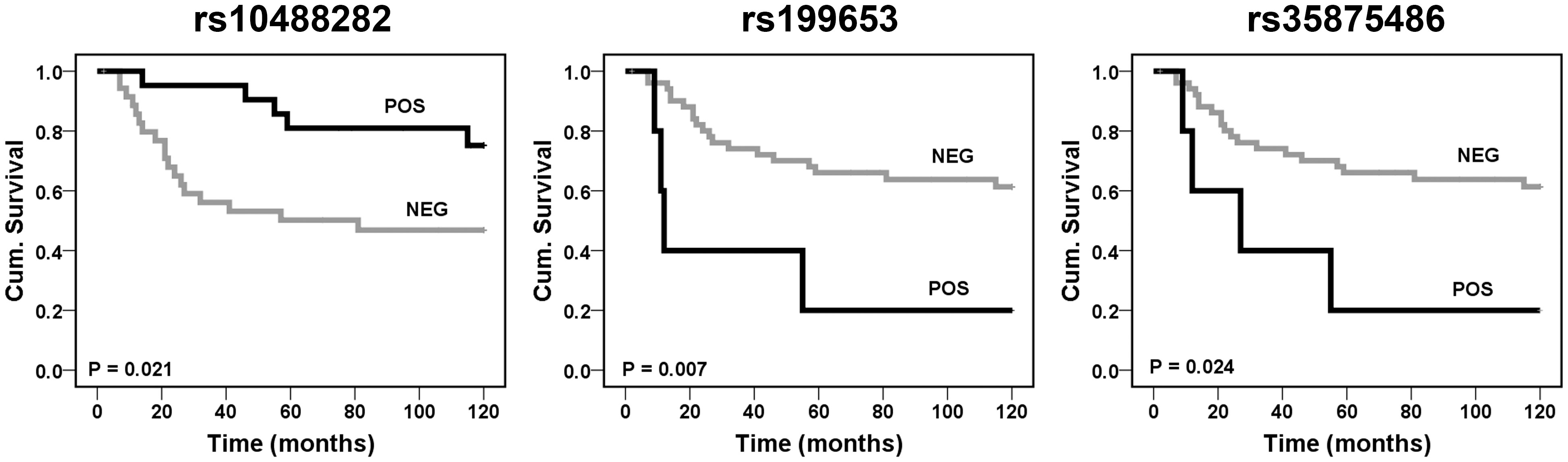
Figure 1 Prognostic impact of the IGF2BP3 rs10488282, rs199653, and rs35875486 variants according to Kaplan-Meier curves and log-rank test. EWS patients were classified for the presence (POS) or absence (NEG) of the variant. Overall survival (OVS) was considered.
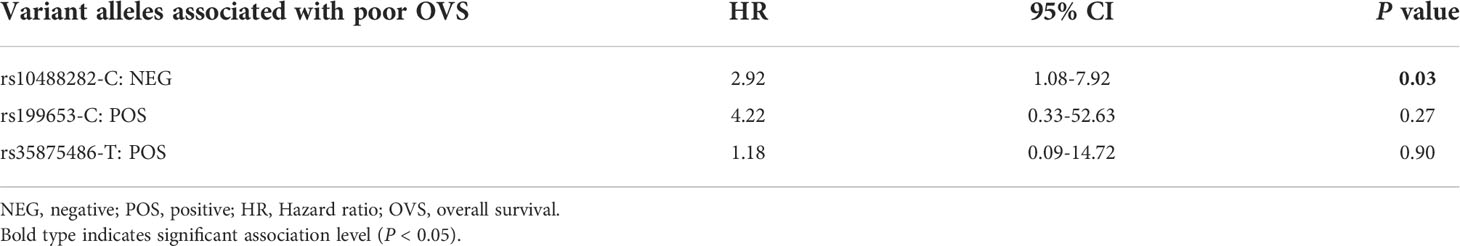
Table 5 Cox proportional-hazards regression multivariate analysis for IGF2BP3 SNPs associated with OVS after univariate analysis in the dataset of 58 EWS patients.
Susceptibility to the development of sporadic tumors is based on a complex interplay that includes various genetic and environmental factors whose degree of influence depends on the type of cancer. In pediatric cancer etiology, the genetic contribution is predominant over the environmental one. In EWS, the peak of incidence in the second decade of life draws attention to genetic predisposition rather than to environmental repercussion for the disease onset. The identification of EWS predisposing genetic factors can lead to clinical benefits for patients, highlighting new oncogenic pathways that may be useful either for the molecular diagnosis or for better therapy. Besides wide-scale approaches, an alternative option to identify germinal predisposing factors is to deeply investigate genes already known to be involved in the biology of cancer disease.
Based on this approach, our study considers IGF2BP3 and SENCR as candidate genes for searching susceptibility genetic factors to EWS. To the best of our knowledge, this is the first study linking germline genetic variants of IGF2BP3 and of its putative modulator SENCR to the risk of EWS development, further supporting the concept that heritable factors can influence susceptibility to EWS (11–18).
Nominal level of significance in pairwise association analysis was obtained with three IGF2BP3 SNPs (rs12700421, rs13242065 and rs112316332). The polymorphisms rs13242065 and rs112316332 were located in a gene region where multipoint association analysis provided evidence of association with different haplotypes. This region, bounded by rs58201821 and rs6953027, spans 29 Kbp across 5’-UTR and the second intron of the gene. According to ENCODE Registry of candidate cis-Regulatory Elements (cCREs) hosted in UCSC Genome Browser on Human GRCh38/hg38 Assembly, such region includes two cCREs showing a promoter-like signature proximal to a transcription start site (EH38E2540316 and EH38E2540292), and several predicted proximal and distal enhancers, suggesting for a potential regulatory function. The relevance of the region spanning across 5’-UTR and the second intron of IGF2BP3 for EWS predisposition was proved also when patients were stratified for sex (rs34033684) or age (rs13242065). Our finding corroborates the hypothesis that susceptibility factors act differently in females than in males and may influence the age of EWS occurrence. In addition to IGF2BP3 polymorphisms, our study also highlighted a potential value for SENCR genetic variants in EWS predisposition. All the four polymorphisms evaluated for SENCR were found to be significantly associated with a different risk to develop EWS and should be considered as inherited susceptibility factors of the disease. Although genetic variants in lncRNAs have been implicated as potential biomarkers in prediction of complex diseases (32), the genetic association between lncRNAs and EWS has yet to be explored. While the rs4526784 maps in the second exon of SENCR gene (http://genome.ucsc.edu/index.html) and may act by affecting the lncRNA sequence, the rs10893909, rs11221437 and rs12420823 map on the first intron of the gene and very likely influence EWS susceptibility in an indirect manner. For example, the rs10893909 and rs11221437 are located in regulatory regions annotated as proximal enhancer-like signature in ENCODE (EH38E1581272 and EH38E1581271, respectively) while according to the JASPAR database of transcription factor binding profiles (33) both the two variants disrupt transcription factor (TF) binding motifs. In particular, the rs10893909 variant was reported to disrupt a transcription factor binding motif with predicted affinity for several TFs, including NRF1 and KLF15 that were shown to cooperate with EWS-FLI1 (34). Likewise, the rs11221437 modifies a transcription factor binding motif with predicted affinity for CTCF, a TF involved in many cellular processes including the regulation of the transcriptional state-dependent 3D organization of the chromatin (35).
In addition, we demonstrate for the first time that three allelic variants of IGF2BP3 may affect EWS patient’s outcome. Particularly the absence of the C allele at rs10488282 SNP was confirmed as an independent factor of prognosis at multivariate analysis, being associated with a poor survival for patients with localized EWS. Although mechanistic studies are needed to explain this observation, our findings support the hypothesis that genetic variants in the IGF2BP3 gene may significantly affect the progression of EWS. Considering the limits related to the low number of patients here considered and the rarity of the tumor, we offer this evidence to the scientific community for more extensive validation studies. Comprehensive genomic and epigenomic profiling has revealed that epigenetic factors likely play a critical role in EWS initiation and progression (5). RNA-binding proteins, along with microRNAs and lncRNAs, which dictate the entire RNA life cycle from alternative splicing to nuclear export, transcript storage, stabilization, subcellular localization and degradation [for a review, please consider (36)], may thus represent major regulators of tumor onset and progression. Over the past few years, studies have increasingly documented the contribution of IGF2BP3 to fundamental processes in cancer biology, such as cell growth, migration, and the response to drugs. Indeed, many tumor types upregulate IGF2BP3 compared to normal tissues but very limited information regarding the molecular regulatory mechanisms responsible for human IGF2BP3 expression is available [for a review see (21)]. Here we focused on SENCR, a gene coding for a lncRNA, recently found to play a critical role in the proliferation and migration of vascular smooth muscle cells (37), which may influence gene expression through multiple mechanisms, including interaction with RNA-binding proteins. The role and mechanism of action of the lncSENCR in malignant tumors remains largely unexplored. Our study supports deeper investigation on this lncRNA as a factor influencing cancer susceptibility.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical committee approval was obtained from the Comitato Etico di Area Vasta Emilia Centro (Codice CE AVEC 505/2019/Sper/IOR). The study was conducted in accordance with the Declaration of Helsinki ethical guidelines, and patient informed consent for research use of biobanking material was obtained.
MM, CZ, and KS: conception and design. MM, CM, LS, AP, PDS, CF, and MP: acquisition and analysis of data. MM, CZ, and KS: drafting the manuscript. MM and KS: study supervision. All authors contributed to the article and approved the submitted version.
The research leading to these results has received funding from AIRC under IG 2019 — ID. 22805 project — P.I. Scotlandi Katia, and from Ricerca Fondamentale Orientata (RFO, University of Bologna) to Zucchini Cinzia and Martinelli Marcella. The materials presented and views expressed herein are the responsibility of the authors only. The sponsor takes no responsibility for any use of the information presented herein. None of the funders played a role in study design; in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the paper for publication.
We thank the patients and their family for supporting this study. We thank Dr. Vilma Mantovani and Dr. Carlotta Cristalli for their technical support during experimental procedure of genotyping at CRBA (Applied Biomedical Research Center, S. Orsola-Malpighi Polyclinic, Bologna, Italy). We thank Dr. Marika Sciandra (Laboratory of Experimental Oncology, IRCCS Istituto Ortopedico Rizzoli, Bologna, Italy) for her technical support. The authors are grateful to Muscolo Skeletal Tumor Biobank-Biobanca dei Tumori Muscoloscheletrici (Biotum)—member of the CRB-IOR—which provided us the biological samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.968884/full#supplementary-material
1. Scotlandi K, Hattinger CM, Pellegrini E, Gambarotti M, Serra M. Genomics and therapeutic vulnerabilities of primary bone tumors. Cells (2020) 9(4), 968. doi: 10.3390/cells9040968
2. Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discovery (2014) 4(11):1326–41. doi: 10.1158/2159-8290.CD-13-1037
3. Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discovery (2014) 4(11):1342–53. doi: 10.1158/2159-8290.CD-14-0622
4. Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PloS Genet (2014) 10(7):e1004475. doi: 10.1371/journal.pgen.1004475
5. Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, et al. Ewing Sarcoma. Nat Rev Dis Primers. (2018) 4(1):5. doi: 10.1038/s41572-018-0003-x
6. Worch J, Cyrus J, Goldsby R, Matthay KK, Neuhaus J, DuBois SG. Racial differences in the incidence of mesenchymal tumors associated with EWSR1 translocation. Cancer Epidemiol Biomarkers Prev (2011) 20(3):449–53. doi: 10.1158/1055-9965.EPI-10-1170
7. Hutter RV, Francis KC, Foote FW Jr. Ewing's sarcoma in siblings: Report of the second known occurrence. Am J Surg (1964) 107:598–603. doi: 10.1016/0002-9610(64)90328-9
8. Joyce MJ, Harmon DC, Mankin HJ, Suit HD, Schiller AL, Truman JT. Ewing's sarcoma in female siblings. A clinical report and review of the literature. Cancer (1984) 53(9):1959–62. doi: 10.1002/1097-0142(19840501)53:9<1959::aid-cncr2820530926>3.0.co;2-9
9. Mc CL, Dockerty MB, Ghormley RK. Ewing's sarcoma. Cancer (1952) 5(1):85–99. doi: 10.1002/1097-0142(195201)5:1<85::aid-cncr2820050111>3.0.co;2-t
10. Ji J, Hemminki K. Familial risk for histology-specific bone cancers: An updated study in Sweden. Eur J Cancer. (2006) 42(14):2343–9. doi: 10.1016/j.ejca.2005.11.043
11. Silva DS, Sawitzki FR, De Toni EC, Graebin P, Picanco JB, Abujamra AL, et al. Ewing's sarcoma: Analysis of single nucleotide polymorphism in the EWS gene. Gene (2012) 509(2):263–6. doi: 10.1016/j.gene.2012.08.012
12. Martinelli M, Parra A, Scapoli L, De Sanctis P, Chiadini V, Hattinger C, et al. CD99 polymorphisms significantly influence the probability to develop Ewing sarcoma in earlier age and patient disease progression. Oncotarget (2016) 7(47):77958–67. doi: 10.18632/oncotarget.12862
13. Thurow HS, Hartwig FP, Alho CS, Silva DS, Roesler R, Abujamra AL, et al. Ewing Sarcoma: influence of TP53 Arg72Pro and MDM2 T309G SNPs. Mol Biol Rep (2013) 40(8):4929–34. doi: 10.1007/s11033-013-2593-4
14. Wang J, Zhou Y, Feng D, Yang H, Li F, Cao Q, et al. CD86 +1057G/A polymorphism and susceptibility to ewing's sarcoma: A case-control study. DNA Cell Biol (2012) 31(4):537–40. doi: 10.1089/dna.2011.1370
15. Zhang C, Hou WH, Ding XX, Wang X, Zhao H, Han XW, et al. Association of cytotoxic T-lymphocyte antigen-4 polymorphisms with malignant bone tumor risk: A meta-analysis. Asian Pac J Cancer Prev (2016) 17(8):3785–91.
16. Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet (2012) 44(3):323–7. doi: 10.1038/ng.1085
17. Machiela MJ, Grunewald TGP, Surdez D, Reynaud S, Mirabeau O, Karlins E, et al. Genome-wide association study identifies multiple new loci associated with Ewing sarcoma susceptibility. Nat Commun (2018) 9(1):3184. doi: 10.1038/s41467-018-05537-2
18. Grunewald TG, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud MM, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet (2015) 47(9):1073–8. doi: 10.1038/ng.3363
19. Monument MJ, Johnson KM, McIlvaine E, Abegglen L, Watkins WS, Jorde LB, et al. Clinical and biochemical function of polymorphic NR0B1 GGAA-microsatellites in Ewing sarcoma: A report from the children's oncology group. PloS One (2014) 9(8):e104378. doi: 10.1371/journal.pone.0104378
20. Manara MC, Pasello M, Scotlandi K. CD99: A cell surface protein with an oncojanus role in tumors. Genes (Basel). (2018) 9(3), 159. doi: 10.3390/genes9030159
21. Mancarella C, Scotlandi K. IGF2BP3 from physiology to cancer: Novel discoveries, unsolved issues, and future perspectives. Front Cell Dev Biol (2019) 7:363. doi: 10.3389/fcell.2019.00363
22. Mancarella C, Pasello M, Manara MC, Toracchio L, Sciandra EF, Picci P, et al. Insulin-like growth factor 2 mRNA-binding protein 3 influences sensitivity to anti-IGF system agents through the translational regulation of IGF1R. Front Endocrinol (Lausanne). (2018) 9:178. doi: 10.3389/fendo.2018.00178
23. Mancarella C, Pasello M, Ventura S, Grilli A, Calzolari L, Toracchio L, et al. Insulin-like growth factor 2 mRNA-binding protein 3 is a novel post-transcriptional regulator of Ewing sarcoma malignancy. Clin Cancer Res (2018) 24(15):3704–16. doi: 10.1158/1078-0432.CCR-17-2602
24. Mancarella C, Caldoni G, Ribolsi I, Parra A, Manara MC, Mercurio AM, et al. Insulin-like growth factor 2 mRNA-binding protein 3 modulates aggressiveness of Ewing sarcoma by regulating the CD164-CXCR4 axis. Front Oncol (2020) 10:994. doi: 10.3389/fonc.2020.00994
25. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol (2014) 34(6):1249–59. doi: 10.1161/ATVBAHA.114.303240
26. Ferrari S, Sundby Hall K, Luksch R, Tienghi A, Wiebe T, Fagioli F, et al. Nonmetastatic Ewing family tumors: high-dose chemotherapy with stem cell rescue in poor responder patients. results of the Italian sarcoma Group/Scandinavian sarcoma group III protocol. Ann Oncol (2011) 22(5):1221–7. doi: 10.1093/annonc/mdq573
27. Bernstein M, Kovar H, Paulussen M, Randall RL, Schuck A, Teot LA, et al. Ewing's sarcoma family of tumors: Current management. Oncologist (2006) 11(5):503–19. doi: 10.1634/theoncologist.11-5-503
28. de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet (2005) 37(11):1217–23. doi: 10.1038/ng1669
29. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet (2007) 81(3):559–75. doi: 10.1086/519795
30. Bradburn MJ, Clark TG, Love SB, Altman DG. Survival analysis part III: multivariate data analysis – choosing a model and assessing its adequacy and fit. Br J Cancer. (2003) 89(4):605–11. doi: 10.1038/sj.bjc.6601120
31. Stahl M, Ranft A, Paulussen M, Bolling T, Vieth V, Bielack S, et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. (2011) 57(4):549–53. doi: 10.1002/pbc.23040
32. Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. (2013) 108(12):2419–25. doi: 10.1038/bjc.2013.233
33. Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res (2022) 50(D1):D165–D73. doi: 10.1093/nar/gkab1113
34. Kovar H. Downstream EWS/FLI1 - upstream ewing's sarcoma. Genome Med (2010) 2(1):8. doi: 10.1186/gm129
35. Xiang JF, Corces VG. Regulation of 3D chromatin organization by CTCF. Curr Opin Genet Dev (2021) 67:33–40. doi: 10.1016/j.gde.2020.10.005
36. Coppin L, Leclerc J, Vincent A, Porchet N, Pigny P. Messenger RNA life-cycle in cancer cells: Emerging role of conventional and non-conventional RNA-binding proteins? Int J Mol Sci (2018) 19(3), 650. doi: 10.3390/ijms19030650
Keywords: Ewing sarcoma, polymorphisms, IGF2BP3, SENCR, cancer predisposition
Citation: Martinelli M, Mancarella C, Scapoli L, Palmieri A, De Sanctis P, Ferrari C, Pasello M, Zucchini C and Scotlandi K (2022) Polymorphic variants of IGF2BP3 and SENCR have an impact on predisposition and/or progression of Ewing sarcoma. Front. Oncol. 12:968884. doi: 10.3389/fonc.2022.968884
Received: 14 June 2022; Accepted: 26 September 2022;
Published: 21 October 2022.
Edited by:
Luisa Lanfrancone, European Institute of Oncology (IEO), ItalyCopyright © 2022 Martinelli, Mancarella, Scapoli, Palmieri, De Sanctis, Ferrari, Pasello, Zucchini and Scotlandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcella Martinelli, bWFyY2VsbGEubWFydGluZWxsaUB1bmliby5pdA==; Katia Scotlandi, a2F0aWEuc2NvdGxhbmRpQGlvci5pdA==
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.