- Department of Radiation Medicine, Georgetown University Hospital, Washington, DC, United States
Purpose: To investigate DNA double strand breaks (DSBs) induced by therapeutic proton beams in plateau and Bragg peak to demonstrate DSB induction due to the higher LET in the Bragg peak.
Materials and Methods: pUC19 plasmid DNA samples were irradiated to doses of 1000 and 3000 Gy on a Mevion S250i proton system with a monoenergetic, 110 MeV, proton beam at depths of 2 and 9.4 cm, corresponding to a position on the plateau and distal Bragg peak of the beam, respectively. The irradiated DNA samples were imaged by atomic force microscopy for visualization of individual DNA molecules, either broken or intact, and quantification of the DNA fragment length distributions for each of the irradiated samples. Percentage of the broken DNA and average number of DSBs per DNA molecule were obtained.
Results: Compared to irradiation effects in the plateau region, DNA irradiated at the Bragg peak sustained more breakage at the same dose, yielding more short DNA fragments and higher numbers of DSB per DNA molecule.
Conclusion: The higher LET of proton beams at the Bragg peak results in more densely distributed DNA DSBs, which supports an underlying mechanism for the increased cell killing by protons at the Bragg peak.
Introduction
The development of dedicated clinical proton technology has accelerated proton radiotherapy adoption and renewed interest in understanding the fundamental physical and biological properties of proton interactions with living tissues and cells. The RBE variation along the proton Bragg peak, especially towards the distal falloff of the peak has been of particular interest and clinical relevance (1–4). Practical considerations have led to the adoption of an average value of 1.1 for the biological effectiveness of protons relative to photons, regardless of proton energy or depth of beam penetration. However, recent in vitro studies of cell survival have demonstrated a continuous increase in RBE along a proton path and a rapid increase in RBE in the region of the Bragg peak, reaching the highest value of 1.7 (5–8). Furthermore, reported clinical observations have been interpreted to support a non-constant RBE (9, 10). Unexpected brainstem injuries of patients receiving proton brain radiation have been attributed to the effect of increased RBE at the distal end of the proton beam (11, 12). Based on laboratory and pre-clinical data, models have been formulated for calculation of proton RBE as a function of dose, energy and depth (13, 14), and the RBE variations have been incorporated into radiation dose distribution calculations (15).
In addition to cell survival studies, RBE effects have also been investigated by measurements of proton induced DNA DSB. Belli etal. (16) measured DSB in V79 cells by low energy protons using low speed sedimentation technique. They observed a linear correlation between DSB yield and dose but not with LET. Campa etal. (17) measured DSB induced by 0.84 MeV, low energy protons using gel electrophoresis and also reported a linear response with dose. More recently Chaudhary etal. (18) investigated DNA damage and repair along a 60 MeV proton beam path using the 53BP1 foci formation assay and found significant persistence of foci at the distal end of the proton SOBP, suggesting more complex DNA DSB induction by the higher-LET protons. Cuaron etal. (19) and Keta etal. (20) also studied DNA damage using the γH2AX assay by therapeutic proton beam in plateau and distal edge of the SOBP, and found persistently higher γH2AX signal at the distal edge.
Using a plasmid model system, Vyšín etal. (21) studied both direct and indirect damage of DNA by low energy proton irradiation using the agarose gel electrophoresis assay. Using a mathematic model to fit the electrophoresis profile, they derived the number of DSBs per Mbp. The DNA samples were placed in the plateau region and therefore no comparisons were made between the effect of Bragg peak and that of the plateau.
Atomic Force Microscopy (AFM) is an established imaging technique capable of atomic resolution for solid state materials and nanometer resolution for soft biomolecules (22). AFM has been used to image individual DNA molecules and DNA fragments following exposure to neutron, electron, and other ion irradiations for determination of DNA DSBs (23–25). In this investigation, we applied AFM imaging to quantify proton induced DNA breaks using the same plasmid DNA model system (pUC19) that we have used in previous studies at various positions along a proton beam. Compared to other biological assays for DNA strand break measurement, AFM offers the unique ability to measure individual, short DNA fragments of a few nanometers in length and it does not require mathematical models for calculation of DSBs, which can introduce biases due to assumptions in the model. As a result, this approach permits accurate, high-resolution quantification of densely distributed DSBs, and therefore is suited to measure DSBs by high-LET radiations. We demonstrate enhanced DNA breakage at the proton Bragg peak in comparison to the plateau regions at the same doses. These observations are interpreted as direct evidence of the increased LET effects on DNA DSB induction and are consistent with the reported RBE increase for cell killing at the Bragg peaks.
Materials and methods
a. DNA sample preparation
PUC19 plasmid DNA (purchased from New England Biolab in Beverley, MA) in its original concentration of 1 mg/ml in buffer (10 mM Tris-HCl, 1 mM EDTA) was diluted to 5 ng/µl in buffer consisting of 10 mM HEPES and 1 mM MgCl2.
b. Proton irradiation
Proton irradiation of the pUC19 DNA was performed on a Mevion S250i proton system at the Proton Center of Medstar Georgetown University Hospital. The Mevion S250i is a compact, single room clinical proton system with highest energy of 227 MeV. A 50-ton super-conducting synchrocyclotron which is directly mounted on the treatment gantry produces proton beams to yield a dose rate of 2 Gy/min at iso center in a 10 cm3 water volume. Proton scanning is facilitated by a single magnet, dual coil beam scanning apparatus that permits scanning in X- and Y-direction at a scanning speed of 10 m/s. Energy modulation is accomplished with use of an energy selector consisting of 18 Lexan plates of various thicknesses. Insertion of plates of proper thicknesses into the beam reduces the 227 MeV proton beam from the accelerator to desired energies, ranging from 227 MeV to 0 MeV, permitting treatment from skin surface to a depth of 32 cm. The proton spots are further trimmed by a dynamic Adaptive Aperture on the field periphery to reduce beam penumbra. The entire beam monitoring and modulating devices are housed in the treatment nozzle of less than 2 m in length (26).
To facilitate irradiation of the DNA samples at specified depths along the path of proton beam, a 1-cm diameter and 1-mm depth well was drilled in a 2-cm thick, 30cm x 30cm solid water plate at its center. The well can hold 70 µl liquid. A single spot, 110 MeV proton beam was chosen to irradiate the liquid DNA samples at a water equivalent depth of 2 and 9.4 cm, respectively, corresponding to a point on the flat plateau and the distal 50% Bragg peak position. Selection of a single spot beam for irradiation reflects consideration of the proton system to produce high doses in the kGy range for our DNA DSB measurement technique (24). The sigma of the 110 MeV beam is 14.7 mm at the water surface using this setup, and gradually increases with depth in water to reach a maximum of roughly 18 mm at the Bragg peak position. When aligned to the center of the 10-mm diameter well, the 110 MeV single spot beam ensures dose uniformity of 93% in the DNA containing well, taking into consideration a potential 1-mm positioning uncertainty. In the depth direction, when positioned at the Bragg peak with a 1-mm positioning uncertainty, the dose variation in the 1 mm-deep well is less than 5% in the plateau and 10% in the distal Bragg peak.
Using an anterior beam and water equivalent solid water plates, the position of the DNA containing chamber on the 110 MeV beam path is determined by the thickness of the plates, which was calculated using the commercial Raystation treatment planning system and verified by dose measurements using a calibrated PPC05 parallel plate chamber. Table 1 shows calculated doses at 2 cm depth on the plateau and at 9.4 cm depth at 50% distal Bragg peak. Also shown are the monitor units required to deliver 1000 Gy at the two positions and the corresponding LET values.
c. AFM imaging and measurement of DNA fragments
The irradiated DNA samples were extracted from the well using a 100 µl pipette and stored in Eppendorf tubes at –200C until use. As a control, an unirradiated DNA sample was subjected to the same storage process. In preparation for AFM imaging, a 2 µl DNA containing buffer was deposited on freshly cleaved mica, followed by a gentle rinse with 600 µl distilled/deionized water, and dried in a gentle flow of nitrogen gas. The AFM images were obtained using a Bruker NanoScope-8 model, operated in ScanAsys mode at a scanning frequency of 1 Hz and scanning area of 2 x 2 µm2.
Measurements of fragment lengths were obtained using a commercial software package, the FemtoScan. Construction of DNA fragment length distributions, from which the number of DSB per DNA, the number of DSB per broken DNA and the spatial distribution of DSB on a DNA molecule follow procedures and formula previously established (24, 25)
Results
Figure 1 shows sample AFM images of unirradiated (1a), irradiated to 1000 Gy at 2 (1b-1) and 9.4 cm (1b-2) and 3000 Gy at 2 (1c-1) and 8.9 cm (1c-2) depths. Unirradiated plasmid DNA molecules remained mostly intact, in circular or slightly supercoiled conformation, while plasmids irradiated to 1000 and 3000 Gy yielded progressively more broken DNA molecules in response to dose. Furthermore, Figures 1b-1, 1b-2 show that irradiation to a dose of 1000 Gy but at two different depths have yielded different amounts of DNA fragmentation. Similarly, as shown in 1c-1 and 1c-2, 3000 Gy at 2 and 8.9 cm yield significantly different amounts of DNA fragmentation, with a more pronounced fragmentation than that for 1000 Gy.
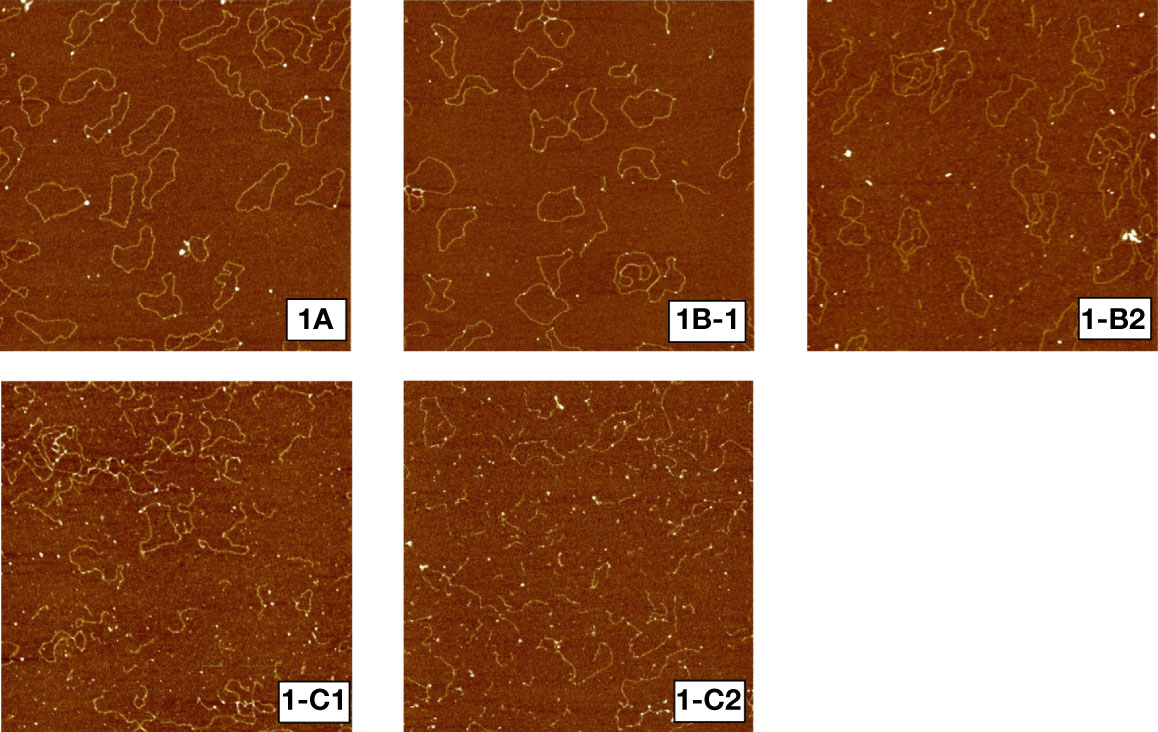
Figure 1 Sample AFM images of pUC19 plasmid DNA. 1a: unirradiated, 1b-1: irradiated to 1000 Gy at 2cm depth, 1b-2: irradiated to 1000 Gy at 9.4 cm depth, 1c-1: irradiated to 3000 Gy at 2 cm depth, 1c-2: irradiated to 3000 Gy at 9.4 cm depth.
Figure 2 shows the fragment size distributions at 1000 and 3000 Gy at the two different depths. Both doses yielded greater quantities of unbroken DNA at 2 cm depth than at 9.4 cm depth. For the 1000 Gy irradiated DNA samples, the size distributions of broken fragments at the two depths are largely similar but with somewhat more fragments in the 0-100 nm region at 9.4 cm depth; in the meantime, more than 80% of the DNA remains intact. However, there are marked differences for the 3000 Gy irradiated samples as demonstrated by a pronounced increase in the number of short DNA fragments in the 50-200 nm region at 9.4 cm depth. Furthermore, less DNA remained intact at 9.4 cm than at 2 cm.
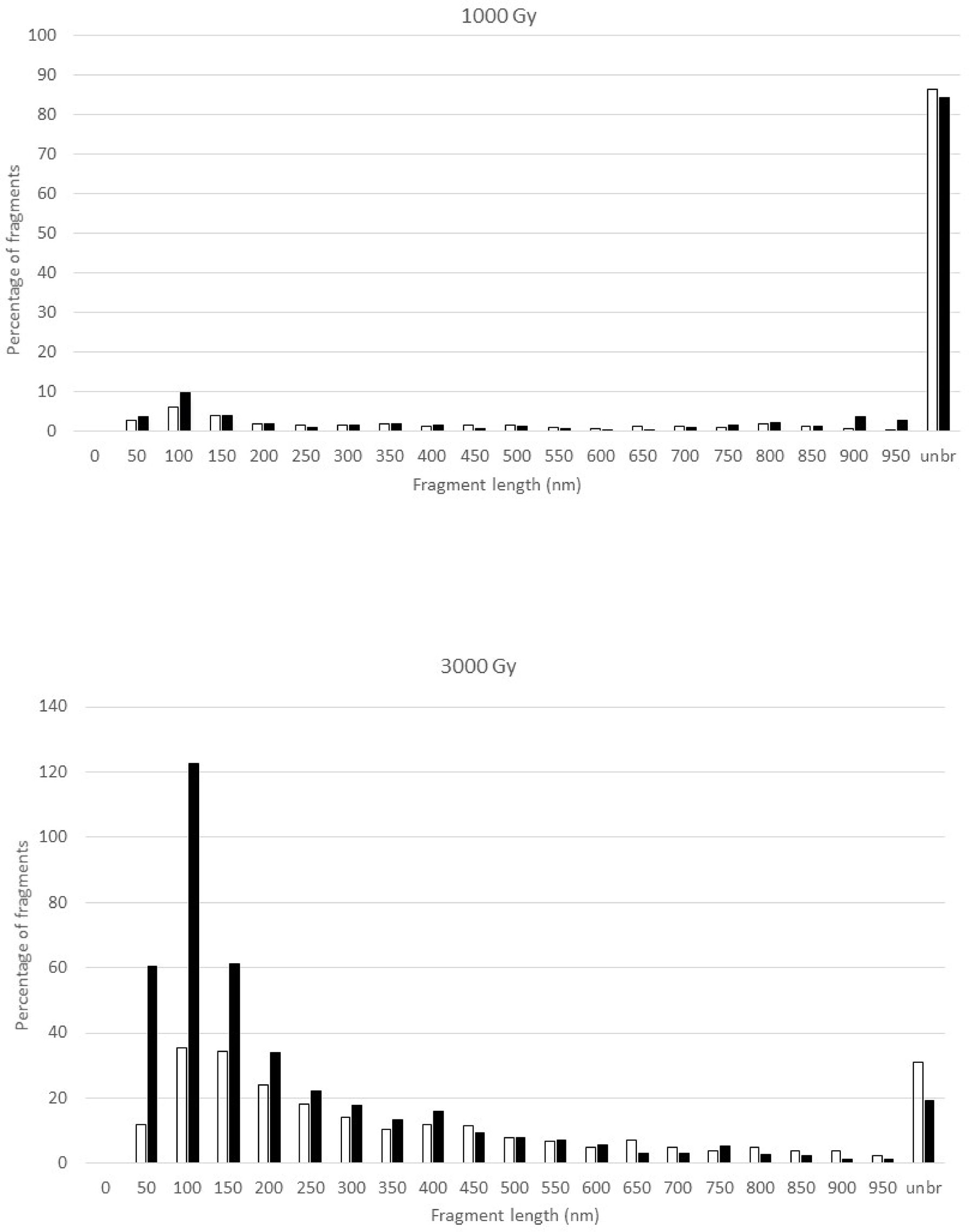
Figure 2 Fragment size distribution of the pUC19 plasmid DNA after exposure to 1000 Gy (upper panel) and 3000 Gy (lower panel) proton irradiation in the plateau (2-cm depth, open bar) and at the peak (9.4-cm depth, solid bar) of the 110 MeV Pristine proton bream. Where the number of fragments in each length bin is normalized to the total number of DNA molecules that include both broken DNA and intact DNA, which results in the percentage of fragments greater than 100 for the 3000 Gy irradiated DNA samples. The Y-axis, the percentage of fragments, is the number of DNA fragments measured in a specific length bin, i.e., 0-50 nm, divided by the total number of DNA molecules measured for a sample, and multiplied by 100.
From the fragment length distributions of Figure 2, we can further calculate the percentage of broken DNA molecules and the average number of DSB per DNA molecule using the formula derived in 24.
Figure 3 shows the percentage of broken DNA at 1000 and 3000 Gy at 2 and 9.4 cm depths. Increase in dose from 1000 to 3000 Gy results in a substantial increase in the fraction of DNA molecules that are broken at both depths. However, a more important observation is that at both 1000 and 3000 Gy, the broken fraction is always higher at 9.4 cm than at 2 cm depth, and the difference increases when the dose is higher.
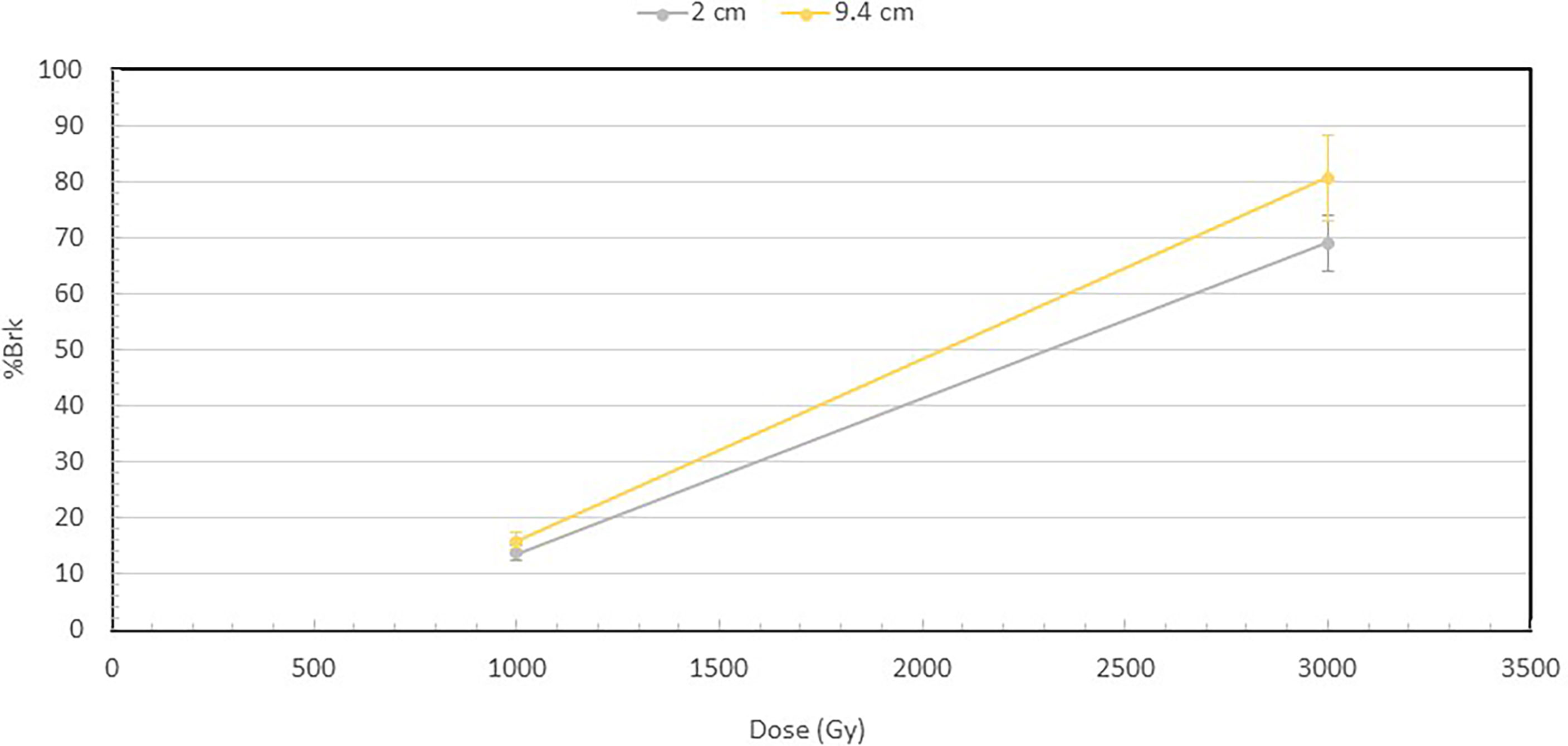
Figure 3 Percentage of broken DNA molecules at the depths of 2 and 9.4 cm for the 1000 and 3000 Gy irradiated DNA. At 1000 Gy, 13.9% of the DNA were broken at 2 cm, while 15.7% at 9.4 cm. At 3000 Gy, 69% of DNA were broken at 2 cm and 80.6% at 9.4 cm. The fraction of broken DNA is the ratio of the number of broken DNA to that of the total number of DNA, which is the sum of both broken DNA and intact DNA. The number of broken DNA is calculated by summing the lengths of the broken DNA fragments and then dividing by the length of an intact DNA.
Figure 4 shows the number of measured DSB per DNA as a function of the LET values at 2 and 9.4 cm depths for both the 1000 and 3000 Gy irradiated DNA samples. At 2 cm depth, the LET is 1.11 keV/µm, but at 9.4 cm depth the LET is substantially higher at 22.6 keV/µm. As shown, the DSB per DNA at both 1000 and 3000 Gy is less than 1 at 2 cm depth. However, at the 9.4 cm depth, the number of DSB per DNA has increased to 2.22 and 3.97, respectively, for the 1000 and 3000 Gy irradiated DNA samples, showing a large dependence on LET.
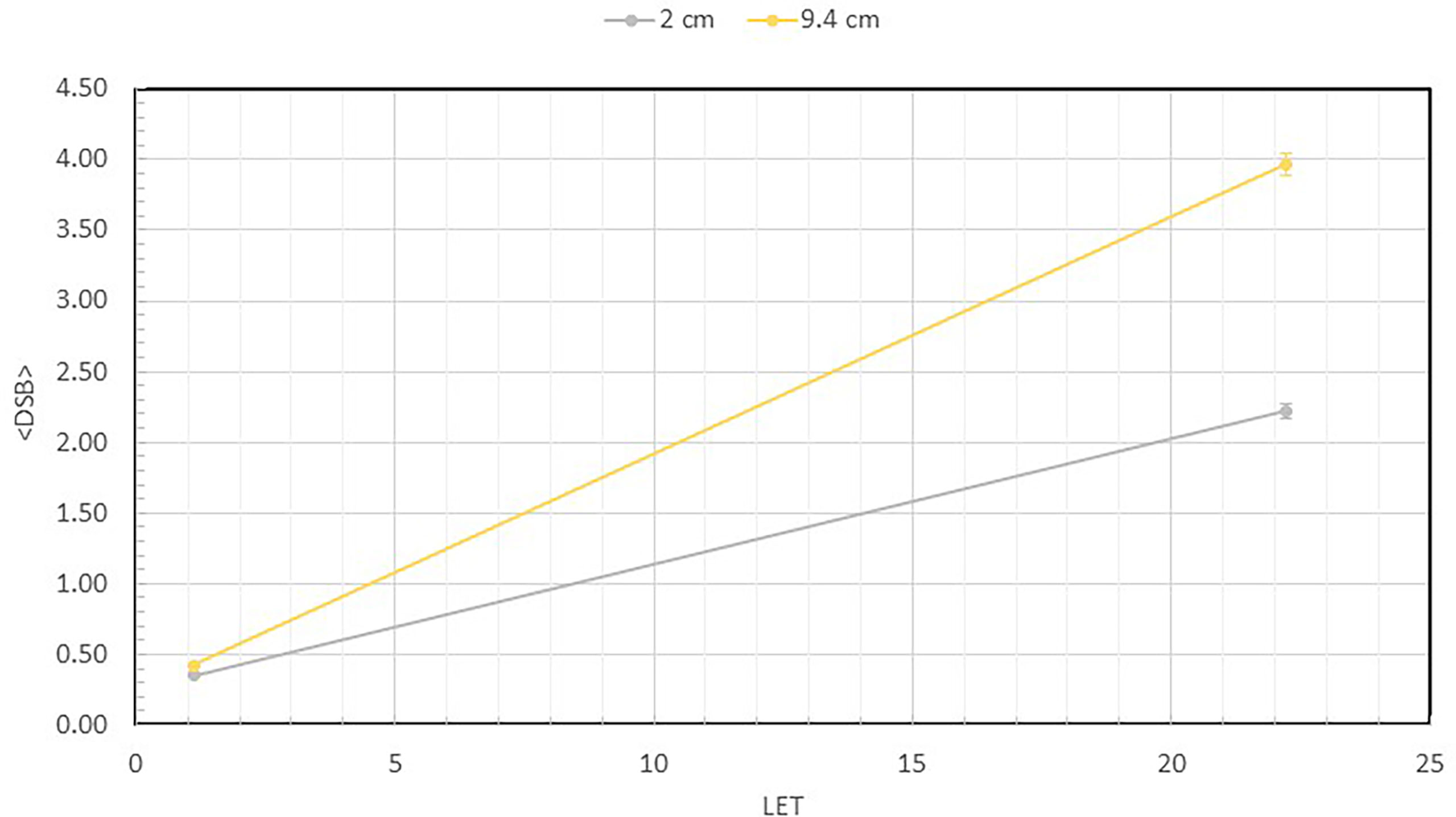
Figure 4 Average number of DSBs per DNA at 2 ad 9.4 cm for both 1000 Gy and 3000 Gy. They are plotted as a function of LET, which shows value of 1.1 keV/µm at 2 cm and 22.6 keV/µm at 9.4 cm depth. This quantity is calculated as the total number of broken DNA fragments divided by the total number of DNA molecules described in Figure 3 for each sample.
Discussion
In this study we investigate critical effects of proton radiation on cells with a focus on potential variation of DNA DSB induction at positions of the Bragg peak. Theoretical calculations have shown a rapid increase in LET along proton beam paths, especially in the Bragg peak region (3). The effects on cell survival have been demonstrated by several investigating groups (5, 27). The increased cell killing capacity of the proton beam in the Bragg peak region, especially towards the end of its range, has significant clinical implications and warrants careful consideration in clinical application of proton irradiation.
The fragmentation distribution profiles shown in Figure 2 show a dose response. This is consistent with what we have observed with other types of radiation. In our previous work, we observed a difference in DNA fragment distribution profiles resulting from the low-LET photon and high-LET neutron irradiation, which is demonstrated by a profile shift towards shorter DNA fragments by neutron radiation (24). This was interpretated as the result of clustered DNA damage by the higher-LET effects of neutrons. In this work with proton irradiation, we observed a significant increase in the number of short DNA fragments less than 250 nm shown in Figure 2 for the DNA sample irradiated at the depth of 9.4 cm as compared to the sample irradiated at 2 cm depth at the same dose of 3000 Gy. We interpret this observation to reflect the enhanced LET effect on DNA fragmentation by proton particles with much higher LET at the Bragg peak.
High-LET radiations produce more densely and clustered DNA damage than low-LET radiation. The clustered DNA damage, which produces more short DNA fragments, is difficult to repair by the cells repair mechanism and leads to greater cellular lethality (28, 29). The significantly enhanced short DNA fragments generation at the Bragg peak measured in this study provides firsthand evidence of clustered DNA damage by therapeutic protons in the Bragg peak region and provides a mechanistic support to the previously reported data of enhanced cell killing at the Bragg peak.
From the fragment size distributions in Figure 2, we calculated the fraction of broken DNA molecules irradiated at 2 cm and 9.4 cm depth to further illustrate the differential DNA damaging effects of protons along the Bragg peak as shown in Figure 3. While there is only a small difference in the number of broken DNAs at the two depths for the 1000 Gy irradiated DNA samples, a substantial difference becomes apparent following exposure of DNA to 3000 Gy irradiation, with 69% of plasmids broken at the 2 cm and 80.6% at the 9.4 cm depth.
To directly quantify the DSB induction, we calculated the average number of DSBs per DNA molecule from the fragment size distribution data using equations from a previous publication (24), and plotted the data as a function of LET. As shown in Figure 4, when irradiated to 1000 Gy, the average DSB per DNA molecule are 0.35 and 0.42, respectively, at 2 and 9.4 cm depth. However, when irradiated to 3000 Gy, a substantial increase is seen to reach 2.22 and 3.97, respectively, showing a much greater number of DSB per DNA at the 9.4 cm depth. We calculated the LET of the 110 MeV beam used in our experiments to be 1.1 keV/µm at 2 cm depth and a substantially higher value of 22.6 keV/µm at 9.4 cm depth. The clear dependence of DNA DSBs on LET is a demonstration of the enhanced DNA damaging effect of the higher LET proton particles at the Bragg peak.
Both Monte Carlo simulation of DNA DSB production (30, 31) and our previous work on AFM measurements have provided evidence for the enhanced short DNA fragment production by high-LET radiations. Here, applying the same AFM measurement technique, we determined DNA DSB induction by clinical protons on their path in the plateau and Bragg peak at the same dose, demonstrating an unambiguous capacity of higher LET protons to generate more DSB.
The direct visualization of individual DNA molecules by AFM offers us the possibility of counting and measuring each DNA fragment with an uncertainty of about 20 nm (24) permitting counting and length measurement of DNA fragments individually. This unique ability of the AFM offers insights into radiation induced DNA DSB induction that are difficult to accomplish by other DNA damage assays.
Proton radiotherapy offers a distinct dosimetric advantage over photon radiotherapy in that proton particles traversing tissues have a unique dose deposition pattern exemplified by a largely flat dose-depth correspondence until the end of their paths where the dose increases rapidly to reach a maximum, followed by a subsequent rapid falloff and stopping of all proton particles. Tissue beyond the particle range will receive no dose at all for a complete sparing. The finite range and distal tissue sparing of protons is the primary reason for enthusiastic adoption of proton radiotherapy. However, it was only recently that the potential biological and clinical impact of the higher LET at the Bragg peak, which is inherent to proton and other heavy charged particle beams, began to be explored. The generic RBE value of current clinical proton beams of 1.1 anywhere along a proton beam path was largely based on in vitro cell survival data where cell irradiations were performed almost exclusively in the plateau region of the proton depth dose curve (3, 14). Recent investigations have demonstrated a variable RBE with clinical implications.
It is accepted that cell killing correlates to unrepaired DNA DSBs, and the effectiveness of radiation in cell killing is due to its ability to inflict DSB (32–35). High-LET radiations generate more complex DSBs that are more difficult to repair than low-LET radiation (28, 36, 37). Our data on DSB production by clinical proton beams provide direct evidence of variable DSB induction along a proton beam path to support the mechanism underlying the variable RBE for cell survival.
This investigation reports the first experimental evidence of DNA DSB induction variation along the path of a clinical proton beam and shows images of resultant broken DNA strands using a plasmid model system. These data provide support for a mechanistic understanding of the enhanced cell killing at the Bragg peak. The increase in short DNA fragment generation at the Bragg peak and the more densely distributed DNA DSB are consistent with reported RBE values.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
CF performed AFM imaging of the DNA samples, counted and measured DNA fragment lengths, assisted with data analysis. YC participated in radiation delivery and assisted with data analyses. JG assisted with DNA fragment measurement. TM and YY assisted with radiation delivery and DNA fragment measurement. AD assisted with manuscript preparation. DP conceived and supervised the project execution and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Britten RA, Nazaryan V, Davis LK, Klein SB, Nichiporov D, Mendonca MS, et al. Variations in the RBE for cell killing along the depth-dose profile of a modulated proton therapy beam. Radiat Res (2013) 179(1):21–8. doi: 10.1667/RR2737.1
2. Gerweck LE, Kozin SV. Relative biological effectiveness of proton beams in clinical therapy. Radiother. Oncol (1999) 50(2):135–42. doi: 10.1016/S0167-8140(98)00092-9
3. Paganetti H, Niemierko A, Ancukiewicz M, Gerweck LE, Goitein M, Loeffler JS, et al. Relative biological effectiveness (RBE) values for proton beam therapy. Int J Radiat Oncol Biol Phys (2002) 53(2):407–21. doi: 10.1016/S0360-3016(02)02754-2
4. Sørensen BS, Bassler N, Nielsen S, Horsman MR, Grzanka L, Spejlborg H, et al. Relative biological effectiveness (RBE) and distal edge effects of proton radiation on early damage in vivo. Acta Oncol (2017) 56(11):1387–91. doi: 10.1080/0284186X.2017.1351621
5. Chaudhary P, Marshall TI, Perozziello FM, Manti L, Currell FJ, Hanton F, et al. Relative biological effectiveness variation along monoenergetic and modulated Bragg peaks of a 62-MeV therapeutic proton beam: a preclinical assessment. Int J Radiat Oncol Biol Phys (2014) 90(1):27–35. doi: 10.1016/j.ijrobp.2014.05.010
6. Howard ME, Beltran C, Anderson S, Tseung WC, Sarkaria JN, Herman MG. Investigating dependencies of relative biological effectiveness for proton therapy in cancer cells. Int J Particle Ther (2017) 4(3):12–22. doi: 10.14338/IJPT-17-00031.1
7. Schuff J, Policastro L, Duran H, Kreiner A, Mazal A, Molinari B, et al. Relative biological effectiveness measurements of low energy proton and lithium beams on tumor cells. Nucl Instruments Methods Phys Res Section B: Beam Interact Mater. Atoms (2002) 187(3):345–53. doi: 10.1016/S0168-583X(01)01136-3
8. Zhang X, Lin SH, Fang B, Gillin M, Mohan R, Chang JY. Therapy-resistant cancer stem cells have differing sensitivity to photon versus proton beam radiation. J Thorac Oncol (2013) 8(12):1484–91. doi: 10.1097/JTO.0b013e3182a5fdcb
9. Bahn E, Bauer J, Harrabi S, Herfarth K, Debus J, Alber M. Late contrast enhancing brain lesions in proton-treated patients with low-grade glioma: clinical evidence for increased periventricular sensitivity and variable RBE. Int J Radiat Oncol Biol Phys (2020) 107(3):571–8. doi: 10.1016/j.ijrobp.2020.03.013
10. Niemierko A, Schuemann J, Niyazi M, Giantsoudi D, Maquilan G, Shih HA, et al. Brain necrosis in adult patients after proton therapy: Is there evidence for dependency on linear energy transfer? Int J Radiat Oncol Biol Phys (2021) 109(1):109–19. doi: 10.1016/j.ijrobp.2020.08.058
11. Indelicato DJ, Flampouri S, Rotondo RL, Bradley JA, Morris CG, Aldana PR, et al. Incidence and dosimetric parameters of pediatric brainstem toxicity following proton therapy. Acta Oncol (2014) 53(10):1298–304. doi: 10.3109/0284186X.2014.957414
12. Peeler CR, Mirkovic D, Titt U, Blanchard P, Gunther JR, Mahajan A, et al. Clinical evidence of variable proton biological effectiveness in pediatric patients treated for ependymoma. Radiother. Oncol (2016) 121(3):395–401. doi: 10.1016/j.radonc.2016.11.001
13. McNamara AL, Schuemann J, Paganetti H. A phenomenological relative biological effectiveness (RBE) model for proton therapy based on all published in vitro cell survival data. Phys Med Biol (2015) 60(21):8399. doi: 10.1118/1.4925192
14. Paganetti H. Relative biological effectiveness (RBE) values for proton beam therapy. variations as a function of biological endpoint, dose, and linear energy transfer. Phys Med Biol (2014) 59(22):R419. doi: 10.1088/00319-9155/59/22/R419
15. Paganetti H. Significance and implementation of RBE variations in proton beam therapy. Technol Cancer Res Treat (2003) 2(5):413–26. doi: 10.1177/153303460300200506
16. Belli M, Cera F, Cherubini R, Ianzini F, Moschini G, Sapora O, et al. DNA Double-strand breaks induced by low energy protons in V79 cells. Int J Radiat Biol (1994) 65(5):529–36. doi: 10.1080/09553009414550621
17. Campa A, Ballarini F, Belli M, Cherubini R, Dini V, Esposito G, et al. DNA DSB induced in human cells by charged particles and gamma rays: experimental results and theoretical approaches. Int J Radiat Biol (2005) 81(11):841–54. doi: 10.1080/09553000500530888
18. Chaudhary P, Marshall TI, Currell FJ, Kacperek A, Schettino G, Prise KM. Variations in the processing of DNA double-strand breaks along 60-MeV therapeutic proton beams. Int J Radiat Oncol Biol Phys (2016) 95(1):86–94. doi: 10.1016/j.ijrobp.2015.07.2279
19. Cuaron JJ, Chang C, Lovelock M, Higginson DS, Mah D, Cahlon O, et al. Exponential increase in relative biological effectiveness along distal edge of a proton Bragg peak as measured by deoxyribonucleic acid double-strand breaks. Int J Radiat Oncol Biol Phys (2016) 95(1):62–9. doi: 10.1016/j.ijrobp.2016.02.018
20. Keta O, Petković V, Cirrone P, Petringa G, Cuttone G, Sakata D, et al. DNA Double-strand breaks in cancer cells as a function of proton linear energy transfer and its variation in time. Int J Radiat Biol (2021) 97(9):1229–40. doi: 10.1080/09553002.2021.1948140
21. Vyšín L, Pachnerová Brabcová K, Štěpán V, Moretto-Capelle P, Bugler B, Legube G, et al. Proton-induced direct and indirect damage of plasmid DNA. Radiat Environ Biophys. (2015) 54(3):343–52. doi: 10.1007/s00411-015-0605-6
22. Pang D, Thierry AR, Dritschilo A. DNA Studies using atomic force microscopy: capabilities for measurement of short DNA fragments. Front Mol Biosci (2015) 2:1. doi: 10.3389/fmolb.2015.00001
23. Gudowska-Nowak E, Psonka-Antonczyk K, Weron K, Elsasser T, Taucher-Scholz G. Distribution of DNA fragment sizes after irradiation with ions. Eur Phys J E Soft Matter (2009) 30(3):317–24. doi: 10.1140/epje/i2009-10522-7
24. Pang D, Berman B, Chasovskikh S, Rodgers J, Dritschilo A. Investigation of neutron-induced damage in DNA by atomic force microscopy: experimental evidence of clustered DNA lesions. Radiat Res (1998) 150(6):612–8. doi: 10.2307/3579883
25. Pang D, Rodgers JE, Berman BL, Chasovskikh S, Dritschilo A. Spatial distribution of radiation-induced double-strand breaks in plasmid DNA as resolved by atomic force microscopy. Radiat Res (2005) 164(6):755–65. doi: 10.1667/RR3425.1
26. Kang M, Pang D. Commissioning and beam characterization of the first gantry-mounted accelerator pencil beam scanning proton system. Med Phys (2020) 47(8):3496–510. doi: 10.1002/mp.13972
27. Matsumoto Y, Matsuura T, Wada M, Egashira Y, Nishio T, Furusawa Y. Enhanced radiobiological effects at the distal end of a clinical proton beam: in vitro study. J Radiat Res (2014) 55(4):816–22. doi: 10.1093/jrr/rrt230
28. Nickoloff JA, Sharma N, Taylor L. Clustered DNA double-strand breaks: biological effects and relevance to cancer radiotherapy. Genes (2020) 11(1):99. doi: 10.3390/genes11010099
29. Pang D, Winters TA, Jung M, Purkayastha S, Cavalli LR, Chasovkikh S, et al. Radiation-generated short DNA fragments may perturb non-homologous end-joining and induce genomic instability. J Radiat Res (2011) 52(3):309–19. doi: 10.1269/jrr.10147
30. Alloni D, Campa A, Belli M, Esposito G, Facoetti A, Friedland W, et al. A Monte Carlo study of the radiation quality dependence of DNA fragmentation spectra. Radiat Res (2010) 173(3):263–71. doi: 10.1667/RR1957.1
31. Friedland W, Dingfelder M, Kundrát P, Jacob P. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat Res. Fundamental Mol Mech Mutagenesis (2011) 711(1-2):28–40. doi: 10.1016/j.mrfmmm.2011.01.003
32. Blöcher D, Pohlit W. DNA Double strand breaks in Ehrlich ascites tumour cells at low doses of X-rays. II. can cell death be attributed to double strand breaks? Int J Radiat Biol Relat. Stud Phys. Chem Med (1982) 42(3):329–38. doi: 10.1080/09553008214551241
33. Frankenberg-Schwager M, Frankenberg D. DNA Double-strand breaks: their repair and relationship to cell killing in yeast. Int J Radiat Biol (1990) 58(4):569–75. doi: 10.1080/09553009014551931
34. Frankenberg D, Frankenberg-Schwager M, Blöcher D, Harbich R. Evidence for DNA double-strand breaks as the critical lesions in yeast cells irradiated with sparsely or densely ionizing radiation under oxic or anoxic conditions. Radiat Res (1981) 88(3):524–32. doi: 10.2307/3575641
35. Ostashevsky J. A model relating cell survival to DNA fragment loss and unrepaired double-strand breaks. Radiat Res (1989) 118(3):437–66. doi: 10.2307/3577405
36. Goodhead DT. Mechanisms for the biological effectiveness of high-LET radiations. J Radiat Res (1999) 40(SUPPL):S1–S13. doi: 10.1269/jrr.40.s1
Keywords: AFM, proton, Bragg peak, DNA, DSB
Citation: Frame CM, Chen Y, Gagnon J, Yuan Y, Ma T, Dritschilo A and Pang D (2022) Proton induced DNA double strand breaks at the Bragg peak: Evidence of enhanced LET effect. Front. Oncol. 12:930393. doi: 10.3389/fonc.2022.930393
Received: 27 April 2022; Accepted: 18 July 2022;
Published: 05 August 2022.
Edited by:
Walter Tinganelli, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Anthony J. Davis, University of Texas Southwestern Medical Center, United StatesYizheng Chen, Stanford University, United States
Copyright © 2022 Frame, Chen, Gagnon, Yuan, Ma, Dritschilo and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dalong Pang, ZGFsb25nLlBhbmdAZ3VuZXQuZ2VvcmdldG93bi5lZHU=
 Cara M. Frame
Cara M. Frame Yu Chen
Yu Chen Jonathan Gagnon
Jonathan Gagnon Anatoly Dritschilo
Anatoly Dritschilo Dalong Pang
Dalong Pang