- Department of Gastrointestinal Surgery, Jinhua Hospital of Zhejiang University, Jinhua, China
Gastric cancer (GC) is one of the leading causes of cancer mortality worldwide. Numerous studies have shown that the gastric microbiota can contribute to the occurrence and development of GC by generating harmful microbial metabolites, suggesting the possibility of discovering biomarkers. Metabolomics has emerged as an advanced promising analytical method for the analysis of microbiota-derived metabolites, which have greatly accelerated our understanding of host-microbiota metabolic interactions in GC. In this review, we briefly compiled recent research progress on the changes of gastric microbiota and its metabolites associated with GC. And we further explored the application of metabolomics and gastric microbiome association analysis in the diagnosis, prevention and treatment of GC.
Introduction
Gastric cancer (GC) is the fourth most common cancer in the world, and its cancer-related mortality rate ranks second in the world (1). According to statistics, GC was responsible for over 1,089,103 new cases and 768,793 deaths in 2020 (2). Currently, there is no effective treatment for the disease, and the lag in the diagnosis of early GC is a major cause of high mortality in cancer patients. Endoscopy is now widely used for early screening, but not only does this method involve invasive procedures, its accuracy depends on the experience of the endoscopist and pathologist, and its economics are still questionable (3). Therefore, it is of great significance to study non-invasive and specific biomarkers for early screening, diagnosis and treatment of GC.
The main risk factors of GC include Helicobacter pylori infection, smoking, dietary factors, etc (4). H. pylori infection is widely recognized as a high risk factor for the development of GC (5). In many developing countries, the H. pylori infection rate exceeds 90% (6). Almost all cases of GC are associated with H. pylori (7). It was previously believed that the highly acidic environment in the stomach is not suitable for bacterial growth in addition to H. pylori. However, with the advancement of sequencing technology, it has proven that the stomach is inhabited by a robust microbiota (4) and changes in gastric microbes may be a possible cause of GC. Studies have found that microbial diversity is significantly lower in GC patients compared with those with superficial gastritis (8, 9) and GC is associated with increased microbial diversity and richness (10, 11).
Microbial metabolites have been shown to play important roles in cancer initiation or progression (12–14). In recent years, with the the development of metabolomics technology, metabolomics has been used to characterize the metabolic perturbation and identify potential biomarkers in various cancers (15). Combining metagenomics and metabolomics may further help us understand the relationship of microbial dysbiosis and harmful metabolites to gastric carcinogenesis, providing new ideas for us to understand the mechanism of GC (16). This review summarizes the research progress on GC-related alterations in gut microbiota and its metabolites, the application of metabolomics and gut microbiome in the diagnosis, prevention, and treatment of GC, and discusses current challenges and future directions.
Microbiome: an overview
The human body is colonized with a large and complex microbial community, and the sum of its genes is called the human microbiome. The performance of the human microbiome is influenced by a variety of environmental and physiological changes, including age, gender, diet, and more. Studies (17–19) found human microorganisms are closely related to a variety of diseases such as infectious diseases, obesity, diabetes, liver disease, coronary heart disease and tumors, etc. They form a symbiotic relationship with the host during the coevolution process, and play an important role in regulating the host’s digestion, absorption, metabolism, and immunity (20, 21). Among them, the human digestive tract is a huge microbial reservoir, and the total number of cells is 10 times that of the total number of human cells, and the number of genes contained in it is 150 times that of the total human genome (22). The digestive tract contains a large spectrum of pathogenic commensal bacteria. These microorganisms exist in the gut in a symbiotic form in a healthy state, and maintain the health of the body together with the host; in an unhealthy state, their diversity and abundance change abnormally, leading to the occurrence and development of various diseases, such as esophageal reflux, gastritis, pseudomembranous colitis (23) and other digestive tract diseases, and tumor diseases such as liver cancer (24) and gastric cancer (25). Therefore, the gastrointestinal microbiota is considered as a potential therapeutic target for various disease interventions. For example, studies have shown that fecal transplantation is significantly effective in improving symptoms of recurrent diarrhea due to Clostridium difficile infection (23). At present, the research on the mechanism of human microorganisms in the occurrence and development of various diseases is still in its infancy. Although the international research on the relationship between human microecology and diseases is becoming more and more intense, there are many key technologies and problems in the field of human microecology research that need to be further explored. Herein, we reviewed the research progress on the relationship between gastrointestinal microecology and GC. In Table 1 we summarize the changes in microbial diversity in gastric cancer.
Methods for studying the microbiota
The traditional method for detecting gastrointestinal microbiota is mainly to quantitatively analyze the microflora by counting the number of viable bacterial clones in gastric mucosa, gastric juice, and intestinal contents. In recent years, with the development of molecular microecology research and the application of related technologies, the detection level of gastrointestinal microecological microflora has been significantly improved. The main molecular technique for studying microbiota expression is DNA amplification of hypervariable regions using a polymerase chain reaction (PCR) (29). Next generation sequencing (NGS) can realize large-scale parallel sequencing of multiple genes and can fundamentally solve the practical problems of difficult diagnosis caused by the heterogeneity of single-gene inheritance, multiple genes, and complex phenotypes. NGS advances the study of the human microbiome and helps understand the association between microbiome imbalances and disease phenotypes (29).
Diet and GC
Diet is thought to influence the development or progression of GC, possibly through complex metabolic and immune pathways. Recent microbiome studies suggest that dysbiosis of the microbiota may be a key risk factor for the development of GC (25). High-fat dietary components, like meat and snacks, have been found to increase the abundance of bile-tolerant microbes and decrease metabolizing plant polysaccharides bacterial levels, which may induce gastrointestinal carcinogenesis (30, 31). Consuming preserved foods can lead to high salt intake, which directly damages the stomach lining and increases the formation of nitroso compounds that significantly increase the risk of GC (32). At the same time, a high-salt diet can increase the risk of H. pylori infection, and the synergistic effect of the two pathogens can further increase the risk of GC occurrence and development (33). A study showed that a healthy dietary pattern characterized by the consumption of vegetables, fruits may reduce the risk of GC (34). Fresh vegetables contain various types of antioxidants that act as protective agents, potentially ameliorating the effects of microbial imbalances. Several antioxidant-related nutrients present in fruits also play a key role in the prevention of GC (35, 36). Dairy products containing probiotics may reduce the risk of various types of gastrointestinal cancers by modulating immune parameters (37). It reduces the levels of several cancer-related biomarkers resulting from microbial and metabolic imbalances, while increasing the production of IFN-γ, which has anticancer effects (38). Therefore, dairy products, fresh vegetables and fruits should be included in the daily diet to reduce the risk of GC.
The Mechanisms of GC mediated by gastrointestinal microbiota
A dysbiosis of the microbiota occurs when the composition of bacterial species and the number of harmful bacteria changes, thereby promoting the development of GC (25). At present, the complex micro-ecosystem in the stomach has attracted the attention of many researchers. Bik (39) et al. found through gene sequencing that there are 128 phylotypes in the gastric microflora, belonging to 8 phyla, of which the dominant microflora accounts for 5 phyla, namely Bacteroidetes, Firmicutes, Fusobacterium, and Nephronomyces and Proteobacteria. H. pylori can colonize the human gastric mucosa, leading to dysbiosis in the gastrointestinal tract, resulting in chronic active gastritis. It can further cause peptic ulcer or malignant gastric epithelial mucosal lesions, affecting the immune function of gastric mucosal epithelial cells (40, 41). Maldonado-Contreras A (42) et al. analyzed the gastric microflora of H. pylori-positive patients and showed that H. pylori infection increased Proteobacteria, Helicobacter and Acidobacteria, while reducing Actinomyces, Bacteroidetes and Firmicutes, significantly changing the bacterial abundance in the stomach. Wang (26) et al. found that the abundance of Armatmonadetes, Chloroflexi, Elusimicrobia, Nitrospirae, Planctomycetes, Verrucomicrobia, and WS3 decreased sequentially from CG, IM, IN to GC. Actinobacteria, Bacteroides, Fusobacterium, SR1 and TM7 were more abundant in IN and GC. At the community level, the proportions of Gram-positive and anaerobic bacteria were higher in IN and GC than in other tissue types, while the proportions of aerobic and facultative anaerobes were significantly reduced in GC, suggesting that these bacteria may play a role in the process of intestinal metaplasia and GC. Below we discuss some of the microbiota associated with GC.
H. pylori is a Gram-negative, microaerophilic pathogen that colonizes the human gastric mucosa, and is closely related to the occurrence of chronic gastritis, peptic ulcer, and GC (43). The World Health Organization has classified it as a class I carcinogen Factors (44). H. pylori is involved in the occurrence and development of GC through pathogenic virulence factors such as vacuolar toxin-associated protein A (VacA), cytotoxin-associated protein (CagA), urease, adhesion factor, blood group antigen-binding adhesin gene, lipopolysaccharide, and inflammation and immune response after infection, etc (45). Lertpiriyapong (46) et al. studied the H. pylori INS-GAS mouse model and found that IL-11, TGF-β and cancer-related gene expression increased, which promote the development of GC. Hayashi (47) et al. showed that CagA secreted by H. pylori can up-regulate the expression of c-MYC, DNMT3B, EZH2, down-regulate the expression of miR-26a and miR-101, and decrease the expression of let-7, thereby activating the Ras pathway and increasing Ras expression, thus participating in the occurrence and development of GC. Cheng (48) et al. applied genome-wide methylation analysis and found that, In human H. pylori-related GC tissues, H. pylori can induce the methylation of FoxD3 and inhibit the activation of cell death regulators CYFIP2 and RARB, thereby promoting the proliferation and invasion of GC cells.
Enterococcus faecalis induce intracellular production of oxidative phosphorylation-independent ROS while disrupting the mitochondrial genome in gastric cells. The bacteria also induce a pro-inflammatory response driven by NF-κB, one of the key transcription factors in cancer-related inflammation. It also impairs the DNA damage response and cell cycle-controlled gene expression, induces mitochondrial DNA instability, and promotes tumor formation. E. faecalis infection also resulted in decreased expression of several genes involved in DNA repair, such as MMR gene expression (49).
Escherichia coli can release cytotoxins such as colibacins, which cause DNA damage (50). At the same time, it can also induce inflammation, destroy gastrointestinal mucosal cells, and promote the production of GC (51).
Fusobacterium nucleatum is one of the enriched strains in the GC microbiota. It can directly act on host cells and affect the expression of cancer marker genes, thereby promoting the occurrence of cancer (52). In addition, Fusobacterium nucleatum can also secrete endotoxin to inhibit the immune function of the body and generate an inflammatory microenvironment (53). Hsieh (27) et al. studied the bacterial species associated with gastric epithelium in 11 GC patients and found that Fusobacterium nucleatum was abundantly enriched in GC patients, and the gastric microbes of most GC patients were different from those of noncancerous gastric disease patients. The experimenter operating characteristic curve analysis showed that the sensitivity of Fusobacterium nucleatum combined with Clostridium colicanis and Fusobacterium canifelinum in the diagnosis of GC was 100%, and the specificity was about 70%. This suggests that Fusobacterium nucleatum may be highly correlated with GC. In addition, Abed (54) et al.found that Gal-GalNAc antigen was highly expressed in GC tissue. Fusobacterium nucleatum, as a cancer-promoting biological factor, can be enriched to the lesion through the specific interaction of Fap2 with Gal-GalNAc antigen on the tumor surface, and lead to the expression of MUC2 and TNF-α in cancer cells (55). According to the above studies, it is speculated that Fusobacterium nucleatum may be involved in the development of GC and can be used as a potential biomarker for monitoring GC.
A significant increase in the relative abundance of lactic acid-producing bacteria (Lactococcus and Lactobacillus) was observed in GC patients (10). While Lactobacillus species are commonly used as probiotics and are thought to be beneficial to the host, elevated lactate levels can be very harmful in the context of cancer. Lactic acid can act as an energy source for tumor cells, inducing glycolysis leads to increased ATP supply, also can promote inflammation and stimulate tumor angiogenesis. In addition, Lertpiriyapong (46) et al. found that Lactobacillus can accelerate the development of H. pylori-associated gastritis into intraepithelial neoplasia in the H. pylori INS-GAS mouse model.
Wang (28) et al. reported that the phylum Nitrospirae was present in all patients with GC but completely absent in patients with chronic gastritis. Notably, several members of the Nitrospirae phylum are known to play a role in the metabolism of nitrates and nitrites (56). It is known that the consumption of nitrates is a significant risk factor for the development of GC, and it is plausible that these bacteria may increase cancer risk.
At present, the application of probiotics and their metabolites in the treatment of gastrointestinal diseases has attracted more and more attention. Probiotics are a class of active microorganisms that are beneficial to the host. They colonize the human gastrointestinal tract and reproductive system, and can produce exact health effects and improve the host’s micro-ecological balance. Studies (57, 58) have shown that probiotics and their metabolites have inhibitory effects on H. pylori, which can inhibit the colonization and growth of H. pylori in gastric mucosal epithelium, reduce H. pylori activity, and kill H. pylori by destroying the cell wall. Fermented milk formed by the fermentation of Propionibacterium freudenreichii can promote the apoptosis of human GC cell line HGT-1, and can induce typical apoptosis processes, including chromosome aggregation, apoptotic body formation, and cell apoptosis, et al. And the fermented milk can enhance the cytotoxicity of the GC chemotherapeutic drug camptothecin. Lactobacillus casei extract can inhibit the proliferation of GC cell line KAT03 and induce its apoptosis by inactivating the NF-κB promoter activity. Further molecular mechanism study found that Lactobacillus casei extract can reduce the expression of NF-κB and I-κB, and then some molecules in the mTOR signaling pathway such as PI3K, Akt and p70S6 kinase phosphorylation decrease, which promotes the occurrence of apoptosis (59). A study by Orlando (60) et al. found that the cytoplasmic extract of Lactobacillus rhamnosus strain GG could significantly inhibit the proliferation of GC cell HGC-27 strain and colon cancer cell DLD-1 strain, indicating that Lactobacillus mainly relies on cytoplasm to exert its anti-tumor proliferation effect. Mahkonen (61) et al. applied Lactobacillus and Bifidobacterium to stimulate primary human GC cells AGS and metastatic human GC cells NCI-N87, and detected cyclooxygenase-1 (COX-1), COX-2, COX-1-IR expression. The results showed that the expression of COX-1 increased after Lactobacillus stimulated NCI-N87 cells, while the expression of COX-1, COX-2 and COX-1-IR did not change significantly after Bifidobacterium stimulated AGS and NCI-N87 cells, suggesting that Lactobacillus can inhibit the growth of metastatic GC cells by inducing the production of cytoprotective COX-1.
Metabolism and GC
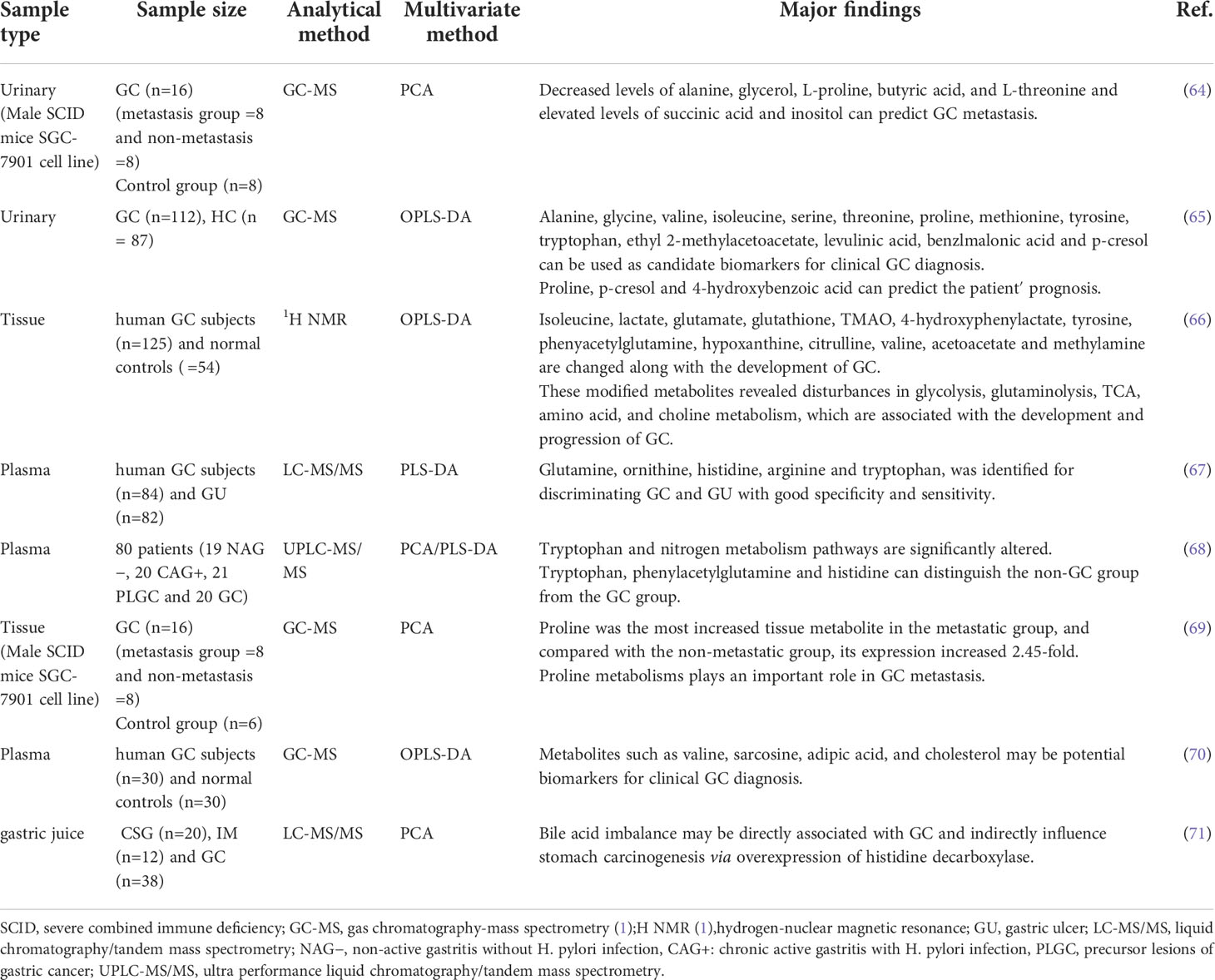
Gastrointestinal microbes play an important role in human health and disease. The metabolic functions of gastrointestinal microbes can be considered as contributing factors for disease development, and their bioactive substances have important effects on the physiological and pathological processes of the host. With the rise of metabolomics, significant progress has been made in understanding the relationship between metabolic regulation and cancer. Extensive studies have shown that metabolic disturbances are one of the hallmarks of cancer (62) and are intricately linked to tumorigenesis and cancer immune escape (20). To understand this connection, We must first understand the metabolic changes in GC and the mechanisms behind these changes (63). In Table 2 we summarize the altered metabolites in different sample in GC.
Glucose metabolism
The “Warburg effect” was first proposed in 1956, and subsequent studies have shown that, compared with control group, the concentration of lactate continued to increase in GC group (64–66), and glucose was significantly depleted, proving that cancer cells increase glucose uptake and obtain energy through glycolysis to meet the energy requirements for maintaining their rapid growth and proliferation (66). At the same time, lactic acid is the final product of glycolysis, and accumulated lactate modulates the activity of proteases that break down the extracellular matrix. These proteases can produce peptides and amino acids that can be used for energy production (72). The acidotic microenvironment also contributes to the formation of cancer blood vessels to meet the nutrients required for tumor invasion and metastasis (73). Based on metabolomic studies, it was found that the level of glucose involved in glycolysis decreased in both the tissues and plasma of GC patients and SGC-7901 tumor-bearing mice, so the glycolytic state may be of great significance for the early diagnosis of GC (66, 74, 75). Studies have reported that in non-diabetic conditions, blood glucose levels are positively correlated with cancer mortality, and the potential mortality of cancer patients with glucose intolerance is also increased (76). Hyperglycemia in patients with both diabetes and GC can promote the proliferation of GC cells and reduce the sensitivity to chemotherapeutic drugs (77). Not only that, glycolysis transcriptional regulators and glycolysis-related proteins are significantly correlated with the prognosis of cancer patients, so glycolysis status may also be a potential biomarker for prognosis (78).
Amino acid metabolism
The tumor microenvironment is far from an ideal cell growth environment, and nutrients such as amino acids can be used as energy sources for tumors to affect the occurrence and development of tumors (79). Many cancer cell lines cannot survive without glutamine (80). Glutamine is produced by fermentation of glutamate-producing bacteria, and it is required for anabolic growth of mammalian cells because of its ability to control protein translation (81). Moreover, reprogramming of glutamine metabolism further promotes proliferative and metabolic responses regulated by the oncogenic transcription factor c-MYC (16, 82). In addition, studies (67, 68) have found that patients with GC have lower levels of tryptophan, which may be due to up-regulation of the expression of tryptophan metabolizing enzymes indoleamine 2,3-dioxygenase 2 and tryptophan dioxygenase (68). This alteration not only promotes cancer progression, but also affects tumor immune regulation. Chen (69) et al. used the human GC cell line SGC-7901 to establish a metastatic and non-metastatic animal model of GC. They found that proline was the most increased tissue metabolite in the metastatic group, and compared with the non-metastatic group, its expression increased 2.45-fold. Elevated proline may be due to the degradation of extracellular matrix and collagen in the microenvironment (83). And they point out that proline metabolism may play an important role in metastasis. However, further functional and clinical sample analysis of the metabolic pathways is needed to demonstrate their role in GC metastasis.
Lipid metabolism
Gut microbes regulate dietary lipid composition, digestion, and absorption, potentially altering intestinal lipoprotein formation. Lipids mainly include fats, phospholipids and sterol. Disorders of lipid metabolism may affect the proliferation and differentiation of tumor cells and accelerate the occurrence and development of cancer70. Song et al. (84) found that serum cholesterol and some fatty acid levels in patients with GC were significantly reduced. It is suggested that GC cells may consume a large amount of fatty acids to meet the needs of cell membrane synthesis and energy production. The survival of cancer cells in the human body depends on lipids, and accumulated lipid droplets are found in various cancer microenvironments (70), so lipid droplets are expected as effective targets for blocking tumor growth (85), and fatty acid metabolism-related proteins may also become diagnostic markers for early GC (86, 87). In addition, GC is prone to omental metastasis, and fatty acid oxidation is regulated by omental adipocytes. However, fatty acid oxidation is enhanced in GC patients, which promotes omental metastasis of GC (88, 89). Fatty acid oxidation also plays an important role in mesenchymal stem cell-mediated chemoresistance in patients with GC (90). Study has shown that fatty acid oxidation inhibitors combined with chemotherapeutic drugs can improve the chemoresistance of patients (91). Notably, the level of O-acetylcarnitine, which increases fatty acid β-oxidation, tends to decrease as early GC progresses to advanced stages (92). This seems to explain the significant reductions in 9-hexadecenoic acid, cis-vaccenic acid, arachidonic acid, hexadecanoic acid and 3-hydroxybutanoic acid in stage III/IV GC tissue samples (70). However, this difference needs to be further elucidated with larger samples and different analytical methods. In addition, patients with advanced GC are often accompanied by cachexia. Study has found that chronic inflammation mediated by TNF and IL-6 may promote the occurrence of cachexia in patients with GC. In the early stage of cachexia, serum TNF is positively correlated with serum free fatty acid (FFA). In early and advanced cancer cachexia, serum IL6 and FFA were also significantly positively correlated (90), indicating that cachexia may be associated with lipid metabolism, but the specific mechanism is still unclear.
Nucleotide metabolism
The rapid proliferation and differentiation of tumor cells can lead to abnormal nucleotide synthesis and catabolism. Nucleotides are related to energy metabolism, mainly in the form of ATP and GTP. At the same time, nucleotide synthesis can ensure the timely replication of DNA. These are all necessary for tumor cell proliferation and key elements of cancer metabolism (16, 93, 94). Studies (75, 84, 95) have found that compounds involved in nucleotide metabolism, such as adenine, xanthine, and inosine are increased in GC patients or animal models, which can be used as biomarkers for early diagnosis of GC. Nucleotide catabolism is characterized by hyperuric acid or hyperurate in GC patients (64, 96). Some purine compounds, such as hypoxanthine and guanosine, increased the accumulation of urea and uric acid in the urine of GC patients, further indicating abnormal nucleotide metabolism in GC patients (97, 98). In addition, nucleotide-related proteins, such as nicotinamide nucleotide transhydrogenase, are also involved in the growth and metastasis of GC (99).
Other Metabolism
Bile acids are products of cholesterol catabolism in the liver. Bile acids are known to be involved in the pathological mechanism of gastric carcinogenesis. Lee (71) et al. found that the metabolism of cholic acid to deoxycholic acid was statistically different on the basis of the progression of chronic superficial gastritis to GC, suggesting that bile acid imbalance may be directly related to GC. Bile acids are cell-surface G protein-coupled receptor 1 (TGR5) ligands that regulate intestinal barrier formation and inflammation-driven immune dysfunction. Studies have shown that TGR5 is overexpressed in gastrointestinal adenocarcinomas and is associated with poor prognosis in GC patients (100). In addition, the bile acid receptor Farnesoid X receptor (FXR) is associated with the expression of Caudal type homeobox 2 (CDX2) and Mucin 2 (MUC2), which can lead to gastrointestinal metaplasia (101). Bile acids induce upregulation of Egr-1 and oncogenes through MAPK signaling in GC cells. Egr-1 has been implicated in biological processes including inflammation, cell proliferation, cell differentiation and cancer progression (102). Primary bile acids can also increase the expression levels of c-MYC and c-Jun genes through MAPK signaling, which are involved in gastric carcinogenesis and progression (103, 104). Continued exposure of gastric epithelial cells to primary bile acids may be a factor in gastric carcinogenesis. Furthermore, studies have shown that the signaling of hydrophobic bile acids is mediated through PKC activation and COX-2 induction, which leads to increased cell invasion. Moreover, by perturbing the bile acid pool, ursodeoxycholic acid (UDCA) was able to attenuate chenodeoxycholic acid CD-induced PGE2 synthesis and tumor invasiveness without affecting COX-2 expression (105).
TMAO is formed by choline, the precursor of trimethylamine (TMA), through the combined action of gut microbes and the liver. Changes in TMAO levels are closely related to gut microbe disturbances (106). Studies have shown that urinary TMAO can be used as a predictor of GC (107).
Nitrite is a precursor of carcinogenic nitroso compounds, and studies have found that the GC microbiota has a nitrate reductase function that promotes the reduction of nitrate to nitrite (55).
The microbiome-metabolomics interplay in GC
The possible role of gut microbiota and metabolomics in cancer prevention and treatment has received extensive attention (108). Studies (109) have found that eradication of H. pylori may play an important role in preventing the occurrence and malignant progression of GC. In addition, various probiotics are widely used in daily life, which can protect the gastric mucosa and enhance the immune response. Kim (107) et al. used (1)H NMR studies to find metabolites related to the tricarboxylic acid (TCA) cycle, such as TMAO, TMA, 3-indolyl sulfate, hippurate, citrate and 2-oxoglutarate can be considered as a therapeutic target to enhance the efficacy of ADR. Tumor Microenvironment(TME) highly affects the metabolism of cancer cells. It provides nutrients to tumor cells and prepares the surrounding environment for proliferation, local invasion and metastasis. The microbiota, through its metabolites, can shape the TME to influence tumor development (110). However, studies investigating the interaction between the microbiome and metabolome in GC are limited. Only a few reports have highlighted the association between the microbiome and metabolome in GC. Coker (8) et al. found the functional changes in the GC microbiomes included significantly increased representation of predicted KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways involved in nucleotide metabolism, carbohydrate digestion and absorption and bacterial ion channels compared with other disease stages. The disturbances in the gut microbiota lead to the production of harmful metabolites, such as acetaldehyde, secondary bile acids, and glucuronic acid, which induce DNA damage and promote gastric carcinogenesis. A study (110) found that polyamines (putrescine, spermidine and spermine) play an important role in cell proliferation and are required to maintain cell growth in both pre-tumor and tumor tissues. Probiotics can affect polyamine metabolism by affecting polyamine metabolism and act as an antineoplastic agent in the stomach. For example, lyophilized and sonicated preparations of Lactobacillus brevis CD2 inhibit arginine-dependent polyamine synthesis, thereby inducing associated apoptosis (111). Dai (112) et al. studied the interaction between gastric microbiota and metabolites in GC, and conducted an association analysis between different genera and different categories of metabolites. They found the metabolome profiles of the GC tumor tissues were strongly influenced by Helicobacter, Lactobacillus, and other microorganisms, which might promote GC development, suggesting that combined analysis of microbiota metabolites with microbiota bacteria may serve as promising diagnostic biomarkers for GC.
Conclusions
GC is one of the most malignant tumors in the world, although its pathogenesis is still unclear, but with the advent of omics studies, there have been encouraging findings. The development of GC is not only affected by the presence of intestinal microflora, but also by metabolic changes. Gastrointestinal microflora, especially some special bacteria, affect the development of GC through metabolic and structural changes, which can provide research data for establishing preventive strategies for GC and determining its pathogenic mechanism. Although metabolomics has been able to explain the biology of GC to a certain extent, it is easily affected by background interference during the detection process, and sometimes cannot fully explain the biological process. Therefore, the comprehensive application of multi-omics methods is required (113). However, studies on the correlation between the microbiome and metabolome in GC are still very limited. Therefore, large-scale studies, including retrospective and prospective studies, are still needed in the future to reveal the impact of the microbiome and metabolomics in the GC microenvironment and how their complex interactions affect the occurrence and development of tumors, and to screen out GC diagnostic markers with high sensitivity and specificity for the diagnosis and prognosis of GC.
Author contributions
LCZ, ZB, ZZW: Manuscript writing, literature research, editing the manuscript. GDJ: Manuscript writing and final approval of manuscript. All authors have read and approved the manuscript.
Funding
The study was supported by grants from the Foundation of the Science and Technology Department of Jinhua (No 2021-3-066) and Jinhua Municipal Central Hospital Foundation (No JY2020-5-03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China. CA Cancer J Clin (2015 2016) 66:115–32. doi: 10.3322/caac.21338
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Correa P. Gastric cancer: overview. Gastroenterol Clin North Am (2013) 42:211–7. doi: 10.1016/j.gtc.2013.01.002
4. Kim J, Cho YA, Choi WJ, Jeong SH. Gene-diet interactions in gastric cancer risk: a systematic review. World J Gastroenterol (2014) 20(28):9600–10. doi: 10.3748/wjg.v20.i28.9600
5. Peek RM, Kuipers EJ. Gained in translation: the importance of biologically relevant models of helicobacter pylori-induced gastric cancer. Gut (2012) 61(1):2–3. doi: 10.1136/gutjnl-2011-301342
6. Khalilpour A, Kazemzadeh-narbat M, Tamayol A, Oklu R, Khademhosseini A. Biomarkers and diagnostic tools for detection of helicobacter pylori. Appl Microbiol Biotechnol (2016) 100:4723–34. doi: 10.1007/s00253-016-7495-7
7. Chen Y, Segers S, Blaser MJ. Association between helicobacter pylori and mortality in the NHANES III study. Gut (2013) 62(9):1262–9. doi: 10.1136/gutjnl-2012-303018
8. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut (2018) 67:1024–32. doi: 10.1136/gutjnl-2017-314281
9. Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, et al. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharm Ther (2020) 51:770–80. doi: 10.1111/apt.15675
10. Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep (2017) 7:15957. doi: 10.1038/s41598-017-16289-2
11. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut (2018) 67(2):226–36. doi: 10.1136/gutjnl-2017-314205
12. Fan X, Jin Y, Chen G, Ma X, Zhang L. Gut microbiota dysbiosis drives the development of colorectal cancer. Digestion (2021) 102:508–15. doi: 10.1159/000508328
13. Dalal N, Jalandra R, Bayal N, Yadav AK, Harshulika, Sharma M, et al. Gut microbiota-derived metabolites in CRC progression and causation. J Cancer Res Clin Oncol (2021) 147:3141–55. doi: 10.1007/s00432-021-03729-w
14. Zhang W, An Y, Qin X, Wu X, Wang X, Hou H, et al. Gut microbiota-derived metabolites in colorectal cancer: The bad and the challenges. Front Oncol (2021) 11:739648. doi: 10.3389/fonc.2021.739648
15. Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discovery (2002) 1:153–61. doi: 10.1038/nrd728
16. Xiao S, Zhou L. Gastric cancer: Metabolic and metabolomics perspectives (Review). Int J Oncol (2017) 51(1):5–17. doi: 10.3892/ijo.2017.4000
17. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (2005) 307(5717):1915–20. doi: 10.1126/science.1104816
18. Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature (2012) 482(7384):179–85. doi: 10.1038/nature10809
19. Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol (2012) 9(10):577–89. doi: 10.1038/nrgastro.2012.156
20. Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol (2008) 16(3):107–14. doi: 10.1016/j.tim.2007.12.008
21. Tlaskalová-Hogenová H, Štěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol (2011) 8:110–20. doi: 10.1038/cmi.2010.67
22. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
23. Kassam Z, Lee CH, Yuan YH, Richard H. Fecal microbiota transplantation for clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol (2013) 108(4):500–8. doi: 10.1038/ajg.2013.59
24. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell (2012) 21(4):504—516. doi: 10.1016/j.ccr.2012.02.007
25. Stewart OA, Wu F, Chen Yu. The role of gastric microbiota in gastric cancer. Gut Microbes (2020) 11(5):1220–30.doi: 10.1080/19490976.2020.1762520
26. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the gastric mucosal microbiome associated with histological stages of gastric carcinogenesis. Front Microbiol (2020) 11:997. doi: 10.3389/fmicb.2020.00997
27. Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep (2018) 8(1):158. doi: 10.1038/s41598-017-18596-0
28. Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol (2016) 28(3):261–6. doi: 10.1097/MEG.0000000000000542
29. Al-Ansari MM, AlMalki RH, Dahabiyeh LA, Abdel AM. Metabolomics-microbiome crosstalk in the breast cancer microenvironment. Metabolites (2021) 11(11):758. doi: 10.3390/metabo11110758
30. Tong Y, Gao H, Qi Q, Liu X, Li J, Gao J, et al. High fat diet, gut microbiome and gastrointestinal cancer. Theranostics (2021) 11:5889. doi: 10.7150/thno.56157
31. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA. Linking long-term dietary patterns with gut microbial enterotypes. Science (2011) 334:105–8. doi: 10.1126/science.1208344
32. Wang JB, Fan JH, Dawsey S, Sinha D, Freedman ND, Taylor PR, et al. Dietary components and risk of total, cancer and cardiovascular disease mortality in the linxian nutrition intervention trials cohort in China. Sci Rep (2016) 6:22619. doi: 10.1038/srep22619
33. Raei N, Behrouz B, Zahri S, Latifi-Navid S. Helicobacter pylori infection and dietary factors act synergistically to promote gastric cancer. Asian Pac J Cancer Prev (2016) 17(3):917–21.doi: 10.7314/APJCP.2016.17.3.917
34. Bahmanyar S, Ye W. Dietary patterns and risk of squamous-cell carcinoma and adenocarcinoma of the esophagus and adenocarcinoma of the gastric cardia: A population-based case-control study in Sweden. Nutr Cancer (2006) 54:171–8. doi: 10.1207/s15327914nc5402_3
35. Hoang BV, Lee J, Choi IJ, Kim YW, Ryu KW, Kim J. Effect of dietary vitamin c on gastric cancer risk in the Korean population. World J Gastroenterol (2016) 22:6257.doi: 10.3748/wjg.v22.i27.6257
36. Kim JH, Lee J, Choi IJ, Kim YI, Kwon O, Kim H, et al. Dietary carotenoids intake and the risk of gastric cancer: A case-control study in Korea. Nutrients (2018) 10:1031.doi: 10.3390/nu10081031
37. Hori T, Matsuda K, Oishi K. Probiotics: A dietary factor to modulate the gut microbiome, host immune system, and gut–brain interaction. Microorganisms (2020) 8:1401.doi: 10.3390/microorganisms8091401
38. Zheng P, Zeng B, Zhou C, Liu M, Fang M, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry (2016) 21:786–96. doi: 10.1038/mp.2016.44
39. Bik E, Eckburg P, Gill S, Nelson K, Purdom E, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA (2006) 103(3):732–7. doi: 10.1073/pnas.0506655103
40. Sibony M, Jones NL. Recent advances in helicobacter pylori pathogenesis. Curr Opin Gastroen (2012) 28(1):30–5. doi: 10.1097/MOG.0b013e32834dda51
41. Conteduca V, Sansonno D, Lauletta G, Russi S, Ingravallo G, Dammacco F. H. pylori infection and gastric cancer: state of the art (review). Int J Oncol (2013) 42(1):5–18. doi: 10.3892/ijo.2012.1701
42. Maldonado-Contreras A, Goldfarb KC, Godoyvitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to helicobacter pylori status. ISME J (2011) 5(4):574–9. doi: 10.1038/ismej.2010.149
43. Malfertheiner P, Megrau D, O'Morain C. Management of helicobacter pylori infection-the maastricht v/florence consensus report. Gut (2017) 66:6–30. doi: 10.1136/gutjnl-2016-312288
44. Wang F, Meng W, Wang B, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett (2014) 345(2):196–202. doi: 10.1016/j.canlet.2013.08.016
45. Sun X, Zhang M, El-Zataari M, Owyang SY, Eaton KA, Liu M, et al. TLR2 mediates helicobacter pylori-induced tolerogenic immune response in mice. PloS One (2013) 8(9):e74595. doi: 10.1371/journal.pone.0074595
46. Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, et al. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut (2014) 63(1):54–63. doi: 10.1136/gutjnl-2013-305178
47. Hayashi Y, Tsujii M, Wang J, Kondo J, Akasaka T, Jin Y, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in helicobacter pylori-related carcinogenesis. Gut (2013) 62(11):1536–46. doi: 10.1136/gutjnl-2011-301625
48. Cheng AS, Li MS, Kang W, Cheng VY, Chou JL, Lau SS, et al. Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology (2013) 144(1):122–33. doi: 10.1053/j.gastro.2012.10.002
49. Strickertsson JA, Desler C, Martin-Bertelsen T, Dantas AM, Wadstrøm T, Winther O, et al. Enterococcus faecalis infection causes inflammation, intracellular oxphos-independent ROS production, and DNA damage in human gastric cancer cells. PloS One (2013) 8(4):e63147. doi: 10.1371/journal.pone.0063147
50. Sunny HW, Thomas NY, Chun YW, Jun Y. Clinical applications of gut microbiota in cancer biology. Semin Cancer Biol (2019) 55:28–36. doi: 10.1016/j.semcancer.2018.05.003
51. Lu YT, Liu HY, Yang K, Meng YJ, Yang LK, Liu OY, et al. A comprehensive update: gastrointestinal microflora, gastric cancer and gastric premalignant condition, and intervention by traditional Chinese medicine. J Zhejiang Univ Sci B (2022) 23(1):1–18. doi: 10.1631/jzus.B2100182
52. Rubinstein MR, Wang X, Liu W, Hao YJ, Cai GF, Han YP. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
53. Brennan CA, Garrett WS. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol (2016) 70:395–411. doi: 10.1146/annurev-micro-102215-095513
54. Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe (2016) 20(2):215–25. doi: 10.1016/j.chom.2016.07.006
55. Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, et al. Fap2 of fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun (2015) 83(3):1104–13. doi: 10.1128/IAI.02838-14
56. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of e. coli in the inflamed gut. Science (2013) 339(6120):708–11. doi: 10.1126/science.1232467
57. Parvez S, Malik KA, Kang SA, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol (2006) 100(6):1171–85. doi: 10.1111/j.1365-2672.2006.02963.x
58. Gotteland M, Brunser O, Cruchet S. Systematic review: are probiotics useful in controlling gastric colonization by helicobacter pylori? Aliment Pharmacol Ther (2006) 23(8):1077–86. doi: 10.1111/j.1365-2036.2006.02868.x
59. Cousin FJ, Jouan-Lanhouet S, Dimanche-Boitrel MT, Corcos L, Jan G. Milk fermented by propionibacterium freudenreichii induces apoptosis of HGT-1 human gastric cancer cells. PloS One (2012) 7(3):e31892. doi: 10.1371/journal.pone.0031892
60. Orlando A, Refolo MG, Messa C, Amati L, Lavermicocca P, Guerra V, et al. Antiproliferative and proapoptotic effects of viable or heat-killed lactobacillus paracasei IMPC2.1 and lactobacillus rhamnosus GG in HGC-27 gastric and DLD-1 colon cell lines. Nutr Cancer (2012) 64(7):1103–11. doi: 10.1080/01635581.2012.717676
61. Mahkonen A, Putaala H, Mustonen H, Rautonen N, Puolakkainen P. Lactobacillus acidophilus 74-2 and butyrate induce cyclooxygenase (COX)-1 expression in gastric cancer cells. Immunopharmacol Immunotoxicol (2008) 30(3):503–18. doi: 10.1080/08923970802135229
62. Beger RD. A review of applications of metabolomics in cancer. Metabolites (2013) 3(3):552–74. doi: 10.3390/metabo3030552
63. Li JV, Swann J, Marchesi JR. Biology of the microbiome 2: Metabolic role. Gastroenterol Clin North Am (2017) 46:37–47. doi: 10.1016/j.gtc.2016.09.006
64. Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, et al. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol (2011) 17:727–34. doi: 10.3748/wjg.v17.i6.727
65. Chen Y, Zhang J, Guo L, Liu L, Wen J, Xu L, et al. A characteristic biosignature for discrimination of gastric cancer from healthy population by high throughput GC-MS analysis. Oncotarget (2016) 7:87496–510. doi: 10.18632/oncotarget.11754
66. Wang HJ, Zhang HL, Deng PC, Liu CQ, Li DD, Jie H, et al. Tissue metabolic profiling of human gastric cancer assessed by 1H NMR. BMC Cancer (2016) 16(1):371. doi: 10.1186/s12885-016-2356-4
67. Jing F, Hu X, Cao Y, Xu M, Wang Y, Jing Y, et al. Discriminating gastric cancer and gastric ulcer using human plasma amino acid metabolic profile. IUBMB Life (2018) 70(6):553–62. doi: 10.1002/iub.1748
68. Lario S, Ramírez-Lázaro MJ, Sanjuan-Herráez D, Brunet-Vega A, Pericay C, Gombau L, et al. Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci Rep (2017) 7(1):17774. doi: 10.1038/s41598-017-17921-x
69. Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol (2010) 16(46):5874–80. doi: 10.3748/wjg.v16.i46.5874
70. Mikami H, Kimura O, Yamamoto H, Kikuchi S, Nakamura Y, Ando T, et al. A multicentre clinical validation of AminoIndex cancer Screening(AICS). Sci Rep (2019) 9(1):13831. doi: 10.1038/s41598-019-50304-y
71. Lee W, Um J, Hwang B, Lee Y, Chung B, Hong J. Assessing the progression of gastric cancer via profiling of histamine, histidine, and bile acids in gastric juice using LC-MS/MS. J Steroid Biochem Mol Biol (2019) 197:105539. doi: 10.1016/j.jsbmb.2019.105539
72. Gatenby RA, Gillies RJ. : Why do cancers have high aerobic glycolysis? Nat Rev Cancer (2005) 4:891–9. doi: 10.1038/nrc1478
73. Dhup S, Dadhich RK, Porporato PE. And sonveaux p: Multiple biological activities of lactic acid in cancer: Influences on tumor growth, angiogenesis and metastasis. Curr Pharm Des (2012) 18:1319–30. doi: 10.2174/138161212799504902
74. Zhang H, Cui L, Liu W, Wang Z, Ye Y, Li X, et al. 1H NMR metabolic profiling of gastric cancer patients with lymph node metastasis. Metabolomics (2018) 14(4):47. doi: 10.1007/s11306-017-1293-9
75. Nie S, Zhao Y, Qiu X, Wang W, Yao Y, Yi M, et al. Metabolomic study on nude mice models of gastric cancer treated with modified Si jun zi tang via HILIC UHPLC-Q-TOF/ MS analysis. Evid Based Complement Alternat Med (2019) 2019:3817879. doi: 10.1155/2019/3817879
76. Tang Z, Li L, Tang Y, Xie D, Wu K, Wei W, et al. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci (2018) 109(8):2590–8. doi: 10.1111/cas.13691
77. Kakehi E, Kotani K, Nakamura T, Takeshima T, Kajii E. Non-diabetic glucose levels and cancer mortality: A literature review. Curr Diabetes Rev (2018) 14(5):434–45. doi: 10.2174/1573399813666170711142035
78. Zhao W, Chen R, Zhao M, Li L, Fan L, Che X, et al. High glucose promotes gastric cancer chemoresistance in vivo and in vitro. Mol Med Rep (2015) 12(1):843–50. doi: 10.3892/mmr.2015.3522
79. Weljie AM, Jirik FR. Hypoxia-induced metabolic shifts in cancer cells: Moving beyond the warburg effect. Int J Biochem Cell Biol (2011) 43:981–9. doi: 10.1016/j.biocel.2010.08.009
80. Hensley CT, Wasti AT. And DeBerardinis RJ: Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J Clin Invest (2013) 123:3678–84. doi: 10.1172/JCI69600
81. Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell (2009) 136:521–34. doi: 10.1016/j.cell.2008.11.044
82. Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA (2012) 109:8983–8. doi: 10.1073/pnas.1203244109
83. Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids (2008) 35:681–90. doi: 10.1007/s00726-008-0063-4
84. Song H, Peng JS, Yao DS, Yang ZL, Liu HL, Zeng YK, et al. Serum metabolic profiling of human gastric cancer based on gas chromatography/ mass spectrometry. Braz J Med Biol Res (2012) 45(1):78–85. doi: 10.1590/S0100-879X2011007500158
85. Petan T, Jarc E, Jusovic M. Lipid droplets in cancer: Guardians of fat in a stressful world. Molecules (2018) 23(8):1941. doi: 10.3390/molecules23081941
86. Wu H, Han Y, Sillke Y, Deng H, Siddiqui S, Treese C, et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol Med (2019) 11(11):e10698. doi: 10.15252/emmm.201910698
87. Jiang Z, Shen H, Tang B, Yu Q, Ji X, Wang L. Quantitative proteomic analysis reveals that proteins required for fatty acid metabolism may serve as diagnostic markers for gastric cancer. Clin Chim Acta (2017) 464:148–54. doi: 10.1016/j.cca.2016.11.032
88. Tsuboi K. 2-hydroxylated fatty acids as candidates of novel drugs to promote chemosensitivity of gastric cancer. EBioMedicine (2019) 41:19–20. doi: 10.1016/j.ebiom.2019.02.029
89. Tan Y, Lin K, Zhao Y, Wu Q, Chen D, Wang J, et al. Adipocytes fuel gastric cancer omental metastasis via PITPNC1-mediated fatty acid metabolic reprogramming. Theranostics (2018) 8(19):5452–68. doi: 10.7150/thno.28219
90. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q, et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene (2019) 38(23):4637–54. doi: 10.1038/s41388-019-0747-0
91. Lee JS, Kim SH, Lee S, Kang JH, Lee SH, Cheong JH, et al. Gastric cancer depends on aldehyde dehydrogenase 3A1 for fatty acid oxidation. Sci Rep (2019) 9(1):16313. doi: 10.1038/s41598-019-52814-1
92. Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ, Kwak JM, et al. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR-based metabolomic profiling. Ann Surg Oncol (2014) 21:S736–42. doi: 10.1245/s10434-014-3886-0
93. Han J, Meng Q, Shen L, Wu G. Interleukin-6 induces fat loss in cancer cachexia by promoting white adipose tissue lipolysis and browning. Lipids Health Dis (2018) 17(1):14. doi: 10.1186/s12944-018-0657-0
94. Shuvalov O, Petukhov A, Daks A, Fedorova O, Vasileva E, Barlev N. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget (2017) 8(14):23955–77. doi: 10.18632/oncotarget.15053
95. Tsai K, Yeh TS, Wu RC, Lai YC, Chiang MH, Lu KY, et al. Metabolomic alterations and chromosomal instability status in gastric cancer. World J Gastroenterol (2018) 24(33):3760–9. doi: 10.3748/wjg.v24.i33.3760
96. Yu L, Aa J, Xu J, Sun M, Qian S, Cheng L, et al. : Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. J Gastroenterol Hepatol (2011) 26:1290–7. doi: 10.1111/j.1440-1746.2011.06724.x
97. Liang Q, Wang C, Li B. Metabolomic analysis using liquid chromatography/ mass spectrometry for gastric cancer. Appl Biochem Biotechnol (2015) 176(8):2170–84. doi: 10.1007/s12010-015-1706-z
98. Wu H, Xue R, Tang Z, Deng C, Liu T, Zeng H, et al. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem (2010) 396:1385–95. doi: 10.1007/s00216-009-3317-4
99. Tsuchiya A, Nishizaki T. Anticancer effect of adenosine on gastric cancer via diverse signaling pathways. World J Gastroenterol (2015) 21(39):10931–5. doi: 10.3748/wjg.v21.i39.10931
100. Cao W, Tian W, Hong J, Li D, Tavares R, Noble L, et al. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol (2013) 304:G322–7. doi: 10.1152/ajpgi.00263.2012
101. Yu JH, Zheng JB, Qi J, Yang K, Wu YH, Wang K, et al. Bile acids promote gastric intestinal metaplasia by upregulating CDX2 and MUC2 expression via the FXR/NF-κB signalling pathway. Int J Oncol (2019) 54:879–92. doi: 10.3892/ijo.2019.4692
102. Lee S-M, Park MS, Park S-Y, Choi Y-D, Chung JO, et al. Primary bile acid activates egr-1 expression through the MAPK signaling pathway in gastric cancer. Mol Med Rep (2022) 25(4):129. doi: 10.3892/mmr.2022.12646
103. Shibata W, Maeda S, Hikiba Y, Yanai A, Sakamoto K, Nakagawa H, et al. C-jun NH2-terminal kinase 1 is a critical regulator for the development of gastric cancer in mice. Cancer Res (2008) 68:5031–9. doi: 10.1158/0008-5472.CAN-07-6332
104. Chen R, Masuo K, Yogo A, Yokoyama S, Sugiyama A, Seno H, et al. SNAIL regulates gastric carcinogenesis through CCN3 and NEFL. Carcinogenesis (2021) 42:190–201. doi: 10.1093/carcin/bgaa133
105. Wu Y-C, Chiu C-F, Hsueh C-T, Hsueh C-T. The role of bile acids in cellular invasiveness of gastric cancer. Cancer Cell Int (2018) 18:75. doi: 10.1186/s12935-018-0569-0
106. Grifin LE, Djuric Z, Angiletta CJ, Mitchell CM, Baugh ME, Davy KP, et al. A Mediterranean diet does not alter plasma trimethylamine n-oxide concentrations in healthy adults at risk for colon cancer. Food Funct (2019) 10(4):2138–47. doi: 10.1039/C9FO00333A
107. Kim KB, Yang JY, Kwack SJ, Kim HS, Ryu DH, Kim YJ, et al. Potential metabolomic biomarkers for evaluation of adriamycin efficacy using a urinary 1H-NMR spectroscopy. J Appl Toxicol (2013) 33:1251–9. doi: 10.1002/jat.2778
108. Matson V, Chervin CS, Gajewski TF. Cancer and the microbiome-influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology (2021) 160:600–13. doi: 10.1053/j.gastro.2020.11.041
109. Talebi BAA, Taghvaei T, Mobarez AM, Vaira G, Vaira D. High correlation of babA 2-positive strains of helicobacter pylori with the presence of gastric cancer. Intern Emerg Med (2013) 8(6):497–501. doi: 10.1007/s11739-011-0631-6
110. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci (2020) 21(16):5786. doi: 10.3390/ijms21165786
111. Russo F, Linsalata M, Orlando A. Probiotics against neoplastic transformation of gastric mucosa: Effects on cell proliferation and polyamine metabolism. World J Gastroenterol (2014) 20(37):13258–72. doi: 10.3748/wjg.v20.i37.13258
112. Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis (2021) 12(12):1104. doi: 10.1038/s41419-021-04396-y
Keywords: gastric cancer, microbiota, metabolite, metabolomics, association analysis
Citation: Lei C, Gong D, Zhuang B and Zhang Z (2022) Alterations in the gastric microbiota and metabolites in gastric cancer: An update review. Front. Oncol. 12:960281. doi: 10.3389/fonc.2022.960281
Received: 02 June 2022; Accepted: 29 July 2022;
Published: 23 August 2022.
Edited by:
Carlos Fernandez-Pena, St. Jude Children’s Research Hospital, United StatesReviewed by:
Enoch Luis, National Council of Science and Technology (CONACYT), MexicoKarolina Kaźmierczak-Siedlecka, Medical University of Gdansk, Poland
Copyright © 2022 Lei, Gong, Zhuang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daojun Gong, Z2RqNzEwNEAxNjMuY29t
 Changzhen Lei
Changzhen Lei Daojun Gong*
Daojun Gong*