- 1P.Herzen Moscow Oncology Research Institute (MORI), Moscow, Russia
- 2Peoples’ Friendship University of Russia, Moscow, Russia
- 3Ulyanovsk Oncology Center, Ulyanovsk, Russia
Lung cancer is a disease with a unique genetic pattern and is occasionally related to hereditary syndromes such as Lynch, Louis–Bar, and Li–Fraumeni. In some patients, germinal mutations may be discovered in combination with somatic alterations. For instance, Li–Fraumeni syndrome often reveals a mixture of TP53 and EGFR mutations. The development of new target therapies necessitates an extensive search for new pathogenic mutations. In this article, we present a rare case report of lung cancer, requiring a pneumonectomy, in three sibling brothers.
Introduction
Lung cancer is a major socioeconomic threat to modern society and is recognized to play a significant role in morbidity and mortality, even in the most developed countries (1). One of the best-known risk factors is smoking, along with pollution.
In recent years, DNA profiling of malignant diseases has become increasingly popular and widely used in clinical practice. Investigation of pathological mutations may provide an accurate treatment plan, including target therapy, thus having been a cornerstone in modern oncology. Key mutations for lung cancer are EGFR, BRAF, and KRAS and alterations in PDL-1, ALK, and ROS1 expression. The current clinical guidelines covering the appropriate administration of immunotherapy and tyrosine kinase inhibitors were devised with consideration of target gene mutations (2).
Germinal mutations are not as common in lung cancer as they are in other tumors, but they are more regularly accompanied with family history. It may be a part of the Lynch, Louis–Bar, or Li–Fraumeni syndromes (3). Given the connection with the latter, germinal mutations in p53 and EGFR are commonly diagnosed (4, 5). To elaborate an explicit and personalized management plan, it is imperative to consider all available options on an extended tumor board with the participation of a geneticist.
Case history
Three male patients, sibling brothers, were independently diagnosed with lung cancer in the left lung. Patient A (56 years old, the oldest one), patient B (63 years old), and patient C (58 years old) were assessed in 2008, 2016, and 2018, respectively. Each of them was referred to the Regional Cancer Centre in Ulyanovsk for further diagnosis and management.
Presentation
All patients were admitted with productive cough, associated with occasional bloody expectoration in patients A and B. They had neither significant comorbidities nor environmental or occupational hazards.
Objective findings
CT scan showed a peripheral 7-cm lesion of the left lower lobe in patient A, a perihilar lesion of the left upper lobe and a tumor with N1 lymph node involvement in patient B, and a centrally located lesion with hypermetabolism of para-aortic lymph nodes with an SUVmax of 7.34 in patient C (Figure 1). In all three cases, a preoperative endoscopic biopsy was performed, which revealed squamous cell carcinoma in patient A and lung adenocarcinoma in patients B and C.
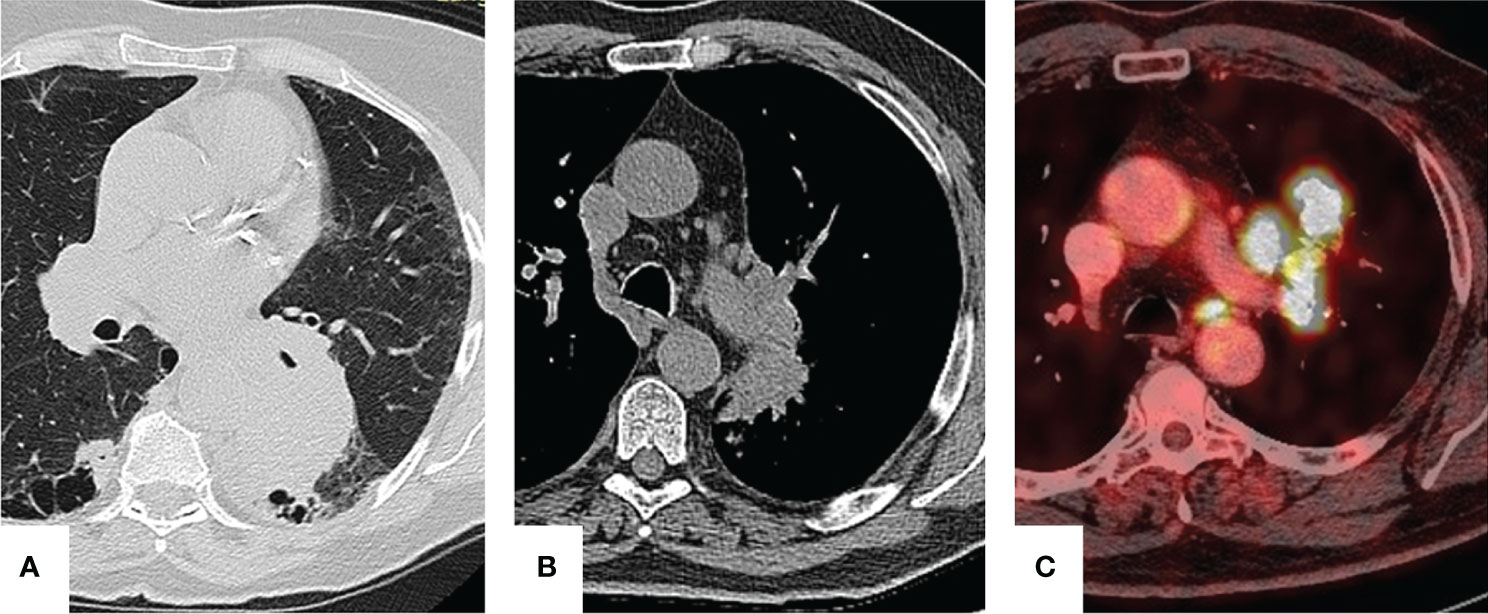
Figure 1 Preoperative imaging. Patient A: a 7-cm lesion is seen in the left lower lobe. Patient B: perihilar lesion with infiltration of bronchopulmonary lymph nodes. Patient C: para-aortic lymph node hypermetabolism.
Diagnosis and management
All patients successfully underwent left-sided pneumonectomy. To downstage the tumor in patient A and because of mediastinal lymph node involvement in patient C, neoadjuvant chemotherapy consisting of etoposide + cisplatin for four cycles was performed.
Postoperative histology showed pT4N0M0, G2, stage IIIa, in patient A; pT2bN1M0, stage IIb, in patient B; and pT2aN2M0, stage IIIa, in patient C. Patients B and C received adjuvant chemotherapy.
After the blood relationship between the patients was revealed, they were referred to a geneticist. Somatic mutations in EGFR, BRAF, KRAS, and PIK3CA genes were ordered first and showed no evidence of presence in any of them. Because of serious concerns about familial history, we evaluated the CHEK2 gene, responsible for the development of different types of cancer, but no mutation was identified. After that, an analysis of microsatellite instability (MSI) was performed, provided by TrueMark MSI Assay (Applied Biosystems, USA) (Tables 1, 2).

Table 2 MSI status associated with functional alteration of the mismatch repair system in patients B and C.
Furthermore, in patient A, a complex molecular genetic testing was completed with the usage of a broad genetic panel: APC, ATM, AXIN2, BARD1, BLM, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDKN2A, CHEK2, DICER1, EPCAM, GALNT12, GREM1, MEN1, MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, NBN, NF1, NTHL1, PALB2, PMS2, POLD1, POLE, PTCH1, PTCH2, PTEN, RAD51C, RAD51D, RET, SMAD4, STK11, SUFU, TP53, TSC1, TSC2, VHL, and WT1. For the naming of the revealed variations, we utilized the nomenclature from http://varnomen.hgvs.org/.
Data processing was carried out with an automated algorithm, comprised of translation alignment of Genome Reference Consortium Human Build 38 (GRCh38), post-processing of alignment, and detection of variants and quality filter, along with the annotation of revealed variants of all known transcripts for each gene from RefSeq based on the usage of the pathogenic potential of substitution prediction (SIFT, PolyPhen2-HDIV, PolyPhen2-HVAR, MutationTaster, MetaSVM) and evolutionary conservative position calculations by PhyloP and PhastCons.
For an appraisal of population prevalence, we adopted samples from the Genome Aggregation Database (gnomAD), Exome Aggregation Consortium (ExAC), 1000 Genomes, and NHLBI Exome Sequencing Project (ESP6500). The classification of nucleotide sequence was accomplished according to a technical standard of next-generation sequencing (NGS), American College of Medical Genetics and Genomics (ACMG) (Table 3).
After amplification and extensive sequencing of lymphocyte’s DNA, we revealed the germinal missense mutation NM_000264.5(PTCH1):c.3941C>T (p.Pro1314Leu, rs1400282737, COSM9550521) in a heterozygous state.
The mutation NM_000264.5(PTCH1):c.3941C>T in the gene PTCH1 is registered in ClinVar and COSMIC databases as a pathologically relevant variant (score 1.00), associated with a high risk of different malignant tumors (Figure 2).
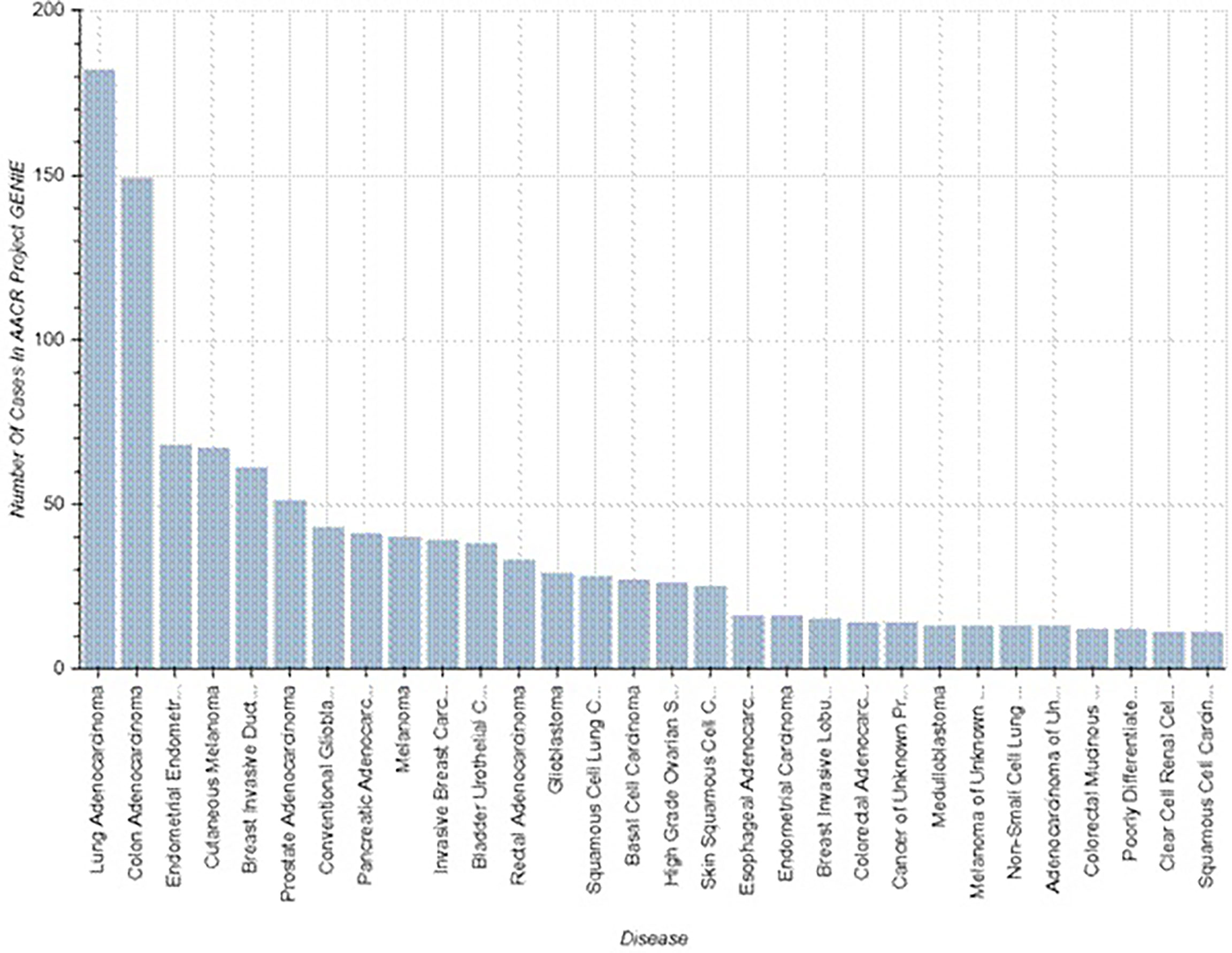
Figure 2 Known cases of malignancy associated with PTCH1 mutations (https://www.mycancergenome.org/content/alteration/ptch1-mutation/).
Discussion
According to GLOBOCAN (2021), lung cancer includes 11.6% of all malignant tumors worldwide and is associated with a large amount of 1-year lethality. Despite significant advances in treatment, it is still a remarkably fatal disease with an average rate of 48.4% of those who die within a year since diagnosis (1).
Genetics, environment, and length of affection may influence the possibility and timing of familial lung cancer cases. A meta-analysis, conducted by The International Lung Cancer Consortium (ILCCO) in 2021, revealed a 1.5-fold increased rate of lung cancer in relatives in the first degree (6). The same findings were shown by Cannon-Albright LA (2019) and Loiola de Alencar (2020), which even stated a two-fold increased rate (7, 8). Ang et al. (2020) appraised high-risk factors of lung cancer, such as Asian race compared to non-Asians, age below 50, smoking, and individuals whose two or more relatives are affected (7). This evidence entails a comprehensive study of genetic factors to determine the optimal pathways for diagnosis, management, and subsequent follow-up.
At the present time, over 754 genes are correlated with lung cancer (8). To establish target genes, DNA diagnostics with polymerase chain reaction are widely implicated. More recently, modern approaches such as NGS have been introduced. The technology is used to determine the order of nucleotides in entire genomes or targeted regions of DNA.
The common pathway for DNA analysis in lung cancer is straightforward. In the case of adenocarcinoma (even in dimorphic combination with squamous cell carcinoma), a molecular genetic assay of EGFR mutations (18–21 exons), BRAF, V600E, ALK, and ROS1 is recommended. For negative or unknown results, further investigation must be proceeded with PDL-1 testing (2).
For a correct perception of the results and to deliberate the familial prepossession, a discussion with a geneticist is advocated. If a connection with one of the genetic syndromes is suspected, a comprehensive DNA testing must be performed.
The PTCH1 gene encodes the patched homolog 1 protein. PTCH1, a 12-pass transmembrane protein, encompasses two large extracellular loops and two large intracellular loops. The PTCH1 protein is one of the membranous receptors involved in Hedgehog signaling (9). Hedgehog signaling is important for embryonic development and tumorigenesis.
PTCH1 is altered in 2.76% of non-small cell lung carcinoma patients with PTCH1 mutation present in 2.56% of all non-small cell lung carcinoma patients (10). The available data on lung cancer features associated with PTCH1 mutation are limited, and no previous study has focused on the long-term survival of these patients. However, it was found that patients with breast cancer and PTCH1 mutation had more metastasis in the lungs and worse recurrence-free survival (11). The features of lung cancer associated with PTCH1 mutation remain to be investigated. In view of the poor prognosis in patient A, a regular follow-up should be provided for life.
It is believed that one of the most important universal causes of cancer development is genomic instability. DNA mismatch repair deficiency leads to microsatellite instability and occurs in 15% of colorectal cancers, leading to the ineffectiveness of standard 5-fluorouracil-based chemotherapy (12).
The MSI-H rate in lung adenocarcinoma has been shown to be rare (0.8%) (13). Patients B and C had MSI-high status; therefore, nivolumab or pembrolizumab can be administered in case of progression.
Overall follow-up comprised 11 years for patient A, 5 years for patient B, and 2 years for patient C with no evidence of recurrence. In case of tumor progression, the thorough genetic testing performed in this study may facilitate the selection of an appropriate treatment option.
Conclusion
This rare observation of familial NSCLC indicates the necessity of a scrupulous analysis of genealogy and elaborate genetic testing. Genetic counseling is mandatory in patients with a familial history of malignancy and the usage of broad panels is advised.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Hertsen Moscow Oncology Research Institute Ethics Committee (#039-B, 23.06.2022). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
AK and OP contributed to the conception and design of the study. EZ and LL organized the database. ET and OA wrote the manuscript. All authors contributed to manuscript revision and read and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw (2022) 20(5):497–530. doi: 10.6004/jnccn.2022.0025
3. Benusiglio PR, Fallet V, Sanchis-Borja M, Coulet F, Cadranel J. Lung cancer is also a hereditary disease. Eur Respir Rev (2021) 30(162):210045. doi: 10.1183/16000617.0045-2021
4. Barbosa MVR, Cordeiro de Lima VC, Formiga MN, Andrade de Paula CA, Torrezan GT, Carraro DM, et al. High prevalence of EGFR mutations in lung adenocarcinomas from Brazilian patients harboring the TP53 p.R337H variant. Clin Lung Cancer (2020) 21:e37–44. doi: 10.1016/j.cllc.2019.11.012
5. Mezquita L, Jové M, Nadal E, Kfoury M, Morán T, Ricordel C, et al. High prevalence of somatic oncogenic driver alterations in patients with NSCLC and Li-fraumeni syndrome. J Thorac Oncol (2020) 15:1232–9. doi: 10.1016/j.jtho.2020.03.005
6. Cotu ML, Liu M, Bonassi S, Neri M, Schwartz AG, Christiani DC, et al. Increased the risk of lung cancer in individuals with a family history of the disease: a pooled analysis from the international lung cancer consortium. Eur J Cancer (2012) 48:1957–68. doi: 10.1016/j.ejca.2012.01.038
7. Ang L, Chan C, Yau WP, Seow WJ. Association between family history of lung cancer and lung cancer risk: a systematic review and meta-analysis. Lung Cancer (2020) 148:129–37. doi: 10.1016/j.lungcan.2020.08.012
8. Wang CY, Chang YC, Kuo YL, Lee KT, Chen PS, Cheung CHA, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet (2008) 40(12):1407–9. doi: 10.1038/ng.273
9. Harvey MC, Fleet A, Okolowsky N, Hamel PA. Distinct effects of the mesenchymal dysplasia gene variant of murine patched-1 protein on canonical and non-canonical hedgehog signaling pathways. J Biol Chem (2014) 289(15):10939–49. doi: 10.1074/jbc.M113.514844
10. The AACR Project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov (2017) 7(8):818–31. doi: 10.1158/2159-8290.CD-17-0151
11. Wang CY, Chang YC, Kuo YL, Lee KT, Chen PS, Cheung CHA, et al. Mutation of the PTCH1 gene predicts recurrence of breast cancer. Sci Rep (2019) 9:16359. doi: 10.1038/s41598-019-52617-4
12. Devaud N, Gallinger S. Chemotherapy of MMR-deficient colorectal cancer. Fam Cancer (2013) 12(2):301–6. doi: 10.1007/s10689-013-9633-z
Keywords: lung cancer, familial cancer, genome sequencing, pneumonectomy, immunotherapy
Citation: Kaprin A, Pikin O, Ryabov A, Aleksandrov O, Toneev E, Lubchenko L and Zelenova E (2022) Case Report: A rare case of familial lung cancer requiring pneumonectomy in three male siblings. Front. Oncol. 12:947210. doi: 10.3389/fonc.2022.947210
Received: 18 May 2022; Accepted: 06 July 2022;
Published: 02 August 2022.
Edited by:
Xiao Zhu, Guangdong Medical University, ChinaReviewed by:
Mengmeng Huang, Boston Children’s Hospital and Harvard Medical School, United StatesYanhan Ren, Rosalind Franklin University of Medicine and Science, United States
Copyright © 2022 Kaprin, Pikin, Ryabov, Aleksandrov, Toneev, Lubchenko and Zelenova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oleg Aleksandrov, b2xlZy5hbGV4YW5kcm92QGljbG91ZC5jb20=
 Andrey Kaprin1,2
Andrey Kaprin1,2 Oleg Aleksandrov
Oleg Aleksandrov
