
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 31 August 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.966981
This article is part of the Research TopicCirculating Molecular Biomarkers: Next-Generation Tools for Monitoring Minimal Residual Disease in Cancer PatientsView all 11 articles
 Xiaomei Yi1,2
Xiaomei Yi1,2 Jie Chen1,2
Jie Chen1,2 Defa Huang1,2
Defa Huang1,2 Shuo Feng3
Shuo Feng3 Tong Yang1,2
Tong Yang1,2 Zhengzhe Li1,2
Zhengzhe Li1,2 Xiaoxing Wang1,2
Xiaoxing Wang1,2 Minghong Zhao1,2
Minghong Zhao1,2 Jiyang Wu1,2
Jiyang Wu1,2 Tianyu Zhong1,2*
Tianyu Zhong1,2*Exosomes are a heterogeneous subset of extracellular vesicles (EVs) that biogenesis from endosomes. Besides, exosomes contain a variety of molecular cargoes including proteins, lipids and nucleic acids, which play a key role in the mechanism of exosome formation. Meanwhile, exosomes are involved with physiological and pathological conditions. The molecular profile of exosomes reflects the type and pathophysiological status of the originating cells so could potentially be exploited for diagnostic of cancer. This review aims to describe important molecular cargoes involved in exosome biogenesis. In addition, we highlight exogenous factors, especially autophagy, hypoxia and pharmacology, that regulate the release of exosomes and their corresponding cargoes. Particularly, we also emphasize exosome molecular cargoes as potential biomarkers in liquid biopsy for diagnosis of cancer.
Extracellular vesicles (EVs) are secreted from almost all cell types (1), and widely distributed in various body fluids, such as urine (2), blood (3), milk (4), saliva (5), cerebrospinal fluid (6), amniotic fluid (7) and semen (8), can transmit information between cells and participate in many physiological and pathological processes. It is known that the extraction and isolation of exosomes from different body fluids are mainly achieved by ultracentrifugation, ultrafiltration, sedimentation, density gradient centrifugation, immune-capture, precipitation and commercial reagents(Table 1). Exosomes are bi-layered lipid vesicles produced by the endosomal pathway, a subset of EVs with a diameter of 30-150nm (36, 37). However, due to the limitations of the isolation method, we usually define the particles less than 200nm in diameter are exosomes. Therefore, the International Society of Extracellular Vesicle (ISEV) statement in the Minimum Information on Extracellular Vesicle Research 2018 (MISEV2018) recommends the use of “EVs” as a general term (36). In this review, EVs mainly refer to exosomes without special instructions.
Exosomes are present in biological fluids as a form of intercellular communication to transport proteins, lipids, nucleic acids, and metabolites to the pericellular environment (38, 39). Exosome biogenesis are tightly regulated, possibly by interactions with different effectors (40, 41), which mainly involved with ESCRT-dependent and ESCRT-independent mechanisms (42). Exosome biogenesis begins in the endocytic pathway, where the plasma membrane invagination packages cell membrane proteins and some extracellular components together to form the early endosomes (EEs) (43, 44). After that, EEs exchange substances with other organelles, or further mature into late endosomes (LEs), and the late endosomal membrane invaginate to form multiple vesicles (MVBs) containing luminal vesicles (ILVs). Next, MVBs bind to lysosomes or autophagosomes for degradation, or they are transported to the plasma membrane through the cytoskeleton and microtubule network, which then efflux to form exosomes (Figure 1A) (45–47). Interestingly, exosomal cargo molecules (Figure 1B) (proteins, lipids, and nucleic acids) regulate the whole process (42, 45, 48, 49). For example, tetraspanin proteins (e. g.: CD9, CD63, CD81, CD82), major histocompatibility complex (MHC) molecules, heat shock proteins (HSPs), endosomal sorting complex (ESCRT) proteins (e. g. Alix, TSG101), Rab proteins, actin, soluble N-acetamide sensitive factor attachment proteins (SNAREs) are the major participating proteins (50–53). Similarly, lipid components such as ceramide, cholesterol, phosphatidic acid, phosphatidylinositol 3-phosphate, phosphatidylinositol-3, 5-diphosphate, and sphingosine 1-phosphate are also involved in the process (54–57). A summary of the molecular cargoes associated with exosome biogenesis process is presented in Table 2.
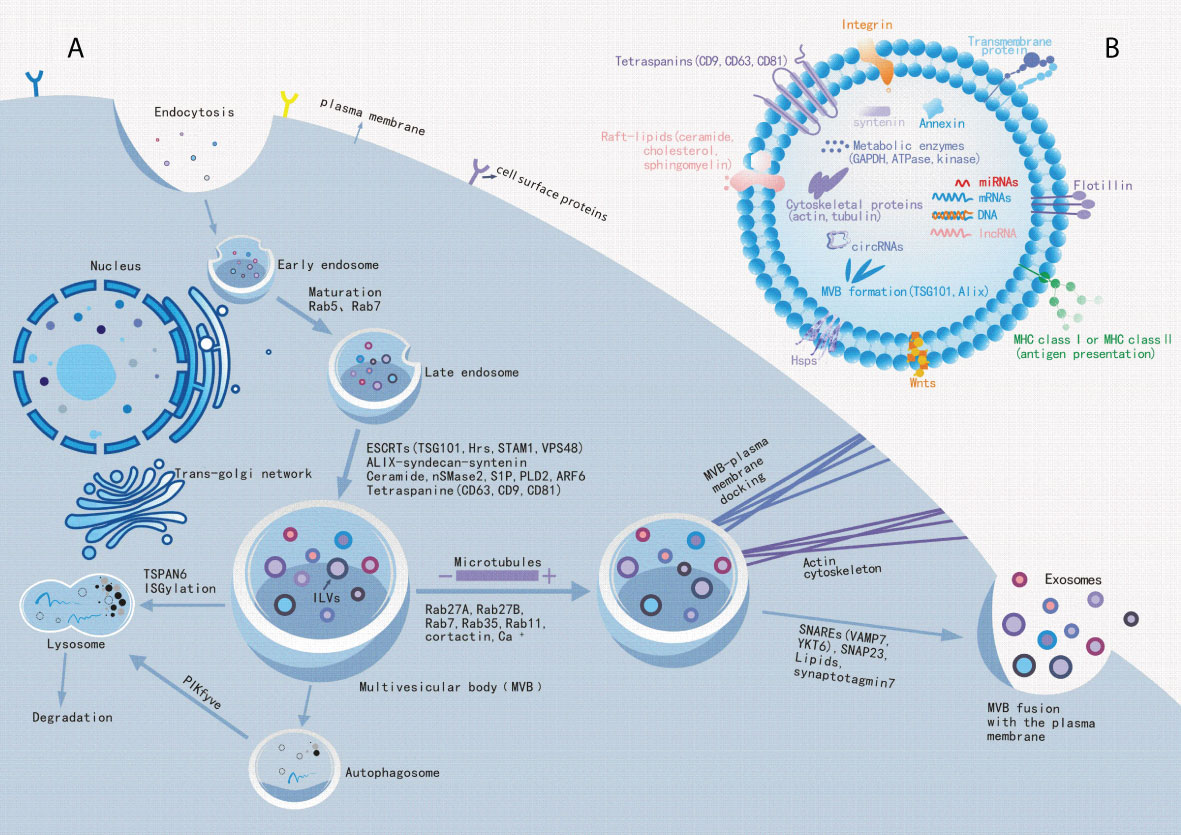
Figure 1 Exosome biogenesis. (A): Schematic diagram of the molecular mechanisms of exosome biogenesis. Extracellular components, such as proteins, lipids, nucleic acids and small molecules, can enter cells with cell surface proteins through endocytosis and plasma membrane invagination. Under endocytosis, it leads to the formation of early endosomes and late endosomes, which bud out into multiple vesicles (MVBs) containing luminal vesicles (ILVs). Some molecules, such as ESCRT proteins (ALIX, TSG101, etc.), lipids and tetraspanin proteins mediate this process. Subsequently, MVBs will fuse to lysosomes or autophagosomes to accelerate their degradation to inhibit exosome release, or MVBs transported along the cytoskeleton and microtubule network to the plasma membrane after maturation, where it can fuse with the plasma membrane and release exosomes into extracellular space. Among these, Rabs, Actin and SNARE proteins are involved in exosome release.(B): Exosome biomarkers. Exosomal luminal cargoes are mainly composed of proteins, lipids, nucleic acids, and other metabolites that can function in the recipient cells. Among these, CD9, CD63, CD81, flotillin, and Annexin can be used as exosome biomarkers.
Initial studies suggested that exosomes were some waste materials excreted by cells in order to maintain homeostasis (138). Recent reports suggest that exosomes are capable of material transport and information transfer between cells, thereby mediating many physiological and pathological processes (51, 72, 139, 140). Furthermore, these small vesicles are involved in immunomodulation and intercellular communication (141), and mediate the disease progression of cancer (142), cardiovascular disease (143–145), metabolic disease (146), degenerative change (147) and autoimmunity (148). It is currently believed that the key to exosomes biological functions lies in their molecular cargoes, including proteins, lipids, and nucleic acids. For example, phosphatidylinositol glycan-1 (GPC1) is a cell surface proteoglycan rich in cancer cell-derived exosomes, and Melo et al. (120) identified that GPC1 has the potential for early detection of pancreatic cancer lesions to promote the possibility of curative surgical treatment (120) found that CRC cell-derived exosomal HSPC111 protein promotes pre-metastatic niche formation and CRC liver metastases (CRLM) via reprogramming lipid metabolism in cancer-associated fibroblasts (CAFs), which implicate HSPC111 may be a potential therapeutic target for preventing CRLM (149). In addition, phosphatidylserine, cholesterol and ceramide are also play key roles in exosome formation, which affect cargo sorting, signaling and exosomes structure (150, 151). MicroRNAs(miRNAs) are one of the most abundant RNA species in exosomes, and miRNAs play roles in various biological processes such as exocytosis and exosome-mediated cellular communication (73, 152). For example, Fu et al. (153) found that exosomes content miR-98-5p inhibits the progression of pancreatic ductal adenocarcinoma(PDAC) by targeting MAPK signaling (153). In addition, microarray profiles identified that miR-106a-5p and miR-19b-3p were remarkably overexpressed in the serum exosomes of patients with gastric cancer(GC). Notably, integrating the two miRNAs could identify GC patients among healthy volunteers with a 0.814 area under the curve (AUC) value, which was higher than that obtained using CEA or AFP (154). Of note, the parental information of these exosomes may differ significantly between healthy people and patients, making some molecular cargoes in exosomes potentially as specific biomarkers of cancer. Importantly, the ability to selectively control the release of exosomes in pathological situations without compromising their role as essential components in physiological situations would make exosomes have promising clinical applications in disease diagnosis, treatment and prognosis. In this review, we conclude the role of exosomes molecular cargoes in their biogenesis. We also underline the potential mechanisms by which autophagy, hypoxia and pharmacology exogenous factors affect exosome release. And summarize the key roles of exosome molecular cargoes play in cancer diagnosis. Furthermore, we discuss the challenges and potential applications of exosomes research.
The biogenesis of exosomes is influenced by a variety of extrinsic factors in addition to the molecular correlation of the above-mentioned cargoes. A greater understanding of the underlying mechanisms that influence exosome release factors could provide new targets for disease diagnosis and treatment. The potential mechanisms by which autophagy, hypoxia, and pharmacological factors affect exosome release are presented below.
Autophagy is a process that causes the degradation of cellular material at the lysosome. Autophagosomes can fuse with MVBs or directly with lysosomes to degrade cargoes (155). It was found that autophagy-related proteins, such as ATG5 and ATG16L1, affects exosome release process. For example, Abdulrahma et al. reported that when the autophagy protein ATG5 was knocked down, it greatly promoted the release of prion protein (PRNP) exosomes (156). Recently, Zheng et al. demonstrated that sulforaphane inhibits autophagy and induces exosome release via regulating mTOR/TEF3 (157). In addition, Guo et al. showed that ATG16L1 and ATG5 autophagy proteins protected MVBs from lysosomal degradation and thus facilitated the fusion of MVBs with the plasma membrane to facilitate exosome release. Conversely, silencing of ATG16L1 and ATG5 decreased exosome release, probably due to the ability of ATG5 to separate ATP6V1E1 from V1V0-ATPase, thereby inhibiting MVBs acidification and facilitating exosome release (158). Crucially, Keller et al. identified that ATG proteins promoted exosome release through a lysosomal non-dependent pathway, i.e. secretory autophagy, which in turn excreted bacterial toxin receptors from the membrane surface in the form of exosomes, assisting host cells to resist toxin damage and enhancing the antimicrobial response of the organism (159). These studies all suggest that autophagy may play a specific role to affect exosome release.
Hypoxia may affect exosome release through hypoxia-inducible factors (HIF), Rab-GTPases, NF-κB and four transmembrane protein signaling pathways, but the specific mechanisms involved remains unclear (160). Hypoxia-inducible factor (HIF) is a major component of the hypoxia-related signaling pathway that directly or indirectly regulates the process of exosome release. Recently, it has been reported that HIF mediates endocytosis mainly by increasing the expression of glucose transporter protein (GLUT-1), transferrin receptor and epidermal growth factor receptor (EGFR), which in turn induces exosome release (161). It was found that the increased release of exosomes from rat proximal renal tubular cells (RPTC) (162) and breast cancer cells (163) in hypoxic environment was mainly mediated by HIF-1α. In particular, hypoxia can cause glycolysis and lactate accumulation. Ban et al. demonstrated that exosome markers such as CD9, CD63, and HSP70 expression increased under acidic conditions and were more conducive to exosome release, whereas exosomal proteins and exosomal RNA were not detected in alkaline environments and exosome release was reduced (164). Wang et al. demonstrated that hypoxia increased the number of exosomes released from colorectal cancer cells compared to hyperoxic conditions (165). On the other hand, hypoxia not only alters exosome size, sorting mechanisms and exosome uptake and binding capacity in the tumor microenvironment, but also impacts exosome-mediated tumor biological functions (166). Interestingly, different hypoxic conditions, such as duration and severity of hypoxia, can have dramatically variable impacts on the amount and content of exosomes released by different cell types (167)
Nowadays, utilizing exosome as nanomaterials for drug delivery is of great interest to researchers. Notably, drugs may have a dramatic impact on drug repositioning and as potential novel anticancer agents by affecting certain molecules in the exosome release process. However, there are no drugs available to control the production of harmful exosomes in tumor cells (168). PH and Ca2+ are required for exosome release. Amiloride is a drug that inhibits Na +/H + exchange pump and Na +/Ca 2+ channels, and Savina et al. demonstrated that it reduced exosome release (169). Importantly, amiloride inhibits ceramide formation by indirectly inhibiting acid sphingomyelinase (aSMase), which in turn inhibits exosome release (170). Similarly, promethazine, a tricyclic antidepressant, has been found to reduce exosome release through inhibition of aSMase activity in the prostate cancer cell line PC3 by Kosgodage et al. (171). Metformin is the first-line drug for the treatment of type 2 diabetes, which increases insulin sensitivity and reduces fat synthesis (172). Recently, Liao et al. have demonstrated that metformin promotes the fusion of MVBs with the plasma membrane through autophagy and thus increased exosome release from mesenchymal stem cells (MSCs), which improved their therapeutic effect on senescent cells (173). In addition, metformin may promote exosome release to regulate stress by increasing the production of reactive oxygen species in tumor cells (174). Gao al.demonstrated that all-trans retinoic acid suppressed GES-1 cell proliferation induced by exosomes from patients with precancerous lesions by arresting the cell cycle in S-phase (175). Therefore, these drugs may act by acting on certain molecules released from exosomes, promoting exosome release may be a protective method against drug stress conditions to eliminate cellular damage.
Ticagrelor is a purinergic drug, it has been widely used in patients with acute coronary syndrome (ACS) and myocardial infarction (176). Existing studies have reported that ticagrelor enhanced the release of cell-derived exosomes from the anti-hypoxic cardiac group by increasing cell proliferation in vitro (177). In addition, extracellular vesicles derived from cardiomyocytes pretreated with ticagrelor have a protective effect on hyperglycemic cardiomyocytes by attenuating oxidative and endoplasmic reticulum stress (178). Recently, Kulshreshtha et al. confirmed that simvastatin, a HMG CoA inhibitor, mediates exosome release by altering MVBs transport and that its mediated reduction in monocyte-derived exosome secretion is protective in vitro model of atherosclerosis (179). Likewise, exosomes derived from mesenchymal stem cells (MSCs) pretreated with atorvastatin (ATV) dramatically enhanced the efficacy of treatment of acute myocardial infarction (AMI), possibly by enhancing endothelial cell function through paracrine mechanisms (180). It was also found that extracellular vesicles of cannabis with high cannabidiol (CBD) content induce anticancer signaling in human hepatocellular carcinoma (181).
Notably, Zhang et al. reported that neutral sphingomyelinase inhibitor (Manumycin A) and ketoconazole had no effect on exosomes released from normal cells, but affected exosomes released from tumor cells, which is crucial for disease treatment (182). It remains to be further investigated whether this can be mediated by the influence of proto-oncogenes and/or oncogenes in the tumor cells or by other factors. Considering that most of the experiments were performed on tumors, it remains to be further explored how these drugs affect the cancer phenotype by influencing the exosome release process and thus the cancer phenotype. Furthermore, we need to be aware that drugs have certain side effects. In the future, there is also need to focus on what doses of these drugs should be used to reach specific sites of cancer in a particular way to inhibit or promote exosome release as a form of cancer treatment.
In addition, other factors such as food compounds (183, 184), temperature (185, 186), radiotherapy (187) and chemotherapy (188, 189)affect intercellular communication mechanisms by mediating exosome release process, which allows exosomes to perform different functions and then contributes to the diagnosis and treatment of diseases.
Exosome components indicate the biological state of the initiating cells and reflect the health status of the organs. Recently, more and more studies have shown that EVs contents can be applied in the diagnosis of various diseases (13, 190–193). This section summarizes the biomarkers that may become clinically common diseases in several major classes of molecular cargoes.
Messenger RNA (mRNA) is a single-stranded ribonucleic acid that carries genetic information and can guide protein synthesis. mRNA is not only an important exosome cargo, but also acts as a functional modulator in cancer cell-derived exosome processes (194). In order to study the diagnostic performance of circulating exosomal messenger RNA (emRNA) and tissue mRNA in prostate cancer (PCa) patients, Ji et al. (195) demonstrated circulating emRNA is more advantageous as a diagnostic biomarker in PCa patients. Recipient operating characteristic curve (ROC) analysis indicated that the AUC value of circulating emRNA in PCa screening and diagnosis was 0.948 and 0.851 respectively. Furthermore, the six molecules in emRNA including CDC42, IL32, MAX, NCF2, PDGFA and SRSF2 were upregulated in the screening and diagnosis of PCa patients compared to healthy controls (195). Similarly, Shephard et al. (88) said that serum-derived EV-mRNA has great potential for the differential diagnosis of prostate cancer. Among these, increased serum-derived EV-mRNA CTGF molecule or decreased EV-mRNA CAV1 molecule were closely associated with the rate of disease progression, and the AUC values of CTGF and CAV1 were 0.8600 and 0.8100 respectively. However, serum PSA could not predict disease progression, suggesting that EV-mRNA CTGF and CAV1 are superior to PSA in predicting disease progression (88). Another study proved that mRNA index of membrane matrix type 1 metalloproteinase (MT1-MMP) was significantly up-regulated in gastric cancer (GC) patients, with an AUC of 0.788, sensitivity of 63.9% and specificity of 87.1%, while the AUC value of serum CEA was only 0.655. Meanwhile, the combined exosomes diagnosis of mRNA(MT1-MMP) and CEA (AUC=0.821) was significantly better than the detection of mRNA (MT1-MMP) or CEA separately in identifying GC patients. In addition, it has been shown that exosomal epithelial growth factor receptor (EGFR) mRNA may be a potential predictor of glioblastoma (196). Serum exosome mRNA(MT1-MMP) was significantly associated with tumor differentiation, depth of invasion, lymphatic metastasis, distal metastasis and TNM stage (89). In brief, these studies show that exosomal mRNAs may have the potential to act as cancer biomarkers, but their specificity for the disease should be further investigated.
MiRNA is a class of small endogenous noncoding RNA composed of 18-24 nucleotides, and the miRNA that delivered to the recipient cells can regulate various gene expression by preventing translation and inducing mRNA degradation (197). In addition, Exosomal miRNAs are more stable than free miRNAs as they are protected from degradation owing to RNase activity in biofluids (198). Recent studies have revealed that exosomal miRNAs may serve as potential biomarkers in certain cancers. For example, Yang et al. (104) found that exosomal miR-423-5p level was highly expressed in gastric cancer (GC) patients serum, and the AUC values of exosomal miR-423-5p, serum CEA and CA-199 were 0.763, 0.596 and 0.607 respectively (104). Notably, the combined detection of miRNAs can improve diagnostic accuracy. Huang et al. (199) found that six miRNAs were significantly higher expressed in serum exosomes of GC patients, whose AUC values were 0.627 (miR-10b-5p), 0.652 (miR-132-3p), 0.637 (miR-185-5p), 0.683 (miR-195-5p), 0.637 (miR-20a-3p) and 0.652 (miR-296-5p). At the same time, the AUC of the combined detection of the six miRNAs was 0.703, significantly improved the diagnostic accuracy of GC patients (199). Another study showed that the AUC values of serum exosomal miR-19b-3p and miR-106a-5p were 0.813 and 0.806 respectively. The AUC of their combined diagnosis was 0.826 (154). Similarly, in urinary exosomes from patients with renal clear cell carcinoma (ccRCC), different combinations of miRNAs, including miR-126-3p + miR-449a, miR-126-3p + miR-34b-5p, miR-126-3p + miR-486-5p, miR-25-3p + miR-34b-5p, miR-34b-5p, miR-2 b-5p-34 b-5p and miR-150-5 p + miR-126-3p have been reported to be potential diagnostic biomarkers in ccRCC patients. The sensitivities of these six combinations were 60.6%, 67.3%, 52.9%, 73.1%, 74%, and 61.5% respectively. Accordingly, specificities were 100%, 82.8%, 95.8%, 79.3%, 72.4%, and 82.8%, respectively. Furthermore, the targets of these miRNAs may be related to cell cycle regulation, tumorigenesis and angiogenesis (200). Muramatsu-Maekawa et al. (201) stated that miRNA-4525 in serum EVs is significantly higher expression in patients with advanced renal cell carcinoma (RCC) (201). Initially, serum exosomal miR-17-5p and miR-21 levels were considered as potential biomarkers for the differentiation of primary adenocarcinoma (PC). The mean levels of miR-17-5p and miR-21 were significantly higher in PC patients than in healthy controls (HPs) and non-PC groups, and the AUC values for miR-17-5p and miR-21 were 0.887 and 0.897 respectively, and the sensitivity and specificity of miR-17-5p were 72.7% and 92.6%, and 95.5% and 81.5% for miR-21 respectively (93). Subsequently, serum exosomal miRNAs (including miR-1246, miR-4644, miR-3976, and miR-4306) were also proposed as potential diagnostic biomarkers for pancreatic cancer (202). Notably, Manterola et al. (203) found that serum exosomal miR-320 and miR-574-3p were significantly higher expression in patients with glioblastoma multiforme (GBM) as compared with healthy controls, and ROC curve analysis indicated AUC for exosomal miR-320 and miR-574-3p of 0.720 and 0.738 respectively (203). In conclusion, exosomal miRNAs may be regarded as potential biomarkers of diseases.
In addition to miRNAs, exosomal lncRNAs are also attractive as potential diagnostic biomarkers. Long noncoding RNA (lncRNA) exists in the nucleus or cytoplasm, and they can interact with DNA, RNA, or proteins (204). Several studies have shown that exosomal lncRNAs may have the potential to act as biomarkers for cancer diagnosis. For example, plasma expression of lncUEGC1 was significantly higher in gastric cancer (GC) patients of stage I or II, and plasma exosomal lncUEGC1 (AUC =0.8760) was significantly superior to serum CEA (AUC = 0.6614). This suggests that exosomal lncUEGC1 may be a highly potential sensitive biomarker in early gastric cancer diagnosis (107). In addition, serum exosomal lncRNA HOTTIP was found to be a potential diagnostic index for gastric cancer patients. The ROC curve indicated that HOTTIP had high diagnostic value with an AUC value of 0.827 and higher diagnostic power than CEA, CA19-9 and CA72-4 (AUC values of 0.653, 0.685 and 0.639, respectively). It’s important that HOTTIP expression level was significantly correlated with the depth of invasion and TNM stage in gastric cancer (108). Another study confirmed that circulating exosomal long noncoding RNA-GC1 (lncRNA-GC1) expression could distinguish early gastric cancer patients and healthy controls, and ROC curve indicated that better exosomal lncRNA-GC1 (AUC=0.9033) compared to serum CEA, CA72-4 and CA19-9 (AUC values of 0.5987,0.6816 and 0.6482, respectively) (109). In addition, LINC00152 was also significantly elevated in the plasma exosomes of gastric cancer patients. Elevated exosomal LINC00152 was considered as a potential diagnostic indicator of gastric cancer with an AUC value of 0.657 (205). Similarly, Xiao et al. (110) demonstrated that lncRNA CCAT1 was significantly higher in serum EVs in gastric cancer patients than in healthy controls, chronic gastritis or dysplasia, with EVs lncRNA CCAT1 having an AUC of 0.890, sensitivity of 79.6%, specificity of 92.6%, while EVs lncRNA CCAT1 and embryo antibody combinations of 0.910 of 80.5% and 92.6% respectively. Moreover, EVs lncRNA CCAT1 may promote gastric cancer cells proliferation, migration and invasion through c-Myc or Bmi-1 upmodulation (110).
Circular RNA (circRNA) is a class of noncoding RNA, mainly produced by pre-mRNA splicing. In contrast to miRNA, circRNA is abnormally stable, conserved and has cells or tissue-specific expression pattern (206). Exosomal circRNAs are anti-degradative, and its secretion into the extracellular environment can be used for many biological applications. Importantly, exosomal circRNAs may serve as novel diagnostic biomarkers. For example, Shao et al. (115) found that the expression of plasma exosomal hsa_circ_0065149 was significantly reduced in gastric cancer patients compared with healthy cohort, suggesting that reduced hsa_circ_0065149 is a potential diagnostic biomarker for gastric cancer (AUC=0.640) (115). Similarly, Xie et al. (116) found significant higher serum circSHKBP1 level in gastric cancer patients with a sharp decrease in exosomal circSHKBP1 after surgical resection of the tumor (116). A previous study in plasma EVs from breast cancer patients proved that nine circRNAs (including hsa_circ_0002190, hsa_circ_0007177, hsa_circ_0000642, hsa_circ_0001439, hsa_circ_0001417, hsa_circ_0005552, hsa_circ_0001073, hsa_circ_0000267 and hsa_circ_04004) combinations display maximum AUC values, and the AUC is 0.83 (207). In cholangiocarcinoma, circ-0000284 was significantly elevated in cholangiocarcinoma cell lines, its tissues and plasma exosomes, and higher expression of circ-0000284 promoted the migration, invasion and proliferation capacity of cholangiocarcinoma cells in vitro and in vivo (208). Therefore, the exosomal circ-0000284 could be used as a potential metastatic diagnostic biomarker. Circulating exosomal hsa-circ-0004771 was significantly upregulated in colorectal cancer (CRC) patients and AUC values of hsa-circ-0004771 were 0.59, 0.86 and 0.88 in differentiating between intercancer, stage I/II and CRC patients and healthy controls respectively, suggesting that hsa-circ-0004771 could serve as a new potential diagnostic biomarker for CRC patients (209). Moreover, exosomal circRNAs in serum and urine have the potential to act as diagnostic biomarker for idiopathic membranous nephropathy (IMN) (210). In short, these studies suggest that exosomal circRNAs have the possibility of act as biomarkers for disease diagnosis. However, whether its expression levels are specific for different disease and tumor subtypes remains to be further investigated.
In addition to nucleic acids, exosomal proteins have been found to act as potential biomarkers for diseases. Because exosomes contain multiple protein molecules that reflect the characteristics of its parental cells (211). Exosomal proteins have been found in different body fluids (including serum, plasma, urine, saliva and cerebrospinal fluid) and may have the potential to serve as biomarkers for cancer diagnosis. For example, the cell surface proteoglycan Glypican-1 (GPC1), a member of the heparan sulfate proteoglycan family, is a widespread cell surface protein (212). It has been suggested that GPC1-positive exosomal was highly expressed in the serum of pancreatic cancer patients, and the diagnostic power of the exosomal protein GPC1 (AUC = 1.0) was significantly better than CA19-9 (AUC =0.739) in distinguishing pancreatic cancer patients from healthy controls. CA19-9 serum levels cannot distinguish patients with intraductal papillary mucinous tumors (PCPL) from healthy controls, while GPC1-positive serum exosomal had 100% sensitivity and specificity in all stages of pancreatic cancer (e. g.: cancer in situ, stage I, and stage II-IV) (120). Similarly, the exosomal protein GPC1 expression was significantly increased in both plasma and tissue samples of colorectal cancer (CRC) patients, and both normalized after surgical treatment (213). Another study indicated that the downregulation of serum exosomal Gastrokine 1 (GKN1) protein may be a valid diagnostic biomarker in gastric cancer patients (129).
Recently, the proteomic analysis of extracellular vesicles and granules (EVP) from 426 human samples derived from tissue explants (TE), plasma and other body fluids by Hoshino et al. (214). They confirmed that CD63 and flotillins were heterogeneous in plasma and tissue EVP. And Leucine-rich repeat protein 26 (LRRC26), ATP-dependent translocase ABCB1 (ABCB1), Bile salt export pump (ABCB11), Adhesion G protein-coupled receptor G6 (ADGRG6), Desmosomes-1 (DSC1), Desmoglein-1 (DSG1), Keratin and Plasminogen-like protein B (PLGLB1) were present only in plasma-derived EVP in patients with pancreatic cancer (PaCa), absent or extremely low expression in tumor tissue (TT) and adjacent normal tissue (AT) -derived EVP. This suggests that these proteins have the potential to act as characteristic tumor-associated EVP proteins. In addition, they said that EVP proteins can distinguish between cancer in the early stages of pancreatic cancer (PaCa) and lung adenocarcinoma (Luca) patients (214). It is interesting that, by proteomic analysis of Sun et al. (215), Annexin family members (Annexin A1, A2, A3, A5, A6, A11), Nitrogen permease regulator 2-like protein(NPRL2), Carcinoembryonic antigen-related cell adhesion molecule 1(CEACAM1), Mucin 1(MUC1), Prominin-1 (PROM1), Histone H4 (HIST1H4A) and Tumor necrosis factor alpha-induced protein 3 (TNFAIP3) were associated with lung cancer, which is helpful in lung cancer diagnosis (215). The expression levels of plasma exosomal Tim-3 and Galectin-9 protein molecules were significantly increased in non-small-cell lung cancer (NSCLC) patients, as compared with healthy controls. It’s important that exosomal Tim-3 and Galectin-9 expression levels were positively correlated with clinicopathological features such as patient age, tumor size, distant metastasis and cancer stage. Moreover, exosomal Tim-3 is also associated with lymph node metastasis. Therefore, exosomal Tim-3 and Galectin-9 may serve as potential biomarkers for the clinical application of NSCLC (216). All of these findings suggest that exosomal proteins have the potential to serve as biomarkers for disease diagnosis. In the future, we still need to focus on the expression levels of specific proteins in a certain disease.
Lipid molecules in exosomes are mainly used to maintain their external morphology. It has been reported that lipid molecules in EVs can not only protect nucleic acids and protein contents from harmful stimuli in the extracellular environment, but also exert bioactive functions to participate in tumor biological processes as signaling molecules (217, 218). It has been shown that lipid molecules in exosomes can also be used as potential biomarkers in cancer patients (136, 219–222). Among them, the expression levels of phosphatidylcholine(PC), phosphatidylethanolamine (PE),phosphatidylinositol(PI),sphingomyelin(SM),ceramide(Cer) and cholesterol are various in difference diseases (150, 223–225).
Previously, Skotland et al. (223) pointed out that urinary exosomal lipid molecules (such as phosphoresterdylserine and lactoceramide) have potential as biomarkers in prostate cancer (134). Subsequently, Brzozowski et al. (226) performed lipid analysis in exosomes released from non-tumorigenic (RWPE1), tumorigenic (NB26) and metastatic (PC-3) prostate cell lines, and they found significant differences in lipid species abundance in cells of these three different prostate species. The abundance of Diacylglycerol (DG) and Triacylglycerol (TG) species were reduced in both the NB26 and PC-3 cell lines EVs as compared to the EVs in the RWPE1 cell line. However, in contrast to EVs in the RWPE1 cell line, EVs in the NB2 and PC-3 cell lines were rich in glycerophospholipids, while Cer and SM species do not differ much among the three cell lines (226). In addition, Exosomal lipid components have been detected in Hepatocellular Carcinoma (HepG2/C3a and Huh7 cells) (227), Melanoma (B16-F10 cells) (228), Glioblastoma (U87 cells) (229)and Pancreatic cancer (AsPC-1 cells) (230). Recently, Glover et al. (135) stated that the content of exosomal lipid molecules such as glycerophospholipids, glycerolips, and sterols is reduced in the urine of patients with hereditary-trypsinaemia (135). Overexpression of exosomal lipid molecules such as acid sphingolipase in the cerebrospinal fluid of multiple sclerosis(MS) patients is strongly associated with disease severity, creating new opportunities for the diagnosis and treatment of the disease (137). Furthermore, sphingomyelin, derived from EVs in tumor cells, promotes endothelial cell migration and angiogenesis during tumor growth and metastasis (231). To sum up, this suggests that the great potential of EVs lipid molecules for cancer diagnostic biomarkers.
In conclusion, exosomal nucleic acids, proteins and lipid molecular cargoes in different body fluids have broad application prospects as cancer diagnostic biomarkers (Table 3). Previous researches have shown that exosomal molecular cargoes are differentially expressed in body fluids, and exosomal molecular cargoes with higher AUC values may effectively distinguish cancer patients from healthy individuals (232–234). It is worth noting that the combined detection of multiple potential exosome molecular cargoes may provide a rapid, reliable and non-invasive aid to the diagnosis of diseases. In addition, that exosomes used as diagnostic biomarkers also requires consideration of all preanalytical variables associated with sample collection, such as whole blood (or other biofluid) treatment, hemolysis interference, and other contaminant interference (235). In the future, we should also focus on large-scale preparation and standardized protocols for exosomes analysis, and need advanced techniques to minimize contaminants in the samples as well.
In this review, we illustrate that exosomal molecular cargoes participate in exosome biogenesis, which is a complex process that may vary in cargoes or cellular origin. In addition, the regulation of exosome biogenesis processes involves the coordination of many different molecular cargoes and signaling mechanisms, mainly dominated by ESCRT-dependent, lipid raft and tetraspanin protein mechanisms, and Rab proteins further assists cargo sorting and exosome release. Notably, this cargo molecules interact with each other to mainly mediate exosome biogenesis by regulating the negative curvature of the cell membrane (236). So far, ESCRT and ceramide pathways are established for exosome biogenesis.
Furthermore, exosomes and their molecular cargoes are elaborated as effective tools for the diagnosis of cancer. Although tissue biopsy is still the gold standard for tumor diagnosis, but it is invasive. An ideal diagnostic approach for cancer should accurately detect tumor-specific biomarkers using non-invasive techniques at the pre-metastatic stage (237). Most of the current molecules used as tumor diagnostic biomarkers are based on detecting the higher expression molecules above the threshold in healthy individuals. For instance, PSA and CEA serve as diagnostic biomarkers for prostate cancer and gastrointestinal cancer respectively, and these biomarkers are significantly elevated only at tumor progression state (238). Since exosomes are present in most body fluids and their stability properties, and the molecular cargoes carried by exosomes reflects the genetic or signaling changes in the cancer cells of origin. If it would be detected earlier as biomarkers, so as to achieve a means of treating the disease, it would make exosomes potentially replace invasive biopsies as cancer diagnostic biomarkers of important clinical significance (239, 240).
Understanding the process of exosome biogenesis is an important part of the research and physiological significance of exosomes function, especially for disease diagnosis, treatment, and prognosis. Controlling exosome generation in pathological states may serve as a therapeutic opportunity to reduce tumorigenesis. However, it is still challenging to investigate the whole mechanism of exosome biogenesis. Because the exosome formation pathway may be different according to different cell types, some specific molecules will participate in multiple processes, leading to the exact mechanism of action of many molecules is not clear, for which their heterogeneity may be a disadvantage of their use as biomarkers. It is worth noting that most studies in the field of exosomes are conducted in vitro, and the laboratory culture conditions or technical methods also affect the biological characteristics of exosomes (241). Therefore, special attention should also be paid to the methods of exosomes extraction used in each study. How to promote the yield and purity of exosomes is a top priority, which has been a bottleneck limiting their translational applications. Recent studies have shown that appropriate combinations of several methods for extracting and purifying exosomes can effectively improve the above problems, and how to integrate them for optimum results remains to be further investigated. More work needs to be done in the future to elucidate the role of exosomes in diseases progression, with particular attention to the precise mechanisms by which exosome biogenesis pathways influence cellular function. The questions will be raised such as, will different biogenesis pathways produce vesicles with different or similar functions? Will there be any correlation between vesicles produced by this biogenesis pathways? This will be useful for treatments involving the pathological mechanisms of exosomes. Understanding the physiological effects and how they can be induced into pathological factors is crucial when developing new therapeutic strategies.
XY searched for literature and wrote the first draft of this article,JC revised the manuscript and developed the main content of this manuscript. SF provided great help for polishing the manuscript. DH, TY, ZL, XW, MZ, and JW were involved in edited the manuscript. TZ supervised the project and contributed to the revision of the final manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Key R&D Planning Project of Jiangxi Science and Technology Commission, China (No. 20203BBGL73126).
We thank Yaojiang Que edited the figure, Zhigang Li reviewed the manuscript and polished the grammar.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gandham S, Su X, Wood J, Nocera AL, Alli SC, Milane L, et al. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol (2020) 38:1066–98. doi: 10.1016/j.tibtech.2020.05.012
2. Barreiro K, Dwivedi OP, Valkonen S, Groop PH, Tuomi T, Holthofer H, et al. Urinary extracellular vesicles: Assessment of pre-analytical variables and development of a quality control with focus on transcriptomic biomarker research. J extracellular vesicles (2021) 10:e12158. doi: 10.1002/jev2.12158
3. Dutta S, Hornung S, Kruayatidee A, Maina KN, Del Rosario I, Paul KC, et al. α-synuclein in blood exosomes immunoprecipitated using neuronal and oligodendroglial markers distinguishes parkinson's disease from multiple system atrophy. Acta neuropathologica (2021) 142:495–511. doi: 10.1007/s00401-021-02324-0
4. Yan C, Chen J, Wang C, Yuan M, Kang Y, Wu Z, et al. Milk exosomes-mediated miR-31-5p delivery accelerates diabetic wound healing through promoting angiogenesis. Drug delivery (2022) 29:214–28. doi: 10.1080/10717544.2021.2023699
5. Li K, Lin Y, Luo Y, Xiong X, Wang L, Durante K, et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer (2022) 21:21. doi: 10.1186/s12943-022-01499-8
6. Li Y, Gu J, Mao Y, Wang X, Li Z, Xu X, et al. Cerebrospinal fluid extracellular vesicles with distinct properties in autoimmune encephalitis and herpes simplex encephalitis. Mol Neurobiol (2022) 59:2441–55. doi: 10.1007/s12035-021-02705-2
7. Li P, Lu X, Hu J, Dai M, Yan J, Tan H, et al. Human amniotic fluid derived-exosomes alleviate hypoxic encephalopathy by enhancing angiogenesis in neonatal mice after hypoxia. Neurosci Lett (2022) 768:136361. doi: 10.1016/j.neulet.2021.136361
8. Su Q, Zhang Y, Cui Z, Chang S, Zhao P. Semen-derived exosomes mediate immune escape and transmission of reticuloendotheliosis virus. Front Immunol (2021) 12:735280. doi: 10.3389/fimmu.2021.735280
9. Sadik N, Cruz L, Gurtner A, Rodosthenous RS, Dusoswa SA, Ziegler O, et al. Extracellular RNAs: A new awareness of old perspectives. Methods Mol Biol (Clifton N.J.) (2018) 1740:1–15. doi: 10.1007/978-1-4939-7652-2_1
10. Markowska A, Pendergrast RS, Pendergrast JS, Pendergrast PS. A novel method for the isolation of extracellular vesicles and RNA from urine. J circulating Biomarkers (2017) 6:1849454417712666. doi: 10.1177/1849454417712666
11. Merchant ML, Rood IM, Deegens JKJ, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol (2017) 13:731–49. doi: 10.1038/nrneph.2017.148
12. Vitorino R, Ferreira R, Guedes S, Amado F, Thongboonkerd V. What can urinary exosomes tell us? Cell Mol Life Sci CMLS (2021) 78:3265–83. doi: 10.1007/s00018-020-03739-w
13. Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer (2022) 21:56. doi: 10.1186/s12943-022-01509-9
14. Burkova EE, Sedykh SE, Nevinsky GA. Human placenta exosomes: Biogenesis, isolation, composition, and prospects for use in diagnostics. Int J Mol Sci (2021) 22. doi: 10.3390/ijms22042158
15. Yamashita T, Takahashi Y, Nishikawa M, Takakura Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur J pharmaceutics biopharmaceutics Off J Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V (2016) 98:1–8. doi: 10.1016/j.ejpb.2015.10.017
16. Li S, Yi M, Dong B, Tan X, Luo S, Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer (2021) 148:2640–51. doi: 10.1002/ijc.33386
17. Wang YT, Shi T, Srivastava S, Kagan J, Liu T, Rodland KD. Proteomic analysis of exosomes for discovery of protein biomarkers for prostate and bladder cancer. Cancers (2020) 12. doi: 10.3390/cancers12092335
18. Wijenayake S, Eisha S, Tawhidi Z, Pitino MA, Steele MA, Fleming AS, et al. Comparison of methods for pre-processing, exosome isolation, and RNA extraction in unpasteurized bovine and human milk. PLoS One (2021) 16:e0257633. doi: 10.1371/journal.pone.0257633
19. Sidhom K, Obi PO, Saleem A. A review of exosomal isolation methods: Is size exclusion chromatography the best option? Int J Mol Sci (2020) 21. doi: 10.3390/ijms21186466
20. Sedykh S, Kuleshova A, Nevinsky G. Milk exosomes: Perspective agents for anticancer drug delivery. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21186646
21. Vaswani K, Koh YQ, Almughlliq FB, Peiris HN, Mitchell MD. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod Biol (2017) 17:341–8. doi: 10.1016/j.repbio.2017.09.007
22. Cheshmi B, Cheshomi H. Salivary exosomes: properties, medical applications, and isolation methods. Mol Biol Rep (2020) 47:6295–307. doi: 10.1007/s11033-020-05659-1
23. Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: Emerging roles in systemic disease. Int J Biol Sci (2018) 14:633–43. doi: 10.7150/ijbs.25018
24. Cheng J, Nonaka T, Wong DTW. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials (Basel Switzerland) (2019) 12. doi: 10.3390/ma12040654
25. Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Korvala J, Salo T, Sormunen R, et al. Human saliva-derived exosomes: comparing methods of isolation. J Histochem Cytochem Off J Histochem Soc (2015) 63:181–9. doi: 10.1369/0022155414564219
26. Tan YJ, Wong BYX, Vaidyanathan R, Sreejith S, Chia SY, Kandiah N, et al. Altered cerebrospinal fluid exosomal microRNA levels in young-onset alzheimer's disease and frontotemporal dementia. J Alzheimer's Dis Rep (2021) 5:805–13. doi: 10.3233/adr-210311
27. Thompson AG, Gray E, Mager I, Fischer R, Thézénas ML, Charles PD, et al. UFLC-derived CSF extracellular vesicle origin and proteome. Proteomics (2018) 18:e1800257. doi: 10.1002/pmic.201800257
28. Yao YF, Qu MW, Li GC, Zhang FB, Rui HC. Circulating exosomal miRNAs as diagnostic biomarkers in parkinson's disease. Eur Rev Med Pharmacol Sci (2018) 22:5278–83. doi: 10.26355/eurrev_201808_15727
29. Hou X, Gong X, Zhang L, Li T, Yuan H, Xie Y, et al. Identification of a potential exosomal biomarker in spinocerebellar ataxia type 3/Machado-Joseph disease. Epigenomics (2019) 11:1037–56. doi: 10.2217/epi-2019-0081
30. Thompson AG, Gray E, Mäger I, Thézénas ML, Charles PD, Talbot K, et al. CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin Proteomics (2020) 17:31. doi: 10.1186/s12014-020-09294-7
31. Ding K, Yu L, Huang Z, Zheng H, Yang X, Tian T, et al. [Differential expression profile of miRNAs in amniotic fluid exosomes from fetuses with down syndrome]. Nan fang yi ke da xue xue bao = J South Med Univ (2022) 42:293–9. doi: 10.12122/j.issn.1673-4254.2022.02.18
32. Tang GY, Yu P, Zhang C, Deng HY, Lu MX, Le JH. The neuropeptide-related HERC5/TAC1 interactions may be associated with the dysregulation of lncRNA GAS5 expression in gestational diabetes mellitus exosomes. Dis Markers (2022) 2022:8075285. doi: 10.1155/2022/8075285
33. Baskaran S, Panner Selvam MK, Agarwal A. Exosomes of male reproduction. Adv Clin Chem (2020) 95:149–63. doi: 10.1016/bs.acc.2019.08.004
34. Yang C, Guo WB, Zhang WS, Bian J, Yang JK, Qi T, et al. [Extraction and identification of semen-derived exosomes using PEG6000]. Nan fang yi ke da xue xue bao = J South Med Univ (2016) 36:1531–5.
35. Mercadal M, Herrero C, López-Rodrigo O, Castells M, de la Fuente A, Vigués F, et al. Impact of extracellular vesicle isolation methods on downstream mirna analysis in semen: A comparative study. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21175949
36. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J extracellular vesicles (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750
37. Sasaki R, Kanda T, Yokosuka O, Kato N, Matsuoka S, Moriyama M. Exosomes and hepatocellular carcinoma: From bench to bedside. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20061406
38. Gurunathan S, Kang MH, Qasim M, Khan K, Kim JH. Biogenesis, membrane trafficking, functions, and next generation nanotherapeutics medicine of extracellular vesicles. Int J nanomedicine (2021) 16:3357–83. doi: 10.2147/ijn.S310357
39. Modani S, Tomar D, Tangirala S, Sriram A, Mehra NK, Kumar R, et al. An updated review on exosomes: biosynthesis to clinical applications. J Drug Targeting (2021) 29:925–40. doi: 10.1080/1061186x.2021.1894436
40. Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol (2015) 35:69–77. doi: 10.1016/j.ceb.2015.04.013
41. Huang D, Chen J, Hu D, Xie F, Yang T, Li Z, et al. Advances in biological function and clinical application of small extracellular vesicle membrane proteins. Front Oncol (2021) 11:675940. doi: 10.3389/fonc.2021.675940
42. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci CMLS (2018) 75:193–208. doi: 10.1007/s00018-017-2595-9
43. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Sci (New York N.Y.) (2020) 367. doi: 10.1126/science.aau6977
44. Huotari J, Helenius A. Endosome maturation. EMBO J (2011) 30:3481–500. doi: 10.1038/emboj.2011.286
45. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem (2019) 88:487–514. doi: 10.1146/annurev-biochem-013118-111902
46. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev (2018) 118:1917–50. doi: 10.1021/acs.chemrev.7b00534
47. Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer (2022) 21:54. doi: 10.1186/s12943-022-01525-9
48. Bebelman MP, Smit MJ, Pegtel DM, Baglio SR. Biogenesis and function of extracellular vesicles in cancer. Pharmacol Ther (2018) 188:1–11. doi: 10.1016/j.pharmthera.2018.02.013
49. Bestard-Escalas J, Maimó-Barceló A, Lopez DH, Reigada R, Guardiola-Serrano F, Ramos-Vivas J, et al. Common and differential traits of the membrane lipidome of colon cancer cell lines and their secreted vesicles: Impact on studies using cell lines. Cancers (2020) 12. doi: 10.3390/cancers12051293
50. Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol (2014) 5:442. doi: 10.3389/fimmu.2014.00442
51. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics (2010) 73:1907–20. doi: 10.1016/j.jprot.2010.06.006
52. van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem (2006) 140:13–21. doi: 10.1093/jb/mvj128
53. Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis (2010) 16:34–8. doi: 10.1111/j.1601-0825.2009.01604.x
54. Zhang C, Ji Q, Yang Y, Li Q, Wang Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol Cancer Res Treat (2018) 17:1533033818763450. doi: 10.1177/1533033818763450
55. Bestard-Escalas J, Reigada R, Reyes J, de la Torre P, Liebisch G, Barceló-Coblijn G. Fatty acid unsaturation degree of plasma exosomes in colorectal cancer patients: A promising biomarker. Int J Mol Sci (2021) 22. doi: 10.3390/ijms22105060
56. Melero-Fernandez de Mera RM, Villaseñor A, Rojo D, Carrión-Navarro J, Gradillas A, Ayuso-Sacido A, et al. Ceramide composition in exosomes for characterization of glioblastoma stem-like cell phenotypes. Front Oncol (2021) 11:788100. doi: 10.3389/fonc.2021.788100
57. Lin M, Liao W, Dong M, Zhu R, Xiao J, Sun T, et al. Exosomal neutral sphingomyelinase 1 suppresses hepatocellular carcinoma via decreasing the ratio of sphingomyelin/ceramide. FEBS J (2018) 285:3835–48. doi: 10.1111/febs.14635
58. Zhao L, Gu C, Gan Y, Shao L, Chen H, Zhu H. Exosome-mediated siRNA delivery to suppress postoperative breast cancer metastasis. J Controlled release Off J Controlled Release Soc (2020) 318:1–15. doi: 10.1016/j.jconrel.2019.12.005
59. Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol (2012) 44:1574–84. doi: 10.1016/j.biocel.2012.06.018
60. Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol (2003) 19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609
61. Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Martín S, Ursa A, Sánchez-Madrid F, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem (2013) 288:11649–61. doi: 10.1074/jbc.M112.445304
62. Lynch S, Santos SG, Campbell EC, Nimmo AM, Botting C, Prescott A, et al. Novel MHC class I structures on exosomes. J Immunol (Baltimore Md. 1950) (2009) 183:1884–91. doi: 10.4049/jimmunol.0900798
63. Gauvreau ME, Côté MH, Bourgeois-Daigneault MC, Rivard LD, Xiu F, Brunet A, et al. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic (Copenhagen Denmark) (2009) 10:1518–27. doi: 10.1111/j.1600-0854.2009.00948.x
64. Caruso Bavisotto C, Cappello F, Macario AJL, Conway de Macario E, Logozzi M, Fais S, et al. Exosomal HSP60: a potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev Mol diagnostics (2017) 17:815–22. doi: 10.1080/14737159.2017.1356230
65. Lauwers E, Wang YC, Gallardo R, Van der Kant R, Michiels E, Swerts J, et al. Hsp90 mediates membrane deformation and exosome release. Mol Cell (2018) 71:689–702.e689. doi: 10.1016/j.molcel.2018.07.016
66. Larios J, Mercier V, Roux A, Gruenberg J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol (2020) 219. doi: 10.1083/jcb.201904113
67. Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood cells molecules Dis (2005) 35:398–403. doi: 10.1016/j.bcmd.2005.08.005
68. Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol (Baltimore Md. 1950) (2001) 166:7309–18. doi: 10.4049/jimmunol.166.12.7309
69. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol (2010) 12:19–30; sup pp 11-13. doi: 10.1038/ncb2000
70. Auger C, Brunel A, Darbas T, Akil H, Perraud A, Bégaud G, et al. Extracellular vesicle measurements with nanoparticle tracking analysis: A different appreciation of up and down secretion. Int J Mol Sci (2022) 23. doi: 10.3390/ijms23042310
71. Fukuda M. Regulation of secretory vesicle traffic by rab small GTPases. Cell Mol Life Sci CMLS (2008) 65:2801–13. doi: 10.1007/s00018-008-8351-4
72. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9
73. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell bioscience (2019) 9:19. doi: 10.1186/s13578-019-0282-2
74. Verweij FJ, Bebelman MP, Jimenez CR, Garcia-Vallejo JJ, Janssen H, Neefjes J, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol (2018) 217:1129–42. doi: 10.1083/jcb.201703206
75. Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun (2017) 8:14041. doi: 10.1038/ncomms14041
76. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Sci (New York N.Y.) (2008) 319:1244–7. doi: 10.1126/science.1153124
77. Menck K, Sönmezer C, Worst TS, Schulz M, Dihazi GH, Streit F, et al. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J extracellular vesicles (2017) 6:1378056. doi: 10.1080/20013078.2017.1378056
78. Inuzuka T, Inokawa A, Chen C, Kizu K, Narita H, Shibata H, et al. ALG-2-interacting tubby-like protein superfamily member PLSCR3 is secreted by an exosomal pathway and taken up by recipient cultured cells. Bioscience Rep (2013) 33:e00026. doi: 10.1042/bsr20120123
79. Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie (2007) 89:205–12. doi: 10.1016/j.biochi.2006.10.014
80. Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 glued and late endosome positioning. J Cell Biol (2009) 185:1209–25. doi: 10.1083/jcb.200811005
81. Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem (2009) 284:34211–22. doi: 10.1074/jbc.M109.041152
82. Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, et al. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry (2005) 44:2097–102. doi: 10.1021/bi0478502
83. Kooijman EE, Chupin V, de Kruijff B, Burger KN. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic (Copenhagen Denmark) (2003) 4:162–74. doi: 10.1034/j.1600-0854.2003.00086.x
84. Morel E, Chamoun Z, Lasiecka ZM, Chan RB, Williamson RL, Vetanovetz C, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun (2013) 4:2250. doi: 10.1038/ncomms3250
85. Raiborg C, Schink KO, Stenmark H. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J (2013) 280:2730–42. doi: 10.1111/febs.12116
86. Hessvik NP, Øverbye A, Brech A, Torgersen ML, Jakobsen IS, Sandvig K, et al. PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell Mol Life Sci CMLS (2016) 73:4717–37. doi: 10.1007/s00018-016-2309-8
87. Kajimoto T, Okada T, Miya S, Zhang L, Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat Commun (2013) 4:2712. doi: 10.1038/ncomms3712
88. Shephard AP, Giles P, Mbengue M, Alraies A, Spary LK, Kynaston H, et al. Stroma-derived extracellular vesicle mRNA signatures inform histological nature of prostate cancer. J extracellular vesicles (2021) 10:e12150. doi: 10.1002/jev2.12150
89. Dong Z, Sun X, Xu J, Han X, Xing Z, Wang D, et al. Serum membrane type 1-matrix metalloproteinase (MT1-MMP) mRNA protected by exosomes as a potential biomarker for gastric cancer. Med Sci monitor Int Med J Exp Clin Res (2019) 25:7770–83. doi: 10.12659/msm.918486
90. Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med (2018) 56:479–84. doi: 10.1515/cclm-2017-0327
91. Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. OncoTargets Ther (2016) 9:139–48. doi: 10.2147/ott.S95565
92. Rodríguez M, Bajo-Santos C, Hessvik NP, Lorenz S, Fromm B, Berge V, et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer (2017) 16:156. doi: 10.1186/s12943-017-0726-4
93. Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol (2013) 11:219. doi: 10.1186/1477-7819-11-219
94. Xu YF, Hannafon BN, Zhao YD, Postier RG, Ding WQ. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget (2017) 8:77028–40. doi: 10.18632/oncotarget.20332
95. Machida T, Tomofuji T, Maruyama T, Yoneda T, Ekuni D, Azuma T, et al. miR−1246 and miR−4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep (2016) 36:2375–81. doi: 10.3892/or.2016.5021
96. Xu Y, Xu L, Zheng J, Geng L, Zhao S. MiR-101 inhibits ovarian carcinogenesis by repressing the expression of brain-derived neurotrophic factor. FEBS Open Bio (2017) 7:1258–66. doi: 10.1002/2211-5463.12257
97. Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol (2019) 25:1890–8. doi: 10.3748/wjg.v25.i15.1890
98. Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surgeons (2019) 19:3250–62. doi: 10.1111/ajt.15490
99. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med (2018) 7:1670–9. doi: 10.1002/cam4.1390
100. Min L, Chen L, Liu S, Yu Y, Guo Q, Li P, et al. Loss of circulating exosomal miR-92b is a novel biomarker of colorectal cancer at early stage. Int J Med Sci (2019) 16:1231–7. doi: 10.7150/ijms.34540
101. Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, et al. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer (2020) 11:630–7. doi: 10.7150/jca.33022
102. Zhong Y, Ding X, Bian Y, Wang J, Zhou W, Wang X, et al. Discovery and validation of extracellular vesicle-associated miRNAs as noninvasive detection biomarkers for early-stage non-small-cell lung cancer. Mol Oncol (2021) 15:2439–52. doi: 10.1002/1878-0261.12889
103. Zhou L, Wang W, Wang F, Yang S, Hu J, Lu B, et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol Cancer (2021) 20:57. doi: 10.1186/s12943-021-01352-4
104. Yang H, Fu H, Wang B, Zhang X, Mao J, Li X, et al. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol carcinogenesis (2018) 57:1223–36. doi: 10.1002/mc.22838
105. Wei S, Peng L, Yang J, Sang H, Jin D, Li X, et al. Exosomal transfer of miR-15b-3p enhances tumorigenesis and malignant transformation through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric cancer. J Exp Clin Cancer Res CR (2020) 39:32. doi: 10.1186/s13046-019-1511-6
106. Liu J, Yoo J, Ho JY, Jung Y, Lee S, Hur SY, et al. Plasma-derived exosomal miR-4732-5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J Ovarian Res (2021) 14:59. doi: 10.1186/s13048-021-00814-z
107. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer (2018) 17:84. doi: 10.1186/s12943-018-0834-9
108. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer (2018) 17:68. doi: 10.1186/s12943-018-0817-x
109. Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: A multiphase study. JAMA Surg (2020) 155:572–9. doi: 10.1001/jamasurg.2020.1133
110. Xiao K, Dong Z, Wang D, Liu M, Ding J, Chen W, et al. Clinical value of lncRNA CCAT1 in serum extracellular vesicles as a potential biomarker for gastric cancer. Oncol Lett (2021) 21:447. doi: 10.3892/ol.2021.12708
111. Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer (2017) 16:143. doi: 10.1186/s12943-017-0714-8
112. Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer (2018) 17:143. doi: 10.1186/s12943-018-0880-3
113. Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer epidemiology Biomarkers Prev Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol (2018) 27:710–6. doi: 10.1158/1055-9965.Epi-17-0770
114. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer (2018) 17:74. doi: 10.1186/s12943-018-0822-0
115. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res POR (2020) 26:1475–82. doi: 10.1007/s12253-019-00716-y
116. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer (2020) 19:112. doi: 10.1186/s12943-020-01208-3
117. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer (2018) 17:137. doi: 10.1186/s12943-018-0888-8
118. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer (2020) 19:101. doi: 10.1186/s12943-020-01221-6
119. Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer (2022) 21:49. doi: 10.1186/s12943-021-01471-y
120. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature (2015) 523:177–82. doi: 10.1038/nature14581
121. Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One (2012) 7:e46737. doi: 10.1371/journal.pone.0046737
122. Li S, Zhao Y, Chen W, Yin L, Zhu J, Zhang H, et al. Exosomal ephrinA2 derived from serum as a potential biomarker for prostate cancer. J Cancer (2018) 9:2659–65. doi: 10.7150/jca.25201
123. Szajnik M, Derbis M, Lach M, Patalas P, Michalak M, Drzewiecka H, et al. Exosomes in plasma of patients with ovarian carcinoma: Potential biomarkers of tumor progression and response to therapy. Gynecol obstetrics (Sunnyvale Calif.) (2013) (Suppl 4):3. doi: 10.4172/2161-0932.S4-003
124. Wei P, Wu F, Kang B, Sun X, Heskia F, Pachot A, et al. Plasma extracellular vesicles detected by single molecule array technology as a liquid biopsy for colorectal cancer. J extracellular vesicles (2020) 9:1809765. doi: 10.1080/20013078.2020.1809765
125. Fu H, Yang H, Zhang X, Wang B, Mao J, Li X, et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res CR (2018) 37:162. doi: 10.1186/s13046-018-0825-0
126. Pan D, Chen J, Feng C, Wu W, Wang Y, Tong J, et al. Preferential localization of MUC1 glycoprotein in exosomes secreted by non-small cell lung carcinoma cells. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20020323
127. Moon PG, Lee JE, Cho YE, Lee SJ, Jung JH, Chae YS, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res an Off J Am Assoc Cancer Res (2016) 22:1757–66. doi: 10.1158/1078-0432.Ccr-15-0654
128. Moon PG, Lee JE, Cho YE, Lee SJ, Chae YS, Jung JH, et al. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget (2016) 7:40189–99. doi: 10.18632/oncotarget.9561
129. Yoon JH, Ham IH, Kim O, Ashktorab H, Smoot DT, Nam SW, et al. Gastrokine 1 protein is a potential theragnostic target for gastric cancer. Gastric Cancer Off J Int Gastric Cancer Assoc Japanese Gastric Cancer Assoc (2018) 21:956–67. doi: 10.1007/s10120-018-0828-8
130. Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Della Mina P, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst (2013) 9:1220–33. doi: 10.1039/c3mb25582d
131. Tian F, Zhang S, Liu C, Han Z, Liu Y, Deng J, et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat Commun (2021) 12:2536. doi: 10.1038/s41467-021-22913-7
132. Pang Y, Shi J, Yang X, Wang C, Sun Z, Xiao R. Personalized detection of circling exosomal PD-L1 based on Fe(3)O(4)@TiO(2) isolation and SERS immunoassay. Biosensors bioelectronics (2020) 148:111800. doi: 10.1016/j.bios.2019.111800
133. Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat Biomed Eng (2019) 3:438–51. doi: 10.1038/s41551-019-0356-9
134. Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer (Oxford Engl 1990) (2017) 70:122–32. doi: 10.1016/j.ejca.2016.10.011
135. Glover SC, Nouri MZ, Tuna KM, Mendoza Alvarez LB, Ryan LK, Shirley JF, et al. Lipidomic analysis of urinary exosomes from hereditary α-tryptasemia patients and healthy volunteers. FASEB bioAdvances (2019) 1:624–38. doi: 10.1096/fba.2019-00030
136. Tao L, Zhou J, Yuan C, Zhang L, Li D, Si D, et al. Metabolomics identifies serum and exosomes metabolite markers of pancreatic cancer. Metabolomics Off J Metabolomic Soc (2019) 15:86. doi: 10.1007/s11306-019-1550-1
137. Pieragostino D, Cicalini I, Lanuti P, Ercolino E, di Ioia M, Zucchelli M, et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of multiple sclerosis patients. Sci Rep (2018) 8:3071. doi: 10.1038/s41598-018-21497-5
138. Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells (2019) 8. doi: 10.3390/cells8040307
139. Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta (2014) 1841:108–20. doi: 10.1016/j.bbalip.2013.10.004
140. Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett (2020) 476:13–22. doi: 10.1016/j.canlet.2020.01.033
141. Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J extracellular vesicles (2015) 4:27066. doi: 10.3402/jev.v4.27066
142. Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol (2018) 8:66. doi: 10.3389/fonc.2018.00066
143. Barile L, Cervio E, Lionetti V, Milano G, Ciullo A, Biemmi V, et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: role of pregnancy-associated plasma protein-a. Cardiovasc Res (2018) 114:992–1005. doi: 10.1093/cvr/cvy055
144. Terrasini N, Lionetti V. Exosomes in critical illness. Crit Care Med (2017) 45:1054–60. doi: 10.1097/ccm.0000000000002328
145. Carrozzo A, Casieri V, Di Silvestre D, Brambilla F, De Nitto E, Sardaro N, et al. Plasma exosomes characterization reveals a perioperative protein signature in older patients undergoing different types of on-pump cardiac surgery. GeroScience (2021) 43:773–89. doi: 10.1007/s11357-020-00223-y
146. Chang W, Wang J. Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cells (2019) 8. doi: 10.3390/cells8080853
147. Elashiry M, Elashiry MM, Elsayed R, Rajendran M, Auersvald C, Zeitoun R, et al. Dendritic cell derived exosomes loaded with immunoregulatory cargo reprogram local immune responses and inhibit degenerative bone disease in vivo. J extracellular vesicles (2020) 9:1795362. doi: 10.1080/20013078.2020.1795362
148. Riazifar M, Mohammadi MR, Pone EJ, Yeri A, Lässer C, Segaliny AI, et al. Stem cell-derived exosomes as nanotherapeutics for autoimmune and neurodegenerative disorders. ACS nano (2019) 13:6670–88. doi: 10.1021/acsnano.9b01004
149. Zhang C, Wang XY, Zhang P, He TC, Han JH, Zhang R, et al. Cancer-derived exosomal HSPC111 promotes colorectal cancer liver metastasis by reprogramming lipid metabolism in cancer-associated fibroblasts. Cell Death Dis (2022) 13:57. doi: 10.1038/s41419-022-04506-4
150. Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res (2019) 60:9–18. doi: 10.1194/jlr.R084343
151. Skryabin GO, Komelkov AV, Savelyeva EE, Tchevkina EM. Lipid rafts in exosome biogenesis. Biochem Biokhimiia (2020) 85:177–91. doi: 10.1134/s0006297920020054
152. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics (2013) 14:319. doi: 10.1186/1471-2164-14-319
153. Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, et al. Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res CR (2018) 37:130. doi: 10.1186/s13046-018-0807-2
154. Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun (2017) 493:1322–8. doi: 10.1016/j.bbrc.2017.10.003
155. Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, et al. Lysosomal exocytosis, exosome release and secretory autophagy: The autophagic- and endo-lysosomal systems go extracellular. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21072576
156. Abdulrahman BA, Abdelaziz DH, Schatzl HM. Autophagy regulates exosomal release of prions in neuronal cells. J Biol Chem (2018) 293:8956–68. doi: 10.1074/jbc.RA117.000713
157. Zheng K, Ma J, Wang Y, He Z, Deng K. Sulforaphane inhibits autophagy and induces exosome-mediated paracrine senescence via regulating mTOR/TFE3. Mol Nutr Food Res (2020) 64:e1901231. doi: 10.1002/mnfr.201901231
158. Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, et al. Atg5 disassociates the V(1)V(0)-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell (2017) 43:716–30.e717. doi: 10.1016/j.devcel.2017.11.018
159. Keller MD, Ching KL, Liang FX, Dhabaria A, Tam K, Ueberheide BM, et al. Decoy exosomes provide protection against bacterial toxins. Nature (2020) 579:260–4. doi: 10.1038/s41586-020-2066-6
160. Jafari R, Rahbarghazi R, Ahmadi M, Hassanpour M, Rezaie J. Hypoxic exosomes orchestrate tumorigenesis: molecular mechanisms and therapeutic implications. J Trans Med (2020) 18:474. doi: 10.1186/s12967-020-02662-9
161. Choudhry H, Harris AL. Advances in hypoxia-inducible factor biology. Cell Metab (2018) 27:281–98. doi: 10.1016/j.cmet.2017.10.005
162. Zhang W, Zhou X, Yao Q, Liu Y, Zhang H, Dong Z. HIF-1-mediated production of exosomes during hypoxia is protective in renal tubular cells. American journal of physiology. Renal Physiol (2017) 313:F906–f913. doi: 10.1152/ajprenal.00178.2017
163. King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer (2012) 12:421. doi: 10.1186/1471-2407-12-421
164. Ban JJ, Lee M, Im W, Kim M. Low pH increases the yield of exosome isolation. Biochem Biophys Res Commun (2015) 461:76–9. doi: 10.1016/j.bbrc.2015.03.172
165. Wang Y, Yin K, Tian J, Xia X, Ma J, Tang X, et al. Granulocytic myeloid-derived suppressor cells promote the stemness of colorectal cancer cells through exosomal S100A9. Advanced Sci (Weinheim Baden-Wurttemberg Germany) (2019) 6:1901278. doi: 10.1002/advs.201901278
166. He G, Peng X, Wei S, Yang S, Li X, Huang M, et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol Cancer (2022) 21:19. doi: 10.1186/s12943-021-01440-5
167. Duan P, Tan J, Miao Y, Zhang Q. Potential role of exosomes in the pathophysiology, diagnosis, and treatment of hypoxic diseases. Am J Trans Res (2019) 11:1184–201.
168. Datta A, Kim H, McGee L, Johnson AE, Talwar S, Marugan J, et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci Rep (2018) 8:8161. doi: 10.1038/s41598-018-26411-7
169. Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem (2003) 278:20083–90. doi: 10.1074/jbc.M301642200
170. Serrano D, Bhowmick T, Chadha R, Garnacho C, Muro S. Intercellular adhesion molecule 1 engagement modulates sphingomyelinase and ceramide, supporting uptake of drug carriers by the vascular endothelium. Arteriosclerosis thrombosis Vasc Biol (2012) 32:1178–85. doi: 10.1161/atvbaha.111.244186
171. Kosgodage US, Trindade RP, Thompson PR, Inal JM, Lange S. Chloramidine/Bisindolylmaleimide-I-Mediated inhibition of exosome and microvesicle release and enhanced efficacy of cancer chemotherapy. Int J Mol Sci (2017) 18. doi: 10.3390/ijms18051007
173. Liao Z, Li S, Lu S, Liu H, Li G, Ma L, et al. Metformin facilitates mesenchymal stem cell-derived extracellular nanovesicles release and optimizes therapeutic efficacy in intervertebral disc degeneration. Biomaterials (2021) 274:120850. doi: 10.1016/j.biomaterials.2021.120850
174. Mogavero A, Maiorana MV, Zanutto S, Varinelli L, Bozzi F, Belfiore A, et al. Metformin transiently inhibits colorectal cancer cell proliferation as a result of either AMPK activation or increased ROS production. Sci Rep (2017) 7:15992. doi: 10.1038/s41598-017-16149-z
175. Gao L, He Y, Wang K, Wang C, Wu H, Hu A, et al. All-trans retinoic acid suppressed GES-1 cell proliferation induced by exosomes from patients with precancerous lesions by arresting the cell cycle in s-phase. Eur J Cancer Prev Off J Eur Cancer Prev Organisation (ECP) (2021) 30:113–9. doi: 10.1097/cej.0000000000000571
176. Wernly B, Erlinge D, Pernow J, Zhou Z. Ticagrelor: a cardiometabolic drug targeting erythrocyte-mediated purinergic signaling? American journal of physiology. Heart Circulatory Physiol (2021) 320:H90–h94. doi: 10.1152/ajpheart.00570.2020
177. Casieri V, Matteucci M, Pasanisi EM, Papa A, Barile L, Fritsche-Danielson R, et al. Ticagrelor enhances release of anti-hypoxic cardiac progenitor cell-derived exosomes through increasing cell proliferation In vitro. Sci Rep (2020) 10:2494. doi: 10.1038/s41598-020-59225-7
178. Bitirim CV, Ozer ZB, Aydos D, Genc K, Demirsoy S, Akcali KC, et al. Cardioprotective effect of extracellular vesicles derived from ticagrelor-pretreated cardiomyocyte on hyperglycemic cardiomyocytes through alleviation of oxidative and endoplasmic reticulum stress. Sci Rep (2022) 12:5651. doi: 10.1038/s41598-022-09627-6
179. Kulshreshtha A, Singh S, Ahmad M, Khanna K, Ahmad T, Agrawal A, et al. Simvastatin mediates inhibition of exosome synthesis, localization and secretion via multicomponent interventions. Sci Rep (2019) 9:16373. doi: 10.1038/s41598-019-52765-7
180. Huang P, Wang L, Li Q, Tian X, Xu J, Xu J, et al. Atorvastatin enhances the therapeutic efficacy of mesenchymal stem cells-derived exosomes in acute myocardial infarction via up-regulating long non-coding RNA H19. Cardiovasc Res (2020) 116:353–67. doi: 10.1093/cvr/cvz139
181. Tajik T, Baghaei K, Moghadam VE, Farrokhi N, Salami SA. Extracellular vesicles of cannabis with high CBD content induce anticancer signaling in human hepatocellular carcinoma. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie (2022) 152:113209. doi: 10.1016/j.biopha.2022.113209
182. Zhang H, Lu J, Liu J, Zhang G, Lu A. Advances in the discovery of exosome inhibitors in cancer. J Enzyme inhibition medicinal Chem (2020) 35:1322–30. doi: 10.1080/14756366.2020.1754814
183. Lionetti V, Tuana BS, Casieri V, Parikh M, Pierce GN. Importance of functional food compounds in cardioprotection through action on the epigenome. Eur Heart J (2019) 40:575–82. doi: 10.1093/eurheartj/ehy597
184. Mantilla-Escalante DC, López de Las Hazas MC, Crespo MC, Martín-Hernández R, Tomé-Carneiro J, Del Pozo-Acebo L, et al. Mediterranean Diet enriched in extra-virgin olive oil or nuts modulates circulating exosomal non-coding RNAs. Eur J Nutr (2021) 60:4279–93. doi: 10.1007/s00394-021-02594-0
185. Otsuka K, Yamamoto Y, Ochiya T. Uncovering temperature-dependent extracellular vesicle secretion in breast cancer. J extracellular vesicles (2020) 10:e12049. doi: 10.1002/jev2.12049
186. Cheng Y, Zeng Q, Han Q, Xia W. Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell (2019) 10:295–9. doi: 10.1007/s13238-018-0529-4
187. Mutschelknaus L, Peters C, Winkler K, Yentrapalli R, Heider T, Atkinson MJ, et al. Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. PLoS One (2016) 11:e0152213. doi: 10.1371/journal.pone.0152213
188. Ab Razak NS, Ab Mutalib NS, Mohtar MA, Abu N. Impact of chemotherapy on extracellular vesicles: Understanding the chemo-EVs. Front Oncol (2019) 9:1113. doi: 10.3389/fonc.2019.01113
189. Avisar A, Cohen M, Brenner B, Bronshtein T, Machluf M, Bar-Sela G, et al. Extracellular vesicles reflect the efficacy of wheatgrass juice supplement in colon cancer patients during adjuvant chemotherapy. Front Oncol (2020) 10:1659. doi: 10.3389/fonc.2020.01659
190. Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol Off J Eur Soc Med Oncol (2021) 32:466–77. doi: 10.1016/j.annonc.2021.01.074
191. Sahoo S, Adamiak M, Mathiyalagan P, Kenneweg F, Kafert-Kasting S, Thum T. Therapeutic and diagnostic translation of extracellular vesicles in cardiovascular diseases: Roadmap to the clinic. Circulation (2021) 143:1426–49. doi: 10.1161/circulationaha.120.049254
192. Kugeratski FG, Hodge K, Lilla S, McAndrews KM, Zhou X, Hwang RF, et al. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat Cell Biol (2021) 23:631–41. doi: 10.1038/s41556-021-00693-y
193. Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji JF, et al. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer (2021) 20:99. doi: 10.1186/s12943-021-01396-6
194. Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C, et al. The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer (2018) 17:75. doi: 10.1186/s12943-018-0823-z
195. Ji J, Chen R, Zhao L, Xu Y, Cao Z, Xu H, et al. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol Cancer (2021) 20:58. doi: 10.1186/s12943-021-01349-z
196. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol (2008) 10:1470–6. doi: 10.1038/ncb1800
197. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer (2006) 6:259–69. doi: 10.1038/nrc1840
198. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci United States America (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
199. Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, et al. Six serum-based miRNAs as potential diagnostic biomarkers for gastric cancer. Cancer epidemiology Biomarkers Prev Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol (2017) 26:188–96. doi: 10.1158/1055-9965.Epi-16-0607
200. Butz H, Nofech-Mozes R, Ding Q, Khella HWZ, Szabó PM, Jewett M, et al. Exosomal MicroRNAs are diagnostic biomarkers and can mediate cell-cell communication in renal cell carcinoma. Eur Urol Focus (2016) 2:210–8. doi: 10.1016/j.euf.2015.11.006
201. Muramatsu-Maekawa Y, Kawakami K, Fujita Y, Takai M, Kato D, Nakane K, et al. Profiling of serum extracellular vesicles reveals miRNA-4525 as a potential biomarker for advanced renal cell carcinoma. Cancer Genomics Proteomics (2021) 18:253–9. doi: 10.21873/cgp.20256
202. Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer (2015) 136:2616–27. doi: 10.1002/ijc.29324
203. Manterola L, Guruceaga E, Gállego Pérez-Larraya J, González-Huarriz M, Jauregui P, Tejada S, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-oncology (2014) 16:520–7. doi: 10.1093/neuonc/not218
204. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
205. Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2015) 36:2007–12. doi: 10.1007/s13277-014-2807-y
206. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer (2019) 18:116. doi: 10.1186/s12943-019-1041-z
207. Lin L, Cai GX, Zhai XM, Yang XX, Li M, Li K, et al. Plasma-derived extracellular vesicles circular RNAs serve as biomarkers for breast cancer diagnosis. Front Oncol (2021) 11:752651. doi: 10.3389/fonc.2021.752651
208. Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (London Engl 1979) (2019) 133:1935–53. doi: 10.1042/cs20190589
209. Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet (2019) 10:1096. doi: 10.3389/fgene.2019.01096
210. Ma H, Xu Y, Zhang R, Guo B, Zhang S, Zhang X. Differential expression study of circular RNAs in exosomes from serum and urine in patients with idiopathic membranous nephropathy. Arch Med Sci AMS (2019) 15:738–53. doi: 10.5114/aoms.2019.84690
211. Mirzaei H, Sahebkar A, Jaafari MR, Goodarzi M, Mirzaei HR. Diagnostic and therapeutic potential of exosomes in cancer: The beginning of a new tale? J Cell Physiol (2017) 232:3251–60. doi: 10.1002/jcp.25739
212. Wang S, Qiu Y, Bai B. The expression, regulation, and biomarker potential of glypican-1 in cancer. Front Oncol (2019) 9:614. doi: 10.3389/fonc.2019.00614
213. Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med (2017) 21:838–47. doi: 10.1111/jcmm.12941
214. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell (2020) 182:1044–61.e1018. doi: 10.1016/j.cell.2020.07.009
215. Sun Y, Xia Z, Shang Z, Sun K, Niu X, Qian L, et al. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci Rep (2016) 6:24669. doi: 10.1038/srep24669
216. Gao J, Qiu X, Li X, Fan H, Zhang F, Lv T, et al. Expression profiles and clinical value of plasma exosomal Tim-3 and galectin-9 in non-small cell lung cancer. Biochem Biophys Res Commun (2018) 498:409–15. doi: 10.1016/j.bbrc.2018.02.114
217. Simbari F, McCaskill J, Coakley G, Millar M, Maizels RM, Fabriás G, et al. Plasmalogen enrichment in exosomes secreted by a nematode parasite versus those derived from its mouse host: implications for exosome stability and biology. J extracellular vesicles (2016) 5:30741. doi: 10.3402/jev.v5.30741
218. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta (2012) 1820:940–8. doi: 10.1016/j.bbagen.2012.03.017
219. Beloribi S, Ristorcelli E, Breuzard G, Silvy F, Bertrand-Michel J, Beraud E, et al. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLoS One (2012) 7:e47480. doi: 10.1371/journal.pone.0047480
220. Beloribi-Djefaflia S, Siret C, Lombardo D. Exosomal lipids induce human pancreatic tumoral MiaPaCa-2 cells resistance through the CXCR4-SDF-1α signaling axis. Oncoscience (2015) 2:15–30. doi: 10.18632/oncoscience.96
221. Wu Q, Ishikawa T, Sirianni R, Tang H, McDonald JG, Yuhanna IS, et al. 27-hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep (2013) 5:637–45. doi: 10.1016/j.celrep.2013.10.006
222. Roberg-Larsen H, Lund K, Seterdal KE, Solheim S, Vehus T, Solberg N, et al. Mass spectrometric detection of 27-hydroxycholesterol in breast cancer exosomes. J Steroid Biochem Mol Biol (2017) 169:22–8. doi: 10.1016/j.jsbmb.2016.02.006
223. Skotland T, Sandvig K, Llorente A. Lipids in exosomes: Current knowledge and the way forward. Prog Lipid Res (2017) 66:30–41. doi: 10.1016/j.plipres.2017.03.001
224. Donoso-Quezada J, Ayala-Mar S, González-Valdez J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic (Copenhagen Denmark) (2021) 22:204–20. doi: 10.1111/tra.12803
225. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell communication Signaling CCS (2021) 19:47. doi: 10.1186/s12964-021-00730-1
226. Brzozowski JS, Jankowski H, Bond DR, McCague SB, Munro BR, Predebon MJ, et al. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis (2018) 17:211. doi: 10.1186/s12944-018-0854-x
227. Chapuy-Regaud S, Dubois M, Plisson-Chastang C, Bonnefois T, Lhomme S, Bertrand-Michel J, et al. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie (2017) 141:70–9. doi: 10.1016/j.biochi.2017.05.003
228. Sancho-Albero M, Jarne C, Savirón M, Martín-Duque P, Membrado L, Cebolla VL, et al. High-performance thin-layer chromatography-Densitometry-Tandem ESI-MS to evaluate phospholipid content in exosomes of cancer cells. Int J Mol Sci (2022) 23. doi: 10.3390/ijms23031150
229. Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J extracellular vesicles (2016) 5:32570. doi: 10.3402/jev.v5.32570
230. Linton SS, Abraham T, Liao J, Clawson GA, Butler PJ, Fox T, et al. Tumor-promoting effects of pancreatic cancer cell exosomes on THP-1-derived macrophages. PLoS One (2018) 13:e0206759. doi: 10.1371/journal.pone.0206759
231. Kim CW, Lee HM, Lee TH, Kang C, Kleinman HK, Gho YS. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res (2002) 62:6312–7.
232. Jiang C, Hopfner F, Berg D, Hu MT, Pilotto A, Borroni B, et al. Validation of α-synuclein in L1CAM-immunocaptured exosomes as a biomarker for the stratification of parkinsonian syndromes. Movement Disord Off J Movement Disord Soc (2021) 36:2663–9. doi: 10.1002/mds.28591
233. Huang C, Pan L, Shen X, Tian H, Guo L, Zhang Z, et al. Hsp16.3 of mycobacterium tuberculosis in exosomes as a biomarker of tuberculosis. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol (2021) 40:2427–30. doi: 10.1007/s10096-021-04246-x
234. Liu Y, Li Y, Zang J, Zhang T, Li Y, Tan Z, et al. CircOGDH is a penumbra biomarker and therapeutic target in acute ischemic stroke. Circ Res (2022) 130:907–24. doi: 10.1161/circresaha.121.319412
235. Sharma R, Huang X, Brekken RA, Schroit AJ. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br J Cancer (2017) 117:545–52. doi: 10.1038/bjc.2017.183
236. Cesselli D, Parisse P, Aleksova A, Veneziano C, Cervellin C, Zanello A, et al. Extracellular vesicles: How drug and pathology interfere with their biogenesis and function. Front Physiol (2018) 9:1394. doi: 10.3389/fphys.2018.01394
237. Drabovich AP, Martínez-Morillo E, Diamandis EP. Toward an integrated pipeline for protein biomarker development. Biochim Biophys Acta (2015) 1854:677–86. doi: 10.1016/j.bbapap.2014.09.006
238. Špilak A, Brachner A, Kegler U, Neuhaus W, Noehammer C. Implications and pitfalls for cancer diagnostics exploiting extracellular vesicles. Advanced Drug delivery Rev (2021) 175:113819. doi: 10.1016/j.addr.2021.05.029
239. Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics. Clin Appl (2015) 9:358–67. doi: 10.1002/prca.201400114
240. Rak J. Extracellular vesicles - biomarkers and effectors of the cellular interactome in cancer. Front Pharmacol (2013) 4:21. doi: 10.3389/fphar.2013.00021
Keywords: exosomes, extracellular vesicles (EVs), biogenesis, exogenous factors, release, molecular cargoes, diagnostics, biomarkers
Citation: Yi X, Chen J, Huang D, Feng S, Yang T, Li Z, Wang X, Zhao M, Wu J and Zhong T (2022) Current perspectives on clinical use of exosomes as novel biomarkers for cancer diagnosis. Front. Oncol. 12:966981. doi: 10.3389/fonc.2022.966981
Received: 12 June 2022; Accepted: 01 August 2022;
Published: 31 August 2022.
Edited by:
Mehdi Jaymand, Kermanshah University of Medical Sciences, IranReviewed by:
Saravanakumar Marimuthu, University of Nebraska Medical Center, United StatesCopyright © 2022 Yi, Chen, Huang, Feng, Yang, Li, Wang, Zhao, Wu and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianyu Zhong, emhvbmd0aWFueXVAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.