- 1Hematopathology, Oman Medical Specialty Board, Muscat, Oman
- 2Hematology Department, Sultan Qaboos University Hospital, Muscat, Oman
- 3Internal Medicine, Oman Medical Specialty Board, Muscat, Oman
- 4College of Medicine and Health Sciences, Sultan Qaboos University, Muscat, Oman
This is a systematic review and meta-analysis evaluating the prognostic significance of epigenetic mutations on the overall survival (OS) in Acute Myeloid Leukemia (AML). We searched for studies evaluating epigenetic mutations in AML (up to November 2018) in PubMed, Trip database and Cochrane library. Hazard ratio (HR) of outcomes were extracted, and random-effects model was used to pool the results. A total of 10,002 citations were retrieved from the search strategy; 42 articles were identified for the meta-analysis (ASXL1 = 7, TET2 = 8, DNMT3A = 12, IDH =15), with fair to good-quality studies. The pooled HR was 1.88 (95% CI: 1.49−2.36) for ASXL1 mutation, 1.39 (95% CI: 1.18−1.63) for TET2 mutation, 1.35 (95% CI 1.16-1.56) for DNMT3a and 1.54 (95% CI: 1.15-2.06) for IDH mutation. However, there was a substantial heterogeneity in the DNMT3a and IDH studies. In conclusion epigenetic mutations in ASXL1, TET2, DNMT3a and IDH adversely impact OS in patients with AML albeit with considerable heterogeneity and possibly publication bias. Further studies are required to address these limitations.
Introduction
Acute myeloid leukemia (AML) is characterized by the uncontrolled proliferation of myeloid blast cells in the bone marrow and peripheral blood (1). Breakthroughs in the past have contributed to our understanding of the genetic failures and the changing biology in the myeloid cells that underlie the initiation and progression of the disease (2). With the application of global DNA sequencing, several recurrent gene mutations have been identified, which has led to improvements in prognostication and molecular characterization within these subsets (3).
It is now recognized that genetic and epigenetic modifications are similarly important in the pathogenesis of AML (2). Epigenetic modification refers to variability in gene expression without underlying genetic changes (4, 5). DNA methylation and histone modifications are the well-known molecular epigenetic mechanisms studied in cancer biology (4). These epigenetics affect gene expression leading to leukemogenesis through silencing tumor suppressors and activation of oncogenes (3). Multiple studies integrating epigenetic modifiers like DNMT3a, IDH1, IDH2, TET2, ASXL1 and EZH2 and clinical outcomes in AML patients identified mutations as markers prognostic stratification (1). In addition to prognostic significance, since these alterations do not change the DNA sequences and are pharmacologically reversible, they have been regarded as optimal targets for what is now known as epigenetic therapy (2).
Several studies and reviews assessed the prognostic significance of these mutations in AML patients; however, the results are widely variable. In a study performed by Ravandi et al. (6) and colleagues, IDH mutation showed no impact on response to therapy nor Overall Survival (OS). However, Shunichiro Yamaguchi and colleagues (7) found IDH mutations are associated with poor prognosis. A meta-analysis performed by Qingyu Xu and colleagues (8) included thirty-three studies and concluded that IDH1 is associated with poor prognosis and IDH2 is associated with good prognosis. Similarly, studies assessing the impact of DNMT3A, TET2, ASXL1 and EZH2 showed conflicting results. Thus, it’s necessary to perform a systematic review and meta-analysis to clarify the prognostic significance of these mutations in AML patients. The rationale of our study is to evaluate the impact of epigenetic mutations in AML patients with the inclusion of recent publications and larger sample size.
Methods
Search strategy
We performed a literature search on several electronic databases, including PubMed (https://pubmed.ncbi.nlm.nih.gov), Trip database (https://www.tripdatabase.com) and Cochrane library (https://www.cochranelibrary.com) up to November 2018. We used various medical subject headings (MeSH terms) and free text search like mutations, epigenetic, acute myeloid leukemia, acute myeloblastic leukemia, acute myelocytic leukemia and AML.
Study selection
We included adult and pediatric AML studies that received any kind of therapy and were tested for epigenetic mutations (DNMT3a, IDH1, IDH2, TET2, ASXL1 and EZH2). In addition, observational and experimental studies were included. We excluded studies not published in the English language or those not reporting outcomes of interest. Two reviewers (RR and FB) independently screened all citations. Unrelated articles and duplicate publications were excluded after the title and abstract screening. Full-text articles were obtained from the remaining articles and were reviewed carefully for eligibility. Any disagreement between the two reviewers was resolved by discussion or involving a third reviewer (MK).
Data abstraction
Data were extracted using a common data collection form which was designed specifically for this study. Two reviewers abstracted the data independently and subsequently compared the results. Any disagreement was resolved by discussion and consensuses. The main variables extracted were patient characteristics, type of therapy, cytogenetics and molecular features, Hazards ratio (HR) and 95% Confidence Interval (CI) for the OS. In all studies that did not report HR or CI, we used Parmar’s methods to estimate the HR from the available data (9). Furthermore, corresponding authors were contacted for missing data.
Risk of bias assessment
The two reviewers assessed the quality of included studies independently using Newcastle-Ottawa-Scale (NOS) for observational studies. The NOS assessment tool included three main elements related to selection, comparability and outcome. A study can be awarded as good, fair or poor quality based on the maximum score. Any discrepancy between the reviewers was resolved by discussion. Cohen kappa coefficient (k) was calculated to assess the agreement.
Analysis and data synthesis
All statistical analyses were performed using R Program version 3.1.2 (R Core Team (2020); R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/). Hazards ratio was used to assess the prognostic impact of epigenetic mutations compared to wild type. The random-effects model was used to pool the results for the HR of OS. Pooled HR less than 1 indicated better outcome among mutated patients. A p-value of 0.05 or less is considered statistical significance. The heterogeneity among studies was assessed by Cochran I2 and Chi-squared test. I-square (I2) < 30%, 30%–50%, 50%–75%, and >75% were defined as low, moderate, substantial, and considerable heterogeneity, respectively. No ethical approval was required for this study as all data were abstracted from published papers.
Results
Study selection
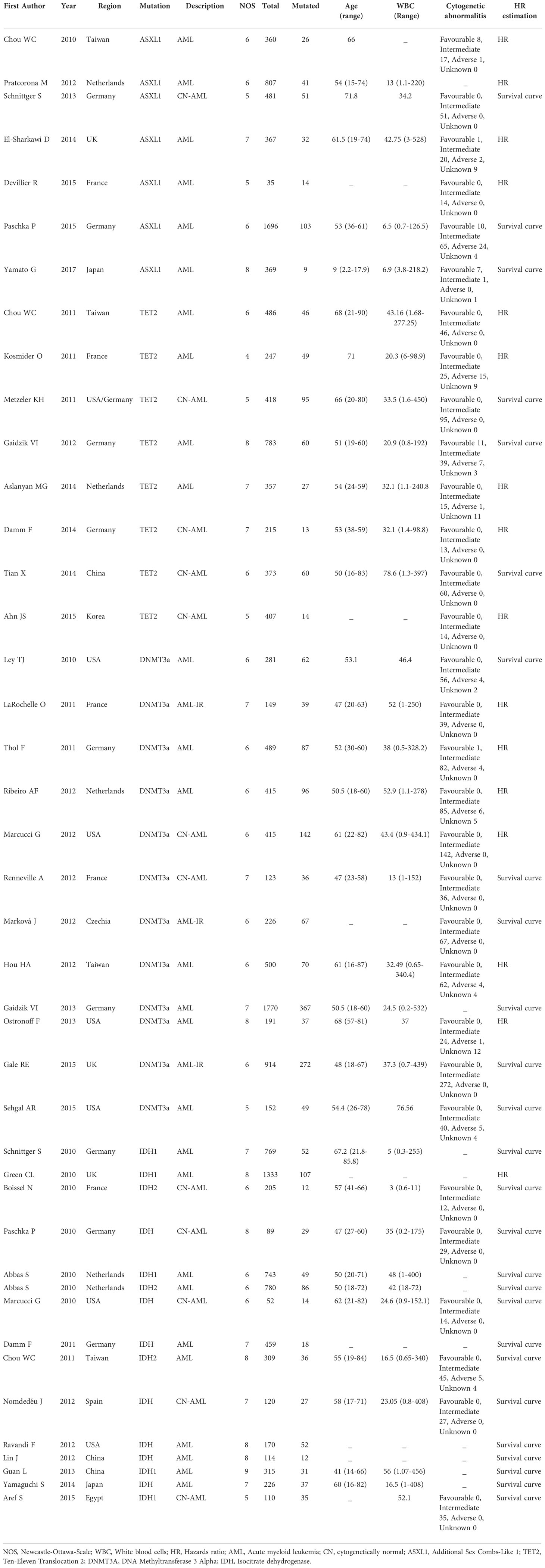
The procedure of study selection is presented in Figure 1. Initially, 10,002 citations were retrieved from the database search. After title screening, 9862 articles were excluded either because not assessing epigenetic mutations, not AML patients or duplicates. We screened the remaining 140 articles. Of these, 49 were excluded after abstract screening for the following reasons: 14 full text not available, ten review articles/case reports and 25 not reporting the outcome of interest. Full-text screening further excluded 49 articles due to the absence of survival analysis or incomplete data. Finally, 42 articles were identified for the systematic review which met the full inclusion criteria. Of these, seven articles were for ASXL1 mutation (10–16), eight for TET2 (17–24), twelve for DNMT3A (25–36) and fifteen for IDH mutation (6, 7, 37–48).
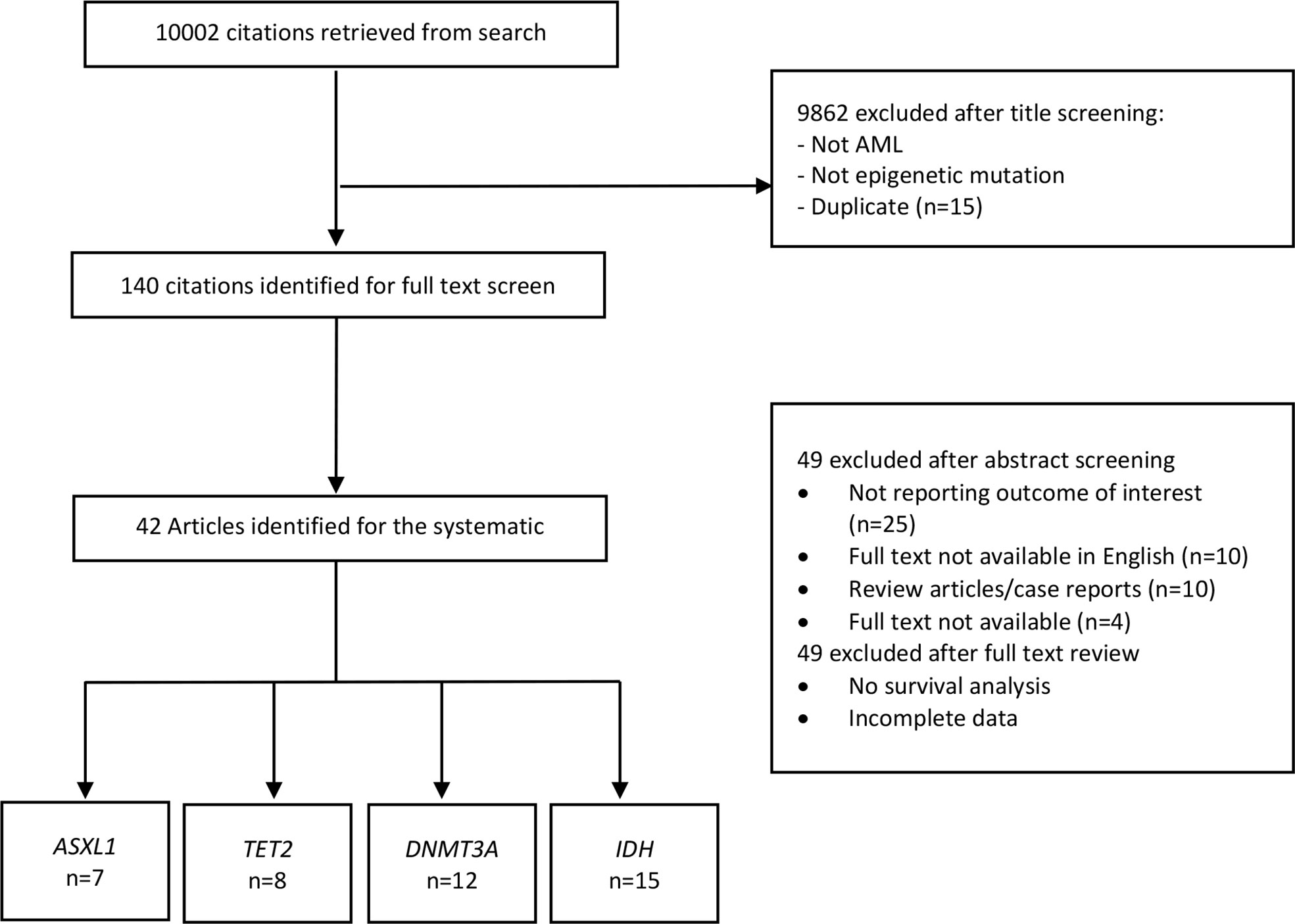
Figure 1 Study selection for the systematic review and meta-analysis. AML, Acute myeloid leukemia; ASXL1, Additional Sex Combs-Like 1; TET2, Ten-Eleven Translocation 2; DNMT3A, DNA Methyltransferase 3 Alpha; IDH, Isocitrate dehydrogenase.
Study characteristics and risk of bias
Table 1 presents the characteristics of the 42 included studies. For ASXL1 mutation articles, five were from Europe and two from Asia. The total number of patients included was 4115 patients; among these, 276 harbored the mutation. One article was for pediatric patients, and the remaining six articles were for adult patients. Eight articles were for TET2 mutation, with a total number of patients 3286 among these 364 had TET2 mutation. Three studies were from Asia, four from Europe and one from Europe and the USA. Finally, for DNMT3A mutation papers total of 12 articles were included, seven from Europe, four from the USA and one from Asia, with a total number of patients 5555 and 1324 with the mutation.
Furthermore, IDH mutation papers were 15 articles. The total number of patients was 5794, and mutated was 597. Eight papers were from Europe, four from Asia, two from the USA and one from Africa. NOS for all 42 included studies ranged from fair to good quality. Cohen kappa coefficient (k) ranged from 0 - 1, with the lowest agreement in exposure and duration of follow-up and highest agreement in confounders.
The details of chemotherapy protocol used in each publication are summarized in Supplementary Material. Majority received intensive cytarabine and anthracycline based chemotherapy. However a subset of frail patients received less intensive chemotherapy, palliative or best supportive care.
Overall survival
As shown in Figure 2, ASXL1 mutation was associated with worse overall survival (HR, 1.88; 95% CI, 1.49 – 2.36, P 0.1316; heterogeneity: I-squared 42.6%). However, this was statistically not significant and was associated with moderate heterogeneity. In addition, the funnel plot showed asymmetry suggesting publication bias or a small study effect.
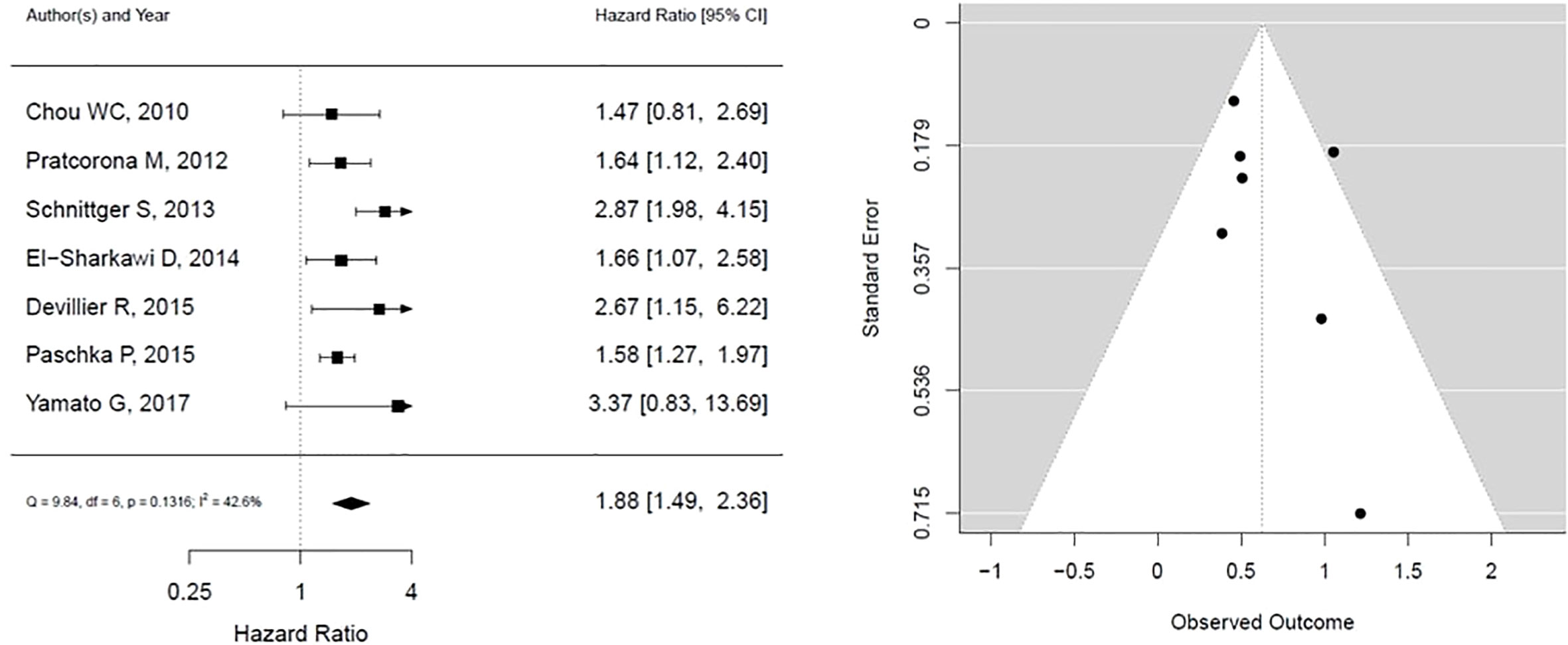
Figure 2 Forest plot and funnel plot of the HR for overall survival in ASXL1 mutation. The first author and year of publication is provided for each study. The hazards ratio (boxes) with 95% confidence intervals (CI, horizontal lines) were calculated, the pooled hazards ratio (diamond) was estimated using random effect model. The P value for comparing heterogeneity between subgroups was calculated using I-squared. ASXL1, Additional Sex Combs-Like 1.
The results for TET2 mutation as presented in Figure 3. It shows that TET2 is associated with statistically insignificant worse overall survival with low heterogeneity (HR, 1.39; 95% CI, 1.18 – 1.63, P 0.1675; heterogeneity: I-squared 27.6%). The funnel plot is symmetrical, suggesting no publication bias.
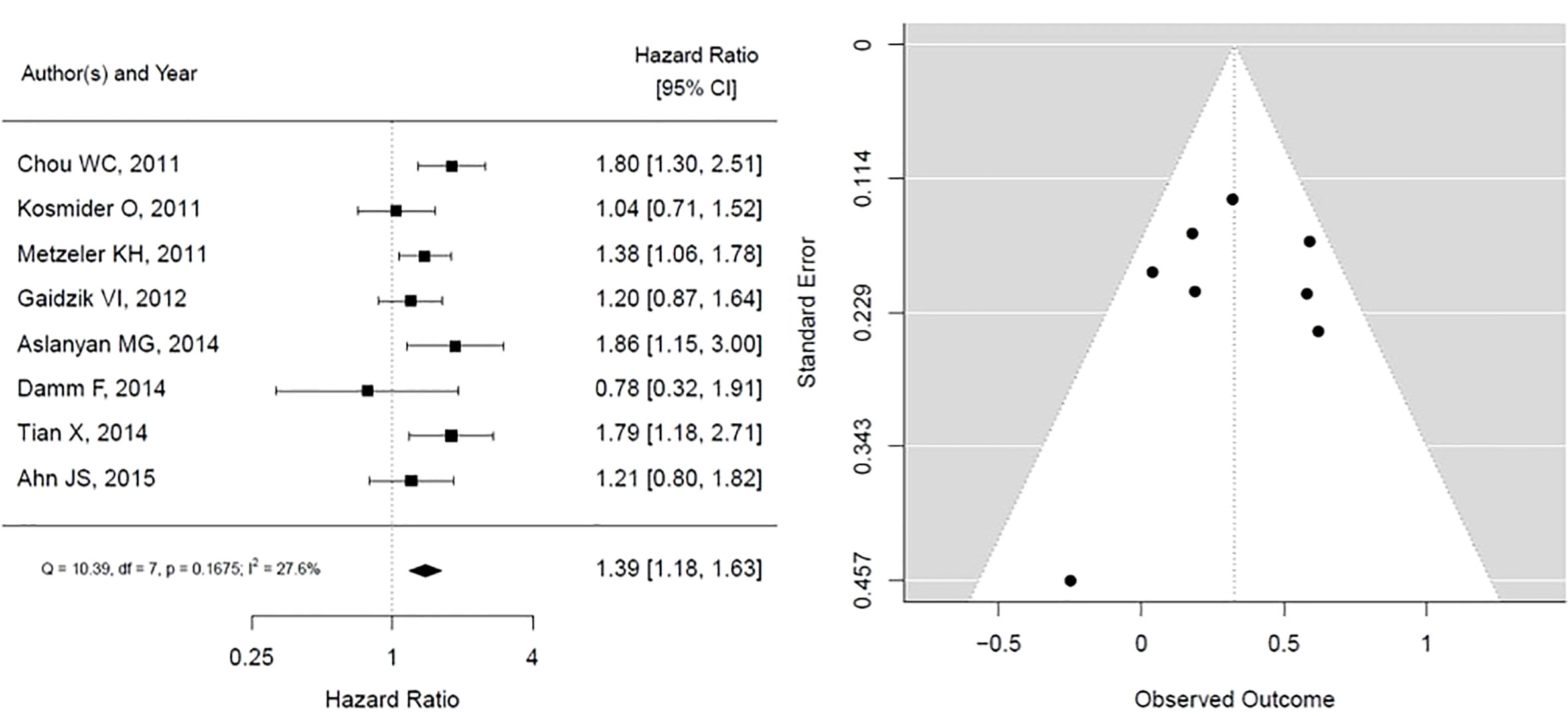
Figure 3 Forest plot and funnel plot of the HR for overall survival in TET2 mutation. The first author and year of publication is provided for each study. The hazards ratio (boxes) with 95% confidence intervals (CI, horizontal lines) were calculated, the pooled hazards ratio (diamond) was estimated using random effect model. The P value for comparing heterogeneity between subgroups was calculated using I-squared. TET2, Ten-Eleven Translocation 2.
For DNMT3A mutations, the forest plot is shown in Figure 4, the pooled HR is associated with worse overall survival, and the results are statistically significant, however with substantial heterogeneity (HR, 1.35; 95% CI, 1.16 – 1.56, P < 0.05; heterogeneity: I-squared 71.0%). In addition, the funnel plot is suggestive of publication bias, given the asymmetry as assessed visually.
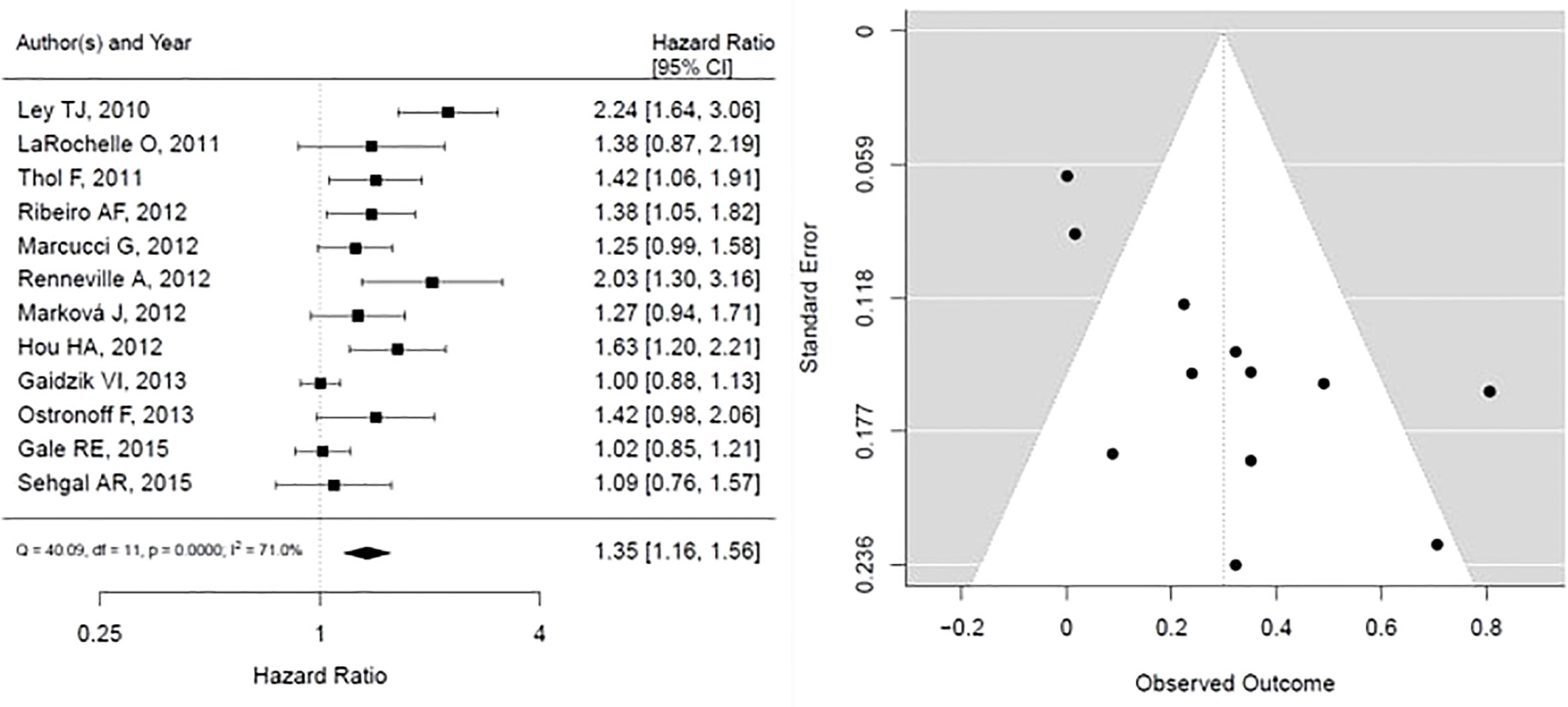
Figure 4 Forest plot and funnel plot of the HR for overall survival in DNMT3a mutation. The first author and year of publication is provided for each study. The hazards ratio (boxes) with 95% confidence intervals (CI, horizontal lines) were calculated, the pooled hazards ratio (diamond) was estimated using random effect model. The P value for comparing heterogeneity between subgroups was calculated using I-squared. DNMT3A, DNA Methyltransferase 3 Alpha.
As shown in Figure 5, IDH mutation is associated with statistically significant worse OS, however with considerable heterogeneity in results (HR, 1.54; 95% CI, 1.15 – 2.06, P <0.05; heterogeneity: I-squared 84.8%). The funnel plot presented is not suggestive of publication bias.
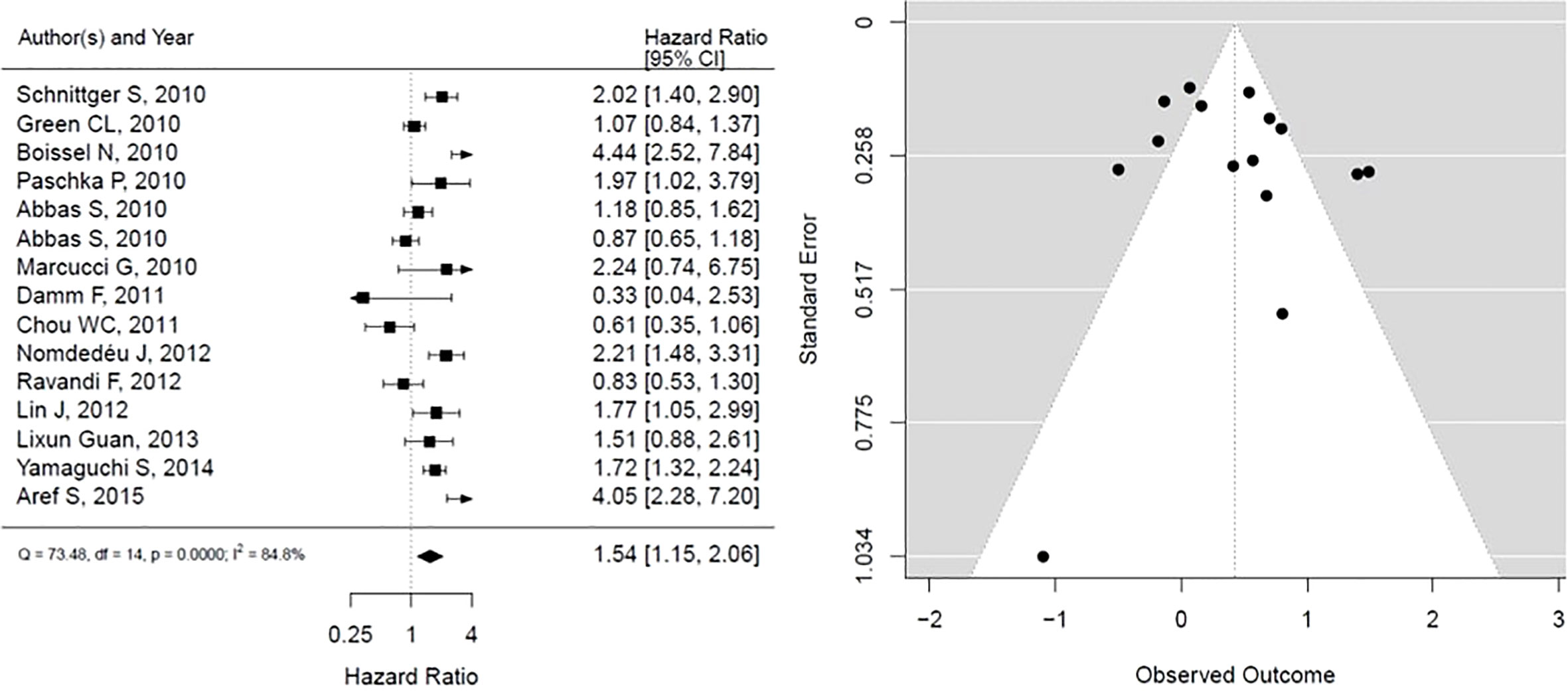
Figure 5 Forest plot and funnel plot of the HR for overall survival in IDH mutation. The first author and year of publication is provided for each study. The hazards ratio (boxes) with 95% confidence intervals (CI, horizontal lines) were calculated, the pooled hazards ratio (diamond) was estimated using random effect model. The P value for comparing heterogeneity between subgroups was calculated using I-squared. IDH, Isocitrate dehydrogenase.
Discussion
In this systematic review and meta-analysis, we found that all epigenetic modifiers mutations (ASXL1, TET2, DNMT3A and IDH) are associated with worse OS in patients with AML based on fair to good-quality studies. However, there was substantial heterogeneity for IDH and DNMT3A mutation studies, respectively.
For ASXL1 mutation, the results revealed a prominent worse overall survival among AML patients. This was consistent among all the included studies. Shivarov V. and his colleagues (49) reported a similar outcome based on the assessment of six large trials and a total of 3311 adult patients. Since there was a single study from the pediatric population, the results cannot be applied to this population. An adverse prognosis was also observed with TET2 mutations, consistent with what Wang R. and his colleagues (50) reported. DNMT3A mutation showed a significantly worse prognosis than the wild type, consistent with what is reported in the literature (51, 52).
Similarly, our data on IDH mutation is reported to show an adverse effect on prognosis. Moreover, we found out that these mutations were frequently found in the intermediate-risk group of the international prognostic scoring system, as shown in Table 1. They were also associated with older patient age and higher presenting white blood cell count.
Although there was substantial heterogeneity in DNMT3A and IDH studies, this could be explained by the year of the publication, age of the patients and cytogenetic risk profile. There has been improvement in supportive care over the last few years, and with the emergence of new targeted therapies, the OS of these patients slightly improved. Jakobsen and colleagues conducted a large population based registry study and they concluded a significant temporal overall survival improvement among patient with AML since 2000. This was particularly seen among the patients aged between 50-75 years where they got curative chemotherapy and option of allogenic stem cell transplant was offered in some cases (53). Furthermore, many other factors affect response to therapy and OS, especially the presence of FLT3 mutation. Publication bias was suggested among ASXL1 and DNMT3A mutation probably; studies with no effect were not published. This was a major limitation to our estimate of the outcome of these mutations.
Our study has several limitations. First, the lack of data from many studies and the high possibility of publication bias could impact the outcome. Second, we did not perform subgroup analysis to assess the impact of the year of the publication to explain the heterogeneity, as older publications probably had worse outcomes. Finally, we did not consider individual patient data, and our analysis was based on cumulative data from different studies.
In conclusion, this meta-analysis revealed that ASXL1, TET2, DNMT3A and IDH mutations had an adverse effect on the survival of AML patients albeit with considerable heterogeneity and possibly publication bias. Further studies are required to address these limitations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Datasets are available on request from the corresponding author.
Author contributions
MA-K suggested the idea, optimized the search strategy, performed the analysis and reviewed and edited the manuscript. FA-B performed the search, extracted the information and drafted the manuscript. RA-R extracted the data and gave final approval. ZA-H and BA-A gave the final approval of the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
Would like to thank Oman Medical Specialty Board (OMSB) library for providing full text articles.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.967657/full#supplementary-material
Abbreviations
AML, Acute myeloid leukemia; ASXL1, Additional Sex Combs-Like 1; TET2, Ten-Eleven Translocation 2; DNMT3A, DNA Methyltransferase 3 Alpha; IDH, Isocitrate dehydrogenase; FLT3, Fms Related Receptor Tyrosine Kinase 3; OS, Overall survival; HR, Hazards Ratio; WBC, White Blood Cells.
References
1. Abdel-Wahab O, Levine RL. Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood. (2013) 121(18):3563–72. doi: 10.1182/blood-2013-01-451781
2. Plass C, Oakes C, Blum W, Marcucci G. Epigenetics in acute myeloid leukemia. Semin Oncol (2008) 35(4):378–87. doi: 10.1053/j.seminoncol.2008.04.008
3. Larsson CA, Cote G, Quintás-Cardama A. The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome. Mol Cancer Res (2013) 11(8):815–27. doi: 10.1158/1541-7786.MCR-12-0695
4. Yamazaki J, Issa J-PJ. Epigenetic aspects of MDS and its molecular targeted therapy. Int J Hematol (2013) 97(2):175–82. doi: 10.1007/s12185-012-1197-4
5. Gutierrez SE, Romero-Oliva FA. Epigenetic changes: a common theme in acute myelogenous leukemogenesis. J Hematol Oncol (2013) 6(1):57. doi: 10.1186/1756-8722-6-57
6. Ravandi F, Patel K, Luthra R, Faderl S, Konopleva M, Kadia T, et al. Prognostic significance of alterations in IDH enzyme isoforms in patients with AML treated with high-dose cytarabine and idarubicin: IDH mutations and SNP in AML. Cancer. (2012) 118(10):2665–73. doi: 10.1002/cncr.26580
7. Yamaguchi S, Iwanaga E, Tokunaga K, Nanri T, Shimomura T, Suzushima H, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol (2014) 92(6):471–7. doi: 10.1111/ejh.12271
8. Xu Q, Li Y, Lv N, Jing Y, Xu Y, Li Y, et al. Correlation between isocitrate dehydrogenase gene aberrations and prognosis of patients with acute myeloid leukemia: A systematic review and meta-analysis. Clin Cancer Res (2017) 23(15):4511–22. doi: 10.1158/1078-0432.CCR-16-2628
9. Parmar MKB, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statist Med (1998) 17(24):2815–34. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8
10. Chou W-C, Huang H-H, Hou H-A, Chen C-Y, Tang J-L, Yao M, et al. Distinct clinical and biological features of de novo acute myeloid leukemia with additional sex comb-like 1 (ASXL1) mutations. Blood. (2010) 116(20):4086–94. doi: 10.1182/blood-2010-05-283291
11. Pratcorona M, Abbas S, Sanders MA, Koenders JE, Kavelaars FG, Erpelinck-Verschueren CAJ, et al. Acquired mutations in ASXL1 in acute myeloid leukemia: Prevalence and prognostic value. Haematologica. (2012) 97(3):388–92. doi: 10.3324/haematol.2011.051532
12. Schnittger S, Eder C, Jeromin S, Alpermann T, Fasan A, Grossmann V, et al. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. (2013) 27(1):82–91. doi: 10.1038/leu.2012.262
13. El-Sharkawi D, Ali A, Evans CM, Hills RK, Burnett AK, Linch DC, et al. ASXL1 mutations are infrequent in young patients with primary acute myeloid leukemia and their detection has a limited role in therapeutic risk stratification. Leukemia Lymphoma. (2014) 55(6):1326–31. doi: 10.3109/10428194.2013.833332
14. Devillier R, Mansat-De Mas V, Gelsi-Boyer V, Demur C, Murati A, Corre J, et al. Role of ASXL1 and TP53 mutations in the molecular classification and prognosis of acute myeloid leukemias with myelodysplasia-related changes. Oncotarget. (2015) 6(10):8388–96. doi: 10.18632/oncotarget.3460
15. Paschka P, Schlenk RF, Gaidzik VI, Herzig JK, Aulitzky T, Bullinger L, et al. ASXL1 mutations in younger adult patients with acute myeloid leukemia: A study by the German-Austrian acute myeloid leukemia study group. Haematologica. (2015) 100(3):324–30. doi: 10.3324/haematol.2014.114157
16. Yamato G, Shiba N, Yoshida K, Shiraishi Y, Hara Y, Ohki K, et al. ASXL2 mutations are frequently found in pediatric AML patients with t(8;21)/ RUNX1-RUNX1T1 and associated with a better prognosis. Genes Chromosomes Cancer. (2017) 56(5):382–93. doi: 10.1002/gcc.22443
17. Chou W-C, Chou S-C, Liu C-Y, Chen C-Y, Hou H-A, Kuo Y-Y, et al. TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood. (2011) 118(14):3803–10. doi: 10.1182/blood-2011-02-339747
18. Kosmider O, Delabesse E, Mas VM-D, Cornillet-Lefebvre P, Blanchet O, Delmer A, et al. TET2 mutations in secondary acute myeloid leukemias: A French retrospective study. Haematologica. (2011) 96(7):1059–63. doi: 10.3324/haematol.2011.040840
19. Metzeler KH, Maharry K, Radmacher MD, Mrózek K, Margeson D, Becker H, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: A cancer and leukemia group b study. JCO. (2011) 29(10):1373–81. doi: 10.1200/JCO.2010.32.7742
20. Gaidzik VI, Paschka P, Späth D, Habdank M, Köhne C-H, Germing U, et al. TET2 mutations in acute myeloid leukemia (AML): Results from a comprehensive genetic and clinical analysis of the AML study group. JCO. (2012) 30(12):1350–7. doi: 10.1200/JCO.2011.39.2886
21. Aslanyan MG, Kroeze LI, Langemeijer SMC, Koorenhof-Scheele TN, Massop M, van Hoogen P, et al. Clinical and biological impact of TET2 mutations and expression in younger adult AML patients treated within the EORTC/GIMEMA AML-12 clinical trial. Ann Hematol (2014) 93(8):1401–12. doi: 10.1007/s00277-014-2055-7
22. Damm F, Markus B, Thol F, Morgan M, Göhring G, Schlegelberger B, et al. TET2 mutations in cytogenetically normal acute myeloid leukemia: Clinical implications and evolutionary patterns: TET2 MUTATIONS IN AML. Genes Chromosomes Cancer. (2014) 53(10):824–32. doi: 10.1002/gcc.22191
23. Tian X, Xu Y, Yin J, Tian H, Chen S, Wu D, et al. TET2 gene mutation is unfavorable prognostic factor in cytogenetically normal acute myeloid leukemia patients with NPM1+ and FLT3-ITD– mutations. Int J Hematol (2014) 100(1):96–104. doi: 10.1007/s12185-014-1595-x
24. Ahn J-S, Kim H-J, Kim Y-K, Jung S-H, Yang D-H, Lee J-J, et al. Adverse prognostic effect of homozygous TET2 mutation on the relapse risk of acute myeloid leukemia in patients of normal karyotype. Haematologica. (2015) 100(9):e351–3. doi: 10.3324/haematol.2015.126227
25. Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med (2010) 363(25):2424–33. doi: 10.1056/NEJMoa1005143
26. LaRochelle O, Bertoli S, Vergez F, Sarry J-E, Mansat-De Mas V, Dobbelstein S, et al. Do AML patients with DNMT3A exon 23 mutations benefit from idarubicin as compared to daunorubicin? a single center experience. Oncotarget. (2011) 2(11):850–61. doi: 10.18632/oncotarget.347
27. Thol F, Damm F, Lüdeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. JCO. (2011) 29(21):2889–96. doi: 10.1200/JCO.2011.35.4894
28. Ribeiro AFT, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, et al. Mutant DNMT3A: A marker of poor prognosis in acute myeloid leukemia. Blood. (2012) 119(24):5824–31. doi: 10.1182/blood-2011-07-367961
29. Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrózek K, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. JCO. (2012) 30(7):742–50. doi: 10.1200/JCO.2011.39.2092
30. Renneville A, Boissel N, Nibourel O, Berthon C, Helevaut N, Gardin C, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: A study by the acute leukemia French association. Leukemia. (2012) 26(6):1247–54. doi: 10.1038/leu.2011.382
31. Marková J, Michková P, Burčková K, Březinová J, Michalová K, Dohnalová A, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia: DNMT3A mutations in patients with AML. Eur J Haematology. (2012) 88(2):128–35. doi: 10.1111/j.1600-0609.2011.01716.x
32. Hou H-A, Kuo Y-Y, Liu C-Y, Chou W-C, Lee MC, Chen C-Y, et al. DNMT3A mutations in acute myeloid leukemia: Stability during disease evolution and clinical implications. Myeloid neoplasia (2012) 119(2):11. doi: 10.1182/blood-2011-07-369934
33. Gaidzik VI, Schlenk RF, Paschka P, Stölzle A, Späth D, Kuendgen A, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: Results of the AML study group (AMLSG). Blood. (2013) 121(23):4769–77. doi: 10.1182/blood-2012-10-461624
34. Ostronoff F, Othus M, Ho PA, Kutny M, Geraghty DE, Petersdorf SH, et al. Mutations in the DNMT3A exon 23 independently predict poor outcome in older patients with acute myeloid leukemia: A SWOG report. Leukemia. (2013) 27(1):238–41. doi: 10.1038/leu.2012.168
35. Gale RE, Lamb K, Allen C, El-Sharkawi D, Stowe C, Jenkinson S, et al. Simpson’s paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. JCO. (2015) 33(18):2072–83. doi: 10.1200/JCO.2014.59.2022
36. Sehgal AR, Gimotty PA, Zhao J, Hsu J-M, Daber R, Morrissette JD, et al. DNMT3A mutational status affects the results of dose-escalated induction therapy in acute myelogenous leukemia. Clin Cancer Res (2015) 21(7):1614–20. doi: 10.1158/1078-0432.CCR-14-0327
37. Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. (2010) 116(25):5486–96. doi: 10.1182/blood-2010-02-267955
38. Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. (2010) 116(15):2779–82. doi: 10.1182/blood-2010-02-270926
39. Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: A study by the acute leukemia French association group. JCO. (2010) 28(23):3717–23. doi: 10.1200/JCO.2010.28.2285
40. Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. JCO. (2010) 28(22):3636–43. doi: 10.1200/JCO.2010.28.3762
41. Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. (2010) 116(12):2122–6. doi: 10.1182/blood-2009-11-250878
42. Marcucci G, Maharry K, Wu Y-Z, Radmacher MD, Mrózek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within De novo cytogenetically normal acute myeloid leukemia: A cancer and leukemia group b study. JCO. (2010) 28(14):2348–55. doi: 10.1200/JCO.2009.27.3730
43. Damm F, Thol F, Hollink I, Zimmermann M, Reinhardt K, van den Heuvel-Eibrink MM, et al. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: A study of the AML–BFM and DCOG study groups. Leukemia. (2011) 25(11):1704–10. doi: 10.1038/leu.2011.142
44. Chou W-C, Lei W-C, Ko B-S, Hou H-A, Chen C-Y, Tang J-L, et al. The prognostic impact and stability of isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. (2011) 25(2):246–53. doi: 10.1038/leu.2010.267
45. Nomdedéu J, Hoyos M, Carricondo M, Esteve J, Bussaglia E, Estivill C, et al. Adverse impact of IDH1 and IDH2 mutations in primary AML: Experience of the Spanish CETLAM group. Leukemia Res (2012) 36(8):990–7. doi: 10.1016/j.leukres.2012.03.019
46. Lin J, Yao D, Qian J, Chen Q, Qian W, Li Y, et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol (2012) 91(4):519–25. doi: 10.1007/s00277-011-1352-7
47. Guan L, Gao L, Wang L, Li M, Yin Y, Yu L, et al. The frequency and clinical significance of IDH1 mutations in Chinese acute myeloid leukemia patients. Konopleva M editor. PLoS One (2013) 8(12):e83334. doi: 10.1371/journal.pone.0083334
48. Aref S, Kamel Areida ES, Abdel Aaal MF, Adam OM, El-Ghonemy MS, El-Baiomy MA, et al. Prevalence and clinical effect of IDH1 and IDH2 mutations among cytogenetically normal acute myeloid leukemia patients. Clin Lymphoma Myeloma Leukemia. (2015) 15(9):550–5. doi: 10.1016/j.clml.2015.05.009
49. Shivarov V, Gueorguieva R, Ivanova M, Tiu RV. ASXL1 mutations define a subgroup of patients with acute myeloid leukemia with distinct gene expression profile and poor prognosis: A meta-analysis of 3311 adult patients with acute myeloid leukemia. Leukemia Lymphoma. (2015) 56(6):1881–3. doi: 10.3109/10428194.2014.974596
50. Wang R, Gao X, Yu L. The prognostic impact of tet oncogene family member 2 mutations in patients with acute myeloid leukemia: A systematic-review and meta-analysis. BMC Cancer. (2019) 19(1):389. doi: 10.1186/s12885-019-5602-8
51. Tie R, Zhang T, Fu H, Wang L, Wang Y, He Y, et al. Association between DNMT3A mutations and prognosis of adults with De novo acute myeloid leukemia: A systematic review and meta-analysis. Eaves CJ editor. PLoS One (2014) 9(6):e93353. doi: 10.1371/journal.pone.0093353
52. Shivarov V, Gueorguieva R, Stoimenov A, Tiu R. DNMT3A mutation is a poor prognosis biomarker in AML: Results of a meta-analysis of 4500 AML patients. Leukemia Res (2013) 37(11):1445–50. doi: 10.1016/j.leukres.2013.07.032
53. Hjort Jakobsen L, Stidsholt Roug A, Kiesbye Øvlisen A, Werenberg Marcher C, Beier Ommen H, Theilgaard-Mönch K, et al. Temporal changes in survival among adult patients with acute myeloid leukaemia in the period 2000–2016: A Danish population-based study. Br J Haematology. (2020) 193(3):482–7. doi: 10.1111/bjh.17213
Keywords: AML, epigenetics, mutation, ASXL1, Dnmt3a, IDH, TET2, review – systematic
Citation: Al-Bulushi F, Al-Riyami R, Al-Housni Z, Al-Abri B and Al-Khabori M (2022) Impact of mutations in epigenetic modifiers in acute myeloid leukemia: A systematic review and meta-analysis. Front. Oncol. 12:967657. doi: 10.3389/fonc.2022.967657
Received: 13 June 2022; Accepted: 11 October 2022;
Published: 28 November 2022.
Edited by:
Mohamed A. Yassin, Hamad Medical Corporation, QatarReviewed by:
Jatinder K. Lamba, University of Florida, United StatesHonar Cherif, Hamad Medical Corporation, Qatar
Copyright © 2022 Al-Bulushi, Al-Riyami, Al-Housni, Al-Abri and Al-Khabori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Murtadha Al-Khabori, a2hhYm9yaUBzcXUuZWR1Lm9t; bWtraGFib3JpQGdtYWlsLmNvbQ==
 Fatma Al-Bulushi
Fatma Al-Bulushi Rahma Al-Riyami3
Rahma Al-Riyami3 Zainab Al-Housni
Zainab Al-Housni Murtadha Al-Khabori
Murtadha Al-Khabori