- 1Department of Computer Science, School of Informatics, Xiamen University, Xiamen, China
- 2School of Science and Technology, Hong Kong Metropolitan University, Hong Kong, Hong Kong SAR, China
Breast cancer is the most common cause of cancer death in women. Early screening and treatment can effectively improve the success rate of treatment. Ultrasound imaging technology, as the preferred modality for breast cancer screening, provides an essential reference for early diagnosis. Existing computer-aided ultrasound imaging diagnostic techniques mainly rely on the selected key frames for breast cancer lesion diagnosis. In this paper, we first collected and annotated a dataset of ultrasound video sequences of 268 cases of breast lesions. Moreover, we propose a contrastive learning–guided multi-meta attention network (CLMAN) by combining a deformed feature extraction module and a multi-meta attention module to address breast lesion diagnosis in ultrasound sequence. The proposed feature extraction module can autonomously acquire key information of the feature map in the spatial dimension, whereas the designed multi-meta attention module is dedicated to effective information aggregation in the temporal dimension. In addition, we utilize a contrast learning strategy to alleviate the problem of high imaging variability within ultrasound lesion videos. The experimental results on our collected dataset show that our CLMAN significantly outperforms existing advanced methods for video classification.
1 Introduction
According to the World Cancer Report (1), the number of new cases of breast cancer has reached 2.26 million worldwide in 2020, and breast cancer accounts for about 6.9% of all deaths from cancer worldwide, ranking fifth. Early detection and timely treatment can effectively improve the survival prognosis of breast cancer patients, prolong their survival years, and improve the people’s living standards. Because it is non-invasive, inexpensive, safe, and free of ionizing radiation, ultrasound imaging is currently the most commonly used technique for the early detection of breast lesions. However, ultrasound imaging provides low-quality imaging, mainly because interference from the ultrasound reflective wavefront causes speckle noise on imaging. During the acquisition or examination of a breast sequence, the operator usually needs to apply processing such as filtering, adjusting brightness levels, and scaling the image to improve the quality of ultrasound imaging, whereas interpreting ultrasound imaging usually requires an experienced and well-trained radiologist. However, in some cases, the breast lesion in the ultrasound imaging is ambiguous, and even experienced radiologists are unable to accurately determine its benignity or malignancy [in medical practice, BI-RADS 1–3 are usually considered benign, BI-RADS 4 for suspicious malignant, and BI-RADS 5–6 for malignant (2)].
Computer-assisted technology has provided new ideas for the diagnosis of breast lesions by ultrasound imaging. With the help of computer-aided diagnosis, the operation-dependent impact of ultrasound imaging can be minimized. At the same time, computer-aided diagnosis can also reduce the workload of radiologists. Most of the existing computer-aided diagnostic techniques analyze a single frame (key frames) in the video sequence of pathology acquisition. Although it helps to reduce the computer diagnostic time, it also reveals two significant problems: first, it is challenging to select typical key frames representing pathology samples; second, too much pathology diagnostic information is lost in the video sequence. The field of benign and malignant classification for breast lesions by ultrasound video sequences is in urgent need of research.
Therefore, we propose an automatic diagnosis model for ultrasound sequences, which uses deep learning methods to achieve high accuracy in classification recognition to assist medical diagnosis tasks. The designed diagnostic model weighs spatial dimensional information through the non-local module, on the one hand, and adaptive and fine-grained attention weight scoring for each feature dimension of each frame through the multi-meta attention module, on the other hand, focusing on the key information in the samples in a self-learning manner. This approach can accept samples of different sequence lengths and make full use of the potential connections between frames in the sample by weighting and aggregating the features of each frame through the aggregation module to improve the accuracy of diagnosis.
The contributions of this work can be summarized as follows: a) We develop a new network for learning video-level classification of breast lesions. b) We collected an ultrasound video dataset (268 sequences) for breast lesion classification. c) A deformed feature extraction module is proposed to facilitate high-quality deep feature representation, whereas a multi-meta attention module is developed to acquire key feature information at the video level adaptively. d) The experimental results show that our network achieves a new state-of-the-art performance in the breast ultrasound lesion classification task on our collected dataset.
2 Related work
2.1 Breast ultrasound classification
Classification of breast lesion pathology is a primary task in computer-aided diagnosis projects. Researchers working on breast ultrasound-related topics have proposed a number of effective deep learning schemes. Han et al. (3) used deep convolutional networks pre-trained on grayscale nature images to discriminate between benign and malignant. Although the lesion regions of interest used in this scheme were all provided by radiologists, this study demonstrated that breast lesion features extracted by deep learning–based networks can achieve comparable classification performance to hand-designed feature methods. To further avoid the potential missing effects that result from manual intervention in the region of interest selection, Cheng et al. (4) proposed the utilization of an unsupervised stacked denoising auto-encoder to extract high-level feature representations for breast lesion imaging with supervised fine-tuning training. Diagnosis models constructed in a deep learning manner usually require a large amount of training data to achieve significant classification results. However, because most cases are benign, the imbalance of medical data makes it particularly difficult to collect sufficient training samples. To alleviate the problem of model underfitting due to data scarcity, Fujioka et al. (5) and Pan et al. (6) started to use generative adversarial networks to simulate and enhance breast ultrasound sample data. The synthesized images will be further used for the training of convolutional neural networks. The semi-automatic classification model proposed by Bocchi et al. (7) is an outstanding early work to study breast lesion classification based on ultrasound video sequence data. In their proposed method, each imaging frame of the video is independently classified as benign or malignant after semi-automatic segmentation and morphological feature extraction. Subsequently, the classification results of all frames of the video are integrated to obtain reliable video-level results. This scheme results in a substantial improvement in the correct classification rate compared with the results of a single-image frame. At the same time, the uncertainty of classification judgments for certain frames reflects the clinical situation that lesions may present different characteristic manifestations when viewed from different viewpoints.
2.2 Contrastive learning
Traditional supervised learning methods rely heavily on a large amount of labeled training data available. In addition to the expensive labeling cost, this approach is also vulnerable to generalization error, spurious correlations, adversarial attaches, etc. (8). More and more studies start to find new ways out and start to learn feature representation by self-supervised learning. Contrastive learning is a discriminative approach, which aims to group similar samples closer together and dissimilar samples as far away from each other as possible. For computer vision tasks, methods such as MoCo (9), SwAV (10), and SimCLR (11) have produced comparable results to the state-of-the-art supervised methods in ImageNet (12) dataset. He et al. (9) proposed the momentum contrast method for unsupervised visual representation learning, which trains visual representation by constructing dynamic dictionaries with queueing and moving average encoders to match with encoded queries encoder. Compared with the direct comparison of features in general contrast learning, Caron et al. (10) save computational overhead by clustering data and computing online for different enhancements of the same image. Chen et al. (11) save computational overhead by incremental image augmentation and by feature representation and introducing a learnable linear transformation between the feature representation and contrast loss, further substantially improving the quality of the learned feature.
2.3 Attention mechanism
In the field of image classification, the attention mechanism is used to extract key regions and recognize images by spatial invariance. The STN (Spatial Transformer Networks) proposed by Jaderberg et al. (13) effectively addresses the insensitivity of convolutional networks to different viewpoints of the same thing through the attention mechanism. Wang et al. (14) proposed the non-local model to apply the self-attention mechanism to the computer vision tasks. For an input feature image, each pixel value is derived from the weighted average of other pixel features. SENet (15) proposed the squeeze-and-excitation module, which enhances important channels and suppresses invalid channels by automatically learning the importance of different channel features, thus improving model accuracy and reducing computational effort and complexity. Woo et al. (16) propose CBAM (Convolutional Block Attention Module) based on SENet. It extends the attentional dimension from focusing on the channel dimension to the spatial dimension.
3 Method
Figure 1 shows the schematic illustration of the designed contrastive learning–guided multi-meta attention network (CLMAN). The network determines the input breast ultrasound sequence as benign or malignant, as well as the predicted score given to that. CLMAN consists of two main modules: a feature extraction module and a multi-meta attention module. The feature extraction module performs self-supervision training on the breast ultrasound video dataset by the contrast learning method before the formal training to the learn high-quality feature extraction patterns. For a given breast ultrasound sequence containing T frames, CLMAN first performs feature extraction on each frame by a pretrained feature extraction module to obtain independently encoded high-level feature vectors. Subsequently, the high-level feature vectors are aggregated for each frame in the multi-meta attention module. The module performs adaptive and fine-grained weight scoring along each feature dimension of each frame to form a compact and differentiated representation of breast lesions. Finally, the aggregated video-level feature vectors are used to determine the pathology of breast lesions by a linear classifier.
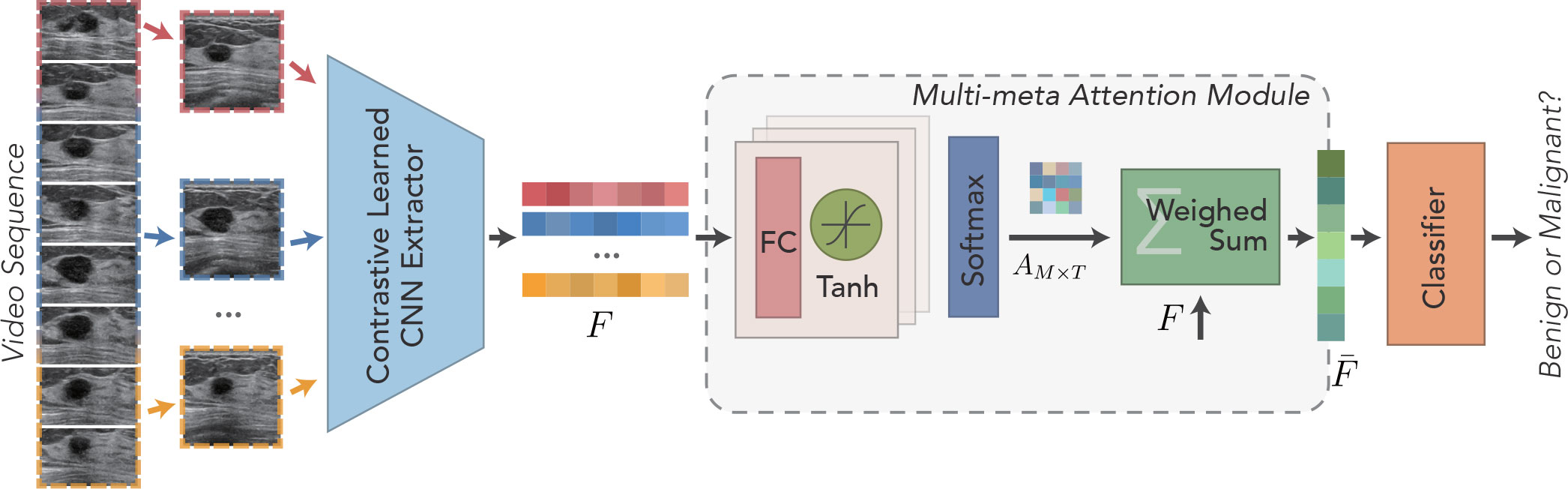
Figure 1 Schematic illustration of the developed Contrastive Learning guided Multi-meta Attention Network (CLMAN) for breast lesion classification in ultrasound sequence.
3.1 Deformed feature extraction module
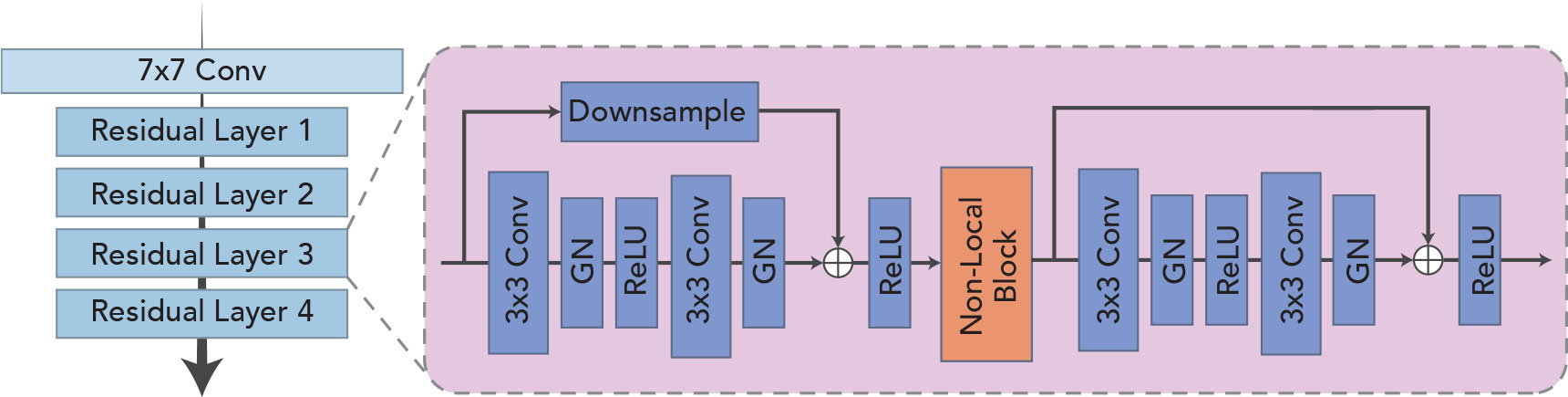
As shown in Figure 2, the feature extraction module is designed to extract features in a sequence and obtain a high-quality feature encoding vector for each frame, which is used for downstream tasks. The module is based on ResNet-18 (17) because the residual structure adopted effectively solves the problem of model degradation due to its depth, and the constant mapping also enhances the information transfer between the upper and lower layers. Because of the inherent multi-frame nature of a sequence, video classification tasks often take smaller batch sizes. Although the amount of training data per batch is sufficient in terms of the number of images, the general batch normalization approach may not be applicable when the model goes normalization because of the high similarity of pixel feature distribution across frames within the same video. In view of this, the group normalization (18) is used in each bottleneck structure in the basic feature extraction module to guarantee the stability of the distribution of the input features. For the problems of low quality and poor contrast of ultrasound imaging, it is especially important to focus on critical regions and suppress invalid regions effectively. The non-local (14) module is introduced and placed in the third and fourth stages of the feature extraction module for capturing spatially distant relationships. It focuses on the correlation between larger objects when the model level is shallow and pays more attention to the correlation between smaller objects when the model level is deep.
Suppose a breast ultrasound sequence V = {vt|t∈[0,T]} , where vt denotes the tth frame and T denotes the index of frames in the sequence. The feature extraction module Θ(·) extracts features from each frame to obtain the high-quality feature coding vector F = {ft|t∈[0,T]} for the whole sequence, which is given by
3.2 Contrast learning strategy
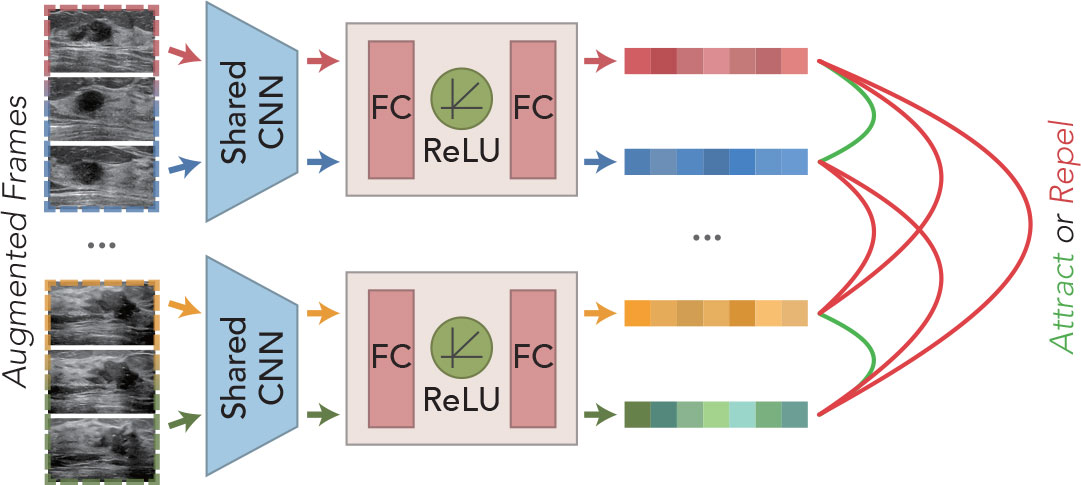
Breast ultrasound tumors tend to be characterized by large intraclass disparities and small interclass disparities in visual presentation. Moreover, the cross-sectional visualization of lesions presented at different stages within the same sequence often varies greatly. How to identify the diversity of different cross-sections of the same lesion is the basis for the correct classification of multi-frame sequences. Inspired by SimCLR (11), we borrowed this method of learning different data augmentation of the same image as positive samples together with negative samples composed of other images to train to determine the proximity of two features and applied it to video data, as shown in Figure 3.
For any N sequence clips, M frames are selected randomly as training samples, and then, the augmented training samples are extracted by the feature extraction network to obtain the high-level feature vectors. The extracted features are cascaded through MLP layers to obtain a tighter feature representation for the model to learn a better similarity representation. Assuming that the training samples provided for learning are , the final feature representation can be obtained by the following:
where τ denotes data augmentation. In the data augmentation stage, we mainly adopt the random combination of flip, crop, scale, modulation of brightness, contrast, and elastic transformation to increase the diversity of sample data.
3.3 Multi-meta attention module
The multi-meta attention module is applied to aggregate high-level feature vectors across frames of video to provide a compact and differentiated representation of mammary nodules. The module adaptively weighs all frames at a fine-grained level along each feature dimension, leveraging the valuable or discriminatory parts of each frame to facilitate commonality recognition without easily discarding or trivializing low-quality frames as the previous approaches have done. The feature extraction module trained by the contrast learning strategy is used to extract feature representations for each frame of the original sequence, denoted as follows:
where Ft denotes the tth frame feature vector with m dimensions.
As shown in Figure 1, a cascading attention module is applied to each frame feature to capture the attention representation better. Each attention module consists of a filter and an activation layer, which are cascaded to perform nonlinear feature learning:
where the fully connected layer is used as the filter and the Tanh function is used as the activation layer σ(·) for nonlinearly transformation. When l = 1, is defined as Ft. For the obtained attention vectors of each frame, the attention linear weights corresponding to each of the Ft channels are obtained by Softmax operation:
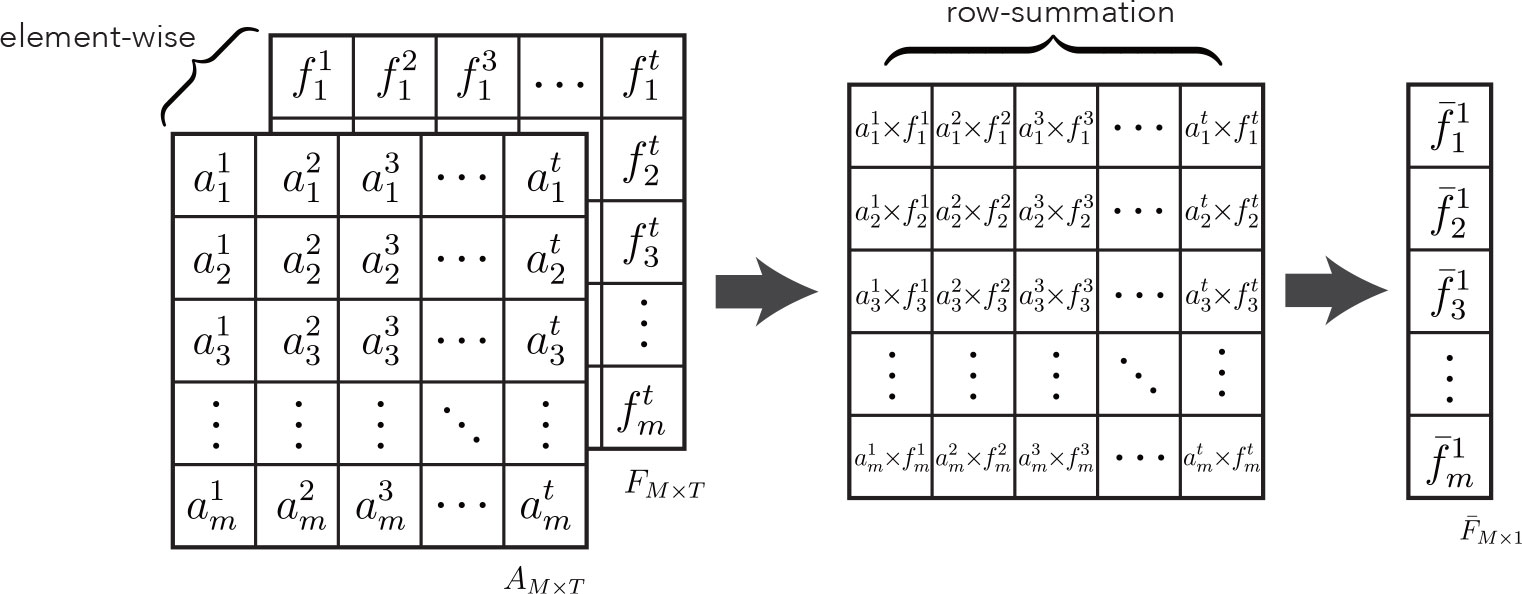
The final aggregated feature is computed by multiplying the attention weights by the cumulative sum of the feature vectors, as shown in Figure 4. The specific aggregation operation can be expressed as follows:
where ⊙ denotes the matrix bitwise product. This aggregation module can weigh the importance of features at the dimensional level. Theoretically, it can achieve the best aggregation with good training. CLMAN uses a fair treatment of each frame of information to maximize the use of any of its valuable local features to facilitate the recognition of lesion sequence. Meanwhile, it is worth noting that the formula degrades to average pooling when each item in the attention matrix A is equal, and of course, the same formula also applies to maximum pooling in extreme cases.
In addition, using this module of mid-term aggregation of features allows the linear classifier to process sequence clips of arbitrary length, whereas the aggregation results have the same vector dimension as the individual features Ft and the order remain constant, i.e., this aggregation module is insensitive to sequence order and temporal information and is generalizable to ultrasound sequence clips of arbitrary start and end points. The module’s parameters can be obtained by the standard backpropagation and gradient descent for supervised learning.
4 Experiments
4.1 Dataset
To evaluate the effectiveness of the developed network, we collected a dynamic breast ultrasound video sequences dataset with 268 videos, of which 152 sequences are malignant and 116 sequences are benign. All sequences are acquired by GE Healthcare equipment (Chicago, IL, USA), with L12-5 ultrasound probe and sampling frequency of 12 MHz, supported by the Xiamen University Xiang’an Hospital. A total of 107 of these sequences are randomly selected as the test set (about 40% of the total data volume), and the rest of the sequences are used as the training set. Data for both the training and test sets are obtained from cases of patients aged 20 years and older with definite benign or malignant pathological findings (BI-RADS categories 3 to 5) of breast lesions as determined by ultrasound.
4.2 Evaluation metrics
The six widely used metrics are utilized for quantitatively comparing different breast lesion ultrasound sequence classification methods. These are accuracy, average precision (AP), sensitivity, specificity, F1 score, and area under curve (AUC).
4.3 Implementation details
Our network is implemented on PyTorch (19) and trained using a SGD (Stochastic Gradient Descent) (20) with 320 epochs, an initial learning rate of 1 × 10−4, a momentum of 0.9, and a weight decay of 5 × 10−4. The sample length T is set to 16, whereas cross-entropy loss is set as the loss function. The whole architecture is trained on one GeForce RTX 2080 Ti GPU, and each GPU has a batch size of 8. In the contrast learning phase, NT-Xent (11) is used as the loss function, and the LARS (21) optimizer is used to train the model in the pre-task with 8,192 epochs, an initial learning rate of 9 × 10−3, and a weight decay of 1 × 10−6. The batch size here is set to 64. The learning rate is adjusted using the Cosine Annealing (22).
4.4 Ablation study
4.4.1 Effectiveness of deformed extraction modules
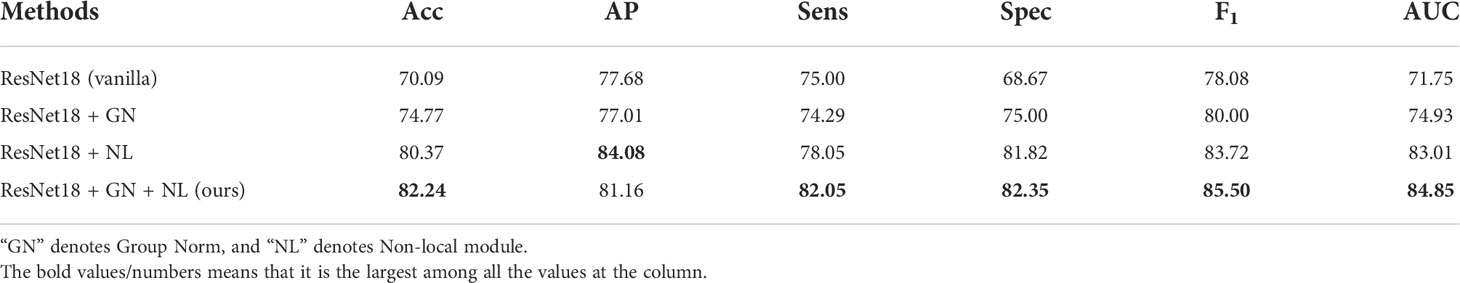
We establish separate control groups based on ResNet-18 and compare the use of different components on the classification performance. As shown in Table 1, “ResNet18 (vanilla)” indicates the most primitive ResNet-18 architecture, “GN” denotes Group Norm, and “NL” denotes non-local module. To avoid the effect of the contrast learning strategy, none of the four settings in Table 1 use that strategy. Compared with the plain ResNet-18 architecture, the feature extraction module with group norm has 4.68%, 6.33%, 1.92% and 3.18% improvement in accuracy, specificity, F1, and AUC, respectively. Meanwhile, the feature extraction module with the non-local module shows a steady increase in all six metrics, with 10.28% increase in accuracy, 6.4% increase in AP, 3.05% increase in sensitivity, 13.15% increase in specificity, 5.64% increase in F1, and 11.26% increase in AUC. The feature extraction module with the group norm and non-local module achieves the average best performance, with accuracy of 82.24%, AP of 81.16%, sensitivity of 82.08%, specificity of 82.35%, F1 of 85.50%, and AUC of 84.85%. It indicates that using the group norm and non-local module for the feature extraction module to obtain high-quality deep features has a certain facilitation effect.
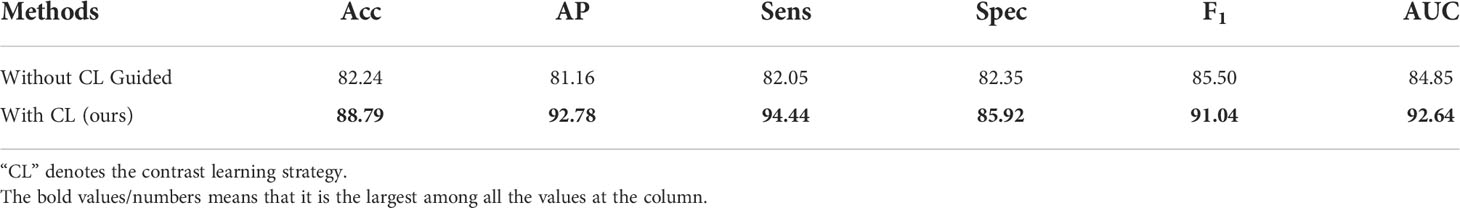
4.4.2 Effectiveness of contrast learning strategy
The feature extraction module used by our network is pre-trained by a contrast learning strategy to effectively identify different geometric patterns of the same lesion under the same sequence imaging before formally training. Table 2 verifies the impact of the contrast learning strategy, which is denoted as “CL”, on the model performance. The experiments show that the performance of the CLMAN decreases when the contrast learning strategy is removed. Specifically, accuracy, AP, sensitivity, specificity, F1, and AUC decreased by 6.55%, 11.62%, 12.39%, 3.57%, 5.54%, and 7.79%, respectively. It suggests that the contrast learning strategy can effectively alleviate the problem of large intraclass differences in the visual presentation of ultrasound lesions.
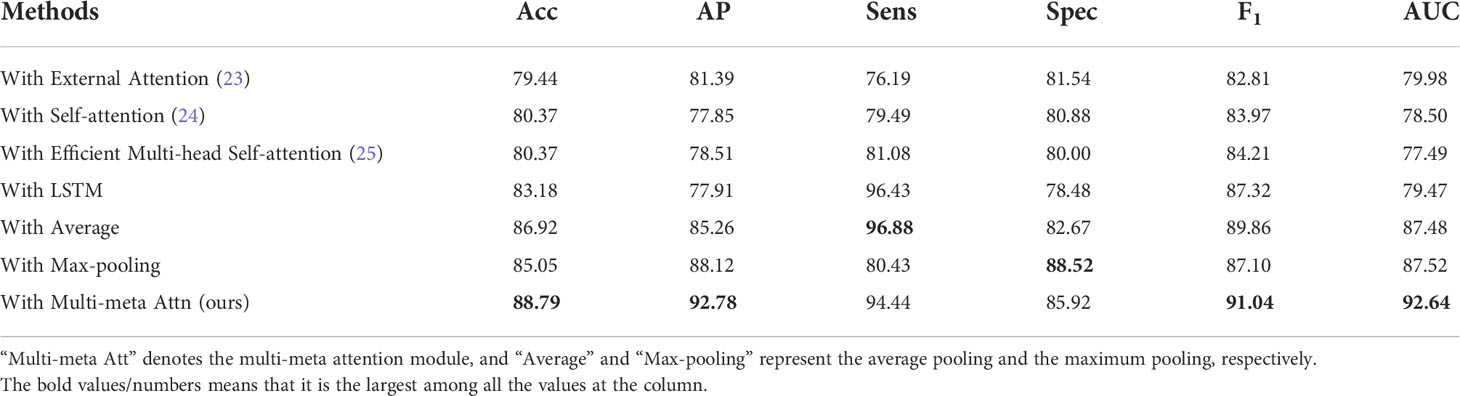
4.4.3 Effectiveness of multi-meta attention module
We conduct ablation experiments of multi-meta attention modules on the CLMAN model. First, the experiment considers the degenerate version of our multi-attention module, i.e., average pooling, as well as the extreme case of the maximum pooling and then compares them. Second, the LSTM (Long Short Term Memory) methods for long sequence feature capture are also compared in this experiment. In addition, we also compared attention modules proposed by other studies to demonstrate the advantage of the multi-meta attention module in video tasks. As shown in Table 3, “Multi-meta Att” denotes the multi-meta attention module, and “Average” and “Max-pooling” represent the degenerate average pooling and the extreme maximum pooling, respectively. According to Table 3 the long sequence feature capture capability of LSTM is not fully applicable to ultrasound video imaging aggregation. The proposed classic attention modules that have often been effective in the past do not seem to be up to our video task. Meanwhile, the simple average pooling and maximum pooling methods achieved the best in terms of sensitivity or specificity, but the other metrics were not satisfactory. The proposed multi-meta attention scheme shows a 1.87% improvement in accuracy, 4.66% improvement in AP, 3.94% improvement in F1, and 5.12% improvement in AUC, with a stronger comprehensive capability. It indicates that the model has different fine-grained trade-offs for each part of the features, whereas such weights are learnable, and the simple and crude average pooling and maximum pooling approaches limit this adaptive capability.
4.5 Comparisons with state of the arts
To demonstrate the effectiveness and feasibility of the designed CLMAN model, Table 4 selects from five papers nine existing methods commonly used to handle video classification task for comparison, including R3D (23), Times Former (27), MC3 (23), P3D (24), R(2 + 1)D (23), TIN(Res18, Res34, Res50)(29), and TSM (25). For providing a fair comparison, we obtain the classification results of all compared methods by exploiting their public implementations or by implementing them. We train these networks on our dataset and only set the batch size and epoch number to the same as ours.
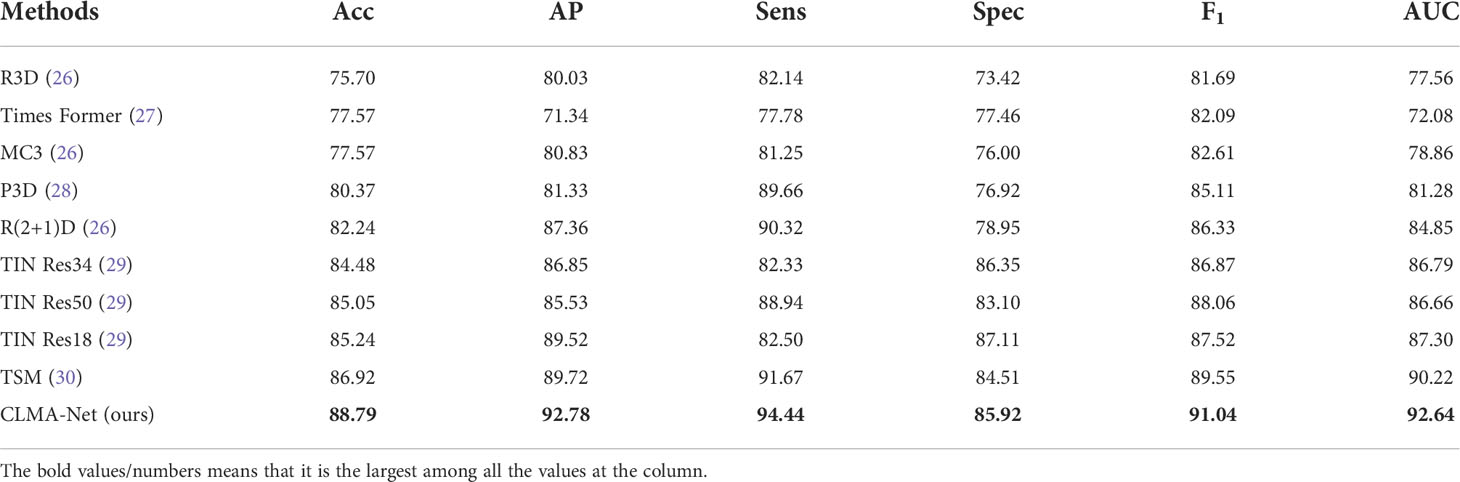
Table 4 Quantitative comparisons of our network and compared methods on the collected ultrasound sequence dataset.
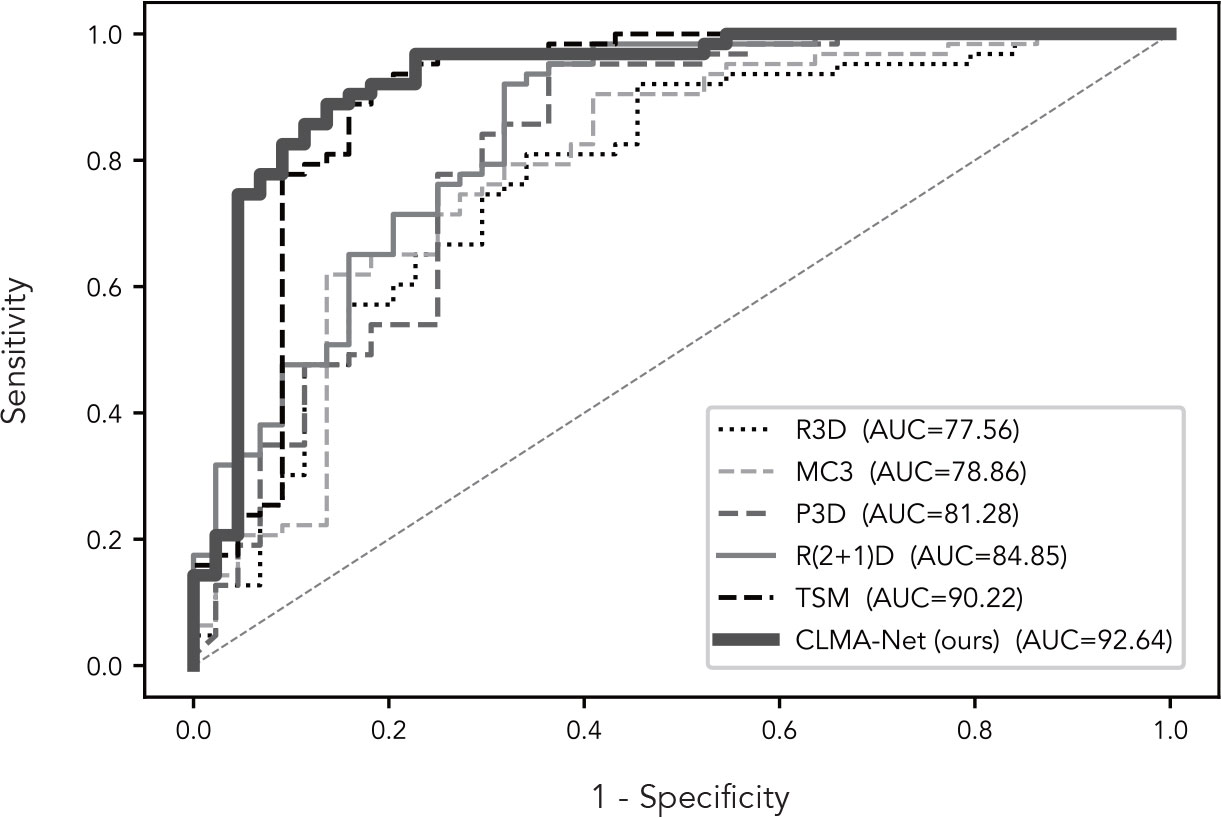
CLMAN performs on par with the best of the methods compared and even better suited for video-level classification tasks of breast ultrasound sequence, with accuracy improved by 1.87%, AP improved by 3.06%, sensitivity improved by 2.77%, specificity by 1.44%, F1 improved by 1.49%, and AUC improved by 2.42%.
More visually, Figure 5 shows the ROC curves of CLMAN with the above five of the nine methods. The performance of R3D, MC3, P3D, and R(2 + 1)D is similar, and the AUC remains around 80%, whereas the area of TSM and CLMAN is comparable, both exceeding 90%.
5 Conclusion
In this paper, we first collected 268 video sequences constituting a video dataset for breast ultrasound classification. Moreover, we propose a CLMAN for lesion diagnosis of ultrasound breast sequences in arbitrary length. Our approach is able to learn the attention weights of each feature dimension adaptively and autonomously in both spatial and temporal dimensions while using a contrast learning predecessor task to effectively address several challenges of the ultrasound video sequence classification problem. Experimental results on the collected dataset show that our network achieves superior diagnostic performance for breast lesions than the state-of-the-art video classification methods.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
XH and SH participated in the design of the study. XH and SH collected the data. ZL and XH performed the statistical analysis. ZL and XH wrote the manuscript. ZL, M-TC, and LW revised the manuscript. FW makes its contributions to prepare and revise the response letter for revised paper,and provide a research grant to cover the publication fee. All authors contributed to the article and agreed to the submitted version.
Acknowledgments
This work was supported by the Research Grants Council of Hong Kong (Project: UGC/FDS16/M08/18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wild C, Weiderpass E, Stewart BW. World cancer report: cancer research for cancer prevention. France: IARC Press (2020).
2. Asbeutah AM, Karmani N, Asbeutah AA, Echreshzadeh YA, AlMajran AA, Al-Khalifah KH. Comparison of digital breast tomosynthesis and digital mammography for detection of breast cancer in kuwaiti women. Med Principles Pract (2019) 28:10–5. doi: 10.1159/000495753
3. Han S, Kang HK, Jeong JY, Park MH, Kim W, Bang WC, et al. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys Med Biol (2017) 62:7714–28. doi: 10.1088/1361-6560/aa82ec
4. Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in us images and pulmonary nodules in ct scans. Sci Rep (2016) 6:1–13. doi: 10.1038/srep24454
5. Fujioka T, Kubota K, Mori M, Kikuchi Y, Katsuta L, Kimura M, et al. Efficient anomaly detection with generative adversarial network for breast ultrasound imaging. Diagnostics (2020) 10:456. doi: 10.3390/diagnostics10070456
6. Pang T, Wong JHD, Ng WL, Chan CS. Semi-supervised gan-based radiomics model for data augmentation in breast ultrasound mass classification. Comput Methods Programs Biomed (2021) 203:106018. doi: 10.1016/j.cmpb.2021.106018
7. Bocchi L, Gritti F, Manfredi C, Giannotti E, Nori J. Semiautomated breast cancer classification from ultrasound video, in: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). Piscataway, NJ: IEEE (2012). p. 1112–5.
8. Liu X, Zhang F, Hou Z, Mian L, Wang Z, Zhang J, et al. Self-supervised learning: Generative or contrastive, in: IEEE Transactions on Knowledge and Data Engineering. Piscataway, NJ: IEEE (2021).
9. He K, Fan H, Wu Y, Xie S, Girshick R. Momentum contrast for unsupervised visual representation learning, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. Piscataway, NJ: IEEE (2020). p. 9729–38.
10. Caron M, Misra I, Mairal J, Goyal P, Bojanowski P, Joulin A. Unsupervised learning of visual features by contrasting cluster assignments. (2020) arXiv preprint (La Jolla: Neural information processing systems foundation). arXiv:2006.09882.
11. Chen T, Kornblith S, Norouzi M, Hinton G. (2020). A simple framework for contrastive learning of visual representations, in: International conference on machine learning (PMLR), New York: PMLR. pp. 1597–607.
12. Deng J, Dong W, Socher R, LJ L, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database, in: 2009 IEEE conference on computer vision and pattern recognition (Ieee). Piscataway, NJ: IEEE (2009). p. 248–55.
13. Jaderberg M, Simonyan K, Zisserman A. Spatial transformer networks. Adv Neural Inf Process Syst (2015) 28:2017–25.
14. Wang X, Girshick R, Gupta A, He K. Non-local neural networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition. (Piscataway, NJ: IEEE) (2018). p. 7794–803.
15. Hu J, Shen L, Sun G. Squeeze-and-excitation networks, in: Proceedings of the IEEE conference on computer vision and pattern recognition. (Piscataway, NJ: IEEE) (2018). p. 7132–41.
16. Woo S, Park J, Lee JY, Kweon IS. in: Cbam: Convolutional block attention module. Proceedings of the European conference on computer vision (ECCV). New York: Springer (2018). p. 3–19.
17. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition. New York: Springer (2016). p. 770–8.
18. Wu Y, He K. Group normalization, in: Proceedings of the European conference on computer vision (ECCV). New York: Springer (2018). p. 3–19.
19. Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: An imperative style, high-performance deep learning library. Adv Neural Inf Process Syst (2019) 32:8026–37.
20. Robbins H, Monro S. A stochastic approximation method. Ann Math Stat (1951) 22:400–7. doi: 10.1214/aoms/1177729586
21. You Y, Gitman I, Ginsburg B. Large Batch training of convolutional networks. (2017). arXiv preprint arXiv:1708.03888.
22. Loshchilov I, Hutter F. Sgdr: Stochastic gradient descent with warm restarts. (2016). arXiv preprint (San Diego: International Conference on Learning Representations). arXiv:1608.03983.
23. Tran D, Wang H, Torresani L, Ray J, LeCun Y, Paluri M. A closer look at spatiotemporal convolutions for action recognition, in: Proceedings of the IEEE conference on Computer Vision and Pattern Recognition. Piscataway, NJ: IEEE (2018). p. 6450–9.
24. Qiu Z, Yao T, Mei T. Learning spatio-temporal representation with pseudo-3d residual networks, in: Proceedings of the IEEE International Conference on Computer Vision. Piscataway, NJ: IEEE (2017). p. 5533–41.
25. Lin J, Gan C, Han S. Tsm: Temporal shift module for efficient video understanding, in: Proceedings of the IEEE/CVF International Conference on Computer Vision. Piscataway, NJ: IEEE (2019). p. 7083–93.
26. Tran D, Wang H, Torresani L, Ray J, LeCun Y, Paluri M. A closer look at spatiotemporal convolutions for action recognition. Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (2018), 6450–9.
27. Bertasius G, Wang H, Torresani L. Is space-time attention all you need for video understanding? 325 Proceedings of the International Conference on Machine Learning (ICML) (2021), 2(3):4.
28. Qiu Z, Yao T, Mei T. Learning spatio-temporal representation with pseudo-3d residual networks. Proceedings of the IEEE International Conference on Computer Vision (2017), 5533–41.
29. Shao H, Qian S, Liu Y. Temporal interlacing network. Proceedings of the AAAI Conference on Artificial Intelligence (2020), Vol. 34, No. 07, 11966–73.
Keywords: ultrasound sequence, video classification, breast lesion, contrastive learning, multi-meta attention network
Citation: Huang X, Lin Z, Huang S, Wang FL, Chan M-T and Wang L (2022) Contrastive learning–guided multi-meta attention network for breast ultrasound video diagnosis. Front. Oncol. 12:952457. doi: 10.3389/fonc.2022.952457
Received: 25 May 2022; Accepted: 18 July 2022;
Published: 24 October 2022.
Edited by:
Guang Yang, Imperial College London, United KingdomReviewed by:
Ping Li, Hong Kong Polytechnic University, Hong Kong SAR, ChinaQiang Zhang, Dalian University of Technology, China
Qiong Wang, Shenzhen Institutes of Advanced Technology (CAS), China
Copyright © 2022 Huang, Lin, Huang, Wang, Chan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moon-Tong Chan, dG10Y2hhbkBoa211LmVkdS5oaw==
 Xiaoyang Huang1
Xiaoyang Huang1 Zhi Lin
Zhi Lin Shaohui Huang
Shaohui Huang Moon-Tong Chan
Moon-Tong Chan Liansheng Wang
Liansheng Wang