- 1Department of Pathology, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Pathology, Eastern Hepatobilliary Surgery Hospital, The Second Military Medical University, Shanghai, China
Ferroptosis is an iron-dependent programmed cell death related to the biological process of many kinds of tumors. Long noncoding RNAs (LncRNA) have been found to play essential roles in the tumor, and their functions in the ferroptosis of tumor cells have been partially discovered. However, there is no summary of ferroptosis-related LncRNA and its functions in melanoma. In the present study, we aim to explore the expression profile of ferroptosis-related LncRNA genes and their value in melanoma prognosis by bioinformatics analysis. The expression of ferroptosis-related gene (FRG) from melanoma clinical data was extracted based on the Cancer Genome Atlas (TCGA) database. By screening the RNA expression data of 472 cases of melanoma and 810 cases of normal skin, eighteen ferroptosis-related differential genes were found to be related to the overall survival rate. Furthermore, 384 ferroptosis-related LncRNAs were discovered through constructing the mRNA-LncRNA co-expression network, and ten of them were found with prognostic significance in melanoma by multivariate Cox analysis. Risk assessment showed that the high expression of LncRNA00520 is associated with poor prognosis, while the increased expression of the other LncRNA is beneficial to the prognosis of patients with melanoma. From univariate and multivariate Cox regression analysis, there were ten ferroptosis-related LncRNA risk models towards to be significant independent prognostic factors for patients with melanoma and valuable predictive factors for overall survival (OS)(P<0.05). The ROC curve further suggested that the risk score has relatively reliable predictive ability (AUC=0.718). The protein level of ferroptosis-related genes was verified by the HPA database and IHC test, leading to the discovery that the expressions of ALOX5, PEBP1, ACSL4, and ZEB1 proteins up-regulated in tumor tissues, and existed differences between tumor tissues and normal tissues. In conclusion, we identified ten ferroptosis-related LncRNA and constructed a prognosis model base.
Introduction
Although melanoma is not the most common malignant tumor in skin cancer, it is the leading cause of skin cancer-related death (1, 2), The occurrence and development of melanoma are related to various complex factors, including lifestyle, living environment, genetic and epigenetic changes. Melanoma has significant heterogeneity which would be a major determinant of invasion and metastasis, and its occurrence is closely related to the aberrant regulation of many genes and signal pathways. Increasing evidences showed that many genes involved the development and progression of melanoma, most recurrent mutation is BRAFV600E, which hyperactivates the mitogen-activated protein kinase (MAPK) pathway (3) followed by NRAS mutation (4) and NF1 mutations (5). Among the regulatory pathways implicated in melanoma, MAPK pathway is most common, others include PI3K pathway, Wingless type MMTV integration site (Wnt) signaling (6). Although the treatment of melanoma has been greatly improved in recent years, the prognosis of tumor patients remains poor due to individual differences. The effect of standard chemotherapy on patients with progressive disease is mainly limited, with only 5-10% effective rate (7) and a 27% 5-year survival rate of metastatic melanoma (8). The acquisition of melanoma treatment strategies depends on an accurate understanding of the exact mechanism of the occurrence and development of melanoma. The search for sensitive and accurate biomarkers will better predict the survival and prognosis of patients with melanoma and improve the therapeutic effect.
As well known, cancer cells, including melanoma cells, have an anti-apoptotic property and exhibit malignancy. Ferroptosis is an iron-dependent cell death dependent on the intracellular accumulation of reactive oxygen species (ROS), distinct from apoptosis and autophagy (9). As a non-apoptotic modality of cell death, ferroptosis, is defined as an iron-dependent regulated necrosis that is caused by massive lipid peroxidation-mediated membrane damage featured by a necrosis-like morphological change (10). It is a ROS-dependent form of cell death associated with iron accumulation and lipid peroxidation, involving a series of genes and protein expression including Acyl-CoA synthetase long-chain family member 4 (ACSL4) (11, 12). It determines cell fates through immune and inflammatory reactions. As a form of inflammatory cell death, ferroptosis can release lipid oxidation products to activate NF-κB pathway during tissue injury or tumor therapy.
Increasing evidence indicated that ferroptosis plays an essential role in cancer (12). Dysregulation of iron metabolism is a risk factor for cancer and can promote tumor growth. Therefore, understanding the mechanism of ferroptosis in cancer is of great significance for developing new methods of diagnosis and treatment. Furthermore, long non-coding RNA (Long non-coding RNAs, lncRNAs) is a subset of RNA molecule with more than 200 nucleotidess and lack of protein-coding ability. lncRNAs interact with mRNAs, DNA, proteins and microRNAs (miRNAs) and modulating epigenetic, transcriptional, post-transcriptional, translational and post-translational events of gene expression in mutlitlevels (13). with four interactional ways such as signal, decoy, guide and scaffold (14). LncRNA performs its functions mainly through lncRNA-protein interaction, lncRNA-ceRNA network, lncRNA-miRNA-mRNA silencing, binding to DNA in cis and binding to promoter regions of encoding genes (15). LncRNA participates in the occurrence and development of many kinds of tumors (16–19). Cancer therapy such as chemotherapy could be modulated by LncRNAs (20). so LncRNAs can be used as diagnostic, therapeutic and prognostic markers in cancers (21, 22).
Although the study of ferroptosis is a relatively new field with fast development, it is still a lack of studies on ferroptosis-related LncRNA genes in melanoma. Thus, this study is aimed to explore the expression profile of ferroptosis-related LncRNA and their value in the prognosis of melanoma by bioinformatics analysis. Meanwhile, using the HPA protein database, we probed into the proteins linked to ferroptosis-related LncRNA genes.
Methods
Data collection
RNA sequence (tumor) data and corresponding clinical information of 1416 skin melanoma patients was extracted from TCGA database. The gene expression profiles were normalized by “Limma” R software installation package. The gene expression of normal skin tissue was extracted from the GTEX data sets from UCXC database. Then the intersection of the genes of TCGA and GTEX data sets was made and merged. Both TCGA data and UCXC data are public, thus the study exempts the approval of local ethics committees.
Identification of ferroptosis-related genes
The differentially expressed genes in tumor and non-tumor tissues in the TCGA cohort were screened by “LIMMA” R software package. The overall survival rate (OS) was analyzed by univariate Cox analysis to screen ferroptosis-related genes with prognostic value. Subsequently, heat map visualization, PPI (Protein-Protein Interaction Networks, https://cn.string-db.org/) network interaction and co-expression correlation analysis on these genes were performed.
Construction and verification of ferroptosis-related lncRNAs
We constructed ferroptosis-related mRNA-lncRNA co-expression network via using ferroptosis-related genes with prognostic value according to the correlation coefficient > 0.3 and P < 0.01. Then we identified 384 ferroptosis-related lncRNA using “LIMMA” R software package for Pearson correlation analysis.
Development of ferroptosis-related lncRNA prognostic signatures
We conducted a univariate Cox proportional hazard analysis according to the criteria of P < 0.01. Then, we carried out multivariate Cox analysis using “survival” R package to establish the optimal prognostic risk model based on the risk score (coef (lncRNA1) × expr(lncRNA1) + coef(lncRNA2) × expr(lncRNA2) +… + coef (lncRNAn) × expr(lncRNAn). Coef (LncRNAn) is defined as the lncRNAs coefficient related to survival. ExpR (LncRNAn) is defined as the expression of lncRNAs. According to the median risk score, melanoma patients in TCGA were divided into high-risk group and low-risk group. Kaplan-Meier survival analysis used survival and Servicer R package to estimate the survival difference between the two groups.
Independent prognostic analysis and ROC curve drawing
In order to evaluate the relationship between survival and clinicopathological factors and risk score, univariate and multivariate Cox regression analysis was performed using the SurvivalR software package. The time-dependent receiver operating characteristic (ROC) curve was drawn by the Survival ROC R software package, and the prediction accuracy of different clinicopathological factors and risk scores on survival time was estimated.
Gene enrichment analysis
Gene set enrichment analysis (GSEA) was used for functional annotation. GSEA (https://www.gSea-msigdb.Org/gsia/index.jsp) is a powerful analytical method to explain the whole genome expression profile, and detect the relationship between several cancer-related pathways, metabolic pathways, transcriptional procedures and stress responses in biological processes. P < 0.05 was considered to be statistically significant.
HPA protein database
HPA (human protein atlas) protein database (https://www.proteinatlas.org/) uses transcriptome and proteomics techniques to study protein expression in different human tissues and organs at RNA and protein levels. Up to now, more than 26000 kinds of antibodies have been collected in HPA. The protein expression information in HPA was from the immunohistochemistry (IHC) staining and confirmed by professionals with high accuracy and credibility. Therefore, we use the HPA database to verify the protein level of ferroptosis-related genes.
Immunohistochemistry analysis of clinical samples
Immunohistochemical studies were performed on formalin-fixed paraffin-embedded (FFPE) tissues, and used monoclonal antibodies including anti-ALOX5 (EP6072, dilution 1:100, Abcam), Anti-PEBP1 (EPR2875Y, dilution 1:250, Abcam), Anti-ACSL4 (EPR8640, dilution 1:500, Abcam) and Anti-ZEB1 (EPR17375, dilution 1:1000, Abcam), which was operated on BENCHMARK automatic immunohistochemical instrument. Positive and negative controls were carried out according to the manufacturers’ recommendations. Immunoreactivity result was recorded as negative without tumor cell expression, conversely as positive.
Statistical analysis
All statistical analyses were carried out using R software (version 4.0.3). Normally and non-normally distributed variables were analyzed using the unpaired student’s t-test and the Wilcoxon test, respectively. The co-expression networks of ferroptosis-related lncRNAs-mRNAs with prognostic value were established. Based on FDR, Benjamini-Hochberg method was used to identify the differently expressed lncRNAs.
Results
Identification of prognostic factors related to ferroptosis in TCGA cohort
We uncovered the differentially expressed ferroptosis-related genes in tumor tissues and paracancerous tissues. Univariate Cox regression analysis identified 18 OS-associated genes (Figure 1A). The genes were clustered to generate a heat map,and the map showed CHAC1, P-EBP1, NQO1, HSPB1, ABCC1, ACSL4, SQLE, ALOX12, FANCD2 are highly expressed, and they are related to the poor prognosis of melanoma patients. In contrast, ALOX5, CISD1, ATP5MC3, NFS1, EMC2, ZEB1 are highly expressed in normal tissues, and they are related to the good prognosis of melanoma patients (Figure 1B). Protein interaction network analysis showed that ACACA was a central gene (Figure 1C), and the correlation between genes was shown in Figure 1D.
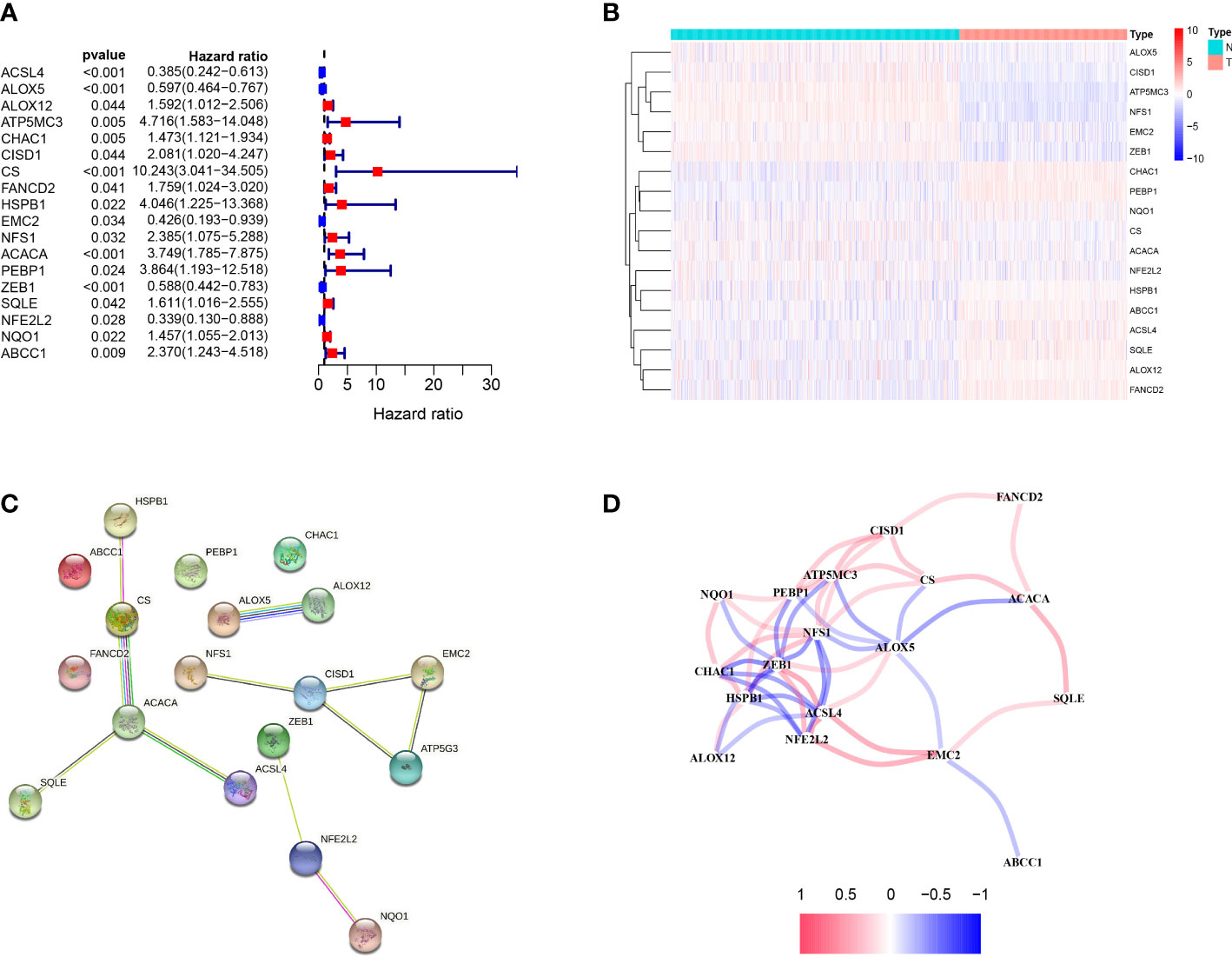
Figure 1 Identification of candidate ferroptosis-related genes in TCGA cohort. (A) Forest plot of univariate Cox regression analysis. (B) The expression of 18 genes in cancer tissues. (C) The interaction between 18 candidate genes in the PPI network. (D) The correlation network of candidate genes. The correlation coefficients are represented by different colors.
Identification of ferroptosis-related lncRNA with important prognostic value In melanoma
A total of 384 ferroptosis-related LncRNAs were obtained by constructing a co-expression network of 18 ferroptosis-related encoding genes (mRNA) with prognostic value. COX regression analysis showed that 46 ferroptosis-related LncRNAs were significantly associated with TCGA melanoma patients (P <0.01), 37 of which were low risk (risk ratio: HR <1), 9 as high-risk (risk ratio: HR> 1). Multivariate Cox analysis further screened 10 prognostic ferroptosis-related LncRNAs. These 10 LncRNAs form an optimal pre-rear risk model of ferroptosis-related LncRNA. As shown in Figures 2A, B, we also established 10 visualized ferroptosis-related LncRNAs-mRNA co-expression networks with prognostic value.
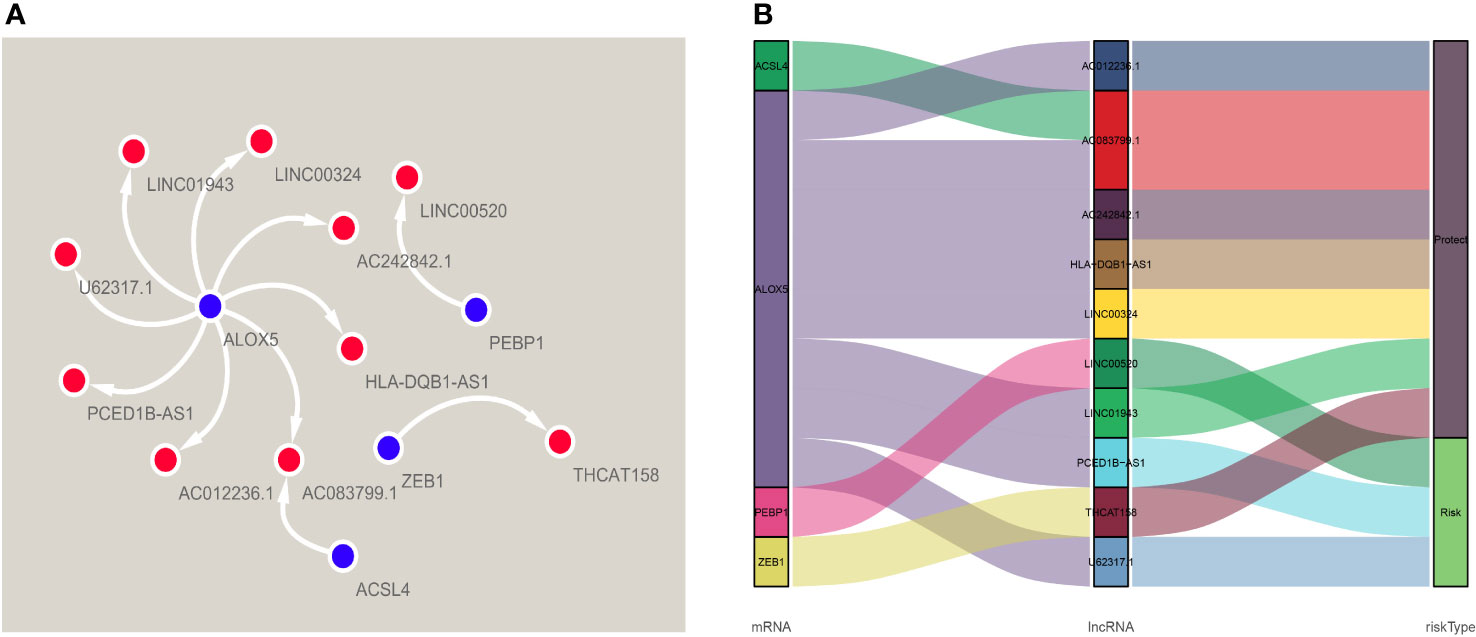
Figure 2 A co-expression network of four ferroptosis-related-lncRNAs-mRNAs with predictive value. (A) The co-expression relationship between ferroptosis genes and lncRNA, visualized by Cytoscape. (B) The co-expression of iron death gene and lncRNA and the correlation of risk value, visualized by Sankey diagram.
According to the risk score formula and the calculated median risk score, we divided melanoma patients into high-risk and low-risk groups. In this study, we drew ten lncRNA survival curves with prognostic value. Despite that LncRNA00520 represents the poor prognosis of patients with melanoma, the high expression of the other several LncRNAs was beneficial to the prognosis of patients with melanoma (Figure 3).
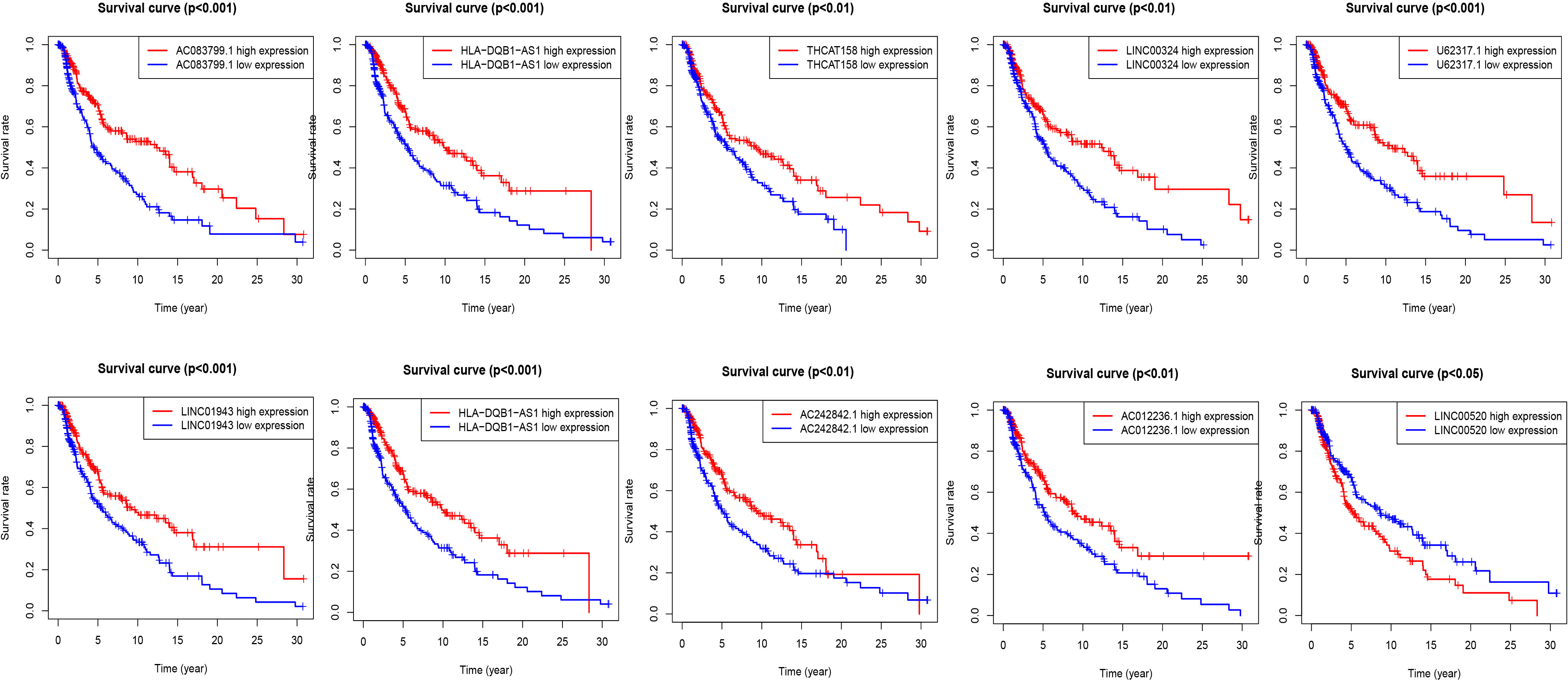
Figure 3 Kaplan-Meier survival curves of lncRNAs that predict the prognosis of melanoma in the TCGA data set. Ten ferroptosis-related-LncRNA were found to be independent prognostic factors for melanoma patients. Nine of them (THCAT158, AC083799.1, AC012236.1, LINC01943, HLA-DQB1-AS1, LINC00324, AC242842.1, PCED1B-AS1, U62317.1) are favorable prognostic factors for melanoma patients, and one of them (LINC00520) is an unfavorable prognostic factor for melanoma.
Kaplan-Meier survival analysis demonstrated that the overall survival rate (OS) of the high-risk group was worse than that in the low-risk group (p <0.001) (Figure 4A). The risk score had a specific predictive value for patients’ prognoses. We use a risk curve and scatter plot to describe the risk score and related survival status of patients with melanoma. The results showed that mortality depended on the risk score (Figures 4B, C). The heat map showed that the expression of LINC00520 was highly expressed in the high-risk group, while the expression of AC012236.1, LINC01943, U62317.1, AC242842.1, HLA−DQB1−AS1, LINC00324, PCED1B−AS1, THCAT158, AC083799.1 was separately up-regulated in the low-risk group (Figure 4D). Therefore, the above studies identified ten LncRNAs associated with ferroptosis, which is significant for the melanoma prognosis.
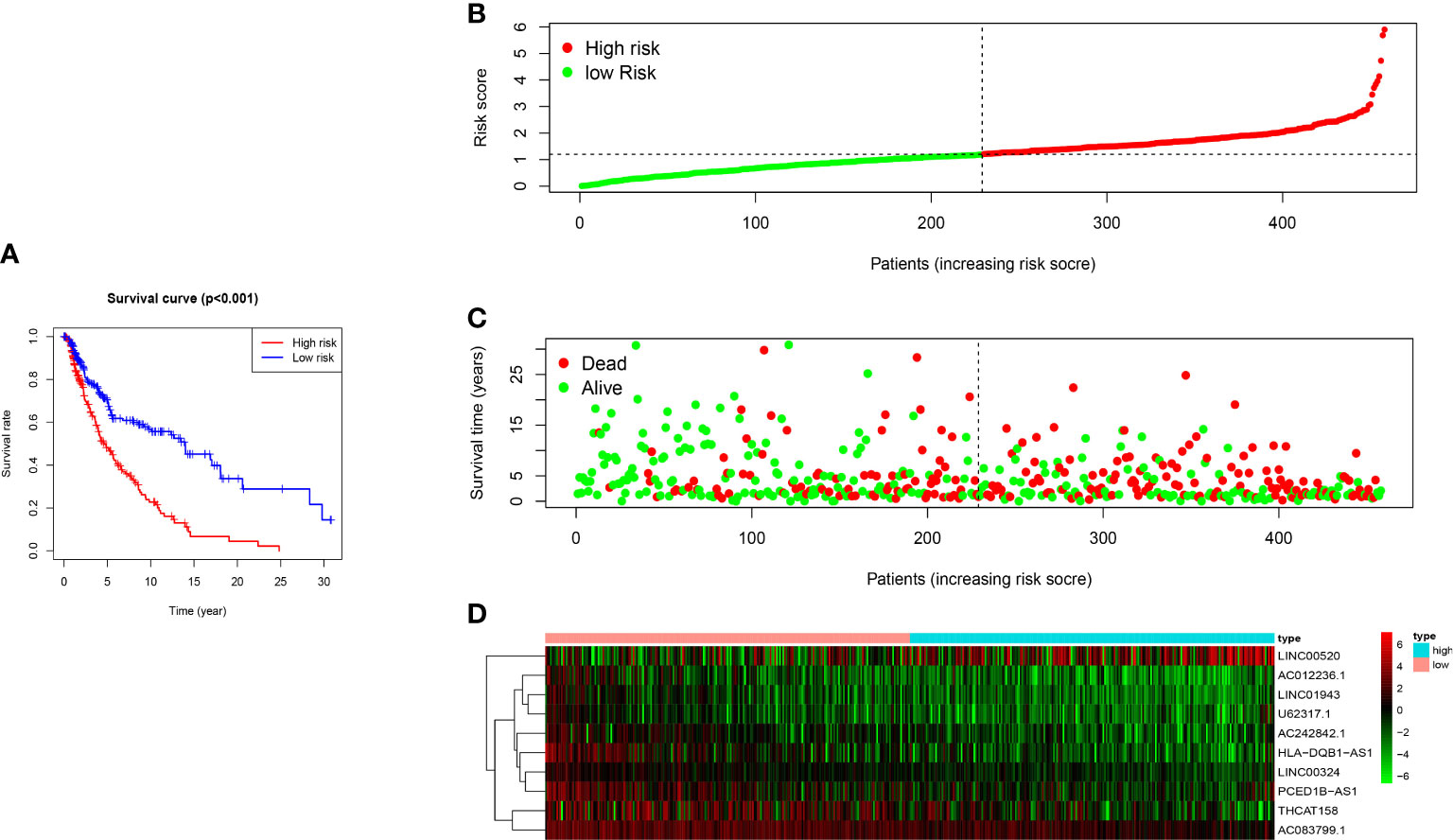
Figure 4 The prognostic value of 10 ferroptosis-related-lncRNA risk models in the TCGA cohort. (A) Kaplan-Meier survival analysis of high-risk and low-risk populations based on risk model and median risk score. (B) Risk curve based on the risk score of each sample. (C) Scatter plot based on the survival status of each sample. The green dot and the red dot represent survival and death, respectively. (D) The heat map shows the expression levels of ferroptosis-related-LncRNA in the high-risk and low-risk groups.
Evaluation of risk model using 10 ferroptosis-related lncRNA as independent prognostic factors in patients with melanoma
We performed univariate and multivariate Cox regression analysis to evaluate whether the above ten ferroptosis-related LncRNA risk models are independent prognostic factors for patients with melanoma. In univariate COX regression analysis (Figure 5A), the risk score and 95%CI (HR) were 1.876 and 1.598-2.202 (P < 0.001), respectively, and in multivariate COX regression analysis (Figure 5B), they were 1.825 and 1.541-2.162 (P < 0.001), respectively. It is suggested that the risk models of 10 ferroptosis-related lncRNA are the most significant prognostic factors for melanoma. In order to evaluate the sensitivity and specificity of risk score in predicting the prognosis of melanoma patients, the area under the ROC curve (AUC) of the risk score was calculated (Figure 5C). The AUC value of the risk score was 0.718, indicating that the prognosis risk model of 10 melanoma iron death related lnc RNAs was quite reliable. Therefore, 10 risk models of iron death related lncRNA are important independent prognostic factors for melanoma cancer patients.
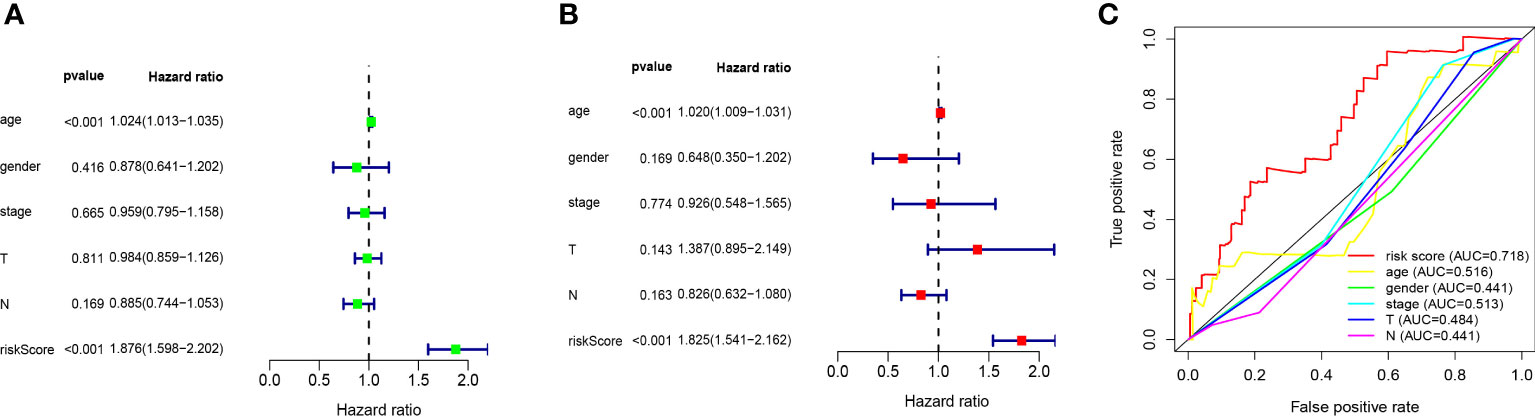
Figure 5 Evaluation of ten ferroptosis-related-lncRNA prognostic risk models in melanoma. (A) Univariate and (B), multivariate Cox regression analysis of risk model score and clinical characteristics on prognosis. (C) The risk model score and AUC of clinical characteristics were calculated according to the ROC curve. Clinical features include age, sex, stage and TNM stage.
GSEA enrichment analysis
Functional pathway enrichment is based on Gene Set Enrichment Analysis (GSEA). The results revealed that most of the ten ferroptosis-related lincRNAs correlate with the metabolism signaling pathway. In the high expression group, ferroptosis-related lincRNAs regulated the glyoxylate and dicarboxylate metabolism signaling pathway, oxidative phosphorylation signaling pathway, and pentose phosphate pathway. As meanwhile, in the low expression group, they trend to affect the JAK-STAT signaling pathway, nod like receptor signaling_pathway, and toll-like receptor signaling pathway (Figure 6, Table 1).
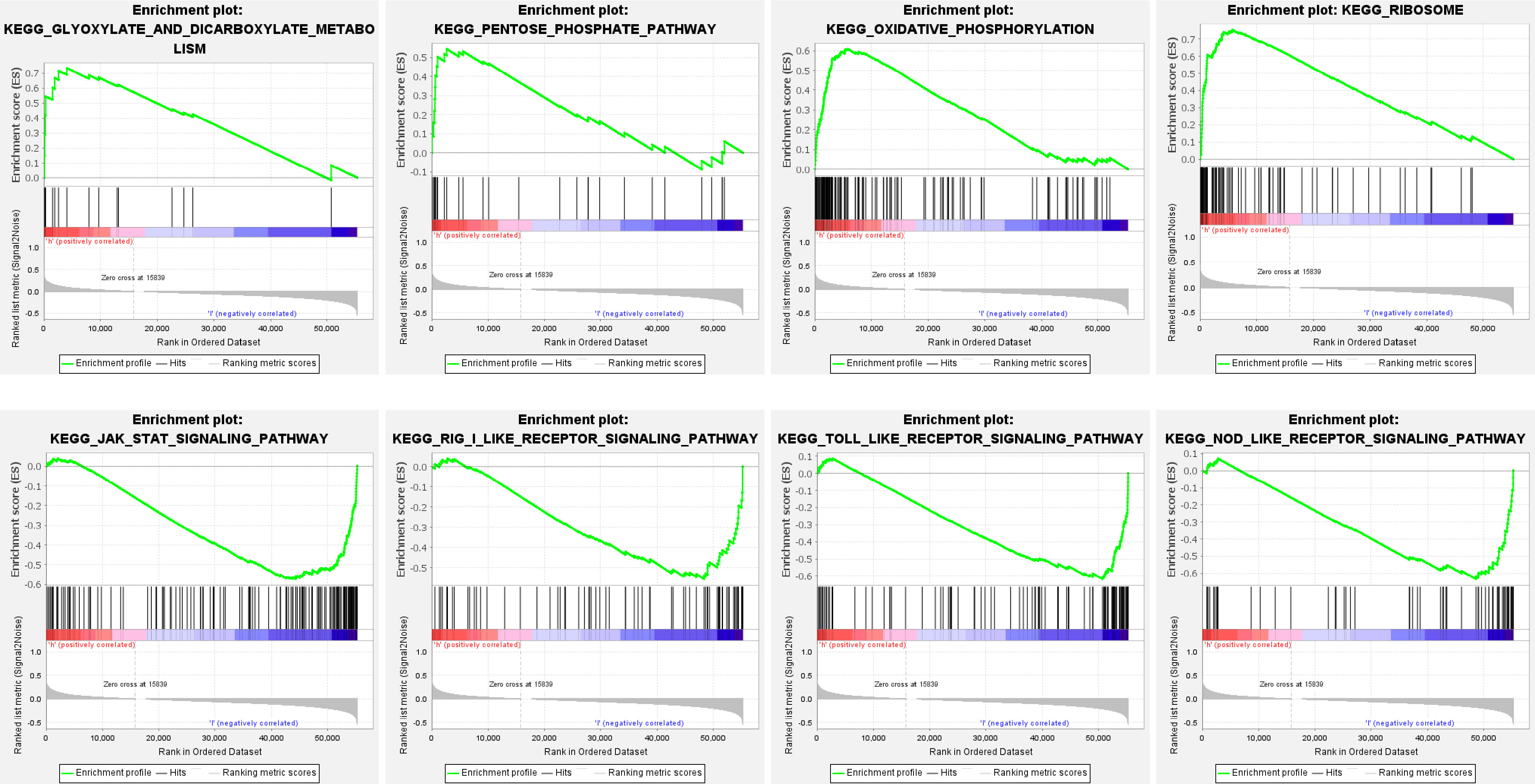
Figure 6 Functional enrichment analysis of ten ferroptosis-related-lncRNA risk models based on GSEA.
HPA and immunohistochemistry (IHC) verification
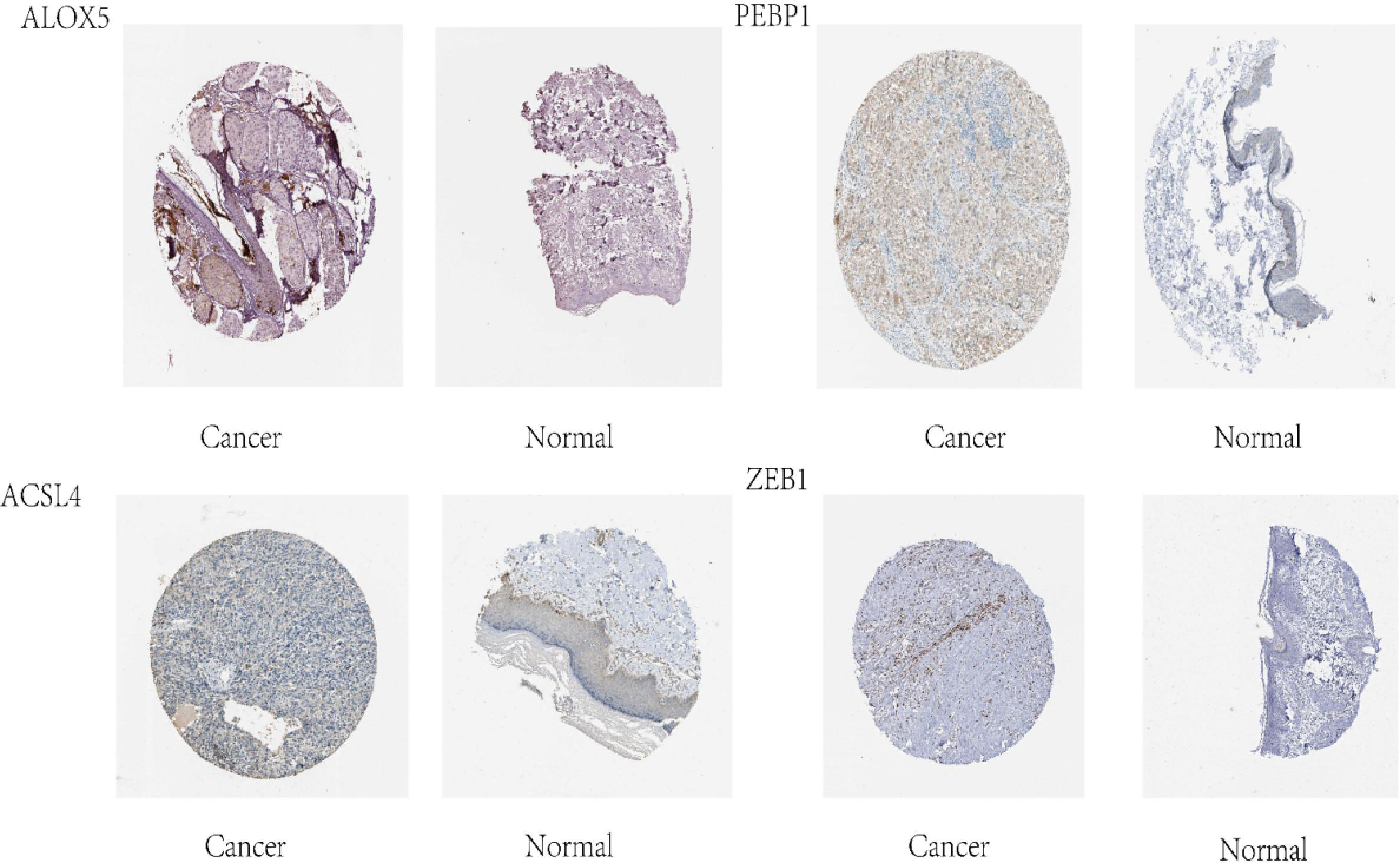
We used the human protein map (http://www.proteinatlas.org) database to verify four ferroptosis-related genes. The results showed that all four ferroptosis-related genes were expressed in varying degrees in melanoma, as shown in Figure 7. We observed expressions of ferroptosis related proteins including ALOX5, PEBP1, ACSL4 and ZEB1 in melanoma, which showed a strong cytoplasmic staining (Figure 8).
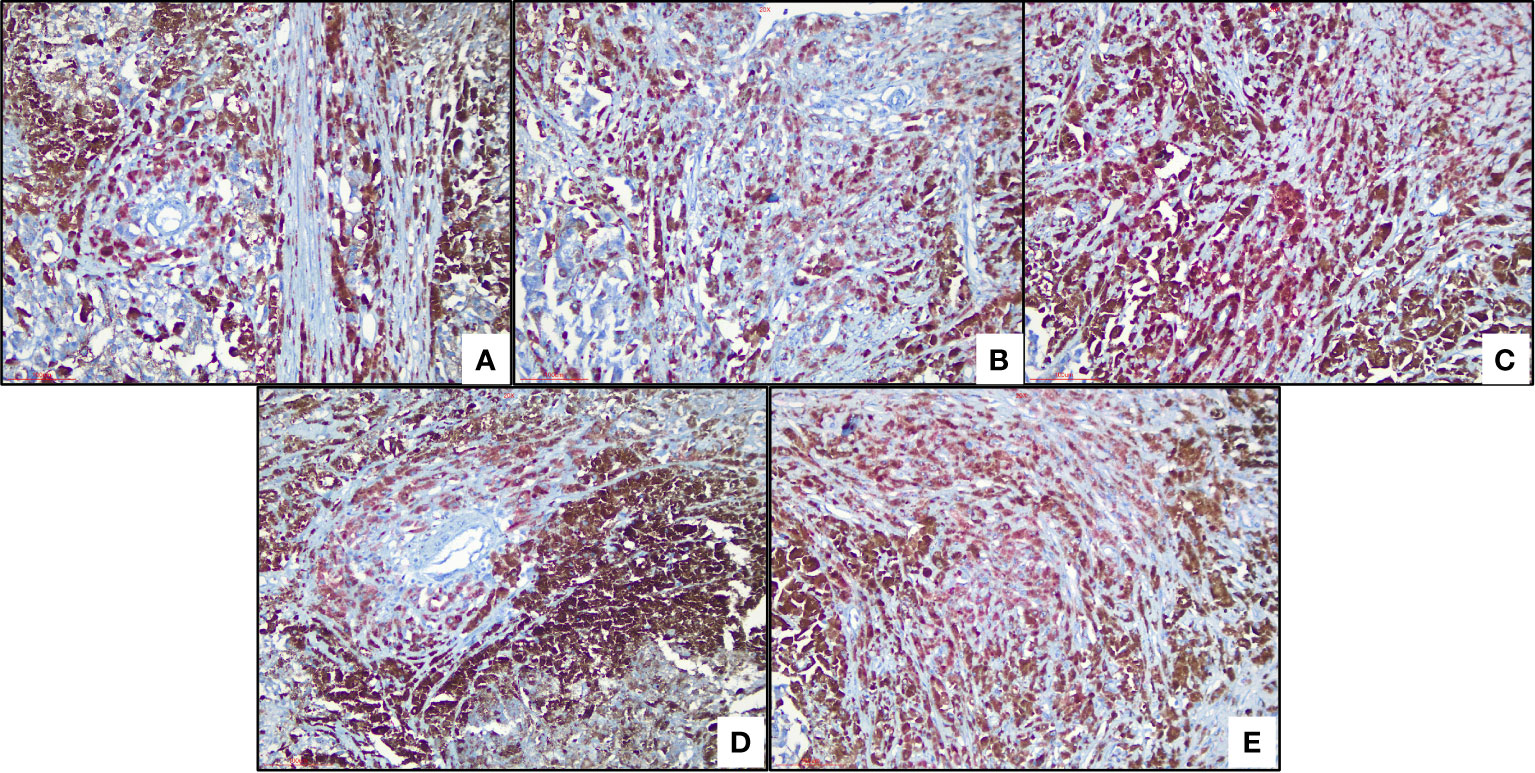
Figure 8 Immunohistochemical analysis of ferroptosis-related proteins (A) Cytoplastic expression of ALOX5 (red color means positive) (B) Cytoplastic expression of PEBP1 (red color means positive). (C) Expression of ACSL4, (red color means positive) (D) ZEB1 (red) and (E) ZEB1 (red) stain the cytoplasm of tumor cells.
Discussion
In recent years, the etiology and pathogenesis of melanoma are constantly being studied to expand its therapeutic areas. Ferroptosis is a newly discovered form of cell death that is different from apoptosis, typical necrosis and autophagy (9). The ferroptosis does not depend on the caspase activity and receptor-interacting protein 1 (RIPK1) kinase activity, but depends on the iron-dependent accumulation of lipid hydrogen peroxide to a lethal level (10). The increasing evidence indicated that ferroptosis plays a vital role in many kinds of tumor progression and prognosis (23–25). including melanoma. Luo et al. (26) reported the ferroptosis-suppressing effect of miR-137. However, many critical issues such as the interconnection between ferroptosis and other regulators of gene expression remain unsolved. This study successfully obtained the expression profile of ferroptosis-related genes in melanoma and identified eighteen ferroptosis-related genes with prognostic value by survival analysis. Through the analysis of mRNA LncRNA co-expression network, we constructed novel, well-performing risk prediction models based on the 10 LncRNA for melanoma patients.
As well known, LncRNA affects the target gene expression through epigenetic modification and transcriptional regulation. It is reported that LncRNA is abnormally expressed and related to the malignant progression of melanoma (27, 28). LncRNA-mRNA co-expression analysis is a standard method for identifying potential target genes of lncRNA and exploring the biological function of lncRNA in diseases. In this study, based on the ferroptosis-related mRNA-lncRNA co-expression network, we identified 384 ferroptosis-related lncRNA and obtained an optimal risk model including 10 LncRNAs (THCAT158, AC083799.1, AC012236.1, LINC01943, HLA-DQB1-AS1, LINC00324, AC242842.1, LINC00520, PCED1B-AS1, U62317.1). Furthermore, we found LINC00520 is an adverse prognostic factor for melanoma by multivariate Cox analysis, LINC00520 was reported to play roles in the occurrence and development of a variety of solid tumors, such as breast cancer (29, 30). However, LINC00520 is a new oncogene in melanoma, of which the exact role and molecular mechanism in malignant melanoma remain unclear. Mei XL et al (31) has found that LINC00520 inhibits the growth and metastasis of skin squamous cell carcinoma. Luan W et al. found that LICN00520 is highly expressed and promotes melanoma cells’ the proliferation, invasion, and migration. It is a risk factor for the prognosis of melanoma patients and might be used as a survival biomarker or therapeutic target for melanoma (32). In this study, survival analysis revealed that LINC00520 was highly expressed in melanoma in the high-risk group, suggesting that the prognosis of this group was poor, but the conclusion still needs to be further verified.
GO enrichment analysis revealed that the ferroptosis-related LncRNA enrichment regions differed between the high expression and the low expression groups. In the high expression group, ferroptosis-related LncRNA is closely related to glyoxylate and dicarboxylate metabolism, oxidative phosphorylation and so on. While the low expression group mainly focused on pathways such as JAK-STAT signaling, NOD-like receptor signaling pathway, RIG-I-like receptor signaling pathway and Toll-like receptor signaling.
In addition, ten lncRNA were analyzed with the human protein database HPA (http://www.proteinatlas.org), and four ferroptosis-related proteins were obtained to understand further the protein function ALOX5, PEBP1, ACSL4 and ZEB1. Moreover, IHC verification in subsequent clinical specimens showed that these ferroptosis-related proteins were highly expressed in melanoma compared with normal tissues. ALOX5, belonging to the Human lipoxygenases (LOX) family (33), is a newly discovered ELF3 target gene, and related to host immune response. It has been proved that ALOX5 can stimulate the expression of oncogenes and promote mitosis and chemotaxis in various types of cancer cells (34–36). It may also promote tumorigenesis, angiogenesis and tumor cell invasion by regulating tumor cell oxidation, arachidonic acid metabolism, carcinogenic signaling pathway and inflammation (37). Although a previous study has shown an association between ALOX5 with poor outcome of tumor (36), there are no studies indicating association of ALOX5 with the prognosis of melanoma, we herein show for the first time that ALOX5 is associated with the prognosis of melanoma.
Moreover, phosphatidylethanolamine binding protein 1 (PEBP1) is a conserved protein, isolated initially based on its ability to bind phospholipids (38). PEBP1 acts as an endogenous inhibitor of the RAF-MEK-ERK pathway by directly blocking the activation of RAF-1 proto-oncogene serine/threonine protein kinase (RAF1) (39). This concurrent activation of autophagy protects cells from ferroptosis death and the release of mitochondrial DNA. The mechanism through which LncRNA regulates PEBP1 is unknown. Our results suggest that PEBP1 may affect the prognosis of melanoma by LncRNA.
Acyl-CoA synthetase long-chain family member 4 (ACSL4) is a vital isoenzyme in the metabolism of polyunsaturated fatty acids(polyunsaturated fatty acids, PUFA), which determines the sensitivity of ferroptosis. ACSL4 disorders are associated with a variety of malignant tumors, including gastric (40), breast (41) and liver cancer (42, 43). Previous studies showed ACSL4 displayed both tumor-promoting and tumor-suppressive functions in different tumor types, while the role of ACSL4 in melanoma was still elusive. Our results provides new insights of ACSL4 into the mechanism of melanoma prognosis and provide a theoretical basis for developing new therapeutic strategies and drugs in the future.
Zinc-fingerE-box-bindinghomeobox1 (ZEB1) is a transcriptional regulator of epithelial-mesenchymal transformation of (EMT) (44, 45). Consisting of two zinc finger clusters and a central homologous domain that binds to DNA (46). The study of Li L shows that ZEB1 is overexpressed in hepatocellular carcinoma, which promotes tumor cell migration, invasion, and metastasis through EMT (47). Besides, ZEB1 reverses the more primitive state of melanoma (a neural crest stem cell (NCSC)) into a more differentiated state and reduces drug resistance to related treatments (48). Our results revealed the correlation between the ZEB1 expression and melanoma prognosis and suggested that targeting ZEB1 might be a potential method for treating melanoma.
Conclusion
In the present study, we systematically employed bioinformatics to analyze the ferroptosis-related differential genes in melanoma and identified independent prognostic differential genes. Through the association of ferroptosis-related mRNA-LncRNA, an effective prognostic model based on the risk score was successfully constructed. Our results provided a direction for prognosis assessment in melanoma patients, and the four ferroptosis-related LncRNA might be used as new biomarkers for melanoma.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
YR and JZ carried out the studies, participated in collecting data, and drafted the manuscript. HZ and WD performed the statistical analysis and participated in its design. QL provided help and advice on administration. YR, JZ, HZ, and QL wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Project of biobank (No.YBKB201911) from Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine and the Cross disciplinary Research Projects of Ninth People’s Hospital, Shanghai JiaoTong university School of Medicine (JYJC202106).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rajanna S, Rastogi I, Wojdyla L, Furo H, Kulesza A, Lin L, et al. Current molecularly targeting therapies in NSCLC and melanoma. Anticancer Agents Med Chem (2015) 15(7):856–68. doi: 10.2174/1871520615666150202100130
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin (2017) 67(1):7–30. doi: 10.3322/caac.21387
3. Wellbrock C. Hurlstone a BRAF as therapeutic target in melanoma. Biochem Pharmacol (2010) 80(5):561–7. doi: 10.1016/j.bcp.2010.03.019
4. Goydos JS, Mann B, Kim HJ, Gabriel EM, Alsina J, Germino FJ, et al. Detection of b-RAF and n-RAS mutations in human melanoma. J Am Coll Surg (2005) 200(3):362–70. doi: 10.1016/j.jamcollsurg.2004.10.032
5. Krauthammer M, Kong Y, Bacchiocchi A, Evans P, Pornputtapong N, Wu C, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet (2015) 47(9):996–1002. doi: 10.1038/ng.3361
6. Motwani J, Eccles MR. Genetic and genomic pathways of melanoma development, invasion and metastasis. Genes (Basel) (2021) 12(10):1543. doi: 10.3390/genes12101543
7. Gupta A, Gomes F, Lorigan P. The role for chemotherapy in the modern management of melanoma. Melanoma Manage (2017) 4(2):125–36. doi: 10.2217/mmt-2017-0003
8. Allais BS, Beatson M, Wang H, Shahbazi S, Bijelic L, Jang S, et al. Five-year survival in patients with nodular and superficial spreading melanomas in the US population. J Am Acad Dermatol (2021) 84(4):1015–22. doi: 10.1016/j.jaad.2020.11.047
9. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell (2012) 149(5):1060–72. doi: 10.1016/j.cell.2012.03.042
10. Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell (2017) 171(2):273–85. doi: 10.1016/j.cell.2017.09.021
11. Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun (2016) 478(3):1338–43. doi: 10.1016/j.bbrc.2016.08.124
12. Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell (2014) 156(1-2):317–31. doi: 10.1016/j.cell.2013.12.010
13. Zhang X, Wang W, Zhu W, Dong J, Cheng Y, Yin Z, et al. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int J Mol Sci (2019) 20(22):5573. doi: 10.3390/ijms20225573
14. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell (2011) 43(6):904–14. doi: 10.1016/j.molcel.2011.08.018
15. Hu Q, Ma H, Chen H, Zhang Z, Xue Q. LncRNA in tumorigenesis of non-small-cell lung cancer: From bench to bedside. Cell Death Discov (2022) 8(1):359. doi: 10.1038/s41420-022-01157-4
16. Wu S, Liu B, Zhang Y, Hong R, Liu S, Xiang T, et al. Long non-coding RNA LEISA promotes progression of lung adenocarcinoma via enhancing interaction between STAT3 and IL-6 promoter. Oncogene (2021) 40(19):3449–59. doi: 10.1038/s41388-021-01769-7
17. Ni X, Ding Y, Yuan H, Shao J, Yan Y, Guo R, et al. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Prolif (2020) 53(1):e12723. doi: 10.1111/cpr.12723
18. Song W, Zheng C, Liu M, Xu Y, Qian Y, Zhang Z, et al. TRERNA1 upregulation mediated by HBx promotes sorafenib resistance and cell proliferation in HCC via targeting NRAS by sponging miR-22-3p. Mol Ther (2021) 29(8):2601–16. doi: 10.1016/j.ymthe.2021.04.011
19. Abolghasemi M, Tehrani SS, Yousefi T, Karimian A, Mahmoodpoor A, Ghamari A, et al. Critical roles of long noncoding RNAs in breast cancer. J Cell Physiol (2020) 235(6):5059–71. doi: 10.1002/jcp.29442
20. Ashrafizaveh S, Ashrafizadeh M, Zarrabi A, Husmandi K, Zabolian A, Shahinozzaman M, et al. Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett (2021) 508:104–14. doi: 10.1016/j.canlet.2021.03.018
21. Peña-Flores JA, Bermúdez M, Ramos-Payán R, Villegas-Mercado CE, Soto-Barreras U, Muela-Campos D, et al. Emerging role of lncRNAs in drug resistance mechanisms in head and neck squamous cell carcinoma. Front Oncol (2022) 12:965628. doi: 10.3389/fonc.2022.965628
22. Liu J, Geng R, Zhong Z, Zhang Y, Ni S, Liu W, et al. N1-Methyladenosine-Related lncRNAs are potential biomarkers for predicting prognosis and immune response in uterine corpus endometrial carcinoma. Oxid Med Cell Longev (2022) 2022:2754836. doi: 10.1155/2022/2754836
23. Shi Z, Zhang L, Zheng J, Sun H, Shao C. Ferroptosis: Biochemistry and biology in cancers. Front Oncol (2021) 11:579286. doi: 10.3389/fonc.2021.579286
24. Ye Y, Dai Q, Li S, He J, Qi H. A novel defined risk signature of the ferroptosis-related genes for predicting the prognosis of ovarian cancer. Front Mol Biosci (2021) 8:645845. doi: 10.3389/fmolb.2021.645845
25. Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater (2019) 31(51):e1904197. doi: 10.1002/adma.201904197
26. Luo M, Wu L, Zhang K, Wang H, Zhang T, Gutierrez L, et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ (2018) 25(8):1457–72. doi: 10.1038/s41418-017-0053-8
27. Luan W, Ding Y, Ma S, Ruan H, Wang J, Lu F. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis (2019) 10(11):855. doi: 10.1038/s41419-019-2090-3
28. Safa A, Gholipour M, Dinger ME, Taheri M, Ghafouri-Fard S. The critical roles of lncRNAs in the pathogenesis of melanoma. Exp Mol Pathol (2020) 117:104558. doi: 10.1016/j.yexmp.2020.104558
29. Guo Q, Xu L, Peng R, Ma Y, Wang Y, Chong F, et al. Characterization of lncRNA LINC00520 and functional polymorphisms associated with breast cancer susceptibility in Chinese han population. Cancer Med (2020) 9(6):2252–68. doi: 10.1002/cam4.2893
30. Chen X, Chen H, Liu M, Xiong J, Song Z. Long noncoding RNA LINC00520 accelerates lung adenocarcinoma progression via miR-1252-5p/FOXR2 pathway. Hum Cell (2021) 34(2):478–90. doi: 10.1007/s13577-020-00478-9
31. Mei XL, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J (Engl) (2019) 132(4):454–65. doi: 10.1097/cm9.0000000000000070
32. Luan W, Ding Y, Yuan H, Ma S, Ruan H, Wang J, et al. Long non-coding RNA LINC00520 promotes the proliferation and metastasis of malignant melanoma by inducing the miR-125b-5p/EIF5A2 axis. J Exp Clin Cancer Res (2020) 39(1):96. doi: 10.1186/s13046-020-01599-7
33. Wang Y, Wang W, Sanidad KZ, Shih PA, Zhao X, Zhang G. Eicosanoid signaling in carcinogenesis of colorectal cancer. Cancer Metastasis Rev (2018) 37(2-3):257–67. doi: 10.1007/s10555-018-9739-8
34. Orafaie A, Matin MM, Sadeghian H. The importance of 15-lipoxygenase inhibitors in cancer treatment. Cancer Metastasis Rev (2018) 37(2-3):397–408. doi: 10.1007/s10555-018-9738-9
35. Zhu L, Yang F, Wang L, Dong L, Huang Z, Wang G, et al. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int (2021) 21(1):124. doi: 10.1186/s12935-021-01821-2
36. Moore GY, Pidgeon GP. Cross-talk between cancer cells and the tumour microenvironment: The role of the 5-lipoxygenase pathway. Int J Mol Sci (2017) 18(2):236. doi: 10.3390/ijms18020236
37. Merchant N, Bhaskar L, Momin S, Sujatha P, Reddy ABM, Nagaraju GP. 5-lipoxygenase: Its involvement in gastrointestinal malignancies. Crit Rev Oncol Hematol (2018) 127:50–5. doi: 10.1016/j.critrevonc.2018.05.012
38. Bernier I, Tresca JP, Jollès P. Ligand-binding studies with a 23 kDa protein purified from bovine brain cytosol. Biochim Biophys Acta (1986) 871(1):19–23. doi: 10.1016/0167-4838(86)90128-7
39. Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, et al. Suppression of raf-1 kinase activity and MAP kinase signalling by RKIP. Nature (1999) 401(6749):173–7. doi: 10.1038/43686
40. Ye X, Zhang Y, Wang X, Li Y, Gao Y. Tumor-suppressive functions of long-chain acyl-CoA synthetase 4 in gastric cancer. IUBMB Life (2016) 68(4):320–7. doi: 10.1002/iub.1486
41. Belkaid A, Ouellette RJ, Surette ME. 17β-estradiol-induced ACSL4 protein expression promotes an invasive phenotype in estrogen receptor positive mammary carcinoma cells. Carcinogenesis. (2017) 38(4):402–10. doi: 10.1093/carcin/bgx020
42. Chen J, Ding C, Chen Y, Hu W, Lu Y, Wu W, et al. ACSL4 promotes hepatocellular carcinoma progression via c-myc stability mediated by ERK/FBW7/c-myc axis. Oncogenesis (2020) 9(4):42. doi: 10.1038/s41389-020-0226-z
43. Chen J, Ding C, Chen Y, Hu W, Yu C, Peng C, et al. ACSL4 reprograms fatty acid metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway. Cancer Lett (2021) 502:154–65. doi: 10.1016/j.canlet.2020.12.019
44. Dong X, Han Y, Zhang E, Wang Y, Zhang P, Wang C, et al. Tumor suppressor DCAF15 inhibits epithelial-mesenchymal transition by targeting ZEB1 for proteasomal degradation in hepatocellular carcinoma. Aging (Albany NY). (2021) 13(7):10603–18. doi: 10.18632/aging.202823
45. Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle (2015) 14(4):481–7. doi: 10.1080/15384101.2015.1006048
46. Han L, Chen W, Xia Y, Song Y, Zhao Z, Cheng H, et al. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger e-box binding homeobox 1. Am J Transl Res (2018) 10(4):1172–83.
47. Li L, Zheng YL, Jiang C, Fang S, Zeng TT, Zhu YH, et al. HN1L-mediated transcriptional axis AP-2γ/METTL13/TCF3-ZEB1 drives tumor growth and metastasis in hepatocellular carcinoma. Cell Death Differ (2019) 26(11):2268–83. doi: 10.1038/s41418-019-0301-1
Keywords: ferroptosis, melanoma, prognosis model, lncRNA, bioinformatics analysis
Citation: Rao Y, Zhu J, Zheng H, Dong W and Lin Q (2022) A novel melanoma prognostic model based on the ferroptosis-related long non-coding RNA. Front. Oncol. 12:929960. doi: 10.3389/fonc.2022.929960
Received: 27 April 2022; Accepted: 23 September 2022;
Published: 12 October 2022.
Edited by:
Cristina Barbagallo, University of Catania, ItalyReviewed by:
Milad Ashrafizadeh, Sabancı University, TurkeyAmin Safa, Complutense University of Madrid, Spain
Copyright © 2022 Rao, Zhu, Zheng, Dong and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingyuan Lin, bGlucWluZ3l1YW4yMDA3QDE2My5jb20=
†These authors have contributed equally to this work
 Yamin Rao
Yamin Rao Jinchao Zhu
Jinchao Zhu Haiyan Zheng
Haiyan Zheng Wei Dong
Wei Dong Qingyuan Lin
Qingyuan Lin